Abstract
The mammalian heart is derived from multiple cell lineages; however, our understanding of when and how the diverse cardiac cell types arise is limited. We mapped the origin of the embryonic mouse heart at single-cell resolution using a combination of transcriptomic, imaging, and genetic lineage labelling approaches. This provided a transcriptional and anatomic definition of cardiac progenitor types. Furthermore, it revealed a cardiac progenitor pool that is anatomically and transcriptionally distinct from currently known cardiac progenitors. Besides contributing to cardiomyocytes, these cells also represent the earliest progenitor of the epicardium, a source of trophic factors and cells during cardiac development and injury. This study provides detailed insights into the formation of early cardiac cell types, with particular relevance to the development of cell-based cardiac regenerative therapies.
The heart is formed from several lineages, which give rise to the various cardiac cell types. The earliest mesodermal progenitors of cardiomyocytes emerge from the primitive streak, starting at approximately Embryonic Day (E)6.5 in the mouse (1–3). These cells migrate laterally across the egg cylinder to occupy an anterior position, initially forming the cardiac crescent before subsequently forming the linear heart tube (LHT) (4) (Fig. 1A). There are at least two distinct types of mesodermal cardiac progenitors, the First and Second Heart Fields, defined broadly on the basis of marker genes expressed in different but overlapping anatomical locations (5). FHF cells occupy a cranio-lateral region and, together with early cardiomyocytes, form the cardiac crescent. The SHF represents a wider domain, dorsal and medial to the cardiac crescent. FHF cells show limited proliferation, instead rapidly differentiating into cardiomyocytes in order to lay down a primitive contractile apparatus (6) (Fig. 1A). In contrast, SHF cells are more proliferative, contributing to the growth of the heart before the cells differentiate. Given the overlap in expression of FHF and SHF markers, particularly with SHF cells upregulating FHF markers as they differentiate into cardiomyocytes, there is some debate about whether FHF and SHF correspond to fundamentally distinct cell types, or whether they represent different states along a shared differentiation trajectory from a common progenitor towards cardiomyocytes.
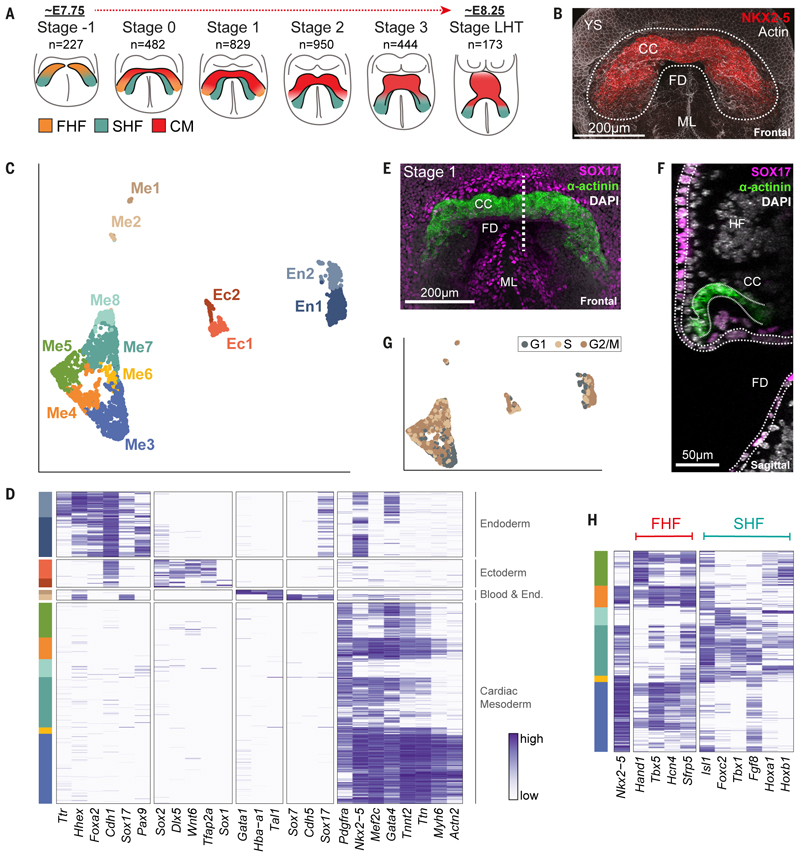
Fig. 1. Single-cell resolution spatiotemporal analysis of the forming murine heart.
A, Schematic of crescent and linear heart tube stages collected for scRNA-seq. n = number of single cells that passed quality control. FHF, first heart field; SHF, second heart field; CM, cardiomyocytes. B, Maximum intensity projection (MIP) of a stage 1 cardiac crescent region, marked by NKX2-5 protein expression (Movie S1). The dotted region highlights the area micro-dissected for collecting cells. CC, cardiac crescent; FD, foregut diverticulum; YS, yolk sac; ML, midline. C, UMAP plot of all cells that passed quality control (n = 3,105) computed from highly variable genes. Cells with similar transcriptional profiles were clustered into 12 different groups, as indicated by the different colors. D, Heatmap showing the expression of well-characterized marker genes. Cells (rows) are ordered first by cluster and then by hierarchical clustering. Each gene’s normalized log expression levels are regularized to be within [0,1]. E, MIP of representative embryo showing SOX17 and sarcomeric α-actinin protein (Movie S2). This serves as an example of the approach used to anatomically localize in the intact embryo the cell types identified in the scRNA-seq dataset. The dotted line represents the location of the section shown in F. F, Sagittal section of the embryo in E. Dotted lines outline the endoderm (SOX17+) and cardiac crescent (α-actinin+). See also Figure S3. HF, headfold. G, UMAP plot as in C but colored based on the inferred cell cycle stage of each cell. H, Heatmap of cardiac related clusters showing the expression of genes associated with the first (FHF) and second (SHF) heart fields.
Retrospective clonal analyses support the idea that there are two separate lineages that contribute to the cardiomyocytes of the heart. The ‘First Lineage’ contributes primarily to the atria and left ventricles, whereas the ‘Second Lineage’ contributes primarily to the atria, right ventricle, and outflow tract (5,7). These two lineages are assumed to correspond to the FHF and SHF respectively. However, it is unclear if the difference in fate of these two lineages is because of fundamental differences in their potential, or due only to differences in their embryonic location during early cardiac morphogenesis.
Cells from outside the traditional heart fields also contribute to the heart. Notably, the proepicardium gives rise to the epicardium, the outer layer of cells of the vertebrate heart. The epicardium generates several cardiac cell types including vascular smooth muscle cells and fibroblasts, in addition to providing important paracrine signals. It has attracted considerable attention as a possible source of cells and trophic factors in the context of repair after injury (8,9). The proepicardium is initially situated at the inflow end of the looping heart tube at approximately E9.0, but its embryonic origin remains unclear (10–12).
Single cell transcriptomics offers a powerful approach to characterize the various cell types of the embryonic heart and generate hypotheses about their lineal origins and fates. Several recent studies have used this approach to characterize cardiogenesis, focusing on sub-populations of cells isolated using pre-defined genes (13–16). Here, we perform a spatially resolved transcriptomic analysis of a micro-dissected region of the anterior embryo, collecting cells from the region corresponding to the embryonic heart over the time-course of cardiac crescent formation and LHT development.
Single-cell analysis of heart formation
We sampled cells, using manual micro-dissection to isolate the anterior cardiac region of mouse embryos, from early crescent to linear heart tube stages (Figure 1A, B, Figure S1A and Movie S1). Our collection was highly time-resolved, covering six defined stages spanning approximately 12 hours of development (roughly E7.75 to E8.25; Figure S1A) (17). Following quality control, the transcriptomes of 3,105 cells were retained for further analysis (Figure S2A; Methods). After identifying genes with highly variable expression, we performed unsupervised clustering to determine the different cell types within our dataset. This revealed a total of 12 distinct clusters (Figure 1C, D), representing mesodermal cell types (including cardiac related (Me3-8) that represented a majority (66.1%) of the cells (18), endothelial (Me2) and blood (Me1)) (13,19), overlying endoderm (including definitive (En1) and yolk sac (En2)) (20) and adjacent ectoderm (surface/amnion (Ec1) and neurectoderm (Ec2)) (21). We experimentally validated cluster identity using whole mount immunofluorescence for cluster-specific combinations of marker genes and single-cell resolution imaging (Figure 1E, F, Figures S3 and S4 and Movie S2). As a user-friendly community resource, we have created a web-interface to interrogate these data, accessible at https://marionilab.cruk.cam.ac.uk/heartAtlas/.
Me3 represented the most differentiated cardiomyocytes within our dataset, showing elevated expression of contractile related genes Myh6, Actn2 and Ttn (Figure 1D). Me3 started to emerge between stages 0 to 1 and increased over time (Figure S1B), consistent with the formation of cardiomyocytes and onset of contractile activity at stage 1 (6). Me3 also showed reduced proliferative capacity in comparison to other clusters, with a higher proportion of cells in G1 (Figure 1G and Figure S1C).
Me4 and Me6 were characterized by strong expression of cardiac progenitor markers, but intermediate to low levels of contractile genes (Figure 1D) and therefore represented differentiating cardiac progenitors. Me5, Me7, and Me8 represented the least differentiated populations based on the low expression of contractile and cardiac physiology related genes (Figure 1D).
Transcriptional and spatial identification of cardiac progenitor subtypes
To characterize these cardiac related clusters (Me3 to Me8) in more detail, we analyzed the expression of common FHF and SHF markers (Figure 1H). The cardiac progenitor marker Nkx2-5 was expressed in all these clusters except Me5 and Me8. Me4 had the highest combined expression of canonical FHF markers such as Tbx5, Nkx2-5, Hcn4 and Sfrp5, although some of these were also found in Me3, Me5 and to a lesser extent, Me7. Me6 and Me7 showed a distinct SHF-like signature (Figure 1H), although again, some SHF markers were also seen in Me3 and Me5.
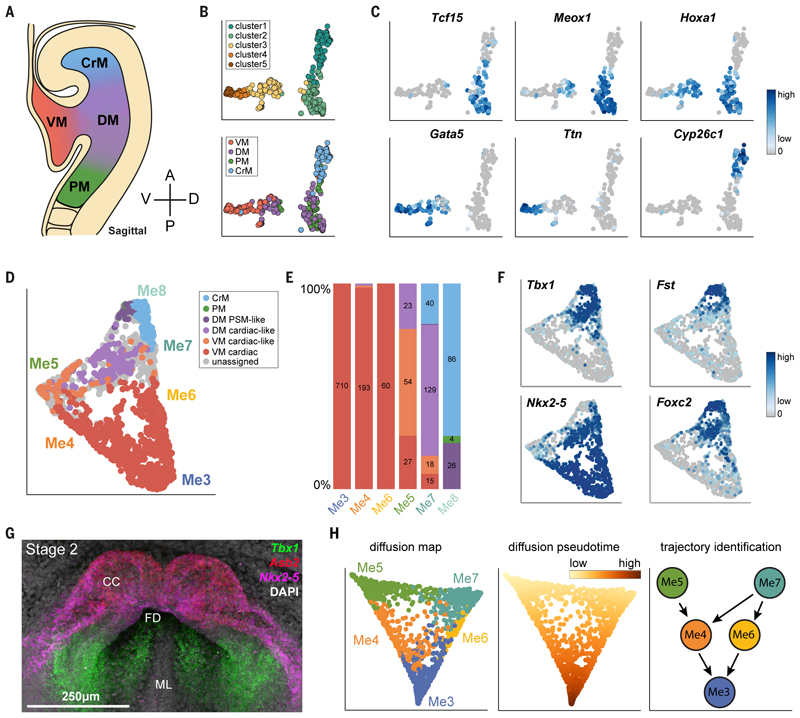
The FHF and SHF are defined not only based on markers but also on their anatomical location. Hence, we next confirmed our assignment of clusters to a FHF or SHF identity by generating a spatially resolved scRNA-seq reference dataset from four anatomically defined regions of mesoderm in stage 3 crescent embryos (~E8.0). We manually micro-dissected and sequenced cells from the ventral mesoderm containing the cardiac crescent (VM), abutting dorsal mesoderm (DM), cranial mesoderm (CrM), and paraxial mesoderm (PM) (Figure 2A). We generated an average of 70 high-quality single-cell transcriptomes for each of these reference regions, which together clustered into five different subpopulations (clusters 1 to 5) that were dispersed to varying degrees across the four regions (Figure 2B and Figure S5A). Cluster 1 expressed the cranial mesoderm marker Cyp26c1 (22) (Figure 2C) and was almost exclusively derived from this reference region. Cluster 2 represented paraxial mesoderm (Tcf15 and Meox1) (23,24) and was composed of cells from this region as well as from the dorsal mesoderm. Clusters 3, 4 and 5 expressed cardiac markers such as Ttn and Gata5 (25) and were made up predominantly of cells from the VM and DM (Figure 2B, C). VM cells could be categorized as more mature, differentiated cardiac cells (cluster 5), and less mature ‘cardiac-like’ cells (clusters 3 and 4) (Figure 2B and Figure S5A). Similarly, DM cells were split across ‘cardiac-like’ and ‘PM-like’ (clusters 3 and 2 respectively; Figure 2B). Hoxa1, which marks cells from the DM, highlights the pronounced transcriptional heterogeneity within this region, being split across clusters 2 and 3 (Figure 2B, C).
Fig. 2. Classification of cells from clusters Me3-Me8 based on an anatomically-resolved mesoderm reference dataset.
A, Schematic indicating the four regions of mesoderm micro-dissected to generate the reference dataset. Single cells from each region were isolated and used for scRNA-seq. B, UMAP plot of reference cells that pass quality control, colored by their anatomical origin (top) or clustering based on their transcriptional profiles (bottom). These cells were used to build a random forest classifier. C, Expression of genes that are important for the random forest, distinguishing the different types of cells. D, UMAP plot of clusters Me3-Me8 as in Figure 1C, but colored based on the class assigned by the random forest classifier. Cells in grey cannot be confidently assigned to a single class. E, Barplot indicating the proportion of cells in each cluster assigned to the different classes.
Numbers indicate the number of cells. F, UMAP plots showing the expression of genes used to identify clusters Me7 and Me8. G, Maximum intensity projection of a stage 2 crescent showing expression of Nkx2-5 and Tbx1 using Hybridization Chain Reaction (Supplementary Movie 3). H, Diffusion map of cells from the cardiac mesoderm meta-cluster (Me3-Me7, n = 1,772); the first two diffusion components are shown. On the left, cells are colored by their cluster annotation as in Fig. 1C. In the middle, the color gradient indicates diffusion pseudotime measurements for each cell. On the right, schematic diagram summarizing the trajectories identified, linking the progenitor clusters Me5 and Me7 to the mature cardiomyocytes of Me3 (see also Figure S9). Color scales in C and F represent log2 normalized counts.
To use these reference cells to guide spatial localization of the cells assigned to clusters Me3-8, we trained a random forest classifier using the combined information from the dissection labels and clustering analysis (Methods; Figure 2B and Figure S5A, B). Upon application of this classifier to the main dataset, the majority of cells in clusters Me3-6 were assigned to the VM (Figure 2D, E), which corresponds broadly to the cardiac crescent. Consistent with the differences in maturity observed between Me5 and Me3/4/6, the former matched ‘cardiac-like’ VM cells while the latter mapped to the more mature cardiac VM (Figure 2D, E). The majority of the cells from Me7 mapped to ‘cardiac-like’ DM reference cells whereas Me8 mapped predominantly to CrM but also PM-like DM (Figure 2D, E). In particular, although Me7 and Me8 were both characterized by high Tbx1 and Foxc2, they could be distinguished by expression of Nkx2-5 (high in Me7, low in Me8) and Fst (low in Me7, high in Me8) (Figure 2F, G, Figure S6-S8, Movie S3-S5 and S7). This allowed us to localize Me7 to areas of mesoderm medial and dorsal to the crescent, consistent with it representing cells from the SHF.
Intermediate states in cardiomyocyte differentiation are spatially localized across development
We next employed a diffusion map approach to infer differentiation trajectories between clusters Me3-7, excluding Me8 due to its CrM and PM-like identity. This analysis revealed two distinct trajectories, both leading to the differentiated cardiomyocytes of Me3 (Figure 2H and Figure S9). One trajectory connected Me7 to Me3 via two distinct intermediates, Me6 and Me4. The second trajectory linked Me5 to Me3 via Me4 (Figure 2H and Figure S9A-C).
The trajectory linking Me5, Me4 and Me3 is consistent with a FHF-like differentiation trajectory. Although Me4 had a broad FHF transcriptional signature, Me5 did not fit with straightforward categorization as it expressed some FHF markers (Hand1 and Tbx5), but significantly lower levels of the canonical cardiac progenitor marker Nkx2-5 (Figure 1D, H). Therefore, to further characterize these clusters, we determined their precise embryonic locations in 3-D at different stages using the combinatorial expression of multiple markers (Figure S10) and single-cell resolution volume imaging.
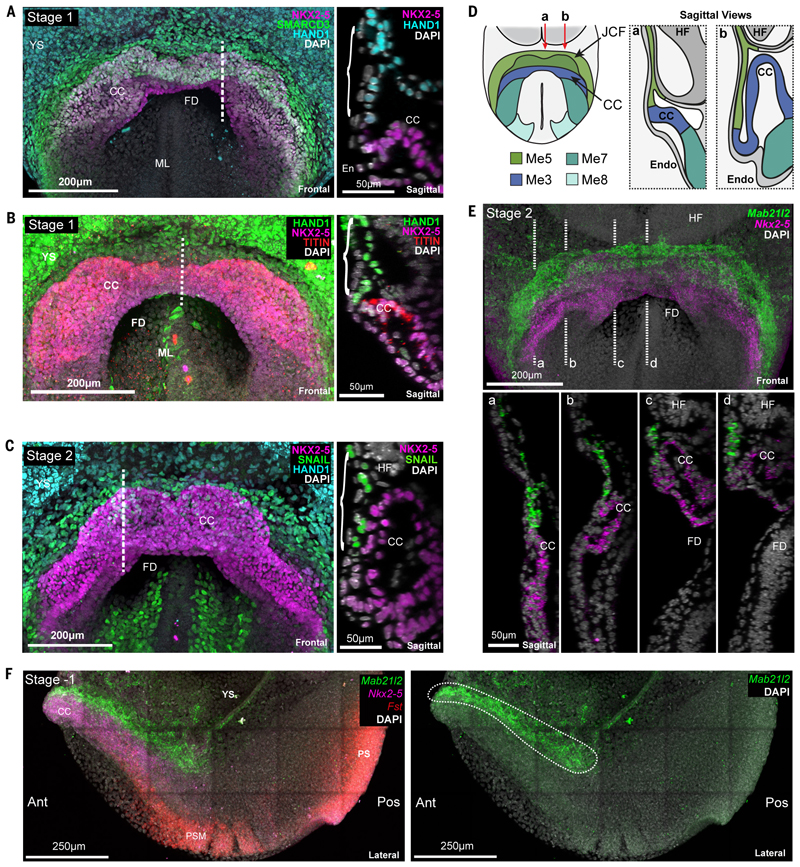
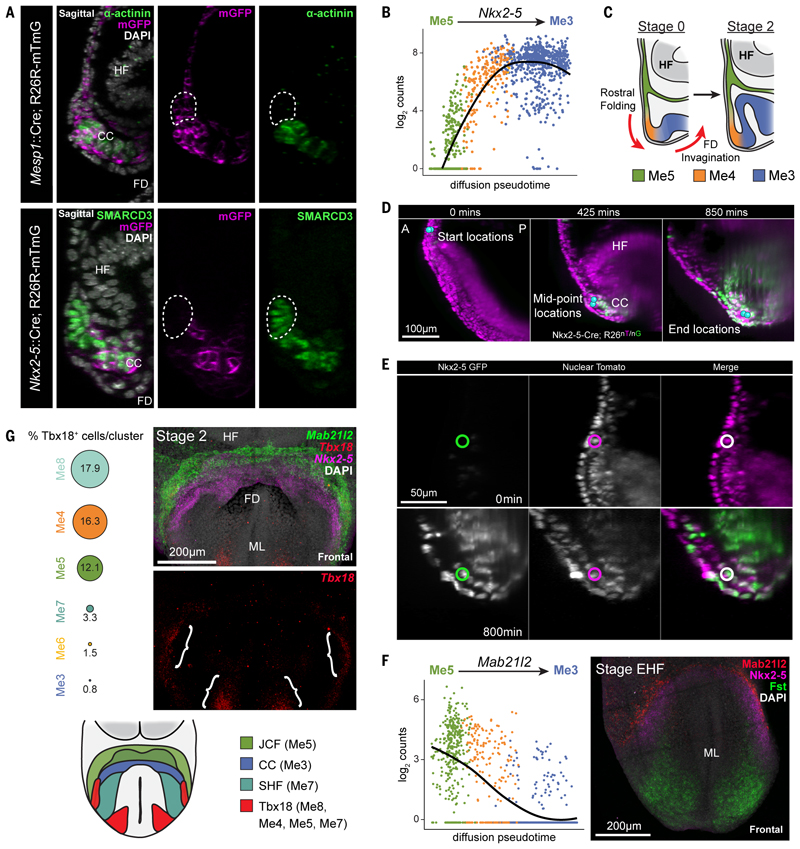
Me5 cells are Hand1 and Snai1 positive but also Nkx2-5 negative. Using these and other markers such as Smarcd3 and Tbx5, we were able to locate Me5 at the rostral border of the cardiac crescent, as a discrete group of cells at the confluence of extra-embryonic and splanchnic mesoderm, corresponding anatomically with the primitive septum transversum (26) (Figure 3A-C and Figure S11). The splanchnic mesoderm here is a monolayer continuous with the extra-embryonic mesoderm of the yolk-sac. Within this sheet, at stage 1 and earlier, Me5 was rostrally positioned with respect to the more caudal NKX2-5 and TITIN positive cardiac progenitors (Figure 3B). Later, at stage 2, rostral folding causes this mesoderm to buckle, such that Me5 is now sandwiched between the endoderm and the more dorsal layer of NKX2-5 positive cardiac progenitors, while maintaining its relative positions within the plane of the mesodermal sheet (Figure 3C and D). Using our scRNA-seq data we identified Mab21l2 as a marker of Me5 (Figure 3E, 3F, Figure S8B, 12, Movie S6 and S7). Expression of Mab21l2 confirmed the location of Me5 at the rostral border of the crescent prior to rostral folding, and between the endoderm and crescent during folding (Figure 3E and Figure S12).
Fig. 3. Identification of a transcriptionally and anatomically distinct cell type with cardiac progenitor like properties.
A-C, Maximum intensity projections (MIP) of immunostained embryos revealing anatomical localization of Me5 (brackets). Dotted line indicates the location of the sagittal section shown at right in each panel. YS, yolk sac; CC, cardiac crescent; FD, foregut diverticulum; ML, midline; En, endoderm; HF, headfold. D, Schematic highlighting the localization of Me5 (Juxta cardiac field (JCF)) and Me7/8, relative to the myocardium of the cardiac crescent (Me3). E, MIP of hybridization chain reaction (HCR) staining of an embryo at stage 2 revealing the distinct anatomical localization of JCF cells marked by Mab21l2 (Movie S6). Dotted lines (a-d) correspond to sagittal sections shown below. F, Left, MIP of lateral view of HCR on a stage -1 embryo showing distinct expression domains of Mab21l2, Nkx2-5 and Fst (Movie S7). Ant, anterior; Pos, posterior; PS, primitive streak; YS, yolk sac; PM, paraxial mesoderm. Right panel shows only Mab21l2 expression highlighting location of Me5 cells (white dotted region).
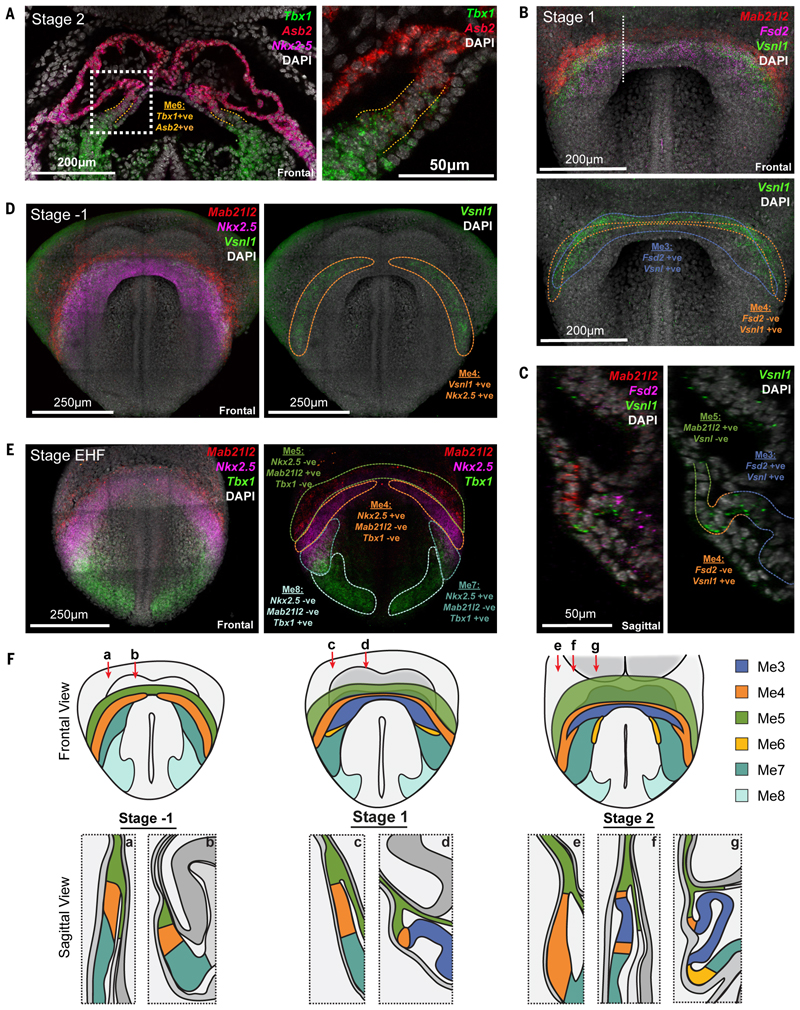
Me6, characterized by overlapping expression of Asb2 and Tbx1, represented a small population of cells restricted to the medial-dorsal interface between Me7 and the cardiac crescent (Figure 4A and F, Figure S13 and Movie S3), consistent with it representing a transitional state during the differentiation of SHF progenitors (Me7) into cardiomyocytes (Me3).
Fig. 4. Anatomical identification of transition states during cardiomyocyte differentiation.
A, Coronal section of a hybridization chain reaction (HCR) stained stage 2 embryo, showing the anatomical location of Me6 (dotted orange lines) marked by co-expression of Tbx1 and Asb2. Dotted white box represents region of higher magnification at right. B, Maximum intensity projection (MIP) of HCR on stage 1 embryo showing expression of Mab21l2, Vsnl1 and Fsd2 to identify cluster Me4. Colored dotted lines correspond to Me3 and Me4 (Movie S8). White dotted line shows location of section in C. C, Sagittal section of embryo in B highlighting locations of Me3, Me4 and Me5. D, MIP of HCR on stage -1 embryo showing Mab21l2, Vsnl1 and Nkx2-5, to identify cluster Me4 (yellow dotted line) (Movie S11). E, MIP of an early headfold (EHF) stage embryo, prior to crescent formation, showing expression of Mab21l2, Tbx1 and Nkx2-5, to identify clusters Me4, Me5, Me7 and Me8. Colored dotted lines outline corresponding regions. F, Schematic highlighting the anatomical location of all mesoderm transcriptional states identified at multiple stages of heart formation. Red arrows represent the location of sagittal sectional views.
Me4, identified by expression of Vsnl1 and absence of Fsd2, localized at stage 1 to a narrow arc within the cardiac crescent, at the interface between Me5 and Me3 (Figure 4B-C, 4F, Figures S14, S15 and Movies S8-S10). At stage -1, prior to formation of cardiomyocytes (Me3), Me4 formed the earliest arcs of the cardiac crescent, just caudal to Me5 (Figure 4D, 4F, Figure S16, S17 and Movie S11), consistent with it representing the FHF. The discrete anatomical locations of these transcriptional clusters highlights the spatially graded maturation of cardiomyocytes in this region. The Me4 state could be detected as late as the LHT stage (Figure S14C).
Our diffusion analysis indicated that Me7 (SHF) could contribute to Me3 (cardiomyocytes) via not only Me6, a transient transcriptional state specific to the SHF trajectory, but also via Me4, that captures the FHF transcriptional state. Consistent with this, we detected Me4 cells not only at the interface between Me5 and Me3 (above) but also at the interface of Me7 and Me3 (Figure 4E, F and Figure S16C, S17C and D). In addition, Me3, defined by the expression of Fsd2, could be separated into two subgroups based on the presence or absence of the Me4 marker Vsnl1. Imaging revealed that the Vsnl1 positive cardiomyocytes of Me3 were located in the ventral crescent, adjacent to Me4, whilst the Vsnl1 negative cardiomyocytes were located more medial-dorsally, adjacent to Me7 and Me6 (Figure 4C and Figure S14D, E). Given the different anatomical locations and transcriptional profiles of SHF intermediates, the branched trajectory of SHF progenitors possibly reflects the early separation of anterior and posterior SHF.
The consensus FHF transcriptional signature and anatomical location of Me4 within the crescent indicate that it represents the FHF. In contrast, the Nkx2-5 negative Me5 corresponds to progenitors of the FHF. Hand1 has been considered a FHF marker due to in situ hybridization using chromogenic detection that appears to label the cardiac crescent27. However, our single-cell resolution imaging shows that HAND1 in fact marks Me5, which is distinct from the FHF and cardiac crescent (Figure 3A-C); due to its ventral position overlying the crescent, volume imaging is required to distinguish the two. Given the proximity of Me5 to the cardiac crescent, we named it the Juxta-Cardiac Field (JCF).
The JCF represents a cardiac progenitor population
To determine if the JCF, similar to other cardiomyocyte progenitors, is derived from cells that expressed Mesp1, we used a Mesp1-Cre (3) driver line. This verified that the JCF arises from Mesp1 positive mesoderm (Figure 5A). Similarly, using an Nkx2-5-Cre (27) driver line showed that JCF cells had never previously expressed Nkx2-5 (Figure 5A).
Fig. 5. The JCF represents a cardiac progenitor population.
A, Top, sagittal section of Mesp1-Cre; R26R-mTmG embryos immunostained for α-actinin and GFP (n=6). Bottom, section of Nkx2-5-Cre; R26R-mTmG embryos immunostained for SMARCD3 and GFP (n=4). Dotted regions highlight the JCF, revealing it is derived from Mesp1, but not Nkx2-5, expressing cells. HF; headfold; FD, foregut diverticulum; CC, cardiac crescent. B, Nkx2-5 expression levels plotted against the diffusion pseudotime of all cells consistent with the trajectory from Me5 to Me3. Points are colored based on their assigned cluster. Nkx2-5 is upregulated during the transition from JCF towards cardiomyocytes. The line is the quadratic local linear fit of the expression levels as a function of pseudotime. C, Schematic diagrams highlighting the relative positions of Me3-Me5 during rostral folding and the transition from stage 0 to stage 2 cardiac crescents. D,E, Frames from time-lapse movie of an E7.5 Nkx2-5-Cre; R26-nTnG embryo imaged over a 16-hour period with light-sheet microscopy. D, JCF cells migrated from a more rostral position towards the developing crescent (blue spheres = representative tracked cells). A, anterior; P, posterior. E, Nkx2-5 was upregulated by JCF cells, as shown by an increase in the nuclear expression of GFP. Circles indicate the same cell tracked across time, shown in different channels. Representative cells also shown in Movies S12 and S13. F, On the left, Mab21l2 gene expression levels are plotted against the diffusion pseudotime of all cells consistent with the trajectory from Me5 to Me3. Points are colored based on their assigned cluster. Mab21l2 expression is downregulated during the transition from the JCF towards cardiomyocytes. At right, maximum intensity projection (MIP) of HCR on an early headfold stage (EHF) embryo, prior to crescent formation, revealing the early rostral expression of Mab21l2 in JCF progenitors proximal to Nkx2-5 and Fst expression. 3D rendering of expression is shown in Movie S14. ML, midline. G, Tbx18 is expressed in multiple cell clusters. The percentages of Tbx18-positive cells from each cluster are shown as proportionally sized circles. At right, MIP of HCR for Tbx18, Mab21l2 and Nkx2-5 in a stage 2 embryo, highlighting location of Tbx18 expression (brackets) (Movie S6). Schematic at bottom, highlights Tbx18 expression domains in the anterior crescent region of the developing embryo.
Supporting a differentiation trajectory from the JCF to cardiomyocytes, the cardiac progenitor gene Nkx2-5 increased in expression along pseudotime as cells transitioned from JCF to Me3 (Figure 5B). We therefore tracked JCF cells in embryos where expression of Nkx2-5 could be monitored using nuclear-eGFP fluorescence (28,29) using single-cell resolution light-sheet microscopy (30). Confirming the diffusion map trajectory, we found that JCF cells activated expression of Nkx2-5:GFP as they moved towards the cardiac crescent during rostral folding (Figure 5C-E and two representative cells in Movie S12 and S13).
The Me5 marker Mab21l2 is expressed rostral to the cardiac crescent and is down-regulated during cardiomyocyte differentiation (Figure 5F). Prior to crescent formation, at early headfold stages(31), Mab21l2 was detected rostral to an Nkx2-5-positive domain, at the boundary between embryonic and extra-embryonic mesoderm (Figure 5F, Figure S18B and Movie S14). At early pre-headfold stages, we could not detect Mab21l2, but found Nkx2-5 expression in the anterior mesoderm (Figure S18C). This suggests that the Me5 population forms after other cardiac progenitor populations.
Our transcriptomic data showed that Me8, Me4 and a small subset of Me5 expressed Tbx18 (Figure 5G). A Tbx18 positive (and Nkx2-5, Isl1 double negative) population of caudal-lateral mesoderm can give rise to sinus venosus myocardium (32). To test whether the JCF and sinus venosus precursors might overlap, we assayed for Tbx18 and Mab21l2 coexpression by in situ Hybridization Chain Reaction (Figure 5G and Figure S19). In contrast to Mab21l2, Tbx18 was not detected until stage 1, with weak expression in two separate domains, a region of the lateral crescent and medial paraxial mesoderm, corresponding with Me8 (Figure 5G). In contrast, Mab21l2, was expressed earlier, at early headfold stages, rostrally and in a significantly larger region covering both medial and lateral regions of mesoderm (Figure S19B). Tbx18 was never detected in the rostral-medial JCF, indicating that the JCF is a spatially and transcriptionally distinct progenitor population from the sinus venosus progenitors (Figure 5G).
Juxta cardiac field progenitors contribute to cardiomyocytes and epicardium
In order to test our diffusion trajectory results indicating the JCF gives rise to cardiomyocytes, we knocked CreERT2 into the Mab2l12 locus in a homozygous R26R-YFP reporter background (33). At E8.0, nuclear localized CreERT2 was found only in the juxta-cardiac field and was not detected in cardiomyocytes (Figure S20B).
To specifically label the JCF, we administered a single low dose of tamoxifen to pregnant dams at 6.5dpc then assessed the fate of labelled cells at several subsequent time-points (Figure 6A). This ensured that tamoxifen was available when Mab2l12 first begins to mark the JCF around E7.5 while minimizing the possibility of labelling cells outside the JCF, or at subsequent stages. This protocol allowed us to stringently assess the fate of the JCF but likely underrepresents the contribution of this progenitor pool due to the early, limiting, dose of tamoxifen.
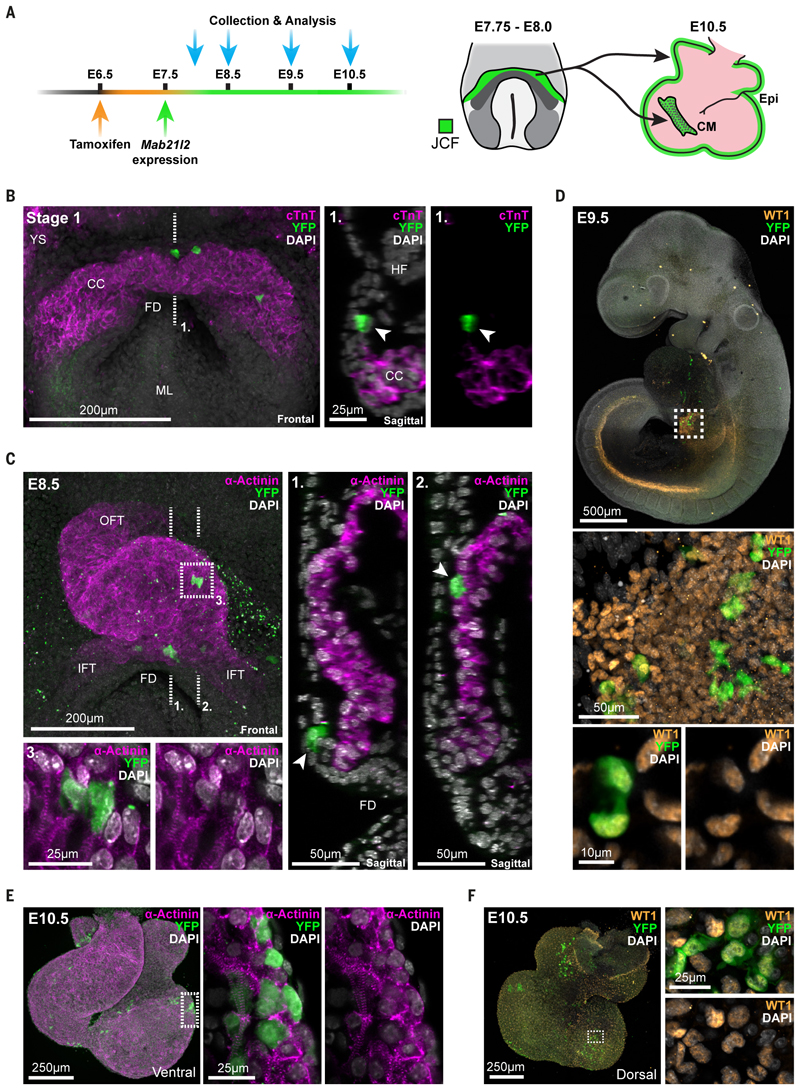
Fig. 6. Juxta-cardiac field progenitors contribute to cardiomyocytes and epicardium.
A, Schematic showing the experimental design of lineage labelling experiments using a Mab21l2- iCreERT2 transgenic mouse line. At right, a schematic highlighting that labelling of the juxta-cardiac field (JCF) between E7.75 and E8.0 leads to labelling of both cardiomyocytes (CM) and the epicardium (Epi) at E10.5. B, Maximum intensity projection (MIP) of a stage 1 Mab21l2- iCreERT2; R26R-YFP embryo immunostained for cardiac troponin T (cTnT) and YFP. The dotted line represents the location of the sagittal section at right. YFP positive cells (arrows) are located in the JCF and do not express cardiomyocyte marker cTnT. CC, cardiac crescent; FD, foregut diverticulum; YS, yolk sac; ML, midline; HF, headfold. C, MIP of a looping heart tube from an E8.5 Mab21l2-iCreERT2; R26R-YFP embryo immunostained for α-actinin and YFP. Dotted lines show locations of sagittal sections at right. Arrows highlight the location of recombined YFP cells within the myocardium (2.) and maintained within the JCF progenitor region (1.). The dotted box (3.) shows zoomed in view of JCF derived cardiomyocytes with sarcomeric striations of α-actinin. OFT, outflow tract; IFT, inflow tract. D, MIP of an E9.5 embryo immunostained for WT1 and YFP. WT1 marks both the nephrogenic cord and proepicardium (boxed). Below are high magnification views of the proepicardium highlighting the coexpression of YFP and WT1. E and F, Immunostaining of E10.5 hearts showing co-expression of α-actinin (E) and WT1 (F) with YFP positive cells, highlighting the dual potential of the JCF. Dotted boxes represent zoomed in regions. Dorsal and ventral views shown.
We first tested specificity of labelling by looking in E8.0 embryos. Fewer than half of Mab2l12- CreERT2 transgenic embryos showed any labelled cells (19 out of 45 embryos) consistent with the limiting dose of tamoxifen. YFP positive cells could be found only in the JCF at early crescent stages and did not express markers of maturing FHF cells such as sarcomeric α-actinin or cardiac Troponin T (cTnT) (Figure 6B and Figure S20B-D). On average, five YFP positive cells could be detected per embryo between stage 1 and LHT (Figure S20A), highlighting the low level of recombination. These data confirmed that the new Mab21l2-iCreERT2 transgenic line specifically targeted JCF cells and would allow us to investigate their fate.
We next analyzed the early looping heart tube at E8.5. Consistent with time-lapse results (Figure 5D) this revealed that JCF cells contribute to cardiomyocytes within the linear heart tube, as verified by sarcomeric striations of alpha-Actinin in YFP positive cells (Figure 6C and Table S1). Additionally, the JCF was maintained throughout these stages as YFP positive non-cardiomyocyte cells were still present in the JCF domain, which by this stage was positioned by rostral folding between the cardiac tube and endoderm (Figure 6C).
JCF cells, in addition to forming cardiomyocytes, also contributed to the proepicardium at E9.5. Whole mount immunofluorescence for WT1, a proepicardium and epicardium marker, revealed colocalization with YFP cells (Figure 6D and Table S2). Consistent with this, at E10.5 we found YFP positive cells in both the epicardium and myocardium. The latter cells were found in the left ventricle, atrioventricular canal and atria, but never in the right ventricle or outflow tract (Figure 6E, F, Table S3 and S4). The direct contribution of the JCF to myocardium, as opposed to via differentiation of later formed epicardial cells, is evident from the JCF derived myocardial cells observed already at E8.5, before the formation of the epicardium (Figure 6C, Table S1 and S2). These data together establish the juxta-cardiac field as a progenitor pool of cardiomyocytes and the proepicardium.
Our spatially and temporally-resolved single-cell anatomical and transcriptional analysis provides a multifaceted resource for understanding how the heart forms. It also provides a spatial and transcriptomic reference definition of the FHF, SHF, and intermediate states to cardiomyocytes. Given that the transcriptional state of a cell is labile, but scRNA-seq offers only a snapshot of its transcriptional state, one has to be cautious in using ‘marker’ genes to make inferences regarding lineage (past history, often with the implicit assumption that a shared transcriptional state implies a shared history) and fate (the future descendants). This is particularly evident in Me4 that represents a FHF transcriptional state, but through which cells of two different origins (JCF or SHF) can transition in forming cardiomyocytes. It also highlights the importance of using independent approaches such as genetic fate mapping to determine the fate of cells.
Our analyses revealed a progenitor pool, the JCF, that can contribute to cardiomyocytes via a FHF transcriptional state, as well as to the proepicardium. The JCF is located at the rostral confluence of the embryonic and extra-embryonic mesoderm, and may represent a subset of the forming septum transversum. Although the JCF can contribute to the FHF, it is unlikely to be the sole progenitor. It is currently unclear if the JCF is a pool of unipotent progenitors of cardiomyocytes and proepicardium, or contains bipotent cells capable of giving rise to both. Future clonal analyses are required to address this question. The significance of the JCF is supported by a previous study which showed that loss of function of the JCF marker Mab21l2 leads to early embryonic lethality due to a reduction in myocardium and proepicardium (34). Our work further indicates there are likely multiple populations contributing to the proepicardium, the JCF representing an early population, and the Tbx18 positive population of paraxial mesoderm (10–12), a later population.
By identifying the juxta-cardiac field, our work widens the cardiac progenitor region, identifies the earliest progenitors of the proepicardium and provides new insights into the lineal origins of cardiac cell types. This, combined with the detailed characterization of these cell types at the transcriptomic level, will contribute to a better understanding of the origin of congenital cardiac defects as well as provide basic insights for informing regenerative approaches to treating heart disease.
Materials and Methods
All animal experiments complied with the UK Animals (Scientific Procedures) Act 1986, approved by the local Biological Services Ethical Review Process and were performed under UK Home Office project licenses PPL 30/3420 and PCB8EF1B4. Anterior embryonic regions were dissected manually, dissociated to single cells and processed to obtain single-cell transcriptomes. Single cell analyses, whole mount immunohistochemistry, in situ Hybridization Chain Reaction, volume imaging, light-sheet live microscopy, 2C-HR-CRISPR based genetic engineering and genetic lineage labelling are described in detail in the Supplementary Methods.
Supplementary Material
Acknowledgements
We thank S. Meilhac, M. Mommersteeg, P. Riley, R. Sambasivan and M. Torres for valuable discussions and comments. We acknowledge the Single Cell Genomics Core Facility at the Wellcome Sanger Institute for processing SMART-seq2 libraries. SS is a member of the British Heart Foundation (BHF) Oxford Centre of Regenerative Medicine and BHF Oxford Centre of Research Excellence. DNA encoding tdTomato was kindly provided by R. Y. Tsien (HHMI/UCSD). We thank N. Ashley at the MRC Weatherall Institute of Molecular Medicine single cell facility for his advice on single cell workflows and the Micron Advanced Bioimaging Unit (supported by Wellcome Awards 091911/B/10/Z and 107457/Z/15/Z and BBSRC ALERT13 Award BB/L014750/1). XI-S is currently affiliated with Computational Biology, Functional Genomics, GSK Medicines Research Centre, Gunnels Wood Road, Stevenage, SG1 2NY, UK.
Funding
This work was funded by Wellcome Awards 108438/Z/15/Z and 105031/C/14/Z (SS and JCM), Wellcome Senior Investigator Award 103788/Z/14/Z (SS), BHF Immediate Fellowship FS/18/24/33424 (RT) and core funding from the European Molecular Biology Laboratory and Cancer Research UK (JCM) and the Howard Hughes Medical Institute (PJK).
Footnotes
Author Contributions: RT, XI-S, JM and SS conceived the study, interpreted the data and wrote the manuscript. RT conducted all whole mount immunohistochemistry, in situ hybridization experiments, imaging and lineage analysis. XI-S performed all computational analyses. RT, SS and AM dissected and processed embryos for single-cell library preparation. KM and PK performed all light-sheet experiments. TvdB and AS performed initial scRNA-seq data analyses. SJ and JG generated the Mab21l2-iCreERT2 transgenic line. PK, JM and SS supervised the study. All authors read and approved the final manuscript. Authors declare no competing interests.
Data and materials availability
The Mab2l12-CreERT2 mouse line is available from SS under a material transfer agreement with the University of Oxford. Raw sequencing data has beendeposited in the European Nucleotide Archive database under study PRJEB14363 (sample accession identifiers provided in Extended Data 3), and ArrayExpress under accession E-MTAB- 7403. The raw count matrices for the unbiased and reference datasets are provided in Data S1 and S2, respectively. Associated sample metadata is available in Extended Data 3 and 4, respectively. Raw and processed data are also available as R data objects from https://content.cruk.cam.ac.uk/jmlab/mouseEmbryonicHeartAtlas/. All code used for data analysis, figure generation and the shiny app is available at https://github.com/MarioniLab/mouseHeart_2020.
References
- 1.Lawson Ka. Fate mapping the mouse embryo. Int J Dev Biol. 1999;43:773–775. [PubMed] [Google Scholar]
- 2.Parameswaran M, Tam PPL. Regionalisation of cell fate and morphogenetic movement of the mesoderm during mouse gastrulation. Dev Genet. 1995;17:16–28. doi: 10.1002/dvg.1020170104. [DOI] [PubMed] [Google Scholar]
- 3.Saga Y, et al. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 4.Meilhac SM, Buckingham ME. The deployment of cell lineages that form the mammalian heart. Nat Rev Cardiol. 2018;15:705–724. doi: 10.1038/s41569-018-0086-9. [DOI] [PubMed] [Google Scholar]
- 5.Lescroart F, et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat Cell Biol. 2014 doi: 10.1038/ncb3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyser RCV, et al. Calcium handling precedes cardiac differentiation to initiate the first heartbeat. Elife. 2016;5 doi: 10.7554/eLife.17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell. 2004 doi: 10.1016/S1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- 8.Cao J, Poss KD. The epicardium as a hub for heart regeneration. Nature Reviews Cardiology. 2018 doi: 10.1038/s41569-018-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simões FC, Riley PR. The ontogeny, activation and function of the epicardium during heart development and regeneration. Development (Cambridge) 2018 doi: 10.1242/dev.155994. [DOI] [PubMed] [Google Scholar]
- 10.Mommersteeg MTM, et al. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res. 2010 doi: 10.1093/cvr/cvq033. [DOI] [PubMed] [Google Scholar]
- 11.Schlueter J, Brand T. Subpopulation of proepicardial cells is derived from the somatic mesoderm in the chick embryo. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.113.301347. [DOI] [PubMed] [Google Scholar]
- 12.Bressan M, Liu G, Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science (80-) 2013 doi: 10.1126/science.1232877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G, et al. Transcriptomic Profiling Maps Anatomically Patterned Subpopulations among Single Embryonic Cardiac Cells. Dev Cell. 2016 doi: 10.1016/j.devcel.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lescroart F, et al. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science (80-) 2018 doi: 10.1126/science.aao4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Soysa TY, et al. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental anomalies. Nature. 2019 doi: 10.1038/s41586-019-1414-x. ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong H, et al. Single-Cell Transcriptomics Reveals Chemotaxis-Mediated Intraorgan Crosstalk during Cardiogenesis. Circ Res. 2019 doi: 10.1161/CIRCRESAHA.119.315243. [DOI] [PubMed] [Google Scholar]
- 17.Tyser RCV, Srinivas S. The First Heartbeat—Origin of Cardiac Contractile Activity. Cold Spring Harb Perspect Biol. 2019:a037135. doi: 10.1101/cshperspect.a037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paige SL, Plonowska K, Xu A, Wu SM. Molecular regulation of cardiomyocyte differentiation. 2015 doi: 10.1161/CIRCRESAHA.116.302752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Soysa TY, et al. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature. 2019 doi: 10.1038/s41586-019-1414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowotschin S, et al. The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature. 2019 doi: 10.1038/s41586-019-1127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pijuan-Sala B, et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. 2019;566:490–495. doi: 10.1038/s41586-019-0933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Holland PWH, Takahashi T. Gene profiling of head mesoderm in early zebrafish development: Insights into the evolution of cranial mesoderm. Evodevo. 2019 doi: 10.1186/s13227-019-0128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess R, Rawls A, Brown D, Bradley A, Olson EN. Requirement of the paraxis gene for somite formation and musculoskeletal patterning. Nature. 1996;384:570–3. doi: 10.1038/384570a0. [DOI] [PubMed] [Google Scholar]
- 24.Candia AF, et al. Mox-1 and Mox-2 define a novel homeobox gene subfamily and are differentially expressed during early mesodermal patterning in mouse embryos. Development. 1992 doi: 10.1242/dev.116.4.1123. [DOI] [PubMed] [Google Scholar]
- 25.Reiter JF, et al. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunwoodie SL, Rodriguez TA, Beddington RSP. Msg1 and mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mech Dev. 1998 doi: 10.1016/S0925-4773(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 27.Moses KA, Demayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001 doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 28.Stanley EG, et al. Efficient cre-mediated deletion in cardiac progenitor cells conferred by a 3’UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int J Dev Biol. 2002 doi: 10.1387/ijdb.12141429. [DOI] [PubMed] [Google Scholar]
- 29.Prigge JR, et al. Nuclear double-fluorescent reporter for in vivo and ex vivo analyses of biological transitions in mouse nuclei. Mamm Genome. 2013 doi: 10.1007/s00335-013-9469-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDole K, et al. In Toto Imaging and Reconstruction of Post-Implantation Mouse Development at the Single-Cell Level. Cell. 2018;175:859–876. doi: 10.1016/j.cell.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 31.Lawson KA, Wilson V. Kaufman’s Atlas of Mouse Development Supplement. 2016. A Revised Staging of Mouse Development Before Organogenesis. [DOI] [Google Scholar]
- 32.Christoffels VM, et al. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res. 2006 doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- 33.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001 doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito Y, Kojima T, Takahashi N. Mab21l2 is essential for embryonic heart and liver development. PLoS One. 2012 doi: 10.1371/journal.pone.0032991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Mab2l12-CreERT2 mouse line is available from SS under a material transfer agreement with the University of Oxford. Raw sequencing data has beendeposited in the European Nucleotide Archive database under study PRJEB14363 (sample accession identifiers provided in Extended Data 3), and ArrayExpress under accession E-MTAB- 7403. The raw count matrices for the unbiased and reference datasets are provided in Data S1 and S2, respectively. Associated sample metadata is available in Extended Data 3 and 4, respectively. Raw and processed data are also available as R data objects from https://content.cruk.cam.ac.uk/jmlab/mouseEmbryonicHeartAtlas/. All code used for data analysis, figure generation and the shiny app is available at https://github.com/MarioniLab/mouseHeart_2020.