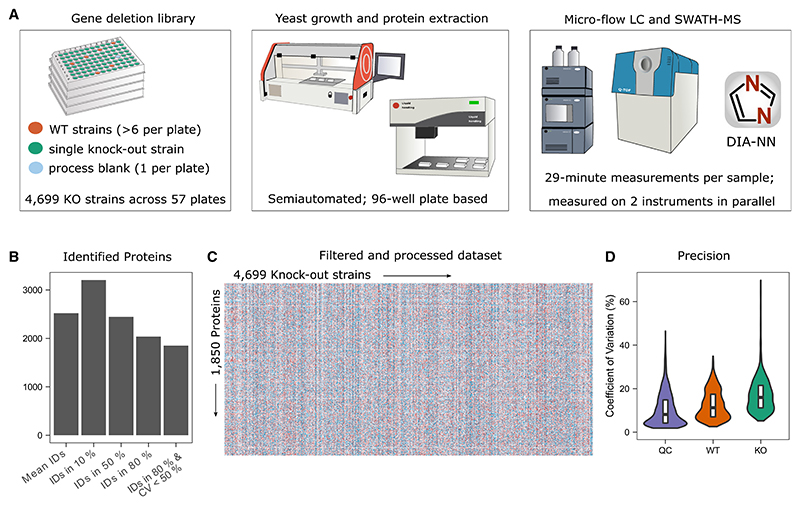
Figure 1. Quantitative proteomes for the genome-scale yeast gene-deletion collection.
(A) Experimental setup (STAR Methods).
(B) Protein identification numbers as mean per sample (2,520), identified in 10% of the samples (3,205), identified in 50% of the samples (2,445), identified in 80% of the samples (2,036), and identified in 80% of the WT samples with CV <50% (filtered dataset as described in STAR Methods) (1,850). All values were calculated for samples that passed the quality control (QC) thresholds.
(C) The filtered quantitative data are shown as a heatmap with 1,850 unique proteins measured across the 4,699 KOs, containing 8,693,150 protein quantities.
(D) The coefficients of variation (CVs; in %) were calculated for each protein and are shown for pooled yeast digest samples (QC, n = 389), whole-process control samples (WT, n = 388), and KO samples (KO, n = 4,699). Median CV values are 8.1% across the technical replicates of a pooled digest, 11.3% across the biological replicates of the wild-type strain, and 16.2% across the KO library. CVs were calculated on the filtered dataset and are shown from 0% to 70% (see Figure S1B for all data points).