Abstract
Compelling evidence for the existence of somatic stem cells in the heart of different mammalian species has been provided by numerous groups; however, so far it has not been possible to maintain these cells as self-renewing and phenotypically stable clonal cell lines in vitro. Thus, we sought to identify a surrogate stem cell niche for the isolation and persistent maintenance of stable clonal cardiovascular progenitor cell lines, enabling us to study the mechanism of self-renewal and differentiation in these cells. Using postnatal murine hearts with a selectable marker as the stem cell source and embryonic stem cells and leukemia inhibitory factor (LIF)-secreting fibroblasts as a surrogate niche, we succeeded in the isolation of stable clonal cardiovascular progenitor cell lines. These cell lines self-renew in an LIF-dependent manner. They express both stemness transcription factors Oct4, Sox2, and Nanog and early myocardial transcription factors Nkx2.5, GATA4, and Isl-1 at the same time. Upon LIF deprivation, they exclusively differentiate to functional cardiomyocytes and endothelial and smooth muscle cells, suggesting that these cells are mesodermal intermediates already committed to the cardiogenic lineage. Cardiovascular progenitor cell lines can be maintained for at least 149 passages over 7 years without phenotypic changes, in the presence of LIF-secreting fibroblasts. Isolation of wild-type cardiovascular progenitor cell lines from adolescent and old mice has finally demonstrated the general feasibility of this strategy for the isolation of phenotypically stable somatic stem cell lines.
Keywords: Somatic stem cells, Cardiovascular progenitor cells, Clonal cell lines, Persistent self-renewal, Leukemia inhibitory factor, Cardiomyogenesis, Restricted differentiation potential, Cardiomyocytes, Vascular endothelial cells, Smooth muscle cells
Abbreviations used in this paper
- CBs
cardiac bodies
- CMCs
cardiomyocytes
- cTnT
cardiac troponin T
- CVPCs
cardiovascular progenitor cells
- EBs
embryoid bodies
- ESCs
embryonic stem cells
- ETCs
endothelial cells
- LIF
leukemia inhibitory factor
- MHC
myosin heavy chain
- PBS
phosphate-buffered saline
- SMA
smooth muscle actin
- SMCs
smooth muscle cells
- Tk
thymidine kinase
- vWF
von Willebrand factor
Introduction
Heart stem cells or cardiovascular progenitor cells (CVPCs) [Scott, 2012; Taubenschmid and Weitzer, 2012] have been identified in neonatal [Porrello et al., 2011] and adult hearts of different species [Beltrami et al., 2001; Messina et al., 2004; Anversa et al., 2007b; Kajstura et al., 2008; Laugwitz et al., 2008; Wu et al., 2008; Bergmann et al., 2009; Hansson et al., 2009; Kajstura et al., 2010]. Within the heart, CVPCs have been identified in epicardial tissue [Limana et al., 2007], ventricular tissue [Galvez et al., 2008] (there referred to as mesoangioblasts), and heart auricles [Gambini et al., 2010]. In vitro they have regularly been found in cardiospheres [Messina et al., 2004; Smith et al., 2007; Davis et al., 2009]. For their identification in situ and for their isolation by preparative fluorescence-activated cell sorting, surface markers such as stem cell antigen-1 (Sca1) [Matsuura et al., 2004; Tateishi et al., 2007; Smits et al., 2009], the drug extruder MDR-1 [Oh et al., 2003], the vascular endothelial growth factor receptor Flk1 [Kattman et al., 2006; Yang et al., 2008], the stem cell factor receptor c-Kit [Beltrami et al., 2003; Bearzi et al., 2007, 2009; Tallini et al., 2009; Gambini et al., 2010], and the transcription factor Islet-1 (Isl-1) [Laugwitz et al., 2005; Moretti et al., 2006; Laugwitz et al., 2008] have been used. In vitro CVPCs differentiate only to cardiomyocytes (CMCs), vascular endothelial cells (ETCs), and smooth muscle cells (SMCs) [Srivastava and Ivey, 2006; Bearzi et al., 2007; Oyama et al., 2007; Wu et al., 2008; Smits et al., 2009]. Introduced into the hearts of mice adjacent to infarcted areas, CVPC populations contribute to the regeneration of the diseased heart to variable extents [Beltrami et al., 2003; Bearzi et al., 2007; Martin-Puig et al., 2008; Davis et al., 2009].
Stem cells and progenitor cells are supposed to exist in niches, where they remain in an undifferentiated and quasi dormant state until external signals stimulate differentiation to specific somatic cells. The myocardium possesses interstitial structures with the architectural organization of specific stem cell niches [Urbanek et al., 2006], particularly located in the atria and in the subepicardium of the ventricles [Popescu et al., 2009; Gherghiceanu and Popescu, 2010; Kuhn and Wu, 2010]. Many niche cells express surface receptors such as N-cadherin or α4β1 integrins found on fibroblasts [Moore and Lemischka, 2006]. Cardiac fibroblasts are abundant in the interstitial space of the heart [Oyama et al., 2007] and express growth factors supporting self-renewal. Notably, CVPCs proliferate in newborn mice as long as leukemia inhibitory factor (LIF) expression is detectable in the myocardium [Robertson et al., 1993]. However, when removed from the niche, and particularly at low cell densities, somatic stem cells in most instances succumb to anoikis, an apoptosis-like process [Frisch and Francis, 1994; Yu et al., 2012]. In contrast, in embryonic stem cell (ESC) cultures a niche-like environment composed of ESC-derived fibroblasts and ESCs themselves maintain the stem cell status by a complex interplay between growth factors secreted from fibroblasts [Bendall et al., 2007], and LIF [Niwa et al., 2009] and do not undergo apoptosis. Likewise, primitive mesodermal cells establish their own progenitor cell niche by a Brachyury maintained autoregulatory mechanism [Martin and Kimelman, 2010], and cardiospheres also seem to establish a niche-like environment [Anversa et al., 2007a; Li et al., 2010].
From these observations we conclude that a temporal high stem cell density at the beginning of the CVPC isolation procedure could foster the survival of CVPCs, and the presence of LIF together with fibroblast-derived growth factors could promote and maintain the self-renewal of CVPCs in vitro.
To test this hypothesis, we designed an in vitro niche composed of LIF-secreting SNL76/7 fibroblasts and murine ESCs. ESCs should support cardiac stem cell survival during the initial phase of isolation and propagation. Once a critical mass of CVPCs is generated, which allows self-supportive self-renewal of CVPCs, ESCs will be removed by drug selection. Then it should be possible to maintain CVPCs exclusively in the presence of LIF-secreting fibroblasts similar to ESC culture.
Here we describe a robust and simple method for the isolation and unlimited culture of clonal self-renewing CVPC lines from newborn and adult mouse hearts, characterize these cell lines, and demonstrate that they can reproducibly differentiate to functional CMCs, ETCs, and SMCs both in monolayer cultures and in embryoid body (EB)-like cardiac bodies (CBs).
Materials and Methods
Ethics Statement
Mice from which hearts were obtained to isolate the new monoclonal CVPC lines were maintained and sacrificed humanely according to the Austrian animal welfare law. According to the Austrian animal welfare law [Bundesgesetz über den Schutz der Tiere (Tierschutzgesetz – TSchG), Art. 2 §6 Abs. 3, BGBl. I Nr. 118/2004 zuletzt geändert durch BGBl. I Nr. 80/2010] no ethical review or permit is required to breed and sacrifice mice at academic institutions such as the Medical University of Vienna.
Isolation and Persistent Culture of CVPCs and ESCs
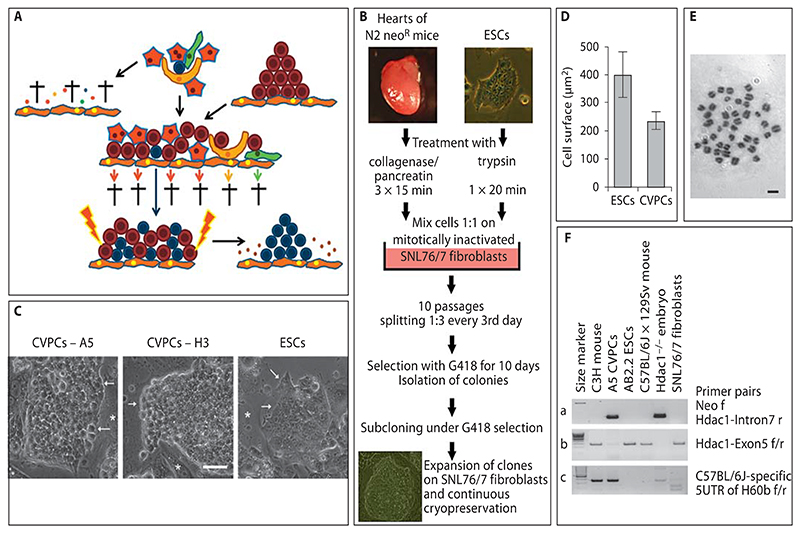
Hearts were obtained from 2-day-old outbred C57BL/6J × 129Sv mice with a neomycin resistance cassette inserted into one allele of the Hdac1 locus [Lagger et al., 2002] and from 2-, 22-, and 180-day-old wild-type Balb/C mice, respectively. Hearts were placed in ice-cold phosphate-buffered saline (PBS), thoroughly rinsed with PBS, and cut into very small pieces. Residual blood cells were removed by centrifugation at 1,000 rpm at room temperature for 1 min and the pelleted tissue was suspended in 1 ml of PBS containing 0.6 mg pancreatin (Gibco) and 0.5 mg collagenase II (Worthington) and incubated at 37 °C on a shaker at 800 rpm for 15 min twice. The supernatant of the first incubation period was stored on ice and combined with the second one. After centrifugation, cells were suspended in M15Hy medium composed of high glucose DMEM containing 2 mmol/l glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, 0.1 mmol/β-mercaptoethanol, and 15% (v/v) fetal bovine serum (Hyclone SH30070.30, lot ASK30724). Cardiac cells (5 × 105) were mixed with 5 × 105 AB2.2 ESCs [Soriano et al., 1991] and plated into 1 well of a 24-well plate coated with mitotically inactivated SNL76/7 fibroblasts as feeder cells [McMahon and Bradley, 1990; Soriano et al., 1991]. Balb/C heart cells were mixed with the transgenic ESC line Tag3 containing 2 Thymidine kinase suicide genes from human herpes simplex virus type I and a neomycin resistance cassette [Höllrigl et al., 2001]. Putative stem cells were enriched by a modified 3T3 protocol for 10 passages, comprising 3-day culture with a daily change of 2 ml M15Hy medium and splitting of the cells 1:3 every third day onto new feeder cells. After 10 passages the cell population was subjected to selection with 180 μg/ml G418 (Gibco) for 10 days. Cells from Balb/C mice were selected 2 times with 2 mmol/l gancyclovir for 14 days and individual clones were counter selected for G418 sensitivity. C57BL/6J × 129Sv clones surviving G418 selection were expanded and seeded at clonal density on feeder cells under continuing G418 selection. Similarly, Balb/C CVPC clones were seeded at clonal density on feeder cells under continuing gancyclovir selection. Individual colonies were expanded to clonal cell lines which were frozen at increasing passages.
Clonal CVPC lines were continuously maintained at high density in M15Hy on mitomycin C-inactivated feeder cells prepared from SNL76/7 fibroblasts in the same way as AB2.2, W4 [Lauss et al., 2005], 663 [Lagger et al., 2002], and Tag3 ESCs [Höllrigl et al., 2001]. To determine the cell fate at a low cell density and in the absence of feeder cells and LIF, CVPCs were plated at 1 × 104 cells/cm2 on gelatine-coated cell culture plates in M15Hy and monitored for 8 days. Stocks of C57BL/6J × 129Sv CVPC clones were frozen up to passage 149, and Balb/C CVPC clones up to passage 113.
Differentiation of CVPCs
To allow differentiation of cells in high-density monolayer cultures, CVPCs were seeded at 3 × 104 cells/cm2 on gelatine-coated tissue culture plates. Starting after 5 days 70% of the M15Hy medium was changed daily. Development of rhythmically contracting CMCs, aperiodically and very slowly contracting SMCs, and ETCs were monitored and quantified using phase-contrast illumination on an Olympus CK2 microscope.
To generate CBs, which are EB-like aggregates made from CVPCs, cells were grown to confluence on feeder cells and split 1:2 on the day before aggregation. The next day CVPCs were dissociated with Trypsin and suspended in M15Si. M15Si has a composition identical to that of M15Hy, with the exception of using a different fetal bovine serum (Sigma F7524). Feeder cells were removed by adsorption on gelatine-coated tissue culture plates at 37 °C for 45 min [Bader et al., 2000], and CVPC aggregates were generated the same way as EBs [Bader et al., 2001]. Shortly, suspended CVPCs were diluted to 4.5 × 104 cells/ml M15Si and 90 ± 6 drops (20 μl) were placed on the lids of bacterial grade 10-cm plates. Hanging drops were incubated over a layer of sterile water to compensate increased vapor pressure of the drops at 37 °C for 5 days. Then aggregates were washed from the lid into gelatinecoated tissue culture plates with 6 ml M15Si, evenly distributed, and cultured using a feeding protocol exactly as described for EBs [Bader et al., 2000; Stary et al., 2005]. LIF (purified from LIF expressing SF9 cells) 100 ng/ml, activin A (#338-AC; R&D Systems) 20 ng/ml, and retinoic acid (#R2625; Sigma) 5 × 10–7 mol/l were added to the medium at the time intervals indicated in the legends to fig. 2 and 3 and in the online suppl. fig. S5. Development of CMCs, SMCs, and ETCs were monitored for up to 108 days. Cardiomyogenesis was monitored daily and quantified by determining the percentage of aggregates with CMCs. Photomicrographs were taken with an Olympus E330 digital camera on an Olympus CK2 microscope under phase contrast illumination.
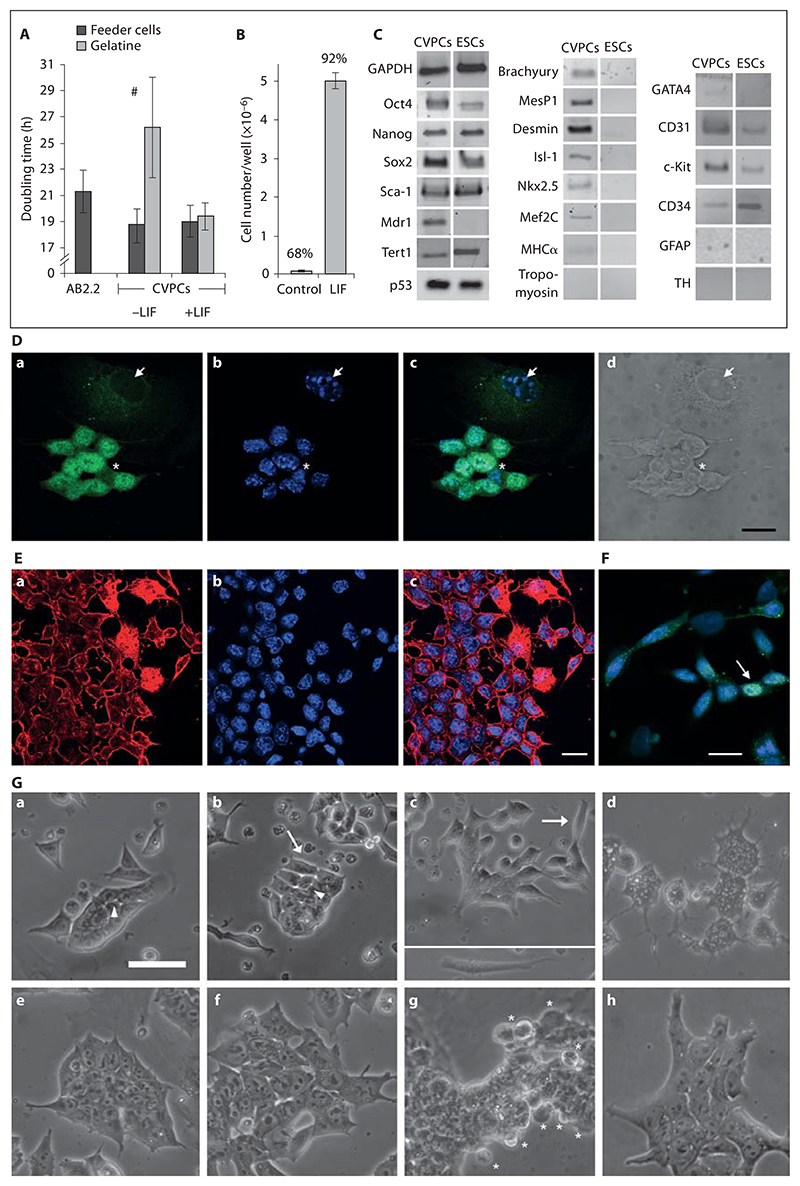
Fig. 2. Characterization of clonal CVPC lines.
A Doubling time of A5, H3, and G3 CVPC clones on SNL76/7 feeder cells and on gelatine without (–) and with (+) 100 ng/ml LIF. AB2.2 = ESCs. Error bars in all figures indicate the standard deviation. # p < 0.05. B Long-term self-renewal of A5 CVPCs in the absence of SNL76/7 feeder cells and in the presence of 100 ng/ml LIF. CVPCs were cultured on gelatine-coated plates for 30 days and cell numbers were determined in 3 plates in parallel. The data above the bars indicates the percentage of living cells. p < 0.001. C Semiquantitative RT-PCR analysis of genes expressed in CVPCs and ESCs. The cycle number was chosen for each pair of primers so that the signal was not saturated. GAPDH, loading control. D Localization of Oct4 proteins in CVPCs by immunofluorescence microscopy. Anti-Oct4 staining with FITC-conjugated secondary antibodies (a), DAPI staining of nuclei (b), merged images (c), and differential interference contrast (d). Arrows indicate Oct4-negative nuclei of feeder cells. Asterisks indicate very rare CVPCs with little Oct4 protein in the nucleus. E Localization of Brachyury protein in undifferentiated CVPCs by immunofluorescence microscopy. Anti-Brachyury staining with TR-conjugated secondary antibodies (a), DAPI staining of nuclei (b), and merged images (c). F Immunolocalization of anti-Brachyury with FITC-conjugated secondary antibodies and DAPI-stained DNA (blue) (merged images). The arrow indicates very rare CVPCs with Brachyury localized in the nucleus. Scale bars = 20 μm. G Morphological changes of CVPCs induced in the absence of LIF and feeder cells. Phase-contrast images of CVPCs (a–d) and ESCs (e–h) 1 day (a, e), 2 days (b, f), 3 days (c, g), and 4 days (d, h) after plating. Arrows, indicate elongating CVPCs at different stages of development. Inset in c: typical elongated migrating cell. Arrowheads point at bright areas indicating decreased cell-cell contact. Asterisks indicate developing primitive endoderm cells at the rim of ESC colonies [Bader et al., 2001; Rula et al., 2007]. Scale bar = 50 μm.
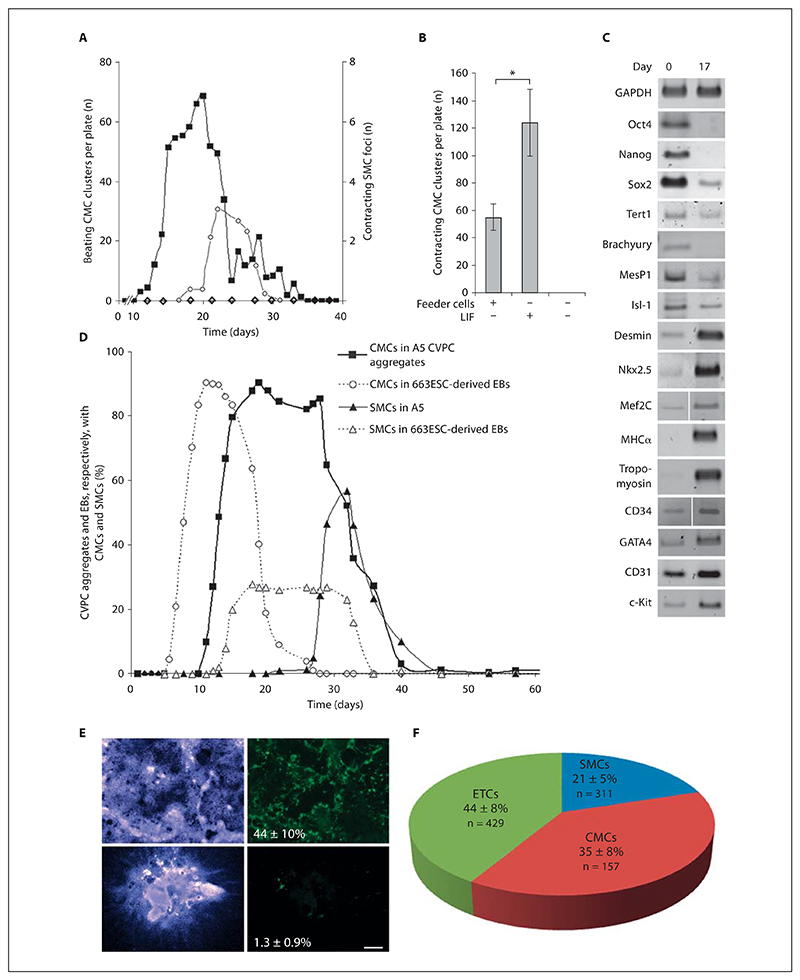
Fig. 3. Differentiation of CVPCs.
A Differentiation of CVPCs in monolayers. Cells were seeded on gelatine-coated tissue culture plates in the absence of SNL76/7 feeder cells and medium was partially replaced only when consumed. Development of CMCs (▪) and SMCs (○). AB2.2 ESC monolayer cultures (◊) gave rise neither to CMCs nor to SMCs. Data are means of 10 experiments for CMCs and 3 experiments for SMCs. B LIF enhances the myocardial differentiation potential of CVPCs. Cells were propagated for 30 days in the presence of SNL76/7 feeder cells and in the presence or absence of LIF and then subjected to differentiation as described in A. Error bars indicate the standard deviation. * p < 0.05. C Change of expression levels of stemness, mesodermal, myocardial, and endodermal genes in differentiating CVPC aggregates (CBs) between days 0 and 17. D Development of CMCs and SMCs in CBs. CVPCs were aggregated in hanging drop cultures for 5 days and plated on gelatine-coated culture plates, and the development of rhythmically contracting CMCs and occasionally and very slowly contracting SMCs was quantified daily. Dashed lines represent data from identically treated 663 ESC-derived EBs. Data are means of 8 triplicate experiments. Number of CBs checked daily: 26 ± 3. E Topology and magnitude of cardiomyogenesis in CBs and W4 ESC-derived EBs, respectively. CBs (top) and EBs (bottom) were fixed 3 days after the first CMCs started to contract. Nuclei (left, blue) and cTnT-positive CMCs (right, green). Values represent the percentage of the cTnT-positive area relating to the DAPI-stained area. n = 3 in each case. Scale bar = 500 μm. F Percentage of cTnT-positive CMCs, vWF- and CD31-positive vascular ETCs, and SMA-positive SMCs in CBs. n = Total number of counted cells.
Genotyping CVPCs by PCR and Semiquantitative RT-PCR
Genomic DNA and mRNA were isolated from self-renewing CVPCs at passages 6–46, from AB2.2 and Tag3 ESCs, from SNL76/7 fibroblasts, and from differentiating CBs at day 13, at the beginning of CMC contraction, and at day 17, when all CBs had developed a maximum number of contracting CMCs. Before isolation of DNA and mRNA, SNL76/7 feeder cells were removed from ESCs and CVPCs by adsorption to gelatine-coated tissue culture plates. Further, genomic DNA was isolated from Balb/C, C3H, and outbred C57BL/6J × 129Sv mice. mRNA was isolated with a Qiagen RNeasy kit.
CVPCs were genotyped in order to proof their origin from the donor mouse, and the absence of cell fusion with ESCs and mitotically inactivated SNL76/7 feeder cells, respectively, by PCR with primer pairs specific for the mutant Hdac1 allele on chromosome 4, present in C57BL/6J × 129Sv mice [Lagger et al., 2002] and in the CVPC lines generated by positive selection. Primer pairs are listed in the online suppl. table 6. Additionally, strain-specific primer pairs for inverted and inserted DNA elements between the H60a, H60b, and H60c loci on chromosome 10 [Zhang et al., 2011] were employed which allow distinguishing of the C57BL/6J mouse-derived alleles from 129Sv ESCs alleles. The origin of the He2, He22, and HeA CVPC lines from Balb/C mice was demonstrated by the presence of the wild-type allele of the sPlA2 gene on chromosome 4 [Kennedy et al., 1995] which is mutated in the 129Sv strain, and the absence of the mutant Desmin allele on chromosome 1 [Höllrigl et al., 2001] which is only present in Tag3 ESCs. The presence of the wild-type Hprt gene on the X chromosome in CVPC lines was used to demonstrate the absence of the mutant Hprt allele [McMahon and Bradley, 1990] present only in SNL76/7 fibroblasts. To provide further evidence making transdifferentiation and fusion of cells involved in the isolation procedure very unlikely, the absence of the Sry gene located on the Y chromosome was used to demonstrate that CVPCs were also isolated from female donor mice in the presence of male ESCs and SNL76/7 feeder cells. PCR was performed with Pfu DNA polymerase (EP8051 Fermentas) to saturation of the signals in order to exclude any false-negative results.
cDNA was synthesized with RevertAid M-MuLV Reverse Transcriptase (EP0441; Fermentas). Semiquantitative RT-PCR was performed with Taq polymerase (EP0402; Fermentas) and primers as indicated in figures 2–4, 6 and listed in the online suppl. table S7. Experiments were performed 1 time with all 11 CVPC clones and at least 2 times with 6 clones (A5, G3, H3, He2, He22, and HeA). The number of cycles for each pair of primers was carefully determined by several preliminary experiments and chosen so that none of the obtained signals were saturated.
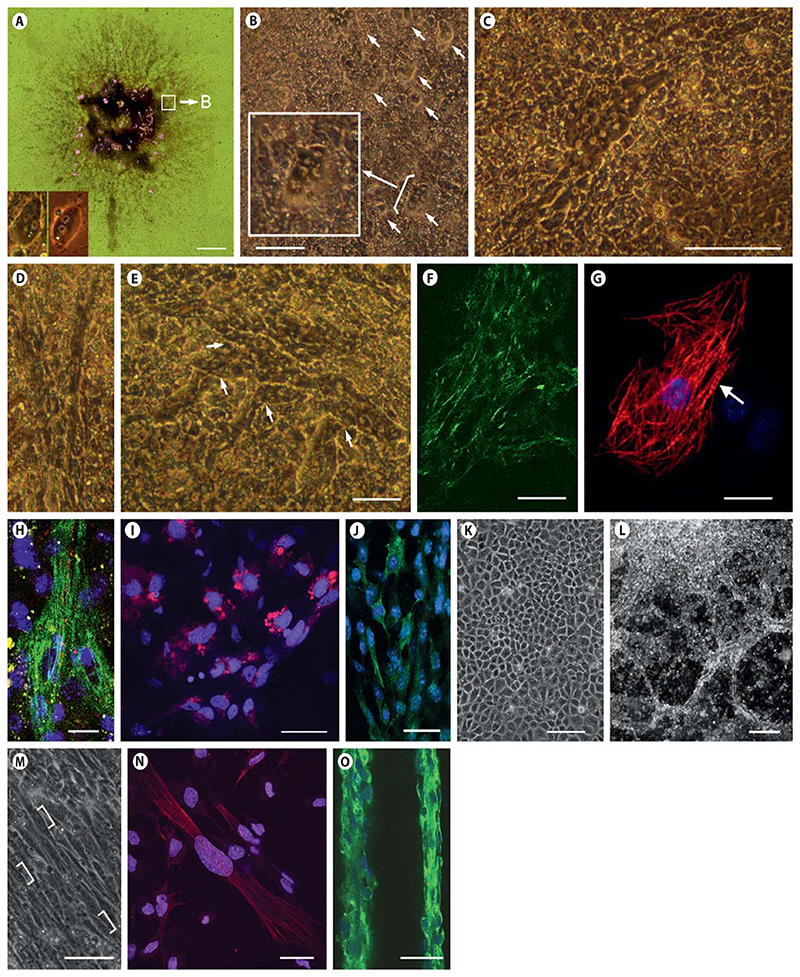
Fig. 4. Development and morphology of CMCs, ETCs, and SMCs in CBs.
A Composed bright-field image (18 images) of a CB at day 13 after aggregation. Rhythmically contracting areas are highlighted in magenta. Insets: left, CMC derived from CVPCs; right, primary CMCs from a 2-day-old mouse heart. B CMCs start to contract as single cells or as small aggregates of 1–6 cells. Inset: magnification of a small aggregate of CMCs. Note the typical gray cytoplasm and bright nuclei of CMCs. C Single contracting CMCs proliferate and form patches composed of dozens of cells. D Elongated bundles of discrete CMCs. E Branching networks of CMC bundles. F Indirect immunofluorescence staining of large arrays of CMCs at day 16 with anti-cTnT (green). G Single CMC at day 22 stained for cTnT (red); the arrow indicates myofibrils with a sarcomeric substructure. Nuclei stained with DAPI (blue). H Connexin 43 staining (red) indicating cell-cell interaction between 2 adjacent CMCs. cTnT (green); nuclei (blue). ETCs at day 11 stained with CD31 antibody (red) (I) and with anti-vWF antibody (green) (J). K Epithelial cluster of ETCs at day 17. L Dark-field image of cord-like networks of ETCs forming vascular-like strings at day 36. M SMCs start to form at day 13 and occasionally contract very slowly once they are in a parallel alignment. Brackets indicate elongated nuclei typical for SMCs. N Elongating SMCs stained with anti-SMA specific antibodies (red). Nuclei (blue). O Large clusters of SMCs aligned in vessel-like structures contracted occasionally between days 20 and 25, stained with SMA-specific antibodies (green). Nuclei (blue). B–E, K, L Phase-contrast images. Scale bars = 1 mm (A), 50 μm (B, D, E, M, N), 100 μm (C, F, K, O), 10 μm (G, H), 20 μm (I, J), and 200 μm (L).
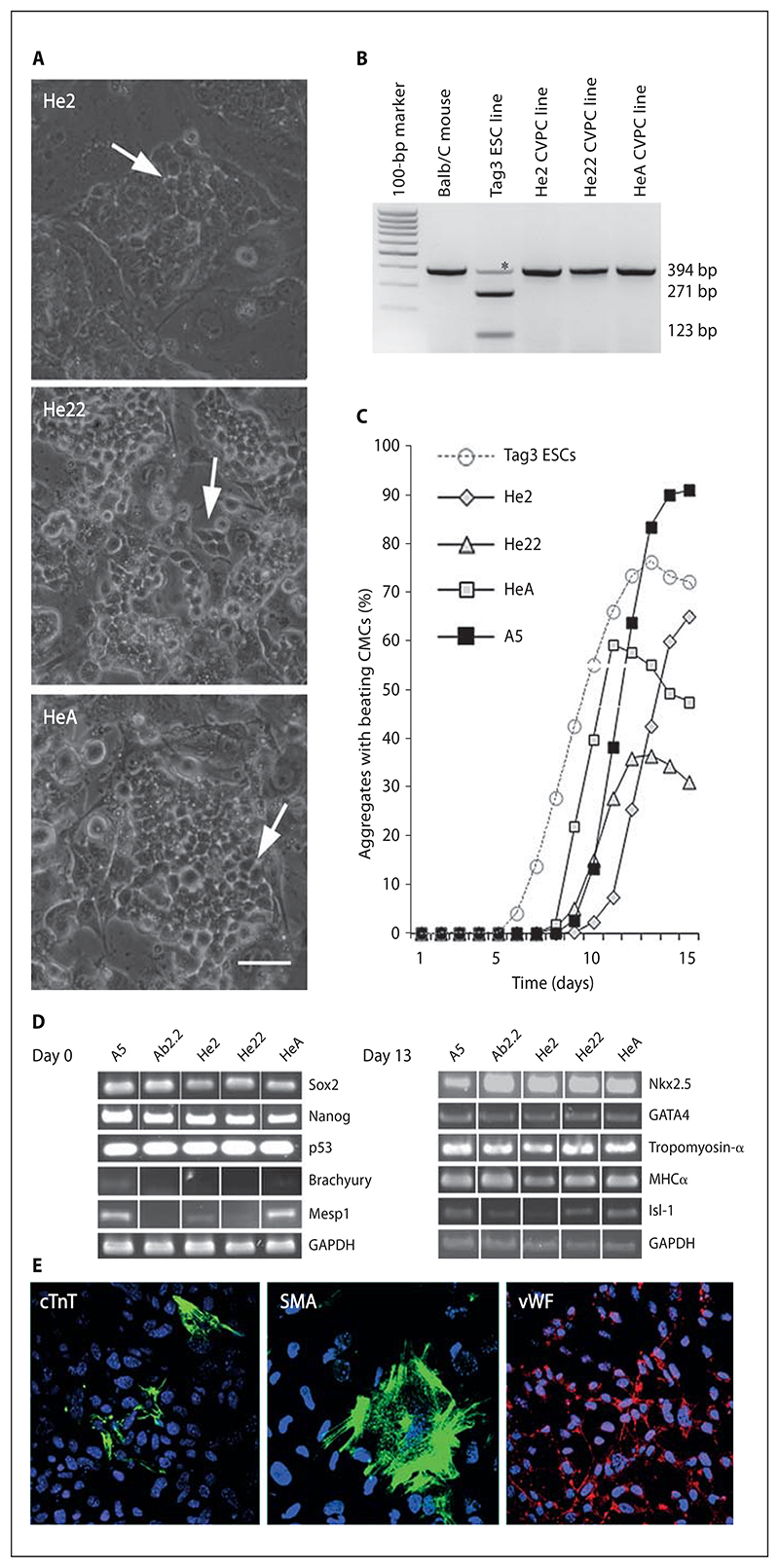
Fig. 6. CVPCs from wild-type Balb/C mouse hearts isolated by negative selection.
A Morphology of CVPC colonies from clones He2, He22, and HeA. Phase contrast images. Arrows indicate areas within colonies with a rather loose arrangement and reduced cell-cell contact between CVPCs. Scale bar = 50 μm. B Genotyping of CVPCs. BamHI digestion of a PCR fragment generated from the sPlA2 gene on chromosome 4 which is wild type in the Balb/C mice and CVPCs derived from this strain but mutated and containing a BamHI restriction site in the 129Sv Tag3 ESC line. Asterisks denote unspecific amplicons present in all strains which migrate a little faster than the 394-bp amplicon after very long electrophoresis. C Development of CMCs in CBs. CVPC lines He2, He22, HeA, and A5 as controls were aggregated in hanging drop cultures for 5 days and plated on gelatine-coated culture plates. Development of rhythmically contracting CMCs in CBs was quantified daily. Dashed lines indicate data from identically treated ESC-derived EBs. Data are means of 3 triplicate experiments. Number of EBs and CBs checked daily: 20. D Semiquantitative RT-PCR analysis of genes expressed in undifferentiated (day 0) and differentiated (day 13) CVPCs. GAPDH, loading control. E Indirect immunofluorescence staining of CBs at day 13. CMCs were detected with anti-cTnT (green, left), SMCs with anti-SMA (green, middle), and ETC with anti-vWF antibodies (red, right). Nuclei were stained with DAPI (blue). Scale bar = 50 μm.
Confocal Immunofluorescence Microscopy
Aggregated and differentiating CVPCs (so-called CBs) were processed and stained exactly as described for EBs [Bader et al., 2001]. CBs and EBs were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature, permeabilized with 0.15% saponin in PBS, and stained with antibodies against cardiac troponin T (cTnT) (#MS-295, 1:200; Thermo Scientific), connexin-43 (C6219, 1:4,000; Sigma), smooth muscle actin (SMA) (#A2547, 1:800; Sigma), von Willebrand factor (vWF) (F3520, 1:200; Sigma), P-Ser21/9-glycogen synthase kinase 3 α/β (#9331, 1:1,000; Cell Signaling), Oct4 (#SC5279, 1:100; Santa Cruz Biotechnology, and Cell #2840, 1:200; Signaling Technology), and CD31 (#555445, 1:50 and 1:5; BD Pharmingen), for 60 min, and consecutively with FITC- and TR-conjugated secondary antibodies (#711-095-152, 1:200; #711-095-151, 1:200; #711-075-152, 1:200; Dianova) for 60 min. Alternatively, isolated cells transferred to coverslips were fixed in 96% ethanol at –20°C for 20 min, dried, and blocked with 1% BSA in PBS for 10 min. Nuclear DNA was stained with DAPI.
The number of cTnT-, CD31-, vWF-, and SMA-positive cells was determined on 6 photomicrographs of different EBs and CBs, respectively, in 2 independent experiments, and put in relation to the total number of cells with DAPI-stained nuclei on these photomicrographs. Photomicrographs were taken with a Zeiss LSM 510 confocal microscope.
Electrophysiological Analysis of CMCs
The standard whole-cell patch-clamp recording technique and the voltage-clamp mode to record voltage-gated Na+ and L-type Ca2+ channel currents have been previously described in detail [Kuzmenkin et al., 2009]. Briefly, after aggregation of A5 CVPCs, beating areas were cut on day 23 and dissociated with Trypsin, and cells were placed on gelatin-coated coverslips. Cells were incubated for 36–72 h prior to the experiments. Electrophysiological recordings were performed using an EPC 9 amplifier, and cell membrane capacitance was determined online using pulse acquisition software (HEKA, Lambrecht, Germany). SigmaPlot (SPSS Inc., Chicago, Ill., USA), CorelDraw (Corel GmbH, Germany) on day 25 ± 1, and Origin (Microcal Software, Northampton, Mass., USA) programs were used for display and analysis of recordings. For a detailed description of the current- and voltage-clamp mode please see online supplementary material (www.karger.com/doi/10.1159/000345804).
Analysis of Smooth Muscle Contraction
CVPCs and ESCs were aggregated and spontaneous contraction of SMCs was monitored on days 21 and 22 after aggregation for 10 min. Then, 1 μmol/l Angiotensin II (A9525; Sigma-Al-drich) was added to the medium and contraction was again monitored for 10 min. After that, 1 μmol/l losartan (61188; Fluka) was added to block the angiotensin II receptor and measurements were repeated. After washout of both angiotensin II and losartan, CBs and EBs, respectively, were incubated for 30 min in fresh medium at 37 °C and then again 1 μmol/l Angiotensin II was added and contraction was monitored for 10 min.
Statistical Analysis
Data are presented as arithmetic means ± standard deviation. Statistical significance was evaluated using a one-sample Student’s t test and values of p < 0.05 were considered statistically significant.
For additional methods please see online supplementary material.
Results
Isolation of CVPC with the Help of ESCs and LIF-Secreting Fibroblasts
The initial question addressed in this study was whether we can identify and employ conditions mimicking a somatic stem cell niche which allow the clonal isolation and persistent maintenance of a CVPC line from the hearts of newborn mice. The strategy we developed (fig. 1A) was based, firstly, on the generally accepted notion that in vivo CVPCs are very low in number and in a quasi dormant state embedded in a stem cell niche of the heart [Urbanek et al., 2006]. Secondly, these stem cells are supposed to be ready to turn on the self-renewal machinery and cell division upon external stimuli such as elevated levels of LIF. Thirdly, hearts (at least those from young animals) do express detectable quantities of LIF [Robertson et al., 1993], and fourthly, LIF has a well-established role in the self-renewal of murine ESCs, germ cells, and somatic stem cells [Ohlstein et al., 2004; Davies and Fuller, 2008]. Thus, we used LIF-expressing neomycin-resistant SNL76/7 fibroblasts as one component of an artificial in vitro niche to stimulate self-renewal and to allow isolation of putative stem cells of the heart. However, explanting a cardiac cell population on these feeder cells did not give rise to any self-renewing clonal cell lines because all cells died over time. Thus, based on the well-known fact that ESCs confer signals supporting survival and self-renewal to neighboring cells in physical contact with them, we additionally used ESCs, as a second cellular component of the in vitro niche, to foster the survival of the supposedly low number of CVPCs during the early phase of isolation. We assumed that once a critical number of self-renewing CVPCs is reached, they can survive by a self-supportive autocrine stimulation of proliferation in the absence of ESCs.
Fig. 1. Isolation procedure and phenotype of clonal CVPC lines.
A Rationale for the isolation strategy. Putative CVPCs (blue) are embedded between hypothetical niche cells (yellow), cardiac fibroblasts (green), and CMCs (red) in the heart (top of the illustration). All other cell types such as ETCs and SMCs were omitted for clarity reasons. If the heart tissue is dissociated and cells are explanted on fibroblasts (orange), all cell types die during long-term culture (arrow to the left). Small dots are fragments of dead cells. If cells are cultured in the presence of ESCs (brown) and in the presence of LIF-secreting feeder cells (orange) (middle of the illustration), all cell types except the CVPCs die or are diluted out during frequent passaging (lower part of the illustration). After CVPCs have reached a critical number allowing self-supportive self-renewal, removal of ESCs by drug selection should give rise to pure populations of continuously proliferating CVPCs (arrow to the right). B Workflow for the isolation of CVPCs. Triturated heart cells (5 × 105) and ESCs were mixed and cultured on mitotically inactivated SNL76/7 feeder cells for 10 passages. Then ESCs were selectively removed by addition of G418 and colonies of CVPCs were subcloned, expanded, and frozen. C Morphology of CVPC colonies from clones A5 and H3 and from ESCs. Phase-contrast images. Arrows indicate discontinued rough borders of CVPC colonies with a rather loose arrangement of cells (bright space between cells), and tightly sealed borders of very densely packed ESC colonies, respectively. Asterisks indicate feeder cells. Scale bar = 50 μm. D Size of CVPCs and ESCs 3 days after plating on gelatine-coated culture plates. Error bars indicate the standard deviation. Number of cells = 55 each. p < 0.05. E Chromosome spread from an A5 CVPC demonstrating a diploid karyotype. Giemsa stain, bright-field image. Scale bar = 10 μm. F PCR analysis of DNA from A5 CVPCs, from mouse strains C3H, outbred wild-type C57BL/6J × 129Sv, and outbred Hdac1–/– C57BL/6J × 129Sv (Hdac1–/– embryo as a positive control), and from AB2.2 ESCs and SNL76/7 fibroblasts with a primer pair specific for the neomycin resistance cassette and intron 7 of the Hdac1 gene (a), exon 5 of the Hdac1 gene (b), and a C57BL/6J-specific region in the H60b gene (c). Left lanes, 100-bp size standard; thick lane = 500 bps.
To test the feasibility of this strategy, we mixed 5 × 105 heart cells from the whole hearts of 7 newborn mice carrying a neomycin resistance gene for positive selection [Lagger et al., 2002] with an equal number of ESCs and subjected this mixture to a modified 3T3 protocol on mitotically inactivated LIF-secreting SNL76/7 fibroblasts to selectively enrich CVPCs (fig. 1B). The modified 3T3 protocol included growth of cells at a high density for 3 days, and splitting of cells irrespective of cell number 1:3 for only 10 passages (online suppl. fig. S1A; table S1). For a more detailed description of the border conditions and a mathematical model describing the influence of the modified 3T3 protocol on cell proliferation, please see online supplementary material. Assuming that higher densities of CVPCs are able to autonomously support self-renewal, ESCs were selectively killed with 180 μg/ml G418 after passage 10. The first time, 24 out of 314 surviving cell colonies were picked, expanded, and subcloned by limiting dilution under continuous selection with G418. This experiment was repeated 4 times, finally giving rise to 11 clonal cell lines which were used for further analysis. Successfully established clonal cell lines were maintained in the logarithmic growth phase at subconfluent density on feeder cells and repeatedly frozen in liquid nitrogen over a period of 7 years without any visible karyotypic and phenotypic change. Repeating the isolation procedure, we were able to identify very small round cells resembling CVPCs which were clearly distinguishable from ESCs as early as at passage 3 (day 9), when somatic cells of the heart were diluted out and/or had started to die (online suppl. fig. S1B). The presence of CVPC-like cells as early as 9 days after the extraction of cells from the heart suggests that CVPC are not a product of the isolation procedure but are genuine endogenous cells of the heart.
To substantiate the distinct nature of CVPCs and to demonstrate their cardiac origin, a detailed phenotypic and genetic analysis was performed. Already during the cloning procedure it became obvious that the morphology of CVPCs differs significantly from that of ESCs. CVPCs had a more globular shape than ESCs (fig. 1C), did not adhere to each other as tightly as ESCs, and were significantly smaller than ESCs (fig. 1D). The latter already suggests that they are not tetraploid cell fusion products generated during the isolation procedure. Fusion of ESCs with somatic cells results in significantly larger nuclei or heterokaryons [Ying et al., 2002] which were never observed in any CVPC line (fig. 1C, 2D, G; online suppl. fig. S3). Additionally, karyotyping of 11 clonal CVPC lines at different passages (fig. 1E; online suppl. table S2) demonstrated a stable diploid karyotype which makes it very unlikely that all CVPC lines were fusion products of somatic heart cells and ESCs. The remaining possibility of pseudo-diploidy caused by reductive mitosis of tetraploid cells is extremely unlikely because: (i) this progress is very slow and requires several cell divisions and thus cannot proceed during the modified 3T3 protocol (online suppl. fig. S1A); (ii) it is very inefficient and thus would always lead to a cell population where the majority remains tetraploid, and this is not the case in CVPCs (online suppl. table S2), and most importantly, (iii) it has been demonstrated that reductive mitosis requires the absence of p53 [Vitale et al., 2010], and CVPC lines definitely express p53 (fig. 2C). Genetic analysis of CVPCs by PCR with a primer pair specific for the neomycin resistance cassette and intron 7 of the Hdac1 gene demonstrated the presence of the unique Hdac1-Neomycin transgene in A5 CVPCs which can only stem from the donor Hdac1+/– C57BL/6J × 129Sv mouse strain [Lagger et al., 2002] and not from AB2.2 ESCs and SNL76/7 fibroblasts used to isolate CVPCs. Amplification of exon 5 of the Hdac1 gene was used as a positive control. Amplification of C57BL/6J-specific regions from the H60b and H60c loci [Zhang et al., 2011] demonstrated that A5 CVPCs do indeed contain alleles from the donor mouse strain which are present neither in AB2.2 ESCs nor in SNL76/7 fibroblasts (fig. 1F). The absence of the mutant Hprt locus, present only in SNL76/7 cells [Thomas and Capecchi, 1987] (online suppl. fig. S1C), provided clear evidence that CVPCs had been neither derived from SNL76/7 fibroblasts nor generated by fusion with these cells. Additionally, we performed an LINE-1 repetitive DNA element polymorphism analysis and found a much closer relationship of CVPC clones to the genotype of the donor mouse line than to AB2.2 ESCs or SNL76/7 fibro-blasts (online suppl. table S3). Finally, numerical modeling of the isolation procedure (online suppl. fig. S1A; table S1) also suggests that the probability that CVPCs have been generated by (i) cell fusion followed by reductive mitosis, (ii) transdifferentiation of ESCs to CVPCs induced by somatic heart cells, and (iii) transdifferentiation of somatic heart cells to CVPCs induced by ESCs becomes null after 9 days of the isolation procedure. This period of 9 days is definitely too short for any of these processes to proceed. In conclusion, CVPCs indeed originated from the mouse heart, and ESCs together with LIF-expressing feeder cells mimic a stem cell niche in a way that allows isolation and maintenance of stable CVPC lines.
LIF Is Sufficient for the Maintenance of CVPCs as Self-Renewing Cell Lines
To test whether LIF secreted by the SNL76/7 fibroblasts is sufficient to maintain proliferation of CVPCs, we first determined the doubling time of CVPCs in the presence and absence of feeder cells and in parallel, in the presence of 100 ng/ml LIF. In the presence of feeder cells the doubling time of CVPCs was shorter than that of ESCs (fig. 2A), but it was increased by 7 h in the absence of feeder cells. Addition of LIF restored the short doubling time observed in the absence of feeder cells but did not influence the doubling time in the presence of feeder cells. To demonstrate that LIF is sufficient for continuous self-renewal and maintenance of the differentiation potential of CVPCs, we cultured CVPCs at a subconfluent density (105 cells/cm2) for 30 days (10 passages) in LIF-containing medium in the absence of feeder cells and determined the number of cells per well and the percentage of living cells, respectively (fig. 2B). LIF supported long-term self-renewal and significantly increased the viability of CVPCs under high-density culture conditions. In the presence of LIF-secreting feeder cells the clonally derived cell lines A5, H3, B5, and G3 maintained their unlimited self-renewing capacity, karyotype, morphology, and tissue-specific differentiation potential (see next paragraph) for 149 (A5), 60 (H3), and 21 passages (B5 and G3), respectively.
Next we investigated the expression of transcription factor genes important for continuous self-renewal, stem cell-specific genes, and markers for mesodermal and ectodermal cells (fig. 2C). All CVPC subclones expressed Oct4, Nanog, Sox2, and Sca1 in the presence of LIF-secreting fibroblasts but differed from ESCs by expressing the ABCG2 transporter Mdr1 and lower levels of Tert1. Surprisingly, and in sharp contrast to ESCs, CVPCs also expressed the early mesodermal markers Brachyury and Mesp1, the muscle cell marker Desmin, and myocardial transcription factors Isl-1, Nkx2.5, and Mef2C in the presence of LIF-secreting feeder cells. However, the definitive myocardial genes MHC and Tropomyosin were barely or not detectable. Expression of endothelial markers GATA4, CD31, and c-Kit was somewhat increased in CVPCs, but CD34 was decreased when compared to ESCs. The ectodermal markers Glial fibrillary acidic protein and Tyrosine hydroxylase were not detected in CVPCs. All 11 clones expressed comparable amounts of Oct 4, Nanog, Sox2, and GATA4, but early mesodermal and myocardial transcription factors were expressed more differently (online suppl. fig. S2). Immunofluorescence microscopy demonstrated the nuclear localization of Oct4 protein (fig. 2D), and similar to ESCs, but with a lower frequency, CVPCs displayed an inhomogeneous expression of Oct4 (online suppl. fig. S3). Brachyury protein localized mainly in the cytoplasm and as distinct foci in the nucleus of most CVPCs (fig. 2E). Brachyury was upregulated in a few apparently differentiating cells; however, only in less than 1% of undifferentiated CVPCs was it enriched in the nucleus (fig. 2F), which very much resembles the behavior of Tbx5 [Stirnimann et al., 2010], which is similar to Brachyury efficiently transported out of the nucleus in undifferentiated cells. Notably, in ESCs less than 0.1%, if any, of the cells were positive for Brachyury.
In addition to the different expression pattern of ESCs and CVPCs, a completely different behavior of CVPCs was observed in the absence of feeder cells and LIF. At a low density (102–103 cells/cm2), culture of CVPCs in the absence of feeder cells was not possible because CVPCs succumbed to cell death (fig. 2G). In the first 5 days CVPCs underwent morphological changes different from those observed in ESC cultures. They elongated and started to migrate and finally generated a lot of intracytoplasmic vesicles before they died. In sharp contrast, ESCs maintained their cellular integrity and only cells at the edges of the stem cell colonies started to differentiate to primitive endoderm as previously described [Bader et al., 2001]. This hints at a reduced capacity of CVPCs, compared to ESCs, to autonomously maintain self-renewal.
LIF Is Required to Maintain the Differentiation Potential of CVPCs
To investigate the differentiation potential of CVPCs, feeder cells were removed and CVPCs were transferred to gelatine-coated culture plates and seeded at a density of 4 × 104 cells/cm2. Within 6–7 days, endothelium forming ETCs became visible and after 11 days spontaneous differentiation of CVPCs gave rise to rhythmically contracting CMCs. Finally, after day 18, SMCs began to contract slowly and aperiodically (fig. 3A; online suppl. table S4). Other cell types could never be observed. Notably, ESCs maintained under identical conditions never gave rise to a single cluster of ETCs, CMCs, or SMCs.
Next we wanted to know whether LIF is sufficient for maintenance of the differentiation potential of CVPCs to CMCs, because LIF promotes early cardiomyogenesis [Bader et al., 2000]. Therefore, CVPCs were propagated at subconfluent density (1.2 × 105 cells/cm2) in the presence of 100 ng/ml LIF in the absence of feeder cells for 30 days and then subjected to differentiation on gelatine-coated plates. Preceding maintenance of CVPCs in the presence of LIF doubled the number of CMC clusters under differ-entiation-promoting conditions, as compared to maintenance on feeder cells (fig. 3B). In the absence of LIF, the myocardial differentiation potential of surviving CVPCs was totally lost. These results demonstrate that LIF sustains the disposition of CVPCs to differentiate to CMCs.
To confirm the specific differentiation potential of CVPCs on the molecular level, semiquantitative RT-PCR was performed with cDNAs from undifferentiated and differentiated CVPCs. In differentiated CVPCs Oct4, Nanog, and Brachyury were completely downregulated and Sox2, Tert1, MesP1, and Isl-1 were partially down-regulated (fig. 3C). Downregulation of Brachyury, MespP1, and Isl-1, in combination with upregulation of the expression of Desmin, Nkx2.5, GATA4, Mef2C, MHC, and Tropomyosin is consistent with molecular events of cardiac cell differentiation observed in vivo. Increased expression of the cell surface glycoprotein CD34, platelet endothelial cell adhesion molecule-1 (PECAM-1 or CD31), and the stem cell growth factor receptor c-Kit in differentiated CVPCs (fig. 2C), as well as the formation of vessel-like structures in areas with densely packed ETCs (fig. 4K, L; online suppl. fig. S4A–E), and Weibel-Palade-like bodies in the cytoplasm of ETCs (fig. 4I; online suppl. fig. S4F, G) suggest that a significant percentage of differentiated CVPCs were ETCs.
Aggregation of CVPC Facilitates Efficient and Reproducible Differentiation to Only CMCs, ETCs and SMCs
To test more rigorously whether CVPCs develop into all lineages of somatic cells, as ESCs do, or are somehow restricted in their differentiation potential, CVPCs were aggregated to CBs using the protocol developed to generate EBs from ESCs [Bader et al., 2000]. In ESC-derived EBs, differentiating cells give rise to derivatives of all 3 germ layers [Lanza, 2004] and extra-embryonic endoderm [Bader et al., 2001]. When aggregated in hanging drop cultures, CVPCs did not give rise to extra-embryonic primitive endoderm and its derivatives, parietal and visceral endoderm [Bader et al., 2001], and never developed a two-fold axis typically observed in EBs [Fuchs et al., 2012]. They rather spontaneously differentiated to contracting CMCs (fig. 3D). First rhythmically contracting CMCs (online suppl. video 1) became apparent on day 11 ± 8 h, developed to large networks of synchronously contracting CMCs in 94 ± 6% of CBs (online suppl. video 2), and contracted rhythmically for another 30 days. In sharp contrast, cardiomyogenesis in ESC-derived EBs began on day 7.5 ± 10 h [Hofner et al., 2007] in 86 ± 6% of EBs and rhythmical contraction lasted only for 18 ± 44 days. Likewise, SMC development in CBs required more time than in EBs (fig. 3D). Most importantly, no shift in this phenotype was observed in the 3 cell lines tested for up to 149 passages. Immunostaining of CMCs in CBs and EBs with cTnT antibodies and counting of cTnT-positive cells and DAPI-stained nuclei, respectively, demonstrated that CBs contain 44 ± 10% CMCs, which is 34 times more CMCs than usually observed in EBs (1.3 ± 0.9%) (fig. 3E). Immunostaining of CBs, in order to detect and quantify cTnT-positive CMCs, vWF- and CD31-positive ETCs, and SMA-positive SMCs (fig. 4I, J, M; online suppl. fig. S4D–G, S5F) demonstrated that ETCs are the most abundant cell types, followed by CMCs and a significantly smaller percentage of SMCs (fig. 3F).
Once attached to culture plates, CBs started to spread with radial symmetry and the first CMCs appeared stochastically dispersed between day 11 and day 13 (fig. 4A, highlighted areas). These first rhythmically contracting cells and primary CMCs from newborn mice had an indistinguishable morphology (fig. 4A, insets). Within 1–2 days CBs became densely infused by an increasing number of clustered and rhythmically contracting CMCs. Single contracting CMCs further proliferated to small patches (fig. 4B), then elongated but did not fuse (fig. 4C), and finally formed long fibers composed of numerous contracting nonfused CMCs (fig. 4D). These fibers soon branched (fig. 4E) and formed a dense interwoven network of synchronously and rhythmically contracting cTnT-positive CMCs (fig. 4F). CMCs displayed sarcomeres and well-developed myofibrils (fig. 4G) and connected to neighboring CMCs by Connexin 43-positive gap junctions (fig. 4H).
To test whether CVPCs can be induced to give rise to cell types of the ectodermal and endodermal lineage as well, we added retinoic acid and Activin A, respectively, to CBs. Stimulation of neuronal differentiation with 5 × 10–7 mol/l retinoic acid [Kim et al., 2009], and endoderm differentiation with 20 ng/ml Activin A [Sulzbacher et al., 2009], never led to the development of any ectodermal or endodermal cell type in CBs (online suppl. fig. S5), whereas in control EBs the numbers of β3 Tubulin-positive neurons (online suppl. fig. S5B, S6B) and FoxA2-positive endodermal cells (online suppl. fig. S5E) significantly increased. Addition of retinoic acid to CBs did not induce expression of Glial fibrillary acidic protein and Tyrosine hydroxylase but significantly increased the expression of both genes in EBs and neurospheres (online suppl. fig. S5C). Induction of other mesodermal cells such as skeletal myoblasts followed by myotube formation under low serum concentrations in EBs [Diel et al., 2008] (online suppl. fig. S6C, D) never gave rise to any such cell types in CBs (online suppl. fig. S6A). Likewise, we could not detect a single erythropoietic cell in several hundreds of CBs or a single Hemoglobin-containing cell out of 2 × 107 cells stained with benzidine (online suppl. fig. S6F, G). Restriction of the differentiation potential of CVPCs to the cardiac lineage strongly suggests that CVPCs are not blood-derived precursor cells [Cesselli et al., 2009].
ETCs became apparent already between days 6 and 8 of CB development, expressed CD31 (fig. 4I) and vWF (fig. 4J; online suppl. fig. S5F), and, later on, formed endothelial sheets and vessel-like structures (fig. 4K, L; online suppl. fig. S4A–E). Aperiodic and slowly contracting SMA-positive SMCs (online suppl. video 3) developed less frequently and much later than ETCs and CMCs (fig. 4M, N) and were sometimes associated to vessel-like structures (fig. 4O; online suppl. fig. S4D, E).
CVPCs Give Rise to Functional Pacemaker, Atrial, and Ventricular CMCs
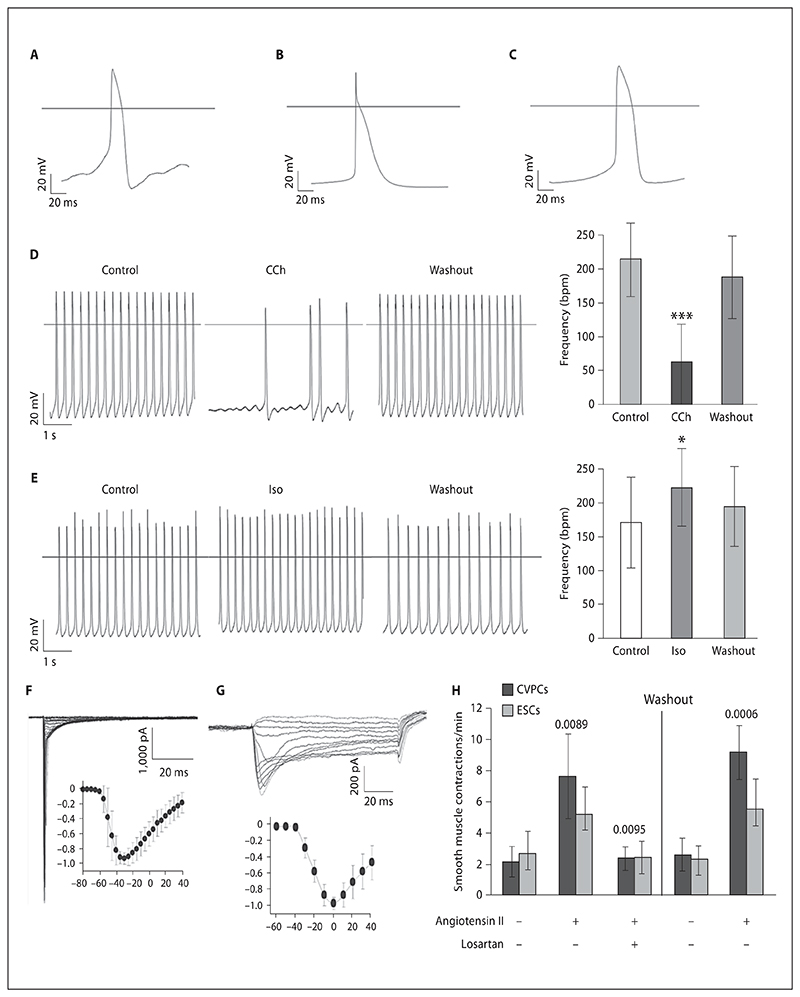
CMCs in CBs interconnected and formed large synchronously and rhythmically contracting networks. To test whether the molecular requirements for electromechanical coupling are expressed in CVPC-derived CMCs, an electrophysiological characterization of CMCs was performed. Spontaneous action potentials of CVPC-derived CMCs demonstrated almost adult phenotypes of pacemaker, atrial, and ventricular CMCs at a ratio of 1:1:1 (fig. 5A–C; online suppl. table S5). CMCs responded to muscarinic and β-adrenergic stimulation (fig. 5D, E). Whole-cell voltage-clamp experiments detected currents through voltage-gated Na+ (fig. 5F) and L-type Ca2+ channels (fig. 5G). The typical current-voltage relationships (fig. 5F, G, insets) matched to already functional but not fully matured primary CMCs.
Fig. 5. Electrophysiological characteristics of CVPC-derived CMCs and angiotensin II dependent contraction of SMCs.
A–C CMCs with pacemaker-, atrial-, or ventricular-like action potentials. Effects of muscarinic (D) and β-adrenergic (E) stimulations of CMCs. Negative chronotropic effect of 1 μmol/l carbachol (CCh) (D) and positive chronotropic effect of 1 μmol/l isoproterenol (Iso) (E). Representative experiments (left), and impact on beating frequency (right). Number of cells used for the analysis: carbachol, 14; isoproterenol, 12. * p < 0.05, *** p < 0.001. Functional expression of cardiac voltage-gated sodium channels (F) and L-type Ca2+ channels (G). Insets: current amplitudes normalized to their maxima and plotted against test voltages. H Angiotensin II increases the contraction rates of SMCs. CVPCs and ESCs were aggregated and SMCs were monitored for spontaneous contractions for 10 min on days 21 and 22 after aggregation. Then 1 μmol/l angiotensin II was added to the plates and contraction was again monitored for 10 min. After that 1 μmol/l losartan was added to block the angiotensin II receptor and measurements were repeated. After washout of both angiotensin II and losartan, CBs and EBs were incubated for 30 min at 37°C and then 1 μmol/l angiotensin II was again added. Number of experiments = 5 with 2 CVPC lines. Error bars indicate the standard deviation. p values are given for CVPC-derived SMC contraction rates in relation to the temporally preceding values.
CVPC-Derived SMCs Respond to Angiotensin II
Spontaneous contraction of large areas of parallel aligned SMCs can be observed in CBs between days 18 and 40 after aggregation (fig. 3D). The basal frequency of SMC contraction could be significantly increased within less than 5 s by addition of 1 μmol/l Angiotensin II to the medium, both in CBs and in EBs (fig. 5H; online suppl. videos 4, 6). The Angiotensin II-mediated increase in the contraction rate could be reduced back to basal levels by addition of 1 μmol/l of the competitive inhibitor losartan (online suppl. videos 5, 7). The response of CVPC-derived SMCs to Angiotensin II was much stronger than that of ESC-derived SMCs, suggesting a more advanced developmental stage of CVPC-derived SMCs.
The physiological response of CMCs and SMCs, and the vessel-like structures formed by ETCs, together with the expression pattern and immunostaining demonstrated that in vitro self-renewing CVPCs can reproducibly give rise to functional ETCs, SMCs, and CMCs.
Isolation of CVPCs from Wild-Type Mice
To demonstrate that the new strategy for CVPC isolation can also be used to isolate CVPCs from organs of wild-type animals of advanced age, we isolated hearts from 2-(mixed female and male), 22-(male), and 180-day-old (female) wild-type Balb/C mice. Triturated heart cells were mixed with an equal number of Tag3 ESCs [Höllrigl et al., 2001]. This ESC line contains 2 thymidine kinase (Tk) suicide genes from human herpes simplex virus type I and a neomycin resistance cassette for counter selection, inserted into exon 1 of the Desmin gene. After subjecting this mixture of cells to the same modified 3T3 protocol, used to isolate the neomycin-resistant CVPCs (fig. 1B), wild-type CVPCs were selected for the absence of the Tk suicide genes in the presence of 2 mmol/l gan-cyclovir for 14 days. Clones were isolated, expanded, and subjected to a second selection with gancyclovir for 14 days, and in parallel they were tested for G418 sensitivity to exclude any false Tk-negative clones.
In contrast to the positive selection procedure, where 314 primary clones finally gave rise to 11 independent clonal CVPC lines from 500,000 primary heart cells, negative selection resulted in only 4 CVPC lines from hearts of 2-day-old mice (He2), 2 CVPC lines from hearts of 22-day-old mice (He22), and 1 CVPC line from 180-day-old mice (HeA). These CVPCs had the same morphology (fig. 6A) as neomycin-resistant C57BL/6J × 129Sv CVPCs (fig. 1C), their doubling times ranged from 16.5 to 20.3 h, and 90–94% of the cells of all clones were euploid (online suppl. table S2). Demonstration that these CVPC lines contain only the donor Balb/C mouse-derived wild-type alleles of the sPlA2 gene [Kennedy et al., 1995] (fig. 6B) excluded that they were generated by cell fusion because the 129Sv strain-derived Tag3 ESC line has a point mutation within exon 3 of the sPlA2 gene which generates a new BamHI restriction site. Additionally, 2 of the 7 cell lines were female, demonstrating unambiguously that they were derived neither from the male Tag3 ESC line nor from the male SNL76/6 cell line (online suppl. fig. S1C). This result was corroborated by the absence of the Desmin-TkTk-Neo-mycin resistance transgene, unique for Tag3 ESCs, and the absence of the neomycin resistance cassette in the Hprt locus, unique for SNL76/7 fibroblasts, in all CVPC lines isolated from Balb/C mice (online suppl. fig. S1C). These CVPC lines maintained their phenotype for up to 113 passages. Unlike ESCs, and similar to A5 CVPCs, all wild-type CVPC lines exclusively differentiated to CMCs, SMCs, and ETCs (online suppl. table S4). Cardiomyogenesis in wild-type CBs was again delayed when compared to cardiomyogenesis in ESC-derived EBs (fig. 3D, 6C). Self-renewing wild-type CVPCs expressed stemness transcription factors Sox2 and Nanog but barely expressed Brachyury and weakly expressed Mesp1 (fig. 6D). Upon differentiation in CBs, they expressed transcription factors Nkx2.5, GATA4, and Isl-1 (fig. 6D) and structural CMC-specific genes tropomyosin, MHC, and cTnT (fig. 6E). Similar to the CVPC lines established by positive selection, they also expressed SMA and vWF in discrete areas of the CBs (fig. 6E). These data together suggest that these CVPC lines, independently isolated from the Balb/C mouse strain, have properties very similar to those of the neomycin-resistant CVPC lines from the transgenic C57BL/6J × 129Sv mouse strain, and demonstrate that CVPCs can be isolated from donors of different age and sex without the requirement of a selectable marker.
Discussion
The Isolation of Clonal CVPC Lines
Here we could demonstrate that CVPCs can be reproducibly isolated from murine hearts and that they can be maintained on LIF-secreting feeder cells as stable clonal cell lines without any phenotypic chance for more than 149 passages over a period of 7 years. A major concern against the isolation procedure was related to the origin of CVPCs. Coculture of 2 cell lines (although one of them was mitotically inactivated) with a undefined population of heart cells opens the theoretical possibility of cell fusion, transdifferentiation, or reprogramming as the decisive mechanism by which CVPCs could have been generated rather than isolated. The chance that these cell lines emerged by phenotypic conversion of some heart cells or ESCs during the isolation procedure can be excluded by statistical means and by experimental data published by others. Briefly, the mathematical model (for a detailed description see online suppl. material) suggests that fusion of any somatic heart cell with an ESC, followed by reductive mitosis, is practically impossible (p << 10–10) because of the low numbers of heart cells present during the isolation procedure and the relatively short time required to isolate CVPCs. In vivo, donor blood stem cell-derived diploid CMCs and liver cells can be identified in recipient solid organ tissue within 9 and 13 days, respectively, a period far too short for fusion followed by reductive mitosis and gain of a stable somatic cell phenotype [Orlic et al., 2001a, b; Korbling et al., 2002]. The latter in vivo data, together with the fact that reductive mitosis requires the absence of p53 [Vitale et al., 2010], which is not the case in CVPCs, make fusion followed by reductive mitosis even more unlikely [Korbling and Estrov, 2003]. Further, fusion of a somatic cell with an ESC would result in dominance of the ESC phenotype [Ying et al., 2002], and fusion of a CMC with a fibroblast would result in a lack of dominance of the cardiac phenotype [Evans et al., 1994], both of which are not the case for CVPCs.
Likewise, the border conditions of the isolation procedure make it extremely unlikely that CVPCs have been artificially generated by direct transdifferentiation or reprogramming of any somatic cell, followed by the establishment of a stable phenotype unique for cardiogenic mesoderm [Leri et al., 2005]. Transdifferentiation or reprogramming occurs with a very low frequency [Ieda et al., 2010; Peran et al., 2010] and has so far exclusively resulted in a meta-stable cardiac phenotype [Condorelli et al., 2001; Acquistapace et al., 2011] but not in stable clones of continuously self-renewing CVPC lines with a constant cardiogenic differentiation potential.
Another limitation seen to blur the origin of CVPCs is the notion that they might have been generated from bone marrow stem cells present in the coronary vessels. Apart from thorough purging of the hearts before CVPC isolation, CVPCs never gave rise to a single hematopoietic cell. They expressed several transcription factor genes typical for committed mesoderm and myocardial cells not found in bone marrow-derived stem cells, and did not require any paracrine stimuli or cell coculture to induce differentiation to CMCs as necessary for bone marrow-derived stem cells. Together, these facts make it extremely unlikely that CVPCs stem from multipotent progenitor cells present in the peripheral blood [Cesselli et al., 2009] in acute coronary syndromes [Wojakowski et al., 2010].
The Unique Features of CVPCs
Several features distinguish CVPCs from ESCs and from primary heart cells, respectively. Self-renewing CVPCs have a shorter doubling time, are significantly smaller than ESCs, exhibit reduced cell-cell interaction in clonal colonies on feeder cells, and are more prone to cell death in the absence of feeder cells. CVPCs continuously self-renew on LIF-secreting feeder cells for up to 450 days, which is close to the life span of a mouse, whereas primary heart cells, placed on feeder cells, divide at most a few times and then start to die or to dedifferentiate to fibroblast-like cells. In sharp contrast to ESCs, which require aggregation to proceed with development [Bader et al., 2001], CVPCs in monolayer cultures spontaneously differentiate to cardiac cell types in the absence of LIF. They give rise exclusively to ETCs, CMCs, and SMCs, whereas ESCs undergo commitment and differentiation to cell types of all 3 germ layers in a gastrulation-like process [Fuchs et al., 2012]. In the presence of strong stimuli like activin A and retinoic acid, CVPCs do not differentiate to definitive endoderm and neurons, respectively, whereas EBs develop large quantities of definitive endodermal and ectodermal cells. CMCs derived from CVPCs exhibit pacemaker-like, atrial, and ventricular action potential types at an equal ratio, contrarily to CMCs derived from ESCs, in which the atrial action potential type predominates [Kuzmenkin et al., 2009]. Similarly, in CVPC-derived CMCs the action potential length indicates a more mature phenotype as usually found in ESC-derived CMCs, and densities of Na+ and L-type Ca2+ currents in CMCs comply with a more advanced developmental state than observed in ESC- and induced pluripotent cell-derived CMCs. Finally, self-renewing CVPCs express stage-specific transcription factor genes not expressed in ESCs which suggests that CVPCs are already committed to the cardiac lineage. These unique features of CVPCs fit those of most other cardiac stem cell populations isolated by preparative FACS and distinguish them in a significant manner from ESCs and from induced or reprogrammed pluripotent cells.
The Differentiation Potential of CVPCs
The spontaneous and restricted differentiation of CVPCs to CMCs, ETCs, and SMCs suggests that the developmental stage of CVPCs is somewhere downstream from the segregation of the hematopoietic lineage from primitive mesoderm [Kattman et al., 2006] and that they are already exclusively committed to the cardiac lineage. ETCs always appear as the first somatic cell type and constitute the major part of differentiated cells in CBs which express CD31, CD34, and vWF. CMCs are the second most prominent cell type which becomes visible 5–6 days after the first ETCs appear. The upregulation of Nkx2.5, but not of Isl-1, in developing CVPC-derived CMC pre-cursors might indicate that these cells contribute to, or stem from, the primary heart field lineage [Laugwitz et al., 2008; Martin-Puig et al., 2008]. Robust and long-lasting rhythmic contraction of CMC networks, propagation of the pulse over 2–3 mm in CBs, and connexin 43-positive CMC-CMC contacts make it likely that these CMCs can participate in tissue formation and that they might support the function of the myocardium. SMCs are the third and least prominent cell type which becomes visible and they respond to angiotensin II in a physiological manner and form vessel-like structures in conjunction with ETCs.
Simultaneous expression of stemness, mesodermal, and myocardial transcription factor genes in self-renewing CVPCs and the presence of both Oct4 and Brachyury protein in nuclei of CVPCs may explain the preparedness of CVPCs to spontaneously differentiate to cardiac cells and suggests a regulatory network in which stemness and myocardial transcription factors balance self-renewal and differentiation. It remains to be demonstrated how these transcription factors interact and facilitate the transition from self-renewal to differentiation, in vitro and in vivo. Understanding this regulatory network in CVPCs will shed new light on the persisting problem of how self-renewal is suppressed for the sake of tumor prevention in vivo, and how somatic stem cells are continuously or temporarily stimulated to contribute to the functional maintenance of an organ throughout adulthood and perhaps also after injury.
The Relation to Other Cardiac Stem Cell Populations and Plasticity of CVPC Lines
A major question awaiting an answer concerns the nature and location of stem cell niches within the heart. In conjunction with this problem, it would be very helpful to know whether only one type of CVPC exists in the heart or more types exist which are located in different niches. Likewise, the relation of the different cardiac stem cell populations previously described to the CVPC lines described in this study is still an unsolved issue. Cardiac stem cells have been characterized by the presence of proteins like Sca1, c-Kit, Mdr1, or the transcription factor Isl-1, and those genes are all expressed in our clonal CVPC lines. Distribution of stem cells in the heart is most likely developmentally and spatially restricted [Pesce et al., 2011]. These facts make it possible that expression of one or more of these factors in stem cell lines varies depending on the time of isolation, on the developmental and metabolic stage of the tissue, and on the local niche from which they were isolated [Wu et al., 2008]. Adding to this diversity, stem cells most likely have a significant degree of plasticity which may allow a temporal shift in the expression pattern during the isolation procedure [Taubenschmid and Weitzer, 2012]. Thus, we argue that to some extent a reversible phenotypic shift of CVPCs during isolation and maintenance in an in vitro niche cannot be excluded, as this process cannot be excluded for any cell lines established in vitro in the past. However, due to the plasticity of CVPCs this phenotypic shift should be reversible, as long as CVPCs remain self-renewing and are placed in the proper niche. This phenomenon has been demonstrated for the conversion of ESCs to primitive ectodermal cells and vice versa [Pelton et al., 2002; Rathjen and Rathjen, 2003; Johnson et al., 2008]. Experimental testing of this hypothesis will become possible once several clonal CVPC lines have been isolated under different conditions and maintained self-renewing in a defined in vitro niche by different investigators.
Conclusion
These data suggest that CVPCs are genuine self-renewing and committed mesodermal precursors of cardiac cells with a restricted differentiation potential directed towards the cardiac lineage. Furthermore, these experiments demonstrate for the first time a feasible, simple, and low-cost isolation procedure for somatic stem cells from hearts of different ages which do not depend on the presence of not-so-specific surface proteins used to isolate CVPCs by fluorescence-activated cell sorting, and which thus may be utilized and modified for the isolation of stem cells from other organs and species in the future. Particularly, isolation of CVPCs from adult mice opens the possibility to obtain somatic stem cell lines from human biopsies by similar strategies. Stable and clonal somatic stem cell lines from animals and humans may also become useful for drug discovery and predictive toxicology. Most important for basic science is that the feasibility of a persistent in vitro culture of somatic stem cells without imposition of any phenotypic shift provides an important foundation and a new model for future basic studies shedding light on the niche-dependent molecular and cellular mechanisms balancing the self-renewal, dormancy, limited expansion, and eventually temporal differentiation of CVPCs.
Supplementary Material
Acknowledgements
We thank Dr. Allan Bradley for the AB2.2 ESCs and SNL76/7 fibroblasts which we have used now for 17 years in our lab without any phenotypical change, and Brigitte Gundacker, Guoxing Xu, Wolfgang Weber, and Nikola Vidovic for technical assistance.
This work was supported by funds from the Austrian Science Fund (FWF), grant P18659, the Herzfelder’sche Familienstiftung, and the Hochschuljubiläumsstiftung der Stadt Wien, grants H-2174/2007, H-1249/2009, and H-2302/2011 to G.W.
References
- Acquistapace A, Bru T, Lesault PF, Figeac F, Coudert AE, le Coz O, Christov C, Baudin X, Auber F, Yiou R, Dubois-Rande JL, et al. Human mesenchymal stem cells reprogram adult cardiomyocytes toward a progenitor-like state through partial cell fusion and mitochondria transfer. Stem Cells. 2011;29:812–824. doi: 10.1002/stem.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anversa P, Kajstura J, Leri A. If I can stop one heart from breaking. Circulation. 2007a;115:829–832. doi: 10.1161/CIRCULATIONAHA.106.682195. [DOI] [PubMed] [Google Scholar]
- Anversa P, Leri A, Rota M, Hosoda T, Bearzi C, Urbanek K, Kajstura J, Bolli R. Concise review: stem cells, myocardial regeneration, and methodological artifacts. Stem Cells. 2007b;25:589–601. doi: 10.1634/stemcells.2006-0623. [DOI] [PubMed] [Google Scholar]
- Bader A, Al-Dubai H, Weitzer G. Leukemia inhibitory factor modulates cardio-genesis in embryoid bodies in opposite fashions. Circ Res. 2000;86:787–794. doi: 10.1161/01.res.86.7.787. [DOI] [PubMed] [Google Scholar]
- Bader A, Gruss A, Hollrigl A, Al-Dubai H, Capetanaki Y, Weitzer G. Paracrine promotion of cardiomyogenesis in embryoid bodies by LIF modulated endoderm. Differentiation. 2001;68:31–43. doi: 10.1046/j.1432-0436.2001.068001031.x. [DOI] [PubMed] [Google Scholar]
- Bearzi C, Leri A, Lo Monaco F, Rota M, Gonzalez A, Hosoda T, Pepe M, Qanud K, Ojaimi C, Bardelli S, D’Amario D, et al. Identification of a coronary vascular progenitor cell in the human heart. Proc Natl Acad Sci USA. 2009;106:15885–15890. doi: 10.1073/pnas.0907622106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bearzi C, Rota M, Hosoda T, Tillmanns J, Nascimbene A, De Angelis A, Yasuzawa-Amano S, Trofimova I, Siggins RW, Lecapitaine N, Cascapera S, et al. Human cardiac stem cells. Proc Natl Acad Sci USA. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- Beltrami AP, Urbanek K, Kajstura J, Yan SM, Finato N, Bussani R, Nadal-Ginard B, Silvestri F, Leri A, Beltrami CA, Anversa P. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Stewart MH, Menendez P, George D, Vijayaragavan K, Werbowetski-Ogilvie T, Ramos-Mejia V, Rouleau A, Yang J, Bosse M, Lajoie G, et al. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448:1015–1021. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesselli D, Beltrami AP, Rigo S, Bergamin N, Verardo R, Piazza S, Klaric E, Fanin R, Toffoletto B, Marzinotto S, Mariuzzi L, et al. Multipotent progenitor cells are present in human peripheral blood. Circ Res. 2009;104:1225–1234. doi: 10.1161/CIRCRESAHA.109.195859. [DOI] [PubMed] [Google Scholar]
- Condorelli G, Borello U, De Angelis L, Latronico M, Sirabella D, Coletta M, Galli R, Balconi G, Follenzi A, Frati G, Cusella De Angelis MG, et al. Cardiomyocytes induce endothelial cells to trans-differentiate into cardiac muscle: implications for myocardium regeneration. Proc Natl Acad Sci USA. 2001;98:10733–10738. doi: 10.1073/pnas.191217898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies EL, Fuller MT. Regulation of self-renewal and differentiation in adult stem cell lineages: lessons from the Drosophila male germ line. Cold Spring Harb Symp Quant Biol. 2008;73:137–145. doi: 10.1101/sqb.2008.73.063. [DOI] [PubMed] [Google Scholar]
- Davis DR, Zhang Y, Smith RR, Cheng K, Terrovitis J, Malliaras K, Li TS, White A, Makkar R, Marban E. Validation of the cardiosphere method to culture cardiac progenitor cells from myocardial tissue. PLoS One. 2009;4:e7195. doi: 10.1371/journal.pone.0007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diel P, Baadners D, Schlupmann K, Velders M, Schwarz JP. C2C12 myoblastoma cell differentiation and proliferation is stimulated by androgens and associated with a modulation of myostatin and Pax7 expression. J Mol Endocrinol. 2008;40:231–241. doi: 10.1677/JME-07-0175. [DOI] [PubMed] [Google Scholar]
- Evans SM, Tai LJ, Tan VP, Newton CB, Chien KR. Heterokaryons of cardiac myocytes and fibroblasts reveal the lack of dominance of the cardiac muscle phenotype. Mol Cell Biol. 1994;14:4269–4279. doi: 10.1128/mcb.14.6.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs C, Scheinast M, Pasteiner W, Lagger S, Hofner M, Höllrigl A, Schultheis M, Weitzer G. Self-organization phenomena in embryonic stem cell-derived embryoid bodies: axis formation and breaking of symmetry during cardiomyogenesis. Cells Tissues Organs. 2012;195:377–391. doi: 10.1159/000328712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez BG, Sampaolesi M, Barbuti A, Crespi A, Covarello D, Brunelli S, Dellavalle A, Crippa S, Balconi G, Cuccovillo I, Molla F, et al. Cardiac mesoangioblasts are committed, self-renewable progenitors, associated with small vessels of juvenile mouse ventricle. Cell Death Differ. 2008;15:1417–1428. doi: 10.1038/cdd.2008.75. [DOI] [PubMed] [Google Scholar]
- Gambini E, Pompilio G, Biondi A, Alamanni F, Capogrossi MC, Agrifoglio M, Pesce M. C-kit+ cardiac progenitors exhibit mesenchymal markers and preferential cardiovascular commitment. Cardiovasc Res. 2010;89(2):362–373. doi: 10.1093/cvr/cvq292. [DOI] [PubMed] [Google Scholar]
- Gherghiceanu M, Popescu LM. cardiomyocyte precursors and telocytes in epicardial stem cell niche: electron microscope images. J Cell Mol Med. 2010;14:871–877. doi: 10.1111/j.1582-4934.2010.01060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson EM, Lindsay ME, Chien KR. Regeneration next: toward heart stem cell therapeutics. Cell Stem Cell. 2009;5:364–377. doi: 10.1016/j.stem.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Hofner M, Puz S, Stary M, Weitzer G. Desmin stimulates differentiation of cardiomyocytes and upregulation of brachyury and nkx2.5. Differentiation. 2007;75:605–615. doi: 10.1111/j.1432-0436.2007.00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höllrigl A, Hergovich A, Görzer I, Bader A, Ellersdorfer G, Habegger K, Hammer E, Enzinger S, Capetanaki Y, Weitzer G. High-throughput site-directed mutagenesis in ES cells. Biochem Biophys Res Commun. 2001;289:329–336. doi: 10.1006/bbrc.2001.5980. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BV, Shindo N, Rathjen PD, Rathjen J, Keough RA. Understanding pluripotency – how embryonic stem cells keep their options open. Mol Hum Reprod. 2008;14:513–520. doi: 10.1093/molehr/gan048. [DOI] [PubMed] [Google Scholar]
- Kajstura J, Urbanek K, Perl S, Hosoda T, Zheng H, Ogorek B, Ferreira-Martins J, Goichberg P, Rondon-Clavo C, Sanada F, Rota M, et al. Cardiomyogenesis in the adult human heart. Circ Res. 2010;107:305–315. doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kajstura J, Urbanek K, Rota M, Bearzi C, Hosoda T, Bolli R, Anversa P, Leri A. Cardiac stem cells and myocardial disease. J Mol Cell Cardiol. 2008;45:505–513. doi: 10.1016/j.yjmcc.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kennedy BP, Payette P, Mudgett J, Vadas P, Pruzanski W, Kwan M, Tang C, Rancourt DE, Cromlish WA. A natural disruption of the secretory group II phospholipase A2 gene in inbred mouse strains. J Biol Chem. 1995;270:22378–22385. doi: 10.1074/jbc.270.38.22378. [DOI] [PubMed] [Google Scholar]
- Kim M, Habiba A, Doherty JM, Mills JC, Mercer RW, Huettner JE. Regulation of mouse embryonic stem cell neural differentiation by retinoic acid. Dev Biol. 2009;328:456–471. doi: 10.1016/j.ydbio.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbling M, Estrov Z. Adult stem cells for tissue repair – a new therapeutic concept? N Engl J Med. 2003;349:570–582. doi: 10.1056/NEJMra022361. [DOI] [PubMed] [Google Scholar]
- Korbling M, Katz RL, Khanna A, Ruifrok AC, Rondon G, Albitar M, Champlin RE, Estrov Z. Hepatocytes and epithelial cells of donor origin in recipients of peripheral-blood stem cells. N Engl J Med. 2002;346:738–746. doi: 10.1056/NEJMoa3461002. [DOI] [PubMed] [Google Scholar]
- Kuhn EN, Wu SM. Origin of cardiac progenitor cells in the developing and postnatal heart. J Cell Physiol. 2010;225:321–325. doi: 10.1002/jcp.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmenkin A, Liang H, Xu G, Pfannkuche K, Eichhorn H, Fatima A, Luo H, Saric T, Wernig M, Jaenisch R, Hescheler J. Functional characterization of cardiomyocytes derived from murine induced pluripotent stem cells in vitro. FASEB J. 2009;23:4168–4180. doi: 10.1096/fj.08-128546. [DOI] [PubMed] [Google Scholar]
- Lagger G, O’Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza R. Handbook of Stem Cells. Elsevier; Amsterdam: 2004. [Google Scholar]
- Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135(2):193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, Platoshyn O, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauss M, Stary M, Tischler J, Egger G, Puz S, Bader-Allmer A, Seiser C, Weitzer G. Single inner cell masses yield embryonic stem cell lines differing in lifr expression and their developmental potential. Biochem Biophys Res Commun. 2005;331:1577–1586. doi: 10.1016/j.bbrc.2005.04.068. [DOI] [PubMed] [Google Scholar]
- Leri A, Kajstura J, Anversa P. Identity deception: not a crime for a stem cell. Physiology (Bethesda) 2005;20:162–168. doi: 10.1152/physiol.00005.2005. [DOI] [PubMed] [Google Scholar]
- Li TS, Cheng K, Lee ST, Matsushita S, Davis D, Malliaras K, Zhang Y, Matsushita N, Smith RR, Marban E. Cardiospheres recapitulate a niche-like microenvironment rich in stemness and cell-matrix interactions, rationalizing their enhanced functional potency for myocardial repair. Stem Cells. 2010;28:2088–2098. doi: 10.1002/stem.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limana F, Zacheo A, Mocini D, Mangoni A, Borsellino G, Diamantini A, De Mori R, Battistini L, Vigna E, Santini M, Loiaconi V, et al. Identification of myocardial and vascular precursor cells in human and mouse epicardium. Circ Res. 2007;101:1255–1265. doi: 10.1161/CIRCRESAHA.107.150755. [DOI] [PubMed] [Google Scholar]
- Martin BL, Kimelman D. Brachyury establishes the embryonic mesodermal progenitor niche. Genes Dev. 2010;24:2778–2783. doi: 10.1101/gad.1962910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2:320–331. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Matsuura K, Nagai T, Nishigaki N, Oyama T, Nishi J, Wada H, Sano M, Toko H, Akazawa H, Sato T, Nakaya H, et al. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J Biol Chem. 2004;279:11384–11391. doi: 10.1074/jbc.M310822200. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, Fiordaliso F, Salio M, Battaglia M, Latronico MV, Coletta M, Vivarelli E, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, Garry DJ, Entman ML, et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci USA. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Kai T, Decotto E, Spradling A. The stem cell niche: theme and variations. Curr Opin Cell Biol. 2004;16:693–699. doi: 10.1016/j.ceb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li B, Pickel J, McKay R, Nadal-Ginard B, Bodine DM, Leri A, et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001a;410:701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- Orlic D, Kajstura J, Chimenti S, Limana F, Jakoniuk I, Quaini F, Nadal Ginard B, Bodine DM, Leri A, Anversa P. Mobilized bone marrow cells repair the infarcted heart, improving function and survival. Proc Natl Acad Sci USA. 2001b;98:10344–10349. doi: 10.1073/pnas.181177898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Nagai T, Wada H, Naito AT, Matsuura K, Iwanaga K, Takahashi T, Goto M, Mikami Y, Yasuda N, Akazawa H, et al. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. J Cell Biol. 2007;176:329–341. doi: 10.1083/jcb.200603014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton TA, Sharma S, Schulz TC, Rathjen J, Rathjen PD. Transient pluripotent cell populations during primitive ectoderm formation: correlation of in vivo and in vitro pluripotent cell development. J Cell Sci. 2002;115:329–339. doi: 10.1242/jcs.115.2.329. [DOI] [PubMed] [Google Scholar]
- Peran M, Marchal JA, Lopez E, Jimenez-Navarro M, Boulaiz H, Rodriguez-Serrano F, Carrillo E, Sanchez-Espin G, de Teresa E, Tosh D, Aranega A. Human cardiac tissue induces transdifferentiation of adult stem cells towards cardiomyocytes. Cytotherapy. 2010;12:332–337. doi: 10.3109/14653240903548202. [DOI] [PubMed] [Google Scholar]
- Pesce M, Burba I, Gambini E, Prandi F, Pompilio G, Capogrossi MC. Endothelial and cardiac progenitors: boosting, conditioning and (re)programming for cardiovascular repair. Pharmacol Ther. 2011;129:50–61. doi: 10.1016/j.pharmthera.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Popescu LM, Gherghiceanu M, Manole CG, Faussone-Pellegrini MS. Cardiac renewing: interstitial Cajal-like cells nurse cardiomyocyte progenitors in epicardial stem cell niches. J Cell Mol Med. 2009;13:866–886. doi: 10.1111/j.1582-4934.2009.00758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen J, Rathjen PD. Lineage specific differentiation of mouse ES cells: formation and differentiation of early primitive ectoderm-like (EPL) cells. Methods Enzymol. 2003;365:3–25. doi: 10.1016/s0076-6879(03)65001-9. [DOI] [PubMed] [Google Scholar]
- Robertson M, Chambers I, Rathjen P, Nichols J, Smith A. Expression of alternative forms of differentiation inhibiting activity (DIA/LIF) during murine embryogenesis and in neonatal and adult tissues. Dev Genet. 1993;14:165–173. doi: 10.1002/dvg.1020140303. [DOI] [PubMed] [Google Scholar]
- Rula ME, Cai KQ, Moore R, Yang DH, Staub CM, Capo-Chichi CD, Jablonski SA, Howe PH, Smith ER, Xu XX. Cell autonomous sorting and surface positioning in the formation of primitive endoderm in embryoid bodies. Genesis. 2007;45:327–338. doi: 10.1002/dvg.20298. [DOI] [PubMed] [Google Scholar]
- Scott IC. Life before nkx2.5: cardiovascular progenitor cells: embryonic origins and development. Curr Top Dev Biol. 2012;100:1–31. doi: 10.1016/B978-0-12-387786-4.00001-4. [DOI] [PubMed] [Google Scholar]
- Smith RR, Barile L, Cho HC, Leppo MK, Hare JM, Messina E, Giacomello A, Abraham MR, Marban E. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- Smits AM, van Vliet P, Metz CH, Korfage T, Sluijter JP, Doevendans PA, Goumans MJ. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nat Protoc. 2009;4:232–243. doi: 10.1038/nprot.2008.229. [DOI] [PubMed] [Google Scholar]
- Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Ivey KN. Potential of stem-cell-based therapies for heart disease. Nature. 2006;441:1097–1099. doi: 10.1038/nature04961. [DOI] [PubMed] [Google Scholar]
- Stary M, Pasteiner W, Summer A, Hrdina A, Eger A, Weitzer G. Parietal endoderm secreted SPARC promotes early cardiomyogenesis in vitro. Exp Cell Res. 2005;310:331–341. doi: 10.1016/j.yexcr.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Stirnimann CU, Ptchelkine D, Grimm C, Muller CW. Structural basis of TBX5-DNA recognition: the T-box domain in its DNA-bound and -unbound form. J Mol Biol. 2010;400:71–81. doi: 10.1016/j.jmb.2010.04.052. [DOI] [PubMed] [Google Scholar]
- Sulzbacher S, Schroeder IS, Truong TT, Wobus AM. Activin A-induced differentiation of embryonic stem cells into endoderm and pancreatic progenitors – the influence of differentiation factors and culture conditions. Stem Cell Rev. 2009;5:159–173. doi: 10.1007/s12015-009-9061-5. [DOI] [PubMed] [Google Scholar]
- Tallini YN, Greene KS, Craven M, Spealman A, Breitbach M, Smith J, Fisher PJ, Steffey M, Hesse M, Doran RM, Woods A, et al. c-kit expression identifies cardiovascular precursors in the neonatal heart. Proc Natl Acad Sci USA. 2009;106:1808–1813. doi: 10.1073/pnas.0808920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Ashihara E, Takehara N, Nomura T, Honsho S, Nakagami T, Morikawa S, Takahashi T, Ueyama T, Matsubara H, Oh H. Clonally amplified cardiac stem cells are regulated by Sca-1 signaling for efficient cardiovascular regeneration. J Cell Sci. 2007;120:1791–1800. doi: 10.1242/jcs.006122. [DOI] [PubMed] [Google Scholar]
- Taubenschmid J, Weitzer G. Mechanisms of cardiogenesis in cardiovascular progenitor cells. Int Rev Cell Mol Biol. 2012;293:195–267. doi: 10.1016/B978-0-12-394304-0.00012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Urbanek K, Cesselli D, Rota M, Nascimbene A, De Angelis A, Hosoda T, Bearzi C, Boni A, Bolli R, Kajstura J, Anversa P, et al. Stem cell niches in the adult mouse heart. Proc Natl Acad Sci USA. 2006;103:9226–9231. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale I, Senovilla L, Jemaa M, Michaud M, Galluzzi L, Kepp O, Nanty L, Criollo A, Rello-Varona S, Manic G, Metivier D, et al. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 2010;29:1272–1284. doi: 10.1038/emboj.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojakowski W, Ratajczak MZ, Tendera M. Mobilization of very small embryonic-like stem cells in acute coronary syndromes and stroke. Herz. 2010;35:467–472. doi: 10.1007/s00059-010-3389-0. [DOI] [PubMed] [Google Scholar]
- Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132:537–543. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Evans EP, Smith AG. Changing potency by spontaneous fusion. Nature. 2002;416:545–548. doi: 10.1038/nature729. [DOI] [PubMed] [Google Scholar]
- Yu X, Cohen DM, Chen CS. miR-125b is an adhesion-regulated microRNA that protects mesenchymal stem cells from anoikis. Stem Cells. 2012;30:956–964. doi: 10.1002/stem.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hardamon C, Sagoe B, Ngolab J, Bui JD. Studies of the H60a locus in C57BL/6 and 129/Sv mouse strains identify the H60a 3′UTR as a regulator of H60a expression. Mol Immunol. 2011;48:539–545. doi: 10.1016/j.molimm.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.