Abstract
Ancestral Coast Salish societies in the Pacific Northwest kept long-haired “woolly” dogs that were bred and cared for over millennia. However, the dog wool-weaving tradition declined during the 19th century, and the population was lost. Here, we analyze genomic and isotopic data from a preserved woolly dog pelt, “Mutton”, collected in 1859. Mutton is the only known example of an Indigenous North American dog with dominant pre-colonial ancestry postdating the onset of settler colonialism. We identify candidate genetic variants potentially linked with their unique woolly phenotype. We integrate these data with interviews from Coast Salish Elders, Knowledge Keepers, and weavers about shared traditional knowledge and memories surrounding woolly dogs, their importance within Coast Salish societies, and how colonial policies led directly to their disappearance.
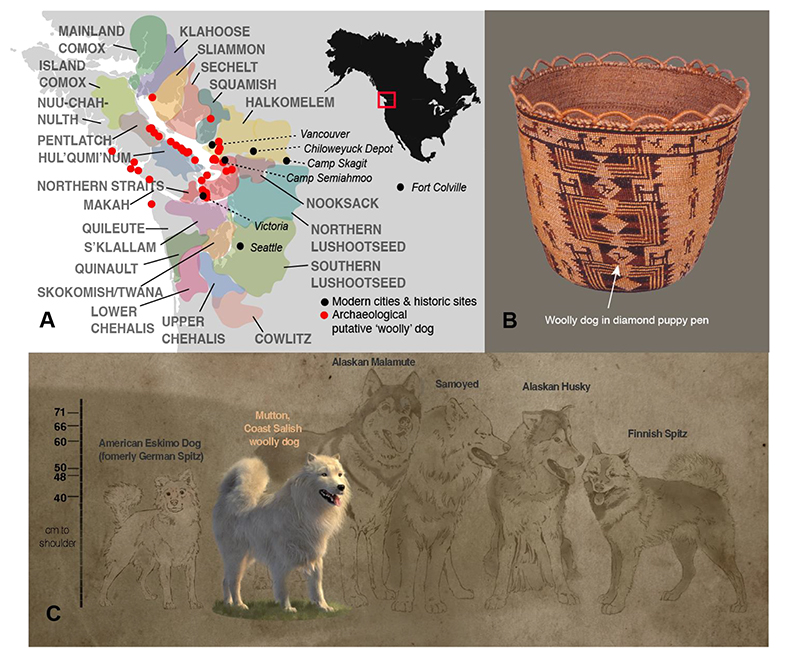
Dogs were introduced to the Americas from Eurasia via northwestern North America ~15,000 years ago, and have been ubiquitous in Indigenous societies of the Pacific Northwest (PNW) for millennia (1–4). Coast Salish peoples in the Salish Sea region (Fig. 1A) kept multiple different types of dogs: hunting dogs, village dogs, and “woolly dogs” with a thick woolen undercoat that was shorn for weaving (4, 5). Dog wool blankets, often blended with mountain goat wool, waterfowl down, and plant fibers like fireweed and cattail fluff, were prestigious cultural belongings (6–8). Woolly dogs, known as sqwemá:y, ske’-ha, sqwǝméy̓, sqwbaý, and QebeO in some Coast Salish languages (9), were emblems of some communities, as depicted in a 19th century Skokomish/Twana basket (Fig. 1B (10)).
Figure 1. Domestic dogs in the culture and society of Indigenous Coast Salish peoples.
1A. Coast Salish ancestral lands include the inner coastal waterways of Salish Sea in southwest British Columbia and Washington State. Archaeological woolly dog data are from (2). Distribution of the Coast Salish languages in the 19th century as indicated by colored areas. The map is modified from https://commons.wikimedia.org/wiki/File:Coast_Salish_language_map.svg and licensed under CC BY-SA 4.0. 1B. Woven Skokomish/Twana basket with woolly dog iconography, depicted with upturned tails. Woolly dog puppies are inside pens represented by diamond shapes (10) (courtesy of Burke Museum, Catalog number #1-507). 1C. Forensic reconstruction of a woolly dog based on Mutton’s pelt measurements and archaeological remains (9). Sketches of Arctic and spitz dog breeds are shown for scale and comparison of appearance, and do not imply a genetic relationship.
The first comprehensive book on Salish weaving (11) scrutinized most Coast Salish woven blankets in museums around the world, questioning if any contained primarily dog wool, and disputing the fiber’s spinnability. More recent proteomic analysis of 19th century blankets confirmed the use of dog wool in Coast Salish weaving (12). In addition, zooarchaeological remains thought to be from woolly dogs have been found in dozens of archaeological sites in Coast Salish territories beginning ~5,000 years before present (BP) (2, 4) (Fig. 1A). The last Coast Salish woolly dogs likely lived in the late 19th/early 20th centuries (5, 13). Later photographs and records referring to woolly dogs extend into the 20th century, but these examples likely reflect mixed ancestry or non-Indigenous breeds (9).
The decline in dog wool weaving has previously been attributed to the proliferation of machine-made blankets by British and American trading companies in the early 19th century (11, 13). However, this explanation ignores the cultural importance of woolly dogs, as reflected through their enduring use by weavers, particularly for high status items like regalia (7, 14). Given their role in Coast Salish societies, it is unlikely that the entire dog wool tradition would have been abandoned simply because of the ready availability of imported textiles. Further, this explanation ignores weavers’ efforts to maintain culturally relevant practices in the face of settler colonialism. The use of blankets and robes served not only a functional purpose, but also a spiritually protective role in Coast Salish cultures. Wearing a ceremonial blanket was spiritually transformative since it intertwined the creator of the blanket, the wearer, and the community (13–15).
The only known pelt of an extinct Coast Salish woolly dog is of “Mutton”, a dog cared for by naturalist and ethnographer George Gibbs during the Northwest Boundary Survey (1857-1862). According to Gibbs’s field journal and Smithsonian ledgers (USNM A4401-A4425), Mutton became ill and died in late 1859 (9, 15). His pelt and lower leg bones are housed at the Smithsonian Institution (USNM 4762) (Figs. S2, S4).
Here, we combine genomic analysis, ethnographic research, stable isotope and zooarchaeological analysis, and archival records to investigate this iconic dog’s history, including ancestry, the genetic underpinnings of woolliness, and their ultimate decline. We sequenced Mutton’s nuclear genome to a mean 3.4x depth of coverage and, for comparison, a non-woolly village dog (Figs. S3, S5) from the nearby Semiahmoo Bay region to low coverage (0.05x; “SB dog” hereafter, USNM 3512; collected 1858). For additional genomic context, we increased the coverage of an ancient dog from Port au Choix, Newfoundland (AL3194; 4,020 cal BP) (3), from 1.9x to 11.9x, and sequenced the genome of an ancient dog from Teshekpuk Lake, Alaska (ALAS_015; 3,763 BP; 1.23x), three modern coyotes, and 59 modern dogs representing 21 breeds (DataS1). We also undertook δ13C and δ15N stable isotope analysis of Mutton and the SB dog to test for substantial differences in their dietary life histories. Finally, we interviewed seven Coast Salish Elders, Knowledge Keepers, and wool weavers about family histories and traditional knowledge surrounding woolly dogs to provide a cultural framework for interpreting the genomic analyses (9). The interviewees span several Coast Salish communities, including Stó:lō, Squamish, Snuneymuxw, and Musqueam Nations in British Columbia (BC) and Suquamish, and Skokomish/Twana in Washington.
Woolly dog origins
Throughout northwestern North America there are numerous oral histories and origin stories involving the woolly dog. Skokomish/Twana Elder, Michael Pavel, reports that in a former time, when all beings including woolly dogs were recognized as relatives, all were ‘people’ and were family. High-status Qw’ó:ntl’an women are an example of those who trace their lineages from the woolly dog at a time when all beings were one family (16). According to Pavel: “…And out of [the origin story], [woolly dogs] were given the gift of the wool, and they were able to teach the women how to gather the wool, how to process the wool, how to spin the wool, and how to weave with the wool” (9).
Early colonial explorers and scholars speculated that woolly dogs originated in Japan (17) or were recently introduced to the Coast Salish by Dene from their homelands in northern boreal Canada (18). However, zooarchaeological remains of morphologically distinct dogs in Coast Salish territories suggest woolly dog husbandry was present for ~5,000 years before European colonization (2, 4). Furthermore, longstanding oral histories and traditional knowledge hold that woolly dogs have been part of Coast Salish society for millennia (9).
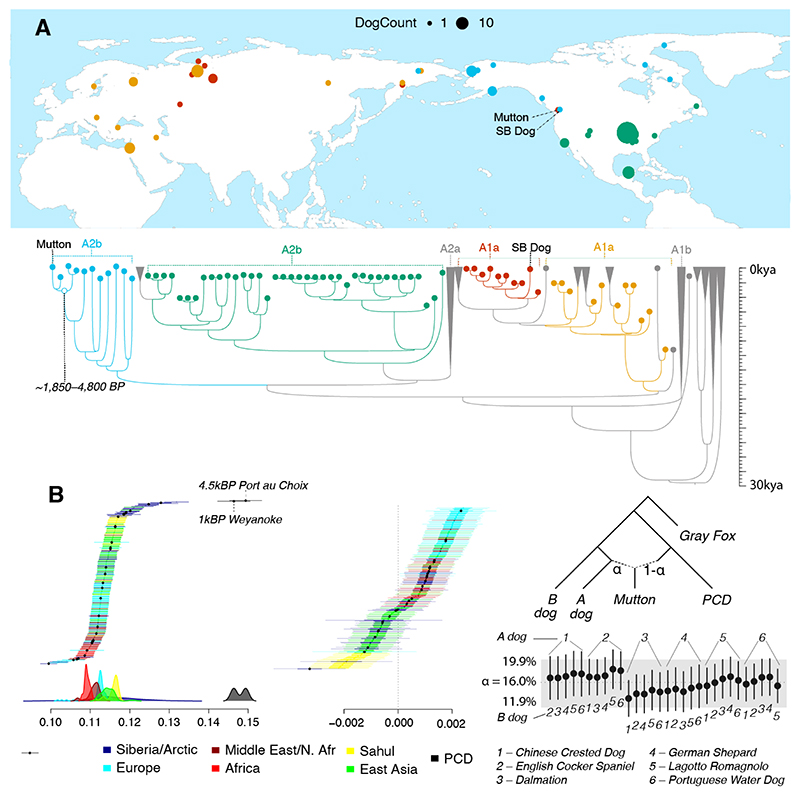
To test whether Mutton has pre-colonial or settler dog ancestry, we first compared his mitochondrial genome to 207 ancient and modern dogs from a global sampling. Mutton carries the A2b mtDNA haplotype, which emerged after dogs initially arrived from Eurasia (3). Most of this mtDNA lineage of so-called pre-colonial dogs (PCDs) disappeared after European colonization (3, 19, 20). Mutton’s nearest mtDNA neighbor is an ancient dog (PRD10, ~1,500 BP) from Prince Rupert Harbour, BC (Figs. 2A, S16). PRD10 is the only archaeological dog from the PNW in the mtDNA dataset, and this similarity reflects the deep roots of Mutton’s maternal ancestry in the region. A pair of modern and ancient (~620 BP) dogs from Alaska form a sister clade of the Mutton-PRD10 grouping, further underscoring the long-term maternal population structure in northwestern North America. In contrast, the SB dog carries an A1a haplotype, similar to most modern European dogs, and the most common present-day haplotype worldwide (64 out of 207 dogs in our analysis) (21).
Figure 2. Genetic ancestry of woolly dogs.
2A. mtDNA tree of 207 dogs with A2b (Mutton) and A1a (SB Dog) haplotypes expanded. Map points correspond to colored tree tips for the most similar archaeological and historic dog mtDNAs, highlighting the subclades of interest and the broader haplotypes. Samples used are listed in DataS1. 2B. Outgroup-f3 statistics (f3(GrayFox; Mutton, B) or estimation of shared drift between Mutton and 229 other dogs reveals that Mutton has highest similarity to PCDs. Black point estimates indicate ancient genomes. 2C. D-statistics (((PCD, Mutton), Test Dog), Gray Fox) consistent with gene flow into Mutton’s background, with European breeds appearing the most likely contributors to Mutton’s non-PCD ancestry. 2D. f4-ratio tests (f4(A, Out; Mutton, AL3194-PortauChoix): f4(A, Out; B, AL3194-PortauChoix)) to estimate the proportion of European settler dog ancestry in Mutton’s background using six modern European breeds as proxies for Mutton’s European ancestry component.
To place a timeframe on the divergence of Mutton’s maternal lineage, we performed a molecular clock analysis on the mitochondrial phylogeny (DataS1). The results suggest a mitochondrial common ancestor estimated between 4,776 and 1,853 years BP for the subclade containing Mutton, PRD10, and the two Alaskan dogs (95% highest posterior density; Figs. 2A, S16). Although we are limited by the analysis of a single individual, this timing is generally consistent with the increasing occurrence of small sized ‘woolly’ dog zooarchaeological remains in the regions surrounding the Salish Sea (2).
To assess Mutton’s nuclear ancestry, we analyzed 217 globally distributed ancient and modern dogs. Outgroup-f3 statistics reveal that Mutton carries substantially greater shared genetic drift with PCDs than with any other dogs, specifically, archaeological remains of a dog from Port au Choix, Newfoundland (4,020 cal BP), and from Weyanoke Old Town, Virginia (~1,000 BP) (Figs. 2B, S17). Since Mutton lived after European colonization and waves of pre-colonial dog introductions (3, 21), we tested for gene flow from introduced lineages using D-statistics. We found that European breeds yielded strongly positive D-statistics, indicating that Mutton’s non-PCD ancestry most likely stemmed from introduced European dogs (Fig. 2C).
To refine these results, we used f4-ratio tests with six modern European breeds (Chinese Crested dog, English Cocker Spaniel, Dalmatian, German Shepherd, Lagotto Romagnolo, and Portuguese Water Dog), estimating that Mutton had 84% PCD and 16% European ancestry (11.9%–19.9% 2 SE range; Fig. 2D). The f4-ratio test may slightly over-estimate Mutton’s European ancestry if the true contributor of this ancestry was equally related (an outgroup) to the two European breeds in the tests. However, estimates across all permutations are broadly consistent (Figs. 2D, S18), suggesting European ancestry roughly on the order of one great-grandparent in Mutton’s background. In contrast, outgroup-f3 statistics indicate that the contemporaneous SB dog appears highly admixed, showing greatest similarity to ancient dogs from Siberia and Alaska (Fig. S17). The distribution of PCD vs. European ancestry tracts in Mutton can provide some additional insight into the timing of admixture. Although this method is imprecise due to recent admixture and the scarcity of PCD source population data, we estimate that Mutton’s European admixture occurred 10.8±4.9 generations before (1 SE). Assuming a three-year generation time, this analysis suggests admixture ~32 years before Mutton’s birth, consistent with post-colonial admixture (9).
To test for dietary differences between Mutton and the SB dog, we performed stable isotope analysis of δ13C and δ15N on bone collagen and hair keratin. The SB dog has high δ13C and δ15N values similar to archaeological dogs from the PNW (22), indicating a traditional marine-based diet (Figs. S13-S14). Mutton’s isotope values reveal a more terrestrial and C3-rich diet, likely reflecting Mutton’s life and travels with Gibbs from an early age (Figs. S14-B,C, S15, (9)).
The persistence of a high proportion of post-colonial PCD ancestry may reflect concerted efforts by Coast Salish peoples to maintain the breed against the pressure of gene flow from non-native dogs. Mutton lived near the end of traditional woolly dog husbandry (5, 9, 13). Although he had mixed ancestry, Mutton’s background is dominated by PCD ancestors, compared to the contemporaneous SB dog. This may indicate careful reproductive management to maintain woolly dogs’ unique genetic makeup and phenotype until their decline. Mutton’s fraction of European ancestry also highlights the turbulent cultural moment when Mutton lived and illustrates how interbreeding with settler-introduced dogs could have threatened the survival of woolly dogs.
The influence of people on the woolly dog genome
Woolly dogs were treated as beloved extended family members. According to Debra qwasen Sparrow, a Musqueam Master weaver, her grandfather [Ed Sparrow, (1898-1998)] told her “every village had [woolly dogs], that they were like gold because they were mixed with the mountain goat and then rove and spun” (9). Dogs also comprised a form of wealth and status for Coast Salish women, who carefully managed the dogs to maintain their woolly coats, isolating them on islands or in pens to strictly manage their breeding (9, 17, 23). Often island names reflect their connection with dogs, such as sqwiqwmi’ (“Little Dog”) village on Cameron Island in Nanaimo, Snuneymuxw territory, British Columbia. The prevention of interbreeding wool dogs with hunting or village dogs was critical for maintaining their unique hair characteristics: soft guard hairs with an unusually long crimpy undercoat (Fig. S2), which was highly spinnable and made warm blanket yarn. These management practices likely contributed to Mutton’s PCD ancestry long after the onset of settler colonialism.
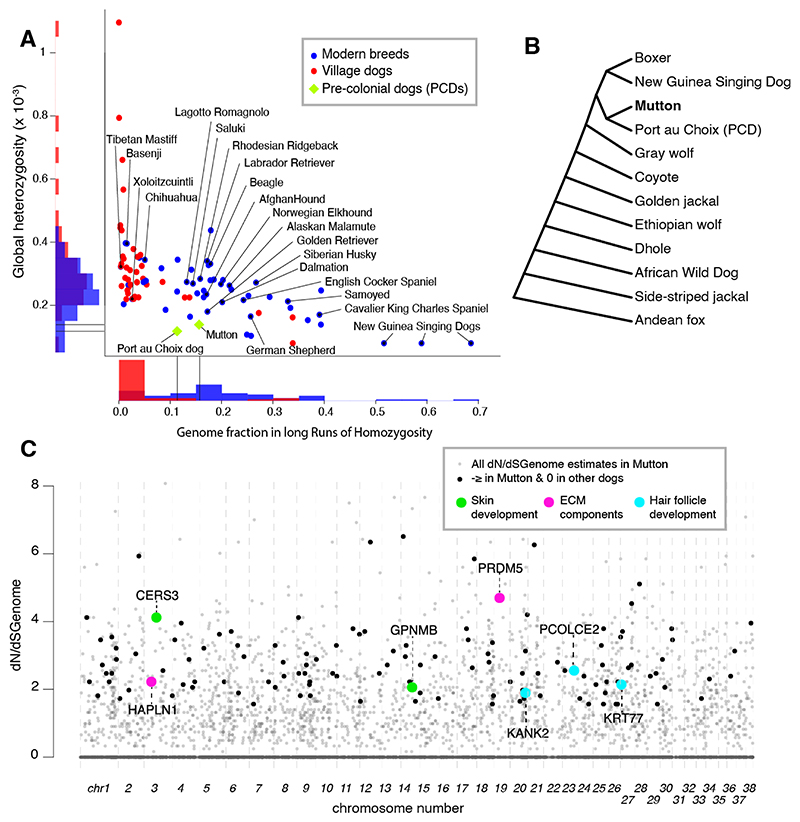
Long-term husbandry for woolly hair likely limited woolly dogs’ effective population size, which would be reflected in nucleotide diversity and thus in Mutton’s heterozygosity. We found that Mutton’s heterozygosity is in the lowest range of living breeds (n=51) and village dogs (n=42) downsampled to the same coverage (Fig. 3A). Additionally, runs of homozygosity (ROH) better reflect recent demography than global heterozygosity. Using an ROH method optimized for low coverage (9, 24), we estimate that 15.7% of Mutton’s genome is in ROH of 2.5Mbp or greater, again in the range of modern breeds. The ancient Port au Choix dog also has low genomic heterozygosity and 11.3% ROH, so Mutton’s low heterozygosity may partly reflect shared demographic history from a small PCD founding population (Fig. 3A). Because of recent European admixture, Mutton’s genome is inevitably more heterozygous than his recent woolly dog ancestors.
Figure 3. Genomic outcomes of management and selection.
3A. Global heterozygosity and long runs of homozygosity over transversions in Mutton compared to modern dogs and the ancient Port au Choix dog. All dogs have been downsampled to Mutton’s coverage level for analysis. 3B. Tree schematic used in dN/dS analysis to identify genes under selection in Mutton compared to other canids. Branching order after (50). dN/dS estimates were done separately including one of the four dogs plus all other canids. Genes with elevated dN/dSGenome values in multiple dogs could reflect more ancient shared selection before the separation of the woolly dog lineage. Therefore, likely candidates for selection in woolly dogs were conservatively assessed where dN/dSGenome>1.5 in Mutton (9), but dN = 0 in the other three dogs, including one PCD. 3C. Genes with an excess of non-synonymous mutations in Mutton. Black points are the 125 selection candidates on the basis of dN/dSgenome ≥1.5 in Mutton but dN=0 in three other dogs including one PCD (9). Several genes with high dN/dSgenome in Mutton (shown in gray) are excluded as selection candidates because they carry at least one non-synonymous mutation in other dogs. This approach is designed to conservatively highlight genes where selection is more likely specific to Mutton’s lineage rather than during dog domestication or in the common ancestors of PCDs. Candidate genes discussed in text are indicated.
To search for evidence of genetic mechanisms for woolliness, we used maximum likelihood-based estimation of the enrichment of non-synonymous mutations (dN/dS) observed within Mutton’s coding regions (9). We evaluated 11,112 genes with sufficient sequence coverage for all dogs and outgroups (DataS1), and restricted selection candidate identification to genes with elevated dN/dS in Mutton but lacking any non-synonymous mutations in three other dogs, including one PCD (Fig. 3B). Although power to detect selection is fundamentally limited with only a single genome, we identified a candidate set of genes with high lineage-specific dN/dS values. We identified 125 genes as candidates for positive selection in woolly dogs (DataS2). Among these, 28 have plausible links to hair growth and follicle regeneration based on a model of the hair growth cycle (Fig. S12), and are associated with cell replication, proliferation, the formation of extracellular matrix components, vascularization, and related processes (25–31) (Fig. 3C, DataS3).
Candidate selection genes in Mutton include KANK2, a steroid signaling regulator responsible for hereditary diseases of the hair shaft in humans (32). A unique non-synonymous mutation in Mutton lies in the adjacent amino acid to the KANK2 mutation causing a “woolly” hair phenotype in humans (32). KRT77 is a member of the keratin gene family responsible for the structural integrity of cells in the epithelium and hair follicles. Mutations in keratin genes are linked to curly hair phenotype in other dogs, rats, and mice (31), woolly hair and hereditary hair loss in humans (26, 30), and multiple KRT genes underwent selection in woolly mammoths (25). CERS3, PRDM5, HAPLN1 are associated with maintaining the integrity of the skin or connective tissue in humans (27, 28). GPNMB is involved in multiple cellular functions in the epidermis, potentially mediating pigmentation (29). We also manually evaluated 15 specific variants from previous literature linked with hair characteristics in living dog breeds (DataS4). Apart from a widespread FGF5 mutation conferring long hair (33, 34), Mutton showed the ancestral allele in all cases with data present (DataS4), illustrating the independent origins of woolly dogs’ unique phenotype.
The impact of colonialism on the iconic breed’s disappearance
Woolly dogs’ decline throughout the 19th century is not fully understood. The narrative that the influx of trade blankets into the region led to the abandonment of woolly dog husbandry oversimplifies a complex scenario. By 1857 (a year before Mutton’s birth) in Sto:lo territory, where Mutton was most likely acquired, the settler population consisted of only a few dozen permanent settlers at Fort Langley (35, 36). The following year, more than 33,000 miners arrived at present-day British Columbia during the 1858 Fraser River Gold Rush. This large-scale migration set off conflicts between miners, colonial governments, and Indigenous peoples. Meanwhile, Indigenous populations declined by an estimated two-thirds between 1830 and 1882 (37). Smallpox epidemics—almost one every generation from the 1700s to 1862 (38)—are estimated to have killed more than 90% of Indigenous people in some villages across BC (38), along with steady depopulation due to other introduced diseases such as mumps, tuberculosis, and influenza (37).
Survival of woolly dogs depended upon the survival of their caretakers. In addition to disease, expanding colonialism increased cultural upheaval, displacement of Indigenous peoples, and a diminished capacity to manage the breed. Policies targeted Indigenous governance and inherent rights, resulting in the deliberate disenfranchisement and criminalization of Indigenous cultural practices (39). Indigenous women, the caretakers of woolly dogs and weaving knowledge, were specifically targeted. Missionization efforts reduced women’s roles in society, and legislation such as the Indian Act (1876) explicitly prohibited women from participating in local governance, denied women basic property rights, and restricted their movement (39). In the 20th century, transference of cultural knowledge was further disrupted by mandatory residential schooling designed to remove children from their families and suppress culture (40).
Through these compounding waves of colonialism, the transmission of important knowledge relating to the husbandry of the woolly dog, processing the hair, spinning, and weaving was interrupted. Stó:lō Elder Rena Point Bolton, 95 years old in 2022, recalls how Th’etsimiya, her great-grandmother, had kept woolly dogs, but was forced to give them up: “They were told they couldn’t do their cultural things. There was the police, the Indian Agent and the priests. The dogs were not allowed. She had to get rid of the dogs.” (9). The dogs represented high status and traditional practices that threatened British and later Canadian dominion, and as such were removed via policies of assimilation (40–42). The weaving traditions were not completely lost, as many cultural teachings and types of expertise were carried on in secret. Bolton said: “Our people were not allowed to spin on shxwqáqelets [traditional spindle whorls]. They could spin on a European one but not on the shxwqáqelets. They couldn’t use their looms, and they would take them out and burn them or they would give them to museums or collectors…The generation that was there when the Europeans came and colonized us, that’s where it ended, and there [were] just a few people who went underground. And my grandmother and my mother were two of them.” (9).
A growing body of research demonstrates how peoples of the PNW cared for and managed their ancestral lands, cultivating diverse and highly localized plants and marine foods (43–45). Woolly dogs may have also been similarly localized and diverse. We focus on Coast Salish dogs, but non-Salish peoples in the PNW also kept woolly dogs. For example, Nuu-chah-nulth peoples of western Vancouver Island kept a different wool dog that were reportedly bigger and had coats of different colors including brown, spotted, black, grey, or white (46–48). These differences could be population-specific, or they could be a result of widespread phenotypic diversity, as noted by explorers in the 18th and 19th centuries (17), reflecting trade among the different Indigenous communities.
Weaving and woolly dogs are intertwined in Coast Salish culture and society, which cannot be separated from the long-time management of their ancestral homelands. Weavers, artists, and Elders continue to promote the renewal of traditional or customary weaving knowledge and practices. Artist Eliot Kwulasultun White-Hill (Snuneymuxw) said (9): “It starts to unravel, in a way, people’s understanding of us as a hunter gatherer society… Our relationship with the woolly dogs, our relationship with the camas patches and the clam beds, the way that we tended the land and tended the forests… these all show the systems in place that are far more complex than what people take for granted about Coast Salish culture.”
Supplementary Material
1 sentence summary.
A 19th century dog genome and Traditional Knowledge illuminate the life, history, and decline of Coast Salish woolly dogs
Acknowledgements
We wish to express our deep gratitude to the Honorable Steven Point, Grand Chief and Dr. Gwen Point of the Stó:lō Nation for giving us permission and encouragement for this research. Thanks to Candace Wellman for her role in re-discovering Mutton, assistance with history of the area, and photographs. We raise our hands in thanks to all people within the Coast Salish communities who have graciously shared their time and knowledge to realize this project, specifically: Xweliqwiya Rena Point Bolton (Stó:lō Nation); Danielle Morsette (Suquamish/Shxwhá:y Village); Eliot Kwulasultun White (Snuneymuxw First Nation); Sulqwan Philomena Williams (Cowichan); Violet Snu’Meethia Elliott (Snuneymuxw); Tracy Sesemiya Williams Skwxwú7mesh Úxwumixw (Squamish Nation); Andrea Fritz, Norris family (Lyacksun); Tillie Jones (Tulalip); Tami Hohn (Puyallup); q́wat́ǝlǝmu Nancy Bob (Lummi). Interviews were carried out under Institutional Review Board and Research Ethics Board approvals from the Smithsonian Institution (Human Subjects Protocol #HS220007) and Vancouver Island University (#101410), with informed consent including explicit opt-in permissions to reprint quotations with personal attribution. Computations performed for this paper were conducted on the Smithsonian High Performance Cluster, Smithsonian Institution: https://doi.org/10.25572/SIHPC, and the Leibniz Supercomputing Centre (LRZ). Portions of the laboratory work were conducted in and with the support of the Laboratories for Analytical Biology (L.A.B.) facilities of the National Museum of Natural History. Thanks to Tom Gilbert for funding the processing/sequencing of AL3194, John Ososky for specimen handling assistance, and Ludovic Orlando and Sierra Harding for providing helpful comments on the manuscript.
Funding
Research was supported by SI funds to LK. ATL, H-LL, and CS were supported by Smithsonian postdoctoral fellowships. Funding for stable isotope analysis provided by Smithsonian Museum Conservation Institute federal and trust funds. PS was supported by EMBO, the Vallee Foundation, the European Research Council (grant no. 852558), the Wellcome Trust (217223/Z/19/Z), and Francis Crick Institute core funding (FC001595) from Cancer Research UK, the Medical Research Council, and the Wellcome Trust. VG was supported by an SSHRC-IG.
Footnotes
Author contributions: Conceptualization: ATL, LH-K, LK; Methodology: ATL, LK, H-LL, LH-K, SGA, CS, CAMF, KC; Investigation: ATL, LK, CS, SGA, H-LL, MTRH, LH-K, JH, IM, GK, TRF, M-HSS, SG, LF, AB, AC, AH; Formal analysis: ATL, LK, CS, CAMF, SGA, DWGS, AH; Visualization: ATL, LK, CS, KC, MH, GK, IM; Resources: LK, MTRH, VG, BNS, IM, EAO; Funding acquisition: LK, PS, LD; Supervision: LK, LH-K; Writing – original draft: ATL, LK, LH-K; Writing – review & editing: all authors.
Competing interests: All authors declare there are no competing interests.
Data availability
Genomic sequencing data for Mutton, SB dog, the Port au Choix dog (AL3194), and ALAS_015 are available for non-commercial use via NCBI SRA Project Accession PRJNA1005336 and BioSample Accessions SAMN36985984-SAMN36985987. The SRA Project Accession for the modern coyote from Wyoming is PRJNA734649. Stable isotope data are available (49). All other public genomic data sources are provided in DataS1.
References and Notes
- 1.Fedje D, Mackie Q, McLaren D, Wigen B, Southon J. Karst caves in Haida Gwaii: Archaeology and paleontology at the Pleistocene-Holocene transition. Quat Sci Rev. 2021;272:107221 [Google Scholar]
- 2.McKechnie I, Moss ML, Crockford SJ. Domestic dogs and wild canids on the Northwest Coast of North America: Animal husbandry in a region without agriculture? Journal of Anthropological Archaeology. 2020;60:101209 [Google Scholar]
- 3.Ní Leathlobhair M, Perri AR, Irving-Pease EK, Witt KE, Linderholm A, Haile J, Lebrasseur O, Ameen C, Blick J, Boyko AR, Brace S, et al. The evolutionary history of dogs in the Americas. Science. 2018;361:81–85. doi: 10.1126/science.aao4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crockford SJ. Osteometry of Makah and Coast Salish Dogs. Archaeology Press, Simon Fraser University; 1997. [Google Scholar]
- 5.Schulting R. The Hair of the Dog: The Identification of a Coast Salish Dog-Hair Blanket from Yale, British Columbia. Canadian Journal of Archaeology / Journal Canadien d’Archéologie. 1994;18:57–76. [Google Scholar]
- 6.Dall WH, Gibbs G. Tribes of the Extreme Northwest, and Tribes of Western Washington and Northwestern Oregon. I. Cosimo Classics; 1877. [Google Scholar]
- 7.Suttles W. In: Indian Art Traditions of the Northwest Coast. Carlson RL, editor. Archaeology Press, Simon Fraser University; Burnaby, B.C: 1982. Productivity and its Constraints: A Coast Salish Case; p. 70. [Google Scholar]
- 8.Barnett HG. The Coast Salish of British Columbia vol 4 of Monographs : Studies in anthropology. University of Oregon; Eugene, OR: 1955. [Google Scholar]
- 9.see Supplementary Materials
- 10.Burke museum basketry exhibition. Burke Museum. (available at https://www.burkemuseum.org/static/baskets/idgame/dreport.html)
- 11.Gustafson P. Salish Weaving. Douglas & McIntyre; Seattle, WA: 1980. [Google Scholar]
- 12.Solazzo C, Heald S, Ballard MW, Ashford DA, DePriest PT, Koestler RJ, Collins MJ. Proteomics and Coast Salish blankets: a tale of shaggy dogs? Antiquity. 2011;85:1418–1432. [Google Scholar]
- 13.Barsh RL, Jones JM, Suttles W. In: Snyder LM, Moore EA, editors. History, ethnography, and archaeology of the Coast Salish woolly-dog; Proceedings of the 9th Conference of the International Council of Archaeozoology, Durham, August 2002; Oxford, OX1 1HN. 2006. pp. 2–11. [Google Scholar]
- 14.Tepper LH, George J, Joseph W. Salish blankets. University of Nebraska Press; Lincoln, NE: 2017. [Google Scholar]
- 15.Gibbs G. Journal, Northwest Boundary Survey. 1859:1857–1862. doi: 10.5962/bhl.title.97030. [DOI] [Google Scholar]
- 16.Carlson KT. In: A Stó:Lō-Coast Salish Historical Atlas. Carlson K, McHalsie AJ, editors. Douglas & McIntyre, Sto:lo Nation; Seattle, WA;Chilliwack, B.C;Vancouver: 2001. Expressions of Cultural Identity; p. 25. [Google Scholar]
- 17.Lord JK. The naturalist in Vancouver Island and British Columbia. R. Bentley, London: 1866. [Google Scholar]
- 18.Howay FW. The Dog’s Hair Blankets of the Coast Salish. The Washington Historical Quarterly. 1918;9:83–92. [Google Scholar]
- 19.Bergström A, Frantz L, Schmidt R, Ersmark E, Lebrasseur O, Girdland-Flink L, Lin AT, Storå J, Sjögren K-G, Anthony D, Antipina E, Amiri S, et al. Origins and genetic legacy of prehistoric dogs. Science. 2020;370:557–564. doi: 10.1126/science.aba9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castroviejo-Fisher S, Skoglund P, Valadez R, Vilà C, Leonard JA. Vanishing native American dog lineages. BMC Evol Biol. 2011;11:73. doi: 10.1186/1471-2148-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ameen C, Feuerborn TR, Brown SK, Linderholm A, Hulme-Beaman A, Lebrasseur O, Sinding M-HS, Lounsberry ZT, Lin AT, Appelt M, Bachmann L, et al. Specialized sledge dogs accompanied Inuit dispersal across the North American Arctic. Proc Biol Sci. 2019;286:20191929. doi: 10.1098/rspb.2019.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hillis D, McKechnie I, Guiry E, St Claire DE, Darimont CT. Ancient dog diets on the Pacific Northwest Coast: zooarchaeological and stable isotope modelling evidence from Tseshaht territory and beyond. Sci Rep. 2020;10:15630. doi: 10.1038/s41598-020-71574-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eells M, Castile GP. The Indians of Puget Sound: The Notebooks of Myron Eells. 1985. (No Title). (available at https://cir.nii.ac.jp/crid/1130282269923522048)
- 24.Daly KG, Mattiangeli V, Hare AJ, Davoudi H, Fathi H, Doost SB, Amiri S, Khazaeli R, Decruyenaere D, Nokandeh J, Richter T, et al. Herded and hunted goat genomes from the dawn of domestication in the Zagros Mountains. Proc Natl Acad Sci U S A. 2021;118:e2100901118. doi: 10.1073/pnas.2100901118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Díez-Del-Molino D, Dehasque M, Chacón-Duque JC, Pečnerová P, Tikhonov A, Protopopov A, Plotnikov V, Kanellidou F, Nikolskiy P, Mortensen P, Danilov GK, Vartanyan S, Gilbert MTP, et al. Genomics of adaptive evolution in the woolly mammoth. Curr Biol. 2023 doi: 10.1016/j.cub.2023.03.084. [DOI] [PubMed] [Google Scholar]
- 26.Shimomura Y, Wajid M, Petukhova L, Kurban M, Christiano AM. Autosomal-dominant woolly hair resulting from disruption of keratin 74 (KRT74), a potential determinant of human hair texture. Am J Hum Genet. 2010;86:632–638. doi: 10.1016/j.ajhg.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radner FPW, Marrakchi S, Kirchmeier P, Kim G-J, Ribierre F, Kamoun B, Abid L, Leipoldt M, Turki H, Schempp W, Heilig R, et al. Mutations in CERS3 cause autosomal recessive congenital ichthyosis in humans. PLoS Genet. 2013;9:e1003536. doi: 10.1371/journal.pgen.1003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burkitt Wright EMM, Spencer HL, Daly SB, Manson FDC, Zeef LAH, Urquhart J, Zoppi N, Bonshek R, Tosounidis I, Mohan M, Madden C, et al. Mutations in PRDM5 in brittle cornea syndrome identify a pathway regulating extracellular matrix development and maintenance. Am J Hum Genet. 2011;89:346. doi: 10.1016/j.ajhg.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biswas KB, Takahashi A, Mizutani Y, Takayama S, Ishitsuka A, Yang L, Yang F, Iddamalgoda A, Katayama I, Inoue S. GPNMB is expressed in human epidermal keratinocytes but disappears in the vitiligo lesional skin. Sci Rep. 2020;10:4930. doi: 10.1038/s41598-020-61931-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasif N, Naqvi SKU-H, Basit S, Ali N, Ansar M, Ahmad W. Novel mutations in the keratin-74 (KRT74) gene underlie autosomal dominant woolly hair/hypotrichosis in Pakistani families. Hum Genet. 2011;129:419–424. doi: 10.1007/s00439-010-0938-9. [DOI] [PubMed] [Google Scholar]
- 31.Harel S, Christiano AM. Keratin 71 mutations: from water dogs to woolly hair. J Invest Dermatol. 2012;132:2315–2317. doi: 10.1038/jid.2012.291. [DOI] [PubMed] [Google Scholar]
- 32.Ramot Y, Molho-Pessach V, Meir T, Alper-Pinus R, Siam I, Tams S, Babay S, Zlotogorski A. Mutation in KANK2, encoding a sequestering protein for steroid receptor coactivators, causes keratoderma and woolly hair. J Med Genet. 2014;51:388–394. doi: 10.1136/jmedgenet-2014-102346. [DOI] [PubMed] [Google Scholar]
- 33.Dierks C, Mömke S, Philipp U, Distl O. Allelic heterogeneity of FGF5 mutations causes the long-hair phenotype in dogs. Anim Genet. 2013;44:425–431. doi: 10.1111/age.12010. [DOI] [PubMed] [Google Scholar]
- 34.Cadieu E, Neff MW, Quignon P, Walsh K, Chase K, Parker HG, Vonholdt BM, Rhue A, Boyko A, Byers A, Wong A, et al. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326:150–153. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibson JR. Farming the Frontier: The Agricultural Opening of the Oregon Country. University of British Columbia Press; Vancouver: 1985. pp. 1786–1846. [Google Scholar]
- 36.Carlson K. In: A Stó:Lō-Coast Salish Historical Atlas. Carlson K, McHalsie AJ, editors. Douglas & McIntyre, Sto:lo Nation; Seattle, WA;Chilliwack, B.C;Vancouver: 2001. The Numbers Game; pp. 76–83. [Google Scholar]
- 37.Carlson KT. In: A Stó:Lō-Coast Salish Historical Atlas. Carlson K, McHalsie AJ, editors. Douglas & McIntyre, Sto:lo Nation; Seattle, WA;Chilliwack, B.C;Vancouver: 2001. The Fraser River Gold Rush, 1858; pp. 92–93. [Google Scholar]
- 38.Boyd R. Smallpox in the Pacific Northwest: the first epidemics. BC Studies: The British Columbian Quarterly. 1994:5–40. [Google Scholar]
- 39.Lawrence B. Indians and others : mixed-blood urban Native peoples and indigenous nationhood. University of Nebraska Press; Lincoln, NE: 2004. [Google Scholar]
- 40.Hanson E, Gamez D, Manuel A. The Residential School System. Indigenous Foundations. 2020. (available at https://indigenousfoundations.arts.ubc.ca/the_residential_school_system/)
- 41.Fisher R. Contact and Conflict: Indian-European Relations in British Columbia. 2nd edition. UBC Press; 1992. pp. 1774–1890. [Google Scholar]
- 42.Bohaker H. Makúk: A new history of aboriginal-white relations. West Hist Q. 2009;40:509. [Google Scholar]
- 43.Armstrong CG, Earnshaw J, McAlvay AC. Coupled archaeological and ecological analyses reveal ancient cultivation and land use in Nuchatlaht (Nuu-chah-nulth) territories, Pacific Northwest. J Archaeol Sci. 2022;143:105611 [Google Scholar]
- 44.Lepofsky D, Toniello G, Earnshaw J, Roberts C, Wilson L, Rowell K, Holmes K. Ancient anthropogenic clam gardens of the northwest coast expand clam habitat. Ecosystems. 2021;24:248–260. [Google Scholar]
- 45.Turner N. Ancient pathways, ancestral knowledge: ethnobotany and ecological wisdom of indigenous peoples of northwestern North America. Vol. 74. McGill-Queen’s Press-MQUP; 2014. [Google Scholar]
- 46.Forrest JT, Kane P, Harper JR. Paul Kane’s frontier: Including wanderings of an artist among the Indians of north America. West Hist Q. 1972;3:79. [Google Scholar]
- 47.Swan JG. The Indians of Cape Flattery: At the Entrance to the Strait of Fuca, Washington Territory. Smithsonian Institution; 1868. [Google Scholar]
- 48.Smith CH. Lizars WH, editor. The Natural History of Dogs: Canidae Or Genus Canis of Authors : Including Also the Genera Hyaena and Proteles. 1839 [Google Scholar]
- 49.Stantis C. stantis/Coast-Salish-wool-dogs-isotopes: v0.2-alpha (v0.2-alpha) Zenodo; 2023. [DOI] [Google Scholar]
- 50.Perri AR, Mitchell KJ, Mouton A, Hulme-Beaman A, Haile J, Jamieson A, Meachen J, Lin AT, Schubert BW, Ameen C, Antipina EE, et al. Dire wolves were the last of an ancient New World canid lineage. Nature. 2021;591:87–91. doi: 10.1038/s41586-020-03082-x. [DOI] [PubMed] [Google Scholar]
- 51.Galloway BD. Dictionary of Upriver Halkomelem vol 1 of University of California Publications in Linguistics. Berkeley and Los Angeles, California: University of California Press; 2009. [Google Scholar]
- 52.Waterman TT. Indian notes and monographs. Museum of the American Indian, Heye Foundation; New York: 1973. Notes on the ethnology of the Indians of Puget Sound. Miscellaneous. [Google Scholar]
- 53.Keddie G. Prehistoric dogs of BC: Wolves in Sheep Clothing. The Midden. 1993;25 [Google Scholar]
- 54.Galloway BD. Phonology, morphology, and classified word list for the Samish dialect of Straits Salish. University of Ottawa Press; Ottawa, ON, Canada: 1990. [Google Scholar]
- 55.Drachman D. T Skokomish Indian, Ed. Skokomish Indian Tribe; 2020. [Google Scholar]
- 56.Kane P. Wanderings of an artist among the Indians of North America: from Canada to Vancouver’s Island and Oregon through the Hudson’s Bay Company’s territory and back again. Vol. 35931. Longman, Brown, Green, Longmans and Roberts; London: 1859. [Google Scholar]
- 57.Leechman JD. Portraits. Adams, Mrs. Mary and her dog, Jumbo. Small fishing canoe made by Jack Adams, of Port Madison Reservation (1920) 1920. Mar, (available at https://cdm16118.contentdm.oclc.org/digital/collection/p16118coll33/id/79/rec/7)
- 58.Waterman TT, Coffin G. Types of canoes on Puget Sound. Museum of the American Indian, Heye Foundation; New York: 1920. [Google Scholar]
- 59.Leechman D. Fleece-Bearing Dogs. Nature Magazine. 1929;14:177. [Google Scholar]
- 60.Jenness D. Woolly dog 02. 1935 doi: 10.58066/5apq-1n64. [DOI] [Google Scholar]
- 61.Jenness D. Woolly dog 01. 1935 doi: 10.58066/tj37-2m64. [DOI] [Google Scholar]
- 62.Allen GM. Dogs of the American Aborigines. Vol. 4. Museum of Comparative Zoology; Cambridge, MA: 1920. [Google Scholar]
- 63.Jenness D. W A Newcombe - correspondence, SERIES A. 1936 [Google Scholar]
- 64.Kennerly CBR. Natural History - Northwest Boundary Survey Zoology Box 1 Folder 6 (1857-1858) (available at https://siarchives.si.edu/collections/fbr_item_modsi1045)
- 65.Baker M. Survey of the Northwestern Boundary of the United States. Government Printing Office; Washington: 1900. pp. 1857–1861. [Google Scholar]
- 66.Miller BG. Be of Good Mind: Essays on the Coast Salish. UBC Press; 2011. [Google Scholar]
- 67.Baird SF. Record Unit 7002 Box 26. pp. 1833–1889. (available at https://siarchives.si.edu/collections/siris_arc_217202)
- 68.Baird SF. General Directions for Collecting and Preserving Objects of Natural History. 1848 [Google Scholar]
- 69.Tuck JA. An Archaic Cemetery at Port Au Choix, Newfoundland. Am Antiq. 1971;36:343–358. [Google Scholar]
- 70.Tuck JA. An Archaic Indian Cemetery in Newfoundland. Sci Am. 1970;222:112–122. [Google Scholar]
- 71.Renouf MAP. Palaeoeskimo seal hunters at Port au Choix, northwestern Newfoundland. Newfoundland Studies. 1993;9:185–212. [Google Scholar]
- 72.Guiry EJ, Grimes V. Domestic dog (Canis familiaris) diets among coastal Late Archaic groups of northeastern North America: A case study for the canine surrogacy approach. Journal of Anthropological Archaeology. 2013;32:732–745. [Google Scholar]
- 73.Tuck JA. Ancient People of Port Au Choix: The Excavation of an Archaic Indian Cemetery in Newfoundland. Institute of Social and Economic Research, Memorial University of Newfoundland; 1976. [Google Scholar]
- 74.Stanton DWG, Alberti F, Plotnikov V, Androsov S, Grigoriev S, Fedorov S, Kosintsev P, Nagel D, Vartanyan S, Barnes I, Barnett R, et al. Early Pleistocene origin and extensive intra-species diversity of the extinct cave lion. Sci Rep. 2020;10:12621. doi: 10.1038/s41598-020-69474-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bergström A, Stanton DWG, Taron UH, Frantz L, Sinding M-HS, Ersmark E, Pfrengle S, Cassatt-Johnstone M, Lebrasseur O, Girdland-Flink L, Fernandes DM, Ollivier M, et al. Grey wolf genomic history reveals a dual ancestry of dogs. Nature. 2022;607:313–320. doi: 10.1038/s41586-022-04824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fulton TL, Shapiro B. Setting up an ancient DNA laboratory. Methods Mol Biol. 2019;1963:1–13. doi: 10.1007/978-1-4939-9176-1_1. [DOI] [PubMed] [Google Scholar]
- 77.Schroeder H, Damgaard Pde Barros, Allentoft ME. Pretreatment: Improving endogenous ancient DNA yields using a simple enzymatic predigestion step. Methods Mol Biol. 2019;1963:21–24. doi: 10.1007/978-1-4939-9176-1_3. [DOI] [PubMed] [Google Scholar]
- 78.Dabney J, Knapp M, Glocke I, Gansauge M-T, Weihmann A, Nickel B, Valdiosera C, García N, Pääbo S, Arsuaga J-L, Meyer M. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci U S A. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kapp JD, Green RE, Shapiro B. A Fast and Efficient Single-stranded Genomic Library Preparation Method Optimized for Ancient DNA. J Hered. 2021;112:241–249. doi: 10.1093/jhered/esab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ersmark E, Orlando L, Sandoval-Castellanos E, Barnes I, Barnett R, Stuart A, Lister A, Dalén L. Population demography and genetic diversity in the Pleistocene cave lion. Open Quat. 2015;1:4. [Google Scholar]
- 82.Mak SST, Gopalakrishnan S, Geng C, Liu S, Sinding M-HS, Kuderna LFK, Zhang W, Fu S, Vieira FG, Bocherens H, Fedorov S, et al. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience. 2017;6:1–13. doi: 10.1093/gigascience/gix049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 84.Dutrow EV, Serpell JA, Ostrander EA. Domestic dog lineages reveal genetic drivers of behavioral diversification. Cell. 2022;185:4737–4755.:e18. doi: 10.1016/j.cell.2022.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dussex N, Stanton DWG, Sigeman H, Ericson PGP, Gill J, Fisher DC, Protopopov AV, Herridge VL, Plotnikov V, Hansson B, Dalén L. Biomolecular analyses reveal the age, sex and species identity of a near-intact Pleistocene bird carcass. Commun Biol. 2020;3:84. doi: 10.1038/s42003-020-0806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Palkopoulou E, Mallick S, Skoglund P, Enk J, Rohland N, Li H, Omrak A, Vartanyan S, Poinar H, Reich D, Dalén L. Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr Biol. 2015;25:1395–1400. doi: 10.1016/j.cub.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN] 2013. (available at http://arxiv.org/abs/1303.3997)
- 92.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, Shakir K, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017 doi: 10.1101/201178. [DOI] [Google Scholar]
- 94.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, Johnson PLF, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 96.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bouckaert R, Vaughan TG, Barido-Sottani J, Fourment M, Gavryushkina A, Heled J, Jones G, De Maio N, Matschiner M, Mendes FK, Ogilvie HA, et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Molak M, Suchard MA, Ho SYW, Beilman DW, Shapiro B. Empirical calibrated radiocarbon sampler: a tool for incorporating radiocarbon-date and calibration error into B ayesian phylogenetic analyses of ancient DNA. Mol Ecol Resour. 2015;15:81–86. doi: 10.1111/1755-0998.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Feuerborn TR, Carmagnini A, Losey RJ, Nomokonova T, Askeyev A, Askeyev I, Askeyev O, Antipina EE, Appelt M, Bachura OP, Beglane F, et al. Modern Siberian dog ancestry was shaped by several thousand years of Eurasian-wide trade and human dispersal. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2100338118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Narasimhan VM, Patterson N, Moorjani P, Rohland N, Bernardos R, Mallick S, Lazaridis I, Nakatsuka N, Olalde I, Lipson M, Kim AM, et al. The formation of human populations in South and Central Asia. Science. 2019;365:eaat7487. doi: 10.1126/science.aat7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marciniak S, Mughal MR, Godfrey LR, Bankoff RJ, Randrianatoandro H, Crowley BE, Bergey CM, Muldoon KM, Randrianasy J, Raharivololona BM, Schuster SC, et al. Evolutionary and phylogenetic insights from a nuclear genome sequence of the extinct, giant, “subfossil” koala lemur Megaladapis edwardsi. Proc Natl Acad Sci U S A. 2021;118:e2022117118. doi: 10.1073/pnas.2022117118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perry GH, Reeves D, Melsted P, Ratan A, Miller W, Michelini K, Louis EE, Jr, Pritchard JK, Mason CE, Gilad Y. A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol Evol. 2012;4:126–135. doi: 10.1093/gbe/evr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grote S. GOfuncR: Gene ontology enrichment using FUNC R package version 1.20.0. 2023 doi: 10.18129/B9.bioc.GOfuncR. [DOI] [Google Scholar]
- 110.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 111.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update. Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shimomura Y, Wajid M, Ishii Y, Shapiro L, Petukhova L, Gordon D, Christiano AM. Disruption of P2RY5, an orphan G protein–coupled receptor, underlies autosomal recessive woolly hair. Nat Genet. 2008;40:335–339. doi: 10.1038/ng.100. [DOI] [PubMed] [Google Scholar]
- 113.Khan GM, Hassan N, Khan N, Humayun M, Khan K, Khaliq S, Rehman FU, Ahmed S, Shah K, Khan SA, Muhammad N, et al. Biallelic mutations in the LPAR 6 gene causing autosomal recessive wooly hair/hypotrichosis phenotype in five Pakistani families. Int J Dermatol. 2019;58:946–952. doi: 10.1111/ijd.14480. [DOI] [PubMed] [Google Scholar]
- 114.Romano M-T, Tafazzoli A, Mattern M, Sivalingam S, Wolf S, Rupp A, Thiele H, Altmüller J, Nürnberg P, Ellwanger J, Gambon R, Baumer A, et al. Bi-allelic mutations in LSS, encoding lanosterol synthase, cause autosomal-recessive hypotrichosis simplex. Am J Hum Genet. 2018;103:777–785. doi: 10.1016/j.ajhg.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shah K, Basit S, Ali G, Ramzan K, Ansar M, Ahmad W. A novel homozygous frameshift variant in the C3orf52 gene underlying isolated hair loss in a consanguineous family. Eur J Dermatol. 2021;31:409–411. doi: 10.1684/ejd.2021.4053. [DOI] [PubMed] [Google Scholar]
- 116.Tariq M, Azhar A, Baig SM, Dahl N, Klar J. A novel mutation in the Lipase H gene underlies autosomal recessive hypotrichosis and woolly hair. Sci Rep. 2012;2:730. doi: 10.1038/srep00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Törnqvist G, Sandberg A, Hägglund AC, Carlsson L. Cyclic expression of lhx2 regulates hair formation. PLoS Genet. 2010;6:e1000904. doi: 10.1371/journal.pgen.1000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harshuk-Shabso S, Dressler H, Niehrs C, Aamar E, Enshell-Seijffers D. Fgf and Wnt signaling interaction in the mesenchymal niche regulates the murine hair cycle clock. Nat Commun. 2020;11:5114. doi: 10.1038/s41467-020-18643-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lim X, Tan SH, Yu KL, Lim SBH, Nusse R. Axin2 marks quiescent hair follicle bulge stem cells that are maintained by autocrine Wnt/β-catenin signaling. Proc Natl Acad Sci U S A. 2016;113:E1498–505. doi: 10.1073/pnas.1601599113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tobin DJ, Gunin A, Magerl M, Handijski B, Paus R. Plasticity and cytokinetic dynamics of the hair follicle mesenchyme: implications for hair growth control. J Invest Dermatol. 2003;120:895–904. doi: 10.1046/j.1523-1747.2003.12237.x. [DOI] [PubMed] [Google Scholar]
- 121.Hoover E, Alhajj M, Flores JL. Physiology, Hair. StatPearls Publishing; 2022. [PubMed] [Google Scholar]
- 122.Alonso L, Fuchs E. The hair cycle. J Cell Sci. 2006;119:391–393. doi: 10.1242/jcs.02793. [DOI] [PubMed] [Google Scholar]
- 123.Peter CJ, Saito A, Hasegawa Y, Tanaka Y, Nagpal M, Perez G, Alway E, Espeso-Gil S, Fayyad T, Ratner C, Dincer A, et al. In vivo epigenetic editing of Sema6a promoter reverses transcallosal dysconnectivity caused by C11orf46/Arl14ep risk gene. Nat Commun. 2019;10:4112. doi: 10.1038/s41467-019-12013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 125.Lokaj M, Kösling SK, Koerner C, Lange SM, van Beersum SEC, van Reeuwijk J, Roepman R, Horn N, Ueffing M, Boldt K, Wittinghofer A. The interaction of CCDC104/BARTL1 with Arl3 and implications for ciliary function. Structure. 2015;23:2122–2132. doi: 10.1016/j.str.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee Y-J, Ho S-R, Graves JD, Xiao Y, Huang S, Lin W-C. CGRRF1, a growth suppressor, regulates EGFR ubiquitination in breast cancer. Breast Cancer Res. 2019;21:134. doi: 10.1186/s13058-019-1212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu Z, Xiang Y, Sun G. The KCTD family of proteins: structure, function, disease relevance. Cell Biosci. 2013;3:45. doi: 10.1186/2045-3701-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chauhan S, Zheng X, Tan YY, Tay B-H, Lim S, Venkatesh B, Kaldis P. Evolution of the Cdk-activator Speedy/RINGO in vertebrates. Cell Mol Life Sci. 2012;69:3835–3850. doi: 10.1007/s00018-012-1050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ghelli Luserna di Rorà A, Cerchione C, Martinelli G, Simonetti G. A WEE1 family business: regulation of mitosis, cancer progression, and therapeutic target. J Hematol Oncol. 2020;13:126. doi: 10.1186/s13045-020-00959-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mecham RP. Overview of extracellular matrix. Curr Protoc Cell Biol. 2012 doi: 10.1002/0471143030.cb1001s57. Chapter 10. [DOI] [PubMed] [Google Scholar]
- 131.Donati G, Proserpio V, Lichtenberger BM, Natsuga K, Sinclair R, Fujiwara H, Watt FM. Epidermal Wnt/β-catenin signaling regulates adipocyte differentiation via secretion of adipogenic factors. Proc Natl Acad Sci U S A. 2014;111:E1501–E1509. doi: 10.1073/pnas.1312880111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tsukada S, Iwai M, Nishiu J, Itoh M, Tomoike H, Horiuchi M, Nakamura Y, Tanaka T. Inhibition of experimental intimal thickening in mice lacking a novel G-protein-coupled receptor. Circulation. 2003;107:313–319. doi: 10.1161/01.cir.0000043804.29963.b4. [DOI] [PubMed] [Google Scholar]
- 133.Guo L, Zhang H, Hou Y, Wei T, Liu J. Plasmalemma vesicle-associated protein: A crucial component of vascular homeostasis. Exp Ther Med. 2016;12:1639–1644. doi: 10.3892/etm.2016.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xu Y, Rong J, Zhang Z. The emerging role of angiotensinogen in cardiovascular diseases. J Cell Physiol. 2021;236:68–78. doi: 10.1002/jcp.29889. [DOI] [PubMed] [Google Scholar]
- 135.Kaetzel MA, Chan HC, Dubinsky WP, Dedman JR, Nelson DJ. A role for annexin IV in epithelial cell function. Inhibition of calcium-activated chloride conductance. J Biol Chem. 1994;269:5297–5302. [PubMed] [Google Scholar]
- 136.Kreft S, Klatt AR, Straßburger J, Pöschl E, Flower RJ, Eming S, Reutelingsperger C, Brisson A, Brachvogel B. Skin wound repair is not altered in the absence of endogenous AnxA1 or AnxA5, but pharmacological concentrations of AnxA4 and AnxA5 inhibit wound hemostasis. Cells Tissues Organs. 2016;201:287–298. doi: 10.1159/000445106. [DOI] [PubMed] [Google Scholar]
- 137.Cieslak M, Reissmann M, Hofreiter M, Ludwig A. Colours of domestication. Biol Rev Camb Philos Soc. 2011;86:885–899. doi: 10.1111/j.1469-185X.2011.00177.x. [DOI] [PubMed] [Google Scholar]
- 138.Schmutz SM, Berryere TG, Ellinwood NM, Kerns JA, Barsh GS. MC1R studies in dogs with melanistic mask or brindle patterns. J Hered. 2003;94:69–73. doi: 10.1093/jhered/esg014. [DOI] [PubMed] [Google Scholar]
- 139.Dürig N, Letko A, Lepori V, Hadji Rasouliha S, Loechel R, Kehl A, Lohi H, Mauri N, Dietrich J, Wiedmer M, Jagannathan V, et al. Two MC1R loss-of-function alleles in cream-coloured Australian Cattle Dogs and white Huskies. Anim Genet. 2018;49:284–290. doi: 10.1111/age.12660. [DOI] [PubMed] [Google Scholar]
- 140.Hédan B, Cadieu E, Botherel N, Dufaure de Citres C, Letko A, Rimbault M, Jagannathan V, Derrien T, Schmutz S, Leeb T, André C. Identification of a missense variant in MFSD12 involved in dilution of phaeomelanin leading to white or cream coat color in dogs. Genes (Basel) 2019;10:386. doi: 10.3390/genes10050386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Drögemüller C, Philipp U, Haase B, Günzel-Apel AR, Leeb T. A noncoding melanophilin gene (MLPH) SNP at the splice donor of exon 1 represents a candidate causal mutation for coat color dilution in dogs. J Hered. 2007;98:468–473. doi: 10.1093/jhered/esm021. [DOI] [PubMed] [Google Scholar]
- 142.Bauer A, Kehl A, Jagannathan V, Leeb T. A novel MLPH variant in dogs with coat colour dilution. Anim Genet. 2018;49:94–97. doi: 10.1111/age.12632. [DOI] [PubMed] [Google Scholar]
- 143.Van Buren SL, Minor KM, Grahn RA, Mickelson JR, Grahn JC, Malvick J, Colangelo JR, Mueller E, Kuehnlein P, Kehl A. A third MLPH variant causing coat color dilution in dogs. Genes (Basel) 2020;11:639. doi: 10.3390/genes11060639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Renaud G, Hanghøj K, Korneliussen TS, Willerslev E, Orlando L. Joint Estimates of Heterozygosity and Runs of Homozygosity for Modern and Ancient Samples. Genetics. 2019;212:587–614. doi: 10.1534/genetics.119.302057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pacheco C, Stronen AV, Jędrzejewska B, Plis K, Okhlopkov IM, Mamaev NV, Drovetski SV, Godinho R. Demography and evolutionary history of grey wolf populations around the Bering Strait. Mol Ecol. 2022;31:4851–4865. doi: 10.1111/mec.16613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Katzenberg MA, Lovell NC. Stable isotope variation in pathological bone. Int J Osteoarchaeol. 1999;9:316–324. [Google Scholar]
- 147.Longin R. New method of collagen extraction for radiocarbon dating. Nature. 1971;230:241–242. doi: 10.1038/230241a0. [DOI] [PubMed] [Google Scholar]
- 148.Brown TA, Nelson DE, Vogel JS, Southon JR. Improved Collagen Extraction by Modified Longin Method. Radiocarbon. 1988;30:171–177. [Google Scholar]
- 149.Harris J, Anderson S. Letters from the 49th Parallel, 1857–1873: Selected Correspondence of Joseph Harris and Samuel Anderson. Champlain Society; Toronto: 2013. Introduction; pp. xiii–cviii. [Google Scholar]
- 150.Pesticides. National Museum of the American Indian. (available at https://americanindian.si.edu/explore/collections/conservation/pesticides)
- 151.Ambrose SH. Preparation and characterization of bone and tooth collagen for isotopic analysis. J Archaeol Sci. 1990;17:431–451. [Google Scholar]
- 152.Ambrose SH, Norr L. On Stable Isotopic Data and Prehistoric Subsistence in the Soconusco Region. Curr Anthropol. 1992;33:401–404. [Google Scholar]
- 153.O’Connell TC, Hedges REM, Healey MA, Simpson AHRW. Isotopic Comparison of Hair, Nail and Bone: Modern Analyses. J Archaeol Sci. 2001;28:1247–1255. [Google Scholar]
- 154.McManus-Fry E, Knecht R, Dobney K, Richards MP, Britton K. Dog-human dietary relationships in Yup’ik western Alaska: The stable isotope and zooarchaeological evidence from pre-contact Nunalleq. Journal of Archaeological Science: Reports. 2018;17:964–972. [Google Scholar]
- 155.Guiry E, Royle TCA, Matson RG, Ward H, Weir T, Waber N, Brown TJ, Hunt BPV, Price MHH, Finney BP, Kaeriyama M, et al. Differentiating salmonid migratory ecotypes through stable isotope analysis of collagen: Archaeological and ecological applications. PLoS One. 2020;15:e0232180. doi: 10.1371/journal.pone.0232180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sharp Z. Principles of Stable Isotope Geochemistry. 2nd edition. 2017. (available at https://digitalrepository.unm.edu/unm_oer/1/?fref=gc&dti=175833885799280) [Google Scholar]
- 157.Keegan WF, DeNiro MJ. Stable Carbon- and Nitrogen-Isotope Ratios of Bone Collagen Used to Study Coral-Reef and Terrestrial Components of Prehistoric Bahamian Diet. Am Antiq. 1988;53:320–336. [Google Scholar]
- 158.Minagawa M, Wada E. Stepwise enrichment of 15N along food chains: Further evidence and the relation between δ15N and animal age. Geochim Cosmochim Acta. 1984;48:1135–1140. [Google Scholar]
- 159.DeNiro MJ, Epstein S. Influence of diet on the distribution of carbon isotopes in animals. Geochim Cosmochim Acta. 1978;42:495–506. [Google Scholar]
- 160.Hoefs J. Stable Isotope Geochemistry. Springer International Publishing; [Google Scholar]
- 161.Smith K, Rennie MJ. New approaches and recent results concerning human-tissue collagen synthesis. Curr Opin Clin Nutr Metab Care. 2007;10:582–590. doi: 10.1097/MCO.0b013e328285d858. [DOI] [PubMed] [Google Scholar]
- 162.Jackson CI. Letters from the 49th Parallel, 1857-1873: Selected Correspondence of Joseph Harris and Samuel Anderson. Vol. 63. Champlain Society; Toronto: 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic sequencing data for Mutton, SB dog, the Port au Choix dog (AL3194), and ALAS_015 are available for non-commercial use via NCBI SRA Project Accession PRJNA1005336 and BioSample Accessions SAMN36985984-SAMN36985987. The SRA Project Accession for the modern coyote from Wyoming is PRJNA734649. Stable isotope data are available (49). All other public genomic data sources are provided in DataS1.