Abstract
Plant genomes are characterized by large and complex gene families that often result in similar and partially overlapping functions. This genetic redundancy severely hampers current efforts to uncover novel phenotypes, delaying basic genetic research and breeding programs. Here, we describe the development and validation of Multi-Knock, a genome-scale CRISPR toolbox that overcomes functional redundancy in Arabidopsis by simultaneously targeting multiple gene-family members, thus identifying genetically hidden components. We computationally designed 59,129 optimal single guide RNAs (sgRNAs) that each target 2 to 10 genes within a family at once. Furthermore, partitioning the library into ten sub-libraries directed towards a different functional group allows flexible and targeted genetic screens. From the 5,635 sgRNAs targeting the plant transportome, we generated over 3,500 independent Arabidopsis lines that allowed us to identify and characterize the first known cytokinin tonoplast-localized transporters in plants. With the ability to overcome functional redundancy in plants at the genome-scale level, the developed strategy can be readily deployed by scientists and breeders for basic research and to expedite breeding efforts.
Introduction
Plant genomics research and breeding programs rely on variation, be it natural, induced, or introduced. Phenotypic variation has been the basis for the identification of novel traits and their introduction into robust elite cultivars. Genetic variation has been expanded over the years by introducing natural variation and by creating random mutagenized lines by treatment with physical (e.g., radiation), chemical (e.g., ethyl methanesulfonate), or biological (e.g., T-DNA insertion or gene silencing) mutagens1–7. These approaches have greatly facilitated and accelerated progress in plant functional genomics and breeding programs over the past several decades.
Comprehensive genetic studies and large-scale genome sequencing projects have shown that it is challenging to uncover many phenotypes due to widespread genetic redundancy in plant genomes. A recurrent history of whole-genome duplications and numerous local duplications of smaller scale over the course of plant evolution has resulted in large gene families of similar sequences and partially overlapping functions. On average, 64.5% of plant genes are part of paralogous gene families, ranging from 45.5% in the moss Physcomitrella patens to 84.4% in the apple Malus domestica8. Given that ancient and fast-evolving paralogs are not easily detected due to sequence divergence, these percentages are likely underestimates. In Arabidopsis (Arabidopsis thaliana), the paralogous gene content, genes belonging to families with at least two members, vary from 63% to 78% depending on the methodological procedures applied2,8. It is speculated that single-copy genes are involved in the maintenance of genome integrity and organelle function, whereas multi-copy genes encode proteins involved in signaling, transport, and metabolism9. Therefore, in many cases mutating multiple gene family members is required to uncover "hidden" phenotypes associated with gene functions10.
Approaches creating random mutations for forward-genetics (e.g., alkylating agents and T-DNA lines) cannot overcome the limitations of genetic redundancy by simultaneously targeting multiple homologous genes in a single mutant line3,11. In recent years, significant progress has been made using genome-scale RNA interference methods and artificial microRNA (amiRNA) collections2,6,7. However, these methods do not work well in several important crops and generally reduce gene expression rather than cause complete knockout phenotypes12.
Recently, the CRISPR/Cas9 system (Clustered Regularly Interspaced Short Palindromic Repeats), involving CRISPR repeat-spacer arrays and Cas proteins, has been used to build large knockout mutant libraries for forward-genetic screens and analysis of gene functions and regulation. This system represents a tremendous breakthrough for generating targeted mutations both in terms of simplicity and efficiency13,14. In the past few years, studies have demonstrated the feasibility of CRISPR-based knockout collections in rice (Oryza sativa L.), maize (Zea mays) and tomato (Solanum lycopersicum)12,15–22. However, thus far, CRISPR/Cas9 has not been used on a genome-scale level to simultaneously target multiple potentially redundant genes in plants or other eukaryotes.
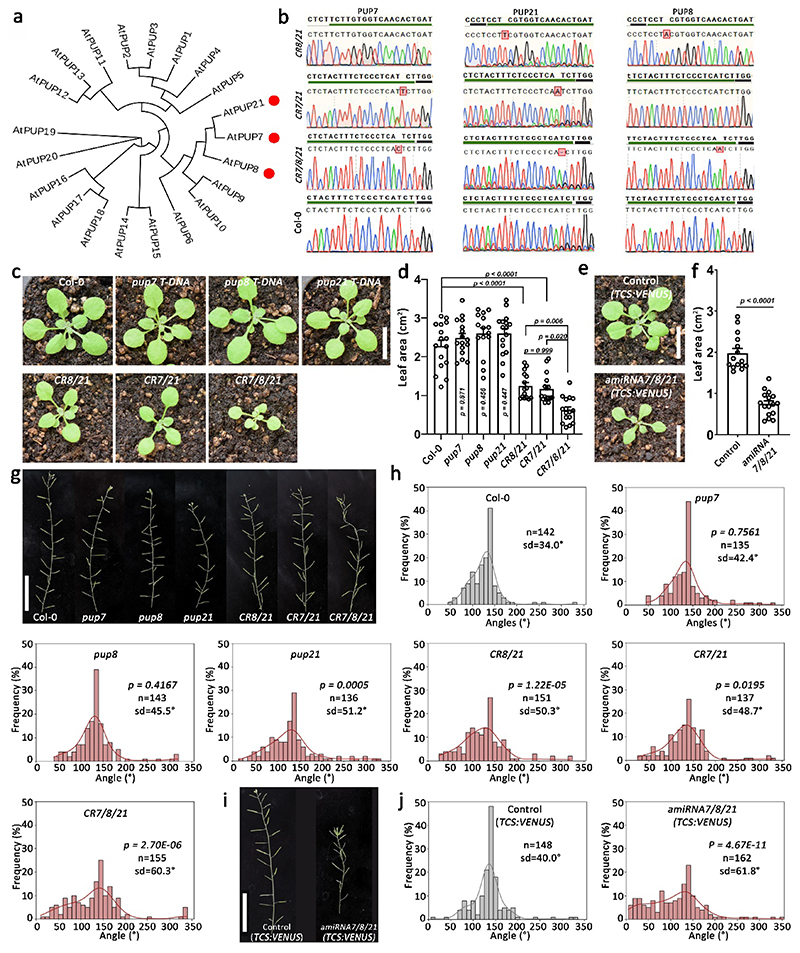
Here, we describe the development and validation of Multi-Knock, a novel genetic approach in plants that combines forward-genetics with dynamically targeted genome-scale CRISPR/Cas9 tools to address the problem of masked phenotypic variation due to functional redundancy (Fig. 1). A total of 59,129 multi-targeted sgRNAs, divided into 10 functional sub-libraries targeting 16,152 genes in Arabidopsis, were designed, synthesized, and cloned into a genome-editing intronized Cas9 vector23. From this collection, 5,635 sgRNAs targeting 1,123 of the 1,327 transporters (TRP) in Arabidopsis were cloned into four different Cas9 vectors generating independent CRISPR libraries, wherein each sgRNA was designed to target closely related homologues within sub-clades in transporter families. A proof of concept forward-genetic screen using over 3,500 CRISPR lines targeting the plant transportome recovered multiple known phenotypes in Arabidopsis, demonstrating the validity of the approach. Moreover, our screen allowed us to uncover novel transporters whose function has been hidden due to genetic redundancy. Specifically, we identified a homologous subfamily of three previously unstudied genes with partially overlapping function, PUP7, PUP21, and PUP8. We discovered that while all three proteins biochemically function as cytokinin transporters, PUP8 localizes to the plasma membrane, while PUP7 and PUP21 are localized to the tonoplast. We show that these proteins regulate meristem size, phyllotaxis, and plant growth, revealing complex redundant activity within this sub-family and providing a demonstration of the power of the Multi-Knock approach to discover new biological functions.
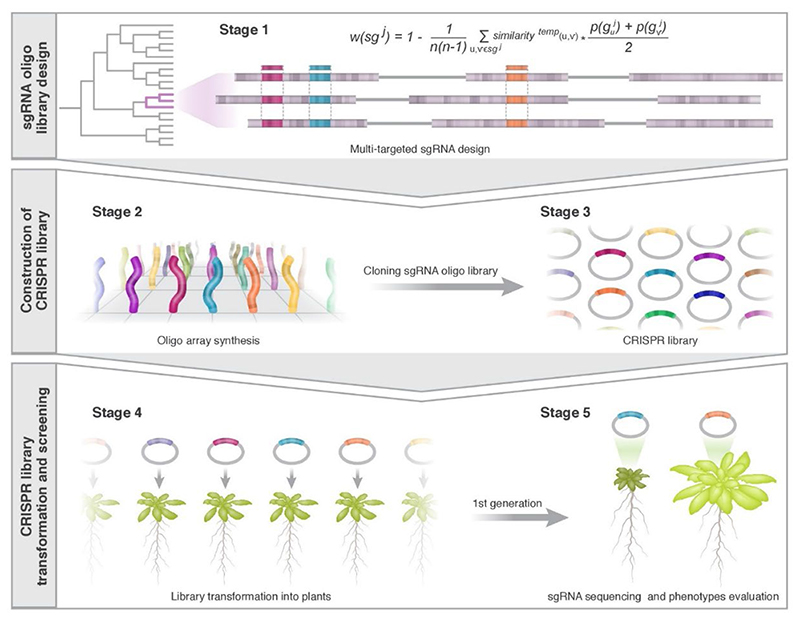
Fig. 1. Overview of the Multi-Knock, genome-scale, multi-targeted CRISPR platform.
Stage 1: Multi-targeted sgRNAs were designed to target multiple genes (coding sequences) from the same family. The Arabidopsis genome was clustered into gene families and multiple sgRNAs were designed to target each node using the CRISPys algorithm. Stages 2 and 3: sgRNA sub-library sequences were synthesized, amplified, and cloned into CRISPR/Cas9 vectors. Stage 4: The pooled CRISPR library was introduced into Agrobacterium and transformed into Arabidopsis to generate stable lines. Each plant expresses a single sgRNA, targeting a clade of 2 to 10 genes from the same family. Stage 5: A phenotypic forward genetic screen was conducted. Candidate lines were genotyped for sgRNAs and targets.
Results
Design of a multi-targeted, CRISPR-based, genome-scale genetic toolbox
The high similarity in coding sequences within plant gene families often results in complete, partial, or conditional functional redundancy, leading to substantial phenotypic buffering. To construct a genome-scale library of sgRNAs that would potentially target multiple members from the same family, all gene families in the Arabidopsis genome (TAIR10), encompassing 27,416 protein-coding genes, were downloaded from the PLAZA 3.0 plant comparative genomics database24. Following the filtration of mitochondrial and chloroplast genes, as well as singletons (i.e., genes without any family members), 21,798 genes remained, belonging to 3,892 families of size 2 or more. We then designed a set of sgRNAs that would optimally target multiple members of each gene family while accounting for the similarity among family members (Fig. 2a,b). Specifically, a phylogenetic reconstruction strategy was used to hierarchically organize each family into a tree structure, such that a homologous subgroup of genes that are more closely related are placed closer to each other on the tree. The optimal sgRNAs that could most efficiently target multiple members of each subgroup were designed using the CRISPys algorithm25. Since CRISPys could potentially design the same sgRNAs for different subgroups of the same family, we considered only one occurrence of each sgRNA (Fig. 2c,d). This procedure resulted in a total of 2,183,722 sgRNAs. Next, we removed sgRNAs that targeted only a single gene with high efficiency, resulting in 1,101,799 sgRNAs. We then removed sgRNAs with potential high off-target activity towards unintended Arabidopsis coding regions and filtered sgRNAs with overlapping targets. This resulted in a total of 59,129 sgRNAs targeting 16,152 genes (~74% of all protein-coding genes that belong to families) (Fig. 3a, Extended Data 1). Of the 59,129 sgRNAs, 98.7% target two to five genes; the rest target six to ten genes (Fig. 3b). This procedure thus created a library of sgRNAs where every sgRNA targets multiple genes, and every gene is targeted by multiple sgRNAs (Fig. 3c, Supplementary Figure 1).
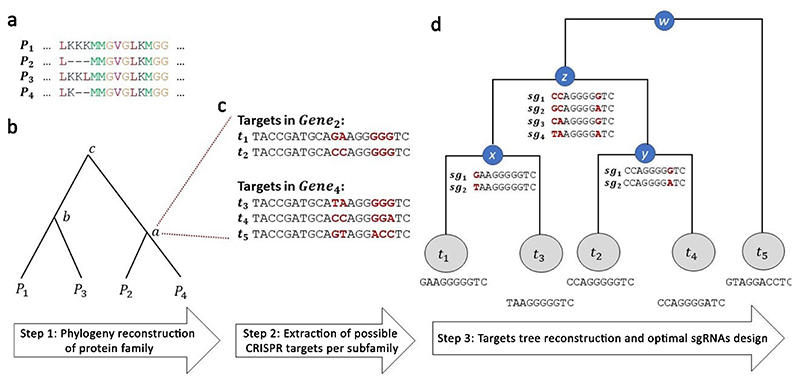
Fig. 2. An overview of sgRNA design strategy for gene families.
a, For each gene family, the multiple alignments of the respective protein sequences is computed. P stands for protein, and letters indicate amino acids. b, A phylogenetic tree is constructed based on the sequence similarity of the protein sequences. Optimal sgRNAs for each subgroup of genes, which are induced by internal nodes in the tree (marked by lowercase letters a-c), are then designed. c, For each subfamily of genes, and illustrated here for node a, all potential CRISPR target sites are extracted. In this case, the subfamily induced by node a includes two genes (g2 and g4, encoding for proteins P2 and P4, respectively). Typically, each gene contains dozens of possible targets. For simplicity, only five targets are presented. Nucleotide positions that are identical in all targets are colored in black, while the polymorphic sites are colored in red and are in bold type. d, A tree of the target sites is constructed based on sequence similarity among the targets while accounting for CRISPR-specific characteristics. sgRNA candidates are constructed for each internal node, where all combinations of the polymorphic sites are considered (marked in red), and the ones with the highest editing efficacy to target the considered subgroup of genes are chosen. For simplicity, only a few candidates (denoted by si) are shown for each internal node. Assuming that the cutoff of the number of polymorphic sites k is 4, the search of sgRNA candidates stops at node z. In practice, k was set to 12 polymorphic sites.
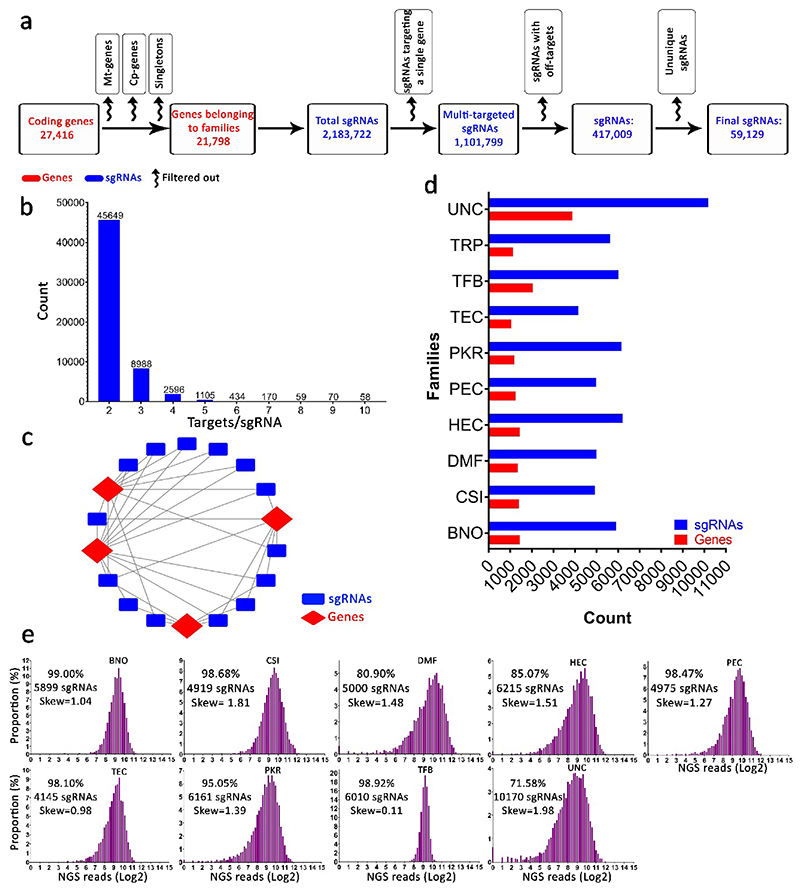
Fig. 3. Multi-targeted genome-scale sgRNA design and construction.
a, Schematic illustration of the computational workflow used to design the Multi-Knock sgRNA library. A filtering process yielded a selection of 59,129 sgRNAs targeting 16,152 genes (~74% of all coding genes belonging to families). The red color represents coding genes, blue color represents sgRNAs, and curved arrows represent filtering steps. Abbreviations: Mt-genes, mitochondrial genes; Cp-genes, chloroplast genes; Singletons, genes that do not belong to a family. b, Histogram showing the number of genes targeted by individual sgRNAs. c, Representative sgRNA-target network in the CRISPR library. Genes are targeted by multiple sgRNAs, and sgRNAs target multiple genes. d, Total number of sgRNAs and target genes in each functional sub-library. e, Deep-sequencing data of sgRNAs in individual sub-libraries. Columns indicate the distribution of sgRNAs. Coverage is indicated for each group. TRP (transporters); PKR (protein kinases, protein phosphatases, receptors, and their ligands); TFB (transcription factors and other RNA and DNA binding proteins); BNO (proteins binding small molecules); CSI (proteins that form or interact with protein complexes including stabilizing factors); HEC (hydrolytic enzymes); TEC (metabolic enzymes and enzymes that catalyze transfer reactions); PEC (catalytically active proteins, mainly enzymes); DMF (proteins with diverse functional annotations not found in the other categories); and UNC (proteins of unknown function or cannot be inferred).
Construction of multi-targeted CRISPR sub-libraries for specific functional groups
In an effort to increase the flexibility of the Multi-Knock library and enable targeted forward-genetics screens, the 59,129 sgRNAs were classified into 10 groups according to the protein functions of their putative target genes2, thus creating the following ten sub-libraries: transporters (TRP: 1,123 genes and 5,635 sgRNAs); protein kinases, protein phosphatases, receptors, and their ligands (PKR: 1,190 genes and 6,161 sgRNAs); transcription factors and other RNA and DNA binding proteins (TFB: 2,042 genes and 6,010 sgRNAs); proteins binding small molecules (BNO: 1,443 genes and 5,899 sgRNAs); proteins that form or interact with protein complexes including stabilizing factors (CSI: 1,399 genes and 4,919 sgRNAs); hydrolytic enzymes (enzyme classification [EC] class 3), excluding protein phosphatases (HEC: 1,438 genes and 6,215 sgRNAs); metabolic enzymes and enzymes (EC class2) that catalyze transfer reactions (TEC: 1,041 genes and 4,145 sgRNAs); catalytically active proteins, mainly enzymes (PEC: 1,252 genes and 4,975 sgRNAs); proteins with diverse functional annotations not found in the other categories (DMF: 1,343 genes and 5,000 sgRNAs); and proteins of unknown function or cannot be inferred (UNC: 3,881 genes and 10,170 sgRNAs) (Fig. 3d, Supplementary Table 1).
To facilitate the creation of the sub-libraries, adaptors of 38 to 47 nucleotides in length were added that were unique to each sub-library (Supplementary Table 2). We amplified each sub-library using primers complementary to the specific adaptors and used the Golden Gate method to clone the sgRNA sub-libraries into the intronized zCas9 vector (pRPS5A:zCas9i). The intronized Cas9 has 13 introns integrated into the maize codon-optimized Cas9; these introns have a significant positive effect on Cas9 genome editing efficiency in Arabidopsis23.
More than 2.0 x 105 clones of each sub-library (average coverage of 20-48× per sgRNA) growing on the selection plates were harvested, and plasmid DNA from each sub-library was isolated. In order to evaluate library quality, each sub-library was deep sequenced in a 150 paired-end mode (PE150). An ideal sgRNA library should have < 0.5% undetected guides, and a skew ratio of less than 10 26. The sequencing data showed that more than 98% of the designed sgRNAs in our libraries were present, with the exception of sgRNAs in four sub-libraries (PKR, DMF, HEC, and UNC) that exhibited lower coverage percentages (95.05%, 80.90%, 85.07%, and 71.58% coverage, respectively) (Fig. 3e). Importantly, the sgRNAs frequencies in the sub-libraries showed a narrow bell-shaped distribution (skew < 2) (Fig. 3e), indicating that no individual sgRNA were overly enriched. In addition, the effect of sgRNA cross-contamination during library construction was evaluated using deep sequencing analysis. The data indicated that all sub-libraries are highly specific (~99.9%), with the exception of the TEC sublibrary that showed significant amplification (35%) of sgRNAs from CSI sublibrary (Supplementary Figure 2). With the exception of the TEC sublibrary, all quality control analyses indicate that the Multi-Knock CRISPR sub-libraries are ready to be used in plants for functional analysis.
Multi-targeted transportome analysis
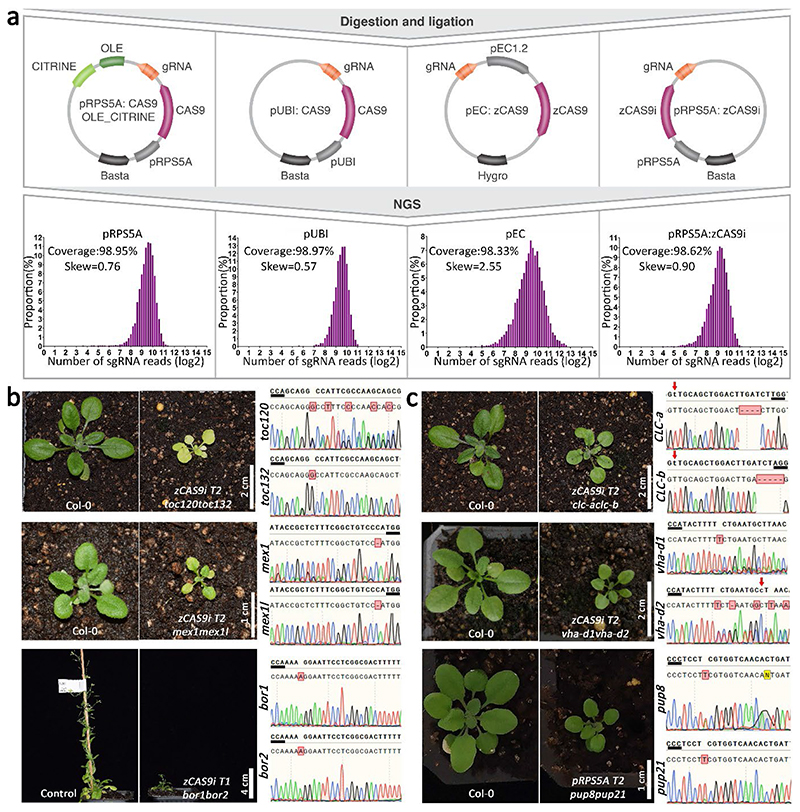
To demonstrate that the Multi-Knock approach overcomes redundancy in forward-genetics screens in planta, we chose to focus on the plant transportome using the TRP sub-library. Transporter families in plants are generally large (e.g., 136 ABC members and 53 NPF members) and relatively uncharacterized genetically27. To expand the functional utility of our tool, we cloned the 5,635 sgRNA sequences into four different Cas9 vectors to create independent TRP-sub-libraries, varying in their Cas9 type, the promoter driving the Cas9, and resistance in plants: pRPS5A:zCas9i library described above, which results in high Cas9 genome-editing activity in Arabidopsis23; pRPS5A:Cas9 with OLE:CITRINE that carries BASTA resistance and allows selection of Cas9 in seeds using a fluorescent Citrine protein28 (Supplementary Figure 3); the commonly used pUBI:Cas9 also imparts BASTA resistantance29,30; and pEC:Cas9 that carries kanamycin resistance and allows mutation specifically in the egg cells to avoid somatic mutations31. The four sub-libraries were cloned and deep-sequenced to evaluate sgRNA coverage and frequency. Coverage was higher than 98%, with a Gaussian distribution for all four libraries (skew < 3) (Fig. 4a).
Fig. 4. Transportome-specific Multi-Knock screen.
a, To create independent sub-libraries, 5,635 sgRNAs, each targeting 2 to 10 transporters from the same family, were amplified and cloned into four different Cas9 vectors to create pRPS5A:Cas9 (OLE:CITRINE), pUBI:Cas9, pEC:Cas9, and pRPS5A:zCas9i sub-libraries. Graphs show coverage and frequency based on next-generation sequencing of the four sub-libraries. The four libraries were transformed into Col-0 plants yielding 3,500 transgenic T1 plants. b, Photographs show representative phenotypes of TRP Multi-Knock proof-of-concept lines. From top to bottom are Col-0 and plant expressing sgRNA targeting toc120 and toc132 (scale bar = 2 cm), Col-0 and plant expressing sgRNA targeting mex1 and mex1l (scale bar = 1 cm), and control DR5:VENUS plant and the T1 plant harboring sgRNA targeting bor1 and bor2 (scale bar = 4 cm).
Chromatograms show the types of mutations. Red arrows indicate the mismatches between sgRNA and target sequence. PAM is marked with a black underline. c, Images show lines with abnormal phenotypes that had not previously been described: from top to bottom adjacent to Col-0 control are plants expressing sgRNA targeting clc-a and clc-b (scale bar = 2 cm), vha-d1 and vhad-2 (scale bar = 2 cm), and pup8 and pup21 (scale bar = 1 cm). Chromatograms show the type of mutations. Red arrows indicate the mismatches between sgRNA and target sequence. PAM is marked with a black underline.
At least 2.2 x 105 Agrobacterium tumefaciens clones of each TRP-sub-library were harvested and transformed into Arabidopsis Col-0 plants yielding about 3,500 transgenic T1 plants (pUBI:Cas9, 500 lines; pEC:Cas9, 500 lines; pRPS5A:Cas9 OLE:CITRINE, 500 lines; and pRPS5A:zCas9i 2,000 lines). To increase on-target mutagenesis in plants, pUBI:Cas9, pEC:Cas9, and pRPS5A:zCas9i T1 plants were subjected to repeated mild heat stress as previously described with slight modifications32. 2,000 T1 lines were collected individually for the pRPS5A:zCas9i library. pUBI:Cas9, pEC:Cas9, and pRPS5A:Cas9 OLE:CITRINE libraries were each collected in bulks of 10 plants. 1,200 independent T2 lines generated from the pRPS5A:zCas9i TRP sub-library were screened in soil at 22°C under long-day conditions (16-h light and 8-h dark) using a high-throughput phenomics system, evaluating photosynthesis-related parameters, thermal imaging, plant color, and shoot morphology, Importantly, the screen recovered previously reported phenotypes of mutants affected in transporters. For example, we isolated two independent lines with pale, bleached, and small-size shoots. Extracting DNA, amplifying the sgRNA cassette, and sequencing revealed that they harbor the same sgRNA sequence, putatively targeting TOC132 and TOC120 (Translocon Outer Complex proteins) (Fig. 4b, Supplementary Figure 4a). Sanger sequencing of TOC132 and TOC120 revealed that frameshift mutations occurred at the sgRNA target sites in these two genes (Fig. 4b, Supplementary Figure 4b). The similar phenotypes we observed in two independent lines indeed mimicked the toc132,toc120 double mutant phenotype that was previously characterized33. In addition, we identified phenotypes driven by two different sgRNAs targeting two maltose transporters (MEX1 and MEX1-Like). The mex1,mex1l CRISPR double mutants phenotype was enhanced compared to the previously described mex1 single mutant34 (Fig. 4b, Supplementary Figure 5). Plants targeting genes encoding two boron transporters (BOR1 and BOR2) were genotyped as double bor1,bor2 knockouts and had growth inhibition phenotypes (Fig. 4b), likely enhancing the bor1-1 mutant-plants35. Unlike all other lines described here which were validated in T2 or T3 generations, the T1 generation bor1,bor2 knockouts were sterile and did not produce seeds. Sequensing the sgRNA and their putative target genes of additional lines showed edited DNA events in the majority of the targeting sites (Supplementary Figure 6).
Many of the phenotypes we observed were driven by previously undescribed genes, For example, plants expressing a single sgRNA resulted in deletions in clc-a, clc-b (Chloride Channels), or vha-d1, vha-d2 (Vacuolar-type H + -ATPases) or pup8, pup21 (Purin Permeases), all showing smaller rosette size than Col-0 plants (Fig. 4c). Notably, while some of the mutations were homozygous, TOC120, VHA-D2, and PUP8 showed superimposed sequencing data. Chromatogram sequence deconvolution showed that PUP8 is a biallelic mutation (+T/+A) (Fig. 4). The data for TOC120 and VHA-D2 is less clear and could point to biallelic or heterozygous mutations. At this stage, we do not know whether the phenotypes are a result of an on-target activity, and further genetic validation is needed to rule out off-target effects. Such genetic validation was carried out below for the PUP candidates. Notably, the Multi-Knock seed collection we generated here is available to the community for any type of forward-genetic screen. Together, the results demonstrate the strength of the Multi-Knock strategy in exposing novel mutant phenotypes.
Multi-Knock revealed partially redundant tonoplast-localized PUP cytokinin transporters
As noted above, the Multi-Knock transportome-scale screen identified a shoot growth inhibition phenotype caused by PUP8 and PUP21 loss-of-function (Fig. 4c). The two unstudied proteins are members of the PUP family, which consists of 21 genes (Fig. 5a). Most of the genes in the PUP Arabidopsis family have not been characterized36, but PUP14 reportedly encodes for a plasma membrane cytokinin transporter37. In addition to plasma membrane-localized PUP1437, PUP1 and PUP2 were also identified as cytokinin transporters in Arabidopsis38,39. In rice, OsPUP1 and OsPUP7 were shown to localize on the endoplasmic reticulum (ER), while OsPUP4 was localized to the plasma membrane40,41. Cytokinins are plant hormones essential for meristem maintenance and additional physiological and developmental processes, such as cell division, lateral root formation, leaf senescence, embryo development and adaptive responses to heat and drought stresses42–44. Because cytokinin biosynthesis, catalyzed by isopentenyl-transferases, does not occur throughout the plant but is limited to certain tissues, cytokinins are translocated through the plant by diffusion and/or through active transport mechanisms36,45. There is a complete genetic linkage between the PUP7, PUP21, and PUP8 genes, and phylogenetic analysis of PUPs in Arabidopsis showed that these three genes form a monophyletic clade (Fig. 5a). Similar to PUP8 and PUP21, the function of PUP7 is unknown. To characterize the activity of PUP7, PUP21, and PUP8, we isolated single PUP7, PUP21, and PUP8 T-DNA homozygous lines. The single pup7 (SALK_084103) and pup8 (SALK_137526) mutants showed no morphological differences compared to Col-0. The pup21 (GABI_288E11) mutant also did not show a phenotype in the vegetative stage compared to Col-0, and presented only a mild plant height phenotype after bolting (Supplementary Figure 7). To validate the potentially redundant on-target activity of PUP7, PUP21, and PUP8 as revealed by the PUP8 and PUP21 loss-of-function line (Fig. 4c), we cloned a multiplexed CRISPR construct targeting PUP7, PUP21, and PUP8 and obtained double (CRISPR7/21) and triple (CRISPR7/8/21) mutants. Mutations were validated in generations T4 (CRISPR7/8/21) or T3 (CRISPR7/21). Both CRISPR7/21 and CRISPR7/8/21 showed frameshift mutations in their targets (Fig. 5b) that exhibited a small rosette size (Fig. 5c,d). The phenotype of the triple mutant (CRISPR7/8/21) was enhanced compared to the CRISPR7/21 double mutant and to the CRISPR8/21 double mutant recovered from the Multi-Knock screen (Fig. 5 c,d), indicating that PUP7, PUP21, and PUP8 redundantly regulate shoot growth. To further validate the on-target activity of PUP7, PUP21, and PUP8 we generated a PUP7, PUP21, and PUP8 multi-targeted amiRNA line (amiRNA7/8/21). amiRNA7/8/21 showed reduced expression of PUP7, PUP21, and PUP8 (Supplementary Figure 8). In agreement with the CRISPR7/8/21 triple mutant, the amiRNA7/8/21 line also exhibited a small rosette size (Fig. 5e,f).
Fig. 5. PUP7, 8, and 21 redundantly regulate shoot growth and phyllotaxis.
a, Phylogenetic tree of Arabidopsis PUP family based on amino acid sequences. Red dots indicate proteins coded by putative CR7/8/21 target genes. b, Chromatograms showing the types of mutations in the CR8/21 (T3 generation), CR7/21 (T3 generation) and CR7/8/21 (T4 generation) lines as identified by sequencing. CR8/21 and CR7/21 stand for CRISPR double mutant pup8/21 and pup7/21, respectively; CR7/8/21 stands for CRISPR triple mutant pup7/8/21. PAM is underlined in black; 20-bp sgRNA is underlined in green. The sgRNA in line CR8/21 was not designed to target PUP7 as it does not contain a respective PAM sequence. c, Shown are representative images of 18-day-old WT (Col-0), the T-DNA single pup7, pup8, pup21 mutants, the double pup8pup21 (CR8/21) (T3 generation), double pup7, pup21 (CR7/21) (T3 generation), and the triple pup7pup8pup21 CRISPR mutant (CR7/8/21) (T4 generation). Scale bar = 1 cm. d, Quantification of genotypes shown in (c). Shown are means (±SE). p value in ordinary one-way ANOVA is indicated for each analysis, Col-0, pup7. pup21, CR7/21 and CR7/8/21: n=16; pup8 and CR8/21: n=15. e, Shown are representative images of 18-day-old Control (TCS:VENUS) and amiRNA7/8/21 (TCS:VENUS background). amiRNA7/8/21 stands for amiRNA knockdown PUP7/8/21. Scale bar = 1 cm. f, Quantification of genotypes shown in (e). Shown are means (±SE). p value two tailed t test is indicated. n = 15 (Control); n = 16 (amiRNA7/8/21). g, Phyllotaxis patterns in inflorescences stem of wild-type (Col-0), single T-DNA insertion mutants, CR8/21, CR7/21 and CR7/8/21. Scale bar = 5 cm. h, Silique divergence angle distribution in inflorescences of Col-0, pup single mutants, CR8/21. CR7/21 and CR7/8/21. P-value, n number and standard deviation (sd) are indicated for each analysis. P-value was extracted using Fligner-Killeen test for equality of variance. i, Phyllotaxis patterns in inflorescence stem of control (TCS:VENUS) and amiRNA7/8/21 mutant. amiRNA7/8/21 stands for amiRNA triple PUP7/8/21 knockdown. Scale bar = 5 cm. j, Distribution of divergence angle frequencies between successive siliques in control and amiRNA7/8/21 stems. p value Fligner-Killeen test for equality of variance is indicated for each analysis.
Cytokinin response was previously shown to regulate the spatial distribution of lateral organs along the stem or phyllotaxis46. We found a significant perturbed phyllotaxis phenotype with an increase in the occurrence of abnormal angles between consecutive organs in CRISPR7/8/21 and amiRNA7/8/21 lines (Fig. 5g-j). These results suggest that PUP7, PUP21, and PUP8 redundantly regulate shoot growth and phyllotaxis.
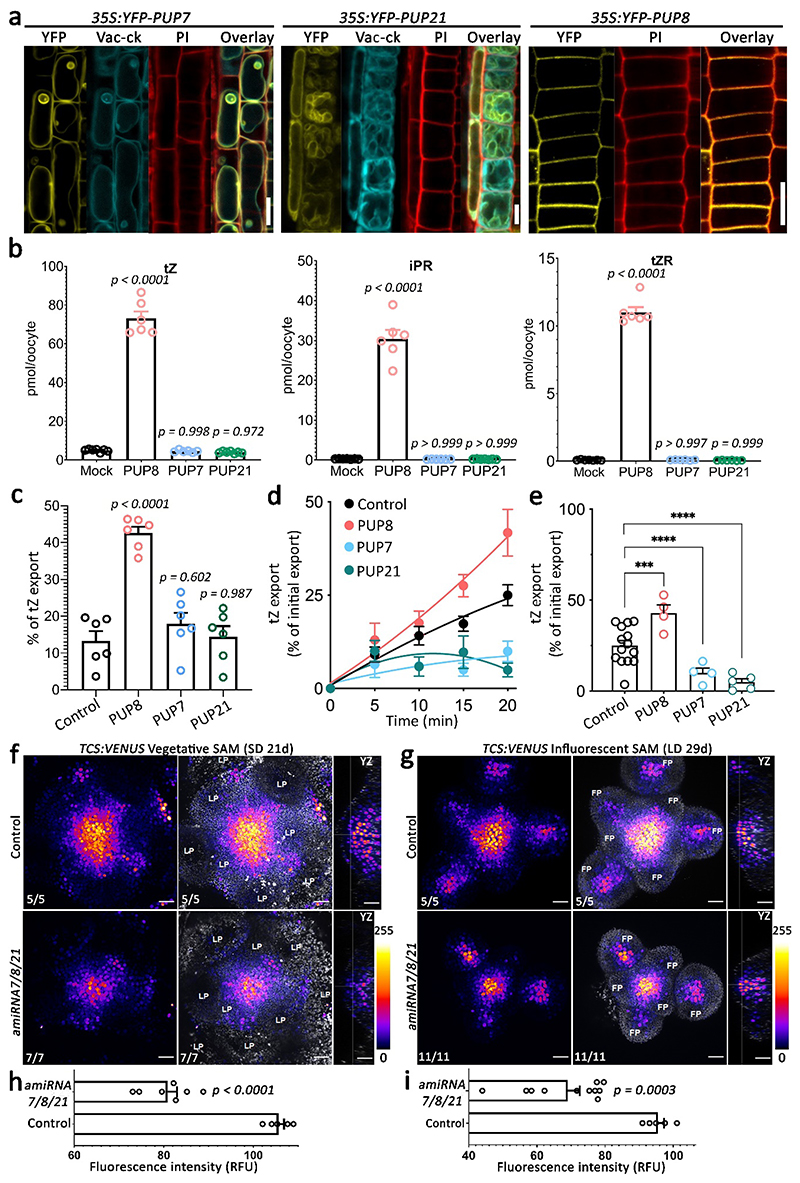
To understand how PUP7, PUP8, and PUP21 function in cellular transport, we generated stable transgenic plants that express PUP7, PUP8 and PUP21 fused with YFP under the control of the cauliflower mosaic virus 35S promoter and evaluated the subcellular localization of these proteins in the root meristem epidermis cells. PUP7 and PUP21 were localized to tonoplasts as indicated by co-localization with the vac-ck tonoplast marker47, whereas PUP8 localized to the plasma membrane (Fig. 6a). We then performed transport assays with the 3 proteins in Xenopus laevis oocytes using the cytokinin trans-zeatin (tZ). Based on standard import assays and injection-based export assays, PUP8 was shown to be capable of bidirectional cytokinin transport along the concentration gradient of cytokinin (Fig. 6b,c, Supplementary Figure 9). The absence of transport activity of PUP7 and PUP21 in oocytes, which do not have vacuoles, likely results from their mis-localization. In order to back up the oocyte data in a plant system, we expressed all three PUP transporters in tobacco protoplasts and quantified tZ export after diffusion-based loading. Expression of PUP7 and PUP21 resulted in significantly reduced cellular tZ export, while PUP8 expression resulted in enhanced tZ export (Fig. 6d,e). In order to demonstrate substrate specificity, we performed competition experiments for PUP8 that allowed as a putative tZ exporter for direct competition of tZ uptake into microsomal fractions. Addition of an access of cold tZ significantly reduced transport activity of radiolabelled tZ (Supplementary Figure 10).
Fig. 6. PUP7, PUP8, and PUP21 regulate cytokinin transport and shoot meristem.
a, Root meristem confocal microscopy images of 35S:YFP-PUPs localization. YFP (yellow) was used to label PUPs, Vac-ck (cyan) was used as a tonoplast marker, and propidium iodide (PI) (red) was used to stain the cell wall. Scale bars = 10 μm. The experiments were independently repeated three times with similar results. b, Standard import Xenopus laevis oocytes assays using the indicated cytokinin froms. 60 min transport assay in 100 μM cytokinins at pH = 5.5. n = 7 (Mock), n = 6 (PUP8 and PUP7), n = 7 (PUP21 tZ and iPR), n = 6 (PUP21 tZR). Shown are means (±SE). p values were determined by One-way ANOVA. c, Injection-based export assay of PUPs in Xenopus laevis oocytes. n = 6 ± SE, p value two-tailed t-test is indicated for each analysis, d, e, tZ export from tobacco protoplasts as percentage of initial export as a function of time (d) and at 20 minutes (e). n = 14 (Control), n = 4 (PUP8 and PUP7), n = 5 (PUP21). Shown are means (±SE). p values were determined by One-way ANOVA. Box plots represent 25th-75th percentiles; whiskers represent min-max (with all points), central bands in the box plots show the medians. f-i, TCS-Venus intensity in control plants (TCS:VENUS) compared to amiR7/8/21 at (f) vegetative and (g) inflorescence stages. amiRNA7/8/21 stands for amiRNA triple knockdown PUP7/8/21. Optical longitudinal sections of the meristem (YZ direction) are shown. LP: leave primordia, FP: floral primordia. Scale bars = 100 μm. The color scale at the right indicates TCS expression. Quantification of TCS-Venus intensity in vegetative (h) and inflorescence (i) stages. Control: n = 5 in (h, i); amiRNA7/8/21: n = 7 in (h), n = 11 in (i). Shown are means (±SE). p value two tailed t test is indicated for each analysis.
To evaluate whether these PUP proteins are implicated in cytokinin transport and signaling, we monitored the shoot apical meristem cytokinin response in Col-0 and amiRNA7/8/21 plants using the synthetic cytokinin-inducible reporter TCS:Venus48. A severely reduced TCS signal was detected in amiRNA7/8/21 vegetative and inflorescence shoot apical meristems compared to controls (Fig. 6f-i). The TCS domain at the center of the SAM and in flowers was also much narrower in amiRNA7/8/21 compared to wild type (Fig. 6f,g), thus demonstrating also a change in the spatial distribution of cytokinin responses in the SAM. Overall, these results indicated that tonoplast-localized PUP7 and PUP21 can act as vacuolar cytokinin importers, while plasma membrane-localized PUP8 is required for cytokinin export. This cytokinin transport activity is required to establish the spatial pattern of cytokinin signaling within the SAM, thus regulating shoot growth and phyllotaxis. However, since directional transport activity of PUP8, and possibly of PUP7 and PUP21, depends on cytoplasmic (or apoplastic) cytokinin levels, it is possible that their transport directionality might change spatially. Taken together, our discovery of these novel cytokinin transporters was made possible through the simultaneous targeting of the multiple PUP genes.
Discussion
The large number of gene families in Arabidopsis results in high levels of functional redundancy49. In recent years, genome-scale amiRNA collections have been developed in Arabidopsis and used for forward-genetic screening to identify hidden phenotypes masked by redundant homologous genes2,7. However, this strategy generally results in incomplete knockout phenotypes. The CRISPR/Cas9 system is a simple, effective method for generating targeted heritable mutations in the genome and has recently enabled large-scale knockout mutant libraries of single genes to be generated for forward-genetic screens in mammalian50,51 and plant systems15–19. An important advantage of the CRISPR/Cas9 method is its capacity to simultaneously target multiple genes, whether they are genetically linked or not. In this study, we developed a novel genome-scale approach with the ability to simultaneously target several genes within the same gene family and applied it in Arabidopsis. This forward-genetic strategy overcomes functional redundancy and enables flexible screening, ranging from a specific functional subgroup to the entire genome. We reported here on six lines with reproducible phenotypes and validated the respective editing of the target genes. It is important to note that we have observed dozens of novel phenotypes (i.e., by screening for differences in shoot size, color, shape and photosynthesis-related parameters) that we have not followed up with detailed experiments. As the intronized-CAS9 efficiency is above 70% 23,52, we project that the rate-limiting step is not the library or the intronized-CAS9 efficiency, but rather the genetic screen setup and cost-effective genotyping strategies. Overall, the approach we developed and the library we constructed should allow a broad spectrum of functional screens to be readily carried out, thereby significantly impacting current genetic analyses in plants.
The present strategy constructed the Multi-Knock tool using a single sgRNA design to optimally target multiple genes. We expect that with the anticipated progress in the field, future developments of the Multi-Knock approach could utilize sgRNA multiplexing. While constructing a genome-scale sgRNA multiplexed CRISPR library is at present technically challenging (currently limited by the base-pare length of pooled oligo libraries containg thouesends of sgRNAs), a breakthrough in the field will increase the overall targeting efficiency and coverage. In addition, the current Multi-Knock tool is designed to target redundancy resulting from multiple proteins with highly similar sequences carrying overlapping activity and having highly similar coding sequences. However, Multi-Knock will not reveal redundancy resulting from overlapping pathways (diverse gene families with overlapping transcriptional or biochemical activities). The development of a multiplexing Multi-Knock library would naturally uncover redundant pathways, by allocating each sgRNA to target a different set of (one or more) homologous proteins.
As a proof-of-concept that Multi-Knock can identify novel gene functions, we identified the first cytokinin tonoplast transporters in plants. There are discrepancies in published data concerning the subcellular localization of cytokinin receptors and the cellular site of cytokinin perception. In different studies, cytokinin receptors have been shown to localize to the endoplasmic reticulum membrane53–56 and to the plasma membrane37. Recently, two independent groups described multiple sites of cytokinin perception at plasma and endoplasmic reticulum membranes57,58. We speculate that the tonoplast localization of PUP7 and PUP21 may facilitate the movement of cytokinins into vacuoles, thereby lowering the availability of cytokinins in the cytoplasm and/or the ER. However, it is important to note that injection or diffusion-based tZ loading might promote PUP-mediated tZ transport from high to low concentration in both Xenopus and tobacco, respectively. Under in vivo conditions, it is possible that PUP transport directionalities may also shift, depending on local cytokinin concentrations or electrochemical gradient changes.
The direction of both uni- and symporters was previously shown to be reversible in vitro depending on the direction of the electrochemical gradient of their substrate and/or symported ion59–62. For example, PUP8 and the SWEET (SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTER) transporters were both identified using import-based screens but were shown to be bidirectional in Xenopus oocytes61,62. Lastly, PUP8 was shown to possess phlorizin transport capability via an import-based screen of > 600 Arabidopsis transporters and also shown to be bidirectional. Phlorizin is a dihydrochalcone abundantly found in apples, which indicates that PUP8 might have acquired additional substrates in plants61. It is thus intriguing to speculate that dual-directional PUPs activity in different cell types, possibly among shoot apical meristem zones, regulates cytokinin response by mediating sub-cellular cytokinin homeostasis. The tonoplast-dependent cytokinin transport may also have an impact on cell-to-cell cytokinin movement. Such coordination among a network of plasma membrane and tonoplast transporters might determine the cytokinin fluxes within the cell and within tissues. Therefore, the tonoplast localization of PUP7 and PUP21 identified here adds another level of regulation to the response mediated by endoplasmic reticulum-localized cytokinin receptors.
Using the Multi-Knock strategy, we demonstrated partially redundant functions of PUP8, PUP7, and PUP21 in cytokinin transport either across the plasma membrane or into the vacuole, which would both lead to reduced cytoplasmic cytokinin levels. In all cases, the genes are genetically linked and functionally redundant, emphasizing the power and need of Multi-Knock as a genome-scale multi-targeted approach. It is common that evolutionary conserved transporter families in plants share redundant activities and are genetically linked, suggesting recent gene-duplication events. For example, in five ABCB auxin transporters (ABCB15-22) of the same phylogenetic branch, their simultaneous knockout using Cas9 exposed their redundant activity63. Additional examples are the genetically linked and redundant activities of ABCB6 and ABCB20 in auxin transport7 and of ABCG17 and ABCG18 in ABA transport64. We predict that future genetic screens, utilizing approaches as the one developed here, will expose the activities of many genetically linked and redundant genes.
Saturated screens allows to observe the same phenotype in multiple independent lines, creating multiple alleles and partially omitting false positive candidate lines. Here, we screened ~1,200 independent T2 lines from the CRISPR TRP library, which contains 5,635 sgRNAs, meaning there is a very low chance of identifying multiple independent lines containing sgRNAs that target the same genes. However, we successfully isolated independent lines for toc120,toc132, and mex1,mex1l, likely because of their strong visible phenotypes. One way to tackle this issue is to screen for high-throughput phenotypes as carried out in the mammalian field using cell culture51. However, such screens do not benefit from the strength of screening in planta. Another way one can isolate multiple events and screen a X10 covorage, saturated population, is to bulk 10 or 100 lines together (for example, 56K lines covering 5,635 sgRNAs). This allows rapid screening and increases the chance of identifying the same sgRNA multiple times. At the same time, this approach has drawbacks as it is harder to pick up on minor phenotypes. An alternative approach to tackle the issue of saturated screens is to generate smaller libraries that cover specific groups of limited size, thereby increasing the chance of observing multiple lines directed towards the same set of genes.
Following successful phenotyping and genotyping of Multi-Knock T2 plants, just as in any other genetic approach (e.g., use of alkylating agents, T-DNA, amiRNA), it is critical to validate that the phenotype is indeed driven by the specific mutation. Ursache et al., reported that an Agrobacteria mix, containing multiple constructs, could result in the integration of more than one construct per plant52. Furthermore, Jacobs et al., reported that transient expression of sgRNAs in tomato might cause CRISPR mediated mutations in respective target gene15, both leading to possible off-target effects. One way to address these obstacles and reveal possible on/off-targets is to use the SMAP multiplex PCR amplicon tool to genotype putative target sites65. Addination methods to validate on-target activity may include complementation lines to demonstrate phenotype rescue, or generate multiple independent mutant lines (such as a combination of T-DNA lines or, in cases of genetic linkage, sgRNAs or amiRNA) that present the same phenotype.
Here we presented the Multi-Knock tool and validated its use for gene function discovery in Arabidopsis. This genome-scale multi-targeted mutagenesis system may also be applied to other plant species and utilized for next-generation breeding programs to uncover hidden genetic variations. However, in contrast to flower-dip transformation in Arabidopsis, large-scale Agrobacterium-mediated plant transformations in crops is still a bottleneck due to low transformation efficiency and the requirement for labor-intensive tissue culture. Thus, once transformation efficiency is enhanced, for example, using sgRNA delivery by viral vectors66,67 or nanoparticle-based carriers68,69, we foresee that the Multi-Knock approach could be readily employed in many other plant species. Notably, the Multi-Knock tool requires high-quality information on gene space. While high-quality sequenced genomes of most important crops, and dozens of other plants, are already available, this could be an issue for less studied plant species. A possible alternative in such cases is to extract the gene-space via deep sequencing of the transcriptome, albeit limiting the off-target search to coding regions only.
Materials and Methods
Plant material and growth conditions
All Arabidopsis plants were derived from the Columbia ecotype and grown in dedicated growth rooms under long-day conditions (16 h light/ 8 h dark) at 22 °C. Arabidopsis Col-0 plants were transformed using Agrobacterium strains (GV3101) by the flower dip method70.
Multi-targeted sgRNA design
All 9,350 gene families in the Arabidopsis thaliana genome, encompassing 27,416 genes, were downloaded from the PLAZA 3.0 plant comparative genomics database24. Genes belonging to the mitochondrial and chloroplast genomes were filtered out, as well as families with a single family member, leaving 3,892 families of size 2 or more that together encompassed 21,798 genes. The CRISPys algorithm was then applied to each family while accounting for the homologous relationships within each family. Specifically, given a family of genes, a gene tree was reconstructed using a hierarchical clustering algorithm71, which clusters the genes according to their sequence similarity. The SΩ design strategy of CRISPys was then recursively applied to each subgroup induced by the gene tree to find the optimal sgRNAs for targeting the desired subfamily. CRISPys was applied using the CFD (Cutting Frequency Determination) score72 as the scoring function with targeting efficacy threshold of Ω = 0.55 and k = 12 as the threshold for the number of polymorphic sites. The number of sgRNAs per each subgroup of genes in a given gene tree was limited to 200. The potential sgRNA targets were allowed only for the first two-thirds of the coding sequence. Since CRISPys could assign the same sgRNAs for different subgroups of homologous genes, where one subgroup is a subset of the other one (for example, assuming that {𝑔1, 𝑔2, 𝑔3} is a subset of homologous genes, and s is an sgRNA that targets this subgroup of genes, the same sgRNA s can also be found for {𝑔1, 𝑔2}). we considered only one occurrence of the sgRNA.
For each remaining sgRNA, a genome-wide off-target detection was applied. In the context of gene-family cleavage, an off-target is defined as a potential genomic target that is outside the specified gene family, while on-targets are nuclear targets that reside within the family, even though some mismatches may occur between them and the examined sgRNA. To this end, given a specified sgRNA, the Burrows-Wheeler Aligner (BWA)73 was applied to the Arabidopsis thaliana genome (PLAZA v3) to identify potential nuclear hits. BWA was executed with the command "bwa aln", with the following parameters: -N, -l 20, -i 0, -n 5, -o 0, -d 3, -k 4, -M 0, -O 1000000, -E 0, thus allowing searching for targets with at most four mismatches and no gaps. Only hits that reside within protein-coding exons were considered off-targets. A potential sgRNA was filtered if it was inferred to cleave an off-target with a CFD score higher than 0.33. We then applied an additional filtering procedure, where we tested the remained sgRNAs for overlapping target regions. A given sgRNA was removed if all its targets overlapped with those of a second potential sgRNA, and the CFD scores of most of these targets were lower. A sgRNA s1 is defined to overlap with sgRNA s2 if the positions of all its targets overlap with those of s2 in at least 10% of the aligned region (i.e.. 2 bp).
CRISPR/Cas9 vectors
To generate the pRPS5A:Cas9 OLE:CITRINE plasmid, Site-Directed Mutagenesis (NEB-E0554S), was used to eliminate the 3 BsaI sites within the OLE:CITRINE sequence, using the following primers: Fwd-ATGGGCCGAGACAGGGACCAGTACCAGATGTCCGGAC Rev-CATCGGGTACTGGTCCCTGCCGATGATATCGTGATGG. The BsaI sites are required for the Golden gate CRISPR library cloning. Next, OLE:CITRINE was cut and ligated from pJET into pRPS5A:Cas9 vector using MluI and BamHI restriction enzymes. pUBI:Cas9 was generated as described previously29. pRPS5A:zCAS9i (Addgene ID: AGM55261)23 and pEC:Cas9 (Addgene ID: pHEE401)31 were purchased from Addgene.
Construction of Multi-Knock, multi-targeted CRISPR libraries
The 20-nucleotide sgRNA target sites were appended with the specific adaptors and BsaI sites (Supplementary Table 2). Synthesis of the 59,129 DNA oligonucleotides (total yield: 500 ng) corresponding to the sgRNAs was performed by Twist Bioscience. A stock solution of oligo pool was prepared by resuspending in 10 mM Tris buffer, pH 8.0 to a concentration of at least 20 ng/μl. The single-stranded oligonucleotide pool was converted to double-stranded DNA by PCR using the high-fidelity Phusion polymerase (NEB) using 12 to 15 cycles of PCR to avoid introducing PCR bias. PCR was conducted using the following conditions: 98 °C for 30 s; 15 cycles of 98 °C for 30 s, 60 °C for 30 s, and 72 °C for 15 s; and a final extension at 72 °C for 10 min. For each family pool, about 6 tubes of 50 μl-volume amplification reactions with a total of 15 ng single-stranded oligonucleotide pool as a template and the specific primers for adaptors (Supplementary Table 3) were used, and the PCR products were purified with a NucleoSpin Gel and PCR clean up Kit (Macherey-Nagel).
The purified DNA products were digested with BsaI restriction enzyme and ligated into the desired Cas9 expression constructs using the Golden Gate cloning method. Golden Gate assembly was performed as follows: 35 cycles of 37 °C for 5 min and 16 °C for 5 min; 50 °C for 20 min; and 80 °C for 20 min. Four 20-μl ligation reactions were combined, and 20 bacterial transformations were carried out using 4 μl of ligation reaction and 50 μl Top10 chemically competent E. coli per transformation according to the manufacturer’s instructions. The 20 transformations were combined and plated onto seven LB agar plates (145 x 20 mm, Greiner Bio-one) supplemented with the relevant antibiotics. Colonies were validated using colony PCR and Sanger sequencing individually, then bacteria from all plates were scraped off and combined. The plasmid DNA was purified with a Plasmid Maxi kit (Qiagen) to produce the CRISPR libraries. In order to verify these plasmid pools, PCR products amplified with the primers listed in Supplementary Table 4 from the CRISPR libraries were sequenced on an Illumina NovaSeq 6000 with the PE150 mode. Eight 50 μl amplification reactions using the high-fidelity Phusion DNA Polymerase (NEB) were set up. PCR was conducted using the following conditions: 98 °C for 30 s; 30 cycles of 98 °C for 30 s, 60 °C for 30 s, and 72 °C for 15 s; and a final extension at 72 °C for 10 min. PCR products were purified with a NucleoSpin Gel and PCR clean-up Kit (Macherey-Nagel). At least 1.5 μg PCR products per each sub-library were sent to Novogene for deep-sequencing.
The number of reads per sgRNA sequence was quantified from the raw sequencing data using the Biopython package in the Python programing language. A list of sgRNAs comprising the library was loaded into python. A loop was performed where each one of the sgRNAs in the list was compared against the raw deep-sequencing data in FASTA using the Biopython package and quantified the number of reads where the specific sgRNA appeared. The sgRNA and its read number were written into a new data frame. CRISPR libraires are available upon request.
Generation of four transportome CRISPR libraries
The four transportome CRISPR plasmids were transformed into Agrobacterium tumefaciens strain GV3101 using electroporation. In brief, for each library, around 20 tubes of GV3101 competent cells (80 μl) were incubated on ice with ~1 μg plasmid in each tube for 5 min and electroporated using a MicroPulser (Bio-Rad Laboratories; 2.2 kV, 5.9 ms). Immediately after electroporation, 700 μl LB medium was added, and samples were shaken for 1.5-2 h at 28 °C. Agrobacterium was then plated on LB agar plates (145 x 20 mm, Greiner Bio-one) containing the relevant antibiotics for 2 days at 28 °C in the dark. Each Agrobacterium transportome CRISPR library was transformed into six trays of Arabidopsis Col-0 plants. T1 Seeds were collected in bulk. After transformant plant selection, transgenic plants for each transportome CRISPR library were propagated, and T2 seeds were collected. CRISPR libraires are available upon request.
Arabidopsis transformation and heat-shock treatment
The Agrobacterium colonies from all plates were scraped off and added into 1 L LB medium with 25 μg/ml gentamycin, 25 μg/ml rifampicin, and vector-specific antibiotic, followed by incubation at 28 °C for 16-24 hours. Agrobacterium was harvested by centrifugation for 10 min at 5,500 rpm, the supernatant was discarded, and the bacteria pellet was resuspended in ~400 ml inoculation medium containing 0.5 x MS (Duchefa Biochemie), 5.0% sucrose, and 0.05% Tween-20 (Sigma-Aldrich). Arabidopsis flowers were then sprayed with the bacterial solution. After spraying, plants were kept in the dark overnight and grown until siliques ripened and dried. T1 seeds were collected in bulk. The T1 seeds of the pEC:zCas9 library were sown on MS media containing hygromycin (25 μg/ml) for the transformant plant selection, whereas the T1 seeds of the other three transportome CRISPR libraries were sown on soil and sprayed with BASTA for selection at the age of 2 weeks. T1 transgenic plants were subjected to repeated heat stress treatments as previously described with slight modifications32 (with the exception of pRPS5A:Cas9 OLE:CITRINE). The plants that were subjected to heat stress were treated as follows: After resistance selection and 4 days of acclimation to the soil, the seedlings were transferred to growth chambers at 32 °C for 24 h, followed by a 48 h recovery at 22 °C (3-day period). This heat stress cycle was performed four times during the vegetative phase of growth. The plants were then grown at 22 °C from that point on.
CRISPR/CAS9 and amiRNA cloning
The 20 nt protospacer (CTCTACTTTCTCCCTCATCT) was picked to target PUP7 (AT4G18197), PUP8 (AT4G18195) and PUP21 (AT4G18205) at once. The oligos (FW: attgCTCTACTTTCTCCCTCATCT; REV: aaacAGATGAGGGAGAAAGTAGAG) were annealed and cloned into the pRPS5A:zCAS9i (Addgene: AGM55261) using the Golden Gate cloning method. In brief, the oligos were incubated at 95°C for 5 mins and cooled at RT for 20 mins. The annealed oligos and the pRPS5A:zCAS9i were added in the following reaction (20 μl): 3μl of annealed oligos; ~150 ng of CAS9 vector; 1 μl T4 ligase (400,000 units/ml, NEB); 1 μl BsaI-HF v2 (20,000 units/ml, NEB); Cutsmart buffer (NEB) and T4 ligase buffer (NEB). Golden Gate assembly was performed as follows: 35 cycles of 37 °C for 5 min and 16 °C for 5 min; 50 °C for 20 min; and 80 °C for 20 min. 1/10 of the reaction was transformed into E.coli DH5α.
To generate the 35S:amiRNA-PUP7/8/21 vector, the amiRNA319 backbone sequence with miR targeting PUP7, PUP8 and PUP21 (MiR-sense: TATCATGGAAAACTGTCACTG) was synthesized by Syntezza Bioscience Ltd. and cloned into the pH2GW7 destination vector using the Gateway system.
Genotyping
To identify the sgRNA of transgenic plants, genomic DNA from young leaf tissue was extracted by grinding 1-2 leaves into 400 μl Extraction Buffer (200 mM Tris-HCl, pH 8.0, 250 mM NaCl, 25 mM EDTA, and 0.5% SDS). After 1-min centrifugation at 14,500 g, 300 μl supernatant was transferred to a new Eppendorf tube and mixed with 300 μl isopropanol, followed by centrifugation for 10 min at maximum speed. The supernatant was removed and the DNA pellets were washed with 70% ethanol and then resuspended in 50 μl of water. The PCR amplified using the primers listed in Supplementary Table 4 and 5 was identified using Sanger sequencing.
T-DNA lines for the single mutants, listed in Supplementary Table 6, were ordered from Gabi Kat (https://www.gabi-kat.de) and The Arabidopsis Information Resource (https://www.arabidopsis.org/). Primers for the T-DNA genotyping were designed using the T-DNA Primer Design Tool powered by Genome Express Browser Server (http://signal.salk.edu/tdnaprimers.2.html). Homozygous mutants were selected by PCR performed with primers listed in Supplementary Table 6.
Phenomics
Morphological and photosynthesis parameters were analyzed with the PlantScreenTM Phenotyping System, Photon Systems Instruments (PSI), Czech Republic. Plants were sowed in PSI standard pots and imaged at day 25.
35S:YFP-PUPs cloning
PUP7 genomic DNA, PUP8-CDS and PUP21-CDS were amplified with Phusion High-fidelity Polymerase (NEB) using the primers list in Supplementary Table 7. PUP7 genomic sequence with intron, PUP8, and PUP21 coding regions was cloned into pENTER/D-TOPO (Invitrogen K2400), verified by sequencing, and subsequently cloned into the binary destination vector (pH7WGY2) using LR Gateway reaction (Invitrogen 11791). p35S:YFP-PUP7, p35S:YFP-PUP8, and p35S:YFP-PUP21 were generated using the pH7WGY2 vector and were selected using spectinomycin in Escherichia coli and hygromycin in plants.
Microscopy imaging
Seedlings were stained in 10 mg L–1 propidium iodide (PI) for 5 min and rinsed in water for 30 s. Confocal microscopy was performed using a Zeiss LSM780 inverted confocal microscope equipped with a 20×/0.8 M27 objective lens. YFP and CFP were excited using an argon-ion laser, whereas PI was excited using a diode laser. Emissions were detected sequentially with ZEN 3.6 (blue edition) to prevent crosstalk between fluorophores. YFP was excited at 514 nm, CFP at 458 nm, and PI at 561 nm. Fluorescence emission was measured at 517-561 nm (YFP), 523-552 (CITRINE), 463-517 nm (CFP), and 588-718 nm (PI).
Expression and cytokinin transport assays in Xenopus oocytes
Coding DNA sequences of PUP7, PUP8 and PUP21 were cloned into Xenopus laevis oocyte expression vector pNB1u using USER cloning technique as described previously74. DNA template for in vitro transcription was generated by PCR using Phusion High-Fidelity DNA Polymerase (NEB) using forward primer (5'-AATTAACCCTCACTAAAGGGTTGTAATACGACTCACTATAGGG-3’) and reverse primer (5'-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTATACTCAAGCTAGCCTCGAG-3’). The PCR products were purified using the E.Z.N.A. Cycle Pure Kit (Omega Bio-tek). Capped complementary RNA (cRNA) was in vitro transcribed using the mMessage mMachine T7 Kit (Invitrogen AM1344) following the manufacturer’s instructions.
X. laevis oocytes (stage V or VI) were purchased from Ecocyte Bioscience (Germany). Oocytes were injected with 15 ng of PUP7, PUP8, or PUP21 cRNA (or 50.6 nl nuclease-free water as mock control) using a Drummond NANOJECT II (Drummond Scientific Company, Broomall Pennsylvania). The injected oocytes were incubated for 3 days at 16°C in a kulori buffer (90 mM NaCl, 1 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 5 mM Hepes pH 7.4) supplemented with gentamycin (100 μg/ml).
Three days after cRNA injection, the injection-based export assay was performed. Oocytes were injected with 23nl of 2mM trans-zeatin (tZ) and then oocytes were incubated in a group of 3 oocytes in 150 ul kulori buffer (90 mM NaCl, 1 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM MES pH 5.5) for 150 min. After 150 min, the oocytes and their respective external medium were harvested separately for intracellular and extracellular tZ quantification, respectively. The samples were homogenized with 50% methanol and then stored at -20°C overnight. Subsequently, the extracts were spun down at 15,000 𝑔 for 15 min at 4°C and the supernatant was diluted with water, filtered through a 0.22 μm polyvinylidene difluoride–based filter plate (MSGVN2250, Merck Millipore), and analyzed by analytical liquid chromatography coupled to mass spectrometry (LC-MS/MS). Export of tZ was calculated as follows: ((amount of tZ in the medium at time t=150 min)/(amount of tZ in the oocyte at time t=150 min + amount of tZ in the medium at time t=150 mm))*100%.
Uptake assay was performed as described previously75, with some modifications. A mixture of three types of cytokinin (trans-zeatin (tZ), trans-zeatin riboside (tZR), and isopentenyl adenosine (iPR)) with a final concentration of each type at 10 or 100 μM were used. Three days after cRNA injection, oocytes were preincubated in kulori buffer (pH 5.5 or pH 8.5) for five minutes, and then the oocytes were incubated in 10 or 100 μM of cytokinin mix (tZ, tZR and iPR) containing kulori buffer (pH 5.5 or pH 8.5) for 60 min. Oocytes were then washed four times in kulori buffer (pH 5.5 or pH 8.5) and homogenized with 50 % methanol. Subsequent extraction and oocyte sample preparation was performed as described above.
Cytokinin quantification by LC-MS/MS
Samples were 100-fold diluted with deionized water and subjected to analysis by liquid chromatography coupled to mass spectrometry. Chromatography was performed on an Advance UHPLC system (Bruker, Bremen, Germany). Separation was achieved on a Kinetex 1.7u XB-C18 column (100 x 2.1 mm, 1.7 μm, 100 Å, Phenomenex, Torrance, CA, USA). Formic acid (0.05%) in water and acetonitrile (supplied with 0.05% formic acid) were employed as mobile phases A and B, respectively. The elution profile was: 0-0.1 min, 5% B; 0.1-1.0 min, 5-45 % B; 1.0-3.0 min 45-100 % B, 3.0-3.5 min 100 % B, 3.5-3.55 min, 100-5 % B and 3.55-4.7 min 5 % B. The mobile phase flow rate was 400 μl min-1. The column temperature was maintained at 40°C. The liquid chromatography was coupled to an EVOQ Elite TripleQuadrupole mass spectrometer (Bruker, Bremen, Germany) equipped with an electrospray ion source (ESI). The instrument parameters were optimized by infusion experiments with pure standards. The ion spray voltage was maintained at +5000 V in positive mode. Cone temperature was set to 350 °C and cone gas to 20 psi. The heated probe temperature was set to 250 °C and probe gas flow to 50 psi. Nebulizing gas was set to 60 psi and collision gas to 1.6 mTorr. Nitrogen was used as a probe and nebulizing gas and argon as collision gas. The active exhaust was constantly on. Multiple reaction monitoring (MRM) was used to monitor analyte precursor ion → fragment ion transitions. MRM transitions were adapted from literature76. Detailed values for mass transitions and references are listed in Supplementary Table 8. Both Q1 and Q3 quadrupoles were maintained at unit resolution. Bruker MS Workstation software (Version 8.2.1, Bruker, Bremen, Germany) was used for data acquisition and processing. Linearity in ionization efficiencies were verified by analyzing dilution series.
Tobacco cytokinin transport assays
For protoplast export experiments, 35S:YFP:PUPs were transiently expressed in N. benthamiana leaf tissues by Agrobacterium tumefaciens-mediated transfection, protoplasts were prepared and [14C]tZ and [3H]BA export was quantified simultaneously as described previously77.
For microsomal uptake experiments, 35S:YFP:PUP8 was expressed in N. benthamiana leaf tissue by Agrobacterium tumefaciens-mediated transfection, and microsomes were prepared and assayed as described previously37. In short, 14C-labelled tZ was added to 300 μg of 25 microsomes in the presence of 5 mM ATP to yield a final concentration of 1 μM [14C]tZ. For substrate competition assays, unlabelled substrate was included in the transport buffer at a 100-fold excess. After 10 s of incubation at 20°C, aliquots of 100 μl were vacuum-filtered and objected to scintillation counting. Means and standard error of means of at least four independent experiments with four technical replicates each are represented.
Measurements of TCS:Venus responses
Leaf or floral primordia were removed with the help of a fine forceps (Swiss No.5) under a Nikon SMZ 745 stereoscope. Dissected shoot apices were inserted into Apex Culture Medium (1/2 x MS medium (Duchefa), 1% sucrose, 1% agarose, 2 mM MES (Sigma), 1x vitamin solution (myo-Inositol 100 mg/L, nicotinic acid 1 mg/L, pyridoxine hydrochloride 1 mg/L, thiamine hydrochloride 10 mg/L, glycine 2 mg/L), 250 nM N6-Benzyladenine)). Confocal images were obtained with a Zeiss LSM 700 microscope equipped with a water-dipping lens (W Plan-Apochromat 40x/1.0 DIC). Venus and auto-fluorescence (chlorophyll A) were excited by both 488 nm laser with emission range at 300-550 nm and 664-800 nm, respectively. Images were processed and analyzed using ImageJ (1.53e) software.
Phylogenetic tree
A phylogenetic tree of Arabidopsis PUP family members, based on protein sequences, was constructed using Phylogeny.fr (http://www.phylogeny.fr/)78 with “one-click” mode. The previously unreported PUP9 protein (AT4G18220), a close paralog of PUP10, was identified and added to the phylogenetic analysis (Fig. 5a).
Measurements of rosette area and silique divergence angles
Rosette area of individual plants was measured by ImageJ (1.53e). Angles separating successive siliques on the main inflorescence stem were quantified using a protractor as previously described79. The divergence angle was measured between the insertion points of two successive floral pedicels. Phyllotaxy orientation can be either clockwise or anticlockwise.
Statistical analysis and reproducibility
Two-tailed Student’s t-test and one-way analysis of variance (ANOVA) with Tukey test were performed to determine the statistical significance of differences between samples. GraphPad Prism 8.0 (www.graphpad.com) software was used for statistical analysis and graphing. All experiments were independently reproduced two or three times.
Supplementary Material
Acknowledgments
We thank Bruno Müller (University of Zurich, Switzerland) for sharing TCS:VENUS seeds.
Funding
This work was supported by grants from the Israel Science Foundation (2378/19 and 3419/20 to E.S.), the Human Frontier Science Program (HFSP—RGY0075/2015 and HFSP—LIY000540/2020 to E.S., H.H.N.-E. and Z.M.B.), Danmarks Grundforskningsfond (DNRF99 to H.H.N.-E.), the European Research Council (757683-RobustHormoneTrans to E.S.), the PBC postdoctoral fellowship (to Y.H.), and by the Swiss National Funds (31003A-165877/1 to M.G.).
Footnotes
Author contributions
Y.H. and E.S. conceived and designed the study and wrote the manuscript. Y.H. performed the research. P.P. assisted in cloning the Multi-Knock libraries and with PUP genetics. O.P. cloned and characterized the PUP amiRNA and PUP reporter lines. A.S., G.H. and O.C. carried out the Multi-Knock sgRNA library bioinformatics design and analysis. Z.M.B. performed transport assays in the Xenopus oocytes. J.B. carried out the Multi-Knock sgRNA library deep-sequencing analysis. S.B. cloned the OLE:CITRINE Cas9 vector. B.S. carried out the TCS:VENUS assays. L.C. performed the tobacco transport assays. C.C. performed the LC-MS analysis. Y.Z. assisted in the PUP genes discovery. M.T. assisted in the library screen. D.W assisted in the Multi-Knock sgRNA library design. A.A. provided the UBI:Cas9 vector. H.H.N.-E., M.G., T.V., and I. M. designed and supervised the work and edited the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests: A US Provisional Patent Application (No. 63/329,506) on the Multi-Knock system described in this study has been filed. The authors declare that they have no competing interests.
Data and materials availability
All the data supporting the findings of this study are available within the article and the Supplementary Materials.
Code availability
All source codes used to generate the library are available in GitHUb (https://github.com/anatshafir1/sgRNA_filtering_procedure)
References
- 1.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 2.Hauser F, et al. A genomic-scale artificial MicroRNA library as a tool to investigate the functionally redundant gene space in arabidopsis. Plant Cell. 2013;25:2848–2863. doi: 10.1105/tpc.113.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry IM, et al. Efficient genome-wide detection and cataloging of EMS-induced mutations using Exome capture and next-generation sequencing. Plant Cell. 2014;26:1382–1397. doi: 10.1105/tpc.113.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tal I, et al. The Arabidopsis NPF3 protein is a GA transporter. Nat Commun. 2016;7:11486. doi: 10.1038/ncomms11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang X, et al. A Single Transcript CRISPR-Cas9 System for Efficient Genome Editing in Plants. Mol Plant. 2016;9:1088–1091. doi: 10.1016/j.molp.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, et al. Construction of a genomewide RNAi mutant library in rice. Plant Biotechnol J. 2013;11:997–1005. doi: 10.1111/pbi.12093. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, et al. A transportome-scale amiRNA-based screen identifies redundant roles of Arabidopsis ABCB6 and ABCB20 in auxin transport. Nat Commun. 2018;9:4204. doi: 10.1038/s41467-018-06410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panchy N, Lehti-Shiu M, Shiu SH. Evolution of gene duplication in plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, et al. Gene duplicability of core genes is highly consistent across all angiosperms. Plant Cell. 2015;28:326–344. doi: 10.1105/tpc.15.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rensing SA. Gene duplication as a driver of plant morphogenetic evolution. Curr Opin Plant Biol. 2014;17:43–48. doi: 10.1016/j.pbi.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Lu X, et al. Gene-Indexed Mutations in Maize. Mol Plant. 2018;11:496–504. doi: 10.1016/j.molp.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Gaillochet C, Develtere W, Jacobs TB. CRISPR screens in plants: Approaches. guidelines. and future prospects. Plant Cell. 2021;33:794–813. doi: 10.1093/plcell/koab099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu Rev Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- 14.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs TB, Zhang N, Patel D, Martin GB. Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol. 2017;174:2023–2037. doi: 10.1104/pp.17.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K, et al. A FLASH pipeline for arrayed CRISPR library construction and the gene function discovery of rice receptor-like kinases. Mol Plant. 2021;15:243–257. doi: 10.1016/j.molp.2021.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Liu HJ, et al. High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell. 2020;32:1397–1413. doi: 10.1105/tpc.19.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y, et al. Genome-wide Targeted Mutagenesis in Rice Using the CRISPR/Cas9 System. Mol Plant. 2017;10:1242–1245. doi: 10.1016/j.molp.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Meng X, et al. Construction of a Genome-Wide Mutant Library in Rice Using CRISPR/Cas9. Mol Plant. 2017;10:1238–1241. doi: 10.1016/j.molp.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Bai M, et al. Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol J. 2020;18:721–731. doi: 10.1111/pbi.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramadan M, et al. Efficient CRISPR/Cas9 mediated Pooled-sgRNAs assembly accelerates targeting multiple genes related to male sterility in cotton. Plant Methods. 2021;17:1–13. doi: 10.1186/s13007-021-00712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenzo CD, et al. BREEDIT: a multiplex genome editing strategy to improve complex quantitative traits in maize. Plant Cell. 2022 doi: 10.1093/plcell/koac243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grützner R, et al. High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Commun. 2021;2:1–15. doi: 10.1016/j.xplc.2020.100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proost S, et al. PLAZA 3.0: An access point for plant comparative genomics. Nucleic Acids Res. 2015;43:D974–D981. doi: 10.1093/nar/gku986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyams G, et al. CRISPys: Optimal sgRNA Design for Editing Multiple Members of a Gene Family Using the CRISPR System. J Mol Biol. 2018;430:2184–2195. doi: 10.1016/j.jmb.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 26.Joung J, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang J, et al. Plant ABC Transporters. Arab B. 2011;9:e0153. doi: 10.1199/tab.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsutsui H, Higashiyama T. PKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol. 2017;58:46–56. doi: 10.1093/pcp/pcw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sussholz O, Pizarro L, Schuster S, Avni A. SlRLK-like is a malectin-like domain protein affecting localization and abundance of LeEIX2 receptor resulting in suppression of EIX-induced immune responses. Plant J. 2020;104:1369–1381. doi: 10.1111/tpj.15006. [DOI] [PubMed] [Google Scholar]
- 30.Fauser F, Schiml S, Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014;79:348–359. doi: 10.1111/tpj.12554. [DOI] [PubMed] [Google Scholar]
- 31.Wang ZP, et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:1–12. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeBlanc C, et al. Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 2018;93:377–386. doi: 10.1111/tpj.13782. [DOI] [PubMed] [Google Scholar]
- 33.Kubis S, et al. Functional specialization amongst the arabidopsis Toc159 Family of chloroplast protein import receptors. Plant Cell. 2004;16:2059–2077. doi: 10.1105/tpc.104.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niittylä T, et al. A Previously Unknown Maltose Transporter Essential for Starch Degradation in Leaves. Science. 2004;303:87–89. doi: 10.1126/science.1091811. [DOI] [PubMed] [Google Scholar]
- 35.Takano J, et al. Arabidopsis boron transporter for xylem loading. Nature. 2002;420:337–340. doi: 10.1038/nature01139. [DOI] [PubMed] [Google Scholar]
- 36.Kang J, Lee Y, Sakakibara H, Martinoia E. Cytokinin Transporters: GO and STOP in Signaling. Trends Plant Sci. 2017;22:455–461. doi: 10.1016/j.tplants.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Zürcher E, Liu J, Di Donato M, Geisler M, Müller B. Plant development regulated by cytokinin sinks. Science. 2016;353:1027–1030. doi: 10.1126/science.aaf7254. [DOI] [PubMed] [Google Scholar]
- 38.Bürkle L, et al. Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 2003;34:13–26. doi: 10.1046/j.1365-313x.2003.01700.x. [DOI] [PubMed] [Google Scholar]
- 39.Gillissen B, et al. A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell. 2000;12:291–300. doi: 10.1105/tpc.12.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao Y, et al. Endoplasmic Reticulum-Localized PURINE PERMEASE1 Regulates Plant Height and Grain Weight by Modulating Cytokinin Distribution in Rice. Front Plant Sci. 2020;11:1–12. doi: 10.3389/fpls.2020.618560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao Y, et al. Big Grain3. encoding a purine permease. regulates grain size via modulating cytokinin transport in rice. J Integr Plant Biol. 2019;61:581–597. doi: 10.1111/jipb.12727. [DOI] [PubMed] [Google Scholar]
- 42.Perilli S, Moubayidin L, Sabatini S. The molecular basis of cytokinin function. Curr Opin Plant Biol. 2010;13:21–26. doi: 10.1016/j.pbi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 43.Wybouw B, De Rybel B. Cytokinin-A Developing Story. Trends Plant Sci. 2019;24:177–185. doi: 10.1016/j.tplants.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012;17:172–179. doi: 10.1016/j.tplants.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 45.Sakakibara H. Cytokinins: Activity. biosynthesis. and translocation. Annu Rev Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- 46.Besnard F, et al. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature. 2014;505:417–421. doi: 10.1038/nature12791. [DOI] [PubMed] [Google Scholar]
- 47.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 48.Bruno M, Jen S. Cytokinin and auxin interplay in root stem-cell specification during early embryogenesis. Nature. 2008;4:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Malley RC, Ecker JR. Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 2010;61:928–940. doi: 10.1111/j.1365-313X.2010.04119.x. [DOI] [PubMed] [Google Scholar]
- 50.Park RJ, et al. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat Genet. 2017;49:193–203. doi: 10.1038/ng.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ursache R, Fujita S, Dénervaud V, Geldner N. Combined fluorescent seed selection and multiplex CRISPR/Cas9 assembly for fast generation of multiple Arabidopsis mutants Robertas. Plant methods. 2021;17:111. doi: 10.1186/s13007-021-00811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caesar K, et al. Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. J Exp Bot. 2011;62:5571–5580. doi: 10.1093/jxb/err238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wulfetange K, et al. The cytokinin receptors of arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 2011;156:1808–1818. doi: 10.1104/pp.111.180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lomin SN, Yonekura-Sakakibara K, Romanov GA, Sakakibara H. Ligand-binding properties and subcellular localization of maize cytokinin receptors. J Exp Bot. 2011;62:5149–5159. doi: 10.1093/jxb/err220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding W, et al. Isolation. characterization and transcriptome analysis of a cytokinin receptor mutant osckt1 in rice. Front Plant Sci. 2017;8:1–13. doi: 10.3389/fpls.2017.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Antoniadi I, et al. Cell-surface receptors enable perception of extracellular cytokinins. Nat Commun. 2020;11:4284. doi: 10.1038/s41467-020-17700-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubiasová K, et al. Cytokinin fluoroprobe reveals multiple sites of cytokinin perception at plasma membrane and endoplasmic reticulum. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-17949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carpaneto A, et al. Phloem-localized. proton-coupled sucrose carrier ZmSUT1 mediates sucrose efflux under the control of the sucrose gradient and the proton motive force. J Biol Chem. 2005;280:21437–21443. doi: 10.1074/jbc.M501785200. [DOI] [PubMed] [Google Scholar]
- 60.Léran S, et al. Arabidopsis NRT1.1 is a bidirectional transporter involved in root-to-shoot Nitrate translocation. Mol Plant. 2013;6:1984–1987. doi: 10.1093/mp/sst068. [DOI] [PubMed] [Google Scholar]
- 61.Mussa Belew Z, et al. Identification and characterization of phlorizin transporter from Arabidopsis 1 thaliana and its application for phlorizin production in Saccharomyces cerevisiae. bioRxiv. 2020 [Google Scholar]
- 62.Chen LQ, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J, et al. ABCB-mediated shootward auxin transport feeds into the root clock. 2023:1–16. doi: 10.15252/embr.202256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, et al. ABA homeostasis and long-distance translocation are redundantly regulated by ABCG ABA importers. Sci Adv. 2021;7:1–18. doi: 10.1126/sciadv.abf6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Develtere W, et al. SMAP design: A multiplex PCR amplicon and gRNA design tool to screen for natural and CRISPR-induced genetic variation. bioRxiv. 2022 doi: 10.1093/nar/gkad036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ellison EE, et al. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat Plants. 2020;6:620–624. doi: 10.1038/s41477-020-0670-y. [DOI] [PubMed] [Google Scholar]
- 67.Wang M, et al. Gene Targeting by Homology-Directed Repair in Rice Using a GeminivirusBased CRISPR/Cas9 System. Mol Plant. 2017;10:1007–1010. doi: 10.1016/j.molp.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 68.Martin-Ortigosa S, et al. Mesoporous silica nanoparticle-mediated intracellular cre protein delivery for maize genome editing via loxP site excision. Plant Physiol. 2014;164:537–547. doi: 10.1104/pp.113.233650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitter N, et al. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat Plants. 2017;3 doi: 10.1038/nplants.2016.207. [DOI] [PubMed] [Google Scholar]
- 70.Clough SJ, Bent AF. Floral dip : a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. 1999;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 71.Gronau I, Moran S. Optimal implementations of UPGMA and other common clustering algorithms. Inf Process Lett. 2007;104:205–210. [Google Scholar]
- 72.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nour-Eldin HH, Hansen BG, Nørholm MHH, Jensen JK, Halkier BA. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jørgensen M, Crocoll C, Halkier B, Nour-Eldin H. Uptake Assays in Xenopus laevis Oocytes Using Liquid Chromatography-mass Spectrometry to Detect Transport Activity. Bio-Protocol. 2017;7:1–13. doi: 10.21769/BioProtoc.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ionescu IA, et al. Transcriptome and metabolite changes during hydrogen cyanamide-induced floral bud break in sweet cherry. Front Plant Sci. 2017;8:1–17. doi: 10.3389/fpls.2017.01233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jarzyniak K, et al. Early stages of legume-rhizobia symbiosis are controlled by ABCG-mediated transport of active cytokinins. Nat Plants. 2021;7 doi: 10.1038/s41477-021-00873-6. [DOI] [PubMed] [Google Scholar]
- 78.Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prasad K, et al. Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr Biol. 2011;21:1123–1128. doi: 10.1016/j.cub.2011.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting the findings of this study are available within the article and the Supplementary Materials.
All source codes used to generate the library are available in GitHUb (https://github.com/anatshafir1/sgRNA_filtering_procedure)