Summary
Doublet microtubules of eukaryotic cilia and flagella are made up of a complete A- and an incomplete B-tubule that are fused together. Of the two fusion points, the outer junction is made of tripartite tubulin connections, while the inner junction contains non-tubulin elements. The latter includes flagellar-associated protein 20 (FAP20) and Parkin co-regulated gene protein (PACRG) that together link the A- and B-tubule at the inner junction. While structures of doublet microtubules reveal molecular details, their assembly is poorly understood. In this study, we purified recombinant FAP20 and characterized its effects on microtubule dynamics. We use in vitro reconstitution and cryo-electron microscopy to show that FAP20 recruits free tubulin to the existing microtubule lattice. Our cryo-electron microscopy reconstruction of microtubule:FAP20:tubulin complex reveals the mode of tubulin recruitment by FAP20 onto microtubules, providing insights into assembly steps of B-tubule closure during doublet microtubule formation.
Graphical abstract.
Introduction
Biological motility of eukaryotes mediated by cilia and flagella beating involves dynein-based sliding of axonemal doublet microtubules, which are conserved across organisms from unicellular ciliates and flagellates to multicellular eukaryotes.1–3 Defects in assembly of the doublet microtubule are associated with structural deformities and impairment of ciliary function implicated in many ciliopathies.4,5 The doublet microtubules are made of a 13-protofilament A-tubule, which resembles cytoplasmic microtubules and an additional, semi-circular B-tubule (with 10 protofilaments) fused to the A-tubule.6 Advances in cryo-electron microscopy (cryo-EM) and tomography have led to determination of several structures of an intact doublet microtubule from cilia/flagella.7–12 These models have revealed intricate details about the doublet microtubule architecture, various microtubule inner proteins decorating the A- and B-tubule as well as the outer and inner junctions between the A- and B-tubule. The outer junction is entirely mediated by tubulin components that form a tripartite lateral interaction between A- and B-tubule protofilaments.7,8,11,13 The assembly of outer junction has been demonstrated in vitro in presence of excess salts as well as in the absence of tubulin carboxy-terminal tails.14,15 However, the partially assembled B-tubule in this intermediate does not fuse with the A-tubule and presumably requires proteins of the inner junction to form the complete doublet microtubule.1,7 The doublet microtubule inner junction had long been suspected to be mediated via non-tubulin subunits1 and the components were later identified from high-resolution structures.7,8 An alternating set of flagellar-associated protein 20 (FAP20) and Parkin co-regulated gene (PACRG) protein units forms the core of the inner junction7,8,16 and recent high-resolution cryo-EM structures reveal additional protein subunits such as FAP45, 52, 106, 126, and 276 further strengthening the inner junction.7
Mutational studies in Chlamydomonas reveal that although PACRG and FAP20 proteins are not essential for the formation of flagella, their null mutants show abnormalities in the structure of axonemes (absence of beak structures and inner dynein arms) and severe defects in motility of the organisms.16–18 Similar effects are also observed in the case of depletion of a homolog of FAP20 in Paramecium, known as Bug22.19 Bug22 mutants in Drosophila show abnormalities associated with ciliopathies, neuronal defects, and impairment of sperm differentiation.20 FAP20 plays an important role in motile cilia function in zebrafish with body curvature and cardiac or gut symmetry defects in the FAP20 null and knockdown mutants.21 Recently, advances in genome sequencing and analysis have helped identify mutations in FAP20 gene linked to inherited retinal dystrophies in human patients.21,22
Interestingly, overlapping roles for both FAP20 and PACRG have been revealed in non-motile ciliary functions of behavioral learning, locomotion, lifespan, and size determination in C. elegans with structural defects (incomplete B-tubules and ruptured inner junctions) in the axonemes of the mutants.21,23 The absence of either PACRG or FAP20 causes slower sliding velocities with respect to dynein in Chlamydomonas but the effect on the double mutant is not cumulative.16 These observations suggest that PACRG and FAP20 play a role in the later stages of the inner junction assembly and contribute toward maintaining the structural integrity and functional aspects of the axoneme. So far, in vitro studies of inner junction proteins have been limited to experiments with recombinant PACRG co-purified with meiosis expressed gene 1 product (MEIG1),24,25 and there is little biochemical evidence to show the direct role of FAP20 in doublet formation and maintenance. To address this, we have purified a recombinant version of FAP20 and characterized its biochemical properties toward microtubules using in vitro reconstitution studies. Our studies with light and electron microscopy show that FAP20 recruits tubulin to the microtubule lattice independent of GTP. The reconstitution of FAP20 binding to a growing microtubule allowed us to identify flexible interactions in the microtubule:FAP20:tubulin complex and propose a model for inner junction assembly.
Results
FAP20 binding stabilizes the microtubule lattice
The properties of FAP20 binding to microtubules in vitro have not been reported so far, and to study this, we cloned, expressed, and purified a recombinant version of full-length FAP20 from Chlamydomonas reinhardtii by nickel-histidine affinity chromatography (See STAR Methods for details, Figure S1A). First, we performed co-pelleting assays with purified FAP20 and microtubules polymerized from goat brain tubulin in the presence of different nucleotides (Figures S1B and S1C). We found that FAP20 binds to all the microtubules in vitro, suggesting that FAP20 is insensitive to minor lattice perturbations due to different nucleotide states26 (Figure S1C). To confirm the interaction of FAP20 with microtubules, we performed cryo-EM experiments using purified FAP20 and GMPCPP-stabilized microtubules. The micrographs showed microtubules with projections of hair-like structures on the surface corresponding to bound FAP20 (Figure S1D).
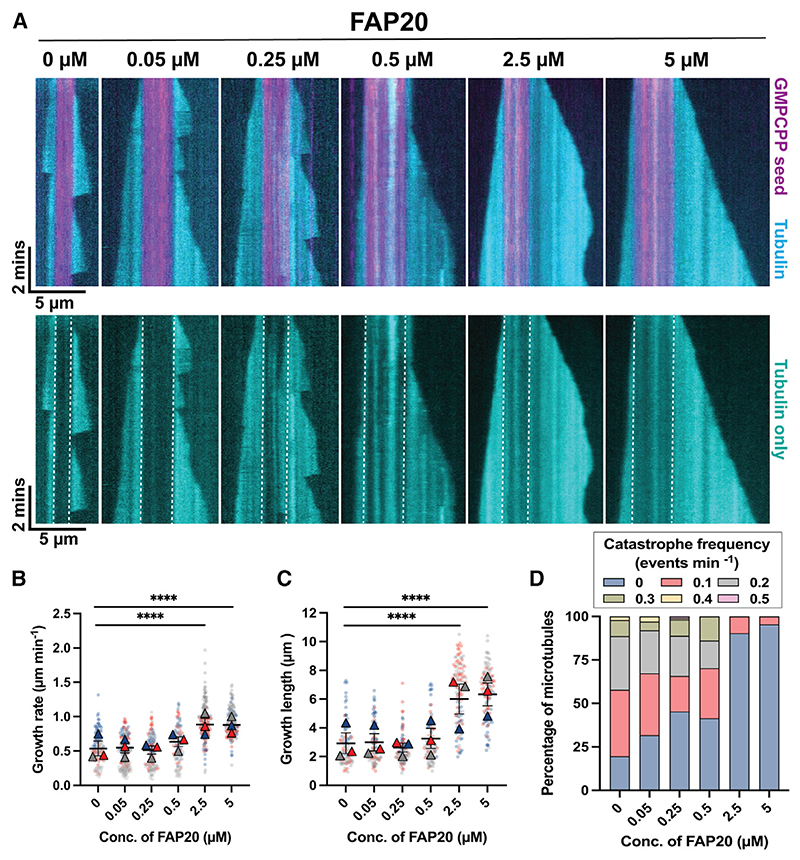
To assess whether FAP20 affects microtubule growth and stability, we then carried out microtubule growth assays in the presence of FAP20 (See STAR Methods for details, Figure 1A, Videos S1 and S2). From the kymograph analysis of total internal reflection fluorescence (TIRF) images, we found that at concentrations of 0.05, 0.25, and 0.5 μM, FAP20 did not induce any significant effect on the growth rate and growth length of the microtubules (Figures 1B, 1C, S2A, and S2B). However, the growth rates and growth lengths of microtubules increased significantly in the presence of 2.5 and 5 μM FAP20. The microtubule growth rates in the absence and presence of 2.5 and 5 μM FAP20 were 0.54, 0.89, and 0.88 μm min−1, respectively (Figure 1B). The microtubule growth lengths recorded were 2.93 μm in the absence of FAP20, 6.00 and 6.32 μm in the presence of 2.5 and 5 μM FAP20, respectively (Figure 1C). Another striking observation apparent from the kymographs was the reduced number of catastrophe events as the concentration of FAP20 was increased in the microtubule growth assays (Figure 1A). Quantification of the frequency of catastrophe events showed a marked decrease in a concentration-dependent manner. In the absence of FAP20, and with 0.05 and 0.25 μM FAP20, 80.4%, 68.3%, and 54.66% of the analyzed microtubules exhibited regular catastrophe events, respectively (Figures 1D and S2C). At higher concentrations of FAP20 (2.5 and 5 μM), almost no catastrophe events could be observed leading to the formation of long microtubules (Figures 1A, 1D, and S2D). Thus, the recombinant FAP20 protein makes stabilizing interactions with the microtubule lattice increasing the growth rate, reducing the number of catastrophe events, and eventually preserving the growing microtubule.
Figure 1. Microtubule dynamics in the presence of FAP20.
(A) Representative kymographs showing microtubule dynamics in the presence of different concentrations of FAP20. The upper panels show an overlay of the GMPCPP seed channel (magenta) and Cy5-labelled tubulin channel (cyan). Lower panels show the corresponding Cy5-labelled tubulin channel only. Dotted white lines in the lower panels denote the ends of GMPCPP-stabilized seeds.
(B and C) SuperPlots of microtubule plus-end growth rates and growth lengths in the presence of 10 μM tubulin and increasing concentrations of FAP20, as indicated. Average values from three independent experiments are shown as colored triangles and individual values from each experimental set are shown as circles in their respective muted colors. Error bars represent the standard error of the mean from three independent experiments. (N = 135: 0 μM FAP20, 130: 0.05 μM FAP20, 137: 0.25 μM FAP20, 118: 0.5 μM FAP20, 164: 2.5 μM FAP20, 150: 5 μM FAP20 for growth rate measurements and N = 97: 0 μM FAP20,101: 0.05 μM FAP20, 108: 0.25 μM FAP20, 94: 0.5 μM FAP20, 125: 2.5 μM FAP20, 110: 5 μM FAP20 for growth length measurements. ****p < 0.0001, Kruskal-Wallis test followed by Dunn’s multiple comparisons test).
(D) Frequency plot showing the percentage of microtubules undergoing different catastrophe frequencies at each FAP20 concentration. Data are obtained from three individual experiments (N = 97: 0 μM FAP20, 101: 0.05 μM FAP20, 108: 0.25 μM FAP20, 94: 0.5 μM FAP20, 125: 2.5 μM FAP20, 110: 5 μM FAP20). For individual values see Figure S2 and Data S1.
FAP20 recruits free tubulin to microtubule lattice
In our microtubule growth assays, we observed fluorescence signal from Cy5-labelled free tubulin accumulating at the existing microtubule lattice, both GMMPCP seeds as well as the newly polymerized microtubules (Figure 2A and Video S1). The tubulin accumulation on the existing microtubule lattice appeared sparse at 0.25 and 0.5 μM FAP20 and more pronounced at 2.5 and 5 μM FAP20 concentrations used in the assay (Figures 1A, Videos S1, and S2). We reasoned that FAP20 promotes the lateral association of free tubulin dimers to the microtubule lattice and investigated the role of GTP in this phenomenon. Under the same conditions without GTP, similar Cy5-labelled free tubulin accumulation was observed (Figure 2B and Video S3) suggesting that the FAP20 protein is the basic unit that links the microtubule lattice to the soluble tubulin dimer. While we observed different patterns of tubulin association with the GMPCPP seeds in presence of GTP (Figures S3A–S3C), the pattern of arrangement was found to be regular and continuous in the case of tubulin recruitment by 5 μM FAP20 when no GTP was present (Figures S3D and S3E).
Figure 2. FAP20-mediated recruitment of tubulin to the microtubule lattice.
(A) Representative TIRF images of GTP microtubules grown from GMPCPP seeds (magenta) in the presence of 10 μM tubulin (cyan) with and without 5 μM FAP20 at different time points, as indicated.
(B) Representative TIRF images of GMPCPP seeds (magenta) in the presence of 10 μM tubulin (cyan) with and without 5 μM FAP20 in the absence of GTP at different time points, as indicated.
(C) Mean fluorescence intensity of Cy5-labelled free tubulin along the GMPCPP seed with time for 10 min in the presence of 10 μM tubulin (light blue), +GTP (pink), +5 μM FAP20 (dark blue), and +5 μM FAP20 with GTP (red). Error bars represent the standard error of mean for values combined from three independent experiments.
(D) Fluorescence intensity line scans of Cy5-labelled free tubulin along the length of a microtubule in the presence of 10 μM tubulin (pink) and GTP, + 5 μM FAP20 without GTP (dark blue), +0.5 μM FAP20 with GTP (red, dashed line), + 5 μM FAP20 with GTP (red) at the end of 10 min. The vertical black lines in each curve represent the ends of the GMPCPP seeds for the respective experiments.
(E) Differences in final (T = 10 min) and initial (T = 0 min) normalized fluorescence intensity of Cy5-labelled free tubulin recruited per length (μm) of GMPCPP seed are shown in presence of 10 μM tubulin (light blue), +GTP (pink), +5 μM FAP20 (dark blue), and +5 μM FAP20 with GTP (red). Values from each experimental set are represented as different symbols and error bars represent the standard error of mean for values combined from three independent experiments (N = 5 for each experiment).
To gain further understanding of the tubulin recruitment pattern, we measured the fluorescence signal associated with bound tubulin dimers over time (Figure 2C) and the length of the microtubule (Figure 2D). We observed a uniform increase in tubulin recruitment over time on the GMPCPP-stabilized seeds, with increased recruitment in the presence of GTP compared to its absence (Figure 2C). This effect was also evident from intensity analysis over the length of the microtubule, where we observed a several fold increase in fluorescence intensity represented by the area under the curve corresponding to the GMPCPP seed only in the presence of GTP (Figure 2D). Here, the tubulin recruitment on existing microtubules showed a graded response ranging from sparse tubulin recruitment at 0.25 and 0.5 μM FAP20 concentrations to the entire length of GMPCPP seed being covered with free tubulin (Cy5-labelled tubulin fluorescence) at 5 μM FAP20 (Figures 2D, S3F, S3G, Videos S1, and S2). Quantification of the difference between initial and final normalized intensities of recruited tubulin to the GMPCPP seed in the presence of 5 μM FAP20 showed a large distribution of values (Figure 2E) suggesting no directed pattern of recruitment in the presence of GTP. The FAP20-mediated tubulin recruitment occurs rapidly as the reaction is set up and the initial stages could not be captured using TIRF imaging (Figures 2A, 2B, Videos S1, and S2). This is particularly evident in the absence of GTP, where the difference in normalized fluorescence intensities between the initial and final time points of the assay is negligible as compared to in the presence of GTP (Figure 2E). The tubulin recruitment saturates almost immediately in the absence of GTP, while in its presence the recruited tubulin dimers act as nucleation points for a steady-state lateral association. In summary, our TIRF imaging experiments have uncovered a unique property of FAP20, where it mediates binding of a tubulin dimer to the microtubule lattice in a GTP-independent manner.
FAP20 mediates flexible interaction of microtubule lattice with tubulin dimers as well as protofilaments
We next performed cryo-EM analysis to understand the FAP20-mediated tubulin recruitment onto the microtubule lattice. Samples were prepared by adapting the steps derived from our TIRF assay on transmission electron microscopy grids, which were then imaged in an electron microscope (See STAR Methods for details). In the absence of FAP20, we observed undecorated GMPCPP microtubules with a diameter between 25 and 30 nm (Figure 3A). In the presence of GTP and FAP20, we observed microtubule-like structures with twice the diameter ~50 nm (Figure 3B), indicating tubulin accumulation as observed in the TIRF images (Figure 2A). Analysis of the raw and filtered images as well as the power spectra suggests the presence of additional layers of protofilaments associated with each other on a conventional microtubule lattice structure (Figure S4A). Attempts to obtain a consensus high-resolution 3D reconstruction of this structure failed due to the heterogeneity of the sample and the complexity of the arrangement in the higher-order structure (Figure S4B). However, a low-resolution structure could be determined, which shows multiple stacks of protofilaments with a hollow center similar to the diameter of a conventional microtubule (Figure S4B).
Figure 3. Ultrastructural analysis of FAP20-mediated tubulin recruitment.
(A–C) Representative cryo-electron micrographs showing GMPCPP microtubules (A), +FAP20 and 10 μM tubulin with GTP (B), and +FAP20 and 10 μM tubulin (C). Inset: Zoomed in version of a section of microtubule outlined by a white box in the micrographs. Right: Longitudinal sections of a single microtubule segment with their Fourier filtered images to highlight the differences.
(D) Line scan plot of intensity distribution along the decorated region of microtubule+FAP20 and tubulin selected by the red dotted line in (C).
(E) Straightened segments of micrographs showing heterogeneous FAP20+tubulin ring/spiral formation along the microtubule lattice in the absence of GTP.
To obtain a more homogeneous sample, we then turned our attention to FAP20-mediated tubulin recruitment in the absence of GTP. In the absence of GTP, the electron micrographs showed the presence of the microtubule lattice (20–30 nm) decorated with rings and/or spirals (Figure 3C). From the line scan analysis of the filtered images (Figure 3D) and the layer lines in the power spectra of the helical segments (Figure S4A), a 10–11 nm regular interval of ring/spiral structures could be observed. Since the in vitro reconstitution system contains only microtubule, free tubulin, and FAP20, and the TIRF images show tubulin recruitment on existing microtubule lattice, we deduce that the rings/spirals observed here are higher-order structures composed of microtubule, FAP20, and tubulin dimers. The interaction between FAP20, tubulin dimers, and microtubule, however, appears to be flexible as the rings/spirals were found at different angles with respect to the microtubule axis and the vertical spacing between the rings/spirals, depending on the region analyzed might actually range around 11 nm (Figure 3E). Hence, we conclude that FAP20 mediates flexible interactions between the microtubule lattice and tubulin dimers in the absence of GTP. This association is retained with protofilaments when GTP is added to the sample.
Comparison of cryo-EM reconstruction of microtubule-FAP20-tubulin complex with doublet inner junction suggests conformational changes in assembly pathway
From the TIRF experiments and electron micrographs, the microtubule:FAP20:tubulin structure (in the absence of GTP) appeared to be less heterogeneous compared to microtubule: FAP20:tubulin:GTP complex (Figures 2E, 3B and 3C). Therefore, we attempted 3D reconstruction of microtubule:FAP20:tubulin complex to deduce its architecture and possible molecular interactions (See STAR Methods for details and Figure S5A). A low-resolution C1 reconstruction of the complex revealed that it is made up of three concentric layers (Figures 4A, S5B and Table 1). Our microtubule co-pelleting and cryo-EM experiments with purified FAP20 and microtubules demonstrated that FAP20 binds directly to the microtubule lattice. Further, our in vitro reconstitution TIRF and cryo-EM experiments unequivocally show the recruitment of tubulin by FAP20. Therefore in our 3D reconstruction, we assigned the innermost layer as microtubule, the intermediate layer as FAP20 bound to microtubule lattice, and the outermost rings as recruited tubulin (Figure 4A). Our 3D reconstruction is comparable to previously determined structures of similar microtubule-MAP-tubulin complexes28–30 that have ring-like structures featuring tubulin dimers (Figure 4A). To improve the resolution of the complex, we performed signal subtraction for the different layers and refined them individually (Figure S5A). While refinement of the microtubule lattice yielded a 3D reconstruction with sufficient distinction between alpha- and beta-tubulins, the particles with subtraction focused on the FAP20 layer and outer tubulin layer did not yield EM maps with defined structures for the intermediate layer and outer rings/spirals (FAP20 and tubulin dimer unit), possibly because of heterogeneity and flexibility in these layers (Figure S5A). In particular, the FAP20 layer was very poorly resolved and did not yield any discernible features that could aid in fitting the model of FAP20 obtained from cryo-EM studies on the isolated doublet.7,8 While the reconstructed maps corresponding to the outermost layer could accommodate only tubulin dimers with no additional space for FAP20, at this resolution, we cannot completely exclude the presence of FAP20 in this layer (Figure 4B). The horizontal protofilament along the ring/spiral showed a better fit into the density compared to the vertical alignment of tubulin dimers along the microtubule axis (Figure 4B). However, due to the highly heterogeneous nature of this layer (Figure 3E), the exact position, orientation, and interaction between tubulin dimers within the layer could not be accurately modeled.
Figure 4. Cryo-EM map and 3D reconstruction of microtubule: FAP20: tubulin complex.
(A) Surface representation of C1 reconstruction of microtubule: FAP20: tubulin complex obtained in the absence of GTP is shown in gray. Left: Cross section of the 3D reconstruction showing three concentric layers, representing the 14 protofilament microtubule (inner), FAP20 (middle), and tubulin dimers (outer). Right: Lateral views of the 3D reconstruction rotated by 180° along the vertical axis of the microtubule to show the rings/spirals of FAP20: tubulin complex and absence of break in the rings/spirals corresponding to the microtubule seam.
(B) Vertical versus horizontal protofilament arrangement model of tubulin dimers in the electron density map of microtubule:FAP20:tubulin complex from (A). Cartoon representations of atomic models of alpha- and beta-tubulin (obtained from AlphaFold2 prediction) are shown in green and blue, respectively. Height of a tubulin dimer and the approximate vertical distance of a tubulin spiral are indicated.
(C) Cross section of 3D reconstruction of microtubule:FAP20:tubulin complex from (A) shown in the top left corner with a zoomed in version out-lined in black. The electron density map corresponding to three protofilaments of the microtubule (inner) layer was fitted with three repeats of a model of FAP20 (red) bound to tubulin dimer (green/blue) obtained from AlphaFold2 prediction. The distance between the microtubule and outer tubulin layer is indicated with a double-sided arrow.
Table 1. Data collection statistics.
| Name | C1 reconstruction of microtubule: FAP20: tubulin complex (without GTP) |
|---|---|
| Microscope | Titan Krios G3-X-FEG |
| Voltage | 300 kV |
| Detector and mode | Falcon III detector and linear mode |
| Pixel size (Å) | 1.78 |
| Defocus range (μm) | −2.0 to −3.2 |
| Software used | EPU |
| Total electron dose (electrons/Å2) | 39.55 |
| Dose rate (electrons/pixel/s) | 62.5 |
| Exposure time (s) | 2 |
| Frames per movie | 20 |
| Number of micrographs | 1467 |
| Initial number of particles | 56145 |
| Final number of particles | 24621 |
| Resolution (Å)a | 11.6 |
| EMDB ID | EMD-34836 |
Resolution corresponding to gold-standard Fourier shell correlation (FSC) value of 0.143 between half maps.
In order to obtain a composite model of the microtubule:-FAP20:tubulin interaction, we attempted AlphaFold231,32 multimer-based structure prediction of complex of FAP20 and tubulin dimer and compared the predicted models with structures of the fully assembled doublet determined by cryo-EM. The AlphaFold2 model of FAP20 bound to a tubulin dimer showed high confidence (pTM+ipTM = 0.91) and was similar to the interaction of FAP20 with alpha-tubulin from A1 protofilament in the atomic models of the doublet (Figures S5C and S5D). The distinct spacing of FAP20-mediated tubulin rings in the structure, high confidence values (pTM+ipTM between 0.89 and 0.91) for top 5 models obtained from AlphaFold 2 (Figure S5D), and buried surface area (865.2 Å2) suggest binding of FAP20 to alpha-tubulin in the microtubule lattice (similar to A1 protofilament in the doublet microtubule). We fit the tubulin part of the model of FAP20 bound to the tubulin dimer (from AlphaFold2) in the electron density map (C1 reconstruction) of the microtubule (Figure 4C). Examination of the model and map revealed that the distance between the microtubule lattice and outer layer was 6 nm with no strong linking connections in the electron density map between the FAP20 and the microtubule or tubulin layers, indicating high flexibility of the interactions. The predicted atomic model of FAP20 associated with the fitted tubulin dimer aligned with the map corresponding to the FAP20 layer but showed space to accommodate a modified version of FAP20 or another molecule of FAP20 between the microtubule and outer tubulin layers. The interaction between FAP20 and tubulin dimers of the outer helix could not be analyzed due to insufficient resolution of the density to accurately fit the tubulin dimers. However, analysis of other interfaces of FAP20 with tubulin including that in the B-tubule in the doublet structure (PDB ID: 6u428) and in C2 microtubule of the ciliary central pair (PDB ID:7SOM27) gave low values of buried surface areas (FAP20: alpha-tubulin-135.2 Å2, FAP20: beta-tubulin-262.4 Å2 in doublet, and FAP20: beta-tubulin-184.7 Å2 in central pair) suggesting weak interactions between FAP20 and tubulin dimers of the outer helix. Hence, this structure might represent an intermediate in the formation of the inner junction which upon interaction with other proteins is stabilized to form the connection observed in the doublet microtubule.
In summary, we have purified FAP20 and characterized its interaction with microtubules and tubulin dimers. Using biochemistry, TIRF, and electron microscopy imaging, we thus conclude that FAP20 mediates flexible interactions between microtubule and tubulin dimers possibly by undergoing conformational changes or oligomerization.
Discussion
The architecture of the doublet microtubule inner junction is fundamentally different from that of the outer junction and is chiefly composed of FAP20 and PACRG repeating units connecting the B-tubule to the A-tubule.16 Previous studies on the inner junction have involved analysis of isolated doublet microtubules,7,8 Chlamydomonas FAP20 and PACRG mutants,16,17 and purified recombinant PACRG in complex with MEIG125; however, the properties of FAP20 have remained largely uncharacterized. In this study, we recombinantly purified FAP20 and characterized its role in mediating the inner junction connections using TIRF and electron microscopy. Microtubule growth and dynamic assays in the presence of FAP20 revealed an increase in microtubule growth rate and length at high concentrations along with a stark reduction in the frequency of observed catastrophe events. Similar stabilizing lattice interactions are often observed in case of microtubule inner proteins (MIPs) associated with the doublet microtubules. Chlamydomonas mutants of MIPs, FAP45, and FAP52 show higher depolymerization rates of the B-tubule compared to the wild type.33 Turbidity studies with recombinant FAP85, a MIP associated with the A-tubule, show reduced microtubule depolymerization rates at lower temperatures.34 Since FAP20 binds uniformly over the microtubule lattice in vitro, we conclude that this mode of microtubule polymer protection is unique to FAP20 among the microtubule doublet-associated proteins that have been characterized so far.
In addition to the microtubule stabilizing role of FAP20, our experiments revealed another salient property of FAP20, where it recruits free tubulin to the existing microtubule polymer. Based on the cryo-EM structure of the ciliary doublet,8 1 FAP20 molecule binds to one helical turn of the doublet (13 + 10 tubulin dimers) leading to a stoichiometry of approximately 1 FAP20 per 23 tubulin dimers. While the observed tubulin recruitment in our assays is most striking at concentrations of 2.5 and 5 μM FAP20, it is also evident at lower concentrations of 0.25 and 0.5 μM FAP20 (Figure 1A). The lower concentration range of 0.25 and 0.5 μM FAP20 corresponds to stoichiometric ratios of 1 FAP20 per 40 and 20 tubulin dimers, respectively, demonstrating the physiological relevance of our reconstitution studies. Recruitment of free tubulin to the existing polymer has been reported in the microtubule repair mechanism and occurs without the aid of any microtubule-associated proteins (MAPs).35 PACRG-mediated tubulin recruitment has also been reported recently and the tubulin has been shown to accumulate as distinct puncta at comparable concentrations.25 In our dynamic assays, FAP20-mediated tubulin recruitment along the GMPCPP seed gives a speckled appearance at concentrations of 0.25 and 0.5 μM FAP20 and a more thorough coating of the microtubule lattice at 2.5 and 5 μM concentrations of FAP20 (Figures 2A, 2D, S3F, S3G, Videos S1, and S2). The concentration of tubulin used in our assay (10 μM) eliminates any tubulin quenching effect due to its lateral association and an overall increase in growth rate and length of microtubules is observed at concentrations of 2.5 and 5 μM FAP20 (Figures 1B, 1C, and S2D).
Through cryo-EM studies, we further show that FAP20 mediates flexible interactions with tubulin dimers in the absence of GTP as well as larger assemblies of protofilaments when GTP is added (Figures 3B and 3C). The conformational heterogeneity observed in the cryo-EM 3D reconstructions can also be attributed to the different modes of tubulin binding that FAP20 adopts at the inner junction of A- and B-tubule of doublet microtubules and C2 microtubule of the central pair of ciliary apparatus.8,27 The higher-order structure of microtubule:FAP20:tubulin obtained in the presence of GTP appears similar to the “double walled microtubules” nucleated in the presence of polycations, peptides, or histones.36,37 In the presence of GTP, the microtubule:FAP20:tubulin complex acts as a nucleation point for the addition of more protofilaments or polymerization of tubulin dimers leading to the formation of puncta or thick higher-order structures manifesting in a steady increase of fluorescence along the GMPCPP seed over time (Figures 2C, Videos S1, and S2). The tubulin rings/spirals connected by FAP20 to the microtubule lattice obtained in the absence of GTP resemble similar tubulin dimer decorations in the microtubule structures determined in complex with depolymerizing kinesin-13, Ska1, and APC proteins.28–30 The periodicity of FAP20 interaction with the microtubule lattice as evident by the spacing between the FAP20-tubulin rings/spirals was found to be around 11 nm which is slightly higher than the 8 nm periodicity of FAP20 binding in the doublet microtubule.7,8,16 This difference can be attributed to a stagger of tubulin dimers in the outer ring as compared to the vertical axis of the microtubule due to FAP20 binding flexibly to alpha-tubulin leading to different helical symmetries for the microtubule and outer ring (Figure 4B). In contrast, kinesin-13 binds between alpha- and beta-tubulin and the spirals formed of kinesin-13 and tubulin follow the helical path of the tubulin heterodimer in the microtubule lattice.29,38 FAP20 mutations were identified in human patients with inherited retinal dystrophy cluster in the binding site of FAP20 with alpha-tubulin further highlighting the importance of this interaction.21
While the absence of FAP20 affects the beak structures in axonemes, it does not appear to be critical for other associated structures of the axoneme such as radial spokes, outer and inner dynein arms, and the dynein regulatory complex.17 High-resolution structures of the doublet microtubule suggest that FAP20 interacts with many inner junction proteins including PACRG, FAP52, FAP276, FAP106, FAP126, and FAP45.7,8 Extensive comparative analysis of expression patterns of these interacting proteins is lacking but studies point toward the relative independence of FAP20 expression. FAP52 mutants do not show a decrease in levels of FAP20 or PACRG7 and reports have shown reduced levels of PACRG and Rib72 in FAP20 mutants,17 but not vice versa,16 suggesting their recruitment to be sequential steps in a pathway. Since mutants of FAP20 or PACRG do not have incomplete B-tubules,16 it is likely that both these proteins play a role in the later stages of the doublet microtubule assembly after it has been initiated by the other proteins.33 However, the absence of FAP20 in Chlamydomonas leads to flaying of isolated axonemes upon the addition of ATP, indicating its importance in the stability of axonemes.17
Although more studies are required to identify the sequence of ciliary proteins and intermediate steps in organizing the doublet microtubule, based on the previously described inferences, we provide a basic model of assembly of the inner junction (Figure 5). The assembly of the B-tubule is initiated at the outer junction and stabilized by multiple proteins.7,11,15 As the B-tubule extends from the A-tubule, several MIPs lining the B-tubule and ribbon of the doublet (FAP52, FAP276, FAP106, FAP126, and FAP45) act synergistically via tubulin posttranslational modifications and aid in addition of requisite protofilaments for reaching the inner junction site.7,8,13 Mutations in these proteins are associated with ciliary defects in structure and function,39–42 emphasizing their role in cilia formation and maintenance. Upon a regulatory cue at this stage, FAP20, PACRG, possibly MEIG1, and similar such proteins depending on the organism might bind in different combinations to the partial doublet structure; it is still not clear if FAP20 and PACRG exist as or form heterodimers.7,16,25 Although we cannot rule out the role of a marker protein (between A13 and A1) such as FAP276 or tubulin posttranslational modifications at the carboxy-termini of B-tubules in recruiting FAP20 and PACRG to the inner junction,7 our TIRF microscopy and structural data suggest that FAP20 is capable of binding to the entire microtubule surface and not a single protofilament. This FAP20 binding, however, is stochastic and transient and can be cooperatively stabilized by PACRG or another accessory protein. At places along the inner junction where both FAP20 and PACRG make secure interactions between the A- and B-tubule, a stable intermediate is formed (Figure 5) which then propagates along the junction stabilizing it. Further studies with purified FAP20 and PACRG will be needed to show the combined effects of their binding and tubulin recruitment to the microtubule lattice. Finally, while higher resolution structures of in vitro reconstituted complexes at different stages of doublet assembly will be required to understand and validate the assembly mechanism in detail, the purification and reconstitution experiments with FAP20 in this study lay the foundation and provide a step closer towards polymerizing doublet microtubules in vitro.
Figure 5. Model for assembly of inner junction and closure of doublet microtubule.
Schematic model illustrating the steps of formation of the inner junction. Cross sections are shown on the top with the corresponding lateral views depicted below. Partial stabilization of B-tubule with proteins implicated in vital contacts at the inner junction (left) followed by the stochastic formation of a nascent inner junction in the presence of FAP20 and PACRG along the joining points of B- and A-tubule (middle) and finally the propagation of this stable inner junction segment along the longitudinal axis leading to the formation of the complete inner junction and the closure of B-tubule of the doublet microtubule (right). Density maps generated using the doublet microtubule model (PDB: 6u428) in chimera are depicted as a surface representation with alpha- and beta-tubulins from the A-tubule shown in green and blue, respectively, FAP20, PACRG, and MEIG1 in red, yellow, and brown, respectively, and alpha- and beta-tubulin from the B-tubule in light and dark gray, respectively. The proteins of the inner junction portrayed here include FAP45 (pink), FAP52 (peach), FAP106 (light green), FAP126 (beige), FAP276 (light blue), and RIB72 (salmon). The different possible combinations of FAP20, PACRG, and MEIG1 interactions along the microtubule lattice are shown in white dashed circles/ovals, while the stable inner junction segment is outlined in a white dashed rectangle.
Star★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| One Shot™ TOP10 Chemically Competent E. coli cells | Invitrogen™ | Cat# C404010 |
| Rosetta (DE3) competent cells | Novagen | Cat# 70954 |
| Chemicals, peptides, and recombinant proteins | ||
| Restriction enzymes BamH1 and Nde1 | New England Biolabs | BamH1: Cat# R0136 Nde1: Cat# R0111 |
| Carbenicillin disodium salt | Thermo Fisher Scientific | Cat# 10177012 |
| Chloramphenicol | Sigma-Aldrich | Cat# C0378 |
| IPTG | Sigma-Aldrich | Cat# I6758 |
| Phenylmethanesulfonyl fluoride (PMSF) | Sigma-Aldrich | Cat# P7626 |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail tablets |
Roche | Cat# 11836170001 |
| PIPES | Sigma-Aldrich | Cat# P6757 |
| Adenosine 5′-triphosphate disodium salt hydrate (ATP) | Sigma-Aldrich | Cat# A2383 |
| Guanosine 5′-triphosphate sodium salt hydrate (GTP) | Sigma-Aldrich | Cat# G8877 |
| Guanosin-5′-[(α,β)-methyleno]triphosphate, Sodium salt (GMPCPP) | Jena Bioscience | Cat# NU-405 |
| Guanosine 5′-[γ-thio]triphosphate tetralithium salt (GTPγS) | Sigma-Aldrich | Cat# 8634 |
| Paclitaxel, Taxus sp. | Sigma-Aldrich | Cat# 580555 |
| β-casein from bovine milk | Sigma-Aldrich | Cat# C6905 |
| Methyl cellulose | Sigma-Aldrich | Cat# M0262 |
| EZ-Link™ Sulfo-NHS-Biotin | Thermo Scientific™ | Cat# 21326 (Discontinued) |
| Alexa Fluor™ 647 NHS Ester (Succinimidyl Ester) | Invitrogen™ | Cat# A37573 |
| Alexa Fluor™ 568 NHS Ester (Succinimidyl Ester) | Invitrogen™ | Cat# A20003 |
| Bovine Serum Albumin, Biotinylated | Pierce™ | Cat# 29130 |
| Streptavidin | Invitrogen™ | Cat# 434302 |
| Protocatechuate 3,4-Dioxygenase (PCD) from Pseudomonas sp. | Sigma-Aldrich | Cat# P8279 |
| 3,4-Dihydroxybenzoic acid (Protocatechuic acid-PCA) | Sigma-Aldrich | Cat# 37580 |
| (±)-6-Hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (Trolox) | Sigma-Aldrich | Cat# 238813 |
| Goat brain tubulin | Purified according to Castoldi and Popov43 | N/A |
| Critical commercial assays | ||
| Gibson Assembly® Master Mix | New England Biolabs | Cat# E2611 |
| Deposited data | ||
| Cryo-EM structure of 14 protofilament undecorated GMPCPP microtubule | Zhang et al.26 | EMD 7973; PDB 6dpu |
| Cryo-EM structure of natively decorated ciliary doublet microtubule | Ma et al.8 | PDB 6u42 |
| Crystal structure of PACRG-MEIG1 complex | Khan et al.25 | PDB 6ndu |
| Cryo-EM structure of microtubule:FAP20:tubulin complex in absence of GTP | This study | EMD 34836 |
| Oligonucleotides | ||
|
pET17b CTer His Cr.CFAP20 FP TAActttaagaaggagatatacatATGTTCAAGAACGCC TTCCAATCCG |
This study | N/A |
|
pET17b CTer His Cr.CFAP20 RP CAGGTccttggaagaggacttcgagCGACTTCTGGATC GGCAGGAAG |
This study | N/A |
| Recombinant DNA | ||
| Chlamydomonas reinhardtii FAP20 cloned into pET17b | This study | N/A |
| Human kinesin-1 (1-324 G234A) cloned in pET17b | Rice et al.44 | N/A |
| Software and algorithms | ||
| Fiji | Schindelin et al.45 | https://imagej.net/software/fiji/ |
| Unblur | Grant and Grigorieff.46 | https://grigoriefflab.umassmed.edu/unblur_summovie |
| MotionCor2 | Zheng et al.47 | https://emcore.ucsf.edu/ucsf-software |
| GCTF | Zhang48 | https://www2.mrc-lmb.cam.ac.uk/download/gctf/ |
| RELION v3.1 | Scheres.49,50 | https://relion.readthedocs.io/en/release-3.1/ |
| COOT | Emsley et al.54 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Chimera | Goddard et al.,55 Pettersen et al.56 | https://www.cgl.ucsf.edu/chimera/ |
| ChimeraX | Goddard et al.,57 Pettersen et al.58 | https://www.cgl.ucsf.edu/chimerax/ |
| AlphaFold2 multimer | Evans et al.,31 Jumper et al.32 | https://github.com/deepmind/alphafold |
| GraphPad Prism Version 9 | GraphPad Software, Dotmatics | https://www.graphpad.com |
| Other | ||
| 5ml and 1ml His-Trap™ high performance column | Cytiva | Cat# 17524801 and 29051021 |
| 1ml HiTrap™ Q HP column | Cytiva | Cat# 29051325 |
| QUANTIFOIL® R1.2/1.3 on Au 300 mesh grids | Quantifoil | Cat# N1-C14nAu30-01 |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Minhajuddin Sirajuddin (minhaj@instem.res.in).
Materials availability
Expression plasmids used in this study are available upon request from the lead contact. This study did not generate new unique reagents.
Experimental Model and Study Participant Details
Bacterial strains
One Shot™ TOP10 Chemically Competent E. coli cells were used for cloning of FAP20 into pET17b and were cultured in Luria-Bertani broth containing 100μg/ml of carbenicillin at 37°C. Rosetta™ (DE3) E. coli cells transformed with plasmid pET17b were cultured in Terrific Broth containing 100μg/ml of carbenicillin and 25μg/ml of chloramphenicol at 37°C.
Method Details
Purification of proteins
Tubulin was purified by a series of polymerization and depolymerization steps from the goat brain using a standard protocol.43 The coding sequence for FAP20 was amplified from C. reinhardtii cDNA and cloned into pET17b vector between Nde1 and Kpn1 restriction sites with a carboxy-terminal deca-histidine tag. The FAP20 pET17b vector was transformed into Rosetta E. coli cells and a single transformed colony was used for inoculation in 25 ml of Luria Bertani broth for overnight growth as primary inoculum. 25 ml of this primary inoculum was added to 1L of Terrific Broth medium containing 100μg/ml of carbenicillin and 25μg/ml of chloramphenicol and incubated at 37°C for 4-6 hours. 0.5 mM IPTG was added for induction of 10X His tagged FAP20 and the culture was left under shaking conditions at 150 rpm, 25°C overnight. The next day, the cells were harvested and resuspended in a buffer containing 50mM HEPES pH 7.5, 500mM KCl, 10 mM imidazole, 5mM β mercaptoethanol, 100mM PMSF and EDTA free Protease Inhibitor Cocktail tablets (Roche, Cat No.11836170001) using a manual dounce. The resuspended cells were lysed by sonication followed by centrifugation to remove cell debris. The clarified lysate was loaded at a flow rate of 0.3 ml/min onto a Ni-NTA column pre-equilibrated with the above buffer. The loaded column was washed with 5 column volumes of wash buffer containing 50mM HEPES pH 7.5, 250 mM KCl, 40 mM imidazole, 1 mM ATP, 2 mM MgCl2 and 5 mM β mercaptoethanol to remove contaminating proteins. Column bound proteins were then eluted using a gradient of 40 mM to 350 mM imidazole in 50mM HEPES pH 7.5, 5mM β mercaptoethanol buffer. The protein that eluted at a concentration of around 200mM imidazole corresponded to FAP20 and was passed through Q column pre-equilibrated with buffer containing 50mM HEPES pH 7.5, 100 mM KCl and 5 mM β mercaptoethanol to remove other contaminants. We observed that FAP20 did not bind to Q column, however, the flowthrough was devoid of other contaminating proteins. The Q column flowthrough and wash were then passed through a Ni-NTA column again and eluted in a smaller volume, which was then concentrated using a centricon with 10 kDa molecular weight cut off. Imidazole was removed by 2 rounds of dilution and concentration with buffer containing no imidazole. The purity of the protein was confirmed by the SDS PAGE which showed a single band at around 20 kDa. Small aliquots (5-10 μL) of proteins were snap frozen in liquid nitrogen and stored at -80°C. From the structures of doublet microtubule and ciliary C1 apparatus,8,27 we confirmed that the carboxy-terminus of FAP20 is not involved in its interactions with tubulins and hence we have left the histidine tag positioned at the carboxy-terminus intact.
Microtubule co-pelleting assay
Purified goat brain tubulin (20 μM) was polymerized in BRB80 buffer (80mM PIPES, 1mM MgCl2, 1mM EGTA, pH 6.8 set with KOH) with 2 mM GTP followed by the addition of 20 μM taxol after 30 minutes and incubated at 37°C for 2-3 hours. Similarly, microtubules were also polymerized with a slowly hydrolysable GTP analog, 0.4 mM GMPCPP but without taxol. Microtubules supplemented with 2mM GTPγS were incubated at 37°C overnight in BRB80 buffer. 5μM FAP20 was incubated with polymerized microtubules for 20 minutes at room temperature followed by centrifugation in a TLA100 rotor at 279,000 x g for 10min at 35°C layered over a 60% glycerol cushion (60mL 100% glycerol, 20mL dd-water, 20mL 5X BRB80). The pellet was dissolved in 40uL of warm BRB80 and was analyzed along with the supernatant on a 10% SDS-PAGE. 10 μM human kinesin-1 (1-324 G234A)44 was incubated with microtubules for 20 minutes at room temperature and pelleted down following the same protocol. The comparative densitometric analysis of FAP20, kinesin and tubulin were done using Fiji software.45
In vitro reconstitution
1.5 mm coverslips were cleaned by sonication in acetone and stored in 80 % ethanol. Short GMPCPP microtubule seeds (spiked with biotinylated tubulin) were prepared by polymerization at 37°C for 30 minutes, spun on a warm 50% BRB80 sucrose cushion and resuspended in warm (37°C) BRB80 buffer. These seeds were attached by means of BSA-biotin-streptavidin linkage onto the coverslips in a flow chamber. Briefly the chamber (~10μL) was filled with solutions of BSA-Biotin (2mg/ml), followed by 2x washes with BRB80 buffer, then streptavidin (1mg/ml), followed by 2x washes with BRB80 buffer supplemented with casein (62μg/ml) and then the biotinylated GMPCPP microtubule seeds. Assay mix consisting of BRB80 buffer supplemented with β-casein (62μg/ml), 10mM β -mercaptoethanol, 2mM GTP, 0.1% methylcellulose and oxygen scavenging system (2.5mM PCA, 50nM PCD and 20nM Trolox) was prepared in ice. To a 20μl aliquot of this mix, 10uM tubulin (spiked with 647-Alexa labelled tubulin) and FAP20 was added and centrifuged to remove aggregates. The range of concentrations of FAP20 (0.05μM to 5μM) used was designed to include the physiological ratio of FAP20: tubulin found in ciliary doublets (1 FAP20 : 23 tubulin dimers)7,8 and the stoichiometric ratio of bound FAP20 to tubulin (0.25:1) obtained from microtubule co-pelleting experiments (Figure S1C). Dilutions of FAP20 were prepared in BRB80 buffer such that the volume of FAP20 added to the assay mix was kept constant. In case of 0nM FAP20, an equivalent volume of assay buffer was added instead of FAP20. The supernatant was warmed to 25°C, flowed into the flow chamber, and was immediately visualized using a Nikon TIRF-2 system at 25°C. Image sequences were collected at 561 and 640nm excitation wavelengths with an exposure time of 100 milliseconds at an interval of 2 seconds for 10 minutes.
Sample preparation and data collection for cryo-EM
Quantifoil gold holey carbon grids R 1.2/1.3 were glow discharged in a GloQube Glow Discharge (Quorum Technologies) system for 90 seconds. Sample application and vitrification was carried out in a Vitrobot Mark IV (ThermoFisher Scientific) set to 25°C and 100% humidity using a Whatmann No. 1 filter paper for blotting.
50μM tubulin was polymerized with 1mM GMPCPP at 37°C for 2 hours, spun on warm 50% BRB80 sucrose cushion and resuspended in warm BRB80 buffer. A mix of 10μM tubulin and 10μM FAP20 in BRB80 buffer or BRB80 buffer supplemented with 2mM GTP was prepared in ice and centrifuged at 4°C to remove aggregates. The supernatant was kept in ice and warmed only prior to application on the grids. 1μL of the GMPCPP microtubule seeds was applied on the grid mounted in the Vitrobot. After a brief incubation of 10-20 seconds to allow for the adsorption of the seeds onto the grid, 3μL of pre-warmed FAP20-tubulin mix was applied to the same grid. The grids were then blotted after an incubation time of 1 minute with a blot force of 0 and blot time of 3 seconds before plunging into liquid ethane. For microtubule + FAP20 sample, 1 μL of GMPCPP microtubules and 3μL of 7.5μM FAP20 were incubated at room temperature for a minute before application on grid, blotting and plunge freezing. The frozen grids were transferred to an Autogrid under liquid nitrogen conditions and mounted into a Titan Krios (FEI) operating at 300keV. Data collection was carried out using the automated pipeline of EPU software system with the Falcon III detector in integration mode at the National Cryo-EM facility, Bangalore. Images were collected at a magnification of 47,000x resulting in a pixel size of 1.78Å and defocus range between -2 to -3.2 μm. Each image had a total electron flux of 62.15 electrons/pixel/sec resulting in a total dose of 39.55 electrons/Å2 and 20 movie frames were stored.
Cryo-EM image processing
Images from multiple datasets were manually inspected for the presence of contaminating ice and filament quality and poor-quality micrographs were discarded. Motion correction of movie frames was carried out using Unblur46 or MotionCor247 and CTF estimation was done on dose-weighted motion corrected summed images using GCTF.48 Summed images were used for the generation of power spectra in Fiji software. All subsequent image processing steps were carried out in Relion v3.1.49,50 Helical segments were manually picked using Relion’s helical picker51 and 2x binned particles were extracted using a box size of ~780Å and overlapping inter-box distance of 82 Å. Microtubule segments containing different protofilament numbers were segregated using supervised 3D classification involving comparison with references representing microtubules made of 11 to 16 protofilaments.52 The 14 protofilament microtubules formed the major fraction of the population and were selected for further processing. In case of microtubule:FAP20:tubulin with GTP dataset, this subset of unbinned 14 protofilament containing microtubule segments was used for several runs of 3D refinement using a combination of cylinder and low-pass filtered microtubule as references, with and without imposing helical symmetry and in absence and presence of soft masks but a well resolved consensus structure could not be obtained (Figure S4B). This could be because of high levels of uncontrolled heterogeneity in the sample that could not be segregated by 3D classification. For the microtubule:FAP20:tubulin (without GTP) dataset, the set of unbinned 14 protofilament microtubules was used for 3D refinement with helical symmetry (twist=-25°, rise=9Å) using a cylinder with inner and outer diameters of 18 and 55 nm respectively as reference. The obtained model was again refined with imposed helical symmetry for 14pf microtubules (twist=-25°, rise=8.5Å) and a soft mask extending between 35 to 60 nm and containing 30% of the filament to improve the alignment of the outer tubulin rings. This model was then used for 3D refinement without symmetry starting with a sampling rate of 1.8 degrees to give the C1 reconstruction (Figure 4A). Examination of the obtained structure revealed that the different layers of microtubule, FAP20 and outer tubulin rings possibly followed different helical parameters leading to improved features in parts of the reconstruction depending on the focus and masks applied. In order to improve the reconstruction of each layer, signal subtraction was applied to the particles using customised masks focussing on each concentric layer. Refinement of subtracted particles containing only the microtubule was carried out using GMPCPP microtubule (EMD 797353) low pass filtered to 15Å as the reference. For subtracted particles focused on FAP20 and outer tubulin ring layers, 3D classification with and without symmetry was attempted to solve the problem of heterogeneity but was unsuccessful. The subtracted particles for each layer (FAP20 and outer tubulin helix) were then independently refined using suitable references and masks (Figure S5A). The data collection and refinement statistics are provided in Table 1.
Since the resolution of the maps obtained was not sufficiently high enough to build atomic models unambiguously, AlphaFold2 was used to obtain atomic models that could be fitted into the electron maps. The models obtained were docked into the map using Chimera’s ‘Fit in Map’ option. Superposition of atomic models was carried out in Coot54 and images were made using Chimera55,56 and ChimeraX.57,58
AlphaFold2 prediction and analysis
Protein sequences for goat brain tubulin (Uniprot IDs A0A452F3L8 for alpha-tubulin and A0A452FIJ2 for beta-tubulin) or C. reinhardtii tubulin (Uniprot ID P09204 for alpha-tubulin and Uniprot ID P04690 for beta-tubulin) and FAP20 (Uniprot ID A8IU92) were used as inputs for locally installed AlphaFold2 multimer.31,32 The model with the highest confidence calculated as a combination of pTM (predicted TM-score) and ipTM (interface predicted TM-score) was used for fitting and analyses.
Quantification and Statistical Analysis
The TIRF imaging data shown in Figures 1B–1D, 2C–2E, and S2A–S2C were analyzed with Fiji software45 including drift correction when necessary, using StackReg59 rigid body transformation. Microtubule growth rates (Figures 1B and S2A), growth lengths (Figures 1C and S2B) and catastrophe frequencies (Figures 1D and S2C) were calculated from kymographs plotted from single isolated microtubules manually selected in the maximum intensity projections of the movie frames. FAP20 together with tubulin can cause aggregation at higher concentrations of FAP20 (2.5μM and above) at the assay temperature. Efforts were taken to avoid microtubules with external aggregates along the growing lattice for analysis of growth and other parameters of microtubule dynamics. Growth length refers to the total length of the newly polymerized segment of the microtubule for the duration of the experiment. In the case of higher concentrations of FAP20 (2.5 and 5μM) catastrophe events were considered when microtubules shortened by at least a third of their growth length. Intensity measurements were made in Fiji over time (Figure 2C) or microtubule length (Figure 2D) for manually selected single isolated microtubules. The intensity in Figure 2E was normalized by subtracting intensity values over the same selected region between microtubules and an adjacent area in the field of view. For each condition, data were analyzed from 3 experiments in which independently purified batches of FAP20 were used and graphs were plotted using GraphPad Prism version 9. SuperPlots were generated as described earlier.60
Supplementary Material
In brief.
Bangera et al. purified and characterized a recombinant version of FAP20, an essential ciliary doublet microtubule inner junction protein. In vitro reconstitution assays and cryo-EM analysis revealed the ability of FAP20 to recruit tubulin to the microtubule lattice, providing critical insights into the assembly of axonemal doublet microtubules during ciliogenesis.
Highlights.
In vitro characterization of ciliary doublet microtubule inner junction protein, FAP20
FAP20 recruits tubulin dimers to the microtubule lattice
Cryo-EM structure of microtubule:FAP20:tubulin complex
Interactions between FAP20 and tubulin are highly flexible
Acknowledgments
The authors acknowledge the National Cryo-EM facility at Bangalore Life Sciences Cluster (BLiSc), the funding by the B-life grant from the Department of Biotechnology (DBT/PR12422/MED/31/287/2014) and K. R. Vinothkumar (National Centre for Biological Sciences, India) for the help in EM data collection. M.B. would like to thank David Houldershaw and Birkbeck IT facility for computational support. We thank K. R. Vinothkumar, Sudarshan Gadadhar, and Aakash Mukhopadhyay for their comments on the manuscript. M.B. acknowledges the SERB-NPDF fellowship (PDF/2016/002682). M.S. acknowledges funding support from inStem core grants from the Department of Biotechnology, India, DBT/Wellcome Trust India Alliance Intermediate Fellowship (IA/I/14/2/501533), EMBO Young Investigator Programme award, CEFI-PRA (5703-1) from the Department of Science and Technology, SERB-EMR grant (CRG/2019/003246) and DBT-BIRAC (BT/PR40389/COT/142/6/2020) grant.
Footnotes
Author contributions
M.B. and M.S. conceived the project. M.B. purified FAP20, performed TIRF and cryo-EM work, analyzed the data, and wrote the paper. S.P. established the protocol for purification of FAP20. A.D. performed the co-sedimentation assays described in Figures S1B and S1C. M.S. acquired the funding, supervised the work, and wrote the paper.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Data and code availability
The cryo-EM density map for C1 reconstruction of microtubule:FAP20:tubulin complex without GTP is deposited in the Electron Microscopy Data bank with deposition number EMD-34836. This paper does not report original code. Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Nicastro D, Fu X, Heuser T, Tso A, Porter ME, Linck RW. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc Natl Acad Sci USA. 2011;108:E845–E853. doi: 10.1073/pnas.1106178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sui H, Downing KH. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature. 2006;442:475–478. doi: 10.1038/nature04816. [DOI] [PubMed] [Google Scholar]
- 3.Viswanadha R, Sale WS, Porter ME. Ciliary Motility: Regulation of Axonemal Dynein Motors. Cold Spring Harbor Perspect Biol. 2017;22:a018325. doi: 10.1101/cshperspect.a018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 5.Oh EC, Katsanis N. Cilia in vertebrate development and disease. Development. 2012;139:443–448. doi: 10.1242/dev.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satir P, Christensen ST. Overview of Structure and Function of Mammalian Cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 7.Khalifa AAZ, Ichikawa M, Dai D, Kubo S, Black CS, Peri K, McAlear TS, Veyron S, Yang SK, Vargas J, et al. The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications. Elife. 2020;9:e52760. doi: 10.7554/eLife.52760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma M, Stoyanova M, Mihaela G, Dutcher SK, Brown A, Zhang R. Structure of the Decorated Ciliary Doublet Microtubule. Cell. 2019;27 doi: 10.1016/j.cell.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gui M, Farley H, Anujan P, Anderson JR, Maxwell DW, Whitchurch JB, Botsch JJ, Qiu T, Meleppattu S, Singh SK, et al. De novo identification of mammalian ciliary motility proteins using cryo-EM. Cell. 2021;184:5791–5806.:e19. doi: 10.1016/j.cell.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung MR, Roelofs MC, Chiozzi RZ, Hevler JF, Heck AJR, Zeev-Ben-Mordehai T. Unraveling the intricate microtubule inner protein networks that reinforce mammalian sperm flagella. bioRxiv. 2022 doi: 10.1101/2022.09.29.510157. [DOI] [Google Scholar]
- 11.Kubo S, Black CS, Joachimiak E, Yang SK, Legal T, Peri K, Khalifa AAZ, Ghanaeian A, McCafferty CL, Valente-Paterno M, et al. Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins. Nat Commun. 2023;14:2168. doi: 10.1038/s41467-023-37868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Greenan GA, Shiozaki M, Liu Y, Skinner WM, Zhao X, Zhao S, Yan R, Yu Z, Lishko PV, et al. In situ cryo-electron tomography reveals the asymmetric architecture of mammalian sperm axonemes. Nat Struct Mol Biol. 2023;30:360–369. doi: 10.1038/s41594-022-00861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichikawa M, Liu D, Kastritis PL, Basu K, Hsu TC, Yang S, Bui KH. Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins. Nat Commun. 2017;8:15035. doi: 10.1038/ncomms15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Euteneuer U, McIntosh JR. Polarity of midbody and phragmoplast microtubules. J Cell Biol. 1980;87:509–515. doi: 10.1083/jcb.87.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt-Cernohorska M, Zhernov I, Steib E, Le Guennec M, Achek R, Borgers S, Demurtas D, Mouawad L, Lansky Z, Hamel V, Guichard P. Flagellar microtubule doublet assembly in vitro reveals a regulatory role of tubulin C-terminal tails. Science. 2019;363:285–288. doi: 10.1126/science.aav2567. [DOI] [PubMed] [Google Scholar]
- 16.Dymek EE, Lin J, Fu G, Porter ME, Nicastro D, Smith EF. PACRG and FAP20 form the inner junction of axonemal doublet microtubules and regulate ciliary motility. MBoC. 2019;30:1805–1816. doi: 10.1091/mbc.E19-01-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanagisawa Ha, Mathis G, Oda T, Hirono M, Richey EA, Ishikawa H, Marshall WF, Kikkawa M, Qin H. FAP20 is an inner junction protein of doublet microtubules essential for both the planar asymmetrical waveform and stability of flagella in Chlamydomonas. MBoC. 2014;25:1472–1483. doi: 10.1091/mbc.e13-08-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng D, Cao M, Oda T, Pan J, Pan J. The conserved ciliary protein Bug22 controls planar beating of Chlamydomonas flagella. J Cell Sci. 2014;127:281–287. doi: 10.1242/jcs.140723. [DOI] [PubMed] [Google Scholar]
- 19.Laligné C, Klotz C, de Loubresse NG, Lemullois M, Hori M, Laurent FX, Papon JF, Louis B, Cohen J, Koll F. Bug22p, a Conserved Centrosomal/Ciliary Protein Also Present in Higher Plants, Is Required for an Effective Ciliary Stroke in Paramecium. Eukaryot Cell. 2010;9:645–655. doi: 10.1128/EC.00368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendes Maia T, Gogendeau D, Pennetier C, Janke C, Basto R. Bug22 influences cilium morphology and the post-translational modification of ciliary microtubules. Biol Open. 2014;3:138–151. doi: 10.1242/bio.20146577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chrystal PW, Lambacher NJ, Doucette LP, Bellingham J, Schiff ER, Noel NCL, Li C, Tsiropoulou S, Casey GA, Zhai Y, et al. The inner junction protein CFAP20 functions in motile and non-motile cilia and is critical for vision. Nat Commun. 2022;13:6595. doi: 10.1038/s41467-022-33820-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González-Del Pozo M, Fernández-Suárez E, Bravo-Gil N, Méndez-Vidal C, Martín-Sánchez M, Rodríguez-de la Rua E, Ramos-Jiménez M, Morillo-Sánchez MJ, Borrego S, Antiñolo G. A comprehensive WGS-based pipeline for the identification of new candidate genes in inherited retinal dystrophies. npj Genom Med. 2022;7:17. doi: 10.1038/s41525-022-00286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loucks CM, Bialas NJ, Dekkers MPJ, Walker DS, Grundy LJ, Li C, Inglis PN, Kida K, Schafer WR, Blacque OE, et al. PACRG, a protein linked to ciliary motility, mediates cellular signaling. MBoC. 2016;27:2133–2144. doi: 10.1091/mbc.E15-07-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda T. Parkin-co-regulated gene PACRG product interacts with tubulin and microtubules. FEBS Lett. 2008;582:1413–1418. doi: 10.1016/j.febslet.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 25.Khan N, Pelletier D, McAlear TS, Croteau N, Veyron S, Bayne AN, Black C, Ichikawa M, Khalifa AAZ, Chaaban S, et al. Crystal structure of human PACRG in complex with MEIG1 reveals roles in axoneme formation and tubulin binding. Structure. 2021;29:572–586.:e6. doi: 10.1016/j.str.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gui M, Wang X, Dutcher SK, Brown A, Zhang R. Ciliary central apparatus structure reveals mechanisms of microtubule patterning. Nat Struct Mol Biol. 2022;29:483–492. doi: 10.1038/s41594-022-00770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monda JK, Whitney IP, Tarasovetc EV, Wilson-Kubalek E, Milligan RA, Grishchuk EL, Cheeseman IM. Microtubule Tip Tracking by the Spindle and Kinetochore Protein Ska1 Requires Diverse Tubulin-Interacting Surfaces. Curr Biol. 2017;27:3666–3675.:e6. doi: 10.1016/j.cub.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benoit MPMH, Asenjo AB, Sosa H. Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s. Nat Commun. 2018;9:1662. doi: 10.1038/s41467-018-04044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serre L, Delaroche J, Vinit A, Schoehn G, Denarier E, Fourest-Lieuvin A, Arnal I. The mitotic role of adenomatous polyposis coli requires its bilateral interaction with tubulin and microtubules. J Cell Sci. 2023;136:jcs260152. doi: 10.1242/jcs.260152. [DOI] [PubMed] [Google Scholar]
- 31.Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv. 2022 doi: 10.1101/2021.10.04.463034. [DOI] [Google Scholar]
- 32.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owa M, Uchihashi T, Yanagisawa HA, Yamano T, Iguchi H, Fukuzawa H, Wakabayashi KI, Ando T, Kikkawa M. Inner lumen proteins stabilize doublet microtubules in cilia and flagella. Nat Commun. 2019;10:1143. doi: 10.1038/s41467-019-09051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirima J, Oiwa K. Flagellar-associated Protein FAP85 Is a Microtubule Inner Protein That Stabilizes Microtubules. Cell Struct Funct. 2018;43:1–14. doi: 10.1247/csf.17023. [DOI] [PubMed] [Google Scholar]
- 35.Schaedel L, John K, Gaillard J, Nachury MV, Blanchoin L, Théry M. Microtubules self-repair in response to mechanical stress. Nat Mater. 2015;14:1156–1163. doi: 10.1038/nmat4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erickson HP, Voter WA. Polycation-induced assembly of purified tubulin. Proc Natl Acad Sci USA. 1976;73:2813–2817. doi: 10.1073/pnas.73.8.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unger E, Böhm KJ, Vater W. Structural diversity and dynamics of microtubules and polymorphic tubulin assemblies. Electron Microsc Rev. 1990;3:355–395. doi: 10.1016/0892-0354(90)90007-F. [DOI] [PubMed] [Google Scholar]
- 38.Tan D, Asenjo AB, Mennella V, Sharp DJ, Sosa H. Kinesin-13s form rings around microtubules. J Cell Biol. 2006;175:25–31. doi: 10.1083/jcb.200605194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gegg M, Böttcher A, Burtscher I, Hasenoeder S, Van Campenhout C, Aichler M, Walch A, Grant SGN, Lickert H. Flattop regulates basal body docking and positioning in mono-and multiciliated cells. Elife. 2014;3:e03842. doi: 10.7554/eLife.03842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jungnickel MK, Sutton KA, Baker MA, Cohen MG, Sanderson MJ, Florman HM. The flagellar protein Enkurin is required for mouse sperm motility and for transport through the female reproductive tract. Biol Reprod. 2018;99:789–797. doi: 10.1093/biolre/ioy105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.TaShma A, Perles Z, Yaacov B, Werner M, Frumkin A, Rein AJJT, Elpeleg O. A human laterality disorder associated with a homozygous WDR16 deletion. Eur J Hum Genet. 2015;23:1262–1265. doi: 10.1038/ejhg.2014.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dougherty GW, Mizuno K, Nöthe-Menchen T, Ikawa Y, Boldt K, Ta-Shma A, Aprea I, Minegishi K, Pang Y-P, Pennekamp P, et al. CFAP45 deficiency causes situs abnormalities and asthenospermia by disrupting an axonemal adenine nucleotide homeostasis module. Nat Commun. 2020;11:5520. doi: 10.1038/s41467-020-19113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- 44.Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 45.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. Elife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheres SHW. A Bayesian View on Cryo-EM Structure Determination. J Mol Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cook AD, Manka SW, Wang S, Moores CA, Atherton J. A microtubule RELION-based pipeline for cryo-EM image processing. J Struct Biol. 2020;209:107402. doi: 10.1016/j.jsb.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang R, LaFrance B, Nogales E. Separating the effects of nucleotide and EB binding on microtubule structure. Proc Natl Acad Sci USA. 2018;115:E6191–E6200. doi: 10.1073/pnas.1802637115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 56.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. UCSF ChimeraX: Meeting modern challenges in visualization and analysis: UCSF ChimeraX Visualization System. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. UCSF ChimeraX : Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 60.Lord SJ, Velle KB, Mullins RD, Fritz-Laylin LK. SuperPlots: Communicating reproducibility and variability in cell biology. J Cell Biol. 2020;219:e202001064. doi: 10.1083/jcb.202001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density map for C1 reconstruction of microtubule:FAP20:tubulin complex without GTP is deposited in the Electron Microscopy Data bank with deposition number EMD-34836. This paper does not report original code. Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.