Abstract
Background
Genetic defects in components of inflammasomes can cause autoinflammation. Biallelic loss-of-function mutations in dipeptidyl peptidase 9 (DPP9), a negative regulator of the NLRP1 and CARD8 inflammasomes, have recently been shown to cause an inborn error of immunity characterized by pancytopenia, skin manifestations, and increased susceptibility to infections.
Objective
We sought to study the molecular basis of autoinflammation in a patient with severe infancy-onset hyperinflammation associated with signs of fulminant hemophagocytic lymphohistiocytosis.
Methods
Using heterologous cell models as well as patient cells, we performed genetic, immunologic, and molecular investigations to identify the genetic cause and to assess the impact of the identified mutation on inflammasome activation.
Results
The patient exhibited pancytopenia with decreased neutrophils and T, B, and natural killer cells, and markedly elevated levels of lactate dehydrogenase, ferritin, soluble IL-2 receptor, and triglycerides. In addition, serum levels of IL-1β and IL-18 were massively increased, consistent with inflammasome activation. Genetic analysis revealed a previously undescribed de novo mutation in DPP9 (c.755G>C, p.Arg252Pro) affecting a highly conserved amino acid residue. The mutation led to destabilization of the DPP9 protein as shown in transiently transfected HEK293T cells and in patient-derived induced pluripotent stem cells. Using functional inflammasome assays in HEK293T cells, we demonstrated that mutant DPP9 failed to restrain the NLRP1 and CARD8 inflammasomes, resulting in constitutive inflammasome activation. These findings suggest that the Arg252Pro DPP9 mutation acts in a dominant-negative manner.
Conclusions
A de novo mutation in DPP9 leads to severe infancy-onset autoinflammation because of unleashed inflammasome activation.
Keywords: Inborn error of immunity, hemophagocytic lymphohistiocytosis, autoinflammation, inflammasome, DPP9, NLRP1, CARD8, proinflammatory cytokines, IL-18, IL-1β
Introduction
Inflammasomes are cytosolic multimeric signaling complexes that integrate non-self and damage recognition with inflammatory responses1–3. Inflammasome assembly is initiated by certain sensor proteins that recognize pathogen-derived or endogenous danger signals, resulting in the recruitment and activation of pro-caspase-1. Most inflammasomes use the adaptor apoptosis-associated speck-like protein containing a CARD (ASC), which serves to amplify signaling in a threshold-dependent manner. Activated caspase-1 cleaves a restricted number of substrates, including the proinflammatory cytokines IL-1β and IL-18 that are rendered bioactive on processing, as well as the pore-forming molecule gasdermin D (GSDMD), which triggers pyroptotic cell death1–3. Given that inadvertent inflammasome engagement can lead to potentially detrimental inflammatory responses and cell death, inflammasome activation needs to be tightly regulated. Indeed, aberrant inflammasome activation due to mutations in the inflammasome sensors, NLRP3 (NLR family pyrin domain containing 3), NLRC4 (NLR family CARD domain containing 4), NLRP1 (NLR family pyrin domain containing 1), and pyrin, underlies a spectrum of autoinflammatory diseases4–6. Most of these mutations constitute gain-of-function mutations that result in the activation of these sensors in the absence of cognate triggers. At the same time, certain inflammasome sensors are maintained in an inactive state by a number of regulatory proteins. One such regulatory protein is dipeptidyl peptidase 9 (DPP9), encoded by the DPP9 gene. DPP9 functions as an N-terminal dipeptide post-proline-cleaving serine protease that specifically restrains activation of the NLRP1 and CARD8 (caspase recruitment domain family member 8) inflammasomes7–11. Recently, biallelic loss-of-function mutations in DPP9 have been described in 4 patients from 3 families with increased susceptibility to infection, recurrent fever, and skin manifestations12. We report the first case of a de novo monoallelic dominant-negative mutation in DPP9 leading to unleashed inflammasome activation in an infant with hemophagocytic lymphohistiocytosis (HLH)-like autoinflammation.
Results and Discussion
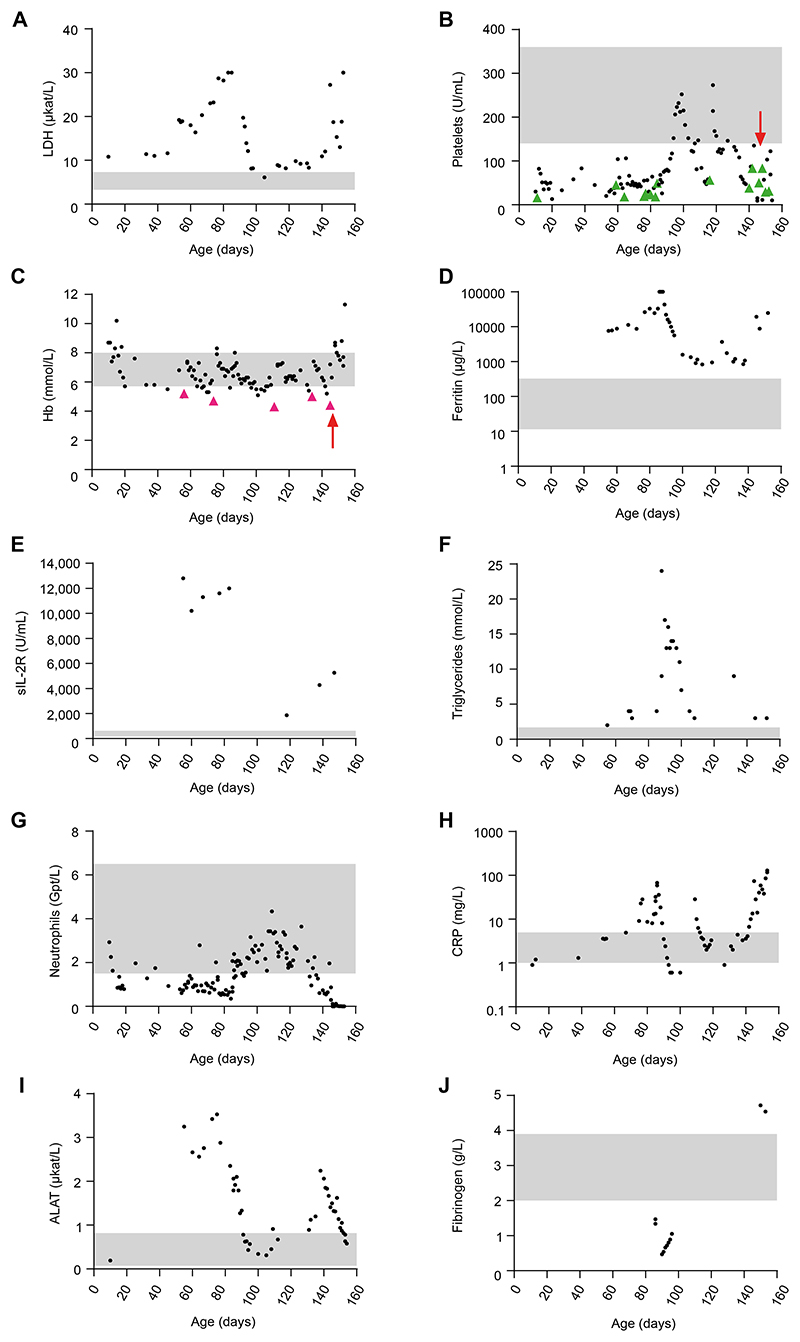
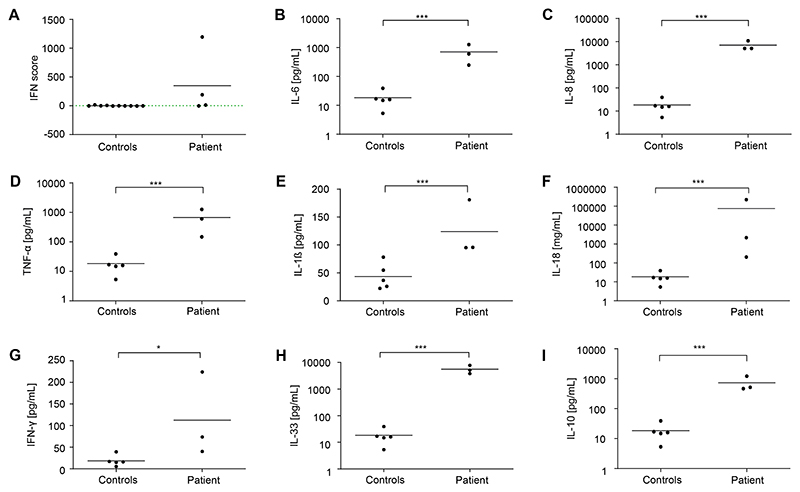
The patient was born at term to nonconsanguineous healthy parents of German origin following an uneventful pregnancy. At the age of 12 days, he was found to have hematomas on the head, neck, and toes, as well as a grade 1 intraventricular hemorrhage due to thrombocytopenia (16 platelets/nL), which was treated with platelet transfusion. The patient and his mother tested negative for antiplatelet antibodies. At the age of 2 months, the infant presented with trilinear pancytopenia and markedly elevated lactate dehydrogenase (LDH) (Fig 1; see also Table S1), requiring repeated platelet and erythrocyte transfusions. A bone marrow analysis revealed increased erythropoiesis and megakaryopoiesis, as well as left-shifted granulopoiesis, without signs of malignancy, excluding primary bone marrow failure. The patient initially showed mild splenomegaly that progressed to pronounced hepatosplenomegaly over the following weeks, along with elevated liver enzymes and massively increased serum levels of ferritin, soluble IL-2 receptor, and triglycerides (Fig 1). Immunophenotyping revealed markedly reduced neutrophils and B, natural killer, and T cells, in particular CD31CD81 cytotoxic T cells (Fig 1 and Table S1). The combination of fever, hepatosplenomegaly, pancytopenia, elevated inflammatory markers, hyperferritinemia, hypertriglyceridemia, hypofibrinogenemia, and increased soluble IL-2 receptor was suggestive of HLH. All test results for viral, bacterial, or fungal infections were negative. Perforin expression and degranulation of natural killer cells after IL-2 stimulation, as well as HLA-DR expression of CD41 and CD81 T cells, were unremarkable, arguing against known primary causes of HLH. Panel sequencing did not reveal disease-causing mutations in genes causing primary bone marrow failure, immunodeficiency, autoinflammatory syndromes, or primary HLH. The patient was started on methylprednisolone (2 mg/kg/d, intravenously) followed by intravenous immunoglobulins (1 g/kg/d) without improvement. Because of persistent fever and neutropenia, antibiotic therapies with piperacillin/tazobactam and fluconazole were given until microbiologic test results came back negative. Treatment with anakinra (2-4 mg/kg/d) over 1 week was also ineffective, and methylprednisolone (10 mg/kg/d) was restarted for 2 days, followed by prednisolone (2 mg/kg/d). During placement of a central venous catheter, the boy experienced a bleeding episode with volume loss and transient renal failure. Because of a strong interferon signature in blood (Fig 2A), ruxolitinib (0.5 mg/kg/d) was added to the prednisolone treatment. This regimen resulted in a transient increase in platelets and neutrophils, along with a decrease in ferritin and triglycerides. However, 2 weeks later, the patient experienced a flare, possibly because of a parainfluenza infection. A cytokine panel revealed strongly elevated serum levels of proinflammatory cytokines, including IL-6, IL-8, TNF-a, IFN-g, IL-1β, and IL-18 (Fig 2). In view of refractory pancytopenia and hyperinflammation in the 5-month-old infant, a clinical decision was made to proceed with allogeneic hematopoietic stem cell transplantation. During treosulfan-based reduced-toxicity conditioning, the patient developed staphylococcus sepsis and reactivation of parainfluenza with hyperinflammatory flare and massive capillary leakage, which led to hyperkalemia and cardiac arrest on day 6 post–hematopoietic stem cell transplantation.
Figure 1. Laboratory findings.
A-J, Blood parameters of the patient taken at the indicated day of life. The gray shaded areas mark the normal range for each parameter. B and C, Green and purple arrowheads indicate platelet and erythrocyte transfusions, respectively, and the red arrows mark allogeneic hematopoietic stem cell transplantation. ALAT, Alanine-aminotransferase; CRP, C-reactive protein; Hb, hemoglobin; sIL-2R, soluble IL-2 receptor.
Figure 2. Interferon signature and serum cytokines.
A, IFN signature in patient PBMCs measured at different time points. An IFN score of 12.49 (dashed green line) indicates the median IFN score of 10 healthy controls + 2 SD. B-I, Cytokine levels in patient sera collected at different time points, compared with 5 healthy controls. Data are presented as mean of 3 technical replicates. The Mann-Whitney U test was used. ***P < .001; *P < .05.
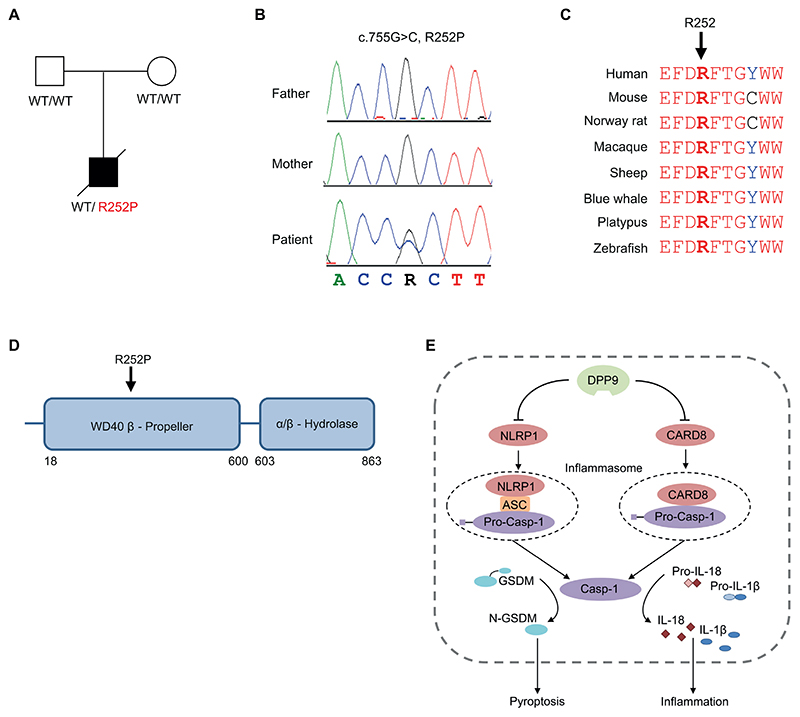
Trio whole-exome sequencing revealed a heterozygous mutation in DPP9 (c.755G>C, R252P) in the patient, which was not detected in his parents (Fig 3A and B). This variant was not reported in the gnomAD database and is predicted to impair function (see Fig S1). Multiple sequence alignment revealed complete conservation of amino acid residue 252 of the DPP9 protein across different species (Fig 3C). DPP9 encodes DPP9, which restrains formation of the NLPR1 and CARD8 inflammasomes (Fig 3D and E) by forming a ternary complex with either 2 CARD8 or 2 NLRP1 molecules, a unique feature of these sensors8–11. This keeps the C-terminal UPA (conserved in UNC5, PIDD, and ankyrin)-CARD fragments of NLRP1 or CARD8, which are required for inflammasome activation, in check. Given the critical negative regulatory role of DPP9 in inflammasome activation, we hypothesized that the autoinflammatory symptoms observed in our patient were caused by an impairment of the DPP9 function.
Figure 3. Identification of a de novo DPP9 mutation.
A, Pedigree of the family. B, Electropherograms showing a heterozygous DPP9 mutation in the patient, which is absent in his healthy parents. C, Multiple sequence alignment showing high conservation of R252 within the DPP9 protein. D, Schematic of domain architecture of DPP9 with the R252P mutation indicated. E, Model depicting the negative regulatory function of DPP9 on the NRLP1 and CARD8 inflammasomes.
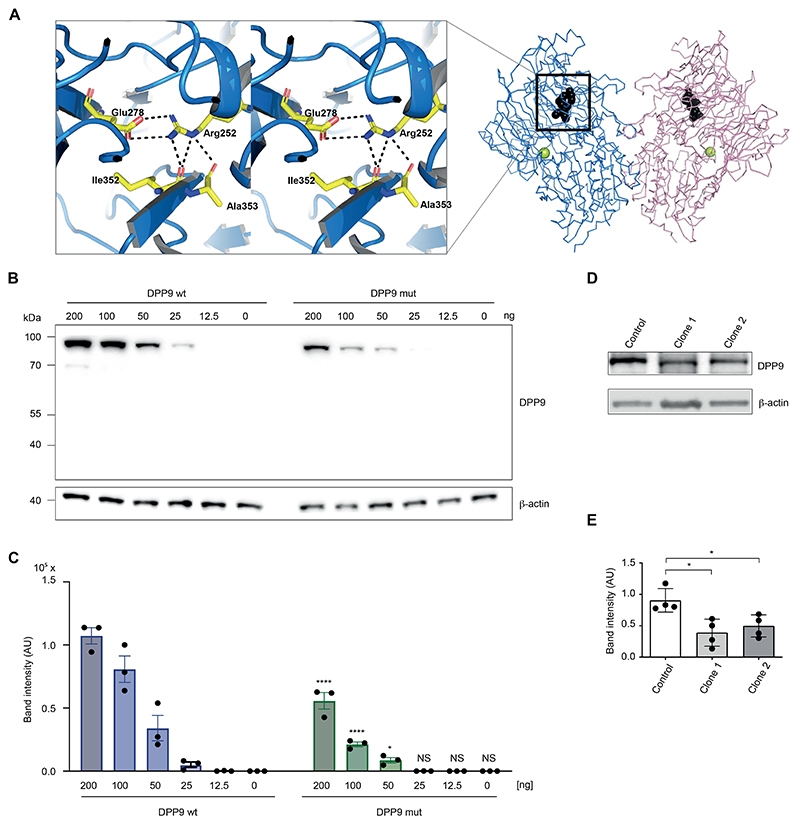
To explore whether the R252P mutation could have an impact on a critical structural aspect of DPP9, we investigated this residue in the structural model of DPP9. The protein segment 242-258, which includes 2 (E248 and E249) of the 4 glutamic acids, forms a pocket and surface cleft by wrapping around the Arg252 side chain (Fig 4A). It is lined with mostly hydrophobic amino acid residues, Ala242, Ile246, Phe250, Phe253, and Trp257, except Glu278. The Arg252 side chain forms a salt bridge with Glu278 and is engaged in hydrogen bonds with carbonyl oxygens of Ile352, Ala353, and Thr254 (Fig 4A). Proline is unable to establish these interactions and may be a nucleus of instability, affecting the local environment and the molecule as a whole. We therefore assessed the impact of the R252P mutation on DPP9 protein abundance. Transient transfection of wild-type and mutant DPP9 at decreasing concentrations in HEK293T cells revealed greatly reduced protein levels of mutant DPP9, with the 2 highest expression levels (200 and 100 ng) studied reaching only 52% and 26%, respectively, of wild-type DPP9 protein (Fig 4B and C). DPP9 expression was also reduced in patient derived induced pluripotent stem cells (iPSCs) (Fig 4D and E), which were generated because of the unavailability of blood cells for functional studies, confirming that R252P compromises DPP9 protein stability in patient cells.
Figure 4. Impact of the DPP9 mutation R252P on protein structure and stability.
A, Model of DPP9 focusing on the R252 pocket (PDB: 6EOR, 6EOQ, 6QZV7). Left, The plot displays the Arg252 pocket in stereo. Right, A chain trace of the DPP9 dimer is shown with Arg252 (black). The position of the active site Ser730 is indicated (yellow sphere). B, HEK293T cells were transiently transfected with plasmids encoding for wild-type DPP9 (wt) or the DPP9 mutant (R252P) at the indicated concentrations. Expression levels were determined 24 hours later via immunoblotting. One representative immunoblot of 3 independent experiments is depicted. C, The band intensities of 3 independent experiments were quantified and summarized as mean ± SEM. A 2-way ANOVA was performed followed by the Šidák multiple comparisons test. ****P < .0001; *P < .05. D, One representative immunoblot of 4 independent experiments showing DPP9 expression in patient-derived iPSCs compared with a control iPSC line. β-actin served as loading control. E, The band intensities, normalized to β-actin, of 4 independent experiments were quantified and summarized as mean values. The Mann-Whitney U test was used. *P < .05. AU, Arbitrary unit; NS, not significant.
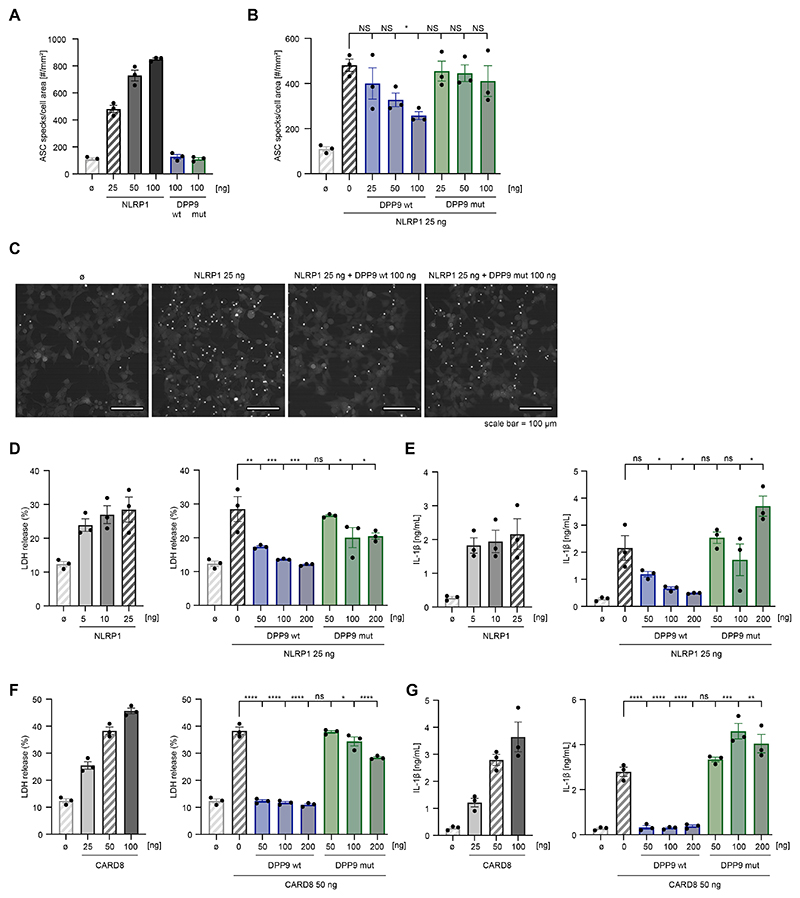
We next investigated the impact of DPP9 R252P on its capacity to restrain NLRP1 inflammasome activation using functional inflammasome assays in HEK293T cells stably expressing ASC with a red fluorescent protein (ASC-RFP) tag. Transient expression of inflammasome sensors in these cells results in the formation of an ASC speck (also known as pyroptosome), which serves as the signaling hub of the inflammasome complex. This phenomenon can be visualized by fluorescence microscopy and can serve as a quantitative readout of inflammasome activation. Consistent with this notion, expressing low amounts of NLRP1 already led to a considerable increase in ASC speck formation in cells, whereas the expression of wild-type or mutant DPP9 on their own had no effect (Fig 5A). To study whether DPP9 restrains NLRP1 activity to trigger inflammasome activation, we coexpressed increasing concentrations of DPP9 together with NLRP1 and studied the impact on ASC speck formation (Fig 5B and C). This resulted in decreased levels of ASC speck formation, reaching about half of the activity at the highest amount of DPP9 being expressed. However, when coexpressing increasing amounts of the mutated DPP9 construct (R252P), the ability of NLRP1 to induce ASC speck formation was not inhibited. This is in line with the critical inhibitory scaffold formed by DPP9 to restrain NLRP1 inflammasome activation10,11. We further determined pyroptosis and IL-1β secretion as a function of NLRP1 in HEK293T cells additionally transfected with NLRP1, ASC, caspase-1, pro-IL-1β and GSDMD. Transient transfection of increasing amounts of NLRP1 led to LDH release or IL-1β secretion in a dose-dependent manner, consistent with pyroptotic cell death and IL-1β maturation induced by inflammasome activation (Fig 5D and E). Activation of the NRLP1 was strongly inhibited by coexpression of wild-type DPP9, confirming the inhibitory effect of DPP9 on inflammasome activation (Fig 5D and E). In contrast, mutant DPP9 was unable to inhibit inflammasome activation or pyroptosis induced by NLRP1, as shown by persistent and strongly enhanced LDH release and IL-1β secretion (Fig 5D and E).
Figure 5. DPP9 R252P fails to inhibit NLRP1- and CARD8-dependent inflammasome activation.
A-C, HEK293T cells expressing fluorescently tagged ASC were transfected with the indicated plasmids or left untreated (B). After 24 hours, cells were imaged to quantify ASC speck formation. A and B, The number of ASC specks was quantified per cell area. Data are presented as mean ± SEM of 3 independent experiments. A 1-way ANOVA was performed followed by the Dunnett multiple comparisons test: *P < .05. Note that certain control conditions are shown twice for each panel, and the corresponding bars are shaded. C, Representative fields of view of 1 representative experiment of 3 independent experiments conducted as in Fig 5B. Scale bar, 100 mm. D-G, HEK293T cells were transfected with inflammasome components and indicated amounts of NLRP1 or CARD8 and/ or wild-type or mutant (R252P) DPP9 for 24 hours and treated with 4 mM Val-boroPro for 4 hours. Cytotoxicity was determined by LDH assay and IL-1β release was measured by ELISA. Data are presented as mean ± SEM of 3 independent experiments. A 1-way ANOVA was performed followed by the Dunnett multiple comparisons test. ****P < .0001; ***P < .001; **P < .01; *P < .05. Note that certain control conditions are shown twice for each panel, and the corresponding bars are shaded. NS, Not significant.
NLRP1 is highly expressed in keratinocytes, and gain-of-function NLRP1 mutations cause autoinflammation with widespread skin dyskeratosis or respiratory papillomatosis13,14. However, our patient exhibited prominent systemic autoinflammation accompanied by massive elevation of serum LDH and panlymphopenia without any skin involvement. Unlike NLRP1, CARD8 is highly expressed in T cells and activation of the CARD8 inflammasome has been shown to cause T-cell pyroptosis15,16 suggesting that uncontrolled CARD8 inflammasome activity may have accounted for pyroptotic T-cell demise with subsequent LDH release. We therefore also examined the ability of mutant DPP9 to restrain pyroptosis induced by CARD8 in HEK293T cells. Doing so revealed similar results to the ones obtained with NLRP1: overexpression of CARD8 led to a dose-dependent increase in LDH release and IL-1β secretion and this could be strongly blunted by coexpressing wild-type, but not mutant, DPP9 (Fig 5F and G). Taken together, these findings indicate that R252P cannot restrain NLRP1 and CARD8 inflammasome activity. Although no variants in genes known to cause auto-inflammation had been detected in the patient, we cannot fully exclude that variants in yet unknown genes contribute to the disease phenotype. Parents of patients with biallelic loss-of-function mutations in DPP9 are healthy heterozygous mutation carriers12. This suggests that the heterozygous de novo DPP9 mutation described in our patient acts in a dominant-negative manner. To address this hypothesis, we transduced N/TERT-1 keratinocytes that are functional for the NLRP1 inflammasome with either a wild-type DPP9 construct, the R252P variant, or green fluorescent protein (GFP). All constructs were tagged with hemagglutinin (HA). Immunoblot analysis for the HA tag revealed that the heterologous constructs were expressed, whereas the DPP9 R252P variant displayed lower expression levels (see Fig S2A). Probing for DPP9 was unable to distinguish endogenous from heterologously expressed DPP9, yet the expression level for the DPP9 R252P transduced cells was lower compared with the wild-type variant (Fig S2A). Stimulating these cells with Val-boroPro to trigger NLRP1 inflammasome activation revealed that the DPP9 R252P transduced cells released more IL-1β than did GFP or DPP9 wild-type transduced cells (Fig S2B). Together, these results indicated that the expression of the DPP9 R252P variant exerted a dominant-negative effect on endogenous DPP9, thereby lowering the threshold for NLRP1 inflammasome activation. Thus, unleashed inflammasome activation and pyroptosis in the patient—presumably triggered by viral infections such as parainfluenza—resulted in the clinical presentation of hyperinflammation due to excessive and self-perpetuating cytokine release. The associated massive IL-18 secretion may explain why the patient did not respond to anakinra and suggests that an IL-18–targeting treatment may have been a potentially more effective therapeutic approach. In summary, we describe a de novo dominant-negative R252P mutation in DPP9 that impedes negative regulation of inflammasome activation, leading to severe early-onset autoinflammation with HLH-like features.
Methods
Study approval
Written informed consent was obtained from the parents of the patient. The study was conducted in accordance with the Declaration of Helsinki and with approval by the Ethics Committee of the Medical Faculty, Technische Universität Dresden.
Whole-exome sequencing
Exome libraries were generated using the Illumina DNA Prep with Enrichment, IDT for Illumina DNA/RNA UD Indexes, and Human Core Exome Probes (TWIST Bioscience, South San Francisco, Calif) and sequenced using the NovaSeq 6000 S1 Reagent Kit on an Illumina NovaSeq 6000 (Illumina, Berlin, Germany). Data analysis was performed using the varvis genomics platform (Limbus Medical Technologies, Rostock, Germany) for annotation of single-nucleotide variants and insertions/deletions. Sequences were mapped to the human reference genome (GRCh37/UCSC hg19).
Sanger sequencing
Genomic DNA flanking the DPP9 (NM_139159) mutation was amplified by PCR using gene-specific primers (Eurofins MWG Operon, Ebersberg, Germany; DPP9: for-CTCACCCCTTGTTGATGCTCTC, rev-ACATGCAGCTCAGAGGGC) and sequenced in both directions using the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, Mass) on a 3130xl Genetic Analyzer (Applied Biosystems). Data were analyzed using the Vector NTI software (Life Technologies, Waltham, Mass).
Generation of iPSCs
PBMCs were reprogrammed using the CytoTune-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific, Waltham, Mass) and individual clones were expanded on human embryonic stem cell-qualified Matrigel (Corning, Kaiserslautern, Germany) in mTeSR1 medium (Stem Cell Technologies, Cologne, Germany) as previously described17. Clones were tested for absence of Sendai virus RNA 8 weeks after transduction by RT-PCR using primer sequences recommended in the CytoTune-iPS 2.0 Sendai Reprogramming Kit manual. Karyotyping of 2 clones by Giemsa banding was analyzed at the Institute of Human Genetics, Jena University, Germany, as previously described17 and it revealed an intact 46,XY[cp20] chromosome set.
Cytokine analysis
Plasma was taken from blood collected in heparin tubes by centrifugation (2000 rpm) at 4°C for 20 minutes. Cytokines were measured using the LEGENDplex Human Inflammation Panel 1 (BioLegend, Koblenz, Germany) according to the manufacturer’s instructions. Data were collected on a fluorescence-activated cell sorting flow cytometer (LSRII, Becton Dickinson, Heidelberg, Germany) and analyzed with the LEGENDplex Data Analysis V8.1 software (BioLegend).
Quantitative real-time RT-PCR
Total RNA was extracted from PBMCs or from lysed whole blood using the ReliaPrep RNA Cell Miniprep System (Promega, Walldorf, Germany) followed by DNase I digestion. RNA was reverse-transcribed using the GoScript Reverse Transcription System (Promega). Gene expression was determined by quantitative real-time RT-PCR using the Taqman Universal PCRMaster Mix (Applied Biosystems) on an ABI7300 and normalized to glyceraldehyde-3-phosphate dehydrogenase (for-GAAGGTGAAGGTCGGAGTC, rev-GAAGATGGTGATGGGATTTC, and FAM-CAAGCTTCCCGTTCTCAGCC-TAMRA) and hypoxanthine phosphoribosyltransferase 1 (Hs02800695_m1; Thermo Fisher Scientific) expression. For calibration, a calibrator cDNA was included in each assay. Target genes were analyzed using predesigned TaqMan probes (Thermo Fisher Scientific) for IFI27 (Hs01086373_g1), IFI44 (Hs00951349_m1), IFI44L (Hs00915292_m1), IFIT1 (Hs01675197_m1), ISG15 (Hs01921425_s1), RSAD2 (Hs01057264_m1), and SIGLEC1 (Hs00988063_m1). The IFN score was calculated as previously described18.
Western blot analysis
HEK293T cells were seeded at 2 x 105 cells per well in a 96-well tissue culture plate. The following day, the cells were transfected with plasmids encoding the DPP9 wild type or the DPP9 R252P mutant with indicated concentrations (200 ng -12.5 ng) using GeneJuice (0.5 μL/well; Merck, Darmstadt, Germany) according to the manufacturer’s instructions. After 24 hours, cells were collected and washed 3 times with PBS. Pellets were lysed in 1 x Lämmli Lysis buffer and boiled for 5 minutes at 95°C. Samples were separated on denaturing and reducing TRIS-glycine SDS-PAGE (Thermo Fisher Scientific), blotted onto 0.2-μm nitrocellulose membranes (10600004; Cytiva, Marlborough, Mass), blocked in 3% milk, and incubated with indicated primary and corresponding secondary antibodies. Primary antibodies were used 1:1000 in 3% milk: mouse–anti-DPP9 (MAB5419-SP; R&D Systems, Minneapolis, Minn) and anti–b-actin horseradish peroxidase (sc-47778; Santa Cruz Biotechnology, Dallas, Tex).
iPSCs were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM TRIS-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM sodium orthovanadate, and 20 mM sodium fluoride) supplemented with 1 x Complete Protease Inhibitor Cocktail, 1 x PhosSTOP phosphatase inhibitors (Roche, Mannheim, Germany), and DNase I (Qiagen, Hilden, Germany). Protein concentration was determined using a BCA Kit (Thermo Fisher Scientific). Lysates were resolved in a 4% to 12% NuPAGE Bis-TRIS gel under reducing and denaturing conditions and blotted onto a nitrocellulose membrane (BA83, Sigma-Aldrich). Membranes were blocked in 5% dry milk and probed using the following antibodies: anti-DPP9 (ab42080, 1:1000; Abcam, Cambridge, UK) and anti–β-actin (A5316, 1:5000; Sigma-Aldrich, Taufkirchen, Germany). Immunoreactive signals were detected by chemiluminescence using the Lumi-Light PLUS (Roche).
Cloning
The short isoform of DPP9 (Uniprot identifier no. Q86TI2-1) was subcloned into pFUGW_blast. The DPP9 R252P point mutation was introduced via site-directed mutagenesis using Gibson assembly. Additional constructs with an N-terminal HA tag to wild-type and mutant DPP9 were cloned into pFUGW_blast using Gibson assembly.
Cell culture
HEK293T cells were cultured in Dulbecco modified Eagle medium (high-glucose, 41965062; Thermo Fisher Scientific) supplemented with 10% FCS (10270106, Thermo Fisher Scientific), 1% penicillin-streptomycin (15140163, Thermo Fisher Scientific), and 1 mM sodium pyruvate (11360088, Thermo Fisher Scientific). N/TERT-1 cells (human immortalized keratinocytes previously shown to be functional for NLRP1 [a gift from J. Rheinwald]) were cultured in a 1:2 mixture of Ham’s F12 (11765054, Thermo Fisher Scientific) and Dulbecco modified Eagle medium (high-glucose, no glutamine, no calcium; 21068028, Thermo Fisher Scientific). This mixture was supplemented with 1% nonessential amino acids (11140050, Thermo Fisher Scientific), 0.5% EpiLife defined growth supplement (S0125, Thermo Fisher Scientific), 25 μg/mL bovine pituitary extract (13028014, Thermo Fisher Scientific), 20 ng/mL epidermal growth factor (Max Planck Institute of Biochemistry CoFa, Munich, Germany), 10 mM HEPES (H0887, Sigma-Al-drich), 2 mM GlutaMAX (35050061, Thermo Fisher Scientific), 0.1 mM CaCl2 (C7902, Sigma-Aldrich), and 1% penicillin-streptomycin.
Lentiviral transduction
For the generation of lentiviruses, HEK293T cells were seeded with 1.2 x 106 cells per well in a 6-well tissue culture plate. The following day, cells were transfected with the HA-tagged constructs in the lentiviral pFUGW_blast vector and respective helper plasmids pMDLg/pRRE, pRSV-rev, and pCMV-VSV-G using PEI Max (Polysciences, Hirschberg an der Bergstrasse, Germany) as transfection reagent. After 48 hours, lentiviral supernatant was harvested and filtered using a 0.45 μm pore filter to remove any remaining cellular debris. N/TERT-1 cells were transduced in a 1:2 mixture of viral supernatant and fresh N/TERT-1 media. Transduced N/TERT-1 cells were selected 48 hours after transduction with medium containing 5 μg/mL blasticidin (A1113903; Thermo Fisher Scientific).
ASC speck assay
HEK293T cells expressing ASC-RFP were seeded at
1.5 x 105 cells per well in an Ibidi 96-well glass-bottom black plate (ibidi, Gräfelfing Germany). The following day, cells were transfected with plasmids encoding the human wild-type (DPP-WT) or mutant DPP9 (DPP-R252P) and/or human NLRP1 with the indicated concentrations (25 ng -250 ng) using GeneJuice (0.75 μL/well; Merck) according to the manufacturer’s instructions. After 24 hours, cells were imaged using the SYNENTEC Cellavista Cell Imager (SYNENTEC, Elmshorn, Germany) using an Olympus 20x/0.50 objective (Olympus, Hamburg, Germany). Total cell area was analyzed from brightfield images, and the number of ASC-RFP specks was quantified using an optimized “Count Fluo Dots” processing function.
Overexpression and inflammasome activation assays
For these experiments, HEK293T cells were seeded at 2 x 105 cells per well in a 96-well tissue culture plate. For the overexpression, cells were transfected the following day with plasmids encoding the DPP9 wild-type or DPP9 mutants (DPP9-R252P) with indicated concentrations using GeneJuice (0.5 μL/ well; Merck) according to the manufacturer’s instructions. DNA concentrations were kept constant across all conditions by stuffer DNA. After 24 hours, cells were collected and prepared for immunoblotting. For the inflammasome activation assays, cells were transfected with inflammasome components (5 ng GSDMD, 20 ng caspase-1, 2 ng ASC, and 25 ng pro-IL-1β) and indicated amounts of NLRP1 or CARD8 and DPP9 wild-type or DPP9 mutant with indicated concentrations using GeneJuice (0.75 μL/ well; Merck). After 24 hours, supernatant was removed and cells were stimulated with 4 μM Val-boroPro (also called Talabostat mesylate; B3941, ApexBio, Houston, Tex) for 4 hours.
Stimulation assay
For stimulation experiments of N/TERT-1, transgenic N/TERT-1 cells were seeded at 0.5 x 105 cells per well in a 96-well tissue culture plate. Cells were stimulated with different concentrations of Val-boroPro (0-0.5 μM, B3941; ApexBio) for 24 hours.
Cytotoxicity assay
LDH release was analyzed using the CyQUANT LDH Cytotoxicity Assay (C20300; Thermo Fisher Scientific) according to the manufacturer’s instructions. Results are presented relative to a lysis control from the same experiment with the subtraction of only medium as background.
ELISA
hIL-1β (557953; BD Biosciences, Franklin Lakes, NJ) ELISA was performed according to the supplier’s protocol.
Statistical analysis
Statistical significance was determined using the 2-tailed Student t test, Mann-Whitney U test, and ANOVA followed by post hoc tests using Microsoft Excel and GraphPad Prism (Version 9.5.1; GraphPad Software, Boston, Mass). P values less than .05 were considered to be statistically significant. Data are presented as mean ± SD or mean ± SEM as indicated.
Supplementary Material
A, PolyPhen-2 report. B, Mutation Taster prediction. The annotation in PolyPhen-2 and Mutation Taster (c.842G>C, R281P) refers to the longer isoform 2 of DPP9 (ENST00000262960.14) that has 31 additional N-terminal amino acids compared with the physiological isoform 1 (ENST00000593973.2).
A and B, N/TERT-1 keratinocytes were transduced with GFP, DPP9 wild type (wt), or DPP9 R252P (all with an N-terminal HA tag). A, Expression of these constructs was analyzed via immunoblotting, probing for DPP9 (top panel), HA (middle panel), and β-actin (bottom panel). One representative immunoblot of 3 independent experiments is depicted. B, Cells were stimulated with the indicated doses of Val-boroPro (VbP) for 24 hours and subsequently IL-1β release was measured by ELISA. NS, Not significant. Data are presented as mean ± SEM of 3 independent experiments. A 2-way ANOVA was performed followed by the Dunnett multiple comparisons test. ****P < .0001; **P < .01.
Acknowledgments
We thank the patient and his family for participation in the study. We also acknowledge the assistance of the Stem Cell Engineering Facility of the Center for Regenerative Therapies Dresden, Technische Universität Dresden.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (grant nos. CRC237 369799452/B21 to M.L.-K., CRC237 369799452/A06 to C.W., CRC237 369799452/A27 to V.H., CRC1054 210592381/B17 to V.H., and CRC1160 256073931/A01 to S.E.), the Federal Ministry of Education and Research (grant nos. BMBF GAIN 01GM2206C to M.L.-K. and BMBF GAIN 01GM2206A to S.E.), Rosemarie-Germscheid Stiftung (to C.S.), and the European Research Council grant ERC-2020-ADG ENGINES (grant no. 101018672 to V.H.).
Footnotes
Disclosure of Potential Conflict of Interest
The authors declare that they have no relevant conflicts of interest.
References
- 1.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–20. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 2.Guo H, Callaway JB, Ting JPY. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–87. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taabazuing CY, Griswold AR, Bachovchin DA. The NLRP1 and CARD8 inflammasomes. Immunol Rev. 2020;297:13–25. doi: 10.1111/imr.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harapas CR, Steiner A, Davidson S, Masters SL. An update on autoinflammatory diseases: inflammasomopathies. Curr Rheumatol Rep. 2018;20:40. doi: 10.1007/s11926-018-0750-4. [DOI] [PubMed] [Google Scholar]
- 5.Romberg N, Vogel TP, Canna SW. NLRC4 inflammasomopathies. Curr Opin Allergy Clin Immunol. 2017;17:398–404. doi: 10.1097/ACI.0000000000000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnappauf O, Chae JJ, Kastner DL, Aksentijevich I. The pyrin inflammasome in health and disease. Front Immunol. 2019;10:1745. doi: 10.3389/fimmu.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross B, Krapp S, Augustin M, Kierfersauer R, Arciniega M, Geiss-Friedlander R, et al. Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer. Proc Natl Acad Sci U S A. 2018;115:E1437–45. doi: 10.1073/pnas.1717565115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong FL, Robinson K, Teo DET, Tan K-Y, Lim C, Harapas CR, et al. Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. J Biol Chem. 2018;293:18864–78. doi: 10.1074/jbc.RA118.004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharif H, Hollingsworth LR, Griswold AR, Hsiao JC, Wang Q, Bachovchin DA, et al. Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment. Immunity. 2021;54:1392–404.:e10. doi: 10.1016/j.immuni.2021.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollingsworth LR, Sharif H, Griswold AR, Fontana P, Mintseris J, Dagbay KB, et al. DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation. Nature. 2021;592:778–83. doi: 10.1038/s41586-021-03350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M, Zhang X, Toh GA, Gong Q, Wang J, Han Z, et al. Structural and biochemical mechanisms of NLRP1 inhibition by DPP9. Nature. 2021;592:773–7. doi: 10.1038/s41586-021-03320-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harapas CR, Robinson KS, Lay K, Wong J, Moreno Traspas R, Nabavizadeh N, et al. DPP9 deficiency: an inflammasomopathy that can be rescued by lowering NLRP1/IL-1 signaling. Sci Immunol. 2022;7:eabi4611. doi: 10.1126/sciimmunol.abi4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong FL, Mamaï O, Sborgi L, Boussofara L, Hopkins R, Robinson K, et al. Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell. 2016;167:187–202.:e17. doi: 10.1016/j.cell.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Drutman SB, Haerynck F, Zhong FL, Hum D, Hernandez NJ, Belkaya S, et al. Homozygous NLRP1 gain-of-function mutation in siblings with a syndromic form of recurrent respiratory papillomatosis. Proc Natl Acad Sci U S A. 2019;116:19055–63. doi: 10.1073/pnas.1906184116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linder A, Bauernfried S, Cheng Y, Albanese M, Jung C, Keppler OT, et al. CARD8 inflammasome activation triggers pyroptosis in human T cells. EMBO J. 2020;39:e105071. doi: 10.15252/embj.2020105071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson DC, Okondo MC, Orth EL, Rao SD, Huang HC, Ball DP, et al. DPP8/9 inhibitors activate the CARD8 inflammasome in resting lymphocytes. Cell Death Dis. 2020;11:628. doi: 10.1038/s41419-020-02865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hänchen V, Kretschmer S, Wolf C, Engel K, Khattak S, Neumann K, et al. Generation of induced pluripotent stem cell lines from three patients with Aicardi-Goutières syndrome type 5 due to biallelic SAMDH1 mutations. Stem Cell Res. 2022;64:102912. doi: 10.1016/j.scr.2022.102912. [DOI] [PubMed] [Google Scholar]
- 18.Wolf C, Brück N, Koss S, Griep C, Kirschfink M, Palm-Beden K, et al. Janus kinase inhibition in complement component 1 deficiency. J Allergy Clin Immunol. 2020;146:1439–42.:e5. doi: 10.1016/j.jaci.2020.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, PolyPhen-2 report. B, Mutation Taster prediction. The annotation in PolyPhen-2 and Mutation Taster (c.842G>C, R281P) refers to the longer isoform 2 of DPP9 (ENST00000262960.14) that has 31 additional N-terminal amino acids compared with the physiological isoform 1 (ENST00000593973.2).
A and B, N/TERT-1 keratinocytes were transduced with GFP, DPP9 wild type (wt), or DPP9 R252P (all with an N-terminal HA tag). A, Expression of these constructs was analyzed via immunoblotting, probing for DPP9 (top panel), HA (middle panel), and β-actin (bottom panel). One representative immunoblot of 3 independent experiments is depicted. B, Cells were stimulated with the indicated doses of Val-boroPro (VbP) for 24 hours and subsequently IL-1β release was measured by ELISA. NS, Not significant. Data are presented as mean ± SEM of 3 independent experiments. A 2-way ANOVA was performed followed by the Dunnett multiple comparisons test. ****P < .0001; **P < .01.