Abstract
Due to medical data privacy regulations, it is often infeasible to collect and share patient data in a centralised data lake. This poses challenges for training machine learning algorithms, such as deep convolutional networks, which often require large numbers of diverse training examples. Federated learning sidesteps this difficulty by bringing code to the patient data owners and only sharing intermediate model training updates among them. Although a high-accuracy model could be achieved by appropriately aggregating these model updates, the model shared could indirectly leak the local training examples. In this paper, we investigate the feasibility of applying differential-privacy techniques to protect the patient data in a federated learning setup. We implement and evaluate practical federated learning systems for brain tumour segmentation on the BraTS dataset. The experimental results show that there is a trade-off between model performance and privacy protection costs.
1. Introduction
Deep Neural Networks (DNN) have shown promising results in various medical applications, but highly depend on the amount and the diversity of training data [10]. In the context of medical imaging, this is particularly challenging since the required training data may not be available in a single institution due to the low incidence rate of some pathologies and limited numbers of patients. At the same time, it is often infeasible to collect and share patient data in a centralised data lake due to medical data privacy regulations.
One recent method that tackles this problem is Federated Learning (FL) [6,8]: it allows collaborative and decentralised training of DNNs without sharing the patient data. Each node trains its own local model and, periodically, submits it to a parameter server. The server accumulates and aggregates the individual contributions to yield a global model, which is then shared with all nodes. It should be noted that the training data remains private to each node and is never shared during the learning process. Only the model’s trainable weights or updates are shared, thus keeping patient data private. Consequently, FL succinctly sidesteps many of the data security challenges by leaving the data where they are and enables multi-institutional collaboration.
Although FL can provide a high level of security in terms of privacy, it is still vulnerable to misuse such as reconstructions of the training examples by model inversion. One effective countermeasure is to inject noise to each node’s training process, distort the updates and limit the granularity of information shared among them [1,9]. However, existing privacy-preserving research only focuses on general machine learning benchmarks such as MNIST, and uses vanilla stochastic gradient descent algorithms.
In this work, we implement and evaluate practical federated learning systems for brain tumour segmentation. Throughout a series of experiments on the BraTS 2018 data, we demonstrate the feasibility of privacy-preserving techniques. Our primary contributions are: (1) implement and evaluate, to the best of our knowledge, the first privacy-preserving federated learning system for medical image analysis; (2) compare and contrast various aspects of federated averaging algorithms for handling momentum-based optimisation and imbalanced training nodes; (3) empirically study the sparse vector technique for a strong differential privacy guarantee.
2. Method
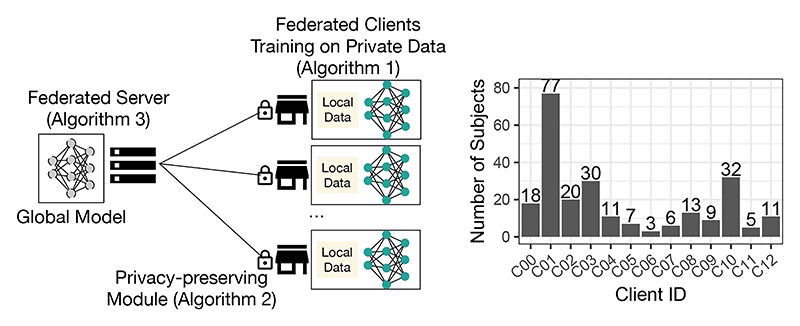
We study FL systems based on a client-server architecture (illustrated in Fig. 1 (left)) implementing the federated averaging algorithm [6]. In this configuration, a centralised server maintains a global DNN model and coordinates clients’ local stochastic gradient descent (SGD) updates. This section presents the client-side model training procedure, the server-side model aggregation procedure, and the privacy-preserving module deployed on the client-side.
Fig. 1.
Left: illustration of the federated learning system; right: distribution of the training subjects (N = 242) across the participating federated clients (K = 13) studied in this paper.
2.1. Client-side model training
We assume each federated client has a fixed local dataset and reasonable computational resources to run mini-batch SGD updates. The clients also share the same DNN structure and loss functions. The proposed training procedure is listed in Algorithm 1. At federated round t, the local model is initialised by reading global model parameters W(t) from the server, and is updated to W(l,t) by running multiple iterations of SGD. After a fixed number of iterations N(local), the model difference ΔW(t) is shared with the aggregation server.
Algorithm 1. Federated learning: client-side training at federated round t.
Require: local training data , num_local_epochs
Require: learning rate η, decay rates β1, β2, small constant ϵ
Require: loss function ℓ defined on training pairs (x, y) parameterised by W
1: procedure local_training(global model W(t))
2: Set initial local model: W(0,t) ← W(t)
3: Initialise momentum terms: m(0) ← 0, v(0) ← 0
4: Compute number of local iterations: N(local) ← Nc · num_local_epochs
5: for l ← 1 … N(local) do ⊳ Training with Adam optimiser
6: Sample a training batch: ℬ(l) ~ 𝒟
7: Compute gradient: g(l) ← ∇ℓ (ℬ(l); W(l−1,t))
8: Compute 1st moment: m(l) ← β1 · m(l−1) + (1 − β1) · g(l)
9: Compute 2nd moment: v(l) ← β2 · v(l−1) + (1 − β2) ·g(l) · g(l)
10: Compute bias-corrected learning rate:
11: Update local model:
12: end for
13: Compute federated gradient: ΔW(t) ← W(l,t) − W(0,t)
14: ← PRIVACY_PRESERVING(ΔW(t))
15: return and N(local) ⊳ Upload to server
16: end procedure
DNNs for medical image are often trained with a momentum-based SGD. Introducing the momentum terms takes the previous SGD steps into account when computing the current one. It can help accelerate the training and reduce oscillation. We explore the choices of design for handling these terms in FL. In the proposed Algorithm 1 (exemplified with Adam optimiser [4]), we re-initialise each client’s momentums at the beginning of each federated round (denoted as m. restart). Since local model parameters are initialised from the global ones, which aggregated information from other clients, the restarting operation effectively clears the clients’ local states that could interfere the training process. This is empirically compared with (a) clients maintaining a set of local momentum variables without sharing; denoted as baseline m. (b) treating the momentum variables as a part of the model, i.e., the variables are updated locally and aggregated by the server (denoted as m. aggregation). Although m. aggregation is theoretically plausible [11], it requires the momentums to be released to the server. This increases both communication overheads and data security risks.
2.2. Client-side privacy-preserving module
The client-side is designed to have full control over which data to share and local training data never leave the client’s site. Still, model inversion attacks such as [3] can potentially extract sensitive patient data from the update or the model W(t) during federated training. We adopt a selective parameter update [9] and the sparse vector technique (SVT) [5] to provide strong protection against indirect data leakage.
Algorithm 2. Federated learning: client-side differential privacy module.
Require: privacy budgets for gradient query, threshold, and answer ε1, ε2, ε3
Require: sensitivity s, gradient bound and threshold γ, τ, proportion to release Q
Require: number of local training iterations N(local)
1: procedure privacy_preserving(ΔW)
2: Normalise by iterations: ΔW ← ΔW/N(local)
3: Compute number of parameters to share: q ← Q · size(ΔW)
4: Track parameters to release: ← empty set
5: Compute a noisy threshold:
6: while size do
7: Randomly draw a gradient component wi from ΔW
8: if then
9: Compute a noisy answer:
10: Release the answer: append wi to
11: end if
12: end while
13: Undo normalisation:
14: return
15: end procedure
Selective parameter sharing
The full model at the end of a client-side training process might have over-fitted and memorised local training examples. Sharing this model poses risks of revealing the training data. Selective parameter sharing methods limit the amount of information that a client shares. This is achieved by (1) only uploading a fraction of : component wi of will be shared iif abs(wi) is greater than a threshold ; (2) further replacing by clipping the values to a fixed range [−γ, γ]. Here abs(x) denotes the absolute value of x; is chosen by computing the percentile of abs(); γ is independent of specific training data and can be chosen via a small publicly available validation set before training. Gradient clipping is also applied, which is a widely-used method, acting as a model regulariser to prevent over-fitting.
Differential privacy module
The selective parameter sharing can be further improved by having a strong differential privacy guarantee using SVT. The procedure of selecting and sharing distorted components of wi is described in Algorithm 2. Intuitively, instead of simply thresholding abs () and sharing its components wi, every sharing wi is controlled by the Laplacian mechanism. This is implemented by first comparing a clipped and noisy version of abs(wi) with a noisy threshold τ(t) + Lap(s/ε2) (Line 8, Algorithm 2), and then only sharing a noisy answer clip(wi + Lap(qs/ε3), γ), if the thresholding condition is satisfied. Here Lap(x) denotes a random variable sampled from the Laplace distribution parameterised by x; clip(x, γ) denotes clipping of x to be in the range of [−γ, γ]; s denotes the sensitivity of the federated gradient which is bounded by γ in this case [9]. The selection procedure is repeated until q fraction of is released. This procedure satisfies (ε1 + ε2 + ε3)-differential privacy [5].
Algorithm 3. Federated learning: server-side aggregation of T rounds.
Require: num_federated_rounds
1: procedure AGGREGATING
2: Initialise global model: W(0)
3: for t ← 1 … T do
4: for client k ←1 … K do ⊳ Run in parallel
5: Send W(t−1) to client k
6: Receive from client’s LOCAL_TRAINING(W(t−1))
7: end for
8:
9: end for
10: return W(t)
11: end procedure
2.3. Server-side model aggregation
The server distributes a global model and receives synchronised updates from all clients at each federated round (Algorithm 3). Different clients may have different numbers of local iterations at round t, thus the contributions from the clients could be SGD updates at different training speeds. It is important to require an N(local) from the clients, and weight the contributions when aggregating them (Line 8, Algorithm 3). In the case of partial model sharing, utilising the sparse property of to reduce the communication overheads is left for future work.
3. Experiments
This section describes the experimental setup, including the common hyper-parameters used for each FL system.
Data preparation
The BraTS 2018 dataset [2] contains multi-parametric pre-operative MRI scans of 285 subjects with brain tumours. Each subject was scanned with four modalities, i.e. (1) T1-weighted, (2) T1-weighted with contrast enhancement, (3) T2-weighted, and (4) T2 fluid-attenuated inversion recovery (T2-FLAIR). Each subject was associated with voxel-level annotations of “whole tumour”, “tumour core”, and “enhancing tumour”. For details of the imaging and annotation protocols, we refer the readers to Bakas et al. [2]. The dataset was previously used for benchmarking machine learning algorithms and is publicly available. We use it to evaluate the FL algorithms on the multi-modal and multiclass segmentation task. For the client-side local training, we adapted the state-of-the-art training pipeline originally designed for data-centralised training [7] and implemented as a part of the NVIDIA Clara Train SDK3.
To test the generalisation ability across the subjects, we randomly split the dataset into a model training set (N = 242 subjects) and a held-out test set (N = 43 subjects). The scans were collected from thirteen institutions with different equipment and imaging protocols, and thus heterogeneous image feature distributions. To make our federated setup realistic, we further stratified the training set into thirteen disjoint subsets, according to where the image data were originated and assigned each to a federated client. The setup is challenging for FL algorithms, because (1) each client only processes data from a single institution, which potentially suffers from more severe domain-shift and over-fitting issues compared with a data-centralised training; (2) it reflects the highly imbalanced nature of the dataset (shown in Fig. 1).
Federated model setup
The evaluation of the FL procedures is perpendicular to the choice of convolutional network architectures. Without loss of generality, we chose the segmentation backbone of [7] as the underlying federated model and used the same set of local training hyperparameters for all experiments: the input image window size of the network was 224 × 224 × 128 voxels, and spatial dropout ratio of the first convolutional layer was 0.2. Similarly to [7], we minimised a soft Dice loss using Adam [4] with a learning rate of 10−4, batch size of 1, β1 of 0.9, β2 of 0.999, and ℓ2 weight decay coefficient of 10−5. For all federated training, we set the number of federated rounds to 300 with two local epochs per federated round. A local epoch is defined as every client “sees” its local training examples exactly once. At the beginning of each epoch, data were shuffled locally for each client. For a comparison of model convergences, we also train a data-centralised baseline for 600 epochs.
In terms of computational costs, the segmentation model has about 1.2 × 106 parameters; a training iteration with an NVIDIA Tesla V100 GPU took 0.85 s.
Evaluation metrics
We measure the segmentation performance of the models on the held-out test set using mean-class Dice score averaged over the three types of tumour regions and all testing subjects. For the FL systems, we report the performance of the global model shared among the federated clients.
Privacy-preserving setup
The selective parameter updates module has two system parameters: fraction of the model q and the gradient clipping value γ. We report model performance by varying both. For differential privacy, we fixed γ to 10−4, the sensitivity s to 2γ, and ε2 to according to [5]. The model performance by varying q, ε1, and ε3 are reported in the next section.
4. Results
Federated vs. data-centralised training
The FL systems are compared with the data-centralised training in Fig. 2 (left). The proposed FL procedure can achieve a comparable segmentation performance without sharing clients’ data. In terms of training time, the data-centralised model converged at about 300 training epochs, FL training at about 600. In our experiments, an epoch of data-centralised training (N = 242) with an NVIDIA Tesla V100 GPU takes 0.85s × 242 = 205.70s per epoch. The FL training time was determined by the slowest client (N = 77), which takes 0.85s × 77 = 65.45s plus small overheads for client-server communication.
Fig. 2. Comparison of segmentation performance on the test set with (left): FL vs. non-FL training, and (right): partial model sharing.
Momentum restarting and weighted averaging
Fig. 2 (left) also compares variants of the FL procedure. For the treatment of momentum variables, restarting them at each federated round outperforms all the other variants. This suggests (1) each client maintaining an independent set of momentum variables slows down the convergence of the federated model; (2) averaging the momentum variables across clients improved the convergence speed over baseline m., but still gave a worse global model than the data-centralised model. On the server-side, weighted averaging of the model parameters outperforms the simple model averaging (i.e. ). This suggests that the weighted version can handle imbalanced numbers of iterations across the clients.
Partial model sharing
Fig. 2 (right) compares partial model sharing by varying the fraction of the model to share and the gradient clipping values. The figure suggests that sharing larger proportions of models can achieve better performance. Partial model sharing does not affect the model convergence speed and the performance decrease can be almost negligible when only 40% of the full model is shared among the clients. Clipping of the gradient can, sometimes, improve the model performance. However, the value needs to be carefully tuned.
Differential privacy module
The model performances by varying differential privacy (DP) parameters are shown in Fig. 3. As expected, there is a trade-off between DP protection and model performance. Sharing 10% model showed better performance than sharing 40% under the same DP setup. This is due to the fact that the overall privacy costs ε are jointly defined by the amount of noise added and the number of parameters shared during training. By fixing the per-parameter DP costs, sharing fewer variables has less overall DP costs and thus better model performance.
Fig. 3. Comparison of segmentation models (ave. mean-class Dice score) by varying the privacy parameters: percentage of partial models, ε1, and ε3.
5. Conclusion
We propose a federated learning system for brain tumour segmentation. We studied various practical aspects of the federated model sharing with an emphasis on preserving patient data privacy. While a strong differential privacy guarantee is provided, the privacy cost allocation is conservative. In the future, we will explore differentially private SGD (e.g. [1]) for medical image analysis tasks.
Acknowledgements
We thank Rong Ou at NVIDIA for the helpful discussions. The research was supported by the Wellcome/EPSRC Centre for Medical Engineering (WT203148/Z/16/Z), the Wellcome Flagship Programme (WT213038/Z/18/Z), the UKRI funded London Medical Imaging and AI centre for Value-based Healthcare, and the NIHR Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnotes
References
- 1.Abadi M, et al. Deep Learning with Differential Privacy; SIGSAC Conference on Computer and Communications Security; 2016. pp. 308–318. [Google Scholar]
- 2.Bakas S, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arxiv. 2018:1811.02629 [Google Scholar]
- 3.Hitaj B, Ateniese G, Perez-Cruz F. Deep models under the GAN: information leakage from collaborative deep learning; SIGSAC Conference on Computer and Communications Security; 2017. pp. 603–618. [Google Scholar]
- 4.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv. 2014:1412.6980 [Google Scholar]
- 5.Lyu M, Su D, Li N. Understanding the sparse vector technique for differential privacy. Proceedings of the VLDB Endowment. 2017;10(6):637–648. [Google Scholar]
- 6.McMahan B, et al. Communication efficient learning of deep networks from decentralized data. Artificial Intelligence and Statistics. 2017:1273–1282. [Google Scholar]
- 7.Myronenko A. 3D MRI brain tumor segmentation using autoencoder regularization; MICCAI Brainlesion Workshop; 2018. pp. 311–320. [Google Scholar]
- 8.Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S. Multi-institutional deep learning modeling without sharing patient data: A feasibility study on brain tumor segmentation; MICCAI Brainlesion Workshop; 2018. pp. 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shokri R, Shmatikov V. Privacy-Preserving Deep Learning; SIGSAC Conference on Computer and Communications Security; 2015. pp. 1310–1321. [Google Scholar]
- 10.Sun C, Shrivastava A, Singh S, Gupta A. Revisiting unreasonable effectiveness of data in deep learning era. ICCV. 2017 [Google Scholar]
- 11.Yu H, Jin R, Yang S. On the linear speedup analysis of communication efficient momentum SGD for distributed non-convex optimization. ICML. 2019 [Google Scholar]