Abstract
Expression of the transcription factor brachyury (TBXT) is normally restricted to the embryo and its silencing is epigenetically regulated. TBXT promotes mesenchymal transition in a subset of common carcinomas, and in chordoma, a rare cancer showing notochordal differentiation, TBXT acts as a putative oncogene. We hypothesized that TBXT expression is controlled through epigenetic inhibition to promote chordoma cell death. Screening of five human chordoma cell lines revealed that pharmacological inhibition of the histone 3 lysine 27 demethylases KDM6A (UTX) and KDM6B (JMJD3) leads to cell death. This effect was phenocopied by dual genetic inactivation of KDM6A/B using CRISPR/Cas9. Inhibition of KDM6A/B with a novel compound KDOBA67 led to a genome-wide increase in repressive H3K27me3 marks with concomitant reduction in active H3K27ac, H3K9ac, and H3K4me3 marks. TBXT was a KDM6A/B target gene, and chromatin changes at TBXT following KDOBA67 treatment were associated with a reduction in TBXT protein levels in all models tested, including primary patient-derived cultures. In all models tested, KDOBA67 treatment downregulated expression of a network of transcription factors critical for chordoma survival and upregulated pathways dominated by ATF4-driven stress and pro-apoptotic responses. Blocking the AFT4 stress response did not prevent suppression of TBXT and induction of cell death, but ectopic overexpression of TBXT increased viability, therefore implicating TBXT as a potential therapeutic target of H3K27 demethylase inhibitors in chordoma. Our work highlights how knowledge of normal processes in fetal development can provide insight into tumorigenesis and identify novel therapeutic approaches.
Introduction
Normal embryonic development requires a finely co-ordinated process of temporal and spatial gene expression which is controlled through epigenetic mechanisms (1,2). In addition, aberrant epigenetic mechanisms also have been implicated in the pathogenesis of a diverse range of malignancies (3). During vertebrate development, mesoderm specification is tightly regulated by the prototypical T-box transcription factor T (TBXT, also known as T, or brachyury). TBXT expression is exquisitely orchestrated through caudo-cranial morphogenic gradients, controlled through fibroblast growth factor and activin signalling, and ultimately transforms the nascent dorsal mesoderm into the rudimentary axial skeleton, the notochord (4,5). TBXT is silenced in the human fetus at approximately 12 weeks of development and consequently, the notochord recedes prenatally (6).
In embryonic stem cells, and mesoderm-derived differentiating adipocytes, regulation of TBXT gene expression is achieved through epigenetic regulatory mechanisms. These include the recruitment of histone deacetylase 1 (HDAC1) (7), CpG island methylation (8,9) and the action of histone lysine demethylases, in particular lysine demethylase 6A (KDM6A, also known as ubiquitously transcribed X chromosome tetratricopeptide repeat protein (UTX)) and lysine demethylase 6B (KDM6B, Jumonji domain containing 3 (JMJD3)) (hereafter referred to as KDM6A/B) (10,11) which act on tri-methylated histone H3 lysine 27 (H3K27me3). H3K27me3 marks are deposited by polycomb repressive complex 2 (PRC2), inhibits H3K27 acetylation (12,13) and recruits the canonical form of polycomb repressive complex 1 (PRC1) to maintain developmental regulator genes in a repressed state (14,15).
TBXT expression is seen in chordoma, a rare cancer of the axial skeleton showing notochordal differentiation, in the benign notochordal cell tumour - the postulated precursor of chordoma, and in hemangioblastoma (16,17). Lower levels of TBXT are also detected in a proportion of tumours of epithelial origin (lung, prostate and breast cancer), where TBXT promotes tumour progression and epithelial-mesenchymal transition (18). TBXT is expressed at high levels in chordomas (19) and several lines of evidence demonstrate that it plays a pivotal role in the development of this disease (20); namely germline tandem duplication of TBXT is associated with familial chordoma (21) and similar somatic changes are seen in sporadic tumours (22). Furthermore, TBXT regulates an oncogenic transcriptional network (20), silencing of which causes growth arrest in chordoma cell lines (23,24). However, the key mechanism(s) that regulate the expression of TBXT in chordoma have not been reported and cannot be accounted for solely by somatic copy number gains which are observed only in up to 27% of sporadic cases (22). Hence, we postulated that persistent expression of TBXT in chordoma results from disruption of the physiological epigenetic process(es) that control its expression in development, and that such pathways could be exploited for targeted therapy.
An increasing number of therapeutic approaches targeting epigenetic pathways have shown promising results in cancer, for example in acute myeloid leukemia (25), multiple myeloma (26), brain tumours (27), and neuroblastoma (28). More recently, chordoma has been added to this list with experiments revealing an association of TBXT with an active super-enhancer which can be therapeutically exploited by inhibiting cyclin dependent kinases with roles in transcriptional regulation, such as CDK7/12/13, via downregulation of TBXT expression (29).
Individuals with chordoma have a median of 7 years survival, and they have seen little benefit from genomic investigations seeking tractable therapeutic targets such as protein kinases (22). Herein, we used a focused library of mechanistically defined epigenetic inhibitor tool compounds with the aim of identifying vulnerable signalling pathways that could be exploited for the development of novel treatments for chordoma.
Materials and Methods
Cell lines and primary cultures
Five human chordoma cell lines, U-CH1, U-CH2, U-CH7, MUG-Chor and UM-Chor were cultured as described previously (30). All chordoma cell lines derive from sacral tumours except UM-Chor which was generated from a clival chordoma (www.chordomafoundation.org). U2OS (ATCC® HTB96™, ATCC, VA, USA), an osteosarcoma cell line that lacked expression of TBXT and was used as a control, was cultured according to ATCC guidelines. Cells lines were quality controlled by short-tandem-repeat (STR) analysis (DNA Diagnostic Centre, London, UK) (last report in Table S1) and were tested regularly to ensure that they were mycoplasma-free.
Fresh sacral chordoma samples were obtained with written informed consent from patients being treated at the Royal National Orthopaedic Hospital (RNOH), Stanmore, United Kingdom. Tumour diagnoses were made according to the WHO classification (16). Human investigations were performed after approval by an institutional review board: ethical approval was obtained from the Cambridgeshire 2 Research Ethics Service (reference 09/H0308/165). The studies were conducted in accordance with the Declaration of Helsinki ethical guidelines. Fresh chordoma samples were obtained within 30 minutes of being removed from the patient; the sample were washed in PBS supplemented with antibiotics (Primocin™, 100 μg/ml, Invivogen, Tolouse, France), minced and incubated with Collagenase II (100 mg / ml, 17101-015, Thermo Fisher Scientific) for one hour. The digested tissue samples was then passed through a 70 μM cell strainer, centrifuged and then resuspended in chordoma medium (as described in reference (30)). On reaching confluence, cells were passaged once and then plated at equal density for KDOBA67 treatment. The assay was repeated with 4 independent donors, each with at least 3 biological replicates.
Screening of small molecule inhibitors, compound synthesis and compound treatments
To screen small molecules inhibitors, human chordoma cell lines (U-CH1, U-CH2, U-CH7, MUG-Chor and UM-Chor) were seeded at a density of 5000 cells per well in a 96 well plate overnight. Cells were treated with either a compound or vehicle control (0.1 % DMSO) for 3-6 days (a list of compounds and concentrations used is available Dataset S1). Cell viability was measured using Presto Blue Cell Viability Reagent (Thermo Fisher Scientific, Loughborough, UK). Viability was normalised to vehicle controls within the same plate. Each assay was repeated three times, using 3 replicates and the mean decrease in viability calculated. For IC50 determination, cells were plated as above and treated with halving the dilution of the compounds with the highest concentration being 50 μM. Viability was measured by Presto Blue Cell Viability Reagent (Thermo Fisher Scientific) as above. IC50 was calculated by fitting 7-point dose response data using Prism version 5. Quoted values represent the mean of 3 independent experiments, each with 3 replicates.
In follow-up experiments chordoma cell lines were treated with compounds at the IC80: GSK-J4 (5 μM, Cayman Chemicals, Ann Arbor, USA), KDOBA67 (5 μM) and ISRIB (250 nM, Sigma Aldrich, St. Louis, MO, USA) for 48 hours (or for 72 hours in apoptosis and cell cycle experiments only) unless otherwise specified. Primary chordoma cultures were treated with KDOBA67 (10 μM) for 6 days. DMSO was used at the same concentration as vehicle control (0.1 %).
Detailed methods regarding the synthesis of the KDOBA67 compound, IC50 determination of GSK-J1 analogs, structure determination of UTY in complex with KDOBA67 (protein crystallisation, X-ray data collection and structure determination) are available in SI Materials and Methods and Figure S1A-C, Table S2-S3.
Apoptosis and cell cycle studies
Experiments were performed on an LSR Fortessa™ (Becton Dickinson, USA) running FACSDiva Software version 6 with 104 events recorded for each sample. Cell cycle analysis. Propidium Iodide (PI) staining: Sub-confluent cultures of cells were treated with compounds at the IC50 concentration for between 47 and 72 hours. Cells were harvested, washed once in phosphate buffer saline (PBS, Thermo Fisher Scientific) and counted. 2 x105 cells were fixed in 70% ethanol in PBS on ice for 30 minutes. Fixed cells were centrifuged at 3000 rpm for 5 minutes and washed with PBS. To ensure that only DNA was stained the pellet was treated with RibonucleaseA (100 μg/ml in PBS, Thermo Fisher Scientific) and subsequently stained with PI solution in PBS (50 μg/ml, Sigma-Aldrich) at room temperature in the dark for 30 minutes prior to analysis. Apoptosis Detection: Apoptosis was determined by detecting phosphatidylserine by APC-conjugated Annexin V using the APC Annexin V Apoptosis Detection Kit with PI (Biolegend, CA, USA). Briefly, following compound treatment as above, cells were harvested, washed once in PBS and 2 x105 cells resuspended in 250 μl of binding buffer containing 5 μL Annexin V-APC and 10 μl PI solution. Cells were incubated in the dark for 15 minutes before being analysed. Each assay was repeated 3 times, each with 3 replicates.
Western blotting, qPCR, immunofluorescence
Western blotting and qPCRs were performed as described previously (30). Antibodies and primers are listed in Table S4 and Table S5 respectively. For the analysis of the nascent unprocessed RNA, equal amounts of total RNA were retrotranscribed using the Superscript-IV RT cDNA synthesis Kit (Thermo Scientific) and qPCR was carried out using specific primers designed to amplify the first intronic regions of the transcript of interest. No amplification was observed in control reactions lacking RT.
Immunofluorescence of primary chordoma cells was performed as described previously (31) using antibodies listed in Table S4.
CRISPR/Cas9 knockdown
Target guide sequences validated against KDM6A, KDM6B and TBXT were designed as previously published (cfr Table S5). Guides were cloned in a CRISPR/Cas9 lentiviral backbone containing a Puromycin and a Blasticidin resistance cassette, respectively, as described previously (32). LentiCRISPR v2-Blast (Addgene plasmid # 83480) was a gift from Mohan Babu. LentiCRISPR v2 (Addgene plasmid # 52961) was a gift from Feng Zhang. The empty vectors were used as a non-targeting control. Lentiviral particles were produced as described previously (31). Chordoma cells were transduced with a MOI of 1 and subjected to antibiotic selection (Puromycin, sc-108071, Insight Biotechnology LTD, at 2 μg / ml for UCH1 and UM-Chor, 4 μg / ml for MUG-Chor and Blasticidine, A1113903, Thermo Fisher Scientific at 10 μg / ml for UCH1 and UM-Chor, 20 μg / ml for MUG-Chor) using constructs in a sequential manner. Each knockout experiment was repeated twice for each three chordoma cell lines, with at least four replicates per condition per assay. Genome targeting efficiency was assessed in a cleavage assay using the T7 Endonuclease I according to manufacturer’s instructions (M0302, New England Biolabs, USA) or by Sanger Sequencing of PCR-amplified DNA of targeted cells. Cell proliferation was assessed 72 hours after the transduction of the second CRISPR construct by Presto Blue Viability Assay (Thermo Fisher Scientific) according to manufacturer’s instruction. Each assay was repeated twice using at least five replicates. Viability was normalized to vehicle controls within the same plate.
TBXT overexpression
The sequence of the TBXT transcript (TBXT-201 ENST00000296946.6, Ensembl) corrected for the G177D variant and in frame to an HA-tag was cloned into a pHIV-dTomato lentiviral vector (a gift from Bryan Welm, Addgene plasmid # 21374), as described in Figure S7K. The empty vector was used as control (EV). Lentiviral particles were produced as described previously (31). Chordoma cells were seeded on 96 well collagen-coated plates and transduced with a MOI of 10 with the addition of Polybrene 5μg/ml, which gave a positivity of infection of >95 % as assessed by the expression of dTomato 72 hours post infection. UCH1 cells were treated with 5uM KDOBA67 and cell proliferation assessed as described above. For western blot, cells were seeded in collagen-coated 6 wells plates.
Chromatin Immunoprecipitation (ChIP), ChIP-Rx and library preparation
ChIP was performed as described in SI Materials and Methods. ChIP-Rx was performed as ChIP with the exception that a fixed ratio of Drosophila S2 cells (20 % of chordoma cells) was spiked in prior to fixation to allow for normalization. Libraries were prepared using the NEBNext Ultra 2 DNA Library Preparation Kit (New England Biolabs, MA, USA) and sequenced on a Nextseq500 (Illumina, CA, USA).
RNA-seq library preparation
Cells were lysed in Trizol and total RNA extracted using the Direct-zol kit (Zymo Research, CA, USA) including an on-column DNA digest. Poly(A) RNA was selected using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs) and a first strand library prepared using NEBNext Ultra Directional RNA Library Prep Kit (New England Biolabs) and sequenced on a Nextseq 500 (Illumina).
Next generation sequencing data processing
Computational pipelines were used by calling scripts from the CGAT toolkit to analyze the next generation sequencing data (https://github.com/cgat-developers) (33). RNA-seq. Briefly, FASTQ files were assessed for quality using FASTQC, aligned to GRCh37 and a concatenated genome consisting of GRCh37 using the default options of HISAT2 (34). Differential gene expression analysis was performed using DESeq2 using cell type and treatment as factors in the model. Genes were considered to be differentially regulated based on log2 fold change and p-value < 0.05. A list of genes differentially expressed in response to KDOBA67 (average for MUG-Chor and UM-Chor) is provided in Dataset S2. ChIP-Rx. ChIP-Rx data were aligned to a concatenated genome sequence of human (GRCh37-hg19) and spike-in of drosophila (BDGP5.25) using bowtie2 (35) with --very-sensitive option. Only uniquely mapped reads and properly-paired reads (for paired-end data) were used for the downstream analysis. PCR duplicates were removed using samtools (http://www.htslib.org/). The peak calling was performed using SICER (36) (FDR = 0.05) and filtered for blacklisted regions in the human genome. Peaks present in all the replicates in each condition (control and treatment) of a histone mark were used for further analysis. A peak set was created from the merged peaks between the two conditions and used for differential binding analysis for each histone mark. Read counts were obtained for individual replicates for the peak set using DiffBind and differential binding analysis was performed using DESeq2 in R. Spike-in calibration for differential binding analysis was performed utilizing methods described in (37). The metaplots and heatmaps were generated with computeMatrix and plotProfile/plotHeatmap functions from deeptools (https://deeptools.readthedocs.io/en/develop/). Genome coverage tracks were obtained using MACS2 pileup function (38) and visualized with Gviz and biomaRt in R. Differentially bound H3K27me3 peaks were overlapped with ±1Kb of the TSS; genes with a log2fold change >1.25 were used for enrichment analysis using gprofiler2 in R.
DNA Methylation Analysis
DNA was extracted from either formalin-fixed paraffin-embedded (FFPE) or fresh frozen tumours using QIAamp DNA Kits (Qiagen) according to the manufacturer’s instructions. Between 0.5ug and 1ug DNA was used for the Infinium Methylation EPIC array or 450K Infinium Human Methylation array (Illumina). Data were assembled using BeadStudio (Illumina) and processed using Minfi1 employing the functional normalisation protocol. Beta values were downloaded from the Cancer Genome Atlas for cancers other than chordoma.
Statistics
Statistical parameters including the exact value of n, precision measures (mean ± SD) and statistical significance are reported in the Figures. Data were judged to be statistically significant when p < 0.05 by two-tailed Student’s t test performed in GraphPad PRISM 5.0; asterisks denote statistical significance as calculated by Student’s t test (*p ≤0.05, **p ≤0.01, ***p ≤0.001, ****p ≤0.0001).
Definition of experimental replicates
‘Independent experiment’ refers to an experiment that has been performed multiple times (e.g. the same experiment was performed on different days, using different primary patient samples). ‘Replicates’ refers to biological replicates and represent multiple tests within the same experiment (i.e. multiple wells exposed to the drug). ‘Technical repeats’ of the same replicate (e.g. multiple qPCR reads of the same replicate) have been averaged and are not used as independently values in the statistical analysis.
Results
Inhibition of H3K27 demethylases reduces chordoma cell viability
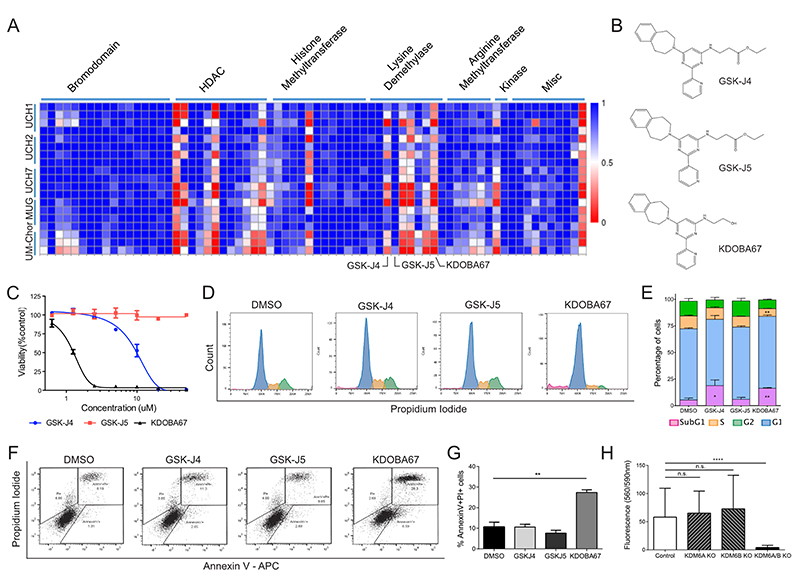
We screened a library of tool compounds targeting proteins involved in chromatin biology including epigenetic readers, writers and erasers, against five chordoma cell lines (www.chordomafoundation.com) (Dataset S1) (39). A resazurin-based viability assay was used as the primary anti-proliferative readout (Figure 1A). Decreased cell viability was observed in all cell lines in response to HDAC inhibitors, as previously reported (40). Broad-spectrum inhibitors of the Jumonji-type of lysine demethylases were found to reduce cell viability, as was the more specific inhibitor of KDM6A/B GSK-J4 (41) (Figure 1A-B). Decreased viability was also observed with KDOBA67, a novel cell-permeable hydroxyl derivative of GSK-J4, which unlike its parental compound, does not require activation by intracellular esterases (Figure 1A-B and Figure S1A-C). KDOBA67 displayed in vitro inhibitory activity in the low micromolar range with IC50 values of 2-5 μM in various chordoma cell lines (Figure 1C) leading to induction of apoptosis (Figure 1D-G and Figure S2A-F). As expected, the regio-isomer control compound GSK-J5 did not display anti-proliferative effects (Figure 1A-G and Figure S2A-F).
Figure 1. Focused epigenetic library screening identifies H3K27 lysine demethylases as potential therapeutic target in chordoma.
(A) Screening of 90 small molecule probes with validated activity against enzymes involved in chromatin biology on 5 chordoma cell lines. Each row represents a separate replicate and each column an individual compound. Columns have been grouped together by inhibitor class (c.f. also to Dataset S1). Values plotted as fractional viability compared to vehicle (DMSO) control. (B) Molecular structures of GSK-J4, GSK-J5 and KDOBA67. (C) Dose response curves for GSK-J4, GSK-J5 and KDOBA67 in UM-Chor. (D-G) Cell cycle changes (D-E) and cell death analysis assessed by AnnexinV-PI staining (F-G) in response to the compounds after 72 hours of treatment. Representative histogram/dot plots and quantification of 3 independent experiments, with 3 replicates per condition (c.f. also Figure S2). (H) Viability measured by Alamar blue of UCH1 following CRISPR/Cas9 editing of KDM6A/B or both. Mean of 2 independent experiments, with at least 6 replicates per condition per experiment. *p ≤0.05, **p ≤0.01, ***p ≤0.001, ****p ≤0.0001.
To confirm that the inhibitory growth effects with the tool compounds were due to on-target inhibition of KDM6A/B, we used CRIPSR/Cas9 to introduce nonsense mutations in both genes (Figure S3A-B). Targeting KDM6A/B independently in UCH1 and UM-Chor cells, in which both genes are intact, was insufficient to reduce cell viability, whereas editing both genes simultaneously resulted in reduction in cell viability and proliferation (Figure 1H and Figure S3C-E). The on-target effect was further supported by the reduction in viability following inactivation of KDM6A only in the MUG-Chor cell line, which contains a biallelic deletion of KDM6B (Figure S3F-H).
Inhibition of H3K27 demethylases targets critical pathways for chordoma cell survival
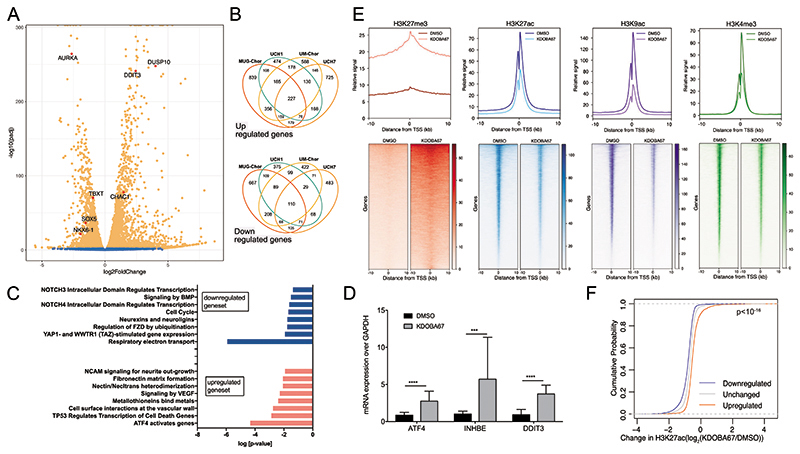
The transcriptional profile of the chordoma cell lines UCH1, UCH7, MUG-Chor and UM-Chor (Dataset S2 and Figure S4A-E) treated with inhibitors of KDM6A/B revealed significant changes in gene expression (Figure 2A-B). Alteration in the expression of 371 genes was identified across all chordoma cell lines and the affected pathways were linked to chordoma biology (20), and relate to cell movement, adhesion, extracellular matrix organisation, cell contact and signalling (Figure 2C). This analysis also revealed enrichment in categories such as apoptosis, mitochondrial impairment, cell cycle arrest and mitosis with identification of key genes such as aurora kinase A (AURK), dual-specific phosphatase 10 (DUSP10), activating transcription factor 4 (ATF4), DNA damage inducible transcript 3 (DDIT3) and the ATF4/DDIT3-target gene ChaC glutathione specific gamma-glutamylcyclotransferase 1 (CHAC1) (Figure 2A-C and Figure S4B-E), consistent with the anti-proliferative results of H3K27 demethylase inhibitors (Figure 1). Treatment of primary chordoma cultures with KDOBA67 revealed a similar transcriptional response to that seen in chordoma cell lines, including increases in ATF4/DDIT3-target genes (Figure 2D).
Figure 2. Epigenomic and transcriptomic profiling in response to H3K27 lysine demethylase inhibitors in chordoma cell lines.
(A) Volcano plot summarising differential expression in chordoma cells (UM-Chor) following treatment with KDOBA67 for 48 hours. Blue, not significant; Orange, adjusted P-value (Padj) <0.05. Average of 3 biological replicates. Results for UCH1, MUG-Chor and UCH7 in Figure S4 and Dataset S2. TBXT expression is reduced across chordoma cell lines (log2-fold changes -2.1 (MUG-Chor), -1.2 (UCH1), -0.96 (UM-Chor), -1.5 (UCH7) (Padj < 0.01)). (B) Venn diagrams showing the numbers of upregulated (top) and downregulated (bottom) genes in 4 chordoma cell lines; an average of 3 biological replicates per cell line. (C) Reactome analysis for UCH1, UCH7, UM-Chor and MUG-Chor in response to KDOBA67. (D) Upregulation of ATF4 and ATF4-target genes (inhibin subunit beta E, INHBE and DDIT3) in patient-derived primary chordoma cultures as assessed by qPCR (4 independent primary samples, at least 3 biological replicates per condition). (E) Metagene plots (top) and heatmaps (bottom) of ChIP-Rx peaks of H3K27me3, H3K27ac, H3K9ac and H3K4me3 in UCH1 cells treated with KDOBA67 or DMSO treatment for 48 hours. The signal represents the average of 2 or 3 biological replicates per condition. Principal component analysis (PCA) plots of single replicates is shown in Figure S4F, H-J. (F) Cumulative distribution frequency of the change in H3K27ac occupancy in UCH1 cells in response to KDOBA67 versus DMSO control as assessed by ChIP-Rx, at the promoter of genes which are upregulated, downregulated or unchanged in response to KDOBA67 as assessed by RNA-sequencing. The three distributions are statistically different one from another by Kolmogorov Smirnov test (p<10-16). *p ≤0.05, **p ≤0.01, ***p ≤0.001, ****p ≤0.0001.
H3K27 demethylase inhibition leads to genome-wide alteration of histone marks
We next investigated the mechanism by which KDOBA67 alters cellular gene expression. Using ChIP-sequencing, normalised with an exogenous reference genome (ChIP-Rx), we found that treatment of UCH1 cells with KDOBA67 increased H3K27me3 at transcription start sites (TSS) and gene bodies across the genome after 48 hours (Figure 2E and Figure S4F). Pathway analysis revealed that differentially bound H3K27me3 peaks were at genes involved in chordoma cell biology, related to extracellular matrix organisation, cell contact and signalling (Figure S4G and Dataset S3). ChIP-Rx also revealed a corresponding global decrease in H3K27ac at TSS (Figure 2E and Figure S4H). To explore the wider effect of the increase in H3K27me3 on chromatin structure, we measured changes in the activating histone marks H3K9ac and H3K4me3 following KDOBA67 treatment and found a global decrease at TSS, similar to that seen for H3K27ac (Figure 2E and Figure S4I-J). We then asked how the global loss of active histone marks in response to KDOBA67 treatment relates to the changes in gene expression. Plotting H3K27ac at the TSS of differentially expressed genes revealed that downregulated genes exhibited the greatest loss of H3K27ac (Figure 2F). Thus, we conclude that inhibition of H3K27 demethylation leads to depletion of H3K27ac and inhibition of gene expression.
We next examined the effect on H3K4me3. As reported previously for GSK-J4 (42) we found that KDOBA67 exerts some inhibitory activity in non-cell-based assays against recombinantly expressed and purified KDM5 (JARID1) enzymes, the principal H3K4me3 demethylases (Table S2). The decrease in H3K4me3 levels observed following treatment with KDOBA67 (Figure 2E) is in stark contrast to the previously reported global increase in H3K4me3 following treatment with specific H3K4me3 demethylase inhibitors (43,44) and indicates that the phenotype observed in our experiments is not an off-target effect against KDM5 enzymes.
H3K27 demethylase inhibition alters chromatin state at TBXT and inhibits its expression
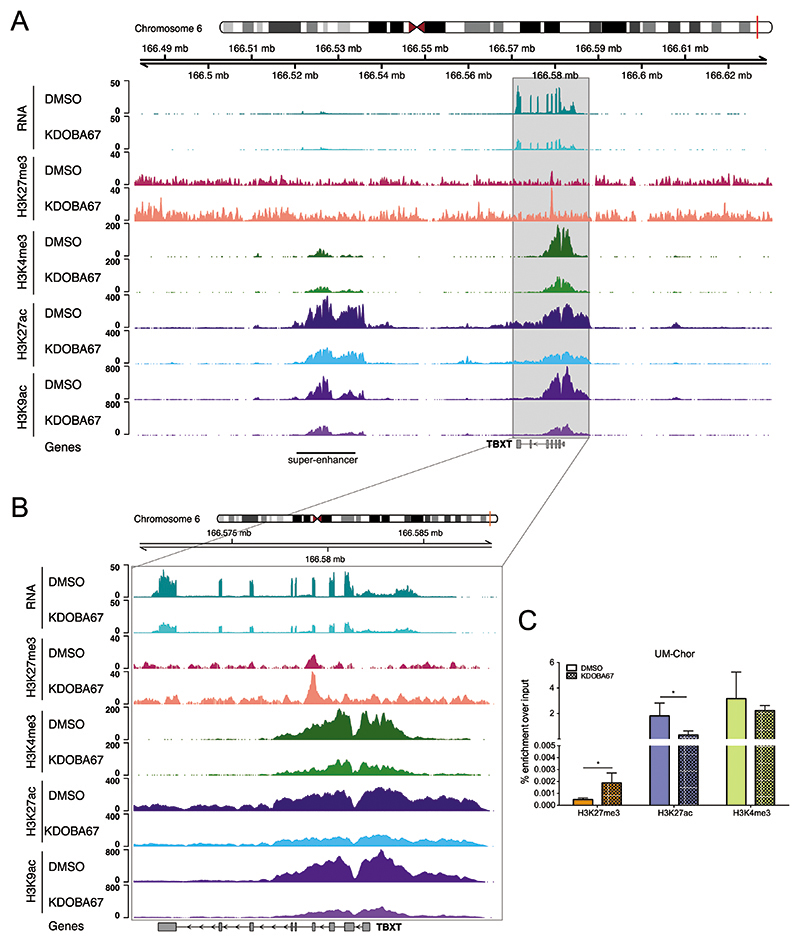
Given the central role of TBXT in the pathogenesis of chordoma (20,23), in addition to the evidence that silencing H3K27 demethylases in development represses TBXT expression (10,11), we next asked whether inhibition of KDM6A/B suppressed TBXT in chordoma cell lines. Treatment with KDOBA67 led to an increase in H3K27me3 and a decrease in H3K27ac, H3K9ac and H3K4me3 at the TBXT gene locus in the UCH1 cell line (Figure 3A-B), which was associated with significantly reduced expression of TBXT as measured by RNA-seq (Figure 3A-B, top pair of tracks and Dataset S2). These chromatin alterations were confirmed in UM-Chor cells using ChIP-qPCR (Figure 3C and Figure S4K-M). H3K27ac was also significantly reduced at the recently identified TBXT-associated super-enhancer and active regulatory region (29) (Figure 3A and Figure S5A-B).
Figure 3. Inhibition of H3K27 lysine demethylase leads to chromatin changes around TBXT in chordoma.
(A) RNA expression (top pair of tracks), H3K27me3, H3K4me3, H3K27ac and H3K9ac modifications at the TBXT locus in UCH1 cells treated with KDOBA67 (light colours) or DMSO (dark colours) for 48 hours. An increase in the level of H3K27me3 over the gene locus is observed with a concurrent decrease in the levels of H3K27ac, H3K9ac and H3K4me3 at the TBXT promoter and super-enhancer (labelled). Tracks show the average of replicates. (B) As (A) but with an expanded view of the TBXT gene. (C) H3K27me3, H3K27ac and H3K4me3 enrichment at the TBXT promoter (-2Kb from the TSS) in UM-Chor cells treated with KDOBA67 or DMSO for 48hours, assessed by ChIP-qPCR. Results from 4 biological replicates per condition. ChIP-qPCR data for positive control sites are shown in Figure S4K-M.
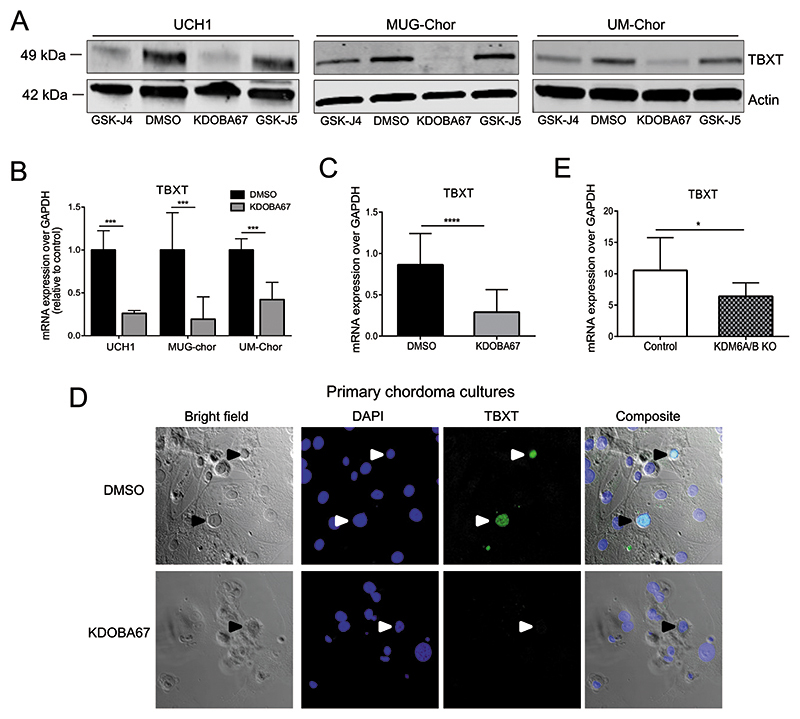
Western blot and qRT-PCR analyses confirmed that treatment with either GSK-J4 or KDOBA67, but not with the control compound GSK-J5, also reduced TBXT expression in two other chordoma cell lines (Figure 4A-B), including MUG-Chor cells that harbour multiple copies of TBXT (www.chordomafoundation.org). The relevance of KDM6A/B inhibition for disease was further supported by the reduction in cell proliferation (Figure 4C) and TBXT expression (Figure 4D) following in vitro treatment of patient-derived primary chordoma samples with KDOBA67. The reduction of TBXT expression was reproduced by CRISPR/Cas9-mediated knock-out of KDM6A/B in both UCH1 and UM-Chor cell lines, demonstrating this to be a direct effect of the compound on these enzymes (Figure 4E and Figure S5C).
Figure 4. Inhibition of H3K27 lysine demethylase leads to inactivation of TBXT in chordoma.
(A) Protein expression of TBXT in chordoma cell lines, following treatment with KDOBA67 for 48 hours as assessed by western blotting. Beta actin is used as an endogenous control. Representative blots of 2 independent experiments. (B) TBXT expression in UCH1, MUG-Chor and UM-Chor following KDOBA67 treatment for 48 hours as assessed by qRT-PCR, in 3 independent experiments with 3 biological replicates per condition per cell line. (C) TBXT transcript level is reduced in primary chordoma cultures treated with KDOBA67 for six days, as assessed by qPCR. N=4 primary samples, at least three replicates per condition per sample. (D) Cell death and reduction in expression of TBXT (harrow heads) as shown by immunofluorescence in KDOBA67- and DMSO-treated patient-derived chordoma cultures for 6 days. TBXT-positive chordoma cells are interspersed between tumour-derived stromal cells. TBXT/green, DAPI/blue. 40X magnification. Representative pictures from one sample, experiment performed on 2 samples. (E) Expression of TBXT is reduced in UCH1 following double KO of KDM6A/B, assessed by qPCR. Quantification of two independent experiments, with two biological replicates per condition per experiment. *p ≤0.05, **p ≤0.01, ***p ≤0.001, ****p ≤0.0001.
Histone modifications directly control TBXT expression in chordoma
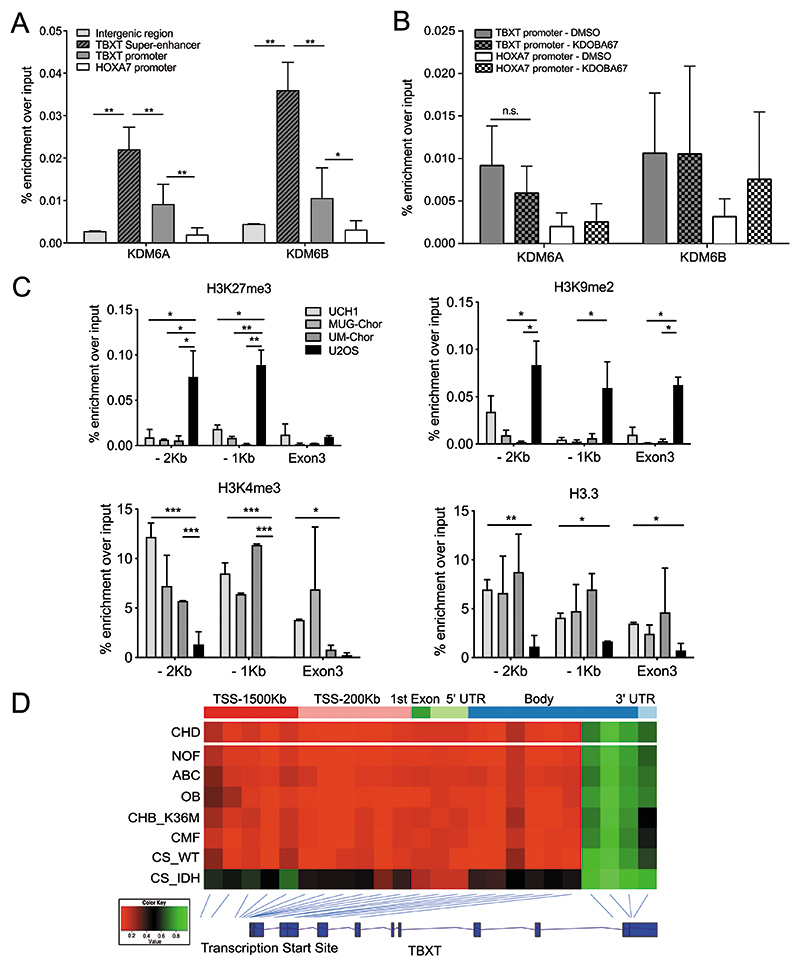
Next, we tested if KDM6A/B directly target the TBXT locus. ChIP-qPCR revealed binding of both KDM6A and KDM6B at the TBXT super-enhancer and promoter regions in the UCH1 cell line (Figure 5A). In contrast, lower levels of KDM6A/B were found at the promoter of homeobox A7 (HOXA7), a known polycomb target (Figure 5A). The binding of KDM6A/B was not altered at either the TBXT or HOXA7 promoter regions in response to KDOBA67 treatment implying that this drug specifically acts to inhibit the catalytic activity of demethylases rather than altering their recruitment to chromatin (Figure 5B).
Figure 5. Histone modifiers epigenetically control TBXT expression in chordoma.
(A) ChIP-qPCR of KDM6A and KDM6B occupancy in UCH1 cells at the TBXT super-enhancer (indicated in Figure 3a), TBXT and HOXA7 promoter (-1Kb from TSS) and a control intergenic region (300Kb downstream of TBXT). Four biological replicates per condition. (B) ChIP-qPCR of KDM6A and KDM6B at the TBXT or HOXA7 promoter (-1Kb from TSS) in UCH1 cells treated with KDOBA67 or DMSO for 48 hours. Two biological replicates per condition per experiment, two independent experiments. (C) ChIP-PCR of H3K27me3 and H3K9me3 (top), H3K4me3 and H3.3 (bottom) in three chordoma cell lines (UCH1, MUG-Chor, UM-Chor) and the osteosarcoma cell line U2OS. Two independent experiments, with three biological replicates per condition per experiment. (D) The DNA at the promoter region of TBXT is hypomethylated in all primary human chordomas (n=35) and in other mesenchymal tumours not associated with the expression of TBXT. From top: chordoma, non-ossifying fibroma (n=12), aneurysmal bone cysts (n=9), osteoblastoma (n=12), chondroblastoma harbouring H3.3-K36M mutation (n=17), chondromyxoid fibroma (n=25), chondrosarcoma WT (n=2), chondrosarcoma-harbouring an IDH mutant (n=3). The average beta value for each probe is plotted with the position of the probe shown relative to the TBXT gene body. *p ≤0.05, **p ≤0.01, ***p ≤0.001, ****p ≤0.0001.
We next asked whether variations in H3K27me3 and other histone marks could explain the difference in expression of TBXT in chordoma cells and other mesoderm-derived cancer cells in which physiological silencing of TBXT occurs during development. ChIP-qPCR revealed that the repressive marks H3K27me3 and H3K9me2 were enriched at the TBXT promoter region in the osteosarcoma U2OS cell line but not in the chordoma lines tested (Figure 5C), consistent with the expression of TBXT in chordoma but not in osteosarcoma. Reciprocally, enrichment of H3K4me3 and H3.3, both associated with active gene transcription, was restricted to chordoma cells (Figure 5C).
We next investigated whether DNA methylation contributes to the regulation of TBXT expression in chordomas in view of the evidence that it cooperates with histone modifications to regulate TBXT expression in development (8). We found low levels of DNA methylation in the promoter region and at the TSS of TBXT in primary chordoma tumours and also in bone tumours that do not express TBXT (Figure 5D and Figure S6). Taken together, these data support the concept that TBXT expression is regulated by histone modification and not by DNA promoter methylation.
H3K27 demethylases inhibition targets TBXT and induces chordoma cell death
In addition to suppression of TBXT in response to treatment with KDOBA67, we noted activation of an ATF4 stress-induced pathway (45) (Figure 2). Below we provide four lines of evidence showing that the antiproliferative effect mediated by KDM6 inhibitors is largely related to the reduction in TBXT expression rather than activation of the ATF4 pathway.
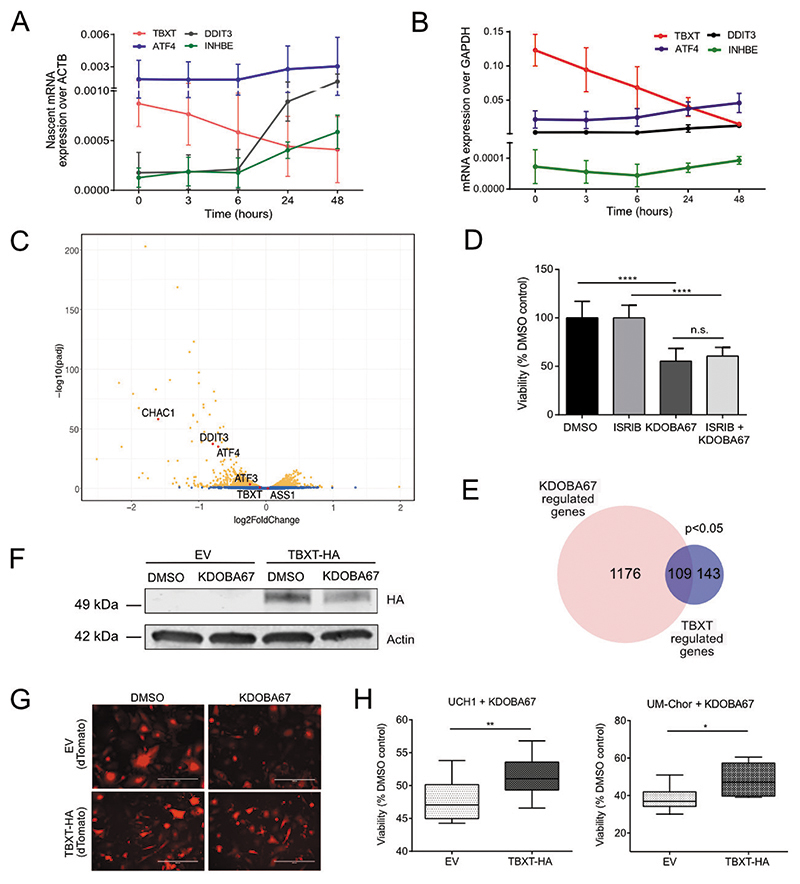
First, having established that TBXT is a KDM6A/B target gene (Figure 5A-B), we asked whether the reduction in TBXT gene expression was a direct consequence of KDM6A/B inhibition or an event secondary to induced metabolic stress. Measuring the effect of KDOBA67 on nascent gene transcription we found that the TBXT pre-mRNA was reduced as early as 3 hours after exposure to KDOBA67 (Figure 6A). Furthermore, time course experiments in three chordoma cell lines demonstrated that TBXT expression decreased prior to upregulation of genes associated with the ATF4 response (Figure 6B and Figure S7A-B).
Figure 6. Epigenetic regulation of TBXT occurs independently of an ATF4-stress response and is key to the lethality induced by H3K27 demethylase inhibitors.
(A-B) ATF4, DDIT3, INHBE and TBXT regulation over a time course in response to KDOBA67 in UCH1, by qPCR of nascent RNA (A) and total RNA (B). Two independent experiments, three replicates per condition each experiment. Results for total RNA for UM-Chor and MUG-Chor are reported in Figure S7. (C) Volcano plot summarising differential gene expression by RNA-seq in UCH1 cells treated with KDOBA67 for 48 hours in the presence or absence of ISRIB. Blue, not significant; Orange, Padj <0.05. Three biological replicates. (D) Proliferation of UCH1 cells in response to treatment with KDOBA67 for 48 hours in the presence or absence of ISRIB. Three independent experiments, three replicates per condition per experiment. (E) Venn diagram showing the number of genes regulated by KDOBA67 treatment in UCH1 cells (by RNA-seq, Figure 2) that overlap with genes previously identified as direct targets of TBXT in UCH1 cells using ChIP-seq in (20). (F) Overexpression of TBXT-HA in UCH1 cells is not affected by KDOBA67 treatment: similar levels of HA by western blot in cells treated with KDOBA67 or DMSO. (G) Overexpression of TBXT-HA or control EV in UCH1 cells is achieved in >95% of cells: expression of dTomato by fluorescence microscopy of EV and TBXT-HA cells treated with KDOBA67 or DMSO (representative images, scale bar 400μm). (H) Overexpression of TBXT-HA mitigates the reduction in cell viability in response to treatment with KDOBA67 for four days in UCH1 and UM-Chor. 4 replicates per condition in three (UCH1) and two (UM-Chor) independent experiments. *p ≤0.05, **p ≤0.01, ***p ≤0.001, ****p ≤0.0001.
Second, we treated chordoma cells with ISRIB, a specific compound that inhibits the induction of the ATF4 response by interfering with eIF2α phosphorylation (46). ISRIB treatment significantly reduced the expression of ATF4 and ATF4-target genes (INHBE and DDIT3) but did not prevent KDOBA67-induced downregulation of TBXT (Figure 6C and Figure S7C-E) nor the reduction in cell proliferation (Figure 6D and Figure S7F-G).
Third, analysis of the transcriptional profile of UCH1 in response to KDOBA67 treatment revealed that, together with TBXT, genes encoding transcription factors directly targeted by TBXT, such as the SRY-Box transcription factor 5 (SOX5) (20,47), or predicted to be targeted by TBXT, such as PBX homeobox 1 (PBX1) and NK homeobox 6-1 (NKX6-1), were downregulated (Figure 2A and Figure S4B-E). Furthermore, experimentally-validated TBXT target genes (20) were significantly enriched among those genes altered in response to KDOBA67 (Figure 6E), indicating that KDM6A/B inhibition results in the repression of a TBXT-controlled transcriptional network. Indeed, the antiproliferative effect of KDM6 inhibition is also phenocopied by CRISPR/Cas9-mediated knock-out of TBXT (Figure S7H-J).
Finally, to determine whether repression of TBXT expression was necessary for KDOBA67-induced cell death, we overexpressed haemagglutinin-tagged TBXT (TBXT-HA) in the human chordoma cell line UCH1 under a strong promoter which, unlike the endogenous regulatory elements, was not sensitive to KDM6A/B inhibition (Figure 6F-G and Figure S7K-L). We found that ectopic expression of TBXT-HA partially rescued KDOBA67-induced cell death (Figure 6G-H).
Discussion
The number of small molecule inhibitors that have been found to induce significant anti-proliferative effects in chordoma cells is limited (29,30,48). Here, we show that H3K27 demethylase inhibitors suppress cell viability and TBXT expression in multiple human chordoma cell lines as well as patient-derived primary chordoma samples. This is in striking contrast to previous studies which reported that EGFR inhibitors decrease chordoma cell viability to a variable degree across only some chordoma cell lines and reduction of TBXT expression was only seen in response to a single compound, afatinib (30,48,49).
Our finding that KDM6A/B inhibition increases H3K27me3 at the TBXT locus demonstrates that it is feasible to block the function of TBXT in chordoma cells by manipulating its chromatin environment. Such a strategy has been demonstrated successfully for oncogenes in other malignancies, for example repression of c-Myc with bromodomain and extra terminal domain (BET) protein inhibitors such as JQ1 in multiple myeloma (26), thereby circumventing the challenge of directly inhibiting transcription factors, which are often considered “undruggable” (50). Moreover, our previous work on human immune cell types such as macrophages, NK or T-cells (28,39,51) demonstrates reversible anti-proliferative and anti-inflammatory effects upon GSK-J4 inhibition without signs of cell death, suggesting selective pro-apoptotic effects on cancer cells. This supports the concept that H3K27 demethylase inhibitors could potentially be used in the treatment of more common cancers which show a dependency on TBXT expression (18). However, KDM6 inhibitor tool compounds such as GSK-J4 and KDOBA67 have been developed as ex vivo tool compounds (41) and require significant chemical optimisation to reach adequate pharmacokinetic and metabolic properties to justify in vivo studies. Although GSK-J1 is a potent in vitro tool compound, its carboxylate structure significantly impairs cellular uptake. The ethyl ester derivative GSK-J4 requires intracellular esterases for release of GSK-J1: this allows cellular uptake in most cell culture experiments but its in vivo use is significantly hampered by the activity of unspecific serum hydrolases. This renders the compound metabolically unstable, requiring unsuitably high dosing in order to reach sufficient in vivo exposure levels.
H3K27me3 is deposited at promoter regions by PRC2 and allows recruitment of canonical CBX-containing forms of PRC1, and further recruitment of PRC2, thus reinforcing polycomb activity at target sites (14,15). Ectopic expression of KDM6B has previously been shown to lead to a marked decrease of H3K27me3 and delocalization of Polycomb proteins in vivo (52), consistent with our observations that KDM6A/B inhibition causes a global increase in H3K27me3. The increase in H3K27me3 across the genome, even at genes at which H3K27me3 is not normally detected, is consistent with the chromatin sampling model for PRC2 recruitment (53). The identified concomitant global decrease in H3K27ac is consistent with the known antagonistic relationship between H3K27 methylation and acetylation (12,13). Previous work suggests that the Mixed Lineage Leukemia/ Complex of Protein-Associated with Set1 (MLL/COMPASS)-like complex 4 (MLL4), which contains KDM6A, is involved in enhancer H3K4 monomethylation and H3K27 acetylation through the recruitment of the acetyltransferase p300 in murine ESCs (54). MLL4 also establishes H3K4me3 and activates super-enhancers at tumour suppressor genes (55) and at genes that drive cellular differentiation (56). Although the role of KDM6A in enhancer activation might not solely rely on its enzymatic function, its presence in MLL4 may explain the global reduction in H3K4me3 and H3K27ac levels in chordoma cells following loss of KDM6A function, a phenomenon reported also in pancreatic cancer (57).
Our study provides new insights into the function of H3K27 demethylases in tumour biology. KDM6 enzymes can act in a cancer type-specific manner as oncogenes or tumour suppressors, but only mutations in KDM6A have thus far been identified. KDM6 enzymes are ubiquitously expressed, suggesting ‘housekeeping’ functions in maintaining H3K27 methylation levels. In addition KDM6B is highly and transiently induced by various stimuli (such as cytokines or growth factors) to modulate the temporal expression of key genes in lineage development and inflammation (58). We show here that inhibition/knockdown of both KDM6 enzymes is necessary to obtain an anti-proliferative effect, suggesting a cooperative role of KDM6 enzymes in the installation of global methylation patterns.
This research reveals that treatment with H3K27 demethylases inhibitors alters a network of genes similar to those altered by knocking-down TBXT in chordoma cells, including genes involved in the cell cycle, production of extracellular matrix, growth factor and cytokine secretion, which are necessary for the survival of chordoma cells (20) and pertinent to notochord formation (20,30). Moreover, in keeping with our previous studies and that of others (28,39,51), we describe the activation of an ATF4- and DDIT3-driven stress response, thereby identifying this pathway as an additional possible therapeutic option for chordoma (45). However, our data suggest that the effect of H3K27me3 demethylase inhibitors on chordoma cell growth is primarily mediated by suppression of TBXT expression rather than activation of the ATF4 metabolic stress response. Our finding that the effect of KDOBA67 treatment on cell death was not fully reversed by TBXT over-expression is likely to be explained by the substantial number of genes, which, in addition to TBXT, are also repressed in response to KDOBA67. These genes include both direct and/or indirect targets of TBXT.
In conclusion, our study shows that inactivation of H3K27 demethylases alters the expression of genes critical to the survival of chordoma cancer cells and opens the door to further studies investigating how KDM6A/B can be exploited as potential therapeutic targets.
Supplementary Material
Statement of significance.
Pharmacological inhibition of H3K27-demethylases in human chordoma cells promotes epigenetic silencing of oncogenic TBXT, alters gene networks critical to survival, and represents a potential novel therapy.
Acknowledgements
Funding for this project was received from the Chordoma Foundation (AMF, UO), Chordoma UK (AMF), Sarcoma UK (AMF, UO), Bone Cancer Research Trust (AC, LC, AMF, UO) Rosetrees Trust, CRUK (A23900, UO), the LEAN program grant of the Leducq Foundation (UO), the Oxford NIHR Biomedical Research Centre, Skeletal Cancer Action Trust (AMF), a RNOH NHS R&D grant (AMF), and the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007-2013) (UO) under REA grant agreement n° [609305]. AMF is a National Institute for Health Research (NIHR) senior investigator. NP is a CRUK clinician scientist (Grant no:18387). LC, RJ, AMF and NP are supported by NIHR, UCLH Biomedical Research Centre and the Cancer Research Experimental Cancer Centre. GK is supported by the CRUK UCL Centre and an MRC grant to RGJ (MR/R001413/1).
We thank Carina Gileadi and Kannan Velupillai for performing crystallisation experiments with KDOBA67 and UTY. We thank Dr Ivana Bjedov and Dr Victoria Martinez Miguel, UCL Cancer Institute, for providing S2 Drosophila cells and Donna Magsumbol for technical support. We thank Professor Paolo Salomoni, Dr Andy Feber, Dr Teresa Sposito, Dr Stanimir Dulev, Dr Christopher Steele, Dr Manuel Beltran Nebot and Dr Javier Herrero for their valuable discussion and input, and UCL Cancer Institute Core Facilities, the Biobank Team at the RNOH, and the healthcare workers who care for the patients. We thank the patients for the generous donation of their material without which this research would not have been feasible.
Footnotes
Conflict of interest statement
The authors declare no potential conflicts of interest.
Data availability
Data are deposited in the National Center for Biotechnology Information GEO database: RNA-seq and ChIP-Rx sequencing experiments under accession number GSE120214, DNA methylation array data under accession number GSE119462.
Coordinates and structure factors for the UTY:KDOBA67 ligand complex are deposited with RCSB under PDB ID 5A1L.
References
- 1.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mallo M, Alonso CR. The regulation of Hox gene expression during animal development. Development. 2013;140:3951–63. doi: 10.1242/dev.068346. [DOI] [PubMed] [Google Scholar]
- 3.Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357 doi: 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerdan C, McIntyre BAS, Mechael R, Levadoux-Martin M, Yang J, Lee JB, et al. Activin A Promotes Hematopoietic Fated Mesoderm Development Through Upregulation of Brachyury in Human Embryonic Stem Cells. Stem Cells Dev. 2012;21:2866–77. doi: 10.1089/scd.2012.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–9. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- 6.Risbud MV, Shapiro IM. Notochordal cells in the adult intervertebral disc: New perspective on an old question. Crit Rev Eukaryot Gene Expr. 2011;21:29–41. doi: 10.1615/critreveukargeneexpr.v21.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyu J, Jho E, Lu W. Smek promotes histone deacetylation to suppress transcription of Wnt target gene brachyury in pluripotent embryonic stem cells. Cell Res. 2011;21:911–21. doi: 10.1038/cr.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dansranjavin T, Krehl S, Mueller T, Mueller LP, Schmoll HJ, Dammann RH. The role of promoter CpG methylation in the epigenetic control of stem cell related genes during differentiation. Cell Cycle. 2009;8:916–24. doi: 10.4161/cc.8.6.7934. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt CS, Bultmann S, Meilinger D, Zacher B, Tresch A, Maier KC, et al. Global DNA hypomethylation prevents consolidation of differentiation programs and allows reversion to the embryonic stem cell state. PLoS One. 2012;7:e52629. doi: 10.1371/journal.pone.0052629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naruse C, Shibata S, Tamura M, Kawaguchi T, Abe K, Sugihara K, et al. New insights into the role of Jmjd3 and Utx in axial skeletal formation in mice. FASEB J. 2017;31:2252–66. doi: 10.1096/fj.201600642R. [DOI] [PubMed] [Google Scholar]
- 11.Morales Torres C, Laugesen A, Helin K. Utx Is Required for Proper Induction of Ectoderm and Mesoderm during Differentiation of Embryonic Stem Cells. PLoS One. 2013;8:1–15. doi: 10.1371/journal.pone.0060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38:4958–69. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavarone E, Barbieri CM, Pasini D. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat Commun Springer US; 2019;10:1–16. doi: 10.1038/s41467-019-09624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16:803–10. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 16.Editorial Board. Notochordal tumours. IARC; Lyon, France: 2020. WHO classification of soft tissue and bone tumours; pp. 449–457. [Google Scholar]
- 17.Vujovic S, Henderson S, Presneau N, Odell E, Jacques TS, Tirabosco R, et al. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol. 2006;209:157–65. doi: 10.1002/path.1969. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton DH, David JM, Dominguez C, Palena C. Development of Cancer Vaccines Targeting Brachyury, a Transcription Factor Associated with Tumor Epithelial-Mesenchymal Transition. Cells Tissues Organs. 2017 doi: 10.1159/000446495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miettinen M, Wang Z, Lasota J, Heery C, Schlom J, Palena C. Nuclear Brachyury Expression Is Consistent in Chordoma, Common in Germ Cell Tumors and Small Cell Carcinomas, and Rare in Other Carcinomas and Sarcomas: An Immunohistochemical Study of 5229 Cases. Am J Surg Pathol. 2015;39:1305–12. doi: 10.1097/PAS.0000000000000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson AC, Pillay N, Henderson S, Presneau N, Tirabosco R, Halai D, et al. An integrated functional genomics approach identifies the regulatory network directed by brachyury (T) in chordoma. J Pathol. 2012;228:274–85. doi: 10.1002/path.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang XR, Ng D, Alcorta DA, Liebsch NJ, Sheridan E, Li S, et al. T (brachyury) gene duplication confers major susceptibility to familial chordoma. Nat Genet. 2009;41:1176–8. doi: 10.1038/ng.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarpey PS, Behjati S, Young MD, Martincorena I, Alexandrov LB, Farndon SJ, et al. The driver landscape of sporadic chordoma. Nat Commun. 2017;8:890. doi: 10.1038/s41467-017-01026-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Presneau N, Shalaby A, Ye H, Pillay N, Halai D, Idowu B, et al. Role of the transcription factor T (brachyury) in the pathogenesis of sporadic chordoma: a genetic and functional-based study. J Pathol. 2011;223:327–35. doi: 10.1002/path.2816. [DOI] [PubMed] [Google Scholar]
- 24.Hsu W, Mohyeldin A, Shah SR, ap Rhys CM, Johnson LF, Sedora-Roman NI, et al. Generation of chordoma cell line JHC7 and the identification of Brachyury as a novel molecular target. J Neurosurg. 2011;115:760–9. doi: 10.3171/2011.5.JNS11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Momparler RL, Bouchard J, Onetto N, Rivard GE. 5-AZA-2’-deoxycytidine therapy in patients with acute leukemia inhibits DNA methylation. Leuk Res. 1984 doi: 10.1016/0145-2126(84)90141-3. [DOI] [PubMed] [Google Scholar]
- 26.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014:1394–6. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lochmann TL, Powell KM, Ham J, Floros KV, Heisey DAR, Kurupi RIJ, et al. Targeted inhibition of histone H3K27 demethylation is effective in high-risk neuroblastoma. Sci Transl Med. 2018;10:1–13. doi: 10.1126/scitranslmed.aao4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharifnia T, Wawer MJ, Chen T, Huang QY, Weir BA, Sizemore A, et al. Small-molecule targeting of brachyury transcription factor addiction in chordoma. Nat Med. 2019;25:292–300. doi: 10.1038/s41591-018-0312-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheipl S, Barnard M, Cottone L, Jorgensen M, Drewry DH, Zuercher WJ, et al. EGFR inhibitors identified as a potential treatment for chordoma in a focused compound screen. J Pathol. 2016;239:320–34. doi: 10.1002/path.4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lampada A, O’Prey J, Szabadkai G, Ryan KM, Hochhauser D, Salomoni P. MTORC1-independent autophagy regulates receptor tyrosine kinase phosphorylation in colorectal cancer cells via an mTORC2-mediated mechanism. Cell Death Differ. 2017;24:1045–62. doi: 10.1038/cdd.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014 doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sims D, Ilott NE, Sansom SN, Sudbery IM, Johnson JS, Fawcett KA, et al. Genome analysis CGAT : computational genomics analysis toolkit. 2014;30:1–2. doi: 10.1093/bioinformatics/btt756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–67. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead and Steven L Salzberg. Bowtie2. Nat Methods. 2013;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–8. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fursova NA, Blackledge NP, Nakayama M, Ito S, Koseki Y, Farcas AM, et al. Synergy between Variant PRC1 Complexes Defines Polycomb-Mediated Gene Repression. Mol Cell. 2019;74:1020–1036.:e8. doi: 10.1016/j.molcel.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9 doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cribbs A, Hookway ES, Wells G, Lindow M, Obad S, Oerum H, et al. Inhibition of histone H3K27 demethylases selectively modulates inflammatory phenotypes of natural killer cells. J Biol Chem. 2018;293:2422–37. doi: 10.1074/jbc.RA117.000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee DH, Zhang Y, Kassam AB, Park MJ, Gardner P, Prevedello D, et al. Combined PDGFR and HDAC Inhibition Overcomes PTEN Disruption in Chordoma. PLoS One. 2015;10:e0134426. doi: 10.1371/journal.pone.0134426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruidenier L, Chung C, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–8. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinemann B, Nielsen JM, Hudlebusch HR, Lees MJ, Larsen DV, Boesen T, et al. Inhibition of demethylases by GSK-J1/J4. Nature. 2014;514:E1–2. doi: 10.1038/nature13688. [DOI] [PubMed] [Google Scholar]
- 43.Johansson C, Velupillai S, Tumber A, Szykowska A, Hookway ES, Nowak RP, et al. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol. 2016;12:78–80. doi: 10.1038/nchembio.2087. [DOI] [PubMed] [Google Scholar]
- 44.Tumber A, Nuzzi A, Hookway ES, Hatch SB, Velupillai S, Johansson C, et al. Potent and Selective KDM5 Inhibitor Stops Cellular Demethylation of H3K4me3 at Transcription Start Sites and Proliferation of MM1S Myeloma Cells. Cell Chem Biol. 2017;138:981–92. doi: 10.1016/j.chembiol.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singleton DC, Harris AL. Targeting the ATF4 pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:1189–202. doi: 10.1517/14728222.2012.728207. [DOI] [PubMed] [Google Scholar]
- 46.Sidrauski C, McGeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife. 2015;4 doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smits P. Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development. 2003 doi: 10.1242/dev.00331. [DOI] [PubMed] [Google Scholar]
- 48.Magnaghi P, Salom B, Cozzi L, Amboldi N, Ballinari D, Tamborini E, et al. Afatinib is a new therapeutic approach in chordoma with a unique ability to target EGFR and Brachyury. Mol Cancer Ther. 2017 doi: 10.1158/1535-7163.MCT-17-0324. [DOI] [PubMed] [Google Scholar]
- 49.Shalaby A, Presneau N, Ye H, Halai D, Berisha F, Idowu B, et al. The role of epidermal growth factor receptor in chordoma pathogenesis: a potential therapeutic target. J Pathol. 2011;223:336–46. doi: 10.1002/path.2818. [DOI] [PubMed] [Google Scholar]
- 50.Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the “undruggable” cancer targets. Nat Rev Cancer. 2017;17:502–8. doi: 10.1038/nrc.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cribbs AP, Terlecki-Zaniewicz S, Philpott M, Baardman J, Ahern D, Lindow M, et al. Histone H3K27me3 demethylases regulate human Th17 cell development and effector functions by impacting on metabolism. Proc Natl Acad Sci. 2020:1–11. doi: 10.1073/pnas.1919893117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 53.Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. Chromatin Sampling-An Emerging Perspective on Targeting Polycomb Repressor Proteins. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang SP, Tang Z, Chen CW, Shimada M, Koche RP, Wang LH, et al. A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription. Mol Cell. 2017;67:308–321. doi: 10.1016/j.molcel.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dhar SS, Zhao D, Lin T, Gu B, Pal K, Wu SJ, et al. MLL4 Is Required to Maintain Broad H3K4me3 Peaks and Super-Enhancers at Tumor Suppressor Genes. Mol Cell. 2018;70:825–841.:e6. doi: 10.1016/j.molcel.2018.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dhar SS, Lee SH, Kan PY, Voigt P, Ma L, Shi X, et al. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012;26:2749–62. doi: 10.1101/gad.203356.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe S, Shimada S, Akiyama Y, Ishikawa Y, Ogura T, Ogawa K, et al. Loss of KDM6A characterizes a poor prognostic subtype of human pancreatic cancer and potentiates HDAC inhibitor lethality. Int J Cancer. 2019;145:192–205. doi: 10.1002/ijc.32072. [DOI] [PubMed] [Google Scholar]
- 58.Johansson C, Tumber A, Che K, Cain P, Nowak R, Gileadi C, et al. The roles of Jumonji-type oxygenases in human disease. Epigenomics. 2014;6:89–120. doi: 10.2217/epi.13.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are deposited in the National Center for Biotechnology Information GEO database: RNA-seq and ChIP-Rx sequencing experiments under accession number GSE120214, DNA methylation array data under accession number GSE119462.
Coordinates and structure factors for the UTY:KDOBA67 ligand complex are deposited with RCSB under PDB ID 5A1L.