Scientists found a new miRNA involved in macrophage responses, paving the way for an improved management of inflammatory diseases.
Abstract
Unbalanced immune responses to pathogens can be life-threatening although the underlying regulatory mechanisms remain unknown. Here, we show a hypoxia-inducible factor 1α–dependent microRNA (miR)–210 up-regulation in monocytes and macrophages upon pathogen interaction. MiR-210 knockout in the hematopoietic lineage or in monocytes/macrophages mitigated the symptoms of endotoxemia, bacteremia, sepsis, and parasitosis, limiting the cytokine storm, organ damage/dysfunction, pathogen spreading, and lethality. Similarly, pharmacologic miR-210 inhibition improved the survival of septic mice. Mechanistically, miR-210 induction in activated macrophages supported a switch toward a proinflammatory state by lessening mitochondria respiration in favor of glycolysis, partly achieved by downmodulating the iron-sulfur cluster assembly enzyme ISCU. In humans, augmented miR-210 levels in circulating monocytes correlated with the incidence of sepsis, while serum levels of monocyte/macrophage-derived miR-210 were associated with sepsis mortality. Together, our data identify miR-210 as a fine-tuning regulator of macrophage metabolism and inflammatory responses, suggesting miR-210–based therapeutic and diagnostic strategies.
INTRODUCTION
The inflammatory response to the entry of pathogens is a complex and multistep process that needs to be tightly regulated since a trade-off between removal of the harmful stimulus and self-preservation must be established to return to tissue homeostasis (1). Notably, an unbalanced host immune response to infections can also culminate in sepsis, a deadly, life-threatening pathological syndrome (2) that can also be secondary to other diseases such as cancer (3).
Innate immune cells, most importantly macrophages, initiate a type 1 inflammatory response to infections by sensing the presence of invading pathogens (namely, bacteria, viruses, and parasites) via pattern recognition receptors, that recognize pathogen-associated molecular patterns such as lipopolysaccharide (LPS) (4). Following their recognition, monocytes extravasate and differentiate into macrophages, starting to produce effector molecules including chemokines and cytokines [e.g., interferon-γ (IFN-γ), interleukin-6 (IL-6), IL-1β, and tumor necrosis factor–α (TNF-α)] that activate other immune cell populations and endothelial cells (1). To fuel the higher biomass and the increased energy demands as well as to produce anti- or proinflammatory metabolites (e.g., itaconate or succinate, respectively), activated immune cells rewire their metabolism (5, 6). For example, it is known that LPS-stimulated macrophages increase tremendously their glucose consumption rate at the expense of oxidative metabolism (7). However, the molecular mechanisms underlying this functional switch are not yet completely unraveled, and it remains an open question whether metabolic changes occurring upon macrophage activation are epistatic to their phenotypic rewiring. These are important issues since deciphering these processes might pave the way toward the development of new therapeutic strategies.
MicroRNAs (miRs) are small, single-stranded noncoding RNA molecules that mediate posttranscriptional repression by targeting specific mRNAs in a sequence-dependent manner (8). MiRs, for instance, miR-155 and miR-146a, have been well recognized for their important contribution to inflammatory responses (9). However, miRs are only recently emerging as mediators of metabolic rewiring in immune cells (8, 10).
Here, we characterize miR-210, classically studied in conditions of low oxygen tension such as cancer and ischemia (11, 12) and for this reason renamed hypoxia-miR (13, 14) as an important executor of the switch to classical (M1) activation and, consequently, of the inflammatory responses in macrophages. Several metabolic targets such as the integral components of the ETC (electron transport chain) NDUFA4 (15), succinate dehydrogenase complex subunit D (15), cytochrome c oxidase 10 (16), and proteins involved in the synthesis and maturation of cofactors essential for the ETC such as ISCU (iron-sulfur cluster assembly enzyme) (16, 17) have been identified mostly in cancer cells. In contrast, although an in vitro study argues in favor of miR-210 as a potential negative regulator of proinflammatory cytokines (18), there are no data investigating its involvement in the metabolic rewiring of macrophages, how this is linked to their phenotypic switch, and how the entire cascade is relevant for disease outcome. By using genetic and pharmacologic approaches to target miR-210 in mice, we show the metabolic and immunological implications of miR-210 in macrophages in the context of several infectious/inflammatory disease models. Moreover, we provide a proof of evidence that miR-210 might be a suitable target for the treatment and management of these diseases. Last, we tentatively transfer this knowledge to the human setting by studying miR-210 in cultured human macrophages and correlating miR-210 levels in circulating monocytes and serum of patients with sepsis.
RESULTS
MiR-210 deletion in the hematopoietic lineage confers protection against sepsis and parasitic infections
We started by assessing the functional relevance of miR-210 expression in the immune system in vivo during various inflammatory/infectious events. For this purpose, we reconstituted lethally irradiated C57BL/6 wild-type (WT) recipient mice with bone marrow cells derived from either miR-210 WT or knockout (KO) donors. Both reconstituted WT and KO chimeras (WT➔WT and miR-210 KO➔WT, respectively) displayed normal blood counts when compared to control mice (table S1).
We then applied a single-shot intraperitoneal LPS injection mimicking an endotoxin-septic shock. Here, we found that loss of miR-210 expression in the hematopoietic lineage was protective against LPS-induced endotoxemia and substantially enhanced the survival of miR-210 KO chimeras (Fig. 1A). A less severe response to this systemic inflammation was further supported by a milder drop in body temperature (Fig. 1B).
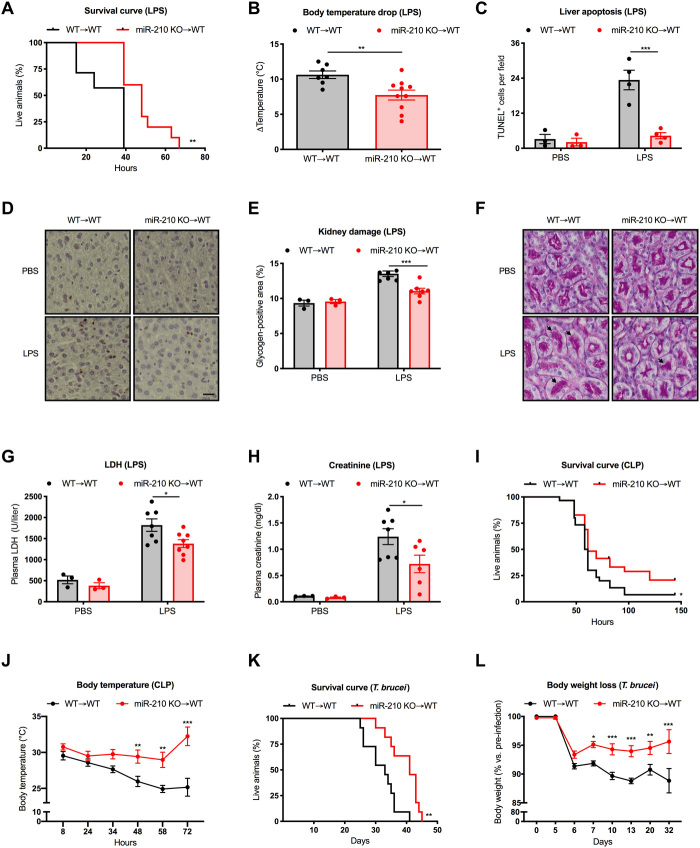
Fig. 1. MiR-210 deletion in the hematopoietic lineage increases survival and mitigates the symptoms of inflammatory diseases.
(A and B) Survival (A) and delta body temperature (°C) (B) of WT➔WT (n = 7) and miR-210 KO➔WT (n = 10) mice injected intraperitoneally with LPS. (C and D) Quantification (C) and representative images (D) of liver apoptosis assessed by TUNEL staining of WT➔WT and miR-210 KO➔WT (PBS = 3 and 3; LPS = 4 and 4). (E and F) Quantification (E) and representative images (F) of glycogen-positive areas in kidneys on periodic acid–Schiff (PAS)–stained kidney sections from WT➔WT and miR-210 KO➔WT mice (PBS = 3 and 3; LPS = 7 and 7). Arrows indicate PAS-positive areas of renal tubules. (G and H) Concentration of plasma LDH (PBS = 3 and 3; LPS = 7 and 8) and creatinine (PBS = 3 and 3; LPS = 7 and 6) in WT➔WT and miR-210 KO➔WT mice. (I) Survival of WT➔WT (n = 30) and miR-210 KO➔WT (n = 29) mice subjected to CLP. (J) Representative body temperature (°C) of WT➔WT (n = 20) and miR-210 KO➔WT (n = 20) mice subjected to CLP. (K) Survival of WT➔WT (n = 11) and miR-210 KO➔WT (n = 11) mice infected with Trypanosoma brucei parasites. (L) Body weight loss (% versus pre-infection) of WT➔WT (n = 16) and miR-210 KO➔WT (n = 15) mice infected with T. brucei parasite. Temperature, histology, and plasma analysis (B to H) were assessed 18 hours after LPS injection. Data show a representative of two (A, B, and K) or a pool of three (I) or two (J) independent experiments. P value was assessed by Log-rank (Mantel-Cox) test (A, I, and K); two-tailed, unpaired, Student’s t test (B); and two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test (C, E, G, H, J, and L). Statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001; graphs show means ± SEM. Scale bar, 100 μm.
Mortality in LPS-induced endotoxemia is generally due to the overwhelming inflammatory response that leads to a progressive organ damage, ultimately resulting in organ dysfunction. Following LPS challenge, livers from miR-210 KO chimeras exhibited fewer apoptotic hepatocytes compared to controls, as assessed by TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) staining on liver sections (Fig. 1, C and D). Similarly, the histological analysis of kidneys revealed a reduced glycogen deposition after LPS challenge in KO chimeras pointing to milder renal damage in LPS-challenged miR-210 KO chimeras (Fig. 1, E and F). In line with these results, we also observed reduced lactate dehydrogenase (LDH) and creatinine plasma levels in miR-210 KO mice indicating a better preservation of liver and kidney functionality, respectively (Fig. 1, G and H).
To evaluate the role of miR-210 in a more clinically relevant model, we adopted the cecal ligation and puncture (CLP) protocol as an experimental model for polymicrobial sepsis (19). Consistently, also in this model, we observed an increased survival of KO versus WT chimeras, associated with a milder drop in body temperature, further supporting the protective function against septic shock conferred by hematopoietic miR-210 depletion (Fig. 1, I and J).
Both LPS-induced endotoxemia and CLP models are fast and acute disease models. We aimed to understand whether miR-210 deficiency could affect the progression and/or outcome of a disease that leads to a chronic state. In addition, we questioned whether miR-210 could be involved in parasite-mediated infectious diseases and not only during the course of bacterial infections. Therefore, we opted for a parasite disease model and specifically for an infection with Trypanosoma brucei, an extracellular protozoan that, in humans, leads to African trypanosomiasis (HAT), better known as sleeping sickness (20). To evaluate the functional role of miR-210 in this model, we intraperitoneally injected the T. brucei AnTat1.1E strain in WT and miR-210 KO chimeras and we followed the survival as well as the main clinical disease symptoms (20). Although miR-210 KO chimeras displayed only mild differences in parasitemia levels during T. brucei infection at days 5 and 20 (fig. S1A), miR-210 ablation in the hematopoietic lineage conferred disease protection and increased survival (Fig. 1K). This disease protection consisted in a milder body weight loss as well as a less severe anemia during the chronic stage of the disease (Fig. 1L and fig. S1B), two critical immunopathological features associated with HAT. Overall, these data demonstrate that miR-210 deletion in the immune system is able to protect the organism against endotoxemia and parasitic infections, both characterized by a proinflammatory immune response.
MiR-210 is up-regulated in monocytes and macrophages in response to pathogens
To understand which immune cell type was mostly involved in our phenotype, we analyzed miR-210 expression levels among different immune cell populations (B cells, T cells, neutrophils, and macrophages) isolated from peritoneal lavages at baseline and after LPS administration. Notably, macrophages were among the immune cells with the highest miR-210 levels at baseline and the only cell type that showed an induction upon LPS challenge (Fig. 2A). At baseline, the number of neutrophils was too low to reliably assess miR-210 levels (Fig. 2A). The representation of different immune cell populations in the peritoneal fluid from chimeric WT versus miR-210 KO mice before or after LPS challenge did not change (fig. S2, A and B). Consistently, circulating monocytes in the blood displayed both the highest basal levels and the strongest up-regulation of miR-210 in response to a challenge with LPS (Fig. 2B). We further confirmed the up-regulation of miR-210 in macrophages and circulating monocytes isolated, upon in vivo exposure to LPS, either by selective adhesion of peritoneal macrophages to nontreated culture dishes or by CD115+ magnetic beads, respectively (fig. S2, C and D). All these expression data suggest a possible role of miR-210 in monocyte/macrophage responses during LPS-induced endotoxemia.
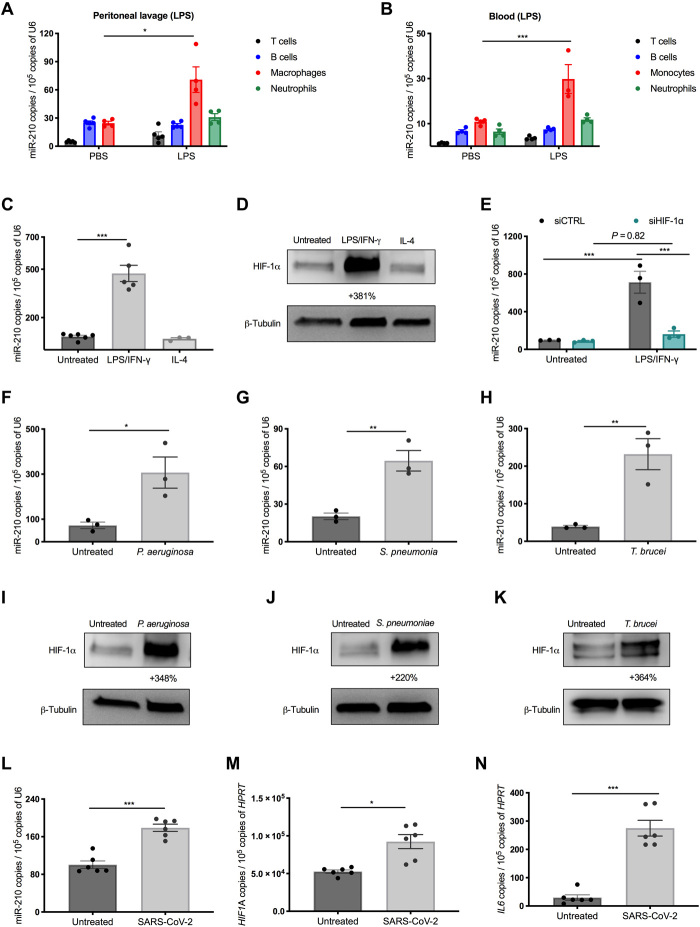
Fig. 2. MiR-210 is up-regulated both in vivo and in vitro in response to pathogens.
(A and B) MiR-210 levels [quantitative real-time PCR (qRT-PCR)] in CD45+ sorted peritoneal (A) and circulating blood cells (B) 18 hours after intraperitoneal injection of LPS or phosphate-buffered saline (PBS): T cells (TCR β+), B cells (B220+), neutrophils (F4/80−, Ly6G+), macrophages (Ly6G−, F4/80+), or monocytes (Ly6G−, CD115+). (C and D) MiR-210 levels (qRT-PCR) (C) and hypoxia-inducible factor 1α (HIF-1α) protein (Western blot) (D) in bone marrow–derived macrophages (BMDMs), untreated or after LPS/IFN-γ or IL-4 stimulation. The percentage attached to the blot indicates densitometric changes. (E) MiR-210 levels (qRT-PCR) in BMDMs untreated or after 24-hour LPS/IFN-γ stimulation. BMDMs were electroporated 24 hours before LPS/IFN-γ treatment with small interfering RNAs (siRNAs) against HIF-1α or a control sequence. (F to K) MiR-210 levels (qRT-PCR) (F to H) and HIF-1α protein (Western blot) (I to K) in BMDMs cocultured with either heat-killed Pseudomonas aeruginosa (F and I) or heat-killed Streptococcus pneumoniae (G and J) or stimulated with lysate from T. brucei (H and K). The percentage attached to the blot indicates densitometric changes. (L to N) MiR-210 (L), HIF1A (M), and IL6 (N) mRNA levels (qRT-PCR) in human monocyte-derived macrophages (hMDMs) untreated or exposed for 12 hours to the SARS-CoV-2 spike protein. For all the in vitro analyses, unless specified in the legend (E and L to N), the treatment (C, D, F to K) was 44 hours. Data show a pool (A, C, and L to N) or a representative (B) of two independent experiments. P value was assessed by multiple Student’s t test (A); two-way ANOVA with Sidak’s (B) and Tukey’s (E) multiple comparisons test; one-way ANOVA with Dunnett’s multiple comparison test (C); and two-tailed, unpaired, Student’s t test (F to H and L to N). Statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001; graphs show means ± SEM.
To better investigate miR-210 regulation in macrophages, we moved to an in vitro system. We found that miR-210 levels were up-regulated in bone marrow–derived macrophages (BMDMs) after stimulation with bacterial/proinflammatory (M1) stimuli, consisting in the combination of LPS and IFN-γ but not in response to LPS or IFN-γ alone (Fig. 2C and fig. S2E). In contrast, anti-inflammatory stimuli such as IL-4 did not significantly affect miR-210 levels (Fig. 2C). A similar induction of miR-210 expression was observed in peritoneal macrophages in response to LPS/IFN-γ, but not after exposure to anti-inflammatory cytokines such as IL-4 or IL-13 (fig. S2F). Although the increased miR-210 levels by the combination of LPS/IFN-γ stimulation may seem to contradict the in vivo evidence showing miR-210 induction by LPS alone, it is well documented that IFN-γ is acutely released by the host upon LPS challenge (21).
The miR-210 gene locus is characterized by a hypoxia-response element, a promoter region recognized and bound by the transcription factor hypoxia-inducible factor 1α (HIF-1α), and consequently, HIF-1α works as a paramount miR-210 regulator (13, 14). Despite the name, HIF-1α can be regulated by many other factors besides low oxygen tension such as inflammatory stimuli (22). Consistent with the literature and with the expression pattern of miR-210 (23, 24), we found that HIF-1α was synergistically stabilized by the combination of LPS and IFN-γ, but much less by each of them alone and not at all by IL-4 (Fig. 2D and fig. S2G). In support of the involvement of HIF-1α in the induction of miR-210, silencing of HIF-1α by small interfering RNAs (siRNAs) abolished miR-210 up-regulation in BMDMs upon LPS/IFN-γ stimulation (Fig. 2E and fig. S2H).
MiR-210 was up-regulated in BMDMs not only by the Gram-negative bacteria component LPS but also by exposure to heat-inactivated Gram-negative bacteria, such as Pseudomonas aeruginosa, heat-killed Gram-positive Streptococcus pneumoniae [leading cause of human sepsis (25)], and T. brucei lysates (Fig. 2, F to H). All of these pathogenic stimuli promoted HIF-1α accumulation as well (Fig. 2, I to K). Recent findings show that HIF-1α is also stabilized in monocytes in response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (26). We found that in vitro culture of human monocyte-derived macrophages (hMDMs) exposed to the spike protein of SARS-CoV-2, together with increasing transcript levels of HIF1A, up-regulates miR-210 and proinflammatory but not anti-inflammatory cytokines (Fig. 2, L to N, and fig. S2I). Therefore, miR-210 is specifically induced in monocytes and macrophages upon their interaction with (components of) both Gram-positive and Gram-negative bacteria, T. brucei, and SARS-CoV-2, most likely in an HIF-1α–dependent manner.
Macrophage-specific or systemic targeting of miR-210 is sufficient to protect mice from endotoxic shock and infections
On the basis of our in vivo and in vitro findings, showing that miR-210 expression is strongly regulated in macrophages upon pathogen interaction, we reasoned to specifically delete miR-210 expression in macrophages in vivo to evaluate the functional role of miR-210 in these cells. To this aim, we crossed miR-210 floxed mice with an inducible, macrophage-specific CSF1R:Cre-ERT (iCre) obtaining miR-210 ΔM0 (iCre-positive mice) mice and controls (iCre-negative mice)—a strategy that, in comparison to the bone marrow chimera approach, circumvents possible procedural side effects. Tamoxifen administration effectively resulted in locus recombination (Fig. 3A). Then, we challenged these mice with a one-shot peritoneal LPS injection. Loss of miR-210 selectively in macrophages was sufficient to protect mice from the septic shock induced by the Gram-negative component LPS (Fig. 3B). Then, we tested the in vivo relevance of miR-210 during Gram-positive infection of S. pneumoniae. As a systemic readout of inflammation, we measured the body temperature before and after intratracheal injection of a high dose of S. pneumoniae, which substantially dropped in control but not in miR-210 ΔM0 mice (Fig. 3C). Compared to controls, miR-210 ΔM0 mice showed reduced counts of bacterial CFUs (i.e., colony-forming units) in the lung and in the blood (Fig. 3D). These data suggest that miR-210 depletion in macrophages prevents the deleterious effects of a septic shock and, at the same time, it retains improved pathogen control.
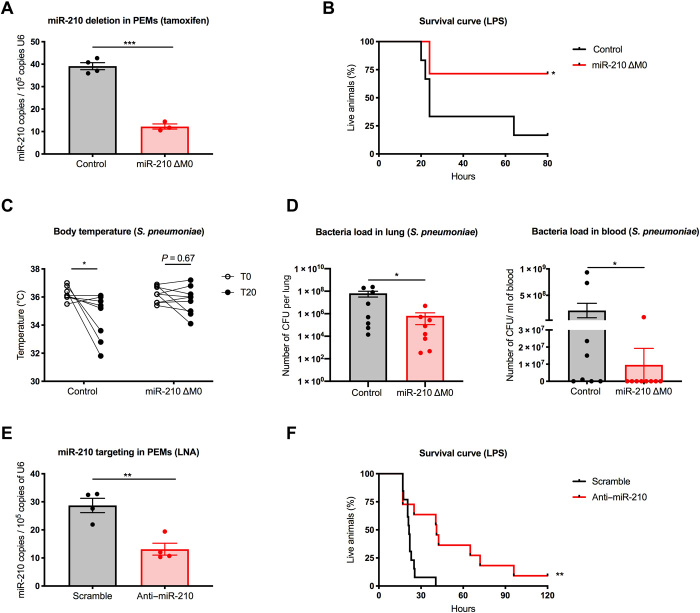
Fig. 3. Inducible deletion in macrophages or systemic pharmacologic inhibition of miR-210 offers a therapeutic advantage in response to pathogens.
(A) MiR-210 levels (qRT-PCR) in sorted peritoneal macrophages (CD45+, Ly6G−, and F4/80+) from control (n = 4) and miR-210 ΔM0 (n = 3) mice 5 days after tamoxifen administration. (B) Survival of control (n = 6) and miR-210 ΔM0 (n = 7) mice injected intraperitoneally with LPS. (C) Body temperature in control (n = 8) and miR-210 ΔM0 (n = 9) mice at baseline (T0) and 20 hours after S. pneumoniae infection (T20). (D) Viable bacterial counts recovered from homogenized lung (left) or whole blood (right) quantified as CFU from control (n = 8) and miR-210 ΔM0 (n = 9) mice 20 hours after S. pneumoniae infection. (E) MiR-210 levels (qRT-PCR) in sorted peritoneal macrophages (CD45+, Ly6G−, and F4/80+) after 62 hours of a single intravenous administration of scramble (n = 4) or anti-miR-210 LNA (n = 4). (F) Survival of C57BL/6 mice injected intravenously with scramble LNA (n = 11) or anti-miR-210 LNA (n = 13) 48 hours before intraperitoneal LPS injection. Data show a representative (B) or a pool (C, D, and F) of two independent experiments. P value was assessed by two-tailed, unpaired, Student’s t test (A and E); Log-rank (Mantel-Cox) test (B and F); two-way RM ANOVA with Sidak’s multiple comparisons test (C); and Mann-Whitney U test (D). Statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001; graphs show means ± SEM.
From a therapeutic point of view, a macrophage-specific approach is very challenging. However, noncoding RNAs can be blocked by the systemic delivery of complementary antisense sequences of locked nucleic acids (LNAs), which are modified RNA structures with increased stability against enzymatic degradation (27). Therefore, we tested the effectiveness of LNA-mediated miR-210 inhibition in the LPS-induced endotoxemia model. Down-regulation of miR-210 by the LNA was confirmed with quantitative real-time PCR (qRT-PCR) on sorted peritoneal macrophages (Fig. 3E). Systemic inhibition of miR-210, obtained by intravenous injection of the LNAs in C57BL/6 WT mice 48 hours before LPS challenge, resulted into a much longer survival compared to control mice that were treated with a scrambled LNA sequence (Fig. 3F). These data pave the way toward miR-210–based targeting approaches for the management of sepsis and other infectious diseases.
MiR-210 orchestrates the glycolytic switch of M1 macrophages
While both anti-inflammatory (M2-polarized) and resting/nonactivated (M0) macrophages rely more on oxidative phosphorylation (OXPHOS), proinflammatory (M1) macrophages respond to external stimuli such as LPS by increasing glycolysis at the expense of OXPHOS (6, 7, 28). However, it is not well known how the latter modulation is controlled. When stimulated with LPS/IFN-γ, WT BMDMs increased glucose utilization as highlighted by a strong augmentation in glycolysis (Fig. 4, A to C). Yet, the increase in glycolysis upon treatment was significantly milder in miR-210 KO macrophages as assessed both by radioactive assays and by mass spectrometry (MS) analysis of intracellular and extracellular lactate accumulation (Fig. 4, A to C). No difference was observed in the untreated condition (Fig. 4, A to C).
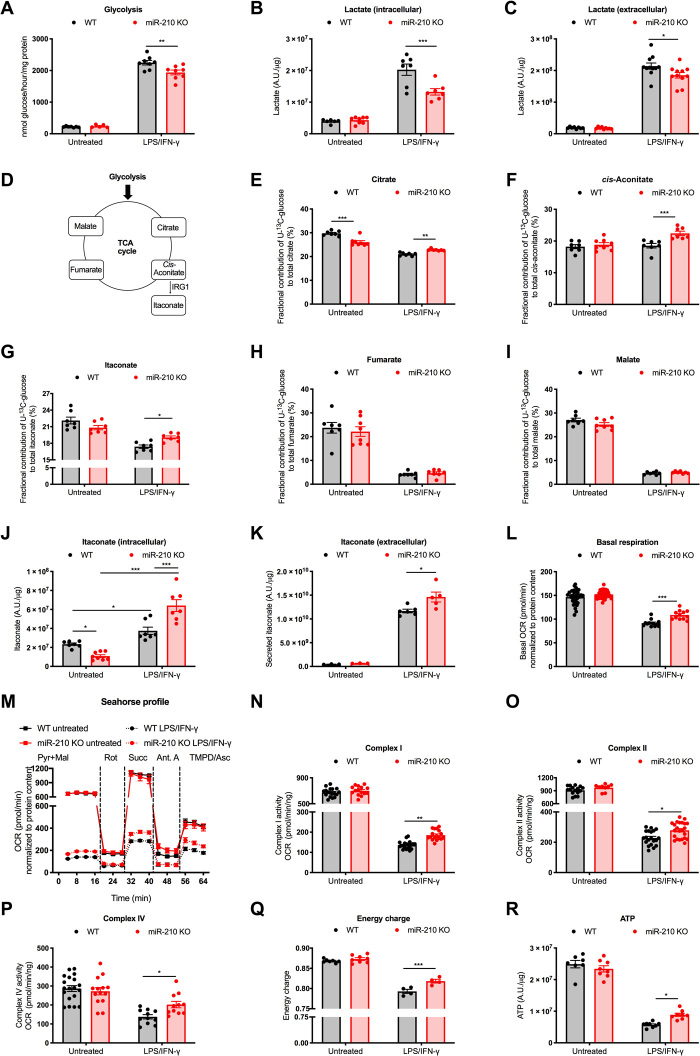
Fig. 4. MiR-210 deletion in macrophages reduces glycolysis while enhancing oxidative metabolism in response to LPS/IFN-γ.
(A) Glycolysis (3H-Glucose) of WT and miR-210 KO BMDMs untreated or after 18-hour LPS/IFN-γ stimulation. (B and C) Intracellular (B) and extracellular (C) lactate (liquid chromatography–mass spectrometry, LC-MS) from WT and miR-210 KO BMDMs untreated or after 24-hour LPS/IFN-γ. (D) Scheme of the TCA cycle and itaconate production. (E to I) Fractional contribution (LC-MS) of U-13C-glucose into different TCA cycle intermediates and derivates: citrate (E), cis-aconitate (F), itaconate (G), fumarate (H), and malate (I) in WT and miR-210 KO BMDMs, untreated or after 24-hour LPS/IFN-γ. (J and K) Intracellular (J) and extracellular (K) itaconate (LC-MS) from WT and miR-210 KO BMDMs untreated or after 24-hour (J) or 48-hour LPS/IFN-γ (K). (L) Basal respiration (seahorse) of WT and miR-210 KO BMDMs, untreated or after 24-hour LPS/IFN-γ. (M to P) A representative oxygen consumption rate (OCR) curve (M) and activity of complex I (N), complex II (O), or complex IV (P) (seahorse) on permeabilized WT and miR-210 KO BMDMs untreated or after 42-hour LPS/IFN-γ in the presence of pyruvate (Pyr) and malate (Mal). Rotenone, succinate, antimycin A, or TMPD + ascorbate was injected at the indicated time point. Complex activities in (N) to (P) were calculated by OCR subtractions. (Q and R) Energy charge (Q) and adenosine triphosphate (ATP) levels (R) (LC-MS) of WT and miR-210 KO BMDMs untreated or after 24-hour LPS/IFN-γ. Data were normalized by protein content (A to C and J to R). Data show a pool of two (A, B, E to K, and M to R) or three (C) or a representative of two (L) independent experiments. P value was assessed by two-way ANOVA with Sidak’s multiple comparisons test (A to R). Statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001; graphs show means ± SEM. A.U., arbitrary units.
When analyzing the fractional contribution of U-13C-glucose to the intermediates of the tricarboxylic acid (TCA) cycle (Fig. 4D), we noticed that 13C labeling of citrate, cis-aconitate, and itaconate, the latter an anti-inflammatory and antimicrobial metabolite (29), was higher in LPS/INF-γ–stimulated miR-210 KO versus WT macrophages (Fig. 4, E to G). Instead, the fractional contribution of U-13C-glucose to fumarate and malate was dropping comparably in the two genotypes (Fig. 4, H and I), consistent with a broken TCA cycle in M1-like macrophages (7). All of these data argue in favor of enhanced glucose utilization into itaconate production through decarboxylation of cis-aconitate. Abundance of itaconate in response to LPS/INF-γ stimulation was increased in WT BMDMs (30), and this increase was even stronger in the absence of miR-210 (Fig. 4J). MS analysis of culture supernatants from LPS/INF-γ–stimulated BMDMs revealed that itaconate was also secreted (Fig. 4K), as also shown before (31). In accordance to the increased production of itaconate, its secretion was also enhanced in miR-210 KO versus WT macrophages (Fig. 4K).
M1-activated macrophages display reduced OXPHOS (6, 7). Consistently, the basal respiration of BMDMs upon administration of LPS/IFN-γ was strongly reduced (Fig. 4L). However, in treated miR-210 KO macrophages, this drop was less severe (Fig. 4L). To better investigate the involvement of the different components of the ETC, we analyzed the oxygen consumption rate (OCR) specifically related to complex I (assessed as the basal OCR in medium containing pyruvate and malate), complex II (assessed by succinate-related OCR, complex II substrate, in the presence of rotenone, complex I inhibitor), and complex IV (assessed by TMPD and ascorbate-related OCR, artificial complex IV substrates, in the presence of the complex III inhibitor antimycin A) on permeabilized macrophages (Fig. 4, M to P). In this setting, untreated WT and miR-210 KO macrophages did not display any significant difference in any of the OCR parameters. However, following LPS/IFN-γ stimulation, the activity of complexes I, II, and IV was reduced in WT BMDMs but significantly less miR-210 KO macrophages (Fig. 4, M to P). As expected (32), LPS/IFN-γ treatment led to a drop of the energy charge (Fig. 4Q), reflected by a reduction in the overall balance of adenosine triphosphate (ATP) (Fig. 4R). Nevertheless, this drop was less pronounced in miR-210 KO versus WT macrophages supporting the increased engagement of the ETC at the expense of glycolysis. Overall, our data argue that miR-210 induction by LPS/IFN-γ is relevant to partly turn off OXPHOS and oxidative metabolism in favor of a pro-glycolytic rewiring, typical of M1 macrophages.
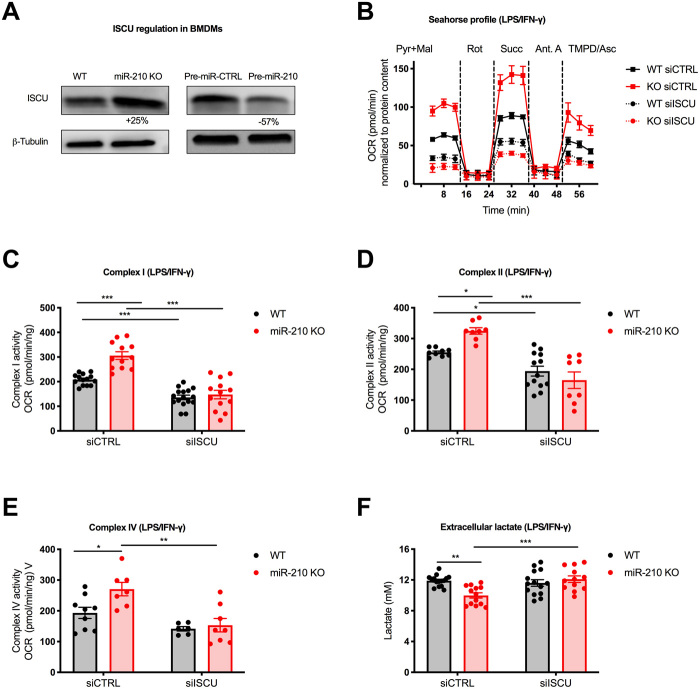
The iron-sulfur (Fe-S) cluster scaffold protein ISCU was shown to be one of the miR-210 targets in cancer and endothelial cells (16, 33). ISCU is involved in both the synthesis and maturation of 2Fe-2S and 4Fe-4S clusters (34). Notably, the ETC has 12 different Fe-S clusters that play an important role in the electron transfer. Therefore, we hypothesized that ISCU could be implicated in the orchestration of the metabolic regulations observed in miR-210 KO BMDMs. Western blot analysis detected increased ISCU protein levels in miR-210 KO versus WT macrophages (Fig. 5A, left). Accordingly, overexpression of miR-210 led to a reduction of the protein level (Fig. 5A, right). We then tested whether the differences in OXPHOS could be reverted by ISCU silencing. Macrophages were silenced by electroporation with either a siRNA control (siCTRL) or a mix of two siRNAs for ISCU. As expected, miR-210 KO macrophages treated with siCTRL displayed an increase in OCR levels for complexes I, II, and IV upon LPS/IFN-γ (Fig. 5, B to E). Silencing of ISCU equalized the OCR of complexes I, II, and IV in miR-210 WT and KO macrophages, diminishing the activity of these complexes down to the same levels in both genotypes (Fig. 5, B to E). Moreover, silencing of ISCU increased the glycolytic flux (measured as lactate release) in miR-210 KO macrophages, equalizing it to the WT levels (Fig. 5F). Together, these data suggest that the metabolic reprogramming of macrophages dictated by miR-210–mediated modulation of ISCU mitigates OXPHOS and, indirectly, favors the glycolytic switch observed in M1-polarized macrophages.
Fig. 5. ISCU modulation by miR-210 mediates the metabolic rewiring in LPS/IFN-γ–stimulated macrophages.
(A) ISCU protein levels (Western blot) in WT and miR-210 KO BMDMs (left) and in WT BMDMs electroporated with pre-control (pre-miR-CTRL) or miR-210 precursor (pre-miR-210) (right). The percentage attached to the blot indicates densitometric changes. (B to E) A representative OCR curve (B) and activity of complex I (C), complex II (D), or complex IV (E) (seahorse) on permeabilized WT and miR-210 KO BMDMs after 42-hour LPS/IFN-γ stimulation in the presence of pyruvate (Pyr) and malate (Mal). Rotenone, succinate, antimycin A, or TMPD + ascorbate was injected at the indicated time point. Complex activities in (C) to (E) were calculated by OCR subtractions. BMDMs were electroporated 24 hours before LPS/IFN-γ treatment with two siRNAs against ISCU or a control sequence. (F) Extracellular lactate concentration (colorimetric assay) in the medium of WT and miR-210 KO BMDMs, after 24-hour LPS/IFN-γ. BMDMs were electroporated 48 hours before LPS/IFN-γ treatment with two siRNAs against ISCU or a control sequence. Data were normalized by protein content (B to F). Data show a representative of two (A) or a pool of two (B to E) or four (F) independent experiments. P value was assessed by two-way ANOVA with Tukey’s multiple comparisons test (A to F). Statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001; graphs show means ± SEM.
MiR-210 enhances M1 responses in macrophages
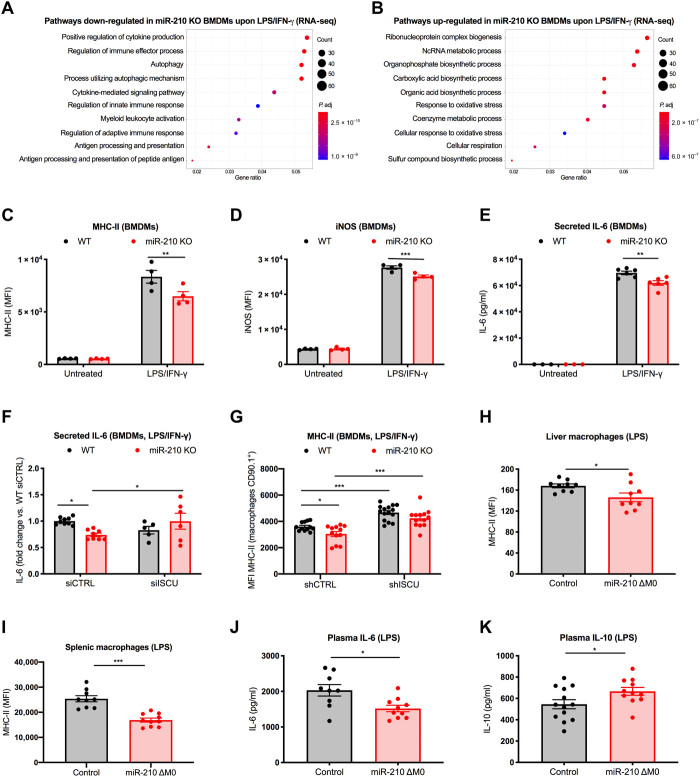
To evaluate the impact of miR-210 on the macrophage phenotype, we performed a genome-wide messenger RNA profiling of either WT or miR-210 KO BMDMs, either untreated or treated with LPS/IFN-γ. In the untreated condition, we identified only 220 differentially expressed genes between WT and miR-210 KO macrophages. On the other hand, 2477 genes were differentially expressed upon LPS/IFN-γ treatment, of which 1301 were down-regulated and 1176 were up-regulated in miR-210 KO macrophages compared to WT macrophages, suggesting an involvement of miR-210 in response to pathogenic stimuli rather than at baseline. Gene ontology analysis revealed that M1-related pathways were less activated in miR-210 KO versus WT macrophages as highlighted by their appearance in the list of the differentially down-regulated genes in the treated condition. Examples of these M1 pathways are the positive regulation of cytokine production, autophagy, cytokine-mediated signaling pathways, antigen processing and presentation, or terms related to general cell activation such as regulation of immune effector processes and regulation of innate immune responses (Fig. 6A). Conversely, gene ontology analysis on the differentially up-regulated genes in the treated condition highlighted cellular respiration and sulfur compound biosynthetic processes among the top 10 enriched pathways in miR-210 KO versus WT BMDMs, supporting the relevance of miR-210 in orchestrating the metabolic reprogramming of macrophages (Fig. 6B).
Fig. 6. MiR-210 deletion in macrophages counters inflammatory pathways and activation markers in response to endotoxin.
(A and B) Top 10 GO terms enriched, based on gene ratio, for significantly down-regulated (A) and up-regulated (B) genes in miR-210 KO versus WT BMDMs after 42-hour LPS/IFN-γ stimulation. Color of the dots indicates adjusted P value. (C and D) MFI of major histocompatibility complex class II (MHC-II) (C) and inducible nitric oxide synthase (iNOS) (D) of WT and miR-210 KO BMDMs, untreated or after 24-hour LPS/IFN-γ. (E) IL-6 concentration (MSD) in the medium of WT and miR-210 KO BMDMs, untreated or after 12-hour LPS/IFN-γ. (F) Fold changes of IL-6 [enzyme-linked immunosorbent assay (ELISA)] in the medium of WT and miR-210 KO BMDMs after 24-hour LPS/IFN-γ. BMDMs were electroporated 48 hours before LPS/IFN-γ treatment with 2 siRNAs against ISCU or a control sequence. (G) MFI of MHC-II of WT and miR-210 KO BMDMs after 24-hour LPS/IFN-γ. Macrophages were transduced with a short hairpin RNA against ISCU or a control sequence. (H) MFI of MHC-II in liver macrophages (CD45+, CD11b+, Ly6G−, and F4/80+) from control (n = 9) and miR-210 ΔM0 (n = 9) mice. (I) MFI of MHC-II in spleen macrophages (CD45+, CD11b+, Ly6G−, and MHC-II+) from control (n = 9) and miR-210 ΔM0 (n = 10) mice. (J and K) IL-6 and IL-10 concentration (ELISA) in the plasma of control (n = 9 to 13) and miR-210 ΔM0 (n = 10 to 11) mice. In vivo analysis was performed 18 hours after sublethal LPS intraperitoneal injection (H to K). Data show a representative of three (C and D) or a pool of two (E and H to K), three (F), or five (G) independent experiments. P value was assessed by two-way ANOVA with Sidak’s multiple comparisons test (C to G) and two-tailed, unpaired, Student’s t test (H to K). Statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001; graphs show means ± SEM.
When looking at differential responses to treatment via proteomics, results were consistent: IFN-γ–mediated signaling pathways, cell activation, response to cytokines, and immune effector processes were less represented in treated versus untreated miR-210 KO macrophages, compared to what was seen in WT BMDMs in response to LPS/IFN-γ (fig. S3A). The most up-regulated terms were instead related to protein localization, which might underline a trend toward an anti-inflammatory, M2 state, as further examined in the discussion (fig. S3B). Overall, these findings suggest that miR-210 induction by LPS/IFN-γ enables a skewing of macrophages toward a functional and metabolic, inflammatory (M1-like) phenotype that exacerbates their responses to pathogenic stimuli.
In line with the omics data, classical M1 features (7, 35), such as major histocompatibility complex class II (MHC-II) expression, inducible nitric oxide synthase (iNOS), and IL-6 secretion (Fig. 6, C to E), were less promoted in miR-210 KO than in WT macrophages upon in vitro treatment with LPS/IFN-γ. ISCU silencing increased IL-6 release by miR-210 KO macrophages to the same levels as in WT macrophages along with increased MHC-II expression in both WT and miR-210 KO macrophages (Fig. 6, F and G). These data underscore the idea that ISCU loss of function can revert, at least in part, the inflammatory response controlled by miR-210 induction.
In vivo flow cytrometry analysis and enzyme-linked immunosorbent assay (ELISA) revealed, respectively, lower MHC-II expression in macrophages from both liver and spleen and reduced plasma levels of IL-6 and TNF-α in LPS-treated macrophage-specific miR-210 KO versus control mice (at a time point when the disease state was similar as assessed by body temperature) (Fig. 6, H to J, and fig. S3C). As a further sign of reduced inflammation, plasma levels of the anti-inflammatory cytokine IL-10 were increased in miR-210 KO versus control mice (Fig. 6K). MiR-210 deletion in macrophages did not alter their infiltration into the liver (fig. S3D) while neutrophil accumulation in response to LPS was reduced, the latter being another sign of a milder inflammatory response (fig. S3E).
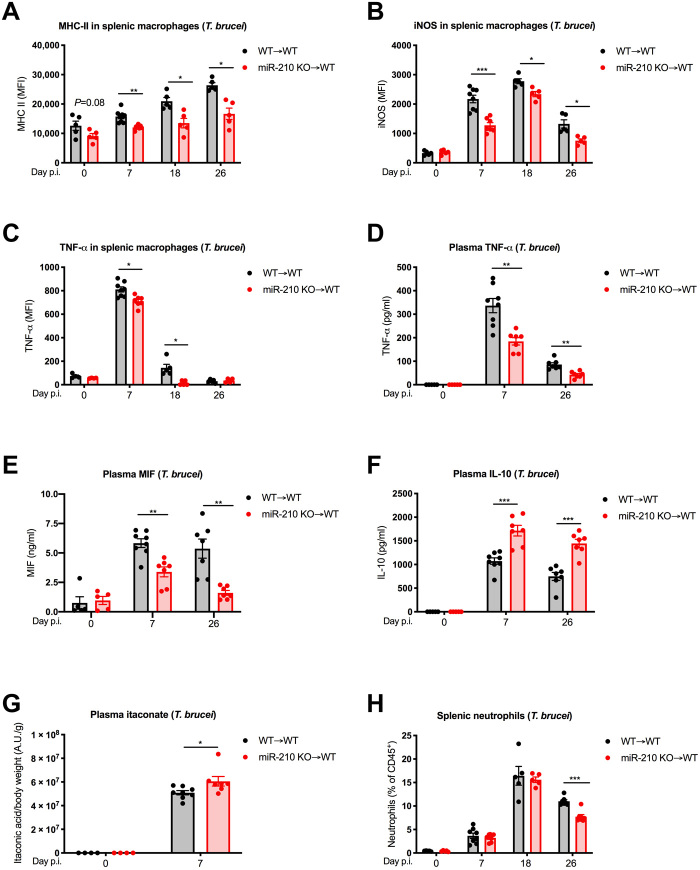
Several signs of reduced inflammation were also found during the course of T. brucei infection. Splenic macrophages from miR-210 KO chimeras displayed a lower induction or no induction of the proinflammatory markers MHC-II, iNOS, and TNF-α, as assessed by FACS both in the acute (day 7) and in the late (days 18 and 26) stage of the infection (Fig. 7, A to C). Along the same line, serum levels of TNF-α and macrophage migration inhibitory factor (MIF), prominent inflammatory mediators during T. brucei infection (36, 37), were lower in miR-210 KO versus WT chimeras (Fig. 7, D and E). Conversely, the anti-inflammatory cytokine IL-10 showed significantly higher plasma levels in miR-210 KO chimeras (Fig. 7F). Moreover, in the acute stage of the disease, the plasma levels of the antimicrobial and anti-inflammatory metabolite itaconate were found to be higher in miR-210 KO versus WT chimeras (Fig. 7G). Despite a change in phenotype, post-infection macrophage numbers in the spleen of miR-210 KO chimeras were following the same trend as in control animals during the infection (fig. S3F). Aligned with a faster resolution of the inflammatory response, neutrophilia was reduced in miR-210 KO versus WT chimeras at the late stage (i.e., day 26 after infection) but not at the early stage after infection (i.e., until day 18 after infection) (Fig. 7H). Overall, miR-210 deletion in macrophages leads to a generally reduced inflammatory response because of the incomplete skewing toward the M1 status, overall explaining the increased survival to endotoxin-mediated septic shock and T. brucei infection.
Fig. 7. MiR-210 deletion reduces macrophage type 1 responses and overall inflammation during T. brucei infection.
(A to C) MFI from WT➔WT and miR-210 KO➔WT of MHC-II (day 0, n = 5 and 5; day 7, n = 8 and 7; day 18, n = 5 and 5; day 26, n = 5 and 5) (A), iNOS (day 0, n = 5 and 5; day 7, n = 8 and 7; day 18, n = 5 and 5; day 26, n = 5 and 5) (B), and TNF-α (day 0, n = 4 and 4; day 7, n = 8 and 7; day 18, n = 5 and 5; day 26, n = 5 and 4) (C) in splenic macrophages (CD45+, CD11b+, Ly6G−, F4/80+, and MHC-II+) before (day 0) or 7, 18, and 26 days after T. brucei infection. (D to F) TNF-α (D), MIF (E), and IL-10 (F) concentration (ELISA) in the plasma of WT➔WT and miR-210 KO➔WT chimeric mice before (day 0) or 7 and 26 days after T. brucei infection (day 0, n = 5 and 5; day 7, n = 8 and 7; day 26, n = 7 and 7). (G) Itaconate levels (LC-MS) in the plasma of WT➔WT and miR-210 KO➔WT chimeric mice before or 7 days after T. brucei infection (day 0, n = 4 and 4; day 7, n = 8 and 7). (H) Percentage (FACS) of splenic neutrophils (CD45+, CD11b+, and Ly6G+) of WT➔WT and miR-210 KO➔WT chimeric mice before (day 0) or 6, 18, and 26 days after T. brucei infection (day 0, n = 4 and 4; day 7, n = 8 and 7; day 18, n = 5 and 5; day 26, n = 7 and 7). Data are representative of two independent experiments (A to F and H). P value was assessed by multiple Student’s t test (A to F and H) and by two-way ANOVA with Sidak’s multiple comparisons test (G). Statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001; graphs show means ± SEM.
Regulation of miR-210 in human monocytes and macrophages might represent a diagnostic and prognostic biomarker for sepsis
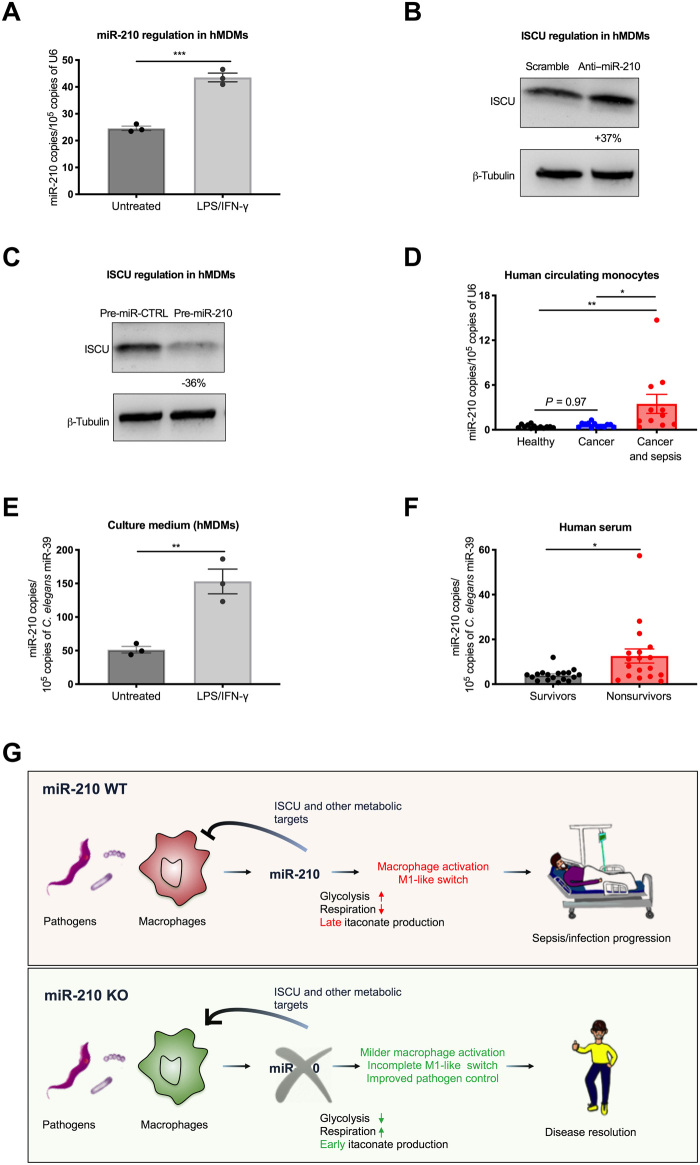
In an attempt to investigate the relevance of our mouse data in a human setting, we first differentiated blood-circulating monocytes isolated from healthy volunteers into proinflammatory hMDMs (38, 39). Real-time analysis on hMDMs showed a significant up-regulation of miR-210 upon LPS/IFN-γ treatment, phenocopying what was observed in murine macrophages (Fig. 8A). Subsequently, we silenced miR-210 levels by the transfection of hMDMs with an anti-miR-210 LNA observing an increased level of ISCU protein (Fig. 8B). Conversely, transfection with the miR-210 precursor resulted in a reduction of ISCU at the protein level, compared to a relative precursor control (Fig. 8C).
Fig. 8. MiR-210 is up-regulated and released by human macrophages and circulating monocytes upon pathogenic stimuli.
(A) MiR-210 levels (qRT-PCR) in hMDMs untreated or after 24-hour LPS/IFN-γ stimulation. (B and C) ISCU protein levels (Western blot) in hMDMs transfected with anti–miR-210 or scramble LNAs (B), and miR-210 precursor (pre-miR-210) or relative control (pre-miR-CTRL) (C). The percentage attached to the blot indicates densitometric changes. (D) MiR-210 levels (qRT-PCR) in circulating CD14+ monocytes isolated from the blood of healthy volunteers (n = 13) and cancer (cancer patients with no sepsis, n = 13) and oncological patients with sepsis (cancer patients with sepsis, n = 11). (E) MiR-210 levels (qRT-PCR) in the conditioned medium from hMDMs, untreated or after 24-hour LPS/IFN-γ. (F) Circulating miR-210 levels (qRT-PCR) in serum of patients with sepsis who will survive (survivors, n = 18) or die (nonsurvivors, n = 18). (G) Following pathogen interaction, both circulating monocytes and macrophages up-regulate miR-210. MiR-210 represses ISCU and (probably) other target genes contributing to the metabolic rewiring of macrophages toward glycolysis, required for the inflammatory burst, at the expense of mitochondrial respiration and itaconate production, ultimately leading to an exacerbated inflammatory response detrimental for the organism. In contrast, blocking miR-210 in macrophages, pharmacologically or genetically, unleashes the expression of ISCU and potentially other targets, impeding a full glycolytic-dependent M1-like activation state and displaying a milder proinflammatory response but earlier itaconate production, favoring pathogen control and altogether improving disease outcome. Data are representative of two independent experiments (A to C and E). P value was assessed by two-tailed, unpaired, Student’s t test (A, B, E, and F) and one-way ANOVA with Tukey’s multiple comparisons (D). Statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001; graphs show means ± SEM.
Since the up-regulation of miR-210 was already observed in mouse monocytes after LPS challenge, we measured miR-210 expression in human blood-circulating monocytes as well and assessed whether its levels were correlating with sepsis. In this regard, we isolated circulating monocytes from the blood of patients with sepsis recruited in the Oncology Unit (“cancer and sepsis”), cancer patients with no sign of sepsis (“cancer”), and healthy volunteers (“healthy”) (table S2). MiR-210 levels in circulating monocytes were much higher in oncological patients with sepsis compared to either cancer patient controls or healthy volunteers, but there was almost no difference between cancer only and healthy conditions (Fig. 8D). This result offers a “proof of concept” that monocyte-derived miR-210 might represent a diagnostic biomarker for human sepsis, which might discriminate for other inflammatory conditions such as cancer.
Soluble markers of sepsis are so far missing. It has been recently demonstrated that miR-210 can be found in the exosomes of a transformed cell line of macrophage origin (40). Therefore, we checked whether the levels of miR-210 in the medium mirrored its expression in macrophages. Here, we found that miR-210 up-regulation in response to the treatment of BMDMs with LPS/IFN-γ led to a similar increase of miR-210 in the culture medium (fig. S4A). Similarly, miR-210 levels were up-regulated in the plasma of WT mice in response to LPS (fig. S4B), but this induction was prevented when challenging miR-210 ΔM0 mice (fig. S4C), suggesting an important contribution of monocyte/macrophage in plasma miR-210 levels upon inflammatory/infectious stimuli.
Mirroring the mouse scenario, miR-210 was also detected in the culture medium of hMDMs upon LPS/IFN-γ treatment, leading to enhanced levels of secreted miR-210 in the culture medium (Fig. 8E). Encouraged by these data, we checked circulating miR-210 levels in a different cohort of patients who were admitted at the intensive care unit (ICU) with sepsis (table S3). Here, we found that miR-210 levels in serum correlated with disease outcome, being strongly up-regulated in the nonsurvivors (Fig. 8F). Although larger cohorts and prospective studies are warranted, these data argue that circulating miR-210 might represent a prognostic soluble biomarker in human sepsis.
DISCUSSION
Macrophage activation results in the prompt up- and down-regulation of a wide pattern of genes and metabolic pathways to mount a proper immune response to infections (4, 5). This entire process, still not entirely understood, must be tightly regulated since its loss of control can lead to deadly inflammatory conditions such as sepsis (35).
Here, we show that macrophages up-regulate miR-210 in response to Gram-negative or Gram-positive bacteria, parasitic T. brucei infection, and SARS-CoV-2 (or, at least, to some of their components) through HIF-1α stabilization, suggesting the involvement of miR-210 in a wide range of host-pathogen interactions. The relevance of miR-210 in different diseases ranging from endotoxemia to bacteria (polymicrobial and S. pneumoniae) and parasite (T. brucei) infections was further confirmed by our in vivo data. While IFN-γ and LPS binding to, respectively, IFN-γR and Toll-like receptor 4 (TLR4) leads to synergic HIF-1α accumulation through JAK/STAT1 (Janus kinase/signal transducer and activator of transcription 1) (23, 41) and NF-κB (nuclear factor κB) activation (42), previous literature suggests that the S. pneumoniae surface lipoprotein diacylglycerol transferase (43) and CpG motifs in the trypanosomal genomic DNA (44) may account for a similar effect via TLR2 and TLR9, respectively. It is likely that the relevance and abundance of the TLR family members in macrophages and monocytes (4) confer the selectivity of miR-210 up-regulation in these specific immune cell types.
Our in vitro analysis reveals that miR-210 up-regulation enables the effectiveness of the glycolytic switch imposed by LPS through the efficient shutdown of cell respiration. This effect was greatly achieved by the down-regulation of ISCU by miR-210 as proven by the observation that ISCU silencing in miR-210 KO macrophages reduced the OCR and, indirectly, increased glycolysis, thus re-establishing metabolic features of WT macrophages upon LPS/INF-γ stimulation. A possibility is that a reduction in NO and an increase in IL-10 levels associated with the phenotypic skewing of miR-210 KO macrophages can install a positive feedback loop that, indirectly, facilitates OXPHOS (45, 46). A previous study has shown that the shift from OXPHOS to aerobic glycolysis is a crucial mechanism for the initial activation of innate immunity in patients with sepsis (47). In addition to OXPHOS regulation, ISCU modulation by miR-210 might partly explain the improved pathogen clearance displayed in vivo by macrophage-specific miR-210–deficient mice. Previous results have already shown that repression of ISCU by miR-210 in endothelial cells reduces the activity of aconitase, another Fe-S cluster protein (33). Here, we argue that itaconate, a potent antimicrobial and anti-inflammatory intracellular and secreted metabolite (7, 29, 48, 49), is promptly and highly induced in activated miR-210–deficient macrophages via the enhanced channeling of glucose carbons down to the immune-responsive gene 1 (Irg1) pathway. Itaconate production by cis-aconitate-decarboxylase (the enzyme codified by the gene Irg1) may then contribute not only to the protection against the immunopathology of several infectious diseases observed in our in vivo model but also to the enhanced pathogen control observed in the S. pneumoniae and, to less extent, in the T. brucei infection models. We cannot exclude the idea that other macrophage-related mechanisms such as pathogen killing, digestion, and phagocytosis might be involved (50). Consistent with the fact that miR-210 controls other metabolic genes, e.g., NDUFA4 (15) and PDK1 (51), and also affects metabolic and nonmetabolic pathways, e.g., AKT/mTOR (52) and NF-κB signaling (18, 53), when silencing ISCU in miR-210–deficient macrophages, we could observe reversion of some, but not all, the immunological features such as increased IL-6 and MHC-II levels. This highlights the complex network of genes and pathways undermined by miRs and the impossibility to consider miR-210 as epistatic to ISCU only.
Following pathogen interaction, both circulating monocytes and macrophages up-regulate miR-210, which “imprints” macrophages toward an M1 activation state. Subsequently, these activated macrophages continue to exacerbate their proinflammatory response in a feed-forward loop as long as they remain in contact with the pathogen, thus decreasing the possibility of the organism to recover from the disease (Fig. 8G). Our in vitro proteomic and transcriptomic approaches as well as flow cytometric and ELISA analysis, both in vitro and in vivo, have consistently revealed that depletion of miR-210 mitigates this inflammatory, type 1 response upon activation, accompanied by an overt metabolic change in macrophages. At the same time, miR-210 ablation does not entirely impair the M1-type response and, therefore, infection does not take over (as we show in experimental CLP, or in case of T. brucei and S. pneumoniae infection) and, in contrast, pathogen control is even improved. On the other hand, miR-210 deletion does not seem to enforce the critical pathways characterizing an anti-inflammatory M2 response. Only one branch of the proteomic analysis highlighted terms related to protein localization. This could be intuitively associated to chaperone pathways, ER stress, and unfolded protein responses, which are more commonly associated to M2 macrophage polarization (54).
Overall, our findings suggest that pathogen-macrophage interaction imposes a deemed glycolytic switch that is sustained by HIF-1α accumulation (55, 56). More in detail, HIF-1α plays a dual role in macrophages: It induces glycolytic gene expression, e.g., LDH, PDK, HK2, PFK1, and G6PD (55), and, as we show, represses oxidative metabolism through the induction of miR-210, altogether permitting glycolysis that is instrumental for the inflammatory burst. This entire cascade is essential for macrophage responses determining disease outcome. Besides our in vivo phenotypic data, this model is reinforced by the observation that inhibition of glycolytic genes in macrophages such as PKM2 confers resistance to endotoxemia by affecting inflammatory responses (57, 58). Consistently, loss of function (56, 59) or gain of function (60) for HIF-1α have been proven to, respectively, mitigate and enhance macrophage-mediated inflammatory responses.
On one hand, all our data reinforce and complement the role of miR-210 in regulating oxidative metabolism and, in turn, glycolysis consistently with previous reports in cancer cells (15–17) or in muscle tissue during ischemia (12). On the other hand, they highlight different functional consequences of miR-210 modulation that are clearly cell and context/disease dependent (53, 61, 62) revealing its involvement in infectious diseases and sepsis.
At this point in time, we are not able to exclude an exocrine or paracrine mechanism of miR-210. MiR-210 was detectable in the extracellular milieu of LPS-stimulated murine or human macrophages and in the plasma of both septic mice and patients. Released miR-210, free or in exosomes, might regulate (metabolic) gene expression of neighboring cells as shown, for miR-210 (40) and other miRs (8, 63, 64) in different contexts.
Since miR-210 might be a suitable biomarker or therapeutic target, our study could also have social and medical impact. It is estimated that sepsis accounts for about 11 million deaths in 2017 (65) and, accordingly with WHO reports, lower respiratory infections are still the fourth global cause of death. Currently, the identification and monitoring of sepsis is still extremely challenging (66) and no targeted therapies are available in clinical practice (67). To render the clinical scenario even more complex, sepsis can occur in patients with multiple comorbidities (66). For example, it is estimated that almost 10% of cancer deaths are associated with sepsis (3). In our pilot study, miR-210 levels are specifically increased in monocytes isolated from patients with septic cancer but not in those with only cancer, suggesting its potential diagnostic value as a discriminant biomarker. In a different cohort of patients, individuals that exhibited the highest miR-210 serum levels are the ones that ultimately will succumb because of sepsis. Overall, these findings could be exploited for on-time medical intervention or to steer decisions in terms of treatment follow-up/discontinuation, thus increasing the possibility for the patient to survive and preventing costly procedures at the ICU. In addition, the implementation of a theragnostic approach would allow stratifying the patients based on miR-210 expression levels to select those patients who may benefit more from miR-210 inhibition. We show that a single injection of anti–miR-210 LNA strongly impairs miR-210 expression in macrophages, which is associated with an increase in survival after endotoxemia. The therapeutic benefit of an LNA against miR-122 is currently tested in a clinical trial for the treatment of hepatitis C virus infections (27), underlying the feasibility of this approach and translational aspect of our data.
Several studies have indicated that the overwhelming inflammatory response to SARS-CoV-2 infection dictates disease severity and survival hypothesizing that a mechanism of viral sepsis could occur (68). In this regard, a strong hyperinflammatory response of monocytes/macrophages was found in the severe cases of patients with COVID-19 (coronavirus disease 2019) (69). More recently, the increased cytokine production in monocytes was found to be mediated by a glycolytic switch orchestrated by HIF-1α (26). In view of our observation that miR-210 is induced in macrophages upon exposure to the SARS-CoV-2 spike protein, one could argue that miR-210 takes part in this HIF-1α–mediated metabolic reprogramming, suggesting the possible involvement of this miR in the pathogenicity of the disease (as we have shown for the other infectious/septic models) that could be further investigated. In addition, in our hands, induction of miR-210 upon macrophage interaction with the spike protein of SARS-CoV-2 alone is able to trigger inflammatory (but not anti-inflammatory) responses, which altogether is aligned with our model.
In conclusion, we identify miR-210 as a nongenetic immunoregulator, an “inflammo-miR,” induced in response to invading pathogens to enable a specific metabolic reprogramming required for macrophage activation and for their switch toward a proinflammatory, M1 phenotype. At large, these findings provide insights for the treatment, detection, and management of several diseases characterized by the over-reaction of the immune system to the entry of viruses, bacteria, and parasites.
MATERIALS AND METHODS
Mice
C57BL/6 miR-210 floxed mice were obtained in collaboration with M. Ivan (70) (University of Indiana, USA). Briefly, miR-210 floxed mice were crossed to Gata-1 Cre mice to constitutively delete the MiR210 locus, obtaining miR-210 KO mice. Alternatively, miR-210 floxed mice were intercrossed with the tamoxifen-inducible, macrophage-specific CSFR1:Cre-ERT (iCre) deleter mouse line (a gift of J. W. Pollard, University of Edinburgh, UK) obtaining miR-210 ΔM0 (iCre-positive mice) mice and controls (iCre-negative mice). In the latter model, the deletion of miR-210 was induced by daily tamoxifen (T5648, Sigma-Aldrich) administration (40 mg/kg) by gavage (for the LPS-induced endotoxin model) or intraperitoneally (for the S. pneumoniae infection model) in 100 μl of a solution containing 87.5% sunflower oil and 12.5% EtOH for 5 days. Moreover, there was an additional administration at day 0 and 18 hours after LPS injection to induce deletion of miR-210 in newly recruited macrophages. Control mice were treated with tamoxifen according to the same protocol. In all the experiments, littermate controls were used. Housing and all experimental animal procedures were approved by the Institutional Animal Care and Research Advisory Committee of the KU Leuven. For all experiments with T. brucei, maintenance and care of the mice complied with the European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes guidelines (CETS n° 123) and were approved by the Ethical Committee for Animal Experiments at the Vrije Universiteit Brussel (permit numbers: 14-220-05).
Bone marrow chimera generation and hematological analysis
C56BL/6 recipient mice were sublethally irradiated with 9.5 Gy (gray) and subsequently received up to 1 × 107 bone marrow cells from WT and miR-210 KO mice via tail vein injections to generate WT and miR-210 KO chimeric mice. Reconstitution of the bone marrow was allowed for 6 weeks. Red and white blood cell count was determined using a hemocytometer on peripheral blood collected in heparin with capillary pipettes by retro-orbital bleeding.
LPS-induced endotoxemia
Eight- to 12-week-old mice were injected with a single shot of LPS Escherichia coli 0111:B4 (20 to 27.5 mg/kg; L2630, Sigma-Aldrich). Mice were continuously monitored for survival. Rectal body temperature was monitored after 18 hours. Blood was taken via retro-orbital puncture and combined with an anticoagulant, e.g., lithium-heparin. Plasma preparation for LDH and creatinine analysis was obtained by centrifugation of the blood for 10 min at 2000g (4°C) and afterwards sent to the clinical laboratory of UZ Leuven, which performed the analysis.
In vivo (and in vitro) downmodulation of miR-210 was carried out as previously described (12). Briefly, 200 μl of phosphate-buffered saline (PBS)–diluted LNA oligonucleotides (12 mg/kg) against miR-210 (anti–miR-210) or SCR control sequence was injected into the tail vein; 15-mer LNA-enhanced sequences with complete phosphothioate backbone were used: anti-miR-210, GCTGTCACACGCACA; SCR, CGTCTAGCCACCTAG (in vivo LNA-miR inhibitors; Exiqon). The injection of LNA oligonucleotides was performed 48 hours before LPS injection.
Cecal ligation and puncture
The CLP procedure was performed according to the general guidelines (19). Briefly, mice were anesthetized by isoflurane inhalation, and a 1-cm incision was made in the abdomen after which the cecum was exposed and ligated. Subsequently, two punctures were made in the cecum with a 21-G needle for induction of a lethal CLP. The abdominal musculature and skin were closed with running sutures and metallic clips, respectively. After 10 and 24 hours from the surgery, mice were injected intraperitoneally with an antibiotic cocktail containing ceftriaxone (25 mg/kg; Sigma-Aldrich) and metronidazole (12.5 mg/kg; Sigma-Aldrich) dissolved in 200 μl of PBS. Rectal body temperature was monitored.
T. brucei infection
Clonal pleomorphic T. brucei AnTat 1.1E parasites were a gift from N. Van Meirvenne (Institute for Tropical Medicine, Belgium) and stored at −80°C. Mice were intraperitoneally infected with 5 × 103 AnTat1.1E trypanosomes. Parasite and red blood cell (RBC) numbers in blood were determined via a hemocytometer by tail cut (2.5 ml of blood in 500 ml of RPMI). Anemia was expressed as percentage of RBCs remaining in infected mice compared to that of noninfected mice.
S. pneumoniae infection
Mice were anesthetized intraperitoneally with ketamine (60 mg/kg, Ketavet, Zoetis) and medetomidine hydrochloride (0.8 mg/kg; Domitor, Orion Pharma). Each mouse received 107 CFUs of S. pneumoniae serotype 2 D39 strain via intratracheal instillation followed by subcutaneous injection of atimepazole hydrochloride (0.8 mg/kg; Antisedan, Orion Pharma) 30 min after infection. Twenty hours after infection, the animals were euthanized, and blood was obtained from the abdominal artery. The left lung was harvested and homogenized under sterile conditions by using a tissue homogenizer (Bullet Blender) and lysing tubes (Precellys). Tenfold serial dilutions were plated from blood and left lung homogenate into blood agar plates and grown overnight at 37°C and 5% CO2, and CFU counts were determined.
Histology and immunostaining
Livers and kidneys from septic mice were dissected, paraffin-embedded, and sectioned at 8-μm thickness. For TUNEL stainings, liver sections were stained using the ApoTag Plus Peroxidase In Situ Apoptosis Detection Kit (Chemicon) according to the manufacturer’s directions. Hematoxylin was used as a nuclear stain. Apoptosis was quantified as TUNEL-positive cells per optical field. Eight fields (20×) per liver were selected in a blinded fashion for quantification. For periodic acid–Schiff stainings, kidney sections were incubated for 10 min in periodic acid solution (2 g of H5IO6 in 200 ml of AD) after deparaffinization. After rinsing several times in tap water for 3 min, sections were incubated in Schiff’s reagent (Prosan) for 20 min and washed for 10 min under tap water. Last, sections were incubated in Harris’s hematoxylin (BDH) for 10 s and again rinsed under tap water for 10 min. The last steps were dehydration and mounting. Staining was performed to determine the glycogen-positive area in the kidney. Eight fields (20×) per kidney were selected in a blinded fashion for quantification.
Flow cytometry
To proceed with the FACS staining, a single-cell suspension was obtained from spleen and liver, as previously described (20). Cells in the peritoneal cavity were isolated by lavage as indicated in the section below, while peripheral blood was collected with heparinized capillary pipettes by retro-orbital bleeding. For the blood, and when required, RBC lysis was performed using Hybri-Max (Sigma-Aldrich). Cells were resuspended and counted in FACS buffer [PBS containing 2% fetal bovine serum (FBS) and 2 mM EDTA]. Subsequently, cells were incubated for 15 min with purified rat anti-mouse Fc blocking monoclonal antibody (Clone 2.4G2, BD-Pharmingen). The surface staining was performed, in the dark, at 4°C for 30 min using the following antibodies: viability dye (Invitrogen), anti-CD45 (30-F11, eBioscience), anti-CD11b (M1/70, eBioscience), anti-F4/80 (BM8, eBioscience), anti-MHC-II (M5/114.15-12, eBioscience), anti-Ly6C (AL-21, BD Pharmingen), anti-Ly6G (1A8, BD Pharmingen), anti-CD115 (AFS98, BioLegend), anti-TCRb (H57-597, BD Biosciences), anti-CD45/B220 (RA3-6B2, BD Biosciences), and anti-CD90.1 (Thy-1.1) (OX-7, BioLegend). After surface staining, cells were washed once with FACS buffer and resuspended in FACS buffer for FACS analysis. For iNOS intracellular staining in vitro on BMDMs, after surface staining (see before) and one washed with FACS buffer, cells were resuspended and incubated, in the dark, at 4°C for 30 min in Fix/Perm buffer (00-5523, eBioscience), prepared according to the manufacturer’s instructions. After one wash with permeabilization buffer (00-5523, eBioscience), cells were stained in permeabilization buffer overnight with anti-NOS2 (CXNFT, Thermo Fisher Scientific). For intracellular staining for iNOS or TNF-α on cells isolated from the spleen, cells were cultured in RPMI-medium + 5% fetal calf serum with brefeldin A (BD Bioscience). Four hours later, cells were washed with FACS buffer and subjected to surface staining (see before). After washing with FACS buffer, the cells were incubated, in the dark, at 4°C for 30 min in Fix/Perm buffer (00-5523, eBioscience), prepared according to the manufacturer’s instructions. After one wash with permeabilization buffer (00-5523, eBioscience), cells were stained in permeabilization buffer with anaphase-promoting complex (APC)–conjugated anti–TNF-α (MP6-XT22, BioLegend) or unlabeled rabbit iNOS (M19, BD Biosciences) and APC-conjugated anti-rabbit immunoglobulin G (eBioscience) for 30 min at 4°C in the dark. Cells were subsequently washed in permeabilization buffer and resuspended in cold permeabilization buffer for further FACS analysis. Cells were sorted by a FACSAria III (BD Biosciences) or acquired on a FACSVerse or FACSCanto II (BD Biosciences). Data analysis was performed using FlowJo software.
Peritoneal lavage
Mice were euthanized and 6 ml of PBS was injected into the peritoneal cavity using a 27-G needle. The abdomen was softly rubbed for 1 to 2 min and disinfected with 70% ethanol. Peritoneal cells were collected by inserting a syringe via a 24-G needle. For in vitro analysis on peritoneal macrophages, retrieved cells were counted and seeded in nontreated culture dishes non–tissue culture–treated dishes for the respected experiments. One hour after, cells were vigorously washed three times with PBS to enrich for peritoneal macrophages. For in vitro experiments, peritoneal macrophages were stimulated for the reported amount of time with LPS (100 ng/ml), IFN-γ (20 ng/ml), IL-4 (10 ng/ml), or IL-13 (10 ng/ml) in the different indicated combinations. For flow cytometry analysis, retrieved cells were counted and resuspended in FACS buffer (PBS containing 2% FBS and 2 mM EDTA) and further processed for flow cytometry.
Bone marrow–derived macrophages
Macrophages were derived from bone marrow precursors as described before (71). Briefly, bone marrow precursors (up to 10 × 106 cells) were cultured in a volume of 6 ml in a 10-cm petri dish (nontreated culture dishes, bacterial grade) in Dulbecco’s modified Eagle’s medium (DMEM) high glucose (41965039, Gibco) supplemented with 20% FBS, 30% L929 conditioned medium as a source of M-CSF, 25 mM Hepes, 2 mM l-glutamine, and Pen/Strep. After 3 days of culture, an additional 3 ml of differentiation medium was added. At day 7, macrophages were detached with ice-cold PBS. The obtained cells were a uniform population of macrophages as assessed by FACS, using the pan-macrophage–specific marker F4/80. For in vitro experiments, BMDMs were stimulated for the reported amount of time with LPS (100 ng/ml), IFN-γ (20 ng/ml), IL-4 (10 ng/ml), and T. brucei lysate (5 μg per 1 × 106 of BMDMs) in the different indicated combinations. For coculture experiments with heat-killed bacteria, a ratio of 1:10 (macrophage:bacteria) was used. BMDMs were cultured in DMEM high glucose (41965039, Gibco), 10% FBS, 2 mM l-glutamine, and Pen/Strep.
Generation of heat-killed P. aeruginosa and S. pneumoniae and T. brucei lysates
Both P. aeruginosa PAO-1 strain and S. pneumoniae serotype 2 D39 strain were exponentially grown in nutrient broth (Oxoid) at 37°C in the case of P. aeruginosa and brain/heart infusion broth (Oxoid) supplemented with 20% of heat-inactivated FBS (Gibco) at 37°C, 5% CO2. Heat killing was achieved by immersion of the bacteria in a water bath for 1 hour at 80°C. For the in vitro stimulation of cells with parasite lysate, parasites were isolated as described. Briefly, mice with a systemic T. brucei (AnTat1.1E) parasitemia were exsanguinated and parasites were purified from heparinized blood by diethylaminoethyl-cellulose (DE-52, Whatman). Parasites were collected in the flow-through and centrifuged for 10 min at 805g (Eppendorf 5810R centrifuge) and washed two times with PBS. Last, the pellet was resuspended in 1 to 2 ml of PBS. The lysate was prepared by three repetitive freeze-thawing cycles (−80°C, 37°C). The concentration of the lysate was determined spectrophotometrically (Nanodrop) following Prosep-Remtox treatment (Immunosource, Schilde, Belgium), and the samples were confirmed to be LPS free by using the Limulus Amebocyte Lysate Kinetic-QCL Kit (Cambrex, East Rutherford, NJ, USA) in accordance with the manufacturer’s instructions.
Spike-in SARS-CoV-2 stimulation
hMDMs differentiated with M-CSF were cultured in RPMI medium with SARS-CoV-2 (2019-nCoV) Spike protein (S1 + S2 ECD-His Recombinant Protein; Sino Biological, 40589-V08B1) at a concentration of 2 μg/ml.
RNA extraction, reverse transcription, and RT-qPCR
Cells were lysed and total RNA was isolated according to the manufacturer’s instructions using the Cell and Plant Kit (no. 300110, Exiqon) or miRNeasy Mini Kit (217004, Qiagen). Before RNA isolation from human serum or mouse plasma samples [blood collected in EDTA and centrifuged 10 min at 2000 rpm (4°C)] or culture media [5 min, 300g (4°C)], 10 fmol of C. elegans miR cel-miR-39 spiked-in (21960, Qiagen) was added to 20 μl of each sample, as previously described (72) and used for normalization. Reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (4366597, Applied Biosystems) and the specific RT primers from the miR assays according to the manufacturer’s instructions. Reverse transcription to cDNA was performed with the SuperScript III First Strand cDNA Synthesis Kit (18080051, Thermo Fisher Scientific) according to the manufacturer’s protocol. The cDNA, primer/probe mix, and TaqMan Fast Universal PCR Master Mix were prepared in a volume of 10 μl according to the manufacturer’s instructions (Applied Biosystems). TaqMan primers/assays used are hsa-miR-210-3p (000512, Thermo Fisher Scientific), U6-snRNA (001973, Thermo Fisher Scientific), and cel-miR-39 (000200, Thermo Fisher Scientific), while for gene detection, pre-made assays were purchased from IDT (HPRT, Hs.PT.58v.45621572; HIF1A, Hs.PT.56a.534274.g; IL6, Hs.PT.58.40226675; TNFA, Hs.PT.58.45380900; and IL10, Hs.PT.58.2807216). Samples were loaded into an optical 96-well Fast Thermal Cycling plate (Applied Biosystems) and quantitative real-time PCR (qRT-PCR) was performed using an ABI Prism 7500 Fast Real-Time PCR System (Applied Biosystems).
Electroporation for siRNA and precursor miR-210 delivery
Briefly, 5.6 × 106 BMDMs were resuspended in 750 μl of Opti-MEM and were electroporated (250 V, 950 μF, ∞ Ω) with 120 pmol of siRNA HIF-1α (mm.Ri.Hif1a.13.1 IDT), scrambled negative control (51-01-19-09, IDT), with 60 pmol of two siRNAs for ISCU (mm.Ri.Iscu.13.1 and mm.Ri.Iscu.13.2 IDT), or with 40 nM (hMDMs) or 150 nM (BMDMs) of Pre-miR miR-210 miR Precursor (PM10516, Thermo Fisher Scientific) or Pre-miR miR Precursor Negative Control (AM17100, Thermo Fisher Scientific).
shRNA-mediated silencing
A short hairpin RNA (shRNA) sequence for Iscu (CGTCATGAAACTGCAGATCCA) was subcloned in a lentiviral pLKO3 Thy1.1 vector backbone, and an shRNA sequence for LacZ was used as control (GTTCCGTCATAGCGATAACGA). Amplified viral particles were purified and titrated, and subsequently used for transduction. Briefly, 2 × 106 bone marrow cells in a six-well plate were transduced at day 5 of differentiation in DMEM high glucose (41965039, Gibco) supplemented with 20% FBS, 2 mM l-glutamine, and Pen/Strep with polybrene (16 mg/ml) overnight. Then, the medium was replaced with DMEM high glucose (41965039, Gibco) supplemented with 20% FBS, 30% L929 conditioned medium as a source of M-CSF, 25 mM Hepes, 2 mM l-glutamine, and Pen/Strep until day 8. The obtained cells were a uniform population of macrophages as assessed by FACS, using the pan-macrophage–specific marker F4/80. Last, CD90.1 surface expression was used to gate the transduced cells.
Protein extraction and immunoblot
Whole-cell protein extraction was performed using radioimmunoprecipitation assay extraction buffer (20 mM tris-HCl, 150 mM NaCl, 1% Triton X-100, 10% glycerol, and 5 mM EDTA) supplemented with Complete Mini protease inhibitor (Roche) and PhosSTOP Phosphatase Inhibitor (Roche). Proteins (15 to 50 μg) were separated by Mini-PROTEAN TGX Stain-Free Precast Gels (4568094, Bio-Rad) and transferred to nitrocellulose membrane Trans-Blot Turbo Midi 0.2-μm Nitrocellulose (no. 1704159, Bio-Rad) using the Trans-Blot Turbo Transfer System (Bio-Rad). Nonspecific binding was blocked in PBS with 0.05% Tween 20 (TBST) containing 5% of bovine serum albumin (BSA). The following antibodies were used: HIF-1α (C-term) (1006421, Cayman Chemical Company), ISCU (14812-1-AP, Proteintech), anti–β-tubulin antibody–Loading Control horseradish peroxidase (HRP) (ab21058, Abcam), anti-Vinculin (V9131, Sigma-Aldrich), and appropriate HRP-conjugated secondary antibodies (Santa Cruz). Signal was visualized by enhanced chemiluminescent reagents (ECL, Invitrogen) or West Femto by Thermo Fisher Scientific according to the manufacturer’s instructions and acquired by a LAS 4000 charge-coupled device camera with ImageQuant software (GE Healthcare).
Seahorse experiment
OCR was measured by using the Seahorse XF96 plate reader. Basal respiration was assessed in BMDMs (7.5 × 104 cells per well) incubated in a nonbuffered DMEM (D5030; Sigma-Aldrich) supplemented with 10 mM glucose and 2 mM glutamine. To assess complexes’ activity, BMDMs (7.5 × 104 cells per well) were first permeabilized with 1 nM Seahorse XF Plasma Membrane Permeabilizer (no. 102504-100; Agilent) and respiration was stimulated by adding 10 mM pyruvate, 5 mM malate, and 2 mM ADP. After the baseline scan, 100 nM rotenone was then added to inhibit complex I and 10 mM succinate was injected to establish complex II respiration. Last, 4 μM antimycin A was added to inhibit complex III and 100 μM TMPD +10 mM ascorbate was injected as a complex IV substrate. OCR data were then normalized according to protein content in each experimental well.
Radioactive metabolic assays
Radioactive metabolic analysis was performed as previously described (71). Briefly, for glycolysis, BMDMs were incubated for 2 hours in M199 with 10% FBS containing 0.4 μCi/ml [5-3H]-d-glucose (PerkinElmer). Subsequently, supernatant was transferred into glass vials sealed with rubber stoppers. 3H2O was captured in hanging wells containing a Whatman paper soaked with H2O over a period of 48 hours at 37°C to reach saturation. The radioactivity in the paper was determined by liquid scintillation counting.
Metabolite analysis by LC-MS/MS
Metabolites were extracted from WT and miR-210 KO BMDMs (2 × 106 BMDMs per well in a six-well plate) cultured in DMEM (A1443001, Gibco) supplemented with 10% dialyzed FBS, 2 mM glutamine, and 5 mM [U-13C]-d-glucose (Cambridge Isotope Laboratories). The extraction was performed using 80% methanol. After 5 min of incubation, cells were scraped and collected in a new tube. Following a centrifugation at 20,000g for 10 min at 4°C, the supernatant was transferred to a new vial for MS analysis. Pellet was used for protein quantification. For the plasma, 300 μl of 80% methanol was added to 100 μl of sample. For analysis on condition medium, 900 μl of 80% methanol was added to 100 μl of sample. After overnight storage at −80°C, samples were centrifuged at 20,000g for 15 min at 4°C and the supernatant was transferred to a new vial for MS analysis. Metabolite measurements were performed using a Dionex UltiMate 3000 LC System (Thermo Fisher Scientific) coupled to a Q Exactive Orbitrap MS (Thermo Fisher Scientific) operated in negative mode. Practically, 10 μl of sample was injected on an iHILIC-Fusion (P) column (HILICON). The gradient started with 10% of solvent B (10 mM Na-acetate in mqH2O, pH 9.3) and 90% solvent A (acetonitrile) and remained at 10% B until 2 min after injection. Next, a linear gradient to 80% B was carried out until 20 min. The gradient was kept at 80% B for 3 min followed by a decrease to 40% B. At 27 min, the gradient returned to 10% B. The chromatography was stopped at 35 min. The flow was kept constant at 200 μl/min. The MS operated in full scan (range, 70 to 1050) and negative mode using a spray voltage of 3.2 kV, a capillary temperature of 320°C, sheath gas at 10.0, and auxiliary gas at 5.0. AGC target was set at 3e6 using a resolution of 70,000. Data collection was performed using the Xcalibur software (Thermo Fisher Scientific) while data analysis was performed with the software El Maven-Polly (Elucidata). Raw abundance values were normalized by protein content. The fractional contribution (FC), which is the fractional percentage of each of the labeled isotopolog out of the total amount of metabolite, was obtained by using the software El Maven-Polly (Elucidata), which is mostly based on the formula FC = (−ni = 0 i × mi)/(n × −ni = 0 mi), where n is the number of C atoms in the metabolite, i denotes the isotopologs, and m indicates the abundance of an isotopolog. Energy charge was calculated as the following using the normalized raw abundancy: (ATP + ADP)/(ATP + ADP + AMP).
Extracellular lactate
BMDMs WT and miR-210 KO BMDMs (1 × 106 BMDMs per well in a six-well plate) were stimulated for 24 hours with LPS/IFN-γ. BMDMs were electroporated 48 hours before LPS/IFN-γ treatment with two siRNAs against ISCU or a control sequence. Lactate was measured in 2 μl of cultured medium by a commercial enzymatic colorimetric lactate assay kit (1001330, SPINREACT) according to the manufacturer’s instructions.
Cytokine detection
Secretion of IL-6 was analyzed using an electrochemiluminescent detection assay on a SECTOR3000 (MesoScale Discovery/MSD) with consecutive analysis using the Discovery Workbench 4.0. software (MSD) or with mouse IL-6 Quantikine ELISA kit (M6000B, R&D Systems) according to the manufacturer’s instructions. TNF-α and MIF levels were analyzed using a commercial mouse TNF-α DuoSet (DY410, R&D Systems), mouse MIF DuoSet (DY1978, R&D Systems), and murine IL-10 DuoSet (DY217, R&D Systems) ELISA kit, respectively, according to the manufacturers’ recommendations.
RNA sequencing and analysis
RNA was extracted from BMDMs isolated from WT and miR-210 KO mice, treated with LPS/IFN-γ or not treated. Per sample, an amount of 300 ng of total RNA was used as input. Sequencing libraries were prepared with the QIAseq FastSelect –rRNA HMR Kit, according to the manufacturer’s protocol. Sequence libraries of each sample were lastly equimolarly pooled and sequenced on a NextSeq 500 in a 75-cycle run. The enriched GO terms of the Biological Processes tree in the DEG (LPS/IFN-γ and not treated, up-regulated and down-regulated separately) were calculated using the enrichGO function of clusterProfiler R package v. 3.12 on R Studio v. 3.6.1 and 4.0.2. The resulting GOs were also “simplified” using the simplify function from clusterProfiler, to remove redundant GO terms. The resulting GOs were plotted using the dotplot function of clusterProfiler.
Proteomics
Proteins were extracted from BMDMs isolated from WT and miR-210 KO mice, treated with LPS/IFN-γ or untreated. All the samples were run on liquid chromatography–tandem MS (LC-MS/MS) and searched together using the MaxQuant algorithm (version 1.6.1.0) with default search settings including a false discovery rate (FDR) set at 5% on both the peptide and protein level. Spectra were searched against the mouse proteins in the SWISS-PROT database (database release version of September 2017 containing 16,931 mouse protein sequences) (www.uniprot.org). To reveal proteins of which the expression level was significantly regulated between the different conditions, sample groups were defined on the basis of treatment (untreated versus LPS/IFN-γ) and cell type (WT versus miR-210 KO), and a two-way analysis of variance (ANOVA) test was performed to compare the intensities of the proteins in the treatment group with the cell-type group. For each protein, this test calculated a P value (actually −log P value) for treatment, a P value for cell type, and a P value for the interaction between treatment and cell type. Overall, 2022 proteins with a P value < 0.05 in at least one of these three conditions were considered to be significantly regulated. Subsequently, using the −log10 (P value) for the interaction term from the two-way ANOVA as metrics (with a positive or negative sign according to whether the protein abundance was higher or lower in the KO condition, respectively), gene set enrichment analysis (73) on GO biological process gene sets was carried out. Significant terms were then selected on the basis of an FDR cutoff value of 0.05.
Human monocyte isolation
Circulating human monocytes were isolated from whole blood with CD14 Dynabeads (Invitrogen, catalog no. 11149D) according to the manufacturer’s instructions. Briefly, 10 ml of whole blood was collected in EDTA tubes and transferred to a 15-ml Falcon tube containing 65 μl of CD14 Dynabeads resuspended in isolation buffer [PBS, 0.1% BSA, and 2 mM EDTA (pH 7.4)] for a 20-min incubation on a carousel at 4°C. After, the tube was placed in a magnet for 2 min and supernatant was removed. After the removal of the tube from the magnet, 1 ml of isolation buffer was used to resuspend before transfer to a new clean tube. Washes were performed three more times before counting. Cells were centrifuged 10 min at 300g. Pellets were resuspended in 350 μl of lysis buffer (Exiqon, 1% β-mercaptoethanol) and stored at −80°C. The clinical study was conducted according to the Declaration of Helsinki, and ethical committee approval was obtained from the ethical committee of the Antwerp University Hospital (B300201837008) and, for healthy volunteers, from UZ/KU Leuven (S62927). Informed consent was obtained from all individual participants in the study.
Human monocyte-derived macrophages
Briefly, as previously described (39), circulating monocytes were isolated from healthy blood donor buffy coats by two-step gradient centrifugation using Ficoll combined with a CD14 MicroBeads (Miltenyi Biotec Inc.) magnetic isolation step (following manufacturer instructions). Last, monocytes were seeded in RPMI 10% FBS, 2 mM l-glutamine, and Pen/Strep in the presence of either recombinant human granulocyte M-CSF (GM-CSF) (100 ng/ml; 300-03, PeproTech) or recombinant human M-CSF (20 ng/ml; 300-25, PeproTech). Then, 50% of fresh volume with GM-CSF was added at day 6. Differentiated hMDMs were cultured in RPMI 1640, 10% FBS, 2 mM l-glutamine, and Pen/Strep. For in vitro experiments with miR-210 modulation, 40 nM precursor miR-210 or anti–miR-210 and relative controls were transfected using Lipofectamine MessengerMAX Transfection Reagent (LMRNA003, Thermo Fisher Scientific) in RPMI 10% FBS, 2 mM l-glutamine, and Pen/Strep supplemented with GM-CSF following manufacturer instructions.
Serum samples of ICU patients with sepsis
This was a secondary analysis of the randomized controlled EPaNIC trial that investigated the impact of early parenteral nutrition. The study protocol and consent forms were approved by the institutional ethical review board (ML4190). The detailed study protocol and primary results have been published elsewhere (74). For this study, 18 nonsurviving patients with sepsis were age- and gender-matched to 18 surviving patients with sepsis.
Statistical analysis
Data entry and all analyses were performed in a blinded fashion. All statistical analyses were performed using GraphPad Prism software. Statistical significance was calculated by unpaired t test, one-way ANOVA, multiple t test using two-sided, two-sample t tests with Holm-Sidak correction, two-way ANOVA followed by Tukey’s or Sidak’s multiple comparisons test, or Mann-Whitney U test as indicated in the figure legends, with P < 0.05 considered statistically significant. Sample sizes for all experiments were chosen on the basis of previous experiences. Detection of mathematical outliers was performed using the Grubbs’ test in GraphPad. Independent experiments were pooled and analyzed together whenever possible as detailed in figure legends. All graphs show mean values ± SEM.
Acknowledgments
We thank the VIB Proteomics Core Facility, Ghent for the proteomics measurements and T. Maia for the pathway analysis; the Metabolomics Core Facility, VIB for metabolic measurements; and L. Archetti, P. Ortelli, C. Guilbaud, C. Merlin, and P. Coelho for technical support. Funding: M.M. was supported by an ERC Consolidator grant (ImmunoFit, no. 773208) and FWO (1501215N) and received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 766214. D.T. was supported by AIRC (IG2017-20258), Fondazione Cassa di Risparmio Torino CRT (2018.1311), and Ricerca Sanitaria Finalizzata (RF-2016-02361048). M.I. was supported by NIH/NCI RO1 grant (CA155332) and acknowledges the Center for Genomics and Bioinformatics (Indiana University) for RNA seq analysis. A.-T.H. was supported by Fonds Wetenschappelijk Onderzoek (FWO) (1297115N and 1501215N). F.M. was supported by the Italian Ministry of Health (“Ricerca Corrente” and “5x1000”), by AFM-Telethon (no. 23054), and by Telethon Foundation (GGP19035A). C.R.-D. was supported by Fonds Wetenschappelijk Onderzoek (Flemish Research Foundation) grant 1108919N. F.C. was supported by the personal fellowship from the Emmanuel Van Der Schueren kom op tegen kanker (K.B.O. nr. 0442.528.054). A.S.M. was supported by Wellcome Trust Postdoctoral Research Training Fellowship for Clinicians (110086/Z/15/Z). S.R.W. was principally supported by a Wellcome Trust Clinical Fellowship award (209220). Moreover, the University of Edinburgh is a charitable body, registered in Scotland, with registration number SC005336. Author contributions: F.V. performed experimental design, all experiments, and data acquisition and interpretation, and wrote the manuscript. F.C. performed in vitro metabolic assays and helped in vivo experiments. B.S. performed all the T. brucei experiments. A.-T.H. performed histological and in vivo experiments. F.C., R.T., and C.R.-D. performed in vitro assays and helped in FACS experiments. J.V.A., H.P., and E.S. collected and supported the analysis of human monocytes species. A.S.M., M.A.S.-G., and S.R.W. performed and helped in the interpretation of S. pneumoniae experiments. J.V. and C.L. performed and helped in the interpretation of CLP experiments. A.G. and C.I. analyzed RNA-seq data. F.O., D.L., F.M., and J.A.V.G. supported with critical suggestion and edits in the writing. L.L. and G.V.d.B. provided human plasma samples. O.F. and C.C. supported with metabolic assays and critical suggestions. B.G. performed all the mass spectrometry measurements. M.I. generated and provided floxed miR-210 mice as well as the global KO strain, and importantly contributed to the RNA-seq data generation. M.M. and D.T. performed experimental design and data analysis, conducted scientific direction, and wrote the manuscript. Competing interests: M.M. and A.-T.H. are inventors on a patent related to this work filed by VIB and KU Leuven (n° WO 2018/065390, filing date: 3 October 2017, published 12 April 2018), now pending as EP3523433 and US 16/332.575. Inventorship of F.V. is under evaluation. The other authors declare that they have no competing interests. Data and materials availability: RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) data repository, with accession number GSF2057 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171270).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/19/eabf0466/DC1
REFERENCES AND NOTES
- 1.Chaplin D. D., Overview of the immune response. J. Allergy Clin. Immunol. 125, S3–S23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Poll T., van de Veerdonk F. L., Scicluna B. P., Netea M. G., The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol. 17, 407–420 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Williams M. D., Braun L., Cooper L. M., Johnston J., Weiss R. V., Qualy R. L., Linde-Zwirble W., Hospitalized cancer patients with severe sepsis: Analysis of incidence, mortality, and associated costs of care. Crit. Care 8, R291–R298 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi O., Akira S., Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Pearce E. J., Pearce E. L., Driving immunity: All roads lead to metabolism. Nat. Rev. Immunol. 18, 81–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riera-Domingo C., Audigé A., Granja S., Cheng W.-C., Ho P.-C., Baltazar F., Stockmann C., Mazzone M., Immunity, hypoxia, and metabolism—The Ménage à Trois of cancer: Implications for immunotherapy. Physiol. Rev. 100, 1–102 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Viola A., Munari F., Sánchez-Rodríguez R., Scolaro T., Castegna A., The metabolic signature of macrophage responses. Front. Immunol. 10, 1462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Virga F., Quirico L., Cucinelli S., Mazzone M., Taverna D., Orso F., MicroRNA-mediated metabolic shaping of the tumor microenvironment. Cancer 13, 127 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connell R. M., Rao D. S., Baltimore D., MicroRNA regulation of inflammatory responses. Annu. Rev. Immunol. 30, 295–312 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Yao Q., Song Z., Wang B., Zhang J., Emerging roles of microRNAs in the metabolic control of immune cells. Cancer Lett. 433, 10–17 (2018). [DOI] [PubMed] [Google Scholar]
- 11.M. Ivan, X. Huang, miR-210: Fine-tuning the hypoxic response, in Tumor Microenvironment and Cellular Stress, C. Koumenis, E. Hammond, A. Giaccia, Eds. (Advances in Experimental Medicine and Biology, Springer New York, 2014), vol. 772, pp. 205–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaccagnini G., Maimone B., Di Stefano V., Fasanaro P., Greco S., Perfetti A., Capogrossi M. C., Gaetano C., Martelli F., Hypoxia-induced miR-210 modulates tissue response to acute peripheral ischemia. Antioxid. Redox Signal. 21, 1177–1188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan Y. C., Banerjee J., Choi S. Y., Sen C. K., miR-210: The master hypoxamir. Microcirculation 19, 215–223 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulshreshtha R., Ferracin M., Wojcik S. E., Garzon R., Alder H., Agosto-Perez F. J., Davuluri R., Liu C.-G., Croce C. M., Negrini M., Calin G. A., Ivan M., A MicroRNA signature of hypoxia. Mol. Cell. Biol. 27, 1859–1867 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puisségur M.-P., Mazure N. M., Bertero T., Pradelli L., Grosso S., Robbe-Sermesant K., Maurin T., Lebrigand K., Cardinaud B., Hofman V., Fourre S., Magnone V., Ricci J. E., Pouysségur J., Gounon P., Hofman P., Barbry P., Mari B., miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 18, 465–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z., Li Y., Zhang H., Huang P., Luthra R., Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 29, 4362–4368 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Ullmann P., Qureshi-Baig K., Rodriguez F., Ginolhac A., Nonnenmacher Y., Ternes D., Weiler J., Gäbler K., Bahlawane C., Hiller K., Haan S., Letellier E., Hypoxia-responsive miR-210 promotes self-renewal capacity of colon tumor-initiating cells by repressing ISCU and by inducing lactate production. Oncotarget 7, 65454–65470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi J., Qiao Y., Wang P., Li S., Zhao W., Gao C., microRNA-210 negatively regulates LPS-induced production of proinflammatory cytokines by targeting NF-κB1 in murine macrophages. FEBS Lett. 586, 1201–1207 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Dejager L., Pinheiro I., Dejonckheere E., Libert C., Cecal ligation and puncture: The gold standard model for polymicrobial sepsis? Trends Microbiol. 19, 198–208 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Stijlemans B., De Baetselier P., Magez S., Van Ginderachter J. A., De Trez C., African trypanosomiasis-associated anemia: The contribution of the interplay between parasites and the mononuclear phagocyte system. Front. Immunol. 9, 218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varma T. K., Lin C. Y., Toliver-Kinsky T. E., Sherwood E. R., Endotoxin-induced gamma interferon production: Contributing cell types and key regulatory factors. Clin. Vaccine Immunol. 9, 530–543 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda N., O’Dea E. L., Doedens A., Kim J., Weidemann A., Stockmann C., Asagiri M., Simon M. C., Hoffmann A., Johnson R. S., Differential activation and antagonistic function of HIF-α isoforms in macrophages are essential for NO homeostasis. Genes Dev. 24, 491–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parra-Izquierdo I., Castaños-Mollor I., López J., Gómez C., San Román J. A., Sánchez Crespo M., García-Rodríguez C., Lipopolysaccharide and interferon-γ team up to activate HIF-1α via STAT1 in normoxia and exhibit sex differences in human aortic valve interstitial cells. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 2168–2179 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Yeh Y.-H., Hsiao H.-F., Yeh Y.-C., Chen T.-W., Li T.-K., Inflammatory interferon activates HIF-1α-mediated epithelial-to-mesenchymal transition via PI3K/AKT/mTOR pathway. J. Exp. Clin. Cancer Res. 37, 70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiser J. N., Ferreira D. M., Paton J. C., Streptococcus pneumoniae: Transmission, colonization and invasion. Nat. Rev. Microbiol. 16, 355–367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Codo A. C., Davanzo G. G., de B. Monteiro L., de Souza G. F., Muraro S. P., Virgilio-da-Silva J. V., Prodonoff J. S., Carregari V. C., de Biagi Junior C. A. O., Crunfli F., Restrepo J. L. J., Vendramini P. H., Reis-de-Oliveira G., dos Santos K. B., Toledo-Teixeira D. A., Parise P. L., Martini M. C., Marques R. E., Carmo H. R., Borin A., Coimbra L. D., Boldrini V. O., Brunetti N. S., Vieira A. S., Mansour E., Ulaf R. G., Bernardes A. F., Nunes T. A., Ribeiro L. C., Palma A. C., Agrela M. V., Moretti M. L., Sposito A. C., Pereira F. B., Velloso L. A., Vinolo M. A. R., Damasio A., Proença-Módena J. L., Carvalho R. F., Mori M. A., Martins-de-Souza D., Nakaya H. I., Farias A. S., Moraes-Vieira P. M., Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/glycolysis-dependent axis. Cell Metab. 32, 498–499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rupaimoole R., Slack F. J., MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Diskin C., Pålsson-McDermott E. M., Metabolic modulation in macrophage effector function. Front. Immunol. 9, 270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordes T., Michelucci A., Hiller K., Itaconic acid: The surprising role of an industrial compound as a mammalian antimicrobial metabolite. Annu. Rev. Nutr. 35, 451–473 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Lampropoulou V., Sergushichev A., Bambouskova M., Nair S., Vincent E. E., Loginicheva E., Cervantes-Barragan L., Ma X., Huang S. C.-C., Griss T., Weinheimer C. J., Khader S., Randolph G. J., Pearce E. J., Jones R. G., Diwan A., Diamond M. S., Artyomov M. N., Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strelko C. L., Lu W., Dufort F. J., Seyfried T. N., Chiles T. C., Rabinowitz J. D., Roberts M. F., Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 133, 16386–16389 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mills E. L., Kelly B., Logan A., Costa A. S. H., Varma M., Bryant C. E., Tourlomousis P., Däbritz J. H. M., Gottlieb E., Latorre I., Corr S. C., McManus G., Ryan D., Jacobs H. T., Szibor M., Xavier R. J., Braun T., Frezza C., Murphy M. P., O’Neill L. A., Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470.e13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan S. Y., Zhang Y.-Y., Hemann C., Mahoney C. E., Zweier J. L., Loscalzo J., MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 10, 273–284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouault T. A., Biogenesis of iron-sulfur clusters in mammalian cells: New insights and relevance to human disease. Dis. Model. Mech. 5, 155–164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sica A., Mantovani A., Macrophage plasticity and polarization: In vivo veritas. J. Clin. Invest. 122, 787–795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magez S., Radwanska M., Beschin A., Sekikawa K., De Baetselier P., Tumor necrosis factor alpha is a key mediator in the regulation of experimental Trypanosoma brucei infections. Infect. Immun. 67, 3128–3132 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stijlemans B., Leng L., Brys L., Sparkes A., Vansintjan L., Caljon G., Raes G., Van Den Abbeele J., Van Ginderachter J. A., Beschin A., Bucala R., De Baetselier P., MIF contributes to Trypanosoma brucei associated immunopathogenicity development. PLOS Pathog. 10, e1004414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lacey D. C., Achuthan A., Fleetwood A. J., Dinh H., Roiniotis J., Scholz G. M., Chang M. W., Beckman S. K., Cook A. D., Hamilton J. A., Defining GM-CSF– and macrophage-CSF–dependent macrophage responses by in vitro models. J. Immunol. 188, 5752–5765 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Palmieri E. M., Menga A., Martín-Pérez R., Quinto A., Riera-Domingo C., De Tullio G., Hooper D. C., Lamers W. H., Ghesquière B., McVicar D. W., Guarini A., Mazzone M., Castegna A., Pharmacologic or genetic targeting of glutamine synthetase skews macrophages toward an M1-like phenotype and inhibits tumor metastasis. Cell Rep. 20, 1654–1666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian F., Tang P., Sun Z., Zhang R., Zhu D., He J., Liao J., Wan Q., Shen J., miR-210 in exosomes derived from macrophages under high glucose promotes mouse diabetic obesity pathogenesis by suppressing NDUFA4 expression. J. Diabetes Res. 2020, 6894684 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spirig R., Djafarzadeh S., Regueira T., Shaw S. G., von Garnier C., Takala J., Jakob S. M., Rieben R., Lepper P. M., Effects of TLR agonists on the hypoxia-regulated transcription factor HIF-1α and dendritic cell maturation under normoxic conditions. PLOS ONE 5, e10983 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rius J., Guma M., Schachtrup C., Akassoglou K., Zinkernagel A. S., Nizet V., Johnson R. S., Haddad G. G., Karin M., NF-κB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1α. Nature 453, 807–811 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomlinson G., Chimalapati S., Pollard T., Lapp T., Cohen J., Camberlein E., Stafford S., Periselneris J., Aldridge C., Vollmer W., Picard C., Casanova J.-L., Noursadeghi M., Brown J., TLR-mediated inflammatory responses to Streptococcus pneumoniae are highly dependent on surface expression of bacterial lipoproteins. J. Immunol. 193, 3736–3745 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mabille D., Caljon G., Inflammation following trypanosome infection and persistence in the skin. Curr. Opin. Immunol. 66, 65–73 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Ip W. K. E., Hoshi N., Shouval D. S., Snapper S., Medzhitov R., Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van den Bossche J., Baardman J., Otto N. A., van der Velden S., Neele A. E., van den Berg S. M., Luque-Martin R., Chen H.-J., Boshuizen M. C. S., Ahmed M., Hoeksema M. A., de Vos A. F., de Winther M. P. J., Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Rep. 17, 684–696 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Cheng S.-C., Scicluna B. P., Arts R. J. W., Gresnigt M. S., Lachmandas E., Giamarellos-Bourboulis E. J., Kox M., Manjeri G. R., Wagenaars J. A. L., Cremer O. L., Leentjens J., van der Meer A. J., van de Veerdonk F. L., Bonten M. J., Schultz M. J., Willems P. H. G. M., Pickkers P., Joosten L. A. B., van der Poll T., Netea M. G., Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat. Immunol. 17, 406–413 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Hooftman A., Angiari S., Hester S., Corcoran S. E., Runtsch M. C., Ling C., Ruzek M. C., Slivka P. F., McGettrick A. F., Banahan K., Hughes M. M., Irvine A. D., Fischer R., O’Neill L. A. J., The immunomodulatory metabolite itaconate modifies NLRP3 and inhibits inflammasome activation. Cell Metab. 32, 468–478.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michelucci A., Cordes T., Ghelfi J., Pailot A., Reiling N., Goldmann O., Binz T., Wegner A., Tallam A., Rausell A., Buttini M., Linster C. L., Medina E., Balling R., Hiller K., Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. U.S.A. 110, 7820–7825 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frank B., Marcu A., de Oliveira Almeida Petersen A. L., Weber H., Stigloher C., Mottram J. C., Scholz C. J., Schurigt U., Autophagic digestion of Leishmania major by host macrophages is associated with differential expression of BNIP3, CTSE, and the miRNAs miR-101c, miR-129, and miR-210. Parasit. Vectors 8, 404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y., Yang C., Zhang L., Yang P., MicroRNA-210 induces endothelial cell apoptosis by directly targeting PDK1 in the setting of atherosclerosis. Cell. Mol. Biol. Lett. 22, 3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu W., Jiang D., Gong F., Huang Y., Luo Y., Rong Y., Wang J., Ge X., Ji C., Fan J., Cai W., miR-210-5p promotes epithelial–mesenchymal transition by inhibiting PIK3R5 thereby activating oncogenic autophagy in osteosarcoma cells. Cell Death Dis. 11, 93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang D., Cao X., Li J., Zhao G., MiR-210 inhibits NF-κB signaling pathway by targeting DR6 in osteoarthritis. Sci. Rep. 5, 12775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oh J., Riek A. E., Weng S., Petty M., Kim D., Colonna M., Cella M., Bernal-Mizrachi C., Endoplasmic reticulum stress controls M2 macrophage differentiation and foam cell formation. J. Biol. Chem. 287, 11629–11641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corcoran S. E., O’Neill L. A. J., HIF1α and metabolic reprogramming in inflammation. J. Clin. Invest. 126, 3699–3707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cramer T., Yamanishi Y., Clausen B. E., Förster I., Pawlinski R., Mackman N., Haase V. H., Jaenisch R., Corr M., Nizet V., Firestein G. S., Gerber H. P., Ferrara N., Johnson R. S., HIF-1α is essential for myeloid cell-mediated inflammation. Cell 112, 645–657 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie M., Yu Y., Kang R., Zhu S., Yang L., Zeng L., Sun X., Yang M., Billiar T. R., Wang H., Cao L., Jiang J., Tang D., PKM2-dependent glycolysis promotes NLRP3 and AIM2 inflammasome activation. Nat. Commun. 7, 13280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang L., Xie M., Yang M., Yu Y., Zhu S., Hou W., Kang R., Lotze M. T., Billiar T. R., Wang H., Cao L., Tang D., PKM2 regulates the Warburg effect and promotes HMGB1 release in sepsis. Nat. Commun. 5, 4436 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peyssonnaux C., Cejudo-Martin P., Doedens A., Zinkernagel A. S., Johnson R. S., Nizet V., Cutting edge: Essential role of hypoxia inducible factor-1α in development of lipopolysaccharide-induced sepsis. J. Immunol. 178, 7516–7519 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Schneider M., Van Geyte K., Fraisl P., Kiss J., Aragonés J., Mazzone M., Mairbäurl H., De Bock K., Jeoung N. H., Mollenhauer M., Georgiadou M., Bishop T., Roncal C., Sutherland A., Jordan B., Gallez B., Weitz J., Harris R. A., Maxwell P., Baes M., Ratcliffe P., Carmeliet P., Loss or silencing of the PHD1 prolyl hydroxylase protects livers of mice against ischemia/reperfusion injury. Gastroenterology 138, 1143–1154.e2 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Kopriva S. E., Chiasson V. L., Mitchell B. M., Chatterjee P., TLR3-induced placental miR-210 down-regulates the STAT6/interleukin-4 pathway. PLOS ONE 8, e67760 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noman M. Z., Janji B., Hu S., Wu J. C., Martelli F., Bronte V., Chouaib S., Tumor-promoting effects of myeloid-derived suppressor cells are potentiated by hypoxia-induced expression of miR-210. Cancer Res. 75, 3771–3787 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Orso F., Quirico L., Dettori D., Coppo R., Virga F., Ferreira L. C., Paoletti C., Baruffaldi D., Penna E., Taverna D., Role of miRNAs in tumor and endothelial cell interactions during tumor progression. Semin. Cancer Biol. 60, 214–224 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Squadrito M. L., Baer C., Burdet F., Maderna C., Gilfillan G. D., Lyle R., Ibberson M., De Palma M., Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 8, 1432–1446 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Rudd K. E., Johnson S. C., Agesa K. M., Shackelford K. A., Tsoi D., Kievlan D. R., Colombara D. V., Ikuta K. S., Kissoon N., Finfer S., Fleischmann-Struzek C., Machado F. R., Reinhart K. K., Rowan K., Seymour C. W., Watson R. S., West T. E., Marinho F., Hay S. I., Lozano R., Lopez A. D., Angus D. C., Murray C. J. L., Naghavi M., Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 395, 200–211 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vincent J.-L., The clinical challenge of sepsis identification and monitoring. PLOS Med. 13, e1002022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marshall J. C., Why have clinical trials in sepsis failed? Trends Mol. Med. 20, 195–203 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Li H., Liu L., Zhang D., Xu J., Dai H., Tang N., Su X., Cao B., SARS-CoV-2 and viral sepsis: Observations and hypotheses. Lancet 395, 1517–1520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Merad M., Martin J. C., Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 20, 355–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mantel C. R., O’Leary H. A., Chitteti B. R., Huang X., Cooper S., Hangoc G., Brustovetsky N., Srour E. F., Lee M. R., Messina-Graham S., Haas D. M., Falah N., Kapur R., Pelus L. M., Bardeesy N., Fitamant J., Ivan M., Kim K.-S., Broxmeyer H. E., Enhancing hematopoietic stem cell transplantation efficacy by mitigating oxygen shock. Cell 161, 1553–1565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wenes M., Shang M., Di Matteo M., Goveia J., Martín-Pérez R., Serneels J., Prenen H., Ghesquière B., Carmeliet P., Mazzone M., Macrophage metabolism controls tumor blood vessel morphogenesis and metastasis. Cell Metab. 24, 701–715 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Mitchell P. S., Parkin R. K., Kroh E. M., Fritz B. R., Wyman S. K., Pogosova-Agadjanyan E. L., Peterson A., Noteboom J., O’Briant K. C., Allen A., Lin D. W., Urban N., Drescher C. W., Knudsen B. S., Stirewalt D. L., Gentleman R., Vessella R. L., Nelson P. S., Martin D. B., Tewari M., Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Casaer M. P., Mesotten D., Hermans G., Wouters P. J., Schetz M., Meyfroidt G., Van Cromphaut S., Ingels C., Meersseman P., Muller J., Vlasselaers D., Debaveye Y., Desmet L., Dubois J., Van Assche A., Vanderheyden S., Wilmer A., Van den Berghe G., Early versus late parenteral nutrition in critically Ill adults. N. Engl. J. Med. 365, 506–517 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/19/eabf0466/DC1