Abstract
The mechanisms by which infectious hepatitis C virus (HCV) particles are assembled and released from infected cells remain poorly characterized. In this regard, many other enveloped viruses, notably human immunodeficiency virus type 1, have been shown to utilize the host vacuolar protein sorting machinery (also known as the endosomal sorting complex required for transport; ESCRT) to traffic through the cell and effect the membrane rearrangements required for the formation of enveloped particles. We postulated that this might also apply to HCV. To test this hypothesis, we established a method of conditional virus-like particle assembly involving trans-complementation of an envelope-deleted JFH-1 genome using plasmid transfection. This system reliably produced virus particles that were infectious and could be enumerated easily by focus-forming assay in Huh7 cells. Following co-transfection with plasmids expressing various dominant-negative forms of either components of the ESCRT-III complex or Vps4 (the AAA ATPase that recycles the ESCRT complexes), a reduction in particle production was seen. No significant effect was observed after co-transfection of dominant-negative ESCRT-I or Alix, an ESCRT associated protein. Dominant-negative Vps4 or ESCRT-III components had no effect on either virus genome replication or the accumulation of intracellular infectious particles. These data were confirmed using cell culture infectious HCV and we conclude that HCV requires late components of the ESCRT pathway for release of infectious virus particles.
Introduction
Hepatitis C virus (HCV) represents a major global health burden. An estimated 170 million people are chronically infected worldwide and a significant proportion of these will go on to develop end stage liver disease. Current drug treatments are poorly tolerated and are only effective in approximately 50% of individuals. There is, therefore, a pressing need for a greater understanding of the molecular mechanisms involved in HCV infection in order for novel drug targets to be identified.
HCV is the prototype member of the genus Hepacivirus in the family Flaviviridae. It is a single-stranded, positive-sense RNA virus whose genome encodes a single ORF. This is translated in a cap-independent manner from an internal ribosome entry site (IRES) located in the 5′ untranslated region, into a single polyprotein of approximately 3000 aa. The polyprotein is proteolytically cleaved by host and viral proteases to form 10 mature proteins; the structural proteins (core, E1 and E2), a viroporin p7 and the non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B).
Although recent advances in the development of tissue culture systems for the study of the HCV life cycle (Lindenbach et al., 2005; Wakita et al., 2005; Zhong et al., 2005) have defined a critical role for lipid droplets in virus assembly (Miyanari et al., 2007), the precise mechanism by which infectious HCV particles are assembled and released remains poorly characterized. In particular, it has not been established how nascent HCV particles acquire a lipid bilayer membrane (lipid droplets are surrounded by a single lipid layer). In this regard, it has been well documented that many enveloped viruses utilize aspects of the host endosomal pathway to acquire a membrane and effect assembly and release following membrane scission. Central to the endosomal pathway is the multivesicular body (MVB), which is generated by inward budding of numerous intraluminal vesicles, a process that is analogous to the mechanism whereby viruses acquire their envelope (for review see McDonald & Martin-Serrano, 2009). The sorting of cargo into MVBs is co-ordinated by several multi-protein complexes collectively termed the endosomal sorting complexes required for transport (ESCRTs). Although the precise workings of the ESCRT machinery remain to be fully characterized, it is generally thought that four ESCRT complexes (0, I, II and III) act sequentially to incorporate ubiquitinated target proteins into the intraluminal vesicle within the MVB (for reviews see Hurley & Emr, 2006; Teis et al., 2009). When the process is complete, the ESCRT complexes are dissociated from the cytosolic side of the endosomal membrane and recycled by the AAA ATPase, Vps4. In the absence of Vps4 activity, the ESCRT complex cannot function and aberrant endosomes are formed. Numerous enveloped RNA viruses such as human immunodeficiency virus type 1 (HIV-1) (Garrus et al., 2001), Ebola virus (Martin-Serrano et al., 2001) and avian sarcoma virus (ASV) (Pincetic et al., 2008) and, more recently, enveloped DNA viruses including hepatitis B virus (HBV) (Lambert et al., 2007) and herpes simplex virus 1 (HSV-1) (Crump et al., 2007) have been shown to utilize ESCRT complexes during virion morphogenesis. In most cases viruses interact with the ESCRT machinery via late (L) domains, short motifs normally found in capsid or matrix proteins, which act as docking sites for ESCRT components. When mutated, the loss of the L-domain causes defective assembly, whereby separation of host and virus fails during the budding process (Bieniasz, 2006). Three canonical L-domains have been identified in viruses: PT/SAP, YPXL and PPXY, which have been shown to interact with TSG101, Alix and the Nedd4-like E3 ubiquitin ligases, respectively (Demirov & Freed, 2004). More recently, a fourth L-domain (FPIV) was described in a paramyxovirus but there is as yet no information about its binding partner (Schmitt et al., 2005).
Where ESCRT complex function is inhibited, viral particle production is compromised. We postulated that the ESCRT complex may also be involved in the HCV life cycle and that inhibiting ESCRT function would reduce HCV viral particle production. In vitro manipulation of fundamental pathways such as the ESCRT system is extremely toxic to cells and therefore cannot be easily achieved using stable cell lines that constitutively express dominant-negative (DN) ESCRT proteins. We therefore investigated whether a transient trans-complementation method could be used to assess the role of ESCRT components in HCV virus assembly and release. Trans-complementation systems have been developed for a number of positive-strand RNA viruses, such as West Nile (Hanna et al., 2005), Sindbis (Bredenbeek et al., 1993) and Kunjin (Khromykh et al., 1998; Harvey et al., 2004) viruses, but it is only recently that analogous systems have been reported for HCV (Ishii et al., 2008; Steinmann et al., 2008; Adair et al., 2009). In this study, we complemented a JFH-1 derivative lacking the E1 and E2 glycoproteins by transfection of a plasmid expressing the relevant structural proteins, either in the presence or absence of plasmids expressing DN derivatives of various ESCRT proteins. This system was advantageous in that the effects of ESCRT disruption were limited to virus-producing cells, although we also qualified our findings using fully infectious HCV. We describe the use of this methodology to demonstrate the requirement for a functional ESCRT complex during the release of HCV virus particles.
Results
Provision of the HCV envelope proteins in trans enables efficient virus particle production
We developed a conditional assembly system (JFH-1/ΔE1E2) based on transfection of Huh7 cells with a JFH-1 derivative containing an internal deletion of 351 aa spanning the envelope glycoproteins. This deletion renders the virus incapable of producing infectious virus particles, yet it retains the ability to undergo productive genome replication in Huh7 cells (Wakita et al., 2005). To restore particle production, E1, E2 and p7 were supplied in trans in the form of a rescue plasmid pcDNA-E1-p7(J4). As subsequent experiments would involve co-transfection of pcDNA-E1-p7(J4) with green fluorescent protein (GFP)-tagged ESCRT protein expression constructs, pEGFP was co-transfected as a control for non-specific effects of GFP expression on particle production. Huh7 cells were electroporated with JFH-1/ΔE1E2 RNA and subsequently transfected with plasmids by lipofection.
Seventy-two hours after electroporation, protein expression in producer cells was verified by Western blot and virus production in culture supernatants was measured by focus-forming assay (Fig. 1a). In the absence of the pcDNA-E1-p7(J4) rescue plasmid, no virus production was detectable. A titration of plasmid input showed that optimal virus production was obtained when 2 μg pcDNA-E1-p7(J4) was transfected (data not shown), giving a mean titre of 1.1×103 focus-forming units (f.f.u.) ml−1 (Fig. 1a); this is equivalent to titres achieved in packaging cell lines (Steinmann et al., 2008). In the context of GFP co-expression, the mean titre was 6.4×102 f.f.u. ml−1. Titres achieved via trans-complementation were lower than those seen for full-length JFH-1 (7.5×104 f.f.u. ml−1), most likely reflecting the fact that only a fraction of the cell population was transfected with the envelope protein expression plasmid. Nevertheless, these findings confirmed that infectious viruses could be reproducibly generated by trans-complementation and that titres were sufficiently high to discern the effects of manipulating the endosomal pathway in subsequent experiments.
Fig. 1. Validation of a trans-complementation assay using JFH-1/ΔE1E2.
(a) Huh7 cells were electroporated with full-length JFH-1 RNA, the non-replicating mutant (GND) or JFH-1/ΔE1E2 (ΔE1E2) alone (bars 1–3). For cells electroporated with JFH-1/ΔE1E2 RNA, they were subsequently transfected with either pcDNA3.1 (pcDNA), the rescue plasmid pcDNA-E1-p7(J4) (pE1p7), or pcDNA-E1-p7(J4) and pEGFP (bars 4–6). Clarified cell supernatants were titrated by focus-forming assay on naïve Huh7 cells. Error bars indicate SEM. Lysates from the producer cells above were analysed by Western blotting with the indicated antibodies; the lanes correlate to the bars in the top panel. (b) DN ESCRT mutants do not affect HCV genome replication. Huh7 cells were electroporated with SGR-Luc-JFH-1 RNA and subsequently transfected with plasmids encoding a panel of wild-type and DN ESCRT proteins, or GFP alone. At 48 h, cells were lysed and levels of replication were compared by luciferase assay. RLU, Relative light units.
To confirm the activity of various DN ESCRT components, their effects on HIV-1 particle production was verified by co-transfection of both HEK 293T and Huh7 cells with the NL43 proviral clone of HIV-1 alongside ESCRT components (data not shown). As expected, DN Tsg101, charged multivesicular body protein (CHMP) 4 and Vps4 reduced levels of extracellular infectious virus and p24 secretion approximately 10-fold in both producer cell types, whereas wild-type controls or DN Alix (apoptosis-linked gene-2 interacting protein X) did not. It was also important to verify that DN ESCRT components did not interfere with aspects of the virus life cycle distinct from particle production. To this end, Huh7 cells were electroporated with a subgenomic HCV replicon, SGR-Luc-JFH-1 (Targett-Adams & McLauchlan, 2005), prior to transfection with plasmids expressing the various ESCRT components. RNA replication was assessed by luciferase assay and, as shown in Fig. 1(b), none of the constructs had any significant effect on HCV genome replication.
Inhibition of Vps4 function blocks HCV particle production
Vps4, a member of the AAA ATPase family, is an essential protein for cargo sorting and MVB formation. Vps4 dissociates ESCRT complexes from the MVB membrane and allows them to recycle for further rounds of vesicle formation. A DN ATPase-defective mutant of Vps4, in which the active site glutamic acid has been mutated to glutamine, leads to defective MVB sorting and formation of aberrant endosomes in BHK cells (Bishop & Woodman, 2000) and also in Huh7 cells. Production of infectious particles of HIV-1 (Garrus et al., 2001), HSV-1 (Crump et al., 2007), HBV (Lambert et al., 2007) and ASV (Pincetic et al., 2008) is inhibited upon expression of the Vps4DN protein. The wild-type or DN forms of Vps4, tagged with GFP (Vps4WT–GFP or Vps4DN–GFP) were transfected into Huh7 cells, the distribution of Vps4DN–GFP was distinct from Vps4WT (Fig. 2a), with a concentration in large cytoplasmic punctae consistent with aberrant endosomes. Vps4WT–GFP or Vps4DN–GFP plasmids were then co-transfected with pcDNA-E1-p7(J4) into Huh7 cells containing JFH-1/ΔE1E2 RNA. A control transfection using pcDNA-E1-p7(J4) and pEGFP (GFP) was included to measure particle production in the absence of exogenous Vps4 expression. Virus titre was quantified by focus-forming assay and expressed as a percentage of the GFP control (Fig. 2b). Transfection of Vps4WT–GFP did not significantly alter particle production (98% of the number of particles produced in the presence of GFP). However, in the presence of Vps4DN–GFP, the extracellular virus titre was reduced approximately 20-fold (5% of GFP control levels). To ascertain whether inhibition of extracellular particle production was dependent on the level of Vps4DN expression, increasing amounts of Vps4DN–GFP DNA were transfected, from 50 ng to 1 μg, resulting in a proportionate increase in protein expression (Fig. 2c). Even the lowest level of Vps4DN–GFP expression resulted in a greater than fivefold reduction (18% of GFP control levels), and extracellular particle levels were further diminished with increased Vps4DN–GFP expression, indicating that the DN effect was both potent and dose-dependent. Western blot analysis of lysates from producer cells transfected with the lowest (50 ng) and highest (1 μg) amounts of Vps4DN–GFP DNA confirmed that expression of the HCV proteins was unaffected by Vps4DN–GFP expression (Fig. 2d), consistent with the observation in Fig. 1(b) that RNA replication was unaffected, and supporting the notion that the effect of Vps4DN was specific to extracellular particle production. These results demonstrate that recycling of ESCRT complex components by Vps4 and a functional endosomal pathway are critical to the production of extracellular HCV particles.
Fig. 2.
Vps4 DN mutant inhibits HCV infectious particle production. Huh7 cells were electroporated with JFH-1/ΔE1E2 RNA and subsequently transfected with pcDNA-E1-p7(J4) and plasmids expressing GFP-tagged Vps4 wild-type (Vps4WT–GFP), DN (VpsDN–GFP) or a pEGFP-only control (GFP). At 72 h, supernatants were removed and used to inoculate naïve Huh7 cells. (a) Huh7 cells transfected with Vps4WT–GFP or Vps4DN–GFP were observed using a fluorescent microscope. In both wild-type- and DN-transfected cells, E2 is seen to localize at the ER; however, the cytoplasmic distribution is altered in the presence of Vps4DN–GFP. (b) Infectious particles were enumerated by focus-forming assay. Data were normalized to the titre produced in the presence of GFP only (100%). (c) A range of amounts of Vps4DN–GFP DNA (50 ng–1 μg) were transfected into JFH-1/ΔE1E2 RNA electroporated Huh7 cells. Infectious particles were enumerated by focus-forming assay. Data were normalized to the titre produced in the presence of GFP only (100%). (d) Lysates from the producer cells transfected with 50 ng (lanes 1 and 3) or 1 μg (lanes 2 and 4) pEGFP (lane 5) or mock-transfected (lane 6) were analysed by Western blotting with the indicated antibodies. All error bars indicate SEM.
Tsg101 and Alix are not required for efficient HCV particle production
The first virus shown to utilize ESCRT complex components during assembly, HIV-1, enters the endosomal pathway via interactions between late domains found in the Gag structural protein and either the ESCRT-I component Tsg101 (Garrus et al., 2001) or the ESCRT-I/III associated protein, Alix (Demirov & Freed, 2004). Accordingly, HIV-1 viral particle production is significantly reduced when Tsg101 is disrupted, whilst disrupting Alix function can be compensated for by Tsg101 interactions (McDonald & Martin-Serrano, 2009).
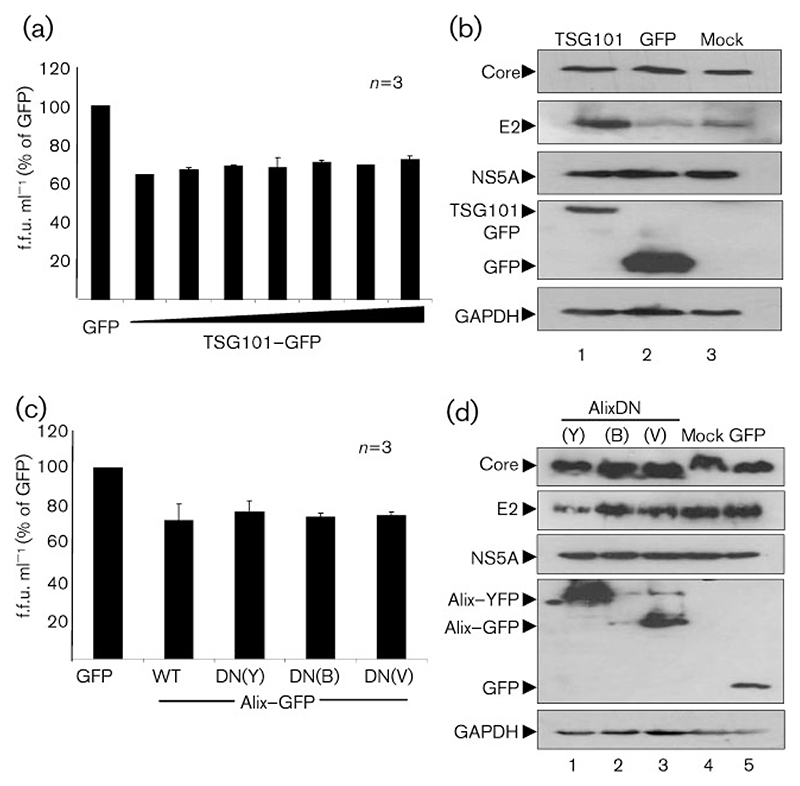
Although sequence analysis failed to identify any canonical late domains in any HCV proteins (data not shown), we investigated whether HCV similarly recruited the ESCRT machinery via interaction with Tsg101 or Alix. To this end, a DN mutant of Tsg101 tagged with GFP (TSG101DN–GFP) that blocks ESCRT recruitment via PT/SAP motifs, was used to assess the importance of Tsg101 for HCV particle production. Consistent with the lack of canonical L domains, expression of TSG101DN–GFP had no significant effect on particle production (Fig. 3a, b). Although a modest reduction was seen in the presence of TSG101DN–GFP (64–73% of GFP control levels), no dose-dependent relationship with expression levels was observed, meaning that the low level of disruption is likely due to non-specific effects associated with overexpression.
Fig. 3.
Tsg101 or Alix DN mutants do not inhibit HCV infectious particle production. Huh7 cells were electroporated with JFH-1/ΔE1E2 RNA and subsequently transfected with pcDNA-E1-p7(J4) and plasmids expressing Tsg101DN–GFP (a, b), wild-type Alix (AlixWT) or one of three DN Alix constructs [AlixDN(Y)–YFP, AlixDN(Bro)–GFP or AlixDN(V)–GFP] or mock and GFP controls (c, d). Infectious particles were enumerated by focus-forming assay (a, c). Data were normalized to the titre produced in the presence of GFP only (100%). Lysates from the producer cells were analysed by Western blotting with the indicated antibodies (b, d). All error bars indicate SEM.
Alix acts as a bridge between ESCRT-I and ESCRT-III (Hurley & Emr, 2006). Three expression constructs of Alix were obtained: AlixDN(Y)–YFP lacks the first half of the Bro1 domain, which is the binding site for the ESCRT-III component CHMP4; AlixDN(Bro)–GFP is the wild-type Bro1 domain in isolation and so will bind CHMP4 but not to YPXL motifs (and therefore may or may not be dominant-negative); and AlixDN(V)–GFP is the wild-type V domain in isolation and so will bind to YPXL motifs but is unable to interact with the remainder of the ESCRT machinery, and therefore inhibits any YPXL motifs from interacting with wild-type Alix (Munshi et al., 2007). A wild-type Alix plasmid (AlixWT) was transfected as a further control. As seen for Tsg101, however, expression of neither the wild-type nor the DN forms of Alix showed any significant effect upon particle production (Fig. 3c, d). These results suggest that recruitment of the ESCRT complexes via PT/SAP or YPXL interaction with Tsg101 or Alix, respectively, are not required for HCV particle production.
Inhibition of ESCRT-III function blocks HCV particle production
ESCRT-III comprises numerous CHMPs – 2A, 2B, 3, 4A, 4B, 4C and 6 – as well as the associated proteins CHMP1A, 1B, 5 and 7, which act in concert to mediate the formation of the intraluminal vesicles in the MVB. The CHMPs are also known to be involved in membrane scission activities alongside Vps4 and can facilitate scission in the absence of ESCRT-I and -II (Wollert et al., 2009).
To generate DN versions of the CHMPs (CHMP-DN) they were cloned as yellow fluorescent protein (YFP) fusion proteins such that the YFP moiety was C-terminal (Pawliczek & Crump, 2009), this prevents the C terminus-mediated interaction of the CHMPs with Vps4 (Obita et al., 2007; Stuchell-Brereton et al., 2007) and therefore perturbs ESCRT complex recycling. These constructs were then introduced into the trans-complementation system, as described above, to determine if they were able to inhibit HCV particle production. As shown in Fig. 4(a), each of the CHMP-DN constructs dramatically reduced extracellular particle production (to between 1 and 15% of GFP control levels). To assess whether this effect was dose-dependent, increasing amounts of the construct expressing the most effective inhibitor of HCV particle production, CHMP4B–YFP, were transfected into Huh7 cells containing JFH-1/ΔE1E2 RNA (Fig. 4b). This experiment confirmed that expression of CHMP4B–YFP reduced extracellular particle production in a dose-dependent fashion. Western blot analysis of lysates from cells transfected with either low (50 ng) or high (1 μg) amounts of CHMP-DN constructs confirmed that expression of the HCV proteins was not affected by CHMP-DN expression (Fig. 4c). We conclude that production of extracellular HCV particles requires the action of the ESCRT-III complex.
Fig. 4.
DN CHMP mutants inhibit HCV infectious particle production. Huh7 cells were electroporated with JFH-1/ΔE1E2 RNA and subsequently transfected with pcDNA-E1-p7(J4) and plasmids expressing DN CHMPs (as GFP fusions). Infectious particles were enumerated by focus-forming assay (a, b). In (b), a range of CHMP4B–GFP plasmid concentrations (50 ng–1 μg) was transfected. Data were normalized to the titre produced in the presence of GFP only (100%). Lysates from the producer cells were analysed by Western blotting with the indicated antibodies (c). All error bars indicate SEM.
Inhibition of ESCRT function does not block the accumulation of intracellular infectious HCV particles
Although our data demonstrate clearly that DN Vps4 or CHMPs were able block the production of extracellular infectious HCV particles, it was not clear whether this block occurred at the stage of assembly of intracellular infectious particles or during their release to the extracellular environment. To test this, intracellular infectious particles were quantified following lysis of transfected cells by repetitive freeze–thaw (Hughes et al., 2009). As shown in Fig. 5(a), none of the DN ESCRT components had any significant effect on the accumulation of intracellular infectious particles. Appropriate expression of the ESCRT components and HCV viral proteins was demonstrated by Western blot (Fig. 5b). We conclude that ESCRT function is not required for the assembly of intracellular infectious HCV particles, but plays a role in the release of these particles from infected cells.
Fig. 5.
DN ESCRT mutants have no effect on the production of intracellular virus particles. Huh7 cells were electroporated with JFH-1/ΔE1E2 RNA and subsequently transfected with pcDNA-E1-p7(J4) and plasmids encoding a panel of wild-type and DN ESCRT proteins. Intracellular contents were liberated by repeated freeze–thaw cycles and infectious particles were quantified by focus-forming assay (a). Data were normalized to the titre produced in the presence of GFP only (100%). Lysates from the producer cells were analysed by Western blotting with the indicated antibodies (b). All error bars indicate SEM.
Release of full-length infectious HCV is dependent on Vps4 and ESCRT-III function
As discussed above, we used a trans-complementation system to facilitate the analysis of the role of the ESCRT complex in HCV particle assembly. We considered it important to verify that the results obtained were replicated in the context of the full-length, tissue-culture-infectious virus. To test this, we utilized the J6/JFH-1 chimeric virus (Jc1), as in our hands this gave significantly higher titres than JFH-1 (Griffin et al., 2008). Huh7 cells were infected with Jc1 and subsequently transfected with the appropriate ESCRT constructs as described above. Titres of both intra- and extracellular virus were determined at 96 h post-infection (p.i.). As shown in Fig. 6(a), TSG101DN and AlixDN expression resulted in a modest but significant reduction in released virus consistent with the non-specific effect observed in the context of the trans-complementation assay (Fig. 3). Furthermore, as observed in the trans-complementation assay (Figs 2 and 4), both CHMP4BDN and Vps4DN expression resulted in a fivefold reduction in released virus (Fig. 6a). Additionally, none of the ESCRT constructs had any significant effect on the intracellular virus titres (Fig. 6b), further confirming that ESCRT function is required for release of infectious HCV particles, but not for their assembly.
Fig. 6.
Inhibition of Vps4 and ESCRT-III blocks release of virus particles from Huh7 cells infected with full-length HCV. Huh7 cells were infected with Jc1 at an m.o.i. of 0.5 and subsequently transfected with plasmids encoding a panel of WT and DN ESCRT proteins. Infectious extracellular and intracellular particles were quantified by focus-forming assay (a, b). Extracellular results were normalized to titres produced in the presence of GFP only (100%). Lysates from the producer cells were analysed by Western blotting with the indicated antibodies (c). All error bars indicate SEM.
Discussion
Our data clearly demonstrate that the release of HCV particles requires late stages in the ESCRT pathway, as DN mutants of either ESCRT-III complex components (CHMPs) or the AAA ATPase Vps4 inhibited production of extracellular infectious virus particles. In contrast with these results, we found no evidence for a role of either ESCRT-I or Alix in the HCV life cycle. These results are similar to recent findings for HSV-1, which has also been shown to be dependent on ESCRT-III and Vps4, but not ESCRT-I or Alix (Pawliczek & Crump, 2009). The observation that DN mutants of all the individual CHMPs tested had a dramatic effect on particle production correlates with the observation that an ordered assembly of the CHMP proteins in ESCRT-III is required for appropriate MVB sorting (Teis et al., 2008).
One explanation of these data might be that DN CHMPs or Vps4 have a general cytotoxic effect by disrupting normal endosome formation. However, the observation that transient expression of the DN Vps4 mutant did not affect production of either influenza virus (Bruce et al., 2009) or respiratory syncytial virus (Utley et al., 2008), whereas it had a marked effect on production of HSV-1 particles (Crump et al., 2007), argues against this explanation. The latter observations suggest that expression of this protein, although undoubtedly detrimental to cell function, does not result in gross perturbation of endosome formation to the extent that all enveloped virus production is impaired.
How might HCV interact with the ESCRT system? For other enveloped viruses, the direct interactions of viral proteins with components of the ESCRT machinery via L-domains has been well characterized (for review see McDonald & Martin-Serrano, 2009). Consistent with the lack of a requirement for Tsg101 or Alix in HCV particle production, sequence analysis of HCV proteins did not reveal the presence of any canonical L-domains. However, the lack of identifiable L-domains does not necessarily mean that there are no potential associations with ESCRT proteins, as it is clear that the interactions between viruses and the ESCRT system are varied and complex. For example, viruses containing the PPxY motif are able to recruit Nedd4L and subsequently co-opt the ESCRT machinery in an as yet uncharacterized manner. Furthermore, despite not containing the PPxY motif, HIV-1 Gag protein deficient in Tsg101- and Alix-binding L-domains, can be rescued by overexpression of Nedd4L (Chung et al., 2008; Usami et al., 2008), highlighting the inter-changeability and flexibility inherent in L-domains. Thus, it is feasible that HCV gains entry to the ESCRT machinery by an as yet uncharacterized interaction, in a manner that is Tsg101- and Alix-independent. In this regard, it should be noted that two HCV proteins with roles in virus assembly – core and NS2 – are ubiquitinated (Shirakura et al., 2007; Suzuki et al., 2001; Welbourn et al., 2009), providing possible mechanisms by which HCV proteins might be recruited to the ESCRT complex.
What might be the role of the ESCRT machinery in HCV virus production? It is generally thought that the role of the ESCRT machinery in enveloped virus production is limited to the processes of membrane deformation and scission – key events in both virus budding and production of intraluminal MVB vesicles. However, our data clearly show that ESCRT perturbation does not block assembly of intracellular infectious virus. We propose instead, that the ESCRT system is involved in either trafficking of complete nascent HCV particles through (and out of) the cell, or possibly the scission event whereby infectious particles are released from cellular membranes.
One explanation for our findings is the observation made several years ago that Vps4DN-expressing cells demonstrated impaired cholesterol trafficking (Bishop & Woodman, 2000), suggesting that defects in the ESCRT system might disrupt normal cellular lipid metabolism. Lipids are known to play a vital role in HCV particle production; a number of HCV proteins are known to associate with lipid droplets, notably core (Boulant et al., 2006) and NS5A (Miyanari et al., 2007), and these organelles play a key, albeit uncharacterized, role in virus production (Miyanari et al., 2007; Huang et al., 2007; Ye, 2007). It is possible therefore that ESCRT function might be required to transport nascent virus particles from the site of assembly (assumed to be proximal to lipid droplets) to the cell surface.
In conclusion, this study is the first to show a requirement for the ESCRT machinery in the HCV life cycle. Further investigation of the mechanisms of this interaction will help clarify the role of the ESCRT complex in HCV assembly release and could pave the way for future therapeutic interventions.
Methods
Cell culture and cell lines
Huh7 cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma) supplemented with 10% fetal calf serum (FCS), 1% non-essential amino acids, 20 mM HEPES-KOH (pH 7.4) and 1% penicillin/streptomycin (complete DMEM).
Plasmid construction and transfection
pcDNA-E1-p7(J4) containing the coding sequence for E1-E2-p7 of the genotype J4 infectious clone, pJ4CV6LS (Yanagi et al., 1998), has been described previously (Griffin et al., 2005). Generation of the CHMP and Alix DN constructs was described recently by Pawliczek & Crump (2009). pcDNA3-myc-ALIX (referred to throughout as AlixWT) was provided by Harald Stenmark (University of Oslo). Wild-type (WT) and dominant-negative (DN) GFP-tagged Vps4 plasmids (Vps4WT–GFP and Vps4DN–GFP) were obtained from Philip Woodman (University of Manchester) (Bishop & Woodman, 2000). The DN Tsg101 plasmid pCR3.1-Tsg101(1-157)–GFP (TSG101DN–GFP) and the DN Alix pCr3.1-Alix–YFP (AlixDN–YFP) were obtained from Juan Martin-Serrano (Kings College, London) (Martin-Serrano et al., 2003).
In vitro transcription and electroporation of RNA
T7 in vitro transcripts were made from linearized DNA templates of the JFH1 subgenomic replicon (SGR-luc-JFH-1) (Targett-Adams & McLauchlan, 2005) or the non-replicating GND mutant (Wakita et al., 2005), using an Ambion T7 megascript kit. JFH-1/ΔE1E2 and Jc1 were not linearized as they contain a T7 terminator sequence. To electroporate Huh7 cells with RNA, subconfluent cells were trypsinised, washed with diethyl pyrocarbonate (DEPC)-treated PBS, then resuspended at 107 cells ml−1 in DEPC-treated PBS. Cells (8×106; 400 μl) were mixed with 10 μg RNA in a 4 mm cuvette, then pulsed in a Bio-Rad Gene Pulser II at 975 μF, 270 V. Cells were resuspended in 12 ml DMEM, then 2 ml of this was added to each well of a six-well plate.
Trans-complementation
Ten hours after electroporation with JFH-1/ΔE1E2 RNA, medium was removed and producer cells were washed with complete DMEM. Cells were then transfected with the appropriate plasmids by lipofection using Lipofectin (according to the manufacturer’s instructions). Overall DNA input was kept constant by supplementing with an appropriate quantity of pcDNA3.1 vector. Transfection mixes were removed after 12 h and cells were incubated at 37 °C in complete DMEM for a further 50 h. For quantification of extracellular virus particle production, cell culture supernatant was removed, clarified by centrifugation, then infectivity was determined by focus-forming assay as described by Griffin et al. (2008). To determine intracellular virus particle production, cells were resuspended in 50 μl PBS and lysed by repetitive freeze–thaw action (five times); lysates were clarified by centrifugation at 2800 g in a microcentrifuge for 5 min, and infectivity was determined by focus-forming assay.
Quantification of particle production using the infectious Jc1 isolate
Concentrated virus inoculum was prepared by electroporation of cells with Jc1 RNA and collection of the media from repeated passages for 2 weeks. Virus was concentrated by centrifugation of supernatants at 150 000 g through a 20% sucrose cushion; this was resuspended in PBS and titrated by focus-forming assay. To infect Huh7 cells with concentrated Jc1, 5×105 cells were seeded in a six-well plate and left to settle overnight. Jc1 (at an m.o.i. of 0.5 in complete DMEM) was added, and removed at 10 h. Cells were then transfected with plasmids by lipofection. Transfection mixes were removed after 12 h and cells were incubated at 37 °C in complete DMEM for a further 74 h. Extracellular and intracellular virus infectious particle production was quantified as described above.
Transient HCV replication assays using luciferase reporter genomes
Ten hours after electroporation of in vitro-transcribed RNA, medium was removed and cells were washed with complete DMEM. Cells were then transfected with the appropriate plasmids by lipofection. Transfection mixes were removed after 12 h and cells were incubated at 37 °C in complete DMEM for a further 26 h. Cells were washed in PBS, lysed in 200 μl passive lysis buffer (Promega) and briefly centrifuged to pellet insoluble material. A luciferase assay was performed using a luminometer (BMG Labtech) and the appropriate reagent (Promega). Assays were performed in duplicate.
Western blot analysis
For protein analysis, cells were washed twice with PBS, lysed in 40 μl lysis buffer [50 mM Tris/HCl (pH 8.0), 140 mM NaCl, 0.1% SDS, 0.5% NP-40, supplemented with EDTA-free complete protease inhibitor cocktail (Roche)], then mixed with 15 μl 4 × Laemmli buffer. Lysates were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore) using a semi-dry blotting apparatus (Bio-Rad). After blocking with 5% dried skimmed milk in TBS-T [150 mM NaCl, 50 mM Tris/HCl (pH 7.5), 0.1% Tween-20], primary and secondary antibodies (diluted in 5% dried skimmed milk in TBS-T) were added sequentially with TBS-T washing after each step. Blots were visualized using chemiluminescence and autoradiography.
Indirect immunofluorescence
In addition to focus-forming assays, immunofluorescence (IF) was also used to verify expression of the ESCRT and HCV proteins. Cells were fixed in 4% (w/v) paraformaldehyde/PBS for 20 min at room temperature, washed in PBS, then permeabilized with 0.2% Triton X-100 (v/v) in PBS at room temperature for 10 min. Primary antibody (diluted in 10% v/v; FCS/PBS) was applied directly and detected using appropriate Alexa-fluor-conjugated secondary antibodies (Invitrogen Molecular Probes) diluted in 10% FCS in PBS at 1/500.
Antibodies
Proteins were detected using the primary antibodies α-E2 (AP33; courtesy of Genetech and Arvind Patel, MRC Virology Unit, Glasgow) at 1 : 1000 (Western blot) or 1 : 500 (IF), α-core (#308; gift from John McLauchlan, MRC Virology unit, Glasgow) at 1 : 3000, polyclonal α-NS5A (Macdonald et al., 2003) at 1 : 10000 (Western blot) or 1 : 4000 (IF), α-GAPDH-6C5 (Abcam) at 1 : 30000 and α-GFP at 1 : 10000 (gift from Frank von Kuppeveld, Radboud University, Netherlands).
Acknowledgements
We are grateful to the following people for provision of experimental materials: Takaji Wakita for the pJFH-1 constructs, Ralf Bartenschlager for the JFH-1/ΔE1E2 and Jc1 constructs, Juan Martin-Serrano for the Tsg101DN–GFP and AlixDN–YFP constructs, Philip Woodman for the Vps4WT–GFP and Vps4DN–GFP constructs, Harald Stenmark for the AlixWT construct, John McLauchlan for the anti-core antibody and the SGR-Luc-JFH-1 constructs, Frank von Kuppeveld for the GFP antibody, and Genetech and Arvind Patel for the E2 (AP33) antibody. L. C. was funded by the Leeds Teaching Hospitals Trust, S. D. C. G. is the recipient of a Medical Research Council New Investigator Award (G0700124) and C. M. C. is a Royal Society University Research Fellow. We thank Jamel Mankouri for helpful advice and discussions during this study. Work in the M. H. laboratory is also supported by the Medical Research Council and the Wellcome Trust.
References
- Adair R, Patel AH, Corless L, Griffin S, Rowlands DJ, McCormick CJ. Expression of hepatitis C virus (HCV) structural proteins in trans facilitates encapsidation and transmission of HCV subgenomic RNA. J Gen Virol. 2009;90:833–842. doi: 10.1099/vir.2008.006049-0. [DOI] [PubMed] [Google Scholar]
- Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Bishop N, Woodman P. ATPase-defective mammalian VPS4 localises to aberrant endosomes and impairs cholesterol trafficking. Mol Biol Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Montserret R, Hope RG, Ratinier M, Targett-Adams P, Lavergne JP, Penin F, McLauchlan J. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J Biol Chem. 2006;281:22236–22247. doi: 10.1074/jbc.M601031200. [DOI] [PubMed] [Google Scholar]
- Bredenbeek PJ, Frolov I, Rice CM, Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce EA, Medcalf L, Crump CM, Noton SL, Stuart AD, Wise HM, Elton D, Bowers K, Digard P. Budding of filamentous and non-filamentous influenza A virus occurs via a Vps4 and Vps28-independent pathway. Virology. 2009;390:268–278. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Chung HY, Morita E, von Schwedler U, Muller B, Kräusslich HG, Sundquist WI. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump CM, Yates C, Minson T. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J Virol. 2007;81:7380–7387. doi: 10.1128/JVI.00222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pormillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, et al. TSG101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Griffin S, Clarke D, McCormick C, Rowlands D, Harris M. Signal peptide cleavage and internal targeting signals direct the hepatitis C virus p7 protein to distinct intracellular membranes. J Virol. 2005;79:15525–15536. doi: 10.1128/JVI.79.24.15525-15536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin S, StGelais C, Owsianka AM, Patel AH, Rowlands D, Harris M. Genotype-dependent sensitivity of hepatitis C virus to inhibitors of the p7 ion channel. Hepatology. 2008;48:1779–1790. doi: 10.1002/hep.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna SL, Pierson TC, Sanchez MD, Ahmed AA, Murtadha MM, Doms RW. N-linked glycosylation of West Nile virus envelope proteins influences particle assembly and infectivity. J Virol. 2005;79:13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey TJ, Liu WJ, Wang XJ, Linedale R, Jacobs M, Davidson A, Le TT, Anraku I, Suhrbier A, et al. Tetracycline-inducible packaging cell line for production of Flavivirus replicon particles. J Virol. 2004;78:531–539. doi: 10.1128/JVI.78.1.531-538.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci U S A. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M, Griffin S, Harris M. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J Gen Virol. 2009;90:1329–1334. doi: 10.1099/vir.0.009332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane trafficking network. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Murakami K, Hmwe SS, Bin Z, Li J, Shirakura M, Morikawa K, Suzuki R, Miyamura T, et al. Trans-encapsidation of hepatitis C virus subgenomic replicon RNA with viral structure proteins. Biochem Biophys Res Commun. 2008;371:446–450. doi: 10.1016/j.bbrc.2008.04.110. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Varnavski AN, Westaway EG. Encapsidation of the Flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J Virol. 1998;72:5967–5977. doi: 10.1128/jvi.72.7.5967-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C, Döring T, Prange R. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J Virol. 2007;81:9050–9060. doi: 10.1128/JVI.00479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Macdonald A, Crowder K, Street A, McCormick C, Saksela K, Harris M. The hepatitis C virus NS5A protein inhibits activating protein-1 (AP1) function by perturbing Ras–ERK pathway signalling. J Biol Chem. 2003;278:17775–17784. doi: 10.1074/jbc.M210900200. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz P. Role of ESCRT-I in retroviral budding. J Virol. 2003;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B, Martin-Serrano J. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J Cell Sci. 2009;122:2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J Biol Chem. 2007;282:3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Pawliczek T, Crump CM. Herpes simplex virus type-1 production requires a functional ESCRT-III complex, but is independent of TSG101 and/or ALIX expression. J Virol. 2009;83:11254–11264. doi: 10.1128/JVI.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincetic A, Medina G, Carter C, Leis J. Avian sarcoma virus and the human immunodeficiency virus, type 1 use different subsets of ESCRT proteins to facilitate the budding process. J Biol Chem. 2008;283:29822–29830. doi: 10.1074/jbc.M804157200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol. 2005;79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakura M, Murakami K, Ichimura T, Suzuki R, Shimoji T, Fukuda K, Abe K, Sato S, Fukasawa M, et al. E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. J Virol. 2007;81:1174–1185. doi: 10.1128/JVI.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann E, Brohm C, Kallis S, Bartenschlager R, Pietschmann T. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus like particles. J Virol. 2008;82:7034–7046. doi: 10.1128/JVI.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Tamura K, Li J, Ishii K, Matsuura Y, Miyamura T, Suzuki T. Ubiquitin-mediated degradation of hepatitis C virus core protein is regulated by processing at its carboxyl terminus. Virology. 2001;280:301–309. doi: 10.1006/viro.2000.0785. [DOI] [PubMed] [Google Scholar]
- Targett Adams P, McLauchlan J. Development and characterisation of a transient replication assay for the genotype 2a hepatitis C virus subgenomic replicon. J Gen Virol. 2005;86:3075–3080. doi: 10.1099/vir.0.81334-0. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr S. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr S. SnapShot: The ESCRT Machinery. Cell. 2009;137:182.:e1. doi: 10.1016/j.cell.2009.03.027. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Gottlinger HG. Efficient and specific release of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol. 2008;82:4898–4907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley TJ, Ducharme NA, Varthakavi V, Shepherd BE, Santangelo PJ, Lindquist ME, Goldenring JR, Crowe JE., Jr Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11–FIP2. Proc Natl Acad Sci U S A. 2008;105:10209–10214. doi: 10.1073/pnas.0712144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welbourn S, Jirasko V, Breton V, Reiss S, Penin F, Bartenschlager R, Pause A. Investigation of a role for lysine residues in non-structural proteins 2 and 2/3 of the hepatitis C virus for their degradation and virus assembly. J Gen Virol. 2009;90:1071–1080. doi: 10.1099/vir.0.009944-0. [DOI] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi M, St Claire M, Shapiro M, Emerson SU, Purcell RH, Bukh J. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- Ye J. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 2007;3:e108. doi: 10.1371/journal.ppat.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng C, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]