Abstract
Biallelic variants in SUMF1 are associated with multiple sulfatase deficiency (MSD), a rare lysosomal storage disorder typically diagnosed in early infancy or childhood, marked by severe neurodegeneration and early mortality. We present clinical and molecular characterisation of three unrelated patients aged 13 to 58 years with milder clinical manifestations due to SUMF1 disease variants, including two adult patients presenting with apparent non-syndromic retinal dystrophy. Whole genome sequencing identified biallelic SUMF1 variants in all three patients; patient 1 homozygous for a complex allele c.[290G>T;293T>A]; p.[(Gly97Val);Val98Glu)], patient 2 homozygous for c.866A>G; p.(Tyr289Cys), and patient 3 compound heterozygous for c.726-1G>C and p.(Tyr289Cys). Electroretinography indicated a rod-cone dystrophy with additional possible inner retinal dysfunction in all three patients. Biochemical studies confirmed reduced, but not absent, sulfatase enzyme activity in the absence of extra-ocular disease (patient 1) or only mild systemic disease (patients 2,3). These cases are suggestive that non-null SUMF1 genotypes can cause an attenuated clinical phenotype, including retinal dystrophy without systemic complications, in adulthood.
Keywords: multiple sulfatase deficiency, retinal dystrophies, MSD, SUMF1, lysosomal storage disorder
Introduction
The SUMF1 (Sulfatase-Modifying Factor 1) gene encodes for the formylglycine-generating enzyme (FGE), which plays a vital role in post-translational modification and catalytic activation of all 17 cellular sulfatase enzymes in humans. These enzymes are essential for multiple cellular functions including hormone regulation, cellular degradation, and modulation of signalling pathways (1). FGE converts a highly-conserved cysteine residue within the catalytic domain of all sulfatase enzymes into Cα-formylglycine, a step necessary for sulfatase enzyme activation (1,2).
Biallelic variants in SUMF1 lead to multiple sulfatase deficiency (MSD, MIM #272200), an extremely rare autosomal recessive multisystem lysosomal storage disorder (LSD), with only approximately 150 affected individuals reported to date (3). In MSD, impairment of FGE function leads to insufficient activation of all sulfatases to varying degrees, with each deficiency contributing to specific clinical manifestations. Ocular features are present in 50-80% of patients with MSD, and include corneal clouding, optic atrophy and retinal dystrophy (3).
This study presents a clinical, biochemical and molecular characterisation of three patients with an attenuated MSD disease phenotype resulting from biallelic SUMF1 variants, including an adult individual with non-syndromic retinal dystrophy, representing the mildest phenotypes associated with SUMF1 gene dysfunction reported to date.
Materials and methods
This study adhered to the tenets of the Declaration of Helsinki and received relevant local research ethics approval. Written informed consent was obtained from all patients and their relatives participating. A comprehensive clinical history was obtained, and in-depth phenotyping, including ophthalmic, systemic and biochemical assessments, was conducted where possible. Whole genome sequencing (WGS) was performed for all three patients. Further details on electrophysiology, biochemical assessments, and genetic testing can be found in the supplemental information.
Results
Clinical presentation
Patient 1 (GC15080) is a 37-year-old Asian Pakistani male, who first noticed a reduction in central vision at the age of 13 years, and was diagnosed with a retinal dystrophy aged 15 years. He suffers from from occasional backache and sciatica but is otherwise fit and well. Parental consanguinity was noted, but there is no family history of vision problems.
Patient 2 (GC19954) is a 59-year-old British Caucasian female diagnosed with retinal dystrophy in her early 40s after experiencing difficulty with reading. She also has glaucoma and Fuch’s corneal endothelial dystrophy. Her medical history includes a deep vein thrombosis and a transient ischaemic attack diagnosed at age 47 and 54 years respectively.
Patient 3 is a 13-year-old British Caucasian male with a complex medical history (see Table 1). His mother and three older siblings have all been diagnosed with Ehlers-Danlos syndrome, and two older siblings have also been diagnosed with autism. While visual symptoms were not initially apparent, the patient reported problems with night and peripheral vision on direct inquiry.
Table 1. Clinical and biochemical studies for patients 1-3.
| Patient 1 (GC15080) | Patient 2 (GC19954) | Patient 3 | |
|---|---|---|---|
| Sex, ethnicity | Male, Asian Pakistani | Female, Caucasian | Male, Caucasian |
| SUMF1 genotype | c. 290G>T; p.(Gly97Val) c.293T>A ; p.(Val98Glu) Homozygous for both variants | c.866A>G ; p.(Tyr289Cys) Homozygous | c.866A>G ; p.(Tyr289Cys) c.726-1G>C Compound heterozygous |
| Presenting symptom (age, y) | Reduced central vision (13) | Reduced reading vision (40s) | Multisystem (childhood) |
| Age at last examination (y) | 37 | 58 | 13 |
| Ocular features | |||
| BCVA (Snellen) | HM both eyes | 3/60 both eyes | 6/7.5 both eyes |
| Cornea | Clear | Endothelial guttata | Clear |
| Lens | Clear | Mild cataracts | Clear |
| Fundus | Central macular atrophy | Mild pigmentary changes at the central macula | Mild RPE mottling at posterior pole |
| Auto-fluorescence | Posterior pole hyper autofluorescence with central mottled hypo autofluorescence | Posterior pole hyper autofluorescence with central mottled hypo autofluorescence | Narrow perimacularring of hyperautofluorescence |
| OCT | Outer retinal disruption | Outer retinal disruption | Perifoveal loss of outer segment structure with a “bulls-ey” appearance |
| ERG | Severe generalised rod photoreceptor dysfunction with mild cone system involvement and severe macular involvement. Reduced DA3 & DA10 ERG b:a ratios (additional inner retinal rod system dysfunction) |
Severe generalised rod/cone photoreceptor dysfunction with severe macular involvement. Reduced DA3, DA10 and LA3 ERG b:a ratios and (additional inner retinal dysfunction) |
LA ERGs normal. DA10 ERG a-wave borderline. Reduced DA3 & DA10 ERG b:a ratios (inner retinal rod system dysfunction) |
| Other | - | Glaucoma, FCED | - |
| Non-ocular features | |||
| Cardiac evaluation | Not available | Echocardiography: LVH with normal systolic function, grade 1 diastolic dysfunction | Normal echocardiography |
| Abdominal evaluation | No organomegaly | Mild fatty liver and gallstones | Small kidneys, mildly enlarged liver with no intrahepatic lesions |
| Skeletal survey | No dysostosis multiplex | No dysostosis multiplex | Scoliosis and subtle thoracolumbar kyphosis. No dysostosis multiplex |
| Neuroimaging | No leukodystrophy, incidental pituitary microadenoma under investigation | Small vessel disease | Mild prominence of perivascular spaces and mild ventriculomegaly involving the lateral ventricles. No emergent features of MSD |
| Other features | - | - | Autism, oral aversion and feeding difficulties, short stature, recurrent ear and throat infections, mild OSA, hypermobility and POTS (EDS type 3) |
| Biochemical testing | |||
| Arylsulphatase A | ↓ ↓ | ↓ ↓ | ↓ ↓ |
| Heparin Sulphamidase | ↓ | ↓ | ↓ ↓ |
| Iduronate Sulphatase |
↓ | Normal | Normal |
| Galactose-6-sulphatase | Normal | Normal | Normal |
| N-acetyl glucosamine-6-sulphatase | Not available | Not available | ↓ ↓ |
| Urinary glycosaminoglycan | Presence of chondroitin sulphate, heparan sulphate and trace dermatan sulphate | Normal | Presence of chondroitin sulphate, heparan sulphate and dermatan sulphate |
Abbreviations: BCVA, best corrected visual acuity; DA, dark-adapted; ECG, electrocardiogram; EDS, Ehlers-Danlos syndrome; ERG, electroretinogram; FCED, Fuchs corneal endothelial dystrophy; LA, light-adapted; LVH, left ventricular hypertrophy; MSD, multiple sulfatase deficiency; OCT, optical coherence tomography, OSA, obstructive sleep apnoea, POTS, postural orthostatic hypotension syndrome, RPE, retinal pigment epithelium; y, year; ↓, reduced ; ↓↓, significantly reduced
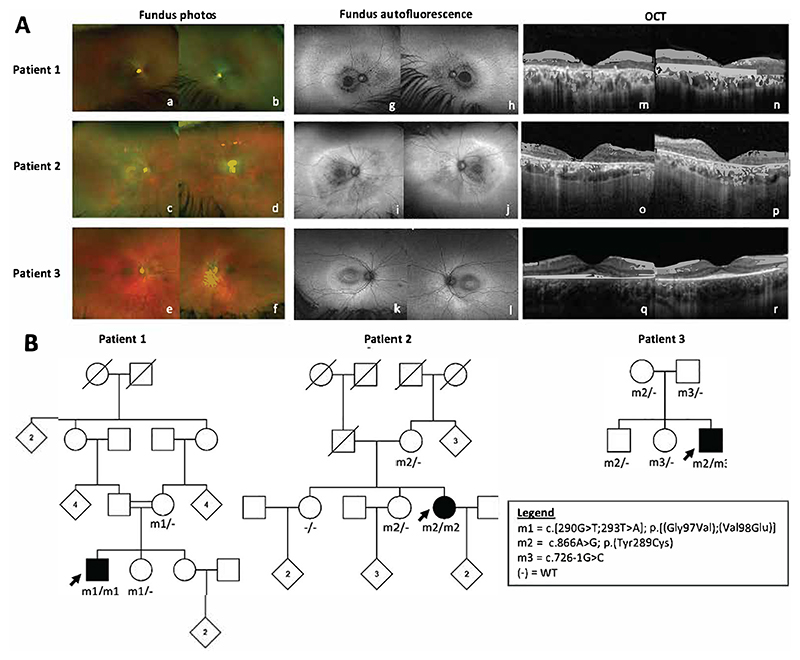
All three patients had a symmetrical retinal dystrophy with macular involvement on ophthalmological examination (Figure 1A). The electroretinogram (ERG) in patients 1 and 2 was indicative of a rod-cone dystrophy with additional inner retinal dysfunction, and with severe macular involvement. The ERG in patient 3 was consistent with severe and selective rod system dysfunction (Supplemental Information).
Figure 1. Clinical findings and family pedigrees for patients 1-3.
(A) Optos pseudocolour fundus photographs (a-f), Optos fundus autofluorescence (g-l) and Spectralis optical coherence tomography (OCT) images (m-r) for patients 1-3 with biallelic SUMF1 variants and retinal dystrophy (B) Family pedigrees showing genotypes and familial segregation where available.
A summary of ocular and systemic findings for all three patients is provided in Table 1.
Genetic results
For patients 1 and 2, WGS and subsequent virtual gene panel analysis excluded pathogenic genotypes in known inherited retinal disease (IRD) genes. Further analysis focused on prioritising rare protein-altering variants with a population frequency of <0.01 in control databases (Genome Aggregation Database; gnomAD v4.0.0, and 100kGP), and the only plausible candidate disease variants identified were located within the SUMF1 gene. Patient
1 was homozygous for a complex allele (NM_182760) c.[290G>T;293T>A]; p.[(Gly97Val);(Val98Glu)], whilst patient 2 was homozygous for missense variant c.866A>G; p.(Tyr289Cys).
WGS in patient 3 identified compound heterozygosity for SUMF1 c.726-1G>C and c.866A>G; p.(Tyr289Cys).
Segregation analysis in all three patients is consistent with the variants being inherited in trans (Figure 1B)
Table 2 provides an overview of population frequency and in silico predictions for all SUMF1 variants reported in this study, none of which have been previously reported in the literature.
Table 2. SUMF1 genetic data for patients 1-3.
| Patient | Ethnicity | Variant (nucleotide) | Variant (protein) | Genotype | gnomAD MAF | In silico predictions † | ClinVar (ID) | ||
|---|---|---|---|---|---|---|---|---|---|
| Revel | AlphaMissense | SpliceAI | |||||||
| 1(GC15080) | Asian Pakistani | c. 290G>T | p.(Gly97Val) | Hom | Absent | Pathogenic (0.928) | Likely pathogenic (0.8002) | Benign (0.01) | Absent |
| c.293T>A | p.(Val98Glu) | Hom | Absent | Uncertain (0.53) | Likely benign (0.1167) | Benign (0) | Absent | ||
| 2 (GC19954) | Caucasian | c.866A>G | p.(Tyr289Cys) | Hom | 0.00001847 | Pathogenic (0.862) | Likely benign (0.299) | Benign (0) | Uncertain significance (1306425) |
| 3 | Caucasian | c.866A>G | p.(Tyr289Cys) | Het | |||||
| c.726-1G>C | - | Het | 0.000003601 | NA | NA | Splice-altering (0.97) | Likely pathogenic (1067238) | ||
REVEL is an ensemble score based on 13 individual scores for predicting the pathogenicity of missense variants. AlphaMissense scores can be interpreted as the approximate probability of a variant being clinically pathogenic. SpliceAI delta scores can be interpreted as the probability that the variant affects splicing at any position within a +/-500bp window around it. Scores for REVEL, AlphaMissense and SpliceAl range from 0 to 1, with higher scores indicating a higher probability of the variant being damaging or having a splice altering effect.
Abbreviations: gnomAD, Genome Aggregation Database v4.0.0; Het, heterozygous, Hom, homozygous; MAF, minor allele frequency
Due to markedly low leucocyte arylsulfatase A (ASA) levels in all three patients, the genomic data was scrutinized for the ASA pseudodeficiency allele ARSA (NM_001085427) c.[1055C>G; *96A>G]; p.[(Asn352Ser);(?)]. Patient 1 was heterozygous for this allele, whilst patients 2 and 3 did not carry this allele.
Discussion
This study describes three individuals with an attenuated systemic phenotype, including non-syndromic retinal dystrophy, due to biallelic variants in SUMF1. SUMF1 variants are typically associated with MSD, marked by severe progressive neurological deterioration and early mortality (4).
In recent years, there is growing recognition that genes initially associated with severe LSDs, such as neuronal ceroid lipofuscinosis (CLN3 and MFSD8) and mucopolysaccharidosis type IIIC (HGSNAT), can also cause non-syndromic retinal dystrophy (5–7). For individual 1 reported in this study, comprehensive clinical evaluations at age 37 years have not identified any extraocular features associated with MSD. This individual with non-syndromic retinitis pigmentosa therefore represents the mildest SUMF1-associated phenotype to date.
SUMF1 variants are typically associated with a variable disease spectrum determined by the severity of FGE protein instability and its residual catalytic ability (4,8). Most SUMF1 variants identified in individuals with MSD are missense variants that likely permit residual enzyme activity, and it has been noted that all MSD patients have reduced but measurable levels of sulfatase activity (9). It is therefore believed that MSD is largely caused by hypomorphic SUMF1 variants; indeed, biallelic loss of function alleles in SUMF1 have only very rarely been reported in cases of hydrops fetalis, and appear incompatible with life (10,11).
Clinical and biochemical analyses support our hypothesis that the novel missense p.(Tyr289Cys) and p.[(Gly97Val);(Val98Glu)] alleles identified in this study are particularly mild alleles, resulting in a limited ocular phenotype (p.[(Gly97Val);(Val98Glu)]; patient 1) or an attenuated MSD phenotype (p.(Tyr289Cys); patients 2 and 3). The biochemical evidence shows reduced enzyme activity across several sulfatases in all three patients, although these levels are not as markedly low as typically seen in classic MSD presentations. Notably, the p.(Tyr289Cys) variant exhibits significant differences in predicted effect according to Revel and AlphaMissense scores; Revel predicts pathogenicity, whilst AlphaMissense suggests it is likely benign. AlphaMissense uniquely incorporates AlphaFold data, which is not included in the 13 tools used to calculate the Revel ensembl score, highlighting the potential for AlphaMissense to provide a more accurate reflection of the variant’s moderate effect. Further detailed structural and biochemical studies investigating the impact of these SUMF1 variants on FGE subcellular localisation, intracellular retention, enzymatic activity, and protein stability would be highly beneficial in understanding the molecular mechanisms through which these hypomorphic variants contribute to milder clinical presentations (12).
Sumf1 knock-out mice have complete deficiency of sulfatase enzyme activity, and exhibit a high mortality rate in the first weeks of life (13). Hypomorphic Sumf1 mice instead show a predominant retinal phenotype with a reduction in rod and cone a- and b-wave on ERG testing, and a reduction in retinal outer nuclear layer thickness consistent with rod and cone degeneration (14). Retinal dystrophies have been described in patients with single sulfatase deficiencies including MPS type II (IDS), III-A (SGSH), III-D (GNS), IV-A (GALNS), X (ARSK) as well as Usher syndrome type IV (ARSG) (15–20), and the retinopathy identified in patients 1-3 are likely due to the resulting deficiencies in one or more of these sulfatases. Within the retina, it is thought that impairment of sulfatase enzyme activity leads to the progressive accumulation of glycosaminoglycans in the retinal pigment epithelial cells as well as the inter-photoreceptor matrix, leading to progressive photoreceptor loss (21). However, the precise pathophysiological mechanisms underlying the selective retinal involvement remains unclear.
These cases reveal the existence of attenuated forms of MSD, expanding the phenotypic spectrum associated with SUMF1 variants, and highlighting a novel association with non-syndromic retinal dystrophy. The retinal phenotype is characterised by posterior pole autofluorescence changes and outer retinal disruption in the macular region, with ERGs consistent with either a rod-cone dystrophy with evidence of additional inner retinal dysfunction, or severe rod system dysfunction.
Classical forms of LSD typically exhibit significantly reduced enzyme activity, while attenuated forms as observed in our case series may retain residual or near-normal levels, limiting the diagnostic utility of biochemical tests. Recognising that attenuated forms of LSD may present with milder symptoms and atypical blood or imaging findings is essential. SUMF1 and other LSD genes linked to retinal phenotypes should be incorporated into IRD gene panels for accurate molecular diagnoses, enabling timely referral to metabolic specialists for comprehensive evaluations, management of systemic associations, and facilitating access to promising therapeutic approaches that could improve patient outcomes (22).
Supplementary Material
Acknowledgements
We are grateful to Dr Derek Burke, Chief Biomedical Scientist at the Enzyme Laboratory, Great Ormond Street Hospital, for his invaluable expertise in interpreting the enzyme results in this paper.
Supported by the Wellcome Trust (grant nos.: 206619/Z/17/Z [OM] and 205174/Z/16/Z [MM]); Fight for Sight UK (Early Career Investigator Award [grant no. 5045/46, GA]); the National Institute of Health Research Biomedical Research Centre (NIHR-BRC) at Moorfields Eye Hospital and UCL Institute of Ophthalmology (grant no NIHR203322, AGR) and Great Ormond Street; and Steven and Elizabeth Archer in memory of Marion Woods (GA).
This research was made possible through access to data in the National Genomic Research Library via the Genomics England Research Environment.
The funding organisations had no role in the design or conduct of this research. The views expressed are those of the author(s) and not necessarily those of the funding organisations, NHS, NIHR or the Department of Health.
Footnotes
Conflicts of interest: none declared.
Author contributions
Conception and design: SL, ARW
Data collection: All authors
Analysis and interpretation: All authors
Writing – original draft preparation: SL, OAM, GA, ARW
Writing – review and editing: All authors
Supervision and overall responsibility: OM, GA, ARW
Data availability statement
Further details of the genome sequencing data presented in the study are available via direct contact with the corresponding author.
References
- 1.Landgrebe J, Dierks T, Schmidt B, von Figura K. The human SUMF1 gene, required for posttranslational sulfatase modification, defines a new gene family which is conserved from pro-to eukaryotes. Gene. 2003 Oct 16;316:47–56. doi: 10.1016/s0378-1119(03)00746-7. [DOI] [PubMed] [Google Scholar]
- 2.Indika NLR, Uçar KT, Law EC, Senarathne UD, Doery JCG, Stepien KM. Genetic Syndromes. Springer International Publishing; Cham: 2023. Multiple Sulfatase Deficiency (MSD) pp. 1–6. [Internet]. Available from: https://link.springer.com/10.1007/978-3-319-66816-1_1774-1. [Google Scholar]
- 3.Schittkowski MP, Naxer S, Elabbasy M, et al. Multiple Sulfatase Deficiency from an Ophthalmologist’s Perspective-Case Report and Literature Review. Children (Basel) 2023 Mar 21;10(3) doi: 10.3390/children10030595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adang LA, Schlotawa L, Groeschel S, et al. Natural history of multiple sulfatase deficiency: Retrospective phenotyping and functional variant analysis to characterize an ultra-rare disease. J Inherit Metab Dis. 2020 Nov;43(6):1298–309. doi: 10.1002/jimd.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F, Wang H, Tuan HF, et al. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet. 2014 Mar;133(3):331–45. doi: 10.1007/s00439-013-1381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roosing S, van den Born LI, Sangermano R, et al. Mutations in MFSD8, encoding a lysosomal membrane protein, are associated with nonsyndromic autosomal recessive macular dystrophy. Ophthalmology. 2015 Jan;122(1):170–9. doi: 10.1016/j.ophtha.2014.07.040. [DOI] [PubMed] [Google Scholar]
- 7.Schiff ER, Daich Varela M, Robson AG, et al. A genetic and clinical study of individuals with nonsyndromic retinopathy consequent upon sequence variants in HGSNAT, the gene associated with Sanfilippo C mucopolysaccharidosis. Am J Med Genet C Semin Med Genet. 2020 Sep;184(3):631–43. doi: 10.1002/ajmg.c.31822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlotawa L, Preiskorn J, Ahrens-Nicklas R, et al. A systematic review and meta-analysis of published cases reveals the natural disease history in multiple sulfatase deficiency. J Inherit Metab Dis. 2020 Nov;43(6):1288–97. doi: 10.1002/jimd.12282. [DOI] [PubMed] [Google Scholar]
- 9.Annunziata I, Bouchè V, Lombardi A, Settembre C, Ballabio A. Multiple sulfatase deficiency is due to hypomorphic mutations of the SUMF1 gene. Hum Mutat. 2007 Sep;28(9):928. doi: 10.1002/humu.9504. [DOI] [PubMed] [Google Scholar]
- 10.Al-Kouatly HB, Makhamreh MM, Rice SM, et al. High diagnosis rate for nonimmune hydrops fetalis with prenatal clinical exome from the Hydrops-Yielding Diagnostic Results of Prenatal Sequencing (HYDROPS) Study. Genet Med. 2021 Jul;23(7):1325–33. doi: 10.1038/s41436-021-01121-0. [DOI] [PubMed] [Google Scholar]
- 11.Schlotawa L, Dierks T, Christoph S, et al. Severe neonatal multiple sulfatase deficiency presenting with hydrops fetalis in a preterm birth patient. JIMD Rep. 2019 Sep;49(1):48–52. doi: 10.1002/jmd2.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlotawa L, Steinfeld R, von Figura K, Dierks T, Gärtner J. Molecular analysis of SUMF1 mutations: stability and residual activity of mutant formylglycine-generating enzyme determine disease severity in multiple sulfatase deficiency. Hum Mutat. 2008 Jan;29(1):205. doi: 10.1002/humu.9515. [DOI] [PubMed] [Google Scholar]
- 13.Settembre C, Annunziata I, Spampanato C, et al. Systemic inflammation and neurodegeneration in a mouse model of multiple sulfatase deficiency. Proc Natl Acad Sci U S A. 2007 Mar 13;104(11):4506–11. doi: 10.1073/pnas.0700382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorrentino NC, Presa M, Attanasio S, et al. New mouse models with hypomorphic SUMF1 variants mimic attenuated forms of multiple sulfatase deficiency. J Inherit Metab Dis. 2023 Mar;46(2):335–47. doi: 10.1002/jimd.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verheyen S, Blatterer J, Speicher MR, et al. Novel subtype of mucopolysaccharidosis caused by arylsulfatase K (ARSK) deficiency. J Med Genet. 2022 Oct;59(10):957–64. doi: 10.1136/jmedgenet-2021-108061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khateb S, Kowalewski B, Bedoni N, et al. A homozygous founder missense variant in arylsulfatase G abolishes its enzymatic activity causing atypical Usher syndrome in humans. Genet Med. 2018 Sep;20(9):1004–12. doi: 10.1038/gim.2017.227. [DOI] [PubMed] [Google Scholar]
- 17.Hendriksz CJ, Al-Jawad M, Berger KI, et al. Clinical overview and treatment options for non-skeletal manifestations of mucopolysaccharidosis type IVA. J Inherit Metab Dis. 2013 Mar;36(2):309–22. doi: 10.1007/s10545-012-9459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkin J, Kerr NC, Byrd KW, Ward JC, Iannaccone A. Characterization of a Case of Pigmentary Retinopathy in Sanfilippo Syndrome Type IIIA Associated with Compound Heterozygous Mutations in the SGSH Gene. Ophthalmic Genet. 2016 Jun;37(2):217–27. doi: 10.3109/13816810.2015.1028647. [DOI] [PubMed] [Google Scholar]
- 19.Liang F, Audo I, Sahel JA, Paques M. Retinal degeneration in mucopolysaccharidose type II. Graefes Arch Clin Exp Ophthalmol. 2013 Jul;251(7):1871–2. doi: 10.1007/s00417-012-2215-1. [DOI] [PubMed] [Google Scholar]
- 20.Nijmeijer SCM, van den Born LI, Kievit AJA, et al. The attenuated end of the phenotypic spectrum in MPS III: from late-onset stable cognitive impairment to a non-neuronopathic phenotype. Orphanet J Rare Dis. 2019 Nov 12;14(1):249. doi: 10.1186/s13023-019-1232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashworth JL, Biswas S, Wraith E, Lloyd IC. Mucopolysaccharidoses and the eye. Surv Ophthalmol. 2006;51(1):1–17. doi: 10.1016/j.survophthal.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Fernández-Pereira C, Millán-Tejado B, Gallardo-Gómez M, et al. Therapeutic Approaches in Lysosomal Storage Diseases. Biomolecules. 2021 Nov 26;11(12) doi: 10.3390/biom11121775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further details of the genome sequencing data presented in the study are available via direct contact with the corresponding author.