Abstract
The fungal genus Armillaria contains necrotrophic pathogens and some of the largest terrestrial organisms that cause tremendous losses in diverse ecosystems, yet, how they evolved pathogenicity in a clade of dominantly non-pathogenic wood-degraders remains elusive. Here, we show that Armillaria species, in addition to gene duplications and de novo gene origins, acquired at least 1,025 genes via 124 horizontal gene transfer (HGT) events, primarily from Ascomycota. HGT might have affected plant biomass-degrading and virulence abilities of Armillaria, and provides an explanation for their unusual, soft rot-like wood decay strategy. Combined multi-species expression data revealed extensive regulation of horizontally acquired and wood-decay related genes, putative virulence factors as well as two novel conserved pathogenicity-induced small secreted proteins, which induced necrosis in planta. Overall, this study details how evolution knitted together horizontally and vertically inherited genes in complex adaptive traits of plant biomass degradation and pathogenicity in important fungal pathogens.
Plant pathogenic fungi cause significant economic losses worldwide in a wide variety of plant species, including forest trees. Among tree pathogens, the genus Armillaria (Basidiomycota, Agaricales, Physalacriaceae) stands out as one of the most important in temperate systems, responsible for great losses in both natural and planted stands of woody plants1–3. At the genus-level they are known to cause ‘Armillaria root-rot disease’3,4. The most common pathogenic species A. mellea sensu lato has been reported to infect >500 plant species1 and is solely responsible for up to 40% annual loss of vinegrape in California5. The gymnosperm-specific A. ostoyae is responsible for considerable losses in conifer forests1.
Armillaria species evolved a range of features exceptional or rare among fungi, which have conceivably all emerged in its most recent common ancestor (MRCA). These include a very low mutation rate, extreme longevity and immense colony sizes (>2,500 years, >900 hectares1–3), diploidy, bioluminescence, specialized underground structures known as rhizomorphs1, and potential to facilitate atmospheric N2 fixation6. Perhaps the most economically important aspect of this genus is the ability to infect and kill woody plants1,3. Most Armillaria species are broad host range necrotrophs1,3. After infection through the roots, they colonize and kill the cambium of mostly weakened trees, causing the death of the plant and enabling the fungus to transition to its necrotrophic phase1,7. Despite a well-documented epidemiology and etiology2,7,8, molecular aspects of the infection process are poorly known. Recent studies and genome sequencing efforts highlighted certain secondary metabolites, plant cell wall degrading enzymes (PCWDEs), chitin-binding proteins and expanded protein-coding repertoires enriched in putative pathogenicity-related genes, among others1,6,9. It is likely that these previous findings only cover a fraction of the virulence genes of Armillaria, leaving much of its pathogenic arsenal yet to be characterized.
As a result, it is unknown how infection models developed based on better-studied necrotrophic fungi (e.g. Sclerotinia sclerotiorum10, or Colletotrichum spp.11), are applicable for Armillaria, if at all, and what traits the latter evolved for plant infection. In other necrotrophs, broad roles of tissue acidification, tolerance towards and detoxification of plant secondary metabolites and reactive oxygen bursts12, secretion of diverse PCWDEs11,13 and effectors14 have been established as key assets of infection. However, given that most relatives of Armillaria are non-pathogenic, it can be inferred that it evolved necrotrophy independently, which might have resulted in unique infection mechanisms, our understanding of which remains limited.
Here, we sequenced eight new genomes and report transcriptome data from new in planta and in vitro pathosystems, enabled us to explore genome evolution and key aspects of the necrotrophic lifestyle of Armillaria spp. We infer that gene duplications, genus-specific gene families and horizontal gene transfer have shaped the genomic toolkit available for plant infection and biomass degradation. RNA-Seq data from six experiments and four Armillaria species, including new in planta time series and fresh tree stem invasion experiments allowed us to decipher gene expression patterns specific for these processes in virulent and non-virulent strains. Experimental validation of predicted pathogenicity-induced small secreted proteins revealed potential conserved virulence factors in Armillaria. Overall, our phylogenomic and gene expression studies elucidate how vertically and horizontally acquired genes became integrated into complex adaptive traits of a globally important fungal group.
New Armillaria genomes
We report the high-quality annotated de novo genomes of eight Armillaria species (Figure 1, Supplementary Table 1). The new genomes were assembled to 33-864 scaffolds comprising 40-79 Mbp haploid size, with 12,228 - 19,984 predicted gene models and BUSCO (fungi) completeness of 97.7-99.7% .
Figure 1. Genome statistics and reconstruction of ancestral genome sizes for 15 Armillaria species and 5 Physalacriaceae outgroups.
Numbers at nodes represent ancestral proteome sizes in the Physalacriaceae tree. Purple circles correspond to sizes for each node in the tree (for gene gains and losses at each node, and for the complete species tree, see Extended Data Fig. 1A). Blue circles represent BUSCO scores for each species. For genome sizes, darker regions show TE sizes (Mbp) from the primary scaffolds, and lighter color shows the genome assembly sizes in Mbp (for TE categories, see Extended Data Fig. 1B). For proteome sizes, darker color shows proteins with no known functional domains (unannotated proteins). Carbohydrate active enzymes darker color shows plant cell wall degrading enzymes, lighter color shows other CAZymes. Secretomes and small secreted proteins - darker color shows unannotated proteins, lighter color shows proteins with known functional domains (for general genome statistics of all 66 species in Dataset 1, see Supplementary Table 1). Clade names are based on Koch et al, 201741.
We sampled all major clades recognized currently15, including the Northern Hemisphere , the Australasian/Southern American , African and melleoid clades (Figure 1). We also included 2 species from subgenus Desarmillaria (A. tabescens and A. ectypa), of which the moss-associated A. ectypa had the smallest genome among Armillaria spp., as well as Guyanagaster necrorhizus6, which is the sister genus of Armillaria15.
Genetic innovations in Armillaria clade
To analyze the genomic innovations associated with the emergence of Armillaria, we used genomes of 15 Armillaria species and 5 outgroups from the Physalacriaceae. We combined these with other Agaricales exhibiting a range of lifestyles, resulting in a dataset of 66 species (Supplementary Table 1).
Reconstruction of genome-wide gene gain/loss patterns revealed genome expansion in Armillaria. We estimated 9,929 ancestral genes at the root node, suggesting an early origin for most genes (Extended Data Fig. 1A). The Armillaria genus showed a net genome expansion with 18,662 protein-coding genes inferred for the most recent common ancestor (mrca) of Armillaria (2,913 duplications, 189 losses), as opposed to 15,938 for that of Armillaria and Guyanagaster and 18,155 for the mrca of Armillaria and Hymenopellis (Figure 1, Extended Data Fig. 1). The mrca of the Northern Hemisphere clade was inferred to have further expanded to 23,756 genes . These data indicate that the large protein coding repertoires of Armillaria spp. can be explained by genus-specific gene duplications, as suggested before9.
The 2,913 duplications in the mrca of Armillaria happened in a total of 1,473 orthogroups (OGs). Gene ontology (GO) and InterPro enrichment analyses showed significant overrepresentation of 55 molecular functions, 18 biological processes and 3 cellular component terms ( p <0.05) (Extended Data Fig. 2) and 733 InterPro terms (Supplementary Table 2). These included functions related to plant biomass utilization, such as pectin-degradation (pectate lyases, pectin lyases, esterases, GH28), cellulose binding, and putative extracellular and aromatic compound breakdown (e.g. intradiol ring-cleavage dioxygenase, multicopper oxidases) (Supplementary Table 2, Extended Data Fig. 2). Duplicated genes were also enriched in putative pathogenesis related gene families, including genes encoding deuterolysins, aspartic peptidases, chitin deacetylases, and SCP-like Golgi-associated pathogenesis-related proteins. Ceratoplatanins and LysM domains, which were reported to assist infection in pathogenic fungi16,17 were also enriched in Armillaria.
Small secreted proteins (SSPs), which include cysteine-rich proteins involved in, among others, host colonization18, were found in 270-507 copies in Armillaria, with A. fuscipes having the fewest and A. luteobubalina having the most. Of these, 45-57% had no known functional domains, which we hereafter refer to as unannotated SSPs. Typically this latter class of SSPs are called candidate secreted effector proteins in pathogenic fungi11,19.
Gene families that arose within and are conserved in most Armillaria spp. may be particularly relevant for explaining Armillaria-specific innovations. We found 212 such families (Supplementary Table 2, Supplementary Fig. 1); of which 116 consisted of proteins with no known functional annotations. The remaining orthogroups were dominated by F-box domains, Leucine-rich repeats, Cytochrome P450s, Zinc-finger C2H2 type transcription factors, protein kinases and other fast evolving gene families.
Armillaria spp. are among the few bioluminescent fungi. Ke et al. reported the luciferase gene cluster in Armillaria, comprising five genes20. We found the luciferase cluster to be conserved and highly syntenic in Armillaria genomes (Supplementary Fig. 2), however, the cluster was missing in other Physalacriaceae, suggesting it was lost in those, or gained in the common ancestor of Armillaria and Guyanagaster.
Plant biomass degradation by Armillaria
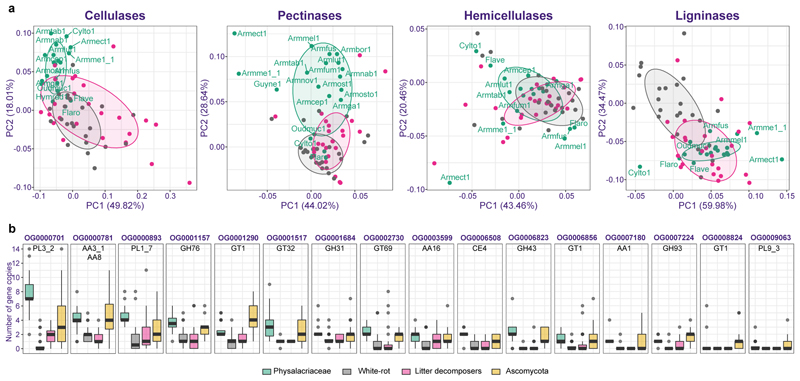
Armillaria species are reported to be facultative necrotrophs that first kill the host, then utilize its biomass during the saprotrophic phase. Using the new genomes we analyzed the wood decay strategy of Armillaria, relative to other fungi based on their PCWDE gene repertoires. Similar to white rot fungi and necrotrophs21, Armillaria species possess the complete enzymatic repertoire for degrading woody plant biomass (Figure 1). We generated phylogenetic PCAs for Armillaria species, other Physalacriaceae, white rot and litter decomposer species (Supplementary Table 1) based on PCWDE copy numbers acting on cellulose, hemicellulose, pectin and lignin. Cellulases and pectinases clearly separated Armillaria spp. and other Physalacriaceae from white rot and litter decomposer fungi (Figure 2, Extended Data Fig. 3), whereas hemicellulase and ligninase PCA-s grouped them together (Figure 2, Supplementary Fig. 3, Supplementary Fig. 4, Supplementary Fig. 5). Cellulase loading factors indicate that this separation was mainly driven by expansins, the AA16, AA8, and AA3_1, GH1 and GH45 families. In line with loading factors, Armillaria and other Physalacriaceae have more AA3_1, GH1 and GH45 genes than white rot and litter decomposer fungi (Supplementary Fig. 6). On the other hand, they were depleted in CBM1 and GH5_5 with an average of 12 and 1.5 genes respectively, while white rotters and litter decomposers have 20-60 CBM1 and 4-10 GH5_5 genes. Notably, a few brown rot and ectomycorrhizal fungi had more gene copies of CBM1 and GH5_5 than Armillaria (Supplementary Fig. 6). The GH44 family, which is specific to Basidiomycota22, is missing in the Physalacriaceae and Armillaria, indicating that in specific cases gene losses also drove trophic mode evolution of these fungi towards an Ascomycota-like lifestyle (see below).
Figure 2. Plant biomass degradation related genes in Armillaria.
a) Phylogenetic PCAs for PCWDE gene families. Species abbreviations are shown only for Physalacriaceae species (all species names and PCA loadings are given in Extended Data Fig. 3 and Supplementary Table 3). b) Boxplot of copy numbers of 16 CAZy orthogroups co-enriched in Physalacriaceae (n=20) and in Ascomycota (n=21) with respect to white rot (n=23) and litter decomposer fungi (n=24). The box plots shows the median and interquartile range, with the upper whiskers extending to the largest value from the 75th percentile, and lower whiskers extending to the smallest value from the 25th percentile. Scale limits for the boxplot were set to 14, losing one sample point (Conioc1 in OG0000781 with 18 genes). Lifestyles of the species used are denoted by color.
In the pectinase PCA, the highest loading family was CBM67 , which is enriched in Armillaria species but absent in most white rot species in our dataset (Supplementary Fig. 7). CBM67s are frequently associated with GH78 and PL1 proteins and are putatively binding rhamnose, a pectin component23. The PL1 family is present in all Physalacriaceae species, but is depleted or missing in many white rotters and litter decomposers (Supplementary Fig. 7). Other pectin-acting families, such as GH28, GH53, GH88, CE8, and CE12, were present in higher numbers in Armillaria and other Physalacriaceae than in white rot and litter decomposer fungi, whereas PL1_7, PL26, PL3_2, and PL4_1 were abundant in Armillaria but absent in most other decomposer species.
These analyses portray Armillaria and the Physalacriaceae as versatile wood decayers that are distinct from white rotters, despite previous classifications as such3,9,24. This is consistent with microscopy, chemical and transcriptomic analyses25,26, which indicated that their decay is similar to soft rot27,28, a decay mode known only in the Ascomycota. This apparent discrepancy prompted us to systematically look for similarities between the Physalacriaceae and the Ascomycota. We found 16 PCWDE orthogroups that were significantly overrepresented in both groups with respect to white rotters and litter decomposers (BH-corrected p-value >0.05, Fisher’s exact test, Supplementary Table 3, Figure 2). These included several of the high-loading families from the PCAs, as well as other PCWDEs acting on cellulose (AA3_1, AA8, CBM1), pectin (PL3_2, PL1_7, PL9_3), cellulose/chitin (AA16), and hemicellulose (GH31, GH43, GH93, CE4). These families could either be the result of co-expansion in both the Ascomycota and the Physalacriaceae or represent horizontal gene transfer (HGT) events. Blast searches with Armillaria GH28 genes suggested the latter scenario to be more likely, which led us to systematically evaluate the role of HGTs.
Widespread horizontal transfer of genes from Ascomycota
To identify horizontally transferred (HT) genes, we first screened candidates based on the Alien index (AI)29 across a broad set of 942 species of fungi, plants and bacteria (Supplementary Table 1, Dataset3, Supplementary Table 4). Then, we validated each candidate HT gene by phylogenetic analyses (≥70% bootstrap in maximum likelihood gene trees) and in-depth similarity searches (UniRef100 database) (see Methods; gene trees available at Figshare - https://figshare.com/articles/dataset/Gene_trees/22730534, Supplementary Table 1, Dataset3). Overall we recovered 124 strongly supported horizontal transfer events (Figure 3A) into Physalacriaceae, corresponding to 1,387 individual genes, in 110 orthogroups. We identified 37-107 HT genes per species in Physalacriaceae, with H. radiata having the most and Cylindrobasidium torrendii the fewest. Among Armillaria, A. tabescens had the highest estimated number of HT genes (83), and G. necrorhizus the lowest (51) (Figure 3A, Supplementary Table 4).
Figure 3. Horizontal gene transfers into Armillaria and the Physalacriaceae family.
a) Barplot showing the number of phylogenetically validated HT genes identified in each genome. b) Schematic representation of donor and recipient relationships for HT events after phylogenetic validation. Size of the arrows are proportional to the number of HGT events inferred from all the nodes belonging to a specific donor group. The number of events for each donor group are listed along the arrows. c) Violin plot showing expression dynamics of phylogenetically validated HT genes and vertically transferred (VT) genes. X-axis shows the sample names and the y-axis shows log2 FPKMs (For expression dynamics of HT and VT genes in developmental dataset, see Extended Data Fig. 4).
Multiple internal nodes of the Physalacriaceae tree were identified as putative recipients. 104 HGT events were confidently associated with Ascomycota as donors, in particular the Sordariomycetes and Dothideomycetes (34 and 31 events, respectively). We find that ~40% of HT genes have undergone duplications, indicating that they were likely integrated into the life history of Physalacriaceae. Expression levels of HT genes support their functionality (Figure 3C, Extended Data Fig. 4).
Among phylogenetically validated HT genes, we found 164 CAZymes (Supplementary Table 4), of which 117 belonged to families co-enriched in Ascomycota and Physalacriaceae (see above), suggesting that the co-enrichment signal was likely created by HGT. In addition to CAZymes, HGT affected intradiol ring-cleavage dioxygenases, CAP domain proteins, Pyr1-like SCP domains, as well as cytochrome P450, peptidases, transporters and transcription factors (Supplementary Table 4).
Taken together, we find that horizontal transfer affected several gene families associated with wood-decay (e.g. AA3_1, GH43, PL3_2), as well as plant-fungal interactions (e.g. CAP domain proteins, peptidases), which suggests that it might have shaped the plant biomass degrading and pathogenic abilities of the genus. Given that Physalacriaceae species have been reported to cause soft-rot-like decay which is classically restricted to the Ascomycota30, we hypothesize that HGT contributed to the evolution of plant biomass degrading ability of Armillaria. We speculate that the large number of putative HGT events from Sordario- and Dothideomycetes might stem from the extensive contact of Armillaria ssp. with plant pathogens in these classes and/or their peculiar niche and longevity. Their rhizomorphs can reach several meters in length and can contact numerous other soil microbes, perhaps providing time windows for gene exchange. Additionally, other idiosyncrasies of Armillaria, such as diploidy could also be factors promoting HGT.
Gene expression profile in plant colonization and wood decay
Armillaria enters the host through the roots, colonizes and kills the cambium, leading to death of the plant and the onset of the necrotrophic phase1. Molecular aspects associated with this process are hardly known, with most information available on the wood-decaying phase9,26. To obtain a molecular perspective on these strategies we produced new RNA-Seq data for two in vitro pathosystems and re-analyzed data from 2 published studies on wood-decay as well as rhizomorph and fruiting body development9,26. New data were generated from an in planta time-series experiment of A. luteobubalina infecting Eucalyptus grandis seedlings (Extended Data Fig. 5A) and for in vitro fresh tree stem invasion assays with highly and less-virulent isolates of both A. ostoyae and A. borealis (Extended Data Fig. 5B), emulating the cambium-killing phase of the fungus.
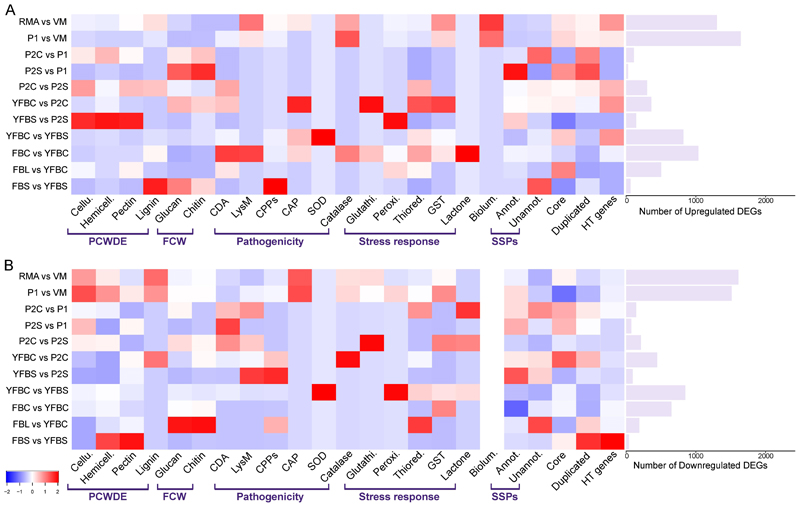
We independently analyzed differentially expressed gene (DEG) lists from each experiment and calculated DEG enrichment ratios in each of the 24 gene groups we defined (see Methods).
These ratios reflect how enriched DEGs are in a given gene group in a given experiment and are shown as heatmap on Figure 4 (and Extended Data Fig. 6). The different experiments showed distinct enrichment patterns. Cellulose-, hemicellulose-, pectin- and lignin-related PCWDE genes were enriched in stem invasion and wood-decay experiments, as expected9,31. On fresh stems, pectinase encoding genes were most dominant, possibly enabling the fungus to break carbohydrate - lignin bonds and spread between the bark and the sapwood. At the same time PCWDEs were depleted during the infection phase in the in planta time series experiment, indicating that A. luteobubalina did not induce these genes during the infection phase. This is consistent with most necrotrophic fungi expressing a limited set of PCWDEs for plant penetration and a larger battery of enzymes during necrotrophy11,13. Other virulence-related genes include cerato-platanins16, which were enriched at 24h and 48h in A. luteobubalina and in stem invasion by A. borealis . HT genes showed an enrichment in wood-decay and stem-invasion experiments. Bioluminescence genes were enriched in root-invading mycelium and rhizomorph (A. ostoyae) samples. SSPs, expanded, novel and stress-related genes were enriched in various fruiting body developmental stages (Extended Data Fig. 6). Genes related to oxidative stress were upregulated in various stages of plant infection and in stem invasion assays (Figure 4, Extended Data Fig. 7). Specifically, genes encoding superoxide dismutases, catalases and members of the glutathione system and ergothioneine pathway were enriched among DEGs in the in planta invasion assays, whereas genes encoding glutathione-S-transferases and catalases were enriched in wood-decay experiments.
Figure 4. Enrichment of differentially expressed genes of wood-decay, pathogenicity, stress-response and other gene families in 6 RNA-Seq datasets.
The heatmap shows enrichment ratios for 24 gene groups (x-axis) from aggregated differential gene expression data across 6 experiments (a - upregulated, b - downregulated genes). Enrichment ratios were calculated by considering the number of DEGs in a given gene group in the given sample, all DEGs in that sample as well as all genes in the genome and all genes in that gene group. The total number of DEGs in each experiment is shown as a barplot at right. In the heatmaps, warmer colors mean higher enrichment ratios (for a complete list of enrichment ratios Supplementary Table 5).
Figure 4 also shows species- and strain-specific enrichment patterns among up- and downregulated genes. For example, the virulent A. borealis A6 strain had a unique enrichment of pathogenicity-related LysM proteins and cerato-platanins, which was not seen in the less-virulent A4 strain or in A. ostoyae. Nevertheless, in both A. borealis and A. ostoyae, the virulent strains (A6 and C18, respectively) showed more enrichment across most gene groups tested. The species- and strain-specific enrichment patterns also suggest that each species mount a different response under the examined conditions, which might correlate with differences in lifestyle or virulence.
Early stages of live host colonization by Armillaria
To understand early stages of Armillaria infection, we analyzed gene expression of A. luteobubalina colonizing E. grandis roots across five time points (pre-symbiosis, 24hrs, 48hrs, 1 week and 2 weeks) in four biological replicates and compared these to non-symbiotic samples (Supplementary Fig. 8, Supplementary Table 5). SuperSeq32 revealed that we could detect 77-94% of differentially expressed genes (DEGs) in A. luteobubalina across different timepoints, whereas, in E. grandis, we could variably detect 25-70% of DEGs (Supplementary Fig. 9). DEGs were arranged into modules based on expression similarity using the Short Time-series Expression Miner (STEM)33. This yielded eight and four significant modules in the fungus and plant, respectively. Five fungal modules showed gradual increase in expression through time (Supplementary Fig. 10). Genes in these were enriched for GO terms often associated with pathogenic interactions34, with the strongest signal for (oxidative) stress (Supplementary Fig. 10, Extended Data Fig. 7). We identified A. luteobubalina DEGs related to four pathogenic processes: host immune suppression, oxidative stress, detoxification, and cytotoxicity (e.g. cerato-platanins).
Genes encoding glutathione-S-transferases were almost uniformly upregulated in plant-associated samples, compared to free-living mycelium, whereas other genes related to oxidative stress response (e.g. superoxide dismutases) were mostly upregulated at 1 and 2 weeks (Extended Data Fig. 7). Toxin efflux systems, which are involved in the tolerance against plant secondary and defense metabolites (e.g. phytoalexins, phytoanticipins), have been reported to be transcriptionally regulated in necrotrophs35. Membrane transporters, cytochrome P450 monooxygenases and laccases, which are often associated with detoxification or plant metabolites35,36, showed extensive regulation during the experiment (Extended Data Fig. 7). On the other hand, we did not observe extensive regulation of PCWDEs (Figure 4).
We observed an upregulation of HT genes at certain stages of in planta colonization. Of the 78 A. luteobubalina HT genes , 29 were differentially expressed at some time point (Supplementary Table 5). We identified two horizontally acquired CAP domain/pathogenicity-related-1 (PR-1) genes which were strongly induced across the time-series and three LysM domain proteins (CBM50) that were upregulated at 24 and 48h. Both PR-1 and LysM genes are known to be involved in fungal pathogenicity, by facilitating the transport of fatty acids and sterols37 and masking the presence of chitin residues from the plant immune system, respectively38.
The four significant expression modules of E. grandis were enriched in defense-related terms, including chitinase activity, chitin binding, defense response to fungus, defense response to bacterium (Supplementary Fig. 11), suggesting an activation of plant immunity. A finer level analysis of the plant response identified 95 DEGs up-regulated across the whole time course and 59 DEGs repressed in the same series (Supplementary Fig. 12). Within the up-regulated genes we found enrichment for phosphoprotein phosphatase activity due to up-regulation of PP2 family genes related to effector immunity and ABA-JA cross-talk39. Other GO terms enriched in this gene set included unfolded protein binding/chaperone activity, heat shock and jasmonic acid response due to PR4 and MYB108 gene induction40. Of the core repressed genes, we found GO enrichment for copper ion binding proteins, peroxidase activity, carboxylic acid binding, and lipid transfer. Reduction of copper ion binding proteins, peroxidase activity and lipid transfer would, likely, lead to a delayed hypersensitive response as copper ions are necessary for ethylene production and repression of the ABA biosynthesis41 while peroxidase activity directly leads to plant cell death and the lipid-associated genes encode DIR-like proteins responsible for long-range immune signaling42. The genes associated to carboxylic acid binding and hydrolase activity, meanwhile, likely affect pathogen nutrition more directly as the repressed genes associated to the former are involved in biosynthesis of L-serine, lack of which would reduce the nutrition value of E. grandis tissues, while the latter are involved in detoxification (i.e. cyanoalanine nitrilase) which could lead to a toxic build-up of 3-cyano-L-alanine thereby inhibiting pathogen growth43.
Pathogenicity-induced SSPs
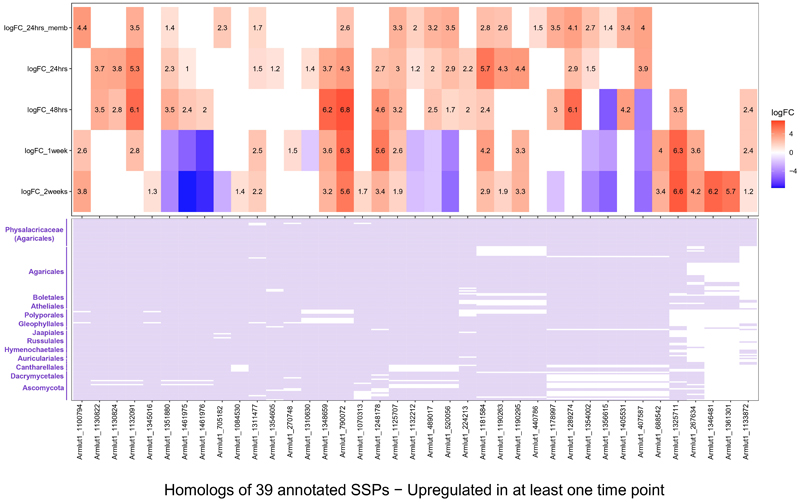
Phytopathogenic fungi often utilize effectors that suppress or manipulate host defense responses14,44. As many effectors are short proteins, we scrutinized the 39 and 38 annotated and unannotated SSPs, respectively, of A. luteobubalina that were upregulated in at least one time point (Figure 5 and Extended Data Fig. 8). Annotated SSPs with highest fold changes (FC) included Pry1-like, SCP- or CAP-, Ricin-B-like lectin- and LysM domain containing genes (Supplementary Table 6). Two unannotated SSPs (1165297 and 1348401) had the highest fold change values among all genes, so we selected these for experimental validation and call them pathogenicity-induced SSPs (PiSSPs, Figure 5A, red arrows). Gene 1348401 had a peak expression at 48hrs whereas 1165297 at 1 week. The encoded proteins contain no known conserved domain and have no predicted function.
Figure 5. Pathogenicity-induced SSPs (PiSSPs) of A. luteobubalina induce cell death in host plants.
a) Heatmap shows log2 fold changes for SSPs significantly upregulated in at least one time point. Red shows higher and blue depicts lower logFC, followed by presence/absence matrix of homologs of unannotated SSPs in 131 species (Dataset 2). The two experimentally validated PiSSPs are shown by red arrows. b) Transient transformation of the non-host N. benthamiana and the host E. grandis with negative controls (empty vector; Pisolithus microsporus proteins Pismi_683611 and Pismi_689601), with a positive control (BAX) or with an in planta expression vector encoding Armlut_1165297 or Armlut_1348401. As further controls, frameshift versions of the A. luteobubalina sequences were included (Armlut_1165297_fs or Armlut_1348401_fs).
We cloned both PiSSPs (1165297 and 1348401) and expressed them transiently in the non-host Nicotiana benthamiana and the host E. grandis (Figure 5B). The positive control expression of BAX led to cell death in N. benthamiana as well as in E. grandis. The expression of the two A. luteobubalina PiSSPs in E. grandis leaves, likewise, led to rapid cell death (Figure 5B), indicating a specific interaction with plant tissue. This cell death phenotype was not seen in heterologous expression of these proteins with an induced frameshift mutation, nor in leaves expressing secreted proteins from mutualistic fungi45, nor from empty vector transformants. These data indicate that these A. luteobubalina PiSSPs can cause necrotic lesions in host plant tissue.
Orthology searches revealed that the two PiSSPs were surprisingly conserved (Figure 5, Extended Data Fig. 9, Extended Data Fig. 10). Homologs of 1348401 are present throughout the Agaricomycetes and the family strongly expanded in Armillaria, with on average 7 genes per species compared to 1.8 in other Agaricales (Extended Data Fig. 9). The non-pathogenic A. ectypa had the fewest genes (3) in this family. The evolutionary conservation of these two proteins outside Armillaria is somewhat surprising and is in contrast with observations on other (mostly Ascomycota) effectors, which are believed to evolve fast and are often species-specific18,19. Whether they trigger cell death via direct (as true effectors) or indirect routes remains to be established. Nevertheless, these data raise the possibility that Armillaria use specific mechanisms for necrotrophy, as postulated by the gene-for-gene model14.
Potential virulence factors in Armillaria spp
In vitro invasion of freshly cut conifer stems were used to explore gene expression in highly and less-virulent isolates46,47 of the conifer-specific A. ostoyae and A. borealis (Extended Data Fig. 5). These experiments emulated cambium colonization and killing, and virulent strains consistently colonized the stems faster. We performed differential gene expression analyses between control and invasive mycelia as well as between virulent and less-virulent strains of the same species (Supplementary Table 5). According to SuperSeq32 we detected 89.4-98.6% of all DEGs (Supplementary Fig. 9).
To uncover cambium colonization strategies, we scrutinized genes upregulated in invasive mycelia, relative to control mycelia (Supplementary Note 1). We identified upregulation of PCWDEs, in particular pectinases, as a major transcriptional response to fresh stems in both species. Enrichment ratios confirmed that pectin-related genes are induced in the largest proportions, followed by hemicellulases, cellulases and lignin-related genes (Figure 4). Horizontally transferred intradiol ring-cleavage dioxygenases were also upregulated in these experiments.
We also aimed to identify genes encoding potential virulence factors by comparing highly and less-virulent isolates (Supplementary Note 1). Based on manual curation and sequence-based comparison of upregulated genes against PHI-base, we identified 303 and 56 candidate virulence factor genes from A. borealis and A. ostoyae (Supplementary Table 6). We further determined the secreted proteins to predict potential plant interacting factors. In A. borealis, 40 secreted proteins, including 9 SSPs, 5 CAZymes, 2 peptidases, 2 intradiol ring-cleavage dioxygenases and one hydrophobin-related gene were identified. In A. ostoyae we found 18 genes, including 3 SSPs, 5 CAZymes and 1-1 genes representing hydrophobins, intradiol ring-cleavage dioxygenases and tyrosinases (Supplementary Table 6). Notably, highly virulent strains of both species upregulated a pair of polygalacturonase (GH28) and cellulase (GH3) genes, both homologous to virulence effectors reported from Ascomycota (Supplementary Table 6).
Discussion
The genus Armillaria is a globally important group of primarily tree pathogens that evolved a range of unique or rare traits, from extreme colony sizes to bioluminescence and represent an independent origin of broad host-range necrotrophy. In search of the genetic underpinnings of the unique biology of these fungi, in this study we combined new genomes with transcriptomic profiling of in planta and in vitro pathosystems. Comparative genomic analyses support the view that Armillaria spp. have an expanded protein-coding repertoire and possess a complete set of CAZymes for degrading lignocellulosic plant biomass (including lignin)3,9,24,48. However, we also detected a considerable influx of Ascomycete genes through HGT which, based on the expression data, seem to have influenced both the plant biomass degrading ability and pathogenic attributes of Armillaria. Previous studies showed that wood-decay by Armillaria is reminiscent of soft-rot26–28,49, a lifestyle known only in Ascomycota. Congruently, our phylogenetic PCA analyses separated Armillaria from white rot species, which is paradoxical given that white rot is the dominant wood-decay strategy in the Agaricales31. Alien CAZyme genes acquired through HGT from Ascomycota may explain this controversy and establish HGT as a source of novelty in the evolution of fungal plant biomass degrading systems in the Basidiomycota.
Armillaria are necrotrophic pathogens that first kill, then feed on their hosts1,9. Our data provided the first insights into the molecular mechanisms of early- and late stages of colonization of host plants. We detected genes related to four major necrotrophic processes: host immune evasion and suppression, oxidative stress, detoxification, and cytotoxicity, as well as specific regulation of horizontally transferred genes. The presence of pathogenicity-induced small-secreted proteins and the lack of broad plant biomass degrading CAZyme gene upregulation during early colonization supports the emerging view that, instead of a non-specific, brute-force attack with a broad battery of CAZymes, necrotrophs use specific effectors for modulating the plant immune system14. The expression patterns of the two PiSSPs are consistent with the gene-for-gene theory of pathogenicity, which has traditionally been discussed for (hemi-)biotrophs50, but may apply to necrotrophs as well14. Determining whether the two PiSSPs are truly specific effectors will require uncovering the mechanisms of their action, especially in light of their broad conservation, but these data provide an entry point into investigating fine details of Armillaria - plant interactions. In contrast to early-stage infection, cambium colonization was characterized by a broad upregulation of plant cell wall degrading CAZymes. Comparisons of low and highly-virulent strains in these assays further provided insights into genes associated with virulence, such as certain polygalacturonase- or cellulase-encoding genes.
Overall, this study revealed the molecular underpinnings of the lifestyle of a group of widespread pathogens of woody plants, which, combined with existing and emerging experimental tools will facilitate research on the molecular mechanisms of plant colonization. We propose that genome evolution in Armillaria relied on combined effects of multiple types of genetic innovation, including HGT, and that genes gained early during the evolution of the genus were integrated into cellular regulatory networks of plant biomass degradation and pathogenicity.
Methods
Strains for Sequencing, Assembly and Annotation
Armillaria strains (Supplementary Table 1) were inoculated on Malt Extract Agar (MEA) and incubated at 25°C in the dark for 7-10 days. Identity of each strain was confirmed by amplifying and sequencing the fungal internal transcribed spacer (ITS1-5,8S-ITS2) region and bacterial contamination was checked using universal 16S primers (Integrated DNA Technologies). Strains were inoculated in Malt Extract Broth in 500ml Erlenmeyer flasks and incubated at 25oC in the dark for 4-5 weeks until a substantial quantity of fungal biomass was grown. Fungal mass was stored at -80°C until DNA and RNA extraction. Prior to nucleic acid extraction, fungal tissues were homogenized using liquid nitrogen in mortar and pestle. DNA extraction was performed using the Blood & Cell Culture DNA Maxi Kit (Qiagen Inc.) and RNA extraction was performed using the RNeasy Midi Kit (Qiagen Inc.) as per manufacturer's instructions. Using the DNA, we again confirmed strain identity by sequencing the ITS region and nucleic acid quantity was measured using Qubit (ThermoFisher) according to the manufacturer’s instructions.
Genomic and transcriptomic library preparation, sequencing, assembly and annotation
For A. nabsnona, A. mellea, and A. ectypa, 5 ug of genomic DNA was sheared to approximately 15-20 kb using Megaruptor3 (Diagenode). The sheared DNA was treated with DNA Prep to remove single-stranded ends, and DNA damage repair mix followed by end repair, A-tail and ligation of PacBio overhang adapters using SMRTbell Express Template Prep 2.0 Kit (Pacific Biosciences). The final libraries were size selected with BluePippin (Sage Science) at 10 kb cutoff size. For A. borealis, A. fumosa, A. novae-zelandiae, and A. tabescens, 5 ug of genomic DNA was left unsheared due to marginal HMW DNA quality. The unsheared gDNA was treated with exonuclease to remove single-stranded ends and DNA damage repair mix followed by end repair and ligation of blunt adapters using SMRTbell Template Prep Kit 1.0 (Pacific Biosciences). All libraries were purified with AMPure PB beads.
The PacBio Sequencing primers were then annealed to the SMRTbell template libraries and sequencing polymerase was bound to them using Sequel Binding kit 2.0 (A. nabsnona, A. mellea, A. tabescens, and A. ectypa), 2.1 (A. borealis), or 3.0 (A. fumosa and A. novae-zelandiae). The prepared SMRTbell template libraries were then sequenced on a Pacific Biosciences' Sequel sequencer using v3 sequencing primer, 1M v2 (A. borealis, A. nabsnona, A. mellea, A. tabescens, and A. ectypa) or v3 (A. fumosa, and A. novae-zelandiae) SMRT cells, and Version 2.1 (A. borealis, A. nabsnona, A. mellea, A. tabescens, and A. ectypa) or 3.0 (A. fumosa and A. novae-zelandiae) sequencing chemistry with 1x360 & 1x600 sequencing movie run times.
Filtered subread data were processed to remove artifacts and assembled together with Falcon (https://github.com/PacificBiosciences/FALCON) version 1.8.8 (A. ectypa, A. nabsnona, A. tabescens, and A. borealis) or version pb-assembly 0.0.2, falcon-kit 1.2.3, pypeflow 2.1.0 (A. mellea, A. novae-zelandiae, and A. fumosa) to generate an initial assembly. Mitochondrial genomes were assembled separately from the Falcon pre-assembled reads (preads) using an in-house tool (assemblemito.sh (A. ectypa, A. nabsnona, A. tabescens, A. borealis, and A. mellea) or assemblemito.py (A. fumosa and A. novae-zelandiae)), used to filter the preads, and polished with Arrow version SMRTLink v5.0.1.9578 (A. ectypa and A. nabsnona), v5.1.0.26412 (A. tabescens and A. borealis), v6.0.0.47841 (A. mellea) or v7.0.1.66975 (A. novae-zelandiae and A. fumosa) (https://github.com/PacificBiosciences/GenomicConsensus). A secondary Falcon assembly was generated using the mitochondria-filtered preads, improved with finisherSC51 version 2.0 (A. ectypa and A. nabsnona) or 2.1 (A. fumosa), and polished with Arrow version SMRTLink v5.0.1.9578 (A. ectypa and A. nabsnona), v5.1.0.26412 (A. tabescens and A. borealis), v6.0.0.47841 (A. mellea), or v7.0.1.66975 (A. novae-zelandiae and A. fumosa). Contigs less than 1000 bp were excluded.
Stranded cDNA libraries were generated using the Illumina Truseq Stranded mRNA Library Prep kit. mRNA was purified from 200 ng (A. nabsnona) or 1ug (all other species) of total RNA using magnetic beads containing poly-T oligos. For A. nabsnona, mRNA was fragmented using divalent cations and high temperature. The fragmented RNA was reverse transcribed using random hexamers and SSII (Invitrogen) followed by second strand synthesis. For all other species, mRNA was fragmented and reverse transcribed using random hexamers and SSII (Invitrogen) followed by second strand synthesis. The fragmented cDNA was treated with end-pair, A-tailing, adapter ligation, and 10 (A. nabsnona) or 8 (all other species) cycles of PCR.
All prepared libraries were quantified using KAPA Biosystems' next-generation sequencing library qPCR kit and run on a Roche LightCycler 480 real-time PCR instrument. Sequencing was performed on the Illumina NovaSeq sequencer using NovaSeq XP V1 reagent kits, S4 flowcell (A. fumosa, A. novae-zelandiae, and A. mellea) or on the Illumina HiSeq2500 sequencer using TruSeq paired-end cluster kits v4 and TruSeq SBS sequencing kits v4 (A. nabsnona, A. borealis, A. tabescens and A. ectypa), following a 2x150 indexed run recipe.
Raw reads were evaluated with BBDuk (https://sourceforge.net/projects/bbmap/) for artifact sequence by kmer matching (kmer=25), allowing 1 mismatch and detected artifacts were trimmed from the 3' end of the reads. RNA spike-in reads, PhiX reads, and reads containing any Ns were removed. Quality trimming was performed using the phred trimming method set at Q6. Following trimming, reads under the length threshold were removed (minimum length 25 bases or 1/3 of the original read length - whichever is longer). Filtered reads were assembled into consensus sequences using Trinity v. 2.3.252 with the --normalize_reads and --jaccard_clip options.
Genomes were annotated with the support of their corresponding transcriptomes using the JGI Annotation Pipeline. Assemblies and annotations are available from the JGI Genome Portal MycoCosm (https://mycocosm.jgi.doe.gov)53,54.
Taxon sampling and functional annotations
We used 3 datasets in this study. Dataset1 comprises 66 species (64 Agaricales, 2 Boletales outgroups), which were used to analyze gene family evolution in Armillaria (Supplementary Table 1: Dataset1). This was extended into a phylogenetically more diverse Dataset2, which was used to perform gene copy number analysis of wood-decay gene repertoires and comprised 131 fungal species from both Basidiomycota and Ascomycota, including species with various lifestyles such as white-rotters, brown-rotters, litter-decomposers, ECM fungi, pathogens, and species with uncertain life history strategies (Supplementary Table 1: Dataset2). Dataset3 was used to identify horizontally transferred genes and comprised 942 species, including species ranging from fungi, bacteria as well as plants (Supplementary Table 1: Dataset3). All the species used in these datasets are published and publicly available sequences from JGI Mycocosm (except a few that were used with Author’s permission). The version numbers of the genomes are provided in Supplementary Table1 for reference. For TE classification, Repeatmasker v4.0.355 was run (with parameters -nolow -no_is -s) on the Armillaria and Physalacriaceae outgroup species genome assemblies using a database composed of the Repbase repeat library 25.0356 and additional rDNA sequences from MycoCosm. For genomes identified as diploid, only primary scaffolds were considered. For A. fuscipes, F. velutipes, and F. rossica, only scaffolds > 1000bp were considered. Proteomes of all the species used in this manuscript were functionally annotated by using InterPro (IPR) Scan v.5.46-81.057. Secreted and small secreted (at least 300 amino acids) proteins were identified as described previously58.The luciferase cluster and its synteny were identified in Armillaria species and 5 Physalacriaceae outgroups as described previously20. Orthologous groups from these genomes were inferred using OrthoFinder v2.5.459,60 and synteny around the luciferase cluster was visualized based on ordering and orienting scaffolds using the Armillaria ostoyae C18/9 genome and using genoPlotR61 package. The hispidin synthase and cytochrome P450 gene were previously missing and re-annotated on the scaffold of NODE_104435 in Armillaria mellea DSM 3731 using predictions of Armillaria mellea ELDO17 v1.0.
Genome evolution in Armillaria spp
Reconstruction of genome wide gene family evolution was performed using Dataset1. Predicted protein sequences were clustered into orthogroups using OrthoFinder v2.3.159,60 with default settings. For the species tree, multiple sequence alignment for 81 single copy orthogroups (SCOGs) were inferred using MAFFT v7.453 (-auto)62, followed by removal of gapped regions from the alignments using TrimAl v1.263 with a gap threshold of 0.9. Trimmed alignments were then concatenated into a supermatix using an in-house R-script. The species tree was reconstructed in RAxML v8.2.1264 with the option of rapid bootstrap analysis and search for the best-scoring ML tree under the PROTGAMMAWAG model of protein evolution. The tree was rooted using FigTree and is provided in Supplementary Table 1.
We analyzed the genome-wide duplications and losses across gene families using the COMPARE method65 which uses reconciled gene trees to perform Dollo parsimony mapping and ortholog coding to tell apart duplications from speciation events. To obtain the reconciled gene trees for COMPARE, we first inferred multiple sequence alignments for the Orthofinder orthogroups with at least 4 proteins using MAFFT v7.453 (-auto)62. Aligned clusters were trimmed to remove spurious regions using TrimAL v1.263 with -strict parameter. Clusters for which TrimAL resulted in blank alignments (due to -strict parameter) were used in their non-trimmed form. In total, 16,936 trimmed and 819 non-trimmed clusters were used in RAxML v8.2.1264 for gene tree inference, followed by Shimodaira-Hasegawa (sh)-like branch support calculation also in RAxML under the PROTGAMMAWAG substitution model. The 17,755 gene trees with their SH-like support values, along with the species tree were used for rerooting, followed by gene tree species tree reconciliation in Notung v2.9 run in batch mode with edge weight threshold set to 70. Reconciled gene trees were used to reconstruct the gene duplication/loss scenarios in orthogroups along the species tree using the COMPARE pipeline (available at https://github.com/laszlognagy/COMPARE). For each gene family, the number of gains, losses, net gains (sum of gains and losses), and ancestral copy numbers were obtained and their summary was mapped back onto the species tree. GO terms significantly enriched (p-values < 0.05) among the duplications at Armillaria MRCA were identified by topGO66 using the weight01 algorithm and Fisher testing.
Analyses of CAZymes
Dataset2 was used to analyze where Physalacriaceae (including Armillaria) fits in terms of their wood-decay repertoires. Annotation of genes encoding Carbohydrate Active enzymes (CAZymes) for the required species were downloaded from JGI Mycocosm. Species whose CAZymes were not present in JGI Mycocosm were annotated using the CAZy annotation pipeline67. Based on previously published reports, annotated CAZymes were further categorized based on their putative plant cell wall degrading preferences (Supplementary Table 3) into those acting on cellulose, hemicellulose, pectin and lignin. We subjected the proteomes of these 131 species to OrthoFinder v2.3.159,60 with default settings, giving us a total of 41,205 orthogroups. For inferring the species tree, we generated SCOGs using a set of in-house R scripts which gave us 548 orthogroups in which the duplications were caused by gains at species levels (or terminal gains). From these we omitted proteins showing less similarity based on amino acid distance against the other members of the cluster. In this step we used at least 40% of all species for each cluster. Finally, the resulting 514 SCOGs were concatenated (min 60 length of AA, and at least 66 species) into one supermatrix and followed by species tree inference in IQ-Tree v1.6.1268.
We generated phylogenetic PCA using the phyl.pca69 function from Phytools70 for Physalacriaceae along with white rot and litter decomposer fungi. We compare Armillaria and other Physalacriaceae to only white rot and litter decomposer fungi, because including brown rot and mycorrhizal species, which have huge differences in gene content, reduced the resolution of the plots. We provided the function with a gene copy number matrix normalized according to proteome sizes for each substrate category and the above described ML species tree (subsetted to white rot, litter decomposer and Physalacriaceae species) as inputs. Independent contrasts were calculated under the Brownian motion model and the parameter mode=“cov”. The substrate-wise gene copy numbers and their resulting phylogenetic PCA loadings are provided in Supplementary Table 3. We also plotted their proteome-size normalized copy numbers into barplots for visualization of copy numbers across the phylogeny (Supplementary Fig. 6, Supplementary Fig. 3, Supplementary Fig. 7, Supplementary Fig. 4, Supplementary Fig. 5).
We further investigated if Physalacriaceae members have CAZymes shared with Ascomycota. For checking this hypothesis, we first identified the CAZy orthogroups from the 131 species, by selecting orthogroups with at least 50% of the proteins annotated as CAZymes, and at least 5 proteins. This gave us 401 CAZy orthogroups which were used to perform Fisher’s exact test-based enrichment for identifying co-enriched CAZy families in Physalacriaceae and Ascomycota with respect to white rot and litter decomposers (Supplementary Table 3).
Analyses of horizontally transferred genes
In order to check if Physalacriaceae species are more similar to Ascomycota than expected by chance, which could either come from horizontal gene transfer or long retained ancestral genes, we compiled the Dataset3. This comprised a taxonomically diverse dataset with a total of 942 species, ranging from 110 fungi from Basidiomycota (including 20 Physalacriaceae), 741 from Ascomycota, 26 from Mucoromycota, 10 from Zoopagomycota, 13 from Chytridiomycota and 17 other early-diverging fungi as well as 15 bacterial and 10 plant species.
We identified candidate horizontally transferred (HT) genes using the Alien index (AI), for which we ran MMseqs (easy-search, e-value 0.001)71 using all Physalacriaceae proteomes merged together as our query, against a database of proteomes of all the other species as our subject. Using an in-house R script, we parsed the output to retain only the top hits (based on e-values) for each taxonomic group. AI scores were calculated as [log10(best hit to species within the group lineage + 1x10-200) - log10(best hit to species outside the group lineage + 1x10-200)]72.
Further, for phylogenetic validation of HT events, we used a subset of 329 species from Dataset3 based on AI of top HT donors. In this case, we restricted our donor list to major HT contributors only, viz. Ascomycota, followed by Mucoromycota and Zoopagomycota as outgroups. The proteomes of these 329 species were subjected to OrthoFinder v2.5.459,60 clustering with default settings. From these, we fished out the orthogroups with at least one AI-based HT gene and also the orthogroups containing CAZymes. Further, we retained orthogroups having at least one Physalacriaceae and one Ascomycota/Mucoromycota/Zoopagomycotina species, resulting in a total of 675 orthogroups.
Proteins from these 675 orthogroups were aligned using MAFFT v7.453 (-auto)62 TrimAL v1.2 (-strict)63. Trimmed alignments were used to infer gene trees using IQ-Tree v1.6.1268 with ultrafast bootstrap (1000 replicates) under the WAG+G substitution model. Using an in-house R-script, we identified potential HT events by extracting clades from the gene trees based on support values (>70%) and taxon occupancy (receiver clade with >70% Physalacriacae species, donor clade with >70% Ascomycota, and finally sister clade with >70% Ascomycota to ensure the direction of gene transfer). Physalacriceae proteins from these putative HT clades, were used as a query against the UniRef100 database73 from the UniProt Reference Cluster using MMSeqs easy-search, e-value 0.001)71. To remove the low sequence similarity matches and false hits, we retained hits with percent identity and bidirectional coverage of more than 45%. We parsed the filtered hits to retain only the top 100 hits (based on percent identity) for each query, and the taxonomic category contributing predominantly among the top 100 hits was assigned as the putative donor. Further, we classified the assigned donor into two categories: Type A, where the top 100 hits were dominated and confidently associated with Ascomycota, and Type B, where the top 100 hits were dominated by non-Agaricomycetes species, however ambiguously distributed among different taxa (Supplementary Table 4).
Live plant and stem invasion pathosystems
To understand gene expression in Armillaria and its host during the early stages of host infection, we performed live in planta assays between Eucalyptus grandis and Armillaria luteobubalina (Extended Data Fig. 5B). A. luteobubalina was cultured onto half-strength modified Melin–Norkrans (MMN) media (pH 5.5; 1 g l-1 glucose) and grown for one month in the dark. E. grandis seeds were sterilized for 10 minutes in 30% hydrogen peroxide (H2O2, v/v) and germinated on 1% (w/v) water agar one month (25°C; 16 h light cycle). Four weeks before contact with the fungus, seedlings were transferred onto half-strength MMN medium (pH 5.5; 1 g l-1 glucose) and grown at 22–30°C night/day temperature with a 16 h light cycle. Once the E. grandis seedlings were two months old and the fungal cultures had grown for one month, the plants were separated into one of three treatment categories: (1) fungal-free controls whereby the seedlings were transferred onto new half-strength MMN medium; (2) ‘pre-symbiosis’ which involved the transfer of seedlings onto new half-strength MMN medium in indirect contact with fungal mycelium for 24 h by separating the two organisms by a permeable cellophane membrane; (3) ‘physical contact’ seedlings were transferred onto new half-strength MMN medium and then placed into direct contact with the active growing edge of a fungal colony and then samples were harvested at 24 h, 48 h, 1 week, and 2 week post-contact. These plates were then closed using micropore tape to allow for gas exchange with the external environment. Four biological replicates per treatment and timepoint were generated and harvested at the described timepoint into liquid nitrogen and stored at -80°C until RNA extraction.
For stem-invasion assays, high and low virulent isolates of conifer-specific A. ostoyae (C18 - highly virulent, C2 - low virulent), and A. borealis (A6 - highly virulent, A4 - low virulent)46,47 were maintained in the dark on Petri dishes on RS medium (40g malt extract, 20g dextrose, 5g bacto peptone, 19g agar / 1L) at 24°C and 4°C, respectively (Extended Data Fig. 5/A). Before the start of the experiment, fresh cultures of all isolates were set up. The system for growing subcortical mycelial fans in the laboratory consisted of plastic jars containing a layer of moistened and inoculated RSTO medium9. After approximately 10 days of incubation in dark at 24°C, 10cm long, freshly cut stem segments of Norway spruce were placed on top of this growing mycelial lawn, allowing Armillaria to invade the stem segments. The timing and effectiveness of the advancing invasive mycelial fans were followed by their arrival in small monitoring "windows" (1 x 1.5 cm) cut into the bark halfway up to the top of the stems. The samples were harvested soon after the mycelial front line appeared in the cutouts. Armillaria vegetative mycelia collected from the jars without spruce stems were used as controls.
RNA isolation and sequencing
For in planta assays, live tissues, frozen samples were harvested and extracted using the ISOLATE II Plant miRNA kit (Bioline, Sydney, Australia) as per the manufacturer's instructions. Following extraction, the RNA samples were sequenced at the Beijing Genome Institute (BGI). For the stem invasion assays, sections of the mycelial fan that were collected from under the bark of infected colonized stems were frozen in liquid nitrogen and stored at -80°C. RNA was isolated from the frozen samples using the RNeasy Plant Midi Kit (Qiagen Inc.) according to the manufacturer’s protocol. Prior to the RNA isolation, fungal tissues were homogenized with the help of liquid nitrogen, mortar and pestle. RNA quantity was measured using Qubit (ThermoFisher) according to the manufacturer’s protocol. Biological triplicates were analyzed for all sample types. The libraries for Illumina sequencing were prepared using NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB, Ipswitch, MA, USA). Briefly, 100 ng RNA was enriched using RiboCop rRNA Depletion Kits (Lexogen, Austria). Thereafter, the RNA was fragmented, end prepped and adapter-ligated. Finally, the libraries were amplified according to the manufacturer’s instructions. The quality of the libraries was checked on Agilent 4200 TapeSation System using D1000 Screen Tape (Agilent Technologies, Palo Alto, CA, USA), the quantity was measured on Qubit 3.0. Illumina sequencing was performed on the NovaSeq 6000 instrument (Illumina, San Diego, CA, USA) with 2 × 151 run configuration. Raw RNA-Seq reads were aligned against the A. ostoyae (NCBI genome GCA_900157425.1 version 2) and A. borealis (JGI: Armillaria borealis FPL87.14 v1.0) genomes using STAR v2.7.5a74. After alignment, the level of expression was estimated using RSEM v1.3.175.
Analysis of expression data
For the stem invasion assay, the estimated counts matrix was used for differential expression analysis using Limma-Voom76. Prior to running the differential expression analysis, counts were normalized using TMM (trimmed mean of M values) using edgeR v3.38.177 and lowly expressed genes (cpm ≤ 10) were filtered. Differential gene expression analysis for the in-planta pathosystem data were carried out as in our previous study26.
In both analyses, genes with at least two-fold expression change and FDR value ≤0.05 were considered significant. Multidimensional scaling (“plotMDS” function in edgeR) was applied to visualize gene expression profiles. Expression heatmaps were generated by hierarchically clustering based on average linkage of FPKM values with heatmap.2 function from gplots package
To check reliability of both expression data, we used the superSeq package32 which uses a subsampling approach to simulate and predict the number of differentially expressed genes at lower read depths and random sampling points from the original dataset. These subsampled reads are then extrapolated to predict the relationship between statistical power and read depth.
Genes from the time-series in planta assay from both fungal and the plant side were clustered into co-expression based modules using STEM v1.3.1133. GO terms significantly enriched (p-values < 0.05) among each of the STEM modules were identified by topGO66, using the weight01 algorithm and Fisher testing.
Enrichment ratios for specific gene families in each gene expression dataset were calculated using the fisher.test function in R which uses the formula (AD)/(BC), where A represents the number of DEGs in the gene family of interest, B represents the total number of genes in that specific gene family, C represents the total number of DEGs excluding the gene family of interest, and D represents the total number of proteins excluding the DEGs, the gene family of interest, and the total genes in that gene family. These enrichment or odds ratio for different gene families in each experiment were visualized using the heatmap.2 function from the gplots package. Expression heatmaps for different gene families in the six datasets are available in Figshare (https://figshare.com/articles/figure/Gene_expression_heatmaps/22778477?file=40472333).
Experimental validation of Pathogenicity-induced SSPs
Aliquots of RNA extracted from the in planta pathosystem assay (6 timepoints, 24 samples) were pooled and used for generation of full length cDNA using the Tetro cDNA synthesis kit (Bioline) according to manufacturer’s instructions and using only the oligo-dT primer. Two A. luteobubalina HWK02 v1.0 SSP sequences were amplified using the KAPA HiFi Hotstart Readymix (Roche) according the manufacturer’s instructions using the following primers: Armlut1_1165297(5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTTYCARYTNYTNTTY GCNGC-3’ and 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTGTTACTCTTCTAGCAGCTTCACGTC-3’) and Armlut_1348401(5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTTGTTCAGTTTCTTC CTCTTCTACC-3’ and 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTGCTARTCNACRCANGTNGTNSWDATRTCNG -3’). As negative controls, we cloned these same genes with a single nucleotide frameshift using the primers Armlut1_1165297_fs (5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTYCARYTNYTNTTYGCNGC) and Armlut1_1348401_fs (5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTGTTCAGTTTCTTCCTCTTCTACC) with the same reverse primer as per the sense constructs. As a further set of negative cell death transformation controls, we also cloned two SSPs lacking the secretion domain from the mutualistic ectomycorrhizal fungus Pisolithus microcarpus isolate SI14 to act as negative controls for the induction of cell death. These were Pismi_683611 and Pismi_689601, effector proteins that do not cause visible plant damage when expressed in planta (Pismi_68361145; Pismi_68960178). Successful amplification was verified using gel electrophoresis, products were gel purified using the Wizard SV Gel and PCR Cleanup kit (Promega). As a positive control for cell death, we synthesized the full length protein apoptosis regulator BAX isoform 1 from Mus musculus (NP_031553.1; MDGSGEQLGSGGPTSSEQIMKTGAFLLQGFIQDRAGRMAGETPELTLEQPPQDASTKKLSECL RRIGDELDSNMELQRMIADVDTDSPREVFFRVAADMFADGNFNWGRVVALFYFASKLVLKALC TKVPELIRTIMGWTLDFLRERLLVWIQDQGGWEGLLSYFGTPTWQTVTIFVAGVLTASLTIWKKM G; Twist Biosciences) with codon usage optimised for in planta expression. All of these constructs were ligated into pDONR221-b1b4 (Invitrogen) using BP ligase (Invitrogen). Ligations were transformed into E. coli and positive colonies were selected based on PCR screening. Positive bacterial colonies were grown overnight in LB + kanamycin (50 mg/L) and plasmids purified using the PureLink HiPure Plasmid Miniprep kit. Gene inserts were sequence verified using Sanger sequencing at the Hawkesbury Institute for the Environment sequencing platform. Plasmids containing the proper gene insert were used in an LR ligation with pBiFCt-2in1destination vector with an empty cassette ligated into the b2b3 position. Similarly, as a control for in planta transformations, a version of pBIFCt-2in1 with empty cassettes was generated as an empty vector control. Screening procedures following were as per with pDONR221. Positive plasmids were purified and transformed into Agrobacterium tumefaciens GV3101. Expression in N. benthamiana was performed as per Plett et al45. Similarly, young leaves of E. grandis were agroinfiltrated using the same solutions as for N. benthamiana and put into growth chambers at 25°C constant temperature (16hr light/8hr dark). As opposed to N. benthamiana leaves, E. grandis leaves were more recalcitrant to infiltration without damaging the leaves, therefore the bacteria was only introduced into a circle the size of the syringe bore. Both plant systems were left until visible symptoms developed in the positive control and the leaves photographed at the site of infiltration. Leaves from three separate plants were infiltrated for each plant model system and representative images presented.
Extended Data
Supplementary Material
Reporting Summary.
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Acknowledgments
We acknowledge support by the 'Momentum' program of the Hungarian Academy of Sciences (contract no. LP2019-13/2019 to LGN) the European Research Council (grant no. 758161 to LGN) as well as the Eotvos Lorand Research Network (SA-109/2021). GS acknowledges support by the Hungarian National Research, Development, and Innovation Office (GINOP-2.3.2-15-2016-00052). The work (proposals: 10.46936/10.25585/60001060 and 10.46936/10.25585/60001019) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231. The research was performed in collaboration with the Genomics and Bioinformatics Core Facility at the Szentágothai Research Centre of the University of Pécs. Ian Hood and Pam Taylor (Scion Research, New Zealand Forest Research Institute Ltd) kindly provided the A. nova-zealandiae 2840 strain. Daniel Lindner (Forest Products Laboratory, USA) kindly shared strains of A. borealis and A. ectypa for sequencing. We appreciate the permission of Gregory Bonito for using the genome of Flagelloscypha sp.
Footnotes
Author contributions
NS, LGN, JP and GS conceived the study. NS, BI, JW-B, ZM, KLP and JP carried out laboratory experiments, including DNA/RNA isolation for genome and transcriptome sequencing. NS, ZM, BI, H-MK, SK, ED, BB, BH, MV, SC, IJT, JS, LGN carried out data analysis. ED and BH annotated CAZymes for the genomes not available in JGI Mycocosm. NS, JS, ZM, SK and LGN analyzed HGT events. SA, MM, AL, BA, JP, AP, KB, KL, MK, MY, RR and IGV performed genome sequencing, assembly and annotation. JP and KLP performed PiSSP experimental validation. KLF and GDF contributed strains to the genome sequencing. CBH contributed genomic data. LGN, NS, JP, FMM, JS, SK, GS and DR wrote the manuscript. All authors reviewed, checked and approved the manuscript.
Competing Interests Statement
The authors declare no competing interests.
Data Availability
New genomic assemblies and annotation generated in this study are deposited under the 1000 Fungal Genome Project at JGI Mycocosm (https://mycocosm.jgi.doe.gov/Armillaria/Armillaria.info.html) and at DDBJ/EMBL/GenBank under the accession numbers PRJNA463936, PRJNA500536, PRJNA500837, PRJNA519860, PRJNA519861, PRJNA571622, PRJNA677793, PRJNA677794. New RNA-seq datasets used in this study are deposited in the NCBI’s Gene Expression Omnibus (GEO) Archive at https://www.ncbi.nlm.nih.gov/geo/. Accession number for the in-planta assay between A. luteobubalina and E. grandis is PRJNA975488, or GSE233220. For the stem-invasion assay, the accession numbers are PRJNA972908 for A. ostoyae and PRJNA972989 for A. borealis.
Phylogenetically validated gene trees and gene expression heatmaps for various gene families for the six RNA-Seq datasets used in this study can be found in the Figshare repository at https://figshare.com/articles/dataset/Gene_trees/22730534 and https://figshare.com/articles/figure/Gene_expression_heatmaps/22778477?file=40472333respectively.
Code availability
Codes associated with the data analyses and visualization are available at https://github.com/nehasahu486/Armillaria-phylogenomics/tree/main.
References
- 1.Baumgartner K, Coetzee MPA, Hoffmeister D. Secrets of the subterranean pathosystem of Armillaria: Subterranean pathosystem of Armillaria. Mol Plant Pathol. 2011;12:515–534. doi: 10.1111/j.1364-3703.2010.00693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinzelmann R, et al. Latest advances and future perspectives in Armillaria research. Can J Plant Pathol. 2019;41:1–23. [Google Scholar]
- 3.Sipos G, Anderson JB, Nagy LG. Armillaria . Curr Biol. 2018;28:PR297-R298. doi: 10.1016/j.cub.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 4.Coetzee M, Wingfield B, Wingfield M. Armillaria Root-Rot Pathogens: Species Boundaries and Global Distribution. Pathogens. 2018;7:83. doi: 10.3390/pathogens7040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgartner K. Root Collar Excavation for Postinfection Control of Armillaria Root Disease of Grapevine. Plant Dis. 2004;88:1235–1240. doi: 10.1094/PDIS.2004.88.11.1235. [DOI] [PubMed] [Google Scholar]
- 6.Koch RA, et al. Symbiotic nitrogen fixation in the reproductive structures of a basidiomycete fungus. Curr Biol. 2021;31:3905–3914.:e6. doi: 10.1016/j.cub.2021.06.033. [DOI] [PubMed] [Google Scholar]
- 7.Ford KL, Henricot B, Baumgartner K, Bailey AM, Foster GD. A faster inoculation assay for Armillaria using herbaceous plants. J Hortic Sci Biotechnol. 2017;92:39–47. [Google Scholar]
- 8.Devkota P, Hammerschmidt R. The infection process of Armillaria mellea and Armillaria solidipes. Physiol Mol Plant Pathol. 2020;112:101543 [Google Scholar]
- 9.Sipos G, et al. Genome expansion and lineage-specific genetic innovations in the forest pathogenic fungi Armillaria. Nat Ecol Evol. 2017;1:1931–1941. doi: 10.1038/s41559-017-0347-8. [DOI] [PubMed] [Google Scholar]
- 10.Liang X, Rollins JA. Mechanisms of Broad Host Range Necrotrophic Pathogenesis in Sclerotinia sclerotiorum. Phytopathology®. 2018;108:1128–1140. doi: 10.1094/PHYTO-06-18-0197-RVW. [DOI] [PubMed] [Google Scholar]
- 11.O’Connell RJ, et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet. 2012;44:1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newman TE, Derbyshire MC. The Evolutionary and Molecular Features of Broad Host-Range Necrotrophy in Plant Pathogenic Fungi. Front Plant Sci. 2020;11:591733. doi: 10.3389/fpls.2020.591733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson Å, et al. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. 2012;194:1001–1013. doi: 10.1111/j.1469-8137.2012.04128.x. [DOI] [PubMed] [Google Scholar]
- 14.Shao D, Smith DL, Kabbage M, Roth MG. Effectors of Plant Necrotrophic Fungi. Front Plant Sci. 2021;12:687713. doi: 10.3389/fpls.2021.687713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch RA, Wilson AW, Séné O, Henkel TW, Aime MC. Resolved phylogeny and biogeography of the root pathogen Armillaria and its gasteroid relative, Guyanagaster. BMC Evol Biol. 2017;17 doi: 10.1186/s12862-017-0877-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baccelli I. Cerato-platanin family proteins: one function for multiple biological roles? Front Plant Sci. 2015;5 doi: 10.3389/fpls.2014.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muraosa Y, Toyotome T, Yahiro M, Kamei K. Characterisation of novel-cell-wall LysM-domain proteins LdpA and LdpB from the human pathogenic fungus Aspergillus fumigatus. Sci Rep. 2019;9:3345. doi: 10.1038/s41598-019-40039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plett JM, Plett KL. Leveraging genomics to understand the broader role of fungal small secreted proteins in niche colonization and nutrition. ISME Commun. 2022;2:49. doi: 10.1038/s43705-022-00139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo Presti L, et al. Fungal Effectors and Plant Susceptibility. Annu Rev Plant Biol. 2015;66:513–545. doi: 10.1146/annurev-arplant-043014-114623. [DOI] [PubMed] [Google Scholar]
- 20.Ke H-M, et al. Mycena genomes resolve the evolution of fungal bioluminescence. Proc Natl Acad Sci. 2020;117:31267–31277. doi: 10.1073/pnas.2010761117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy LG, et al. Genetic Bases of Fungal White Rot Wood Decay Predicted by Phylogenomic Analysis of Correlated Gene-Phenotype Evolution. Mol Biol Evol. 2017;34:35–44. doi: 10.1093/molbev/msw238. [DOI] [PubMed] [Google Scholar]
- 22.Sun P, et al. Fungal glycoside hydrolase family 44 xyloglucanases are restricted to the phylum Basidiomycota and show a distinct xyloglucan cleavage pattern. iScience. 2022;25:103666. doi: 10.1016/j.isci.2021.103666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Resl P, et al. Large differences in carbohydrate degradation and transport potential among lichen fungal symbionts. Nat Commun. 2022;13:2634. doi: 10.1038/s41467-022-30218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins C, et al. Genomic and Proteomic Dissection of the Ubiquitous Plant Pathogen, Armillaria mellea: Toward a New Infection Model System. J Proteome Res. 2013;12:2552–2570. doi: 10.1021/pr301131t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daniel G, Volc J, Nilsson T. Soft rot and multiple T-branching by the basidiomycete Oudemansiella mucida. Mycol Res. 1992;96:49–54. [Google Scholar]
- 26.Sahu N, et al. Hallmarks of Basidiomycete Soft-and White-Rot in Wood-Decay-Omics Data of Two Armillaria Species. Microorganisms. 2021;9:149. doi: 10.3390/microorganisms9010149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell WG. The chemistry of the white rots of wood. Biochem J. 1931;25:2023–2027. doi: 10.1042/bj0252023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarze FWMR. Wood decay under the microscope. Fungal Biol Rev. 2007;21:133–170. [Google Scholar]
- 29.Gladyshev EA, Meselson M, Arkhipova IR. Massive Horizontal Gene Transfer in Bdelloid Rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 30.Worrall JJ, Anagnost SE, Zabel RA. Comparison of Wood Decay among Diverse Lignicolous Fungi. Mycologia. 1997;89:199. [Google Scholar]
- 31.Floudas D, et al. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genet Biol. 2015;76:78–92. doi: 10.1016/j.fgb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bass AJ, Robinson DG, Storey JD. Determining sufficient sequencing depth in RNA-Seq differential expression studies. 2019 doi: 10.1101/635623. [DOI] [Google Scholar]
- 33.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bautista D, et al. Comprehensive Time-Series Analysis of the Gene Expression Profile in a Susceptible Cultivar of Tree Tomato (Solanum betaceum) During the Infection of Phytophthora betacei. Front Plant Sci. 2021;12:730251. doi: 10.3389/fpls.2021.730251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westrick NM, Smith DL, Kabbage M. Disarming the Host: Detoxification of Plant Defense Compounds During Fungal Necrotrophy. Front Plant Sci. 2021;12:651–716. doi: 10.3389/fpls.2021.651716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lah L, et al. The versatility of the fungal cytochrome P450 monooxygenase system is instrumental in xenobiotic detoxification: Fungal P450 systems in xenobiotic detoxification. Mol Microbiol. 2011;81:1374–1389. doi: 10.1111/j.1365-2958.2011.07772.x. [DOI] [PubMed] [Google Scholar]
- 37.Darwiche R, et al. Plant pathogenesis-related proteins of the cacao fungal pathogen Moniliophthora perniciosa differ in their lipid-binding specificities. J Biol Chem. 2017;292:20558–20569. doi: 10.1074/jbc.M117.811398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao F, et al. Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat Plants. 2019;5:1167–1176. doi: 10.1038/s41477-019-0527-4. [DOI] [PubMed] [Google Scholar]
- 39.Saito N, et al. Roles of RCN1 Regulatory A Subunit of Protein Phosphatase 2A, in Methyl Jasmonate Signaling and Signal Crosstalk between Methyl Jasmonate and Abscisic Acid. Plant Cell Physiol. 2008;49:1396–1401. doi: 10.1093/pcp/pcn106. [DOI] [PubMed] [Google Scholar]
- 40.Cui F, Brosché M, Sipari N, Tang S, Overmyer K. Regulation of ABA dependent wound induced spreading cell death by MYB 108. New Phytol. 2013;200:634–640. doi: 10.1111/nph.12456. [DOI] [PubMed] [Google Scholar]
- 41.Liu H, et al. Copper ions suppress abscisic acid biosynthesis to enhance defence against Phytophthora infestans in potato. Mol Plant Pathol. 2020;21:636–651. doi: 10.1111/mpp.12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature. 2002;419:399–403. doi: 10.1038/nature00962. [DOI] [PubMed] [Google Scholar]
- 43.O’Leary B, Preston GM, Sweetlove LJ. Increased β-Cyanoalanine Nitrilase Activity Improves Cyanide Tolerance and Assimilation in Arabidopsis. Mol Plant. 2014;7:231–243. doi: 10.1093/mp/sst110. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka S, Kahmann R. Cell wall-associated effectors of plant-colonizing fungi. Mycologia. 2021;113:247–260. doi: 10.1080/00275514.2020.1831293. [DOI] [PubMed] [Google Scholar]
- 45.Plett JM, et al. Mycorrhizal effector PaMiSSP10b alters polyamine biosynthesis in Eucalyptus root cells and promotes root colonization. New Phytol. 2020;228:712–727. doi: 10.1111/nph.16759. [DOI] [PubMed] [Google Scholar]
- 46.Heinzelmann R, Prospero S, Rigling D. Virulence and Stump Colonization Ability of Armillaria borealis on Norway Spruce Seedlings in Comparison to Sympatric Armillaria Species. Plant Dis. 2017;101:470–479. doi: 10.1094/PDIS-06-16-0933-RE. [DOI] [PubMed] [Google Scholar]
- 47.Prospero S, Holdenrieder O, Rigling D. Comparison of the virulence of Armillaria cepistipes and Armillaria ostoyae on four Norway spruce provenances. For Pathol. 2004;34:1–14. [Google Scholar]
- 48.Collins C, et al. Proteomic Characterization of Armillaria mellea Reveals Oxidative Stress Response Mechanisms and Altered Secondary Metabolism Profiles. Microorganisms. 2017;5:60. doi: 10.3390/microorganisms5030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell WG. The chemistry of the white rots of wood. Biochem J. 1932;26:1829–1838. doi: 10.1042/bj0261829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flor HH. Current Status of the Gene-For-Gene Concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 51.Lam K-K, LaButti K, Khalak A, Tse D. FinisherSC: a repeat-aware tool for upgrading de novo assembly using long reads. Bioinformatics. 2015;31:3207–3209. doi: 10.1093/bioinformatics/btv280. [DOI] [PubMed] [Google Scholar]
- 52.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grigoriev IV, et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuo A, Bushnell B, Grigoriev IV. Fungal Genomics. Vol. 70. Elsevier; 2014. pp. 1–52. (Advances in Botanical Research). [Google Scholar]
- 55.Smit AFA, Hubley R, Green P. RepeatMasker Open-40. 2015:2013–2015. [Google Scholar]
- 56.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Almási É, et al. Comparative genomics reveals unique wood-decay strategies and fruiting body development in the Schizophyllaceae. New Phytol. 2019;224:902–915. doi: 10.1111/nph.16032. [DOI] [PubMed] [Google Scholar]
- 59.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guy L, Roat Kultima J, Andersson SGE. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagy LG, et al. Latent homology and convergent regulatory evolution underlies the repeated emergence of yeasts. Nat Commun. 2014;5:4471. doi: 10.1038/ncomms5471. [DOI] [PubMed] [Google Scholar]
- 66.Alexa A, Rahenfuhrer J. topGO: Enrichement Analysis for Gene Ontology. 2016 [Google Scholar]
- 67.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Revell LJ. Size-Correction And Principal Components For Interspecific Comparative Studies. Evolution. 2009;63:3258–3268. doi: 10.1111/j.1558-5646.2009.00804.x. [DOI] [PubMed] [Google Scholar]
- 70.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things): phytools: R package. Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- 71.Steinegger M, Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. 2017;35:1026–1028. doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 72.Alexander WG, Wisecaver JH, Rokas A, Hittinger C. T Horizontally acquired genes in early-diverging pathogenic fungi enable the use of host nucleosides and nucleotides. Proc Natl Acad Sci. 2016;113:4116–4121. doi: 10.1073/pnas.1517242113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suzek BE, et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Plett JM, et al. Speciation Underpinned by Unexpected Molecular Diversity in the Mycorrhizal Fungal Genus Pisolithus. Mol Biol Evol. 2023;40:msad045. doi: 10.1093/molbev/msad045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New genomic assemblies and annotation generated in this study are deposited under the 1000 Fungal Genome Project at JGI Mycocosm (https://mycocosm.jgi.doe.gov/Armillaria/Armillaria.info.html) and at DDBJ/EMBL/GenBank under the accession numbers PRJNA463936, PRJNA500536, PRJNA500837, PRJNA519860, PRJNA519861, PRJNA571622, PRJNA677793, PRJNA677794. New RNA-seq datasets used in this study are deposited in the NCBI’s Gene Expression Omnibus (GEO) Archive at https://www.ncbi.nlm.nih.gov/geo/. Accession number for the in-planta assay between A. luteobubalina and E. grandis is PRJNA975488, or GSE233220. For the stem-invasion assay, the accession numbers are PRJNA972908 for A. ostoyae and PRJNA972989 for A. borealis.
Phylogenetically validated gene trees and gene expression heatmaps for various gene families for the six RNA-Seq datasets used in this study can be found in the Figshare repository at https://figshare.com/articles/dataset/Gene_trees/22730534 and https://figshare.com/articles/figure/Gene_expression_heatmaps/22778477?file=40472333respectively.
Codes associated with the data analyses and visualization are available at https://github.com/nehasahu486/Armillaria-phylogenomics/tree/main.