Abstract
Since the turn of the millenium, RNA-based control of gene expression has added an extra dimension to the central dogma of molecular biology. Still, the roles of Mycobacterium tuberculosis regulatory RNAs, and the proteins that facilitate their functions remain elusive, although there can be no doubt that RNA biology plays a central role in the baterium’s adaptation to its many host environments. In this review we have presented examples from model organisms and from M. tuberculosis to showcase the abundance and versatility of regulatory RNA, in order to emphasize the importance of these ‘fine-tuners’ of gene expression.
Keywords: Mycobacterium tuberculosis, post-transcriptional regulation, regulatory RNA, small RNA, riboswitch, riboregulation
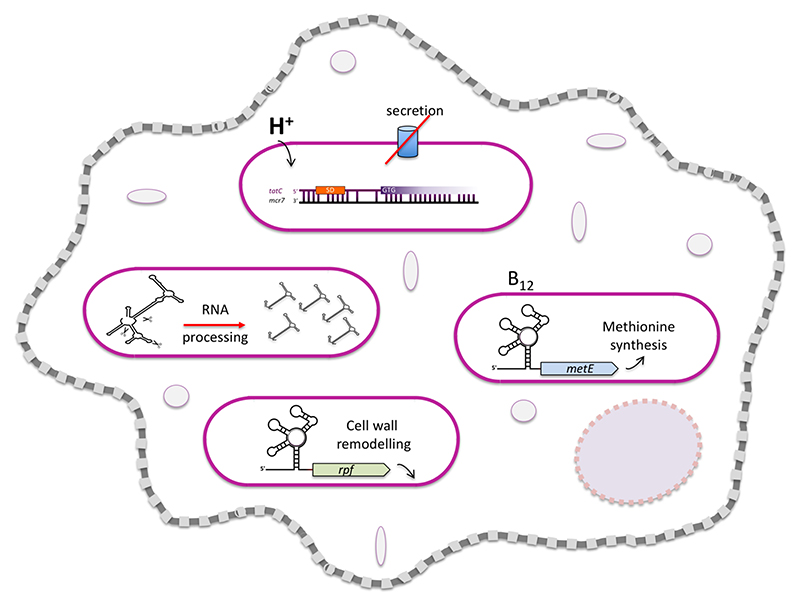
Graphical abstract. aspects of M. tuberculosis regulatory RNA discussed in this review.
Introduction
Bacterial gene expression consists of two tightly coupled processes, transcription and translation. A detailed, systematic and molecular to global characterisation of how these processes are regulated in pathogens is critical for development and improvement of disease interventions. While the regulation of transcription initiation is chiefly protein-based and orchestrated by sigma- and transcription factors, post-transcriptional regulation is to a large extent influenced by regulatory RNA species or ‘riboregulators’.
Until the turn of the millenium, riboregulators were few and far between; their discovery had been serendipituous, and in particular small regulatory RNAs (sRNAs) were mostly associated with plasmid copy number control, phages and transposable elements, reviewed in (Wagner et al., 2002). Two publications from 2001 signalled a change in attitude towards regulatory RNA with systematic searches for intergenic sRNAs based on cDNA cloning, tiling arrays and bioinformatics (Argaman et al., 2001, Wassarman et al., 2001). This was soon followed by the identification and characterisation of a number of highly conserved cis-regulatory elements, e.g. (Grundy et al., 2002, Mironov et al., 2002, Nahvi et al., 2002, Winkler et al., 2002). The trend continued at an accelerated pace with the advent of Deep sequencing methods, including variations such as dRNA-seq, ribo-seq, term-seq and grad-seq providing unbiased and comprehensive mapping of global transcriptomes e.g (Ingolia et al., 2009, Sharma et al., 2010, Dar et al., 2016, Smirnov et al., 2016). Currently, a plethora of riboregulators with steadily increasing complexity and functionality has been revealed in a substantial number of prokaryotic species, reviewed in (Wagner & Romby, 2015), and by now, the number of riboregulators is likely to equal or exceed the number of transcription factors in many bacteria (Ishihama, 2010, Rau et al., 2015, Holmqvist & Wagner, 2017, Smirnov et al., 2017). The more we uncover, the clearer it becomes that just like the processes of transcription and translation are tightly coupled, the functions of their regulators are interwoven and part of each other’s regulons (Arnvig & Young, 2010, Beisel & Storz, 2010, Lee & Gottesman, 2016).
RNA-based control of gene expression
The question ‘why regulatory RNA?’ keeps emerging in scientific discussions. There are several answers to this question that to some extent dependent on the type of element. Judging by their evolutionary conservation, some regulatory RNAs, including certain riboswitches are very old and possibly remnants from the RNA world and thus predate proteinaceous regulators (McCown et al., 2017). This is not the case for sRNAs, which have evolved later than other non-coding RNAs (ncRNAs) (Peer & Margalit, 2014). Many sRNAs are induced by stress and their synthesis is faster and less costly than transcription and translation of a proteinaceous transcription factor, two qualities that may be of significance during stress. In addition, one might argue from first principles that RNA is the obvious interaction partner for RNA - because basepairing is less complicated than evolving RNA-protein interactions. New sRNAs are continuously emerging as a result of single nucleotide polymorphisms (SNPs) leading to spurious promoters e.g. (Rose et al., 2013), and probably continuously evolving, as sRNA-mRNA interactions can be easily modulated by a few SNPs adjusting their basepairing (Updegrove et al., 2015). Whether these SNPs become fixed depends on the resulting fitness gain or -loss.
Several reports indicate that sRNA regulators have different kinetic properties than protein regulators and that (sRNAs as well as asRNAs) may play a role in suppressing transcriptional noise (Levine & Hwa, 2008, Lasa et al., 2011, Holmqvist & Wagner, 2017).
Finally, it is important to keep in mind that regulatory RNAs operate not instead of, but in addition to, conventional protein-based regulation thereby significantly expanding the number of available components and the complexity of cellular regulatory network.
M. tuberculosis differs from model organisms
M. tuberculosis is subject to multiple stresses and environments through the course of transmission, infection, immune response attack, dormancy and resuscitation, and surviving these changes requires extensive rewiring of its gene expression programme. Regrettably, protein-centric gene expression analysis still provides the main point of reference for M. tuberculosis. Our knowledge about mycobacterial riboregulation to a large extent still remains ‘dark matter’, although there is currently no doubt that this is an important aspect of M. tuberculosis’s intracellular life. Most of our knowledge about bacterial riboregulation originates from studies of the Gram-negative Escherichia coli and Salmonella typhimurium, and the Gram-positive Listeria monocytogenes, Bacillus subtilis and Staphylococcus aureus. These model organisms have provided extensive and novel insights into the structure, function and mechanisms of different types of regulatory RNA. In addition to integrating ‘ribo-knowledge’ into our understanding of how M. tuberculosis gene expression is controlled, we may ask whether it has anything to offer to the already vast knowledgebase of RNA biology or whether it is all about finding a cure? Should we simply rely on extrapolating findings from significantly more tractable model organisms instead of sweating in the Cat 3 lab playing catch up? The answer is a resounding ‘No!’ and the reasons will hopefully be clearer after reading this review. In here we provide a brief overview of different types of riboregulation, showcasing pertinent examples from model organisms as well as M. tuberculosis. Due to the rapidly expanding field of regulatory RNA, we are not able to provide a comprehensive overview, and we apologize in advance for the ommission of our colleagues’ work that has not been included due to space constraints. We refer to more extensive reviews on riboswitches e.g. (Serganov & Nudler, 2013, Sherwood & Henkin, 2016, Quereda & Cossart, 2017) and on sRNAs e.g. (Updegrove et al., 2015, Wagner & Romby, 2015, Smirnov et al., 2017). Our key aim is to highlight and further the appreciation of riboregulators as important operators in the regulation of gene expression in M. tuberculosis.
Cis- and trans-, non-coding and regulatory RNAs
In the early days of the era of riboregulation, classifications were relatively simple. However, as we learn more about the origins and the functions of the different types of regulatory RNA, the boundaries between cis-regulatory elements, trans-acting RNA, and cis- and trans-encoded regulatory RNAs and have become increasingly blurred. In BOX 1 we offer a short list of definitions, bearing in mind that there may be overlaps between these.
Box1.
Non-coding RNA (ncRNA) is a transcript or part of a transcript that does not encode protein or peptide; it may or may not be regulatory. Conversely, there are several examples of regulatory RNAs that are coding.
Cis-regulatory elements are part of the RNAs they regulate, and they include RNA leaders (5’ of both coding and ncRNA) and 3’ untranslated regions (UTRs), both of which may be a source of sRNAs.
Trans-acting RNAs, as opposed to cis-regulatory elements, refer to RNAs that are not part of the transcripts they regulate; they include antisense RNA (asRNA) and small regulatory RNA ($RNA).
sRNAs comprise a small number of protein binding RNAs, and a large number of basepairing RNAs, which again are divided into cis- or trans-encoded.
asRNAs are encoded opposite their targets and sizes vary significantly.
Dual function sRNAs are transcripts that act both as riboregulators and as templates for protein synthesis.
RNA chaperones and ribonucleases
As riboregulators adopt more prominent roles, the proteins that regulate their expression, stability and/or degradation have attracted more attention as well. Two classes of proteins important for riboregulator function are the RNA chaperones and Ribonucleases (RNases).
RNA chaperones can have a profound influence on the effect that riboregulators can exert, both as molecular matchmakers, but also due to their role in modulating the stability of these regulators, reviewed in (Vogel & Luisi, 2011). For several years, the widely conserved RNA chaperone, Hfq was considered the facilitator of sRNA-mRNA interactions, but different species rely on Hfq to different extents. For example, several sRNAs in the otherwise highly Hfq-dependent S. typhimurium and E. coli do not bind Hfq; there are conflicting reports on the requirement for Hfq in Gram-positive bacteria, and certain species including Helicobacter pylori and M. tuberculosis do not encode an Hfq homologue at all (Jousselin et al., 2009, Arnvig & Young, 2012, Oliva et al., 2015).
Recently, the new global RNA chaperone ProQ was discovered by a combination of density gradient fractionation with mass spectrometry- (of proteins) and and RNA-seq analysis of each fraction (Grad-seq) (Smirnov et al., 2016). Similar to Hfq, there is no ProQ homologue in M. tuberculosis, but its identification does rekindle the question as to whether M. tuberculosis sRNAs require a matchmaker and if so, how does it work?
A third contender as a general RNA chaperone with a wide range of targets is Cold Shock Protein A (CspA), which belongs to a large family of Csp’s, and this protein does have a highly expressed homologue in M. tuberculosis (Arnvig et al., 2011, Caballero et al., 2018). CspA is an RNA binding protein that facilitates the melting of secondary RNA structures, which are otherwised stabilised at low temperatures (Jiang et al., 1997). Although well-expressed at all times, the E. coli cspA mRNA is itself regulated by a thermoswitch that leads to increased translation at low temperatures (Giuliodori et al., 2010). Whether M. tuberculosis CspA ia regulated in a similar manner remains to be seen, but certainly an RNA chaperone that specialises in melting highly structured RNA would seem appropriate for M. tuberculosis.
Degrading RNA
The abundance of a transcript is a carefully controlled balance between synthesis and degadation, and hence, ribonucleases (RNases) are crucial players in riboregulation.
In addition to encoding an unprecedented number of type II toxins (Ramage et al., 2009, Sala et al., 2014), M. tuberculosis (and other mycobacteria) encode a curious complement of RNases, with elements from both Gram negative (E. coli) and Gram positive (B. subtilis) species. A comprehensive review of the similarities and differences can be found in (Durand et al., 2015), and we will only mention a few important corner stones. Mycobacteria contain functional homologues of both RNase E and RNase J, while RNase E is absent from B. subtilis and RNase J is absent from E. coli (Even et al., 2005, Taverniti et al., 2011, Durand et al., 2015).
The endonucleolytic activity of E. coli RNase E as well as the exonucleolytic activity of B. subtilis RNase J are both sensitive to the phosphorylation state of the 5’ nucleotide of their substrates, i.e. they both have a strong preference for mono-phosphorylated transcripts as substrates, and the same ‘rules’ are likely to apply to the M. tuberculosis enzymes (Mackie, 1998, Koslover et al., 2008, Li de la Sierra-Gallay et al., 2008, Mathy et al., 2010, Taverniti et al., 2011). RNA 5’ monophosphates can be generated either by endonucleolytic cleavage of a transcript or by the removal of a pyrophosphate group from the 5’ nucleotide by the action of the Nudix hydrolase, RppH (RNA pyrophosphohydrolase) (Deana et al., 2008). So far, there are no reports of RppH homologues in mycobacteria, although there are some indications that such an enzyme exists (Moores et al., 2017). Regardless of the enzymes involved and the pathways employed, the degradation of RNA is as important as its synthesis for managing resources and ensuring appropriate execution of the cell’s gene expression programme.
RNA 5’ leaders
RNA leaders serve as hubs for post-transcriptional regulation, which in many cases involves some means of controlling ribosome entry. In its most basic form, the RNA leader is short and simply provides a Shine-Dalgarno (SD) sequence for binding of the 30S ribosomal subunit (Shine & Dalgarno, 1974). Translation efficiency can be modulated by changing the SD sequence to be more or less complimentary to the 16S sequence or by altering the spacing between the SD and the start codon (Vellanoweth & Rabinowitz, 1992). Certain (longer) RNA leaders have the ability to switch between mutually exclusive conformations that are either permissive or non-permissive for downstream gene expression (transcription or translation), and these are referred to as RNA switches. The switching between alternative RNA structures can be triggered by changes in temperature, pH, metal ions and metabolites, but also by RNA and proteins (Babitzke et al., 2009, Nechooshtan et al., 2009, Ferre-D’Amare & Winkler, 2011, Kortmann & Narberhaus, 2012, Sherwood & Henkin, 2016, McCown et al., 2017). Moreover, the leader is often targeted by sRNAs with a variety of outcomes.
Protein binding 5’ leaders
Many 5’ leaders regulate downstream gene expression by binding specific proteins reviewed in (Babitzke et al., 2009). This is particularly evident in the case of ribosome biosynthesis, which represents a major drain on cellular resources, and therefore has to be tightly regulated. The expression of many ribosomal protein (r-protein) operons is regulated by direct binding of one or more r-proteins encoded in these operons to their cognate mRNA leaders, usually proximal to the translation initiation region (TIR) typically blocking ribosome entry. This has been extensively characterised in E. coli and examples include r-proteins S4, S8, L20, and a complex of L10/L12 (Babitzke et al., 2009). The highly conserved organisation of many of these genes between E. coli and other bacteria including M. tuberculosis suggests that these feedback mechanisms are also conserved (Arnvig et al., 2011).
In parallel, the transcription of ribosomal RNA (rRNA), is likewise heavily regulated by the 5’ leader, which is removed by nucleolytic cleavage from the nascent RNA to generate the mature transcript (Deutscher, 2009). Studies in E. coli have elucidated how binding of the antitermination factor NusB and the r-protein S10 to the rRNA leader nucleates a conformational change in the transcription elongation complex that leads to an increase in elongation rate and processivity, i.e. antitermination e.g. (Greive et al., 2005). Similar mechanisms are likely to occur in M. tuberculosis, although a specific role for the NusB/E heterodimer has not been demonstrated. However, M. tuberculosis has contributed to the antitermination story via its NusA protein and one of the earliest investigations on mycobacterial regulatory RNA. The M. tuberculosis NusA lacks the C-terminal domain that masks part of the RNA-binding domain in its E. coli counterpart (Gopal et al., 2001). This in turn facilitated the identification of a highly specific interaction between the KH domains of NusA and the antitermination site of the M. tuberculosis rRNA leader, an interaction that was also shown to affect RNAP processivity (Arnvig et al., 2004, Beuth et al., 2005). It remains to be seen how NusA in this context changes from a pausing/termination factor to an antitermination factor.
Riboswitches, leaders sensing metabolites
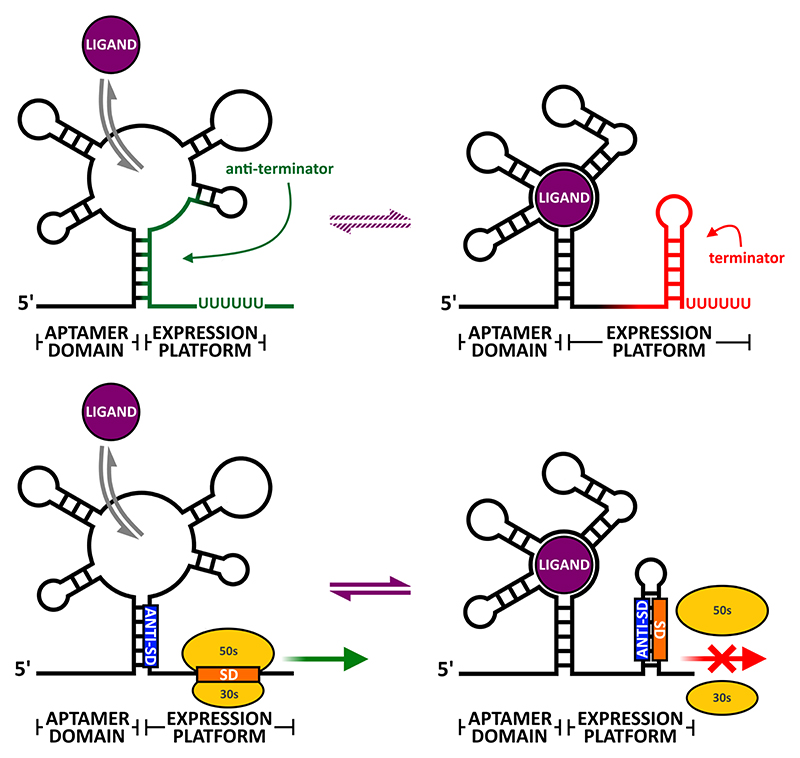
The term ‘riboswitch’ refers to a subset of RNA switches that sense changes in the concentration of metabolites or ions without the aid of accessory proteins, and these currently make up the largest class of RNA switches (Serganov & Nudler, 2013, Sherwood & Henkin, 2016, McCown et al., 2017). Riboswitch aptamers sense and interact with a variety of small molecules including nucleotides, amino acids and enzyme co-factors with high affinity and specificity, making them potentially ideal drug targets (Sherwood & Henkin, 2016, Dersch et al., 2017). In many cases these ligands are synthesized by enzymes encoded by the genes that are controlled by the riboswitches itself, thereby implementing feedback regulation (Nudler & Mironov, 2004). The expression platform executes the regulatory output triggered by the presence or absence of ligand binding. If ligand binding leads to reduced expression, the switch is classified as an ‘Off’ switch (Fig. 1); conversely, if ligand binding leads to increased expression, the switch is classified as an ‘On’ switch.
Fig. 1.
Riboswitch architecture. Top panel illustrates how a ligand induces transcriptional termination in a transcriptionally controlled ‘Off’ switch; panel below illustrates a translational ‘Off’ switch.
Similar to other leader-based regulators, many riboswitches function by blocking/unblocking ribosome entry. Whether the default (i.e. ligand free) conformation is ‘On’ or ‘Off’ depends on the individual switch, but in all cases ligand binding to the aptamer domain induces the alternative conformation within the expression platform.
Other riboswitches are based on intrinsic (i.e. factor-independent) termination/antitermination, which sets them apart from other leader-based modes of regulation as access to the SD is not involved. Moreover, while the two conformers of a translationally regulated riboswitch may exist in a dynamic equilibrium a transcriptionally regulated riboswitch cannot, since both transcription termination and readthrough are irreversible events. This adds additional kinetic requirements to transcriptional riboswitches, as the decision between one or the other conformer has to be made after transcribing the aptamer domain and sensing of a cognate ligand, but before reaching the end of the expression platform. This may require the RNAP to pause at specific and functionally critical positions within the riboswitch to allow for correct co-transcriptional folding of the RNA (Steinert et al., 2017).
A curious characteristic of some riboswitches is that conserved aptamer domains, recognising identical ligands, may be associated with different expression platforms in different species, and in some cases even within the same species. For example, the B. subtilis RFN element, which senses Flavin Mononucleotide (FMN) uses SD sequestration within the ypaA riboflavin transporter mRNA, but transcriptional termination in the ribDEAHT mRNA, encoding a series of FMN biosynthetic enzymes (Winkler et al., 2002). It is still unclear exactly where and when a translational expression platform is more or less advantageous than a transcriptional expression platform. While the latter requires ongoing RNA synthesis, the former could in theory regulate the translation of extant transcripts provided that these are relatively stable.
Riboswitches in M. tuberculosis
The vitamin B12-sensing riboswitch
Currently only a single metabolite-sensing riboswitch has been experimentally validated in M. tuberculosis, although several have been predicted by sequence homology and covariance analyses (Warner et al., 2007, Nawrocki et al., 2015). This is the cobalamine or B12-sensing riboswitch upstream of the metE gene, encoding a B12-independent methionine synthase. In the presence of B12, expression of the MetE enzyme, which catalyses the conversion of homocysteine to methionine, is repressed, making this an ‘Off’ switch. In M. tuberculosis H37Rv, this reaction is instead carried out by the B12-dependent isozyme, encoded by metH. This means that in the absence of B12, metE is required, while in the presence of B12, metH is required, due to riboswitch-mediated repression of metE. However, this gene has been partially disrupted in M. tuberculosis CDC1551 with the result that this strain of M. tuberculosis has a severe growth defect in the presence of vitamin B12 (Warner et al., 2007).
A second B12-sensing rioswitch is located in the 5’ leader of the PPE2-cobQ1-cobU operon. PPE2 (Rv0256c) belongs to the family of proteins sharing proline-proline-glutamate (PPE) N-terminal motifs that were identified in the M. tuberculosis genome sequence, many of which are found on the cell surface of M. tuberculosis. PPE2 was originally predicted to be a vitamin B12 transporter (Rodionov et al., 2003, Vitreschak et al., 2003). However, a more recent study demonstrated that Rv1819c, an ABC transporter is the ‘sole corrinoid transporter’ responsible for vitamin B12 uptake in M. tuberculosis under standard in vitro growth conditions (Gopinath et al., 2013). Expression of Rv1819c is not controlled by a B12 riboswitch, and the exact function of PPE2 remains obscure, although the presence of the riboswitch and the cobQ1-cobU genes does suggest a role in B12 uptake/metabolism.
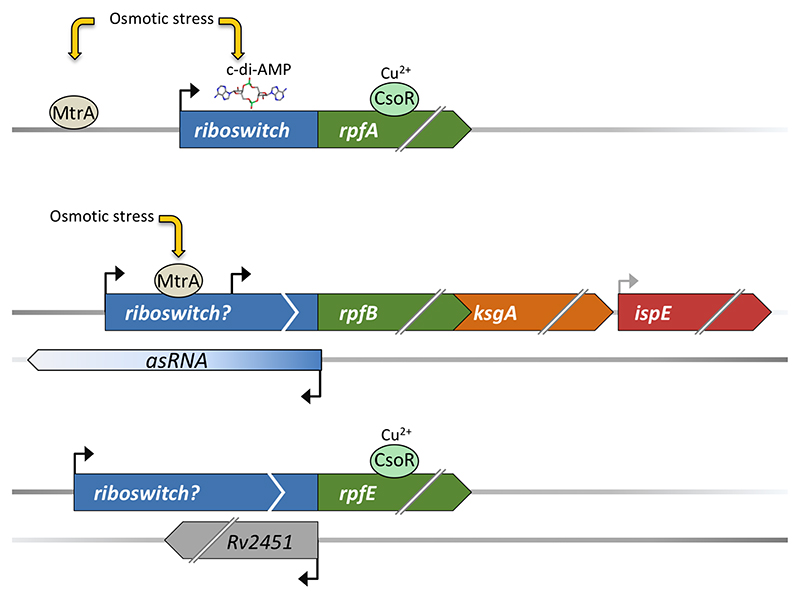
A Cyclic-di-AMP sensing riboswitch regulates rpfA expression
M. tuberculosis encodes five so-called resuscitation promoting factors (RpfA-E). These are cell wall remodelling enzymes critical for the transition between dormancy and resuscitation (Chao & Rubin, 2010, Kana & Mizrahi, 2010, Mukamolova et al., 2010, Turapov et al., 2014). Precise control of Rpf expression is vital as these enzymes are potentially lethal for M. tuberculosis itself, and multiple, at times shared signals converge in the control of rpf transcription (Fig. 2). The rpfA 5’ leader is 272 nucleotides in length and harbours a homologue of the ydaO aptamer domain (Block et al., 2010, Arnvig & Young, 2012). Identified almost a decade before its cognate ligand, cyclic di-AMP (c-di-AMP), the ydaO riboswitch regulates genes associated with cell wall metabolism and osmotic stress in a wide range of bacteria (Barrick et al., 2004, Nelson et al., 2013). The prolonged pursuit for the correct ligand illustrates the difficulty of identifying some riboswitch ligands, even after an element has been characterised. A similar element has been identified in the 5’ leader of Streptomyces coelicolor rpfA mRNA, where it has been shown to control expression of RpfA in a c-di-AMP-dependent manner (St-Onge et al., 2015, St-Onge & Elliot, 2017). Due to the close relationship between S. coelicolor and M. tuberculosis, we expect the M. tuberculosis ydaO homologue may also respond to c-di-AMP. Curiously, unlike the B. subtilis element, there are no apparent intrinsic terminators, i.e. a stable stem-loop followed by a poly-U tail, associated with neither the Streptomyces nor the M. tuberculosis riboswitch, suggesting a different expression platform (Nelson et al., 2013, St-Onge & Elliot, 2017) (J. Green and G. Mukamolova personal communication).
Fig. 2.
Control of rpf expression in M. tuberculosis. The figure illustrates how different, sometimes shared, transcriptional regulators contribute to rpf regulation in addition to long 5’ leaders, which in the case of rpfA harbours a riboswitch with a known ligand (c-d-AMP), in rpfB a riboswitch candidate, with unknown ligand and in rpfE, a so far entirely uncharacterised element.
It remains to be seen how this element affects M. tuberculosis pathogenesis, but adaptation to changing osmolarity does play an important role in M. tuberculosis’s lifestyle as well as in phenotypic drug tolerance (Larrouy-Maumus et al., 2016). Moreover, while the rpfA CDS is highly polymorphic in M. bovis (Amadio et al., 2005), the ydao element is 100% conserved between M. tuberculosis and Mycobacterium bovis, suggesting an important role for this riboswitch.
A novel riboswitch candidate regulating expression of rpfB
Remarkably, three of the five rpf mRNAs (encoding RpfA, B and E), have extensive 5’ leaders of more than 100 nucleotides in length (Arnvig et al., 2011, Cortes et al., 2013), suggestive of post-transcriptional regulation; the Rpfs encoded by the same three genes are critical players for Rpf-mediated phenomena such as resuscitation of dormant mycobacteria, growth on solid medium and resistance to detergents (Kana & Mizrahi, 2010).
An RNA switch without a known ligand may be considered a riboswitch candidate (Meyer et al., 2011). Similar to rpfA, the 176-nucleotide 5’ leader of the rpfB mRNA harbours an RNA switch (or riboswitch candidate), and like rpfA, identification of the rpfB element precedes identification of its ligand. Unlike ydaO however, the rpfB switch has a recognisable intrinsic terminator structure, and also unlike ydaO, the rpfB switch appears to be restricted to a small subset of pathogenic mycobacteria (Schwenk et al., 2018). By extensive genetic and biochemical analysis, this switch has been shown to control rpfB transcription via an intrinsic terminator located immediately upstream of the TTG start codon, which was experimentally re-annotated in the same study. The rpfB switch regulates a tri-cistronic operon, which also encodes the methyltransferase KsgA, crucial for ribosome biogenesis and IspE, essential for early steps in M. tuberculosis cell wall synthesis (Connolly et al., 2008, Schwenk et al., 2018).
This arrangement provides an intriguing, regulatory link between riboswitch co-ordinated resuscitation from dormancy, ribosome maturation and cell wall synthesis. Moreover, as the operon represents two classical drug targets, i.e. cell wall synthesis and ribosome function under one regulatory roof, it is tempting to speculate that this riboswitch candidate may represent a new target for anti-tuberculosis drug development. Identification of the cognate ligand will undoubtedly provide novel insights into coordinated regulation of macromolecular synthesis as well as post-transcriptional regulation of gene expression in M. tuberculosis.
A potential RNA switch regulating rpfE
Little is known about the regulation of rpfE expression other than it is induced by chloride, and it is not yet clear if this effect is transcriptional or post-transcriptional (Tan et al., 2013). TSS mapping indicates that the rpfE 5’ leader is at least 251 nucleotides in length, and overlaps the divergently transcribed Rv2451 (of unknown function). Similar to the rpfB leader, the rpfE leader harbours the potential to form a stem-loop followed by a poly-U tail close to the TIR. However, the rpfE poly-U tail is short with only three uridine residues, which may be insufficient to confer intrinsic termination without the support of additional factors/ligands.
Across bacterial species it is clear that only a fraction of riboswitches has been identified to date, and rare (i.e. not broadly conserved) riboswitches are unlikely to be identified by genome alignments. Novel, more experimental approaches are required to tackle this conservation bias. One such approach is Term-seq, which provides a genome-wide display of RNA 3’ ends facilitating the identification of conditional terminators and potential novel riboswitches (Dar et al., 2016). Finally, it is worth mentioning in this context that transcriptionally terminated riboswitches can act in trans as sRNAs, thus blurring the boundaries cis-regulatory elements and trans-acting RNA (Loh et al., 2009).
Trans-acting RNAs
In contrast to cis-regulatory elements, asRNAs and sRNAs are not part of the transcript they regulate and may therefore be considered ‘trans-acting’ (Lease & Belfort, 2000, Loh et al., 2009). This class of transcripts include a small number of protein binding RNAs, and a very large number of basepairing RNAs, which again are divided into cis- or trans-encoded. Due to space constraints we will focus on basepairing RNAs in this review.
Cis-encoded RNAs are transcripts encoded opposite their target mRNAs (i.e.’true’ asRNAs). These transcripts have perfect complementarity to their mRNA targets, suggesting that the resulting hybrids are ideal RNase III substrates. asRNAs can be of varying sizes from <100 nucleotides to several kb, and they are likely to have different modes of action depending on their size and location.
Many, smaller asRNAs are encoded opposite the TIR of their mRNA targets, where they function in a manner similar to trans-encoded sRNAs, by blocking ribosome entry and translation. An important class of such small asRNAs are those associated with type I toxin-antitoxin (TA) systems (Brantl & Jahn, 2015). Curiously however, while there is an abundance of Type II/protein based TA systems in M. tuberculosis, so far no Type I systems have been identified.
Longer asRNAs can be several hundred nucleotides long and in a few cases even several kb (Arnvig et al., 2011, Lasa et al., 2011, Sesto et al., 2013). An example is the asRNA covering Rv2817-2816c, encoding Cas1 and Cas2, respectively in the M. tuberculosis CRISPR locus, and while this transcript is relatively abundant, there is very little expression of the coding strand under standard in vitro growth conditions, suggesting an inverse correlation in abundance between sense and antisense (Arnvig et al., 2011). The function of these asRNAs is still debated, but pervasive antisense transcription may suppress sense transcriptional noise via transcriptional (RNAP) interferences and/or RNase III mediated cleavage of hybridised sense-antisense transcripts (Lasa et al., 2011).
The 5’ leaders of divergently transcribed genes or 3’ UTRs of convergently transcribed genes, can also act as asRNA on mRNAs transcribed from the opposing strand, once more blurring the boundaries between cis-regulatory elements and trans-acting RNA. This phenomenon was first observed in L. monocytogenes (Toledo-Arana et al., 2009) where it has since led to the to the ‘Excludon’ concept, coined by Pascale Cossart’s group. The excludon specifically refers to ‘an unusually long asRNA that spans divergent genes or operons with related or opposing functions (Sesto et al., 2013).
In M. tuberculosis, converging 3’ UTRs make a significant contribution to the overall antisense transcriptome, and these show a striking enrichment of genes associated with cell wall functions (Arnvig et al., 2011, Cortes et al., 2013). Future studies on gene function and expression should reveal if an excludon mechanism is employed in M. tuberculosis.
Finally, some cis-encoded sRNAs also have the potential to act as trans-encoded sRNAs on mRNA targets with similar sequences as the primary targets e.g. (Arnvig & Young, 2009, Jager et al., 2012).
Trans-encoded sRNAs are encoded in different genomic locations to their targets. The majority of these transcripts are induced by stress and therefore often associated with pathogen adaptation to hostile host environments. In the early days of sRNA identification, searches for sRNAs focused on intergenic regions, and hence this class of regulators were perceived to originate primarily from disinct promoters within these regions, e.g. (Argaman et al., 2001, Wassarman et al., 2001, Arnvig & Young, 2009, Dichiara et al., 2010). However, with the accumulation of data from RNA-seq based methods, it has become evident that many sRNAs are in fact derived from mRNAs. As already mentioned, transcriptionally attenuated leaders can act as sRNAs (Loh et al., 2009); and mRNA 3’ UTRs are avid sRNA generators either from processing or from internal promoters (Chao et al., 2012, Chao et al., 2017).
Mode of action
Unlike the interaction between cis-encoded (as)RNAs and their targets, the interaction between trans-encoded sRNAs and their targets proceeds via limited basepairing apart from a short ‘seed sequence’, which means that in many cases, trans-encoded sRNAs depend on an RNA chaperone to facilitate the interaction with their targets (Vogel & Luisi, 2011). In addtion to the seed sequence, most sRNAs contain another characteristic feature, which is an intrinsic terminator critical for the interaction with the RNA chaperone Hfq (Otaka et al., 2011, Morita et al., 2017). The limited complementarity also means that prediction of targets can be challenging, and several algorithms have been developed to facilitate this, e.g. TargetRNA2, (Kery et al., 2014) and CopraRNA (Wright et al., 2013). Moreover, a number of experimental approaches have been developed, e.g. RIL-seq, which exploits the proximity of sRNAs to mRNA targets on Hfq (Melamed et al., 2016) or MAPS (pull-downs with MS2-tagged sRNAs), which does not require a protein (Lalaouna et al., 2017). Both predictive and experimental approaches require further validation, in particular in an organism such as M. tuberculosis, where little remains known about sRNA targets. Individual sRNAs can both repress and increase expression of genes in their regulons, depending on the location of the target region.
Repressing interactions
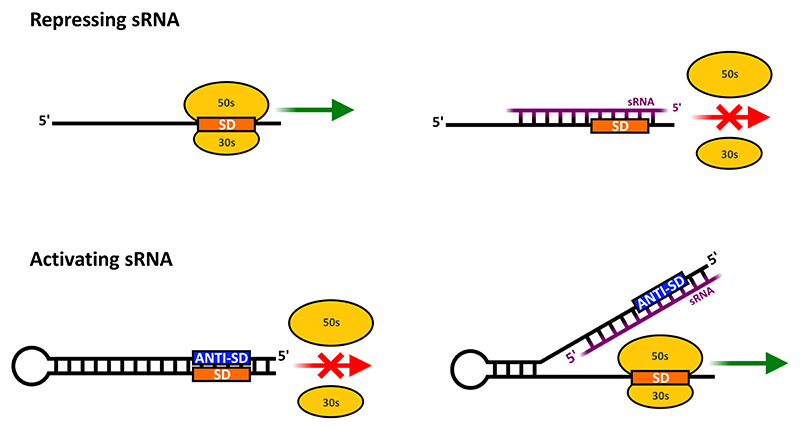
The most commonly known mode of action for trans-encoded sRNAs is repression of translation by blocking the TIR, often followed by mRNA degradation (Fig. 3), reviewed in (Wagner & Romby, 2015).
Fig. 3.
Basic sRNA modes of action. Top half illustrates how an sRNA (cis- or trans-encoded) can block ribosome entry and translation. Bottom panel illustrates how an sRNA can activate translation by an anti-antisense mechanism; in this situation the mRNA leader itself blocks translation, by masking the TIR, but an sRNA can interact with the leader to unmask the TIR.
If the TIR is located early within a multi-cistronic operon, this block may also lead to Rho-dependent termination of transcription further downstream (i.e. polarity), (Bossi et al., 2012). The interaction can also take place downstream of the TIR, several codons into the coding region of the mRNA (Pfeiffer et al., 2009). This may be a means of regulating the many leaderless transcripts in M. tuberculosis (Cortes et al., 2013).
Activating interactions
sRNA-mRNA interaction can also lead to increased translation either by direct stabilisation of the mRNA, by unmasking of the TIR and/or by interfering with Rho-dependent termination, reviewed in (Papenfort & Vanderpool, 2015).
An example of direct stabilisation has been observed in Salmonella, where the RydC sRNA blocks an RNase E cleavage site in the cfa1 mRNA. This interaction leads to stabilisation of the mRNA even in the absence of translation (Frohlich et al., 2013).
A somewhat more sophisticated means of activation involves a so-called ‘anti-antisense’ mechanism (Majdalani et al., 1998). In this situation, the leader of the target mRNA contains an auto-inhibitory secondary structure that masks the TIR, and which can be unmasked sRNA binding. A well-characterised example is the E. coli rpoS mRNA, which encodes the stationary phase sigma factor, Sigma38 (Battesti et al., 2011). The rpoS mRNA harbours a 567-nucleotide 5’ leader, which blocks its own TIR (Majdalani et al., 1998, Peng et al., 2014). Upon binding of one of three sRNAs (DsrA, RprA, ArcZ) to the inhibitory region, the SD sequence and start codon are unmasked via the anti-antisense mechanism to permit translation (Battesti et al., 2011).
Recently, it was shown that the same three sRNAs in addition to unmasking the rpoS mRNA TIR, could also inhibit Rho-dependent termination of rpoS transcription in E. coli by masking one or more Rho binding sites in the rpoS leader, thus making the sRNA activating effect two-pronged. The authors argued that this novel sRNA-regulated antitermination is likely to be widespread in long leaders (Sedlyarova et al., 2016).
To summarise, sRNAs can both repress or promote translation initiation, and repress or promote Rho-dependent termination of transcription. Moreover, the effect of an sRNA can be greatly enhanced if the mRNA target encodes a regulator such as a sigma or a transcription factor. An overview of different regulatory networks, and their evolution can be found in (Beisel & Storz, 2010, Peer & Margalit, 2014).
M. tuberculosis sRNAs
In spite of several M. tuberculosis sRNAs being identified and mapped, and their expression patterns investigated e.g. (Arnvig & Young, 2009, Dichiara et al., 2010, Arnvig et al., 2011, Miotto et al., 2012), only few, including MTS2823, ncRv12659, DrrS and Mcr7, have been functionally characterised in any greater detail (Arnvig et al., 2011, Houghton et al., 2013, Solans et al., 2014, Moores et al., 2017). Like their counterparts in model organisms, M. tuberculosis sRNAs are often stress induced and some are highly abundant during infection. The evolutionary conservation of M. tuberculosis small RNAs is subject to considerable variation. Some sRNAs, such as ncRv12659 are specific for a subset of M. tuberculosis strains (Houghton et al., 2013), some are found throughout species of the M. tuberculosis complex, some a little further afield including non-tuberculous, pathogenic mycobacteria and a few M. tuberculosis sRNAs are conserved in Mycobacterium smegmatis and other Actinomycetes e.g. (Arnvig & Young, 2009, Dichiara et al., 2010, Haning et al., 2014). Many M. tuberculosis sRNAs are highly structured, in part due to the high GC content of the bacterium. Furthermore, by comparing results from 5’ and 3’ RACE, RNA-seq, northern blotting and RNA structure prediction, it is evident that many M. tuberculosis sRNAs do not contain conventional intrinsic terminator structures. For some time this lack of conventional terminators was attributed to the presence of so-called I-shaped terminators, i.e. stem-loop structures without a poly-U tail (Mitra et al., 2008). However, more recently, RNA-seq and in vitro transcription experiments using M. bovis RNA polymerase, have demonstrated that in most cases this type of structure is not sufficient for termination of transcription in vivo or in vitro (Arnvig et al., 2011, Czyz et al., 2014). This in turn suggests that many sRNA 3’ termini may be generated by processing in M. tuberculosis, setting them apart from the well-known Hfq-dependent sRNAs that require a poly-U tail to function (Otaka et al., 2011). The predicted processing also suggests that some sRNAs may exist as different isoforms, as is the case for the DosR regulated sRNA, DrrS (Moores et al., 2017).
The 108-nucleotide DrrS was first identified by RNA-seq and shown to accumulate to high levels during chronic mouse infection (Arnvig et al., 2011). Recently it was shown that DrrS expression is induced by DosR, but it is a combination of DosR-dependent induction and the unrivalled stability of DrrS that determines the overall levels (Moores et al., 2017).
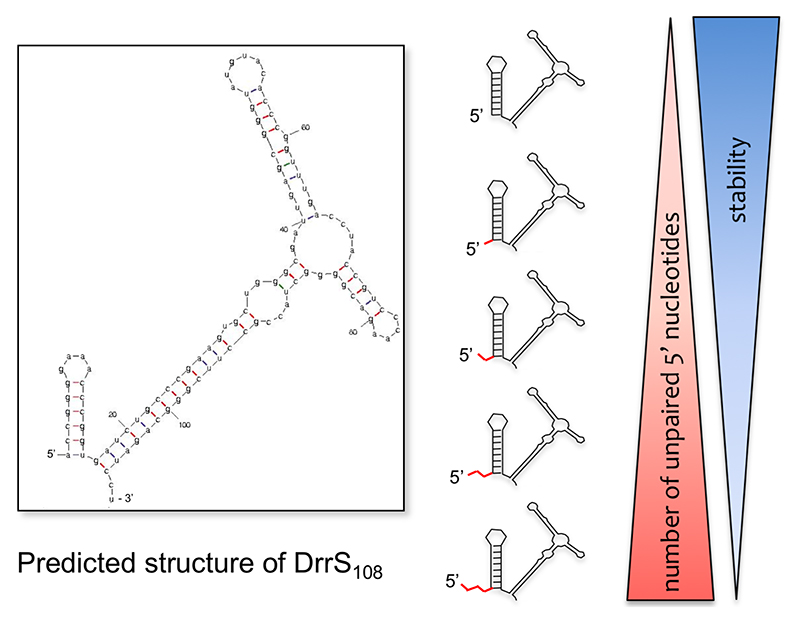
DrrS has a half-life in the order of several hours due to a stable stem-loop structure at its 5’ end. The addition of two or more unpaired nucleotides 5’ of this stem-loop, reduces stability significantly, suggesting the involvement of a mycobacterial RppH homologue (Fig. 4). Moreover, this structure increases expression of a lacZ reporter when added to the 5’ end of its mRNA, suggesting that it represents a general stabilising feature (Moores et al., 2017). In addition to elucidating how RNA stabiity may be modulated in M. tuberculosis, DrrS provides insights into sRNA processing. DrrS is transcribed as a longer (>300 nucleotide) precursor, (DrrS+) that is rapidly (in M. tuberculosis terms) processed to the shorter, stable 108-nucleotide sRNA (DrrS108).
Fig. 4.
Stability of DrrS. Large image shows the predicted structure of DrrS108, while the schematic representation illustrates how the number of unpaired nucleotides 5’ are inversely correlated to transcript stability.
While DrrS+ levels peak in early stationary phase, DrrS108 accumulates continuously for at least three weeks into stationary phase (Moores et al., 2017). The substantial difference in size and maximum expression between DrrS and DrrS+ implies that the longer isoform may play a different role than the the shorter isoform. Apart from shedding light on RNA processing and stability, the DrrS example also highlights the importance of thoroughly characterising multiple aspects of an sRNA before defining its regulon. The application of Term-seq to define M. tuberculosis 3’ ends on a global scale (Dar et al., 2016), is likely to be hugely informative at this stage.
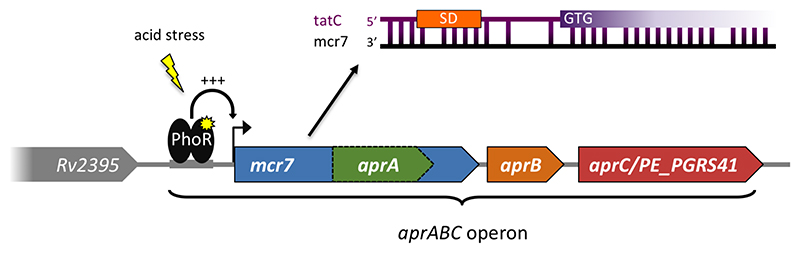
The best characterised M. tuberculosis sRNA in terms of biological role is Mcr7. This sRNA was first identified as a 350-400 nucleotide transcript by cloning and sequencing of M. bovis BCG cDNA, and in the same study predicted by sequence homology to be conserved throughout the M. tuberculosis complex (Dichiara et al., 2010). RNA-seq later confirmed high expression in M. tuberculosis H37Rv (Arnvig et al., 2011). Mcr7 is encoded downstream of Rv2395 and according to TSS mapping, a single promoter drives transcription in the region downstream of Rv2395 and into PE_PGRS41 (Cortes et al., 2013), suggesting that Mcr7 is (part of) the 5’ leader of the latter. However, there is more to this locus than a PE_PGRS protein with a long 5’ leader.
In 2011 David Russell’s group reported the characterisation of the PhoPR-dependent aprABC (Acid and Phagosome Regulated) locus encoding the conserved hypothetical proteins, AprA and AprB, as well as PE_PGRS41 (AprC) (Abramovitch et al., 2011). The aprA coding region lies entirely within the boundaries of Mcr7 (62 basepairs downstream of the annotated TSS), with AprB and AprC encoded downstream of Mcr7 (Fig.5).
Fig. 5.
The mcr7/aprABC locus in M. tuberculosis. The figure illustrates the elements associated with the PhoP/R regulated operon with the ncRNA Mcr7, which contains an open reading frame encoding the acid inducible AprA, and the proposed interaction between Mcr7 and the tatC mRNA.
The proteins have not yet been experimentally validated in M. tuberculosis, but aprA does have a likely ribosome binding site upstream of its start codon. Moreover, the recombinant protein has been expressed and purified in E. coli, suggesting this is a bona fide, stable protein (Abramovitch et al., 2011). Was this then an indication that Mcr7 had been wrongly annotated as an sRNA?
In 2014 the mcr7/aprABC promoter was identified as one of the major targets of PhoR (Solans et al., 2014). Based on this finding and the assumption that Mcr7 was a post-transcriptional regulator of gene expression, the authors used in silico prediction to identify putative targets of Mcr7, one of which was the tatC mRNA. More specifically positions -16 to +19 relative to the annotated GTG start of the tatC mRNA are targeted by the central portion (nucleotides 119 to 151) of Mcr7, i.e. well within the coding region of aprA. The prediction suggests that PhoP/R dependent expression of Mcr7 represses the translation of TatC resulting in reduced secretion of TAT-dependent proteins, which was supported by proteomics on culture supernatants on M. tuberculosis wildtype and phoP mutant. This study therefore strongly supports the notion of Mcr7 being an sRNA that represses translation of TatC, thereby changing the secretome and modifying the host-pathogen interface (Solans et al., 2014).
So, although AprA has not yet been identified in M. tuberculosis and a direct interaction between Mcr7 and tatC mRNA has not been experimentally validated, it appears that this sRNA is a prime candidate for a dual function sRNA in M. tuberculosis. As there are no additional TSS in this operon, it also suggests that the 5’ end of Mcr7 may regulate aprA expression via an as yet uncharacterised post-transcriptional mechanism. If all these elements really represents their annotated functions, this operon represents a complex arrangement of a 5’ leader that acts as a trans-encoded dual function sRNA.
Concluding remarks
Pathogen survival depends on constant monitoring of, and adaptation to, a range of host environments, an adaptation that sometimes requires rapid and drastic changes in gene expression. This is most efficiently achieved by multi-pronged approaches combining several layers of control, such as transcriptional, post-transcriptional and post-translational regulation. A comprehensive insight into all of these mechanisms is necessary to fully understand how a pathogen interacts with its host, and more importantly, how we might exploit this to our own advantage. Whether the aim is drug discovery or vaccine development, a thorough understanding of the basic molecular mechanisms of the pathogen in question is fundamental.
In this review we have illustrated (i) how riboregulators work, (ii) argued why riboregulation should be considered by the M. tuberculosis community, and (iii) why M. tuberculosis should be considered by the RNA community. Although some general rules may apply, riboregulation is still full of surprises, and M. tuberculosis is different; with its high GC content (>65%), abundance of leader-less mRNA, distinct complement of RNases and lack of Hfq and ProQ chaperones. In summary, M. tuberculosis has the potential to greatly advance our knowledge of RNA based control of gene expression.
Acknowledgements
We thank Galina Mukamolova and Finn Werner for critical reading of the manuscript.
Funding
This work was supported by The Medical Research Council New Investigator Award for KBA [grant number MR/L018519/1].
References
- Abramovitch RB, Rohde KH, Hsu FF, Russell DG. aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Molecular microbiology. 2011;80:678–694. doi: 10.1111/j.1365-2958.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadio A, Romano MI, Bigi F, Etchechoury I, Kubica T, Niemann S, Cataldi A, Caimi K. Identification and characterization of genomic variations between Mycobacterium bovis and M. tuberculosis H37Rv. Journal of clinical microbiology. 2005;43:2481–2484. doi: 10.1128/JCM.43.5.2481-2484.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Arnvig KB, Young DB. Identification of small RNAs in Mycobacterium tuberculosis. Molecular microbiology. 2009;73:397–408. doi: 10.1111/j.1365-2958.2009.06777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnvig KB, Young DB. Regulation of pathogen metabolism by small RNA. Drug Discov Today Dis Mech. 2010;7:e19–e24. [Google Scholar]
- Arnvig KB, Young DB. Regulatory RNAs in Mycobacterium tuberculosis. RNA biology. 2012 [Google Scholar]
- Arnvig KB, Pennell S, Gopal B, Colston MJ. A high-affinity interaction between NusA and the rrn nut site in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8325–8330. doi: 10.1073/pnas.0401287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnvig KB, Comas I, Thomson NR, et al. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002342. doi: 10.1371/journal.ppat.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Baker CS, Romeo T. Regulation of translation initiation by RNA binding proteins. Annual review of microbiology. 2009;63:27–44. doi: 10.1146/annurev.micro.091208.073514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annual review of microbiology. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS microbiology reviews. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 2005;24:3576–3587. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block KF, Hammond MC, Breaker RR. Evidence for widespread gene control function by the ydaO riboswitch candidate. Journal of bacteriology. 2010;192:3983–3989. doi: 10.1128/JB.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L, Schwartz A, Guillemardet B, Boudvillain M, Figueroa-Bossi N. A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes & development. 2012;26:1864–1873. doi: 10.1101/gad.195412.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S, Jahn N. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS microbiology reviews. 2015;39:413–427. doi: 10.1093/femsre/fuv003. [DOI] [PubMed] [Google Scholar]
- Caballero CJ, Menendez-Gil P, Catalan-Moreno A, et al. The regulon of the RNA chaperone CspA and its auto-regulation in Staphylococcus aureus. Nucleic acids research. 2018 doi: 10.1093/nar/gkx1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MC, Rubin EJ. Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annual review of microbiology. 2010;64:293–311. doi: 10.1146/annurev.micro.112408.134043. [DOI] [PubMed] [Google Scholar]
- Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3’ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Li L, Girodat D, et al. In Vivo Cleavage Map Illuminates the Central Role of RNase E in Coding and Non-coding RNA Pathways. Molecular cell. 2017;65:39–51. doi: 10.1016/j.molcel.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, Aebersold R, Young DB. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013;5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyz A, Mooney RA, Iaconi A, Landick R. Mycobacterial RNA polymerase requires a U-tract at intrinsic terminators and is aided by NusG at suboptimal terminators. mBio. 2014;5:e00931. doi: 10.1128/mBio.00931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar D, Shamir M, Mellin JR, Koutero M, Stern-Ginossar N, Cossart P, Sorek R. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science. 2016;352:aad9822. doi: 10.1126/science.aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5’ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- Dersch P, Khan MA, Muhlen S, Gorke B. Roles of Regulatory RNAs for Antibiotic Resistance in Bacteria and Their Potential Value as Novel Drug Targets. Frontiers in microbiology. 2017;8:803. doi: 10.3389/fmicb.2017.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci. 2009;85:369–391. doi: 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- Dichiara JM, Contreras-Martinez LM, Livny J, Smith D, McDonough KA, Belfort M. Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis. Nucleic acids research. 2010 doi: 10.1093/nar/gkq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Tomasini A, Braun F, Condon C, Romby P. sRNA and mRNA turnover in Gram-positive bacteria. FEMS microbiology reviews. 2015;39:316–330. doi: 10.1093/femsre/fuv007. [DOI] [PubMed] [Google Scholar]
- Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic acids research. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D’Amare AR, Winkler WC. The roles of metal ions in regulation by riboswitches. Met Ions Life Sci. 2011;9:141–173. doi: 10.1039/9781849732512-00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich KS, Papenfort K, Fekete A, Vogel J. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J. 2013;32:2963–2979. doi: 10.1038/emboj.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliodori AM, Di Pietro F, Marzi S, Masquida B, Wagner R, Romby P, Gualerzi CO, Pon CL. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Molecular cell. 2010;37:21–33. doi: 10.1016/j.molcel.2009.11.033. [DOI] [PubMed] [Google Scholar]
- Gopal B, Haire LF, Gamblin SJ, Dodson EJ, Lane AN, Papavinasasundaram KG, Colston MJ, Dodson G. Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 A resolution. Journal of molecular biology. 2001;314:1087–1095. doi: 10.1006/jmbi.2000.5144. [DOI] [PubMed] [Google Scholar]
- Gopinath K, Venclovas C, Ioerger TR, Sacchettini JC, McKinney JD, Mizrahi V, Warner DF. A vitamin B12 transporter in Mycobacterium tuberculosis. Open biology. 2013;3:120175. doi: 10.1098/rsob.120175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greive SJ, Lins AF, von Hippel PH. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. The Journal of biological chemistry. 2005;280:36397–36408. doi: 10.1074/jbc.M507146200. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haning K, Cho SH, Contreras LM. Small RNAs in mycobacteria: an unfolding story. Front Cell Infect Microbiol. 2014;4:96. doi: 10.3389/fcimb.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E, Wagner EGH. Impact of bacterial sRNAs in stress responses. Biochemical Society transactions. 2017;45:1203–1212. doi: 10.1042/BST20160363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton J, Cortes T, Schubert OT, Rose G, Rodgers A, De Ste Croix M, Aebersold R, Young DB, Arnvig KB. A small RNA encoded in the Rv2660c locus of Mycobacterium tuberculosis is induced during starvation and infection. PloS one. 2013;8:e80047. doi: 10.1371/journal.pone.0080047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS microbiology reviews. 2010;34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- Jager D, Pernitzsch SR, Richter AS, Backofen R, Sharma CM, Schmitz RA. An archaeal sRNA targeting cis- and trans-encoded mRNAs via two distinct domains. Nucleic acids research. 2012;40:10964–10979. doi: 10.1093/nar/gks847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. The Journal of biological chemistry. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- Jousselin A, Metzinger L, Felden B. On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends in microbiology. 2009;17:399–405. doi: 10.1016/j.tim.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Kana BD, Mizrahi V. Resuscitation-promoting factors as lytic enzymes for bacterial growth and signaling. FEMS immunology and medical microbiology. 2010;58:39–50. doi: 10.1111/j.1574-695X.2009.00606.x. [DOI] [PubMed] [Google Scholar]
- Kery MB, Feldman M, Livny J, Tjaden B. TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic acids research. 2014;42:W124–129. doi: 10.1093/nar/gku317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J, Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nature reviews Microbiology. 2012;10:255–265. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- Koslover DJ, Callaghan AJ, Marcaida MJ, Garman EF, Martick M, Scott WG, Luisi BF. The crystal structure of the Escherichia coli RNase E apoprotein and a mechanism for RNA degradation. Structure. 2008;16:1238–1244. doi: 10.1016/j.str.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalaouna D, Prevost K, Eyraud A, Masse E. Identification of unknown RNA partners using MAPS. Methods. 2017;117:28–34. doi: 10.1016/j.ymeth.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Larrouy-Maumus G, Marino LB, Madduri AV, Ragan TJ, Hunt DM, Bassano L, Gutierrez MG, Moody DB, Pavan FR, de Carvalho LP. Cell-Envelope Remodeling as a Determinant of Phenotypic Antibacterial Tolerance in Mycobacterium tuberculosis. ACS Infect Dis. 2016;2:352–360. doi: 10.1021/acsinfecdis.5b00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I, Toledo-Arana A, Dobin A, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1113521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease RA, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Gottesman S. sRNA roles in regulating transcriptional regulators: Lrp and SoxS regulation by sRNAs. Nucleic acids research. 2016;44:6907–6923. doi: 10.1093/nar/gkw358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine E, Hwa T. Small RNAs establish gene expression thresholds. Current opinion in microbiology. 2008;11:574–579. doi: 10.1016/j.mib.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase J. Nat Struct Mol Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Mackie GA. Ribonuclease E is a 5’-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathy N, Hebert A, Mervelet P, Benard L, Dorleans A, Li de la Sierra-Gallay I, Noirot P, Putzer H, Condon C. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Molecular microbiology. 2010;75:489–498. doi: 10.1111/j.1365-2958.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. Riboswitch Diversity and Distribution. RNA. 2017 doi: 10.1261/rna.061234.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed S, Peer A, Faigenbaum-Romm R, Gatt YE, Reiss N, Bar A, Altuvia Y, Argaman L, Margalit H. Global Mapping of Small RNA-Target Interactions in Bacteria. Molecular cell. 2016;63:884–897. doi: 10.1016/j.molcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MM, Hammond MC, Salinas Y, Roth A, Sudarsan N, Breaker RR. Challenges of ligand identification for riboswitch candidates. RNA biology. 2011;8:5–10. doi: 10.4161/rna.8.1.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto P, Forti F, Ambrosi A, Pellin D, Veiga DF, Balazsi G, Gennaro ML, Di Serio C, Ghisotti D, Cirillo DM. Genome-wide discovery of small RNAs in Mycobacterium tuberculosis. PloS one. 2012;7:e51950. doi: 10.1371/journal.pone.0051950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- Mitra A, Angamuthu K, Nagaraja V. Genome-wide analysis of the intrinsic terminators of transcription across the genus Mycobacterium. Tuberculosis (Edinb) 2008;88:566–575. doi: 10.1016/j.tube.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Moores A, Riesco AB, Schwenk S, Arnvig KB. Expression, maturation and turnover of DrrS, an unusually stable, DosR regulated small RNA in Mycobacterium tuberculosis. PloS one. 2017;12:e0174079. doi: 10.1371/journal.pone.0174079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Nishino R, Aiba H. Role of the terminator hairpin in the biogenesis of functional Hfq-binding sRNAs. RNA. 2017;23:1419–1431. doi: 10.1261/rna.060756.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. Resuscitation-promoting factors reveal an occult population of tubercle Bacilli in Sputum. Am J Respir Crit Care Med. 2010;181:174–180. doi: 10.1164/rccm.200905-0661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chemistry & biology. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Burge SW, Bateman A, et al. Rfam 12.0: updates to the RNA families database. Nucleic acids research. 2015;43:D130–137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes & development. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nature chemical biology. 2013 doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Oliva G, Sahr T, Buchrieser C. Small RNAs, 5’ UTR elements and RNA-binding proteins in intracellular bacteria: impact on metabolism and virulence. FEMS microbiology reviews. 2015;39:331–349. doi: 10.1093/femsre/fuv022. [DOI] [PubMed] [Google Scholar]
- Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13059–13064. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Vanderpool CK. Target activation by regulatory RNAs in bacteria. FEMS microbiology reviews. 2015;39:362–378. doi: 10.1093/femsre/fuv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer A, Margalit H. Evolutionary patterns of Escherichia coli small RNAs and their regulatory interactions. RNA. 2014;20:994–1003. doi: 10.1261/rna.043133.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Soper TJ, Woodson SA. Positional effects of AAN motifs in rpoS regulation by sRNAs and Hfq. Journal of molecular biology. 2014;426:275–285. doi: 10.1016/j.jmb.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- Quereda JJ, Cossart P. Regulating Bacterial Virulence with RNA. Annual review of microbiology. 2017;71:263–280. doi: 10.1146/annurev-micro-030117-020335. [DOI] [PubMed] [Google Scholar]
- Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS genetics. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau MH, Bojanovic K, Nielsen AT, Long KS. Differential expression of small RNAs under chemical stress and fed-batch fermentation in E. coli. BMC genomics. 2015;16:1051. doi: 10.1186/s12864-015-2231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. The Journal of biological chemistry. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- Rose G, Cortes T, Comas I, Coscolla M, Gagneux S, Young DB. Mapping of genotype-phenotype diversity amongst clinical isolates of Mycobacterium tuberculosis by sequence-based transcriptional profiling. Genome Biology and Evolution. 2013 doi: 10.1093/gbe/evt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala A, Bordes P, Genevaux P. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 2014;6:1002–1020. doi: 10.3390/toxins6031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk S, Moores A, Nobeli I, McHugh TD, Arnvig KB. Nucleic acids research. 2018. Cell-wall synthesis and ribosome maturation are co-regulated by an RNA switch in Mycobacterium tuberculosis. press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlyarova N, Shamovsky I, Bharati BK, Epshtein V, Chen J, Gottesman S, Schroeder R, Nudler E. sRNA-Mediated Control of Transcription Termination in E. coli. Cell. 2016;167:111–121.:e113. doi: 10.1016/j.cell.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P. The excludon: a new concept in bacterial antisense RNA-mediated gene regulation. Nature reviews Microbiology. 2013;11:75–82. doi: 10.1038/nrmicro2934. [DOI] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Sherwood AV, Henkin TM. Riboswitch-Mediated Gene Regulation: Novel RNA Architectures Dictate Gene Expression Responses. Annual review of microbiology. 2016;70:361–374. doi: 10.1146/annurev-micro-091014-104306. [DOI] [PubMed] [Google Scholar]
- Shine J, Dalgarno L. The 3’-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Schneider C, Hor J, Vogel J. Discovery of new RNA classes and global RNA-binding proteins. Current opinion in microbiology. 2017;39:152–160. doi: 10.1016/j.mib.2017.11.016. [DOI] [PubMed] [Google Scholar]
- Smirnov A, Forstner KU, Holmqvist E, Otto A, Gunster R, Becher D, Reinhardt R, Vogel J. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:11591–11596. doi: 10.1073/pnas.1609981113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solans L, Gonzalo-Asensio J, Sala C, Benjak A, Uplekar S, Rougemont J, Guilhot C, Malaga W, Martin C, Cole ST. The PhoP-dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004183. doi: 10.1371/journal.ppat.1004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Onge RJ, Elliot MA. Regulation of a muralytic enzyme-encoding gene by two non-coding RNAs. RNA biology. 2017 doi: 10.1080/15476286.2017.1338241. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Onge RJ, Haiser HJ, Yousef MR, Sherwood E, Tschowri N, Al-Bassam M, Elliot MA. Nucleotide second messenger-mediated regulation of a muralytic enzyme in Streptomyces. Molecular microbiology. 2015;96:779–795. doi: 10.1111/mmi.12971. [DOI] [PubMed] [Google Scholar]
- Steinert H, Sochor F, Wacker A, et al. Pausing guides RNA folding to populate transiently stable RNA structures for riboswitch-based transcription regulation. Elife. 2017;6 doi: 10.7554/eLife.21297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Sukumar N, Abramovitch RB, Parish T, Russell DG. Mycobacterium tuberculosis responds to chloride and pH as synergistic cues to the immune status of its host cell. PLoS Pathog. 2013;9:e1003282. doi: 10.1371/journal.ppat.1003282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverniti V, Forti F, Ghisotti D, Putzer H. Mycobacterium smegmatis RNase J is a 5’-3’ exo-/endoribonuclease and both RNase J and RNase E are involved in ribosomal RNA maturation. Molecular microbiology. 2011;82:1260–1276. doi: 10.1111/j.1365-2958.2011.07888.x. [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- Turapov O, Glenn S, Kana B, Makarov V, Andrew PW, Mukamolova GV. The in vivo environment accelerates generation of Resuscitation-promoting factor-dependent mycobacteria. AJRCCM. 2014 doi: 10.1164/rccm.201407-1289LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegrove TB, Shabalina SA, Storz G. How do base-pairing small RNAs evolve? FEMS microbiology reviews. 2015;39:379–391. doi: 10.1093/femsre/fuv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellanoweth RL, Rabinowitz JC. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Molecular microbiology. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA. 2003;9:1084–1097. doi: 10.1261/rna.5710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nature reviews Microbiology. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EG, Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv Genet. 2015;90:133–208. doi: 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Wagner EG, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- Warner DF, Savvi S, Mizrahi V, Dawes SS. A riboswitch regulates expression of the coenzyme B12-independent methionine synthase in Mycobacterium tuberculosis: implications for differential methionine synthase function in strains H37Rv and CDC1551. Journal of bacteriology. 2007;189:3655–3659. doi: 10.1128/JB.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes & development. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, Backofen R, Georg J. Comparative genomics boosts target prediction for bacterial small RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E3487–3496. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]