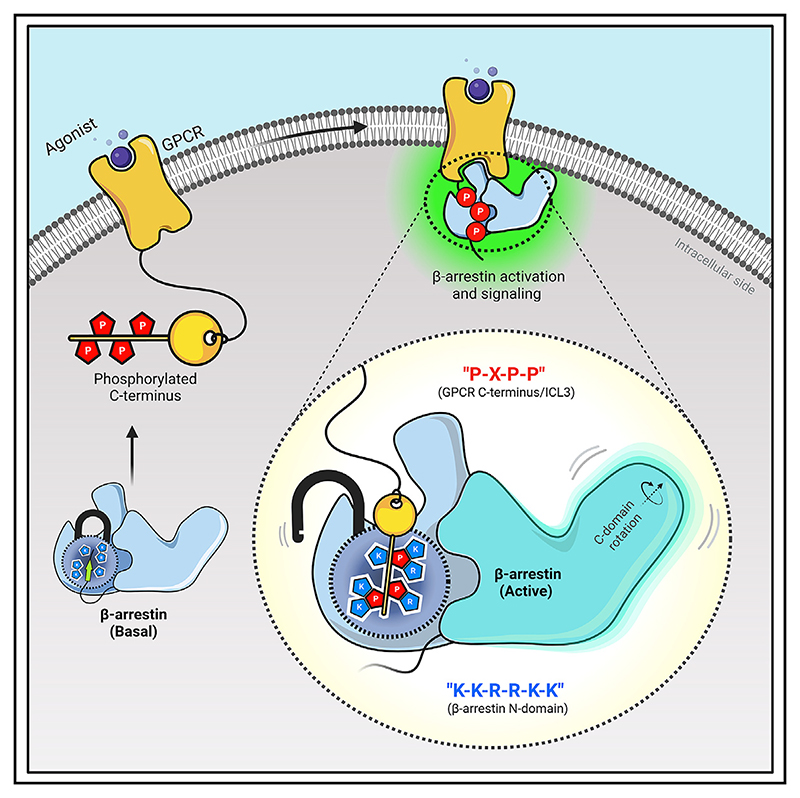
Summary
Agonist-induced GPCR phosphorylation is a key determinant for the binding and activation of β-arrestins (βarrs). However, it is not entirely clear how different GPCRs harboring divergent phosphorylation patterns impart converging active conformation on βarrs leading to broadly conserved functional responses such as desensitization, endocytosis, and signaling. Here, we present multiple cryo-EM structures of activated βarrs in complex with distinct phosphorylation patterns derived from the carboxyl terminus of different GPCRs. These structures help identify a P-X-P-P type phosphorylation motif in GPCRs that interacts with a spatially organized K-K-R-R-K-K sequence in the N-domain of βarrs. Sequence analysis of the human GPCRome reveals the presence of this phosphorylation pattern in a large number of receptors, and its contribution in βarr activation is demonstrated by targeted mutagenesis experiments combined with an intra-body-based conformational sensor. Taken together, our findings provide important structural insights into the ability of distinct GPCRs to activate βarrs through a significantly conserved mechanism.
Graphical abstract.
Introduction
G protein-coupled receptors (GPCRs) are typically characterized by their conserved seven transmembrane (7TM) architecture and agonist-induced coupling to heterotrimeric G-proteins and β-arrestins (βarrs).1 Of these, βarrs are multifunctional cytosolic proteins critically involved in regulating the signaling and trafficking of GPCRs.2–4 Their interaction with GPCRs involves a major contribution from receptor phosphorylation, which not only drives the affinity of receptor-βarr interaction but also imparts functionally competent active conformation in βarrs driving downstream functional outcomes5–12 (Figure 1A). Additional interaction of βarrs with the receptor transmembrane core and membrane lipid bilayer induces further structural changes as visualized using biophysical and direct structural approaches,13–20 which also fine-tune the functional capabilities of βarrs and possibly spatiotemporal aspects of their regulatory mechanisms.21,22 Despite a poorly conserved primary sequence of GPCRs in terms of the number and spatial positioning of the putative phosphorylation sites, the near-universal nature of βarr interaction, ensuing signaling and regulatory responses, remains to be fully understood at the molecular level. Moreover, differential receptor phosphorylation by different subtypes of GPCR kinases (GRKs), and possibly other kinases, in cell type-specific manner adds further complexity to GPCR-βarr binding modalities and context-specific functional specialization.23–29
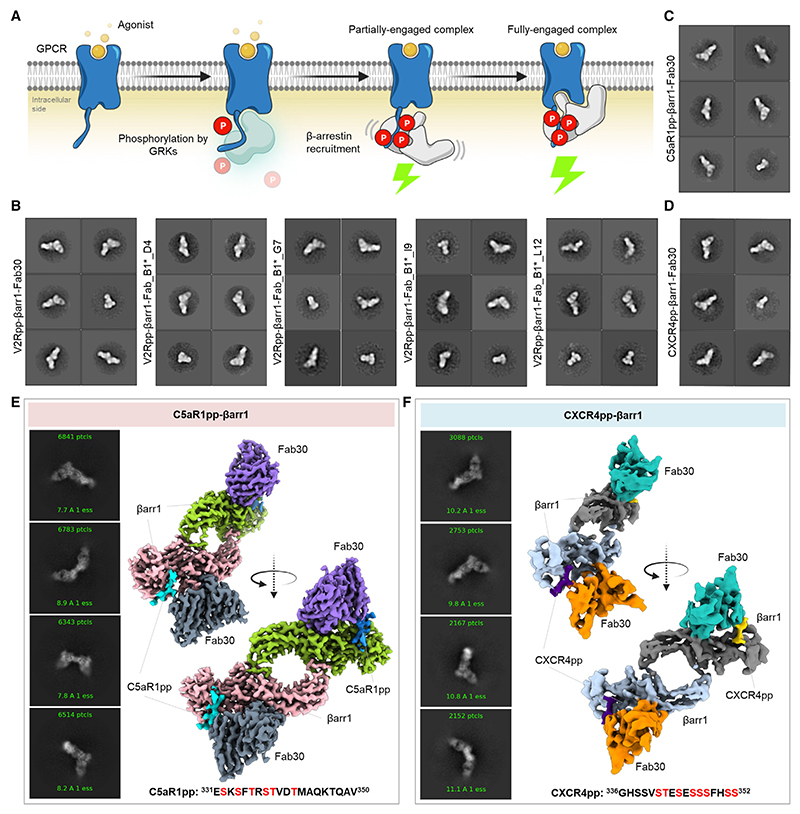
Figure 1. Reconstitution and structure determination of C5aR1pp/CXCR4pp-βarr1 complexes.
(A) Agonist stimulation of GPCRs leads to receptor phosphorylation by GPCR kinases (GRKs) followed by the recruitment and activation of βarrs governed through the phosphorylated residues and activated receptor core.
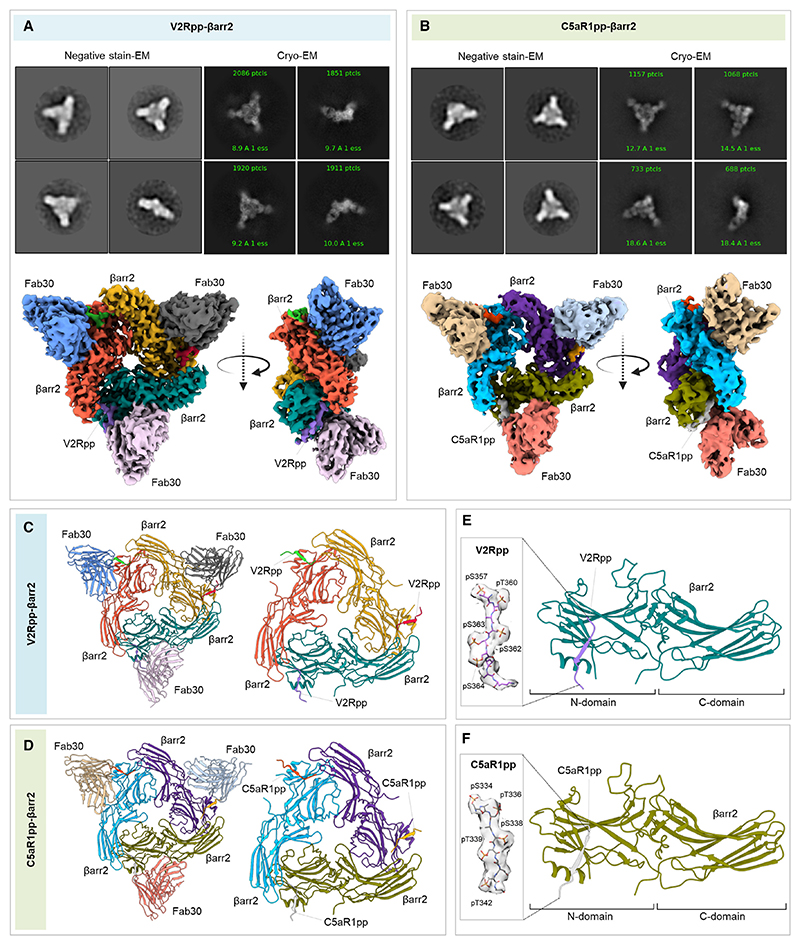
(B) Negative-staining EM-based 2D class averages of V2Rpp-βarr1 complexes stabilized by Fab30, Fab_B1*_D4, Fab_B1*_G7, Fab_B1*_I9, and Fab_B1*_L12, respectively.
(C and D) Negative-staining EM-based 2D class averages of C5aR1pp-βarr1-Fab30 and CXCR4pp-βarr1-Fab30 complexes, respectively.
(E and F) Selected 2D class averages and surface representation of C5aR1pp-βarr1-Fab30 and CXCR4pp-βarr1-Fab30 structures, respectively, determined by cryo-EM. The missing Fab30 constant domain densities were truncated during local refinement. See also Figures S1–S3 and Table 1.
There are two isoforms of βarrs namely, βarr1 and 2, also known as Arrestin2 and 3, respectively, and despite a high degree of sequence and structural similarity, they often exhibit a significant diversity in their functional contribution toward GPCR signaling and regulation.30 There are only a very few structures of βarr1 in active conformation, either in complex with receptor-derived phosphopeptides31,32 or in complex with agonist-bound, phosphorylated GPCRs.15–20 The structural coverage for βarr2 is even more sparse with the structural snap-shots limited to either a complex with CXC chemokine receptor subtype 7 (CXCR7)-derived phosphopeptide33 or in IP6-bound state.34 Although these structures provide useful information about βarrs’ interaction with the receptors, there is only limited information available about the phosphorylation patterns either due to chimeric constructs used in these studies or the lack of visualization of multiple phosphorylation sites resulting from insufficient structural resolution. Thus, the quest to decipher precise molecular details of convergent βarr activation by GPCRs harboring different phosphorylation patterns remains open. This represents a major knowledge gap in our current understanding of GPCR signaling and regulatory paradigms governing and fine-tuning signal transduction through this versatile class of receptors.
In this backdrop, here we present cryogenic-electron microscopy (cryo-EM) structures of full-length βarr1 and 2 activated by defined phosphorylation patterns encoded in the form of phosphopeptides, which are derived from three different GPCRs, namely, the complement C5a receptor subtype 1 (C5aR1), the CXC chemokine receptor subtype 4 (CXCR4), and the vasopressin receptor subtype 2 (V2R). These structural snapshots reveal a P-X-P-P type pattern of phosphorylation in GPCRs that engages a K-K-R-R-K-K sequence in the N-domain of βarrs leading to βarr activation. Interestingly, a large repertoire of GPCRs encodes the P-X-P-P motif either in their carboxyl terminus or in the 3rd intracellular loop (ICL3), suggesting a broad implication of this activation mechanism. We further validate the contribution of the P-X-P-P motif with respect to βarr activation in cellular context for several GPCRs using an intrabody-based conformational biosensor and structure-guided mutagenesis studies. Collectively, our data help identify the presence of the P-X-P-P motif in GPCRs and uncover the molecular basis of its ability to activate βarrs.
Results
Although cryo-EM has been used to determine the structures of GPCR-βarr1 complexes,15–19 all the previous structures of βarrs in basal state,35–38 bound to phosphopeptides31–33 or IP634 have been determined using X-ray crystallography. Therefore, in order to test the feasibility of structure determination of βarrs in complex with different phosphorylation patterns encoded in GPCR phosphopeptides by cryo-EM, we first reconstituted V2Rpp-βarr1 complex together with a set of conformationally selective antigen-binding fragments (Fabs), which recognize activated βarr1.39,40 Subsequently, we analyzed these complexes using negative-staining single particle EM, which revealed monodisperse particle distribution and 2D class averages where the densities of βarr1 and Fabs were clearly discernible (Figures 1B and S1A). Based on these observations, we synthesized and characterized a set of phosphopeptides corresponding to the carboxyl terminus of the C5aR1 and the CXCR4, and assessed their ability to activate βarr1, measured in terms of Fab30 reactivity (Figures S1B–S1E). We identified the phosphorylation patterns from C5aR1 (C5aR1pp2; referred to as C5aR1pp hereon) and CXCR4 (CXCR4pp2; referred to as CXCR4pp hereon), which elicited maximal Fab30 reactivity as a measure of βarr1 activation (Figures S1B–S1E). Subsequently, we reconstituted C5aR1pp-βarr1-Fab30 and CXCR4pp-βarr1-Fab30 complexes, validated their monodispersity and architecture using negative-staining EM (Figures 1C, 1D, S1F, and S1G), and subjected these complexes to cryo-EM. We successfully determined the structures of C5aR1pp-βarr1-Fab30 and CXCR4pp-βarr1-Fab30 complexes at global resolutions of 3.26 and 4.45 Å, respectively (Figures 1E, 1F, and S2).
βarr1 structures in complex with C5aR1 and CXCR4 phosphorylation patterns
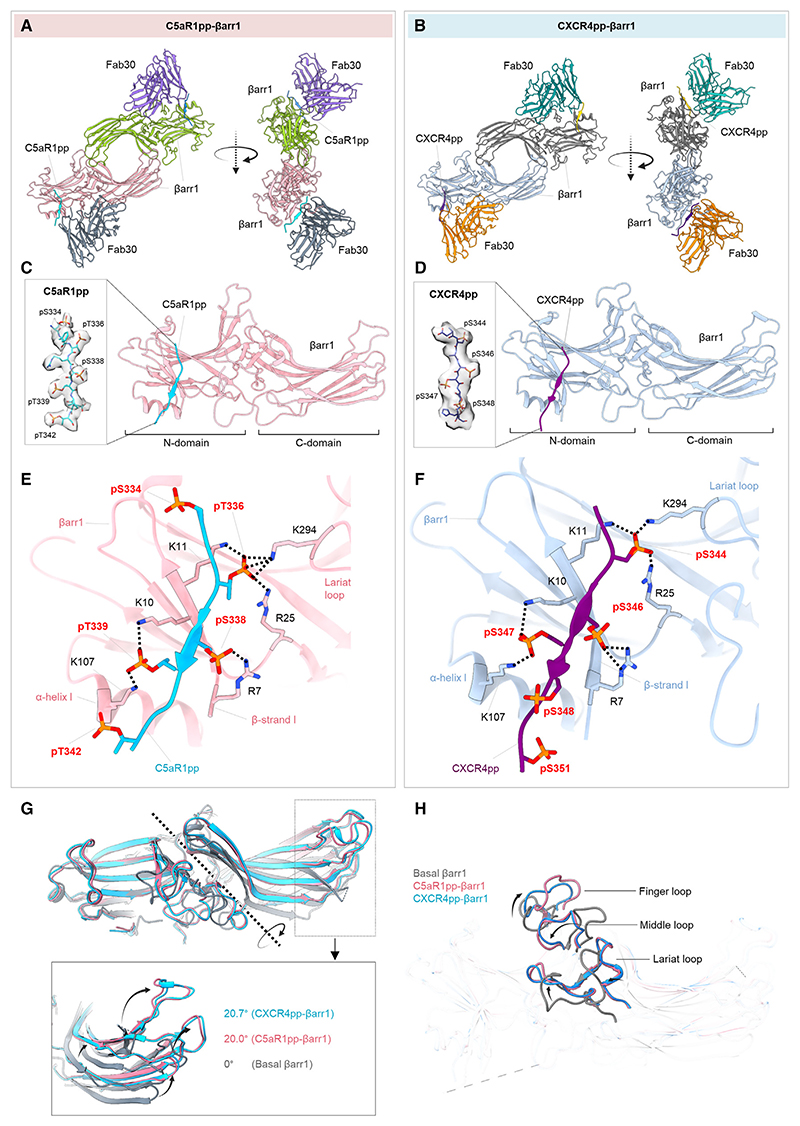
Both structures revealed a dimeric arrangement with the two βarr1 protomers making contacts through the C-edge loops and finger loops (Figures 2A and 2B). βarr1 protomers exhibit nearly identical overall structures with each other (root-mean-square deviation [RMSD] < 0.5 Å) with clear densities of the phosphopeptides visible in the EM map (Figures S3A and S3B). C5aR1pp and CXCR4pp are positioned in a positively charged groove on the N-domain of βarr1 (Figures 2C and 2D), and the phosphate moieties make extensive contacts with Arg/Lys residues at the binding interface (Figures 2E and 2F). Interestingly, three phosphate groups arranged in a P-X-P-P type pattern, where P is a phosphorylated residue and X is any other amino acid, in both the phosphopeptides engage a nearly identical set of Lys/Arg residues in βarr1 (Figures 2E and 2F). Specifically, the pT336-R337-pS338-pT339 pattern in C5aR1pp and pS344-E345-pS346-pS347 pattern in CXCR4pp engages K294-K11-R25-R7-K10-K107 in βarr1. The other phosphate groups present in the phosphopeptides are either not involved in direct contact or sparsely linked with Arg/Lys or positioned outside the binding groove. The N- and C-domains of βarr1 exhibit an inter-domain rotation of approximately 20° when compared with the basal state of βarr1, in both structures, which is a hallmark of βarr activation upon binding of phosphorylated GPCRs31 (Figure 2G). Moreover, the three major loops in βarr1 namely, the finger, middle, and lariat loop also exhibit significant reorientation upon binding of C5aR1pp and CXCR4pp compared with the basal state, although their positioning is almost identical between the two structures (Figure 2H). Finally, the three-element interaction and polar-core network in βarr1 are also disrupted upon binding to C5aR1pp and CXCR4pp when compared with the basal state structure through the displacement of the carboxyl terminus of βarr1 from the N-domain and repositioning of the lariat loop through the interaction of K294 with a phosphate moiety (Figures S4A and S4B). These structural features and interaction interface are analogous to that observed in the V2Rpp-βarr1-Fab30 crystal structure determined previously,31 although the primary sequence and phosphorylation patterns encoded by C5aR1pp and CXCR4pp are distinct from V2Rpp (Figure S4C).
Figure 2. Overall structures and key structural features of C5aR1pp/CXCR4pp-βarr1 complexes.
(A and B) Overall structures of C5aR1pp-βarr1-Fab30 and CXCR4pp-βarr1-Fab30 complexes shown with ribbon representation. The constant domains of Fab30 were masked out during refinement.
(C and D) Structure of individual C5aR1pp-βarr1 and CXCR4pp-βarr1 complex protomers shown as ribbon representation to indicate the binding of phosphopeptides on the N-domain of βarr1. Cryo-EM densities of corresponding phosphopeptides have been provided in insets.
(E and F) Stabilizing charge-charge interactions of C5aR1pp and CXCR4pp with the N-domain groove residues of βarr1 indicated as dotted lines. pS and pT refer to phospho-Ser and phospho-Thr residues, respectively.
(G) Inter-domain rotation in βarr1 upon binding of C5aR1pp (pink) and CXCR4pp (blue) is compared with the basal conformation of βarr1 determined previously (PDB: 1G4M, gray).
(H) Superimposition of C5aR1pp- and CXCR4pp-bound βarr1 structures with the basal conformation of βarr1 (PDB: 1G4M, gray) indicating the repositioning of finger, middle, and lariat loops upon βarr1 activation. See also Figure S4.
Structure-guided engineering yields structures of activated βarr2
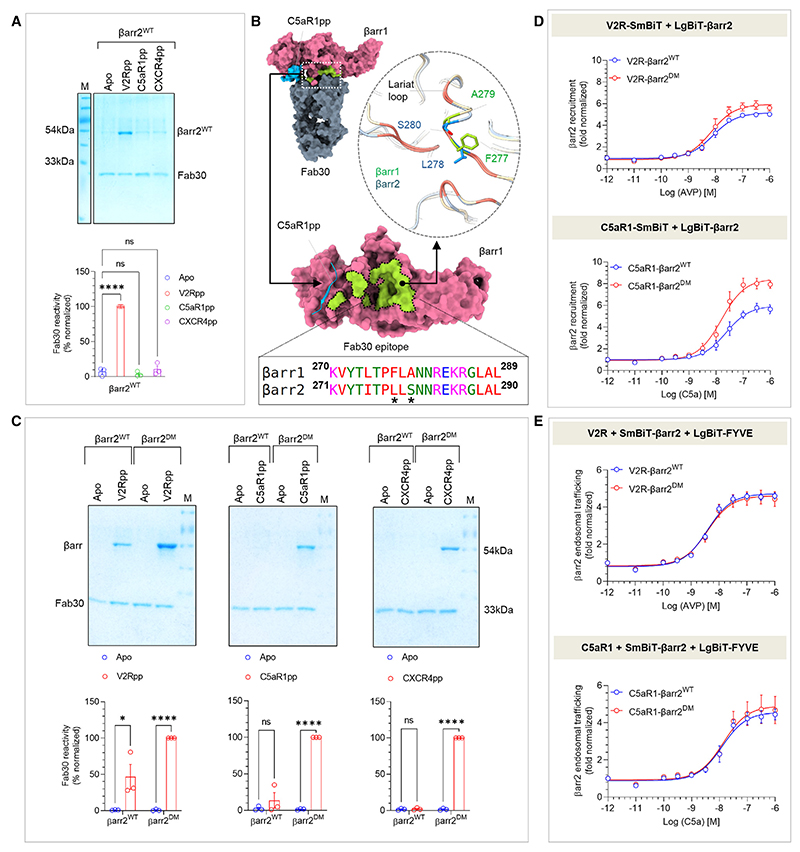
As mentioned earlier, the structural coverage of active βarrs, either in complex with full GPCRs or GPCR-derived phosphopeptides, is limited primarily to βarr1. Activated structures of the other isoform i.e., βarr2 are represented only by an IP6-bound crystal structure34 and a complex of truncated βarr2 with a phosphopeptide derived from a βarr-biased seven-transmembrane receptor (7TMR) (CXCR7).33 Therefore, we set out to reconstitute and determine the structure of βarr2 in complex with the phosphopeptides derived from different receptors, i.e., C5aR1pp and CXCR4pp using cryo-EM. Surprisingly, however, we did not observe a significant Fab30 reactivity to C5aR1pp/CXCR4pp-bound βarr2 while it robustly recognized the V2Rpp-βarr2 complex (Figure 3A). Therefore, we analyzed the Fab30 interaction interface on the C5aR1pp-bound βarr1 structure to identify a potential reason for the lack of Fab30 reactivity with βarr2. Interestingly, we observed that Fab30 epitope is conserved between βarr1 and 2 with the exception of two residues, i.e., instead of F277 and A279 as in βarr1, βarr2 contains L278 and S280 in the corresponding positions, respectively (Figure 3B). Therefore, we generated a βarr2 double mutant, referred to as βarr2DM, and tested its reactivity to Fab30 upon activation by distinct phosphopeptides. In line with our hypothesis, we observed a robust interaction of Fab30 with C5aR1pp-βarr2DM and CXCR4pp-βarr2DM complex, and we also noticed that the interaction of Fab30 with V2Rpp-βarr2DM was further enhanced compared with βarr2WT (Figure 3C). We also confirmed that βarr2DM exhibits a similar pattern of interaction as βarr2WT with V2R and C5aR1 in cellular context (Figure 3D) and shows near-identical endosomal trafficking as βarr2WT upon the stimulation of V2R and C5aR1 (Figure 3E). The receptor surface expression was assessed by whole-cell-based surface ELISA assay (Figure S5A). Thus, βarr2DM provides us with an excellent handle to reconstitute stable complexes with receptor phosphopeptides suitable for cryo-EM. In fact, we successfully managed to reconstitute monodisperse V2Rpp-βarr2DM-Fab30 and C5aR1pp-βarr2DM-Fab30 complexes (Figures S5C and S5I) and determine their cryo-EM structures at 3.96 and 4.33 Å resolution, respectively (Figures 4A, 4B, S5D–S5H, and S5J–S5N). In order to simplify the discussion, we refer to βarr2DM as βarr2 from here onward unless specified otherwise.
Figure 3. Generation and characterization of βarr2DM for structure determination.
(A) Fab30 reactivity to C5aR1pp and CXCR4pp activated βarr2WT was measured by co-immunoprecipitation (coIP) assay. C5aR1pp and CXCR4pp activated βarr2WT were not recognized by Fab30 (top). Densitometry-based quantification of the coIP data is presented (bottom) (mean ± SEM; n = 3; normalized with respect to V2Rpp signal as 100%; one-way ANOVA, Dunnett’s multiple comparisons test). The exact p values are as follows: Apo vs. V2Rpp (p ≤ 0.0001), Apo vs. C5aR1pp (p = 0.7719), Apo vs. CXCR4pp (p = 0.7899) (****p < 0.0001; ns, non-significant).
(B) Comparison of the epitope region of Fab30 in βarr1 with βarr2 reveals that instead of F277 and A279 as in βarr1, βarr2 contains L278 and S280 in corresponding positions (indicated with an asterisk).
(C) CoIP assay showing the reactivity of Fab30 toward activated βarr2DM upon binding of C5aR1pp and CXCR4pp (top). Densitometry-based quantification is presented (bottom) (mean ± SEM; n = 3; normalized with respect to Fab30 reactivity toward activated βarr2DM treated as 100%; two-way ANOVA, Šídák’s multiple comparisons test.) The exact p values are as follows: for βarr2WT: Apo vs. V2Rpp (p = 0.018), Apo vs. C5aR1pp (p = 0.3369), Apo vs. CXCR4pp (p = 0.9338); for βarr2DM: Apo vs. V2Rpp (p < 0.0001), Apo vs. C5aR1pp (p < 0.0001), Apo vs. CXCR4pp (p < 0.0001) (*p < 0.05, ****p < 0.0001).
(D) A side-by-side comparison of agonist-induced βarr2WT and βarr2DM recruitment to V2R (top) and C5aR1 (bottom) in the NanoBiT assay (Receptor-SmBiT+LgBiT-βarr2) (mean ± SEM; n = 4; normalized as fold over basal).
(E) A side-by-side comparison of βarr2WT and βarr2DM endosomal trafficking in response to agonist (arginine vasopressin peptide [AVP] for V2R and C5a for C5aR1) in the NanoBiT assay (Receptor+SmBiT-βarr2+LgBiT-FYVE) (mean ± SEM; n = 5; normalized as fold over basal). See also Figure S5.
Figure 4. Structures of V2Rpp/C5aR1pp-βarr2 complexes.
(A and B) Overall cryo-EM structures of V2Rpp-βarr2-Fab30 (left) and C5aR1pp-βarr2-Fab30 complexes (right), respectively, in a trimeric assembly with βarr2 and Fab30 molecules colored as individual units. Front and side views of the trimeric complex EM maps have been shown with βarr2 molecules in blue, olive green, and purple; and Fab30 molecules in beige, red, and gray, respectively. The constant domain densities of Fab30 were removed during local refinement and not observed in the final cryo-EM maps.
(C and D) Overall trimeric arrangement of V2Rpp/C5aR1pp-βarr2 complexes in the cryo-EM structures shown here as cartoon representation (left). Domain organization of the βarr2 molecules in trimeric assembly without Fab30 shown as cartoon representation (right).
(E and F) Structure of individual βarr2 protomers in V2Rpp/C5aR1pp-βarr2 complexes showing the binding of phosphopeptides on the N-domain of βarr2. Coulombic densities of corresponding phosphopeptides have been provided in insets. See also Figures S3 and S5 and Table 1.
Structures of βarr2 in complex with V2Rpp and C5aR1pp
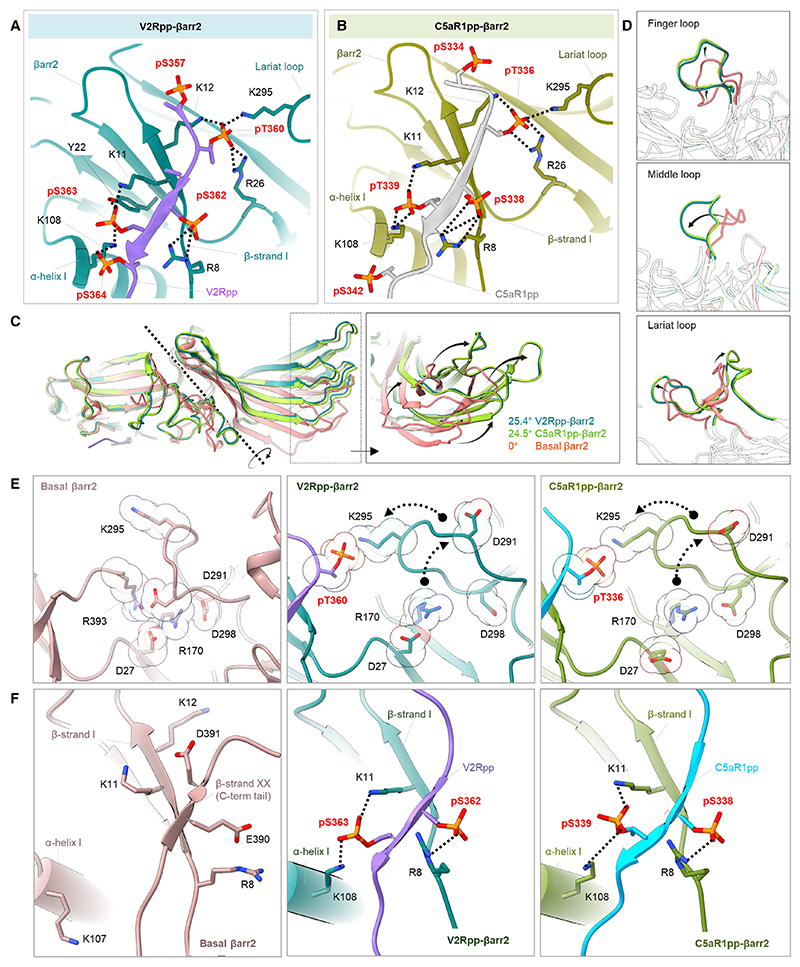
The V2Rpp-βarr2 and C5aR1pp-βarr2 structures exhibited a trimeric assembly of βarr2 with the individual protomers arranged through N- to C-domain contacts (Figures 4A–4D and S7; Table S2). The overall structural features of the individual protomers in each structure were nearly identical as reflected by low RMSD (<0.5 Å) with the phosphopeptide densities clearly visible in the EM maps (Figures S3C and S3D). Like βarr1 structures, V2Rpp and C5aR1pp are positioned in a positively charged groove on the N-domain of βarr2 (Figures 4E and 4F), and the phosphate moieties make extensive contacts with Arg/ Lys residues at the binding interface (Figures 5A and 5B). Remarkably, we observed that three phosphate groups arranged in a P-X-P-P pattern in the phosphopeptides, engage an analogous set of Lys/Arg residues as in βarr1 (Figures 5A and 5B). Specifically, pT360-A361-pS362-pS363 in V2Rpp and pT336-R337-pS338-pT339 in C5aR1pp engage K295-K12-R26-R8-K11-K108 in βarr2 (Figures 5A and 5B). Similar to βarr1 structures, the other phosphate groups present in the phosphopeptides are either not involved in direct contact with Arg/Lys or positioned outside the binding groove. The N- and C-domains of βarr2 exhibit an inter-domain rotation of approximately 25° when compared with the basal state of βarr2 in both the structures, which is relatively higher from that observed in phosphopeptide-bound βarr1 (Figure 5C). Moreover, the three major loops in βarr2 namely, the finger, middle, and lariat loop also exhibit significant reorientation upon binding of V2Rpp and C5aR1pp compared with the basal state, although their positioning is almost identical between the two structures (Figure 5D). Finally, the three-element interaction and polar-core network in βarr2 are also disrupted upon binding to V2Rpp and C5aR1pp when compared with the basal state structure through the displacement of the carboxyl terminus of βarr2 from the N-domain and repositioning of the lariat loop through the interaction of K295 with a phosphate moiety (Figures 5E and 5F).
Figure 5. Active conformations of phosphopeptide-bound βarr2.
(A and B) Extensive charge-charge interactions between the phosphate residues in V2Rpp/C5aR1pp with Lys/Arg in the N-domain (represented as black dotted lines) stabilize the V2Rpp and C5aR1pp into the N-domain groove of βarr2.
(C) V2Rpp (dark green) and C5aR1pp (light green) activated βarr2 structures were superimposed with the basal state of βarr2 (PDB: 3P2D, orange), and inter-domain rotations were calculated.
(D) Conformational changes observed in the finger (top), middle (middle), and lariat loops (bottom) in the activated βarr2 compared with the basal state crystal structure of βarr2.
(E) Polar-core environment in basal βarr2 (PDB: 3P2D, left) and disruption of polar-core interactions upon binding of V2Rpp (middle) and C5aR1pp to βarr2 (right).
(F) Three-element interaction network consisting of βarr2 C-terminal b-strand XX, α-helix I, and β -strand I in the basal state of βarr2 (left). Binding of the phosphopeptides V2Rpp and C5aR1pp to βarr2 results in the displacement of the β-strand XX, and engages the phosphopeptide V2Rpp (middle) and C5aR1pp (right) into the N-domain groove of βarr2 through hydrogen bonds and polar interactions. See also Figure S6.
Identification of a key phosphorylation motif driving βarr activation
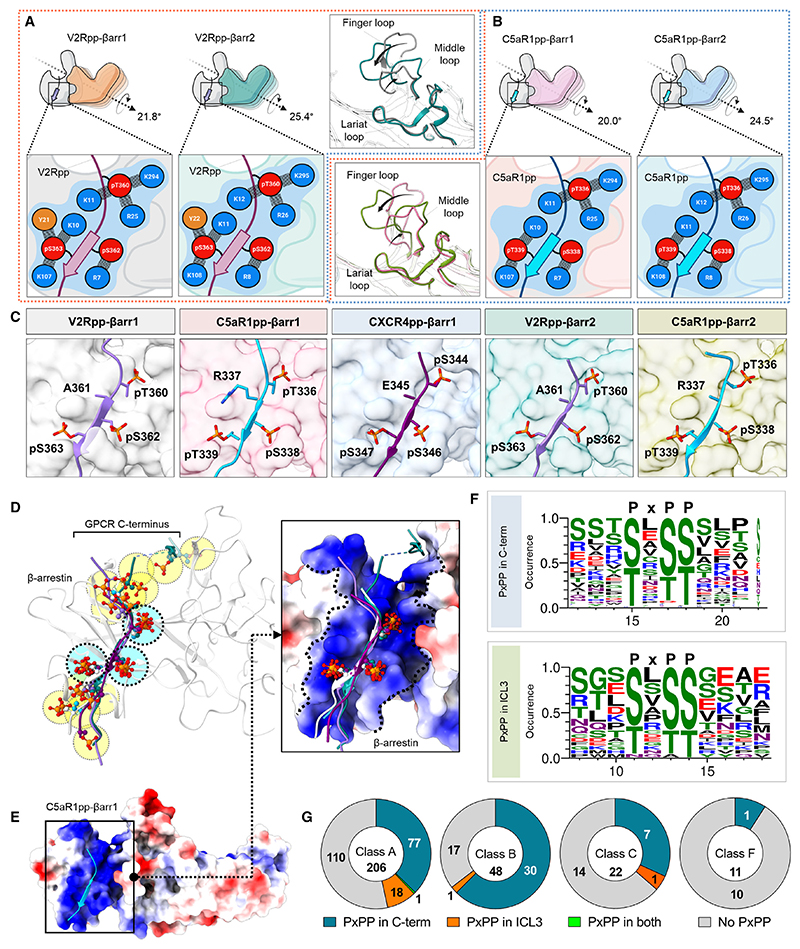
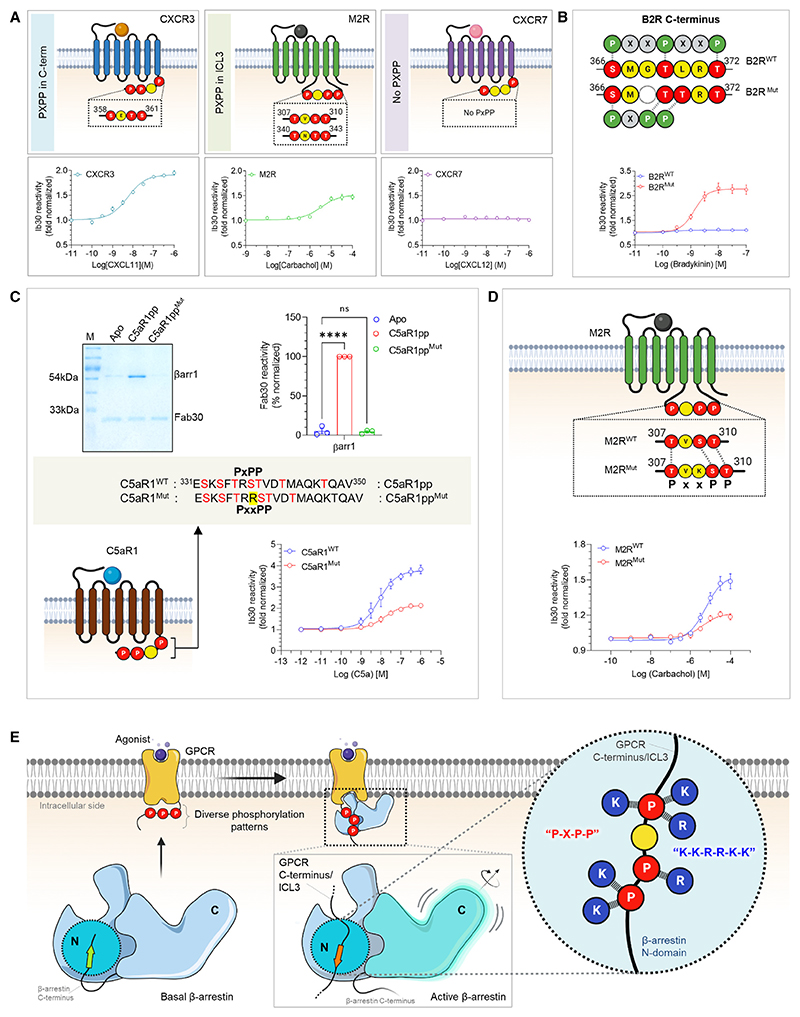
As mentioned earlier, the analysis of these structural snapshots in terms of phosphorylation sites revealed a P-X-P-P type pattern with nearly identical interactions with analogous residues in βarr1 and 2 (Figures 6A–6C). Therefore, we analyzed the primary sequence of all non-olfactory and non-orphan GPCRs in their carboxyl terminus and ICL3 to identify the occurrence of the P-X-P-P pattern in these receptors (Table S1). We observed that a large set of GPCRs harbored this motif in their carboxyl terminus sequence and several receptors also included it in their ICL3 (Figures 6F and 6G). In order to validate the functional contribution of this motif in GPCR-induced βarr activation, we employed Ib30-based conformational biosensor in cellular context using three different receptors, which possess P-X-P-P motif either in their C terminus (CXCR3), ICL3 (M2R), or lack it (CXCR7). At the level of βarr1 conformation, the Ib30 sensor reports the degree (>15°) of inter-domain rotation as a proxy of βarr activation upon its interaction with activated and phosphorylated receptors.40,41–43 In agreement with our hypothesis, we observed robust reactivity of Ib30 sensor with βarr1 for CXCR3 and M2R but not for CXCR7 although CXCR7 is capable of recruiting βarr1 upon agonist stimulation44–46 (Figure 7A). We note however that the lack of Ib30 reactivity for CXCR7 does not indicate the absence of βarr1 activation but instead, a different active conformation that is not recognized by the Ib30 sensor. Surface expression of the receptors was confirmed in these assays using the whole-cell ELISA assay (Figure S6B). To further corroborate these findings, we used two different receptors namely, the Bradykinin receptor subtype 2 (B2R) and C5aR1 for structure-based targeted mutagenesis to probe gain of function and loss of function, respectively, in terms of Ib30 reactivity pattern. As presented in Figure 7B, the activation of the wild-type B2R fails to induce an interaction between βarr1 and Ib30 as it lacks a P-X-P-P motif in its C terminus, although B2R is capable of recruiting βarrs.43,47 Similar to CXCR7, the lack of Ib30 reactivity for B2R likely suggests a distinct activated conformation in βarr1 that is not efficiently recognized by Ib30 sensor. Interestingly however, reconstitution of the P-X-P-P motif in B2R by double mutation (ΔG368+L370T) results in robust Ib30 reactivity upon agonist stimulation (Figure 7B). Along the same lines, a mutant version of C5aR1pp, where the P-X-P-P motif is disrupted by the insertion of an additional arginine residue between pT336 and pS338, completely loses the ability to induce a conformation that is recognizable by Fab30 (Figure 7C). Moreover, the corresponding mutation in C5aR1 also leads to a dramatic loss of Ib30 reactivity in cellular context (Figure 7C). Finally, we also disrupted a P-X-P-P motif present in the ICL3 of M2R by site-directed mutagenesis and measured the reactivity of Ib30 upon agonist stimulation (Figure 7D). We observed that similar to C5aR1, the disruption of the P-X-P-P motif in M2R also results in a marked decrease in Ib30 reactivity (Figure 7D). Taken together these data establish that the P-X-P-P phosphorylation pattern, when present in GPCRs, serves as a critical determinant for interaction and activation of βarrs (Figure 7E). In these experiments, the wild-type and mutant receptors were expressed at comparable levels as assessed in the surface expression assay (Figures S6C–S6E).
Figure 6. Identification of a key phosphorylation motif in GPCRs driving βarr activation.
(A) Comparison of the V2Rpp-bound βarr1 and βarr2 structures reveals similar interactions of V2Rpp with both isoforms of βarrs although a slightly higher inter-domain rotation is observed in βarr2 (left). A schematic representation of the interface network between negatively charged phospho-residues (red) and positively charged residues (blue) of βarrs is shown (below, zoomed-in box). Although the lariat loops of the two structures align well, significant deviations can be observed for the finger and middle loops (right, inset box).
(B) Comparative analysis of C5aR1pp-bound βarr1 and βarr2 structures uncover similar interactions of C5aR1pp with both βarr isoforms, but again, a higher inter-domain rotation is observed for βarr2. A similar representation of the interface between negatively charged phospho-residues (red) and positively charged residues (blue) of βarrs is shown (below, zoomed-in box).
(C) In all the structures of phosphopeptide-bound βarrs, a conserved motif can be observed with respect to three phospho-residues, referred to as the P-X-P-P motif, where “P” is a phospho-Ser/Thr and “X” can be any other residue.
(D) Superposition of V2Rpp-βarr1 (PDB 4JQI), V2Rpp-βarr2, C5aR1pp-βarr1, CXCR4pp-βarr1, and C5aR1pp-βarr2 shows conservation of phosphates corresponding to P-X-P-P position, whereas other phosphates are distributed throughout the phosphopeptides.
(E) Superposition of phosphopeptides on C5aR1pp-βarr1 reveals the conserved phospho-residues on positively charged cleft present on βarrs’ N-domain. βarr is shown as Coulombic-charged surface here.
(F) A sequence alignment of the C-terminal tail and ICL3 residues of non-olfactory and non-orphan class A receptors reveal the consensus sequence, “P-X-P-P” required for activation of βarrs. The consensus sequence logo was generated with the WEBLOGO tool48 and sequence alignment was performed with Kalign.49 A stretch of 11 amino acid residues has been shown for better representation.
(G) Proportions of GPCRs of class A, B, C, and F having P-X-P-P motif in C terminus or ICL3 have been represented as pie charts. See also Figure S6 and Table S1.
Figure 7. A key phosphorylation motif for βarr activation.
(A) NanoBiT-based assay for assessing Ib30 reactivity to CXCR3 (left), M2R (middle), and CXCR7 (right) activated βarr1 (Receptor+SmBiT-βarr1+LgBiT-Ib30) (mean ± SEM; n = 3; normalized as fold over basal).
(B) Deletion of G368 and substitution of L370 to Thr in B2R engineers the P-X-P-P (referred to as B2RMut) and results in gain of function in terms of Ib30 reactivity as measured using the NanoBiT assay (Receptor+SmBiT-βarr1+LgBiT-Ib30) (mean ± SEM; n = 3; normalized as fold over basal).
(C) Addition of an extra Arg between positions 336 and 337 in C5aR1pp to disrupt the P-X-P-P (referred to as C5aR1ppMut) leads to a near-complete loss of Fab30 (top) reactivity as measured in coIP assay (mean ± SEM; n = 3; densitometry-based data normalized with respect to C5aR1pp signal as 100%; one-way ANOVA, Dunnett’s multiple comparisons test). The exact p values are as follows: Apo vs. C5aR1pp2 (p < 0.0001), Apo vs. C5aR1pp4 (p = 0.9160) (****p < 0.0001, ns, non-significant). Corresponding mutation in C5aR1 to disrupt the P-X-P-P (referred to as C5aR1Mut) results in a dramatic decrease in Ib30 reactivity (bottom) as measured using the NanoBiT assay (mean ± SEM; n = 5; normalized as fold over basal).
(D) Insertion of a Lys residue between positions 308 and 309 to disrupt the P-X-P-P pattern in ICL3 of M2R (referred to as M2RMut) shows reduced Ib30 reactivity compared with the wild type (M2RWT) as measured using the NanoBiT assay (mean ± SEM; n = 4; normalized as fold over basal).
(E) Schematic representation summarizing the identification of a phosphorylation motif in GPCRs that drives βarr activation. See also Figure S6.
Discussion
Understanding the contribution of specific phosphorylation patterns in GPCRs for βarr recruitment, activation, and functional outcomes has been a key focus area in the field of GPCR biology, and several previous studies have provided interesting insights into this. For example, the bar-code hypothesis suggests differential contribution of distinct GPCR phosphorylation patterns in inducing specific conformations in βarrs that are linked to corresponding functional outcomes.28,50 Taking lead from the rhodopsin-arrestin fusion protein structure determined by X-ray free electron laser (XFEL), a previous study identified and validated PXPXXP and PXXPXXP type phosphorylation codes in GPCRs that appear to be critical for βarr recruitment.51,52 Sub-sequently, another study used peptide array and biophysical approaches to identify and propose a framework of the “key,” “inhibitory,” and “modulatory” phosphorylation sites in GPCRs that influence βarr binding, activation, and global conformation.53 Additionally, we have also previously explored the functional contribution of phosphorylation codes in several GPCRs to establish a link between different codes and ERK1/2 mitogen-activated protein (MAP) kinase activation using biochemical and cellular experiments.43 This study now visualizes the molecular interaction of multiple GPCR phosphorylation patterns with βarrs and identifies a P-X-P-P motif that plays a crucial role in βarr binding and activation when present in the interacting receptor.
The structural snapshots determined here reveal that the P-X-P-P phosphorylation pattern simultaneously engages multiple elements in βarrs that are typically responsible for maintaining the inactive conformation in order to facilitate βarr activation. It is intriguing that the P-X-P-P phosphorylation pattern from different GPCR phospho-peptides utilizes a conserved set of interactions and docking interface on both isoforms of βarrs (Figures 6D and 6E). This conserved binding interface and corresponding interactions ensure the displacement of βarrs’ C terminus from the N-domain and repositioning of the lariat loop, leading to the release of the two major “breaks” on βarr activation namely, the three-element interaction and the polar-core network10,11,54 (Figure S6A). It is important to note that some GPCRs tested here such as B2R and CXCR7, which lack the P-X-P-P motif, do not exhibit Ib30 reactivity although they are capable of recruiting βarrs in functionally competent conformation as reported previously.43–45,47 Taken together, these data indicate that although P-X-P-P motif is not always essential for βarr binding, however, when present in GPCRs, it contributes critically in βarr interaction, and imparts an active conformation on βarrs through a structurally conserved mechanism. On the other hand, in the absence of the P-X-P-P motif, the receptors likely induce a distinct active conformation in βarrs that is also capable of directing functional responses. These scenarios underscore the structural diversity engrained in the GPCR-βarr system, which orchestrates their functional diversity and versatility. The P-X-P-P motif, when present in GPCRs, appears to be sufficient to activate βarrs as measured using Ib30 sensor; however, additional phosphorylation sites may also contribute to improve the affinity of GPCR-βarr interaction and exert modulatory activities as proposed earlier.52,53 Moreover, several GPCRs harbor multiple copies of P-X-P-P motif in their carboxyl terminus and ICL3 (Table S2), similar to PXPXXP and PXXPXXP codes as reported earlier.52 While it is tempting to speculate that different P-X-P-P motif may be utilized in a context-dependent fashion such as cell type-specific, GRK-specific, and ligand-specific manner, further studies are essential to systematically probe these interesting possibilities.
An intriguing observation in these structural snapshots is novel dimer and trimer assemblies of βarrs. Although βarrs have a strong propensity to adopt different oligomeric states,55–57 the dimer and trimer interfaces observed here differ significantly from previously reported interfaces34,56,57 (Figure S7). The overall buried surface area in dimer and trimer assemblies are approximately 1,500 and 4,500 Å2, respectively, suggesting a robust and stable oligomeric arrangement. The two protomers in C5aR1pp- and CXCR4pp-bound βarr1 interface with each other through multiple hydrogen bonds, salt bridges, and non-bonded contacts in a manner where the C-edge loop residues of one protomer are positioned into the central crest of the other protomer in the proximity of the finger loop. An analogous set of interactions are also involved in the trimer arrangement of βarr2 in complex with V2Rpp and C5aR1pp. Interestingly, a previous crystal structure of βarr2 in complex with IP6 also shows a trimeric arrangement although the trimer interface is different from that observed here in phosphopeptide-bound conformations.34 A comprehensive map of dimer and trimer interface with residue-level contacts is listed in Table S2. Considering the functional multiplicity of βarrs in terms of distinct signaling and regulatory outcomes and receptor-specific responses,58 it is possible that distinct oligomeric interfaces in βarrs may be a modular mechanism to fine-tune the functional contributions by providing distinct possibilities for adaptable protein-protein interaction interfaces for binding partners. Nonetheless, the biological implications of the oligomeric assemblies observed here for activated βarrs remain to be explored further in future studies. The comparison of V2Rpp- and C5aR1pp-bound βarr1 and 2 reveal a significantly higher inter-domain rotation in βarr2 compared with βarr1 as hypothesized earlier based on cellular and biochemical studies,40 and this may provide a plausible explanation for functional differences between the βarr isoforms as observed for multiple GPCRs.30 However, structural visualization of GPCR-βarr2 complexes would be required to validate this possibility.
In summary, guided by the structural snapshots, we identify and experimentally validate a P-X-P-P motif in GPCRs that imparts an active conformation on βarrs by engaging a conserved network of interacting residues in their N-domain. Our study therefore provides an important missing link in our current conceptual framework of GPCR-mediated βarr activation and paves the way to decipher the structural and functional diversity encoded in GPCR signaling and regulatory paradigms. We also note that a companion manuscript presenting the crystal structures of βarr1 in complex with phosphopeptides derived from the chemokine receptor CCR5 converges to similar conclusions about a critical contribution of the P-X-P-P motif in GPCRs on βarr interaction and activation.59
Limitations of the study
The structural snapshots presented here involve isolated phosphopeptides with defined phosphorylation patterns without the transmembrane core of the receptors. As the interaction of receptor transmembrane core is also important for generating fully engaged GPCR-βarr complexes,60–63 it is possible that βarrs exhibit additional conformational changes in complex with activated and phosphorylated receptors. However, the ability of the P-X-P-P motif, when present in GPCRs, to engage the K-K-R-R-K-K pattern in βarrs is likely to be maintained and guide βarr activation even in the context of full-length receptors. Additionally, we cannot rule out the possibility that for some receptors, a functional P-X-P-P motif may not be generated in cellular context despite having a suitable primary sequence because all the phosphorylatable residues may not undergo efficient phosphorylation.
Star+Methods
Detailed methods are provided in the online version of this paper and include the following:
-
-
○
Lead contact
-
○
Materials availability
-
○
Data and code availability
-
○
-
EXPERIMENTAL MODEL AND SUBJECT DETAILS
-
○
Human cell lines
-
○
Insect cells
-
○
-
-
○
General reagents, plasmids, and cell culture
-
○
Expression and purification of βarrs
-
○
Expression and purification of Fabs
-
○
Co-immunoprecipitation assay
-
○
Reconstitution of phosphopeptide-βarr-Fab complexes
-
○
Negative-staining EM
-
○
Cryo-EM sample preparation and data acquisition
-
○
Cryo-EM data processing and model building
-
○
Model building and refinement
-
○
NanoBiT assay for βarr2WT and βarr2DM recruitment
-
○
NanoBiT assay for βarr trafficking
-
○
NanoBiT assay for Ib30 reactivity B Receptor surface expression
-
○
Star+Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal ANTI-FLAG M2-HRP antibody | Sigma-Aldrich | Cat# A8592; RRID: AB_439702 |
| Chemicals, peptides, and recombinant proteins | ||
| TRIS | SRL | Cat# 71033 |
| HEPES | SRL | Cat# 63732 |
| NaCl | SRL | Cat# 41721 |
| EDTA | SRL | Cat# 12070 |
| Phenylmethanesulfonyl Fluoride (PMSF) | SRL | Cat# 84375 (84375) |
| Benzamidine Hydrochloride | SRL | Cat# 93014 (0248255) |
| Lysozyme | SRL | Cat# 45822 |
| Glycerol | SRL | Cat# 77453 |
| Dithiothreitol | HiMedia | Cat# MB070 |
| Lauryl Maltose Neopentyl Glycol (MNG) | Anatrace | Cat# NG310, CAS no.1257852-96-2 |
| Paraformaldehyde (PFA) | Sigma Aldrich | Cat# P6148, CAS no. 30525-89-4 |
| Poly-D-lysine | Sigma Aldrich | Cat# P0899 |
| TMB (Tetramethylbenzidine) | Thermo Fisher Scientific | Cat# 34028 |
| Janus Green B | Sigma Aldrich | Cat# 201677 |
| PEI (Polyethylenimine) | Polysciences | Cat# 23966 |
| Bovine Serum Albumin, BSA | SRL | Cat# 83803 (0140105) |
| HBSS - Hank’s Balanced Salt Solution | Thermo Fisher Scientific | Cat# 14065 |
| GIBCO Fetal Bovine Serum | Thermo Fisher Scientific | Cat# 10270-106 |
| DMEM | Cellclone | Cat# CC3004 |
| Phosphate-buffered saline (PBS) | Sigma Aldrich | Cat# D1283 |
| GIBCO Penicillin-Streptomycin | Thermo Fisher Scientific | Cat# 15140122 |
| Coelenterazine | Goldbio | Cat# CZ05 |
| Glyco-diosgenin (GDN) | Anatrace | GDN101 |
| Cholesteryl Hemisuccinate | Sigma | C6512 |
| Coomassie briliiant Blue | SRL | Cat# 64222 |
| Uranyl formate | Polysciences | Cat# 24762-1 |
| Recombinant rat β-arrestin1 | Purified | N/A |
| Recombinant bovine β-arrestin2 | Purified | N/A |
| C5aR1pp1 | Chemically synthesized | N/A |
| C5aR1pp2 | Chemically synthesized | N/A |
| C5aR1pp3 | Chemically synthesized | N/A |
| V2Rpp | Chemically synthesized | N/A |
| CXCR4pp1 | Chemically synthesized | N/A |
| CXCR4pp2 | Chemically synthesized | N/A |
| CXCR4pp3 | Chemically synthesized | N/A |
| CXCR4pp4 | Chemically synthesized | N/A |
| Recombinant human C5a | Purified | N/A |
| Bradykinin | Genscript | N/A |
| Arginine Vasopressin Peptide (AVP) | Genscript | N/A |
| Uranyl formate | Polysciences | Cat# 24762-1 |
| Formvar/carbon coated 300 mesh copper grids |
PELCO (Ted Pella) | Cat# 01753-F |
| Critical commercial assays | ||
| Site directed mutagenesis kit | NEB | Cat# E0554 |
| NanoBiT assay | Promega | N/A |
| Deposited data | ||
| C5aR1pp-βarr1-Fab30 | This study | PDB: 8GO8, EMD-34173 |
| V2Rpp-βarr2-Fab30 | This study | PDB: 8GOC, EMD-34175 |
| C5aR1pp-βarr2-Fab30 | This study | PDB: 8GOO, EMD-34178 |
| CXCR4pp-βarr1-Fab30 | This study | PDB: 8GP3, EMD-34188 |
| C5aR1pp-βarr1-Fab30-Local-refine | This study | PDB: 8I0N, EMD-35104 |
| V2Rpp-βarr2-Fab30-Local-refine | This study | PDB: 8I10, EMD-35115 |
| C5aR1pp-βarr2-Fab30-Local-refine | This study | PDB: 8I0Z, EMD-35114 |
| CXCR4pp-βarr1-Fab30-Local-refine | This study | PDB: 8I0Q, EMD-35106 |
| Experimental models: Cell lines | ||
| Human: HEK293 | ATCC | Cat# CRL-3216 |
| Oligonucleotides | ||
| C5aR1_R-insertion SDM primer Forward: CGCTCCACAGTGGACACTATGG |
This study | N/A |
| C5aR1_R-insertion SDM primer Reverse: GCGCGTGAATGACTTGCT |
This study | N/A |
| B2Rmut Forward: AACACGGACCTCCATCTCCGTG |
This study | N/A |
| B2RmutReverse GTCATGGAGTTCTCCATCTGAATGGG |
This study | N/A |
| M2Rmut_Forward: AAGTCTACTTCACTGGGCCAC |
This study | N/A |
| M2Rmut_Reverse: GACGGTGTTTTCGTCCTG |
This study | N/A |
| β-arrestin2 Double mutant (βarr2DM) Fw: ggcgAACAACCGTGAAAAACGTG |
This study | N/A |
| β-arrestin2 Double mutant (βarr2DM) Rv: aggaaCGGGGTGATGGTGTAAAC |
This study | N/A |
| Recombinant DNA | ||
| PcDNA_3.1 (empty vector) | Dr. Arun K Shukla | N/A |
| pcDNA3.1_V2R-WT | Dr. Arun K Shukla | N/A |
| pcDNA3.1_C5aR1-WT | Dr. Arun K Shukla | N/A |
| pcDNA3.1_CXCR3-WT | Dr. Arun K Shukla | N/A |
| pcDNA3.1_CXCR7-WT | Dr. Arun K Shukla | N/A |
| pcDNA3.1_B2R-WT | Dr. Arun K Shukla | N/A |
| pcDNA3.1_M2R-WT | Dr. Arun K Shukla | N/A |
| pcDNA3.1_C5aR1-mutTRRST | This study | N/A |
| pcDNA3.1_B2R-mutΔG368/L370T | Dr. Arun K Shukla | N/A |
| pcDNA3.1_M2R-mutTVKST | This study | N/A |
| pCAGGS_V2R-WT-SmBiT | Dr. Arun K Shukla | N/A |
| pCAGGS _C5aR1-SmBiT | Dr. Arun K Shukla | N/A |
| pCAGGS_SmBiT-βarr2WT | Dr. Asuka Inoue | N/A |
| pCAGGS_SmBiT-βarr2DM | This study | N/A |
| pCAGGS_LgBiT-βarr2WT | Dr. Asuka Inoue | N/A |
| pCAGGS_LgBiT-βarr2DM | This study | N/A |
| pCAGGS_LgBiT-FYVE | Dr. Asuka Inoue | N/A |
| pCAGGS_LgBiT-Ib30 | Dr. Asuka Inoue | N/A |
| Software and algorithms | ||
| UCSF Chimera | Pettersen et al.64 | https://www.cgl.ucsf.edu/chimera/ |
| UCSF Chimera X | Pettersen et al.65 | https://www.rbvi.ucsf.edu/chimerax/ |
| COOT | Emsley et al.66 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| cryoSPARC | Punjani et al.67 | https://cryosparc.com/ |
| PDBsum | Laskowski et al.68 | http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/ |
| Phenix | Liebschner et al.69 | https://www.phenix-online.org/ |
| PyMol | Schrodinger | https://pymol.org/2/ |
| Prism 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Relion3.1.2 | Zivanov et al.70 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page |
| SerialEM | Mastronarde71 | https://bio3d.colorado.edu/SerialEM/ |
| Graphpad Prism 9 | GraphPad Software, San Diego, California USA | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | Schneider et al.72 | https://imagej.nih.gov/ij/download.html |
Resource Availability
Lead contact
Further information and requests for reagents should be addressed to the lead contact, Dr. Arun K. Shukla (arshukla@iitk.ac.in).
Materials availability
Reagents described in this manuscript are available upon reasonable request from the lead contact.
Experimental Model and Subject Details
Human cell lines
HEK-293 cells were purchased from ATCC for all the cellular experiments performed in the study. The cell line was examined frequently under the microscope for proper morphology, but they were not authenticated. They were cultured in DMEM with fetal bovine serum (FBS) at 37°C in 5% CO2. In this study, any stable, knockout, or knockdown cell lines were not generated, and the details of previously generated cell lines are referenced in the manuscript.
Insect cells
Sf9 cells were obtained from Expression systems, and they were routinely monitored under the microscope for proper morphology. These cells were maintained in a shaker incubator at 27°C with 135rpm shaking, and sub-cultured in protein-free insect cell medium purchased from Expression Systems.
Method Details
General reagents, plasmids, and cell culture
Most of the general reagents were purchased from Sigma Aldrich unless mentioned otherwise. Dulbecco’s Modified Eagle’s Medium (DMEM), Dulbecco’s Phosphate buffer saline (PBS), Fetal-Bovine Serum (FBS), Trypsin-EDTA, Hank’s balanced salt solution (HBSS), and penicillin-streptomycin solution were purchased from Thermo Fisher Scientific. HEK-293 cells were obtained from ATCC and maintained in DMEM (Gibco, Cat no. 12800-017) supplemented with 10% FBS (Gibco, Cat no. 10270-106) and 100U ml-1 penicillin (Gibco, Cat no. 15140122) and 100μg ml-1 streptomycin (Gibco, Cat no. 15140-122) at 37°C under 5% CO2. The cDNA coding region for the mentioned receptors namely, V2R, C5aR1, B2R, M2R, CXCR3, and CXCR7 were cloned in pcDNA3.1 consist of HA signal sequence followed by FLAG tag at the N-terminus of the receptor. The mutants generated for the study are as follows: deletion of G368 and a substitution L370T in B2R; insertion of an Arg residue between R337 and S338 in C5aR1; insertion of a Lys residue between V308 and S309 in M2R; using Q5 site-directed mutagenesis kit (NEB, Cat. no. E0554S). For the NanoBiT assay, receptors harboring SmBiT at the C-terminus were generated by subcloning in the lab, and other constructs have been described previously.41,43,73 All the constructs were verified by DNA sequencing (Macrogen).
Expression and purification of βarrs
Full length rat βarr1, βarr2WT and bovine βarr2DM were cloned into pGEX-4T3 vector with thrombin cleavage site between GST tag and βarr. Similar protocol was followed for purifying all three forms of βarr. βarrs were expressed in E. coli BL21 cells and grown in Terrific broth media supplemented with 100μg ml-1 ampicillin. A primary culture of 50ml volume was inoculated with an isolated colony from freshly transformed LB-amp plate. Primary culture was grown till a cell optical density at 600nm (OD600) of 0.8-1 and further inoculated into a secondary culture of TB-Amp of 1.5L volume till OD600 0.8-1. The expression of βarrs was then induced with 25μM IPTG concentration and cells were allowed to grow till 16h at 18°C. Cultures were harvested and stored at -80°C until further use. Harvested pellets were of 12-15g in mass.
For purification, cells were lysed by sonication in lysis buffer; 25mM Tris, pH 8.5, 150mM NaCl, 1mM PMSF (phenylmethylsulfonyl fluoride), 2mM Benzamidine, 1mM EDTA (Ethylenediaminetetraacetic acid), 5% Glycerol, 2mM Dithiothreitol (DTT) and 1mg ml-1 Lysozyme. The lysate was centrifuged at 18,000-20,000rpm at 4°C and supernatant was allowed to bind to Glutathione resin (GS resin) (Glutathione Sepharose™ 4 Fast Flow, GE Healthcare Cat. no. 17-5132-02) in a batch binding mode for overnight at 4°C. GS-resin bound GST-βarr was transferred into Econo columns (Biorad, Cat. no. 7372512) and washed rigorously with wash buffer (25mM Tris, pH 8.5, 150mM NaCl, 2mM DTT and 0.02% n-dodecyl-β-D-maltopyranoside (DDM). Afterward, on-column cleavage was set up by adding thrombin to 1:1 resin:buffer slurry at room temperature for 2h. βarrs were then eluted with gravity flow and further with buffer 25mM Tris, pH 8.5, 350mM NaCl and 0.02% DDM and 2mM DTT. Eluted proteins were concentrated and further purified on a HiLoad 16/600 Superdex column in buffer 25mM Tris, pH 8.5, 350mM NaCl, 2mM DTT and 0.02% DDM. Fractions corresponding to pure βarr were flash frozen with 10% glycerol and stored at -80°C until further use.
Expression and purification of Fabs
A similar protocol for expression and purification was followed for all the Fabs and they were purified as previously mentioned.39 Briefly, Fabs were expressed in the periplasmic fraction of E. coli M55244 cells (ATCC) and purified using Protein L resin (GE Healthcare Cat. no. 17547802) with gravity flow affinity chromatography. Cells transformed with Fab plasmid were grown in 50ml 2xYT media and allowed to grow overnight at 30°C. 1L 2xYT media was inoculated with 5% volume of initial inoculum and grown for an additional 8h at 30°C. Cells were collected and resuspended in an equal volume of CRAP medium supplemented with 100μg ml-1 ampicillin, and grown further for 16h at 30°C. For purification, cells were lysed in lysis buffer (50mM HEPES-Na+, pH 8.0, 0.5M NaCl, 0.5% (v/v) Triton X-100, 0.5mM MgCl2) by sonication. Cell lysate was heated in a 65°C water bath for 30min and cooled immediately on ice for 5min. Lysate was centrifuged at 20,000rpm and passed through pre-equilibrated Protein L resin packed gravity flow affinity columns. Binding was performed at room temperature and beads were washed extensively with wash buffer (50mM HEPES-Na+, pH 8.0, 0.5M NaCl). Fabs were eluted with 100mM acetic acid into tubes containing 10% volume of 1M HEPES, pH 8.0 for neutralization. Eluted samples were desalted into buffer (20mM HEPES-Na+, pH 8.0, 0.1M NaCl) using a pre-packed PD-10 column (GE Healthcare Cat. no. 17085101). Purified Fabs were flash-frozen and stored at -80°C supplemented with 10% (v/v) glycerol until further use.
Co-immunoprecipitation assay
For co-immunoprecipitation assay, 2.5μg of β-arrestins were incubated with different phosphopeptides at 10-fold molar excess in binding buffers (20mM HEPES, pH 7.4, 150mM NaCl) for 1h at room temperature for activation. Post peptide-induced activation, 5μg Fab30 was added, and reaction was incubated for an additional 1h at room temperature. After 1h, 25μl of pre-equilibrated protein L beads (Capto™ L resin, GE Healthcare Cat. no. 17547802) was added and reaction was incubated for 90min at room temperature, followed by five washes with binding buffer containing 0.01% LMNG. Bound protein was eluted with 2X SDS loading buffer and 15μl sample was analyzed on 12% SDS-PAGE. For statistical analyses, protein bands were quantified using ImageJ software suite72 and the values were plotted using GraphPad Prism software v 9.5. The data were normalized with respect to their respective experimental control and appropriate statistical analyses were performed as indicated in the corresponding figure legend.
Reconstitution of phosphopeptide-βarr-Fab complexes
A previously published protocol was followed for complex purification with minor modifications.31 Briefly, βarrs were activated with corresponding phosphopeptides at a 1:3 molar ratios of βarr:phosphopeptide for 30-40min at room temperature. Respective Fabs were added to the phosphopeptide-βarr mixture at 1:1.5 molar ratio of βarr:Fab and incubated for 1h at room temperature. To remove excess Fabs, the phosphopeptide-βarr-Fab complexes were concentrated with 30,000 MWCO concentrators (Vivaspin, Cytiva Cat. no. 28932361) and injected into Superose 6 Increase 10/300 GL (Cytiva Cat. no. 29091596) gel-filtration column. Fractions were further analyzed on SDS-PAGE and selected fractions were pooled and concentrated for structural studies.
Negative-staining EM
Complex formation, homogeneity, and particle quality of the samples were judged through negative staining of the samples prior to data collection under cryogenic conditions for high resolution reconstructions. Negative staining of the samples was performed with uranyl formate in accordance with the previously published protocols.74 For imaging, 3.5μl of the samples were dispensed on glow discharged carbon/formvar coated 300 mesh Cu grids (PELCO, Ted Pella) and allowed to adsorb for 1min, followed by blotting off the sample using a filter paper. The grid was then touched on a first drop of freshly prepared 0.75% (w/v) uranyl formate stain and immediately blotted off, followed by staining for 30sec on a second drop of stain. Imaging of the negatively stained samples were performed on a FEI Tecnai G2 12 Twin TEM (LaB6) operating at 120kV and equipped with a Gatan CCD camera (4k x 4k) at 30,000x magnification. Data processing of the collected micrographs for the individual samples were performed with Relion 3.1.2.70 Approximately 10,000 particles were autopicked using the gaussian blob picker within Relion and the extracted particles were subjected to reference free 2D classification.
Cryo-EM sample preparation and data acquisition
Quantifoil holey carbon grids (Cu or Au, R2/1 or R2/2) were glow discharged for 45sec with a Glocube glow discharge system (Quorum technologies Ltd, UK). 3μl of the complex was dispensed on the glow discharged grid, blotted for 3sec with a Whatman paper filter no. 1 at 10°C and maintained at 90% humidity and then plunge frozen into liquid ethane (−180 °C) using a Leica GP plunger (Leica Microsystems, Austria).
For C5aR1pp-βarr1-Fab30 complex, cryo-EM data collection was performed on R2/2 Cu 300 mesh grid using a Titan Krios electron microscope (Thermofisher Scientific, USA) operating at 300kV equipped with the Gatan Energy Filter. Movies were recorded in counting mode with a Gatan K2 Summit DED (Gatan, USA) using the automated SerialEM software71 at a nominal magnification of 165,000x and a pixel size of 0.82Å at sample level. 6,212 movie stacks consisting of 40 frames were recorded over a defocus range of 0.5 to 2.5μm with a total dose of 49e-/A2 and total exposure time of 5sec.
For CXCR4pp-βarr1-Fab30 complex, cryo-EM data collection was performed on R2/2 Cu 300 mesh grid using a Glacios electron microscope (Thermofisher Scientific, USA) operating at 200kV. Movies were recorded in counting mode with a Gatan K3 DED (Gatan, USA) using the automated SerialEM software at a nominal magnification of 46,000x and a pixel size of 0.878Å at sample level. 5,637 movie stacks consisting of 40 frames were recorded over a defocus range of 0.5 to 2.5μm with a total dose of 49.3e-/A2 and total exposure time of 2.9sec.
For V2Rpp-βarr2-Fab30 complex, cryo-EM data collection was performed on R2/2 Au 200 mesh grid using a Titan Krios electron microscope (Thermofisher Scientific, USA) operating at 300kV equipped with the Gatan Energy Filter. Movies were recorded in counting mode with a Gatan K2 Summit DED (Gatan, USA) using the automated SerialEM software at a nominal magnification of 165,000x and a pixel size of 0.82Å at sample level. 9,720 movie stacks consisting of 40 frames were recorded over a defocus range of 0.5 to 2.5μm with a total dose of 48.7e-/A2 and total exposure time of 4sec.
For C5aR1pp-βarr2-Fab30 complex, cryo-EM data collection was performed on R2/2 Cu 300 mesh grid using a Glacios electron microscope (Thermofisher Scientific, USA) operating at 200kV. Movies were recorded in counting mode with a Gatan K3 DED (Gatan, USA) using the automated SerialEM software at a nominal magnification of 46,000x and a pixel size of 0.878Å at sample level. 8,614 movie stacks consisting of 40 frames were recorded over a defocus range of 0.5 to 2.5μm with a total dose of 51e-/A2 and total exposure time of 3sec.
Cryo-EM data processing and model building
All image processing steps were performed in cryoSPARC version 3.3.267 unless otherwise stated. In brief, for the C5aR1pp-βarr1-Fab30 complex, 6,212 movie stacks were subjected to patch motion correction (multi), followed by CTF refinement with patch CTF multi. 5,790 motion corrected micrographs with CTF fit resolution better than 4.5Å were selected for further processing. 4,304,237 particle projections were automatically picked with blob picker, extracted with a box size of 480pixels and fourier cropped to 64pixels. The particle stack so obtained was subjected to multiple rounds of 2D classification. The class averages with clear secondary structural features were selected and re-extracted with a box size of 480pixels and fourier cropped to 256pixels resulting in a pixel size of 1.5375Å. 295,922 re-extracted particles were then subjected to Ab-initio reconstruction and 3D classification/Heterogeneous refinement with C1 symmetry yielding 4 models. 80,437 particles corresponding to a dimer and containing 47.9% of the total particles were subjected to non-uniform refinement with C2 symmetry to yield a map with an estimated resolution of 3.41Å (voxel size of 1.5375Å). Further, local refinement was performed by masking out the variable domains of Fab30 resulting into an estimated resolution of 3.26Å. Local resolution of all reconstructions was estimated using the Blocres within cryoSPARC version 3.3.2.
For the CXCR4pp-βarr1-Fab30 complex data set, 5,637 movies were motion corrected using a patch of 5x5 patch within patch motion correction (multi). Following CTF estimation, 5,236 motion corrected micrographs with CTF fit resolution better than 6Å were curated for further processing. 3,236,193 particles were automatically picked using the blob-picker sub-program and subsequently extracted with a box size of 480pixels and fourier cropped to 64pixels. The extracted particles were subjected to several rounds of 2D classification to remove junk particles. 104,707 particles corresponding to the clean class averages were selected, re-extracted with a box size of 480pixels and fourier cropped to 256pixels (pixel size of 1.65) and used to produce two ab-initio models. The particles corresponding to the two ab-initio models were subjected to heterogenous refinement/3D classification which produced a 3D class with clear dimeric conformation and a particle count of 86,525. This particle set was re-extracted with full box size of 480pixels (pixel size of 0.878) and subjected to non-uniform refinement with C2 symmetry which converge to a map with 4.81Å resolution as estimated using the gold standard Fourier Shell Correlation (GFSC) using the 0.143 criterion. Local refinement was performed by masking out the variable domains of Fab30 which resulted into an estimated resolution of 4.45Å.
For the V2Rpp-βarr2-Fab30 complex, 9,720 movies were motion corrected with 5x5 patches followed by CTF estimation with patch CTF (multi). Following CTF refinement, 8,295 movies with CTF fit resolution better than 4.5Å were used for further processing. Particle picking from the curated micrographs was performed automatically with the blob picker sub-program to obtain an initial stack of 2,444,407 particles. The particles were then extracted with a box size of 512pixels and fourier cropped to a box size of 64pixels. The extracted particles were subjected to several rounds of reference free 2D classification. 2D class averages with evident secondary features containing 161,436 particles were extracted with a box size of 512pixels and fourier cropped to a box size of 256 (pixel size of 1.64). This sub-set of particles was used for ab-initio reconstruction and subsequent rounds of 3D/Heterogeneous classification with C1 symmetry to obtain 2 models. 92,018 particles corresponding to a trimer were re-extracted with full box size of 512pixels which refined to an overall resolution of 4.18Å (voxel size of 0.82Å) with NU refinement (C3 symmetry) according to the gold standard Fourier shell correlation (FSC) criterion of 0.143. Subsequently, local refinement was performed on βarr and the variable domains of Fab30 and resulted into an estimated resolution of 3.96Å.
For the C5aR1pp-βarr2-Fab30 complex data set, 8,614 movies were motion corrected using patch motion correction (multi) and subsequent CTF estimation was performed through patch CTF (multi). 8,157 micrographs with CTF fit resolution better than 6Å were curated for particle picking using the blob picker sub-program. 4,012,616 particles were automatically picked and extracted with a box size of 512pixels and fourier cropped to 64pixels. Reasonable class averages after several rounds of reference free 2D classification yielded a particle set containing 54,193 particle projections, which was re-extracted with a box size of 512pixels and fourier cropped to 360pixels (pixel size of 1.2487Å) for subsequent used for ab-initio reconstruction generating two ab-initio models. Following heterogenous refinement/3D classification, the 3D class with evident features of a trimer and containing 38,206 particles was subjected to non-uniform refinement with C3 symmetry to yield a reconstruction at 4.41Å (final voxel size of 1.2487Å) as determined by gold standard Fourier Shell Correlation (FSC) using the 0.143 criterion. Local refinement was performed masking out the variable domains of Fab30 which improved the resolution to 4.33Å.
Model building and refinement
Coordinates from a previously solved V2Rpp bound βarr1 structure (PDB 4JQI) was used to dock the model into the EM density map of C5aR1pp-βarr1-Fab30 using Chimera.64 The EM map was then used for manual rebuilding of the βarr1 residues and placing the phosphopeptide in COOT.66 The rebuilt model was subjected to real space refinement in Phenix69 to obtain a model with 97.23% of the residues in most favored region and 2.77% in the allowed region of the Ramachandran plot.
The protomeric structure from the IP6-βarr2 (PDB 5TV1) complex solved in a previous study was used as an initial model to dock into the density map of V2Rpp-βarr2-Fab30 complex and regenerate the trimeric complex with C3 symmetry. The rigid body fitted trimeric model and the phosphopeptides were then rebuilt manually into the EM density map. The rebuilt trimeric coordinates with the phosphopeptides were subsequently subjected to real space refinement in Phenix to reach a final model with 95.05% in the favored region and 4.76% in the allowed region of the Ramachandran plot.
For model building into the 4.33Å C5aR1pp-βarr2-Fab30 coulombic map, the co-ordinates corresponding to V2Rpp peptide were deleted from the trimeric co-ordinates of V2Rpp-βarr2-Fab30 complex (PDB 8GOC), and the resulting model was docked into the EM map in Chimera. The “all atom refine” sub-module within the “refine” module in COOT was used for initial fitting of the model into the EM map, followed by manual rebuilding of the phosphopeptides. Multiple rounds of Phenix real space refinement combined with iterative model building yielded a model with 94.9% of the residues residing in the most favored region of the Ramachandran plot.
The dimeric co-ordinates from the cryo-EM structure of C5aR1pp-βarr1-Fab30 (PDB 8GO8) without the phosphopeptide was used as an initial model to dock into the CXCR4pp-βarr1-Fab30 EM map using Chimera. The docked model along with the coulombic map were imported into COOT and the model was subjected to “all atom refine” for fitting the atoms into the density. The phosphopeptide was manually built into the density to yield a complete model, which was subsequently used to refine the model against the EM map with Phenix real space refinement. The final refined model had 96.62% residues in the most favored regions and 3.38% in the allowed regions of the Ramachandran plot. For model building into the locally refined maps, coordinates of the individual full-length structures were used to dock into the corresponding maps followed by iterative rounds of manual adjustments in COOT and real space refinement in Phenix.
All the refined models were validated using “Comprehensive Validation (cryo-EM)” sub-module in Phenix. 3D reconstruction and model refinement statistics for both full and local-refined structures are provided as Table 1. Figures in the manuscript have been prepared with Chimera64 and ChimeraX65 software. Domain rotation analysis was performed with PyMOL.75 The interaction interface of βarr oligomers in the cryo-EM structures were identified using PDBSum.68
Table 1. Cryo-EM data collection, processing, and refinement statistics, related to Figures 1, 4, S2, and S5.
| C5aR1pp- βarr1-Fab30 |
C5aR1pp-βarr1- Fab30 (local refined) |
V2Rpp- βarr2-Fab30 |
V2Rpp- βarr2-Fab30 (local refined) |
C5aR1pp- βarr2-Fab30 |
C5aR1pp-βarr2- Fab30 (local refined) |
CXCR4pp- βarr1-Fab30 |
CXCR4pp-βarr1- Fab30 (local refined) |
|
|---|---|---|---|---|---|---|---|---|
| Code | PDB:8GO8 EMD 34173 |
PDB: 8I0N EMD 35104 |
PDB: 8GOC EMD 34175 |
PDB: 8I10 EMD 35115 |
PDB: 8GOO EMD 34178 |
PDB: 8I0Z EMD 35114 |
PDB: 8GP3 EMD 34188 |
PDB: 8I0Q EMD 35106 |
| Microscope | Titan Krios | Titan Krios | Titan Krios | Titan Krios | Glacios | Glacios | Glacios | Glacios |
| Camera | GIF/K2 | GIF/K2 | GIF/K2 | GIF/K2 | Gatan K3 | Gatan K3 | Gatan K3 | Gatan K3 |
| Magnification | 165,000 | 165,000 | 165,000 | 165,000 | 46,000 | 46,000 | 46,000 | 46,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 | 200 | 200 | 200 | 200 |
| Defocus range (μm) | 0.5–2.5 | 0.5–2.5 | 0.5–2.5 | 0.5–2.5 | 0.5–2.5 | 0.5–2.5 | 0.5–2.5 | 0.5–2.5 |
| Exposure time (s) | 5 | 5 | 4 | 4 | 3 | 3 | 2.9 | 2.9 |
| Total dose (e−/Ǻ2) | 49 | 49 | 48.7 | 48.7 | 51 | 51 | 49.38 | 49.38 |
| Number of frames | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 40 |
| Pixel size (Ǻ) | 0.82 | 0.82 | 0.82 | 0.82 | 0.878 | 0.878 | 0.878 | 0.878 |
| Micrographs (no.) | 6,212 | 6,212 | 9,720 | 9,720 | 8,614 | 8,614 | 5,637 | 5,637 |
| Initial particles (no.) | 4,304,237 | 4,304,237 | 2,444,407 | 2,444,407 | 4,012,616 | 4,012,616 | 3,236,193 | 3,236,193 |
| Symmetry imposed | C2 | C2 | C3 | C3 | C3 | C3 | C2 | C2 |
| Final particles (no.) | 80,437 | 80,437 | 92,018 | 92,018 | 38,206 | 38,206 | 86,525 | 86,525 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution (Ǻ) | 3.41 | 3.26 | 4.18 | 3.96 | 4.41 | 4.33 | 4.81 | 4.45 |
| Refinement | ||||||||
| Initial code (PDB) | 4JQI | 8GO8 | 5TV1 | 8GOC | 8GOC | 8GOO | 8GO8 | 8GP3 |
| Model resolution (Ǻ) | 3.5 | 3.4 | 4.7 | 4.1 | 4.7 | 4.7 | 5.3 | 5.7 |
| FSC threshold | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Model composition | ||||||||
| Non-hydrogen atoms | 11,519 | 9,141 | 16,942 | 13,205 | 16,966 | 13,163 | 11,494 | 8,842 |
| Protein residues | 1,522 | 1,190 | 2,223 | 1,729 | 2,223 | 1,725 | 1,522 | 1,192 |
| Ligand atoms | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RMSD | ||||||||
| Bond length (Ǻ) | 0.004 | 0.005 | 0.003 | 0.007 | 0.004 | 0.006 | 0.005 | 0.004 |
| Bond angle (°) | 0.942 | 1.001 | 0.716 | 1.256 | 0.665 | 1.128 | 0.982 | 0.967 |
| Validation | ||||||||
| Favored (%) | 97.23 | 95.43 | 95.05 | 93.63 | 94.90 | 93.24 | 96.62 | 94.92 |
| Allowed (%) | 2.77 | 4.57 | 4.76 | 6.37 | 4.96 | 6.76 | 3.38 | 5.08 |
| Disallowed (%) | 0 | 0 | 0.18 | 0 | 0.14 | 0 | 0 | 0 |
| MolProbity score | 1.41 | 1.46 | 1.97 | 1.88 | 1.91 | 1.77 | 1.77 | 1.85 |
| Clash score | 5.23 | 3.49 | 13.07 | 8.60 | 10.83 | 6.12 | 10.81 | 9.44 |
NanoBiT assay for βarr2WT and βarr2DM recruitment
βarr2WT and βarr2DM recruitment downstream of V2R and C5aR1 in response to AVP and C5a, respectively, was measured using NanoBiT (Enzyme linked complementation-based assay) assay following the protocol described earlier.76 Receptor constructs were tagged with SmBiT at the carboxyl-terminus, and βarr2 constructs were N-terminally tagged with LgBiT. Briefly, HEK-293 cells were transfected with indicated receptor constructs (3.57#x03BC;g) and βarr2 (βarr2WT/DM) constructs (3.5μg) using polyethylenimine (PEI) linear (Polysciences, Cat. no. 19850) at a ratio of 1:3 (DNA:PEI linear). After 16-18h of transfection, cells were trypsinized, harvested, and resuspended in assay buffer (1XHBSS, 5mM HEPES, pH 7.4, 0.01% BSA) containing 10μM coelenterazine (GoldBio, Cat. no. CZ05). Resuspended cells were seeded in a white flat bottom 96-well plate (100μl well-1). After 2h of incubation (90min at 37°C and 30min at room temperature), basal luminescence was recorded using a multimode plate reader (FLUOstar Omega, BMG Lab-tech). Later, cells were stimulated with varying doses of indicated ligands followed by measurement of luminescence signal for 20 cycles. For data analysis, ligand induced change in signals were taken and normalized with the lowest ligand dose luminescence value, and fold normalized data was plotted using nonlinear regression three-parameter sigmoidal concentration-response curve in GraphPad Prism v 9.5 software.
NanoBiT assay for βarr trafficking
Agonist-induced βarr2WT and βarr2DM endosomal trafficking downstream of the receptors mentioned above was studied using NanoBiT assay as described in the recruitment experiment. The only exception from the recruitment assay was that the receptor constructs were not tagged with SmBiT, but rather βarr2 (βarr2WT/DM) and FYVE constructs N-terminally fused with SmBiT and LgBiT respectively were used for enzyme complementation. For each experiment, 3μg of indicated receptors, 2μg of SmBiT-βarr2WT/DM, and 5μg of LgBiT-FYVE were used. Fold normalized change in signals were plotted using nonlinear regression three-parameter sigmoidal concentration-response curve in GraphPad Prism v 9.5 software.
NanoBiT assay for Ib30 reactivity
To assess Ib30 reactivity in response to an agonist for the mentioned receptors, NanoBiT assay was used following the same protocol as discussed in the βarr2WT and βarr2DM recruitment assay.77 For enzyme complementation, N-terminally SmBiT fused βarr1 and N-terminally LgBiT fused Ib30 were used. For transfection, 3μg receptor except for M2R, CXCR7 (5μg), and CXCR3 (7μg), 2μg SmBiT-βarr1, and 5μg LgBiT-Ib30 were used. Transfected cells were stimulated with varying doses of respective ligands (mentioned in corresponding figures). Fold normalized change in luminescence were plotted using nonlinear regression three/four-parameter sigmoidal concentration-response curve in GraphPad Prism v 9.5 software.
Receptor surface expression
Receptor surface expression in various assays was measured using a previously described whole cell-based surface ELISA assay.78 To study the surface expression of the receptor, cells transfected with a particular receptor were seeded into a 0.01% poly-D-Lysine pre-coated 24-well plate at a density of 2x105 cells well-1. Post 24h of seeding cells were washed once with ice-cold 1XTBS, fixed with 4% PFA (w/v in 1XTBS) on ice for 20min, washed again three times with 1XTBS, and blocked with 1% BSA (prepared in 1XTBS) at room temperature for 1.5h. Afterward, cells were incubated with anti-FLAG M2-HRP antibody at 1:5000 dilution (Sigma, Cat. no. A8592) for 1.5h, which was followed by three washes in 1% BSA. Subsequently, incubated with TMB-ELISA substrate (Thermo Fisher Scientific, Cat. no. 34028) until a light blue color appeared. To quench the reaction, 100μl of the colored solution was transferred to another 96-well plate containing 100μl of 1M H2SO4, and the absorbance was measured at 450nm. Afterward, the TMB substrate was removed, washed twice with 1XTBS, and incubated with 0.2% (w/v) Janus Green (Sigma; Cat. no. 201677) for 15min at room temperature. Later, cells were washed with water to remove the excess stain, followed by the addition of 800μl of 0.5N HCl in each well. Thereupon, the colored solution was transferred to a 96-well plate for measuring the absorbance at 595nm. The signal intensity was normalized by calculating the ratio of A450/A595 values followed by quantifying fold increase with respect to the A450/A595nm value of negative control (mock transfection) and plotted using the GraphPad Prism v 9.5.
Quantification And Statistical Analysis
GraphPad Prism v9.5 was used to plot and analyze all the functional data presented in this manuscript, and all the relevant details such as number of replicates, data normalization, mean±sem, and statistical analyses are mentioned in the corresponding figure legends.
Highlights.
Cryo-EM structure determination of β-arrestins in complex with GPCR phosphopeptides
Identification of P-X-P-P motif in GPCRs for β-arrestin interaction and activation
Experimental validation of P-X-P-P motif using mutagenesis and conformational sensor
Discovery of a significantly conserved β-arrestin activation mechanism by GPCRs
In brief.
Maharana et al. determine multiple structures of activated β-arrestins in complex with the carboxyl terminus phosphopeptides of different GPCRs using cryo-EM. Based on these structural snapshots, they discover and experimentally validate a significantly conserved phosphorylation motif in GPCRs that drives β-arrestin interaction and activation.
Acknowledgments
Research in A.K.S.’s laboratory is supported by the Senior Fellowship of the DBT Wellcome Trust India Alliance (IA/S/20/1/504916) awarded to A.K.S., Science and Engineering Research Board (SPR/2020/000408 and IPA/2020/000405), Council of Scientific and Industrial Research (37(1730)/19/EMR-II), Indian Council of Medical Research (F.NO.52/15/2020/BIO/BMS), the Young Scientist Award from Lady Tata Memorial Trust, and IIT Kanpur. We thank A. Ranjan, M. Chaturvedi, and H. Dwivedi-Agnihotri for their help with the characterization of the phosphopeptides; M. Ganguly for assisting with GPCR sequence analysis; E. Ghosh for initial characterization of βarr2DM; and A. Dalal and N. Zaidi for helping with the functional assays on M2R. Cryo-EM was performed at the BioEM lab of the Biozentrum at the University of Basel, and we thank Carola Alampi and David Kalbermatter for their excellent technical assistance.
Footnotes
Author Contributions
J.M. and M.K.Y. prepared and characterized the βarr complexes. J.M. performed negative-staining EM with R.B. and processed the cryo-EM data with R.B. P.S. carried out all the functional assays related to βarr2DM characterization and Ib30 sensor assay. M.K.Y. purified βarrs and carried out the coIP experiments with V.S. and Sayantan Saha. Shirsha Saha contributed to the functional characterization of βarr2DM. M.C. screened the samples and collected cryo-EM data. A.K.S. supervised and managed the overall project. All authors contributed to data analysis, interpretation, and manuscript writing.
Declaration of Interests
The authors declare no competing interests.
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Data and code availability
All three-dimensional cryo-EM density maps, coordinates for the atomic models and local-refined maps generated in this study have been deposited and are publicly available as of the date of publication. Accession numbers (EMDB and PDB IDs) are listed in the key resources table. Original gel images have been deposited to Mendeley data, and they are publicly available after publication. The DOI is listed in the key resources table.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Molecular mechanism of beta-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol. 2012;52:179–197. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shenoy SK, Lefkowitz RJ. Seven-transmembrane receptor signaling through beta-arrestin. Sci STKE. 2005;2005:cm10. doi: 10.1126/stke.2005/308/cm10. [DOI] [PubMed] [Google Scholar]
- 4.Pierce KL, Lefkowitz RJ. Classical and new roles of beta-arrestins in the regulation of G-protein-coupled receptors. Nat Rev Neurosci. 2001;2:727–733. doi: 10.1038/35094577. [DOI] [PubMed] [Google Scholar]
- 5.Maharana J, Banerjee R, Yadav MK, Sarma P, Shukla AK. Emerging structural insights into GPCR-beta-arrestin interaction and functional outcomes. Curr Opin Struct Biol. 2022;75:102406. doi: 10.1016/j.sbi.2022.102406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranjan R, Dwivedi H, Baidya M, Kumar M, Shukla AK. Novel structural insights into GPCR-beta-arrestin interaction and signaling. Trends Cell Biol. 2017;27:851–862. doi: 10.1016/j.tcb.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Ahn S, Shenoy SK, Luttrell LM, Lefkowitz RJ. SnapShot: beta-arrestin functions. Cell. 2020;182:1362.:e1. doi: 10.1016/j.cell.2020.07.034. [DOI] [PubMed] [Google Scholar]
- 8.Shenoy SK, Lefkowitz RJ. beta-arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol Sci. 2011;32:521–533. doi: 10.1016/j.tips.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang DS, Tian X, Benovic JL. Role of beta-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Curr Opin Cell Biol. 2014;27:63–71. doi: 10.1016/j.ceb.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seyedabadi M, Gharghabi M, Gurevich EV, Gurevich VV. Receptor-arrestin interactions: the GPCR perspective. Biomolecules. 2021;11:218. doi: 10.3390/biom11020218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurevich VV, Gurevich EV. The structural basis of the arrestin binding to GPCRs. Mol Cell Endocrinol. 2019;484:34–41. doi: 10.1016/j.mce.2019.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurevich VV, Gurevich EV. Overview of different mechanisms of arrestin-mediated signaling. Curr Protoc Pharmacol. 2014;67:2.10.11–12.10.19. doi: 10.1002/0471141755.ph0210s67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiraishi Y, Kofuku Y, Ueda T, Pandey S, Dwivedi-Agnihotri H, Shukla AK, Shimada I. Biphasic activation of beta-arrestin 1 upon interaction with a GPCR revealed by methyl-TROSY NMR. Nat Commun. 2021;12:7158. doi: 10.1038/s41467-021-27482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shukla AK, Westfield GH, Xiao K, Reis RI, Huang LY, Tripathi-Shukla P, Qian J, Li S, Blanc A, Oleskie AN, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bous J, Fouillen A, Orcel H, Trapani S, Cong X, Fontanel S, Saint-Paul J, Lai-Kee-Him J, Urbach S, Sibille N, et al. Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex. Sci Adv. 2022;8:eabo7761. doi: 10.1126/sciadv.abo7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, Warne T, Nehmé R, Pandey S, Dwivedi-Agnihotri H, Chaturvedi M, Edwards PC, García-Nafría J, Leslie AGW, Shukla AK, Tate CG. Molecular basis of beta-arrestin coupling to formoterol-bound beta(1)-adrenoceptor. Nature. 2020;583:862–866. doi: 10.1038/s41586-020-2419-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W, Masureel M, Qu Q, Janetzko J, Inoue A, Kato HE, Robertson MJ, Nguyen KC, Glenn JS, Skiniotis G, Kobilka BK. Structure of the neurotensin receptor 1 in complex with beta-arrestin 1. Nature. 2020;579:303–308. doi: 10.1038/s41586-020-1953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staus DP, Hu H, Robertson MJ, Kleinhenz ALW, Wingler LM, Capel WD, Latorraca NR, Lefkowitz RJ, Skiniotis G. Structure of the M2 muscarinic receptor-beta-arrestin complex in a lipid nanodisc. Nature. 2020;579:297–302. doi: 10.1038/s41586-020-1954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin W, Li Z, Jin M, Yin YL, de Waal PW, Pal K, Yin Y, Gao X, He Y, Gao J, et al. A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Res. 2019;29:971–983. doi: 10.1038/s41422-019-0256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao C, Barros-Álvarez X, Zhang S, Kim K, Dämgen MA, Panova O, Suomivuori CM, Fay JF, Zhong X, Krumm BE, et al. Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD. Neuron. 2022;110:3154–3167.:e7. doi: 10.1016/j.neuron.2022.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latorraca NR, Wang JK, Bauer B, Townshend RJL, Hollingsworth SA, Olivieri JE, Xu HE, Sommer ME, Dror RO. Molecular mechanism of GPCR-mediated arrestin activation. Nature. 2018;557:452–456. doi: 10.1038/s41586-018-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichel K, Jullié D, Barsi-Rhyne B, Latorraca NR, Masureel M, Sibarita JB, Dror RO, von Zastrow M. Catalytic activation of beta-arrestin by GPCRs. Nature. 2018;557:381–386. doi: 10.1038/s41586-018-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reiter E, Lefkowitz RJ. GRKs and beta-arrestins: roles in receptor silencing, trafficking and signaling. Trends Endocrinol Metab. 2006;17:159–165. doi: 10.1016/j.tem.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Prihandoko R, Bradley SJ, Tobin AB, Butcher AJ. Determination of GPCR phosphorylation status: establishing a phosphorylation barcode. Curr Protoc Pharmacol. 2015;69:2.13.1–2.13.26. doi: 10.1002/0471141755.ph0213s69. [DOI] [PubMed] [Google Scholar]
- 25.Tobin AB. G-protein-coupled receptor phosphorylation: where, when and by whom. Br J Pharmacol. 2008;153:S167–S176. doi: 10.1038/sj.bjp.0707662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobin AB, Butcher AJ, Kong KC. Location, location, location…site-specific GPCR phosphorylation offers a mechanism for cell-type-specific signalling. Trends Pharmacol Sci. 2008;29:413–420. doi: 10.1016/j.tips.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, Yang F, Zhang D, Liu Z, Lin A, Liu C, Xiao P, Yu X, Sun JP. Phosphorylation of G protein-coupled receptors: from the barcode hypothesis to the flute model. Mol Pharmacol. 2017;92:201–210. doi: 10.1124/mol.116.107839. [DOI] [PubMed] [Google Scholar]
- 28.Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, Strachan RT, Huang TY, Bressler EA, Hara MR, et al. Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Sci Signal. 2011;4:ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q, Tesmer JJG. G protein-coupled receptor interactions with arrestins and GPCR kinases: the unresolved issue of signal bias. J Biol Chem. 2022;298:102279. doi: 10.1016/j.jbc.2022.102279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srivastava A, Gupta B, Gupta C, Shukla AK. Emerging functional divergence of beta-arrestin isoforms in GPCR function. Trends Endocrinol Metab. 2015;26:628–642. doi: 10.1016/j.tem.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, Staus DP, Hilger D, Uysal S, Huang LY, et al. Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature. 2013;497:137–141. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He QT, Xiao P, Huang SM, Jia YL, Zhu ZL, Lin JY, Yang F, Tao XN, Zhao RJ, Gao FY, et al. Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2. Nat Commun. 2021;12:2396. doi: 10.1038/s41467-021-22731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min K, Yoon HJ, Park JY, Baidya M, Dwivedi-Agnihotri H, Maharana J, Chaturvedi M, Chung KY, Shukla AK, Lee HH. Crystal structure of beta-arrestin 2 in complex with CXCR7 phosphopeptide. Structure. 2020;28:1014–1023.:e4. doi: 10.1016/j.str.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Q, Perry NA, Vishnivetskiy SA, Berndt S, Gilbert NC, Zhuo Y, Singh PK, Tholen J, Ohi MD, Gurevich EV, et al. Structural basis of arrestin-3 activation and signaling. Nat Commun. 2017;8:1427. doi: 10.1038/s41467-017-01218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation. Structure. 2001;9:869–880. doi: 10.1016/S0969-2126(01)00644-X. [DOI] [PubMed] [Google Scholar]
- 36.Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J Mol Biol. 2011;406:467–478. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milano SK, Pace HC, Kim YM, Brenner C, Benovic JL. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–3328. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]
- 38.Kang DS, Kern RC, Puthenveedu MA, von Zastrow M, Williams JC, Benovic JL. Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking. J Biol Chem. 2009;284:29860–29872. doi: 10.1074/jbc.M109.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh E, Srivastava A, Baidya M, Kumari P, Dwivedi H, Nidhi K, Ranjan R, Dogra S, Koide A, Yadav PN, et al. A synthetic intrabody-based selective and generic inhibitor of GPCR endocytosis. Nat Nanotechnol. 2017;12:1190–1198. doi: 10.1038/nnano.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh E, Dwivedi H, Baidya M, Srivastava A, Kumari P, Stepniewski T, Kim HR, Lee MH, van Gastel J, Chaturvedi M, et al. Conformational sensors and domain swapping reveal structural and functional differences between beta-arrestin isoforms. Cell Rep. 2019;28:3287–3299.:e6. doi: 10.1016/j.celrep.2019.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baidya M, Chaturvedi M, Dwivedi-Agnihotri H, Ranjan A, Devost D, Namkung Y, Stepniewski TM, Pandey S, Baruah M, Panigrahi B, et al. Allosteric modulation of GPCR-induced beta-arrestin trafficking and signaling by a synthetic intrabody. Nat Commun. 2022;13:4634. doi: 10.1038/s41467-022-32386-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dwivedi-Agnihotri H, Chaturvedi M, Baidya M, Stepniewski TM, Pandey S, Maharana J, Srivastava A, Caengprasath N, Hanyaloglu AC, Selent J, Shukla AK. Distinct phosphorylation sites in a prototypical GPCR differently orchestrate beta-arrestin interaction, trafficking, and signaling. Sci Adv. 2020;6:eabb8368. doi: 10.1126/sciadv.abb8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baidya M, Kumari P, Dwivedi-Agnihotri H, Pandey S, Chaturvedi M, Stepniewski TM, Kawakami K, Cao Y, Laporte SA, Selent J, et al. Key phosphorylation sites in GPCRs orchestrate the contribution of beta-arrestin in ERK1/2 activation. EMBO Rep. 2020;21:e49886. doi: 10.15252/embr.201949886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajagopal S, Kim J, Ahn S, Craig S, Lam CM, Gerard NP, Gerard C, Lefkowitz RJ. Beta-arrestin-but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc Natl Acad Sci USA. 2010;107:628–632. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen HT, Reyes-Alcaraz A, Yong HJ, Nguyen LP, Park HK, Inoue A, Lee CS, Seong JY, Hwang JI. CXCR7: a beta-arrestin-biased receptor that potentiates cell migration and recruits beta-arrestin2 exclusively through Gbetagamma subunits and GRK2. Cell Biosci. 2020;10:134. doi: 10.1186/s13578-020-00497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reyes-Alcaraz A, Lee YN, Yun S, Hwang JI, Seong JY. Conformational signatures in beta-arrestin2 reveal natural biased agonism at a G-protein-coupled receptor. Commun Biol. 2018;1:128. doi: 10.1038/s42003-018-0134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmerman B, Simaan M, Akoume MY, i N, Chevallier S, Séguéla P, Laporte SA. Role of ssarrestins in bradykinin B2 receptor-mediated signalling. Cell Signal. 2011;23:648–659. doi: 10.1016/j.cellsig.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 48.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lassmann T. Kalign 3: multiple sequence alignment of large data sets. Bioinformatics. 2019;36:1928–1929. doi: 10.1093/bioinformatics/btz795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shukla AK, Violin JD, Whalen EJ, Gesty-Palmer D, Shenoy SK, Lefkowitz RJ. Distinct conformational changes in beta-arrestin report biased agonism at seven-transmembrane receptors. Proc Natl Acad Sci USA. 2008;105:9988–9993. doi: 10.1073/pnas.0804246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou XE, He Y, de Waal PW, Gao X, Kang Y, Van Eps N, Yin Y, Pal K, Goswami D, White TA, et al. Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell. 2017;170:457–469.:e13. doi: 10.1016/j.cell.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer D, Damberger FF, Samarasimhareddy M, Feldmueller M, Vuckovic Z, Flock T, Bauer B, Mutt E, Zosel F, Allain FHT, et al. Distinct G protein-coupled receptor phosphorylation motifs modulate arrestin affinity and activation and global conformation. Nat Commun. 2019;10:1261. doi: 10.1038/s41467-019-09204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 55.Gurevich VV, Gurevich EV. Solo vs. Chorus: monomers and Oligomers of arrestin Proteins. Int J Mol Sci. 2022;23:7253. doi: 10.3390/ijms23137253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Q, Zhuo Y, Kim M, Hanson SM, Francis DJ, Vishnivetskiy SA, Altenbach C, Klug CS, Hubbell WL, Gurevich VV. Self-association of arrestin family members. Handb Exp Pharmacol. 2014;219:205–223. doi: 10.1007/978-3-642-41199-1_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Q, Zhuo Y, Sharma P, Perez I, Francis DJ, Chakravarthy S, Vishnivetskiy SA, Berndt S, Hanson SM, Zhan X, et al. An eight amino acid Segment Controls Oligomerization and Preferred Conformation of the two Non-visual arrestins. J Mol Biol. 2021;433:166790. doi: 10.1016/j.jmb.2020.166790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gurevich VV, Gurevich EV. Plethora of functions packed into 45 kDa arrestins: biological implications and possible therapeutic strategies. Cell Mol Life Sci. 2019;76:4413–4421. doi: 10.1007/s00018-019-03272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isaikina P, P I, Jakob RP, Sarma P, Ranjan A, Baruah M, Panwalkar V, Maier T, Shukla AK, Grzesiek S. A key GPCR phosphorylation motif discovered in arrestin2•CCR5 phosphopeptide complexes. Mol Cell. 2022;83 doi: 10.1016/j.molcel.2023.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Kumari P, Srivastava A, Banerjee R, Ghosh E, Gupta P, Ranjan R, Chen X, Gupta B, Gupta C, Jaiman D, Shukla AK. Functional competence of a partially engaged GPCR-beta-arrestin complex. Nat Commun. 2016;7:13416. doi: 10.1038/ncomms13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumari P, Srivastava A, Ghosh E, Ranjan R, Dogra S, Yadav PN, Shukla AK. Core engagement with beta-arrestin is dispensable for agonist-induced vasopressin receptor endocytosis and ERK activation. Mol Biol Cell. 2017;28:1003–1010. doi: 10.1091/mbc.E16-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cahill TJ, 3rd, Thomsen AR, Tarrasch JT, Plouffe B, Nguyen AH, Yang F, Huang LY, Kahsai AW, Bassoni DL, Gavino BJ, et al. Distinct conformations of GPCR-beta-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc Natl Acad Sci USA. 2017;114:2562–2567. doi: 10.1073/pnas.1701529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asher WB, Terry DS, Gregorio GGA, Kahsai AW, Borgia A, Xie B, Modak A, Zhu Y, Jang W, Govindaraju A, et al. GPCR-mediated beta-arrestin activation deconvoluted with single-molecule precision. Cell. 2022;185:1661–1675.:e16. doi: 10.1016/j.cell.2022.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 68.Laskowski RA, Jabłońska J, Pravda L, Vařeková RS, Thornton JM. PDBsum: structural summaries of PDB entries. Protein Sci. 2018;27:129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zivanov J, Nakane T, Scheres SHW. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020;7:253–267. doi: 10.1107/S2052252520000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 72.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baidya M, Kumari P, Dwivedi-Agnihotri H, Pandey S, Sokrat B, Sposini S, Chaturvedi M, Srivastava A, Roy D, Hanyaloglu AC, et al. Genetically encoded intrabody sensors report the interaction and trafficking of beta-arrestin 1 upon activation of G-protein-coupled receptors. J Biol Chem. 2020;295:10153–10167. doi: 10.1074/jbc.RA120.013470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peisley A, Skiniotis G. 2D projection analysis of GPCR complexes by negative stain electron microscopy. Methods Mol Biol. 2015;1335:29–38. doi: 10.1007/978-1-4939-2914-6_3. [DOI] [PubMed] [Google Scholar]
- 75.Schrödinger L, DeLano W. PyMOL. 2020. http://www.pymol.org/pymol.
- 76.Kawakami K, Yanagawa M, Hiratsuka S, Yoshida M, Ono Y, Hiroshima M, Ueda M, Aoki J, Sako Y, Inoue A. Heterotrimeric Gq proteins act as a switch for GRK5/6 selectivity underlying beta-arrestin transducer bias. Nat Commun. 2022;13:487. doi: 10.1038/s41467-022-28056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dwivedi-Agnihotri H, Sarma P, Deeksha S, Kawakami K, Inoue A, Shukla AK. An intrabody sensor to monitor conformational activation of beta-arrestins. Methods Cell Biol. 2022;169:267–278. doi: 10.1016/bs.mcb.2021.12.023. [DOI] [PubMed] [Google Scholar]
- 78.Pandey S, Roy D, Shukla AK. Measuring surface expression and endocytosis of GPCRs using whole-cell ELISA. Methods Cell Biol. 2019;149:131–140. doi: 10.1016/bs.mcb.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All three-dimensional cryo-EM density maps, coordinates for the atomic models and local-refined maps generated in this study have been deposited and are publicly available as of the date of publication. Accession numbers (EMDB and PDB IDs) are listed in the key resources table. Original gel images have been deposited to Mendeley data, and they are publicly available after publication. The DOI is listed in the key resources table.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.