Abstract
Background
The effects of inhaled corticosteroids (ICS) on healthy airways are poorly defined.
Objectives
To delineate the effects of ICS on gene expression in healthy airways, without confounding caused by changes in disease-related genes and disease-related alterations in ICS-responsiveness.
Methods
Randomised open-label bronchoscopy study of high dose ICS therapy in 30 healthy adult volunteers randomised 2:1 to i) fluticasone propionate 500 mcg bd daily or, ii) no treatment, for 4 weeks. Laboratory staff were blinded to allocation. Biopsies and brushings were analysed by immunohistochemistry, bulk RNA sequencing, DNA methylation array and metagenomics.
Results
ICS induced small between-group differences in blood and lamina propria eosinophil numbers, but not in other immunopathological features, blood neutrophils, FeNO, FEV1, microbiome or DNA methylation. ICS treatment upregulated 72 genes in brushings and 53 genes in biopsies, and downregulated 82 genes in brushings and 416 genes in biopsies. The most downregulated genes in both tissues were canonical markers of type-2 inflammation (FCER1A, CPA3, IL33, CLEC10A, SERPINB10 and CCR5), T cell-mediated adaptive immunity (TARP, TRBC1, TRBC2, PTPN22, TRAC, CD2, CD8A, HLA-DQB2, CD96, PTPN7), B cell immunity (CD20, immunoglobulin heavy and light chains), and innate immunity, including CD48, Hobit, RANTES, Langerin and GFI1. An IL-17-dependent gene signature was not upregulated by ICS.
Conclusions
In healthy airways, 4-week ICS exposure reduces gene expression related to both innate and adaptive immunity, and reduces markers of type-2 inflammation. This implies that homeostasis in health involves tonic type-2 signalling in the airway mucosa, which is exquisitely sensitive to ICS.
Registered at ClincialTrials.gov: NCT02476825
Keywords: asthma, health, inhaled corticosteroids, bronchial biopsy, transcriptome, epigenetics, microbiome
Abbreviations
- ASM
airway smooth muscle
- FeNO
fraction of exhaled nitric oxide
- FEV1
forced expiratory volume
- ICS
inhaled corticosteroid
- IQR
interquartile range
- MBP
major basic protein
- MCT
tryptase only mast cell
- MCTC
tryptase and chymase mast cell
- PG
prostaglandin
- RBM
reticular basement membrane
- RTU
ready-to-use
- SD
standard deviation
- T2
Type-2 cytokines (IL-4, IL-5, IL-13)
Introduction
Inhaled corticosteroids (ICS) are the cornerstone of asthma treatment. They attenuate eosinophilic airway inflammation1,2, improve lung function, and reduce asthma symptoms, exacerbations, and mortality3. However, their use is also associated with an increased risk of pneumonia4. Corticosteroids modulate the expression of many molecular pathways at the level of gene transcription, through direct upregulation of anti-inflammatory molecules and β-adrenoceptors (transactivation), and suppression of pro-inflammatory genes, either through direct DNA-binding or via inhibition of pro-inflammatory transcription factor binding (transrepression)5. In people with severe asthma, ICS are relatively ineffective even at high doses, but the mechanisms behind this corticosteroid insensitivity are poorly understood and likely multi-factorial6,7.
When considering the underlying molecular pathways driving both severe asthma and relative corticosteroid insensitivity, it is unclear to what extent this is driven by pathways that are not responsive to corticosteroids, as opposed to inhibition of corticosteroid signalling. Gene expression profiling in asthmatic epithelial bronchial brushings and bronchial biopsies has identified several molecular pathways present in subgroups of patients with mild, moderate and severe asthma8,9. Approximately 50-80% of people with steroid-naïve “mild” asthma demonstrate evidence of blood or airway eosinophilia, with a concomitant increase in the fraction of exhaled nitric oxide (FeNO)10,11. This phenotype is characterised by increased expression of an airway gene expression signature driven by IL-4 and IL-138,9,12,13; tissue eosinophilia is also dependent on IL-514. Together these cytokines are described as Th2 or type-2 cytokines (T2). T2 expression and the accompanying eosinophilia are suppressed by ICS in mild asthma15. In severe asthma, a persistent T2 gene signature is evident in about 25% of patients, suggesting corticosteroid insensitivity8,9. In addition, in severe asthma, about 25% of patients have evidence of an IL-17-dependent gene signature, which is seen only in people on ICS8,9, and mutually exclusive with the T2 signature. It is therefore not clear whether this IL-17 activity represents an independent corticosteroid-insensitive pathway driving severe asthma or a consequence of ICS therapy. Approximately 50% of people with severe asthma have neither a T2- or IL-17-dependent airway gene signature, and the mechanisms driving their persistent disordered airway physiology remains unknown. So, while gene expression profiling provides insight into the abnormalities present in severe asthmatic airways, the multiple effects of ICS on airway gene expression make it difficult to disentangle the changes due to the disease as opposed to the treatment, and whether corticosteroid activity is inhibited or not.
The effects of ICS on healthy airways are poorly defined. We hypothesise that gene expression data in severe asthma will be more interpretable if we can delineate and thus allow for the effects of high dose ICS therapy. We have therefore performed a randomised open label bronchoscopy study of high dose ICS therapy in healthy adult volunteers, with the aim of understanding transcriptional consequences of ICS therapy without the confounding effects due to disease-related processes.
Methods
Detailed methods are provided in the online data supplement.
Ethics and consent
This prospective study was approved by the East Midlands-Leicester Central Research Ethics Committee (reference:15/EM/0313) and registered at clinicaltrials.gov (NCT02476825). Participants gave written informed consent.
Participant population
Healthy volunteers aged 18-65 were eligible, were current non-smokers with <10 pack year smoking history, and had no prior history or clinical evidence of lower respiratory disease with normal spirometry. Participants with a history of rhinitis were required to have a PC20 methacholine >16 mg/ml.
Study design
This was a randomised, open-labelled, bronchoscopy study designed to assess the effects of 4 weeks treatment with fluticasone propionate on airway gene expression and cellularity in healthy adult volunteers. The primary endpoint was the corticosteroid-inducible gene expression pattern in healthy airways. Secondary endpoints included the relative change from baseline in airway cellularity.
30 participants were randomised by a blinded investigator (MR) in a 2:1 ratio to one of two study groups: i) fluticasone propionate 500 mcg b.i.d. via Accuhaler (Diskus) daily for 4 weeks (n=20), or ii) no treatment (observation) for 4 weeks (n=10). Bronchoscopy was performed at baseline prior to the start of treatment/observation, and at the end of week 4. Genentech and Leicester laboratory support staff were blinded to treatment allocation.
To ensure there were sufficient data for analysis, if a subject withdrew before completion of the study, a further subject(s) was randomised after the first 30 randomisations until a total of 30 subjects had completed the study.
Bronchoscopy
Subjects underwent bronchoscopy conducted according to British Thoracic Society guidelines16. Mucosal biopsies and brushes were collected from 2nd-5th generation bronchi under direct vision as per study procedure manual.
Tissue processing, immunohistochemistry and assessment of immunopathology
Please see the online supplement.
All pathological data were assessed by an observer blinded to the identity and treatment allocations of the participants.
RNA sequencing
Please see the online supplement.
Bisulphite conversion and DNA methylation arrays, DNA methylation data quality control and normalization, Differential DNA methylation analysis, Expression quantitative trait methylation (eQTM) analysis
Please see the online supplement.
Microbiota Sequence Data Generation, Processing, and Analysis
Please see the online supplement.
Transcriptomic analysis
Sequences in fastq files (in single and pair ends) were aligned using STAR aligner (version 2.7.1a) to the human reference genome GRCh38; R package 17Rsubread was employed for quantification of reads assigned to genes.
Raw count pre-processing, normalisation and differential gene expression analysis was performed using R, packaged edgeR (for pre-processing and gene expression filtering) and gene expression analysis with DESeq2, and limma. Gene lists of differentially expressed genes and over-representation analysis of gene pathways/categories were produced from DESeq2 and limma results; volcano plots were generated with log2 fold changes and adjusted p values resulting from moderated t tests in limma17,18.
Statistical analysis
Basic summary statistical analysis was performed using GraphPad Prism version 7.03 (GraphPad Software, San Diego). Parametric and non-parametric data are presented as mean (standard deviation [SD]) and median (interquartile range [IQR]) respectively unless otherwise stated.
Results
Clinical characteristics
We recruited 44 healthy participants. 32 proceeded to bronchoscopy but one patient withdrew from the study after the first bronchoscopy. 31 completed 2 bronchoscopies, but one was subsequently withdrawn due to both <80% medication adherence and an intercurrent asymptomatic bronchitis evident at the 2nd bronchoscopy (with bronchial wash samples positive for a non-Covid coronavirus and Staphylococcus aureus). The clinical characteristics of the 30 participants completing the study are shown in table 1.
Table 1. Demographics of the study participants.
| Healthy – ICS (n=20) |
Healthy – observation (n=10) |
p value† | |
|---|---|---|---|
| Age - years | 38 (22-52) | 24 (22-33) | 0.2 |
| Sex - M/F | 8/12 | 7/3 | 0.2 |
| BMI (kg/m2) | 24.7 ± 3.7 | 24.4 ± 2.7 | 0.8 |
| Ethnicity Caucasian (%) |
80 | 70 | 0.7 |
| Atopic (%) | 25 | 50 | 0.2 |
| Ex smoker (%) | 20 | 10 | 0.6 |
| Smoking (pack years) |
0.0 (0.0-0.0) | 0.0 (0.0-0.0) | 0.4 |
| FEV1 Pre BD (L) | 3.32 ± 0.79 | 4.0 ± 0.93 | 0.047 |
| FEV1 Pre BD (% predicted) |
103 ± 11 | 100 ± 8 | 0.5 |
| FEV1/FVC (%) | 82.1 ± 4.6 | 82.4 ± 3.3 | 0.9 |
| FeNO (ppb) | 16 (11-25) | 18 (15-29) | 0.4 |
| Blood eosinophils at screening (×109/L) |
0.11 (0.06-0.15) | 0.11 (0.08-0.15) | 0.8 |
Continuous variables are presented as mean ± SD or median (interquartile range). BD, bronchodilator; BMI, body mass index; FVC, forced vital capacity; FeNO, fractional exhaled nitric oxide; FEV1, forced expiratory volume in 1 second; ICS, inhaled corticosteroids.
All tests for continuous variables are t-test or Mann Whitney U. For categorical variables, a Fischer’s exact test was used.
*P<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Effects of ICS on biomarker and physiological measurements
There was a significant increase in blood eosinophil numbers after 4 weeks in the observation group compared to people using ICS, but this was not related to atopic status (Supplementary figure E1A). There were no significant between-group differences for changes in blood neutrophils, FeNO or FEV1 (Supplementary figure E1B-D).
Effects of ICS on airway inflammatory and structural cells
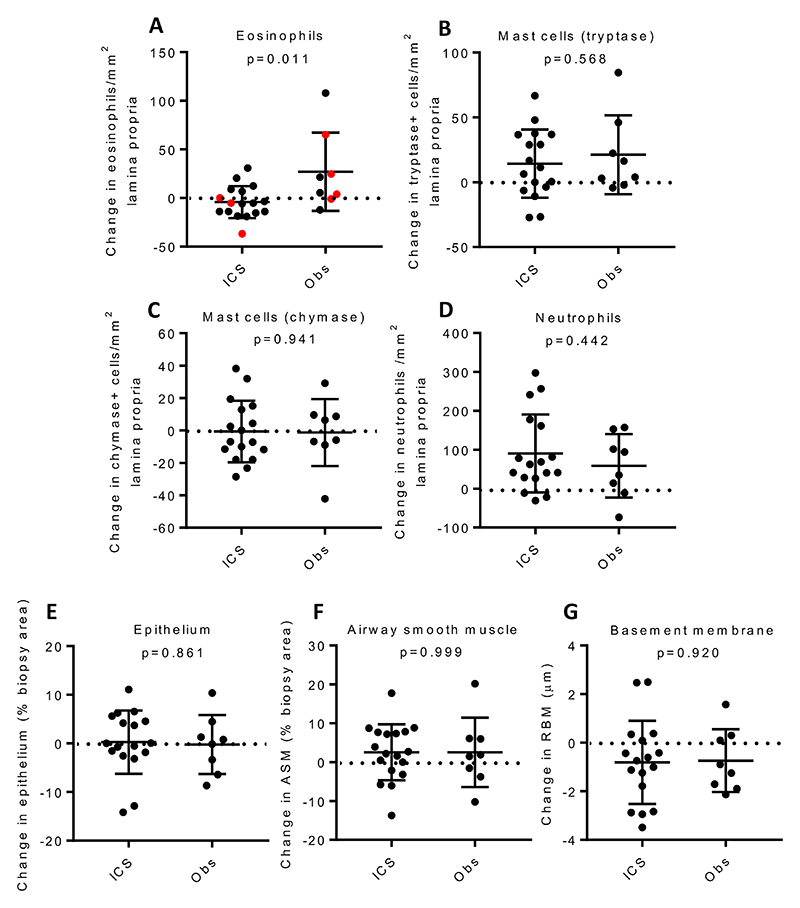
Suitable paired samples for immunohistochemical analysis of the lamina propria were available from 17 participants receiving ICS and 8 undergoing observation (Figure 1 and Supplementary Figure E2). Although there was no significant change in lamina propria eosinophil counts within either group from the 1st to 2nd bronchoscopy, there was a significant between-group difference in the changes (p=0.01), due to a non-significant increase in the observation group (Figure 1A). There was no correlation between the change in blood eosinophils versus tissue eosinophils in the observation group (rs= -0.024, p=0.97). Comparing the changes in measurements from the 1st to 2nd bronchoscopy for the ICS versus the observation group, there were no significant differences between treatment groups for lamina propria neutrophil, tryptase+ or chymase+ mast cell counts, airway smooth muscle (ASM) and epithelial area expressed as a percentage of biopsy area, or reticular basement membrane depth (Figure 1B-G)(Supplementary Figure E2). We performed additional cell deconvolution analysis on bronchial brushings and bronchial biopsy transcriptomic data to infer changes in cellular composition (supplementary methods). There was an increase in club cells (FDR p=0.02) and we confirmed a suppression of the innate and adaptive immune responses by a marked decrease in type 2 dendritic cells (FDR p=0.02) and plasma cells (FDR p=3×10-9) after 4 weeks of ICS (Supplementary Figure E3 and E4) and by GSEA an associated decrease in transcriptional activity of T, B, NK, and dendritic cell genesets and a basophil/mast cell geneset (Supplementary Table E8).
Figure 1.
Immunohistochemical analysis of the lamina propria biopsies showing cell counts and remodelling features on participants with available paired data, before and after 4 weeks treatment with inhaled fluticasone or without treatment. The change in numbers of A) eosinophils, with atopic participants shown in red, B) tryptase-positive mast cells, C) chymase-positive mast cells, or D) neutrophils, expressed in absolute counts/mm2. Changes in area of E) epithelium or F) airway smooth muscle (ASM), expressed as a percentage of biopsy area or of G) reticular basement membrane (RBM) thickness. Horizontal bars represent mean (SD)(mast cells, neutrophils, ASM, epithelium, RBM) or median (IQR) (eosinophils), analysed by unpaired t test or Mann Whitney U respectively. Obs represents the observation group.
Effects of ICS on airway gene expression
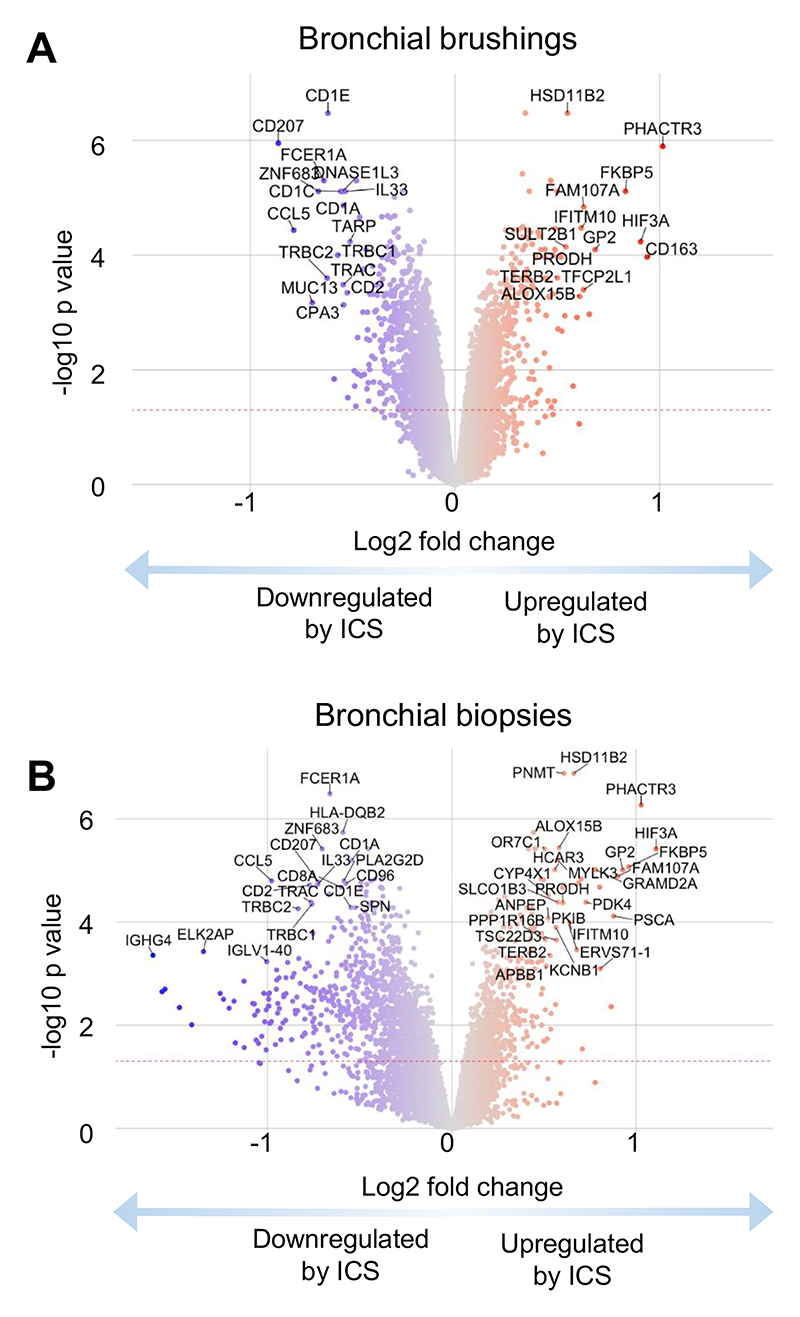
Suitable paired brush and biopsy samples were available for 15 and 20 participants respectively receiving ICS. We observed significant differential expression of genes amongst participants at week 4 compared with baseline in the ICS-treatment group, with upregulation of 72 genes in brushings and 53 genes in biopsies, and downregulation of 82 genes in brushings and 416 genes in biopsies (Figure 2, Supplementary Tables E1-E4, Supplementary Figures E5, E6). Amongst participants in the observation-only group there were no significant changes in gene expression observed between baseline and week 4 (Supplementary Figure E7). There was a close correlation between epithelial brush and bronchial biopsy gene expression, with 20 genes common to the top 24 most significantly differentially upregulated genes in both airway compartments (Table 2) and 20 genes common to the top 41 most significantly downregulated genes in both compartments (Table 3).
Figure 2.
Changes in gene expression measured by RNAseq in response to 4 weeks treatment with inhaled fluticasone. Log2 fold changes and statistical tests calculated using moderated t tests on vsn normalized log2(count per million) values in Limma. A) Bronchial brush volcano plot. B) Bronchial biopsy volcano plot.
Table 2. Top upregulated differentially expressed genes.
| Bronchial brushes | Bronchial biopsies | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Baseline read count |
Log2 fold change | FDR P value | Baseline read count | Log2 fold change | FDR P value |
Mean of FDR P values | Comments and aliases | Functional role |
| PHACTR3 | 497 | 3.59 | 1.43E-41 | 325 | 3.28 | 4.29E-38 | 2.14E-38 | Phosphatase And Actin Regulator 3; associated with the nuclear scaffold in proliferating cells. | Cytoskeletal changes |
| HSD11B2 | 468 | 1.89 | 3.22E-41 | 392 | 1.81 | 9.88E-37 | 4.94E-37 | Hydroxysteroid 11-β Dehydrogenase 2: converts cortisol to cortisone, prevents mineralocorticoid receptor activation | Steroid metabolism |
| FKBP5 | 3336 | 1.88 | 2.17E-27 | 5029 | 1.67 | 3.14E-20 | 1.57E-20 | Regulates corticosteroid sensitivity; asthma susceptibilty gene. | Steroid metabolism |
| FAM107A | 3486 | 1.45 | 6.14E-22 | 3657 | 1.60 | 4.02E-17 | 2.01E-17 | Family With Sequence Similarity 107 Member A; role in actin and microtubule organisation. | Cytoskeletal changes |
| HIF3A | 584 | 3.18 | 4.42E-17 | 990 | 2.67 | 5.35E-21 | 2.21E-17 | Hypoxia Inducible Factor 3 Subunit α; canonical sensor of hypoxia. | Hypoxic sensing |
| MYLK3 | 476 | 1.10 | 1.19E-40 | 490 | 1.47 | 6.54E-17 | 3.27E-17 | Myosin Light Chain Kinase 3; Immune response to CCR3 signaling in eosinophils, regulates smooth muscle contracti | Smooth muscle/immune |
| GP2 | 341 | 2.48 | 1.26E-15 | 286 | 3.12 | 9.81E-28 | 6.29E-16 | Glycoprotein 2; binds pathogens such as enterobacteria. | Innate immune |
| SULT2B1 | 1706 | 1.38 | 3.05E-17 | 1199 | 1.44 | 2.01E-15 | 1.02E-15 | Hydroxysteroid Sulfotransferase 2; catalyses the sulfate conjugation hormones. | Steroid metabolism |
| GRAMD2A | 2452 | 1.15 | 5.37E-24 | 2309 | 1.38 | 2.69E-15 | 1.35E-15 | GRAM Domain Containing 2A; organization of endoplasmic reticulum-plasma membrane contact sites. | Cellular metabolism |
| SYT8 | 503 | 1.12 | 2.93E-15 | 470 | 1.13 | 2.37E-22 | 1.46E-15 | Synaptotagmin 8; regulate exocytosis of hormones and in intracellular organelles. | Steroid metabolism |
| SLCO1B3 | 124 | 1.94 | 1.69E-17 | 111 | 2.27 | 5.11E-14 | 2.55E-14 | Solute Carrier Organic Anion Transporter 1B3; transports conjugated steroids, leukotrienes, prostaglandins. | Steroid metabolism |
| HCAR2 | 4673 | 1.10 | 9.06E-20 | 3361 | 1.22 | 1.15E-13 | 5.73E-14 | Niacin Receptor 1; receptor for niacin and mediates neutrophil apoptosis and anti-inflammatory effect of butyrate | Anti-inflammatory |
| PSCA | 23905 | 1.21 | 9.32E-14 | 13008 | 1.87 | 3.83E-14 | 6.58E-14 | Prostate stem cell antigen; may regulate cell proliferation. | Celll proliferation |
| TFCP2L1 | 3228 | 1.38 | 2.76E-13 | 4849 | 1.40 | 1.64E-16 | 1.38E-13 | Transcription corepressor activity, and probably Wnt/β-catenin signaling. | Celll proliferation |
| PRODH | 5268 | 1.20 | 5.53E-14 | 4754 | 1.10 | 8.22E-13 | 4.39E-13 | Proline Dehydrogenase 1; catalyses proline catabolism. | Cellular metabolism |
| TSC22D3 | 7099 | 1.12 | 1.19E-17 | 8967 | 1.14 | 1.55E-12 | 7.73E-13 | Glucocorticoid-Induced Leucine Zipper; mediates anti-inflammatory effects of glucocorticoids in macrophages. | Anti-inflammatory |
| IFITM10 | 1872 | 1.55 | 4.46E-22 | 1816 | 1.23 | 3.71E-11 | 1.86E-11 | IFITM family functions include controlling cell proliferation, and promoting homotypic cell adhesion. | Celll proliferation |
| ANPEP | 2159 | 1.16 | 4.30E-14 | 1707 | 1.02 | 6.28E-11 | 3.14E-11 | Aminopeptidase M, CD13; broad specificity aminopeptidase, promotes angiogenesis, and cell growth. | Celll proliferation |
| KCNB1 | 602 | 1.03 | 1.83E-15 | 662 | 1.28 | 6.63E-10 | 3.31E-10 | Potassium Voltage-Gated Channel Subfamily B Member 1; neurotransmission and regulation of exocytosis. | Cell signalling |
| GNMT | 704 | 1.28 | 2.24099E-12 | 548 | 1.10 | 1.97E-07 | 9.84E-08 | Glycine N-Methyltransferase; regulation of methyl group metabolism. | Cellular metabolism |
Top 20 upregulated genes common to both epithelial brushes and bronchial biopsies occurring in the top 24 differentially upregulated genes when ranked by FDR p value. Table ranks genes by arithmetic mean of FDR p values across both tissues.
Table 3. Top downregulated differentially expressed genes.
| Bronchial brushes | Bronchial biopsies | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Baseline read count | Log2 fold change | FDR P value | Baseline read count | Log2 fold change | FDR P value | Mean of FDR P values | Comments and aliases | Functional role |
| FCER1A | 144 | -2.78 | 2.15E-24 | 228 | -2.25 | 6.93E-29 | 1.08E-24 | High affinity IgE receptor; expressed on airway mast cells and dendritic cells. | Type 2 immunity |
| ZNF683 | 136 | -3.33 | 2.31E-23 | 131 | -3.06 | 1.32E-20 | 6.62E-21 | Hobit; transcription factor promoting tissue residency in innate and adaptive lymphocytes. | Adaptive immunity |
| CD207 | 169 | -3.78 | 3.05E-23 | 144 | -2.90 | 4.24E-18 | 2.12E-18 | Langerin; C-type lectin with mannose binding specificity, inolved in nonclassical antigen-processing. | Antigen presentation |
| CCL5 | 1133 | -2.39 | 1.21E-15 | 1233 | -2.15 | 2.67E-16 | 7.37E-16 | RANTES; chemoattractant for monocytes, memory T helper cells and eosinophils. | Adaptive immunity |
| IL33 | 3885 | -1.20 | 2.18E-22 | 6985 | -1.24 | 3.70E-15 | 1.85E-15 | Interleukin 33; airway epithelial cell alarmin which acts via ST2 to promote type-2 responses. | Alarmin |
| CD96 | 253 | -1.62 | 1.31E-12 | 399 | -1.64 | 3.49E-14 | 6.71E-13 | TACTILE (T cell activation, increased late expression); involved in adhesion of T and NK cells to target cells. | Cell mediated immunity |
| TARP | 100 | -2.50 | 7.30E-15 | 112 | -1.90 | 3.18E-10 | 1.59E-10 | TCR Gamma Alternate Reading Frame Protein. | T cell adaptive immunity |
| TRBC1 | 353 | -2.33 | 3.39E-10 | 540 | -2.07 | 3.65E-13 | 1.70E-10 | T cell receptor Beta constant chain 1. | T cell adaptive immunity |
| TRBC2 | 463 | -2.41 | 7.16E-10 | 741 | -2.07 | 2.98E-13 | 3.58E-10 | T cell receptor Beta constant chain 2. | T cell adaptive immunity |
| PTPN22 | 165 | -1.26 | 1.43E-09 | 243 | -1.52 | 5.76E-11 | 7.42E-10 | Protein Tyrosine Phosphatase Non-Receptor Type 22. Regulates Treg frequency and T cell motility. | T cell adaptive immunity |
| TRAC | 539 | -1.92 | 3.37E-09 | 764 | -1.84 | 4.48E-13 | 1.68E-09 | T cell receptor Alpha constant chain. | T cell adaptive immunity |
| CPA3 | 228 | -2.26 | 7.00E-09 | 913 | -1.98 | 1.51E-09 | 4.26E-09 | Carboxypeptidase A3: mast cell-specific pepridase located in secretory granules. | Type 2 immunity |
| CD2 | 566 | -1.87 | 1.23E-08 | 732 | -1.87 | 9.60E-14 | 6.14E-09 | Lymphocyte-Function Antigen-2; T and NK cell marker and co-stimulatory molecule. | T cell adaptive immunity |
| CLEC10A | 135 | -1.71 | 1.82E-08 | 199 | -2.20 | 4.39E-14 | 9.11E-09 | C-Type Lectin Domain Containing 10A; CD301: marker for alternatively activated macrophages. | Type 2 immunity |
| SERPINB10 | 287 | -1.18 | 1.82E-08 | 350 | -1.42 | 1.25E-12 | 9.12E-09 | Serpin Family B Member 10; inhibits Th2 cell apoptosis, highly upregulated on airway epithelium by IL-13. | Type 2 immunity |
| CD8A | 516 | -1.55 | 6.40E-07 | 611 | -1.52 | 5.11E-14 | 3.20E-07 | T cell co-receptor for MHCI restriction. | T cell adaptive immunity |
| GFI1 | 133 | -1.06 | 1.73E-06 | 177 | -1.23 | 2.46E-10 | 8.64E-07 | Growth Factor Independent 1 Transcriptional Repressor; involved in haematopoiesis, especially of neutrophils. | Innate immunity |
| HLA-DQB2 | 798 | -1.01 | 9.52E-06 | 640 | -1.51 | 4.63E-18 | 4.76E-06 | Major histocompatibility complex, class II, DQ beta 2 | T cell adaptive immunity |
| CCR5 | 295 | -1.42 | 9.97E-05 | 426 | -1.91 | 1.11E-12 | 4.99E-05 | Ligands include CCL5 (RANTES), MCP-2, MIP-1a, MIP-1b. Chemotactic for T2–induced eosinophils. | Type 2 immunity |
| PTPN7 | 248 | -1.32 | 0.00033 | 312 | -1.64 | 2.36E-10 | 1.67E-04 | Protein Tyrosine Phosphatase Non-Receptor 7; regulation of TCR signaling, early response gene in lymphokine stimulated | T cell adaptive immunity |
Top 20 downregulated genes common to both epithelial brushes and bronchial biopsies occurring in the top 41 biopsy differentially downregulated genes when ranked by FDR p value. Table ranks genes by arithmetic mean of FDR p values across both tissues.
The most significantly upregulated genes were predominantly those involved in steroid metabolism (HSD11B2, FKBP5, SULT2B1, SYT8, and SLCO1B3), cellular proliferation (PSCA, TFCP2L1, IFITM10, ANPEP), cellular metabolism (GRAMD2A, PRODH, GNMT) and cytoskeletal changes (PHACTR3, FAM107A). By contrast genes which were most significantly downregulated in both brushings and biopsies were key components of T2-driven inflammation (FCER1A, CPA3, IL33, CLEC10A, SERPINB10 and CCR5) and T cell-mediated adaptive immunity (TARP, TRBC1, TRBC2, PTPN22, TRAC, CD2, CD8A, HLA-DQB2, CD96, PTPN7) and (Table 3). All other top 20 common downregulated genes were involved with innate or adaptive immunity, including the transcription factor Hobit (ZNF683) which promotes lymphocyte tissue residency, the chemokine RANTES (CCL5), the antigen presentation-associated molecule Langerin (CD207), and the growth factor GFI1 which is involved in haematopoiesis, especially of neutrophils. In addition, there was downregulation of genes associated with B cell function and immumunoglobulin production (CD20/79, most heavy and variable light chains for IgA, IgG and IgM, JCHAIN), protective innate immunity (e.g. CD48, CD163), mast cell proteases (TPSB1, TPSAB1, CPA3), the beta chain of the high affinity IgE receptor (MS4A2), and prostaglandin D2 synthase (PTGDS1).
Consistent with these findings, pathway analysis of bronchial brushings with Reactome showed strong ICS-related downregulation of innate and adaptive pathways, including ‘Immunoregulatory interactions between a lymphoid and non-lymphoid cell’, five pathways related to TCR signaling, the immunological synapse or co-stimulation, with weaker signals for ‘Generation of second messenger molecules’, ‘PD-1 signaling’, and ‘Chemokine receptors bind chemokines’, showing the potent ability of ICS to suppress local adaptive T cell immunity (Supplementary Figures E8, Supplementary Table E5). Parallel analysis of the bronchial biopsies again showed effects on lymphoid – non-lymphoid interactions and TCR signaling, but also strong suppression of ‘Extracellular matrix organization’, ‘Cell surface interactions at the vascular wall’ and ‘Integrin cell surface interactions’, suggesting the potential for suppression of inflammatory cell recruitment to the mucosa (Supplementary Figure E9, Supplementary Table E6). ICS did not upregulate the IL-17-dependent gene signature identified previously in people with moderate-severe asthma (Supplementary Figure E10).
Next we compared our set of differentially upregulated genes with a set of 26 genes previously reported as induced by 10 weeks of inhaled fluticasone in bronchial brushings from participants with mild asthma19. Geneset enrichment analysis profiles showed very close agreement between genesets (Supplementary Figure E11), particularly for brushings, which were directly comparable between studies, showing very similar ICS-induced gene induction in health and mild asthma. Similar downregulation of type 2 immune-related genes was observed both irrespective of atopic status (Supplementary Table E7).
To analyse the effects of corticosteroid treatment on specific structural cells we used geneset enrichment analysis to compare our findings with publicly available data obtained from fibroblast cell lines, primary smooth muscle cells and primary respiratory epithelium treated in vitro with corticosteroids. We found no enrichment for a signature obtained from a cortisol-treated human lung fibroblast cell line, but observed strong enrichment for signatures obtained from primary human airway smooth muscle cells treated with fluticasone propionate, and primary human airway epithelial cells treated in vitro with budesonide (details in Supplementary Results, Supplementary Table E8 and Supplementary Figure E12). Together these suggest 4 weeks of ICS have direct effects on airway epithelial cells and airway smooth muscle cells. Although there were minimal direct effects on fibroblasts, it is possible ICS in vivo might modulate other cells to alter the function of fibroblasts indirectly.
To determine whether ICS have differential effects on specific epithelial cell types we used the proportions of 5 different epithelial and structural cell types (multiciliated, basal resting, peribronchial fibroblasts, club and suprabasal), estimated using MuSiC deconvolution, to estimate differential gene expression between treatment conditions according to cell type, using TOAST20 in R. Results are presented in Supplementary table E9.
Effects of ICS on the airway microbiome
Minimal effects of ICS treatment were observed on the airway microbiome (details in Supplementary Results and Supplementary Figure E13).
Effects of ICS on airway DNA methylation
There were minimal effects of ICS treatment on DNA methylation (details in Supplementary Results and Supplementary Figure E14).
Discussion
In this open label, randomised study in healthy adult volunteers, we have defined genes that are altered directly by medium-term (4 weeks), twice daily high-dose ICS therapy independent of the confounding effect of asthma pathophysiology. Changes in airway cellularity were minimal between ICS treatment and no treatment, and there were no measurable biologically significant effects on the airway microbiome or DNA methylation.
By contrast, we observed widespread changes in gene expression following ICS use in these healthy individuals, which predominantly comprised downregulated genes. A striking finding was that the most significantly downregulated genes statistically were canonical markers of T2-driven inflammation. Notably the most highly downregulated gene, in both airway brushes, which predominantly sample the airway epithelium, and mucosal biopsies, was FCER1A which encodes the α chain of the high affinity IgE receptor, a key effector component of mast cell activation by allergens and of T2 immunity, which is highly upregulated in asthma 21,22. In addition, there was downregulation of genes encoding mast cell tryptases (TPSB1, TPSAB1) and carboxypeptidase A3 (CPA3), the beta chain of the high affinity IgE receptor (MS4A2), and prostaglandin D2 synthase (PTGDS1). We did not see a reduction in mast cell numbers in the airway mucosa of these healthy individuals, so the predominant effect of ICS appears to be on gene transcription, rather than mast cell survival, but nevertheless suggests that ICS have a potentially important dampening effect on mast cell function. In biopsies, there was also downregulation of the important mast cell chemoattractants CXCL10 and CXCL11 which promote mast cell migration to the ASM in mild steroid-naive asthma through the airway mast cell chemokine receptor CXCR323. In people with severe asthma using high dose ICS +/- oral corticosteroids, mast cells are not increased within the ASM 24,25, suggesting suppression of mast cell chemoattractants from ASM may remain corticosteroid-sensitive in severe disease. This is in contrast to the release of mast cell-derived proteases and autacoids which demonstrate ongoing release in severe disease24,26.
Other highly downregulated T2-related genes included IL33, an airway epithelial cell alarmin which acts to promote initiation of T2 responses27, CLEC10A which is a marker for alternatively activated macrophages28, SERPINB10 which is highly upregulated on airway epithelium by IL-13 and inhibits Th2 cell apoptosis, and CCR5 whose ligands include CCL5 (RANTES), a chemoattractant for eosinophils and mast cells. Moreover PTGS1, which encodes cyclooxygenase-1 was the 7th most downregulated gene in brushings. This enzyme converts arachidonate to prostaglandins and its dysregulation or inhibition is implicated in salicylate-sensitive asthma and sino-nasal eosinophilic inflammation29. Similarly in biopsies, periostin (POSTN), an IL-13-induced epithelial gene associated with T2 high asthma30, was also downregulated.
Taken together, the ability of ICS to downregulate this extensive set of T2-related genes implies that homeostasis in health involves a low level of tonic T2 signalling in the airway mucosa that is very sensitive to ICS. This would be consistent with the observation that IL-4- and IL-5-positive cells are present in healthy airways31, as are mast cells and dendritic cells expressing FcεRIα32. Constitutive T2 signalling is also evident in primary epithelial cell cultures grown at air-liquid interface, where we previously observed that STAT6-dependent genes including POSTN, CLCA1 and SERPINB2 were repressed by NFκB-dependent cytokine stimulation or dexamethasone33. Thus differentiated bronchial epithelial cells have a low level of tonic STAT6 dependent signalling in the absence of exogenous IL-4 or IL-13. However, FeNO was not reduced by ICS in these healthy volunteers, and NOS2, which regulates FeNO production, was not altered by ICS therapy, suggesting tonic T2 signalling is below the threshold required for pathological FeNO generation.
The role of low-level, tonic, constitutive T2 signalling in the airways is uncertain but likely beneficial for tissue homeostasis. T2 immunity exhibits many host-protective functions, including maintaining metabolic homeostasis, suppressing excessive T1 inflammation, maintenance of barrier defence and regulation of tissue regeneration34–37. For example, IL-33 is pleiotropic and can promote type 2 inflammation but in other contexts it can be immunoregulatory27, and preserves epithelial integrity during influenza infection in a mouse model38,39. Thus the effects of type 2 immunity may be context specific and affected by the cellular source and concomitant inflammatory milieu, and at steady state may promote airway epithelial barrier function. Inhibition of homeostatic tonic T2 signalling might therefore have deleterious effects which may have an impact clinically, and might explain an enhanced propensity to proteobacterial colonisation or infection, as was observed in the MEX40 and RASP-UK41 studies amongst participants with very low T2 biomarkers.
In addition to the inhibition of T2-driven genes, there was marked suppression of molecules involved in both protective innate immunity (e.g. CD48, CEACAM5) and adaptive immunity, with suppression of genes related to dendritic cells (CD207), T cells (e.g. CD2/3/6/8/96, TRBC1/2, TRAC), and B cell function (e,g, CD20/79, most heavy and variable light chains for IgA, IgG and IgM, JCHAIN). This potential impairment of innate, cell-mediated and antibody-dependent immunity likely explains the reproducible dose-dependent increased pneumonia risk in people with asthma and COPD who are using ICS4. A subgroup of people with asthma also become colonised with certain fungi42, and it is possible that this is also a consequence of the mucosal immunosuppression observed here.
An important aim of this study was to provide information on the activity of ICS in healthy airways, to remove the confounding that might occur due to disease-related changes in gene expression or inherent ICS responsiveness, which should facilitate analyses of gene expression changes in asthma. Studies of severe asthma to-date43 have attempted to account for the effects of ICS based on gene expression changes derived from in vitro cell cultures, from an interventional study of gene expression changes in people administered ICS for chronic obstructive pulmonary disease, and from one previous intervention study which used microarrays to assess acute airway transcriptional consequences 6 hours after a single inhaled dose of budesonide (1600 μg) in 12 healthy, steroid-naïve men44. Our findings are complementary to the previous budesonide study in that some differentially regulated genes are common to our two studies (including upregulation of FKBP5, ZBTB16, PHACTR3, TSC22D3 and downregulation of CD207, FCER1A, IL33), but there is a striking difference in the overall consequences of a single dose versus 4 weeks twice daily ICS use. The predominant effect of a single large acute dose of ICS was upregulation of genes (transactivation), with 68 genes upregulated and only 28 downregulated. Many of the upregulated genes were proinflammatory including growth factors, chemokines, chemokine receptors, cytokines, growth factors, and coagulation factors. By comparison in our chronic high dose exposure study only 53 genes were upregulated in biopsies, whilst 416 genes were downregulated (transrepression). This highlights the critical and often overlooked importance of the complex temporal dynamics of corticosteroid effects45.
Early during an acute infectious or traumatic challenge a surge in endogenous corticosteroids will tend to enhance protective inflammatory and procoagulant responses, but as time passes these responses will attenuate and slower but potent transrepressive effects will dominate to curtail uncontrolled inflammatory cell recruitment and activation, to prevent bystander tissue damage, and to direct resolution of tissue homeostasis. It is this latter situation which is of greatest clinical relevance to long term asthma management. Of note, comparing our data to the study by Woodruff12, where people with mild asthma received inhaled placebo or fluticasone propionate (500 μg) twice daily for 8 weeks, there was a strong correlation between the studies for ICS-inducible genes, suggesting that in mild asthma at least, ICS-dependent transactivation is preserved.
When considering previous studies of gene expression in severe asthma, an IL-17-dependent gene signature is expressed in a subset of people, and mutually exclusive with a T2 gene signature8,9. This IL-17-dependent signature was only seen in people using ICS, but it has been unclear whether this is disease- or treatment-related, and whether it is harmful or potentially protective. We did not see upregulation of this IL-17 signature after 4 weeks of ICS treatment in this study, thus it seems most likely a feature of disease. Similarly, there is upregulation of CEACAM family members in severe asthma, notably the IL-13-dependent gene CEACAM546,47, and CEACAM6 which was increased on both the epithelium and neutrophils in bronchial biopsies46. Here we found that CEACAM6 expression did not change with ICS treatment, while CEACAM5 expression was reduced (tables E2, E4), in keeping with the inhibition of other T2-related genes. Therefore the upregulation of these CEACAMs in severe asthma appears to be a feature of the disease rather than treatment. In the recent U-BIOPRED bronchoscopy study25, ICS-inducible genes such as FKBP5 were upregulated in severe asthma, suggesting that their participants were adherent to treatment, and importantly, that ICS transactivation appears to be preserved in severe asthma, but many pathological pathways that should be sensitive to transrepression by ICS such as T2 signalling are not responsive in a subset of patients8,9,24.
There are some limitations to our work. Firstly the effects of ICS in healthy airways at 4 weeks, while likely representative of the steady state in long term therapy, might not be fully representative of longer term therapy. However it is not reasonable to ask healthy volunteers to take ICS for a year, and adherence would likely wane. Secondly our analyses use bulk sequencing of airway brushes and biopsies, and ICS effects are likely to be highly cell-specific, so future studies using spatial sequencing, from central and peripheral airways as well as nasal tissues and peripheral blood would be informative. For example, as we did not enumerate all of the cell types implicated in the transcriptomic analyses, we cannot conclude whether the changes in T- and B-lymphocyte related gene expression are due to transcriptional changes within lymphocytes, changes in the relative proportions of lymphocytes in the samples, or a combination of the two.
In summary, we defined genes altered directly by ICS therapy without confounding by disease. We provide evidence that IL-17-dependent signalling in asthma is disease-dependent rather than ICS-dependent, and demonstrate downregulation of canonical markers of T2 inflammation, implying that homeostasis in health involves tonic T2 signalling in the airway mucosa, which is exquisitely sensitive to ICS. There was also broad suppression of innate and adaptive immunity, in keeping with known immunosuppressive effects of corticosteroids.
Supplementary Material
Acknowledgements
The authors are grateful to all the participants who volunteered and to the clinical and research teams at all the participating centres. For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. The authors vouch for the integrity and completeness of the data and the fidelity of the trial to the protocol. This study has been conducted in accordance with the Declaration of Helsinki and any applicable regulatory requirements.
Funding sources
This work was supported by an investigator-led grant from Genentech to the University Hospitals of Leicester NHS Trust, and supported in part by the National Institute for Health Research (NIHR) Leicester Biomedical Research Centre (Respiratory). EM is supported by Asthma+Lung UK (WADR22\100015). TSCH is supported by a Wellcome Trust Fellowship (211050/Z/18/z). RLC is supported by a University of Nottingham Anne McLaren Fellowship. All authors had full access to the data in the study and accept responsibility to submit for publication. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnotes
Author conflict of interest statement
EM-none.
TSCH - TSCH has received grants from Pfizer Inc., the University of Oxford, the Wellcome Trust, the Medical Research Council, Asthma UK+Lung, the NIHR Oxford Biomedical Research Centre, Sensyne Health, and Kymab during the conduct of the study; and personal fees from AZ Pieris, and Kymab outside the submitted work.
MR – none.
LK – none.
FAS – none.
PR – none.
RC – RC is supported by an Anne McLaren Fellowship.
BH-none.
CDA – CDA is an employee of Genentech.
JLM – none.
MSK – none.
SS – SS has received consulting fees and honoraria from AstraZeneca, GSK, Roche, CSL Behring, Chiesi, Areteia Therapeutics, ERT Medical, Medscape, European Respiratory Society.
JSM – JSM is an employee of Genentech.
JRA – JRA is a former employee of Genentech and named inventor on granted and pending patents related to asthma diagnosis and treatment.
DFC – DFC is an employee of Genentech, and is an inventor of filed and approved patents for the treatment and diagnosis of chronic lung diseases. As an employee of Genentech, DFC holds Roche shares and stock options.
PB - PB has received research funding from Genentech via the University Hospitals of Leicester NHS Trust; consultancy for Genentech via the University of Leicester. Support to attend scientific meetings from Chiesi, GSK and Sanofi-Regeneron, and one honorarium from AstraZeneca.
Author contributions
All authors reviewed the data and contributed to its interpretation, edited the manuscript, and approved the final submitted version.
Specific contributions:
EM: Analysed the RNA seq data and drafted the manuscript
TSCH: Contributed to data analysis and interpretation, and drafting of the manuscript.
MR: Analysed the RNA seq data.
LC and FAS: Performed immunohistochemistry, and quantified and collated the data.
PR: performed and analysed DNA methylation studies
RLC: Designed, supervised and analysed DNA methylation studies.
BH: Managed the study, contributed to patient recruitment, collated the demographic data.
CDA: Contributed to bronchoscopy protocol design.
JLM and MSK performed DNA methylation studies.
SS: Analysed the RNA seq data.
JRA: Conceived and designed the study.
DC: Conceived and designed the study, analysed RNA seq data, drafted the manuscript.
PB: Conceived and designed the study, wrote the study protocol and ethical application, analysed the data, drafted the manuscript, performed all bronchoscopies. Chief investigator.
Data sharing
The data analysed and presented in this study are available from the corresponding author on reasonable request, providing the request meets local ethical and research governance criteria after publication. Patient-level data will be anonymised. The RNA Sequencing data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE242048. All microbiome sequence data is available from the European Genome-Phenome Archive (https://ega-archive.org/, EGAS00001007538).
References
- 1.Laitinen LA, Laitinen A, Haahtela T. A comparative study of the effects of an inhaled corticosteroid, budesonide, and a beta 2-agonist, terbutaline, on airway inflammation in newly diagnosed asthma: a randomized, double-blind, parallel-group controlled trial. J Allergy Clin Immunol. 1992;90(1):32–42. doi: 10.1016/s0091-6749(06)80008-4. [DOI] [PubMed] [Google Scholar]
- 2.Djukanovic R, Wilson JW, Britten KM, et al. Effect of an inhaled corticosteroid on airway inflammation and symptoms in asthma. Am Rev Respir Dis. 1992;145(3):669–674. doi: 10.1164/ajrccm/145.3.669. [DOI] [PubMed] [Google Scholar]
- 3.Suissa S, Ernst P, Benayoun S, Baltzan M, Cai B. Low-dose inhaled corticosteroids and the prevention of death from asthma. N Engl J Med. 2000;343(5):332–336. doi: 10.1056/NEJM200008033430504. [DOI] [PubMed] [Google Scholar]
- 4.McKeever T, Harrison TW, Hubbard R, Shaw D. Inhaled corticosteroids and the risk of pneumonia in people with asthma: a case-control study. Chest. 2013;144(6):1788–1794. doi: 10.1378/chest.13-0871. [DOI] [PubMed] [Google Scholar]
- 5.Jaffuel D, Demoly P, Gougat C, et al. Transcriptional potencies of inhaled glucocorticoids. Am J Respir Crit Care Med. 2000;162(1):57–63. doi: 10.1164/ajrccm.162.1.9901006. [DOI] [PubMed] [Google Scholar]
- 6.Chachi L, Shikotra A, Duffy SM, et al. Functional KCa3.1 channels regulate steroid insensitivity in bronchial smooth muscle cells. J Immunol. 2013;191(5):2624–2636. doi: 10.4049/jimmunol.1300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alzahrani A, Hussain A, Alhadian F, et al. Potential Role of Mast Cells in Regulating Corticosteroid Insensitivity in Severe Asthma. Adv Exp Med Biol. 2021;1303:1–12. doi: 10.1007/978-3-030-63046-1_1. [DOI] [PubMed] [Google Scholar]
- 8.Choy DF, Hart KM, Borthwick LA, et al. TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Sci Transl Med. 2015;7(301):301ra129. doi: 10.1126/scitranslmed.aab3142. [DOI] [PubMed] [Google Scholar]
- 9.Ostling J, van Geest M, Schofield JPR, et al. IL-17-high asthma with features of a psoriasis immunophenotype. J Allergy Clin Immunol. 2019;144(5):1198–1213. doi: 10.1016/j.jaci.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Berry MA, Morgan A, Shaw DE, et al. Pathological features and inhaled corticosteroid response of eosinophilic and non-eosinophilic asthma. Thorax. 2007 doi: 10.1136/thx.2006.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGrath KW, Icitovic N, Boushey HA, et al. A large subgroup of mild-to-moderate asthma is persistently noneosinophilic. Am J Respir Crit Care Med. 2012;185(6):612–619. doi: 10.1164/rccm.201109-1640OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodruff PG, Modrek B, Choy DF, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180(5):388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choy DF, Modrek B, Abbas AR, et al. Gene Expression Patterns of Th2 Inflammation and Intercellular Communication in Asthmatic Airways. J Immunol. 2010 doi: 10.4049/jimmunol.1002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flood-Page P, Menzies-Gow A, Phipps S, et al. Anti-IL-5 treatment reduces deposition of ECM proteins in the bronchial subepithelial basement membrane of mild atopic asthmatics. J Clin Invest. 2003;112(7):1029–1036. doi: 10.1172/JCI17974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallin A, Sandstrom T, Cioppa GD, Holgate S, Wilson S. The effects of regular inhaled formoterol and budesonide on preformed Th-2 cytokines in mild asthmatics. Respir Med. 2002;96(12):1021–1025. doi: 10.1053/rmed.2002.1388. [DOI] [PubMed] [Google Scholar]
- 16.Du Rand IA, Blaikley J, Booton R, et al. British Thoracic Society guideline for diagnostic flexible bronchoscopy in adults: accredited by NICE. Thorax. 2013;68(Suppl 1):i1–i44. doi: 10.1136/thoraxjnl-2013-203618. [DOI] [PubMed] [Google Scholar]
- 17.Foundation TR. The R Project for Statistical Computing. 2023. [Accessed 23/05/2023]. http://www.r-project.org/index.html .
- 18.Reimers M, Carey VJ. Bioconductor: an open source framework for bioinformatics and computational biology. Methods Enzymol. 2006;411:119–134. doi: 10.1016/S0076-6879(06)11008-3. [DOI] [PubMed] [Google Scholar]
- 19.Woodruff PG, Boushey HA, Dolganov GM, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104(40):15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Wu H. TOAST: improving reference-free cell composition estimation by cross-cell type differential analysis. Genome Biol. 2019;20(1):190. doi: 10.1186/s13059-019-1778-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singhania A, Wallington JC, Smith CG, et al. Multitissue Transcriptomics Delineates the Diversity of Airway T Cell Functions in Asthma. Am J Respir Cell Mol Biol. 2018;58(2):261–270. [Google Scholar]
- 22.Holgate ST. Pathogenesis of asthma. Clin Exp Allergy. 2008;38(6):872–897. doi: 10.1111/j.1365-2222.2008.02971.x. [DOI] [PubMed] [Google Scholar]
- 23.Brightling CE, Ammit AJ, Kaur D, et al. The CXCL10/CXCR3 axis mediates human lung mast cell migration to asthmatic airway smooth muscle. Am J Respir Crit Care Med. 2005;171:1103–1108. doi: 10.1164/rccm.200409-1220OC. [DOI] [PubMed] [Google Scholar]
- 24.Khalfaoui L, Symon FA, Couillard S, et al. Airway remodelling rather than cellular infiltration characterizes both type2 cytokine biomarker-high and -low severe asthma. Allergy. 2022 doi: 10.1111/all.15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson SJ, Ward JA, Sousa AR, et al. Severe asthma exists despite suppressed tissue inflammation: findings of the U-BIOPRED study. Eur Respir J. 2016;48(5):1307–1319. doi: 10.1183/13993003.01129-2016. [DOI] [PubMed] [Google Scholar]
- 26.Hinks TS, Zhou X, Staples KJ, et al. Innate and adaptive T cells in asthmatic patients: Relationship to severity and disease mechanisms. J Allergy Clin Immunol. 2015;136(2):323–333. doi: 10.1016/j.jaci.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung LY, Tanaka Y, Herbine K, et al. Cellular context of IL-33 expression dictates impact on anti-helminth immunity. Sci Immunol. 2020;5(53) doi: 10.1126/sciimmunol.abc6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staples KJ, Hinks TS, Ward JA, Gunn V, Smith C, Djukanovic R. Phenotypic characterization of lung macrophages in asthmatic patients: overexpression of CCL17. J Allergy Clin Immunol. 2012;130(6):1404–1412.:e1407. doi: 10.1016/j.jaci.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietruszewska W, Fendler W, Podwysocka M, et al. Expression of Transcript Variants of PTGS1 and PTGS2 Genes among Patients with Chronic Rhinosinusitis with Nasal Polyps. Diagnostics (Basel) 2021;11(1) doi: 10.3390/diagnostics11010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia G, Erickson RW, Choy DF, et al. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. J Allergy Clin Immunol. 2012;130(3):647–654. doi: 10.1016/j.jaci.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradding P, Roberts JA, Britten KM, et al. Interleukin-4, -5, and -6 and tumor necrosis factor-alpha in normal and asthmatic airways: evidence for the human mast cell as a source of these cytokines. Am J Respir Cell Mol Biol. 1994;10(5):471–480. doi: 10.1165/ajrcmb.10.5.8179909. [DOI] [PubMed] [Google Scholar]
- 32.Tunon-De-Lara JM, Redington AE, Bradding P, et al. Dendritic cells in normal and asthmatic airways: expression of the alpha subunit of the high affinity immunoglobulin E receptor (Fc epsilon RI-alpha) Clin Exp Allergy. 1996;26(6):648–655. [PubMed] [Google Scholar]
- 33.Choy DF, Hart KM, Borthwick LA, et al. TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Sci Transl Med. 2015;7(301):301ra129. doi: 10.1126/scitranslmed.aab3142. [DOI] [PubMed] [Google Scholar]
- 34.Gieseck RL, 3rd, Wilson MS, Wynn TA. Type 2 immunity in tissue repair and fibrosis. Nat Rev Immunol. 2018;18(1):62–76. doi: 10.1038/nri.2017.90. [DOI] [PubMed] [Google Scholar]
- 35.Van Dyken SJ, Locksley RM. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol. 2013;31:317–343. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goh YP, Henderson NC, Heredia JE, et al. Eosinophils secrete IL-4 to facilitate liver regeneration. Proc Natl Acad Sci U S A. 2013;110(24):9914–9919. doi: 10.1073/pnas.1304046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mesnil C, Raulier S, Paulissen G, et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest. 2016;126(9):3279–3295. doi: 10.1172/JCI85664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monticelli LA, Sonnenberg GF, Abt MC, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12(11):1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dagher R, Copenhaver AM, Besnard V, et al. IL-33-ST2 axis regulates myeloid cell differentiation and activation enabling effective club cell regeneration. Nature communications. 2020;11(1):4786. doi: 10.1038/s41467-020-18466-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDowell PJ, Diver S, Yang F, et al. The inflammatory profile of exacerbations in patients with severe refractory eosinophilic asthma receiving mepolizumab (the MEX study): a prospective observational study. Lancet Respir Med. 2021;9(10):1174–1184. doi: 10.1016/S2213-2600(21)00004-7. [DOI] [PubMed] [Google Scholar]
- 41.Diver S, Haldar K, McDowell PJ, et al. Relationship between inflammatory status and microbial composition in severe asthma and during exacerbation. Allergy. 2022;77(11):3362–3376. doi: 10.1111/all.15425. [DOI] [PubMed] [Google Scholar]
- 42.Agbetile J, Fairs A, Desai D, et al. Isolation of filamentous fungi from sputum in asthma is associated with reduced post-bronchodilator FEV1. Clin Exp Allergy. 2012;42(5):782–791. doi: 10.1111/j.1365-2222.2012.03987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ginebaugh SP, Hagner M, Ray A, et al. Bronchial epithelial cell transcriptional responses to inhaled corticosteroids dictate severe asthmatic outcomes. J Allergy Clin Immunol. 2023 doi: 10.1016/j.jaci.2023.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leigh R, Mostafa MM, King EM, et al. An inhaled dose of budesonide induces genes involved in transcription and signaling in the human airways: enhancement of anti- and proinflammatory effector genes. Pharmacol Res Perspect. 2016;4(4):e00243. doi: 10.1002/prp2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stavreva DA, Coulon A, Baek S, et al. Dynamics of chromatin accessibility and long-range interactions in response to glucocorticoid pulsing. Genome Res. 2015;25(6):845–857. doi: 10.1101/gr.184168.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shikotra A, Choy DF, Siddiqui S, et al. A CEACAM6-High Airway Neutrophil Phenotype and CEACAM6-High Epithelial Cells Are Features of Severe Asthma. J Immunol. 2017;198(8):3307–3317. doi: 10.4049/jimmunol.1600606. [DOI] [PubMed] [Google Scholar]
- 47.Mumby S, Kermani NZ, Garnett JP, et al. CEACAM5 is an IL-13-regulated epithelial gene that mediates transcription in type-2 (T2) high severe asthma. Allergy. 2022;77(11):3463–3466. doi: 10.1111/all.15465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analysed and presented in this study are available from the corresponding author on reasonable request, providing the request meets local ethical and research governance criteria after publication. Patient-level data will be anonymised. The RNA Sequencing data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE242048. All microbiome sequence data is available from the European Genome-Phenome Archive (https://ega-archive.org/, EGAS00001007538).