Abstract
Intestinal inflammation shifts microbiota composition and metabolism. How the host monitors and responds to such changes remains unclear. Here, we describe a protective mechanism by which mucosal-associated invariant T (MAIT) cells detect microbiota metabolites produced upon intestinal inflammation and promote tissue repair. At steady state, MAIT ligands derived from the riboflavin biosynthesis pathway were produced by aerotolerant bacteria residing in the colonic mucosa. Experimental colitis triggered luminal expansion of riboflavin-producing bacteria, leading to increased production of MAIT ligands. Modulation of intestinal oxygen levels suggested a role for oxygen in inducing MAIT ligand production. MAIT ligands produced in the colon rapidly crossed the intestinal barrier and activated MAIT cells, which expressed tissue-repair genes and produced barrier-promoting mediators during colitis. Mice lacking MAIT cells were more susceptible to colitis and colitis-driven colorectal cancer. Thus, MAIT cells are sensitive to a bacterial metabolic pathway indicative of intestinal inflammation.
Introduction
Many human diseases are associated with changes in the composition and activity of intestinal microbiota. At steady state, the host controls critical nutrient availability to allow the growth of beneficial microbes and contain the expansion of potentially harmful bacteria. The abundance of electron acceptors, such as oxygen and nitrates in the colonic lumen, has emerged as a key ecological parameter that shapes microbiota composition (1). At steady state, oxygen coming from the blood is consumed by the colonic epithelium, effectively maintaining a hypoxic luminal environment that selects for fermentative, strict anaerobic bacteria such as Firmicutes and Bacteroidetes (2). By contrast, the availability of oxygen in the lumen (dysanaerobiosis) enables bacterial respiration, a metabolism that yields more energy than fermentation, and fuels the expansion of facultative anaerobes such as Enterobacteriaceae (1). Dysanaerobiosis represents a major driver of dysbiosis and underlies many pathological situations, such as inflammatory bowel diseases (IBDs), enteric infections, and colorectal cancer (CRC) (3). Whether the host monitors microbial metabolites indicative of intestinal dysbiosis remains unclear.
Mucosal-associated invariant T (MAIT) cells are innate-like T cells that recognize microbial metabolites presented by the major histocompatibility complex–related molecule MR1 and are abundant in liver and mucosal tissues (10 to 40% of T cells in the liver and 1 to 10% in the gut) (4). The particular tissue location of MAIT cells suggests a key role in the surveillance of microbiota-derived ligands produced by the intestinal microbiota (5). MAIT ligands are derived from the riboflavin precursor 5-amino-6-(D-ribitylamino)uracil (5-A-RU), which reacts with methylglyoxal or glyoxal to generate the potent antigens 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) and 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil (5-OE-RU). 5-A-RU is synthesized by the microbial enzyme RibD, which is strictly necessary for the production of MAIT ligands in both Gram-positive and Gram-negative bacteria (6, 7). The amount of bacterial MAIT ligands produced is proportional to the amount of riboflavin produced (8), suggesting that MAIT cells may become activated through the T cell receptor (TCR) in the context of strong microbial riboflavin production. However, the rules governing riboflavin production by the microbiota are unknown. Riboflavin is the precursor of flavin mono-nucleotide and flavin adenine dinucleotide (FAD), two major coenzymes involved in many redox reactions owing to their ability to shuttle electrons from donor to acceptor molecules. Riboflavin-derived FAD is an essential component of complex II in the respiratory electron transport chain (9). Flavins are used also by anaerobic bacteria such as Faecalibacterium prausnitzii to reduce oxygen and grow in contact with air (10). Thus, intestinal dysanaerobiosis may increase riboflavin needs and lead to MAIT ligand synthesis by the microbiota.
In support of this hypothesis, MAIT cells are activated in pathologies associated with intestinal dysanaerobiosis, such as IBD (11, 12) and CRC (13–15). The intestinal microbiota of patients with IBD (16, 17) and CRC (18) harbors increased abundance of bacteria capable of riboflavin synthesis, raising the possibility that MAIT cell activation in these patients results from the production of riboflavin adduct ligands by the dysbiotic microbiota. TCR stimulation of MAIT cells drives expression of tissue-repair genes (19–21). Skin wound healing occurs more slowly in Mr1−/− mice, which lack MAIT cells, compared with wild-type (WT) mice, thereby demonstrating a role for MAIT cells in promoting tissue repair (22, 23). In an in vitro wound healing assay, MAIT cells promoted the growth of a human intestinal epithelial cell line in a TCR-dependent manner (19), suggesting that antigenic stimulation of MAIT cells may support growth of the intestinal epithelium. Yet, the exact role of MAIT cells in IBD is unknown.
In this study, we used a metatranscriptomic approach coupled with a highly specific MAIT ligand quantification assay to investigate the effect of intestinal inflammation on the production of riboflavin and MAIT ligands in the gut. We found that MAIT ligands were synthesized by aerotolerant bacteria that bloom in the gut lumen during experimental colitis. The subsequent MAIT cell activation leads to the production of anti-inflammatory and tissue-repair mediators. Mice lacking MAIT cells were more susceptible to chronic colitis and colitis-induced CRC, strongly suggesting that MAIT cells reduce intestinal inflammation.
Results
Bacterial production of MAIT ligands in the colonic mucosa
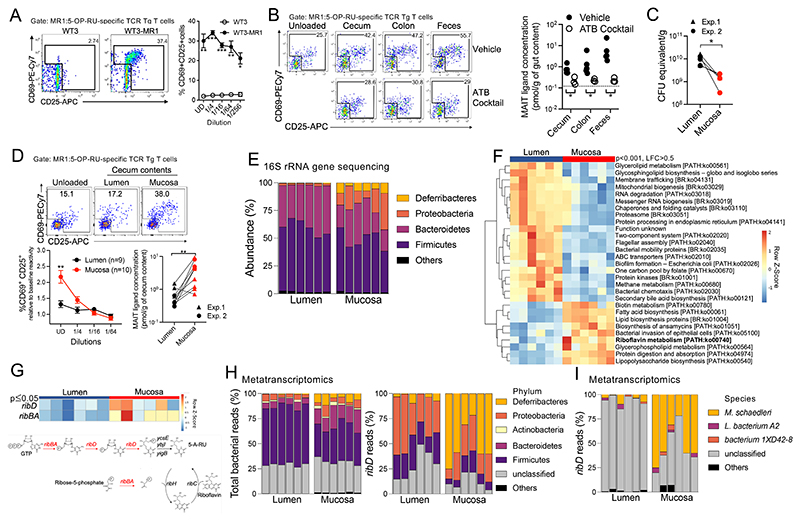
To quantify MAIT ligands in the large intestine, we adapted a sensitive in vitro biological assay (7) based on the activation of monospecific TCR transgenic (Tg) T cells (24) recognizing the canonical MAIT ligand 5-OP-RU. The mouse embryonic fibroblast cell line WT3, engineered to overexpress MR1 (25), was used as MAIT antigen-presenting cells. WT3-MR1 cells loaded with synthetic 5-OP-RU efficiently induced CD25 and CD69 at the surface of tetramer+ 5-OP-RU–specific TCR Tg T cells after overnight coculture (fig. S1, A and B). To assess the presence of MAIT ligands in the intestine, colon contents from a B6 mouse were solubilized in culture medium, passed through a 0.22-μm filter to eliminate bacteria, and incubated with WT3 cells (which express low levels of MR1) or WT3-MR1 cells. The antigen-loaded cells were then washed and incubated overnight with TCR Tg T cells. 5-OP-RU–specific TCR Tg T cells expressed CD25 and CD69 after coculture with WT3-MR1 cells but not WT3 cells, indicating cognate activation by MR1-restricted MAIT ligands (Fig. 1A). Using a standard curve of synthetic 5-OP-RU dilutions, the concentration of MAIT ligands was estimated at 0.8 pmol/g of cecum contents, 1.8 pmol/g of colon contents, and 7 pmol/g of feces (Fig. 1B). Both fungi and bacteria can produce riboflavin-derived MAIT ligands (6, 26). MAIT ligands were depleted from intestinal contents when mice were treated with broad-spectrum antibiotics (Fig. 1B) but not with the antifungal drug fluconazole (fig. S1C), indicating that MAIT ligands were mostly produced by the commensal bacteria.
Fig. 1. Bacterial production of MAIT ligands in the colonic mucosa.
(A) activation of MR1:5-OP-RU–specific TCR Tg T cells after coculture with WT3 or WT3-MR1 cells pulsed with colon luminal contents at the indicated dilutions (n = 3 mice per group; data pooled from N = 2). *P < 0.05, **P < 0.01, and ***P < 0.001 by unpaired multiple t tests with a false discovery rate (FDR) = 1%. UD, undiluted. (B) left, MR1:5-OP-RU–specific TCR Tg T cell activation after coculture with WT3-MR1 cells pulsed with filtered luminal contents from control or antibiotic-treated mice. Right, MAIT ligand concentrations expressed as 5-OP-RU equivalent (n = 3 to 5 mice per group; representative of N = 2). the dotted line indicates TCR Tg T cell activation in the absence of intestinal contents. *P < 0.05 by unpaired multiple Mann-Whitney U tests with an FDR = 1%. (C) Estimation of cecal bacterial numbers by qPcR amplification of the 16S rRna gene (data pooled from N = 2). *P < 0.05 by paired Wilcoxon test. (D) top, MR1:5-OP-RU–specific TCR Tg T cell activation after coculture with WT3-MR1 cells pulsed with lumen or mucosal cecal contents. Bottom left, relative expression of CD25 and CD69 by MR1:5-OP-RU–specific TCR Tg T cells in the indicated conditions (means ± SEMs, n = 9 or 10 mice; data pooled from N = 2). **P < 0.01 by paired multiple t tests with an FDR = 1%. Bottom right: Mait ligand concentrations expressed as 5-OP-RU equivalent. **P < 0.01 by paired Wilcoxon test. (E) abundance of the indicated phyla in the cecum. columns indicate individual mice. (F) Differentially expressed metabolic pathways between the lumen and mucosal bacterial communities [Padj < 0.001 by Mann-Whitney U test, log fold change (LFC) > 0.5]. (G) top, riboflavin biosynthetic pathway gene expression in the cecal lumen and mucosa (Padj ≤ 0.05 by Mann-Whitney U test). Bottom, scheme of the riboflavin biosynthesis pathway. (H) taxonomy of total (left) and ribD (right) bacterial transcripts in the lumen and mucosa. (I) Species-level taxonomy of ribD transcripts in the lumen and mucosa.
The large intestine presents ecological niches populated by microbial communities with distinct metabolic requirements (27), which could affect MAIT ligand production. In particular, oxygen arriving from the blood partially diffuses through the colonic epithelium into the lumen, creating a steep oxygen gradient from the mucosal surface to the lumen that shapes microbial communities (28). We evaluated how the radial partitioning of the gut microbiota affects MAIT ligand production. Whereas mucosal scrapes contained fewer bacteria (Fig. 1C), they were nonetheless more potent than lumen samples at activating 5-OP-RU–specific TCR Tg T cells (Fig. 1D), indicating stronger MAIT ligand production in the mucosal side compared with the lumen. To explore the mechanisms underlying increased MAIT ligand production in the mucosa, we sequenced the 16S ribosomal RNA (rRNA) genes and used PICRUSt (29) to estimate the metabolic potential of the lumen and mucosa-associated bacterial communities.
Mucosal communities were enriched in Proteobacteria (families Oxalobacteraceae and Enterobacteriaceae) and Deferribacteres (family Deferribacteraceae) (Fig. 1E, fig. S1D, and table S1), consistent with a previous report (30). In agreement with higher MAIT ligand production in the mucosa, PICRUSt predicted an enrichment in the riboflavin biosynthetic pathway in the mucosa as compared with the lumen (fig. S1E). The oxidative phosphorylation and glutathione metabolisms were also enriched in the mucosa (fig. S1E), consistent with higher oxygenation of the mucosa than the lumen.
To directly quantify the transcriptional activity of the riboflavin biosynthetic pathway, we next sequenced the non-rRNAs from both the mucosa and the lumen. Metatranscriptomic analyses identified several bacterial genes (fig. S2) and pathways (Fig. 1F) modulated between the two locations and revealed higher expression of the riboflavin biosynthesis pathway in the mucosa as compared with the lumen (Fig. 1F). The gene ribD, which controls production of the MAIT ligand precursor 5-A-RU, was overexpressed in the mucosa (Fig. 1G), consistent with active synthesis of MAIT ligands. Thus, the activity of the riboflavin biosynthesis pathway and the concentration of MAIT ligands vary in the distinct ecological niches of the gut and are higher in mucosal layers than in the lumen.
To identify the bacterial taxa contributing to MAIT ligand biosynthesis in the gut, we analyzed the taxonomy of ribD transcripts. In the lumen, ribD was mostly expressed by Proteobacteria and Firmicutes (Fig. 1H and fig. S1F). At the family level, the Proteobacteria were identified as Desulfovibrionaceae, whereas the Firmicutes could not be identified (fig. S1F). In the mucosa, ribD reads were primarily expressed by Deferribacteres (family Deferribacteraceae) and Proteobacteria (family Desulfovibrionaceae). Both taxa represented minor components of the total metatranscriptome (Fig. 1H), suggesting that MAIT ligand production may be dominated by a few strong producers. At the species level, ribD reads from the mucosa were mapped to Mucispirillum schaedleri (Fig. 1I), the only member of the Deferribacteres living in the mammalian intestine. M. schaedleri is found in most mammals, including humans, and resides in crypts and mucus layers (30) owing to its ability to scavenge oxygen and resist oxidative stress (31). In the mucosa, M. schaedleri expressed a catalase, which protects against oxidative damage by reactive oxygen species, and the high-affinity cbb3-type cytochrome c oxidase, which can be used either for protection from O2 stress or for microaerobic respiration (fig. S1G) (31). Thus, the strong riboflavin biosynthesis pathway activity in the mucosa may reflect needs for riboflavin in microbiota living in an oxygen-exposed environment.
Dysanaerobiosis drives microbiota production of MAIT ligands
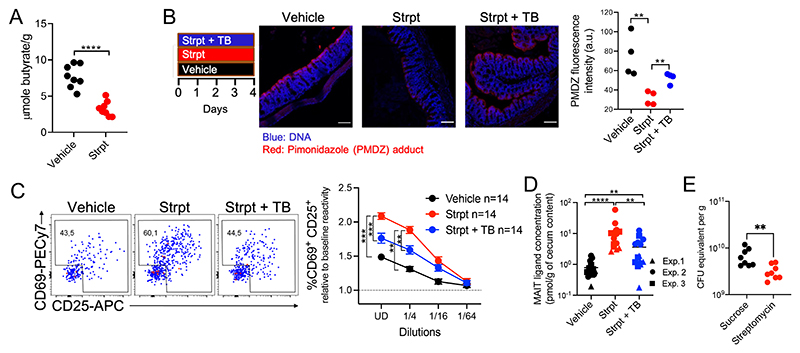
In neonates compared with adults, intestinal hypoxia is not yet established, and the colon presents a higher abundance of facultative anaerobes such as Enterobacteriaceae (22, 32, 33). To assess MAIT ligand production by the microbiota before hypoxia formation, MAIT ligands were quantified in colon contents from 2-week-old and 3-month-old mice. MAIT ligands were more abundant in the colons of neonates compared with those of adults, consistent with a link between intestinal oxygenation and MAIT ligand production in vivo (fig. S3A). In adults, intestinal hypoxia is maintained by epithelial cells through the β-oxidation of the microbial fermentation product butyrate (2, 34). Colon epithelial oxygenation can be modulated through the streptomycin-mediated depletion of butyrate-producing bacteria (34, 35). To increase the oxygen concentration in the gut, adult mice were treated with streptomycin, resulting in reduced butyrate concentrations in the cecum (Fig. 2A). To quantify hypoxia in the colon, we used the exogenous oxygen-sensitive dye pimonidazole (PMDZ), which is retained in tissues at oxygen concentrations of <1% (35). PMDZ was retained at the surface of colonocytes in nontreated mice (fig. S3B), confirming epithelial hypoxia. Streptomycin treatment reduced PMDZ staining (Fig. 2B), indicating increased colon oxygenation in these mice. Loss of hypoxia led to increased production of MAIT ligands, with concentrations increased 10-fold in the cecum (Fig. 2, C and D) despite lower bacterial loads (Fig. 2E). We then supplemented streptomycin-treated mice with tributyrin (TB), a butyrate precursor, which has been used to restore epithelial hypoxia upon streptomycin treatment (34, 35). TB supplementation partially restored epithelial hypoxia (Fig. 2B) and partially reduced the concentration of MAIT ligands in the cecum (Fig. 2, C and D). Thus, MAIT ligand production follows the oxygen concentration in the colon.
Fig. 2. Dysanaerobiosis drives microbiota production of MAIT ligands.
(A) cecal butyrate concentrations in control (vehicle) and streptomycin (strpt)–treated mice. n = 8 mice per group, N = 1. ****P < 0.0001 by unpaired Student’s t test. (B) left, PMDZ adduct (red) and DaPi (blue) staining at colon epithelial surfaces (each dot summarizes data from one mouse). White bars represent 100 μm. Representative of N = 2. **P < 0.01 by unpaired Student’s t test. (C) MR1:5-OP-RU–specific TCR Tg T cell activation after coculture with WT3-MR1 cells pulsed with cecal contents from the indicated mice (n = 14 mice per group; data pooled from N = 3). ***P < 0.001, **P < 0.01 by unpaired multiple t tests with FDR = 1%. (D) MAIT ligand concentrations expressed as 5-OP-RU equivalent (n = 14 mice per group, data pooled from N = 3). **P < 0.01 and ****P < 0.0001 by unpaired Mann-Whitney U tests. (E) qPCR-based estimation of cecal bacterial numbers in the indicated mice. n = 8 mice per group, N = 1. **P < 0.01 by unpaired Mann–Whitney U test.
The oxygen-induced production of MAIT ligands can result from shifts in microbiota composition favoring the outgrowth of strong MAIT ligand producers or from the response of individual species to oxygen. A previous study reported reduced MAIT ligand production by Escherichia coli grown aerobically as compared with anaerobically (36). To further evaluate how oxygen exposure affects MAIT ligand production at the species level, four species belonging to the main intestinal phyla (Bacteroides thetaiotaomicron, E. coli, Clostridium perfringens, and Clostridioides difficile) and competent for riboflavin biosynthesis (37) were grown anaerobically before exposure to air for 1 hour. Oxygen exposure did not induce MAIT ligand synthesis (fig. S3C), suggesting that these individual bacterial responses to oxygen are not a major source of MAIT ligands during dysanaerobiosis.
Intestinal inflammation triggers riboflavin and MAIT ligand production by the microbiota
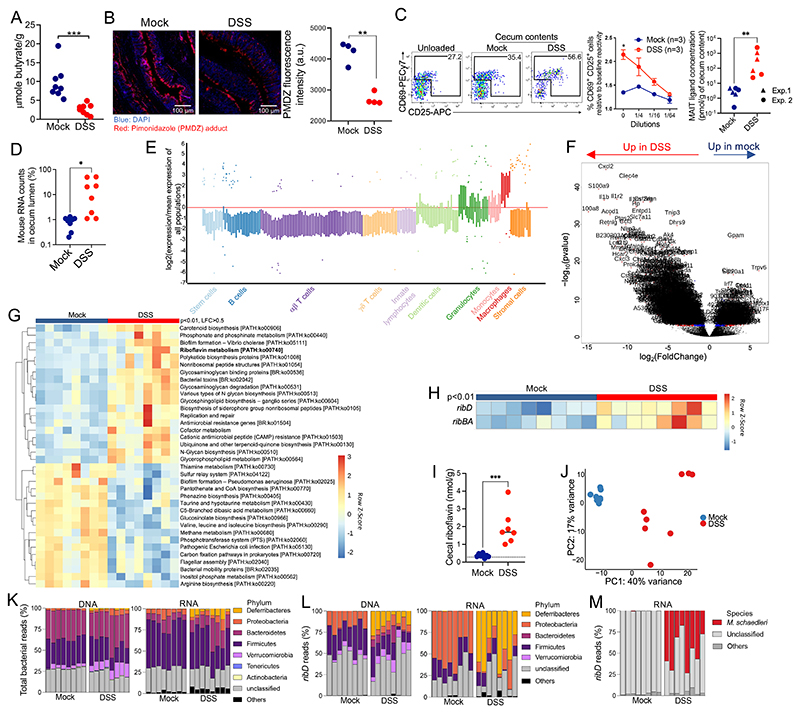
Because oxygenation of the colon lumen induces shifts in microbiota composition during intestinal inflammation (38), we hypothesized that MAIT ligands could be overproduced by the microbiota during colitis. We used dextran salt sulfate (DSS) to induce colonic epithelium injuries and inflammation (fig. S4A). Colitis was associated with a reduced butyrate concentration in the cecum (Fig. 3A) and loss of PMDZ staining in the colon, indicating reduced epithelial hypoxia (Fig. 3B). Accordingly, MAIT ligand concentrations were increased in the ceca of DSS-treated mice (Fig. 3C), indicating overproduction of MAIT ligands by the microbiota.
Fig. 3. Intestinal inflammation triggers riboflavin and MAIT ligand production by the microbiota.
(A) cecal butyrate concentrations in control (mock) and DSS-treated mice. n = 8 mice, N = 1. ***P < 0.001 by unpaired Student’s t test. (B) left, PMDZ adduct (red) and DNA (blue) staining in the colon. White bars represent 100 μm. Right, quantification of PMDZ staining at colon epithelium surfaces (each dot summarizes data from one mouse. Data representative of N = 2. **P < 0.01 by unpaired Student’s t test. (C) left, MR1:5-OP-RU–specific TCR Tg T cell activation after coculture with WT3-MR1 cells pulsed with cecal contents from control or DSS-treated mice (means ± SEMs, n = 3 mice per group, representative of N = 2). *P < 0.05 by unpaired multiple t tests with an FDR = 1%. Right, Mait ligand concentrations expressed as 5-OP-RU equivalent (n = 6 mice per group, data pooled from N = 2). **P < 0.01 by unpaired Mann-Whitney U test. (D) Percentage of mouse transcripts in ceca from the indicated mice. *P < 0.05 by unpaired Mann-Whitney U test. (E) Predicted cell-type enrichment in the cecum lumen upon DSS treatment (see Materials and Methods). (F) Differentially expressed mouse genes in the cecum lumen upon DSS treatment. (G) Bacterial metabolic pathways differentially expressed between control and DSS-treated mice (Padj < 0.01 by Mann-Whitney U test, LFC > 0.5). (H) Differentially expressed riboflavin biosynthetic pathway transcripts between control and DSS-treated mice (Padj < 0.01 by Mann-Whitney U test). (I) cecal riboflavin concentration (n = 7 or 8 mice per group, representative of N = 2). the dotted line indicates the plasma riboflavin concentration at steady state. ***P < 0.001 by unpaired Student’s t test. (J) PCA of cecal DNA reads at the family level in control and DSS-treated mice. (K) Phylum-level taxonomy of total bacterial DNA (left) or RNA (right) reads from the cecum. (L) Phylum-level taxonomy of ribD DNA (left) or RNA (right) reads from the cecum. (M) Species-level taxonomy of ribD transcripts.
We then sequenced the cecal RNA and DNA contents to better define the bacterial communities and functions associated with MAIT ligand production during colitis. Metagenomic and metatranscriptomic analyses revealed the presence of mouse DNA and RNA in the cecum lumen during colitis (Fig. 3D and fig. S4B). The expressed genes matched granulocyte and phagocyte transcriptional signatures (Fig. 3E), suggesting that innate immune cells crossed the inflamed intestinal epithelium and were released into the lumen. We identified the up-regulated mouse and bacterial genes to determine how intestinal inflammation shaped host-microbiota cross-talk. Multiple mouse genes involved in antibacterial defense (Nos2, Lcn2, and antimicrobial peptides S100a8 and S100a9), innate immune responses (Nlrp3, Il1a, and Il1b), and oxidative stress resistance (glutathione reductase Gsr, Slc7a11, and Dusp1) (Fig. 3F; fig. S4, C and D; and table S2A) were up-regulated in the cecum lumen upon colitis, consistent with increased inflammation and oxidative stress. In response, bacterial genes involved in resistance to antimicrobial peptides and to oxidative stress (catalase-peroxidase, methionine sulfoxide reductase, catalase, and cytochrome-c peroxidase, among others) were up-regulated in colitis (Fig. 3G, fig. S4E, and table S2B), whereas genes involved in butyrate metabolism were down-regulated (fig. S4F). The genes of the riboflavin biosynthetic pathway were also up-regulated in bacteria during colitis (Fig. 3G), specifically ribBA and ribD, which control 5-A-RU production (Fig. 3H). Quantification of riboflavin itself revealed increased concentrations in the ceca of DSS-treated mice as compared with control mice, well above plasma levels (39), confirming increased activity of this metabolic pathway in the gut during colitis (Fig. 3I).
Next, we analyzed the taxonomy of the microbiota at the DNA and RNA levels to determine the microbial composition and transcriptional activity. Principal components analysis (PCA) of bacterial DNA reads clearly distinguished mock- and DSS-treated mice (Fig. 3J), indicating shifts in the microbiota composition induced by the treatment. Bacterial families enriched at the DNA level were usually also enriched at the RNA level, suggesting that most families were transcriptionally active (fig. S4G). In particular, Deferribacteres (family Deferribacteraceae) were a minor component of the microbiota at steady state but were enriched at the DNA and RNA level in DSS-treated mice (Fig. 3K and fig. S4H), indicating that this phylum was expanded and transcriptionally active during colitis, in line with earlier reports (31, 40, 41). Entero-bacteriaceae were also enriched in DSS-treated mice at both DNA and RNA levels (table S2, C and D). We then analyzed the ribD transcripts to identify bacteria contributing to MAIT ligand production during colitis. Whereas steady-state ribD expression was dominated by Proteobacteria (family Desulfovibrionaceae), ribD was mostly expressed by Deferribacteres (family Deferribacteraceae) during colitis (Fig. 3L and fig. S4I). M. schaedleri alone contributed 47% of the ribD expressed in colitis (Fig. 3M), suggesting that this species is a major producer of MAIT ligands during colitis. ribD reads from Bacteroides (family Bacteroidaceae) and Verrucomicrobia (family Akkermansiaceae) were detected at the DNA level (Fig. 3L and fig. S4I) but not the RNA level, suggesting transcriptional regulation of ribD in vivo.
To assess the robustness of our findings, we reanalyzed a published metatranscriptomic dataset from fecal bacteria upon Helicobacter hepaticus–induced colitis (42). Although the colitis model and sampling locations were different, the riboflavin pathway genes were also upregulated (fig. S4J). All of the genes controlling 5-A-RU production, including ribD, were more expressed during colitis (Padj < 0.01, fig. S4J). PCA of riboflavin biosynthesis gene expression readily distinguished mice before and after colitis induction (fig. S4J), which suggests that the riboflavin biosynthesis pathway activity can be used as a colitis marker across experimental models.
MAIT ligands cross the intestinal barrier and activate MAIT cells in various tissues
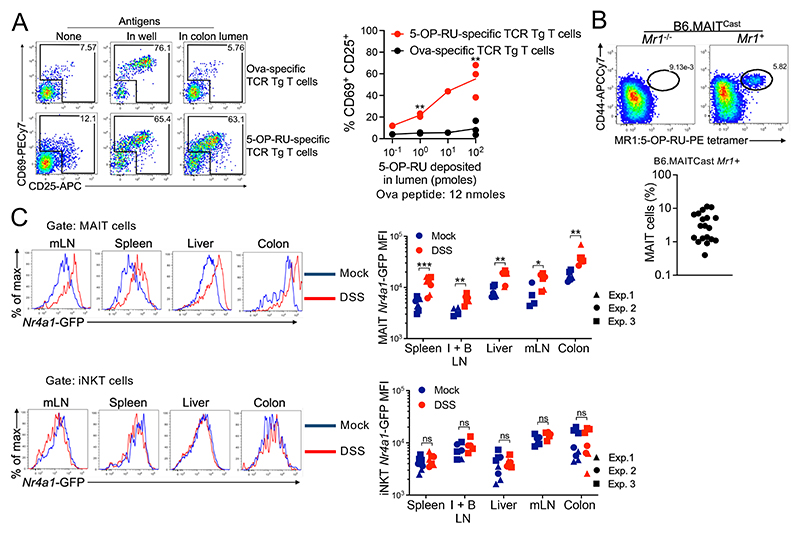
MAIT ligands produced in the lumen may diffuse throughout the body because MAIT ligands can rapidly cross the mouse skin (43). To assess whether the intestinal epithelium is permeable to 5-OP-RU, a mixture of antigenic peptide (OVA323–339) and 5-OP-RU metabolite was deposited into the lumen of intestinal explants from untreated mice and incubated on top of antigen-presenting cell lines (fig. S5A). WT3-MR1 cells were used for 5-OP-RU presentation, whereas mutuDC cells were used for Ova presentation (44). After 10 min of incubation, colon explants were removed, and antigen-presenting cells were incubated with 5-OP-RU–specific and Ova-specific TCR Tg T cells to evaluate the presence of these ligands (fig. S5A). 5-OP-RU–specific, but not Ova-specific, TCR Tg T cells became activated (Fig. 4A), which indicated rapid and efficient transfer of 5-OP-RU across the whole intestinal wall ex vivo. This rapid passage of 5-OP-RU through the gut wall was observed at a physiological dose (≤10 pmol in the lumen). Given the hydrophilic nature of 5-OP-RU, this result suggests the presence of an unknown transporter expressed by intestinal epithelial cells.
Fig. 4. MAIT ligands cross the intestinal barrier and are presented to MAIT cells in various tissues.
(A) TCR Tg T cell activation in the indicated conditions. Data were pooled from N = 2. **P < 0.01 by paired multiple t tests with FDR = 1%. (B) identification of Mait cells from the colon lamina propria of B6-MAITCast mice. Pooled data from N = 3. (C) Nr4a1-GFP expression in MAIT (top) and iNKT (bottom) cells. Data pooled from N = 3. ns, P > 0.05, *P < 0.05, **P < 0.01, and ***P < 0.001 by unpaired multiple Mann-Whitney tests with FDR = 1%.
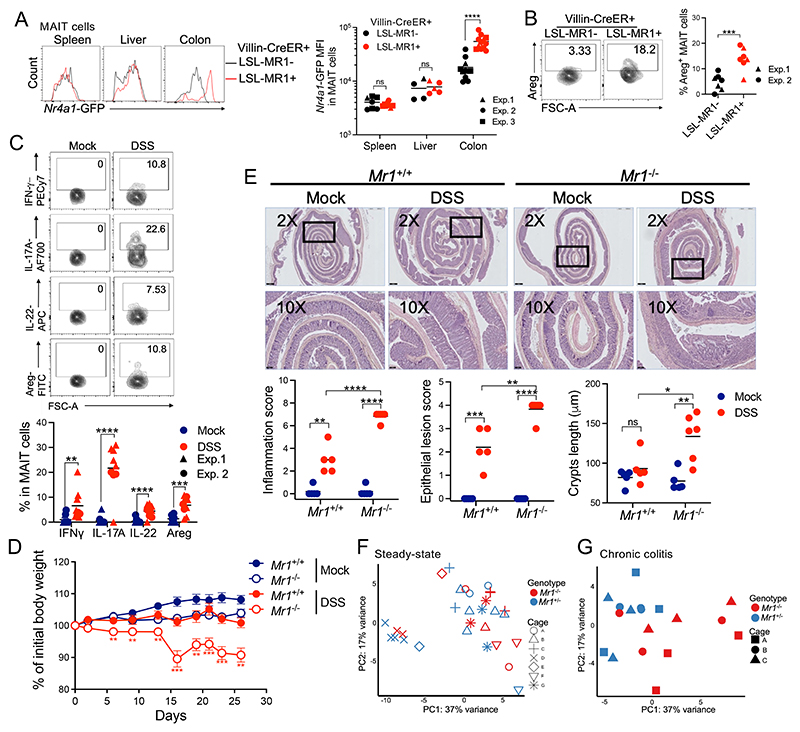
To test whether MAIT cells sense ligands in vivo, we used B6-MAITCast mice, which present an increased frequency of MAIT cells owing to a more frequent rearrangement of the MAIT TCRα chain (45), crossed with the Nr4a1–green fluorescent protein (GFP) reporter mouse in which TCR signaling drives GFP expression (46). We confirmed that TCR, but not interleukin-12 (IL-12) and IL-18 stimulation, controls Nr4a1-GFP expression in MAIT cells (fig. S5B). MAIT cells identified using the MR1:5-OP-RU tetramer (see gating strategy in fig. S5C) represented 0.5 to 10% of T cells in the colonic lamina propria in B6-MAITCast mice (Fig. 4B). At steady state, MAIT cells expressed Nr4a1-GFP in the colons, mesenteric lymph nodes (mLNs), and livers (fig. S5D), consistent with some ligand production in the colon and transport of these ligands through the lymph and the blood. DSS-induced colitis increased Nr4a1-GFP expression in MAIT cells from the colon and also from the mLN, liver, inguinal and brachial LN, and spleen (Fig. 4C and S5E), indicating TCR-mediated activation across peripheral tissues. By contrast, intestinal inflammation had no effect on Nr4a1-GFP expression in invariant natural killer T (iNKT) cells (Fig. 4C). Colitis was also associated with increased numbers of MAIT cells in the colon (fig. S5F). Thus, colitis drives expression of the riboflavin pathway in microbiota, resulting in MAIT ligand production and in MAIT cell activation.
Mice lacking MAIT cells are more susceptible to chronic colitis
TCR stimulation of MAIT cells in vitro triggers expression of tissue-repair mediators (19–21), including the growth factor amphiregulin (Areg), which induces differentiation of intestinal epithelial cells and protects against intestinal damage (47). To assess the effect of TCR signaling on intestinal MAIT cells in vivo, we generated a conditional MR1 Tg mouse (Lox-STOP-Lox-MR1, subsequently referred to as LSL-MR1) for Cre-mediated overexpression of MR1 (fig. S6A). This strain was validated by crossing with CD11c-Cre and confirming MR1 overexpression in dendritic cells (fig. S6B). The LSL-MR1 mouse was then crossed with the villin-CreER mouse for inducible expression in intestinal epithelial cells (48). Tg MR1 expression in intestinal epithelial cells had no effect on microbiota production of MAIT ligands (fig. S6C) but elicited strong TCR signaling in MAIT cells from the colon, but not the spleen and liver, as expected (Fig. 5A). Thus, the LSL-MR1 x villin-CreER strain can help isolate the effect of TCR stimulation on colonic MAIT cells. TCR signaling in colonic MAIT cells was sufficient to induce the production of Areg in vivo (Fig. 5B), consistent with previous in vitro observations (19–21) and suggesting that TCR stimulation of MAIT cells may promote epithelial repair.
Fig. 5. MAIT cells reduce colitis severity.
(A) Nr4a1-GFP expression by Mait cells from the indicated mice. Both LSL-MR1+ and LSL-MR1− mice carried the villin-CreER Tg and received tamoxifen. Pooled data from N = 3. ****P < 0.0001 by unpaired Mann-Whitney U test. (B) areg expression by colonic Mait cells from the indicated mice. n = 7 mice per group. Data pooled from N = 2. ***P < 0.001 by unpaired Student’s t test. (C) cytokine production by colonic MAIT cells from control mice and from mice with chronic colitis. n = 9 to 11 mice per group. Data were pooled from N = 2. **P < 0.01, ***P < 0.001, and ****P < 0.0001 by unpaired Student’s t tests. (D) Mouse body weight after mock or chronic DSS treatment. n = 12 or 13 mice per group; data pooled from N = 3. **P < 0.01 and ***P < 0.001 between DSS-treated Mr1+/+ and Mr1−/− mice, by unpaired multiple t tests with FDR = 1%. (E) top, H&E staining of colon from the indicated condition. Representative 2× and 10× magnifications are shown. Black bars represent 500 μm (top) and 100 μm (bottom). Bottom, investigator-blinded pathology assessments in the indicated conditions. n = 5 to 7 mice per group. Data representative of N = 3. ns, P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by unpaired Student’s t tests. (F and G) PCA of bacterial families in Mr1+/− and Mr1−/− B6-MAITCast mice either untreated (F) or upon chronic DSS-induced colitis (G). Symbols indicate the cage of origin.
To investigate the role of MAIT cells in colitis, we turned to a chronic DSS model, which mimics intestinal inflammatory flares and can provide insights into adaptive immune responses to colitis (49). To this end, B6-MAITCast mice on a Mr1+/+ or Mr1−/− background [hence lacking MAIT cells (45)] were cohoused to allow for sharing of microbiota (49) and then subjected to successive cycles of DSS treatments followed by a recovery phase (fig. S6D). At the end of the last recovery phase, MAIT cells from colitic mice spontaneously produced interferon-γ (IFN-γ), IL-17A, IL-22, and Areg in the colon (Fig. 5C). IFN-γ promotes mucus secretion by goblet cells (50), where- as IL-17A and IL-22 stimulate secretion of antimicrobial peptides and expression of tight-junction proteins in epithelial cells (51), thereby reinforcing the epithelial barrier. To determine the role of MAIT cells in chronic colitis, colons from Mr1+/+ and Mr1−/− mice were scored for inflammation and histopathology upon chronic DSS treatment. Mr1−/− mice experienced more severe weight loss than Mr1+/+ counterparts upon chronic colitis (Fig. 5D). The presence of MAIT cells was associated with reduced inflammatory cell infiltrates, improved epithelial architecture, reduced hyperplasia (Fig. 5E), and reduced splenomegaly (fig. S6E). The numbers of iNKT and γδ T cells remained unchanged in the thymus and peripheral tissues in the absence of Mr1 (fig. S6F), ruling out a potential compensatory expansion of these cells in the absence of MAIT cells. Thus, MAIT cells likely protect against chronic intestinal inflammation.
Previous studies reported dysbiosis in Mr1−/− mice at steady state (52, 53), which may contribute to the observed enhanced pathology in these mice during chronic colitis. To characterize the effect of Mr1 on the intestinal microbiota, fecal samples from cohoused, nontreated Mr1+ and Mr1−/− B6-MAITCast mice were compared by 16S rRNA sequencing (rRNA-seq). We could not detect any bacterial family with changed abundance (Padj < 0.01) between the two groups (table S3A). Samples clustered together by cage of origin rather than by mouse genotype (Fig. 5F), indicating that the microbiota was not affected by Mr1 at steady state. Thus, enhanced pathology in Mr1−/− mice is likely independent of the preexisting microbiota composition. To determine whether MAIT cells affect the microbiota composition upon colitis, samples from cohoused Mr1+ and Mr1−/− B6-MAITCast mice were analyzed by 16S rRNA-seq after chronic colitis induction. Five bacterial families were differentially abundant (Padj < 0.01) between Mr1+ and Mr1−/− mice with chronic colitis (table S3B), and mice clustered by genotype rather than by cage of origin (Fig. 5G). The families Bacteroidaceae and Enterobacteriaceae, which expand during colitis [table S2D and (1)], were further expanded in Mr1−/− mice as compared with Mr1+ controls (table S3B) consistent with Mr1−/− mice suffering worse colitis than Mr1+ mice. Thus, the lack of MAIT cells during chronic colitis is associated with an expansion of bacterial families such as Enterobacteriaceae, likely as a consequence of increased pathology.
In an attempt to demonstrate a direct protective effect of MAIT cells on DSS-induced colitis, we transferred in vitro–expanded MAIT cells into CD3ε−/− mice. Unexpectedly, CD3ε−/− mice were not protected against chronic colitis (fig. S6, G and H). This negative result does not dismiss a protective effect of MAIT cells because technical caveats are numerous: a possible modification of MAIT cells during in vitro expansion, a low number of cells reaching the colon, or potentially incorrect localization of MAIT cells in the colon. The microbiota may also be modified in CD3ε−/− mice. Moreover, the protective effect of MAIT cells may rely on triggering other T cells that are lacking in CD3e−/− mice or may be counteracted by an increase in other immune cells compensating for the lack of T cells. Because Mr1+ are less susceptible to colitis than MR1−/− mice (Fig. 5D), this negative result could also suggest that the protective effect of MAIT cells might be indirect.
Mice lacking MAIT cells are more susceptible to inflammation-induced CRC
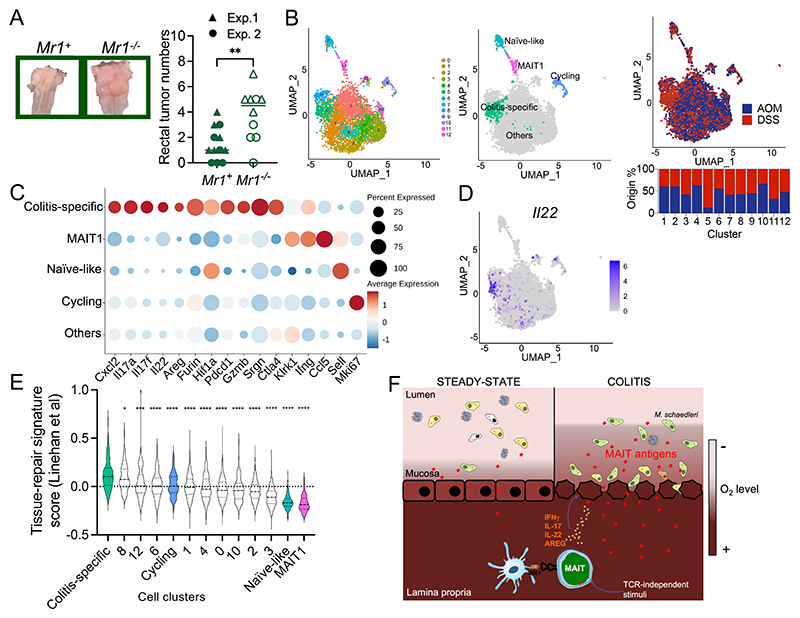
Chronic intestinal inflammation is a risk factor for CRC. To define the role of MAIT cells in colitis-induced CRC, colorectal tumors were induced in Mr1+ and Mr1−/− B6-MAITCast mice using the azoxymethane (AOM) + DSS model, in which administration of the carcinogen AOM is followed by three phases of DSS-induced colitis. Again, Mr1+ and Mr1−/− mice were cohoused to normalize the microbiota before and during the experiment. Colorectal tumors were enumerated 60 to 80 days after the last DSS cycle (fig. S7A). Only mice receiving both AOM and DSS developed colonic tumors. Mr1−/− mice developed higher numbers of rectal tumors than Mr1+ mice, strongly suggesting that MAIT cells alleviate inflammation-driven colonic tumorigenesis (Fig. 6A). The presence of colorectal tumors was not associated with increased frequencies or numbers of MAIT cells in the colon (fig. S7B) or with the production of MAIT ligands (fig. S7C). TCR signaling was not increased in MAIT cells from tumor-bearing mice (fig. S7D), indicating that MAIT cells were not activated by microbiota-derived ligands at the time of euthanasia (i.e., 60 to 80 days after the last DSS cycle) in CRC-bearing mice. Furthermore, MAIT cells did not provide protection against colorectal tumor development in an orthotopic model (fig. S7, E and F), suggesting that the MAIT cell protective effect in the colitis-associated CRC model was exerted by decreasing intestinal inflammation.
Fig. 6. MAIT cells protect against inflammation-induced CRC.
(A) Left, representative images of the rectum after inflammation-induced CRC. Right, rectal tumor numbers in the indicated mice. Pooled data from N = 2. **P < 0.01 by unpaired Student’s t test. (B) UMAP of colonic MAIT cells isolated from control (AOM) or colitic (DSS) mice analyzed by single-cell RNA-seq. The relative contribution of MAIT cells from control and colitic mice to each cluster is indicated. (C) Expression of selected genes by MAIT cells from the indicated UMAP clusters. (D) Il22 expression by single-cell RNA-seq in colonic MAIT cells. (E) Tissue-repair gene signature expression in the indicated clusters. the dashed bar represents the median score. *P < 0.05, ***P < 0.001, and ****P < 0.0001 by unpaired Student’s t tests. (F) Schematic summary of the main findings of the study.
To define the transcriptional response of MAIT cells to chronic colitis, colonic MAIT cells from control (AOM alone) or colitic (DSS-treated) mice were fluorescence-activated cell sorting (FACS)–sorted and analyzed by single-cell RNA-seq. After quality control and filtering, 2903 and 2584 MAIT cells from control and colitic mice, respectively, were retained and integrated for downstream analyses. Uniform Mani-fold Approximation and Projection (UMAP) identified 13 clusters of cells (Fig. 6B). Genes differentially expressed in each cell cluster were identified (table S4) and displayed in a heatmap (fig. S8A). Cell clusters were then labeled according to the expression of known marker genes (fig. S8A). The analysis identified a small subset of naïve-like cells [expressing Sell (encoding CD62L), Ccr7, and Klf2] (cluster #7) (fig. S8A and table S3), MAIT1 cells (expressing Tbx21, Ifng, and Ccl5) (cluster #11), and cycling cells (expressing proliferation genes such as Mki67) (cluster #9) (Fig. 6C and fig. S8A). Except for the naïve-like and MAIT1 cells, all cells expressed a MAIT17 program, which was blended with a MAIT1 program in some cells (fig. S8B). Partition of the cells according to their origin revealed a cluster of MAIT cells uniquely present upon chronic intestinal inflammation (Fig. 6B, cluster 5) and characterized by expression of anti-inflammatory (Tgfb1 and Ctla4) and cytotoxicity genes (Gzmb and Srgn) (Fig. 6C, fig. S8A, and table S3). MAIT cells from the colitis-specific cluster were also characterized by the expression of the key effector cytokines Il17a and Il22, with well-documented beneficial effects on the intestinal epithelium during colitis (Fig. 6, C and D) (51, 54–57). These MAIT cells also expressed a gene signature (described in table S5) previously associated with tissue-repair functions (58) (Fig. 6E). In particular, MAIT cells in colitis overexpressed the transcriptional regulator Hif1a (Fig. 6C and table S4), whose expression protects against colitis (59), and Furin (Fig. 6C and table S4), which encodes a convertase essential for the maturation of anti-inflammatory pro-proteins such as transforming growth factor–β1 (TGF-β1) (60). Furin controls the protective function of regulatory T cells in the adoptive transfer model of colitis (60), suggesting that Furin expression by MAIT cells may contribute to their protective functions. Thus, MAIT cells in the inflamed colon express anti-inflammatory and tissue-repair genes, and mice lacking MAIT cells are more susceptible to chronic colitis and colitis-induced CRC, strongly arguing for a protective role of MAIT cells against colonic inflammation.
Discussion
Microbiota-derived MAIT ligands control MAIT cell development and functions (22, 43), yet the bacterial species involved and the factors controlling MAIT ligand production in the intestinal ecosystem are poorly defined. Here, we used metatranscriptomics coupled to a sensitive bioassay to quantify MAIT ligands in distinct ecological niches of the large intestine. At steady state, the genes of the riboflavin biosynthetic pathway were expressed by mucosa-associated bacteria, and MAIT ligands were produced in the colon mucosa. MAIT ligands were also abundantly synthesized in the colon lumens of mice before weaning, during a developmental time window critical for MAIT cell seeding of the skin (22). Colonic hypoxia is only established at weaning, and mucosa-associated bacterial communities in adults are profoundly shaped by the oxygen emanating from the colon epithelium, suggesting a link between oxygen availability and microbiota synthesis of MAIT ligands. In line with this hypothesis, disruption of intestinal hypoxia using streptomycin treatment increased production of MAIT ligands, whereas restoration of hypoxia using TB reduced MAIT ligand production. Exposure of individual intestinal bacterial species to oxygen did not induce MAIT ligand production, suggesting that oxygen-induced community shifts, rather than individual bacterial responses, caused MAIT ligand production during dysanaerobiosis. Thus, oxygen may represent a key ecological factor explaining the temporally and spatially segregated production of MAIT ligands observed in the colon.
The production of MAIT ligands during dysanaerobiosis appears particularly relevant in the context of IBD, a condition associated with increased oxygenation of the colon (1, 2). Experimental colitis was indeed associated with increased oxygen availability in the colon lumen and with the expansion of aerotolerant and facultative anaerobic bacteria. Bacterial genes involved in resistance to oxidative stress were induced by colitis, together with the genes of the riboflavin biosynthetic pathway. Increased transcription of rib genes translated into increased production of riboflavin and MAIT ligands upon DSS treatment. Although 85% of all bacterial species harbor the genes encoding a full riboflavin biosynthesis pathway (61), only a handful of species expressed ribD in vivo. In addition, the increased transcription of ribD in mucosa and during colitis was mainly driven by a single species (M. schaedleri), which resided in the mucosa at steady state and bloomed during inflammation. These observations support the hypothesis that MAIT ligands are primarily produced by expanded aerotolerant bacteria during dysanaerobiosis. M. schaedleri can trigger colitis in immunodeficient hosts (62) but also impart protection against Salmonella infections (63). Whether these functions involve the antigen-driven activation of MAIT cells remains to be determined.
Given the ability of MAIT ligands to rapidly cross the intestinal epithelium, MAIT cells may detect the earliest events of dysbiosis and provide an immediate effector response (Fig. 6F). Intriguingly, colitis triggered TCR signaling in MAIT cells from all the studied tissues, including non–gut-draining lymph nodes, consistent with the ability of 5-OP-RU to travel across the body (43). MAIT cell activation in distant tissues may represent a systemic surveillance strategy of intestinal inflammation, the implications of which remain to be investigated. Because MAIT cell transcriptional features are shaped by their tissue of residence (64), body-wide TCR activation as seen during colitis may result in local production of mediators tuned to each tissue (65).
In patients with IBD, several groups observed decreased MAIT cell frequencies in blood coupled with activation and recruitment of MAIT cells in inflamed lesions (11, 66), which correlated with disease activity (66, 67). Because MAIT cells produce cytokines that can be either deleterious [tumor necrosis factor–α (TNF-α) (67)] or beneficial [IL-17A and IL-22 (11, 66, 67)] in patients with IBD, their role in the progression of the disease was unclear in the absence of experimental manipulation. Our data establish that the presence of MAIT cells is associated with reduced intestinal epithelial damage during chronic colitis and reduced development of colitis-associated colorectal tumors. Chronic colitis induced the expression of a tissue-repair transcriptional program in MAIT cells, in good agreement with the proposed link between MAIT cell TCR stimulation and tissue-repair functions (19, 21). These results are in line with previous studies reporting weakened intestinal barrier function in Mr1−/− mice (68, 69) and support a key role for MAIT cells in promoting epithelial barrier integrity. The exact contribution of each MAIT-derived effector molecule remains to be determined. Our results contradict a previous study reporting worse pathology in Mr1+/+ mice as compared with Mr1−/− mice in the oxazolone model of colitis, in which inflammation is triggered by immunization against microbiota antigens (69). The apparent discrepancy may derive from the models used (acute in the case of oxazolone colitis versus chronic in our model). In addition, interpretation of the results from Yasutomi et al. (69) is complicated by the use of a lower oxazolone dose for some of the control mice, which were more susceptible to colitis because they originated from commercial vendors, whereas Mr1−/− mice were produced in house. Thus, colitis severity in that study may be primarily determined by the microbiota instead of by MAIT cells. In conclusion, we highlight a specific function of MAIT cells in the colon: monitoring the activity of a microbial metabolic pathway indicative of intestinal inflammation to provide anti-inflammatory and tissue-repair mediators in return.
Materials and Methods
Study design
Before inclusion in experimental groups, mice from different litters were cohoused or cage contents were mixed for at least 2 weeks to normalize the microbiota. Individual mice were randomly assigned to experimental groups. The assessment of colitis and CRC severity was performed by scientists blinded to mouse genotypes (Mr1+ or Mr1−/−), which remained concealed until pathology measurements were completed. The number of independent experiments performed, N, is indicated in the figure legends.
Mice
C57BL/6J (B6) and B6-CD45.1 mice were purchased from Charles River Laboratories. iVα19 and iVα19 Vβ8 TCR Tg Cα−/− B6 mice, WT, and Mr1−/− B6-MAITCast mice have been described (24, 45, 70). Nr4a1-eGFP mice (46) were imported from the MMRRC (UC Davis). OTII mice (71) were maintained on an RAG−/− background. All mice were bred and maintained on a standard chow diet in specific pathogen–free conditions at Institut Curie. All experiments were performed according to national and international guidelines and were approved by the Institut Curie Ethic committee (APAFIS #25203-2020042318109119, #24245-2020021921558370, #IOI68-2017060909448377, and #26939-2020082515024782 v1). To generate the Lox-stop-Lox-MR1 (LSL-MR1) Tg mouse, a DNA segment containing a stop cassette flanked with loxP sites upstream of the murine Mr1 gene was prepared by digestion of the pOD2G-LS CAGGS [7279 base pairs (bp)] plasmid (72) (a gift from J. J. Moon, Massachusetts General Hospital) with the enzymes HindIII and SpeI allowing the insertion of a full-length MR1 cDNA. Efficient induction of MR1 protein expression under the control of the chicken actin promoter was validated in human embryonic kidney 293 cells cotransfected with a Cre expression vector. The construct was then injected into pronuclei of fertilized C57BL6/N oocytes. Founders were crossed with Tg mice bearing either a CD11c-Cre transgene (73) or a tamoxifen-dependent Cre recombinase expressed under the control of the villin promoter (48).
Antibiotic treatments, colitis induction, and tamoxifen administration
Female mice aged 6 to 10 weeks at the start of the experiments were used. For microbiota depletion, B6/J mice were given four antibiotics [vancomycin (0.5 g/liter, Mylan), ampicillin (10 g/liter, Coophavet), metronidazole (1 g/liter, B Braun), and neomycin (1 g/liter, Coophavet)] in drinking water supplemented with 2% sucrose for 7 days. Fresh antibiotic solutions were provided after 3 days of treatment. Streptomycin (2.5 mg/ml, Sigma-Aldrich, S9137) was provided for 4 days in drinking water supplemented with 2% sucrose. When indicated, streptomycin-treated mice also received TB (5 g/kg) (Sigma-Aldrich, T8626) by oral gavage each day for 4 days. For fungal depletion, fluconazole (0.5 mg/ml, Sigma-Aldrich) was provided in the drinking water for 7 days.
Acute colitis was induced in B6/J mice by adding 2% (w/v) DSS (molecular weight: 36,000 to 50,000 Da; MP Biomedicals) to the drinking water for 7 days or until the mice had lost 15% of their initial weight. For modeling chronic colitis, B6-MAITCast mice received successive 5-day DSS treatments, each with increasing DSS concentrations, followed by 4 days with water only. Cohoused Mr1+/+ and Mr1−/− mice received three cycles of DSS before clinical pathology assessment. Pathology assessment was performed by an investigator blinded to the mouse genotypes. Mr1+/+ mice received four or five cycles until the mice had lost weight for 2 consecutive weeks before phenotypic analyses of MAIT cells. Control groups (mock) received only water. For inducible MR1 overexpression, all mice were injected intraperitoneally with 2 mg of tamoxifen (T5648, Sigma-Aldrich) in a corn oil–ethanol mixture once per day for 3 consecutive days. Tissues were collected 24 hours after the last injection.
CRC models
The AOM/DSS model (74) was used as a model of colitis-induced CRC. Male and female Mr1+ and Mr1−/− B6-MAITCast mice aged at least 12 weeks were cohoused for at least 2 weeks before the start of the experiment to normalize the microbiota. The mice received a single intraperitoneal injection of saline or of azoxymethane in saline (10 mg/kg, Sigma-Aldrich, A5486-25MG). Mice were then treated three times with AOM DSS (2.5%) in the drinking water for 4 days, followed by 17 days on water. The first DSS treatment started 4 days after AOM injection, and mice were euthanized 110 to 130 days later. Colonic and rectal tumors were counted by an investigator blind to the mouse genotypes. For orthotopic CRC development, cohoused female B6-MAITCast Mr1+ or Mr1−/− mice aged 12 to 18 weeks were anesthetized and implanted intrarectally with 2 × 106 MC38 cells. Colorectal tumors were counted 11 days later by an investigator blinded to the mouse genotypes.
Cell isolation
Spleens were mashed over a 40-μm cell strainer to create single-cell suspensions followed by red blood cell lysis. Livers were mashed over a 100-μm cell strainer and centrifuged over Histopaque 1077 (Sigma-Aldrich) layers to isolate the lymphocyte-containing fraction. Colons were opened longitudinally and cut into approximately 0.5-cm pieces. Dissociation of epithelial cells was performed by incubation with constant stirring in Hanks’ balanced salt solution (HBSS) without Ca/Mg (Life Technologies) containing 5 mM EDTA (5 mM; Thermo Fisher Scientific), dithiothreitol (1 mM; Euromedex), and 5% fetal calf serum (FCS) twice for 20 min at 37°C. After each step, samples were vortexed and the epithelial fraction discarded. Tissue fragments were then washed in HBSS, and enzymatic digestion was performed in CO2-independent medium (Life Technologies) containing collagenase D (1 mg/ml; Roche), Liberase TM (0.17 U/ml; Roche), and deoxyribonuclease (DNase I, 100 μg/ml; Roche) on a shaker for 30 min at 37°C. Colon fragments were then dissociated with a gentleMACS dissociator according to the manufacturer’s instructions (Miltenyi). Lymphocytes were collected at the interface of a 40%/80% Percoll gradient (GE Healthcare).
Tetramers and flow cytometry
CD1d tetramers loaded with PBS-57 and conjugated to BV421, and MR1 tetramers loaded with 5-OP-RU (75) and conjugated to phycoerythrin were provided by the National Institutes of Health (NIH) tetramer core facility (Emory University). Cell suspensions prepared from various tissues were stained in phosphate-buffered saline (PBS), 2% FCS, and 2 mM EDTA complemented with rat anti-mouse CD16/CD32 antibody (clone 2.4G2 produced in house) to block nonspecific binding to the Fcγ receptor. Tetramer staining was performed at room temperature for 30 min. Surface staining was performed at 4°C with fluorescently labeled antibodies and fixable viability dye eFluor780 (eBioscience, Hatfield, UK). MR1:5-OP-RU–specific TCR Tg T cells were identified as shown in fig. S1A. MAIT and iNKT cells were identified as shown in fig. S5C. A list of the antibodies and reagents used in the study is provided in table S6.
Fixation and permeabilization were performed for intranuclear staining (FoxP3 staining kit; eBioscience) and intracytoplasmic staining (Cytofix/Cytoperm kit; BD Biosciences) according to the manufacturer’s instructions. For intracytoplasmic staining, cells were cultured for 3 hours at 37°C in complete RPMI 1640 in the presence of GolgiPlug (1:1000; BD). Cultured cells were subsequently stained for surface antigens as above. Cells were fixed and permeabilized, washed, and incubated for 30 min at 4°C with anti–IL-17 (TC11-18H), –IL-22 (IL22JOP), and –IFN-γ (XMG1.2). Cells were then stained overnight with anti–Areg-biotin (BAF989) followed by fluorescein isothiocyanate– or BV605-conjugated streptavidin. In some experiments, non-fixed dead cells were stained with 4′,6-diamidino-2-phenylindole (DAPI)–staining solution and directly analyzed by flow cytometry. Cells were acquired on Cytoflex (Beckman), LSRII (BD Biosciences), or Fortessa (Becton Dickinson), and the results were analyzed with FlowJo V10.2 (Treestar).
Single-cell RNA-seq
Mice injected with AOM or treated with DSS (nine mice per group) were euthanized 53 days after AOM injection or 3 days after the end of the third DSS treatment. Colon MAIT cells were identified as live CD11c−B220−CD19−TCRb+CD44+MR1:5-OP-RU tetramer+ and FACS-sorted into PBS containing 0.04% sterile bovine serum albumin (Sigma-Aldrich) using the BD FACSAriaIII. Sorted MAIT cells were counted and processed for 3′ v3 single-cell RNA-seq according to manufacturer’s instructions (10X Genomics). Reads were aligned to the mm10 genome using CellRanger (6.0.0) and analyzed by Seurat (4.1.0) package in R (4.1.0). Contaminating cells (B lymphocytes, macrophages, and epithelial cells) and cells with high mitochondrial content were filtered out. Data were normalized using default parameters. The 2000 (AOM) and 3000 (DSS) most variable features were selected. Both datasets were integrated using default parameters without the SC transform option. Reduction of dimensions was performed using 30 principal components and a resolution of 0.8. Differential expression (DE) analysis was performed on assay RNA using the classical DE function FindAllMarkers(), but using logistic regression and taking into account batch effect in the logistic regression model. To compute a tissue-repair score, we aggregated the expression of the following genes [(58) and table S5]: Hbegf, Csf2, Mmp25, Cxcl10, Areg, Jag1, Tnf, Pdgfb, Tgfb3, Cxcl2, Furin, Il1b, Tgfb1, Hif1a, Thbs1, Vegfb, Hmgb1, Ptges2, and Disp1.
5-OP-RU preparation
5-A-RU was synthesized as described (7, 76). For preparation of 5-OP-RU, 5-A-RU was incubated with a fourfold excess of methylglyoxal (Sigma-Aldrich) for 10 min at room temperature.
In vitro validation of the Nr4a1-eGFP mouse
Splenic T cells from a Tg Nr4a1-eGFP mouse were isolated by negative cell enrichment using the Magnisort mouse T cell enrichment kit (Thermo Fisher Scientific). Isolated cells were cultured at a density of 20 × 106/ml in complete RPMI 1640 and stimulated with either recombinant mouse IL-12 (10 ng/ml) and IL-18 (12.5 ng/ml) or with 10 nM 5-OP-RU in the presence of the murine embryonic fibroblast cell line WT3 engineered to overexpress MR1 (25) (WT3-MR1 cells, 5 × 106/ml) for 16 hours at 37°C. GolgiPlug was added during the final 6 hours of culture. MAIT cells were then analyzed for Nr4a1-GFP expression and IFN-γ production.
In vitro detection of MAIT ligands
To detect intestinal MAIT ligands, 100 mg of luminal contents was collected from the cecum or colon. Feces were collected from cage floors immediately after defecation. Samples were weighed, dissolved in complete RPMI 1640 medium (RMPI without folic acid supplemented with 10% FCS, penicillin, and streptomycin), and kept on ice. Tubes were vortexed to homogenize the mixture and centrifuged at maximum speed for 1 min at 4°C. Supernatants were filtered through a 0.22-μm–pore-size filter and loaded at different dilutions on 105 WT3-MR1 cells for 2 hours and 30 min at 37°C. WT3-MR1 cells, a mouse embryonic fibroblast cell line engineered to overexpress MR1, have been described elsewhere (25). WT3-MR1 cells were then washed and cocultured overnight at a 1:1 ratio with total splenocytes from a Vα19 TCR Tg Cα−/− or Vα19Vβ8 TCR Tg Cα−/− mouse. TCR Tg splenocytes were also incubated with unloaded WT3-MR1 cells to measure background activation. Cells were then harvested and stained for CD25 and CD69 expression on 5-OP-RU:MR1–specific T cells. The resulting activation was internally normalized to the unloaded condition (unloaded WT-MR1 cells). In each experiment, MAIT ligand concentrations were estimated using a standard curve prepared with serial dilutions of synthetic 5-OP-RU. For comparing lumen and mucosal samples, 50 mg of luminal contents and 50 mg of mucosal scrapings were sampled by scraping biomass from intestinal walls with a sterile scalpel. Samples were then processed as described for luminal contents. For comparing 2-week-old and 3-month-old samples, 15 mg of colon contents was harvested from both groups and processed as described above for luminal contents.
Intestinal permeability to 5-OP-RU
Seventy-five microliters of PBS containing 12 nmol of OVA 323-339 peptide (Invivogen) mixed with the indicated amount of 5-OP-RU (0.1 to 100 pmol) was introduced into the lumens of freshly isolated colons from healthy mice using a p200 pipette. Colon segments were then immersed in complete RPMI 1640 medium at 37°C in a well on top of mixed MutuDC (44) and WT3-MR1 cells for OVA and 5-OP-RU presentation, respectively. As a positive control, ligand solutions were also added directly on top of mixed antigen-presenting cells. Colon segments were removed from the wells after 10 min of incubation. For the measurement of antigen presentation, splenocytes from a Vα19 TCR Tg Cα−/− mouse and from an OTII-Rag2−/− mouse were cultured overnight with antigen-presenting cells at, respectively, 1:1 and 1:10 ratios. 5-OP-RU–specific and Ova-specific splenocytes were identified as MR1:5-OP-RU tetramer+ and Vα2+ T cells, respectively, and analyzed for surface expression of CD25 and CD69.
Intestinal sample collection for nucleic acid extraction
Immediately after euthanasia, luminal contents and mucosal scrapings were collected from the cecum. Cecal contents were resuspended in RLT lysis buffer (Qiagen) supplemented with acid-washed glass beads (Sigma-Aldrich). Bacteria were lysed using a Fast Prep bead beater (MP Biomedicals) (three times 1 min at 6.5 m/s). Total DNA and RNA were purified using the AllPrep DNA/RNA Kit (Qiagen). On-column DNA digestion was performed before RNA elution.
16S rRNA gene PCR amplification and sequencing
16S rRNA gene sequencing was performed in house or at the University of Minnesota Genomics Center with similar results. In-house library preparation was performed as described (77). Briefly, a single polymerase chain reaction (PCR) step was used for amplification of the V3-V4 region of the 16S rRNA gene and for dual-index labeling of each sample. PCR amplification was performed using the Accuprime Pfx DNA polymerase (Thermo Fisher Scientific). PCR products were purified (AMPure XP beads, Beckman Coulter) and sequenced on MiSeq (Illumina, USA) to generate paired-end 300-bp reads.
To study the effect of Mr1 on the microbiota at steady state, the V3-V4 region was amplified (using the Accuprime Pfx DNA polymerase) from DNA isolated from the feces of cohoused Mr1+/− and Mr1−/− B6-MAITCast mice. PCR products were purified on agarose gels (1.5%) using the Qiaquick gel extraction kit (Qiagen). After gel extraction, sequencing was performed on MiSeq (Illumina, USA) to generate paired-end 300-bp reads.
Bioinformatic analysis of 16S rRNA-seq data
Adapter sequences were trimmed from the raw fastq files using Cutadapt v 2.10 (78). Reads were further corrected for known sequencing error using SPAdes (v3.14.1 – PMID: 32559359) and then merged using PEAR (v0.9.11 – 24142950). Amplicon sequence variants (ASVs) were identified using a Vsearch pipeline (v2.21.1 – 27781170) designed to dereplicate (--derep_prefix –minuquesize 2) and cluster (--unoise3) the merged reads and to check for chimeras (uchime3_denovo). Taxonomic classification of ASVs was performed using the classifier from the RDPTools suite (v2.13 – 24288368). Representative ASV sequences were taxonomically assigned using the RDP classifier with an SAB score ≥ 0.5. Downstream analyses and visualization were performed in the R environment v 4.0.4 (R Core Team 2019). Metabolic pathway abundance in each sample was predicted using PICRUSt2 (79), and differentially abundant pathways were identified using LEfSe (80).
Metatranscriptomic and metagenomic data generation
For metatranscriptomic analyses, total RNA was prepared from the cecum mucosa and lumens of six nontreated mice and from the cecum lumens of eight untreated mice and eight mice treated with 2.5% DSS for 7 days followed by 2 days on water. DNA was also isolated from the cecum lumens of the same mock- and DSS-treated mice for paired metagenomic analyses (see below for metagenomic library preparation). Sequencing libraries were prepared from DNase-treated total RNA using the Illumina Stranded Total RNA Prep Ligation with Ribo-Zero Plus kit. Indexed libraries were sequenced (SR100) on a NovaSeq (Illumina). A total of 23 to 80 million reads (mean 54.4 ± 15.3 million) were obtained for each library.
For metagenomic analyses, sequencing libraries were prepared from cecum lumen DNA using the KAPA HyperPrep Library PCR-Free kit (Roche). Indexed libraries were sequenced (SR100) on a NovaSeq (Illumina). A total of 5.4 to 71 million reads (mean 42 ± 22 million) were obtained for each library.
Metatranscriptomic and metagenomic data analysis
Single-end reads were quality checked and trimmed using TrimGalore v0.6.6 (https://github.com/FelixKrueger/TrimGalore). Host reads were identified by aligning metatranscriptomes and metagenomes against the mouse genome (http://ftp.ensembl.org/pub/release-102/fasta/mus_musculus/dna/Mus_musculus.GRCm38.dna.primary_assembly.fa.gz) using STAR v2.7.7 (81). Ribosomal reads were removed from metatranscriptomic datasets using sortmerna v4.2.0 (82). Last, the remaining nonhost nonribosomal reads were mapped against the iMGMC mouse gene catalog (83) using bwa v0.7.17-r1188 with default parameters. Read counts aligned to the iMGMC gene catalog were obtained from bam files processed with featureCounts v2.0.1 (84). Only genes with at least 50 total reads were selected for downstream analyses. For analysis of microbial gene expression, reads were binned by KEGG categories at the level of metabolic pathways or at the level of genes. The binned features (either pathways or genes) were then used as input in DESeq2 (85) to identify differentially abundant features, which were displayed in heatmaps using the pheatmap R package v 1.0.12. The same analysis pipeline was applied to the published metatranscriptomic dataset (42) stored under the accession number E-MTAB-4082 and downloaded from EBI ENA. To predict which mouse cells were present in the cecum during DSS treatment, the top 200 mouse transcripts with increased abundance in the ceca of DSS-treated mice were used as input in the Immgen Microarray v1.
Estimation of bacterial number in intestinal samples
Total bacterial numbers from intestinal samples were estimated by TaqMan quantitative PCR (qPCR) quantification of bacterial 16S rRNA (LightCycler 480 Instrument II, Roche). The universal 16S rRNA gene primers and probe sequences used in this study were as follows (86): 16S Forward: 5′TGGAGCATGTGGTTTAATTCGA3′. 16S Reverse: 5′TGCGGGACTTAACCCAACA3′. Probe: 5′CACGAG CTGACGACA(AG)CCATGCA3′. A standard curve was built using E. coli MG1655 DNA to convert the threshold cycle (Ct) values into colony-forming unit equivalents.
SCFA analysis
Short-chain fatty acid (SCFA) analysis was carried out as described previously (87), with slight modifications. Samples were water-extracted, and proteins were precipitated with phosphotungstic acid. A volume of 0.3 μl of the supernatant fraction was injected into a gas-liquid chromatograph (Agilent 6890; Les Ulis, France) equipped with a split-splitless injector, a flame-ionization detector, and a capillary polyethylene glycol column (15 m by 0.53 mm, 0.5 μm). The carrier gas (H2) flow rate was 10 ml/min, and inlet, column, and detector temperatures were 200°, 100°, and 240°C, respectively. Between each sample injection, the column was cleaned by increasing the temperature from 100° to 180°C (20°C/min) followed by holding at 180°C for 2 min before returning to 100°C for the next analysis. 2-Ethylbutyrate was used as an internal standard. Samples were analyzed in duplicate. Data were collected and peaks integrated with the OpenLab CDS Chemstation Edition software (Agilent, Les Ulis, France).
Riboflavin dosage
Cecum contents from eight untreated mice and seven mice treated with 2.5% DSS for 7 days were harvested, weighed, and dissolved in water. After centrifugation (14,000 rpm, 5 min, 4°C), supernatants were passed through a 0.22-μm filter and analyzed by reverse-phase high-performance liquid chromatography following a method adapted from Lopez-Anaya et al. (88). Briefly, samples were diluted 1:20 in distilled water and then 1:2 in acetonitrile. Diluted samples were vortexed for 1 min followed by centrifugation (4000 rpm, 10 min, 12°C). Supernatants were transferred into glass tubes and mixed with 2 ml of chloroform followed by vortexing for 1 min. Samples were centrifugated (4000 rpm, 10 min, 12°C), and 20 ml of supernatant was injected on a Uptisphere C18 ODB-15QK 3 m 4.6 by 150 mm column (Interchim) with isocratic elution mode at 1 ml/min. The mobile phase contained 700 ml of buffer (1.35 g of potassium phosphate and 1 g of ammonium acetate in 700 ml of water) and 300 ml of acetonitrile (potassium phosphate 10 mM and ammonium acetate 12.3 mM). Riboflavin was detected by fluorescence after excitation at 445 nm and detection at 530 nm. The retention time of riboflavin was about 8 min.
Detection of intestinal hypoxia
PMDZ (Sigma-Aldrich) was administered at 60 mg/kg by intraperitoneal injection 1 hour before euthanasia. Colon samples were fixed in formalin solution 10% neutral buffer (Sigma-Aldrich). Paraffin-embedded sections were blocked with protein block (Dako, X0909) and treated with avidin/biotin blocking reagents (Dako, X0590) followed by overnight incubation at 4°C with either biotinylated or purified mouse monoclonal anti-PMDZ antibody (Hypoxyprobe-1) at 1.2 μg/ml according to the manufacturer’s instructions. Sections stained with purified anti-PMDZ primary antibody were then incubated for 30 min with biotinylated anti-mouse immunoglobulin G secondary antibody (Vectastain). Negative control sections were treated identically except that primary antibody was substituted with control antibody (BD) for overnight incubation. All sections were then incubated for 30 min with Alexa546-conjugated streptavidin (Thermo Fisher Scientific). Tissues were counterstained with DAPI. PMDZ staining was quantified using Fiji and plotted as fluorescence intensity per 400 μm2.
Individual bacterial cultures
All bacterial strains were first grown anaerobically (5% H2, 5% CO2, 90% N2). C. difficile 630Δerm (89), C. perfringens SM101 (90), and E. coli MG1655 (91) were grown in brain heart infusion (BHI, Difco), and B. thetaiotaomicron VPI-5482 (92) (collection of S. Rabot, INRAE, Jouy en Josas, France) was grown in M17 (Difco) supplemented with glucose 0.5% and hemin 5 μM (Sigma-Aldrich). An overnight liquid preculture was diluted in 10 ml of fresh medium (1/1000 for E. coli and C. perfringens and 1/50 for the other strains) and grown anaerobically until late exponential growth phase. After 3 hours for E. coli and C. perfringens, 8 hours for B. thetaiotaomicron, and 6 hours for C. difficile, cultures were distributed in 24-well plates, 1 ml per well, and placed in air at 37°C. Culture supernatants were harvested after 0 and 60 min of air exposure and fast-frozen in liquid nitrogen. MAIT ligands were quantified in 0.22-μm filtered supernatants using the MAIT ligand quantification bioassay.
Assessment of intestinal pathophysiology
In chronic DSS experiments, the spleen and colon were isolated from each mouse. Individual spleen tissues were weighed. The entire colon was fixed in formalin solution 10% neutral buffer, embedded in paraffin, and stained with hematoxylin and eosin (H&E). H&E-stained sections of the colon were analyzed for colitis severity in an investigator-blinded manner. Epithelial damage (architectural lesion), inflammatory cell infiltration, and crypt length were scored using the histopathological scoring system described previously (93).
Statistical analyses
Statistical analyses and graphical representations related to the microbiome pipeline were conducted in R (v 4.0.4) (R Core Team 2019) with the R package ggplot2 v 3.2.0. All other statistical analyses were performed with Prism software (GraphPad). Two-tailed P values were determined using Wilcoxon’s and Mann-Whitney’s tests for paired and nonpaired samples, as appropriate. A false discovery rate of 1% was calculated for multiple t tests and multiple Mann-Whitney’s tests using the two-stage step-up method of Benjamini, Krieger, and Yekutieli.
Supplementary Material
Acknowledgments
We thank M. Garcia and V. Dangles-Marie, the mouse facility technicians, and the flow cytometry core at Institut Curie. We thank F. El Marjou and C. Jouhanneau for the generation of the LSL-MR1 transgenic mouse and S. Robine for the villin-CreER mouse. We thank the ICGex NGS platform of the Institut Curie for technical help with single-cell RNA-seq and metagenomic experiments. ICGex is supported by the grants ANR10EQPX03 (Equipex) and ANR10INBS0908 (France Génomique Consortium) from the Agence Nationale de la Recherche (‘Investissements d’Avenir’ program), by the ITMO-Cancer Aviesann (Plan Cancer III), and by the SiRIC-Curie program (INCa-DGOS-465 and INCa-DGOS-Inserm_12554). We are grateful to the INRAE MIGALE bioinformatics facility for providing help with computing and storage resources. We thank D. Halpern (INRAE, Jouy-en-Josas, France) for expertise and technical help with B. thetaiotaomicron. We thank J.J. Moon (Massachusetts General Hospital) for providing the LSL-MR1 backbone. We thank N. Manel and B. Chassaing for critical reading of the manuscript. We thank the NIH tetramer core facility (Emory University) for providing CD1d and MR1 tetramers. The MR1:5-OP-RU tetramer technology was developed jointly by J. McCluskey, J. Rossjohn, and D. Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne. The sequencing data generated in this study are available on the EBI ENA through the accession number PRJEB58263.
Funding
This study was funded by the Institut National de la Santé et de la Recherche Médicale (O.L. and F.L.), Institut Curie (O.L.), Agence Nationale de la Recherche [JCJCANR-19-CE15-0002-01 MAIT (F.L.), MicrobMAIT ANR-23-CE15-0036-01 (F.L.), MAIT ANR-16-CE15-0020-01 (O.L.), diabMAIT ANR-17-CE14-0002-02 (O.L.), MAITrepair ANR-20-CE15-0028-01 (O.L.), ANR-10-IDEX-0001-02 PSL (O.L.)], European Research Council (ERC-2019-AdG-885435) (O.L.), Chaire de recherche from Rennes Métropole (22C0451) (F.L.), and Fondation pour la Recherche Médicale (FDT202304016434) (M.F.).
Footnotes
Author contributions: Y.E.M., M.F., K.F., C.M., T.D., M.S., V.P., L.C., S.R., and F.L. conducted the experiments with the assistance of L.P., H.B., A.D., B.J., C.P., E.P., and A.d.H. L.C. and R.R. provided 5-OP-RU. M. Mestdagh, M. Maurin, H.B., and A.V. conducted computational analysis with the assistance of F.L. F.L. wrote the paper with input from all other authors. F.L. and O.L. supervised the work.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability
16S rRNA amplicon sequencing data, metatranscriptomic, metagenomic, and single-cell RNA-seq data are available on the EBI ENA under the accession number PRJEB58263. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Raw data are provided in data file S1. Mouse strains and cell lines can be provided upon signing a material transfer agreement with O.L. and F.L.
References and Notes
- 1.Lee J-Y, Tsolis RM, Bäumler AJ. The microbiome and gut homeostasis. Science. 2022;377:eabp9960. doi: 10.1126/science.abp9960. [DOI] [PubMed] [Google Scholar]
- 2.Litvak Y, Byndloss MX, Bäumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018;362:eaat9076. doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byndloss MX, Bäumler AJ. The germ-organ theory of non-communicable diseases. Nat Rev Microbiol. 2018;16:103–110. doi: 10.1038/nrmicro.2017.158. [DOI] [PubMed] [Google Scholar]
- 4.Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, Milder M, Le Bourhis L, Soudais C, Treiner E, Lantz O. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 5.Legoux F, Salou M, Lantz O. MAIT cell development and functions: The microbial connection. Immunity. 2020;53:710–723. doi: 10.1016/j.immuni.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Corbett AJ, Eckle SBG, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, Williamson NA, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 7.Soudais C, Samassa F, Sarkis M, Le Bourhis L, Bessoles S, Blanot D, Hervé M, Schmidt F, Mengin-Lecreulx D, Lantz O. In vitro and in vivo analysis of the Gram-negative bacteria-derived riboflavin precursor derivatives activating mouse MAIT cells. J Immunol. 2015;194:4641–4649. doi: 10.4049/jimmunol.1403224. [DOI] [PubMed] [Google Scholar]
- 8.Tastan C, Karhan E, Zhou W, Fleming E, Voigt AY, Yao X, Wang L, Horne M, Placek L, Kozhaya L, Oh J, et al. Tuning of human MAIT cell activation by commensal bacteria species and MR1-dependent T-cell presentation. Mucosal Immunol. 2018;11:1591–1605. doi: 10.1038/s41385-018-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HJ, Winge DR. Emerging concepts in the flavinylation of succinate dehydrogenase. Biochim Biophys Acta. 2013;1827:627–636. doi: 10.1016/j.bbabio.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan MT, Duncan SH, Stams AJM, van Dijl JM, Flint HJ, Harmsen HJM. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 2012;6:1578–1585. doi: 10.1038/ismej.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serriari N-E, Eoche M, Lamotte L, Lion J, Fumery M, Marcelo P, Chatelain D, Barre A, Nguyen-Khac E, Lantz O, Dupas J-L, et al. Innate mucosal-associated invariant T (MAIT) cells are activated in inflammatory bowel diseases. Clin Exp Immunol. 2014;176:266–274. doi: 10.1111/cei.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haga K, Chiba A, Shibuya T, Osada T, Ishikawa D, Kodani T, Nomura O, Watanabe S, Miyake S. MAIT cells are activated and accumulated in the inflamed mucosa of ulcerative colitis. J Gastroenterol Hepatol. 2016;31:965–972. doi: 10.1111/jgh.13242. [DOI] [PubMed] [Google Scholar]
- 13.Ling L, Lin Y, Zheng W, Hong S, Tang X, Zhao P, Li M, Ni J, Li C, Wang L, Jiang Y. Circulating and tumor-infiltrating mucosal associated invariant T (MAIT) cells in colorectal cancer patients. Sci Rep. 2016;6:20358. doi: 10.1038/srep20358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zabijak L, Attencourt C, Guignant C, Chatelain D, Marcelo P, Marolleau J-P, Treiner E. Increased tumor infiltration by mucosal-associated invariant T cells correlates with poor survival in colorectal cancer patients. Cancer Immunol Immunother. 2015;64:1601–1608. doi: 10.1007/s00262-015-1764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Simoni Y, Becht E, Loh CY, Li N, Lachance D, Koo S-L, Lim TP, Tan EKW, Mathew R, Nguyen A, et al. Human tumor-infiltrating MAIT cells display hallmarks of bacterial antigen recognition in colorectal cancer. Cell Rep Med. 2020;1:100039. doi: 10.1016/j.xcrm.2020.100039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, Su L, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 19.Leng T, Akther HD, Hackstein C-P, Powell K, King T, Friedrich M, Christoforidou Z, McCuaig S, Neyazi M, Arancibia-Cárcamo CV, Hagel J, et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. 2019;28:3077–3091.:e5. doi: 10.1016/j.celrep.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamichhane R, Schneider M, de la Harpe SM, Harrop TWR, Hannaway RF, Dearden PK, Kirman JR, Tyndall JDA, Vernall AJ, Ussher JE. TCR- or cytokine-activated CD8+ mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep. 2019;28:3061–3076.:e5. doi: 10.1016/j.celrep.2019.08.054. [DOI] [PubMed] [Google Scholar]
- 21.Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, Meehan BS, Kostenko L, Turner SJ, Corbett AJ, Chen Z, et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. 2019;28:3249–3262.:e5. doi: 10.1016/j.celrep.2019.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han S-J, Chen YE, Li K, Farhat S, Weckel A, Krishnamurthy SR, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019;366:eaax6624. doi: 10.1126/science.aax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.du Halgouet A, Darbois A, Alkobtawi M, Mestdagh M, Alphonse A, Premel V, Yvorra T, Colombeau L, Rodriguez R, Zaiss D, El Morr Y, et al. Role of MR1-driven signals and amphiregulin on the recruitment and repair function of MAIT cells during skin wound healing. Immunity. 2023;56:78–92.:e6. doi: 10.1016/j.immuni.2022.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, Premel V, Devys A, Moura IC, Tilloy F, Cherif S, et al. Stepwise development of MAIT cells in mouse and human. PLOS Biol. 2009;7:e54. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S, Martin E, Kim S, Yu L, Soudais C, Fremont DH, Lantz O, Hansen TH. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci USA. 2009;106:8290–8295. doi: 10.1073/pnas.0903196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Bourhis L, Martin E, Péguille I, Guihot A, Froux N, Coré M, Lévy E, Dusseaux M, Meyssonnier V, Premel V, Ngo C, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11:701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 27.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, Thom SR, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063.:e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaborin A, Penalver Bernabe B, Keskey R, Sangwan N, Hyoju S, Gottel N, Gilbert JA, Zaborina O, Alverdy JC. Spatial compartmentalization of the microbiome between the lumen and crypts is lost in the murine cecum following the process of surgery, including overnight fasting and exposure to antibiotics. mSystems. 2020;5:e00377–e00320. doi: 10.1128/mSystems.00377-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loy A, Pfann C, Steinberger M, Hanson B, Herp S, Brugiroux S, Gomes Neto JC, Boekschoten MV, Schwab C, Urich T, Ramer-Tait AE, et al. Lifestyle and horizontal gene transfer-mediated evolution of Mucispirillum schaedleri, a core member of the murine gut microbiota. mSystems. 2017;2:e00171–16. doi: 10.1128/mSystems.00171-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes KR, Schofield Z, Dalby MJ, Caim S, Chalklen L, Bernuzzi F, Alcon-Giner C, Le Gall G, Watson AJM, Hall LJ. The early life microbiota protects neonatal mice from pathological small intestinal epithelial cell shedding. FASEB J. 2020;34:7075–7088. doi: 10.1096/fj.202000042R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035s–1045s. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 34.Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Bäumler AJ. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmaler M, Colone A, Spagnuolo J, Zimmermann M, Lepore M, Kalinichenko A, Bhatia S, Cottier F, Rutishauser T, Pavelka N, Egli A, et al. Modulation of bacterial metabolism by the microenvironment controls MAIT cell stimulation. Mucosal Immunol. 2018;11:1060–1070. doi: 10.1038/s41385-018-0020-9. [DOI] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigottier-Gois L. Dysbiosis in inflammatory bowel diseases: The oxygen hypothesis. ISME J. 2013;7:1256–1261. doi: 10.1038/ismej.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimatsu H, Yonezawa A, Yamanishi K, Yao Y, Sugano K, Nakagawa S, Imai S, Omura T, Nakagawa T, Yano I, Masuda S, et al. Disruption of Slc52a3 gene causes neonatal lethality with riboflavin deficiency in mice. Sci Rep. 2016;6:27557. doi: 10.1038/srep27557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, Artis D, Bushman F. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect Immun. 2009;77:4668–4678. doi: 10.1128/IAI.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry D, Schwab C, Milinovich G, Reichert J, Ben Mahfoudh K, Decker T, Engel M, Hai B, Hainzl E, Heider S, Kenner L, et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012;6:2091–2106. doi: 10.1038/ismej.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ilott NE, Bollrath J, Danne C, Schiering C, Shale M, Adelmann K, Krausgruber T, Sims D, Powrie F. Defining the microbial transcriptional response to colitis through integrated host and microbiome profiling. ISME J. 2016;10:2389–2404. doi: 10.1038/ismej.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, Procopio E, Salou M, Gilet J, Ryffel B, Balvay A, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. 2019;366:494–499. doi: 10.1126/science.aaw2719. [DOI] [PubMed] [Google Scholar]
- 44.Fuertes Marraco SA, Grosjean F, Duval A, Rosa M, Lavanchy C, Ashok D, Haller S, Otten LA, Steiner Q-G, Descombes P, Luber CA, et al. Novel murine dendritic cell lines: A powerful auxiliary tool for dendritic cell research. Front Immunol. 2012;3:331. doi: 10.3389/fimmu.2012.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui Y, Franciszkiewicz K, Mburu YK, Mondot S, Le Bourhis L, Premel V, Martin E, Kachaner A, Duban L, Ingersoll MA, Rabot S, et al. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J Clin Invest. 2015;125:4171–4185. doi: 10.1172/JCI82424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zikherman J, Parameswaran R, Weiss A. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature. 2012;489:160–164. doi: 10.1038/nature11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monticelli LA, Osborne LC, Noti M, Tran SV, Zaiss DMW, Artis D. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc Natl Acad Sci USA. 2015;112:10762–10767. doi: 10.1073/pnas.1509070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.el Marjou F, Janssen K-P, Chang BH-J, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 49.Wirtz S, Popp V, Kindermann M, Gerlach K, Weigmann B, Fichtner-Feigl S, Neurath MF. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12:1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- 50.Klose CSN, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, Göppert N, Croxford AL, Waisman A, Tanriver Y, Diefenbach A. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 51.Liang SC, Tan X-Y, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varelias A, Bunting MD, Ormerod KL, Koyama M, Olver SD, Straube J, Kuns RD, Robb RJ, Henden AS, Cooper L, Lachner N, et al. Recipient mucosal-associated invariant T cells control GVHD within the colon. J Clin Invest. 2018;128:1919–1936. doi: 10.1172/JCI91646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith AD, Foss ED, Zhang I, Hastie JL, Giordano NP, Gasparyan L, VinhNguyen LP, Schubert AM, Prasad D, McMichael HL, Sun J, et al. Microbiota of MR1 deficient mice confer resistance against Clostridium difficile infection. PLOS ONE. 2019;14:e0223025. doi: 10.1371/journal.pone.0223025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JS, Tato CM, Joyce-Shaikh B, Gulen MF, Cayatte C, Chen Y, Blumenschein WM, Judo M, Ayanoglu G, McClanahan TK, Li X, et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity. 2015;43:727–738. doi: 10.1016/j.immuni.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song X, Dai D, He X, Zhu S, Yao Y, Gao H, Wang J, Qu F, Qiu J, Wang H, Li X, et al. Growth factor FGF2 cooperates with interleukin-17 to repair intestinal epithelial damage. Immunity. 2015;43:488–501. doi: 10.1016/j.immuni.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 56.Maxwell JR, Zhang Y, Brown WA, Smith CL, Byrne FR, Fiorino M, Stevens E, Bigler J, Davis JA, Rottman JB, Budelsky AL, et al. Differential roles for interleukin-23 and interleukin-17 in intestinal immunoregulation. Immunity. 2015;43:739–750. doi: 10.1016/j.immuni.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 57.Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Linehan JL, Harrison OJ, Han S-J, Byrd AL, Vujkovic-Cvijin I, Villarino AV, Sen SK, Shaik J, Smelkinson M, Tamoutounour S, Collins N, et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. 2018;172:784–796.:e18. doi: 10.1016/j.cell.2017.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karhausen J, Furuta GT, Tomaszewski JE, Johnson RS, Colgan SP, Haase VH. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Invest. 2004;114:1098–1106. doi: 10.1172/JCI21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pesu M, Watford WT, Wei L, Xu L, Fuss I, Strober W, Andersson J, Shevach EM, Quezado M, Bouladoux N, Roebroek A, et al. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature. 2008;455:246–250. doi: 10.1038/nature07210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mondot S, Boudinot P, Lantz O. MAIT, MR1, microbes and riboflavin: A paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules? Immunogenetics. 2016;68:537–548. doi: 10.1007/s00251-016-0927-9. [DOI] [PubMed] [Google Scholar]
- 62.Caruso R, Mathes T, Martens EC, Kamada N, Nusrat A, Inohara N, Núñez G. A specific gene-microbe interaction drives the development of Crohn’s disease-like colitis in mice. Sci Immunol. 2019;4:eaaw4341. doi: 10.1126/sciimmunol.aaw4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herp S, Brugiroux S, Garzetti D, Ring D, Jochum LM, Beutler M, Eberl C, Hussain S, Walter S, Gerlach RG, Ruscheweyh HJ, et al. Mucispirillum schaedleri antagonizes salmonella virulence to protect mice against colitis. Cell Host Microbe. 2019;25:681–694.:e8. doi: 10.1016/j.chom.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 64.Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, Richer W, Goubet A-G, Daviaud C, Menger L, Procopio E, et al. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med. 2019;216:133–151. doi: 10.1084/jem.20181483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ataide MA, Knöpper K, Cruz de Casas P, Ugur M, Eickhoff S, Zou M, Shaikh H, Trivedi A, Grafen A, Yang T, Prinz I, et al. Lymphatic migration of unconventional T cells promotes site-specific immunity in distinct lymph nodes. Immunity. 2022;55:1813–1828.:e9. doi: 10.1016/j.immuni.2022.07.019. [DOI] [PubMed] [Google Scholar]
- 66.Haga K, Chiba A, Shibuya T, Osada T, Ishikawa D, Kodani T, Nomura O, Watanabe S, Miyake S. MAIT cells are activated and accumulated in the inflamed mucosa of ulcerative colitis. J Gastroenterol Hepatol. 2016;31:965–972. doi: 10.1111/jgh.13242. [DOI] [PubMed] [Google Scholar]
- 67.Tominaga K, Yamagiwa S, Setsu T, Kimura N, Honda H, Kamimura H, Honda Y, Takamura M, Yokoyama J, Suzuki K, Wakai T, et al. Possible involvement of mucosal-associated invariant T cells in the progression of inflammatory bowel diseases. Biomed Res. 2017;38:111–121. doi: 10.2220/biomedres.38.111. [DOI] [PubMed] [Google Scholar]
- 68.Rouxel O, Da Silva J, Beaudoin L, Nel I, Tard C, Cagninacci L, Kiaf B, Oshima M, Diedisheim M, Salou M, Corbett A, et al. Cytotoxic and regulatory roles of mucosal-associated invariant T cells in type 1 diabetes. Nat Immunol. 2017;18:1321–1331. doi: 10.1038/ni.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yasutomi Y, Chiba A, Haga K, Murayama G, Makiyama A, Kuga T, Watanabe M, Okamoto R, Nagahara A, Nagaishi T, Miyake S. Activated mucosal-associated invariant T cells have a pathogenic role in a murine model of inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2022;13:81–93. doi: 10.1016/j.jcmgh.2021.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 71.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Z, Legoux FP, Vaughan SW, Moon JJ. Opposing peripheral fates of tissue-restricted self antigen-specific conventional and regulatory CD4+ T cells. Eur J Immunol. 2020;50:63–72. doi: 10.1002/eji.201948180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8-dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanaka T, Kohno H, Suzuki R, Yamada Y, Sugie S, Mori H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 2003;94:965–973. doi: 10.1111/j.1349-7006.2003.tb01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SBG, Meehan B, Chen Z, Whittle B, Liu L, Fairlie DP, Goodnow CC, et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med. 2015;212:1095–1108. doi: 10.1084/jem.20142110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yvorra T, Steinmetz A, Retailleau P, Lantz O, Schmidt F. Synthesis, biological evaluation and molecular modelling of new potent clickable analogues of 5-OP-RU for their use as chemical probes for the study of MAIT cell biology. Eur J Med Chem. 2021;211:113066. doi: 10.1016/j.ejmech.2020.113066. [DOI] [PubMed] [Google Scholar]
- 77.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. [Google Scholar]
- 79.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kopylova E, Noé L, Touzet H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 83.Lesker TR, Durairaj AC, Gálvez EJC, Lagkouvardos I, Baines JF, Clavel T, Sczyrba A, McHardy AC, Strowig T. An integrated metagenome catalog reveals new insights into the murine gut microbiome. Cell Rep. 2020;30:2909–2922.:e6. doi: 10.1016/j.celrep.2020.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 85.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nonnenmacher C, Dalpke A, Mutters R, Heeg K. Quantitative detection of periodontopathogens by real-time PCR. J Microbiol Methods. 2004;59:117–125. doi: 10.1016/j.mimet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 87.Lan A, Bruneau A, Philippe C, Rochet V, Rouault A, Hervé C, Roland N, Rabot S, Jan G. Survival and metabolic activity of selected strains of Propionibacterium freudenreichii in the gastrointestinal tract of human microbiota-associated rats. Br J Nutr. 2007;97:714–724. doi: 10.1017/S0007114507433001. [DOI] [PubMed] [Google Scholar]
- 88.Lopez-Anaya A, Mayersohn M. Quantification of riboflavin, riboflavin 5′-phosphate and flavin adenine dinucleotide in plasma and urine by high-performance liquid chromatography. J Chromatogr. 1987;423:105–113. doi: 10.1016/0378-4347(87)80332-8. [DOI] [PubMed] [Google Scholar]
- 89.Hussain HA, Roberts AP, Mullany P. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J Med Microbiol. 2005;54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
- 90.Zhao Y, Melville SB. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens . J Bacteriol. 1998;180:136–142. doi: 10.1128/jb.180.1.136-142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 92.Cato EP, Johnson JL. Reinstatement of species rank for Bacteroides fragilis, B. ovatus, B. distasonis, B. thetaiotaomicron, and B. vulgatus: Designation of neotype strains for Bacteroides fragilis (Veillon and Zuber) Castellani and Chalmers and Bacteroides thetaiotaomicron (Distaso) Castellani and Chalmers. Int J Syst Bacteriol. 1976;26:230–237. [Google Scholar]
- 93.Laroui H, Ingersoll SA, Liu HC, Baker MT, Ayyadurai S, Charania MA, Laroui F, Yan Y, Sitaraman SV, Merlin D. Dextran sodium sulfate (DSS) induces colitis in mice by forming nano-lipocomplexes with medium-chain-length fatty acids in the colon. PLOS ONE. 2012;7:e32084. doi: 10.1371/journal.pone.0032084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA amplicon sequencing data, metatranscriptomic, metagenomic, and single-cell RNA-seq data are available on the EBI ENA under the accession number PRJEB58263. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Raw data are provided in data file S1. Mouse strains and cell lines can be provided upon signing a material transfer agreement with O.L. and F.L.