Abstract
In the complex realm of bacterial infections, particularly those caused by Staphylococcus aureus (S. aureus), macrophages play a pivotal role in orchestrating the immune response. During the initial stages of infection, the monocytes give rise to macrophages with a pro-inflammatory (M1 type) behaviour, engulfing and neutralizing the invading pathogens. However, under the sustained influence of S. aureus infection, monocytes can undergo a transition into an anti-inflammatory M2 state (pro-infection) rather than the M1 state (anti-infection), thereby compromising effective infection control. Therefore, it is necessary to develop a strategy that would preserve the pro-inflammatory functions of macrophages, in a safe and controlled manner. For this, we focused on harnessing the potential of S. aureus-derived ghost cells (GCs) which are non-live empty envelopes of bacterial cells, but with the antigenic determinants intact. Through a unique Lugol’s-iodine treatment, we generated GCs and characterization of these GCs using gel electrophoresis, FTIR, flow cytometry, TEM, and SEM confirmed their structural integrity. Following this, we assessed the extend of cellular association of the GCs with RAW267.4 macrophages, and observed an immediate interaction between the two, as evident from the flowcytometry and microscopy studies. We then performed macrophage polarisation on a human monocyte-macrophage model cell line, THP-1. Our findings revealed that GCs effectively activated macrophages, and promoted a pro-inflammatory polarisation with the expression of M1 differentiation markers (CD86, TNFα, IL-1β, IL-6, IL-12) evaluated through both qPCR and ELISA. Interestingly an intermediary expression of M2 markers viz., CD206 and IL-10 was also observed, but was over-ruled by the enhanced expression of M1 markers at a later time point. Overall, our study introduces a novel approach utilizing GCs to guide naïve macrophages towards M1 subtypes, thereby potentiating immune responses during microbial infections. This innovative strategy can modulate macrophage function, ultimately improving outcomes in S. aureus infections and beyond.
Keywords: S. aureus, Macrophages, Inflammation, Cytokines, Bacterial ghosts, Infection
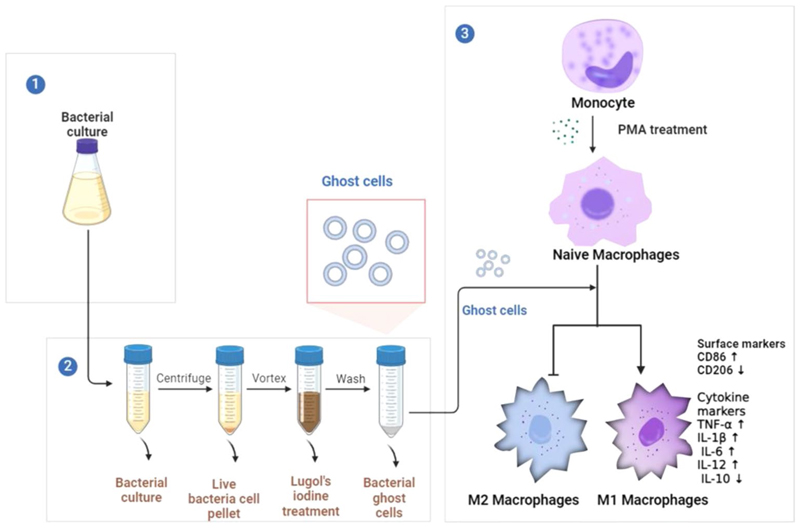
Graphical Abstract.
The present study devised a rapid Lugol’s iodine-based protocol to obtain structurally intact bacterial ghost cells (GCs), which effectively induced pro-inflammatory macrophage polarization, upregulating M1 markers and cytokines, while downregulating IL-10, an anti-inflammatory cytokine. This suggested a promising therapeutic strategy for combatting bacterial infections via modulating macrophage immune responses
Introduction
Macrophages, are a class of versatile immune cells that exhibit remarkable plasticity, transitioning between distinct functional phenotypes in response to cues from their microenvironment. The classical pro-inflammatory (M1) and anti-inflammatory (M2) macrophage subtypes represent two ends of this spectrum, each with unique roles in host defence, tissue repair, and immunoregulation (Yunna et al. 2020). The interplay between macrophage sub-types significantly shapes infection outcomes. The pro-inflammatory M1 macrophages, with heightened cytokine secretion and reactive oxygen species (ROS) production, execute robust antimicrobial responses through enhanced phagocytosis, antigen presentation, and adaptive immune activation (Jain et al. 2019). Conversely, the M2 macrophages contribute to tissue repair, but excessive M2 polarization during infection can inadvertently under-mine the ability of the host to combat pathogens, thereby, facilitating pathogen evasion and enhancing the potential for establishment of chronic infection (Ravichandran et al. 2021; Strizova et al. 2023). Macrophages can actively engulf S. aureus in their planktonic state while the biofilms formed by S. aureus avoid conventional recognition by Toll-like receptors (TLRs) and modify the immune response to infection. Therefore, this interaction between S. aureus during the later stages of established infections skews macrophage polarization towards the M2 type. Also, the biofilms formed by S. aureus could attenuate the production of pro-inflammatory mediators which leads to bacterial persistence and macrophage killing (Thurlow et al. 2011). An example for such events happens in chronic rhinosinusitis associated with S. aureus, where, an increased count of the M2 phenotype is observed leading to an impaired immune response with a decreased phagocytic ability (Krysko et al. 2011).
Exploring deliberate modulation of monocytes/naive macrophages to differentiate into the M1 subtype (utilizing cytokines, small molecules, and other biological agents) has lately shown potential as a strategy to fortify the host defence system (Mohamed Elashiry et al. 2021). By exposing macrophages to external stimuli, researchers have attempted to enhance the secretion of cytokines, such as tumor necrosis factor-alpha and interleukin-12, along with heightened production of reactive oxygen species (ROS) and other supportive pro-inflammatory cytokines by the macrophages (Varotto-Boccazzi et al. 2020). This orchestrated shift, primes the macrophages for an intensified antimicrobial response, encompassing augmented phagocytosis, efficient antigen presentation, and activation of adaptive immune pathways.
However, the M1 differentiation strategies are often bothered by their non-specificity, thereby leading to undesirable immune-activation as well as associated immunotoxicity (Sica and Mantovani 2012). At this juncture, it is prudent to evoke the fact that macrophages are notable for their abundant expression of pattern recognition receptors (PRRs) and therefore, using PRR-agonists would be an ideal approach to design macrophage-specific therapies, especially to manifest M1 differentiation. Activation via the PRRs can trigger downstream signalling pathways, leading to the activation of NF-κB, MAPKs, and transcription factors, that drive the expression of pro-inflammatory cytokines like IL-1β, TNF-α, and IL-6, promoting the M1 phenotype (Wang et al. 2014; Li and Wu 2021; Kolliniati et al. 2022).
The present study demonstrates the potential of hollow bacterial structures called bacterial ghosts in polarizing naïve macrophages to the M1 subtype. Bacterial ghosts are structures devoid of the genetic content, while preserving the natural reservoir of PRR-agonists on their surface, thereby equipped with the potential to interact with immune cells, especially the macrophages. In this work, we developed ghost cells of Staphylococcus aureus (S. aureus) (henceforth mentioned as GC) via a rapid technique using Lugol’s iodine, following which the GCs were validated for their size, structure, status of genetic content and surface chemical integrity using electron microscopic and spectroscopic techniques as well as electrophoresis and flow cytometry. We then interrogated the cytocompatibility and cellular association (encompassing both binding and uptake) of the GCs with RAW 264.7 macrophage cells. For elucidating the differentiation of naïve macrophage (M0) we used THP-1-derived M0 and and performed a time-kinetic protein expression analyses of the cytokines viz., TNFα, IL-1β, IL-6, IL-12 and IL-10 using ELISA, while the assessment of gene expression for CD86, CD206, and IL-12 was done through qPCR. By focusing on the capacity of S. aureus GCs to modulate macrophage differentiation towards the M1 subtype, we unveil a potential avenue for precise immunomodulation, and also provide novel insights into the development of cost-effective and scalable therapeutic strategy that harnesses the potential of bacterial ghost cells for infectious diseases.
Materials and methods
Bacteria and cell culture
Nutrient broth (Himedia), Nutrient agar (Himedia), Lugol’s iodine (Sigma), Phosphate buffered saline (Sigma), PMA (Phorbol 12-myristate 13-acetate, Sigma), FBS (Fetal Bovine Serum, Gibco-Invitrogen), RPMI medium (Gibco-Invitrogen), Trypsin (Sigma).
The Staphylococcus aureus strain NCIM 2127 was acquired from the National Collection of Industrial Micro-organisms (NCIM), India and was cultured in nutrient broth maintained at 37 °C within a shaking incubator operating at 200 rpm to ensure sufficient aeration. The bacterial growth was monitored by assessing the optical density at 600 nm (OD600) until it reached a value of 1.0, at which point the culture was deemed suitable for subsequent experimental procedures.
The cell lines viz., RAW 264.7 macrophages and THP-1 monocytes were purchased from the National Centre for Cell Sciences (NCCS), India and the American Type Culture Collection (ATCC), USA respectively and were cultured in RPMI medium supplemented with 2 mM l-glutamine, 2.5 g/L glucose, 1 mM sodium pyruvate, 10 mM HEPES, 0.05 mM 2-mercaptoethanol, 1000/100 mg/mL streptomycin / penicillin, and 10% Fetal Bovine Serum (Invitrogen), at 37 °C and 5% CO2 and maintained at a density between 0.1 and 1 × 106 cells / mL. The cells were fed and subcultured twice a week and they were used before reaching the passage 20 in culture.
GC preparation and viability tests
Staphylococcus aureus was subjected to systematic treatments involving varying concentrations and exposure durations of Lugol’s iodine. The experiment employed a range of Lugol’s iodine concentrations (0.05%, 0.1%, 0.15%, 0.2%, 0.25%, and 0.5%) prepared in Luria-Bertani (LB) broth. A consistent volume of the bacterial culture with an OD = 1, was introduced into individual tubes containing the respective Lugol’s iodine + LB broth solution. Subsequently, the samples were vortexed and incubated at ambient temperature for 5 min. A 100 µL volume of each sample was spread-plated onto nutrient agar plates under sterile conditions and were then subjected to a 24 h incubation. Enumeration of resultant bacterial colonies on the agar plates, conducted post-incubation, facilitated a comprehensive and comparative analysis of bacterial proliferation across varying iodine concentrations and exposure durations. The following equation was used to calculate the percentage of metabolically active cells;
Additionally, a scaling-up procedure was executed and the centrifugally purified samples were lyophilized to remove moisture, allowing subsequent quantification of dry weight. To enhance the yield of GCs, we scaled up the protocol to a 50 ml culture that was subjected to 0.25% iodine for a 5 min incubation period, maintaining a volume ratio of iodine-to-culture at 1:10 v/v. In a separate experimental setup, a similar approach was followed with a 100 ml bacterial culture for both 5 and 10 m ins incubation intervals while maintaining the same iodine-to-culture v/v ratio. Following the incubation, the samples were centrifuged @10,000 rpm at 4 °C to obtain the respective pellets which was resuspended into 1 ml of sterile saline and checked for viability (data provided as supplementary fig. S2d). Post-incubation, thorough removal of iodine was accomplished through multiple centrifugal washes using MilliQ water, resulting in the production of ghost cells displaying minimal residual iodine content.
Characterization of the bacterial ghosts
Field emission scanning electron microscopy (FE-SEM)
To further elucidate the morphological characteristics, both untreated bacterial cells and the GCs were fixed using 1.25% glutaraldehyde at room temperature for 15 min, followed by an overnight incubation at 4 °C. The fixative was then thoroughly removed through repeated washing with double distilled water. To facilitate gradual dehydration, the samples were subjected to an ethanol gradient of 20%, 40%, 60%, 80%, and 100%, for 10 min each at 4 °C. Following this, the samples were loaded onto copper grids, gold sputtered and visualised under FE-SEM (Carl Zeiss - Sigma). The images were further analysed using the software SMARTSEM.
Transmission electron microscope
The GC samples were fixed and dehydrated as mentioned above and were loaded onto copper grids and stained with uranyl acetate and lead citrate. The grids (10 random sections per treatment) were examined using the transmission electron microscope.
Gel electrophoresis and Fourier transform infra-red (FTIR) spectroscopy
To understand the status of the genomic DNA in the GCs, we first extracted the DNA using a commercially available kit following the protocols of the manufacturer (HiMedia, India). The GC pellets and untreated S. aureus cells were processed separately to extract their genomic DNA. The concentration and purity of extracted DNA were determined using a UV spectrophotometer (Nano-drop, USA).
Agarose gel electrophoresis was employed to visually assess the extracted DNA samples. A 1% agarose gel enriched with ethidium bromide was prepared and equivalent volumes of extracted DNA samples obtained from both untreated bacterial cells and GC pellets were meticulously loaded onto the gel matrix. Electrophoretic separation was conducted at a voltage of 100 V for a duration of 10 min, followed by a subsequent voltage of 80 V for 1 h. Visualization of the gel was achieved using a UV gel documentation system and a DNA ladder consisting of 1 kilobase pair (kbp) marker (Takara) was incorporated onto the gel as reference.
FTIR spectroscopic analysis, conducted using the Nicolet™ iS50 FTIR Spectrometer (Thermo Scientific, USA), aimed to assess the chemical composition of the prepared GCs. The analysis encompassed a spectral range from 4000 to 500 cm− 1. To facilitate accurate measurements, the freeze-dried samples were uniformly blended with KBr, serving as the reference material, before the scanning process was initiated. Subsequently, the obtained spectra were smoothened using OMNIC 8.0 software, this smoothing step aids in reducing noise and enhancing the clarity of spectral features.
Fluorescent-tagging of GC
The preparation of fluorescent-GC was achieved by labelling them with Wheat Germ Agglutinin (WGA)-conjugated Alexa Fluor 488 (Invitrogen). Briefly, the purified GCs were resuspended in different concentrations of WGA-Alexa Fluor 488 conjugate (1–5 µg/ml in PBS) and incubated for time points of 1, 2 and 3 h at room temperature, under dark conditions. After the incubation time, the suspension was washed thoroughly and fixed using 4% paraformaldehyde by gently mixing and incubating for 60 min at room temperature or at 4 °C. After fixing the sample, they were washed thoroughly with PBS to remove the fixative and was visualised under a fluorescent microscope (Nikon eclipse Ti 2).
Flow cytometry
To further validate the formation of GCs, the forward-scatter (to understand size difference) and the side-scatter (to understand granularity difference) was measured using flow cytometry (BD FACS Celesta™ Cell Analyzer). For this, the GCs and S. aureus samples were stained with WGA Alexa Fluor 488 as mentioned above and were resuspended in flowcytometry buffer (BD Biosciences, CA, USA), following which the analysis was performed and data were analyzed with a FlowJo™ software (BD Biosciences, CA, USA).
Cytotoxicity
Cytotoxicity was evaluated through the MTT assay (Sigma-Aldrich), with some modifications in the established procedures (Seidl and Zinkernagel 2013). In brief, RAW 264.7 cells (2 × 105 cells/mL) were incubated with varying concentrations of S. aureus or GCs for 24 h at varying dosages (0.5–2.5 µg/ml). After the incubation period, the test medium was removed and the cell bed was rinsed with PBS following which MTT solution (0.1 mg/mL) was added to the wells and incubated for 4 h at 37 °C. After the incubation, the dark purple formazan crystals were dissolved in DMSO and the absorbance was recorded at 570 nm using a plate reader (BioTek, Germany). The experiments were conducted in triplicate and the results were compared with untreated cells to calculate the percentage viability of the cells.
Cellular interactions of GCs with macrophages: flow cytometry and microscopy
RAW 264.7 cells were cultured in a 6-well plate at a seeding density of 5 × 105 cells per well in RPMI medium. After overnight incubation, the cells were incubated with fluorescent labelled GCs for 30 min to evaluate particle interaction. After incubation, the cells were washed with PBS, trypsinised, centrifuged at 1500 rpm for 5 mins and resuspended in flowcytometry buffer. The differences in fluorescence were measured on the flow cytometer capturing 20,000 events per sample. The results were expressed as the percentage of fluorescence-positive cells. Furthermore, to visualize the interactions, RAW 264.7 cells (seeded on an acid-etched coverslips) were incubated with trypan blue-stained GCs for 30 min. Subsequently, the cells were rinsed thrice with PBS and fixed with glutaraldehyde and were subjected to imaging using phase-contrast microscopy. Fluorescent microscopy was performed after similarly incubating the cells with fluorescent-GCs.
Monocyte-macrophage differentiation and GC treatment
THP-1 human monocytic cells were subjected to macrophage differentiation using 100nM PMA for 2 days. The resulting naive macrophages were then allowed a rest period of 3 days after which they were passaged and seeded onto culture plates at a density of 2 × 105 cells/well. These cells were then exposed to a treatment with the GC units at a concentration of 100 ng/ml for a duration of 24 h. After the 24 h incubation period, the wells were washed with PBS and pellets were harvested after trypsinisation followed by centrifugation at 3000 rpm for 10 min at 20 °C, and were then taken for total RNA isolation preceding qPCR.
Cytotoxicity, total RNA isolation and quantitative RT-PCR (qRT-PCR)
A cytotoxicity experiment was carried on the THP-1 cells prior to qRT-PCR experiments. For this, the protocol mentioned above for RAW 264.7 cells was repeated. A dose-dependent study was performed at the rage of 0.1–2.0 µg/ml. Lipopolysaccharide (LPS) treated cells were used as control along with the untreated cells. Total RNA extraction was carried out using the RNeasy plus mini kit (Qiagen, GmbH). Subsequently, complementary DNA (cDNA) synthesis was performed using the SuperScript III reverse transcriptase kit along with random hexamers (Invitrogen, Carlsbad, CA) in a 20 µL total volume. For gene expression analysis, SYBR Green RT-PCR Master Mix from TaKaRa (Japan) was employed, and the quantification of fold change was done through the 2−ΔΔCT method, in a real-time PCR system (Applied Biosystem-7300). All experimental procedures were done in three independent sets of triplicates. The following are the primers used: CD86: F-5′-AGCCTTATCGGAAATGATCCAGT-3′, R-5′-GGCCTTGTAGACACCTTGGT-3′; CD206: F-5’-ACACAAACTGGGGGA AAGGTT-3’, R-5’-TCAAGGAAGGGTCGGATCG-3’; Glyceraldehyde-3-phosphate dehydrogenase (GAPDH): F-5′-CGACCACTTTGTCAAGCTCA − 3′, R-5′-AGGGG AGATTCAGTGTGGTG − 3′. The primers were obtained from Merck, India.
Enzyme-linked immunosorbent assay (ELISA)
The THP-1-derived macrophages were subjected to treatment similarly as mentioned for gene expression study, following which, the supernatants were collected and cytokine quantification was performed as per the manufacturers’ protocols in a Tecan plate reader. For cytokine quantification, specific ELISA kits were used for Tumor necrosis factor-α (TNFα) (Invitrogen, USA), Interleukin-6 (IL-6) (Invitrogen, USA), Interleukin-1β (IL-1β) (Abcam, USA), Interleukin-12 (IL-12p70) (RnDsystems, USA), and Inter-leukin-10 (IL-10) (RnDsystems, USA). All experiments were performed in three individual sets of triplicates.
Statistical analysis
The data underwent analysis of variance (ANOVA), followed by post hoc Tukey’s test using GraphPad Prism, CA. Results are expressed as mean ± SEM, with statistical significance determined by p-values below 0.05.
Results and discussion
Preparation and characterization of GCs
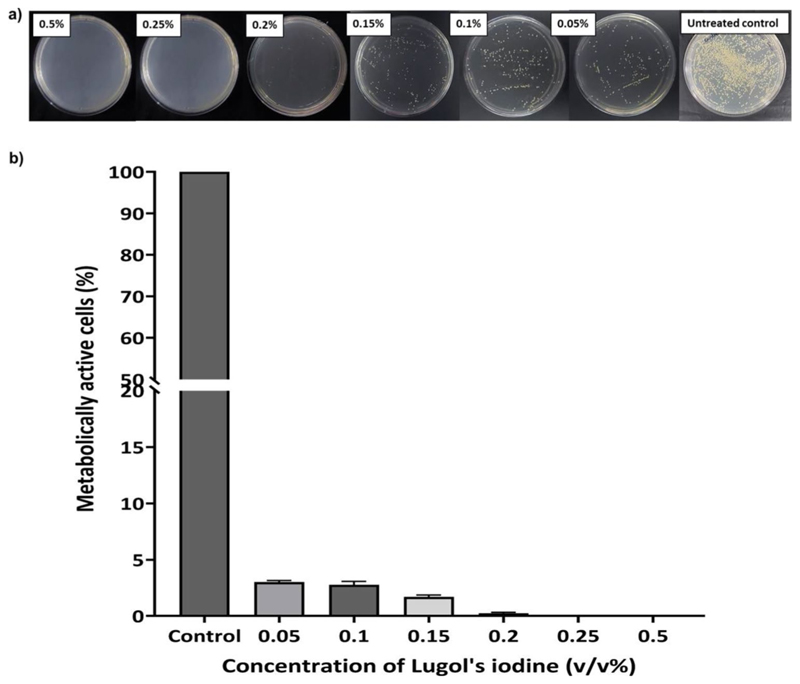
In our pursuit to illustrate ghost cell-driven macrophage polarization, we initially embarked upon the development of a feasible protocol for the generation of GCs. Often, the creation of structurally-intact ghost cells from gram-positive bacteria have posed formidable challenges, primarily attributing to the inherent complexity of perforating their robust outer cell wall. We had previously reported a scalable alkaline-medium culture method for deriving intact ghost cells from S. aureus, which is a gram-positive bacterium (Narayanan et al. 2024). This method, though was competent in yielding structurally sturdy ghost cells, it required overnight culturing and was time-consuming. Therefore, in this study, we have optimised a rapid and efficient protocol using very-low concentrations of Lugol’s iodine for obtaining GCs. This study also sought to surmount existing limitations associated with chemically-driven protocols, which often compromises the structural integrity of these entities, rendering the cells excessively porous and morphologically distorted. The preservation of both the structural and functional integrity of the GCs was deemed indispensable, as it reinforced their potential utility as versatile platforms for immunotherapy, drug delivery, and vaccine. To begin with, we initially determined the Minimum Inhibitory Concentration (MIC) of Lugol’s iodine against Staphylococcus aureus NCIM 2127 using the two-fold broth dilution method (Vinod et al. 2015). The bacterium was subjected to a time- and concentration-dependent treatment with Lugol’s iodine which is a solution containing iodine and potassium iodide, and is commonly used as a disinfectant. The bacterial samples, growth synchronised at OD = 1 were allowed to incubate in the varying concentrations of Lugol’s iodine at room temperature for 5 min after which, the samples were spread-plated and incubated for 24 h to check for any colony regrowth. Notably, we observed no colony formation for iodine concentrations from 0.25 to 1%, while concentrations below 0.25% showed colony revival (Fig. 1). We also conducted the S. aureus viability tests viz., mannitol salt agar (MSA) test and catalase test to validate the above data and compared it with untreated control sample (Supplementary fig. S1a and S2b). The MSA test revealed a characteristic colour change from red to yellow in the medium containing untreated samples, indicative of S. aureus viability, while no colour change was observed in the medium containing 0.25% Iodine-treated samples. For the catalase test, live S. aureus exhibited bubble formation upon application of 15% H2O2 on a glass slide, signifying catalase positivity. In contrast, no bubble formation was observed following ghost cell inoculation. Consequently, we choose 0.25% of iodine as optimal for ghost cell generation based on these validations. We next assessed the presence of iodine content in the ghost cells after their purification from the preparation pot. For this, a method based on the formation of a blue starch-iodine complex was employed, as described by (Sulistyarti et al. 2015). Maintaining a constant concentration of starch, we subsequently determined whether iodine was completely removed through repeated washing steps. Before iodine treatment, the bacterial pellet exhibited a creamish-white colour, which transitioned to brown upon iodine treatment. As the washing steps progressed, the pellet gradually lost the brown tint, indicative of iodine removal and assumed a white color by the end of the fifth centrifugal wash. Upon conducting the starch-iodine assay using the supernatants after each centrifugation, we observed a decrease in iodine content with increasing number of centrifugations,as was evident from the reduction in blue colour formation in the assay mix (Supplementary fig. S2e).
Fig. 1.
(a) Nutrient agar plates representing colony forming units of bacterial cultures treated with Lugol’s iodine concentrations of 0.5%, 0.25%, 0.2%, 0.15%, 0.1%, and 0.05% respectively along with untreated control plate (b) Plot representing percentage of metabolically active cells vs. iodine concentration determined from the colony forming units
Subsequently, to quantify the average dry weight of GCs obtained from a single preparation, we subjected the GC wet pellets to lyophilization (Supplementary fig. S1d). The untreated control exhibited an average dry weight of 23 mg/mL, while the GCs displayed a notable reduction in dry weight, amounting to 13 mg/mL. Additionally, we estimated the number of bacterial units (in the control and GC samples) by unit counting using a hemocytometer and observed no significant variation in unit counts between the untreated control and GC sample. This suggested that the decrease in dry weight is likely attributed to the loss of internal cellular constituents rather than differences in unit counts between the untreated and treated samples.
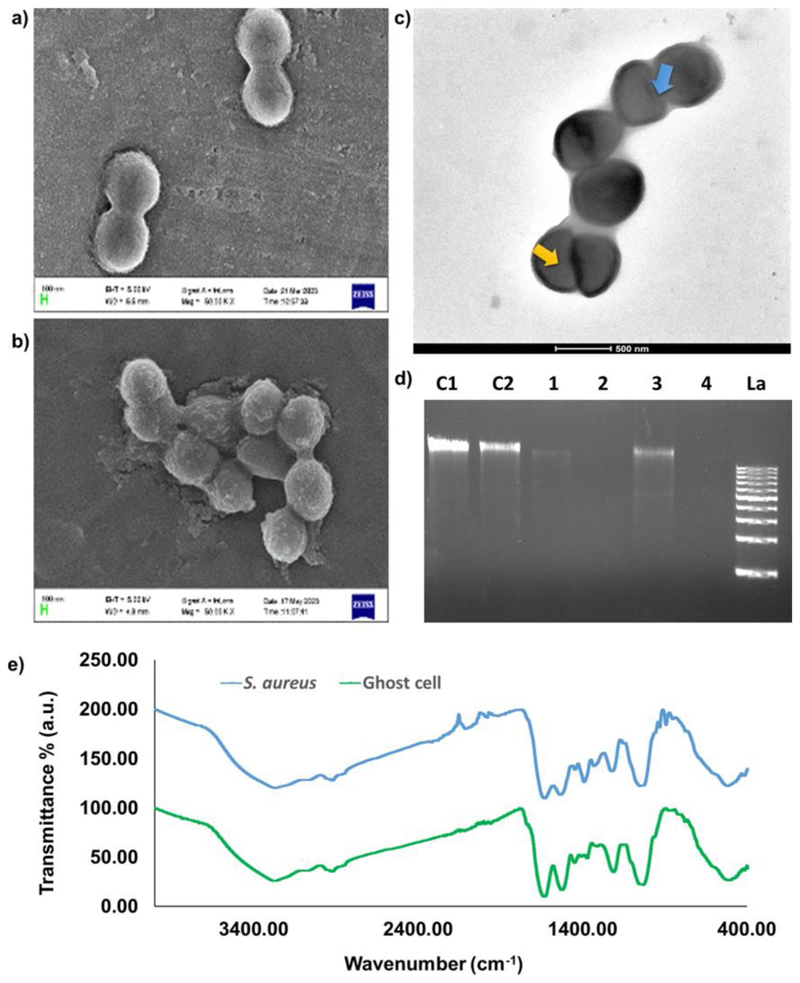
Following GC preparation and purification, the initial investigation aimed to discern the structural characteristics of the ghost cells, involving the utilization of FE-SEM and TEM. As depicted in Fig. 2a and b, FE-SEM images revealed a slightly reduced size for the ghost cells compared to the parent bacterial cells. This size variation might be linked to a loss of turgidity due to the expulsion of cytoplasm and the nucleoid content. Further insights about the morphology and intracellular status were gained through TEM imaging (Fig. 2c). Earlier reports on live S. aureus TEM imaging have demonstrated the presence of highly dense interiors due to the availability of the cytoplasm and the nucleoid centre (Kim et al. 2007; Shu et al. 2020). Our TEM analysis highlighted noticeable intra-cellular vacancies in the GCs, indicated by less dense centres due to the unavailability of the cytoplasmic content. However, the outer wall integrity was preserved and was noticeably denser than the intracellular compartment. Notably, the absence of tunnel-like structures in the obtained GCs diverges from earlier findings, potentially due to transient pore formation induced by the ultra-low iodine treatment. In our case, we found small notch-like structures on the GCs indicative of pores which plausibly underwent self-sealing owing to cell wall rearrangement.
Fig. 2.
Scanning electron microscopic images of bacterial cells (a) control (live S. aureus) and (b) treated (S. aureus ghost cells) with observable size reduction under 10,000 X magnification. (c) Transmission electron microscopic images of ghost cells with decreased internal density (indicated by yellow arrow), increased transparency and intact cell wall. On the surface of the ghost cell small notches could also be seen (indicated by blue arrow). (d) Agarose gel electrophoresis image showing the reduction in DNA content in ghost cell samples compared to the respective control from two different culture volumes and time points (C1-untreated 100 mL, C2-untreated 50 mL, 1–50 mL culture plus 5 min iodine treatment, 2–50 mL culture plus 10 min iodine treatment, 3-100 mL culture plus 5 min iodine treatment, 4-100 mL culture plus 10 min iodine treatment, La-DNA ladder 1 kb). (e) FT-IR spectra of lyophilized samples of untreated bacterial cells (S. aureus) and treated ghost cells with characteristic peaks between the range 4000 cm−1 to 400 cm−1
We further validated the ghost cell formation by checking the DNA integrity of the GCs and compared them with that of the parent cells via agarose gel electrophoresis. Figure 2d illustrates distinct outcomes, where DNA extracted from untreated control samples of varying culture volumes, specifically 100 and 50 µl (Lane C1 and C2, respectively), as well as the DNA ladder (Lane La), displayed clear and intact DNA bands. Conversely, GC samples obtained after 5 and 10 min of iodine incubation in 50 µl of culture (Lane 1 and 2, respectively), and samples obtained after similar incubation times in 100 µl of culture (Lane 3 and 4, respectively), exhibited no discernible DNA bands, indicative of the absence of nucleic acids within their intracellular space. Interestingly, consistently lower DNA content was observed after 10 min of iodine incubation across both culture volumes, indicating the effects of iodine incubation time on GC generation. Overall, the data specified a substantial reduction in genomic DNA content within the GC pellets compared to the untreated S. aureus cells, corroborating with the less dense cores that were observed in the GCs during TEM analysis.
For assessing the status of PAMPs on the GCs and reaf-firming the absence of DNA, we employed FTIR spectroscopy (Fig. 2e). PAMPs such as peptidoglycan, lipoteichoic acid, and lipoproteins are major constituents of the S. aureus cell wall and membrane and therefore our primary focus was in identifying their characteristic spectral signatures. The spectral peaks centred at 1635 cm−1 for S. aureus and 1639 cm−1 for GCs correspond to Amide-I, while those at 1539 cm−1 for S. aureus and 1532 cm−1 for GCs aligns to Amide-II which are specific for peptidoglycan (Hayles et al. 2024). The peaks ranging from 1500 –1200 cm−1, encompasses a combination of signals arising from the bending vibrations of fatty acids and proteins. The bands responsible for > CH2 and -CH3 bending modes in Amide-III contribute to this region, providing additional insights into the characteristics of the fatty acid segment (Zarnowiec et al. 2015). Peaks observed at 2927 cm−1 for GCs and 2926 cm−1 for the parent organism signify the vibration of -CH3 groups, characteristic of alkane groups predominantly found within lipids that form a part of lipoproteins (Marseno et al. 2014). The spectral range spanning 900 to 700 cm−1 exhibited subtle yet distinctive patterns associated with aromatic ring structures found in amino acids and nucleic acids. This region serves as the characteristic peak for discriminating specific bacteria. Furthermore, specific peaks observed only in the untreated bacterial sample, notably at 934.17 cm−1 and 895.28 cm−1, corresponding to vibrations attributed to the sugar-phosphate chain (Banyay et al. 2003)and the deoxy ring (Zhang et al. 2016), both in nucleic acids, respectively, were absent in the GC sample. This strongly implied the absence of DNA, aligning coherently with the validation obtained through the TEM and agarose gel electrophoresis analyses. These findings collectively supported the structural characterization of the GCs while affirming their lack of genetic material.
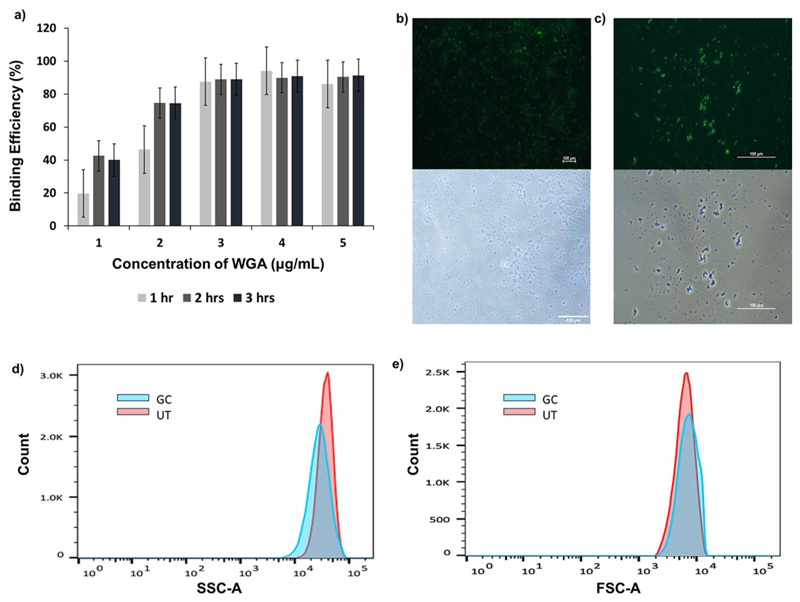
In our next pursuit, we first undertook an optimization process to facilitate effective conjugation of WGA Alexa Fluor 488 fluorophore to the parent and ghost cells for the ease of conducting flowcytometry. WGA, a plant lectin renowned for its specificity toward N-acetyl-D-glucos-amine (NAG) present in peptidoglycan (Hsu et al. 2016), complemented our investigations. To create fluorescent live cells and ghost cells, varied concentrations of WGA-Alexa Fluor 488 conjugate (ranging from 1 to 5 µg/ml) and diverse incubation time points (1, 2, and 3 h) were meticulously assessed at room temperature, with the experimental samples shielded from light throughout the procedure. Subsequent spectrofluorimetric analysis of the supernatants post-incubation revealed that an optimal fluorophore conjugation for both the parent and the ghost cells was achieved at a concentration of 3 µg/ml with an incubation period of 1 h (Fig. 3a). Any further increase in fluorophore concentration or incubation time did not result in significant enhancement in fluorescent tagging. The consistency observed in optimal conjugation conditions implies that the surface characteristics of the GCs did not undergo alterations in the NAG moiety, thus facilitating an unhindered WGA-NAG interaction. This assurance of consistent reactivity between WGA and the NAG component further reinforces the stability and structural integrity of the surface components of the generated GCs as observed under the fluorescent microscope (Fig. 3b and c).
Fig. 3.
(a) Comparison of the binding efficiency of WGA-Alexa Flour 488 stain using a spectrofluorometer with concentrations ranging from 1 to 5 µg/mL at three different time points (1 h, 2 h, and 3 h), quantified in terms of percentage intensity in the supernatant. (b) Fluorescence microscopy images of WGA Alexa Flour 488-stained untreated cells (S. aureus) and (c) GC samples under 20X magnification. Lower panel of (b) and (c) show bright field images of the respective regions. (d) and (e) are side scatter and forward scatter plots obtained for assessing granularity and size respectively of ghost cells in comparison to live bacterial cells
Utilizing the aforementioned fluorescent-tagged GCs and parent cells we performed the SSC analyses which revealed distinct patterns, as illustrated in the provided figure (Fig. 3d). Specifically, in the SSC analysis, indicative of cellular granularity, notable differences were observed where the GCs exhibited a reduced level of granularity compared to their parental counterpart S. aureus, suggesting a discernible loss of internal components consequential to the ghost cell treatment. FSC (forward scatter) analysis of the sample was also performed in which, the size reduction is clearly visible when comparison with live S. aureus (Fig. 3e). A similar fashion of validation was recently reported by Hjelm et al., where they used flowcytometry to study the granularity of ghost cells (Hjelm et al. 2015).
Cytotoxicity analysis of GCs on macrophages
Cytotoxicity assessment was performed utilizing the MTT assay, entailing the evaluation of Raw 264.7 macrophage viability in response to varying concentrations of untreated S. aureus or GCs in RPMI media (Supplementary Fig. S3a). During a 24-hr exposure period, four distinct concentrations (0.5, 1, 1.5, and 2.5 µg/µL) were administered to the cells. Notably, at 0.5 µg/µL, GCs exhibited maximal cell viability, reaching an impressive 98%, while untreated S. aureus demonstrated a viability of 80% at the same concentration. In contrast, the lowest cell viability was recorded at 2.5 µg/µL, with GCs showing 89% viability while untreated S. aureus exhibiting a significantly lower 60%, indicating a pronounced divergence in outcomes compared to other concentrations. S. aureus, a recognized pathogenic bacterium, exhibits active proliferation and infectivity in its live state, leading to pronounced deleterious effects on the cultured environment, including the induction of cell death. This phenomenon was conspicuously manifested by the turbidity observed in the macrophage cultures treated with parent cells (live S. aureus), indicating a substantial impact on the culture due to the vigorous growth and infectious activity of the pathogenic bacteria. Therefore, our cytotoxicity results revealed that the survival of the S. aureus-infected macro-phages was severely affected compared to those treated with the GCs at the dose, indicative of the inertness of the GCs even at higher concentrations.
Cellular interaction of GCs
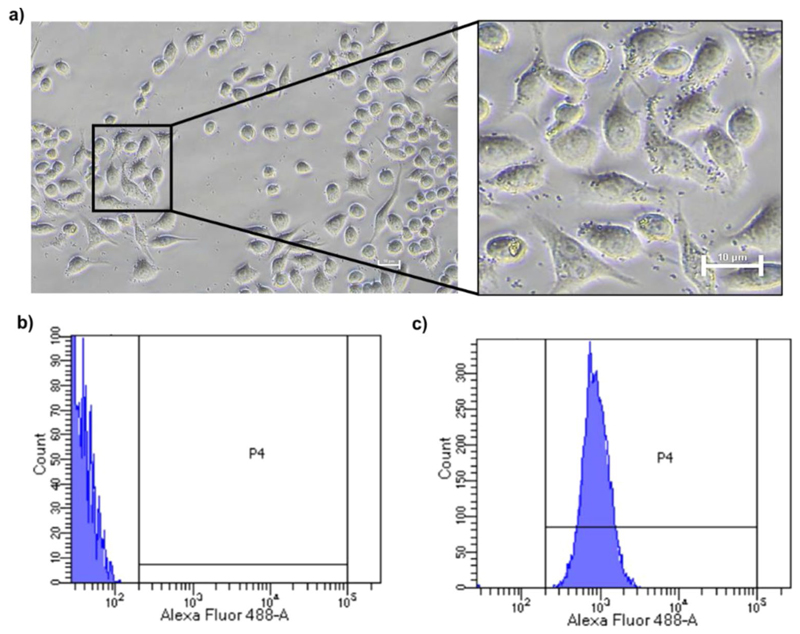
To visualize the cellular localization of GCs in macro-phages, a phase-contrast microscopy approach was initially employed. Pre-staining of GCs with trypan blue facilitated the tracking of GC localization. After a 30-minute incubation of RAW264.7 macrophages with 0.5 µg/µL GCs, microscopic examination revealed distinct localization of GCs around the membrane and cytoplasmic regions of the macrophages, manifesting as small, rounded, blue granules (Fig. 4a). This observation provided insights into the interaction process and spatial distribution (encompassing surface binding and internalization) of GCs in the macrophages.
Fig. 4. Interaction of GCs with RAW 264.7 macrophages.
(a) Phase contrast microscopy images of GC-internalized macrophages after 30 min incubation. Outset shows magnified image of GC localization in the cells. (b) Macrophages before co-incubation and (c) Macrophages co-incubated with Alexa Flour 488-labeled bacterial ghost cells following a 30 min incubation period
Subsequently, flow cytometry and fluorescent microscopy experiments were conducted to validate the association of the GCs with RAW 264.7 macrophages, utilizing 0.5 µg/µL of WGA-FITC labelled GCs. The assessment revealed that over 90% of the cells exhibited fluorescence within a brief 30 min incubation (Fig. 4b and c), notably prompting to a high particle interaction efficiency in macrophages. Similar observations were made when the fluorescent-GC incubated macrophages were visualised under the fluorescent microscope after 30 min of incubation (Supplementary Fig S5). The data showed fluorescence signals in, and around the membrane of the macrophages possibly indicating the binding and initial uptake within the stipulated time period. Macrophages are antigen presenting cells that phagocytose bacteria and portray immediate uptake of foreign materials often within a matter of minutes (Stuart et al. 2005; Gustafson et al. 2015). While the swift uptake is often considered a challenge in conventional drug delivery systems where drugs may be quickly internalized and potentially lost, our study highlights a beneficial aspect of this phenomenon. In our case, the immediate internalization of GCs by macrophages proves advantageous for several critical immunological processes. The rapid internalization of GCs would allow macrophages to process and present bacterial components, triggering a cascade of events that includes cytokine release. The cytokine release, in turn, could contribute to the activation of immune cells and the expansion of the immune response. Importantly, this immediate response could set the stage for efficient communication with the adaptive immune system, ultimately enhancing the overall immunomodulatory capacity of the GCs.
Assessment of gene expression via qPCR
The pro-inflammatory M1 subtype of macrophages is distinguished by its capacity for anti-microbial cytokine production (Mamilos et al. 2023), playing a pivotal role in eradicating pathogens within infected tissues. This elimination is orchestrated through various mechanisms, including the induction of cell toxicity, necrosis, apoptosis, and the recruitment of other inflammatory cells such as neutrophils and leukomonocytes, thereby orchestrating a robust immune response against invading pathogens (Ni et al. 2023). In pursuit of a macrophage-mediated approach for infection control, we sought to explore the potential of GCs in programming naïve macrophages to adopt the pro-inflammatory M1 subtype. To achieve this, we focused on investigating the human monocytic cell line THP-1-derived naïve macrophages, obtained through the culture of THP-1 cells in medium supplemented with 100 nM PMA. The observation following the suspension culture of THP-1 cells revealed a morphological transition from rounded suspension cells to elongated adherent forms (Supplementary Fig S3b and S3c) by the conclusion of the resting phase. These adherent macrophage cultures were subsequently designated as naïve macrophages (M0). These cells were first used to evaluate the dose-dependent effect of GC and lipopolysaccharide (LPS) using the MTT cell viability assay (Supplementary Fig S3d). LPS was chosen as a comparison point as it is a classical inducer of M1 differentiation in macrophages and we intended to use it as a positive control for the protein and gene expression studies to ensure reliability and accuracy of our experimental outcomes.
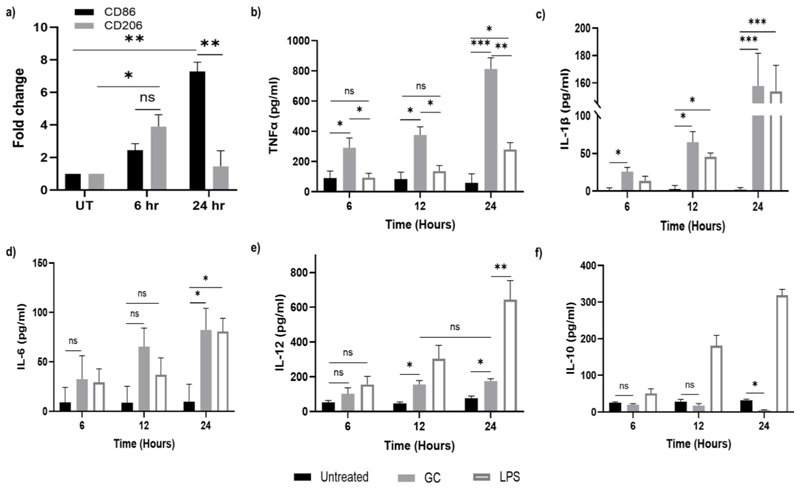
For the subsequent protein and gene expression studies, the M0 macrophages were subjected to treatment with GCs at a chosen concentration of 100 ng/mL. This concentration was chosen to ensure an effective response without inducing undue stress or potential adverse effects on the macrophages. Subsequent to the stimulation period, well contents were collected, and centrifugation was performed to obtain cell-free supernatant and cell pellet for ELISA and qPCR analyses, respectively. First, the gene expression of M1 (CD86) and M2 (CD206) markers were analysed. A significant 7.2-fold increase in the CD86 expression (p < 0.005) was observed compared to untreated control cells after 24 h of GC-treatment of M0. However, in the case of CD206 expression, though an increase in expression was found at 6 h, the fold change nearly reduced to the untreated level and was significantly lower than the expression of CD86 (p < 0.005). Notably, the expression levels of CD206 were initially higher, suggesting a potential dominance or prevalence of the M2 marker during the early stages. However, over time, there was a discernible increase in the expression of CD86, indicating a dynamic shift in cellular subtype (Fig. 5a). These findings suggest that GCs may selectively induce the expression of CD86 on naïve macrophages. Notably, the augmented expression of CD86 in antigen-presenting cells such as macrophages, assumes significance in the amplification of pro-inflammatory factors, given its role as a costimulatory molecule for the B7 receptor on T-cells (Parker 2018).
Fig. 5.
(a) Gene expression levels of CD86 and CD206, markers for M1 and M2 -subtypes respectively performed using qPCR after incubation with 100 ng/ml of GC. The results were normalized and are shown as fold changes relative to gene expression in untreated control cells. Quantification of cytokines (b) Quantification of TNF-α in GC or LPS- treated (positive control) supernatants compared to untreated control (negative control) at different time intervals performed via sandwich ELISA. (c) IL-1β, (d) IL-6, (e) IL-12p70 and (f) IL-10 in supernatants of 100 ng/ml GC or LPS -treated naïve macrophages in a time-dependent manner via sandwich ELISA. Data represented as mean ± SD from 3 independent experiments. p-values: * <0.05, **<0.01, *** <0.005
Quantification of cytokines via ELISA
In the pursuit of deciphering the immunomodulatory impact of GCs on M0 macrophages, the quantification of key cytokines, namely TNF-α, IL-1β, IL-6, and IL-12, was undertaken through highly sensitive and specific ELISA experiments. These cytokines are classical pro-inflammatory cytokines that represent as specific markers for the M1 macrophage subtype. The quantification, as mentioned earlier, was done using the cell-free supernatant of the treated cells and was compared with two controls viz., cells cultured in medium only (negative control) and cells cultured with equivalent concentration of LPS containing medium (positive control). The quantitative assessment of TNF-α levels in the supernatant of GC-treated M0 macrophages exhibited a discernible and steady escalation, with fold increases of up to 16-fold, during the critical 12–24 h post-treatment interval as opposed to the untreated control group. Intriguingly, comparative analysis between distinct time points unveiled a statistically significant temporal surge in TNF-α secretion from 6 h (p < 0.05) to 12 h (p < 0.05) and 24 h (p < 0.005) intervals (Fig. 5b). This temporal disparity is indicative of potential activation cascades and intricate signalling pathways triggered by GCs within this timeframe. Existing research posits that TLR activation instigates the onset of TNF-α production, fostering an autocrine loop that amplifies the activation of M1 macrophages (Parameswaran and Patial 2010). Interestingly, an equivalent concentration of LPS was ineffective in triggering the production of TNF-α at all test intervals, though the levels were greater than untreated control.
Considering that GCs is a TLR-agonist reservoir with several PAMP molecules available for TLR activation, while LPS is a singular PAMP molecule that specifically binds only to TLR-4, it is reasonable to expect a temporally regulated pattern in the release of this crucial pro-inflammatory cytokine. This anticipation stems from the fact that macrophages possess multiple TLR subsets and the intricate interplay between GCs and macrophages may influence the timing and dynamics of pro-inflammatory cytokine release.
Subsequent to our investigations, we quantified the pro-inflammatory cytokine IL-1β, widely acknowledged as a hallmark of the M1 macrophage subtype (Chen et al. 2023). Discernible IL-1β expression following GC treatment manifested significance as early as the 6-hr time point (p < 0.05) in comparison to untreated controls. This significance intensified by the 24-hr time point (p < 0.005) (Fig. 5c). Notably, the IL-1β expression pattern closely mirrored that of TNFα, albeit at concentrations lower than the latter. Extant literature suggests that IL-1β and TNFα are capable of upregulating CD86 expression (Saito et al. 2006), a proposition substantiated in our study where we observed a higher fold-change in CD86 gene expression after 24 h of GC-treatment. This coherence in the expression patterns of pro-inflammatory cytokines and associated immune markers accentuates the molecular events orchestrated by GC treatment, emphasizing the potential impact on macrophage polarization and activation states.
Next, our focus extended to IL-6 which is yet another marker of M1 macrophage subtype (Chen et al. 2022). Notably, the analysis of IL-6 in the cell-free supernatant revealed a delayed expression, becoming statistically significant only following 24 h of GC incubation. This temporal pattern aligns with prior reports indicating that the induction of IL-6 necessitates an initial priming with TNFα and IL-1β which are the most potent inducers of IL-6 expression, mainly regulated at the transcriptional level(Furia et al. 2010; Lopez-Castejon and Brough 2011). Consequently, the observed delay in IL-6 expression within our experimental context may reasonably be attributed to this inherent lag associated with the prerequisite priming phase (Fig. 5d). Interestingly, both GC and an equivalent concentration of LPS exhibited comparable gene expression profiles at 24 h, indicating a parallel response in the examined condition. This congruence was consistent with the observed patterns for the previously investigated cytokine, IL-1β.
We next assessed IL-12p70 expression in the GC-treated cells for similar time points as above. Its primary function is to bolster cell-mediated immunity by activating Th1 cells as well as cytolytic cells such as the cytotoxic T-cells and the natural killer cells (Ullrich et al. 2020). This cytokine also acts synergistically with TNFα and other proinflammatory cytokines in amplifying the production of IFN-γ (Arango Duque and Descoteaux 2014). Our results show a discernible increase in expression for IL-12 at both 12 and 24 h post GC-treatment, with no significant cytokine release at the early hours. Earlier investigations have identified IL-1β as an IL-12-inducing agent (Wesa and Galy 2001). Hence, we posit that the noteworthy upregulation of IL-12 observed 12 h post GC treatment may be intricately regulated by the early expression of IL-1β, a correlation evident in our experimental data. Interestingly, the IL-12 expression pattern following LPS treatment revealed a markedly elevated expression at the 24-hr time point, signifying a robust and prolonged pro-inflammatory response (Fig. 5e).
Conversely, when scrutinizing IL-10, an anti-inflammatory cytokine and a marker associated with the M2 macrophage phenotype, a noteworthy deviation from the observed trends for other cytokines emerged. In contrast to the upregulation witnessed for pro-inflammatory markers, IL-10 exhibited a decrement in expression over time, demonstrating a diminishing trend. Interestingly, the untreated naïve macrophages exhibited a basal level of IL-10 release, a phenomenon that persisted even at the 24 h time point. In contrast to this sustained expression, macrophages subjected to GC-treatment displayed a temporal decrease in IL-10 expression and was noted to be significant at 24 h (Fig. 5f). This observed diminution starkly contrasted with the expression profile exhibited by their LPS-treated counterparts where the IL-10 expression was accentuated, exhibiting a notable increase in IL-10 over time. This was in agreement with earlier reports which suggested LPS to be an inducer for IL-10 expression.
Notably, the observed reduction in IL-10 levels in the GC-treated cells was of significance and aligns with prior literature suggesting TNFα as a potential down-regulator of IL-10 expression. Earlier reports alluded to TNFα blockade as a strategy aimed at enhancing IL-10 expression, providing further context to the observed trend in our study (Evans et al. 2014). In accordance with our earlier findings of elevated TNFα expression in GC-treated cells, we postulate that TNFα likely played a modulatory role in the observed reduction of IL-10 expression, favouring a proinflammatory milieu. Conversely, the lower levels of TNFα in LPS-treated cells may not have exerted a comparable influence on diminishing IL-10 expression. The mechanistic insights gleaned from this observation contributed to our understanding of the complex immunomodulatory effects associated with the GCs, shedding light on potential therapeutic strategies aimed at fine-tuning the delicate balance between pro- and anti-inflammatory cytokine expression. Our observations also suggest the initial manifestation of an M1/M2 chimeric phenotype in GC-treated cells, characterized by concurrent presence of IL-12 and IL-10, which subsequently transitions towards a predominant M1 phenotype over time. We also conducted an assessment of the intracellular ROS generation in THP-1 macrophages as well as RAW264.7 cells after treatment with GCs for pre-determined concentrations and time intervals. Our findings, illustrated in Supplementary Figure S4 a indicate that GCs did not induce significant ROS production in RAW 264.7 macrophages compared to untreated control samples. We believe that the lack of significant ROS production rendered the GCs more compatible with the macrophages as was also evident from our cytotoxicity results. We also conducted the DCFH-DA assay on PMA-induced THP1 macrophages for concentrations 100 and 500 ng/ml for incubation tie intervals of 1 and 24 h Supplementary Figure S4b The results indicated to us that ROS production induced by the GCs at 1 h was very insignificant when compared to the ROS levels in the untreated cells. However, after 24 h we saw mild changes in the ROS levels for both the test concentrations. For our gene and protein expression studies also, we had used 100 ng/ml of GC and we had found that investigated gene and protein expression levels were enhanced only by 24 h wherein the ROS levels have also increased. Therefore, we sense an association between ROS and the concerned molecular changes induced by the GCs, though further validations are recommended.
Conclusion
In conclusion, our study successfully established a rapid and efficient protocol for the generation of intact Staphylococcus aureus ghost cells (GCs), and demonstrated their potential in transforming naïve macrophages to the M1 pro-inflammatory subtype which is implicated in effective infection management by the immune system. Our investigations into GC-mediated macrophage polarization revealed a notable upregulation of pro-inflammatory markers, such as TNF-α, IL-1β, IL-6, and IL-12, indicative of M1 subtype activation as well as M1-specific CD86 biomarker expression. Additionally, our observations revealed the emergence of an M1/M2 chimeric phenotype during the initial hours following GC-treatment, indicating a gradual but consistent transition towards the M1 phenotype. This effect was accompanied by a reduction in the anti-inflammatory cytokine IL-10, suggesting a shift toward a pro-inflammatory milieu. Furthermore, our results propose a mechanistic link between TNF-α and IL-10 expression, providing insights into the delicate balance of pro- and anti-inflammatory responses.
Overall, our results emphasize the potential of GCs as a valid therapeutic platform for modulating the pro-inflammatory transition of macrophages, a phenomenon that is otherwise opposed by the invading pathogens during the realm of an infection. Our study opens way to further research on macrophage-mediated therapeutic strategies to mitigate bacterial infections which are highly complex and manipulative. Non-live entities like GCs can serve as effective adjuvants for antibiotic-therapies, and may hold significant implications for advancing the field of immunotherapy, offering new insights into combatting microbial infections and enhancing treatment efficacy in the face of evolving antibiotic resistance. Although our study shows encouraging findings, the future prospects can be extended to include studies on the impact of GC-triggered macrophages on live bacteria as well as deciphering any possibilities of direct bactericidal activity of the GCs on live bacteria. Also, microbicidal mechanisms and the effect of GCs on already polarised macrophages would provide valuable support to their potential therapeutic applications.
Supplementary Material
Highlights.
A simplistic protocol developed for obtaining ghost cells (GC) from S. aureus.
High cellular interaction of GCs demonstrated in macrophages within short incubation time intervals.
GCs induced M1 macrophage polarization, upregulating markers like CD86 and cytokines TNF-α, IL-1β, IL-6, and IL-12.
Selective M1 polarization demonstrated by GCs evident from the downregulation the anti-inflammatory cytokine IL-10.
Novel approach for immunotherapy in S. aureus infections via pro-inflammatory (M1) polarization of macrophages.
Acknowledgements
We express our sincere gratitude to the DBT-Wellcome Trust India Alliance for the research funds and the Department of Biotechnology, CUSAT for the infrastructure. We would like to thank Mr. Bins KC for technical support during flowcytometry analyses, Department of Physics-CUSAT for FE-SEM analyses, School of Marine Sciences, CUSAT for spectrofluorimetric analyses and fluorescent microscopy, electron microscope facility-Indian Institute of Science, Bengaluru for TEM analyses, and Sophisticated Test and Instrumentation Centre (STIC), CUSAT for FTIR analysis.
Declarations
Author contributions
S.N. conceptualized and designed the study, and performed data analyses. A.P and A.I.M contributed to experimental work, data analyses and manuscript writing. A.P and S.N edited the manuscript. P.K.V and S.G.B provided critical feedback during the execution of the work and while manuscript writing. All authors approved the final version for submission.
Competing interests The authors declare no competing interests.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Data availability
No datasets were generated or analysed during the current study.
References
- Arango Duque G, Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol. 2014;5 doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyay M, Sarkar M, Gräslund A. A library of IR bands of nucleic acids in solution. Biophys Chem. 2003;104:477–488. doi: 10.1016/S0301-4622(03)00035-8. [DOI] [PubMed] [Google Scholar]
- Chen X, Dou J, Fu Z, et al. Macrophage M1 polarization mediated via the IL-6/STAT3 pathway contributes to apical periodontitis induced by Porphyromonas gingivalis. J Appl Oral Sci. 2022;30 doi: 10.1590/1678-7757-2022-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Saeed AFUH, Liu Q, et al. Macrophages in immuno-regulation and therapeutics. Signal Transduct Target Ther. 2023;8:207. doi: 10.1038/s41392-023-01452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans HG, Roostalu U, Walter GJ, et al. TNF-α blockade induces IL-10 expression in human CD4 + T cells. Nat Commun. 2014;5:3199. doi: 10.1038/ncomms4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furia A, Confalone E, D’Alessio G. IL-6 induction by TNFα and IL-1β in an osteoblast-like Cell Line. Int J Biomedical Sci. 2010;6:135–140. doi: 10.59566/IJBS.2010.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson HH, Holt-Casper D, Grainger DW, Ghandehari H. Nanoparticle uptake: the phagocyte problem. Nano Today. 2015;10:487–510. doi: 10.1016/j.nantod.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles A, Bright R, Nguyen NH, et al. Staphylococcus aureus surface attachment selectively influences tolerance against charged antibiotics. Acta Biomater. 2024;175:369–381. doi: 10.1016/j.actbio.2023.12.029. [DOI] [PubMed] [Google Scholar]
- Hjelm A, Söderström B, Vikström D, et al. Autotransporter-based Antigen Display in bacterial ghosts. Appl Environ Microbiol. 2015;81:726–735. doi: 10.1128/AEM.02733-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-P, Meng X, VanNieuwenhze MS. Methods for visualization of peptidoglycan biosynthesis. 2016:3–48. [Google Scholar]
- Jain N, Moeller J, Vogel V. Mechanobiology of macrophages: how physical factors coregulate macrophage plasticity and Phagocytosis. Annu Rev Biomed Eng. 2019;21:267–297. doi: 10.1146/annurev-bioeng-062117-121224. [DOI] [PubMed] [Google Scholar]
- Kim Y, Farrah S, Baney RH. Membrane damage of bacteria by silanols treatment. Electron J Biotechnol. 2007;10 doi: 10.2225/vol10-issue2-fulltext-7. 0-0. [DOI] [Google Scholar]
- Kolliniati O, Ieronymaki E, Vergadi E, Tsatsanis C. Metabolic regulation of macrophage activation. J Innate Immun. 2022;14:51–68. doi: 10.1159/000516780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysko O, Holtappels G, Zhang N, et al. Alternatively activated macrophages and impaired phagocytosis of S. Aureus in chronic rhinosinusitis. Allergy. 2011;66:396–403. doi: 10.1111/j.1398-9995.2010.02498.x. [DOI] [PubMed] [Google Scholar]
- Li D, Wu M. Pattern recognition receptors in health and diseases. Signal Transduct Target Ther. 2021;6:291. doi: 10.1038/s41392-021-00687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Castejon G, Brough D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011;22:189–195. doi: 10.1016/j.cytogfr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamilos A, Winter L, Schmitt VH, et al. Macrophages: from simple phagocyte to an Integrative Regulatory Cell for Inflammation and tissue Regeneration—A review of the literature. Cells. 2023;12:276. doi: 10.3390/cells12020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marseno DW, Haryanti P, Adiseno B, Haryadi H. Synthesis and characterization of hydroxypropylcellulose from oil palm empty fruit bunches (Elaeis guineensis Jacq) Indonesian Food Nutr Progress. 2014;13:24. doi: 10.22146/jifnp.112. [DOI] [Google Scholar]
- Mohamed Elashiry M, Tian F, Elashiry M, et al. Enterococcus faecalis shifts macrophage polarization toward M1-like phenotype with an altered cytokine profile. J Oral Microbiol. 2021;13 doi: 10.1080/20002297.2020.1868152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan S, Baburajan AP, Muhammad M, et al. Demonstrating the immunostimulatory and cytokine-augmentation effects of bacterial ghosts on natural killer cells and Caenorhabditis Elegans. Biotechnol Bioeng. 2024;121:959–970. doi: 10.1002/bit.28619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni R, Jiang L, Zhang C, et al. Biologic mechanisms of macrophage phenotypes responding to infection and the Novel therapies to moderate inflammation. Int J Mol Sci. 2023;24:8358. doi: 10.3390/ijms24098358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran N, Patial S. Tumor necrosis Factor-α signaling in macrophages. Crit Rev Eukaryot Gene Expr. 2010;20:87–103. doi: 10.1615/CritRevEukarGeneExpr.v20.i2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D. CD80/CD86 signaling contributes to the proinflammatory response of Staphylococcus aureus in the airway. Cytokine. 2018;107:130–136. doi: 10.1016/j.cyto.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran S, BS B, A B, et al. Knock-down of a regulatory barcode shifts macrophage polarization destination from M1 to M2 and increases pathogen burden upon S. Aureus infection. bioRxiv. 2021 doi: 10.1101/2021.10.19.464946. 2021-10 bioRxiv. [DOI] [Google Scholar]
- Saito Y, Yanagawa Y, Kikuchi K, et al. Low-dose lipopolysaccharide modifies the production of IL-12 by Dendritic Cells in response to various cytokines. J Clin Experimental Hematopathology. 2006;46:31–36. doi: 10.3960/jslrt.46.31. [DOI] [PubMed] [Google Scholar]
- Seidl K, Zinkernagel AS. The MTT assay is a rapid and reliable quantitative method to assess Staphylococcus aureus induced endothelial cell damage. J Microbiol Methods. 2013;92:307–309. doi: 10.1016/j.mimet.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Shu Q, Wei T, Lu H, et al. Mannosylerythritol lipids: dual inhibitory modes against Staphylococcus aureus through membrane-mediated apoptosis and biofilm disruption. Appl Microbiol Biotechnol. 2020;104:5053–5064. doi: 10.1007/s00253-020-10561-8. [DOI] [PubMed] [Google Scholar]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strizova Z, Benesova I, Bartolini R, et al. M1/M2 macrophages and their overlaps – myth or reality? Clin Sci. 2023;137:1067–1093. doi: 10.1042/CS20220531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Deng J, Silver JM, et al. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol. 2005;170:477–485. doi: 10.1083/jcb.200501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulistyarti H, Atikah A, Fardiyah Q, et al. A simple and safe Spectrophotometric Method for Iodide determination. Makara J Sci. 2015;19 doi: 10.7454/mss.v19i2.4736. [DOI] [Google Scholar]
- Thurlow LR, Hanke ML, Fritz T, et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186:6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich KA-M, Schulze L, Lou, Paap E-M, et al. Immunology of IL-12: an update on functional activities and implications for disease. EXCLI J. 2020;19:1563–1589. doi: 10.17179/excli2020-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto-Boccazzi I, Epis S, Arnoldi I, et al. Boosting immunity to treat parasitic infections: Asaia bacteria expressing a protein from Wolbachia determine M1 macrophage activation and killing of Leishmania protozoans. Pharmacol Res. 2020;161:105288. doi: 10.1016/j.phrs.2020.105288. [DOI] [PubMed] [Google Scholar]
- Vinod N, Oh S, Park HJ, et al. Generation of a Novel Staphylococcus aureus ghost vaccine and examination of its immunogenicity against virulent challenge in rats. Infect Immun. 2015;83:2957–2965. doi: 10.1128/IAI.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage M1â€M2 polarization balance. Front Immunol. 2014;5 doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesa AK, Galy A. IL-1β induces dendritic cells to produce IL-12. Int Immunol. 2001;13:1053–1061. doi: 10.1093/intimm/13.8.1053. [DOI] [PubMed] [Google Scholar]
- Yunna C, Mengru H, Lei W, Weidong C. Macrophage M1/M2 polarization. Eur J Pharmacol. 2020;877:173090. doi: 10.1016/j.ejphar.2020.173090. [DOI] [PubMed] [Google Scholar]
- Zarnowiec P, Lechowicz L, Czerwonka G, Kaca W. Fourier Transform Infrared Spectroscopy (FTIR) as a Tool for the identification and differentiation of pathogenic Bacteria. Curr Med Chem. 2015;22:1710–1718. doi: 10.2174/0929867322666150311152800. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Ma R, Tian Y, et al. Sterilization efficiency of a Novel Electrochemical disinfectant against Staphylococcus aureus. Environ Sci Technol. 2016;50:3184–3192. doi: 10.1021/acs.est.5b05108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.