Abstract
Plant genome sequences catalogue genes and the genetic elements that regulate their expression. Such inventories further research aims as diverse as mapping the molecular basis of trait diversity in domesticated plants or inquiries into the origin of evolutionary innovations in flowering plants millions of years ago. The transformative technological progress of DNA sequencing in the past two decades has enabled researchers to sequence ever more genomes with greater ease. Pangenomes — complete sequences of multiple individuals of a species or higher taxonomic unit — have now entered the geneticists’ toolkit. The genomes of crop plants and their wild relatives are being studied with translational applications in breeding in mind. But pangenomes are applicable also in ecological and evolutionary studies, as they help classify and monitor biodiversity across the tree of life, deepen our understanding of how plant species diverged, and show how plants adapt to changing environments or new selection pressures exerted by human beings.
Introduction
Genomes comprise the entirety of genic and non-genic sequences of an organism. Comparisons between the genome sequences of different individuals of the same species have revealed a high extent of intraspecies variation, which ranges from single nucleotide changes and small insertions or deletions (indels) to large-scale structural variation; some individuals lack entire genes that are present in others, or the linear order of genetic elements can differ between members of the same species1. By creating genome sequence assemblies for a representative set of individuals from a species of interest, researchers can catalogue and characterize the genetic diversity within and between species; decipher the role of structural changes in evolutionary processes, such as speciation, adaptation, domestication or polyploidization; and investigate the genetic basis of phenotypic variation.
Pangenomics (pan, from the Greek word meaning whole) seeks to capture the full spectrum of genetic variation within a species through the assembly and comparative analysis of genome sequences from multiple individuals. In bacterial genomics, where the term originated, the pangenome is often defined as the full complement of genes present in the members of a species2, with the core genome consisting of genes that are present in all or nearly all individuals and the accessory genome including genes that are variably present among different individuals or strains. In eukaryotes such as animals and plants, allowance must be made for non-coding sequences, most of which are derived from repetitive elements. Although ‘pangenome’ in an all-encompassing sense is defined as the full genomic content of a species, terms such as “pangenome reference”3 or simply “pangenome”4 often denote a collection of genome sequences belonging to one species or higher taxonomic group and the computational and logistical infrastructure5 appertaining to it (Figure 1).
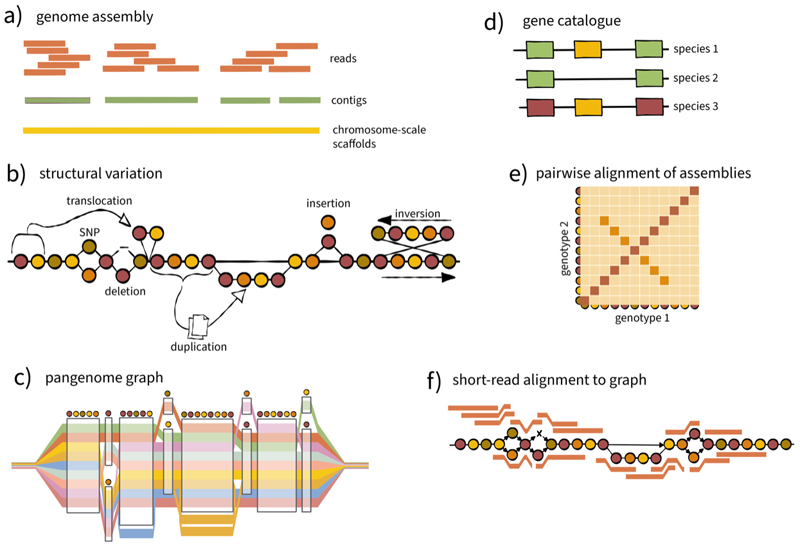
Figure 1. Pangenomics: assembly and comparison of genome sequences.
(a) Sequence reads, these days mostly long (>15 kb) reads, are assembled into contigs, which are arranged into chromosome-scale scaffolds (pseudomolecules) with the help of genetic and physical linkage information (dashed lines). (b) Comparisons of sequence assemblies reveal the full spectrum of sequence variation in the assembled genomes. (c) Pangenome graphs are computational representations of the assemblies and the differences between them. In this example, colour bands represent genomes as paths through the pangenome graph. Graphs with single base pair resolution are still challenging to construct at the whole-genome level. (d) A gene-centric view reduces complexity as do (e) pairwise alignments of genome sequences. (f) Short-read data (red bars), which is used for population-scale resequencing, can be integrated with pangenomes, for example, by aligning them to pangenome graphs.
Genome sequence assemblies are central to pangenomics. The first genome sequence assemblies were ‘drafts’, split into thousands of sequence contigs and often without a linear order6. As genome sequencing used to be a costly undertaking, its applications long relied on a single ‘high-quality’ genome sequence per species that was at chromosome scale, contiguous and often constructed with much effort. Many more genomes were only re-sequenced with comparatively cheap short-read sequencing technologies, with short reads then aligned to one or more reference genomes for variant detection7–9. But short reads cannot accurately represent repetitive sequences, which abound especially in plant genomes, and fail to resolve balanced structural variants (SVs), such as inversions or inter-chromosomal translocations10–12 (Figure 1). Advances in DNA sequencing technologies, in particular the development of long-read sequencing, has made genome assembly easier and faster and its products more contiguous and complete13,14,15,16. Gap-free sequences have been generated for the chromosomes of several eukaryotes, from plants11 to humans17,18. Piecing together such telomere-to-telomere assemblies remains challenging, owing in large part to the presence of homogeneous repeats19,20. But the routinely applicable techniques that underpin primary contig assembly and subsequent scaffolding yield in silico representations of chromosomes that, despite their occasional lack of completeness or positional assignment, are sufficiently informative to extract most biological information21.
In agriculture, pangenomes of crop species promise to advance crop improvement22–24 by identifying genetic variation underlying the expression of desirable genetic traits, which in turn might help breed improved crop varieties. Over the past decade, crop pangenomes have been constructed for cereals, legumes, vegetables, fruit trees and tuber-bearing crops (Table 1). Although a success story going full circle from the identification of a SV to creating a new crop variety has yet to emerge, crop pangenomes have shown the impact of structural variation on crop evolution. Recently, the application of pangenomes has moved beyond domesticated plants to their wild progenitors and more distant relatives, highlighting their utility for ecological and evolutionary studies. Pangenomes are on their way to replace short-read reference genomes as the preferred inventories of sequence variation, with tree-of-life projects, such as the Earth BioGenome Project, the Darwin Tree of Life project and the 10,000 Plant Genome Project (10KP), aiming to sequence and compare as many genomes as possible to help capture and preserve biodiversity.
Table 1. Plant pangenome studies.
| Species | Common name | Genome size (Mbp) | n | Sequencing technology | Year | Ref |
|---|---|---|---|---|---|---|
| Glycine soja | Wild soybean | 1,000 | 7 | Illumina | 2014 | 28 |
| Medicago truncatula | Barrel medic | 400 | 15 | Illumina | 2017 | 183 |
| Brachypodium distachyon | Purple false brome | 250 | 54 | Illumina | 2017 | 184 |
| Oryza spp. | Rice wild relatives | 400-500 | 13 | Illumina | 2018 | 82 |
| Oryza sativa and O. rufipogon | Asian and common wild rice | 400 | 66 | Illumina | 2018 | 185 |
| Solanum lycopersicum | Tomato | 950 | 72 5 | Illumina | 2019 | 4 |
| Solanum lycopersicum | Tomato | 950 | 10 0 | Oxford Nanopore | 2020 | 152 |
| Oryza sativa | Asian rice | 400 | 12 | PacBio | 2020 | 186 |
| Brassica napus | Rapeseed | 1,100 | 8 | PacBio | 2020 | 187 |
| Hordeum vulgare | Barley | 5,000 | 20 | Illumina | 2020 | 40 |
| Glycine max and G. soja | Soybean | 1,100 | 29 | PacBio | 2020 | 188 |
| Arabidopsis thaliana | Thale cress | 135 | 8 | PacBio | 2020 | 189 |
| Triticum aestivum | Bread wheat | 15,000 | 15 | Illumina | 2020 | 39 |
| Oryza sativa | Rice | 400 | 33 | PacBio | 2021 | 190 |
| Zea mays | Maize | 2,100 | 26 | PacBio | 2021 | 49 |
| Sorghum bicolor | Sorghum | 800 | 16 | PacBio | 2021 | 191 |
| Raphanus spp. | Radish | 500 | 11 | PacBio | 2021 | 192 |
| Cucumis sativus | Cucumber | 350 | 12 | PacBio | 2022 | 193 |
| Solanum lycopersicum | Tomato | 950 | 32 | PacBio (HiFi) | 2022 | 60 |
| Solanum spp. | Potato wild relatives | 800 | 44 | PacBio (HiFi) | 2022 | 83 |
| Solanum tuberosum | Potato | 800 | 6 | Illumina Oxford Nanopore | 2022 | 194 |
| Glycine spp. | Soybean wild relatives | 1,100 | 26 | PacBio | 2022 | 195 |
| Gossypium spp. | Cotton | 750–2,500 | 7 | Oxford Nanopore | 2022 | 196 |
| Vigna unguiculata | Cowpea | 650 | 7 | PacBio | 2023 | 197 |
| Arabidopsis thaliana | Thale cress | 135 | 38 | PacBio (HiFi) | 2022 | 198 |
| Pennisetum glaucum | Pearl millet | 1,700 | 10 | PacBio (HiFi) | 2023 | 199 |
| Zea mays | Maize | 2,100 | 26 | PacBio | 2023 | 200 |
| Citrus spp. | Orange | 217-419 | 12 | PacBio Oxford Nanopore | 2023 | 201 |
| Solanum spp. | Tomato and relatives | 770-1,200 | 13 | PacBio | 2023 | 43 |
| Setaria italica | Foxtail millet | 430 | 11 0 | PacBio | 2023 | 202 |
| Capsicum spp. | Chili peppers | 3000–4,100 | 11 | PacBio (HiFi) | 2023 | 203 |
| Arabidopsis thaliana | Thale cress | 135 | 72 | PacBio (HiFi) Oxford Nanopore | 2023 | 204 |
In this Review, we take stock of the progress of plant pangenomics over the past decade. We first provide an overview of the application of pangenomes to crop plants, before discussing the role pangenomes can play in the conservation of biodiversity and how they further evolutionary and biodiversity research. Finally, we outline future developments for the field.
Applications of pangenomes in crop plants
The publication of the first genome sequence of a crop, that of rice in 200525, ushered in a step-change in the speed of genetic research in that species26, largely owing to easier and faster gene mapping27. As we move beyond single reference genomes, complete genome sequences for multiple individuals are used to catalogue sequence diversity in crop plants. One of the first reports that can lay claim to being a plant pangenome paper was published in 201428. In that study, Li et al. assembled draft genome sequences of seven accessions of Glycine soja, the wild progenitor of domesticated soy bean. Plant genetic resources29, which comprise traditional landraces and wild progenitors, are potential sources of beneficial genes and alleles, for example, those conferring resistance against disease, that are absent from modern elite varieties. Pangenomes can assist in realizing this potential by more effectively linking sequence variation to phenotypes deployed in breeding programmes. In this section, we describe three applications of pangenomes in crop plant genetics: mapping or selecting for beneficial alleles; the generation of inventories of resistance genes (also known as R genes); and the study of crop-wild relatives (Figure 2).
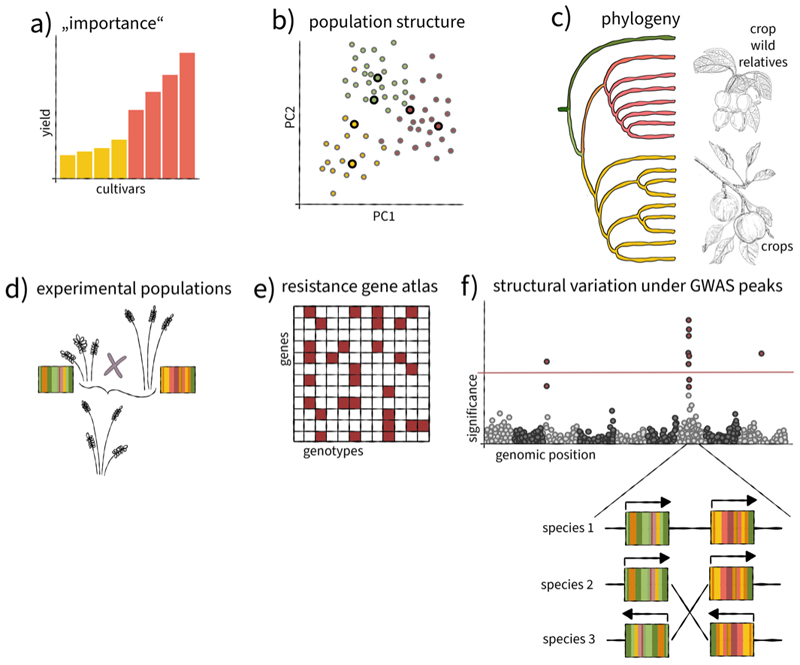
Figure 2. Pangenomics in crop plants.
Most pangenome studies to date have focused on crops. The varieties under investigation are selected based on different criteria.(a) Cultivars of great ‘importance’ include those that are widely grown or used in genetic research. (b) Surveys of population structure enable the selection of core sets that represent with a limited number of samples genetic diversity in a given crop as best as possible. The diversity space of species is often represented in principal component analysis (PCA). Population structure is reflected in clusters (shown in different colours) that correspond to geographic origins or infraspecific taxonomy. (c) Crop-wild relatives (wild progenitors and more distant relatives) are studied because they broaden allelic diversity in cultivated varieties. (d) Pangenomes have diverse applications in crop genetics. Genome sequences of the parents of experimental population assist in mapping traits to single genetic factors (coloured bar). (d). (e) Catalogues of resistance genes enrich the toolkit of plant pathology and may be represented in matrices that record the presence (blue square) or absence (grey square) of genes in the sequenced individuals. (f) Thanks to genome sequences, geneticists can include structural variants in their search for causal polymorphisms under GWAS peaks.
Genetic mapping and selection of variants associated with desired traits
Genetic mapping refers to the process of identifying and understanding the genetic basis of specific traits within a population, often with the aim of improving those traits through selective breeding. In crop genetics, mapping employs molecular markers — predominantly based on single-nucleotide polymorphism (SNPs) or indels — to establish causal relationships between discrete genetic elements and variation in breeding-relevant phenotypes such as seed traits, yield or disease resistance30,31. Molecular markers targeting SNPs and indels are the most amenable to rapid genotyping and hence effective at delineating genomic regions of interest in experimental or natural populations. But the search for candidate genes needs to take into account SVs, which have also been associated with phenotypes relevant to breeding progress. For example, spring wheats harbour whole-gene deletions at the VERNALIZATION2 locus, which means that, in contrast to winter wheats, they do not require prolonged exposure to low temperatures to flower32 and can thus be planted in spring. Another example is the locus Mla of barley, which confers resistance to the fungal disease powdery mildew and is a hotspot of copy-number variation33. As a third example, a 13-Mb inversion in the maize genome that originated in a wild relative has a possible role in environmental adaptation, although functional studies are hampered by the fact that it is inherited as a single haplotype block34,35. These three examples imply that SVs are not only associated with agronomic traits but may exert a direct influence on such traits.
Genome sequences are crucial in identifying and characterizing structural variation. For example, genome sequences of resistant donor varieties of wheat and barley have facilitated mapping of resistance genes36–38, and pangenomes of tomato have shed light on the role of regulatory variation in this crop (Box 1). However, to date, whole-genome assembly remains a costly undertaking, which is why crop pangenome projects must balance different interests when genotypes are selected for sequencing. Researchers may choose to focus on either an ‘important’ crop variety, that is, a genotype that many farmers grow or researchers work with39; a diversity panel that aims, with a ‘core set’ of predetermined size, to represent as much of the genetic diversity of a specific crop as feasible40,41; or crop-wild relatives, which are used as a means to broaden genetic diversity but often still lack sequence assemblies42,43 (Figure 2). As more genomes are assembled, crop pangenomes might in the future turn into ‘haplotype catalogues’, in which researchers can look up the genome sequence of their variety of interest5. When choosing which crop varieties to work with, one might select genotypes that are amenable to genetic transformation44–47, those of parents of experimental populations38,48,49 or genotypes that carry beneficial genes or haplotypes of interest50. Maize serves as a good example, given that genome sequences for many maize varieties have been published in the past 5 years, each with different aims. A pangenome of 25 maize lines represents global diversity49, whereas other studies have reported on single varieties: a parent of a widely used mutant population51; a parent of a mapping population18,52; or a tropical maize line that proved helpful in the mapping of yield-related traits53.
Box 1. Pangenomics in tomato and potato.
Tomato and potato, two species in the genus Solanum, are the world’s most important vegetable and tuber crop, respectively205. Because of their economic importance and – at least in tomato – tractable genetics, the genomes of both species have been studied intensely. Three tomato pangenomes4,60,152 and a super-pangenome43 of the crop and its wild relatives have been published to date. Thanks to the depth and breadth of genetic research in tomato, links between classic mutants and structural variants (SVs) became evident as soon as pangenomes were available. Li et al.43 sequenced 13 genomes of cultivated tomato and its wild relatives and compiled a catalogue of structural variants. One of these was a 244-bp deletion in the cytochrome P450 gene Sgal12g015720 that was found in all cultivated tomatoes but only in 22% of wild forms (see the figure, left panel). Transgenic overexpression of the gene had higher yield in a laboratory setting (see the figure, right panel), making the gene a potential target for future breeding efforts. A variant in the sequence inventory of Gao et al.4 was a substitution in the promotor of a gene involved in the synthesis of flavour compounds. Alonge et al.152 observed that SVs have a widespread impact on gene expression and dissected the haplotype structure at loci affecting the weight and aroma of fruits. Zhou et al.60 genotyped SVs with the help of a pangenome graph to improve genome-wide association mapping. A recurring theme is the link between structural variation and gene expression, which in turn modulates gene regulatory networks. A systematic inquiry of gene-by-gene through induced regulatory and genic variation in pairs of putatively interacting gene has been proposed as a strategy to investigate dosage-dependent regulatory interactions in crops155.
The potato haploid genome size is similar to that of tomato, but genetics and genomics approaches in the crop are more difficult to implement because of autotetraploidy and clonal propagation. Haplotype-resolved genome assembly became feasible only with accurate long reads206,207. An ambitious research programme aims to turn potato into an inbred seed crop to speed up genetic gains and set up hybrid breeding208,209. Wild relatives play a crucial role in that effort. Several Solanum species are sources of self-compatibility. Single genetic factors involved in the transition from outcrossing to selfing have been isolated210,211. As of now, one pangenome study of tetraploid cultivars and two on diploid relatives have been published. Hoopes et al.194 established the technical feasibility of pangenomics in autopolyploids and studied gene expression in an allele-specific manner. Tang et al. compared the resistance gene repertoire of wild potato and identified a tuber identity gene83. Wu et al.42 focused on deleterious variants, which are a barrier to an inbred potato: harmful variants are masked by functional alleles at the same locus in heterozygous genomes but may be lethal in inbred lines. Genome-assisted selection can rid the genome of individual loci, but the mutational load across the entire genome requires a phylogenomic approach, as chosen by Wu et al.42. They assembled the genomes of 87 Solanaceous species and 5 outgroup taxa and inferred from multiple sequence alignments evolutionarily constrained sites that do not tolerate amino acid exchanges in wild relatives but are affected by putatively deleterious variants in the crop. When included in genomic prediction models, the prediction accuracy for yield grew by an astonishing 25%42.
ToC blurb
Plant pangenomes have had a transformative impact on crop enhancement, biodiversity conservation and evolutionary research. This Review delves into the application of pangenomes for understanding trait diversity, aiding breeding, biodiversity classification and monitoring, and illuminating evolutionary innovations.
Inventories of SVs are useful beyond genetic mapping. Genomic selection is a breeding technique that predicts phenotypes from genome-wide marker profiles54. Rather than linking any single genetic variant to phenotypic variation, the statistical models underlying genomic selection are premised on evolutionary models that posit that quantitative traits, such as yield or yield components, are controlled by many loci of small effect55,56. Even when only a few thousand markers are used, the accuracy of genomic selection matches that of phenotypic selection, that is, the evaluation and selection of individuals based on observable traits57. Still, the inclusion of SVs can improve prediction accuracy. Linked SNPs have proved to be but incomplete proxies of SVs58. Models that take into account pangenome data are better at imputing sparse genotyping data59 and predicting phenotypes60 (Box 1).
Disease resistance gene atlases
Resistance breeding involves the development of crops that are more resilient to factors that can limit their productivity, such as diseases, pests or environmental stresses. Various methods are used for this approach: crossbreeding different plant varieties relies on natural variation to integrate resistance traits; marker-assisted selection uses genetic markers to efficiently identify desired traits; and genetic engineering directly modifies or inserts genes into a plant genome to confer resistance. Developing crops with inherent resistance reduces the reliance on chemical pesticides and fungicides, thereby promoting more sustainable agricultural practices and food security, as resistant crops are more likely to maintain their yield potential in adverse conditions. However, many plant pathogens, including viruses, bacteria and fungi, evolve rapidly due to large populations and short generation times. Resistance conferred through genes introduced by breeders is often overcome in a matter of years by newly evolved pathogen strains61. Complete knowledge of the resistance gene repertoire of a crop can help breeders find novel sources of resistance and combine them to achieve durable resistance. 62–64 For example, a resistance gene atlas has been proposed in wheat62, an effort that entails, among other things, assembling the sequences of all wheat resistance genes50.
Before whole-genome assembly became economical for a large number of samples, researchers focussed on one class of resistance genes. For example, the nucleotide-binding leucine-rich repeat (NLR) genes are a multifarious class of resistance genes that guard vast swathes of the green kingdom against a host of pathogens65 and, for that reason, are especially well-researched66. Thanks to conserved gene structure and sequencing, capture approaches using oligonucleotide baits as were employed in wheat are a cost-effective means of sequencing many NLR genes at once. Sequence-wide inventories of NLR genes have been compiled for the model plant Arabidopsis thaliana67, tomato and wheat wild relatives68. In future, targeted resistance genes enrichment sequencing may be eschewed in favour of whole-genome sequencing. An ‘NLRome’, that is, a pangenome limited to a single gene family, is defined by more than mere sequence content. Resistance genes are found in clusters that evolve rapidly owing to frequent unequal crossing over69. What matters beyond the mere presence or absence of genes is the number and arrangement in a given genome of often virtually identical copies of NLR genes33. The structural and functional annotation of R gene homologues is an active research field70, and the availability of more reference-quality genome annotations will help to annotate newly assembled genomes. As computational methods improve, including for the prediction of protein structures such as AlphaFold2 (ref. 71), it may become possible to model molecular interactions and design targeted interventions to respond to rapidly evolving pathogens72. Pangenomes may also tell us about the evolutionary origins and patterns of structural variation also in other types of resistance genes73 and similarly complex loci where duplication is common, such as metabolic gene clusters74 or storage proteins75.
Crop-wild relatives and ‘super-pangenomes’
Crosses between crops and their wild relatives are sources of variation that breeders are keen to exploit29,76. Crop-wild introgressions harbour wild-derived (or ‘alien’) chromatin in an otherwise elite background and have been successfully deployed in many crops, among them wheat77 and tomato78. A classification scheme by Harlan and de Wet79 divides the wild relatives of a given crop into primary, secondary and tertiary tiers, or ‘gene pools’, according to how easily they can be crossed with the cultivated form. This gene pool hierarchy affords a natural order — from most to least amenable to inter-specific crosses — in which crop wild relatives may be prioritized for genome sequencing. Extending pangenomics to higher taxonomic levels presents few conceptual hurdles, even though an entire taxonomic group is studied rather than one species. Analysis methods may differ according to whether sequence variants are fixed between reproductively isolated species or segregate in mutually interfertile populations connected by gene flow80. Moreover, owing to the rapid turnover of repetitive sequences, sequence alignments are often confined only to genes and conserved regulatory elements. But the bare sequence assembly of the genomes of ten wheat wild relatives is not much harder than a similar feat with ten wheat cultivars.
Hence, crop wild relatives have been first among the targets of “super-pangenomics”81, a moniker given to the comparative analysis of genome sequences at taxonomic levels above the species. Genome sequence assemblies of 13 wild relatives of rice in the genus Oryza have shed light on the evolutionary dynamics of genes and repetitive elements in that taxon82. By scouring variation in 46 genomes of potato and its wild relatives, one study identified a gene involved in the development of tubers, storage organs that have made Solanum species targets for domestication83 (Box 1). As genomes of ever more crop wild relatives84–86 are being reported, inquiries into the evolutionary origins of crops and translational applications in introgression breeding will benefit.
The role of pangenomes in biodiversity research
The degradation of ecosystems and global warming threaten species richness87,88 and ecosystem services, such as freshwater availability, temperature regulation and carbon sequestration89. Pangenomics, including the sequencing of purposefully chosen genomes90,91, can help to counteract the environmental fallout of human economic activity, including agriculture92, by helping ecologists monitor and mitigate biodiversity losses93,94, thereby supporting conservation efforts, sustainable agriculture and ecosystem management.
Digitizing living libraries
Conservation genomics is the application of genetic sequencing to understand, catalogue and safeguard biodiversity. Such biodiversity may be sampled in situ, from herbaria95 or genebanks of cultivated and wild plants29. Examples of ‘plant genetic resources’, as they are termed by conservationists and breeders, are seeds stored in genebanks around the world and wild plants in the Amazon rainforest awaiting botanists’ collection missions. Genebanks are structured collections of plant materials (‘germplasm’) associated with searchable and curated data records (so-called ‘accessions’). As such, genebanks lend themselves well to systematic sequencing96. A non-exhaustive list of crop species that have been the focus of genebank genomics include rice97, maize98, wheat99, barley100 and chili peppers101. These activities yield “molecular passport records”96 that provide information about the structure and representativeness of collections and can help monitor seed identity in the future. DNA sequences can be complemented with other types of data, such as historic field observations and molecular phenotypes, including seed metabolites, transcript abundances or epigenetic profiles, to serve as ‘digital twins’ of genebank accessions. These surveys of genetic diversity, stored in easily accessible biorepositories, support informed choices on which germplasm to sequence in pangenomics projects. In contrast to short-read sequencing, long-read sequencing does not yet scale to thousands of samples stored in genebanks. Moreover, the genomes of some plant species, for example outcrossing and polyploid taxa, remain difficult to assemble with the latest technologies and can require expensive supporting evidence, such as genetic maps102,103, which are more time-consuming to construct than sequence assemblies. Hence, a judicious mix of short-read and long-read sequencing at appropriate depths of coverage is needed to maximize knowledge gain per unit of currency in genebank genomics efforts.
A tiered strategy for pangenomics
The method of choice for genebank genomics is reduced representation sequencing, also known as genotyping-by-sequencing (GBS) or restriction site associated DNA markers sequencing (RADseq)8. In contrast to other marker systems, such as SNP arrays, sequencing does not require prior knowledge on the patterns of diversity104 in a species or larger group of taxa and works reasonably well in the absence of even reference genome sequences105. Tens of thousands of accessions can thus be genotyped. The high levels of duplication in genebanks100,106 mean indiscriminate whole-genome sequencing will become cost-effective only if, and when, future drops in sequencing costs obliterate the gap between reduced representation and whole-genome sequencing. Those employing sequence-based genotyping often restrict their attention to SNPs, and for a good reason. When no alternatives were around, short-reads were used to discover and genotype SVs7. Of late, we have come to realize the extent to which anything but the most accurate of long reads compromises our ability fully to grasp the spectrum of structural variation12,107. Conclusions drawn from short-read data may have been premature. Chromosome-scale sequences are now being assembled on the scale of dozens to hundreds of individuals per species. The resultant catalogues of variants, running the gamut from SNPs, short indels, to genic copy number variants to inversions or translocations of large chunks of chromatin, underpin the genotyping of SVs in short-read data of a wider set of germplasm5. Allelic states of SVs can be inferred by either linked SNPs108 or k-mers, short oligonucleotide sequences whose copy numbers are indicative of those of the underlying sequence variation68,109. More sophisticated approaches employing alignments of reads to genome graphs to call variants are being developed3,9,107. A design of a pangenomics projects that strikes a reasonable balance between sequencing depth and broad taxon sampling can be visualized as a pyramid at whose tip sit genome sequences of a select few and whose base is short-read genotyping of many accessions (Figure 3). It is hoped that, thanks to technological progress, long-read sequencing will percolate all the way to the foundation10.
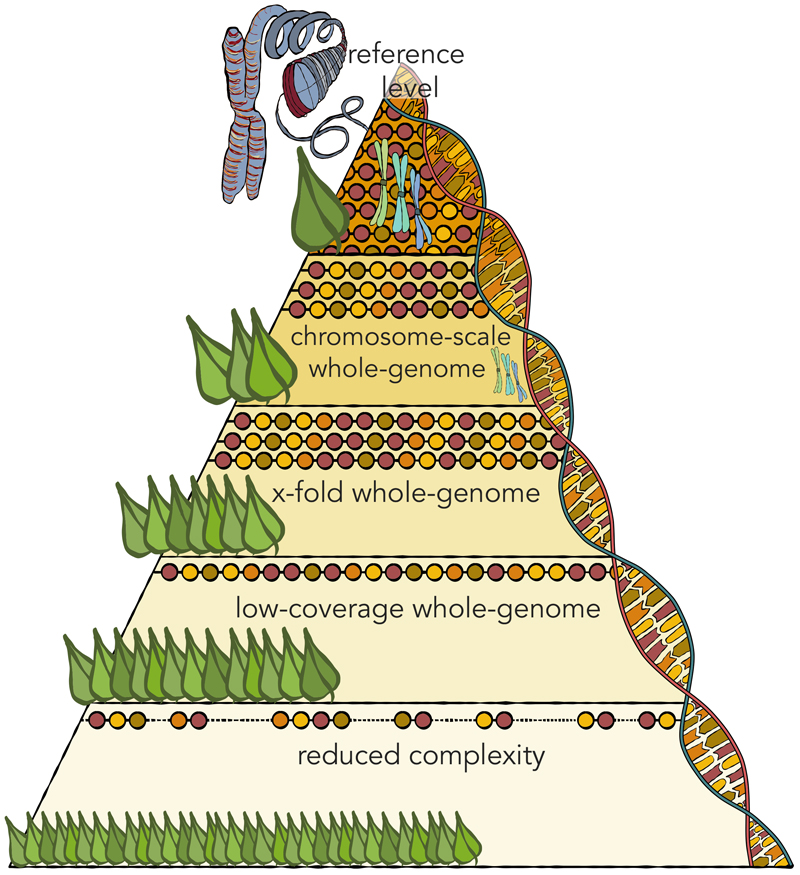
Figure 3. A tiered strategy for pangenomics.
Different sequence strategies (level of the pyramid) are suitable for different panel size (represented by leaf numbers). Reduced representation sequencing is done on as many genotypes, sampled in situ or from genebank collection, as possible. Representative coresets, sequenced to ever greater depth, are selected for different applications. Low-coverage (1- to 5-fold coverage) short-read whole genome sequencing aided by imputation is useful for genome-wide association scans and for genotyping known SVs. High-coverage (> 10-fold for inbred, > 30-fold for heterozygous genomes) short-read sequencing underpins selection scans, haplotype definition and demographic analyses. Genome assemblies based on long-read sequencing and chromosome-scale mapping catalogue the full spectrum of structural variation. Potentially extraordinary effort will be expended on a small number of genotypes to close gaps in difficult-to-assemble regions such as long tandem repeat arrays and centromeres to obtain telomere-to-telomere (T2T) assemblies. As technology progresses, the pyramid may turn into a cube and long-read sequencing may be employed in the bottom layers as well.
Pangenomes in evolutionary research
Full genome sequences have enabled phylogenomics or “big-data phylogenetics”110 where high-throughput sequencing data and increasingly whole-genome sequences are used to construct and refine phylogenetic trees. Similar to their role in crop plant research, genome sequences are useful tools for evolutionary biologists as they help map discrete genetic factors that underlie evolutionary innovations or are driving speciation.
Pangenomes in taxonomy and phylogenetics
Taxonomists and phylogeneticists name species and represent their evolutionary relationships in phylogenetic trees. But drawing boundaries between species can be difficult. Full genome sequences of entire taxonomic groups can improve the robustness of phylogenetic inference by obtaining consensus trees across many genes111, or help explain, when there is no consensus, discrepancies that arise, for example, from hybridization or incomplete lineage sorting (the persistence of segregating variants inherited from a common ancestor)112. Tree-of-life genome projects aim to sequence all forms of life. This ambitious goal requires taxonomic or geographic circumscription to achieve logistical viability in the short term. For example, the Earth Biogenome Project limits itself to eukaryotes113. Nested therein, the Darwin Tree of Life Project114 focusses on the British Isles. Other geographically circumscribed efforts target other regions of the world, for example, Europe115 or California116. The 1000 Plants Initiative reported assemblies of the vegetative transcriptomes of 1,124 plants species sensu latu, including green plants (Chloroplastida), glaucophytes and red algae117. These data illustrate and confirm hallmarks of land plant evolution, such as repeated whole-genome duplications and expansions of gene families. Even with abundant gene sequences and broad taxon sampling, some discordant phylogenies remain unresolved, possibly because of rapid speciation millions of years ago117. The successor to the 1000 Plants Initiative is the 10KP (10,000 Plants) Genome Sequencing Project, whose aim is to sequence representative genomes of embryophytes and green algae118. Complementary to these taxonomically comprehensive efforts, reduced representation sequencing has resolved several more recent branches of the plant tree of life, such as Hordeum119, Triticum120 and Crocus121. It is hoped that pangenomes will serve the same purpose in the future. Pangenomic studies will aid in defining species boundaries and will be pivotal in assessing the diversity and relatedness of different populations and subspecies.
Revisions to the tree of life springing from genomics may not only move about nodes and redraw edges, but also question the very nature of the tree; that is, the tree of life would be more appropriately named the graph of life, for some of its nodes have more than one parent122. Horizontal gene transfer set off the evolution of eukaryotes123 and, as genome sequences have revealed, happened repeatedly in the evolution of land plants84,124,125. Un-tree-like structures may also arise from polyploidization — the coalescence of two parental species’ genomes in one nucleus — or homoploid hybrid speciation, whereby diverged, but not yet fully reproductively isolated parents mate and their hybrid offspring evolve into a thriving species in their own right. Polyploid plants are common, both on our plates126 and in our laboratories127. How frequent is homoploid speciation and how can it be reconciled with the homogenizing effects of gene flow counteracting speciation is an open question128. Genomics has helped answer research questions on polyploidy using RFLP markers129 and has served that purpose ever since130. Of later provenance are inferences, from genome assemblies and resequencing data, for example, about the homoploid hybrid origin of mind shade131 and chestnut trees132. Genome sequencing can help map barriers to gene flow and thus illuminate the mechanisms of incipient speciation. Research is underway on the importance (or lack thereof) of islands of speciation133. These discrete genomic regions of elevated differentiation between taxa may be related to reproductive isolation. Alternatively, such patterns may arise from processes other than population divergence such as linked selection or heterogeneous recombination landscapes134. The latter pattern may stem from inversions, SVs that flip around large chunks of chromatin and have been for decades known to impede crossovers. Catalogues of polymorphic inversions are a by-product of pangenomes40, and such inventories have helped dissect the role of these rearrangements in species such as barley40 and sunflower135. In summary, the information on the full complement of genes and their arrangements in different species afforded by pangenomes expands our ability to resolve phylogenetic trees and to understand sources of discrepancy arising in tress as a result of gene flow between closely related species.
Pangenomes reveal evolutionary innovations
The same genetic methods136 that have been pioneered and are routinely used by crop scientists — mapping of quantitative trait loci, genome-wide association scans population genomic selection scans — are steadfastly entering the evolutionary biologist’s toolkit137,138 (Figure 4). For example, common garden experiments supplemented by genome sequences have provided cues as to how switchgrass, both a promising bioenergy crop and an important component of the tallgrass prairie, is adapting to climate change139. An enticing prospect is offered by the insights afforded by the comparison of 22 mammalian genome sequences into the convergent evolution of echolocation in such animals140. Similar genomic approaches to study convergent evolution might be adopted to study evolutionarily innovative metabolite profiles in fruit and vegetable crops and their wild relatives, which may be mediated by copy number and presence/absence variation in genes involved in the biosynthesis of such molecules. For example, the sympatric speciation of bee orchids (Ophrys spp.) might be driven by mimicking the scent, through compounds such as alkene hydrocarbons141, and the shape of the pollinating insect142,143. Traditional model plants may also be useful in evolutionary and ecological research. Arabidopsis thaliana, a plant exceptionally well-adapted to the laboratory, also grows in the wild and has become a study object of ecological genomics144. Long before population-scale sequencing had been applied to other plants, 1000 Arabidopsis genomes were sequenced145.
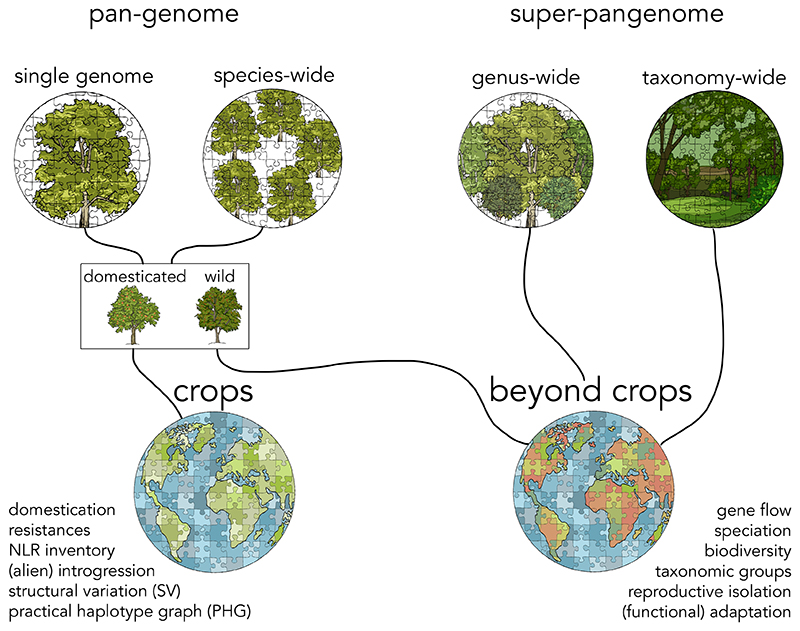
Figure 4. Pangenomics at different taxonomic levels.
Reference sequences can be assembled for the genomes of both wild and domesticated plants. Diversity panels employed in pangenome studies may span different taxonomic levels, from single species to the tree of life. The term ‘super-pangenome’ is a useful shorthand to refer to pangenomics beyond the species level. Analysis methods differ according to whether the observed genomic variants segregate in a population of interfertile individuals or represent fixed differences between reproductively isolated species. Broadly speaking, intraspecific diversity fuels genetic mapping and breeding, whereas super-pangenomes hold answers to taxonomic and evolutionary questions. At higher taxonomic levels, taxon sampling cannot but look beyond crops, as the species that farmers attend to are in a minority.
Pangenomes can help delineate adaptive changes and evolutionary processes by revealing the acquisition or loss of genes during the evolutionary history of a lineage. Beyond speciation, another evolutionary process whose study benefits from genome sequences is domestication, which results from rapid adaptation to new habitats. Owing to the selective breeding of plants (and animals) for desirable attributes, a suite of traits are commonly observed in diverse domesticated species, a phenomenon that is referred to as the domestication syndrome146–148. Arduous genetic mapping and laboursome sequencing were required to identify the insertion of a transposable element as the causal genetic variant underlying the reduced tillering observed in domesticated maize compared with its wild progenitor149,150. Arguably, genome sequences would make an analogous task easier these days. As more genomes of crops and their wild relatives are sequenced, more links between crop evolution and specific SVs will emerge. For example, research in tomato has established the role of structural variation in tomato breeding and its link to gene regulatory interactions151,152 (Box 1).
In addition to genetic factors, which are comprehensively encapsulated in pangenomes, epigenetic information must be considered to understand environmental adaptation153,154. This is part of a wider research agenda that no longer considers only the actions of genes but also their interactions155. Gene regulatory networks are influenced by and, in turn, influence the abundance of genic transcripts and various classes of non-coding RNAs, DNA methylation levels, histone modifications and chromatin accessibility156. Pan-epigenomes, that is, epigenetic profiles of several individuals of a species have been collected in cereal crops49,157. Epigenomes of long-lived organisms such as forest trees might help us understand how they cope with climate change158. Epigenetic mechanisms may underlie plasticity in growth and development, which in turn may enable plants better to respond to environmental stress factors158. Pan-epigenomics is still in its infancy, but similar to genome assembly this field has benefited from faster and cheaper whole-genome sequencing and the development of new protocols such as assay for transposase-accessible chromatin using sequencing (ATAC-seq) to map chromatin accessiblity159 and DNA affinity purification sequencing (DAP-seq) to map transcription-factor binding sites160. When applied to diverse species, these techniques are bound to lead to new insights into the molecular facets of biodiversity161.
Future perspectives
Sequencing a human genome cost US$100,000 in 2009 and US$1,000 in 2019. Another drop by three orders of magnitude would make genome sequencing no costlier than genotyping it with a set of PCR markers. Genomicists have generated widely applicable resources for breeders, evolutionary biologists and developmental geneticists. As we widen our gaze to find new uses for our now mature tools, we should be aware of what might constrain pangenomics other than the per gigabase price of sequencing.
Future methodological challenges
Challenges in logistics may overshadow those in the lab, although both aspects are interwoven. Genotyping DNA samples by their thousands is now considerably cheaper, but not much easier and faster, than it was 10 years ago. As long-read sequencing scales to large germplasm collections or the tree-of-life’s foliage162, taxon and tissue sampling163,164, isolation and quality control of high-molecular weight DNA10,165, preparation of multiplexed sequencing libraries166,167 and data management and archiving167 will become increasingly challenging. Approaches that can extract nucleic acids from myriad seeds into digital sequences in a matter of weeks are needed, as are improved analysis methods that can compare thousands of whole-genome sequences in a reference-free manner. The assembly and comparison of genome sequences is particularly challenging in plants owing to the large size and repeat-rich nature of their genomes resulting from polyploidy168. Haplotype phasing, the assignment of sequence to parental haplotypes169, is required in heterozygous and autopolyploid plants. The development of better alignment algorithms would enable the comparison of many genomes at base-level resolution170. Pangenome graphs have emerged in the last couple of years as the data structure most suited to storing and analysing multiple genome sequences171. They hold the promise of greater accuracy in various downstream applications such as variant calling, transcript abundance quantification and the resolution of structurally complex loci3. But there is an obstacle to the widespread adoption of pangenome graphs in plants: at the moment, these graphs test the limits of computational infrastructures, even if they operate on only a few dozen human-sized genomes172. Another open question is how pangenome graphs, which are now geared towards multiples genomes of a single species, can be generalized to genome sequences of entire genera, where divergence is higher and alignment rates are lower.
Pangenomes as community resources
A key aspect of reference genome and later pangenome projects has been the compilation of community resources and the provision of an ancillary infrastructure to facilitate access, such as genome browsers and repositories for bulk download173,174. With genome sequencing about to become a quotidian effort, genome assembly often occurs in the pursuit of narrowly circumscribed research projects without consideration of long-term resourcing. However, accessibility in the long term is in part what democratizing DNA sequencing175 is about. Submitting genome assemblies to public sequence archives enables later synthesis and collation of genome sequences to derive knowledge from a comparative outlook176 or simply curation to host all genome sequences of a species in one community hub177. As of now, such efforts have been few and far between and are possibly hampered by the diversity of applied sequencing strategies. As the speed of progress relents and best practices settle in, these endeavours will gain in prominence. Procedures for depositing sequence assemblies and their underlying raw data are well established by the repositories that are members of the International Nucleotide Sequence Database Collaboration178. Standards recommendation for phylogenomic sequencing initiatives, including minimum quality standards for assemblies and annotation, have been proposed by the Earth Biogenome Project179.
Access to biological diversity
Genome researchers are accustomed to unfettered access to sequence data with no other obligations than to cite research articles written by data depositors. By contrast, property rights to plant genetic material are governed by a legal framework of bewildering complexity (or so it may seem to the uninitiated). Access to plant genetic resources is governed by international covenants, among the best known of which are the Convention on Biological Diversity, the International Treaty on Plant Genetic Resources for Food and Agriculture, and the Nagoya Protocol on access and benefit sharing180. Today’s debates revolve around Digital Sequence Information (DSI), a concept that defies easy definition181. In any event, genome sequences fall well under the DSI extended purview. Many scientists wish for sequence data to remain publicly accessible, and practical solutions have been proposed to keep international sequence archives accessible while ensuring the equitable distribution of proceeds among stakeholders182.
Conclusions
Surveys of structural variation have taught us that, to understand the full extent of sequence diversity of a species, we need to compare many individual genome sequences. However, pangenomics remains in its infancy; although sequencing technologies and analysis methods are improving at a rapid pace, and most crop plants have reference genomes, few have pangenomes. Pangenomes of higher taxonomic units may become foundational community resources that help to better appreciate the role of evolutionary processes. After crop genome sequencing, wild relatives are the next frontier in agricultural genomics. A democratization of pangenomes driven by cheaper DNA sequencing and easier-to-use computational methods is underway. We eagerly await the outcomes of sequencing the tree of life.
Acknowledgements
The authors’ research activities in barley and its wild relatives are supported by grants from, respectively, the German Federal Ministry of Education and Research (BMBF, SHAPE-P3, 031B1302A to N.S. and M.M.) and the European Research Council (Starting Grant TRANSFER, action number 949873 to M.M.). MJ’s work on faba bean genomics is funded by the Leibniz Association (REPLACE, J118/2021).
Footnotes
Author contributions
The authors contributed equally to all aspects of the manuscript.
Competing interests
The authors declare no competing interests.
Related links
Earth BioGenome Project https://www.earthbiogenome.org/
Darwin Tree of Life https://www.darwintreeoflife.org/
10,000 Plant Genome Project https://db.cngb.org/10kp/
Peer review information
Nature Reviews Genetics thanks the anonymous reviewers for their contribution to the peer review of this work.
References
- 1.Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell. 2005;17:343–360. doi: 10.1105/tpc.104.025627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao W-W, et al. A draft human pangenome reference. Nature. 2023;617:312–324. doi: 10.1038/s41586-023-05896-x. [This study showcases how pangenome graphs work in a case study of a model primate] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao L, et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nature Genetics. 2019;51:1044–1051. doi: 10.1038/s41588-019-0410-2. [DOI] [PubMed] [Google Scholar]
- 5.Jayakodi M, Schreiber M, Stein N, Mascher M. Building pan-genome infrastructures for crop plants and their use in association genetics. DNA Research. 2021;28:dsaa030. doi: 10.1093/dnares/dsaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proceedings of the National Academy of Sciences. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nature Reviews Genetics. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey JW, et al. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nature Reviews Genetics. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- 9.Hickey G, et al. Genotyping structural variants in pangenome graphs using the vg toolkit. Genome Biology. 2020;21:35. doi: 10.1186/s13059-020-1941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Coster W, Weissensteiner MH, Sedlazeck FJ. Towards population-scale long-read sequencing. Nature Reviews Genetics. 2021;22:572–587. doi: 10.1038/s41576-021-00367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson ND, et al. Variant calling and benchmarking in an era of complete human genome sequences. Nature Reviews Genetics. 2023;24:464–483. doi: 10.1038/s41576-023-00590-0. [DOI] [PubMed] [Google Scholar]
- 12.Mahmoud M, et al. Utility of long-read sequencing for All of Us. bioRxiv. 2023:2023.2001.2023.525236. doi: 10.1101/2023.01.23.525236. [DOI] [Google Scholar]
- 13.Sedlazeck FJ, Lee H, Darby CA, Schatz MC. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nature Reviews Genetics. 2018;19:329–346. doi: 10.1038/s41576-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 14.Wenger AM, et al. Highly-accurate long-read sequencing improves variant detection and assembly of a human genome. bioRxiv. 2019:519025. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolmogorov M, et al. Scalable Nanopore sequencing of human genomes provides a comprehensive view of haplotype-resolved variation and methylation. Nature Methods. 2023 doi: 10.1038/s41592-023-01993-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Method of the Year 2022: long-read sequencing. Nature Methods. 2023;20:1. doi: 10.1038/s41592-022-01759-x. [DOI] [PubMed] [Google Scholar]
- 17.Miga KH, et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020;585:79–84. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, et al. A complete telomere-to-telomere assembly of the maize genome. Nature Genetics. 2023;55:1221–1231. doi: 10.1038/s41588-023-01419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabanal FA, et al. Pushing the limits of HiFi assemblies reveals centromere diversity between two Arabidopsis thaliana genomes. Nucleic Acids Research. 2022;50:12309–12327. doi: 10.1093/nar/gkac1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navrátilová P, et al. Prospects of telomere-to-telomere assembly in barley: Analysis of sequence gaps in the MorexV3 reference genome. Plant Biotechnology Journal. 2022;20:1373–1386. doi: 10.1111/pbi.13816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whibley A, Kelley JL, Narum SR. The changing face of genome assemblies: Guidance on achieving high-quality reference genomes. Molecular Ecology Resources. 2021;21:641–652. doi: 10.1111/1755-0998.13312. [DOI] [PubMed] [Google Scholar]
- 22.Tay Fernandez CG, et al. Pangenomes as a resource to accelerate breeding of under-utilised crop species. International Journal of Molecular Sciences. 2022;23:2671. doi: 10.3390/ijms23052671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman MA, He Y, Zhou M. Beyond a reference genome: pangenomes and population genomics of underutilized and orphan crops for future food and nutrition security. New Phytologist. 2022;234:1583–1597. doi: 10.1111/nph.18021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Della Coletta R, Qiu Y, Ou S, Hufford MB, Hirsch CN. How the pan-genome is changing crop genomics and improvement. Genome Biology. 2021;22:3. doi: 10.1186/s13059-020-02224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki T, International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, Shang L, Zhu Q-H, Fan L, Guo L. Twenty years of plant genome sequencing: achievements and challenges. Trends in Plant Science. 2022;27:391–401. doi: 10.1016/j.tplants.2021.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Jackson SA. Rice: The First Crop Genome. Rice. 2016;9:14. doi: 10.1186/s12284-016-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y-h, et al. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nature Biotechnology. 2014;32:1045–1052. doi: 10.1038/nbt.2979. [DOI] [PubMed] [Google Scholar]
- 29.Frankel OH. Genetic conservation: our evolutionary responsibility. Genetics. 1974;78:53–65. doi: 10.1093/genetics/78.1.53. [This classic paper set the conceptual and moral agenda for the discipline of conservation genetics] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanksley SD. Mapping polygenes. Annual review of genetics. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. [DOI] [PubMed] [Google Scholar]
- 31.Semagn K, Bjørnstad Å, Ndjiondjop M. Principles, requirements and prospects of genetic mapping in plants. African Journal of Biotechnology. 2006;5 [Google Scholar]
- 32.Yan L, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bettgenhaeuser J, et al. The barley immune receptor Mla recognizes multiple pathogens and contributes to host range dynamics. Nature Communications. 2021;12:6915. doi: 10.1038/s41467-021-27288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crow T, et al. Gene regulatory effects of a large chromosomal inversion in highland maize. PLoS Genet. 2020;16:e1009213. doi: 10.1371/journal.pgen.1009213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romero Navarro JA, et al. A study of allelic diversity underlying flowering-time adaptation in maize landraces. Nature Genetics. 2017;49:476–480. doi: 10.1038/ng.3784. [DOI] [PubMed] [Google Scholar]
- 36.Thind AK, et al. Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nat Biotechnol. 2017;35:793–796. doi: 10.1038/nbt.3877. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, et al. An unusual tandem kinase fusion protein confers leaf rust resistance in wheat. Nature Genetics. 2023;55:914–920. doi: 10.1038/s41588-023-01401-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Athiyannan N, et al. Long-read genome sequencing of bread wheat facilitates disease resistance gene cloning. Nature Genetics. 2022;54:227–231. doi: 10.1038/s41588-022-01022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walkowiak S, et al. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020;588 doi: 10.1038/s41586-020-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jayakodi M, et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature. 2020;588:284–289. doi: 10.1038/s41586-020-2947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranallo-Benavidez TR, et al. Optimized sample selection for cost-efficient long-read population sequencing. Genome Research. 2021;31:910–918. doi: 10.1101/gr.264879.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, et al. Phylogenomic discovery of deleterious mutations facilitates hybrid potato breeding. Cell. 2023;186:2313–2328.:e2315. doi: 10.1016/j.cell.2023.04.008. [DOI] [PubMed] [Google Scholar]
- 43.Li N, et al. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nat Genet. 2023;55:852–860. doi: 10.1038/s41588-023-01340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain R, et al. Genome sequence of the model rice variety KitaakeX. BMC Genomics. 2019;20:905. doi: 10.1186/s12864-019-6262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato K, et al. Chromosome-scale genome assembly of the transformation-amenable common wheat cultivar ‘Fielder’. DNA Research. 2021;28 doi: 10.1093/dnares/dsab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreiber M, et al. A Genome Assembly of the Barley ‘Transformation Reference’ Cultivar Golden Promise. G3 Genes|Genomes|Genetics. 2020;10:1823–1827. doi: 10.1534/g3.119.401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin G, et al. Chromosome-level genome assembly of a regenerable maize inbred line A188. Genome Biology. 2021;22:175. doi: 10.1186/s13059-021-02396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, et al. Long-read genome sequencing accelerated the cloning of <em>Pm69</em> by resolving the complexity of a rapidly evolving resistance gene cluster in wheat. bioRxiv. 2022:2022.2010.2014.512294. doi: 10.1016/j.xplc.2023.100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hufford MB, et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science. 2021;373:655–662. doi: 10.1126/science.abg5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kale SM, et al. A catalogue of resistance gene homologs and a chromosome-scale reference sequence support resistance gene mapping in winter wheat. Plant Biotechnology Journal. 2022;20:1730–1742. doi: 10.1111/pbi.13843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Springer NM, et al. The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat Genet. 2018;50:1282–1288. doi: 10.1038/s41588-018-0158-0. [DOI] [PubMed] [Google Scholar]
- 52.Sun S, et al. Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nature Genetics. 2018;50:1289–1295. doi: 10.1038/s41588-018-0182-0. [DOI] [PubMed] [Google Scholar]
- 53.Yang N, et al. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nature Genetics. 2019;51:1052–1059. doi: 10.1038/s41588-019-0427-6. [DOI] [PubMed] [Google Scholar]
- 54.Crossa J, et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends in Plant Science. 2017;22:961–975. doi: 10.1016/j.tplants.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 55.Bernardo R. Bandwagons I, too, have known. Theor Appl Genet. 2016;129:2323–2332. doi: 10.1007/s00122-016-2772-5. [DOI] [PubMed] [Google Scholar]
- 56.Barton NH, Etheridge AM, Véber A. The infinitesimal model: Definition, derivation, and implications. Theoretical Population Biology. 2017;118:50–73. doi: 10.1016/j.tpb.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 57.Zhang H, Yin L, Wang M, Yuan X, Liu X. Factors Affecting the Accuracy of Genomic Selection for Agricultural Economic Traits in Maize, Cattle, and Pig Populations. Front Genet. 2019;10:189. doi: 10.3389/fgene.2019.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu F, et al. High-resolution genetic mapping of maize pan-genome sequence anchors. Nature Communications. 2015;6:6914. doi: 10.1038/ncomms7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bradbury PJ, et al. The Practical Haplotype Graph, a platform for storing and using pangenomes for imputation. Bioinformatics. 2022;38:3698–3702. doi: 10.1093/bioinformatics/btac410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Y, et al. Graph pangenome captures missing heritability and empowers tomato breeding. Nature. 2022;606:527–534. doi: 10.1038/s41586-022-04808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wulff BBH, Krattinger SG. The long road to engineering durable disease resistance in wheat. Current Opinion in Biotechnology. 2022;73:270–275. doi: 10.1016/j.copbio.2021.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Hafeez AN, et al. Creation and judicious application of a wheat resistance gene atlas. Mol Plant. 2021;14:1053–1070. doi: 10.1016/j.molp.2021.05.014. [This study provides a good example of resistance gene sequencing for crop improvement] [DOI] [PubMed] [Google Scholar]
- 63.Jupe F, et al. Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 2013;76:530–544. doi: 10.1111/tpj.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dracatos PM, Lu J, Sánchez-Martín J, Wulff BBH. Resistance that stacks up: engineering rust and mildew disease control in the cereal crops wheat and barley. Plant Biotechnology Journal. doi: 10.1111/pbi.14106. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamborski J, Krasileva KV. Evolution of Plant NLRs: From Natural History to Precise Modifications. Annual Review of Plant Biology. 2020;71:355–378. doi: 10.1146/annurev-arplant-081519-035901. [DOI] [PubMed] [Google Scholar]
- 66.Kourelis J, van der Hoorn RAL. Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell. 2018;30:285–299. doi: 10.1105/tpc.17.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van de Weyer AL, et al. A Species-Wide Inventory of NLR Genes and Alleles in Arabidopsis thaliana. Cell. 2019;178:1260–1272.:e1214. doi: 10.1016/j.cell.2019.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arora S, et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat Biotechnol. 2019;37:139–143. doi: 10.1038/s41587-018-0007-9. [DOI] [PubMed] [Google Scholar]
- 69.Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 70.Steuernagel B, et al. The NLR-Annotator Tool Enables Annotation of the Intracellular Immune Receptor Repertoire1 [OPEN] Plant Physiology. 2020;183:468–482. doi: 10.1104/pp.19.01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Förderer A, et al. A wheat resistosome defines common principles of immune receptor channels. Nature. 2022;610:532–539. doi: 10.1038/s41586-022-05231-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bayer PE, et al. Variation in abundance of predicted resistance genes in the Brassica oleracea pangenome. Plant Biotechnology Journal. 2019;17:789–800. doi: 10.1111/pbi.13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fan P, et al. Evolution of a plant gene cluster in Solanaceae and emergence of metabolic diversity. eLife. 2020;9:e56717. doi: 10.7554/eLife.56717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Halstead-Nussloch G, et al. Multiple wheat genomes reveal novel gli-2 sublocus location and variation of celiac disease epitopes in duplicated α-gliadin genes. Frontiers in Plant Science. 2021;12:715985. doi: 10.3389/fpls.2021.715985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flint-Garcia S, Feldmann MJ, Dempewolf H, Morrell PL, Ross-Ibarra J. Diamonds in the not-so-rough: Wild relative diversity hidden in crop genomes. PLOS Biology. 2023;21:e3002235. doi: 10.1371/journal.pbio.3002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao L, et al. The Aegilops ventricosa 2NvS segment in bread wheat: cytology, genomics and breeding. Theoretical and Applied Genetics. 2021;134:529–542. doi: 10.1007/s00122-020-03712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Rengs WMJ, et al. A chromosome scale tomato genome built from complementary PacBio and Nanopore sequences alone reveals extensive linkage drag during breeding. The Plant Journal. 2022;110:572–588. doi: 10.1111/tpj.15690. [DOI] [PubMed] [Google Scholar]
- 79.Harlan JR, de Wet JMJ. Toward a Rational Classification of Cultivated Plants. TAXON. 1971;20:509–517. doi: 10.2307/1218252. %U https://onlinelibrary.wiley.com/doi/abs/10.2307/1218252. [A classic paper that may inform which wild plant species to prioritize for today’s pangenomics from the perspective of the agricultural geneticist] [DOI] [Google Scholar]
- 80.Kryazhimskiy S, Plotkin JB. The Population Genetics of dN/dS. PLOS Genetics. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khan AW, et al. Super-Pangenome by Integrating the Wild Side of a Species for Accelerated Crop Improvement. Trends Plant Sci. 2019 doi: 10.1016/j.tplants.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stein JC, et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nature Genetics. 2018;50:285–296. doi: 10.1038/s41588-018-0040-0. [DOI] [PubMed] [Google Scholar]
- 83.Tang D, et al. Genome evolution and diversity of wild and cultivated potatoes. Nature. 2022;606:535–541. doi: 10.1038/s41586-022-04822-x. [This study showcases the seemingly straight-forward implementation of what 3 years ago was beyond reach: genus-wide pangenomics] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang H, et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science. 2020;368 doi: 10.1126/science.aba5435. [DOI] [PubMed] [Google Scholar]
- 85.Yu G, et al. Aegilops sharonensis genome-assisted identification of stem rust resistance gene Sr62. Nature Communications. 2022;13:1607. doi: 10.1038/s41467-022-29132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang X, et al. A chromosome-scale genome assembly of Dasypyrum villosum provides insights into its application as a broad-spectrum disease resistance resource for wheat improvement. Mol Plant. 2023;16:432–451. doi: 10.1016/j.molp.2022.12.021. [DOI] [PubMed] [Google Scholar]
- 87.Niskanen AKJ, Niittynen P, Aalto J, Väre H, Luoto M. Lost at high latitudes: Arctic and endemic plants under threat as climate warms. Diversity and Distributions. 2019;25:809–821. doi: 10.1111/ddi.12889. [DOI] [Google Scholar]
- 88.Lippmann R, Babben S, Menger A, Delker C, Quint M. Development of Wild and Cultivated Plants under Global Warming Conditions. Current Biology. 2019;29:R1326–R1338. doi: 10.1016/j.cub.2019.10.016. [DOI] [PubMed] [Google Scholar]
- 89.Campbell JL, et al. Forest and Freshwater Ecosystem Responses to Climate Change and Variability at US LTER Sites. BioScience. 2022;72:851–870. doi: 10.1093/biosci/biab124. [DOI] [Google Scholar]
- 90.Formenti G, et al. The era of reference genomes in conservation genomics. Trends in Ecology & Evolution. 2022;37:197–202. doi: 10.1016/j.tree.2021.11.008. [DOI] [PubMed] [Google Scholar]
- 91.Theissinger K, et al. How genomics can help biodiversity conservation. Trends in Genetics. 2023;39:545–559. doi: 10.1016/j.tig.2023.01.005. [DOI] [PubMed] [Google Scholar]
- 92.Poore J, Nemecek T. Reducing food’s environmental impacts through producers and consumers. Science. 2018;360:987–992. doi: 10.1126/science.aaq0216. [DOI] [PubMed] [Google Scholar]
- 93.Anderson CB. Biodiversity monitoring, earth observations and the ecology of scale. Ecology Letters. 2018;21:1572–1585. doi: 10.1111/ele.13106. [DOI] [PubMed] [Google Scholar]
- 94.Cordier T, et al. Ecosystems monitoring powered by environmental genomics: A review of current strategies with an implementation roadmap. Molecular Ecology. 2021;30:2937–2958. doi: 10.1111/mec.15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bieker VC, Martin MD. Implications and future prospects for evolutionary analyses of DNA in historical herbarium collections. Botany Letters. 2018;165:409–418. doi: 10.1080/23818107.2018.1458651. [DOI] [Google Scholar]
- 96.Mascher M, et al. Genebank genomics bridges the gap between the conservation of crop diversity and plant breeding. Nat Genet. 2019;51:1076–1081. doi: 10.1038/s41588-019-0443-6. [DOI] [PubMed] [Google Scholar]
- 97.Wang W, et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Romay MC, et al. Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biology. 2013;14:R55. doi: 10.1186/gb-2013-14-6-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sansaloni C, et al. Diversity analysis of 80,000 wheat accessions reveals consequences and opportunities of selection footprints. Nature Communications. 2020;11:4572. doi: 10.1038/s41467-020-18404-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Milner SG, et al. Genebank genomics highlights the diversity of a global barley collection. Nat Genet. 2019;51:319–326. doi: 10.1038/s41588-018-0266-x. [DOI] [PubMed] [Google Scholar]
- 101.Tripodi P, et al. Global range expansion history of pepper (Capsicum spp.) revealed by over 10,000 genebank accessions. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2104315118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun H, et al. Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. bioRxiv. 2021:2021.2005.2015.444292. doi: 10.1038/s41588-022-01015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Q, et al. Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nature Genetics. 2022;54:885–896. doi: 10.1038/s41588-022-01084-1. [DOI] [PubMed] [Google Scholar]
- 104.Darrier B, et al. A Comparison of Mainstream Genotyping Platforms for the Evaluation and Use of Barley Genetic Resources. Frontiers in Plant Science. 2019;10 doi: 10.3389/fpls.2019.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Leggett RM, MacLean D. Reference-free SNP detection: dealing with the data deluge. BMC Genomics. 2014;15:S10. doi: 10.1186/1471-2164-15-S4-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van Hintum TJL, Visser DL. Duplication within and between germplasm collections. Genetic Resources and Crop Evolution. 1995;42:135–145. doi: 10.1007/BF02539517. [DOI] [Google Scholar]
- 107.Kirsche M, et al. Jasmine and Iris: population-scale structural variant comparison and analysis. Nature Methods. 2023 doi: 10.1038/s41592-022-01753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hehir-Kwa JY, et al. A high-quality human reference panel reveals the complexity and distribution of genomic structural variants. Nat Commun. 2016;7:12989. doi: 10.1038/ncomms12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Voichek Y, Weigel D. Identifying genetic variants underlying phenotypic variation in plants without complete genomes. Nat Genet. 2020;52:534–540. doi: 10.1038/s41588-020-0612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Young AD, Gillung JP. Phylogenomics — principles, opportunities and pitfalls of big-data phylogenetics. Systematic Entomology. 2020;45:225–247. doi: 10.1111/syen.12406. [DOI] [Google Scholar]
- 111.Huelsenbeck JP, Bull JJ, Cunningham CW. Combining data in phylogenetic analysis. Trends Ecol Evol. 1996;11:152–158. doi: 10.1016/0169-5347(96)10006-9. [DOI] [PubMed] [Google Scholar]
- 112.Solís-Lemus C, Yang M, Ané C. Inconsistency of Species Tree Methods under Gene Flow. Systematic Biology. 2016;65:843–851. doi: 10.1093/sysbio/syw030. [DOI] [PubMed] [Google Scholar]
- 113.Lewin HA, et al. Earth BioGenome Project: Sequencing life for the future of life. Proceedings of the National Academy of Sciences. 2018;115:4325–4333. doi: 10.1073/pnas.1720115115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Darwin Tree of Life Project Consortium. Sequence locally, think globally: The Darwin Tree of Life Project. Proceedings of the National Academy of Sciences. 2022;119:e2115642118. doi: 10.1073/pnas.2115642118. [This paper reviews the practicalities of tree-of-life projects] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mazzoni CJ, Ciofi C, Waterhouse RM. Biodiversity: an atlas of European reference genomes. Nature. 2023;619:252. doi: 10.1038/d41586-023-02229-w. [DOI] [PubMed] [Google Scholar]
- 116.Shaffer HB, et al. Landscape Genomics to Enable Conservation Actions: The California Conservation Genomics Project. Journal of Heredity. 2022;113:577–588. doi: 10.1093/jhered/esac020. [DOI] [PubMed] [Google Scholar]
- 117.Leebens-Mack JH, et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679–685. doi: 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheng S, et al. 10KP: A phylodiverse genome sequencing plan. GigaScience. 2018;7 doi: 10.1093/gigascience/giy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brassac J, Blattner FR. Species-Level Phylogeny and Polyploid Relationships in Hordeum (Poaceae) Inferred by Next-Generation Sequencing and In Silico Cloning of Multiple Nuclear Loci. Syst Biol. 2015;64:792–808. doi: 10.1093/sysbio/syv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Glémin S, et al. Pervasive hybridizations in the history of wheat relatives. Sci Adv. 2019;5:eaav9188. doi: 10.1126/sciadv.aav9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nemati Z, Harpke D, Gemicioglu A, Kerndorff H, Blattner FR. Saffron (Crocus sativus) is an autotriploid that evolved in Attica (Greece) from wild Crocus cartwrightianus. Molecular Phylogenetics and Evolution. 2019;136:14–20. doi: 10.1016/j.ympev.2019.03.022. [DOI] [PubMed] [Google Scholar]
- 122.Doolittle WF. Phylogenetic Classification and the Universal Tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 123.McInerney J, Pisani D, O’Connell MJ. The ring of life hypothesis for eukaryote origins is supported by multiple kinds of data. Philosophical Transactions of the Royal Society B: Biological Sciences. 2015;370:20140323. doi: 10.1098/rstb.2014.0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ma J, et al. Major episodes of horizontal gene transfer drove the evolution of land plants. Molecular Plant. 2022;15:857–871. doi: 10.1016/j.molp.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 125.Mahelka V, et al. Multiple horizontal transfers of nuclear ribosomal genes between phylogenetically distinct grass lineages. Proceedings of the National Academy of Sciences. 2017;114:1726–1731. doi: 10.1073/pnas.1613375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Udall JA, Wendel JF. Polyploidy and Crop Improvement. Crop Science. 2006;46 doi: 10.2135/cropsci2006.07.0489tpg. S-3-S14. [DOI] [Google Scholar]
- 127.Soltis PS, Marchant DB, Van de Peer Y, Soltis DE. Polyploidy and genome evolution in plants. Current Opinion in Genetics & Development. 2015;35:119–125. doi: 10.1016/j.gde.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 128.Schumer M, Rosenthal GG, Andolfatto P. How Common is Homoploid Hybrid Speciation? Evolution. 2014;68:1553–1560. doi: 10.1111/evo.12399. [DOI] [PubMed] [Google Scholar]
- 129.Dvorak J, McGuire PE, Cassidy B. Apparent sources of the A genomes of wheats inferred from polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome. 1988;30:680–689. doi: 10.1139/g88-115. [DOI] [Google Scholar]
- 130.Ramírez-González RH, et al. The transcriptional landscape of polyploid wheat. Science. 2018;361 doi: 10.1126/science.aar6089. [DOI] [PubMed] [Google Scholar]
- 131.Wang Z, et al. Genomic evidence for homoploid hybrid speciation between ancestors of two different genera. Nature Communications. 2022;13:1987. doi: 10.1038/s41467-022-29643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sun Y, Lu Z, Zhu X, Ma H. Genomic basis of homoploid hybrid speciation within chestnut trees. Nature Communications. 2020;11:3375. doi: 10.1038/s41467-020-17111-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wolf JBW, Ellegren H. Making sense of genomic islands of differentiation in light of speciation. Nature Reviews Genetics. 2017;18:87–100. doi: 10.1038/nrg.2016.133. [DOI] [PubMed] [Google Scholar]
- 134.Burri R, et al. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 2015;25:1656–1665. doi: 10.1101/gr.196485.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang K, Andrew RL, Owens GL, Ostevik KL, Rieseberg LH. Multiple chromosomal inversions contribute to adaptive divergence of a dune sunflower ecotype. Molecular Ecology. 2020;29:2535–2549. doi: 10.1111/mec.15428. [DOI] [PubMed] [Google Scholar]
- 136.Ross-Ibarra J, Morrell PL, Gaut BS. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc Natl Acad Sci U S A. 2007;104(Suppl 1):8641–8648. doi: 10.1073/pnas.0700643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schreiber M, Stein N, Mascher M. Genomic approaches for studying crop evolution. Genome Biology. 2018;19:140. doi: 10.1186/s13059-018-1528-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Savolainen O, Lascoux M, Merilä J. Ecological genomics of local adaptation. Nature Reviews Genetics. 2013;14:807–820. doi: 10.1038/nrg3522. [DOI] [PubMed] [Google Scholar]
- 139.Lovell JT, et al. Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass. Nature. 2021;590:438–444. doi: 10.1038/s41586-020-03127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Parker J, et al. Genome-wide signatures of convergent evolution in echolocating mammals. Nature. 2013;502:228–231. doi: 10.1038/nature12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schlüter PM, et al. Stearoyl-acyl carrier protein desaturases are associated with floral isolation in sexually deceptive orchids. Proceedings of the National Academy of Sciences. 2011;108:5696–5701. doi: 10.1073/pnas.1013313108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Breitkopf H, Onstein RE, Cafasso D, Schlüter PM, Cozzolino S. Multiple shifts to different pollinators fuelled rapid diversification in sexually deceptive Ophrys orchids. New Phytologist. 2015;207:377–389. doi: 10.1111/nph.13219. [DOI] [PubMed] [Google Scholar]
- 143.Baguette M, Bertrand JAM, Stevens VM, Schatz B. Why are there so many bee-orchid species? Adaptive radiation by intra-specific competition for mnesic pollinators. Biological Reviews. 2020;95:1630–1663. doi: 10.1111/brv.12633. [DOI] [PubMed] [Google Scholar]
- 144.Weigel D. Natural Variation in Arabidopsis: From Molecular Genetics to Ecological Genomics Plant Physiology. 2011;158:2–22. doi: 10.1104/pp.111.189845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Alonso-Blanco C, et al. 1,135 Genomes Reveal the Global Pattern of Polymorphism in <em>Arabidopsis thaliana</em>. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.De Wet JMJ, Harlan JR. Weeds and Domesticates: Evolution in the man-made habitat. Economic Botany. 1975;29:99–108. doi: 10.1007/BF02863309. [DOI] [Google Scholar]
- 147.Hammer K. Das Domestikationssyndrom. Die Kulturpflanze. 1984;32:11–34. doi: 10.1007/BF02098682. [DOI] [Google Scholar]
- 148.Wilkins AS, Wrangham RW, Fitch WT. The “domestication syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics. 2014;197:795–808. doi: 10.1534/genetics.114.165423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Clark RM, Wagler TN, Quijada P, Doebley J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- 150.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 151.Soyk S, et al. Bypassing Negative Epistasis on Yield in Tomato Imposed by a Domestication Gene. Cell. 2017;169:1142–1155.:e1112. doi: 10.1016/j.cell.2017.04.032. [DOI] [PubMed] [Google Scholar]
- 152.Alonge M, et al. Major Impacts of Widespread Structural Variation on Gene Expression and Crop Improvement in Tomato. Cell. 2020;182:145–161.:e123. doi: 10.1016/j.cell.2020.05.021. [A tour de force of pangenomics and classical genetics] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.McGuigan K, Hoffmann AA, Sgrò CM. How is epigenetics predicted to contribute to climate change adaptation? What evidence do we need? Philos Trans R Soc Lond B Biol Sci. 2021;376:20200119. doi: 10.1098/rstb.2020.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gao Y, et al. Complementary genomic and epigenomic adaptation to environmental heterogeneity. Molecular Ecology. 2022;31:3598–3612. doi: 10.1111/mec.16500. [DOI] [PubMed] [Google Scholar]
- 155.Soyk S, Benoit M, Lippman ZB. New Horizons for Dissecting Epistasis in Crop Quantitative Trait Variation. Annual Review of Genetics. 2020;54:287–307. doi: 10.1146/annurev-genet-050720-122916. [This review includes a proposal on how to harness induced structural variation in the study of gene-by-gene interactions] [DOI] [PubMed] [Google Scholar]
- 156.Lloyd JPB, Lister R. Epigenome plasticity in plants. Nature Reviews Genetics. 2022;23:55–68. doi: 10.1038/s41576-021-00407-y. [DOI] [PubMed] [Google Scholar]
- 157.Zhao L, et al. Integrative analysis of reference epigenomes in 20 rice varieties. Nature Communications. 2020;11:2658. doi: 10.1038/s41467-020-16457-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bräutigam K, et al. Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecology and Evolution. 2013;3:399–415. doi: 10.1002/ece3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.O’Malley Ronan C, et al. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 2016;165:1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Greally JM. Population Epigenetics. Curr Opin Syst Biol. 2017;1:84–89. doi: 10.1016/j.coisb.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Walker A. Adding genomic ‘foliage’ to the tree of life. Nature Reviews Microbiology. 2014;12:78. doi: 10.1038/nrmicro3203. [DOI] [PubMed] [Google Scholar]
- 163.McKain MR, Johnson MG, Uribe-Convers S, Eaton D, Yang Y. Practical considerations for plant phylogenomics. Applications in Plant Sciences. 2018;6:e1038. doi: 10.1002/aps3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kress WJ, et al. Green plant genomes: What we know in an era of rapidly expanding opportunities. Proceedings of the National Academy of Sciences. 2022;119:e2115640118. doi: 10.1073/pnas.2115640118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Trigodet F, et al. High molecular weight DNA extraction strategies for long-read sequencing of complex metagenomes. Molecular Ecology Resources. 2022;22:1786–1802. doi: 10.1111/1755-0998.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Al’Khafaji AM, et al. High-throughput RNA isoform sequencing using programmable cDNA concatenation. bioRxiv. 2021:2021.2010.2001.462818. doi: 10.1101/2021.10.01.462818. [DOI] [Google Scholar]
- 167.Cai Z-F, et al. Long amplicon HiFi sequencing for mitochondrial DNA genomes. Molecular Ecology Resources. doi: 10.1111/1755-0998.13765. n/a. [DOI] [PubMed] [Google Scholar]
- 168.Heslop-Harrison JSP, Schwarzacher T, Liu Q. Polyploidy: its consequences and enabling role in plant diversification and evolution. Ann Bot. 2023;131:1–10. doi: 10.1093/aob/mcac132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Klau GW, Marschall T. Springer International Publishing; pp. 50–63. [Google Scholar]
- 170.Song B, Buckler ES, Stitzer MC. New whole-genome alignment tools are needed for tapping into plant diversity. Trends in Plant Science. doi: 10.1016/j.tplants.2023.08.013. [DOI] [PubMed] [Google Scholar]
- 171.Eizenga JM, et al. Pangenome Graphs. Annual Review of Genomics and Human Genetics. 2020;21:139–162. doi: 10.1146/annurev-genom-120219-080406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Andreace F, Lechat P, Dufresne Y, Chikhi R. Construction and representation of human pangenome graphs. bioRxiv. 2023:2023.2006.2002.542089. doi: 10.1101/2023.06.02.542089. [DOI] [Google Scholar]
- 173.Yates AD, et al. Ensembl Genomes 2022: an expanding genome resource for non-vertebrates. Nucleic acids research. 2022;50:D996–D1003. doi: 10.1093/nar/gkab1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Contreras-Moreira B, et al. In: Plant Bioinformatics: Methods and Protocols. Edwards David., editor. Springer US; 2022. pp. 27–55. [Google Scholar]
- 175.Democratizing sequencing. Nature Methods. 2005;2:633. doi: 10.1038/nmeth0905-633. [DOI] [PubMed] [Google Scholar]
- 176.Gui S, et al. A pan-Zea genome map for enhancing maize improvement. Genome Biology. 2022;23:178. doi: 10.1186/s13059-022-02742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Portwood JL, II, et al. MaizeGDB 2018: the maize multi-genome genetics and genomics database. Nucleic Acids Research. 2018;47:D1146–D1154. doi: 10.1093/nar/gky1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Arita M, Karsch-Mizrachi I, Cochrane G, on behalf of the International Nucleotide Sequence Database, C The international nucleotide sequence database collaboration. Nucleic Acids Research. 2021;49:D121–D124. doi: 10.1093/nar/gkaa967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lawniczak MKN, et al. Standards recommendations for the Earth BioGenome Project. Proceedings of the National Academy of Sciences. 2022;119:e2115639118. doi: 10.1073/pnas.2115639118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Marden E, Sackville Hamilton R, Halewood M, McCouch S. International agreements and the plant genetics research community: A guide to practice. Proceedings of the National Academy of Sciences. 2023;120:e2205773119. doi: 10.1073/pnas.2205773119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Watanabe ME. The Nagoya Protocol: The Conundrum of Defining Digital Sequence Information. BioScience. 2019;69:480. doi: 10.1093/biosci/biz034. [DOI] [Google Scholar]
- 182.Scholz AH, et al. Multilateral benefit-sharing from digital sequence information will support both science and biodiversity conservation. Nature Communications. 2022;13:1086. doi: 10.1038/s41467-022-28594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhou P, et al. Exploring structural variation and gene family architecture with De Novo assemblies of 15 Medicago genomes. BMC Genomics. 2017;18:261. doi: 10.1186/s12864-017-3654-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gordon SP, et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nature Communications. 2017;8:2184. doi: 10.1038/s41467-017-02292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhao Q, et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nature Genetics. 2018;50:278–284. doi: 10.1038/s41588-018-0041-z. [DOI] [PubMed] [Google Scholar]
- 186.Zhou Y, et al. A platinum standard pan-genome resource that represents the population structure of Asian rice. Scientific Data. 2020;7:113. doi: 10.1038/s41597-020-0438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Song J-M, et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nature Plants. 2020;6:34–45. doi: 10.1038/s41477-019-0577-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu Y, et al. Pan-Genome of Wild and Cultivated Soybeans. Cell. 2020;182:162–176.:e113. doi: 10.1016/j.cell.2020.05.023. [DOI] [PubMed] [Google Scholar]
- 189.Campoy JA, et al. Chromosome-level and haplotype-resolved genome assembly enabled by high-throughput single-cell sequencing of gamete genomes. bioRxiv. 2020:2020.2004.2024.060046. doi: 10.1186/s13059-020-02235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Qin P, et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell. 2021;184:3542–3558.:e3516. doi: 10.1016/j.cell.2021.04.046. [DOI] [PubMed] [Google Scholar]
- 191.Tao Y, et al. Extensive variation within the pan-genome of cultivated and wild sorghum. Nature Plants. 2021;7:766–773. doi: 10.1038/s41477-021-00925-x. [DOI] [PubMed] [Google Scholar]
- 192.Zhang X, et al. Pan-genome of Raphanus highlights genetic variation and introgression among domesticated, wild, and weedy radishes. Molecular Plant. 2021;14:2032–2055. doi: 10.1016/j.molp.2021.08.005. [DOI] [PubMed] [Google Scholar]
- 193.Li H, et al. Graph-based pan-genome reveals structural and sequence variations related to agronomic traits and domestication in cucumber. Nature Communications. 2022;13:682. doi: 10.1038/s41467-022-28362-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hoopes G, et al. Phased, chromosome-scale genome assemblies of tetraploid potato reveal a complex genome, transcriptome, and predicted proteome landscape underpinning genetic diversity. Molecular Plant. 2022;15:520–536. doi: 10.1016/j.molp.2022.01.003. [DOI] [PubMed] [Google Scholar]
- 195.Zhuang Y, et al. Phylogenomics of the genus Glycine sheds light on polyploid evolution and life-strategy transition. Nature Plants. 2022;8:233–244. doi: 10.1038/s41477-022-01102-4. [DOI] [PubMed] [Google Scholar]
- 196.Wang M, et al. Genomic innovation and regulatory rewiring during evolution of the cotton genus Gossypium. Nature Genetics. 2022;54:1959–1971. doi: 10.1038/s41588-022-01237-2. [DOI] [PubMed] [Google Scholar]
- 197.Liang Q, et al. A view of the pan-genome of domesticated Cowpea (Vigna unguiculata [L.] Walp. The Plant Genome. 2023:e20319. doi: 10.1002/tpg2.20319. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kang M, et al. The pan-genome and local adaptation of <em>Arabidopsis thaliana</em>. bioRxiv. 2022:2022.2012.2018.520013. doi: 10.1101/2022.12.18.520013. [DOI] [Google Scholar]
- 199.Yan H, et al. Pangenomic analysis identifies structural variation associated with heat tolerance in pearl millet. Nature Genetics. 2023;55:507–518. doi: 10.1038/s41588-023-01302-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang B, et al. De novo genome assembly and analyses of 12 founder inbred lines provide insights into maize heterosis. Nature Genetics. 2023;55:312–323. doi: 10.1038/s41588-022-01283-w. [DOI] [PubMed] [Google Scholar]
- 201.Huang Y, et al. Pangenome analysis provides insight into the evolution of the orange subfamily and a key gene for citric acid accumulation in citrus fruits. Nature Genetics. 2023 doi: 10.1038/s41588-023-01516-6. [DOI] [PubMed] [Google Scholar]
- 202.He Q, et al. A graph-based genome and pan-genome variation of the model plant Setaria. Nature Genetics. 2023;55:1232–1242. doi: 10.1038/s41588-023-01423-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liu F, et al. Genomes of cultivated and wild Capsicum species provide insights into pepper domestication and population differentiation. Nature Communications. 2023;14:5487. doi: 10.1038/s41467-023-41251-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lian Q, et al. A pan-genome of 72 Arabidopsis thaliana accessions reveals a conserved genome structure throughout the global species range. 2023 doi: 10.1038/s41588-024-01715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.FAO. Agricultural production statistics 2000–2020. 2022.
- 206.Bao Z, et al. Genome architecture and tetrasomic inheritance of autotetraploid potato. Molecular Plant. 2022;15:1211–1226. doi: 10.1016/j.molp.2022.06.009. [DOI] [PubMed] [Google Scholar]
- 207.Sun H, et al. Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. Nature Genetics. 2022;54:342–348. doi: 10.1038/s41588-022-01015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jansky SH, et al. Reinventing potato as a diploid inbred line–based crop. Crop Science. 2016;56:1412–1422. [Google Scholar]
- 209.Zhang C, et al. Genome design of hybrid potato. Cell. 2021;184:3873–3883.:e3812. doi: 10.1016/j.cell.2021.06.006. [DOI] [PubMed] [Google Scholar]
- 210.Ye M, et al. Generation of self-compatible diploid potato by knockout of S-RNase. Nat Plants. 2018;4:651–654. doi: 10.1038/s41477-018-0218-6. [DOI] [PubMed] [Google Scholar]
- 211.Ma L, et al. A nonS-locus F-box gene breaks self-incompatibility in diploid potatoes. Nat Commun. 2021;12:4142. doi: 10.1038/s41467-021-24266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]