Summary
Signal transduction pathways stimulated by secreted growth factors are tightly regulated at multiple levels between the cell surface and the nucleus. The trafficking of cell surface receptors is emerging as a key step for regulating appropriate cellular responses, with perturbations in this process contributing to human diseases, including cancer. For receptors recognizing ligands of the transforming growth factor β (TGF-β) family, little is known about how trafficking is regulated or how this shapes signaling dynamics. Here, using whole genome small interfering RNA (siRNA) screens, we have identified the ESCRT (endosomal sorting complex required for transport) machinery as a crucial determinant of signal duration. Downregulation of ESCRT components increases the outputs of TGF-β signaling and sensitizes cells to low doses of ligand in their microenvironment. This sensitization drives an epithelial-to-mesenchymal transition (EMT) in response to low doses of ligand, and we demonstrate a link between downregulation of the ESCRT machinery and cancer survival.
Graphic abstract.
Introduction
Cell communication, mediated via signal transduction pathways, underpins both embryonic development and adult tissue homeostasis, and deregulation of these pathways is the underlying cause of many human diseases, a prominent example being cancer (Weber et al., 2016). Cell signaling is initiated by the binding of cytokines and growth factors to cell surface receptors that then transmit signals to the nucleus to induce new programs of gene expression that result in changes in cell behavior. Intracellular trafficking and turnover of the cell surface receptors are key determinants of the amplitude of the response and dictate the manner in which the signal is modulated over time (Le Roy and Wrana, 2005; Schmid, 2017; Zwang and Yarden, 2009). For ligands of the transforming growth factor β (TGF-β) family, signaling dynamics have been shown to be crucial for determining how cells respond to ligand stimulation (Nicolás and Hill, 2003; Vizán et al., 2013), but we know little about how TGF-β receptor trafficking or turnover is regulated.
Members of the TGF-β family regulate a wide range of cellular processes including cell growth, migration, adhesion, differentiation, and apoptosis, and as a result, are essential for orchestrating embryonic development and regulating tissue homeostasis (Miller and Hill, 2016). The TGF-β family ligands activate signaling by binding to serine/threonine kinase receptors at the cell surface. These receptors are functionally categorized into two classes, type I and type II, and the canonical TGF-β ligands (TGF-β1, TGF-β2, and TGF-β3) use TGFBR1 and TGFBR2 as their type I receptor and type II receptor, respectively (Feng and Derynck, 2005). The ligand brings the two types of receptors together, which allows the constitutively active kinase domain of the type II receptor to phosphorylate an intracellular glycine- and serinerich (GS) domain of the type I receptor (Feng and Derynck, 2005). This activates the type I receptor and provides a binding site for the downstream substrates of the pathway, the receptor-regulated SMADs (R-SMADs), which are phosphorylated at their extreme C termini. Phosphorylated SMADs (PSMADs) form complexes with SMAD4, which accumulate in the nucleus and regulate the transcription of a battery of genes in conjunction with other co-factors. SMAD phosphorylation is therefore a direct readout of receptor activity (Massagué, 2012). The family has traditionally been divided into two branches, whereby the TGF-βs, activins, and NODAL induce SMAD2 and SMAD3 phosphorylation, and the bone morphogenetic proteins (BMPs) and growth and differentiation factors (GDFs) induce SMAD1, SMAD5, and SMAD9 phosphorylation, although some cross-talk between these branches also occurs (Grönroos et al., 2012; Ramachandran et al., 2018).
The TGF-β receptors are localized to the basolateral membranes in polarized cells, and short sequences have been identified in both TGFBR1 and TGFBR2 that are responsible for this targeting (Yin et al., 2013, 2017). Moreover, both receptors are constantly internalized and recycled in the absence and presence of ligand (Di Guglielmo et al., 2003; Mitchell et al., 2004). However, their route of internalization remains controversial. One study reported that receptors internalize uniquely into clathrin-coated pits and subsequently into EEA1-positive endosomes for active signaling and via caveolin-marked vesicles for degradation (Di Guglielmo et al., 2003). Others have suggested that while SMAD signaling occurs in clathrin-coated pits and in clathrin-mediated endosomal compartments, non-SMAD signaling, such as TGF-β-induced activation of ERK MAPK signaling and PI3K-AKT signaling occurs uniquely in the caveolar compartment (Budi et al., 2017). Interestingly, recent work has indicated that the two routes of internalization may merge, since TGF-β receptors were tracked to endosomal vesicles that were positive for both EEA1 and caveolin (He et al., 2015). It also remains unclear whether internalization is absolutely necessary for signaling to occur, with different cell lines and methods of tagging receptors leading to conflicting conclusions (Budi et al., 2017; Di Guglielmo et al., 2003; Lu et al., 2002).
Once internalized, a proportion of receptors appear to be re-cycled to the cell surface in a RAB11-dependent manner, while the remainder are ubiquitinated and targeted for degradation (Mitchell et al., 2004). A number of E3 ubiquitin ligases have been identified as playing a role in this process including SMURF1 (Kavsak et al., 2000), SMURF2 (Ogunjimi et al., 2005), WWP1 (Komuro et al., 2004), and NEDD4L (Kuratomi et al., 2005). Deubiquitinating enzymes have also been identified that reverse this process and increase receptor stability, such as USP15 (Eichhorn et al., 2012), UCHL5 (UCH37) (Wicks et al., 2005), and USP4 (Zhang et al., 2012). Mathematical modeling with experimental validation has demonstrated that the lifetime of activated receptors dictates how TGF-β signaling intensity is modulated over time and thus how cells respond to continuous ligand stimulation (Vizán et al., 2013). However, the pathway of degradation subsequent to ubiquitination for the TGF-β receptors has never been identified.
Derailed endocytosis is emerging as a driver in a number of human diseases, with cancer being the most prominent (Mellman and Yarden, 2013; Mosesson et al., 2008). Multiple stages in internalization, trafficking, recycling, and degradation of cell surface receptors can become deregulated during the process of carcinogenesis, leading to aberrant signaling that can promote tumor growth and dissemination (Tomas et al., 2014). Therefore, understanding how perturbations in receptor trafficking alter signaling, and how the downstream effects that result from these alterations change cell behavior in response to external factors is critical for understanding the link between receptor trafficking and disease states. TGF-β signaling mediates a range of tumor-suppressive effects including growth arrest and apoptosis but also plays an important tumor-promoting role, acting as an immune suppressor and a pro-angiogenic factor. In addition, by inducing an epithelial-to-mesenchymal transition (EMT) in tumor cells, TGF-β can promote invasion and metastasis, among other effects (David and Massagué, 2018).
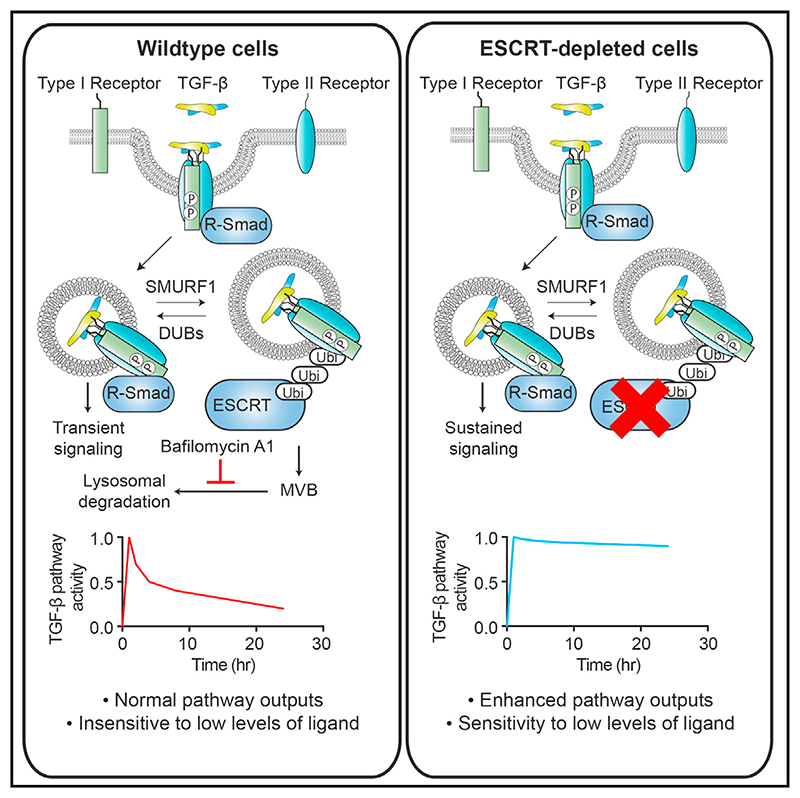
In this study, we performed a series of whole genome loss-of-function screens to find regulators of TGF-β signaling duration. These screens identified the ESCRT (endosomal sorting complexes required for transport) pathway as the degradation route of the TGF-β receptors and hence essential for limiting signal duration. Knockdown of ESCRT components led to persistent pathway activity as measured by R-SMAD phosphorylation, as well as to upregulation of the downstream outputs of TGF-β signaling, including its ability to drive growth arrest and EMT. Furthermore, these results demonstrate that downregulation of ESCRT components sensitizes cells to TGF-β in their extracellular environment, which we reveal could be important in tumorigenesis.
Results
Whole Genome Screening Identifies ESCRT Components as Key Regulators of TGF-β Signaling Dynamics
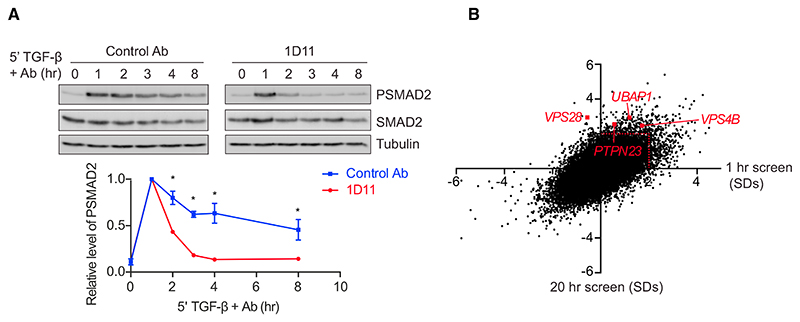
Upon acute exposure of cells to TGF-β, levels of PSMAD2 increase for around 1 hr before attenuating to lower levels at later time points (Figure S1A). These signaling dynamics are seen with a range of TGF-β concentrations, and we have subsequently used 2 ng/mL for our experiments as an example of a saturating dose. These characteristic signaling dynamics are the result of the TGF-β receptors becoming depleted from the cell surface within 5–10 min of ligand exposure, rendering the cells refractory to further acute stimulation (Vizán et al., 2013). We first sought to understand whether the internalized receptors continue to signal in these conditions from intracellular compartments and used the well-characterized human keratinocyte cell line, HaCaT, for these experiments (Vizán et al., 2013). HaCaT cells were treated for 5 min with TGF-β to internalize receptors, then external ligand was removed with a TGF-β neutralizing antibody (1D11) or cells were incubated with an isotype-matched control antibody (Nam et al., 2008). With the neutralizing antibody, signaling persisted for between 1 and 2 hr, despite no new signaling being initiated from the cell surface (Figure 1A). This indicates that receptors signal for 1–2 hr from internal compartments.
Figure 1. A Whole Genome Screen for Regulators of TGF-β Signaling Dynamics.
(A) HaCaT cells were treated for 5 min with TGF-β, followed by the TGF-β neutralizing antibody 1D11 or an isotype-matched control (Control Ab) for the times indicated. Levels of PSMAD2, SMAD2, and tubulin as a loading control were assayed by western blotting. Quantifications are the normalized average ± SD of three independent experiments. *p < 0.05.
(B) A scatterplot showing changes in levels of nuclear SMAD2/3 for every hit in a whole genome siRNA screen analyzed after 1 hr (x axis) or 20 hr (y axis), expressed as SDs from the average of the screen. The red dotted line indicates 2 SDs, the significance threshold for these screens. Four components of the ESCRT machinery, VPS28, UBAP1, VPS4B, and PTPN23 are indicated.
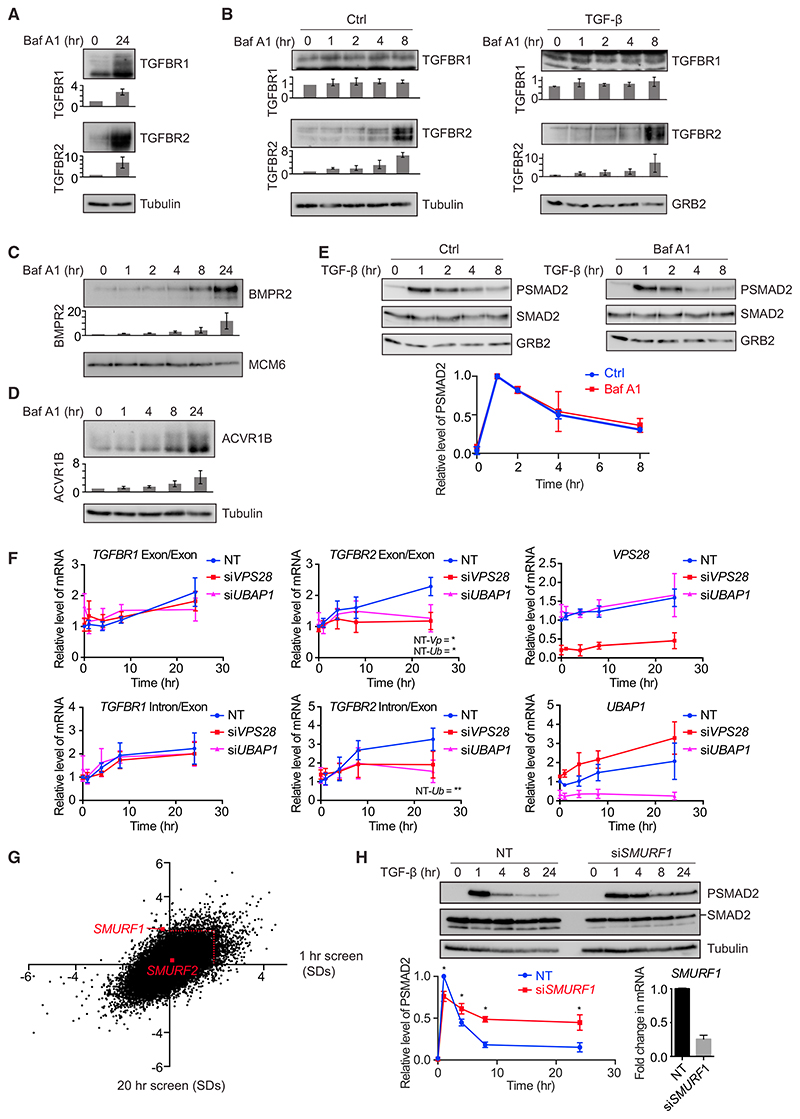
To determine how receptor trafficking is controlled, we performed a series of whole genome small interfering RNA (siRNA) screens for regulators of TGF-β signaling. We were particularly interested in identifying regulators that changed the dynamics of the TGF-β response. HaCaT cells were transfected with SMARTpools comprising four individual siRNA oligonucleotides targeting each gene in the human genome. Screens were performed after two different time points of TGF-β treatment, 1 hr and 20 hr, and the accumulation of endogenous SMAD2/3 in the nucleus was assayed by immunofluorescence using automated microscopy to capture images. We confirmed that SMAD2/3 accumulated in the nucleus in response to TGF-β after 1 hr and this response was attenuated after 20 hr (Figure S1B). As a positive control, we showed that nuclear SMAD2/3 accumulation was inhibited by knockdown of the type I TGF-β receptor, TGFBR1, at both the 1 hr and 20 hr time points (Figure S1B). siRNAs that led to a significant decrease in cell number (>2 SDs from the screen average) were excluded from further analysis. We chose to focus on hits whose knockdown led to normal induction of signaling after 1 hr, but to a persistence of signaling over 20 hr, to identify components that potentially regulated the trafficking of activated receptors (Figure S1C). A significant hit was considered one that exhibited an increase in nuclear SMAD2/3 levels of >2 SDs relative to the average of the screen after 20 hr of TGF-β treatment, with no significant change in nuclear SMAD2/3 after 1 hr of TGF-β treatment (Figure 1B). For a full list of hits, see Table S1. Among the hits in this category were several components of the ESCRT machinery, namely VPS28, UBAP1, PTPN23, and VPS4B (Figures 1B and S1D). The ESCRT machinery is composed of four protein assemblies (ESCRT-0, -I, -II, and –III). The ESCRT-0 and -I complexes, with the help of ESCRT-II, recognizes ubiquitinated cargo, which is then incorporated by ESCRT-III into intra-luminal vesicles (ILVs) by invagination of the endosomal limiting membrane (Clague et al., 2012; Schmidt and Teis, 2012; Szymanska et al., 2018; Wollert and Hurley, 2010). This leads to the generation of the multi-vesicular body (MVB), a structure that fuses with lysosomes to degrade cargo (Futter et al., 1996). VPS28 and UBAP1 are members of ESCRT-I, PTPN23 is an accessory protein that links ESCRT-I to ESCRT-III, and VPS4B associates with ESCRT-III and is required for ESCRT disassembly and recycling (Ali et al., 2013; Lata et al., 2008; Szymanska et al., 2018).
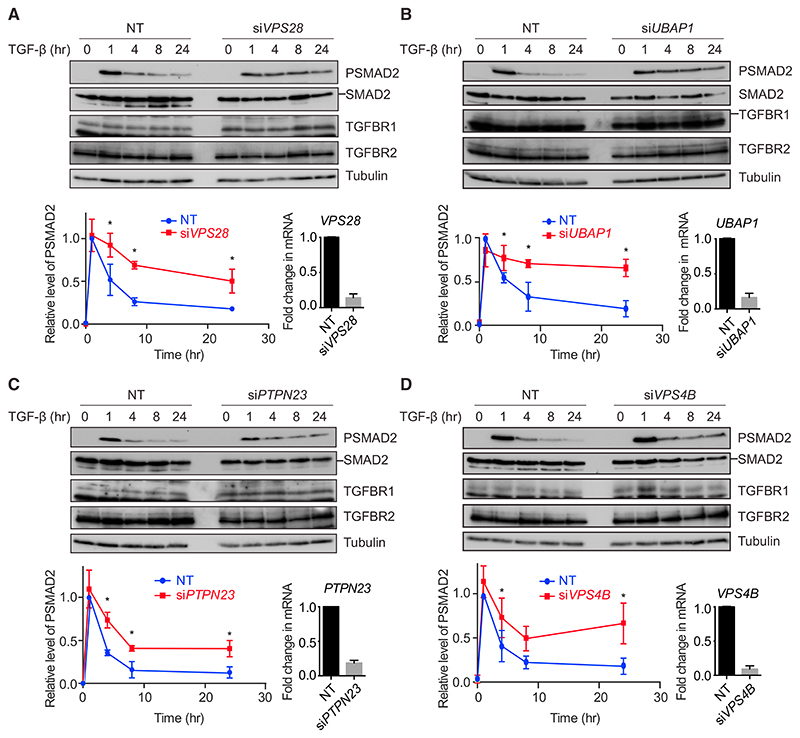
To determine the impact of knocking down these components on signaling dynamics, time courses of TGF-β treatment were performed in HaCaT cells in the presence or absence of siRNAs against these targets. Knocking down VPS28, UBAP1, PTPN23, or VPS4B led to a persistence in SMAD2 phosphorylation over a time course of 24 hr, compared to the attenuation in signal seen with non-targeting (NT) siRNA controls (Figure 2). Crucially, the acute induction of PSMAD2 was unaffected by ESCRT knockdown, suggesting that the major impact of losing ESCRT components is on signaling duration. In all cases, no significant upregulation in total levels of the TGF-β receptors TGFBR1 and TGFBR2 was seen (Figure 2). To ensure that siRNA pools were not dominated by individual oligonucleotides, pools were deconvoluted, and we demonstrated that each individual member of the pools targeting, for example, VPS28 and UBAP1, had the same impact on TGF-β signaling (Figures S2A and S2B). Knockdown of ESCRT components also led to a persistence in SMAD3 phosphorylation after TGF-β treatment (Figures S2C and S2D). Importantly, upregulation of SMAD2 phosphorylation upon TGF-β stimulation with ESCRT knockdown occurred at a range of TGF-β doses from 0.1 ng/mL to 2 ng/mL, and ESCRT knockdown additionally clearly increased basal levels of PSMAD2 (Figures S2E and S2F).
Figure 2. Knockdown of ESCRT Components Leads to a Persistence in SMAD2 Phosphorylation upon TGF-β Treatment.
(A–D) HaCaT cells were transfected with non-targeting (NT) control siRNAs or siRNAs targeting VPS28 (A), UBAP1 (B), PTPN23 (C), or VPS4B (D)and stimulated with TGF-β for the times indicated. Levels of PSMAD2, SMAD2/3, TGFBR1, TGFBR2, and tubulin as a loading control were assayed by western blot. Quantifications are the normalized average ± SD of three independent experiments. *p < 0.05. The extent of knockdown was assessed by qPCR, and the normalized average ± SD from the same three independent experiments is shown bottom right.
The ESCRT Machinery Determines the Dynamics of BMP and Activin Signaling
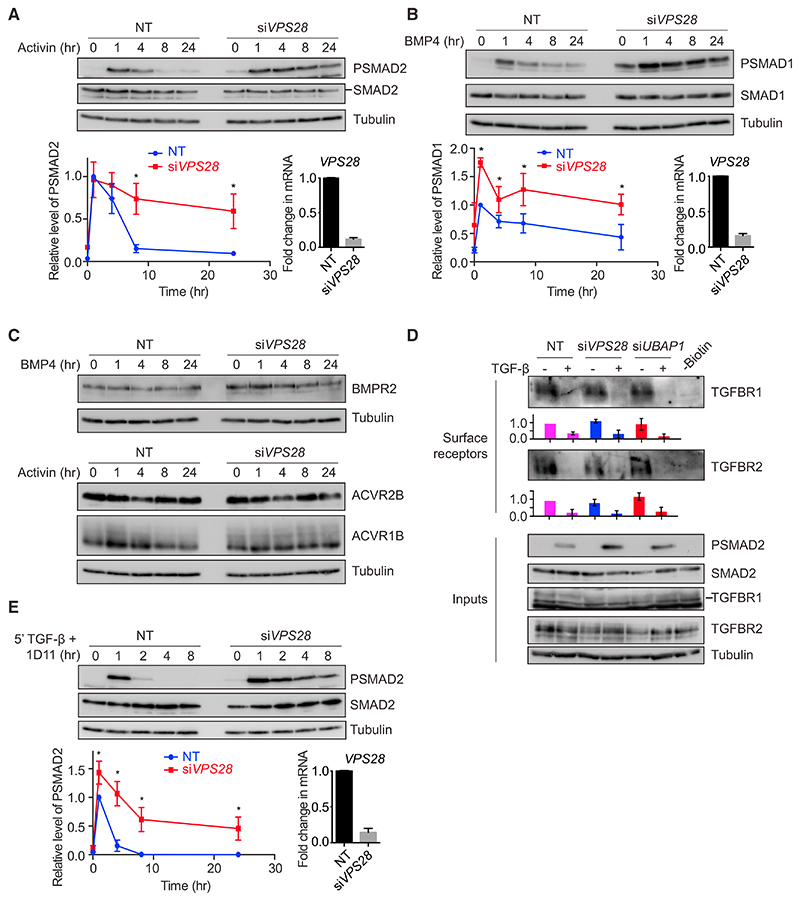
We next extended our findings to signaling by other members of the TGF-β family. Stimulation of HaCaTs with activin led to transient SMAD2 phosphorylation, which peaked after 1 hr before rapidly declining over the next 8 hr (Figure 3A). With knockdown of VPS28, this response became persistent, indicating that the ESCRT machinery also shapes the dynamics of the cellular response to activin. In contrast to the response to activin and TGF-β, exposure of HaCaTs to BMP4 led to persistent SMAD1 phosphorylation (Figure 3B). When VPS28 was knocked down, this response became elevated across the entire time course of BMP4 stimulation, including the baseline. In both cases, as with TGF-β, the levels of receptors were not changed upon ESCRT knockdown (Figure 3C).
Figure 3. The Effects of ESCRT Knockdown Are Not Limited to the TGF-β Pathway and Result in Sustained Signaling from Internal Compartments.
(A–C) HaCaT cells were transfected with non-targeting (NT) control siRNAs or siRNAs targeting VPS28 and stimulated with activin (A and C) or BMP4 (B and C) for the times indicated. Levels of PSMAD2, SMAD2/3, PSMAD1, SMAD1, BMPR2, ACVR1B, ACVR2B, and tubulin as a loading control were assayed by western blot. Quantifications are the normalized average ± SD of three independent experiments. Quantifications are normalized to NT untreated samples. *p < 0.05. The extent of knockdown was assessed by qPCR, and the normalized average ± SD from the same three independent experiments is shown bottom right.
(D) HaCaT cells were transfected with NT control siRNAs or siRNAs targeting VPS28 or UBAP1 and stimulated with TGF-β as indicated for 4 hr. A surface biotinylation assay was performed, and surface levels of TGFBR1 or TGFBR2, or total levels of TGFBR1, TGFBR2, PSMAD2, SMAD2/3, or tubulin as a loading control were assayed by western blot. Quantifications are the average ± SD of four independent experiments and are normalized to untreated NT samples.
(E) HaCaT cells transfected with NT siRNAs or siRNAs targeting VPS28 were treated for 5 min with TGF-β, followed by the neutralizing antibody 1D11 for the times indicated. Levels of PSMAD2, SMAD2/3, and tubulin as a loading control were assayed by western blot. Quantifications and knockdown controls were as in (A) and (B).
These results demonstrate that the ESCRT machinery limits the amplitude and duration of signaling in response to multiple TGF-β family ligands.
Depletion of the ESCRT Machinery Traps Activated Receptors in Internal Compartments
As mentioned above, the ESCRT machinery functions to target ubiquitinated cell surface receptors for degradation (Szymanska et al., 2018). We therefore investigated whether inhibition of this pathway blocked the internalization of the TGF-β receptors from the cell surface or serves to trap activated receptors in intracellular compartments. TGFBR1 and TGFBR2 are both almost completely depleted from the surface of HaCaTs after 4 hr of exposure to TGF-β (Vizán et al., 2013). This depletion was not inhibited by knockdown of VPS28 or UBAP1, suggesting that receptor internalization is not affected in these conditions (Figure 3D). As previously indicated, when cells were stimulated for 5 min with TGF-β, followed by a neutralizing antibody over a time course of 8 hr, receptors continued to signal from internal compartments for 1–2 hr (Figure 3E). After VPS28 knockdown, signaling persisted from internal compartments for the entire duration of an 8-hr time course, suggesting that activated signaling receptors were indeed trapped in internal compartments (Figure 3E). To address this more directly, we looked for co-localization between endosomal markers and TGFBR1. As there are no receptor antibodies suitable for immunostaining, we made use of a cell line stably expressing HA-tagged TGFBR1 (Figure S3). The type I receptor clearly co-localized with endogenous EEA1, which marks early endosomes, and with the ESCRT-0 component HGS (also called HRS) (Figures S3B and S3C). Quantitation revealed that approximately one-third of HA-marked puncta co-localized with endosome/ESCRT components in both the absence and presence of TGF-β (Figure S3D). We also attempted to look at co-localization between endosomal markers and TGFBR1 when ESCRT components were knocked down. However, in these conditions, we observed an expansion of the endosomes, as has been seen by others (Doyotte et al., 2005), making any changes in co-localization difficult to interpret (data not shown).
TGF-β Receptors Are Degraded via the Lysosome and This Requires SMURF1
The results so far suggest that the TGF-β receptors are targeted for degradation by the ESCRT machinery. Cargo recognized by ESCRT components is degraded in the lysosome. To investigate whether this was true of the TGF-β receptors, cells were treated with bafilomycin A1, which inhibits the fusion of MVBs with the lysosome (Bowman et al., 1988). Cellular pools of both TGFBR1 and TGFBR2 accumulated after 24 hr of bafilomycin A1 treatment, demonstrating that both receptors are indeed degraded in the lysosome (Figure 4A). Interestingly, TGFBR2 accumulated after only 8 hr of bafilomycin treatment, while TGFBR1 required up to 24 hr before accumulation was detectable, and this occurred both in the absence or presence of TGF-β (Figures 4A and 4B). Consistent with our finding that activin and BMP signaling was also regulated by the ESCRT machinery, we observed that the BMP type II receptor BMPR2 accumulated with bafilomycin A1 treatment, as did the activin type I receptor ACVR1B (Figures 4C and 4D). Receptors in MVBs generally do not contact their cytoplasmic substrates (Tomas et al., 2014), and consistent with this, the attenuation in PSMAD2 levels seen in response to TGF-β was not affected by bafilomycin A1 treatment (Figure 4E).
Figure 4. ESCRT Components Are Responsible for Trafficking the Receptors to MVBs.
(A–D) HaCaT cells were treated with bafilomycin A1 (Baf A1) and/or TGF-β for the times indicated. Levels of TGFBR1, TGFBR2 (A and B), BMPR2 (C), or ACVR1B
(D) were assayed by western blot together with a loading control (tubulin, GRB2, or MCM6). Quantifications are the average ± SD of three independent experiments, normalized to untreated samples.
(E) HaCaT cells were treated or not with bafilomycin A1 and TGF-β for the times indicated. Levels of PSMAD2, SMAD2/3, or GRB2 as a loading control were assayed by western blot. Quantifications are the normalized average ± SD of three independent experiments.
(F) HaCaT cells were transfected with non-targeting (NT) control siRNAs or siRNAs targeting VPS28 or UBAP1 and stimulated with TGF-β for the times indicated. Expression levels of the genes indicated were assayed by qPCR and are the average ± SD of three independent experiments, normalized to levels in the NT untreated sample. Exon/Exon indicates mature mRNA; Intron/Exon indicates nascent mRNA. NT-Vp indicates the p value when values are compared between NT and siVPS28 samples after 24 hr of treatment, and NT-Ub indicates the same for NT and siUBAP1. *p < 0.05, **p < 0.005.
(G) A scatterplot showing changes in levels of nuclear SMAD2/3 for every hit in a whole genome siRNA screen analyzed after 1 hr (x axis) or 20 hr (y axis), expressed as SDs from the average of the screen. The red dotted line indicates 2 SDs, the significance threshold for these screens. SMURF1 and SMURF2 are indicated.
(H) HaCaT cells were transfected with non-targeting (NT) control siRNAs or siRNAs targeting SMURF1 and stimulated with TGF-β for the times indicated. Levels of PSMAD2, SMAD2/3, and tubulin as a loading control were assayed by western blot. Quantifications are the normalized average ± SD of three independent experiments. *p < 0.05. The extent of knockdown was assessed by qPCR, and the normalized average ± SD from the same three independent experiments is shown bottom right.
Both bafilomycin A1 treatment and knockdown of the ESCRT machinery prevent trafficking of the receptors to the lysosomes, but there are key differences in the effects these treatments have on the signaling pathway and receptor levels. In the case of bafilomycin A1, receptors accumulate in the MVBs, most rapidly in the case of TGFBR2, but signaling is not sustained. When ESCRT components are knocked down, however, signaling is sustained, indicating that in this case the receptors are trapped in an internal compartment upstream of the MVB, likely the signaling endosome. This led us to ask why we could not detect receptor accumulation when ESCRT components were depleted. In wild-type cells, we have previously noted that receptor degradation rates are matched by synthesis rates, keeping levels of receptors stable (Vizán et al., 2013), and suggesting that a feedback mechanism may be operating. We hypothesized therefore that when the ESCRT machinery is depleted such a feedback mechanism might result in reduced rates of receptor synthesis if receptor degradation was inhibited. We would expect this to be most obvious for TGFBR2, as it is turned over much faster than TGFBR1 (Vizán et al., 2013). Consistent with this idea, we demonstrated, using a cycloheximide chase assay, that VPS28 knockdown resulted in increased stability of both TGFBR1 and TGFBR2 in unstimulated cells (Figure S4A). Moreover, VPS28 or UBAP1 knockdown also resulted in a decreased rate of accumulation of TGFBR2, as read out by both nascent and mature mRNA levels (Figure 4F).
Ubiquitination controls the trafficking routes of activated receptors after internalization and may govern the efficiency of recycling from endosomes to the plasma membrane versus lysosomal sorting through the MVB pathway (Clague et al., 2012). A number of E3 ubiquitin ligases have previously been demonstrated to be capable of ubiquitylating the TGF-β receptors, but definitive proof as to which is responsible for targeting the receptors for degradation has been lacking (Budi et al., 2017). We therefore examined our screen hits and searched for siRNAs targeting E3 ubiquitin ligases whose activity led to significantly prolonged signaling compared with the control NT siRNAs. SMURF1, an E3 ubiquitin ligase that has been associated with TGF-β receptor ubiquitination, was one of the most significant hits from the screen, while the closely related E3 ubiquitin ligase, SMURF2, which has also been suggested to play the same role (Kavsak et al., 2000), had no effect on the dynamics of SMAD2/3 nuclear localization (Figure 4G). This result was confirmed by examining images from the screen (Figure S4B) and performing time courses of SMAD2 phosphorylation in the absence and presence of SMURF1 or SMURF2 knockdown (Figures 4H and S4C–S4E). Furthermore, we showed that knockdown of SMURF1 also stabilized TGFBR1 and TGFBR2 in a cycloheximide chase assay (Figure S4F).
Prolonged TGF-β Signaling Results in Enhanced Target Gene Expression
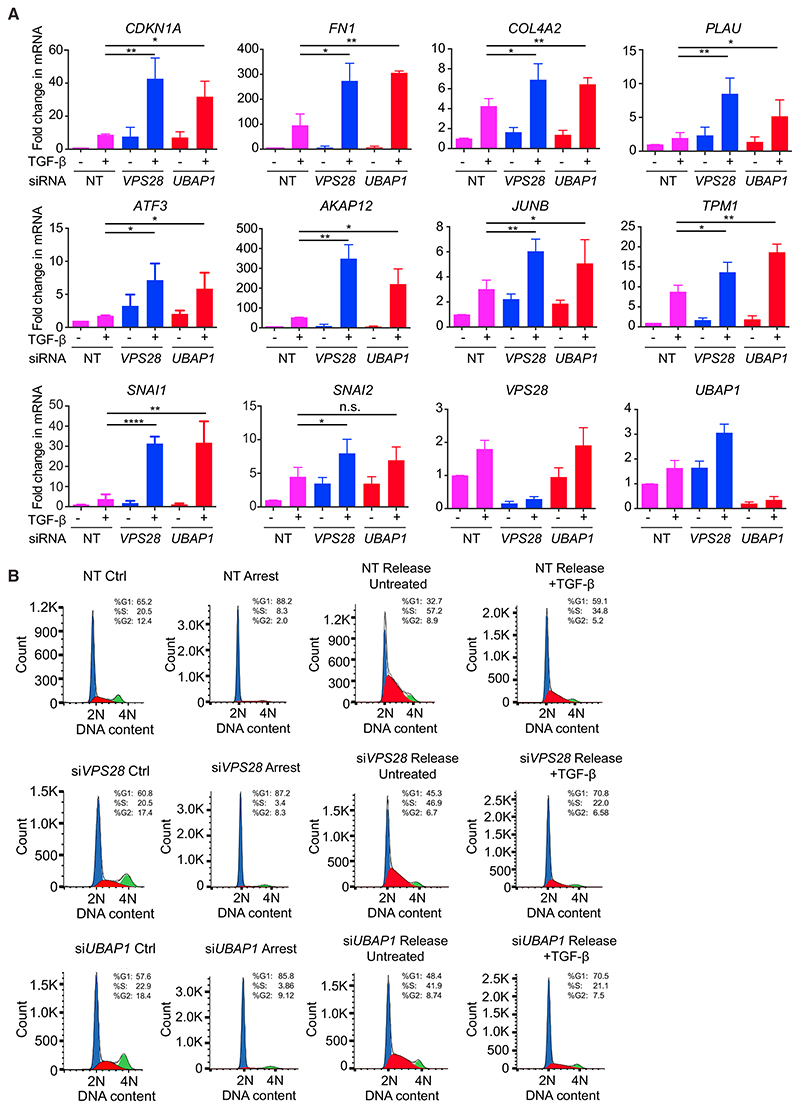
We have clearly demonstrated that knockdown of components of the ESCRT machinery prolongs the cellular response to TGF-β stimulation by blocking the degradation in the lysosome of actively signaling receptors. We next investigated whether this persistence in SMAD2 and SMAD3 phosphorylation is translated into an enhancement of the downstream biological outputs of TGF-β signaling. TGF-β drives the expression of a plethora of target genes, with varying dynamic patterns. Knockdown of VPS28 or UBAP1 led to a dramatic upregulation of the expression of a range of target genes in response to 24 hr of TGF-β stimulation (Figure 5A). These included several genes encoding transcriptional regulators (JUNB, ATF3), proteins responsible for driving the deposition, and degradation of extracellular matrix (ECM) components (PLAU, COL4A2, FN1), as well as proteins involved in limiting cell proliferation (CDKN1A). Finally, genes encoding drivers of TGF-β-mediated EMT, such as SNAI1 (previously called SNAIL) and SNAI2 (previously called SLUG), were also upregulated upon TGF-β stimulation when VPS28 or UBAP1 were knocked down as compared to controls. By contrast, the expression of some target genes was not altered by VPS28 or UBAP1 knockdown (Figure S5A). This is not entirely unexpected, as we have recently demonstrated that neither SMAD2 phosphorylation nor PSMAD2 chromatin association directly correlates with the kinetics of target gene expression (Coda et al., 2017).
Figure 5. A Failure to Attenuate SMAD2 Phosphorylation Results in the Upregulation of Downstream TGF-β Responses.
(A) HaCaT cells were transfected with non-targeting (NT) control siRNAs, siRNAs targeting VPS28 or UBAP1, and stimulated with TGF-β for 24 hr. Expression levels of the genes indicated were assayed by qPCR and are the average ± SD of three independent experiments, normalized to levels in the NT untreated sample. *p < 0.05, **p < 0.005, ****p < 0.00005. n.s., not significant.
(B) HaCaT cells were transfected with non-targeting (NT) control siRNAs, or siRNAs targeting VPS28 or UBAP1. They were serum starved for 24 hr to arrest the cell cycle (Arrest), then transferred into full serum media for 20 hr in the absence (Release Ctrl) or presence (Release +TGF-β) of 0.5 ng/mL TGF-β. Control (Ctrl) cells were siRNA-transfected cells growing in full media. After treatments, cells were fixed, cell-cycle analysis was performed, and cells were assigned to a phase of the cell cycle. Blue, G1; red, S; green, G2/M. % of cells in each phase of the cell cycle is given. The x axis indicates DNA content measured by propidium iodide fluorescence.
Prolonged TGF-β Signaling Results in Enhanced TGF-β Responses
To confirm that an upregulation in gene expression translates into alterations in cell behavior, we first performed cell-cycle analysis in HaCaT cells. The release of HaCaT cells arrested in G0 phase by serum starvation is potently inhibited by 2 ng/mL TGF-β (Levy and Hill, 2005). A lower dose of TGF-β (0.5 ng/mL) partially inhibited the release of cells into the cell cycle, as evidenced by the higher number of cells in G0/G1 phase, and lower number of cells in G2/S phase, compared to a full release in the absence of TGF-β (Figures 5B, S5B, and S5C). This partial inhibition was enhanced with VPS28 or UBAP1 knockdown, resulting in a higher proportion of cells in G0/G1 phase and a lower proportion of cells in G2/S phase compared to NT siRNA controls in each condition (Figures 5B, S5D, and S5E). In addition, the release of cells from G0 in the absence of TGF-β was also inhibited with knockdown of ESCRT components, presumably due to the increase in basal SMAD2 phosphorylation (Figures 5B, S2E, S2F, S5D, and S5E).
To further address the impact of ESCRT knockdown on cell behavior, we turned to another key TGF-β-induced response, EMT (Miettinen et al., 1994). Importantly, we have recently shown in two different mouse mammary epithelial cell lines, NMuMG and EpRas, that both TGF-β-induced SMAD2 and also non-canonical SMAD1/5 phosphorylation are essential for a full EMT (Ramachandran et al., 2018). We chose to focus on the ESCRT-I component, VPS28, for these experiments as a representative of the ESCRT machinery and first deconvoluted an siRNA SMARTpool against Vps28 to confirm that all the siRNAs acted as expected. This identified two siRNAs that led to the greatest extent of knockdown (siRNAs 1 and 3), thus a pool of these two siRNAs was used for all further experiments (Figure S5F). Knockdown of VPS28 in NMuMG cells led to a persistence in SMAD2 phosphorylation over a TGF-β time course relative to induction after 1 hr (Figure S5G). In addition, we observed a striking persistence of SMAD1 phosphorylation, which is normally induced only transiently by TGF-β, over the same period (Figure S5H). A similar effect on both SMAD1 and SMAD2 phosphorylation was seen in EpRas cells (see Figure S7A).
EMT in NMuMGs is accompanied by cell death, and we used a CellTiter-Glo assay to test the effect of knocking down VPS28 on TGF-β-mediated cell death. This showed that TGF-β drives a dose-dependent decrease in viable cell number, with the effect of TGF-β enhanced with VPS28 knockdown at every dose of ligand tested, relative to the NT controls (Figure S6A).
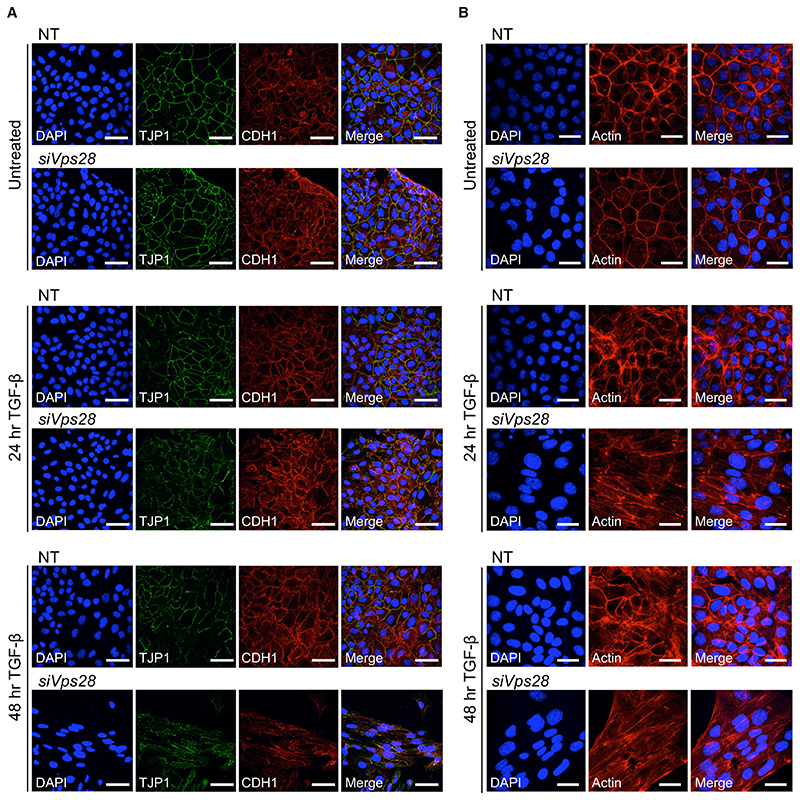
We next investigated the effects of depletion of the ESCRT machinery on EMT itself. Hallmarks of EMT include the formation of actin stress fibers, as well as the downregulation and delocalization from the plasma membrane of epithelial markers such as TJP1 (formerly known as ZO-1) and CDH1 (formerly known as E-cadherin), which contribute to the tight junctions and adherens junctions, respectively, (Miettinen et al., 1994). These effects are readily detectable after treatment of NMuMGs for 48 hr with a saturating dose (2 ng/mL) of TGF-β (Figures S6B and S6C) (Ramachandran et al., 2018). At a sub-saturating dose of TGF-β (0.5 ng/mL), there was only very minimal evidence of an EMT occurring in NT controls after either 24 hr or 48 hr, as TJP1 and CDH1 remained localized at the cell surface (Figure 6A). With VPS28 knockdown, however, we observed a partial delocalization of epithelial markers after 24 hr of treatment and a more substantial delocalization and downregulation of both TJP1 and CDH1 after 48 hr (Figure 6A). In addition, cells appeared to elongate after 48 hr of low dose TGF-β with VPS28 knockdown and stress fibers were induced, as evidenced by staining with phalloidin (Figure 6B). These results indicate that knocking down VPS28 leads to a more sustained TGF-β response in NMuMGs, and this sustained response drives EMT after exposure to a low dose of TGF-β, which is not by itself sufficient to promote EMT. We additionally confirmed that the enhanced EMT observed when VPS28 was knocked down was still dependent on TGF-β receptors, thus ruling out the possibility that it was due to a distinct signaling pathway that might be strengthened when ESCRT components are depleted (Figures S6D and S6E).
Figure 6. ESCRT Knockdown Allows TGF-β to Drive EMT at Low Doses.
(A) NMuMG cells were transfected with non-targeting (NT) control siRNAs or siRNAs targeting Vps28 and treated or not with 0.5 ng/mL TGF-β for the times indicated. Cells were fixed and stained for TJP1, CDH1, and DAPI to mark nuclei and imaged by confocal microscopy. Scale bar, 50 μm.
(B) As in (A) except that cells were stained for actin using phalloidin. Scale bar, 30 μm.
To confirm that the sensitization of cells to low dose TGF-β was not just a feature of NMuMG cells, we performed similar experiments in EpRas cells, which undergo EMT in a more extended time frame (7 days). As with the NMuMGs, we showed that 0.5 ng/mL TGF-β was not sufficient to induce EMT in control cells, but could do so in cells knocked down for VPS28 (Figures S7B and S7C). Furthermore, this EMT was accompanied by a dramatic upregulation in ACTA2 (previously called alpha smooth muscle actin [α-SMA]) (Valcourt et al., 2005) and transcriptional repressors known to be drivers of EMT, SNAI1, SNAI2, ZEB1, and ZEB2 (Katsuno et al., 2013) (Figures S7D and S7E).
Although we noted in both cell lines that SMAD2 phosphorylation was sustained with VPS28 knockdown, we observed a more dramatic effect on SMAD1 phosphorylation (Figures S5H and S7A). This suggested that the prolonged PSMAD1 may play a more dominant role in sensitizing these cells to TGF-β-induced EMT than the PSMAD2 (Figures S5G and S7A). To address this hypothesis directly, we investigated whether prolonging PSMAD1 signaling in NMuMG cells by additionally treating the cells with BMP4 (Ramachandran et al., 2018) could potentiate TGF-β-induced EMT. Using the phalloidin staining of actin as a readout for EMT, we observed that BMP signaling enhanced the ability of a low dose of TGF-β to induce stress fibers after 48 hr (Figure S6F). The effect of BMP4 was less obvious when a saturating dose of TGF-β was used, as prominent stress fibers were already observed in this condition (Figure S6F). We also used the upregulation of ACTA2 as a readout of EMT. Again, it was evident that the combined effect of TGF-β and BMP4 was greater than TGF-β alone (Figure S6G). These results strongly suggest that prolonged PSMAD1 signaling, as occurs when ESCRT components are depleted, enhances TGF-β-induced EMT.
Downregulation of ESCRT Components Correlates with Poor Survival in Multiple Cancer Contexts
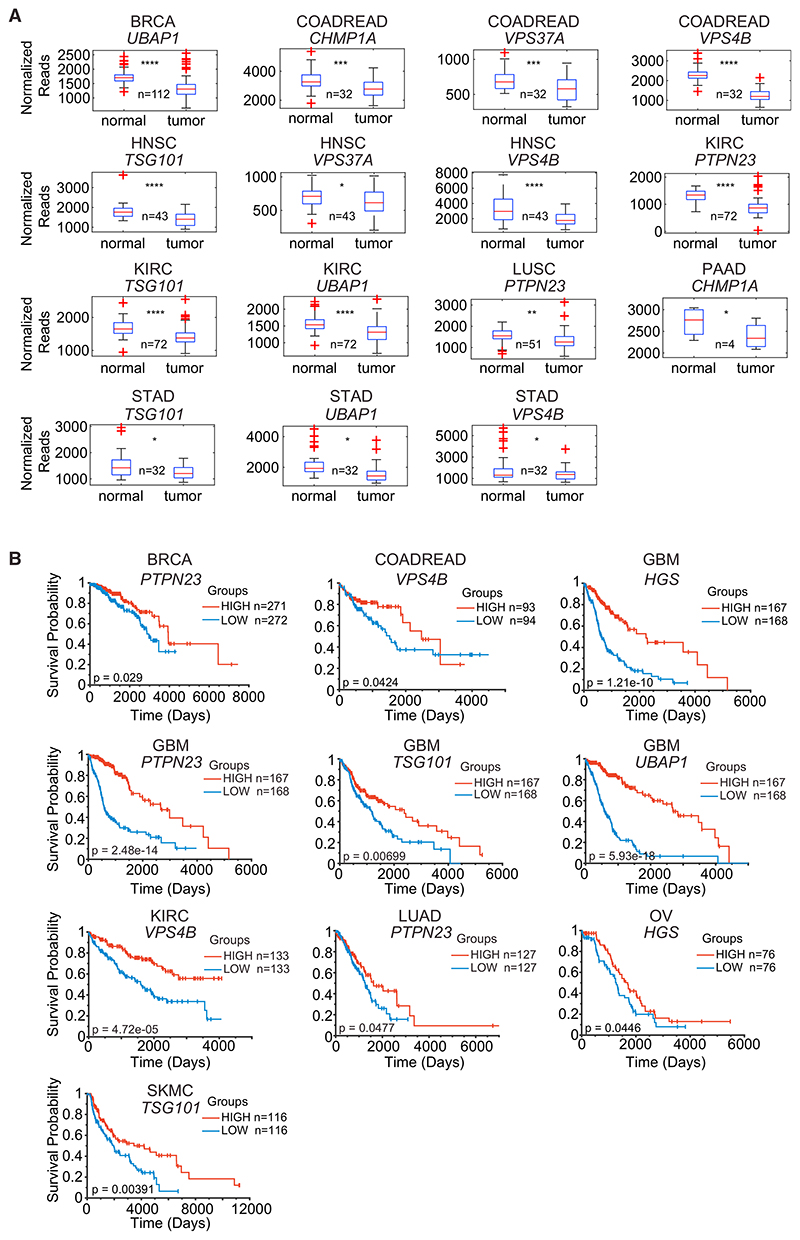
EMT is a key step in tumorigenesis that contributes to a migratory phenotype, acquisition of stem cell properties, and resistance to chemotherapeutic agents (Ye and Weinberg, 2015). We have shown that knockdown of ESCRT components in two different cell systems confers an increased sensitivity to TGF-β-induced EMT. We therefore postulated that losing expression of ESCRT components in cancer might lead to a more invasive and aggressive tumor, if TGF-β was present in the micro-environment. Indeed, loss of PTPN23 has previously been linked to more invasive cancers (Lin et al., 2011; Manteghi et al., 2016). By mining The Cancer Genome Atlas (TCGA) data, we found evidence for downregulation of the ESCRT components we have identified in our screen, as well as other ESCRT components like VPS37A, CHMP1A, and TSG101, in multiple tumor types (Figure 7A). Note, though, that this analysis could be complicated in the case of VPS4B as it resides close to SMAD4 on chromosome 18 and, in some cancers, they are co-deleted (McDonald et al., 2017). Importantly, we also found that downregulation of ESCRT components was associated with poorer survival in multiple different tumor contexts (Figure 7B). It is thus possible that an enhanced response to TGF-β drives more invasive, and thus more metastatic, cancers in these instances.
Figure 7. Reduced Expression of ESCRT Components in Tumors Correlates with Poorer Prognosis.
(A) RSEM-normalized TCGA RNA-seq expression datasets were analyzed for expression of ESCRT components in matched tumor and normal samples. Box and whisker plots are shown, where the central line indicates the sample median, box edges represent quartiles, and whiskers extend to the most extreme data points not considered outliers. Outliers are indicated with red crosses. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.00005.
(B) RSEM-normalized TCGA RNA-seq expression datasets were analyzed for expression of ESCRT components. The top and bottom quantile expression ranked values for selected genes were plotted as Kaplan-Meier curves. p values are shown.
BRCA, breast invasive carcinoma; COADREAD, colorectal adenocarcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma.
Discussion
In this paper, we have identified the ESCRT pathway as the degradative route for the TGF-β receptors and thus crucial for determining the lifetime of TGF-β receptors and hence signal duration. We have shown that components of the ESCRT machinery act to limit the extent of TGF-β signaling by trafficking activated TGF-β receptors for degradation in the lysosome. Our whole genome screens identified two components of the ESCRT-I complex that traffics ubiquitinated cargos to the endosomes (VPS28 and UBAP1), a protein that couples ESCRT-I to ESCRT-III (PTPN23) and another component that is important for ESCRT disassembly and recycling (VPS4B). Knockdown of these ESCRT components not only leads to prolonged signaling, but also to enhanced transcriptional responses, and moreover, confers on cells increased sensitivity to low levels of TGF-β for inducing growth arrest and EMT. The latter result raises the intriguing possibility that a reduction in ESCRT activity, as we have shown to occur in the context of cancer, could sensitize tumor cells to the tumor-promoting activity of TGF-β produced by the stroma. Indeed, we have demonstrated that downregulation of ESCRT components correlates with poorer survival of cancer patients for a number of different tumors.
We have demonstrated that TGF-β signaling is prolonged with knockdown of ESCRT components despite receptors internalizing normally. As this also occurs when new signaling from the cell surface is blocked with a TGF-β neutralizing antibody, we conclude that in these conditions TGF-β receptors continue to signal from internal endosomal compartments. Consistent with this idea, we have also directly demonstrated that knockdown of ESCRT components stabilizes both TGF-β receptors. The issue of whether the receptors signal at the plasma membrane or from internal vesicles has been the source of considerable debate, with some studies concluding that all signaling occurs at the cell surface (Hagemann et al., 2009; Lu et al., 2002), while others have shown that internalization is required for signaling (Di Guglielmo et al., 2003; Jullien and Gurdon, 2005). Although our study does not determine whether receptor internalization is a prerequisite for signaling, it strongly suggests that signaling occurs from endosomes in the normal lifetime of the TGF-β receptors. This fits well with the proposal that the endosomal FYVE domain-containing proteins, SARA (also called ZFYVE9) and endofin (also called ZFYVE16), aid recruitment of R-SMADs to the activated receptors for phosphorylation (Shi et al., 2007; Tsukazaki et al., 1998). Interestingly, the ESCRT component, PTPN23, has very recently been shown to interact with SARA and endofin, thus preventing PTPN23’s interaction with the ESCRT-III component CHMP4 (Gahloth et al., 2017). This suggests that PTPN23 could have a positive role in TGF-β signaling mediated via its association with SARA, in addition to its negative role mediating ESCRT-III recruitment and trafficking of the receptors to the MVBs. Our findings that depletion of PTPN23 results in sustained TGF-β signaling suggest that the PTPN23 interaction with ESCRT-III is the dominant one.
We have demonstrated that the sustained TGF-β signaling we observe when components of the ESCRT machinery are depleted is translated into sustained TGF-β-induced transcriptional responses and also into an increased sensitivity to low levels of TGF-β for induction of growth arrest or EMT. Although this sort of phenomenon has been observed for some other signaling pathways, such as cytokine receptors activating the nuclear factor κB (NF-κB) pathway (Mamińska et al., 2016), this correlation has not necessarily held true for all signaling pathways. For the EGF receptors, depletion of ESCRT components caused retention of EGFR in endosomes, and increased activation of EGFR and its downstream kinases (e.g., MEK1/2 and ERK1/2) but had virtually no effect on the overall profile and amplitude of the EGF-induced transcriptional response (Brankatschk et al., 2012). These authors could only observe an effect on the EGF-induced transcriptional program if the receptors were retained at the cell surface. One reason that the retention of the active TGF-β receptors on endosomes might result in enhanced transcriptional responses may be due to the nucleo-cytoplasmic shuttling behavior of the SMADs. During signaling, R-SMADs and SMAD4 constantly shuttle between the cytoplasm and the nucleus, with the R-SMADs cycling between being phosphorylated by active receptor in the cytoplasm and dephosphorylated by nuclear phosphatases (for review, see Schmierer and Hill, 2007). There are no amplification steps in this pathway, and thus, through the shuttling mechanism, the R-SMADs constantly monitor receptor activity. This mechanism ensures that as long as receptors are active and the kinase domains are available to cytoplasmic R-SMADs, a pool of activated SMAD complexes is maintained in the nucleus. Prolonging the presence of activated receptors in endosomes by depleting ESCRT components and observing the resulting enhancement of transcriptional responses serves as an important demonstration of this mechanism.
We have confirmed that inhibition of the E3 ubiquitin ligase SMURF1 also prolongs signaling from the TGF-β receptors, in line with its previously suggested role in targeting the receptors for degradation (Ebisawa et al., 2001). Interestingly, knockdown of SMURF2, which has previously also been implicated in ubiquitinating the TGF-β receptors and targeting them for degradation, had no impact on SMAD phosphorylation dynamics. This corroborates work performed with SMURF2 knockout mouse embryonic fibroblasts, which showed that knockout of SMURF2 did not affect receptor turnover or PSMAD2 dynamics (Tang et al., 2011). It will now be important to identify the ubiquitination sites on the receptors, as alterations in these residues would be predicted to abrogate receptor degradation and lead to a persistence in signaling in a similar manner to ESCRT knockdown.
Deregulation of the ESCRT complexes (Mattissek and Teis, 2014), and derailed endocytosis more generally (Mosesson et al., 2008), are emerging as common features in diseases such as cancer. Our results implicate a failure to downregulate TGF-β receptors as essential for driving an EMT in response to low doses of ligand. Thus, downregulation of ESCRT components in cancer could lead to a more invasive tumor phenotype in the presence of low levels of TGF-β. Indeed, haploinsuffciency in PTPN23 has been linked to poor prognosis in cancer (Manteghi et al., 2016), and suppression of the same protein has been to shown to drive invasive behaviors in mammary epithelial cells (Lin et al., 2011). The effects of PTPN23 haploin-suffciency have been ascribed to several different signaling pathways, including integrin and PDGF signaling (Gingras et al., 2017). In the case of VPS4B, downregulation in breast tumors was correlated with increased levels of EGFR (Lin et al., 2012). Given our results, it is an intriguing possibility that in cancers showing loss or depletion of ESCRT components, prolonged, upregulated TGF-β signaling could also play a driving role in tumorigenesis. We have previously shown that in the continuous presence of TGF-β, which is commonly found in the micro-environment of tumors (Pickup et al., 2013), cells have a counterintuitively low level of SMAD2 phosphorylation due to the rapid depletion of cell surface receptors (Vizán et al., 2013). How tumor cells in this situation would be able respond to high levels of TGF-β, and induce invasive behaviors, is unclear. Here, we provide one potential mechanism. By downregulating components of the ESCRT machinery, cells could escape from the low levels of signaling imposed by receptor depletion and undergo an EMT or other TGF-β-mediated tumor promoting cell behaviors. The induction of an EMT can be a critical step in tumor invasion and metastasis (Kalluri and Weinberg, 2009), and thus the regulation of ESCRT components is likely to be important for understanding TGF-β-driven tumor dissemination.
Star★Methods
Detailed methods are provided in the online version of this paper and include the following:
-
EXPERIMENTAL MODEL AND SUBJECT DETAILS
-
○
Cell lines
-
○
-
-
○
Generation of TGFBR1 CRISPR/Cas9 knockout line
-
○
siRNA transfection and ligand treatment time courses
-
○
Whole genome siRNA screen
-
○
Cell lysis and western blotting
-
○
Surface biotinylation assays
-
○
Cell cycle analysis
-
○
Immunofluorescence
-
○
Image acquisition and analysis
-
○
qPCR
-
○
TCGA analysis
-
○
-
QUANTIFICATION AND STATISTICAL ANALYSIS
-
○
Quantifying western blots
-
○
Statistical analysis
-
○
Star★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-phospho-SMAD2 (Western) | Cell Signaling Technology | Cat# 3108; RRID: AB_490941 |
| Anti-SMAD2/3 (IF and Western) | BD Biosciences | Cat# 610843; RRID: AB_398162 |
| Anti-phospho-SMAD1/5 (Western) | Cell Signaling Technology | Cat# 13820; RRID: AB_2493181 |
| Anti-SMAD1 (Western) | Invitrogen | Cat# 38-5400; RRID: AB_2533373 |
| Anti-phospho-SMAD3 (Western) | Cell Signaling Technology | Cat# 9520; RRID: AB_2193207 |
| Anti-SMAD3 (Western) | Abcam | Cat# 40854; RRID: AB_777979 |
| Anti-ACVR1B (Western) | LSBio | Cat# LS-B2695 |
| Anti-ACVR2B (Western) | Aviva Systems Biology | Cat# ARP45041; RRID: AB_10644782 |
| Anti-BMPR2 (Western) | BD Biosciences | Cat# 612292; RRID: AB_399609 |
| Anti-UBAP1 (Western) | Proteintech | Cat# 12385-1; RRID: AB_2211886 |
| Anti-SMURF1 (Western) | Santa Cruz | Cat# sc-25510; RRID: AB_2302385 |
| Anti-SMURF2 (Western) | Cell Signaling Technology | Cat# 12024 |
| Anti-TGFBR1 (Western) | Santa Cruz | Cat# sc-398 RRID: AB_632493 |
| Anti-TGFBR2 (Western) | Santa Cruz | Cat# sc-17792; RRID: AB_628349 |
| Anti-ACTA2 (Western) | Sigma Aldrich | Cat# A5228; RRID: AB_262054 |
| Anti-GRB2 (Western) | BD Biosciences | Cat# 610112; RRID: AB_397518 |
| Anti-MCM6 (Western) | Santa Cruz | Cat# sc-9843; RRID: AB_2142543 |
| Anti-Tubulin (Western) | Abcam | Cat# ab6160; RRID: AB_305328 |
| Anti-Actin (Western) | Sigma Aldrich | Cat# A3853; RRID AB_262137 |
| Anti-TJP1 (IF) | Invitrogen | Cat# 61-7300; RRID: AB_2533938 |
| Anti-CDH1 (IF) | BD Biosciences | Cat# 610181; RRID: AB_397580 |
| Anti-HA (IF) | Sigma Aldrich | Cat# 11867423001; RRID: AB_10094468 |
| Anti-EEA1 (IF) | BD Biosciences | Cat# 610456; RRID: AB_397829 |
| Anti-HGS (IF) | Enzo Life Sciences | Cat# ALX-804-382; RRID: AB_2118912 |
| Anti-Rabbit Alexa Fluor 488 (IF) | ThermoFisher Scientific | Cat# A-21206; RRID: AB_2535792 |
| Anti-Mouse Alexa Fluor 488 (IF) | ThermoFisher Scientific | Cat# A-11001; RRID: AB_2534069 |
| Anti-Mouse Alexa Fluor 594 (IF) | ThermoFisher Scientific | Cat# A-21203; RRID: AB_2535789 |
| Anti-Rabbit Alexa Fluor 594 (IF) | ThermoFisher Scientific | Cat# A-21244; RRID: AB_10562581 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SB-431542 | Tocris | Cat# 1614 |
| Rhodamine-Phalloidin | ThermoFisher Scientific | Cat# R415 |
| Human recombinant TGFβ1 | Peprotech | Cat# 100-21 |
| Human recombinant BMP4 | Peprotech | Cat# 120-05ET |
| Human recombinant Activin A | Peprotech | Cat# 120-14 |
| cOmplete, EDTA-free Protease Inhibitor Cocktail | Sigma Aldrich | Cat# 000000011873580001 |
| Bafilomycin A1 | Merck Millipore | Cat# 196000 |
| EZ-Link Sulfo-NHS-Biotin | ThermoFisher Scientific | Cat# 21217 |
| Pierce NeutrAvidin Agarose | ThermoFisher Scientific | Cat# 29200 |
| Cycloheximide | Sigma Aldrich | Cat# C7698 |
| 1D11 | Gift from Lalage Wakefield (NCI, Bethesda) | N/A |
| 13C4 | Gift from Lalage Wakefield (NCI, Bethesda) | N/A |
| INTERFERin | Polyplus | Cat# 409-10 |
| PowerUp SYBR Green Master Mix | ThermoFisher Scientific | Cat# A25742 |
| TRIzol | ThermoFisher Scientific | Cat# 15596026 |
| DAPI | Sigma Aldrich | Cat# 10236276001 |
| Critical Commercial Assays | ||
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat# G7570 |
| Experimental Models: Cell lines | ||
| HaCaT cells, human | Francis Crick Institute Cell Services | N/A |
| EpRas cells, mouse | Gift from Martin Oft and Hartmut Beug (IMP, Vienna) | N/A |
| MDA-MB-231 cells, human | ECACC/HPA culture collection | Cat# 92020424; RRID: CVCL_0062 |
| MDA-MB-231 HA-TGFBR1 cells, human | This paper | N/A |
| NMuMG cells, mouse | ATCC | Cat# CRL-1636; RRID: CVCL_0075 |
| NMuMG KO TGFBR1 | This paper | N/A |
| Oligonucleotides | ||
| See Table S2 for oligonucleotides | N/A | N/A |
| See Table S2 for siRNAs | N/A | N/A |
| TGFBR1 guide RNA: GGTGAATGACAGTGCGGTTA | This paper | N/A |
| Recombinant DNA | ||
| HA-TGFBR1 | Kavsak et al., 2000 | N/A |
| pSUPER.retro.puro | OligoEngine | Cat# VEC-pRT-0002 |
| pSpCas9(BB)-2A-GFP (PX458) | Ran et al., 2013 | Addgene Cat# 48138 |
| Software and Algorithms | ||
| FIJI (ImageJ) | https://imagej.net/Fiji/Downloads | N/A |
| MATLAB (Version R2016b) | https://uk.mathworks.com | N/A |
| FIREHOSE | https://gdac.broadinstitute.org/ | N/A |
| FlowJo 10 | FlowJo | N/A |
| HCS Studio Cell Analysis Software | ThermoFisher Scientific | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Caroline Hill (caroline.hill@crick.ac.uk).
Experimental Model and Subject Details
Cell lines
All cell lines were maintained at 37°C and 10% CO2 and have been certified mycoplasma negative by the Francis Crick Institute Cell Services. HaCaTs and MDA-MB-231 cells have been authenticated by short tandem repeat profiling, while NMuMG and EpRas cells had species confirmation by the Francis Crick Institute Cell Services. The identity of all cell lines was also authenticated by confirming that their responses to ligands and their phenotype were consistent with published history. Cell line details are as follows:
HaCaT, human keratinocyte, male, maintained in Dulbecco’s modified Eagle’s medium (DMEM) + 10% fetal bovine serum (FBS) + 1% penicillin/streptomycin (Pen/Strep).
MDA-MB-231, human triple negative breast cancer, female, maintained in DMEM + 10% FBS + 1% Pen/Strep.
MDA-MB-231 HA-TGFBR1, human triple negative breast cancer, female, stably transfected with HA-TGFBR1, maintained in DMEM + 10% FBS + 1% Pen/Strep + 1 μg/mL Puromycin.
EpRas, mouse mammary epithelial, female, maintained in DMEM + 10% FBS + 1% Pen/Strep.
NMuMG, mouse mammary epithelial, female, maintained in DMEM + 10% FBS + 1% Pen/Strep + 10 μg/mL insulin.
NMuMG KO TGFBR1, mouse mammary epithelial, female, maintained in DMEM + 10% FBS + 1% Pen/Strep + 10 μg/mL insulin.
Method Details
Generation of TGFBR1 CRISPR/Cas9 knockout line
From the wild-type NMuMG cells, a parental clone was selected that expressed robust junctional markers (TJP1 and CDH1) and underwent an efficient EMT in response to TGF-β. A guide RNA(see Key Resources Table) targeting the intracellular domain of TGFBR1 was expressed from the plasmid pSpCas9(BB)–2A-GFP (PX458) (Ran et al., 2013) and used to knock out TGFBR1. NMuMG parental clone cells were transfected with this plasmid, sorted for GFP expression, plated as single cells in 96-well plates and screened by sequencing to verify mutations in TGFBR1. Loss of TGFBR1 was confirmed by western blotting.
siRNA transfection and ligand treatment time courses
For siRNA experiments, cells were plated, and 24 hr later transfected with 50 nM (HaCaTs and EpRas) or 30 nM (NMuMG) siRNA (Dharmacon) with 8 μL INTERFERin (Polyplus) and 200 μL Opti-MEM (ThermoFisher Scientific) for a 6-well plate in fresh media. Experiments were performed 72 hr after siRNA transfection. For EMT assays in NMuMGs, cells were transfected with siRNAs for 24 hr, followed by TGF-β for the time indicated. For EMT assays in EpRas, cells were reverse transfected with siRNAs for 24 hr, followed by TGF-β for 72 hr. Cells were then split, re-transfected with siRNAs, and treated with TGF-β for a further 96 hr. A full list of siRNAs used is given in Table S2.
Ligands and reagents were used at the following concentrations, and re-constituted in the following buffers: TGF-β1 (Peprotech, 4.4 mM HCl/0.1% BSA), 2 ng/mL except where indicated otherwise; BMP4 (Peprotech, 4.4 mM HCl/0.1% BSA), 20 ng/mL; Activin (Peprotech, 4.4 mM HCl/0.1% BSA), 20 ng/mL; Bafilomycin A1 (Merck Millipore, DMSO), 50 nM; SB-431542 (Tocris, DMSO), 10 μM; cycloheximide (Sigma Aldrich, water), 20 μg/mL. The TGF-β neutralizing antibody, 1D11, and isotype-matched IgG1 monoclonal control antibody raised against Shigella toxin (13C4) were as described (Nam et al., 2008), and used at 30 μg/mL. All stimulations were performed in 10% FCS.
Whole genome siRNA screen
A Dharmacon (GE Healthcare) siGENOME siRNA library covering the human genome was plated out into Greiner Sensoplate glassbottom multiwell 384/1536 well plates (ThermoFisher Scientific) at a final concentration of 37.5 nM. Control siRNAs, ON-TARGET Non-targeting siRNA or siGENOME RISC-free siRNA (GE Healthcare), were also plated at the same concentration. For 384-well plate screens, 0.1 μL of INTERFERin (PolyPlus) was added per well, and 2000 HaCaT cells per well were then plated. For the 1536-well plate format, 0.025 μL of INTERFERin was used per well, and 500 HaCaTs were plated. After 72 hr, cells were stimulated with TGF-β for 1 hr or 20 hr. The whole screen was performed in triplicate, with each replicate on a separate plate. Cells were fixed and stained using an anti-SMAD2/3 antibody (BD Biosciences, Cat. # 610843) and with DAPI, then imaged using an ArrayScan XTI Live High Content microscope (Cellomics). Results were analyzed using HCS Studio Cell Analysis Software (ThermoFisher Scientific). Nuclear and cytoplasmic boundaries were delineated using DAPI as a nuclear marker and fitting a concentric shape around this to model the cytosol, and the relative level of nuclear and cytoplasmic SMAD2/3 determined for each hit. Cells with a very bright DAPI signal were excluded, as they were considered likely to be dividing or dying cells. Hits that led to a reduction in cell number of greater than 2 SD from the average of the screen were excluded from further analysis. Hits with a less than 2 SD increase in nuclear SMAD2/3 in the 1 hr screen and greater than 2 SD increase in nuclear SMAD2/3 in the 20 hr screen were taken forward for analysis. See Table S1 for a list of these hits.
Cell lysis and western blotting
Whole cell extracts were prepared as previously described (Inman et al., 2002). Briefly, cells were lysed in D0.4 lysis buffer (20 mM HEPES pH 7.5, 10% Glycerol, 0.4 M KCl, 0.4% Triton X-100,10 mM EGTA, 5 mM EDTA, 1X protease inhibitors (Roche), 25 mM NaF), sonicated, centrifuged, and supernatants retained. Cells were then analyzed by western blotting as previous described (Inman et al., 2002). Samples were loaded onto standard 15% SDS polyacrylamide gels. After electrophoresis, proteins were transferred to PVDF membrane (Millipore) and immunoblotted using standard techniques. Western blots were visualized on film or using an ImageQuant LAS 4000 mini (GE Healthcare) and quantified with ImageJ. Blots were normalized to densitometry measurements in control cells after 1 hr of treatment except where indicated. The following antibodies were used for western blotting: anti-PSMAD2 (Cell Signaling Technology, Cat. # 3108), anti-SMAD2/3 (BD Biosciences, Cat. # 610843), anti-PSMAD1/5 (Cell Signaling Technology, Cat. # 13820), anti-SMAD1 (Invitrogen, Cat. # 38-5400), anti-PSMAD3 (Cell Signaling Technology, Cat. # 9520), anti-SMAD3 (Abcam Cat. # 40854), anti-ACVR1B (LSBio, Cat. # LS-B2695), anti-ACVR2B (Aviva Systems Biology, Cat. # ARP45041), anti-BMPR2 (BD Biosciences, Cat. # 612292), anti-UBAP1 (Proteintech, Cat. # 12385-1), anti-GRB2 (BD Biosciences, Cat. # 610112), anti-TGFBR1 (Santa Cruz, Cat. # sc-398), anti-TGFBR2 (Santa Cruz, Cat. # 17792), anti-MCM6 (Santa Cruz, Cat. # sc-9843), anti-Tubulin (Abcam, Cat. # Ab6160), anti-Actin (Sigma Aldrich, Cat # A3853), anti-SMURF1 (Santa Cruz, Cat. # sc-25510), anti-SMURF2 (Cell Signaling Technology, Cat. # 12024) and anti-ACTA2 (Sigma Aldrich, Cat# A5228).
Surface biotinylation assays
Surface biotinylation assays were as previously described (Vizán et al., 2013). Briefly, cells were labeled with EZ-Link Sulfo-NHS-Biotin in PBS-CM (0.8 mM MgCl2, 1 mM CaCl2, PBS) at 4°C for 40 mins, labeling quenched with 20 mM Tris-HCl pH 7.5 in PBS and then cells lysed in lysis buffer (1% Triton X-100, 0.1% SDS, 10 mM EDTA, 1X protease inhibitors). Samples were normalized for protein levels, inputs taken, and biotin-labeled proteins pulled down with NeutrAvidin Agarose resin (ThermoFisher Scientific) overnight. Beads were washed with lysis buffer three times and samples eluted before analysis by western blotting.
Cell cycle analysis
Cell cycle analysis was performed as previously described (Levy and Hill, 2005). Cells were seeded, and the next day transferred to DMEM with 0.1% serum and transfected with siRNAs as described. For arrested samples, after 72 hr, cells were washed twice with PBS, then trypsinized, pelleted, and fixed in ice-cold 70% ethanol. The remaining cells were transferred to DMEM with 10% FBS for 22 hr in the presence or absence of 0.5 ng/mL TGF-β, and processed as for the arrested samples. Control samples were plated for the same amount of time, but not serum starved. Cells were then treated with 50 μLof 100 μg/mL Ribonuclease to degrade RNAs. 200 μL of 50 μg/mL propidium iodide (PI) was then added to stain DNA. PI incorporation was analyzed with a cytometer using a 610/20-Yellow filter. Cell debris and cell doublets were gated out, and different phases of the cell cycle were fitted to the resulting histogram of PI incorporation using a Dean-Jett-Fox model with FlowJo 10 software (FlowJo). For quantifications, the fold change in the number of cells in each phase of the cell cycle relative to arrested (Figures S5B and S5C) or non-targeting siRNA transfected (Figures S5D and S5E) cells was determined, and averaged across three independent experiments.
Immunofluorescence
Immunofluorescence experiments were performed as previously described (Nicolás and Hill, 2003). Cells were fixed for 20 mins at −20°C in methanol/acetone (1:1) for TJP1/CDH1 staining, and for 10 mins at room temperature in 4% formaldehyde for phalloidin, HA, HGS and EEA1 staining. After fixation, cells were washed three times in 0.3% Triton X-100/PBS, blocked for 30 mins in 0.3% Triton X-100/0.3% BSA/10% FBS and then stained with primary antibody for 1 hr. Cells were then washed a further three times and stained with secondary antibody, with DAPI to counterstain nuclei. F-actin was detected using rhodamine-phalloidin (Sigma Aldrich) in place of the secondary antibody. The following primary antibodies were used for immunofluorescence experiments: anti-TJP1 (Invitrogen, Cat. # 61-7300), anti-CDH1 (BD Biosciences, Cat. # 610181), anti-HA (Sigma Aldrich, Cat. # 11867423001), anti-EEA1 (BD Biosciences, Cat. # 610456) and anti-HGS (Enzo Life Sciences, Cat. # ALX-804-382). The following secondary antibodies were used for immunofluorescence experiments: Anti-Rabbit Alexa Fluor 488 (ThermoFisher Scientific, Cat. # A-21206), Anti-Mouse Alexa Fluor 488 (ThermoFisher Scientific, Cat. # A-11001), Anti-Mouse Alexa Fluor 594 (ThermoFisher Scientific, Cat. # A-21203), Anti-Rabbit Alexa Fluor 594 (ThermoFisher Scientific, Cat. # A-21244).
Image acquisition and analysis
All imaging was performed using ZEISS LSM710 or 780 upright confocal microscopes. Coverslips were mounted on slides using Mowiol (Calbiochem, #475904) and imaged with 63x/1.40 or 40x/1.3 Oil Plan-Apochromat lenses. In all cases, scale bars are indicated on images. For EMT assays, Z stacks were acquired for all channels and maximal intensity projections are shown, while for co-localization assays single slices were taken. All images were processed and analyzed in ImageJ. For co-localization assays, images were thresholded uniformly to remove background staining. The percentage of red pixels that overlapped with green pixels was then quantified for 20 individual cells in each condition.
qPCR
qPCR was performed as previously described (Grönroos et al., 2012). For primer sequences see Table S2. In brief, mRNA was extracted using TRIzol (ThermoFisher Scientific) and cDNA synthesis was performed on 1 μg mRNA using Affnity Script (QIAGEN), both according to the manufacturer’s instructions. qPCRs were performed using Fast SYBR Green Master mix (ThermoFisher Scientific) on an ABI 7500 Fast (Applied Biosystems) thermocycler. Technical replicates for each condition were taken and experiments were repeated three times. Calculations were performed using the ΔΔCt method, and levels of mRNA are expressed as fold change relative to untreated control cells. Means ± SD from at least three independent experiments are shown. Results were analyzed using GraphPad Prism 7 software and statistics were performed on these data using a Student’s t test.
TCGA analysis
RSEM normalized TCGA RNA-seq expression datasets were downloaded from Firehose (https://gdac.broadinstitute.org/). Box-and-whisker plots were generated from matched tumor and normal data using MATLAB (Version R2016b), using the standard Boxplot function. For Kaplan-Meier curves the top and bottom quantile expression ranked values for selected genes were plotted using the ‘MatSurv’ package (version 1.0.0.0) in MATLAB (https://uk.mathworks.com/matlabcentral/fileexchange/64582-matsurv). Statistics were performed on these data using a t test.
Quantification and Statistical Analysis
Quantifying western blots
Quantification of western blots was performed by densitometry measurements of each lane using ImageJ software, and was normalized to the relative densitometry of the loading control from the same blot. In each case, quantifications were normalized to the control samples stimulated for 1 hr, except where otherwise indicated in the Figure legends.
Statistical analysis
In all cases except Figure 7, the data shown is the mean and SD of at least three independent experiments, except where the presence of a representative experiment is indicated in the Figure legends. The exact number of repeated experiments is indicated in the Figure legend in each case. Statistics were performed using a Student’s t test in GraphPad Prism 7 software, Correction for multiple testing was used if several points in a time course were compared by t test. In this case, statistical significance was determined in GraphPad Prism 7 using the Holm-Sidak method, with alpha = 0.05. Each pair of samples was analyzed individually, without assuming a consistent SD.
For the TCGA analysis, the number of samples in each dataset is indicated in the Figure, where n = the number of matched tumor and normal pairs for Figure 7A and the number of samples in each of the high and low expression datasets for Figure 7B. Analysis was performed in MATLAB and in boxplots, the central line indicates the sample median, box edges represent quartiles and whiskers extend to the most extreme data points not considered outliers. Outliers are indicated with red crosses. An outlier is defined as a value that is more than 1.5 times the interquartile range (w = 1.5) away from the top or bottom of the box, calculated as greater than q3 + w × (q3 – q1) or less than q1 – w × (q3 – q1).
In all cases, significance was defined as a p value of less than 0.05, with the exact degree of significance indicated by asterisks within the Figures. * = p < 0.05, ** = p < 0.005, *** = p < 0.0005, **** = p < 0.00005.
Supplementary Material
Highlights.
The TGF-β receptors are targeted for degradation by the ESCRT machinery
Depletion of ESCRT components traps signaling receptors in internal compartments
This leads to upregulation of transcriptional and phenotypic outputs of the pathway
ESCRT inhibition sensitizes cells to low levels of TGF-β in their micro-environment
Acknowledgements
We thank Lalage Wakefield for providing the TGF-β neutralizing antibody and the isotype-matched control, and the late Anita Roberts for the HA-TGFBR1 expression plasmid. We are grateful to the Francis Crick Institute Light Microscopy and Flow Cytometry facilities and to the Genomics Equipment Park. We thank Sabine Reichert for analyzing the data for Figure 7, Silvia Benito for her contribution to the screens, Anassuya Ramachandran for generating the TGFBR1 null NMuMG cell line, and Pedro Vizán for generating the MDA-MB-231 HA-TGFBR1 stable cell line. We thank Jeremy Carlton and all the members of the Hill lab for useful comments on the manuscript. This work was supported by the Francis Crick Institute that receives its core funding from Cancer Research UK (FC001095), the UK Medical Research Council (FC001095), and the Wellcome Trust (FC001095).
Footnotes
Author contributions
D.S.J.M. and C.S.H. conceived the study. D.D., I.G., M.J., R.E.S., and M.H. designed and performed the whole genome screens. D.S.J.M., R.D.B., and P.C. designed, performed, and analyzed all the experiments and D.S.J.M. and C.S.H. wrote the paper.
Declaration of Interests
The authors declare no competing interests.
References
- Ali N, Zhang L, Taylor S, Mironov A, Urbé S, Woodman P. Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr Biol. 2013;23:453–461. doi: 10.1016/j.cub.2013.02.033. [DOI] [PubMed] [Google Scholar]
- Bowman EJ, Siebers A, Altendorf K. Bafilomycins: a class of inhibitors of membraneATPasesfrom microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brankatschk B, Wichert SP, Johnson SD, Schaad O, Rossner MJ, Gruenberg J. Regulation ofthe EGFtranscriptional response byendo-cytic sorting. Sci Signal. 2012;5:ra21. doi: 10.1126/scisignal.2002351. [DOI] [PubMed] [Google Scholar]
- Budi EH, Duan D, Derynck R. Transforming growth factor-ß receptors and Smads: regulatory complexity and functional versatility. Trends Cell Biol. 2017;27:658–672. doi: 10.1016/j.tcb.2017.04.005. [DOI] [PubMed] [Google Scholar]
- Clague MJ, Liu H, Urbé S. Governance ofendocytic trafficking and signaling by reversible ubiquitylation. Dev Cell. 2012;23:457–467. doi: 10.1016/j.devcel.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Coda DM, Gaarenstroom T, East P, Patel H, Miller DSJ, Lobley A, Matthews N, Stewart A, Hill CS. Distinct modes of SMAD2 chromatin binding and remodeling shape the transcriptional response to NODAL/Activin signaling. eLife. 2017;6:e22474. doi: 10.7554/eLife.22474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Massagué J. Contextual determinants ofTGFß action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19:419–435. doi: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-ß receptor signalling and turnover. Nat Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- Doyotte A, Russell MR, Hopkins CR, Woodman PG. Depletion of TSG101 forms a mammalian “Class E” compartment: a multicisternal early endosome with multiple sorting defects. J Cell Sci. 2005;118:3003–3017. doi: 10.1242/jcs.02421. [DOI] [PubMed] [Google Scholar]
- Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-ß type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- Eichhorn PJ, Rodón L, González-Juncá A, Dirac A, Gili M, Martinez-Saez E, Aura C, Barba I, Peg V, Prat A, et al. USP15 stabilizes TGF-ß receptor I and promotes oncogenesis through the activation of TGF-ß signaling in glioblastoma. Nat Med. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in tgf-ß signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Futter CE, Pearse A, Hewlett LJ, Hopkins CR. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahloth D, Levy C, Walker L, Wunderley L, Mould AP, Taylor S, Woodman P, Tabernero L. Structural basis for specific interaction of TGFß signaling regulators SARA/endofin with HD-PTP. Structure. 2017;25:1011–1024. doi: 10.1016/j.str.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras MC, Kazan JM, Pause A. Role of ESCRT component HD-PTP/PTPN23 in cancer. Biochem Soc Trans. 2017;45:845–854. doi: 10.1042/BST20160332. [DOI] [PubMed] [Google Scholar]
- Grönroos E, Kingston IJ, Ramachandran A, Randall RA, Vizan P, Hill CS. Transforming growth factor ß inhibits bone morphogenetic protein-induced transcription through novel phosphorylated Smad1/5-Smad3 complexes. Mol Cell Biol. 2012;32:2904–2916. doi: 10.1128/MCB.00231-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann AI, Xu X, Nentwich O, Hyvonen M, Smith JC. Rab5-mediated endocytosis of activin is not required for gene activation or long-range signalling in Xenopus. Development. 2009;136:2803–2813. doi: 10.1242/dev.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Yan X, Li N, Dang S, Xu L, Zhao B, Li Z, Lv Z, Fang X, Zhang Y, Chen YG. Internalization of the TGF-ß typeI receptorinto cav-eolin-1 and EEA1 double-positive early endosomes. Cell Res. 2015;25:738–752. doi: 10.1038/cr.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman GJ, Nicolás FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-ß superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Jullien J, Gurdon J. Morphogen gradient interpretation byaregu-lated trafficking step during ligand-receptor transduction. Genes Dev. 2005;19:2682–2694. doi: 10.1101/gad.341605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno Y, Lamouille S, Derynck R. TGF-ß signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol. 2013;25:76–84. doi: 10.1097/CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF ß receptorfordegradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K, Miyazawa K. Negative regulation of transforming growth factor-ß (TGF-ß) signaling by WW domain-containing protein 1 (WWP1) Oncogene. 2004;23:6914–6923. doi: 10.1038/sj.onc.1207885. [DOI] [PubMed] [Google Scholar]
- Kuratomi G, Komuro A, Goto K, Shinozaki M, Miyazawa K, Miyazono K, Imamura T. NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-ß (transforming growth factor-ß) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-ß type I receptor. Biochem J. 2005;386:461–470. doi: 10.1042/BJ20040738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weis-senhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy C, Wrana JL. Clathrin-and non-clathrin-mediated endo-cytic regulation of cell signalling. Nat Rev Mol Cell Biol. 2005;6:112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor ß (TGF-ß) target genes and distinguishes TGF-ß-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol. 2005;25:8108–8125. doi: 10.1128/MCB.25.18.8108-8125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Aranda V, Muthuswamy SK, Tonks NK. Identification of PTPN23 as a novel regulator of cell invasion in mammary epithelial cells from a loss-of-function screen ofthe ‘PTP-ome’. Genes Dev. 2011;25:1412–1425. doi: 10.1101/gad.2018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Li X, Chen JL, Sun X, Cooper FN, Chen YR, Zhang W, Chung Y, Li A, Cheng CT, et al. Identification of an AAA ATPase VPS4B-dependent pathway that modulates epidermal growth factor receptor abundance and signaling during hypoxia. Mol Cell Biol. 2012;32:1124–1138. doi: 10.1128/MCB.06053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Murray JT, Luo W, Li H, Wu X, Xu H, Backer JM, Chen YG. Transforming growth factor ß activates Smad2 inthe absence of receptor endocytosis. J Biol Chem. 2002;277:29363–29368. doi: 10.1074/jbc.M203495200. [DOI] [PubMed] [Google Scholar]
- Mamińska A, Bartosik A, Banach-Orłowska M, Pilecka I, Jastrzebski K, Zdzalik-Bielecka D, Castanon I, Poulain M, Neyen C, Wolinska-Nizioł L, et al. ESCRT proteins restrict constitutive NF-κB signaling bytrafficking cytokine receptors. Sci Signal. 2016;9:ra8. doi: 10.1126/scisignal.aad0848. [DOI] [PubMed] [Google Scholar]
- Manteghi S, Gingras MC, Kharitidi D, Galarneau L, Marques M, Yan M, Cencic R, Robert F, Paquet M, Witcher M, et al. Haploinsufficiency of the ESCRT component HD-PTP predisposes to cancer. Cell Rep. 2016;15:1893–1900. doi: 10.1016/j.celrep.2016.04.076. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGFß signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattissek C, Teis D. The role of the endosomal sorting complexes required for transport (ESCRT) in tumorigenesis. Mol Membr Biol. 2014;31:111–119. doi: 10.3109/09687688.2014.894210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald ER, 3rd, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, Belur D, Castelletti D, Frias E, Gampa K, et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell. 2017;170:577–592. doi: 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Mellman I, Yarden Y. Endocytosis and cancer. Cold Spring Harb Perspect Biol. 2013;5:a016949. doi: 10.1101/cshperspect.a016949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-ß induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DSJ, Hill CS. In: Encyclopedia of Cell Biology. Bradshaw RA, Stahl PD, editors. Elsevier; 2016. TGF-ß superfamily signalling; pp. 37–50. [Google Scholar]
- Mitchell H, Choudhury A, Pagano RE, Leof EB. Liganddependent and-independent transforming growth factor-ß receptor recycling regulated by clathrin-mediated endocytosis and Rab11. Mol. Biol Cell. 2004;15:4166–4178. doi: 10.1091/mbc.E04-03-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- Nam JS, Terabe M, Mamura M, Kang MJ, Chae H, Stuelten C, Kohn E, Tang B, Sabzevari H, Anver MR, et al. An anti-transforming growth factor ß antibody suppresses metastasis via cooperative effects on multiple cell compartments. Cancer Res. 2008;68:3835–3843. doi: 10.1158/0008-5472.CAN-08-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolás FJ, Hill CS. Attenuation of the TGF-ß-Smad signaling pathway in pancreatic tumor cells confers resistance to TGF-ß-induced growth arrest. Oncogene. 2003;22:3698–3711. doi: 10.1038/sj.onc.1206420. [DOI] [PubMed] [Google Scholar]
- Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, Rasmussen RK, Seet BT, Sicheri F, Wrana JL. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19:297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Pickup M, Novitskiy S, Moses HL. The roles of TGFß in the tumour microenvironment. Nat Rev Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A, Vizán P, Das D, Chakravarty P, Vogt J, Rogers KW, Müller P, Hinck AP, Sapkota GP, Hill CS. TGF-ß uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition. eLife. 2018;7:e31756. doi: 10.7554/eLife.31756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL. Reciprocal regulation of signaling and endocytosis: Implications for the evolving cancer cell. J Cell Biol. 2017;216:2623–2632. doi: 10.1083/jcb.201705017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Teis D. The ESCRT machinery. Curr Biol. 2012;22:R116–R120. doi: 10.1016/j.cub.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer B, Hill CS. TGFß-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- Shi W, Chang C, Nie S, Xie S, Wan M, Cao X. Endofin acts as a Smad anchor for receptor activation in BMP signaling. J Cell Sci. 2007;120:1216–1224. doi: 10.1242/jcs.03400. [DOI] [PubMed] [Google Scholar]
- Szymanska E, Budick-Harmelin N, Miaczynska M. Endosomal “sort” of signaling control: The role of ESCRT machinery in regulation of receptor-mediated signaling pathways. Semin Cell Dev Biol. 2018;74:11–20. doi: 10.1016/j.semcdb.2017.08.012. [DOI] [PubMed] [Google Scholar]
- Tang LY, Yamashita M, Coussens NP, Tang Y, Wang X, Li C, Deng CX, Cheng SY, Zhang YE. Ablation of Smurf2 reveals an inhibition in TGF-ß signalling through multiple mono-ubiquitination of Smad3. EMBO J. 2011;30:4777–4789. doi: 10.1038/emboj.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFß receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-ß and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;16:1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizán P, Miller DSJ, Gori I, Das D, Schmierer B, Hill CS. Controlling long-term signaling: receptor dynamics determine attenuation and refractory behavior of the TGF-ß pathway. Sci Signal. 2013;6:ra106. doi: 10.1126/scisignal.2004416. [DOI] [PubMed] [Google Scholar]
- Weber JD, Yarden Y, Dustin M, Sassoon DA, Ezekowitz A. In: Encyclopedia of Cell Biology. Bradshaw RA, Stahl PD, editors. Elsevier; 2016. Functional cell biology: an overview; pp. 1–7. [Google Scholar]
- Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Dijke PT, Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-ß signalling. Oncogene. 2005;24:8080–8084. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Murphy SJ, Wilkes MC, Ji Y, Leof EB. Retromer maintains basolateral distribution of the type II TGF-ß receptor via the recycling endosome. Mol Biol Cell. 2013;24:2285–2298. doi: 10.1091/mbc.E13-02-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Kang JH, Andrianifahanana M, Wang Y, Jung MY, Hernandez DM, Leof EB. Basolateral delivery of the type I transforming growth factor ß receptor is mediated by a dominant-acting cytoplasmic motif. Mol Biol Cell. 2017;28:2701–2711. doi: 10.1091/mbc.E17-05-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, Huang H, Sheppard KA, Porter JA, Lu CX, ten Dijke P. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-ß type I receptor. Nat Cell Biol. 2012;14:717–726. doi: 10.1038/ncb2522. [DOI] [PubMed] [Google Scholar]
- Zwang Y, Yarden Y. Systems biology of growth factor-induced receptor endocytosis. Traffic. 2009;10:349–363. doi: 10.1111/j.1600-0854.2008.00870.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.