Abstract
The threat of bacteria resistant to multiple antibiotics poses a major public health problem requiring immediate and coordinated action worldwide. While infectious pathogens have become increasingly resistant to commercially available drugs, antibiotic discovery programs in major pharmaceutical companies have produced no new antibiotic scaffolds in forty years. As a result, new strategies must be sought to obtain a steady supply of novel scaffolds capable of countering the spread of resistance. The bacterial ribosome is a major target for antimicrobials and is inhibited by more than half of the antibiotics used today. Recent studies showing that the ribosome is a target for several classes of ribosomally synthesized antimicrobial peptides point to ribosome-targeting peptides as a promising source of antibiotic scaffolds. In this perspective, we revisit the current paradigm of antibiotic discovery by proposing that the bacterial ribosome can be used both as a target and as a tool for the production and selection of peptide-based antimicrobials. Turning the ribosome into a high-throughput platform for the directed evolution of peptide-based antibiotics could be achieved in different ways. One possibility would be to use a combination of state-of-the-art microfluidics and genetic reprogramming techniques, which we will review briefly. If successful, this strategy has the potential to produce new classes of antibiotics for treating multidrug-resistant pathogens.
Antibiotics are the workhorses of modern medicine, but their misuse has favored the emergence of resistant bacterial strains that reduce the effectiveness of our drug arsenal over time. Strains resistant to multiple antibiotics are particularly worrying, as they can cause infections that are difficult to treat and seriously compromise clinical outcomes. The fight against antibiotic resistance is a complex problem that requires coordinated action on many levels, from the way antibiotics are prescribed to the development of new drugs. This last point is essential because a common tactic used by physicians to treat resistant pathogens is to switch a patient to a class of antibiotics with an unrelated molecular scaffold, a different mode of action and, importantly, a distinct mechanism of resistance. Without an effective discovery platform to deliver a steady supply of new antibiotic scaffolds, it will be increasingly difficult to contain the spread of resistance 1–3.
This perspective focuses on antimicrobials that inhibit the bacterial ribosome, the complex molecular machine responsible for producing the cellular proteome. Given its central role, the bacterial ribosome is a target for more than half of the antibiotics available today 4, 5. The majority of these are based on molecular scaffolds that date back to the golden era of antibiotic discovery (1950-1960), when soil-dwelling bacteria were mined extensively for antimicrobial compounds. As resistance against these early antibiotics began to spread in the 1960s, new scaffolds also became more difficult to find and the focus shifted to replicating and expanding antibiotic scaffolds from the golden era through synthetic chemistry. Derivatives produced during this period showed greater potency and improved pharmacokinetic parameters, were more effective against resistant strains and had fewer side effects than their predecessors. Unfortunately, efforts since the 1990s to identify new antibiotic scaffolds by high-throughput screening of large compound libraries or by rational drug design have failed to deliver inhibitors with the necessary physical and chemical properties to either target a broad spectrum of pathogens or to effectively penetrate Gram-negative bacteria 2. As a result, drug discovery programs have not yielded a new class of ribosome targeting antibiotic in the past forty years and novel strategies are needed to find molecules that can successfully reach and block their cellular target.
Recent studies showing that ribosomally synthesized antimicrobial peptides are potent inhibitors of the bacterial ribosome point to peptide-based antibiotics as a possible way forward 6–13. Long dismissed by the pharmaceutical industry, peptides are increasingly considered as drug candidates14, 15. They are easy to synthesize chemically, tend to show greater target selectivity and fewer side effects compared to conventional small molecules (MW < 500 Da) and, once they have been suitably modified, offer the potential for better bioavailability, membrane permeability and metabolic stability relative to protein therapeutics (MW > 5000 Da). In addition, peptides can be synthesized biologically by the ribosome, simply by encoding the desired amino acid sequence in a DNA template. This property makes peptides stand out relative to other antimicrobials and offers us a unique opportunity to revisit the antibiotic discovery paradigm that has prevailed for more than half a century.
Our basic premise is simple: the ribosome should not only be considered a target for antibiotics, but also a discovery platform for peptide-based antibiotics. Instead of screening large libraries of synthetic compounds or natural products for antimicrobial activity, we could use the ribosome to produce complex peptide libraries in vitro and select those peptides that are most effective at reaching and blocking ribosomes within bacterial cells. This could be achieved through a directed evolution approach, a well-established strategy that mimics the natural selection process to evolve proteins or nucleic acids with a set of desired properties. Directed evolution relies on similar principles as Darwinian selection: genetic diversity giving rise to varied phenotypes, a strong phenotype-genotype linkage to ensure the recovery of desired genotypes and amplification of the selected genotypes. Although the implementation of these basic principles could vary significantly, considerations such as the quality and complexity of the input library and the sensitivity and specificity of the selection procedure are the key to ensuring success. Directed evolution has been used to discover or improve therapeutic peptides 16, proteins 17 and aptamers 18. Its remarkable success was rewarded with the 2018 Nobel Prize in Chemistry. Yet, with one possible exception19, it has never been applied to the search for novel antimicrobials to block the ribosome. Here, we will review recent biochemical advances that uphold the validity of developing ribosome-targeting peptides as antibiotic scaffolds and that suggest how this could actually be achieved using directed evolution. The general approach outlined in this perspective could be implemented in a number of different ways, but we hope that its underlying principle – that the ribosome can be used both as a production and a selection tool to discover new peptide-based antibiotics – will drive further efforts to replenish our dwindling collection of antimicrobials.
Ribosomally-Synthesized Peptides are Promising Antibiotic Scaffolds
The ribosome is a validated antibiotic target
The ribosome is an extremely complex and sophisticated macromolecular machine, composed of multiple functional sites and moving parts. In bacteria, the ribosome consists of two subunits composed of about two-thirds of ribosomal RNA (rRNA) and one-third of protein. The small 30S subunit contains the decoding center, where the anticodon of an aminoacylated transfer RNA (tRNA) recognizes its cognate codon on the messenger RNA (mRNA), effectively translating the 4-nucleotide language used to store genetic information into the 20-amino acid language of proteins. The large 50S subunit includes the peptidyl transferase center (PTC), where amino acids are added one at a time to the elongating polypeptide, and the nascent polypeptide exit tunnel, a long cavity through which newly synthesized proteins transit on their way out of the ribosome. Protein synthesis is a dynamic process that requires additional protein factors to facilitate the initiation, elongation, release and recycling steps that make up the translation cycle 5. During the course of translation, factors come in and out of the ribosome, subunits move relative to each other and the ribosome inches along the mRNA, one codon at a time, in a process known as translocation.
Just like the machines we use in every day life, the ribosome has weak points that can be targeted to block the activity of the entire complex. Small molecules that efficiently block the bacterial ribosome, but fail to inhibit or reach our cytoplasmic and mitochondrial ribosomes, account for a large portion of commonly used antibiotics. Different antibiotics target different steps of the translation cycle, either by binding to various functional sites on the ribosome or by preventing factors from binding to it. The mechanisms of action of these antibiotics and the means by which bacteria become resistant to them have been reviewed extensively 5 and will not be addressed here. Instead, we will focus on peptides that block the ribosome when they are added extraneously (in trans) or that inhibit the ribosome that is translating them while they are still attached to a ribosome-bound tRNA (in cis). While the former are obvious candidates for further development as antimicrobials, the latter share enough common points with certain ribosome-targeting antibiotics to deserve a mention here.
Antimicrobial peptides block the ribosome in trans
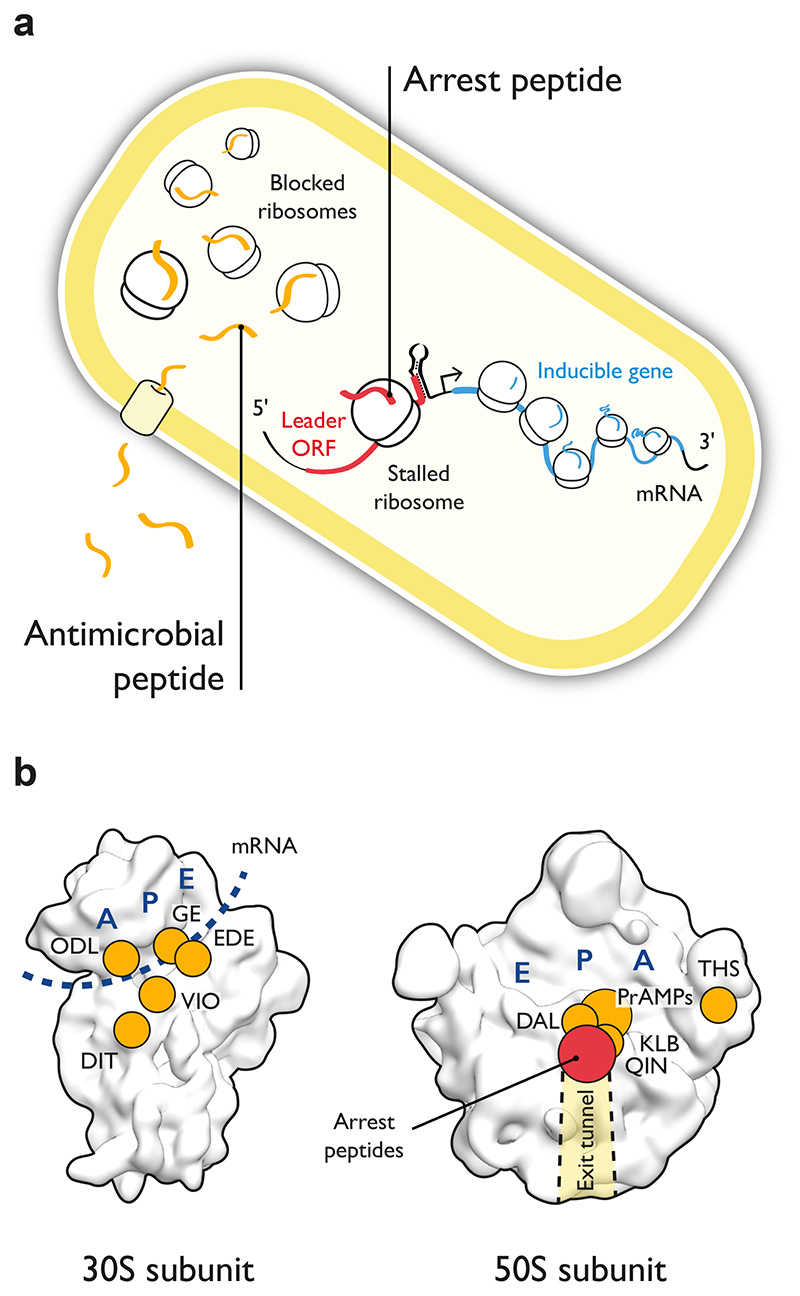
Several classes of naturally occurring ribosome-targeting antimicrobial peptides exist, based on either linear or cyclic peptide backbones (Figure 1a) 20. These peptides can be classified into three groups: (i) peptides that are produced by the ribosome, such as the recently characterized proline-rich antimicrobial peptides (PrAMPs) 21; (ii) ribosomally synthesized and post-translationally modified peptides (RiPPs) 22 like klebsazolicin or thiostrepton; or (iii) peptides that are synthesized by non-ribosomal peptide synthetases (NRPSs) 23, such as edeine or GE81112. Peptides that affect the 30S subunit target various steps of the translation cycle, including initiation (edeine 24, GE81112 25), mRNA decoding (odilorhabdins 26) or tRNA translocation (GE82832/dityromycin 27, viomycin and capreomycin 28, 29) (Figure 1b). Peptide-based antibiotics that affect the large subunit do so by targeting the PTC or the nascent polypeptide exit tunnel in order to block different processes, namely the transition from the initiation to the elongation phase of translation (Type-I PrAMPs like oncocin8, 11, 13 or Bac-7 7, 9, 12), the elongation phase (streptogramins A and B 30–33, klebsazolicin 34) or the termination phase (Type-II PrAMPs like apidaecin 6) (Figure 1b). Some peptide antibiotics, like thiostrepton, block translation by preventing the binding of key factors to the ribosome 35. With the exception of streptogramins and the anti-tuberculosis drugs capreomycin and the now-retired viomycin, peptide-based ribosomal antibiotics have yet to be used clinically as antibiotics 20.
Figure 1. Antimicrobial peptides and arrest peptides are trans- and cis-acting inhibitors of bacterial translation, respectively.
(a) Antimicrobials peptides (left part, orange) are produced by the host immune response of eukaryotes and must cross the bacterial membrane to inhibit the translational machinery in trans. Arrest peptides (center, red) are cis-acting inhibitors of the translational machinery that regulate the expression of inducible genes in bacteria and in eukaryotes. Inhibition in cis results from interactions between an arrest peptide in its nascent state and components of the large ribosomal subunit. (b) Surface representation of the E. coli 30S and 50S ribosomal subunits (PDB: 4ybb 96) showing the sites of action of various ribosome-targeting antimicrobial peptides (orange) and of arrest peptides (red). Abbreviations for antimicrobial peptides are DAL (Dalfopristin), DIT (Dityromycin), EDE (Edeine), GE (GE81112), KLB (Klebsazolicin), ODL (Odilorhabdin), PrAMPs (Proline-rich antimicrobial peptides), QIN (Quinupristin), THS (Thiostrepton), VIO (Viomycin). The A, P and E tRNA binding sites are in dark blue and the path of the mRNA on the 30S subunit is indicated with a dark blue dotted line. The nascent polypeptide exit tunnel is indicated with a shaded area.
Ribosomally synthesized antimicrobial peptides in particular are a promising source of antibiotic scaffolds, as it should be possible to produce them in an in vitro translation system at a high enough concentration to measure the inhibitory effect that they exert upon their own synthesis. Using a ribosome to produce a peptide that has the capacity to inhibit it may seem counter-intuitive at first. However, it is important to remember that this peptide cannot act on the same ribosome that is translating it and that it will only become inhibitory once a sufficient peptide concentration is reached in the in vitro translation reaction. For example, the insect-derived PrAMP Onc112 blocks luciferase production in an Escherichia coli (E. coli) in vitro translation system with a half-inhibitory concentration (IC50) 13. This value is one order of magnitude smaller than the ~10 μM of peptide that can theoretically be obtained using a commercial PURExpress system, if we assume that each ribosome in the reaction translates a minimum of 5 mRNA molecules and the concentration of ribosomes is 2 μM 36. Significantly, recent data from our group show that recombinant Onc112 can inhibit its own synthesis once a sufficient amount of peptide has been produced by this type of in vitro translation system (to be published). This means that a directed evolution strategy could be devised in which a collection of peptides is translated in vitro and a selection procedure ensures that only those peptides that inhibit translation with a given IC50 are retained. Similarly, peptides produced in vitro could accumulate at a sufficient concentration to block bacterial cell growth, as suggested by the minimum inhibitory concentration (MIC) of 10 μM measured for Onc112 13. This means that assays could be devised that test the ability of peptides produced by a cell-free translation system to reach their ribosomal target inside growing bacterial cells.
For a directed evolution strategy to succeed, the genetic diversity of the input DNA or mRNA library is critical. On the one hand, a library that is too focused on a particular region of sequence space might compromise our ability to discover truly novel inhibitory peptides. On the other hand, the selection method could fail to yield any hits if the library is too complex and variants with the desired phenotype only occur at a very low frequency. The availability of known antimicrobial peptide sequences makes it possible to prepare focused libraries comprised of millions of peptide variants differing by only a few amino acids, in addition to using truly random peptide-encoding libraries. Focused libraries based on insect or mammalian-derived PrAMPs like Onc112 8, 11, 13 or Bac-7 7, 9, 12, respectively, could help identify shortened variants with enhanced activity against the ribosomes of many or perhaps even just a single bacterial species, including resistant strains. Indeed, the high degree of conservation of the bacterial ribosome makes it a prime target for broad-spectrum antibiotics, but small sequence variations among ribosomes from different species could be exploited to develop species-specific translation inhibitors. The choice of selection scheme is important as well and care must be taken that the variants identified not only display high inhibitory activity against the ribosome in vitro (inhibitory peptides), but also retain their ability to cross the bacterial membrane and block cell growth (antimicrobial peptides). Additional selection schemes to isolate protease-resistant variants could also be established in order to produce antibiotic scaffolds that are suitable for further development. Establishing reliable protocols in future directed evolution strategies will be the key to ensuring success.
Arrest peptides inhibit translation in cis
An intriguing but so far unexploited source of translation inhibition is that of ribosome-arresting peptides, or arrest peptides for short (Figure 1a) 37–39. During their synthesis by the ribosome, arrest peptides make extensive interactions with the nascent polypeptide exit tunnel that force them to remain trapped inside the large ribosomal subunit (Figure 1b). This in turn results in the inactivation and stalling of the ribosome that was translating the arrest peptide on the mRNA. Translation inhibition mediated by arrest peptides is dependent on the nascent amino acid sequence and sometimes requires the presence of a low molecular weight ligand, with the ribosome and arrest peptide effectively acting as a metabolite sensor 37, 39, 40. As a result, ribosome stalling induced by arrest peptides can regulate the expression of other genes on the same mRNA through diverse mechanisms, both in bacteria and in eukaryotes. Biological phenomena that rely on arrest peptides in bacteria include the induction of drug-resistance genes by antibiotics of the macrolide family (e.g. Erm leader peptides) 41–45, the sensing of soluble tryptophan by a ribosome-associated TnaC peptide 46, 47, targeting of the expression of the SecA pre-protein translocase to the cell membrane by the nascent SecM polypeptide 48–50, the expression of the YidC2 membrane insertase by the MifM peptide 51–53 and the regulation of SecDF2 in low-salinity environments by the arrest peptide VemP 54. Arrest can occur during the elongation phase of translation by inhibiting peptide bond formation or aminoacyl-tRNA accommodation, or during termination by preventing peptide release from the ribosome 39, 46. It may also take place at a clearly defined location on the mRNA or at multiple sites within the same sequence 51.
The nascent polypeptide exit tunnel of the ribosome measures ~80 Å in length and has a diameter of 10-20 Å in bacteria. Several classes of antibiotics 5, natural antimicrobial peptides 20 and all arrest peptides characterized to date 37, 38 exert their effects in the upper and central sections of this tunnel, where the PTC and the exit tunnel constriction formed by the extensions of ribosomal proteins uL22 and uL4 are located. As a result, the significant structural and functional overlap between these three classes of molecules suggests that arrest peptides could help inspire new peptide-based translation inhibitors that target the exit tunnel of bacterial ribosomes. Like ribosome-targeting antimicrobial peptides, known arrest peptides are short. Only a few conserved amino acids within a stretch of 10-30 residues appear to be needed to induce translational arrest. However, one significant difference exists between antimicrobial peptides that target the ribosomal exit tunnel and arrest peptides. The latter exert their inhibitory effects in cis, while they are still covalently linked to a tRNA molecule bound to the P-site of the ribosome. The additional affinity for the ribosome provided by the tRNA moiety is likely to be partly responsible for the ability of arrest peptides to stably interact with the ribosomal exit tunnel and no ribosomal inhibition has been reported to date for an arrest peptide provided in trans. Nevertheless, focused libraries encoding variants of arrest peptides (in particular ligand-independent ones, such as enhanced SecM variants 55) could provide a suitable input for a directed evolution scheme in which peptides that gain the ability to inhibit the ribosome in trans are selected. Although there is no evidence at present that arrest peptides can successfully be converted into trans-inhibitors of translation, this option is certainly worth exploring.
Establishing an Effective Selection Procedure for Ribosome-Targeting Peptides
A strong phenotype-genotype linkage is the cornerstone of directed evolution
Known peptides that target translation interact with the ribosome using 4-20 amino acids 20. Given the set of 20 standard amino acids, the number of possible 10-amino acid peptides produced by the ribosome (and hence starting with formyl-methionine) is ~5x1011, a number greatly exceeding, for example, the >450,000 compounds made available for screening by the seven established pharmaceutical partners of the European Lead Factory 56, 57. As a result, the limiting factor in accessing this molecular diversity will be our ability to develop suitable assays to select peptides with the highest inhibitory activity against the ribosome and the best cell-penetrating ability and intracellular stability. While choosing a suitable peptide-encoding library is important, the most critical requirement to ensure a successful directed evolution experiment is to achieve a strong linkage between the desired phenotype and the genotype that has given rise to it. Possible selection strategies based on existing technologies are discussed below, both for trans and cis peptide inhibitors of translation.
Compartmentalization can be used for high-throughput selection of trans-acting peptides
Directed evolution techniques such as ribosome display 58 or mRNA display 59 have been used successfully to identify proteins or peptides with a high binding affinity for a molecular target of interest. In both cases, the link between phenotype and genotype is established directly at the molecular level, be it through the formation of a stable ribosome nascent chain complex carrying both the peptide of interest (phenotype) and the mRNA that encodes it (genotype) 58 or through the covalent linkage of the nascent peptide and its mRNA 59. While these approaches are effective at selecting proteins or peptides that bind to the exposed surface of a macromolecular target, ribosome-targeting inhibitory peptides present additional challenges. Indeed, the peptides under selection in a ribosome display or mRNA display experiment are attached to a 2.5 MDa ribosome or to a long mRNA molecule, respectively. This could lead to non-specific interactions between these molecules and the selection target (the bacterial ribosome) or, conversely, to the loss of potential binders that are unable to access buried active sites on the target. How then, might one proceed in the absence of a direct physical linkage between the peptide and the mRNA that encodes it?
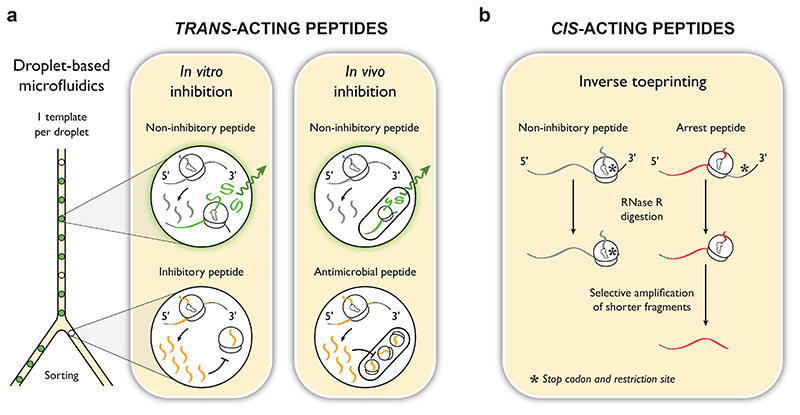
An elegant way to circumvent this limitation would be to compartmentalize the in vitro translation reaction 60 using droplet-based microfluidics 61, 62 (Figure 2a). Indeed, several recent studies have shown that picoliter-sized droplets can be used as individual reaction chambers for high-throughput analyses of libraries with therapeutic uses 63, 64. Unlike conventional screening that relies on microplates, droplet-based microfluidics offer a relatively inexpensive way of ensuring high-throughput, making it possible to study >107 mutants per experiment. In our proposed scheme, each droplet would contain multiple copies of a unique peptide-encoding sequence, an expression cassette for a reporter protein and all the molecular components necessary to perform in vitro translation. The reporter protein would provide a means to monitor expression levels within each droplet and would thus reflect the inhibitory activity of the peptides produced. The most promising peptides could later be specifically selected by means of microfluidic sorting systems 65. In this way, templates encoding the peptides of interest would be pooled and analyzed by next-generation sequencing, leading to the large-scale identification of trans-acting protein synthesis inhibitors, including peptides that directly block the ribosome. An efficient counter-selection procedure would be needed to eliminate transcription inhibitors and peptides that target components of the translational machinery other than the ribosome, such as translation factors or aminoacyl-tRNA synthetases. An additional selection system could then be devised to isolate ribosome-targeting peptides with antimicrobial activity, by placing the expression reporter cassette inside bacteria 66–68 contained within the droplets or within liposomes that mimic the bacterial cell wall 69. This would be effective as a secondary strategy to eliminate peptides that display inhibitory activity in vitro but fail to inhibit bacterial cell growth.
Figure 2. Possible selection schemes for identifying trans- or cis-acting peptides that inhibit bacterial translation.
(a) Approaches for the identification of trans-acting peptides capable of in vitro translation inhibition or in vivo cell growth inhibition (orange). The proposed microfluidic strategy allows the sorting of picoliter-sized droplets based on the level of expression of a fluorescent reporter protein (green). Each droplet contains multiple copies of a unique peptide variant. For the in vitro inhibition approach, the expression level of the reporter protein will be reduced inside droplets that contain inhibitory concentrations of a trans-acting inhibitory peptide, leading to low fluorescence. In contrast, drops that contain non-inhibitory peptides will show strong fluorescence. For the in vivo inhibition approach, the same principle applies, but the reporter protein is expressed inside bacterial cells contained within the droplets. (b) Identification of cis-acting peptides (red) by inverse toeprinting 70. On the left, ribosomes translating a non-inhibitory peptide reach the stop codon and remain stalled on the mRNA due to the omission of release factor-2 from the in vitro translation reaction. On the right, translation is inhibited during the elongation step due to the presence of an arrest sequence. This yields shorter RNA fragments after RNase R digestion, which can be specifically amplified and sequenced.
High-throughput inverse toeprinting enables selection of cis-acting peptides
As reported above, arrest peptides are a promising albeit immature source of translation inhibitors. A method to identify new ligand-independent arrest peptides would yield cis-acting molecules with the potential to be evolved into trans-inhibitory peptides. To this end, our group has developed inverse toeprinting, a method to map the position of a stalled ribosome on the mRNA with codon resolution, while protecting the entire peptide-encoding sequence up to the point of stalling 70 (Figure 2b). This contrasts with ribosome profiling 71, which only generates a short ribosome-protected footprint and therefore loses sequence information for a majority of the coding region. Like ribosome profiling, however, inverse toeprinting is characterized by a strong phenotype-genotype linkage, made possible by the inherent stability of stalled ribosome nascent chain complexes. The use of next-generation sequencing as readout and the specificity of inverse toeprinting make it suitable for incorporation into directed evolution schemes, as evidenced by the ability of a single cycle of inverse toeprinting to identify variants of existing drug-dependent arrest peptides with specificities for different antibiotics 70. Inverse toeprinting allows >1012 sequences to be analyzed in parallel in a single experiment and thus might have a reasonable chance of finding inhibitory peptides within a random pool of sequences. This could be beneficial by providing additional templates as input for the microfluidics-based approach, provided that the resulting cis-acting peptides could eventually be made to work in trans.
Expanding the Chemical Diversity of Ribosome-Targeting Peptides
Non-proteinogenic groups make peptides more drug-like
Peptides that inhibit the bacterial ribosome in trans must also possess drug-like properties to become viable antibiotic scaffolds, such as the capacity to cross biological membranes, good oral bioavailability, low toxicity against eukaryotic cells and sufficient stability in a physiological environment. Despite their huge potential as pharmaceutical compounds, most peptides are not considered good drug candidates in their natural, unmodified state, due to their limited stability and short half-life in vivo, a very low membrane permeability and negligible oral bioavailability 72, 73. Drawing inspiration from small natural macrocyclic peptides produced via ribosome-independent pathways (e.g. cyclosporin A, vancomycin, actinomycin D…), a strategy aimed at overcoming these limitations consists in adding non-canonical chemical groups to the peptides of interest 74, 75. Indeed, the incorporation of residues capable of inducing the formation of macrocycles, complemented with N-methylation, D-amino acids or residues harboring bulky side chains have shown great promise for the design of new antibiotics, as their chemical properties can confer rigidity and protease resistance, help cross biological membranes and increase the affinity for their target 76. Significant efforts have been made over the past decade to introduce these non-canonical groups into peptide backbones of interest in a site-specific manner. Within the context of this perspective, we have chosen to focus on the co-translational incorporation of non-proteinogenic side chains and backbones into a ribosomally synthetized peptide by in vitro genetic code reprogramming.
In vitro genetic code reprogramming
Genetic code reprogramming is the artificial reassignment of codons to non-canonical residues in order to allow the ribosome-based synthesis of polypeptides that incorporate these unnatural moieties (Figure 3). Various exotic groups have been incorporated at the N-terminus of proteins using a standard bacterial in vitro translation system, including a large selection of unnatural L-amino acids, residues to induce peptide cyclization77, N-methyl amino acids78, β-amino acids79, D-amino acids80, 81 and even helical aromatic foldamers82. Incorporation at internal positions of ribosomally synthesized proteins has also been achieved for many of these groups, with efforts focused over the past decade on increasing the efficiency of this process for ‘difficult’ or bulky non-canonical groups. However, the incorporation of a single or consecutive residues with a backbone chemistry differing from that of standard L-amino acids remains a significant challenge in the field 83–86, though the recent development of Ribo-T 87 and splinted orthogonal ribosome systems 88 suggest that this aim is now within reach. Genetic reprogramming can be achieved in vitro or in vivo, but in the context of discovering new antibiotics, we will focus exclusively on one of the most promising in vitro approaches: the flexizyme technology. Complementary information about in vivo and other in vitro approaches can be found in several excellent reviews 76, 89, 90.
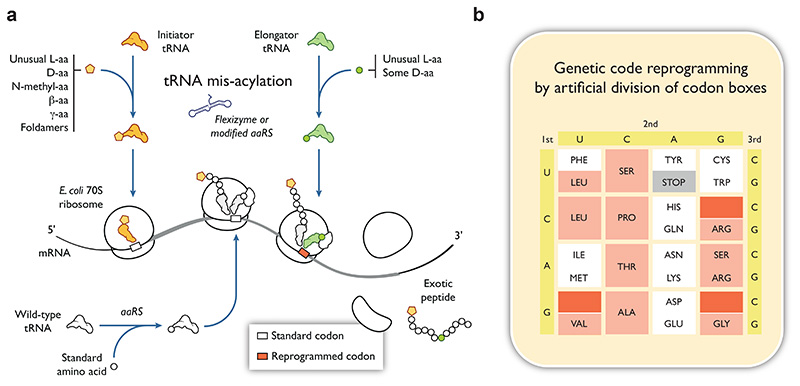
Figure 3. In vitro genetic code reprogramming to extend the chemical diversity of ribosomally synthetized peptides.
(a) Flexizyme-based mis-acylation of initiator and elongator tRNAs (yellow and green respectively) with diverse non-proteinogenic groups (top). Addition of the acyl-tRNA to an in vitro bacterial translation system allows the synthesis of peptides with both proteinogenic (white) and non-proteinogenic groups at their N-terminus (orange) or at internal positions (green). (b) Genetic code expansion strategies 94, 95 permit the reallocation of some codons to non-proteinogenic groups without compromising the presence of natural amino acids in the final genetic code. For the ‘artificial division of codon boxes’ approach shown here, codons that have been successfully reprogrammed are shown in red, whereas redundant codons that could theoretically be reprogrammed are shown in salmon.
The first step of genetic reprogramming is the mis-acylation of a tRNA molecule with an unusual amino acid or group of interest (Figure 3a). This can be accomplished in a relatively easy and flexible way using small RNA enzymes called flexizymes 91 or, traditionally, using mutated amino acyl-tRNA synthetases (aaRSs) with more permissive active sites and/or altered editing sites90. A distinct advantage of the flexizyme technology over aaRSs-based approaches is its applicability to virtually any non-canonical amino acid. Three types of flexizyme are typically used – eFx, aFx and dFx – that act upon amino acid substrates previously activated with specific ester-based leaving groups 91, 92. The only sequence element in the tRNA needed for recognition by flexizymes is an intact 3’ CCA end. This flexibility ensures that a wide range of substrates can be acylated onto any tRNA molecule, irrespective of its anticodon. Thus, more than 300 non-canonical amino acids, exotic monomers or short polymers (N-alkyl, N-acyl, D-amino acids, β-amino acids, γ-amino acids, macrocyclic peptides, aromatic-helix folding foldamers hybrids) have been attached to the 3’ end of tRNAs since flexizymes were first introduced 77, 81, 82, 92.
The second step of genetic reprogramming is the pairing of the mis-acylated tRNA to its cognate codon in the mRNA and the subsequent incorporation of the unusual moiety it carries into the nascent polypeptide chain. This can be achieved thanks to a fully reconstituted and tunable bacterial in vitro translation system comprising mRNA, mis-acylated-tRNA, ribosomes, translation factors and all other necessary substrates and components of the translational machinery93. While the flexizyme technology has considerably extended the array of non-canonical groups that can be charged onto tRNAs, recent efforts have sought to expand the number of codons that can be reassigned in a single translation reaction. Owing to the redundancy of the genetic code, the reallocation of a given codon to an unusual amino acid limits the sequence space available, as the amino acid that is encoded naturally by this codon must be omitted from the reaction. To overcome this, codons must artificially be reassigned to increase the total number of codons available for genetic reprogramming.
Two strategies have been developed to address the issue of codon reassignment (Figure 3b). The ‘artificial division of codon boxes’ strategy reduces the redundancy of codon assignment by replacing all wild-type tRNAs with 32 in vitro transcribed tRNAs bearing SNN anticodons (S=G or C; N = U, C, A or G) 94. Among these, tRNAs with GNN anticodons can decode NNY (Y=C or T) codons independently from tRNAs with CNN anticodons from the same codon box, which recognize NNG codons. After the 20 standard amino acids have been accounted for, the redundancy of the genetic code leaves 11 codons vacant that could theoretically be reprogrammed. Three such codons have successfully been reallocated to date, enabling the synthesis of a linear peptide containing 20 natural amino acids and 3 reassigned N-methyl-amino acids, as well as a macrocyclic N-methyl peptide. The ‘codon table duplication’ strategy makes use of orthogonal ribosome/tRNA pairs that do not cross-react with the wild-type translational machinery. This is made possible by introducing mutations in the A- and P-sites of the orthogonal ribosome that allow it to interact with orthogonal tRNAs bearing a C75G mutation 95. Introducing the orthogonal tRNA/ribosome pair into an in vitro translation system containing a single mRNA yields two distinct peptides in similar ratios: one translated by the wild-type ribosome and one translated by the orthogonal ribosome, which uses the parallel genetic code. Used in combination with the flexizyme technology, these approaches make in vitro genetic reprogramming a powerful tool to introduce chemical and functional diversity into directed evolution experiments aimed at identifying linear or cyclic peptide-based antibiotic scaffolds to target the ribosome.
Turning the Ribosome Into an Antibiotic Discovery Platform
A possible way forward
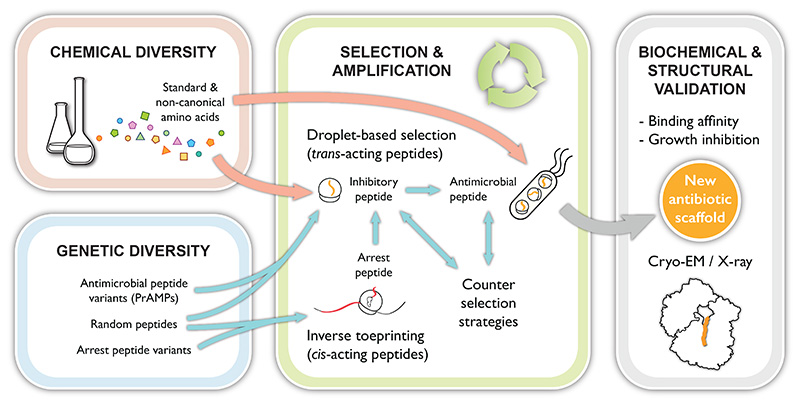
Various strategies could foreseeably be adopted to use the ribosome as a production and selection platform for peptides with the potential to become new ribosome-targeting antibiotic scaffolds. The strategy that we favor is outlined in Figure 4. In brief, peptide-encoding template libraries will be used to drive the expression of short peptides in a compartmentalized in vitro translation system, such that no more than a single peptide variant is expressed in each compartment. The choice of methodology for compartmentalization will be important, with droplet-based microfluidics providing an attractive solution in terms of consistency, reproducibility and ability to sort different phenotypes. Different readouts could be used to determine whether the peptides produced in each drop inhibit translation, an obvious solution being the use of a fluorescent reporter protein. Sorting of the droplets would yield a pool of templates encoding potential inhibitory peptides, which could be enriched through further cycles of selection and amplification. An additional selection strategy could be used to retain peptides that display antimicrobial properties against live bacterial cells, which will again require the use of compartmentalization to link phenotype to genotype. It is important to note that this approach could foreseeably lead to the identification of peptides that inhibit transcription, components of the translation machinery other than the ribosome (translation factors, aminoacyl-tRNA synthetases) or enzymes tasked with replenishing the in vitro translation system with energy (ATP, GTP). Different counter-selection schemes may thus need to be developed to identify and remove these unwanted peptides. Eventually, repeated cycles of selection and counter-selection would ensure that the most active peptide variants are retained. In parallel, cis-acting arrest peptides could be identified by inverse toeprinting and further mutated to act in trans using the directed evolution scheme described above. Directed evolution experiments could either make use of focused libraries of known antimicrobial peptides or arrest peptides variants as input or could be initiated with truly random peptide-encoding libraries. Finally, the use of genetic reprogramming to introduce chemical and molecular diversity into the peptides produced could yield modified linear or macrocyclic molecules with the desired properties for cellular uptake, oral bioavailability and in vivo stability. Implementing this or any other strategy will require new methods to be developed and many hurdles to be overcome, but ensuring a steady supply of novel antibiotic scaffolds for the generations to come is certainly worth the effort.
Figure 4. Proposed strategy for the discovery of new peptide-based antibiotic scaffolds that target the ribosome.
Acknowledgments
The authors thank A.-X. van der Stel for productive discussions and reading of the manuscript.
Funding
J.C. was supported by a grant form the Aquitaine Regional Council (2015-1R30104 – 00005037). A.M. is the recipient of an EMBO Long-Term Fellowship. C.A.I. is an EMBO Young Investigator and received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant Agreement No. 724040).
Notes
The authors declare no competing financial interest.
References
- 1.Braine T. Race against time to develop new antibiotics. Bulletin of the World Health Organization. 2011;89:88–89. doi: 10.2471/BLT.11.030211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 3.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 4.Wilson DN. The A-Z of bacterial translation inhibitors. Critical reviews in biochemistry and molecular biology. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 5.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nature reviews Microbiology. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 6.Florin T, Maracci C, Graf M, Karki P, Klepacki D, Berninghausen O, Beckmann R, Vazquez-Laslop N, Wilson DN, Rodnina MV, Mankin AS. An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nature structural & molecular biology. 2017;24:752–757. doi: 10.1038/nsmb.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gagnon MG, Roy RN, Lomakin IB, Florin T, Mankin AS, Steitz TA. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krizsan A, Volke D, Weinert S, Sträter N, Knappe D, Hoffmann R. Insect‐Derived Proline‐Rich Antimicrobial Peptides Kill Bacteria by Inhibiting Bacterial Protein Translation at the 70 S Ribosome. Angewandte Chemie International Edition. 2014;53:12236–12239. doi: 10.1002/anie.201407145. [DOI] [PubMed] [Google Scholar]
- 9.Mardirossian M, Grzela R, Giglione C, Meinnel T, Gennaro R, Mergaert P, Scocchi M. The host antimicrobial peptide Bac71-35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chemistry & biology. 2014;21:1639–1647. doi: 10.1016/j.chembiol.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Mardirossian M, Pérébaskine N, Benincasa M, Gambato S, Hofmann S, Huter P, Müller C, Hilpert K, Innis CA, Tossi A, Wilson DN. The dolphin proline-rich antimicrobial peptide Tur1A inhibits protein synthesis by targeting the bacterial ribosome. Cell Chemical Biology. 2018 doi: 10.1016/j.chembiol.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy RN, Lomakin IB, Gagnon MG, Steitz TA. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nature structural & molecular biology. 2015;22:466–469. doi: 10.1038/nsmb.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seefeldt AC, Graf M, Perebaskine N, Nguyen F, Arenz S, Mardirossian M, Scocchi M, Wilson DN, Innis CA. Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkv1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seefeldt AC, Nguyen F, Antunes S, Perebaskine N, Graf M, Arenz S, Inampudi KK, Douat C, Guichard G, Wilson DN, Innis CA. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nature structural & molecular biology. 2015;22:470–475. doi: 10.1038/nsmb.3034. [DOI] [PubMed] [Google Scholar]
- 14.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chemical biology & drug design. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 15.Kaspar AA, Reichert JM. Future directions for peptide therapeutics development. Drug Discov Today. 2013;18:807–817. doi: 10.1016/j.drudis.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Passioura T, Katoh T, Goto Y, Suga H. Selection-based discovery of druglike macrocyclic peptides. Annual review of biochemistry. 2014;83:727–752. doi: 10.1146/annurev-biochem-060713-035456. [DOI] [PubMed] [Google Scholar]
- 17.Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 18.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 19.Llano-Sotelo B, Klepacki D, Mankin AS. Selection of small peptides, inhibitors of translation. Journal of molecular biology. 2009;391:813–819. doi: 10.1016/j.jmb.2009.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polikanov YS, Aleksashin NA, Beckert B, Wilson DN. The Mechanisms of Action of Ribosome-Targeting Peptide Antibiotics. Front Mol Biosci. 2018;5:48. doi: 10.3389/fmolb.2018.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graf M, Mardirossian M, Nguyen F, Seefeldt AC, Guichard G, Scocchi M, Innis CA, Wilson DN. Proline-rich antimicrobial peptides targeting protein synthesis. Nat Prod Rep. 2017;34:702–711. doi: 10.1039/c7np00020k. [DOI] [PubMed] [Google Scholar]
- 22.Hudson GA, Mitchell DA. RiPP antibiotics: biosynthesis and engineering potential. Curr Opin Microbiol. 2018;45:61–69. doi: 10.1016/j.mib.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reimer JM, Haque AS, Tarry MJ, Schmeing TM. Piecing together nonribosomal peptide synthesis. Current opinion in structural biology. 2018;49:104–113. doi: 10.1016/j.sbi.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, Kalpaxis D, Nierhaus KH. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding. Molecular cell. 2004;13:113–124. doi: 10.1016/s1097-2765(04)00002-4. [DOI] [PubMed] [Google Scholar]
- 25.Fabbretti A, Schedlbauer A, Brandi L, Kaminishi T, Giuliodori AM, Garofalo R, Ochoa-Lizarralde B, Takemoto C, Yokoyama S, Connell SR, Gualerzi CO, et al. Inhibition of translation initiation complex formation by GE81112 unravels a 16S rRNA structural switch involved in P-site decoding. Proc Natl Acad Sci U S A. 2016;113:E2286–2295. doi: 10.1073/pnas.1521156113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantel L, Florin T, Dobosz-Bartoszek M, Racine E, Sarciaux M, Serri M, Houard J, Campagne JM, de Figueiredo RM, Midrier C, Gaudriault S, et al. Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site. Molecular cell. 2018;70:83–94.:e87. doi: 10.1016/j.molcel.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Brandi L, Maffioli S, Donadio S, Quaglia F, Sette M, Milon P, Gualerzi CO, Fabbretti A. Structural and functional characterization of the bacterial translocation inhibitor GE82832. FEBS Lett. 2012;586:3373–3378. doi: 10.1016/j.febslet.2012.07.040. [DOI] [PubMed] [Google Scholar]
- 28.Liou YF, Tanaka N. Dual actions of viomycin on the ribosomal functions. Biochem Biophys Res Commun. 1976;71:477–483. doi: 10.1016/0006-291x(76)90812-3. [DOI] [PubMed] [Google Scholar]
- 29.Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nature structural & molecular biology. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. Journal of molecular biology. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 31.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods in enzymology. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 32.Noeske J, Huang J, Olivier NB, Giacobbe RA, Zambrowski M, Cate JH. Synergy of streptogramin antibiotics occurs independently of their effects on translation. Antimicrobial agents and chemotherapy. 2014;58:5269–5279. doi: 10.1128/AAC.03389-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterman IA, Khabibullina NF, Komarova ES, Kasatsky P, Kartsev VG, Bogdanov AA, Dontsova OA, Konevega AL, Sergiev PV, Polikanov YS. Madumycin II inhibits peptide bond formation by forcing the peptidyl transferase center into an inactive state. Nucleic Acids Res. 2017;45:7507–7514. doi: 10.1093/nar/gkx413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metelev M, Osterman IA, Ghilarov D, Khabibullina NF, Yakimov A, Shabalin K, Utkina I, Travin DY, Komarova ES, Serebryakova M, Artamonova T, et al. Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel. Nat Chem Biol. 2017;13:1129–1136. doi: 10.1038/nchembio.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Molecular cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 36.Tuckey C, Asahara H, Zhou Y, Chong S. Protein synthesis using a reconstituted cell-free system. Curr Protoc Mol Biol. 2014;108:16 31 11–22. doi: 10.1002/0471142727.mb1631s108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito K, Chiba S. Arrest peptides: cis-acting modulators of translation. Annu Rev Biochem. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 38.Wilson DN, Arenz S, Beckmann R. Translation regulation via nascent polypeptide-mediated ribosome stalling. Current opinion in structural biology. 2016;37:123–133. doi: 10.1016/j.sbi.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Wilson DN, Beckmann R. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Current opinion in structural biology. 2011;21:274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Seip B, Innis CA. How Widespread is Metabolite Sensing by Ribosome-Arresting Nascent Peptides? Journal of molecular biology. 2016;428:2217–2227. doi: 10.1016/j.jmb.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 41.Gryczan TJ, Grandi G, Hahn J, Grandi R, Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980;8:6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horinouchi S, Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980;77:7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narayanan CS, Dubnau D. Demonstration of erythromycin-dependent stalling of ribosomes on the ermC leader transcript. The Journal of biological chemistry. 1987;262:1766–1771. [PubMed] [Google Scholar]
- 44.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Molecular microbiology. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 45.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Molecular cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 46.Gong F, Ito K, Nakamura Y, Yanofsky C. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro) Proc Natl Acad Sci U S A. 2001;98:8997–9001. doi: 10.1073/pnas.171299298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gong F, Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 48.Murakami A, Nakatogawa H, Ito K. Translation arrest of SecM is essential for the basal and regulated expression of SecA. Proc Natl Acad Sci U S A. 2004;101:12330–12335. doi: 10.1073/pnas.0404907101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakatogawa H, Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Molecular cell. 2001;7:185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 50.Nakatogawa H, Murakami A, Mori H, Ito K. SecM facilitates translocase function of SecA by localizing its biosynthesis. Genes & development. 2005;19:436–444. doi: 10.1101/gad.1259505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiba S, Ito K. Multisite ribosomal stalling: a unique mode of regulatory nascent chain action revealed for MifM. Molecular cell. 2012;47:863–872. doi: 10.1016/j.molcel.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 52.Chiba S, Kanamori T, Ueda T, Akiyama Y, Pogliano K, Ito K. Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proc Natl Acad Sci U S A. 2011;108:6073–6078. doi: 10.1073/pnas.1018343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiba S, Lamsa A, Pogliano K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 2009;28:3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishii E, Chiba S, Hashimoto N, Kojima S, Homma M, Ito K, Akiyama Y, Mori H. Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria. Proc Natl Acad Sci U S A. 2015;112:E5513–5522. doi: 10.1073/pnas.1513001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cymer F, Hedman R, Ismail N, von Heijne G. Exploration of the arrest peptide sequence space reveals arrest-enhanced variants. The Journal of biological chemistry. 2015;290:10208–10215. doi: 10.1074/jbc.M115.641555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Besnard J, Jones PS, Hopkins AL, Pannifer AD. The Joint European Compound Library: boosting precompetitive research. Drug Discov Today. 2015;20:181–186. doi: 10.1016/j.drudis.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 57.Karawajczyk A, Giordanetto F, Benningshof J, Hamza D, Kalliokoski T, Pouwer K, Morgentin R, Nelson A, Muller G, Piechot A, Tzalis D. Expansion of chemical space for collaborative lead generation and drug discovery: the European Lead Factory Perspective. Drug Discov Today. 2015;20:1310–1316. doi: 10.1016/j.drudis.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 58.Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci U S A. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci U S A. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tawfik DS, Griffiths AD. Man-made cell-like compartments for molecular evolution. Nature biotechnology. 1998;16:652. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- 61.Fallah-Araghi A, Baret J-C, Ryckelynck M, Griffiths AD. A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab on a Chip. 2012;12:882–891. doi: 10.1039/c2lc21035e. [DOI] [PubMed] [Google Scholar]
- 62.Sharma S, Srisa-Art M, Scott S, Asthana A, Cass A. Droplet-based microfluidics. Methods in molecular biology. 2013;949:207–230. doi: 10.1007/978-1-62703-134-9_15. [DOI] [PubMed] [Google Scholar]
- 63.Gielen F, Colin P-Y, Mair P, Hollfelder F. Protein engineering. Springer; 2018. Ultrahigh-throughput screening of single-cell lysates for directed evolution and functional metagenomics; pp. 297–309. [DOI] [PubMed] [Google Scholar]
- 64.Miller OJ, El Harrak A, Mangeat T, Baret J-C, Frenz L, El Debs B, Mayot E, Samuels ML, Rooney EK, Dieu P. High-resolution dose–response screening using droplet-based microfluidics. Proceedings of the National Academy of Sciences. 2012;109:378–383. doi: 10.1073/pnas.1113324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baret J-C, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab on a Chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 66.Armbrecht L, Gabernet G, Kurth F, Hiss JA, Schneider G, Dittrich PS. Characterisation of anticancer peptides at the single-cell level. Lab on a Chip. 2017;17:2933–2940. doi: 10.1039/c7lc00505a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Churski K, Kaminski TS, Jakiela S, Kamysz W, Baranska-Rybak W, Weibel DB, Garstecki P. Rapid screening of antibiotic toxicity in an automated microdroplet system. Lab on a Chip. 2012;12:1629–1637. doi: 10.1039/c2lc21284f. [DOI] [PubMed] [Google Scholar]
- 68.Mahler L, Tovar M, Weber T, Brandes S, Rudolph MM, Ehgartner J, Mayr T, Figge MT, Roth M, Zang E. Enhanced and homogeneous oxygen availability during incubation of microfluidic droplets. RSC Advances. 2015;5:101871–101878. [Google Scholar]
- 69.Deshpande S, Birnie A, Dekker C. On-chip density-based purification of liposomes. Biomicrofluidics. 2017;11:034106. doi: 10.1063/1.4983174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seip B, Sacheau G, Dupuy D, Innis CA. Ribosomal stalling landscapes revealed by high-throughput inverse toeprinting of mRNA libraries. Life Science Alliance. 2018;1 doi: 10.26508/lsa.201800148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di L. Strategic approaches to optimizing peptide ADME properties. The AAPS journal. 2015;17:134–143. doi: 10.1208/s12248-014-9687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yin N, Brimble M, Harris P, Wen J. Enhancing the oral bioavailability of peptide drugs by using chemical modification and other approaches. Med Chem. 2014;4:763–769. [Google Scholar]
- 74.Driggers EM, Hale SP, Lee J, Terrett NK. The exploration of macrocycles for drug discovery—an underexploited structural class. Nature Reviews Drug Discovery. 2008;7:608. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]
- 75.Nielsen DS, Shepherd NE, Xu W, Lucke AJ, Stoermer MJ, Fairlie DP. Orally absorbed cyclic peptides. Chemical reviews. 2017;117:8094–8128. doi: 10.1021/acs.chemrev.6b00838. [DOI] [PubMed] [Google Scholar]
- 76.Passioura T, Suga H. Reprogramming the genetic code in vitro. Trends in biochemical sciences. 2014;39:400–408. doi: 10.1016/j.tibs.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 77.Goto Y, Ohta A, Sako Y, Yamagishi Y, Murakami H, Suga H. Reprogramming the translation initiation for the synthesis of physiologically stable cyclic peptides. ACS chemical biology. 2008;3:120–129. doi: 10.1021/cb700233t. [DOI] [PubMed] [Google Scholar]
- 78.Subtelny AO, Hartman MC, Szostak JW. Ribosomal synthesis of N-methyl peptides. Journal of the American Chemical Society. 2008;130:6131–6136. doi: 10.1021/ja710016v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maini R, Nguyen DT, Chen S, Dedkova LM, Chowdhury SR, Alcala-Torano R, Hecht SM. Incorporation of beta-amino acids into dihydrofolate reductase by ribosomes having modifications in the peptidyltransferase center. Bioorg Med Chem. 2013;21:1088–1096. doi: 10.1016/j.bmc.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 80.Dedkova LM, Fahmi NE, Golovine SY, Hecht SM. Enhanced D-amino acid incorporation into protein by modified ribosomes. Journal of the American Chemical Society. 2003;125:6616–6617. doi: 10.1021/ja035141q. [DOI] [PubMed] [Google Scholar]
- 81.Goto Y, Murakami H, Suga H. Initiating translation with D-amino acids. Rna. 2008;14:1390–1398. doi: 10.1261/rna.1020708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rogers JM, Kwon S, Dawson SJ, Mandal PK, Suga H, Huc I. Ribosomal synthesis and folding of peptide-helical aromatic foldamer hybrids. Nature chemistry. 2018;10:405. doi: 10.1038/s41557-018-0007-x. [DOI] [PubMed] [Google Scholar]
- 83.Achenbach J, Jahnz M, Bethge L, Paal K, Jung M, Schuster M, Albrecht R, Jarosch F, Nierhaus KH, Klussmann S. Outwitting EF-Tu and the ribosome: translation with d-amino acids. Nucleic acids research. 2015;43:5687–5698. doi: 10.1093/nar/gkv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fujino T, Goto Y, Suga H, Murakami H. Reevaluation of the D-amino acid compatibility with the elongation event in translation. Journal of the American Chemical Society. 2013;135:1830–1837. doi: 10.1021/ja309570x. [DOI] [PubMed] [Google Scholar]
- 85.Fujino T, Goto Y, Suga H, Murakami H. Ribosomal synthesis of peptides with multiple β-amino acids. Journal of the American Chemical Society. 2016;138:1962–1969. doi: 10.1021/jacs.5b12482. [DOI] [PubMed] [Google Scholar]
- 86.Katoh T, Tajima K, Suga H. Consecutive elongation of D-amino acids in translation. Cell chemical biology. 2017;24:46–54. doi: 10.1016/j.chembiol.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 87.Orelle C, Carlson ED, Szal T, Florin T, Jewett MC, Mankin AS. Protein synthesis by ribosomes with tethered subunits. Nature. 2015;524:119–124. doi: 10.1038/nature14862. [DOI] [PubMed] [Google Scholar]
- 88.Fried SD, Schmied WH, Uttamapinant C, Chin JW. Ribosome Subunit Stapling for Orthogonal Translation in E. coli. Angew Chem Int Ed Engl. 2015;54:12791–12794. doi: 10.1002/anie.201506311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chin JW. Expanding and reprogramming the genetic code. Nature. 2017;550:53. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 90.Mukai T, Lajoie MJ, Englert M, Soll D. Rewriting the Genetic Code. Annu Rev Microbiol. 2017;71:557–577. doi: 10.1146/annurev-micro-090816-093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goto Y, Katoh T, Suga H. Flexizymes for genetic code reprogramming. Nature protocols. 2011;6:779. doi: 10.1038/nprot.2011.331. [DOI] [PubMed] [Google Scholar]
- 92.Passioura T, Suga H. Flexizyme‐Mediated Genetic Reprogramming As a Tool for Noncanonical Peptide Synthesis and Drug Discovery. Chemistry–A European Journal. 2013;19:6530–6536. doi: 10.1002/chem.201300247. [DOI] [PubMed] [Google Scholar]
- 93.Hipolito CJ, Suga H. Ribosomal production and in vitro selection of natural product-like peptidomimetics: the FIT and RaPID systems. Curr Opin Chem Biol. 2012;16:196–203. doi: 10.1016/j.cbpa.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 94.Iwane Y, Hitomi A, Murakami H, Katoh T, Goto Y, Suga H. Expanding the amino acid repertoire of ribosomal polypeptide synthesis via the artificial division of codon boxes. Nature chemistry. 2016;8:317. doi: 10.1038/nchem.2446. [DOI] [PubMed] [Google Scholar]
- 95.Terasaka N, Hayashi G, Katoh T, Suga H. An orthogonal ribosome-tRNA pair via engineering of the peptidyl transferase center. Nature chemical biology. 2014;10:555. doi: 10.1038/nchembio.1549. [DOI] [PubMed] [Google Scholar]
- 96.Noeske J, Wasserman MR, Terry DS, Altman RB, Blanchard SC, Cate JH. High-resolution structure of the Escherichia coli ribosome. Nature structural & molecular biology. 2015;22:336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]