Summary
Intrinsically disordered domains represent attractive therapeutic targets because they play key roles in cancer as well as in neurodegenerative and infectious diseases. They are however considered undruggable because they do not form stable binding pockets for small molecules and therefore have not been prioritized in drug discovery. Under physiological solution conditions many biomedically relevant intrinsically disordered proteins undergo phase separation processes leading to the formation of mesoscopic highly dynamic assemblies, generally known as biomolecular condensates, that define environments that can be quite different from the solutions surrounding them. In what follows we review key recent findings in this area and show how biomolecular condensation can offer opportunities for modulating the activities of intrinsically disordered targets.
Introduction
Protein domains that do not fold into well-defined structures are said to be intrinsically disordered [1]. The conformations of this class of domains cannot be represented by a single structure and are best depicted by conformational ensembles that describe their structural heterogeneity [2]. The widespread nature of intrinsic disorder and the often important functions of intrinsically disordered domains challenge our understanding of how protein sequences encode biological functions. It also represents a challenge for the field of drug discovery because the tools used to target globular domains with small molecules may not entirely suit intrinsically disordered ones [3].
This class of domains can have a propensity to phase separate into biomolecular condensates often formed by liquid liquid phase separation [4,5]. This phenomenon leads to the formation of dynamic mesoscopic assemblies, stabilized by a large number of weak transient non-covalent interactions, that are liquid and generate unique chemical environments [6]. Our understanding of how biomolecular condensation may allow specific functions to emerge is likely still incomplete; it is nevertheless already clear that they can act as reservoirs of primed inactive protein [7], as scaffolds to facilitate protein protein interactions [8,9] and as molecular sieves to regulate molecular traffic through biological membranes [10,11], among other functions [12].
It is thus natural that this phenomenon has raised substantial interest in the field of drug discovery, as evidenced by the foundation of a number of biotechnology companies with this focus [13]. Indeed, understanding how small molecules partition in biomolecular condensates and whether they can be used to modify the composition, stabilities, rates of formation and physical properties of these assemblies holds substantial promise for challenging indications [14] and may allow using small molecules to modify the activity of therapeutic targets currently considered undruggable.
The free energy landscape of intrinsically disordered proteins
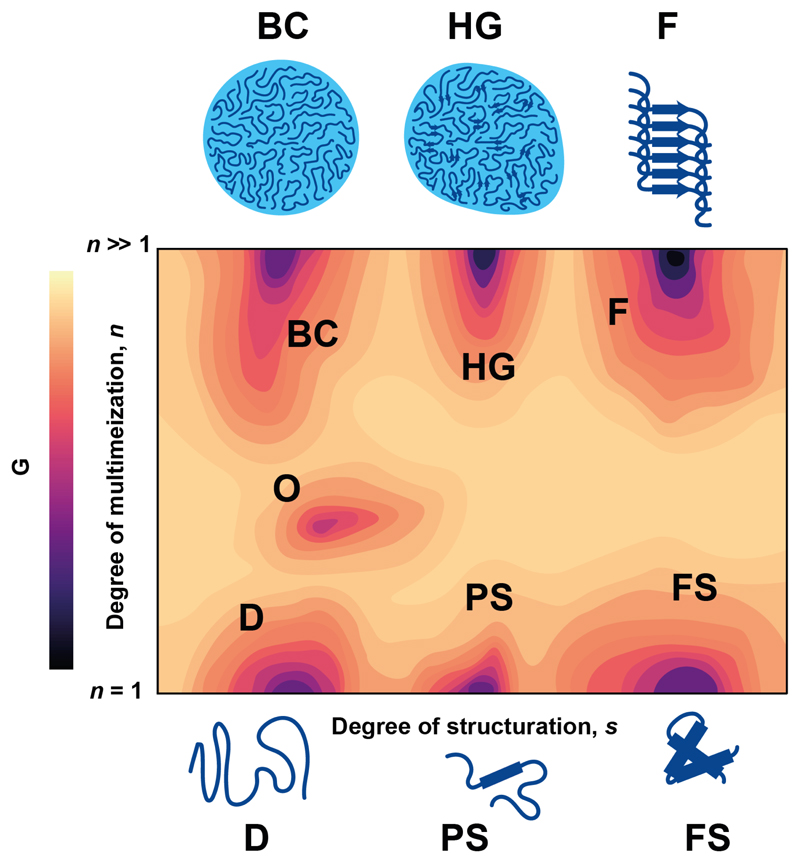
Free energy landscapes are useful to describe the conformational properties of intrinsically disordered proteins and of the multimeric assemblies that they can form [15]. They represent the free energy of the protein as a function of its conformation and are usually plotted in 3D as contour plots. The free energy is plotted in the z axis and the conformational space available to the protein in the xy plane, projected on two structural descriptors such as, for intrinsically disordered proteins, the degrees of structuration, s, and multimerization, n (Fig. 1). Stable states correspond to free energy minima and the frequency of transitions between them is given by the height of the free energy barriers.
Figure 1.
Generic free energy landscape of an intrinsically disordered protein with a propensity to condensate where the free energy of a protein molecule (G) is represented as a function of its degree of structuration (s) and multimerization (n). The letters represent the different minima that may be populated by such molecule, that are illustrated by representative conformations and include highly disordered state (D), partially and fully structures states (PS, FS), an oligomeric state (O), a biomolecular condensate (BC), a hydrogel (HG) and a fibril (F). Changes in the free energy of any state caused by interaction with a small molecule can lead to population shifts or changes in kinetic stability that can be used to alter the propensity of the protein to interact with a binding partner, form condensates or form fibrillar aggregates as shown in Figure 2.
The landscape expected for an intrinsically disordered protein undergoing biomolecular condensation is shown in Figure 1. In the region of the landscape corresponding to no multimerization (n=1) the minima correspond to the various states that may be populated by monomeric intrinsically disordered proteins, such as disordered (D), partially (PS) and fully structured (FS) states. Since they have similar free energies and the barriers connecting them are low, the structural properties of the intrinsically disordered protein can abruptly change upon population shifts caused for example by changes in solution conditions [16], post-translational modifications [17] or by interactions with other molecules [18,19].
In the region corresponding to multimers (n ≫1) minima may correspond to biomolecular condensates (BCs), glassy solids or hydrogels (HGs) and fibrils (Fs). Intrinsically disordered proteins that form condensates by liquid-liquid phase separation are thought to remain disordered [20,21] and can diffuse across the liquid-liquid interface and in the bulk of the condensate. Upon fibrillization, by contrast, protein molecules change conformation to form quaternary structures stabilized by intermolecular hydrogen bonds (F) in which they occupy permanent positions [22]. Hydrogels are in an intermediate situation both in terms of structuration and dynamism [12].
Contrary to what is the case for monomeric intrinsically disordered proteins, the stable states of biomolecular condensates can have quite different thermodynamic stabilities. Biomolecular condensates formed by liquid-liquid phase separation are in fact thought to be kinetically but not thermodynamically stable relative to fibrillization. Indeed amyloid fibrils represent the most thermodynamically stable state available to protein sequences and their formation is in general irreversible [23]. The barriers connecting such states are high due to their relatively high density and the polymeric nature of their components [24].
Small molecules can reshape the energy landscapes of intrinsically disordered proteins that form biomolecular condensates
At equilibrium the populations of the various states accessible to a protein depend on their relative free energies. Decreasing the free energy of a specific state, for example by selectively targeting it with a small molecule, can be used to reshape the energy landscape for therapeutic intervention [3]. For a simple two-state system such as a globular protein in exchange with the corresponding unfolded state, for example, targeting the globular state with small molecule chemical chaperones stabilizes the protein against proteolytic degradation and aggregation by decreasing the population of the unfolded protein that is a precursor of both processes [25,26].
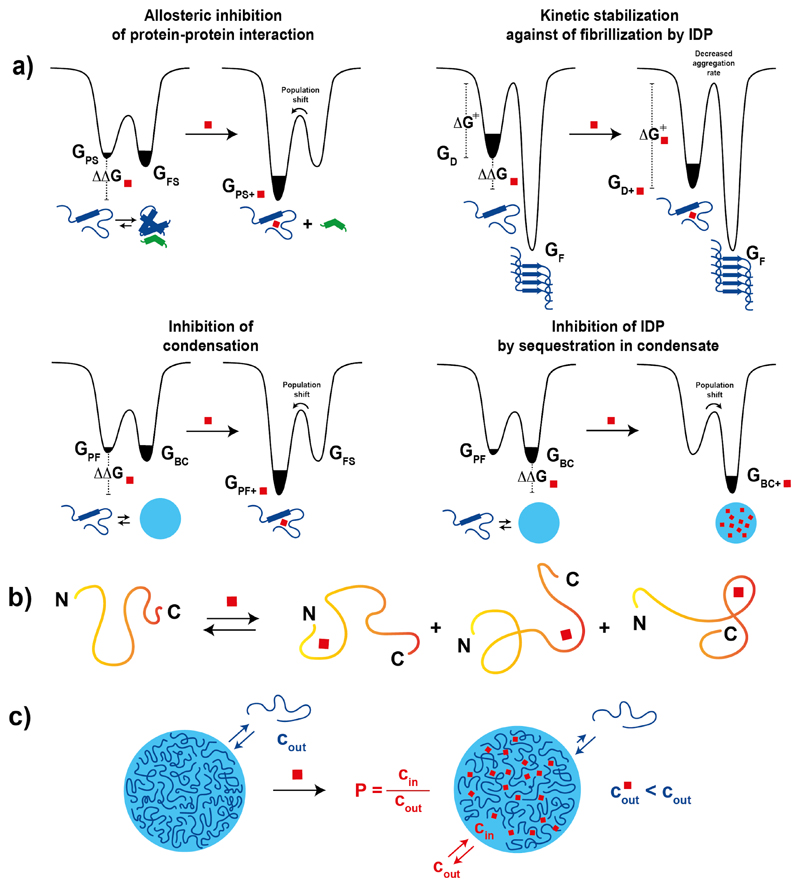
In a generalization of this mechanism of action, targeting a specific state of an intrinsically disordered protein, including multimeric states, with small molecules can be useful for different therapeutic purposes, as shown in Figure 2a. For intrinsically disordered proteins that fold upon interaction with a binding partner, for instance, small molecules that stabilize an alternate, binding-incompetent conformation will act as allosteric inhibitors of the interaction [27]. For intrinsically disordered proteins that can phase separate into biomolecular condensates stabilizing the monomer state with small molecules can inhibit the condensation process [28], whereas stabilizing the condensate will have the opposite effect, thus inhibiting the functions of the monomeric protein. Finally, although it is unlikely that small molecule binding to an intrinsically disordered protein will abolish amyloid formation, it can kinetically stabilize the monomer and thus decrease its aggregation propensity [29]. Given that these processes play key roles in the biological functions of intrinsically disordered proteins [4], as well as in disease [30], it is necessary to develop tools, both conceptual and technical, allowing us to direct small molecules to all the relevant states that they populate.
Figure 2.
a) Energy landscape of an intrinsically disordered protein before and after small molecule binding illustrating how it can inhibit protein-protein interactions, provide kinetic stability against fibril formation as well as cause population shifts that promote or suppress biomolecular condensation. b) Schematic illustration of a generalized mechanism for the interaction between small molecules and intrinsically disordered proteins derived from both experimental and computational studies. c) Schematic representation of a biomolecular condensate, of the exchange of protein molecules from the biomolecular condensate to the surrounding solution and of the effect of small molecule binding to an intrinsically disordered protein undergoing biomolecular condensation according to the polyphasic linkage framework.
We direct the reader to other reviews to gain a good understanding of the state of the art in targeting small molecules to orthosteric and allosteric sites in fully structured states of intrinsically disordered proteins (FS) [31] and to sites of both primary and secondary nucleation in amyloid fibrils (F) [32]. Although targeting such states, even when they exist at low population, can allow modulating structural and functional properties of intrinsically disordered proteins, we here focus our attention on the challenging and relatively unexplored goals of directing small molecules to their disordered and partially structured states (D, PF) (Fig. 2a,b) as well as to biomolecular condensates (Fig. 2c).
Targeting disordered states with small molecules
Targeting intrinsically disordered regions with small molecules represents a formidable challenge for drug discovery [33–35]. These proteins do not display stable, well-defined binding pockets and as a consequence they are generally considered undruggable therapeutic targets and thus not prioritized by the pharmaceutical industry. Despite this, due to their potential as targets for highly relevant disease areas, including neurodegeneration and oncology [36], a number of drug discovery programs have explored this enticing prospect in different ways, with promising results (Table 1).
Table 1. Small molecule inhibitors of disordered states of proteins with proven in vivo efficacy.
| Target | Disease family | Compound | Identification | In vivo efficacy | Clinical trials (Phase) | Structure | Ref. |
|---|---|---|---|---|---|---|---|
| ɑ-synuclein | Parkinson’s disease | Fasudil | Drug repurposing | Improved motor and cognitive functions at 10 and 30 mg/kg | Approved for use in humans but not assayed for Parkinson’s disease |
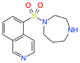
|
[37] |
| Aβ | Alzheimer’s disease | SEN1576 | Rational design | Reduced deficits in in vivo long-term potentiation and memory at 0.3 and 1 mg/kg | - |
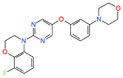
|
[38] |
| AR | Castration resistant prostate cancer | EPI-7170 | Second generation lead compound from a phenotypic screen | Tumor growth inhibition at 25 mg/kg | NCT044212221 (1) |
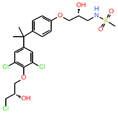
|
[39] |
| c-Myc | Cancer | MYCMI-6 | Cell-based protein interaction screen | Apoptosis induction and reduction of tumor cell proliferation and microvascularity at 20 mg/kg | - |
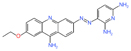
|
[40] |
| EN4 | Covalent ligand screen | Tumor growth inhibition at 50 mg/kg | - |
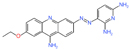
|
[41] | ||
| EWS-FLI1 | Ewing’s sarcoma | TK216 | Rational design | Tumor growth inhibition at 100 mg/kg |
NCT02657005 (1) |
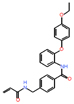
|
[42] |
| NUPR1 | Pancreatic adenocarcinoma | ZZW-115 | Ligand-based design | Tumor growth arrested at a 5 mg/kg | - |
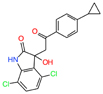
|
[43] |
| PTP1B | Diabetes, obesity | MSI-1436 | Serendipity | Suppresses appetite, reduces body weight and improves plasma insulin at 5-10 mg/kg | Completed: NCT00806338 NCT00606112 NCT00509132 Discontinued: NCT02524951 (1) |
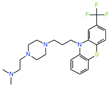
|
[44, 45] |
| Tau | Alzheimer’s disease | TRx0237 | In vitro aggregation assay | Reduces the brain atrophy rate at 4 mg/kg twice a day. |
NCT03446001 (1) |
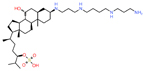
|
[46, 47] |
Clinical trial of EPI-7386 (structure not available).
As structure-based drug discovery cannot in principle be used to target intrinsically disordered proteins one approach is to rely on screens of inhibitors of protein-protein interactions and on phenotypic screens. Examples of this for oncology include the discovery of a small molecule that targets the intrinsically disordered region of p53 interacting with MDM-2 [48], of direct binders of c-Myc that inhibit its interaction with Max [40,49–52], of a small molecule targeting EWS-FLI1 that inhibits its interaction with RNA helicase A [53,54], of an allosteric inhibitor of PTP1B that targets its intrinsically disordered domain [44] and of a direct inhibitor of the transactivation domain of the androgen receptor (AR) [55,56].
Nuclear magnetic resonance (NMR) is a powerful tool to study weak interactions and has recently been used to identify small molecules that bind to protein p27 [19,57]. Although the affinity was weak, in the mM range, the authors were able to show how titrating the inhibitor destabilized the interaction between p27 and Cdk2/cyclin A [57], establishing a proof of concept. Identifying small molecules stabilizing the structural properties of intrinsically disordered proteins against thermal denaturation has also been used for screening, leading to a compound interacting with NUPR1 that after optimization has shown promising anticancer activity [43,58].
Although, on average, intrinsically disordered proteins are devoid of the structural features associated with druggability, they may transiently populate collapsed/structured conformations that instead may be druggable. The situation is reminiscent of the formation of cryptic binding pockets in globular proteins [59] and, as in this case, molecular simulations can help to reveal the relevant conformations, which can then subsequently be studied with tools of structure-based drug discovery such as molecular docking. In its first implementations this approach was used to investigate the druggability of the intrinsically disordered proteins Aβ42 [60], α-synuclein [61] and has more recently been used to discover inhibitors for c-Myc, MBD2 and p53 with some success [62–64].
Studying in detail how intrinsically disordered proteins interact with small molecule inhibitors identified by screening can help to unveil the intermolecular interactions that stabilize the complexes, the nature of the conformational changes that the small molecule can induce in the protein and thus the molecular basis for selectivity. Several academic laboratories have focused their attention on inhibitors identified by two-hybrid screening to inhibit the interaction between c-Myc and Max by targeting c-Myc [49]. By using biophysical techniques such as NMR and circular dichroism (CD) it was found that different inhibitors appeared to recognize independently different motifs with partial helical secondary structure [65,66] suggesting that small molecules can be targeted to specific intrinsically disordered sequences.
Similarly, a study of the interaction between a small molecule inhibitor, EPI-001, and the transactivation domain of AR by using solution NMR showed how this small molecule interacts with a sub-domain formed by three partially folded helices (Tau-5) but not with a similar one formed by two such helices (Tau-1), also supporting the idea that it is possible to target small molecules to intrinsically disordered proteins with some degree of selectivity. In this specific case it was found that this small molecule did not interact with the three partially helical sequences independently, suggesting the formation of a binding pocket involving residues found in at least two of them [67].
Although small molecule binding can change the conformation of intrinsically disordered targets [65] an analysis of the biophysical properties of these complexes clearly indicates that they do not lose their disordered character upon binding, precluding the obtention of a structure of the complex by using conventional structural biology methods. In this scenario molecular simulations represent powerful tools to complement the information obtained experimentally. The most studied system is again that formed by c-Myc and its small molecule inhibitors and the results obtained by different laboratories [68–70] consistently indicate that the inhibitors do not have a single binding pose, that they can interact with different motifs in the disordered target, in different conformations, and that they have a moderate effect on its conformational ensemble (Fig. 2b); the study of a similar system such as p27 led to equivalent results [71]. A very recent investigation of the binding of fasudil and 49 of its analogs to α-synuclein [37] showed good agreement with the results of NMR experiments, highlighting that molecular simulations may be useful in the future to guide the optimization of hits for drug discovery [72].
Covalent inhibition can be attractive for intrinsically disordered proteins because it can alleviate weak affinity issues. It requires the presence of a nucleophilic side chain in the target, typically a Cys or Lys, and of a warhead moiety in the inhibitor, typically an electrophilic group that may be generated in situ [73]. The modification of the structure of the intrinsically disordered target produced by covalent modification can be easily detected by mass spectrometry and lead to target inhibition by the mechanisms shown in Figure 2a. In addition it can directly inhibit the target if it modifies a residue in a motif mediating its interaction with a binding partner. This orthosteric mechanism of inhibition is less plausible with non-covalent inhibitors due to the non-druggable nature of the extended conformations involved in protein-protein interactions. A number of small molecule inhibitors of intrinsically disordered proteins appear to act as covalent inhibitors such as oleocanthal, that inhibits tau aggregation [74], baicalein, that inhibits α-synuclein aggregation [75], EPI-001, that binds to transactivation domain of AR [56] and nimbolide, that inhibits RNF114, a E3 ubiquitin ligase, and therefore stabilizes its substrates [76]. Very recently a systematic search for covalent inhibitors of the interaction between c-myc and Max lead to the identification of EN4, a compound bearing an acrylamide warhead, that reacted with some selectivity with Cys 171 in c-Myc and showed promising antiproliferative properties [41].
Targeting biomolecular condensates with small molecules
One defining feature of biomolecular condensates produced by liquid-liquid separation that differentiates them from conventional protein aggregates such as amyloid fibrils is their liquid character (Fig. 2c) [4]. Similarly to what is the case in the liquid-liquid extraction procedures used in the chemistry laboratory to isolate reaction products, small drug-like molecules can distribute between specific condensates and the surrounding solution depending on their partition coefficient P (Fig. 2c) [6,77]. In doing so they may change the properties of the condensate in ways that could be used for drug discovery in various disease areas such as neurodegeneration, [24] oncology [77] and infectious diseases [30,78].
Like intrinsically disordered proteins, biomolecular condensates are unconventional drug targets, out of the reach of conventional structure-based drug discovery tools. In this scenario cell-based or phenotypic assays may be again used to identify small molecules that appear to target them [79]. In an example of this approach a library of drug-like molecules was screened for their ability to dissolve stress granules by using a cell-based multiparametric imaging assay. The resulting hits, lipoamide and lipoic acid, could potentially be used to treat amyotrophic lateral sclerosis (ALS) after further development. This currently incurable disease can be caused by mutations in intrinsically disordered proteins such as FUS [80] or TDP-43 [81] that accelerate liquid to solid transitions in stress granules [82].
In these initiatives it is important to characterize in as much detail as possible the mechanism by which small molecules modulate the relevant phase transition. This will be crucial to investigate whether the approach can be generalized to other relevant targets, to design robust assays amenable to high throughput mode and, finally and most importantly, to guide the optimization of the structure of the small molecules to maximize potency. This is a highly challenging endeavour, however, due to the difficulties associated with reproducing in vitro key properties of biomolecular condensates. Despite recent progress, enumerating their components, and their stoichiometries, for example, is non-trivial [83], as is reproducing the highly dynamic character that they can have in cells due to energy consumption processes [84].
In considering what the effect of small molecule partitioning in biomolecular condensates will be, it is relevant to invoke the phenomenon called polyphasic linkage, that links the phase separation properties of a specific protein with its interaction with ligands [85–87]. Specifically, when the strength of the interaction between a ligand and a protein is not the same in the various phases that the protein may form, addition of the ligand will change the phase diagram. Polyphasic linkage, similarly to allostery, does not put forward a specific mechanism for the correlation but it is plausible that it occurs due to differences in the strength of intermolecular interactions in the various phases, that can have both energetic and entropic causes as revealed by both experiments and simulations [87–89] and to changes in molecular structure upon phase separation [90] among other factors [14,82].
Outlook
Intrinsically disordered proteins and biomolecular condensates represent unconventional therapeutic targets, which will require the development of new concepts and new tools for drug discovery. Phenotypic or high-throughput screens have led to the identification of a number of small molecules that appear to target these protein states, which has led to great interest in understanding their detailed mechanisms of action. The results from the limited number of studies available clearly indicate that the interaction between small molecules and both intrinsically disordered proteins and biomolecular condensates has specific features that should be taken into consideration in designing screening assays, choosing libraries for screening and, especially, in optimizing the chemical structures of hits and leads during drug development.
As far as intrinsically disordered proteins are concerned much of the emphasis has been put on applying the tools of structure-based drug discovery, in an ensemble fashion, for virtual screening. Although intuitive, this approach assumes that drug action relies on the establishment of highly specific interactions between the drug and a druggable conformation of the target identified by clustering a conformational ensemble. It appears, however, that small molecules bind to disordered targets in different poses and conformations, by establishing different non-covalent interactions. As a consequence, the effect of changes in the chemical structure of hits and leads on the stability of the complex and the structure of the target can only be rationalized by considering the energy landscape of the monomeric disordered protein (Fig. 1B).
Even in cases where the interaction between small molecules and intrinsically disordered targets can be modeled for drug development, for example by using advanced sampling techniques of molecular simulations [71], the optimized small molecules are likely to interact with intrinsically disordered targets with weak affinity. Indeed the size of small molecules precludes by definition the establishment of a large number of simultaneous interactions with the target, or allows it only with a substantial entropic penalty upon partial folding of the target. It has been proposed that in certain cases the interaction between small molecules and disordered proteins could increase the entropy of the intrinsically disordered target, perhaps by releasing long range interactions [91] but it remains to be seen whether this mechanism is compatible with selective targeting [29].
It is becoming increasingly clear that, for a given intrinsically disordered protein forming biomolecular condensates, collapse and biomolecular condensation are favoured by similar solution conditions because they are stabilized by equivalent interactions, that are intramolecular in one case and intermolecular in the other [92] (Fig. 2c). Since the collapsed conformations of disordered proteins are those easiest to target with small drug-like molecules i.e. more druggable, it is possible that biomolecular condensates produced by liquid-liquid phase separation feature a high density of binding sites for small molecules with the appropriate functionalities. As we gain a more detailed understanding of the specific interactions that stabilize the biomolecular condensates representing therapeutic targets [93–96] we will be in a better position to design and optimize the structures of small molecules targeting these assemblies to modify their properties in ways that are useful for drug discovery.
Highlights.
Intrinsically disordered domains are challenging therapeutic targets because they do not form stable binding pockets for small molecules
Small molecules that bind intrinsically disordered domains typically interact with different motifs, in different poses and with low to moderate affinity and selectivity
Biomolecular condensates generate environments that can selectively accumulate small drug-like molecules
Targeting monomeric proteins and biomolecular condensates with small molecules can lead to populations shifts that alter protein phase equilibria
Acknowledgements
M.B. acknowledges a PhD fellowship within the PREBIST programme of the Barcelona Institute for Science and Technology (BIST), that has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 754558. M.F. acknowledges a PhD fellowship awarded by “la Caixa” foundation in the 2015 call of the International Doctoral Fellowships Programme “la Caixa” - Severo Ochoa. X.S. acknowledges funding from AGAUR (2017 SGR 324), MINECO (BIO2015-70092-R) and the European Research Council (CONCERT, contract number 648201). IRB Barcelona is the recipient of a Severo Ochoa Award of Excellence from MINECO (Government of Spain).
References
- 1.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fenwick RB, Esteban-Martín S, Salvatella X. Understanding biomolecular motion, recognition, and allostery by use of conformational ensembles. Eur Biophys J. 2011;40:1339–1355. doi: 10.1007/s00249-011-0754-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heller GT, Bonomi M, Vendruscolo M. Structural Ensemble Modulation upon Small-Molecule Binding to Disordered Proteins. J Mol Biol. 2018;430:2288–2292. doi: 10.1016/j.jmb.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [*The authors provide a clear and comprehensive review of how liquid-liquid phase separation leads to the formation of cellular assemblies with key biological functions.] [DOI] [PubMed] [Google Scholar]
- 5.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nott TJ, Craggs TD, Baldwin AJ. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat Chem. 2016;8:569–575. doi: 10.1038/nchem.2519. [*This work shows that molecules can exchange between the surrounding solution and the interior of biomolecular condensates, where they experience very different solution conditions that can cause them to undergo conformational changes.] [DOI] [PubMed] [Google Scholar]
- 7.Guillén-Boixet J, Buzon V, Salvatella X, Méndez R. CPEB4 is regulated during cell cycle by ERK2/Cdk1-mediated phosphorylation and its assembly into liquid-like droplets. eLife Sciences. 2016;5:e19298. doi: 10.7554/eLife.19298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell. 2017;169:1066–1077.:e10. doi: 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 9.Mitrea DM, Cika JA, Stanley CB, Nourse A, Onuchic PL, Banerjee PR, Phillips AH, Park C-G, Deniz AA, Kriwacki RW. Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat Commun. 2018;9:842. doi: 10.1038/s41467-018-03255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemke EA. The Multiple Faces of Disordered Nucleoporins. J Mol Biol. 2016;428:2011–2024. doi: 10.1016/j.jmb.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt HB, Görlich D. Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem Sci. 2016;41:46–61. doi: 10.1016/j.tibs.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357 doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 13.Dolgin E. Drug startups coalesce around condensates. Nat Biotechnol. 2021 doi: 10.1038/s41587-021-00828-4. [DOI] [PubMed] [Google Scholar]
- 14.Wheeler RJ. Therapeutics-how to treat phase separation-associated diseases. Emerg Top Life Sci. 2020 doi: 10.1042/ETLS20190176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobson CM, Sali A, Karplus M. Protein folding: A perspective from theory and experiment. Angew Chem Int Ed. 1998;37:868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 16.Marsh JA, Forman-Kay JD. Sequence determinants of compaction in intrinsically disordered proteins. Biophys J. 2010;98:2383–2390. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519:106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 18.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ban D, Iconaru LI, Ramanathan A, Zuo J, Kriwacki RW. A small molecule causes a population shift in the conformational landscape of an intrinsically disordered protein. J Am Chem Soc. 2017;139:13692–13700. doi: 10.1021/jacs.7b01380. [*This work shows how nuclear magnetic resonance can be used to describe the thermodynamics and kinetics of the population shift process caused by the binding of a small molecule to an intrinsically disordered protein, p27.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [*The authors show by using nuclear magnetic resonance in solution that phase-separated FUS has similar conformational properties in the phase separated compartment and in the surrounding, that is that condensation does not necessarily involve conformational changes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brady JP, Farber PJ, Sekhar A, Lin Y-H, Huang R, Bah A, Nott TJ, Chan HS, Baldwin AJ, Forman-Kay JD, et al. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc Natl Acad Sci U S A. 2017;114:E8194–E8203. doi: 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin AJ, Knowles TPJ, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas SL, Waudby CA, Mossuto MF, Meehan S, Gras SL, et al. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc. 2011;133:14160–14163. doi: 10.1021/ja2017703. [DOI] [PubMed] [Google Scholar]
- 24.Mathieu C, Pappu RV, Taylor JP. Beyond aggregation: Pathological phase transitions in neurodegenerative disease. Science. 2020;370:56–60. doi: 10.1126/science.abb8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connelly S, Choi S, Johnson SM, Kelly JW, Wilson IA. Structure-based design of kinetic stabilizers that ameliorate the transthyretin amyloidoses. Curr Opin Struct Biol. 2010;20:54–62. doi: 10.1016/j.sbi.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makley LN, McMenimen KA, DeVree BT, Goldman JW, McGlasson BN, Rajagopal P, Dunyak BM, McQuade TJ, Thompson AD, Sunahara R, et al. Pharmacological chaperone for α-crystallin partially restores transparency in cataract models. Science. 2015;350:674–677. doi: 10.1126/science.aac9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Follis AV, Hammoudeh DI, Daab AT, Metallo SJ. Small-molecule perturbation of competing interactions between c-Myc and Max. Bioorg Med Chem Lett. 2009;19:807–810. doi: 10.1016/j.bmcl.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, Hyman AA. ATP as a biological hydrotrope. Science. 2017;356:753–756. doi: 10.1126/science.aaf6846. [**The authors show that physiological concentrations of ATP, that acts as biological hydrotrope, strongly decreases the propensity of a range of intrinsically disordered proteins to phase separate into biomolecular condensates. This work illustrates how stabilizing monomeric intrinsically disordered proteins can efficiently prevent biomolecular condensation.] [DOI] [PubMed] [Google Scholar]
- 29.Heller GT, Aprile FA, Michaels TCT, Limbocker R, Perni M, Ruggeri FS, Mannini B, Löhr T, Bonomi M, Camilloni C, et al. Small-molecule sequestration of amyloid-β as a drug discovery strategy for Alzheimer’s disease. Science Advances. 2020;6:eabb5924. doi: 10.1126/sciadv.abb5924. [The authors show how the interaction of a small molecule with an intrinsically disordered protein can strongly decrease its propensity to aggregate into amyloid fibrils by kinetically stabilizing the monomeric state.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alberti S, Dormann D. Liquid-Liquid Phase Separation in Disease. Annu Rev Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [*The authors review published evidence that biomolecular condensates play key roles in disease classes of high biomedical interest and therefore represent therapeutic targets of great promise.] [DOI] [PubMed] [Google Scholar]
- 31.Kalyaanamoorthy S, Chen Y-PP. Structure-based drug design to augment hit discovery. Drug Discov Today. 2011;16:831–839. doi: 10.1016/j.drudis.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Arosio P, Vendruscolo M, Dobson CM, Knowles TPJ. Chemical kinetics for drug discovery to combat protein aggregation diseases. Trends Pharmacol Sci. 2014;35:127–135. doi: 10.1016/j.tips.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Santofimia-Castaño P, Rizzuti B, Xia Y, Abian O, Peng L, Velázquez-Campoy A, Neira JL, Iovanna J. Targeting intrinsically disordered proteins involved in cancer. Cell Mol Life Sci. 2020;77:1695–1707. doi: 10.1007/s00018-019-03347-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruan H, Sun Q, Zhang W, Liu Y, Lai L. Targeting intrinsically disordered proteins at the edge of chaos. Drug Discov Today. 2019;24:217–227. doi: 10.1016/j.drudis.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 35.Tsafou K, Tiwari PB, Forman-Kay JD, Metallo SJ, Toretsky JA. Targeting Intrinsically Disordered Transcription Factors: Changing the Paradigm. J Mol Biol. 2018;430:2321–2341. doi: 10.1016/j.jmb.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 36.Bushweller JH. Targeting transcription factors in cancer - from undruggable to reality. Nat Rev Cancer. 2019;19:611–624. doi: 10.1038/s41568-019-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatenhorst L, Eckermann K, Dambeck V, Fonseca-Ornelas L, Walle H, Lopes da Fonseca T, Koch JC, Becker S, Tönges L, Bähr M, et al. Fasudil attenuates aggregation of α-synuclein in models of Parkinson’s disease. Acta Neuropathol Commun. 2016;4:39. doi: 10.1186/s40478-016-0310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Hare E, Scopes DIC, Kim E-M, Palmer P, Spanswick D, McMahon B, Amijee H, Nerou E, Treherne JM, Jeggo R. Novel 5-aryloxypyrimidine SEN1576 as a candidate for the treatment of Alzheimer’s disease. Int J Neuropsychopharmacol. 2014;17:117–126. doi: 10.1017/S1461145713000886. [DOI] [PubMed] [Google Scholar]
- 39.Banuelos CA, Ito Y, Obst JK, Mawji NR, Wang J, Hirayama Y, Leung JK, Tam T, Tien AH, Andersen RJ, et al. Ralaniten Sensitizes Enzalutamide-Resistant Prostate Cancer to Ionizing Radiation in Prostate Cancer Cells that Express Androgen Receptor Splice Variants. Cancers. 2020;12 doi: 10.3390/cancers12071991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castell A, Yan Q, Fawkner K, Hydbring P, Zhang F, Verschut V, Franco M, Zakaria SM, Bazzar W, Goodwin J, et al. A selective high affinity MYC-binding compound inhibits MYC:MAX interaction and MYC-dependent tumor cell proliferation. Sci Rep. 2018;8:10064. doi: 10.1038/s41598-018-28107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boike L, Cioffi AG, Majewski FC, Co J, Henning NJ, Jones MD, Liu G, McKenna JM, Tallarico JA, Schirle M, et al. Discovery of a Functional Covalent Ligand Targeting an Intrinsically Disordered Cysteine within MYC. Cell Chem Biol. 2020 doi: 10.1016/j.chembiol.2020.09.001. [*This work shows how covalent inhibition is a valid approach for targeting intrinsically disordered proteins with small molecules. The authors identify an electrophilic compound that reacts with a cysteine side chain in MYC and decreases the transcriptional activity of this transcription factor, which is an important therapeutic target in oncology.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spriano F, Chung EYL, Gaudio E, Tarantelli C, Cascione L, Napoli S, Jessen K, Carrassa L, Priebe V, Sartori G, et al. The ETS Inhibitors YK-4-279 and TK-216 Are Novel Antilymphoma Agents. Clin Cancer Res. 2019;25:5167–5176. doi: 10.1158/1078-0432.CCR-18-2718. [DOI] [PubMed] [Google Scholar]
- 43.Santofimia-Castaño P, Xia Y, Lan W, Zhou Z, Huang C, Peng L, Soubeyran P, Velázquez-Campoy A, Abián O, Rizzuti B, et al. Ligand-based design identifies a potent NUPR1 inhibitor exerting anticancer activity via necroptosis. J Clin Invest. 2019;129:2500–2513. doi: 10.1172/JCI127223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnan N, Koveal D, Miller DH, Xue B, Akshinthala SD, Kragelj J, Jensen MR, Gauss C-M, Page R, Blackledge M, et al. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat Chem Biol. 2014;10:558–566. doi: 10.1038/nchembio.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lantz KA, Hart SGE, Planey SL, Roitman MF, Ruiz-White IA, Wolfe HR, McLane MP. Inhibition of PTP1B by trodusquemine (MSI-1436) causes fat-specific weight loss in diet-induced obese mice. Obesity. 2010;18:1516–1523. doi: 10.1038/oby.2009.444. [DOI] [PubMed] [Google Scholar]
- 46.Harrington CR, Storey JMD, Clunas S, Harrington KA, Horsley D, Ishaq A, Kemp SJ, Larch CP, Marshall C, Nicoll SL, et al. Cellular Models of Aggregation-dependent Template-directed Proteolysis to Characterize Tau Aggregation Inhibitors for Treatment of Alzheimer Disease. J Biol Chem. 2015;290:10862–10875. doi: 10.1074/jbc.M114.616029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melis V, Magbagbeolu M, Rickard JE, Horsley D, Davidson K, Harrington KA, Goatman K, Goatman EA, Deiana S, Close SP, et al. Effects of oxidized and reduced forms of methylthioninium in two transgenic mouse tauopathy models. Behav Pharmacol. 2015;26:353–368. doi: 10.1097/FBP.0000000000000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LGGC, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 49.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 50.Kiessling A, Sperl B, Hollis A, Eick D, Berg T. Selective inhibition of c-Myc/Max dimerization and DNA binding by small molecules. Chem Biol. 2006;13:745–751. doi: 10.1016/j.chembiol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 51.Choi SH, Mahankali M, Lee SJ, Hull M, Petrassi HM, Chatterjee AK, Schultz PG, Jones KA, Shen W. Targeted Disruption of Myc-Max Oncoprotein Complex by a Small Molecule. ACS Chem Biol. 2017;12:2715–2719. doi: 10.1021/acschembio.7b00799. [DOI] [PubMed] [Google Scholar]
- 52.Hart JR, Garner AL, Yu J, Ito Y, Sun M, Ueno L, Rhee J-K, Baksh MM, Stefan E, Hartl M, et al. Inhibitor of MYC identified in a Kröhnke pyridine library. Proc Natl Acad Sci U S A. 2014;111:12556–12561. doi: 10.1073/pnas.1319488111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erkizan HV, Kong Y, Merchant M, Schlottmann S, Barber-Rotenberg JS, Yuan L, Abaan OD, Chou T-H, Dakshanamurthy S, Brown ML, et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing's sarcoma. Nat Med. 2009;15:750–756. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zöllner SK, Selvanathan SP, Graham GT, Commins RMT, Hong SH, Moseley E, Parks S, Haladyna JN, Erkizan HV, Dirksen U, et al. Inhibition of the oncogenic fusion protein EWS-FLI1 causes G2-M cell cycle arrest and enhanced vincristine sensitivity in Ewing's sarcoma. Sci Signal. 2017;10 doi: 10.1126/scisignal.aam8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung J-K, Watt K, Tam T, Yang YC, Banuelos CA, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell. 2010;17:535–546. doi: 10.1016/j.ccr.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 56.Myung J-K, Bañuelos CA, Fernandez JG, Mawji NR, Wang J, Tien AH, Yang YC, Tavakoli I, Haile S, Watt K, et al. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest. 2013;123:2948–2960. doi: 10.1172/JCI66398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iconaru LI, Ban D, Bharatham K, Ramanathan A, Zhang W, Shelat AA, Zuo J, Kriwacki RW. Discovery of Small Molecules that Inhibit the Disordered Protein, p27(Kip1) Sci Rep. 2015;5:15686. doi: 10.1038/srep15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neira JL, Bintz J, Arruebo M, Rizzuti B, Bonacci T, Vega S, Lanas A, Velázquez-Campoy A, Iovanna JL, Abián O. Identification of a Drug Targeting an Intrinsically Disordered Protein Involved in Pancreatic Adenocarcinoma. Sci Rep. 2017;7:39732. doi: 10.1038/srep39732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vajda S, Beglov D, Wakefield AE, Egbert M, Whitty A. Cryptic binding sites on proteins: definition, detection, and druggability. Curr Opin Chem Biol. 2018;44:1–8. doi: 10.1016/j.cbpa.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu M, De Simone A, Schenk D, Toth G, Dobson CM, Vendruscolo M. Identification of small-molecule binding pockets in the soluble monomeric form of the Aβ42 peptide. J Chem Phys. 2013;139:035101. doi: 10.1063/1.4811831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tóth G, Gardai SJ, Zago W, Bertoncini CW, Cremades N, Roy SL, Tambe MA, Rochet J-C, Galvagnion C, Skibinski G, et al. Targeting the intrinsically disordered structural ensemble of α-synuclein by small molecules as a potential therapeutic strategy for Parkinson's disease. PLoS One. 2014;9:e87133. doi: 10.1371/journal.pone.0087133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu C, Niu X, Jin F, Liu Z, Jin C, Lai L. Structure-based Inhibitor Design for the Intrinsically Disordered Protein c-Myc. Sci Rep. 2016;6:22298. doi: 10.1038/srep22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruan H, Yu C, Niu X, Zhang W, Liu H, Chen L, Xiong R, Sun Q, Jin C, Liu Y, et al. Computational strategy for intrinsically disordered protein ligand design leads to the discovery of p53 transactivation domain I binding compounds that activate the p53 pathway. Chem Sci. 2020 doi: 10.1039/d0sc04670a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim MY, Na I, Kim JS, Son SH, Choi S, Lee SE, Kim J-H, Jang K, Alterovitz G, Chen Y, et al. Rational discovery of antimetastatic agents targeting the intrinsically disordered region of MBD2. Sci Adv. 2019;5:eaav9810. doi: 10.1126/sciadv.aav9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem Biol. 2008;15:1149–1155. doi: 10.1016/j.chembiol.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 66.Hammoudeh DI, Follis AV, Prochownik EV, Metallo SJ. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J Am Chem Soc. 2009;131:7390–7401. doi: 10.1021/ja900616b. [DOI] [PubMed] [Google Scholar]
- 67.De Mol E, Fenwick RB, Phang CTW, Buzón V, Szulc E, de la Fuente A, Escobedo A, García J, Bertoncini CW, Estébanez-Perpiñá E, et al. EPI-001, A Compound Active against Castration-Resistant Prostate Cancer, Targets Transactivation Unit 5 of the Androgen Receptor. ACS Chem Biol. 2016;11:2499–2505. doi: 10.1021/acschembio.6b00182. [*The authors show how it is possible to target with small molecules a partially folded sub-domain of an intrinsically disordered protein with some selectivity. EPI-001, the small molecule studies in this work, showed efficacy in a clinical trial for castration resistant prostate cancer.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michel J, Cuchillo R. The Impact of Small Molecule Binding on the Energy Landscape of the Intrinsically Disordered Protein C-Myc. PLoS One. 2012;7:e41070. doi: 10.1371/journal.pone.0041070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin F, Yu C, Lai L, Liu Z. Ligand clouds around protein clouds: a scenario of ligand binding with intrinsically disordered proteins. PLoS Comput Biol. 2013;9:e1003249. doi: 10.1371/journal.pcbi.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heller GT, Aprile FA, Bonomi M, Camilloni C, De Simone A, Vendruscolo M. Sequence Specificity in the Entropy-Driven Binding of a Small Molecule and a Disordered Peptide. J Mol Biol. 2017;429:2772–2779. doi: 10.1016/j.jmb.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 71.Herrera-Nieto P, Pérez A, De Fabritiis G. Small Molecule Modulation of Intrinsically Disordered Proteins Using Molecular Dynamics Simulations. J Chem Inf Model. 2020;60:5003–5010. doi: 10.1021/acs.jcim.0c00381. [DOI] [PubMed] [Google Scholar]
- 72.Robustelli P, Ibanez-de-Opakua A, Campbell-Bezat C, Giordanetto F, Becker S, Zweckstetter M, Pan AC, Shaw DE. Molecular basis of small-molecule binding to α-synuclein. Cold Spring Harbor Laboratory. 2021 doi: 10.1021/jacs.1c07591. [**This preprint exemplifies that molecular simulations of the interaction between small molecules and intrinsically disordered proteins can reproduce the results obtained by solution nuclear magnetic resonance and rationalize, to some extent, structure-activity relationships.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gersch M, Kreuzer J, Sieber SA. Electrophilic natural products and their biological targets. Nat Prod Rep. 2012;29:659–682. doi: 10.1039/c2np20012k. [DOI] [PubMed] [Google Scholar]
- 74.Li W, Sperry JB, Crowe A, Trojanowski JQ, Smith AB, 3rd, Lee VM-Y. Inhibition of tau fibrillization by oleocanthal via reaction with the amino groups of tau. J Neurochem. 2009;110:1339–1351. doi: 10.1111/j.1471-4159.2009.06224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu M, Rajamani S, Kaylor J, Han S, Zhou F, Fink AL. The Flavonoid Baicalein Inhibits Fibrillation of α-Synuclein and Disaggregates Existing Fibrils. J Biol Chem. 2004;279:26846–26857. doi: 10.1074/jbc.M403129200. [DOI] [PubMed] [Google Scholar]
- 76.Spradlin JN, Hu X, Ward CC, Brittain SM, Jones MD, Ou L, To M, Proudfoot A, Ornelas E, Woldegiorgis M, et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat Chem Biol. 2019;15:747–755. doi: 10.1038/s41589-019-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall'Agnese A, Oksuz O, Henninger JE, Shrinivas K, Sabari BR, et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386–1392. doi: 10.1126/science.aaz4427. [**This work shows how nuclear condensates generate environments where cancer therapeutics accumulate, proposes that such accumulation plays a role in their mode of action and establishes a link between the intermolecular interactions that stabilize such condensates and the chemical structures of the drugs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmid M, Speiseder T, Dobner T, Gonzalez RA. DNA virus replication compartments. J Virol. 2014;88:1404–1420. doi: 10.1128/JVI.02046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang MY, Markmiller S, Vu AQ, Javaherian A, Dowdle WE, Jolivet P, Bushway PJ, Castello NA, Baral A, Chan MY, et al. Small-Molecule Modulation of TDP-43 Recruitment to Stress Granules Prevents Persistent TDP-43 Accumulation in ALS/FTD. Neuron. 2019;103:802–819.:e11. doi: 10.1016/j.neuron.2019.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 81.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wheeler RJ, Lee HO, Poser I, Pal A, Doeleman T. Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. BioRxiv. 2019 [*This preprint represents pioneering work aiming at screening libraries of drug-like small molecules for their ability to dissolve stress granules associated with the development of ALS. Although the authors could not firmly establish the mode of action of the hits identified, this work represents an important proof of concept for this field.] [Google Scholar]
- 83.Ditlev JA, Case LB, Rosen MK. Who's In and Who's Out-Compositional Control of Biomolecular Condensates. J Mol Biol. 2018;430:4666–4684. doi: 10.1016/j.jmb.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zwicker D, Seyboldt R, Weber CA, Hyman AA, Julicher F. Growth and division of active droplets provides a model for protocells. Nat Phys. 2016;13:408. [Google Scholar]
- 85.Wyman J, Gill SJ. Ligand-linked phase changes in a biological system: applications to sickle cell hemoglobin. Proc Natl Acad Sci U S A. 1980;77:5239–5242. doi: 10.1073/pnas.77.9.5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Posey AE, Ruff KM, Harmon TS, Crick SL, Li A, Diamond MI, Pappu RV. Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J Biol Chem. 2018;293:3734–3746. doi: 10.1074/jbc.RA117.000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruff KM, Dar F, Pappu RV. Ligand Effects on Phase Separation of Multivalent Macromolecules. Cold Spring Harbor Laboratory. 2020 doi: 10.1073/pnas.2017184118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin Y-H, Song J, Forman-Kay JD, Chan HS. Random-phase-approximation theory for sequence-dependent, biologically functional liquid-liquid phase separation of intrinsically disordered proteins. J Mol Liq. 2017;228:176–193. [Google Scholar]
- 89.Babinchak WM, Dumm BK, Venus S, Boyko S, Putnam AA, Jankowsky E, Surewicz WK. Small molecules as potent biphasic modulators of protein liquid-liquid phase separation. Nat Commun. 2020;11:5574. doi: 10.1038/s41467-020-19211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peran I, Mittag T. Molecular structure in biomolecular condensates. Curr Opin Struct Biol. 2020;60:17–26. doi: 10.1016/j.sbi.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heller GT, Sormanni P, Vendruscolo M. Targeting disordered proteins with small molecules using entropy. Trends Biochem Sci. 2015;40:491–496. doi: 10.1016/j.tibs.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 92.Zeng X, Holehouse AS, Chilkoti A, Mittag T, Pappu RV. Connecting Coil-to-Globule Transitions to Full Phase Diagrams for Intrinsically Disordered Proteins. Biophys J. 2020;119:402–418. doi: 10.1016/j.bpj.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang J, Choi J-M, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell. 2018;174:688–699.:e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, Mittag T. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science. 2020;367:694–699. doi: 10.1126/science.aaw8653. [**In this work, by using biophysical methods and molecular simulations, the authors show the importance of interactions between aromatic residues for the propensity of intrinsically disordered proteins to phase separate into biomolecular condensates.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nguemaha V, Zhou H-X. Liquid-Liquid Phase Separation of Patchy Particles Illuminates Diverse Effects of Regulatory Components on Protein Droplet Formation. Sci Rep. 2018;8:6728. doi: 10.1038/s41598-018-25132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]