Abstract
Background
Alzheimer’s disease (AD)-related neuropathological changes can occur decades before clinical symptoms. We aimed to investigate whether neurodevelopment and/or neurodegeneration affects the risk of AD, through reducing structural brain reserve and/or increasing brain atrophy, respectively.
Methods
We used bidirectional two-sample Mendelian randomisation to estimate the effects between genetic liability to AD and global and regional cortical thickness, estimated total intracranial volume, volume of subcortical structures and total white matter in 37 680 participants aged 8−81 years across 5 independent cohorts (Adolescent Brain Cognitive Development, Generation R, IMAGEN, Avon Longitudinal Study of Parents and Children and UK Biobank). We also examined the effects of global and regional cortical thickness and subcortical volumes from the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) Consortium on AD risk in up to 37 741 participants.
Results
Our findings show that AD risk alleles have an age-dependent effect on a range of cortical and subcortical brain measures that starts in mid-life, in non-clinical populations. Evidence for such effects across childhood and young adulthood is weak. Some of the identified structures are not typically implicated in AD, such as those in the striatum (eg, thalamus), with consistent effects from childhood to late adulthood. There was little evidence to suggest brain morphology alters AD risk.
Conclusions
Genetic liability to AD is likely to affect risk of AD primarily through mechanisms affecting indicators of brain morphology in later life, rather than structural brain reserve. Future studies with repeated measures are required for a better understanding and certainty of the mechanisms at play.
Introduction
The earliest Alzheimer’s disease (AD)-related histopathological changes are typically observed within the medial temporal lobes and disperse throughout the frontal, parietal and temporal neocortices and subcortical regions by the time a clinical diagnosis of AD is made.1 Amyloid-β accumulation in the brain may be apparent 20 years before the appearance of clinical symptoms.2 Hence, the integration of biological data prior to the onset of clinical symptoms is crucial in understanding the aetiology, timing and progression of the disease, and for the development of more efficient strategies for early detection and screening of individuals for AD risk.
It has been argued that variability in AD risk may be mediated through both morphology (‘brain reserve’) and/or functional capacity to compensate for pathology (‘cognitive reserve’),3 which may operate independently or synergistically. Consequently, it has been hypothesised that genetic risk for AD may be mediated through determining the underlying brain reserve of an individual.4–6 Furthermore, the relationship between brain structures and AD may be bidirectional, as genes associated with brain morphology (such as thickness and surface area) have been shown to be involved in neurogenesis.7
Genetic instruments allow for the identification of factors that modify disease risk, establish effects of prodromal disease and can help us discover biomarkers that predict disease. Genome-wide association studies (GWAS)8–11 for AD have identified approximately 30 single nucleotide polymorphisms (SNPs), each with a modest effect on the risk of AD, apart from the ε4 genotype in the APOE gene, whereby carriers may have up to 12-fold increased risk.11 Previous studies examining the association of genetic liability to AD with brain morphology have typically used polygenic risk scores (PRS) at liberal thresholds, which can increase bias due to horizontal pleiotropy.6 12 13 They also have small sample sizes, as genetic and neuroimaging data are rarely available in combination. Furthermore, SNPs associated with brain structure have been discovered using larger sample sizes7 14 than previous neuroimaging studies,15 16 allowing for the bidirectional investigation of the causal effects of structural brain measures on risk of AD, using Mendelian randomisation (MR). MR is a form of instrumental variable analysis which uses SNPs as instruments for exposures to estimate lifetime effects of phenotypes on disease risk (and vice versa).17
We investigated how genetic liability to AD affects brain morphology across the life course (from ages 8 to 81 years) using two-sample MR. This approach examines whether AD genetic susceptibility affects brain development or degeneration. Using two-sample MR, we also investigated whether brain morphology has a causal effect on the risk of AD, to establish whether greater thickness/volume provides a protective effect against advancing neuropathology and thus, reduces risk of an AD diagnosis (‘brain reserve’ hypothesis).
Materials and Methods
Data
Alzheimer’s disease GWAS
We extracted SNPs from the largest GWAS of clinically diagnosed AD,9 which identified 27 SNPs to be associated with AD risk in participants of European ancestry. For each SNP, we used the effect estimates from the stage with the largest sample size (n=82 771 to 94 437 participants).
Brain structure GWAS
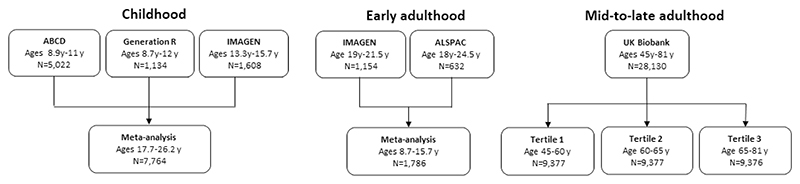
We used GWAS of brain structures (average thickness of 34 gyral-based cortical regions of interest, mean thickness, estimated total intracranial volume (eTIV), 9 subcortical volumes and the total volume of white matter) conducted within different cohorts across the life course. Regional thickness has been used to differentiate between mild cognitive impairment and individuals with AD with excellent accuracy, specificity and reproducibility across independent cohorts.18 We conducted all GWAS described, except for the GWAS in the ENIGMA consortium which has been previously published.7 14 19 20 GWAS for regional cortical thickness and subcortical volumes were adjusted for global cortical thickness and eTIV, respectively. For the peripubertal period, we used Generation R,21 22 the Adolescent Brain Cognitive Development study (ABCD)23 24 and IMAGEN.25 For early adulthood, we meta-analysed the Avon Longitudinal Study of Parents and Children (ALSPAC)26–28 and the second wave of data collection for the IMAGEN study.25 For adulthood, we used the UK Biobank (UKB)29 and stratified the sample into three equal-sized age tertiles, to examine age-specific effects (figures 1 and 2 and table 1). Finally, we used summary data from ENIGMA7 14 (n=37 741 participants, age range 3.4−91.4 years), which includes the first release of UKB imaging data. All GWAS in the analyses were conducted in participants of European ancestry. Details of the cohorts, including the genotyping and neuroimaging procedures, are provided in online supplemental tables 1 and 2, respectively.
Figure 1.
Study cohorts in the age-stratified analysis of genetic liability to Alzheimer’s disease on brain morphology. ABCD, Adolescent Brain Cognitive Development; ALSPAC Avon Longitudinal Study of Parents and Children; y, years.
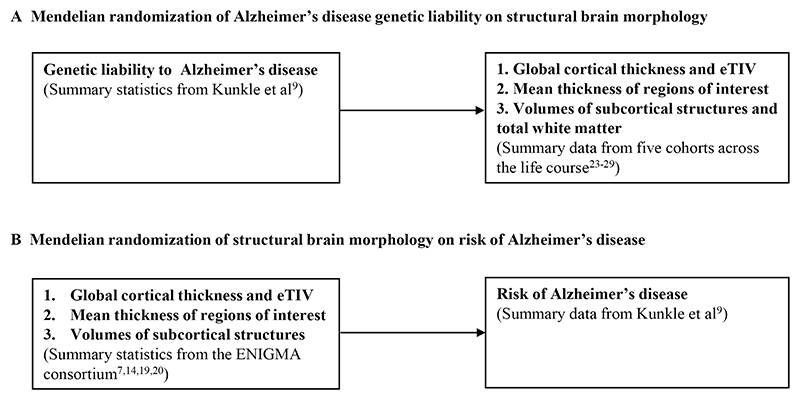
Figure 2. Study design for examining the bidirectional effects between Alzheimer’s disease and brain morphology.
(A) Mendelian randomisation of Alzheimer’s disease genetic liability on structural brain morphology. (B) Mendelian randomisation of structural brain morphology on risk of Alzheimer’s disease. eTIV, estimated total intracranial volume.
Table 1. Descriptive statistics of the cohorts used in the analysis.
| Cohort | N | Number of Alzheimer’s disease SNPs | F-statistic for Alzheimer’s disease SNPs | Mean age (SD) | Age range | % female |
|---|---|---|---|---|---|---|
| Childhood | ||||||
| ABCD | 5022 | 25 | 223.28 | 9.91 (0.6) | 8.9−11 | 52.6 |
| Generation R | 1134 | 23 | 239.34 | 10.2 (0.6) | 8.9−12 | 49.2 |
| IMAGEN | 1151−1154 | 23 | 224.67 | 14.4 (0.4) | 13.3−15.7 | 50.6 |
| Early adulthood | ||||||
| ALSPAC | 358−632 | 25 | 231.7 | 20.5 (1.6) | 18−24.5 | 22.4 |
| IMAGEN | 1577−1608 | 23 | 224.7 | 26.2 (0.7) | 17.7−26.2 | 51.3 |
| Adulthood | ||||||
| UK Biobank tertile 1 | 9377 | 24 | 231.5 | 55 (3.4) | 45−60 | 57.0 |
| UK Biobank tertile 2 | 9377 | 24 | 231.5 | 64.3 (2.2) | 60−68 | 53.7 |
| UK Biobank tertile 3 | 9376 | 24 | 231.5 | 72.0 (2.9) | 68−81 | 46.0 |
ABCD, Adolescent Brain Cognitive Development; ALSPAC, Avon Longitudinal Study of Parents and Children; SNP, single nucleotide polymorphism.
Statistical analyses
Estimating the causal effect of genetic liability to Alzheimer’s disease on brain structures
Two-sample Mendelian randomisation
Two-sample MR is an extension of MR,30 where the SNP effects on the exposure and on the outcome are extracted from separate GWAS studies. To examine the effects of genetic liability to AD on structural brain measures, we extracted SNPs strongly associated with AD (p≤5×10−8).9 Where SNPs were not available, we used proxy SNPs (r2>0.80). SNPs were clumped using r2>0.001 and a physical distance of 10 000 kb. We also included rs7412 and rs423958 to tag the APOE ε4 allele. We used 23−25 SNPs as instruments for AD, the number varying by availability within each cohort (table 1). We harmonised the AD and brain structure GWAS in IMAGEN, Generation R, ABCD, ALSPAC and the UKB (online supplemental methods).
We then employed univariable MR to estimate the effect of the AD SNPs on 9 subcortical volumes and the 34 cortical regions defined by the Desikan-Killiany atlas31 (as well as total volume of white matter where available) within each cohort. We used a random-effects inverse-variance weighted (IVW) regression analysis, which assumes no directional horizontal pleiotropy17 and used the F-statistic as a measure of instrument strength.32 All effect estimates reflect SD changes in the outcome per doubling of genetic liability to AD.33 Using the metagen function,34 we applied random-effects models to meta-analyse the effects of the AD SNPs on structural brain measures for the three peri-pubertal cohorts: IMAGEN, ABCD and Generation R (figures 1 and 2). To examine how the age-level covariate was associated with the causal effect estimates across the three age-stratified tertiles of UKB, we extracted a p for trend across these groups, using the meta regress command in STATA V.1635 and using the mean age of each tertile as the exposure. Sample sizes differed by brain structure due to quality control and missing data.
Estimating the causal effect of brain structures on risk of Alzheimer’s disease
Two-sample Mendelian randomisation
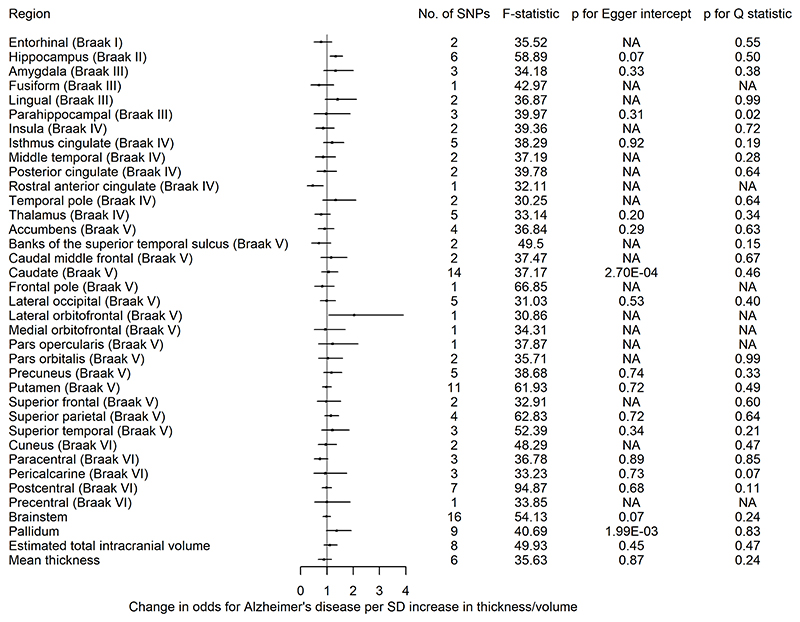
Using the ENIGMA GWAS,7 14 19 20 we extracted SNPs associated with eight subcortical volumes and the thickness of the regions of interest as defined by the Desikan-Killiany atlas.31 The same parameters and harmonisation methods were used as in the previous analysis. Again, we employed univariable MR to examine the causal effects of each brain structure on risk of AD using a random-effects IVW regression. All effect estimates represent an OR for AD per SD increase in thickness or volume. There is overlap between ENIGMA and some of the individual-level cohorts. However, it has been shown that sample overlap results in little bias in the presence of strong instruments (ie, F>10).36
Sensitivity analyses
We conducted sensitivity analyses to examine for potential violation of key MR assumptions. For MR to generate unbiased causal effect estimates, each genetic variant that is used as an instrumental variable must satisfy three assumptions: (1) that it is associated with the exposure (relevance assumption), (2) that it is not associated with the outcome through a confounding pathway (exchangeability assumption) and (3) is only associated with the outcome through the exposure (exclusion restriction assumption). IVW regression assumes no horizontal pleiotropy and provides unbiased causal effect estimates only when there is balanced or no horizontal pleiotropy. We compared estimates from IVW with those from Egger regression,37 38 weighted median39 and weighted mode,40 which relax this assumption. Heterogeneity in the causal estimates was assessed using Cochran’s Q-statistic.37 Furthermore, to exclude the possibility that the SNPs used to proxy for AD are instruments for brain structures and vice versa, we performed a directionality (Steiger) test.41 Where the hypothesised direction was false, we performed sensitivity analyses removing SNPs explaining more variance in the outcome than the exposure (details in online supplemental methods). Lastly, we excluded the two SNPs in the APOE locus from the AD instrument, to investigate whether the effects observed are primarily driven by the variants in the APOE gene. This study involves evaluating global patterns of effect estimates; hence, we focus on effect size and precision.42 43 Adjusted p values, controlling for the false discovery rate are in provided online supplemental tables 4, 8, 9 and 11.
Results
We used bidirectional two-sample MR30 to first examine the effect of genetic liability to AD (p≤5×10−8) on global and regional cortical thickness, eTIV, volumes of subcortical structures. We also included total white matter as an outcome where available. To boost the statistical power of the smaller childhood cohorts, we meta-analysed the causal effect estimates across ABCD,23 24 Generation R44 45 and IMAGEN25 (aged 8−16 years). For early adulthood, we used participants selected for neuroimaging in ALSPAC substudies46 (aged 18−24.5 years). For mid-to-late adulthood, we stratified the UKB population into three age tertiles: 45−60 years, 60−68 years and 68−81 years. In total, we used 23−25 independent AD SNPs from the largest GWAS of clinically diagnosed AD,9 depending how many were available in each cohort used (table 1). Second, we examined the causal effects of brain morphology on AD risk, using genetic instruments for each brain structure from the ENIGMA consortium GWAS. A summary of our study design is presented in figures 1 and 2.
Of the 34 cortical regions and 10 subcortical structures examined, there was evidence to suggest that genetic liability to AD has an age-dependent effect on the thickness and volume of these measures, respectively, across mid-to-late adulthood, but the evidence for such effects in childhood through young adulthood is weak. When we examined the causal effects of the thickness of 27 cortical regions (ie, those regions with genetic variants at 5×10−8), we found little evidence of an effect of greater thickness on risk of AD. We only found evidence that hippocampal volume and thickness of lateral orbitofrontal and rostral anterior cingulate cortices affected AD risk. An overview of the findings is shown in table 2.
Table 2. Summary of main findings.
| Exposure | Outcome | Timepoint | Direction |
|---|---|---|---|
| Genetic liability to Alzheimer’s disease | Caudal anterior cingulate Thalamus | Childhood | ↓ |
| ↓ | |||
| Thalamus | Early adulthood | ↓ | |
| Cuneus | Adulthood (45−60 years) | ↑ | |
| Inferior temporal | ↑ | ||
| Accumbens | Adulthood (60−68 years) | ↓ | |
| Caudal middle frontal | ↓ | ||
| Caudate | ↓ | ||
| Putamen | ↓ | ||
| Thalamus | ↓ | ||
| Accumbens* | Adulthood (68−81 years) | ↓ | |
| Amygdala* | ↓ | ||
| Caudate | ↓ | ||
| Entorhinal* | ↓ | ||
| Fusiform | ↓ | ||
| Hippocampus* | ↓ | ||
| Inferior temporal | ↓ | ||
| Lateral occipital | ↑ | ||
| Lateral ventricles | ↑ | ||
| Middle temporal* | ↓ | ||
| Parahippocampal | ↓ | ||
| Pericalcarine | ↑ | ||
| Postcentral | ↑ | ||
| Precentral | ↑ | ||
| Superior parietal | ↑ | ||
| Thalamus* | ↓ | ||
| Transverse temporal | ↑ | ||
| Hippocampus | Alzheimer’s disease | Across the life course (summary data) |
↑ |
Only analyses where 95% CIs show some evidence of association are displayed.
Indicates p<0.05 following correction for multiple testing.
Causal effects of genetic liability to Alzheimer’s disease on brain structures
Childhood
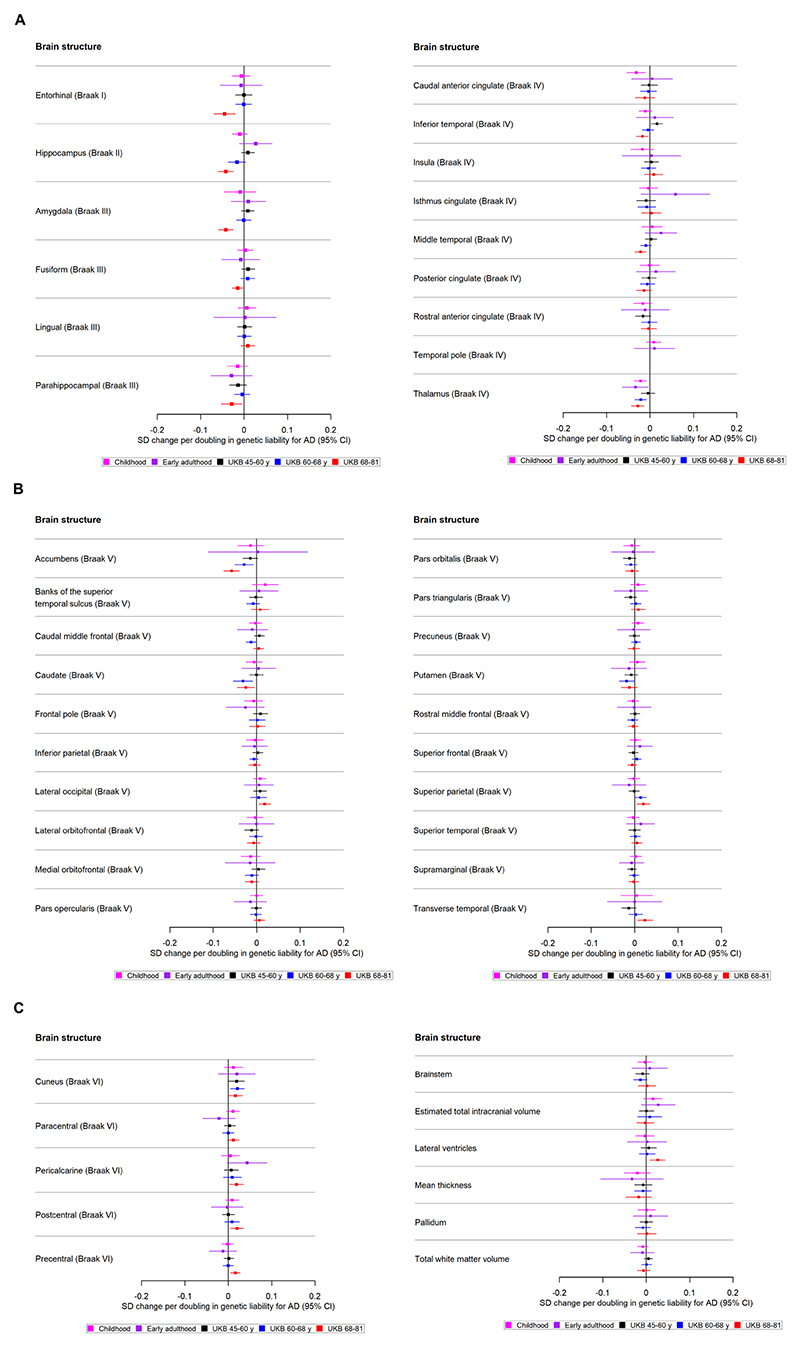
Only weak evidence supported the association between genetic liability to AD and cortical thickness or subcortical volumes in school-aged children. A doubling in odds of genetic liability to AD was associated with a −0.02 SD (95% CI −0.04 to −0.01) smaller volume of the thalamus (Braak stage IV) (figure 3A) and −0.03 SD (95% CI −0.05 to −0.01) lower thickness of the caudal anterior cingulate (Braak stage IV) (figure 3A, online supplemental tables 1–4).
Figure 3.
(A) The causal effects of genetic liability to AD on brain structures in Braak stages I−IV at different ages across the life course (see figure 3B for structures in Braak stage V and figure 3C for Braak stage VI). The childhood cohorts include meta-analysed effects of three peri-pubertal cohorts: ABCD, GEN R and IMAGEN. The early adulthood cohort includes ALSPAC and the later adulthood cohorts include UKB. Effect estimates for cortical regions and subcortical structures represent SD changes in thickness and volume. Cortical regions were adjusted for mean thickness and subcortical volumes were adjusted for estimated intracranial volume. Where an effect estimate is missing, that structural measure was not available in that cohort. (B) The causal effects of genetic liability to AD on brain structures in Braak stages V at different ages across the life course (see figure 3C for structures in Braak stage VI). The childhood cohorts include meta-analysed effects of three peri-pubertal cohorts: ABCD, GEN R and IMAGEN. The early adulthood cohort includes ALSPAC and the later adulthood cohorts include UKB. Effect estimates for cortical regions and subcortical structures represent SD changes in thickness and volume. Cortical regions were adjusted for mean thickness and subcortical volumes were adjusted for estimated intracranial volume. Where an effect estimate is missing, that structural measure was not available in that cohort. (C) The causal effects of genetic liability to AD on brain structures in Braak stage VI, and those not included in Braak staging, at different ages across the life course. The childhood cohorts include meta-analysed effects of three peri-pubertal cohorts: ABCD, GEN R and IMAGEN. The early adulthood cohort includes ALSPAC and the later adulthood cohorts include UKB. Effect estimates for cortical regions and subcortical structures represent SD changes in thickness and volume. Cortical regions were adjusted for mean thickness, subcortical structures and volume of cerebral white matter were adjusted for estimated intracranial volume. Where an effect estimate is missing, that structural measure was not available in that cohort. ABCD, Adolescent Brain Cognitive Development; AD, Alzheimer’s disease; ALSPAC, Avon Longitudinal Study of Parents and Children; GEN R, Generation R; UKB, UK Biobank.
Early adulthood
There was little evidence to suggest that a higher genetic liability to AD is associated with cortical regions and subcortical structures. However, a doubling in odds of genetic liability to AD was weakly associated with a −0.03 lower thalamic volume (Braak stage IV) (figure 3A, online supplemental table 8) (SD−0.03; 95% CI −0.06 to −0.004).
Mid-life to late life
We identified evidence of an age-dependent effect of AD genetic liability on smaller volume of the hippocampus (Braak stage II), accumbens (Braak stage II), amygdala (Braak stage II) and thalamus (Braak stage IV) (p for trend across age tertiles for each respective structure: 1.32×10−5, 0.001, 0.02 and 0.03; online supplemental tables 9 and 10). Furthermore, we found evidence of age-dependent effect of AD genetic liability on lower thickness of the inferior temporal and middle temporal cortices (p for trend across age tertiles=0.001 and p=0.009, respectively; Braak stage IV, figure 3A). A doubling in odds of genetic liability to AD, for example, was associated with 0.02 SD (95% CI −0.04 to −0.01) lower thickness in the middle temporal cortex for participants of aged 68−81 years and a trend in the same direction was observed for participants aged 60−68 years. On the contrary, for the superior and transverse temporal cortices (Braak stage V, figure 3B), we identified AD genetic liability to be associated with greater thickness (p for trend across age tertiles=0.03 and p=0.003, respectively).
We also identified effects which did not show clear age-dependent associations. Within the youngest UKB participants aged 45−60 years, a higher genetic liability to AD was associated with a greater thickness in the cuneus. In participants aged 60−68 years, a higher genetic liability to AD was associated with a lower volume in the caudate (Braak stage V, figure 3B), and putamen (Braak stage V, figure 3B). In participants aged 68−81 years, a doubling in odds of genetic liability to AD was associated with 0.05 SD (95% CI 0.07 to 0.02) lower thickness in the entorhinal cortex (Braak stage I), fusiform and parahippocampal cortices (Braak stage III, figure 3A). Additionally, AD genetic liability was associated with a thicker pericalcarine, postcentral, precentral cortex and a larger volume in the lateral ventricles (Braak stage VI, figure 3C).
Causal effects of brain morphology on risk of Alzheimer’s disease
We found little evidence of causal effects for the global measures of thickness and eTIV on AD risk (online supplemental table 11). However, of the eight subcortical structures examined, we observed that a 1 SD increase hippocampal volume, instrumented by six SNPs, increased AD risk on average by 33% (95% CI 1.11 to 1.59). A 1 SD increase in the thickness of the lateral orbitofrontal cortex increased AD risk (OR 2.74; 95% CI 1.08 to 6.93), while a 1 SD higher thickness in the rostral anterior cingulate cortex decreased AD risk (OR 0.40; 95% CI 0.19 to 0.83) (figure 4). However, for these two structures, we have one instrument and could not perform sensitivity analyses for assessing heterogeneity or pleiotropy.
Figure 4.
The causal effects of genetic predisposition to higher thickness and volume of cortical, subcortical and white matter measures, respectively on risk for AD. This figure shows the change in OR for AD per SD change in thickness and volume of cortical, subcortical structures, respectively. Effects for lateral ventricles is missing due to inability in obtaining access to summary statistics. The F-statistic is a measure of instrument strength. AD, Alzheimer’s disease; NA, not available.
Sensitivity analyses
Detailed results of analyses examining potential pleiotropy are provided in online supplemental tables 1–18. The evidence of a causal effect of genetic liability to AD on the caudal anterior cingulate in peri-pubertal childhood was consistent across pleiotropy-robust methods (SD −0.03; 95% CI −0.06 to −0.004 in MR-Egger and SD −0.03; 95% CI −0.05 to −0.01 per doubling in odds of genetic liability to AD for weighted mode). The association with thalamic volume in school-aged children and early adulthood were consistent across most of the pleiotropy-robust methods (online supplemental tables 4 and 8). In the analysis of AD genetic liability on brain structures in UKB, the magnitude of effect sizes for the MR-Egger, weighted median and mode were consistent with the IVW estimates for all brain structures (online supplemental table 9). In the MR analysis of brain structures on AD risk, the observed detrimental effect of a larger hippocampus on AD was consistent across pleiotropy-robust methods (online supplemental tables 11 and 18).
When we removed the APOE SNPs from our analyses in the peri-pubertal childhood cohort meta-analysis, the effect observed for AD liability on thalamic volume and the thickness of the caudal anterior cingulate cortex attenuated to the null (online supplemental table 19). The effect observed for AD liability on thalamic volume in early adulthood also attenuated to the null (online supplemental table 20). In UKB analyses, the associations with regional cortical thickness and subcortical structures largely remained (online supplemental table 21).
The directionality test indicated that, on average, the instruments for AD explained more variance in AD than they did in the brain structures in UKB (online supplemental table 22). The directionality test for SNPs associated with the hippocampus, lateral occipital and rostral anterior cingulate cortices on AD showed that they explain more variance in these structures than they do in AD risk (online supplemental table 23).
Discussion
Our findings suggest that AD risk alleles have an age-dependent effect on a range of cortical and subcortical brain measures across mid-to-late adulthood, but we found little evidence for such effects in childhood and early adulthood, with the exception of an observed effect of AD genetic liability on thalamic volume and the thickness of the caudal anterior cingulate. These results therefore suggest that genetic liability to AD operates largely through causing changes in brain morphology in later life (eg, potential neurodegeneration), rather than initial brain reserve. In the age-stratified analysis of UKB participants, a higher AD genetic liability was associated with an age-dependent decrease in the thickness of the middle temporal, inferior temporal cortices as well as volume of structures such as the hippocampus, accumbens and thalamus. Some effects were only apparent in the oldest participants (aged 68−81 years), such as the decrease in the thickness of the fusiform, entorhinal and parahippocampal cortices and the volume of the amygdala. When SNPs in the APOE gene region were removed, effects across all structures largely remained but as expected, became less precise. In the reverse direction, there was little evidence that the thickness and volume of cortical and subcortical structures, respectively, affected the risk of AD, except for a greater hippocampal volume increasing risk.
In adults, genetic liability to AD was associated with regions known to show significant atrophy early in disease progression, such as the entorhinal,47–49 inferior, middle temporal and para-hippocampal cortices,50 as well as the hippocampus.51 Change in hippocampal volume is an important imaging phenotype to define preclinical stages of AD, where atrophy predicts conversion from mild cognitive impairment to AD.52 We observed a trend of a higher genetic liability to AD being associated with a smaller hippocampus in the younger participants, of ages 45−68 years. Additionally, there was strong evidence of an effect of genetic liability on a lower hippocampal volume in the oldest age participants (aged 68−81 years), using genetic instruments both including and excluding the APOE locus. A study also using the UKB identified strong evidence of an effect of the AD PRS (p≤5×10−8) and hippocampal subfield volumes in older individuals (aged 63−80 years), which was driven by SNPs in the APOE locus.51 Contrary to previous PRS studies,12 13 we found weak evidence that genetic liability to AD was associated with a lower hippocampal volume in childhood. However, in comparison with the stringent threshold we used in our study (p<5×10− 8), these studies used liberal p-value thresholds for SNP inclusion (p≤0.132 and p≤0.0001) (increasing risk of bias due to horizontal pleiotropy).12 13
The focus of previous PRS studies with brain MRI data on the hippocampus and the neocortex can be attributed to their well-recognised role in cognition and episodic memory.53 54 However, there are other structures that are relevant for cognition that are less well studied in relation to genetic liability to AD,55 such as the thalamus. The medial temporal lobe connects to thalamic nuclei and the retrosplenial cortex, constituting the hippocampal-diencephalic system, whose integrity is important for normal episodic memory.56 In our study, we found the earliest, most robust effects of genetic liability to AD in the thalamus as early as childhood (aged 8−14 years) and in the caudate and accumbens from 60 years of age. A study investigating how the APOE genotype changes whole-brain large-scale structural networks in subjects with mild cognitive impairment,57 found APOE ε4 carriers showed pronounced atrophy in specific regions such as the thalamus and the hippocampus, both of which had strong structural covariance association with the left caudate nucleus. Furthermore, a longitudinal brain imaging study examining the effects of the APOE ε4 genotype found evidence of differences between carriers/non-carriers in rates of amyloid-β plaque accumulation across the adult lifespan only in the caudate at age 56 years and the putamen at age 63 years.58 APOE ε4 carriers showed accelerated rates of amyloid-beta deposition in the entorhinal cortex at age 68 years. We observed that the oldest participants (aged 68−81 years) with higher genetic liability to AD showed, on average, lower entorhinal thickness.
Like other studies, we also found causal effects of genetic liability to AD on larger thickness in the lateral occipital, which is consistent with two previous studies in healthy individuals where APOE ε4 carriers have a thicker occipital cortex in comparison with normal controls.59 60 The thickening of certain brain regions has been speculated to reflect brain swelling in response to glial activation in preclinical AD stages.61
Genetic liability to AD is hypothesised to affect brain structures through influencing neurodevelopment, resulting in structural differences in the brain which may increase tolerance to pathology (ie, altering brain reserve and increasing the age of disease onset), or by changing the rates or mechanisms of neurodegeneration.3 We observe an age-dependent decrease in the volume of structures such as the thalamus, caudate and accumbens in UKB participants. However, a longitudinal analysis would be required to test the variable neurodegeneration hypothesis and such a conclusion cannot be extrapolated from findings in cross-sectional data (as in our analyses). Walhovd et al examined the association between AD PRS and hippocampal volume in 1181 cognitively healthy people with a wide age range (4−95 years).4 They identified an effect of a higher AD PRS on reduced hippocampal volumes in young adults, which was consistent across age groups, suggesting the AD PRS results in an earlier onset of brain ageing instead of accelerated ageing through variable neurodegeneration. The MR of brain morphology on AD in our study provides little support for the notion that brain structure alterations change the risk for AD, except for a larger hippocampal volume increasing the risk for AD, which is contrary to most existing research.4 62 63 This hippocampus finding from our MR study may be due to chance, or due to the small number of SNPs used (n=6). It is unlikely that these effects are a result of pathways independent of hippocampal volume (ie, horizontal pleiotropy), as the MR estimators which relax the assumptions about instrument validity are consistent with the IVW method, and there was little evidence of heterogeneity or pleiotropy in the causal effect estimates. Although we found little evidence of effects of brain morphology on AD risk, we observed that AD genetic liability influenced the volume of the thalamus from childhood to adulthood, which suggests that initial thalamic reserve could potentially play a role in AD risk. However, given that this structure is not typically implicated in the earliest AD-related brain atrophy, it is possible that this is a chance finding reflecting variability around the null. The caudal anterior cingulate was observed to be associated with genetic effects in childhood but not in adulthood. However, a recent recall-by-genotype study also reported an effect in this region in adults of the PROTECT cohort.64 In summary, the fewer effects observed in childhood and early adulthood compared with those later in the life course may be due to developmental noise, or a greater effect of genetic variation on more biological pathways in older individuals. It is also possible that genetic effects become more pronounced later in the life course due to the accumulation of gene-environment interactions and/or potential epigenetic mechanisms. Future studies should seek to replicate this in large independent samples with repeated measures when more data become available.
The MR method requires that genetic variants must fulfil three key assumptions to be considered valid instrumental variables: (1) that the genetic variants are strongly associated with the exposure (relevance assumption), (2) that there is no confounding of the genetic instrument − outcome association (eg, by population stratification, or dynastic effects; the independence assumption), and (3) that the genetic instruments affect the outcome only through the exposure (exclusion restriction assumption). Only the first assumption can be tested with the use of statistical parameters indicating instrument strength (variants associated with the trait at genome-wide significance and/or F-statistics in our analyses >10). The independence and exclusion restriction assumptions are not testable but are falsifiable with sensitivity analyses. We adjusted our GWAS for ancestry-informative principal components to control for population stratification. We were unable to account for dynastic effects in this study, but future within-family MR study designs should look to examine this. For the exclusion-restriction assumption, sensitivity analyses were performed to examine potential bias due to horizontal pleiotropy. That said, several brain measures had too few genetic proxies for pleiotropy sensitivity analyses to be performed and hence, these results should be considered with caution.
While previous studies have examined whether genetic liability to AD is associated with specific structural brain measures, our study is the first to examine these in such large samples, using an exploratory approach from childhood to old age. One of the main strengths of our study is that genetic variants are subject to little measurement error, contrary to observational neuro-imaging phenotypes, and can serve as unconfounded indicators of particular traits values.17 Furthermore, using aggregate PRS precludes the examination of key potential sources of bias such as horizontal pleiotropy, which we have examined in detail here. We examined regions that have not been shown to be vulnerable to AD pathology, allowing us to discover novel regions affected by genetic liability to AD, such as the caudate. The large modern biobanks with neuroimaging and genetic data allowed us to recreate to the best of our ability a pseudo-longitudinal cohort. The precision of age-dependent dose effects suggest that our results are unlikely to be due to chance or other forms of bias. However, for studies such as ALSPAC, participants were selected for imaging for (1) a case-control study of psychotic experiences, (2) recall-by-genotype for schizophrenia, (3) testosterone study, making the ALSPAC sample unrepresentative of the general population. Another limitation is that different Free-surfer versions were used across cohorts. However, we allowed for this technical variation using random-effects meta-analyses. Although we applied multiple correction strategies controlling the false discovery rate, our findings were consistent across multiple cohorts. Finally, the participants in our analyses were of European ancestry and the findings may not be generalisable to other populations.
Our study shows that genetic liability to AD is associated with age-dependent changes in brain morphology in non-clinical populations, starting as early as 60 years of age, potentially highlighting the earliest phenotypic manifestations of the disease and the optimal timing for intervention with any potential neuroprotective therapy. Brain imaging to detect AD focuses on hippo-campal, whole brain and parietal volume. The findings of our study highlight the importance of brain volume in other regions — notably the striatum — for AD. The analysis of these regions could be incorporated into early diagnosis imaging analysis algorithms for clinical use. The lack of evidence to support an effect of brain morphology on AD suggests that genetic liability to AD affects biological pathways leading to neurodegeneration rather than neurodevelopment. Future research should aim to use a longitudinal design and integrate their findings with biological and clinical data.
Supplementary Material
What is Already Known on this Topic
-
⇒
Little is known about the dynamic interplay between brain morphology and Alzheimer’s disease throughout the life course.
-
⇒
Most prior research has predominantly focused on overall brain structure metrics, such as estimated total intracranial volume, mean thickness and total surface area.
What This Study Adds
-
⇒
This is the first bidirectional Mendelian randomisation study to assess the effects between Alzheimer’s disease, and both global and regional measurements of cortical thickness, estimated total intracranial volume, total white matter and subcortical structure volumes, using different cohorts spanning the life course.
How this Study Might Affect Research, Practice or Policy
-
⇒
Brain morphology is likely to play a role in changing the risk of Alzheimer’s disease through neurodegenerative pathways such as a loss of brain matter, rather than neurodevelopmental pathways such as building brain reserve.
-
⇒
Future research should focus efforts on using different measures of structural and functional brain morphology, starting in mid-adulthood.
Acknowledgements
Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive Development (ABCD) Study (https://abcdstudy.org), held in the NIMH Data Archive (NDA). This is a multisite, longitudinal study designed to recruit >10 000 children aged 9−10 years and follow them over 10 years into early adulthood. The ABCD study is supported by the National Institutes of Health (NIH) and additional federal partners under award numbers U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, U24DA041147. A full list of supporters is available at https://abcdstudy.org/federal-partners.html. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/consortium_members/. ABCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in analysis or writing of this report. The ABCD data repository grows and changes over time. The ABCD data used in this report came from NIMH Data Archive Digital Object Identifier (10.15154/1503209). We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and Wellcome (grant ref: 217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. This publication is the work of RK-L and ELA and NMD will serve as guarantors for the contents of this paper. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). The ALSPAC-Testosterone study was funded by the NIH, USA (R01MH085772 to TP). The ALSPAC-PE study was funded by a grant from the UK Medical Research Council (G0901885). ASD was also supported by the NIH Research Biomedical Research Centre at the South London and Maudsley Hospital Foundation NHS Trust and the IoPPN, King’s College London. The ALSPAC SCZ-RbG study was funded by grant MR/K004360/1 from the Medical Research Council (MRC) titled: ‘Behavioural and neurophysiological effects of schizophrenia risk genes: a multi-locus, pathway based approach’ and by the MRC Centre for Neuropsychiatric Genetics and Genomics (G0800509) and the NIHR Bristol Biomedical Research Centre. IMAGEN is supported by the European Union-funded FP6 Integrated Project IMAGEN (reinforcement-related behaviour in normal brain function and psychopathology) (LSHM-CT-2007-037286), the Horizon 2020 funded ERC Advanced Grant ‘STRATIFY’ (brain network-based stratification of reinforcement-related disorders) (695313), Human Brain Project (HBP SGA 2, 785907 and HBP SGA 3, 945539), the Medical Research Council Grant ‘c-VEDA’ (Consortium on Vulnerability to Externalising Disorders and Addictions) (MR/N000390/1), the National Institute of Health (NIH) (R01DA049238, a decentralised macro and micro gene-by-environment interaction analysis of substance use behaviour and its brain biomarkers), the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, the Bundesministeriumfür Bildung und Forschung (BMBF grants 01GS08152; 01EV0711; Forschungsnetz AERIAL 01EE1406A, 01EE1406B; Forschungsnetz IMAC-Mind 01GL1745B), the Deutsche Forschungsgemeinschaft (DFG grants SM 80/7-2, SFB 940, TRR 265, NE 1383/14-1), the Medical Research Foundation and Medical Research Council (grants MR/R00465X/1 and MR/S020306/1), the NIH funded ENIGMA (grants 5U54EB020403-05 and 1R56AG058854-01). Further support was provided by grants from the ANR (ANR-12-SAMA-0004, AAPG2019 — GeBra), the Eranet Neuron (AF12-NEUR0008-01 — WM2NA and ANR-18-NEUR00002-01 — ADORe), the Fondation de France (00081242), the Fondation pour la Recherche Médicale (DPA20140629802), the Mission Interministérielle de Lutte-contre-les-Drogues-et-les-Conduites-Addictives (MILDECA), the Assistance-Publique-Hôpitaux-de-Paris and INSERM (interface grant), Paris Sud University IDEX 2012, the Fondation de l’Avenir (grant AP-RM-17-013), the Fédération pour la Recherche sur le Cerveau; the NIH, Science Foundation Ireland (16/ERCD/3797), USA (Axon, Testosterone and Mental Health during Adolescence; RO1 MH085772-01A1) and by NIH Consortium grant U54 EB020403, supported by a cross-NIH alliance that funds Big Data to Knowledge Centres of Excellence. The UK Biobank data used in this work were obtained from UK Biobank Data Application 48970. We thank UK Biobank for making the data available, and to all UK Biobank study participants, who generously donated their time to make this resource possible. We thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients and their families. The i-Select chips was funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence programme investment for the future) DISTALZ grant, INSERM, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD/PERADES was supported by the Medical Research Council (grant no. 503480), Alzheimer’s Research UK (grant no. 503176), the Wellcome Trust (grant no. 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant no. 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01-AG-12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC-10-196728.
Funding
RK-L was supported by a Wellcome Trust PhD studentship (grant ref: 215193/Z18/Z). ELA is supported by a fellowship from the UK Medical Research Council (MR/P014437/1). The Medical Research Council (MRC) and the University of Bristol support the MRC Integrative Epidemiology Unit (MC_UU_00011/1). NMD is supported by a Norwegian Research Council Grant number 295989. YB-S receives grant funding from the following: MRC, Wellcome Trust, NIHER, Templeton Foundation, Parkinson’s UK, HQIP, Versus Arthritis, Dunhill Medical Trust, Gatsby Foundation, Kidney Research UK. LDH is funded by a Career Development Award from the UK Medical Research Council (MR/M020894/1). EW is funded by the European Union’s Horizon 2020 research and innovation programme (grant no. 848158) and by CLOSER, who was funded by the Economic and Social Research Council (ESRC) and the MRC between 2012 and 2017. Its initial 5-year grant has since been extended to March 2021 by the ESRC (grant reference: ES/K000357/1). TW is funded by Netherlands Organization for Health Research and Development (ZonMw) TOP project number 91211021. This research was also supported by contract R01-HL105756-07 from the National Heart, Lung and Blood Institute (NHLBI). EC is supported by grants with the Above and Beyond Charity for research optimising sleep to slow dementia progression and improve quality of life and the Alzheimer’s Research UK on research about the long-term memory testing to predict the presence of Alzheimer’s disease pathology from BRACE charity. CC is supported by the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement no. 707404 and grant agreement no. 848158 (EarlyCause Project). The work of Henning Tiemeier is supported by Netherlands Organization for Health Research and Development (ZonMW: 016.VICI.170.200). GH is supported by the Wellcome Trust and Royal Society (208806/Z/17/Z). AW is supported by Wellcome Trust PhD Grant, BRACE Pilot grant (co-applicant). SD is supported by the MRC. AB has received two grants from the National Children’s Hospital Foundation — Tallaght (Ireland), a grant from the Health Research Board (Ireland), a grant from the Irish Research Council and a grant from the European Union Horizon 2020 Programme (MSCA-ITN). All funds paid to Trinity College Dublin. HF is supported by the Coviddrug German Research Foundation Perpain German Ministry of Education and Research. TP is supported by the Canadian Institutes of Health research. MS is supported by the Deutsche Forschungsgemeinschaft, grant numbrs 186318919, 178833530 and 402170461. HW is supported by EU Horizon 2020: GA101016127; GA 777084, European Research Commission: GA 695313, BMBF (German Ministery for Health and Education): 01ZX1909C, 01EE1407G, 01ZX1614B, DFG (German Research Foundation): SFB 940; TRR 256/1; GRK 2386; WA 1539/9-1; WA 1539/11-1.
Footnotes
Contributors RK-L, ELA, NMD, LDH and YB-S designed, conceptualised and interpreted results of the study. RK-L and BX performed the statistical analyses. EC, EW, TS, TW and AW provided advice regarding structural brain measures. GH provided advice and feedback on the implementation of software. TS, AB, SD, HF, AG, HG, PG, AH, RB, J-LM, M-LPM, EA, FN, DPO, TP, LP, SM, JHF, MS, HW, RW, GS, and JW worked on the processing and provision of IMAGEN data. All authors provided valuable feedback and comments on the manuscript. ELA and NMD will serve as guarantors for the contents of this paper.
Disclaimer This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators.
Competing interests TB served in an advisory or consultancy role for Lundbeck, Medice, Neurim Pharmaceuticals, Oberberg GmbH, Shire. He received conference support or speaker’s fee by Lilly, Medice, Novartis and Shire. He has been involved in clinical trials conducted by Shire & Viforpharma. He received royalties from Hogrefe, Kohlhammer, CIP Medien, Oxford University Press. The present work is unrelated to the above grants and relationships. LP served in an advisory or consultancy role for Roche and Viforpharm and received speaker’s fee by Shire. She received royalties from Hogrefe, Kohlhammer and Schattauer. The present work is unrelated to the above grants and relationships.
Patient consent for publication Not applicable.
Ethics approval UK Biobank is approved by the National Health Service National Research Ethics Service (ref 11/NW/0382; UK Biobank application number 48970). Ethics approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees and informed consent for the use of data collected via questionnaires and clinics was obtained from participants. In Generation R, all study protocols and measurements assessed in each wave of data collection were approved by the Medical Ethical Committee (MEC 198.782/2001/31) of the Erasmus MC, University Medical Center Rotterdam. The IMAGEN study was approved by the institutional ethics committee of Kings College London, University of Nottingham, Trinity College Dublin, University of Heidelberg, Technische Universität Dresden, Commissariat á l Energie Atomique et aux Energies Alternatives and University Medical Center at the University of Hamburg in accordance with the Declaration of Helsinki. The UCSD IRB approved all data collection protocols for ABCD. IRB number: 160091. All analyses in this study used de-identified data, therefore no additional IRB approval was required. Participants gave informed consent to participate in the study before taking part.
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement
Data may be obtained from a third party and are not publicly available. The ENIGMA consortium MRI summary measures from genetic association analyses of estimated total intracranial volume, subcortical structures as well as cortical thickness were requested online. The ABCD study data are openly available to qualified researchers for free (https://nda.nih.gov/abcd/request-access). Requests for Generation R data should be directed towards the management team of the Generation R study (secretariaat.genr@erasmusmc.nl), which has a protocol of approving data requests. For access to IMAGEN data, researchers may submit a request to the IMAGEN consortium (https://imagen-europe.com/resources/imagen-project-proposal/). ALSPAC details and data descriptions are available on their website (www.bristol.ac.uk/alspac/researchers/access), where applications for individual-level data can be made (managed access). UK Biobank data are available through a procedure described on their website (http://www.ukbiobank.ac.uk/using-the-resource/). The UCSD IRB approved all data collection protocols for ABCD. IRB number: 160091. In Generation R, all study protocols and measurements assessed in each wave of data collection were approved by the Medical Ethical Committee (MEC 198.782/2001/31) of the Erasmus MC, University Medical Center Rotterdam. The IMAGEN study was approved by the institutional ethics committee of Kings College London, University of Nottingham, Trinity College Dublin, University of Heidelberg, Technische Universität Dresden, Commissariat á l Energie Atomique et aux Energies Alternatives, and University Medical Center at the University of Hamburg in accordance with the Declaration of Helsinki. Ethics approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees and informed consent for the use of data collected via questionnaires and clinics was obtained from participants. UK Biobank is approved by the National Health Service National Research Ethics Service (ref 11/NW/0382; UK Biobank application number 48970). All analyses in this study used de-identified data, therefore no additional IRB approval was required. All necessary patient/participant consent has been obtained. Code is available at https://github.com/rskl92/AD_BRAIN_BIDIRECTIONAL_MR.
References
- 1.Yang J, Pan P, Song W, et al. Voxelwise meta-analysis of gray matter anomalies in Alzheimer’s disease and mild cognitive impairment using anatomic likelihood estimation. J Neurol Sci. 2012;316:21–9. doi: 10.1016/j.jns.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Bateman RJ, Xiong C, Benzinger TLS, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern Y. Cognitive reserve in ageing and Alzheimer’s disease. Lancet Neurol. 2012;11:1006–12. doi: 10.1016/S1474-4422(12)70191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walhovd KB, Fjell AM, Sørensen Ø, et al. Genetic risk for alzheimer disease predicts hippocampal volume through the human lifespan. Neurol Genet. 2020;6:e506. doi: 10.1212/NXG.0000000000000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mormino EC, Sperling RA, Holmes AJ, et al. Polygenic risk of alzheimer disease is associated with early- and late-life processes. Neurology. 2016;87:481–8. doi: 10.1212/WNL.0000000000002922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabuncu MR, Buckner RL, Smoller JW, et al. The association between a polygenic alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex. 2012;22:2653–61. doi: 10.1093/cercor/bhr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grasby KL, Jahanshad N, Painter JN, et al. The genetic architecture of the human cerebral cortex. Science. 2020;367:eaay6690. doi: 10.1126/science.aay6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51:404–13. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and Implicates Aβ, Tau, immunity and lipid processing. Nat Genet. 2019;51:414–30. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 12.Axelrud LK, Santoro ML, Pine DS, et al. Polygenic risk score for Alzheimer’s disease: implications for memory performance and hippocampal volumes in early life. Am J Psychiatry. 2018;175:555–63. doi: 10.1176/appi.ajp.2017.17050529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foley SF, Tansey KE, Caseras X, et al. Multimodal brain imaging reveals structural differences in Alzheimer’s disease polygenic risk carriers: a study in healthy young adults. Biol Psychiatry. 2017;81:154–61. doi: 10.1016/j.biopsych.2016.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satizabal CL, Adams HHH, Hibar DP, et al. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet. 2019;51:1624–36. doi: 10.1038/s41588-019-0511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hibar DP, Stein JL, Renteria ME, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–9. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott LT, Sharp K, Alfaro-Almagro F, et al. Genome-wide association studies of brain imaging phenotypes in UK biobank. Nature. 2018;562:210–6. doi: 10.1038/s41586-018-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Human Molecular Genetics. 2014;23:R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desikan RS, Cabral HJ, Hess CP, et al. Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer’s disease. Brain. 2009;132:2048–57. doi: 10.1093/brain/awp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams HHH, Hibar DP, Chouraki V, et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. 2016;19:1569–82. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hibar DP, Adams HHH, Jahanshad N, et al. Novel genetic Loci associated with hippocampal volume. Nat Commun. 2017;8:13624. doi: 10.1038/ncomms13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White T, El Marroun H, Nijs I, et al. Pediatric population-based neuroimaging and the generation R study: the intersection of developmental neuroscience and epidemiology. Eur J Epidemiol. 2013;28:99–111. doi: 10.1007/s10654-013-9768-0. [DOI] [PubMed] [Google Scholar]
- 22.Muetzel RL, Mulder RH, Lamballais S, et al. Frequent bullying involvement and brain morphology in children. Front Psychiatry. 2019;10:696. doi: 10.3389/fpsyt.2019.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casey BJ, Cannonier T, Conley MI, et al. The Adolescent Brain Cognitive Development (ABCD) study: imaging acquisition across 21 sites. Dev Cogn Neurosci. 2018;32:43–54. doi: 10.1016/j.dcn.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagler DJ, Hatton S, Cornejo MD, et al. Image processing and analysis methods for the adolescent brain cognitive development study. Neuroimage. 2019;202 doi: 10.1016/j.neuroimage.2019.116091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumann G, Loth E, Banaschewski T, et al. The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry. 2010;15:1128–39. doi: 10.1038/mp.2010.4. [DOI] [PubMed] [Google Scholar]
- 26.Boyd A, Golding J, Macleod J, et al. Cohort profile: the ‘children of the 90S’-The index offspring of the avon longitudinal study of parents and children. Int J Epidemiol. 2013;42:111–27. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser A, Macdonald-Wallis C, Tilling K, et al. Cohort profile: the avon longitudinal study of parents and children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Northstone K, Lewcock M, Groom A, et al. The Avon longitudinal study of parents and children (ALSPAC): an update on the enrolled sample of index children in 2019. Wellcome Open Res. 2019;4:51. doi: 10.12688/wellcomeopenres.15132.1. [version 1; peer review: 2 approved] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller KL, Alfaro-Almagro F, Bangerter NK, et al. Multimodal population brain imaging in the UK biobank prospective epidemiological study. Nat Neurosci. 2016;19:1523–36. doi: 10.1038/nn.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawlor DA. Commentary: two-sample mendelian randomization: opportunities and challenges. Int J Epidemiol. 2016;45:908–15. doi: 10.1093/ije/dyw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–80. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 32.Burgess S, Thompson SG, CRP CHD Genetics Collaboration Avoiding bias from weak instruments in mendelian randomization studies. Int J Epidemiol. 2011;40:755–64. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 33.Burgess S, Labrecque JA. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur J Epidemiol. 2018;33:947–52. doi: 10.1007/s10654-018-0424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarzer G. Meta: an R package for meta-analysis. R News. 2015;7:40–5. doi: 10.1007/978-3-319-21416-0. [DOI] [Google Scholar]
- 35.Stata press. STATA statistical software: release 16. 2019 [Preprint] [Google Scholar]
- 36.Sadreev II, Elsworth BL, Mitchell RE, et al. Navigating sample overlap, winner’s curse and weak instrument bias in mendelian randomization studies using the UK biobank. medRxiv. 2021 doi: 10.1101/2021.06.28.21259622. [Preprint] [DOI] [Google Scholar]
- 37.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. 2015;44:512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowden J, Davey Smith G, Haycock PC, et al. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985–98. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wasserstein RL, Lazar NA. The ASA statement on p-Values: context, process, and purpose. Am Stat. 2016;70:129–33. doi: 10.1080/00031305.2016.1154108. [DOI] [Google Scholar]
- 43.Sterne JA, Davey Smith G. Sifting the evidence—what’s wrong with significance tests? BMJ. 2001;322:226–31. doi: 10.1136/bmj.322.7280.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White T, Muetzel RL, El Marroun H, et al. Paediatric population neuroimaging and the generation R study: the second wave. Eur J Epidemiol. 2018;33:99–125. doi: 10.1007/s10654-017-0319-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White T, Jansen PR, Muetzel RL, et al. Automated quality assessment of structural magnetic resonance images in children: comparison with visual inspection and surface-based reconstruction. Hum Brain Mapp. 2018;39:1218–31. doi: 10.1002/hbm.23911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp TH, McBride NS, Howell AE, et al. Population neuroimaging: generation of a comprehensive data resource within the ALSPAC pregnancy and birth cohort. Wellcome Open Res. 2020;5:203. doi: 10.12688/wellcomeopenres.16060.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desikan RS, Schork AJ, Wang Y, et al. Polygenic overlap between C-reactive protein, plasma lipids, and alzheimer disease. Circulation. 2015;131:2061–9. doi: 10.1161/CIRCULATIONAHA.115.015489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Killiany RJ, Hyman BT, Gomez-Isla T, et al. MRI measures of entorhinal cortex vs hippocampus in preclinical AD. Neurology. 2002;58:1188–96. doi: 10.1212/wnl.58.8.1188. [DOI] [PubMed] [Google Scholar]
- 49.Tan CH, Bonham LW, Fan CC, et al. Polygenic hazard score, amyloid deposition and alzheimer’s neurodegeneration. Brain. 2019;142:460–70. doi: 10.1093/brain/awy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Köhler S, Black SE, Sinden M, et al. Memory impairments associated with hippocampal versus parahippocampal-gyrus atrophy: an MR volumetry study in Alzheimer’s disease. Neuropsychologia. 1998;36:901–14. doi: 10.1016/s0028-3932(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 51.Foo H, Thalamuthu A, Jiang J, et al. Associations between Alzheimer’s disease polygenic risk scores and hippocampal subfieldvolumes in 17,161 UK biobank participants. Neurobiol Aging. 2021;98:108–15. doi: 10.1016/j.neurobiolaging.2020.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Macdonald KE, Bartlett JW, Leung KK, et al. The value of hippocampal and temporal horn volumes and rates of change in predicting future conversion to ad. Alzheimer Dis Assoc Disord. 2013;27:168–73. doi: 10.1097/WAD.0b013e318260a79a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Squire LR, Stark CEL, Clark RE. The medial temporal lobe. Annu Rev Neurosci. 2004;27:279–306. doi: 10.1146/annurev.neuro.27.070203.144130. [DOI] [PubMed] [Google Scholar]
- 54.Diana RA, Yonelinas AP, Ranganath C. Imaging recollection and familiarity in the medial temporal lobe: a three-component model. Trends Cogn Sci. 2007;11:379–86. doi: 10.1016/j.tics.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Aggleton JP, Pralus A, Nelson AJD, et al. Thalamic pathology and memory loss in early Alzheimer’s disease: moving the focus from the medial temporal lobe to papez circuit. Brain. 2016;139:1877–90. doi: 10.1093/brain/aww083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Opitz B, Friederici AD. Interactions of the hippocampal system and the Prefrontal cortex in learning language-like rules. Neuroimage. 2003;19:1730–7. doi: 10.1016/s1053-8119(03)00170-8. [DOI] [PubMed] [Google Scholar]
- 57.Novellino F, López ME, Vaccaro MG, et al. Association between hippocampus, thalamus, and caudate in mild cognitive impairment APOEε4 carriers: a structural covariance MRI study. Front Neurol. 2019;10:1303. doi: 10.3389/fneur.2019.01303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mishra S, Blazey TM, Holtzman DM, et al. Longitudinal brain imaging in preclinical alzheimer disease: impact of APOE E4 genotype. Brain. 2018;141:1828–39. doi: 10.1093/brain/awy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Espeseth T, Westlye LT, Fjell AM, et al. Accelerated age-related cortical thinning in healthy carriers of apolipoprotein E E4. Neurobiol Aging. 2008;29:329–40. doi: 10.1016/j.neurobiolaging.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 60.Espeseth T, Westlye LT, Walhovd KB, et al. Apolipoprotein E E4-related thickening of the cerebral cortex modulates selective attention. Neurobiol Aging. 2012;33:304–322. doi: 10.1016/j.neurobiolaging.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 61.Calsolaro V, Edison P. Neuroinflammation in Alzheimer’s disease: current evidence and future directions. Alzheimers Dement. 2016;12:719–32. doi: 10.1016/j.jalz.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 62.Stern Y, Barnes CA, Grady C, et al. Brain reserve, cognitive reserve, compensation, and maintenance: operationalization, validity, and mechanisms of cognitive resilience HHS public access. Neurobiol Aging. 2019;83:124–9. doi: 10.1016/j.neurobiolaging.2019.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonner-Jackson A, Mahmoud S, Miller J, et al. Verbal and non-verbal memory and hippocampal volumes in a memory clinic population. Alzheimers Res Ther. 2015;7:61. doi: 10.1186/s13195-015-0147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lancaster T, Creese B, Escott-Price V, et al. Proof-of-concept recall-by-genotype study of extremely low and high alzheimer’s polygenic risk reveals autobiographical deficits and cingulate cortex correlates. Alzheimers Res Ther. 2023;15:213. doi: 10.1186/s13195-023-01362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data may be obtained from a third party and are not publicly available. The ENIGMA consortium MRI summary measures from genetic association analyses of estimated total intracranial volume, subcortical structures as well as cortical thickness were requested online. The ABCD study data are openly available to qualified researchers for free (https://nda.nih.gov/abcd/request-access). Requests for Generation R data should be directed towards the management team of the Generation R study (secretariaat.genr@erasmusmc.nl), which has a protocol of approving data requests. For access to IMAGEN data, researchers may submit a request to the IMAGEN consortium (https://imagen-europe.com/resources/imagen-project-proposal/). ALSPAC details and data descriptions are available on their website (www.bristol.ac.uk/alspac/researchers/access), where applications for individual-level data can be made (managed access). UK Biobank data are available through a procedure described on their website (http://www.ukbiobank.ac.uk/using-the-resource/). The UCSD IRB approved all data collection protocols for ABCD. IRB number: 160091. In Generation R, all study protocols and measurements assessed in each wave of data collection were approved by the Medical Ethical Committee (MEC 198.782/2001/31) of the Erasmus MC, University Medical Center Rotterdam. The IMAGEN study was approved by the institutional ethics committee of Kings College London, University of Nottingham, Trinity College Dublin, University of Heidelberg, Technische Universität Dresden, Commissariat á l Energie Atomique et aux Energies Alternatives, and University Medical Center at the University of Hamburg in accordance with the Declaration of Helsinki. Ethics approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees and informed consent for the use of data collected via questionnaires and clinics was obtained from participants. UK Biobank is approved by the National Health Service National Research Ethics Service (ref 11/NW/0382; UK Biobank application number 48970). All analyses in this study used de-identified data, therefore no additional IRB approval was required. All necessary patient/participant consent has been obtained. Code is available at https://github.com/rskl92/AD_BRAIN_BIDIRECTIONAL_MR.