Summary
RNA viruses have evolved elaborate strategies for protection of their genomes, including 5’ capping. However, so far no RNA 5’ cap was identified for hepatitis C virus (HCV) 1,2, which causes chronic infection, liver cirrhosis and cancer3. Here, we demonstrate that the cellular metabolite flavin adenine dinucleotide (FAD) is used as noncanonical initiating nucleotide by the viral RNA-dependent RNA polymerase resulting in a 5’FAD cap on the HCV RNA. The HCV FAD capping frequency is ~75 %, which is the highest observed for any RNA metabolite cap across all kingdoms of life4–8. FAD capping is conserved among HCV isolates for the replication intermediate negative strand and partially for the positive strand. It is also observed in vivo on HCV RNA isolated from patient sera and the liver and plasma of a human liver chimeric mouse model. Furthermore, we show that 5’FAD capping protects RNA from RIG-I mediated innate immune recognition but does not stabilize the HCV RNA. These results establish capping with cellular metabolites as a novel viral RNA capping strategy, which could be used by other viruses and affect viral treatment outcomes and persistence of infection.
Introduction
More than 30 years after its discovery, HCV remains a serious health burden associated with ~60 million chronically infected individuals and ~300,000 annual deaths3. HCV is a member of the Hepacivirus genus within the Flaviviridae family and has six epidemiologically important genotypes. The ~9.6 kb positive-strand RNA genome, HCV(+), contains 5’ and 3’ untranslated regions (UTRs) and a single open reading frame, encoding a polyprotein that is cleaved into 10 mature viral proteins3. The NS5B RNA-dependent RNA polymerase is key for viral RNA replication and binds to the 3’ end of the HCV(+) RNA to produce the reverse complementary negative strand, HCV(−)1. Initiation of replication occurs de novo9,10 with the priming and the incoming nucleotides base pairing with the two 3’ terminal positions of the HCV RNA template to form the first bond. HCV(+) terminates with a 3’ uridine (U) for all strains, and cell culture propagation of strains with other 3’ terminal nucleotides leads to rapid reversion to U11,12, suggesting that the use of adenosine (A) as the priming nucleotide is required for production of HCV(–). The 5’ nucleotide of HCV(+) is A or guanosine (G) for all strains, and 5’ mutagenesis of intergenotypic recombinants based on the robustly replicating genotype 2a JFH1 strain13 leads to rapid reversion to its native 5’ A, indicating selective pressure for initiation of replication with A also for HCV(+)14,15. Initiation of HCV RNA replication with nucleoside triphosphates results in 5’ triphosphate (5´ppp) termini2, which can trigger anti-viral responses through activation of RIG-I like receptors (RLRs) 16 and make the HCV RNA susceptible to dephosphorylation to 5’ monophosphate (5’p) and subsequent degradation by 5’ exonucleases2,17,18. Whereas the HCV(+) 5’ terminus can be protected by proximal binding of cellular miRNA-1221,19,20, no viral strategy for protection of the HCV(–) 5’ terminus has been identified.
Following the initial discovery of RNA 5’ capping with metabolic dinucleotides, such as nicotinamide adenine dinucleotide (NAD+) and 3’-dephospho-Coenzyme A in E. coli21,22, NAD+ capped RNAs have been identified in both prokaryotes and eukaryotes4–8,23. This type of capping is the result of NAD+ being used as non-canonical initiating nucleotide for transcription by DNA-dependent RNA polymerases4,23–25. Mass spectrometry (MS)-based analysis of RNA 5’ caps has confirmed the presence of metabolite caps on mRNA from bacteria, yeast and human cells26. In human cells, metabolite caps constitute ~0.27 % of all caps identified and the abundance correlates with the relative abundance of the metabolites26, indicating that metabolite initiation largely is a stochastic event. A low average frequency of FAD capping has been observed for human mRNA (0.02 %)26 and for short RNAs27, but to this point FAD capping at any significant frequency has not been reported for any single specific RNA.
In this study, we demonstrate that HCV(–), and for some strains also HCV(+), is FAD capped at high frequency (~75 %) as a result of NS5B-mediated de novo initiation of replication with FAD. Moreover, we find that HCV FAD capping occurs in vivo and that the 5’FAD cap protects RNA from cell-intrinsic innate immune recognition. The HCV FAD cap is the first example of viral 5’ metabolite capping, which could be widespread and provide opportunities for novel therapeutic intervention strategies.
Results
HCV RNA is 5’ capped with FAD
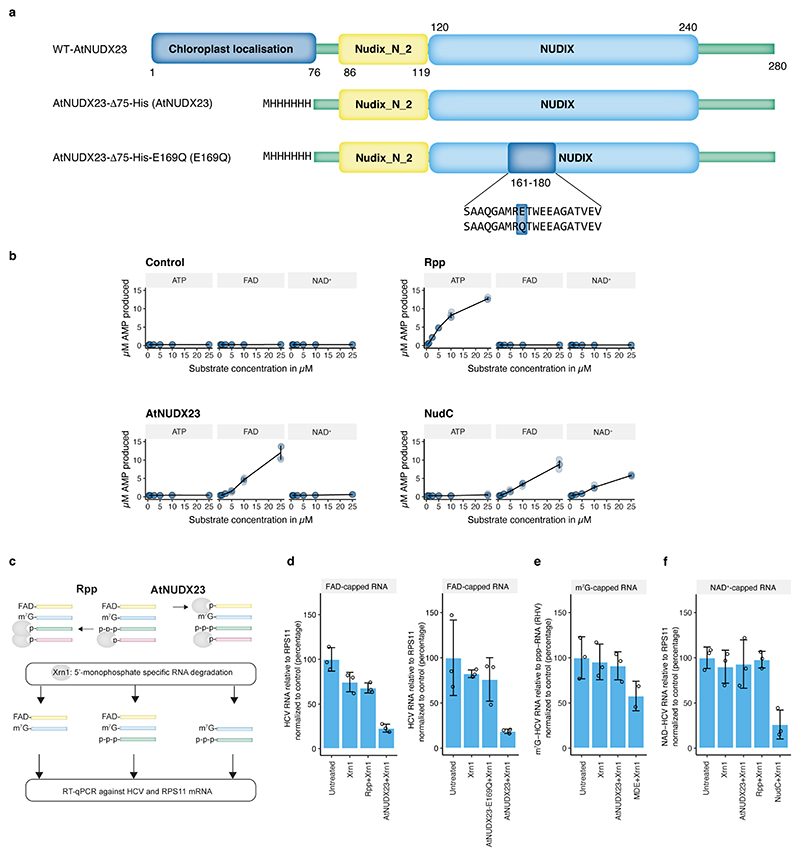
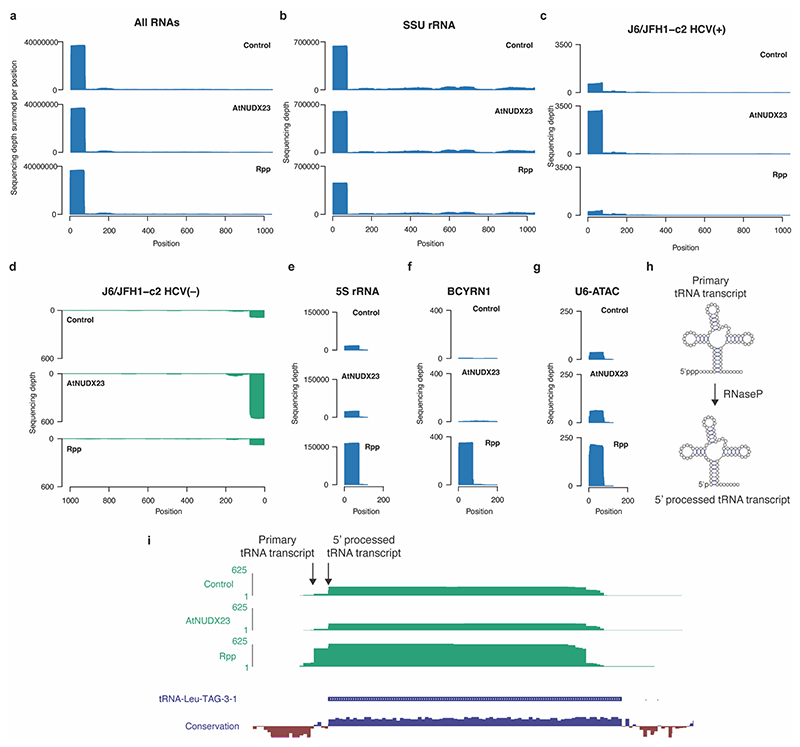
The ubiquitous metabolic cofactor FAD is required for HCV RNA replication in cell culture28, but the underlying mechanism remains unknown. We therefore hypothesized that HCV could be using FAD to initiate replication, leading to 5’ capped viral RNA strands. To investigate the chemical nature of HCV 5’ ends, we purified recombinant Arabidopsis Nudix pyrophosphohydrolase 23 (AtNUDX23), which is specific for FAD29 (Extended Data Fig. 1a-b). We confirmed cleavage also when FAD is present as an RNA 5’ cap and the absence of activity on NAD+, ATP or N7-Methylguanosine 5′-triphosphate (m7GpppN) capped RNA (Fig. 1a-b, Extended Data Fig. 1c-f). We then applied the CapZyme-seq method, which has previously been used to detect NAD+ caps25, with AtNUDX23 or RNA 5’ polyphosphatase (Rpp), which specifically cleaves RNA 5’ppp ends (Fig. 1c). As expected, most reads from total RNA isolated from Huh7.5 hepatoma cells infected with the HCV genotype 2a J6/JFH1 recombinant variant c230 (J6/JFH1 from here onwards) mapped to the 5’ termini of RNAs (Extended Data Fig. 2a).
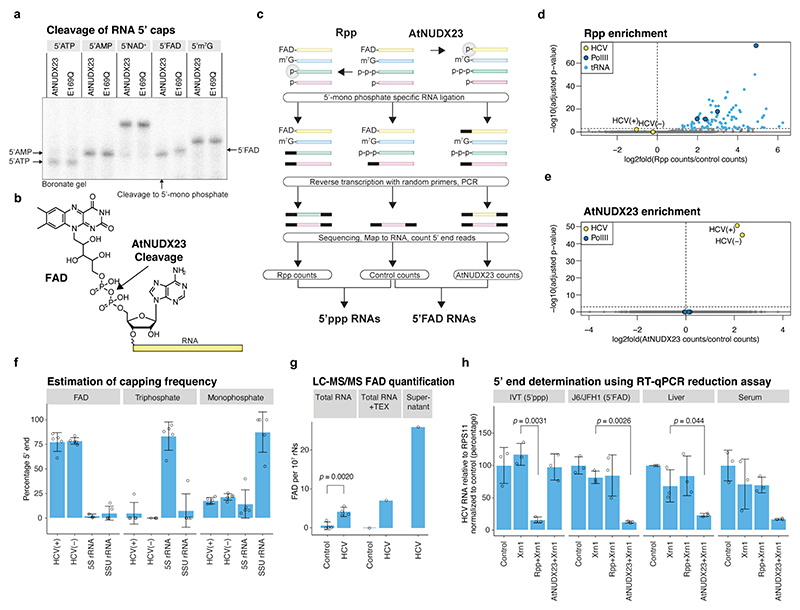
Fig. 1 |. Demonstration of HCV FAD capping.
(a) AtNUDX23 specificity against different 5’caps on a 40 nt RNA. Reactions were analysed on a boronate gel, which affects the migration of RNAs depending on their 5’ cap. E169Q is a catalytically inactive AtNUDX23 mutant. (b) AtNUDX23 cleavage of FAD-capped RNA. (c) Schematic representation of the CapZyme-seq method. (d) Identification of 5’ppp RNAs from J6/JFH1 infected Huh7.5 cells. Volcano plot showing DeSeq2 adjusted p-values versus log2(fold change) for reads mapping to the 5’ termini of individual RNAs (n=5 biological replicates). Rpp treated samples are compared to control samples. (e) Identification of 5’FAD capped RNAs. As in (d), but AtNUDX23 treated samples are compared to control samples. (f) Estimation of the percentage of FAD, tri- and mono-phosphate 5’ termini for J6/JFH1(+) and (−) RNA, 5S rRNA and SSU rRNA (n=5 biological replicates). The analysis assumes stoichiometric conversion to monophosphate by Rpp and AtNUDX23. (g) LC-MS/MS FAD quantification using stable isotope-labelled internal standards for FAD and normalization to ribonucleotide content. RNA samples were isolated from J6/JFH1 infected and control Huh7.5 cells (n=4 biological replicates) and single samples of TEX treated J6/JFH1 infected and control Huh7.5 cells plus ultrafiltration supernatant from HCV infected Huh7.5 cells. TEX: Terminator 5’ exonuclease. (h) RT-qPCR reduction assay showing HCV RNA levels after treatment with Xrn1 5’ exonuclease (5’p specific reduction), Rpp + Xrn1 (5’ppp reduction) or AtNUDX23 + Xrn1 (5’FAD reduction). The following samples were compared: in vitro transcribed 5’ppp J6/JFH1 (IVT), RNA extracted from J6/JFH1 infected Huh7.5 cells (J6/JFH1), and liver and plasma from a J6/JFH1A876P infected human liver chimeric uPA-SCID mouse (n=3 independent replicates). The p-values in (g) and (h) are calculated using one-sided Welch’s unequal variances t-test. Data are presented as mean +/− SD. For gel source data, see Supplementary Figure 1.
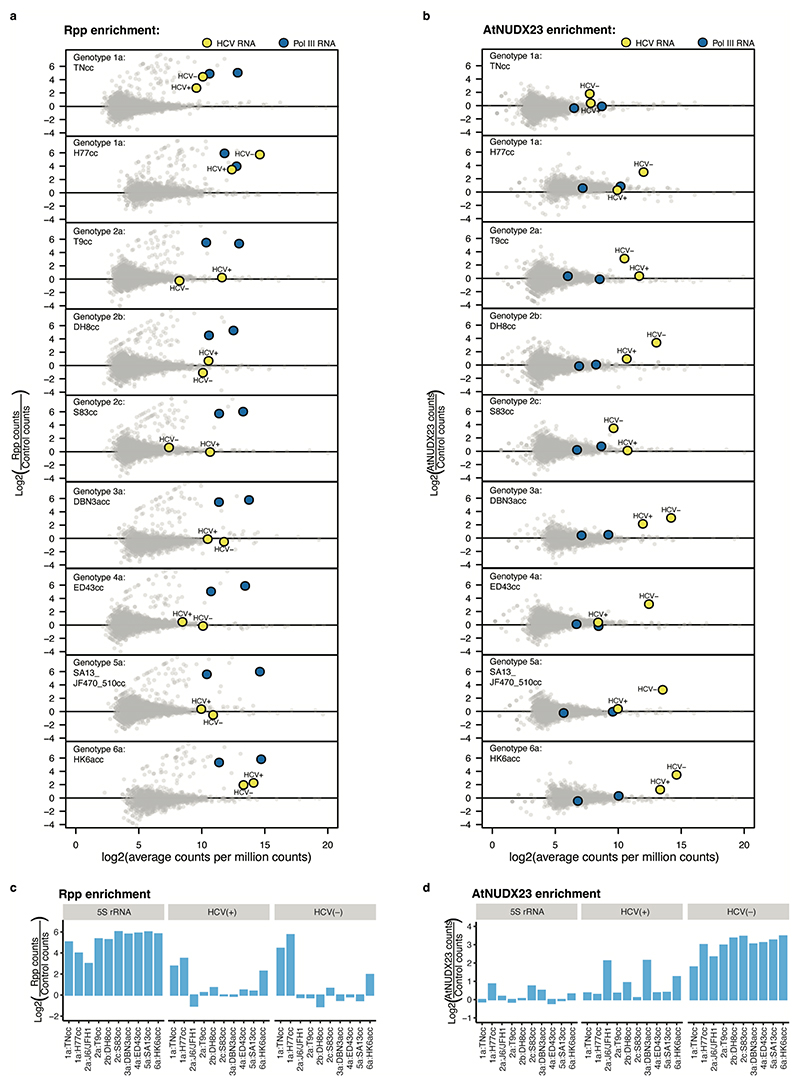
Rpp treatment led to significant enrichment of 117 known endogenous 5’ppp RNAs, including 5S rRNA, BCYRN-1 ncRNA, U6-ATAC snRNA and primary tRNA transcripts, but not HCV(+) or HCV(–) (Fig. 1d and Extended Data Fig. 2). In contrast, HCV(+) and HCV(–) were significantly enriched in the AtNUDX23 treated libraries (Fig. 1e and Extended Data Fig. 2c-d), showing that the J6/JFH1 HCV prototype culture strain is 5’FAD capped in vitro. We estimated the 5’ termini of HCV(+) and HCV(−) to be ~75 % FAD capped with the remaining ~25 % being 5’p, whereas the 5S rRNA and SSU rRNA as expected predominantly had 5’ppp and 5’p, respectively (Fig. 1f).
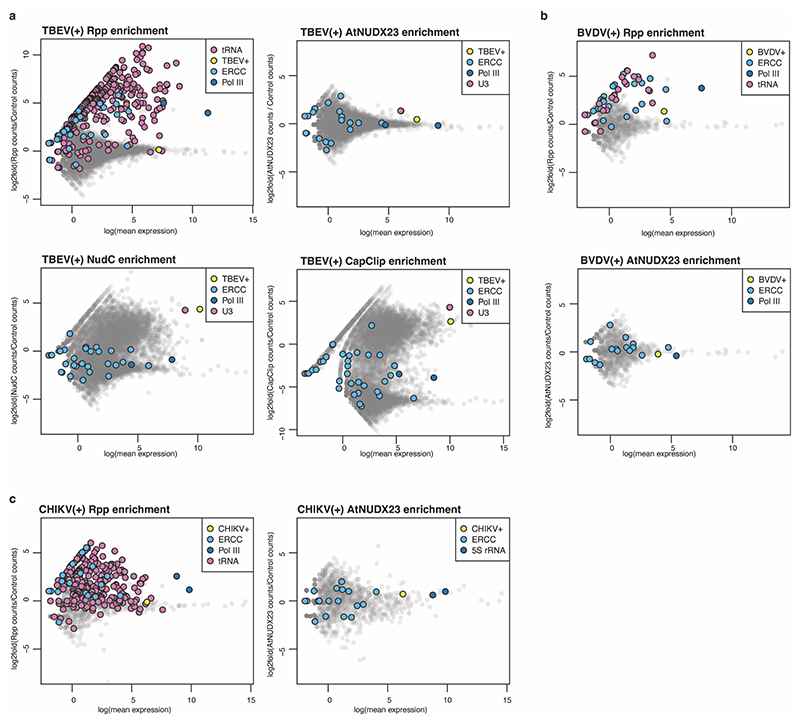
For comparison, we analysed the genomic RNA of the pestivirus bovine viral diarrhea virus (BVDV) and the flavivirus tick-borne encephalitis virus (TBEV), both from the Flaviviridae family, as well as chikungunya virus (CHIKV), an alphavirus of the Togaviridae family. We did not observe FAD capping of any of these viral genomes, whereas we confirmed the m7GpppN cap of the TBEV genome using NudC and CapClip enzymes for CapZyme-seq enrichment (Extended Data Fig. 3). We did not detect any FAD-capped cellular RNAs in any of the four cell lines studied (Fig. 1e, and Extended Data Fig. 3).
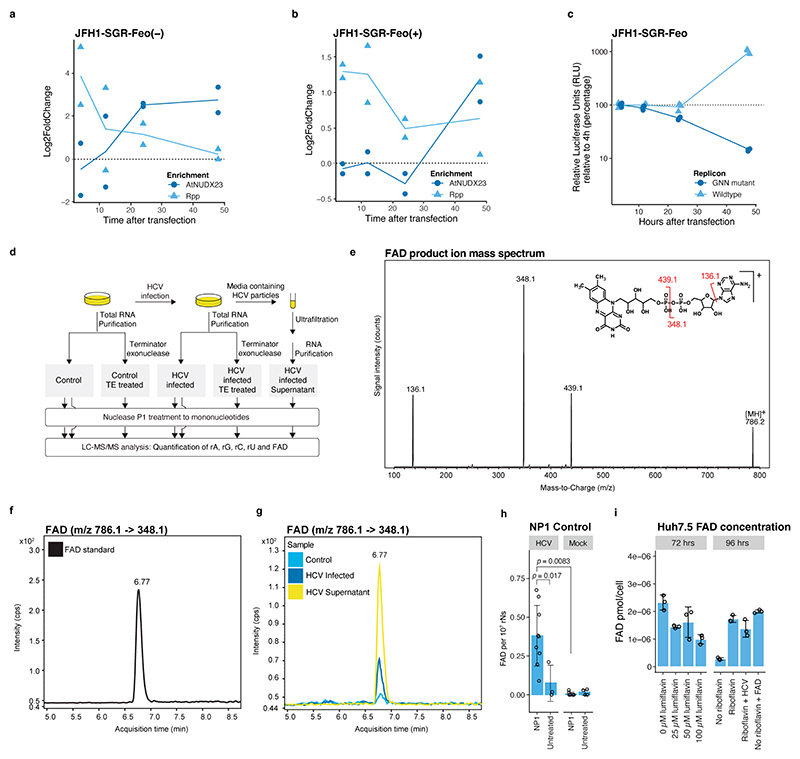
To test that HCV becomes FAD capped upon replication, we transfected in vitro transcribed 5’ppp HCV(+) RNA of the sub-genomic reporter replicon (SGR), JFH1-SGR-Feo, into Huh7.5 cells. Initiation of replication at 24-48 hrs correlated with a switch from HCV 5’ppp to 5’FAD detectable at 24 hrs for HCV(–) and 48 hrs for HCV(+) (Extended Data Fig. 4a-c). As an independent validation of FAD capping, we digested total RNA from J6/JFH1 infected Huh7.5 cells and quantified the released FAD using LC-MS/MS. We consistently detected FAD in HCV infected, but not in uninfected or non-nuclease treated samples (Fig. 1g and Extended Data Fig. 4d-h). Partial removal of rRNA with 5’ monophosphate specific exonuclease led to an increase in relative FAD concentration and enrichment for viral particles by ultrafiltration of supernatant containing a high-titer JFH1-replicase recombinant13 further increased FAD concentration by ~5-fold (Fig. 1g), whereas HCV infection did not increase the intracellular free FAD levels (Extended Data Fig. 4i). To verify FAD capping in vivo, we used the human liver chimeric uPA-SCID mouse model31, and extracted RNA from liver and plasma during long-term infection with an in vivo adapted J6/JFH1 strain13. As the low-abundance in vivo HCV RNA species led to reduced depth in CapZyme-seq assays, we developed an RT-qPCR reduction assay (Extended Data Fig. 1c) and confirmed FAD capping for J6/JFH1 RNA in vivo (Fig. 1h). Taken together, these findings demonstrate that the 5’ termini of the genotype 2a HCV(+) and HCV(−) are FAD capped at high frequency in vitro and in vivo.
FAD capping across the HCV genotypes
To investigate the conservation of HCV FAD capping, we applied CapZyme-seq to RNA isolated from Huh7.5 cells infected with culture adapted recombinants of genotypes 1-613,32 (Fig. 2a). Among strains with HCV(+) 5’G, the 2b and 6a genotype strains changed to 5’A during replication in culture, whereas only genotype 1a strains retained their 5’G (Fig. 2b). Accordingly, except for HCV(+) of genotype 1a, the 5’ ends of HCV(+) and (–) of all strains would be compatible with FAD capping. In agreement with this, we observed AtNUDX23 dependent enrichment of HCV(–) for all genotypes and of HCV(+) for genotypes 2a, 3a and 6a (Fig. 2c, Extended Data Fig. 5). For the genotype 1a strains, we observed strong Rpp enrichment and no AtNUDX23 enrichment of HCV(+), suggesting that their RNAs predominantly initiate with 5’ppp. In addition, genotype 1a and 6a strains also showed Rpp dependent 5’ enrichment of HCV(–), indicating that these genotypes have a mixture of 5’FAD and 5’ppp termini (Fig. 2c, Extended Data Fig. 5). To substantiate these findings, we applied the RT-qPCR reduction assay to plasma samples from patients having ongoing infections with HCV genotypes 1-3 and confirmed 5’FAD capping of genotype 2a and 3a HCV(+) and 5’ppp termini on genotype 1a HCV(+) (Fig. 2d).
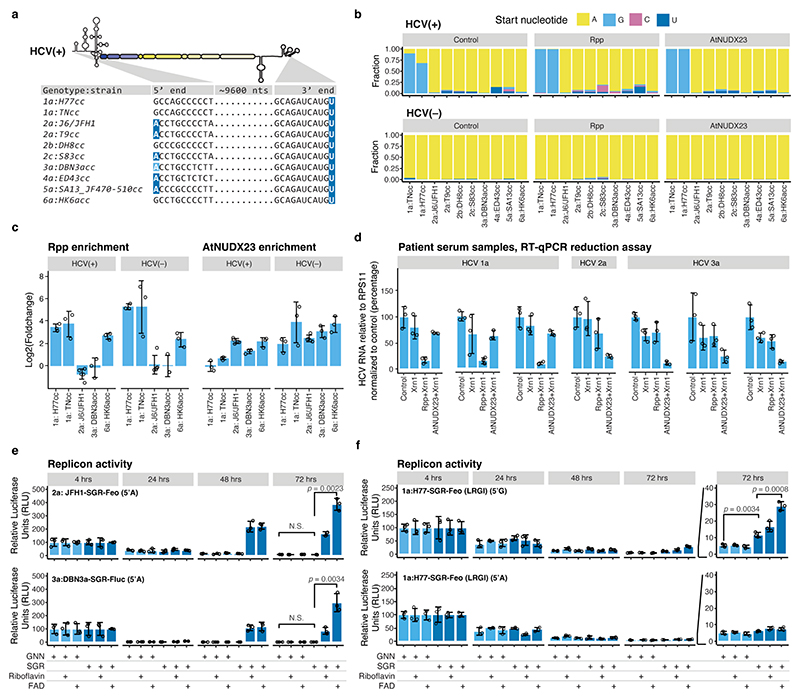
Fig. 2 |. FAD capping for HCV strains of genotypes 1-6.
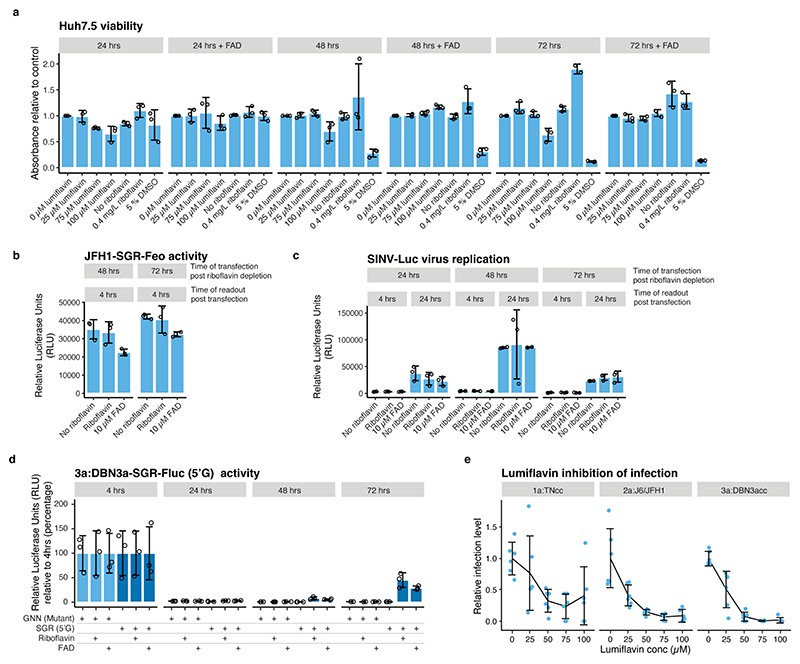
(a) Alignment showing the 5’ and 3’ termini of the HCV strains used in this study. Residues compatible with FAD capping are highlighted in dark blue. Genotype 3a isolates in general, as well as DBN, have 5’A compatible with FAD capping, however, the DBN3acc clone used had a non-authentic 5’G that reverts in culture (light blue). (b) Observed 5’ nucleotides in control, Rpp and AtNUDX23 enriched CapZyme-seq libraries for the HCV(+) and (–) RNAs after propagation of the indicated strains in Huh7.5 cell culture. (c) Enrichment of 5’ terminal reads for HCV(+) and (–) RNAs. Shown are DESeq2 log2FoldChange observed in Rpp (5’ppp, left) and AtNUDX23 (5’FAD, right) enriched CapZyme-seq libraries from RNA isolated from Huh7.5 cells infected with indicated HCV strains. Error bars show the DESeq2 standard error of the log2FoldChange estimate, n=3 biological replicates, except J6/JFH1 with n=5. (d) RT-qPCR reduction assays on RNA isolated from plasma samples taken from patients infected with HCV genotypes 1-3 (technical triplicate for each sample). (e) Replication of wildtype (SGR) and corresponding non-replicating mutant (GNN) of the JFH1-SGR-Feo and DBN3a-SGR-Fluc replicons in Huh7.5 cells grown in riboflavin-depleted media shown as relative luciferase units (RLU) relative to 4 hrs at the indicated time points. (f) Replication of H77-SGR-Feo (LRGI) replicon with authentic 5’G (top) or engineered 5’A (bottom) presented as in panel (e). Transient replication levels are generally lower for H77-SGR-Feo (LRGI) than for JFH1-SGR-Feo and DBN3a-SGR-Fluc. The p-values in (d-f) are calculated using one-sided Welch’s unequal variances t-test. Data are presented as mean +/− SD (n=3 biological replicates); N.S. are non-significant differences. Riboflavin (0.4 mg/L) or FAD (10 μM) was used for reconstitution in (e-f).
Next, we assessed the effect of depletion of the FAD precursor, riboflavin, on viral replication. In agreement with previous results28, we found that replication of the JFH1 genotype 2a strain required FAD, as no replication of JFH1-SGR-Feo was observed upon riboflavin depletion, whereas replication was rescued by supplementing exogenous riboflavin or FAD (Fig. 2e).
Riboflavin depleted cells maintained the ability to support HCV IRES mediated translation and replication of the unrelated Sindbis virus (SINV), showing that the inhibition of HCV replication was not an effect of decreased HCV translation or a general inability to sustain viral replication (Extended Data Fig. 6a-c). As expected from the high 5’FAD capping levels observed for genotype 3, replication of a DBN-SGR genotype 3a replicon also had an absolute requirement for FAD (Fig. 2e) and mutagenesis to 5’G resulted in reversion to 5’A and delayed replication (Extended Data Fig. 6d). In contrast, an H77-SGR genotype 1a replicon showed a strict requirement for 5’G and reduced but active replication in riboflavin depleted media, which was enhanced upon FAD supplementation (Fig. 2f), consistent with partial 5’FAD capping on HCV(−) but not HCV(+). Furthermore, treatment with the riboflavin uptake inhibitor, lumiflavin, inhibited infection with J6/JFH1 (2a) and DBN3acc (3a), whereas TNcc (1a) infection was only partially attenuated (Extended Data Fig. 6a and 6e). These results show that HCV genotype 2a and 3a strains are strictly dependent on FAD, whereas genotype 1a is less dependent, consistent with a lower level of FAD capping.
HCV initiates replication with FAD
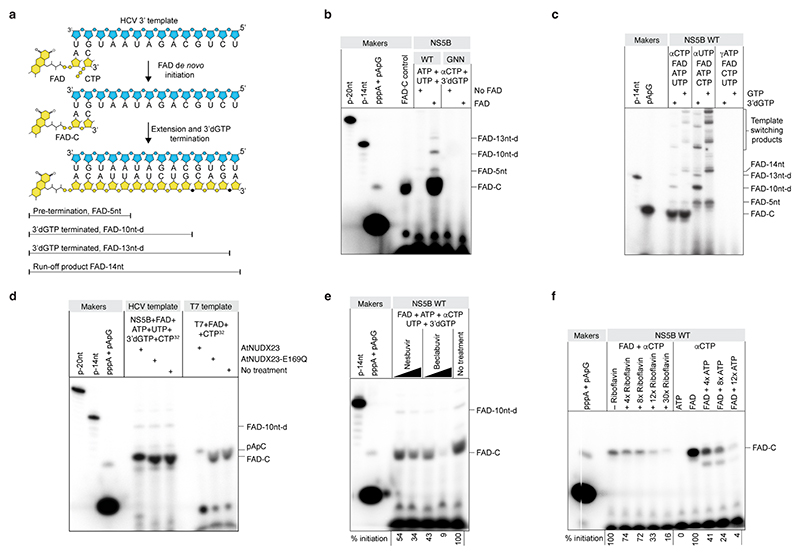
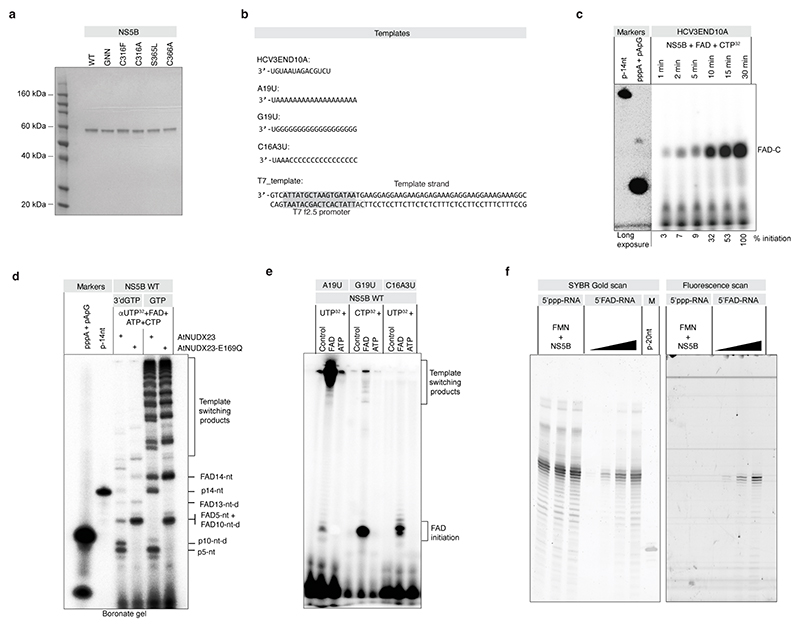
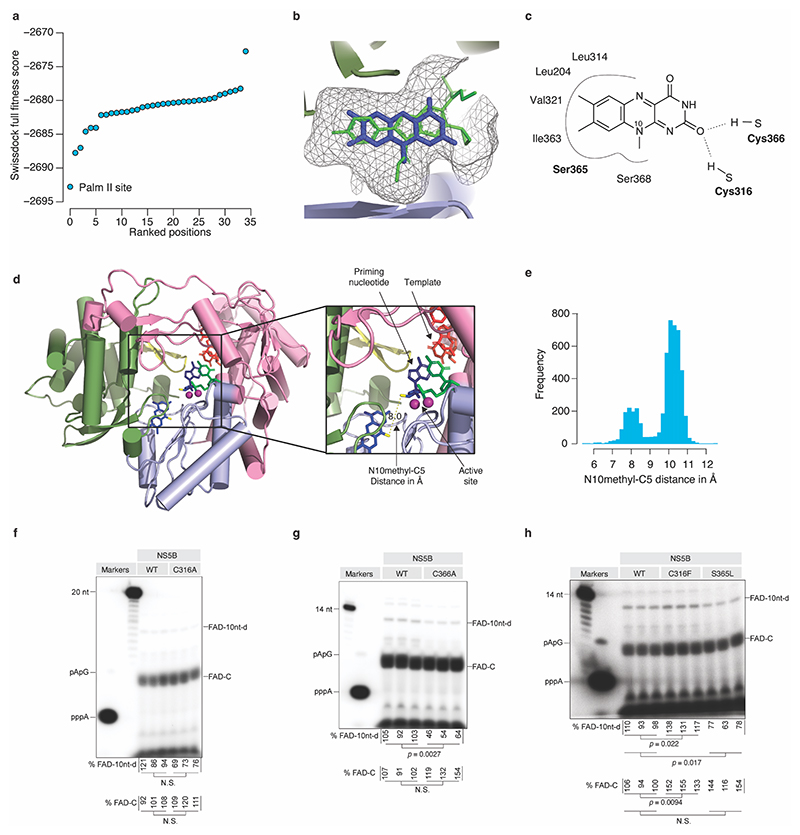
To assess the ability of the HCV NS5B polymerase to initiate replication with FAD, we performed in vitro replication initiation assays using recombinant JFH1 NS5B and an RNA template based on the conserved 3’ terminal sequence of HCV(+) (Fig. 3a, Extended Data Fig. 7a-b). With ATP in the reaction, we did not observe any replication products, whereas inclusion of FAD as the priming nucleotide resulted in efficient de novo initiation of replication and production of an initiation product consisting of FAD linked to CMP (FAD–C) as well as a 5 nt pre-termination product and the expected extension products terminated by incorporation of 3’deoxy-GTP (3’dGTP) (Fig. 3a-c). The FAD–C initiation product accumulated independently of ATP and GTP in a time-dependent manner (Extended Data Fig. 7c) and FAD incorporation in the initiation and extension products was confirmed by AtNUDX23 treatment (Fig. 3d and Extended Data Fig. 7d). FAD-dependent de novo initiation was also observed for other RNA templates (Extended Data Fig. 7e) and was inhibited by treatment with the allosteric NS5B inhibitors nesbuvir and beclabuvir (Fig. 3e). Incubation of 5’ppp RNA with recombinant NS5B and flavin mononucleotide (FMN) did not result in 5’FAD capping, supporting that 5’FAD capping of HCV RNA depends on NS5B de novo initiation with FAD and not post-initiation capping of 5’ppp RNA (Extended Data Fig. 7f). De novo FAD initiation was sensitive to increasing doses of riboflavin or ATP (Fig. 3f), indicating that the nucleotide and riboflavin parts of FAD both contribute to NS5B binding. In silico docking suggested binding of riboflavin to the NS5B palm II site hydrophobic pocket, which binds inhibitors of the benzofuran class, such as nesbuvir33, and is located in a position compatible with the ADP moiety of FAD occupying the active site (Extended Data Fig. 8a-e). Among four NS5B polymerase mutants predicted to affect FAD binding in the palm II pocket, C316F, S365L and C366A significantly affected FAD extension but not initiation, whereas RNA synthesis was unaffected for the C316A mutant (Extended Data Fig. 8f-h). Taken together, these results demonstrate that JFH1 NS5B selectively initiates replication with FAD as the priming nucleotide and implicates residues C316, S365 and C366 in extension of FAD initiated products.
Fig. 3 |. The HCV NS5B RNA-dependent RNA polymerase initiates replication with FAD.
(a) Schematic of expected products of in vitro replication initiation reactions with recombinant HCV JFH1 NS5B polymerase and a 14-nt HCV(+) 3’ UTR template using 3’d-GTP to terminate extension at 10 (or 13) nts. (b) NS5B, wild-type (WT) or catalytically dead (GNN) de novo replication initiation assay with FAD and labeling with αCTP. (c) NS5B replication initiation assay as in (b) but comparing GTP to 3’dGTP to directly visualize the 10 and 13 nt 3’dGTP terminated extension bands and the 14 nt GTP run-off band (left two lanes). To focus labeling on extension products and avoid initiation products, the same reactions were done with γUTP (middle two lanes). To demonstrate the lack of ATP primed initiation, the same reactions were done with γATP (right two lanes). (d) NS5B replication initiation assay as in (b) but using AtNUDX23 to demonstrate presence of FAD in the initiation product. Cleavage of T7 produced FAD-C is shown for comparison. Because of higher negative charge, FAD-C migrates faster than pApC. (e) Inhibition of FAD replication initiation by HCV NS5B inhibitors, nesbuvir and beclabuvir, at 0.1 and 1 mM concentrations. FAD initiation signal is quantified. (f) Effect of riboflavin or ATP on FAD de novo initiation. FAD initiation signal is quantified. To focus analysis on the FAD initiation product, no UTP was included in the competition reactions preventing extension. For all in vitro replication reactions, products were radioactively labeled as indicated and resolved with 18 % denaturing PAGE. For gel source data, see Supplementary Figure 1.
FAD capped RNA evades immune sensing
To investigate the function of the HCV FAD cap, we first addressed its impact on RNA stability. We observed no difference in the stability of 5’ppp and 5’FAD capped in vitro transcribed non-replicating JFH1-SGR-Feo-GNN RNA delivered by electroporation, which both had half-lives of ~1 hour (Fig. 4a). Next, we infected Huh7.5 cells with the HK6acc strain, which has a mixture of 5’ppp and 5’FAD termini on both strands, and inhibited replication with the allosteric NS5B inhibitor beclabuvir before performing CapZyme-seq. Compared to the electroporated in vitro transcribed RNA, viral RNA from infection was stabilized with half-lives of ~10 hrs for HCV(−) and ~5 hrs for HCV(+), but again, no difference in stability was observed between the 5’ppp and 5’FAD sub-populations of viral RNA (Fig. 4b). Thus, compared to 5’ppp, 5´FAD does not increase the stability of the HCV RNA.
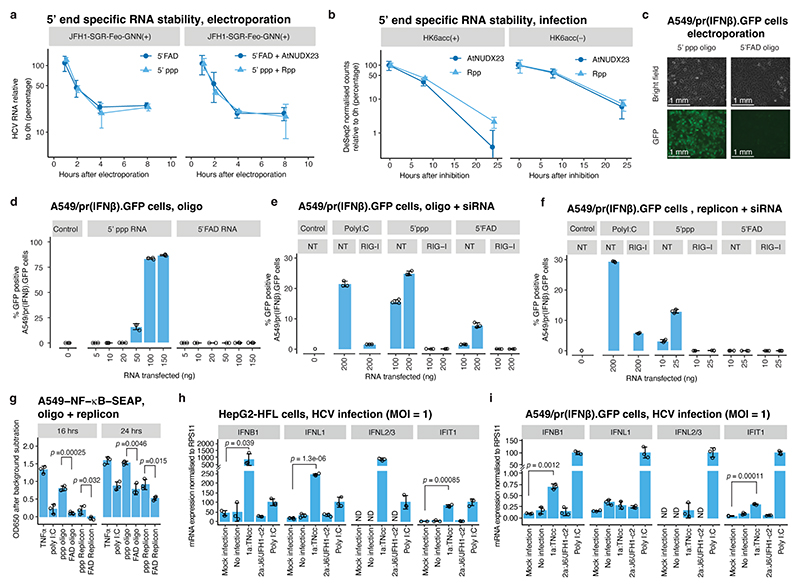
Fig. 4 |. Functional implications of 5’FAD capping.
(a) Stability of 5’ppp and 5’FAD capped in vitro transcribed non-replicating JFH1-SGR-Feo-GNN RNA determined by RT-qPCR after electroporation into Huh7.5 cells. Controls treated with Rpp or AtNUDX23 to generate 5’p are shown for comparison. (b) Stability of 5’ppp and 5’FAD sub-populations of HK6acc (+) and (–) RNA after inhibition of replication by the NS5B inhibitor, beclabuvir, was quantified using CapZyme-seq with Rpp (5’ppp) or AtNUDX23 (5’FAD) enrichment. The 5’ end counts were normalized between libraries and control sample counts subtracted. (c) 5’ppp or 5’ FAD in vitro transcribed RNA oligos were transfected into A549/pr(IFNβ).GFP cells containing a GFP reporter under control of the IFN-β promoter. Images of bright field and GFP channel 48 hrs after transfection are shown. (d) Transfection of RNA oligos as in (c) but with increasing doses and analysis using flow-cytometry. (e) Transfection of RNA oligos as in (d) but in cells pretreated with siRNAs targeting RIG-I or no target (NT). Flow-cytometry was performed 24 hrs after oligo transfection. (f) Transfection as in (e) but using JFH1-SGR-Feo-GNN replicon RNA. (g) Transfection of RNA oligos (150 ng), JFH1-SGR-Feo-GNN RNA (25 ng) or poly I:C (25 ng) into A549-NF-κB-SEAP reporter cells or stimulation with 5ng/mL TNF-α. OD650 was determined at indicated timepoints. (h) mRNA expression levels for IFNB1, IFNL1, IFNL2/3 and IFIT1 18 hrs post infection of HepG2-HFL cells with MOI=1 of indicated HCV strains shown relative to poly I:C induced levels (n=3). P-values compared to mock are shown for samples significantly different from both controls (non-treated and mock supernatant concentrated in parallel with virus stocks). (i) mRNA expression levels as in (h) but in A549/pr(IFNβ).GFP cells 24 hours post infection. ND: Not detected. (a-i) Data are presented as mean +/− SD (n=3 biological replicates.). The p-values in (g-i) are calculated using one-sided Welch’s unequal variances t-test.
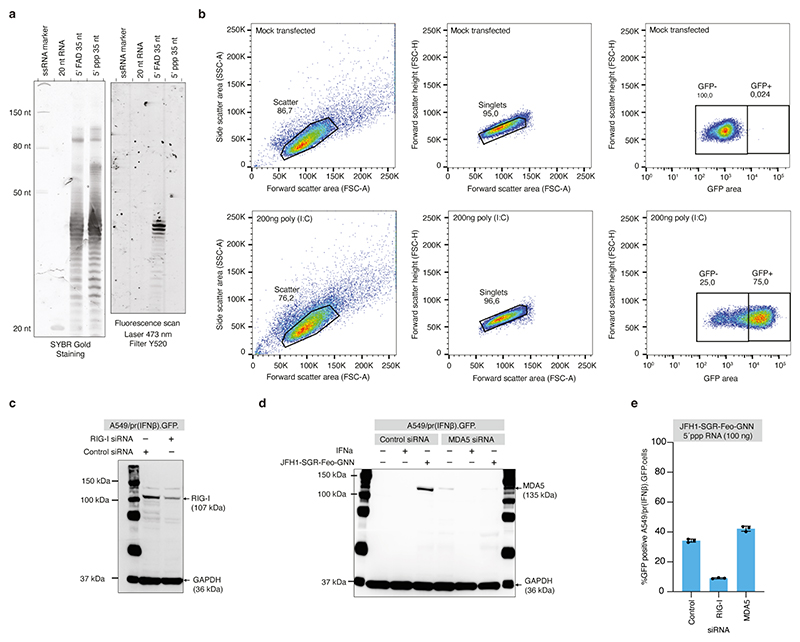
Protection from cell-intrinsic innate immune recognition could constitute another potential role of 5’FAD capping16. RIG-I is the main cytoplasmic sensor of 5’ppp RNA and its activation leads to signalling through MAVS followed by IRF3 mediated induction of type I and III interferon (IFN) or NF-κB mediated induction of inflammatory pathways34. To assess the role of 5’FAD, we transfected in vitro transcribed 5’ppp or 5’FAD capped RNA oligos (Extended Data Fig. 9a) into A549 cells expressing GFP under the control of an IRF3 dependent IFN-β promoter35,36. We detected strong dose-dependent activation by 5’ppp RNA but not by 5’FAD capped RNA (Fig. 4c-d, Extended Data Fig. 9b). The innate immune induction by 5’ppp RNA oligos or 5’ppp JFH1-SGR-Feo-GNN RNA was completely prevented by RIG-I knock-down (Fig. 4e-f and Extended Data Fig. 9c). Knock-down of MDA5, another important RLR not implicated in 5’ppp sensing, did not reduce innate immune activation irrespective of the JFH1-SGR RNA 5’ end (Extended Data Fig. 9d-e), suggesting that MDA5 is not an important sensor of HCV RNA.
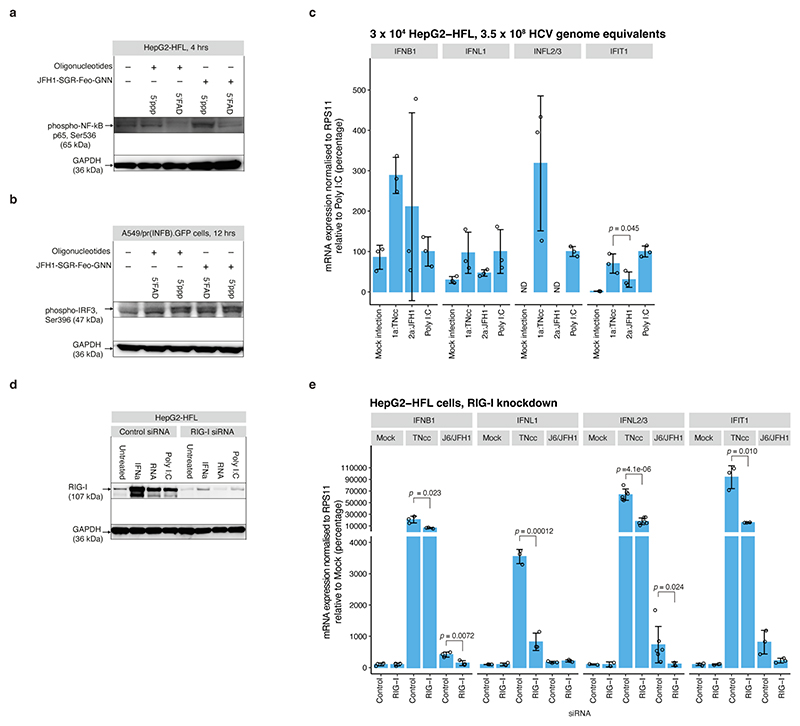
5’ppp RNA also induced stronger secreted embryonic alkaline phosphatase (SEAP) expression from NF-κB driven A549 reporter cells compared to 5’FAD RNA (Fig. 4g). These observations were supported by minimal 5’FAD RNA induced activation of IRF3 and the NF-κB p65 subunit compared to that of 5’ppp RNA, as demonstrated by immunoblotting for protein phosphorylation (Extended Data Fig. 10a-b).
RLR signalling is inhibited by HCV NS3-4A mediated cleavage of MAVS37–39, but this requires active replication and is activated after ~18 hrs post infection40. To study 5’FAD mediated evasion of innate immune sensing during HCV infection, we therefore compared genotype 1a (TNcc) and 2a (J6/JFH1), having 5’ppp and 5’FAD genomic HCV(+) termini, respectively, at early time points in HCV permissive HepG2-HFL cells41 (18 hrs) and in A549/pr(IFNβ).GFP cells, which supports HCV entry but not replication42 (24 hrs). At a multiplicity of infection (MOI) = 1, genotype 1a but not 2a infection consistently induced the innate response genes IFNB1, IFNL1, IFNL2/3, and IFIT1 (Fig. 4h-i). Compared to J6/JFH1, TNcc has lower specific infectivity13 and the TNcc inoculum used accordingly contained 23-fold more viral RNA. To control for a potential influence of this, we increased the J6/JFH1 inoculum to match the RNA titers of TNcc, and also under these conditions, dramatically favouring J6/JFH1 infection, we observed more efficient innate immune induction by TNcc (Extended Data Fig. 10c).
Furthermore, the TNcc induced IFNB1, IFNL1, IFNL2/3, and IFIT1 expression was reduced during RIG-I knock-down (Extended Data Fig. 10d-e), indicating that the observed genotype difference depends on detection of genotype 1a 5’ppp termini. These data support a role for 5’FAD capping in evasion of innate immunity early in HCV infection and possibly at other points of infection.
Discussion
Here, we demonstrate that HCV RNA is 5’FAD capped through non-canonical initiation of replication with FAD. Some HCV strains, including the prototype genotype 2a culture strain J6/JFH1, are 5’FAD capped with high frequency on both RNA strands, whereas other strains are only 5’FAD capped on the negative strand. This finding indicates a biological function of the HCV FAD cap in vivo, consistent with validation of FAD capping for genotype 2a and 3a virus circulating in patient plasma and genotype 2a in the human liver chimeric uPA-SCID mouse model. The demonstration of HCV RNA 5’FAD capping rationalizes the previously unexplained dependence of HCV replication on FAD28. Further, HCV 5’FAD RNA capping could be a determinant for the requirement of the HCV genome to terminate in 3’U11,12 and for some HCV genomes to start with 5’A11,14,15. HCV RNA was previously considered to have 5’ppp termini based on data from a single study investigating HCV(+) of a genotype 1a recombinant2. Since FAD capping was not investigated in that study, the detected 5’ppp RNA may either represent a minor subpopulation or be a result of the genotype 1a sequence present in the studied recombinant.
RNA viruses, such as HCV, evolve under strong selective pressure, suggesting that FAD capping is a functional adaptation conferring a selective advantage to the virus. We show that the HCV JFH1 NS5B polymerase initiates replication efficiently with FAD as the priming nucleotide and not with ATP, which is consistent with the observed absence of 5’ppp and the ~75 % FAD capping of the J6/JFH strain in cell culture. In fact, ATP or riboflavin addition inhibited formation of the FAD–C initiation product, indicating that ATP works as a competitive inhibitor for FAD initiation intracellularly, where ATP is 500-1000 fold more abundant than FAD43. Thus, FAD and ATP availability could regulate HCV replication initiation efficiency in vivo and hence may contribute to viral long-term persistence by restricting replication. Intriguingly, oxidative stress induced by HCV infection is known to reduce viral replication via the membrane anchor of NS5B44 and it is possible that the different redox states of FAD also affect replication initiation. The molecular mechanisms responsible for selective NS5B de novo initiation with FAD remains to be further characterized.
Another selective advantage of the HCV 5’FAD cap could be protection against immune responses, especially during early steps of the viral life cycle where the HCV RNA is not protected inside replication vesicles and RIG-I signalling is intact34. Accordingly, 5’FAD capping of in vitro transcribed RNA that were transfected into cells resulted in evasion of innate immune sensing and reduced RIG-I mediated signalling through IRF3 and NF-κB. Moreover, infection with genotype 1a, having an unprotected 5’ppp RNA genome terminus, stimulated expression of innate immune response genes at early timepoints, whereas infection with the 5’FAD capped genotype 2a did not. Thus, the 5’ FAD cap could serve as an independent evasion mechanism early in infection and complement NS3-4A induced MAVS cleavage37–39 during active replication. This would be consistent with the finding that the effect of RLR knock-down on HCV JFH1 replication is minor or absent18, in contrast to the strong RLR activation towards HCV RNA produced by in vitro transcription16. Binding of miR-122 to the HCV(+) 5’ end19 may serve as an alternative strategy for RLR protection for genotypes without the HCV(+) 5’FAD cap. Consistent with this, HCV(+) RNA of genotype 1, and of some strains of genotype 6, can form 4 supplementary base pairs with miR-122, compared to only 3 for other genotypes20.
For HCV(−), some level of 5’FAD capping is observed for all genotypes consistent with the lack of miRNA protection. Nonetheless, since HCV JFH1 replication is dependent on FAD in Huh7.5 cells, which are deficient in the RIG-I pathway45, the 5’FAD cap must have functional roles other than those related to immune evasion; the FAD requirement for JFH1 NS5B replication initiation constitutes one such role. In contrast, 5’FAD capping did not stabilize HCV RNA beyond that of 5’ppp. This could be the result of the viral RNA being secluded in replication vesicles or complementary miR-122 mediated protection of HCV(+) against 5’ pyrophosphatases18,20,46. In addition, the HCV FAD cap may be removed by the decapping exonuclease DXO/DOM3Z, which has been shown to limit HCV replication18 and remove 5’FAD caps from RNA27, or alternatively by the 5’ exonuclease, Xrn1, which also has some activity against 5’FAD capped RNA47.
We demonstrate that HCV RNAs are 5’FAD capped at high frequency (~75 %), which is the highest observed for any metabolite capped RNA across all kingdoms of life4–8,23. The overall FAD capping frequency observed for human mRNA is low (0.02 %) and appears to be a result of stochastic initiation of transcription26. We did not identify any human RNAs with a significant level of FAD capping, supporting that FAD capping of cellular mRNAs is stochastic and low frequency. This indicates that 5’FAD RNA caps could provide a virus-specific target for therapeutic intervention. Furthermore, HCV FAD capping is likely to influence viral persistence and the treatment outcomes of NS5B directed inhibitors and drugs interacting with innate immunity.
The ability of the HCV RNA-dependent RNA polymerase to initiate replication with FAD implies that other RNA viruses could also utilize cellular metabolites for de novo initiation of replication and RNA 5’ capping. Candidate viruses include other hepaci-, pegi- and flaviviruses of the Flaviviridae family, as well as members of the Orthomyxo-, Paramyxo- and Rhabdoviridae families. For example, all genomes of the hepaci-, pegi- and flaviviruses terminate with 3’ U, and for flaviviruses, selective pressure on this residue has been demonstrated48,49. Conversely, metabolite capping appears unlikely on negative strands for RNA viruses with 3’ poly-A tails (e.g., Picornaviridae, Togaviridae) and on positive strands for those with 5’ covalently linked uridylated viral proteins (e.g., Picornaviridae) 50. It will be important to establish the extent of viral RNA metabolite capping, especially for intermediate RNA strands, for which the chemical nature of RNA termini is mostly unknown. For HCV, further studies are needed to elucidate the structural and mechanistic details of selective FAD de novo initiation of replication by NS5B and how 5’FAD RNA capping influences viral propagation in vivo and pathogenesis in the human host.
Methods
Cell culture and virus infections
The human hepatoma cell lines Huh7.551 and HepG2-HFL41 were maintained in DMEM supplemented with 10 % fetal bovine serum (FBS) and in EMEM supplemented with 10 % FBS and 5 μg/mL blasticidin, respectively. The adenocarcinomic human alveolar basal epithelial A549/pr(IFNβ).GFP cell line was maintained in DMEM supplemented with 10 % FBS35,36. A549-Dual cells (Invivogen) were maintained in DMEM supplemented with 10 % FBS, 10 μg/mL blasticidin and 100 μg/mL zeocin. Human SH-SY5Y neuroblastoma cells (ATCC) were maintained in DMEM/F12 (1:1) + GlutaMax-I supplemented with 10 % FBS. Human TIG3 fibroblast cells were maintained in DMEM/F12 (1:1) + GlutaMax-I supplemented with 10 % FBS. Madin-Darby bovine kidney (MDBK, ATCC) cells were maintained in DMEM with 10 % horse serum. All cell lines tested negative for mycoplasma contamination. SH-SY5Y was authenticated by ATCC, whereas the other cell lines were not authenticated.
For infection with HCV strains TNcc (1a) 52, H77cc (1a) 53, J6/JFH1-c2 (2a) 30, T9cc (2a) 54, DH8cc (2b) 55, S83cc (2c) 54, DBN3acc (3a) 56, ED43cc (4a) 32, SA13 (5a) 57 or HK6acc (6a) 58, Huh7.5 cells were seeded one day prior. Cells were sub-cultured every 2-3 days and monitored for viral spread by immunostaining using NS5A monoclonal antibody59 (9E10, Cell Essentials) and Core antibody (C7-50, Enzo Life Sciences Inc) until > 90 % of cells were HCV antigen positive. Infection with tick-borne encephalitis virus (TBEV) of the Neudoerfl strain60 was done in SH-SY5Y cells for 48 hrs at MOI=0.1. Infection with chikungunya virus (CHIKV) of the La Reunion strain61 was done in TIG3 fibroblasts for 24 hrs at MOI=10 or 48 hrs at MOI=0.1.
Infection with bovine viral diarrhea virus (BVDV) of the NADL strain62, with changes to the ORF as described63, was done in MDBK cells for 24 hrs at MOI=0.005. For RNA extraction, supernatants were removed, cells were washed with PBS and lysed with Trizol for RNA extraction. Trizol RNA extraction was followed by purification with RNA Clean & Concentrator kit-5 (Zymo Research, R1013) using manufacturer’s instructions. DNase I digestion (Qiagen) was carried out followed by additional purification with RNA Clean & Concentrator kit-5. RNA quantity and integrity were assessed using NanoDrop One (Thermo Fisher Scientific) and Agilent 2100 Bioanalyzer.
J6/JFH1 infection of human liver chimeric mouse
Mice with humanized chimeric livers were generated as described previously64, according to the animal study proposals (2017-15-0201-01288 and 2021-15-0201-01063) approved by the Danish national animal use and care committee. Male and female Alb-uPA mice were maintained in a CB-17/Icr-Prkdcscid/scid/Rj (SCID) background with ad libitum access to food (SAFE D03, SAFE Complete Care Competence, Rosenheim, Germany) and water. The animals were housed by gender (except for breed-ers) in Innovive IVC caging containing wood chip bedding, shelters, nesting material and biting sticks on a 12-hour light dark cycle at 21 °C and 55 % humidity. All experimentation was conducted during light cycle adhering to 3R principles and with procedures consistent with affirmative response to the ARRIVE 10 questionnaire. Engraftment of human hepatocytes (Lonza) was confirmed 7 weeks after intra-splenic hepatocyte injection by human serum albumin levels using an enzyme linked immunosorbent assay (Bethyl Laboratories) and infection was subsequently done via intraperitoneal injection of 2x104 focus forming units (FFU) of J6/JFH1A876P64. Infection with HCV was evaluated at day 10 post infection by TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher) using primers previously described65 on viral RNA isolated from blood samples collected by temporal vein puncture and purified by Trizol LS reagent (Invitrogen). Sampling was performed every 6-10 days until day 60 post infection. Peak titers of 3.65x107 genome equivalents per mL (GE/mL) were observed 18-days post infection and titers consistently exceeded 1x106 GE/mL throughout follow-up. Human liver nodules collected from mice engrafted with human hepatocytes were collected following euthanasia at day 100 (corresponding serum titer of 1.07x106 GE/mL) and stored in RNA later (Sigma) until use. Total RNA was prepared by homogenization in Trizol reagent (Invitrogen) using a MagNA Lyser (Roche). Trizol RNA extraction was followed by purification with RNA Clean & Concentrator kit-25 (Zymo research, R1017), using manufacturer’s instructions. The sample size (one mouse) was chosen based on availability. No randomisation or blinding was used.
Generation of HCV enriched supernatant for LC-MS/MS
HCV enriched supernatant was generated as previously described66. Briefly, Huh7.5 cells were seeded in DMEM supplemented with 10 % FBS, 100 units/mL of penicillin and 100 μg/mL streptomycin (P/S) solution (Sigma) in triple-layer cell culture flasks the day before infection. Cells were infected at MOI=0.003 with the adapted SA13/JFH1 recombinant67, followed by transfer to a 10-layer EasyFill Cell Factory (Nunc) the next day. When 80 % of cells were HCV NS5A-positive based on immunostaining of a parallel culture, the medium was removed, and the cells were washed with 1x PBS. The cells were maintained in AEM (Gibco) supplemented with P/S solution. Every 2-3 days, supernatant was collected and replaced for a total of five 0.8 L harvests. Pooled supernatant was filtered with 5 μm and 0.65 μm capsule filters (Sartopure PP3, Sartorius) and concentrated 325-fold by ultrafiltration (MiniKros sampler 500 K mPES, Repligen). RNA from 1 mL of concentrated supernatant was extracted using Trizol LS reagent (Invitrogen) and purified with RNA Clean & Concentrator kit-5 following manufacturer’s instructions. RNA quantity was assessed using Qubit RNA High Sensitivity assay kit (ThermoFisher Scientific).
Human samples
Patients with chronic hepatitis C (CHC) were identified from the Danish Database for Hepatitis B and C (DANHEP), the Danish nationwide database with patients referred with CHC (positive for HCV-RNA ≥ 6 months) 68,69. DANHEP was approved by the Regional Scientific Ethical Committee (Capital Region) (protocol AHH-2017-103), and informed written consent was provided by all participants. Three patients with HCV genotype (GT) 1a, one patient with GT 2a and three patients with GT 3a with an HCV RNA titer >300.000 IU/ml were selected for this study. None of the patients were co-infected with hepatitis B virus. One patient was HIV-positive, received antiretroviral treatment and had undetectable viral load. Viral RNA was isolated from plasma by Trizol LS reagent (Invitrogen) followed by purification with RNA Clean & Concentrator kit-25 (Zymo research, R1017) according to manufacturer’s instructions.
Expression and purification of AtNUDX23, AtNUDX23-E169Q and NudC enzymes
The protein coding sequences for AtNUDX23 and NudC were used for gene synthesis and cloning into pET30(a) and pET28(+), respectively, by GenScript Biotech. For AtNUDX23 and AtNUDX23-E169Q, the chloroplast targeting N-terminal was deleted70, and a codon optimized sequence was used. The plasmids were transformed by heat shock into Escherichia coli Rosetta (DE3) competent cells. The cell cultures were grown at 37 °C to OD600=0.6 in LB media supplemented with 50 μg/mL kanamycin and 25 μg/mL chloramphenicol (for AtNUDX23 and AtNUDX23-E169Q) or 50 μg/mL kanamycin (for NudC) before induction with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 18 °C overnight. Cells were harvested and washed with ice-cold binding buffer A (5 mM imidazole, 20 mM Tris-HCl (pH 7.9), 500 mM NaCl) followed by lysis using French press in ice-cold binding buffer B (5 mM imidazole, 20 mM Tris-HCl (pH 7.9), 500 mM NaCl, 400 mM urea, 0.5 % Triton-X 100). Cell lysates were centrifuged at 9-10,000x g for 40 min at 4 °C and the supernatant was passed through a 0.45 μm filter. The cleared supernatant was applied to ProBond nickel-chelating resin (ThermoFisher Scientific) pre-washed with binding buffer B, and incubated for 1 hr at 4 °C with slow mixing. The resin was washed twice with 10 resin volumes of binding buffer B and twice with 10 volumes of binding buffer A. Wash buffers contained 60 mM imidazole instead of 5 mM. Protein was eluted with 6 volumes of elution buffer containing 20 mM Tris-HCl (pH 7.9), 500 mM NaCl and 0.5 M (for NudC) or 1 M (for AtNUDX23) imidazole. The buffer was exchanged into the storage buffer (25 mM Tris-HCl (pH 8.0), 300 mM NaCl, 5 % glycerol) using PD Minitrap G-10 columns (GE Healthcare) for AtNUDX23 and AtNUDX23-E169Q and D-tube™ Dialyzer Maxi tubes for NudC. Protein quality and quantity were assessed using SDS-PAGE and Bradford protein assay.
Purification of NS5B polymerases
JFH-1 NS5B with a deletion of the sequence coding for the C-terminal 21 amino acids (NS5BD21) was cloned from the full-length HCV J6/JFH159 into a Champion pET303/CT-His plasmid with a C-terminal His6 tag (Thermo Scientific). The sequence was amplified with NS5B-F and NS5B-R primers (Table S1) using Q5 Hot Start High-Fidelity DNA polymerase (New England Biolabs). The amplified NS5BD21sequence and the pET303 plasmid were digested with XhoI and XbaI restriction enzymes (New England Biolabs) and ligated using the Rapid DNA Ligation Kit (Thermo Scientific). The ligation product was transformed by heat shock into XL10-Gold ultracompetent cells (NEB) following the manufacturer’s instructions. The complete NS5B sequence of the NS5BD21- 4 pET303 plasmid obtained with the Sigma-Aldrich midiprep kit was confirmed by Sanger sequencing (Macrogen Europe). Mutations were introduced in the NS5BD21- 4 pET303 plasmid using a megaprimer approach with primers described in Table S1 at a concentration of 10 μM, 50 ng of the NS5BD21- 4 pET303 plasmid, and the Q5 High-Fidelity 2X Master Mix reagent, with the following PCR conditions: 98°C for 30 seconds; followed by 35 cycles of 98°C for 10 seconds, 71°C for 15 seconds, and 72°C for 1 minute; followed by a final extension at 72°C for 5 minutes. PCR products were gel purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research), and 200 ng of the amplicon were used as primer in a second PCR reaction (megaprimer reaction) with 50 ng of the NS5BD21-4 pET303 plasmid conducted at 98°C for 30 seconds; followed by 20 cycles of 98°C for 10 seconds, 48°C for 1 minute, and 72°C for 20 minutes, followed by a final extension at 72°C for 20 minutes. PCR products were treated overnight with DpnI (NEB). Obtaining of plasmids and confirmation of their NS5B sequence was performed as described above. Plasmids were transformed by heat shock into E. coli Rosetta 2(DE3) competent cells. A starter culture was grown in 2X YT medium supplemented with ampicillin (50 μg/mL) and chloramphenicol (34 μg/mL) at 37 °C to OD600=0.5-1.0. The final expression culture was inoculated with the starter culture and grown in Overnight Express Instant TB autoinduction medium (Novagen) supplemented with ampicillin (50 μg/mL) and chloramphenicol (34 μg/mL) at 22 °C for 24 hrs. The cells were pelleted at 4,000x g for 20 min at 4 °C and resuspended in buffer containing 50 mM Na2HPO4 (pH 8.0), 500 mM NaCl, 0.05 % n-octyl-b-D-glucoside, 20 mM imidazole, 4 mM MgCl2 and 1 U/mL DNase I. The cells were lysed using The cells were lysed using a French pressure cell press and thereafter centrifuged at 20,000x g for 60 min at 4 °C and the supernatant was passed through a 0.45 μm filter. The cleared supernatant was loaded onto a HisTrap HP purification column (Cytiva) on an ÄKTA Go purification system and washed with 50% binding buffer (50 mM Na2HPO4 (pH 8.0), 500 mM NaCl, 0.05 % n-octyl-b-D-glucoside, 20 mM imidazole) and 50% elution buffer (50 mM Na2HPO4 (pH 8.0), 500 mM NaCl, 0.05 % n-octyl-b-D-glucoside, 250 mM imidazole). Protein was eluted with 100% elution buffer. The fractions were analysed on an SDS-PAGE gel. Fractions with NS5B were pooled and buffer exchanged using PD10 Desalting Columns (Cytiva) into 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1 mM EDTA, 1 mM DTT and 5% glycerol. Protein purity and concentration were assessed using SDS-PAGE and the Bradford protein assay, respectively. The purified protein was stored in aliquots at -80 °C.
Assessment of AtNUDX23, NudC and Rpp enzymatic activity
Activity and specificity of AtNUDX23, NudC and Rpp (Lucigen, RP8092H) were determined with AMP-Glo assay (Promega, V5011) by measuring concentration of AMP generated by these proteins in the presence of different substrates (ATP, FAD and NAD+). Substrate concentrations were varied between 0.25 and 25 μM; protein concentrations were kept at 1 U/μl for Rpp, and 1 μM for AtNUDX23 and NudC. Reactions were carried out in quadruplicates. For reactions with Rpp, protein and substrate were incubated for 30 min at 37 °C in 1x Rpp buffer containing 50 mM HEPES-KOH (pH 7.5), 0.1 M NaCl, 1 mM EDTA, 0.1% β-mercaptoethanol, 0.01% Triton X-100. For reactions with AtNUDX23 and NudC, protein and substrate were incubated for 15 min at 37 °C in 1x NUDX buffer containing 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, and 1 mM DTT. All protein-substrate reactions were followed by 1 hr room temperature incubation with AMP-Glo Reagent I solution and 1 hr room temperature incubation with AMP detection solution as per manufacturer’s instructions. Luminescence was detected using Synergy HTX multi-mode reader (BioTek). An AMP standard curve was used to calculate AMP concentrations from luminescence measurements.
AtNUDX23 RNA decapping gel assay
AtNUDX23 RNA cleavage specificity was assessed with RNA decapping activity assays71 that compare AtNUDX23 ability to cleave RNA with different 5’ caps. Radioactively labelled 40 nt RNA 5’ capped with FAD, NAD, m7G, ATP and AMP were synthesized as previously described71 except with polymerase and buffer from MEGAscript T7 transcription kit (Invitrogen, AM1334). DNA template was prepared by annealing two DNA oligonucleotides (T7f2.5-40n_AG_F and T7f2.5-40n_AG_R, Supplementary Table 1) that included T7φ2.5 promoter followed by transcribed region of 40 nt with a single adenosine at the transcription start site. Radioactively labelled 5’ capped RNA was incubated with either 1 μM AtNUDX23 or 1 μM AtNUDX23-E169Q for 15 min at 37 °C in 1x NUDX buffer containing 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, and 1 mM DTT. Cleavage reactions were terminated by addition of equal volume of Gel loading buffer II (Ambion), resolved with a 7 M urea, 0.2 % 3’-acrylamidophenylboronic acid, 10 % PAGE and visualized using a PhosphorImager.
CapZyme-seq
1.25 μg of RNA supplemented with 2 μl of 1:100 diluted ERCC Spike-in mix (SIRV-Set 3, Lexogen, 051.01) was used in each enzymatic reaction (i.e., AtNUDX23, NudC, CapClip, Rpp and no enzyme control). To decrease the amount of ribosomal RNA in experiments with HCV strains (except for J6/JFH1) and TBEV, total RNA was treated with Terminator 5’-phosphate-dependent Exonuclease (Lucigen, TER51020), as previously described72, prior to enzymatic reactions. To convert 5’FAD capped RNA to 5’monophosphate RNA, RNA was treated with 1 μM AtNUDX23 in 1x NUDX buffer at 37 °C for 15 min. For reactions with NudC, RNA was treated with 1 μM NudC in 1x NUDX buffer at 37 °C for 15 min. To convert 5’m7G capped RNA to 5’-monophosphate RNA, RNA was treated with 5 U of Cap-Clip Acid Pyrophosphatase (CELLSCRIPT, C-CC15011H) in 1x Cap-Clip Acid Pyrophosphatase Reaction Buffer at 37 °C for 30 min. In parallel, RNA was incubated in 1x NUDX buffer without addition of any enzyme. Enzymatic reactions were stopped by adding EDTA to 16.7 mM and purified with RNA Clean & Concentrator kit-5 following manufacturer’s instructions. To convert 5’-triphosphate RNA to 5’-monophosphate RNA, RNA was treated with 1 U/μl of Rpp in 1x Rpp buffer supplemented with 1 U/μl of RiboLock RNase inhibitor (Thermo Fisher Scientific) for 30 min at 37 °C followed by purification with RNA Clean & Concentrator kit-5, as per manufacturer’s instructions. RNA generated from each enzymatic reaction was heat denatured in the presence of 10 μM 5’_ RNA_adapter_oligo (Table S1) at 65 °C for 5 min and stored on ice. The denaturation was followed by 5’ adapter ligation reaction using 0.5 U/μl T4 RNA ligase (Thermo Fisher Scientific) in 1x T4 RNA ligase buffer (Thermo Fisher Scientific) supplemented with BSA (1 mg/mL) and DMSO (10 % (v/v)). These reactions were incubated at 37 °C for 2 hrs and purified using RNAClean XP beads (Beckman Coulter) in a sample:beads ratio of 1:1.8, as per manufacturer’s instructions. 5’-adapter-ligated products were heat denatured in the presence of 0.5 μM reverse transcription primer (Table S1) at 65 °C for 5 min and stored on ice. Reverse transcription reactions were performed in 1x PrimeScript buffer (Takara Bio) with 5 U/μl of PrimeScript reverse transcriptase (Takara Bio) and dNTPs (1 mM each) by incubation at 30 °C for 10 min, 42 °C for 60 min and 70 °C for 15 min. The reactions were purified using AMPure XP beads (Beckman Coulter) in a sample:beads ratio of 1:1.8, as per manufacturer’s instructions. cDNA was amplified using 0.04 U/μl Phusion High-Fidelity DNA Polymerase (NEB, m0530) and 0.5 μM PCR_forward and PCR_reverse_index_# primers (Table S1) in 1x HF buffer (NEB) with 0.2 mM dNTPs. The reactions were denatured for 3 min at 98 °C followed by 12 cycles of 98 °C for 80 sec, 68 °C for 30 sec and 72 °C for 30 sec. The reactions were purified twice using AMPure XP beads (Beckman Coulter) in a sample:beads ratio of 1:1 as per manufacturer’s instructions. Quantity and quality of the libraries were assessed using Qubit 2.0 Fluorometer and Agilent 2100 Bioanalyzer. Single read sequencing with a read length of 75 bp was carried out on Illumina NextSeq550 using NextSeq500/550 High Output Kit v2.5.
Sequencing data analysis
Sequencing data was trimmed for adapter sequences and reads shorter than 15 nt using cutadapt (cutadapt -a AGATCGGAAGAGCACACGTCT -q 20 -m 15) 73. The RNAs expressed in the sample were identified by pseudomapping of the trimmed reads to the entire human transcriptome (Ensembl v96, GRCh38), or in case of BVDV infected MDBK cells, the bovine transcriptome (Ensembl v101, bosTau9) using Kallisto (kallisto quant --single -l 200 -s 30) 74. The transcripts with an expression level higher than 2 TPM were used to make a Bowtie2 index together with sequences for human or bovine ncRNAs (miRNA, tRNA, snRNA, snoRNA). The fasta files used for the analysis is available at https://doi.org/10.17894/ucph.c8675313-45c6-4a90-b4ab-c65abffd3f38. Subsequently, the sequencing reads were mapped against the index using bowtie2 (bowtie2 --norc --very-sensitive) 75. For analysis of coverage at tRNA loci, sequencing reads were mapped to the human genome sequence (hg19). The featureCounts function from the Subread package were used to obtain the 5’ termini counts for the first 100 nts for each transcript76. For the analysis of sequencing depth, the Samtools depth tool were used (samtools depth -a -H -m 0) 77. The nucleotide distribution at the 5’ end of reads mapping precisely to the end of HCV RNAs were extracted from sam files using a custom script. Example code for the processing of the sequencing data is available at https://github.com/jeppevinther/CapZyme. For plotting and statistical analysis, count and depth files were imported into R78 and plotted using base-R and ggplot279, and the final figures were assembled using Adobe Illustrator. Statistical analysis of the sequencing experiments was performed using the DESeq2’s Wald significance tests using a multi-factor design including the sample information as a term in the design formula (paired test) 80. For technical replicates, counts were merged before statistical analysis using the collapseReplicates function from DESeq2. To estimate FAD, 5’ppp and 5’p fractions on the ends of RNAs, the counts in each library were normalized to the combined counts obtained for the mitochondrial and cytoplasmic LSU and SSU rRNAs and the efficiency of Rpp and AtNUDX23 was assumed to be 100%. The percentage for 5’ppp and 5’FAD capping for each RNA was estimated by subtracting the normalized count of the control sample from the normalized count of the Rpp or AtNUDX23 sample and dividing by the sum of the normalized counts observed in all three samples. In cases where the normalized counts from the Rpp or AtNUDX23 treated samples were lower than the normalized Control counts, the difference was set to zero. For the analysis of HK6acc RNA stability, counts were obtained using the mapping/counting strategy described above. Libraries for each time point were normalized for sequencing depth using DeSeq2 and the control samples normalized counts was subtracted from Rpp and AtNUDX23 normalized counts, respectively.
The resulting values were normalized to the mean value obtained at time point 0. For the time course analysis of 5’ capping for JFH1-SGR-Feo replicon81, counts were obtained using the mapping/counting strategy described above. The obtained counts were analysed using DESeq2 for each time point using a multi-factor design including the sample information as a term in the design formula and comparing the control samples with Rpp and AtNUDX23 samples, respectively. The obtained DESeq2 Log2FoldChange and the associated standard error at different time points were used for plotting. All sequence data and the FeatureCounts output files are available at NCBI GEO under the accession GSE180956.
Quantification of FAD using LC-MS/MS
50 μg (total RNA samples), 7 μg (Terminator exonuclease treated samples) or the RNA output of ultrafiltration HCV supernatant prep was added to 0.15 pmol 13C5-FAD internal standard (Medical Isotopes, Inc., Pelham, US) and hydrolyzed to ribonucleotides by nuclease P1 (Sigma, see figure legends for enzyme concentration) in 10 mM ammonium acetate (pH 6.0) and 1 mM ZnCl2 at 40 °C for 1 h for the analysis of FAD (enzyme omitted for control samples). The pH was then adjusted with ammonium bicarbonate pH 7.5 to final concentration 50 mM to increase FAD stability. An aliquot was hydrolyzed by 20 U benzonase (Santa Cruz Biotech), 0.2 U nuclease P1 (Sigma) and 0.1 U alkaline phosphatase from E. coli (Sigma) in 10 mM ammonium acetate (pH 6.0) and 1 mM MgCl2 at 40 °C for 1 hr for the analysis of unmodified ribonucleosides. The hydrolysates were mixed with 3 volumes of acetonitrile and centrifuged (16,000x g, 30 min, 4 °C). The supernatants were lyophilized and dissolved in 8 mM ammonium bicarbonate (pH 7.0) for LC-MS/MS analysis of FAD and unmodified ribonucleosides.
Chromatographic separation of FAD was performed using an Agilent 1290 Infinity II UHPLC system with a ZORBAX RRHD Eclipse Plus C18 150 x 2.1 mm ID (1.8 μm) column (Agilent Technologies). The mobile phase was a gradient of buffer A 8 mM ammonium bicarbonate (pH 7.0) and B (methanol) starting with 0.5 min of 5 % B, then 2.5 min of 5-15 % B, 6 min of 15-95 % B, and finally 4 min re-equilibration with 5 % B at flow rate 0.2 mL/min. Unmodified ribonucleosides were chromatographed isocratically with water:methanol:formic acid 80:20:0.1 % at 0.25 mL/min. Mass spectrometric detection was performed using an Agilent 6495 Triple Quadrupole system operating in positive electrospray ionization mode, monitoring the mass transitions m/z 786.1-348.1 (FAD, CE 20), 786.1-136.1 (FAD, CE 44), m/z 791.1-353.1 (13C5-FAD, CE 20), m/z 268.1-268.1 (A), 284.1-152.1 (G), 244.1-112.1 (C), and 245.1-113.1 (U). The significance of FAD detection in the HCV infected samples compared to control samples was calculated using the one-sided Welch’s unequal variances t-test, as implemented in R78.
NS5B de novo replication initiation assay
10 μl de novo replication reactions contained 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 5 mM DTT, 0.3 μM or 0.18 μM NS5B, 10 μM RNA template (IDT), 1 mM of indicated initiating nucleotide (ATP or FAD, Sigma-Aldrich), 1 mM 3’dGTP (Jena Biosciences) or GTP (Sigma, Aldrich) and 1 mM of elongating nucleotide (CTP and UTP, Sigma Aldrich) supplemented with 0.5 μl or 1 μl of indicated α32P-labeled or γ32P-labeled nucleotide (3,000 Ci/mmol, Perkin Elmer). For the reactions with mutant NS5B proteins, visualized in each gel are reactions with equal concentrations of mutant and WT NS5B polymerases. Reactions with nesbuvir (HCV-796) and beclabuvir (both from Acme Biosciences) contained 0.1 mM and 1 mM of the antiviral. For the reactions showing time-dependent formation of the initiation product, ATP, 3’dGTP and UTP were omitted. For the reactions showing the effect of riboflavin and ATP on FAD de novo initiation, FAD concentration was reduced to 0.25 mM and 3’dGTP and UTP were omitted. In addition, either riboflavin (at 1 mM, 2 mM, 3 mM and 7.5 mM) or ATP (at 1 mM, 2 mM or 3 mM) were added. Assembled reactions were incubated at 30 °C for 1 hour and were terminated by extraction with equal volume of phenol-chloroform or by addition of equal volume of Gel loading buffer II (Ambion). 20 nt RNA, 14 nt RNA (both from IDT) and ApG dinucleotide (Jena Bioscience) were 5’ radioactively labelled with T4 polynucleotide kinase (ThermoFisher Scientific) following manufacturer’s instructions and used as size markers. For the FAD-C size maker, the dinucleotide was synthesized with a T7 in vitro transcription reaction as previously described71 except using polymerase and buffer from MEGAscript T7 transcription kit (Invitrogen, AM1334). DNA template was generated by annealing oligonucleotides (T7f2.5-40n_AC_F and T7f2.5-40n_AC_R, Supplementary Table 1), specifically designed to produce FAD-C dinucleotide. In the template, the T7φ2.5 promoter is followed by the sequence ACT at the transcription start site and the underlined adenosine is the only adenosine in the entire transcribed sequence. FAD-C dinucleotide is obtained by including FAD and α32P-labelled CTP in the T7 transcription reaction. RNA products along with size markers were resolved using 7 M urea, 18 % PAGE. Gels were visualized using a PhosphorImager and analysed with ImageQuantTL software. Percentage extension was calculated relative to the CTP band (loading control) and the average of the NS5B WT extension products. Percentage initiation was calculated relative to the CTP band and the FAD only initiation product, except for experiments showing time-dependent formation of the FAD initiation product, where % initiation is relative to the 30 min initiation product.
NS5B de novo replication initiation assay followed by AtNUDX23 enzyme treatment
10 μl de novo replication reactions containing 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 5 mM DTT, 0.3 μM NS5B, 10 μM HCV3END10A RNA template (IDT), 1 mM ATP (Sigma-Aldrich), 1 mM FAD (Sigma-Aldrich), 1 mM 3’dGTP (Jena Biosciences) or GTP (Sigma, Aldrich) and 1 mM CTP (Sigma Aldrich) were supplemented with 0.5 μl or 1 μL of α32P-labeled CTP or UTP (3,000 Ci/mmol, Perkin Elmer). To demonstrate presence of FAD in the replication initiation product, the reactions were incubated for 1 hr at 30 °C followed by treatment with AtNUDX23 enzyme, AtNUDX23-E169Q enzyme or water (negative control) by adding 10 μl containing 1 μM enzyme or water in 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, and 1 mM DTT. After 15 min incubation at 37 °C, the reactions were terminated by addition of equal volume of Gel loading buffer II (Ambion). To demonstrate presence of FAD in the replication elongation products, the reactions were incubated for 1 hr at 30 °C followed by purification with microspin G-25 columns (Merck) using manufacturer’s instructions. Purified reactions were incubated with 3 μM of AtNUDX23 enzyme, AtNUDX23-E169Q enzyme or water (negative control) for 30 min at 37 °C in 1x NUDX buffer containing 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, and 1 mM DTT. 10 μl of radioactively labelled FAD-C dinucleotide generated as described above were treated with AtNUDX23 enzyme, AtNUDX23-E169Q enzyme or water (negative control) by adding 10 μl containing 1 μM enzyme or water in 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, and 1 mM DTT. After 15 min incubation at 37 °C, these reactions were terminated by addition of equal volume of Gel loading buffer II (Ambion). 20 nt RNA, 14 nt RNA (both from IDT) and ApG dinucleotide (Jena Bioscience) were 5’ radioactively labelled with T4 polynucleotide kinase (ThermoFisher Scientific) following manufacturer’s instructions and used as size markers. The cleavage products were resolved using either 7 M urea, 18 % PAGE, or 7 M urea, 18 % PAGE, 0.2 % 3-(Acrylamido)phenylboronic acid (Sigma) (labelled as Boronate gel) and visualized using a PhosphorImager.
Production of 5’FAD capped RNA
35 nt FAD-capped RNA was generated using a template of annealed oligonucleotides containing a T7 Aφ2.5 promoter (CAGTAATACGACTCACTATT) followed by a 35 nt sequence (AGGGAAGCGGGCATGCGGCCAGCCATAGCCGATCA)82. In vitro transcription reaction was performed with MEGAscript T7 transcription kit (Invitrogen, AM1334) and contained 150 ng of template; 6.5 mM each of CTP, UTP, GTP; 1.5 mM ATP and 6 mM FAD. A control reaction containing 6.5 mM each of ATP, CTP, UTP and GTP was carried out in parallel. Following 6 hrs incubation at 37 °C, the template was removed by treatment with 0.1 U/μl TURBO DNase (Invitrogen AM1907) for 15 min at 37 °C and purification with RNA Clean & Concentrator kit-5 (Zymo Research). The resulting in vitro transcribed 5´ppp and 5´FAD RNA was quantified by Nanodrop and Qubit HS RNA kit (ThermoFisher Scientific) and resolved on a 7 M urea, 18 % PAGE gel together with a 20 nt RNA marker (IDT) and low range ssRNA ladder (NEB). To visualize FAD capped RNA, the gel was scanned using a Typhoon FLA 7000 scanner (ex. 473 nm, em. Y520 filter). SYBR GOLD staining (ThermoFisher Scientific) was used to visualize all of the nucleic acids in the sample.
For production of 5´FAD- and NAD-capped HCV RNA, we engineered pJFH1-SGR-Feo-GNN to include a T7 Aφ2.5 promoter (TAATACGACTCACTATT). The pJFH1-SGR-Feo-GNN-T7 Aφ2.5 plasmid was linearized using XbaI. For production of 5´m7G-HCV RNA, pJFH1-SGR-Feo-GNN plasmid was linearized using XbaI. For production of 5´ppp-capped Rat Hepacivirus (RHV) RNA, pRHV-rn-1plasmid83 was linearized using MluI. The in vitro transcription was performed with MEGAscript T7 transcription kit (Invitrogen, AM1334) in buffer containing 1 μg of template; 6.5 mM each of CTP, UTP, GTP; 1.5 mM ATP and 6 mM of either FAD, NAD or m7G. A control reaction with 6.5 mM each of ATP, CTP, UTP and GTP was carried out in parallel to generate a 5’ppp RNA. Following 2 hrs incubation at 30 °C, the template was removed by treatment with 0.1 U/μl TURBO DNase (Invitrogen AM1907) for 15 min at 37 °C. To control for free FAD co-purification, 6 mM of FAD was added to the control reaction after DNAse treatment. Unincorporated nucleotides were removed with a G-50 column (Roche, 11274015001) followed by purification with RNA Clean & Concentrator kit-25 (Zymo Research). The resulting in vitro transcribed 5´ppp, 5´FAD, 5´ NAD and 5´m7G RNA was quantified by Nanodrop and Qubit HS RNA kit (ThermoFisher Scientific) and resolved on a 1 % agarose, 3.7 % formaldehyde denaturing gel. To determine the percent of 5´metabolite-capping, RNA was subjected to the RT-qPCR reduction assay described below.
NS5B capping activity assay
To determine if NS5B uses FMN for generation of 5’FAD-cap in 5’ppp RNA, 1.5 μg of in vitro transcribed 5’ppp RNA (35 nt) was incubated with 0.3 μM NS5B and 1 mM FMN (Sigma-Aldrich) in 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 5 mM DTT for 1 hr at 30 °C. The reactions were purified with Oligo Clean & Concentrator kit (Zymo Research). The products were resolved using 7 M urea, 18 % PAGE gel. The in vitro transcribed 5’FAD-capped RNA (1.5 μg, 0.75 μg, 0.15 μg and 0.015 μg) was used as a control for FAD-cap visualization and 20 nt RNA was used as a size marker. To visualize FAD capped RNA, the gel was scanned using a Typhoon FLA 7000 scanner (ex. 473 nm, em. Y520 filter). SYBR GOLD staining (ThermoFisher Scientific) was used to visualize all of the nucleic acids in the sample.
Replication assays
The JFH1-SGR-Feo81, DBN3a-SGR-Fluc (with 5’G and adaptive mutations84), DBN3a-SGR-Fluc-GNN and H77-SGR-Feo (P1496L/K1691R/E1726G/S2204I; LRGI for simplicity85) replicons were previously described. Here, we generated H77-SGR-Feo (LRGI) construct by InFusion cloning and mutagenesis from H77/SG-Feo (L+8) 86 and H/SG-Neo (L+I)87 constructs, as the published construct could not be obtained. By similar methods, we generated JFH1-SGR-Feo-GNN, H77-SGR-Feo(LRGI)-GNN, H77-SGR-Feo(LRGI)-5’A, DBN3a-SGR-Fluc-GNN, and DBN3a-SGR-Fluc-5’A constructs. For 5’A constructs, an upstream G, which is part of the T7 promoter was preserved; this leads to in vitro transcribed RNA having an additional upstream 5’ G, which is known to rapidly be deleted upon HCV replication15,88. The pToto1101/SINV-Luc plasmid for generation of infectious reporter Sindbis virus (SINV) was previously described89.
The JFH1-SGR-Feo and DBN3a-SGR-Fluc plasmids were linearized using XbaI, H77-SGR-Feo (LRGI) using HpaI and pToto1101/SINV-Luc using ScaI. HCV replicon RNA in vitro transcription was performed using the T7 RiboMAX Express Large Scale RNA Production System (Promega P1320). SINV-Luc m7G-capped RNA was generated using the mMESSAGE mMACHINE SP6 Transcription kit (Invitrogen, AM1340). The resulting in vitro transcribed RNAs were treated with RQ1 DNase I (Promega, M6101) and purified with RNA Clean & Concentrator kit-25 (Zymo research, R1017). For cell culture, riboflavin-depleted media (rdRPMI) was prepared by dissolving RPMI-1640 Medium without L-alanine, L-glutamine, folic acid, riboflavin, Culture Media powder (MyBioSource, MBS653004) in milliQ water and supplemented with 1 mg/L of folic acid (Sigma, F8758), 2 mM of L-analyl-L-glutamine dipeptide (Gibco, 35050061) and 2 g/L of sodium bicarbonate. Riboflavin-depleted FBS (rdFBS) was obtained by dialysis against 1.2 M NaCl solution through a 3.5 MWCO dialysis cassette (ThermoFisher, 87726) at 4 °C for 96 hrs. For transfection, 5x104 Huh7.5 cells were seeded in triplicate into 6-well plates and incubated for 24 hrs at 37 °C. Cells were washed twice with PBS and rdRPMI + 10 % rdFBS either with or without 0.4 mg/L of riboflavin was added for 24 hrs at 37 °C and 5 % CO2. Cells were transfected with a mix of 1 μg of purified replicon RNA and 1 μL of Lipofectamine 2000 (Invitrogen 11668027) in rdRPMI supplemented with 3 % rdFBS for 4 hrs at 37 °C. Transfection media was removed, and cells were incubated at 37 °C with rdRPMI + 10 % rdFBS with or without supplementation with 0.4 mg/L riboflavin or 10 μM FAD. Cells were then incubated for 24, 48 or 72 hrs at 37 °C and 5 % CO2. Luciferase expression in cell lysates was measured using Luciferase Assay System (Promega E1500) on a CLARIOStar Plus microplate reader.
RT-qPCR reduction assay
1 μg of total RNA was subjected to two enzymatic steps, 1) Rpp, AtNUDX23, AtNUDX23-E169Q, NudC, mRNA decapping enzyme (MDE) (NEB M0608S) or mock, and 2) Xrn1 or mock. For samples with lower RNA quantity, such as that derived from plasma, the extracted RNA was spiked into 1 μg total RNA from Huh7.5 mock cells. To convert 5´ppp to 5´p, RNA was treated with 1 U/μl of Rpp in 20 μl of 1x Rpp buffer for 30 min at 37 °C followed by purification with RNA Clean & Concentrator kit-5 (Zymo Research). To convert 5´FAD to 5´p, RNA was treated with 1 μM of AtNUDX23 in 50 μl of 1x NUDX buffer supplemented with 1 mM DTT for 15 min at 37 °C followed by purification with RNA Clean & Concentrator kit-5. To convert 5´NAD to 5’ p, RNA was treated with 1 μM of NudC in 50 μl of 1x NUDX buffer supplemented with 1 mM DTT for 15 min at 37 °C followed by purification with RNA Clean & Concentrator kit-5. To convert 5´m7G to 5´p, RNA was treated with 0.25 U/μl of MDE in 20 μl of 1x mRNA decapping enzyme reaction buffer for 30 min at 37 °C followed by purification with RNA Clean & Concentrator kit-5. For 5´p RNA degradation, RNA from enzymatic reactions was treated with 1 U/μl of Xrn1 (NEB M0338L) in 20 μl of 1x NEB buffer 3 supplemented with 1 U/μl of Murine RNase Inhibitor (NEB M0314) for 60 min at 37 °C followed by purification with RNA Clean & Concentrator kit-5.
Quantification of RNA was performed using RT-qPCR with TS-O-01528, TS-O-01529 and TS-O-01530 primers recognizing HCV RNA; TS-O-00176, TS-O-00177 and TS-O-00178 primers recognizing mRNA encoding human ribosomal protein S11 (RPS11); TS-O-01544, TS-O-01545 and TS-O-01546 primers recognizing RHV RNA; or TS-O-01516, TS-O-01517; and TS-O-01518 recognizing mouse RPS11 mRNA (Table S1). The RT-qPCR reactions were assembled using TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems 4444432) and thermal cycling was performed with the following conditions: 55 °C for 10 min; 95 °C for 3 min; 45 cycles of 95 °C for 15 s, and 58 °C for 30 s; and 40 °C for 1 min. Statistical analysis is based on the one-sided Welch’s unequal variances t-test as implemented in R.
Inhibition of HCV infection by lumiflavin
To monitor the effect of lumiflavin on HCV infection, Huh7.5 cells were infected with TNcc (1a), J6/JFH1-c2 (2a) or DBN3acc (3a) virus titrated to yield 100-200 FFU/well after 72 hrs. Huh7.5 cells were seeded in triplicate at a density of 2x104 cells/well on a flat bottom 96-well plate coated with poly-L-lysine and cultured with 100 μL of DMEM supplemented with 10 % FBS at 37 °C 5 % CO2. 24 hrs post seeding cells were infected in medium supplemented with 0 μM, 25 μM, 50 μM, 75 μM and 100 μM lumiflavin (Cayman) with or without 10 μM FAD. Plates were then incubated for 72 hrs. After incubation, the plates were fixed with methanol, washed, and incubated overnight with the primary antibody anti-NS5A 9E10 combined with Core antibody (C7-50, Enzo Life Sciences Inc). Plates were then washed and incubated overnight with the secondary antibody anti-mouse conjugated HRP (1:500, Cytiva NA931-1ML). After incubation and washing, the plates were developed using a DAB staining kit according to the manufacturer’s instructions (Immunologic). The number of FFU were then automatically determined with an ImmunoSpot Series 5 Analyzer with customized software (CTL). FFU/well were normalized to the 0 μM lumiflavin control.
Cell viability assay
To monitor the effects of lumiflavin addition and riboflavin depletion on cell viability, we used mitochondrial function as a surrogate marker using XTT reduction to measure viability. Briefly, Huh7.5 cells were seeded at a density of 2x104 cells/well in a flat bottom 96-well plate and cultured in 100 μL of DMEM supplemented with 10 % FBS at 37 °C 5% CO2. 24 hrs post seeding, cells were supplemented with 0 μM, 24 μM, 50 μM, 75 μM and 100 μM lumiflavin (Cayman) + DMEM + 10 % FBS. Alternatively, DMEM was replaced with rdRPMI + 10 % rdFBS with or without supplementation with 0.4 mg/L riboflavin or 10 μM FAD. Cells were then incubated for 24, 48, 72 or 96 hrs at 37 °C 5 % CO2. After each time point, medium was replaced with FluoroBrite DMEM (Gibco) supplemented with 7.67 μg/mL PMS (Sigma) and 300 μg/mL XTT (Thermofisher) and incubated at 37 °C 5 % CO2 for 2 hrs. After incubation, absorbance was measured at 670 and 450 nm using a plate reader (MWG AG). For each plate, a blank value was measured that was subtracted from each sample on the plate.
Intracellular FAD quantification
To monitor the amount of intracellular FAD, cells were subjected to regular culture conditions with increasing concentrations of lumiflavin or to rdRPMI, and intracellular FAD was quantified. Briefly, Huh7.5 cells were seeded at a density of 3x105 cells/well in a 6-well plate and cultured with 2 mL of DMEM supplemented with 10 % FBS at 37 °C 5 % CO2. 24 hrs post seeding, cells were supplemented with 0, 24, 50 and 100 μM lumiflavin (Cayman) + DMEM + 10 % FBS. Alternatively, DMEM was replaced with rdRPMI + 10 % rdFBS with or without supplementation with 0.4 mg/L riboflavin or 10 μM FAD. Cells were then incubated for 72 or 96 hrs at 37 °C 5 % CO2. After incubation, intracellular FAD was quantified using a FAD Colorimetric/Fluorometric Assay Kit (BioVision) according to the manufacturer’s instructions.
Briefly, cells were washed twice in cold PBS, counted, lysed and deproteinated using a PCA deproteination kit (BioVision). Deproteinated samples were centrifuged at 12,000x g for 5 min and the supernatant was used to quantify intracellular FAD using a 535/587 nm band pass filter in a plate reader (Clarion). Concentration was determined by interpolation with a standard curve included in the run. For each plate, a blank value was measured that was subtracted from each sample on the plate.
Lumiflavin docking and modelling of the de novo initiation complex
Docking of the oxidized lumiflavin (PDB:LFN) was performed using SWISSDOCK90 on the crystal structure of HCV NS5B polymerase (PDB:3FQL) after removing the nesbuvir inhibitor. The pose search was performed in a cubic box of 2.5 nm size centered in the position of atom C14 of nesbuvir (PDB:79Z). The docking algorithm returned 256 poses, grouped into 36 structurally divergent positions. To model the de novo initiation complex containing the 3’ HCV(+) sequence, adenosine and CMP and lumiflavin, the ideal A-form double stranded RNA with sequence UGU was created using the nucleic acid builder91. To locate adenosine and CMP in the active site, the G-C base-pair in the double stranded RNA was superposed to the A-UDP base-pair in the primed initiation complex (PDB:4WTM), which contains a CC primer basepaired to a 3’-GGA template and an incoming UDP92. To locate lumiflavin, the HCV polymerase (PDB:3FQL) containing the docked lumiflavin was then superposed to the 4WTM structure using PyMOL93. All experimental structures containing FAD were retrieved from the Protein Data Bank as of September 2020, resulting in 4,928 conformations. For each FAD structure, the distance between atoms C1’ and C5B in FAD was calculated using PyMOL.
HK6acc RNA stability experiments
Huh7.5 cells in triplicates were infected with 1 mL of HK6acc containing supernatant58. Subsequently, cells were transferred to T80 culture flasks and sub-cultured 2-3 days, with a small portion of cells seeded on chamber slides. Slides were fixed and HCV antigen positive cells detected by immunostaining using NS5A monoclonal antibody (9E10)59 and Core antibody (C7-50, Enzo Life Sciences Inc). When > 90 % of cells were HCV antigen positive, cells were plated on 6-well plates for beclabuvir treatment at 10x EC50 (341 nM)58. Supernatants were removed and cells were collected at 0, 8, and 24 hrs. Cells were washed with 1x PBS and lysed with 1 mL of Trizol, collected and stored at -80 °C until use. Trizol RNA extraction was followed by DNase I digestion and purification with RNA Clean & Concentrator kit-5 using manufacturer’s instructions. CapZyme-Seq was performed as described above using Rpp and AtNUDX23 for enrichment.
HCV RNA stability experiments by electroporation
To measure RNA stability, 6x106 Huh7.5 cells in 400 μL cold cytomix (120 mM KC1; 0.15 mM CaCl2; 10 mM K2HPO4/KH2PO4, pH 7.6; 25 mM HEPES, pH 7.6; 2 mM EGTA, pH 7.6; 5mM MgCl2, pH adjusted with KOH; 2mM ATP and 5mM glutathione) were electroporated in triplicates with 1 μg of either 5´ FAD-, 5´ ppp-or 5´p-HCV RNA. 5´p-HCV RNA was derived from 5´FAD- and 5´ppp treated with AtNUDX23 or Rpp, respectively. Cells were electroporated using 4 mm cuvettes at infinite resistance, 270 V and 950 μF using a BioRad GenePulser XCell (BioRad). Electroporated cells were resuspended in 15 mL of DMEM + 10 % FBS and 2 ml per time point were plated in 6-well plates for RNA extraction at 0, 1, 2, 4, 8, 24 and 48 hrs post-electroporation. The extracted RNA was analysed by RT-qPCR as described above.
A549 reporter cell experiments
For transfection of RNA, 1.5x105 A549/pr(IFNβ).GFP cells42 producing GFP under an IRF3 dependent promoter, or A549-Dual cells (Invivogen) producing SEAP under an NF-κB dependent promoter, were seeded in triplicates into 6-well plates and incubated for 24 hrs at 37 °C. Cells were transfected with indicated amounts of either 5´ppp RNA or 5´FAD RNA and 1 μL of Lipofectamine 2000 in DMEM supplemented with 3 % FBS for 4 hrs at 37 °C. Cells transfected with indicated amounts of poly (I:C) (Sigma P1530) or with TNF-α added to the culture medium were used as a positive control. Mock transfections were carried out in parallel. After removal of the transfection media, cells were incubated in DMEM supplemented with 10 % FBS for 24 or 48 hrs at 37 °C. For flow cytometry, cells were washed with PBS, treated with trypsin, harvested with PBS supplemented with 2 % FBS and 2 mM EDTA and pelleted by centrifugation at 350 x g for 5 min at 4 °C. Single cell suspension was prepared using PBS supplemented with 2 % FBS and 2 mM EDTA, and filtered through a 40 μm cell strainer. Cells were analysed on a FACSMelody (BD Biosciences), and a minimum of 50,000 events were recorded. Data was analysed using the FlowJo program, and the percentage of GFP positive cells was determined. For SEAP measurements, the OD650 of cell culture media was measured according to manufacturer’s instructions.
For RIGI and MDA5 knockdown, 8x106 A549/pr(IFNβ).GFP cells in 400 μL cold cytomix were first electroporated with 60 pmol of either RIGI siRNA (ThermoFisher Scientific s223615), MDA5 siRNA (ThermoFisher Scientific s34499) or siGENOME Non-Targeting siRNA #5 (Horizon discovery D-001210-05-05), as described above. Electroporated cells were resuspended in 30 ml of DMEM + 10 % FBS, plated in 15-cm dishes and incubated at 37 °C and 5 % CO2.
Two days post first electroporation, 8x106 A549/pr(IFNβ).GFP cells were electroporated again with 60 pmol of the same siRNA, as described previously. Electroporated cells were resuspended in 40 mL of DMEM + 10 % FBS, and 2 mL plated in 6-well plates and incubated 24 hrs at 37 °C and 5 % CO2. One day post second siRNA electroporation, cells were transfected in triplicate with indicated amounts of 5´ppp or 5´FAD RNA and 1 μL of Lipofectamine 2000 in DMEM supplemented with 3 % FBS for 4 hrs at 37 °C. To avoid unintended 5’ppp derived signal, 5´FAD capped JFH1-SGR-Feo-GNN RNA was treated with Rpp as described above prior to transfection. Cells transfected with 200 ng of poly (I:C) (Sigma P1530) were used as a positive control. Mock transfections were carried out in parallel. After removal of the transfection media, cells were incubated in DMEM + 10 % FBS for 24 hrs at 37 °C and 5 % CO2. FACS was performed 24 hrs post-transfection, as described above.
Western blots
Total protein was harvested in RIPA lysis buffer (Santa Cruz sc-24948) and quantified using the Pierce BCA Protein Assay Kit (ThermoFisher Scientific 23225). The protein was resuspended in an LDS sample buffer, subjected to denaturing electrophoresis, and transferred to a nitrocellulose membrane. The membrane was probed with the primary antibody (rabbit anti-RIG-I [EPR18629] (1:1,000; Abcam ab180675), rabbit anti-MDA5 (D74E4) (1:1,000; Cell Signaling 5321T), rabbit anti-phospho-IRF3 (Ser396) (4D4G) (1:1,000; Cell Signaling 4947), or rabbit anti-phospho-NF-κB p65 (Ser536) (93H1) (1:1,000; Cell Signaling 3033) antibody) and the control rabbit anti-GAPDH [EPR16891] (1:10,000; Abcam ab181602) primary antibody. Subsequently, the blot was probed with the secondary antibody, anti-rabbit conjugated with horseradish peroxidase antibody (1:10,000; Abcam ab97051), visualized using enhanced chemiluminescence (ThermoFisher Scientific 34579 or 34094) and imaged with the ChemiDoc Touch Imaging System (BioRad).
Innate immune response during HCV infection
TNcc (1a) and J6/JFH1-c2 (2a) stocks were generated by infecting Huh7.5 cells as described above. When > 90 % of cells were HCV antigen positive, supernatants were collected, and HCV infectivity titers were determined as FFU/mL for normalization during concentration. Mock and HCV-containing supernatants were concentrated in parallel by centrifugal ultrafiltration with a 100 kDa MWCO membrane (Millipore UFC910024) at 3,500 x g for 25 min at 4 °C and HCV infectivity and RNA titers were determined as FFU/mL and GE/mL, respectively. For infection, 3x104 HepG2-HFL or A549/pr(IFNβ).GFP cells were seeded into 48-well plates in EMEM supplemented with 10 % FBS and 5 μg/mL blasticidin or DMEM supplemented with 10 % FBS, respectively. Cells were incubated overnight at 37 °C and infected with TNcc (1a) or J6/JFH1-c2 (2a) in triplicates with either 6.98x108 GE/mL (adjusted to 100 μL of concentrated supernatant in 500 μL of media), or at MOI=1 (adjusted to 30 μL of concentrated supernatant in 500 μL of media), corresponding to 1.07x109 and 4.57x107 GE/mL, respectively. Cells transfected with 25 ng of poly (I:C) (Sigma P1530) were used as a positive control and incubations with either mock concentrated supernatant or non-concentrated media were carried out in parallel. Cells were incubated for 18 or 24 hrs at 37 °C and lysed with Trizol for RNA extraction.
For RIGI knockdown, 8x106 HepG2-HFL or A549/pr(IFNβ).GFP cells in 400 μL cold cytomix were first electroporated with 60 pmol of either RIGI siRNA (ThermoFisher Scientific s223615), or siGENOME Non-Targeting siRNA #5 (Horizon discovery D-001210-05-05), as described above. Electroporated cells were resuspended in 30 mL of DMEM with 15 % FBS, plated in 15-cm dishes and incubated at 37 °C, 5 % CO2. Two days post initial electroporation, 8x106 of electroporated cells were electroporated again with 60 pmol of the same siRNA, as described previously. 3x104 of electroporated cells resuspended in DMEM and 20 % FBS were plated into 48-well plates and incubated for 24 hrs at 37 °C, 5 % CO2. One day post second siRNA electroporation, cells were infected in triplicates with either TNcc (1a) or J6/JFH1-c2 (2a) at MOI=1. Cells transfected with 25 ng of poly (I:C) (Sigma P1530) were used as a positive control. Cell incubations with either mock concentrated supernatant or non-concentrated media were carried out in parallel. Cells were incubated for 18 or 24 hrs at 37 °C and lysed with Trizol for RNA extraction. Trizol RNA extraction was followed by RNA purification and on-column DNase I digestion using RNA Clean & Concentrator kit-5, using manufacturer’s instructions. To measure levels of innate response genes, RNA was subjected to the RT-qPCR with TS-O-01693 and TS-O-01694 primers recognizing IFNL1; TS-O-01695 and TS-O-01696 primers- IFNL2/3; TS-O-01697 and TS-O-01698 primers- IFNB1; TS-O-01699 and TS-O-01700 primers-IFIT1, and TS-O-00176, TS-O-00177 primers -RPS11 used for normalization (Table S1). The RT-qPCR reactions were assembled using iTaq Universal SYBR Green One-Step Kit (BioRad 1725150) and thermal cycling was performed with the following conditions: 50 °C for 10 min; 95 °C for 1 min; 40 cycles of 95 °C for 15 s, and 60 °C for 60 s. Statistical analysis is based on the one-sided Welch’s unequal variances t-test as implemented in R.
Display of sequencing depth in UCSC genome browser
The reads obtained from one of the J6/JFH1 infected Huh7.5 cell samples were mapped to the human genome sequence (GRCh37/hg19) using bowtie2 with the (--very-sensitive) setting. The resulting bam files were converted to wig files for display in the UCSC genomes browser94 as described at https://github.com/jeppevinther/CapZyme. A UCSC session showing the Rpp, AtNUDX23 and control sequencing depth across GRCh37/hg19 is available here:http://www.genome.ucsc.edu/s/vinther/hg19_CapZyme%2Dseq_tRNA%2Dleu%2DTAG.
Statistics and reproducibility
Unless mentioned otherwise, experiments were not repeated. All repeated experiments showed similar results.
Extended Data
Extended Data Fig.1 |. Specificity of enzymes used for CapZyme-seq enrichment.
(a) Schematic representation of the AtNUDX23 and AtNUDX23-E169Q proteins used in this study. For both, the N-terminal 75 residues were truncated to remove the chloroplast localisation signal present in the WT protein. (b) AMP production in the presence of increasing concentrations of ATP, FAD and NAD+ for the indicated enzymes and the no enzyme control (n=4, independent replicates). Data are presented as mean +/− SD. (c) Strategy for the RT-qPCR reduction assay. (d) RT-qPCR reduction assay testing activity of AtNUDX23 and Rpp against in vitro transcribed FAD-capped RNA (left) and showing the lack of activity of the AtNUDX23-E169Q protein (right). (e) RT-qPCR reduction assay testing activity of AtNUDX23 and mRNA decapping enzyme (MDE) against in vitro transcribed m7G-capped RNA. (f) RT-qPCR reduction assay testing activity of AtNUDX23, Rpp and NudC against in vitro transcribed NAD+-capped RNA. For (d-f), data are presented as mean +/− SD, n=3 independent replicates.
Extended Data Fig.2 |. CapZyme-seq reads map to 5’ termini of RNAs.
Normalized sequencing depth plots for CapZyme-seq libraries: no enzyme control, AtNUDX23 and Rpp enrichment. Reads were mapped to human noncoding RNAs plus the most highly expressed mRNAs (> 2 TPM) in Huh7.5 cells. The normalized sequencing depth is shown for (a) all RNAs combined, (b) SSU rRNA, (c) J6/JFH1-c2 HCV(+) RNA, (d) J6/JFH1-c2 HCV(–) RNA, (e) 5S rRNA, (f) BCYRN1 RNA and (g) U6-ATAC RNA. (h) Schematic representation of tRNA processing: RNase P mediates processing of 5’ppp termini of primary tRNAs to 5’p mature tRNAs. (i) Normalized CapZyme-seq sequencing depth for tRNA-Leu-TAG-3-1 is shown for no enzyme control, AtNUDX23- and Rpp enrichment (top panels). Base wise conservation (phyloP score) for 100 vertebrate species and gene annotation (both obtained from UCSC genome browser) are shown in the bottom panels. Conserved positions have positive phyloP scores (blue). For browsing of the CapZyme-seq data at other human genomic loci, the following UCSC genome session can be used: http://www.genome.ucsc.edu/s/vinther/hg19_CapZyme%2Dseq_tRNA%2DLeu%2DTAG
Extended Data Fig.3 |. CapZyme-seq analysis of TBEV, BVDV and CHIKV infected cells.
Mean-difference plots showing fold changes (log2) as a function of average abundance (log) for reads in 5’ termini of individual RNA molecules. (a) CapZyme-seq analysis of RNA isolated from SH-SY5Y neuroblastoma cells infected with TBEV using Rpp, AtNUDX23, NudC and CapClip for enrichment. NudC has broad specificity, including cleavage of FAD, NAD+ and m7GpppN; Cap-Clip Acid Pyrophosphatase hydrolyzes m7GpppN (n=3 biological replicates). (b) CapZyme-seq analysis of RNA isolated from MDBK bovine kidney cells infected with BVDV using Rpp and AtNUDX23 for enrichment (n=3 biological replicates). (c) CapZyme-seq analysis of RNA isolated from TIG3 fibroblast cells infected with CHIKV using Rpp and AtNUDX23 for enrichment (n=2 biological replicates).
Extended Data Fig.4 |. Time course analysis of 5’ capping for JFH1-SGR-Feo replicon and LC-MS/MS detection of RNA 5’FAD caps.
(a) Enrichment of 5’ terminal reads for JFH1-SGR-Feo (–) at the indicated time points. DESeq2 log2(fold change) values were calculated by comparison of enzyme treatment (AtNUDX23 in dark blue and Rpp in light blue) to no enzyme control libraries. The 4 and 12 hrs time points for JFH1-SGR-Feo (–) are associated with uncertainty, because of low counts. (n=2 biological replicates; mean is represented by the line). (b) As in (a) but for JFH1-SGR-Feo (+). (c) Analysis of viral replication at different time points. Relative luciferase units (RLU) show HCV IRES mediated translation, and replication levels can be deduced by comparing JFH1-SGR-Feo to JFH1-SGR-Feo-GNN (catalytically dead replicon control) at the indicated time points after transfection (n=3 biological replicates). (d) Sample preparation for FAD detection using LC-MS/MS. (e) FAD product ion mass spectrum. Insert shows the structure of FAD with the detected ions indicated. (f) LC-MS/MS chromatogram showing the detection of the FAD control. (g) LC-MS/MS chromatogram showing the detection of FAD in a control Huh7.5 mock-infected sample (light blue), intracellular RNA from HCV infected Huh7.5 cells (dark blue) and in HCV particles concentrated from supernatant (yellow). (h) LC-MS/MS FAD quantification. FAD concentration based on internal stable isotope-labelled standards for FAD and normalization to ribonucleotide content. RNA was isolated from J6/JFH1 infected or control Huh7.5 cells (mock) and treated with or without Nuclease P1. Due to different lot activities, lower concentration of Nuclease P1 (0.006 U/μg total RNA) was used here, compared to the experiment presented in Fig. 1g (0.12 U/μg total RNA), resulting in lower FAD/rNs ratios. The p-values are calculated using one-sided Welch’s unequal variances t-test. Data are presented as mean +/− SD (from left to right n = 9, 3, 9 and 6 biological replicates). (i) Intracellular FAD levels in Huh7.5 cells with and without riboflavin and HCV infection or after lumiflavin treatment. Cells were cultured with or without supplementation of riboflavin (0.4 mg/L) or FAD (10 μM) or were treated with increasing lumiflavin concentrations. HCV indicates infection with J6/JFH1. Fluorescence measurements were used for FAD quantification; shown values for FAD pmol/cell were calculated using a standard curve. Data are presented as mean +/− SD (n=3 biological replicates).
Extended Data Fig.5 |. Identification of 5’ capping for different HCV strains.
CapZyme-seq analysis of RNA isolated from Huh7.5 cells infected with the indicated HCV genotypes/strains. Mean-difference plots show log2 (fold changes) as a function of log2 (average abundance) for reads mapping to the 5’ termini of individual RNA molecules. Enrichment with Rpp (a) and AtNUDX23 (b) was based on comparison of enzyme-treated libraries to no enzyme control libraries. The log2 (fold values) observed for HCV(+), HCV(–) and 5S rRNA (5’ppp) for the different genotypes/strains are summarized in (c) for Rpp and (d) for AtNUDX23. Data is derived from a single biological replicate. Main Fig. 2c shows replicate experiments for selected strains.
Extended Data Fig.6 |. The effect of riboflavin depletion and lumiflavin treatment on cellular viability and HCV genotype-specific dependency on FAD.
(a) Cell viability after lumiflavin treatment or riboflavin depletion. (b) Measurement of translational activity at 4 hrs after transfection of JFH1-SGR-Feo RNA with and without 0.4 mg/L riboflavin and 10μM FAD. Transfection at several time points after riboflavin depletion was included to exclude that differences in cell viability affected HCV IRES-mediated translation. (c) Measurement of Sindbis virus (SINV) replication after transfection of in vitro transcribed RNA from infectious SINV Toto-1101/Luc reporter virus with and without 0.4 mg/L riboflavin and 10 μM FAD. Transfection at several time points after riboflavin depletion was included to exclude that differences in cell viability affected SINV replication. (d) Replication of DBN3a-SGR(5’G) and GNN (non-replicating mutant) in Huh7.5 cells grown in riboflavin-depleted media. DBN3a-SGR-Fluc(5’G) had partially reverted to 5’A at the 72 hrs time point. Conditions with riboflavin (0.4 mg/L) and FAD (10 μM) included are indicated. Replication was measured in relative luciferase units (RLU) at the indicated time points and quantified relative to 4 hrs. (e) Inhibition of HCV infection by lumiflavin. Infection was measured in focus forming units relative to the untreated control obtained after infection with HCV TNcc (1a), J6/JFH1 (2a) or DBN3acc (3a) strains. For all panels, data are presented as mean +/− SD, n=3 biological replicates.
Extended Data Fig.7 |. FAD dependent replication initiation by HCV NS5B.
(a) Coomassie stained SDS-PAGE of the HCV NS5B WT, GNN, C316F, C316A, S365L and C366A recombinant proteins. (b) RNA templates used for in vitro replication reactions. (c) Time-dependent formation of the initiation product. HCV NS5B polymerase was incubated with HCV3END10A RNA in the presence of FAD and CTP, complemented with α 32P-labeled CTP for visualization. Reactions were terminated at the indicated time points. Products were resolved with 18 % denaturing PAGE together with 5’ radioactively labelled markers. (d) The replication extension products contain FAD. NS5B polymerase was incubated with HCV3END10A RNA in the presence of FAD, ATP, CTP, UTP (complemented with α-32P-labeled UTP) and either GTP or 3’dGTP. After 1 hr incubation, the reactions were treated with AtNUDX23 or AtNUDX23-E169Q. The products were resolved with 18 % denaturing 0.2 % boronate PAGE. The replication reactions subjected to AtNUDX23 or AtNUDX23-E169Q treatment are identical to the α-32P-labeled UTP reactions separated on a regular PAGE gel in main Fig. 3c. The presence of boronate retards FAD-capped RNAs compared to uncapped RNAs due to the relatively stable diol complexes formed between the gel derived boronyl groups and naturally present diols of the RNA 3’ end and of FAD. Accordingly, both 5’ FAD capping and 3’ deoxy termination of extension products will affect migration in the boronate gel. Products were annotated by comparison between the 3’dGTP/GTP conditions, AtNUDX23/AtNUDX23-E169Q conditions and the migration observed on the regular PAGE gel (main Fig. 3c). (e) FAD promotes replication initiation on different RNA templates. Indicated RNA templates were incubated with HCV NS5B polymerase. ATP, FAD or control was added as initiating nucleotide. Elongating UTP or CTP nucleotides were supplemented with α32P-labeled UTP or CTP, respectively. The resulting products were resolved with 18 % denaturing PAGE. (f) Lack of HCV NS5B mediated post-initiation FAD capping. To exclude that NS5B mediated 5’FAD capping could happen post-initiation on a 5’ppp initiated RNA, 5’ppp RNA was incubated with FMN in the presence of NS5B polymerase. RNA was resolved with 18 % PAGE and visualised by nucleic acid staining (left) and with fluorescent signal indicative of the 5’FAD cap (right). Increasing concentrations of 5’FAD capped RNA generated with T7 polymerase were used as markers on the same gel. For gel source data, see Supplementary Figure 1.
Extended Data Fig.8 |. Prediction model of FAD − HCV NS5B interaction.
(a) In silico docking analysis of FAD into HCV NS5B using SwissDock. Full fitness scores for the top 36 positions obtained by docking of lumiflavin into HCV NS5B are shown. (b) Close-up view of the palm II binding pocket demonstrating the overlap between the nesbuvir (green) binding site and the putative binding site of the lumiflavin moiety of FAD (blue). The docking was based on (PDB: 3FQL) with the nesbuvir molecule removed. (c) Predicted contacts between NS5B and lumiflavin when docked in the palm II site of NS5B. (d) Predicted lumiflavin binding site modelled into the structure of the NS5B primed initiation complex (PDB:4WTM) colored by subdomains: fingers (pink), palm (light blue), thumb (pale green) and beta-loop (yellow). The HCV(+) 3’ end sequence was modeled into the position of the template (red). An adenosine (blue) and CMP (green) were modeled into the position of the priming and incoming nucleotides, respectively. The close-up view shows the active site with two localized Mn2+ ions in violet. The direct distance between the lumiflavin N10 methyl and the ribose C5 is indicated. (e) Observed FAD N10 methyl-C5 distances from 4,928 crystal structures of proteins containing a FAD cofactor. (f-h) Triplicate analysis (independent replicates) of de novo initiation with FAD for the indicated NS5B mutants using the HCV3END10A template with the FAD extension and initiation signal quantified below the gels. The p-values are calculated using two-sided Welch’s unequal variances t-test. N.S. equals p-val > 0.05. For gel source data, see Supplementary Figure 1.
Extended Data Fig.9 |. RNA stimulation of innate immune responses.
(a) Analysis of the in vitro transcribed RNAs used in cell-intrinsic innate immune recognition experiments (main Fig. 4c-e and g, and Extended Data Fig. 10a-b). RNA was resolved with 18 % PAGE and visualised by nucleic acid staining (left) or with fluorescent signal from the 5’FAD cap (right). (b) Gating strategy for flow cytometry experiments. Mock- and poly (I:C) transfected A549/pr(IFNβ).GFP reporter cells were used to set gates. Live cells were SSC-A:FSC-A gated, followed by FSC-H:FSC-A gating to select single cells. Single cells were then GFP(–) or GFP(+) gated. (c) Immunoblot for RIG-I after control or RIGI siRNA mediated knock-down in A549/pr(IFNβ).GFP reporter cells. The RIG-I specific band is indicated. (d) Immunoblot for MDA5 after control or MDA5 siRNA mediated knock-down in A549/pr(IFNβ).GFP reporter cells and induction of MDA5 expression by transfection with 5´ppp JFH1-SGR-Feo-GNN RNA. 50 IU/mL IFN-a 2a, is shown for comparison; this concentration was too low to induce MDA5 expression at the shown exposure level. (e) Analysis of innate immune activation upon transfection of JFH1-SGR-Feo-GNN replicon RNA into A549/pr(IFNβ).GFP cells after knock-down of RIG-I or MDA5. Data are presented as mean +/− SD, n=3 biological replicates. For gel source data, see Supplementary Figure 1.
Extended Data Fig.10 |. RNA stimulation of innate immune responses.
(a) Immunoblot for phosphorylated NF-kB subunit p65 in HepG2-HFL cells 4 hrs after stimulation with 5’ppp or 5’FAD as indicated. (b) Immunoblot for phosphorylated IRF3 in A549/pr(IFNβ).GFP cells 12 hrs after stimulation with 5’ppp or 5’FAD as indicated. The upper band, consistent with the size of p-IRF3 appears only after stimulation. Note the different loading order compared to panel (a). (c) mRNA expression levels for IFNB1, IFNL1, IFNL2/3 and IFIT1 18 hrs post infection of HepG2-HFL cells with equal RNA titers of indicated HCV strains shown relative to polyI:C induced levels. Data represent mean +/− SD (n=3 biological replicates). ND: Not detected. (d) Immunoblot for RIG-I after control or RIGI siRNA mediated knock-down in HepG2-HFL cells and induction of RIG-I expression by transfection of 100 ng in vitro transcribed 5´ppp J6/JFH1 RNA, 100 ng Poly I:C or treatment with 500 U/mL IFN-a 2a. (e) mRNA expression levels for IFNB1, IFNL1, IFNL2/3 and IFIT1 18 hrs post infection of HepG2-HFL cells with indicated HCV strains shown relative to levels obtained for Mock infected control siRNA treated cells. Data represent mean +/− SD (n=3 biological replicates). Prior to infection, cells were transfected with control siRNA or RIGI targeting siRNA. The p-values in (c) and (e) are calculated using one-sided Welch’s unequal variances t-test by comparing to the mock infection samples. For gel source data, see Supplementary Figure 1.
Supplementary Material
Acknowledgments
We thank Dr. Charles M. Rice (Rockefeller University) and Dr. Jan Christiansen (University of Copenhagen) for critical comments; members of the authors’ laboratories for helpful discussion; Dr. Konstantinos Kokkonos, Lotte Mikkelsen and Nele Brinkmann for assistance with reagents; Dr. Matthew Evans (Mount Sinai School of Medicine) for HepG2-HFL cells; Dr. Catherine Adamson (University of St Andrews) for A549/pr(IFNβ).GFP cells; Dr. Philip Meuleman (University of Ghent) for help with establishing the human liver chimeric mouse model; and the BIO-UCPH sequencing and biocomputing core facilities for access to necessary infrastructure. Mass spectrometry analyses were performed by the Proteomics and Modomics Experimental Core (PROMEC), Norwegian University of Science and Technology, which is a member of the National Network of Advanced Proteomics Infrastructure (NAPI), Norway. This work was supported by Independent Research Fund Denmark grant DFF-6110-00350 (JB, JV) and DFF-9039-00380B (SR, JB); European Research Council (ERC) Starting Grant 802899 (TKHS); Novo Nordisk Foundation Distinguished Investigator grant NNF19OC0054518 (JB), NNF18OC0052354 (JV), NNF19OC0055462 (JB), and NNF19OC0058443 (TKHS); Carlsberg Foundation Young Researcher Fellowship (TKHS), Research Infrastructure grant CF18-1075; Innovation Fund Denmark grant 1046-00020B (JMG); Danish Agency for Science and Higher Education grant 0237-00005B (SR, JB); Candys Foundation grant 2017-248 (AO, JMG, JB); PROMEC is funded by The Central Norway Regional Health Authority and the Research Council of Norway INFRASTRUKTUR-program (295910) (CBV).
Footnotes
Author contributions:
Conceptualization: JB, TKHS, JV (lead); Funding acquisition: CBV, JMG, SR, JB, TKHS, JV; Experiments: CapZyme-Seq (AVS, HSL, KMA and AA-C), LC-MS/MS (CBV and AK), protein expression (AVS, GI, AP-C, JERG and NWL), human liver chimeric mouse infection (KH), patient samples (CS and NW), virus infection and cell assays (LRRR, HSL, RC, EJ, LVP, LN and NF), NS5B polymerase expression and in vitro replication experiments (AVS, LAR, CF-A and GI), NS5B modelling (SB), RT-qPCR reduction assay (LRRR and HSL), HCV RNA half-life (LRRR and LVP), innate immune response assays (LRRR and EJ), and high-titer viral supernatant (AO), Project administration: AVS, JB, TKHS, JV (lead); Supervision: JMG, SR, JB, TKHS, JV; Visualization: JV; Writing − original draft: AVS, LRRR, JB, TKHS, JV (lead); All authors reviewed and edited the manuscript.
Competing interests: Authors declare no competing interests.
Data availability
The sequencing data and the count files obtained for the different experiments are available in NCBI GEO with the accession GSE180956 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE180956). Fasta files used for mapping is available at: https://doi.org/10.17894/ucph.c8675313-45c6-4a90-b4ab-c65abffd3f38. Data used for plotting of the main figures 1, 2 and 4 are available as Source Data 1-3.
Code availability
Analysis of the CapZyme-seq data is based on existing tools and is described in detail at https://github.com/jeppevinther/CapZyme. Additional information about the code used for the analyses described in this paper is available from the corresponding author (JV) upon request.
References
- 1.Li Y, Yamane D, Masaki T, Lemon SM. The yin and yang of hepatitis C: synthesis and decay of hepatitis C virus RNA. Nat Rev Microbiol. 2015;13:544–558. doi: 10.1038/nrmicro3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Masaki T, Yamane D, McGivern DR, Lemon SM. Competing and noncompeting activities of miR-122 and the 5’ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1881–1886. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukh J. The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J Hepatol. 2016;65:S2–S21. doi: 10.1016/j.jhep.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 4.Cahova H, Winz ML, Hofer K, Nubel G, Jaschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015;519:374–377. doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- 5.Jiao X, et al. 5’ End Nicotinamide Adenine Dinucleotide Cap in Human Cells Promotes RNA Decay through DXO-Mediated deNADding. Cell. 2017;168:1015–1027.:e1010. doi: 10.1016/j.cell.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walters RW, et al. Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:480–485. doi: 10.1073/pnas.1619369114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird JG, et al. Highly efficient 5’ capping of mitochondrial RNA with NAD(+) and NADH by yeast and human mitochondrial RNA polymerase. Elife. 2018;7 doi: 10.7554/eLife.42179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, et al. NAD(+)-capped RNAs are widespread in the Arabidopsis transcriptome and can probably be translated. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:12094–12102. doi: 10.1073/pnas.1903682116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo G, et al. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol. 2000;74:851–863. doi: 10.1128/jvi.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong W, Uss AS, Ferrari E, Lau JY, Hong Z. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol. 2000;74:2017–2022. doi: 10.1128/jvi.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Z, Liang TJ, Luo G. Effects of Mutations of the Initiation Nucleotides on Hepatitis C Virus RNA Replication in the Cell. Journal of Virology. 2004;78:3633–3643. doi: 10.1128/JVI.78.7.3633-3643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi M, Lemon SM. Structure-function analysis of the 3’ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA. 2003;9:331–345. doi: 10.1261/rna.2144203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramirez S, Bukh J. Current status and future development of infectious cell-culture models for the major genotypes of hepatitis C virus: Essential tools in testing of antivirals and emerging vaccine strategies. Antiviral Res. 2018;158:264–287. doi: 10.1016/j.antiviral.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 14.Li YP, Ramirez S, Gottwein JM, Bukh J. Non-genotype-specific role of the hepatitis C virus 5’ untranslated region in virus production and in inhibition by interferon. Virology. 2011;421:222–234. doi: 10.1016/j.virol.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5’ UTR. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4991–4996. doi: 10.1073/pnas.1016606108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sedano CD, Sarnow P. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe. 2014;16:257–264. doi: 10.1016/j.chom.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amador-Canizares Y, Bernier A, Wilson JA, Sagan SM. miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5’ end from the cellular pyrophosphatases, DOM3Z and DUSP11. Nucleic acids research. 2018;46:5139–5158. doi: 10.1093/nar/gky273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 20.Gebert LFR, Law M, MacRae IJ. A structured RNA motif locks Argonaute2:miR-122 onto the 5’ end of the HCV genome. Nat Commun. 2021;12:6836. doi: 10.1038/s41467-021-27177-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat Chem Biol. 2009;5:879–881. doi: 10.1038/nchembio.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowtoniuk WE, Shen Y, Heemstra JM, Agarwal I, Liu DR. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7768–7773. doi: 10.1073/pnas.0900528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frindert J, et al. Identification, Biosynthesis, and Decapping of NAD-Capped RNAs in B. subtilis. Cell Rep. 2018;24:1890–1901.:e1898. doi: 10.1016/j.celrep.2018.07.047. [DOI] [PubMed] [Google Scholar]
- 24.Julius C, Yuzenkova Y. Bacterial RNA polymerase caps RNA with various cofactors and cell wall precursors. Nucleic acids research. 2017;45:8282–8290. doi: 10.1093/nar/gkx452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vvedenskaya IO, et al. CapZyme-Seq Comprehensively Defines Promoter-Sequence Determinants for RNA 5’ Capping with NAD. Molecular cell. 2018;70:553–564.:e559. doi: 10.1016/j.molcel.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, et al. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic acids research. 2019;47:e130. doi: 10.1093/nar/gkz751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doamekpor SK, et al. DXO/Rai1 enzymes remove 5’-end FAD and dephospho-CoA caps on RNAs. Nucleic acids research. 2020;48:6136–6148. doi: 10.1093/nar/gkaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marceau CD, et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruta T, et al. An Arabidopsis FAD pyrophosphohydrolase, AtNUDX23, is involved in flavin homeostasis. Plant Cell Physiol. 2012;53:1106–1116. doi: 10.1093/pcp/pcs054. [DOI] [PubMed] [Google Scholar]
- 30.Catanese MT, et al. Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission. J Virol. 2013;87:8282–8293. doi: 10.1128/JVI.01102-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercer DF, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 32.Pham LV, et al. HCV genome-wide analysis for development of efficient culture systems and unravelling of antiviral resistance in genotype 4. Gut. 2022;71:627–642. doi: 10.1136/gutjnl-2020-323585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howe AY, et al. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob Agents Chemother. 2008;52:3327–3338. doi: 10.1128/AAC.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwerk J, Negash A, Savan R, Gale M., Jr Innate Immunity in Hepatitis C Virus Infection. Cold Spring Harb Perspect Med. 2021;11 doi: 10.1101/cshperspect.a036988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasou A, et al. Modular cell-based platform for high throughput identification of compounds that inhibit a viral interferon antagonist of choice. Antiviral Res. 2018;150:79–92. doi: 10.1016/j.antiviral.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S, et al. Heterocellular induction of interferon by negative-sense RNA viruses. Virology. 2010;407:247–255. doi: 10.1016/j.virol.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 38.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loo YM, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narbus CM, et al. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J Virol. 2011;85:12087–12092. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang W, et al. Correlation of the tight junction-like distribution of Claudin-1 to the cellular tropism of hepatitis C virus. The Journal of biological chemistry. 2008;283:8643–8653. doi: 10.1074/jbc.M709824200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park JO, et al. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat Chem Biol. 2016;12:482–489. doi: 10.1038/nchembio.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamane D, et al. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med. 2014;20:927–935. doi: 10.1038/nm.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sumpter R, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kincaid RP, Lam VL, Chirayil RP, Randall G, Sullivan CS. RNA triphosphatase DUSP11 enables exonuclease XRN-mediated restriction of hepatitis C virus. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:8197–8202. doi: 10.1073/pnas.1802326115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma S, et al. Identification of a novel deFADding activity in human, yeast and bacterial 5’ to 3’ exoribonucleases. Nucleic acids research. 2022;50:8807–8817. doi: 10.1093/nar/gkac617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tilgner M, Shi PY. Structure and function of the 3’ terminal six nucleotides of the west nile virus genome in viral replication. J Virol. 2004;78:8159–8171. doi: 10.1128/JVI.78.15.8159-8171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teramoto T, et al. Genome 3’-end repair in dengue virus type 2. RNA. 2008;14:2645–2656. doi: 10.1261/rna.1051208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lescar J, Canard B. RNA-dependent RNA polymerases from flaviviruses and Picornaviridae. Curr Opin Struct Biol. 2009;19:759–767. doi: 10.1016/j.sbi.2009.10.011. [DOI] [PubMed] [Google Scholar]
References for Methods
- 51.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li YP, et al. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19757–19762. doi: 10.1073/pnas.1218260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li YP, Ramirez S, Mikkelsen L, Bukh J. Efficient infectious cell culture systems of the hepatitis C virus (HCV) prototype strains HCV-1 and H77. J Virol. 2015;89:811–823. doi: 10.1128/JVI.02877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramirez S, et al. Cell Culture Studies of the Efficacy and Barrier to Resistance of Sofosbuvir-Velpatasvir and Glecaprevir-Pibrentasvir against Hepatitis C Virus Genotypes 2a, 2b, and 2c. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.01888-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramirez S, et al. Highly efficient infectious cell culture of three hepatitis C virus genotype 2b strains and sensitivity to lead protease, nonstructural protein 5A, and polymerase inhibitors. Hepatology. 2014;59:395–407. doi: 10.1002/hep.26660. [DOI] [PubMed] [Google Scholar]
- 56.Ramirez S, Mikkelsen LS, Gottwein JM, Bukh J. Robust HCV Genotype 3a Infectious Cell Culture System Permits Identification of Escape Variants With Resistance to Sofosbuvir. Gastroenterology. 2016;151:973–985.:e972. doi: 10.1053/j.gastro.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 57.Humes D, et al. Recombinant hepatitis C virus genotype 5a infectious cell culture systems expressing minimal JFH1 NS5B sequences permit polymerase inhibitor studies. Virology. 2018;522:177–192. doi: 10.1016/j.virol.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 58.Pham LV, et al. HCV Genotype 6a Escape From and Resistance to Velpatasvir, Pibrentasvir, and Sofosbuvir in Robust Infectious Cell Culture Models. Gastroenterology. 2018;154:2194–2208.:e2112. doi: 10.1053/j.gastro.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 59.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 60.Mandl CW, Ecker M, Holzmann H, Kunz C, Heinz FX. Infectious cDNA clones of tick-borne encephalitis virus European subtype prototypic strain Neudoerfl and high virulence strain Hypr. J Gen Virol. 1997;78(Pt 5):1049–1057. doi: 10.1099/0022-1317-78-5-1049. [DOI] [PubMed] [Google Scholar]
- 61.Tsetsarkin K, et al. Infectious clones of Chikungunya virus (La Reunion isolate) for vector competence studies. Vector Borne Zoonotic Dis. 2006;6:325–337. doi: 10.1089/vbz.2006.6.325. [DOI] [PubMed] [Google Scholar]
- 62.Mendez E, Ruggli N, Collett MS, Rice CM. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J Virol. 1998;72:4737–4745. doi: 10.1128/jvi.72.6.4737-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheel TK, et al. A Broad RNA Virus Survey Reveals Both miRNA Dependence and Functional Sequestration. Cell Host Microbe. 2016;19:409–423. doi: 10.1016/j.chom.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prentoe J, et al. HVR1-mediated antibody evasion of highly infectious in vivo adapted HCV in humanised mice. Gut. 2016;65:1988–1997. doi: 10.1136/gutjnl-2015-310300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Engle RE, Russell RS, Purcell RH, Bukh J. Development of a TaqMan assay for the six major genotypes of hepatitis C virus: comparison with commercial assays. J Med Virol. 2008;80:72–79. doi: 10.1002/jmv.21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lothert K, et al. Development of a downstream process for the production of an inactivated whole hepatitis C virus vaccine. Sci Rep. 2020;10:16261. doi: 10.1038/s41598-020-72328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mathiesen CK, et al. Adaptive Mutations Enhance Assembly and Cell-to-Cell Transmission of a High-Titer Hepatitis C Virus Genotype 5a Core-NS2 JFH1-Based Recombinant. J Virol. 2015;89:7758–7775. doi: 10.1128/JVI.00039-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sølund C, et al. Direct acting antiviral treatment of chronic hepatitis C in Denmark: factors associated with and barriers to treatment initiation. Scand J Gastroenterol. 2018;53:849–856. doi: 10.1080/00365521.2018.1467963. [DOI] [PubMed] [Google Scholar]
- 69.Weis N, Thomsen RW. The Danish Database for Hepatitis B and C. Ugeskr Laeger. 2012;174:2521. [PubMed] [Google Scholar]
- 70.Ogawa T, et al. Molecular characterization of organelle-type Nudix hydrolases in Arabidopsis. Plant Physiol. 2008;148:1412–1424. doi: 10.1104/pp.108.128413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh Y, Bird JG. A gel electrophoresis-based assay for measuring enzymatic RNA decapping activity. Methods in enzymology. 2022;675:323–350. doi: 10.1016/bs.mie.2022.07.014. [DOI] [PubMed] [Google Scholar]
- 72.Poulsen LD, Vinther J. RNA-Seq for Bacterial Gene Expression. Curr Protoc Nucleic Acid Chem. 2018;73:e55. doi: 10.1002/cpnc.55. [DOI] [PubMed] [Google Scholar]
- 73.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011;17:3. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 74.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 75.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–U354. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 77.Danecek P, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Core_Team., R. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2020. R version 4.0.2. [Google Scholar]
- 79.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2016. [Google Scholar]
- 80.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saeed M, et al. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob Agents Chemother. 2012;56:5365–5373. doi: 10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coleman TM, Wang G, Huang F. Superior 5’ homogeneity of RNA from ATP-initiated transcription under the T7 phi 2.5 promoter. Nucleic acids research. 2004;32:e14. doi: 10.1093/nar/gnh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trivedi S, et al. Viral persistence, liver disease, and host response in a hepatitis C-like virus rat model. Hepatology. 2018;68:435–448. doi: 10.1002/hep.29494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ward JC, et al. Insights into the unique characteristics of hepatitis C virus genotype 3 revealed by development of a robust sub-genomic DBN3a replicon. J Gen Virol. 2020;101:1182–1190. doi: 10.1099/jgv.0.001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Voitenleitner C, Bechtel J, Arfsten A, Hamatake R. Hepatitis C genotype 1a replicon improved through introduction of fitness mutations. Biotechniques. 2012;52:273–275. doi: 10.2144/000113841. [DOI] [PubMed] [Google Scholar]
- 86.Kinge Wose, et al. Hepatitis C virus genotype 5a subgenomic replicons for evaluation of direct-acting antiviral agents. Antimicrob Agents Chemother. 2014;58:5386–5394. doi: 10.1128/AAC.03534-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J Virol. 2003;77:3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gottwein JM, et al. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J Virol. 2010;84:5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bick MJ, et al. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grosdidier A, Zoete V, Michielin O. Fast docking using the CHARMM force field with EADock DSS. Journal of computational chemistry. 2011;32:2149–2159. doi: 10.1002/jcc.21797. [DOI] [PubMed] [Google Scholar]
- 91.Macke TJ, Case DA. Molecular Modeling of Nucleic Acids Vol 682 ACS Symposium Series; American Chemical Society; 1997. pp. 379–393. [Google Scholar]
- 92.Appleby TC, et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 93.Schrödinger L. The PyMOL Molecular Graphics System. 2022 Version 2.4.2. [Google Scholar]
- 94.Kent WJ, et al. The human genome browser at UCSC. Genome research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data and the count files obtained for the different experiments are available in NCBI GEO with the accession GSE180956 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE180956). Fasta files used for mapping is available at: https://doi.org/10.17894/ucph.c8675313-45c6-4a90-b4ab-c65abffd3f38. Data used for plotting of the main figures 1, 2 and 4 are available as Source Data 1-3.
Code availability
Analysis of the CapZyme-seq data is based on existing tools and is described in detail at https://github.com/jeppevinther/CapZyme. Additional information about the code used for the analyses described in this paper is available from the corresponding author (JV) upon request.
Analysis of the CapZyme-seq data is based on existing tools and is described in detail at https://github.com/jeppevinther/CapZyme. Additional information about the code used for the analyses described in this paper is available from the corresponding author (JV) upon request.