Abstract
Chimeric-antigen receptor (CAR) T cells have emerged as a powerful treatment option for patients with B cell malignancies but have yet to achieve success in treating acute myeloid leukemia (AML) due to a lack of safe targets. Here, we leveraged an atlas of publicly available RNA sequencing data of over 500,000 single cells from 15 AML patients and nine healthy human tissues for prediction of target antigens that are expressed on malignant cells but lacking on healthy cells, including T cells. Aided by this, high-resolution, single-cell expression approach, we computationally identify colony-stimulating factor 1 receptor (CSF1R) and cluster of differentiation 86 (CD86) as targets for CAR-T cell therapy in AML. Functional validation of these established CAR-T cells shows robust in vitro and in vivo efficacy in cell line and patient-derived AML models with minimal off-target toxicity towards relevant healthy human tissues. This provides strong rationale for further clinical development.
Keywords: Single-cell RNA-sequencing, machine learning, chimeric antigen receptor, adoptive T cell therapy, acute myeloid leukemia, colony-stimulating factor 1 receptor, CSF1R, cluster of differentiation 86, CD86
Chimeric antigen receptor (CAR) T cells are patient-derived effector cells, genetically engineered to therapeutically target a specific epitope on malignant cells1. CAR-T cells targeting the B cell lineage antigens CD19 or B cell maturation antigen (BCMA) have shown clinical efficacy in heavily pretreated patients suffering from different B cell malignancies such as B cell lymphoma, B cell acute lymphoblastic leukemia and multiple myeloma2–4. However, CAR-T cells aiming at non-B cell-associated epitopes have yet to show similar response rates5. For instance, in myeloid malignancies such as acute myeloid leukemia (AML), common target structures are often co-expressed on vital tissues, such as endothelial cells or hematopoietic stem and progenitor cells (HSPC), increasing the risk for on-target off-tumor toxicity6,7. Identifying safe target structures is thus pivotal to translate the vast potential of CAR-T cell therapy to myeloid neoplasms.
AML is the most common acute leukemia in adults and its molecular heterogeneity has complicated the successful development of novel therapeutic agents8. Despite upfront curative intent in most patients with combinatorial chemotherapy, disease relapse is frequent, occurring in over 50 % of treated patients9. Upon relapse, allogeneic hematopoietic stem cell transplantation (allo-HSCT) remains the only curative approach, but even then, long-term survival probabilities are below 20 %. Therefore, innovative treatment options represent a high unmet medical need. Currently, CAR-T cells targeting AML-associated target antigens CD33 and anti-interleukin 3 receptor subunit alpha (IL3RA, CD123) are undergoing clinical investigation. Due to preclinical evidence of off-tumor toxicity towards HSPC, most clinical trials are evaluating the potential of anti-CD123 or anti-CD33 CAR-T cells as a bridge-to-transplant regimen before allo-HSCT. Early reports of these trials have only shown limited therapeutic efficacy10–12. Yet, more complete results of these clinical studies in AML are eagerly awaited. In the meanwhile, other targets such as CD70, C-type lectin-like molecule-1 (CLL-1), fms-like tyrosine kinase-3 (FLT3), CD44 variant 6 (CD44v6), sialic acid-binding Ig-like lectin (Siglec)-6 or CD117 have been tested in preclinical studies as alternative CAR targets13–17. Yet, clinical validation is pending and expression profiles of most of the targets raise at least some uncertainties as to their clinical safety and efficacy.
Often, newly developed CAR-T cells are directed against target structures, which have already been used for antibody therapy. In contrast, unbiased de novo target screenings for CAR-T cell therapy have only rarely been conducted18. In addition, until recently, off-tumor antigen projections could only leverage bulk sequencing data - entirely missing detailed information about cell type-specific target antigen expression patterns. Conveniently, the revolution in single-cell technologies in the last decade has generated massive single-cell expression data that provide precise information about the transcriptomic anatomy of healthy and malignant cells19, a yet mostly untapped resource for therapeutic development, at least in the context of de novo antigen predictions and CAR-T cell development. These advancements allow in-depth on- and off-tumor antigen prediction20, offering unique insights into healthy and malignant cells at an unmatched resolution.
We thus developed a single-cell RNA sequencing (scRNA-seq)-based approach, specifically tailored to identify promising antigens for CAR-T cell therapy on a discovery AML cohort of 15 patients21. We generated a transcriptomic atlas from publicly available datasets, consisting of over 28,000 healthy and malignant bone marrow cells from these patients, and additionally over 500,000 healthy cells from nine of the most vital human tissues. We screened this data for cell surface antigens expressed on malignant cells with minimal co-expression on healthy cells including T cells. With rigorous cutoffs, we identified two, so far unrecognized targets for CAR-T cells in AML: colony-stimulating factor 1 receptor (CSF1R) and cluster of differentiation 86 (CD86). We developed CAR-T cells against both targets and tested their efficacy in vitro and in vivo in cell lines and patient-derived models including primary AML blasts. We assessed their safety in vitro using advanced primary cell cultures for target-expressing cell types, demonstrating better discriminatory capacity than established anti-CD33 CAR-T cells. In addition, we used several in vivo models to mitigate safety concerns. Our results illustrate the translational potential of an unbiased scRNA-seq-based screening approach and lay the basis for clinical development of our CAR candidates.
Results
Development of a scRNA-seq-based screening algorithm
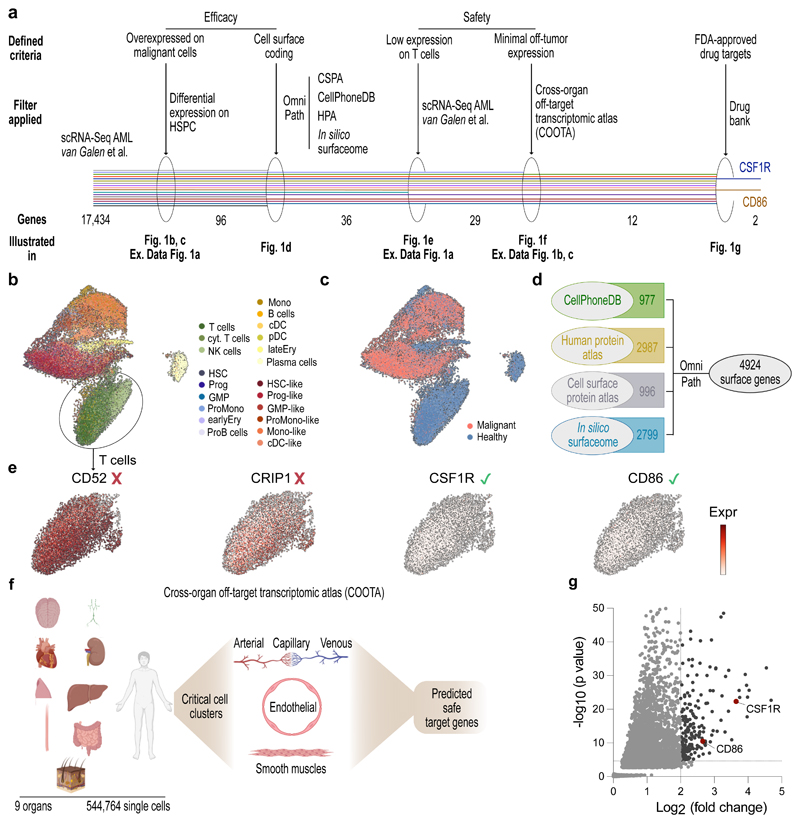
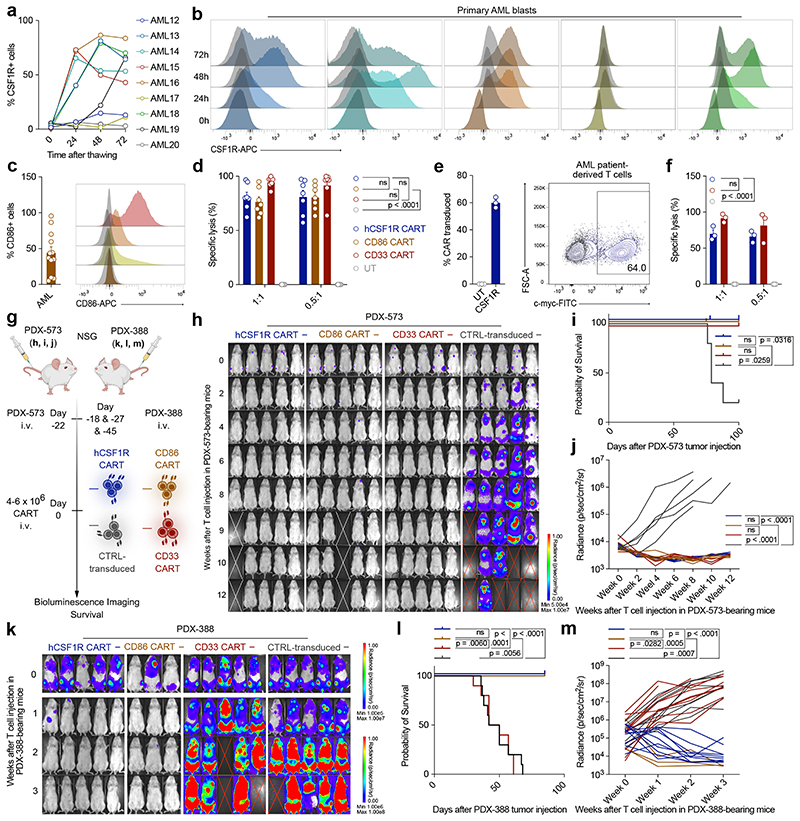
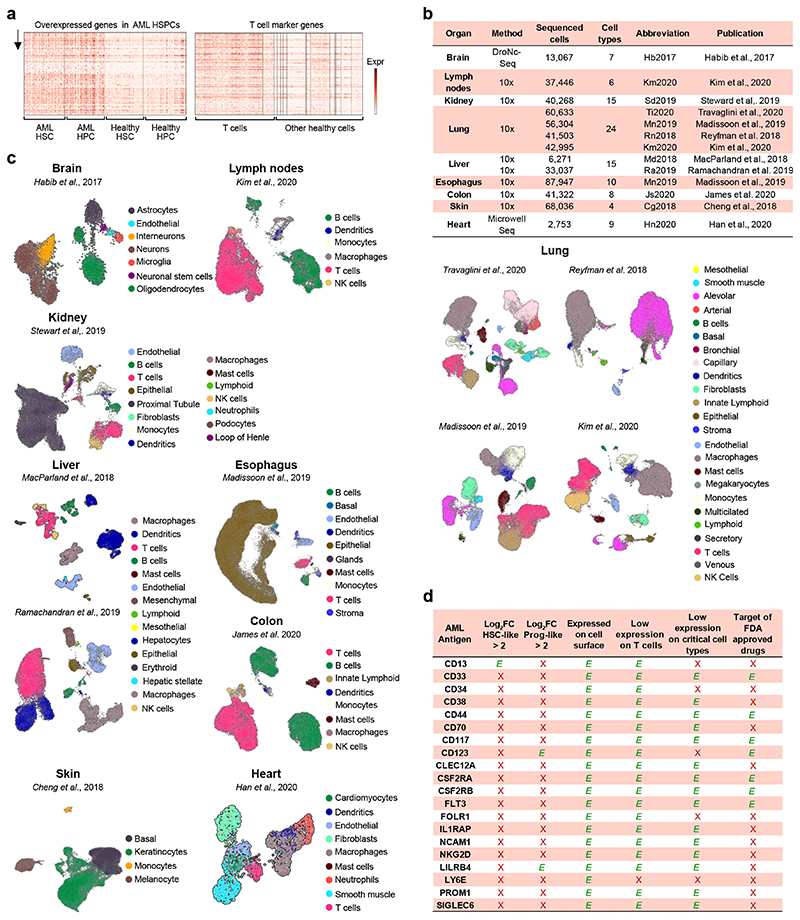
We created an unbiased scRNA-seq-based discovery approach for identification of CAR targets. To ensure CAR efficacy, a suitable candidate is 1) overexpressed in malignant cells and 2) located on the cell surface. In terms of CAR safety, the candidate should 3) not be expressed on T cells, and 4) show minimal expression across vital, healthy tissues (Fig. 1a). Applying our approach to AML, we used publicly available scRNA-seq data of 15 AML patients21. From these, a total of 28,404 sequenced healthy and malignant bone marrow cells passed quality control (Fig. 1b, c, see methods section for a detailed description of quality control steps). For maximal CAR efficacy, we sought to identify candidates with high expression on malignant HSPC-like cells (herein termed hematopoietic stem cell (HSC)-like and Progenitor (Prog)-like) compared to their healthy counterparts. Differential gene expression analyses between malignant and healthy HSPC revealed 96 genes that were strongly overexpressed in HSPC-like cells and used for further downstream analyses (Extended Data Fig. 1a).
Figure 1. A single-cell RNA Seq-based screening approach identifies CSF1R and CD86 as potential CAR targets in AML.
(a) Workflow of computational CAR target antigen identification by stepwise evaluation against a set of criteria for an ideal and effective CAR target antigen. The decreasing number of screened AML target genes are shown on the bottom. CSPA: Cell surface protein atlas; HPA: Human protein atlas. (b, c) UMAP showing 28,404 healthy and malignant cells from data of 15 previously published AML patients harboring 15 different mutations21. Normalized gene expression values were log-transformed. Cell annotations were provided by the authors. (d) Summary of databases used to identify cell surface coding genes. (e) Quantification of T cell expression of newly identified targets. Red cross: Targets with high expression on T cells, which were excluded from further analyses. Green check: No significant expression on T cells. (f) Harmonization of 11 scRNA-Seq datasets from nine healthy human tissues into a cross-organ off-target transcriptomic atlas (COOTA) consisting of 544,764 cells. A detailed summary of all used datasets is provided in Extended Data Fig. 1b. Targets highly expressed in non-immune cell lineages or on cell types in direct proximity to infused T cells (critical cell clusters: arterial, capillary, venous, endothelial and smooth muscle cells) were excluded from further analysis (g) Volcano plot showing the remaining two target antigens with their respective FDR-adjusted log10 p-value and log2 fold changes from differential expression analysis between malignant HSPC-like and healthy HSPC using a t-test with overestimated variance. Dotted lines indicate applied thresholds at log2fc=2 and p-value=0.01.
To identify candidates accessible for CAR-T cells on the target cell surface, we used OmniPath22, a large-scale molecular database, to integrate data of multiple resources23–26 into a comprehensive human surface-gene library of 4924 genes (Fig. 1d). Of the 96 genes overexpressed in HSPC-like cells, 36 were present in this library. Genes which passed all previous filters but showed high expression on T cells (e.g. CD52 and CRIP1) were excluded from further analysis (Fig. 1e).
To minimize on-target off-tumor effects, we processed and harmonized 11 scRNA-seq datasets from nine healthy human tissues (brain, lung, lymph nodes, heart, skin, liver, kidney, colon and esophagus) into a massive cross-organ off-target transcriptomic atlas (COOTA) consisting of over 500,000 single healthy cells (Fig. 1f)27–37. A detailed summary of all datasets used for COOTA is given in Extended Data Fig. 1b and 1c. Targets highly expressed in vital non-immune cell lineages or on cell types of tissues in direct proximity to infused T cells (i.e. endothelium, arteries, veins, bronchial vessels, capillary and smooth muscle cells) were excluded from further analyses (Fig. 1f).
Using this stringent and rigorous approach, 12 potential candidates for CAR development remained. Interestingly, most of the so far described CAR targets for AML (n = 20) failed the thresholds of our analyses at different levels (Extended Data Fig. 1d). For example, prototypic AML antigens CD33 and CD123 did not fulfill our strict criteria of overexpression on malignant HSPC (see methods for applied thresholds), most likely due to expression of both antigens on healthy HSPC. In addition, CD123 had high expression levels across endothelial and various lung cell types (see Fig. 2d for detailed analysis).
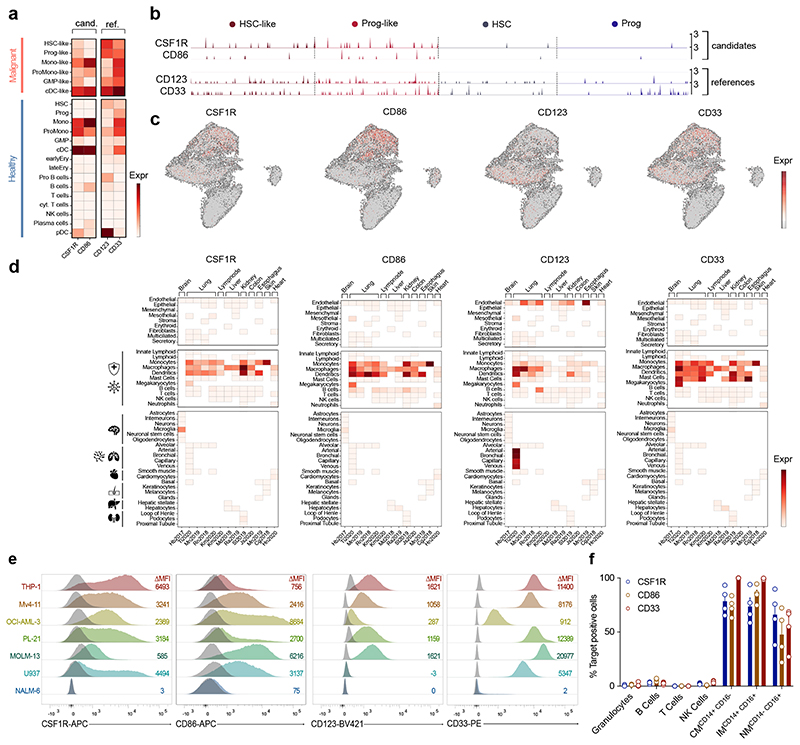
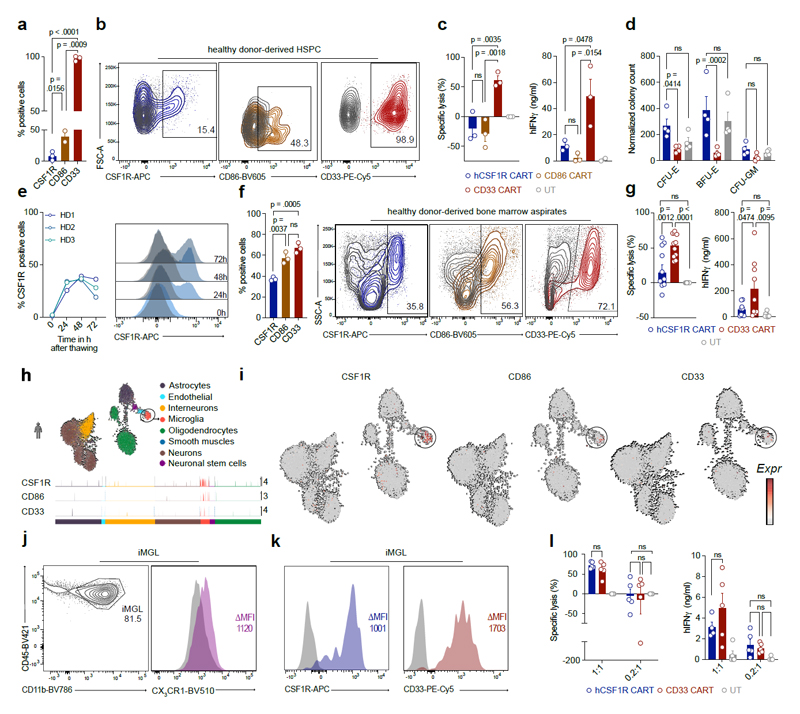
Figure 2. CSF1R and CD86 are preferentially expressed on malignant HSPC-like compared to healthy HSPC and off-tumor expression is restricted to infiltrating or tissue-resident immune cells.
(a) Expression of target and reference genes (CD123, CD33) on single healthy and malignant cell types. Normalized expression values were log-transformed and scaled to unit variance. (b) Expression of CSF1R and CD86 target genes in malignant (HSC-like, Prog-like; left) and healthy (HSC, Prog; right) stem cells. For visualization purposes, normalized expression values of healthy HSPC and a random subsample of malignant HSPC were log-transformed and scaled to unit variance. Each peak corresponds to a cell, peak height indicates expression intensity. (c) Expression of CSF1R and CD86 target genes in healthy and malignant cells of 15 AML patients. Normalized gene expression values were log-transformed and visualized in a UMAP embedding. (d) Single-cell cross-organ off-target transcriptomic atlas screening for target (CSF1R, CD86) and reference (CD123, CD33) genes. Single-cell transcriptomic atlas consists of a total of 544,764 sequenced cells comprising 9 different organs. Each field represents the mean expression value per cluster. Blank fields indicate cell types not present in a study. (e) Representative flow cytometric images of target gene expression on a panel of six different AML cell lines or NALM-6 control cells. Staining for target antigens was carried out at least twice. (f) Expression of target antigens on human immune cell population quantified by flow cytometry. Data are mean ± s.e.m. of four different donors. CM, classical monocytes; IM, intermediate monocytes; NM, non-classical monocytes.
To further optimize the safety profile of newly developed CAR-T cells, we reasoned that, if targeted therapies for any of the 12 identified candidates have already been approved by the FDA, the risk for unexpected, severe on-target off-tumor toxicities of newly developed CAR-T cells will be minimized. In addition, this could shorten the time span and decrease regulatory hurdles for translation of newly developed CAR-T cells into clinical routine, as safety of target-directed therapies was previously demonstrated. Thus, we used an accessible database of all monitored FDA-approved drugs containing information on the interaction, pharmacology and chemical structures of drugs and drug targets38. Indeed, we identified two targets, CD86 and CSF1R, which have already undergone clinical investigation (Fig. 1g). To the best of our knowledge, neither anti-CD86- nor anti-CSF1R CAR-T cells have been implicated for CAR-T cell therapy in AML. We thus decided to further investigate their potential.
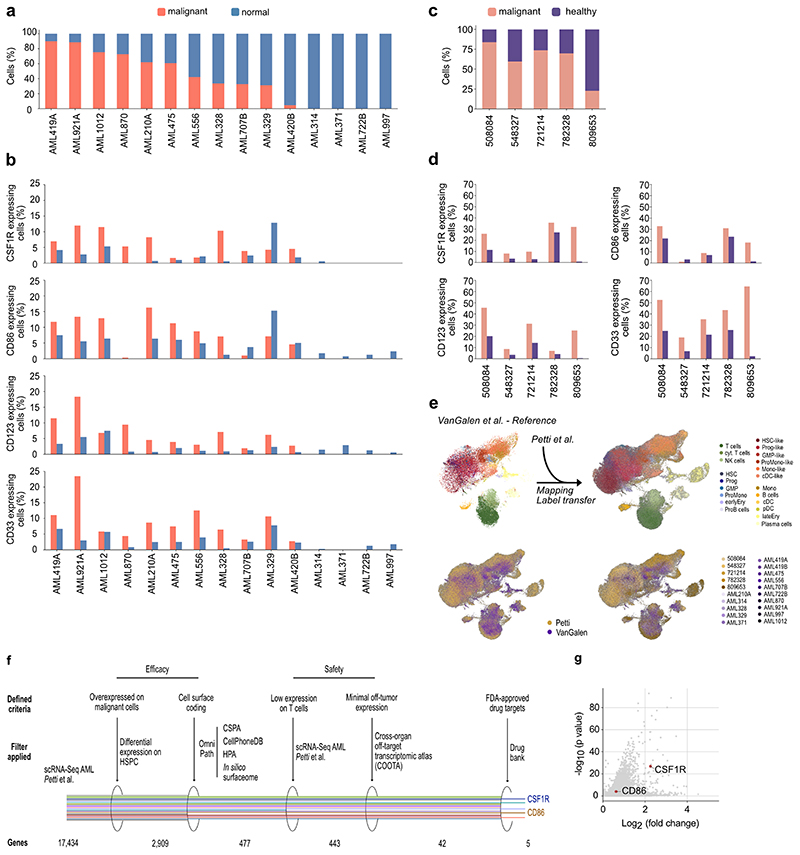
Both antigens were highly expressed across malignant cells in 100 % of the AML patients with captured malignant blasts (11 out of 15, Extended Data Fig. 2a, b), despite the heterogeneous molecular profile of the patient collective (see van Galen et al.21 for patient characteristics).
To ensure the validity of our analyses and to better reflect the cytogenetic diversity of AML as a disease, we next set out to further increase the size of our patient cohort. Thus, we obtained a second publicly available scRNA-seq dataset of five additional AML patients39 (Extended Data Fig. 2c). For the cross validation of our computational target identification approach, we used scANVI, a semi-supervised variational auto-encoder40 to map the data from Petti et al. (query) onto a newly generated reference map of van Galen et al. (Extended Data Fig. 2e). In line with the results above, CSF1R and CD86 were preferentially expressed on malignant cells compared to healthy hematopoietic cells (Extended Data Fig. 2d). Next, upon extending our target identification approach to these five additional AML patients (Fig. 1a), both CSF1R and CD86 were again identified as suitable target antigens for CAR therapy in this second AML cohort (Extended Data Fig. 2f, g). In summary, using two independent single-cell AML cohorts consisting of a total of 20 patients, we identified CSF1R and CD86 as potential CAR targets for AML therapy.
On- and off-tumor expression analysis of CSF1R and CD86
Next, we benchmarked the two target antigens CSF1R and CD86 to the reference genes CD123 and CD33 to ease interpretation of receptor expression on a transcriptomic level (Fig. 2a - c). CSF1R was expressed on all six malignant cell clusters, but was the highest on monocytic (Mono)-like or conventional dendritic cell (cDC)-like clusters. CD86 was most strongly expressed on Mono-like, pro-monocytic (ProMono)-like and cDC-like clusters (Fig. 2a). In terms of expression in malignant HSPC clusters, CSF1R expression was higher than CD86, albeit lower than CD123 and CD33 reference genes (Fig. 2a, b). In contrast, CD123 or CD33 were detected in healthy HSC and progenitors, while both CSF1R and CD86 were only minimally expressed among these cells (Fig. 2b). Visualized in an UMAP embedding, the expression profiles of CSF1R and CD86 were very comparable to CD123 and CD33 reference genes (Fig. 2c).
COOTA analysis revealed target antigen expression mainly in immune cells of myeloid origin (monocytes, macrophages and dendritic cells), similar to the peripheral expression profile of CD33 (Fig. 2d). Both, CSF1R and CD86 were not highly expressed on epithelial or stromal cells (Fig. 2d, top panel). In organ specific cell clusters (Fig. 2d, bottom panel), expression was restricted to microglia cells in the brain as described in literature41. We next sought to assess expression of the target antigens on a protein level. We carried out primary screening using a panel of six different human AML cell lines (THP-1, Mv4-11, OCI-AML-3, PL-21, MOLM-13, U937) and B cell malignant NALM-6 cells as negative staining control (Fig. 2e). CSF1R and CD86 were detected on all screened AML cell lines. CD123 and CD33 expression was measured as reference (Fig. 2e). Given the similar expression profile on mature, healthy immune cells of our targets to CD33, we decided to use CD33 as the main control for all subsequent experiments.
To validate the transcriptomic profiles predicted by COOTA, we assessed receptor expression on peripheral blood immune cells from healthy human donors of each candidate antigen using multicolor flow cytometry (Fig. 2f). In accordance with our transcriptomic prediction, expression of CSF1R and CD86 was mainly restricted to monocytic cell populations with no expression on granulocytes or T cells (Fig. 2f).
mCSF1R CAR-T cells do not cause toxicity in mice
Despite the stringent thresholds set by our approach and our in-depth off-tumor antigen projection, expression patterns of CSF1R and CD86 were still broader than those of candidates in clinical use (CD19 and BCMA), which are almost entirely confined to B cells or B cell subsets11. Therefore, we first tested the safety of the developed anti-target CAR-T cells in fully immunocompetent syngeneic mice models. To ensure similar target expression in mouse and human, we compared expression of candidates in different organs using available bulk sequencing data (Fig. 3a). CSF1R showed higher expression in organs of both mouse and human, while CD86 was only detected in the spleen. Also, in line with our COOTA prediction, CSF1R is known to be expressed on microglia42, raising additional safety concerns. ScRNA-seq analysis of archived mouse brain tissue27 confirmed expression of CSF1R on microglia and similar expression patterns on tissue resident myeloid cells (Fig. 3b).
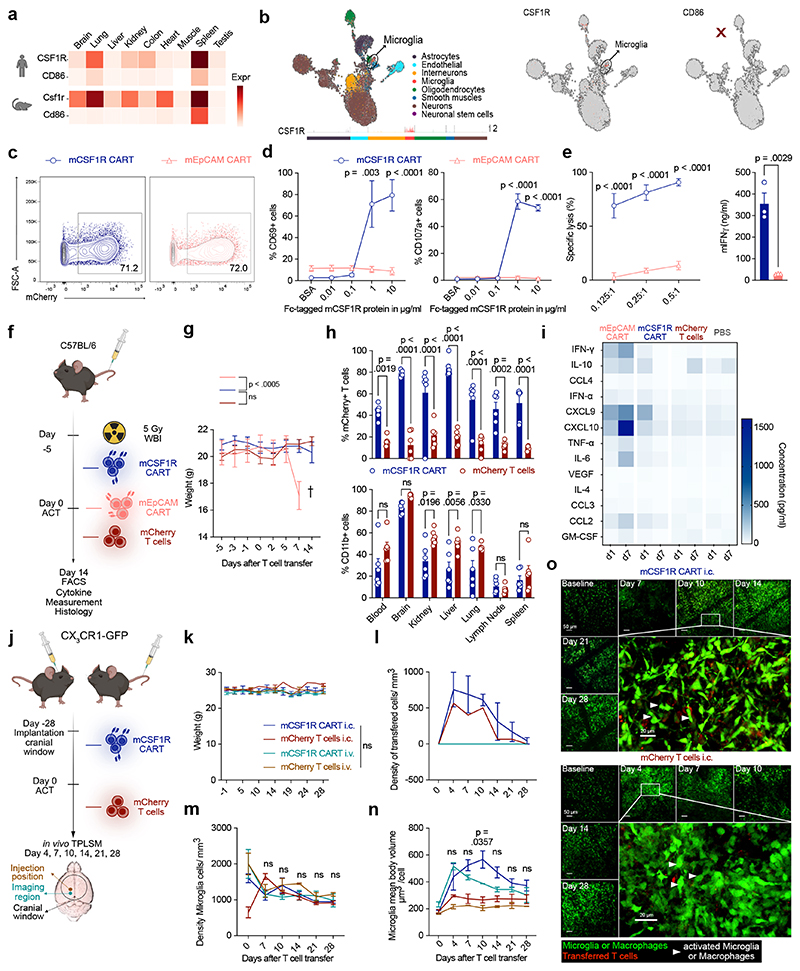
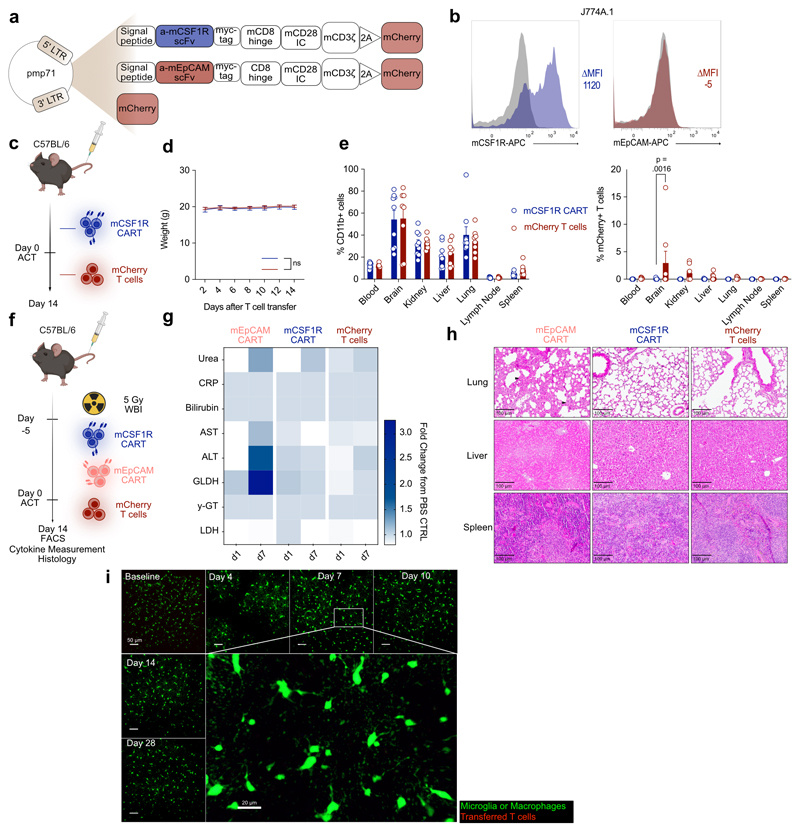
Figure 3. mCSF1R CART do not cause toxicity in mice.
(a) Target expression (transcripts per million) across organs in human (top) or mouse (bottom) quantified using bulk RNAseq. (b) Target expression in single mouse brain cells. UMAP embedding of sequenced brain cells (left). Each peak corresponds to a cell, peak height indicates expression intensity. Normalized, log-transformed antigen expression per cell type (right). (c) Construct expression on transduced primary murine T cells. (d) Activation of mCSF1R or mEpCAM CART after incubation with plate-bound mCSF1R measured by flow cytometry. (e) mCSF1R or mEpCAM CART co-cultured with J774A.1-Luc+ for 48 hours. Cell lysis quantified by BLI (left). Secretion of IFNγ quantified by ELISA (right). (d, e) Data are mean ± s.e.m of three independent experiments. (e) Right: statistical significance calculated with unpaired t test. (f) Treatment schedule for in vivo toxicity assessment of mCSF1R CART. WBI, whole body irradiation. (g) Weight curves of mice treated with 3 x 106 mCSF1R CART (n = 9) or mCherry T cells (n = 11). 6 x 106 mEpCAM CART (n = 5) were transferred as a toxicity control. Error bars indicate s.e.m. (h) Quantification of mCherry+ T cells of parent population (left; parent population: CD3+-CD8+ cells), or CD11b+ cells (right) by flow cytometry. Data are mean ± s.e.m. of n = 6 mice. Shown statistical significance applies to d7. (i) Serum cytokine levels one (d1) or seven (d7) days after ACT. Cytokine levels measured with LEGENDplex™. n = 3 mice. Statistically significant increases in serum cytokine levels (mEpCAM versus mCSF1R CART or mCherry T cell-treated mice) at d7: IFN-γ (p = 0.0371), CXCL9 (p = 0.0096) and CXCL10 (p < 0.0001). (j)Treatment regimen to assess neurological toxicity in CX3CR1-GFP reporter mice. (k) Weight curves of mice after intracranial (i.c. – 2 x 105) or intravenous (i.v. – 3 x 106) injection of mCSF1R CART or mCherry T cells. Quantification of transferred T cells (l) or microglia (m) by TPLSM. (m) Indicated p-values apply to comparison between all groups. (n) Mean body volume of microglia (MBV). P-values of comparison mCSF1R CAR i.c. versus mCherry T cells i.c. (o) Representative maximum intensity projection of microglia, macrophages (green) in CX3CR1-GFP mice after i.c. injection mCSF1R CART (red, top) or mCherry T cells (red, bottom). White Arrowhead: Microglia and macrophages with higher mean density and MBV. Depth from brain surface: 0 - 100 μm. Scale bars: 50 μm and 20 μm. (k, m, n) Data are mean ± s.e.m. mCSF1R CART: i.c., n = 5; i.v., n = 4; mCherry T cells: i.c./ i.v. n = 2. (l) Data are mean ± s.e.m. mCSF1R CART: i.c., n = 3; i.v., n = 4; mCherry T cells: i.c. one control mouse.
For all panels, if not otherwise indicated, statistical significance was calculated using two-way ANOVA with Šidák multiple comparison correction.
Given the above, we decided to use CSF1R to model potential off-target toxicity in mice. We sequenced an anti-murine CSF1R (mCSF1R) antibody-producing hybridoma and designed second generation anti-mCSF1R CAR-T cells (mCSF1R CART; Extended Data Fig. 3a). Murine anti-EpCAM CAR (mEpCAM CART) or mCherry-transduced T cells were used as a negative controls for all experiments (Extended Data Fig. 3a). mCSF1R CAR construct could be efficiently transduced into primary murine T cells (Fig. 3c). mCSF1R CART were dose-dependently activated through Fc-immobilized, recombinant murine CSF1R protein, as seen by upregulation of activation marker CD69 (Fig. 3d, left) and cell surface exposure of degranulation marker CD107a (Fig. 3d, right) compared to mEpCAM CART.
To further validate functionality of the developed mCSF1R CART, we investigated killing capacity towards mCSF1R-expressing cell lines. Therefore, we selected murine reticulum cell sarcoma cell line J774A.1, which expresses mCSF1R43. Using flow cytometry, we verified expression of mCSF1R on J774A.1 cells, while mEpCAM was not detected (Extended Data Fig. 3b). Co-culturing mCSF1R or mEpCAM CART with J774A.1 tumor cells demonstrated efficient lysis of J774A.1 tumor cells by mCSF1R CART (Fig. 3e, left). As a mark of selective activation, high amounts IFNγ were secreted by mCSF1R CART (Fig. 3e, right).
Next, we used in vivo experiments to assess the risk for on-target toxicities. Initially, mCSF1R CART or controls were injected intravenously into healthy C57Bl/6 mice with limited engraftment (Extended Data Fig. 3c - e). To enhance persistence of the T cells, mice were next pre-conditioned using whole body irradiation (WBI, 5 Gy) 5 days prior to adoptive cell transfer (ACT) of mCSF1R CAR-T cells (Fig. 3f). High counts of mEpCAM CART and mCherry T cells were used as positive and negative controls, respectively. Following transfer of T cells, we did not detect a measurable change of weight of mCSF1R CART-treated mice as a sensitive surrogate for toxicities in mice (Fig. 3g). In comparison, as described in literature44, mEpCAM CART-treated mice rapidly lost weight one week after ACT (Fig. 3g). On day 7 (d7), when mEpCAM-treated mice reached the predefined abortion criteria, organs were harvested for subsequent analyses. Remaining mCSF1R CART or mCherry T cell-treated mice were sacrificed two weeks after ACT and organ-derived cell suspensions were analyzed by flow cytometry. We detected higher percentages of mCSF1R CART in all organs compared to mCherry-transduced T cells, indicative of better persistence – or antigen-dependent proliferation - of mCSF1R CART (Fig. 3h, top). We observed lower numbers of tissue-resident, CD11b+ cells in the kidney, liver and lung but not in other analyzed organs (Fig. 3h, bottom), most likely due to on-target effects of mCSF1R CART. Multiplex serum cytokine measurements on day 1 or day 7 after ACT revealed no differences in cytokine levels on d7 between mCSF1R CART- and mCherry T cell- or PBS-treated mice (Fig. 3i). In contrast, high levels of proinflammatory cytokines such as IFNγ, CXCL9 or CXCL10 were detected in the serum of mice that received the mEpCAM CART (Fig. 3i). Similarly, serum levels of clinically used markers of organ damage (e.g. urea, bilirubin, liver enzymes) were elevated in mice treated with mEpCAM CART, but not in mice that received mCSF1R CART or mCherry T cells (Extended Data Fig. 3g). Finally, we carried out histopathological analysis of organs with known high expression of CSF1R. mCSF1R CART-treated mice did not exhibit any signs of organ damage in hematoxylin and eosin (H&E)-stained lungs, livers or spleens (Extended Data Fig. 3h). Notably, as previously reported44, lungs of mEpCAM CART-treated mice showed thickening of the alveolar epithelium indicative of on-target off-tumor toxicities of the transferred mEpCAM CART (Extended Data Fig. 3h).
To investigate homing and killing potential of CAR-T cells to the brain compartment, we made use of CX3CR1-GFP reporter mice, which enable direct visualization of CAR-microglia interaction by 2-photon laser scanning microscopy (TPLSM). After implantation of cranial windows, mCSF1R CART or mCherry-transduced T cells were either intravenously or intracranially implanted into CX3CR1-GFP reporter mice (Fig. 3j - o). T cell-microglia interactions, change in microglia morphology, and reduction of overall microglia counts were monitored using TPLSM for a total of 28 days (Fig. 3l - o). Again, we observed no changes in weight or behavior across all treatment groups (Fig. 3k). We detected high T cell numbers following intracranial implantation of mCSF1R CART or mCherry T cells (Fig. 3l, o). These numbers gradually declined over the course of 28 days, regardless of whether mice were implanted with mCSF1R CART or mCherry T cells. At day 28, no transferred T cells could be detected in any group (Fig. 3l, o). Furthermore, microglia numbers did not substantially differ between any of the groups (Fig. 3m, o). Following intracranial implantation of T cells, mean body volume of microglia increased, most likely due to activation of the cells45 (Fig. 3n, o). This activation was most pronounced in mice injected with mCSF1R CART (Fig. 3n, o). However, by day 28, signs of microglia activation diminished in all groups (Fig. 3n, o). After intravenous injection of T cells, we detected neither mCSF1R CART nor mCherry control T cells in the brain and observed no signs of microglia activation or depletion (Fig. 3k - o, Extended Data Fig. 3i). Our results suggest that, despite expression of target antigens on tissue-resident immune cells in different organs as well as on microglia, there were no relevant safety signals that would prevent further therapeutic development.
Anti-human CART exhibit high potency in AML xenograft models
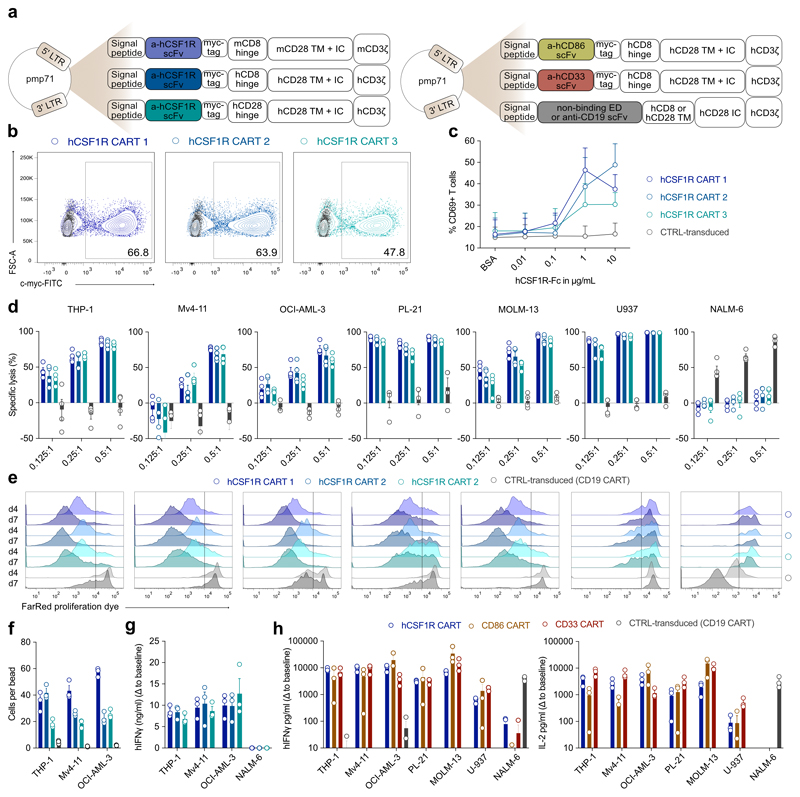
After proving the safety of CSF1R in various syngeneic mouse models, we next aimed to validate the targets in human models. We cloned an anti-human CSF1R binding scFv into the pre-existing anti-mCSF1R CAR backbone, which allows direct cross-comparisons of CAR-T cell activation thresholds of both anti-murine and anti-human CAR-T cells in mice and men. In addition, we created two fully human anti-CSF1R CAR constructs, harboring either a CD8 or CD28 hinge domain (hCSF1R CART 1-3; Extended Data Fig. 4a, left). First, we extensively cross-compared the functionality of the different anti-hCSF1R CAR constructs. All constructs could be efficiently introduced into primary human T cells (Extended Data Fig. 4b) and were dose-dependently activated by recombinant plate-bound hCSF1R protein (Extended Data Fig. 4c). CAR products efficiently lysed all six human AML cell lines tested, but not antigen-negative NALM-6 cells (Extended Data Fig. 4d). Constructs harboring CD8 hinge domains showed a tendency for higher lytic potency at lower effector to target (E:T) cell ratios (Extended Data Fig. 4d). To evaluate antigen-specific proliferation, we co-cultured CSF1R CART with AML cell lines for 4 or 7 days. All CSF1R CART showed antigen-specific, time-dependent proliferation (Extended Data Fig. 4e). Absolute quantification of T cell numbers revealed a more robust expansion of CD8 hinge-based anti-CSF1R CAR constructs (Extended Data Fig. 4f). All CSF1R CART secreted high amounts of IFNγ upon co-culture with THP-1, Mv4-11, or OCI-AML-3 AML cell lines, but not when co-cultured with NALM-6 control cells (Extended Data Fig. 4g). Building on these results, we decided to further proceed with CSF1R CAR-T cells harboring a CD8 hinge domain (hCSF1R CART 1, herein named hCSF1R CART).
Constructs for human CD86 CAR-T cells (CD86 CART) and human CD33 CAR-T control cells (CD33 CART) were similarly designed (Extended Data Fig. 4a, right). All CAR-T cell products could be efficiently introduced into primary human T cells (Fig. 4a). To validate functionality of CD86 CART and to compare sensitivity thresholds of both newly developed therapeutics, both CAR-T cells were incubated with their respective plate-bound antigens (Fig. 4b). Activation of CD86 CART was already observed at very low concentrations of target protein (0.01 μg/ml). In comparison, hCSF1R CART required concentrations of 1 μg/ml or higher (Fig. 4b). We co-cultured all CART with AML cell lines and assessed both specific lysis of AML cells and antigen-dependent proliferation (Fig. 4c, d). hCSF1R and CD86 CART efficiently lysed all six AML cell lines, comparable to CD33 CART (Fig. 4c) and proliferated to a similar extent (Fig. 4d). CD19 CART were used as control-transduced cells.
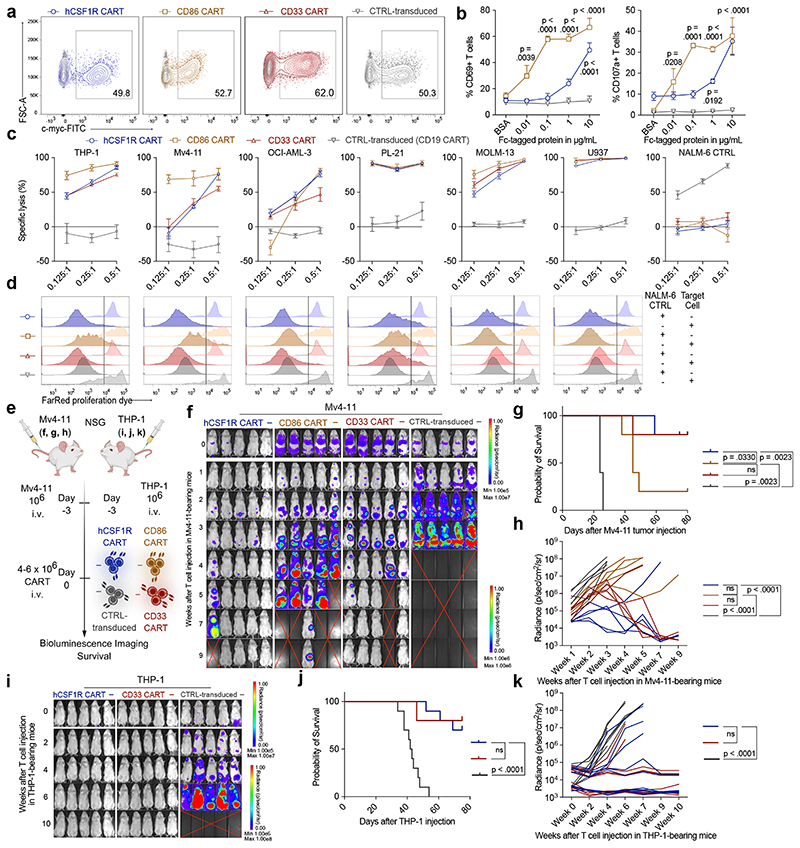
Figure 4. Anti-target CAR-T cells are functional and efficiently lyse AML cell lines in vitro and in vivo.
(a) Representative flow cytometric images of construct expression on primary human T cells. (b) Activation of hCSF1R or CD86 CART after incubation with plate-bound hCSF1R or hCD86 protein was quantified using flow cytometry. Data are mean ± s.e.m of three different donors. (c) hCSF1R or CD86 CART were co-cultured with luc+ target antigen expressing AML tumor cell lines or antigen negative NALM-6 control cells expressing luciferase for 48 hours at indicated E:T ratios. CD33 and CD19 CART (CTRL-transduced) were used as positive or negative controls, respectively. Cell lysis was quantified by BLI. Data are mean ± s.e.m of three different donors. (d) Proliferation dye-labeled CART were co-cultured with above indicated cell lines for 7 days at a E:T ratio of 0.5:1. One representative image of three different donors is shown. (e) Schematic of treatment scheme used for in vivo experiments. BLI images (f) survival curves (g) and quantification of tumor-burden (h) in Mv4-11-tumor bearing mice after treatment with different CART. (f - h) n = 5 mice per group. (i - k) BLI images (i), survival curves (j) and quantification of tumor burden (k) of THP-1-bearing mice treated with hCSF1R CART or control-transduced T cells. n = 10 mice per group. Shown is pooled data ± s.e.m. from two independent experiments. (f, i) Red cross, mice succumbed to disease. For all experiments, statistical significance was calculated using two-way ANOVA with Šidák multiple comparison correction. For Kaplan-Meier-Curves, statistical significance was calculated with log-rank test.
To prove in vivo efficacy of newly developed CART, we injected NOD-SCID IL-2Ry-null (NSG) mice with a lethal dose of Mv4-11 AML cells and treated them with hCSF1R, CD86, CD33 or control CART (Fig. 4e). We monitored tumor progression using bioluminescence imaging (BLI). Both hCSF1R and CD86 CART eliminated Mv4-11 tumor burden in vivo (Fig. 4f - h). To deliver in vivo proof for another AML model, we injected a lethal dose of THP-1 cells intravenously into NSG mice and again treated them with CSF1R, CD33 or control CART (Fig. 4e). Again, hCSF1R CART efficiently controlled experimental leukemia with similar complete remission (CR) rates as CD33 CART (hCSF1R CART: CR in 7 of 10; CD33 CART: CR in 8 of 10), with overall survival up to 80 days after tumor cell injection (Fig. 4i - k). In summary, we were able to demonstrate both in vitro and in vivo efficacy of newly developed anti-hCSF1R and anti-CD86 CART towards a large panel of human AML cell lines.
CSF1R and CD86 CART are effective in primary patient models
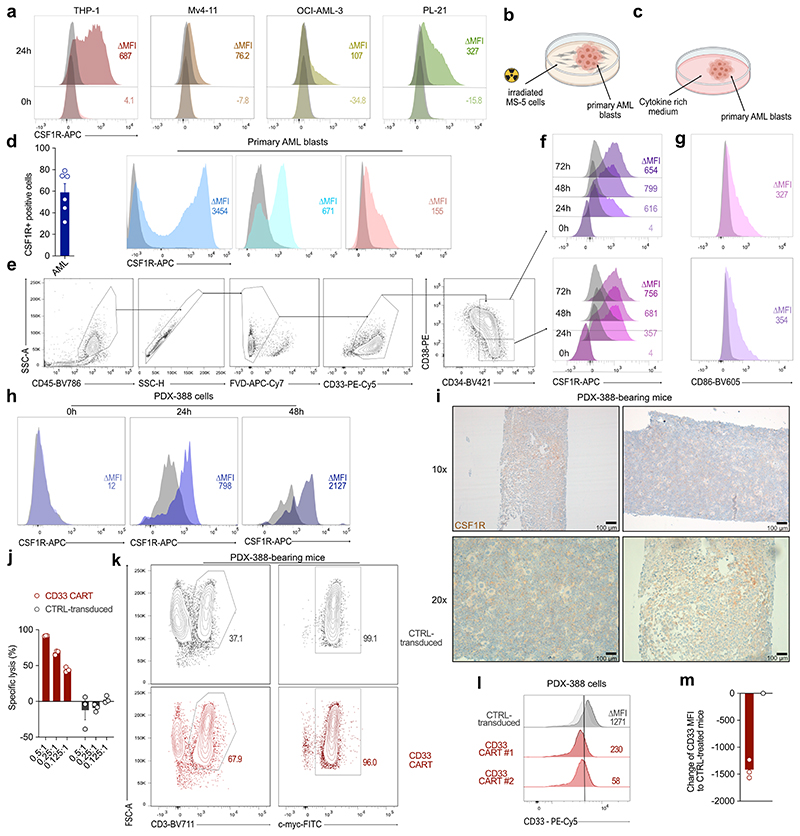
We next assessed receptor expression in primary AML samples. Until now, CSF1R expression on primary AML blasts was thought to be restricted to “AML supportive cells” or only to mature leukemic cells46. Indeed, when analyzing surface CSF1R expression on frozen bone marrow samples immediately after thawing, we could not detect any measurable receptor expression using flow cytometry (Fig. 5a, b). However, when primary AML cells were co-cultured on MS-5 murine bone marrow stromal cells (Extended Data Fig. 5b), we observed a strong, time-dependent increase of CSF1R expression (Fig. 5a, b). We hypothesized that these discrepancies in measurable surface CSF1R expression were most likely due to receptor downmodulation during the freezing and thawing process. To probe this, we analyzed receptor expression on AML cell lines after freeze-thaw cycles. Similar to the results seen on primary AML blasts, CSF1R was undetectable directly after thawing but regained high expression after 24 to 48 hours of culture (Extended Data Fig. 5a). To further exclude any cell culture artifacts, we analyzed surface receptor expression on primary AML blasts after culturing in cytokine-rich media47 (Extended Data Fig. 5c). Again, CSF1R was highly expressed on malignant primary AML blasts after culture (Extended Data Fig. 5d). We also confirmed expression of CD86 on primary AML blasts (Fig. 5c).
Figure 5. CSF1R and CD86 are readily detected on primary AML samples and hCSF1R CART show efficient lysis of primary AML samples in vitro and in vivo.
(a) Expression of CSF1R following thawing of primary AML samples over 72 h. Each line represents one patient. (b) Representative histograms of CSF1R (colored) expression on primary AML samples over time in comparison to isotype control (grey). (c) Expression of CD86 on primary AML samples. Each dot represents one patient. Left: Percentage positive cells gated to isotype. Right: Representative histograms of four different patients. Data are mean ± s.e.m of eleven different primary AML samples. (d) hCSF1R, CD86 or CD33 CART or untransduced T cells were co-cultured with primary AML samples for 72 hours. Specific lysis was assessed using flow cytometry. Data are mean ± s.e.m of seven different primary AML samples. Indicated p-values apply to E:T ratio 0.5:1. (e) hCSF1R CAR construct transduced into T cells of AML patients. Left: Transduction efficiency of AML patient-derived CAR-T cells. Right: Representative flow cytometric image. (f) Patient-derived CART or untransduced T cells were co-cultured with primary AML samples of the same donor. Experiments were carried out as outlined in (d). (e, f) Data are mean ± s.e.m of three different autologous donors. (g) Summary of treatment scheme used for in vivo experiments. (h - j) BLI images (h), survival curves (i) and BLI quantification of tumor-burden (j) of PDX-573 tumor-bearing mice injected with 6 x 106 hCSF1R, CD33 CART or control-transduced T cells (n = 5 mice per group). (j) P-values calculated at week 8. (h) White cross, censored mice; red cross, mice succumbed to disease. (k - m) BLI images (k), survival curves (l) and BLI quantification of tumor-burden (m)of PDX-388 tumor-bearing mice injected with 6 x 106 hCSF1R, CD86, CD33 CART, control-transduced T cells or PBS. (n = 3 - 10 mice per group). CD86 CART treatment was carried out separately. For all experiments statistical significance was calculated using two-way ANOVA with Šidák multiple comparison correction. For Kaplan-Meier-Curves, statistical significance was calculated with log-rank test.
Our single-cell gene expression analysis revealed lower expression of CSF1R and CD86 on malignant HSPC compared to CD123 and CD33 reference genes. Thus, we analyzed the protein expression of CSF1R and CD86 on malignant HSPC-like cells (Extended Data Fig. 5e – g). Both, CSF1R and CD86 were expressed on malignant HSPC-like cells, showing no differences in expression between these cell types (Extended Data Fig. 5f, g),illustrating the conserved expression of target antigens on these cells.
Next, we co-cultured primary AML samples with CAR-T cells and determined specific lysis by flow cytometry. hCSF1R and CD86 CART specifically lysed primary AML samples comparable to CD33 CART at low E:T ratios (Fig. 5d). In order to reflect the genetic heterogeneity of AML, a total of seven different primary AML specimens with differing cytogenetics were used for in vitro assays. To probe whether new anti-target CART can be introduced into T cells derived from AML patients, we transduced anti-hCSF1R CAR constructs into T cells of patients suffering from AML (Fig. 5e). Patient-derived hCSF1R CART were then co-cultured with autologous primary AML blasts resulting in potent lysis of primary samples (Fig. 5f).
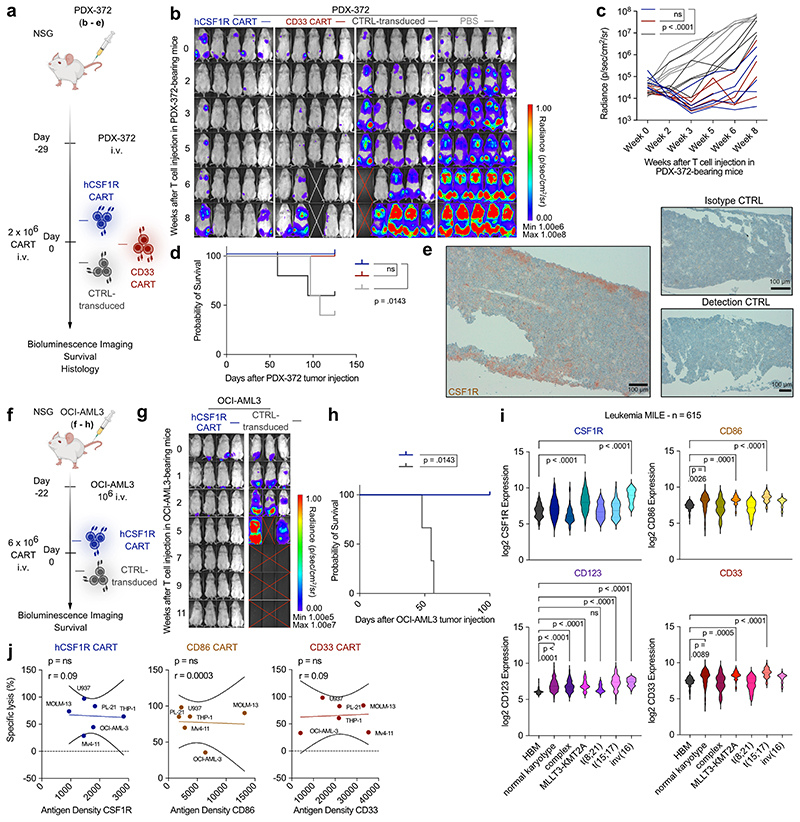
To prove efficacy of hCSF1R and CD86 CART in more relevant in vivo models, we transplanted cytogenetically distinct patient-derived xenograft models (PDX) models48 into mice and treated with the respective CART. First, we selected PDX-573, a model that was derived of a relapsed AML patient with high-risk cytogenetics (European LeukemiaNet (ELN) 2017 – adverse prognosis, see Supplementary Table 1b for detailed characteristics). Three weeks later, we injected hCSF1R, CD86, CD33 CART or CTRL-transduced T cells (Fig. 5g - j). All CART were highly effective, inhibiting tumor outgrowth in all treated mice ( 5 of 5, Fig. 5h - j). Next, we tested the efficacy of hCSF1R, CD86 and CD33 CART in PDX-388, derived of an initial diagnosis AML patient with KMT2A rearrangement (ELN 2017 – adverse prognosis) (Fig. 5k - m). Notably, expression of CSF1R on PDX-388 samples mimicked the above-described pattern: Following thawing of the cells, CSF1R was not expressed on PDX-388 cells, but was detectable after at least 24 hours of in vitro culture (Extended Data Fig. 5h), and also in vivo in bone marrow sections of control-treated PDX-388 mice (Extended Data Fig. 5i). hCSF1R and CD86 CART induced sustained remission in all treated mice over a period of 85 days (CR in 10 of 10 for CSF1R CART, 3 of 3 for CD86 CART) (Fig. 5k - m).
Interestingly, in this model, CD33 CART completely failed to control tumor growth in all mice (CR 0 of 10). We excluded manufacturing failure of CD33 CART in vitro (Extended Data Fig. 5j). Furthermore, in a separate cohort, we verified that CD33 CART were present in the circulation of treated mice (Extended Data Fig. 5k, left) and expressed the CAR on the cell surface (Extended Data Fig. 5k, right). Ex vivo flow cytometric measurement of CD33 on PDX-388 blasts revealed a strong decrease of CD33 surface expression on PDX cells of mice treated with CD33 CART compared to CTRL-transduced T cells (Extended Data Fig. 5l, m). Failure of CD33 CART to control tumor burden was thus most likely due to downregulation of surface CD33 expression on PDX-388 blasts. However, the detailed biological mechanism remains elusive and still requires further characterization.
To unambiguously validate the potential of hCSF1R CART in vivo, we used a third PDX model (PDX-372; Extended Data Fig. 6a - e) as well as third cell line xenograft model (OCI-AML3, Extended Data Fig. 6f - h). PDX-372 samples were again derived from a patient with relapsed AML with high-risk cytogenetics and TP53 mutation (Supplementary Table 1b). In addition, to create more challenging models, we transferred reduced numbers of CART into PDX-372-bearing mice (Extended Data Fig. 6a). hCSF1R CART stunted AML growth in 3 of 5 mice. Detected BLI signal did not vary between hCSF1R and CD33 CART (Extended Data Fig. 6b, c). As previously described for PDX-388, immunohistochemical analysis revealed high expression of CSF1R on PDX cells in vivo (Extended Data Fig. 6e). hCSF1R CART transferred into OCI-AML3-tumor bearing mice were similarly effective (Extended Data Fig. 6f - h).
To gain a better understanding of the expression patterns of CSF1R and CD86 in the complex molecular landscape of AML and of potentially differing expression patterns in different AML subtypes, we used a published large-scale dataset (Leukemia MILE study) and analyzed the gene expression of CSF1R and CD86 in comparison to CD123 and CD33 reference genes. Similar to CD33, CSF1R and CD86 were broadly expressed in different subtypes, with highest expression observed in KMT2A::MLLT3 (MLL::AF9), t(15;17) and inv(16)-mutated AML (Extended Data Fig. 6i).
Given the comprehensive panel of different in vitro and in vivo models used throughout our studies, we next sought to investigate whether we can determine an antigen threshold for effective CAR-T cell therapy in AML. However, we did not observe correlations between antigen site density measured with flow cytometry and lysis capacity of CAR-T cells for any of the tested antigens (Extended Data Fig. 6j).
In summary, using three different, cytogenetically distinct PDX-models, as well as three cell line xenograft models, we were able to provide strong evidence of functionality of newly developed anti-target CAR-T cells in vitro and in vivo.
Toxicity analyses of anti-human CSF1R and CD86 CART
After having verified the expression on malignant AML cells, we next evaluated target antigen expression on CD34+ HSPC. Using flow cytometry, we demonstrated lower expression of CSF1R and CD86 than CD33 on healthy HSPC (Fig. 6a, b). To directly assess toxicity towards HSPC, we co-cultured enriched bone marrow-derived CD34+ cells with hCSF1R, CD86, CD33 CART or untransduced T cells for 24 hours (Fig. 6c). CD34+ HSPC were exclusively lysed by CD33 CART (Fig. 6c, left). Also, CD33 CART secreted more IFNγ into co-culture supernatant than hCSF1R or CD86 CART (Fig. 6c, right). To further validate these results, we performed conventional colony-forming unit (CFU) assays. Colony counts of CFU-E and BFU-E were higher when HSPC were co-cultured with hCSF1R CART than with CD33 CART, indicative of better survival of stem cells in the presence of hCSF1R CART (Fig. 6d). Importantly, colony counts of HSPC co-cultured with either hCSF1R CART or untransduced T cells did not vary (Fig. 6d).
Figure 6. hCSF1R CART show better discriminatory capacity towards healthy human hematopoietic cells than CD33 CART.
(a) Target expression on MACS-enriched, bone marrow-derived CD34+ HSPC. Data are mean ± s.e.m of two to three independent, pooled HSPC donors. (b) Representative flow cytometric image of target expression on HSPC. (c) CSF1R, CD86 or CD33 CART or untransduced T cells were co-cultured with HSPC for 24 hours at an E:T ratio of 2:1. Lysis of HSPC was quantified by flow cytometry (left). IFNγ secretion measured by ELISA (right). (d) CSF1R, CD33 CART or untransduced T cells were co-cultured with HSPC for 24 hours at an E:T ratio of 2:1 and CFU assay was performed. Colony count was quantified after 14 days. (c, d) Data are mean ± s.e.m. from three (c) or four (d) different donors. (e) CSF1R expression on healthy donor-derived bone marrow aspirates (HD). Left: Percentage CSF1R positive cells gated to isotype. Right: Representative histograms of CSF1R expression on HD. (f) Quantified target expression on HD. Left: Percentage positive cells gated to isotype. Right: Representative flow cytometric image. (e, f) Data are mean ± s.e.m. from three different donors. (g) hCSF1R, CD33 CART or untransduced T cells were co-cultured with HD for 72 hours at the indicated E:T ratios. Left: Off-tumor lysis of CART assessed by flow cytometry. Right: Activation of T cells quantified by IFNγ secretion. Data are mean ± s.e.m. from 11 different samples. (h, i) Quantification of log-transformed normalized target expression in 13,067 single human brain cells. Each peak corresponds to a cell, peak height indicates expression intensity. (i) UMAP plot illustrating expression patterns of CSF1R, CD86 and CD33 in human brain cells. (j) Phenotype of human iPSC-derived microglia-like cells (iMGL). (k) Representative histograms of CSF1R and CD33 expression on iMGL. (l) hCSF1R CART, CD33 or untransduced T cells were co-cultured with iMGL for 24 hours at indicated E:T ratios. Lysis of iMGL was quantified by flow cytometry (left). T cell activation quantified by ELISA (right). Data are mean ± s.e.m. from five T cell donors. For all experiments statistical significance was calculated using two-way ANOVA with Šidák multiple comparison correction.
Next, we analyzed expression of target antigens on samples of healthy human bone marrow donors (HD samples) (Fig. 6e – f). Again, surface CSF1R expression could only be detected after at least 24 hours of culture (Fig. 6e), but its expression remained lower than that of CD86 or CD33 (Fig. 6f). Co-cultures of hCSF1R, CD33 CART or untransduced T cells with HD samples revealed higher lysis of healthy bone marrow samples (Fig. 6g, left) and increased secretion of IFNγ by CD33 CART (Fig. 6g, right). Both, lysis of HD samples and IFNγ secretion did not differ between hCSF1R CART and untransduced T cells (Fig. 6g).
ScRNA-seq analysis of single human brain cells confirmed expression of CSF1R on microglia (Fig. 6h, i). On a single-cell level, CSF1R showed higher expression on microglia than CD86 or CD33 (Fig. 6h, i). To model toxicity of CAR-T cells towards human microglia, we generated iPSC-derived human microglia-like cells (iMGL)49,50 and verified their phenotype (Fig. 6j). Both CSF1R and CD33 were highly expressed on iMGL (Fig. 6k). Co-cultures of human iMGL with either CSF1R, CD33 CART or untransduced T cells demonstrated lysis of human iMGL by both CAR at high E:T ratios of 1:1 (Fig. 6l, left). At more physiological E:T ratios (0.2:1), neither CSF1R nor CD33 CART were able to lyse human iMGL, consistent with our in vivo data (Fig. 6l, left). IFNγ release mimicked the results obtained from flow cytometric analysis (Fig. 6l, right). In summary, our data suggest a superior discriminative capacity towards healthy hematopoiesis of our newly developed CAR-T cells compared to CD33 CART and indicate that microglia might not be a relevant off-tumor target of anti-CSF1R CAR-T cells.
Discussion
We developed an unbiased scRNA-seq approach for de novo target identification and in-depth, high-resolution off-tumor mapping across multiple tissues, specifically tailored to predict potential candidates for CAR-T cell therapy. Applying our approach to AML, we identified two target antigens: CSF1R and CD86. Extensive in vitro and in vivo validation revealed broad expression on AML blasts, strong and durable treatment responses of newly developed CAR-T cells in vitro and in vivo, and minimal toxicities towards relevant healthy cells and tissue.
For primary target screening, we leveraged single-cell sequencing data of 15 primary AML specimens with differing cytogenetic properties21. In addition, we validated the obtained results in an independent cohort of five additional AML patients39. The top hits of the present study were reliably found overexpressed in large bulk sequencing AML cohorts (n = 615). Given the highly complex molecular landscape of AML, rare AML subtypes might still not be fully represented in our analyses. Despite this limitation, our study clearly demonstrates the translational potential of unbiased, scRNA-seq-based screening approaches and provides proof-of-principle of the whole spectrum of scRNA-seq-guided drug development spanning from computational target identification to thorough preclinical investigation of newly developed CAR-T cells.
CSF1R has been previously implicated as a target for small molecule inhibition in AML46. However, its expression was thought to be restricted to a small subset of AML supportive cells in certain patients, while the majority of patient blasts are regarded as antigen-negative51. Using various techniques, including transcriptomic analysis, flow cytometry, immunohistochemistry and comprehensive functional investigation of CSF1R-directed CAR-T cell therapy, we were able to confirm high CSF1R expression on AML blasts. These reported ambiguities of CSF1R expression on malignant AML blasts, encourage the use of unbiased, RNA-based screening algorithms for target identification and prioritization, as methodological or biological confounders can easily mask protein expression analysis. Nevertheless, it is crucial to bear in mind that scRNA-seq-centered strategies come with their own limitations (e.g. the zero or dropout problem of singe-cell gene expression52) and in any case require protein validation.
CD86 is expressed on malignant AML blasts and high receptor expression is associated with significantly shortened overall survival of AML patients53,54, but to the best of our knowledge, CD86 has never been explored as a target for (immuno-) therapy of cancer. The expression of CD86 is not limited to AML, but has also been reported in numerous B cell malignancies55. As such, the use of CD86 CART promises not only treatment options for AML, but may also be applicable for a variety of other hematological diseases such as multiple myeloma56 and childhood B-cell precursor acute lymphoblastic leukemia57. Nevertheless, CD86 is also expressed on healthy macrophages and dendritic cells58–60 and might increase the risk of immunosuppression and ensuing severe infection. However, CTLA-4 fusion proteins such as abatacept, targeting both CD80 and CD86, have received approval by the FDA and are clinically used for the treatment of autoinflammatory disorders61. In clinical studies, abatacept was generally well tolerated61.
For both, CSF1R and CD86, the measured antigen site densities were rather low, especially in comparison CD33 expression, which was high. Yet, despite our extensive functional validation, we did not observe marked differences between CSF1R and CD86 CART in comparison to established CD33 CART. Along these lines, we did not observe correlation between lysis capacity of CAR-T cells and site density of the respective target antigen. To a certain extent these findings are in line with recent reports observed for anti-mesothelin CAR-T cells in solid tumors62. Several factors such as affinity and binding properties of the used scFv and conformation of the target antigen can positively or negatively influence these CAR – tumor cell interactions. Ultimately, while high target antigen expression undoubtedly increases killing efficacy, our data suggests that in some cases functional cross-comparison might help to identify promising target antigens, despite, on first glance rather low antigen expression.
Similar to previous results in AML18, we were not able to identify target antigens with expression limited to a single immune cell lineage, as is the case for CD19 or BCMA in B cell malignancies. However, expression of our prime candidates is limited to immune cells of myeloid origin (monocytes, tissue-resident macrophages and dendritic cells) with minimal detection on stem or progenitor cells. Thus, our candidates could bear the advantage of clinical application without the risk for severe bone marrow toxicity, which is a current concern of AML-targeted treatments10. It should be noted however, that to date, the clinical outcomes of off-tumor gene expression on HSPC remains elusive. Along these lines, precise projection of off-tumor antigen expression is one of the central objectives of our single-cell approach, since unwanted toxicity may be inferred from high transcriptomic off-target antigen expression20,63. Yet, as outlined above, the risk of severe adverse effects caused by off-tumor activity of CAR-T cells is not fully understood and different outcomes have been reported64. As such, the latest trials evaluating the safety of CD123 CART did not show sustained cytopenia64. However, in most anti-CD123 CAR trials currently conducted, patients eventually received allo-HSCT, which presumably eradicated CAR-T cells. Of note, the development of fatal cytokine release syndrome and capillary leak syndrome following CD123 CART infusion, potentially due to off-target expression of CD123 on small vessels, has been reported7. Altogether, current clinical evidence does not support a clear definition of the critical cell types as well expression thresholds which would preclude the development of CAR-T cells against a certain target to avoid unmanageable toxicities. In any case, in the long run, detailed knowledge of off-tumor expression will allow vigilant monitoring of “high-risk off-tumor organs" in clinical trials and might enable rapid side effect-mitigating treatments. Similarly, clinical lessons from anti-CD19 or anti-BCMA CAR-T cell therapy deem lineage-restricted expression patterns as highly desirable, providing further strong evidence for the use of single-cell technologies for de novo target identification, as these technologies might be able to aid the search for unrecognized target antigens with minimal off-tumor expression in healthy tissues.
Many of the currently investigated CAR targets in AML failed the thresholds of overexpression on malignant HSPC compared to their healthy counterpart in our analyses. Herein, to a certain extent, our data contradicts with publications from our colleagues13,65. Sauer et al. for example illustrated the higher expression of CD70 in bone marrow biopsies of AML patients, compared to bone marrow samples of healthy donors using immunohistochemistry13. This discrepancy is most likely due to our restrictive analyses, in which we have chosen rather high cutoff criteria to ensure maximal safety of identified target antigens. Dynamic adjustment of these thresholds might yield different results and many of the previously identified target antigens (e.g. CD123, CD33, CD70, FLT3, CLL-1, CD44v6) will most likely be of aid to improve patient care of refractory or relapsed AML patients. Nonetheless, our data clearly demonstrates the value of CSF1R and CD86 as targets for CAR-T cell therapy in AML and, especially considering the complex molecular landscape of AML and its highly diverse subsets, are expected to be valuable additions to the immunotherapeutic repertoire in AML.
Unsurprisingly, CSF1R was expressed on microglia, which share a common monocytic precursor, as also known for CD3366. Clinical investigation of the so far only CSF1R-directed monoclonal antibody did not reveal neurotoxicity as a concern when depleting CSF1R positive cells from the periphery67. However, given the different mode of action of cellular versus antibody-based therapies, these results might not be directly transferable to anti-CSF1R-CAR therapy. In addition, CAR-T cells are known to be able to cross the blood-brain-barrier already at steady-state68,69 and peak levels of pro-inflammatory cytokines further increase permeability of this tightly regulated barrier70,71. Because of these considerations, we rigorously tested the possibility for neurotoxicity in numerous models. These models included fully syngenic mice models, in which we implanted large quantities of CAR-T cells directly into mouse brains. Yet, we did not observe any signs of neurotoxicity. Nevertheless, future clinical validations will need to include well-designed protocols to vigilantly detect any signs of neurotoxicity.
Our results highlight the potential of using unbiased, high-resolution, single-cell transcriptomic data for target selection and drug development. Leveraging this data and the appropriate high dimensional analyses as standard operating procedures promises to improve safety and efficacy of newly engineered CAR-T cells and enables identification of new target structures for targeted immunotherapy in malignant disorders.
Methods
Single-cell transcriptome analysis
All preprocessing and analyses steps of scRNA-seq data were run in python 3 using Scanpy72 v.1.4.6 to 1.9.1 and anndata73 v.0.7.1 to 0.8.0 unless otherwise stated. All scRNA-seq figures were plotted using matplotlib and seaborn.
Preprocessing publicly available scRNA-seq data of healthy and AML cells
For healthy donors and AML patients, we obtained raw, annotated count data of healthy bone marrow cells from two resources. (i) The data from van Galen et al.21 were downloaded from GEO (GSE116256). Here, we excluded patient AML916, as this is a mixed AML phenotype expressing markers of stem cells, myeloid, T and B lineages. (ii) For the scRNA-seq data of Petti et al.39, we obtained raw count data from https://zenodo.org/record/3345981.
For the data from GSE116256, barcodes were filtered for each sample for high-quality cells based on the total distributions of UMI counts and genes. For exact threshold values, see provided notebook. Cells with a fraction of mitochondria-encoded genes over 20 % were excluded. Barcodes that could not be confidentially assigned to either healthy or tumor cells were discarded. Genes detected in less than 20 cells were excluded from further analyses. The resulting count matrix was used for normalization. UMI counts of each cell were normalized using the SCRAN algorithm as implemented in the R-based package74,75. Briefly, size factors were estimated by preliminary clustering of the data using the Louvain algorithm implemented in Scanpy (tl.louvain) with a resolution of 0.5 before running ComputeSumFactors (min.mean = 0.1). The estimated size factors were then used for cell normalization. Finally, the data was log-transformed (log(count+1)).
Feature selection and visualization in a low dimensional embedding
The top 4000 variable genes were identified based on normalized dispersion as described previously76 using Scanpy’s pp.highly_variable_genes with flavor=cell_ranger. Briefly, genes were ordered along their mean expression in several bins and selected according to their highest variance-to-mean ratio. To efficiently capture the underlying data structure in two dimensions, principal component analysis (PCA) dimension reduction was carried out by computing 15 principal components on highly variable genes using Scanpy’s pp.pca. To account for technical batches, harmony77 was used to integrate data from the respective patients. Next, a neighborhood graph was computed on the first 50 harmony-adjusted principal components using Scanpy’s pp.neighbors with 15X neighbors. For 2D visualization, embedding the neighborhood graph via UMAP78 was done by running Scanpy’s tl.umap with an effective minimum distance between embedded points of 0.5.
Differential gene expression of AML HSPC and T cell marker gene identification
Enriched gene expression in T cells were identified by comparing the mean expression of healthy T cells to the mean expression of all other healthy cell types using a t-test with overestimated variance as adopted in Scanpy in the tl.rank_genes_groups function. Testing was performed on the log-transformed normalized data to account for differences in sequencing depth between samples. Upregulated genes with FDR-adjusted p ≤ 0.01 and a log fold change > 0 were considered for target antigen filtering. Characteristic gene signatures of AML HSPC were identified by performing separate differential expression analysis of AML HSC-like and HPC-like cells against their healthy equivalents, respectively. Genes that were expressed in at least 2% of all cells with log fold change > 2 and FDR-adjusted p ≤ 0.01 were defined as enriched marker genes.
Harmonizing public databases to obtain surface-protein coding genes
To obtain genes coding for proteins on the cell surface, we used OmniPath22, a large scale molecular database to access data from (1) the Mass Spectrometry based Cell Surface Protein Atlas23, (2) CellPhoneDB25, a repository of curated receptors, ligands and their interactions, (3) the machine learning-based In silico human surfaceome24 and (4) the Human Protein Atlas26 v20.1 (https://www.proteinatlas.org/). Permissive integration of all datasets was critical, as cell surface expression showed strong variabilities between databases. Consequently, union of these databases were used for all subsequent analyses.
Targets of FDA approved drugs
To identify genes that code for druggable proteins, we utilized DrugBank38, a database containing information on the interaction, pharmacology and chemical structures of drugs and drug targets. We defined druggable genes as targets with known pharmacological action of FDA-approved drugs.
Cross-organ off-target transcriptomic atlas (COOTA)
To quantify off-target effects, we analyzed and combined a total of 11 scRNA-seq datasets across 9 healthy tissues27–37. Raw annotated scRNA-seq data from the respective studies 27,29,31,33–37 were obtained using the python-based data repository sfaira79. To quantitatively analyze the expression of possible CAR-T cell therapy targets across healthy tissues, comparable preprocessing steps were carried out for each dataset separately, which involved removing low-quality cells and lowly expressed genes (see provided notebook for exact thresholds), cell count normalization using scran, selecting highly variable genes based on normalized dispersion and visualizing the cells in a two-dimensional UMAP embedding as described above. For the lung datasets of Travaglini, Madissoon and Reyfman30–32, we used publicly available data with cell annotations deriving from a study integrating multiple single-cell RNA-seq datasets80. To account for technical batches along the respective samples, batch balanced k nearest neighbors81 were calculated for the datasets of Travaglini, Madissoon, Reyfman, Ramachandran, James, Cheng and Han30–32,34–37. Finally, processed and annotated count matrices were concatenated on union variables using Scanpy’s concatenate with join = outer and the resulting matrix was used for target antigen filtering.
Genes were excluded if they were expressed in over 2% of cells of a critical cell cluster (endothelial, arterial, bronchial, capillary, venous and smooth muscle cells). A single-gene-based count matrix with barcodes x datasets was created and used for plotting mean expression values across cell types.
Reference mapping and label transfer
Raw count data from Petti et al.39 were filtered to obtain high-quality cells. Cells with less than 10 expressed genes as well as genes detected in less than 3 cells were excluded from further analyses. Barcodes with a fraction of mitochondria-encoded genes over 10 % and a fraction of ribosomal genes over 50 % were excluded. For reference mapping and label transfer, we used scANVI40 a semi-supervised variational auto-encoder model to leverage the cell type knowledge from the data from GSE116256 (reference) to infer the states of cells from Petti et al. (query). Briefly, we trained the SCVI Model was trained on the reference data with 2 hidden layers and a dropout rate of 0.2 (for the exact parameters, see provided notebook). Next, we initialized the scanVI model from the pretrained scVI model, before training the scANVI model for 20 epochs with 100 samples per label. Afterwords, we created a new query model instance before training the query data with a weight_decay of 0 for 100 epochs. The latent representation and the label predictions were obtained using get_latent_representation() and predict(), respectively. Finally, we computed a neighborhood graph on with 15 neighbors using the scANVI representation, before embedding the graph using UMAP as mentioned before.
Bulk expression data
The data used for the bulk analyses of CSF1R and CD86 expression in human and mouse tissues were obtained from the GTEX portal82 (E-MTAB-5214) and Merkin et al.83 (E-MTAB-2801) via the Expression atlas84 (https://www.ebi.ac.uk/gxa/home).
Cell lines
Human AML cell lines THP-1, MV4-11, OCI-AML-3, PL-21, MOLM13, U937, NALM-6 were purchased from ATCC (USA). All cell lines were cultured in RPMI containing 20% FBS, 2 mM L-Glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Murine J774A.1 cell line was provided by Peter Düwell, Institute of Innate Immunity, University Hospital, Bonn. Cells were cultured in DMEM containing 10 % FBS, 2 mM L-Glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. All cells were grown at 37°C in a humidified incubator with 5% CO2. Short tandem repeat (STR) profiling was used to verify identity of human cell lines. Cells were negatively tested for mycoplasma contamination using polymerase chain reaction (PCR). All cell lines were lentivirally transduced with a pCDH-EF1a-eFly-eGFP plasmid85. After transduction, enhanced green-fluorescent protein (eGFP) positive cells were single-cell sorted using a BD FACSAria™ III Cell Sorter and expression of firefly luciferase (fLuc) was verified using Bio-Glo™ Luciferase Assay System. Cells were frozen in medium containing 90% FCS and 10% DMSO and stored at -80°C or in liquid nitrogen for long-term storage. Generation of PDX cells was previously described48. Anti-mouse c-fms (CD115)-producing Hybridoma RCB4486 was acquired from the RIKEN Bio-Resource Research Center86.
AML blast isolation and culture
Primary AML blasts or healthy donor control samples (HD) were obtained from the bone marrow or peripheral blood of patients suffering from AML after written informed consent in accordance with the Declaration of Helsinki and approval by the Institutional Review Board of the Ludwig-Maximilians Universität (Munich, Germany) or the Ethics committee of the medical association of Hamburg. Bone marrow aspirates were enriched for AML blasts either through density centrifugation or lysis of red blood cells using osmotic gradient solutions and frozen in liquid nitrogen. Prior to T cell-based assay, bone marrow aspirates were thawed and T cells were depleted using a CD3 positive selection kit (StemCell Technologies).
Primary AML samples were either cultured in IMDM basal medium supplemented with 15% BIT 9500 serum substitute and beta-Mercaptoethanol (10-4 M), 100 ng/ml SCF, 50 ng/ml FLT3-Ligand, 20 ng/ml G-CSF, 20 ng/ml IL-3, 1 μM UM729 and 500 nM SR129 or alternatively in alpha-MEM supplemented with 12.5% horse serum, 12,5% FCS, 1% penicillin/ streptomycin, 1% L-glutamine, G-CSF, IL-3, TPO and 2-Mercaptoethanol on irradiated MS-5 (murine bone marrow stromal cells) for co-culture experiments87–89. Co-Cultures with primary AML samples were carried out after a three-day pre-culture of the thawed AML blasts89.
Flow cytometry
Flow cytometric analysis was carried out using a BD FACSCanto™, a BD LSRFortessa™ II or Beckman Coulter CytoFLEX. All staining steps for identified AML target antigens were conducted on ice. Cells were centrifuged at 200 – 400 g for 5 min at 4°C in a pre-cooled centrifuge. For staining of primary AML blasts and AML cell lines a maximum of 106 cells were counted and transferred to a U-bottom 96-well plate. Cells were washed twice with ice-cold phosphate-based saline (PBS) containing 2% FBS and incubated for 15 min with 5 μl of human TrueStain FcX™. CSF1R was stained for 30 min in the dark either using unconjugated anti-human m-CSF-R/CD115 antibody (R&D, Clone 61701) or mouse IgG1 isotype as control (R&D Systems, Clone 11711), followed by secondary staining with AlexaFluor® 647 rat anti-mouse IgG (H+L) antibody (Jackson ImmunoResearch, USA) or alternatively after incubation with biotinylated recombinant CSF-1 protein (Sino Biological, China) followed by secondary staining with Streptavidin-APC. Staining for CD86 was conducted with anti-human CD86 antibody (clone IT2.2)
Dead cells were excluded after staining with a fixable viability dye (eFluor™ 780, eBioscience, USA) in all experiments. Quantification of absolute cell counts was carried out by using Count Bright™ Absolute Counting Beads (Thermo Fisher Scientific, USA).
Primary AML blasts were identified using anti-human CD45 (clone: HI30) and anti-human CD33 (clone P67.6; WM53, Invitrogen/eBioscience). Staining with anti-human CD34 (clone 561) and anti-CD38 (clone: clone HB-7) were included for gating of leukemia-initiating cells. CAR expression was quantified using anti-c-myc FITC (clone SH1-26E7.1.6; Miltenyi Biotec, Germany). CAR activation was measured using anti-murine or anti-human CD69 (murine - clone: H1.2F3, human – clone: FN50), CD107a (murine – clone: 1D4B, BD Biosciences, human – clone: H4A3), PD-1 (human – clone: PD-1, EH12.2H7). Anti-CD2 (human - clone RPA-2.10), anti-CD3 (murine – clone: 145-2C11, human - clone: UCHT1, HIT3a) anti-CD4 (murine – clone: GK1.5, human - clone: OKT4) and anti-CD8 (murine – clone: 53-6.7, human – clone SK1, HIT8aa) antibodies were used to gate on T cells. Gating on tissue-resident immune cells was carried using anti-mouse CD45 antibody (clone: 30-F11) and anti-mouse/human CD11b (clone: M1/70). Preparation of organs for flow cytometric analysis was carried out as recently described90.
All antibodies and reagents were purchased from BioLegend, USA, unless otherwise specified.
Absolute quantification of antigen densities per molecule were carried using BD Biosciences Quantibrite™ Phycoerythrin (PE)-Beads according to manufacture’s instruction.
All flow cytometric stainings were carried out at a dilution of 1:50. For flow cytometric quantification of absolute molecule count per cell dilutions of 1:20 were used.
Generation of CAR constructs
All CAR constructs contained myc tags to readily detect CAR expression. Anti-CSF1R single chain fragment variable (scFv) was designed based on the patented sequence of anti-CSF1R heavy and light chain variable domains of the anti-CSF1R antibody clone 2F11-e7 (EP 2 510 010 B1). Anti-human CD86 scFv was derived of 3D1 (US 2002/0176855 A1). Anti-CD19 CAR-T cells were designed based on anti-CD19-CAR-FMC63-28Z CAR-T cells (WO2015187528A1). h-P67.7 scFv was used for anti-CD33 CAR-T cells. Murine anti-CSF1R scFv (clone AFS98) was derived from anti-mouse c-fms-producing Hybridoma described above. Murine CAR constructs contained a mCherry fluorescence tag, separated from the CAR construct via a 2A sequence. Constructs were either created using conventional cloning techniques or codon-optimized and cloned into pMP71 retroviral vectors using commercial cloning services (Twist Bioscience, USA).
Retroviral murine and human T cell transduction
For virus production, retroviral pMP71 vectors carrying the sequence of the relevant receptor were stably expressed in packaging cell lines 293Vec-Galv, 293Vec-Eco and 293Vec-RD11490,91,92–94. Human T cells were isolated from healthy donor PBMC using density gradient centrifugation, enriched for T cells using anti-CD3 microbeads (Miltenyi Biotec, Germany) and stimulated for 48 hours prior to retroviral transduction with human T-Activator CD3/CD28 Dynabeads® (Life Technologies, Germany). Human T cells were expanded in human T cell medium (hTCM) containing 2.5% human serum, 2 mM L-Glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin, 1% Sodium Pyruvate and 1% non-essential amino acids supplemented with recombinant human IL-2 (PeproTech, USA, Novartis, Switzerland) and IL-15 (PeproTech USA, Miltenyi Biotech, Germany). Murine T cells were derived from splenocytes, activated using anti-CD3 and anti-CD28 antibodies and transduced in murine T cell medium (10% FBS, 2 mM L-Glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin, 1% Sodium Pyruvate and 0.5% HEPES) supplemented with IL-2 and mouse T-Activator CD3/CD28 Dynabeads® (Life Technologies, Germany) as described. Following retroviral transduction, murine T cells were expanded in medium containing human IL-15 (PeproTech, USA). For all experiments comparing different CAR-T cells, transduction efficiency was adjusted to the lowest measured efficiency of the respective construct.
Animal experiments
Animal experiments were approved by the local regulatory agency (Regierung von Oberbayern, ROB) and carried out in accordance with guidelines and regulations implemented by the ROB. Animals were housed in specific-pathogen-free facilities. C57BL/6, BALB/c and NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ were purchased from Janvier (St Berthevin, France), Charles River Laboratories (Sulzfeld, Germany) or bred at the local facilities. CX3CR1-GFP reporter mice were bred at local facilities. Mice were held in facilities with a 12-hours dark/light cycle including a 30 min twilight phase of 30 minutes at noise levels below 50 dBA. Air velocity was held below 0.2 m/s. Air humidity in the facilities were between 45 – 60% and average temperature was held between 20 to 22 °C.
Patient-derived xenograft (PDX) models AML-573, AML-388 and AML-372 were genetically modified to express fLuc48. For BLI, mice were anesthetized using an isoflurane-oxygen mixture (1.5 – 2.5%) following intraperitoneal (i.p.) injection of BLI substrate (Xenolight D Luciferin potassium salt, Perkin Elmer, USA) into each mouse according to manufacturer’s protocol. In vivo Imaging System Platform Lumina X5 (IVIS, PerkinElmer, USA) was used to measure BLI signal.
Xenograft models using THP-1, MV4-11 or OCI-AML3 cell lines or PDX models were established by i.v. injection. T cells were transferred at indicated times and numbers. Mice that had to be removed from animal experiments due to non-tumor related toxicities (e.g. did not have measurable BLI signal at the exclusion timepoint) were censored. Censored mice are indicated in the respective Kaplan-Meier-Curves and are marked with a white cross in the BLI images.
Surgical procedures and stereotactic implantation
Preparation of chronic cranial windows using microsurgical implantation and stereotactic CAR-T cell injection was performed as previously described92. After the mouse was deeply anesthetized with intraperitoneal injection of MMF (Midazolam 5 mg/kg, Medetomidin 0.05 mg/kg and Fentanyl 0.5 mg/kg), skin was cut off and the periosteum was removed. After marking the cortical area of interest, a 5.5 mm circular part of the cranium was removed using a sterile carbon steel microdrill. Dura mater was separated from leptomeninges using forceps and removed to prevent dural fibrosis. Sterile round cover glasses and tailored rings were attached to the cranial bone with acrylic dental glue. To prevent postsurgical astroglial or microglial activation affecting tumor growth, stereotactic implantation of CAR-T cells was performed at least two weeks after cranial window implantation. For stereotactic implantation of CAR-T cells, 2 x 105 transduced T cells were resuspended in 1 – 2 μl PBS and injected at predefined coordinates (1 mm lateral and 2 mm posterior to the bregma at an intraparenchymal depth of 1.5 mm). Perioperative care included daily recording of weight and neurological scores.
Two-photon laser scanning microscopy (TPLSM)
TPLSM was performed using a Multiphoton TrimScope I system (LaVision Biotec) connected to an upright Olympus microscope attired with a MaiTai Laser (690 to 1.040 nm, Spectra Physics) and a 20× water immersion objective (numerical aperture 0.95, Olympus). Single images were acquired from different depth, depending on different regions, with z-interval of and μm. nm was used as an excitation wavelength with 1024 × 1024 pixels and detected by PMTs (G6780-20, Hamamatsu). Mice were anesthetized by isoflurane and maintained with a constant flow from 0.8% to 2.0% (as low as possible according to the physical condition of the mouse). After original images were acquired using Imspector Pro, Bitplane Imaris Software was used for further analysis. To obtain high-quality images, brightness, contrast or color balance were regulated manually for the whole images.
Immunohistochemistry
CSF1R staining on human in mouse xenograft bone marrow samples was performed using anti-human CSF1R primary antibody (Cell Signaling, rabbit monoclonal, clone E4T8Z, Cat. No.: 28917). Samples were formalin-fixed, decalcified in EDTA and paraffin-embedded. Heat-mediated epitope retrieval was accomplished using Epitope Retrieval Solution pH8 (Novocastra, RE7116). Slides were incubated in primary antibody for 60 min at RT at a dilution of 1:180. Biotinylated anti-rabbit IgG secondary antibody (Vector, Cat. No.: BA-1000) and Streptavidin-HRP Reagent (Novocastra, Cat. No.: RE 7104) was used for antibody detection. Finally, slides were stained with DAB+ (Agilent Technologies, Cat. No.: K3468) and counterstained with Hematoxylin Gill`s Formula (Vector, Cat. No.: H-3401).
T cell stimulation assay using plate-bound recombinant protein
96-well, half area, flat bottom, polystyrene plates (Corning, USA) were coated overnight at 4°C with fc-tagged recombinant protein diluted in 100 μl PBS at the indicated concentrations. On the next day, plates were washed with PBS and blocked with 2% BSA dissolved in PBS for 30 min. After another washing step, 50,000 T cells resuspended in hTCM without cytokines were added. T cell activation was assessed using flow cytometry after 24 hours of incubation.
Cytotoxicity assays
For co-culture experiments, 30,000 to 50,000 human AML cells were plated in a flat-bottom 96-well plate. Tumor cells were co-cultured with T cells at the indicated E:T ratio for 48 hours in hTCM without supplements unless otherwise specified. Killing was assessed using either Bio-Glo™ Luciferase Assay System (Promega Corporation, USA) according to the manufacturer’s protocol or flow cytometry. Specific lysis was calculated after normalization to control conditions. Co-cultures of primary AML blasts or healthy bone marrow cells and CAR-T cells were performed in the conditions outlined above. Killing was quantified with flow cytometry.
Proliferation assays
Prior to co-cultures (E:T ratio 0:5:1), T cells were stained using CellTrace™ Far Red Cell Proliferation Kit (ThermoFisher Scientific, USA) according to the manufacturer’s instructions. Trace dilution was measured by flow cytometry at day 7.
Cytokine measurements
Cytokine levels in co-culture supernatants were analyzed using BD Bioscience (IFNγ and IL-2) according to the manufacturer´s protocol. LEGENDplex™ mouse cytokine release syndrome panel (Biolegend, USA) was used to analyze serum cytokine levels of mice. Procedures were carried out as described by the manufacturer.
Hematopoietic stem cell co-cultures
Human CD34+ bone-marrow or cord blood-derived hematopoietic stem cells (HSC) were acquired from Stemcell Technologies. Healthy human bone marrow samples (HD) were obtained from patients undergoing hip replacement surgery at the University Hospital of the LMU, Munich. All cells were collected after informed consent in accordance with the Declaration of Helsinki. HSC were thawed in a pre-warmed water bath at 37°C. Directly after thawing, cells were expanded using StemSpan II Medium (Stemcell Technologies, Canada), supplemented with serum-free nutrient supply and UM729 small molecule inhibitor. Flow cytometric analysis of HSC or co-cultures of HSC and CAR-T cells were conducted after a total expansion phase of 7 days. Fresh expansion medium was added on day 3. 30,000 HSC were co-cultured with T cells at indicated E:T ratios for 24 hours prior to flow cytometric analysis. Co-cultures of HD and T cells were performed as the co-cultures of primary human AML blasts (see above).
Generation of iPSC-derived human microglia like cells (iMGL)
Human iMGL were generated as previously described49,50. In brief, human iPSC cell lines were differentiated to hematopoietic progenitors (iHPCs) using STEMdiff Hematopoietic Kit (Stemcell Technologies, Canada). Following successful development to iHPC, cells were grown in serum-free iMGL-differentiation medium, containing CSF-1, IL-34 and TGFβ for at least 12 days. Cells were then harvested and used for in vitro co-cultures with CAR-T cells. Co-cultures of iMGL and CAR-T cells were carried out as described above.
Software and statistical analysis
Flow cytometric data was obtained in the BD FACSDiva™ or Beckman Coulter software. G*Power software v3.1 was used to calculate group size of animal experiments. Luminescence and absorbance was measured with Mithras Reader using MicroWin2000 software. Flow cytometric data were analyzed using FlowJo V10.3 to V10.8.1 software. ImageJ/Fiji and Imaris (Bitplane AG) were used for analysis of TPLSM images. Radiance calculation of BLI images was performed using Living Image 4.4 (PerkinElmer, USA). All statistical analyses were performed using GraphPad Prism software V9.2.0 to V9.5.0 (San Diego, CA, USA).
Extended Data
Extended Data Figure 1. Summary of the cross-organ off-target transcriptomic atlas (COOTA).
(a) Top 100 overexpressed genes in AML HSPC (left) and healthy T cells (right) from differential expression analysis. Normalized expression values were logarithmized and scaled to unit variance. (b) Overview of 11 scRNA-seq datasets of various healthy tissues used to quantify off-target antigen expression. (c) UMAP plots of 11 scRNA-seq datasets with colors highlighting clustering into respective cell types. Cell annotations were provided by the authors of the respective studies. (d) Current CAR targets in AML were cross-referenced to filters used for the single cell-based target screening approach. OE HSC-/Prog-like: overexpressed on HSC-/Prog-like cells with log fold change > 2 and FDR-adjusted p ≤ 0.01, using a t-test with overestimated variance. Red cross: Antigen did not fulfill the respective threshold or criteria. Green check: Thresholds or criteria were passed.
Extended Data Figure 2. CSF1R and CD86 are consistently expressed across multiple patients with differing molecular subtypes.
(a, c) Amount of malignant and normal cells per AML patient of van Galen et al. (a) or Petti et al. (c). (b, d) Percentage of malignant and normal cells expressing target genes CSF1R, CD86 and reference genes CD123, CD33 for each sequenced AML patient of van Galen et al. (b) or Petti et al. (d). (e) Data from van Galen et al.21 was used as a reference (top left) to map cells from Petti et al.28 (top right) using scANVI29. UMAP representation showing the mapped query and reference data together (bottom). (f) Computational CAR target antigen identification using the mapped dataset of Petti et al. by stepwise evaluation against a set of criteria for an ideal and effective CAR target antigen. The decreasing number of screened AML target genes are shown on the bottom. CSPA: Cell surface protein atlas; HPA: Human protein atlas. (g) Volcano plot showing CD86 and CSF1R target genes with their respective FDR-adjusted log10 p-value and log2 fold changes from differential expression analysis between malignant HSPC-like and healthy HSPC using a t-test with overestimated variance.
Extended Data Figure 3. mCSF1R CART do not persist in immunocompetent mice.
(a) Construct design of mCSF1R or mEpCAM CART or mCherry T cells. (b) Representative histograms of mCSF1R or mEpCAM expression on J774A.1 cells. Staining was carried out twice. (c) Summary of treatment schedule for in vivo toxicity assessment of mCSF1R CART. (d) Mean weight curves of mice treated with 3 x 106 mCSF1R CART or mCherry T cells. n = 10 mice per group. Error bars indicate s.e.m. (e) Quantification of tissue-resident CD11b+ cells (left) or mCherry+ T cells of parent population (right, parent population: CD3 and CD8 positive cells) in different organs by flow cytometry. Data are mean ± s.e.m. of n = 10 mice. (d, e) Statistical significance was calculated using two-way ANOVA with Šidák multiple comparison correction. (f) Scheme of treatment schedule for in vivo toxicity assessment of mCSF1R CART. WBI, whole body irradiation. Mice were treated with 3 x 106 mCSF1R CART or mCherry T cells per mouse. 6 x 106 mEpCAM CART were transferred as a positive control. (g) Serum levels of indicated markers one (d1) or seven (d7) days after ACT. Depicted is the fold change of serum levels of the indicated groups from the PBS-treated control group. CRP, C-reactive protein; AST, aspartate aminotransferase; ALT, alanine aminotransferase; GLDH, glutamate dehydrogenase; γ-GT, gamma-glutamyl-transferase; LDH, lactate dehydrogenase. n = 3 mice per group. Statistically significant increases in the serum of mEpCAM CART treated mice compared to mCSF1R CART or control-treated mice at day 7 were observed for GLDH (p < 0.0001). (h) Histopathological analysis of indicated organs after treatment with mEpCAM or mCSF1R CART or mCherry T cells. Representative images of n = 3 mice per group. Signs of organ damage are indicated in the picture: black arrowhead: thickening of alveolar epithel. (i) Representative maximum intensity projection of microglia (green) and macrophages (green) in CX3CR1-GFP mice on days 0 (baseline), 4, 7, 10, 14, 21 and 28 after intravenous injection of 107 mCSF1R CART (red). Depth from brain surface: 0-100 μm. Scale bars as depicted: 50 μm and 20 μm. mCSF1R CART: i.c. injection, n = 5 mice; i.v. injection, n = 3 mice; mCherry T cells: i.c./ i.v. injection = 2 mice.
Extended Data Figure 4. Assessment of different hCSF1R CAR constructs in vitro.
(a) Construct design of all anti-human constructs used throughout the course of the study. (b) Representative flow cytometric images of construct expression on primary human T cells. (c) Activation of different hCSF1R CART after incubation with plate-bound hCSF1R protein quantified by flow cytometry. (d) T cells expressing different hCSF1R CAR constructs were co-cultured with luc+ target antigen expressing AML tumor cell lines or antigen negative NALM-6 control cells for 48 hours at the indicated E:T ratios. Cell lysis was quantified by BLI. (e) Proliferation dye-labeled hCSF1R CART were co-cultured with respective cell lines for 4 or 7 days at a E:T ratio of 0.5:1. Proliferation was subsequently assessed by trace dilution. One representative image of three different donors is shown. (f) Bead quantified T cell numbers. (g) Secretion of IFNγ by T cells transduced with different hCSF1R CAR constructs after co-culture with AML cell lines. (h) Secretion of IFNγ (left) or IL-2 (right) of hCSF1R, CD86 or control CART in co-culture with AML cell lines. (c, d, f - h) Data are mean ± s.e.m. of three independent donors.
LTR, long terminal repeat; scFv, single chain fragment variable; TM, transmembrane, IC, intracellular, ED, extracellular domain, CTRL-transduced, control-transduced.
Extended Data Figure 5. CSF1R is highly expressed on primary AML blasts.
(a) Representative histograms of CSF1R expression on AML cell lines after freeze and thaw cycles. (b, c) Schematic of culture methods used to cultivate primary AML samples throughout the course of the study. (d) Expression of CSF1R on primary AML samples after 24 to 48 hours of culture in cytokine rich medium. Left: Percentage positive cells gated to isotype. Each dot represents different primary AML samples. Right: Representative flow cytometric images from three different AML samples. Data are mean ± s.e.m. from six different donors. (e) Gating strategy to identify CD34+ CD38± malignant HSPC. (f, g) Expression of target antigens (CSF1R, f; CD86, g) on malignant HPC (top) and HSC (bottom). (h) Expression of CSF1R on PDX-388 sample at indicated time points after thawing. (i) IHC staining of human CSF1R in the bone marrow of control-treated PDX-388-bearing mice. Shown are two representative pictures (right, left) in two different magnifications (top 10x, bottom 20x). (j) Left: CD33 CART used for i.v. injection into PDX-388-bearing mice (Fig. 5 k - m) were co-cultured ex vivo with luc+ Mv4-11 tumor cells for 48 hours at indicated E:T ratios. Specific lysis was quantified by BLI. Shown is mean ± s.e.m. of three biological replicates. Experiment was carried out twice. (k) Representative flow cytometric image of percentage of CD3 positive T cells (left) and percentage of CAR (c-myc) positive T cells (right) in the blood of PDX-388-bearing mice. (l, m) Ex vivo CD33 expression on PDX-388 AML blasts in the bone marrow after treatment with CD33 CART or CTRL-transduced T cells (CD19 CART) measured by flow cytometry. Depicted are representative histograms (l) or the change of CD33-PE-Cy5 MFI in CD33 CART treated mice compared to CTRL-transduced treated mice (m). (m) Data are mean ± s.e.m. from n = 3 mice injected with CD33 CART compared to CTRL-transduced mouse. (k - m) n = 3 mice injected with CD33 CART), n = 1 mouse injected with CTRL-transuced T cells.
Extended Data Figure 6. hCSF1R CAR T cells are effective in vivo.
(a) Treatment scheme used for PDX-372 model. (b - d) BLI images (b), BLI quantification of tumor-burden (c) and survival curves (d) of PDX-372 tumor-bearing mice injected with 2 x 106 hCSF1R, CD33 CART or control-transduced T cells (n = 5 mice per group). (b) White cross, censored mice; red cross, mice succumbed to disease. (e) IHC staining of human CSF1R in the bone marrow of control-treated PDX-372-bearing mice. Left: IHC for human CSF1R. Right: Isotype (top) and detection system control (bottom) for CSF1R IHC staining. (f) Schematic of treatment scheme used for OCI-AML3 cell line xenograft model. (g, h) BLI images (g) and survival curves (h) of OCI-AML3 tumor-bearing mice injected with 6 x 106 hCSF1R CART or control-transduced T cells (n = 3 - 4 mice per group). (g) Red cross, mice succumbed to disease. (a - h) Statistical significance was calculated using two-way ANOVA with Sidak multiple comparison correction. (i) log2 expression of CSF1R and CD86 target antigens or CD123 and CD33 controls in bulk RNA-sequencing dataset of the Leukemia MILE study (n = 615 different patients). HBM, healthy bone marrow. Data was obtained from bloodspot.eu. Dashed line represents the median, dotted line the interquartile ranges. Statistical significance was calculated using ordinary one-way ANOVA with Sidak multiple comparison correction. (j) Simple linear regression analysis of in vitro lysis of CAR T cells and target antigen density of the indicated AML cell line measured by flow cytometry. r = spearman correlation coefficient, p = p-value. Three independent antigen density measurements were used to perform regression analysis. For Kaplan-Meier-Curves, statistical significance was calculated with log-rank test.
Supplementary Material
Acknowledgements
This study was supported by the Förderprogramm für Forschung und Lehre der Medizinischen Fakultät der LMU (grant number 1138 to A.G.), the Bavarian Cancer Research Center (BZKF) (A.G.), the Deutsche Forschungsgemeinschaft (DFG, grant number GO 3823/1-1 to A.G.; grant number: KO5055-2-1 and KO5055/3-1 to S.K.), the international doctoral program ‘i-Target: immunotargeting of cancer’ (funded by the Elite Network of Bavaria; to S. K. and S.E.), the Melanoma Research Alliance (grant number 409510 to S. K.), Marie Sklodowska-Curie Training Network for the Immunotherapy of Cancer (IMMUTRAIN) (funded by the Horizon 2020 programme of the European Union; to S.E. and S. K.), Marie Sklodowska-Curie Training Network for Optimizing Adoptive T Cell Therapy of Cancer (funded by the Horizon 2020 programme of the European Union; grant 955575 to S. K.), Else Kröner-Fresenius-Stiftung (to S. K.), German Cancer Aid (to S.K.), the Wilhelm-Sander-Stiftung (to S. K.), Ernst Jung Stiftung (to S. K.), Institutional Strategy LMUexcellent of LMU Munich (within the framework of the German Excellence Initiative; to S.E. and S. K.), the Go-Bio-Initiative (to S. K.), the m4-Award of the Bavarian Ministry for Economical Affairs (to S. E. and S. K.), Bundesministerium für Bildung und Forschung (to S.E. and S. K.), European Research Council (Starting Grant 756017 and PoC Grant 101100460 to S. K.), Deutsche Forschungsgemeinschaft (DFG; to S. K.), by the SFB-TRR 338/1 2021–452881907 (to S. K.), Fritz-Bender Foundation (to S. K.), Deutsche José Carreras Leukämie Stiftung (to S. K.), the Bayerische Forschungsstiftung (BAYCELLator to S.K) and Hector Foundation (to S. K.), Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy – ID 390857198, to E. B. and D. P.) and Alzheimer’s Association (to D. P.). M.T. is funded by the Volkswagen Foundation (project OntoTime). S.S. was supported by the Else Kröner-Fresenius Clinician Scientist Program Cancer Immunotherapy, the Munich Clinician Scientist Program (MCSP) and the DKTK School of Oncology. C.M. has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement number 866411). Cytometry data were obtained in the Core Facility Flow Cytometry of the University Hospital, LMU Munich using a BD LSRFortessa™ II or Beckman Coulter CytoFLEX.
In vivo imaging device was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – INST 409/231-1.
We acknowledge Life Science Editors for their editing services. Figure illustrations were created with Biorender.com under a paid subscription.
Footnotes
Author contributions
A.G., M.T., R.G., S.L., L.R., V.I., D.B., M.B., S.D., K.M., T.X. D.D., F.M., S. R., A. S., H.S., Ö.U., V.K., C.T., E.C., S.N., T.S, T.L, S.S., P.J.M., J.D., M.S., B.L.C., R.B., N.R., F.R., M.N., performed or assisted with the experiments, A.G., M.T., R.G., S.L., L.R., S. D., K. M., T.X., E.B., and T.H. analyzed the data and supported the project. S.R., B.V., J.W., F.M., W.F., C.K., D.P., I.J. and L.v.B. provided critical reagents, S.K., C.M. and S.E. supervised the project and acquired the funding. A.G., M.T., S.L., T.H., D.M., D.P., L.v.B, S.E, M. S., C.M. and S.K designed the experiments. A.G., M.T., C.M. and S. K. wrote the manuscript.
All authors critically read and approved the final manuscript.
Competing interests
Parts of this work have been performed for the doctoral thesis of M.T., R.G., D.D. and S.D., at Technische Universität München and Ludwig -Maximilians-Universität München. A.G., M.T., R.G., S.L., S.E., C.M. and S.K. submitted patent applications related to this work filed by the Ludwig-Maximilians-Universität München the University Hospital of the LMU Munich or the Helmholtz Centre Munich. S.K. has received honoraria from TCR2 Inc, Novartis, BMS and GSK. S.K. and S.E. are inventors of several patents in the field of immuno-oncology. S.K. and S.E. received license fees from TCR2 Inc and Carina Biotech. A.G. received research support from Tabby Therapeutics for work unrelated to the manuscript. S.K. and S.E. received research support from TCR2 Inc. and Arcus Bioscience for work unrelated to the manuscript.
F. M. received support for meeting attendance from Servier, AbbVie, Incyte, Gilead, Jazz Pharmaceuticals, Novartis, Teva, Pfizer, Amgen; received support for medical writing from Servier; received research grant from Apis Technologies, Daiichi Sankyo and received speaker honoraria from Servier, Jazz Pharmaceuticals and AbbVie.
W.F. received payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Novartis, Abbvie, Pfizer, Amgen. W.F. received support for attending meetings and/or travel: Amgen, Gilead, Jazz Pharmaceuticals, Servier, Daiichi Sankyo. W.F. participates on a Data Safety Monitoring Board or Advisory Board: Amgen, ARIAD/Incyte, Pfizer, Novartis, Jazz Pharmaceuticals, Morphosys, Abbvie, Celgene, Stemline, Clinigen.
The remaining authors declare no competing interests.
Life Sciences Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
Data of publicly available scRNA-seq can be found via the accession numbers or the provided link of the respective studies: GSE11625621, https://zenodo.org/record/334598139, GSE7158527, https://www.gtexportal.org/home/datasets, GSE13435537, https://www.gutcellatlas.org/36, GSE13190728, GSE11546933, https://www.tissuestabilitycellatlas.org/31,GSE13610334, https://nupulmonary.org/resources/32, https://www.kidneycellatlas.org/29, https://www.synapse.org/#!Synapse:syn21041850/files/30, EGAS0000100292735.
Data of publicly available bulk sequencing data can be found via the accession numbers: E-MTAB-521482, E-MTAB-280183 via the Expression atlas (https://www.ebi.ac.uk/gxa/home). The surface-gene library was obtained by integrating publicly available data23–26 using OmniPath22 (https://omnipathdb.org/). Targets of FDA-approved drugs were obtained using DrugBank (https://go.drugbank.com/).
All reagents and biological material will be made available upon reasonable request to the authors given the agreement by the providing institution.
Code Availability
Python scripts for replicating the figures from the scRNA-seq as jupyter notebooks can be found in following Github repository: (https://github.com/marrlab/CAR_T_Targetidentification) 74.
Count matrices of processed scRNA-seq data will be made available upon reasonable request.
References
- 1.June CH, Sadelain M. Chimeric Antigen Receptor Therapy. New England Journal of Medicine. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maude SL, et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N Engl J Med. 2018;378:439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster SJ, et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N Engl J Med. 2019;380:45–56. doi: 10.1056/NEJMoa1804980. [DOI] [PubMed] [Google Scholar]
- 4.Raje N, et al. Anti-BCMA CAR T-Cell Therapy bb2121 in Relapsed or Refractory Multiple Myeloma. N Engl J Med. 2019;380:1726–1737. doi: 10.1056/NEJMoa1817226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lesch S, et al. Determinants of response and resistance to CAR T cell therapy. Semin Cancer Biol. 2020;65:80–90. doi: 10.1016/j.semcancer.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Lamers CH, et al. Treatment of metastatic renal cell carcinoma with CAIX CAR-engineered T cells: clinical evaluation and management of on-target toxicity. Mol Ther. 2013;21:904–912. doi: 10.1038/mt.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummins KD, Gill S. Will CAR T cell therapy have a role in AML? Promises and pitfalls. Semin Hematol. 2019;56:155–163. doi: 10.1053/j.seminhematol.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thol F, Ganser A. Treatment of Relapsed Acute Myeloid Leukemia. Curr Treat Options Oncol. 2020;21:66. doi: 10.1007/s11864-020-00765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummins KD, Gill S. Chimeric antigen receptor T-cell therapy for acute myeloid leukemia: how close to reality? Haematologica. 2019;104:1302–1308. doi: 10.3324/haematol.2018.208751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacKay M, et al. The therapeutic landscape for cells engineered with chimeric antigen receptors. Nat Biotechnol. 2020;38:233–244. doi: 10.1038/s41587-019-0329-2. [DOI] [PubMed] [Google Scholar]
- 12.Tambaro FP, et al. Autologous CD33-CAR-T cells for treatment of relapsed/refractory acute myelogenous leukemia. Leukemia. 2021;35:3282–3286. doi: 10.1038/s41375-021-01232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauer T, et al. CD70-specific CAR T-cells have potent activity against Acute Myeloid Leukemia (AML) without HSC toxicity. Blood. 2021 doi: 10.1182/blood.2020008221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jetani H, et al. Siglec-6 is a novel target for CAR T-cell therapy in acute myeloid leukemia (AML) Blood. 2021 doi: 10.1182/blood.2020009192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myburgh R, et al. Anti-human CD117 CAR T-cells efficiently eliminate healthy and malignant CD117-expressing hematopoietic cells. Leukemia. 2020;34:2688–2703. doi: 10.1038/s41375-020-0818-9. [DOI] [PubMed] [Google Scholar]
- 16.Tashiro H, et al. Treatment of Acute Myeloid Leukemia with T Cells Expressing Chimeric Antigen Receptors Directed to C-type Lectin-like Molecule 1. Mol Ther. 2017;25:2202–2213. doi: 10.1016/j.ymthe.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casucci M, et al. CD44v6-targeted T cells mediate potent antitumor effects against acute myeloid leukemia and multiple myeloma. Blood. 2013;122:3461–3472. doi: 10.1182/blood-2013-04-493361. [DOI] [PubMed] [Google Scholar]
- 18.Perna F, et al. Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML. Cancer Cell. 2017;32:506–519.:e505. doi: 10.1016/j.ccell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suva ML, Tirosh I. Single-Cell RNA Sequencing in Cancer: Lessons Learned and Emerging Challenges. Mol Cell. 2019;75:7–12. doi: 10.1016/j.molcel.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Jing Y, et al. Expression of chimeric antigen receptor therapy targets detected by single-cell sequencing of normal cells may contribute to off-tumor toxicity. Cancer Cell. 2021;39:1558–1559. doi: 10.1016/j.ccell.2021.09.016. [DOI] [PubMed] [Google Scholar]
- 21.van Galen P, et al. Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell. 2019;176:1265–1281.:e1224. doi: 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turei D, et al. Integrated intra-and intercellular signaling knowledge for multicellular omics analysis. Mol Syst Biol. 2021;17:e9923. doi: 10.15252/msb.20209923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bausch-Fluck D, et al. A mass spectrometric-derived cell surface protein atlas. PLoS One. 2015;10:e0121314. doi: 10.1371/journal.pone.0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bausch-Fluck D, et al. The in silico human surfaceome. Proc Natl Acad Sci U S A. 2018;115:E10988–E10997. doi: 10.1073/pnas.1808790115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multisubunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 26.Thul PJ, et al. A subcellular map of the human proteome. Science. 2017;356 doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 27.Habib N, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim N, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020;11:2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart BJ, et al. Spatiotemporal immune zonation of the human kidney. Science. 2019;365:1461–1466. doi: 10.1126/science.aat5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Travaglini KJ, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. 2020;587:619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madissoon E, et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 2019;21:1. doi: 10.1186/s13059-019-1906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reyfman PA, et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am J Respir Crit Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacParland SA, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramachandran P, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng JB, et al. Transcriptional Programming of Normal and Inflamed Human Epidermis at Single-Cell Resolution. Cell Rep. 2018;25:871–883. doi: 10.1016/j.celrep.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James KR, et al. Distinct microbial and immune niches of the human colon. Nat Immunol. 2020;21:343–353. doi: 10.1038/s41590-020-0602-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han X, et al. Construction of a human cell landscape at single-cell level. Nature. 2020;581:303–309. doi: 10.1038/s41586-020-2157-4. [DOI] [PubMed] [Google Scholar]
- 38.Wishart DS, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petti AA, et al. A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat Commun. 2019;10:3660. doi: 10.1038/s41467-019-11591-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu C, et al. Probabilistic harmonization and annotation of single-cell transcriptomics data with deep generative models. Mol Syst Biol. 2021;17:e9620. doi: 10.15252/msb.20209620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurga AM, Paleczna M, Kuter KZ. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front Cell Neurosci. 2020;14:198. doi: 10.3389/fncel.2020.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erblich B, Zhu L, Etgen AM, Dobrenis K, Pollard JW. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One. 2011;6:e26317. doi: 10.1371/journal.pone.0026317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chihara T, et al. IL-34 and M-CSF share the receptor Fms but are not identical in biological activity and signal activation. Cell Death Differ. 2010;17:1917–1927. doi: 10.1038/cdd.2010.60. [DOI] [PubMed] [Google Scholar]
- 44.Qin D, et al. Potential lung attack and lethality generated by EpCAM-specific CAR-T cells in immunocompetent mouse models. Oncoimmunology. 2020;9:1806009. doi: 10.1080/2162402X.2020.1806009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nayak D, Roth TL, McGavern DB. Microglia development and function. Annu Rev Immunol. 2014;32:367–402. doi: 10.1146/annurev-immunol-032713-120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards DKt, et al. CSF1R inhibitors exhibit antitumor activity in acute myeloid leukemia by blocking paracrine signals from support cells. Blood. 2019;133:588–599. doi: 10.1182/blood-2018-03-838946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pabst C, et al. Identification of small molecules that support human leukemia stem cell activity ex vivo. Nat Methods. 2014;11:436–442. doi: 10.1038/nmeth.2847. [DOI] [PubMed] [Google Scholar]
- 48.Vick B, et al. An advanced preclinical mouse model for acute myeloid leukemia using patients’ cells of various genetic subgroups and in vivo bioluminescence imaging. PLoS One. 2015;10:e0120925. doi: 10.1371/journal.pone.0120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McQuade A, et al. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol Neurodegener. 2018;13:67. doi: 10.1186/s13024-018-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reifschneider A, et al. Loss of TREM2 rescues hyperactivation of microglia, but not lysosomal deficits and neurotoxicity in models of progranulin deficiency. EMBO J. 2022;41:e109108. doi: 10.15252/embj.2021109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sletta KY, Castells O, Gjertsen BT. Colony Stimulating Factor 1 Receptor in Acute Myeloid Leukemia. Front Oncol. 2021;11:654817. doi: 10.3389/fonc.2021.654817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haque A, Engel J, Teichmann SA, Lonnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura H, et al. Expression of functional B7-H2 and B7.2 costimulatory molecules and their prognostic implications in de novo acute myeloid leukemia. Clin Cancer Res. 2005;11:5708–5717. doi: 10.1158/1078-0432.CCR-04-2672. [DOI] [PubMed] [Google Scholar]
- 54.Re F, et al. Expression of CD86 in acute myelogenous leukemia is a marker of dendritic/monocytic lineage. Exp Hematol. 2002;30:126–134. doi: 10.1016/s0301-472x(01)00768-8. [DOI] [PubMed] [Google Scholar]
- 55.Zheng Z, et al. Expression patterns of costimulatory molecules on cells derived from human hematological malignancies. J Exp Clin Cancer Res. 1998;17:251–258. [PubMed] [Google Scholar]
- 56.Gavile CM, et al. CD86 regulates myeloma cell survival. Blood Adv. 2017;1:2307–2319. doi: 10.1182/bloodadvances.2017011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sedek L, et al. Differential expression of CD73 CD86and CD304 in normal vs.leukemic B-cell precursors and their utility as stable minimal residual disease markers in childhood B-cell precursor acute lymphoblastic leukemia. J Immunol Methods. 2019;475:112429. doi: 10.1016/j.jim.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 58.Guinan EC, Gribben JG, Boussiotis VA, Freeman GJ, Nadler LM. Pivotal role of the B7:CD28 pathway in transplantation tolerance and tumor immunity. Blood. 1994;84:3261–3282. [PubMed] [Google Scholar]
- 59.Zhou LJ, Tedder TF. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Proc Natl Acad Sci U S A. 1996;93:2588–2592. doi: 10.1073/pnas.93.6.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smyth C, et al. Identification of a dynamic intracellular reservoir of CD86 protein in peripheral blood monocytes that is not associated with the Golgi complex. J Immunol. 1998;160:5390–5396. [PubMed] [Google Scholar]
- 61.Blair HA, Deeks ED. Abatacept: A Review in Rheumatoid Arthritis. Drugs. 2017;77:1221–1233. doi: 10.1007/s40265-017-0775-4. [DOI] [PubMed] [Google Scholar]
- 62.Adusumilli PS, et al. A Phase I Trial of Regional Mesothelin-Targeted CAR T-cell Therapy in Patients with Malignant Pleural Disease, in Combination with the Anti-PD-1 Agent Pembrolizumab. Cancer Discov. 2021;11:2748–2763. doi: 10.1158/2159-8290.CD-21-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parker KR, et al. Single-Cell Analyses Identify Brain Mural Cells Expressing CD19 as Potential Off-Tumor Targets for CAR-T Immunotherapies. Cell. 2020;183:126–142.:e117. doi: 10.1016/j.cell.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Majzner RG, Mackall CL. Clinical lessons learned from the first leg of the CAR T cell journey. Nat Med. 2019;25:1341–1355. doi: 10.1038/s41591-019-0564-6. [DOI] [PubMed] [Google Scholar]
- 65.Jetani H, et al. CAR T-cells targeting FLT3 have potent activity against FLT3-ITD+ AML and act synergistically with the FLT3-inhibitor crenolanib. Leukemia. 2018;32:1168–1179. doi: 10.1038/s41375-018-0009-0. [DOI] [PubMed] [Google Scholar]
- 66.Griciuc A, et al. TREM2 Acts Downstream of CD33 in Modulating Microglial Pathology in Alzheimer’s Disease. Neuron. 2019;103:820–835.:e827. doi: 10.1016/j.neuron.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cassier PA, et al. CSF1R inhibition with emactuzumab in locally advanced diffuse-type tenosynovial giant cell tumours of the soft tissue: a dose-escalation and dose-expansion phase 1 study. Lancet Oncol. 2015;16:949–956. doi: 10.1016/S1470-2045(15)00132-1. [DOI] [PubMed] [Google Scholar]
- 68.O’Rourke DM, et al. A single dose of peripherally infused EGFRvIII-directed CAR-T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gust J, et al. Endothelial Activation and Blood-Brain Barrier Disruption in Neurotoxicity after Adoptive Immunotherapy with CD19 CAR-T Cells. Cancer Discov. 2017;7:1404–1419. doi: 10.1158/2159-8290.CD-17-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sterner RM, et al. GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances CAR-T cell function in xenografts. Blood. 2019;133:697–709. doi: 10.1182/blood-2018-10-881722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan AHJ, Vinanica N, Campana D. Chimeric antigen receptor-T cells with cytokine neutralizing capacity. Blood Adv. 2020;4:1419–1431. doi: 10.1182/bloodadvances.2019001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Virshup I, Rybakov S, Theis FJ, Angerer P, Wolf FA. anndata: Annotated data. bioRxiv. 2021:2021.2012.2016.473007. doi: 10.1101/2021.12.16.473007. [DOI] [Google Scholar]
- 74.Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lun AT, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. 2018. arXiv:1802.03426. https://ui.adsabs.harvard.edu/abs/2018arXiv180203426M .
- 79.Fischer DS, et al. Sfaira accelerates data and model reuse in single cell genomics. Genome Biol. 2021;22:248. doi: 10.1186/s13059-021-02452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muus C, et al. Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat Med. 2021;27:546–559. doi: 10.1038/s41591-020-01227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Polanski K, et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2020;36:964–965. doi: 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Consortium GT Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Papatheodorou I, et al. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 2020;48:D77–D83. doi: 10.1093/nar/gkz947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sandhofer N, et al. Dual PI3K/mTOR inhibition shows antileukemic activity in MLL-rearranged acute myeloid leukemia. Leukemia. 2015;29:828–838. doi: 10.1038/leu.2014.305. [DOI] [PubMed] [Google Scholar]
- 86.Sudo T, et al. Functional hierarchy of c-kit and c-fms in intramarrow production of CFU-M. Oncogene. 1995;11:2469–2476. [PubMed] [Google Scholar]
- 87.Benmebarek MR, et al. A modular and controllable T cell therapy platform for acute myeloid leukemia. Leukemia. 2021;35:2243–2257. doi: 10.1038/s41375-020-01109-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Gosliga D, et al. Establishing long-term cultures with self-renewing acute myeloid leukemia stem/progenitor cells. Exp Hematol. 2007;35:1538–1549. doi: 10.1016/j.exphem.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 89.Herrmann M, et al. Bifunctional PD-1 x alphaCD3 x alphaCD33 fusion protein reverses adaptive immune escape in acute myeloid leukemia. Blood. 2018;132:2484–2494. doi: 10.1182/blood-2018-05-849802. [DOI] [PubMed] [Google Scholar]
- 90.Lesch S, et al. T cells armed with C-X-C chemokine receptor type 6 enhance adoptive cell therapy for pancreatic tumours. Nature Biomedical Engineering. 2021 doi: 10.1038/s41551-021-00737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ghani K, et al. Efficient human hematopoietic cell transduction using RD114-and GALV-pseudotyped retroviral vectors produced in suspension and serum-free media. Hum Gene Ther. 2009;20:966–974. doi: 10.1089/hum.2009.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mulazzani M, et al. Long-term in vivo microscopy of CAR T cell dynamics during eradication of CNS lymphoma in mice. Proc Natl Acad Sci U S A. 2019;116:24275–24284. doi: 10.1073/pnas.1903854116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data of publicly available scRNA-seq can be found via the accession numbers or the provided link of the respective studies: GSE11625621, https://zenodo.org/record/334598139, GSE7158527, https://www.gtexportal.org/home/datasets, GSE13435537, https://www.gutcellatlas.org/36, GSE13190728, GSE11546933, https://www.tissuestabilitycellatlas.org/31,GSE13610334, https://nupulmonary.org/resources/32, https://www.kidneycellatlas.org/29, https://www.synapse.org/#!Synapse:syn21041850/files/30, EGAS0000100292735.
Data of publicly available bulk sequencing data can be found via the accession numbers: E-MTAB-521482, E-MTAB-280183 via the Expression atlas (https://www.ebi.ac.uk/gxa/home). The surface-gene library was obtained by integrating publicly available data23–26 using OmniPath22 (https://omnipathdb.org/). Targets of FDA-approved drugs were obtained using DrugBank (https://go.drugbank.com/).
All reagents and biological material will be made available upon reasonable request to the authors given the agreement by the providing institution.
Python scripts for replicating the figures from the scRNA-seq as jupyter notebooks can be found in following Github repository: (https://github.com/marrlab/CAR_T_Targetidentification) 74.
Count matrices of processed scRNA-seq data will be made available upon reasonable request.