Abstract
In female mammals, one of the two X chromosomes becomes inactivated during development by X-chromosome inactivation (XCI). Although Polycomb repressive complex (PRC) 1 and PRC2 have both been implicated in gene silencing, their exact roles in XCI during in vivo development have remained elusive. To this end, we have studied mouse embryos lacking either PRC1 or PRC2. Here we demonstrate that the loss of either PRC has a substantial impact on maintenance of gene silencing on the inactive X chromosome (Xi) in extra-embryonic tissues, with overlapping yet different genes affected, indicating potentially independent roles of the two complexes. Importantly, a lack of PRC1 does not affect PRC2/H3K27me3 accumulation and a lack of PRC2 does not impact PRC1/H2AK119ub1 accumulation on the Xi. Thus PRC1 and PRC2 contribute independently to the maintenance of XCI in early post-implantation extra-embryonic lineages, revealing that both Polycomb complexes can be directly involved and differently deployed in XCI.
In mammals, differences in X-linked gene dosage between the sexes are compensated for by X-chromosome inactivation (XCI)1 that takes place during female embryogenesis. In the mouse, XCI happens in two waves2, a first wave of imprinted paternal XCI that initiates at the two- to four-cell stage and is maintained in extra-embryonic tissues, and a second wave of random paternal or maternal XCI that initiates at around embryonic day (E)5.0 in the inner cell mass, which gives rise to the epiblast/embryo proper. The X-inactive specific transcript (Xist) non-coding RNA that is transcribed from the Xist gene is both necessary and sufficient to trigger gene silencing during XCI3,4. Downstream epigenetic changes ensure that gene repression can be initiated on the Xist-coated future inactive X chromosome (Xi) and that this gene repression can be maintained on the Xi at subsequent developmental stages. The two waves of XCI differ in the extent and nature of these epigenetic modifications. Polycomb repressive complexes (PRCs) accumulate on the Xi during both waves of XCI. CpG islands (CGIs) are target DNA elements for both PRC1 and PRC2 (ref. 5), and this may contribute to PRC-mediated silencing of CGI-associated genes. CGIs can also be cytosine-methylated, and this is a characteristic of the Xi, although it occurs mainly in the embryo proper, during the second wave of XCI6 and is thought to be key to the highly stable silencing found in somatic cells7. In cells of extra-embryonic tissues such as the yolk sac, CGIs on the Xi remain hypomethylated2,6. Although gene silencing is slightly less stable in extra-embryonic tissues presumably partly because of CGI hypomethylation on the Xi, the inactive state is still globally maintained, implying that other epigenetic factors, such as Polycomb, are indeed important for Xi maintenance
When Xist RNA initially coats the future Xi, it recruits, via its A-repeat region, factors such as SPEN that mediate the initiation of gene silencing8–12. Xist coating also leads to the recruitment of both PRC1 and PRC2 (refs. 13–16), via the BC repeats of the Xist RNA. Interestingly, human XIST has been recently reported to recruit PRC1 and PRC2 via different domains of XIST RNA17, although it is not known whether this is also the case for mouse Xist. There are at least six PRC1 subcomplexes in mammals, characterized partly by different PCGF1–6 co-factor proteins, but all PRC1 subcomplexes contain RING1A/B, the two E3 ubiquitin ligases that catalyse monoubiquitination of H2A Lysine 119 (H2AK119ub1) (refs. 18,19). Mammalian PRC2 complex contains the core factors EED, SUZ12 and EZH1 or EZH2, with the catalytic subunit of EZH1/2 catalysing the trimethylation of H3 Lysine 27 (H3K27me3) (ref. 20). Both H2AK119ub1 and H3K27me3 marks become enriched on the Xi during XCI13–16. Previous analyses of Eed mutant mice revealed that, in the absence of Eed, the silencing of an X-linked green fluorescent protein transgene was not properly maintained in extra-embryonic tissues at post-implantation stages, from E5.5 to E7.5 (ref. 21). However, whether endogenous X-linked genes are also affected in a PRC2 mutant situation has remained unanswered. More recently, the PRC1 subcomplex PCGF3/5-PRC1, which is recruited directly by Xist RNA22, was found to be implicated in XCI on the basis of studies in embryonic stem cells (ESCs) and embryos22–24. Using an ESC model that recapitulates the second, random wave of XCI, it was shown that PCGF3/5-PRC1 is important at least to initiate Xist-mediated gene silencing on an autosome, although its implication in stable maintenance of gene silencing was not studied23,24. Importantly, it was also shown that PCGF3/5-PRC1, probably through RING1A/B-mediated H2A ubiquitination activity, regulates PRC2/H3K27me3 on the Xi at the initiation of XCI23,24. The above studies have implicated both PRC1 and PRC2 in XCI in vitro and in certain in vivo contexts; however, their respective roles and possible redundancy, especially for the maintenance of endogenous X-linked gene silencing in XCI in vivo, have remained poorly understood. In this study, we analysed mice that are mutated for the core subunits of each complex, PRC1 or PRC2, to uncover their relative implications in the maintenance of XCI. Our data indicate that both PRC1 and PRC2 are critical for XCI and that they independently contribute to maintaining the silencing of the Xi in extra-embryonic lineages in vivo.
Role of PRC1 in H2AK119ub1 and H3K27me3 accumulation on the Xi in E7.5 and E8.5 post-implantation mouse embryos
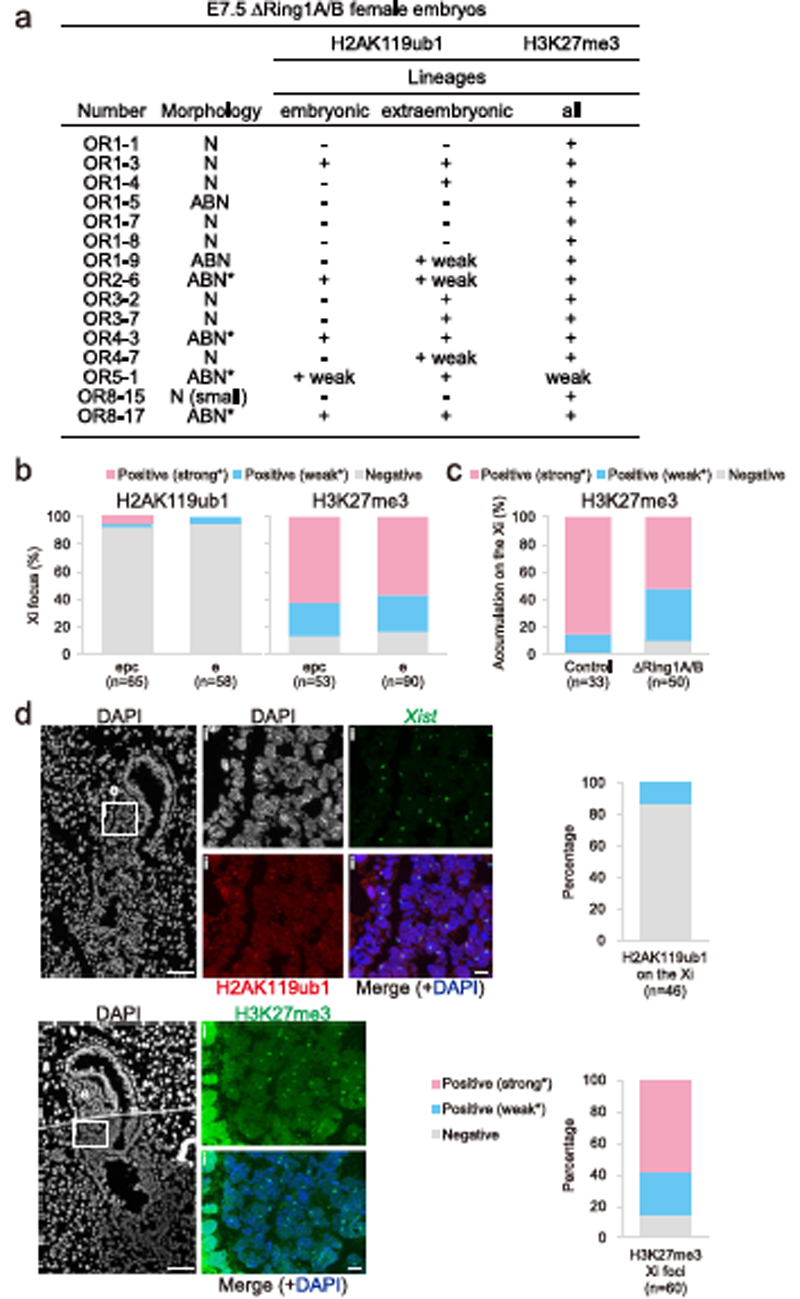
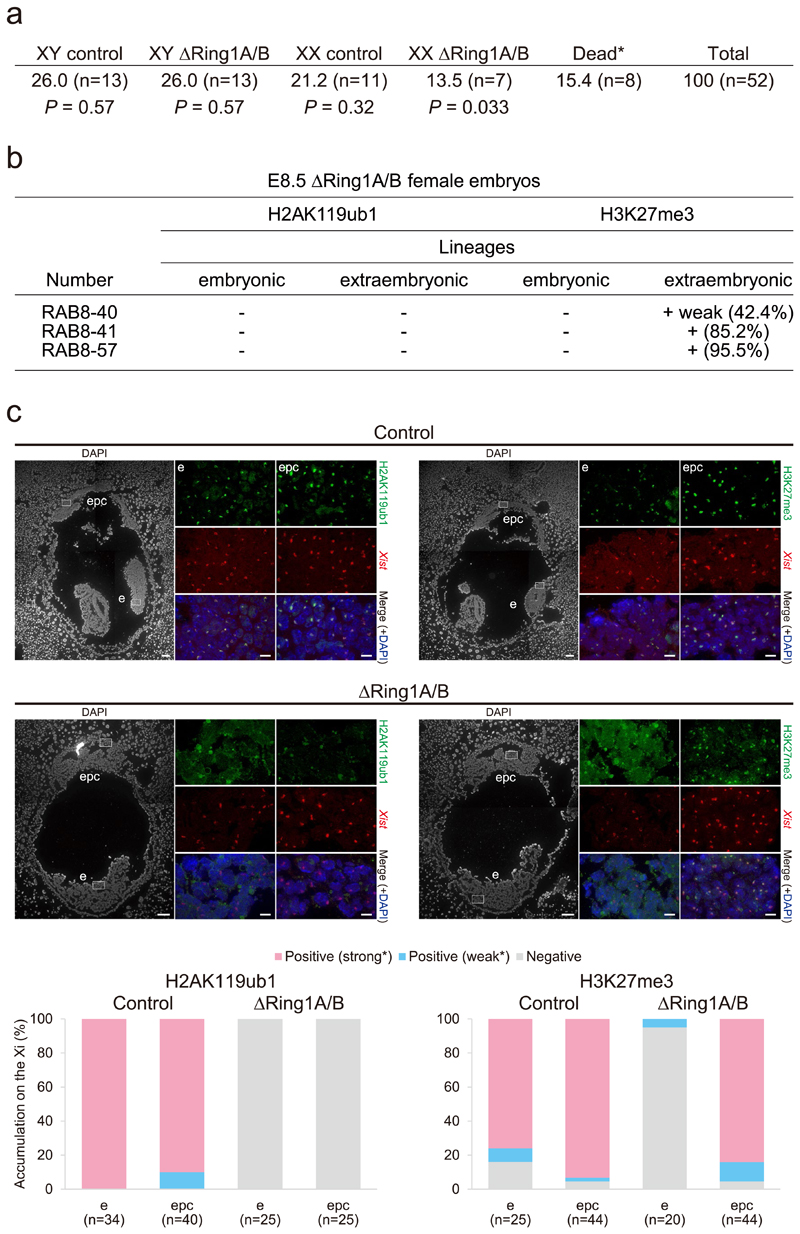
First, to examine the in vivo role of PRC1 in XCI, we analysed embryos from conditional PRC1 knockout mice that had been generated previously25. We used mice bearing a constitutively mutated Ring1A allele and a conditionally inactivated Ring1B allele. Using this mouse model, Ring1B is mutated upon tamoxifen-mediated activation of CreERT2, which creates PRC1-null (∆Ring1A/B) mice, thus allowing us to assess the role of PRC1 in XCI. Tamoxifen was injected at E5.5, and embryos were recovered at E7.5 (Fig. 1a). During this time window, imprinted XCI is already well established in extra-embryonic tissues and random XCI has just been established in the embryo proper26,27. We obtained a total of 15 female ∆Ring1A/B embryos from 68 analysed embryos (for genotyping, see Methods; Extended Data Fig. 1a). H2AK119ub1 accumulation on the Xi was assessed by immunofluorescence (IF) (Fig. 1b and Extended Data Fig. 1b) combined with Xist RNA fluorescence in situ hybridization (FISH) (Extended Data Fig. 1d), on all E7.5 embryo sections. H2AK119ub1 is normally highly enriched on the Xi at this stage of embryogenesis. Five of the 15 ∆Ring1A/B female embryos completely lacked H2AK119ub1 Xi enrichment in all embryonic and extra-embryonic lineages at E7.5 2 days post-deletion induction. The remaining ten embryos retained some H2AK119ub1, suggesting that the mark is progressively lost (Fig. 1b and Extended Data Fig. 1a–d). Regardless of the H2AK119ub1 Xi-accumulation status, all 15 ∆Ring1A/B embryos, including those lacking H2AK119ub1 on the Xi, displayed H3K27me3 accumulation on the Xi similar to their wild-type (WT) littermates. This suggested that H3K27me3 accumulation on the Xi at this stage of development might be independent of PRC1 (Fig. 1b and Extended Data Fig. 1a–d). This would contrast with previous reports that H3K27me3 accumulation on the Xi is dependent on PRC1/H2AK119ub1 in ES cells and peri-implantation embryos22–24. To assess whether Xi accumulation of H3K27me3 might be lost at a later stage in ∆Ring1A/B embryos, we examined ∆Ring1A/B embryos at E8.5. A substantially lower number of ∆Ring1A/B female embryos could be obtained, suggesting that they are dying at this stage (13.5%, P = 0.033; Extended Data Fig. 2a). Nevertheless, three of these mutant female embryos could be analysed using immuno-RNA FISH (Extended Data Fig. 2b,c). In control E8.5 females (n = 2), we could detect both H2AK119ub1 and H3K27me3 accumulation on the Xi in both embryonic and extra-embryonic tissues. In contrast, in all three ∆Ring1A/B E8.5 female embryos, no H2AK119ub1 accumulation could be detected on the Xi throughout the whole embryo, indicating that PRC1-mediated H2AK119ub1 was completely depleted from the Xi at this stage. On the other hand, comparable levels of H3K27me3 accumulation on the Xi were found in extra-embryonic ectoplacental cones (EPCs) in all three ∆Ring1A/B mutant female embryos (control 95.5% versus ∆Ring1A/B 95.5%) (Extended Data Fig. 2b,c). These data support the idea that deposition of H3K27me3 on the Xi is not dependent on PRC1/H2AK119ub1 at this post-implantation stage, at least in extra-embryonic tissues. However, in the embryonic tissues of these ∆Ring1A/B E8.5, H3K27me3 on the Xi could no longer be seen (control 84.0% versus ∆Ring1A/B 5.0%) (Extended Data Fig. 2b,c). It should be noted that at E8.5 the embryonic tissues of ∆Ring1A/B embryos are severely affected, with many small and abnormal nuclei. Nevertheless, the lack of H3K27me3 on the Xi in embryonic tissues of ∆Ring1A/B mutants is consistent with the previous study that showed H3K27me3 accumulation is lost in Pcgf3/5 mutant mouse embryos23.
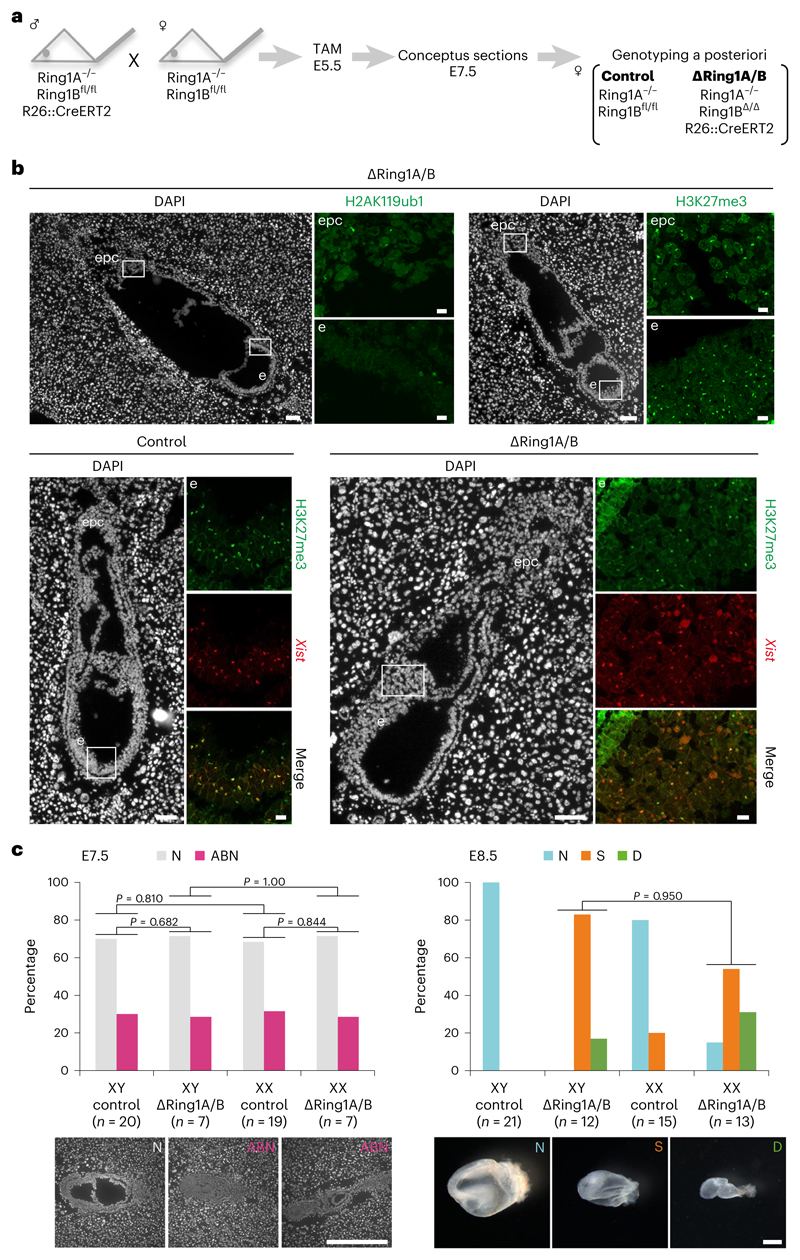
Fig. 1. Deletion of Ring1A/B (PRC1) does not impact H3K27me3 accumulation on the Xi in E7.5 embryos.
a, Schematic representation of ΔRing1A/B mouse deletion strategy. Tamoxifen (TAM) was injected intraperitoneally to pregnant mother at E5.5 for both control (without R26::CreERT2) and ΔRing1A/B (bearing R26::CreERT2). Embryos were recovered at E7.5 for the experiments. For more details, see Methods. b, ΔRing1A/B E7.5 embryos are deprived of H2AK119ub1, but not H3K27me3 on the Xi. Analyses on longitudinal cryosections; boxed regions are shown with higher magnification. Top: ΔRing1A/B E7.5 (OR1-1) embryo studied by IF for two histone marks, H2AK119ub1 (left) and H3K27me3 (right); consecutive sections; lack of H2AK119ub1 accumulation on the Xi and H3K27me3 accumulation on the Xi in most cells. For quantification, see Extended Data Fig. 1b. Four independent embryos were examined, and all showed similar results. Bottom: immuno-RNA FISH (H3K27me3 and Xist) for control (left) and another ΔRing1A/B (OR3-2) E7.5 embryos (right); similar accumulation of H3K27me3 on the Xi. For quantification, see Extended Data Fig. 1c. Eight independent embryos were examined, and all showed similar results. epc, ectoplacental cone; e, embryo proper. Scale bars: 100 μm (whole embryos) and 10 μm (enlarged images). c, Morphology of E7.5 and E8.5 control and ΔRing1A/B male and female embryos. Left: E7.5 embryos on sections; N, normal; ABN, abnormal. Scale bar, 500 μm. Sum of six independent litters. Right: E8.5 embryos in the yolk sac; N, normal; S, small; D, dying. Scale bar, 500 μm. Sum of nine independent litters. P values were calculated using one-sided binomial test.
Role of PRC1 in post-implantation development of female embryos
The impact of PRC1 depletion on female mouse development was examined at E7.5 and E8.5. At E8.5, evidence of female ∆Ring1A/B embryo lethality was found (Extended Data Fig. 2a). However, both male and female ∆Ring1A/B E8.5 embryos were small (>50% female, >80% male) or dying (>30% female, >10% male) compared with controls indicating a general role of PRC1 in the development of both sexes (Fig. 1c). At E7.5, male and female ∆Ring1A/B and control embryos were indistinguishable, on the basis of whole embryo sections. Mild abnormalities, probably due to tamoxifen injections, were observed in both ∆Ring1A/B and control embryos at E7.5 (Fig. 1c and Extended Data Fig. 1a). These data suggest that PRC1 depletion starting at E5.5 has a detrimental impact on embryogenesis between E7.5 and E8.5 in both sexes, with an earlier impact on viability of females compared with males, consistent with an essential role for PRC1 in XCI.
Role of PRC1 in XCI maintenance in E7.5 post-implantation embryo tissues
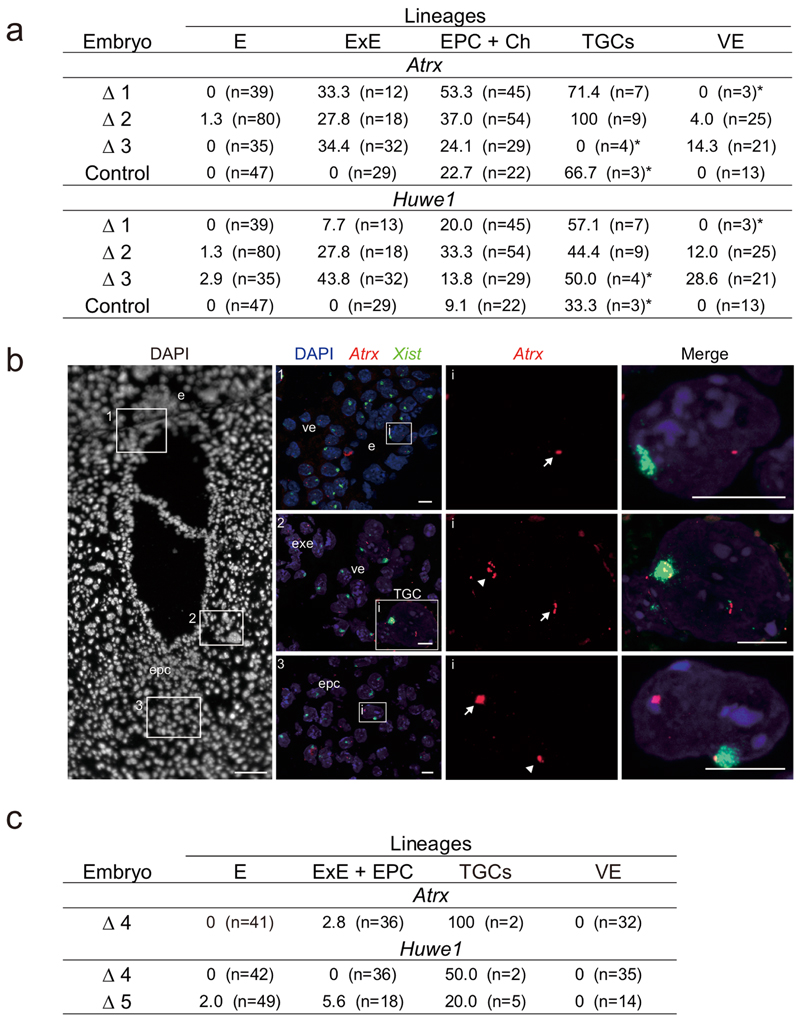
To assess the potential XCI defects in ∆Ring1A/B embryos, we investigated gene activity on the Xi in different lineages of ∆Ring1A/B and control embryos at E7.5, just before the abnormal morphologies in ∆Ring1A/B female embryos. At E7.5, random XCI in embryonic lineages is complete, and imprinted XCI in extra-embryonic lineages is already in full maintenance mode26,27. We analysed the expression of X-linked Atrx and Huwe1 by nascent transcript RNA FISH, together with Xist RNA. Both genes are silent on the Xi in most extra-embryonic and embryonic cells at this stage of development, with only a minor degree of escape from XCI in some cell types28. Three ∆Ring1A/B female E7.5 embryos showing total depletion for H2AK119ub1 were compared with one control female retaining H2AK119ub1 accumulation on the Xi (Fig. 2a and (Extended Data Fig. 3a,b). In control females, no escape from XCI (Atrx, 0%; Huwe1, 0%) was found in the epiblast/embryo proper, extra-embryonic ectoderm or visceral endoderm, and only a modest degree of escape (Atrx, 22.7%; Huwe1, 9.1%) was found in the EPC and the chorion (Fig. 2a and Extended Data Fig. 3a,b), consistent with our previous study in WT E7.5 embryos28. In contrast, in ∆Ring1A/B mutant embryos, we found a striking increase in cells showing biallelic Atrx and Huwe1 expression in all extra-embryonic lineages (extra-embryonic ectoderm, EPC + chorion, and visceral endoderm): Atrx (31.8%, 38.2% and 6.1%, respectively) and Huwe1 (26.4%, 22.4% and 13.5%, respectively) (Fig. 2a, Extended Data Fig. 3a, b). No Xi expression of Atrx and Huwe1 was observed in the embryo proper of ∆Ring1A/B embryos. Importantly, ∆Ring1A/B female mutants displaying incomplete loss of H2AK119ub1 on the Xi showed less Atrx and Huwe1 derepression from the Xi, pointing to the role of PRC1/H2AK119ub1 levels in maintaining Xi gene silencing (Extended Data Fig. 3c). In summary, loss of PRC1 activity and associated H2AK119ub1 results in Xi derepression of Atrx and Huwe1 in extra-embryonic lineages, but not in embryonic lineages.
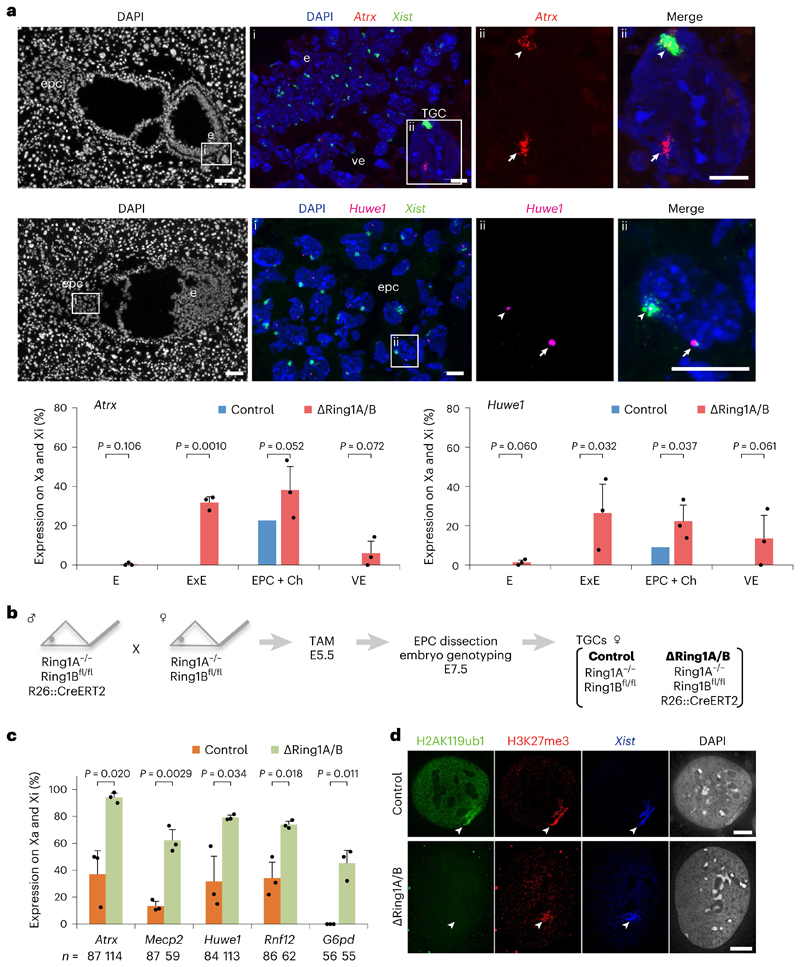
Fig. 2. Deletion of Ring1A/B (PRC1) results in loss of gene silencing on the Xi in extra-embryonic lineages.
a, X-linked gene expression on the Xi in extraembryonic tissues upon Ring1A/B deletion. Consecutive sections of ΔRing1A/B E7.5 embryo (Δ2) showing transcripts of two X-linked genes (Atrx and Huwe1) and Xist by RNA FISH. Top: Atrx and Xist. (i) Boxed area showing embryo proper (e), visceral endoderm (ve) and TGC. TGC has endoreplicated genome DNA and shows multi-spot RNA FISH signal (here, Atrx). (ii) Boxed region in i showing a TGC with biallelic Atrx expression (Xa and Xi). Middle: Huwe1 and Xist. (i) Boxed area showing ectoplacental cone (epc). (ii) Boxed region in i showing a cell with biallelic Huwe1 expression (Xa and Xi). Three independent embryos (Δ1: OR1–7, Δ2: OR1–8 and Δ3: OR8–15) were examined, and all showed similar results. Δ1 is shown in Extended Data Fig. 3b. Bottom: quantification of Atrx and Huwe1 biallelic expression (Xa and Xi) in three ΔRing1A/B E7.5 embryos totally deprived of H2AK119ub1 on the Xi (Δ1, Δ2 and Δ3). Mean (percentage of nuclei) ± standard deviation (s.d.) from three independent embryos. P values were calculated using two-sided t-test. For the number of analysed cells, see Extended Data Fig. 3a. Scale bars: 100 μm (whole embryos) and 10 μm (enlarged images). Arrowheads: Xist-coated Xi. Arrows: Xa. ExE: extra-embryonic ectoderm. Ch: chorion. b, Schematic of the experiment to generate TGCs. Embryos were recovered at E7.5 after intraperitoneal tamoxifen (TAM) injection to pregnant mother at E5.5. EPC was isolated and cultivated for 3 or 4 days to derive TGCs (Methods). c, Deletion of Ring1A/B induces escape of X-linked genes in TGCs. Percentage of cells showing biallelic expression (Xa and Xi) of five X-linked genes in control and ΔRing1A/B TGCs. Mean (percentage of nuclei) ± s.d.; n indicates number of TGCs from three experiments. The expected ratio and variance of biallelic expression in the control and knockout were calculated assuming a binomial distribution, and the differences between them were evaluated by two-sided binomial test. d, Immuno-RNA FISH showing lack of H2AK119ub1 but accumulation of H3K27me3 on the Xi in ΔRing1A/B TGC. Arrowheads: Xist-coated Xi. Scale bar, 10 μm. For quantification, see Extended Data Fig. 4b.
Role of PRC1 in XCI maintenance in E7.5 EPC-derived TGCs
In light of the above results, we wished to explore how X-linked gene dosage is affected in trophoblast giant cells (TGCs) given the critical role of TGCs for normal development29–31. We observed an increased frequency of Atrx and Huwe1 expression on the Xi in TGCs of ∆Ring1A/B E7.5 embryos (Extended Data Fig. 3a). Given the very limited number of TGCs in single E7.5 embryo sections, we derived TGCs from EPCs from control and ∆Ring1A/B embryos using short-term cultures28,32 (Fig. 2b). Several X-linked genes Atrx, Mecp2, Huwe1, Rnf12 and G6pd were examined by RNA FISH (Fig. 2c). All were found to have higher frequencies of escape (Xi expression in addition to Xa) in ∆Ring1A/B-derived TGCs compared with controls (Fig. 2c). Importantly, G6pd, which normally never displays expression from the Xi in TGCs28, exhibited a very high rate of expression from the Xi (>40%) in ∆Ring1A/B TGCs (Fig. 2c). Thus, loss of PRC1 has a major impact on maintenance of Xi silencing for all genes tested.
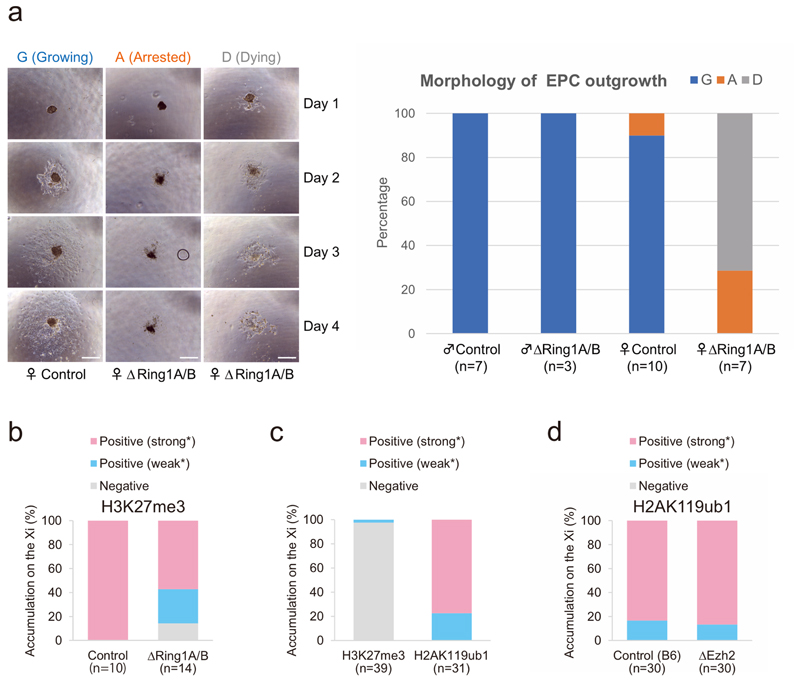
Importantly, female ∆Ring1A/B TGCs showed severely reduced growth and high rates of cell death compared with male ∆Ring1A/B mutant TGCs, consistent with the negative impact of impaired XCI on female mutant TGCs. This probably contributes to the female-biased embryonic lethality described above (Fig. 1c and Extended Data Figs. 2a and 4a).
We also investigated the impact of PRC1 loss on H2AK119ub1 and PRC2-associated H3K27me3 on the Xi in these EPC-derived TGCs. As in the embryo sections, ∆Ring1A/B TGCs robustly retained H3K27me3 accumulation on their Xi (control 100% versus ∆Ring1A/B 85.7%), despite the lack of H2AK119ub1 (Fig. 2d and Extended Data Fig. 4b). It should be noted that the TGCs were examined 5–6 days following tamoxifen induction and deletion of Ring1A/B. Thus, deletion of PRC1 does not appear to directly impact PRC2/H3K27me3 accumulation on the Xi in TGCs but nevertheless results in a pronounced loss of gene silencing of Xi genes in this developmentally critical extra-embryonic lineage, for both implantation and placenta formation.
Role of PRC1 in chromosome-wide maintenance of XCI in E7.5 EPCs
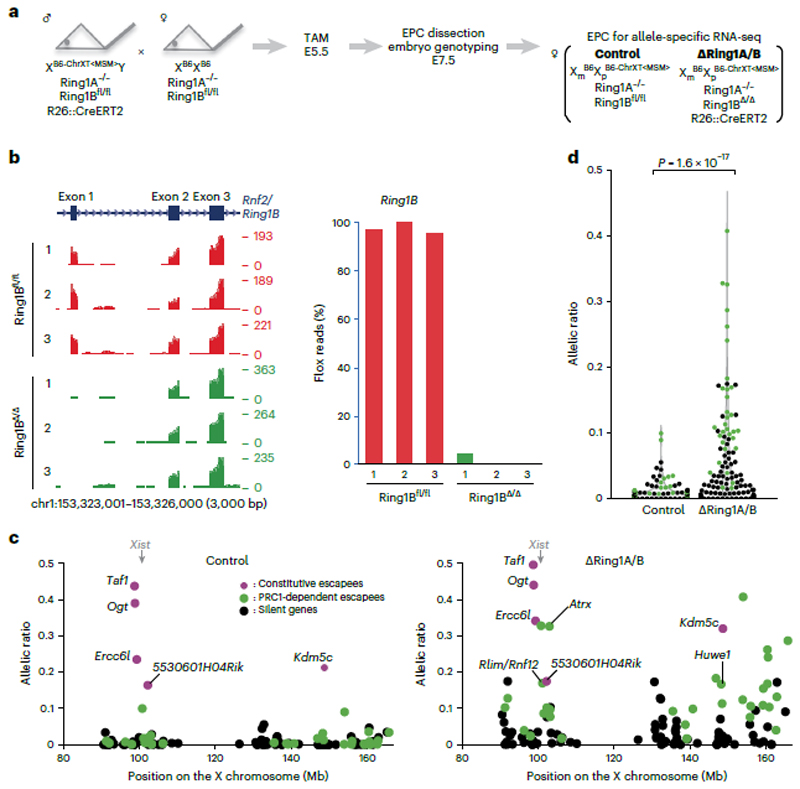
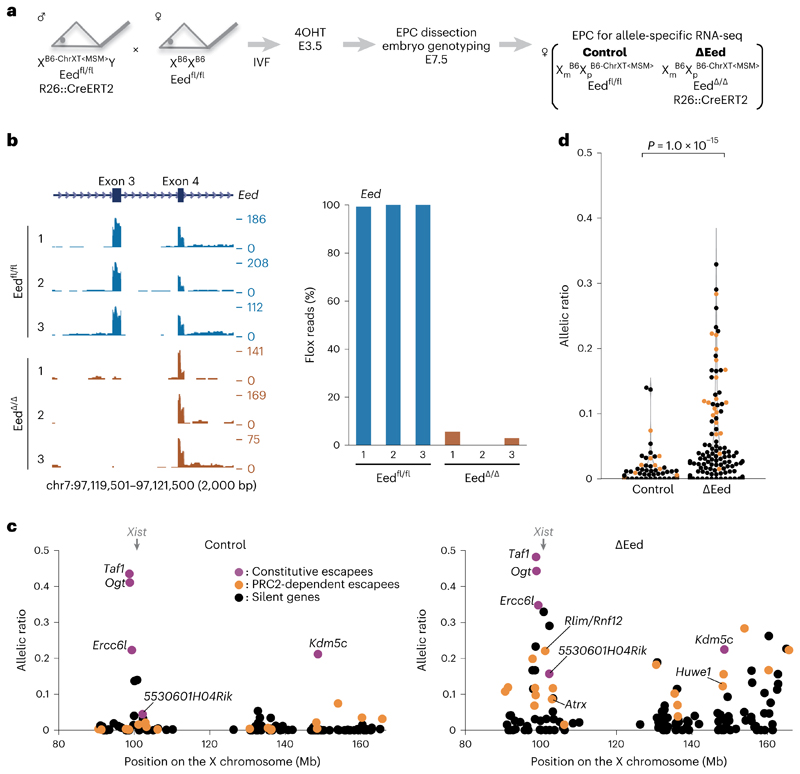
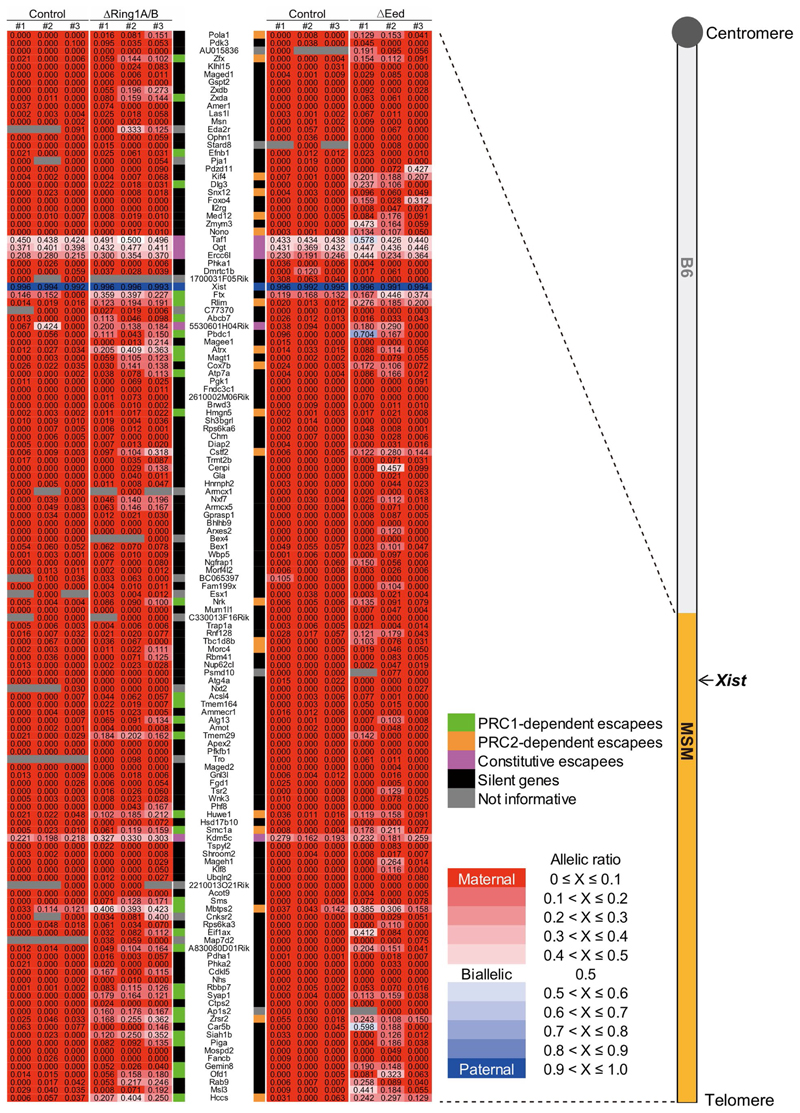
To examine the impact of PRC1 loss on XCI maintenance more globally, beyond the few genes examined by RNA FISH (Fig. 2a,c), we performed allele-specific RNA sequencing (RNA-seq) analysis on E7.5 EPCs from control and ∆Ring1A/B mutant embryos. Mice with polymorphic X chromosomes: B6 (C57BL/6, Mus musculus domesticus) and MSM/Ms (Mus musculus molossinus) were used to distinguish active and inactive X-linked alleles on the basis of the high frequency of single nucleotide polymorphisms (SNPs) and strain-specific insertions/deletions (indels). We used a subconsomic B6/MSM hybrid mouse strain, with telomeric X region of MSM/Ms-origin and centromeric X region (and autosomes) of B6-origin (B6-ChrXTMSM) 33 (Fig. 3a and Extended Data Fig. 5). Males with the B6-ChrXTMSM were crossed with B6 females such that resulting female embryos would display imprinted XCI of the paternally inherited B6-ChrXTMSM in extra-embryonic tissues of both control and ∆Ring1A/B mutant embryos (Fig. 3a). RNA-seq analysis was performed on E7.5 EPCs derived from three independent control and ∆Ring1A/B mutant embryos. Loss of Ring1B messenger RNA was confirmed in ∆Ring1A/B EPCs (Fig. 3b). We obtained informative allele-specific reads, which we converted to allelic ratios (see Methods), for 122 expressed X-linked genes (for full set of processed data, see Supplementary Table 1). Five known constitutive escape genes (Taf, Ogt, Ercc6l, 5530601H04Rik and Kdm5c) showed unchanged levels of biallelic expression in ∆Ring1A/B EPCs compared to controls, as expected (Fig. 3c, magenta circles). Of the 122 informative X-linked genes, aside from the 5 escapees and Xist, 116 genes were found to be monoallelically expressed and silent on the Xi in control females (Fig. 3c left, green and black circles, and Extended Data Fig. 5). However, in the ∆Ring1A/B EPCs, we found that a total of 32 genes (27.6%) showed significant derepression on the Xi (Fig. 3c right, green circles, and Extended Data Fig. 5). Many other informative genes (60/84; 71.4%) also showed some degree of Xi derepression in at least one of the mutant EPCs, although when averaged across three replicates, they were no longer significantly derepressed and so have been categorized as ‘silent’ (Fig. 3c, d, black circles, and Extended Data Fig. 5).
Fig. 3. Deletion of Ring1A/B (PRC1) results in loss of chromosome-wide gene silencing on the Xi in EPC.
a Schematic of allele-specific RNA-seq using ΔRing1A/B conditional knockout mouse. Embryos were recovered at E7.5 after intraperitoneal tamoxifen (TAM) injection to pregnant mother at E5.5. EPC, which retains imprinted XCI where B6-ChrXTMSM is dominantly silenced, was isolated and applied for RNA-seq library preparation. b, Genome browser plots showing allele-specific RNA-seq reads of three controls and three ΔRing1A/B EPCs at Ring1B locus. The first coding exon including ATG start codon is deleted upon CreERT2 activation. c, Allele-specific RNA-seq shows chromosome-wide gene escape from XCI upon PRC1 deletion. A total of 122 informative genes including Xist and 5 constitutive escapees (magenta), 32 significant escapees upon PRC1 deletion (green) and 84 silenced genes even in the absence of PRC1 (black) are shown in both control and ΔRing1A/B EPCs. Average of three independent EPCs. P values were calculated using one-sided Student’s t-test and are presented in Supplementary Table 2. d, Total change of allelic ratio upon PRC1 deletion. Allelic ratios of each X-linked gene in control and ΔRing1A/B E7.5 EPCs. A total of 116 informative genes including 32 significant escapees upon PRC1 deletion (green) and 84 silenced genes even in the absence of PRC1 (black) are shown in both control and ΔRing1A/B EPCs. Average of three independent EPCs. P values were calculated using one-sided Brunner–Munzel test.
Overall, these allele-specific RNA-seq (Fig. 3c) and nascent RNA FISH (Fig. 2a,c) analyses reveal that substantial Xi re-activation occurs in ∆Ring1A/B female extra-embryonic lineages, independently of H3K27me3 on the Xi, demonstrating that PRC1 plays a key role in XCI maintenance.
Role of PRC2 in H3K27me3 and H2AK119ub1 accumulation on the Xi in post-implantation female mouse embryos
To assess whether PRC2 also has a role in XCI maintenance or whether PRC1 is the main player, we investigated the in vivo impact of depleting PRC2 on the maintenance of XCI at similar stages in embryos as well as in EPC-derived TGCs32. We first looked at embryos conditionally ablated for Ezh2, the main catalytic component of PRC2, using a similar conditional deletion approach to that for Ring1A/B above (Fig. 1a). We used a floxed allele flanking exons that code for the catalytic SET domain of EZH2 protein34 (Fig. 4a). Deletion of the floxed Ezh2 exons was induced by injection of tamoxifen at E5.5 to mediate activation of CreERT2. Unlike ∆Ring1A/B embryos, which show defects at E8.5 but not at E7.5, female ∆Ezh2 embryos already showed substantial developmental delay and defects at E7.5, with normal morphology at E6.5 (Fig. 4b). Immuno-RNA FISH analysis of embryo sections at different stages (from E6.0 to E8.5; E6.5 is shown here in Fig. 4b and Extended Data Fig. 4c) allowed us to ascertain that the Ezh2 deletion had indeed occurred, based on loss of H3K27me3 staining. At E6.0 (seven embryos) and E6.5 (five embryos), all female embryos displayed normal morphology despite the absence of H3K27me3 staining on the Xi. By E7.5, the number of aborted embryos was high (nine embryos for four litters) and the few ∆Ezh2 embryos (n = 3) recovered from these pregnancies had a highly abnormal morphology. By E8.5, no ∆Ezh2 embryos could be recovered at all.
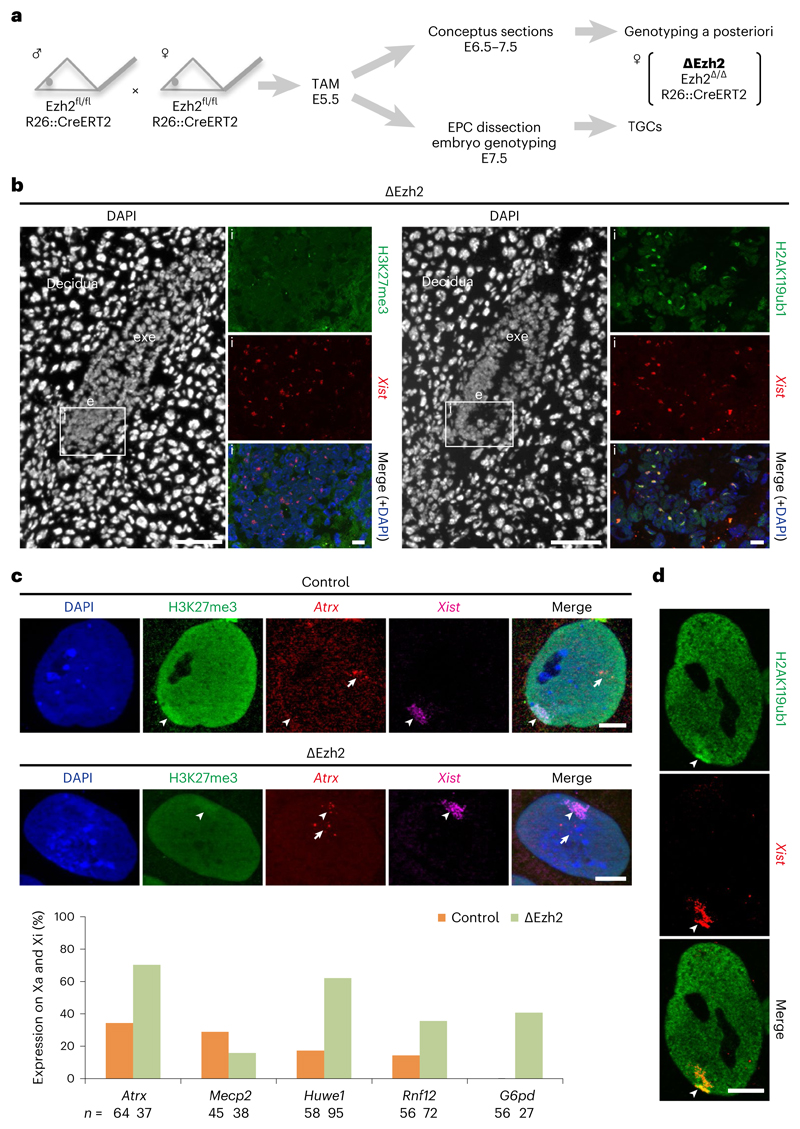
Fig. 4. Deletion of Ezh2 (PRC2) results in loss of gene silencing on the Xi in TGCs without affecting H2AK119ub1 accumulation.
a Schematic representation of ΔEzh2 experiments. Tamoxifen (TAM) was injected same as ΔRing1A/B experiments (Fig. 1a). Embryos were recovered at E6.5 or E7.5 and used for either cryosectioning or TGC derivation. For more details, see Methods. b, ΔEzh2 E6.5 embryos are deprived of H3K27me3, but not H2AK119ub1 on the Xi. Immuno-RNA FISH for two histone marks, H3K27me3 (left) or H2AK119ub1 (right), and Xist; consecutive sections; (i) boxed area showing the embryo proper (e); lack of H3K27me3 but accumulation of H2AK119ub1 shown here for the embryo proper. exe, extra-embryonic ectoderm. Scale bars: 100 μm (whole embryos) and 10 μm (enlarged images). For quantification, see Extended Data Fig. 4c. Two independent embryos for both control and ΔEzh2 were examined, and all showed similar results. c, Deletion of Ezh2 leads to aberrant expression of X-linked genes from the Xi in TGCs. Upper part shows immuno-RNA FISH for H3K27me3, Xist and an X-linked gene Atrx. Top: example of control TGC showing H3K27me3 accumulation on the Xi and monoallelic expression of Atrx on the Xa. Bottom: example of ΔEzh2 TGC showing strong decrease of H3K27me3 accumulation on the Xi and biallelic expression of Atrx on both Xa and Xi. Arrowheads: Xist-coated Xi. Arrows: Xa. Scale bar, 10 μm. Lower part shows percentage of cells showing biallelic expression of five X-linked genes on both Xa and Xi in ΔEzh2 and control TGCs (from WT B6D2F1). n indicates number of TGCs from two experiments. d, Deletion of Ezh2 does not impact H2AK119ub1 accumulation on the Xi in TGCs. Immuno-RNA FISH showing accumulation of H2AK119ub1 on the Xi in ΔEzh2 TGC. Arrowheads: Xist-coated Xi. Scale bar, 10 μm. For quantification, see Extended Data Fig. 4d. Thirty control TGCs derived from a single control embryo and 30 ΔEzh2 TGCs derived from a single ΔEzh2 embryo were analysed.
Immuno-RNA FISH analysis of the E6.5 sections revealed that all five ∆Ezh2 embryos lacked H3K27me3 on the Xi in both embryonic and extra-embryonic lineages (Fig. 4b and Extended Data Fig. 4c), while H2AK119ub1 was still enriched on the Xi in all lineages (Fig. 4b and Extended Data Fig. 4c). This suggests that PRC1 recruitment to the Xi may be independent of PRC2/H3K27me3 at this stage of mouse development.
Role of PRC2 in XCI maintenance in E7.5 EPC-derived TGCs
As many ∆Ezh2 embryos showed aberrant morphology and degeneration at E7.5, we could not analyse XCI status at this stage, unlike for the PRC1 mutants above. Therefore, we investigated XCI status in E7.5 EPC-derived TGCs (∆Ezh2 TGCs) from PRC2 mutant embryos (Fig. 4a,c). These ∆Ezh2 female TGCs grew similarly to control TGCs. Immuno-RNA FISH analysis revealed that Xist RNA accumulation appeared normal, but there was reduced or no H3K27me3 staining on the Xi in ∆Ezh2 TGCs compared with controls (Fig. 4c). However, H2AK119ub1 was still robustly enriched on the Xi in these ∆Ezh2 TGCs (Fig. 4d and Extended Data Fig. 4d). We also analysed expression of five X-linked genes, Atrx, Mecp2, Huwe1, Rnf12 and G6pd, alongside Xist RNA and H3K27me3 on control and ∆Ezh2 TGCs. Some of these five X-linked genes normally show a mild degree of escape from XCI in TGCs at this stage in WT cells28. Four out of the five X-linked genes examined by RNA FISH revealed Xi derepression in the absence of PRC2 (Fig. 4c), very similar to the impact seen in the absence of PRC1 in ∆Ring1A/B mutant female TGCs (Fig. 2c). In particular, G6pd, which is normally completely repressed on the Xi in control TGCs, was clearly active on the Xi in 40.7% of ∆Ezh2 TGCs.
Taken together, these results indicate that, in TGCs derived from PRC2 mutants at E7.5, Xi recruitment of PRC1/H2AK119ub1 is not affected but nevertheless some X-linked genes are re-expressed, indicating an important role of PRC2 in maintaining X-linked gene silencing on the Xi in extra-embryonic cells.
Role of PRC2 in chromosome-wide maintenance of XCI in E7.5 EPCs
As the above findings were based on RNA FISH for only a few genes, we wished to assess the impact of PRC2 mutation on the Xi in a chromosome-wide manner. Allele-specific RNA-seq was therefore performed on a PRC2 mutant, similarly to the design for ∆Ring1A/B EPCs used above (Fig. 3a). As ∆Ezh2 embryos show a severe embryonic lethality phenotype at E7.5, we instead used Eed conditional knockout mouse35. EED is another core component of PRC2, and its inactivation results in rapid loss of PRC2 activity35. In contrast to ∆Ezh2 embryos, ∆Eed embryos display a milder morphological phenotype. We induced Eed knockout at an earlier stage (E3.5) to ensure complete depletion and collected EPCs from E7.5 embryos (Fig. 5a). We confirmed efficient Eed deletion at E7.5, 4 days after knockout induction (Fig. 5b). Allele-specific RNA-seq analyses of three control and three ∆Eed female E7.5 EPCs revealed 132 informative X-linked genes (for full set of processed data, see Supplementary Table 1). After removing 5 constitutive escapees (Taf, Ogt, Ercc6l, 5530601H04Rik and Kdm5c) and Xist, we found 19 (15.1%) out of 126 X-linked genes in ∆Eed female E7.5 EPCs to be significantly derepressed from the Xi, indicating an important role(s) for PRC2 in the maintenance of X-linked gene silencing in EPCs (Fig. 5c right, orange circles, and Extended Data Fig. 5). Similarly to the situation for ∆Ring1A/B female EPCs, many other X-linked genes showed some degree of derepression in at least one ∆Eed EPC (78/107, 72.9%) but were classified as silenced due to variations in the degree of escape detected for such genes between replicates (Fig. 5c,d, black circles, and Extended Data Fig. 5).
Fig. 5. Deletion of Eed (PRC2) results in loss of chromosome-wide gene silencing on the Xi in EPC.
a Schematic of allele-specific RNA-seq using ÄEed conditional knockout mouse. Embryos were recovered at E7.5 after 4-hydroxytamoxifen (4OHT) treatment of blastocysts obtained from in vitro fertilization (IVF) at E3.5 (Methods). EPC was isolated and applied for RNA-seq library preparation. b, Genome browser plots showing allele-specific RNA-seq reads of three controls and three ÄEed EPCs at Eed locus. The third coding exon is deleted upon CreERT2 activation. c, Allele-specific RNA-seq shows chromosomewide gene escape from XCI upon PRC2 deletion. A total of 132 informative genes including Xist and 5 constitutive escapees (magenta), 19 significant escapees upon PRC2 deletion (orange) and 107 silenced genes even in the absence of PRC2 (black) are shown in both control and ÄEed EPCs. Average of three independent EPCs. P values were calculated using one-sided Student’s t-test, and each P value is shown in Supplementary Table 3. d, Total change of allelic ratio upon PRC2 deletion. Allelic ratios of each X-linked gene in control and ÄEed E7.5 EPCs. A total of 126 informative genes including 19 significant escapees upon PRC2 deletion (orange) and 107 silenced genes even in the absence of PRC2 (black) are shown in both control and ÄEed EPCs. P values were calculated using one-sided Brunner–Munzel test.
In summary, RNA FISH and allele-specific RNA-seq on PRC2 knockout female embryos reveal that PRC2 also has an important role in maintaining X-linked gene silencing on the Xi, and this appears to be independent of PRC1, at least in extra-embryonic tissues.
A substantial but partial overlap between PRC1- and PRC2-dependent escapees
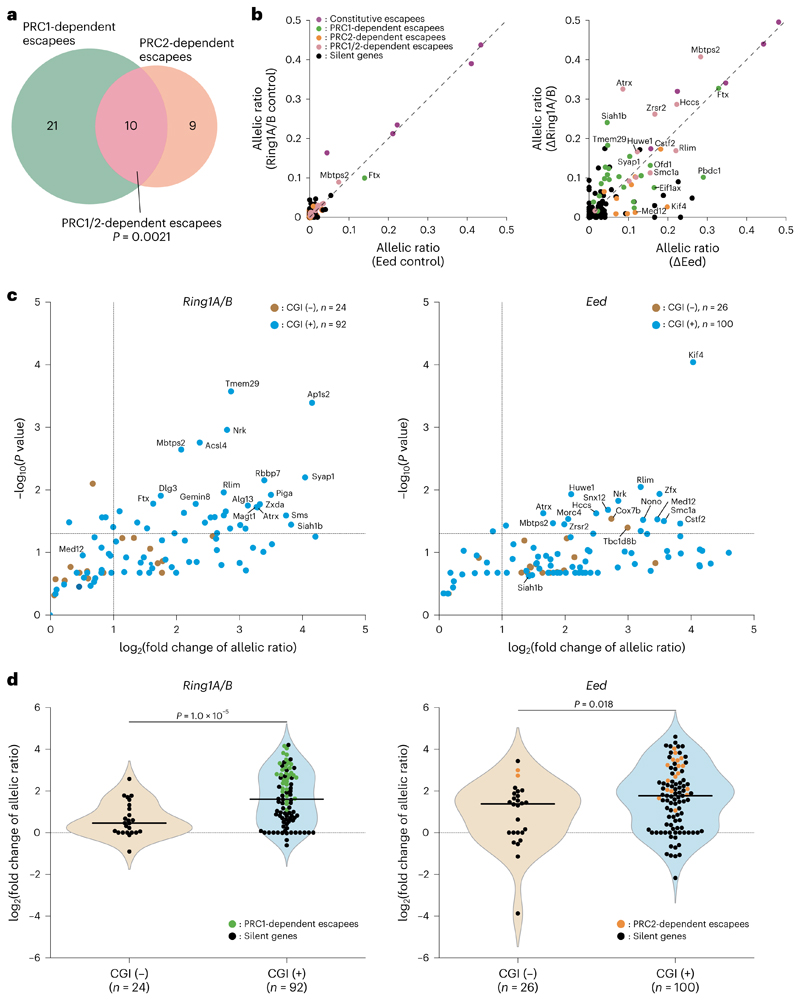
We compared the degree of overlap between PRC1-dependent (Fig. 3c) and PRC2-dependent escapees (Fig. 5c) from the allele-specific RNA-seq analyses in EPCs. A substantial overlap was seen between the two datasets (10 out of 31 PRC1-dependent escapees and 19 PRC2-dependent escapees, P = 0.0021), indicating that repression of these X-linked genes depends on both PRC1 and PRC2 (Fig. 6a,b, Extended Data Fig. 5 and Supplementary Tables 2–4). However, most X-linked genes such as Siah1b (fold change 14.1, P = 0.036 in ∆Ring1A/B; fold change 2.7, P = 0.24 in ∆Eed) and Med12 (fold change 1.4, P = 0.11 in ∆Ring1A/B; fold change 11.0, P = 0.030 in ∆Eed) are preferentially derepressed by PRC1 and PRC2 knockout, respectively, suggesting that their silencing is controlled by a single PRC (Fig. 6b, Extended Data Fig. 5 and Supplementary Tables 2–4). These results indicate that, although both PRC1 and PRC2 have a substantially overlapping role in maintaining Xi gene silencing, each complex also appears to affect a different range of X-linked genes.
Fig. 6. PRC1-dependent escapees are enriched with CGI at the promoter.
a, A Venn diagram showing the degree of overlap between PRC1-dependent escapees (N = 31) and PRC2-dependent escapees (N = 19) in 112 commonly informative X-linked genes. Average of three independent EPCs. Ap1s2 was omitted from the analysis as it was not informative in the Eed dataset. Detailed information is indicated in Supplementary Table 4. P value was calculated using chi-square test. b, Correlation of PRC1- and PRC2-dependent escapees. Allelic ratio of each informative X-linked gene was plotted. A total of 117 informative genes including 5 constitutive escapees (magenta), 21 PRC1-dependent escapees (green), 9 PRC2-dependent escapees (orange), 10 PRC1/2-dependent escapees (pink) and 72 silenced genes (black) are shown in both Ring1A/B and Eed control EPCs (left) and ΔRing1A/B and ΔEed EPCs (right). Average of three independent EPCs. Detailed information is indicated in Supplementary Tables 1 and 4. c, PRC1-dependent escapees are significantly enriched with promoter-associated CGI. Left: distribution of 32 PRC1-dependent escapees and 84 silent genes. All 32 PRC1-dependent escapees have CGI at their promoter (P = 6.9 × 10−4, chi-square test). For processed data, see Supplementary Table 2. Right: distribution of 19 PRC2-dependent escapees and 107 silent genes. Seventeen out of 19 PRC2-dependent escapees have CGI at their promoter (P = 0.24, chi-square test). For processed data, see Supplementary Table 3. P values for each gene were calculated using one-sided Student’s t-test and converted to −log10 values (see also Supplementary Tables 2 and 3). d, Correlation of promoter-associated CGI and degree of escape. Left: a total of 116 informative genes analysed in ΔRing1A/B mutant EPCs including 32 PRC1-dependent escapees (green circles) and 84 silent genes (black circles). For processed data, see Supplementary Table 2. Right: a total of 126 informative genes analysed in ΔEed mutant EPCs including 19 PRC2-dependent escapees (orange circles) and 107 silent genes (black circles). For processed data, see Supplementary Table 3. P values were calculated using two-sided Brunner–Munzel test. In c and d, each dot represents an average of three independent experiments. To all allelic ratio values, 1% dispersion (0.01) was added to calculate the fold change of allelic ratio (Methods). CGI (−) and CGI (+): genes without and with CGI at promoter (±2 kb from transcription start site), respectively.
Role of CGIs in PRC1-mediated gene silencing on the Xi
As CGIs have been proposed as genomic target elements of both PRC1 and PRC2 (refs. 5,36), we assessed this feature for both PRC1-dependent (Fig. 3c) and PRC2-dependent escapees (Fig. 5c) in EPCs. We found that all 32 PRC1-sensitive genes have a CGI at their promoter, which represents a significant enrichment (P = 6.9 × 10−4) compared with the total population (92/116, 79.3%) (Fig. 6c, left). These 92 CGI-associated X-linked genes have substantially higher expression from the Xi compared with other informative X-linked genes with no CGI (24/116 genes, 20.7%) (P = 1.0 × 10−5) in ∆Ring1A/B mutant EPCs (Fig. 6d, left), suggesting an important role for CGIs in PRC1-mediated maintenance of Xi gene silencing. In the case of the 19 significant PRC2-dependent escapees shown in Fig. 5c, 17 (89.5%) have a CGI at their promoter; however, the degree of CGI enrichment was not significant (P = 0.24) compared with the total 126 informative genes (100/126, 79.4%) (Fig. 6c, right). Furthermore, in ∆Eed mutants, informative genes without a promoter-associated CGI (N = 26) tend to be derepressed, similarly to genes with CGIs (N = 100) (Fig. 6d, right). Therefore, PRC2 appears to play a role in maintaining repression of X-linked genes with or without a CGI (Fig. 6d, right), unlike the more CGI-associated role of PRC1 (Fig. 6d, left).
Discussion
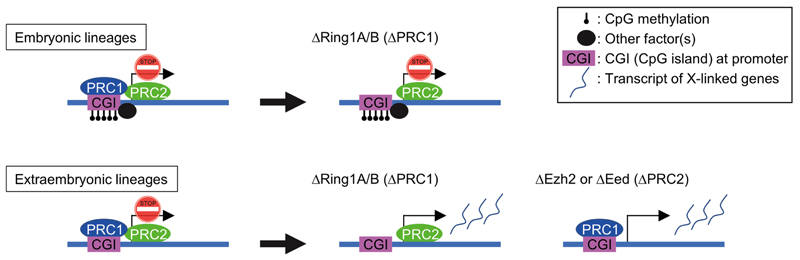
In this study, we have analysed the in vivo roles of PRCs in the maintenance of XCI in post-implantation embryos, using conditional knockout mouse mutants for either PRC1 (Ring1A/B) or PRC2 (Ezh2 or Eed). Our results using combined immunostaining and RNA FISH on whole embryo cryosections, as well as EPC-derived TGCs, have revealed that the accumulation of H2AK119ub1 and H3K27me3 on the Xi, mediated by PRC1 and PRC2 respectively, appear to be independent of each other at least at this stage of development, and that both pathways contribute to maintaining X-linked gene silencing on the Xi in extra-embryonic tissues. Furthermore, we demonstrated that both PRC1- and PRC2-dependent gene silencing on the Xi in extra-embryonic lineages is broadly distributed, affecting multiple genes across the X chromosome, on the basis of allele-specific RNA-seq analyses in mutant EPCs. The critical role of both PRC1 and PRC2 complexes for XCI in extra-embryonic tissues, contrasts with the embryo proper, where a lack of PRC1 or PRC2 does not appear to affect gene silencing on the Xi. This is presumably due to the Xi role played by other epigenetic marks, such as DNA methylation, which is present at CGIs on the Xi in embryonic tissues, but not in most extra-embryonic lineages6.
Our transcriptomic analysis in PRC1 and PRC2 mutant extra-embryonic tissues also demonstrated that sensitivity to PRC1 or PRC2 loss differs substantially among most Xi genes (Fig. 6a,b), implying limited redundancy in PRC1 and PRC2 functions for X-linked gene silencing at this stage. The fact that promoter CGIs tend to be strongly enriched in PRC1-dependent escapees but less so in PRC2-dependent escapees is consistent with likely CGI-mediated targeting of PRC1 to target genes in general5,36. This implies that CGI-associated PRC1 may be critical for maintenance of silencing of some genes on the Xi while PRC2 targets other genes via a different mechanism (Extended Data Fig. 6) at least during this developmental time window.
The different PRC1 and PRC2 effects on the Xi that we observed at E7.5 in our study differ at first sight with more overlapping effects in a recent study by Andergassen et al.37. However, this probably reflects the different experimental setups and time windows examined (Supplementary Discussion).
Our study demonstrates that PRC1/H2AK119ub1 and PRC2/H3K27me3 histone modifications occur independently of each other on the Xi at the post-implantation stages of development examined here. PRC2-mediated H3K27me3 still accumulates on the Xi in the absence of PRC1/H2AK119ub1 in all lineages of E7.5 embryos, implying that PRC1 is not essential for PRC2 propagation at this stage. Consistent with this, H3K27me3 accumulation on the Xi was still observed in ∆Ring1A/B mutants at least in extra-embryonic tissues at E8.5. These results seem to be in contrast to a previous study, where embryonic deletion of non-canonical PRC1 factors Pcgf3/5 resulted in loss of PRC2/H3K27me3 on the Xi23. This apparent discrepancy might be explained in at least two ways. First, once XCI is established and enters into maintenance phase, PRC2 may no longer be dependent on PRC1 for its propagation, via the PRC2 accessory factor JARID2 that can bind to H2AK119ub1 (refs. 38,39). This may be the case for the Xi at E5.5–8.5 in extraembryonic lineages and explain why the removal of PRC1 does not affect PRC2 in our study. Second, the kinetics of loss/removal of H2AK119ub1 and H3K27me3 from the Xi may differ in different cell lineages. Upon acute removal of RING1A/B, H2AK119ub1 has been reported to be lost much more rapidly than H3K27me3 from chromatin in mouse ES cells40. H3K27me3 is thought to be lost by dilution during chromatin replication at each cell division much more slowly in the absence of PRC2 (refs. 40,41). At E3.5–7.5, examined in Almeida et al., cells may divide more frequently than E5.5–7.5 and the deficiency of PCGF3/5-PRC1 at this earlier stage may accelerate the removal of H3K27me3 from the Xi chromatin23. Nevertheless, the fact that several rounds of divisions must occur during the 3 days of post-deletion even in extra-embryonic cells, suggests that there is PRC1-independent accumulation of PRC2 on the Xi in extra-embryonic lineages.
Here we provide evidence that both PRC1 and PRC2 act independently and both have essential roles in the maintenance of XCI, at least in extra-embryonic lineages. The general models for PRC1 and PRC2 inter-dependent functions on the Xi have mainly focused on earlier phases of XCI: Xist RNA recruits PRC1 (PCGF3/5 subcomplex), and this in turn enables PRC2 recruitment22,23. Once XCI is established and enters the maintenance phase, PRC1 and PRC2 propagation on the Xi may occur independently of each other and may have independent roles in maintaining X-linked gene repression with varying degree of sensitivity for different X-linked genes. We propose that Polycomb complexes PRC1 and PRC2 each preserve the Xi from harmful transcriptional re-activation in extra-embryonic tissues, and that this is crucial in the absence of CGI methylation, the other epigenetic system that ensures stable Xi silencing in embryonic tissues. Thus, the molecular mechanisms underlying Polycomb-mediated maintenance of gene silencing in XCI may differ for the PRC1 and PRC2 machineries, potentially opening up exciting new avenues to explore the diversity of Polycomb-mediated epigenetic memory mechanisms.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41556-022-01047-y.
Methods
Mouse mating and embryo obtention
All animal experiments were carried out according to the in-house guidelines for care and use of laboratory animals of RIKEN, Yokohama Institute, Japan and Institut Curie, Paris, France. Eight-to-16-week-old female mice and 10-to-24-week-old male mice were used in all experiments. The mice were bred in 12 h light/12 h dark condition, 21–25 °C and 40–60% humidity. All mice used for the experiments were randomly chosen from multiple mouse colonies. Ring1A null and Ring1B conditional alleles have been described previously16,25. To generate conditional PRC1 knockout mice, males with CreERT2 (Ring1A−/−, Ring1Bfl/fl, Rosa26::CreERT2) and females without CreERT2 (Ring1A−/−, Ring1Bfl/fl) were mated. Generation of these mice (Ring1A−/−, Ring1Bfl/fl, Rosa26::CreERT2) was described previously16. Conditional deletion of Ring1B was carried out by 100–150 µl of 15 mg ml−1 tamoxifen (Sigma-Aldrich) intraperitoneal injection to the pregnant mother at E5.5. The embryos were collected at E7.5 and applied for cryosectioning or TGC derivation.
Mice carrying subconsomic B6-ChrXTMSM X chromosome where telomeric half of B6 Mus musculus domesticus X chromosome was replaced with MSM/Ms Mus musculus molossinus X chromosome were obtained from National Institute of Genetics in Japan.
Ezh2fl/fl, Rosa26::CreERT2 mice were kindly given by R. Margueron34. Ezh2 deletion was induced upon injection of tamoxifen as described above.
Eed conditional allele has been described previously35. Briefly, the coding exon 3 of Eed was floxed with two loxP sites, and Cre-mediated excision produced an in-frame EED mutation that lacks a part of WD40 repeats and functionally null EED protein. Conditional deletion of Eed was performed as described previously23 with several modifications. In vitro fertilized eggs were grown in KSOM medium until blastocyst stage and treated with 4-hydroxytamoxifen (Sigma-Aldrich, final concentration 1.6 µM) for 10 h and transplanted into E3.5 foster mothers.
Genotyping of mice and embryos
Genomic DNA for mouse genotyping was obtained from mouse tail biopsies. Genotyping of ∆Ring1A/B or ∆Ezh2 mouse embryos was performed with genomic DNA, which was obtained by either scratching the embryonic parts from the frozen E7.5 sections after immuno-RNA FISH analysis (∆Ring1A/B and ∆Ezh2) or collecting yolk sac, embryonic and extra-embryonic ectoderm regions subsequently to the dissection of the EPC for TGC derivation (∆Ezh2). Genotyping of ∆Ring1A/B or ∆Eed mouse embryos was performed with complementary DNA. Total RNA of ∆Ring1A/B or ∆Eed E7.5 embryos was obtained using AllPrep DNA/RNA Micro Kit (QIAGEN) by collecting yolk sac, embryonic and extra-embryonic ectoderm regions subsequently to the dissection of the EPC for TGC derivation (∆Ring1A/B) and allele-specific RNA-seq (∆Ring1A/B and ∆Eed) and reverse-transcribed with SuperScript VILO (Thermo Fisher Scientific). qPCR was performed with KOD SYBR qPCR Mix (TOYOBO). Genotyping primers are listed in Supplementary Table 5.
Derivation of TGCs in culture
To generate TGCs in short-term culture, EPCs were isolated from E7.5 embryos as previously described28,32. Individual EPCs were grown on glass coverslips in four-well multidishes, containing 0.5 ml medium (RPMI 1640, 15% foetal calf serum, 0.1 mM 2-mercaptoethanol and antibiotics) at 37 °C in 5% CO2 for 3–4 days.
Cryosections
Conceptuses (whole decidua containing an embryo) were collected at E6.5, E7.5 and E8.5, embedded in OCT compound (Tissue-Tek), frozen in liquid N2, then cut using a CM 1950 and CM3050 cryostats (Leica) into 8 µm sections32.
RNA FISH
Embryonic cryostat sections on slides and cultured TGCs on coverslips were directly fixed in 3% paraformaldehyde for 10 min at room temperature. After washing three times with PBS, they were permeabilized on ice for 5 min in PBS, 0.5% Triton X-100 and 2 mM vanadyl-ribonucleoside complex (New England Biolabs) and washed three times with PBS and stored in 70% ethanol at −20 °C. The stored samples were dehydrated with 1× 80%, 1× 95% and 2× 100% ethanol for 5 min each, dried out and hybridized with RNA FISH probes in a humid chamber at 37 °C overnight. RNA FISH probes were prepared with Nick Translation Kit (Abbott) according to the manufacturer’s instructions, purified by ethanol precipitation and dissociated in hybridization buffer (50% formamide, 2× saline sodium citrate, 10% dextran sulphate, 1 mg ml−1 BSA and 10 mM vanadyl-ribonucleoside complex). After the probe hybridization, the samples were washed three times at 42 °C with 50% formamide in 2× SSC for 7 min, three times at 42 °C with 2× SSC for 5 min, two times at room temperature with 2× SSC for 5 min, and mounted with VECTASHIELD mounting medium with DAPI (Vector laboratories).
We used the p510 plasmid28 to detect Xist and the following gene-specific BAC and fosmid probes:
RP23-265D6 (Atrx), RP24-157H12 (Huwe1), G135P601977A8 (Mecp2), RP24-240J16 (Rnf12) and RP23-13D21 (G6pd). They were labelled with Spectrum Green-dUTP (Vysis), Spectrum Red-dUTP (Vysis) or Cy5-dUTP (Dutscher) by nick translation.
IF and immuno-RNA FISH
To perform IF, embryo sections and TGCs were directly fixed and permeabilized as described above for RNA FISH, then blocked with 1% BSA in PBS for 15 min at room temperature. The samples were incubated for 40 min at room temperature with primary antibodies followed by washing five times with PBS for 5 min each. The samples were incubated with secondary antibodies for 40 min at room temperature, then washed five times with PBS for 5 min each. Primary and secondary antibodies were diluted with 1% BSA, 0.2 U µl −1 SUPERase-In (Thermo Fisher Scientific) in PBS. After the final wash, the samples were mounted with VECTASHIELD mounting medium with DAPI.
For immuno-RNA FISH, IF was performed before RNA FISH as described above. After the final wash of IF, the samples were post-fixed in 3% paraformaldehyde for 10 min at room temperature, then washed with PBS three times and with 2× SSC twice, and RNA FISH was performed as described above. Three monoclonal antibodies were used in this study: an anti-H3K27me3 antibody (7B11, 1:200, mouse mAb, a gift from R. Margueron; MABI0323, 1:200, mouse mAb, Takara) and an anti-H2AK119ub1 antibody (8240S, 1:50, rabbit mAb, Cell Signaling). Four secondary antibodies were used in this study: goat anti-mouse IgG Alexa568 (#A11031, 1:500, Thermo Fisher Scientific), goat anti-mouse IgG Alexa488 (#A11029, 1:800, Thermo Fisher Scientific), goat anti-rabbit IgG Alexa488 (#A11034, 1:800, Thermo Fisher Scientific) and goat anti-rabbit IgG Alexa647 (#A21044, 1:800, Thermo Fisher Scientific).
Imaging and quantification
All imaging data were analysed by at least two independent persons. All fluorescent images were acquired using Apotome (Zeiss) and Deltavision (GE Healthcare) and analysed using Adobe Photoshop (v10.0.1). Quantification of each image was performed as follows.
For immunostained sections, nuclei were selected upon DAPI staining and those with accumulation as a focus of a given histone mark were recorded. For sections upon immunostaining combined with RNA FISH, nuclei with a Xist domain were selected. Those exhibiting an accumulation of a given histone mark at the Xist domain (co-localization) were recorded. Three categories were defned: two positive, either strong (a compact and dense staining) or weak (a loose and relaxed staining) and one negative.
Allele-specific RNA-seq
E7.5 EPCs were surgically isolated, and their total RNAs were purified using AllPrep DNA/RNA Micro Kit (QIAGEN). Embryonic and extra-embryonic ectoderm regions of the same E7.5 embryo were used for genotyping by RT–qPCR. RNA-seq libraries were prepared with SMARTer Stranded Total RNA-seq Kit - Pico Input Mammalian (Takara Bio) and analysed by HiSeq 2500 and NovaSeq 6000 DNA sequencer (Illumina) with 20–60 million 100 bp paired-end reads. Allele-specific RNA-seq analysis was performed according to Sakata et al. 31, by constructing the B6-ChrXTMSM specific reference. To prepare a set of the SNPs and indels (referred to as variants) of the MsM strain, MsM genomic reads (NCBI Sequence Read Archive accession number DRX050256) were assembled into scaftigs using fermi v1.1-r751 (ref. 42) with option ‘-k 100’. Those scaftigs were aligned to a reference genome (UCSC mm9), including unlocalized/unplaced scaffolds and one unit of ribosomal DNA repeats (GenBank accession number BK000964.1) as decoys, using bwa mem v0.7.10 (ref. 43) with option ‘-D 0 -c 10000 -w 1000’. Scaftigs with low mapping quality (MAPQ <40) or overlapping satellite repeats (annotated as GSAT_MM and SYNREP_NM in the UCSC RepeatMasker track) were filtered out to avoid misidentification of variants. Using outputs of samtools v0.1.19 mpileup44, we called MsM variants satisfying the following criteria: (1) a depth of scaftigs ≥1 and ≤3; (2) all bases among scaftigs were consistent when multiple scaftigs were overlapped; (3) indel length ≤40 bp. We constructed the MsM-specific genome by incorporating the MsM variants into the B6 genome. To determine the boundary between B6 and MSM alleles in B6-ChrXTMSM, ChIP–seq reads derived from testes of male B6-ChrXTMSM strain (NCBI Sequence Read Archive accession number DRP001325) were mapped to B6/MsM diploid genomes using bwa samse v0.7.10 (ref. 45) as described46. As a result of visual inspection of allele-specific reads (MAPQ ≥16), alleles at rs261303783 and rs236638645 (dbSNP database ID) in B6-ChrXTMSM were B6 and MsM, respectively. The pseudoautosomal region was adapted as previously described47. We constructed B6-ChrXTMSM reference from mm9 genome by incorporating the MsM variant in mm9 chrX:85995615-166407703.
Paired-end reads were trimmed to remove any portion of the SMARTer adaptor sequences and the first two bases of read 1 (an error-prone stretch caused by random hexamer priming during library preparation) using cutadapt v2.1 (ref. 48), FLASH2 v2.2.00 (ref. 49) and seqtk v1.3 (https://github.com/lh3/seqtk). We filtered out RNA-seq reads derived from ribosomal RNA and mitochondrial transcripts by mapping to 45S RNA, 5S RNA and mitochondrial DNA (GenBank accession numbers BK000964.1 and NR_030686.1; UCSC mm9 chrM). The filtered reads were mapped to the custom reference, including the strain-specific genomes and sequences spanning splice junctions on the basis of the RefSeq genes for all possible alleles, as single-end reads using bwa samse v0.7.10. The coordinates of hits mapping to splice junctions and/or MsM sequences were converted to mm9 genomic coordinates using samtools v0.1.19, bedtools v2.28.0 (ref. 50) and UCSC genome browser utilities v379 ref. 51), and redundancy in the coordinates for each read was removed while retaining their allelic information. For each gene, the number of B6 (maternal) and MsM (paternal) allele-specific reads was counted while ignoring variants overlapping multiple hits in order to control for mapping bias caused by differences in uniquely mappable positions among strain-specific genomes52 and ratios of expression from paternal alleles were calculated. Allelic ratio shows the degree of escape from imprinted XCI. Allelic ratios 0 and 0.5 indicate complete silencing by XCI and complete escape from XCI, respectively. We considered genes with more than ten allele-specific reads in all six EPCs as informative for each ∆Ring1A/B and ∆Eed experiments. On initial analysis, we noticed that Tmsb4x and Tsix showed aberrant data due to an error-prone variant and incompleteness of strand specificity, respectively (Supplementary Information). Therefore, those two genes were omitted from further analysis. To estimate gene expression level as the fragments per kilobase of exon per million fragments mapped (FPKM) in a non-allelic manner, the alignments including both unique and multiple hits (the number of best hits ≤20) were processed using Cufflinks v2.2.1 cuffdiff53 with option ‘–library-type ff-firststrand–multi-read-correct’.
Statistics and reproducibility
All statistical calculations were done by using Python SciPy library software. In allele-specific RNA-seq analysis, significantly derepressed genes upon PRC1 or PRC2 deletion were identified using one-sided Student’s t-test between paternal expression in control and that in ∆Ring1A/B or ∆Eed EPCs (P < 0.05) under assumption of normal distribution examined using F-test (P > 0.05). Mean paternal expression for each gene was calculated and derepression of total consomic regions was evaluated using Brunner–Munzel non-parametric test. To calculate fold change of allelic ratio in the mutants compared with its controls, 1% dispersion (0.01) was added to each allelic ratio value to avoid divergence in fold change. At least two biological replicates were analysed in all experiments in this study.
Sample size
All experiments were performed with at least two distinct biological replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Replicates of E7.5 ΔRing1A/B female embryos.
a Features of all fifteen E7.5 ΔRing1A/B female embryos. Morphologies of embryos: N: normal, ABN: abnormal E7.5 morphology or (*) small E6.5-like morphology. H2AK119ub1 accumulation status on the Xi for embryonic and/or extraembryonic lineages. H3K27me3 accumulation status on the Xi for all lineages: embryonic and extraembryonic. 15 out of 27 female embryos were ΔRing1A/B among 68 embryos analysed. b, Quantification of H2AK119ub1 and H3K27me3 Xi focus expression in nuclei from sections in Fig. 1b (upper part). Both embryo (e) and ectoplacental cone (epc) nuclei from ΔRing1A/B E7.5 (OR1-1) embryos are analysed as positive (strong* or weak* expression as an Xi focus) or negative. Percentage and number of cells analysed are given for both histone marks. c, Quantification of H3K27me3 Xi accumulation in nuclei from sections in Fig. 1b (lower part). Results are given for control and ΔRing1A/B E7.5 female embryo (OR3-2). All nuclei are analysed as positive (strong* or weak* expression as an Xi focus) or negative. Percentage and number of cells analysed are given. d, Another example of ΔRing1A/B E7.5 female embryo (OR1–9) showing lack of H2AK119ub1 and presence of H3K27me3 on the Xi on sections. Upper part: General view of the embryo with DAPI staining is shown on the left. i: higher magnification of embryonic region (e) analyzed by immuno-RNA FISH for H2AK119ub1 and Xist. Quantification of H2AK119ub1 Xi accumulation in nuclei from the represented section. Lower part: Consecutive section. General view of the whole embryo with DAPI staining is shown on the left. i: higher magnification of embryonic region (e) immunostained for H3K27me3. Note; because the DAPI image of this cryosection was accidently lost, a consecutive section was DAPI stained and displayed here. Quantification of H3K27me3 Xi foci expression in nuclei from the represented section. All nuclei are analysed as positive (strong* or weak* expression as an Xi focus) or negative. Percentage and number of cells analysed are given for both histone marks. Scale bars: 100 μm for the whole embryo and 10 μm for the enlarged images indicated with white rectangles.
Extended Data Fig. 2. ΔRing1A/B E8.5 embryos are deprived of H2AK119ub1, but not H3K27me3 on the Xi in extraembryonic tissues.
a Female specific lethality upon deletion of Ring1A/B at E8.5. E8.5 control and ΔRing1A/B male and female embryos were analysed. Percentage and number are given. A significantly lower than expected number of ΔRing1A/B female embryos at E8.5 (13.5% instead of 25%, P = 0.033) and an increased number of empty deciduae (15.4%), suggesting female ΔRing1A/B embryo lethality at this stage. *empty decidua that is, dead embryo. Under the assumption that all genotypes are observed in 25% of the cases, one-sided binomial distribution was used to calculate p-values less than the expected number of embryos. b, H2AK119ub1/H3K27me3 accumulation status on the Xi analysed on sections for three E8.5 ΔRing1A/B female embryos. Percentage of H3K27me3-positive cells on the Xi is given. c, ΔRing1A/B E8.5 embryos are deprived of H2AK119ub1, but not H3K27me3 on the Xi in extraembryonic lineages, for example epc. Analyses on longitudinal sections; boxed regions (e and epc) are shown with higher magnification. Upper part: control embryo studied by immuno-RNA FISH for two histone marks, H2AK119ub1 (left), H3K27me3 (right) and Xist; consecutive sections. Lower part: ΔRing1A/B female embryo (RAB8–57). Quantifications of H2AK119ub1 and H3K27me3 Xi accumulation in nuclei from these sections are shown below. All nuclei are analysed as positive (strong* or weak* expression as an Xi focus) or negative. Percentage and numbers of cells analyzed are given. e: embryo proper, epc: ectoplacental cone. Scale bars: 100 μm for the whole embryos and 10 μm for the enlarged images.
Extended Data Fig. 3. Replicates of E7.5 ΔRing1A/B female embryos analyzed by RNA FISH.
a Quantification of Atrx and Huwe1 biallelic expression on both Xa and Xi in embryonic and extraembryonic lineages in three ΔRing1A/B E7.5 embryos totally deprived of H2AK119ub1 accumulation on the Xi: Δ1, Δ2 and Δ3 (OR1–7, OR1–8 and OR8–15, respectively) and in one control (OR2–17). Percentage and numbers of cells analyzed for each lineage including TGCs. *Not significant due to low number of analyzed cells. b, Another example of ΔRing1A/B female embryo (Δ1, OR1–7) showing escape of Atrx in extraembryonic tissues. Three different regions are shown. 1 (ve and e), i: higher magnification showing a cell from the embryo proper in which Atrx is monoallelically expressed from the Xa. 2 (exe, ve and TGC), i: higher magnification of a TGC in which Atrx is biallelically expressed on both Xa and Xi; 3 (epc), i: higher magnification showing a cell from epc in which Atrx is biallelically expressed on both Xa and Xi. Arrowheads: Xist-coated Xi. Arrows: Xa. Scale bars: 100 μm for DAPI staining of the whole embryos and 10 μm for the enlarged images indicated with white rectangles. e; embryo proper, epc; ectoplacental cone, ve; visceral endoderm, exe; extraembryonic ectoderm, TGC; trophoblast giant cell. Three independent embryos (Δ1: OR1–7, Δ2: OR1–8 and Δ3: OR8–15) were examined and all showed similar results. Δ2 (OR1–8) is shown in Fig. 2a. c, H2AK119ub1 level on the Xi in extraembryonic lineages correlates with the degree of derepression on the Xi. Quantification of Atrx and Huwe1 biallelic expression on both Xa and Xi in embryonic and extraembryonic lineages in two ΔRing1A/B E7.5 embryos which partially lost H2AK119ub1 accumulation on the Xi in extraembryonic lineages: Δ4 and Δ5 (OR1–4 and OR1–9 respectively; see Extended Data Fig. 1a). Percentage and numbers of cells analyzed for each lineage including TGCs.
Extended Data Fig. 4. Morphology of ΔRing1A/B TGCs derived from E7.5 ectoplacental cone and quantification of H3K27me3 and H2AK119ub1 Xi accumulation.
a Female-specific lethality of TGCs upon Ring1A/B (PRC1) deletion. Morphology of EPC outgrowths, containing TGCs were recorded from days 1 to 4 of culture. Each TGC culture was categorized into three categories, Growing (G), Arrested (A), and Dying (D), based on their morphologies. Representative pictures are shown on the left. Scale bar: 100 μm. Summary chart is shown on the right. ΔRing1A/B female EPC outgrowth showed more severe phenotypes such as Arrested (A) and Dying (D) than male mutants and controls. b, Quantification of H3K27me3 Xi accumulation in control and ΔRing1A/B TGCs illustrated in Fig. 2d. c, Quantification of H3K27me3 and H2AK119ub1 Xi accumulation in nuclei from sections in Fig. 4b. d, Quantification of H2AK119ub1 Xi accumulation in ΔEzh2 TGCs as illustrated in Fig. 4d. b–d, All nuclei are analysed as positive (strong* or weak* expression as an Xi focus) or negative. Percentage and numbers of cells analyzed are given.
Extended Data Fig. 5. Details of allele-specific RNA-seq.
(Right) Schematic of B6-ChrXTMSM chromosome used in this study. Telomeric half of the B6 X chromosome is replaced with MSM/Ms X chromosome. (Left) Allele-specific expression ratio in 3 independent EPCs of Ring1A/B control, ΔRing1A/B, Eed control and ΔEed E7.5 embryos are represented as heat maps. Maternal expression in red and paternal expression in blue. Total 136 informative genes (122 for Ring1A/B and 132 for Eed) including 5 constitutive escapees (magenta), Xist (Blue), significant escapees upon PRC1 deletion (green), significant escapees upon PRC2 deletion (orange), not informative genes (gray) and silenced genes even in the absence of PRC1 or PRC2 (black) are shown. Values are allele-specific expression ratios in each EPC.
Extended Data Fig. 6. A model for the role of Polycomb complexes in the maintenance of XCI.
(Left) X-linked genes in embryonic and extraembryonic lineages on the Xi are silenced by XCI. CGIs on the Xi are heavily methylated in embryonic lineages, but maintained as hypo-methylated in extraembryonic lineages. (Middle) Ring1A/B knockout resulted in a depletion of all PRC1 subcomplexes on the Xi, but PRC2 is still retained on the Xi, at least in extraembryonic lineages. In this situation, PRC2 accumulation on the Xi is retained at E7.5 but lost at E8.5 in embryonic lineages (here E7.5 is shown). (Right) Ezh2 or Eed knockout resulted in a depletion of PRC2 on the Xi, but PRC1 is still retained on the Xi, in extraembryonic lineages. (Middle and Right) In these situations, many of X-linked genes undergo robust reactivation from the Xi only in extraembryonic lineages. In embryonic lineages, however, X-linked genes are still silenced in the absence of PRC1 or PRC2. DNA methylation of CGIs and/or some other factor(s) might compensate a lack of PRC1 or PRC2 to secure a tight silencing of X-linked genes on the Xi in embryonic lineages.
Supplementary Material
Acknowledgements
We thank members of the Heard and Koseki laboratories for helpful discussion; Tim Pollex for critical reading of the manuscript; the mouse facilities in Curie Institute and RIKEN for breeding the mice; D. Reinberg and R. Margueron for antibodies and advice; R. Margueron for providing Ezh2fl/fl, Rosa26::CreERT2 mice. We acknowledge the NGS core facility of the Genome Information Research Center at the Research Institute for Microbial Deseases of Osaka University for support in RNA-seq. This work was funded by the LabEx DEEP (ANR-11-LABX-0044), and an ERC Advanced Investigator award ERC-ADG-2014 671027 to E.H.; Core Research for Evolutional Science and Technology and Precursory Research for Embryonic Science and Technology from the Japan Science and Technology Agency–Agency for Medical Research and Development, and Grants-in-Aid for Scientific Research from Japan Society for the Promotion of Science to Koseki; JPMJPR11SE to O.M., from PRESTO, JST. 17H06426 to K.N. and 18H05532 to C.O., from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT).
Footnotes
Author contributions
O.M., C.C., H.K. and E.H. conceived the main idea of the whole analysis. O.M. and C.C. designed and conducted the experiments. K.N. and C.O. analysed allele-specific RNA-seq data. T.A.E. performed statistical analyses of all data. M.N. designed Eed conditional knockout mouse. F.K., P.D., M.K. and Y.K. bred all mice used in this study. O.M., C.C., F.K. and P.D. prepared frozen sections of embryos. C.C., O.M. and F.K. performed RNA FISH and immuno-RNA FISH on sections. O.M., C.C. and F.K. derived TGCs and performed RNA FISH and immuno-RNA FISH for TGCs. O.M. and C.C. analysed the data and wrote the paper. H.K. and E.H. edited the paper.
Competing interests
The authors declare no competing interests
Additional information
Peer review information Nature Cell Biology thanks Christine Disteche and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Reprints and permissions information is available at http://www.nature.com/reprints
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Data Availability
The allele-specific RNA-seq dataset is publicly available through Gene Expression Omnibus (GEO). The accession number is GSE168140. Raw data of Figs. 3c and 5c are provided in Supplementary Tables 2 and 3, respectively. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
- 1.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Żylicz JJ, Heard E. Molecular mechanisms of facultative heterochromatin formation: an X-chromosome perspective. Annu Rev Biochem. 2020;89:255–282. doi: 10.1146/annurev-biochem-062917-012655. [DOI] [PubMed] [Google Scholar]
- 3.Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 4.Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 5.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lock LF, Takagi N, Martin GR. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 7.Gendrel A-V, et al. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev Cell. 2012;23:265–279. doi: 10.1016/j.devcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu C, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monfort A, et al. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 2015;12:554–561. doi: 10.1016/j.celrep.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moindrot B, et al. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 2015;12:562–572. doi: 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dossin F, et al. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature. 2020;578:455–460. doi: 10.1038/s41586-020-1974-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brockdorff N. Polycomb complexes in X chromosome inactivation. Philos Trans R Soc Lond B. 2017;372:20170021. doi: 10.1098/rstb.2017.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva J, et al. Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 Polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 15.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 16.de Napoles M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Dixon-McDougall T, Brown CJ. Independent domains for recruitment of PRC1 and PRC2 by human XIST. PLoS Genet. 2021;17:e1009123. doi: 10.1371/journal.pgen.1009123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 19.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, et al. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- 22.Pintacuda G, et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish Polycomb-mediated chromosomal silencing. Mol Cell. 2017;68:955–969. doi: 10.1016/j.molcel.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almeida M, et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science. 2017;356:1081–1084. doi: 10.1126/science.aal2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nesterova T, et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat Commun. 2019;10:3129. doi: 10.1038/s41467-019-11171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endoh M, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 26.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 27.Borensztein M, et al. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol. 2017;24:226–233. doi: 10.1038/nsmb.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corbel C, Diabangouaya P, Gendrel AV, Chow JC, Heard E. Unusual chromatin status and organization of the inactive X chromosome in murine trophoblastgiant cells. Development. 2013;140:861–872. doi: 10.1242/dev.087429. [DOI] [PubMed] [Google Scholar]
- 29.Takagi N, Abe K. Detrimental effects of two active X chromosomes on early mouse development. Development. 1990;109:189–201. doi: 10.1242/dev.109.1.189. [DOI] [PubMed] [Google Scholar]
- 30.Mugford JW, Yee D, Magnuson T. Failure of extra-embryonic progenitor maintenance in the absence of dosage compensation. Development. 2012;139:2130–2138. doi: 10.1242/dev.076497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakata Y, et al. Defects in dosage compensation impact global gene regulation in the mouse trophoblast. Development. 2017;144:2784–2797. doi: 10.1242/dev.149138. [DOI] [PubMed] [Google Scholar]
- 32.Corbel C, Heard E. Transcriptional analysis by nascent RNA FISH of in vivo trophoblast giant cells or in vitro short-term cultures of ectoplacental cone explants. J Vis Exp. 2016;114:e54386. doi: 10.3791/54386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takada T, et al. Mouse inter-subspecific consomic strains for genetic dissection of quantitative complex traits. Genome Res. 2008;18:500–508. doi: 10.1101/gr.7175308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su IH, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 35.Sugishita H, et al. Variant PCGF1-PRC1 links PRC2 recruitment with differentiation-associated transcriptional inactivation at target genes. Nat Commun. 2021;12:5341. doi: 10.1038/s41467-021-24894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farcas AM, et al. KDM2B links the Polycomb repressive complex 1 (PRC1) to recognition of CpG islands. eLife. 2012;18:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andergassen D, Smith ZD, Kretzmer H, Rinn JL, Meissner A. Diverse epigenetic mechanisms maintain parental imprints within the embryonic and extraembryonic lineages. Dev Cell. 2021;56:2995–3005. doi: 10.1016/j.devcel.2021.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.da Rocha ST, et al. Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol Cell. 2014;53:301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Cooper S, et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun. 2016;7:13661. doi: 10.1038/ncomms13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dorbrinić P, Szczurek AT, Klose RJ. PRC1 drives Polycomb-mediated gene repression by controlling transcription initiation and burst frequency. Nat Struct Mol Biol. 2021;28:811–824. doi: 10.1038/s41594-021-00661-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jadhav U, et al. Replicational dilution of H3K27me3 in mammalian cells and the role of poised promoters. Mol Cell. 2020;78:141–151. doi: 10.1016/j.molcel.2020.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012;28:1838–1844. doi: 10.1093/bioinformatics/bts280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. [DOI] [Google Scholar]
- 44.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi S, et al. Genome-wide stability of the DNA replication program in single mammalian cells. Nat Genet. 2019;51:529–540. doi: 10.1038/s41588-019-0347-5. [DOI] [PubMed] [Google Scholar]
- 47.Perry J, Palmer S, Gabriel A, Ashworth A. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11:1826–1832. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011;17:10–12. [Google Scholar]
- 49.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kent WJ, et al. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Degner JF, et al. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The allele-specific RNA-seq dataset is publicly available through Gene Expression Omnibus (GEO). The accession number is GSE168140. Raw data of Figs. 3c and 5c are provided in Supplementary Tables 2 and 3, respectively. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.