Abstract
Trillions of bacteria inhabit the mammalian gastrointestinal tract. In the majority of hosts, these symbionts contribute largely to beneficial functions promoting microbe-host homeostasis. However, an increasing number of human diseases is associated with altered microbiota composition and enrichment of certain bacterial species. A well-known example of this is Mucispirillum schaedleri, which has been associated with inflammatory conditions in the intestine. Mucispirillum spp. belong to the phylum Deferribacteres and are prevalent but low abundant members of the rodent, pig and human microbiota. Recently, Mucispirillum schaedleri was causally linked to the development of Crohn’s disease − like colitis in immunodeficient mice. While this study certifies a considerable pathogenic potential, the same organism can also promote health in the immunocompetent host: M. schaedleri protects from Salmonella enterica serovar Typhimurium (S. Tm)-induced colitis by interfering with the expression of the pathogen´s invasion machinery. In this review, we summarize the current knowledge on the mammalian gut symbiont M. schaedleri and its role in intestinal homeostasis and discuss open questions and perspectives for future research.
Keywords: Pathobiont, colitogenic, dysbiosis, microbiome, Crohn´s disease, Ulcerative colitis, inflammation
Ecology of Mucispirillum schaedleri
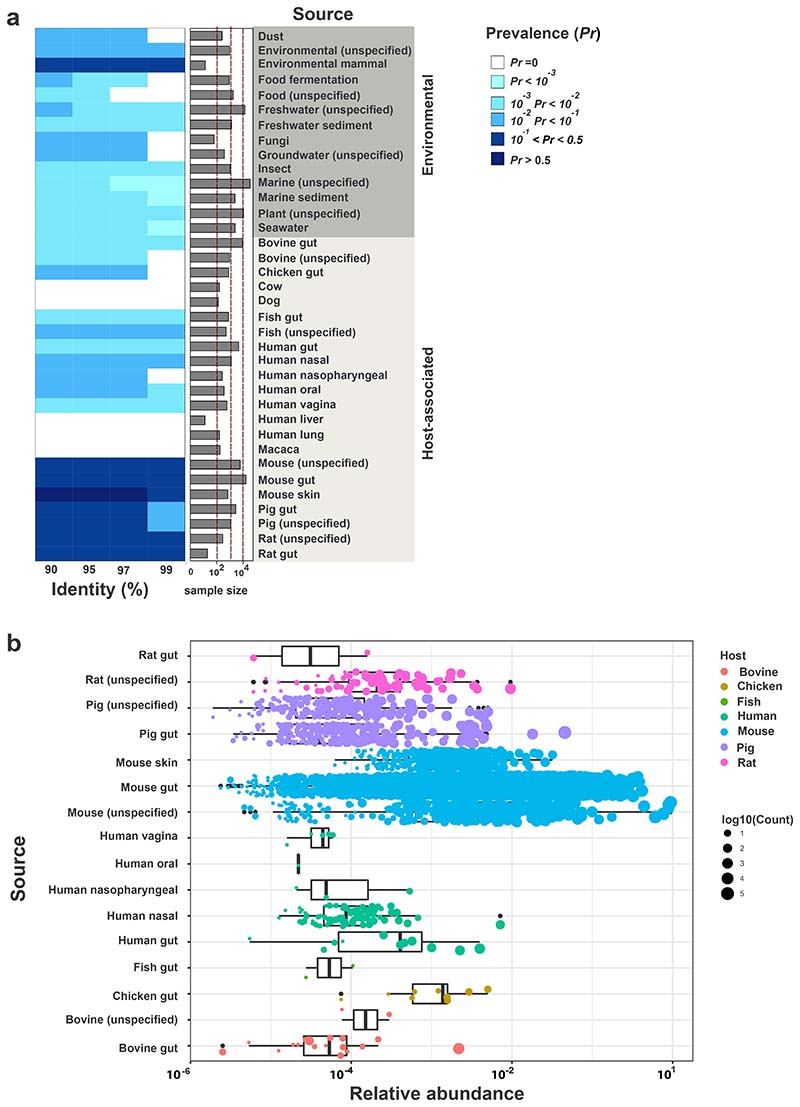
Eubacterial communities in the gut of vertebrates are dominated by the Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria. Low abundance phyla include Fusobacteria, Verrucomicrobia, Spirochaetes and Deferribacteres. Deferribacteres [1], comprise two families, Deferribacteraceae and Calditrichaceae, that mostly group environmental genera (e.g. Calditerrivibrio, Deferribacter, Denitrovibrio, Flexistipes, Geovibrio, Seleniivibrio and Petrothermobacter) living in hot springs, deep-sea or sediments [2–8]. Mucispirillum is, as of today, the only genus of the Deferribacteraceae known to inhabit the vertebrate gastrointestinal tract [9–12]. The type species, Mucispirillum schaedleri HRI I17 was first isolated from intestinal mucus scrapings of laboratory mice [13]. Sequence-based surveys of the mouse gut microbiota have consistently detected this species in mouse intestinal samples. While it shows a low relative abundance in murine feces (˜ 1%), it is enriched in the colonic mucus (relative abundance up to 10%) [14–16]. To gain more insights in the ecology of M. schaedleri, we determined the prevalence of this organism in host-associated and environmental samples by conducting a 16S rRNA gene amplicon-based survey in published datasets contained in IMNGS [17]. Our results are in line with previous reports on the occurrence of M. schaedleri in the intestinal tract of various animals. Compared to environmental samples, a higher prevalence of M. schaedleri was found in host-associated samples, in particular derived from the gut of rodents and pigs (Fig. 1A). Highly similar sequences (>99% identity) were also detected in the gut of cattle, fish and humans, but at lower prevalence. Strikingly, M. schaedleri was detectable in a significant fraction of human nasal samples, at low relative abundance (Fig. 1B). Other environmental samples harbored sequences at 97% identity and below. Interestingly, a fraction of soil and insect gut samples also harbored M. schaedleri sequences.
Figure 1. Mucispirillum spp. prevalence and abundance in host-associated and environmental microbial communities determined from published 16S rRNA amplicon studies using the IMNGS portal.
(A) Prevalence of Mucispirillum schaedleri ASF457 at 90%, 95%, 97% and 99% identity. The y-axis represents the source of the samples, which is organized as “environmental” and “host-associated”. The barplot adjacent to the heatmap represents the sample size from all sources of interest. Prevalence is indicated by different shades of blue. (B) Relative abundance of M. schaedleri ASF457 shown for samples with relative abundance > 0 at 97% identity. The x-axis represents log10-transformed relative abundance values and the y-axis different host-associated sources of interest. The dots within each boxplot represent rel. abundance in individual samples and the size of the dots corresponds to the total number of M. schaedleri ASF457 reads detected in the sample at 97% identity.
Relative abundance of M. schaedleri was only determined for host-associated samples (Fig. 1B). The highest relative abundance was clearly found in mouse-derived specimen, followed by samples from pig, chicken, rat and human gut. The mean relative abundance of M. schaedleri in the human gut was clearly below 0,1%. It was previously shown that the prevalence of M. schaedleri in human stool samples of healthy individuals is low (detected in 10/339 samples). However, mucosal biopsies exhibited a higher prevalence (e.g. detected in 20/27 jejunum, 12/21 cecum and 11/22 ascending colon) in the same study [16, 18]. Likewise, the mean relative abundance of M. schaedleri in human stool samples was 0,02%, while it increased up to 1% in mucosal biopsies. A generally applied procedure in 16S rRNA amplicon sequencing studies is to exclude spurious taxa (<0,25% rel. abundance) [19]. This may explain, why M. schaedleri has long been considered to be absent in the human microbiota [20]. Instead, similar to its location in the murine gastrointestinal tract, the bacterium preferentially colonizes the mucus layer and its relative abundance in stool is rather low [10, 16, 18].
Lifestyle and metabolism
M. schaedleri is a spiral shaped, flagellated, Gram- and obligate anaerobic bacterium [13]. It colonizes the intestinal mucus layer but only harbours a very limited set of genes encoding glycan-degrading enzymes [21]. Instead, M. schaedleri seems to utilize monosaccharides, amino acids or short-chain fatty acids for energy metabolism [21, 22]. M. schaedleri is also enriched at mucosal sites in Anterior gradient 2 protein (Agr2-/-)-deficient mice, which have a poorly developed inner colonic mucus layer [16]. This suggests that mucus is not an important prerequisite for M. schaedleri colonization. The close proximity of the epithelium represents a rather hostile environment where microbes are exposed to reactive oxygen and nitrogen species [23]. M. schaedleri encodes superoxide reductase, catalase, and cytochrome c oxidase genes, which may be important to overcome host defences and colonize mucosal sites [21]. Furthermore, M. schaedleri can perform dissimilatory nitrate reduction to ammonia (DNRA) and addition of nitrate enhances its growth in vitro [21].
Intestinal inflammation and M. schaedleri
A large number of studies have linked Mucispirillum spp. to inflammatory bowel disease (IBD) and a variety of other diseases. The result of our (non-exhaustive) literature search is summarized in Table 1. Several studies in mice reported that Mucispirillum spp. is enriched in intestinal samples under conditions of gut inflammation, including genetic and chemical colitis models and infection (Table 1). Moreover, increase of Mucispirillum spp. has been linked to high-fat diet, drug treatment, stress and diseases like Rheumatoid Arthritis or Parkinson´s Disease (Table 1). The fact that M. schaedleri can respire nitrate, which becomes abundant during inflammatory conditions, may explain blooming of this bacterium in an inflamed environment, as is the case for Enterobacteriaceae [24]. However, so far there is little evidence that M. schaedleri can drive disease in an immunocompetent host background. Of note, M. schaedleri ASF457 is part of the Altered Schaedler Flora (ASF), a low complex consortium of eight different bacterial species [25] widely used in research on microbiota-host interactions [26]. The ASF is considered as “pathobiont-free microbiota”, which, on its own, does not trigger inflammation. This applies to WT mice but also to several immunodeficient lines (Rag1-/-, Rag2-/-, Tlr5-/-) and IBD mouse models (IL10-/-) [27–29] suggesting, that M. schaedleri is not colitogenic within the context of the ASF.
Table 1. Research articles linking Mucispirillum spp. to inflammation and disease.
| Article | Findings regarding Mucispirillum spp. | Associated Disease |
|---|---|---|
| [38] | M. schaedleri is coated by human derived IgA from patients with Crohn's disease or healthy donors. | IBD |
| [39] | Mucispirillum spp. are increased during active DSS colitis in the murine gut and was identified as indicator species for previous DSS induced colitis. | Dextrane-sulfate sodium (DSS)-colitis mouse model / IBD |
| [40] | Mucispirillum was identified as indicator for DSS-colitis in mice. | DSS-colitis mouse model / IBD |
| [41] | During early colonization of germ-free mice with a conventional microbiota, Mucispirillum spp. are overrepresented which coincides with a proinflammatory milieu. | n.a. |
| [42] | Mucispirillum spp. were enriched during active colitis in TRUC mice (T-bet−/− Rag2−/− ulcerative colitis). | TRUC mice (T-bet−/− Rag2−/− ulcerative colitis) |
| [43] | Mucispirillum spp. were found to be a biomarker for spontaneous colitis in an IBD model (A20IEC/myel-KO). | IBD model (A20IEC/myel-KO) |
| [44] | Mucispirillum spp. were associated with the pro-inflammatory microbiota of CD1 d−/− mice. | DSS-colitis mouse model; CD1 d−/− mice |
| [31] | Mucispirillum spp. induce T-cell dependent IgA in the colon and jejunum. | n.a. |
| [45] | Citrobacter rodentium infection decreases Mucispirillum spp. by diminishing its niche during early infection. Mucispirillum spp. can only recover during clearance of C. rodentium. | Pathogen infection |
| [46] | Patients with Parkinson's disease exhibit an increased relative abundance of Mucispirillum spp.. Those patients also showed higher plasma levels of TNF-α and INF-γ indicating a subinflammatory status. | Parkinson’s Disease |
| [47] | P. scandens extract reduced inflammatory cell infiltration in a mouse model which was correlated with reduced abundance of Mucispirillum spp.. | Rheumatoid arthritis |
| [48] | Mucispirillum spp. were positively correlated with SLE symptoms in a mouse model. Mucispirillum spp. abundance decreased after treatment with a glucocorticoid (prednisone). | Systemic lupus erythematosus (SLE) |
| [49] | Cholesterol-lowering atorvastatin and rosuvastatin increase the abundance of Mucispirillum spp. in high-fat diet fed mice. | Metabolic syndrome |
| [50] | Mucispirillum spp. abundance is increased by stress during pregnancy in mice. | Stress |
| [51] | M. schaedleri was enriched in mice facing social stress. | Social Stress |
| [52] | M. schaedleri was positively correlated with body weight in high-fat diet fed mice. | High-fat diet / metabolic syndrome |
In contrast, a recent study established that Mucispirillum schaedleri is causally involved in Crohn’s disease − like colitis in severely immunodeficient mice [30]. Caruso et al. found that doubly deficient Nod2−/− Cybb−/− (but not WT or single deficient) mice develop spontaneous colitis when exposed to a specific microbiota. Induction of inflammation was linked to enrichment of Mucispirillum spp. in this colitogenic microbiota. Transfer of M. schaedleri ASF457 to Nod2−/− Cybb−/− mice triggered colitis, demonstrating that this bacterium indeed is the disease driver. Accumulation of Mucispirillum spp. at mucosal sites in doubly deficient mice suggested that impaired neutrophil recruitment and bacterial killing leads to increased tissue loads of Mucispirillum spp., which then triggers inflammation. Notably, maternal Mucispirillum-specific IgG and IgA protected mice from development of inflammation until weaning and led to increased survival. Further studies reported that M. schaedleri triggers T-cell dependent IgA and IgG responses, suggesting that this organism has the ability of efficient immune priming [31, 32]. Robertson et al. even isolated M. schaedleri from mouse liver samples, suggesting that the species can translocate from the intestinal tract to the hepatobiliary system [13].
Together, this shows that M. schaedleri is a mammalian symbiont with elevated pathogenic potential, which is sufficiently invasive to prime adaptive immune responses in a normal host context. Additionally, in a severely immunocompromised host, the invasive character of M. schaedleri can trigger intestinal inflammatory responses.
M. schaedleri interferes with Salmonella infection
Increasing evidence exists, that the same bacterial strain termed as “pathobiont” could also exert health-promoting functions on its host in a different scenario [33]. Along these lines, we demonstrated a beneficial role of M. schaedleri in the context of non-typhoidal Salmonella enterica serovar Typhimurium (S. Tm) infection [16]. Salmonella is a food-borne pathogen and a major cause of self-limiting gastroenteritis in humans. The intestinal microbiota broadly protects from S. Tm by different mechanisms, including competition for substrates, production of antimicrobial compounds and strengthening of barrier functions [34]. When ingested, Salmonella has to colonize the gut lumen to high levels in order to invade the intestinal epithelium and subsequently induce intestinal inflammation [35]. Invasion into epithelial cells is mediated by employing its major pathogenicity factor, a type 3 secretion system encoded on the Salmonella Pathogenicity Island-1 (SPI1-T3SS) [36]. We found that Agr2-/- mice which exhibit an impaired mucus layer, were protected from S. Tm induced inflammation [16]. This correlated with a vast increase in relative abundance of M. schaedleri at mucosal sites in Agr2-/- but not isobiotic Agr2+/- control mice. Experiments using mice associated with defined microbiota revealed that M. schaedleri cannot block S. Tm colonization but delay the onset of S. Tm colitis by several days. The underlying reason is that S. Tm downregulated its SPI1-T3SS in the presence of M. schaedleri. Since high levels of NO3- promote expression of the SPI1-T3SS, it is conceivable that competition for NO3- is the underlying mechanism how M. schaedleri blocks S. Tm virulence. This study provided the first evidence of an intestinal symbiont which can block virulence functions of a major human enteric pathogen.
Concluding remarks
M. schaedleri is a prevalent symbiont of the gut microbiota of humans and various animals. Due to its low relative abundance in human fecal samples, it has not been noted in the majority of human studies. In mouse models Mucispirillium spp. have been associated with several disease conditions but M. schaedleri can also promote health by interfering with pathogen invasion. For this reason, M. schaedleri may have potential for therapeutic application against human Salmonella infection. Obviously, it has to be clarified, whether M. schaedleri in general or only specific strains are associated with inflammatory, autoimmune and neoplastic diseases. In addition, future work is needed to study the ecology of Mucispirillium spp. in the human and animal gut. 16S rRNA gene amplicon-based survey on published datasets revealed that highly similar sequences (>99% identity) are present in gut samples from humans, mice, rats, other mammals and fish (Fig. 1A). Pigs and chicken show a higher prevalence of less similar sequences (>97% identity), suggesting that there is a higher species diversity. Clearly, a culture collection of Mucispirillium spp. isolates from humans and animals will be of great value for future research, as comparative genomic and functional studies of isolates will be required to understand the precise roles of these symbionts in their different host species.
Methods
One 16S rRNA gene sequence (GeneID: 2558850189) representative for Mucispirillum schaedleri ASF457 genome (NCBI Taxon ID: 1379858) was obtained from the IMG database [37]. In order to identify the prevalence and relative abundance of Mucispirillum schaedleri ASF457 in published 16S rRNA amplicon datasets, we used the IMNGS portal [17] (https://www.imngs.org/16S_rRNA/search/). A list of samples was preselected according to their sources (1) host-associated (for host species, unifying general name and scientific name) and the corresponding body site and (2) environmental sources. The M. schaedleri ASF457 16S rRNA gene sequence (GeneID: 2558850189) was used for the similarity search against these selected lists of samples for 90%, 95%, 97% and 99% identity and minimum length of 100 bp. The prevalence was determined as fraction of M. schaedleri ASF457 positive samples of all samples from the respective group. The relative abundance was determined for M. schaedleri ASF457 positive samples at 97% identity.
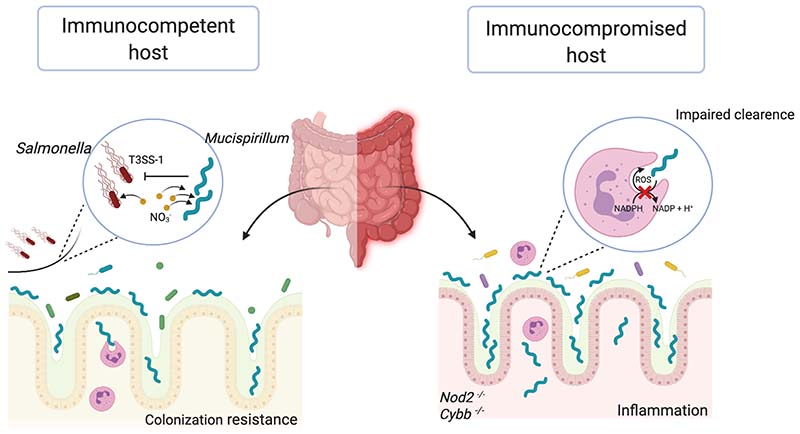
Figure 2. Role of Mucisprillum schaedleri in microbiota-host homeostasis.
Mucispirillum schaedleri, is highly prevalent in the murine gut and enriched at the mucosal surface of the colon. In the immunocompetent host (left side), M. schaedleri can protect from Salmonella enterica serovar Typhimurium (S. Tm)-induced colitis by competing for NO3- and interfering with the expression of the pathogen´s SPI-1 encoded T3SS. In severely immunodeficient Nod2−/− Cybb−/− hosts (right side), which exhibit deficient neutrophil migration and killing functions, higher numbers of M. schaedleri overcome the mucosal barrier and trigger Crohn’s disease − like colitis.
Acknowledgements
Bärbel Stecher acknowledges funding by the German Center of Infection Research (DZIF), the DFG Priority Program SPP1656, the Collaborative Research Center CRC1371 and the European Research Council (grant no. 865615).
References
- 1.Huber H, Stetter KO. Class I. Deferribacteres class. nov. Bergey’s Manual of Systematic Bacteriology. 2001;1:465. [Google Scholar]
- 2.Caccavo F, Coates JD, Rossello-Mora RA, et al. Geovibrio ferrireducens, a phylogenetically distinct dissimilatory Fe(III)-reducing bacterium. Arch Microbiol. 1996;165:370–376. doi: 10.1007/s002030050340. [DOI] [PubMed] [Google Scholar]
- 3.Fiala G, Woese CR, Langworthy TA, et al. Flexistipes sinusarabici, a novel genus and species of eubacteria occurring in the Atlantis II Deep brines of the Red Sea. Arch Microbiol. 1990;154:120–126. doi: 10.1007/BF00423320. [DOI] [Google Scholar]
- 4.Myhr S, Torsvik T. Denitrovibrio acetiphilus, a novel genus and species of dissimilatory nitrate-reducing bacterium isolated from an oil reservoir model column. Int J Syst Evol Microbiol. 2000;50(4):1611–1619. doi: 10.1099/00207713-50-4-1611. [DOI] [PubMed] [Google Scholar]
- 5.Takai K, Kobayashi H, Nealson KH, et al. Deferribacter desulfuricans sp. nov., a novel sulfur-, nitrate- and arsenate-reducing thermophile isolated from a deep-sea hydrothermal vent. Int J SystEvolMicrobiol. 2003;53:839–846. doi: 10.1099/ijs.0.02479-0. [DOI] [PubMed] [Google Scholar]
- 6.Iino T, Nakagawa T, Mori K, et al. Calditerrivibrionitroreducens gen. nov., sp. nov., a thermophilic, nitrate-reducing bacterium isolated from a terrestrial hot spring in Japan. Int J Syst Evol Microbiol. 2008;58:1675–1679. doi: 10.1099/ijs.0.65714-0. [DOI] [PubMed] [Google Scholar]
- 7.Tamazawa S, Mayumi D, Mochimaru H, et al. Petrothermobacter organivorans gen. nov., sp. nov., a thermophilic, strictly anaerobic bacterium of the phylum Deferribacteres isolated from a deep subsurface oil reservoir. Int J Syst Evol Microbiol. 2017;67:3982–3986. doi: 10.1099/ijsem.0.002234. [DOI] [PubMed] [Google Scholar]
- 8.Rauschenbach I, Posternak V, Cantarella P, et al. Seleniivibrio woodruffii gen. nov., sp. nov., a selenate- and arsenate-respiring bacterium in the Deferribacteraceae. Int J Syst Evol Microbiol. 2013;63:3659–3665. doi: 10.1099/ijs.0.043547-0. [DOI] [PubMed] [Google Scholar]
- 9.Rosshart SP, Vassallo BG, Angeletti D, et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171:1015–1028.:e13. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrell L, Wang Y, Antonopoulos D, et al. Standard colonic lavage alters the natural state of mucosal-associated microbiota in the human colon. PLoS One. 2012;7:e32545. doi: 10.1371/journal.pone.0032545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michel AJ, Ward LM, Goffredi SK, et al. The gut of the finch: uniqueness of the gut microbiome of the Galápagos vampire finch. Microbiome. 2018;6:167. doi: 10.1186/s40168-018-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umu ÖCO, Mydland LT, Øverland M, et al. Rapeseed-based diet modulates the imputed functions of gut microbiome in growing-finishing pigs. Sci Rep. 2020;10:9372. doi: 10.1038/s41598-020-66364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson BR, O’Rourke JL, Neilan BA, et al. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol. 2005;55:1199–1204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- 14.Linnenbrink M, Wang J, Hardouin EA, et al. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol. 2013;22:1904–1916. doi: 10.1111/mec.12206. [DOI] [PubMed] [Google Scholar]
- 15.Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A, et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015;16:164–177. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herp S, Brugiroux S, Garzetti D, et al. Mucispirillum schaedleri Antagonizes Salmonella Virulence to Protect Mice against Colitis. Cell Host Microbe. 2019;25:681–694.:e8. doi: 10.1016/j.chom.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Lagkouvardos I, Joseph D, Kapfhammer M, et al. IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep. 2016;6:33721. doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zmora N, Zilberman-Schapira G, Suez J, et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell. 2018;174:1388–1405.:e21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 19.Reitmeier S, Hitch TCA, Fikas N, et al. Handling of spurious sequences affects the outcome of high-throughput 16S rRNA gene amplicon profiling. 2020 doi: 10.1038/s43705-021-00033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krych L, Hansen CHF, Hansen AK, et al. Quantitatively different, yet qualitatively alike: a meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PLoS One. 2013;8:e62578. doi: 10.1371/journal.pone.0062578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loy A, Pfann C, Steinberger M, et al. Lifestyle and Horizontal Gene Transfer-Mediated Evolution of Mucispirillum schaedleri, a Core Member of the Murine Gut Microbiota. mSystems. 2017;2 doi: 10.1128/mSystems.00171-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry D, Stecher B, Schintlmeister A, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci U S A. 2013;110:4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aviello G, Knaus UG. ROS in gastrointestinal inflammation: Rescue Or Sabotage? Br J Pharmacol. 2017;174:1704–1718. doi: 10.1111/bph.13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winter SE, Winter MG, Xavier MN, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orcutt RP, Gianni FJ, Judge RJ. Development of an “altered Schaedler flora” for NCI gnotobiotic rodents. Microecol Ther. 1987;17 [Google Scholar]
- 26.Wymore Brand M, Wannemuehler MJ, Phillips GJ, et al. The Altered Schaedler Flora: Continued Applications of a Defined Murine Microbial Community. ILAR J. 2015;56:169–178. doi: 10.1093/ilar/ilv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomes-Neto JC, Kittana H, Mantz S, et al. A gut pathobiont synergizes with the microbiota to instigate inflammatory disease marked by immunoreactivity against other symbionts but not itself. Sci Rep. 2017;7:17707. doi: 10.1038/s41598-017-18014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolsega S, Basic M, Smoczek A, et al. Composition of the Intestinal Microbiota Determines the Outcome of Virus-Triggered Colitis in Mice. Front Immunol. 2019;10:1708. doi: 10.3389/fimmu.2019.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chassaing B, Gewirtz AT. Mice harboring pathobiont-free microbiota do not develop intestinal inflammation that normally results from an innate immune deficiency. PLoS One. 2018;13:e0195310. doi: 10.1371/journal.pone.0195310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caruso R, Mathes T, Martens EC, et al. A specific gene-microbe interaction drives the development of Crohn’s disease-like colitis in mice. Sci Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunker JJ, Flynn TM, Koval JC, et al. Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity. 2015;43:541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jergens AE, Wilson-Welder JH, Dorn A, et al. Helicobacter bilis triggers persistent immune reactivity to antigens derived from the commensal bacteria in gnotobiotic C3H/HeN mice. Gut. 2006;56:934–940. doi: 10.1136/gut.2006.099242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jochum L, Stecher B. Label or Concept - What Is a Pathobiont? Trends Microbiol. 2020;28:789–792. doi: 10.1016/j.tim.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Rogers AWL, Tsolis RM, Bäumler AJ. Salmonella versus the Microbiome. Microbiol Mol Biol Rev. 2021;85 doi: 10.1128/MMBR.00027-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hapfelmeier S, Stecher B, Barthel M, et al. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol. 2005;174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- 36.Galán JE. Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 37.Chen I-MA, Chu K, Palaniappan K, et al. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res. 2021;49:D751–D763. doi: 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabbert J, Benckert J, Rollenske T, et al. High microbiota reactivity of adult human intestinal IgA requires somatic mutations. J Exp Med. 2020;217 doi: 10.1084/jem.20200275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berry D, Kuzyk O, Rauch I, et al. Intestinal Microbiota Signatures Associated with Inflammation History in Mice Experiencing Recurring Colitis. Front Microbiol. 2015;6:1408. doi: 10.3389/fmicb.2015.01408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berry D, Schwab C, Milinovich G, et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012;6:2091–2106. doi: 10.1038/ismej.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El Aidy S, Derrien M, Aardema R, et al. Transient inflammatory-like state and microbial dysbiosis are pivotal in establishment of mucosal homeostasis during colonisation of germ-free mice. Benef Microbes. 2014;5:67–77. doi: 10.3920/BM2013.0018. [DOI] [PubMed] [Google Scholar]
- 42.Rooks MG, Veiga P, Wardwell-Scott LH, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vereecke L, Vieira-Silva S, Billiet T, et al. A20 controls intestinal homeostasis through cell-specific activities. Nat Commun. 2014;5:5103. doi: 10.1038/ncomms6103. [DOI] [PubMed] [Google Scholar]
- 44.Selvanantham T, Lin Q, Guo CX, et al. NKT Cell-Deficient Mice Harbor an Altered Microbiota That Fuels Intestinal Inflammation during Chemically Induced Colitis. J Immunol. 2016;197:4464–4472. doi: 10.4049/jimmunol.1601410. [DOI] [PubMed] [Google Scholar]
- 45.Belzer C, Gerber GK, Roeselers G, et al. Dynamics of the microbiota in response to host infection. PLoS One. 2014;9:e95534. doi: 10.1371/journal.pone.0095534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin C-H, Chen C-C, Chiang H-L, et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J Neuroinflammation. 2019;16:129. doi: 10.1186/s12974-019-1528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao M, Fu X, Ni Y, et al. Protective effects of Paederia scandens extract on rheumatoid arthritis mouse model by modulating gut microbiota. J Ethnopharmacol. 2018;226:97–104. doi: 10.1016/j.jep.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 48.He Z, Kong X, Shao T, et al. Alterations of the Gut Microbiota Associated With Promoting Efficacy of Prednisone by Bromofuranone in MRL/lpr Mice. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Lee H, An J, et al. Alterations in Gut Microbiota by Statin Therapy and Possible Intermediate Effects on Hyperglycemia and Hyperlipidemia. Front Microbiol. 2019;10:1947. doi: 10.3389/fmicb.2019.01947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jašarević E, Howard CD, Misic AM, et al. Stress during pregnancy alters temporal and spatial dynamics of the maternal and offspring microbiome in a sex-specific manner. Sci Rep. 2017;7:44182. doi: 10.1038/srep44182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Werbner M, Barsheshet Y, Werbner N, et al. Social-Stress-Responsive Microbiota Induces Stimulation of Self-Reactive Effector T Helper Cells. mSystems. 2019;4 doi: 10.1128/mSystems.00292-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ussar S, Griffin NW, Bezy O, et al. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab. 2015;22:516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]