Abstract
Noncovalent interactions are the main driving force in the folding of proteins into a 3D functional structure. Motivated by the wish to reveal the mechanisms of the associated self-assembly processes, scientists are focusing on studying self-assembly processes of short protein segments (peptides). While this research has led to major advances in the understanding of biological and pathological process, only in recent years has the applicative potential of the resulting self-assembled peptide assemblies started to be explored. Here, major advances in the development of biomimetic supramolecular peptide assemblies as coatings, gels, and as electroactive materials, are highlighted. The guiding lines for the design of helical peptides, β strand peptides, as well as surface binding monolayer-forming peptides that can be utilized for a specific function are highlighted. Examples of their applications in diverse immerging applications in, e.g., ecology, biomedicine, and electronics, are described. Taking into account that, in addition to extraordinary design flexibility, these materials are naturally biocompatible and ecologically friendly, and their production is cost effective, the emergence of devices incorporating these biomimetic materials in the market is envisioned in the near future.
Keywords: bioelectronics, hydrogels, nanostructures, peptides, self-assembly
1. Introduction
The formation of ordered nanostructures by molecular selfassembly represents a central theme in nanoscience and nanotechnology.[1] The association of molecular building blocks by a set of non-covalent interactions, including hydrogen bonds, aromatic, electrostatic, and hydrophobic interactions, results in their organization into various architectures at the nanoscale. The simplest structures are nanospheres and 1D structures, such as nanotubes, nanoplates, and fibrils.[2] However, other 2D and 3D structures could also spontaneously emerge from the assembly of one, or more, types of molecular elements into energetically stable supramolecular entities.[3,4] These noncovalent interactions can, for example, lead to the formation of macroscopic structures such as hydrogels with nanoscale order.[5,6]
Biological and bioinspired molecules are especially attractive building blocks for the construction of self-assembled structures.[7,8] This is because of the natural hierarchical organization of biomolecules into functional supramolecular assemblies in living cells, which due to the biocompatibility, water solubility as monomers, the ability to be easily functionalized, and availability of established synthesis procedures is technologically attractive. Among the biological building blocks, proteins and peptides are the most diverse ones. These polyamide structures can be hydrophobic, hydrophilic, or possess mixed water interacting properties that result in amphiphilicity, a central theme in the process of self-organization. Furthermore, the functional side chains of proteins and peptides may facilitate other noncovalent interactions that are essential for the organization into defined 3D architectures. This includes the aromatic amino acids that may be involved in stacking interactions, charged amino acids that allow electrostatic interactions, and polar amino acids that could be involved in intramolecular and intermolecular hydrogen bond networks. Indeed, protein and peptide assemblies are the core of the vast majority of functional elements and molecular machines in nature.
Assemblies of bioinspired and de novo designed peptides exhibit remarkable functional behaviors. They can display structural and mechanical robustness, yet can be controllably and reversibly disassembled. Furthermore, they may also have unique optical, electronic, and other physical properties, making them attractive candidates for technological applications.[9] The self-assembly behavior of peptide nanostructures and immerging properties have been described in several reviews in recent years.[10–13] The aim of this prospective is tohighlight peptide design considerations for tailoring the functional behavior of the peptide assemblies. After a brief background on different self-assembling motifs and peptide folding, we describe the utilization of self-assembling peptides in three very different applications: functional surface coatings, electroactive materials, and hydrogels. We specifically highlight guiding lines for peptide design for each of the discussed applications and overview the functional behavior of the resulting assemblies and devices based on them. This prospective elucidates the extraordinary potential of these biomimetic supramolecular assemblies to become extremely useful in technological applications in the near future.
2. Background-Peptide Folding and Self-Assembly
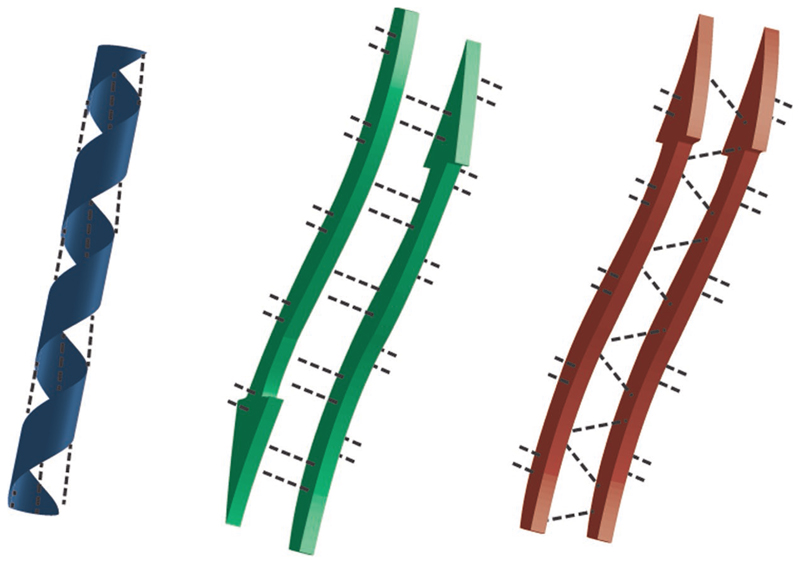
Folding of proteins into functional tertiary structures is governed by self-association of their backbone and amino acid side chains via a variety of noncovalent interactions.[3,14,15] These interactions lead to the formation of subunits organized mainly into helices or sheet-like structures (Figure 1), constituting the secondary structure of the proteins. These self-assembled segments are spatially arranged into the 3D functional structure of the proteins. Understanding the relationships between the primary structures of proteins—the sequence of amino acids covalently bonded into a polymeric chain—and their 3D tertiary structure, and, moreover, its emerging functional behavior, has been, and still is, a major scientific challenge. Nevertheless, the factors governing the folding of its short segments into a secondary structure have been vastly revealed during the years. Folding was found to be dictated by a network of hydrogen bonding interactions formed between a carbonyl oxygen of one amino acid and an amino hydrogen of another amino acid (Figure 1). Intrastrand hydrogen bonding interactions between amine and the oxygen carbonyl of amide bonds that are three or four amino acids apart in the sequence induce helix formation, whereas multiple interstrand hydrogen bonding interactions between amine group in one strand and the oxygen carbonyl group in another strand govern β-pleated sheet formation. The specific arrangement of the hydrogen bond network further dictates the type of helix (e.g., α, 310) or β-sheet (e.g., parallel or antiparallel) formed.
Figure 1. Schematic presentation of peptide assembly into helix and β-sheets. Dashed lines represent hydrogen bonding networks facilitating the assembly process.
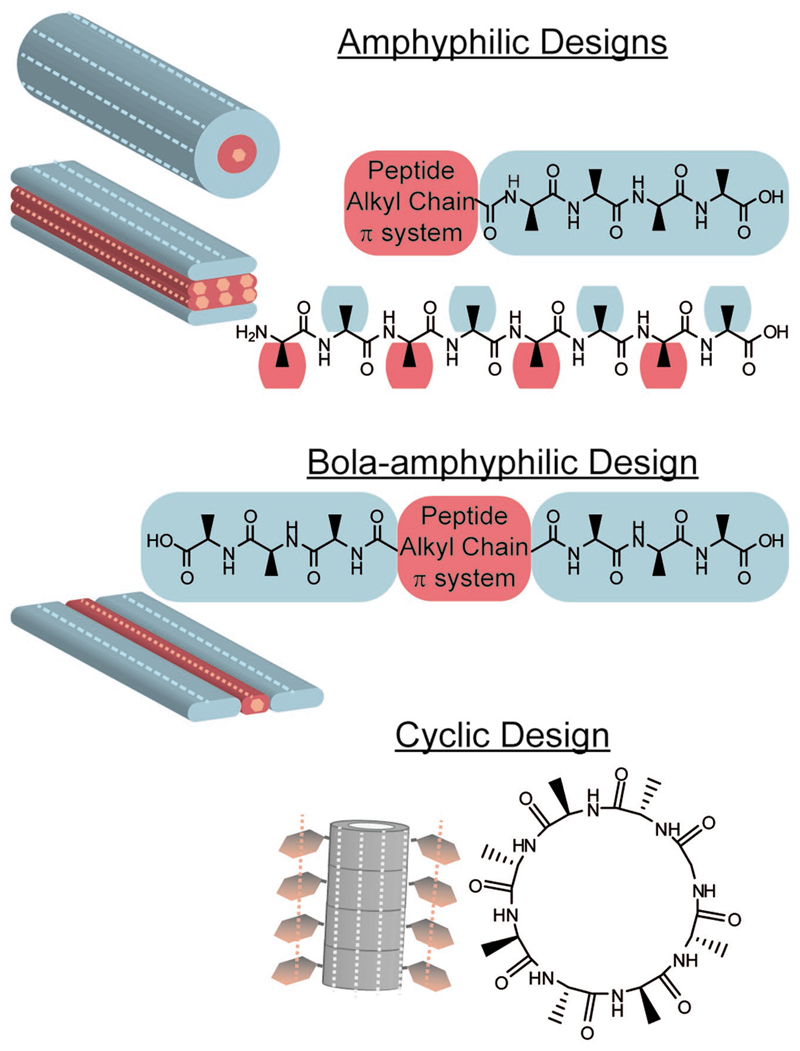
Realization of short sequences of amino acids, namely peptide sequences, which would self-assemble and fold into a well-defined secondary structure,[16–18] and possibly to larger aggregates, sets the grounds for the use of peptides as building blocks of functional materials. Long-range self-assembly of β-pleated sequences into fiber, ribbon, or nanotube architectures has attracted a lot of attention. In general, amphiphilicity is one of the main driving forces of peptide self-assembly and β-sheet folding (Figure 2).[19] In the simplest design, a hydrophobic and a hydrophilic segments are fused to each other forming an amphiphilic peptide.[12] One of the most widely studied β-sheet self-assembling motif is an amphiphilic pentapeptide sequence (KLVFF) derived from the core of amyloid β (Aβ) proteins, which are commonly found in a misfolded β-sheet fibrillary structure in the brain of Alzheimer’s patients.[20–22] The self-assembly of amyloid protein and peptide fibrils and immerging applications have been widely reviewed in recent years.[10,11] Reches and Gazit have found that the dipeptide homomotif, diphenylalanine, of the Aβ core pentapeptide can, by itself, self-assemble into β-sheet arrangement, forming rigid nanotubes.[23] This minimal design has attracted a considerable interest due to its simplicity. It is now well established that many homo- and hetero-dipeptides can self-assemble in a similar fashion, forming diverse nanometric objects.[24–27] This approach was further extended by Gazit and co-workers, recently, to the demonstration of self-assembling dimeric peptide nucleic acid molecules.[28] Furthermore, recently it was demonstrated that even single amino acids can aggregate to form fibrils.[29]
Figure 2.
β-Sheet forming designs. Light blue and red represent designated hydrophilic and hydrophobic segments of the peptide, respectively. Side chains were omitted for clarity. Schematic representation of common self-assembled β-sheet structures is depicted, with dashed light blue and pink lines showing the direction of hydrogen bonds and π-stacking, respectively. Note that the cyclic peptide design is an example from a family of cyclic designs consisting of D,L-α-peptides (the gray color represents unspecified requirement for the nature of side chains).
The understanding of the self-assembly process has led to the realization of other β-sheet forming peptide motifs. For example, a more synthetic approach for the design of selfassembling peptide amphiphiles is based on the conjugation of an aliphatic tail to a short β sheet forming peptide sequence.[12] Alternative amphiphilic designs include bola-amphiphilic sequences with a hydrophobic core appended by two hydrophilic segments at both ends (Figure 2),[30] and alternating amphiphilic sequences that promote bilayer assembly of fibers with hydrophobic core and hydrophilic surfaces.[7,31] Furthermore, cyclic peptide designs that form flat ring conformations upon cyclization, which undergo self-assembly into nanotubes with diameter that is defined by the sequence length, have also been suggested.[32]
Interactions between amino acid side chains dictate and stabilize peptide folding. Furthermore, these interactions govern further folding and aggregation into large supramolecular architectures and the resulting morphology of the peptide assemblies. The specific choice of amino acid sequence also determines the properties of the assemblies. This is a particularly important feature since the pallet of 22 natural amino acids provides an extraordinary flexibility in the design of materials properties. Moreover, since the peptide monomers are prepared by organic chemistry methodologies, synthetic ligands that are not part of the canonical amino acid pallet can be included in the design and further increase the design flexibility.
Since self-assembly is a kinetically controlled process, the assembly environment provides another handle to affect peptide assemblies’ morphology. For example, it was shown that by altering the solvent environment of diphenylalanine, either nanotubes or small colloids could be obtained in a reversible manner.[33] Mixing two variants of diphenylalanine with different termini was shown to enable simultaneous appearance of both morphologies and their intermixing into necklaces like architecture.[34] For longer peptide sequences, e.g., peptides based on the Aβ core sequence, the solvent environment was shown to dictate fiber bundling tendency, and thus, fibers diameter, as well as their rigidity.[35] Recent work has demonstrated that the chirality of fibrils based on alternating amphiphilic peptide with a naphthalene diimide (NDI) side chain could be reversed by tuning the solvent composition.[36] Finally, it should be noted that peptide side chains and termini could interact with other molecules and with materials’ surfaces, enabling the formation of hybrid structures.
The development of peptide models for helical assemblies reflects a much more complicated task, as the stabilization of helical structure requires much longer sequences as compared to the β-sheet ones. In the first demonstration of helical peptide nanostructure assembly, a linear combination of 3-4 heptad repeats (i.e., 21–28 amino acid long peptide sequence) was used to form ordered assemblies.[37] The need for long peptide sequences stems from the fact that stable helical structures require numerous intrastrand hydrogen bonds to stabilize the secondary structure. In addition to hydrogen bonding networks that stabilize the secondary structure within each helix, hydrophobic interaction between side chains of differenthelices provides energetic forces to stabilize the supramolecular arrangement of the networks. Noncoded amino acid building blocks can be used to overcome the building block length limit and stabilize much shorter helical fragments. Specifically, the α-aminoisobutyric acid (Aib), a natural, noncoded amino acid, that is used by microorganisms to secure helical structure for nonribosomal synthesized peptides such as the alamethicin antibiotics produced by the Trichoderma viride fungus,[38] was used to stabilize the structure of a much shorter peptide fragment.[39] The steric hindrance induced by an additional methyl group of the C carbon of the amino acid makes Aib the amino acid with the highest reported helical propensity.[38] Indeed, it was shown that Aib incorporation into a single heptad monomeric peptide results in the formation of supra-helical assemblies, in which helixes from different building blocks interact to form larger ordered structures. This was obtained by helical organization of each of the building blocks, with hydrophobic interactions stabilizing the interactions between building blocks.[39]
Another very important recent extension to the family of helical structures came from the study of functional bacterial amyloids. Landau and co-workers have used crystallography to determine the molecular organization of Staphylococcus aureus PSMα3 toxic fibrils.[40] The researchers have revealed the formation of amphipathic α-helices that are folded to stack perpendicular to the fibril axis into sheets.[40] This organization of cross-α fibrils, as compared to the canonical cross-β organization, represents a newly identified fold for amyloid fibrils and stress the importance of helical fibrillary assemblies also for functional extracellular entities. However, further studies may provide a more minimalistic functional peptide fragments.
3. Peptide Monolayers and Surface Coatings
Controlling the properties of a substrate is of high importance in many research fields and applications. This includes selfcleaning surfaces, sensors, cell engineering, and biomedical devices. Tailoring surface properties can be achieved by patterning the surface and/or binding a coating with different chemical and/or biological moieties. Peptides offer the ability to tailor multiple surface properties by sequence design. In this section, we review recent approaches to modify solid surfaces using peptides and peptide-based assemblies.
3.1. Surface Modification by Peptides Self-Assembled Monolayers
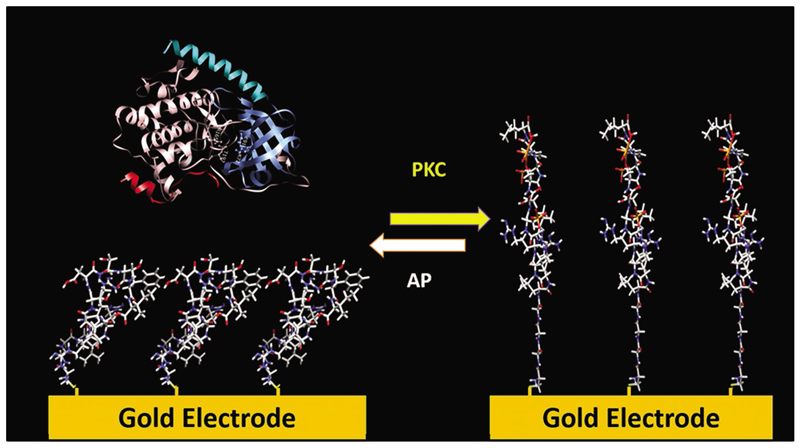
The most common approach for attaching peptides to solid surfaces is using the thiol moiety of the amino acid cysteine to bind a peptide to gold and form a peptide monolayer on the surface.[41] This approach was used, for example, to modify gold surfaces to detect the activity of a family of enzymes, called kinases, which catalyze protein phosphorylation.[42–45] The phosphorylation process affects cell growth, differentiation, and apoptosis, and thus irregular phosphorylation is strongly linked to different diseases such as cancer, diabetes, and Alzheimer.[44] To detect the activity of several kinases, various peptides, which can undergo phosphorylation by different kinases, were immobilized as a well-packed self-assembled monolayer (SAM) on gold electrodes.[42–45] The electrical properties of the electrode were found to depend on minor changes of the electrode surface. Therefore, measuring differences in the electrochemical behavior of the electrode has enabled identifying phosphorylation and dephosphorylation processes that promote changes in the peptides conformation and surface roughness, and disrupt the monolayer order, forming pinholes (Figure 3). Accordingly, the changes in the peptides conformation enable the sensitive and selective detection of kinase activity.[42–45]
Figure 3.
Sensing enzyme catalysis of phoshosphorylation and dephosphorylation processes using SAM of peptides on gold. A dense monolayer of peptides on a gold electrode undergoes phosphorylation that causes disruption of the monolayer order and leads to a change in the surface impedance. Removal of the phosphate groups induces reordering of the monolayer. Figure 3 is based on data in refs.[42–45].
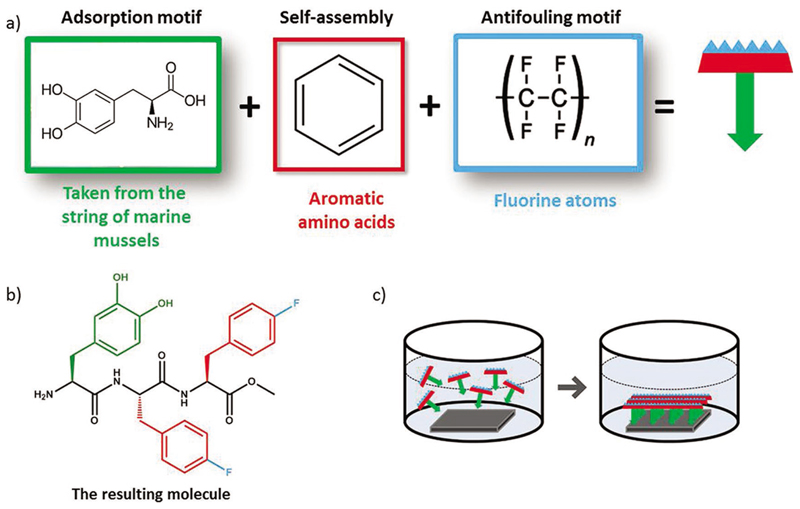
One of the most promising uses of peptide monolayers is as antifouling coating to prevent the adhesion of proteins and organisms to the surface. The nonspecific adsorption of proteins on a surface, and later on, the adsorption and growth of microorganisms to form a biofilm, is a serious problem for the marine industry, water delivery systems, food packaging, and for medical devices.[46–55] Formation of biofilm on medical devices, for example on implants, may cause severe infection that may even result in the removal of the implant. Therefore, there is an acute need to resist bacterial colonization and prevent biofilm formation.[54] Several approaches were studied for this purpose, including the binding of antimicrobial agents to the surface of the implant and the development of antiadhesive coatings.[46] An intriguing approach is the use of antimicrobial peptides (AMPs) coatings to affect bacterial adhesion.[56] AMPs are being produced by the immune system of many organisms and are known for their wide spectrum of activity against bacteria and other pathogens.[53,56,57] The main concerns of using AMPs coatings were that surface-bound AMPs, which are usually used in their soluble form, may be less active once immobilized, and that the most influential factors that affect the activity profile would be the length of the spacer and the amount of target-accessible peptides.[58] Yet, several studies proved the ability to immobilize AMPs to a substrate while maintaining their activity. For instance, the synthetic peptide, melimine, which combines portions from two cationic AMPs, melittin and protamine, was covalently linked to different substrates, to polymeric contact lenses via 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) coupling reaction and to glass via azide linkers that has an additional variable reactive group toward amine group in the peptide sequence. This AMP monolayer indeed reduced the adhesion of tested bacteria.[59,60] Another cationic AMP, Palm–Arg–Arg–NH2, was fixed to silicone substrate through a hydroxysilane group that was coupled to the peptide during synthesis.[61] Other AMP-based coatings were formed on gold and titanium substrates through thiol and silane groups, respectively.[62,63] Immobilizing the AMP cecropin-melittin on gold nanoparticles coated surfaces improved the AMP density on the surface, compared to most of the other studies, and resulted in high antimicrobial activity.[64] The phage display method was used to identify a peptide that binds to titanium, which was used as a linker to bind AMP to a titanium surface.[65,66] Zwitterionic peptides can also prevent biofouling as they strongly hydrate via ionic solvation. These peptides act similarly to the commonly used antifouling polymer poly(ethylene glycol) that achieves its hydration through hydrogen bonding. This hydration layer prevents protein absorption and therefore avoids adhesion of other organisms.[67,68] Peptide sequences of alternating or randomly mixed charged canonical amino acids: glutamic acid, aspartic acid, and lysine, were used to decorate gold surfaces. Protein adsorption assays proved that these peptides present a good antifouling behavior.[69] Recently, a research by Reches and coworkers demonstrated the rational design of a new self-assembled antifouling coating based on a tripeptide (Figure 4).[70] In this novel type of peptide, the amino acid 3,4-dihydroxy-L-phenylalanine (DOPA) is used to anchor the peptide to the surface. DOPA has a key functional role in mussel adhesive proteins (MAPs), the glue proteins of marine mussels.[71,72] Having an adaptive nature, it can adhere to a large variety of surfaces.[71] Two fluorinated phenylalanine appended to the DOPA are used both to promote intermolecular self-assembly and to mimic the fluorine-carbon bond of Teflon, and hence providing a surface with nonsticky, antifouling properties.
Figure 4.
De novo designed peptide based antifouling coating. a) Conceptual design approach: the peptide design includes three functions: adsorption to the surface by a sticky amino acid, self-assembly motif of aromatic amino acids, and antifouling motif by incorporation of fluorine atoms. b) Chemical structure of the peptide NH2–DOPA–(4F)Phe–(4F)Phe–COMe. c) Schematic presentation of spontaneous coating of the surface with a peptide monolayer by a dip-coating process. Figure 4 is based on data in ref. [70].
Promotion of cell adhesion and growth on biomaterials plays a significant role in the success of implants integration. Anchoring cell binding motifs, such as the Arg–Gly–Asp peptide, to the surface can improve the biomaterial properties.[73] The functionalization of titanium surfaces, a material that is widely used in the implant industry,[74] with cell binding peptides by utilizing the DOPA or Cys amino acids as binding element was demonstrated.[73,75]
Aromatic groups can also be exploited for the attachment of peptides onto surfaces. For example, the aromatic rings of tyrosine residues can bind to graphite.[76] Controlling graphite surface wettability through self-assembled peptides was demonstrated by displaying hydrophobic or hydrophilic amino acids on a graphite-binding peptide. In addition, the co-assembly of the mixed peptides (hydrophobic or hydrophilic) in different ratios enabled the formation of a film with tunable wetting properties.[76]
To increase the functionally of surfaces, we should understand better the underlying principles of peptide interactions with solid inorganic or organic surfaces. A better understanding would lead to the discovery of additional functional groups, beyond cysteine and DOPA.[77–79]
3.2. Surface Modification by Self-Assembled Peptide Particles
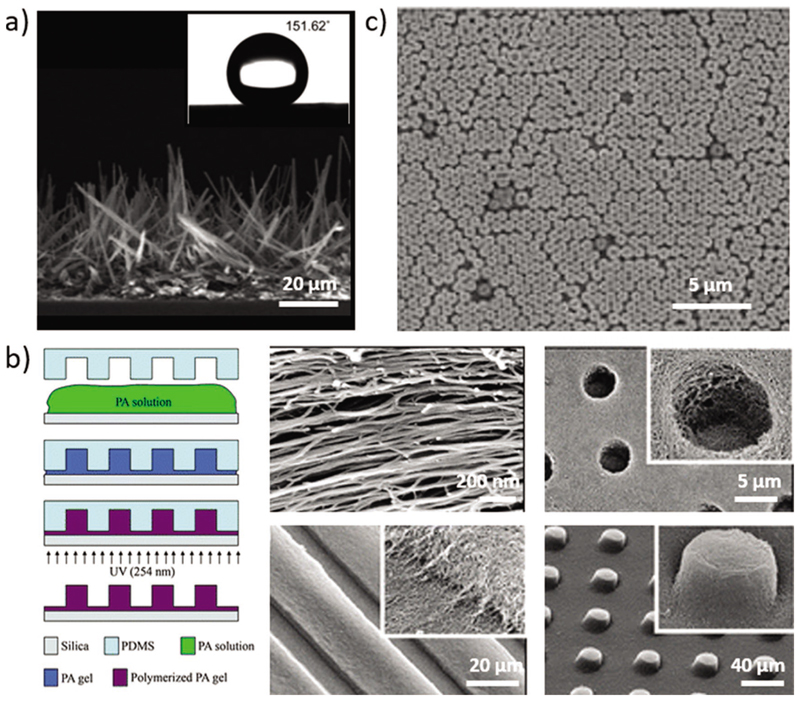
Peptides hold a great promise as biomolecular surface coatings and can provide functionality derived not only from the peptide sequence, but also from its nanoscale assembly. Park and co-workers fabricated self-cleaning superhydrophobic surfaces via the formation of hierarchical nano-microstructures by solidphase self-assembly of diphenylalanine into vertical nanowires (Figure 5a).[80]
Figure 5.
Surface modification by self-assembled peptides particles. a) Peptide nanowires array formed after treating FF with pentafluoroaniline. A water contact angle shown in the inset indicates the formation of a superhydrophobic surface. Reproduced with permission.[80] Copyright 2009, RSC. b) Surface patterning using nanofibers of PAs, including holes, channels, and posts. Reproduced with permission.[82] Copyright 2009, RSC. c) Hexagonal monolayer arrangement of self-assembled Boc–Phe–Phe–Phe bio-nanospheres as fabrication patterning masks. Reproduced with permission.[83] Copyright 2010, Wiley-VCH.
For many technological applications, it is also required to control the placement and orientation of the assemblies in the macroscopic scale, over a large area.[81] Orienting and micropatterning of peptide amphiphiles (PAs), over large area, can be obtained simultaneously to the self-assembly of the peptides to nanofibers or gels (Figure 5b).[81,82] Directing the assemblies within topographically confined areas was obtained by combining top-down methods such as soft lithography and electron-beam lithography together with the self-assembly process. As the peptide monomers that were used contain cell adhesion or neurons outgrowth function, the alignment and micropatterning of the nanometric assemblies were shown to direct cells and control cellular behavior, making this strategy useful and important in regenerative medicine and cell engineering.[81,82]
The self-assembly of the aromatic protected tripeptide Boc–Phe–Phe–Phe into nanospheres led to the development of a novel patterning method (Figure 5c).[83] In this method, the spontaneous packing of self-assembled bio-nanospheres into a hexagonal monolayer serves as a patterning mask for selective deposition and etching.[83]
The modification of surfaces with peptide-based self-assembled particles can also increase the surface area of the substrate. This could be useful for electrochemical detection as the increase in the surface area of the electrode increases the signal.[84,85]
4. Bioinspired Electro Active Materials
The realization of charge transport in peptidic systems has been attracting a substantial interest over the years. This research field is motivated both by the desire to use peptides as simple models to understand charge-transport processes in natural systems,[86–92] and by the possibility to prepare biocompatible electronic materials via self-assembly of small molecules. Progress in the research in these two directions is highlighted below.
4.1. Peptide Molecular Bridges
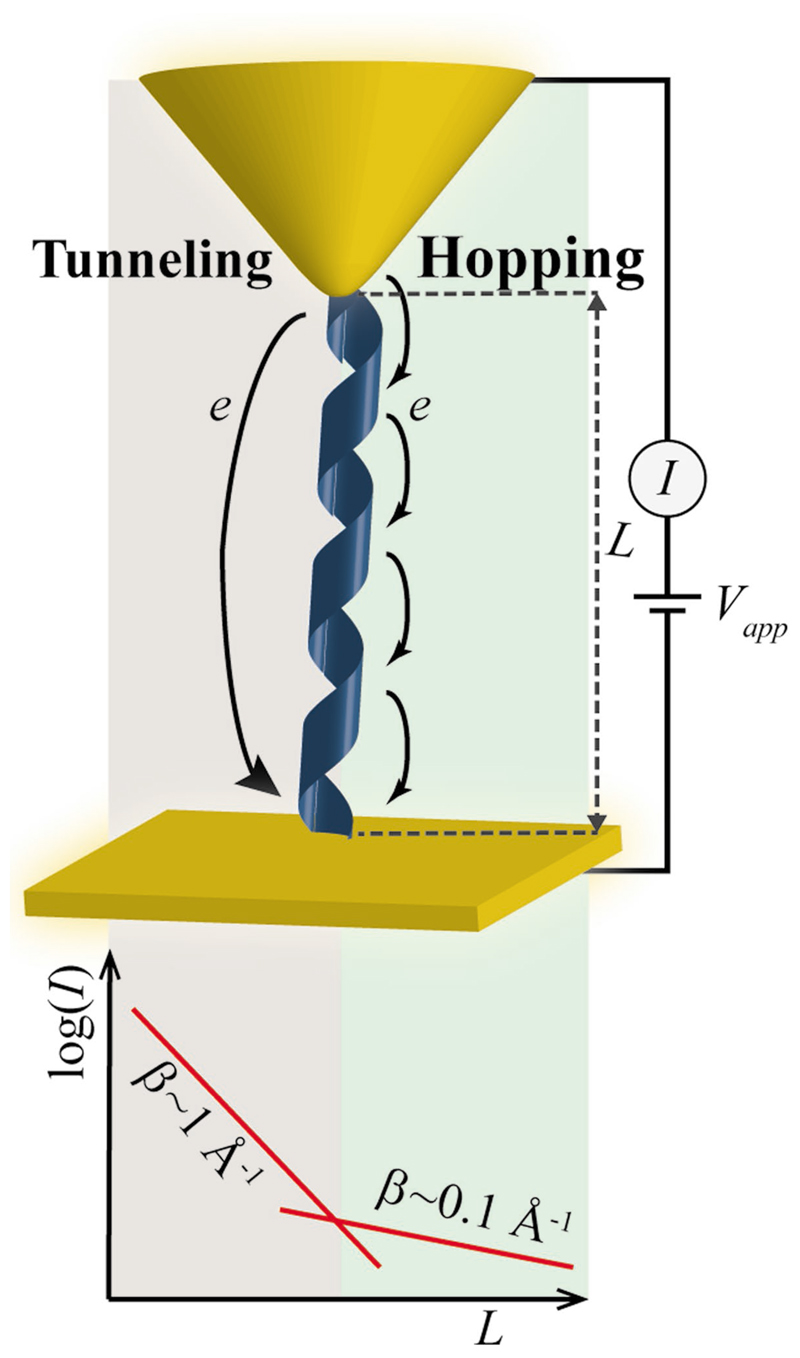
Studies of electron transfer (ET) in peptide molecular junctions have revealed that, in similar to other organic molecular bridges, ET processes in peptides occur either by one-step superexchange mechanism, also known as electron tunneling, or by a multistep hopping mechanism, depending on the length of the molecular bridge (Figure 6). For molecular bridges shorter than ≈1–2 nm, tunneling governs ET processes, giving rise to a strong dependence of the current on the bridge length.[86,93–99] For longer bridges ET is facilitated mainly by the hopping mechanism, and the length dependence becomes less pronounced.[86,93,96] The secondary structure of the peptide molecules is highly important in determining the ET efficiency. For example, for the commonly studied helical peptide molecular junctions the bridge length is much shorter than the extended peptide molecular length, giving rise to ET efficiency higher than expected from the molecule length. Moreover, helical peptides possess a significant inherent dipole that introduces a rectifying behavior to the peptide molecular junctions,[95,100–103] allowing exploitation of peptides as diodes in various switching applications. The ability to control the dimerization of such helical peptides to form parallel or antiparallel coiled-coil protein conformations was used to design and switch the rectification polarity and magnitude of such peptide molecular switches.[104] It was also suggested that the presence of hydrogen bonds may increase the rate of ET.[105] This may explain the prediction that an antiparallel β-sheet peptide assembly would provide a higher ET rate than both α-helix and 310-helix peptides.[106,107]
Figure 6.
ET in peptide molecular bridges. Schematic depiction ofET mechanisms through a helical peptide bridging a gold substrate, and an atomic force microscopy tip (a common experimental setup to study ET through monolayers). The dependence of the current (I) on the length of the molecule (L) is depicted below, demonstrating a crossover between tunneling and hopping dominated ET, depending on the length of the molecule. A crossover point at 1 < L < 2 nm, indicated by a change in the current decay coefficient, β, has been reported. Figure 6 is based on data in refs. [94,95,97,99].
One of the intriguing questions of the ET process is how it is affected by the peptide sequence. In one of the first studies of the effect of side chains on peptide molecular junction ET rate, only a small dependence of ET on the peptide sequence was observed.[108] However, recent studies indicate that ET efficiency does depend on the side chain, with preference to redox active side chains, or ones that can be charged.[97,109] It was further suggested that the efficiency is largely dependent on the overall charge of protonating side chains[97,98] and that it strongly depends on the rotational motion of the peptide chain.[106,110,111]
4.2. Charge Transport through Self-Assembled Peptide Structures
The formation of aromatic stacking upon self-assembly (Figure 2) has motivated studies of long-range ET through selfassembled peptide structures. Efficient ET was observed in supramolecular ET bridges constructed of vertically aligned selfassembled cyclic peptide nanotubes with alternating tryptophan and tyrosine side chains.[112] Despite the short bridge length, ET was found to be dominated by the hopping mechanism. The ability to sustain intermolecular ET has further been exploited in the formation of planner devices, in which a specifically designed peptide—containing a self-assembling motif, an aromatic unit and a surface linking group—self-assembled into a monolayer between two remotely located electrodes.[113] Indeed, ET between the electrodes was detected and monitored by means of current–voltage curves. ET could also be detected for peptide structures self-assembled in solution prior to deposition between the electrodes. One of the simplest examples involves the diphenylalanine nanotubes. Assembly of single diphenylalanine nanotubes, and bundles of them, between electrodes by electrophoresis has resulted in the appearance of current, which, albite being in the range of a few pA, was significantly higher than the background current.[114] Currents in the range of nA were observed for cyclic diphenylalanine peptide nanowires.[26] The conductivity of fibers of a de novo designed phenylalanine rich helix forming peptide was found to be much higher than that of a film of the non-assembled peptide, attesting the importance of self-assembly and the ensued π stacking in promoting the conductivity.[115] It should be noted that such long-range ET is dominated by the hopping process.
As in the case of molecular junctions, amino acid side chains play a critical role in controlling the electronic properties of the peptide assemblies. Theoretical studies have shown that the introduction of aromatic side chains reduce the bandgap of the self-assembled structures, with the largest effect (i.e., smallest bandgap) predicted for peptides incorporating tryptophan.[116,117] An experimental insight to this behavior was manifested by a fivefold higher conductivity of self-assembled diphenylalanine nanodots once replacing one of the phenylalanine in the original sequence with tryptophan.[118]
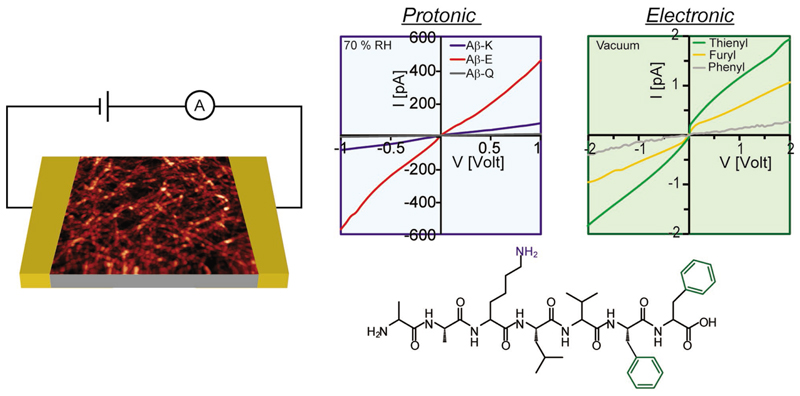
The ability to influence the conductivity by the nature of peptide side chains suggests that the conductivity can be further tuned by the incorporation of nonnatural aromatic groups. One of the first demonstrations of promoting electron delocalization by stacking large aromatic groups was provided using cyclic peptide nanotubes to which NDI groups were embedded by conjugation to lysine.[119–121] Indeed, the replacement of phenyl group of phenylalanine by other aromatic groups, such as NDI, as well as thienyl and furyl, was shown to promote ET in peptide fibrils (Figure 7, right curve).[35,36,122] ET promoting aromatic groups can also be conjugated to the peptide termini,[123–125] or embedded in the backbone in a bola-amphiphilic design (Figure 2).[126–131] A negative reference testifying the significant role of aromatic rings in inducing conductivity of peptide fibrils is the lack of measurable current in a peptide fibrils lacking aromatic amino acids under vacuum conditions.[35,132] Coassembly of donor and acceptor peptide conjugates was achieved either by kinetic control,[133] or by peptide side chain electrostatic interactions.[134] Both self-sorted and coassembled fibers were obtained by kinetic control of selfassembly providing insight into the correlation between energy transport within the assemblies and their structural organization.[133] The formation of electron–donor pairs along peptide fibers was obtained by coassembly of positively and negatively charged peptide amphiphiles to which an electron donor and acceptor were, respectively, conjugated.[134] An improved conductivity was suggested for these fibers.
Figure 7.
Effect of side chain on charge transport. Effects induced by the choice of charged/aromatic side chains are shown at the left/right I–V curves, respectively. Data were collected at 70% relative humidity/vacuum for the left/right panels, respectively. The structure of the “native” Aβ peptide used in these studies is shown below. Schematic depiction of current measurement setup is shown to the left, together with an atomic force microscopy image of the fibers. Reproduced with permission.[122] Copyright 2014, Wiley-VCH.
In addition to the dependence on the sequence, the electronic properties also depend on the assembly configuration, and hence, on the assembly conditions.[35] Generally, it was found that enhancement of the conductivity is obtained by promoting the formation of long nanofilaments with a large persistent length. A fascinating example of environmentally controlled structural effect is the dependence of the conductivity on fibrils’ chirality, which in return can be tuned by addition of cosolvents to the assembly environment to control the different intermolecular interactions.[36] The alignment of the fibrils between the electrodes is also a critical parameter for the conductivity, with the conductivity along the peptide filaments exceeding that in perpendicular to the filaments by an order of magnitudes.[127] Such alignment could be obtained, for example, by forming a wettability gradient on the chip surface.[135]
4.3. Proton Conduction
Self-assembling peptide sequences commonly contain acidic or basic side chains, and their edges remain many times uncapped, and hence, can also add acidic and basic groups to the peptide (Figure 7, bottom). These groups can easily protonate or deprotonate, by this facilitating a route for proton transport. Proton transport could explain the enhancement of the conductivity with increase in the relative humidity of the atmosphere, observed for nanofibrillar films of elastin-related peptide, in which sequence does not include any aromatic unit.[132] Indeed, a current onset was observed in these studies once applying a voltage greater than ≈1.2 V, suggesting water electrolysis as the source of charge carriers in the system. Proton transport could also explain the much higher conductivity measured for Fmoc-L3 nanotube films when measured in air atmosphere compared to vacuum.[123]
As in the case of ET, side chains can modulate the proton transport process. Replacing the basic lysine amino acid with aspartic acid was found to give rise to increase in the conductivity of peptide fibrils (Figure 7, left curve).[136,137] This effect was assigned to both higher concentration and higher mobility of the charge carriers. The peptide fibrils network was found to behave as a dilute proton conducting acid or base for peptides containing acidic or basic side chains, respectively. The uncapped end groups of the peptides used in this study were found to be much less effective in promoting proton conduction. It was recently shown that under high relative humidity conditions proton conduction can dominate the conduction of peptide fibrils to which nonnatural aromatic side chains were introduced in order to enhance electron conductivity.[138] A mixed conductivity of both electrons and protons was found for these peptides under vacuum conditions.[138] It should be emphasized that similar to the case of electron transport, proton conduction also depends on the morphology of the fibril network.[121]
4.4. Piezo- and Ferroelectricity
The formation of crystal structures lacking inversion symmetry by peptide self-assembly, which is related to the chirality of the peptide monomers, opens the way to the use of these materials in piezo micro-electromechanical systems. Indeed, piezoelectric effect was found for diphenylalanine-based nanotubes.[139–143] The piezoelectric coefficient of the diphenylalanine nanotubes was found to be approximately 60 pm V–1, comparable to that of the well-known inorganic piezoelectric material lithium niobate.[139] This value is significantly larger than the 2 pm V–1 value measured for biological tissues.[140] Furthermore, it was shown that a photoirradiation process can be used to switch coercive fields of diphenylalanine microtubes, comprised of bundled nanotubes, and hence, can influence the ferroelectric behavior of the structure.[144] However, the high coercive electric fields of the assemblies limit the polarization switching efficiency.[145] Upon heat treatment above 150 °C, these peptide nanotubes undergo an irreversible phase transition from noncentrosymmetric hexagonal structure to centrosymmetric orthorhombic structure, resulting in the loss of their piezoelectric and ferroelectric behavior.[145,146] Uniform polarization of nanotubes ensemble could be achieved by applying an electric field during the peptide self-assembly process, enabling fabrication of a peptide-based power generator.[147] In recent works, it was shown that the ferroelectric FF nanotubes exhibit a remarkable pyroelectricity farther extending the possibilities of using these peptide assemblies for heat energy harvesting as well as for thermometry applications.[148]
4.5. Applications of Peptides in Electronic Devices
The utilization of peptide nanostructures as active layers of electronic and optoelectronic devices is still in its infancy. In some devices the unique electronic and optoelectronic properties of the structures are exploited, while in other cases the structures were used as the scaffold of the device to aid in optimizing device performance.
Inspired by natural photosynthesis processes, the exploitation of peptides in solar cell applications is probably the most straightforward electro-optic application. Extending the absorption spectrum and photocurrent generation capabilities of peptide incorporating solar cell devices was obtained by appending a dye to the side chain, or one of the ends, of the peptide.[149–152] Monolayers of chromophore-modified peptide helix were found to be good ET antennas in dye-sensitized solar cells.[152] Chromophore-modified peptides were found to generate photocurrent, even when the peptide is physically adsorbed to the surface.[151] The formation of mixed monolayers of helical peptides with opposite dipole direction to which dyes with different excitation wavelengths were coupled enabled switching photocurrent direction by controlling the excitation wavelength.[153] Making use of peptides molecular dipole, the performance of organic solar cells was improved by adjusting their interfaces’ work function using peptide monolayers.[154] It should be noted that the peptide sequence and backbone conformation can have a significant influence on the work function modulation effect.[155,156]
Beyond the use of peptide monolayers, self-assembled peptides were also considered as scaffold organizers in solar cells. For example, the order induced by peptides’ self-assembly was considered to be the reason for improvement in cell performance of porphyrin—fullerene supramolecular composite layers in which porphyrin–peptide oligomers were used.[157] The applicability of peptide–poly-thiophene conjugates for hydrogen evolution applications was recently demonstrated.[158] In these materials, the peptide self-assembly improved electron transfer rate from the photoexcited polythiophene ligand, by this increasing the hydrogen evolution rate. In another example, self-assembled peptide scaffolds have been used to create light-harvesting supramolecular structures that mimic the natural photosynthesis process.[159] To this end, diphenylalanine nanotubes were co-self-assembled with tetra (p-hydroxyphenyl) porphyrins that served as a light-harvesting molecule and platinum nanoparticles that catalyzed electron dissociation. A recent work has demonstrated that porphyrins embedded in amphiphilic peptide nanofibers are packed into linear chains due to the 1D assembly of the peptides.[160] By controlling the porphyrin doping level, exciton diffusion length can be controlled by this influencing the energy transfer efficiency. As in the case of solar cells, the exploitation of selfassembled peptide fibrils as components of electronic devices was demonstrated. A cyclic tri-β peptide nanotubes appended with tetrathiafulvalene (TTF) side chains was used as the p-type layer of a Schottky diode.[161] A transistor active channel made of peptide-aromatic conjugates, e.g., in a bola-amphiphilic design, was also demonstrated.[127,162–164]
Even if charge transport is not directly exploited, peptide nanostructures offer large surface area, controllable wettability properties, and high stability that makes them attractive for use as the dielectric layer in supercapacitors.[165–167] The capacitive nature of peptide nanostructures has allowed the realization of non-volatile memory devices, with peptide nanodots as nanometric charge storage elements.[168]
5. Self-Assembling Hydrogels for Biomedical Applications
Hydrogels are semisolid gelatinous materials that contain mostly water (in some cases more than 99%).[169] Hydrogels frequently exhibit a combination of flexibility, softness, and bio-compatibility that is highly useful for a large number of applications. The predominant water content is highly desirable for biological application both at the molecular level as well as the cellular one. The utilization of hydrogels includes the slow release of drug molecules, tissue engineering and regeneration, and as a general scaffold for the organization of both organic and inorganic structures.
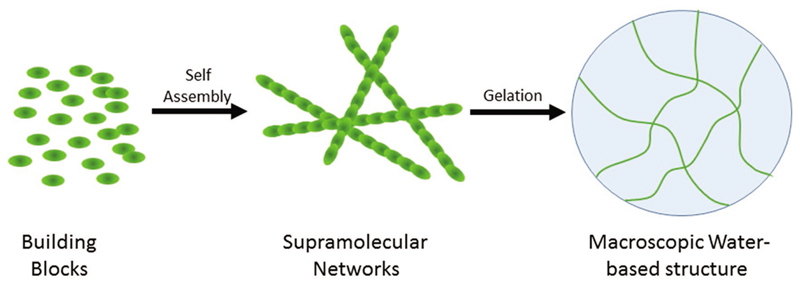
The formation of physical hydrogels is based on the molecular self-assembly of small building blocks that can form a network of fibrous structures at the nanoscale to support the macroscopic structures (Figure 8).[169] The structure of the intact gel is based on a set of noncovalent interactions that together can allow the formation of a rigid and continuous structure. The organization of assembling units into gels could be readily monitored by the transition of the solution from a solution phase into a solid one. The physical properties of the gels are determined by rheology while the internal order of the molecular network is probed by electron microscopy. Environmental scanning electron microscopy is an especially useful technique to determine the organization of hydrogels as the imaging is being done at a wet mode where the integrity of the gel material is being kept. Other physical tools to study the molecular ordering of hydrogels include circular dichroism, Fourier-transform infrared spectroscopy, and fluorescence spectroscopy. The molecular networks of the hydrogelator building blocks result is the capturing of a large number of water molecules to provide a structure that is both rigid but also mainly contain water without the need for cross linking. The main driving forces of the gelation process are hydrophobic interactions, π-π stacking, electrostatic interactions, and the formation of hydrogen bond networks.
Figure 8.
Hydrogel structure and organization. Discrete building blocks self-associate to form a noncovalent supramolecular structures. The binding forces may include aromatic, electrostatic, and hydrophobic interactions. The interaction of the formed supramolecular networks with water molecules in a gelation process results in the attainment of macroscopic semirigid structure that can contain more than 99% water.
Peptides have become very popular building blocks for the fabrication of physical hydrogels due to their inherent biocompatibility and ease of synthesis and modification as discussed above. The first demonstration of the ability of peptide to form ordered hydrogels emerged from the pioneering work of Zhang and co-workers in the 1990s.[170,171] The researchers demonstrated efficient assembly of ionic self-complementary hexadecapeptides into macroscopically ordered physical hydrogels. The investigators also demonstrated the ability of the gel material to serve as a scaffold for the growth of neurite cells, providing the earliest utilization of peptide hydrogels in tissue engineering and regeneration applications.[172]
In spite of the importance of the use of synthetic peptides in the formation of ordered structures, there was a continues need for much shorter peptide that could be produced in large amount and in an economical manner. This was indeed achieved when protected short peptide fragments and even amino acids were studied. The ability of fluorenylmethyloxycarbonyl (Fmoc) modified dipeptides to form ordered hydrogel materials was discovered during studies of protected fragments of a d-Ala–d-Ala dipeptide motif that is found in bacterial cell wall.[173] During the synthesis of d-Ala–d-Ala derivatives, the researchers noticed that the Fmoc-d-Ala–d-Ala protected dipeptide forms hydrogels with high efficiency. It was estimated that one molecule of Fmoc-d-Ala–d-Ala can form a gel with a ratio of about 1–15 000 water molecules. The ability of Fmoc–Phe–Phe to form remarkably rigid hydrogel was later on demonstrated in two independent studies.[174] Ulijn and co-workers had discovered the ability of Fmoc–Phe–Phe to efficiently form hydrogels from the systematic study of Fmoc derivatives,[175] while Gazit and co-workers had identified this building block by the systematic exploration of various capping moieties of the diphenylalanine motif.[176] The convergence of the two independent directions into the identification of the Fmoc–Phe–Phe motif indeed reflects its unique properties as one of the most notable bioorganic hydrogeltaors. As of today, the Fmoc–Phe–Phe is still one of the most studied hydrogelators due its remarkable tendency to form gel with extraordinary mechanical properties and biocompatibility that enable its use as a scaffold in biological systems. Followup studies had demonstrated that not only Fmoc-modified dipeptides, but also Fmoc protected amino acids could form hydrogels with high mechanical rigidity,[177–179] providing another indication for the unmatched gelation potential of the Fmoc moiety and its ability to dictate the assembly of various motifs attached to it.[177–179]
While Fmoc-modified peptides and amino acids, and especially the Fmoc–Phe–Phe dipeptide, were extensively studied, there is a continuous quest for peptide hydrogels that are made by natural, nonmodified peptide building blocks. A systematic computational assessment of the aggregation propensity of all 8000 possible natural tripeptides, led to the discovery that several peptide molecules that combine positively charged lysine and diaromatic motif can form ordered macroscopic hydrogels.[180] Such gelating tripeptides include Lys–Tyr–Phe, Lys–Tyr–Tyr, Lys–Phe–Phe, and Lys–Tyr–Trp. Following the identification of Lys–Tyr–Phe tripeptide as a potent hydrogelator, several analogs of the tripeptide were explored revealing the canonical structural organization as noted above.
6. Conclusions and Future Prospects
The growing interest in the use of functional peptides nano-structures is reflected by the significant number of research works being done in this field. These works include the study of the self-assembly mechanisms for better understanding and controlling the final organized assemblies, as well as expanding the diversity of the morphology and the available functional properties of the peptides assemblies. The combination of interesting and useful properties that arise both from the structural possibilities and the tailored chemical characteristics may contribute in a variety of disciplines including applications of biomedicine, tissue engineering, self-cleaning, detection, and electrical devices, as discussed here. Nevertheless, there are still several open questions, and there is a room for further developments in this field. One of the challenges is the production of new and complex nanostructures that may provide new interesting properties and applications. This requires a better understanding of the relationships between the sequence of the self-assembling peptides and the structure and function of the resulting assemblies. High-resolution characterization techniques and modeling studies should be used for solving open questions regarding the structure of the assemblies. Furthermore, additional work is still required in order to fully exploit the modularity of the peptides and the diversity of their side chains for improving the control over the self-assembly process and the resulting function. Our expectation is that the well-established potential of this field will be materialized in the coming years, and self-assembled peptide nanostructure will be commercially utilized in diverse applications.
Acknowledgements
M.A. is a recipient of the postdoctoral fellowship for women scientists from the Planning and Budgeting Committee, the Council for Higher Education, Israel. S.Y. acknowledges the Clore Foundation for financial support. The authors thank the support by the European Research Council BISON project (E.G.) and N.A. acknowledges support by Grant No. 2015673 from the United States–Israel Binational Science Foundation (BSF), joint grant with the United States National Science Foundation (NSF).
Biographies
 Ehud Gazit is a Professor and Endowed Chair at the Department of Molecular Microbiology and Biotechnology and the Department of Material Science and Engineering, Tel Aviv University. He is also the academic director of the Blavatnik Center for Drug Development. In 2015, he was knighted by the Italian Republic for his service to science and society.
Ehud Gazit is a Professor and Endowed Chair at the Department of Molecular Microbiology and Biotechnology and the Department of Material Science and Engineering, Tel Aviv University. He is also the academic director of the Blavatnik Center for Drug Development. In 2015, he was knighted by the Italian Republic for his service to science and society.
 Meital Reches is an associate professor at the Institute of Chemistry and the Center for Nanoscience and Nanotechnology, the Hebrew University of Jerusalem. She joined the Hebrew University as a faculty member in October 2010. Her research focuses on understanding, controlling and developing biomolecular self-assembly processes, and generating new functional materials. Specifically, her group studies these biomolecular assemblies at the interface with inorganic surfaces.
Meital Reches is an associate professor at the Institute of Chemistry and the Center for Nanoscience and Nanotechnology, the Hebrew University of Jerusalem. She joined the Hebrew University as a faculty member in October 2010. Her research focuses on understanding, controlling and developing biomolecular self-assembly processes, and generating new functional materials. Specifically, her group studies these biomolecular assemblies at the interface with inorganic surfaces.
 Nurit Ashkenasy is an associate professor at the Materials Engineering Department at the Ben-Gurion University of the Negev (BGU), which she has joined in 2006. She is also a member of the BGU’s Ilse Katz Institute for Nanoscale Science and Technology at BGU. Her research interests include engineering of materials based on protein structural motifs for molecular electronics applications.
Nurit Ashkenasy is an associate professor at the Materials Engineering Department at the Ben-Gurion University of the Negev (BGU), which she has joined in 2006. She is also a member of the BGU’s Ilse Katz Institute for Nanoscale Science and Technology at BGU. Her research interests include engineering of materials based on protein structural motifs for molecular electronics applications.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Dr. Moran Amit, Department of Materials Engineering Ben Gurion University of the Negev Beer-Sheva 84105, Israel; Department of Electrical and Computer Engineering, UC San Diego, La Jolla, CA 92093-0407, USA
Sivan Yuran, Institute of Chemistry and The Center for Nanoscience and Nanotechnology, The Hebrew University of Jerusalem, Jerusalem 91904, Israel.
Prof. Ehud Gazit, Department of Molecular Microbiology and Biotechnology, Department of Materials Science and Engineering, Tel Aviv University, Tel Aviv 69978, Israel
Prof. Meital Reches, Institute of Chemistry and The Center for Nanoscience and Nanotechnology, The Hebrew University of Jerusalem, Jerusalem 91904, Israel
Prof. Nurit Ashkenasy, Department of Materials Engineering Ben Gurion University of the Negev Beer-Sheva 84105, Israel
References
- [1].Whitesides G, Mathias J, Seto C. Science. 1991;254:1312. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- [2].Palmer LC, Stupp SI. Acc Chem Res. 2008;41:1674. doi: 10.1021/ar8000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Whitesides GM, Grzybowski B. Science. 2002;295:2418. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- [4].Gazit E. Chem Soc Rev. 2007;36:1263. doi: 10.1039/b605536m. [DOI] [PubMed] [Google Scholar]
- [5].Hoffman AS. Adv Drug Delivery Rev. 2012;64:18. [Google Scholar]
- [6].Fisher SA, Baker AEG, Shoichet MS. J Am Chem Soc. 2017;139:7416. doi: 10.1021/jacs.7b00513. [DOI] [PubMed] [Google Scholar]
- [7].Zhang S. Nat Biotechnol. 2003;21:1171. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- [8].Tseng P, Napier B, Zhao SW, Mitropoulos AN, Applegate MB, Marelli B, Kaplan DL, Omenetto FG. Nat Nanotechnol. 2017;12:474. doi: 10.1038/nnano.2017.4. [DOI] [PubMed] [Google Scholar]
- [9].Drexler KE. Proc Natl Acad Sci USA. 1981;78:5275. doi: 10.1073/pnas.78.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wei G, Su Z, Reynolds NP, Arosio P, Hamley IW, Gazit E, Mezzenga R. Chem Soc Rev. 2017;46:4661. doi: 10.1039/c6cs00542j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Knowles TPJ, Mezzenga R. Adv Mater. 2016;28:6546. doi: 10.1002/adma.201505961. [DOI] [PubMed] [Google Scholar]
- [12].Hendricks MP, Sato K, Palmer LC, Stupp SI. Acc Chem Res. 2017;50:2440. doi: 10.1021/acs.accounts.7b00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ekiz MS, Cinar G, Khalily MA, Guler MO. Nanotechnology. 2016;27 doi: 10.1088/0957-4484/27/40/402002. 402002/1. [DOI] [PubMed] [Google Scholar]
- [14].Zhang SG. Nat Biotechnol. 2003;21:1171. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- [15].Bishop KJM, Wilmer CE, Soh S, Grzybowski BA. Small. 2009;5:1600. doi: 10.1002/smll.200900358. [DOI] [PubMed] [Google Scholar]
- [16].Scanlon S, Aggeli A. Nano Today. 2008;3:22. [Google Scholar]
- [17].Mandal D, Shirazi AN, Parang K. Org Biomol Chem. 2014;12:3544. doi: 10.1039/c4ob00447g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hamley IW. Angew Chem, Int Ed. 2014;53:6866. doi: 10.1002/anie.201310006. [DOI] [PubMed] [Google Scholar]
- [19].Hamley IW. Soft Matter. 2011;7:4122. [Google Scholar]
- [20].Caughey B, Lansbury PT. Annu Rev Neurosci. 2003;26:267. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- [21].Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Nature. 2005;435:773. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ke PC, Sani M-A, Ding F, Kakinen A, Javed I, Separovic F, Davis TP, Mezzenga R. Chem Soc Rev. 2017;46:6492. doi: 10.1039/c7cs00372b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Reches M, Gazit E. Science. 2003;300:625. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- [24].Renliang H, Rongxin S, Wei Q, Jun Z, Zhimin H. Nanotechnology. 2011;22 245609. [Google Scholar]
- [25].Reches M, Gazit E. Nano Lett. 2004;4:581. [Google Scholar]
- [26].Lee JS, Yoon I, Kim J, Ihee H, Kim B, Park CB. Angew Chem, Int Ed. 2011;50:1164. doi: 10.1002/anie.201003446. [DOI] [PubMed] [Google Scholar]
- [27].Maity S, Nir S, Reches M. J Mater Chem B. 2014;2:2583. doi: 10.1039/c3tb21456g. [DOI] [PubMed] [Google Scholar]
- [28].Berger O, Adler-Abramovich L, Levy-Sakin M, Grunwald A, Liebes-Peer Y, Bachar M, Buzhansky L, Mossou E, Forsyth VT, Schwartz T, Ebenstein Y, et al. Nat Nanotechnol. 2015;10:353. doi: 10.1038/nnano.2015.27. [DOI] [PubMed] [Google Scholar]
- [29].Babar DG, Sarkar S. Appl Nanosci. 2017;7:101. [Google Scholar]
- [30].Claussen RC, Rabatic BM, Stupp SI. J Am Chem Soc. 2003;125 doi: 10.1021/ja035882r. 12680. [DOI] [PubMed] [Google Scholar]
- [31].Cui H, Webber MJ, Stupp SI. Biopolymers. 2010;94:1. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chapman R, Danial M, Koh ML, Jolliffe KA, Perrier S. Chem Soc Rev. 2012;47:6023. doi: 10.1039/c2cs35172b. [DOI] [PubMed] [Google Scholar]
- [33].Amdursky N, Molotskii M, Gazit E, Rosenman G. J Am Chem Soc. 2010;132 doi: 10.1021/ja104373e. 15632. [DOI] [PubMed] [Google Scholar]
- [34].Yuran S, Razvag Y, Reches M. ACS Nano. 2012;6:9559. doi: 10.1021/nn302983e. [DOI] [PubMed] [Google Scholar]
- [35].Amit M, Cheng G, Hamley IW, Ashkenasy N. Soft Matter. 2012;8:8690. [Google Scholar]
- [36].Ivnitski D, Amit M, Silberbush O, Atsmon-Raz Y, Nanda J, Cohen-Luria R, Miller Y, Ashkenasy G, Ashkenasy N. Angew Chem, Int Ed. 2016;55:9988. doi: 10.1002/anie.201604833. [DOI] [PubMed] [Google Scholar]
- [37].Woolfson DN. Biopolymers. 2010;94:118. doi: 10.1002/bip.21345. [DOI] [PubMed] [Google Scholar]
- [38].Karle IL, Balaram P. Biochemistry. 1990;29:6747. doi: 10.1021/bi00481a001. [DOI] [PubMed] [Google Scholar]
- [39].Mondal S, Adler-Abramovich L, Lampel A, Bram Y, Lipstman S, Gazit E. Nat Commun. 2015;6 doi: 10.1038/ncomms9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tayeb-Fligelman E, Tabachnikov O, Moshe A, Goldshmidt-Tran O, Sawaya MR, Coquelle N, Colletier J-P, Landau M. Science. 2017;355:831. doi: 10.1126/science.aaf4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zong JY, Cobb SL, Cameron NR. Biomater Sci. 2017;5:872. doi: 10.1039/c7bm00006e. [DOI] [PubMed] [Google Scholar]
- [42].Amit E, Obena R, Wang YT, Zhuravel R, Reyes AJF, Elbaz S, Rotem D, Porath D, Friedler A, Chen YJ, Yitzchaik S. Chem Sci. 2015;6:4756. doi: 10.1039/c5sc00560d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Snir E, Amit E, Friedler A, Yitzchaik S. Biopolymers. 2015;104:515. doi: 10.1002/bip.22653. [DOI] [PubMed] [Google Scholar]
- [44].Snir E, Joore J, Timmerman P, Yitzchaik S. Langmuir. 2011;27 doi: 10.1021/la202247m. 11212. [DOI] [PubMed] [Google Scholar]
- [45].Zhuravel R, Amit E, Elbaz S, Rotem D, Chen YJ, Friedler A, Yitzchaik S, Porath D. Sci Rep. 2016;6 doi: 10.1038/srep36793. 36793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nir S, Reches M. Curr Opin Biotechnol. 2016;39:48. doi: 10.1016/j.copbio.2015.12.012. [DOI] [PubMed] [Google Scholar]
- [47].Lutchmiah K, Verliefde ARD, Roest K, Rietveld LC, Cornelissen ER. Water Res. 2014;58:179. doi: 10.1016/j.watres.2014.03.045. [DOI] [PubMed] [Google Scholar]
- [48].Lejars M, Margaillan A, Bressy C. Chem Rev. 2012;112:4347. doi: 10.1021/cr200350v. [DOI] [PubMed] [Google Scholar]
- [49].Almeida JR, Vasconcelos V. Biotechnol Adv. 2015;33:343. doi: 10.1016/j.biotechadv.2015.01.013. [DOI] [PubMed] [Google Scholar]
- [50].Bazaka K, Jacob MV, Chrzanowski W, Ostrikov K. RSC Adv. 2015;5 48739. [Google Scholar]
- [51].Kirschner CM, Brennan AB. Annu Rev Mater Res. 2012;42:211. [Google Scholar]
- [52].Banerjee I, Pangule RC, Kane RS. Adv Mater. 2011;23:690. doi: 10.1002/adma.201001215. [DOI] [PubMed] [Google Scholar]
- [53].Stern T, Zelinger E, Hayouka Z. Chem Commun. 2016;52:7102. doi: 10.1039/c6cc01438k. [DOI] [PubMed] [Google Scholar]
- [54].Epstein AK, Wong TS, Belisle RA, Boggs EM, Aizenberg J. Proc Natl Acad Sci USA. 2012;109:13182. doi: 10.1073/pnas.1201973109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Leslie DC, Waterhouse A, Berthet JB, Valentin TM, Watters AL, Jain A, Kim P, Hatton BD, Nedder A, Donovan K, Super EH, et al. Nat Biotechnol. 2014;32:1134. doi: 10.1038/nbt.3020. [DOI] [PubMed] [Google Scholar]
- [56].Segev-Zarko L, Saar-Dover R, Brumfeld V, Mangoni ML, Shai Y. Biochem J. 2015;468:259. doi: 10.1042/BJ20141251. [DOI] [PubMed] [Google Scholar]
- [57].Hayouka Z, Chakraborty S, Liu RH, Boersma MD, Weisblum B, Gellman SH. J Am Chem Soc. 2013;135:11748. doi: 10.1021/ja406231b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bagheri M, Beyermann M, Dathe M. Antimicrob Agents Chemother. 2009;53:1132. doi: 10.1128/AAC.01254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Willcox MDP, Hume EBH, Aliwarga Y, Kumar N, Cole N. J Appl Microbiol. 2008;105:1817. doi: 10.1111/j.1365-2672.2008.03942.x. [DOI] [PubMed] [Google Scholar]
- [60].Chen RX, Cole N, Willcox MDP, Park J, Rasul R, Carter E, Kumar N. Biofouling. 2009;25:517. doi: 10.1080/08927010902954207. [DOI] [PubMed] [Google Scholar]
- [61].Pinese C, Jebors S, Echalier C, Licznar-Fajardo P, Garric X, Humblot V, Calers C, Martinez J, Mehdi A, Subra G. Adv Healthcare Mater. 2016;5:3067. doi: 10.1002/adhm.201600757. [DOI] [PubMed] [Google Scholar]
- [62].Humblot V, Yala JF, Thebault P, Boukerma K, Hequet A, Berjeaud JM, Pradier CM. Biomaterials. 2009;30:3503. doi: 10.1016/j.biomaterials.2009.03.025. [DOI] [PubMed] [Google Scholar]
- [63].Godoy-Gallardo M, Mas-Moruno C, Fernandez-Calderon MC, Perez-Giraldo C, Manero JM, Albericio F, Gil FJ, Rodriguez D. Acta Biomater. 2014;10:3522. doi: 10.1016/j.actbio.2014.03.026. [DOI] [PubMed] [Google Scholar]
- [64].Rai A, Pinto S, Evangelista MB, Gil H, Kallip S, Ferreira MGS, Ferreira L. Acta Biomater. 2016;33:64. doi: 10.1016/j.actbio.2016.01.035. [DOI] [PubMed] [Google Scholar]
- [65].Yazici H, O’Neill MB, Kacar T, Wilson BR, Oren EE, Sarikaya M, Tamerler C. ACS Appl Mater Interfaces. 2016;8:5070. doi: 10.1021/acsami.5b03697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Liu Z, Ma S, Duan S, Xuliang D, Sun Y, Zhang X, Xu X, Guan B, Wang C, Hu M, Qi X, et al. ACS Appl Mater Interfaces. 2016;8:5124. doi: 10.1021/acsami.5b11949. [DOI] [PubMed] [Google Scholar]
- [67].Ostuni E, Chapman RG, Holmlin RE, Takayama S, Whitesides GM. Langmuir. 2001;17:5605. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- [68].Harder P, Grunze M, Dahint R, Whitesides GM, Laibinis PE. J Phys Chem B. 1998;102:426. [Google Scholar]
- [69].Chen SF, Cao ZQ, Jiang SY. Biomaterials. 2009;30:5892. doi: 10.1016/j.biomaterials.2009.07.001. [DOI] [PubMed] [Google Scholar]
- [70].Maity S, Nir S, Zada T, Reches M. Chem Commun. 2014;50:11154. doi: 10.1039/c4cc03578j. [DOI] [PubMed] [Google Scholar]
- [71].Lee H, Scherer NF, Messersmith PB. Proc Natl Acad Sci USA. 2006;103:12999. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Waite JH, Tanzer ML. Science. 1981;212:1038. doi: 10.1126/science.212.4498.1038. [DOI] [PubMed] [Google Scholar]
- [73].Mas-Moruno C, Fraioli R, Abericio F, Manero JM, Gil FJ. ACS Appl Mater Interfaces. 2014;6:6525. doi: 10.1021/am5001213. [DOI] [PubMed] [Google Scholar]
- [74].Niinomi M. Mater Sci Eng, A. 1998;243:231. [Google Scholar]
- [75].Pagel M, Hassert R, John T, Braun K, Wiessler M, Abel B, Beck-Sickinger AG. Angew Chem, Int Ed. 2016;55:4826. doi: 10.1002/anie.201511781. [DOI] [PubMed] [Google Scholar]
- [76].Khatayevich D, So CR, Hayamizu Y, Gresswell C, Sarikaya M. Langmuir. 2012;28:8589. doi: 10.1021/la300268d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Razvag Y, Gutkin V, Reches M. Langmuir. 2013;29:10102. doi: 10.1021/la4015866. [DOI] [PubMed] [Google Scholar]
- [78].Das P, Reches M. Nanoscale. 2016;8:15309. doi: 10.1039/c6nr04550b. [DOI] [PubMed] [Google Scholar]
- [79].Maity S, Zanuy D, Razvag Y, Das P, Aleman C, Reches M. Phys Chem Chem Phys. 2015;17:15305. doi: 10.1039/c5cp00088b. [DOI] [PubMed] [Google Scholar]
- [80].Lee JS, Ryu J, Park CB. Soft Matter. 2009;5:2717. [Google Scholar]
- [81].Hung AM, Stupp SI. Nano Lett. 2007;7:1165. doi: 10.1021/nl062835z. [DOI] [PubMed] [Google Scholar]
- [82].Mata A, Hsu L, Capito R, Aparicio C, Henrikson K, Stupp SI. Soft Matter. 2009;5:1228. doi: 10.1039/b819002j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Han TH, Ok T, Kim J, Shin DO, Ihee H, Lee HS, Kim SO. Small. 2010;6:945. doi: 10.1002/smll.200902050. [DOI] [PubMed] [Google Scholar]
- [84].Yemini M, Reches M, Gazit E, Rishpon J. Anal Chem. 2005;77:5155. doi: 10.1021/ac050414g. [DOI] [PubMed] [Google Scholar]
- [85].Yemini M, Reches M, Rishpon J, Gazit E. Nano Lett. 2005;5:183. doi: 10.1021/nl0484189. [DOI] [PubMed] [Google Scholar]
- [86].Shah A, Adhikari B, Martic S, Munir A, Shahzad S, Ahmad K, Kraatz H-B. Chem Soc Rev. 2015;44:1015. doi: 10.1039/c4cs00297k. [DOI] [PubMed] [Google Scholar]
- [87].Sisido M, Tanaka R, Inai Y, Imanishi Y. J Am Chem Soc. 1989;111:6790. [Google Scholar]
- [88].Inai Y, Sisido M, Imanishi Y. J Phys Chem. 1991;95:3847. [Google Scholar]
- [89].Lee H, Faraggi M, Klapper MH. Biochim Biophys Acta. 1992;1159:286. doi: 10.1016/0167-4838(92)90058-l. [DOI] [PubMed] [Google Scholar]
- [90].Antonello S, Formaggio F, Moretto A, Toniolo C, Maran F. J Am Chem Soc. 2003;125:2874. doi: 10.1021/ja029787e. [DOI] [PubMed] [Google Scholar]
- [91].Watanabe J, Morita T, Kimura S. J Phys Chem B. 2005;109 doi: 10.1021/jp051592g. 14416. [DOI] [PubMed] [Google Scholar]
- [92].Dey SK, Long Y-T, Chowdhury S, Sutherland TC, Mandal HS, Kraatz H-B. Langmuir. 2007;23:6475. doi: 10.1021/la070175n. [DOI] [PubMed] [Google Scholar]
- [93].Giese B, Graber M, Cordes M. Curr Opin Chem Biol. 2008;72:755. doi: 10.1016/j.cbpa.2008.08.026. [DOI] [PubMed] [Google Scholar]
- [94].Juhaniewicz J, Sek S. Bioelectrochemistry. 2012;87:21. doi: 10.1016/j.bioelechem.2011.11.013. [DOI] [PubMed] [Google Scholar]
- [95].Pawlowski J, Juhaniewicz J, Tymecka D, Sek S. Langmuir. 2012;28:17287. doi: 10.1021/la302716n. [DOI] [PubMed] [Google Scholar]
- [96].Yu J, Zvarec O, Huang DM, Bissett MA, Scanlon DB, Shapter JG, Abell AD. Chem Commun. 2012;48:1132. doi: 10.1039/c2cc16665h. [DOI] [PubMed] [Google Scholar]
- [97].Sepunaru L, Refaely-Abramson S, Lovrinčić R, Gavrilov Y, Agrawal P, Levy Y, Kronik L, Pecht I, Sheves M, Cahen D. J Am Chem Soc. 2015;137:9617. doi: 10.1021/jacs.5b03933. [DOI] [PubMed] [Google Scholar]
- [98].Xu X, Tao J Am Chem Soc. 2004;126:5370. doi: 10.1021/ja049469a. [DOI] [PubMed] [Google Scholar]
- [99].Sek S, Misicka A, Swiatek K, Maicka E. J Phys Chem B. 2006;770:19671. doi: 10.1021/jp063073z. [DOI] [PubMed] [Google Scholar]
- [100].Kitagawa K, Morita T, Kawasaki M, Kimura S. J Polym Sci A. 2003;41:3493. [Google Scholar]
- [101].Sek S, Tolak A, Misicka A, Palys B, Bilewicz R. J Phys Chem B. 2005;109:18433. doi: 10.1021/jp052157p. [DOI] [PubMed] [Google Scholar]
- [102].Sek S, Swiatek K, Misicka A. J Phys Chem B. 2005;109:23121. doi: 10.1021/jp055709c. [DOI] [PubMed] [Google Scholar]
- [103].Kitagawa K, Morita T, Kimura S. Thin Solid Films. 2006;509:18. [Google Scholar]
- [104].Shlizerman C, Atanassov A, Berkovich I, Ashkenasy G, Ashkenasy N. J Am Chem Soc. 2010;132:5070. doi: 10.1021/ja907902h. [DOI] [PubMed] [Google Scholar]
- [105].Kraatz H-B, Bediako-Amoa I, Gyepi-Garbrah SH, Sutherland TC. J Phys Chem B. 2004;108:20164. [Google Scholar]
- [106].Santhanamoorthi N, Kolandaivel P, Senthilkumar K. J Phys Chem A. 2006;110:11551. doi: 10.1021/jp063069n. [DOI] [PubMed] [Google Scholar]
- [107].Winkler JR, Gray HB. J Biol Inorg Chem. 1997;2:399. [Google Scholar]
- [108].Juhaniewicz J, Sek S. J Electroanal Chem. 2010;649:83. [Google Scholar]
- [109].Guo C, Yu X, Refaely-Abramson S, Sepunaru L, Bendikov T, Pecht I, Kronik L, Vilan A, Sheves M, Cahen D. Proc Natl Acad Sci USA. 2016;113:10785. doi: 10.1073/pnas.1606779113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Schlag EW, Sheu SY, Yang DY, Selzle HL, Lin SH. J Phys Chem B. 2000;104:7790. [Google Scholar]
- [111].Schlag EW, Sheu S-Y, Yang D-Y, Selzle HL, Lin SH. Angew Chem, Int Ed. 2007;46:3196. doi: 10.1002/anie.200601623. [DOI] [PubMed] [Google Scholar]
- [112].Mizrahi M, Zakrassov A, Lerner-Yardeni J, Ashkenasy N. Nanoscale. 2012;4:518. doi: 10.1039/c1nr11068c. [DOI] [PubMed] [Google Scholar]
- [113].Yew SY, Shekhawat G, Wangoo N, Mhaisalkar S, Suri CR, Dravid VP, Lam YM. Nanotechnology. 2011;22 doi: 10.1088/0957-4484/22/21/215606. 215606. [DOI] [PubMed] [Google Scholar]
- [114].Castillo J, Tanzi S, Dimaki M, Svendsen W. Electrophoresis. 2008;29:5026. doi: 10.1002/elps.200800260. [DOI] [PubMed] [Google Scholar]
- [115].Creasey RCG, Shingaya Y, Nakayama T. Mater Chem Phys. 2015;158:52. [Google Scholar]
- [116].Akdim B, Pachter R, Naik RR. Appl Phys Lett. 2015;106 183707. [Google Scholar]
- [117].Okamoto H, Nakanishi T, Nagai Y, Kasahara M, Takeda K. J Am Chem Soc. 2003;125:2756. doi: 10.1021/ja0212720. [DOI] [PubMed] [Google Scholar]
- [118].Amdursky N. Phys Chem Chem Phys. 2013;15:13479. doi: 10.1039/c3cp51748a. [DOI] [PubMed] [Google Scholar]
- [119].Horne WS, Ashkenasy N, Ghadiri MR. Chem - Eur J. 2005;11:1137. doi: 10.1002/chem.200400923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Ashkenasy N, Horne WS, Ghadiri MR. Small. 2006;2:99. doi: 10.1002/smll.200500252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ivnitski D, Amit M, Rubinov B, Cohen-Luria R, Ashkenasy N, Ashkenasy G. Chem Commun. 2014;50:6733. doi: 10.1039/c4cc00717d. [DOI] [PubMed] [Google Scholar]
- [122].Amit M, Ashkenasy N. IsrJ Chem. 2014;54:703. [Google Scholar]
- [123].Xu H, Das AK, Horie M, Shaik MS, Smith AM, Luo Y, Lu X, Collins R, Liem SY, Song A, Popelier PLA, et al. Nanoscale. 2010;2:960. doi: 10.1039/b9nr00233b. [DOI] [PubMed] [Google Scholar]
- [124].Nalluri SKM, Shivarova N, Kanibolotsky AL, Zelzer M, Gupta S, Frederix PWJM, Skabara PJ, Gleskova H, Ulijn RV. Langmuir. 2014;30:12429. doi: 10.1021/la503459y. [DOI] [PubMed] [Google Scholar]
- [125].Sun Y, Jiang L, Schuermann KC, Adriaens W, Zhang L, Boey FYC, De Cola L, Brunsveld L, Chen X. Chem - Eur J. 2011;17:4746. doi: 10.1002/chem.201003760. [DOI] [PubMed] [Google Scholar]
- [126].Diegelmann SR, Gorham JM, Tovar JD. J Am Chem Soc. 2008;130:13840. doi: 10.1021/ja805491d. [DOI] [PubMed] [Google Scholar]
- [127].Wall BD, Diegelmann SR, Zhang S, Dawidczyk TJ, Wilson WL, Katz HE, Mao H-Q, Tovar JD. Adv Mater. 2011;23:5009. doi: 10.1002/adma.201102963. [DOI] [PubMed] [Google Scholar]
- [128].Diegelmann SR, Hartman N, Markovic N, Tovar JD. J Am Chem Soc. 2012;134:2028. doi: 10.1021/ja211539j. [DOI] [PubMed] [Google Scholar]
- [129].Tovar JD. Acc Chem Res. 2013;46:1527. doi: 10.1021/ar3002969. [DOI] [PubMed] [Google Scholar]
- [130].Ardona HAM, Besar K, Togninalli M, Katz HE, Tovar JD. J Mater Chem C. 2015;3:6505. [Google Scholar]
- [131].Ardona HAM, Tovar JD. Bioconjugate Chem. 2015;26:2290. doi: 10.1021/acs.bioconjchem.5b00497. [DOI] [PubMed] [Google Scholar]
- [132].del Mercato LL, Pompa PP, Maruccio G, Della Torre A, Sabella S, Tamburro AM, Cingolani R, Rinaldi R. Proc Natl Acad Sci USA. 2007;104:18019. doi: 10.1073/pnas.0702843104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Ardona HAM, Draper ER, Citossi F, Wallace M, Serpell LC, Adams DJ, Tovar JD. J Am Chem Soc. 2017;139:8685. doi: 10.1021/jacs.7b04006. [DOI] [PubMed] [Google Scholar]
- [134].Khalily MA, Bakan G, Kucukoz B, Topal AE, Karatay A, Yaglioglu HG, Dana A, Guler MO. ACS Nano. 2017;11:6881. doi: 10.1021/acsnano.7b02025. [DOI] [PubMed] [Google Scholar]
- [135].Almohammed S, Oladapo SO, Ryan K, Kholkin AL, Rice JH, Rodriguez BJ. RSC Adv. 2016;6:41809. [Google Scholar]
- [136].Silberbush O, Amit M, Roy S, Ashkenasy N. Adv Funct Mater. 2017;27 [Google Scholar]
- [137].Yardeni JL, Amit M, Ashkenasy G, Ashkenasy N. Nanoscale. 2016;8:2358. doi: 10.1039/c5nr06750b. [DOI] [PubMed] [Google Scholar]
- [138].Amit M, Appel S, Cohen R, Cheng G, Hamley IW, Ashkenasy N. Adv Funct Mater. 2014;24:5873. [Google Scholar]
- [139].Kholkin A, Amdursky N, Bdikin I, Gazit E, Rosenman G. ACS Nano. 2010;4:610. doi: 10.1021/nn901327v. [DOI] [PubMed] [Google Scholar]
- [140].Rosenman G, Beker P, Koren I, Yevnin M, Bank-Srour B, Mishina E, Semin S. J Pept Sci. 2011;17:75. doi: 10.1002/psc.1326. [DOI] [PubMed] [Google Scholar]
- [141].Bosne ED, Heredia A, Kopyl S, Karpinsky DV, Pinto AG, Kholkin AL. Appl Phys Lett. 2013;102 073504. [Google Scholar]
- [142].Nuraeva A, Vasilev S, Vasileva D, Zelenovskiy P, Chezganov D, Esin A, Kopyl S, Romanyuk K, Shur VY, Kholkin AL. Cryst Growth Des. 2016;16:1472. [Google Scholar]
- [143].Ryan K, Beirne J, Redmond G, Kilpatrick JI, Guyonnet J, Buchete N-V, Kholkin AL, Rodriguez BJ. ACS Appl Mater Interfaces. 2015;7:12702. doi: 10.1021/acsami.5b01251. [DOI] [PubMed] [Google Scholar]
- [144].Gan Z, Wu X, Zhu X, Shen J. Angew Chem, Int Ed. 2013;52:2055. doi: 10.1002/anie.201207992. [DOI] [PubMed] [Google Scholar]
- [145].Heredia A, Bdikin I, Kopyl S, Mishina E, Semin S, Sigov A, German K, Bystrov V, Gracio J, Kholkin AL. J Phys D: Appl Phys. 2010;43 462001. [Google Scholar]
- [146].Handelman A, Beker P, Mishina E, Semin S, Amdursky N, Rosenman G. Ferroelectrics. 2012;430:84. [Google Scholar]
- [147].Nguyen V, Zhu R, Jenkins K, Yang RS. Nat Commun. 2016;7 doi: 10.1038/ncomms13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Esin A, Baturin I, Nikitin T, Vasilev S, Salehli F, Shur VY, Kholkin AL. Appl Phys Lett. 2016;109 142902. [Google Scholar]
- [149].Morita T, Kimura S, Kobayashi S, Imanishi Y. J Am Chem Soc. 2000;122:2850. [Google Scholar]
- [150].Gatto E, Stella L, Formaggio F, Toniolo C, Lorenzelli L, Venanzi M. J Pept Sci. 2008;14:184. doi: 10.1002/psc.973. [DOI] [PubMed] [Google Scholar]
- [151].Gatto E, Caruso M, Porchetta A, Toniolo C, Formaggio F, Crisma M, Venanzi M. J Pept Sci. 2011;17:124. doi: 10.1002/psc.1329. [DOI] [PubMed] [Google Scholar]
- [152].Gatto E, Quatela A, Caruso M, Tagliaferro R, De Zotti M, Formaggio F, Toniolo C, Di Carlo A, Venanzi M. ChemPhysChem. 2014;15:64. doi: 10.1002/cphc.201300901. [DOI] [PubMed] [Google Scholar]
- [153].Yasutomi S, Morita T, Imanishi Y, Kimura S. Science. 2004;304:1944. doi: 10.1126/science.1098489. [DOI] [PubMed] [Google Scholar]
- [154].Nie R, Li A, Deng X. J Mater Chem A. 2014;2:6734. [Google Scholar]
- [155].Matmor M, Lengyel GA, Horne WS, Ashkenasy N. Phys Chem Chem Phys. 2017;19:5709. doi: 10.1039/c6cp07198h. [DOI] [PubMed] [Google Scholar]
- [156].Matmor M, Ashkenasy N. J Am Chem Soc. 2012;134:20403. doi: 10.1021/ja3078494. [DOI] [PubMed] [Google Scholar]
- [157].Hasobe T, Saito K, Kamat PV, Troiani V, Qiu H, Solladie N, Kim KS, Park JK, Kim D, D’Souza F, Fukuzumi S. J Mater Chem. 2007;17:4160. [Google Scholar]
- [158].Lu H, Zhou C, Zhou X, Sun H, Bai H, Wang Y, Lv F, Liu L, Ma Y, Wang S. Adv Electron Mater. 2017;3 1700161. [Google Scholar]
- [159].Kim JH, Lee M, Lee JS, Park CB. Angew Chem, Int Ed. 2012;51:517. doi: 10.1002/anie.201103244. [DOI] [PubMed] [Google Scholar]
- [160].Solomon LA, Sykes ME, Wu YA, Schaller RD, Wiederrecht GP, Fry HC. ACS Nano. 2017;11:9112. doi: 10.1021/acsnano.7b03867. [DOI] [PubMed] [Google Scholar]
- [161].Uji H, Kim H, Imai T, Mitani S, Sugiyama J, Kimura S. Biopolymers. 2016;106:275. doi: 10.1002/bip.22850. [DOI] [PubMed] [Google Scholar]
- [162].Sanders AM, Dawidczyk TJ, Katz HE, Tovar JD. ACS Macro Lett. 2012;1:1326. doi: 10.1021/mz3004665. [DOI] [PubMed] [Google Scholar]
- [163].Besar K, Ardona HAM, Tovar JD, Katz HE. ACS Nano. 2015;9:12401. doi: 10.1021/acsnano.5b05752. [DOI] [PubMed] [Google Scholar]
- [164].Sanders AM, Kale TS, Katz HE, Tovar JD. ACS Omega. 2017;2:409. doi: 10.1021/acsomega.6b00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Adler-Abramovich L, Aronov D, Beker P, Yevnin M, Stempler S, Buzhansky L, Rosenman G, Gazit E. Nat Nanotechnol. 2009;4:849. doi: 10.1038/nnano.2009.298. [DOI] [PubMed] [Google Scholar]
- [166].Beker P, Rosenman G. J Mater Res. 2010;25:1661. [Google Scholar]
- [167].Beker P, Koren I, Amdursky N, Gazit E, Rosenman G. J Mater Sci. 2010;45:6374. [Google Scholar]
- [168].Amdursky N, Shalev G, Handelman A, Litsyn S, Natan A, Roizin Y, Rosenwaks Y, Szwarcman D, Rosenman G. APL Mater. 2013;1 062104. [Google Scholar]
- [169].Fichman G, Gazit E. Acta Biomater. 2014;10:1671. doi: 10.1016/j.actbio.2013.08.013. [DOI] [PubMed] [Google Scholar]
- [170].Zhang S, Holmes T, Lockshin C, Rich A. Proc Natl Acad Sci USA. 1993;90:3334. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Zhang S, Holmes TC, DiPersio CM, Hynes RO, Su X, Rich A. Biomaterials. 1995;16:1385. doi: 10.1016/0142-9612(95)96874-y. [DOI] [PubMed] [Google Scholar]
- [172].Holmes TC, de Lacalle S, Su X, Liu G, Rich A, Zhang S. Proc Natl Acad Sci USA. 2000;97:6728. doi: 10.1073/pnas.97.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Zhang Y, Gu H, Yang Z, Xu B. J Am Chem Soc. 2003;125:13680. doi: 10.1021/ja036817k. [DOI] [PubMed] [Google Scholar]
- [174].Görbitz CH. Chem Commun. 2006:2332. doi: 10.1039/b603080g. [DOI] [PubMed] [Google Scholar]
- [175].Jayawarna V, Ali M, Jowitt TA, Miller AF, Saiani A, Gough JE, Ulijn RV. Adv Mater. 2006;18:611. [Google Scholar]
- [176].Mahler A, Reches M, Rechter M, Cohen S, Gazit E. Adv Mater. 2006;18:1365. [Google Scholar]
- [177].Orbach R, Adler-Abramovich L, Zigerson S, Mironi-Harpaz I, Seliktar D, Gazit E. Biomacromolecules. 2009;10:2646. doi: 10.1021/bm900584m. [DOI] [PubMed] [Google Scholar]
- [178].Ryan DM, Anderson SB, Nilsson BL. Soft Matter. 2010;6:3220. [Google Scholar]
- [179].Fichman G, Guterman T, Adler-Abramovich L, Gazit E. CrystEngComm. 2015;17:8105. [Google Scholar]
- [180].Frederix PW, Scott GG, Abul-Haija YM, Kalafatovic D, Pappas CG, Javid N, Hunt NT, Ulijn RV, Tuttle T. Nat Chem. 2015;7:30. doi: 10.1038/nchem.2122. [DOI] [PubMed] [Google Scholar]