Abstract
Proteins and lipids decorated with glycans are found throughout biological entities, playing roles in biological functions and dysfunctions. Current analytical strategies for these glycan-decorated biomolecules, termed glycoconjugates, rely on ensemble averaged methods that do not provide a full view of positions and structures of glycans attached at individual sites in a given molecule, especially for glycoproteins. Here we show single molecule analysis of glycoconjugates by direct imaging of individual glycoconjugate molecules using low-temperature scanning tunneling microscopy. Intact glycoconjugate ions from electrospray are soft-landed on surface for their direct single molecule imaging. The sub-molecular imaging resolution corroborated by quantum mechanical modeling unveils the entire structures and attachment sites of glycans in glycopeptides, glycolipids, N-glycoproteins, and O-glycoproteins densely decorated with glycans.
Glycan (also known as carbohydrate) is one of the four essential organic building blocks found in all forms of life (1–7). Glycans play key roles in cellular functions (7, 8), growth and development (2, 9), identification (2–4), shapes (10, 11), and energy storage (12). In biological systems, glycans are predominantly found attached to other biomolecules such as proteins and lipids. These glycan-decorated biomolecules, termed glycoconjugates, are produced via complex enzymatic glycosylation events – the most common and diverse post translational modification (PTM) that greatly expands the functions of biomolecules (13, 14). The abundance of glycoconjugates in biological systems and their roles in health and disease make them attractive targets in basic and translational research for new therapeutic and diagnostic strategies (1–3, 15, 16). However, despite the ubiquity and importance of glycoconjugates, research to unveil their structure-property relationships has been challenging (17–19).
Glycoconjugates possess extensive structural heterogeneity (i.e. multiple variants of sequence) and structural isomerism (i.e. structures with equal masses), which pose a challenge for today’s analytical methods (17–19). Glycoconjugates are presently analyzed by a combination of chemical labeling, chemoenzymatic digestion, and ensemble averaged methods to indirectly obtain the most likely structures present in a sample (18, 19). Ensemble averaged analysis of structurally heterogeneous and isomeric glycoconjugates however obscures the position and structures of glycans bonded to a biomolecule, particularly for proteins with multiple glycans attached. As a result, insights into the structures of individual molecules are lost with ensemble averaged analysis, which hinders structure-property relationship studies of glycoconjugates. Preventing the loss of structural information for individual molecules requires glycoconjugate molecules to be analyzed at single molecule level.
Here we realize single molecule analysis of glycoconjugates by performing direct, label-free, spatial imaging on individual glycoconjugate molecules. We show that imaging single glycoconjugates at sub-nanometer resolution reveals the primary structure of each molecule, by unveiling how its constituent amino acid, lipid, and monosaccharide subunits connect to one another. As a result, our imaging method establishes glycan sequences at every glycan attachment site in a glycoconjugate by locating every monosaccharide in the glycoconjugate molecule discriminated by its stereoconfiguration and side group. We demonstrate our method for a wide range of glycoconjugates, starting from simple glycopeptides and glycolipids, up to complex glycoproteins with more than 20 attached glycans. Our work shows that single molecule imaging provides a direct access to all glycan structures bonded to the complex glycopeptides, glycolipids, and glycoproteins at single molecule level.
Direct imaging of glycoconjugates soft-landed at a surface
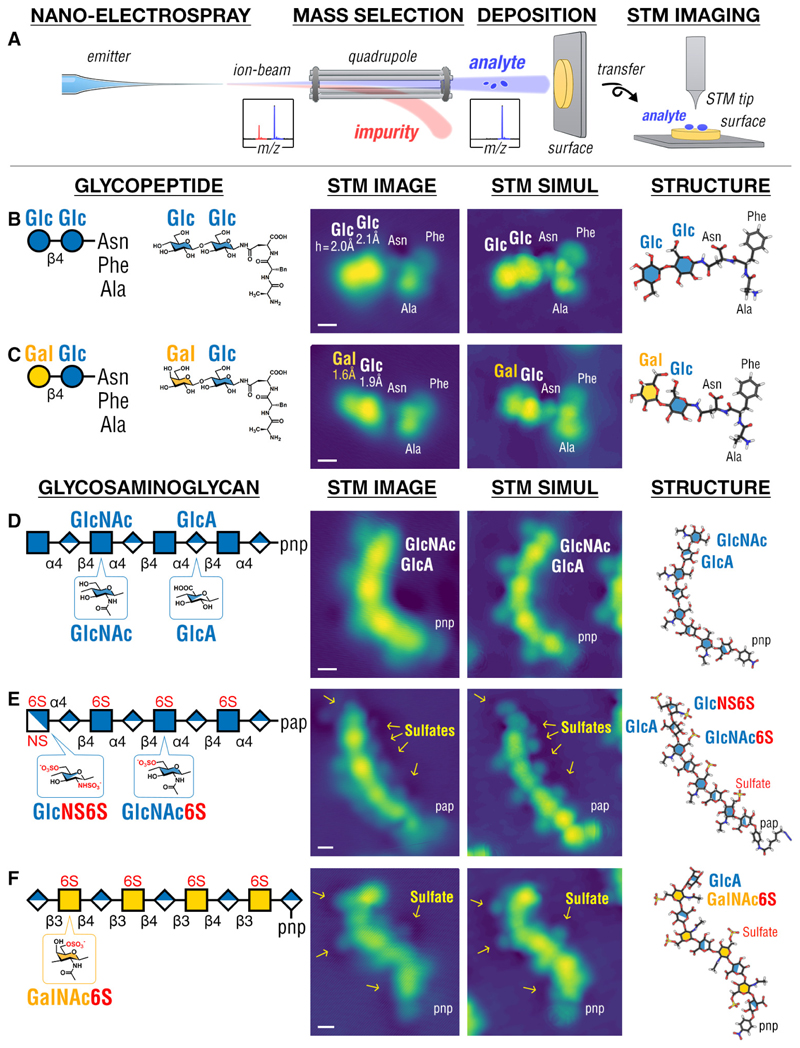
We accomplished direct imaging of single glycoconjugate molecules by combining soft-landing electrospray ion beam deposition (ESIBD) (20, 21) and scanning tunneling microscopy (STM) (see Methods for details). We show STM imaging of single glycoconjugates on Cu-surface at cryogenic temperatures corroborated by Density Functional Theory (DFT) calculations provides direct access to their structural information (Fig. 1). We imaged glycoconjugate ions obtained from nanoelectrospray ionization (nESI) (22) (Fig. S1), whose usage was critical to lower the amount of sample required to ~1 nanomole, given that glycoconjugates, unlike proteins (23, 24) and glycans (25), are more limited in sample quantity. In cases of sulfated molecules (Fig. S2), we deposited the molecules on a more inert Ag-surface to preserve the labile sulfate groups on surface (Fig. S3).
Fig. 1. STM imaging of simple glycoconjugates and glycosaminoglycans (GAGs).
(A) Schematic of the experiment: glycoconjugate or GAG ions generated by nESI were mass-selected, soft-landed intact on surface held at 120 K, and imaged by STM at 11 K (see Methods). STM images of glycopeptides, Glc-Glc-AsnPheAla (B) and Gal-Glc-AsnPheAla (C), reveal the glycan and the peptide domains of the molecule and differentiate each monosaccharide in the glycan domain i.e. glucose (Glc) vs galactose (Gal). Imaging GAGs (D-F) reveals the positions of N-Acetyl groups on all N-acetylglucosamine (GlcNAc) monosaccharides, differentiating them from the glucuronic acid (GlcA) monosaccharides as well as the sulfated GlcNAc6S, GalNAc6S, and GlcNS6S monosaccharides. The GAGs in (D) and (F) are terminated by para-nitrophenyl (pnp), while in (E) the GAG is terminated by para-(6-azidohexanamido)phenyl (pap). STM images were interpreted by STM simulation of molecular structures computed by DFT. Scale bar is 0.5 nm. Glycan icons follow the SNFG standard (56).
We first highlight the capabilities of STM imaging and DFT modeling in characterizing monosaccharide structures of glycans. We imaged two glycopeptides (26) (Fig. 1b,c) composed of a disaccharide (cellobiose, Glcβ1–4Glc, or lactose, Galβ1–4Glc) linked to a tripeptide, and three glycosaminoglycans (GAGs) (Fig. 1d-f). STM imaging of the glycopeptides was found to differentiate the glycan and peptide moieties by their heights: tall/bright for the glycan and low/dim for the peptide (Fig. 1b,c), which allowed the DFT calculations to yield the primary structure of the glycopeptides, resolving the order by which amino acid and monosaccharide subunits in the molecule are connected to one another. In addition, STM imaging was found to discriminate glucose from its epimer, galactose (i.e. they differ only in the stereoconfiguration of their C4-atoms), given that the glucose (h = 2.1 ± 0.2 Å, N = 45) was observed consistently taller than the galactose (h = 1.7 ± 0.3 Å, N = 75). STM imaging was also found to locate and identify side groups and sulfate moieties present in every monosaccharide, as exemplified by the imaging of GAGs (Fig. 1d-f). Each GlcNAc monosaccharide was observed with a dim protrusion corresponding to the N-Acetyl (NAc) moiety, clearly distinct from the GlcA monosaccharides (Fig. 1d). Interestingly, each GlcNAc6S and GalNAc6S monosaccharide was observed with its sulfate moiety (a dim protrusion encircled with a dark ring) on the opposite side from its NAc moiety (a dim protrusion without dark ring) (Fig. 1e,f); whereby GlcNS6S was observed with two sulfate moieties, each appearing as a dim protrusion with a dark ring (Fig. 1e). We further ascertained the STM appearance of sulfate moiety on the same surface as a dim protrusion encircled with a dark ring by imaging simple aryl sulfates (Fig. S3). Our findings show that STM imaging and DFT modeling have sufficient sensitivity and resolution to locate glycans in molecules, and discriminate the constituent monosaccharides based on their stereoconfigurations and side groups.
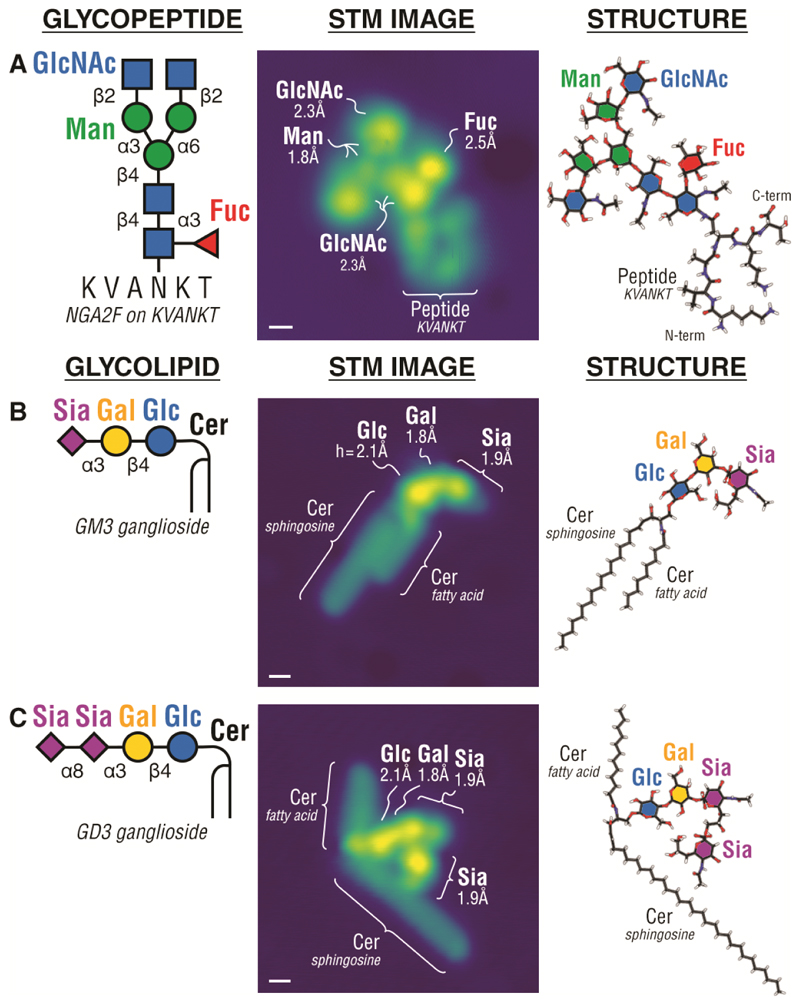
We show the perspectives of our direct single molecule analysis by determining structures of entire glycans present in complex glycopeptides, glycolipids, and glycoproteins at single molecule level. For glycopeptides, we examined an egg yolk sialoglycopeptide derivative (Fig. 2a), which is widely used in biochemical applications (27). For glycolipids, the GM3 and GD3 gangliosides (Fig. 2b,c) were chosen due to their roles as cancer antigens (5, 15). For N-glycoproteins, we chose the widely studied pancreatic RNase B (28). Finally, as a representative for O-glycoproteins, we chose a fragment of human mucin MUC1, one of the most complex glycoproteins in biological systems, which is also overexpressed with aberrant O-glycans in cancer, and a promising cancer biomarker and immunotherapeutic target (29, 30). In all cases, there is a clear height contrast between the bright glycan domain and the dim peptide or lipid domains that establishes their respective primary structures. For the N-glycopeptide (Fig. 2a), the height contrast discriminated the GlcNAc and Fuc monosaccharides (h = 2.2 ± 0.4 Å, N = 132) from the mannose (h = 1.9 ± 0.4 Å, N = 66); while, in the glycolipids (Fig. 2b,c), the height contrast differentiated glucose (one lobe, h = 2.1 ± 0.3 Å, N = 158), galactose (one lobe, h = 1.9 ± 0.2 Å, N = 158), and sialic acid (Neu5Ac) (two or more lobes) from one another. The imaging was found to distinguish glycosidic bonds by the characteristic angle formed between monosaccharides when their pyranose rings adsorb horizontally on surface (experimentally verifiable by their respective heights). For example, the Siaα2–3Gal β1–4Glc in GM3 and GD3 (Sia = Neu5Ac monosaccharide) was observed to form an obtuse 141 ± 22º angle (N = 158), while the Glcβ1–4Glcβ1–4Glc in cellohexaose was observed to form a straight 180 ± 25º angle (N = 204) (31). In addition, the imaging of single glycolipids revealed their molecular conformations (Fig. S4) and allowed discrimination of the ceramide moiety with varied lipid chain lengths (Fig. S5), both of which may provide additional information towards structural studies of lipids. Interestingly, we observed the ‘open’ conformation of the lipid moiety in glycolipids (Fig. 2c and S4), which has been discussed in relation to mechanisms of membrane fusions and protein-membrane interactions (32, 33).
Fig. 2. STM imaging of single glycopeptides and glycolipids.
STM images of an N-glycopeptide (biantennary N-glycan NGA2F on the KVANKT peptide) (A), and glycolipids, GM3 ganglioside (B) and GD3 ganglioside (C), reveal the glycan, peptide, and ceramide (Cer) domains in the respective molecules (Cer consisted of varying length fatty acid chains and sphingosine). STM imaging differentiates individual monosaccharides in the glycoconjugate i.e. glucose (Glc), galactose (Gal), sialic acid (Sia = Neu5Ac), N-acetylglucosamine (GlcNAc), mannose (Man), and fucose (Fuc). STM images were interpreted by structures computed by DFT. Scale bar is 0.5 nm.
Imaging single N- and O-glycoproteins
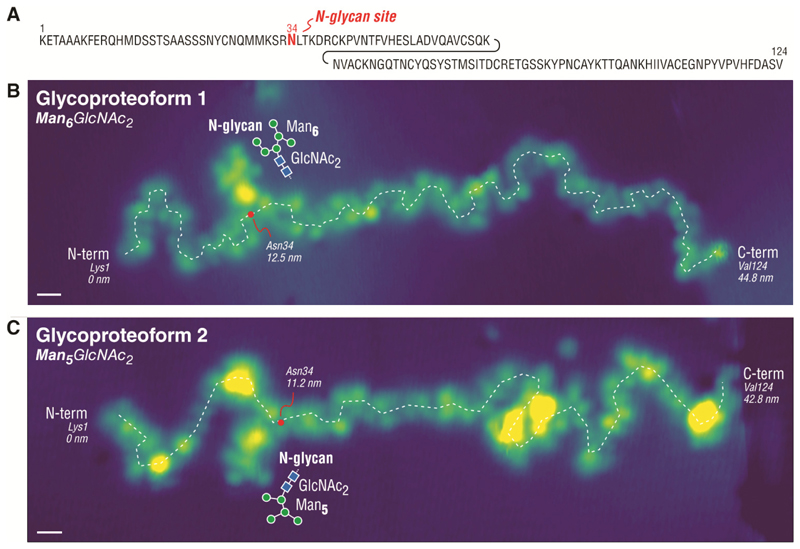
Direct imaging of single a N-glycoprotein, RNase B, revealed the structure and the location of the N-glycan bonded to the protein backbone (28) (Fig. 3). We examined RNase B by imaging individual proteins in their fully unfolded state, which we prepared by exclusively depositing the highly charged protein ions on surface (24) (Fig. S6). Given that RNase B has five glycoproteoforms (28) (each featuring one of five distinct N-glycan structures from Man5GlcNAc2 to Man9GlcNAc2), our single protein imaging allowed the glycoproteoforms of RNase B to be determined one-molecule-at-a-time, as shown in Figure 3b for Man6GlcNAc2 and in Figure 3c for Man5GlcNAc2 – the two most abundant glycoproteoforms of RNase B (Fig. S1g). The STM imaging clearly revealed the glycan attachment site by locating the intersection between the N-glycan and the protein backbone (red dots in Fig. 3b,c). Analysis of 33 individual N-glycoproteins confirmed residue 34 (± 2) as the glycan attachment site, consistent with Asn34 as the known glycan attachment site for RNase B (28).
Fig. 3. STM imaging of single N-glycoproteins.
(A) Sequence of RNase B (124aa) with one N-linked glycan at Asn34. Imaging single unfolded RNase B molecules reveals the N-glycan position along the protein backbone and the N-glycan structure found on individual glycoproteoforms one-molecule-at-a-time, as shown in (B) for Man6GlcNAc2 and in (C) for Man5GlcNAc2. The position of Asn34 is estimated by a red dot along the protein backbone (white dashed line). Scale bar is 1 nm.
To illustrate the full perspective of single glycoprotein imaging, we examined an O-glycoprotein fragment derived from the large mucin MUC1. Mucins are considered one of the last frontiers in glycoanalytics that has remained unexplored to a large extent (34–36), despite their wide importance in mucosal biology and host-pathogen interactions (37). Structural analysis of mucins and their multiple glycosylation sites is challenging due to their enormous size and dense decoration of O-glycans, resulting in heterogeneity and resistance to protease digestion (34–36). Here we show that it is possible to analyze such heavily O-glycosylated proteins one-molecule-at-a-time (Fig. 4) by using the soft deposition and imaging of single glycoproteins on surface (Fig. S7).
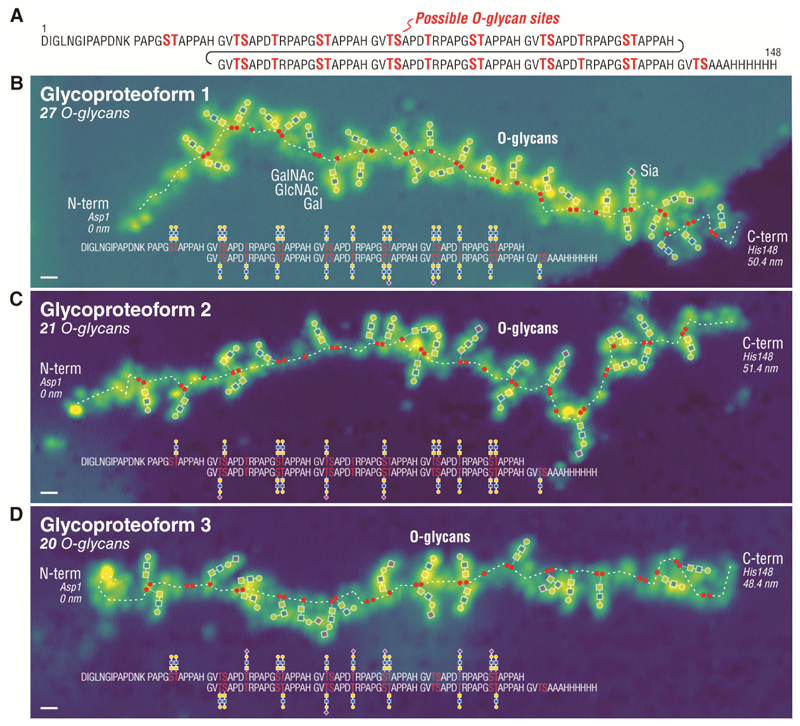
Fig. 4. STM imaging of single O-glycoproteins.
(A) Sequence of MUC1 reporter (148aa) containing 6.5 tandem repeats of 20 amino acids (GVTSAPDTRPAPGSTAPPAH) decorated with O-glycans at the Ser (S) and Thr (T) residues (total of 34 potential O-glycosites). Imaging single MUC1 proteins reveals the number, the structure, and the attachment site of O-glycans decorating the protein, as shown for a glycoproteoform with 27 O-glycans in (B), 21 O-glycans in (C), and 20 O-glycans in (D). The positions of S and T residues are indicated by red dots along the protein backbone (white dashed line). The unannotated STM images are given in Fig. S8. Scale bar is 1 nm.
We imaged a representative fragment of the densely O-glycosylated tandem repeat region of human MUC1 mucin as an O-glycoprotein reporter with relatively homogeneous trisaccharide O-glycans (ie. ‘core 3’: Galβ1-4GlcNAcβ1-3GalNAcα1-O-S/T) (Fig. 4). For this, we employed our recently developed cell-based strategy using genetically glycoengineered HEK293 cells for recombinant production of mucin reporter glycoproteins with custom-designed O-glycosylation (35) (Fig. 4a) (see Methods). The MUC1 O-glycoprotein reporter, featuring 34 potential O-glycosylation sites, was analyzed by intact and bottom-up mass spectrometry, and profiling of released O-glycans, which revealed a relatively homogeneous mixture of O-glycans (mainly ‘core 3’) and number of O-glycans (mainly 19 – 23 glycans) (35, 38) (see also Fig. S1h and Fig. S9 for MUC1 sample used in single molecule imaging experiments).
Imaging single MUC1 O-glycoproteins allows direct observation of the variation in number, structure, and attachment sites of O-glycans on the protein backbone, as shown in examples with 27, 21, and 20 O-glycans (Fig. 4b,c,d). On the individual MUC1 molecules, we found the O-glycans mainly to be the ‘core 3’ trisaccharides with the occasional sialylated ‘core 3’ tetrasaccharide (Siaα2-3Galβ1-4GlcNAcβ1-3GalNAc) in agreement with the glycoprofiling analysis (35, 38) (Fig. S9). Most importantly, the direct imaging of MUC1 clearly revealed the positions of each O-glycan at S and T sites along the protein (red dots in Fig. 4b,c,d) (see Fig. S10 for an example). Analysis of the O-glycan positions on 18 MUC1 O-glycoproteins observed (Table S1) revealed three prevalent patterns of O-glycan distribution on the MUC1 tandem repeats as -TS-T-ST- ; -TS-T-ST- ; and -TS-T-ST- (bold underlined indicates glycosylated, see Table S2). These patterns are largely in agreement with the predicted O-glycosylation sequence of the MUC1 from both in vitro (39) and in vivo (38) enzyme specificity analysis. The O-glycosylation process is a complex event with multiple isoenzymes (polypeptide GalNAc-transferases) each attaching O-glycans at select positions in proteins, and the O-glycosylation of the five possible sites in the MUC1 tandem repeat requires sequential orchestrated action of multiple isoenzymes (39). Further analysis of the STM results (Table S1) revealed on average 3.4 O-glycans per tandem repeat with preferred positions at T in VTSA (87% occupied) and ST in GSTA (78% and 83% occupied respectively) and less preferred positions at S in VTSA (44% occupied) and T in PDTR (55% occupied). These results corroborate our previous studies of the MUC1 reporter protein with ‘core 3’ O-glycans (35, 38), for which we found reduced occupancy of O-glycans at S in VTSA and T in PDTR (38, 40, 41). Direct STM imaging thereby yields detailed snapshots of single molecule glycoproteoforms that can unveil potential interplay between glycosylation at different positions in proteins and the glycan structures that may be assembled at these positions (Table S2). In addition, STM imaging allows direct observation of glycan-glycan interactions dictating the overall shape of the protein backbone. We expect the single molecule analysis approach to be widely applicable to glycoproteins that can be electrosprayed in unfolded states, regardless of size and numbers of attached glycans. In case of increasingly dense glycans causing proteins to unfold incompletely, the STM tip could be used to further unfold the protein to clarify its primary structure (Fig. S11).
Conclusion
Our combination of electrospray deposition and scanning tunnelling microscopy analysis provides an opportunity to look directly at the primary structures of complex glycoconjugates, including glycoproteins with multiple glycans attached. This technology, corroborated by DFT modeling, should enable direct observation of diverse post-translational modifications (PTMs) on biomolecules (42, 43), as well as structures of glycoconjugates that are well beyond today’s analytical capabilities, such as proteoglycans (44), glycoRNAs (45), lipopolysaccharides (46), and carbohydrate vaccines (16). While the present work demonstrates that prior knowledge of the amino acid sequence of the glycoproteins is advantageous to enable interpretation of the STM images, we recognize that STM imaging can still be further improved to identify each amino acid and monosaccharide in a molecule and thus identify single proteins/glycoproteins in complex biological mixtures. These improvements include the use of a functionalized tip to resolve and distinguish covalent bonds in a molecule (47), as well as the use of tunneling spectroscopy (48), nuclear spin detection (49), or optical fingerprinting (50–52) to identify electronic signatures of specific atoms or functional groups in molecules. With these improvements, we expect STM can contribute to identification of unknown glycoproteins or glycolipids, which may ultimately lead to the discovery of individual glycoproteins and glycolipids in a complex cellular mixtures, particularly in the context of glycoproteomics and glycolipidomics studies. Complementing these improvements with automated structure solvers (53–55) or tip preparation will increase the throughput of scanning probe microscopy and create opportunities to solve previously intractable problems in single molecule (bio)analytical chemistry.
Supplementary Material
One sentence summary.
Single molecule imaging of glycan-decorated proteins and lipids unveils all glycan structures bonded to the host molecules
Acknowledgements
Funding
Alexander von Humboldt Foundation (KA, XW)
Baden Württemberg Foundation Project GlycoPepSurf (KA, LS, TZ, KK)
European Research Council Project GlycoX grant 101075996 (KA)
EMBO Postdoctoral Fellowship Program (TJ)
Mizutani Foundation (YN)
Novo Nordisk Foundation grant NNF21OC0071658 (HC)
Lundbeck Foundation grant R223-2016-563 (HC)
Danish National Research Foundation grant DNRF107 (HC)
Novo Nordisk Foundation grant NNF22OC0073736 (RLM)
Carlsberg Foundation grant CF20-0412 (RLM)
Footnotes
Author contributions:
KA, TZ, RLM, KK initiated and supervised the project. KA performed the ESIBD deposition, the STM imaging, the ab initio calculations, and the data analysis. LS, TZ synthesized the glycopeptide standards. TJ, YN, HC, RLM engineered and produced the recombinant MUC1 glycoprotein reporters. XW, SR participated in the initial phase of the project. KA wrote the paper with input from all the authors. All authors contributed to the manuscript.
Competing interests:
Authors declare no competing interests.
Data and materials availability
All data are available in the main text or the supplementary materials. Raw STM images and DFT computed structures are available at the Data Repository of the Max Planck Society (57).
References and Notes
- 1.Hart GW, Copeland RJ. Glycomics Hits the Big Time. Cell. 2010;143:672–676. doi: 10.1016/j.cell.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varki A. Biological roles of glycans. Glycobiology. 2016;27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith BAH, Bertozzi CR. The clinical impact of glycobiology: targeting selectins, Siglecs and mammalian glycans. Nat Rev Drug Discov. 2021;20:217–243. doi: 10.1038/s41573-020-00093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poole J, Day CJ, von Itzstein M, Paton JC, Jennings MP. Glycointeractions in bacterial pathogenesis. Nat Rev Microbiol. 2018;16:440–452. doi: 10.1038/s41579-018-0007-2. [DOI] [PubMed] [Google Scholar]
- 5.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 6.Duan S, Paulson JC. Siglecs as Immune Cell Checkpoints in Disease. Annu Rev Immunol. 2020;38:365–395. doi: 10.1146/annurev-immunol-102419-035900. [DOI] [PubMed] [Google Scholar]
- 7.Ohtsubo K, Marth JD. Glycosylation in Cellular Mechanisms of Health and Disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haltiwanger RS, Lowe JB. Role of Glycosylation in Development. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 10.Cosgrove DJ. Growth of the plant cell wall. Nat Rev Mol Cell Biol. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- 11.Shurer CR, Kuo JC-H, Roberts LM, Gandhi JG, Colville MJ, Enoki TA, Pan H, Su J, Noble JM, Hollander MJ, O’Donnell JP, et al. Physical Principles of Membrane Shape Regulation by the Glycocalyx. Cell. 2019;177:1757–1770.:e21. doi: 10.1016/j.cell.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tena G. Starch synthesis: Seeding the pearl. Nat Plants. 2017;3:17123. doi: 10.1038/nplants.2017.123. [DOI] [PubMed] [Google Scholar]
- 13.Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, Ye Z, Chen Y-H, Schjoldager KT, Steentoft C, Furukawa S, et al. An Atlas of Human Glycosylation Pathways Enables Display of the Human Glycome by Gene Engineered Cells. Mol Cell. 2019;75:394–407.:e5. doi: 10.1016/j.molcel.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moremen KW, Tiemeyer M, V Nairn A. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL, Weiner LM, Matrisian LM. The Prioritization of Cancer Antigens: A National Cancer Institute Pilot Project for the Acceleration of Translational Research. Clin Cancer Res. 2009;15:5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Astronomo RD, Burton DR. Carbohydrate vaccines: developing sweet solutions to sticky situations? Nat Rev Drug Discov. 2010;9:308–324. doi: 10.1038/nrd3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wormald MR, Petrescu AJ, Pao Y-L, Glithero A, Elliott T, Dwek RA. Conformational Studies of Oligosaccharides and Glycopeptides: Complementarity of NMR, X-ray Crystallography, and Molecular Modelling. Chem Rev. 2002;102:371–386. doi: 10.1021/cr990368i. [DOI] [PubMed] [Google Scholar]
- 18.Gray CJ, Migas LG, Barran PE, Pagel K, Seeberger PH, Eyers CE, Boons G-J, Pohl NLB, Compagnon I, Widmalm G, Flitsch SL. Advancing Solutions to the Carbohydrate Sequencing Challenge. J Am Chem Soc. 2019;141:14463–14479. doi: 10.1021/jacs.9b06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagdonaite I, Malaker SA, Polasky DA, Riley NM, Schjoldager K, Vakhrushev SY, Halim A, Aoki-Kinoshita KF, Nesvizhskii AI, Bertozzi CR, Wandall HH, et al. Glycoproteomics. Nat Rev Methods Prim. 2022;2:48. [Google Scholar]
- 20.Grill V, Shen J, Evans C, Cooks RG. Collisions of ions with surfaces at chemically relevant energies: Instrumentation and phenomena. Rev Sci Instrum. 2001;72:3149–3179. [Google Scholar]
- 21.Rauschenbach S, Ternes M, Harnau L, Kern K. Mass Spectrometry as a Preparative Tool for the Surface Science of Large Molecules. Annu Rev Anal Chem. 2016;9:473–498. doi: 10.1146/annurev-anchem-071015-041633. [DOI] [PubMed] [Google Scholar]
- 22.Wilm M, Mann M. Analytical Properties of the Nanoelectrospray Ion Source. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 23.Deng Z, Thontasen N, Malinowski N, Rinke G, Harnau L, Rauschenbach S, Kern K. A Close Look at Proteins: Submolecular Resolution of Two-and Three-Dimensionally Folded Cytochrome c at Surfaces. Nano Lett. 2012;12:2452–2458. doi: 10.1021/nl3005385. [DOI] [PubMed] [Google Scholar]
- 24.Rinke G, Rauschenbach S, Harnau L, Albarghash A, Pauly M, Kern K. Active Conformation Control of Unfolded Proteins by Hyperthermal Collision with a Metal Surface. Nano Lett. 2014;14:5609–5615. doi: 10.1021/nl502122j. [DOI] [PubMed] [Google Scholar]
- 25.Wu X, Delbianco M, Anggara K, Michnowicz T, Pardo-Vargas A, Bharate P, Sen S, Pristl M, Rauschenbach S, Schlickum U, Abb S, Seeberger PH, et al. Imaging Single Glycans. Nature. 2020;582:375–378. doi: 10.1038/s41586-020-2362-1. [DOI] [PubMed] [Google Scholar]
- 26.Sršan L, Ziegler T. Synthesis of new asparagine-based glycopeptides for future scanning tunneling microscopy investigations. Beilstein J Org Chem. 2020;16:888–894. doi: 10.3762/bjoc.16.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Prudden AR, Capicciotti CJ, Bosman GP, Yang J-Y, Chapla DG, Moremen KW, Boons G-J. Streamlining the chemoenzymatic synthesis of complex N-glycans by a stop and go strategy. Nat Chem. 2019;11:161–169. doi: 10.1038/s41557-018-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joao HC, Dwek RA. Effects of glycosylation on protein structure and dynamics in ribonuclease B and some of its individual glycoforms. Eur J Biochem. 1993;218:239–244. doi: 10.1111/j.1432-1033.1993.tb18370.x. [DOI] [PubMed] [Google Scholar]
- 29.Posey AD, Clausen H, June CH. Distinguishing Truncated and Normal MUC1 Glycoform Targeting from Tn-MUC1-Specific CAR T Cells: Specificity Is the Key to Safety. Immunity. 2016;45:947–948. doi: 10.1016/j.immuni.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor-Papadimitriou J, Burchell JM, Graham R, Beatson R. Latest developments in MUC1 immunotherapy. Biochem Soc Trans. 2018;46:659–668. doi: 10.1042/BST20170400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anggara K, Zhu Y, Fittolani G, Yu Y, Tyrikos-Ergas T, Delbianco M, Rauschenbach S, Abb S, Seeberger PH, Kern K. Identifying the Origin of Local Flexibility in a Carbohydrate Polymer. Proc Natl Acad Sci. 2021;113:e2102168118. doi: 10.1073/pnas.2102168118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinnunen PKJ. Fusion of lipid bilayers: a model involving mechanistic connection to HII phase forming lipids. Chem Phys Lipids. 1992;63:251–258. doi: 10.1016/0009-3084(92)90041-m. [DOI] [PubMed] [Google Scholar]
- 33.Corkery RW. The anti-parallel, extended or splayed-chain conformation of amphiphilic lipids. Colloids Surfaces B Biointerfaces. 2002;26:3–20. [Google Scholar]
- 34.Levery SB, Steentoft C, Halim A, Narimatsu Y, Clausen H, Vakhrushev SY. Advances in mass spectrometry driven O-glycoproteomics. Biochim Biophys Acta - Gen Subj. 2015;1850:33–42. doi: 10.1016/j.bbagen.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 35.Nason R, Büll C, Konstantinidi A, Sun L, Ye Z, Halim A, Du W, Sørensen DM, Durbesson F, Furukawa S, Mandel U, et al. Display of the human mucinome with defined O-glycans by gene engineered cells. Nat Commun. 2021;12:4070. doi: 10.1038/s41467-021-24366-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ince D, Lucas TM, Malaker SA. Current strategies for characterization of mucin-domain glycoproteins. Curr Opin Chem Biol. 2022;69:102174. doi: 10.1016/j.cbpa.2022.102174. [DOI] [PubMed] [Google Scholar]
- 37.Hansson GC. Mucins and the Microbiome. Annu Rev Biochem. 2020;89:769–793. doi: 10.1146/annurev-biochem-011520-105053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konstantinidi A, Nason R, Caval T, Sun L, Sørensen DM, Furukawa S, Ye Z, Vincentelli R, Narimatsu Y, Vakhrushev SY, Clausen H. Exploring the glycosylation of mucins by use of O-glycodomain reporters recombinantly expressed in glycoengineered HEK293 cells. J Biol Chem. 2022;298 doi: 10.1016/j.jbc.2022.101784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassan H, Reis CA, Bennett EP, Mirgorodskaya E, Roepstorff P, Hollingsworth MA, Burchell J, Taylor-Papadimitriou J, Clausen H. The Lectin Domain of UDP-N-acetyl-d-galactosamine:PolypeptideN-acetylgalactosaminyltransferase-T4 Directs Its Glycopeptide Specificities*. J Biol Chem. 2000;275:38197–38205. doi: 10.1074/jbc.M005783200. [DOI] [PubMed] [Google Scholar]
- 40.de las Rivas M, Paul Daniel EJ, Coelho H, Lira-Navarrete E, Raich L, Compañón I, Diniz A, Lagartera L, Jiménez-Barbero J, Clausen H, Rovira C, et al. Structural and Mechanistic Insights into the Catalytic-Domain-Mediated Short-Range Glycosylation Preferences of GalNAc-T4. ACS Cent Sci. 2018;4:1274–1290. doi: 10.1021/acscentsci.8b00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coelho H, de las Rivas M, Grosso AS, Diniz A, Soares CO, Francisco RA, Dias JS, Compañon I, Sun L, Narimatsu Y, Vakhrushev SY, et al. Atomic and Specificity Details of Mucin 1 O-Glycosylation Process by Multiple Polypeptide GalNAc-Transferase Isoforms Unveiled by NMR and Molecular Modeling. JACS Au. 2022;2:631–645. doi: 10.1021/jacsau.1c00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- 43.Michaela F, T HB, Mikaela B, Chuan H. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.V Iozzo R, Schaefer L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015;42:11–55. doi: 10.1016/j.matbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, Johnson AG, George BM, Majzoub K, Villalta PW, Carette JE, Bertozzi CR. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell. 2021;184:3109–3124.:e22. doi: 10.1016/j.cell.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raetz CRH, Whitfield C. Lipopolysaccharide Endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gross L, Mohn F, Moll N, Liljeroth P, Meyer G, Leo G, Fabian M, Nikolaj M, Peter L, Gerhard M. The Chemical Structure of a Molecule Resolved by Atomic Force Microscopy. Science. 2009;325:1110–1114. doi: 10.1126/science.1176210. [DOI] [PubMed] [Google Scholar]
- 48.Bian K, Gerber C, Heinrich AJ, Müller DJ, Scheuring S, Jiang Y. Scanning probe microscopy. Nat Rev Methods Prim. 2021;1:36. [Google Scholar]
- 49.Willke P, Yang K, Bae Y, Heinrich AJ, Lutz CP. Magnetic resonance imaging of single atoms on a surface. Nat Phys. 2019;15:1005–1010. [Google Scholar]
- 50.Gutzler R, Garg M, Ast CR, Kuhnke K, Kern K. Light-matter interaction at atomic scales. Nat Rev Phys. 2021;3:441–453. [Google Scholar]
- 51.Zhang R, Zhang Y, Dong ZC, Jiang S, Zhang C, Chen LG, Zhang L, Liao Y, Aizpurua J, Luo Y, Yang JL, et al. Chemical mapping of a single molecule by plasmon-enhanced Raman scattering. Nature. 2013;498:82–86. doi: 10.1038/nature12151. [DOI] [PubMed] [Google Scholar]
- 52.Davies HS, Singh P, Deckert-Gaudig T, Deckert V, Rousseau K, Ridley CE, Dowd SE, Doig AJ, Pudney PDA, Thornton DJ, Blanch EW. Secondary Structure and Glycosylation of Mucus Glycoproteins by Raman Spectroscopies. Anal Chem. 2016;88:11609–11615. doi: 10.1021/acs.analchem.6b03095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alldritt B, Hapala P, Oinonen N, Urtev F, Krejci O, Federici Canova F, Kannala J, Schulz F, Liljeroth P, Foster AS. Automated structure discovery in atomic force microscopy. Sci Adv. 2022;6:eaay6913. doi: 10.1126/sciadv.aay6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rashidi M, Wolkow RA. Autonomous Scanning Probe Microscopy in Situ Tip Conditioning through Machine Learning. ACS Nano. 2018;12:5185–5189. doi: 10.1021/acsnano.8b02208. [DOI] [PubMed] [Google Scholar]
- 55.Choudhary K, Garrity KF, Camp C, V Kalinin S, Vasudevan R, Ziatdinov M, Tavazza F. Computational scanning tunneling microscope image database. Sci Data. 2021;8:57. doi: 10.1038/s41597-021-00824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, Prestegard JJ, et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology. 2015;25:1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anggara K. Data from “Direct observation of glycans bonded to proteins and lipids at single molecule level”. Data Repository of the Max Planck Society. 2023 doi: 10.17617/3.3F5JPU. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bourgoin-Voillard S, Leymarie N, Costello CE. Top-down tandem mass spectrometry on RNase A and B using a Qh/FT-ICR hybrid mass spectrometer. Proteomics. 2014;14:1174–1184. doi: 10.1002/pmic.201300433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Haan N, Narimatsu Y, Koed Møller Aasted M, Larsen ISB, Marinova IN, Dabelsteen S, Vakhrushev SY, Wandall HH. In-Depth Profiling of O-Glycan Isomers in Human Cells Using C18 Nanoliquid Chromatography-Mass Spectrometry and Glycogenomics. Anal Chem. 2022;94:4343–4351. doi: 10.1021/acs.analchem.1c05068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Juraschek R, Dülcks T, Karas M. Nanoelectrospray—more than just a minimized-flow electrospray ionization source. J Am Soc Mass Spectrom. 1999;10:300–308. doi: 10.1016/S1044-0305(98)00157-3. [DOI] [PubMed] [Google Scholar]
- 61.Anggara K, Zhu Y, Delbianco M, Rauschenbach S, Abb S, Seeberger PH, Kern K. Exploring the Molecular Conformation Space by Soft Molecule-Surface Collision. J Am Chem Soc. 2020;142:21420–21427. doi: 10.1021/jacs.0c09933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horcas I, Fernández R, Gómez-Rodríguez JM, Colchero J, Gómez-Herrero J, Baro AM. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev Sci Instrum. 2007;78:013705. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
- 63.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nuñez JR, Anderton CR, Renslow RS. Optimizing colormaps with consideration for color vision deficiency to enable accurate interpretation of scientific data. PLoS One. 2018;13:e0199239. doi: 10.1371/journal.pone.0199239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kresse G, Hafner J. Ab initio molecular dynamics for liquid metals. Phys Rev B. 1993;47:558–561. doi: 10.1103/physrevb.47.558. [DOI] [PubMed] [Google Scholar]
- 66.Kresse G, Furthmüller J. Efficient iterative schemes for ab initio total-energy calculations using a plane-wave basis set. Phys Rev B. 1996;54:11169–11186. doi: 10.1103/physrevb.54.11169. [DOI] [PubMed] [Google Scholar]
- 67.Blöchl PE. Projector augmented-wave method. Phys Rev B. 1994;50:17953–17979. doi: 10.1103/physrevb.50.17953. [DOI] [PubMed] [Google Scholar]
- 68.Kresse G, Joubert D. From ultrasoft pseudopotentials to the projector augmented-wave method. Phys Rev B. 1999;59:1758–1775. [Google Scholar]
- 69.Perdew JP, Burke K, Ernzerhof M. Generalized Gradient Approximation Made Simple. Phys Rev Lett. 1996;77:3865–3868. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 70.Grimme S, Antony J, Ehrlich S, Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys. 2010;132:154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 71.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Momma K, Izumi F. VESTA3 for three-dimensional visualization of crystal, volumetric and morphology data. J Appl Crystallogr. 2011;44:1272–1276. [Google Scholar]
- 73.Tersoff J, Hamann DR. Theory of the scanning tunneling microscope. Phys Rev B. 1985;31:805–813. doi: 10.1103/physrevb.31.805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. Raw STM images and DFT computed structures are available at the Data Repository of the Max Planck Society (57).