Abstract
Increasing evidence highlight the involvement of immune cells in brain activity and its dysfunction. The brain’s immune compartment is a dynamic ensemble of cells that can fluctuate even in naïve animals. However, the dynamics and factors that can affect the composition of immune cells in the naïve brain are largely unknown. Here we examined whether acute sleep deprivation can affect the brain’s immune compartment (parenchyma, meninges and choroid plexus). Using high-dimensional mass cytometry analysis we broadly characterized the effects of short-term sleep deprivation on the immune composition in the mouse brain. We found that after 6 hours of sleep deprivation there was a significant increase in the abundance of B cells in the brain compartment. This effect can be accounted for, at least in part, by the elevated expression of the migration-related receptor, CXCR5, on B cells and its ligand, cxcl13, in the meninges following sleep deprivation. Thus, our study reveals that short-term sleep deprivation affects the brain’s immune compartment, offering a new insight into how sleep disorders can affect brain function and potentially contribute to neurodegeneration and neuroinflammation.
Keywords: Neuroimmunology, sleep deprivation, immunology, immune cells, B cells, brain compartment, CXCR5, cxcl13
Introduction
Many neurodegenerative conditions such as Alzheimer’s disease (AD)1,2, multiple sclerosis (MS)3,4, and Parkinson’s disease (PD)5 are accompanied by disordered sleep. These same conditions are also often characterized by immune dysfunction in the brain and in the periphery6–14. However, the nature of the relationship between sleep, immunity and neurodegeneration remains unknown. Moreover, it is still unclear whether sleep has an effect on the brain’s immune compartment under normal conditions. Uncovering such a connection is important since increasing evidence reveals the central role played by immune cells in brain plasticity15, memory16,17, cognition18 and emotional processes19,20, all of which are also affected by sleep, and especially by sleep deprivation.
Previous studies have demonstrated that sleep affects peripheral immunity21–24. For example, circadian fluctuations and sleep were suggested to induce the migration of lymphocytes between the blood and the lymphatic system25,26, and sleep deprivation has been shown to impair immunological memory formation following hepatitis B vaccination27,28. Notably, the relationship between sleep and immunity is bi-directional, as made evident by sickness behavior and specific induced immune fluctuations, when immune activity alters sleep architecture29–31. For example, when centrally administered, interleukin (IL-1) was shown to increase non-rapid eye movement sleep32,33. This cytokine has been shown to affect neuronal activity in the wake-regulating centers of the hypothalamic preoptic area/basal forebrain34, and consequently increase sleep duration. Additionally, sleep restriction has been associated with increased secretion of proinflammatory cytokines35, such as tumor necrosis factor α (TNF-α) in the blood plasma36, and neurodegeneration and neuroinflammation are commonly characterized by sleep disturbances2,37,38. More specifically, microglia, the brain’s resident immune cell populations, has been proposed as a key player in the pathophysiology of both sleep disorders and neurodegeneration39–42. Thus, understanding how sleep affects the “brain’s immune compartment” may uncover novel sleep-regulatory mechanisms.
The “brain’s immune compartment” includes the composition of immune cells in the brain parenchyma, its surrounding tissues, the choroid plexus and meninges, and the perivascular spaces. It is characterized by resident myeloid cells and peripheral cells, which enter the brain compartment from the blood. These hematopoietic myeloid and lymphoid populations are mostly located in the meninges and choroid plexus43. Under pathological conditions, these cells can also be detected within the brain parenchyma44–47. In recent years, our understanding of the resident and infiltrating immune populations in the brain compartment has been expanded considerably. The emergence of high-dimensional cell characterization techniques such as mass cytometry (CyTOF) and single-cell RNA sequencing, revealed several largely uncharacterized immune populations in the naïve brain43,48–50. Especially interesting among these populations are B cells, which comprise ~10% of the brain’s blood-derived immune populations. Even in peripheral tissues, new evidence highlights the effects of B cells as antigen-presenting cells and cytokine-producing entities, in addition to their well-characterized role in antibody production51. Similarly, new roles for B cells within the brain compartment are constantly being revealed; these cells were shown to play a role in brain myelination processes during development52. Following stroke, analysis of postmortem tissue suggested that B cells reaching the damaged area are associated with the subsequent development of dementia53. These findings are supported by studies conducted in mice where pharmacologic ablation of B-lymphocytes using an anti-CD20 antibody prevented the appearance of delayed cognitive deficits following stroke53. Other studies identified regulatory B cells as immune modulators in stroke54, in autoimmune conditions and under various stressors 55. For example, B cells were found within MS plaques9,56,57 and targeting B cells was shown to attenuate clinical and pathological symptoms of experimental autoimmune encephalomyelitis (EAE)58–61. Psychological stress has been suggested as a modulator of B cell numbers, via glucocorticoids that induce apoptosis of pre-B-cells62. Collectively, these findings contribute to the emerging perspective of the brain as a privileged, yet vibrant immune environment, and has led us to characterize immune cells in the brain compartment following acute sleep deprivation. Nevertheless, the specific role of B cells in the brain’s immune compartment is still largely unknown.
Methods
Mice
Adult (8-10 weeks of age; 20-25 gr) male C57BL/6 mice were used in all experiments. Mice were maintained under Specific-Pathogen-Free (SPF) conditions; four mice were housed in each cage maintained on a 12:12 light cycle (lights on at 07:00) 24±1°C, humidity 30–70%. All experiments were performed in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals. All procedures and protocols were approved by the Technion Administrative Panel of Laboratory Animal Care.
Sleep deprivation
Mice were left to sleep, or sleep-deprived for 6 hours, starting at light onset (ZT0). Sleep deprivation was maintained by gentle handling as shown previously96, to minimize stress responses.
Brain single-cell dissociation
Mice were fully anesthetized (ketamine 80 mg/kg, xylazine 15-20 mg/kg) and perfused with at least 20 ml ice-cold PBS-/- (without Mg+2 and Ca+2; Sigma-Aldrich) through the left ventricle of the heart. Brain tissue was dissociated as previously described43,71,97, with several modifications. Briefly, brains were dissected and placed in PBS-/- on ice. Brain tissue was cut to small pieces, transferred to 7 ml of RPMI-1640 (Sigma-Aldrich) dissociation solution, with collagenase D (0.4 mg/ml; Sigma-Aldrich), DNase I (0.2 mg/ml; Sigma-Aldrich), 1% fetal bovine serum (FBS; Biological Industries), and 1 mM HEPES (Sigma-Aldrich), and mechanically dissociated with a dounce homogenizer. Following incubation (30 min, 37°C. shaking 200 rpm), immune cells were isolated by centrifugation (30 min at 500 g, 18°C) using a 30% Percoll (Sigma-Aldrich, GE Healthcare Bio-Sciences) gradient, comprised of 3 ml stock isotonic Percoll (SIP; 10% X10 PBS-/- diluted in 90% Percoll) and 7 ml of the RPMI-1640 dissociation solution containing the cells. The top myelin layer and supernatant were removed, and the cell pellet was washed once with 10 ml of PBS-/- (7 min at 500 g, 4°C), then re-suspended in 1 ml of staining buffer (PBS-/-, containing 1% bovine serum albumin and 0.05% sodium azide).
Blood extraction
Prior to perfusion, the right atrium of the heart was cut and blood was collected into EDTA coated sterile test tubes, lysed with 10 ml red blood cell lysis buffer for 15 min (BD Biosciences, NJ, US), and then washed twice with staining buffer.
Mass cytometry
For mass cytometry analyses, we pooled six mouse brains for each sample. Pooled cells (3-4·106) from each tissue were incubated with TruStain fcX™ (anti-mouse CD16/32, clone 93; 1:100) for FC blocking, and rhodium-103 (1:2000; Rh; Fluidigm) for live/dead discrimination (20 min, 4°C). Samples were washed twice (5 min, 500 g, 4°C) with staining buffer and then treated for palladium barcoding98 according to the manufactures’ instructions (Cell-ID 20-Plex Pd Barcoding Kit; Fluidigm). Briefly, each sample was incubated in X1 fix buffer (10 min, room temperature; RT), then washed twice with X1 perm buffer, and incubated with a palladium barcode for 30 min, RT. Following barcode incubation, samples were washed twice (5 min, 500 g, 4°C), pooled to a single sample, and stained with the metal-conjugated antibody mix (1 hr, 4°C; a complete list of antibodies is provided in Supplementary Table 1). The pooled sample was washed twice (5 min, 500 g) and fixed in 1.6% freshly made PFA in PBS-/- (1 hr, RT; Sigma-Aldrich), then incubated with iridium-191/193 (1:2000, 20 min, RT; Ir; Fluidigm) for live/dead discrimination, washed in ultrapure H2O (5 min, 500 g) and analyzed on a CyTOF I machine (Fluidigm). Antibodies were conjugated in-house using the MAXPAR reagent (Fluidigm), and the optimal concentration for staining was determined by titration. Internal metal isotope bead standards were added for sample normalization by Matlab as shown previously99, to account for the decline in mean marker intensity over time. Acquired data were uploaded to Cytobank100 for data processing, exclusion of dead cells and normalization beads, and analysis. Immune population in were defined as (Figure 1, Figure S1): resident myeloid cells (CD45lowCD44negCX3CR1+CD11b+), monocytes (CD45highCD11b+Ly6C+), granulocytes (CD45highCD11b+Gr-1+), DCs (CD45highMHC-II+CD11c+), CD4 T cells (CD45highTCR-β+CD4+), CD8 T cells (CD45highTCR-β+CD8+), B cells (CD45highMHC-II+CD19+), NK cells (CD45highTCR-βnegNK1.1+CD49b+).
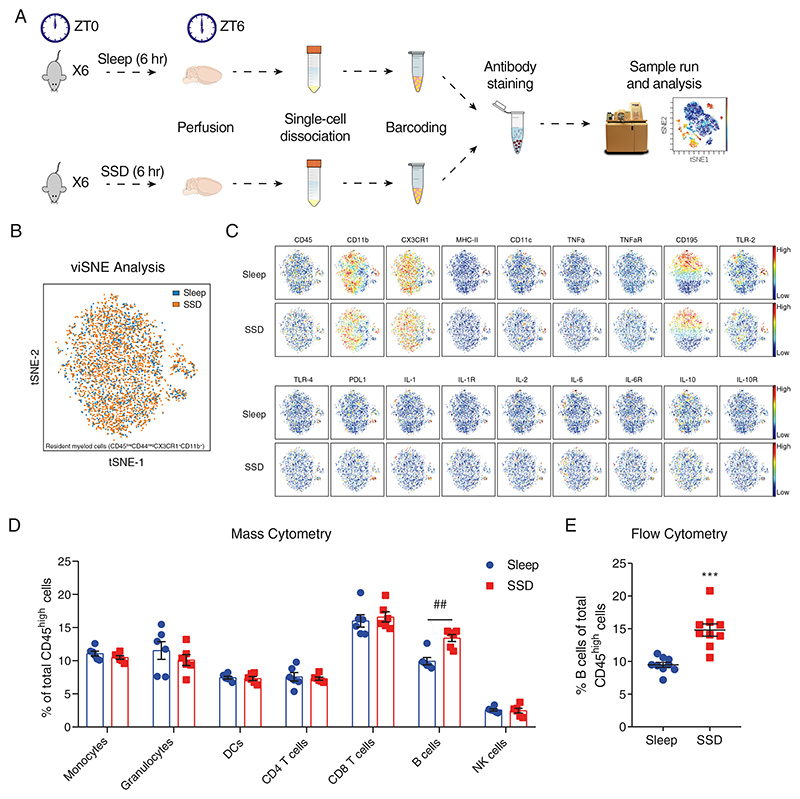
Figure 1. Characterization of immune cells in the brain compartment following short-term sleep deprivation.
(A) Schematic representation of the experimental procedure. Following 6 hours of sleep or sleep deprivation (ZT0 to ZT6), mice were sacrificed and perfused. Each mouse brain was dissociated to a single-cell suspension. Pools of single-cell suspensions from 6 mouse brains per group were barcoded, stained and analyzed using a mass cytometer. (B) viSNE analysis of sleep-control and sleep-deprived resident myeloid cells in an overlaid dot plot, and (C) in a color gradient plot for each marker. (D) The various blood-derived immune cell populations in the brain compartment, following 6 hours of sleep (sleep; left) or 6 hours of sleep deprivation (SSD; right), and the cell subset proportions of each group. Data are presented as mean±sem; multiple t-tests, n=6 pooled specimens of six mice in each sample, corrected p-value ##P<0.002; data of three experiments. (E) Flow cytometry analysis of the percentage of B cells (CD45high/+CD19+MHC-II+) of total CD45high cells in the brain compartment, following 6 hours of sleep (sleep; blue circle) or sleep deprivation (SSD; red square). Data are presented as mean±sem; Student’s t-test, n=9 individual mice; ***P<0.001; representative data of three experiments.
Flow cytometry
Cells were stained with the following antibodies: Pacific Blue or PE/Cy7-conjugated anti-CD19 (6D5, 115523, 115520, Biolegend), Alexa Fluor 488 or Pacific Blue-conjugated anti-MHC-II (M5/114.15.2, 107616,107620, Biolegend), PE or Alexa Fluor 700-conjugated anti-CD45 (30-F11, 103106, 103128, Biolegend), Alexa Fluor 488-conjugated anti-CD40 (HM40-3, 102910, Biolegend), APC-conjugated anti-IgM (1B4B1, 1140-11, Southern Biotech), APC-conjugated anti-IgG (X56, 550874, BD Bioscience), Alexa Fluor 647-conjugated anti-TLR-2 (QA16A01, 153008, Biolegend), APC-conjugated anti-CX3CR1 (SA011F11, 149008, Biolegend), PE-conjugated anti-CD54 (YN1/1.7.4, 116108, Biolegend), PerCP or Brilliant Violet 605-conjugated anti-CD62L (MEL-14, 104430, 104438, Biolegend), PE or Brilliant Violet 510-conjugated anti-CD44 (IM7, 103008, 103043, Biolegend), PE-conjugated anti-CXCR4 (L276F12, 146506, Biolegend), PE-conjugated anti-CXCR5. Zombie NIR™ Fixable Viability (423106, Biolegend) staining was performed for live/dead discrimination according to the manufacturer’s instructions. In each sample, 1·106 cells were incubated (30 min, 4°C) with the antibody mixture in staining buffer (total volume 50 μl, in a 96 well U-shaped plate), then washed twice with staining buffer. Samples were re-suspended in 200 μl of 1% PFA in staining buffer, and analyzed by flow cytometry using CytoFLEX S flow cytometer (Beckman Coulter).
Flow cytometry data analysis
Flow cytometry analysis was performed using FlowJo 10.1r5 software (Tree Star). Pre-gating for double discrimination of cells was performed in each analysis, followed by the selection of live cells. Marker expression level was analyzed by median fluorescence intensity (MFI).
Immunofluorescence
Following perfusion, mice were decapitated, and the skin was removed from the skull. The skull and the brain compartment within it were transferred to 10 ml of 4% PFA in PBS (48 hours, 4°C). Tissues were then transferred to 10 ml of 0.5 M EDTA in PBS-/- for 3-5 days, at room temperature. Following cryoprotection in 30% sucrose in PBS-/- (48 hours, 4°C), tissues were frozen in dry ice. Cryosections of the tissues were sliced at 10 μm thickness and mounted on super-frost slides (Fisherbrand). Tissues were stained with purified anti-CD19 (1:100; 6D5, 115502, Biolegend) and Alexa Fluor 488 AffiniPure Donkey Anti-Rat IgG (1:100; AB_2340684, Jackson), and with PE-conjugated anti-CD45 (1:250; 30-F11, 103106, Biolegend). All images were taken at 40X magnification using an Axio imager M2 microscope (Carl Zeiss Inc. US).
Corticosterone ELISA
Mouse blood was collected in EDTA coated tubes and centrifuged for 10 min at 300×g. Plasma was collected and stored at -80°C until analyzed. Corticosterone levels in plasma were analyzed using Corticosterone ELISA kit (Enzo Life Sciences, US) according to the manufacturer’s instructions.
Quantitative PCR (qPCR) analysis
Brain parenchyma, meninges and choroid plexus were dissected from mice as previously shown101. We have included all meningeal compartments (the dural/arachnoid meninges and the leptomeninges) in our analyses. Tissues were lysed in TRI-Reagent (Sigma) and stored at −80°C overnight. Total RNA was isolated with TRI-Reagent according to the manufacturer’s protocol. The concentration and purity of RNA samples were determined using Take3 Trio Micro-Volume Plate (BioTek, USA). Total RNA (0.1 μg) was reverse transcribed (RT) with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative Real-time PCR analysis was performed on an Applied Biosystems StepOnePlus Real Time PCR System (Foster City, CA) in two independent experiments in duplicates using the Fast SYBR Green Master Mix (Applied Biosystems). Appropriate no-RT and nontemplate controls were included in each 96-well PCR reaction, and dissociation analysis was performed at the end of each run to confirm the specificity of the reaction. Real-time PCR efficiencies were determined for all sets of primers used. The cycle conditions for real-time PCR were 95°C for 20 s, followed by 40 cycles of 95°C for 3 s, 60°C for 30 s and a melt curve stage (95°C for 15 s, 60°C for 1 min and 95°C for 15 s). Relative quantification of gene expression was performed according to the ΔΔ-CT method using StepOne Software 2.3 (Applied Biosystems). The following primers were used: GAPDH forward: 5′-TGAAGCAGGCATCTGAGGG-3′, reverse: 5′-CGAAGGTGGAAGAGTGGGAG-3′; CXCL12 forward: 5′-TGCATCAGTGACGGTAAACCA-3′, reverse: 5′-TCTTCAGCCGTGCAACAATC-3′; CXCL13 forward: 5′-GGCCACGGTATTCTGGAAGC-3′, reverse: 5′-GGGCGTAACTTGAATCCGATCTA-3′.
Statistics
For each mass cytometry experiment, pools of six mouse brains were analyzed as a single sample. Two tailed Student’s t-test was used for the analysis of differences between sleep-control and sleep-deprived mice. When indicated, multiple comparisons correction was performed using Holm-Sidak method; multiple comparisons using one-way ANOVA were performed with Fisher’s LSD test. qPCR data were normalized according to the mean relative expression of the control group and analyzed using One column t-test. Statistics were calculated using GraphPad Prism7. Possible outliers were identified using GraphPad Prism7 ROUT algorithm, based on false discovery rate (FDR) with Q = 5%. One outlier was detected in Figure S5 (indicated in the figure legend) and included in the analysis (no change in the statistical outcome of not significant). Results are reported as mean ± standard error of the mean (sem). Each replicated experiment is a biological repeat, indicated in the text as “representative data of X experiments” so that the presented figure includes the data of one of these repeats. Randomization and blinding were not applied in this study. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications43,102. Data distribution was assumed to be normal, but this was not formally tested.
Results
Sleep is a homeostatic process that affects various physiological mechanisms and has long-term effects on endocrine and metabolic pathways63–66. Thus, manipulations that examine the effects of sleep deprivation inevitably include many contributing factors (e.g., the secretion of stress hormones) that can account for some of the observed changes in physiological and mental processes67–69. Although these accompanying changes are in fact endogenous aspects of sleep deprivation, in this study, to minimize the impact of these alternate processes, we chose to focus on short-term sleep deprivation (SSD). We sleep-deprived mice by gentle handling, which is considered to have minimal effect on stress. Control mice were allowed to sleep at the same time (6 hours Zeitgeber time (ZT) 0 to ZT6). We broadly characterized the complete immunological brain compartment, including the parenchyma, the choroid plexus, and the meninges of mice in both experimental groups (Figure 1A). We used mass cytometry as an initial screening tool to identify possible phenotypic changes in the resident myeloid cells and in blood-derived immune cell populations. To distinguish between resident and blood-derived cells, we used their expression of the extracellular markers CD45 and CD44. Resident myeloid cells were identified by their relatively low or absent expression of these markers (CD45low CD44neg), along with expression of CX3CR1 and CD11b (see Figure S1 for gating strategy). Blood-derived cells were defined by their high levels of CD45 (CD45high). For further identification of each cell population, we used additional cell surface markers in our analyses (Figure S1). We characterized resident myeloid cells of sleep-control and sleep-deprived mice using viSNE analysis70. We used viSNE analysis to describe broad changes in the resident myeloid cell population while preserving the fine details of the single-cell level. However, our analysis did not reveal changes in the proportions of the various resident myeloid cell subsets (Figure 1B) or in the expression of specific cell markers between the groups (Figure 1C, Figure S2A). Even with a more targeted approach, utilizing flow cytometry, which has higher sensitivity compared to mass cytometry and used as an additional validation method, we could detect only limited changes in resident myeloid cells. Most notably, we found a significant decrease in CX3CR1 expression, yet the effect size of this change was relatively small (sleep-control: 1±0.02; SSD: 0.91±0.01; n=4, Student’s t-test, P<0.003; Figure S2B). Thus, we concluded that the resident myeloid cells were largely unaffected by our short-term sleep deprivation.
Next, we examined the blood-derived immune cell populations in the brain compartment of sleep-control and sleep-deprived mice: monocytes, granulocytes, dendritic cells (DCs), CD8 T cells, CD4 T cells, B cells, and natural killer (NK) cells from total blood-derived cells (CD45high; Figure 1D). While we could not detect any changes in the phenotype (Figure S3) or overall number of blood-derived cells between the groups (Figure S4), there was a significant increase in the relative abundance of B cells in the tissue derived from sleep-deprived mice (CD45highMHC-II+CD19+; sleep-control: 9.92±0.56%, SSD: 13.43±0.54%; n=6, multiple t-tests, corrected P<0.002; Figure 1D). We validated this effect and its specificity to B cells using flow cytometry (sleep-control: 9.48±0.39%, SSD: 14.8±0.94%; n=9, Student’s t-test, P<0.001; Figure 1E; Figure S5). While B cell proportion increased following sleep deprivation, there was no specific population whose levels were decreased following sleep deprivation (Figure 1D; Figure 1E; Figure S5). Such overall decrease in the proportion of all other cell types, further highlights the specificity of the effect on B cells. Since B cells are the most abundant population in the mouse blood43, blood contamination in the brain may affect our analyses. We have previously described the identification of blood contamination levels in the brain following perfusion43,71. Using this technique, we have showed that our potential error from blood contamination is approximately 1% for all blood-borne cells in the brain compartment, and 0.25% for B cells43. Given the limited contamination rate, the described effect observed following sleep deprivation, cannot be accounted by contamination from the blood.
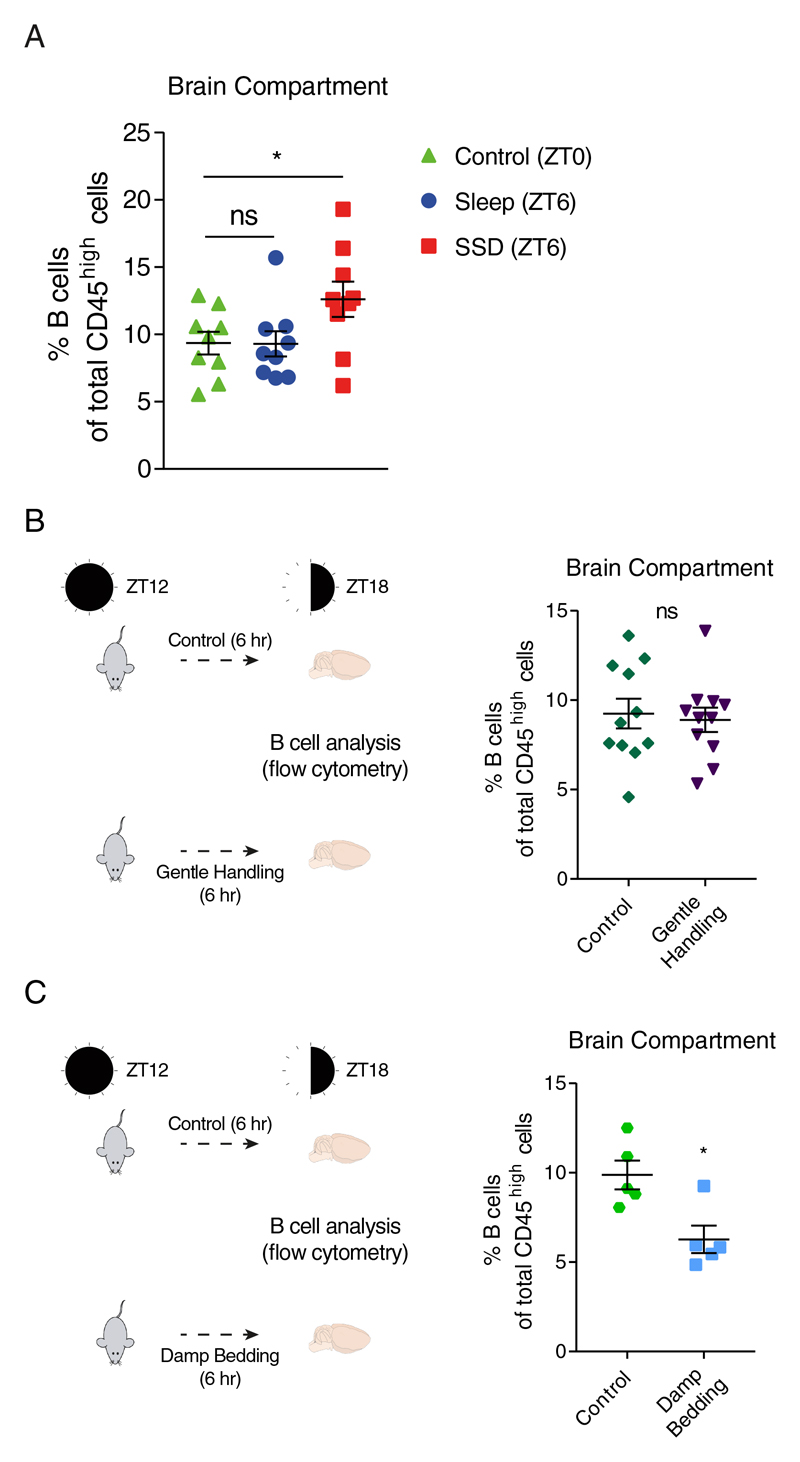
To uncover whether the detected difference in B cell abundance was due to a sleep-induced decrease of B cell numbers or a sleep deprivation induced increase, we compared the proportion of B cells in the sleep-control and sleep-deprived group to the baseline condition, analyzed at ZT0 (control; Figure 2A). There was no difference in the abundance of B cells in the sleep group compared to baseline conditions (ZT0), while in the sleep deprivation group, there was an increase in the percentage of B cells in the brain compartment (control: 9.36±0.84%; sleep: 9.3±0.93%; SSD: 12.62±1.32%; n=9, one-way ANOVA followed by Fisher’s LSD, P<0.04; Figure 2A). Thus, sleep on its own had no effect on the number of B cells, but sleep deprivation increased their abundance in the brain compartment. Moreover, this suggests that the effect on B cells was not dependent on circadian mechanisms as both sleep-control and sleep-deprived mice were analyzed at the same time point.
Figure 2. Comparison of B cell proportions in the brain compartment at baseline conditions (ZT0), and validation of the effects of gentle handling and stress during waking hours (ZT12-ZT18) on B cell proportion in the brain compartment.
Flow cytometry analysis. (A) The percentage of B cells (CD45high/+CD19+MHC-II+) of total CD45high cells at ZT0 (control; green triangle), and following 6 hours of sleep (ZT6; blue circle) or sleep deprivation (SSD; ZT6; red square). Data are presented as mean±sem; one-way ANOVA followed by Fisher’s LSD, n=9 individual mice; *P<0.05; data of two experiments. (B) Mice were housed normally or gently handled for 6 hours (ZT12-ZT18), and the percentage of B cells in the brain compartments of total blood-derived cells was analyzed using flow cytometry. Data are presented as mean±sem; Student’s t-test, n=11 individual mice; P>0.747; data of two experiments. (C) Mice were housed normally or exposed to damp bedding for 6 hours (ZT12-ZT18), and the percentage of B cells in the brain compartments of total blood-derived cells was analyzed using flow cytometry. Data are presented as mean±sem; Student’s t-test, n=5 individual mice; *P<0.05. ns, not significant.
Next, we wanted to address the possibility that the observed increase in B cells abundance was induced by the gentle handling procedure rather than the sleep manipulation. Although gentle handling is considered to be mild intervention that does not induce a significant stress response (validated by corticosterone measurement in our experimental mice; Figure S6), we repeated gentle handling during waking hours (ZT12-ZT18). Under these conditions, we did not observe any effect on B cell percentage (Figure 2B). Moreover, some level of stress is an integral part of sleep deprivation even if it is not made evident by corticosterone levels. Thus, we evaluated the direct effects of stress induced by damp bedding on B cell abundance in the brain. However, when we exposed mice to 6 hours of stress using damp bedding72, there was no increase of B cells in the brain compartment. Rather, mice in the stress group exhibited a significant reduction in B cells in the brain compartment (control: 9.88±0.8%, damp bedding: 6.27±0.77%; n=5, Student’s t-test, P<0.012; Figure 2C), further highlighting the unique effect induced by sleep deprivation. Circadian timing had no significant effect on the proportions of other cell types in the brain compartment (Figure S6B).
Blood-derived immune cells in the brain compartment enter from the blood and under naïve conditions are mainly located in the border areas, the choroid plexus and meninges43. To determine whether sleep deprivation affected the spatial localization of B cells, we performed an immunofluorescence analysis of the brains in the sleep-control and sleep-deprived groups. Sleep deprivation did not induce B cell infiltration to the parenchyma, and all were located only in the border tissues of brains of both groups (Figure S7). It is important to note that given the relatively low abundance of B cells under naïve conditions (i.e., no central nervous system inflammation), these results are mainly qualitative. It was designed to evaluate cell location rather than a numeric comparison in cell number, since quantitative immunofluorescence analysis, with such a low number of cells, has limited power.
To determine whether the cellular profile of B cells was affected by the sleep deprivation, we characterized cell surface markers indicative of specific cell subsets and B cell activation (Figure S8). We found a mild increase in IgG expression (sleep-control: 1±0.08; SSD: 1.21±0.07; n=20; Student’s t-test, P<0.043; Figure S8B), yet there was no significant change in the proportion of IgM+ or IgG+ B cells, or in the relative expression level of IgM on B cells in the brain (Figure S8A, S8B). We additionally found an increase in CD40 expression (sleep-control: 1±0.04; SSD: 1.16±0.04; n=20, Student’s t-test, P<0.004; Figure S8E), a costimulatory membrane protein required for the generation of germinal centers, isotype switching, and sustained antibody production73. Nonetheless, we could not identify a comprehensive change in B cell profile following sleep deprivation (Figure S8F) and the major effect was evidently in the number of these cells. Despite our relatively broad analysis, further studies may provide additional insight on the brain’s B cell population, while extending the characterization of B cell markers, as shown by single-cell RNA sequencing of B cells in the periphery74–76.
The size of a specific immune population can be affected by changes in cell proliferation, death or cell migration. Given the short duration of our manipulation (6 hours), we excluded the possibility of cell proliferation. Flow cytometry analysis using Zombie staining (live/dead discrimination) demonstrated that there was no change in cell viability (Figure S9), suggesting that sleep deprivation induced infiltration and B cell homing towards the brain compartment.
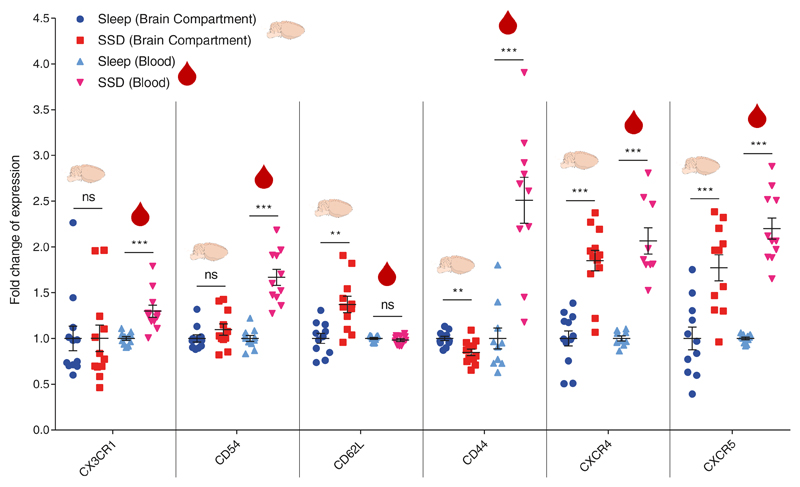
Homing of immune cells to different tissues is regulated by cytokines, chemokines, and their receptors. Therefore, we used flow cytometry to identify changes in migration-related receptors on B cells extracted from the brain compartment or blood of mice in the sleep-control and sleep-deprived groups (Figure 3). CX3CR1 and CD54 on B cells were not affected in the brain compartment, but we detected an increase in their expression on B cells in the blood (CX3CR1: sleep-control:1±0.02, SSD: 1.3±0.07, n=11, Student’s t-test, P<0.001; CD54: sleep-control:1±0.03, SSD: 1.67±0.09, n=11, Student’s t-test, P<0.001). CD62L was elevated in the brain compartment (sleep-control:1±0.05; 1.37±0.09, n=11, Student’s t-test, P<0.003) but not in the blood. CD44 expression decreased in the brain, yet increased in the blood (brain: sleep-control:1±0.0.02, SSD: 0.85±0.04, n=12, Student’s t-test, P<0.002; blood: sleep-control:1±0.11, SSD: 2.51±0.25, n=10, Student’s t-test, P<0.001). By contrast, CXCR4 and CXCR5 were elevated on B cells derived from both the brain compartment and blood of sleep-deprived mice compared to sleep-controls (CXCR4: brain: sleep-control:1±0.08, SSD: 1.85±0.11, n=12, Student’s t-test, P<0.001; blood: sleep-control:1±0.03, SSD: 2.07±0.14, n=9, Student’s t-test, P<0.001; CXCR5: brain: sleep-control:1±0.12, SSD: 1.77±0.14, n=11, Student’s t-test, P<0.001; blood: sleep-control:1±0.01, SSD: 2.2±0.12, n=11, Student’s t-test, P<0.001; Figure 3). The interaction between CXCR4 and its ligand, CXCL12 (also known as stromal cell derived factor 1; SDF-1), regulates the trafficking of all lymphocytes26. This chemokine receptor has been shown to be affected by various neuronal and hormonal signals, such as catecholamines and glucocorticoids77. It has been recently described as a major regulator of circadian-dependent immune cell trafficking, controlling the migration of multiple immune cell types between various tissues26. On the other hand, the interaction between CXCR5 and its ligand, CXCL13 (also known as B cell–attracting chemokine 1; BCA-1), is more specific to B cells78–80 (ImmGen). Based on Allen Brain Atlas, within the brain, cxcl13 is mainly expressed in the meninges81.
Figure 3. Characterization of a B cell migration phenotype in the brain compartment and blood following short-term sleep deprivation.
Flow cytometry analysis. Fold change of expression in migration-related markers (CX3CR1, CD54, CD62L, CD44, CXCR4, CXCR5), in B cells isolated from the brain compartment or in B cells in the blood following 6 hours of sleep (sleep; blue circle) or sleep deprivation (SSD; red square). Fold change of expression was calculated by dividing each value with the mean value of the sleep group for each experiment. Data are presented as mean±sem; Student’s t-test, n=9-12 individual mice; **P<0.01; ***P<0.001; data of two experiments. ns, not significant.
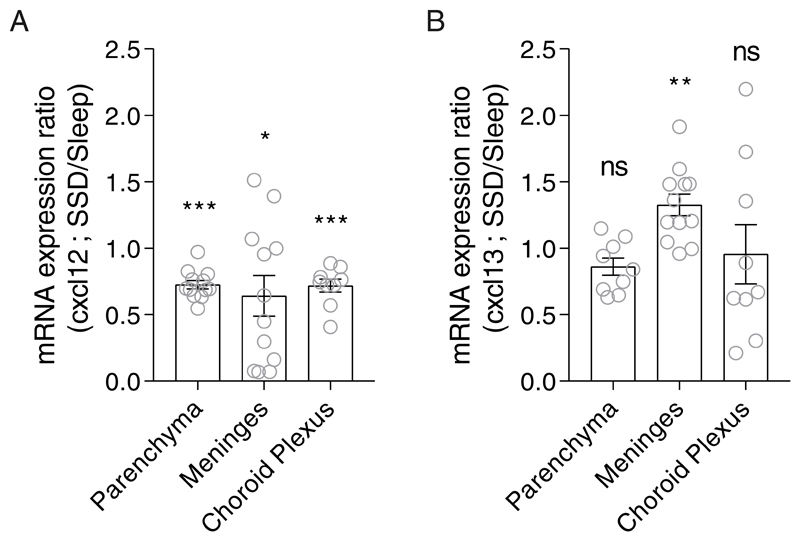
To determine whether the expression of these chemokines in the brain compartment was affected by sleep deprivation, we analyzed their mRNA levels in the meninges, choroid plexus and brain parenchyma. Given that by 6 hours we had already observed an increase in B cell abundance, we analyzed the mRNA levels of the ligands after 2 hours of sleep or sleep deprivation (ZT2). cxcl12 levels decreased in all isolated sites of the brain (meninges, choroid plexus and brain parenchyma; Figure 4A). However, cxcl13 levels were elevated only in the meninges of sleep-deprived mice compared to sleep-controls (SSD/sleep expression ratio: 1.33±0.08, n=12, One sample t-test, P<0.002; Figure 4B). Thus, sleep deprivation induced the increase in CXCR5 on B cells and the elevated expression of cxcl13 in the meninges, which formulates preferential conditions for B cell trafficking to the brain compartment.
Figure 4. Relative mRNA expression of cxcl12 and cxcl13 in the meninges, choroid plexus (CP) and brain parenchyma.
qPCR analyses of (A) cxcl12 and (B) cxcl13 expression levels in the brain parenchyma, meninges, and choroid plexus following 2 hours of sleep deprivation. Relative mRNA expression was calculated by dividing each value in the sleep-deprived group with the mean value of the sleep group for each gene and tissue. Data are presented as mean±sem; One sample t-test, n=9-12 individual mice; *P<0.05; **P<0.01; ***P<0.001; data of three experiments. SSD, sleep-deprived; ns, not significant.
Discussion
The involvement of B cells and other immune cells in brain function, repair or neurodegeneration has become increasingly apparent. However, the factors and the conditions regulating their infiltration and surveillance in the naïve brain, are not fully elucidated. Our study introduces short-term sleep deprivation as such condition. This effect on immune cell trafficking is in line with previous reports that sleep affects the distribution of immune cells between peripheral tissues25. We suggest that the interaction between CXCR5 and its ligand cxcl13 mediated this increase in B cell abundance in the brain compartment following sleep deprivation. However, further studies are required to identify the factors that promote these changes.
In this study, we focused on the immune effects of short term sleep deprivation, which is a common affliction in modern society. Our approach has enabled us to evaluate the immediate effects of sleep deprivation independently from other factors that are associated with longer or chronic sleep deprivation (e.g., stress, metabolic changes). Chronic and prolonged sleep restrictions can alter the homeostatic state of the organism and thus, some factors commonly associated with sleep deprivation may not be apparent 82–84. For example, in some cases no change or even a decrease in plasma corticosterone levels were previously described even though acute sleep deprivation is accompanied by a prominent stress response 85–88. Thus the transition from the acute sleep restricted phase to the chronic one is likely to be evident in the immune compartment of the brain. As many neurodegenerative conditions are accompanied by chronic sleep disruptions 37,89,90, it would be especially interesting to evaluate the immune outcomes of sleep manipulations in specific models of neurodegeneration. Moreover, sleep deprivation can potentially impact various autoimmune conditions in the central nervous system, specifically MS, in which the involvement of B cells has become increasingly evident12,56,57,91,92. Under these conditions, future studies should address how chronic sleep deprivation affects the brain immune compartment of these illnesses. Moreover, the brain is a very complex organ, that holds numerous unique niches, further research may also reveal changes in the spatial localization of the infiltrating immune cells in these various conditions. Such studies will become possible with the emergence of novel analysis techniques allowing us to decipher the immune phenotypes in different brain sites93–95. Thus, our study, demonstrating that acute sleep deprivation affects the immune compartment in the brain, mainly introduces new questions and potentially, an opportunity for intervention.
Supplementary Material
Statement of Significance.
In this study, we show that even after a single and brief deprivation of sleep (6 hours), there is a significant increase in the B cell population in the mouse brain compartment. The recruitment of B cells is especially interesting given the emerging role of B cells in neurodegenerative and neuroinflammatory diseases, brain development and homeostasis. Furthermore, we provide a potential mechanism showing that sleep deprivation induces an increase in the expression levels of the homing receptor CXCR5 on B cells in the brain and in the blood, and a complementary increase in the CXCR5 ligand (cxcl13) in the brain.
Acknowledgments
We thank Prof. Ami Aronheim, Prof. Doron Melamed, Dr. Slava Berger, Maya Schiller and Mariam Amer for their support and helpful discussions, and Shelley Schwarzbaum for editing the paper. We thank Tania Dubovic, Dr. Amir Grau and the Biomedical Core Facility at the Technion Faculty of Medicine for technical support and comments. This work benefited from data assembled by the ImmGene consortium. This study was supported by the Israeli Ministry of Science and Technology, the Adelis Foundation and Prince Center for Neurodegenerative Diseases, A.R. is an International Howard Hughes Medical Institute (HHMI) - Wellcome Trust researcher.
Footnotes
Disclosure statement
Financial Disclosure: none.
Non-financial Disclosure: none.
Author contributions
B.K. designed and carried out the experiments, analyzed and interpreted the results and wrote the manuscript; S.A. contributed to the execution and design of experiments, interpretation of results and the manuscript; H.A-D. carried out the qPCR analyses and contributed to the manuscript; D.F. performed the immunofluorescence staining and acquisition of images; F.H. contributed to the interpretation of results; A.R. contributed to the experimental design, interpretation of results, and revised the manuscript.
References
- 1.Mander BA, Winer JR, Jagust WJ, Walker MP. Sleep: A Novel Mechanistic Pathway, Biomarker, and Treatment Target in the Pathology of Alzheimer’s Disease? Trends Neurosci. 2016;39(8):552–566. doi: 10.1016/j.tins.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peter-Derex L, Yammine P, Bastuji H, Croisile B. Sleep and Alzheimer’s disease. Sleep Med Rev. 2015;19:29–38. doi: 10.1016/j.smrv.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Sahraian MA, Rezaali S, Hosseiny M, Doosti R, Tajik A, Moghadasi AN. Sleep Disorder as a Triggering Factor for Relapse in Multiple Sclerosis. Eur Neurol. 2017;77(5-6):258–261. doi: 10.1159/000470904. [DOI] [PubMed] [Google Scholar]
- 4.Brass SD, Duquette P, Proulx-Therrien J, Auerbach S. Sleep disorders in patients with multiple sclerosis. Sleep Med Rev. 2010;14(2):121–129. doi: 10.1016/j.smrv.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Yeh N-C, Tien K-J, Yang C-M, Wang J-J, Weng S-F. Increased Risk of Parkinson’s Disease in Patients With Obstructive Sleep Apnea. Medicine (Baltimore) 2016;95(2) doi: 10.1097/MD.0000000000002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai KSP, Liu CS, Rau A, et al. Peripheral inflammatory markers in Alzheimer’s disease: a systematic review and meta-analysis of 175 studies. J Neurol Neurosurg Psychiatry. 2017;88(10):876–882. doi: 10.1136/jnnp-2017-316201. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama H, Barger S, Barnum S, et al. Inflammation and Alzheimer’s disease. Neurobiol Aging. 2000;21(3):383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heppner FL, Ransohoff RM, Becher B. Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci. 2015;16(6):358–372. doi: 10.1038/nrn3880. [DOI] [PubMed] [Google Scholar]
- 9.Dendrou CA, Fugger L, Friese MA. Immunopathology of multiple sclerosis. Nat Rev Immunol. 2015;15(9):545–558. doi: 10.1038/nri3871. [DOI] [PubMed] [Google Scholar]
- 10.McGeer PL, McGeer EG. Inflammation and neurodegeneration in Parkinson’s disease. Parkinsonism Relat Disord. 2004;10:S3–S7. doi: 10.1016/j.parkreldis.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Mosley RL, Hutter-Saunders JA, Stone DK, Gendelman HE. Inflammation and adaptive immunity in Parkinson’s disease. [Accessed April 3, 2014];Cold Spring Harb Perspect Med. 2012 2(1) doi: 10.1101/cshperspect.a009381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pikor NB, Prat A, Bar-Or A, Gommerman JL. Meningeal Tertiary Lymphoid Tissues and Multiple Sclerosis: A Gathering Place for Diverse Types of Immune Cells during CNS Autoimmunity. Front Immunol. 2016;6 doi: 10.3389/fimmu.2015.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mesquita SD, Louveau A, Vaccari A, et al. Functional aspects of meningeal lymphatics in ageing and Alzheimer’s disease. Nature. 2018;560(7717):185. doi: 10.1038/s41586-018-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mundt S, Mrdjen D, Utz SG, Greter M, Schreiner B, Becher B. Conventional DCs sample and present myelin antigens in the healthy CNS and allow parenchymal T cell entry to initiate neuroinflammation. Sci Immunol. 2019;4(31):eaau8380. doi: 10.1126/sciimmunol.aau8380. [DOI] [PubMed] [Google Scholar]
- 15.Ziv Y, Ron N, Butovsky O, et al. Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat Neurosci. 2006;9(2):268–275. doi: 10.1038/nn1629. [DOI] [PubMed] [Google Scholar]
- 16.Baruch K, Deczkowska A, David E, et al. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science. 2014;346(6205):89–93. doi: 10.1126/science.1252945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt E-M, Linz B, Diekelmann S, Besedovsky L, Lange T, Born J. Effects of an interleukin-1 receptor antagonist on human sleep, sleep-associated memory consolidation, and blood monocytes. Brain Behav Immun. 2015;47:178–185. doi: 10.1016/j.bbi.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Kipnis J, Gadani S, Derecki NC. Pro-cognitive properties of T cells. Nat Rev Immunol. 2012;12(9):663–669. doi: 10.1038/nri3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herz J, Kipnis J. Bugs and Brain: How Infection Makes You Feel Blue. Immunity. 2016;44(4):718–720. doi: 10.1016/j.immuni.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9(1):46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshall L, Born J. Brain-immune interactions in sleep. Int Rev Neurobiol. 2002;52:93–131. doi: 10.1016/s0074-7742(02)52007-9. [DOI] [PubMed] [Google Scholar]
- 22.Lange T, Dimitrov S, Born J. Effects of sleep and circadian rhythm on the human immune system. Ann N Y Acad Sci. 2010;1193(1):48–59. doi: 10.1111/j.1749-6632.2009.05300.x. [DOI] [PubMed] [Google Scholar]
- 23.Besedovsky L, Ngo H-VV, Dimitrov S, Gassenmaier C, Lehmann R, Born J. Auditory closed-loop stimulation of EEG slow oscillations strengthens sleep and signs of its immune-supportive function. Nat Commun. 2017;8(1):1984. doi: 10.1038/s41467-017-02170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opp MR, Krueger JM. Sleep and Immunity: A Growing Field with Clinical Impact. Brain Behav Immun. 2015;47:1–3. doi: 10.1016/j.bbi.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Besedovsky L, Lange T, Born J. Sleep and immune function. Pflüg Arch - Eur J Physiol. 2011;463(1):121–137. doi: 10.1007/s00424-011-1044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He W, Holtkamp S, Hergenhan SM, et al. Circadian Expression of Migratory Factors Establishes Lineage-Specific Signatures that Guide the Homing of Leukocyte Subsets to Tissues. Immunity. 2018;49(6):1175–1190.:e7. doi: 10.1016/j.immuni.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange T, Dimitrov S, Bollinger T, Diekelmann S, Born J. Sleep after vaccination boosts immunological memory. J Immunol Baltim Md. 2011;187(1):283–290. doi: 10.4049/jimmunol.1100015. 1950. [DOI] [PubMed] [Google Scholar]
- 28.Prather AA, Hall M, Fury JM, et al. Sleep and Antibody Response to Hepatitis B Vaccination. Sleep. 2012;35(8):1063–1069. doi: 10.5665/sleep.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imeri L, Opp MR. How (and why) the immune system makes us sleep. Nat Rev Neurosci. 2009;10(3):199–210. doi: 10.1038/nrn2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Besedovsky L, Lange T, Haack M. The Sleep-Immune Crosstalk in Health and Disease. Physiol Rev. 2019;99(3):1325–1380. doi: 10.1152/physrev.00010.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krueger JM, Clinton JM, Winters BD, et al. In: Progress in Brain Research. Slow Brain Oscillations of Sleep, Resting State and Vigilance. Van Someren EJW, Van Der Werf YD, Roelfsema PR, Mansvelder HD, Lopes Da Silva FH, editors. Vol. 193. Elsevier; 2011. Chapter 3 - Involvement of cytokines in slow wave sleep; pp. 39–47. [DOI] [PubMed] [Google Scholar]
- 32.Krueger JM, Walter J, Dinarello CA, Wolff SM, Chedid L. Sleep-promoting effects of endogenous pyrogen (interleukin-1) Am J Physiol. 1984;246(6 Pt 2):R994–999. doi: 10.1152/ajpregu.1984.246.6.R994. [DOI] [PubMed] [Google Scholar]
- 33.Tobler I, Borbély AA, Schwyzer M, Fontana A. Interleukin-1 derived from astrocytes enhances slow wave activity in sleep EEG of the rat. Eur J Pharmacol. 1984;104(1-2):191–192. doi: 10.1016/0014-2999(84)90391-1. [DOI] [PubMed] [Google Scholar]
- 34.Alam MN, McGinty D, Bashir T, et al. Interleukin-1beta modulates state-dependent discharge activity of preoptic area and basal forebrain neurons: role in sleep regulation. Eur J Neurosci. 2004;20(1):207–216. doi: 10.1111/j.1460-9568.2004.03469.x. [DOI] [PubMed] [Google Scholar]
- 35.Irwin MR. Why Sleep Is Important for Health: A Psychoneuroimmunology Perspective. Annu Rev Psychol. 2015;66(1):143–172. doi: 10.1146/annurev-psych-010213-115205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vgontzas AN, Zoumakis E, Bixler EO, et al. Adverse effects of modest sleep restriction on sleepiness, performance, and inflammatory cytokines. J Clin Endocrinol Metab. 2004;89(5):2119–2126. doi: 10.1210/jc.2003-031562. [DOI] [PubMed] [Google Scholar]
- 37.Musiek ES, Holtzman DM. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science. 2016;354(6315):1004–1008. doi: 10.1126/science.aah4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holth J, Patel T, Holtzman DM. Sleep in Alzheimer’s Disease - Beyond Amyloid. Neurobiol Sleep Circadian Rhythms. 2017;2:4–14. doi: 10.1016/j.nbscr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nadjar A, Wigren H-KM, Tremblay M-E. Roles of Microglial Phagocytosis and Inflammatory Mediators in the Pathophysiology of Sleep Disorders. Front Cell Neurosci. 2017;11 doi: 10.3389/fncel.2017.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q, Wang Y, Feng J, Cao J, Chen B. Intermittent hypoxia from obstructive sleep apnea may cause neuronal impairment and dysfunction in central nervous system: the potential roles played by microglia. Neuropsychiatr Dis Treat. 2013;9:1077–1086. doi: 10.2147/NDT.S49868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Q, Xie X, Fan Y, et al. Phenotypic dysregulation of microglial activation in young offspring rats with maternal sleep deprivation-induced cognitive impairment. Sci Rep. 2015;5:9513. doi: 10.1038/srep09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wisor JP, Schmidt MA, Clegern WC. Evidence for Neuroinflammatory and Microglial Changes in the Cerebral Response to Sleep Loss. Sleep. 2011;34(3):261–272. doi: 10.1093/sleep/34.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korin B, Ben-Shaanan TL, Schiller M, et al. High-dimensional, single-cell characterization of the brain’s immune compartment. Nat Neurosci. 2017;20(9):1300–1309. doi: 10.1038/nn.4610. [DOI] [PubMed] [Google Scholar]
- 44.Ferretti MT, Merlini M, Späni C, et al. T-cell brain infiltration and immature antigen-presenting cells in transgenic models of Alzheimer’s disease-like cerebral amyloidosis. Brain Behav Immun. 2016;54:211–225. doi: 10.1016/j.bbi.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Heneka MT, Kummer MP, Latz E. Innate immune activation in neurodegenerative disease. Nat Rev Immunol. 2014;14(7):463–477. doi: 10.1038/nri3705. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz M, Shechter R. Systemic inflammatory cells fight off neurodegenerative disease. Nat Rev Neurol. 2010;6(7):405–410. doi: 10.1038/nrneurol.2010.71. [DOI] [PubMed] [Google Scholar]
- 47.Keren-Shaul H, Spinrad A, Weiner A, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 2017;169(7):1276–1290.:e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 48.Zeisel A, Muñoz-Manchado AB, Codeluppi S, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 49.Mrdjen D, Pavlovic A, Hartmann FJ, et al. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity. 2018;48(2):380–395.:e6. doi: 10.1016/j.immuni.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Ofengeim D, Giagtzoglou N, Huh D, Zou C, Yuan J. Single-Cell RNA Sequencing: Unraveling the Brain One Cell at a Time. Trends Mol Med. 2017;23(6):563–576. doi: 10.1016/j.molmed.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarlinton D. B cells still front and centre in immunology. Nat Rev Immunol. 2019 January;1 doi: 10.1038/s41577-018-0107-2. [DOI] [PubMed] [Google Scholar]
- 52.Tanabe S, Yamashita T. B-1a lymphocytes promote oligodendrogenesis during brain development. Nat Neurosci. 2018;21(4):506–516. doi: 10.1038/s41593-018-0106-4. [DOI] [PubMed] [Google Scholar]
- 53.Doyle KP, Quach LN, Solé M, et al. B-Lymphocyte-Mediated Delayed Cognitive Impairment following Stroke. J Neurosci. 2015;35(5):2133–2145. doi: 10.1523/JNEUROSCI.4098-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren X, Akiyoshi K, Dziennis S, et al. Regulatory B Cells Limit CNS Inflammation and Neurologic Deficits in Murine Experimental Stroke. J Neurosci. 2011;31(23):8556–8563. doi: 10.1523/JNEUROSCI.1623-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersen BL, Goyal NG, Weiss DM, et al. Cells, cytokines, chemokines, and cancer stress: A biobehavioral study of patients with chronic lymphocytic leukemia. Cancer. 2018;124(15):3240–3248. doi: 10.1002/cncr.31538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michel L, Touil H, Pikor NB, Gommerman JL, Prat A, Bar-Or A. B Cells in the Multiple Sclerosis Central Nervous System: Trafficking and Contribution to CNS-Compartmentalized Inflammation. Front Immunol. 2015;6 doi: 10.3389/fimmu.2015.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stern JNH, Yaari G, Heiden JAV, et al. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci Transl Med. 2014;6(248):248ra107-248ra107. doi: 10.1126/scitranslmed.3008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cross AH, Trotter JL, Lyons J-A. B cells and antibodies in CNS demyelinating disease. J Neuroimmunol. 2001;112(1):1–14. doi: 10.1016/s0165-5728(00)00409-4. [DOI] [PubMed] [Google Scholar]
- 59.Matsushita T, Yanaba K, Bouaziz J-D, Fujimoto M, Tedder TF. Regulatory B cells inhibit EAE initiation in mice while other B cells promote disease progression. J Clin Invest. 2008;118(10):3420–3430. doi: 10.1172/JCI36030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fillatreau S, Sweenie CH, McGeachy MJ, Gray D, Anderton SM. B cells regulate autoimmunity by provision of IL-10. Nat Immunol. 2002;3(10):944–950. doi: 10.1038/ni833. [DOI] [PubMed] [Google Scholar]
- 61.Barr TA, Shen P, Brown S, et al. B cell depletion therapy ameliorates autoimmune disease through ablation of IL-6–producing B cells. J Exp Med. 2012;209(5):1001–1010. doi: 10.1084/jem.20111675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGregor BA, Murphy KM, Albano DL, Ceballos RM. Stress, cortisol, and B lymphocytes: a novel approach to understanding academic stress and immune function. Stress. 2016;19(2):185–191. doi: 10.3109/10253890.2015.1127913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma S, Kavuru M. Sleep and Metabolism: An Overview. Int J Endocrinol. 2010;2010 doi: 10.1155/2010/270832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.AlDabal L, BaHammam AS. Metabolic, endocrine, and immune consequences of sleep deprivation. Open Respir Med J. 2011;5:31. doi: 10.2174/1874306401105010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Briançon-Marjollet A, Weiszenstein M, Henri M, Thomas A, Godin-Ribuot D, Polak J. The impact of sleep disorders on glucose metabolism: endocrine and molecular mechanisms. Diabetol Metab Syndr. 2015;7(1):25. doi: 10.1186/s13098-015-0018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmid SM, Hallschmid M, Schultes B. The metabolic burden of sleep loss. Lancet Diabetes Endocrinol. 2015;3(1):52–62. doi: 10.1016/S2213-8587(14)70012-9. [DOI] [PubMed] [Google Scholar]
- 67.Kushida CA. Sleep Deprivation: Basic Science, Physiology and Behavior. CRC Press; 2016. [Google Scholar]
- 68.Knutson KL, Spiegel K, Penev P, Van Cauter E. The metabolic consequences of sleep deprivation. Sleep Med Rev. 2007;11(3):163–178. doi: 10.1016/j.smrv.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cirelli C, Faraguna U, Tononi G. Changes in brain gene expression after long-term sleep deprivation. J Neurochem. 2006;98(5):1632–1645. doi: 10.1111/j.1471-4159.2006.04058.x. [DOI] [PubMed] [Google Scholar]
- 70.Amir ED, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korin B, Dubovik T, Rolls A. Mass cytometry analysis of immune cells in the brain. Nat Protoc. 2018;13(2):377. doi: 10.1038/nprot.2017.155. [DOI] [PubMed] [Google Scholar]
- 72.Jaggi AS, Bhatia N, Kumar N, Singh N, Anand P, Dhawan R. A review on animal models for screening potential anti-stress agents. Neurol Sci. 2011;32(6):993–1005. doi: 10.1007/s10072-011-0770-6. [DOI] [PubMed] [Google Scholar]
- 73.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229(1) doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andor N, Simonds EF, Czerwinski DK, et al. Single-cell RNA-Seq of follicular lymphoma reveals malignant B-cell types and coexpression of T-cell immune checkpoints. Blood. 2019;133(10):1119–1129. doi: 10.1182/blood-2018-08-862292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen A, Khoo WH, Moran I, Croucher PI, Phan TG. Single Cell RNA Sequencing of Rare Immune Cell Populations. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang L, Dong X, Lee M, Maslov AY, Wang T, Vijg J. Single-cell whole-genome sequencing reveals the functional landscape of somatic mutations in B lymphocytes across the human lifespan. Proc Natl Acad Sci. 2019;116(18):9014–9019. doi: 10.1073/pnas.1902510116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ince LM, Weber J, Scheiermann C. Control of Leukocyte Trafficking by Stress-Associated Hormones. Front Immunol. 2019;9 doi: 10.3389/fimmu.2018.03143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heng TSP, Painter MW, The Immunological Genome Project Consortium et al. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 79.Legler DF, Loetscher M, Roos RS, Clark-Lewis I, Baggiolini M, Moser B. B Cell–attracting Chemokine 1, a Human CXC Chemokine Expressed in Lymphoid Tissues, Selectively Attracts B Lymphocytes via BLR1/CXCR5. J Exp Med. 1998;187(4):655–660. doi: 10.1084/jem.187.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sáez de Guinoa J, Barrio L, Mellado M, Carrasco YR. CXCL13/CXCR5 signaling enhances BCR-triggered B-cell activation by shaping cell dynamics. Blood. 2011;118(6):1560–1569. doi: 10.1182/blood-2011-01-332106. [DOI] [PubMed] [Google Scholar]
- 81.Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 82.Leemburg S, Vyazovskiy VV, Olcese U, Bassetti CL, Tononi G, Cirelli C. Sleep homeostasis in the rat is preserved during chronic sleep restriction. Proc Natl Acad Sci U S A. 2010;107(36):15939–15944. doi: 10.1073/pnas.1002570107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Everson CA. Functional consequences of sustained sleep deprivation in the rat. Behav Brain Res. 1995;69(1):43–54. doi: 10.1016/0166-4328(95)00009-i. [DOI] [PubMed] [Google Scholar]
- 84.Everson CA, Wehr TA. Nutritional and metabolic adaptations to prolonged sleep deprivation in the rat. Am J Physiol-Regul Integr Comp Physiol. 1993;264(2):R376–R387. doi: 10.1152/ajpregu.1993.264.2.R376. [DOI] [PubMed] [Google Scholar]
- 85.Everson CA, Crowley WR. Reductions in circulating anabolic hormones induced by sustained sleep deprivation in rats. Am J Physiol-Endocrinol Metab. 2004;286(6):E1060–E1070. doi: 10.1152/ajpendo.00553.2003. [DOI] [PubMed] [Google Scholar]
- 86.Alexandre C, Latremoliere A, Ferreira A, et al. Decreased alertness due to sleep loss increases pain sensitivity in mice. Nat Med. 2017;23(6):768–774. doi: 10.1038/nm.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Everson CA, Reed HL. Pituitary and peripheral thyroid hormone responses to thyrotropin-releasing hormone during sustained sleep deprivation in freely moving rats. Endocrinology. 1995;136(4):1426–1434. doi: 10.1210/endo.136.4.7895653. [DOI] [PubMed] [Google Scholar]
- 88.Salín-Pascual RJ, Ortega-Soto H, Huerto-Delgadillo L, Camacho-Arroyo I, Roldán-Roldán G, Tamarkin L. The effect of total sleep deprivation on plasma melatonin and cortisol in healthy human volunteers. Sleep. 1988;11(4):362–369. doi: 10.1093/sleep/11.4.362. [DOI] [PubMed] [Google Scholar]
- 89.Abbott SM, Videnovic A. Chronic sleep disturbance and neural injury: links to neurodegenerative disease. Nat Sci Sleep. 2016;8:55–61. doi: 10.2147/NSS.S78947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wulff K, Gatti S, Wettstein JG, Foster RG. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat Rev Neurosci. 2010;11(8):589–599. doi: 10.1038/nrn2868. [DOI] [PubMed] [Google Scholar]
- 91.Milo R. Therapeutic strategies targeting B-cells in multiple sclerosis. Autoimmun Rev. 2016;15(7):714–718. doi: 10.1016/j.autrev.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 92.Wekerle H. B cells in multiple sclerosis. Autoimmunity. 2017;50(1):57–60. doi: 10.1080/08916934.2017.1281914. [DOI] [PubMed] [Google Scholar]
- 93.Hove HV, Martens L, Scheyltjens I, et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci. 2019;22(6):1021. doi: 10.1038/s41593-019-0393-4. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y, Chen K, Sloan SA, et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J Neurosci. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McKenzie AT, Wang M, Hauberg ME, et al. Brain Cell Type Specific Gene Expression and Co-expression Network Architectures. Sci Rep. 2018;8(1):8868. doi: 10.1038/s41598-018-27293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rolls A, Pang WW, Ibarra I, et al. Sleep disruption impairs haematopoietic stem cell transplantation in mice. [Accessed November 11, 2015];Nat Commun. 2015 6 doi: 10.1038/ncomms9516. http://www.nature.com/ncomms/2015/151014/ncomms9516/full/ncomms9516.html . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kokiko-Cochran ON, Saber M, Puntambekar S, et al. Traumatic Brain Injury in hTau Model Mice: Enhanced Acute Macrophage Response and Altered Long-Term Recovery. J Neurotrauma. 2017;35(1):73–84. doi: 10.1089/neu.2017.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zunder ER, Finck R, Behbehani GK, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10(2):316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Finck R, Simonds EF, Jager A, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83A(5):483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kotecha N, Krutzik PO, Irish JM. Current Protocols in Cytometry. John Wiley & Sons, Inc; 2001. [Accessed September 5, 2016]. Web-Based Analysis and Publication of Flow Cytometry Experiments. http://onlinelibrary.wiley.com/doi/10.1002/0471142956.cy1017s53/abstract . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Derecki N, Derecki N, Kipnis J. Mouse meninges isolation for FACS. Protoc Exch. 2014 September; doi: 10.1038/protex.2014.030. [DOI] [Google Scholar]
- 102.Guilliams M, Dutertre C-A, Scott CL, et al. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity. 2016;45(3):669–684. doi: 10.1016/j.immuni.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.