Abstract
To avoid innate sensing and immune control, HIV-1 has to prevent the accumulation of viral cDNA species. Here, we show that the late HIV-1 accessory protein Vpu hijacks DNA repair mechanisms to promote degradation of nuclear viral cDNA in cells that are already productively infected. Vpu achieves this by interacting with RanBP2/RanGAP1*SUMO1/Ubc9 SUMO E3-ligase complexes at the nuclear pore to reprogram promyelocytic leukemia protein nuclear bodies (PML-NB) and to reduce SUMOylation of Bloom syndrome protein (BLM) unleashing end degradation of viral cDNA. Concomitantly, Vpu inhibits RAD52-mediated homologous repair of viral cDNA preventing generation of dead-end 1-LTR circular forms and permitting sustained nucleolytic attack. Our results identify Vpu as a key modulator of the DNA repair machinery. We show that BLM eliminates nuclear HIV-1 cDNA and thereby suppresses immune sensing and proviral hyper-integration. Therapeutic targeting of DNA repair may facilitate induction of antiviral immunity and suppress proviral integration replenishing latent HIV reservoirs.
Introduction
Induction of DNA damage responses (DDRs) and innate immune activation are inevitable consequences of HIV-1 infection. Reverse transcription (RT) converts the single-stranded viral RNA (ssRNA) genome into linear double-stranded DNA (dsDNA). This process is associated with formation of RNA:DNA hybrid intermediates and generation of 1-LTR and 2-LTR circular forms of HIV-1 cDNA that do not support productive infection1. Accumulation of these viral nucleic acids, however, may trigger antiviral immune responses2. To avoid this, HIV-1 uses its capsid core and cellular cofactors to shield its ssRNA genome and RT intermediates from recognition by cytosolic sensors3,4. However, this “cloaking” is imperfect and accumulating non-integrated HIV-1 DNA triggers immune activation and interferon production, unless it is degraded by cellular nucleases, such as TREX15.
HIV-1 also directly modulates DNA repair. Particularly the viral accessory protein R (Vpr) is a well-known modulator of the DDR machinery. Vpr is contained in virions and induces proteasomal degradation of the DNA repair enzymes HLTF, UNG2 and MUS81 immediately after viral entry to prevent restriction of viral cDNA RT products6–8. Following RT and nuclear import, a double-strand break (DSB) is introduced into the host cell genome for integration of the linear viral cDNA9,10. Completion of proviral integration requires non-homologous end joining (NHEJ), i.e. DNA-PKcs, Ku, XRCC4 and Nibrin, and other DDR factors, such as ATM11. Thus, DDRs play key roles in viral replication and HIV-1 needs to prevent accumulation of its nucleic acids to minimize immune activation.
Numerous studies examined how HIV-1 avoids immune sensing and affects DNA repair during early steps of viral replication. In contrast, it is poorly understood how HIV-1 modulates DNA repair in productively infected cells. Here, we used fluorescence-based assays to identified the HIV-1 accessory factor Vpu as key modulator of DNA repair. Vpu is a small, single-pass trans-membrane protein that is only present during the late stages of HIV-1 infection. It prevents superinfection by inducing CD4 degradation12,13 and promotes virus release by antagonizing the restriction factor tetherin14,15. We found that Vpu interacts with RanBP2/RanGAP1*SUMO1/Ubc9 SUMO E3-ligase complexes at the nuclear pore to manipulate functions of PML-NBs and DDR components like RAD52 or BLM. This allows Vpu to suppress superinfection by elimination of newly incoming viral cDNA species by two cooperative mechanisms: (i) inhibition of RAD52-dependent homologous repair and thereby circularization of non-integrated linear viral DNA to maintain susceptibility to nucleolytic attack and (ii) enhancement of DNA processing by BLM to promote degradation of linear viral DNA suppressing innate immune activation and hyper-integration. We also demonstrate that BLM restricts proviral integration of HIV-1. In summary, our results show that Vpu modulates DNA repair to suppress accumulation and sensing of nuclear viral DNA species and suggest a role of BLM in viral latency.
Results
Proviral HIV-1 expression inhibits homologous repair
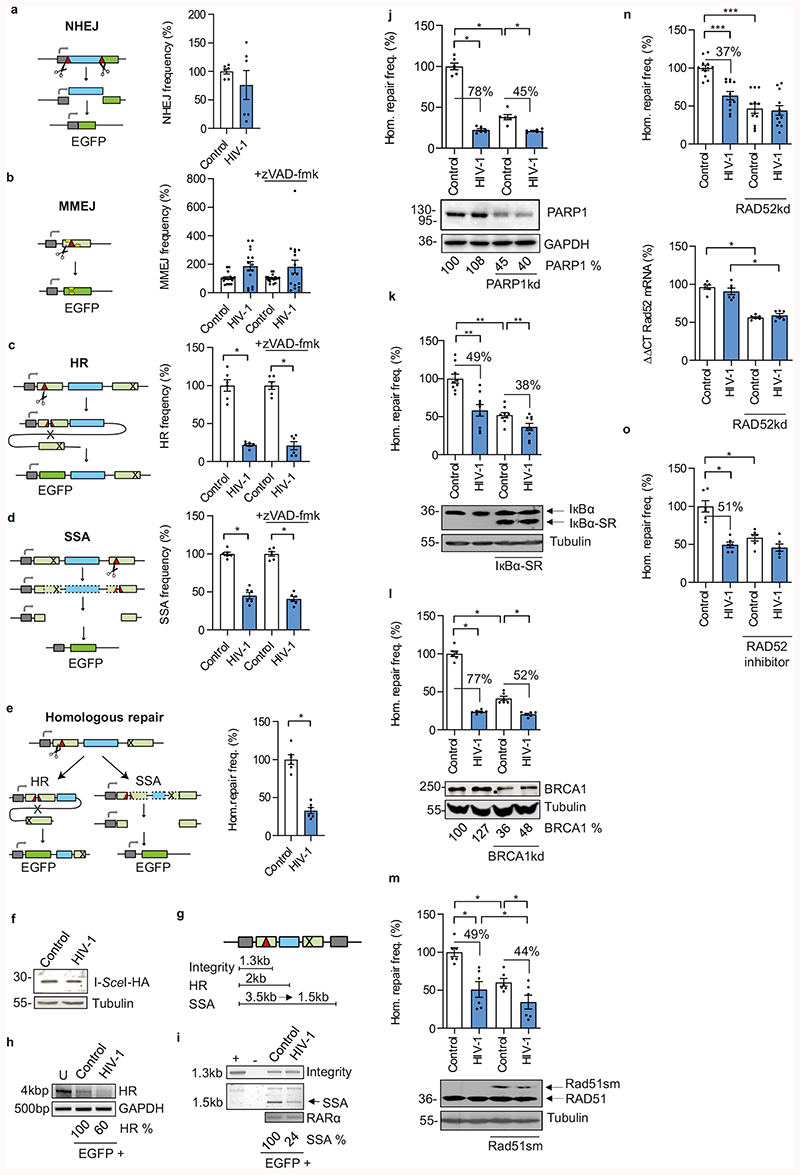
HIV-1 is known to modulate DSB repair, but the underlying mechanisms remained elusive16. We applied enhanced green fluorescent protein (EGFP)-based reporters for specific DSB repair pathways, namely NHEJ, microhomology-mediated end joining (MMEJ) and two types of homologous repair: homologous recombination (HR) and single-strand annealing (SSA)17,18. For assessment of extrachromosomal repair, Jurkat T cells were co-transfected with an HIV-1 NL4-3 IRES mCherry proviral construct (HIV-1 refers to NL4-3 unless specified otherwise), DSB repair substrates and I-Sce I expression vector for targeted cleavage (Fig. 1a-d, left panels). Scoring red and green fluorescent cells allowed to monitor DSB repair in HIV-1 positive cells (Extended Data Fig. 1a). Upon transfection of proviral HIV-1 DNA that bypasses the earliest steps of viral replication from entry to integration, HIV-1 had no effect on NHEJ (Fig. 1a) and or MMEJ (Fig. 1b). In contrast, HR and SSA were strongly reduced (Fig. 1c, d). Addition of pan-caspase inhibitor Z-VAD-FMK reduced the Sub-G1 fraction, i.e. the percentage of apoptotic cells (Extended Data Fig. 1b), but did not alter HIV-1 mediated repair patterns (Fig. 1b-d). Thus, no effects were under (MMEJ) or overestimated (HR, SSA) due to HIV-1 induced apoptosis. Further analyses also excluded indirect effects through cell cycle changes (Extended Data Fig. 1c). HIV-1-dependent inhibition of homologous repair (HR and SSA) was confirmed in the human lymphoblastoid cell line WTK1 (Extended Data Fig. 1d-f).
Fig. 1. Proviral HIV-1 gene expression modulates homologous repair.
a-d, EGFP-based evaluation of extrachromosomal DSB repair. Jurkat T cells were transfected with pHIV-1-NL4-3-env*-IRES-mCherry (HIV-1), pCMV-HA-I-Sce I and reporter plasmids for detection of distinct DSB repair pathways. Samples were treated with zVAD-fmk as indicated. Bars represent means ±SEM, n=2 (a, c, d) or n=5 biologically independent experiments in triplicates (b); *p =0.0313; absolute mean DSB repair frequencies in controls set to 100% were: a, NHEJ 0.1%; b, MMEJ 0.03%; c, HR 0.04%; d, SSA 2% (see also Extended Data Fig. 1a). Kinked arrow, transcriptional promoter; light green bar, mutated EGFP sequence; dark green bar, reconstituted EGFP; blue bar, spacer sequence; red triangle, I-Sce I cleavage site; cross, deletion; yellow bar, 5 bp microhomology; dashed lines, eliminated sequence. e, EGFP-based evaluation of intrachromosomal homologous repair, i.e. HR+SSA frequency (Hom. repair freq.) in WTK1(HR/3’) cells transfected with HIV-1-NL4-3-env*-IRES-mCherry and pCMV-HA-I-Sce I. Bars represent means ±SEM, n=2 biologically independent experiments in triplicates; *p =0.0313 (absolute mean value 0.1% in controls =100%). f, Western blot indicating HA-tagged I-Sce I expression in WTK1(HR/3’). g-i, PCR-based analysis of homologous repair. g, Schematic depiction of PCR products from repair construct in WTK1(HR/3’) cells. h and i, Genomic PCR on FACS sorted WTK1(HR/3’) to determine HR, SSA and construct integrity in EGFP+ cells. +, positive control with HR-EGFP/3’EGFP plasmid; -, negative control without DNA; U, untransfected cells. Numbers indicate HR- and SSA-specific band intensities in the presence versus absence (100%) of HIV-1 normalized to control PCR amplifications in GAPDH or RARα each. j-n, Homologous repair frequency in WTK1(HR/3’) cells co-transfected with pHIV-1-NL4-3-env*-IRES-mCherry, pCMV-HA-I-Sce I and shRNA plasmids to downregulate (kd) PARP1 (j), BRCA1 (l), or RAD52 (n) or expression plasmids for dominant negative IκBα–SR (k), or Rad51sm (m). o, Split samples of WTK1(HR/3’) cells co-transfected with pHIV-1-NL4-3-env*-IRES-mCherry and pCMV-HA-I-Sce I were treated with RAD52 inhibitor 6-Hydroxy-DL-DOPA (5 μM). Bars represent means ±SEM, n=2 (j, l, m), n=3 (k) or n=4 (n) biologically independent experiments in triplicates; * p =0.0313, ** p = 0.039, ***p=0.0005. Controls set to 100% (absolute mean value: 0.1%). Lower panels: Immunoblots with percentages of residual protein levels for knockdowns, stained as indicated. Right panel: to (n), presents RT-PCR to verify RAD52 downregulation; n=3. Two sided Wilcoxon matched-pairs test in c, d, e, j-o.
To examine HIV-1 mediated effects on DSB repair in the chromatin environment, we used the WTK1(HR/3’) cell line19, carrying chromosomally integrated HR-EGFP/3’EGFP substrate (Fig. 1e). Transfection with the HIV-1 IRES mCherry construct decreased homologous repair by ~70% (Fig. 1e) without affecting I-Sce I expression (Fig. 1f). Genomic PCR analysis of sorted EGFP+ WTK1(HR/3’) cells (Fig. 1g) revealed decreases of 40% for HR (Fig. 1h) and of 76% for SSA (Fig. 1i). Analysis of repair construct integrity excluded simple destruction by nucleases (Fig. 1i). HIV-dependent suppression of homologous repair was confirmed in WTK1(Δ/3’) cells with chromosomally integrated ΔEGFP/3’EGFP substrate that allowed to distinguish between HR and SSA in hygromycin B selection medium (Extended Data Fig. 1g)18. Altogether, these results show that proviral HIV-1 gene expression suppresses HR and SSA both extra- and intra-chromosomally.
RAD52 is critical for HIV-1-dependent suppression of homologous repair
Silencing of ATM, ATR, Nibrin, CtIP and Ku70 involved in DDR and end-processing neither diminished homologous repair nor abrogated the HIV-1-mediated decline in WTK1(HR/3’) (Extended Data Fig. 1h-l). Depletion of ATM and ATR even increased basal homologous repair frequencies consistent with de-repression of error-prone pathways like SSA20. In contrast, silencing of the DNA damage sensor PARP1 diminished homologous repair in control cells and reduced the inhibitory effect of HIV-1 (Fig. 1j). Dominant negative interference with NF-κB, which is modulated by HIV-121 and activates DSB repair downstream of PARP122, slightly reduced HIV 1-mediated suppression of homologous repair (Fig. 1k). A similar reduction was observed upon BRCA1 knockdown (Fig. 1l) but not upon dominant negative interference with RAD51 (Fig. 1m). Strikingly, silencing or pharmacological inhibition of the SSA factor RAD52 entirely abolished the inhibitory effect of HIV-1 (Fig. 1n, o). Thus, although PARP1 and BRCA1 are also involved, RAD52 plays a dominant role in HIV 1-dependent suppression of homologous repair.
HIV-1 Vpu inhibits homologous repair and modulates RAD52 function
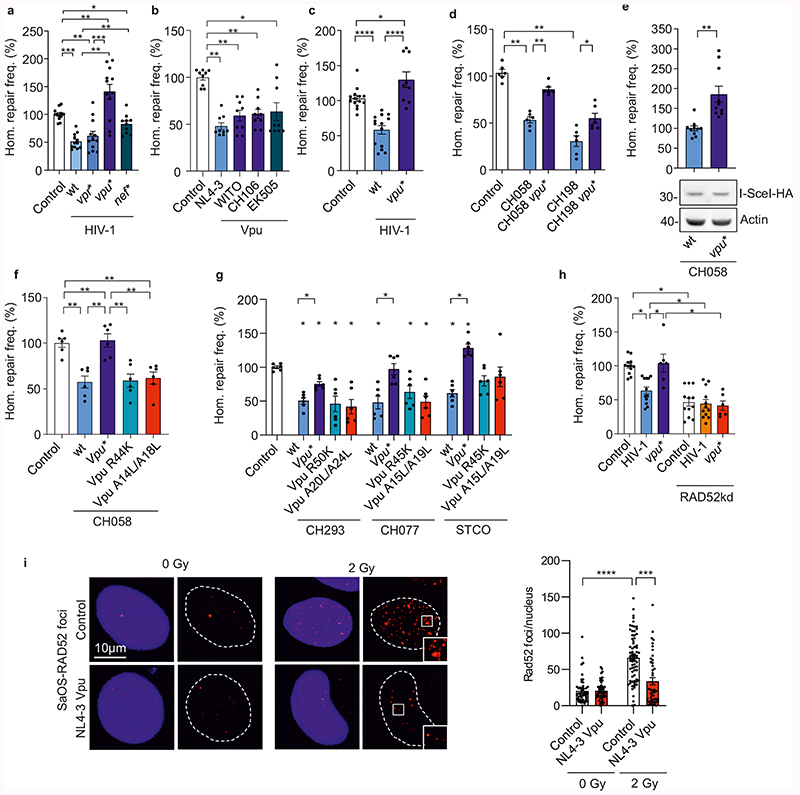
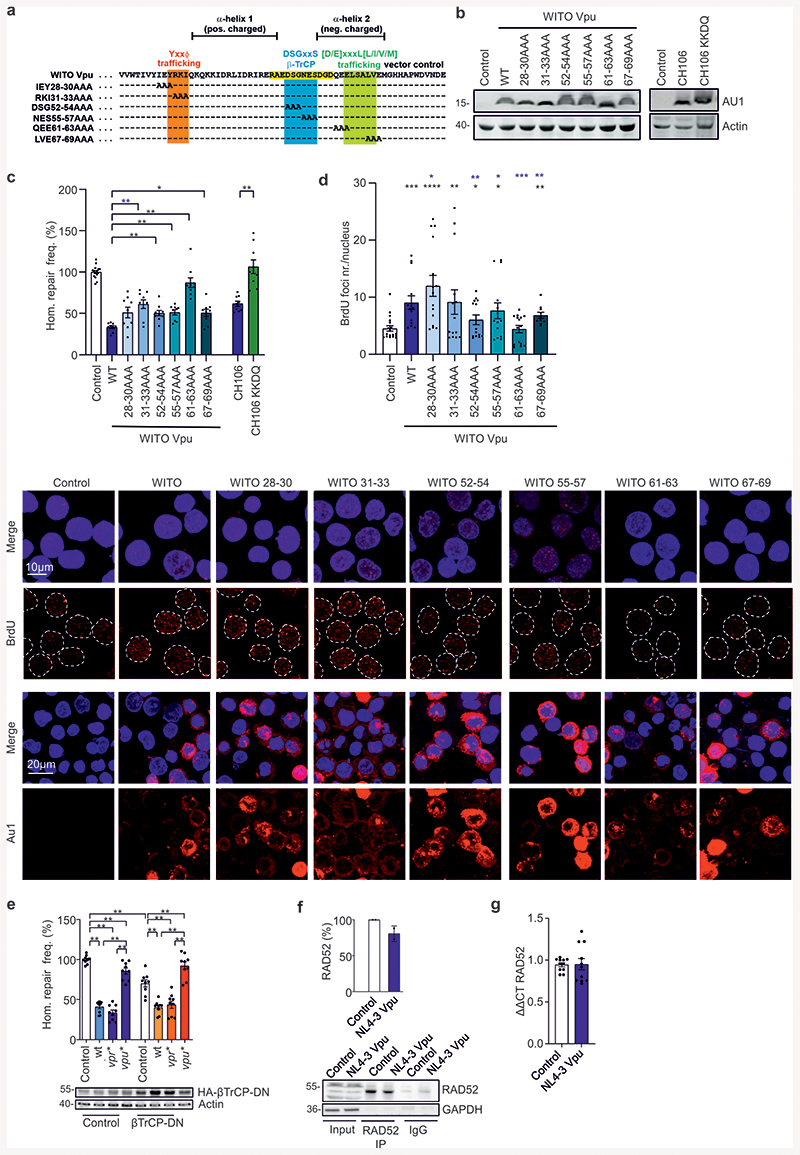
HIV-1 accessory proteins Vpr, Vpu and Nef are known to affect DNA repair and/or NF-κB signaling7,21,23,24. Thus, we examined their involvement in the suppression of homologous repair by HIV-1. Intriguingly, lack of Vpu abrogated the inhibitory effect and even resulted in 1.4-fold enhanced homologous repair in HIV-1-transfected lymphoid cells, while lack of Vpr and Nef had little if any effect (Fig. 2a). Vpu from HIV-1 NL4-3, the transmitted-founder HIV-1 strain WITO, the chronic HIV-1 strain CH106 and the chimpanzee virus SIVcpz EK505 all suppressed homologous repair by ~50% (Fig. 2b). Vpu inhibited homologous repair already at low expression levels (Extended Data Fig. 2a) and was detected at similar levels in cells infected with HIV-1, transfected with proviral constructs, or transfected with Vpu expression vectors (Extended Data Fig. 2b). Vpu did not induce apoptosis or cell cycle changes (Extended Data Fig. 2c) suggesting direct effects on the DNA repair machinery.
Fig. 2. Viral proteins involved in HIV-1 modulated homologous repair.
a-b, Homologous repair frequency in WTK1(HR/3’) cells transfected with (a), wt or mutant pHIV-1-NL4-3-env*-IRES-mCherry and (b), expression plasmids for Vpu and pCMV-HA-I-Sce I. Bars represent means ±SEM of n=4 (a) or n=3 (b) biologically independent experiments in triplicates. Controls set to 100%, (absolute mean value: 0.1%, *p=0.0488, **p=0.002, ***p=0.0005 (a); *p=0.0195, **p=0.0039 (b). c-d, Homologous repair in CD4+ T cells. c, Cells transfected with HR-EGFP/3’EGFP repair plasmid, wt or vpu* pHIV-1-NL4-3-env*-IRES-mCherry constructs and pCMV-HA-I-Sce I. Bars represent means ±SEM, n=3 biologically independent experiments in triplicates, *p=0.0391, ****p=0.0001. d, Cells transfected with HR-EGFP/3’EGFP, wt or vpu* pHIV-1 CH058 or CH198 constructs and pCMV-HA-I-Sce I. Bars represent means ±SEM, n=3 biologically independent experiments in duplicates; *p=0.0313, **p=0.0022; absolute mean value: 0.03%. e, Homologous repair in WTK1(HR/3’) cells first transduced with VSV-G pseudotyped wt or vpu* pHIV-1-CH058-env* IRES-BFP constructs and later transfected with I-Sce I expression plasmid. Bars represent means ±SEM, n=3 biologically independent experiments in triplicates, **p=0.002. The lower panel shows I-Sce I expression. f, Effect of HIV-1 on homologous repair in primary CD4+ T cells. Cells transfected with HIV-1 CH058 proviral constructs (R44K disrupts anti-NF-κB and A14L/A18L anti-tetherin activity), repair reporter and I-Sce I plasmid were analyzed by FACS to evaluate DSB repair frequencies. Bars represent means ±SEM, n=3 biologically independent experiments in duplicates, absolute mean value: 0.3%; **p=0.002. g, Homologous repair in WTK1(HR/3’) cells transfected with wt and mutant HIV-1 CH293, CH077 and STCO constructs were transfected and analyzed as described in panel f. Bars represent means ±SEM, n=2 biologically independent experiments in triplicates, (absolute mean values: CH293 4%, CH077 2%, STCO 2.5%); *p=0.0313. h, Homologous repair in WTK1(HR/3’) cells transfected with wt or vpu* pHIV-1-NL4-3-env*-IRES-mCherry constructs, shRNA targeting RAD52 and pCMV-HA-I-Sce I. Bars represent means ±SEM, n=3 biologically independent experiments conducted in triplicates; *p=0.0313. i, Evaluation of RAD52 foci formation in SaOS cells transfected with NL4-3 Vpu expression or control plasmid. SaOS cells were γ-irradiated (2Gy) or left untreated (0Gy), cultivated for 1h and stained for RAD52. Representative microscopic images are shown. The right panel shows RAD52 foci numbers, bars represent means ±SEM, n=2 biologically independent replicates; ***p=0.0003, ****p<0.0001. Two sided Wilcoxon matched-pairs test in a, b, e, g, h, i. Two-tailed Mann Whitney test in c, d, f.
In agreement with the results obtained in WTK1 cells (Fig. 2a), wild-type (wt) HIV-1 reduced homologous repair in primary CD4+ T cells by ~50% and lack of Vpu disrupted this effect (Fig. 2c). Vpu-dependent suppression of homologous repair was confirmed for HIV-1 CH058 and CH198 containing defective env genes (Fig. 2d and Extended Data Fig. 2d) and was not due to different infection rates (Extended Data Fig. 2d). The inhibitory effect of Vpu on DSB repair was confirmed in HIV-1 infected WTK1 cells (Fig. 2e). Vpu inhibited homologous repair in the context of both env-defective (Fig. 2d) and wt HIV-1 (Extended Data Fig. 2e, left). HIV-1 integration was reported to affect cell survival25. However, evaluation of apoptosis and the cell cycle distribution revealed no significant changes between CD4+ T cells harboring wt or vpu-defective HIV-1 (Extended Data Fig. 2e).
To obtain insights into the mechanism(s) underlying Vpu-mediated inhibition of homologous repair, we analyzed mutant forms of the HIV-1 CH058, CH293, CH077 and STCO infectious molecular clones (IMCs) defective in inhibiting NF-κB or counteracting tetherin26–28. Lack of these Vpu functions did not impair the ability of HIV-1 to inhibit homologous repair in primary CD4+ T cells (Fig. 2f) as well as in WTK1(HR/3’) cells (Fig. 2g). To map critical domains, we examined alanine-substitution mutants of WITO Vpu containing changes in potential trafficking domains29 or the beta-transducin repeat-containing protein (β-TrCP) binding motif30 (Extended Data Fig. 3a) critical for Vpu-mediated degradation of CD431. All mutant Vpus were expressed (Extended Data Fig. 3b) and mutation of QEE61-63AAA affecting an EXXXLV motif in HIV-1 Vpu disrupted its ability to inhibit homologous repair and end processing (Extended Data Figs. 3c, 3d). In comparison, changes in the YxxΦ trafficking and β-TrCP binding motifs did not disrupt these Vpu functions. Mutations in the DSGxxS and EXXXLV motifs in Vpu significantly reduced BrdU foci numbers measured as indicator of DNA end processing (Extended Data Fig. 3d). Addition of a C-terminal ER-retention signal to Vpu disrupted its effect on homologous repair (Extended Data Fig. 3c, compare CH106 KKDQ with CH106) supporting that proper subcellular location of Vpu is critical. Consistent with the mutational analyses, coexpression of a dominant negative mutant of β-TrCP, did not affect suppression of homologous repair by HIV-1 (Extended Data Fig. 3e). Altogether, these results show that the ability of Vpu to inhibit homologous repair is conserved among primary HIV-1 strains and genetically separable from other Vpu activities.
Next, we examined whether Vpu targets RAD52 that was critical for suppression of homologous repair by HIV-1 (Fig. 1n, 1o). Indeed, RAD52 silencing lowered the repair frequency and abolished the effect of Vpu (Fig. 2h). Vpu had little effect on RAD52 protein or mRNA levels (Extended Data Fig. 3f, 3g) but prevented the increase in discrete nuclear RAD52 foci after irradiation of SaOS cells (Fig. 2i). The latter effect was only observed in the absence of irradiation (Fig. 2i) suggesting that Vpu prevents de novo formation of active RAD52 repair centers in response to DNA damage but does not disrupt pre-existing RAD52 foci.
Vpu enhances BLM/EXO1-dependent DNA end-processing
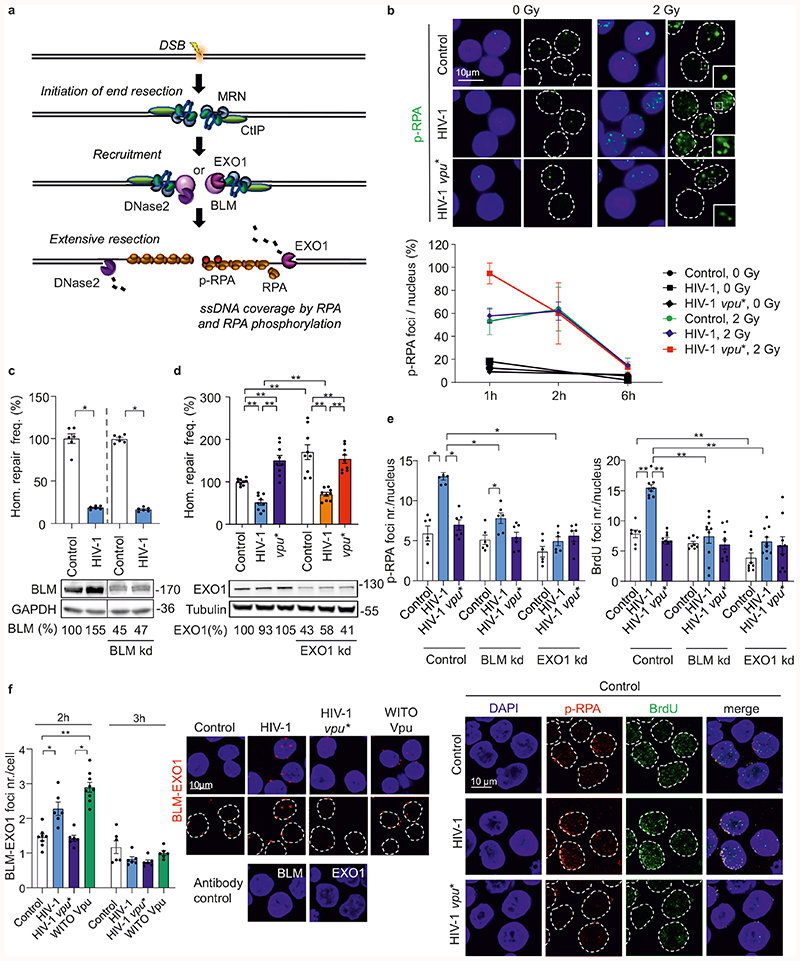
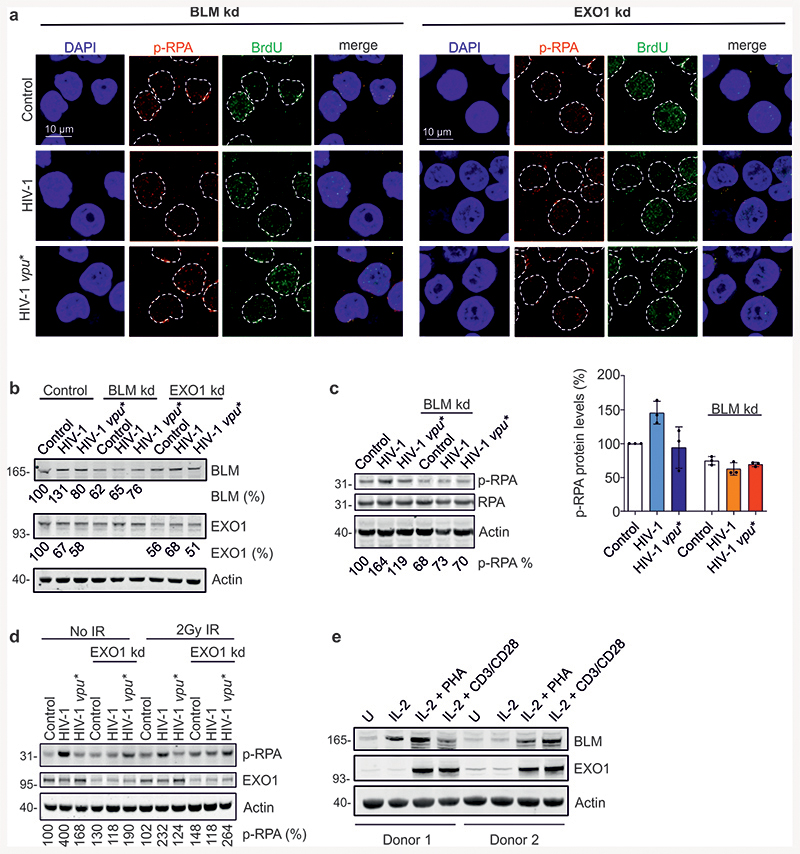
Nucleolytic DNA-end processing generates ssDNA and plays a key role in homologous repair (Fig. 3a)32. Immunofluorescence microscopy revealed that HIV-1 does not affect formation or clearance kinetics of radiation-induced p53-binding protein 1 (53BP1) foci representing indicators of DSBs (Extended Data Fig. 4a). However, HIV-1 enhanced formation of foci containing phosphorylated replication protein A (p-RPA) in a Vpu-dependent manner early (1h) after radiation exposure (Fig. 3b). p-RPA is generated by phosphorylation of the RPA32 subunit upon binding to ssDNA and represents a marker for ssDNA formation33,34 (Fig. 3a). Thus, HIV-1 Vpu enhances ssDNA formation at DSBs and consequently promotes nucleolytic processing of DNA ends.
Fig. 3. HIV-1 regulates BLM mediated DNA end-processing.
a, Overview illustrating the published role of BLM in end processing and the suitability of p-RPA as ssDNA marker. b, Detection of DSB-associated ssDNA by p-RPA. WTK1(HR/3’) cells transfected with wt or vpu* pHIV-1-NL4-3-env*-IRES-mCherry constructs were γ-irradiated (2 Gy) or left untreated (0 Gy), fixed and stained for p-RPA32 S4/8 at the indicated time points. Immunolabeled foci were scored by automated quantification in 150 nuclei each, n=3 biologically independent experiments. Maximum scores were set to 100% (absolute foci numbers were 5), bars represent means ±SD. The upper panel shows representative microscopic images of p-RPA. c-d, Homologous repair frequency in WTK1(HR/3’) cells 48 h after transfection with pHIV-1-NL4-3-env*-IRES-mCherry constructs containing intact or defective vpu genes, pCMV-HA-I-SceI and shRNA to silence BLM (c), or EXO1 (d). Bars represent means ±SEM, n=2 (c) or n=3 (d) biologically independent experiments in triplicates, absolute mean value: 0.1%, *p=0.0313, **p=0.0039. Lower panels depict Western blots to confirm the efficiency of knockdowns (frame indicates signals derived from the same membrane and exposure). e, Evaluation of ssDNA by p-RPA or BrdU. WTK1(HR/3’) cells were cultivated in medium containing 10μg/ml BrdU 24 h prior transfection with wt or vpu* pHIV-1-NL4-3-env*-IRES-mCherry and shRNA targeting BLM or EXO1. Cells were cultivated 48 h when they were γ-irradiated (2 Gy), fixed and stained for p-RPA S4/8 or BrdU in non-denaturing conditions. Foci were scored by automated quantification, each dot (5-9) presenting mean value of foci from one microscopic field containing approximately 30-50 cells, n=3 biologically independent experiments; Bars represent means ±SEM, *p=0.0195, **p=0.0039. Lower panel depicting representative microscopic images. f, PLA to evaluate proximity between BLM-EXO1. WTK1(HR/3’) cells were transfected with 5 μg proviral NL4-3 HIV-1 wt or vpu* or 1 μg WITO Vpu-AU1. 24 h later cells were fixed and PLA was performed. Left, evaluation of BLM-EXO1 PLA foci per cell from 2 independent experiments where each dot (6-9) presents mean value of approximately 30-50 analyzed cells in one microscopic view field; bars represent means ±SEM, *p=0.0313, **p=0.0078. Right, corresponding microscopic images. Two sided Wilcoxon matched-pairs test in c, d, e, f.
To clarify whether Vpu-dependent effects on DNA end-processing and homologous repair are linked, we examined involvement of various DNA repair nucleases and components of end-processing complexes using shRNA interference35. Similar to the MRE11-RAD50-Nibrin (MRN) nuclease complex component Nibrin and CtIP, initiating end resection (Extended Data Fig. 1j, 1k), FEN1, ERCC1, SLX4 and DNase2 did not alter the effect of HIV-1 on homologous repair (Extended Data Fig. 3b). Knockdown of the helicase BLM, coordinating end resection by exonuclease 1 (EXO1), DNase2 (Fig. 3c, 3d)35, and the structure-specific endonuclease subunit SLX4 targeted by Vpr23 (Extended Data Fig. 4c), did also not abrogate HIV-1 mediated suppression of homologous repair. In addition, HIV 1 did not affect expression of various proteins involved in nucleolytic processing (Extended Data Fig. 4d).
To further examine how Vpu promotes DNA end processing, we investigated the impact of BLM and EXO1 on HIV-1 induced p-RPA accumulation. EXO1 is a nuclease involved in long stretch DNA end processing stimulated by interaction with BLM helicase36. Detection of p-RPA and of BrdU under non-denaturing conditions as independent indicators of ssDNA34 revealed that lack of Vpu or silencing of BLM or EXO1 expression eliminate HIV-1 mediated end processing (Fig. 3e and Extended Data Fig. 5a, b). Proximity ligation assays (PLA)37 demonstrated that wt HIV-1 as well as WITO Vpu increased the number of BLM-EXO1 PLA foci in irradiated cells (Fig. 3f), suggesting that Vpu recruits BLM to stimulate EXO1. HIV-1 moderately increased p-RPA but not RPA32 levels in a Vpu, BLM and EXO1 dependent manner (Extended Data Fig. 5c, 5d). Notably, BLM and EXO1 are induced in the main target cells of HIV-1 replication, activated CD4+ T cells (Extended Data Fig. 5e). Collectively, these results show that Vpu promotes processing of DNA ends by BLM and EXO1 independently of its effects on homologous repair.
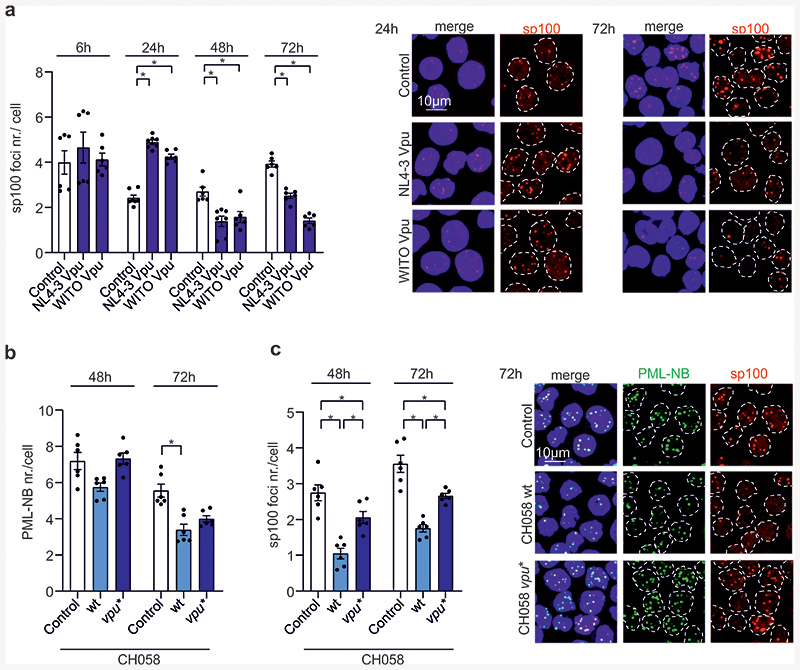
Vpu manipulates PML-NB and modulates SUMOylation
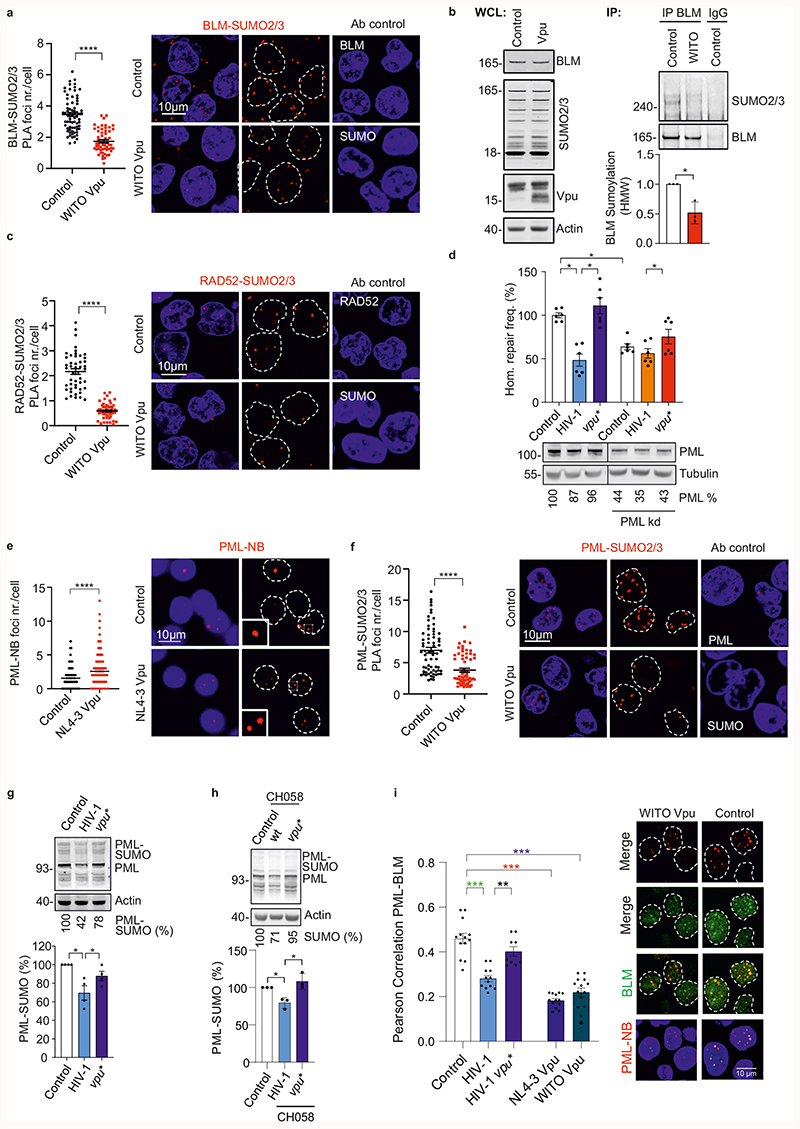
BLM function is governed by SUMOylation, which regulates its intra-nuclear trafficking between DNA damage induced foci and PML-NBs38,39. PLA revealed that Vpu reduces BLM-SUMO2/3 signals by ~50% (Fig. 4a) and immunoprecipitation confirmed reduction of BLM SUMOylation (Fig. 4b). Similarly, Vpu reduced RAD52-SUMO PLA signals by ~70% (Fig. 4c). PML-NBs are hot spots for SUMOylation and harbor many DNA repair proteins, including BLM and RAD5240. Silencing of PML by ~60% reduced homologous repair and abolished further reduction by Vpu in HIV-1 transfected cells (Fig. 4d). This agrees with results showing that PML is important for efficient HR41 and suggested that Vpu might affect PML function to modulate HR. Indeed, we found that Vpu increased PML- or Sp100-stained PML-NB numbers ~2-fold by 24h post-transfection followed by disappearance of a fraction of PML-NBs (Fig. 4e; Extended Data Fig. 6a). This kinetic is reminiscent of PML-NB dispersal and disruption after genotoxic stress40. Marked PML-NB reduction at later time points was confirmed with wt HIV-1 CH058 (Extended Data Fig. 6b, c). In addition, Vpu reduced PML-SUMO2/3 PLA signals by ~50% (Fig. 4f). HIV-1 NL4-3 in WTK1 cells (Fig. 4g) and CH058 in primary CD4+ T cells (Fig. 4h) reduced PML-SUMO levels in a Vpu-dependent manner. Finally, HIV-1 impaired colocalization between BLM and PML-NBs (Fig. 4i) and Vpu was required and sufficient for this effect. Altogether, these data support that Vpu manipulates PML-NBs and mitigates SUMOylation and/or SUMO interactions of BLM and RAD52 to promote end-processing and suppress homologous repair.
Fig. 4. Effects of Vpu on SUMOylation and PML-NBs.
a, PLA for BLM-SUMO. WTK1(HR/3’) cells were transfected with WITO Vpu-AU1 expression or control. Dot plot shows distribution of BLM-SUMO2/3 PLA foci numbers per cell, bars represent means ±SEM, n=3 biologically independent experiments (Control n=72, WITO n=53), ****p<0.0001. Representative microscopic images on the right. b, Immunoprecipitation of endogenous BLM. WTK1(HR/3’) cells transfected as in 4a. The right panel shows BLM SUMOylation values of 3 independent IPs in the presence of Vpu relative to the control IP; bars represent means ±SD, *p=0.0452. c, PLA for RAD52-SUMO2/3. WTK1(HR/3’) cells treated as in 4a. Dot plot shows distribution of RAD52-SUMO2/3 PLA foci numbers per cell, bars represent means ±SEM, n=3 biologically independent experiments (Control n=47, WITO n=55), ****p<0.0001. d, Involvement of PML in HIV-1 regulated homologous repair. WTK1(HR/3’) cells transfected with wt or vpu* pHIV-1-NL4-3-env*-IRES-mCherry constructs plus shRNAs targeting PML. Bars represent means ± SEM, n=2 biologically independent experiments in triplicates, right Western blot for PML knockdown efficiency; *p=0.0313. e, PML staining of WTK1(HR/3’) cells transfected with NL4-3 Vpu expression or control plasmid. Dot plot presents raw foci values and bars represent means ±SEM, n=3 biologically independent experiments (Control n=178, WITO n=196), ****p<0.0001. Representative microscopic images on the right. f, PLA to assess PML SUMOylation. Cells transfected as in panel 4a. Dot plot shows raw foci values and bars represent means ±SEM, n= 3 biologically independent experiments (Control n=67, WITO n=52), ****p<0.0001. Right, representative microscopic images. g, Immunodetection of SUMOylated PML. Western blotting of WTK1(HR/3’) cells transfected with wt or vpu* pHIV-1-NL4-3-env* constructs using a PML-specific antibody. Right: bars represent means ± SEM of 4 independent blots; *p=0.0344. h, Immunodetection of SUMOylated PML in CD4+ T cells. Cells transfected with wt or vpu* pHIV-1-CH058-env*-IRES-BFP were lysed, blotted and blots incubated with antibody specific for SUMOylated PML. Right: bars represent means +/- SEM of PML-SUMO levels from 3 donors, *p=0.0389. i, Pearson correlation analysis between BLM and PML-NB. WTK1(HR/3’) cells transfected as in 4g plus NL4-3 or WITO Vpu. Immunofluorescence performed 24h later. N=4 biologically independent experiments exp, ~100 nuclei analyzed each, bars represent means ± SEM, **p=0.0039, ***p=0.001, ***p=0.0002, ***p=0.0005. Two sided Wilcoxon matched-pairs test in a, c, d, e, f, i. Two sided unpaired t-test in b, g, h.
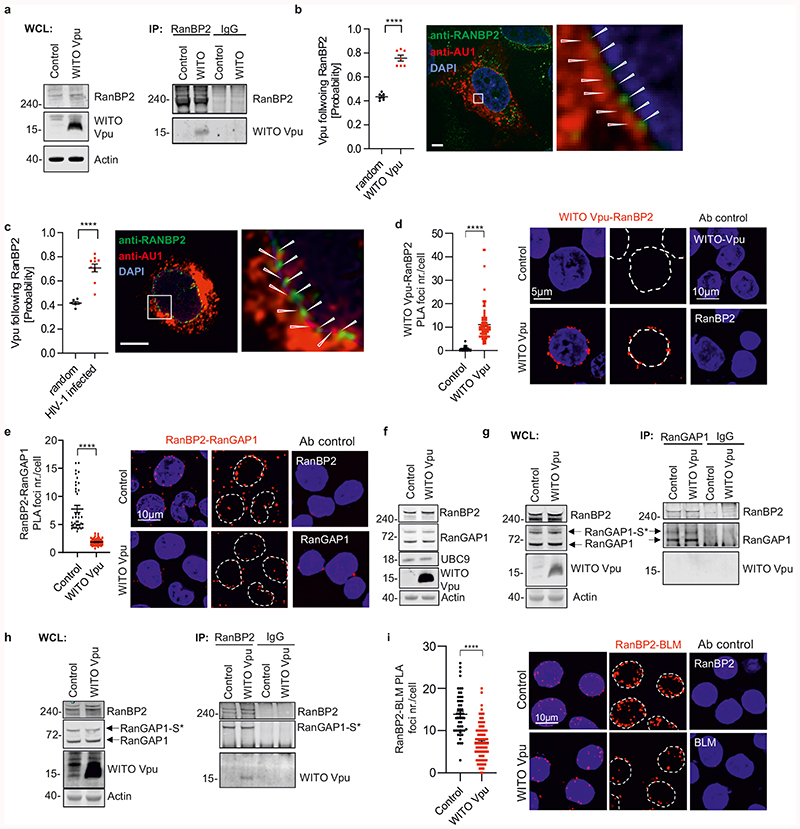
Vpu modulates the RanBP2/RanGAP1*SUMO1/UBC9 SUMO E3-ligase complex
Our findings were surprising because Vpu is known to localize in perinuclear compartments including the trans-Golgi network, but not in the nucleus42. However, subcellular protein fractionation revealed the presence of Vpu in both, the membrane and nuclear fraction (Extended Data Fig. 7a), which agrees with the reported interaction of Vpu with RanBP2 in mass spectrometry analysis43. This nucleoporin (also known as Nup358) forms filaments at the cytoplasmic surface of the nuclear pore that assemble SUMO E3-ligase complexes modulating SUMOylation and function of PML-NB44. In support of an interaction, RanBP2 co-immunoprecipitated Vpu (Fig. 5a). Analysis of HIV-1 infected or transfected SaOS cells (Fig. 5b, Extended Data Fig. 7b, Video 1) and of infected primary CD4+ T cells (Fig. 5c) confirmed the presence of Vpu at the nuclear envelope. Strikingly, Vpu and RanBP2 alternated at the nuclear rim in a non-random pattern (Fig. 5b, 5c). PLA analysis verified that Vpu and endogenous RanBP2 are in close proximity at the nuclear pore (Fig. 5d). Notably, SIVcpz MB897 Vpu showed stronger colocalization with RanBP2 than HIV-1 Vpu in CD4+ T cells and SaOS cells (Extended Data Fig. 7b, 7c and Videos 2 and 3), conceivably because SIVcpz Vpu proteins lack the functional constraints associated with anti-tetherin activity of HIV-1 Vpu at cytoplasmic membranes45.
Fig. 5. Interaction of Vpu with RanBP2/RanGAP1*SUMO1/Ubc9 SUMO E3-ligase complexes.
a, Co-immunoprecipitation of WITO Vpu by RanBP2. WTK1(HR/3’) cells were transfected with WITO Vpu, lysed and RanBP2 was pulled down. b, Confocal microscopic localization analysis of WITO Vpu and RanBP2. SaOS cells were transduced with an HIV-1-NL4-3 construct expressing the AU1-tagged WITO Vpu and lacking the overlap between vpu and env genes. The graph on the left presents the evaluation of the probability of Vpu following RanBP2 calculated from the images such as obtained on the right, n=7, bars represent means ± SEM ****p<0.0001; size bar 5 μm. c, Primary CD4+ T cells were infected with an NL4-3 construct lacking the vpu and env overlap and expressing WITO-AU1. Cells were fixed, were treated as in b. The graph on the left presents the evaluation of the probability of Vpu following RanBP2 calculated as in b, bars represent means ± SEM, n=10. d, PLA assays of WITO Vpu and RanBP2. The assay was performed in WTK1(HR/3’) cells transfected with WITO Vpu-AU1 expression plasmid or empty control. Cells were stained using antibodies against AU1 and RanBP2 plus respective PLA probes. Dot plot presents raw foci values and bars represent means ±SEM derived from two independent experiments (Control n=39, WITO n=90), ****p<0.0001. Right, representative microscopic images. e, RanBP2 and RanGAP1 PLA proximity evaluation in WTK1(HR/3’) cells. Cells transfected as in 5d. Left, evaluation of RanBP2-RanGAP1 PLA foci per cell from 3 independent experiments (Control n=39, WITO n=61), bars represent means ± SEM, ****p<0.0001. Right, representative microscopic images. f, Western blot presenting levels of RanBP2, RanGAP1, UBC9 and WITO-AU1 in WTK1(HR/3’) cells transfected with WITO Vpu-AU1 or empty control. g-h, Immunoprecipitation of RanBP2 or RanGAP1 in WTK1(HR/3’) cells transfected with WITO Vpu-AU1. Cells were lysed and samples incubated with magnetic beads pre-incubated with the designated antibodies overnight. WB Experiments from f-h performed 2 times with similar outcome. i, Proximity analysis between RanBP2 and BLM. WTK1(HR/3’) cells were treated as in panel d. Left, evaluation of RanBP2-BLM PLA foci per cell from 3 independent experiments where in each experiment 50 cells were analyzed. Bars represent means ± SEM, ****p<0.0001. Right, representative microscopic images. Two sided unpaired t- test in b, c. Two sided Wilcoxon matched-pairs test in d, e,i.
RanBP2 filaments form complexes with RanGAP1 and UBC946 that operate as E3 SUMO ligase targeting PML-NBs44. Thus, we analyzed whether Vpu interferes with the RanBP2/RanGAP1*SUMO1/UBC9 complex to manipulate SUMOylation and function of RAD52 and BLM in the nucleus. Vpu reduced the signal for RanBP2/RanGAP1 colocalization in WTK1(HR/3’) cells (Fig. 5e) and in CD4+ T cells (Extended Data Fig. 7d) by ~70%, without changing RanBP2, RanGAP1 and UBC9 expression levels (Fig. 5f). Vpu did not abrogate co-precipitation of RanBP2 and RanGAP1 (Fig. 5g, h) but reduced RanGAP1-SUMO PLA signals at the nuclear membrane (Extended Data Fig. 7e) and nuclear EXO1-SUMO PLA signals (Extended Data Fig. 7f). Notably, Vpu also diminished the number of RanBP2-BLM PLA foci (Fig. 5i). Moreover, SUMOylation of BLM was dependent on both RanBP2 and the E3 SUMO-protein ligase PIAS4 but not on PIAS1 (Extended Data Fig. 8a). In addition, Vpu reduced the levels of SUMO1 in the nucleus but had no effect on the levels of RanBP2 and RanGAP1 at the nuclear membrane (extended Data Fig. 8b). Altogether, these data suggest that Vpu interacts with RanBP2 to modulate the spatial organization of RanBP2/RanGAP1*SUMO1/UBC9 complexes rather than their integrity thereby reducing their SUMOylation activity and consequently affecting PML, RAD52, BLM and EXO1 functions.
Vpu prevents accumulation, sensing and hyper-integration of nuclear viral DNA
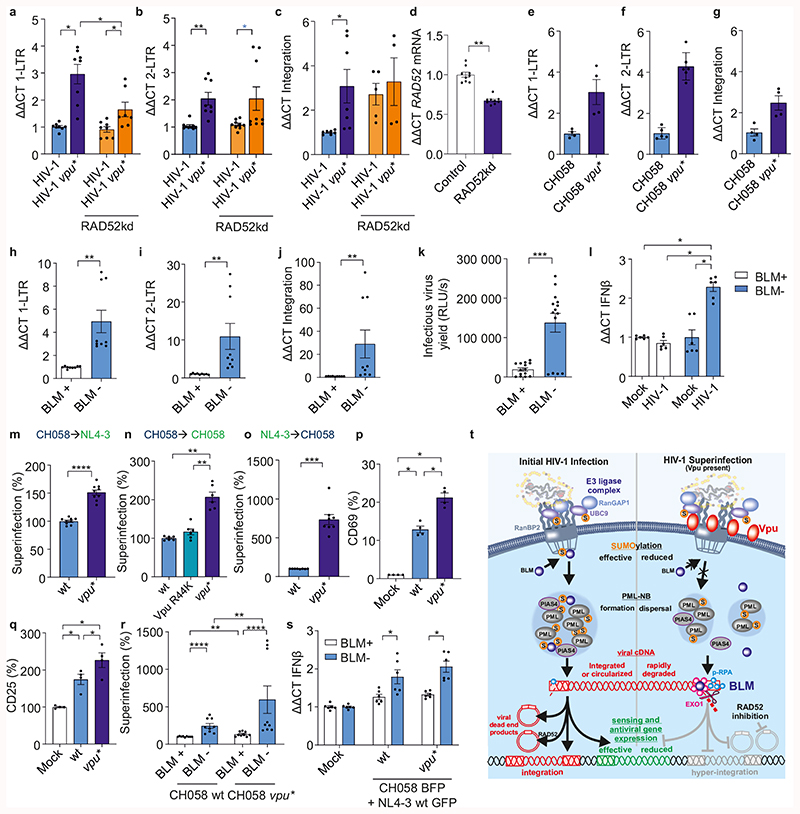
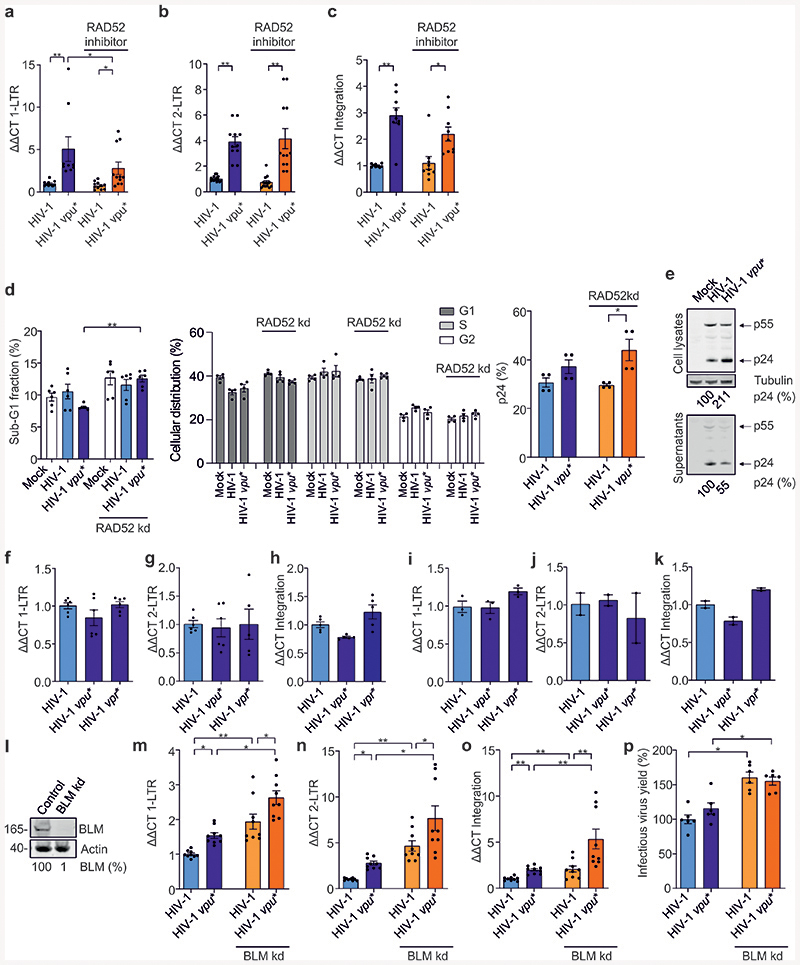
Our data showed that HIV-1 uses Vpu to suppress homologous repair in a RAD52-dependent manner and enhances BLM-mediated DNA end processing. To elucidate the biological significance of these effects, we analyzed the impact of Vpu on nuclear viral cDNA species. Quantitative PCR of HIV-1 infected primary CD4+ T cells revealed that lack of Vpu was associated with 2- to 5-fold increases of 1-LTR and 2-LTR cDNA forms as well as proviral copies (Fig. 6a-c; Extended Data Fig. 9a-c). Silencing or pharmacological inhibition of RAD52 reduced 1-LTR circle numbers in the absence of Vpu (Fig. 6a, Extended Data Fig. 9a), but had no effect on 2-LTR circles (Fig. 6b, Extended Data Fig. 9b). This observation is consistent with the role of homologous repair in 1-LTR and NHEJ in 2-LTR formation47,48. Efficient integration was restored by RAD52 knockdown but not inhibition (Fig. 6c, Extended Data Fig. 9c). Notably, the RAD52 inhibitor 6-Hydroxy-DL-DOPA prevents homologous repair but not ssDNA binding49. RAD52 was also reported to mediate integration of recombinant adeno-associated virus50 but to suppress proviral integration by competition with the integrase for ssDNA51. This might explain why RAD52 knockdown and chemical inhibition differently affected integration. Control experiments showed that neither silencing of RAD52 nor HIV-1 infection markedly affected the cell cycle or apoptosis (Extended Data Fig. 9d). Vpu did not affect viral p24 and p55 Gag expression, although it promoted virion release due to tetherin antagonism (Extended Data Fig. 9e). Vpu-dependent reduction of 1-LTR and 2-LTR circular forms as well as proviral integration were confirmed using the transmitted-founder CH058 HIV-1 strain (Fig. 6e-g). Importantly, the effects of Vpu on nuclear viral DNA species were not observed with env-defective HIV-1 or in the presence of the protease inhibitor Saquinavir (Extended Data Fig. 9f-k). Thus, they required secondary rounds of HIV-1 infection.
Fig. 6. Modulation of viral DNA species and superinfection by Vpu, RAD52 and BLM.
a-c, RT-PCR analysis of indicated viral DNA species in untreated and RAD52 silenced CD4+ T cells infected with wt or vpu* HIV-1. n=4 biologically independent experiments in duplicates, *p=0.0156, **p=0.078. d, RAD52 mRNA levels in T cells transfected with shRNA targeting RAD52. n=3 biologically independent experiments in triplicates, **p=0.0039. e-g, Viral DNA species in CD4+ T cells infected with wt or vpu* HIV-1 CH058. n=2-3 biologically independent experiments in duplicates. h-j, Viral DNA species in BLM deficient patient (BLM-) or reconstituted fibroblasts (BLM+) transduced with VSV-G pseudotyped HIV 1 NL4-3. n=3 biologically independent experiments in triplicates, **p=0.0039. k, Infectious virus yield from BLM- and BLM+ cells. n=5 biologically independent experiments in triplicates, ***p=0.0001. l, IFNβ mRNA levels in BLM- and BLM+ cells +/- VSV-G pseudotyped HIV 1. n=3 biologically independent experiments in triplicates, *p=0.0313. m-o, CD4+ T cells were transduced with VSV-G pseudotyped (m, n) wt, vpu* or R44K HIV-1-CH058-env* or (o) wt or vpu* HIV-1 NL4-3 IRES-BFP constructs. 3d later cells were exposed to VSV-G pseudotyped HIV-1 NL4-3 or CH058 IRES-EGFP constructs and examined by FACS 2d later. Superinfection was calculated as fraction of GFP+BFP+ cells among double positive and GFP+ cells. n=9 (m), n=6 (n) or 8(o) different donors relative to the value obtained for wt CH058 or NL4-3 set to 100%. **p=0.0022, ***p=0.0004, ****p=0.0001. p-q, CD4+ T cells treated as in (m) were stained for (p) CD69 and (q) CD25. Bars represent mean percentages ±SEM of activated CD69+ and CD25+ cells in the superinfected GFP+BFP+ cell population from 4 donors, *p=0.0286. r, Superinfection in BLM- or BLM+ cells treated and analyzed as described in panel 6a. n=3 biologically independent experiments in triplicates, **p=0.0039, ****p=0.0001 (absolute mean value corresponding to 100% for wt: 25%). s, mRNA IFNβ levels in superinfected BLM- or BLM+ cells. n=2 biologically independent experiments in triplicates, *p=0.0313. t, Overview of the role of Vpu as modulator of the DNA repair machinery. See main text for details. In all panels, bars represent means ±SEM. Two-sided Wilcoxon matched-pairs test in a-l, r, s. Two-tailed Mann Whitney test in m-q.
To examine the role of BLM in Vpu-dependent modulation of nuclear viral DNA forms, we transduced fibroblasts derived from a Bloom syndrome patient and the corresponding BLM-reconstituted cells with HIV-1. Both 1-LTR and 2-LTR circles, proviral integration and infectious HIV-1 yield were strongly increased in the absence of BLM (Fig. 6h-k). Similarly, BLM knockdown increased nuclear viral DNA levels, proviral integration and infectious HIV-1 production in Jurkat T cells (Extended Data Fig. 9l-p). HIV-1 induced IFNβ mRNA expression in the absence of BLM but not in its presence (Fig. 6l). Altogether, the results support that BLM reduces HIV-1 induced innate immune activation and proviral integration by promoting degradation of linear viral DNA in the nucleus.
Our results suggested that Vpu might suppress superinfection of HIV-1 independently of CD4 downmodulation by promoting BLM-dependent degradation of unintegrated linear viral DNA. To examine this, we transduced CD4+ T cells with wt or vpu-defective HIV-1-NL4-3-env* IRES-BFP constructs and exposed them to VSV-G pseudotyped env-defective HIV-1 IRES-GFP constructs allowing CD4+-independent viral entry (Extended Data Fig. 10a). Lack of Vpu enhanced superinfection 1.5- and 2.0-fold when the cells were first infected by CH058 and subsequently challenged with NL4-3 or CH058, respectively (Figs. 6m, n; Extended Data Fig. 10b, c). Vpu suppressed superinfection independently of its ability to suppress NF-κB activation (Fig. 6n). Superinfection was even 7.3-fold increased when CD4+ T cells were initially infected with HIV-1 NL4-3 and subsequently exposed to CH058 (Fig. 6o).
Lack of Vpu increased T cell activation in superinfected cells as indicated by upregulated expression of the activation markers CD69 and CD25 in the double positive cell population (Fig. 6p, q). Superinfection was increased in the absence of BLM (Fig. 6r, Extended Data Fig. 10c). Interestingly, however, Vpu still suppressed superinfection in BLM-deficient fibroblasts. A possible reason for this is that Vpu might also affect other RanBP2 dependent processes, such as docking of the HIV-1 preintegration complex to the nuclear pore and nuclear entry52–54. Lack of BLM was associated with significantly increased expression of IFNβ in superinfected cells (Fig. 6s). Thus, BLM eliminates linear viral cDNA species and suppresses innate sensing and proviral integration of HIV-1.
Discussion
Previous studies demonstrated that the HIV-1 accessory protein Vpu inhibits superinfection by downmodulating CD455 and suppresses innate immune activation by inhibiting activation of NF-κB21,27,56. Here, we show that Vpu also prevents superinfection and immune activation by modulating DNA repair. Specifically, Vpu inhibits homologous repair, while stimulating nucleolytic processing of DNA ends to suppress accumulation of nuclear viral cDNA species (summarized in Fig. 6t). To prevent circularization of viral cDNA and permit nucleolytic attack, Vpu manipulates the homologous repair factor RAD52. To stimulate processing of viral DNA ends it activates BLM. Vpu achieves this by interacting with RanBP2 at the nuclear pore to suppress the SUMOylation activity of RanBP2/RanGAP1*SUMO1/UBC9 complexes and consequently PML-NB-mediated SUMOylation of BLM. Thus, Vpu prevents superinfection and immune sensing by synergistic mechanisms including CD4 degradation, suppression of NF-κB activation, inhibition of RAD52, and activation of BLM for degradation of excess viral nuclear cDNA and prevention of proviral hyper-integration (summarized in Supplementary Fig.1).
It may seem surprising that Vpu performs activities reducing all forms of nuclear HIV-1 cDNAs. It must be considered, however, that Vpu is present only during the late stages of the viral replication cycle, when HIV-1 already achieved integration and established productive infection57. In fact, reducing effects on viral DNA species were only observed under conditions allowing multiple rounds of HIV-1 infection and are in line with the emerging role of Vpu as a multi-functional inhibitor of immune activation and viral superinfection. Inhibition of superinfection by reducing CD4 cell surface levels seems important since HIV-1 utilizes three of its gene products (Vpu, Nef and Env) to achieve this58. Nonetheless, CD4 downmodulation does not entirely prevent superinfection13. Thus, increased DNA end processing by Vpu may represent a back-up mechanism preventing superinfection via degradation of linear viral DNA products and consequently proviral hyper-integration. Strikingly, reconstitution of BLM expression in cells from a patient with a truncating BLM mutation resulted in ~30-fold decreased levels of proviral integration (Fig. 6j). Thus, by preventing integration of new proviruses that might become latent BLM may also play a role in the replenishment of latent viral reservoirs.
Another prominent function of Vpu is inhibition of antiviral host gene expression by preventing nuclear translocation of p6521 and by counteracting tetherin that traps virions and acts as immune sensor inducing NF-κB-dependent proinflammatory responses14,59. Prevention of superinfection will also suppress innate sensing because less viral material enters the cell. Increased degradation of nuclear viral DNA will further suppress innate sensing of HIV-1 infection. The exact determinants of Vpu-mediated modulation of DNA repair need further investigation. Our results show, however, that this Vpu function is conserved between HIV-1 and SIVcpz and genetically separable from effects on NF-κB and tetherin.
Vpu is not localized in the nucleus or directly interacting with homologous repair proteins. However, previous data indicate that substantial Vpu fractions are localized in the perinuclear region60 and might bind to RanBP2/Nup35843, a component of the nuclear pore complex and part of a multi-subunit SUMO E3-ligase complex46. RanBP2 has been identified as a HIV-1 dependency factor required for nuclear import of viral DNA43,61. In addition, depletion of RanBP2 compromises SUMOylation and function of PML-NBs44. We found that RanBP2 co-immunoprecipitates Vpu (Fig. 5a, h) and show that Vpu and RanBP2 are located at the nuclear membrane (Figs. 5b, c, Extended Data Fig. 7b, c). Intriguingly, Vpu reduces the number of RanBP2-RanGAP1 foci (Fig. 5e, Extended Data Fig. 7d) and manipulates PML-NBs (Fig. 5e-i; Extended Data Fig. 6a-c). These nuclear organelles are major sites of SUMOylation and colocalize with DSB repair proteins, including RAD52 and BLM40,62. Thus, Vpu might suppress SUMOylation of these DNA repair proteins by altering the SUMO ligase activity of the RanBP2/RanGAP1*SUMO1/UBC9 complex and downstream PML-NB function. Vpu achieves this without disrupting RanBP2/RanGAP1 interaction or altering their expression levels (Figs 5g, h). Instead, Vpu seems to localize between RanBP2 molecules (Figs 5b, c) presumably altering spacing and consequently functionality. Manipulation of PML-NBs and SUMOylation to promote viral replication or immune evasion has already been reported for Simian Virus 40, adeno-, herpes simplex and influenza viruses40,63,64. Notably, proviral integration in close proximity to PML-NBs promotes HIV-1 latency and disruption of these organelles reactivates viral gene expression65. Thus, it will be interesting to further examine whether Vpu modulates HIV-1 latency by affecting PML-NB integrity and function.
Our data show that Vpu downregulates SUMOylation of BLM (Fig. 4a, b) and promotes complex formation of BLM with the nuclease EXO1 (Fig. 3f). Intriguingly, BLM and EXO1 were shown to generate radiation-induced ssDNA fragments that leak into the cytosol of breast cancer cells, where they can be fully digested by TREX166. We show that HIV-1 targets the same factors to prevent accumulation of viral cDNA via manipulation of SUMOylation, which control DNA end resection67. This mechanism may contribute to downregulation of RAD52 foci numbers by Vpu because SUMOylation of BLM seems required for efficient recruitment of RAD52 to DNA damage sites39.
Vpu-mediated activation of BLM is reminiscent of Vpr-induced activation of SLX4, another SUMOylated nuclease co-factor associating with PML-NB and involved in Holliday junction cleavage during HR23,68. Both BLM and SLX4 suppress antiviral IFN expression23 (Fig. 6s). Thus, Vpu and Vpr may cooperate in activating nuclease complexes to suppress innate immune sensing via destruction of excess viral DNA. In contrast to Vpu, however, Vpr is incorporated into viral particles and virion-associated Vpr is sufficient to mediate degradation of DNA repair enzymes7. Thus, Vpr might predominantly prevent accumulation of viral cDNA species during the initial establishment of HIV-1 infection. In contrast, Vpu acts during the late stage of infection and its effects on the DNA repair machinery seem mostly relevant for superinfection events. Notably, evidence suggests that mechanical forces acting on the nucleus during T cell migration in vivo also induce changes including induction of DNA repair and sensing responses69. It will be of interest to further analyze how Vpu and Vpr cooperate to modulate DDRs and to facilitate HIV-1 escape from innate immune sensing.
In conclusion, our data indicate that HIV-1 utilizes Vpu to manipulate RanBP2/RanGAP1 *SUMO1/UBC9 Sumo E3-ligase complexes and consequently PML-NBs and SUMOylation of DNA repair factors for removal of immune-activating and/or dead-end viral DNA products in the nucleus. Vpu activated BLM may synergize with TREX15, located in the cytoplasm, to prevent accumulation of viral DNA species that may serve as pathogen-associated molecular patterns (PAMPs) to trigger innate immune activation and interferon responses. Our discovery of Vpu as modulator of DNA repair pathways thus identifies a higher-level control mechanism exerted by HIV-1 within the network of virus-host circuits affecting DDRs and the antiviral immune defense. Critical players in this control mechanism may represent promising targets for the development of drugs preventing inflammation and premature aging in HIV-1 infected individuals.
Methods
Proviral constructs
The pHIV-1-NL4-3-IRES-mCherry carrying STOP codons in the env, vpu and nef gene were cloned by replacing the IRES-eGFP cassette in the respective GFP reporter viruses55 with an IRES-mCherry cassette. The IRES-mCherry was PCR amplified using primers containing the unique single cutter MluI and XmaI. The vpr-defective pHIV-1-NL4-3-IRES-mCherry construct was created by site directed mutagenesis as described previously70 for the pHIV-1-NL4-3-IRES-mCherry–env* construct. Proviral HIV-1 CH058 constructs were cloned in the pBR322 vector by using the unique restriction sites MluI and NotI flanking the proviral sequence. For the BFP reporter constructs of CH058 the overlapping part of nef and LTR was duplicated and new unique PmeI and SacII restriction sites were introduced via overlap extension PCR. The outer primers overlap with the unique restriction site StuI in the env gene of CH058 and the MluI site after the 3’LTR. After introducing the overlap extension-PCR product in the provirus the new single cutters PmeI and SacII were used to introduce the reporter cassette. The IRES-BFP cassette was PCR amplified using primers containing PmeI and SacII. To generate the env deficient CH058 construct nucleotides 122 to 1371 of the env gene were deleted by using the Q5® Site-Directed Mutagenesis Kit of NEB as recommended by the manufacturer. This kit was also used to introduce point mutations or premature stops in the second and third codon of the vpu gene. A NL4-3 proviral clone without overlapping env and vpu genes was generated via overlap extension PCR as previously described45. During this process, a unique SacII restriction site was introduced upstream of vpu and a unique NcoI site downstream of vpu. The vpu of HIV-1 subtype B WITO was PCR amplified using primers (Supplementary Table 1) containing SacII and NcoI sites and an AU1-Tag at the C-terminus and cloned into this NL4-3 proviral backbone.
Cell culture
Jurkat cells (ATCC), WTK1 (Levy et al. 1968), WTK1(HR/3’)18, K562(Δ/3)18 cells were cultivated in RPMI-1640 medium with 10 % fetal calf serum (FCS), 2 mM L-glutamine, streptomycin, penicillin. The human osteogenic sarcoma cell line SaOS (ATCC) was cultivated in McCoys medium with 10 % FCS, 2 mM L-glutamine, streptomycin, penicillin. Human Embryonic Kidney (HEK) 293T cells (ATCC) and Hela HIV-1 reporter TZM-bl (NIH) cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10 % FCS, 2 mM L-glutamine, streptomycin, penicillin. Fibroblast GMO8505 cells from a patient with Bloom syndrome71 were cultivated in alpha Minimum Essential medium (α-MEM) with 10 % FCS, 2 mM L-glutamine and 350 μg/ml G41872. CD4+ T cells were isolated from healthy donors using a combination of lymphocyte separation medium (Biocoll separating solution, Biochrom) and negative isolation (RosetteSep, Stemcell Technologies). Cells were stimulated with IL-2 (10 ng/ml) and with anti-CD3/CD28 beads (Life Technologies). Cultivation was done in RPMI-1640 medium with 20 % FCS. All cell lines were routinely tested for mycoplasma contamination.
Cell cycle and apoptosis
Cell-cycle and apoptosis results were obtained by FACS analysis on ethanol/acetone fixed and propidium iodide stained cells as previously described73 processed under the conditions of the DSB repair assay.
Transfections to analyze DSB repair
DSB repair was analyzed by the use of the EGFP-based test system as described18. in Jurkat cells, 4x106 in 400ul of DMEM medium without phenol red, were transfected via electroporation (Bio-Rad Laboratories, Hercules) with 10 μg pCMV-HA-I-Sce I, 10 μg pBS/wtEGFP, 5 μg pHIV-1-NL4-3-env*-IRES-mCherry or pBS as control and 10 μg repair substrate EJ5SceGFP17, EJ-EGFP/3’EGFP, HR-EGFP/5’EGFP or 5’EGFP/HR-EGFP. WTK1(HR/3’) cells, 4x106 in 400ul of DMEM medium without phenol red, were electroporated with a DNA mixture consisting of 10 μg pCMV-HA-I-Sce I or 10 μg pBS/wtEGFP, 10 μg pHIV-1-NL4-3-env*-IRES-mCherry or its vpr, vpu and nef STOP variants (vpr*, vpu*, nef*) or 1 μg expression plasmids for different vpu alleles namely pCG-NL4-3 Vpu-AU1, pCG-CH106 Vpu, pCG-WITO Vpu and pCG-EK505 Vpu. For the analysis of the effects of different Vpu mutants on DSB repair WTK1(HR/3’) cells were transfected with 1 μg of WITO Vpu where amino acids from 28-81 were substituted to alanine in triplicates, plus 10 μg pCMV-HA-I-Sce I and 19 μg of pBS. In another experiment WTK1(HR/3’) cells were transfected with 4 μg of proviral transmitted founder viruses CH293, CH077 or STCO wt, vpu* or vpu NF-кB or BST mutants together with 1 μg pCMV-HA-I-Sce I. Primary CD4+ T cells were transfected with a 3 μg DNA mixture of pCMV-HA-I-Sce I, pBS, repair plasmid HR-EGFP/3’EGFP and viral construct: pHIV-1-NL4-3-env*-IRES-mCherry wt or its vpr*, vpu* variants, pCG-NL4-3 Vpu-AU1 expression plasmid; pHIV-1 M subtype B CH058, pHIV-1 M subtype B CH058 vpu*, pXL-TOPO HIV-1 M subtype C CH198, pXL-TOPO HIV-1 M subtype C CH198 vpu*; pHIV-1 M subtype B CH058 env+ wt, vpu* or vpr*; or pHIV-1 M subtype B CH058 env* where Vpu function to inhibit NF-кB or tetherin (BST) were lost. The mix contained all plasmids in a ratio of 1:1:1:1 using Amaxa T-cell nucleofection kit, program U15.
Transfections to analyze DSB repair after gene silencing
To evaluate the role of specific DNA repair proteins in HIV-1 mediated DSB repair, a general DNA mix of 10 μg pCMV-HA-I-Sce I or 10 μg pBS/wtEGFP, 10 μg of pHIV-1-NL4-3-env*-IRES-mCherry or its vpu* variant was supplemented with 40 μg of distinct shRNA or dominant negative protein expression plasmid. The following shRNA and dominant negative protein expression plasmids were used: pRS-PARP1, pRS-EXO1 and pRS-SLX4 mixes of 4 shRNAs each, mixes of pRS-BRCA1-4 and 6, pRS-CtIP-6 and −8, pRS-NBN-4 and −5, pRS-FEN1-0, −7 and −8, pRS-ERCC1-5 and −6, pRS-PIAS1-5, −6 and −7, pRS-PIAS4-7, −8 and −9, pRS-RanBP2 mix of all 4, respectively (all purchased from Origene), pSuper-midDNase2-1 and-374; pcDNA3.0-IкBα-SR22, pSuper-Rad5275; pcDNA3.1-Rad51sm76; pSuper-BLMmi37; pCG-beta-TrCP1 C-HA-IRES-DsRed277; p-Super-PML-a and PML-c mixture78; pSHAG-ATM and pSHAG- ATR79, pSuper-Ku7073.
Evaluation of DSB repair frequency
Following cultivation for 48 h, 50,000-200,000 living cells were examined to determine EGFP-positive and EGFP-negative cells by flow cytometry (FACS Calibur or FACS Canto; BD). Transfected samples were individually corrected using the transfection efficiency of each sample, where pBS was substituted by wtEGFP-expression plasmid in order to correct differences in transfection, transcription, translation, proliferation and lethality. Transfection efficiencies were ranging from 50-80%. Mean values ± SEM of ≥6 measurements were calculated and set to 100 % for controls each.
In situ PLA and immunofluorescence microscopy
WTK1(HR/3’) cells were transfected with 1 μg pCG-WITO Vpu-AU1 or corresponding controls. SaOS cells engaged for immunofluorescence analysis were transfected with 5 μg pCG-NL4-3 Vpu-AU1 expression plasmid or empty control with Amaxa nucleofection kit V, program D24. 24h later cells were taken for the PLA or immunofluorescence microscopy. The in situ PLA was performed as described in Hampp et al., 201637. The following antibodies were used: AU1 Go (Novus Biol; # NB600-452), RanBP2 Rb (Abcam; # ab64276), RanGAP1 Ms (Santa Cruz; # sc-28322), RanGAP1 Ms (Abcam), PML Ms (Abcam ab96051), PML (Bethyl; # A301-167), BLM (Bethyl; # A300-120), BLM Rb (Abcam #2179), RAD52 (Novus Biologicals; # NBP2-58116), SUMO2/3 (Novus Biologicals; # NBP2-34384, NBP1-77163), EXO1 (Abnova; # H00009156-B01P). Imaging was performed using LSM 710 confocal microscope. Quantification of foci was carried out with ImageJ software.
In Vpu localization studies CD4+ T cells were transfected with 0.5 μg pCG-WITO Vpu-AU1 or pCR-XL TOPO-SIVcpz Ptt MB897.2, Amaxa U15 Program, whereas SaOS cells were transfected with 1 μg pCG-WITO Vpu-AU1, Amaxa D24 Program. Vpu localization was analyzed also in SaOS cells infected with HIV-1 constructs where the vpu gene was AU1 tagged and separated from env as described45.
Immunofluorescence experiments were performed 24 h after transfection or infection. Briefly, cells were fixed with 4 % Paraformaldehyde (PFA) and permeabilized with 0.5 % Triton-x. After blocking with BSA, Vpu and RanBP2 were stained with anti-AU1 (Novus Biologicals), anti-RanBP2 (Abcam), p-RPA32 S4/8 (Bethyl, A300-245A), RAD52 (Novus Biologicals), BLM (Bethyl), PML (Bethyl, Abcam), SUMO1 and SUMO2 (Novus Biologicals), RanGAP1 (Abcam), BrdU (Abcam), sp100 (Abcam), 53BP1 (Novus Biologicals). Secondary antibodies conjugated to Alexa Fluor 488 and 647 were used for detection. A confocal microscope (LSM 710, Zeiss) with the corresponding software (LSM 710 Release version 5.5SP1, Zeiss; ImageJ) was used for analysis.
Virus production and infectivity assay
To generate virus stocks, HEK293T cells were transfected with 5 μg pHIV-1-NL4-3-IRES-mCherry wt or vpu* mutant plasmids thereof using a calcium phosphate transfection method55. For the evaluation of 1-LTR, 2-LTR circles and integration HIV-1 NL4-3 without reporter was used. In the case of HIV-1 pseudo-type production, 1 μg of pHIT-G-VSV-G was added to the sample mixture. HEK293T mock samples were transfected only with transfection reagents. Supernatants were harvested 40 h post-transfection. Viral infectivity was determined by infection of TZM-bl reporter cells. 6,000 cells were seeded per well on a 96-well plate and infected with cell culture supernatants. After 3 days, supernatants were removed and a β-galactosidase assay was performed following the manufacturer’s instructions (GalScreen-Applied Bioscience). β-galactosidase activities were quantified as relative light units per second (RLU/s) using an Orion Microplate Luminometer.
Western blotting and antibodies
Cellular pellets were lysed in lysis buffer (50 mM Tris, pH7,4; 150 mM NaCl; 2 mM EGTA; 2 mM EDTA; 25 mM NaF; 25 mM β-glycerophosphate; 0.1 mM NaV; proteinase inhibitor, Roche). To examine the phosphorylation status of proteins, phosphatase inhibitor was added into the lysis buffer (PhosSTOP Phosphatase Inhibitor Cocktail Tablets, Roche). Viral supernatants were centrifuged through 20 % sucrose cushion at 20,800 x g for 90 min at 4°C and lysed in Western blot lysis buffer. Protein concentrations were determined by the BCA™ Protein Assay Kit (Thermo Scientific). Samples were mixed with Protein Sample Loading buffer (Li-COR) with 10 % β-mercaptoethanol, heated at 95°C for 5 min and 60-80 μg of protein were loaded onto 8-15 % SDS-PAGE gels. Electrophoresed protein samples were blotted onto Hybond™-C Extra, Hybond™-P (GE Healthcare) or Immobilon-FL PVDF (Merck Millipore) membranes.
The following antibodies were applied: Actin ab8226 (Abcam), Artemis Rb, ATM 2C1 Ms (Abcam), ATR Ab-2 Rb (Calbiochem), AU1 Ms (Convance), AU1 Go/Rb (Novus Biol.), BLM C16 Gt (Santa Cruz), BLM Rb (Bethyl), BLM Go (Bethyl), BLM Rb (Abcam), BRCA1 Ab-1 Clone MS110 Ms (Calbiochem/Millipore), CtIP T-16 Gt (Santa Cruz), DNase2 Rb (Imgenex), ERRC1 8F1 Ms (BD), EXO1 Rb (GeneTex), EXO1 Ms (Abnova), FEN1 Ms (BD), GAPDH Rat (Biolegend), HA F-7 Ms, HA HAc5 Ms (Abcam), IкBα C-21 Rb (Santa Cruz), Ku70 S5C11 Ms (Abcam), Mre11 Rb (Novus Biologicals), Nbs1 1D7 Ms (Novus Biologicals), PARP1 Rb (Cell Signalling), PCNA Ms (Abcam), PIAS1 (Cell signaling), PIAS4 (Cell signaling), Rad51 H-92 Rb (Santa Cruz), Rad52 F-7 Ms (Santa Cruz), RAD52 (Novus Biologicals), RanBP2 Rb (Abcam), RanGAP1 Rb (Bethyl), RanGAP1 Ms (Abcam), RCC1 (santa Cruz), RPA32 (Bethyl), p-RPA32 S4/S8 Rb (Bethyl), PML PG-M3 Ms (Santa Cruz), PML Rb (Bethyl), PML C7 Ms (Abcam), anti PML-SUMO Rb80, SLX4 Rb (Bethyl), SUMO1 (Ms) and SUMO2 (Ms, Rb) (Novus Biologicals), tubulin DM1A Ms (Abcam) and Ubc9 Ms (Santa Cruz). Secondary mouse, rabbit and goat antibodies were purchased from Rockland or Thermo Fischer. Western blot signals were visualized by Super Signal® West Pico Chemiluminescent Substrate, Super Signal® West Dura Extended Duration Substrate (Thermo Scientific) with the use of Bio-Rad Chemi Doc or Licor Odyssey Scanner (Li-COR Biosciences, US). Values of band intensities for the protein of interest were corrected with the values obtained for the corresponding loading control each.
Subcellular protein fractionation and immunoprecipitation
CD4+ T cells were transfected with pCG-WITO-Vpu-AU1 plasmid, incubated for 24 h when protein fractionation was performed according to the manufacturer’s instructions (Subcellular Protein Fractionation Kit, Thermo Scientific).
Cells used for immunoprecipitation were transfected with 1 μg pCG-WITO Vpu-AU1 or corresponding controls. 24h later samples were lysed with IP lysis buffer (50 mM, Tris pH8, 150 mM NaCl, 1 % NP40, protease inhibitor) or RIPA buffer to demonstrate covalent SUMOylation (150mM NaCl, 1% NP40, 0.5% DOC, 0.1% SDS, 50mM Tris pH 8.0, 25mM NEM) for 30 min on ice or 10 min in the case of RIPA buffer. Lysed samples were centrifuged and incubated for 3 h with Pierce Protein A/G Magnetic beads (#88802) which were pre-incubated over night with antibody (5μg of primary antibody per 10μl of beads per sample).
PCR and quantitative real time PCR (RT-PCR)
WTK1(HR/3’) cells were transfected with 10 μg pCMV-HA-I-Sce I, 10 μg pNL4-3-IRES-mCherry-HIV-1-delta-env and 10 μg pBS and after 48 h were FACS sorted on EGFP positive and EGFP negative cells. Genomic DNA was isolated by the use of Blood and Cell Culture DNA Mini Kit (Qiagen) following the instructions of the manufacturer. Taq PCR Core Kit from Qiagen was used to perform PCR analysis of distinct repair pathways with the use of the following primers: PCR1.1 5’-CCCGCAACCTCCCCTTCTAC-3’ and Sce1.2 5’-ACCTTGAT-GCCGTTCTG-3’ to detect construct integrity as a control; PCR1.1 and PCR1.2 5’-GAACCCGCTC-GTCTGGCTAAG-3’ primers to detect HR; PCR1.1 and PCR2.2 5’-CTGTCTTTAACAAATTG-GACTAATCG3’ to detect SSA. Analysis of 1-LTR, 2-LTR and integrated provirus was done as described61,81. CD4+ T cells were first transfected with 2μg shRNA to knockdown RAD52 or empty control. Following cultivation for 24h cells were infected with wt or vpu- HIV-1 NL4-3 constructs normalized for p24 content. In the case of RAD52 inhibitor, CD4+T cells were pre-treated for 4h with 5μM 6-Hydroxy-DL-DOPA before spinoculation. For detection of 1-LTR the HIV-1 gag (LA1) and envelope (LA15) specific primers were used. As shown by Jacque and Stevenson81, PCR conditions used almost exclusively generate small 1-LTR products while large 2-LTR amplicons are inefficiently generated. Primers for detection of 2-LTRs are binding in the R-U3 region and thereby are specific for 2-LTR circles only. Quantification of viral circles and integrants was normalized to the RNaseP housekeeping gene. ΔΔCT was calculated as described in82. Amplification of specifically 1-LTR and 2-LTR viral cDNA was verified by DNA sequence analysis. Moreover, using RT-PCR cycling conditions as in Munir et al.83 had the same outcome for the evaluation of 1-LTR proving the specificity of our assay.
To determine mRNA levels of IFNβ and RAD52, mRNA was isolated and cDNA was synthesized by the use of the RNase Plus kit, QIAGEN, and PrimeScript, TAKARA, using manufacturer’s instructions. Quantitative PCR was run using Taqman qPCR mix from BioBudget and quantification normalized on the RNaseP housekeeping gene. All primer/probes were purchased from Applied Biosystems, Thermo Fischer.
Statistical Analysis and quantification
All experiments were independently repeated at least twice with similar results. Where statistics were calculated (DSB repair assay, cell cycle and apoptosis, RT-PCR, superinfection, immunofluorescence microscopy, PLA, pattern analysis) at least three replicates were measured in each experiment. Calculations were made using the Prism 7.05 software (GraphPad, La Jolla, CA, USA). Graphical data presentations show mean values and SEM.; asterisks indicate statistical significance (n⩾4), *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Significances for pattern analysis were determined using Mann-Whitney tests. Mean values and SD are shown for WBs; no statistics were calculated for these data, unless otherwise specified.
Extended Data
Extended Data Fig. 1. Primary DSB repair data and controls.
Primary DSB repair data and Role of DNA damage sensors (a) Primary FACS data for EGFP-based evaluation of DSB repair as in Fig. 1d. Evaluation of DSB repair frequency described in Methods section. (b-c) Apoptosis induction and cell cycle distribution in Jurkat cells transfected with pHIV-1-NL4-3-env*-IRES-mCherry (HIV-1), pCMV-HA-I-Sce I plus repair reporter plasmid HR-EGFP/5’EGFP. In parallel, split samples were treated with caspase inhibitor zVAD-fmk. Cells were fixed 48 h post-transfection with ethanol/acetone and stained with propidium iodide for DNA content analysis. Bars represent means ± SEM, n=2 biologically independent experiments in triplicates, *p=0.0313. In c, samples in the same cell cycle phase were statistically analyzed. (d) Extrachromosomal homologous repair frequency (Hom. repair freq.) in WTK1 cells transfected with pHIV-1-NL4-3-env*-IRES-mCherry, pCMV-HA-I-Sce I plus repair plasmid HR-EGFP/3’EGFP for detection of HR and SSA (construct on top). Split samples were treated with zVAD-fmk. Bars represent means ± SEM, n=3 biologically independent experiments in triplicates, *p=0.0391. Control samples transfected with control plasmid set to 100 % (absolute mean value: 2 %). (e-f) Apoptosis induction and cell cycle distribution in WTK1(HR/3’) cells 48 h after transfection with HIV-1 plus pCMV-HA-I-Sce I. Bars represent means ± SEM, n=2 biologically independent experiments in duplicates (f) or triplicates (g). In (f) samples in the same cell cycle phase were statistically analyzed. (g) Intrachromosomal homologous repair frequency in KMV(Δ/3’) cells transfected with HIV-1 plus pCMV-HA-I-Sce I. Split samples were treated with and without Hygromycin B (60 μg/ml) 4 h post-transfection. Bars represent means ± SEM, n=2 biologically independent experiments in triplicates, *p=0.0313, (absolute mean value: 0,2 %). Note the schematic presentation of the chromosomally integrated repair construct ΔEGFP/3’EGFP in KMV(Δ/3’) with a Hygromycin resistance cassette (blue bar) on top. (h-l) Homologous repair frequency in WTK1(HR/3’) cells co-transfected with pHIV-1-NL4-3-env*-IRES-mCherry, pCMV-HA-I-Sce I and shRNA to downregulate ATM (a), ATR (b), Nibrin (c), CtIP (d) and Ku70 (e) and corresponding empty vectors in the controls. Bars represent means ± SEM, n=2 biologically independent experiments in triplicates, *p=0.0313 (absolute mean value: 0.04%). Lower panels show Western blots demonstrating efficiency of knockdowns. Two sided Wilcoxon matched-pairs test in b, d, g, h-l.
Extended Data Fig. 2. Vpu expression in different experimental settings and controls to exclude potentially confounding factors.
Vpu expression in different experimental settings and controls to exclude potentially confounding factors. (a) Vpu titration. Evaluation of DSB repair frequencies in WTK1(HR/3’) cells transfected with increasing amounts of NL4-3 Vpu-AU1 expression plasmid. Bars represent means ± SEM, n=2 biologically independent experiments in triplicates, *p=0.0313. Lower panels show Western blot demonstrating expression efficiency. (b) Expression of WITO Vpu-AU1 in WTK1(HR/3’) cells transduced with pHIV-1-NL4-3 WITO Vpu-AU1 compared to cells transfected with pCG-WITO Vpu-AU1 expression vector or the pHIV-1-NL4-3 WITO Vpu-AU1 proviral construct. In the proviral construct vpu and env were separated and the NL-4-3 vpu replaced by an AU-1-tagged WITO vpu. 24 h post-transfection samples were analyzed by Western blot. (c) Apoptosis and cell cycle analysis in WTK1(HR/3’) cells after transfection with pCG-NL4-3 Vpu-AU1 expression plasmid. Bars represent means ± SEM, n=2 biologically independent experiments in triplicates, statistical analysis was calculated for comparisons between different samples in the same cell cycle phase. (d) Infection rates. Graph presenting mean values ± SD of p24 stained in CD4+ T cells from 3 donors shown in Fig. 3d. Right, primary flow cytometry data. Percentages of p24 positive cells in the live cell population (SSC/FSC gate) were calculated. (e) Homologous repair, apoptosis and cell cycle in primary HIV-1 transfected CD4+ T cells. Cells were transfected with infectious (env+) HIV-1 CH058 wt, vpr* or vpu* constructs, together with repair reporter and meganuclease I-Sce I expression plasmid. 48 h post-transfection samples were examined for repair frequencies or aceton/ethanol fixed and propiduim iodide stained to evaluate cell cycle and apoptosis. Bars represent means ± SEM, n=3 biologically independent experiments in triplicates, *p=0.0152, **p=0.022 (absolute mean value of homologous repair frequency was 0.3%). Two sided Wilcoxon matched-pairs test in a and e.
Extended Data Fig. 3. Analysis of mutant Vpu proteins and evaluation of known cellular Vpu targets.
Analysis of mutant Vpu proteins and evaluation of known cellular Vpu targets. (a) Sequence of WITO Vpu with mutated positions indicated. (b) Western blot depicting expression of different WITO Vpu mutants from (a) and CH106 wt and its KKDQ mutant with the ER retention sequence. Experiment repeated 2 times with similar outcome. (c) Homologous repair in WTK1(HR/3’) cells transfected with different mutants of WITO Vpu or CH106 and the CH106 KKDQ mutant with ER retention sequence. 48 h post transfection cells were analyzed by FACS to evaluate DSB repair frequencies. Bars represent means ± SEM, n=3 or 4 biologically independent experiments in triplicates, *p=0.0273, **p=0078, **p=0.0039 (absolute mean value: 0.7 %). Statistical evaluation shown for the comparison between wt WITO Vpu and the indicated Ala substitution mutants. (d) BrdU assay to analyze DNA processing mediated by different WITO Vpu mutants. WTK1(HR/3’) WITO transfected cells were IR (2 Gy), pre-extracted, fixed and stained against BrdU. Data presents 4 independent experiments where each dot (n=14-18) presents mean value of 30-50 analyzed nuclei from one image view, ± SEM. *p=0.0166, **p=0.012, ***p=0.0085, ****p=0.0001. Black stars presenting comparison to the control, blue stars comparison to WITO Vpu. The lower panel shows representative BrdU and AU1 microscopic images. (e) Homologous repair in WTK1(HR/3’) (β-TrCP). Cells were transfected with wt, vpr* or vpu* pHIV-1-NL4-3-env*-IRES-mCherry constructs together with expression plasmids for I-Sce I and the DN form of β-TrCP. 48 h later cells were harvested for FACS analysis. Bars represent means ± SEM, n=3 biologically independent experiments in triplicates, **p=0.039 (absolute mean value: 0.1%). Lower panels show Western blot demonstrating overexpression of β-TrCP-DN. (f) Immunoprecipitation (IP) of RAD52 1h after γ-irradiation (2Gy) of WTK1(HR/3’) cells transfected 24h before with NL4-3 Vpu expression plasmid or empty vector (Control). Graph presents mean values ± SD from two independent experiments. Note that RAD52 is difficult to detect by Western blotting unless it is concentrated e.g. by IP. Thus, most analyses had to rely on mRNA expression levels. (g) Evaluation of RAD52 mRNA levels in SaOS cells treated as described in the legend Fig. 2i. Bars represent means ± SEM, n=4. Two sided Wilcoxon matched-pairs test in c, d, e, g.
Extended Data Fig. 4. Role of 53BP1 and cellular nucleases in HIV 1-mediated homologous repair regulation.
Role of 53BP1 and cellular nucleases in HIV 1-mediated homologous repair regulation. (a) Detection of DSBs by 53BP1 foci. WTK1(HR/3’) cells transfected with wt or vpu* pHIV-1-NL4-3-env*-IRES-mCherry constructs were γ-irradiated (2Gy) or left untreated (0Gy), fixed and stained for 53BP1 at the indicated time points. Immunolabeled foci were scored by quantification in 150 nuclei each, n=3 biologically independent experiments. Maximum scores were set to 100% (absolute foci numbers for 53BP1: 7), bars represent means ±SD. The left panel shows representative microscopic images of 53BP1 foci. (b) Role of DNase2, ERCC1 and FEN1 nucleases in HIV-1 mediated DNA repair. WTK1(HR/3’) cells were transfected with pHIV-1-NL4-3-env*-IRES-mCherry, I-Sce I-HA and shRNA to silence DNase2, ERCC1 or FEN1. Bars represent means ± SEM, n=2 (DNase2, ERCC10) or n=4 (FEN1) biologically independent experiments in triplicates, *p=0.0313 (absolute mean values: Dnase2 2.2%, ERCC1 0.3% and FEN1 1.0%). Right three graphs presenting RT-PCR analysis to show DNase2, ERCC1 and FEN1 knockdown efficiencies. Bars represent means ± SEM, n=4 biologically independent experiments, two sided unpaired t-test, ***p=0.0001. (c) Homologous repair frequency in WTK1(HR/3’) cells 48 h after transfection with pHIV-1-NL4-3-env*-IRES-mCherry constructs containing intact or defective vpu genes, pCMV-HA-I-Sce I and shRNA to silence SLX4. Bars represent means ± SEM, n=2 biologically independent experiments in triplicates, *p=0.0313 (absolute mean value: 0.1%). (d) Western blot analysis of WTK1(HR/3’) cells 48h after transfection with pHIV-1-NL4-3-env*-IRES-mCherry and pCMV-HA-I-Sce I plasmids. The left panel shows examples of primary data and the right panel mean values ± SD of 3-6 independent experiments after quantification of band intensities using Bio-Rad ChemiDoc. Ttwo sided Wilcoxon matched-pairs test in b (left three panels) and c.
Extended Data Fig. 5. Role of BLM and EXO1 in Vpu-mediated modulation of DNA end processing.
Role of BLM and EXO1 in Vpu-mediated modulation of DNA end processing. (a) Representative images for immunofluorescence analysis of p-RPA and BrdU and (b) knock-down efficiency of BLM and EXO1. (c) p-RPA accumulation as a function of BLM. WTK1(HR/3’) cells were γ-irradiated (2Gy) 48h after transfection with wt or vpu* pHIV-1-NL4-3-env*-IRES-mCherry plus shRNA to silence BLM (BLMkd). 1h post-irradiation, samples were taken for p-RPA32 S4/8 and RPA Western Blot analysis. The right panel shows mean values (±SD) of p-RPA levels obtained in three independent experiments relative to control cells. (d) p-RPA Western blot analysis in WTK1(HR/3’) cells transfected with wt or vpu* pHIV-1-NL4-3-env*-IRES-mCherry, together with shRNA against EXO1. 48 h after transfection cells were irradiated and taken for Western blotting. (e) Expression of BLM and EXO1 in primary CD4+ T cells. CD4+ T cells isolated form two blood donors were either treated with IL-2 alone or stimulated with IL-2/PHA or IL-2 and CD3/CD28 magnetic beads or left untreated (U) for 3d when cells were harvested for Western blot analysis.
Extended Data Fig. 6. Effect of Vpu on PML-NBs. Role of BLM and EXO1 in Vpu-medidated modulation of DNA end processing.
Role of BLM and EXO1 in Vpu-mediated modulation of DNA end processing. (a) Representative images for immunofluorescence analysis of p-RPA and BrdU and (b) knock-down efficiency of BLM and EXO1. (c) p-RPA accumulation as a function of BLM. WTK1(HR/3’) cells were γ-irradiated (2Gy) 48h after transfection with wt or vpu* pHIV-1-NL4-3-env*-IRES-mCherry plus shRNA to silence BLM (BLMkd). 1h post-irradiation, samples were taken for p-RPA32 S4/8 and RPA Western Blot analysis. The right panel shows mean values (±SD) of p-RPA levels obtained in three independent experiments relative to control cells. (d) p-RPA Western blot analysis in WTK1(HR/3’) cells transfected with wt or vpu* pHIV-1-NL4-3-env*-IRES-mCherry, together with shRNA against EXO1. 48 h after transfection cells were irradiated and taken for Western blotting. (e) Expression of BLM and EXO1 in primary CD4+ T cells. CD4+ T cells isolated form two blood donors were either treated with IL-2 alone or stimulated with IL-2/PHA or IL-2 and CD3/CD28 magnetic beads or left untreated (U) for 3d when cells were harvested for Western blot analysis.
Extended Data Fig. 7. Localization of Vpu at the nuclear membrane and effects on RanBP2-RanGAP1 proximity and function.
Localization of Vpu at the nuclear membrane and effects on RanBP2-RanGAP1 proximity and function. (a) Subcellular protein fractionation performed in CD4+ T cells transfected with pCG-WITO Vpu-AU1 expression plasmid or empty control. Experiment conducted 2 times with similar outcome. (b) Confocal microscopic localization analysis of Vpu and RanBP2. SaOS cells were transfected with 1 μg SIVcpz MB897 Vpu-AU1. 24 h later cells were fixed, permeabilized and costained against AU1-tagged Vpu and RanBP2. Graph on left presents the probability of Vpu following RanBP2 calculated from the images such as displayed on the right, bars represent means ± SEM, Two sided unpaired t- test here and in c, n=6, ****p=0.0001. Size bar = 5 μm. (c) Primary CD4+ T cells were transfected with 0.5 μg of a vector expressing MB897 Vpu-AU1. Cells were permeabilized, stained with anti-AU1 and RanBP2 Ab and analyzed by confocal microscopy 24h post-transfection. Graph on left presents the probability of Vpu following RanBP2 calculated from the images such as displayed on the right, bars represent means ±SEM, n=5, ****p=0.0001. Size bar =5 μm. (d) RanBP2 and RanGAP1 PLA in primary CD4+ T cells. Cells were transfected with 0.5 μg WITO Vpu-AU1 expression plasmid or empty vector control. Left, evaluation of RanBP2-RanGAP1 PLA foci per cell from two different donors, n=32, bars represent means ± SEM, two sided Mann-Whitney test, ****p=0.0001. Right, representative microscopic images. (e) PLA to evaluate RanGAP1 and SUMO1 interaction in WTK1(HR/3’) cells transfected with WITO Vpu-AU1 expression plasmid or empty control. Left panel presenting evaluation of RanGAP1-SUMO PLA foci per cell, n=4 biologically independent experiments; two sided Wilcoxon matched-pairs test, mean values ± SEM, right panel presenting microscopic images. (f) PLA assay to evaluate EXO1 and SUMO2/3 interaction in WTK1(HR/3’) cells transfected with WITO Vpu-AU1 expression plasmid or empty control. Left panel presenting evaluation of EXO1-SUMO2/3 foci per cell, n=2 biologically independent experiments, two sided Wilcoxon matched-pairs test (control n= 27, WITO n=20), mean values ± SEM, ****p=0.0001. Right panel presenting microscopic images.
Extended Data Fig. 8. Role of PIAS1, PIAS4 and RanBP2 SUMO ligases in BLM SUMOylation.
Role of PIAS1, PIAS4 and RanBP2 SUMO ligases in BLM SUMOylation. (a) Role of PIAS1, PIAS4 and RanBP2 SUMO ligases in BLM SUMOylation. WTK1(HR/3’) cells were transfected with 1 μg WITO Vpu-AU1 or control plus 3 μg of shRNA to target PIAS1, PIAS4 and RanBP2. 24h later PLA was performed. Left, evaluation of BLM-SUMO2/3 foci per cell, n=3 biologically independent experiments where each dot (8-12) represents a mean from 30-50 cells analyzed from one microscopic view; bars represent means ± SEM, two sided Wilcoxon matched-pairs test, *p=0.0117, **p=0.02, **p=0.0039, **p=0.0078. Right, Western blots presenting knockdown of PIAS1, PIAS4 and RanBP2. Lower panels, representative microscopic images. (b) WTK1(HR/3’) cells were transfected with 1 μg WITO Vpu-AU1 or control and stained against SUMO1, RanBP2 and RanGAP1 24h later. Intensity profiles including the corresponding DAPI (blue) staining in the presence (red) or absence (black) of WITO vpu were quantified using ImageJ. Displayed are the means (full lines) of 10μm profiles of 25 individually quantified and aligned cells including SEM (dotted line). Exemplary confocal images are shown next to the quantifications and the profile indicated by a white line.
Extended Data Fig. 9. Effect of Vpu and RAD52 or BLM on unintegrated viral cDNA, integration and superinfection in primary CD4+ T cells.
Effect of Vpu and RAD52 or BLM on unintegrated viral cDNA, integration and superinfection in primary CD4+ T cells. (a-c) RT-PCR analysis of 1-LTR, 2-LTR circles and integration in CD4+ T cells infected with wt or vpu* pHIV-1-NL4-3 in the presence or absence of a RAD52 inhibitor (5 μM 6-Hydroxy-DL-DOPA) 48 h post infection. Bars represent means ± SEM, n=3 biologically independent experiments in triplicates, (a) *p=0.0156, **p=0.0039, (b) **p=0.0078, (c) *p=0.0273, **p=0.0039. (d) Propidium iodide staining of acetone/ethanol fixed cells and p24 stain of primary CD4+ T cells treated as in Fig. 7 (a-c). n=3, bars represent means ± SEM of 6 (apoptosis) or 4 measurements, **p=0.022. (e) Western blot analysis of p24 protein expression in CD4+ T cells infected with wt or vpu* pHIV-1-NL4-3. Experiment conducted two times with similar outcome. (f-k) RT-PCR analysis of 1-LTR, 2-LTR circles and integration in CD4+ T cells transduced with wt, vpu* or vpr* defective pHIV-1-NL4-3-env* constructs (f-h) or infected with replication-competent pHIV-1-NL4-3 In the presence of the protease inhibitor saquinavir (i-k). Bars represent means ± SEM from 6 (f,g), 5 (h), 3(i), 2(j, k) donors are shown. Analysis done 48 h post-infection/transduction. (l) Western blot demonstrating BLM knockdown efficiency in Jurkat cells. Experiment conducted 2 times with similar outcome. (m-o) RT-PCR analysis of 1-LTR and 2-LTR circles and integration in BLM silenced Jurkat cells infected with wt or vpu* VSV-G pseudotyped pHIV-1-NL4-3. Bars represent means ± SEM, n=3 biologically independent experiments in triplicates, (m) *p=0.0156, **p=0.0039; (n) *p=0.0313, **p=0.0039, (o) **p=0.0039. Analysis done 48 h post-infection. (p) Supernatants from m-o were taken to analyze infectious yields determined by β-galactosidase assay in TZM-bl cells. Bars represent means ± SEM, n=2 biologically independent experiments in triplicates, *p=0.0313. Two sided Wilcoxon matched-pairs test in a-d, m-p.
Extended Data Fig. 10. Effect of Vpu on HIV-1 superinfection in primary CD4+ T cells and BLM deficient cells.
Effect of Vpu on HIV-1 superinfection in primary CD4+ T cells and BLM deficient cells. (a) Schematic presentation of the experimental design to measure and calculate HIV-1 superinfection. (b) Primary flow cytometry data of CD4+ T cells treated as described in panel b; n=6. Calculated percentages of superinfected cells are shown between the dot plot panels. (c) Primary flow cytometry data of BLM+/- cells treated as described in the legend to Fig. 6r.
Supplementary Material
Acknowledgements
We thank Daniela Schloesser for technical help with PLA experiments, Valérie Lallemand-Breitenbach for providing us with the antibody recognizing PML isoforms I -IV, Marina Lusic for support and helpful discussion and Jeremy M. Stark for giving us EJ5SceGFP construct for NHEJ measurements. The Quadro P6000 used for this research project was made available by the NVIDIA Corporation. LW was supported by grants from the German Research Foundation (DFG WI 3099/7, B05 in CRC 1279, the European-German Space Agency (ESA/DLR) and German Ministry of Economy (BMWi, A0-10-IBER-2 funding 50WB1225). FK was supported by the DFG (KI 548/11-1; CRC 1279) and an Advanced ERC investigator grant (Anti-Virome). DS, KMJS and FK are funded by the DFG priority program SPP 1923 and TGH is supported by CRC 1036. KMJS was supported by a Marie Skłodowska-Curie Individual Fellowship from the European Union’s Framework Program for Research and Innovation Horizon 2020 (2014-2020) under the Grant Agreement No. 794803 (“VIAR”) and the DFG (SP1600/4-1).
Footnotes
Author Contributions
Author contributions: M.V. performed and analysed most of the experiments. K.S, L.K., D.H. and D.S. contributed additional assay work, image analysis and experimental expertise. C.S. performed the cloning. M.S. and T.S. contributed assays to analyse PML. N.A. contributed expertise. M.V., K.S., L.W. and F.K. wrote the manuscript. M.V., L.W. and F.K. supervised the study.
Competing interests
The authors declare no competing interests.
Data availability
The data that support the finding of this study are available from the corresponding author upon request. Complete western blot images in the manuscript are provided in the Source Data. Correspondence and requests for materials should be addressed to FK.
References
- 1.Craigie R, Bushman FD. HIV DNA integration. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a006890. a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brégnard C, Benkirane M, Laguette N. DNA damage repair machinery and HIV escape from innate immune sensing. Front Microbiol. 2014;5:176. doi: 10.3389/fmicb.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mamede JI, Cianci GC, Anderson MR, Hope TJ. Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proc Natl Acad Sci U S A. 2017;114:E7169–E7178. doi: 10.1073/pnas.1706245114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Towers GJ, Noursadeghi M. Interactions between HIV-1 and the cell-autonomous innate immune system. Cell Host Microbe. 2014;16:10–18. doi: 10.1016/j.chom.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–13. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn J, et al. HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation. J Biol Chem. 2010;285:37333–41. doi: 10.1074/jbc.M110.133181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hrecka K, et al. HIV-1 and HIV-2 exhibit divergent interactions with HLTF and UNG2 DNA repair proteins. Proc Natl Acad Sci U S A. 2016;113:E3921–30. doi: 10.1073/pnas.1605023113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schröfelbauer B, Yu Q, Zeitlin SG, Landau NR. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J Virol. 2005;79:10978–87. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–37. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 10.Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 11.Daniel R, et al. Evidence that Stable Retroviral Transduction and Cell Survival following DNA Integration Depend on Components of the Nonhomologous End Joining Repair Pathway. J Virol. 2004;78:8573–8581. doi: 10.1128/JVI.78.16.8573-8581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol. 1992;66:7193–200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wildum S, Schindler M, Munch J, Kirchhoff F. Contribution of Vpu, Env, and Nef to CD4 Down-Modulation and Resistance of Human Immunodeficiency Virus Type 1-Infected T Cells to Superinfection. J Virol. 2006;80:8047–8059. doi: 10.1128/JVI.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neil SJD, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. [DOI] [PubMed]
- 15.Van Damme N, et al. The Interferon-Induced Protein BST-2 Restricts HIV-1 Release and Is Downregulated from the Cell Surface by the Viral Vpu Protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan EL, Hollingworth R, Grand RJ. Activation of the DNA Damage Response by RNA Viruses. Biomolecules. 2016;6:2. doi: 10.3390/biom6010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akyuz N, et al. DNA Substrate Dependence of p53-Mediated Regulation of Double-Strand Break Repair. Mol Cell Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Restle A, et al. Dissecting the role of p53 phosphorylation in homologous recombination provides new clues for gain-of-function mutants. Nucleic Acids Res. 2008;36:5362–5375. doi: 10.1093/nar/gkn503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keimling M, et al. Functional characterization connects individual patient mutations in ataxia telangiectasia mutated (ATM) with dysfunction of specific DNA double-strand break-repair signaling pathways. FASEB J. 2011;25:3849–60. doi: 10.1096/fj.11-185546. [DOI] [PubMed] [Google Scholar]
- 21.Sauter D, et al. Differential regulation of NF-κB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep. 2015;10:586–99. doi: 10.1016/j.celrep.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volcic M, et al. NF-κB regulates DNA double-strand break repair in conjunction with BRCA1-CtIP complexes. Nucleic Acids Res. 2012;40:181–195. doi: 10.1093/nar/gkr687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laguette N, et al. Premature activation of the slx4 complex by vpr promotes g2/m arrest and escape from innate immune sensing. Cell. 2014;156:134–145. doi: 10.1016/j.cell.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 24.Hotter D, et al. Primate lentiviruses use at least three alternative strategies to suppress NF-kappaB-mediated immune activation. PLoS Pathog. 2017;13:e1006598. doi: 10.1371/journal.ppat.1006598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper A, et al. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498:376–9. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 26.Kmiec D, et al. Vpu-Mediated Counteraction of Tetherin Is a Major Determinant of HIV-1 Interferon Resistance. MBio. 2016;7:e00934–16. doi: 10.1128/mBio.00934-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langer S, et al. HIV-1 Vpu is a potent transcriptional suppressor of NF-κB-elicited antiviral immune responses. Elife. 2019;8 doi: 10.7554/eLife.41930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada E, et al. Human-Specific Adaptations in Vpu Conferring Anti-tetherin Activity Are Critical for Efficient Early HIV-1 Replication In Vivo. Cell Host Microbe. 2018;23:110–120. doi: 10.1016/j.chom.2017.12.009. e7. [DOI] [PubMed] [Google Scholar]
- 29.Kueck T, Neil SJD. A cytoplasmic tail determinant in HIV-1 vpu mediates targeting of tetherin for endosomal degradation and counteracts interferon-induced restriction. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margottin F, et al. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–74. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 31.Akari H, Bour S, Kao S, Adachi A, S K. The Human Immunodeficiency Virus Type 1 Accessory Protein Vpu Induces Apoptosis by Suppressing the Nuclear Factor κB–dependent Expression of Antiapoptotic Factors. J Exp Med. 2001;194:12991312. doi: 10.1084/jem.194.9.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maréchal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015;25:9–23. doi: 10.1038/cr.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee B, Tomimatsu N, Burma S. Immunofluorescence-based methods to monitor DNA end resection. [DOI] [PMC free article] [PubMed]
- 35.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–52. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. [DOI] [PMC free article] [PubMed]
- 37.Hampp S, et al. DNA damage tolerance pathway involving DNA polymerase? and the tumor suppressor p53 regulates DNA replication fork progression. Proc Natl Acad Sci. 2016;113:E4311–E4319. doi: 10.1073/pnas.1605828113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eladad S, et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- 39.Ouyang KJ, Yagle MK, Matunis MJ, Ellis Na. BLM SUMOylation regulates ssDNA accumulation at stalled replication forks. Front Genet. 2013;4:1–11. doi: 10.3389/fgene.2013.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dellaire G, Bazett-Jones DP. PML nuclear bodies: Dynamic sensors of DNA damage and cellular stress. BioEssays. 2004;26:963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- 41.Vancurova M, et al. PML nuclear bodies are recruited to persistent DNA damage lesions in an RNF168-53BP1 dependent manner and contribute to DNA repair. DNA Repair (Amst) 2019;78:114–127. doi: 10.1016/j.dnarep.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Vigan R, Neil SJD. Separable determinants of subcellular localization and interaction account for the inability of group O HIV-1 Vpu to counteract tetherin. J Virol. 2011;85:9737–48. doi: 10.1128/JVI.00479-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jäger S, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–70. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saitoh N, et al. In situ SUMOylation analysis reveals a modulatory role of RanBP2 in the nuclear rim and PML bodies. Exp Cell Res. 2006;312:1418–1430. doi: 10.1016/j.yexcr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 45.Sauter D, et al. Tetherin-Driven Adaptation of Vpu and Nef Function and the Evolution of Pandemic and Nonpandemic HIV-1 Strains. Cell Host Microbe. 2009;6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werner A, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol Cell. 2012;46:287–98. doi: 10.1016/j.molcel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 47.Kilzer JM, et al. Roles of host cell factors in circularization of retroviral DNA. Virology. 2003;314:460–467. doi: 10.1016/s0042-6822(03)00455-0. [DOI] [PubMed] [Google Scholar]
- 48.Shank PR, et al. Mapping unintegrated avian sarcoma virus DNA: Termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978;15:1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- 49.Chandramouly G, et al. Small-Molecule Disruption of RAD52 Rings as a Mechanism for Precision Medicine in BRCA-Deficient Cancers. Chem Biol. 2015;22:1491–504. doi: 10.1016/j.chembiol.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kan Y, Batada NN, Hendrickson EA. Human somatic cells deficient for RAD52 are impaired for viral integration and compromised for most aspects of homology-directed repair. DNA Repair (Amst) 2017;55:64–75. doi: 10.1016/j.dnarep.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau A, Kanaar R, Jackson SP, O’Connor MJ. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 2004;23:3421–3429. doi: 10.1038/sj.emboj.7600348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaller T, et al. HIV-1 Capsid-Cyclophilin Interactions Determine Nuclear Import Pathway, Integration Targeting and Replication Efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Nunzio F, et al. Human Nucleoporins Promote HIV-1 Docking at the Nuclear Pore, Nuclear Import and Integration. PLoS One. 2012;7:e46037. doi: 10.1371/journal.pone.0046037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang R, Mehla R, Chauhan A. Perturbation of host nuclear membrane component RanBP2 impairs the nuclear import of human immunodeficiency virus −1 preintegration complex (DNA) PLoS One. 2010;5:e15620. doi: 10.1371/journal.pone.0015620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wildum S, Schindler M, Münch J, Kirchhoff F. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J Virol. 2006;80:8047–59. doi: 10.1128/JVI.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil SJD. Innate sensing of HIV-1 assembly by Tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe. 2012;12:633–44. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohammadi P, et al. 24 Hours in the Life of HIV-1 in a T Cell Line. PLoS Pathog. 2013;9:e1003161. doi: 10.1371/journal.ppat.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lama J. The physiological relevance of CD4 receptor down-modulation during HIV infection. Curr HIV Res. 2003;1:167–84. doi: 10.2174/1570162033485276. [DOI] [PubMed] [Google Scholar]
- 59.Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil SJD. Innate Sensing of HIV-1 Assembly by Tetherin Induces NFκB-Dependent Proinflammatory Responses. Cell Host Microbe. 2012;12:633–644. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J Virol. 1990;64:621–9. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.König R, et al. Global Analysis of Host-Pathogen Interactions that Regulate Early-Stage HIV-1 Replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhong S, et al. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]
- 63.Sohn S-Y, Hearing P. Adenovirus regulates sumoylation of Mre11-Rad50-Nbs1 components through a paralog-specific mechanism. J Virol. 2012;86:9656–65. doi: 10.1128/JVI.01273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boichuk S, Hu L, Makielski K, Pandolfi PP, Gjoerup OV. Functional connection between Rad51 and PML in homology-directed repair. PLoS One. 2011;6:e25814. doi: 10.1371/journal.pone.0025814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lusic M, et al. Proximity to PML Nuclear Bodies Regulates HIV-1 Latency in CD4+ T Cells. Cell Host Microbe. 2013;13:665–677. doi: 10.1016/j.chom.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 66.Erdal E, Haider S, Rehwinkel J, Harris AL, McHugh PJ. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 2017;31:353–369. doi: 10.1101/gad.289769.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Himmels S-F, Sartori AA. Controlling DNA-End Resection: An Emerging Task for Ubiquitin and SUMO. Front Genet. 2016;7:152. doi: 10.3389/fgene.2016.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.González-Prieto R, Cuijpers SAG, Luijsterburg MS, van Attikum H, Vertegaal ACO. SUMOylation and PARylation cooperate to recruit and stabilize SLX4 at DNA damage sites. EMBO Rep. 2015;16:512–9. doi: 10.15252/embr.201440017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirby TJ, Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nature Cell Biology. 2018;20:373–381. doi: 10.1038/s41556-018-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vermeire J, et al. HIV Triggers a cGAS-Dependent, Vpu- and Vpr-Regulated Type I Interferon Response in CD4+ T Cells. Cell Rep. 2016;17:413–424. doi: 10.1016/j.celrep.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 71.Pedrazzi G. Direct association of Bloom’s syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 2001 doi: 10.1093/nar/29.21.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pedrazzi G, et al. Direct association of Bloom’s syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 2001;29:4378–86. doi: 10.1093/nar/29.21.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uhl M, et al. Role of SIRT1 in homologous recombination. DNA Repair (Amst) 2010;9:383–93. doi: 10.1016/j.dnarep.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 74.Gole B, Baumann C, Mian E, Ireno CI, Wiesmüller L. Endonuclease G initiates DNA rearrangements at the MLL breakpoint cluster upon replication stress. Oncogene. 2015;34:3391–3401. doi: 10.1038/onc.2014.268. [DOI] [PubMed] [Google Scholar]
- 75.Obermeier K, et al. Heterozygous PALB2 c.1592delT mutation channels DNA double-strand break repair into error-prone pathways in breast cancer patients. Oncogene. 2016;35:3796–3806. doi: 10.1038/onc.2015.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lambert S, Lopez BS. Characterization of mammalian RAD51 double strand break repair using non-lethal dominant-negative forms. EMBO J. 2000;19:3090–9. doi: 10.1093/emboj/19.12.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sauter D, et al. Human Tetherin Exerts Strong Selection Pressure on the HIV-1 Group N Vpu Protein. PLoS Pathog. 2012;8:e1003093. doi: 10.1371/journal.ppat.1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Herzer K, Weyer S, Krammer PH, Galle PR, Hofmann TG. Hepatitis C Virus Core Protein Inhibits Tumor Suppressor Protein Promyelocytic Leukemia Function in Human Hepatoma Cells. 2005. [DOI] [PubMed]
- 79.Collis SJ, Swartz MJ, Nelson WG, DeWeese TL. Enhanced radiation and chemotherapy-mediated cell killing of human cancer cells by small inhibitory RNA silencing of DNA repair factors. Cancer Res. 2003;63:1550–4. [PubMed] [Google Scholar]
- 80.Lallemand-Breitenbach V, et al. Arsenic degrades PML or PML–RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 81.Jacque J-M, Stevenson M. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature. 2006;441:641–645. doi: 10.1038/nature04682. [DOI] [PubMed] [Google Scholar]
- 82.Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 2013;3:71–85. [PMC free article] [PubMed] [Google Scholar]
- 83.Munir S, Thierry S, Subra F, Deprez E, Delelis O. Quantitative analysis of the time-course of viral DNA forms during the HIV-1 life cycle. Retrovirology. 2013;10:87. doi: 10.1186/1742-4690-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the finding of this study are available from the corresponding author upon request. Complete western blot images in the manuscript are provided in the Source Data. Correspondence and requests for materials should be addressed to FK.