Abstract
Whole genome characterizations of crop plants based on ancient DNA have provided unique keys for a better understanding of the evolutionary origins of modern cultivars, the pace and mode of selection underlying their adaptation to new environments and the production of phenotypes of interest. Although forests are among the most biologically rich ecosystems on earth and represent a fundamental resource for human societies, no ancient genome sequences have been generated for trees. This contrasts with the generation of multiple ancient reference genomes for important crops. Here, we sequenced the first ancient tree genomes using two white oak wood remains from Germany dating to the Last Little Ice Age (15th century CE, 7.3X and 4.0X) and one from France dating to the Bronze Age (1700 BCE, 3.4X). We assessed the underlying species and identified one medieval remains as a hybrid between two common oak species (Quercus robur and Q. petraea) and the other two remains as Q. robur. We found that diversity at the global genome level had not changed over time. However, exploratory analyses suggested that a reduction of diversity took place at different time periods. Finally, we determined the timing of leaf unfolding for ancient trees for the first time. The study extends the application of ancient wood beyond the classical proxies of dendroclimatology, dendrochronology, dendroarchaeology and dendroecology, thereby enhancing resolution of inferences on the responses of forest ecosystems to past environmental changes, epidemics and silvicultural practices.
Keywords: first tree paleogenomes, admixture, leaf unfolding timing, Quercus. robur, Q. robur x Q. petraea, Bronze Age, Last Little Ice Age
1. Introduction
The increasing capacity to characterize genomic data from ancient plant remains, such as seeds, leaves and wood material stored in archaeological and herbarium collections has considerably advanced our understanding of the evolutionary, domestication and cultural history of plants (Kistler et al. 2020a). This advancement has unleashed the new research field of plant paleogenomics, with current research focused primarily on crops, including maize, barley, emmer, millet, cotton, sunflower, squash, potato and grapevine (Palmer et al. 2012; Fonseca et al. 2015; Kistler et al. 2015, 2018, 2020b; Ramos-Madrigal et al. 2016, 2019; Vallebueno-Estrada et al. 2016; Swarts et al. 2017; Wales et al. 2018; Smith et al. 2019; Gutaker et al. 2019; Scott et al. 2019). These represent species cultivated by humans for millennia and showing massive agronomical and economical interest today. The ancient plant genomes characterized thus far have solved long-standing debates about the evolutionary origins of modern cultivars, and the tempo and mode of selection underlying their adaptation to new environments and the production of new phenotypes-of-interest (Mascher et al. 2016; Ramos-Madrigal et al. 2016; Vallebueno-Estrada et al. 2016; Swarts et al. 2017; Smith et al. 2019; Scott et al. 2019). In contrast to crops, trees have remained almost entirely overlooked (with notable exceptions; (Lendvay et al. 2017; Wagner et al. 2018)) and the first ancient genome of forest trees remains to be sequenced, despite forests being among the most biologicallyrich and geneticallydiverse ecosystems on the planet and trees providing an essential resource for human societies (Crowther et al. 2015; Brockerhoff et al. 2017).
Characterizing the genome sequences of ancient trees is extremely challenging due to the nature of the sub-fossil (non-fossilized) material available, which largely consists of single cell pollen grains, macrofossils, and inert wood tissues with few living cells; these tissues are also abundant in chemical products interfering with DNA extraction (Rachmayanti et al. 2006, 2009) and enzymatic manipulation (Deguilloux et al. 2002; Rachmayanti et al. 2006, 2009). However, sub-fossil wood remains are ubiquitous and represent key primary sources for dendroclimatology, dendrochronology, dendroarchaeology and dendroecology studies (Domínguez-Delmás 2020; Tegel et al. 2022). These materials also show promising potential for paleogenomic investigations, especially sapwood (outer wood), which contains higher DNA content than heartwood (inner wood) (Lendvay et al. 2017; Wagner et al. 2018). Waterlogged wood remains deposited in suitable microenvironments such as clay sediments on calcareous bedrock or neutral organic sediments are known to preserve plastid and nuclear DNA and to provide sufficient molecular templates for whole-genome characterization through next-generation sequencing technologies (Wagner et al. 2018).
Among European trees, the four deciduous, inter-fertile white oak species Quercus robur, Q. petraea, Q. pubescens and Q. pyrenaica, which are widespread in Western Europe north of the Pyrenees (Roloff et al. 2010), represent a particularly promising model for arboreal plant paleogenomic studies. In particular, Q. robur and Q. petraea stand out among all European wood species as they predominate in temperate forests and have been intensively used as construction timber for dwellings and other essential constructions throughout human history. As a result, white oak wood is very well represented in Holocene lake sediments and soils and is often exceptionally well preserved (Čufar et al. 2010; Aguilera et al. 2011; Tegel et al. 2012; Martinelli 2013; Bernabei et al. 2019). Oaks are known for their long-life spans that can be accurately determined by annual growth rings, provided they are not sprouts resulting from coppicing, a forestry technique that was common throughout history (Billamboz & Martinelli 2015, Roloff et al. 2010). Maturity is reached at 30-40 years of age, but it is difficult to determine when full reproductive potential is reached. Among living oaks, maximum ages of 400 to 1000 years are reached, but very old trees, such as the Birnam Oak from William Shakespeare’s MacBeth (https://www.ancienttreeforum.org.uk/ancient-trees/ancient-tree-sites-to-visit/scotland/birnham-oak/), estimated to be 600 years-old, or populations older than 300 years (Saleh et al. 2022) are rare and geographically restricted. Genomes generated from ancient oak timber therefore provide an invaluable complement to living old-growing oaks, and allow the study of tree populations from very recent to very ancient times. However, all four white oak species show a highly similar wood anatomical structure (Schweingruber 1990). Therefore, when starting from wood, the exact distribution ranges of oaks and potential temporal range shifts, including during global climatic changes and human forest modifications, cannot be reconstructed without DNA characterization.
In oaks, other anatomical features, such as those of the leaves or fruits, do not always allow a clear species assignment. As a result, the delimitation of species within the European white oak complex is the subject of ongoing debate and methodological developments (Rellstab et al. 2016, Neophytou 2014). Indeed, recent genome-wide studies have shown that a large part of the genome of the four species Q. robur, Q. petraea, Q. pubescens and Q. pyrenaica is permeable to interspecific gene flow and only very narrow genomic regions represent species barriers (Leroy et al., 2020b). However, it is unclear since when these species barriers have existed and whether they have changed over time. Time series from ancient DNA can provide clues important for this debate. The narrow species barriers indicate numerous admixture events between species, probably related to their postglacial history. For Q. robur and Q. petraea, it is assumed that the areas that had been released after the last glaciation were initially colonized by Q. robur and that in the course of progressive climatic improvement Q. petraea invaded these forests and pushed Q. robur back to marginal areas or areas less favorable for Q. petraea (Petit et al. 2004). Interestingly, it has been shown that despite these large-scale postglacial migrations and their sessile lifestyle, European white oak shows exceptionally high levels of genomic diversity (Plomion et al. 2018); it can thus be assumed that diversity was maintained throughout history. This does not exclude the possibility that changes took place in genomic sub-regions during numerous moments in time. For example, temperature-induced fluctuating selection resulting in allele frequencies changes in some genomic sub-regions has been observed in three successive oak generations covering the transition from the last Little Ice Age to the modern era (Saleh et al. 2022). Present-day populations have been intensively studied and show a clear population structure as well as examples of recent secondary contacts and adaptive introgression between species (Lepais et al. 2009; Alberto et al. 2010; Leroy et al. 2020a; Degen et al. 2021; Nocchi et al. 2022). Genome-wide population data suggests that secondary contacts took place in the course of the current interglacial period and that they are associated with environmental changes such as temperature and precipitation shifts, since Q. robur is more adapted to cooler and wetter climate whereas Q. petraea prefers a warmer climate and well-drained soils (Leroy et al. 2017, 2020b; Roloff et al. 2010). However, both the timing and number of admixture events remain contentious. In this context, time series of ancient oak genomes would be very valuable for revealing the stability and dynamics of oak population structure as well as potential impacts of agroforestry practices and climate change from the perspective of a few to numerous ancestral generations. This appears particularly interesting given that not only demographic trajectories (Leroy et al. 2017, 2020b) but also pathogen resistance and phenological life history traits (e.g., leaf unfolding timing (Leroy et al. 2020a) can be reconstructed from genome-scale patterns of DNA variation.
In oaks, leaf phenology is one of the most important adaptive traits. The date of leaf unfolding precedes the timing of flowering and contributes to the length of the growing season. Thus, leaf phenology correlates to fitness. Populations surveys and common garden experiments indicate that leaf phenology exhibits strong genetic variation, both within and between populations (Firmat et al., 2017; Girard et al., 2022). Among phenotypic traits observed so far, the date of leaf unfolding shows the highest heritability (h>0.8, Alberto et al. 2011), and strong genetic control has been confirmed by the discovery of numerous QTLs (Derory et al. 2010). The timing of leaf unfolding shows strong genetic clines along temperature driven geographic and temporal gradients that are mirrored by linear relationships of allelic frequencies of their underlying candidate genes (Vitasse et al. 2009; Firmat et al. 2017; Girard et al. 2022, Alberto et al. 2013; Leroy et al. 2020a). In a recent study, 18 Q. petraea populations from different latitudinal and altitudinal gradients, for which leaf folding indices describing the time of leaf unfolding were available were sequenced (Leroy et al, 2020a). Subsequent investigations of population differentiation and genotype-phenotype associations revealed a number of outlier loci that allow populations to be categorized as either early- or late-leaf unfolding (Leroy et al, 2020a). Given these particularly strong genotype-phenotype associations, these SNPs seemed very suitable for testing if phenological signatures can be tracked back in time.
In this study, we sequenced the first ancient oak genomes using two wood remains dating to the last Little Ice Age (15th century CE, Common Era) and one remains sample dating to the Bronze Age (1700 BCE, Before Common Era). First, we assessed if ancient wood remains are suitable for assessing the taxonomic status and the population structure of ancient oaks, which allowed us to identify one medieval sample as a hybrid between Q. robur and Q. petraea and the two other samples as Q. robur. Second, we explored whether genomic diversity has changed over time, by examining changes in different genomic regions; we discovered that global genomic diversity had not changed but that some regions may have undergone changes during different time periods. Finally, we genotyped loci that had previously been shown to be associated with leaf unfolding (Leroy et al, 2020a) and predicted the timing of leaf unfolding phenology of ancient trees for the first time.
2. Material And Methods
2.1. Ancient and Modern Samples
Ancient wood remains were collected from an excavation in the Seine Valley in Northern France in 2017 (Figure 1). The site is located on chalk cliffs at an average altitude of 66 meters and was leveled in the Holocene by flooding and the deposition of silty clay sediments in the floodplain. The wood remains that exhibited good DNA preservation levels (Seine.748) came from a tree of the river plain that was deposited in the sediments of a former channel arm during the Bronze Age and was radiocarbon dated to 1767-1623 cal. BCE (Table S1). Given the small trunk diameter (< 25 cm) and assuming a regular tree growth, we estimate the age to be less than 50 years. An exact age based on dendrochronology is unfortunately not available. To date, climate reconstructions that allow the local climate conditions at that time at that site to be determined are not available. The closest reconstructions come from the Northern French Alps and indicate climate reversals associated with cooler and wetter conditions around 1500 BCE (Magny et al. 2009). We also leveraged the preliminary analysis of 140 ancient wood remains analyzed in a previous study (Wagner et al. 2018),to select two samples from Northern and Southern Germany showing excellent preservation levels (Ham.382: 9.31% endogenous DNA content and Con.548: 16.45%). These samples dated back to the early stage of the Last Little Ice Age (Ham.382: 1415 - 1479 cal. CE, Con.548: 1410 - 1457 cal. CE), a climate context that was significantly colder than today (Luterbacher et al. 2004, 2016; Corona et al. 2010; Weikinn et al. 2017). As estimated based on the number of preserved tree rings, the specimen from Hamburg and Lake Constance were 30 years old at most (Figure S1). However, since coppicing was a widespread forestry technique during the Middle Ages, it cannot be rule out that they were sprouts of older trees. To obtain comparative genome data, we also collected modern leaves and DNA samples as well as previously published sequence data (Schmid-Siegert et al. 2017) from a total of three Q. robur, three Q. petraea as well as one first-generation Q. robur x petraea hybrid (Table S2). The decision to include a hybrid was based on the observation that hybridization between different oak species is very common (Bacilieri et al. 1996; Petit et al. 2004; Lepais et al. 2009; Reutimann et al. 2020) and we could, thus, not exclude the presence of Q. robur x Q. petraea hybrids among the three ancient wood remains analyzed. All modern samples came from sites near the ancient wood remains (Figure 2). Ages of the forests from which the modern samples had been taken ranged between 30 and 100 years (Table S2); the already published genome came from an individual that had been dendrochronologically dated to 234 years in 2012 and sequenced in 2017. Finally, we sequenced one Q. rubra from the section Lobatae, which split from the section Quercus between 47.9 and 54.1 Ma ago (Hipp et al. 2020), as an outgroup individual.
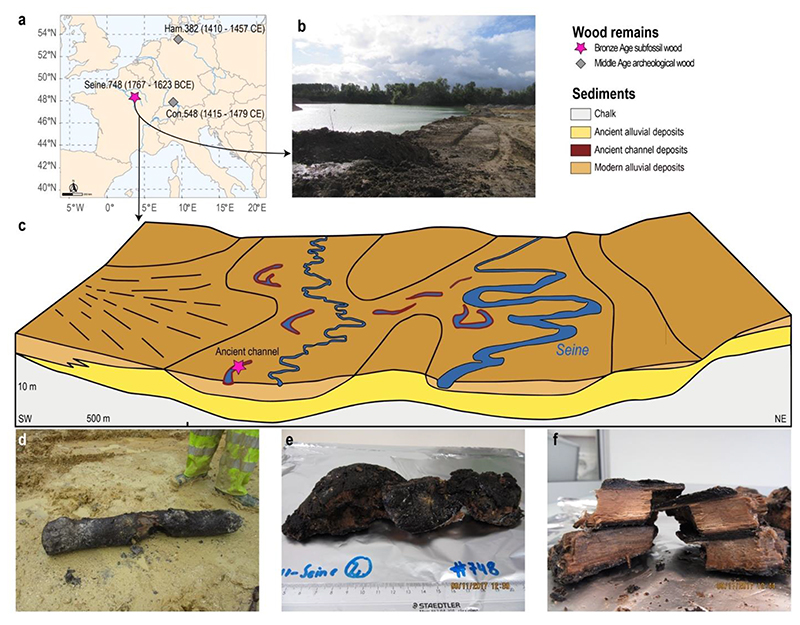
Figure 1. Origin of the ancient waterlogged oak wood remains analyzed in this study.
(a) Site locations of the newly excavated Bronze Age wood remain and the two Medieval wood remains (Wagner et al. 2018). (b) General view of the water-filled excavation site in the Seine Valley. (c) Stratigraphic scheme of the Seine river plain and the Bronze Age wood finding site. The climatic and environmental conditions at this site favored the preservation of DNA in waterlogged wood over millennia. (d) Sub-fossil oak log from the Seine river plain used in the analysis after its recovery from the sediments. (e) Bronze Age wood sample transferred to the ancient DNA laboratory and (f) Detail of its sapwood. Original source (c): www.developpement-durable.gouv.fr, 2017, mod. J. Brenot, S. Poirier and S. Wagner
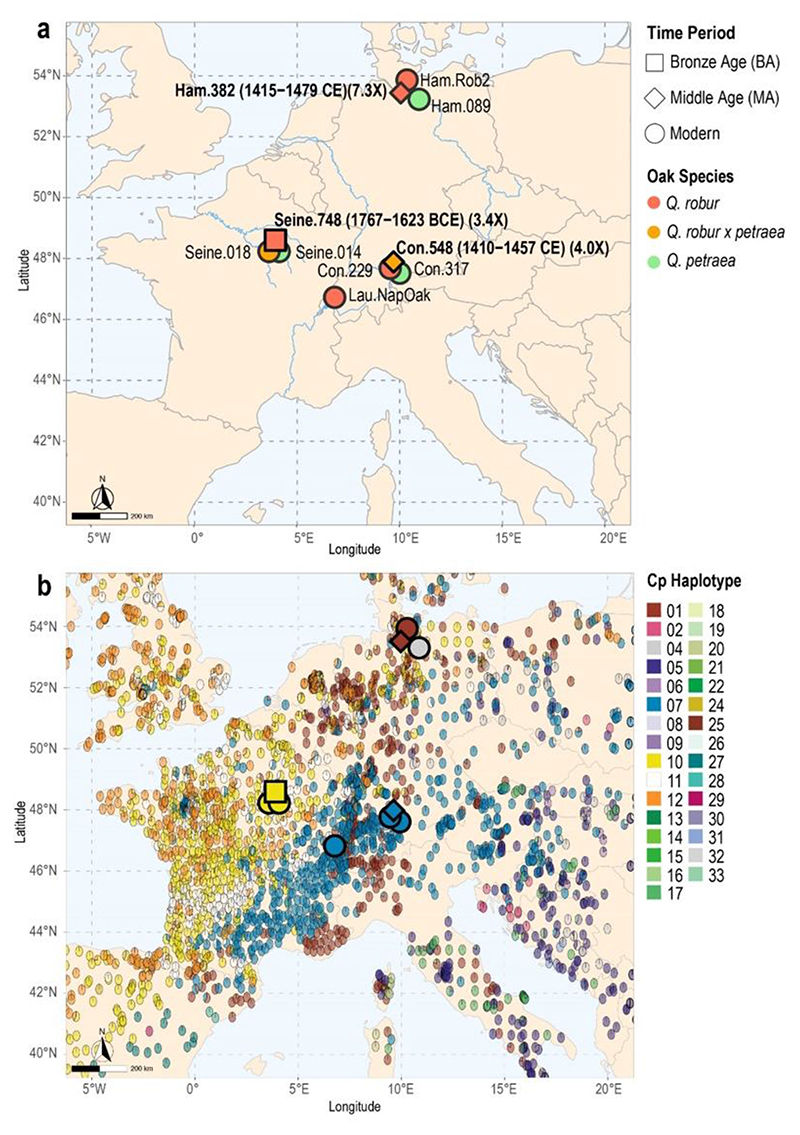
Figure 2. Genome coverage, species assignment and chloroplast haplotypes.
(a) Ancient genomes (bold letters) together with modern genomes analyzed in this study. Calibrated radiocarbon dates and average nuclear coverage for the ancient genomes are shown in brackets. Species assignment of the ancient oaks are based on diagnostic markers and genome-wide data analyses specified in sections 3.3 and 3.4. (b) Haplotypes assigned to ancient and modern oaks against a background of >2000 modern Q. robur and Q. petraea reference populations (source: https://gd2.pierroton.inra.fr/). Haplotype colors correspond to those used in the original publication (Petit et al. 2002).
2.2.1. Ancient DNA
The molecular work on ancient DNA, defined as DNA derived from organic material preserved for more than 200 years under natural conditions (Raxworthy & Smith, 2021), was carried out in the ancient DNA facilities of the Center for Anthropobiology and Genomics of Toulouse (CAGT, France) and the Center for GeoGenetics at the University of Copenhagen (Denmark) and followed strict experimental guidelines. For DNA extraction, genomic library construction and amplification, and DNA sequencing, we used a strategy previously established for ancient wood DNA (Wagner et al. 2018) with slight modifications. DNA extraction involved the following sequential steps for all samples: (1) removal of all external surfaces with a sterile scalpel, (2) crushing a 0.2 cm3 sapwood cuboid using a sterile mortar, (3) pre-digestion of the wood paste at 37°C for 1h in 3 ml lysis buffer consisting of 10 mM Tris-HCl, 10 mM NaCl, 2% w/v SDS, 5 mM CaCl, 2, 2. 5 mM EDTA, 40 mM DTT and 5% proteinase K, (4) removal of the supernatant, (5) digestion of the remaining wood paste at 42°C for 20h in 3 mL of fresh lysis buffer, (6) extraction of the DNA twice in phenol and then once in chloroform, (7) mixing the DNA supernatant with 3 mL Tris-EDTA (1x), (8) concentration on Amicon Ultra 4 (30 kD) filters, and; (9) purification on MinElute PCR purification columns (Qiagen, Hilden, Germany). The pre-digestion step was introduced to increase the proportion of endogenous DNA, following a technique used for animal remains (Damgaard et al. 2015). After initial screening on the new sample Seine.748, DNA libraries were constructed based on USER-treated DNA extracts following Meyer & Kircher (2010) and Gamba et al. (2016). These libraries were subsequently amplified in a 25 μL final reaction volume using 1 unit of AccuPrimeTM Pfx DNA polymerase, 4 to 6 μL DNA library and with an overall concentration of 0.2μM of each primer, including the InPE1.0 primer and a custom PCR primer. While only a single 6bp sequence tag was introduced by PCR in the original protocol (Meyer & Kircher 2010; Gamba et al. 2016), here, two additional 7bp sequence tags were introduced by ligation during DNA library construction (method B, Gaunitz et al. 2018). This triple indexed method was applied for samples Con.548 and Seine.748. For sample Ham.382, libraries that had been constructed based on the original single indexed method were already available for deep sequencing. Indexed DNA libraries were purified with Minelute columns (Qiagen, Hilden, Germany), eluted in 25 μL EB (10 mM Tris-Cl, pH 8.5) + 0.05 % Tween and quantified with the TapeStation 4200 instrument (Agilent Technologies). For contamination monitoring, blank controls were introduced at each laboratory step. Despite undergoing amplification attempts with the maximum number of cycles observed in the entire experimental sets (n=10, 13, 16 for samples 382, 548 and 748, respectively), all blank controls introduced at each step of the experimental workflow (i.e., DNA extraction, library construction and library amplification) revealed negative profiles on the TapeStation. To limit the overall clonality of the sequencing results, one to four library constructs per DNA extract and several amplifications of DNA libraries were carried out with different index primers (Tables S3 and S4). Purified DNA libraries were then pooled and sequenced on Illumina HiSeq2500 (single indexed library) and Illumina HiSeq4000 (triple indexed libraries) instruments at the Danish National High Throughput DNA Sequencing Center and the French National Sequencing Center (Genoscope, Evry) to generate three low-coverage genomes (average fold-of-coverage = 7.3X, 4.0X and 3.4X).
2.2.2. Modern DNA
Modern DNA was extracted from seven samples as described in Saleh et al. 2022 (samples Ham.089, Ham.Rob.2, Seine.014, Seine.018 and Q. rubra) and Rellstab et al. 2016 (samples Con.229 and Con.317) and paired-end libraries were sequenced on an Illumina HiSeq4000 sequencer at the French National Sequencing Center (Genoscope, Evry).
2.3. Read alignment, rescaling and trimming, authenticity assessment
Raw reads were demultiplexed based on index match allowing for 1bp mismatch on each individual internal index. Adapters and sequence tags were removed using AdapterRemoval v.2.2 (Schubert et al. 2016). Trimmed sequencing reads were mapped against the haploid version of the Q. robur reference genome (740 Mb/C: Plomion (2016, 2018) using PALEOMIX v1.2.13.5 (Schubert et al. 2014), with the bwa-back-track algorithm within the Burrows-Wheeler Aligner v.0.7.15 (Li et al. 2009). Seeding was disabled as recommended (Schubert et al. 2012), and alignments with a mapping quality below 25, unmapped reads and PCR duplicates were discarded. Reads were locally realigned around indels using the IndelRealigner procedure from GATK (McKenna et al. 2010). The same procedure was used for mapping against the oak chloroplast genome (160 kb: https://w3.pierroton.inra.fr/QuercusPortal/index.php?p=GENOMIC_SEQ). This alignment strategy was chosen following previous work demonstrating no confounding impact of nuclear organelle insertions (Wagner et al. 2018). The endogenous DNA content was calculated as the proportion of high-quality unique nuclear and chloroplast sequences remaining within the pool of sequences after trimming, quality filtering and removal of PCR duplicates. The distribution of fragment sizes was approximated using the size distribution of the high-quality collapsed read alignments. Ancient DNA authenticity was assessed by analyzing DNA fragmentation and nucleotide misincorporation patterns for nuclear and chloroplast DNA using mapDamage 2.0 (Jónsson et al. 2013; Schubert et al. 2014). To limit biases due to post-mortem DNA damage in downstream analyses, we trimmed and rescaled read alignments following Gaunitz et al.( 2018), and calculated the sequencing error rates per genome using ANGSD v.0.923 (Korneliussen et al. 2014). The same alignment procedure was used when analyzing the sequence data obtained from modern samples, with the exception of the trimming and rescaling procedures.
2.4. Ancient chloroplast haplotype assignment
To expand knowledge of the distribution of geographically structured maternal haplotypes of oak through space and time, we examined 34 commonly-used diagnostic loci for haplotype chloroplast assignment in modern and ancient oaks (Petit et al. 2002; Guichoux & Petit 2014; Wagner et al. 2018). For each sample, alleles of these loci with at least 8-fold depth of coverage and a minimum base quality of 30 were extracted using the PALEOMIX phylo_pipeline (Schubert et al. 2014). The haploid genotypes were determined using a majority rule applied to allele numbers found per locus with a threshold value of 90%, and the haplotypes were subsequently determined by comparison with the haplotype reference (Guichoux & Petit 2014). The haplotypes were then compared with haplotypes found in a range-wide high-resolution reference sample of 2,073 oak populations retrieved from the georeferenced GD2 database (https://gd2.pierroton.inra.fr/gd2).
2.5. Taxonomic assignment of archaeological and subfossil wood samples
Oak species classification is paramount to understanding their evolutionary history as well as for practical applications in forestry, dendroecology, dendroclimatology and dendroarchaeology. We first tested whether a set of 54 near species-diagnostic single nucleotide polymorphisms (SNPs, Leroy et al., 2020b, Kremer et al., in prep.) could be recovered from low coverage shotgun sequencing data and used for species identification. For the sake of clarity, near-diagnostic alleles at single nucleotide polymorphisms (SNPs), were identified thanks to a large whole-genome sequencing effort for the 4 species (Leroy et al. 2020b) and some SNPs exhibiting very large differences among species (usually Δf> 0.80 between one species and the three others). These positions correspond to the extreme of the FST distribution since most of the polymorphisms are shared among these species and the differentiation at neutral markers rarely exceeds 2% (Scotti-Saintagne et al. 2004). The marker set included 8-21 near-diagnostic positions per species and were genotyped using the PALEOMIX phylo_pipeline (minDepth=8, minBase=30). Approximating allele frequencies from a reference dataset of 186 present-day individuals of the four white oak species (Kremer et al. in prep.), we calculated the likelihoods of species membership, integrating the data over the ancient genotypes recovered.
2.6. Population structure and admixture
The following analyses were performed on the full set of chromosomes considering all three ancient genomes and the seven modern genomes. We first reconstructed the phylogenetic relationships between the ancient oaks and modern representatives of the Q. robur and Q. petraea groups, then assessed their genetic ancestry and tested for possible admixture. To avoid the effects of heterogeneous sequencing depths on genotyping results, we first calculated genotype likelihoods taking into account sequencing depths using the software package ANGSD v0.923 (Korneliussen et al. 2014), with filter settings as specified in the Supplementary Materials (Text S1). The phylogenetic reconstruction was carried out using TreeMix (Pickrell & Pritchard 2012), considering all transversion site positions represented across all individuals and showing a minimum genotype probability of 99%. Minimum allele frequency was set to 0.125. We ran TreeMix from 0 to 10 migration edges (for parameter settings and number of retained sites after filtering, see Text S2, Figure 4), determining the optimal number of migration edges using the package OptM (Fitak 2021) and running a total of 1,000 bootstrap replicates for assessing node support. We also ran an unsupervised admixture analysis using NGSadmix (Skotte et al. 2013) to infer the Q. robur / Q. petraea genetic ancestry proportions of the ancient genomes (k=2). A modern first-generation hybrid (Seine.018) was used as positive control and the robustness of the results was assessed through 25 pseudo-replicates including 25% random positions. Finally, we tested whether the tree topology recovered was sufficient to model the species evolutionary history or whether our data provided evidence for gene flow. To achieve this, we calculated D-statistics for the topologies ((Q. robur, Q. petraea), ancient), Q. rubra) and ((Q . robur, ancient), Q. petraea), Q. rubra) using ADMIXTOOLS, as implemented in the R package admixr (Patterson et al. 2012; Petr et al. 2019). The former formally tests for unbalanced proportions of genetic sharedness between the ancient specimen considered and Q. robur or Q. petraea, respectively. Conversely, the latter was implemented to test for an excess of genetic relatedness between the ancient specimen considered and Q. petraea. For computing Z scores, we used a weighted block jackknife, with block size set to 200kb.
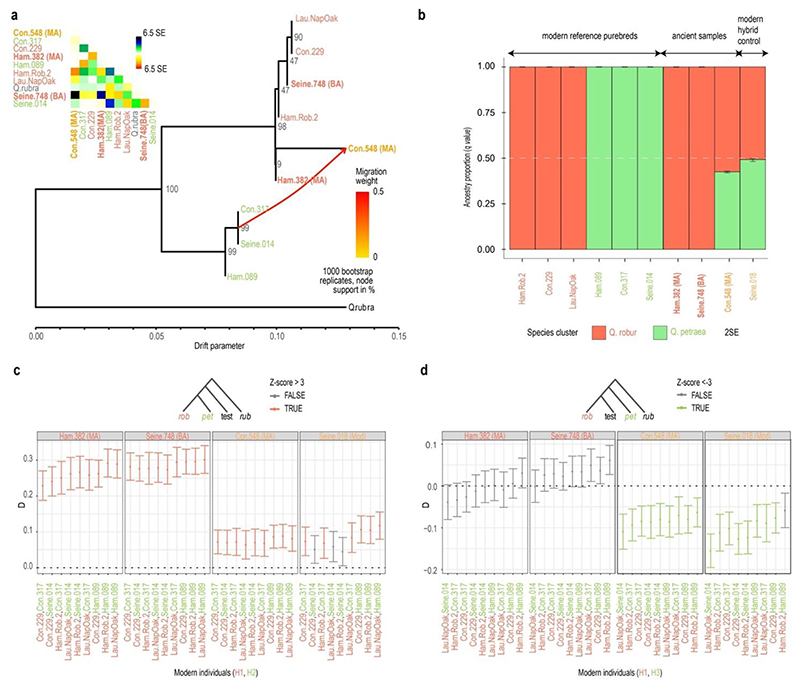
Figure 4. Phylogenetic relationships, gene flow and ancestry components.
(a) TreeMix consensus tree for the most likely topology for 3 ancient and 6 modern oak genomes and corresponding residues (inlet on the upper left). MA: Middle Age, BA: Bronze Age. The analysis was carried out on a total 186,975 transversion sites that were retained for all individuals after quality filtering (i.e., loci with minimum genotype likelihoods of 99% and minimum allele frequency of 0.125). (b) Unsupervised admixture analysis. Each bar shows the genetic ancestries present in a given sample. A modern hybrid was included as a positive control for comparison with the ancient hybrid. (c) D-statistics in the form of (((Q. robur, Q. petraea), test), outgroup) support a significant excess of genetic sharedness of all individuals tested (panel subtitles) and Q. robur. (d) D-statistics in the form of (((Q. robur, test), Q. petraea), outgroup). Configurations showing statistical support for non-null D-statistics are highlighted in color, and provide evidence of an excess of genetic sharedness between the Con.548 sample and Q. petraea, in line with its hybrid status. Similar support was detected in a modern hybrid individual used as positive control (Seine.018). Red fonts are used for Q. robur individuals, green for Q. petraea individuals and orange ones for Q. robur x Q. petraea hybrids.
2.7. Genetic diversity
We next investigated whether the global nucleotide diversities within ancient oak tree populations were as high as those observed within present-day oak tree populations. Since nucleotide diversity estimates are sensitive to genome coverage variation, we first used SAMtools v.1.9 (Li et al. 2009) to downsample all genomes to an equal average depth-of-coverage. This was repeated three times independently to reflect the different coverages of the three ancient genomes. We then estimated Watterson’s theta for transversion sites across the genome using ANGSD (Korneliussen et al. 2014) in a sliding window approach with 50kb non-overlapping windows. Window scores were normalized by their total number of sites (min = 12,500). As a control, individual global heterozygosity estimates at transversion sites were calculated using the same analysis software. The quality filters were the same as before (Text S2).
To assess whether low diversity regions had undergone diversity changes through time, we focused on those genomic windows that had higher nucleotide diversity in a subset of ancient genomes than in modern genomes. This analysis focused on Q. robur, as the only purebred species represented among the ancient samples. Q. robur individuals were grouped temporally and low diversity genomic windows were identified as those common to the 10% quantiles of each theta distribution (i.e., windows of low diversity identified for each individual and overlapping in the 3-4 individuals of the analysis sets specified below). For regions that showed higher nucleotide diversities, we then assessed if genomes were grouped by time period (all modern vs (medieval and Bronze Age)), (all modern and medieval) vs Bronze Age). We also identified checked regions showing low nucleotide diversities across all ancient and modern genomes (all robur), and those showing low nucleotide diversities across all modern and ancient genomes, with the exception of the Bronze Age sample (Seine.748; (all modern and Bronze Age) vs medieval). The annotated genes falling into the detected windows were identified using bedtools (Quinlan & Hall 2010) and the pedunculate oak gene prediction file (Plomion et al. 2018). Functional information was retrieved from Arabidopsis thaliana orthologues using Pathway Studio Plant software (Elsevier) and literature research.
2.8. Ancient phenology prediction
We chose to infer leaf unfolding behavior in the ancient samples analyzed as one of the fundamental climate-dependent processes that is central to the survival of trees. Previously published PoolSeq analyses had identified SNPs associated with leaf unfolding timing and their allele frequencies were estimated in 17 Q. petraea populations from across three geographical gradients (gradient 1: latitudinal gradient reaching from Southern France to Northern Germany; gradient 2: altitudinal gradient in the French Pyrenean valley of Luz with populations growing at average altitudes between 131 and 1,630 m a.s.l., and gradient 3: altitudinal gradient in the French Pyrenean valley of Ossau with populations growing at average altitudes between 259 and 1,614 m a.s.l.) (Leroy et al. 2020a). This study provided SNPs associated with leaf unfolding and subdivided in a leaf unfolding minor (N=825) and a leaf unfolding major (N=30) category (Leroy et al. 2020a). These categories had been based on the levels of differentiation, as assessed from the XtX statistic (Günther & Coop 2013), with the minor category representing SNPs with XtX values above the 0.999 quantile threshold and the main category representing SNPs with XtX values above the 0.99999 quantile threshold (Leroy et al. 2020a). We restricted our analyses to loci represented in all modern and ancient individuals, and generated 20 replicates of pseudo-haploidized genotypes for each individual using ANGSD (same filter settings as above) as an alternative genotyping strategy for low coverage data. We then calculated the likelihoods of belonging to one of the 17 Q. petraea populations from Leroy et al. (2020a) by integrating likelihoods across all loci and using the allele frequencies of these populations. Log-likelihood scores were calculated per population as follows:
where Sk is the kth population, X is the vector representing the sample haplotype and xi is the ith SNP in the sample haplotype. The leaf unfolding behavior of a sample was inferred as that of the population with maximum likelihood. We first used the three present-day Q. petraea genomes that were expected to show late leaf unfolding behavior with respect to Leroy’s Q. petraea reference populations for validation. We then applied our methodology to the modern Q. robur individuals for validation of the transferability of the method from Q. petraea to Q. robur before considering those predictions obtained on ancient individuals as reliable. In order to check if detected similarities extended to other sites, we applied the same procedure to SNPs shown to be correlated to annual mean temperatures and precipitation and published in the same study (Leroy et al. 2020a).
3. Results
3.1. First complete ancient tree genome sequences generated from wood
We characterized three ancient oak genomes from the DNA extracts of three waterlogged sapwood samples (Figure 2a). The oldest sample (Seine.748), newly excavated from the Seine valley, and was dated to the Bronze Age (BA; 1,767-1,623 cal. BCE) and sequenced for the first time in this study. Sequence data from the one DNA library constructed using raw DNA extracts showed typical post-mortem DNA damage, supporting authenticity. This included higher C-to-T (G-to-A) nucleotide mis-incorporation rates at sequence starts (ends), and depurination-driven DNA fragmentation (Figure S2). The endogenous DNA content of the BA sample was sufficient (9.2%) to allow whole-genome characterization using shotgun DNA sequencing at moderate cost. We thus treated the DNA extract with the USER enzymatic mix to reduce the presence of post-mortem DNA damage signature and proceeded with further sequencing on Illumina HiSeq instruments, generating a final genome sequence at an average 3.4-fold coverage. To provide comparative data, a total of three DNA libraries from two late Middle Age samples for which authentic ancient DNA had been recovered in a previous study were subjected to the same experimental procedure, leading to additional ancient genomes at an average 4.0-fold and 7.3-fold coverage (Figure 2a, Table S5). Sequence trimming and base quality rescaling further helped reduce the error rate to minimal levels (Orlando et al. 2015) (0.027 % to 0.048 % per base, Table S6). Finally, we completed our genome panel by sequencing modern DNA from two Q. robur, one Q. robur x petraea and three Q. petraea individuals resulting in six additional high-quality genomes, and mapping of sequencing reads for an additional Q. robur individual (Schmid-Siegert et al. 2017) (Figure 2a, Table S5).
3.2. Ancient chloroplast haplotype assignment
Three chloroplast haplotypes belonging to three different maternal lineages were identified from the ancient sequence data mapped against the chloroplast reference genome (>350-fold) (Figure 2b). The Bronze Age oak (Seine.748) was assigned to haplotype 10, which belongs to so-called lineage B, and is distributed from Spain to the British Isles in present-day populations (Petit et al. 2002). Haplotypes 7 (lineage A, from the Balkans to the Baltic Sea and southern France) and 1 (lineage C, from Italy to Scandinavia) were identified for the medieval samples Con.548 and Ham.382, confirming previous results based on more limited sequence data (Wagner et al. 2018). Overall, all ancient and modern haplotypes matched those haplotypes most commonly found in nearby present-day populations, with the exception of one modern Q. petraea individual from northern Germany (Ham.089), which was assigned to the less abundant haplotype 4 (lineage A).
3.3. Taxonomic assignment of archaeological and subfossil wood samples
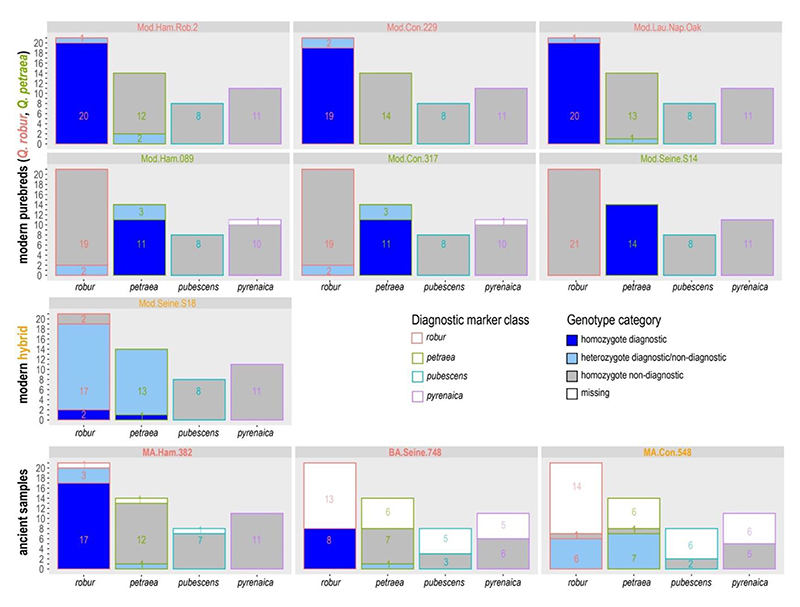
Previous work has identified a total of 54 nuclear loci with maximum differences in allele frequencies among pairs of present-day populations of Quercus robur, Q. petraea, Q. pubescens and Q. pyrenaica (Leroy et al. 2020a, Kremer et al. in prep.). For modern high-coverage genomes, we confirmed that the reference and alternative alleles of these loci were equally represented in the high coverage purebred and hybrid individuals (Figure S3). Although we did not sequence representative of Q. pubescens and Q. pyrenaica at high coverage, we assumed that this should also be the case for these species given the fact that markers have been designed in the same way. These loci could thus provide the basis for a first identification of the species underlying the ancient wood remains analyzed. However, it was unclear whether these loci were represented in low-coverage genomes generated based on ancient wood DNA. We found that most of the 54 loci were represented in all samples (without sequencing depth filter 54, 53 and 50 positions were covered by one or more sequencing reads in the three ancient samples, respectively), but that only some of them were covered by more than 8 reads, as expected given the low sequencing depths (Figure S3, Table S7). We successfully genotyped 20, 25, and 51 positions (minimum depth of coverage = 8X) for samples Con.548, Seine.748 and Ham.382, respectively (Figure 3, Table S7). The alleles observed in all three ancient samples segregate only in present-day robur and petraea populations. This was also the case for the seven modern oak samples of this study for that all positions were covered (Figure 3, Table S8). Both Ham.382 and Seine.748 were primarily characterized by the presence of near-diagnostic robur alleles found in homozygous state (Figure 3), with the exception of respectively one single allele at the heterozygous state. Both of these alleles are found at higher frequencies in present-day Q. petraea populations. Consistently, the likelihood of belonging to Q. robur given the distribution of alleles in present-day populations was considerably higher than the likelihood of belonging to Q. petraea for both samples (Ham.382: Log10((L(robur)/L(petraea)) = 54.9, Seine.748: Log10((L(robur)/L(petraea)) = 27.9). This supports the conclusion that the Ham.382 and Seine.748 samples are derived from Q. robur wood remains. Sample Con.548, however, showed all but one of positions in heterozygous state, and returned similar likelihoods of belonging to Q. robur or Q. petraea (Figure 3Con.548: Log10((L(robur)/L(petraea)) = -6). This suggests this sample is a possible cross-species hybrid, a conclusion which was explored further by inferring the population structure using the full genome data.
Figure 3. Taxonomic assignment based on 54 diagnostic markers.
Genotyping results of diagnostic loci per diagnostic marker class for modern controls and ancient samples. Minimum depth-of-coverage for genotyping was set to 8X; other loci were considered missing. Diagnostic alleles were found only for the marker classes robur and petraea and occurred mostly in homozygous state in purebreds and in heterozygous state in hybrids. Names of Q. robur, Q. petraea and Q. robur x Q. petraea hybrid individuals are written in red, green and orange letters, respectively. Mod: Modern, BA: Bronze Age, MA: Middle Age.
3.4. Population structure and admixture
TreeMix inference including one migration edge was found to best model the evolutionary history of the genome panel considered (Figure S4). It clearly separated the modern representatives of the Q. robur and Q. petraea into distinct phylogenetic clades, as expected (Figures 4a and S5-S7). Consistent with the analyses based on the 54 species informative markers, the medieval Ham.382 and the Bronze Age Seine.748 samples clustered within the Q. robur clade. While sample Con.548 was also placed in the Q. robur clade, it also received massive gene flow from Q. petraea (39.9%), confirming the preliminary assignment as a potential hybrid. Unsupervised admixture analysis provided similar results, with the genomes of both Ham.382 and Seine.748 samples almost exclusively derived from Q. robur ancestry (99.9%±2.3*10-7% and 99.9%±1*10-9%, respectively), and the Con.548 sample consisting of a mixture of Q. robur (57%±0.3%) and Q. petraea (43%±0.3%) genetic ancestries (Figure 4b). These analyses revealed balanced ancestry proportions in the first-generation modern hybrid (51%±0.5% Q. robur, 49%±0.5% Q. petraea), suggesting that our methodology could provide reliable admixture estimates. Finally, D-statistics supported these findings, showing cladal relationships between Ham.382 and Seine.748 samples and present-day Q. robur specimens (Figure 4c), but the presence of an excess of genetic sharedness between the Con.548 sample and present-day Q. petraea relative to Q. robur (Figure 4d). This confirmed its taxonomic assignment as a Q. robur x petraea specimen.
3.5. Genome-wide diversity patterns
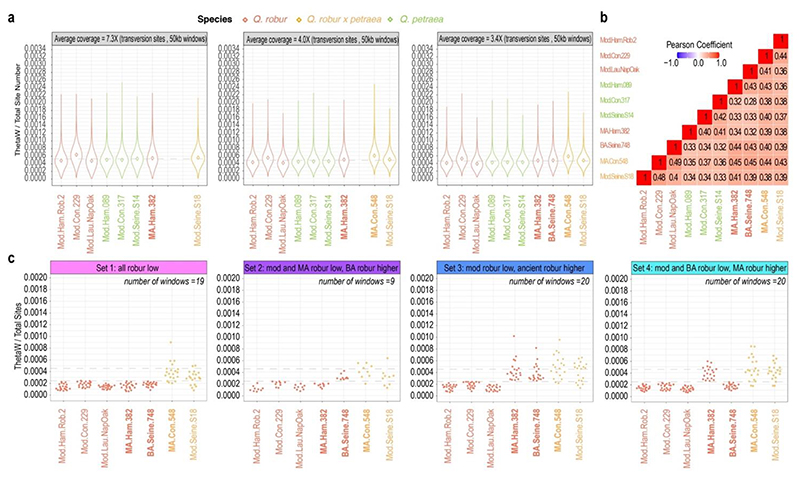
Next, we used our genomic time series to allow a first assessment of the diversity of oaks during the Bronze Age and 15th century CE compared to today. As the probability of identifying genetic variants increases with sequencing efforts, we downsampled the sequence data to the average depth-of-coverage obtained for the least-covered ancient genome (Figure 5a). At the lowest coverage level, we obtained between 11,991 and 13,360 windows with at least 12,500 total sites for each of the samples. The overall distribution of the normalized thetaW diversity found in the two ancient Q. robur samples was largely consistent with that found in present-day populations (Figure 5a, Table S9), suggesting there has been no major demographic shift between the Bronze Age to more recent times, which was also supported by a weakly positive correlation between nucleotide diversities of all samples (Figure 5b). However, given the long generation time of oaks, which reach seed maturity at 30-40 years in closed stands (Roloff et al. 2010), and the low number of generations assumed to have elapsed since the 15th century(N=15-20, when assuming that reproductive potential was reached at seed maturity; N <15 when assuming that full reproductive potential was reached later), this result cannot rule out relatively recent shifts. Consistent with a hybrid status, the medieval Con.548 sample showed slightly inflated thetaW estimates compared to present-day Q. robur and Q. petraea accessions, as well as compared to the other medieval sample (Ham.382; estimates as all others) (Figure 5a). Note that the maximum coverage was 7.3X and therefore not all heterozygous loci can be expected to be taken into account; i.e., values would be higher if high-coverage genomes were analyzed.
Figure 5. Genetic diversity patterns.
(a) Normalized nucleotide diversity for non-overlapping 50kb windows based on transversion sites. Sequence data were downsampled to the average depth-of-coverage of each individual ancient genomes. The dashed line indicates the group median at each coverage level. (b) Covariance of nucleotide diversities between samples (average coverage = 3.4X). (c) Genomic windows characterized by low nucleotide diversity within groups of genomes from different time windows: modern, Middle Age and Bronze Age (set 1), modern and Middle Age vs Bronze Age (set 2), modern vs Middle Age and Bronze Age (set 3), and modern and Bronze Age vs Middle Age (set 4). MA.Con.548 and Mod.Seine.018 hybrids are shown for comparison. The analysis was carried out at the lowest average coverage level (3.4X) and focused on Q. robur samples. The dashed lines indicate the Q. robur group median and the lower 10% quantile.
We next identified those genomic windows showing minimal genetic diversity among modern Q. robur individuals (first 10%-quantile of each of the individual thetaW distributions that overlapped between all individuals of a temporal group); those serve as proxies for genomic regions that have experienced diversity losses. This uncovered sets of a few low diversity windows characterizing four temporal groups: 19 genomic windows exhibited low diversity in all temporal contexts (i.e., potential diversity changes prior to 1,700 BCE as the calibrated age of the oldest sample (Figure 5c, Set 1)); 9 genomic windows showed minimal diversity in modern and medieval genomes, but not in the Bronze Age genome (i.e., potential diversity changes after the Bronze Age but before the Middle Age (Figure 5c, Set 2)); 20 genomic windows displayed minimal diversity in modern genomes but in none of the ancient ones (i.e., candidates for potential changes after the Middle Age (Figure 5c, Set 3)); and 20 windows exhibited minimal diversity today and in the Bronze Age but not during the Middle Age (i.e., potential fluctuating changes (Figure 5c, Set 4)). Each of the detected regions included several Q. robur genes (set 1: N=23, set 2: N=11, set 3: N=8, set 4: N=22), some of which match unique orthologs identified in Arabidopsis thaliana (set 1: N=19, set 2: N=11, set 3: N=7, set 4: N=16) and which have key biological functions involved in plant development, growth, fertility, pathogen defense and drought response (Tables S10-S13, Figure S8). These preliminary results need to be confirmed by additional ancient and modern samples but can provide starting points for studies focusing on the detection of selection signatures using other methods. It is important to stress that ancient tree genomes that can be generated based on abundant ancient wood remains can provide important elements in this context.
3.6. Phenology prediction
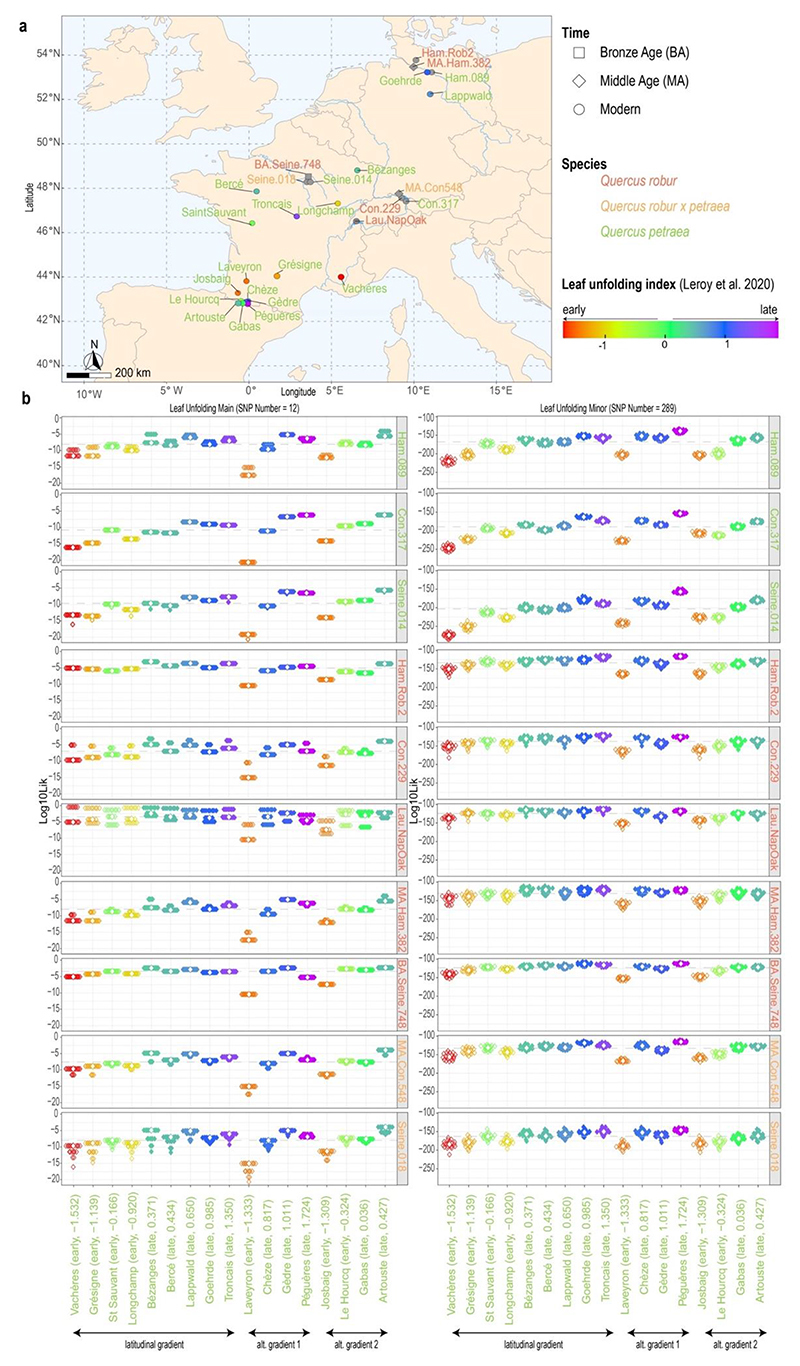
As previous work has revealed a fraction of the genetic variation that is associated with leaf unfolding behavior in modern Q. petraea populations (Figure 6a), we next attempted to assess this phenological trait in the three ancient trees analyzed here. We first validated our approach by predicting the trait for the three modern Q. petraea individuals in our sample, which derived from populations showing late leaf unfolding. Likelihood calculations for the combination of genotypes observed in these individuals returned maximum values for those populations showing late leaf unfolding, especially from northern latitudinal and high altitudinal environments (Figure 6b, 1st to 3rd row), as expected. We next tested whether expected trait values could also be inferred when considering modern specimens from the closely related Q. robur species. Likelihood calculations of the budburst minor category were consistent between replicates, and showed highest scores in Tronçais and Bézanges, Péguères and Gèdre and Artouste (Figure 6b, 4th to 6th row), which represent populations with the expected late leaf unfolding behavior. We thus concluded that our approach could return reliable phenological predictions, despite leveraging allele frequencies in the modern populations of only a single oak species (Q. petraea). Applied to the three ancient samples analyzed here, this method provided higher statistical support for late flushing (Figure 6b, 7th to second last row), suggesting that all three ancient samples likely exhibited late leaf unfolding. As expected similar tendencies for similarities between our samples and the reference populations could be observed when using temperature correlated SNPs, whereas no clear pattern resulted when using loci associated with precipitation (Supplementary Figures S9 and S10). This makes sense given that phenology has been shown to be strongly driven by temperature but less correlated with annual precipitation (Leroy et al. 2020a).
Figure 6. Phenology predictions.
(a) Reference populations with known leaf unfolding behavior and samples of this study. (b) Log10-likelihood of belonging to one of the reference populations. The reference populations are ordered by geographical gradient and within each gradient from earliest to latest leaf unfolding. Classification (early/late) and leaf unfolding index according to Leroy et al. (2020a) are shown in brackets after the population names. The probabilities were calculated for 20 pseudohaploid replicates and for loci that were presented in all samples. Left and right columns correspond to major and minor outlier loci identified by Leroy et al. (2020a). Each point represents one replicate. Open (closed) symbols are used for early (late) flushing populations. Dashed lines indicate median values across all populations within each sample; white diamonds indicate median values across replicates.
4. Discussion
The work described in this study extends the analysis of ancient wood material beyond the classical toolkit of dendroclimatology, dendrochronology, dendroarchaeology and dendroecology, thereby enhancing resolution of the response of forest ecosystems to past environmental changes. To the best of our knowledge, only two previous studies have applied Next Generation Sequencing to wood archaeological and subfossil material (Lendvay et al. 2017; Wagner et al. 2018). The first was limited to amplicon-sequencing data and was aimed at improving ancient DNA recovery from subfossil wood (Lendvay et al. 2017). The second applied the same methodological framework as that described here to 140 ancient oak sapwood samples, but the sequencing efforts resulted in only scattered genome data at extremely low nuclear coverage and focused on chloroplast data. By reporting full genome sequences with average depth-of-coverage between 3.4X and 7.3X, our study thus presents the first ancient genomes retrieved from tree subfossil material. It confirms the suitability of waterlogged sapwood remains for paleogenomic investigation. Interestingly, the oldest sample (Seine.748, c. 1700 BCE, 3.4X) was obtained from a subfossil tree log that was newly excavated from a Bronze Age site in the Seine river valley in Northern French. This is in agreement with a previous contention that water saturated anoxic clay sediments on calcareous bedrock provide favorable environments for DNA preservation in waterlogged contexts (Wagner et al. 2018). Therefore, these environments should be preferentially considered in future paleogenomic studies. Given the abundance of oak remains, and the relatively limited endogenous DNA content of the material investigated here (9.2-16.4%) and in previous studies (0.000343-16.4%, median = 0.02063%, N= 140), future wood paleogenomic research could alternatively take advantage of target capture approaches to generate nuclear datasets across space and time. Such approaches could leverage commercial reagent solutions (e.g., myBaits) or home-made methodologies from the hyRAD family, which were successfully applied to 7.2-ky old silver fir needles (Schmid et al. 2017) and ancient animal bones dating beyond the range of radiocarbon dating (Suchan et al. 2021, 2022). Our study is a first step in the characterization of past forests based on paleogenomes. By continuing this work, it will be possible to map the genomic makeup of whole tree populations through space and time and to address current questions such as the adaptability of long-lived trees to ongoing rapid environmental changes and forest epidemics.
The shotgun sequence data generated in this study was sufficient to characterize three ancient high-quality chloroplast genomes (>350X) that could be assigned to the more than 30 haplotypes identified within modern populations. The three resulting haplotypes correspond to the predominant haplotypes found in modern populations located in the vicinity of the sampling locations. Such congruent patterns of past and present haplotype distributions are consistent with local genetic continuity of maternal haplotypes; however, they do not rule out the existence of movements within each range, as discussed earlier (Wagner et al. 2018). The distribution of maternal lineages in oaks is a very important topic from an evolutionary biology perspective, especially since it is considered to be relatively unchanged despite the numerous environmental changes during the Holocene. The study of haplotype distribution is an important complement to the study of species distribution, as European white oak species share geographically structured haplotypes. Mapping combined haplotype/species data sets across space and time will provide a better understanding of oak populations persisting in different glacial refuge areas, postglacial spread into new regions, population establishment, and gene flow in already established populations.
Species identification based on nuclear diagnostic markers returned clear Q. robur assignments for a medieval sample from Hamburg (Ham.382) and the Bronze Age sample (Seine.748). This was reflected in high likelihood ratios in favor of Q. robur compared to Q. petraea (LL(robur/petraea): 55 and 28 for samples Ham.382 and Seine.748 respectively). In contrast, the likelihood ratio for the sample from Lake Constance (Con.548, LL(robur/petraea): -6) pointed to presumably more balanced proportions of each parental species, indicating the sample as a hybrid, although the number of successfully genotyped diagnostic loci was limited (N=6 and 7 for Q. robur and Q. petraea respectively). Note that no diagnostic alleles for Q. pubescens and Q. pyrenaica species were found in the ancient individuals, ruling out those species as the source of the wood material analyzed. Also, no diagnostic pubescens or pyrenaica alleles were found in any of the modern individuals, which indicates that at these latitudes there were no exchanges, neither ancient nor recent, between temperate species (robur and petraea) and species with a more Mediterranean preference (pubescens and pyrenaica). The current distribution range of Q. pyrenaica does not overlap with the study areas and no Q. pyrenaica alleles were expected. This is not the case for Q. pubescens. The current range covers the study area in southern Germany and the Seine valley, and it would have been conceivable to find Q. pubescens alleles in these places. For instance, Q. pubescens grows in Eastern France along the Rhine river at similar latitudes, with clear imprints of Q. pubescens introgression in Q. petraea (Neophytou 2014). Taking into account the environmental and climate conditions at the three study sites, we would have expected to find Q. robur rather than Q. petraea in all three cases, but given the overlapping ranges of the two species and the anatomical similarity of the wood, it would not have been possible to go beyond the genus level to confirm this assumption and, furthermore, to detect interspecific gene flow. This highlights the complexity of contemporary oak population structure and the importance of paleogenomics for species assignment, a better understanding the origin of the oak population structure and the interplay of the four species in the context of Holocene climate and environmental changes. Additionally, the species assignments made from diagnostic markers were confirmed using the full genome-wide datasets. Combined with chloroplast assignments, our study thus shows that species assignments based on nuclear diagnostic markers can provide a rapid and cost-effective methodology for reliably clarifying the origins of oak wood materials used in dendroclimatology, dendroarchaeology and archaeobotany or even art (Bauch & Eckstein 1981), as well as industry including cooperage (Deguilloux et al. 2004). Since species identification within the white oak section cannot be clarified by paleobotanical records, the migration dynamics of the different species during the post-glacial warming is so far unclear. Our analysis strategy opens up new avenues to track these dynamics and uncover clues to future distributions of these species.
Importantly, both Q. robur and Q. petraea species are common in the region of Lake Constance today and cross-species hybridization between these two species is frequent (Petit et al. 2004; Lepais et al. 2009; Reutimann et al. 2020). The identification of the Con.548 sample as a hybrid showing 58% Q. robur and 42% Q. petraea genetic material suggests that hybridization was already common ~600 years ago. This suggests that both species have been in contact in the Lake Constance region at least since the 15th century. Furthermore, our study reveals similar levels of genetic diversity across all samples, regardless of species (Q. robur, Q. petraea) and age. This is consistent with previous work revealing high nucleotide diversity in modern Q. robur genomes (Plomion et al. 2018) and suggests no global diversity depletion in the recent history of European white oaks. We estimated that at most 15-20 generations have elapsed between the Bronze Age and recent times, and it may be fewer in forests with trees being older than the ones we studied (approx. 30-100 years). We cannot completely rule out that recent shifts or minor ancient shifts have remained undetected but we do not believe that major diversity changes happened since the Bronze Age. This is in agreement with marker-based studies on modern trees that demonstrated intensive pollen-mediated gene flow between geographically distant gene pools that contributed to the maintenance of diversity during recolonization as well as during times when forests have already been established (Gerber et al. 2014, Petit et al. 2004). However, in an exploratory analysis, we identified some genomic regions that may have experienced a reduction of genetic diversity at different time periods. These regions represent candidates that can be scrutinized for signatures of positive selection in future studies. These results are in line with expectations from a three-generation comparison between oak populations with ages of approximately 350, 150 and 50 years (Saleh et al. 2022). This comparison indicated fluctuating selection driven by rapid warming after the Last Little Ice Age and resulting in fluctuating allele frequencies in some genomic regions. Given the long generation times, paleogenomic data sets are a wild card for the study of selection processes in trees because the time scale can be extended from a few to many generations and, moreover, relatively young macroremains can extend the insight provided by relatively old living oak trees that are rare in Europe.
Finally, we took advantage of the known allele contribution to leaf unfolding timing in modern Q. petraea populations to test whether phenological behavior could be inferred in the past. Our approach was able to replicate known leaf unfolding traits in both modern Q. petraea and Q. robur specimens, providing confidence in the traits inferred in ancient individuals. The best likelihood scores supported a proximity with late flushing Q. petraea populations for both the Bronze Age and medieval samples. This suggests a late leaf unfolding phenotype as the most likely, which is consistent with historical climate records and climate models revealing conditions cooler than today during the 15th century (Luterbacher et al. 2004, 2016; Corona et al. 2010; Weikinn et al. 2017). Historical climate conditions at the Bronze Age site are difficult to estimate since no local climate reconstructions are currently available. The geographically closest climatic reconstructions from the northwestern French Alps (Magny et al. 2009) suggest cooler and wetter conditions, which would be consistent with the predicted leaf unfolding behavior of the Seine.748 sample. Apical bud phenology is prone to rapid evolution due to its large genetic variation in natural populations (Alberto et al. 2011), its sharp response to temperature gradients (Firmat et al. 2017), and assortative mating that amplifies evolutionary response (Soularue & Kremer 2014). Thus, if our results are confirmed by additional data, they could lead to reciprocal predictions of ancient climatic conditions, given the genomic diversity of phenology related candidate SNPs. This extends the potential of paleogenomic analyses of ancient wood remains beyond species and lineage assignment, population structure and selection.
Supplementary Material
Acknowledgements
This research was supported by the European Research Council, through the Advanced Grant Project TREEPEACE (#FP7-339728), the Consolidator PEGASUS (#H2020-681605) and French ANR (GENOAK project, 11 - BSV6-009-021). Sequencing was performed at the Center for the Danish National High-Throughput Sequencing Center, Copenhagen, Denmark, the Center for Anthropobiology and Genomics of Toulouse, France, and the French National Sequencing Center (Genoscope), Evry, France. We thank all members from CAGT lab, especially Laure Tonasso and Stéphanie Schiavinato for managing the ancient DNA facilities and running screening experiments on the Illumina Miniseq instrument. We thank Christian Rellstab, Heike Hertel, Bernd Degen, Oliver Nelle, Joachim Koeniger, Uwe Heussner and Jan Vanmoerkerkefor assistance during sampling and providing sampling materials and DNA extracts. We thank Michael Börngen for extracting historical climate information. We thank Pablo Librado, Ludovic Duvaux and Isabelle Lesur for discussion and assistance. We thank the Genotoul Bioinformatics Platform Toulouse Midi-Pyrenees (Bioinfo Genotoul) for providing computing and storage resources. We thank Arnaud Bellec for hosting this project at the CNRGV and Catherine Bastien for coordinating work and scientific feedback.
Footnotes
Author Contributions
S.W., A.K. and L.O. conceived and designed the experiments. S.W. and A.S.O. performed the experiments, with input from L.O. S.W. and L.O. analyzed the data. J.C.L. performed gene annotations together with SW. S.W., A.S.O, J.C.L., T.L., C.L., K.L., J.M.A., S.P., P.W., C.P., A.K. and L.O. contributed with materials/experimental/analysis tools. S.W., A.K and L.O. wrote the manuscript with contributions from A.S.O., C.P. and T.L.
Data Accessibility
All the sequencing data are available for download from the European Nucleotide Archive under Accession no. PRJEB54826.
References
- Aguilera M, Ferrio Díaz JP, Araus Ortega JL, Tarrús J, Voltas Velasco J. Climate at the onset of western Mediterranean agriculture expansion: Evidence from stable isotopes of sub-fossil oak tree rings in Spain. Palaeogeography, Palaeoclimatology, Palaeoecology. 2011;299:541–551. [Google Scholar]
- Alberto F, Bouffier L, Louvet J-M, et al. Adaptive responses for seed and leaf phenology in natural populations of sessile oak along an altitudinal gradient. Journal of Evolutionary Biology. 2011;24:1442–1454. doi: 10.1111/j.1420-9101.2011.02277.x. [DOI] [PubMed] [Google Scholar]
- Alberto FJ, Derory J, Boury C, et al. Imprints of natural selection along environmental gradients in phenology-related genes of Quercus petraea. Genetics. 2013;195:495–512. doi: 10.1534/genetics.113.153783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberto F, Niort J, Derory J, et al. Population differentiation of sessile oak at the altitudinal front of migration in the French Pyrenees. Molecular Ecology. 2010;19:2626–2639. doi: 10.1111/j.1365-294X.2010.04631.x. [DOI] [PubMed] [Google Scholar]
- Bacilieri R, Ducousso A, Petit RJ, Kremer A. Mating system and asymmetric hybridization in a mixed stand of European oaks. Evolution. 1996;50:900–908. doi: 10.1111/j.1558-5646.1996.tb03898.x. [DOI] [PubMed] [Google Scholar]
- Bauch J, Eckstein D. Woodbiological investigations on panels of Rembrandt paintings. Wood Science and Technology. 1981;15:251–263. [Google Scholar]
- Bernabei M, Bontadi J, Rea R, Büntgen U, Tegel W. Dendrochronological evidence for longdistance timber trading in the Roman Empire. PloS One. 2019;14:1–13. doi: 10.1371/journal.pone.0224077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billamboz A. Regional patterns of settlement and woodland developments: Dendroarchaeology in the Neolithic pile-dwellings on Lake Constance (Germany) Vol. 24. The Holocene; 2014. pp. 1–10. [Google Scholar]
- Billamboz A, Martinelli N. In: The end of the lake-dwellings in the Circum-Alpine region. Menotti F, editor. Oxbow books; Oxford, UK, and Philadelphia, PA: 2015. Dendrochronology and Bronze Age pile-dwellings on both sides of the Alps: from chronology to dendrotypoloy highlighting settlement developments and woodland structural changes; pp. 68–84. [Google Scholar]
- Brockerhoff EG, Barbaro L, Castagneyrol B, et al. Forest biodiversity, ecosystem functioning and the provision of ecosystem services. Biodiversity and Conservation. 2017;26:3005–3035. [Google Scholar]
- Corona C, Guiot J, Edouard JL, et al. Millennium-long summer temperature variations in the European Alps as reconstructed from tree rings. Climate of the Past. 2010;6:379–400. [Google Scholar]
- Crowther TW, Glick HB, Covey KR, et al. Mapping tree density at a global scale. Nature. 2015;525:201–205. doi: 10.1038/nature14967. [DOI] [PubMed] [Google Scholar]
- Čufar K, Kromer B, Tolar T, Velušček A. Dating of 4th millennium BC pile-dwellings on Ljubljansko barje, Slovenia. Journal of Archaeological Science. 2010;37:2031–2039. [Google Scholar]
- Damgaard PB, Margaryan A, Schroeder H, et al. Improving access to endogenous DNA in ancient bones and teeth. Scientific Reports. 2015;5:1–12. doi: 10.1038/srep11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degen B, Yanbaev Y, Mader M, et al. Impact of Gene Flow and Introgression on the Range Wide Genetic Structure of Quercus robur (L.) in Europe. Forests. 2021;12:1–17. [Google Scholar]
- Deguilloux M-F, Pemonge M-H, Petit RJ. Novel perspectives in wood certification and forensics: dry wood as a source of DNA. Proceedings of the Royal Society of London. Series B. 2002;269:1039–1046. doi: 10.1098/rspb.2002.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguilloux M-F, Pemonge M-H, Petit RJ. DNA-based control of oak wood geographic origin in the context of the cooperage industry. Annals of Forest Science. 2004;61:97–104. [Google Scholar]
- Derory J, Scotti-Saintagne C, Bertocchi E, et al. Contrasting relationships between the diversity of candidate genes and variation of bud burst in natural and segregating populations of European oaks. Heredity. 2010;104:438–448. doi: 10.1038/hdy.2009.134. [DOI] [PubMed] [Google Scholar]
- Domínguez-Delmás M. Seeing the forest for the trees: New approaches and challenges for dendroarchaeology in the 21st century. Dendrochronologia. 2020;62:1–12. [Google Scholar]
- Firmat C, Delzon S, Louvet J-M, Parmentier J, Kremer A. Evolutionary dynamics of the leaf phenological cycle in an oak metapopulation along an elevation gradient. Journal of Evolutionary Biology. 2017;30:2116–2131. doi: 10.1111/jeb.13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitak RR. OptM: estimating the optimal number of migration edges on population trees using Treemix. Biology Methods & Protocols. 2021;6:1–6. doi: 10.1093/biomethods/bpab017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca RR, Smith BD, Wales N, et al. The origin and evolution of maize in the Southwestern United States. Nature Plants. 2015;1:1–5. doi: 10.1038/nplants.2014.3. [DOI] [PubMed] [Google Scholar]
- Gamba C, Hanghøj K, Gaunitz C, et al. Comparing the performance of three ancient DNA extraction methods for high-throughput sequencing. Molecular Ecology Resources. 2016;16:459–469. doi: 10.1111/1755-0998.12470. [DOI] [PubMed] [Google Scholar]
- Gaunitz C, Fages A, Hanghøj K, et al. Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science. 2018;360:111–114. doi: 10.1126/science.aao3297. [DOI] [PubMed] [Google Scholar]
- Gerber S, Chadœuf J, Gugerli F, Lascoux M, Buiteveld J, Cottrell J, et al. Kremer A. High rates of gene flow by pollen and seed in oak populations across Europe. PloS One. 2014;9(1):e85130. doi: 10.1371/journal.pone.0085130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard Q, Ducousso A, Gramont CB, et al. Provenance variation and seed sourcing for sessile oak (Quercus petraea (Matt.) Liebl.) in France. Annals of Forest Science. 2022;79:1–16. [Google Scholar]
- Guichoux E, Petit RJ. Déclaration d’invention (n° DI-RV-13-00566) auprès de l’INPI : Méthode de traçabilité géographique des bois de chêne. Patent. 2014
- Günther T, Coop G. Robust identification of local adaptation from allele frequencies. Genetics. 2013;195:205–220. doi: 10.1534/genetics.113.152462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutaker RM, Weiß CL, Ellis D, et al. The origins and adaptation of European potatoes reconstructed from historical genomes. Nature Ecology & Evolution. 2019;3:1093–1101. doi: 10.1038/s41559-019-0921-3. [DOI] [PubMed] [Google Scholar]
- Hipp AL, Manos PS, Hahn M, et al. Genomic landscape of the global oak phylogeny. The New Phytologist. 2020;226:1198–1212. doi: 10.1111/nph.16162. [DOI] [PubMed] [Google Scholar]
- Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler L, Bieker VC, Martin MD, et al. Ancient Plant Genomics in Archaeology, Herbaria, and the Environment. Annual Review of Plant Biology. 2020a;71:605–629. doi: 10.1146/annurev-arplant-081519-035837. [DOI] [PubMed] [Google Scholar]
- Kistler L, Maezumi SY, Gregorio de Souza J, et al. Multiproxy evidence highlights a complex evolutionary legacy of maize in South America. Science. 2018;362:1309–1313. doi: 10.1126/science.aav0207. [DOI] [PubMed] [Google Scholar]
- Kistler L, Newsom LA, Ryan TM, et al. Gourds and squashes (Cucurbita spp.) adapted to megafaunal extinction and ecological anachronism through domestication. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:15107–15112. doi: 10.1073/pnas.1516109112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler L, Thakar HB, VanDerwarker AM, et al. Archaeological Central American maize genomes suggest ancient gene flow from South America. Proceedings of the National Academy of Sciences of the United States of America. 2020b;117:33124–33129. doi: 10.1073/pnas.2015560117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay B, Hartmann M, Brodbeck S, et al. Improved recovery of ancient DNA from subfossil wood - application to the world’s oldest Late Glacial pine forest. The New Phytologist. 2017;217:1737–1174. doi: 10.1111/nph.14935. [DOI] [PubMed] [Google Scholar]
- Lepais O, Petit RJ, Guichoux E, et al. Species relative abundance and direction of introgression in oaks. Molecular Ecology. 2009;18:2228–2242. doi: 10.1111/j.1365-294X.2009.04137.x. [DOI] [PubMed] [Google Scholar]
- Leroy T, Louvet J-M, Lalanne C, et al. Adaptive introgression as a driver of local adaptation to climate in European white oaks. The New Phytologist. 2020a;226:1171–1182. doi: 10.1111/nph.16095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy T, Rougemont Q, Dupouey JL, et al. Massive postglacial gene flow between European white oaks uncovered genes underlying species barriers. The New Phytologist. 2020b;226:1183–1197. doi: 10.1111/nph.16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy T, Roux C, Villate L, et al. Extensive recent secondary contacts between four European white oak species. The New Phytologist. 2017;214:865–878. doi: 10.1111/nph.14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luterbacher J, Dietrich D, Xoplaki E, Grosjean M, Wanner H. European seasonal and annual temperature variability, trends, and extremes since 1500. Science. 2004;303:1499–1503. doi: 10.1126/science.1093877. [DOI] [PubMed] [Google Scholar]
- Luterbacher J, Werner JP, Smerdon JE, et al. European summer temperatures since Roman times. Environmental Research Letters. 2016;11:1–12. [Google Scholar]
- Magny M, Peyron O, Gauthier E, et al. Quantitative reconstruction of climatic variations during the Bronze and early Iron ages based on pollen and lake-level data in the NW Alps, France. Quaternary International. 2009;200:102–110. [Google Scholar]
- Martinelli N. Dendro: Chronologie, Typologie, Oekologie, Festschriftfür André Billamboz zum 65 Geburtstag. Freiburg i. Br; Janus Verlag: 2013. Dendrotypology in Italy: The case studies of the pile-dwelling villages Lucone D (Brescia) and Sabbione (Varese) pp. 117–124. [Google Scholar]
- Mascher M, Schuenemann VJ, Davidovich U, et al. Genomic analysis of 6,000-year-old cultivated grain illuminates the domestication history of barley. Nature Genetics. 2016;48:1089–1093. doi: 10.1038/ng.3611. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols. 2010;2010:db.prot5448. doi: 10.1101/pdb.prot5448. 1–7. [DOI] [PubMed] [Google Scholar]
- Neophytou C. Bayesian clustering analyses for genetic assignment and study of hybridization in oaks: effects of asymmetric phylogenies and asymmetric sampling schemes. Tree Genetics & Genomes. 2014;10:273–285. [Google Scholar]
- Nocchi G, Brown N, Coker TLR, et al. Genomic structure and diversity of oak populations in British parklands. Plants People Planet. 2022;4:167–181. [Google Scholar]
- Palmer SA, Clapham AJ, Rose P, et al. Archaeogenomic evidence of punctuated genome evolution in Gossypium. Molecular Biology and Evolution. 2012;29:2031–2038. doi: 10.1093/molbev/mss070. [DOI] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, et al. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A. Hybridization as a mechanism of invasion in oaks. The New Phytologist. 2004;161:151–164. [Google Scholar]
- Petit RJ, Csaikl UM, Bordács S, et al. Chloroplast DNA variation in European white oaks: Phylogeography and patterns of diversity based on data from over 2600 populations. Forest Ecology and Management. 2002;156:5–26. [Google Scholar]
- Petr M, Vernot B, Kelso J. admixr-R package for reproducible analyses using ADMIXTOOLS. Bioinformatics. 2019;35:3194–3195. doi: 10.1093/bioinformatics/btz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genetics. 2012;8:1–17.:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomion C, Aury J-M, Amselem J, et al. Decoding the oak genome: public release of sequence data, assembly, annotation and publication strategies. Molecular Ecology Resources. 2016;16:254–265. doi: 10.1111/1755-0998.12425. [DOI] [PubMed] [Google Scholar]
- Plomion C, Aury J-M, Amselem J, et al. Oak genome reveals facets of long lifespan. Nature Plants. 2018;4:440–452. doi: 10.1038/s41477-018-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmayanti Y, Leinemann L, Gailing O, Finkeldey R. Extraction, amplification and characterization of wood DNA from dipterocarpaceae. Plant Molecular Biology Reporter. 2006;24:45–55. [Google Scholar]
- Rachmayanti Y, Leinemann L, Gailing O, Finkeldey R. DNA from processed and unprocessed wood: factors influencing the isolation success. Forensic Science International Genetics. 2009;3:185–192. doi: 10.1016/j.fsigen.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Ramos-Madrigal J, Runge AKW, Bouby L, et al. Palaeogenomic insights into the origins of French grapevine diversity. Nature Plants. 2019;5:595–603. doi: 10.1038/s41477-019-0437-5. [DOI] [PubMed] [Google Scholar]
- Ramos-Madrigal J, Smith BD, Moreno-Mayar JV, et al. Genome Sequence of a 5,310-Year-Old Maize Cob Provides Insights into the Early Stages of Maize Domestication. Current Biology. 2016;26:3195–3201. doi: 10.1016/j.cub.2016.09.036. [DOI] [PubMed] [Google Scholar]
- Raxworthy CJ, Smith BT. Mining museums for historical DNA: advances and challenges in museomics. Trends in Ecology & Evolution. 2021;36:1049–1060. doi: 10.1016/j.tree.2021.07.009. [DOI] [PubMed] [Google Scholar]
- Rellstab C, Bühler A, Graf R, Folly C, Gugerli F. Using joint multivariate analyses of leaf morphology and molecular-genetic markers for taxon identification in three hybridizing European white oak species (Quercus spp.) Annals of Forest Science. 2016;73:669–679. [Google Scholar]
- Reutimann O, Gugerli F, Rellstab C. A species-discriminatory single-nucleotide polymorphism set reveals maintenance of species integrity in hybridizing European white oaks (Quercus spp.) despite high levels of admixture. Annals of Botany. 2020;125:663–676. doi: 10.1093/aob/mcaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roloff A, Weissgerber H, Lang U, Stimm B, editors. Bäume Mitteleuropas. Wiley-Vch; Weinheim: 2010. [Google Scholar]
- Saleh D, Chen J, Leplé J-C, et al. Genome-wide evolutionary response of European oaks during the Anthropocene. Evolution Letters. 2022;6:4–20. doi: 10.1002/evl3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S, Genevest R, Gobet E, et al. HyRAD-X, a versatile method combining exome capture and RAD sequencing to extract genomic information from ancient DNA (M Gilbert, Ed,) Methods in Ecology and Evolution. 2017;8:1374–1388. [Google Scholar]
- Schmid-Siegert E, Sarkar N, Iseli C, et al. Low number of fixed somatic mutations in a long-lived oak tree. Nature Plants. 2017;3:926–929. doi: 10.1038/s41477-017-0066-9. [DOI] [PubMed] [Google Scholar]
- Schubert M, Ermini L, Der Sarkissian C, et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nature Protocols. 2014;9:1056–1082. doi: 10.1038/nprot.2014.063. [DOI] [PubMed] [Google Scholar]
- Schubert M, Ginolhac A, Lindgreen S, et al. Improving ancient DNA read mapping against modern reference genomes. BMC Genomics. 2012;13:178. doi: 10.1186/1471-2164-13-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Research Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber FH. Microscopic Wood Anatomy. Kommissionsverlag Züricher AG; Zürich: 1990. [Google Scholar]
- Scott MF, Botigué LR, Brace S, et al. A 3,000-year-old Egyptian emmer wheat genome reveals dispersal and domestication history. Nature Plants. 2019;5:1120–1128. doi: 10.1038/s41477-019-0534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti-Saintagne C, Mariette S, Porth I, et al. Genome scanning for interspecific differentiation between two closely related oak species (Quercus robur L. and Q. petraea (Matt.) Liebl.) Genetics. 2004;168:1615–1626. doi: 10.1534/genetics.104.026849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotte L, Korneliussen TS, Albrechtsen A. Estimating individual admixture proportions from next generation sequencing data. Genetics. 2013;195:693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith O, Nicholson WV, Kistler L, et al. A domestication history of dynamic adaptation and genomic deterioration in Sorghum. Nature Plants. 2019;5:369–379. doi: 10.1038/s41477-019-0397-9. [DOI] [PubMed] [Google Scholar]
- Soularue J-P, Kremer A. Evolutionary responses of tree phenology to the combined effects of assortative mating, gene flow and divergent selection. Heredity. 2014;113:485–494. doi: 10.1038/hdy.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchan T, Chauvey L, Poullet M, et al. Assessing the impact of USER-treatment on hyRAD capture applied to ancient DNA. Molecular Ecology Resources. 2022;22:2262–2274. doi: 10.1111/1755-0998.13619. [DOI] [PubMed] [Google Scholar]
- Suchan T, Kusliy MA, Khan N, et al. Performance and automation of ancient DNA capture with RNA hyRAD probes. Molecular Ecology Resources. 2021;22:891–907. doi: 10.1111/1755-0998.13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts K, Gutaker RM, Benz B, et al. Genomic estimation of complex traits reveals ancient maize adaptation to temperate North America. Science. 2017;357:512–515. doi: 10.1126/science.aam9425. [DOI] [PubMed] [Google Scholar]
- Tegel W, Elburg R, Hakelberg D, Stäuble H, Büntgen U. Early Neolithic water wells reveal the world’s oldest wood architecture. PloS One. 2012;7:1–8.:e51374. doi: 10.1371/journal.pone.0051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegel W, Muig B, Skiadaresis G, Vanmoerkerke J, Seim A. Dendroarchaeology in Europe. Frontiers in Ecology and Evolution. 2022;10:1–31. [Google Scholar]
- Vallebueno-Estrada M, Rodríguez-Arévalo I, Rougon-Cardoso A, et al. The earliest maize from San Marcos Tehuacán is a partial domesticate with genomic evidence of inbreeding. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:14151–14156. doi: 10.1073/pnas.1609701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitasse Y, Delzon S, Bresson CC, Michalet R, Kremer A. Altitudinal differentiation in growth and phenology among populations of temperate-zone tree species growing in a common garden. Canadian Journal of Forest Research. 2009;39:1259–1269. [Google Scholar]
- Wagner S, Lagane F, Seguin-Orlando A, et al. High-Throughput DNA sequencing of ancient wood. Molecular Ecology. 2018;27:1138–1154. doi: 10.1111/mec.14514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales N, Akman M, Watson RHB, et al. Ancient DNA reveals the timing and persistence of organellar genetic bottlenecks over 3,000 years of sunflower domestication and improvement. Evolutionary Applications. 2018;12:38–53. doi: 10.1111/eva.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikinn C, Ronniger K, Renner S, et al. Weikinn’sche Quellensammlung zur Witterungsgeschichte Europas (Meteorologischer Teil) Leibniz-Institut für Länderkunde, Sächsische Akademie der Wissenschaften zu Leipzig; Leipzig: 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the sequencing data are available for download from the European Nucleotide Archive under Accession no. PRJEB54826.