Abstract
The p7 protein of hepatitis C virus functions as an ion channel both in vitro and in cell-based assays and is inhibited by amantadine, long alkyl chain imino-sugar derivatives, and amiloride compounds. Future drug design will be greatly aided by information on the stoichiometry and high resolution structure of p7 ion channel complexes. Here, we have refined a bacterial expression system for p7 based on a glutathione S-transferase fusion methodology that circumvents the inherent problems of hydrophobic protein purification and the limitations of chemical synthesis. Rotational averaging and harmonic analysis of transmission electron micrographs of glutathione S-transferase-FLAG-p7 fusion proteins in liposomes revealed a heptameric stoichiometry. The oligomerization of p7 protein was then confirmed by SDS-PAGE and mass spectrometry analysis of pure, concentrated FLAG-p7. The same protein was also confirmed to function as an ion channel in suspended lipid bilayers and was inhibited by amantadine. These data validate this system as a means of generating high resolution structural information on the p7 ion channel complex.
Hepatitis C virus (HCV)5 currently infects over 3% of the world’s population and is the major indicator for liver transplantation in the developed world. Liver disease in the form of cirrhosis, liver failure, or hepatocellular carcinoma arises after many years of persistent virus infection following sub-clinical acute episodes. For this reason intervention is limited to chemotherapeutic treatment of chronic patients. The current regime of pegylated interferon α and ribavirin, while effective against certain viral genotypes, is largely ineffective against the most common HCV genotype 1 (1, 2) viruses making the search for new HCV anti-viral drug targets crucial. A recent meta-analysis of multiple clinical trials where amantadine was included alongside current therapies showed that an improved sustained viral response was achieved in patients that had previously failed to respond to dual therapy (3).
HCV is the prototype member of the Hepacivirus genus of the Flaviviridae family. It is an enveloped virus, and its genome comprises a single-stranded positive sense RNA of ~9.6 kb (4) that is translated from an internal ribosome entry site to yield a single polyprotein. This is cleaved by both host signalases and virus-encoded proteases yielding viral structural proteins and the non-structural proteins (5); the latter form the viral RNA replication complexes on modified cellular membranes and modulate host cell metabolism (6).
The region of the HCV genome between the structural and non-structural regions encodes a 63-amino acid integral membrane protein known as p7 (7), comprising two trans-membrane domains separated by a short basic loop (7, 8). We identified the function of p7 as a cation channel whose activity in suspended lipid bilayers was sensitive to the drug amantadine, providing a potential mechanism for the proposed efficacy of this drug in HCV treatment (9). Others have subsequently identified alternative channel blocking compounds, validating p7 as a potential therapeutic target (10, 11). It is not known whether p7 is a component of the HCV virion, nor at which stage in the replication cycle of HCV it performs its function, although importantly it has been shown as essential for HCV replication in chimpanzees (12) and is required for production of infectious virus in cell culture for the related bovine viral diarrhea virus (13).
p7 is proposed to belong to a family of viral ion channels known as viroporins. These are small integral membrane proteins that homo-oligomerize to form channels that mediate cation fluxes across cellular and viral membranes (14). Precedent for these proteins as therapeutic targets in a clinical setting is made by the M2 protein of influenza A virus (IAV), which is also sensitive to amantadine and related derivatives (15). We showed previously that p7 was able to replace M2 in a functional assay assessing its role in maintaining the pH-sensitive IAV hemagglutinin protein in its receptor binding conformation during cellular export (16). Amantadine inhibited both M2 and p7 in this assay at the same concentration, and we identified the basic loop as critical for p7 function; a fact subsequently confirmed in the context of virus replication in chimpanzees (12).
High resolution structural analysis is a vital component of present day drug design. There have been many structural analyses of the M2 ion channel (17, 18), and the interaction of amantadine with histidine residues in the channel lumen has been modeled (19, 20). M2 is known to form a tetrameric channel (21), and the trans-membrane domain of the protein is sufficient for this interaction to occur; indeed, the trans-membrane domain alone is capable of forming channels in vitro (22), and it is this region in isolation that has been the subject of many studies (23–25). In contrast p7 has two trans-membrane domains that comprise the majority of the protein sequence; thus channel formation is likely to involve almost the entirety of the protein. The length and composition of p7 makes it difficult to synthesize chemically, and those that have pursued this method have found the resulting material to be contaminated with premature termination products (10). We previously described a bacterial expression system for p7 based on a glutathione S-transferase (GST) fusion methodology. The fusion protein was seen to form ring-like structures in liposomes by transmission electron microscopy (TEM) corresponding to the formation of channel structures (9). Both the fusion protein and the subsequently derived near-native p7 protein formed functional ion channels in vitro. Here, we have refined this methodology to increase both yield and purity of an N-terminally FLAG-tagged p7 protein: FLAG-p7. We have used rotational averaging and harmonic analyses of TEM images to determine the precise stoichiometry of channel complexes formed by GST-p7 fusion proteins in a lipidic environment. Oligomerization of this protein was supported by higher order structures observed in subsequent SDS-PAGE and mass spectrometry of concentrated near-native protein. Finally, we confirm that the near-native FLAG-p7 utilized in these studies forms functional ion channels and can be blocked by amantadine. This system should provide the future means for high resolution analysis of the p7 channel complex to facilitate drug design.
Experimental Procedures
Plasmid Design and Construction
A PCR amplimer generated using Vent DNA polymerase (New England Biolabs) comprising the p7 sequence of the HCV genotype 1b J4 infectious clone (26) with appropriate sites in the forward and reverse primers was cloned into pGEX6P-1 (Amersham Biosciences) digested with EcoRI and NotI as previously described (9), generating plasmid pGEX6-FLAG-p7. The pGEX-6 backbone contains a “PreScission” protease (Rhinovirus serotype 14 3C protease, available as a GST fusion from Amersham Biosciences) cleavage site at the C terminus of GST and immediately prior to the polylinker region where the p7 cassette was introduced. The forward primer included a FLAG epitope tag (DYKDDDDK) immediately N-terminal to the p7 sequence and a short linker to increase the distance between the hydrophilic GST and the hydrophobic p7 domains of the fusion protein. The integrity of the plasmids was checked by DNA sequencing. All primer sequences are available on request.
Expression and Purification of GST-FLAG-p7 from Escherichia coli
E. coli strain BL21(DE3) was transformed with pGEX6-FLAG-p7 and grown at 30 °C overnight. Fresh colonies were used to inoculate a 5-ml culture in Luria-Bertani broth supplemented with ampicillin at 100 μg/ml. After overnight growth this was diluted 1/100 into a 2-liter flask containing 400 ml of broth with antibiotic and grown at 30 °C to an A600 of 0.7. Protein expression was induced by the addition of 1 mM isopro-pyl 1-thio-β-D-galactopyranoside, and cultures were grown for a further 4 h. Cells were harvested by centrifugation then resuspended in 30 ml of lysis buffer (phosphate-buffered saline with 1× EDTA-free protease inhibitor mixture (Roche Applied Science)). Cell suspensions were then subjected to two rounds of lysis at 14,000 p.s.i. in a manual fill French Pressure Cell (Thermo Life Sciences). To eliminate contaminating nucleic acids, 200 units of deoxyribonuclease I (Sigma) was added, and the sample was incubated at room temperature for 10 min. Lysates were then stored at 4 °C for immediate use or at −80 °C for long term storage.
To obtain soluble protein, detergent was added to the bacterial lysate (typically 1% (v/v) Triton X-100), and the lysate was centrifuged at 20,000 × g in a Sorvall SLA1500 rotor for 15 min. Supernatant recovered from this centrifugation represented the soluble E. coli fraction. GST fusion proteins recovered from the soluble fraction were purified using a 5-ml GSTrap column (Amersham Biosciences) in accordance with the manufacturer’s instructions. Where appropriate, protease cleavage of GST-FLAG-p7 fusion proteins was conducted while protein was bound to the GSTrap column, eluted in phosphate-buffered saline.
Inclusion bodies were isolated from bacterial cell lysates (typically in 1 ml of lysates for each preparation) by centrifugation at 3000 × g for 15 min. Pellets were then washed with 1/10th original volume of 100 mM ammonium acetate prior to further centrifugation at 3000 × g for 5 min. Subsequent pellets were resuspended in 1× original volume cleavage buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT).
At this point, fusion protein could be solubilized by adding 0.5% w/v N-laurylsarcosine or could be subjected to proteolysis.
Generation and Purification of Cleaved Near-native FLAG-p7
FLAG-p7 was cleaved from the parental fusion protein by the addition of 20 units of PreScission protease (Amersham Biosciences). Cleavage was conducted at 4 °C on a rotating blood mixer for 16 h. The resulting mix was adjusted to 0.5% w/v N-laurylsarcosine to solubilize the cleaved proteins, and the efficiency of cleavage was checked by SDS-PAGE. The resulting mix was then separated, and proteins were purified by high-performance liquid chromatography using a Dionex HPLC system, incorporating an automated fraction collector. The system was controlled using Chromeleon software (Dionex) on a windows-based PC. An ACE 5 C4–300 column (Advanced Chromatography Technologies) was used for analytical HPLC purifications.
Purifications were carried out using a flow rate of 0.5 ml/min. Initially, all purifications were conducted on an analytical scale, with sample injections of 50 μl in volume. Solvent solutions for the mobile phase were: Solution A = 5% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid. Solution B = 80% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid. Solution A was used for equilibration of the column and washing after the injection of sample. Solution B was used for protein elution, introduced over a continuous gradient of 0–100% of solution B. The protein content of eluted solvent was monitored by measuring the absorbance at 280 and 215 nm. Fractions were collected automatically, using a peak threshold, which was typically an absorbance of 150 milliabsorbance units at 215 nm. Protein was then lyophilized in a vacuum drier and resuspended in appropriate buffer, either 100% MeOH, SDS-PAGE Laemmli sample buffer (150 mM Tris-HCl, pH 6.8, 4% SDS w/v, 20% glycerol, 100 mM DTT) or phosphate-buffered saline, and checked by SDS-PAGE and Western blot analysis. Larger scale production was later achieved using a Jupiter C4 300 semipreparative column (Phenomenex) column using the same buffers, with flow rate adjusted for column volume.
Western Analysis and Antibodies
Proteins were typically separated on a 12.5% Tris-glycine polyacrylamide-resolving gel, poured using a mini-protean III system (Bio-Rad). Proteins were transferred in 20% MeOH, 1× Tris-glycine on a Bio-Rad semi-dry electroblotter at 15 V for 50 min onto a polyvinylidene difluoride membrane (Millipore). Filters were blocked in 5% (w/v) fat-free milk in TBS-T (25 mM Tris-HCl, pH 7.5, 137 mM NaCl, 0.1% (v/v) Tween 20) for a minimum of 1 h prior to incubation with antibody. Antibody #1055 was used at 1/1000 dilution in blocking solution and is an affinity-purified rabbit polyclonal anti-peptide antibody, raised against the C-terminal 6 amino acids of the HCV 1b J4 infectious clone p7 sequence: PPRAYA. The peptide was coupled to tuberculin-purified protein derivative via a CGG linker. Antibody #1055 has been described previously (27). The anti-FLAG tag monoclonal mouse antibody, M2, was obtained from Sigma (used at 1/20,000), and the mouse monoclonal anti-GST antibody was from Serotech (used at 1/1,000). Horseradish peroxidase-conjugated goat anti-rabbit or -mouse secondary antibodies were used at 1/5,000 dilution and were obtained from Sigma. Filters were washed for 3 × 10 min in TBS-T following incubation with primary and secondary antibodies, and proteins were visualized using in-house enhanced chemiluminescence (ECL) reagent.
TEM
2 μg of GST-p7 fusion protein was incubated at room temperature for 30 min in the presence or absence of 20 μl of1 mg/ml artificially synthesized PA:PC unilamellar vesicles prepared by extrusion. Both protein and vesicles were taken from stock solutions in phosphate-buffered saline (pH 7.0). 10 μl of sample was then applied directly to Formvar/carbon 400-mesh copper electron microscopy grids (Agar) for 30 s, and negative stain was applied as follows: residual sample was removed by blotting with filter paper, grids were then washed with 10 μl of distilled water for a further 10 s, prior to staining with 10 μl of 2% uranyl acetate for another 30 s. Excess stain was removed by blotting with filter paper. Grids were allowed to air dry prior to examination. Images were captured on a Phillips CM-10 electron microscope using Kodak SO-163 film. Micrographs were taken at ×52,000 or ×73,000 magnification. Negatives were scanned into a digital format using an Imacon Flextight 848 scanner. Digital micrographs were imported into the SPI-DER suite of programs and subjected to single-particle image processing (28). Particle picking was performed manually using the computer mouse to locate the approximate center. Windowed images were brought into mutual alignment by a reference-free strategy. Image classification was performed using K-means clustering (29). Harmonic analysis, incorporating rotational filtering, was conducted by Prof. P. Bullough (University of Sheffield).
Cross-linking of FLAG-p7
Dithiobis succinimidyl propionate (DSP, Pierce) of an appropriate working concentration (diluted from a 10 mM stock in Me2SO) was incubated with FLAG-p7 at a ratio of 20:1 (protein/DSP, v/v). Cross-linking was conducted at 20 °C for 30 min. The reaction was quenched by incubation with 1.5 M Tris-HCl, pH 7.5, at room temperature for 15 min. SDS-PAGE analysis of cross-linked samples was performed under non-reducing conditions.
Slow Crystallization Mass Spectrometry
Cross-linked FLAG-p7 was analyzed by MALDI-TOF mass spectrometry using a TofSpec 2E (Micromass, Manchester, UK) in linear mode. The sample was prepared using the slow crystallization method (30). Briefly, the protein sample (5 μl ofa1 μg/μl preparation in 4% SDS, 5% Me2SO) was thoroughly mixed with 20 μl of matrix solution (10 mg/ml α-cyano-4-hydroxycinnamic acid in formic acid/water/2-propanol (1:2:3, v/v) in a microcentrifuge tube. The tube was left with the lid open at 20 °C for 3 h, to allow protein-doped matrix crystals to form. Scratching the sides of the tube with a pipette tip helped promote crystal growth. The supernatant was removed, and the crystals were washed twice with deionized water. After the final wash the crystals were suspended in 2 μl of deionized water. A portion of the crystal suspension (0.5 μl) was applied to the MALDI target plate and allowed to dry. Samples were calibrated using a suitable standard protein prepared following the slow crystallization protocol.
Black Lipid Membrane Analysis
Lipids L-α-phosphatidylserine (porcine brain, sodium salt) and L-α -phosphatidyletha-nolamine (plant) were acquired from Avanti Polar Lipids. Stock solutions of individual lipids for black lipid membranes (BLMs) were made at 20 mg/ml in chloroform. Two chloroform solutions were then mixed 1:1 (v/v) before being dried to a thin film under nitrogen gas. Lipids were then resuspended by vortexing in n-decane (Sigma) to a final concentration of 40 mg/ml. Lipid preparations were made only on the day of the experiment. Aqueous solutions of 1 M KCl and 10 mM HEPES, pH 7.4, were passed through a 0.22-μm filter before use.
BLM experiments were performed in a polycarbonate cup and chamber. The cup contained a circular aperture of 200 μm in diameter, across which the lipids were painted after being primed with lipid in n-decane and dried under nitrogen gas. The aperture was the only connection between two chambers; cis and trans that each contained 600 μl of buffered salt solutions. Each chamber was connected to a 300-μl, 1 M KCl salt reservoir via a salt bridge (1 M KCl, 3% agar). An Ag/AgCl electrode was placed into each of the salt reservoirs, the trans electrode was connected to the input of a Geneclamp 500B voltage and patch clamp amplifier, and the cis electrode was connected to ground. Potentials were expressed as the trans chamber with respect to cis. FLAG-p7 dissolved in methanol was added to the cis chamber at 1 μM. No stirring mechanism was applied to either chamber.
Bilayer integrity was monitored by capacitance using a BC-200 bilayer capacitance measuring system. A stable capacitance of ≥115 picofarads was used as evidence of a bilayer of sufficient quality to show activity. Background capacitance of the system was 76 picofarads. All bilayers were formed with the amplifier set to the nanoamp range. This was adjusted to picoamp sensitivity after the bilayer (capacitance) was shown to be stable. Recordings were filtered at 200 Hz, and sampling was at 500 Hz. Recordings were made using the software Axoscope 9 (pClamp 9). Statistics, histograms, and representative traces were derived using the associated software Clampfit 9. BLM traces shown were further digitized at 50 Hz in the software using a low pass eight-pole Bessel filter.
Calculations Used in Ion Channel Analysis
Reversal potentials in asymmetrical buffers were calculated using the Nernst equation (Equation 1),
| (Eq. 1) |
where Erev is the reverse potential, R is the gas constant (8.3143 J/mol/degree), T is temperature (298 K), F is the Faraday constant (96,490 J/mol/V), and z is the charge and valency of the transported substance.
A derivation of the Goldman-Hodgkin-Katz equation was used to determine the relative permeabilities of potassium and chloride ions in the BLM system (Equation 2),
| (Eq. 2) |
where PK is the permeability of potassium ions, PCl is the permeability of chloride ions, and Erev is the reverse potential determined by the Nernst equation.
Results
Generation of High Purity GST-p7 Fusions from E. coli
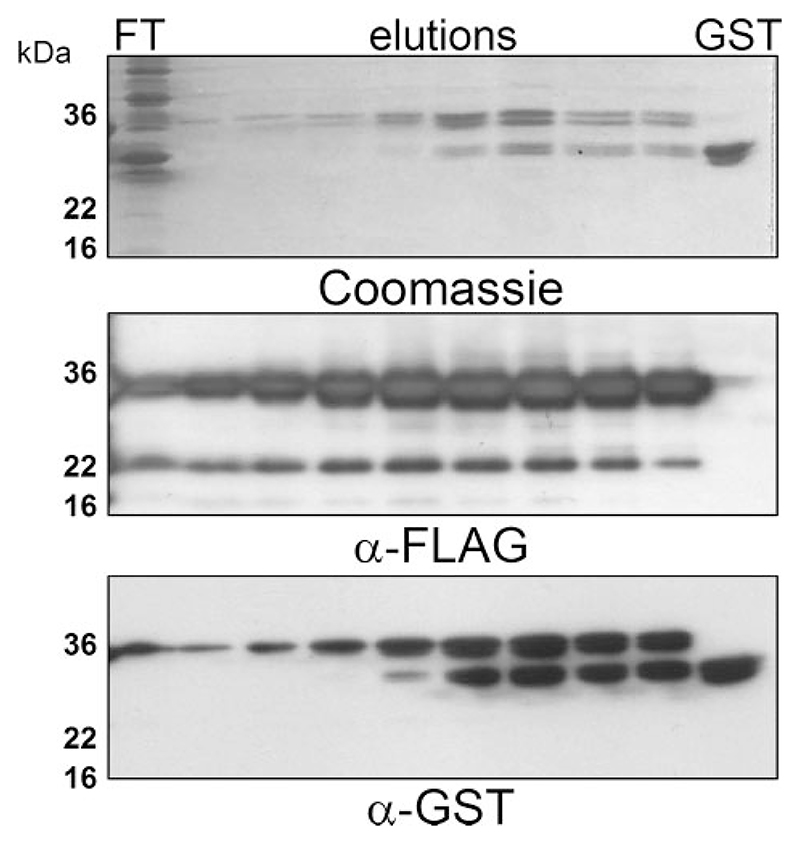
We previously described a GST-p7 fusion-based methodology for the purification of HCV p7 (9). This was based on the observation that fusion to GST rendered a portion of the fusion protein soluble, thereby allowing purification via glutathione-agarose chromatography. Immobilized fusion protein could then be cleaved using thrombin resulting in a near-native HIS6-tagged p7 protein with which we demonstrated the ion channel function of p7. Here we have further developed this methodology, replacing the His6 tag with a FLAG epitope, and incorporating a PreScission protease (Amersham Biosciences) cleavage site in place of a thrombin recognition sequence. The FLAG tag permits improved immunological detection of the protein and Pre-Scission protease avoids issues of protein purity associated with many commercially available thrombin preparations. Preparation of the fusion protein was performed as previously described (9), using Triton X-100 to generate a soluble protein fraction from sonicated bacteria. This fraction was then passed over a glutathione-agarose column and washed, and protein was eluted using reduced glutathione (Fig. 1). A degree of cleavage of the fusion protein occurred during preparations from soluble protein fractions, although the more hydrophobic p7 fusion protein was easily separated from GST by taking the early elutions from the column (Fig. 1). In addition, a band of 22 kDa was always observed. As the 22-kDa band was reactive to the anti-FLAG antibody in Western blots and not to the anti-GST antibody, it was thought to represent an N-terminal truncation of GST-FLAG-p7.
Figure 1. Purification of GST-FLAG-p7 from the soluble E. coli fraction SDS-PAGE.
Analysis of flow-through and elutions from a GSTrap affinity chrosmatography column (Amersham Biosciences). The column was loaded with the soluble fraction of E. coli lysates containing GST-FLAG-p7 after centrifugation at 20,000 X g for 15 min. Bound protein was eluted with 10 mM reduced glutathione. Upper panel: Coomassie staining; middle panel: Western blot using a mouse monoclonal anti FLAG-tag antibody; lower panel: Western blot using a mouse monoclonal anti-GST antibody. FT, flow through; GST, recombinant GST protein control.
GST-FLAG-p7 Forms Heptameric Oligomers in Membranes
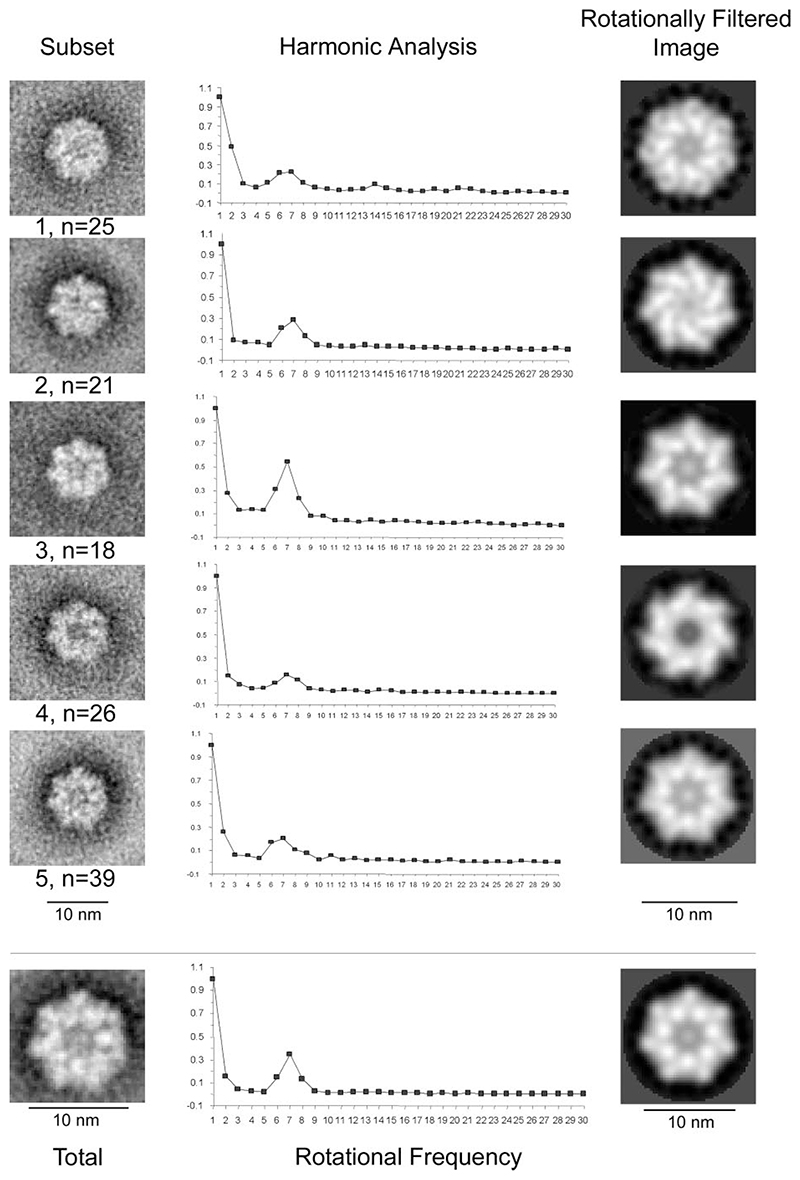
We previously reported the lipid-dependent formation of ring-like structures by GST-p7 fusion proteins in liposomes using TEM and estimated visually that these structures represented hexameric assemblages (9). Immunogold labeling showed that the GST moiety was present on the outside of the rings; the central electron dense core of between 3 and 5 nm was therefore surmised to contain the p7 portion of the fusion protein projecting into the plane of the liposome. To generate definitive data on stoichiometry using TEM images, however, it is necessary to apply computer-based rotational averaging and harmonic analysis to relatively large data sets to resolve individual components of protein structures: individual GST moieties in this instance. Large scale preparation of GST-FLAG-p7 from soluble bacterial protein fractions allowed us to revisit these experiments by using protein of improved concentration and purity, and thereby allowing the generation of several data sets, each containing over 100 individual particles. These data sets were deemed sufficiently large due to the mono-planar orientation of the complex within the liposome (one such representative data set used for Fig. 2; other data sets (not shown) were comparable). GST-FLAG-p7 was incubated with or without artificially synthesized PA:PC unilamellar vesicles (see “Experimental Procedures”) prior to negative staining and visualization. As seen previously for GST-HIS-p7, the presence of lipid promoted either the formation or stability of ring structures by the GST-FLAG-p7 fusion protein; no such structures were visible in the absence of membranes, nor when GST was added to liposomes (9) (data not shown). Data sets of individual complexes were then subjected to rotational symmetry analysis as described (see “Experimental Procedures”). For the data set shown, five image subsets were defined (Fig. 2). Averaged images were generated for each image subset and also for the entire data set (Fig. 2, left-hand panels). Harmonic analysis (Fig. 2, central panels) was performed on each of the averaged images using rotational frequencies of 1–30. In the majority of cases it was determined that 7-fold symmetry was optimal for GST-FLAG-p7, although the harmonic analysis of image subset 1 was unable to distinguish between optimal 6-fold or 7-fold rotational symmetry, as shown by the flat peak in Fig. 2 (top center). Additional resonance peaks for class 1 were observed at rotational frequencies of 14 and 21, however, implying an inherent 7-fold symmetry. The combined agreement between the harmonic analyses for the other image subsets and a global analysis of the complete data set plus other data sets (data not shown), supported the conclusion that GST-FLAG-p7 complexes were, in fact, comprised of heptamers and not hexamers as we previously described (9).
Figure 2. Harmonic analysis and rotational averaging of TEM images depicting GST-FLAG-p7 protein complexes.
GST-FLAG-p7 was incubated with liposomes containing equimolar amounts of PA and PC. Samples were then subjected to negative staining prior to visualization with TEM at ×52,000 magnification. Left, upper panels: averaged images of five GST-FLAG-p7 subsets, as defined using K-means clustering in the SPIDER suite of programs. Left, lower panel: averaged image of an entire data set of GST-FLAG-p7 complexes (n = 129). Center: harmonic analysis of the corresponding averaged image, using rotational frequencies of 1–30. Right: rotationally filtered images of the corresponding averaged image, derived from symmetry indicated by the harmonic analysis.
A rotationally filtered image was generated for each GST-FLAG-p7 subset, based on the results of the harmonic analysis (Fig. 2, right-hand panels). Images depict different conformations of the protein complex that may indicate a switch between different states, or a tolerance for flexibility within the heptameric assembly, although we have not ruled out the possibility that these observations could be due to differences in stain depth and/or penetration. Some of the rotationally filtered images exhibit opposing handedness (Fig. 2, subsets 2 and 4, and possibly subsets 3 and 5), which could represent top and bottom views of the same protein complex. In this eventuality, it is interesting that stain intensity varies at the center of the protein complexes, implying the presence of a funnel-like cavity. That the 7-fold symmetry had not been apparent from the comparatively small data sets obtained in our previous studies was not surprising, because it was not then possible to resolve the individual protein subunits. It is also possible that equivalent complexes to subgroup 1 created the impression of hexamers to the naked eye. The heptameric stoichiometry of these p7 complexes supports the notion that this oligomerization event is mediated by the p7 portion of the fusion protein and is not influenced by the propensity of GST to form homodimers as may have been argued for a hexameric complex.
Generation and Purification of Near-native FLAG-p7 from E. coli Inclusion Bodies
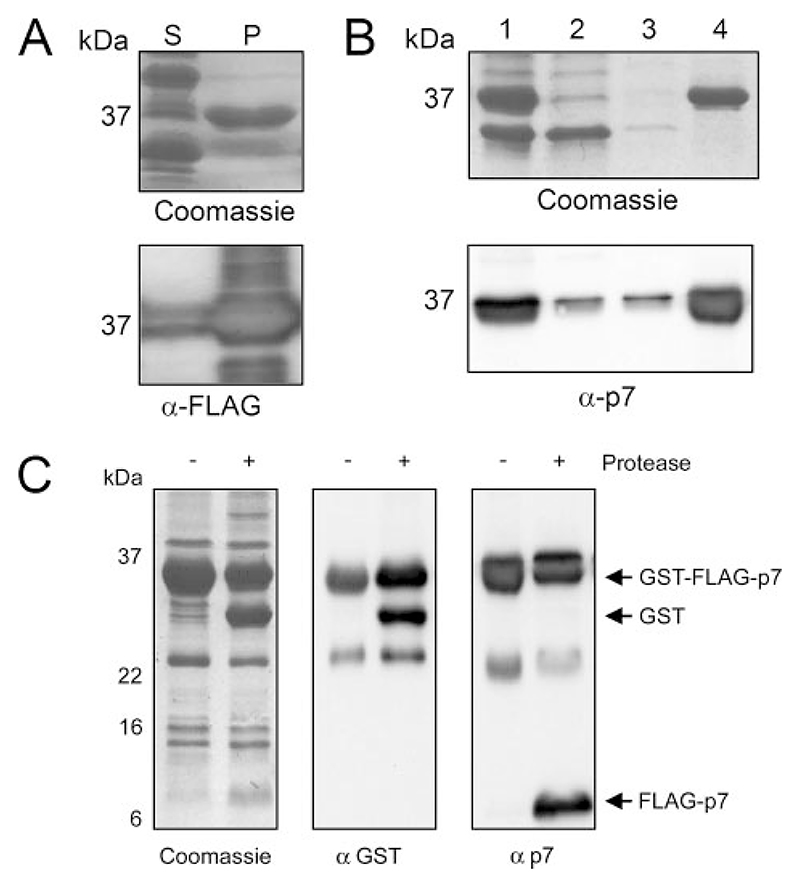
GST-FLAG-p7 generated from the soluble E. coli fraction was sufficient for use in TEM studies and biochemical analysis, although the initial yield of protein was not deemed sufficient to generate FLAG-p7 for prospective structural studies. In addition, it was difficult to maintain the solubility of FLAG-p7 following protease cleavage, which prohibited many affinity-based methods of protein purification that were necessary to remove the residual GST and uncleaved GST-FLAG-p7 from the sample. Upon examination of the insoluble fraction of E. coli-expressing GST-FLAG-p7, it was found that much of the fusion protein was retained in that fraction following the addition of Triton X-100 (Fig. 3A) and was found to remain within inclusion bodies (Fig. 3B). In addition, GST alone, which was always observed in soluble bacterial GST fusion fractions, was absent from inclusion bodies, thus increasing sample purity. These inclusion bodies were therefore separated by centrifugation, washed, and subjected to digestion with PreScission protease (Amersham Biosciences), which resulted in cleavage of ~50% of the protein as determined by Western blot analysis (Fig. 3C). In solubility studies, FLAG-p7 was found to be solubilized by methanol as we reported previously (9), as well as anionic, but not non-ionic detergents (data not shown). Cleaved protein was therefore solubilized by adjusting the cleavage reaction to 0.5% (w/v) N-laurylsarcosine, which was used in preference to SDS due to its lesser propensity to denature protein as well as its solubility at low temperatures. FLAG-p7 was detectable using a specific antibody, #1055 (27), against the p7 C terminus as well as by an anti-FLAG monoclonal antibody to the N terminus, indicating that the near-native p7 protein was intact and did not degrade during the purification process (Figs. 3C and 5, and data not shown).
Figure 3. Isolation of GST-FLAG-p7 from E. coli inclusion bodies.
SDS-PAGE analysis of the methods used to improve the yield and purity of the 36-kDa GST-FLAG-p7 protein from the insoluble E. coli fraction after expression. A, comparison of the soluble and insoluble E. coli fractions after centrifugation at 20,000 × g for 15 min. S, soluble fraction; P, pellet (representing insoluble fraction). Upper, Coomassie staining; lower, Western blot using a mouse monoclonal anti FLAG tag antibody. B, isolation of inclusion bodies from the insoluble E. coli fraction. Lanes: 1, Total E. coli lysate; 2, supernatant following centrifugation at 3,000 × g for 15 min; 3, supernatant of centrifugation at 3,000 × g for 15 min following resuspension of inclusion bodies in 100 mM ammonium acetate; 4, inclusion bodies resuspended in 50 mM Tris-HCl, 150 mM NaCl, pH 7.0. Upper, Coomassie staining; Lower, Western blot using affinity-purified rabbit anti p7 antibody, #1055. C, cleavage of FLAG-p7 from GST-FLAG-p7 using PreScission protease (Amersham Biosciences). Left, Coomassie staining; middle, Western blot using a mouse monoclonal anti GST antibody; right, Western blot using affinity purified rabbit anti-p7 antibody, 1055.
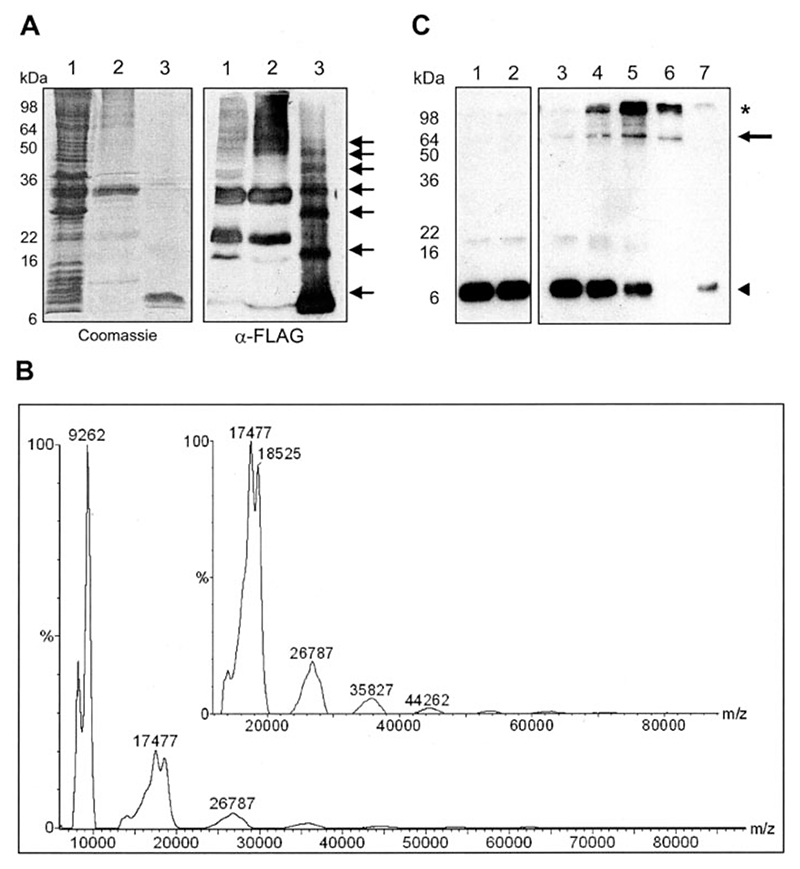
Figure 5. Formation of stable, high molecular weight forms of FLAG-p7.
A, concentrated near-native FLAG-p7, as purified by HPLC, was seen to form seven different molecular weight species by SDS-PAGE. Left, Coomassie staining; right, Western blot. Lanes: 1, GST-FLAG-p7-expressing E. coli lysate; 2, inclusion bodies containing GST-FLAG-p7; 3, protease-released FLAG-p7, HPLC-purified. B, slow crystallization MALDI-TOF mass spectrometry of FLAG-p7, cross-linked with 0.5 mM DSP in 4% SDS. Peaks were calibrated against both low and high molecular weight standards. The inset shows an expanded view of the region from 13 to 80 kDa. Low molecular weight species were calibrated against cytochrome c and high molecular weight species (inset) were calibrated against bovine serum albumin. C, Western blot of a non-reducing SDS-PAGE with a monoclonal anti-FLAG antibody of FLAG p7 from the soluble E. coli fraction. Protein was incubated with the lipid-soluble, thiol-cleavable cross-linking reagent, DSP, at a range of concentrations in the presence of PA:PC liposomes. Lanes: 1, FLAG-p7 only; 2, FLAG-p7 plus Me2SO; 3–6, FLAG-p7 plus DSP (0.01/0.05/0.1/0.5 mM); 7, FLAG-p7 plus 0.5 mM DSP incubated in the presence of 10 mM DTT prior to gel loading. The arrow indicates the FLAG-specific band of apparent molecular mass 64 kDa; the asterisk indicates high molecular weight aggregates. ◭, monomeric p7.
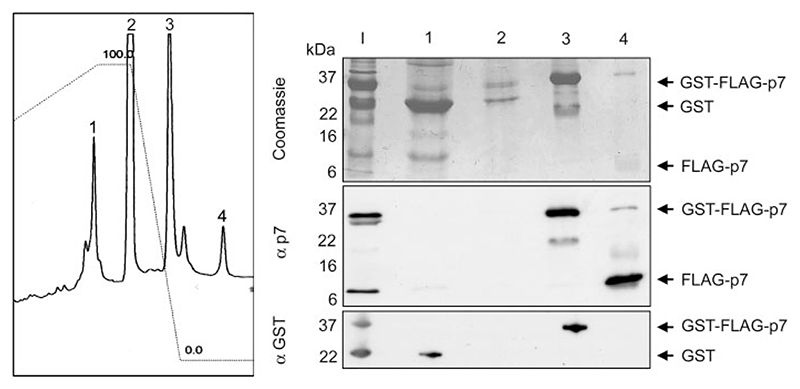
Solubilization of FLAG-p7 permitted its purification away from other components of the cleavage reaction by HPLC using a 0 to 80% acetonitrile gradient on a C4 analytical column. Four discrete peaks were obtained upon elution (Fig. 4, left panel); the first containing pre-dominantly GST, the second mainly N-laurylsarcosine bound to the column, the third comprised uncleaved fusion protein, whereas the last peak, eluted at the highest acetonitrile concentration, contained >95% FLAG-p7 as estimated by Coomassie staining and Western blotting of the four peak fractions using specific antibodies (Fig. 4).
Figure 4. HPLC purification of near-native FLAG-p7.
A C4 HPLC chromatography column was used to purify FLAG-p7 from major contaminants on the basis of hydrophobicity, using a linear acetonitrile gradient. Left, absorbance (215 nm) trace of eluates from an analytical C4 HPLC column. The column was loaded with GST-FLAG-p7 inclusion bodies, cleaved with PreScission protease, and solubilized with 0.5% (w/v) N-laurylsarcosine. Right, SDS-PAGE analysis of HPLC column eluates, with lanes 1–4 corresponding to the peaks on the HPLC trace. Lane I, input sample. Upper, Coomassie staining; middle, Western blot using affinity-purified rabbit anti p7 antibody, 1055; lower, Western blot using a mouse monoclonal anti-GST antibody.
N-Lauryl Sarcosine-solubilized FLAG-p7 Forms Detergentresistant Oligomers
Scaling up the production of FLAG-p7 using a preparative C4 column in place of the analytical column resulted in the production of large quantities of the cleaved FLAG-p7 product; 2.5 mg from 1 liter of bacterial culture. Protein was lyophilized and stored at −20 °C. To check purity, lyophilized protein samples were directly resuspended in Laemmli buffer and subjected to SDS-PAGE and Coomassie staining or Western blot analysis (Fig. 5A). During the course of this study we observed that the FLAG-p7 species does not stain strongly with Coomassie in comparison to other protein species (e.g. GST), we therefore overloaded the SDS-PAGE with a large amount of the purified protein (20 μg per lane) to permit the effective visualization of this species. A minor band migrating slightly faster than the predominant FLAG-p7 species may represent a proteolytic degradation product or may be an artifact of the gel system as we have previously shown that FLAG-p7 expressed in mammalian cells migrates as two distinct species on SDS-PAGE (27). Interestingly, Western blotting revealed a series of higher molecular weight species that were reactive with both the anti-FLAG and anti-p7 antibodies. This observation was consistent with the presence of oligomeric forms of p7. Furthermore, the spacing of the higher molecular weight species appeared to be uniform, indicating that complex formation likely occurs via recruitment of monomers rather than by association of intermediate species, although the latter cannot be ruled out. The oligomers were apparently resistant to the usually fully denaturing SDS concentration present within the buffer (4% w/v) as well as a reducing environment (100 mM DTT) and heating to 100 °C for 1 min prior to loading. In this regard they are reminiscent of oligomers formed by bovine viral diarrhea virus p7 under similar conditions (16).
As SDS-PAGE only provides information about the apparent molecular weight of protein species we sought to use mass spectrometry to confirm the nature and mass of the higher molecular weight oligomeric species. The difficulty of obtaining mass spectrometric signals from protein in 4% SDS, 5% Me2SO was overcome using the slow crystallization method to prepare samples for MALDI-TOF analysis (30). Using this method, the mass of monomeric FLAG-p7 has been determined to within 3 Da of the expected molecular mass: 9059 Da (data not shown). In all cases, lower molecular weight species, deemed to be a minority of truncated FLAG-p7 protein, have been observed.
To stabilize higher molecular weight species prior to MALDI-TOF analysis, purified FLAG-p7 was cross-linked using the lipid-soluble cross-linker DSP at a concentration of 0.5 mM. As can be seen (Fig. 5B) this analysis revealed the presence of uniformly spaced peaks separated by mass differences consistent with FLAG-p7 monomers. As observed in the SDS-PAGE analysis (Fig. 5A), the mass spectrometric data showed that the sample was predominately monomeric, however, peaks up to the equivalent of a heptameric form could be elucidated. Taken together, these observations suggest that anionic detergents such as SDS or N-laurylsarcosine may create a hydrophobic environment mimicking that of a lipid bilayer, thereby facilitating the protein-protein interactions necessary for p7 oligomer formation. However, under the experimental conditions necessary to solubilize FLAG-p7, the protein was not able to form a unique, stable, heptameric species.
Cross-linking of FLAG-p7 Oligomers
Because the Triton X-100-solubilized GST-FLAG-p7 protein was able to form stable heptamers in the presence of liposomes as determined by TEM (Fig. 2), we carried out a further DSP cross-linking analysis of FLAG-p7 obtained from the soluble E. coli fraction, again in the presence of unilamellar liposomes. In this case, an anti-FLAG-reactive protein species was observed at ~64 kDa in Western blots and not at 9 kDa as was typical for the monomeric species (Fig. 5C). This was consistent with FLAG-p7 forming a stable heptameric species. To confirm that the 64-kDa species did indeed represent a cross-linked oligomer, 10 mM DTT was added to cleave the thiol-cleavable cross-linking reagent prior to SDS-PAGE analysis, upon which the intensity of the 64-kDa band was reduced and the 9-kDa monomeric species reappeared.
Characterization of Ion Channels Formed by FLAG-p7 in Suspended Lipid Bilayers
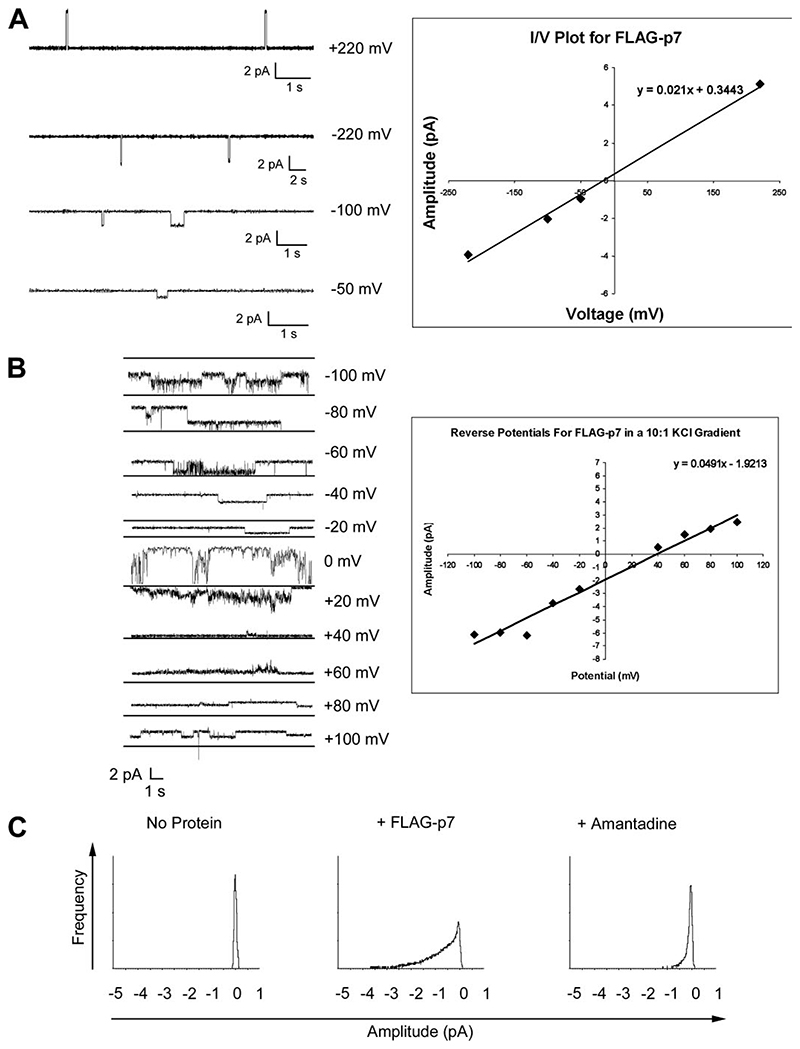
We previously used the BLM system and recombinant His-tagged p7 protein to provide the first description of the p7 ion channel function (9). Since then, synthetic p7 peptides have been characterized giving more detailed electrophysiological information on p7 ion channel complexes (10, 11). Therefore, to validate our system as a means of obtaining structural information on functional p7 ion channels, it was important to show that bacterially expressed FLAG-p7 behaved similarly. Initially, single channel opening events were analyzed under symmetrical buffer conditions in a potassium electrolyte. Under these conditions, the Nernst equation dictates that the linear current (I): voltage (V) ratio (I/V) conductance should pass through zero in an ideal system (Fig. 6A). The linear distribution for FLAG-p7 did indeed pass near to zero under the given conditions indicating that, within the limits of experimental error, FLAG-p7 functioned as a true ion channel with a conductance of 21 picosiemens.
Figure 6. Recombinant FLAG-p7 forms amantadine-sensitive ion channels in BLMs.
BLMs were established by painting an equimolar solution of L-α-phosphatidylserine and L-α-phosphatidylethanolamine in N-decane across a 200-μm aperture separating two 300-μl buffer chambers, each connected to an Ag/AgCl electrode. Recombinant FLAG-p7 was added to the cis buffer chamber, and the current flow was monitored for ion channel activity. A: Left, examples of single ion channel events in symmetrical KCl buffers at variable applied potentials. Right, linear relationship of I/V for FLAG-p7-mediated ion channel events in symmetrical buffer conditions at variable applied potentials, shown on the left. B: determination of the reverse potential for FLAG-p7 in asymmetrical buffer conditions, using a 10:1 KCl gradient. Left, ion channel activity measured at variable applied potentials. Right, linear I/V relationship for FLAG-p7-mediated ion channel data, shown on the left. C, all points histograms of FLAG-p7-mediated ion channel events in the presence of amantadine.
As well as discrete single channel events, FLAG-p7 ion channel activity was also observed to adopt a more erratic “burst activity” pattern; presumably the result of multiple and varied channel opening events, which has also been noted by other investigators using synthetic p7 peptides (10). This pattern was most common in experiments involving asymmetric buffer conditions used to define ion preferences via determination of reverse potentials. By plotting the amplitude of the signal against the potential it is possible to derive a theoretical reverse potential for FLAG-p7 in asymmetrical KCl buffer conditions (Fig. 6B). In deriving this value it should be noted that the values at 0 mV and +20 mV were excluded from the graph due to poor resolution. This analysis defined the reverse potential for FLAG-p7 as +39.13 mV, in good agreement for the value of +35.5 mV obtained using a synthetic p7 peptide (10). We conclude that FLAG-p7 ion channels are indeed cation-selective but also permit the flow of chloride ions, albeit to a far lesser extent: the relative permeability to potassium ions over chloride ions is 7.9:1.
Finally, it was necessary to show that the channels formed by FLAG-p7 could be inhibited by amantadine as had been the case for the HIS-p7 protein used previously (9). This would also demonstrate that different affinity tags placed at the N terminus of the p7 sequence did not affect ion channel function. To assess the effects of amantadine on FLAG-p7 ion channel activity, we analyzed the burst activity of FLAG-p7 over prolonged time periods. The amplitude of current flow was then plotted against frequency of channel opening events as an all-points histogram (Fig. 6C). As expected, in the absence of protein the histogram peaked at 0 pA indicating a low baseline activity in all cases, whereas upon the addition of protein a second peak or shoulder can be seen on the graph reflecting the frequency of channel opening events over time. As each of these experiments was performed with a separate bilayer preparation, the variation in the shape of the graphs upon addition of FLAG-p7 most likely reflects the inherent variability in the composition of the bilayer. After the induction of burst activity at a patched voltage of −100 mV, the voltage was reduced to zero and burst activity continued; presumably as a result of the electrochemical gradient acting upon channels already in the membrane. Upon the addition of amantadine, the curve can be seen to shift back toward the zero point reflecting inhibition of ion channel activity. Others have derived kinetic data for such inhibitor studies based on the inhibitor concentration in solution and the amount of protein added to the system (10). However, we have not performed such calculations, because one cannot determine what proportion of the protein is resident in the bilayers at a given time, nor how many functional complexes are responsible for such activity, even when observing supposed single channel events.
Discussion
This work validates our bacterial expression system as a means of investigating the structure of a functional hepatitis C virus p7 ion channel complex. Production of p7 as a fusion with GST still allows it to fold correctly within membranes promoting interactions with other p7 moieties and the formation of what are presumably ion channel complexes. Computer enhancement of TEM-imaged complexes in liposomes revealed a heptameric stoichiometry that was predominant over homodimeric interactions between the GST portions of the fusion protein.
Near-native FLAG-p7 produced following cleavage of the fusion protein was also seen to form oligomeric complexes by SDS-PAGE under conditions of high protein concentration. In this latter case, oligomers were stable in the presence of high SDS concentrations and a reducing environment. This means that disulfide linkages are unlikely to be involved in the formation of p7 oligomers and that SDS micelles may serve to emulate conditions necessary for hydrophobic protein-protein interactions that potentially mediate oligomerization. The oligomerization of FLAG-p7 in SDS micelles was confirmed by analysis of cross-linked protein using slow crystallization mass spectrometry.
Although under these conditions the higher order oligomers were only present at very low abundance, compared with the monomer and dimer forms of FLAG-p7, this may be explained by the steric constraints imposed by the structure of the oligomer on the cross-linking reaction. DSP will cross-link primary amines of which there are four in each monomer: three near the N terminus (the N terminus itself and two within the FLAG tag) and the fourth within the cytosolic loop. It is possible that in 4% SDS stable cross-linking of successive monomers becomes increasingly unfavorable due to the lack of proximity of the amine groups. However, it appears that in what might be considered more physiological conditions (i.e. in the presence of unilamellar liposomes) the FLAG-p7 could be cross-linked as a stable heptamer, mirroring the data obtained by TEM analysis of GST-FLAG-p7 in liposomes.
Other viroporins studied to date have also been shown to self-associate into oligomeric helical bundles under conditions mimicking that within lipid bilayers. The bitopic influenza A virus (IAV) M2 protein and human immunodeficiency virus, type 1 Vpu protein have been predicted to form tetrameric proton channels and pentameric cation channels, respectively (31, 32). The only other polytopic viroporin studied, picornaviral 2B protein, is also thought to form a tetrameric channel when cross-linked as a maltose-binding protein fusion protein (33), although the propensity of maltose-binding protein to tetramerize could be a contributing factor to this observation. The finding that p7 forms a heptamer is inconsistent with a predominant dimeric nature of GST. However, it agrees with the notion proposed by others (10) that p7 must form at least a pentameric complex to support its observed conductance states. In addition, we previously observed that p7 from bovine viral diarrhea virus appears to form higher molecular weight complexes both in vitro and in cell culture and these were of similar stability to p7 oligomers formed at high protein concentration in this study (16).
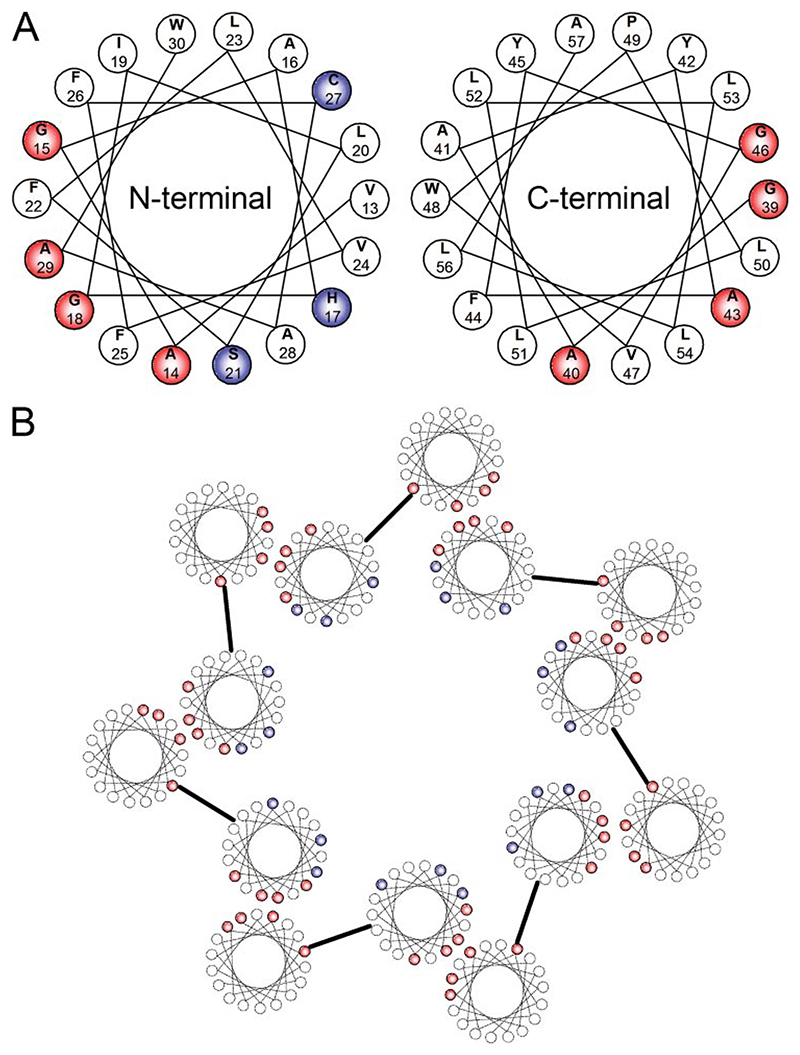
As a bitopic protein, each helix of the IAV M2 oligomer contributes essential residues for ion channel function, i.e. HIS37, to the lumen of the channel. If p7 is to form a similar channel, it is proposed that only one of the two trans-membrane helices will face the channel interior, forming a hydrophilic pore. Helical wheel diagrams show that the N-terminal helix of p7 is likely to be the most amphipathic of the two (Fig. 7A) (8). The hydrophilic residues His, Cys, and Ser could then contribute to the pore interior, because they are all predicted to lie on one side of the N-terminal helix. We previously modeled such a complex based on our initial estimate of hexameric stoichiometry. A revised model for the packing of p7 within the heptamer is proposed, with the N-terminal helix at its center (Fig. 7B).
Figure 7. A model for interactions in the assembly of p7 heptamers.
A, helical wheel diagrams of each of the two predicted helices of p7. Amino acid residues shown in red are small neutral residues that are clustered on one face of each helix, potentially involved in helix-helix interactions. Amino acid residues shown in blue are more hydrophilic residues that contribute to the amphipathicity of the N-terminal helix. Included is histidine 17, which may be similar to an analogous histidine that projects into the lumen of the M2 proton channel of IAV, conferring both ion conductance and amantadine sensitivity. B, proposed model of a heptameric p7 assembly, wherein the small neutral cluster on the N-terminal helix interacts with the analogous cluster on the C-terminal helix, allowing the more hydrophilic residues to reside within the lumen of the pore.
The residues responsible for oligomerization of p7 have not yet been identified, although the study of other ion channels allows the identification of some key residues that should be the target of further investigation. The single cysteine that lies within the N-terminal helix of p7 is unlikely to be involved, because the protonation of a thiol group is not usually permitted in such a dielectric environment as the lipid bilayer. Indeed, oligomers were also observed in this study under reducing conditions. It has been shown for M2, however, that disulfide bonds between adjacent ectodomains help to stabilize the tetrameric complex (31). Asparagine residues located outside of the trans-membrane region have been implicated in oligomerization for Coxsackievirus 2B protein (34). There are two well conserved asparagine residues within the p7 sequence, both toward the N terminus in regions thought to lie outside of the trans-membrane domains (Asn-4 and Asn-9 in the 1b genotype), although these are not present in the genotype 2a JFH-1 sequence, which is able to undergo a complete replication cycle in culture (35–37). Alternatively, a motif resembling a leucine zipper (typically used for oligomerization of soluble proteins) has been identified in the oligomerization of at least one membrane protein (38). p7 has a conserved heptad motif of leucines toward its C terminus in the second trans-membrane α-helix, however, this sequence is not present in the p7 protein of JFH-1.
Most intra-membrane interactions are brought about by hydrophobic interactions that involve direct inter-helix association. In the majority of membrane proteins assessed so far, small neutral residues such as glycine and alanine are implicated in the formation of structurally complementary regions on adjacent helical faces. Two main consensus sequences for hydrophobic inter-helical oligomerization have been identified, Gly-X-X-X-Gly (39), and Gly-Val-X-X-Gly-Val-X-X-Thr (or Ser) (40). Although p7 does not possess either of these motifs, it is tempting to speculate from the helical wheel diagrams that there is a potential hydrophobic binding site on the face of each helix. The N-terminal helix of the J4, genotype 1b p7 sequence has a face comprising residues Ala-14, Gly-15, Gly-18, and Ala-29. The C-terminal helix has a face comprising residues Gly-39, Ala-40, Ala-43, and Gla-46. These residues are highly, although not totally, conserved throughout published HCV sequences (data not shown and Ref. 8). Together, two of the best conserved residues, Ala-14 and Gly-18 make the consensus sequence Ala-X-X-X-Gly, similar to the Gly-X-X-X-Gly motif mentioned previously. In the model proposed these regions would imply a potential hydrophobic interaction between helices 1 and 2 of adjacent p7 monomers within the oligomeric complex, at the same time allowing the polar residues (His, Cys, and Ser of helix 1) to face the lumen of the pore. Production of GST-p7 fusion proteins containing substitution mutants for these and other potentially important residues is ongoing and should reveal functional determinants for ion channel formation.
FLAG-p7 produced in a bacterial system displayed similar electrophysiological characteristics to synthetic p7 peptides used by other investigators. We again showed that amantadine was able to inhibit the flow of ions through the p7 ion channel complex. These findings suggest that bacterial recombinant p7 protein is able to form a complex corresponding to that which likely exists in infected cells, making it ideal for future rational drug design.
Given the existence of p7-like proteins within genomes of other Flaviviridae, e.g. bovine viral diarrhea virus and GB virus B, it seems likely that the function of p7 or its equivalent may be well conserved, even between different virus species, yet sequence differences may contribute to variations in the ion channel complexes formed by different viruses. Indeed, unusually for the highly conserved p7 region of the HCV genome, genotype 2a p7, such as that of JFH-1 (the current method of choice for studying HCV replication in cell culture), is the most sequence-divergent from genotype 1 of all p7 sequences we have assessed to date. It is likely, therefore, that future inhibitors of p7 will have to be tested on multiple genotypes to assess potential clinical efficacy; a purpose for which the bacterial expression system described here for p7 is well suited.
Acknowledgments
We are grateful to Prof. Per Bullough (University of Sheffield) for conducting the harmonic analysis of TEM images, Catherine Botting (University of St. Andrews) for the mass spectrometric analysis, and Malcolm Hunter and Paul Bacchus (University of Leeds) for invaluable help with electrophysiology. We also thank Helen Bright and Tony Carroll (GlaxoSmithKline, Stevenage) for useful discussions.
Footnotes
The abbreviations used are: HCV, hepatitis C virus; BLM, black lipid membrane; DSP, dithiobis succinimidyl propionate; ECL, enhanced chemiluminescence; GST, glutathione S-transferase; IAV, influenza A virus; M2, matrix protein 2; TEM, transmission electron microscopy; Vpu, virus protein U; PA, phosphatidic acid; PC, phosphatidylcholine; DTT, dithiothreitol; HPLC, high-performance liquid chromatography; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight.
References
- 1.Pawlotsky JM. Curr Opin Infect Dis. 2003;16:587–592. doi: 10.1097/00001432-200312000-00012. [DOI] [PubMed] [Google Scholar]
- 2.Simmonds P. Gut. 1997;40:291–293. doi: 10.1136/gut.40.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deltenre P, Henrion J, Canva V, Dharancy S, Texier F, Louvet A, De Maeght S, Paris JC, Mathurin P. J Hepatol. 2004;41:462–473. doi: 10.1016/j.jhep.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Science. 1989;244:359–362. doi: 10.1016/s0168-8278(02)00051-x. [DOI] [PubMed] [Google Scholar]
- 5.Lohmann V, Koch JO, Bartenschlager R. J Hepatol. 1996;24:11–19. [PubMed] [Google Scholar]
- 6.Moradpour D, Gosert R, Egger D, Penin F, Blum HE, Bienz K. Antiviral Res. 2003;60:103–109. doi: 10.1016/j.antiviral.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Lin C, Lindenbach BD, Pragai BM, McCourt DW, Rice CM. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrere-Kremer S, Montpellier-Pala C, Cocquerel L, Wychowski C, Penin F, Dubuisson J. J Virol. 2002;76:3720–3730. doi: 10.1128/JVI.76.8.3720-3730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin SD, Beales LP, Clarke DS, Worsfold O, Evans SD, Jaeger J, Harris MP, Rowlands DJ. FEBS Lett. 2003;535:34–38. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 10.Pavlovic D, Neville DC, Argaud O, Blumberg B, Dwek RA, Fischer WB, Zitzmann N. Proc Natl Acad Sci U S A. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Premkumar A, Wilson L, Ewart GD, Gage PW. FEBS Lett. 2004;557:99–103. doi: 10.1016/s0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- 12.Sakai A, Claire MS, Faulk K, Govindarajan S, Emerson SU, Purcell RH, Bukh J. Proc Natl Acad Sci U S A. 2003;100:11646–11651. doi: 10.1073/pnas.1834545100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harada T, Tautz N, Thiel HJ. J Virol. 2000;74:9498–9506. doi: 10.1128/jvi.74.20.9498-9506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez ME, Carrasco L. FEBS Lett. 2003;552:28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 15.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin SD, Harvey R, Clarke DS, Barclay WS, Harris M, Rowlands DJ. J Gen Virol. 2004;85:451–461. doi: 10.1099/vir.0.19634-0. [DOI] [PubMed] [Google Scholar]
- 17.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. Proc Natl Acad Sci U S A. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Z, Kovacs FA, Wang J, Denny JK, Shekar SC, Quine JR, Cross TA. Biophys J. 2000;79:767–775. doi: 10.1016/S0006-3495(00)76334-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salom D, Hill BR, Lear JD, DeGrado WF. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stouffer AL, Nanda V, Lear JD, DeGrado WF. J Mol Biol. 2005;347:169–179. doi: 10.1016/j.jmb.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 21.Sakaguchi T, Tu Q, Pinto LH, Lamb RA. Proc Natl Acad Sci U S A. 1997;94:5000–5005. doi: 10.1073/pnas.94.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duff KC, Ashley RH. Virology. 1992;190:485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- 23.Pinto LH, Holsinger LJ, Lamb RA. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 24.Tang Y, Zaitseva F, Lamb RA, Pinto LH. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Kim S, Kovacs F, Cross TA. Protein Sci. 2001;10:2241–2250. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanagi M, St Claire M, Shapiro M, Emerson SU, Purcell RH, Bukh J. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 27.Griffin S, Clarke D, McCormick C, Rowlands D, Harris M. J Virol. 2005;79:15525–15536. doi: 10.1128/JVI.79.24.15525-15536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies, Visualization of Biological Molecules in Their Native State. Oxford University Press; Oxford, UK: 2005. [Google Scholar]
- 29.Burgess SA, Walker ML, White HD, Trinick J. J Cell Biol. 1997;139:675–681. doi: 10.1083/jcb.139.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Botting CH. Rapid Commun Mass Spectrom. 2000;14:2030–2033. doi: 10.1002/1097-0231(20001115)14:21<2030::AID-RCM129>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Holsinger LJ, Lamb RA. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 32.Kukol A, Arkin IT. Biophys J. 1999;77:1594–1601. doi: 10.1016/S0006-3495(99)77007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agirre A, Barco A, Carrasco L, Nieva JL. J Biol Chem. 2002;277:40434–40441. doi: 10.1074/jbc.M205393200. [DOI] [PubMed] [Google Scholar]
- 34.de Jong AS, Melchers WJ, Glaudemans DH, Willems PH, van Kuppeveld FJ. J Biol Chem. 2004;279:19924–19935. doi: 10.1074/jbc.M314094200. [DOI] [PubMed] [Google Scholar]
- 35.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 36.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Barten-schlager R, et al. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurezka R, Laage R, Brosig B, Langosch D. J Biol Chem. 1999;274:9265–9270. doi: 10.1074/jbc.274.14.9265. [DOI] [PubMed] [Google Scholar]
- 39.Russ WP, Engelman DM. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 40.Orzaez M, Lukovic D, Abad C, Perez-Paya E, Mingarro I. FEBS Lett. 2005;579:1633–1638. doi: 10.1016/j.febslet.2005.01.078. [DOI] [PubMed] [Google Scholar]