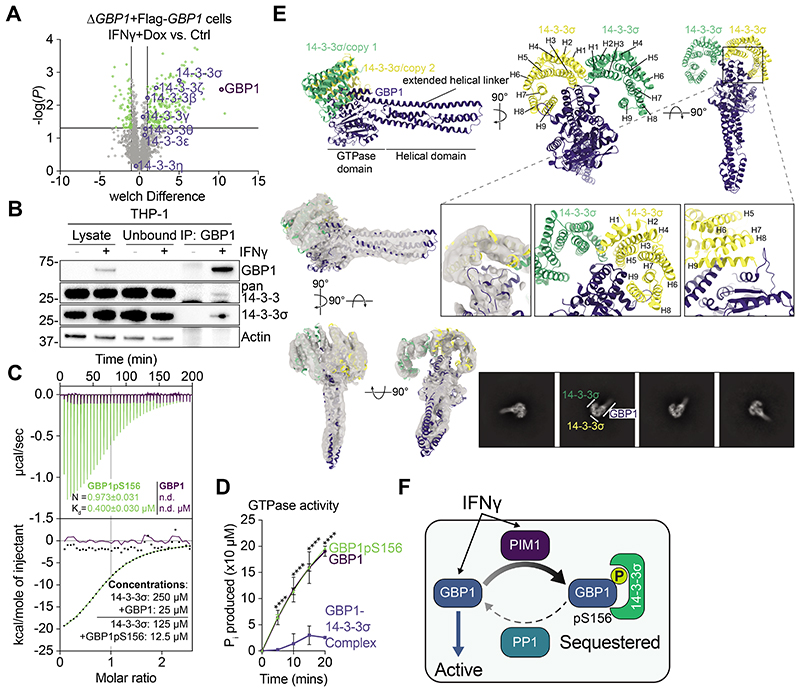
Fig. 3. Phosphorylated GBP1 is bound and inactivated by 14-3-3σ.
(A) Volcano plot of mass spectrometry data analysis of GBP1-interacting proteins obtained following co-immunoprecipitation of Flag-GBP1 from IFNγ (CTRL) or IFNγ+ Dox-treated THP-1ΔGBP1+Flag-GBP1 cells. GBP1-interacting proteins above the significance threshold highlighted in green and 14-3-3 proteins in blue. (B) Immunoblots of co-immunoprecipitation of endogenous GBP1 from THP-1 WT cells treated with IFNγ. (C) Isothermal titration calorimetry determining thermodynamics of GBP1:14-3-3σ complex formation. 14-3-3σ at indicated concentrations was injected in 7 μL aliquots to non-phosphorylated or in vitro phosphorylated GBP1 at indicated concentrations. Determined molar ratio N at equilibrium and dissociation constant Kd as shown in the figure. (D) GTPase activity assay of 2 μM free GBP1, in vitro phosphorylated GBP1 or GBP1:14-3-3σ complex. (E) Top: Overview of the trimeric GBP1:14-3-3σ complex consisting of two 14-3-3 copies (yellow + green) bound to the GBP1 GTPase domain. Middle: insets depict details of GBP1:14-3-3 dimer binding interface. Cryo-EM density is shown in grey with rigid-body docked GBP1 (PDB: 1F5N) and 14-3-3σ (PDB: 1YWT) crystal structures. Bottom: Representative 2D classes with highlighted complex components. (F) Cartoon depicting the observed inhibitory mechanism, in which PIM1 phosphorylates human GBP1 at Ser156 which is subsequently bound by 14-3-3σ. Upon dephosphorylation, GBP1 is liberated and becomes active. Data information: Images in (B) representative of n = 3 experiments. Graph in (A+D) show mean ± SD from n = 3 experiments. Curves in (C) representative of n = 3 experiments and fitted to a model with one set of binding sites. **** P < 0.0001 for indicated comparisons in (D) from 2-way ANOVA following adjustment for multiple comparisons. For gel source data, see Data S5.