Abstract
Purpose of Review
This review summarises recent advances in identifying genetic risk factors for atopic dermatitis (AD) and how these genetic associations are being used to explore the causal relationships between atopic dermatitis and potential risk factors and downstream outcomes.
Recent Findings
A recent large-scale GWAS meta-analysis has identified 91 genetic loci associated with AD. Rare variant studies have also identified new gain- or loss-of-function variants implicated in AD, particularly for FLG and STAT6/JAK1. Finally, there has been a surge in utilising genetic association data to investigate the causal relationships between AD and other traits. Mendelian Randomization studies have found that various metabolites and gut microbiota are causal for AD and have causally implicate AD in the development of alopecia areata, diabetes, vascular dementia and some cancers.
Keywords: Atopic dermatitis, genetics, rare variants, GWAS, Mendelian Randomization
Introduction
It has long been known that atopic dermatitis (AD) is highly heritable [1]. Since FLG was the first gene implicated for AD, the past 15 years has seen advances in genotyping technology and Biobank data collection, which has resulted in studies identifying many genomic loci associated with AD.
New genetic variants identified in GWAS
The largest GWAS meta-analysis for AD was published last year [2**]. In that study we analysed genetic data from >4 million participants to identify 91 genetic loci associated with AD, 32 of these being novel. Downstream analyses of this set of GWAS signals showed enrichment for pathways involved in immune system regulation and activation.
As more genetic associations for AD emerge, it is also becoming clearer how AD is genetically related to other traits. For example, a recent study showed that genes shared between AD and autoimmune disease cluster in Th1, Th2, Th17 pathways [3*]. In our GWAS study we also identified notable genetic correlations with depression/anxiety and gastritis (in addition to the expected allergic traits) [2**].
Rare variants
Whilst GWAS focus on common variants, many studies aim to identify rare variants. FLG has been established as an AD risk factor for a long time, but new variants are still being discovered. A recent step-change in genotyping FLG was achieved using Oxford Nanopore long-read sequencing (as opposed to short-read sequencing), resulting in increased ability to identify copy number variation, more accurate phasing and repeat-specific locations for FLG loss of function variants [4*].
STAT6, previously implicated in AD GWAS [5, 6] has recently gained attention as a source of rare gain-of-function (GOF) variants that result in a severe, early-onset broad allergic disease phenotype. Across five studies, 21 individuals harbouring 11 different STAT6 GOF variants have been identified [7**, 8**, 9**, 10**, 11**]. STAT6 is part of the IL4R/JAK/STAT6 pathway and consistent with this, STAT6 GOF patients have responded well to anti-IL4 antibody dupilumab and JAK-inhibitors. Given this, it has been suggested that patients with severe early-onset allergic disease may benefit from early STAT6 GOF assessment [12].
Rare JAK1 GOF variants have also been identified in families with a broad and severe inflammatory phenotype, including C787F identified in 9 affected individuals of one family [13**]. Another recent study identified 59 individuals with 4 novel JAK1 GOF variants, showing a more common autoimmune and inflammatory disease manifestation than has been reported previously [14**]. Consistent with STAT6, there is preliminary evidence that treatment of these patients with JAK-inhibitors may be especially effective, and warrant early identification and a precision medicine approach to treatment.
A dominant rare variant has also recently been identified in CARD11 in a young patient (of Italian descent) with severe atopic dermatitis, food induced anaphylaxis and hypogammaglobulinemia [15*]. Common variants predicted to affect this gene have also been previously implicated in AD GWAS, but interestingly, only in Japanese individuals [5, 16].
Differences in genetic effects by ancestry
Individuals of non-European descent are currently disproportionately under-represented in genetic studies of AD. Therefore, differences in the genetic aetiology of AD between individuals of different ancestry, could result in large inequalities in terms of available effective treatments that target the underlying causes of disease. This has been seen for cardiovascular disease, where several drugs are less (or in-)effective in people of colour [17].
There is some reassuring evidence from our recent GWAS, that although the index SNPs association may not always replicate between ancestral groups, gene-based or region-based comparisons suggest far fewer differences exist, with only 2 of the 91 associations showing strong evidence of population-specific association (rs4312054 and rs9864845, which appear to be specific to East Asians) [2**].
It has long been reported that there may ethnic differences in the role of FLG in AD, with mutations observed in Europeans being found at much lower frequencies (or entirely absent) in individuals of Asian and African descent [18, 19]. New evidence is starting to fill the gap of knowledge of FLG variants in those of African-descent. Studies in Brazil (with high African ancestry) have investigated the frequency and AD association of FLG variants [20*, 21*] and a US-based study report 20-shared and 10 African ancestry specific (1 novel) FLG variants [22*].
When differences are seen at the variant level, but not at the gene level, this can be because the populations share a causal variant, but this getting tagged differently in the two populations (due to differences in linkage disequilibrium), or the causal variants exist at different frequencies, or different causal mutations have arisen in ancestrally separate groups. The first situation is less of a concern, but the latter two, could result in differences of magnitude in efficacy of drugs that target these genes. Therefore, further careful analysis comparing genetic associations in different population groups is necessary to ensure effective and equitable drug design.
Post-GWAS functional studies
Identification of a SNP, or region associated with a trait in a GWAS requires follow-up to identify the implicated gene (as well as functional mechanism and the relevant cell type). In our recent GWAS, we follow the GWAS with a bioinformatic pipeline to integrate evidence from a wide variety of sources and prioritise genes as the likely causal candidates at each locus.
Several other recent studies have attempted to identify the likely candidate gene from GWAS and validate/functionally characterise these through experimental work. Investigation of one GWAS hit (at 9q21.11) intronic to TJP2, found strong evidence that the GWAS SNP was associated with TJP2 expression, and further investigation suggested that methylation of a nearby CpG site could provide the functional mechanism [23*]. The same group also similarly characterised a genetic association at LOC100294145 with bioinformatic and in vitro analyses [24*]. A new bioinformatic method, maxATAC, which aims to predict transcription factor binding sites using ATAC-Seq data, demonstrated their method in AD [25*]. They found that transcription factors MYB and FOXP1 are predicted to be associated with allele dependent chromatin accessibility at AD loci, adding another dimension to how GWAS loci can be functionally followed up with in silico analyses. Another statistical approach to identify causal genes recently employed in AD is to combine the genetic data with transcriptomic data to directly determine the genes whose expression has a causal effect on AD [26]. This identified four novel genes for AD: AQP3, PDCD1, ADCY3, and DOLPP1.
As a demonstration of how the function of a gene can then be further characterised experimentally, a recent paper has used CRISPR/Cas9 technology to generate FLG knock-out keratinocytes for causal functional characterisation of this gene [27**]. This technology is likely to become more widely used for functional characterisation of implicated genes, though identifying the right model can be a barrier for such investigations.
Genetically informed drug targets
It has been shown that genetic evidence (such as from GWAS) implicating a target gene in the pathogenesis of a disease leads to higher success rate in clinical trials [28*]. The recently approved, Lebrikizumab [29], is supported by genetic associations with IL4, KIF3A and IL13. However, dermatology may have not yet reached the same potential as other disease areas in utilising genetic data for efficient drug discovery [28*] and so it will be interesting to see if over the next few years more genetically supported AD drugs emerge.
Predictive ability of genetic risk factors
The vast majority of genetic associations for AD have very small effect sizes, but can be combined together into a polygenic risk score that captures more genetic risk. Previously a study looking at the predictive ability of genetics for AD, found that using a broad allergy PRS incorporating FLG, the predictive ability is promising (area under the curve (AUC)=0.76, rising to 0.80 with the inclusion of age and sex) [30]. In a recent extension of this, an atopic PRS was found to be associated with paradoxical eczema developing in psoriasis patients treated with biologics, but the predictive ability of this has not yet reached clinical significance (maximum AUC=0.69) [31*].
Digging deeper into the AD phenotype
Following the success in identifying genes implicated in AD susceptibility, attention is now turning to related phenotypes to get a deeper understanding of AD aetiology. Longitudinal clustering methods have been utilised to allow for genetic association testing with longitudinal patterns of allergic disease [32*]. GWAS of related trait IgE has also recently been performed, identifying eight genome-wide significant variants [33*] and whilst FLG genotype has significant impact on AD susceptibility, it has been shown that it does not affect response to dupilumab treatment [34*]. Further refinement of phenotypes will help build a complete picture of how genetic factors influence AD and subsequent progression of the disease.
Utilising genetic data to explore causal relationships
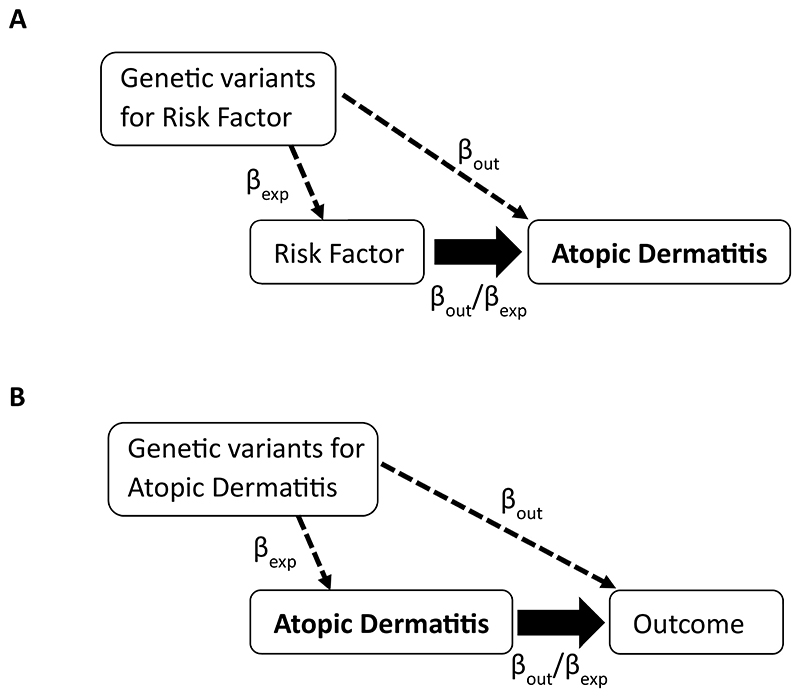
With the increasing availability of large-scale openly accessible AD GWAS data, there has been a surge in utilising this data to investigate causality for many AD epidemiological relationships with an approach called Mendelian Randomization (MR). MR uses genetic proxies for exposures of interest and tests for an association between these genetic proxies and outcomes (figure 1). In this context, GWAS data for AD is combined with GWAS of other traits to determine if AD is causal for certain outcomes, or if particular risk factors have a causal effect on AD. A recent study conducted a systematic review of 30 MR studies in AD [35**], finding evidence for body mass index, gut microbial flora, the IL-18 signalling pathway and gastroesophageal reflux disease as causal risk factors for AD. They also reported that genetic evidence shows that AD is causal for several disease outcomes including heart failure, rheumatoid arthritis and conjunctivitis [35**]. Indicative of the popularity of this statistical technique, in the past 18 months >20 additional AD MR studies have been published. This has included exploring the causal role of metabolites in disease, with one study reporting evidence that increased TNFS14 (and decreased DHA) in the blood are causal risk factors for AD [36*] and another showing that reduced urinary tyrosine [37*] is causal for AD, as well as further studies reporting the causal role of specific gut microbiota [38*, 39*, 40*]. An even greater number of studies have claimed evidence that genetic liability to AD is causal for later health outcomes (Figure 1B), including Type 1 and Type 2 diabetes [41*], basal cell carcinoma and cutaneous squamous cell carcinoma [42*], alopecia areata [43*], vascular dementia [44*], allergic rhinitis [45*] and increased caudate volume [46*], as well as reduced Parkinson’s disease risk [47*]. MR has also been used to refute causal relationships for some observational associations, such as between AD and COVID-19 [48*, 49*]. There are important limitations of this approach to be considered when undertaking or interpreting such studies and STROBE-MR reporting guidelines have been developed to mitigate these [50]. One assumption of MR is that variants do not have a pleiotropic effect on the outcome (i.e. the genetic variants used to proxy the exposure do not affect the outcome through a path independent of the exposure). This is difficult assumption to test, but several methods exist that attempt to estimate and account for pleiotropy. Violation of this assumption can also manifest in situations where an underlying biological impact of the variant affects two diseases independently. This is a particular concern when the causal effect appears to go in both directions, as has been observed for AD and ADHD [51], inflammatory bowel disease [52], asthma [53] and chronic kidney disease [54]. Another way of thinking of this is that the genetic instrument used is not specific for the exposure trait. This should be investigated in studies and Steiger filtering is one approach to avoid this pitfall [55]. Null studies must also be reported carefully, as some MR estimates can have wide confidence intervals, which while do not provide any evidence for a causal effect, often also do not rule out quite large causal effects, as was reported for glaucoma, where confidence intervals for a causal odds ratio of AD on glaucoma ranged from 0.95 to 1.27 [56] and ADHD 0.93 to 1.11 [57]. As larger GWAS become available, statistical power for these research questions will increase. Given the paucity of large GWAS in individuals of non-European ancestry, the current MR studies also suffer from lack of generalisability. However, recently an MR study was able to compare the causal effect of AD on esophageal cancer in Europeans and East Asians, finding causal evidence only in the latter [58].
Figure 1.
Schematic of Mendelian Randomization analysis involving AD (A) Testing for the causal effect of potential risk factors on AD, and (B) Testing for the causal effect of AD on downstream outcomes. In both cases the bold arrow is the causal effect that is being estimated. Genetic risk factors (instruments) for the relevant exposure are selected (and the association recorded, βexp,) and then the association of these instruments is obtained for the outcome (βout). Then the wald ratio [βout/βexp] can be calculated to estimate the causal effect of the exposure on the outcome.
Conclusion
In this paper I have outlined the key recent advances in the genetic aetiology of AD. Whilst the list of genes implicated in the disease has grown substantially, the field now moves into a more challenging phase, where nuance of how genetic associations functionally affect the underlying molecular pathways, and how consistent genetic associations are between individuals of different ancestry is necessary.
Summary.
The past year has seen a huge increase in the genes implicated for AD and in the use of genetics to explore causal relationships. The latter requires caution in implementation and interpretation, but is a promising area of research. In the coming years, increasing the ethnic diversity of AD genetic studies would be very welcome and the translation of current genetic findings into new drugs will be an exciting area of development.
Key points.
Recent large-scale GWAS have identified a large number of genetic variants associated with atopic dermatitis
Functional follow-up studies to characterise these genetic associations, particularly to identify the causal gene, relevant tissue and mechanism of perturbation, are emerging and will be critical to ensure translation of findings.
Genetic studies of diverse cohorts have generally demonstrated that at the gene level there is huge consistency in genetic effects between populations, but at the variant level there can be significant variability.
GWAS resources are now being utilised to undertake Mendelian Randomization studies to investigate causal relationships between AD and potential causes and consequences.
Acknowledgements
none
Financial support
LP receives support from the UK Medical Research Council Integrative Epidemiology Unit at the University of Bristol (MC_UU_00032/01, MC_UU_00032/03), the NIHR Biomedical Research Centre at the University Hospitals Bristol and Weston NHS Foundation Trust and the University of Bristol (NIHR203315), the Leo Foundation (LF-AW_EMEA-23-400257) and the Academy of Medical Sciences Springboard Award, which is supported by the Wellcome Trust, The Government Department for Business, Energy and Industrial Strategy, the Global Challenges Research Fund and the British Heart Foundation (SBF003\1094).
Funding
NIHR, MRC, Leo Foundation, Academy of Medical Sciences (full details in acknowledgements section).
Footnotes
Conflicts of interest: none
References
- [1].Elmose C, Thomsen SF. Twin Studies of Atopic Dermatitis: Interpretations and Applications in the Filaggrin Era. J Allergy. 2015;2015:902359. doi: 10.1155/2015/902359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Budu-Aggrey A, Kilanowski A, Sobczyk MK, et al. European and multi-ancestry genome-wide association meta-analysis of atopic dermatitis highlights importance of systemic immune regulation. Nat Commun. 2023;14:6172. doi: 10.1038/s41467-023-41180-2. [** Large-scale GWAS meta-analysis for AD that identified 91 genetic loci and explored differences between groups of different ancestry] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ahn J, Shin S, Lee GC, et al. Unraveling the link between atopic dermatitis and autoimmune diseases in children: Insights from a large-scale cohort study with 15-year follow-up and shared gene ontology analysis. Allergol Int. 2024;73:243–254. doi: 10.1016/j.alit.2023.12.005. [* Reported that genetic correlation between AD and IBD was predominantly due to shared genetic loci in Th1, Th2 and Th17 pathways] [DOI] [PubMed] [Google Scholar]
- [4].Wong C, Tham CY, Yang L, et al. Nanopore Sequencing Enables Allelic Phasing of FLG Loss-of-Function Variants, Intragenic Copy Number Variation, and Methylation Status in Atopic Dermatitis and Ichthyosis Vulgaris. J Invest Dermatol. 2024 doi: 10.1016/j.jid.2024.01.020. [* Employed new long-range sequencing method which showed improved FLG genotyping in regions that were difficult to genotype with previous technology] [DOI] [PubMed] [Google Scholar]
- [5].Tanaka N, Koido M, Suzuki A, et al. Eight novel susceptibility loci and putative causal variants in atopic dermatitis. J Allergy Clin Immunol. 2021;148:1293–1306. doi: 10.1016/j.jaci.2021.04.019. [DOI] [PubMed] [Google Scholar]
- [6].Sliz E, Huilaja L, Pasanen A, et al. Uniting biobank resources reveals novel genetic pathways modulating susceptibility for atopic dermatitis. J Allergy Clin Immunol. 2022;149:1105–1112.:e1109. doi: 10.1016/j.jaci.2021.07.043. [DOI] [PubMed] [Google Scholar]
- [7].Baris S, Benamar M, Chen Q, et al. Severe allergic dysregulation due to a gain of function mutation in the transcription factor STAT6. J Allergy Clin Immunol. 2023;152:182–194.:e187. doi: 10.1016/j.jaci.2023.01.023. [** One of 5 recent studies that identified GOF variants in STAT6 that cause a broad and severe allergic phenotype] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Minskaia E, Maimaris J, Jenkins P, et al. Autosomal Dominant STAT6 Gain of Function Causes Severe Atopy Associated with Lymphoma. J Clin Immunol. 2023;43:1611–1622. doi: 10.1007/s10875-023-01530-7. [** One of 5 recent studies that identified GOF variants in STAT6 that cause a broad and severe allergic phenotype] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sharma M, Leung D, Momenilandi M, et al. Human germline heterozygous gain-of-function STAT6 variants cause severe allergic disease. J Exp Med. 2023;220 doi: 10.1084/jem.20221755. [** One of 5 recent studies that identified GOF variants in STAT6 that cause a broad and severe allergic phenotype] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Suratannon N, Ittiwut C, Dik WA, et al. A germline STAT6 gain-of-function variant is associated with early-onset allergies. J Allergy Clin Immunol. 2023;151:565–571.:e569. doi: 10.1016/j.jaci.2022.09.028. [** One of 5 recent studies that identified GOF variants in STAT6 that cause a broad and severe allergic phenotype] [DOI] [PubMed] [Google Scholar]
- [11].Takeuchi I, Yanagi K, Takada S, et al. STAT6 gain-of-function variant exacerbates multiple allergic symptoms. J Allergy Clin Immunol. 2023;151:1402–1409.:e1406. doi: 10.1016/j.jaci.2022.12.802. [** One of 5 recent studies that identified GOF variants in STAT6 that cause a broad and severe allergic phenotype] [DOI] [PubMed] [Google Scholar]
- [12].Human germline gain-of-function in STAT6: from severe allergic disease to lymphoma and beyond. Trends Immunol. 2024;45:138–153. doi: 10.1016/j.it.2023.12.003. [DOI] [PubMed] [Google Scholar]
- [13].Fayand A, Hentgen V, Posseme C, et al. Successful treatment of JAK1-associated inflammatory disease. J Allergy Clin Immunol. 2023;152:972–983. doi: 10.1016/j.jaci.2023.06.004. [** Demonstrated that JAK-inhibitors were effective treatment in a family with C787F JAK1 variant] [DOI] [PubMed] [Google Scholar]
- [14].Horesh ME, Martin-Fernandez M, Gruber C, et al. Individuals with JAK1 variants are affected by syndromic features encompassing autoimmunity, atopy, colitis, and dermatitis. J Exp Med. 2024;221 doi: 10.1084/jem.20232387. [** Found 4 novel JAK1 GOF variants in 59 individuals, all with broad autoimmune and inflammatory phenotypes] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Baronio M, Gazzurelli L, Rezzola S, et al. CARD11 dominant negative mutation leads to altered human Natural Killer cell homeostasis. Immunobiology. 2023;228:152381. doi: 10.1016/j.imbio.2023.152381. [* Characterised the p.Glu57Lys CARD11 variant identified in a 12 year old boy with sever AD and allergy] [DOI] [PubMed] [Google Scholar]
- [16].Hirota T, Takahashi A, Kubo M, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- [17].Johnson JA. Ethnic differences in cardiovascular drug response: potential contribution of pharmacogenetics. Circulation. 2008;118:1383–1393. doi: 10.1161/CIRCULATIONAHA.107.704023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chiricozzi A, Maurelli M, Calabrese L, et al. Overview of Atopic Dermatitis in Different Ethnic Groups. J Clin Med. 2023;12 doi: 10.3390/jcm12072701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Margolis DJ, Mitra N, Wubbenhorst B, et al. Association of Filaggrin Loss-of-Function Variants With Race in Children With Atopic Dermatitis. JAMA Dermatol. 2019;155:1269–1276. doi: 10.1001/jamadermatol.2019.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Laczynski CMM, Machado Filho CDS, Miot HA, et al. Prevalence of filaggrin gene polymorphisms (exon-3) in patients with atopic dermatitis in a multiracial Brazilian population. An Bras Dermatol. 2023;98:236–239. doi: 10.1016/j.abd.2022.04.005. [Study investigating the prevalence and AD-associations of FLG mutations in a Brazilian population] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rios R, Magalhães da Silva T, Strina A, et al. Genetic polymorphism (rs6587666) in FLG protects from eczema in admixed Brazilian children population with high African ancestry. Heliyon. 2023;9:e13659. doi: 10.1016/j.heliyon.2023.e13659. [Found the effect of the SNP rs6587666 in FLG was more strongly associated with AD in individuals with higher proportion of African ancestry in a Brazilian cohort] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Virolainen SJ, Satish L, Biagini JM, et al. Filaggrin loss-of-function variants are associated with atopic dermatitis phenotypes in a diverse, early life prospective cohort. JCI Insight. 2024 doi: 10.1172/jci.insight.178258. [* US-based study which reported 20 FLG variants shared between inviduals of European and African ancestry and 10 African ancestry specific FLG variants – 1 of which was novel] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Eliza Lim YY, Yie Sio Y, Say YH, et al. Deep intronic 9q21.11 polymorphism contributes to atopic dermatitis risk through methylation regulated expression of tight junction protein 2. J Investig Allergol Clin Immunol. 2023 doi: 10.18176/jiaci.0978. 0. [* Follow-up of genetic association at 9q21.11 found that this genetic signal affects expression of TJP2 and that methylation of a nearby CpG site may by the mechanism by which the expression of this gene is affected] [DOI] [PubMed] [Google Scholar]
- [24].Teo WY, Lim YYE, Sio YY, et al. Atopic dermatitis-associated genetic variants regulate LOC100294145 expression implicating interleukin-27 production and type 1 interferon signaling. World Allergy Organ J. 2024;17:100869. doi: 10.1016/j.waojou.2023.100869. [* Characterisation of genetic association at 6p21.32 which implicates LOC100294145 and type 1 interferon and IL27 signalling] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cazares TA, Rizvi FW, Iyer B, et al. maxATAC: Genome-scale transcription-factor binding prediction from ATAC-seq with deep neural networks. PLoS Comput Biol. 2023;19:e1010863. doi: 10.1371/journal.pcbi.1010863. [* Bioinformatic method maxATAC for prediction of transcription factor binding sites using ATAC-Seq data, AD is used as a case example, where they found that transcription factors MYB and FOXP1 are predicted to be associated with allele dependent chromatin accessibility at AD loci] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu H, Ke X, Huang W, et al. Multitissue Integrative Analysis Identifies Susceptibility Genes for Atopic Dermatitis. J Invest Dermatol. 2023;143:602–611.:e614. doi: 10.1016/j.jid.2022.09.006. [* Muti-tissue transcriptome-wide association study for AD found evidence causally implicating AQP3, PDCD1, ADCY3, and DOLPP1] [DOI] [PubMed] [Google Scholar]
- [27].Smits JPH, van den Brink NJM, Meesters LD, et al. Investigations into the FLG Null Phenotype: Showcasing the Methodology for CRISPR/Cas9 Editing of Human Keratinocytes. J Invest Dermatol. 2023;143:1520–1528.:e1525. doi: 10.1016/j.jid.2023.02.021. [** CRISPR/Cas9 editing of FLG in keratinocytes demonstrates the causal functional consequences of FLG knock-out] [DOI] [PubMed] [Google Scholar]
- [28].Minikel EV, Painter JL, Dong CC, Nelson MR. Refining the impact of genetic evidence on clinical success. Nature. 2024 doi: 10.1038/s41586-024-07316-0. [* Study showing that targets with genetic evidence have higher success in clinical trials, but that the dermatology field doesn’t have as many genetically-supported targets as other disease areas] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Keam SJ. Lebrikizumab: First Approval. Drugs. 2024;84:347–353. doi: 10.1007/s40265-024-02000-z. [DOI] [PubMed] [Google Scholar]
- [30].Arehart CH, Daya M, Campbell M, et al. Polygenic prediction of atopic dermatitis improves with atopic training and filaggrin factors. J Allergy Clin Immunol. 2022;149:145–155. doi: 10.1016/j.jaci.2021.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Al-Janabi A, Eyre S, Foulkes AC, et al. Atopic Polygenic Risk Score Is Associated with Paradoxical Eczema Developing in Patients with Psoriasis Treated with Biologics. J Invest Dermatol. 2023;143:1470–1478.:e1471. doi: 10.1016/j.jid.2023.01.021. [* Polygenic risk score for AD is associated with paradoxical eczema in psoriasis patients treated with biologics, but not to a clinically useful level] [DOI] [PubMed] [Google Scholar]
- [32].Kilanowski A, Thiering E, Wang G, et al. Allergic disease trajectories up to adolescence: Characteristics, early-life, and genetic determinants. Allergy. 2023;78:836–850. doi: 10.1111/all.15511. [* Longitudinal trajectories of allergic disease are associated with allergic disease PRS] [DOI] [PubMed] [Google Scholar]
- [33].Lu HF, Chou CH, Lin YJ, et al. The genome-wide association study of serum IgE levels demonstrated a shared genetic background in allergic diseases. Clin Immunol. 2024;260:109897. doi: 10.1016/j.clim.2024.109897. [* GWAS of IgE levels identified 8 genome-wide significant variants] [DOI] [PubMed] [Google Scholar]
- [34].Clabbers J, Boesjes C, Spekhorst L, et al. Influence of pathogenic filaggrin variants on dupilumab treatment in atopic dermatitis. J Allergy Clin Immunol. 2024;153:1155–1161.:e1154. doi: 10.1016/j.jaci.2023.12.027. [* FLG variants are not associated with response to dupilumab treatment] [DOI] [PubMed] [Google Scholar]
- [35].Elhage KG, Kranyak A, Jin JQ, et al. Mendelian Randomization Studies in Atopic Dermatitis: A Systematic Review. J Invest Dermatol. 2024;144:1022–1037. doi: 10.1016/j.jid.2023.10.016. [** Systematic review of 30 Mendelian Randomisation studies involving AD] [DOI] [PubMed] [Google Scholar]
- [36].Huang XW, Pang SW, Yang LZ, et al. TNFSF14 mediates the impact of docosahexaenoic acid on atopic dermatitis: a Mendelian randomization study. Eur Rev Med Pharmacol Sci. 2024;28:107–117. doi: 10.26355/eurrev_202401_34896. [* MR study reporting evidence that TNFS14 and DHA are causal risk factors for AD] [DOI] [PubMed] [Google Scholar]
- [37].Tu J, Wen J, Luo Q, et al. Causal relationships of metabolites with allergic diseases: a transethnic Mendelian randomization study. Respir Res. 2024;25:94. doi: 10.1186/s12931-024-02720-6. [* An MR study of the causal effect of metabolites on AD reports evidence of a causal effect of decreased urinary tyrosine on AD] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Feng F, Li R, Tian R, et al. The causal relationship between gut microbiota and immune skin diseases: A bidirectional Mendelian randomization. PLoS One. 2024;19:e0298443. doi: 10.1371/journal.pone.0298443. [* An MR study showing evidence a causal effect of 7 gut microniota genera on AD] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xue Y, Zhang L, Chen Y, et al. Gut microbiota and atopic dermatitis: a two-sample Mendelian randomization study. Front Med (Lausanne) 2023;10:1174331. doi: 10.3389/fmed.2023.1174331. [* An MR study showing evidence that 6 gut microbiota tax causally increase AD and 7 taxa causally decrease AD] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhong Y, Wang F, Meng X, Zhou L. The associations between gut microbiota and inflammatory skin diseases: a bi-directional two-sample Mendelian randomization study. Front Immunol. 2024;15:1297240. doi: 10.3389/fimmu.2024.1297240. [* An MR study showing evidence that gut microbiota family XIII is causally associated with AD and genus Dialister causally reduces AD risk] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lu F, Wu B, Wang Y. Mendelian randomization indicates that atopic dermatitis contributes to the occurrence of diabetes. BMC Med Genomics. 2023;16:132. doi: 10.1186/s12920-023-01575-y. [* An MR study showing evidence that AD causally increase type 1 and type 2 diabetes risk] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Luo M, Zheng Y, Zhuo Q, et al. The causal effects of atopic dermatitis on the risk of skin cancers: A two-sample Mendelian randomization study. J Eur Acad Dermatol Venereol. 2024;38:703–709. doi: 10.1111/jdv.19674. [* An MR study reporting evidence that AD causally effects the risk of basal cell carcinoma and cutaneous squamous cell carcinoma] [DOI] [PubMed] [Google Scholar]
- [43].O’Hagan R, Caldas SA, Correa da Rosa JM, et al. The impact of atopic dermatitis on alopecia areata: A 2-sample Mendelian randomization study. J Am Acad Dermatol. 2023;89:600–602. doi: 10.1016/j.jaad.2023.05.023. [* An MR study showing evidence that AD is causal for increased alopecia areata risk] [DOI] [PubMed] [Google Scholar]
- [44].Wang Y, Wang S, Wu J, et al. Causal Association Between Allergic Diseases and Dementia: Evidence from Multivariate Mendelian Randomization Study. J Alzheimers Dis. 2024;98:505–517. doi: 10.3233/JAD-231091. [* An MR study reporting evidence that AD is causally related to increased risk of vascular dementia] [DOI] [PubMed] [Google Scholar]
- [45].Zhao J, Zhang M, Li Z. Association Between Immune-Related Disease and Allergic Rhinitis: A Two-Sample Mendelian Randomization Study. Am J Rhinol Allergy. 2024;38:31–37. doi: 10.1177/19458924231207131. [* An MR study reporting causal evidence for the role of AD in allergic rhinitis] [DOI] [PubMed] [Google Scholar]
- [46].Chen Y, Cui L, Li H, Gao A. Abnormal brain structure in atopic dermatitis: Evidence from Mendelian randomization study. Skin Res Technol. 2023;29:e13515. doi: 10.1111/srt.13515. [* An MR study reporting evidence for AD causally increasing brian caudate volume] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [47].Zhou T, Wei B, Hu Y, et al. Causal association between atopic dermatitis and Parkinson’s disease: A bidirectional Mendelian randomization study. Brain Behav. 2024;14:e3468. doi: 10.1002/brb3.3468. [* An MR study reporting that gentic evidence suggests that AD causally decreases the risk of Parkinson’s disease] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jin C, Li Y. No causal association between atopic dermatitis and COVID-19 outcomes: A Mendelian randomization study. Skin Res Technol. 2024;30:e13619. doi: 10.1111/srt.13619. [* A large 2-sample MR study of AD and COVID-19 finds no evidence for a causal relationship] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [49].Luo M, Zhu C, Lin J, et al. Bidirectional Mendelian randomization analysis did not indicate a causal relationship between atopic dermatitis and COVID-19. Br J Dermatol. 2023;189:486–488. doi: 10.1093/bjd/ljad208. [* An MR study found no evidence for a causal realtionship between AD and covid-19] [DOI] [PubMed] [Google Scholar]
- [50].Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR Statement. JAMA. 2021;326:1614–1621. doi: 10.1001/jama.2021.18236. [DOI] [PubMed] [Google Scholar]
- [51].Cao S, Zhang Z, Liu L, et al. Causal relationships between atopic dermatitis and psychiatric disorders: a bidirectional two-sample Mendelian randomization study. BMC Psychiatry. 2024;24:16. doi: 10.1186/s12888-023-05478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang L, Wang Y, Wang XE, et al. Causal association between atopic eczema and inflammatory bowel disease: A two-sample bidirectional Mendelian randomization study of the East Asian population. J Dermatol. 2023;50:327–336. doi: 10.1111/1346-8138.16642. [DOI] [PubMed] [Google Scholar]
- [53].Wang X, Huang Y, Li X, et al. The associations between asthma and common comorbidities: a comprehensive Mendelian randomization study. Front Med (Lausanne) 2023;10:1251827. doi: 10.3389/fmed.2023.1251827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang H, Yuan S, Li Y, et al. Atopic dermatitis and chronic kidney disease: a bidirectional Mendelian randomization study. Front Med (Lausanne) 2023;10:1180596. doi: 10.3389/fmed.2023.1180596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27:R195–R208. doi: 10.1093/hmg/ddy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Luan X, Cui X, Fan L, et al. No Evidence of Causal Association Between Atopic Dermatitis and Primary Open-Angle Glaucoma: A Bidirectional Two-Sample Mendelian Randomization Study. Dermatitis. 2024 doi: 10.1089/derm.2023.0380. [DOI] [PubMed] [Google Scholar]
- [57].Ng W, Loh M, Yew YW. Investigating causal relationships between genetically determined increased risk of attention-deficit/hyperactivity disorder (ADHD) and atopic dermatitis (AD): A Mendelian randomization analysis. Exp Dermatol. 2023;32:1468–1475. doi: 10.1111/exd.14851. [DOI] [PubMed] [Google Scholar]
- [58].Liu Y, Gu Y, Zhou J, et al. Mendelian randomization analysis of atopic dermatitis and esophageal cancer in East Asian and European populations. World Allergy Organ J. 2024;17:100868. doi: 10.1016/j.waojou.2023.100868. [DOI] [PMC free article] [PubMed] [Google Scholar]