Abstract
Self-renewing cells of the vertebrate heart have become a major subject of interest in the past decade. However, many researchers had a hard time to argue against the orthodox textbook view that defines the heart as a postmitotic organ. Once the scientific community agreed on the existence of self-renewing cells in the vertebrate heart, their origin was again put on trial when transdifferentiation, dedifferentiation, and reprogramming could no longer be excluded as potential sources of self-renewal in the adult organ. Additionally, the presence of self-renewing pluripotent cells in the peripheral blood challenges the concept of tissue-specific stem and progenitor cells. Leaving these unsolved problems aside, it seems very desirable to learn about the basic biology of this unique cell type. Thus, we shall here paint a picture of cardiovascular progenitor cells including the current knowledge about their origin, basic nature, and the molecular mechanisms guiding proliferation and differentiation into somatic cells of the heart.
Keywords: Stem cell, Cardiovascular progenitor cell, Self-renewal, Cell differentiation, Cardiogenesis, Transcriptional control, Cell plasticity.
1. Introduction
Following the identification of numerous self-renewing cells in vertebrate hearts during the past decade, the assumption emerged that the mammalian heart also has a limited, intrinsic, regenerative potential. Endogenous, self-renewing, and differentiating cells were only recently discovered to contribute to the maintenance, homeostasis, and proper function of the heart throughout the life of an organism. However, clinical approaches show that those cells fail to repair injuries sufficiently after acute myocardial infarction and cannot hinder chronic degeneration of the myocardium. In stark contrast to lower vertebrates, such as fish and newts that are able to respond to cardiac damage by generation of de novo cardiomyogenesis, mammals respond to injury of the heart with scar formation (Ausoni and Sartore, 2009). Interestingly, a continual, limited turnover of human heart cells has been demonstrated elegantly by C14-dating of postmortem heart cells (Bergmann et al., 2009). Most importantly, this study also showed a declining regenerative potential in elderly patients, which may be interpreted as a decrease in the number of self-renewing cells with increasing age.
These dividing heart cells shall be named here cardiovascular progenitor cells for several reasons explained in the next section on the definition of terms and have been identified in situ in the hearts of humans (Bearzi et al., 2009, 2007; Beltrami et al., 2001; Bergmann et al., 2009; Kajstura et al., 2010; Laugwitz et al., 2008; Messina et al., 2004; Smits et al., 2009), mice (Messina et al., 2004; Tallini et al., 2009; Tateishi et al., 2007), rats (Oyama et al., 2007), dogs (Linke et al., 2005), and pigs (Johnston et al., 2009). Cardiovascular progenitor cells have been characterized by the expression of stem cell antigen 1, SCA1 (Matsuura et al., 2004; Smits et al., 2009); Islet-1, ISL1 (Laugwitz et al., 2005; Moretti et al., 2006); the multidrug resistance protein, MDR1 (Oh et al., 2003); and the Stem Cell Factor receptor, cKIT (Bearzi et al., 2009, 2007; Beltrami et al., 2003). They were found mainly in the ventricular and atrial myocardium but also in the epicardial tissue (Limana et al., 2007), cardiospheres (Davis et al., 2009; Messina et al., 2004; Smith et al., 2007), heart auricles (Gambini et al., 2010), and embryonic stem cell-derived embryoid bodies (Kattman et al., 2006, 2011). In most publications, these cells have been uniformly termed cardiac stem cells or progenitor cells, but were also named mesoangioblasts (Galvez et al., 2008) and cardiac side population cells (Oyama et al., 2007).
The renewing cell populations were characterized in vitro by their potential to differentiate into cardiomyocytes, endothelial cells, smooth muscle cells (Bearzi et al., 2007; Oyama et al., 2007; Smits et al., 2009; Srivastava and Ivey, 2006; Wu et al., 2008), and possibly also cardiac fibroblasts (Zeisberg and Kalluri, 2010). These data led to the hypothesis that these somatic cell types in the heart share a common progenitor heritage, possibly a kind of primordial cardiovascular stem cell (Garry and Olson, 2006; Moretti et al., 2006). In mice, such isolated, primary cell populations contributed to the regeneration of the diseased heart to variable extents when injected into the myocardium adjacent to infarcted areas (Bearzi et al., 2007; Beltrami et al., 2003; Davis et al., 2009; Martin-Puig et al., 2008). However, their functional contribution to the heart has not been unambiguously demonstrated so far.
The identification and characterization of cardiovascular progenitor cells has been described in numerous reviews because of the feasible therapeutical potential of these cells in regenerative medicine. Thus, only a few reviews can be mentioned here (Anversa et al., 2007b; Hansson et al., 2009; Kajstura et al., 2008; Laugwitz et al., 2008; Wu et al., 2008), some of them dealing with the transcriptional regulation of the cell cycle (Goetz and Conlon, 2007) and cardiomyogenesis (Bruneau, 2002; Chien et al., 1993; Firulli and Thattaliyath, 2002; Laugwitz et al., 2008), others focusing on the various developmental stages a common progenitor has to run through before complete differentiation into a mature heart cell (Bruneau and Black, 2007; Garry and Olson, 2006; Martin-Puig et al., 2008; Musunuru et al., 2010; Wu et al., 2008).
The existence of tissue-specific self-renewing cells has been challenged by the identification of multipotent progenitor cells present in the peripheral human blood (Cesselli et al., 2009). Possibly, these cells are able to transmigrate through the vessel walls and populate the heart either constantly or after activation through paracrine mechanisms after injury. Likewise, induced reprogramming (Efe et al., 2011; Ieda et al., 2010), transdifferentiation (Takeuchi and Bruneau, 2009), and naturally occurring plasticity of somatic cells (Raff, 2003; Zipori, 2004a) open new questions about the origin and nature of cardiovascular progenitor cells.
Nonetheless, understanding the framework of molecular and cellular mechanisms guiding the instruction and limitation of maintenance, dormancy, self-renewal, and differentiation of cardiovascular progenitor cells in their natural microenvironment or niche is of fundamental biological interest and an indispensable prerequisite for future medical applications. On the way to reach this ultimate goal, numerous obstacles have to be overcome. First of all, there is still no consent about the molecular markers specifying somatic cardiac stem cells and whether they have a significant functional role in cardiovascular progenitor cells. Second, cells described so far differ in their morphology and developmental phenotypes. Third, we still know far too little about the stem cell niche of the heart, the transcriptional network regulating self-renewal of cardiovascular progenitor cells, and the developmental cues guiding differentiation into all cardiac cell types. Finally, but most importantly, we do not know the conditions for in vitro maintenance of isolated cardiovascular progenitor cells to keep them in an indefinite state of self-renewal in conjunction with the preservation of their differentiation potential. This last issue is of major interest and an indispensable prerequisite for studying and comparing molecular mechanisms of self-renewing cells of the heart (Moretti et al., 2007; Smits et al., 2009; Zhang et al., 2009).
Thus, most information obtained so far about the transcriptional regulation and the influence of growth factors, cytokines and small molecules on self-renewal, commitment, and differentiation of cardiovascular progenitor cells has been from genetic model organisms, such as Drosophila melanogaster, Xenopus laevis, Danio rerio, Mus musculum, and embryonic stem cell-derived embryoid bodies from mouse and man. For the sake of clarity, we will not distinguish between mechanisms analyzed in human and mouse stem cells and tissues, first, in order not to complicate the subject and, second, because there is too little data to draw a clear picture of the differences between these species. Consequently, several assumptions had to be made to build this hypothetical model of molecular regulation of cardiomyogenesis in the heart. We presume that the core mechanisms of self-renewal and differentiation are evolutionarily conserved in vertebrates and comparable to those in insects. Further, we suppose that molecular mechanisms responsible for cardiac development during embryogenesis are comparable to those involved in cardiomyogenesis and homeostasis of the adult heart.
Based on these assumptions, we shall review and discuss data concerning the fundamental nature of cardiovascular progenitor cells, their origin within the heart, and how our knowledge about the underlying molecular mechanisms evolved. These insights will foster our understanding of embryonic heart development, homeostasis in the adult heart, also in the context of congenital and acquired heart diseases, and finally on the inevitable decay of the aging heart.
2. Definition of Terms
Since dividing cells residing in the adult heart were discovered not even a decade ago, the definition and use of terms in the literature is still inconsistent. First of all, it is not entirely clear what connotations go along with the use of the terms “cardiac” and “heart” in combination with “cells” in the literature. In most cases, muscle cells of the heart, cardiomyocytes, are meant, regardless of the assumption that the heart might be composed of one or two dozens of different somatic cell types. Among them, cardiomyocytes, interstitial cells or fibroblasts, telocytes or podocyte-like cells, endothelial cells, smooth muscle cells, and all different cell types composing the complex cardiac conduction system can be defined. Going into further detail, one should keep in mind that the heart muscle is composed of different types of cardiomyocytes, such as atrial and ventricular ones. Similarly, cells forming the endocardium, heart valves, epicardium, and pericardium most likely also display different phenotypes. The same applies for the coronary blood vessels and the descending large vessels, where it is still hard to identify a clear border between arteries and veins on one side, and the core myocardium on the other side. Because of this plethora of cell types composing the heart, and the fact that all described self-renewing heart cells give rise to more than one somatic cell type, we will use here, as in the past, the term “cardiovascular” when speaking of cells located in the heart.
Second, there is still a Babylonian confusion concerning the terms “stem,” “stemness,” “progenitor,” and “precursor.” Concomitantly, the lack of knowledge about the differentiation potential of a cell further complicates this issue. To start with the easiest part, we suggest to prefer “progenitor” over “precursor” in regard to a cell placed within a series of cell divisions. A connotation of progenitor is to descend from something living, which fits best when talking about cells, whereas “precursor” is more often used in a chemical or technical sense. To decide whether a cell is a stem cell or a progenitor cell, we should first consider that the term “stem” implies the positioning at the base of a stem, a genealogical tree or a pedigree, giving rise to all descendants. Thus, we would assume this cell type to be totipotent or at least pluripotent. This would fit best to the zygote, the blastomeres, and perhaps to embryonic stem cells, but not to somatic or adult cells that inherit some, however, not all, properties of stemness. The latter ones include here the capability of a cell to self-renew, either indefinitely or for a limited period of time, and the potential to give rise to all types of somatic cells, including germ cells and somatic dividing cells. As self-renewing cells of an adult organ always stem from some predecessors classified in a certain lineage and are neither totipotent nor multipotent, we suggest not to use the terms “somatic stem cells” or “adult stem cells” any longer. Based on these considerations, we shall rather use here and in the future the term “cardiovascular progenitor cell” for a somatic cell in the heart that is capable of self-renewal and displays a certain degree of differentiation potential, being either multipotent or unipotent.
Two other terms that are often used to describe attributes of cardiovascular progenitor cells are “commitment” and “clonogenicity.” At a first glance, it seems simple and straight forward to argue that an organ-specific progenitor cell has to be committed to a certain lineage giving rise to at least some somatic cell types of the organ of origin, but not to those of other organs. Similarly, if progenitor cell are self-renewing they must be clonogenic, as they give rise to at least one identical daughter cell. However, since we see accumulating evidence for natural plasticity of cells and cannot exclude reprogramming of somatic cells as a potential source of progenitor or stem cell, we should not use “commitment” and “clonogenicity” as stringent attributes to define progenitor cells.
Concerning the heart as well as other organs, a future task will be to provide a more accurate definition of the progression of the differentiation process that characterizes the lineage commitment of cardiovascular progenitor cells with the ultimate acquisition of the adult phenotype. However, it remains an open question whether defined and stable cell lines showing a certain developmental stage between the primitive mesoderm and the adult heart can be isolated and characterized. Likewise, it is currently impossible to determine and classify cardiovascular progenitor cells isolated by different groups of scientists, according to their maturity, developmental potential, or dedication. In fact, the existence of distinguishable subpopulations of cardiovascular progenitor cells is still uncertain. Most populations are identical concerning their phenotype, mostly because of their smallness. However, the use of different marker genes to isolate and characterize different cell types by fluorescence-activated cell sorting does not solve this problem. First, no functional relevance of these markers has been described so far, and second, different expression levels of sets of genes, considered as typical for a certain cell type, do not guarantee a different phenotype. This we shall see later when discussing fluctuations of gene expression in cells with a high differentiation potential. Vice versa, diverse expression levels in differentially isolated cardiovascular progenitor cells cannot be taken as evidence for their discrimination and different function. It seems that as long as a certain degree of “stemness” can be attributed to a cell, the expression of responsible genes and functional consequences are inherently linked to some uncertainness. To address this problem properly, a first step would be to culture all differently isolated cardiovascular progenitor cells in an identical and parallel way. Characterization of these cell lines regarding their self-renewal, differentiation potential, and gene expression pattern would help to answer the question, about the existence of only one or several different types of cardiovascular progenitor cells. Consequently, in this review, we shall describe all primary cell populations published so far but will not distinguish between these cell population when discussing the potential and proven influence of transcription factor networks and paracrine signaling on their self-renewal and differentiation.
3. Origin of Cardiovascular Progenitor Cells
3.1. Evolutionary aspects
Inevitably, the heart is considered as the most essential and central organ of the human body as it is absolutely necessary for existence of all higher organisms. However, at the beginning of life, there was no need for heart-like structures and primitive multicellular organisms still do not use circulatory systems. An efficient pump became necessary for the first time when distribution of oxygen became an inevitable prerequisite for the survival of triploblastic organisms (Romer and Parsons, 1977). All along the evolutionary pathways from earliest sponges to humans, the major molecular and cellular mechanisms of heart development only slightly changed (Olson, 2006). The ancestral genes coding for transcription factors that form a network regulating cardiomyogenesis expanded through duplication, refined through modification and subsequential selection. This network was mainly comprised of NKX, MEF, GATA, TBX, and HAND transcription factors. The conserved principal coordination of cardiomyogenesis over millions of years presents the importance of the idea of the heart, even though today’s perfectly working organ only vaguely resembles the ancient contracting tubes of primitive sponges (Bishopric, 2005). Noteworthy, not all metazoans possess a heart. One group of miniaturized animals in the taxon Panarthropoda, the Tardigrades, has lost the heart during evolution and reduction of body size (Schmidt-Rhaesa, 2001).
In evolution, the earliest hearts served as primitive organs in late diploblastic or early triploblastic organisms (Martynova, 1995). A gradually increasing body size of these organisms led to the development of a body cavity, the coelom, or a fluid filled vessel-like structure. Improved nutrient and gas exchange, as well as centralized sexual reproduction allow evolutionary advantage for the newly arisen coelomata (Boero et al., 1998). The gradual specification of this “gastroderm” in diploblasts results in the appearance of a third germ layer, a prototype of mesoderm. Diploblastic jellyfish development was shown to involve the formation of the so-called entocodon, an autonomous tissue layer between the distal ectodermal and the endodermal tissue (Boero et al., 1998). In medusa, this layer is not only separated from the others by an extracellular matrix but also shows expression of muscle-specific forms of Troponin and Myosin heavy chain, distinctive mesoderm-patterning genes, such as Twist and Brachyury, as well as the muscle regulatory proteins MEF2 and SNAIL (Spring et al., 2002). As another prerequisite for a more structured tissue, collagen came up and served as a major component of extracellular matrix of the separating germ layers (Schroder et al., 2000). Interestingly, a protein homologue of cardiac Myotrophin, a factor that stimulates myocyte growth and triggers myocardial hypertrophy in mammalians, was shown to stimulate collagen synthesis in sponges. These early autocrine and paracrine signaling pathways together with the appearance of matrix molecules and the characteristic mesodermal gene expression in primitive diploblastic organisms can be considered as the earliest predisposition of a third germ layer, subsequently allowing myogenesis and muscle-like development (Chen and Fishman, 2000).
The first primitive myocytes appeared before the divergence of Radiata and Bilateria 555 million years ago (Oota and Saitou, 1999). Initially, organized muscle-like cells for early cardiac-like purpose may then have evolved from local dorsal, in insects, or ventral, in vertebrates, excrescences of the primitive foregut (Martynova, 2004). The function of these simple, but already electrically and functionally coupled, contracting structures was the formation of a pumping system for body fluids through the coelom of bilaterian organisms (Moorman and Christoffels, 2003; Simoes-Costa et al., 2005). Later, muscular tubes evolved, squeezing rhythmically and moving blood-like liquid through peristaltic contraction through the body. Invertebrates, such as modern earthworms, still possess these evolutional relicts, some of them having seven pumping tracts regulating their straightforward body fluid circulation system (Avery and Thomas, 1997). Further, most insects still inherit the so-called dorsal vessel which fulfill the function of a primitive heart (Baccetti and Bigliardi, 1969). In D. melanogaster, a pulsating blood vessel already containing valves is formed from heart precursor cells in response to Decapentaplegic, DPP, a Bone Morphogenetic Protein, BMP, homologue, and Wingless, WG, a WNT homologue. Here, the NKX2.5 homologue Tinman is required for cardiac specification (Chen and Schwartz, 1995), resulting in two major cell types: cardioblasts of the contractile tube and flanking pericardial cells (Bodmer, 1995). Additionally, Pannier, a GATA4 homologue, T-box transcription factors, and Hand genes play fundamental roles in heart development of the fruit fly (Olson, 2006). Through modification and specification of this conserved set of genes, the mammalian homologue proteins namely, NKX2.5, TBX5, TBX20, HAND2, and GATA4, nowadays function in a similar manner as the ancient regulators of heart development. Thus, autoregulation of the key transcription factors has induced and stabilized definitive cardiac identity throughout evolution (Bruneau, 2002; Buckingham et al., 2005; Srivastava and Olson, 2000).
As a lineage of the primordial invertebrates slowly morphed through primitive chordates, it became the first fishes about 500 million years ago (Long, 1996). In addition to the extensive modification of the body structure, also the pumping and vessel system developed more complex characteristics in fish. Primary muscle cells diversified into skeletal, cardiac, and smooth muscle cells, ultimately resulting in the formation of atrial, ventricular, and conductive myocytes. Accordingly, a quite primitive organ considered as the heart aroused in fish 500 million years ago (Bishopric, 2005; Moorman and Christoffels, 2003; Romer, 1967; Simoes-Costa et al., 2005). The previously tube-like structure developed into a two-chambered, synchronously pumping heart, composed of an atrium and a ventricle, separated by a rudimentary atrioventricular valve. In this self-contained single circuit circulation in fish, the blood is pumped through the atrium and the ventricle and exits the organ through the conus arteriosus. It receives oxygen at the gills and is then pumped to the organs of the fish for gas, nutrients, and waste exchange before returning back to the atrium.
Together with further development and specialization of primitive hearts, different classes of cardiac and skeletal muscle gene isoforms evolved such as cardiac Actins and Troponin C (Gillis and Tibbits, 2002). The underlying mechanisms such as gene duplication and subsequent change of DNA sequence allow functional specialization not only on the cellular level but in a greater context (Olson, 2006). Novel regulatory transcription factor networks tolerate greater plasticity and adaptability to more specified demands and structural components.
Alongside, the amphibians came up evolving a three-chambered heart, comprising two atria and one ventricle (Simoes-Costa et al., 2005). The atria evolved through physical division of the original atrium or through duplication of an incoming vessel. The formation of the right and the left atrium permits somewhat separation of oxygenated and deoxygenated blood circulation. In reptiles, the heart became almost four-chambered (Wang et al., 2002). An incomplete ventricular division allows double circuit circulation and directed flow of mostly separated oxygenated and deoxygenated blood, necessary for survival in the terrestrial environment.
With the evolution of birds and mammals, 120 million years ago, the division between the left and the right ventricles was completed (Rishniw and Carmel, 1999). In this regard, the boundary between high expression of the T-box transcription factor 5, TBX5, in the left ventricle and low expression in the right ventricle of mammalian hearts marks the line where the septum forms during embryogenesis and divides the ventricles into two parts (Bruneau et al., 1999). The created double circuit circulation then includes total separation of oxygenated and deoxygenated blood as well as separation of the pulmonary and the systemic circulation. Alongside, cardiac shape and functionality developed and led to the formation of chambers, valves, and the conduction system. The latter one was only possible through the evolutionary invention of Connexins, intracellular channel forming proteins, transmitting chemical, and electrical signals (Becker et al., 1998). Whereas only one major type of Connexins can be found in primitive chordata, 20 separate forms developed in mammalia, enabling greater plasticity of complex heart formation. Accelerated communication through these channels allowed rapid, synchronous contraction, and has finally found a climax in the formation of the today’s cardiac conduction system (Myers and Fishman, 2003).
Hand in hand with the first steps of cell differentiation and tremendous changes in heart development, the question of integrity, maintenance, and repair strategies arose. In this context, a distinct, ideally well-controlled number of multipotent, somatic stem cells came up to fulfill all these requirements (Bosch, 2009). Stem cells were shown to keep responsibility of cellular homeostasis, replacing dysfunctional somatic cells, and generating new ones through asymmetric cell division. As the stem cell pool in adult organisms has to comprise a constant cell number, molecular pathways regulating maintenance and differentiation are required for their nonpathological function.
Stem cells were supposed to be at least 800 million years old; they first evolved in the oldest extant metazoan, the sponges (Bosch, 2009). The chemokine network of porifera provides earliest markers for stem cells, mesenchymal stem cell-like proteins, and stem cell maintenance factors such as Noggin and Glia Maturation Factor (Muller et al., 2004b). Likewise, homologues of these genes can still be found in the human genome (Muller et al., 2004a). More primitive organisms, such as molluscs, arthropods, and amphibians, use the mentioned progenitor cell pool to easily reconstruct parts of their heart or other organs via self-renewal or replacement (Martynova, 2004). In contrast to lower vertebrates, mammals respond to injury of the heart with scar formation (Ausoni and Sartore, 2009). As it was recently shown, also humans posses a limited ability of heart renewal; however, their regenerative potential is diminished in comparison to many other lower vertebrates (Beltrami et al., 2003). It is hypothesized that, in long-lived organisms, adult stem cells may serve as a source of restoration of minor injuries, prevent degeneration, and slow down aging during long-time organ function. However, possibly due to avoidance of neoplasia and tumor growth, the mammalian heart is not capable of complete regeneration after acute myocardial injury. This evolutionary adaptation excludes proper replacement of large-scale damaged tissue and makes cardiovascular diseases one of the leading causes of morbidity and mortality in the western word.
3.2. Origin of cardiovascular progenitor cells during embryogenesis
In vertebrates, the cardiovascular system and its central apparatus, the heart, represent the first organ system to become functional long before other parts in the early embryo are discernable. All subsequent events in life depend on the heart’s continuous contractility and ability to pump oxygen and nutrients through the body of higher organisms. During heart development, initial cardiac progenitor cell populations driven by underlying molecular mechanisms guide heart development, from the primitive contractile cardiac tube to gradually more specific structures, including chambers, valves, and the conduction system. In birds and mammals, this finally leads to the formation of a four-chambered heart, such as it can be found in humans. Due to the complex nature of the heart, developmental abnormalities result in congenital heart disease, the most common human birth defect, and may lead to medical consequences such as arrhythmias, cardiomyopathies, hypertrophies, or heart failure.
During embryonic development, the earliest cells showing cardiac fate were confirmed to have their origin in mesodermal tissue derived from the primitive streak that forms during gastrulation (Rawles, 1943; Robb and Tam, 2004). Recently, the T-box transcription factor Eomesodermin was described to be an initial and essential factor for epithelial to mesenchymal transition, marking the earliest formation of cardiac mesoderm through directly activating the basic helix–loop–helix (bHLH), transcription factor Mesoderm Posterior Homolog 1 (MESP1) (Costello et al., 2011; Saga et al., 2000). Accurate temporal and local control of transcription factors such as T-box transcription factor Brachyury and MESP1 is required for successful activation of downstream cardiac signaling and initiation of heart development (Solloway and Harvey, 2003). In one of the evolutionary oldest cardiac-like structure, the dorsal vessel of D. melanogaster, distinct gene expression patterns of developmental genes guide heart development (Harvey, 1996). However, the more complex the contractile organ gets, the more intricate pathways are required for its development. In mammalian cardiogenesis, a much greater diversity is needed to ensure formation and function of the multifaceted protein network in cells adopting cardiac fate (Fishman and Olson, 1997).
Among the most significant pathways, the canonical WNT/βCatenin signaling is absolutely required for embryonic mesoderm formation and thus lays the foundations for heart development during the third week of human embryogenesis (Eisenberg and Eisenberg, 1999). Remarkably, from that time point on, distinct canonical WNTs prohibit further heart formation and their inhibition was shown to induce heart activating factors such as the chemokine receptor CXCR4 (Marvin et al., 2001; Schneider and Mercola, 2001). In contrast, other noncanonical WNTs act through calcium regulation and phosphorylation of c-Jun N-Terminal Kinases, JNK, resulting in activation of cardiac progenitor cells during embryogenesis (Pandur et al., 2002). Further key molecules inducing heart development are the members of the Transforming Growth Factor beta (TGFβ) super-family, including Nodal, Activin, BMP, and Growth Differentiation Factor (GDF; Olson, 2006). These factors act through SMA and Mothers-Against-Dekapentaplegic homologs (SMAD) signaling molecules and were shown to be responsible for early mesendodermal induction. BMP antagonists chordin and noggin prevent downstream signaling of the stated pathways and hence the formation of cardiac mesoderm in inappropriate areas (Schlange et al., 2000). Further, Cdx genes, a family of very early expressed transcription factors regulating the Hox genes, together with retinoic acid signaling were shown to restrict the formation of anterior mesoderm and suppress cardiac development by implementing posterior identity to developing cells (Lengerke et al., 2011).
After and during initial gene expression patterning, specialization, and definition of cardiac progenitor cells, they migrate from the ventral splanchnic or visceral region of the mesoderm to the anterior lateral region of the early embryo to form the cardiac crescent (Tam et al., 1997). Subsequently to these initial molecular and physiological steps essential for the onset of early cardiomyogenesis, inductive endogenous signals, and those of the surrounding tissue induce mesodermal progenitor cell specification to the cardiac lineage (Tam et al., 1997). Utilized signaling for cardiac commitment includes Fibroblast Growth Factor, FGF, BMP, WNT, and Hedge-hog-induced signal transduction. Early cardiac and noncardiac patterning, regional activation, and inhibition of differentiation and signal response require a few irreplaceable key players. Among those, the cardiac transcription factors NKX2.5, TBX5, TBX20 and GATA4 are necessary and guiding for cardiomyogenesis (Gelb, 2004). Upstream of this network, FGF and BMP signaling were shown to stimulate the expression of the homeodomain transcription factor NKX2.5 and, as mentioned previously, activate a number of downstream cardiac transcription factors such as MEF2 and GATA4, finally resulting in the onset of muscle-specific gene expression (Tanaka et al., 2001).
The earliest specified precursor cells commit henceforward irreversibly to the cardiac lineage and begin to differentiate. These primed precursor cells are in a premature state still within a progenitor cell pool named the “first heart field” and can be characterized through the expression of TBX5, HAND1, and the first wave of NKX2.5 (Lengerke et al., 2011). They have cardiac developmental potential when explanted and cultured in vitro (Jacobson and Sater, 1988). After cells of the first heart field eventually build up the primordium, the latter one subsequently fuse to a linear heart tube. The inner layer, the endocardium, is composed of endothelial cells, and the outer layer, the myocardium, of myocardial cells (Buckingham et al., 2005). At this point, the heartbeat is initiated, possibly as a direct result of induction of cTnT and Tropomyosin-4 expression (Nishii et al., 2008).
After elongation of the tubular heart structure, a second population of proliferating progenitors in the pharyngeal mesoderm, lying anterior of the cardiac crescent, is recruited to the poles of the heart tube (Christoffels et al., 2000). These cells of the second heart field are characterized through expression of Insulin gene enhancer protein 1, Islet-1 or ISL1, FGF10, bHLH, transcription factor HAND2, and a second wave of NKX2.5 during embryonic development. Using dyes, retroviral lineage tagging, or LacZ transgenes, the migration of these cells into the cranial part of the previously formed heart tube was revealed. They contribute primary to cardiomyocytes and vascular smooth muscle cells, constructing the outflow tract and the right ventricular myocardium (Moses et al., 2001). According to in vitro analysis, signals from the already existing outflow tract myocardium are sufficient to recruit cells from this second heart field to a myocardial fate (Mjaatvedt et al., 2001), and BMP antagonist Noggin was shown to inhibit the effect (Waldo et al., 2001).
During cardiac looping, where the linear heart tube folds ventrally, the migrating cells of the second heart field present a reserve pool of progenitors before contributing to the developing heart. Along the linear heart tube, recruited cells display diverse molecular behavior and gene expression patterns according to their positions in the looping heart. Asymmetric specification and remodeling of the folded heart tube require especially the embryonic left–right axis accompanied by sided expression of Pituitary Homeobox 2 transcription factor (PITX2), HAND1 and 2, and XIN, an Actin binding protein, among several other factors (Franco and Campione, 2003; Grosskurth et al., 2008; Harvey, 1998; McFadden et al., 2005). Together with the invasion of cells of the second heart field, progenitor cells from a nearby, transient, primitive organ-like structure, called the proepicardium, migrate toward the looping heart. These progenitors give rise to smooth muscle cells, endothelial cells, cardiomyocytes, and cardiac fibroblasts, finally forming the epicardium, the outer layer of the heart tissue, and contributing to the coronary vasculature (Limana et al., 2011). Regulating signals between the endo-, myo-, and epicardium are essential for correct growth and development of cardiac chambers later on. During the process of cardiac looping, the myocardium noticeably expands through invading cells of the second heart field and the heart tube bulges at its outer curvature. Anterior and posterior, as well as dorsal and ventral patterning is required for the primitive precursor structures of the later forming cardiac chambers (Christoffels et al., 2000).
Further specification of different heart tissues requires complete differentiation of cardiovascular progenitor cells and an augmented expression of cardiac-specific proteins such as actins, troponins, tropomyosins, and connexin channel proteins. Extensive remodeling of the internal structures of the heart includes septation and building of valves, guaranteeing the separation, and concurrent connection of the heart chambers. Definitive, coordinated myogenic specialization of restricted cells enables functional, contractile chamber formation.
During the ongoing formation and patterning of the heart, retrograde in vivo tracing of participating cells reveals that progenitors of the first heart field later contribute predominantly to the left ventricle in the adult heart. On the contrary, cells of the second heart field give rise to the outflow tract, the right ventricle, and to the right and left arterial chambers at the venous pole. The T-box transcription factor family plays a key role in establishment of chamber and nonchamber myocardium. TBX5 together with NKX2.5 promotes expression of chamber-specific genes, whereas TBX2 acts antagonistically and inhibits chamber formation (Christoffels et al., 2004). The remaining primary, nonchamber myocardium adopts another distinct fate, partly giving rise to the proximal conduction system, the atrioventricular node and the bundle of His, inflow and outflow vessel myocardium, and fibrotic tissue of the atrioventricular junction (Harvey et al., 2009). In addition to both heart fields and the contributions from the cranial neural crest to the heart (Scholl and Kirby, 2009), recent studies suggest the existence of a proepicardium-derived cardiac progenitor cell population (Zhou et al., 2008a). Cells of this distinct pool of progenitors expressing the WT1 genes separate early from the rest of the developing heart and later form the epicardial layer of the heart and the coronary blood vessels (Perez-Pomares et al., 2002).
Wide-ranging adjustment of the pumping structure, finally leading to cardiac valve formation, complete septation, separation of the oxygenated and deoxygenated blood in the heart, occurrence of the coronary vasculature, the conduction system, and the epicardium, grants the heart its final form. Importantly, cardiogenesis requires a non-negligible amount of plasticity. This is supported by the fact that boundaries between distinct tissues and their flanking regions are not clonally restricted, but dependent on progressively changing signaling gradients.
For now, just the top of the iceberg about the fundamental mechanisms of cardiogenesis during embryonic development has been explored. The intricate dilemma that insights into the generation of life go mostly hand in hand with its destruction is still challenging. Anyway, the understanding about these early processes must be internalized to acquire understanding of similar mechanisms guiding cardiogenesis in cardiovascular progenitor cells in the adult heart. Piece by piece, in single steps, the molecular puzzle of the heart yet has to be solved to give a general idea about our innermost organ.
3.3. Cardiovascular progenitor cells in the adult organism
For many decades, cardiac biology was regarded as a very static field of research as the heart was considered a postmitotic organ without any regenerative potential. The changing came with the discovery of a subpopulation of small, immature, and proliferating myocardial cells in the adult heart. Before that, myocytes were presumed to exist from early embryonic mesodermal development onward until the very old age of men. Although other organs such as bone marrow, liver, skin, brain, skeletal muscle, and pancreas harbor-specific populations of regenerative, proliferating cells (Passier and Mummery, 2003), putative stem cells of the heart remained undetectable. Accordingly, the organ was thought to be composed out of terminally differentiated myocytes without any reproductive potential (Nadal-Ginard et al., 2003). As it was assumed that the actual number of cardiomyocytes in the mammalian heart hits the maximum right after birth and progressively diminishes with age, the only compensatory mechanism after injury of the heart was considered to be hypertrophy of the remaining cells.
The phenomenon that after heart injury the affected organ does not show great ability to self-repair and reconstruct seemed to assure the initial idea of the heart as a postmitotic organ. However, earliest evidence for postnatal cardiac regeneration came from the detection of cycling cells in the fully developed mammalian heart right after myocardial infarction (Anversa and Nadal-Ginard, 2002; Leri et al., 2005). The stated study shows an increased number of immature, mitotic cardiomyocytes in the infracted border zone of the heart. Further, incorporation of bromodeoxyuridine during DNA replication, expression of Ki67 and phosphohistone-H3, and activation of cyclins and cyclin-dependent kinases (CDKs) has been evidential for karyo- and cytokinesis in adult myocytes (Bergmann et al., 2009; Kajstura et al., 2010; Leri et al., 2005). Unfortunately, the outcome of these initial experiments was rather misapprehended as cardiomyocytes were proposed to reenter the cell cycle after terminal differentiation (Anversa and Kajstura, 1998).
Anyway, the search for the concrete source of cycling cells led to the identification of a subpopulation of immature cells in the adult mammalian heart that was able to divide and give rise to new cells of the cardiac lineage (Beltrami et al., 2003). Anversa and colleagues isolated and characterized these small myocytes, according to their expression of the stem cell growth factor receptor tyrosine kinase, cKIT. The isolated cells were considered as bona fide cardiac stem cells that were confirmed to show self-renewing capacity, were clonogenic, and multipotent; moreover, they could differentiate into myocytes, smooth muscle cells, or endothelial cells (Beltrami et al., 2003). In vivo, subsequently to commitment to the myocytes lineage, these stem cells give rise to progenitor cells that divide once or twice before they finally develop into mature, terminally differentiated myocytes (Nadal-Ginard et al., 2003; Torella et al., 2005). The finding of a stem cell source in the adult heart properly explained the previously misinterpreted discovery of cycling cells in the alleged postmitotic organ.
Another source of progenitor cells might come from the epicardium. Thymosin β4 induces the synthesis of Plasminogen Activator Inhibitor 1 (Al-Nedawi et al., 2004) in endothelial cells and has been demonstrated to provide endogenous stem cells from a pool within the epicardium (Smart et al., 2011). After myocardial infarction and intraperitoneal Thymosin β4 administration in mice, these stem cells reactivate the embryonic epicardial gene Wilm’s tumor 1 (Wt1), begin to express ISL1 and NKX2.5, migrate to the infarcted area, and transdifferentiate to functional cardiomyocytes.
The plasticity of pericytes, differentiating into vascular smooth muscle cells (Nehls and Drenckhahn, 1993; Nehls et al., 1992), other mesenchymal cells types, including fibroblasts, osteoblasts, chondrocytes, and adipocytes (Collett and Canfield, 2005), was used to argue, that pericytes may present a source of stem cells in the heart with a niche located along the cardiac microvasculature (Dore-Duffy, 2008), similarly to their function as stem cells in the brain (Bonkowski et al., 2011). Likewise, cardiomyocytes that induce endothelial cells to transdifferentiate into cardiac muscle cells (Condorelli et al., 2001) as well as isolated mesoangioblasts were suggested to contribute to myocardial regeneration (Galvez et al., 2008; Messina et al., 2004). However, it is unclear whether these types of cellular plasticity should be regarded as transdifferentiation between different somatic cell types or a common mesenchymal stem cell. Although pericytes and mesoangioblasts may be of pathophysiological importance or even contribute to tissue regeneration and homeostasis on the adult heart, it is important to remember that the lack of definitive markers do not allow reliable fate mapping of these cells in vivo and in vitro. Hence, conducted experiments are generally confounded by the uncertain origin and identity of pericyte cultures.
These initial, outstanding findings were supported by later studies and numerous independent descriptions of populations of cardiac stem and progenitor cells and their isolation from different mammalian species, including mouse, rat, pig, dog, and human (Laugwitz et al., 2005; Martin et al., 2004; Matsuura et al., 2004; Messina et al., 2004; Oh et al., 2003; Pfister et al., 2005; Tomita et al., 2005). The obtained stem and progenitor cells are proposed to offer the heart a basis for homeostasis, plasticity, and regenerative potential during age-dependent or injury-induced degeneration of cardiac cells. Apart from hypertrophy, the heart was now demonstrated to inherit hyperplasic resource and competence.
However, there still is much discrepancy between and among the various cardiac stem and progenitor cell populations (Garry and Olson, 2006). The diverse cell types were identified with the help of completely different, independent surface markers such as SCA1, ATP-binding cassette transporter, ABCG2, or ISL1 and display 1–2% of total heart cells (Laugwitz et al., 2005; Martin et al., 2004; Oh et al., 2003). Phenotypical and methodical discrepancies between all collections of immature myocytes remain, possibly resulting in the unequal differentiation potential and expression profile of characterized cells. The lineage relation of the isolated subpopulations is still unclear, and the definition of a cardiac stem or progenitor state is ambiguous.
In regard to the fact that many organs only harbor one or two different types of progenitor cells, it is quite doubtful that the heart depends on three or more dissimilar groups of progenitors (Bollini et al., 2011). However, due to the lack of knowledge, it was also suggested that all or some of the described cell populations may present sequential or alternative differentiation stages of only one cell type (Ellison et al., 2007). As existence and the molecular characteristics of quiescent or cycling, undifferentiated or already committed stem, and progenitor cells could not yet be adequately defined, the question remains to be answered.
3.4. Marker of cardiovascular progenitor cells
The originally described and most promising population of cardiac stem cells was isolated according to the expression of cKIT and absence of the expression of Lin genes, a set of eight blood cell markers. So far, the isolated cell type represents the only one to fulfill the requirement of a bona fide stem cell, being self-renewing, clonogenic, and multipotent, giving rise to myocytes, smooth muscle, and vascular cells (Beltrami et al., 2003). When cultured in suspension, the cells were able to form so-called cardiospheres, similar to pseudo-embryoid bodies. Outgrowing cells from these aggregates express marker of myocytes, smooth muscle cells, and endothelial cells (Torella et al., 2005). Moreover, using chemically or genetically tagged cKIT + cells for injection into the border zone of the heart after experimentally inducing myocardial infarction, the labeled cells gave rise to functional myocytes and vascular structures in vivo, hence, partly replacing the infracted zone. Especially, the expression of Connexin 43 and N-cadherin demonstrates electrical and mechanical coupling to the surrounding tissue (Beltrami et al., 2003).
In human, cKIT+ cardiac cells were identified (Quaini et al., 2002; Urbanek et al., 2003) and isolated from adult patients (Smith et al., 2007). These cells were shown to be negative for hematopoietic and endothelial markers but did express the MDR1, which is a glycoprotein of the same family of membrane transporters as SCA1, and ABCG2 (Anversa and Nadal-Ginard, 2002). This fact mainly supports the theory that cKIT+ cardiovascular progenitor cells represent a more original, immature cell population among the various isolated progenitor cell types, with the potential to generate side population cells and SCA1+ cells. Human cKIT+ cells were shown to be able to give rise to cardiomyocytes, endothelial cells, and ventricular smooth muscle cells, indicating their capacity for cardiac regeneration (Bearzi et al., 2007).
Another population of cardiac progenitor cells was isolated from the murine adult heart based on the expression of SCA1+ (Oh et al., 2003). The origin and the molecular identity of SCA1+ cells remain unclear, as distinct groups reported different amount of the expression of marker proteins such as cKIT, TIE2, ANG1, CD31, CD34, or CD45 (Matsuura et al., 2004; Oh et al., 2003). The inconsistent expression profiles of SCA1 cells may correspond to isolation artifacts or dissimilar sources and origins of the gained cells. SCA1+ cells only start to express cardiac transcription factors NKX2.5 and GATA4, structural proteins cTNT, cActin, αMHC, and the Vascular Endothelial Growth Factor Receptor 1, FLT1, upon treatment with oxytocin (Matsuura et al., 2004) or the demethylating agent 5-azacytidine (Oh et al., 2003). However, in further guided development, they formed sarcomeric structures and spontaneously beating clusters. After ischemic reperfusion, SCA+ cells that were injected intravenously into the patient’s abdomen target the injured myocardium through differentiation into cardiomyocytes (Oh et al., 2003). Anyway, the identity of SCA1+ cells as cardiac stem cells and their therapeutic potential have been questioned because the marker gene is not decisive on human cells.
Cells of the so-called side population within the SCA1+ fraction represent another population of cardiac progenitor cells (Martin et al., 2004). They are marked by the expression of ABCG2, that enables the cells to exclude Hoechst and rhodamine dyes (Oh et al., 2003). Side population cells were shown to exist from early embryonic heart development on and persist into the adulthood in various organs such as muscle, liver brain, lung, skin, and finally heart (Montanaro et al., 2003). These cells show stem cell-like properties but prove ability to differentiate into cardiomyocyte lineages only upon treatment with oxytocin or trichostantin A, a histone deacetylase (HDAC) inhibitor (Oyama et al., 2007). When injected into the ischemic heart, side population cells differentiated to endothelial cells and cardiomyocytes (Oyama et al., 2007).
Recent work identified ISL1 expressing cells as cardiovascular progenitor cells in the human adult heart (Laugwitz et al., 2005). They were shown to be remnants from the cardiac primordia, the anterior pharynx, and subsequently the second heart field (Cai et al., 2003). In adult hearts, ISL1+ cells can be found in the outflow tract, the atria, and the right ventricle. They have been characterized as cardiac stem cells, implying the capability to self-renew and expand or differentiate to smooth muscle cells, cardiomyocytes, and endothelial cells (Bu et al., 2009). Approving the theory of ISL+ cells exclusively giving rise to myocytes of the formerly second heart field, no contribution of the subtype to the left ventricle was found. Secluded ISL+ cells from newborn rodents and humans showed potential to give rise to cardiac myocytes both in vivo and in vitro. Electromechanical coupling of in vitro differentiated ISL1+ cells into functional signaling networks indicates the possibility of treatment of conductive system diseases. However, the number of isolated stem cells decreases dramatically during the first weeks after the birth of mammals (Laugwitz et al., 2005), and moreover, in adult humans, no ISL1+ cells could be identified so far.
The epicardium was lately discovered as another potential source of fetal and adult cardiac stem cells (Lepilina et al., 2006). The outermost layer of the heart was shown to harbor multipotent mesenchymal progenitor cells expressing the early epicardial genes Wt1 and Tbx18 during embryonic development and in the injured heart of adult organisms. The according cells were hypothesized to undergo epithelial mesenchymal transition, before forming so-called epicardium-derived mesenchymal cells and finally giving rise to multiple cardiac lineages such as cardiac fibroblasts, vascular smooth muscle cells, endothelial cells, and cardiomyocytes (Limana et al., 2011). The named properties are of particular importance during embryonic cardiogenesis and formation of the coronary vasculature but also in adult heart regeneration. After coronary artery occlusion, restoration of embryonic epicardial genes and developmental programs has been demonstrated to be essential for tissue regeneration (Urbanek et al., 2005). Moreover, in vitro culture of epicardial derived cells allows differentiation into smooth muscle cells, cardiomyocytes, and endothelial cells (Wu et al., 2006a). However, similar to ISL1+ cells, stem cells of the epicardium are mainly abundant during fetal development and merely disappeared in the adult heart (Limana et al., 2007).
Apart from problematic determination of their lineage relation, also the origin of cardiac stem cells yet has to be revealed. Either intrinsic cardiac cells exist in the adult organism since fetal life or cells of extracardiac origin have colonized the myocardium in postnatal life through the circulatory system. The small number of ISL1+ progenitors in the postnatal mammalian heart indicates the existence of remnant cardiac stem cells from embryonic development onward. Moreover, also cKIT+ cells and side population cells have been identified in embryonic and fetal development (Martin et al., 2004; Messina et al., 2004). On the other side, sex-mismatched cardiac and bone marrow transplants (Bayes-Genis et al., 2004; Quaini et al., 2002; Thiele et al., 2004) suggest an extracardiac source of recruited progenitors reconstituting the heart because cells with genetic markers of the donor and recipient, respectively, mix in the transplanted heart. If the bone marrow indeed harbors a source of cardiac stem cells in the adult organism, these cells would present a hitherto unidentified subpopulation of progenitors.
However, the in vitro and in vivo differentiation potential, the phenotype, the molecular identity, and the expression of defined marker genes of these immature cells vary among subpopulations (Di Nardo et al., 2010). Precardiac and cardiac transcription factors such as MESP1, TBX2, and NKX2.5 were shown to be unequally expressed. Further, numerous marker proteins such as SCA1, cKIT, MDR1, NKX2.5, and GATA4 are also abundant in other tissues and organs (Di Nardo et al., 2010). Hence, more definitive markers of the various cell types still need to be termed. Conflicting views of different functional characteristics have to be further investigated, integrated, and united. Especially for clinical reasons, homogeneous, fully characterized cardiovascular progenitor cells will be necessary to avoid decontamination by hyperplastic or tumorigenic cells.
3.5. Cardiac stem cell niche
Stem cells and progenitor cells are supposed to reside in habitats called niches. We use “niche” here as a collective term including connotations such as microenvironmental influences, paracrine and autocrine influences, and cellular contact, as well as the physical parameters influencing cells. They remain there in an undifferentiated and quasi-dormant state until external signals stimulate their commitment and differentiation into specific somatic cell types required for repair and maintenance of the according organ. Consequently, the myocardium possesses interstitial structures with the architectural organization of specific stem cell niches (Urbanek et al., 2006) particularly located in the atria and in a subepicardial region of the ventricles (Gherghiceanu and Popescu, 2010; Kuhn and Wu, 2010; Popescu et al., 2009).
Embryonic stem cells were shown to support themselves to maintain their self-renewing gene expression pattern in the presence of a minimal cocktail of growth factors just by cell–cell contact. In this case, the niche-like environment is composed of embryonic stem cell-derived Fibroblasts and the embryonic stem cells themselves that keep their status by a complex interplay between the Insulin-like Growth Factor 2 (IGF2), basic Fibroblast Growth Factor (bFGF), (Bendall et al., 2007), and Leukemia Inhibitory Factor (LIF) signaling (Niwa et al., 2009). In cardiospheres, as suggested by Anversa et al. (2007a), also niche-like environmental structures were identified (Li et al., 2010). Many of these niche cells express surface receptors such as N-cadherin, or β4α1 Integrins found on fibroblasts (Moore and Lemischka, 2006) in the interstitial space in the heart (Oyama et al., 2007). As the expression of Brachyury in primitive mesoderm is sufficient to establish the embryonic mesodermal stem cell state (Martin and Kimelman, 2010), it was argued here that these cells build up their own niche by supporting each other, without the need for a cellular compartment of different cell types.
In the heart, so far no specific stem cell niche comprising diverse types of supporting cells has been described. However, several cell populations residing in various locations in the heart were suggested to have stem cell potential. Recently, the epicardial stem cell niche has been proposed to contain cardiomyocyte precursors, potentially nursed by telocytes (Gherghiceanu and Popescu, 2010) and interstitial Cajal-like cells (Popescu et al., 2009).
3.6. Cardiovascular progenitor cell descent
Induction and specification of mesoderm commence during gastrulation of triploblastic organisms (Stern, 2004). All mesodermal cells share the expression of Brachyury, the founding member of the T-box family of transcription factors (Herrmann et al., 1990; Kispert and Herrmann, 1993). Its expression is downregulated during commitment and differentiation and upon patterning and specification of nascent mesoderm into derivative tissue (Kispert and Herrmann, 1994). As founders of all mesodermal cell types, the positioning of Brachyury positive cells along the primitive streak, together with distinct gradients of morphogen, predestine them already to a specific developmental fate. To mention just one out of many examples, the gradual expression of the transcription factor Goosecoid significantly influences Brachyury expression and mesodermal patterning (Niehrs et al., 1994) by a reciprocal inhibition of both genes (Artinger et al., 1997).
Brachyury positive, T+, primitive mesodermal cells can be divided into two populations. Those cells expressing the Tyrosine kinase receptor, FLK1, develop into hemangioblasts and consequently give rise to all descendants of the hematopoietic lineage (Kouskoff et al., 2005). The major part of the T+ cells, not expressing FLK1, differentiates into cardiomyocytes. Later, a subpopulation of FLK1 positive cells expressing the Platelet-Derived Growth Factor Receptor α (PDGFRα) was reported to gave rise to a significant number of cardiomyocytes when stimulated with Activin, Nodal, and BMP4 (Kattman et al., 2006, 2011). These sets of data have been obtained from cells generated and isolated from embryonic stem cell-derived embryoid bodies that allow, in contrast to in vivo studies, to manipulate developmental processes by growth factors and to isolate defined subpopulations of cells. Differentiation of embryonic stem cells in combination with preparative FACS can be used to define the biochemical status of a certain cell type expressing a common surface antigen. However, these methods exclude the effects of paracrine signaling on cell proliferation, commitment, and differentiation during cardiomyogenesis in vivo. Small differences in the culture conditions upon differentiation of embryonic stem cells and cardiovascular progenitor cells may easily lead to varying expression levels and thus to contradicting results.
MESP1, another transcription factor marking cardiogenic mesoderm (Kitajima et al., 2000; Saga et al., 1996, 2000, 1999), promotes cardiomyogenic differentiation (David et al., 2008). However, a population of MESP1 negative precursor cells was shown to contribute to parts of the ventricular cardiac conduction system (Kitajima et al., 2006), displaying again the ambivalent situation concerning the specificity of the used markers. Most recently, it has been demonstrated that Brachyury (David et al., 2011) and Eomesodermin (Costello et al., 2011) activate expression of MESP1 providing at least one of the numerous remaining missing links between primitive mesodermal and cardiovascular progenitor cells.
A third transcription factor, the LIM/homeodomain protein, ISL1, was identified in embryonic heart cells (Bu et al., 2009; Cai et al., 2003; Laugwitz et al., 2005; Moretti et al., 2006) that gave rise to cardiomyocytes, endothelial cells, and smooth muscle cells of the second heart field. ISL1 negative precursor cells were shown to be characteristic for the first heart field (Musunuru et al., 2010). In a more detailed hierarchy, Chien and colleagues suggest that ISL1 and FLK1 positive cells gave rise to vascular progenitors and finally to endothelial and vascular smooth muscle cells, whereas ISL1 and NKX2.5 positive precursors develop into smooth muscle cells, cells of the conduction system, and atrial, as well as ventricular cardiomyocytes (Laugwitz et al., 2008). Similar cell types could be derived from human-induced pluripotent stem cells (Moretti et al., 2010).
Anversa and colleagues isolated cKIT positive and lineage negative, Lin−, cells from adult hearts and subpopulations of progenitor cells expressing cKIT together with SCA1 or with both SCA1 and ISL1 and showed their differentiation potential into endothelial, smooth muscle, and cardiomyocyte lineages (Bearzi et al., 2007; Beltrami et al., 2003; Sun et al., 2007). These diverse cell types may represent different stages during cardiac stem cell development and differentiation (Ott et al., 2007). Likewise, a population of cKIT+, SCA1+ cells expressing P-glycoprotein, a member of the multidrug resistance ABC protein family that is found in side population cells, was identified in cardiac tissue (Barile et al., 2007).
cKIT+ progenitor cells were also found in human heart auricles giving rise to mesenchymal stem cells expressing the appropriate markers but no cardiac-specific genes (Aghila Rani et al., 2008; Gambini et al., 2010). These cells had a different phenotype, and when cocultured with cardiomyocytes, differentiated primarily into adipocytes and osteoblasts. As they developed into smooth muscle, endothelial, or cardiomyocytes to a much lesser extent, a noncardiac origin of these progenitors was suggested. In contrast, cKIT and nestin positive cells from postnatal murine hearts readily differentiated into endothelial, smooth muscle, and cardiac muscle cells in vitro and in vivo (Tallini et al., 2009). This study also demonstrates that cKIT expression in postinfarction hearts does not reflect stem cell activity but rather demonstrates infiltration of cKIT positive blood cells. Notably, more than 60% of the CD34 positive blood cells are also positive for cKIT (Reisbach et al., 1993).
To obtain a population of cells with a differentiation potential more restricted to the vasculature, the Vascular Endothelial Growth Factor Receptor 2, KDR, together with cKIT was used to isolate and expand resident coronary vascular progenitor cells from human myocardial samples (Bearzi et al., 2009; Leri et al., 2011). Some of these cells showed self-renewal, were clonogenic, and differentiated predominantly into endothelial and smooth muscle cells. However, as the expression of KDR did not exclude the differentiation into cardiomyocytes, a high degree of plasticity was again apparent. This suggests that most populations isolated with different surface antigens may be reprogrammed during isolation and culture and are not necessarily committed to a certain lineage.
SCA1 was used to isolate a population of cells displaying high telomerase activity, resembling the side population of hematopoietic stem cells (Oh et al., 2004), and cardiac progenitor cells, that were negative for CD34 and CD45 and did not express any endothelium or myocardium specific gene. Comparable cells were also isolated from human hearts (van Vliet et al., 2008) but differentiated into cardiomyocytes only upon exposure to 5-azacytidine (Oh et al., 2004). Later, it was demonstrated that a cardiac-specific side population of cells positive for SCA1, but negative for CD31, gave rise to functional cardiomyocytes when cocultured with adult cardiomyocytes. This suggests the requirement of the intimate contact with cardiac cells and most likely paracrine signaling to adopt a cardiomyocyte phenotype (Pfister et al., 2005). These data demonstrate further, that blood stem cells can be isolated from cardiac tissue, and question any attempts to isolate tissue-specific stem cells. Moreover, the group showed that CD34 and CD45 negative progenitor cells do not have the intrinsic information for differentiation into cardiomyocytes. Although other progenitor cell populations also express SCA1, this marker is most likely inadequate for the enrichment of tissue-specific cardiovascular progenitor cells. Most importantly, similar multipotent progenitor cells can be isolated from peripheral blood after Granulocyte Colony-Stimulating Factor (GCSF) stimulation (Cesselli et al., 2009).
These multipotent progenitor cells are clonogenic, self-renew in vitro for a long time, and, under appropriate environmental conditions, differentiate into derivatives of all three germ layers. They express surface proteins similar to those present in mesenchymal stem cells but are developmentally younger. Their molecular and functional characteristics inevitably resemble human embryonic stem cells rather than any other somatic stem cell type described so far. In these multipotent progenitor cells, the pluripotency-specific transcription factors OCT4, Nanog, SOX2, KLF4, and c-MYC are expressed and the telomerase activity is comparable to that in embryonic stem cells. According to first reports, which still have to be confirmed by independent investigators, these multipotent progenitor cells of the blood migrate to most organs and integrate into the tissue by acquiring the structural and functional identity of the resident cell types. They give rise to endothelial cells of the blood vessels, form hepatocytes in the liver, and most astonishingly, transmigrate through the brain–blood barrier and give rise to neurons of the brain of immunodeficient mice. Finally, to make the current picture even more complex, very small embryonic-like stem cells, so-called VSELs, have been isolated from murine and human hearts (Zhang et al., 2011). This finding adds to the notion that stem cells isolated from a particular tissue or organ may well come from the bone marrow as the only source of stem and progenitor cells in the adult body.
It will be interesting to investigate whether all these cardiovascular progenitor cells are ontologically related to human embryonic stem cells. If they are, we have to consider the mechanisms of stem cells surviving developmental pressure during gastrulation. Possibly, niches in the early eutherian embryo allow the endurance of embryonic stem cells into adulthood, or mature cells find their way back to the embryonic stem cell phenotype similarly to mechanisms first demonstrated by Yamanaka et al. in vitro (Takahashi and Yamanaka, 2006). Further, there may be ways to oscillate from a quiescent and more mature mesenchymal phenotype to a highly proliferative and immature state resembling embryonic stem cells. Similarities to the reversible epithelial-to-mesenchymal transition suggest that the study and comparison of both processes on the molecular level may lead to new fundamental insight into the process of naturally occurring reprogramming of somatic cells.
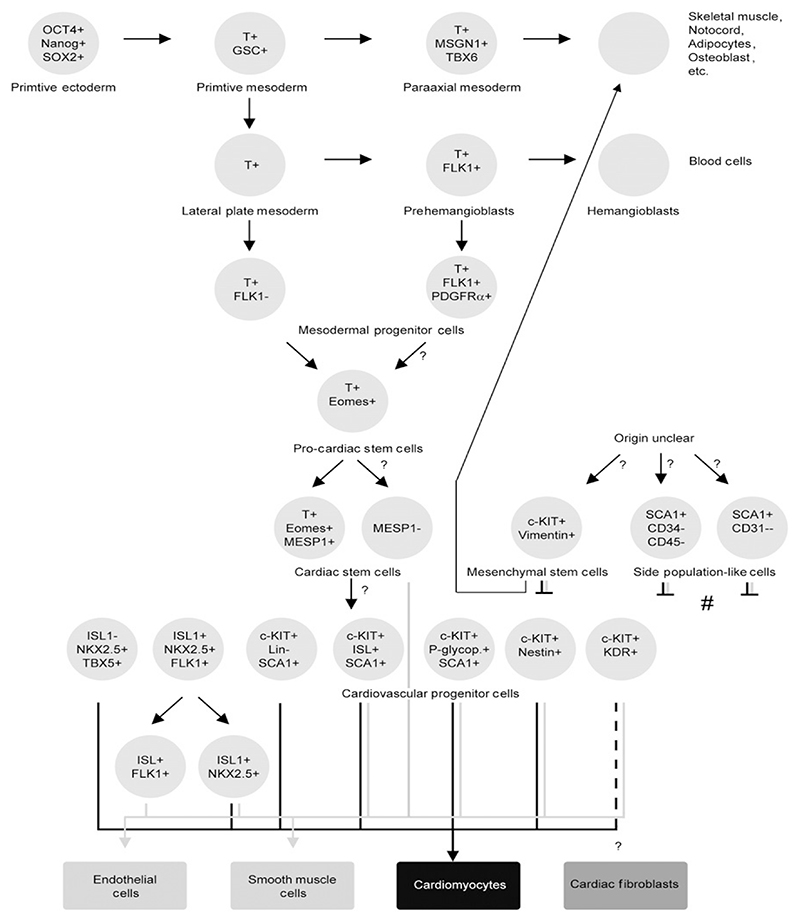
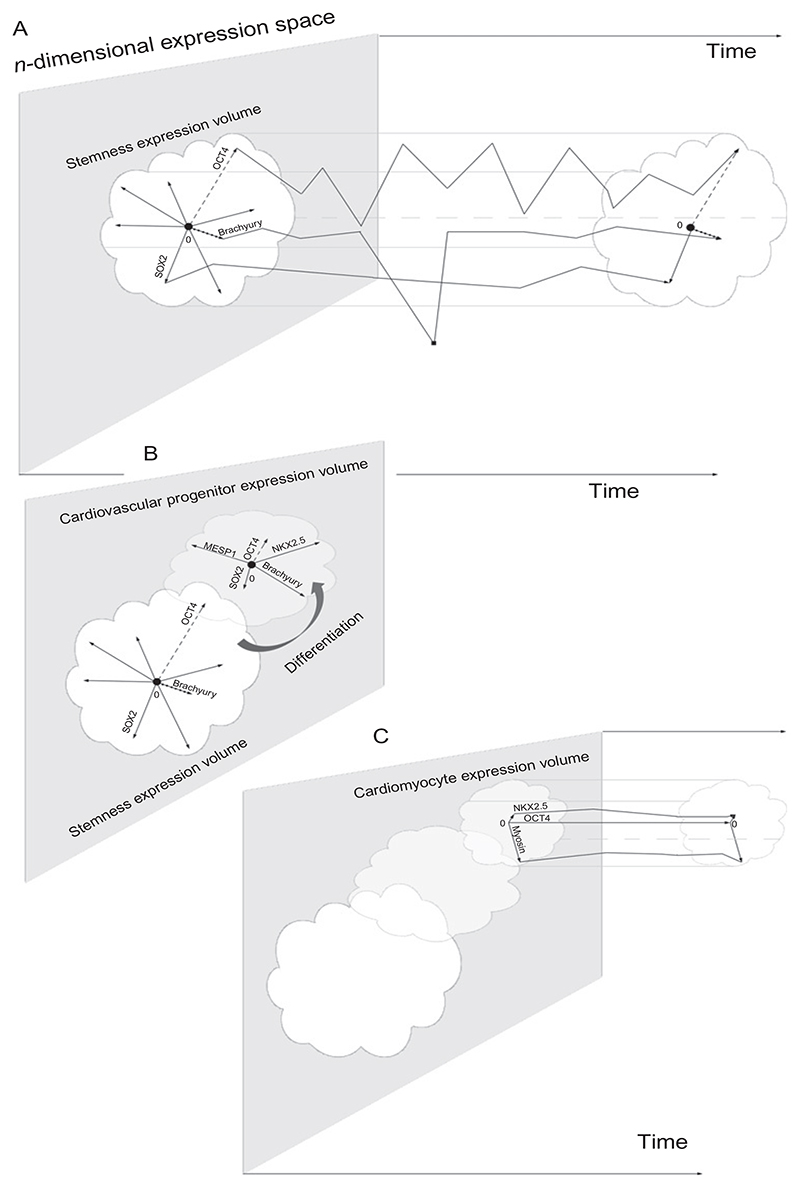
Taken together, this substantial work from many research institutions gave rise to several, however, only partially overlapping models how cardiac cells descend from primitive mesoderm and cardiovascular progenitor cells, respectively. In Fig. 7.1, we try to combine these different models of descent in one illustration. However, we state at the same time that presumably, this static view must be replaced by a more dynamic model displaying constant fluctuation of gene expression, which would better reflect the high plasticity and differentiation potential of all stem and progenitor cells. Thus, this picture may be still far away from reality. Most importantly, it is still unclear if a single type of cardiovascular progenitor cell stands at the root of a pedigree and if a hierarchy composed of different developmental stages of cardiovascular progenitor cells in the heart exists.
Figure 7.1.
Descent of cardiovascular progenitor cells from the primitive mesoderm. Molecules used to identify cell types (circles) are indicated. Rectangles, somatic cardiac cell types. Question marks, marks descent not directly supported by data or simply unknown. Black arrows in the lower part of the cartoon indicate differentiation of progenitor cells to cardiomyocytes, whereas gray lines indicate differentiation to smooth muscle and endothelial cells, respectively. The dashed line reflects only minor contribution to somatic cell type. (#) indicates the two cell types which differentiate to cells of the cardiac lineages only after reprogramming with 5-azacytidine or coculture with cardiomyocytes as feeder layers. Stemness markers: OCT4, Octamer-binding transcription factor 4; Nanog, homeodomain transcription factor; SOX2, sex determining region Y-box 2 transcription factor; mesodermal markers: T, Brachyury; GSC, goosecoid; MSGN, Mesogenin 1, a basic helix–loop–helix transcription factor; FLK1, a receptor for the Vascular Endothelial Growth Factor; PDGFRα, Platelet-Derived Growth Factor Receptor alpha; Eomes, Eomesodermin, a maternal T-box transcription factor; MESP1, Mesoderm Posterior 1 homolog transcription factor; cKIT, stem cell growth factor receptor tyrosine kinase; SCA1, Stem cell antigen 1; Vimentin, a mesenchymal cell-specific type III intermediate filament protein; CD34, 45, and 31, cluster of differentiation antigens; ISL1, a LIM domain transcription factor; NKX2.5, a zinc finger transcription factor; Lin, a set of nine lineage markers of the hematopoietic lineages; P-glyop., P-glycoprotein, an ABC transmembrane transporter; Nestin, a type VI intermediate filament protein, expressed mainly in neuronal cells but also in some stem cells; KDR, Vascular Endothelial Growth Factor Receptor 2; TBX5, T-box transcription factor.
4. Regulation of Cardiogenesis in Cardiovascular Progenitor Cells
4.1. Transcriptional regulation of cardiogenesis
4.1.1. Transcriptional regulation of cardiomyogenesis in cardiovascular progenitor cells
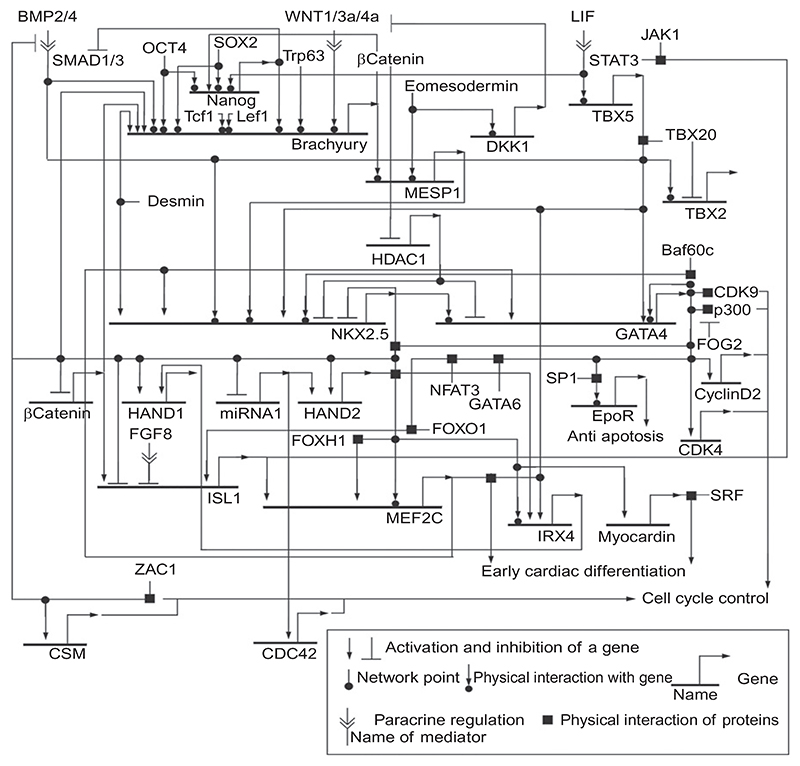
Currently, we do not have the knowledge about a continuous hierarchy of transcription factors regulating the expression of a cell in the primitive mesoderm in a way that it undergoes several defined developmental stages to finally become a cardiomyocyte. We have also very little data on the spatiotemporal interaction of the transcription factors involved in maintenance of the stem cell character and driving cardiomyogenesis in cardiovascular progenitor cells. Thus, we will focus first on two of the best studied transcription factors involved in early cardiomyogenesis the homeobox transcription factor NKX2.5, and the zinc-finger transcription factors GATA4. Both are essential in heart formation during embryogenesis, but neither can initiate cardiomyogenesis on its own in mammalian cardiovascular progenitor cells. Importantly, GATA and NK class proteins are coexpressed in many other tissues, and thus, interaction of these two transcription factors may be crucial for organogenesis. In the case of GATA4 and NKX2.5, it is very likely that they are also important regulators for late events during heart formation (Durocher et al., 1997). Beginning there, we shall try to extend the network of transcriptional regulation, first backward to the rather uncertain beginning in the primitive mesoderm, and second but more securely, toward committed and differentiated cardiomyocytes. Finally, we will try to construct an evidence-based network of physical and genetic interactions of transcription factors as a working model for further investigations.
4.1.2. NKX2.5
NK2 transcription factor related, locus 5, Nkx2.5, or cardiac-specific homeobox gene, Csx (Komuro and Izumo, 1993), codes for a member of the evolutionary highly conserved NK2 homeodomain transcription factor family (Kim and Nirenberg, 1989; Komuro and Izumo, 1993; Lints et al., 1993a,b) which are distantly related to the large Hox gene family. Nk2 genes, where NK stands for Nirenberg and Kim, the authors of the first paper describing these genes in D. melanogaster, are expressed in a tissue-specific manner at different times during mammalian embryogenesis (Harvey, 1996; Harvey et al., 2002). This occurrence suggests versatile roles in commitment and differentiation of cells and patterning of tissues and organs.
In the heart, NKX2.5 is first expressed in the cardiomyogenic progenitor cells during the formation of the lateral plate mesoderm in the late gastrula. NKX2.5 is temporally downregulated during differentiation of cardiomyocytes and then expressed at lower levels in the myocardium throughout the live of an organism. However, on the cellular level expression is likely to significantly vary between different cell types (Gittenberger-de Groot et al., 2007). NKX2.5 is a closely related homolog of Tinman, expressed in D. melanogaster (Bodmer, 1993; Bodmer et al., 1990) where it has an essential function in the development of the cardiac mesoderm (Grow and Krieg, 1998). In D. rerio, NKX2.5 seems to mark the earliest heart field and when ectopically expressed induces the initial but not late steps of cardiomyogenesis (Chen and Fishman, 1996). In mice, NKX2.5 expression starts at embryonic day 7.5 in the paired primordia arising from the splanchnic which builds, by midline fusion, the primitive heart tube (Kasahara et al., 1998). Most likely, NKX2.5 is expressed at low levels much earlier in cardiovascular progenitor cells because in embryoid bodies NKX2.5 expression can be detected on day 4 after initiation of in vitro differentiation (Hofner et al., 2007). Expression before the fusion of the primitive heart tube (Harvey, 1996; Moses et al., 2001) additionally suggests that it is expressed in still proliferating cardiovascular progenitor cells. This notion is heavily supported by data obtained from chicken (Schultheiss et al., 1997, 1995) and X. laevis (Sater and Jacobson, 1989; Sparrow et al., 2000), where cNkx2.5 and XNkx2.5 transcripts accumulate already in midgastrulation during formation of the primitive streak, and can be even detected in the pregastrulation epiblast of chicken embryos (Yatskievych et al., 1997). Nkx2.5-null mutation results in death before embryonic day 11 (Lyons et al., 1995; Tanaka et al., 1999a) and dominant negative mutants of Nkx2.5 negatively affected cardiomyogenesis (Jamali et al., 2001b). Although a rhythmically contracting primitive heart tube is formed in Nkx2.5-null mouse embryos, looping, chamber formation, and trabeculation are severely affected. The fact that NKX2.3 and NKX2.7 can step in for NKX2.5 during early cardiomyogenesis (Fu et al., 1998; Grow and Krieg, 1998; Tu et al., 2009) may explain this results. Otherwise we would have to conclude that NKX2.5 is not essential for cardiomyogenesis in at least a certain subpopulation of cells forming the paired heart primordia. In opposition to this interpretation stands the Nkx2.5 haploinsufficiency, the negative effects of various NKX2.5 point mutations in humans (Benson et al., 1999; Schott et al., 1998; Watanabe et al., 2002), and the fact that NKX2.5 is essential for the commitment of mesodermal cells into the cardiomyogenic lineage in teratocarcinoma cell-derived embryoid bodies (Jamali et al., 2001b).
Recent experiments designed to generate induced cardiovascular progenitor cells from fibroblasts suggest that NKX2.5 is indeed one of the key players for cells to become cardiomyocytes (Efe et al., 2011; Ieda et al., 2010; Takeuchi and Bruneau, 2009). Bruneau and colleagues and Srivastava and colleagues, respectively, demonstrated that ectopic expression of NKX2.5, GATA4, and TBX5 in neonatal murine cardiac fibroblasts is sufficient to convert them to cardiomyocytes. Ding and colleagues demonstrated that ectopic expression of the stemness transcription factors OCT4, KLF4, SOX2, and c-MYC for 4 days in combination with LIF withdrawal, and administration of BMP4, which induces NKX2.5 expression (Jamali et al., 2001a), is sufficient to generate significant numbers of cardiomyocytes from mouse embryonic fibroblasts. The latter demonstrated that this regime led to the upregulation of GATA4, MESP1, and ISL1 expression. Taken together, these data suggest that, in sharp contrast to Tinman in flies, none of the transcription factors identified so far in mammals is sufficient to induce cardiomyogenesis, and that most likely different sets of three or more myocardial transcription factors only suffice to initiate the myocardial differentiation program.
Most importantly, NKX2.5 has also a negative role in early cardiomyogenesis as it downregulates myocardial genes at a very early stage of cardiac induction (Prall et al., 2007). Later in cardiomyogenesis, NKX2.5 can also have detrimental effects as ectopic overexpression of NKX2.5 in mice suppresses the formation of the sinoatrial node (Espinoza-Lewis et al., 2011). Our own results demonstrate that murine cardiovascular progenitor cells temporally downregulate NKX2.5 expression when induced to differentiate in aggregates in vitro (submitted for publication). This phenomenon may be required for proper differentiation of cardiovascular progenitor cells along the cardiac lineage. In contrast, higher expression levels in cardiovascular progenitor cells perhaps maintain the stem cell state and at the same time prevent cardiovascular progenitor cells to escape the cardiomyogenic lineage. Based on the observation of a higher β-galactosidase activity in hearts of homozygous mice with a LacZ gene knocked into the Nkx2.5 locus, replacing the entire coding sequence, than in heterozygous mice (Tanaka et al., 1999b), a negative autoregulatory loop has been suggested for this gene. This model could explain the inhibitory roles and the temporal downregulation of NKX2.5 during cardiomyogenesis in cardiovascular progenitor cells.
At the end of this section, however, there is need for a note of caution; NKX2.5 is also expressed in the anterior endoderm and ectoderm (Schultheiss et al., 1995) and not all cells expressing NKX2.5 in the mesodermal compartment giving rise to the heart become cardiomyocytes (Jacobson and Sater, 1988). Thus, as long as germ layer-specific knockouts of the Nkx2.5, Nkx2.3, and other family members do not exist, we cannot exclude that NKX2.5 expression in these compartments contributes to cardiomyogenesis and cardiovascular progenitor cell homeostasis in vivo, by an unknown paracrine pathway. Finally, its diversified expression pattern definitely prevents NKX2.5 to be used as a specific marker for cardiovascular progenitor cells.
4.1.3. GATA4
The second key player regulating cardiomyogenesis in cardiovascular progenitor cells seems to be GATA4, a member of a zinc-finger transcription factor family with rather diverse functions. The GATA family can be divided into two groups: GATA1–3 are important regulators of hematopoietic stem cell differentiation and diversification and GATA4–6 are expressed in the presumptive cardiac mesoderm (Molkentin, 2000), and in the developing heart (Charron and Nemer, 1999) but are also important for the development of endodermal cell lineages (Afouda et al., 2005). GATA5 seemed to be restricted to the endocardium (Nemer and Nemer, 2002) while GATA4 and GATA6 are expressed in the developing and postnatal myocardium, physically interact with each other (Charron et al., 1999), and transactivate a variety of myocardium-specific genes. Vice versa, expression of several cardiac genes was significantly downregulated in cells lacking GATA4 or GATA6, indicating that these factors are required for the maintenance of the specific genetic program in cardiovascular progenitor cells (Charron et al., 1999). Recently, it has been demonstrated that also GATA4 and GATA5 cooperatively regulate the proliferation of cardiovascular progenitor cells in mouse hearts (Singh et al., 2010). From these data, it may be concluded that GATA4, GATA5, and GATA6 regulate myocardial gene expression by a cooperative pairwise interaction of these transcription factors at the very beginning of cardiomyogenesis in cardiovascular progenitor cells in a rather complex spatiotemporal manner.
GATA4 interacts physically with NKX2.5 (Durocher et al., 1997; Lee et al., 1998), and both factors together but neither alone can induce cardiomyogenesis in mammalian mesoderm (Harvey, 1996). However, the interchangeability of GATA4 and GATA 6 is limited because GATA6 cannot substitute for GATA4 for interaction with NKX2.5 (Durocher et al., 1997) and thus most likely does not induce cardiomyogenesis in the absence of GATA4.
Knockout of the Gata4 gene resulted in embryonic lethality between days E8.5 and 10.5 of embryonic development, but clearly after the formation of the primitive heart tube between days E7.0 and E8.5 (Kuo et al., 1997; Molkentin et al., 1997). Not much surprisingly, targeted deletion of Gata4 in embryonic stem cells disrupted the development of visceral endoderm in embryoid bodies (Soudais et al., 1995). Rescue of Gata4-null embryos by tetraploid aggregation or addition of wild-type extraembryonic endoderm demonstrated that arrest of heart tube formation at day E8 depends on the absence of extraembryonic visceral endoderm, where GATA4 fulfills an indispensable role (Narita et al., 1997; Watt et al., 2004). At the same time, it became evident that GATA4 seems not to influence significantly the gene expression in the myocardium, and its absence does not affect endocardial development and trabeculation of the heart but clearly affects the development of the proepicardium. In contrast, transcription of the Gata4 gene together with Baf60c, coding for a chromatin remodeling complex member (Lickert et al., 2004), proved sufficient to convert mouse posterior mesoderm, normally giving rise to the somites, and extraembryonic mesoderm of the amnion, into cardiovascular progenitor cells (Dixon et al., 2011; Takeuchi and Bruneau, 2009). Notably, in this case, BAF60c mediated the binding of GATA4 to other cardiac genes such as Nkx2.5 which in combination with Tbx5 facilitated the differentiation to cTnT positive cardiomyocytes. Likewise, GATA4 also forms a complex with the CDK9, which contributes to the increased expression of NKX2.5 in differentiating embryonic stem cell-derived cardiovascular progenitor cells (Kaichi et al., 2011). These last two facts suggest that at least in some cells GATA4 acts upstream of NKX2.5.
From the observations that GATA4 alone does not induce cardiomyogenesis, and that its expression is essential in cardiovascular progenitor cells in mammals, we may conclude that only if NKX2.5 together with GATA4 is expressed in a primitive mesodermal cell, the cardiogenic program commences.
4.1.4. Events upstream of NKX2.5 and GATA4
Assuming a hierarchically structured network of transcription factors instructing cells in the primitive mesoderm to become cardiomyocytes, we searched for the earliest event in this hypothetical path. The earliest event we could get hold of was the physical interaction between the transformation-related protein 63, Trp63, a member of the Trp53 transcription factor gene family and the promoter of the Brachyury gene (Cho et al., 2010). More specifically, the isoform DeltaNp63 binds to and activates transcription of the gene. Brachyury is expressed in the primitive mesoderm, and particular in the node, notochord, and posterior mesodermal tissue (Beddington et al., 1992; Tam and Beddington, 1992), which is a major source of instructive signals influencing heart development.
From ChIP-Seq data and studies on transcription factor networks in embryonic stem cells, it became evident that OCT4 and SOX2 are involved in the regulation of Nanog, maintaining self-renewal, and Brachyury, that drives differentiation along the mesodermal lineage into progenitor cells (Thomson et al., 2011).
Thus, already at the time when OCT4 and SOX2 are expressed in stem cells, they actively contribute to lineage decisions after downregulation of Nanog by binding to the Brachyury gene. A regulatory loop wherein Nanog influences Brachyury expression, and vice versa (Sarkar et al., 2011; Suzuki et al., 2006), may well contribute to balance self-renewal and differentiation of cardiovascular progenitor cell. Brachyury is also a direct target of the WNT signaling and βCatenin-activated transcription factors LEF1 and TCF1 activate the Brachyury gene (Yamaguchi et al., 1999). Most surprisingly, the type III intermediate filament protein Desmin contributes to increased Brachyury and NKX2.5 expression when overexpressed in differentiating embryonic stem cells (Hofner et al., 2007; Höllrigl et al., 2007). Since Desmin expression is activated also by MEF2C (Kuisk et al., 1996), the latter one may well contribute to a positive feedback loop by reactivating Brachyury and NKX2.5 expression in the cardiovascular progenitor cell pool, to attenuate differentiation, and to maintain the self-renewal potential.
Brachyury, on his part, binds to and activates Mesp1, and MESP1 is among the earliest proteins expressed in cardiovascular progenitor cells (David et al., 2011). MESP1 and MESP2 are bHLH transcription factors (Bondue and Blanpain, 2010), both expressed in the primitive mesoderm and in cardiovascular progenitor cells (Saga et al., 1996, 1999). Knockout of Mesp1 resulted in severe heart defects and knockout of both genes resulted in death around day E9.5 and the lack of anterior mesoderm including the heart primordia (Saga et al., 2000). MESP1 has been suggested as the key player in vertebrate cardiomyogenesis in an excellent review by Bondue and Blanpain (2010). An alternative paracrine activation of MESP1 in cardiovascular progenitor cells may be mediated by increased OCT4 expression and activation of Sox17 (Stefanovic et al., 2009).
Probably, simultaneously to Brachyury, Eomesodermin, Eomes, another transcription-box transcription factor, also binds to and transactivates Mesp1 (Costello et al., 2011). MESP1, in turn, directly induces Dickkopf 1, DKK1 expression (David et al., 2008), that negatively affects canonical WNT signaling, and thus promotes cardiomyogenesis in cardiovascular progenitor cells. Finally, and most importantly, MESP1 directly activates Nkx2.5 and many other myocardial transcription factors (Bondue et al., 2008) that, in turn, additionally activates transcription of GATA4 and MEF2C.
The expression of the Nkx2.5 gene in cardiovascular progenitor cells is also regulated by SMAD transcription factors and thus by BMP2- and BMP4- induced signal transduction (Liberatore et al., 2002). The proximal promoter of the Nkx2.5 gene contains three consensus SMAD binding sites that confer expression in the cardiac crescent on E7.25. A detailed investigation on the function of different 3′ untranslated regions of the Nkx2.5 gene revealed very complex regional expression patterns (Tanaka et al., 1999b) suggesting the involvement of many transcription factors in the regulation of Nkx2.5 expression. One of these transcription factors is TBX5 that induces Nkx2.5 and accelerates cardiomyogenesis in teratocarcinoma cell-derived embryoid bodies (Hiroi et al., 2001). However, apart from a single paper demonstrating the activation of the Tbx5 gene by STAT3 (Snyder et al., 2010), we do not know how TBX5 fits into the hierarchy of transcription factors described above. It seems still unclear whether TBX5 is upstream or downstream of NKX2.5 and GATA4.
Nonetheless, TBX5, another prominent member of the T-box transcription factor family, plays an important role in cardiogenesis. It is expressed in the cardiac mesoderm and the myocardium of the fetal and adult murine and human heart (Horb and Thomsen, 1999). Later in cardiac development, TBX5 expression becomes rapidly refined and in adulthood only low levels of TBX5 can be detected in both ventricles. Absence of TBX5 caused decreased GATA4 and NKX2.5 expression and affects primarily the ventricle-specific genes Mlc2v, Irx4, and Hey2 (Hatcher et al., 2000). Most importantly, TBX5, allowed complete transdifferentiation of fibroblasts into cardiomyocytes in the presence of GATA4 and BAF60c (Takeuchi and Bruneau, 2009). The factor is necessary to control the length of the embryonic cardiac cell cycle, with depletion leading to cardiac cell cycle arrest in late G1- or early S-phase. Further, this leads to a decrease in cardiac cell number, an alteration in the timing of the cardiac differentiation program, defects in cardiac sarcomere formation, and ultimately, to cardiac programmed cell death (Goetz and Conlon, 2007; Goetz et al., 2006). From these data, we may conclude that TBX5 is an inevitable component of the transcriptional network regulating self-renewal in cardiovascular progenitor cells. Inhibition of TBX5 in Xenopus embryos leads to hypoplasia of cardiac tissues and decreased Nkx2.5 mRNA levels (Horb and Thomsen, 1999) further supporting that Nkx2.5 is a target gene of TBX5.
Taken together these data suggest that TBX5 positions upstream of NKX2.5 and GATA4 in the transcriptional network regulating self-renewal and cardiomyogenesis in cardiovascular progenitor cells. TBX5 physically interacts with TBX20, which, when knocked out, displays a similar phenotype as TBX5 (Brown et al., 2005), and TBX20 directly interacts with NKX2.5, GATA4, and GATA5 in regulation of the gene expression in the developing heart (Stennard et al., 2003). TBX20 represses the expression of TBX2 during cardiomyogenesis (Cai et al., 2005). Tbx2, apart from Brachyury, is so far the only T-box gene which is directly activated by SMADs and thus a target of BMP2/4 signaling in cardiovascular progenitor cells (Shirai et al., 2009; Yamada et al., 2000). Notably, TBX2 may also act antagonistically in cardiomyogenesis (Christoffels et al., 2004), thus constituting a negative feedback loop composed of BMPs and TBX2. All these observations are very similar to effects noticed for NK proteins and suggest a highly redundant function of T-box transcription factors in cardiomyogenesis. However, not all T-box transcription factors can replace or step in for TBX5. For example, TBX1 seems to play only a subordinate role in the diversification of differentiating cells in the second heart field (Liao et al., 2008) and most likely is not directly involved in self-renewal and fate decisions in cardiovascular progenitor cells.
Most of these data hint at a role of TBX5 upstream of NKX2.5 and GATA4. However, the recent generation of induced cardiomyocytes by ectopic expression of GATA4 together with BAF60c (Lickert et al., 2004) suggests that GATA4 together with BAF60c is sufficient to induce the cardiomyogenic program in cardiovascular progenitor cells, and TBX5 only comes then to maintain and further develop differentiation along the myocardial lineage. Last but not least, NKX2.5 expression is also influenced by methylation and acetylation. During the early stage of differentiation, the Nkx2.5 promoter was activated by acetylation of histones H3 and H4 which was accompanied by a significant reduction in HDAC1 expression. Additional suppression of HDAC1 activity stimulated cardiac differentiation accompanied by increased expression of cardiac-specific genes and cell cycle arrest. Accordingly, overexpression of HDAC1 induced the downregulation of GATA4 and NKX2.5 in cardiomyocytes (Liu et al., 2009). Presently, very little is known about epigenetic regulation in cardiovascular progenitor cells (Eilertsen et al., 2008); nonetheless, the fact that cardiovascular progenitor cells have a significantly different and restricted developmental potential as compared to pluripotent embryonic stem cells and cells of the primitive ectoderm clearly demonstrates an epigenetic component that defines the status of cardiovascular progenitor cells.
4.1.5. Target genes and interaction partners of NKX2.5 and GATA4
Physical interaction of GATA4 with NKX2.5 and a positive feedback loop involving both transcription factors makes it very difficult to separate downstream genes which are only targeted by one of these factors. The Nkx2.5 gene contains regulatory sequences in the enhancer which are bound and activated by GATA4 (Lien et al., 1999; Reecy et al., 1999). Downregulation of Nkx2.5 using antisense RNA resulted in a decreased GATA4 expression in populations of human fetal cardiomyocytes (Riazi et al., 2009) corroborating the positive feedback loop between the Nkx2.5 and Gata4 genes (Brown et al., 2004; Jiang et al., 1999; Molkentin et al., 2000; Peterkin et al., 2003). However, GATA factors might also antagonize transcription of Nkx2.5 (Jiang et al., 1999), and the role of NKX2.5 may not always depend on GATA4 because Nkx2.5 also directly activates the Gata6 gene by binding to a cardio-specific enhancer element in its 3′ untranslated region (Molkentin et al., 2000). To additionally complicate the situation, GATA and NK proteins seem to have redundant functions. In D. rerio, for example, Nkx2.7 and NKX2.5 are required for cardiac morphogenesis (Tu et al., 2009). So far, from these data, we cannot solve the problem which transcription factor comes first in a sequence of events committing primitive mesodermal cells to the cardiomyogenic lineage, NKX2.5 or GATA4.
Abnormal expression of transcription factor genes Mef2C and HAND1, among several others, in Nkx2.5-null mouse embryos (Lyons et al., 1995; Tanaka et al., 1999a) suggests them as downstream targets of NKX2.5 of transcriptional regulators activating the cardiomyogenic program in cardiovascular progenitor cells.
Likewise, βCatenin expression was upregulated in the presence of anti-Nkx2.5 miRNA and WNT3 and WNT3a signaling caused the downregulation of HDAC1 which suppresses NKX2.5 and GATA4 expression in cardiovascular progenitor cells (Liu et al., 2009). Simultaneously, upregulation of βCatenin could very well lead to the upregulation of Brachyury in cardiovascular progenitor cells (Yamaguchi et al., 1999). On top of that NKX2.5 also directly suppresses βCatenin expression (Riazi et al., 2009). Altogether, these data suggest that NKX2.5 also influences early cardiomyogenesis by a negative feedback loop involving WNT signaling.
Another target of NKX2.5 and candidate factor involved in transcriptional regulation of cardiomyogenesis in cardiovascular progenitor cells is the Iroquois Homeobox gene 4, Irx4, a transcription factor expressed in the ventricular myocardium on day E7.5 (Bao et al., 1999). It is downregulated in Nkx2.5-null mice and its expression cannot be maintained in Hand2-null mice (Bruneau et al., 2000). In Nkx2.5–Hand2 double-null mice, IRX4 expression was completely abolished (Bruneau et al., 2001; Yamagishi et al., 2001), and we may infer that IRX4 plays a role in late progenitor differentiation downstream of NKX2.5 and HAND2.
The heart- and neural crest derivatives-expressed proteins 1 and 2, HAND1 and HAND2, also named eHAND and dHAND, are evolutionary conserved bHLH, transcription factors (Firulli, 2003) that play crucial and partially redundant roles in cardiac growth, morphogenesis, and gene expression (Hollenberg et al., 1995; Srivastava et al., 1995; Thattaliyath et al., 2002b). In mice, HAND2 is expressed throughout the linear heart tube but HAND1 is expressed only in segments of the linear heart tube giving rise to the conotruncus and left ventricle (Biben and Harvey, 1997; Thomas et al., 1998). D. rerio most likely has only a single Hand gene, closely related to HAND2 (Yelon et al., 2000). Mutation of this Hand gene results in a dramatic reduction in the number of ventricular cardiovascular progenitor cells. HAND1 seems not to be essential for early cardiac development (McFadden et al., 2005), but Hand2-null mice die around day E10.5 from right ventricular hypoplasia and vascular malformations (Srivastava et al., 1997). Expression in the linear heart tube around day E8.0 both in the first and second heart field (Tsuchihashi et al., 2011) suggests that HAND transcription factors are already active in cardiovascular progenitor cells. Later in heart development, they contribute to the proper morphogenesis of the four-chambered heart (Tsuchihashi et al., 2011), and HAND2 directly regulates transcription of the Atrial Natriuretic Factor, Anf, gene (Thattaliyath et al., 2002a). Because NKX2.5 mutants failed to activate HAND2 expression in the early precardiac mesoderm where both factors are usually coexpressed (Yamagishi et al., 2001), we may conclude that Hand2 similar to Hand1 (Tanaka et al., 1999a) is a target gene of NKX2.5.
NKX2.5 also directly binds to the enhancer region of the micro-RNAs 1, miR1 gene and represses expression of miR1 in neonatal cardiomyocytes (Qian et al., 2011). miRNA1 on its part binds and activates expression of the Cdc42 gene (Qian et al., 2011). CDC42 functions as a potent cell cycle regulator in cardiac cells (Maillet et al., 2009), influences cell fate decisions (Brown et al., 2006), and promotes stability of cardiomyocytes (Nagai et al., 2003). From the role of CDC42 in other stem cells, such as hair follicle (Wu et al., 2006b) and neural progenitor cells (Cappello et al., 2006), we may infer that it fulfills similar functions in cardiovascular progenitor cells. Thus, expression of NKX2.5 during differentiation of cardiovascular progenitor cells does not only promote differentiation to cardiomyocytes but also contributes to attenuated cell proliferation.
One of the best characterized downstream targets of NKX2.5 is the Myocyte Enhancer Factor 2C gene, Mef2C, (Potthoff and Olson, 2007; Tanaka et al., 1999a). The four members of the MEF2 family of MADS box transcription factors play critical roles in all muscle cells (Molkentin and Olson, 1996), and neuronal cells (Leifer et al., 1994) and have been shown to be important for various processes involved in differentiation, survival, and apoptosis. In D. melanogaster, the single dMef2 gene is indispensable for cardiac, skeletal, and smooth muscle development (Lilly et al., 1995). Mice lacking Mef2C (Martin et al., 1993) but not other family members were deficient in cardiac looping and died around day E10 (Lin et al., 1997). MEF2C seems to be the first transcription factor in the sequence of events leading to differentiated cardiomyocytes, as it no longer directly contributes to the maintenance and commitment of cardiovascular progenitor cells because heart development commences in Mef2C-null embryos.
Although Mef2C is clearly a target of NKX2.5, it also upregulates NKX2.5 expression and initiates cardiomyogenesis in teratocarcinoma cells (Skerjanc et al., 1998). NKX2.5 and MEF2C interact physically and genetically (Vincentz et al., 2008). Nkx2.5–Mef2C double-null mouse embryos develop only atrial parts of the heart and cells express only markers of the second heart field. Thus, MEF2C and NKX2.5 obligatory synergize in regulation of primary heart field progenitors to ventricular cardiomyocytes. Similar to NKX2.5, MEF2C expression resulted in the upregulation of Brachyury, BMP4, NKX2.5, and GATA4. Activation of a very early player in the chain of events such as Brachyury by the later factor MEF2C suggests that already differentiating cardiomyocytes retain the potential to the cardiovascular progenitor cell-specific program. This may be interpreted as an increased degree of plasticity which allows differentiating and perhaps fully differentiated cardiomyocytes to reenter the proliferative state, and to switch on the “embryonic” transcriptional program typical for cardiovascular progenitor cells. Thus, the “bandwidth” between a cardiovascular progenitor cell and a functional cardiac cell of any type seems to be rather broad.
Additionally, Mef2C is a direct target of FOXH1 which physically and functionally interacts with NKX2.5. FOXH1 mediates a strong SMAD-dependent activation of a TGFβ response element in the Mef2C gene (von Both et al., 2004); Mef2C is also a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development (Dodou et al., 2004). Most importantly, MEF2C physically interacts with TBX5 and in this particular case is indispensable for very early heart development (Ghosh et al., 2009). Thus, it appears that most, if not all, of the early transcription factors in cardiovascular progenitor cells converge in activating the MEF2C gene, which seems to be a prerequisite for the proper building of the contractile apparatus in cardiomyocytes. Just as one example, GATA4 and MEF2C transcription factors control the tissue-specific expression of the cell–cell interaction mediator αTCatenin in cardiomyocytes (Vanpoucke et al., 2004).
NKX2.5 seems to orchestrate the transition between cardiovascular progenitor cell proliferation, cardiac induction, and outflow tract morphogenesis in the second heart field by a negative feedback loop and repression of BMP2/SMAD1 signaling (Prall et al., 2007). Since TGFβ/BMP signaling affects self-renewal of stem cells via a Brachyury- and STAT3-mediated activation of the Nanog gene (Suzuki et al., 2006), and Nanog blocks BMP-induced mesoderm differentiation by binding to and sequestering SMAD1, it seems very likely that NKX2.5 is involved in maintaining self-renewal in cardiovascular progenitor cells, and thus in maintaining the cardiovascular progenitor cell pool in the heart.
As a downstream event following GATA4 expression, the Erythropoietin Receptor gene, EpoR was upregulated in cardiomyocytes. Similar to the situation in erythropoietic cells, where GATA1 together with specific protein 1, SP1, another zinc-finger transcription factor (Rotheneder et al., 1999), activates EpoR (Youssoufian et al., 1993), GATA4 requires SP1 for the upregulation Erythropoietin and to protect cardiomyocytes from apoptosis (Salisch et al., 2011). GATA4 is a direct transcriptional activator of CyclinD2 and Cdk4 (Rojas et al., 2008) supporting a model in which GATA4 controls cardiomyocyte proliferation by coordinately regulating cell cycle genes. GATA4 interacts not only with GATA6 but also with nuclear factor of activated T cell, NFAT3 (Charron et al., 1999), which seems to be important for proper development of cardiac valves and the septum (de la Pompa et al., 1998; Ranger et al., 1998). Finally, GATA4 also interacts and is acetylated by the histone acetyltransferase E1A binding protein, p300, which promotes transcriptional active configurations at promoter and enhancer DNA sequences. This interaction of GATA4 with p300 is inhibited by the protein Friend of GATA 2 (FOG-2; Hirai et al., 2004).
NKX2.5 negatively regulates Isl1 (Prall et al., 2007) which may explain why NKX2.5 positive progenitor cells in the first heart field express TBX5 but not or very little ISL1. Notably, this observation does not fit to the expression of both genes in progenitor cells of the second heart field (Musunuru et al., 2010). In contrast, GATA4 together with the Forkhead box protein O1, FOXO1, activates the Isl1 gene (Kang et al., 2009; Kappen and Salbaum, 2009).
ISL1 is abundant in many different cell types and organs of all three germ layers during embryogenesis and in organs of adult mammals. Nonetheless, ISL1 expression also marks a population of cardiovascular progenitor cells which makes a substantial contribution to the right ventricle, both atria and the outflow tract of the embryonic heart (Cai et al., 2003; Zhou et al., 2008b). At the same time, ISL1 has been described to be expressed both in the first and second heart field (Bu et al., 2009; Ma et al., 2008) and in cardiovascular progenitor cells of embryoid bodies between day 3 and 4.5 and at day 6 in up to 6% of the cells (Moretti et al., 2010). The timing of the expression suggests that ISL1 is involved in very early fate decisions within the cardiomyogenic lineage together with NKX2.5 and other transcription factors, but most likely not in all cardiovascular progenitor cells because at that time the percentage of cells expressing NKX2.5 is much higher (Terami et al., 2004). Similarly, only a subset of progenitor cells in murine hearts expressing ISL1 during embryogenesis become cardiomyocytes (Laugwitz et al., 2005).
ISL1 forms a complex with the Janus Tyrosine Kinase, JAK-1, and the signal transducer and activator of transcription 3, STAT3, but not with STAT1 and STAT5, suggesting specificity of the formation of this ternary complex. ISL1 increases the autophosphorylation kinase activity of JAK1, enhances the DNA binding activity and target gene expression of STAT3 in mouse motor neuron cells (Hao et al., 2005), and increase the STAT3 interaction with p300. Since neurons may be generated from stem cells residing in the subventricular zone in the dentate gyros of the hippocampus, and in the neural tube after injury, we may infer that LIF signaling, that is also active in cardiovascular progenitor cells, may contribute to balance self-renewal and differentiation.
Finally, during anterior–posterior heart patterning, ILS1 is downregulated by retinoic acid via the FGF8 pathway (Sirbu et al., 2008) and is directly regulated by βCatenin (Lin et al., 2007) and thus WNT signaling (Qyang et al., 2007). Similar to NKX2.5, ISL1 directly activates Mef2C and promotes cardiomyogenesis in cardiovascular progenitor cells (Dodou et al., 2004).
A quite different cofactor of NKX2.5 is the zinc-finger transcription factor 1, ZAC1 that is encoded by a maternally imprinted gene. It is strongly expressed in the heart from cardiac crescent stages and shows a chamber-restricted pattern in the looping heart. ZAC1 and NKX2.5 physically associate through zinc fingers 5 and 6 in ZAC1, and the homeodomain in NKX2.5 (Yuasa et al., 2010). Knockout of the Zac1 locus showed only partial lethality and atrial and ventricular septal defects. This phenotype indicates that the major role of ZAC1 lies in progressed heart development and not in specification of cardiovascular progenitor cells. However, ZAC1 is known to regulate many imprinted genes involved in cell proliferation (Varrault et al., 2006). NKX2.5 also transactivates the cardiac-specific isoform of Mov10l1, CSM, a putative RNA helicase (Ueyama et al., 2003b), which may be involved in the maintenance of self-renewal and/or differentiation of cardiovascular progenitor cells. Possibly, NKX2.5 via CSM and ZAC1 plays a role in the maintenance of stem cell self-renewal and temporal induction of reprogramming in the myocardium, because ZAC1 alters the expression of imprinted genes such as Igf2 and directly regulates the Igf2/H19 locus through binding of a shared enhancer. This model fits to fact that IGF2 plays a significant role in cardiovascular progenitor cell proliferation (D’Amario et al., 2011).
The SAP family transcription factor, Myocardin, is a cofactor of the serum response factor (SRF) in the transcriptional program regulating cardiac and smooth muscle cell differentiation, and its gene is transactivated by NKX2.5 (Ueyama et al., 2003a). Physically interacting with SRF (Niu et al., 2007, 2005), myocardin plays an important part in cardiac development. The human and murine Myocardin genes are expressed in vascular and visceral smooth muscle cells and in the heart. Forced expression of Myocardin activates expression of the SM22 alpha, smooth muscle alpha-Actin, and Calponin-h1 in undifferentiated mouse ES cells (Du et al., 2003). SRF also regulates miR1 which targets HAND2 during cardiogenesis (Zhao et al., 2005). These data demonstrate that Myocardin plays an important role in the SRF-dependent transcriptional program that regulates smooth muscle cell and cardiomyocyte development and differentiation contributing to the development of the mammalian heart. Most recently, a different point of view on the transcriptional regulation of cardiomyogenesis has been suggested which adds MEF2A, and SRF to the key players GATA4 and NKX2.5 and sets SRF at the core of the network regulating cardiomyogenesis (Schlesinger et al., 2011).
Assuming that cardiovascular progenitor cells can give rise to all somatic cell types composing the myocardium, endocardium, epicardium, outflow tract, and the conduction system, and that no inescapably committed subpopulations of cardiovascular progenitor cells exist in these compartments, we come here to rather arbitrarily set end and summarize the findings described above in a cartoon showing all possible interactions of transcription factors balancing self-renewal and differentiation in cardiovascular progenitor cells (Fig. 7.2).
Figure 7.2.
Model of a network of the genetic and physical interactions of transcription factors in cardiovascular progenitor cells. This model was inferred from data obtained in vivo at the time when cardiovascular progenitor cells exist and in vitro in comparable model systems such as embryonic stem cells derived embryoid bodies. Notably, this model can only be proven or rejected when stable self-renewing and differentiating cardiovascular progenitor cell lines exist. The acronyms used for genes and proteins are the same as described in Section 4.1.
4.2. Role of micro-RNAs in cardiogenesis
Micro-RNAs or miRNAs have been discovered not long ago and were described to play critical roles in numerous biological processes, including development, cell function, and disease. Among those, also the complex transcriptional network underlying initiation and direction of cardiovascular development was shown to require these small RNAs, as some of them were specifically expressed in cardiac cell types, during cardiomyocyte differentiation and vascularization.
miRNAs are non-coding single-stranded ribonucleic acid molecules that are responsible for the fine-tuning of gene expression. They act through posttranscriptional gene silencing and inhibit the action of their target genes through messenger RNA degradation or translational repression (Bartel, 2004). Far more than 500 miRNAs are encoded in inter- and intragenic regions of the genome (Chaudhuri and Chatterjee, 2007). Briefly, the biogenesis of mature miRNAs initiates with the transcription of the primary miRNA, through DNA polymerase II and III. Still in the nucleus, a protein complex comprising RNase III, Drosha, and a double strand RNA-binding protein, DGCR8, named Pasha in D. melanogaster, then erases the 7-methylguanosin-cap structure and the polyA tail of the primary miRNA to form a precursor miRNA (Han et al., 2004). After nuclear exportation through exportin-5 together with the cofactor Ran-GTP (Zeng and Cullen, 2004), the precursor miRNA is further modified in the cytoplasm. The RNase III, Dicer, cuts the precursor miRNAs into smaller pieces of 17–24 nucleotides, unwinds the RNA, and forms mature single-stranded miRNA molecules (Dykxhoorn et al., 2003). These fragments are then integrated into a ribonucleoprotein complex, miRNP, or RISC for final functionality. The RISC-miRNA combination degrades highly complementary target mRNAs by endonucleatic activity after base pairing. If complementarity is beyond a certain threshold, the complex only binds in the untranslated region of the mRNA and there-with inhibits its translation.
Earliest evidence of the necessity of the miRNA machinery yet in embryonic stem cell maintenance and development demonstrates its ubiquitary importance (Marson et al., 2008). Through interaction with OCT4, Nanog, and SOX2, miRNAs regulate cell cycle genes, pluripotency, early embryonic development, and the differentiation potential of cells (Card et al., 2008; Kanellopoulou et al., 2005). However, by reason of their destructiveness, the expression of miRNAs is highly regulated and cell type specific. Possibly, each one of the detected miRNAs acts on more than 100 different downstream targets in numerous pathways. It is only the combination of synergistic performing miRNAs that enables potent and complex transcriptional regulation as is needed the most in precisely controlled processes such as cardiogenesis. Numerous studies and experiments in recent years confirmed the necessity of these small molecules in early cardiomyogenesis and consequent cardiac cell identity (Hosoda et al., 2011).
The most abundant miRNAs in conjunction with cardiac signaling were shown to be miR-1 and miR-133. Beside the specific expression pattern of the miRNAs in cardiac muscle- and skeletal muscle cells during development, they were also approved to exist in adult human hearts (Chen et al., 2006). The fact that miR-1 and miR-133 are encoded side by side in the genome and that both are generated from a bicistronic precursor transcript proves their common function and cooperation in terms of cardiac development. The cardiac-specific expression of miR-1 and miR-133 is directly regulated by MEF2 and SRF (Zhao et al., 2005).
In early embryonic development, miR-1 and miR-133 first appear in precardiac mesoderm (Ivey et al., 2008). Before that state, both miRNAs are not present and undifferentiated embryonic stem cells do not express them. miR-1 and miR-133 were shown to have antagonistic effects on cardiomyogenesis, as the latter one shows inhibition of differentiation into cardiac mesoderm, whereas miR-1 promotes differentiation of embryonic stem cells toward a cardiac fate (Ivey et al., 2008).
miR-1 is the most abundant miRNA in cardiac cells and has been associated with cardiac hypertrophy (Care et al., 2007), heart development, cardiovascular progenitor cell differentiation, and arrhythmias (Chen et al., 2006). It effects and degrades multiple downstream targets such as Notch ligand Delta-like; it enhances gene activation of MEF2-dependent promoters through repression of HDAC4 (Chen et al., 2006; Kwon et al., 2005) and directly targets Hand2 (Zhao et al., 2005). Besides, NKX2.5 was shown to act via miR-1 on CDC42, and cell cycle progression (Qian et al., 2011) and miR-1 regulate cardiac electrophysiology by controlling the cardiac gap junction protein Connexin 43 and the potassium channel subunit Kir2.1 (Yang et al., 2007). Taken together, miR-1 and miR-133 reciprocally regulate cardiomyocyte proliferation, sarcomeric Actin organization, and cardiac conduction system function.
miR-208 is exclusively expressed in the heart after myocardial injury and is therefore considered as a biomarker for heart damage (Ji et al., 2009). It acts as a regulator of transcriptional repressors of slow muscle fiber genes (van Rooij et al., 2009) and was shown to directly inhibit CDK inhibitor protein p21. miR-499, like miR-208, is encoded in an intron of the MHC gene and similarly upregulated during cardiomyogenesis. In human cardiovascular progenitor cells, miR-499, miR-208, and miR-1 share similar expression patterns and target genes, such as GATA4, MEF2C, and αMHC suggesting that miR-499 plays an important role in cardiomyogenesis (Hosoda et al., 2011).
Another important pair of key regulators of cardiomyogenesis is miR-143 and miR-145. The miRNAs are expressed in cardiac progenitor and cardiac crescent cells and transcribed as a bicistronic element under the transcriptional control of SRF, Myocardin, the Notch intracellular domain N1ICD, and SMAD4. miR-143 and miR-145 regulate vascular smooth muscle proliferation and plasticity, most likely through dose- and time-dependent simulation of TGFβ1. The targeted factors include retinoic acid receptor and ligands, Krüppel-like factors KLF4 and 5, the myocardin-related transcription factor MRTFB, and ELK1 (Davis-Dusenbery et al., 2011).
The miRNA miR-138 was shown to be important in cardiac maturation and cardiac patterning during embryogenesis. It is expressed in the ventricular chambers, regulating the atrioventricular canal gene expression through retinoic acid signal repression (Morton et al., 2008). Further, BMP2/4 controls the miR-17–92 cluster and by this the development of the outflow tract, endothelial cell differentiation, and angiogenesis (Wang et al., 2010). Isl1 and TBX1 are directly repressed by miR-17 (Wang et al., 2010), and miR-206 and miR-29 repress Hdac4 (Winbanks et al., 2011).
Comparison of the various miRNA expression patterns in both heart development and disease suggests similar mechanisms in embryonic development and regeneration of the adult heart. Both myocardial infarction and embryonic heart development support the concept of revitalizing cardiac genes in the adult heart after damage. The great number of different players in the field of heart development and function again shows the importance of the exact regulation of factors involved in this process. The inhibiting mechanism of miRNAs provides an excellent tool for the accurate timing and dosage of the required molecules, but research on that part of cardiogenesis in cardiovascular progenitor cells obviously is in its infancy.
4.3. Autocrine and paracrine regulation of cardiomyogenesis
4.3.1. Influence of growth factors
During early embryogenesis, cardiac tissue develops from the anterior splanchnic mesoderm. In its vicinity, surrounding cell layers such as neuroectoderm and anterior endoderm positively and negatively regulate the initial cardiovascular progenitor cell fate decisions leading to cardiac differentiation of previously undetermined mesodermal cells (Noseda et al., 2011). Mainly, factors secreted from the adjacent endodermal tissue are necessary for the local induction of heart development (Andree et al., 1998; Nascone and Mercola, 1995; Solloway and Harvey, 2003; Wagner and Siddiqui, 2007). The guiding signals include positively acting growth factors such as BMP-2 (Behfar et al., 2002), FGF-8 (Alsan and Schultheiss, 2002), Crescent (Schneider and Mercola, 2001), and WNT11 (Eisenberg and Eisenberg, 1999), as well as negatively acting Chordin (Matsui et al., 2005), Noggin (Choi et al., 2007), Serrate (Rones et al., 2000), and WNT3 and WNT8 (Schneider and Mercola, 2001). Their major role in the process of cardiogenesis is the activation or regulation of a cardiac transcriptional cascade including NKX2.5, MEF2C, and GATA4 (Harvey, 2002).
Most likely, TGFβ and BMPs signaling pathways serve as the most important key regulators in early heart development (Monzen et al., 2002). During embryogenesis, the proximal endodermal tissue secretes BMPs that bind to receptors on the surface of the primitive mesodermal cells and activate SMAD signaling pathways to induce cardiogenesis (Choi et al., 2007; Monzen et al., 2002). BMP2 and 4 are capable of and necessary for cardiac induction in vivo during embryogenesis and in vitro (Andree et al., 1998; Barron et al., 2000; Ladd et al., 1998). SMAD1 and SMAD4 transcription factors directly bind to an evolutionary conserved SMAD-binding site in the enhancer of Nkx2.5 activating its transcription (Behfar et al., 2002; Brown et al., 2004; Liberatore et al., 2002; Lien et al., 2002). A negative feedback loop comprising NKX2.5-mediated repression of BMP2 and related SMADs allows accurate control of the involved factors (Prall et al., 2007). SMAD4 signaling further maintains the expression level of other cardiogenic transcription factors including GATA4 and MEF2C, both required for expression of cardiac-specific proteins and cardiac patterning at the early stage of cardiogenesis (Olson, 2006; Rojas et al., 2005).
On the other hand, it was recently shown that temporal inhibition of BMP2 and 4 signaling, by Noggin or other antagonists, in the still undifferentiated but primed mesodermal cells is crucial for further cardiac differentiation (Choi et al., 2007). This again indicates the critical role of accurate timing and local restriction of cardiogenic signals; however, the underlying mechanism remains rather unclear (Yuasa et al., 2005). Further, the secreted protein acidic and rich in cysteine, SPARC, is released by mesenchymal parietal endoderm and was shown to specifically promote early myocardial cell differentiation in embryoid bodies synergistically with BMP2 and enhances NKX2.5 expression (Stary et al., 2005).
FGF1, 2, 4, and 8 in chickens and FGF8 in mice together with BMP2 were shown to synergistically regulate the induction of cardiac mesoderm during cardiomyogenesis (Alsan and Schultheiss, 2002; Lopez-Sanchez et al., 2002; Lough et al., 1996; Zhu et al., 1996). Early FGF signaling was shown to initially block premature differentiation of cardiovascular progenitor cells in the embryonic heart (Tirosh-Finkel et al., 2010). However, subsequent FGF signals, through the mitogen-activated protein kinase, p38 MAPK, pathway, result in activation of the transcription factor, cAMP Response Element Binding protein, CREB, and the expression of NKX2.5 in precardiac cells (Keren-Politansky et al., 2009).
WNT signaling serves as another important pathway in early cardiogenesis, acting antagonistically during embryogenesis. Noncanonical WNTs, such as WNT11, activate the cardiac transcription factor machinery through phospholipase C or JAK signaling (Eisenberg and Eisenberg, 1999; Pandur et al., 2002). Additionally, repression of the inhibitory WNT3 and WNT8 signals through DKK1 and Crescent very early in cardiogenesis leads to cardiac induction (Marvin et al., 2001). Canonical WNTs usually act through βCatenin signaling and would at that stage hinder cardiogenesis, possibly through directly inhibiting GATA expression.
Through binding of transcriptional activators TCF1, LEF1, and βCatenin in the promoter region of Brachyury, WNT also influences Brachyury gene expression (Arnold et al., 2000) and WNT3 causes downregulation of HDAC1 during early stages of cardiac development. Simultaneously, NKX2.5 expression is induced and cardiac differentiation is promoted (Liu et al., 2009). Additionally, WNT signaling was shown to be regulated by GATA transcription factors (Afouda et al., 2008). Noteworthy, SMAD2 and 4 also interact with LEF1 (Nawshad and Hay, 2003) and thus interfere with WNT/βCatenin signaling pathways.
Paracrine Notch signaling was recently revealed as another critical factor for early heart development. Endocardial Notch1 expression has been shown to be required for myocardial BMP10 expression during ventricular chamber formation (Grego-Bessa et al., 2007). The inhibition of Notch signaling in the second heart field downregulates FGF8 and BMP4 (High et al., 2009) and Notch signaling directly targets Nkx2.5 gene activity through binding of the downstream factors N1ICD and recombining binding protein suppressor of hairless, RBP-Jk, to its promoter region (Boni et al., 2008).
LIF signaling was demonstrated not only maintain self-renewal of embryonic stem cells (Hall et al., 2009; Niwa et al., 1998, 2009; Zandstra et al., 2000) but also to influence differentiation of cardiac progenitor cells (Bader et al., 2000, 2001). JAK2/STAT3 signaling (Foshay et al., 2005) leads to transcriptional activation of Nkx2.5, Gata4, and Tbx5 genes (Snyder et al., 2010) and at the same time induces expression of Nanog.
Various other pathways including retinoic acid and Hedgehog signaling were also shown to be required for the early determination of the cardiac fate. However, only a few growth factors and their apparent impact on heart development were investigated so far. To obtain a full picture of the early activation of cardiac gene expression patterns, more attention must be paid to the mostly overlapping intracellular pathways guiding transcription factor networks required for self-renewal and differentiation of cardiovascular progenitor cells.
4.3.2. Small molecules supporting cardiomyogenesis
Small molecules interfering with signaling pathways that influence cardiomyogenesis may be used in vitro to direct cardiovascular progenitor cells to a defined state and lineage. This could possibly serve as a prerequisite for stem cell therapy and reduce the inherent risk of teratoma formation. Administrable in vivo, small molecules would be less costly and safer than growth factors or cytokines. Their biological activity could be controlled more easily than that of proteins, leading to the avoidance of adverse side effects outside of target tissues. More important for basic science, small molecules present a highly efficient tool to study signaling pathways involved in the regulation of cardiomyogenesis in vitro and in vivo. Large numbers of newly synthesized compounds can be efficiently tested for desired cardiogenic activities by high-throughput screening in differentiating embryonic, patient-specific induced pluripotent, and cardiovascular progenitor cell lines. A set of cardiac-specific reporter genes would reliably indicate lineage specificity.
One of the first small molecules identified accordingly was ascorbic acid or vitamin C. Addition of vitamin C to differentiating murine embryonic stem cells with a cardiac-specific αMHC promoter-driven enhanced green fluorescent protein, inserted into their genome, increased the number of EGFP positive cells. Moreover, administration of vitamin C induced the expression of GATA4 and two of the myofibrillar motor-protein αMHC and βMHC (Takahashi et al., 2003). Notably, this effect could not be mimicked by other antioxidants, suggesting that vitamin C promotes cardiomyogenesis by so far unknown molecular pathway. Likewise, a small sulfonlyhydrazon, named Shz (Sadek et al., 2008), and Cardiogenol C (Wu et al., 2004) could be identified as potent cardiogenic drugs in the murine teratocarcinoma cells which had been stably transfected with the luciferase reporter gene under the control of the cardiac-specific Anf promoter, Cardiogenol induced GATA4 expression in more than 90% of the differentiating cells and more than 50% expressed αMHC. In a rat transplantation model, human mobilized peripheral blood mononuclear cells treated with Shz led to an increased recovery of heart function in the infarcted myocardium (Sadek et al., 2008). However, unambiguous evidence, that these outcome resulted from electro-mechanical coupling of human cardiomyocytes with the remaining healthy rat heart tissue, is so far missing. Further, due to the nature of the assay used, in all these cases, the underlying molecular mechanisms that induce cardiomyogenesis remain elusive. Thus, in alternative approaches small molecules were tested to specifically interfere with signaling pathways such as TGFβ and WNT, which are particularly involved in cardiomyogenesis. Using small molecules to specifically modulate cardiomyogenesis in stem and progenitor cells would be very promising for the in vitro generation of large numbers of differentiated cardiomyocytes. However, due to the universal involvement of TGFβ and WNT signaling in the regulation of development and homeostasis of the entire organism, these molecules are likely to produce threatening side effects when used in vivo.
TGFβ signaling is mediated by heterotetramers of TGFβ type II receptors and activin-linked kinase receptors, ALKs, or Type I receptors. Of those, ALK2 is activated by BMP2 and Activin, the latter ones specifically promoting the differentiation of mesodermal precursor cells into cardiomyocytes in a concentration and time-dependent manner (Andree et al., 1998; Barron et al., 2000; Brand, 2003; Johansson and Wiles, 1995; Kattman et al., 2011; Moore et al., 1998; Tirosh-Finkel et al., 2010; Winnier et al., 1995). Nevertheless, ALK2 can also inhibit Activin signaling by sequestering Activin and BMP2 from their type II receptors (Renlund et al., 2007). Consequently, inhibition of ALK2 activity by the small molecule Dorsomorphin significantly increased the development of cardiomyocytes in embryonic stem cell-derived embryoid bodies (Hao et al., 2008). Interestingly, inhibition takes place in the first 24h of differentiation, long before the mesoderm determination factor Brachyury is expressed. This suggests that Dorsomorphin acts on very early, so far unidentified progenitor cells that later give rise to cardiomyocytes but not to other mesodermal cell types, and that these stem cells respond to the kinase activity of ALK2.
WNT signaling has been demonstrated to influence cardiomyogenesis in a bivalent manner, both during embryogenesis (Klaus et al., 2007) and in embryoid bodies (Gadue et al., 2006). Whereas in the earliest steps of cardiac development, WNT molecules promote cardiomyogenesis, their later activation inhibits cardiomyogenesis (Naito et al., 2006; Paige et al., 2010). In D. rerio, WNT signaling promotes cardiac differentiation before but inhibits heart formation during gastrulation (Ueno et al., 2007). Thus, WNT signaling provides a very good model system for the identification of compounds with cardiomyogenic activity (Willems et al., 2011). Among them, the well-characterized GSK3 inhibitor 6-bromoindirubin-3′ oxime, BIO, was shown to induce cardiomyogenesis through WNT signaling in a very early stage of embryoid body differentiation (Tseng et al., 2006). Controverse data, however, demonstrated that BIO and the other WNT signaling activator CHIR99021 can also be used to maintain LIF-independent self-renewal in embryonic stem cells (Ying et al., 2008). During later stages of cardiomyogenesis, the Tankyrase inhibitor XAV939, which stabilizes Axin and thus inhibits WNT signaling, can robustly induce cardiomyogenesis in embryonic stem cells. Timely administration of XAV939 immediately following the formation of mesoderm progenitor cells promotes cardiomyogenic development at the expense of other mesoderm-derived lineages (Wang et al., 2011). Most importantly, XAV939 even when applied in later stages of development tremendously increases cardiomyogenesis (Wang et al., 2011). These data together with the fact that Tankyrase is also involved in MAPK signal transduction (Chi and Lodish, 2000) and promotes telomere elongation (Smith and de Lange, 2000) suggest a so far undefined reprogramming activity of this compound. Thus, XAV939 may serve as a starting point for chemical reprogramming of somatic cells to cardiomyocytes.
Finally, at least in D. rerio, a very early fate decision in cardiovascular progenitor cells can be affected by activation of FGF/ERK signaling (Molina et al., 2009). (E)-2-benzylidene-3-(cyclohexylamino)-2,3-dihydro-1H-inden-1-one, BCI inhibits the dephosphorylation of ERK1/2 by the dual-specificity phosphatase 6, DUSP6. This leads to increased FGF signaling and to the expansion of the cardiovascular progenitor cell pool, resulting in enlarged hearts at subsequent developmental stages. Notably, this effect was also seen in DUSP6-null mice where increased ERK phosphorylation caused the same deformation (Maillet et al., 2008). This indicates that FGF signaling might be an evolutionarily conserved mechanism that regulates cardiovascular progenitor cell proliferation and fate decision between the endothelial and myocardial lineage.
4.4. Plasticity of cardiovascular progenitor cells
With increasing numbers of studies on the expression levels of genes in established cell lines and large sets of data becoming available from microarray analysis, it became evident that expression of many genes is not constant in otherwise phenotypical identical cell lines (Bhattacharya et al., 2004; Carpenter et al., 2004; Ramalho-Santos et al., 2002; Zeng et al., 2004). It was shown that non-housekeeping genes fluctuate over time (Mansergh et al., 2009) despite the stability of the epigenetic status and phenotype (Rugg-Gunn et al., 2005). This inconsistency was interpreted at this time as genetic instability of the tested cell lines, as a consequence of irreproducible isolation and culture conditions, or as inappropriateness of gene expression analyses. However, since the phenotype of these cell lines remained constant, it became more and more evident that the expression of at least some genes in a given stem cell line is not constant and varies over time.
The set of genes most extensively studied in connection with stem cell properties are the so-called stemness factors in embryonic stem cells, including Oct4, Sox2, and Nanog. Austin Smith’s group firstly discovered stochastically changing OCT4 levels in colonies of embryonic stem cells. A relative high level of OCT4 was shown to drive embryonic stem cell differentiation along the mesendodermal lineage, and low levels of OCT4 give way to the formation of extraembryonic trophectoderm (Niwa et al., 2000). Only a medium level of OCT4 maintains the self-renewal potential of embryonic stem cells. Monitoring the expression of these proteins with appropriate antibodies demonstrated that in a given clone, most cells expressed OCT4, SOX2, and Nanog at different level, varying between cells and over time (Chambers et al., 2007; Silva et al., 2009). This phenomenon became evident also in vivo in the developing inner cell mass of mouse blastocysts (Buehr and Smith, 2003; Nichols et al., 2009). The fluctuation was not restricted to transcription factors as the expression of the LIF-Receptor, an important factor to maintain LIF-induced self-renewal in murine embryonic stem cells (Niwa et al., 2009), also varied within the inner cell mass of murine blastocysts (Lauss et al., 2005). More importantly, subcloning of individual cells with a certain expression pattern of stemness proteins always resulted in heterogeneous cell colonies unevenly expressing OCT4, SOX2, and Nanog (Silva and Smith, 2008; Toyooka et al., 2008). Further, plasticity and variable expression pattern have been extensively described for hematopoietic and mesenchymal stem cells (Raff, 2003).
From these results, we may conclude that most likely all cells display inherent fluctuating or stochastic gene expression. Particularly, stem and progenitor cells may require the increased fluctuation of gene expression to properly respond to external stimuli inducing differentiation. This plasticity may well be the basis for different and increased developmental potentials of progenitor and stem cells. This notion is supported by data demonstrating that fluctuations of OCT4 and SOX2 expression prepare cells for the differentiation along either the mesendodermal or neuroectodermal lineages (Thomson et al., 2011). The higher the differentiation potential of a cell, the more genes fluctuate in their expression level. The less potent these cells are the fewer genes vary in their expression levels. Cardiovascular progenitor cells are supposed to be of mesodermal origin, and we could demonstrate that murine cardiovascular progenitor cells express at the same time the stemness factors OCT4, SOX2, and Nanog, and mesodermal and early myocardial transcription factors such as Brachyury, NKX2.5, and GATA4 (submitted for publication) which would fit to the hypothesis presented above. One could argue that expression of stemness transcription factors guarantees their self-renewal and that the expression of transcription factors responsible for differentiation along the cardiac lineage makes cardiovascular progenitor cells prone to spontaneously develop into cardiomyocytes, endothelial cells, and smooth muscle cells and simultaneously exclude the differentiation to ectodermal and endodermal cell types.
4.4.1. Stochastic fluctuation of gene expression in stem and progenitor cells
Fluctuations in gene expression have been suggested a long time ago by Dov Zipory to describe the differentiation potential and plasticity of mesenchymal stem cells in the bone marrow (Pevsner-Fischer et al., 2011; Zipori, 2004a,b). He suggests a model which includes fluctuations in the expression of different sets of genes even in stem cells displaying the same phenotype. These propositions correspond well with similar observations made in embryonic stem cell clones much later (Chambers et al., 2007; Silva et al., 2009). In his own words, Dov Zipori explained “[…] two types of stem cells […] differ markedly in their gene-expression profiles, but in both, a similar pattern is formed when the ‘stem state’ is adopted […] Such similar ‘organization’ might be formed in each cell by different molecules and could entail post-translational protein modifications, protein degradation, specific localization of molecules within specific cellular compartments or, probably, a combination of the three.” Today, these statements could be easily explained by epigenetic differences found in the regulation of gene expression. He continues with a rather unconventional view of stem cell differentiation by stating “One possibility is that the stem cell is a ‘blank slate’ and that differentiation entails acquisition of different gene-expression capacities,” which at that time was the orthodox view of most scientists involved in that field. However, further he presents his own opinion about the subject by stating “An opposing possibility is that stem cells express many genes at a low level, and that the expression of many of these is reduced during differentiation, with the expression of a small collection of the rest increased to a higher level.” This statement provides an explanation of the commitment of a stem cell while still proliferating and subsequent differentiation into a particular somatic cell type. Further, the model would explain the occurrence of cells, already in an ongoing state of differentiation, comparable to the supposed situation of cardiovascular progenitor cells, adopting a distinct “stem cell state” that allows both limited self-renewal and proliferation (Zipori, 2006).
These findings and ideas can be best illustrated by an n-dimensional expression space, where, within a defined volume, fluctuations in the gene expression level do not change the phenotype. Thus, a minimal volume in this imaginative space would present a certain cellular phenotype, for example, a “stemness expression volume” would be characteristic for stem cells. This idea was inspired by the introduction and description of coarse graining regions in phase space to describe areas of not uniform but indistinguishable values of entropy in the universe (Penrose, 2010). Supposing that the entropy would rise significantly, this part of space would be found in a new, larger coarse-graining region. Similarly, as long as a cell’s expression space remains within the limits of its “stemness expression volume,” this cell will phenotypically be a stem cell. The adopted state is independent of the up- or downregulation of other subsets of genes—and the later implemented differentiation trait. Runaway expression of any gene will be compensated by other gene products remaining in the adequate expression space, thus guaranteeing self-renewal and proliferation of these cells (Fig. 7.3A). However, once a significant number of gene expression levels gain values outside this “stemness volume,” and enter a region defined by a different phenotype, cells will start to differentiate, along a certain lineage led by a distinct number of the expressed genes. Most likely, this event will not be triggered by a single key factor, for which innumerable scientists have searched since developmental biology adopted the tools of molecular biology, but by different finite sets of genes. The direction of the exit out of “stemness expression volume” determines whether ectodermal, mesodermal, or endodermal lineage development follows. These decisions are repeated for each developmentally defined, transiently stable stage, where an intermediate cell type with a defined phenotype can be identified (Fig. 7.3B). This model would also explain how a common mesodermal stem cell, for example, a cardiovascular progenitor cell, with constant fluctuations in the expression of stemness and fate determining transcription factor genes, may function in different roles. Similarly, it would explicate how only small and dynamic changes of gene expression ultimately lead to the commitment and differentiation of cardiovascular progenitor cells to cardiomyocytes, cells of the conduction system, endothelial, and smooth muscle cells (Srivastava and Ivey, 2006). Most importantly, this model would also explain the changes in plasticity as cells differentiate. Following this line of reasoning, we may find the expression space becoming smaller and smaller as a cell differentiates into an inevitably committed and fully differentiated somatic cell type that has lost the differentiation potential of its progenitor or stem cell it originated from (Fig. 7.3C). Consequently, in such a small expression volume, fluctuations of gene expression should decrease and possibly disappear when cells enter senescence. Finally, because of lending this model partly from the description of entropy in space, we may allude to the notion that a living organism at least temporarily and locally diminishes entropy of the system. Reduced fluctuation of gene expression during differentiation at least fits this concept.
Figure 7.3.
(A) Expression of genes in a stem cell in an n-dimensional expression space, where n is the number of genes in a genome. Changes of expression levels over time do not influence the phenotype of a stem cell as long as the majority of genes contributing to the phenotype remain within the stemness expression volume. High fluctuations (e.g., OCT4) or constant expression levels (e.g., SOX2) do not influence the phenotype; however, high fluctuations keep the stem cell prone to differentiation if they increase in number and are biased by external stimuli. Thus, short lasting runaway expression, for example, Brachyury (black rectangle) does not necessarily lead to differentiation. (B) Change in the expression of a sufficiently large number of genes causes differentiation. Different sets of genes may lead to the same phenotype caused by, for example, lower OCT4 and SOX2 levels and higher levels of Brachyury, MESP1, and NKX2.5 in cardiovascular progenitor cells. (C) Reaching the status of a differentiated somatic cell, for example, cardiomyocyte, fluctuations diminish, useless or detrimental genes are downregulated (e.g., Oct4 and Nkx2.5), and genes typical for this cell type are expressed at higher and nearly constant levels (e.g., Myosin) over the lifespan of this cell.
4.4.2. Inherent inhomogeneity of progenitor and stem cell lines
A persisting problem in stem cell biology is that we currently cannot decide whether different types of progenitor cells within an organ exist or if the identified cells are just derivatives of a single cell type with high plasticity that display different phenotypes depending on their location in a niche.
As “plasticity” we understand a significant degree of uncertainty regarding the commitment and future properties of a cell. This also leads to the question about the clonogenicity of stem and progenitor cells. If they show constant changes in the expression of genes regulating self-renewal and differentiation, the concept of monoclonality of a cell line, both in vivo and in vitro, becomes obsolete. From the model presented above, we may expect that generation of clonal cell lines from a population of primary stem cells will give, after as few as one passage, again a heterogeneous pool of cells regarding their quantitative gene expression, but nonetheless maintain their particular phenotype. Subcloning, however, will be no way to escape this stem cell inherent characteristic. The existence of these cells in diverse microenvironments within an organ may add variable external signals either increasing the cell’s inherent fluctuation of gene expression or decreasing and stabilizing the variablility of gene expression in vivo. Consequently, it seems currently too rigorous to unquestionably define stem cells by the expression of a limited set of marker genes.
5. Concluding Remarks
Here, we compiled the current knowledge about cardiovascular progenitor cells of the heart to paint a unified picture of their biology. We gathered information on the origin of these cells during evolution and their supposed localization in the adult mammalian heart. We discussed the current heterogeneity of their features, possible technical reasons, and elaborated from several model organisms a current state of knowledge about the molecular mechanisms which may be responsible for self-renewal and differentiation of cardiovascular progenitor cells to somatic cells of the heart.
Although we tried hard to find all relevant data in the infinite space of the World Wide Web, we are pretty sure that we missed or omitted some considered as important by other scientist. We hope to provide at least some new and perhaps unconventional perspectives from different angles and by this provoke new ideas for future research in the gentle reader. Having said this, we end by pointing out what themes we do not know or understand so far and try to suggest possible strategies for future investigations.
While a considerable number of transcription factors and signaling molecules critical for cardiomyogenesis are known, there are significant gaps in our understanding of molecular and cell biological mechanisms that guide cardiovascular progenitor cells during proliferation, migration, and differentiation processes, from primitive streak to the splanchnic mesoderm and into the different regions of the heart. In particular, we do not know if and how the gearwheels driving self-renewal fit into those driving cardiac differentiation. Interconnected to this complex issue the question rises, whether a reversion of this machinery, in order to accomplish the phenomena of cellular plasticity, and de- and transdifferentiation is possible. Likewise, the borders separating a stem cell from a progenitor and a somatic cell still have to be defined.
Further, we need to identify the critical time windows for the function of genes involved in the specification of cardiovascular progenitor cells. This will provide an important additional dimension for the integration of molecular mechanisms into a unified picture of cell cycle control, self-renewal, and cardiogenesis. Of particular interest is here to define mechanisms that allow self-renewing cells to remain dormant in an organ or tissue until there is a need for them.
Finally, a complex of more theoretical questions that are based on the apparent or real inhomogeneity of cardiovascular progenitor cells in particular, and stem cells in general await answers. Are cardiovascular progenitor cells indeed clonogenic or does a single self-renewing cell always develop into a population of differing progenitor cells in a clone? In other words, is there an inherent stochastic shift between symmetric and asymmetric cell division, perhaps caused by stochastic fluctuations in the expression of genes? These considerations finally culminate in the question whether a developmental process must necessarily represent a hierarchy of distinct developmental stages and associated cell types.
As an experimental approach and in addition to studying molecular mechanisms of self-renewal and cardiac differentiation in cardiovascular progenitor cell lines isolated from different very early bilateria, both protostomia and deuterostomia, it would be very interesting to study the molecular machinery controlling maintenance of self-renewal and differentiation in cardiovascular progenitor cell, in the tiny water bears, the Tardigrades (Schmidt-Rhaesa, 2001), which lost their heart during evolution, and to compare the remnants of the transcriptional network in this group to other groups of the taxon Panarthropoda and all kinds of vertebrates up to the mammalia. This strategy would not only contribute to improve the relationship of taxa within the tree of the animal kingdom but further allow getting additional information on the most basal genes and proteins which stand at the beginning of heart development and apparently regulate the behavior and the features of cardiovascular progenitor cells.
Acknowledgments
We thank Brigitte Gundacker for extensive literature search and Philipp Heher for critical reading of the chapter and helpful discussion. This work was supported by funds from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung, Grants P15303, P11189, and P18659; the Herzfelder´sche Familienstiftung; the Österreichische Nationalbank, Grant 8437; the Austrian Federal Ministry of Science, Grant GZ70.078/0002-Pr/472002; and the Hochschuljubiläumsstiftung der Stadt Wien, Grants H933-2003, H-2174/2007, and H-1249/2009 to G. W.
References
- Afouda BA, Ciau-Uitz A, Patient R. GATA4, 5 and 6 mediate TGFbeta maintenance of endodermal gene expression in Xenopus embryos. Development. 2005;132:763–774. doi: 10.1242/dev.01647. [DOI] [PubMed] [Google Scholar]
- Afouda BA, Martin J, Liu F, Ciau-Uitz A, Patient R, Hoppler S. GATA transcription factors integrate Wnt signalling during heart development. Development. 2008;135:3185–3190. doi: 10.1242/dev.026443. [DOI] [PubMed] [Google Scholar]
- Aghila Rani KG, Jayakumar K, Srinivas G, Nair RR, Kartha CC. Isolation of ckit-positive cardiosphere-forming cells from human atrial biopsy. Asian Cardiovasc Thorac Ann. 2008;16:50–56. doi: 10.1177/021849230801600113. [DOI] [PubMed] [Google Scholar]
- Al-Nedawi KN, Czyz M, Bednarek R, Szemraj J, Swiatkowska M, Cierniewska-Cieslak A, et al. Thymosin beta 4 induces the synthesis of plasminogen activator inhibitor 1 in cultured endothelial cells and increases its extracellular expression. Blood. 2004;103:1319–1324. doi: 10.1182/blood-2003-04-1015. [DOI] [PubMed] [Google Scholar]
- Alsan BH, Schultheiss TM. Regulation of avian cardiogenesis by Fgf8 signaling. Development. 2002;129:1935–1943. doi: 10.1242/dev.129.8.1935. [DOI] [PubMed] [Google Scholar]
- Andree B, Duprez D, Vorbusch B, Arnold HH, Brand T. BMP-2 induces ectopic expression of cardiac lineage markers and interferes with somite formation in chicken embryos. Mech Dev. 1998;70:119–131. doi: 10.1016/s0925-4773(97)00186-x. [DOI] [PubMed] [Google Scholar]
- Anversa P, Kajstura J. Ventricular myocytes are not terminally differentiated in the adult mammalian heart. Circ Res. 1998;83:1–14. doi: 10.1161/01.res.83.1.1. [DOI] [PubMed] [Google Scholar]
- Anversa P, Nadal-Ginard B. Myocyte renewal and ventricular remodelling. Nature. 2002;415:240–243. doi: 10.1038/415240a. [DOI] [PubMed] [Google Scholar]
- Anversa P, Kajstura J, Leri A. If I can stop one heart from breaking. Circulation. 2007a;115:829–832. doi: 10.1161/CIRCULATIONAHA.106.682195. [DOI] [PubMed] [Google Scholar]
- Anversa P, Leri A, Rota M, Hosoda T, Bearzi C, Urbanek K, et al. Concise review: stem cells, myocardial regeneration, and methodological artifacts. Stem Cells. 2007b;25:589–601. doi: 10.1634/stemcells.2006-0623. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Stappert J, Bauer A, Kispert A, Herrmann BG, Kemler R. Brachyury is a target gene of the Wnt/beta-catenin signaling pathway. Mech Dev. 2000;91:249–258. doi: 10.1016/s0925-4773(99)00309-3. [DOI] [PubMed] [Google Scholar]
- Artinger M, Blitz I, Inoue K, Tran U, Cho KW. Interaction of goosecoid and brachyury in Xenopus mesoderm patterning. Mech Dev. 1997;65:187–196. doi: 10.1016/s0925-4773(97)00073-7. [DOI] [PubMed] [Google Scholar]
- Ausoni S, Sartore S. From fish to amphibians to mammals: in search of novel strategies to optimize cardiac regeneration. J Cell Biol. 2009;184:357–364. doi: 10.1083/jcb.200810094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Thomas JH. Feeding and Defecation. 2011/03/18 ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- Baccetti B, Bigliardi E. Studies on the fine structure of the dorsal vessel of arthropods. 1. The "heart" of an orthopteran. Z Zellforsch Mikrosk Anat. 1969;99:13–24. doi: 10.1007/BF00338794. [DOI] [PubMed] [Google Scholar]
- Bader A, Al-Dubai H, Weitzer G. Leukemia inhibitory factor modulates cardiogenesis in embryoid bodies in opposite fashions. Circ Res. 2000;86:787–794. doi: 10.1161/01.res.86.7.787. [DOI] [PubMed] [Google Scholar]
- Bader A, Gruss A, Hollrigl A, Al-Dubai H, Capetanaki Y, Weitzer G. Paracrine promotion of cardiomyogenesis in embryoid bodies by LIF modulated endoderm. Differentiation. 2001;68:31–43. doi: 10.1046/j.1432-0436.2001.068001031.x. [DOI] [PubMed] [Google Scholar]
- Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science. 1999;283:1161–1164. doi: 10.1126/science.283.5405.1161. [DOI] [PubMed] [Google Scholar]
- Barile L, Chimenti I, Gaetani R, Forte E, Miraldi F, Frati G, et al. Cardiac stem cells: isolation, expansion and experimental use for myocardial regeneration. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl. 1):S9–S14. doi: 10.1038/ncpcardio0738. [DOI] [PubMed] [Google Scholar]
- Barron M, Gao M, Lough J. Requirement for BMP and FGF signaling during cardiogenic induction in non-precardiac mesoderm is specific, transient, and cooperative. Dev Dyn. 2000;218:383–393. doi: 10.1002/(SICI)1097-0177(200006)218:2<383::AID-DVDY11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bayes-Genis A, Muniz-Diaz E, Catasus L, Arilla M, Rodriguez C, Sierra J, et al. Cardiac chimerism in recipients of peripheral-blood and bone marrow stem cells. Eur J Heart Fail. 2004;6:399–402. doi: 10.1016/j.ejheart.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Bearzi C, Rota M, Hosoda T, Tillmanns J, Nascimbene A, De Angelis A, et al. Human cardiac stem cells. Proc Natl Acad Sci USA. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearzi C, Leri A, Lo Monaco F, Rota M, Gonzalez A, Hosoda T, et al. Identification of a coronary vascular progenitor cell in the human heart. Proc Natl Acad Sci USA. 2009;106:15885–15890. doi: 10.1073/pnas.0907622106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Becker DL, Cook JE, Davies CS, Evans WH, Gourdie RG. Expression of major gap junction connexin types in the working myocardium of eight chordates. Cell Biol Int. 1998;22:527–543. doi: 10.1006/cbir.1998.0295. [DOI] [PubMed] [Google Scholar]
- Beddington RS, Rashbass P, Wilson V. Brachyury–a gene affecting mouse gastrulation and early organogenesis. Dev Suppl. 1992:157–165. [PubMed] [Google Scholar]
- Behfar A, Zingman LV, Hodgson DM, Rauzier JM, Kane GC, Terzic A, et al. Stem cell differentiation requires a paracrine pathway in the heart. FASEB J. 2002;16:1558–1566. doi: 10.1096/fj.02-0072com. [DOI] [PubMed] [Google Scholar]
- Beltrami AP, Urbanek K, Kajstura J, Yan SM, Finato N, Bussani R, et al. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Stewart MH, Menendez P, George D, Vijayaragavan K, Werbowetski-Ogilvie T, et al. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448:1015–1021. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, et al. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya B, Miura T, Brandenberger R, Mejido J, Luo Y, Yang AX, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- Biben C, Harvey RP. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997;11:1357–1369. doi: 10.1101/gad.11.11.1357. [DOI] [PubMed] [Google Scholar]
- Bishopric NH. Evolution of the heart from bacteria to man. Ann N Y Acad Sci. 2005;1047:13–29. doi: 10.1196/annals.1341.002. [DOI] [PubMed] [Google Scholar]
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- Bodmer R. Heart development in Drosophila and its relationship to vertebrates. Trends Cardiovasc Med. 1995;5:21–28. doi: 10.1016/1050-1738(94)00032-Q. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Jan LY, Jan YN. A new homeobox-containing gene, msh-2, is transiently expressed early during mesoderm formation of Drosophila. Development. 1990;110:661–669. doi: 10.1242/dev.110.3.661. [DOI] [PubMed] [Google Scholar]
- Boero F, Gravili C, Pagliara P, Piraino S, Bouillon J, Schmid V. The cnidarian premises of metazoan evolution: from triploblasty, to coelom formation, to metamery. Ital J Zool. 1998;65:5–9. [Google Scholar]
- Bollini S, Smart N, Riley PR. Resident cardiac progenitor cells: at the heart of regeneration. J Mol Cell Cardiol. 2011;50:296–303. doi: 10.1016/j.yjmcc.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Bondue A, Blanpain C. Mesp1: a key regulator of cardiovascular lineage commitment. Circ Res. 2010;107:1414–1427. doi: 10.1161/CIRCRESAHA.110.227058. [DOI] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Boni A, Urbanek K, Nascimbene A, Hosoda T, Zheng H, Delucchi F, et al. Notch1 regulates the fate of cardiac progenitor cells. Proc Natl Acad Sci USA. 2008;105:15529–15534. doi: 10.1073/pnas.0808357105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bonkowski D, Katyshev V, Balabanov RD, Borisov A, Dore-Duffy P. The CNS microvascular pericyte: pericyte-astrocyte crosstalk in the regulation of tissue survival. Fluids Barriers CNS. 2011;8:8. doi: 10.1186/2045-8118-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TC. Hydra and the evolution of stem cells. Bioessays. 2009;31:478–486. doi: 10.1002/bies.200800183. [DOI] [PubMed] [Google Scholar]
- Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- Brown CO, 3rd, Chi X, Garcia-Gras E, Shirai M, Feng XH, Schwartz RJ. The cardiac determination factor, Nkx2-5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004;279:10659–10669. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
- Brown DD, Martz SN, Binder O, Goetz SC, Price BM, Smith JC, et al. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132:553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Del Re DP, Sussman MA. The Rac and Rho hall of fame: a decade of hypertrophic signaling hits. Circ Res. 2006;98:730–742. doi: 10.1161/01.RES.0000216039.75913.9e. [DOI] [PubMed] [Google Scholar]
- Bruneau BG. Transcriptional regulation of vertebrate cardiac morphogenesis. Circ Res. 2002;90:509–519. doi: 10.1161/01.res.0000013072.51957.b7. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Black BL. The heart’s Da Vinci code: a Renaissance at Keystone. Development. 2007;134:1631–1633. doi: 10.1242/dev.002014. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, et al. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev Biol. 1999;211:100–108. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Bao ZZ, Tanaka M, Schott JJ, Izumo S, Cepko CL, et al. Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2-5 and dHand. Dev Biol. 2000;217:266–277. doi: 10.1006/dbio.1999.9548. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Bao ZZ, Fatkin D, Xavier-Neto J, Georgakopoulos D, Maguire CT, et al. Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol Cell Biol. 2001;21:1730–1736. doi: 10.1128/MCB.21.5.1730-1736.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, et al. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Buehr M, Smith A. Genesis of embryonic stem cells. Philos Trans R Soc Lond B Biol Sci. 2003;358:1397–1402. doi: 10.1098/rstb.2003.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, et al. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Brauninger M, et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Carpenter MK, Rosler ES, Fisk GJ, Brandenberger R, Ares X, Miura T, et al. Properties of four human embryonic stem cell lines maintained in a feeder-free culture system. Dev Dyn. 2004;229:243–258. doi: 10.1002/dvdy.10431. [DOI] [PubMed] [Google Scholar]
- Cesselli D, Beltrami AP, Rigo S, Bergamin N, D’Aurizio F, Verardo R, et al. Multipotent progenitor cells are present in human peripheral blood. Circ Res. 2009;104:1225–1234. doi: 10.1161/CIRCRESAHA.109.195859. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Charron F, Nemer M. GATA transcription factors and cardiac development. Semin Cell Dev Biol. 1999;10:85–91. doi: 10.1006/scdb.1998.0281. [DOI] [PubMed] [Google Scholar]
- Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri K, Chatterjee R. MicroRNA detection and target prediction: integration of computational and experimental approaches. DNA Cell Biol. 2007;26:321–337. doi: 10.1089/dna.2006.0549. [DOI] [PubMed] [Google Scholar]
- Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- Chen JN, Fishman MC. Genetics of heart development. Trends Genet. 2000;16:383–388. doi: 10.1016/s0168-9525(00)02075-8. [DOI] [PubMed] [Google Scholar]
- Chen CY, Schwartz RJ. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NW, Lodish HF. Tankyrase is a Golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J Biol Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- Chien KR, Zhu H, Knowlton KU, Miller-Hance W, van-Bilsen M, O’Brien TX, et al. Transcriptional regulation during cardiac growth and development. Annu Rev Physiol. 1993;55:77–95. doi: 10.1146/annurev.ph.55.030193.000453. [DOI] [PubMed] [Google Scholar]
- Cho MS, Chan IL, Flores ER. DeltaNp63 transcriptionally regulates brachyury, a gene with diverse roles in limb development, tumorigenesis and metastasis. Cell Cycle. 2010;9:2434–2441. doi: 10.4161/cc.9.12.12051. [DOI] [PubMed] [Google Scholar]
- Choi M, Stottmann RW, Yang YP, Meyers EN, Klingensmith J. The bone morphogenetic protein antagonist noggin regulates mammalian cardiac morphogenesis. Circ Res. 2007;100:220–228. doi: 10.1161/01.RES.0000257780.60484.6a. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Habets PE, Franco D, Campione M, de Jong F, Lamers WH, et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol. 2000;223:266–278. doi: 10.1006/dbio.2000.9753. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn. 2004;229:763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- Collett GD, Canfield AE. Angiogenesis and pericytes in the initiation of ectopic calcification. Circ Res. 2005;96:930–938. doi: 10.1161/01.RES.0000163634.51301.0d. [DOI] [PubMed] [Google Scholar]
- Condorelli G, Borello U, De Angelis L, Latronico M, Sirabella D, Coletta M, et al. Cardiomyocytes induce endothelial cells to trans-differentiate into cardiac muscle: implications for myocardium regeneration. Proc Natl Acad Sci USA. 2001;98:10733–10738. doi: 10.1073/pnas.191217898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello I, Pimeisl IM, Drager S, Bikoff EK, Robertson EJ, Arnold SJ. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol. 2011;13:1084–1091. doi: 10.1038/ncb2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amario D, Cabral-Da-Silva MC, Zheng H, Fiorini C, Goichberg P, Steadman E, et al. Insulin-like growth factor-1 receptor identifies a pool of human cardiac stem cells with superior therapeutic potential for myocardial regeneration. Circ Res. 2011;108:1467–1481. doi: 10.1161/CIRCRESAHA.111.240648. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- David R, Brenner C, Stieber J, Schwarz F, Brunner S, Vollmer M, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- David R, Jarsch VB, Schwarz F, Nathan P, Gegg M, Lickert H, et al. Induction of MesP1 by Brachyury(T) generates the common multipotent cardiovascular stem cell. Cardiovasc Res. 2011;92:115–122. doi: 10.1093/cvr/cvr158. [DOI] [PubMed] [Google Scholar]
- Davis DR, Zhang Y, Smith RR, Cheng K, Terrovitis J, Malliaras K, et al. Validation of the cardiosphere method to culture cardiac progenitor cells from myocardial tissue. PLoS One. 2009;4:e7195. doi: 10.1371/journal.pone.0007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Dusenbery BN, Chan MC, Reno KE, Weisman AS, Layne MD, Lagna G, et al. Down-regulation of Kruppel-like Factor-4 (KLF4) by MicroRNA-143/145 Is Critical for Modulation of Vascular Smooth Muscle Cell Phenotype by Transforming Growth Factor-{beta} and Bone Morphogenetic Protein 4. J Biol Chem. 2011;286:28097–28110. doi: 10.1074/jbc.M111.236950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E, et al. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature. 1998;392:182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- Di Nardo P, Forte G, Ahluwalia A, Minieri M. Cardiac progenitor cells: potency and control. J Cell Physiol. 2010;224:590–600. doi: 10.1002/jcp.22165. [DOI] [PubMed] [Google Scholar]
- Dixon JE, Dick E, Rajamohan D, Shakesheff KM, Denning C. Directed differentiation of human embryonic stem cells to interrogate the cardiac gene regulatory network. Mol Ther. 2011;19:1695–1703. doi: 10.1038/mt.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Dore-Duffy P. Pericytes: pluripotent cells of the blood brain barrier. Curr Pharm Des. 2008;14:1581–1593. doi: 10.2174/138161208784705469. [DOI] [PubMed] [Google Scholar]
- Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, et al. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- Eilertsen KJ, Floyd Z, Gimble JM. The epigenetics of adult (somatic) stem cells. Crit Rev Eukaryot Gene Expr. 2008;18:189–206. doi: 10.1615/critreveukargeneexpr.v18.i3.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg CA, Eisenberg LM. WNT11 promotes cardiac tissue formation of early mesoderm. Dev Dyn. 1999;216:45–58. doi: 10.1002/(SICI)1097-0177(199909)216:1<45::AID-DVDY7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Ellison GM, Torella D, Karakikes I, Nadal-Ginard B. Myocyte death and renewal: modern concepts of cardiac cellular homeostasis. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl. 1):S52–S59. doi: 10.1038/ncpcardio0773. [DOI] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Liu H, Sun C, Chen C, Jiao K, Chen Y. Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev Biol. 2011;356:359–369. doi: 10.1016/j.ydbio.2011.05.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli AB. A HANDful of questions: the molecular biology of the heart and neural crest derivatives (HAND)-subclass of basic helix-loop-helix transcription factors. Gene. 2003;312:27–40. doi: 10.1016/s0378-1119(03)00669-3. [DOI] [PubMed] [Google Scholar]
- Firulli AB, Thattaliyath BD. Transcription factors in cardiogenesis: the combinations that unlock the mysteries of the heart. Int Rev Cytol. 2002;214:1–62. doi: 10.1016/s0074-7696(02)14002-2. [DOI] [PubMed] [Google Scholar]
- Fishman MC, Olson EN. Parsing the heart: genetic modules for organ assembly. Cell. 1997;91:153–156. doi: 10.1016/s0092-8674(00)80397-9. [DOI] [PubMed] [Google Scholar]
- Foshay K, Rodriguez G, Hoel B, Narayan J, Gallicano GI. JAK2/STAT3 directs cardiomyogenesis within murine embryonic stem cells in vitro. Stem Cells. 2005;23:530–543. doi: 10.1634/stemcells.2004-0293. [DOI] [PubMed] [Google Scholar]
- Franco D, Campione M. The role of Pitx2 during cardiac development. Linking left-right signaling and congenital heart diseases. Trends Cardiovasc Med. 2003;13:157–163. doi: 10.1016/s1050-1738(03)00039-2. [DOI] [PubMed] [Google Scholar]
- Fu Y, Yan W, Mohun TJ, Evans SM. Vertebrate tinman homologues XNkx2-3 and XNkx2-5 are required for heart formation in a functionally redundant manner. Development. 1998;125:4439–4449. doi: 10.1242/dev.125.22.4439. [DOI] [PubMed] [Google Scholar]
- Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci USA. 2006;103:16806–168011. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez BG, Sampaolesi M, Barbuti A, Crespi A, Covarello D, Brunelli S, et al. Cardiac mesoangioblasts are committed, self-renewable progenitors, associated with small vessels of juvenile mouse ventricle. Cell Death Differ. 2008;15:1417–1428. doi: 10.1038/cdd.2008.75. [DOI] [PubMed] [Google Scholar]
- Gambini E, Pompilio G, Biondi A, Alamanni F, Capogrossi MC, Agrifoglio M, et al. c-kit+ cardiac progenitors exhibit mesenchymal markers and preferential cardiovascular commitment. Cardiovasc Res. 2010;89:362–373. doi: 10.1093/cvr/cvq292. [DOI] [PubMed] [Google Scholar]
- Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. 2006;127:1101–1104. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- Gelb BD. Genetic basis of congenital heart disease. Curr Opin Cardiol. 2004;19:110–115. doi: 10.1097/00001573-200403000-00007. [DOI] [PubMed] [Google Scholar]
- Gherghiceanu M, Popescu LM. Cardiomyocyte precursors and telocytes in epicardial stem cell niche: electron microscope images. J Cell Mol Med. 2010;14:871–877. doi: 10.1111/j.1582-4934.2010.01060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh TK, Song FF, Packham EA, Buxton S, Robinson TE, Ronksley J, et al. Physical interaction between TBX5 and MEF2C is required for early heart development. Mol Cell Biol. 2009;29:2205–2218. doi: 10.1128/MCB.01923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis TE, Tibbits GF. Beating the cold: the functional evolution of troponin C in teleost fish. Comp Biochem Physiol A Mol Integr Physiol. 2002;132:763–772. doi: 10.1016/s1095-6433(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Mahtab EA, Hahurij ND, Wisse LJ, Deruiter MC, Wijffels MC, et al. Nkx2.5-negative myocardium of the posterior heart field and its correlation with podoplanin expression in cells from the developing cardiac pacemaking and conduction system. Anat Rec (Hoboken) 2007;290:115–122. doi: 10.1002/ar.20406. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Conlon FL. Cardiac progenitors and the embryonic cell cycle. Cell Cycle. 2007;6:1974–1981. doi: 10.4161/cc.6.16.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Brown DD, Conlon FL. TBX5 is required for embryonic cardiac cell cycle progression. Development. 2006;133:2575–2584. doi: 10.1242/dev.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskurth SE, Bhattacharya D, Wang Q, Lin JJ. Emergence of Xin demarcates a key innovation in heart evolution. PLoS One. 2008;3:e2857. doi: 10.1371/journal.pone.0002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow MW, Krieg PA. Tinman function is essential for vertebrate heart development: elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, XNkx2-3 and XNkx2-5. Dev Biol. 1998;204:187–196. doi: 10.1006/dbio.1998.9080. [DOI] [PubMed] [Google Scholar]
- Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, et al. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson EM, Lindsay ME, Chien KR. Regeneration next: toward heart stem cell therapeutics. Cell Stem Cell. 2009;5:364–377. doi: 10.1016/j.stem.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Hao A, Novotny-Diermayr V, Bian W, Lin B, Lim CP, Jing N, et al. The LIM/homeodomain protein Islet1 recruits Janus tyrosine kinases and signal transducer and activator of transcription 3 and stimulates their activities. Mol Biol Cell. 2005;16:1569–1583. doi: 10.1091/mbc.E04-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Daleo MA, Murphy CK, Yu PB, Ho JN, Hu J, et al. Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS One. 2008;3:e2904. doi: 10.1371/journal.pone.0002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- Harvey RP. Links in the left/right axial pathway. Cell. 1998;94:273–276. doi: 10.1016/s0092-8674(00)81468-3. [DOI] [PubMed] [Google Scholar]
- Harvey RP. Patterning the vertebrate heart. Nat Rev Genet. 2002;3:544–556. doi: 10.1038/nrg843. [DOI] [PubMed] [Google Scholar]
- Harvey RP, Lai D, Elliott D, Biben C, Solloway M, Prall O, et al. Homeodomain factor Nkx2-5 in heart development and disease. Cold Spring Harb Symp Quant Biol. 2002;67:107–114. doi: 10.1101/sqb.2002.67.107. [DOI] [PubMed] [Google Scholar]
- Harvey RP, Meilhac SM, Buckingham ME. Landmarks and lineages in the developing heart. Circ Res. 2009;104:1235–1237. doi: 10.1161/CIRCRESAHA.109.199729. [DOI] [PubMed] [Google Scholar]
- Hatcher CJ, Goldstein MM, Mah CS, Delia CS, Basson CT. Identification and localization of TBX5 transcription factor during human cardiac morphogenesis. Dev Dyn. 2000;219:90–95. doi: 10.1002/1097-0177(200009)219:1<90::AID-DVDY1033>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Labeit S, Poustka A, King TR, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- High FA, Jain R, Stoller JZ, Antonucci NB, Lu MM, Loomes KM, et al. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue-tissue interactions during outflow tract development. J Clin Invest. 2009;119:1986–1996. doi: 10.1172/JCI38922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M, Ono K, Morimoto T, Kawamura T, Wada H, Kita T, et al. FOG-2 competes with GATA-4 for transcriptional coactivator p300 and represses hypertrophic responses in cardiac myocytes. J Biol Chem. 2004;279:37640–37650. doi: 10.1074/jbc.M401737200. [DOI] [PubMed] [Google Scholar]
- Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, et al. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- Hofner M, Höllrigl A, Puz S, Stary M, Weitzer G. Desmin stimulates differentiation of cardiomyocytes and upregulation of brachyury and nkx2.5. Differentiation. 2007;75:605–615. doi: 10.1111/j.1432-0436.2007.00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höllrigl A, Hofner M, Stary M, Weitzer G. Differentiation of cardiomyocytes requires functional serine residues within the amino-terminal domain of desmin. Differentiation. 2007;75:616–626. doi: 10.1111/j.1432-0436.2007.00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. Tbx5 is essential for heart development. Development. 1999;126:1739–1751. doi: 10.1242/dev.126.8.1739. [DOI] [PubMed] [Google Scholar]
- Hosoda T, Zheng H, Cabral-da-Silva M, Sanada F, Ide-Iwata N, Ogorek B, et al. Human cardiac stem cell differentiation is regulated by a mircrine mechanism. Circulation. 2011;123:1287–1296. doi: 10.1161/CIRCULATIONAHA.110.982918. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson AG, Sater AK. Features of embryonic induction. Development. 1988;104:341–359. doi: 10.1242/dev.104.3.341. [DOI] [PubMed] [Google Scholar]
- Jamali M, Karamboulas C, Rogerson PJ, Skerjanc IS. BMP signaling regulates Nkx2-5 activity during cardiomyogenesis. FEBS Lett. 2001a;509:126–130. doi: 10.1016/s0014-5793(01)03151-9. [DOI] [PubMed] [Google Scholar]
- Jamali M, Rogerson PJ, Wilton S, Skerjanc IS. Nkx2-5 activity is essential for cardiomyogenesis. J Biol Chem. 2001b;276:42252–42258. doi: 10.1074/jbc.M107814200. [DOI] [PubMed] [Google Scholar]
- Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55:1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Drysdale TA, Evans T. A role for GATA-4/5/6 in the regulation of Nkx2.5 expression with implications for patterning of the precardiac field. Dev Biol. 1999;216:57–71. doi: 10.1006/dbio.1999.9469. [DOI] [PubMed] [Google Scholar]
- Johansson BM, Wiles MV. Evidence for involvement of activin A and bone morphogenetic protein 4 in mammalian mesoderm and hematopoietic development. Mol Cell Biol. 1995;15:141–151. doi: 10.1128/mcb.15.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston PV, Sasano T, Mills K, Evers R, Lee ST, Smith RR, et al. Engraftment, differentiation, and functional benefits of autologous cardiosphere-derived cells in porcine ischemic cardiomyopathy. Circulation. 2009;120:1075–1083. doi: 10.1161/CIRCULATIONAHA.108.816058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaichi S, Takaya T, Morimoto T, Sunagawa Y, Kawamura T, Ono K, et al. Cyclin-dependent kinase 9 forms a complex with GATA4 and is involved in the differentiation of mouse ES cells into cardiomyocytes. J Cell Physiol. 2011;226:248–254. doi: 10.1002/jcp.22336. [DOI] [PubMed] [Google Scholar]
- Kajstura J, Urbanek K, Rota M, Bearzi C, Hosoda T, Bolli R, et al. Cardiac stem cells and myocardial disease. J Mol Cell Cardiol. 2008;45:505–513. doi: 10.1016/j.yjmcc.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Kajstura J, Urbanek K, Perl S, Hosoda T, Zheng H, Ogorek B, et al. Cardiomyogenesis in the adult human heart. Circ Res. 2010;107:305–315. doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Nathan E, Xu SM, Tzahor E, Black BL. Isl1 is a direct transcriptional target of Forkhead transcription factors in second-heart-field-derived mesoderm. Dev Biol. 2009;334:513–522. doi: 10.1016/j.ydbio.2009.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappen C, Salbaum JM. Identification of regulatory elements in the Isl1 gene locus. Int J Dev Biol. 2009;53:935–946. doi: 10.1387/ijdb.082819ck. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circ Res. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Keren-Politansky A, Keren A, Bengal E. Neural ectoderm-secreted FGF initiates the expression of Nkx2.5 in cardiac progenitors via a p38 MAPK/CREB pathway. Dev Biol. 2009;335:374–384. doi: 10.1016/j.ydbio.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Kim Y, Nirenberg M. Drosophila NK-homeobox genes. Proc Natl Acad Sci USA. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A, Herrmann BG. The Brachyury gene encodes a novel DNA binding protein. EMBO J. 1993;12:3211–3220. doi: 10.1002/j.1460-2075.1993.tb05990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A, Herrmann BG. Immunohistochemical analysis of the Brachyury protein in wild-type and mutant mouse embryos. Dev Biol. 1994;161:179–193. doi: 10.1006/dbio.1994.1019. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Miyagawa-Tomita S, Inoue T, Kanno J, Saga Y. Mesp1-nonexpressing cells contribute to the ventricular cardiac conduction system. Dev Dyn. 2006;235:395–402. doi: 10.1002/dvdy.20640. [DOI] [PubMed] [Google Scholar]
- Klaus A, Saga Y, Taketo MM, Tzahor E, Birchmeier W. Distinct roles of Wnt/beta-catenin and Bmp signaling during early cardiogenesis. Proc Natl Acad Sci USA. 2007;104:18531–18536. doi: 10.1073/pnas.0703113104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskoff V, Lacaud G, Schwantz S, Fehling HJ, Keller G. Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc Natl Acad Sci USA. 2005;102:13170–13175. doi: 10.1073/pnas.0501672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn EN, Wu SM. Origin of cardiac progenitor cells in the developing and postnatal heart. J Cell Physiol. 2010;225:321–325. doi: 10.1002/jcp.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuisk IR, Li H, Tran D, Capetanaki Y. A single MEF2 site governs desmin transcription in both heart and skeletal muscle during mouse embryogenesis. Dev Biol. 1996;174:1–13. doi: 10.1006/dbio.1996.0046. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci USA. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Yatskievych TA, Antin PB. Regulation of avian cardiac myogenesis by activin/TGFbeta and bone morphogenetic proteins. Dev Biol. 1998;204:407–419. doi: 10.1006/dbio.1998.9094. [DOI] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135:193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- Lauss M, Stary M, Tischler J, Egger G, Puz S, Bader-Allmer A, et al. Single inner cell masses yield embryonic stem cell lines differing in lifr expression and their developmental potential. Biochem Biophys Res Commun. 2005;331:1577–1586. doi: 10.1016/j.bbrc.2005.04.068. [DOI] [PubMed] [Google Scholar]
- Lee Y, Shioi T, Kasahara H, Jobe SM, Wiese RJ, Markham BE, et al. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer D, Golden J, Kowall NW. Myocyte-specific enhancer binding factor 2C expression in human brain development. Neuroscience. 1994;63:1067–1079. doi: 10.1016/0306-4522(94)90573-8. [DOI] [PubMed] [Google Scholar]
- Lengerke C, Wingert R, Beeretz M, Grauer M, Schmidt AG, Konantz M, et al. Interactions between Cdx genes and retinoic acid modulate early cardiogenesis. Dev Biol. 2011;354:134–142. doi: 10.1016/j.ydbio.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Leri A, Kajstura J, Anversa P. Cardiac stem cells and mechanisms of myocardial regeneration. Physiol Rev. 2005;85:1373–1416. doi: 10.1152/physrev.00013.2005. [DOI] [PubMed] [Google Scholar]
- Leri A, Hosoda T, Kajstura J, Anversa P, Rota M. Identification of a coronary stem cell in the human heart. J Mol Med. 2011;10:947–959. doi: 10.1007/s00109-011-0769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TS, Cheng K, Lee ST, Matsushita S, Davis D, Malliaras K, et al. Cardiospheres recapitulate a niche-like microenvironment rich in stemness and cell-matrix interactions, rationalizing their enhanced functional potency for myocardial repair. Stem Cells. 2010;28:2088–2098. doi: 10.1002/stem.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Aggarwal VS, Nowotschin S, Bondarev A, Lipner S, Morrow BE. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev Biol. 2008;316:524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberatore CM, Searcy Schrick RD, Vincent EB, Yutzey KE. Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev Biol. 2002;244:243–256. doi: 10.1006/dbio.2002.0604. [DOI] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Lien CL, Wu C, Mercer B, Webb R, Richardson JA, Olson EN. Control of early cardiac-specific transcription of Nkx2-5 by a GATA-dependent enhancer. Development. 1999;126:75–84. doi: 10.1242/dev.126.1.75. [DOI] [PubMed] [Google Scholar]
- Lien CL, McAnally J, Richardson JA, Olson EN. Cardiac-specific activity of an Nkx2-5 enhancer requires an evolutionarily conserved Smad binding site. Dev Biol. 2002;244:257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Limana F, Zacheo A, Mocini D, Mangoni A, Borsellino G, Diamantini A, et al. Identification of myocardial and vascular precursor cells in human and mouse epicardium. Circ Res. 2007;101:1255–1265. doi: 10.1161/CIRCRESAHA.107.150755. [DOI] [PubMed] [Google Scholar]
- Limana F, Capogrossi MC, Germani A. The epicardium in cardiac repair: from the stem cell view. Pharmacol Ther. 2011;129:82–96. doi: 10.1016/j.pharmthera.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, et al. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci USA. 2007;104:9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke A, Muller P, Nurzynska D, Casarsa C, Torella D, Nascimbene A, et al. Stem cells in the dog heart are self-renewing, clonogenic, and multipotent and regenerate infarcted myocardium, improving cardiac function. Proc Natl Acad Sci USA. 2005;102:8966–8971. doi: 10.1073/pnas.0502678102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993a;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993b;119:969. doi: 10.1242/dev.119.3.969. [DOI] [PubMed] [Google Scholar]
- Liu Z, Li T, Liu Y, Jia Z, Li Y, Zhang C, et al. WNT signaling promotes Nkx2.5 expression and early cardiomyogenesis via downregulation of Hdac1. Biochim Biophys Acta. 2009;1793:300–311. doi: 10.1016/j.bbamcr.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Long JA. The Rise of Fishes: 500 Million Years of Evolution. Johns Hopkins University Press; Baltimore: 1996. [Google Scholar]
- Lopez-Sanchez C, Climent V, Schoenwolf GC, Alvarez IS, Garcia-Martinez V. Induction of cardiogenesis by Hensen’s node and fibroblast growth factors. Cell Tissue Res. 2002;309:237–249. doi: 10.1007/s00441-002-0567-2. [DOI] [PubMed] [Google Scholar]
- Lough J, Barron M, Brogley M, Sugi Y, Bolender DL, Zhu X. Combined BMP-2 and FGF-4, but neither factor alone, induces cardiogenesis in non-precardiac embryonic mesoderm. Dev Biol. 1996;178:198–202. doi: 10.1006/dbio.1996.0211. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Ma Q, Zhou B, Pu WT. Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev Biol. 2008;323:98–104. doi: 10.1016/j.ydbio.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet M, Purcell NH, Sargent MA, York AJ, Bueno OF, Molkentin JD. DUSP6 (MKP3) null mice show enhanced ERK1/2 phosphorylation at baseline and increased myocyte proliferation in the heart affecting disease susceptibility. J Biol Chem. 2008;283:31246–31255. doi: 10.1074/jbc.M806085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet M, Lynch JM, Sanna B, York AJ, Zheng Y, Molkentin JD. Cdc42 is an antihypertrophic molecular switch in the mouse heart. J Clin Invest. 2009;119:3079–3088. doi: 10.1172/JCI37694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansergh FC, Daly CS, Hurley AL, Wride MA, Hunter SM, Evans MJ. Gene expression profiles during early differentiation of mouse embryonic stem cells. BMC Dev Biol. 2009;9:5. doi: 10.1186/1471-213X-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BL, Kimelman D. Brachyury establishes the embryonic mesodermal progenitor niche. Genes Dev. 2010;24:2778–2783. doi: 10.1101/gad.1962910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JF, Schwarz JJ, Olson EN. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc Natl Acad Sci USA. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CM, Meeson AP, Robertson SM, Hawke TJ, Richardson JA, Bates S, et al. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004;265:262–275. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2:320–331. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Martynova MG. Possible cellular mechanisms of heart muscle growth in invertebrates. Ann N Y Acad Sci. 1995;752:149–157. doi: 10.1111/j.1749-6632.1995.tb17418.x. [DOI] [PubMed] [Google Scholar]
- Martynova MG. Proliferation and differentiation processes in the heart muscle elements in different phylogenetic groups. Int Rev Cytol. 2004;235:215–250. doi: 10.1016/S0074-7696(04)35005-9. [DOI] [PubMed] [Google Scholar]
- Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Ikeda K, Nakatani K, Sakabe M, Yamagishi T, Nakanishi T, et al. Induction of initial cardiomyocyte alpha-actin–smooth muscle alpha-actin–in cultured avian pregastrula epiblast: a role for nodal and BMP antagonist. Dev Dyn. 2005;233:1419–1429. doi: 10.1002/dvdy.20477. [DOI] [PubMed] [Google Scholar]
- Matsuura K, Nagai T, Nishigaki N, Oyama T, Nishi J, Wada H, et al. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J Biol Chem. 2004;279:11384–11391. doi: 10.1074/jbc.M310822200. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, Fiordaliso F, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, et al. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- Molina G, Vogt A, Bakan A, Dai W, de Oliveira Queiroz P, Znosko W, et al. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat Chem Biol. 2009;5:680–687. doi: 10.1038/nchembio.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Antos C, Mercer B, Taigen T, Miano JM, Olson EN. Direct activation of a GATA6 cardiac enhancer by Nkx2.5: evidence for a reinforcing regulatory network of Nkx2.5 and GATA transcription factors in the developing heart. Dev Biol. 2000;217:301–309. doi: 10.1006/dbio.1999.9544. [DOI] [PubMed] [Google Scholar]
- Montanaro F, Liadaki K, Volinski J, Flint A, Kunkel LM. Skeletal muscle engraftment potential of adult mouse skin side population cells. Proc Natl Acad Sci USA. 2003;100:9336–9341. doi: 10.1073/pnas.1133179100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzen K, Nagai R, Komuro I. A role for bone morphogenetic protein signaling in cardiomyocyte differentiation. Trends Cardiovasc Med. 2002;12:263–269. doi: 10.1016/s1050-1738(02)00172-x. [DOI] [PubMed] [Google Scholar]
- Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- Moore CS, Mjaatvedt CH, Gearhart JD. Expression and function of activin beta A during mouse cardiac cushion tissue formation. Dev Dyn. 1998;212:548–562. doi: 10.1002/(SICI)1097-0177(199808)212:4<548::AID-AJA8>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Moorman AF, Christoffels VM. Cardiac chamber formation: development, genes, and evolution. Physiol Rev. 2003;83:1223–1267. doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Moretti A, Lam J, Evans SM, Laugwitz KL. Cardiovascular development: towards biomedical applicability : biology of Isl1 (+) cardiac progenitor cells in development and disease. Cell Mol Life Sci. 2007;64:674–682. doi: 10.1007/s00018-007-6520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Bellin M, Jung CB, Thies TM, Takashima Y, Bernshausen A, et al. Mouse and human induced pluripotent stem cells as a source for multipotent Isl1+ cardiovascular progenitors. FASEB J. 2010;24:700–711. doi: 10.1096/fj.09-139477. [DOI] [PubMed] [Google Scholar]
- Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DY, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci USA. 2008;105:17830–17835. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- Muller WE, Wiens M, Adell T, Gamulin V, Schroder HC, Muller IM. Bauplan of urmetazoa: basis for genetic complexity of metazoa. Int Rev Cytol. 2004a;235:53–92. doi: 10.1016/S0074-7696(04)35002-3. [DOI] [PubMed] [Google Scholar]
- Muller WE, Wiens M, Muller IM, Schroder HC. The chemokine networks in sponges: potential roles in morphogenesis, immunity and stem cell formation. Prog Mol Subcell Biol. 2004b;34:103–143. doi: 10.1007/978-3-642-18670-7_5. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Domian IJ, Chien KR. Stem cell models of cardiac development and disease. Annu Rev Cell Dev Biol. 2010;26:667–687. doi: 10.1146/annurev-cellbio-100109-103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers DC, Fishman GI. Molecular and functional maturation of the murine cardiac conduction system. Trends Cardiovasc Med. 2003;13:289–295. doi: 10.1016/s1050-1738(03)00119-1. [DOI] [PubMed] [Google Scholar]
- Nadal-Ginard B, Kajstura J, Leri A, Anversa P. Myocyte death, growth, and regeneration in cardiac hypertrophy and failure. Circ Res. 2003;92:139–150. doi: 10.1161/01.res.0000053618.86362.df. [DOI] [PubMed] [Google Scholar]
- Nagai T, Tanaka-Ishikawa M, Aikawa R, Ishihara H, Zhu W, Yazaki Y, et al. Cdc42 plays a critical role in assembly of sarcomere units in series of cardiac myocytes. Biochem Biophys Res Commun. 2003;305:806–810. doi: 10.1016/s0006-291x(03)00838-6. [DOI] [PubMed] [Google Scholar]
- Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, et al. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci USA. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita N, Bielinska M, Wilson DB. Cardiomyocyte differentiation by GATA-4-deficient embryonic stem cells. Development. 1997;124:3755–3764. doi: 10.1242/dev.124.19.3755. [DOI] [PubMed] [Google Scholar]
- Nascone N, Mercola M. An inductive role for the endoderm in Xenopus cardiogenesis. Development. 1995;121:515–523. doi: 10.1242/dev.121.2.515. [DOI] [PubMed] [Google Scholar]
- Nawshad A, Hay ED. TGFbeta3 signaling activates transcription of the LEF1 gene to induce epithelial mesenchymal transformation during mouse palate development. J Cell Biol. 2003;163:1291–1301. doi: 10.1083/jcb.200306024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls V, Drenckhahn D. The versatility of microvascular pericytes: from mesenchyme to smooth muscle? Histochemistry. 1993;99:1–12. doi: 10.1007/BF00268014. [DOI] [PubMed] [Google Scholar]
- Nehls V, Denzer K, Drenckhahn D. Pericyte involvement in capillary sprouting during angiogenesis in situ. Cell Tissue Res. 1992;270:469–474. doi: 10.1007/BF00645048. [DOI] [PubMed] [Google Scholar]
- Nemer G, Nemer M. Cooperative interaction between GATA5 and NF-ATc regulates endothelial-endocardial differentiation of cardiogenic cells. Development. 2002;129:4045–4055. doi: 10.1242/dev.129.17.4045. [DOI] [PubMed] [Google Scholar]
- Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C, Steinbeisser H, De Robertis EM. Mesodermal patterning by a gradient of the vertebrate homeobox gene goosecoid. Science. 1994;263:817–820. doi: 10.1126/science.7905664. [DOI] [PubMed] [Google Scholar]
- Nishii K, Morimoto S, Minakami R, Miyano Y, Hashizume K, Ohta M, et al. Targeted disruption of the cardiac troponin T gene causes sarcomere disassembly and defects in heartbeat within the early mouse embryo. Dev Biol. 2008;322:65–73. doi: 10.1016/j.ydbio.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Niu Z, Yu W, Zhang SX, Barron M, Belaguli NS, Schneider MD, et al. Conditional mutagenesis of the murine serum response factor gene blocks cardiogenesis and the transcription of downstream gene targets. J Biol Chem. 2005;280:32531–32538. doi: 10.1074/jbc.M501372200. [DOI] [PubMed] [Google Scholar]
- Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol. 2007;19:618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Noseda M, Peterkin T, Simoes FC, Patient R, Schneider MD. Cardiopoietic factors: extracellular signals for cardiac lineage commitment. Circ Res. 2011;108:129–152. doi: 10.1161/CIRCRESAHA.110.223792. [DOI] [PubMed] [Google Scholar]
- Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci USA. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Chi X, Bradfute SB, Mishina Y, Pocius J, Michael LH, et al. Cardiac muscle plasticity in adult and embryo by heart-derived progenitor cells. Ann N Y Acad Sci. 2004;1015:182–189. doi: 10.1196/annals.1302.015. [DOI] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oota S, Saitou N. Phylogenetic relationship of muscle tissues deduced from superimposition of gene trees. Mol Biol Evol. 1999;16:856–867. doi: 10.1093/oxfordjournals.molbev.a026170. [DOI] [PubMed] [Google Scholar]
- Ott HC, Matthiesen TS, Brechtken J, Grindle S, Goh SK, Nelson W, et al. The adult human heart as a source for stem cells: repair strategies with embryonic-like progenitor cells. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl. 1):S27–S39. doi: 10.1038/ncpcardio0771. [DOI] [PubMed] [Google Scholar]
- Oyama T, Nagai T, Wada H, Naito AT, Matsuura K, Iwanaga K, et al. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. J Cell Biol. 2007;176:329–341. doi: 10.1083/jcb.200603014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Osugi T, Afanasiev OK, Pabon L, Reinecke H, Murry CE. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010;5:e11134. doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur P, Lasche M, Eisenberg LM, Kuhl M. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- Passier R, Mummery C. Origin and use of embryonic and adult stem cells in differentiation and tissue repair. Cardiovasc Res. 2003;58:324–335. doi: 10.1016/s0008-6363(02)00770-8. [DOI] [PubMed] [Google Scholar]
- Penrose R. Cycles of Time. The Bodley Head Random House; London: 2010. [Google Scholar]
- Perez-Pomares JM, Phelps A, Sedmerova M, Carmona R, Gonzalez-Iriarte M, Munoz-Chapuli R, et al. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs) Dev Biol. 2002;247:307–326. doi: 10.1006/dbio.2002.0706. [DOI] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. GATA-6 maintains BMP-4 and Nkx2 expression during cardiomyocyte precursor maturation. EMBO J. 2003;22:4260–4273. doi: 10.1093/emboj/cdg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner-Fischer M, Levin S, Zipori D. The origins of mesenchymal stromal cell heterogeneity. Stem Cell Rev. 2011;7:560–568. doi: 10.1007/s12015-011-9229-7. [DOI] [PubMed] [Google Scholar]
- Pfister O, Mouquet F, Jain M, Summer R, Helmes M, Fine A, et al. CD31-but Not CD31+ cardiac side population cells exhibit functional cardiomyogenic differentiation. Circ Res. 2005;97:52–61. doi: 10.1161/01.RES.0000173297.53793.fa. [DOI] [PubMed] [Google Scholar]
- Popescu LM, Gherghiceanu M, Manole CG, Faussone-Pellegrini MS. Cardiac renewing: interstitial Cajal-like cells nurse cardiomyocyte progenitors in epicardial stem cell niches. J Cell Mol Med. 2009;13:866–886. doi: 10.1111/j.1582-4934.2009.00758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Wythe JD, Liu J, Cartry J, Vogler G, Mohapatra B, et al. Tinman/Nkx2-5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J Cell Biol. 2011;193:1181–1196. doi: 10.1083/jcb.201006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaini F, Urbanek K, Beltrami AP, Finato N, Beltrami CA, Nadal Ginard B, et al. Chimerism of the transplanted heart. N Engl J Med. 2002;346:5–15. doi: 10.1056/NEJMoa012081. [DOI] [PubMed] [Google Scholar]
- Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Raff M. Adult stem cell plasticity: fact or artifact? Annu Rev Cell Dev Biol. 2003;19:1–22. doi: 10.1146/annurev.cellbio.19.111301.143037. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T, et al. The transcription factor NF-ATc is essential for cardiac valve formation. Nature. 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- Rawles ME. The heart-forming regions of the early chick blastoderm. Physiol Zool. 1943;16:22–42. [Google Scholar]
- Reecy JM, Li X, Yamada M, DeMayo FJ, Newman CS, Harvey RP, et al. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development. 1999;126:839–849. doi: 10.1242/dev.126.4.839. [DOI] [PubMed] [Google Scholar]
- Reisbach G, Bartke I, Kempkes B, Kostka G, Ellwart J, Birner A, et al. Characterization of hemopoietic cell populations from human cord blood expressing c-kit. Exp Hematol. 1993;21:74–79. [PubMed] [Google Scholar]
- Renlund N, O’Neill FH, Zhang L, Sidis Y, Teixeira J. Activin receptor-like kinase-2 inhibits activin signaling by blocking the binding of activin to its type II receptor. J Endocrinol. 2007;195:95–103. doi: 10.1677/JOE-07-0281. [DOI] [PubMed] [Google Scholar]
- Riazi AM, Takeuchi JK, Hornberger LK, Zaidi SH, Amini F, Coles J, et al. NKX2-5 regulates the expression of beta-catenin and GATA4 in ventricular myocytes. PLoS One. 2009;4:e5698. doi: 10.1371/journal.pone.0005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishniw M, Carmel BP. Atrioventricular valvular insufficiency and congestive heart failure in a carpet python. Aust Vet J. 1999;77:580–583. doi: 10.1111/j.1751-0813.1999.tb13193.x. [DOI] [PubMed] [Google Scholar]
- Robb L, Tam PP. Gastrula organiser and embryonic patterning in the mouse. Semin Cell Dev Biol. 2004;15:543–554. doi: 10.1016/j.semcdb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development. 2005;132:3405–3417. doi: 10.1242/dev.01913. [DOI] [PubMed] [Google Scholar]
- Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer AS. Major steps in vertebrate evolution. Science. 1967;158:1629–1637. doi: 10.1126/science.158.3809.1629. [DOI] [PubMed] [Google Scholar]
- Romer AS, Parsons TS. The Vertebrate Body. Holt-Saunders International; Philadelphia: 1977. [Google Scholar]
- Rones MS, McLaughlin KA, Raffin M, Mercola M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 2000;127:3865–3876. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- Rotheneder H, Geymayer S, Haidweger E. Transcription factors of the Sp1 family: interaction with E2F and regulation of the murine thymidine kinase promoter. J Mol Biol. 1999;293:1005–1015. doi: 10.1006/jmbi.1999.3213. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Epigenetic status of human embryonic stem cells. Nat Genet. 2005;37:585–587. doi: 10.1038/ng1556. [DOI] [PubMed] [Google Scholar]
- Sadek H, Hannack B, Choe E, Wang J, Latif S, Garry MG, et al. Cardiogenic small molecules that enhance myocardial repair by stem cells. Proc Natl Acad Sci USA. 2008;105:6063–6068. doi: 10.1073/pnas.0711507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM. MesP1: a novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122:2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med. 2000;10:345–352. doi: 10.1016/s1050-1738(01)00069-x. [DOI] [PubMed] [Google Scholar]
- Salisch S, Klar M, Thurisch B, Bungert J, Dame C. Gata4 and Sp1 regulate expression of the erythropoietin receptor in cardiomyocytes. J Cell Mol Med. 2011;15:1963–1972. doi: 10.1111/j.1582-4934.2010.01193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Shields B, Davies ML, Muller J, Wakeman JA. Brachyury confers cancer stem cell characteristics on colorectal cancer cells. Int J Cancer Int J Cancer. 2011 doi: 10.1002/ijc.26029. [DOI] [PubMed] [Google Scholar]
- Sater AK, Jacobson AG. The specification of heart mesoderm occurs during gastrulation in Xenopus laevis. Development. 1989;105:821–830. doi: 10.1242/dev.105.4.821. [DOI] [PubMed] [Google Scholar]
- Schlange T, Andree B, Arnold HH, Brand T. BMP2 is required for early heart development during a distinct time period. Mech Dev. 2000;91:259–270. doi: 10.1016/s0925-4773(99)00311-1. [DOI] [PubMed] [Google Scholar]
- Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, et al. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet. 2011;7:e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Rhaesa A. Tardigrades—are they really miniaturized dwarfs? Zool Anz. 2001;240:549–555. [Google Scholar]
- Schneider VA, Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl AM, Kirby ML. Signals controlling neural crest contributions to the heart. Wiley Interdiscip Rev Syst Biol Med. 2009;1:220–227. doi: 10.1002/wsbm.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Schroder HC, Krasko A, Batel R, Skorokhod A, Pahler S, Kruse M, et al. Stimulation of protein (collagen) synthesis in sponge cells by a cardiac myotrophin-related molecule from Suberites domuncula. FASEB J. 2000;14:2022–2031. doi: 10.1096/fj.00-0043com. [DOI] [PubMed] [Google Scholar]
- Schultheiss TM, Xydas S, Lassar AB. Induction of avian cardiac myogenesis by anterior endoderm. Development. 1995;121:4203–4214. doi: 10.1242/dev.121.12.4203. [DOI] [PubMed] [Google Scholar]
- Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;11:451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- Shirai M, Imanaka-Yoshida K, Schneider MD, Schwartz RJ, Morisaki T. T-box 2, a mediator of Bmp-Smad signaling, induced hyaluronan synthase 2 and Tgfbeta2 expression and endocardial cushion formation. Proc Natl Acad Sci USA. 2009;106:18604–18609. doi: 10.1073/pnas.0900635106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa MS, Vasconcelos M, Sampaio AC, Cravo RM, Linhares VL, Hochgreb T, et al. The evolutionary origin of cardiac chambers. Dev Biol. 2005;277:1–15. doi: 10.1016/j.ydbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Singh MK, Li Y, Li S, Cobb RM, Zhou D, Lu MM, et al. Gata4 and Gata5 cooperatively regulate cardiac myocyte proliferation in mice. J Biol Chem. 2010;285:1765–1772. doi: 10.1074/jbc.M109.038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu IO, Zhao X, Duester G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev Dyn. 2008;237:1627–1635. doi: 10.1002/dvdy.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerjanc IS, Petropoulos H, Ridgeway AG, Wilton S. Myocyte enhancer factor 2C and Nkx2-5 up-regulate each other’s expression and initiate cardiomyogenesis in P19 cells. J Biol Chem. 1998;273:34904–34910. doi: 10.1074/jbc.273.52.34904. [DOI] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- Smith RR, Barile L, Cho HC, Leppo MK, Hare JM, Messina E, et al. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- Smits AM, van Vliet P, Metz CH, Korfage T, Sluijter JP, Doevendans PA, et al. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nat Protoc. 2009;4:232–243. doi: 10.1038/nprot.2008.229. [DOI] [PubMed] [Google Scholar]
- Snyder M, Huang XY, Zhang JJ. Stat3 directly controls the expression of Tbx5, Nkx2.5, and GATA4 and is essential for cardiomyocyte differentiation of P19CL6 cells. J Biol Chem. 2010;285:23639–23646. doi: 10.1074/jbc.M110.101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solloway MJ, Harvey RP. Molecular pathways in myocardial development: a stem cell perspective. Cardiovasc Res. 2003;58:264–277. doi: 10.1016/s0008-6363(03)00286-4. [DOI] [PubMed] [Google Scholar]
- Soudais C, Bielinska M, Heikinheimo M, MacArthur CA, Narita N, Saffitz JE, et al. Targeted mutagenesis of the transcription factor GATA-4 gene in mouse embryonic stem cells disrupts visceral endoderm differentiation in vitro. Development. 1995;121:3877–3888. doi: 10.1242/dev.121.11.3877. [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Cai C, Kotecha S, Latinkic B, Cooper B, Towers N, et al. Regulation of the tinman homologues in Xenopus embryos. Dev Biol. 2000;227:65–79. doi: 10.1006/dbio.2000.9891. [DOI] [PubMed] [Google Scholar]
- Spring J, Yanze N, Josch C, Middel AM, Winninger B, Schmid V. Conservation of Brachyury, Mef2, and Snail in the myogenic lineage of jellyfish: a connection to the mesoderm of bilateria. Dev Biol. 2002;244:372–384. doi: 10.1006/dbio.2002.0616. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Ivey KN. Potential of stem-cell-based therapies for heart disease. Nature. 2006;441:1097–1099. doi: 10.1038/nature04961. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Stary M, Pasteiner W, Summer A, Hrdina A, Eger A, Weitzer G. Parietal endoderm secreted SPARC promotes early cardiomyogenesis in vitro. Exp Cell Res. 2005;310:331–341. doi: 10.1016/j.yexcr.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Abboud N, Desilets S, Nury D, Cowan C, Puceat M. Interplay of Oct4 with Sox2 and Sox17: a molecular switch from stem cell pluripotency to specifying a cardiac fate. J Cell Biol. 2009;186:665–673. doi: 10.1083/jcb.200901040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJ, Lai D, et al. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262:206–224. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- Stern CD. Gastrulation: from Cells to Embryo. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2004. [Google Scholar]
- Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, et al. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, et al. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc Natl Acad Sci USA. 2006;103:10294–10299. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Lord B, Schulze PC, Fryer RM, Sarang SS, Gullans SR, et al. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation. 2003;107:1912–1916. doi: 10.1161/01.CIR.0000064899.53876.A3. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini YN, Greene KS, Craven M, Spealman A, Breitbach M, Smith J, et al. c-kit expression identifies cardiovascular precursors in the neonatal heart. Proc Natl Acad Sci USA. 2009;106:1808–1813. doi: 10.1073/pnas.0808920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, Beddington RS. Establishment and organization of germ layers in the gastrulating mouse embryo. Ciba Found Symp. 1992;165:27–49. doi: 10.1002/9780470514221.ch3. [DOI] [PubMed] [Google Scholar]
- Tam PP, Parameswaran M, Kinder SJ, Weinberger RP. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: the role of ingression and tissue movement during gastrulation. Development. 1997;124:1631–1642. doi: 10.1242/dev.124.9.1631. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999a;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Wechsler SB, Lee IW, Yamasaki N, Lawitts JA, Izumo S. Complex modular cis-acting elements regulate expression of the cardiac specifying homeobox gene Csx/Nkx2.5. Development. 1999b;126:1439–1450. doi: 10.1242/dev.126.7.1439. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Schinke M, Liao HS, Yamasaki N, Izumo S. Nkx2.5 and Nkx2.6, homologs of Drosophila tinman, are required for development of the pharynx. Mol Cell Biol. 2001;21:4391–4398. doi: 10.1128/MCB.21.13.4391-4398.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Ashihara E, Takehara N, Nomura T, Honsho S, Nakagami T, et al. Clonally amplified cardiac stem cells are regulated by Sca-1 signaling for efficient cardiovascular regeneration. J Cell Sci. 2007;120:1791–1800. doi: 10.1242/jcs.006122. [DOI] [PubMed] [Google Scholar]
- Terami H, Hidaka K, Katsumata T, Iio A, Morisaki T. Wnt11 facilitates embryonic stem cell differentiation to Nkx2.5-positive cardiomyocytes. Biochem Biophys Res Commun. 2004;325:968–975. doi: 10.1016/j.bbrc.2004.10.103. [DOI] [PubMed] [Google Scholar]
- Thattaliyath BD, Firulli BA, Firulli AB. The basic-helix-loop-helix transcription factor HAND2 directly regulates transcription of the atrial naturetic peptide gene. J Mol Cell Cardiol. 2002a;34:1335–1344. doi: 10.1006/jmcc.2002.2085. [DOI] [PubMed] [Google Scholar]
- Thattaliyath BD, Livi CB, Steinhelper ME, Toney GM, Firulli AB. HAND1 and HAND2 are expressed in the adult-rodent heart and are modulated during cardiac hypertrophy. Biochem Biophys Res Commun. 2002b;297:870–875. doi: 10.1016/s0006-291x(02)02297-0. [DOI] [PubMed] [Google Scholar]
- Thiele J, Varus E, Wickenhauser C, Kvasnicka HM, Lorenzen J, Gramley F, et al. Mixed chimerism of cardiomyocytes and vessels after allogeneic bone marrow and stem-cell transplantation in comparison with cardiac allografts. Transplantation. 2004;77:1902–1905. doi: 10.1097/01.tp.0000127591.34203.8e. [DOI] [PubMed] [Google Scholar]
- Thomas T, Yamagishi H, Overbeek PA, Olson EN, Srivastava D. The bHLH factors, dHAND and eHAND, specify pulmonary and systemic cardiac ventricles independent of left-right sidedness. Dev Biol. 1998;196:228–236. doi: 10.1006/dbio.1998.8849. [DOI] [PubMed] [Google Scholar]
- Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh-Finkel L, Zeisel A, Brodt-Ivenshitz M, Shamai A, Yao Z, Seger R, et al. BMP-mediated inhibition of FGF signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development. 2010;137:2989–3000. doi: 10.1242/dev.051649. [DOI] [PubMed] [Google Scholar]
- Tomita Y, Matsumura K, Wakamatsu Y, Matsuzaki Y, Shibuya I, Kawaguchi H, et al. Cardiac neural crest cells contribute to the dormant multipotent stem cell in the mammalian heart. J Cell Biol. 2005;170:1135–1146. doi: 10.1083/jcb.200504061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torella D, Ellison GM, Nadal-Ginard B, Indolfi C. Cardiac stem and progenitor cell biology for regenerative medicine. Trends Cardiovasc Med. 2005;15:229–236. doi: 10.1016/j.tcm.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Tseng AS, Engel FB, Keating MT. The GSK-3 inhibitor BIO promotes proliferation in mammalian cardiomyocytes. Chem Biol. 2006;13:957–963. doi: 10.1016/j.chembiol.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi T, Maeda J, Shin CH, Ivey KN, Black BL, Olson EN, et al. Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev Biol. 2011;351:62–69. doi: 10.1016/j.ydbio.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu CT, Yang TC, Tsai HJ. Nkx2.7 and Nkx2.5 function redundantly and are required for cardiac morphogenesis of zebrafish embryos. PLoS One. 2009;4:e4249. doi: 10.1371/journal.pone.0004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, et al. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueyama T, Kasahara H, Ishiwata T, Nie Q, Izumo S. Myocardin expression is regulated by Nkx2.5, and its function is required for cardiomyogenesis. Mol Cell Biol. 2003a;23:9222–9232. doi: 10.1128/MCB.23.24.9222-9232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueyama T, Kasahara H, Ishiwata T, Yamasaki N, Izumo S. Csm, a cardiac-specific isoform of the RNA helicase Mov10l1, is regulated by Nkx2.5 in embryonic heart. J Biol Chem. 2003b;278:28750–28757. doi: 10.1074/jbc.M300014200. [DOI] [PubMed] [Google Scholar]
- Urbanek K, Quaini F, Tasca G, Torella D, Castaldo C, Nadal-Ginard B, et al. Intense myocyte formation from cardiac stem cells in human cardiac hypertrophy. Proc Natl Acad Sci USA. 2003;100:10440–10445. doi: 10.1073/pnas.1832855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek K, Rota M, Cascapera S, Bearzi C, Nascimbene A, De Angelis A, et al. Cardiac stem cells possess growth factor-receptor systems that after activation regenerate the infarcted myocardium, improving ventricular function and long-term survival. Circ Res. 2005;97:663–673. doi: 10.1161/01.RES.0000183733.53101.11. [DOI] [PubMed] [Google Scholar]
- Urbanek K, Cesselli D, Rota M, Nascimbene A, De Angelis A, Hosoda T, et al. Stem cell niches in the adult mouse heart. Proc Natl Acad Sci USA. 2006;103:9226–9231. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet P, Roccio M, Smits AM, van Oorschot AA, Metz CH, van Veen TA, et al. Progenitor cells isolated from the human heart: a potential cell source for regenerative therapy. Neth Heart J. 2008;16:163–169. doi: 10.1007/BF03086138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpoucke G, Goossens S, De Craene B, Gilbert B, van Roy F, Berx G. GATA-4 and MEF2C transcription factors control the tissue-specific expression of the alphaT-catenin gene CTNNA3. Nucleic Acids Res. 2004;32:4155–4165. doi: 10.1093/nar/gkh727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, Firulli BA, Conway SJ, Firulli AB. Cooperative interaction of Nkx2.5 and Mef2c transcription factors during heart development. Dev Dyn. 2008;237:3809–3819. doi: 10.1002/dvdy.21803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, et al. Foxh1 is essential for development of the anterior heart field. Dev Cell. 2004;7:331–345. doi: 10.1016/j.devcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Wagner M, Siddiqui MA. Signal transduction in early heart development (I): cardiogenic induction and heart tube formation. Exp Biol Med. 2007;232:852–865. [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, et al. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Wang T, Altimiras J, Axelsson M. Intracardiac flow separation in an in situ perfused heart from Burmese python Python molurus. J Exp Biol. 2002;205:2715–2723. doi: 10.1242/jeb.205.17.2715. [DOI] [PubMed] [Google Scholar]
- Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, et al. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a Micro-RNA-mediated mechanism. Dev Cell. 2010;19:903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hao J, Hong CC. Cardiac induction of embryonic stem cells by a small molecule inhibitor of Wnt/beta-catenin signaling. ACS Chem Biol. 2011;6:192–197. doi: 10.1021/cb100323z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Benson DW, Yano S, Akagi T, Yoshino M, Murray JC. Two novel frameshift mutations in NKX2.5 result in novel features including visceral inversus and sinus venosus type ASD. J Med Genet. 2002;39:807–811. doi: 10.1136/jmg.39.11.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci USA. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, et al. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winbanks CE, Wang B, Beyer C, Koh P, White L, Kantharidis P, et al. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem. 2011;286:13805–13814. doi: 10.1074/jbc.M110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Wu X, Ding S, Ding Q, Gray NS, Schultz PG. Small molecules that induce cardiomyogenesis in embryonic stem cells. J Am Chem Soc. 2004;126:1590–1591. doi: 10.1021/ja038950i. [DOI] [PubMed] [Google Scholar]
- Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006a;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- Wu X, Quondamatteo F, Lefever T, Czuchra A, Meyer H, Chrostek A, et al. Cdc42 controls progenitor cell differentiation and beta-catenin turnover in skin. Genes Dev. 2006b;20:571–585. doi: 10.1101/gad.361406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132:537–543. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Revelli JP, Eichele G, Barron M, Schwartz RJ. Expression of chick Tbx-2, Tbx-3, and Tbx-5 genes during early heart development: evidence for BMP2 induction of Tbx2. Dev Biol. 2000;228:95–105. doi: 10.1006/dbio.2000.9927. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Yamagishi C, Nakagawa O, Harvey RP, Olson EN, Srivastava D. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev Biol. 2001;239:190–203. doi: 10.1006/dbio.2001.0417. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 1999;13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- Yatskievych TA, Ladd AN, Antin PB. Induction of cardiac myogenesis in avian pregastrula epiblast: the role of the hypoblast and activin. Development. 1997;124:2561–2570. doi: 10.1242/dev.124.13.2561. [DOI] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, et al. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H, Longmore G, Neumann D, Yoshimura A, Lodish HF. Structure, function, and activation of the erythropoietin receptor. Blood. 1993;81:2223–2236. [PubMed] [Google Scholar]
- Yuasa S, Itabashi Y, Koshimizu U, Tanaka T, Sugimura K, Kinoshita M, et al. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat Biotechnol. 2005;23:607–611. doi: 10.1038/nbt1093. [DOI] [PubMed] [Google Scholar]
- Yuasa S, Onizuka T, Shimoji K, Ohno Y, Kageyama T, Yoon SH, et al. Zac1 is an essential transcription factor for cardiac morphogenesis. Circ Res. 2010;106:1083–1091. doi: 10.1161/CIRCRESAHA.109.214130. [DOI] [PubMed] [Google Scholar]
- Zandstra PW, Le HV, Daley GQ, Griffith LG, Lauffenburger DA. Leukemia inhibitory factor (LIF) concentration modulates embryonic stem cell self-renewal and differentiation independently of proliferation. Biotechnol Bioeng. 2000;69:607–617. [PubMed] [Google Scholar]
- Zeisberg EM, Kalluri R. Origins of cardiac fibroblasts. Circ Res. 2010;107:1304–1312. doi: 10.1161/CIRCRESAHA.110.231910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Miura T, Luo Y, Bhattacharya B, Condie B, Chen J, et al. Properties of pluripotent human embryonic stem cells BG01 and BG02. Stem Cells. 2004;22:292–312. doi: 10.1634/stemcells.22-3-292. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang CS, Liu YC, Yang XQ, Xiong SH, Wen Y, et al. Isolation, culture and characterization of cardiac progenitor cells derived from human embryonic heart tubes. Cells Tissues Organs. 2009;190:194–208. doi: 10.1159/000204750. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Yang YJ, Qian HY, Wang H, Xu H. Very small embryonic-like stem cells (VSELs)—a new promising candidate for use in cardiac regeneration. Ageing Res Rev. 2011;10:173–177. doi: 10.1016/j.arr.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008a;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, von Gise A, Ma Q, Rivera-Feliciano J, Pu WT. Nkx2-5- and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochem Biophys Res Commun. 2008b;375:450–453. doi: 10.1016/j.bbrc.2008.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Sasse J, McAllister D, Lough J. Evidence that fibroblast growth factors 1 and 4 participate in regulation of cardiogenesis. Dev Dyn. 1996;207:429–438. doi: 10.1002/(SICI)1097-0177(199612)207:4<429::AID-AJA7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Zipori D. Mesenchymal stem cells: harnessing cell plasticity to tissue and organ repair. Blood Cells Mol Dis. 2004a;33:211–215. doi: 10.1016/j.bcmd.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Zipori D. The nature of stem cells: state rather than entity. Nat Rev Genet. 2004b;5:873–878. doi: 10.1038/nrg1475. [DOI] [PubMed] [Google Scholar]
- Zipori D. The stem state: mesenchymal plasticity as a paradigm. Curr Stem Cell Res Ther. 2006;1:95–102. doi: 10.2174/157488806775269133. [DOI] [PubMed] [Google Scholar]