Abstract
Coiled-coil peptides represent the principal building blocks for structure-based design of bionanomaterials. The sequence–structure relationship and precise nanoscale ordering of the coiled-coil helices originate from the knob-into-hole (KIH) packing of side chains. The helical interface stabilized by the KIH interaction is known to have chain lengths ranging from 30 to 1000 residues. Yet the shortest peptide required for oligomerization through KIH assembly is still unknown. Here, we report that through atomic resolution a minimal seven-residue amphipathic helix forms a different type of KIH motif, termed “supramolecular KIH packing”, which confers an exceptional stability to the helical dimers. Significantly, at a low pH, the peptide self-assembles into nanofibers with coiled-coil architecture resembling the natural fibrous proteins. Furthermore, hierarchical ordering of the nanofibers affords lyotropic liquid crystals composed of a shortest natural helical sequence. Thus, this study expands the sequence space for a coiled-coil folding manifold and provides another paradigm for designer nanomaterials from minimal helical sequences.
Keywords: bionanomaterials, lyotropic liquid crystal, helical peptide assembly, X-ray crystallography, nanofibers
Prediction and validation of higher order packing of helical secondary structures are fundamental to understand the natural protein folding and for exploration of such elements as building blocks for functional biomaterials.1–3 Although the design of short peptides with a high propensity to fold into the helical structure is highly explored,4,5 the oligomerization of short helical sequences guided by predetermined side chain packing, a strategy extensively used in de novo protein design,1,2 has not been experimentally validated and has yet to be demonstrated. The well-established and mathematically parametrized side-chain packing of coiled-coil proteins can serve as an excellent model system in this direction.6
Coiled-coil proteins are abundant in nature and found in many biological fibers, including hair, muscles, and skin.7 These proteins are also play important roles in the regulation of gene expressions.8 In dimeric coiled-coil, two α-helices pack with each other by interdigitating the side chains with a well-established geometric architecture termed knob-into-hole (KIH) packing. Subsequently, such KIH packing is established as a core recognition module for all coiled-coil proteins. This packing module was originally proposed by Crick to explain the electron diffraction pattern of the helical fibrous proteins α-keratin, myosin, epidermin, and fibrinogen.6 Most of these proteins usually comprise of amino acid chains that extend well over 100 residues.9 In a pioneering work, Kim and co-workers established that a much shorter sequence derived from the transcriptional activator GCN4 leucine zipper, composed only of 32 amino acids, can form stable dimers in which the interface between adjacent helices is stabilized by KIH packing.10 The high-resolution X-ray crystal structure of this sequence afforded detailed insights on KIH packing and provided an excellent paradigm to study the peptide sequence to structure relationship and the design of nanomaterials.11, 9,12 Consequently, leucine zipper-based peptide modules allowed the successful design of diverse nanostructures, including nanofibers,13,14 nanotubes,15 spheres,16 and sheetlike structures17 with a multitude of functionalities.
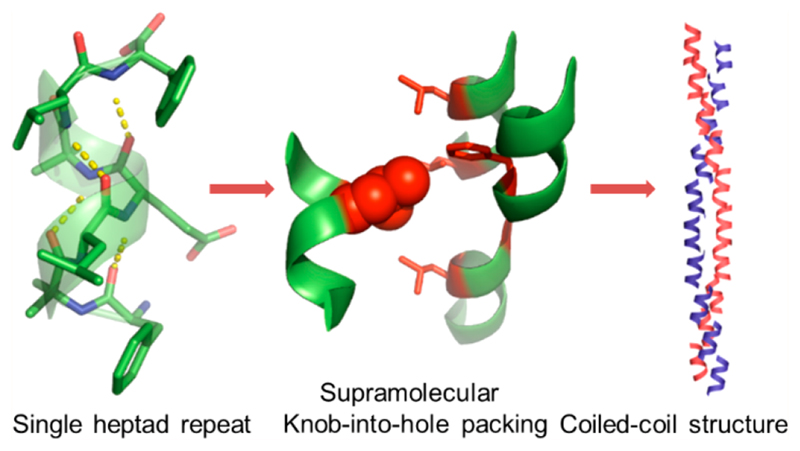
Coiled-coil proteins are composed of recurring sequences of seven amino acids, known as heptad repeats, and the majority these proteins require a minimum of three to four heptad repeats or 21-28 residue peptide chains to stabilize the dimeric helical interface.9,11 Accordingly, until now, all of the coiled-coil peptide-based nanomaterials are composed of chain lengths having more than three heptad repeats. However, a close analysis revealed that a single repeat can accommodate more than one KIH packed side chains from adjacent helices.10, 18,19 These insights prompted us to investigate whether the highly stabilizing KIH packing has the potential to act as an oligomerization module for helical sequences consisting of only seven residues or a single heptad repeat (SHR). Here, we present the atomic resolution (0.8 Å) analysis of a facially amphiphilic helical heptapeptide that gave rise to a continuous chain of dimers in which the interfaces between the adjacent helices are stabilized by KIH interactions. Furthermore, the peptide self-assembled to nanofibers at a low pH and the ensemble showed lyotropic liquid crystalline ordering. The folding of the peptide in the nanofibers was comprehensively analyzed by fiber diffraction, proton nuclear magnetic resonance (1H NMR) spectroscopy, and circular dichroism spectroscopy (CD), revealing super helical coiled-coil architecture from the shortest seven residues sequence.
Results and Discussion
Design and Atomic Level Packing of Minimal Helical Peptide
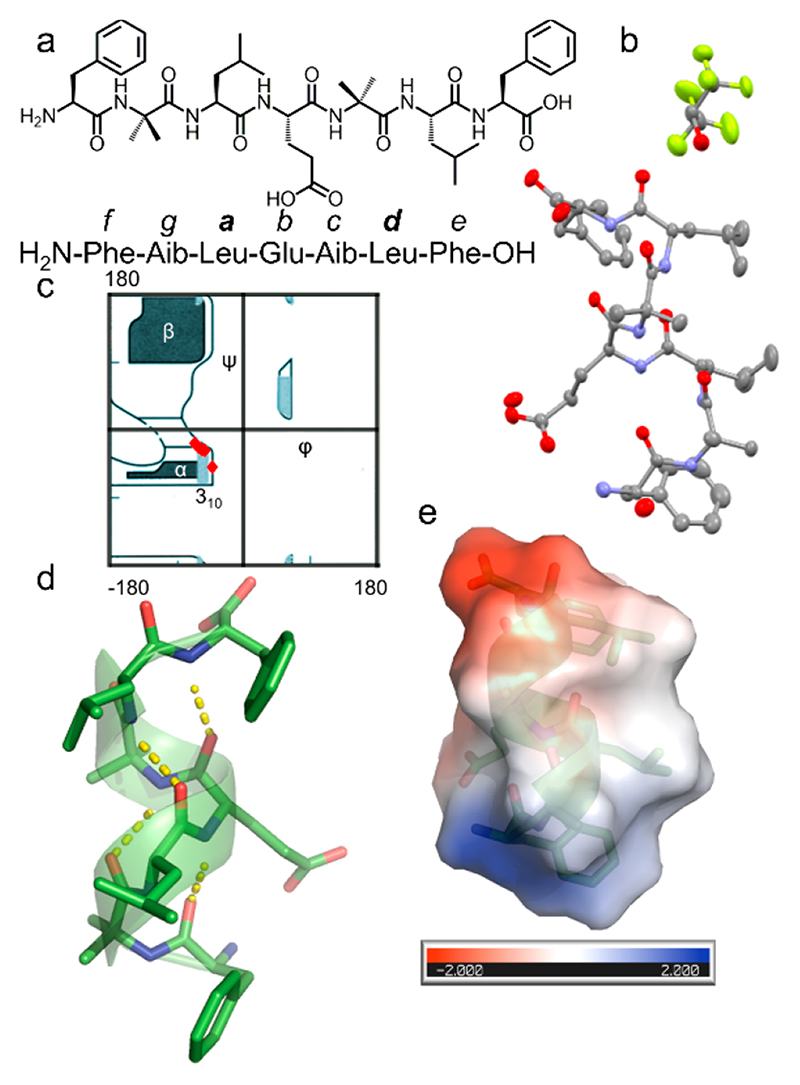
In this study, we aim to design a seven-residue peptide sequence that can self-assemble into a coiled-coil structure in which the helical interface will be stabilized by KIH packing. The seven-residue peptide was designed based on the heptad repeats, which is usually designated as abcdefg, in which the a and d positions are usually occupied by hydrophobic amino acids such as leucine (Leu), valine (Val), and isoleucine (Ile). Following the structural features of the GCN4 leucine zipper, we initially designed several sequences containing either Leu at both the a and d positions or Leu at the a position, phenylalanine (Phe) at the d position, and Phe at both the a and d positions.20 We found that the sequence with only Phe at both the a and d positions produced nanofibers in which the interfaces between the adjacent helices are stabilized by hydrophobic interaction. However, the predominant interactions are π-stacking in nature and not the KIH packing, the fundamental aspect of coiled-coil organization. Also, the optimal geometry for π-stacking forces the Phe to adopt dihedral angles not consistent with the helical region of the Ramachandran plot. This observation prompted us to redesign the sequence in which we again placed Leu at both the a and d positions. At the same time, we retained the Phe because of its high aggregation propensity toward supramolecular assembly. Most importantly, Phe was positioned at the terminus so that it will have minimal impact on the helical folding. The resulting bolaamphiphilic heptapeptide with Leu at the third and sixth positions (corresponding to the a and d position of the leucine zipper heptad repeat) adopts a robust helical conformation but failed to afford a continuous stretch of hydrophobic interface, a crucial aspect of coiled-coil packing.21 From these studies, we inferred that a facially amphiphilic helical structure, as observed in the leucine zipper, has the potential to form a long stretch of hydrophobic interface with the KIH architecture. This analysis led to the design of CCSHR (Figure 1a). Conformationally constrained natural noncoded α-aminoisobutyric acid (Aib) was incorporated into the sequence to induce helicity. An important aspect of CCSHR design was the introduction of glutamic acid (Glu) as it can introduce facial amphiphilicity, a crucial feature of leucine zipper dimerization (Figure 1a).10,13 To validate the design principle, single-crystal X-ray analysis was performed using a CCSHR crystal grown in a phosphate buffer (pH 7.4)-hexafluoroisopropanol (HFIP) (1:1) solvent mixture. The asymmetric unit comprised a single CCSHR molecule and included cocrystallized HFIP and water molecules (Figure 1b). The Ramachandran plot revealed that the peptide adopted torsion angles consistent with right-handed 310 helical structures, with average φ and ψ values of -58.9° and -30.4°, respectively (Figure 1c, Table S1). Further analysis by HELANAL confirmed that the CCSHR helix has a pitch of 5.7 Å and 3.26 residues per turn.22 Most importantly, the peptide formed four intermolecular i and i+3 hydrogen bonds, the maximum number allowed for seven residues peptide (Figure 1d). To the best our knowledge, this represents one of the few crystal structures in which a peptide with free N- and C-termini adopts a canonical helical conformation with ideal H-bonding and dihedral angles. In addition, intramolecular H-bonding between Glu side chain and N-terminal amine further reinforced the helical conformation by end-capping (Figure S1). As initially expected, the organization of Glu side chains and free termini indeed revealed a polar interface along the length of the helices, while the rest of the helical surface was occupied by hydrophobic Leu and Phe residues (Figure 1e). These side chains organizations established the facial amphiphilic nature of CCSHR, similar to GCN4.
Figure 1. Atomistic details of CCSHR.
(a) Chemical structure of CCSHR. The relative position of the residues with respect to the designation of the heptad repeat is also shown. (b) X-ray crystal structure of the molecules present in the asymmetric unit. (c) Ramachandran plot showing the dihedral angles (φ, ψ) of CCSHR (red square) superimposed over ideal secondary structural regions. (d) Representation of a canonical 310 helical structure with four i, i +3 hydrogen bonding. (e) Electrostatic surface potential depicting the segregation of hydrophilic and hydrophobic facet.
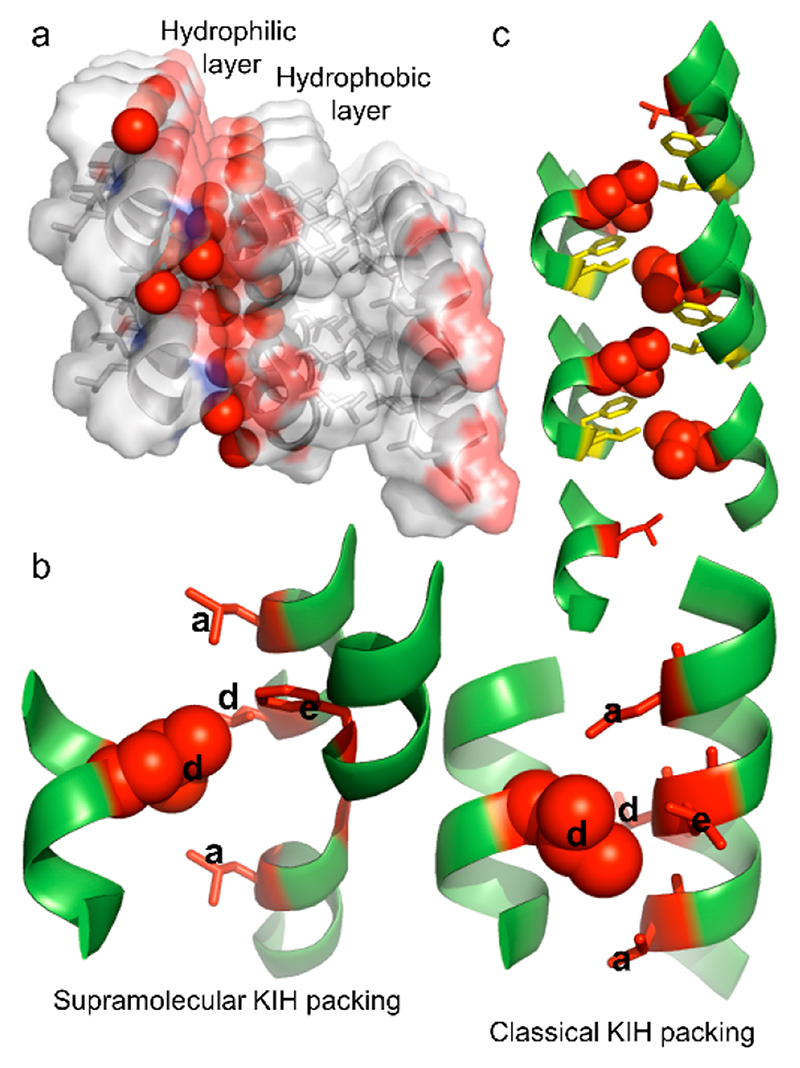
The CCSHR monomers packed on top of each other forming a helical column (Figure S1). As anticipated, the cocrystallized water molecules resided on the hydrophilic faces of the column and formed H-bonding network with both the backbone and the Glu side chain (Figure 2a). The hydrophobic facet of one column interacted with the adjacent column through a packing of side chains, which to the best of our knowledge, has not been previously observed in such short sequences. As shown in Figure 2b, the Leu residue from one helix packed in a hole composed of three Leu and one Phe from the adjacent three helices. This structural arrangement revealed good similarity with KIH packing of the GCN4 peptide (Figure 2b). This is also strongly supported by the superposition of the supra-molecular KIH packing residues with canonical GCN4 KIH motif (Figure S2). In addition, the residues involve in hole formation are located at a, a, d, and e position of the heptad sequence for both CCSHR and GCNR peptide. However, as evident from the Figure 2b, while the GCN4 KIH packing is composed of two helices, the KIH architecture in CCSHR was assembled from four individual helices (Figure 2b). Moreover, unlike coiled-coil packing, the hole-forming residues of CCSHR are not only located on the helical surface but also at the junction between the three helical subunits. Accordingly, we termed this structural arrangement “supramolecular knob-into-hole packing”. Most importantly, the superposition of the residues associated with supramolecular KIH packing and canonical dimeric helices obtained from CC+ database shows very similar geometric architecture despite the supramolecular nature of KIH packing and association of three helical subunits in hole organization in CCSHR (Figure S2).23 The supra-molecular KIH interface propagated along the long axis of the helix, thus forming a continuous stretch of hydrophobic interface (Figure 2c). The voids between dimeric helical columns were occupied by cocrystallized HFIP and afforded further stabilization to the supramolecular KIH motif through hydrophobic forces (Figure 2a, Figure S1). Most strikingly, CCSHR molecules located on top of each other along the helical column did not form any intermolecular head-to-tail H-bonding and the interface between adjacent helices was also devoid of any π-stacking interactions, a predominant driving force in the self-assembly of short peptides,24,25 thus validating the good stabilizing effect conferred by the supramolecular KIH packing (Figure S3). In addition, the helix–helix packing angle in the CCSHR dimer has value of 72°, as opposed to 22° originally proposed by Crick and measured in GCN4 (Figure S3). Taken together, these features indicate a preference for KIH stabilization over other structural parameters and energetic interactions.
Figure 2. Features of CCSHR higher order packing.
(a) Columnar organization of the peptide into an amphiphilic helix. Water molecules (presented as red sphere) concentrated on the hydrophilic face of the helical column, whereas the adjacent helix–helix interface was stabilized by hydrophobic interactions. (b) Atomic details of the hydrophobic face showing supra-molecular KIH packing (left) in which a leucine residue (knob) packed into the hole created by four residues from three adjacent helices, as opposed to the classical KIH architecture of the GCN4 leucine zipper (pdb:2zta). The KIH residues are labeled according to heptad repeat designation. (c) Propagation of KIH packed side chains along the long axis of the helical column revealed a parallel coiled-coil-like organization.
Self-Assembly of CCSHR
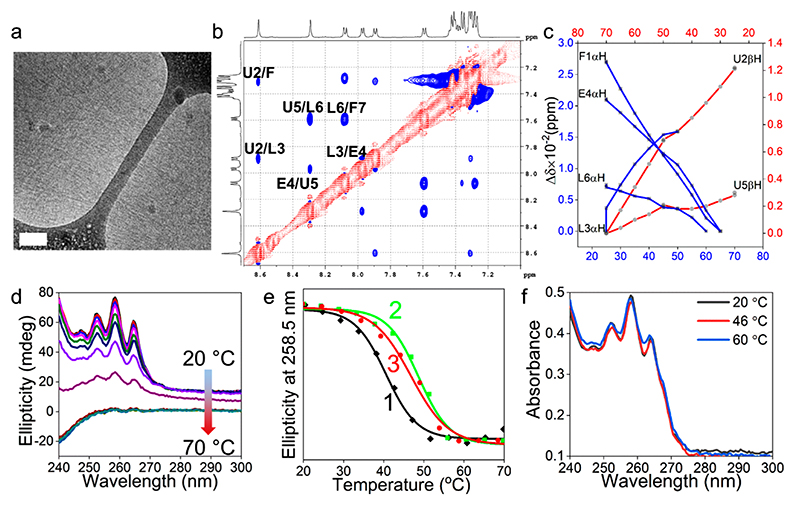
To understand the higher order organization of the peptide in solution, we studied the selfassembly of CCSHR at ~pH 2 in aqueous solution, owing to the low solubility at physiological pH, the conditions for crystallization. As shown in Figure 3a, in water at acidic pH, CCSHR (7 mg mL−1) formed nanofibers of ~2.5–3.5 nm in diameters displaying very high aspect ratios and rodlike features. The conformation of the peptide in the nanofibers was probed by comprehensive 1H NMR spectroscopy as we did not observe any crystallization under these conditions after a prolonged incubation period. Sharp well-dispersed resonances for all of the protons indicated a well-folded peptide. Additional evidence about the nature of the secondary structures were inferred from 2D experiments. Residue specific resonance assignments were established using a combination of TOCSY and NOE. Typical TOCSY assignments of CCSHR residues are depicted in Figure S4. The chemical shift positions of the backbone protons pertaining to different amino acids are shown in Table 1. The partial ROSEY spectrum shown in Figure 3b confirmed the presence of all sequential NiH–Ni+1 (dNN) connectivity, demonstrating the folding of the peptide into a helical conformation and the relatively stronger dNN cross peaks toward the C-terminal region, indicating a more regular helical geometry. This assessment was also supported by the comparable or weaker signals for dαN(CαiH–Ni+1) NOE (Figure S4).26 Additional insight into the backbone dynamics was obtained by temperature-dependent 1H NMR experiments (Figure S4). Three of the residues located at the C-terminus, namely Glu4, Leu6, and Phe7, revealed a temperature coefficient (dδ/dT) consistent with a helical conformation (Table 1, Figure S4).27
Figure 3. CCSHR self-assembly and folding.
(a) Cryo-TEM image of the nanofibers (scale bar 100 nm). (b) NH region ROESY spectrum showing sequential dNN connectivity typical of a helical secondary structure (one letter notation of the amino acids: Phe, F; Aib, U; Leu, L; Glu, E. Residues are numbered in ascending order from N- to C-terminus). (c) Plot of chemical shift variations against temperature for α- and β-protons. (d) Temperature-dependent CD spectra. (e) Thermal unfolding profiles at different peptide concentrations. [(1) Tm = 40 °C (5 mg mL−1); (2) Tm = 48 °C (7 mg mL−1); (3) Tm = 47 °C (8.5 mg mL−1)]. (f) Absorbance vs wavelength profile of nanofibers prepared at 7 mg mL−1 concentration at different temperatures.
Table 1. NMR Chemical Shifts and Temperature Coefficients of Different Protons in CCSHR.
| residues | chemical shifts (ppm) | dδ/dT (ppb/°C) | ||||
|---|---|---|---|---|---|---|
| NH | CαH | CβH | CγH | CδH | ||
| Phe1 | 4.21 | 3.21 | ||||
| Aib2 | 8.61 | 1.35 | 1.09 | −9.6 | ||
| Leu3 | 7.89 | 4.36 | 1.69 | 1.59 | 0.89 | −10.6 |
| Glu4 | 7.97 | 4.18 | 2.02 | 2.48 | −4.3 | |
| Aib5 | 8.29 | 1.41 | −9.4 | |||
| Leu6 | 7.59 | 4.24 | 1.48 | 1.40 | 0.84 | −3.4 |
| Phe7 | 8.08 | 4.67 | 3.07/3.25 | −6.6 | ||
Aib2 and Leu3 showed dδ/dT values typical of solvent-exposed residues as these were not involved in intramolecular i and i+3 H-bonding in the crystal structure. The Aib5 showed higher dδ/dT, inconsistent with helical conformation, but still can adopt helical fold as the allowed dihedral angles are restricted in the helical region of Ramachandran plot. Moreover, the well-resolved resonances also provided atom-by-atom details of the backbone protons at different temperatures. As shown in Figure 3c, overall changes in the chemical shift (Δδ) upon increasing the temperature from 25 to 70 °C were very small with a maximal difference of 27 ppb for Phe1αH and confirmed that the backbone conformation is highly robust and showed little variation with temperatures, consistent with the resistance of Aib-containing peptides to thermal unfolding.28 Furthermore, the C-terminal residues Aib5βH and Leu6αH have lower temperature sensitivity than the N-terminal residues Phe1αH, Aib2βH, and Glu4αH, indicating that the C-terminal helical fold was more robust compared to the N-terminal domain. Thus, cumulatively, the NMR studies confirmed that the CCSHR peptide adopted a rigid helical conformation.
Higher Order Packing of CCSHR in the Nanofibers
The oligomeric state of the CCSHR helices in the nanofibers was probed by measuring the changes in the CD signal at different temperatures arising from the phenylalanine chromophore, as it reports the cumulative nanoscale organization of this residue over the entire nanofiber while the monomer has a negligible CD optical activity. Figure 3d shows the spectra of CCSHR (7 mg mL−1, pH 2) in the region of 240-300 nm, corresponding to the absorbance of phenylalanine, at a temperature range of 20-70 °C. At 20 °C, the spectrum revealed positive ellipticity at 258 nm and shoulders at 252 and 264 nm. The very high signal intensity confirmed the well-ordered super helical organization of the peptide within the nanofibers. Upon increasing the temperature, the optical activity of the solution in the examined range almost completely disappeared. A plot of the temperature vs ellipticity at 258.5 nm demonstrated that the temperature-dependent melting of the nanofibers followed a sigmoidal pathway with a melting temperature (Tm) of 48 °C (Figure 3e). This characteristic confirmed that the thermal unfolding was highly cooperative and the dissociation of the helical oligomer led to melting of the nanofibers super helical packing.29 The Tm values also depended on the peptide concentration (Figure 3e). At 5 and 8.5 mg mL−1, the melting temperatures were 40 and 47 °C, respectively. It is important to mention that the simultaneous measurement of temperature-dependent optical density showed negligible variation over the entire temperature range at all measured concentrations (Figure 3f, Figure S5). The cooperativity arising from the association of a helical peptide composed of only seven amino acids could be attributed to super helical packing of CCSHR. This analysis also supported by the crystal structure of CCSHR in which the column of Phe residues involves in the KIH packing are aligned parallel to the KIH motif. Thus, the supercoiling of the KIH motif observed in the crystal structure will also lead to the helical alignment of Phe residues along the long axis of the nanofibers (Figure S6).
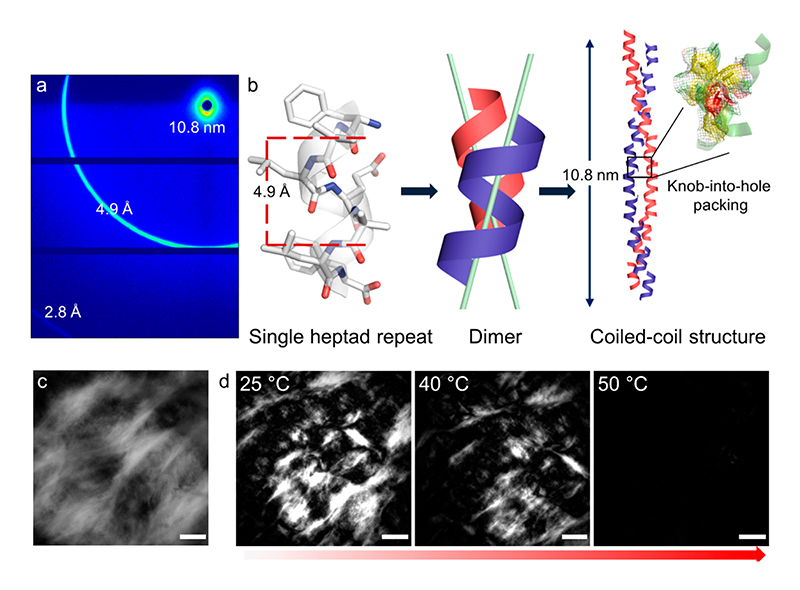
Followed by the evidence of the helical conformation of the CCSHR in the nanofibers and the hierarchical chiroptical organization of phenylalanine residue signaling the super helical packing, we probed the atomic details of the nanoscale ordering of CCSHR in nanofibers by wide-angle X-ray scattering. The diffraction pattern of the partially aligned dried fibers showed well-resolved meridian arch at 4.9 Å and 10.8 nm (Figure 4a). An additional signal at 2.8 Å can be detected, but determination of alignment was not possible (signal falls at detector edge). In accordance with the super helical nature of the nanofibers, the diffraction signal at 4.9 Å can be assigned to the repeats of helical pitches along the long axis of the fibers.30 However, the peak at 4.9 Å is considerably lower than the helical pitch of 5.7 Å observed in CCSHR crystal structure. This can be accounted by considering that the protonation of carboxylic group at low pH introduced additional electrostatic field against helix dipole which can resulted in a change in pitch value. This is further supported by the reported crystal structure of a similar sequence which differs only in one amino acids and showed a helical pitch of only 5.2 Å.20 It is also known that for effective coiled-coiling, as observed in canonical coiled-coil structure but not required for all coiled-coil packing,31 the CCSHR helices must be packed in some angle away form a parallel position which can further reduce the pitch of the helix (for example, a tentative helix crossing angle of 22° will reduced the pitch to 5.2 Å from 5.7 Å).6 Moreover, the supramolecular KIH packing observed in the solid-state crystal structure need to undergo minimal reorganization in solution, in which nanofibers assembly was observed, to allow coiling around supramolecular KIH packing. As mentioned before, such coiling is important to afford the high chiroptical activity observed in CD for phenylalanine chromophore. Most intriguingly, the meridian signal at 10.8 nm can be correlated with the pitch of super helix, demonstrating highly regular packing of the CCSHR coiled-coil along the length of the fibers. This crystalline packing of CCSHR can be attributed to the conformationally constrained nature of the Aib residues, which make the CCSHR helix highly rigid, and consequently, nanofibers have robust packing. On the basis of this analysis and CD and NMR studies, a model of the CCSHR packing in the nanofibers is illustrated in Figure 4b. Furthermore, the exceptional stability of KIH packing implied that the recognition as well as favorable interactions leading to the formation of CCSHR nanofibers was dominated by the interdigitation of the side chains, reminiscent of KIH packing observed in the crystal structure.
Figure 4. Coiled-coil organization and lyotropic liquid crystalline state of CCSHR nanofibers.
(a) Wide-angle X-ray diffraction pattern from partially aligned fibers. All of the reflections are labeled. (b) Schematic illustration of nanofibers assembly. CCSHR has a helical pitch of 4.9 Å and the two helices are inclined away from the parallel position in helical dimers. Propagation of the dimers afforded coiled-coil structure with a super helical pitch of 10.8 nm. (c) Birefringence from lyotropic liquid crystalline phases observed under polarized optical microscopy phases (scale bar 200 μm). (d) Temperature dependent lyotropic to isotropic phase transition (scale bar 500 μm).
Lyotropic Liquid Crystalline Property of the Nano-fiber Assembly
The rigid packing of the CCSHR and the high aspect ratio of the nanofibers can lead to a high excluded volume in solution, and such properties have been the basis for lytropic liquid crystalline ordering of rodlike nanofiber ensembles.32,33 Accordingly, analysis of the viscous solution (10 mg mL−1) with polarized optical microscopy revealed the presence of a crystalline domain in milimter length scale and confirmed that the CCSHR nanofibers had mesogenic properties and formed lyotropic liquid crystals (LC) (Figure 4c).32 Moreover, the cooperative melting of the nanofibers endowed the LC phase with thermotropic characteristics and converted to isotropic phases around 45-50 °C, identical to the Tm calculated based on the CD melting experiment (Figure 4d). It can be envisioned that by modulating the helix–helix interactions it may be possible to control the phase transition temperature.
Conclusion
Until now, design of short peptide-based nanomaterials was dominated by the β-sheet conformation.34 In contrast, we have demonstrated a robust coiled-coil organization of a single heptad repeat or seven amino acid sequence. The single-crystal X-ray crystallography, fiber diffraction, and NMR spectroscopy demonstrated that the dimeric helical interface of the heptad repeats was stabilized by supramolecular KIH packing. The cooperative unfolding of the higher order ensemble indicated the protein-like architecture of this simple peptide assembly. Most importantly, the hierarchical organization of the rodlike super helical nanofibers afforded the formation of lyotropic liquid crystals with predictable thermotropic activities and can be envisaged to find applications in the multifaceted areas related to liquid crystalline materials.35–37 Finally, the short sequence and their modular characteristics may provide another paradigm for functional bionanomaterials design from economically viable helical building blocks with predictable side chain orientation.34
Methods
General Sample Preparation
CCSHR was acquired from Peptron, Inc., located in South Korea. The peptide was dispersed in aqueous solution and cooled in an ice bath. HCl (1 N) was added to the cooled peptide solution to lower the pH ≈ 2 and vortexed for few seconds. A clear solution was obtained in all experimental conditions.
Cryo-transmission Electron Microscopy
Samples were prepared as described in the General Sample Preparation section. Peptide solution (7 mg mL−1, 2.5 μL) was applied on a holey carbon TEM grid (Lacey substrate, 300 mesh) and subsequently blotted with a filter paper and plunged into liquid ethane. The vitrified samples were kept under liquid nitrogen and transferred to a Tecnai G2, FEI TEM using a Gatan workstation and cryoholder. The images were acquired at 98 K with an operating voltage of 120 kV in low electron dose mode.
Crystallization and Structure Determination
Single crystals suitable for X-ray diffraction were grown by slow evaporation of the peptide in hexafluoroisopropanol–phosphate buffer (10 mM, pH 7.4) mixture (1:1) at room temperature, and crystals of diffraction quality were obtained after 2–5 days of sample preparation. Crystals suitable for diffraction were coated with Paratone oil (Hampton Research) and mounted on loops and flash frozen in liquid nitrogen. Singlecrystal X-ray diffraction data measurement was done on a Rigaku XtaLabPro system with CuKα1 (λ = 1.5418 Å) radiation at 100(2) K. Data were collected and processed with CrysAlisPro 1.171.39.22a (Rigaku OD, 2015). The structure was solved by direct methods using SHELXT-2016/4. The structure was refined by full-matrix leastsquares against F2 with SHELXL-2013. The crystallographic data are given in Table S2 (Supporting Information). The X-ray crystallographic coordinates for the peptide have been deposited at the Cambridge Crystallographic Data Centre (CCDC), under deposition no. CCDC 1867195. The structure was illustrated using Pymol (PyMOL).38 APBS (Adaptive Poisson–Boltzmann Solver) have been used to calculate the electrostatic surface potential.39 For this purpose, Aib was mutated with Ala within PyMOL. For HELANAL analysis and to draw the superposition of CCSHR supramolecular KIH packing and canonical coiled-coil packing, Aib was also mutated with Ala within PyMOL. The interhelical angles of CCSHR helices were calculated within the PyMOL interface using the command “AngleBetweenHelices”.
Optical Microscopy
CCSHR was dissolved in aqueous solution as described above at a concentration of 10 mg mL−1. The solution was placed in 1 mm quartz cell and visualized under optical microscope with or without polarizer. The appearance of characteristic birefringence confirmed the formation of lyotropic liquid crystal phase. For temperature-dependent experiments, the sample temperatures were increased by 5 °C starting from 25 °C and equilibrated at the specific temperature for a period of 20 min, and images were recorded at regular time intervals. All of the images were acquired with a Nikon Ti-E inverted motorized microscope equipped with an Okolab objective heater.
NMR Spectroscopy
CCSHR was dissolved at different concentrations in aqueous solution containing 10% D2O according to the conditions described above. D2O was added for field frequency locking on the spectrometer. All NMR spectra were recorded on Bruker (AVANCE III) 500 MHz spectrometers with 5 mm probes at a temperature 278 K and DSS-d6 as external reference. Water signal suppression in all experiments was carried out by excitation sculpting techniques using gradients.40 Two-dimension spectra were recorded in phase sensitive mode using State-TPPI algorithm. For TOCSY41 and NOESY42 experiments, a spectral width of 16 ppm was used in both dimensions. The 2048 and 256 (TOCSY) and 400 (ROESY) time domain points were used in t2 and t1 domains, respectively. A relaxation delay of 1.5 s was used for both of the 2D experiments. Zero filling was done while processing the data to yield a final data set of 2K × 1K points before Fourier transformation. Apodization for time domain data points was done using a π/2 shifted sine bell. A mixing time of 100 and 250 ms was used for TOCSY and NOESY spectra, respectively. Temperature-dependent 1H NMR spectra were recorded in the temperature ranges of 25–70 °C. All NMR data processing was carried out using Bruker TOPSPIN version 3.5 software.
Circular Dichroism Spectroscopy
CCSHR was dissolved at different concentrations in aqueous solution according to the conditions described above. CD spectra in the range of 240–300 nm was recorded on a Chirascan spectrometer in the temperature ranges of 20–70 °C with stepped ramping of 5 °C and the samples were equilibrated for 10 min at the desired temperature before carrying out the CD measurement. The data were recorded in a 2 mm (for 5 mg mL−1) or 1 mm (for 7 mg mL−1 and 8.5 mg mL−1) cuvettes. The background was subtracted from the CD spectra. Simultaneous measurements of absorbance were also performed.
Wide-Angle X-ray Scattering
A 10–20 μL portion of CCSHR solution (10 mg mL−1) was dried between two wax-filled capillaries. The dried partially aligned fibers were vertically stacked in a quartz capillary with a 1 mm diameter. WAXS measurements were performed using an in-house X-ray scattering system with a GeniX (Xenocs) low divergence Cu Kα radiation source (wavelength of 1.54 Å) and a scatterless slits setup.43 Two-dimensional scattering data, with a momentum transfer wave vector (q) range of 0.045–2.34 Å−1 at a sample-to-detector distance of about 185 mm, was collected on a Pilatus 300 K detector (Dectris, Baden-Daettwil, Switzerland). Azimuthal integration was performed using Matlab (MathWorks, Natick, MA)-based procedures (SAXSi). Calibration was performed using silver behenate. The scattering data of the empty capillary was collected as the background and used for subtraction of spurious scattering from the WAXS system itself, for example, Kapton vacuum windows and air gaps.
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.9b04148.
Supporting figures and tables (PDF)
Crystallographic coordinates of CCSHR (CIF)
Acknowledgments
We thank L. Burlaka for help with TEM analysis and members of the Gazit laboratory for helpful discussions. S.M. and V.B. thank the PBC Program for outstanding Postdoctoral Researchers from China and India and Tel Aviv University for scholarships. R.B. acknowledges support from the Israeli Science Foundation (550/15). This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant Agreement No. BISON-694426 to E.G.). We thank S. Rencus-Lazar for her valuable comments and suggestions. We thank S. Raghothama for help with NMR analysis.
Notes
The authors declare no competing financial interest.
References
- 1.Boyken SE, Chen Z, Groves B, Langan RA, Oberdorfer G, Ford A, Gilmore JM, Xu C, DiMaio F, Pereira JH, Sankaran B, et al. De Novo Design of Protein Homo-Oligomers with Modular Hydrogen-Bond Network-Mediated Specificity. Science. 2016;352:680–687. doi: 10.1126/science.aad8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Regan L, DeGrado W. Characterization of a Helical Protein Designed from First Principles. Science. 1988;241:976–978. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- 3.Galloway JM, Senior L, Fletcher JM, Beesley JL, Hodgson LR, Harniman RL, Mantell JM, Coombs J, Rhys GG, Xue W-F, Mosayebi M, et al. Bioinspired Silicification Reveals Structural Detail in Self-Assembled Peptide Cages. ACS Nano. 2018;12:1420–1432. doi: 10.1021/acsnano.7b07785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein M. Stabilized Helical Peptides: Overview of the Technologies and Its Impact on Drug Discovery. Expert Opin Drug Discovery. 2017;12:1117–1125. doi: 10.1080/17460441.2017.1372745. [DOI] [PubMed] [Google Scholar]
- 5.Henchey LK, Jochim AL, Arora PS. Contemporary Strategies for the Stabilization of Peptides in the α-Helical Conformation. Curr Opin Chem Biol. 2008;12:692–697. doi: 10.1016/j.cbpa.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crick FHC. The Packing of α-Helices: Simple Coiled-Coils. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 7.Parry DAD. Fifty Years of Fibrous Protein Research: A Personal Retrospective. J Struct Biol. 2014;186:320–334. doi: 10.1016/j.jsb.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Nikolaev Y, Deillon C, Hoffmann SRK, Bigler L, Friess S, Zenobi R, Pervushin K, Hunziker P, Gutte B. The Leucine Zipper Domains of the Transcription Factors GCN4 and C-Jun Have Ribonuclease Activity. PLoS One. 2010;5:e10765. doi: 10.1371/journal.pone.0010765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robson Marsden H, Kros A. Self-Assembly of Coiled Coils in Synthetic Biology: Inspiration and Progress. Angew Chem, Int Ed. 2010;49:2988–3005. doi: 10.1002/anie.200904943. [DOI] [PubMed] [Google Scholar]
- 10.O’Shea EK, Klemm JD, Kim PS, Alber T. X-Ray Structure of the GCN4 Leucine Zipper, a Two-Stranded, Parallel Coiled Coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 11.Conticello V, Hughes S, Modlin C. Fibrous proteins: structures and mechanisms; Subcellular Biochemistry. Springer International Publishing; Cham: 2017. Biomaterials Made from Coiled-Coil Peptides; pp. 575–600. [DOI] [Google Scholar]
- 12.Al-Ahmady ZS, Al-Jamal WT, Bossche JV, Bui TT, Drake AF, Mason AJ, Kostarelos K. Lipid-Peptide Vesicle Nanoscale Hybrids for Triggered Drug Release by Mild Hyperthermia In Vitro and In Vivo. ACS Nano. 2012;6:9335–9346. doi: 10.1021/nn302148p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong H, Paramonov SE, Hartgerink JD. Self-Assembly of α-Helical Coiled Coil Nanofibers. J Am Chem Soc. 2008;130:13691–13695. doi: 10.1021/ja8037323. [DOI] [PubMed] [Google Scholar]
- 14.Ing NL, Spencer RK, Luong SH, Nguyen HD, Hochbaum AI. Electronic Conductivity in Biomimetic α-Helical Peptide Nanofibers and Gels. ACS Nano. 2018;12:2652–2661. doi: 10.1021/acsnano.7b08756. [DOI] [PubMed] [Google Scholar]
- 15.Egelman EH, Xu C, DiMaio F, Magnotti E, Modlin C, Yu X, Wright E, Baker D, Conticello VP. Structural Plasticity of Helical Nanotubes Based on Coiled-Coil Assemblies. Structure. 2015;23:280–289. doi: 10.1016/j.str.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher JM, Harniman RL, Barnes FRH, Boyle AL, Collins A, Mantell J, Sharp TH, Antognozzi M, Booth PJ, Linden N, Miles MJ, et al. Self-Assembling Cages from Coiled-Coil Peptide Modules. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magnotti EL, Hughes SA, Dillard RS, Wang S, Hough L, Karumbamkandathil A, Lian T, Wall JS, Zuo X, Wright ER, Conticello VP. Self-Assembly of an α-Helical Peptide into a Crystalline Two-Dimensional Nanoporous Framework. J Am Chem Soc. 2016;138:16274–16282. doi: 10.1021/jacs.6b06592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson AR, Wood CW, Burton AJ, Bartlett GJ, Sessions RB, Brady RL, Woolfson DN. Computational Design of Water-Soluble α-Helical Barrels. Science. 2014;346:485–488. doi: 10.1126/science.1257452. [DOI] [PubMed] [Google Scholar]
- 19.Lupas AN, Bassler J, Dunin-Horkawicz S. In: Fibrous Proteins: Structures and Mechanisms. Parry DAD, Squire JM, editors. Springer International Publishing; Cham: 2017. The Structure and Topology of α-Helical Coiled Coils; pp. 95–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mondal S, Adler-Abramovich L, Lampel A, Bram Y, Lipstman S, Gazit E. Formation of Functional Super-Helical Assemblies by Constrained Single Heptad Repeat. Nat Commun. 2015;6 doi: 10.1038/ncomms9615. 8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mondal S, Varenik M, Bloch DN, Atsmon-Raz Y, Jacoby G, Adler-Abramovich L, Shimon LJW, Beck R, Miller Y, Regev O, Gazit E. A Minimal Length Rigid Helical Peptide Motif Allows Rational Design of Modular Surfactants. Nat Commun. 2017;8 doi: 10.1038/ncomms14018. 14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar P, Bansal M. HELANAL-Plus: A Web Server for Analysis of Helix Geometry in Protein Structures. J Biomol Struct Dyn. 2012;30:773–783. doi: 10.1080/07391102.2012.689705. [DOI] [PubMed] [Google Scholar]
- 23.Testa OD, Moutevelis E, Woolfson DN. CC+: A Relational Database of Coiled-Coil Structures. Nucleic Acids Res. 2009;37:D315–322. doi: 10.1093/nar/gkn675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reches M, Gazit E. Casting Metal Nanowires within Discrete Self-Assembled Peptide Nanotubes. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 25.Pellach M, Mondal S, Harlos K, Mance D, Baldus M, Gazit E, Shimon LJW. A Two-Tailed Phosphopeptide Crystallizes to Form a Lamellar Structure. Angew Chem, Int Ed. 2017;56:3252–3255. doi: 10.1002/anie.201609877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijayalakshmi S, Rao RB, Karle IL, Balaram P. Comparison of Helix-Stabilizing Effects of α,α-Dialkyl Glycines with Linear and Cycloalkyl Side Chains. Biopolymers. 2000;53:84–98. doi: 10.1002/(SICI)1097-0282(200001)53:1<84::AID-BIP8>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 27.Andersen NH, Neidigh JW, Harris SM, Lee GM, Liu Z, Tong H. Extracting Information from the Temperature Gradients of Polypeptide NH Chemical Shifts. 1. the Importance of Conformational Averaging. J Am Chem Soc. 1997;119:8547–8561. [Google Scholar]
- 28.Banerjee R, Basu G. Direct Evidence for Alteration of Unfolding Profile of a Helical Peptide by Far-Ultraviolet Circular Dichroism Aromatic Side-Chain Contribution. FEBS Lett. 2002;523:152–156. doi: 10.1016/s0014-5793(02)02974-5. [DOI] [PubMed] [Google Scholar]
- 29.Zolkiewski M, Redowicz MJ, Korn ED, Hammer JA, Ginsburg A. Two-State Thermal Unfolding of a Long Dimeric Coiled-Coil: The Acanthamoeba Myosin II Rod. Biochemistry. 1997;36:7876–7883. doi: 10.1021/bi962947c. [DOI] [PubMed] [Google Scholar]
- 30.Banwell EF, Abelardo ES, Adams DJ, Birchall MA, Corrigan A, Donald AM, Kirkland M, Serpell LC, Butler MF, Woolfson DN. Rational Design and Application of Responsive α-Helical Peptide Hydrogels. Nat Mater. 2009;8:596–600. doi: 10.1038/nmat2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang P-S, Oberdorfer G, Xu C, Pei XY, Nannenga BL, Rogers JM, DiMaio F, Gonen T, Luisi B, Baker D. High Thermodynamic Stability of Parametrically Designed Helical Bundles. Science. 2014;346:481–485. doi: 10.1126/science.1257481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakata M, Zanchetta G, Chapman BD, Jones CD, Cross JO, Pindak R, Bellini T, Clark NA. End-to-End Stacking and Liquid Crystal Condensation of 6- to 20-Base Pair DNA Duplexes. Science. 2007;318:1276–1279. doi: 10.1126/science.1143826. [DOI] [PubMed] [Google Scholar]
- 33.Nyström G, Arcari M, Mezzenga R. Confinement-Induced Liquid Crystalline Transitions in Amyloid Fibril Cholesteric Tactoids. Nat Nanotechnol. 2018;13:330–336. doi: 10.1038/s41565-018-0071-9. [DOI] [PubMed] [Google Scholar]
- 34.Lampel A, McPhee SA, Park H-A, Scott GG, Humagain S, Hekstra DR, Yoo B, Frederix PWJM, Li T-D, Abzalimov RR, Greenbaum SG, et al. Polymeric Peptide Pigments with Sequence-Encoded Properties. Science. 2017;356:1064–1068. doi: 10.1126/science.aal5005. [DOI] [PubMed] [Google Scholar]
- 35.Aranson IS. Harnessing Medium Anisotropy to Control Active Matter. Acc Chem Res. 2018;51:3023–3030. doi: 10.1021/acs.accounts.8b00300. [DOI] [PubMed] [Google Scholar]
- 36.Keber FC, Loiseau E, Sanchez T, DeCamp SJ, Giomi L, Bowick MJ, Marchetti MC, Dogic Z, Bausch AR. Topology and Dynamics of Active Nematic Vesicles. Science. 2014;345:1135–1139. doi: 10.1126/science.1254784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhai J, Fong C, Tran N, Drummond CJ. Non-Lamellar Lyotropic Liquid Crystalline Lipid Nanoparticles for the Next Generation of Nanomedicine. ACS Nano. 2019;13:6178–6206. doi: 10.1021/acsnano.8b07961. [DOI] [PubMed] [Google Scholar]
- 38.PyMOL Molecular Graphics System. Schrödinger, L.L.C; Cambridge, MA: 2015. version 1.8. [Google Scholar]
- 39.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang TL, Shaka AJ. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J Magn Reson Ser A. 1995;112:275–279. [Google Scholar]
- 41.Braunschweiler L, Ernst RR. Coherence Transfer by Isotropic Mixing: Application to Proton Correlation Spectroscopy. J Magn Reson. 1983;53:521–528. [Google Scholar]
- 42.Kumar A, Ernst RR, Wüthrich K. A Two-Dimensional Nuclear Overhauser Enhancement (2D NOE) Experiment for the Elucidation of Complete Proton-Proton Cross-Relaxation Networks in Biological Macromolecules. Biochem Biophys Res Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Beck R, Huang T, Choi MC, Divinagracia M. Scatterless Hybrid Metal-Single-Crystal Slit for Small-Angle X-Ray Scattering and High-Resolution X-Ray Diffraction. J Appl Crystallogr. 2008;41:1134–1139. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.