Abstract
Metastatic breast cancer cells disseminate to organs with a soft microenvironment. Whether and how local tissue mechanical properties influence their response to treatment remains unclear. Here we found that a soft ECM empowers redox homeostasis. Cells cultured on a soft ECM display increased peri-mitochondrial F-actin promoted by Spire1C and Arp2/3 nucleation factors, and increased DRP1- and MIEF1/2-dependent mitochondrial fission. Changes in mitochondrial dynamics lead to increased mtROS production and activate the NRF2 antioxidant transcriptional response, including increased cystine uptake and glutathione metabolism. This retrograde response endows cells with resistance to oxidative stress and ROS-dependent chemotherapy drugs. This is relevant in a mouse model of metastatic breast cancer cells dormant in the lung soft tissue, where inhibition of DRP1 and NRF2 restored cisplatin sensitivity and prevented disseminated cancer cell awakening. We propose that targeting this mitochondrial dynamics- and redox-based mechanotransduction pathway could open avenues to prevent metastatic relapse.
The physical and mechanical properties of the tissue microenvironment, and the ensuing forces to which cells are subjected, are important clues driving cell behavior. Among these, extracellular matrix (ECM) stiffness is a universal signal that controls cell proliferation, differentiation, and death. ECM stiffness is an important regulator of physiological tissue function and contributes to aberrant cell behavior observed in disease 1-4.
Cells measure ECM stiffness by actively developing myosin-dependent contractile forces through the actin cytoskeleton and integrin-mediated adhesions. Downstream of this core system, forces are then translated into biochemical signals to enable a coherent biological response, including the rapid reinforcement of the actomyosin cytoskeleton and of focal adhesions, and the regulation of gene expression 5,6.
In cancer, much attention has been devoted to stiffening of the primary tumor microenvironment, which promotes tumor growth and invasiveness 7. However, when breast cancer cells disseminate to distant organs such as the bone marrow, brain, lung and liver they find themselves in a soft environment, and the consequences of this new mechanical environment for metastatic growth and therapy are unknown 8,9. Given the importance of metabolism for cancer therapy, and the recent observation that ECM mechanical cues regulate metabolism 10, we explored metabolic traits potentially relevant for therapy and regulated by ECM mechanical cues.
Results
ECM mechanotransduction regulates glutathione metabolism
Metabolic profiling previously indicated that MCF10AneoT human transformed mammary epithelial cells (MCF10A-RAS) treated with ROCK (Rho-associated kinase) and MLCK (myosin light chain kinase) inhibitors, which recapitulates the reduced cell contractility observed on a soft ECM, display increased lipid synthesis 11. In the same dataset we also observed a striking increase in both cystine, the disulfide form of cysteine that is transported across the plasma membrane, and of cysteine, occurring as late events (Fig. 1a,b). This increase in cysteine was associated with a significant increase in all the intermediates of the glutathione cycle, where cysteine is used for glutathione synthesis (Fig. 1c, see scheme in Extended Data Fig. 1a). We observed increased cystine uptake also in cells treated with the Non-muscle Myosin Type-2 (NMII) inhibitor Blebbistatin, and in cells cultured on soft or stiff hydrogels (Fig. 1d and Extended Data Fig. 1b). Moreover, similar results were obtained using MCF10A immortalized human mammary epithelial cells and mouse D2.0R metastatic breast cancer cells (Fig. 1e,f), indicating a general effect of ECM stiffness on cysteine metabolism.
Fig. 1. ECM stiffness regulates cystine metabolism and glutathione oxidation.
a-c Levels of intracellular metabolites in MCF10A-RAS cells treated with vehicle (DMSO, n=4 biologically independent samples) or with the Y27632 ROCK inhibitor and ML7 MLCK inhibitor (hereafter YM, n=6 biologically independent samples) as measured by mass spectrometry. Data from one single metabolomics experiment. Welch's two-sample t-tests. d-f, Uptake of FITC-labeled Cystine by flow cytometry in MCF10A-RAS (d), MCF10A (e) and mouse D2.0R cells (f) treated with YM, with the Blebbistatin (BLEBBI) NMII myosin inhibitor for 6 h, or cultured on stiff (E≈15kPa) or soft (E≈0.5kPa) Fibronectin-coated acrylamide hydrogels. Normalized to mean intensity in control (in d n=11 biologically independent samples pooled across three independent experiments for DMSO; n=6 (YM) n=9 (BLEBBI) n= 4 (STIFF) n=6 (SOFT) biologically independent samples pooled across two independent experiments. In e n=5 biologically independent samples pooled across two independent experiments for DMSO, YM and BLEBBI; n=2 (STIFF) and n=4 (SOFT) biologically independent samples pooled across two independent experiments; Dunnet’s tests). g-h, Levels of reduced (GSH, g) and oxidized (GSSG, h) glutathione in MCF10A-RAS cells treated and analyzed as in a. i, Glutathione redox analysis with the cytoplasmic Grx1-roGFP2 genetically-encoded sensor in MCF10A-RAS treated with YM (n=32 (DMSO) n=20 (YM3h) n=29 (YM6h) n=24 (YM24h) cells pooled across two independent experiments; Dunnet’s test). j, Glutathione redox analysis in MCF10A-RAS cells cultured on stiff Matrigel-coated plates (E≈GPa) or soft Matrigel thick gels (E≈2o0Pa) (n=31 (STIFF) n=32 (SOFT) cells pooled across two independent experiments; unpaired two-tailed Student’s t-test).
Data are mean and single points. See Source Data Table 1.
ECM stiffness regulates glutathione redox homeostasis
In cells with reduced ROCK and MLCK activity, reduced glutathione (GSH) levels were initially reduced and subsequently recovered (Fig. 1g), while oxidised glutathione (GSSG) showed a late increase (Fig. 1h). This suggested initial oxidation of GSH, followed by increased GSH synthesis together with continued oxidation. We validated this idea using cytoplasmic and mitochondrial ratiometric glutathione sensors 12, which confirmed an early and sustained shift towards glutathione oxidation in cells with reduced contractility and on a soft ECM (Fig. 1i,j and Extended Data Fig. 1c,d - see below, Fig. 8f,k for similar data in D2.0R in vitro and in vivo). This was independently confirmed by direct glutathione quantification (Extended Data Fig. 1e-g). Collectively, this suggested that cells on a soft ECM increase cystine uptake and glutathione metabolism to achieve redox balance in the face of increased glutathione oxidation.
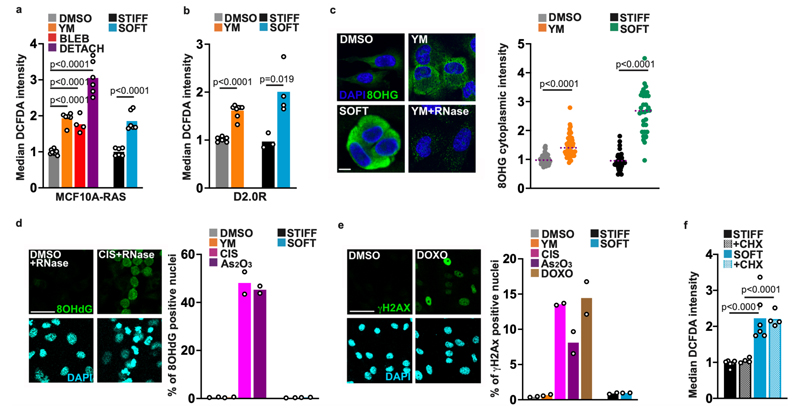
Glutathione oxidation can be due to increased ROS or to decreased NADPH-dependent reduction. We measured the redox state of NADP+/H with an established ratiometric sensor 13, but failed to detect significant steady-state alterations (Extended Data Fig. 2a). In contrast, we observed a consistent increase of general and mitochondrial ROS (mtROS) production in conditions of reduced contractility in multiple cell lines (Fig. 2a,b and Extended Data Fig. 2b-g). ROS increase was detectable as soon as 1 hour after treatment, thus preceding glutathione oxidation and cystine uptake, up to 24 hours (Extended Data Fig. 2h,i). As for the magnitude, ROS production was less in cells with reduced contractility compared to detached cells 14-16 (Fig. 2a) or to cells subjected to oxidative stress (Extended Data Fig. 2b,c). Consistent with increased ROS production, we observed the formation of cytoplasmic 8-hydroxyguanosine (8-OHG) adducts (Fig. 2c). This was not linked to genotoxic damage, as we failed to detect nuclear 8-OHdG or γ-H2AX (Fig. 2d,e). We also detected increased lipid peroxides (Extended Data Fig. 2j-l), and a corresponding increase in 4-hydroxy-2-nonenal (4-HNE) adducts (Extended Data Fig. 2m). By using these in situ read-outs we detected increased ROS in D2.0R cells cultured up to 7 days on a soft ECM, indicating a long-lasting effect in response to ECM stiffness ex vivo and in vivo (see below Fig. 8b,e,j). Finally, ROS induction also occurred in the absence of protein translation, indicating a direct response (Fig. 2f). Thus, altered glutathione metabolism on a soft ECM is associated with increased ROS.
Fig. 2. ECM stiffness regulates reactive-oxygen species (ROS) levels.
a,b, Quantification of ROS by flow cytometry in MCF10A-RAS (a) and D2.0R cells (b), treated with YM, with the Blebbistatin (BLEBBI) NMII myosin inhibitor for 6 h, or cultured on stiff (E≈15kPa) or soft (E≈0.5kPa) Fibronectin-coated acrylamide hydrogels. Cells kept in suspension (DETACH) were included for comparison. Normalized to mean intensity in control (in a n=10 (DMSO) n=6 (YM and DETACH) n=4 (BLEBBI) n=6 (STIFF and SOFT) biologically independent samples pooled across two independent experiments; in b n=6 (DMSO) n=8 (YM) n=4 (STIFF and SOFT) biologically independent samples pooled across two independent experiments; Dunnet’s tests). c, Immunofluorescence of cytoplasmic 8-hydroxy-guanosine (8-OHG) in MCF10A-RAS cells treated and cultured as above. YM+RNase treatment is a control for cytoplasmic nucleic acid adducts. Normalized to mean intensity in control (n=45 (DMSO) n=52 (YM) n= 39 (STIFF) n=33 (SOFT) cells pooled across two independent experiments; unpaired two-tailed Student’s t-tests). d, Immunofluorescence of nuclear 8-hydroxy-deoxy-guanosine (8-OHdG) in MCF10A-RAS treated with YM, cultured on stiff or soft hydrogels, or treated with genotoxic doses of Cisplatin (CIS) and As2O3 (n=4840 (DMSO) n=5195 (YM) n= 4333 (CIS) n=4249 (As2O3) n=2133 (STIFF) n=2344 (SOFT) cells pooled across two independent experiments per condition; Dunnet’s test). e, Immunofluorescence of γH2AX in MCF10A-RAS cells treated as in d. Doxorubicin (DOXO) is a control for DNA damage (n=5873 (DMSO) n=2912 (YM) n= 4015 (CIS) n=3702 (As2O3) n=5378 (DOXO) n=3145 (STIFF) n=2198 (SOFT) cells pooled across two independent experiments per condition; Dunnet’s test). f, Quantification of ROS in MCF10A-RAS treated with cycloheximide (CHX) to inhibit de novo protein synthesis. Normalized to mean intensity in control (n=6 (DMSO and SOFT) n=4 (STIFF+CHX and SOFT+CHX) biologically independent samples pooled across two independent experiments; Dunnet’s test). Images in c,d,e, are representative of at least two independent experiments with similar results. Scale bars, 25 μm, except for c (5 μm). Data are mean and single points. See Source Data Table 2.
ECM stiffness regulates the KEAP1/NRF2 pathway
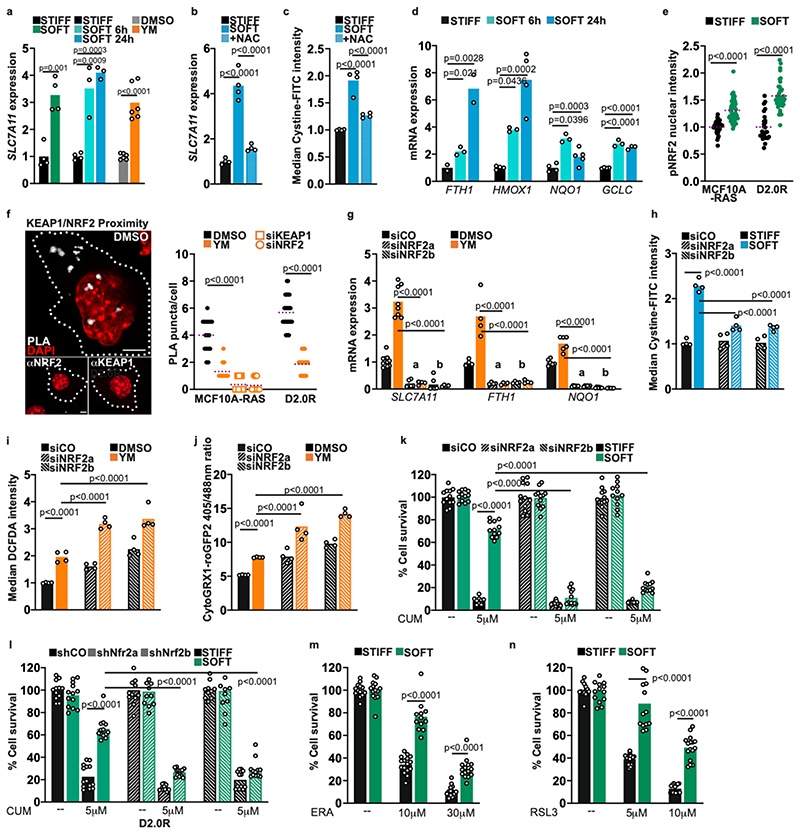
Uptake of extracellular cystine occurs via the system Xc-transporter, encoded by SLC7A11. In our cells, we found that reduced actomyosin contractility upregulated SLC7A11 gene expression (Fig. 3a and Extended Data Fig. 3a-c). This upregulation follows the increase in ROS, but precedes the import of cystine. To establish whether these events were causally linked we treated cells with the N-acetyl-L-cysteine (NAC) antioxidant, which prevented upregulation of SLC7A11 expression (Fig. 3b) and cystine uptake (Fig. 3c). We thus concluded that ROS activates some transcriptional response to induce cystine uptake.
Fig. 3. NRF2 activation on soft ECM potentiates cell resistance to exogenous oxidative stress.
a, qPCR for SLC7A11 in MCF10A-RAS cells cultured on stiff Matrigel-coated plates or soft Matrigel thick gels (green), on stiff (E≈10kPa) or soft (E≈0.5kPa - blue) Fibronectin-coated hydrogels, or treated with YM for 6 h (n=4 (STIFF and SOFT); n=6 (DMSO and YM)). b, qPCR for SLC7A11 in MCF10A-RAS cells cultured on a soft hydrogel and treated with N-acetyl-L-cysteine (NAC) (n=4). c Uptake of FITC-Cystine in MCF10A-RAS cells cultured on a soft hydrogel and treated with NAC. Normalized to mean intensity in control (n=4). d, qPCR for established NRF2 target genes in MCF10A-RAS cells cultured on stiff (E≈50kPa) or soft (E≈0.5kPa) Fibronectin-coated acrylamide hydrogels (n=4). e, Quantification of active nuclear S40-phosphorylated NRF2 in MCF10A-RAS and D2.0R cells cultured on stiff or soft Matrigel. Normalized to mean intensity in control (MCF10A-RAS n=32 (STIFF) n= 50 (SOFT) D2.0R n=27 (STIFF) n= 41 (SOFT)). f, Proximity ligation assays (PLA) between endogenous NRF2 and KEAP1 in MCF10A-RAS cells treated with YM. αNRF2, αKEAP1: a single ab was used. White dotted lines indicate the cell contours. Scale bars = 5 μm. Images are representative of at least two independent experiments with similar results. Knockdown of KEAP1 and NRF2 (siKEAP1, siNRF2) as specificity control (MCF10A-RAS n=28 (DMSO and YM) n= 17 (siKEAP and siNRF2) and D2.0R n=16 (DMSO and YM)). g, qPCR for NRF2 targets in MCF10A-RAS cells transfected with control siRNA (siCO) or two independent NRF2 siRNAs (indicated by a and b), and treated with YM (n=8 and n=6 for SLC7A11 and FTH1, respectively; n=4 for NQO1). h, Uptake of FITC-labeled Cystine in MCF10A-RAS cells transfected with siRNAs and plated on stiff or soft Fibronectin-coated hydrogels. Normalized to mean intensity in control (n=4). i, Quantification of ROS in MCF10A-RAS as in j. Normalized to mean intensity in control (n=4). j, Glutathione redox analysis by flow cytometry in MCF10A-RAS cells transfected and treated as in j (n=4). k,l Cell survival by the resazurin assay in MCF10A-RAS (k) and D2.0R (l) cells with the indicated knockdowns, plated on stiff or soft Matrigel, treated for 48 h with Cumene hydroperoxide (CUM). Mean cell number in controls was set to 100%, (solid black) and all other samples are relative to this (n=12). m,n, Cell survival in MCF10A-RAS cells cultured on stiff or soft Matrigel and treated for 48 h with Erastin (ERA) or RSL3. Normalized to mean cell number in control (n=12).
In a,b,d,g mRNA expression is relative to GAPDH levels normalized to control. ‘n’ refers to the number of biologically independent samples (a-d, g-n) or cells (e, f) analyzed across two independent experiments, except for SLC7A11 and FTH1 in g, where the samples were pooled from three independent experiments. Dunnet’s tests in b-d and f-n, and unpaired two-tailed Student’s t-tests in a and e. Data are mean and single points. See Source Data Table 3.
To identify the mechanism underlying this response, we focused on the notion that SLC7A11 expression is regulated by the NRF2 transcription factor (Nuclear factor erythroid 2 Related Factor 2, or NFE2L2) 17. In the presence of ROS, the NRF2 protein is released from KEAP1 (Kelch-like ECH Associated Protein 1) inhibition and turns on the expression of a general antioxidant program 18,19. To test for NRF2 activation we checked induction of multiple established NRF2 targets by qPCR and found them consistently upregulated across several cell lines when contractility was reduced (Fig. 3d and Extended Data Fig. 3a-h). Among these targets were key enzymes regulating glutathione synthesis, in line with metabolomics results. This was further confirmed by increased endogenous NRF2 nuclear levels (Fig. 3e and Extended Data Fig. 3i) and by reduced interaction of endogenous NRF2 with KEAP1 (Fig. 3f) in conditions of reduced contractility. Importantly, NRF2 was required for upregulation of these target genes (Fig. 3g and Extended Data Fig. 3j-l, and see below Fig. 5m) as well as for increased cystine uptake (Fig. 3h) in cells cultured on a soft ECM, indicating a functional axis.
Fig. 5. ECM stiffness regulates mitochondrial fission through DRP1.
a, Mitochondrial network morphology in MCF10A-RAS cells treated with YM, cultured on stiff or soft Matrigel, or on stiff (E≈10kPa) or soft (E≈0.5kPa) Fibronectin-coated acrylamide hydrogels. FCCP induces the formation of toroidal mitochondria. Scale bars = 5 μm. (n=158 (DMSO) n=242 (YM) n=127 (STIFF) n=118 (SOFT) n=164 (STIFF and SOFT hydrogels)). b, Transmission electron microscope images of mitochondria in MCF10A-RAS cells on stiff or soft Fibronectin-coated hydrogels (n=40). Scale bars = 500 nm. c, Classification of mitochondrial network morphology and mitochondrial fragmentation count (MFC) analysis in MCF10A-RAS cells treated with YM (n=7). Scale bar = 25 μm. d, Endogenous Drpi puncta in MCF10A-RAS cells transfected with mito-RFP and plated on stiff or soft Matrigel substrata (n=35). Scale bar =5 μm. e, Quantification of endogenous puncta of S6i6-phosphorylated DRPi as in d (n=47). f, 3D reconstructions of ER-mitochondrial contacts visualized with the short range (8-10 nm) GFP complementation SPLICs sensor in MCF10A-RAS cells (n=20). Scale bars =10 μm. g, Mitochondrial length in MCF10A-RAS cells plated on soft Matrigel (n=118 (siCO) n=i48 (s1DRP1)). h, Mitochondrial network morphology analysis in MCF10A-RAS cells in response to YM treatment and DRP1 inhibition (n=7). i, Quantification of mtROS in MCF10A-RAS cells plated on stiff or soft Fibronectin-coated acrylamide hydrogels. Where indicated, cells were treated with the MDIVI1 or Drpitor1a DRP1 inhibitors. Normalized to mean intensity in control (n=10 (siCO); all other bars n=4). j, Quantification of mtROS in MCF10A-RAS cells with knockdown of the MIEF1 and MIEF2 (siMIEF1/2), and treated with YM. Normalized to mean intensity in control (n=4 each bar). k,l, qPCR for established NRF2 targets in MCF10A-RAS (k) or D2.oR cells (l) with knockdown of DRP1 and plated on stiff or soft Fibronectin-coated hydrogels. Expression relative to GAPDH normalized to control (MCF10A-RAS: n=6 each bar; D2.0R: n=4 each bar). m. Heatmap of NRF2- and DRP1-dependent genes activated on soft (E≈0.2kPa) compared to stiff (E≈50kPa) Collagen-I-coated acrylamide hydrogels based on RNAseq of D2.oR cells. Each column is an independent biological sample; each line is a single gene. Downregulation and upregulation relative to Z-score (P<0.05) (n=3 independent biological samples from a single experiment each condition). n. Uptake of FITC-Cystine in MCF10A-RAS cells as in i. Normalized to mean intensity in control (siCO n=8; all other bars n=4). o,p, Cell survival in MCF10A-RAS (o) D2.oR (p) plated on stiff or soft Matrigel, and treated for 48 h with Cumene hydroperoxide (CUM). Normalized to mean cell number in Stiff controls (in o n=6 (siCO CUMENE SOFT and siDRP1 VEHICLE STIFF) n=8 all other conditions; in p n=8 (shCO VEHICLE STIFF and shCO CUMENE STIFF) n=6 all other conditions).
Images in a,b,c,d,h are representative of two independent experiments with similar results. ‘n’ refers to number of mitochondria (a,b,g) from 10 cells pooled from two independent experiments; number of cells (d,e,f) across two independent experiments each condition; number of pictures (c,h) pooled from two independent experiments each condition; number of biologically independent samples (i,j,n,o,p) across two independent experiments each bar, except siCO in i and n (four independent experiments), MCF10A RAS in k,l (three independent experiments). Unpaired two-tailed Student’s t-tests in a-e,g,h,j; Dunnet’s tests for f,i,k,l,n,o,p. Data are mean and single points, except for c,g (mean and s.d.). See Source Data Table 5.
Widespread activation of NRF2 target genes on a soft ECM
To expand the generality of our findings, we searched for NRF2 “footprints” 20-23 among genes regulated by ECM stiffness in MCF10A 24, mouse D2.0R 25, human MDA-MB-453 breast cancer cells and primary cultures obtained from the mouse MMTV-PyMT breast cancer model 26 (Extended Data Fig. 4a-d). We also used data on breast cancer patients before and after successful neoadjuvant therapy (Extended Data Fig. 4e) 27, when the stiffness of primary tumors is greatly reduced 28, and a recent microarray of human keloids (Extended Data Fig. 4f) 29, a fibro-proliferative disease caused by stiffening of the dermis 30. In all these experiments we found upregulation of NRF2 targets on a soft ECM, or in the softer tissue condition. As a control, the targets of the mechanosensitive YAP/TAZ and SREBP1/2 factors were coherently regulated 11,31. We also monitored the targets of another ROS-sensitive transcription factor, HSF1 32,33, which we found inhibited in MCF10A cells on a soft ECM (Extended Data Fig. 4g), as in Ref.34. This was however incoherent with ROS, but coherent with mechanically-regulated YAP/TAZ (Extended Data Fig. 4h), in line with Ref.35, suggesting this regulation is uncoupled from ROS. Collectively, these analyses indicate that regulation of NRF2 by the physical properties of the ECM is a conserved process.
A soft ECM empowers cell resistance to oxidative stress
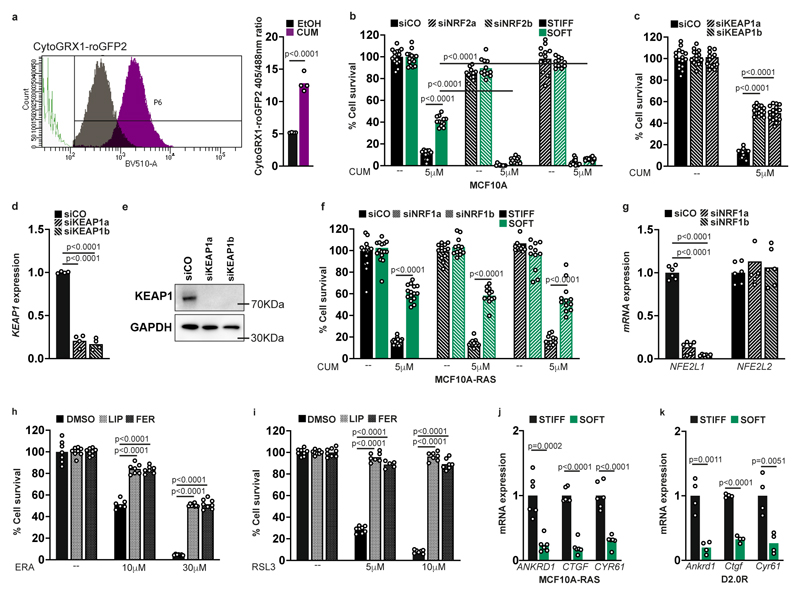
We next addressed the functional consequence of NRF2 activation on a soft ECM. We first tested if this limits production of endogenous toxic ROS. NRF2 knockdown led to higher ROS and higher glutathione oxidation (Fig. 3i,j and Extended Data Fig. 5a), indicating a feedback effect. However, this did not affect cell viability on a soft ECM (Fig. 3k,l and Extended Data Fig. 5b). We then tested the alternative possibility that ROS induce NRF2 to bestow on cells increased tolerance to oxidative stress, a mechanism known as hormesis 36-38. As shown in Fig. 3k,l and Extended Data Fig. 5b, cells on a soft ECM display enhanced survival to exogenous oxidative stress, and this was dependent on NRF2. In line, knockdown of the NRF2 inhibitor KEAP1 enhanced survival on stiff plastics (Extended Data Fig. 5c-e), while NRF1 (Nuclear factor erythroid 2 Related Factor 1 or NFE2L1) had no effect (Extended Data Fig. 5f,g). To further challenge this hypothesis we induced ferroptosis, a form of cell death caused by lipid peroxidation 39-40. As shown in Fig. 3m,n and Extended Data Fig. 5h,i, cells on a soft ECM were also less sensitive to induction of ferroptosis. These results suggest that cells on a soft ECM have increased capacity to resist oxidative stress, which is not observed upon loss of the NRF2 transcription factor.
The antioxidant response to softness depends on mtROS
On a soft ECM we observed increased production of cytoplasmic, mitochondrial and membrane lipid ROS. We thus asked what is the most relevant source of ROS inducing the antioxidant response. For this we inhibited mitochondrial ROS, endoplasmic reticulum (ER) stress and ER-stress-associated ROS, or membrane-associated NADPH oxidases 19. As shown in Fig. 4a-c only MitoTEMPO rescued both cytoplasmic and mitochondrial ROS, suggesting a mitochondrial origin of ROS. Accordingly, treatment of cells cultured on a soft ECM with MitoTEMPO also prevented activation of NRF2 targets (Fig. 4d) and cystine uptake (Fig. 4e). The contribution of other ROS sources remains to be fully determined.
Fig. 4. Mitochondrial ROS (mtROS) initiate the antioxidant response induced by ECM stiffness.
a,b, Quantification of ROS by flow cytometry in MCF10A-RAS cells treated with YM for 3 h (a), or cultured on stiff (E≈10kPa) or soft (E≈0.5kPa) Fibronectin-coated acrylamide hydrogels for 6 h (b), in combination with inhibitors of ROS production. Normalized to mean intensity in control (n=10 (VEHICLE) n=8 (wortmannin) n=6 (TUDCA) all other conditions n=4 for each bar). c, Quantification of mtROS in MCF10A-RAS cells cultured on stiff or soft Fibronectin-coated hydrogels for 6 h. Where indicated, cells were treated with TUDCA or the mitochondrial antioxidant MitoTEMPO (MITO). Normalized to mean intensity in control (n=8 (VEHICLE and MITO); n=4 (TUDCA)). d, qPCR for NRF2 targets in MCF10A-RAS cells cultured as in c 24 h. mRNA expression data are relative to GAPDH levels normalized to control (HMOX1 n=6 (STIFF and SOFT+MITOTEMPO) n=8 (SOFT); NQO1 n=6 (STIFF and SOFT+MITOTEMPO) n=7 (SOFT); GCLC n=4 (STIFF) n=5 (SOFT) n=6 (SOFT+MITOTEMPO)). e, Uptake of FITC-labeled Cystine in MCF10A-RAS cells cultured as in d. Normalized to mean intensity in control (n=4 for each bar). f,g, Oxygen Consumption Rate (OCR) analysis performed with an extracellular flux analyzer on monolayers of MCF10A-RAS (f) D2.0R cells (g) cultured on stiff or soft Matrigel substrata. Oligomycin (OLIGO 0.8 μM), FCCP (900 nM), Rotenone (ROT 1 μM) plus Antimycin A (ANTI A 1 μM) were used to determine basal respiration, ATP-coupled respiration, maximal respiratory capacity and non-mitochondrial oxygen consumption. (MCF10A-RAS n=20 (STIFF and SOFT) D2.0R n=20 (STIFF and SOFT)).
In a-g, ‘n’ refers to the number of biologically independent samples analyzed across two independent experiments, except for VEHICLE and MITO in c (four independent experiments), HMOX1 and NQO1 in d (three independent experiments). Dunnet’s tests in d,e and f-n, and unpaired two-tailed Student’s t-tests in a-c and e-g. Data are mean and single points except from f,g (mean and s.d. - shaded areas). See Source Data Table 4.
Mitochondrial ROS can be produced by multiple mechanisms including changes in oxidative phosphorylation and altered activity of enzymes such as mitochondrial NADPH and monoamine oxidases 19. However, treatment of cells with inhibitors for these enzymes did not prevent ROS production (Fig. 4a,b). Cells cultured on a soft ECM did not display significant changes in basal respiration either (Fig. 4f,g and Extended Data Fig. 5j,k), as also previously found by others 41,42, which suggests the soft matrix does not lead to major defects in oxidative phosphorylation. Finally, NRF2 activity is increased when respiration is enhanced 43,44, but cells of soft ECM displayed reduced maximal respiratory capacity (see Fig. 4f,g), and increased NRF2 (see above). We thus looked for other mitochondria-related mechanisms that increase ROS production.
ECM stiffness regulates mitochondrial fission through DRP1
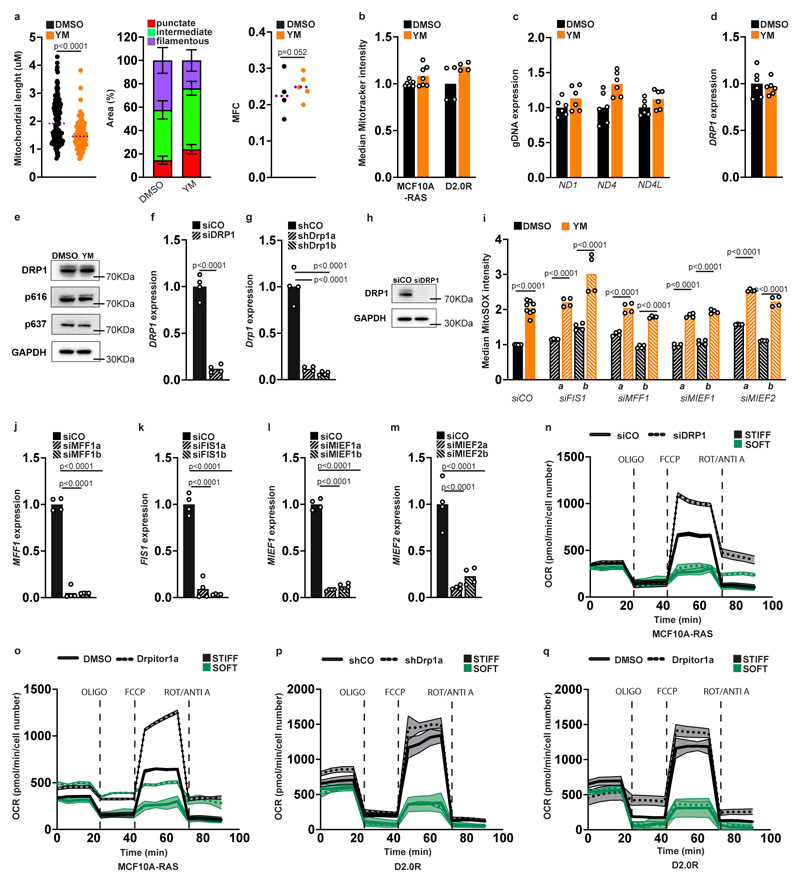
While doing the above experiments we noticed that cells on a soft ECM had an altered mitochondrial network. We then quantified mitochondrial morphology and observed that upon conditions of reduced contractility, cells display fragmented or punctate mitochondria (Fig. 5a,b and Extended Data Fig. 6a) without reduction in overall mitochondria content (Extended Data Fig. 6b,c). We confirmed this observation by an independent quantitative analysis and with an automated image analysis system based on machine learning algorithms 45 (Fig. 5c and Extended Data Fig. 6a).
A shift towards shorter mitochondria can be due to more fission or to fewer fusion events 46. The activity of DRP1 (Dynamin Related Protein 1), the main mitochondrial fission factor, can be visualized by the formation of puncta at mitochondrial sites undergoing fission 47. On a soft ECM we observed an increased number of endogenous DRP1 puncta (Fig. 5d and see below Fig. 6e), as well as of DRP1’s active form which is phosphorylated at serine 616 47 This occurred without changes in DRP1 expression (Extended Data Fig. 6d,e). As an additional read-out of DRP1 activity, we built on the notion that activation of DRP1 leads to reduced ER-mitochondria contacts 48,49. Strikingly, we found a reduced number of shortrange ER-mitochondria contacts on a soft ECM, which was rescued by inhibition of DRP1 with Drpitor1a, a specific inhibitor of DRP1 GTPase activity 45 (Fig. 5f). Finally, inhibition of DRP1 induced the elongation of mitochondria on a soft ECM (Fig. 5g,h), as well as on a stiff ECM as expected, which is overall consistent with increased DRP1 activity and increased mitochondrial fission on a soft ECM.
Fig. 6. ECM stiffness regulates fission via Spire1c- and Arp2/3-dependent peri-mitochondrial F-actin.
a, Left: colocalization of transfected WT but not R62D monomeric mutant Flag-Actin with phalloidin in MCF10A-RAS cells. Centre: PLA (proximity ligation assay) between Flag-Actin and TOMM20. No probes: absence of secondary antibodies (ab). αTOMM20, αFLAG: only one primary ab was used. Cells were treated with YM alone and in combination with LatrunculinA (LATA) or Arp2/3 inhibitor (CK666). Scale bars = 2 μm (n=20). b, PLA between endogenous filamentous actin and TOMM20 in MCF10A-RAS cells treated as in a. αTOMM20; Bio-Phalloidin + αBio: only one primary ab was used as specificity control. Scale bars = 5 μm (n=20). c, Co-localization of F-Actin/mitochondria PLA with DRP1-GFP puncta in MCF10A-RAS cells. Scale bar = 5 μm. d, Quantification of endogenous DRP1 puncta in MCF10A-RAS cells cultured on soft or stiff Matrigel and treated with LatrunculinA (LATA) or Arp2/3 inhibitor (CK666) (n=35). e, Quantification of DRP1-GFP puncta in D2.0R cells treated as in a. Scale bars = 10 μm.
(n=35). f, Mitochondrial length in MCF10A-RAS cells expressing dominant-negative Spire1C isoforms (n=141). g, Quantification of mtROS in MCF10A-RAS cells expressing dominant-negative Spire1C isoforms and plated on stiff or soft hydrogels for 6 h. Normalized to median intensity in the control (n=4).
h, Quantification of mitochondrial ROS in MCF10A-RAS cells depleted of total INF2 (siINF2) or of the mitochondrial splice isoform (siINF2-CAAX), and treated with YM. Normalized to mean intensity in control (n=4). i,j, qPCR in MCF10A-RAS for total (i) or of the mitochondrial splice isoform (j) of INF2. mRNA expression data are relative to GAPDH levels normalized to control (n=4).
Images in a,b,c,e, are representative of at least two independent experiments with similar results. ‘n’ refers to number of cells (a,b,d,e) across two independent experiments each condition; number of mitochondria (f) from 10 cells across two independent experiments each condition; number of biologically independent samples (g-j) across two independent experiments each bar. Unpaired two-tailed Student’s t-tests in a and b; Dunnet’s tests in d-j Data are mean and single points. See Source Data Table 6.
We then probed the functional relevance of DRP1 in producing cell tolerance to oxidative stress on a soft ECM. First, knockdown of DRP1 or treatment with the MDIVI1 and Drpitor1a DRP1 inhibitors, at concentrations ensuring specificity 45,50,51, prevented mtROS production on a soft ECM (Fig. 5i and Extended Data Fig. 6f-h). Second, the same results were achieved by depleting MIEF1 and MIEF2 (Mitochondrial Elongation Factors 1 and 2, also known as MiD49 and MiD51) mitochondrial DRP1 adaptors 52-55 (Fig. 5j and Extended Data Fig. 6i-m). In these experiments mtROS levels were not directly related to oxidative phosphorylation, because a soft ECM reduced maximal respiratory capacity even in cells with inhibited DRP1 (Extended Data Fig. 6n-q). Third, DRP1 was required for NRF2 activation by looking at selected targets (Fig. 5k,l) and at global gene expression (Fig. 5m). Of note, the genes altered by NRF2 and by DRP1 overlap very significantly with those regulated by ECM stiffness (4-fold overrepresented over the random expectation, P=3.6*10^-147 for DRP1 targets, P=2.9*10^-49 for NRF2 targets by Fisher's exact test), confirming a strong link. Finally, inhibition of DRP1 reduced cystine uptake (Fig. 5n) and restored sensitivity to cumene hydroperoxide on a soft ECM (Fig. 5o,p), phenocopying the loss of NRF2 (see above). Collectively, this data indicates that a soft ECM induces mitochondrial fission through activation of DRP1, and shows that DRP1 is instrumental to the regulation of mtROS, NRF2 and the cell’s antioxidant capacity.
ECM stiffness regulates peri-mitochondrial Factin
A remaining question is how ECM stiffness controls mitochondrial morphology. Some data suggests that contractile F-actin bundles can be in a reciprocal equilibrium with other F-actin structures that depend on the Arp2/3 complex 56-60. Interestingly, peri-mitochondrial F-actin and fission also depend on the Arp2/3, as well as on the INF2 (Inverted Formin 2) and Spire1C F-actin nucleation factors 61-63, suggesting a potential crosstalk based on different F-actin pools.
We first developed a method to visualize peri-mitochondrial F-actin specifically, based on proximity-ligation assays (PLA) between F-actin and the mitochondrial outer-membrane protein TOMM20. To capture actin filaments we used two approaches, one based on the incorporation of WT Flag-Actin, but not of the R62D unpolymerized mutant 64 (Fig. 6a), and another based on phalloidin (Fig. 6b). We obtained specific and discrete PLA signals (Fig. 6a,b) which colocalized with DRP1 puncta (Fig. 6c). Strikingly, these F-actin/mitochondria “contacts” were increased upon ROCK/MLCK inhibition, and this was abolished by also inhibiting Factin or Arp2/3 (Fig. 6a,b). F-actin and Arp2/3 were also required for the formation of DRP1 puncta observed upon reduced contractility (Fig. 6d,e). This suggests that when actomyosin is inhibited, the number of F-actin/mitochondria “contacts” is increased, which is causal for DRP1 recruitment and oligomerization.
To address the functional role of peri-mitochondrial F-actin, we then inhibited the INF2 and Spire1C factors. Inhibition of mitochondrial Spire1C rescued mitochondria elongation and mtROS on a soft ECM (Fig. 6f,g), while knockdown of INF2 did not (Fig. 6h-j). This suggests that INF2 and Spire1C do not always work together, consistent with the findings of others 65-67. In aggregate these findings suggest that decreased contractility of the actomyosin cytoskeleton directly or indirectly tips the balance towards formation of peri-mitochondrial F-actin driven by Spire1C and Arp2/3 and this enhances fission.
ECM mechanotransduction regulates chemotherapy resistance
Some chemotherapeutics work by inducing oxidative stress, so that the antioxidant response triggered by NRF2 is known to favor chemotherapy resistance, including on plastics 18,19. To test whether this is regulated in response to ECM stiffness we selected drugs that kill by inducing ROS, such as Cisplatin and Arsenic trioxide (As2O3) (Fig. 7a,b and Extended Data Fig. 7a,b). We found that cells cultured on a soft ECM display increased resistance to these drugs (Fig. 7c,d and Extended Data Fig. 7c,d). As a specificity control, cell death induced by Doxorubicin occurred independently from both ROS and ECM stiffness in our cells (Extended Data Fig. 7e,f). Importantly, inhibition of DRP1 or NRF2 reinstalled cell death on a soft ECM (Fig. 7e-h and Extended Data Fig. 7g,h), indicating that ECM softness increases resistance to ROS-dependent drugs through DRP1 and NRF2.
Fig. 7. A soft ECM increases resistance to Cisplatin and As2O3 chemotherapy through NRF2 and DRP1.
a,b, Cell survival in D2.0R cells treated for 48 h with the indicated concentrations of Cisplatin (a) or As2O3 (b), in combination with N-Acetyl-L-cysteine (NAC) (n=16 all conditions). c,d. Cell survival in D2.0R cells cultured on stiff or soft Matrigel substrata, and treated for 48 h with the indicated concentrations of Cisplatin (c) or As2O3 (d) (in c n=10 (VEHICLE STIFF and SOFT) n=14 all other conditions; in d n=12 (VEHICLE STIFF and SOFT) n=11 (As2O3 STIFF) n=14 all other conditions). e,f. Cell survival in D2.0R cells stably expressing control shRNA or two independent NRF2 shRNAs. Cells were cultured and treated as above (in e n=9 (shNrf2a VEHICLE and shNrf2b VEHICLE) n=11 (shNrf2a SOFT and shNrf2b SOFT) n=16 (shCO CISPLATIN STIFF) n=13 all other conditions; in f n=8 (shNrf2b VEHICLE STIFF and SOFT) n=9 (shCO VEHICLE SOFT and shCO As2O3 STIFF) n=10 (shNrf2b As2O3 STIFF and SOFT) n=11 (shNrf2a VEHICLE STIFF and SOFT, shNrf2a As2O3 STIFF) n=12 (shCO VEHICLE STIFF, shCO As2O3 SOFT and shNrf2a VEHICLE STIFF)). g,h. Cell survival in D2.0R cells stably expressing control shRNA, two independent DRP1 shRNAs, or treated with the Drpitor1a DRP1 inhibitor. Cells were cultured and treated as above (in g n=6 (shDrp1a STIFF CISPLATIN and shDRP1b STIFF CISPLATIN); n=12 (DRPITOR); n=8 all the other conditions; in h n=6 (shCO SOFT As2O3, shDrp1a SOFT As2O3 and shDRP1b SOFT As2O3); n=8 all the other conditions).
All experiments normalized to mean cell number in controls. ‘n’ refers to the number of biologically independent samples across two independent experiments each bar. Data are mean and single points with Dunnet’s tests. See Source Data Table 7.
Fig. 8. A soft metastatic niche protects dormant breast cancer cells from chemotherapy.
a, Immunofluorescence on D2.0R cells cultured on soft Matrigel for 2 (EARLY) or 14 days (LATE). Collagen-I, Phalloidin (F-actin) and nuclear YAP/TAZ are indicative of increased ECM stiffness at 14 days (see Extended Data Fig. 8a). Scale bars = 10 μm. b, secondary ROS adducts in D2.0R cells cultured on stiff Matrigel (24h), soft Matrigel (24h, 48h, 7d) and stiffened soft Matrigel (14d, LATE). Samples normalized to the mean intensity in the controls (8OHG n=63 (STIFF) n=59 (SOFT 24h) n=79 (SOFT 48h) n=109 (SOFT 7d and SOFT 14d); 4HNE n=51 (STIFF) n=60 (SOFT 24h) n=30(SOFT 48h and SOFT 7d) n=60 (SOFT l4d)). c, Cell survival in D2.0R cells cultured as in b for the indicated time-points and then treated for 48 h with Cisplatin l0μM (n=120). d, tissue stiffness by atomic force microscopy on mouse mammary glands, primary D2.0R mammary tumors (1.5 months) and normal lungs, using fresh tissue vibratome sections (n=16 (MAMMARY and LUNG) n= 19 (MAMMARY TUMOR) samplings from 4 sections coming from two mice). e,f, ROS adducts (e) and glutathione redox analysis (f) in D2.0R cells cultured on mouse lung decellularized ECM slices that had been previously incubated with Ribose (RIB), or obtained from fibrotic lungs (FIB) (8OHG n=99 (LUNG ECM and LUNG ECM+RIB) n=148 (LUNG ECM) n=123 (LUNG ECM +FIB); 4HNE n= 99 (LUNG ECM and LUNG ECM+RIB) n=133 (LUNG ECM) n=108 (LUNG ECM+FIB)). g, qPCR in D2.0R cells cultured on plastics, infiltrated into normal or fibrotic (FIB) decellularized lung ECM scaffolds. mRNA expression relative to GAPDH normalized to control (n=8 biologically independent samples pooled across two independent experiments for PLASTIC and LUNG ECM; n=2 biologically independent samples from one experiment for LUNG ECM+FIB). h, tissue stiffness by atomic force microscopy on normal lungs (WT), lungs with early quiescent (14 days, EARLY) and late relapsing (1.5 months, LATE) disseminated D2.0R cells, using frozen tissue microtome sections (n=7 (WT) n=16 (EARLY) n=15 (LATE) sections coming from two mice). i, Immunofluorescence on D2.0R cells disseminated in the mouse lung at early (14 days) and late (1.5 months) time-points. Scale bars = 10 μm. j,k, ROS adducts (j) and glutathione redox analysis (k) in D2.0R cells disseminated in the mouse lung at early and late time-points (in j 8OHG n=25 (EARLY and LATE); 4HNE n=25 (EARLY and LATE; in k n=33 (EARLY and LATE)). l,m. Bioluminescent intravital quantification of lung metastatic burden by GFP/Fluc D2.0R cell lines expressing control or NRF2 shRNAs (l) or DRP1 shRNAs. (m), and treated with Cisplatin at early time-points (see Extended Data Fig. 9a). Total photon flux (photons/s) for each mouse at day 0 (injection) was set to 1, and all other time-points are relative to this (n=6 mice for each condition). In l.m ✝ mice were sacrificed at humane endpoint.
Images in a,i are representative of two independent experiments or mice with similar results Data are mean and single points, except l,m (mean). ‘n’ refers to cells pooled across two independent experiments (b,c,e,f) and cells pooled across three mice (j,k) Unpaired two-tailed Student’s t-tests for (e,f,j,k,l,m), Dunnet’s tests for (b,c,d,h). See Source Data Table 8.
Lung stiffness regulates the ROS/NRF2 axis
D2.0R cells are a model for late-relapsing or “dormant” ER-positive breast cancer metastasis to the lung 68,69. Late relapse of breast cancer remains a clinically unmet problem in patients 70. The transition of metastatic D2.0R cells from dormancy to relapse has been linked to ECM composition: these cells remain quiescent on a basement-membrane (BM) ECM and in the normal lung microenvironment, while they actively grow on a collagenous ECM and in collagen-rich fibrotic lungs. Moreover, if cultured long enough, these cells secrete Collagen-I to self-enable adhesion and resume proliferation 25,71,72.
The amount and cross-linking of Collagen-I are key factors contributing to tissue ECM stiffness. We thus re-examined the dormancy-to-relapse transition of D2.0R cells in light of ECM mechanics (see scheme in Extended Data Fig. 8a). In vitro, cells cultured up to 1 week on a BM ECM display inhibition of the mechanosensitive YAP/TAZ factors, in line with ECM softness; after 2 weeks, however, cells secreted Collagen-I and underwent a morphological shift indicative of increased adhesion, coupled to activation of YAP/TAZ, indicating a stiffened microenvironment (Fig. 8a). Strikingly, this was accompanied by a reduction of ROS markers (Fig. 8b), and by a corresponding restored sensitivity to Cisplatin treatment at 2 weeks (Fig. 8c). This overall suggested a prolonged activation of ROS/NRF2 on a soft ECM, which is inhibited when cells remodel and stiffen the ECM.
We then asked if the lung ECM can be a relevant input to control our phenotypes. Lung elasticity, as measured by atomic force microscopy (AFM), approximated that of soft hydrogels and of the normal mammary gland, while it was softer than a primary tumor induced by D2.0R orthotopic injection into the mammary fat pad (Fig. 8d and Extended Data Fig. 8b). This supports the idea that metastatic dissemination to the lungs occurs towards a softer tissue microenvironment. We then cultured D2.0R cells on slices of decellularized mouse lung ECM 73-75, which increased ROS markers compared to cells on stiffer slices obtained either from fibrotic lungs 76 or by increasing collagen cross-linking in vitro with ribose 77,78 (Fig. 8e,f and Extended Data Fig. 8c,d). Furthermore, cells infiltrated into normal lung ECM display increased NRF2 target genes compared to cells infiltrated into fibrotic lung ECM (Fig. 8g and Extended Data Fig. 8e). This indicates that a physiologically relevant variation of lung ECM stiffness is sufficient to regulate the ROS/NRF2 axis.
Finally, we asked whether this also happened in vivo. For this we induced metastatic dissemination by injecting D2.0R cells in syngeneic female BALB/C mice, and compared normal lungs, lungs bearing early disseminated quiescent cells, and lungs with late outgrowing macroscopic metastases. Using atomic force measurements, Collagen-I expression and YAP/TAZ localization, we found that early disseminated cells are found in a soft microenvironment, while late metastatic outgrowths are stiffer (Fig. 8h,i). Strikingly, this was associated with corresponding higher ROS markers in early disseminated cells (Fig. 8j,k), recapitulating the above in vitro and ex vivo results. Overall, this supports the view that the mechanical properties of the lung ECM regulate the ROS/NRF2 axis in disseminated D2.0R cells.
DRP1 and NRF2 protect metastatic cells from chemotherapy
Dormant metastatic cancer cells are notoriously refractory to chemotherapy 79, which has been ascribed in the past to their proliferative quiescence. Our results instead suggest an active mechanism, i.e. increased antioxidant capacity in response to a soft microenvironment. To verify this idea in vivo we induced metastatic lung dissemination and then treated mice with Cisplatin during the early quiescent phase (see scheme in Extended Data Fig. 9a). Cells expressing control shRNA remained dormant for 6-7 weeks and then resumed proliferation developing into overt metastases, and were marginally sensitive to Cisplatin treatment (Fig. 8l,m). NRF2 knockdown did not affect early metastatic spread, but slowed down cell growth at the metastatic site (Fig. 8l and Extended Data Fig. 9b,c). Most importantly, NRF2 knockdown made quiescent cells vulnerable to Cisplatin treatment, leading to a more effective reduction of cancer cells and preventing relapse up to 28 weeks (Fig. 8l). This was associated with increased cleaved Caspase-3 in cancer cells, indicating the induction of apoptotic cell death (Extended Data Fig. 9d). Finally, this was strikingly recapitulated by knockdown of DRP1 (Fig. 8m and Extended Data Fig. 9e), indicating a key role of mitochondrial fission in regulating chemotherapy sensitivity in vivo. This is in line with a general role of NRF2 in promoting therapy resistance and with general anti-cancer effects of DRP1 and MIEF1/2 inhibition observed by others 18,45,53,80, and is compatible with the view that a soft metastatic niche potentiates breast cancer cell ability to resist chemotherapy by fostering DRP1 and NRF2 activity.
Discussion
Here we identify a link between the physical properties of the ECM and the cell antioxidant homeostasis. When cells are in a soft microenvironment they increase mtROS production, which bestows on cells increased tolerance to oxidative stress via NRF2 (Extended Data Fig. 10). In contrast, when cells are detached from the ECM and unable to generate tension, endogenous ROS production becomes toxic 14-16,81. In this view, this represents a hormetic response by which a physiological level of ECM elasticity maintains a beneficial level of mtROS that entrain cells’ antioxidant metabolism 19,36-38. Scattered elements of this pathway were described in other cells 41,82,83, suggesting widespread conservation. Moreover, the finding that a similar pathway is relevant for neural stem cell commitment in the mouse brain 84, the softest among tissues, suggests this link may have other roles beyond resistance to oxidative stress.
The antioxidant response to ECM elasticity is centered on mitochondrial dynamics, and provides a physiologically-relevant context in which peri-mitochondrial F-actin controls DRP1 activity and mitochondrial fission 61,63,85. The requirement for MIEF1/2, together with the observation that these bear a broad potential for F-actin interaction 55,86, suggest these factors might serve as a specific interface in response to ECM stiffness. Based on the molecular requirements, this fission modality resembles actin-mediated mitochondrial reshuffling 67,87, while it is different from other fission events, including toroidal mitochondria 65,66,88,89. In future, it will be interesting to understand whether mechanosensitive and peri-mitochondrial F-actin pools directly compete for a steady-state amount of actin monomers 58,60, or whether more specific signalling mechanisms are involved. More in general, this provides an example of how organelle structure and dynamics is intimately linked with extracellular forces.
Our results suggest that resistance of disseminated metastatic cells to cisplatin is actively instructed by mechanical signaling at the metastatic niche, and support the view that ECM softness is a double-edged sword in cancer, opposing cell proliferation and invasion, but also favoring chemotherapy resistance. Since the main organs targeted by breast cancer metastasis (the lung, bone marrow, brain and liver) share an elastic modulus in the soft range, it will be interesting to test if the mechanism we here describe also applies to other metastatic sites. In future, these findings may pave the way to testing pharmacological inhibition of NRF2 or DRP1 as a strategy to help eradicate disseminated cells and reduce the risk of metastatic disease relapse in patients.
Methods
Plasmids and Reagents
Cyto-Grx1-roGFP2 (Addgene #64975), mito-Grx1-roGFP2 (Addgene #64977), mito-Clover and mito-RFP (De Stefani), FLAG-Actin and FLAG-R62D-Actin (Posern), iNap1 and iNapC (Yang), deltaKIND and mWH2 dominantnegative Spire1c (Manor), OMM-GFP1-10 and ER-beta11 (Calì). All plasmid preps used for transfection were endotoxin-free and sequence-verified.
Compounds (Source, concentration) were: Y27632 (Axon 1683, 20 microM); ML7 (Sigma I2764, 20 microM), Blebbistatin (Sigma B0560, 20 microM); Cycloheximide (Sigma 247502); Cisplatin (STCB sc-200896); As2O3 (Sigma 202673); Doxorubicin (Sigma D1515); N-Acetyl-L-cysteine (Sigma A7250, 5 mM); MitoTEMPO (Sigma SML0737, 15 microM); Drpitor1a (Stephen Archer, 0.5 microM); MDIVI1 (Sigma M0199, 10 microM); KI696 (MCE HY-101140, 10 microM); TUDCA (SelleckChem S3654, 1milliM); Pargyline (Sigma D026, 2.5 mM); VAS2870 (Cayman 19205, 10 microM); Sulfasalazine (Cayman 15025, 0.5 mM); Erastin (MCE HY-15763); RSL3 (Sigma SML2234); DHEA (Sigma 700087P, 100uM); Diamide (Sigma D3648 50uM), Wortmannin (Sigma W1628 5uM).
Cell culture, substrata and transfections
MCF10A and MCF10AneoT (MCF10A-RAS) human mammary epithelial cells were cultured in DMEM/F12, 5%HS, 10 microg/ml Insulin (Sigma), 20 ng/ml hEGF (Peprotech), 100 ng/ml Cholera Toxin (Sigma), 0.5 microg/ml Hydrocortisone (Sigma). Univocal identification was based on exon PCR sequencing to identify coding point mutations present in MCF10AneoT (MAP3K12 A662D), MCF10AT1 (also known as MCF10AT1k, or MCF1oA MII - PCSK5 M452I) and MCF10CA1a cells (also known as MCF10A MIV - PCSK5 M452I and PIK3CA H1047R) 90. D2.0R mouse metastatic breast cancer cells were cultured in DMEM, 10%FBS. Glutamine was freshly added to a final concentration of 2mM to all media. General media, serum and supplements were from Thermo. Cell lines were cultured without antibiotics and routinely tested with ATCC Universal Mycoplasma Detection Kit to exclude contaminations. Stable MCF10AneoT or D2.0R Grx1-roGFP1 reporter lines were obtained by retroviral infection followed by puromycin selection (2 and 50 microg/ml, respectively). Stable MCF10AneoT iNapC and iNap1 reporter lines were obtained by retroviral infection followed by puromycin selection. D2.0R EGFP/luc cells were obtained by lentiviral infection followed by sorting of light-emitting cells by flow cytometry in the presence of luciferin.
Stiff (E≈15kPa) and soft (E≈0.5kPa) Fibronectin-coated polyacrylamide hydrogels were assembled in-house by standard protocols 91. For selected experiments, we used commercial Collagen-I-coated polyacrylamide hydrogels of E≈50kPa and E≈0.2kPa stiffness (Matrigen). Matrigel (BD) was diluted 2% vol/vol in 1xPBS to coat stiff tissue-culture dishes (E≈GPa), while it was used pure and gelled at 37°C to obtain thick and soft substrata (E≈250Pa).
Cell transfections were carried out with Transit-LT1 (MirusBio), Lipofectamine 3000 (Thermo) or with Lipofectamine RNAi-MAX (Thermo). siRNAs were selected among FlexiTube GeneSolution 4 siRNA sets (Qiagen) and reordered after validation as dTdT-overhanging 19nt RNA duplexes (Thermo). siRNA sequences were: hNRF2 1 CAU UGA UGU UUC UGA UCU A; hNRF2 2 GGU UGA GAC UAC CAU GGU U; hNRF1 1 GAU GGU UCC AGG ACU ACA A; hNRF1 2 GGC AGG AUU CAG CAC AAC A; hKEAP1 1 CCA GCG CCC UGG ACU GUU A; hKEAP1 2 GAG UGU UAC GAC CCA GAU A; hMFF 1 AGU CGA AUU CAG UAC GAA A; hMFF 2 CGC UGA CCU GGA ACA AGG A; hFIS1 1 GGC CAU GAA GAA AGA UGG A; hFIS1 2 GGC UCA AGG AAU ACG AGA A; hMIEF1 1 GGU GCG UCG UGA GAA UCC A; hMIEF1 2 GUA UGA GCG UGA CAA ACA U; hMIEF2 1 GGU GCC UGA UGG UAG GUA U; hMIEF2 2 GCU GAG AGA AGG UGA CUU A; hINF2 1 GGA UCA ACC UGG AGA UCA UCC GC; hINF2 2 GCA GUA CCG CUU CAG CAU UGU CA ; hINF2 CAAX isoform 1 ACA AAG AAA CUG UGU GUG A; hINF2 CAAX isoform 2 CCC UGA UUC UGA UGA UAA U; hDRP1 AAC AAA GUC UCA GUA UUA C; AllStars Negative Control siRNA (Qiagen - the sequence is a proprietary information). shRNAs were selected among 5 different predesigned pLKO-1 vectors (Sigma) stably integrated in D2.0R cells by lentiviral infection and puromycin (50 microg/ml) selection. shRNA sequences were: mNRF2 A TRCN0000054658 CCA AAG CTA GTA TAG CAA TAA; mNRF2 B TRCN0000054659 CTT GAA GTC TTC AGC ATG TTA; mDRP1 A TRCN0000321169 CGG TGG TGC TAG GAT TTG TTA; mDRP1 B TRCN0000012606 GGC AAT TGA GCT AGC GTA TAT; Control TTC TCC GAA CGT GTC ACG T.
Flow Cytometry
Staining for ROS and FITC-cystine uptake were carried out by incubating cells in plate at the end of the experiments with the indicated reagents in full medium w/out phenol red (DCFDA 1 microM 10 min, MitoSOX 1 microM 10 min, C11-Bodipy-581/591 5 microM 15 min, FITC-cystine 5 microM 30 min). After trypsinization and a brief wash in 1xHBSS, quantification was run on live cells in 1xHBSS (Ca- and Mg-free). For detection of stably expressed Grx1-roGFP2 sensor fluorescence ratio, cells were fixed after trypsinization with 4% PFA at 4°C for 7 min. Analysis was carried out on a LSRFortessa X20 cell analyzer (BD). Cells were gated based on SSC-A or FSC-A to eliminate cell debris, and on SSC-W to eliminate cell doublets. DCFDA and FITC-cystine were measured using a FITC-A filter. MitoSOX with a PE-A filter. The MFI (median fluorescence intensity) was measured for each independent biological sample, based on at least 3*104 gated events. For C11-BODIPY-581/591 the signals from both non-oxidized C11 (PE-TexasRed) and oxidized C11 (FITC-A) were acquired, and the ratio of FITC-A to PE-TexasRed was calculated for each sample. For Grx1-roGFP, the signals from both non-oxidized (FITC-A) and oxidized (BV510-A) were acquired, and the ratio of BV510-A to FITC-A was calculated for each sample. See Extended Data Fig. 1b, Extended Data Fig. 2b,c,j, Extended Data Fig. 5a for representative gating schemes.
Survival assays
Cells were seeded in 96-well plates with the indicated coatings (5000 MCF10A-RAS cells/well, 4000 D2.0R cells/well), in parallel with standards in the linear range of detection (8000, 4000, 2000, 1000, 500, 250 cells/well). Where pertinent, siRNA transfection was performed in bulk the day before seeding. 24 hours after seeding, culture medium was renewed by adding the indicated chemicals or the corresponding amounts of vehicle. MCF10A-RAS were in glucose-free medium. After 48 hours of treatment, cells were washed with 1xPBS and incubated with Resazurin (Sigma R7017, 100 microM) in culture medium and incubated at 37°C for 2 h. Absorbance at 544/590 nm was measured on live cells, by checking that the signal was in the temporal linear range. Absolute cell number was then calculated based on the standard curve, after background subtraction (medium without cells). Relative survival was calculated as % of remaining cells compared to control cells (100%).
qPCR
Total RNA was isolated using commercial kits with DNAse treatment (Norgen). cDNA synthesis was carried out with M-MLV Reverse Transcriptase (Thermo) and oligo-dT primers. qPCR reactions were assembled with FastStart SYBR Green Master Mix (Roche) and run on a QuantStudio6 thermal cycler (Thermo). GAPDH expression levels were used to normalize gene expression between samples, based on eff(gene)^-ΔCt(gene) / eff(GAPDH)^-ΔCt(GAPDH), where eff is the primer amplification efficiency experimentally determined from three consecutive five-fold dilutions of one control sample cDNA, centered on the dilution used for all other samples. Gene expression levels for each biological sample was quantified as the mean between three technical replicates.
Primer sequences for human genes: ANKRD1: ACT TGG GCC GGT ATT TCT TC; TGG GCT AGA AGT GTC TTC AGA T. CTGF (CCN2): AGG AGT GGG TGT GTG ACG A; CCA GGC AGT TGG CTC TAA TC. CYR61 (CCN1): CCT TGT GGA CAG CCA GTG TA; ACT TGG GCC GGT ATT TCT TC. DRP1: TCA AGA ACC AAC CAC AGG CA; CAC AAT CTC GCT GTT CCC GA. GAPDH: CTC CTG CAC CAC CAA CTG CT; GGG CCA TCC ACA GTC TTC TG. GCLC: TCC AGG TGA CAT TCC AAG CC; GAA ATC ACT CCC CAG CGA CA. GCLM: CTC CTG CTG TGT GAT GCC A; CTC GTG CGC TTG AAT GTC AG. HMOX-1: AGT CTT CGC CCC TGT CTA CT; GCT GGT GTG TAG GGG ATG AC. NFE2L2: TTC TCC CAA TTC AGC CAG CC; GGG AAT GTC TGC GCC AAA AG. NQO1: GCA CTG ATC GTA CTG GCT CA; CCA CCA CCT CCC ATC CTT TC. SLC7A11: TTT TCT GAG CGG CTA CTG GG; CAG CTG GTA GAG GAG TGT GC. DRP1: TCA AGA ACC AAC CAC AGG CA; CAC AAT CTC GCT GTT CCC GA; NFE2L1: GAT GTG ACA GGC CCA GAC AA; CCA CAT CCT GCT CCT TCT GG; FIS1: CAA GGA GGA ACA GCG GGA TT; TGC CCA CGA GTC CAT CTT TC; MFF: CCA AAC GCT GAC CTG GAA CA; TGT TTT CAG TGC CAG GGG TT; MIEF1 (MID49): GGT TCA TTG ACA GGG CCA CT; GAT CAG TTT CTG CAG GCC CT; MIEF2 (MID51): ATT CGG GGA AAA GGA GCT GG; TCG AAG GTG GAG GAG TCT GT; KEAP1: TCG TCT CCT TTA TGC CGT GG; TGC TGT GAT CAT TCG CCA CT; INF2: GCC AAG AAG AGC CTG AAC CT; TCA ATC TCG TGC TTC TCG GG; HIST1H4A: GGA AGG GTT TGG GTA AGG GG; ACC AGA GAT CCG CTT CAC AC; HIST1H4D: AAT CAC CAA GCC CGC TAT CC; TGA CAG CAT CGC GGA TTA CA; HSP90AA1: CAC GTC TCT GCA TTC CCT GT; GTT GGT CTT GGG TCT GGG TT; EFEMP1: CAT TCC CTC CAA CCC TTC CC; TGA TGC ACA CTT GGT CTG CT; SERPINH1: CCG ACC ACC CCT TCA TCT TC; TCG TCT CGC ATC TTG TCA CC.
Primer sequences for mouse genes: Ankrd1: CTG TGA GGC TGA ACC GCT AT; TCT CCT TGA GGC TGT CGA AT. Ctgf (Ccn2): CTG CCT ACC GAC TGG AAG AC ; CAT TGG TAA CTC GGG TGG AG. Cyr61 (Ccn1): GCT CAG TCA GAA GGC AGA CC ; GTT CTT GGG GAC ACA GAG GA. Drp1: CAG GTG GTG GGA TTG GAG AC; CGG GCA GAC AGT TTT CTT GC. Gapdh: ATC CTG CAC CAC CAA CTG CT ; GGG CCA TCC ACA GTC TTC TG. Gclc: CAG CTG CAG AGG AGT ACA CC; ATA CTG CAT CGG GTG TCC AC. Gclm: GTT CCC AAA TCA GCC CCG AT; GAG CAT GCC ATG TCA ACT GC. Hmox-1: AGC CCC ACC AAG TTC AAA CA; CAT CAC CTG CAG CTC CTC AA. Nfe2l2: AAC AGA ACG GCC CTA AAG CA; CCA GGA CTC ACG GGA ACT TC. Nqo-1: GCG AGA AGA GCC CTG ATT GT; TCG AGT CCT TCA GCT CAC CT. Slc7a11: TGG CTA TCA TCA CAG TGG GC; GCA ACA AAG ATC GGG ACT GC. Drp1: CAG GTG GTG GGA TTG GAG AC; CGG GCA GAC AGT TTT CTT GC.
Immunofluorescence and PLA
Immunofluorescence was performed as in 92-94 with minor modifications: 15 min 1.5 mg/ml glycine in PBS before permeabilization to reduce background; blocking in 10% BSA. Images were acquired sequentially with a Zeiss LSN700, a Leica SP8 or a Leica Stellaris confocal microscope equipped with a CCD camera using ZEN 2 or Leica LASX softwares. Typical acquisition settings for IF were: image size 1024x1024 pixels; acquisition mode xyz; pixel size 0.15mm; image depth 8 bits; acquisition speed 5/10, with average 2; Plan-Apochromat 63x/1.40 oil DIC M27 objective. For multichannel acquisitions, we used a main beam splitter 405/488/555/639. Raw images (saved in .czi or .lif formats) were opened in Fiji v.1.0 for analysis. Figure panels were assembled with Inkscape v.1.1.1.
For mitochondrial morphology analysis, cells were fixed 10 min at 4°C with 4% FA and washed with cold PBS before permeabilization. We measured: the length of mitochondria major axis; mitochondrial fragmentation count (MFC) 80, i.e. the total mitochondrial number divided by the total mitochondrial area in each confocal microscopy image; and the percentage area of punctate, intermediate, or filamentous mitochondria 95.
Quantitative analysis of 8OHG and 4HNE was based on single-plane confocal images. Intensity was calculated in a ROI covering the whole cytoplasm with ImageJ as the mean pixel intensity, normalized to area. Analysis of 8OHdG was based on RNAse-treated cells, and the percentage of positive nuclei. For Grx1-roGFP, a ROI of fixed size randomly chosen in the cytoplasm was measured at 510nm upon excitation at 405nm (oxidized) and 488nm (reduced) for each cell. The ratio between 405/488 was calculated for each cell.
For PLA (Duolink proximity-ligation assays) we followed manufacturer’s instructions (Sigma DUO92102) with minor modifications: cells were fixed in 4% FA for 10min at 4°C; after a brief wash, cells were incubated 15 min with 1.5 mg/ml Glycine in PBS before permeabilization and then blocked 1h at RT with Duolink Blocking Solution (Sigma); primary antibodies were incubated with 2% Goat Serum at 4°C ON. Primary antibodies for F-actin PLA were: TOMM20, FLAG (mouse or rabbit), and biotin-phalloidin (Sigma P8716) together with an anti-biotin antibody.
For Multiplex Immunofluorescence we used the TSA-based Opal method (Akoya Biosciences) on the Leica BOND RX automated immunostainer (Leica Microsystems). FFPE tissue sections were deparaffinized and subjected to heat-induced epitope retrieval (HIER) pretreatments at 97 °C using BOND Epitope Retrieval (ER) Solutions ER1 and ER2 (Leica Biosystems). Tissue sections were blocked with Normal Goat Serum (Vector Laboratories) for 10 minutes before applying primary antibodies. A fluorescent singleplex was carried out for each biomarker to determine the optimal staining conditions and the order in which the primary antibodies would be applied in the multiplex protocol. Primary antibodies were added sequentially on the slides. The HRP-conjugated secondary antibodies goat anti-rabbit (Vector Laboratories), goat antichicken (Abcam) and goat anti-rat (Vector Laboratories) were incubated for 10 minutes. The TSA-conjugated fluorophores Opal 540, 650 and 480 (Akoya Biosciences) were then added for 10 minutes. Slides were rinsed with a washing buffer after each step. Finally, the spectral DAPI (Akoya Biosciences) was used as a nuclear counterstain, and slides were mounted in ProLong Diamond Antifade Mountant (Life Technologies). Multiplex stained slides were imaged using the Mantra Quantitative Pathology Workstation (Akoya Biosciences) at 20X magnification. For each sample, only areas comprising tumor cells were considered. The inForm Image Analysis software (version 2.4.9, Akoya Biosciences) was used to unmix multispectral images using a spectral library built from acquisition of single fluorophore-stained control tissues, and containing fluorophores-emitting spectral peaks. A selection of representative multispectral images was used to train the inForm software to create algorithms to apply in the batch analysis of all acquired multispectral images. Cell density and percentage data were reported as the mean of all acquired fields from the same tissue slide (at least 20 fields at 20X magnification for each stained slide).
Primary antibodies (catalogue number) were: 8OH(d)G (SCBT sc-66036); 4HNE (ABCAM ab48506); NRF2 PS40 (ABCAM ab76026); DRP1 (BD 611738); FLAG mouse monoclonal (Sigma M2); FLAG rabbit monoclonal (CST BK14793); Alexa647-Phalloidin (Thermo A22287); TOMM20 (Thermo STD4-72); Cleaved Caspase-3 (CST 9661S); GFP (Abcam AB13970); CD31 (Optistain DIA310); Collagen-I (Abcam AB34710); KEAP1 used for WB and PLA (STCB sc-365626); NRF2 used for WB (Proteintech 13396-1-AP); NRF2 used for PLA (CST 12712); DRP1p616 for IF and WB (CST#4494); DRP1p637 for WB (CST #4867). Secondary antibodies were: Alexa-488 anti-mouse (Thermo A21202); Alexa-568 anti-rabbit (Thermo A11036); Alexa-568 anti-mouse IgG1 (Thermo A21124); Alexa-647 antimouse IgG2b (A21242).
Oxygen Consumption Rate analysis
An XF24 extracellular flux analyzer (Seahorse Bioscience) was used to detect real-time changes in cellular respiration rates as in 96. Standard XF24 plates were coated with 70ul of 2% Matrigel (stiff), or with 25ul of 100% Matrigel (soft) which was evenly distributed with a micropipette tip before gelling at 37°C. 90000 cells/well MCF10A-RAS or 120000 cells/well D2.0R were seeded 24 hours before performing the measures. Measurements were performed following manufacturer’s instructions with at least 5 biological replicates for each condition in each experiment, and normalized to cell number quantified by DAPI staining after extracellular flux analysis.
Mouse lung decellularized ECM
Procedures on animals were performed in accordance with local approvals and under license released from the UK Home office. Wild-type male adult CD1 mice were euthanized by cervical dislocation in accordance with local animal guidelines and approvals (in the UK, Home Office guidelines under the Animals Scientific Procedures Act 1986). After opening the rib cage, a 2mM EDTA solution was injected through the inferior vena cava to prevent coagulation. Following removal of the thymus, the aortic artery, right superior vena cava, and left superior vena cava were divided. The pulmonary artery was perfused with 2mM EDTA to remove blood in the pulmonary circulation. The heart was dissected and removed. A 24G catheter was inserted into the trachea and secured using 3-0 silk sutures. The trachea was separated right below the cricoid cartilage, and the whole lung was extracted. Perfusion of the lung was confirmed by gently injecting a PBS solution through the cannula.
Decellularization was performed with the intent to preserve the overall lung structure and extracellular-matrix composition, as previously reported 75,97 with minor modifications. Briefly, decellularization reagents were intermittently inflated (for 30 s) and deflated (for 30 s) through the trachea with automatically controlled peristaltic pumps. One cycle of decellularization consisted of 4 series of inflation/deflation with double distilled water, 4 series with 2% Sodium Deoxycholate (Sigma D6750), 8 series with PBS supplemented with 1% p/s, 4 series with 0.00225% DNAse (Sigma D5025) in a 9 g/l NaCl, 1 g/l CaCl2 solution, and 8 series with PBS supplemented with 1% p/s. 9 cycles of decellularization were performed in total.
Decellularized lungs were embedded in Optimal Cutting Temperature (OCT) tissue embedding compound (Electron Microscopy Science) and snap-frozen in dry ice. Samples were cut to a final thickness of 50μm with a cryostat (Thermo A78900003). Slices were placed on 13 mm-diameter poly-lysine coated coverslips. Slices were equilibrated in PBS, sterilised under UV light for 25 min and stored at 4°C until needed. Slices on coverslips were incubated at 4°C with 1xPBS 44 mM NaHCO3 25 mM HEPES, without or with 250 mM Ribose for 5 days to induce non-specific collagen crosslinking.
Bleomycin-induced fibrosis
Animal studies were approved by the University College London Biological Services Ethical Review Committee and licensed under the Animals (Scientific Procedures) Act 1986 (Home Office, London, United Kingdom). The bleomycin model were performed under the UK Home Office Animal Research project license P36565407. Wild-type male age-matched CD1 mice (Charles River Laboratories, body weight: 35-45 g) were randomly divided into bleomycin-treated and sham groups. Anesthesia was maintained by inhalation of 4% isoflurane. 50 μL of either 1.25U/mg bleomycin (Apollo scientific, UK) or PBS were administered endotracheally. 14 days after bleomycin administration, the mice were euthanized. Lungs were harvested for decellularization. The fibrotic mouse lung decellularized ECM were selected based on increased Collagen-I immunofluorescent staining compared to control samples.
Recellularization experiments
Prior to recellularization, decellularized lung ECM from both bleomycin-treated and control mice were sterilised by γ-irradiation. The right and left lungs were separated by ligation of the right main bronchus, and the left lung was used for cell seeding. The right lungs were immediately dried and snap-frozen. The left lung was perfused with D2.0R cells culture medium and equilibrated for 1h at 37 °C and 5% CO2. A single-cell suspension of D2.0R cells (107 cells in 0.5 ml) was injected into the left lobe with a 1 mL-syringe and incubated overnight at 37 °C and 5% CO2 submerged in culture medium. The day after the recellularized lung was perfused with a 1mL-syringe to remove nonadherent cells, and cultured for additional 48 hours. Then, the recellularized lobe was retrieved and the upper airway was removed at the level of the left main bronchus. The lung was dried and snap-frozen. All the lung samples were weighed before snap-freezing and stored at -80 °C. As a control, in parallel to the lung recellularization, 5 × 105 cells were seeded in a 10 cm Petri Dish and cultured in standard conditions. Cells were harvested in Trizol reagent with a pestel homogenizer for RNA extraction. Decellularized lung ECM without infiltrated cells was devoid of tissue RNA.
AFM measurements
Tissue samples were embedded into OCT without prior fixation, and sectioned at the microtome (20 um) onto poly-lysine coated slides, or embedded in low-melting agarose and sectioned at the vibratome (400 um) with similar results. Sections were dried to allow attachment and rehydrated in 1xPBS. All the samples were analyzed by using an Atomic Force Microscope mounted on an Inverted Optical Microscope (XEBio, Park Systems, Korea) to position the AFM tip on the area of interest of the samples 98. Force-displacement curves were collected in 1xPBS at RT using PPP-CONTSCR-10 pyramidal tips mounted on Si3N4 cantilevers with a nominal spring constant of 0.2 N/m precalibrated by the manufacturer (NanoSensors, Neuchatel, Switzerland). Force curves were acquired at indentation rates in the range of 0.3-1 um/s. Young’s modulus was calculated by applying a fit of the Hertz model to the force curve, assuming a Poisson ratio of 0.5. The moduli of at least 3 samples for each condition were calculated as an average over 3-6 sites per sample. Moduli of acrylamide hydrogels of defined stiffness and soft Matrigel substrata were used as positive controls.
In vivo metastasis assay
Animal experiments were performed according to Italian laws, as approved by the University Animal Welfare Commission (OPBA) and authorized by the Ministry of Health (615-2020-PR). D2.0R-EGFP/Luc cells were trypsinized, washed with 1xPBS, and then resuspended before injecting into the tail vein of 6- to 8-week-old female BALB/C mice (100 microl per mouse) using a 26 G needle. D2.0R cells expressing exogenous fluorescent proteins can be used for metastasis assays in immune-competent syngeneic BALB/C mice 99-101. Mice were kept in SPF cages with a limit of 5 mice per cage, with a 12-hour light/dark cycle, ad libitum feeding and drinking, at an average temperature of 19-24°C and with ambient humidity.
For quantification of D2.0R extravasation, 5 × 105 D2.0R-mCherry cells (serving as internal quantitative reference) were injected together with 5 × 105 D2.0R-EGFP/Luc cells expressing the different shRNAs. Three days after injection lungs were cleared of cells remaining in the vessels by postmortem intracardiac perfusion of 50 ml 1xPBS, and fixed in 4% PFA. The ratio of red/green cells was calculated based on quantification of 10 sections for each mouse, whose mean value is a single point indicated in the figure.
For quantification of D2.0R cell growth, 1 × 106 D2.0R-EGFP-Luc cells expressing shRNAs were injected (day 0). Each cell line was injected in n=12 different mice, chosen randomly. At day 7, mice were randomly divided in two groups, and treated by four sequential i.p. (intra peritoneal) injections of 200 microl Cisplatin (6 mg/Kg day 7 - 4 mg/Kg day 14 - 6 mg/Kg day 21 - 4 mg/Kg day 32) 102, or of the corresponding volume of vehicle (1xPBS). Cell growth was monitored by intravital bioluminescence imaging on anesthetized mice at day 0, before each cisplatin injection, and every 11 days thereafter. For this we injected i.p. D-Luciferin solution (PerkinElmer 122799, 200 microl 15 mg/ml in 1xPBS) in anesthetized mice, waited 10 minutes to allow its absorption, and acquired the total photon emission over 60 s. in a ROI spanning the whole chest. Background was measured by using an equivalent ROI positioned over the lower abdomen, which was subtracted to the chest ROI values. Metastatic growth was normalized in each mouse to injected cells, i.e. the total photon emission at day 0 (100%). Mice with relapsing lung metastatic growth, or with growth of cells close to the injection site at late time points, were culled at the first signs of distress.
RNA sequencing
Library generation was based on Quant Seq 3' mRNA-seq Library Prep kit (Lexogen) with oligo-dT priming. Sequencing was performed on NextSeq500 ILLUMINA instruments and based on at least 5 million reads (75bp SE) for each sample. The reads were trimmed using BBDuk (BBMap v. 37.87), with parameters indicated in the Lexogen data analysis protocol. After trimming, reads were aligned to the Mus musculus genome (GRCm38.p6) using STAR (v. 2.7.6a). The gene expression levels were quantified using featureCounts (v. 2.0.1). Genes were sorted removing those that had a total number of counts below 10 in at least 3 samples. All RNA-seq analyses were carried out in the R environment (v. 4.0.0) with Bioconductor (v. 3.7). We computed differential expression analysis using the DESeq2 R package (v. 1.28.1). Normalization to CPM has been performed exploiting the cpm function from the edgeR package (v. 3.32.0).
Statistics and Reproducibility
All data are based on independent experiments with independent biological replicates. Independent experiments were carried out by thawing a new aliquot of cells, deriving from the same original stock. Most results were qualitatively replicated across two different cell lines and with independent techniques providing coherent results. All n values are pooled between independent experiments. No samples or experiments were excluded. Experiments were not blind.
Experimental data are presented as mean and single points or mean and SD, as indicated in the figure legends. Significance tests were unpaired twotailed Student’s t-tests, or unpaired two-tailed Dunnet’s tests for multiple comparisons to a single control. The tests have been performed under the assumption that values follow a normal distribution and have similar variance. Tests were run with Prism v.9 or Excel v.16.55.
Extended Data
Extended Data Fig. 1. ECM stiffness regulates cystine metabolism and glutathione oxidation.
a, A simplified scheme depicting the major metabolic intermediates of the glutathione synthesis pathway, together with the enzymes mediating each reaction. In blue, the intermediates increased in MCF10A-RAS cells cultured in conditions of decreased actomyosin contractility.
b, Representative gating scheme for quantification of FITC-labeled cystine uptake by flow cytometry. Cells without stain were used as negative control (outlined white area). Cells treated with Y27632 and ML7 (YM - orange) display increased uptake compared to DMSO (grey), while treatment with the sulfasalazine (SAS - purple) inhibitor of the cystine transporter inhibits uptake. Scale bar, 50 μm. On the right: quantification of FITC-labeled cystine uptake. Data are mean and single points relative to DMSO (black bar). P-values by Dunnet’s test from a sample size of n=4 biologically independent samples pooled across two independent experiments for each bar.
c,d, Glutathione redox analysis using the mitochondrial mito-Grx1-roGFP2 sensor in MCF10A-RAS cells treated with YM (c), or cultured on stiff or soft Matrigel substrata (d). Data are mean and single cells. P-values by Dunnet’s test or Student’s t-test from a sample size of n=21 in c and n=31 in d cells pooled across two independent experiments for each bar.
e-g, Direct quantification of reduced (GSH, e) and oxidised (GSSH, f) glutathione in extracts from MCF10A-RAS cells treated with YM for 24 h. The ratio of reduced to oxidized glutathione was calculated in g. Data are mean and single points. n=2 independent experiments.
Images in b,c are representative of two independent experiments with similar results. See Source Data Extended Data Fig. 1.
Extended Data Fig. 2. ECM stiffness regulates ROS levels.
a, NADPH redox analysis using the iNap1 sensor in MCF10A-RAS cells treated with YM. Co-treatment with Diamide (DIA) and the G6PD inhibitor DHEA serves as positive control for NADPH depletion. iNapC is a NADP-insensitive and pH-sensitive control sensor (n=35 cells pooled across two independent experiments for each bar; unpaired two-tailed Student’s t-test).
b,c, Representative gating schemes and quantifications of DCFDA (b) and MitoSOX (c) oxidation by flow cytometry. Cells without stain were used as negative control (outlined white area). Cells treated with the positive control cumene hydroperoxide (CUM - purple) display increased uptake compared to DMSO (grey). Median intensity in the controls were set to 1, and other samples are relative to these (n=6 biologically independent samples pooled across three independent experiments for each bar; unpaired two-tailed Student’s t-test).
d-i, Quantification of reactive-oxygen species by the DCFDA and MitoSOX reagents in MCF10A, MCF10A-RAS and D2.0R cells. Median intensity in the controls were set to 1, and other samples are relative to these (in d n=4 (STIFF) and n=5 (all other conditions); in e n=11 (DMSO) n=6 (YM and SOFT) n=5 (BLEBBI) n=8 (STIFF); in f n=4 (STIFF) n=5 (all other conditions); in g n=4 (DMSO and YM) n=6 (STIFF and SOFT); in h n=6 (DMSO, YM6h and YM 24h) n=4 (YM1h and YM 3h); in i n=5 (YM 1h) and n=6 (all other conditions) biologically independent samples pooled across two independent experiments; unpaired two-tailed Student’s t-test or Dunnet’s test).
j, Representative gating scheme and quantification of C11-Bodipy-581/591 lipid peroxidation by flow cytometry. Cells without C11-Bodipy-581/591 were used as negative control (outlined white area). Cells treated with cumene hydroperoxide (CUM - dark purple) or with the GPX4 lipid hydroperoxide glutathione peroxidase inhibitor RSL3 (blue) for 3 h were used as positive controls. Mean intensity in the controls were set to 1, and other samples are relative to these (n=4 biologically independent samples pooled across two independent experiments for each bar; Dunnet’s test).
k, l, Quantification of C11-Bodipy-581/591 lipid peroxidation in MCF10A-RAS and D2.0R cells treated with YM for 3 h or cultured on Fibronectin-coated stiff or soft acrylamide hydrogels. Median intensity in the controls were set to 1, and other samples are relative to these (n=4 biologically independent samples pooled across two independent experiments for each bar; unpaired two-tailed Student’s t-test).
m, Immunofluorescence of 4-hydroxy-2-nonenal (4HNE) lipid peroxidation adducts in MCF10A-RAS cells cultured on stiff or soft Matrigel substrata for 24 h. Images are representative of at two experiments with similar results. Scale bar, 5 μm. Mean intensity in the controls were set to 1, and other samples are relative to these (n=36 (STIFF) n=34 (SOFT) cells pooled across two independent experiments; unpaired two-tailed Student’s t-test).
Data are mean and single points. See Source Data Extended Data Fig. 2.
Extended Data Fig. 3. ECM stiffness regulates NRF2 activity.
a,b, qPCR for established NRF2 target genes in D2.0R and MCF10A cells treated with YM for 6 h (in a FTH1 n=2 (DMSO) n=4 (YM); HMOX1 n=2 (DMSO) n=4 (YM); NQO1 n=2 (DMSO) n=4 (YM); GCLC n=2 (DMSO) n=4 (YM); SLC7A11 n=7 (DMSO) n=5 (YM) independent samples pooled across two independent experiments; in b HMOX1 n=8; FTH1 n=8 (DMSO) n=7 (YM); GCLC n=8 (DMSO) n=6 (YM); GCLM n=8 (DMSO) n=6 (YM); SLC7A11 n=8 (DMSO) n=6 (YM) independent samples pooled across two independent experiments; unpaired two-tailed Student’s t-tests.
c qPCR for established NRF2 target genes in D2.0R cells cultured on Matrigel substrata of different stiffness. (Fth1 n=7(STIFF) n=5 (SOFT); Hmox1 n=4; Nqo1 n=7; Gclm n=4; SLC7A11 n=6 (STIFF) n=5 (SOFT) independent samples pooled across two independent experiments; unpaired two-tailed Student’s t-tests.
d qPCR for established NRF2 target genes in MCF10A-RAS (FTH1 n=4; HMOX1 n=6 (DMSO and YM) n=4 (Ki696); NQO1 n=6 (DMSO and YM) n=4 (ki696); GCLC n=6 (DMSO and YM) n=4 (ki696) independent samples pooled across two independent experiments; unpaired two-tailed Student’s t-tests.
e-g, qPCR for established YAP/TAZ targets in D2.oR (e) and NRF2 and YAP/TAZ target genes in MCF10A-RAS (f,g). YAP/TAZ targets serve as internal positive controls.(in e n=4, in f n=4, in g n=10 samples pooled across two independent experiments for each bars; unpaired two-tailed Student’s t-tests).
h, qPCR for established NRF2 target genes in MCF10A cells cultured on soft (E≈0.2kPa) or stiff (E≈50kPa) CollagenI-coated hydrogels of different stiffness (n=4 samples pooled across two independent experiments for each bars; unpaired two-tailed Student’s t-tests).
i, Quantification of active nuclear S40-phosphorylated NRF2 (pNRF2) intensity from immunofluorescence stainings of MCF10A-RAS and D2.0R cells cultured on stiff or soft Matrigel substrata. KI696 is a KEAP1 inhibitor used as a positive control. Mean levels in the controls were set to 1, and other samples are relative to these (n=59 (DMSO) n=60 (YM and ki696) cells were imaged for each condition, pooled across two independent experiments; unpaired two-tailed Student’s t-tests).
j, qPCR to control for efficient knockdown of NE2L2 (encoding for NRF2) in MCF10A-RAS (n=10 (siCO) n=8 (siNRF2a and siNRF2b) samples pooled across two independent experiments for each bars; unpaired twotailed Student’s t-tests).
k, Immunoblotting for endogenous NRF2 from extracts of MCF10A-RAS cells transfected with the indicated siRNAs. Equal total proteins were loaded in each lane, and GAPDH was used as loading control. Images are representative of two independent experiments with similar results. Unprocessed blots in Source Data Extended Data Fig. 3.
j, qPCR to control for efficient knockdown of NE2L2 in D2.0R cells (n=5 (shCO) n=6 (shNrf2a and shNrf2b) samples pooled across two independent experiments for each bars; unpaired two-tailed Student’s t-tests).
Data are mean and single points. mRNA expression data are relative to GAPDH levels; mean level in the control was set to 1, and other samples are relative to this. See Source Data Extended Data Fig. 3.
Extended Data Fig. 4. Activation of NRF2 gene signatures in response to a soft ECM in published datasets.
a, Gene-Set Enrichment Analysis (GSEA) was performed on genes upregulated by culturing MCF10A cells on stiff 2D Matrigel-coated plates (red) or soft 3D Matrigel gels (blue). NRF2 signatures: KEAP/SFN (normalized enrichment score NES=-1.53 P<0.0001 false discovery rate FDR=0.038), LIST (NES=-1.91 P<0.0001 FDR<0.0001), PANIERI (NES=-1.75 P<0.0001 FDR=0.0006), HER2 (NES=-1.80 P<0.001 FDR=0.0005). YAP/TAZ (NES=2.01 P<0.0001 FDR<0.0001) and SREBP (NES=-1.47 P<0.017 FDR 0.06) serve as positive controls for gene signatures regulated by ECM stiffness.
b, GSEA was performed on genes upregulated by culturing D2.0R cells on stiff Matrigel+Collagen-I gels (red) or soft Matrigel gels (blue). NRF2 signatures: KEAP/SFN (NES=-2.11 P<0.0001 FDR<0.0001), LIST (NES=- 2.11 P<0.0001 FDR<0.0001), HER2 (NES=-1.86 P<0.001 FDR=0.0008). YAP/TAZ (NES=1.64 P=0.004 FDR=0.009) and SREBP (NES=-1.77 P<0.0001 FDR=0.0026) serve as positive controls for gene signatures regulated by ECM stiffness.
c, GSEA was performed on genes upregulated by culturing human MDA-MB-453 breast cancer cells on stiff (red) Fibronectin-coated plates or soft (blue) acrylamide hydrogels. NRF2 signatures: KEAP/SFN (NES=-3.25 P<0.001 FDR<0.001), LIST (NES=-3.60 P<0.001 FDR<0.001), HER2 (NES=-4.50 P<0.001 FDR<0.001). The YAP/TAZ signature (NES=1.71 P=0.025 FDR=0.048 Zhao NES=2.43 P<0.001 FDR<0.001) serves as positive control for genes regulated by ECM stiffness.
d, Heatmap of NRF2 target genes in RNAseq data of mouse MMTV-PyMT breast cancer cells cultured ex vivo on Fibronectin-coated plates, stiff or soft acrylamide hydrogels. Each column is an independent biological sample (n=4 for each condition); each line is a single gene. Expression levels for each gene were normalized relative to the mean expression on plates, which was set to 1 (white). Black and blue indicate downregulation and upregulation (P<0.05), respectively. YAP/TAZ target genes serve as positive controls.
e, GSEA was performed on genes regulated in matched cases of pre-treatment fine-needle aspiration (PRE, red) and the respective post neoadjuvant treatment operative sample (POST, blue) of primary breast cancer patients. Neoadjuvant treatment is known to reduce tumor stiffness. NRF2 signatures: KEAP/SFN (NES=-1.32 P=0.012 FDR=0.096), LIST (NES=-1.34 P=0.11 FDR=0.088), PANIERI (NES=-1.34 P=0.093 FDR=0.087). SREBP (NES=-1.94 P<0.001 FDR<0.001) and YAP/TAZ (NES=2.56 P<0.001 FDR<0.001) serve as positive controls for gene signatures regulated by ECM stiffness.
f, Heatmap of NRF2 and YAP target levels in n=7 patient-matched stiff keloid tissue or soft normal skin. Each column represents -log2(keloid/skin) values for a single patient; each line is a single gene probe; genes ranked according to expression in patient #1. Black and blue indicate downregulation and upregulation (P<0.05), respectively. YAP target genes serve as positive controls.
g, h, qPCR for established HSF1 target genes in MCF10A cells cultured on Fibronectin-coated stiff or soft hydrogels (g), or transfected on plastics with control (siCO) or with YAP/TAZ siRNAs (h). Data are mean and single points. mRNA expression data are relative to GAPDH levels; mean level in the control was set to 1, and other samples are relative to this (n=2 biologically independent samples from one experiment).
See Source Data Extended Data Fig. 4.
Extended Data Fig. 5. ECM stiffness regulates resistance to oxidative stress.
a, Representative gating scheme for quantification of cyto-Grx1-roGFP2 by flow cytometry. Cells without cyto- Grx1-roGFP2 expression were used as negative control (outlined white area). Cells treated with cumene hydroperoxide (CUM, purple) for 10 min. to induce glutathione oxidation were used as positive control (n=4 biologically independent samples pooled across two independent experiments for each bar; unpaired twotailed Student’s t-test).
b, Cell survival by the resazurin assay in MCF10A cells with the indicated knockdowns, plated on stiff or soft Matrigel substrata, and treated for 48 h with Cumene hydroperoxide (CUM). Mean cell number in controls was set to 100%, and all other samples are relative to this (n=12 biologically independent samples pooled across two independent experiments for each bar; Dunnet’s tests).
c, Cell survival of MCF10A-RAS cells transfected with the indicated KEAP1 siRNAs and treated for 48 h with Cumene hydroperoxide (CUM). Mean cell number in Stiff controls was set to 100%, and all other samples are relative to this (n=12 biologically independent samples pooled across two independent experiments for each bar; Dunnet’s test).
d, qPCR to control for efficient knockdown of KEAP1 in MCF10A-RAS. mRNA expression data are relative to GAPDH levels; mean level in the control was set to 1, and other samples are relative to this (n=4 biologically independent samples from two independent experiments; Dunnet’s test)
e, Immunoblotting for endogenous KEAP1 from extracts of MCF10A-RAS cells transfected with the indicated siRNAs. Equal total proteins were loaded in each lane, and GAPDH was used as loading control. Images are representative of two independent experiments with similar results. Unprocessed blots in Source Data Extended Data Fig. 5.
f, Cell survival of MCF10A-RAS cells transfected with the indicated NRF1 siRNA, plated on stiff or soft Matrigel substrata, and treated for 48 h with Cumene hydroperoxide (CUM). Mean cell number in Stiff controls was set to 100%, and all other samples are relative to this (n=12 biologically independent samples pooled across two independent experiments for each bar; unpaired two-tailed Student’s t-tests).
g, qPCR for NFE2L1 (encoding for NRF1) and NFE2L2 (encoding for NRF2) expression levels in MCF10A-RAS transfected with NRF1 siRNAs. mRNA expression data are relative to GAPDH levels; mean level in the control was set to 1, and other samples are relative to this (n=6 biologically independent samples from two independent experiments; Dunnet’s test).
h, i, Cell survival of MCF10A-RAS cells treated for 48 h with Erastin (ERA, g) or RSL3 (h) alone and together with the anti-ferroptosis compounds Liproxstatin-1 (LIP) or Ferrostatin (FER). Mean cell number in Stiff controls was set to 100%, and all other samples are relative to this (in h n=7 (VEHICLE DMSO, ERASTIN 10μM FER, ERASTIN 10μM LIP, ERASTIN 30μM DMSO) n=6 (ERASTIN 10μM DMSO) n=8 (all other conditions) biologically independent samples pooled from a single experiment; in i n=8 (VEHICLE) n=6 (RSL3 5 μ M LIP) n=7 (all other conditions) biologically independent samples pooled from a single experiment; Dunnet’s tests).
j, qPCR for YAP/TAZ targets in MCF10A-RAS cells used as a control for differential stiffness of the Matrigel substrata within the extracellular flux analyzer plates (see Fig. 4f). mRNA expression data are relative to GAPDH levels; mean level in the control was set to 1, and other samples are relative to this (n=6 biologically independent samples from three independent experiments; unpaired two-tailed Student’s t-tests).
k, qPCR for YAP/TAZ targets in D2.0R cells used as a control for differential stiffness of the Matrigel substrata within the extracellular flux analyzer plates (see Fig. 4g). mRNA expression data are relative to GAPDH levels; mean level in the control was set to 1, and other samples are relative to this (n=4 biologically independent samples from two independent experiments; unpaired two-tailed Student’s t-tests).
Data are mean and single points. See Source Data Extended Data Fig. 5.
Extended Data Fig. 6. ECM stiffness regulates mitochondrial fission through DRP1.
a, Quantification of mitochondria length in D2.0R cells transfected with mito-RFP and treated with YM (n=128 (DMSO) n=117(YM) mitochondria pooled across 10 cells per condition in two independent experiments for each bar; unpaired two-tailed Student’s t-tests). The same set of images was analyzed by automated classification of mitochondrial length (centre) and by MFC analysis (right). Data are mean and single mitochondria (left), mean and s.d. (centre) and mean and single pictures (right).
b, Quantification of mitochondria content by MitoTracker staining in MCF10A-RAS and D2.oR cells treated with YM. Median level in the control was set to 1, and other samples are relative to this (n=7 biologically independent samples pooled across three independent experiments for MCF10A-RAS; n=4 biologically independent samples pooled across two independent experiments for D2.0R).
c, Quantification of mitochondrial DNA content in MCF10A-RAS cells treated with YM, based on qPCR for three different mtDNA loci. mtDNA levels are relative to genomic DNA (gDNA) levels; mean level in the control was set to 1, and other samples are relative to this (n=6 biologically independent samples from two independent experiments).
d, qPCR for DRP1 in MCF10A cells treated with YM. mRNA expression data are relative to GAPDH levels; mean level in the control was set to 1, and other samples are relative to this (n=6 biologically independent samples from three independent experiments).
e, Immunoblotting for endogenous total and phosphorylated DRP1 (p616, p637) from extracts of MCF10A-RAS cells treated with ROCK/MLCK inhibitors. Equal total proteins were loaded in each lane, and GAPDH was used as loading control.
f, g. qPCR to control for efficient knockdown of DRP1 in MCF10A-RAS (f) or in D2.0R (g). mRNA expression data are relative to GAPDH levels; mean level in the control was set to 1, and other samples are relative to this (n=4 biologically independent samples from two independent experiments for each bar; unpaired two-tailed Student’s t-test in MCF10A-RAS, Dunnet’s test in D2.0R).
h. Immunoblotting for endogenous DRP1 from extracts of MCF10A-RAS cells transfected with the indicated siRNAs. Equal total proteins were loaded in each lane, and GAPDH was used as loading control.
i. Quantification of mtROS in MCF10A-RAS cells with knockdown of the indicated fission factors, and treated with YM. Median intensity in the control were set to 1, and other samples are relative to these (n=8 biologically independent samples pooled across four independent experiments for siCO bars; n=4 biologically independent samples pooled across two independent experiments for other bars; unpaired two-tailed Student’s t-tests). j-m. qPCR in MCF10A-RAS to control for efficient knockdown of the FIS1, MFF, MIEF1 and MIEF2 fission factors. mRNA expression data are relative to GAPDH levels; mean level in the control was set to 1, and other samples are relative to this (n=4 biologically independent samples from two independent experiments for each bar; Dunnet’s tests).
n. Oxygen Consumption Rate (OCR) analysis performed on monolayers of MCF10A-RAS cells transfected with control (siCO) or DRP1 siRNA and plated on stiff or soft Matrigel substrata (n=20 biologically independent samples pooled across two independent experiments).
o. OCR analysis on cells were treated with Drpitor1a (n=20 biologically independent samples pooled across two independent experiments).
p. OCR analysis performed on monolayers of D2.0R cells stably expressing control (shCo.) or DRP1a shRNA and cultured on stiff or soft Matrigel substrata (n=20 biologically independent samples pooled across two independent experiments).
q. OCR analysis on cells were treated with Drpitor1a (n=20 biologically independent samples pooled across two independent experiments).
Images in e,h are representative of two independent experiments with similar results. Unprocessed blots in Source Data Extended Data Fig. 6. Data are mean and single points, except n-q (mean and s.d. - shaded areas). See Source Data Extended Data Fig. 6.
Extended Data Fig. 7. A soft ECM increases resistance to Cisplatin and As2O3 chemotherapy through NRF2 and DRP1 in MCF10A-RAS cells.
a,b, Cell survival of MCF10A-RAS cells treated for 48 h with the indicated concentrations of Cisplatin (a) or As2O3 (b), in the absence or presence of the antioxidant N-Acetyl-L-cysteine (NAC, 5mM) (in a n=14 (VEHICLE) n=13 (VEHICLE NAC) n=15 (CISPLATIN 5μM) n=16 (CISPLATIN 20 μM) biologically independent samples pooled across two independent experiments; in b n=22 (VEHICLE and As2O3 20 μM) n=19 (VEHICLE NAC) n=21 (As2O3 5μM) biologically independent samples pooled across two independent experiments; Dunnet’s tests).
c,d, Cell survival assay in MCF10A-RAS cells cultured on stiff Matrigel-coated plates or soft Matrigel gels, and treated for 48 h with the indicated concentrations of Cisplatin (c) or As2O3 (d) (n=16 biologically independent samples pooled across two independent experiments for each bar; Dunnet’s test).
e, Cell survival assay in MCF10A-RAS cells treated for 48 h with the indicated concentrations of Doxorubicin (DOXO), in the absence or presence of N-Acetyl-L-cysteine (NAC, 5mM) (n=16 biologically independent samples pooled across two independent experiments for each bar).
f, Cell survival assay in MCF10A-RAS cells cultured on stiff Matrigel-coated plates (black) or soft Matrigel gels (green), and treated for 48 h with the indicated concentrations of Doxorubicin (DOXO) (n=14 biologically independent samples pooled across two independent experiments for each bar).
g, Cell survival of MCF10A-RAS cells transiently transfected as indicated and cultured on stiff Matrigel-coated plates or soft Matrigel gels, and treated for 48 h with the indicated concentration of Cisplatin. (n=8 (siCO and siNRF2b CISPLATIN SOFT) n=6 (all other conditions) biologically independent samples pooled across two independent experiments; Dunnet’s test).
h, Cell survival of MCF10A-RAS cells transiently transfected as indicated, cultured on stiff Matrigel-coated plates or soft Matrigel gels, and treated for 48 h with the indicated concentration of Cisplatin. Where indicated cells were also treated with the DRP1 inhibitor Drpitor1a. (n=14 (siCO) n=12 (siDRP1) n=16 (Drpitor1a) biologically independent samples pooled across two independent experiments; Dunnet’s test).
Data are mean and single points. Mean expression in untreated controls were set to 100%, and all other samples are relative to this. See Source Data Extended Data Fig. 7.
Extended Data Fig. 8. Controls to experiments shown in main Fig. 8.
a, A simplified model of the mechanical conditions used throughout Fig. 8. Cells are seeded in a soft microenvironment (wavy lines: in vitro BM ECM, ex vivo decellularized lung ECM, in vivo lung tissue) or directly in stiff conditions (straight bold lines: lung ECM slices treated with Ribose or derived from fibrotic mice). Cancer cells remodel the ECM, such that they stiffen their microenvironment at late time-points (LATE: 14 days in vitro, 1.5 months in vivo).
b, Quantification of tissue stiffness by atomic force microscopy on mouse mammary glands and normal lungs, using frozen tissue microtome sections. Data are mean and single points (n=4 and n=3 tissue sections from two mice).
c, YAP/TAZ immunofluorescence in D2.0R cells cultured on normal and Ribose-stiffened (RIB) lung ECM slices. Images are representative of two independent experiments with similar results. Scale bars, 25 μm.
d, Collagen-I and YAP/TAZ immunofluorescence in D2.0R cells cultured on normal and fibrosis-stiffened (FIB) lung ECM slices. Images are representative of two independent experiments with similar results. Scale bars, 25 μm.
e, qPCR to control for efficient regulation of YAP/TAZ in D2.0R cells infiltrated into normal or fibrotic (FIB) lung ECM scaffolds. Data are mean and single points; mRNA expression data are relative to GAPDH levels; mean level in the control was set to 1, and other samples are relative to this (n=7 biologically independent samples from two independent experiments for bars 1 and 2; n=2 biologically independent samples from one single experiment for bars 3).
See Source Data Extended Data Fig. 8.
Extended Data Fig. 9. Controls to in vivo experiments shown in main Fig. 8.
a, Experimental set-up to study D2.0R metastatic cell behavior in mice. D2.0R cells expressing GFP and Firefly-luciferase (GFP/Fluc) were injected via the tail vein to induce metastatic dissemination into the lungs and their initial cell number was quantified by intravital bioluminescence imaging (BLI). After letting cells settle for one week, mice were injected i.p. with Cisplatin or with the equivalent volume of vehicle (1xPBS) for four consecutive rounds. Cell growth was monitored after every injection and then every 11 days.
b, GFP/Fluc D2.0R cells expressing the indicated shRNAs were injected via the tail vein mixed at a 1:1 ratio together with RFP D2.0R cells to induce metastatic spread into the lungs. After three days, the ratio of red/green cells in the lung parenchyma was quantified to measure extravasation efficiency. Scale bars, 20 μm. Images are representative of three mice with similar results. Data are mean and single points (n=3 mice for each condition).
c, Intravital imaging of mice with lung metastases from GFP/Fluc D2.0R cells quantified in Fig. 8l, taken at day 90. The rainbow LUT was used to visualize the radiance. Images are representative of six mice with similar results per condition.
d, Quantification of activated Caspase-3 in D2.0R cells disseminated to the mouse lung after treatment with two doses of Cisplatin (see scheme above) (n=2 mice). On the right, an immunofluorescent picture with spectral unmixing showing GFP-positive D2.0R cells (cyan) disseminated into the lung parenchyma and one example of a double-positive Caspase-3/GFP cell (yellow). Tissue sections were also stained with CD31 (magenta) to visualize blood vessels. Scale bar, 20 μm.
e, Intravital imaging of mice with lung metastases from GFP/Fluc D2.0R cells quantified in Fig. 8m, taken at day 75. The rainbow LUT was used to visualize the radiance. Images are representative of six mice with similar results per condition.
Data are mean and single points. See Source Data Extended Data Fig. 9.
Extended Data Fig. 10. Proposed model.
A simplified model that recapitulates the main findings of the manuscript, ordered in time after shifting cells to a soft microenvironment. On a soft ECM, cells develop reduced actomyosin tension, which is associated with increased formation of peri-mitochondrial F-actin, increased DRP1-dependent mitochondrial fission and enhanced mtROS production. mtROS activate a NRF2-dependent antioxidant metabolic response which includes increased cysteine uptake and glutathione synthesis. Functionally, this response has two main outcomes: it keeps redox balance in the face of increased ROS and glutathione oxidation (upward facing arrow), and it results in a better ability of cells to resist exogenous oxidative stress and ROS-dependent chemotherapy (downward facing arrow), compared to cells in a stiff microenvironment. mt is an abbreviation for mitochondria.
Acknowledgements
We are grateful to Diego De Stefani for help with plasmids and advice on toroidal mitochondria; to Luca Scorrano and Margherita Zamberlan for protocols to measure mitochondrial morphology and reagents to study DRP1; to Cecilia Laterza for help with tissue sectioning; to Jessica Weedon who contributed to the setup of the Bleomycin model; to Ildiko Szabo for help with the TEM and for critically reading of the manuscript. Support: Worldwide Cancer Research Grant 21-0156, AIRC Foundation Investigator Grant 21392 and CARIPARO Excellence Grant to S.D. P.R. is supported by a Veronesi Foundation Postdoctoral Fellowship. AIRC Foundation Investigator Grant 2018-ID 2135, AIRC Foundation Investigator Grant 5 per mille 22759, Ministero della Salute RCR-2019-23669115, Ministero della Salute NET-2016-02361632, Istituto Oncologico Veneto to A.R. Giovanni Armenise–Harvard Foundation, ERC Starting Grant (MetEpiStem) to G.M. Canadian Institutes for Health Research (CIHR) Foundation Grant, National Institutes of Health R01-HL071115, 1RC1HL099462, a Tier I Canada Research Chair, the William J. Henderson Foundation to S.L.A. Longfonds (Voorhen Astma Fonds) BREATH Consortium, NIHR GOSH Biomedical Research Centre, the National Institute for Health Research (NIHR-RP-2014-04-046) to P.D.C. F.M. is supported by a GOSH BRC Catalyst Fellowship. University of Padua STARS Consolidator Grant 2019 to T.C. University of Padua TWINING-VELT 2017 Grant to N.E. Fondazione IRP Città della Speranza to A.U.
Footnotes
Author contributions
P.R. carried out experiments and analyzed data with help from N.N. M.A. retrieved raw data for GSEA and performed RNAseq analysis, with support from G.M. A.T. and V.B. performed IHC and helped with mouse experiments, with support from A.R. D.W. performed mitochondrial morphology analysis, C.C.T.H. and S.L.A. provided Drpitor1a and support. F.M. S.S. and T. B. prepared mouse lung decellularized ECM slices and performed lung infiltration experiments, with support from P.D.C. M.G. and A.U. performed AFM measurements, with support from N.E. F.G. helped with SPLICs experiments, with support from T.C. A.R. performed GSH/GSSG quantifications. P.C. and M.M. helped with GSEA analysis. S.D. conceptualized, coordinated and supervised the project, and wrote the manuscript.
Competing interests
The invention of Drpitor1a is the subject of a US Patent Application 20200323829 from D.W. and S.L.A. The remaining authors declare no competing interests.
Data availability
Previously published metabolomics data have been deposited to Figshare database and available at DOI 10.6084/m9.figshare.7338764. RNA sequencing data have been deposited to GEO database as GSE189803 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189803). Source data are provided with this study. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.Min E, Schwartz MA. Translocating transcription factors in fluid shear stress-mediated vascular remodeling and disease. Exp Cell Res. 2019;376:92–97. doi: 10.1016/j.yexcr.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petridou NI, Spiró Z, Heisenberg C-P. Multiscale force sensing in development. Nature Cell Biology. 2017;19:581–588. doi: 10.1038/ncb3524. [DOI] [PubMed] [Google Scholar]
- 3.Tschumperlin DJ, Ligresti G, Hilscher MB, Shah VH. Mechanosensing and fibrosis. J Clin Invest. 2018;128:74–84. doi: 10.1172/JCI93561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vining KH, Mooney DJ. Mechanical forces direct stem cell behaviour in development and regeneration. Nature Reviews Molecular Cell Biology. 2017;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol. 2014;15:802–812. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iskratsch T, Wolfenson H, Sheetz MP. Appreciating force and shape—the rise of mechanotransduction in cell biology. Nat Rev Mol Cell Biol. 2014;15:825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammadi H, Sahai E. Mechanisms and impact of altered tumour mechanics. Nat Cell Biol. 2018;20:766–774. doi: 10.1038/s41556-018-0131-2. [DOI] [PubMed] [Google Scholar]
- 8.Gensbittel V, et al. Mechanical Adaptability of Tumor Cells in Metastasis. Dev Cell. 2021;56:164–179. doi: 10.1016/j.devcel.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Montagner M, Dupont S. Mechanical Forces as Determinants of Disseminated Metastatic Cell Fate. Cells. 2020;9 doi: 10.3390/cells9010250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romani P, Valcarcel-Jimenez L, Frezza C, Dupont S. Crosstalk between mechanotransduction and metabolism. Nature Reviews Molecular Cell Biology. 2020:1–17. doi: 10.1038/s41580-020-00306-w. [DOI] [PubMed] [Google Scholar]
- 11.Romani P, et al. Extracellular matrix mechanical cues regulate lipid metabolism through Lipin-1 and SREBP. Nat Cell Biol. 2019;21:338–347. doi: 10.1038/s41556-018-0270-5. [DOI] [PubMed] [Google Scholar]
- 12.Gutscher M, et al. Real-time imaging of the intracellular glutathione redox potential. Nature Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 13.Tao R, et al. Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat Methods. 2017;14:720–728. doi: 10.1038/nmeth.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawk MA, et al. RIPK1-mediated induction of mitophagy compromises the viability of extracellular-matrix-detached cells. Nat Cell Biol. 2018;20:272–284. doi: 10.1038/s41556-018-0034-2. [DOI] [PubMed] [Google Scholar]
- 15.Jiang L, et al. Reductive carboxylation supports redox homeostasis during anchorageindependent growth. Nature. 2016;532:255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schafer ZT, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki H, et al. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J Biol Chem. 2002;277:44765–44771. doi: 10.1074/jbc.M208704200. [DOI] [PubMed] [Google Scholar]
- 18.Rojo de la Vega M, Chapman E, Zhang DD. NRF2 and the Hallmarks of Cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sies H, Jones DP. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nature Reviews Molecular Cell Biology. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 20.Agyeman AS, et al. Transcriptomic and Proteomic Profiling of KEAP1 Disrupted and Sulforaphane Treated Human Breast Epithelial Cells Reveals Common Expression Profiles. Breast Cancer Res Treat. 2012;132:175–187. doi: 10.1007/s10549-011-1536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends in Biochemical Sciences. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Panieri E, Telkoparan-Akillilar P, Suzen S, Saso L. The NRF2/KEAP1 Axis in the Regulation of Tumor Metabolism: Mechanisms and Therapeutic Perspectives. Biomolecules. 2020;10 doi: 10.3390/biom10050791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero R, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Barrueco R, et al. Inhibition of the autocrine IL-6-JAK2-STAT3-calprotectin axis as targeted therapy for HR-/HER2+ breast cancers. Genes Dev. 2015;29:1631–1648. doi: 10.1101/gad.262642.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun. 2018;9:1944. doi: 10.1038/s41467-018-04070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina SH, et al. Identification of a mechanogenetic link between substrate stiffness and chemotherapeutic response in breast cancer. Biomaterials. 2019;202:1–11. doi: 10.1016/j.biomaterials.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havas KM, et al. Metabolic shifts in residual breast cancer drive tumor recurrence. J Clin Invest. 2017;127:2091–2105. doi: 10.1172/JCI89914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jing H, et al. Early Evaluation of Relative Changes in Tumor Stiffness by Shear Wave Elastography Predicts the Response to Neoadjuvant Chemotherapy in Patients With Breast Cancer. J Ultrasound Med. 2016;35:1619–1627. doi: 10.7863/ultra.15.08052. [DOI] [PubMed] [Google Scholar]
- 29.Hsu C-K, et al. Caveolin-1 Controls Hyperresponsiveness to Mechanical Stimuli and Fibrogenesis-Associated RUNX2 Activation in Keloid Fibroblasts. J Invest Dermatol. 2018;138:208–218. doi: 10.1016/j.jid.2017.05.041. [DOI] [PubMed] [Google Scholar]
- 30.Huang C, Ogawa R. Fibroproliferative disorders and their mechanobiology. Connect Tissue Res. 2012;53:187–196. doi: 10.3109/03008207.2011.642035. [DOI] [PubMed] [Google Scholar]
- 31.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 32.Ahn S-G, Thiele DJ. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003;17:516–528. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendillo ML, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tharp KM, et al. Adhesion-mediated mechanosignaling forces mitohormesis. Cell Metab. 2021;33:1322–1341.:e13. doi: 10.1016/j.cmet.2021.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo M, et al. Heat stress activates YAP/TAZ to induce the heat shock transcriptome. Nat Cell Biol. 2020;22:1447–1459. doi: 10.1038/s41556-020-00602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandel NS. Evolution of Mitochondria as Signaling Organelles. Cell Metabolism. 2015;22:204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Shadel GS, Horvath TL. Mitochondrial ROS Signaling in Organismal Homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yun J, Finkel T. Mitohormesis. Cell Metabolism. 2014;19:757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon SJ, Stockwell BR. The Hallmarks of Ferroptosis. Annual Review of Cancer Biology. 2019;3:35–54. [Google Scholar]
- 40.Ursini F, Maiorino M. Lipid peroxidation and ferroptosis: The role of GSH and GPx4. Free Radic Biol Med. 2020;152:175–185. doi: 10.1016/j.freeradbiomed.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 41.Bertero T, et al. Tumor-Stroma Mechanics Coordinate Amino Acid Availability to Sustain Tumor Growth and Malignancy. Cell Metab. 2019;29:124–140.:e10. doi: 10.1016/j.cmet.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty M, et al. Mechanical Stiffness Controls Dendritic Cell Metabolism and Function. Cell Reports. 2021;34:108609. doi: 10.1016/j.celrep.2020.108609. [DOI] [PubMed] [Google Scholar]
- 43.Khan AUH, et al. Mitochondrial Complex I activity signals antioxidant response through ERK5. Scientific Reports. 2018;8:7420. doi: 10.1038/s41598-018-23884-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santacatterina F, et al. Down-regulation of oxidative phosphorylation in the liver by expression of the ATPase inhibitory factor 1 induces a tumor-promoter metabolic state. Oncotarget. 2015;7:490–508. doi: 10.18632/oncotarget.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu D, et al. Identification of novel dynamin-related protein 1 (Drp1) GTPase inhibitors: Therapeutic potential of Drpitor1 and Drpitor1a in cancer and cardiac ischemia-reperfusion injury. The FASEB Journal. 2020;34:1447–1464. doi: 10.1096/fj.201901467R. [DOI] [PubMed] [Google Scholar]
- 46.Giacomello M, Pyakurel A, Glytsou C, Scorrano L. The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol. 2020;21:204–224. doi: 10.1038/s41580-020-0210-7. [DOI] [PubMed] [Google Scholar]
- 47.Kraus F, Roy K, Pucadyil TJ, Ryan MT. Function and regulation of the divisome for mitochondrial fission. Nature. 2021;590:57–66. doi: 10.1038/s41586-021-03214-x. [DOI] [PubMed] [Google Scholar]
- 48.Cieri D, et al. SPLICS: a split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ. 2018;25:1131–1145. doi: 10.1038/s41418-017-0033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallese F, et al. An expanded palette of improved SPLICS reporters detects multiple organelle contacts in vitro and in vivo. Nature Communications. 2020;11:6069. doi: 10.1038/s41467-020-19892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bordt EA, et al. The Putative Drp1 Inhibitor mdivi-1 Is a Reversible Mitochondrial Complex I Inhibitor that Modulates Reactive Oxygen Species. Developmental Cell. 2017;40:583–594.:e6. doi: 10.1016/j.devcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cassidy-Stone A, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atkins K, Dasgupta A, Chen K-H, Mewburn J, Archer SL. The role of Drp1 adaptor proteins MiD49 and MiD51 in mitochondrial fission: implications for human disease. Clin Sci (Lond) 2016;130:1861–1874. doi: 10.1042/CS20160030. [DOI] [PubMed] [Google Scholar]
- 53.Dasgupta A, et al. An epigenetic increase in mitochondrial fission by MiD49 and MiD51 regulates the cell cycle in cancer: Diagnostic and therapeutic implications. The FASEB Journal. 2020;34:5106–5127. doi: 10.1096/fj.201903117R. [DOI] [PubMed] [Google Scholar]
- 54.Koirala S, et al. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. PNAS. 2013;110:E1342–E1351. doi: 10.1073/pnas.1300855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osellame LD, et al. Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J Cell Sci. 2016;129:2170–2181. doi: 10.1242/jcs.185165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lomakin AJ, et al. Competition for actin between two distinct F-actin networks defines a bistable switch for cell polarization. Nat Cell Biol. 2015;17:1435–1445. doi: 10.1038/ncb3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pocaterra A, et al. Fascin1 empowers YAP mechanotransduction and promotes cholangiocarcinoma development. Commun Biol. 2021;4:1–13. doi: 10.1038/s42003-021-02286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suarez C, Kovar DR. Internetwork competition for monomers governs actin cytoskeleton organization. Nat Rev Mol Cell Biol. 2016;17:799–810. doi: 10.1038/nrm.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Q, Zhang X-F, Pollard TD, Forscher P. Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J Cell Biol. 2012;197:939–956. doi: 10.1083/jcb.201111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carlier M-F, Shekhar S. Global treadmilling coordinates actin turnover and controls the size of actin networks. Nat Rev Mol Cell Biol. 2017;18:389–401. doi: 10.1038/nrm.2016.172. [DOI] [PubMed] [Google Scholar]
- 61.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li S, et al. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J Cell Biol. 2015;208:109–123. doi: 10.1083/jcb.201404050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manor U, et al. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife. 2015;4 doi: 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Posern G, Sotiropoulos A, Treisman R. Mutant Actins Demonstrate a Role for Unpolymerized Actin in Control of Transcription by Serum Response Factor. Mol Biol Cell. 2002;13:4167–4178. doi: 10.1091/mbc.02-05-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Helle SCJ, et al. Mechanical force induces mitochondrial fission. eLife. 2017;6:e30292. doi: 10.7554/eLife.30292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kleele T, et al. Distinct fission signatures predict mitochondrial degradation or biogenesis. Nature. 2021;593:435–439. doi: 10.1038/s41586-021-03510-6. [DOI] [PubMed] [Google Scholar]
- 67.Moore AS, et al. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature. 2021;591:659–664. doi: 10.1038/s41586-021-03309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barkan D, Chambers AF. β1-integrin: a potential therapeutic target in the battle against cancer recurrence. Clin Cancer Res. 2011;17:7219–7223. doi: 10.1158/1078-0432.CCR-11-0642. [DOI] [PubMed] [Google Scholar]
- 69.Montagner M, Sahai E. In vitro Models of Breast Cancer Metastatic Dormancy. Front Cell Dev Biol. 2020;8:37. doi: 10.3389/fcell.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klein CA. Cancer progression and the invisible phase of metastatic colonization. Nature Reviews Cancer. 2020;20:681–694. doi: 10.1038/s41568-020-00300-6. [DOI] [PubMed] [Google Scholar]
- 71.Barkan D, et al. Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Res. 2008;68:6241–6250. doi: 10.1158/0008-5472.CAN-07-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barkan D, et al. Metastatic growth from dormant cells induced by a col-I-enriched fibrotic environment. Cancer Res. 2010;70:5706–5716. doi: 10.1158/0008-5472.CAN-09-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alsafadi HN, et al. Applications and Approaches for Three-Dimensional Precision-Cut Lung Slices. Disease Modeling and Drug Discovery. Am J Respir Cell Mol Biol. 2020;62:681–691. doi: 10.1165/rcmb.2019-0276TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giobbe GG, et al. Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat Commun. 2019;10:5658. doi: 10.1038/s41467-019-13605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maghsoudlou P, et al. Preservation of microarchitecture and angiogenic potential in a pulmonary acellular matrix obtained using intermittent intra-tracheal flow of detergent enzymatic treatment. Biomaterials. 2013;34:6638–6648. doi: 10.1016/j.biomaterials.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu F, et al. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J Cell Biol. 2010;190:693–706. doi: 10.1083/jcb.201004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Girton TS, Oegema TR, Tranquillo RT. Exploiting glycation to stiffen and strengthen tissue equivalents for tissue engineering. J Biomed Mater Res. 1999;46:87–92. doi: 10.1002/(sici)1097-4636(199907)46:1<87::aid-jbm10>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 78.Roy R, Boskey A, Bonassar LJ. Processing of Type I Collagen Gels Using Non-Enzymatic Glycation. J Biomed Mater Res A. 2010;93:843–851. doi: 10.1002/jbm.a.32231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goddard ET, Bozic I, Riddell SR, Ghajar CM. Dormant tumour cells, their niches and the influence of immunity. Nature Cell Biology. 2018;20:1240–1249. doi: 10.1038/s41556-018-0214-0. [DOI] [PubMed] [Google Scholar]
- 80.Rehman J, et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26:2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takahashi N, et al. 3D Culture Models with CRISPR Screens Reveal Hyperactive NRF2 as a Prerequisite for Spheroid Formation via Regulation of Proliferation and Ferroptosis. Molecular Cell. 2020 doi: 10.1016/j.molcel.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen K, Wang Y, Deng X, Guo L, Wu C. Extracellular matrix stiffness regulates mitochondrial dynamics through PINCH-1- and kindlin-2-mediated signalling. Current Research in Cell Biology. 2021;2:100008 [Google Scholar]
- 83.Yang H, et al. Materials Stiffness-Dependent Redox Metabolic Reprogramming of Mesenchymal Stem Cells for Secretome-Based Therapeutic Angiogenesis. Advanced Healthcare Materials. 2019;8:1900929. doi: 10.1002/adhm.201900929. [DOI] [PubMed] [Google Scholar]
- 84.Khacho M, et al. Mitochondrial Dynamics Impacts Stem Cell Identity and Fate Decisions by Regulating a Nuclear Transcriptional Program. Cell Stem Cell. 2016;19:232–247. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 85.Hatch AL, Ji W-K, Merrill RA, Strack S, Higgs HN. Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol Biol Cell. 2016;27:3109–3121. doi: 10.1091/mbc.E16-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Palmer CS, et al. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moore AS, Wong YC, Simpson CL, Holzbaur ELF. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat Commun. 2016;7:12886. doi: 10.1038/ncomms12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X, Hajnóczky G. Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia–reoxygenation stress. Cell Death Differ. 2011;18:1561–1572. doi: 10.1038/cdd.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miyazono Y, et al. Uncoupled mitochondria quickly shorten along their long axis to form indented spheroids, instead of rings, in a fissionindependent manner. Scientific Reports. 2018;8:350. doi: 10.1038/s41598-017-18582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maguire SL, et al. Three-dimensional modelling identifies novel genetic dependencies associated with breast cancer progression in the isogenic MCF10 model. The Journal of Pathology. 2016;240:315–328. doi: 10.1002/path.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pocaterra A, et al. F-actin dynamics regulates mammalian organ growth and cell fate maintenance. J Hepatol. 2019;71:130–142. doi: 10.1016/j.jhep.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 92.Ignesti M, et al. A polydnavirus-encoded ANK protein has a negative impact on steroidogenesis and development. Insect Biochem Mol Biol. 2018;95:26–32. doi: 10.1016/j.ibmb.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 93.Romani P, et al. Dynamin controls extracellular level of Awd/Nme1 metastasis suppressor protein. Naunyn Schmiedebergs Arch Pharmacol. 2016;389:1171–1182. doi: 10.1007/s00210-016-1268-9. [DOI] [PubMed] [Google Scholar]
- 94.Romani P, Duchi S, Gargiulo G, Cavaliere V. Evidence for a novel function of Awd in maintenance of genomic stability. Sci Rep. 2017;7:16820. doi: 10.1038/s41598-017-17217-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen K-H, et al. Epigenetic Dysregulation of the Dynamin-Related Protein 1 Binding Partners MiD49 and MiD51 Increases Mitotic Mitochondrial Fission and Promotes Pulmonary Arterial Hypertension: Mechanistic and Therapeutic Implications. Circulation. 2018;138:287–304. doi: 10.1161/CIRCULATIONAHA.117.031258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Enzo E, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015;34:1349–1370. doi: 10.15252/embj.201490379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hagen CK, et al. High contrast microstructural visualization of natural acellular matrices by means of phase-based x-ray tomography. Sci Rep. 2015;5:18156. doi: 10.1038/srep18156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Urciuolo A, et al. Intravital three-dimensional bioprinting. Nat Biomed Eng. 2020;4:901–915. doi: 10.1038/s41551-020-0568-z. [DOI] [PubMed] [Google Scholar]
- 99.Albrengues J, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science. 2018;361 doi: 10.1126/science.aao4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Montagner M, et al. Crosstalk with lung epithelial cells regulates Sfrp2-mediated latency in breast cancer dissemination. Nat Cell Biol. 2020;22:289–296. doi: 10.1038/s41556-020-0474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shibue T, Brooks MW, Weinberg RA. An integrin-linked machinery of cytoskeletal regulation that enables experimental tumor initiation and metastatic colonization. Cancer Cell. 2013;24:481–498. doi: 10.1016/j.ccr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aston WJ, et al. A systematic investigation of the maximum tolerated dose of cytotoxic chemotherapy with and without supportive care in mice. BMC Cancer. 2017;17 doi: 10.1186/s12885-017-3677-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Previously published metabolomics data have been deposited to Figshare database and available at DOI 10.6084/m9.figshare.7338764. RNA sequencing data have been deposited to GEO database as GSE189803 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189803). Source data are provided with this study. All other data supporting the findings of this study are available from the corresponding author on reasonable request.