Abstract
Background
Preeclampsia is a leading cause of maternal and perinatal morbidity and mortality. However, current understanding of its underlying biological pathways remains limited.
Methods
In this study, we performed a cross-platform proteome- and transcriptome-wide genetic analysis aimed at evaluating the causal relevance of more than 2,000 circulating proteins with preeclampsia, supported by data on expression of over 15,000 genes across 36 tissue leveraging large-scale preeclampsia genetic association data from women of European ancestry.
Results
We demonstrate genetic associations of 18 circulating proteins with preeclampsia (SULT1A1, SH2B3, SERPINE2, RGS18, PZP, NOTUM, METAP1, MANEA, jun−D, GDF15, FGL1, FGF5, FES, APOBR, ANP, ALDH−E2, ADAMTS13, 3MG), among which 11 were either directly or indirectly supported by gene expression data, 9 were supported by Bayesian colocalization analyses, and 5 (SERPINE2, PZP, FGF5, FES and ANP) were supported by all lines of evidence examined. Protein interaction mapping identified potential shared biological pathways through natriuretic peptide signalling, blood pressure regulation, immune tolerance and thrombin activity regulation.
Conclusions
This investigation identified multiple targetable proteins linked to cardiovascular, inflammatory, and coagulation pathways, with SERPINE2, PZP, FGF5, FES and ANP identified as a pivotal proteins with likely causal roles in the development of preeclampsia. The identification of these potential targets may guide the development of targeted therapies for preeclampsia.
Keywords: preeclampsia, proteins, genes, Mendelian randomization, genetics, pathways
Nonstandard Abbreviations and Acronyms
- AMR
Mendelian randomization
- GWAS
Genome wide association study
- pQTL
protein quantitative trait loci
- eQTL
expression quantitative trait loci
- PPH1
posterior probability of hypothesis 1
- PPH2
posterior probability of hypothesis 2
- PPH3
posterior probability of hypothesis 3
- PPH4
posterior probability of hypothesis 4
Introduction
Preeclampsia is a leading cause of maternal and perinatal morbidity and mortality. Current understanding of its underlying biological pathways remains limited. This is a great hindrance to the development of therapies which might prevent its development and downstream consequences on short- and long-term maternal and fetal health. To date, only a handful of drugs have been tested in randomized clinical trials to prevent preeclampsia, and only one therapy, aspirin, has demonstrated efficacy in high-risk women and been translated to clinical recommendations1.
Proteins play a key role in human biology and are the primary targets of most drug therapies. Understanding the causal relevance of proteins and their networks on diseases can aid drug development and repurposing by uncovering the key underlying biological processes, providing rationale for intervening on these proteins with drugs. This can streamline the discovery process and increase the efficiency of drug development.
Mendelian randomization (MR) is a genetic epidemiological method that leverages the natural randomness that is involved in the inheritance of many risk factors, to inform the causal relevance of these risk factors on a specific disease2. In the proteomic setting, the MR paradigm leverages the natural variability in genetic variants encoding proteins, which are potential drug targets, to explore the predicted effects of their perturbation. Since allocation of these genetic variants that determine circulating protein levels occurs randomly through the process of mating and allele assortment at conception, the method resembles randomisation in a clinical trial. In this way, it can be helpful in providing evidence to support causal relevance of a protein on a disease with less potential for issues relating to confounding and reverse causation. Additionally, it can help overcome the limitations of cell or mouse models that may not fully replicate human biological processes. It is therefore a valuable tool in drug discovery: therapies targeting proteins that are supported by evidence from human genetic studies have been shown to have a greater efficacy in trials and are more likely to gain regulatory approval3.
In recent years, there has been an increase in large-scale profiling of the plasma proteome and transcriptome. This has led to the creation of extensive catalogues of genetic variants that influence protein levels and gene transcription across hundreds of tissues within the human body. Combining these two resources allows for in-depth exploration of biological pathways underlying diseases, allowing the examination of the gene expression and downstream proteomic signatures that might influence disease outcomes with a causal role4.
In this study, we performed a proteome-wide genetic analysis aimed at evaluating the causal relevance of more than 1,500 circulating proteins for risk of preeclampsia across two different proteomic platforms, and corroborated and extended these analyses with data on expression of over 15,000 genes across 36 tissue types.
Methods
This study uses publicly available GWAS summary data accessible via the cited sources summarized in Table 1. All included studies had gained ethical approval and participant consent according to individual protocols available at the referenced publications. The study design is summarised in Figure 1, The methods for this study can be found in the Supplementary Methods.5–9
Table 1. Details of data sources utilised in this study.
| Phenotype | Data source | Number of participants | Data link |
|---|---|---|---|
| Preeclampsia | Honigberg et al. (2023)5 | 16,349 cases / 595,135 controls | https://doi.org/10.6084/m9.figshare.22680904.v1 |
| pQTLs | Ferkingstad et al. (2021)6 deCODE |
35,559 | https://www.decode.com/summarydata/ |
| Sun et al. (2023)7 UK Biobank |
34,557 | https://www.synapse.org/#!Synapse:syn53038826/tables/ | |
| eQTLs | GTEx Analysis Release V88,9 (dbGaP Accession phs000424.v8.p2) |
15,201 tissue samples from 838 donors | https://www.gtexportal.org/home/ |
Figure 1.
Flowchart outlining project design and statistical analysis. pQTLs = protein quantitative trait loci, EUR = European
Results
Proteome-wide Mendelian randomization
After instrument selection and harmonization with outcome association data, pQTLs were available for analysis of 1,593 of the 1,881 proteins with cis-pQTLs in deCODE, and 1,991 of the 2,923 proteins in UK Biobank. There was no evidence of weak instruments, with all instruments exceeding the recommended F-statistic threshold of 10. All instrument F-statistics are presented in Supplementary Table 1.
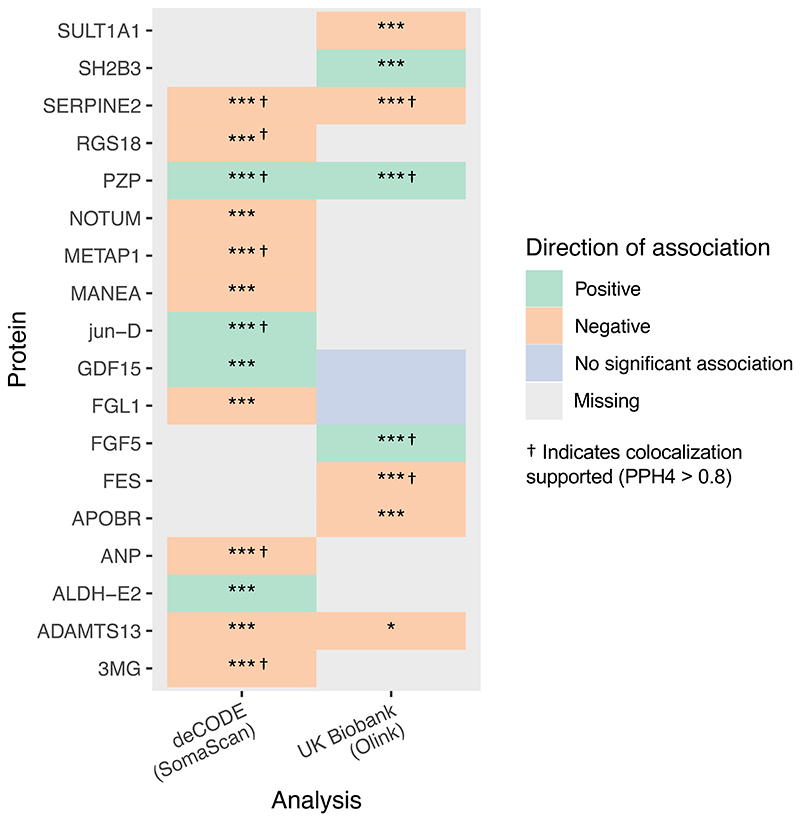
Eighteen proteins had significant genetic associations with preeclampsia. Higher circulating levels of SULT1A1, SERPINE2, RGS18, NOTUM, ?????1, MANEA, FGL1, FES, APOBR, ANP, ADAMTS13 and 3MG were associated with lower risk of preeclampsia. Conversely, higher circulating levels of SH2B3, PZP, jun-D, GDF15, FGF5 and ALDH-E2 were associated with a greater risk of preeclampsia. The results are summarized in Figure 2, and presented in Supplementary Table 2 for deCODE and Supplementary Table 3 for UK Biobank. SERPINE2, PZP, and ADAMTS13 had available pQTLs and consistent associations with preeclampsia across both SOMAscan and Olink datasets.
Figure 2.
Genetic associations of protein levels with preeclampsia, displaying all proteins associated with preeclampsia after correction for multiple testing in the main analysis. Grey fields indicate data was not available for analysis. PPH4 = Posterior probability of shared causal variant, PPH3 = Posterior probability of distinct causal variant. * = nominally significant (p<0.05) ** = p<0.001, *** = significant after Benjamini-Hochberg correction for multiple testing
Sensitivity analyses
Multiple sensitivity analyses were performed to address instrumental variable assumptions, the results of which are summarized in Table 2. Weighted median MR and MR-Egger were not possible for the primary pQTL instruments due to insufficient instruments at genome-wide significance level. After relaxing selection criteria for instruments (p<5×10-6, r2<0.2) and using MR-RAPS, the main analysis was replicated for all proteins except for SULT1A1, NOTUM, MANEA, jun-D, GDF15 (in both deCODE and UK Biobank) and ALDH-E2. After relaxing selection criteria for instruments, MR-Egger and weighted median MR produced consistent results for APOBR, FES, FGF5 and SERPINE2. Sensitivity analyses for ALDH-E2, FGL1, GDF15, MANEA, NOTUM and SULT1A1 were suggestive of horizontal pleiotropy either through a significant MR-Egger intercept (after accounting for multiple testing), or through inconsistency of results when utilising both MR-Egger and weighted median MR methods compared to the main results. The results suggested potential directional pleiotropy for 3MG, ADAMTS14, METAP1, and SH2B3, as the results were consistent on weighted median MR but not on MR-Egger, or vice versa. The analyses could not be performed for ANP, PZP, RGS18 because no additional SNPs were found with the relaxed criteria. The full results are outlined in Supplementary Table 4. Next, we performed a reverse direction MR analysis of significant pQTLs, which did not highlight any evidence of reverse causation (Supplementary Table 5). Finally, we repeated the analyses with a different data source, Zheng et al.’s study, for pQTL instruments4. In this data source, instruments for only 5 of the 18 pQTLs significant on primary analysis were available for validation. The results remained consistent for SERPINE2, PZP and GDF15, but not MANEA or FGL1 (Supplementary Table 6).
Table 2. Summary of proteome-wide, transcriptome-wide and Bayesian genetic colocalization analysis for the proteins significant in the main analysis.
| Protein | Location (GrCh38) | Observational association (PMID) | Consistent in both deCODE and UKB | Consistent on MR-RAPS | Evidence of horizontal pleiotropy | No evidence of bidirectional association | Consistent in Zheng et al pQTLs | Supporting eQTL evidence | Interacting protein eQTL evidence | Colocalization supported |
|---|---|---|---|---|---|---|---|---|---|---|
| SULT1A1 | 16:28,605,196 - 28,614,279 | 29059671 | Unavailable | No | Yes | Yes | Unavailable | Yes | Yes | No |
| SH2B3 | 12:111,405,923 - 111,451,623 | 33239696 | Unavailable | Yes | Some | Yes | Unavailable | No | Yes | No |
| SERPINE2 | 2:223,975,045 - 224,039,318 | - | Yes | Yes | None | Yes | Yes | Yes | Yes | Yes |
| RGS18 | 1:192,158,462 - 192,185,815 | - | Unavailable | Yes | Unavailable | Yes | Unavailable | Yes | Yes | Yes |
| PZP | 12:9,148,840 - 9,208,395 | 37285119; 35843132 | Yes | Yes | Unavailable | Yes | Yes | Unavailable | Unavailable | Yes |
| NOTUM | 17:81,952,507 - 81,961,840 | - | Unavailable | No | Yes | Yes | Unavailable | No | Yes | No |
| METAP1 | 4:98,995,659 - 99,062,809 | - | Unavailable | Yes | Some | Yes | Unavailable | No | No | Yes |
| MANEA | 6:95,577,485 - 95,609,470 | - | Unavailable | No | Yes | Yes | No | Yes | Yes | No |
| jun−D | 19:18,279,694 - 18,281,622 | 24885447 | Unavailable | No | Unavailable | Yes | Unavailable | Unavailable | Unavailable | Yes |
| GDF15 | 19:18,374,731 - 18,389,176 | 36410395 | No | No | Yes | Yes | Yes | No | No | No |
| FGL1 | 8:17,864,380 - 17,910,365 | 36203334 | Unavailable | Yes | Yes | Yes | No | No | No | No |
| FGF5 | 4:80,266,639 - 80,336,680 | 35380267; 33239696 | Unavailable | Yes | None | Yes | Unavailable | Yes | Yes | Yes |
| FES | 15:90,883,695 - 90,895,776 | - | Unavailable | Yes | None | Yes | Unavailable | Yes | Yes | Yes |
| APOBR | 16:28,494,643 - 28,498,964 | - | Unavailable | Yes | None | Yes | Unavailable | Unavailable | No | No |
| ANP | 1:11,845,709 - 11,848,345 | 15511835; 37248299 | Unavailable | Yes | Unavailable | Yes | Unavailable | Unavailable | Yes | Yes |
| ALDH−E2 | 12:111,766,887 - 111,817,532 | - | Unavailable | No | Yes | Yes | Unavailable | Yes | Yes | No |
| ADAMTS13 | 9:133,414,358 - 133,459,402 | 21512165 | Yes | Yes | Some | Yes | Unavailable | Unavailable | No | No |
| 3MG | 16:77,007 - 85,851 | - | Unavailable | Yes | Some | Yes | Unavailable | Yes | Yes | No |
Colocalization
Using Bayesian colocalization analyses, we evaluated the posterior probability of shared causal variants within protein coding regions for the potentially causal pQTL with preeclampsia. The results are presented in in Supplementary Table 7 and Supplementary Table 8 for deCODE and UK Biobank respectively.
In deCODE, results supported colocalization (PPH4 >80%) for SERPINE2 (99.7%), RGS18 (95.2%), PZP (89.8%), METAP1 (92.1%), jun-D (98.1%), ANP (88.4%), and 3MG (90.4%). In UKB, colocalization was supported for SERPINE2 (99.4%), PZP (89.9%), FGF5 (99.7%) and FES (87.9%). The results are presented in Figure 3. In total, 9 of the prioritized proteins were supported by colocalization in one or both datasets.
Figure 3.
Bayesian colocalization analysis evaluating the posterior probability of a shared causal variants influencing protein levels and preeclampsia risk, and probability of colocalization versus non-colocalization conditional on there being a causal variant for both traits [PPH4/(PPH3+PPH4)]. Grey fields indicate data was not available for analysis
Transcriptome-wide Mendelian randomization
In the transcriptome-wide MR, we evaluated the association of gene expression for 103,476 gene/tissue combinations, spanning more than 15,000 genes across 36 different tissues with preeclampsia. Overall, 86 eQTLs displayed at least one association with preeclampsia in at least one tissue that was significant after adjustment for multiple comparisons. These are summarised in Supplementary Figure 1 and Supplementary Table 9.
Direct comparison with significant pQTL results
Comparing the eQTL results with those of the primary pQTL analysis, there was directionally consistent evidence corroborating the pQTL associations in at least one tissue for analysis for SULT1A1, SH2B3, SERPINE2, RGS18, FGF5, FES, ALDH-E2 and 3MG. Among these, ALDH2, FES, MANEA, RGS18, SERPINE2 and SULT1A1 were corroborated using whole blood eQTLs. No tissue-specific eQTL data was available for PZP, jun-D, FGL1, ANP and ADAMTS13. There was no evidence of a directionally consistent association for the remaining pQTLs. Genetically predicted expression of METAP1 in aortic and tibial arterial tissue was significantly associated with preeclampsia, with opposing direction of effect compared to that observed for pQTLs.
Comparison with closely interacting proteins
In addition to looking for eQTL data that directly supports the pQTL findings, we expanded the comparison to also evaluate consistency of associations of eQTLs of the proteins with closest interactions to the significant pQTLs in the main analysis. These were identified as proteins with a STRING interaction score of more than 0.90, with a maximum of 5 proteins included for each significant pQTL. A summary of the identified proteins and the evidence supporting their interaction is provided in Supplementary Table 10.
In these analyses, biologically consistent associations of interacting proteins were noted for ANP, FGF5, SERPINE2, SH2B3 and SULT1A1. For ANP, the clearance receptor NPR3 eQTL was associated with preeclampsia in the opposite direction to the ANP pQTL, which is biologically consistent given its role in clearance of ANP, and a consistent association was found for NPR2 in the tibial artery. There were no additional findings for ADAMTS13, APOBR, FES, FGL1, GDF15, MANEA, METAP1, and RGS18, and no eQTLs relating to interacting proteins were available for 3MG and PZP. The results of the analyses are summarized in Figure 4.
Figure 4.
Genetic associations of gene expression levels with preeclampsia, limited to the proteins identified as significant on the proteome-wide analysis or their closest interacting proteins (maximum of 5 proteins, all with STRING interaction score >0.90). Grey fields indicate data was not available for analysis. * = nominally significant (p<0.05) ** = p<0.001, *** = significant after Benjamini-Hochberg correction for multiple testing
Annotation
Protein function, interactions and druggability
The protein network displaying the 50 top high-confidence interactions (interaction score >0.70) for all 18 proteins is displayed in Supplementary Figure 2. A number of common STRING clusters were identified among the proteins, including ‘Complement and coagulation cascades, and Protein-lipid complex’ (ADAMTS13, APOBR, PZP), ‘Wnt signaling pathway’ (NOTUM, ANP) and ‘Dissolution of Fibrin Clot’ (ADAMTS13, SERPINE2). Full functional annotation for all proteins is provided in Supplementary Table 11.
The results of the druggability evaluation for the significant pQTLs is presented in Supplementary Table 12. A total of 11 of the 18 proteins were classified as druggable (SERPINE2, PZP, NOTUM, METAP1, GDF15, FGL1, FGF5, ANP, ALDH-E2 and ADAMTS13). Among these, three are targets of currently available compounds (METAP1, ANP and ALDH-E2) of which none have safety data relating to pregnancy.
Phenome-wide scanning
The results of phenome-wide scanning for all significant pQTLs are shown in Supplementary Table 13. Among the 18 significant pQTLs, 10 had genome-wide significant associations with other traits. These included blood pressure and cardiovascular disease traits relating to coronary artery disease and heart failure (SH2B3, FGF5, FES, ANP, ALDH-E2), anthropometric traits relating to body mass index, size and adiposity (jun-D, GDF15, APOBR), blood cell traits (SH2B3, FGF5, jun-D, APOBR, ALDH-E2). Instruments for ADAMTS13 were only associated with ADAMTS13 levels in a separate study. FES was additionally associated with birth weight traits. Instruments for SERPINE2 were specifically only associated with platelet traits, as well as PDGF-BB levels, a clinical marker for preeclampsia10.
Discussion
In this study, we aimed to leverage large-scale genetic data to elucidate potential biological pathways contributing to the development of preeclampsia. Overall, genetically-predicted circulating levels of 18 proteins were associated with preeclampsia (SULT1A1, SH2B3, SERPINE2, RGS18, PZP, NOTUM, METAP1, MANEA, jun−D, GDF15, FGL1, FGF5, FES, APOBR, ANP, ALDH−E2, ADAMTS13, 3MG). Among these, 11 were either directly or indirectly supported by gene expression data, and 9 were corroborated by colocalization analyses. Among the studied proteins, 5 were found to have consistent evidence of association and colocalization in all primary and sensitivity analysis (SERPINE2, PZP, FGF5, FES and ANP). Protein interaction mapping identified potential shared biological pathways through natriuretic peptide signalling, blood pressure regulation, immune tolerance and thrombin function.
In this study, lower genetically-predicted levels of SERPINE2 (i.e., serpin family E member 2), a serine protease that directly inhibits thrombin activity, was associated with preeclampsia. Though this association has not been identified before in observational research, the result was consistent across proteomic platforms, and was supported every sensitivity, eQTL and colocalization analysis. In addition, lower genetically-predicted levels of ADAMTS13, a disintegrin and metalloproteinase that modulates thrombin similarly to SERPINE2, were also associated with preeclampsia. Dysregulation of the thrombin pathway plays a well-recognised central role in the pathogenesis of thrombotic thrombocytopenic purpura (TTP), a thrombotic microangiopathy with phenotypic overlap with preeclampsia11. On the whole, these findings suggest deranged thrombin regulation as a key mechanism underlying the development of preeclampsia. This is consistent with the known clinical association between procoagulant conditions such as Factor V Leiden and preeclampsia risk12, with the pathological overlap of preeclampsia with thrombotic microangiopathies13, and with the greater thrombin levels that women with preeclampsia display compared to controls14.
The role of ANP in the maintenance of cardiovascular homeostasis, regulation of natriuresis, vascular remodelling, and its interaction with the renin-angiotensin-aldosterone pathway are well-described15–21. The role of ANP in the development of preeclampsia is less recognised but has been suggested in preclinical models. A previous study by Cui et al.22 demonstrated that pregnant mice lacking either ANP or corin (a cardiac protease that activates ANP, also known as ANP-converting enzyme) exhibited a preeclampsia phenotype. Observational studies have additionally demonstrated that low first trimester levels of the closely related NT-proBNP are associated with greater risk of hypertensive disorders of pregnancy23, a finding corroborated by a recent protein-based MR study24. In our study, lower genetically-predicted ANP levels were associated with preeclampsia. Then, when exploring the association of gene expression of ANP and its 5 strongest associated proteins with preeclampsia, we found that expression of NPR2, a receptor for both ANP and BNP, was associated with higher risk of preeclampsia, whereas greater expression of NPR3, a clearance receptor for ANP, is associated with greater risk of preeclampsia. Finally, colocalization analyses supported that the genetic predisposition to lower ANP and preeclampsia are due to a shared causal variant. On the whole, the combination of these findings support a causal relevance of low pre-pregnancy ANP levels in the development of preeclampsia that warrants its further investigation as a potential therapeutic target.
In the analysis in the UK Biobank, a strong genetic association was found between FGF5 (or fibroblast growth factor 5) and preeclampsia which was corroborated by colocalization. This was also corroborated by gene expression data, where renal cortex FGF5 expression was strongly associated with preeclampsia. FGF5 polymorphisms have been linked with preeclampsia in a Chinese population25 and the FGF5 locus has been consistently prioritised as potentially causal in multiple GWASs of preeclampsia5,26,27 as well as blood pressure traits28–31. Similarly, lower levels of FES, a protein regulating endothelial permeability and leukocyte transmigration, were associated with preeclampsia across all analyses, indicating that FES is the likely causal gene at the previously identified FURIN/FES locus5,32. Together with the additional association of SH2B3, another established blood pressure locus31,33–35, this result supports the hypothesis that at least one of the mechanisms predisposing to preeclampsia is through a shared pathway with hypertension. This is in line with the multitude of observational evidence linking pre-existing hypertension to preeclampsia risk, and conversely, preeclampsia with later life hypertension risk36. The present study extends these prior data by implicating renal physiology in preeclampsia risk and pinpointing an important role for FGF5.
The pathophysiological mechanism underpinning the associations of PZP with preeclampsia is unclear. Higher genetically-predicted levels of circulating maternal PZP were associated with higher risk of preeclampsia. PZP is a broad-spectrum immunosuppressive protein inhibits protease activity and stabilizes misfolded proteins, as well as modulating T helper cell response, through which it is thought to play a part in preventing rejection of the fetus during pregnancy37. Its relevance in the pathophysiology of preeclampsia is not well studied to date, with only one study so far describing lower expression of PZP in preeclamptic placentas38, an association which is opposite in direction to the findings of our study. This association suggests a potential importance of immune regulation and tolerance. Importantly, PZP has previously been highlighted as susceptibility loci in a previous GWAS study32. Overall, our study highlights a likely role of these two proteins in the development of preeclampsia that warrants further investigation.
In this study, we found that higher GDF15 (or growth/differentiation factor 15) protein levels were associated with preeclampsia. GDF15 is a protein that regulates food intake, energy expenditure and body weight in response to metabolic and toxin-induced stresses, and has been implicated as a key factor in the pathogenesis of hyperemesis gravidarum where high fetal protein levels are combined with maternal sensitivity to the hormone due to low intrinsic GDF15 levels39. In the setting of preeclampsia, higher GDF15 levels have been described prior to diagnosis of preeclampsia40, findings that were corroborated in a recent meta-analysis41. However, there was some inconsistency in sensitivity analyses in our study, and we were not able to corroborate this finding using gene expression data and in colocalization analyses. For this reason, its role in preeclampsia remains unclear and remains an important target for future investigation.
This study has a number of key strengths. First, the analysis spans a large scale of cross-platform proteomic and transcriptomic data, which has not been previously leveraged to investigate the biological pathways underlying preeclampsia. This higher coverage compared to any previous study increases the likelihood of uncovering important related mechanisms. In addition to this, we utilised a careful and step-wise approach starting with cross-platform proteomic data, corroborating it with transcriptomic data and colocalization analyses, and linking the results through interaction networks. This provides a comprehensive overview of the results and utilises available tools to help understand how the results of the analyses relate to each other. The use of MR and colocalization analyses helps increase confidence in the causal relevance of explored proteins on preeclampsia, where the results of these both support it. This is an important advantage, as it avoids the issues of confounding and reverse causation that often limit causal inference in biomarker studies of preeclampsia. Finally, the cross-platform approach is an important strength. Current data regarding the sensitivity and specificity of available commercial multiplexed protein assays is has highlighted important shortcomings42. For this reason, when proteome-wide exploration analyses are carried out, validation with both alternative proteomic assays as well as gene expression data are crucial to lend reliability to the results and reassure against off-target results due to poor assay specificity.
Limitations
There are a number of limitations to discuss. First, despite using the largest-to-date studies available, protein and tissue-specific gene expression coverage in the data sources remain incomplete. For example, it can be noted that many of the proteins that have been observationally associated with preeclampsia (e.g., s-Flt1, PLGF, LEP, ENGL) were not available. In addition to this, no data were available for analysis of ANP in the replication analysis using Zheng et al.’s data43, and similarly, there were no instruments for gene expression of the ANP gene, NPPA, in any of the studied tissues. Similar to this, even when instruments were available, where a result was not replicated in the transcriptome-wide MR its eQTL data was sometimes only available in very few tissues (e.g., GDF15, available in only one tissue). For this reason, a non-significant result might simply be due to lack of instrument availability in the ‘correct’ tissue where expression might relate to preeclampsia. Relating to this point, we were unable to analyse eQTL data for placental tissue. This is because, since the placenta is fetal and not maternal tissue, it is representative of fetal gene expression and would therefore require corresponding gene-outcome association data for fetal variants with maternal preeclampsia risk; as opposed to the gene-outcome association data used in this study that reflects the influence maternal variants on maternal preeclampsia risk. Second, in order to avoid potential confounding by population stratification, we restricted the analysis to using only data derived from European ancestry populations. This might limit the generalisability of the findings to populations of other ancestries. Given the lower risk of preeclampsia in women of European ancestry compared to, for example, those of African ancestry, it is imperative for this investigation to be repeated in cohorts of other ancestries once sufficient data is available to do so. Finally, the reliability of MR results is strongly dependent on meeting instrumental variable assumptions. We addressed these in the study design by restriction of ancestry, utilisation of cis-MR to reduce risk of pleiotropy, as well as multiple sensitivity analyses and corroboration using gene expression data as well as colocalization. Nevertheless, even where robust evidence of potential causal relevance was found, validation of results in interventional trials remains the gold standard and cannot be replaced by genetic studies.
Conclusion
In this study, we performed a proteome- and transcriptome-wide genetic analysis aimed at evaluating the causal relevance of thousands of circulating proteins with preeclampsia, and supporting this with data on expression of over 15,000 genes across 36 tissue types. Our investigation identified multiple targetable proteins linked to cardiovascular, inflammatory, and coagulation pathways, with SERPINE2, PZP, FGF5, FES and ANP identified as a pivotal proteins with likely causal roles in the development of preeclampsia. Future studies should focus on evaluating an epidemiological temporal association of these proteins in women before and during pregnancy to corroborate the link with preeclampsia, and if this is confirmed, evaluate the efficacy and safety of intervening in these pathways through animal models and human trials.
Supplementary Material
Acknowledgments
The authors acknowledge all investigators and participants of the studies contributing to the present analyses.
Sources of Funding
This work was supported by core funding from the British Heart Foundation (RG/18/13/33946), NIHR Cambridge Biomedical Research Centre (BRC-1215-20014; NIHR203312) [*], Cambridge BHF Centre of Research Excellence (RE/18/1/34212), BHF Chair Award (CH/12/2/29428). MA is supported by the National Institute for Health Research Academic Clinical Fellowship. PN is supported by the U.S. National Heart, Lung, and Blood Institute (NHLBI R01HL127564, NHGRI U01HG011719). AdM is supported by the Fetal Medicine Foundation (495237). MCH is supported by the U.S. National Heart, Lung, and Blood Institute (NHLBI, K08HL166687) and the American Heart Association (940166, 979465). AB is supported by core funding from the British Heart Foundation (RG/18/13/33946) and NIHR Cambridge Biomedical Research Centre (BRC-1215-20014; NIHR203312) [*]. All other authors have no funding to declare. *The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
Footnotes
Disclosures: P.N. reports research grants from Allelica, Amgen, Apple, Boston Scientific, Genentech / Roche, and Novartis, personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Foresite Labs, Genentech / Roche, GV, HeartFlow, Magnet Biomedicine, Merck, and Novartis, scientific advisory board membership of Esperion Therapeutics, Preciseli, and TenSixteen Bio, scientific co-founder of TenSixteen Bio, equity in MyOme, Preciseli, and TenSixteen Bio, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. MCH reports consulting fees from Comanche Biopharma, advisory board service for Miga Health, and grant support from Genentech, all unrelated to the present work. AB reports institutional grants from AstraZeneca, Bayer, Biogen, BioMarin, Bioverativ, Novartis, Regeneron and Sanofi. All other authors have no disclosures to declare.
References
- 1.Rolnik DL, Wright D, Poon LC, O’Gorman N, Syngelaki A, de Paco Matallana C, Akolekar R, Cicero S, Janga D, Singh M, et al. Aspirin versus Placebo in Pregnancies at High Risk for Preterm Preeclampsia. New England Journal of Medicine. 2017;377:613–622. doi: 10.1056/NEJMoa1704559. [DOI] [PubMed] [Google Scholar]
- 2.Burgess S, Scott RA, Timpson NJ, Smith GD, Thompson SG. Using published data in Mendelian randomization: A blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30:543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 4.Zheng J, Haberland V, Baird D, Walker V, Haycock PC, Hurle MR, Gutteridge A, Erola P, Liu Y, Luo S, et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet. 2020;52:1122–1131. doi: 10.1038/s41588-020-0682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honigberg MC, Truong B, Khan RR, Xiao B, Bhatta L, Vy HMT, Guerrero RF, Schuermans A, Selvaraj MS, Patel AP, et al. Polygenic prediction of preeclampsia and gestational hypertension. Nat Med. 2023;29:1540–1549. doi: 10.1038/s41591-023-02374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL, Gunnarsdottir K, Helgason A, Oddsson A, Halldorsson BV, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet. 2021;53:1712–1721. doi: 10.1038/s41588-021-00978-w. [DOI] [PubMed] [Google Scholar]
- 7.Sun BB, Chiou J, Traylor M, Benner C, Hsu YH, Richardson TG, Surendran P, Mahajan A, Robins C, Vasquez-Grinnell SG, et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023;622:329–338. doi: 10.1038/s41586-023-06592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.GTEx Consortium, Laboratory DA &Coordinating C (LDACC)—Analysis WG, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, NIH/NHGRI, NIH/NIMH, NIH/NIDA, Biospecimen Collection Source Site—NDRI et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu M, Niu Y, Ma K, Leung PCK, Chen ZJ, Wei D, Li Y. Identification of novel first-trimester serum biomarkers for early prediction of preeclampsia. J Transl Med. 2023;21:634. doi: 10.1186/s12967-023-04472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George JN, Nester CM, McIntosh JJ. Syndromes of thrombotic microangiopathy associated with pregnancy. Hematology. 2015;2015:644–648. doi: 10.1182/asheducation-2015.1.644. [DOI] [PubMed] [Google Scholar]
- 12.Dizon-Townson DS, Nelson LM, Easton K, Ward K. The factor V Leiden mutation may predispose women to severe preeclampsia. Am J Obstet Gynecol. 1996;175:902–905. doi: 10.1016/s0002-9378(96)80022-6. [DOI] [PubMed] [Google Scholar]
- 13.Jackson SP, Darbousset R, Schoenwaelder SM. Thromboinflammation: challenges of therapeutically targeting coagulation and other host defense mechanisms. Blood. 2019;133:906–918. doi: 10.1182/blood-2018-11-882993. [DOI] [PubMed] [Google Scholar]
- 14.Rafik Hamad R, Curvers J, Berntorp E, Eriksson MJ, Bremme K. Increased thrombin generation in women with a history of preeclampsia. Thromb Res. 2009;123:580–586. doi: 10.1016/j.thromres.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 15.Bordicchia M, Liu D, Amri EZ, Ailhaud G, Dessì-Fulgheri P, Zhang C, Takahashi N, Sarzani R, Collins S. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J Clin Invest. 2012;122:1022–1036. doi: 10.1172/JCI59701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birkenfeld AL, Budziarek P, Boschmann M, Moro C, Adams F, Franke G, Berlan M, Marques MA, Sweep FCGJ, Luft FC, et al. Atrial natriuretic peptide induces postprandial lipid oxidation in humans. Diabetes. 2008;57:3199–3204. doi: 10.2337/db08-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Souza SC, Chau MDL, Yang Q, Gauthier MS, Clairmont KB, Wu Z, Gromada J, Dole WP. Atrial natriuretic peptide regulates lipid mobilization and oxygen consumption in human adipocytes by activating AMPK. Biochem Biophys Res Commun. 2011;410:398–403. doi: 10.1016/j.bbrc.2011.05.143. [DOI] [PubMed] [Google Scholar]
- 18.Rubattu S, Bigatti G, Evangelista A, Lanzani C, Stanzione R, Zagato L, Manunta P, Marchitti S, Venturelli V, Bianchi G, et al. Association of atrial natriuretic peptide and type a natriuretic peptide receptor gene polymorphisms with left ventricular mass in human essential hypertension. J Am Coll Cardiol. 2006;48:499–505. doi: 10.1016/j.jacc.2005.12.081. [DOI] [PubMed] [Google Scholar]
- 19.Rubattu S, Marchitti S, Bianchi F, Di Castro S, Stanzione R, Cotugno M, Bozzao C, Sciarretta S, Volpe M. The C2238/αANP variant is a negative modulator of both viability and function of coronary artery smooth muscle cells. PLoS One. 2014;9:e113108. doi: 10.1371/journal.pone.0113108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Koller KJ, Lowe DG, Bennett GL, Minamino N, Kangawa K, Matsuo H, Goeddel DV. Selective activation of the B natriuretic peptide receptor by C-type natriuretic peptide (CNP) Science. 1991;252:120–123. doi: 10.1126/science.1672777. [DOI] [PubMed] [Google Scholar]
- 21.Lenz W, Herten M, Gerzer R, Drummer C. Regulation of natriuretic peptide (urodilatin) release in a human kidney cell line. Kidney Int. 1999;55:91–99. doi: 10.1046/j.1523-1755.1999.00242.x. [DOI] [PubMed] [Google Scholar]
- 22.Cui Y, Wang W, Dong N, Lou J, Srinivasan DK, Cheng W, Huang X, Liu M, Fang C, Peng J, et al. Role of corin in trophoblast invasion and uterine spiral artery remodelling in pregnancy. Nature. 2012;484:246–250. doi: 10.1038/nature10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauspurg A, Marsh DJ, McNeil RB, Bairey Merz CN, Greenland P, Straub AC, Rouse CE, Grobman WA, Pemberton VL, Silver RM, et al. Association of N-Terminal Pro–Brain Natriuretic Peptide Concentration in Early Pregnancy With Development of Hypertensive Disorders of Pregnancy and Future Hypertension. JAMA Cardiol. 2022;7:268. doi: 10.1001/jamacardio.2021.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuermans A, Truong B, Ardissino M, Bhukar R, Slob EAW, Nakao T, Dron JS, Small AM, Cho SMJ, Yu Z, et al. Genetic Associations of Circulating Cardiovascular Proteins With Gestational Hypertension and Preeclampsia. JAMA Cardiol. 2024;9:209–220. doi: 10.1001/jamacardio.2023.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin Q, Han Y, Jiang W, Wang J, Luan Y, Ji Q, Sun W. Genetic susceptibility analysis of FGF5 polymorphism to preeclampsia in Chinese Han population. Mol Genet Genomics. 2022;297:791–800. doi: 10.1007/s00438-022-01889-z. [DOI] [PubMed] [Google Scholar]
- 26.Steinthorsdottir V, McGinnis R, Williams NO, Stefansdottir L, Thorleifsson G, Shooter S, Fadista J, Sigurdsson JK, Auro KM, Berezina G, et al. Genetic predisposition to hypertension is associated with preeclampsia in European and Central Asian women. Nat Commun. 2020;11:5976. doi: 10.1038/s41467-020-19733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Changalidis AI, Maksiutenko EM, Barbitoff YA, Tkachenko AA, Vashukova ES, Pachuliia OV, Nasykhova YA, Glotov AS. Aggregation of Genome-Wide Association Data from FinnGen and UK Biobank Replicates Multiple Risk Loci for Pregnancy Complications. Genes (Basel) 2022;13:2255. doi: 10.3390/genes13122255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeuchi F, Akiyama M, Matoba N, Katsuya T, Nakatochi M, Tabara Y, Narita A, Saw WY, Moon S, Spracklen CN, et al. Interethnic analyses of blood pressure loci in populations of East Asian and European descent. Nat Commun. 2018;9:5052. doi: 10.1038/s41467-018-07345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, Ntalla I, Surendran P, Liu C, Cook JP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49:403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X, Wang L, Lin X, Huang J, Charles Gu C, He M, Shen H, He J, Zhu J, Li H, et al. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum Mol Genet. 2015;24:865–874. doi: 10.1093/hmg/ddu478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, Kelly TN, Saleheen D, Lehne B, Leach IM, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;4:1282–1293. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyrmi JS, Kaartokallio T, Lokki AI, Jääskeläinen T, Kortelainen E, Ruotsalainen S, Karjalainen J, Ripatti S, Kivioja A, Laisk T, et al. Genetic Risk Factors Associated With Preeclampsia and Hypertensive Disorders of Pregnancy. JAMA Cardiol. 2023;8:674. doi: 10.1001/jamacardio.2023.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feitosa MF, Kraja AT, Chasman DI, Sung YJ, Winkler TW, Ntalla I, Guo X, Franceschini N, Cheng CY, Sim X, et al. Novel genetic associations for blood pressure identified via gene-alcohol interaction in up to 570K individuals across multiple ancestries. PLoS One. 2018;13:e0198166. doi: 10.1371/journal.pone.0198166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander MR, Hank S, Dale BL, Himmel L, Zhong X, Smart CD, Fehrenbach DJ, Chen Y, Prabakaran N, Tirado B, et al. A Single Nucleotide Polymorphism in SH2B3/LNK Promotes Hypertension Development and Renal Damage. Circ Res. 2022;131:731–747. doi: 10.1161/CIRCRESAHA.121.320625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ardissino M, Slob EAW, Millar O, Reddy RK, Lazzari L, Patel KHK, Ryan D, Johnson MR, Gill D, Ng FS. Maternal Hypertension Increases Risk of Preeclampsia and Low Fetal Birthweight: Genetic Evidence From a Mendelian Randomization Study. Hypertension. 2022;79:588–598. doi: 10.1161/HYPERTENSIONAHA.121.18617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skornicka EL, Kiyatkina N, Weber MC, Tykocinski ML, Koo PH. Pregnancy zone protein is a carrier and modulator of placental protein-14 in T-cell growth and cytokine production. Cell Immunol. 2004;232:144–156. doi: 10.1016/j.cellimm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Löb S, Vattai A, Kuhn C, Mittelberger J, Herbert SL, Wöckel A, Schmoeckel E, Mahner S, Jeschke U. The Pregnancy Zone Protein (PZP) is significantly downregulated in the placenta of preeclampsia and HELLP syndrome patients. J Reprod Immunol. 2022;153:103663. doi: 10.1016/j.jri.2022.103663. [DOI] [PubMed] [Google Scholar]
- 39.Fejzo M, Rocha N, Cimino I, Lockhart SM, Petry CJ, Kay RG, Burling K, Barker P, George AL, Yasara N, et al. GDF15 linked to maternal risk of nausea and vomiting during pregnancy. Nature. 2024;625:760–767. doi: 10.1038/s41586-023-06921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruickshank T, MacDonald TM, Walker SP, Keenan E, Dane K, Middleton A, Kyritsis V, Myers J, Cluver C, Hastie R, et al. Circulating Growth Differentiation Factor 15 Is Increased Preceding Preeclampsia Diagnosis: Implications as a Disease Biomarker. J Am Heart Assoc. 2021;10:e020302. doi: 10.1161/JAHA.120.020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Yang Q. Circulating Growth Differentiation Factor 15 and Preeclampsia: A Meta-Analysis. Horm Metab Res. 2023;55:114–123. doi: 10.1055/a-1956-2961. [DOI] [PubMed] [Google Scholar]
- 42.Katz DH, Robbins JM, Deng S, Tahir UA, Bick AG, Pampana A, Yu Z, Ngo D, Benson MD, Chen ZZ, et al. Proteomic profiling platforms head to head: Leveraging genetics and clinical traits to compare aptamer-and antibody-based methods. Sci Adv. 2022;8:eabm5164. doi: 10.1126/sciadv.abm5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng J, Haberland V, Baird D, Walker V, Haycock PC, Hurle MR, Gutteridge A, Erola P, Liu Y, Luo S, et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet. 2020;52:1122–1131. doi: 10.1038/s41588-020-0682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Core Team. R: A language and environment for statistical computing. Computing. R Foundati; 2021. [Google Scholar]
- 45.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haslam DE, Li J, Dillon ST, Gu X, Cao Y, Zeleznik OA, Sasamoto N, Zhang X, Eliassen AH, Liang L, et al. Stability and reproducibility of proteomic profiles in epidemiological studies: comparing the Olink and SOMAscan platforms. Proteomics. 2022;22:e2100170. doi: 10.1002/pmic.202100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, Janjic N. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol Ther Nucleic Acids. 2014;3:e201. doi: 10.1038/mtna.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39:e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. The Annals of Statistics. 2020;48 doi: 10.1214/19-AOS1866. [DOI] [Google Scholar]
- 53.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, Sarwath H, Thareja G, Wahl A, DeLisle RK, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emilsson V, Ilkov M, Lamb JR, Finkel N, Gudmundsson EF, Pitts R, Hoover H, Gudmundsdottir V, Horman SR, Aspelund T, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361:769–773. doi: 10.1126/science.aaq1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao C, Chen G, Song C, Keefe J, Mendelson M, Huan T, Sun BB, Laser A, Maranville JC, Wu H, et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun. 2018;9:3268. doi: 10.1038/s41467-018-05512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Folkersen L, Fauman E, Sabater-Lleal M, Strawbridge RJ, Frånberg M, Sennblad B, Baldassarre D, Veglia F, Humphries SE, Rauramaa R, et al. Ripatti S, editor. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13:e1006706. doi: 10.1371/journal.pgen.1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zuber V, Grinberg NF, Gill D, Manipur I, Slob EAW, Patel A, Wallace C, Burgess S. Combining evidence from Mendelian randomization and colocalization: Review and comparison of approaches. Am J Hum Genet. 2022;109:767–782. doi: 10.1016/j.ajhg.2022.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ochoa D, Hercules A, Carmona M, Suveges D, Baker J, Malangone C, Lopez I, Miranda A, Cruz-Castillo C, Fumis L, et al. The next-generation Open Targets Platform: reimagined, redesigned, rebuilt. Nucleic Acids Res. 2023;51:D1353–D1359. doi: 10.1093/nar/gkac1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finan C, Gaulton A, Kruger FA, Lumbers RT, Shah T, Engmann J, Galver L, Kelley R, Karlsson A, Santos R, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, Butterworth AS, Staley JR. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35:4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.