Abstract
The mitogen-activated protein kinase (MAPK) p38α is a central component of signaling in inflammation and the immune response and is, therefore, an important drug target. Little is known about the molecular mechanism of its activation by double phosphorylation from MAPK kinases (MAP2Ks), because of the challenge of trapping a transient and dynamic heterokinase complex. We applied a multidisciplinary approach to generate a structural model of p38α in complex with its MAP2K, MKK6, and to understand the activation mechanism. Integrating cryo-electron microscopy with molecular dynamics simulations, hydrogen-deuterium exchange mass spectrometry, and experiments in cells, we demonstrate a dynamic, multistep phosphorylation mechanism, identify catalytically relevant interactions, and show that MAP2K-disordered amino termini determine pathway specificity. Our work captures a fundamental step of cell signaling: a kinase phosphorylating its downstream target kinase.
Mitogen-activated protein kinases (MAPKs) are conserved in all eukaryotes, where they form signaling cascades responding to extracellular stimuli and leading to diverse responses from differentiation to apoptosis. In higher organisms, the MAPK p38α acts in response to stresses such as irradiation, hypoxia, and osmotic shock, and also to signaling from inflammatory cytokines (1). Pathogens, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), often elicit up-regulation of p38α, which can lead to the cytokine storm associated with severe COVID-19 (2, 3). Signals propagate through phosphorylation of successive protein kinases (MAP4K, MAP3K to MAP2K), which eventually phosphorylate and activate a MAPK through double phosphorylation at the TxY motif in the activation loop (A loop), leading to a conformational change (4, 5). When the A loop is phosphorylated, key residues surrounding the adenosine triphosphate (ATP) and Mg2+ ions reorient, stabilizing the activated MAPK. The activated MAPK is then transported to the nucleus, where it modulates gene expression (6–8).
The key role of p38α in inflammation and the implication that aberrant p38α signaling is involved in numerous diseases, such as arthritis and cancer, but also in the response to infection, make it a highly studied drug target (1). Despite initial successes in developing potent compounds that target the kinase nucleotide-binding pocket, many potential therapies have failed in clinical trials because of off-target effects (9). Therefore, a molecular understanding of p38α’s interaction with upstream activators is essential to develop strategies to target allosteric sites, either in p38α or its activators. The structures of individual MAPKs (10–12) and MAP2Ks (13–16) have been extensively studied, and subsequent work has defined the interacting regions between these partners (17–20). The MAP2Ks contain a kinase interaction motif (KIM, or D motif) at the start of their intrinsically disordered N termini, which interacts with an allosteric common docking (CD) site on their target MAPK. The KIM partially ensures specificity between members of the pathway (17); acts allosterically to expose the A loop, preparing the MAPK for activation (21, 22); and enhances the local concentration of kinase and substrate (23). However, there is little structural data on the global interactions between MAPK-cascade components. Beyond the KIM interaction, molecular details of selectivity and activation of a MAPK by its upstream MAP2K remain unknown. Moreover, our knowledge of kinase-kinase interactions, in general, is restricted to homodimers and inactive conformations (24–29).
To further our understanding of how signals are transmitted through the MAPK pathway, we applied a multidisciplinary approach—combining cryo-electron microscopy (cryo-EM) with hydrogen-deuterium exchange mass spectrometry (HDX-MS), small angle x-ray scattering (SAXS), enhanced sampling molecular dynamics (MD) simulations, and cellular assays—to characterize a complex between the MAP kinase p38α (MAPK14) and its activating MAP2K, MKK6 (MAP2K6). We present a detailed molecular model of this highly dynamic and transient interaction between signaling components, providing insights into specificity and multistep catalysis. Our findings open routes to drug development for the MAPK signaling pathway and lead to a better understanding of a crucial step in kinase signaling cascades.
Engineering an active and stable MKK6-p38α complex for structural studies
The MAP kinases are malleable proteins that must adapt to multiple upstream and downstream effectors, as well as phosphorylate a large range of substrates. The proteins therefore adopt multiple conformations, and interactions with effectors and substrates must be transient to maintain signal transmission. To stabilize the MKK6-p38α complex for structural studies, we created a chimera of MKK6, named MKK6DDGRA, in which we replaced the native KIM with that from the Toxoplasma gondii effector protein GRA24 (30), which has a 100-fold higher affinity for p38α but induces the same conformation on p38α as the MKK6 KIM (fig. S1A) (31). We also inserted phosphomimetic mutations in the MKK6 A loop (S207D and T211D) to transform the kinase into a constitutively active form that is ready to phosphorylate p38α (Fig. 1A and fig. S1, B and C). SAXS analysis revealed heterogeneity in the size of the complex formed with p38α (fig. S2A). By screening MKK6DDGRA with a combination of different nucleotide analogs and p38α A-loop mutants, we identified the mutant p38αT180V (one of the phosphosites of the A loop) in combination with the transition-state analog ADP.AlF4- as the complex with the smallest radius of gyration (Rg) (table S1). Because a smaller Rg for a given sample indicates a more compact structure, we reasoned that in this sample, the transition-state analog complex is stably assembled with the single residue available in the A loop (Y182) and should be suitable for structural studies. Subsequent analysis, including HDX-MS, MD, and Bayesian modeling, confirmed that the engineering increases the population of the transphosphorylation-competent state of the complex and that its nature is consistent with that of the wild type (WT).
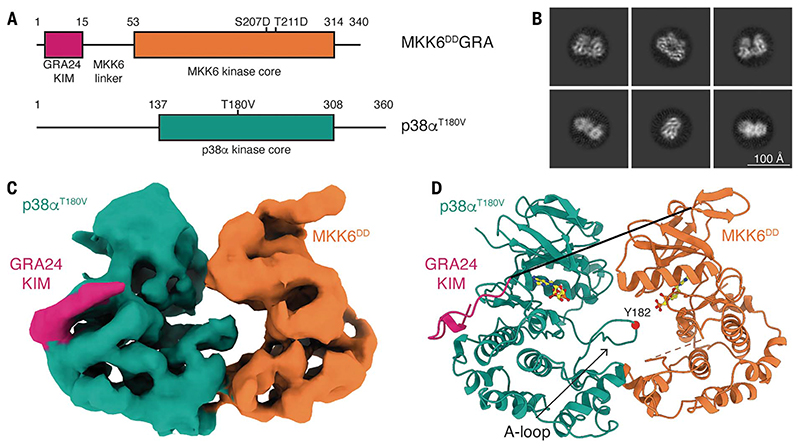
Fig. 1. Structure of the MKK6DDGRA-p38αT180V complex.
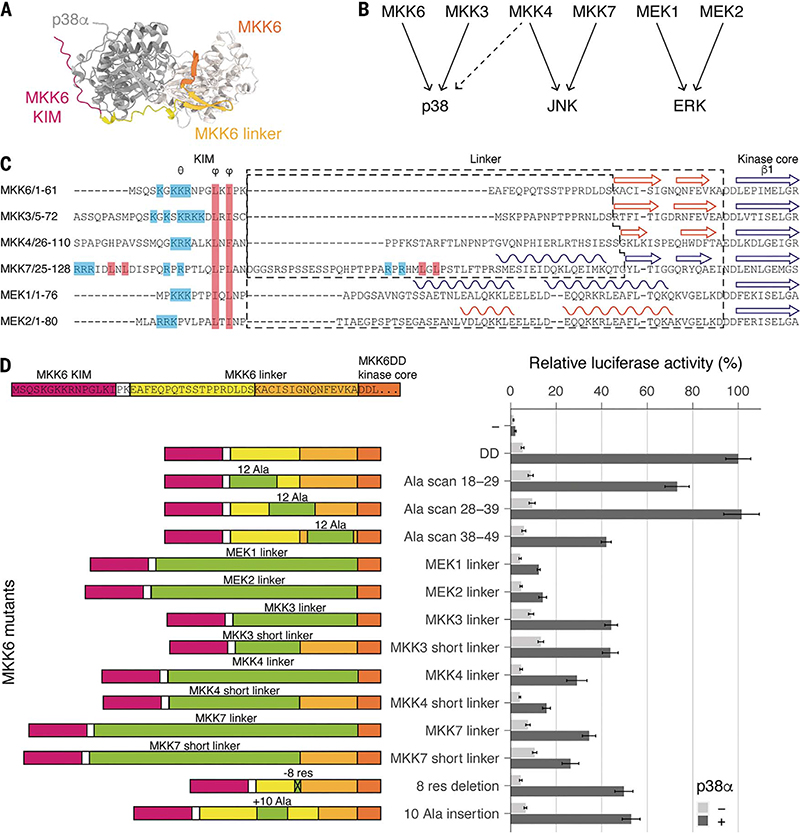
(A) Schematic of the two protein constructs. Key domains and mutations are indicated. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. In the mutants, other amino acids were substituted at certain locations; for example, S207D indicates that serine at position 207 was replaced with aspartic acid. (B) Representative 2D class averages from single-particle cryo-EM analysis of the MKK6DDGRA-p38αT180V complex. Scale bar, 100 Å. (C) Segmented single-particle reconstruction cryo-EM map of the MKK6DDGRA-p38αT180V complex, resolved to 4-Å resolution and colored according to (A). (D) Model of the MKK6DDGRA-p38αT180V complex, showing the overall structure of the complex. MKK6DDGRA-p38αT180V is represented as a cartoon and colored according to (A). AMP-CP nucleotides are represented as balls and sticks (carbon atoms are yellow). The MKK6 linker and A loop are disordered and respectively shown as a full black line and an orange dashed line. The position of the Y182 residue in the p38α A loop is indicated by a red sphere.
Architecture of the MKK6-p38α complex
We imaged the 80-kDa MKK6DDGRA-p38αT180V complex with cryo-EM (Fig. 1). The purified complex was well dispersed and yielded two-dimensional (2D) class averages that showed clear secondary-structure features (Fig. 1B). The selected particles allowed the 3D reconstruction to a nominal resolution of 4 Å (map range 2.7 to 6.5 Å) (Fig. 1C, fig. S3, and movie S1). The final resolution was limited not only by the small size of the complex but also by the highly dynamic nature of the interaction between the kinases, leading to particle heterogeneity. Therefore, very strict selection of particles was necessary to capture this conformation. Multiple rounds of picking—with a combination of Topaz (32), template picking, and 2D classification to eliminate the noisy 2D class averages—led to a good-quality set of particles that could then be further selected to remove particles clearly showing only a single kinase or two loosely connected kinases (these classes were always low resolution). Classification in 3D was then used to select particles corresponding to the heterokinase complex, and further 2D classification was then used to select class averages showing strong features and low background. Subsequent rounds of heterogeneous refinement and quality assessment with 2D classification were then used to differentiate between face-to-face conformations where the A loop was stabilized or disordered.
To build the model of the complex, we docked structures of p38α and MKK6 into the map. Because all available structures of inactive p38α have an unstructured or mobile A loop, we determined the crystal structures of human p38α and of an inactive mutant (p38αK53R), in which the A loops are well ordered in the occluded conformation in order to have a starting point for refinement of the extended conformation (table S2). The available MKK6 structures are not in the active conformation (fig. S4). We therefore modeled the structure from the active conformation of MKK7 (16). Docking of these structures was followed by one round of morphing and real-space refinement. To select the model that best recapitulates the available data, we then computed the cross-correlation between experimental and predicted maps for relevant structures obtained from the ensemble that we derived by combining Bayesian inference with MD simulations and SAXS data (Fig. 1D, fig. S3C, and tables S3 and S4). The reconstruction shows the kinases adopting a face-to-face conformation, with most contact between the C lobes. MKK6 is in the active kinase conformation according to its αC-helix rotated position and in the DFG-in BLAminus conformation according to kinase nomenclature (33) (fig. S4), and density can be observed for nucleotide in its active site (fig. S5). The p38α A loop is ordered and extends toward the MKK6 active site; however, it remains dynamic, limiting the resolution of this region (fig. S5). The p38α active site also binds nucleotide (fig. S5), implying that it is already in an active conformation. MKK6 interacts with the p38α CD site through the GRA24 KIM at the start of its N-terminal extension, which can be traced in the density (Fig. 1C and fig. S5). Most of the linker between the KIM and the MKK6 kinase core, as well as the MKK6 A loop, remains disordered and, therefore, unresolved. AlphaFold2 multimer (34) predicts a similar face-to-face conformation for the MKK6-p38αWT complex, as well as for MKK6DD-p38αWT and MKK6DDGRA-p38αT180V (fig. S6). Superimposing the predicted models to our experimental model shows a high consensus of the interaction. However, relative domain rotation in the prediction leads to differences in the interface of the two kinases (fig. S6A). High confidence in intermolecular contact placement seen in the predicted aligned error (PAE) plots (fig. S6, D to F) supports the overall face-to-face conformation of the complex.
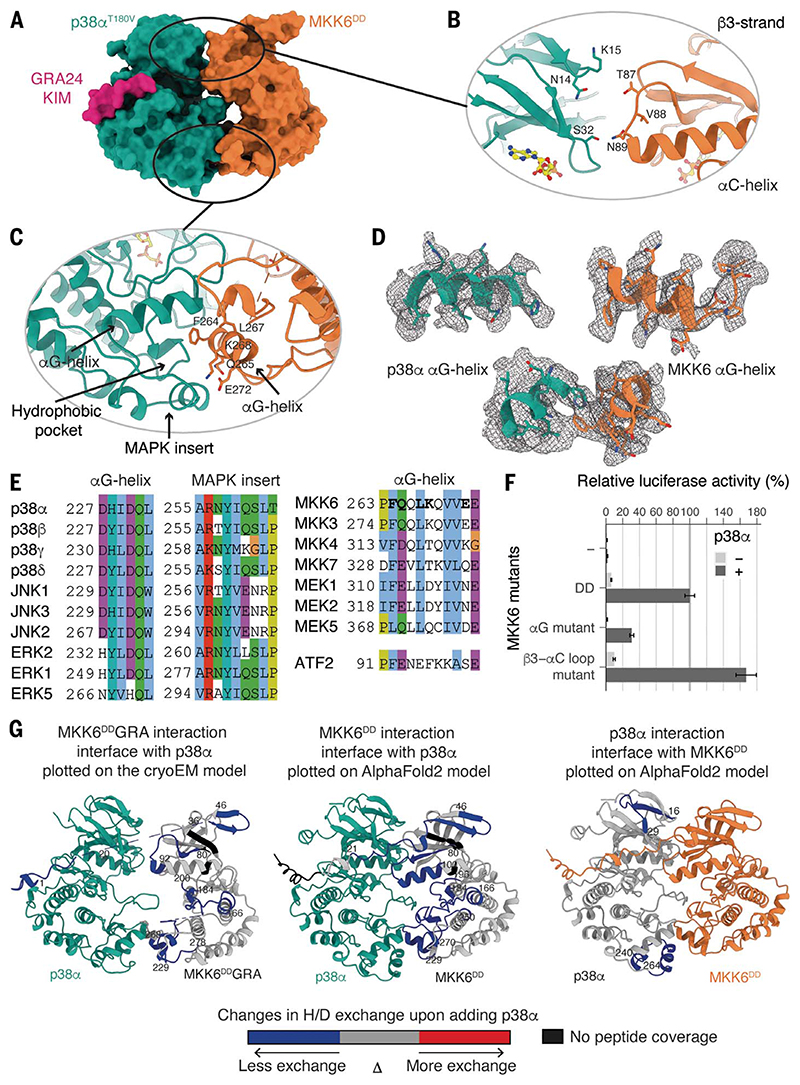
The main kinase core-fold interaction is between the αG helix (residues 262 to 273) of MKK6 and a hydrophobic pocket in p38α, partly formed by the MAPK-specific insert in the C lobe (Fig. 2, A, C, and D, and movie S2), which clamps the MKK6 αG helix upon binding (movie S3). This insert has been described as a lipid-binding site important in regulation (35, 36), and several studies have identified small molecules that target this pocket (37–41) (fig. S7). This site has also been shown to be important in MAPK substrate binding (42, 43) (fig. S7). The exposed residues of this helix, as well as residues lining the MAPK hydrophobic pocket, are highly conserved in the MAP2Ks and MAPKs, respectively, suggesting a common interaction site for the pathways (Fig. 2E). Additionally, there is a potential interaction between the N lobes through the loop between the β3 sheet and αC helix of MKK6 (residues 87 to 89) and β strands 1, 2, and 3 of the N lobe of p38α (Fig. 2B). We validated the observed interactions using HDX-MS (44). In both the MKK6DD (with the wild-type KIM) and chimera MKK6DDGRA, several regions were protected from hydrogen-deuterium exchange in the presence of an excess of p38α: the N terminus, the β3 strand, the A loop, and the αG helix (Fig. 2G, fig. S8, and table S5). The interaction sites identified in solution highly correlate with the cryo-EM model for both the wild-type and chimeric complexes. Although no peptide was detected for MKK6 KIM, the GRA24 KIM was strongly protected upon binding with p38α. In an excess of MKK6DD, only the p38α MAPK insert and a small region of the N lobe are protected from exchange (Fig. 2G and fig. S8). We also evaluated the protection on p38α when we mutated the MKK6 αG-helix residues facing the p38α hydrophobic pocket to alanine (F264A, Q265A, L267A, K268A, and E272A—referred to as the MKK6DD αG-helix mutant). Whereas its interaction with MKK6DD is stable, the p38α MAPK insert is not protected in the presence of the MKK6DD αG-helix mutant, which implies that this mutant cannot interact with p38α in that region (fig. S9). Free-energy calculations of p38α-MKK6 and p38α-MKK6 αG-helix mutant further support the lower stability of the C-lobe interface in the latter (supplementary text and fig. S9). These experiments define the αG-helix-MAPK-insert interaction as the primary site of interaction between the two kinases during phosphorylation of the p38α A loop.
Fig. 2. Interaction interfaces between MKK6 and p38α, distal from the active site.
(A) Surface representation of the MKK6DDGRA-p38αT180V complex. (B) Potential interaction between the N lobes. (C) Interaction between the αG helix of MKK6 and the hydrophobic pocket of p38α. (D) αG helices of p38α and MKK6 in the sharpened Coulomb potential map (black mesh). (E) Sequence alignment of MAPK hydrophobic pockets, MAP2K αG helices, and the p38α substrate ATF2 peptide (Clustal coloring scheme). (F) Luciferase reporter assay to monitor the activity of the p38α signaling pathway in HEK293T cells, showing the ability of MKK6DD mutants to activate p38α (statistical analysis in table S6 and protein expression levels in fig. S10). (G) (Left) MKK6DDGRA and (middle) MKK6DD interaction sites with p38α and (right) p38α interaction sites with MKK6DD, identified with HDX-MS. Regions showing protection upon the addition of p38α or MKK6DD, indicative of interaction, are highlighted in blue.
To further study the importance of these interactions on p38α activation, we used the MKK6DD αG-helix mutant and also mutated the β3-αC loop of the MKK6 N-terminal interaction to alanine residues (T87A, V88A, and N89A, referred to as the MKK6DD β3-αC loop mutant) and read out the effect on p38α signaling in cells using a luciferase reporter assay (Fig. 2F). We transiently transfected human embryonic kidney (HEK)293T cells with p38α and MKK6DD mutants, together with a plasmid encoding firefly luciferase under the control of the activator protein 1 (AP-1) promoter, in which AP-1 activity is stimulated by p38α through several substrates [e.g., activating transcription factor 2 (ATF2); materials and methods]. The MKK6DD αG-helix mutant reduced p38α signaling by 70% when compared with MKK6DD, demonstrating the substantial role that this interaction plays in p38α activation. The MKK6DD β3-αC loop mutant increased signaling to ~170% over MKK6DD. This loop is the site of numerous activating mutations, particularly in cancer, that stabilize the “αC-in” conformation (45, 46). This could explain the observed increase in signaling but does not rule out a role for the p38α N lobe in the stabilization of the active conformation of MKK6 in the face-to-face complex.
Molecular dynamics confirm that the complex is metastable and show how catalytically competent states can be formed
To better understand the details of the heterogeneous structural ensemble of the complex and the mechanism of transphosphorylation of p38α on T180 and Y182 by MKK6, we ran a series of MD simulations, starting with a set of 18 simulations, each 1 μs long (table S7). The simulations started from models derived from the cryo-EM structure: MKK6GRA-p38α (with wild-type A loop) and MKK6-p38α, in which we reverted the GRA24 KIM back to the wildtype sequence.
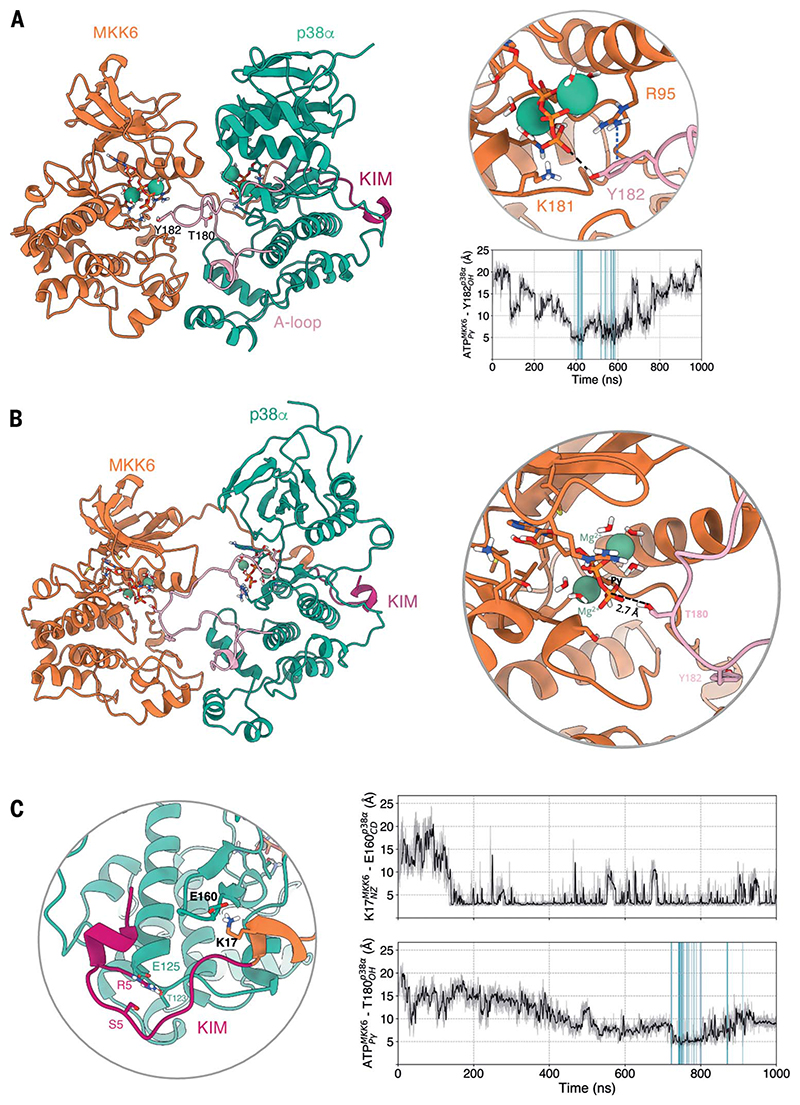
The simulations explored several conformations. In most cases, the contact between the KIM and p38α was retained, and catalytically competent states, in which p38α T180 or Y182 approached the γ-phosphate of MKK6 ATP at a catalytically compatible distance, were observed. Partial detachment of p38α from MKK6 was also observed in some instances. Additionally, the C-lobe interface proved to be important for the stability of the dimer, which is in agreement with our other observations. In all simulations in which the two kinases break apart, this tight hydrogen-bonding network at the C-lobe level is the last point of contact to be lost. In particular, the interaction between MKK6 αG-helix residues K268 and E272 and p38α S261 seems to be one of the main interactions keeping the two kinases in close proximity (fig. S9E). The observed conformational variety is consistent with the SAXS and cryoEM data, which are indicative of the transient nature of the interaction of the two kinases. With respect to the phosphorylation mechanism, in a number of trajectories, either p38α T180 or Y182 approach the ATP γ-phosphate located in the MKK6 active site (47) (Fig. 3, table S8, and movies S4 and S5). In particular, in the simulations of MKK6-p38α, both T180 and Y182 come close to the catalytic site. The tendency of monomeric, unphosphorylated p38α to adopt inactive, sequestered A-loop conformations has been shown in previous studies (48). Thus, the observation that the unphosphorylated A loop of p38α adopts an exposed conformation in multiple instances is indicative of an active role of MKK6 in stabilizing such conformations. The active role of MKK6 is confirmed by the fact that in the simulations in which the MKK6 KIM detaches, leading to the separation of the two kinases, the p38α A loop transitions from an exposed to a sequestered conformation (fig. S11).
Fig. 3. MD simulations show that both p38α Y182 and T180 can approach MKK6 ATP and that a rotated conformation of p38α favors T180 phosphorylation.
(A) Frame extracted from one of the unrestrained MD simulations in which Y182 approaches the γ-phosphate of MKK6 ATP at a catalytically compatible distance (3.8 Å). The p-cation interaction of R95 with Y182 that further stabilizes Y182 close to ATP is shown in a blue dashed line. The frames where p38α Y182 reaches MKK6 ATP at a catalytically compatible distance in this set of simulations are highlighted in the plot of distances over time (blue). (B) Simulation frame in which p38α has rotated around its axis with respect to the cryo-EM structure. (C) Detailed view around the KIM. The frames where T180 reaches MKK6 ATP at a catalytically compatible distance in this set of simulations are highlighted in the plot of distances over time (blue).
A rotated complex might facilitate the phosphorylation of p38α T180
In this initial set of MKK6GRA-p38α simulations, we observe some complexes in which T180 approaches the γ-phosphate of ATP in the catalytic site of MKK6 from an unexpected angle, where the p38α N lobe underwent a 50° to 80° rotation around its axis (Fig. 3B and movie S4). In this conformation, p38α was kept close to MKK6 through the KIM, despite the rotation. Before T180 gets close to the ATP, K17, a key lysine located just after the KIM, forms a salt bridge with p38α E160 that seems to initiate the rotation of p38α (Fig. 3C).
Integration of simulations with SAXS data through Bayesian/maximum-entropy reweighting shows how the kinases associate
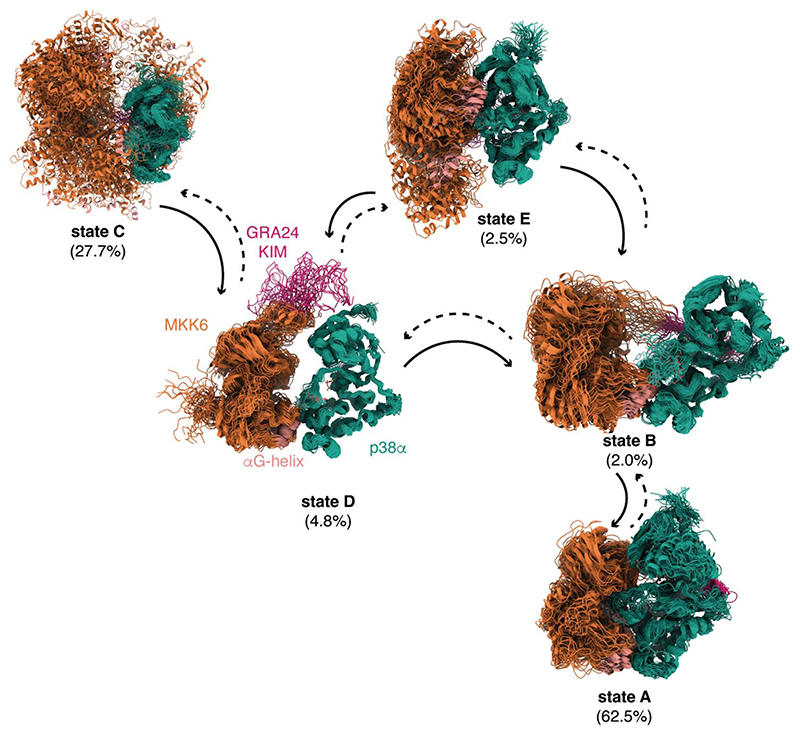
To better characterize intermediate states involved in the formation of the catalytically competent dimer, we extended the conformational sampling of the MD simulations using adaptive Markov state model (MSM) simulations (49, 50) (supplementary material sp. S8 and fig. S13) and used a kinetic-based clustering of the total accumulated MD trajectories to determine the main metastable states. By extending the sampling to more than 18 μs and performing the kinetic clustering, we obtained five macrostates for p38α with MKK6GRA (Fig. 4 and movies S6 to S10). In the case of MKK6GRA-p38α, one of the clusters corresponds to an ensemble that contains conformations where the two kinases are face-to-face, equivalent to the cryo-EM structure (state A, movie S6). In this macrostate, we also see conformations corresponding to the rotated N lobe of p38α, in which T180 is ideally positioned for phosphorylation. The clustering of the face-to-face and rotated conformations within the same metastable state reflects the rapid interconversion from one to the other (few μs). In the second macrostate, the KIM is tightly bound, whereas MKK6 adopts different orientations (state E, movie S7). The third macrostate represents conformations resulting from the detachment and nonspecific binding of the two kinases, in which MKK6 binds to different surfaces of p38α (state C, movie S8). Lastly, we observed two macrostates in which the two kinases are held together through the C-lobe interface and in which the KIM is either bound (state B, movie S9) or explores the N lobe of p38α (state D, movie S10). The connections between the macrostates (Fig. 4) indicate one or more reactive trajectories connecting one state to another. The resulting network shows that the binding of the KIM to its recognition site plays a fundamental role in establishing the specific contacts needed for the formation of the face-to-face dimer.
Fig. 4. Kinetic-based clustering of the accumulated simulations of MKK6GRA-p38α.
Each macrostate shows an overlay of 50 representative conformations sampled proportionally to the equilibrium probability of each microstate in the corresponding macrostate that correlates to different kinetically distinct states. The population of each state as derived from the fitting to the MKK6DDGRA + p38αWT + ADP + AlF4- SAXS curve (fig. S2) is given in parenthesis. The solid arrows indicate the main directions of the observed reactive trajectories connecting the macrostates.
Despite a total sampling time of more than 18 μs, we could not fully converge the kinetics and the population of all the macrostates directly from the simulations, probably thanks to the slow nature of some of the transitions seen in protein-protein associations (50). An exhaustive sampling of complex formation from the detached states would require a prohibitive number of long MD simulations and might still be affected by the quality of the force field. We therefore refined the structural ensemble and validated the clustering results by using the SAXS data and a Bayesian/maximum-entropy approach (51–53). The SAXS profile of each conformer within the clustered MD ensemble was calculated, and the weights associated with each conformer in the ensemble that maximize the agreement with experiments were iteratively determined (fig. S12A). For the refinement of the MKK6GRA-p38α ensemble, we used the SAXS curves of the complex with ADP.AlF4- and with the nonhydrolyzable ATP analog β, γ-methyleneadenosine 5′-triphosphate (AMP-PCP). The reweighted ensemble fits extremely well with the SAXS curve of MKK6DDGRA-p38αT180V with ADP.AlF4- (χ2 = 0.89; movie S15), which showed the most compact conformation (table S1). Reassuringly, all the macrostates from kinetic modeling contribute to the SAXS curve (Fig. 4). State A, which contains conformations equivalent to the cryo-EM structure, is the most populated (62.5%), followed by state C (27.8%), which reflects the transient nature of the complex and the non-specific binding associated with such transient complexes, and then ensemble E (2.5%). From the weights (and the other experiments), we deduce that the latter is the main intermediate for the formation of the face-to-face complex. After the KIM domain is bound, the MD simulations indicate an important role for the hydrophobic patch and a network of hydrogen bonds located at the interface of the C lobes of the two kinases in the formation of the complex. These conclusions are in accordance with the 70% decrease in activity seen in the MKK6DD αG-helix mutant (Fig. 2F), as well as the metadynamics free-energy calculations and the HDX-MS data (fig. S9).
The ensemble reweighted to match the SAXS data obtained from the MKK6DDGRA-p38αWT with the ATP analog AMP-PCP also resulted in a good fit (χ2 = 2.1). The population of the face-to-face cluster A decreases to 1%, whereas that of the detached cluster C and intermediate D increases considerably, to 71.5 and 21.7%, respectively (fig. S12, B and C). Although the populations of the macrostates shift, which is expected because the transition state is not stabilized, the general description of the macrostates remains similar. Within cluster A, the weights of the “rotated” face-to-face dimer increase (fig. S12B). We also sampled the conformational space of the MKK6-p38α with the same adaptive MSM approach. The four macrostates found in the MKK6-p38α complex are equivalent to four of the macrostates found in MKK6GRA-p38α (fig. S13), reflecting the similarity of the dynamics of the two complexes (supplementary text).
The N-terminal extension of MKK6 remains mainly disordered, and its length and secondary-structure elements contribute to pathway specificity
Our structure and macrostate modeling imply that once the KIM is bound, the N-terminal linker plays an important role in conformational sampling, allowing the catalytically competent state to form, a phenomenon that has been observed in other tethering proteins (54). There is a wealth of data on interactions between KIMs and MAPKs, but the role of the remainder of the N terminus is poorly understood (55, 56). In our cryo-EM map, the engineered KIM is clearly visible (Fig. 1C and fig. S5), but it does not appear to interact further with p38α. To investigate whether the N-terminal extension has a role beyond simply associating the kinase to the KIM, we again used the luciferase reporter assay. We produced constructs that scan the region between the KIM and the first consensus β sheet of the kinase core with alanine blocks to determine if there is sequence specificity or direct interaction with p38α (Fig. 5). Our results show that the sequence of the middle region (MKK6DD Ala scan 28 to 39) of the N-terminal linker has no effect on p38α signaling in cellulo. However, the sequence of the linker close to the KIM (MKK6DD Ala scan 18 to 29) and the region close to the kinase core, which comprises the predicted β strands (MKK6DD Ala scan 38 to 49) (Fig. 5) seem to have some importance because their mutation to alanine reduced p38α signaling by 27 and 58%, respectively.
Fig. 5. MKK6 N-terminal extension length and secondary structure define the specificity of p38α activation.
(A) Top view of an MKK6-p38α AlphaFold2 multimer model, for illustrative purposes. (B) Specificity of the MAP2K/MAPK signaling pathways. (C) Sequence alignment of MAP2K N termini. Secondary structure elements are indicated, from experimental structures in blue and AlphaFold2 predictions [predicted local difference distance test (pLDDT) score > 65] in orange. (D) Luciferase reporter assay to monitor the activity of the p38α signaling pathway in HEK293T cells, showing the ability of MKK6DD mutants and chimeras to activate p38α (statistical analysis in table S9 and protein expression levels in fig. S10).
Although KIM sequences are important, they do not sufficiently explain specificity. When the N-terminal linkers of the MAP2Ks are compared, it is found that they differ in their length and secondary-structure elements (16, 57, 58) (Fig. 5, B and C). Could this contribute to specificity between the pathways? When we substituted the MKK6 linker with those from the MAP2Ks of the other pathways, activity was reduced (Fig. 5D). This is most observable with the linkers from MEK1 and 2. The linkers from MAP2Ks that activate p38α also see reduced activity and are not sufficient to rescue the activation, even though the KIM, MAP2K, and MAPK are in the correct combination. Our structure and macrostate models imply that the length of the linker is just sufficient to allow the observed interactions between the kinases, in particular, the MAP2K αG-helix position relative to the MAPK hydrophobic pocket formed by the MAPK insert. By removing or adding 10 residues to the linker region, activity is reduced by ~50%. This implies that the length of the linker, controlled by the number of amino acids and/or by secondary-structure elements, is important in the positioning of the MAP2K for engagement with the MAPK and is finely tuned for MAP kinase pairs. The specificity of a MAP2K is therefore defined by cooperation between the KIM, the linker, and the kinase core folds themselves.
Discussion
Initial docking
The N termini of the MAP2Ks contain a conserved KIM motif that is required for binding to their substrate MAPKs. The first step in activation is KIM binding, after which a wide variety of conformations are observed, showing the fast timescale of association and dissociation (Fig. 4). During the MD simulations, KIM binding induced allosteric changes in p38α, which adopts a prone-to-be-phosphorylated conformation with its A loop extended, exposing the tyrosine and threonine residues, which is in agreement with crystal structures. Our data indicate that the interaction between the MKK6 N terminus and p38α extends beyond the hydrophobic residues of the KIM and is important in the activation process. Although most of the MKK6 N-terminal linker remains disordered, its length seems to be tightly linked to the substrate MAPK, and the presence of some secondary structural elements differs between MAP2Ks, contributing to specificity (Fig. 5C). Our reweighted ensemble, obtained by combining Bayesian inference with adaptive MSM simulations and SAXS data, shows that once the KIM is bound, many conformations are sampled before the engagement of the αG helix (Fig. 4). This sampling is fully consistent with the experiments that show that if the linker is not the correct length, the engagement of the αG helix will be impeded. Once p38α is recruited, some parts of the MKK6 N-terminal linker could participate in the conformational shift toward the MKK6 active conformation, but its main function appears to be to prepare the MAPK A loop and to guide the engagement with the αG helix.
Engagement with substrate
We show that the predominant interaction during activation is mediated by the αG helix of the MAP2K MKK6 and the p38α hydrophobic pocket formed by the αG helix of p38α and the specific MAPK insert. This region is also important in MAPKs for substrate recognition (42) and interaction with scaffold proteins and downstream regulators such as phosphatases (27, 43, 59). The engagement of αG helices seems to be an emerging theme in kinase heterodimers: Only a few structures have been determined, such as the KSR2-MEK1 heterodimer (27) and more recently, the BRAF-MEK1 complex (25), both upstream components of the related extracellular signal-regulated kinase (ERK) MAPK pathway. In both these structures, the αG-helices interaction is very similar, as well as the relative orientation of the kinases. Several studies have shown that small molecules can target this region in p38 isoforms (37–41) and modulate the position of the MAPK-insert helices (fig. S7). However, as the pocket is very deep, the molecules may not extend far enough to disrupt the interactions identified here. There has been intense interest, and some success (60), in developing drugs that target p38α, mostly binding to the nucleotide pocket, but they have been mired by off-target and toxicity effects (9). The molecules that have been identified as binding to the MAPK-insert pocket could be further developed to better disrupt both the MAP2K interaction and that of substrates, which could lead to highly specific p38 inhibitors.
The MKK6 KIM-p38α docking site, the MKK6 N-terminal linker, and the MKK6 αG-helix-p38α hydrophobic pocket interactions are essential for defining the specificity of the kinase-substrate interaction, for positioning the two kinases, and for triggering the necessary changes toward the MKK6 active kinase conformation and positioning the A loop of p38α to be accessible for phosphorylation. Our data demonstrate that altering any of these three components perturbs the activation of downstream signaling.
Freedom of interaction at the catalytic center
All contacts observed between the kinases are distal to the active site, which explains the lack of sequence specificity or conservation in MAPK A loops in the region preceding the TxY motif (58), and our HDX-MS data show no tight interaction between the A loop of p38α and MKK6 (Fig. 2 and fig. S8). Rather than the active site of a classical enzyme, where substrates are perfectly positioned for catalysis, the MAP2K-MAPK complex appears to provide a zone of proximity, allowing the flexible activation loop to move but increasing the probability, or local concentration, of the p38α T180 and Y182 residues to be positioned for nucleophilic attack of the γ-phosphate of ATP by relative positioning of the two kinases. Dual-specificity MAP2Ks are unusual because the two amino acids targeted are considerably different from each other when compared with serine/threonine kinases. In addition, the two substrate-phosphorylation sites are only one residue apart, and the MAP2Ks need to have an active site that can accommodate, effectively, four substrates [(A-loop T-x-Y; T-x-Y; T-x-YP; and TP-x-Y (targeted residue in bold)], all with markedly different sizes and charges. This implies that flexibility in the active site is essential to accommodate such a wide variety of substrates. By not binding any residues of the A loop specifically, various residues and phosphorylation states could be accommodated by sacrificing catalytic efficiency for flexibility.
The MD simulations revealed that the face-to-face architecture of the heterokinase dimer of our model is compatible with the phosphorylation of either T180 or Y182 of the p38α A loop (Fig. 3 and fig. S14). Another conformation, where p38α rotated, emerged and seems to favor the phosphorylation of T180. Kinetic data have shown that the dual phosphorylation of p38α by MKK6 involves a partially processive mechanism in vitro, in which the monophosphorylated intermediates can either dissociate from the enzyme or proceed to the next catalytic step. Wang et al. (61) and others (62) propose that both p38α monophosphorylated forms can be produced by MKK6 catalysis with a preference for Y182, whose phosphorylation is four times as fast as that of T180. With respect to the second catalytic step, the experimentally measured kinetic rates indicate that phosphorylation at Y182 in the first step enhances the catalytic efficiency of MKK6 phosphorylation at T180 in the second step. According to our simulations (Fig. 3 and fig. S14), when the N lobe of p38α undergoes a large conformational change toward the rotated dimer, T180 is in the right position to be phosphorylated, but Y182 is more distant from the catalytic site, which might explain the lower catalytic efficiency of MKK6 observed with kinetic measurements when the second phosphorylation is on Y182. Conversely, Y182 seems to be able to easily approach MKK6 ATP in the face-to-face catalytically competent conformations and then be phosphorylated. Thus, the difference in the experimental kinetic rates measured for the first phosphorylation step may reflect the rotation of the p38α N lobe around its axis, which seems to be involved in the T180 phosphorylation only. The partial processivity of the dual phosphorylation in a cellular context remains an open question (63, 64), but the architecture of the interaction that we observed between MKK6 and p38α, as well as the microstates seen in modeling, seem to be compatible with either a distributive, partially processive or fully processive mechanism.
By combining cryo-EM and MD simulations, together with HDX-MS and structure-driven mutagenesis in cellulo, we describe the range of attributes that lead to MKK6 selectivity and p38α activation. Moreover, we identify critical interfaces for dimer stabilization. In this regard, small molecules that prevent MKK6-p38α complex dimerization could be therapeutically effective in treating diseases resulting from abnormal MKK6-p38α-mediated signaling. What is more, our data describe molecular details of the activation of one protein kinase by another—one of the most fundamental mechanisms in cell signaling.
Materials and methods summary
The details of the methodology are described in the supplementary materials and are summarized here as follows:
Purification of MKK6-p38α
To increase the affinity of the constitutively active MKK6DD mutant (S207D and T211D) for p38α for structural studies, the natural KIM sequence was replaced with the T. gondii effector protein GRA24 KIM sequence (MKK6DDGRA), and a C-terminal Twin-Strep tag (IBA Lifesciences) was added. MKK6DD and MKK6DDGRA were expressed in Sf21 insect cells and purified through streptavidin affinity and size-exclusion chromatography (SEC). Human MAPK p38α constructs were expressed in Escherichia coli with an N-terminal His6 tag and purified through nickel affinity and SEC. The heterokinase complex was prepared by mixing MKK6DDGRA and p38α in a 1:1 molar ratio followed by SEC. The transphosphorylation conformation was then assembled by using the transition-state analog ADP.AlF4- through incubation with 10 mM ADP, 10 mM NH4F, and 1 mM AlCl3 for 30 min before proceeding with biophysical studies.
Cryo-EM grid preparation, imaging, data processing, and modeling
Purified MKK6DDGRA-p38αT180V with ADP. AlF4- was applied to UltraAufoil 1.2/1.3 grids (Quantifoil) and plunge-cooled with a Vitrobot Mark IV (FEI), screened on a FEI Talos Glacios (EMBL Grenoble), and imaged on a FEI Titan Krios (EMBL Heidelberg). Data processing in CryoSPARC (65) yielded a final map with an average resolution of 4.0 Å [Fourier shell correlation (FSC) 0.143], showing the two kinases in a face-to-face conformation with the A loop of p38α extended toward the active site of MKK6.
MD simulations and modeling
The cryo-EM structure was used as the starting conformation of the MD simulations, and bound nucleotides were replaced with ATP. The simulations were set up by using GROMACS v.2021.3 with the DES-Amber force field and TIP4PD water molecules. The PLUMED plugin was used for free-energy calculations. Adaptive MD simulations were performed to further explore the prebound and bound states of the complex. The conformational ensembles were clustered kinetically and refined using an iterative Bayesian/maximum-entropy (iBME) protocol to obtain a structural ensemble that reflects the states captured in the solution SAXS data.
Supplementary Material
Research Article Summary.
Introduction
Mitogen-activated protein kinases (MAPKs) are important signaling proteins found in eukaryotes. They respond to external signals and regulate various cellular processes. One such MAPK is p38α, which plays a crucial role in cell stress, inflammation, and response to infection, including the cytokine storm associated with severe COVID-19 and influenza. Dysregulation of p38α signaling is linked to several diseases, making p38α an important drug target. Understanding the interactions between p38α and its upstream activating MAP2K, MKK6, is essential to devise drug development strategies targeting allosteric sites. The MAP2Ks have a kinase interaction motif (KIM, or D motif) at the beginning of their intrinsically disordered N termini, which interacts with an allosteric docking site on their target MAPK, ensuring some level of specificity. Once engaged, the MAP2K phosphorylates a TxY motif (where T is threonine, Y is tyrosine, and x is any residue) on the MAPK activation loop (A loop), allowing it to adopt an active conformation. Whereas the individual kinases and their interactions through KIM motifs have been extensively studied, the overall interactions between components of the MAP kinase cascades are not yet fully understood. In particular, how specificity is maintained between different MAPK pathways and how MAP2Ks can phosphorylate both tyrosine and threonine residues—which is highly unusual in protein kinases—is unknown.
Rationale
The interaction between the kinases needs to be transient in order to maintain signal transmission. However, this fast interaction time hinders structural studies. We used a multidisciplinary approach—including cryo–electron microscopy (cryo-EM), small-angle x-ray scattering (SAXS), enhanced sampling molecular dynamics (MD) simulations, Bayesian modeling, hydrogen-deuterium exchange mass spectrometry, and cellular assays—to characterize the complex between p38α (MAPK14) and its activating MAP2K, MKK6 (MAP2K6).
Results
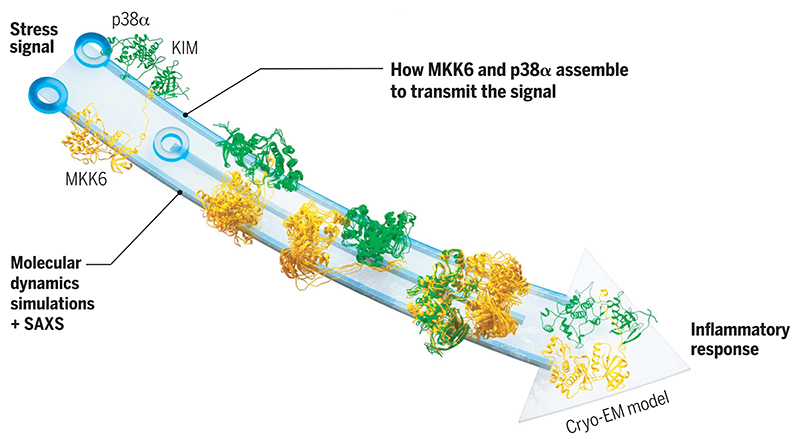
The study provides a detailed molecular model of the dynamic MKK6-p38α complex, shedding light on specificity and multistep catalysis within the MAP kinase pathway. The cryo-EM structure shows that the kinases adopt a face-to-face conformation, with all contact between the kinases distal to the MKK6 active site. The structure reveals two major contacts occurring during complex formation: The MKK6 KIM binds at the expected p38α common docking site, and the MKK6 αG helix engages the p38α C lobe at the so-called lipid-binding site that is specific to MAPKs and has been implicated in regulation of the pathway. MD simulations reveal that the observed conformation facilitates the approach of the A loop of p38α to the active site of MKK6 without compromising the dual specificity of MKK6. The simulations show that both the A-loop threonine and tyrosine can access the active site without specific recognition or binding of the A loop. Adaptive-sampling MD simulations combined with solution SAXS data in a Bayesian/maximum-entropy approach were then used to reconstruct the heterogeneous conformational ensemble and understand how the two kinases assemble and initiate phosphorylation. The populations of the main states captured in this ensemble, as well as the reaction paths connecting them, show the importance of the N terminus of MKK6 and the C lobes of the two kinases in correctly positioning them for phosphorylation. Cellular assays performed with variants of MKK6 N termini revealed that the length and structure of the N-terminal linker are important in determining specificity between MAP2K and MAPK pairs.
Conclusion
Resolving the architecture of a MAP2K activating its target MAPK has identified the interaction sites between the two kinases and allowed us to model the mechanism of activation and discover components that are important in specificity. The N termini of MAP2Ks guide the engagement of specific kinases by being tuned to the correct distance for MAP2K/MAPK pairs. Once bound, rather than acting like a classical enzyme and positioning substrates precisely for catalysis, MKK6 creates a zone of proximity enabling either the tyrosine or threonine to approach the active site, regardless of their state, allowing dual specificity. Through a comprehensive multidisciplinary approach, the study elucidated the architecture and dynamics of the formation of the MKK6-p38α complex. The findings pave the way for targeted drug development and enhance our understanding of essential steps in kinase signaling cascades.
Structure and dynamics of MAP kinase activation.
The MAP2K MKK6 binds its substrate MAPK p38α through a KIM motif and then samples many conformations before engaging at the C-terminal lobes (right). Once bound in the catalytically active conformation, the activation loop of p38α approaches the active site of MKK6, leading to phosphorylation and activation of the MAPK.
Editor’s summary.
Mitogen-activated protein kinases (MAPKs) are a key player in cellular responses to various stimuli in eukaryotes. Signaling cascades occur through a series of upstream kinases, eventually resulting in double phosphorylation of MAPK that occurs in a complex that is transient and dynamic and thus difficult to visualize by traditional structural approaches. Juyoux et al. combined cryo–electron microscopy, biophysical techniques, and molecular dynamics simulations to construct a model of the active complex between the MAPK p38# and its upstream kinase, MKK6. Based on this model, the authors discuss specific interactions, selectivity, and the overall mechanism of p38# activation. These findings will be important for researchers seeking to target MAPKs for drug development. —Michael A. Funk
Acknowledgments
We thank S. Cusack and A. McCarthy (EMBL Grenoble) for encouragement, W. Galej (EMBL Grenoble) for critical reading of the manuscript, and M.-A. Hakimi (IAB, Grenoble) for introducing us to the T. gondii protein GRA24. We acknowledge S. Schneider for her support in using the EM Facility at EMBL Grenoble and thank F. Weiss, W. Hagen, and S. Fromm for excellent data collection at the EMBL Imaging Centre. We thank A. Aubert (EMBL Grenoble) for support in eukaryotic protein expression, the proteomic core facility at EMBL Heidelberg for mass spectrometry, and U. Kapp (ESRF) for assistance in the crystallization of p38α. We acknowledge M. Bonomi (Institut Pasteur, CNRS, France) for discussions and technical support in cross-correlation analysis. K. Lindorff-Larsen, S. Bottaro, and F. Pesce are acknowledged for support in running the BME procedure. This work used the platforms of the Grenoble Instruct-ERIC center (ISBG; UAR 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB), supported by FRISBI (ANR-10-INBS-0005-02) and GRAL, and financed within the Université Grenoble Alpes graduate school (Écoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003). We acknowledge the European Synchrotron Radiation Facility for the provision of synchrotron radiation facilities, and we would like to thank the staff of the ESRF and EMBL Grenoble for assistance and support in using beamlines ID23-1 and ID23-2. SAXS data were collected at beamline B21 at Diamond Light Source, UK. Molecular graphics and analyses were performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases. We acknowledge PRACE and the Swiss National Supercomputing Centre (CSCS) for large supercomputer time allocations on Piz Daint (project IDs: pr126, s1107).
Funding
This project was funded by EMBL (M.W.B.), the Swiss National Science Foundation [project no. 200021_204795] (F.L.G.), and Bridge [project no. 40B2-0_203628] (F.L.G.). P.J. was supported by a EMBL predoctoral fellowship.
Footnotes
Author contributions: Conceptualization: P.J., E.P., F.L.G., and M.W.B. Methodology: P.J., I.G., D.G., J.v.V., M.P., and O.V. Investigation: P.J., I.G., D.G., J.v.V., O.V., M.T., F.L.G., E.P., and M.W.B. Visualization: P.J., I.G., D.G., J.v.V., and O.V. Funding acquisition: F.L.G. and M.W.B. Supervision: F.L.G., E.P., and M.W.B. Writing – original draft: – P.J. and M.W.B. Writing – review and editing: P.J., I.G., D.G., J.v.V., M.P., O.V., M.T., F.L.G., E.P., and M.W.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability
Structure of the MKK6DDGRA-p38αT180V complex: EMPIAR-11203 (raw micrographs; Electron Microscopy Public Image Archive), EMD-15233 (map; Electron Microscopy Data Bank), and PDB-8A8M [coordinates; Protein Data Bank (PDB)]. Crystal structures of apo p38α and p38αK53R: PDB-5ETC and PDB-5ETI (coordinates and structure factors; PDB). The SAXS curves and pair distribution functions are deposited in the Small Angle Scattering Biological Data Bank (access codes: SASBDB-SASDRC6 and SASBDB-SASDRD6). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE partner repository with the dataset identifier PXD040499 and 10.6019/PXD040499. The MD simulations data (trajectories, topology files, and structures of the SAXS-refined ensembles) are deposited in the Yareta repository (66).
References and Notes
- 1.Canovas B, Nebreda AR. Nat Rev Mol Cell Biol. 2021;22:346–366. doi: 10.1038/s41580-020-00322-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouhaddou M, et al. Cell. 2020;182:685–712.:e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battagello DS, et al. Clin Sci. 2020;134:2137–2160. doi: 10.1042/CS20200904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellon S, Fitzgibbon MJ, Fox T, Hsiao H-M, Wilson KP. Structure. 1999;7:1057–1065. doi: 10.1016/s0969-2126(99)80173-7. [DOI] [PubMed] [Google Scholar]
- 5.Taylor SS, Kornev AP. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blüthgen N, et al. FEBS J. 2006;273:895–906. doi: 10.1111/j.1742-4658.2006.05105.x. [DOI] [PubMed] [Google Scholar]
- 7.Ortega F, Acerenza L, Westerhoff HV, Mas F, Cascante M. Proc Natl Acad Sci USA. 2002;99:1170–1175. doi: 10.1073/pnas.022267399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thattai M, van Oudenaarden A. Biophys J. 2002;82:2943–2950. doi: 10.1016/S0006-3495(02)75635-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammaker D, Firestein GS. Ann Rheum Dis. 2010;69:i77–i82. doi: 10.1136/ard.2009.119479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson KP, et al. J Biol Chem. 1996;271:27696–27700. doi: 10.1074/jbc.271.44.27696. [DOI] [PubMed] [Google Scholar]
- 11.Xie X, et al. Structure. 1998;6:983–991. doi: 10.1016/s0969-2126(98)00100-2. [DOI] [PubMed] [Google Scholar]
- 12.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 13.Min X, et al. Structure. 2009;17:96–104. doi: 10.1016/j.str.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto T, et al. J Biochem. 2012;151:541–549. doi: 10.1093/jb/mvs023. [DOI] [PubMed] [Google Scholar]
- 15.Ohren JF, et al. Nat Struct Mol Biol. 2004;11:1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 16.Schröder M, et al. Cell Chem Biol. 2020;27:1285–1295.:e4. doi: 10.1016/j.chembiol.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Garai Á, et al. Sci Signal. 2012;5:ra74. doi: 10.1126/scisignal.2003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanoue T, Adachi M, Moriguchi T, Nishida E. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 19.Peti W, Page R. Protein Sci. 2013;22:1698–1710. doi: 10.1002/pro.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeke A, et al. Mol Syst Biol. 2015;11:837. doi: 10.15252/msb.20156269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou T, Sun L, Humphreys J, Goldsmith EJ. Structure. 2006;14:1011–1019. doi: 10.1016/j.str.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Chang CI, Xu BE, Akella R, Cobb MH, Goldsmith EJ. Mol Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 23.Dyla M, Kjaergaard M. Proc Natl Acad Sci USA. 2020;117:21413–21419. doi: 10.1073/pnas.2006382117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beenstock J, Mooshayef N, Engelberg D. Trends Biochem Sci. 2016;41:938–953. doi: 10.1016/j.tibs.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Park E, et al. Nature. 2019;575:545–550. doi: 10.1038/s41586-019-1660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondo Y, et al. Science. 2019;366:109–115. doi: 10.1126/science.aay0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennan DF, et al. Nature. 2011;472:366–369. doi: 10.1038/nature09860. [DOI] [PubMed] [Google Scholar]
- 28.Sok P, et al. Structure. 2020;28:1101–1113.:e5. doi: 10.1016/j.str.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haar ET, Prabakhar P, Liu X, Lepre C. J Biol Chem. 2007;282:9733–9739. doi: 10.1074/jbc.M611165200. [DOI] [PubMed] [Google Scholar]
- 30.Braun L, et al. J Exp Med. 2013;210:2071–2086. doi: 10.1084/jem.20130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellegrini E, et al. Structure. 2017;25:16–26. doi: 10.1016/j.str.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bepler T, et al. Nat Methods. 2019;16:1153–1160. doi: 10.1038/s41592-019-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modi V, Dunbrack RL., Jr Proc Natl Acad Sci USA. 2019;116:6818–6827. doi: 10.1073/pnas.1814279116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans R, et al. bioRxiv. 2022:2021.10.04.463034. [Preprint] [Google Scholar]
- 35.Diskin R, Engelberg D, Livnah O. J Mol Biol. 2008;375:70–79. doi: 10.1016/j.jmb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Tzarum N, Eisenberg-Domovich Y, Gills JJ, Dennis PA, Livnah O. J Mol Biol. 2012;424:339–353. doi: 10.1016/j.jmb.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Bührmann M, Hardick J, Weisner J, Quambusch L, Rauh D. Angew Chem Int Ed. 2017;56:13232–13236. doi: 10.1002/anie.201706345. [DOI] [PubMed] [Google Scholar]
- 38.Bührmann M, et al. PLOS ONE. 2017;12:e0184627. doi: 10.1371/journal.pone.0184627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Comess KM, et al. ACS Chem Biol. 2011;6:234–244. doi: 10.1021/cb1002619. [DOI] [PubMed] [Google Scholar]
- 40.Laufkötter O, Hu H, Miljković F, Bajorath J. J Med Chem. 2022;65:922–934. doi: 10.1021/acs.jmedchem.0c02076. [DOI] [PubMed] [Google Scholar]
- 41.Zhang XH, et al. FEBS Lett. 2021;595:2570–2592. doi: 10.1002/1873-3468.14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirsch K, et al. Nat Commun. 2020;11:5769. doi: 10.1038/s41467-020-19582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, et al. Nat Commun. 2016;7:10879. doi: 10.1038/ncomms10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.James EI, Murphree TA, Vorauer C, Engen JR, Guttman M. Chem Rev. 2022;122:7562–7623. doi: 10.1021/acs.chemrev.1c00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foster SA, et al. Cancer Cell. 2016;29:477–493. doi: 10.1016/j.ccell.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Zhang B, et al. Mol Genet Genomic Med. 2020;8:e1395. doi: 10.1002/mgg3.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pérez-Gallegos A, Garcia-Viloca M, González-Lafont À, Lluch JM. Phys Chem Chem Phys. 2015;17:3497–3511. doi: 10.1039/c4cp03579h. [DOI] [PubMed] [Google Scholar]
- 48.Kuzmanic A, et al. eLife. 2017;6:e22175. doi: 10.7554/eLife.22175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doerr S, De Fabritiis G. J Chem Theory Comput. 2014;10:2064–2069. doi: 10.1021/ct400919u. [DOI] [PubMed] [Google Scholar]
- 50.Plattner N, Doerr S, De Fabritiis G, Noé F. Nat Chem. 2017;9:1005–1011. doi: 10.1038/nchem.2785. [DOI] [PubMed] [Google Scholar]
- 51.Pesce F, Lindorff-Larsen K. Biophys J. 2021;120:5124–5135. doi: 10.1016/j.bpj.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larsen AH, et al. PLOS Comput Biol. 2020;16:e1007870. doi: 10.1371/journal.pcbi.1007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orioli S, Larsen AH, Bottaro S, Lindorff-Larsen K. Prog Mol Biol Transl Sci. 2020;170:123–176. doi: 10.1016/bs.pmbts.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 54.González-Foutel NS, et al. Nat Struct Mol Biol. 2022;29:781–790. doi: 10.1038/s41594-022-00811-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fleming Y, et al. Biochem J. 2000;352:145–154. [PMC free article] [PubMed] [Google Scholar]
- 56.Tournier C, Whitmarsh AJ, Cavanagh J, Barrett T, Davis RJ. Mol Cell Biol. 1999;19:1569–1581. doi: 10.1128/mcb.19.2.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischmann TO, et al. Biochemistry. 2009;48:2661–2674. doi: 10.1021/bi801898e. [DOI] [PubMed] [Google Scholar]
- 58.Modi V, Dunbrack RL., Jr Sci Rep. 2019;9:19790. doi: 10.1038/s41598-019-56499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang YY, Wu JW, Wang ZX. Sci Signal. 2011;4:ra88. doi: 10.1126/scisignal.2002241. [DOI] [PubMed] [Google Scholar]
- 60.Dominguez C, Powers DA, Tamayo N. Curr Opin Drug Discov Devel. 2005;8:421–430. [PubMed] [Google Scholar]
- 61.Wang YL, et al. FEBS J. 2019;286:1030–1052. doi: 10.1111/febs.14762. [DOI] [PubMed] [Google Scholar]
- 62.Humphreys JM, Piala AT, Akella R, He H, Goldsmith EJ. J Biol Chem. 2013;288:23322–23330. doi: 10.1074/jbc.M113.462101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aoki K, Takahashi K, Kaizu K, Matsuda M. Sci Rep. 2013;3:1541. doi: 10.1038/srep01541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salazar C, Höfer T. FEBS J. 2009;276:3177–3198. doi: 10.1111/j.1742-4658.2009.07027.x. [DOI] [PubMed] [Google Scholar]
- 65.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 66.Gervasio FL, Galdadas I, Gobbo D. Architecture of the MKK6-p38α complex defines the basis of MAPK specificity and activation, Yareta. 2023 doi: 10.26037/yareta:jmnbt6lzkrfrpdcf3m3343smxq. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure of the MKK6DDGRA-p38αT180V complex: EMPIAR-11203 (raw micrographs; Electron Microscopy Public Image Archive), EMD-15233 (map; Electron Microscopy Data Bank), and PDB-8A8M [coordinates; Protein Data Bank (PDB)]. Crystal structures of apo p38α and p38αK53R: PDB-5ETC and PDB-5ETI (coordinates and structure factors; PDB). The SAXS curves and pair distribution functions are deposited in the Small Angle Scattering Biological Data Bank (access codes: SASBDB-SASDRC6 and SASBDB-SASDRD6). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE partner repository with the dataset identifier PXD040499 and 10.6019/PXD040499. The MD simulations data (trajectories, topology files, and structures of the SAXS-refined ensembles) are deposited in the Yareta repository (66).