Abstract
The study of brain development in humans is limited by the lack of tissue samples and suitable in vitro models. Here, we model early human neural tube development using human embryonic stem cells (hESCs) cultured in a microfluidic device. The approach, named microfluidic-controlled stem cell regionalisation (MiSTR), exposes pluripotent stem cells to signalling gradients that mimic developmental patterning. Using a WNT-activating gradient, we generated neural tissue that exhibits progressive caudalisation from forebrain to midbrain to hindbrain, including formation of isthmic organiser characteristics. Single-cell transcriptomics revealed that rostro-caudal organisation was already established at 24 hours of differentiation, and that the first markers of a neural-specific transcription program emerged in the rostral cells at 48 hours. The transcriptomic hallmarks of rostro-caudal organisation recapitulated gene expression patterns of the early rostro-caudal neural plate in mouse embryos. Thus, MiSTR will facilitate research on the factors and processes underlying rostro-caudal neural tube patterning.
The establishment of early neural tube regionalisation is known from model organisms to involve morphogenic gradients along the rostro-caudal and dorso-ventral axes. In the rostro-caudal axis, regionalisation is mediated by a conserved WNT signalling gradient1, which, through unknown mechanisms, leads to the formation of the major neural regions: the forebrain, midbrain, hindbrain and spinal cord. Secondary organising centres subsequently emerge (e.g., the midbrain-hindbrain boundary, MHB), imposing local signalling gradients to sequentially pattern the neural tube into sub-regions2, 3. Following this, dorso-ventral identity is established by gradients perpendicular to the rostro-caudal axis4. Recent studies have modelled dorso-ventral patterning in mouse and human embryonic stem cells (ESCs) through guided self-organisation of neural cysts5, by microfluidic diffusion-based gradient generators6 or by organoid fusion techniques7, 8, 9. In contrast, we know much less about the mechanisms underlying rostro-caudal patterning, and the rostro-caudal neural patterning axis has, to our knowledge, not been modelled in vitro.
We have previously shown that exposure of hESCs to increasing concentrations of a glycogen synthase kinase 3 inhibitor (GSK3i), an activator of the canonical WNT signalling pathway, induces differentiation into progressively caudalised neural fates10. Here, we aimed to recreate the entire cranial rostral-to-caudal neural axis (from forebrain to hindbrain) in a single tissue by exposing hESCs to a linear concentration gradient of GSK3i, thereby mimicking the WNT gradient underlying the rostral-to-caudal neural axis specification in the early embryo1, 3 (Fig. 1a). Our MiSTR system is based on a microfluidic gradient generator relying on sequential diffusive mixing of two inlet media (0% and 100%)11 to obtain a stable and linear gradient of GSK3i (Fig. 1a, b and Supplementary Fig. 1a). The gradient profile in the cell culture chamber was assessed using fluorescein as a surrogate of the GSK3i (CHIR99021), since these molecules have similar diffusion coefficients and Péclet numbers (see Methods and Supplementary Notes).
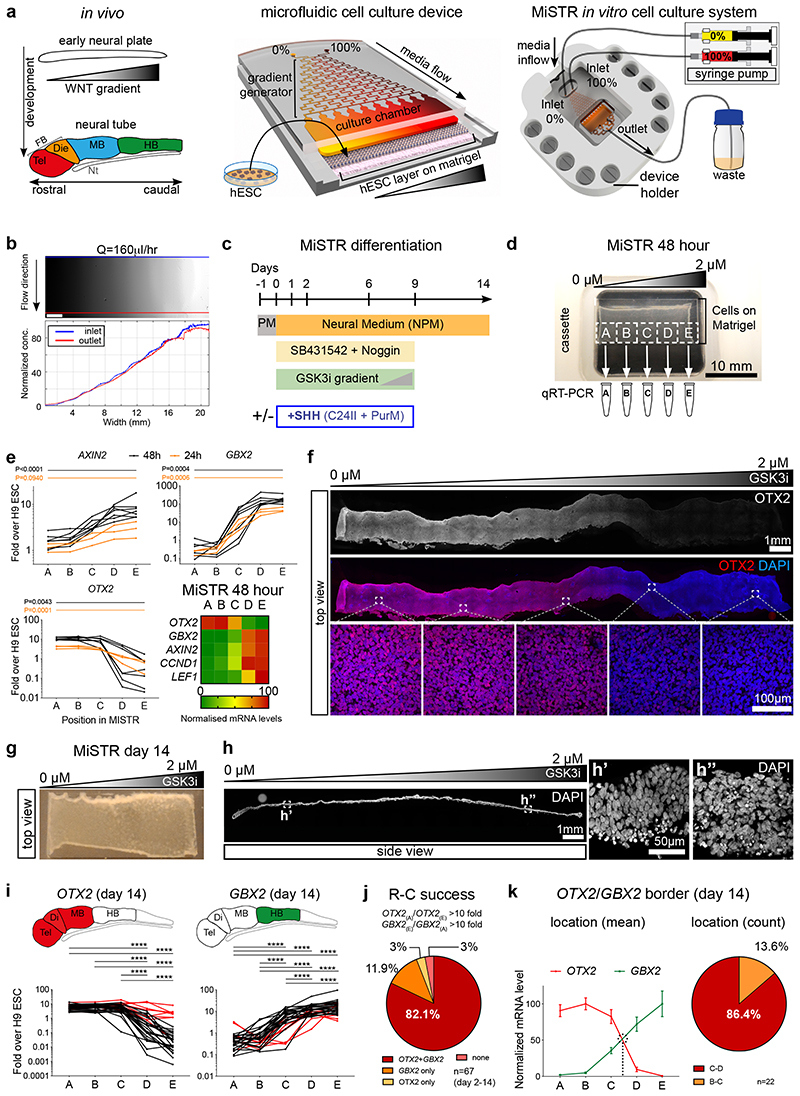
Figure 1. Design of the MiSTR cell culture system and establishment of a WNT signalling gradient in hESC-derived tissue.
(a) The in vivo WNT gradient at the early neural plate stage was mimicked in vitro by a microfluidic gradient generator producing sequential mixing of two media containing 0% and 100% morphogen. The media with the morphogen gradient flows laminarly from the microfluidic channels into the cell culture chamber containing a layer of hESCs seeded onto a bed of Matrigel. Media inflow is controlled by precision syringe pumps, while on the opposite side of cell chamber the used media outflows to a waste container. FB: Forebrain, Tel: telencephalon, Die: diencephalon, MB: midbrain, HB: hindbrain, Nt: Notochord.
(b) Fluorescein gradient (top view), measured across the whole width and length of the cell culture chamber at a flow rate of Q = 160 μl/h; blue line indicates inlet side measurements, and red line outlet side measurements. Scale bars, 2 mm.
(c) Overview of MiSTR differentiation protocol. Ventralising factors (+SHH) were added only when indicated. PM: Pluripotency Medium, PurM: Purmorphamine.
(d) Representative hESCs-derived tissue atop the Matrigel substrate after 48 hours of neural differentiation in the MiSTR device under a GSK3i gradient. The tissue was sub-dissected into 5 samples (A-E) for qRT-PCR analysis.
(e) qRT-PCR analysis along the 5 regions A-E, for the canonical WNT signalling target AXIN2 and of early neural genes (OTX2, rostral; GBX2, caudal), at 24 hours (n = 3 MiSTR tissues) and 48 hours of differentiation (n = 7 MiSTR tissues) under a 0 μM to 2 μM GSK3i gradient. Line graphs represent individual experiments of 24 hours (orange) and 48 hours (black). Heatmap displays normalized data at 48 hours for each gene, plus canonical WNT targets CCND1 and LEF1. 1-way ANOVA followed by Dunn’s (OTX2, GBX2, 48 hours) or Tukey’s (all others) multiple comparison test between all regions – only A to E comparisons are shown. For further comparisons and exact p-values see also Supplementary Fig. 1d and Supplementary Table 2.
(f) Representative whole-mount staining of a strip of 48-hour MiSTR tissue (n=3), showing decreased OTX2 (in red) along the imposed 0 μM to 2 μM GSK3i gradient (shown above).
(g) Representative overview of hESC-derived neural tissue after 14 days of culture in the MiSTR device. Note increased tissue opacity in comparison to 2 day-old MiSTR tissue, as the original monolayer of cells expands in thickness over time.
(h) Representative longitudinal section along the widest axis of MiSTR tissue after 14 days of culture (n=3). Magnified views (h’, h”) show the multicellular layer across the entire culture area, seeded at day 0 as a monolayer of hESCs.
(i) qRT-PCR analysis along the 5 positions, for OTX2 and GBX2, each line representing individual experiments. Red lines indicate experiments which failed the rostral-caudal patterning criterion (reciprocal fold change between regions A and E above 10 fold for both OTX2 and GBX2, see Methods). *p < 0.05; **p < 0.01; ***p < 0.001; **** p<0.0001; Tukey’s (GBX2) or Dunn’s (OTX2) multiple comparison test between all regions, n=32 MiSTR tissues. Schematics above show corresponding expression patterns in the embryo. See Supplementary Table 2 for p-values.
(j) Quantification of MiSTR tissues (from differentiation days d2 through d14) passing to the rostro-caudal patterning criterion.
(k) Normalized mean expression of OTX2 and GBX2 along the A-E regions of 14-day MiSTR tissues which passed the rostro-caudal criterion. Quantification of OTX2/GBX2 border localization, calculated arithmetically for each individual experiment (visualised as dotted line in graph). Data as mean SEM, n=22 MiSTR tissues.
To examine neural regionalisation in the cell culture system, hESCs (H9 cell line) were seeded as a confluent monolayer of cells onto a bed of Matrigel in the device (Supplementary Fig. 1b) and the cells were differentiated under a gradient of GSK3i (0-2 μM) in neural induction medium with dual SMAD inhibition12 (Fig. 1c). Under these conditions the cells robustly formed neural fates without mesodermal or endodermal differentiation (Supplementary Fig. 1c). Upon differentiation, the hESCs formed a thin tissue, which could be sub-dissected into 5 equally sized samples (A-E) for quantitative RT-PCR (qRT-PCR) analysis (Fig. 1d). Analysis at 24-48 hours showed a significant (e.g. 48 hour AXIN2 p<0.0001) progressive increase in expression of WNT target genes from samples A (low GSK3i exposure) to samples E (high GSK3i exposure, Fig. 1e and Supplementary Fig. 1d), indicating that the GSK3i gradient was translated into a WNT signalling gradient. The gradient further induced expression of the caudal marker GBX2 at high GSK3i exposures, whereas OTX2, a rostral marker showed the inverse expression profile (Fig. 1e and Supplementary Fig. 1d). This regionalised profile was corroborated by immunocytochemical staining for OTX2 (Fig. 1f), indicating that already within 24 hours, the hESC tissue layer was undergoing rostral-to-caudal patterning as a response to the microfluidic GSK3i gradient. When subjecting the cells to a gradient of another caudalising agent, retinoic acid (RA), we observed induction of HOXA1 (a transcriptional target of RA13), but absence of an opposing OTX2/GBX2 expression gradient, indicating that WNT but not RA can mimic the early rostro-caudal molecular patterning of the embryo (Supplementary Fig. 1e).
We proceeded to differentiate the GSK3i-patterned cells in MiSTR for 14 days at which point the cells had expanded to produce a tissue of approximately 100 μm thickness (Fig. 1g, h) Again, we consistently observed opposing expression profiles of OTX2 and GBX2 by qRT-PCR, and we defined an experiment as successful and eligible for further analysis when displaying a >10-fold change from A-to-E for both OTX2 and GBX2 (i.e. 82.1% of all 48-hour to 14 day experiments, Fig. 1i, j). At day 14, the boundary between OTX2 and GBX2, i.e. where the midbrain-hindbrain boundary (MHB) arises during development, was consistently located between the C and D regions (86.4% of cases, Fig. 1k). Concomitantly, several key markers of the MHB, including FGF8 and WNT1, showed expression peaks also located to the middle region C (Fig. 2a, b). This was validated by immunostaining showing a distinct OTX2 positive region at the rostral end and increased PAX8 in the middle of the tissue (Fig. 2c). Further qRT-PCR analyses revealed that samples from regions A and B (exposed to low GSK3i) both expressed high levels of forebrain markers OTX2 and FEZF1, whereas only region A showed expression of LHX2 and FOXG1, indicating telencephalic identity in region A and diencephalic identity in region B (Fig. 2d). Samples D and E expressed caudal neural tube markers (GBX2, IRX3) along with several HOX genes, of which the more posterior markers HOXA3 and HOXA5 were restricted to region E (Fig. 2d and Supplementary Fig. 2a). Robustness of the rostro-caudal differentiation axis was confirmed across three additional pluripotent cell lines (Supplementary Fig. 2b).
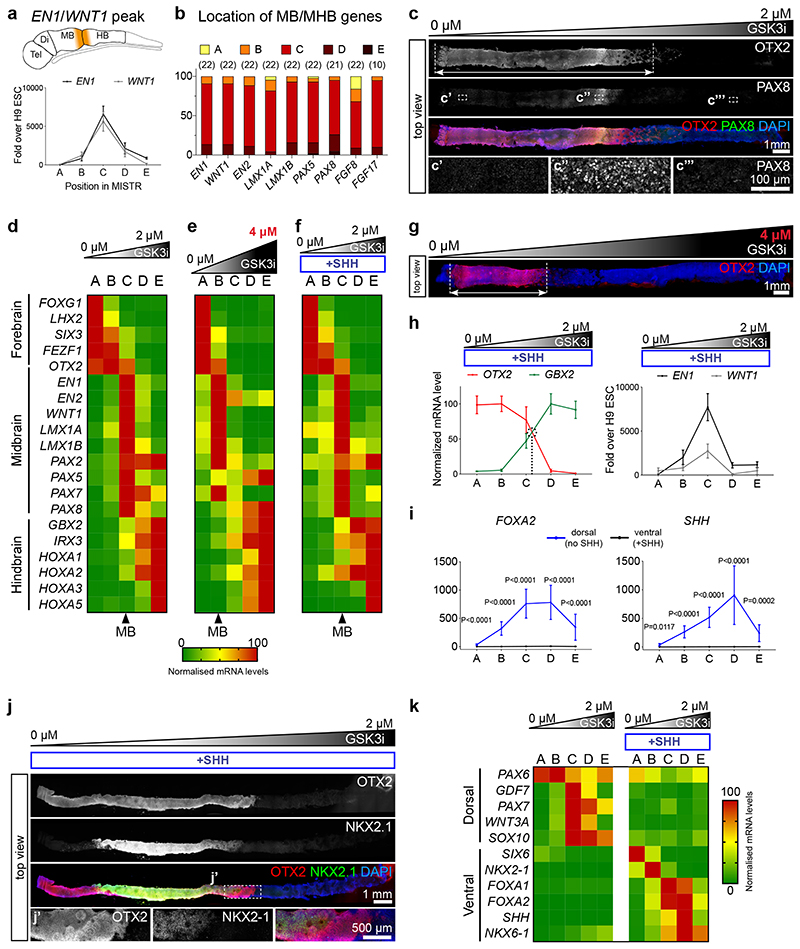
Figure 2. Neural patterning along the rostro-caudal axis in day 14 MiSTR tissue.
(a) qRT-PCR analysis along the 5 regions (A-E), for the MHB markers EN1 and WNT1, peaking at the central C regions (peak regions > 75 percentile values of gene expression, see Methods). Data as mean SEM, n = 22 MiSTR tissues.
(b) Quantification of location of peaked expression for markers of midbrain and MHB. Respective n is shown above.
(c) Representative whole-mount staining of 14-day MiSTR tissue (n=2), generated with a 0 μM to 2 μM GSK3i gradient. OTX2 staining (red) marks the forebrain/midbrain regions (arrow), while increased PAX8 (green) labels the MHB region.
(d) Normalized expression of forebrain, midbrain and hindbrain markers along the A-E regions of MiSTR tissue under a 0 μM to 2 μM GSK3i gradient. Arrowhead below marks the C region, with elevated midbrain (MB) markers. n = 17-24 MiSTR tissues. See Supplementary Fig. 2a.
(e) Normalized expression of forebrain, midbrain and hindbrain markers along the A-E regions of MiSTR tissue under the steeper 0 μM to 4 μM GSK3i gradient (shown above). Note the rostralized location of the midbrain markers in region B (arrowhead). n = 4 MiSTR tissues.
(f) Normalized expression of forebrain, midbrain and hindbrain markers along the A-E regions of ventralized MiSTR tissue (+SHH, SHH agonists added to entire tissue) under a 0 μM to 2 μM GSK3i gradient (shown above). n = 9 MiSTR tissues.
(g) Representative hole-mount staining of 14-day MiSTR tissue (n=2), with a 0 μM to 4 μM GSK3i gradient (shown above). OTX2 staining (red) marks the forebrain/midbrain regions (arrow), compressed at the low GSK3i side, in comparison with the 0 μM to 2 μM gradient. See also Supplementary Fig. 3a.
(h) Normalized mean expression of OTX2 and GBX2 along the 5 regions A-E of ventralized MiSTR (+SHH), and relative expression of the MHB markers EN1 and WNT1. Data as mean SEM, n = 9 MiSTR (+SHH) tissues.
(i) qRT-PCR analysis along the 5 regions A-E of dorsal (no SHH) and ventral (+SHH) MiSTR, for the ventral markers FOXA2 and SHH. 2-way ANOVA, followed by Sidak’s multiple comparison between no SHH and +SHH, for each region. Data as mean SEM, n=12 (SHH) and n = 19 (FOXA2) dorsal MiSTR tissues (no SHH), and n = 9 for ventral MiSTR (+SHH) tissues. See Supplementary Fig. 3b and Supplementary Table 2.
(j) Representative whole-mount staining of ventralised MiSTR tissue (+SHH) (n=3), showing the ventral diencephalic marker NKX2-1 (green) within the OTX2-positive region (red).
(k) Normalized expression of dorsal and ventral markers along the A-E regions of dorsal (no SHH) and ventral (+SHH) MiSTR tissue under a 0 μM to 2 μM GSK3i gradient. n = 9 ventral MiSTR tissues (+SHH), n = 10 to 19 dorsal MiSTR tissues (no SHH). Note expression of roof plate markers PAX7, GDF7, WNT3A and SOX10 in regions C and D. See also Supplementary Fig. 3b and Supplementary Table 2.
To verify that the regionalised neural plate tissue identity in the MiSTR was a direct positional response to the GSK3i level, we increased the upper GSK3i concentration from 2 μM to 4 μM. Under this steeper gradient, the MiSTR patterning profile shifted in a scaled manner, compressing forebrain identities into region A and shifting midbrain/MHB identities from region C to B, while hindbrain markers occupied all remaining regions C-E (Fig. 2e, g and Supplementary Fig. 3a), thereby reinforcing the direct relationship between the rostro-caudal organisation and the steepness of the WNT activation gradient.
We next assessed if MiSTR rostro-caudal regionalisation could be performed under ventralising conditions by differentiating the MiSTR tissue in the presence of ventral agonists, that is, Sonic Hedgehog (SHH) and Purmorphamine (hereafter referred as “+SHH”) across the entire GSK3i gradient. We verified that the ventralised tissue retained its rostro-caudal axis and MHB identity in position C (Fig. 2f, h). In addition, ventralisation induced markers of the ventral neural tube at expected locations, including floor plate markers FOXA1/2 and SHH at mid- and hindbrain locations (Fig. 2i), ventral forebrain markers NKX2-1 and SIX6 at rostral locations and NKX6-1 at caudal locations (Fig. 2k and Supplementary Fig. 3b). Immunostaining of the ventralised tissue revealed that NKX2-1 expression was confined to a region within the medial OTX2+ domain in the presumptive diencephalic part of the tissue (Fig. 2j). In comparison, the non-ventralised MiSTR tissue showed corresponding expression patterns of the dorsal neural tube, including roof plate markers in midbrain and hindbrain regions (Fig. 2k and Supplementary Fig. 3b). To assess if the MiSTR system could be applied for creating regionalised neural tissue with other morphogenic gradients, we subjected hindbrain-patterned tissue to a gradient of RA from day 3 to 14 to mimic the rostral-to-caudal RA gradient of the hindbrain14. Although RA alone did not induce an early rostro-caudal axis in differentiating hESCs (Supplementary Fig. 1e), the combination of an RA gradient with early caudalisation using GSK3i (2 uM), caused progressive induction of more posterior HOX genes along the hindbrain axis (Supplementary Fig. 3c) These data confirmed that rostro-caudal organisation could be obtained in both a dorsal and ventral neural tube context by the simple combination of GSK3i gradient and SHH agonists, whereas hindbrain caudalisation in the MiSTR could be further controlled by a gradient of RA.
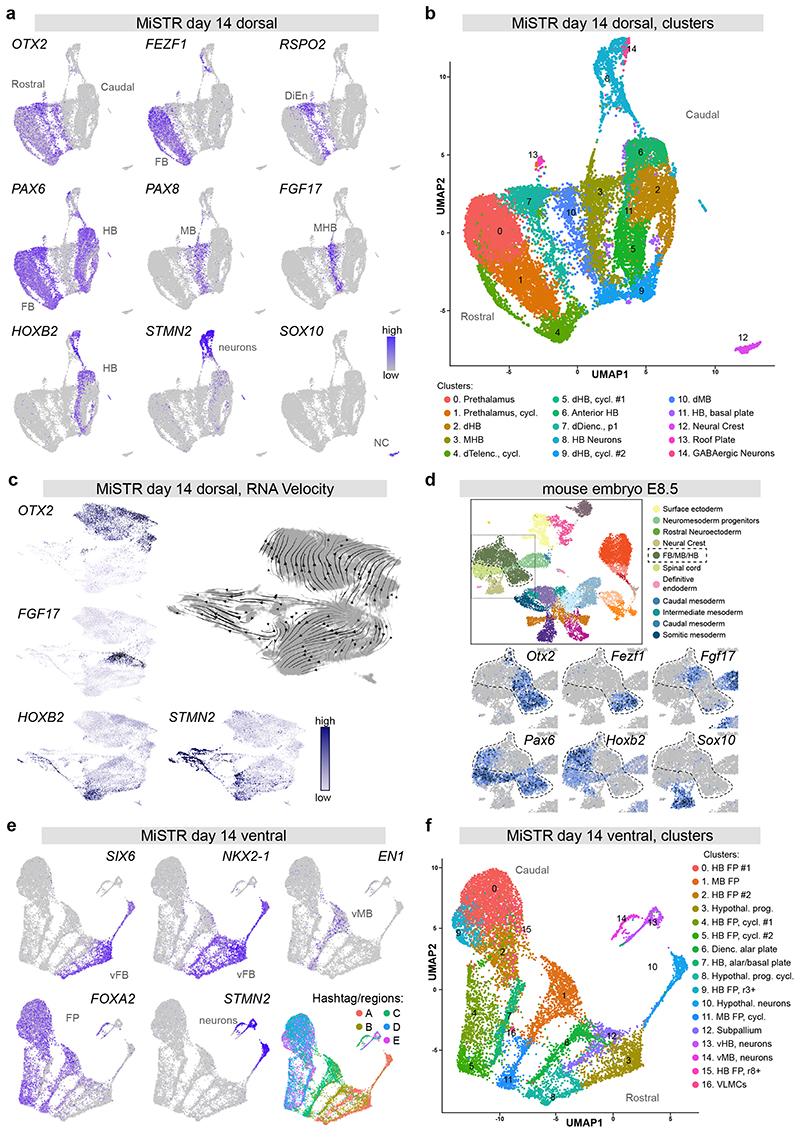
As an unbiased assessment of the full variety of cell types present in the tissue, we performed droplet-based single cell RNA sequencing (scRNAseq) on day 14 dorsal MiSTR tissue. UMAP visualisation of the data showed a remarkable segregation between OTX2+ and OTX2- cells, with FEZF1+ (forebrain), FOXG1+ (telencephalic) and RSPO2+ (diencephalic) clusters present within the OTX2+ population (Fig. 3a, Supplementary Fig. 4a, d and Supplementary Table 1). The OTX2- population on the other hand contained HOXB2+ and GBX2+ (hindbrain) cells (Fig. 3a and Supplementary Fig. 4a, d). At the interface between the OTX2+ and OTX2- cell population appeared two separate clusters (#10 and #3) marked by expression of PAX8 and PAX8/FGF17, indicating the formation of midbrain and MHB cell fates, respectively. These clusters were uniquely negative for PAX6; a gene which in mouse is expressed in all dorsal neuroepithelial tissues except the midbrain and MHB (Fig. 3a and Supplementary Fig. 4b). In line with the crucial organizing role of FGF signalling at the MHB3, the MHB cluster further expressed FGF8 and several markers of active FGF signalling including FGFBP3 (an enhancer of FGF signalling) and the transcriptional targets PTN, SPRY2 and DUSP615 (Supplementary Fig. 4a and Supplementary Table 1). Analysis using the RNA velocity tool16 showed strong opposing RNA velocity trajectories away from the MHB cells (FGF17+), indicating the presence of a molecular boundary effect preventing transitioning between rostral and caudal cell fates (Fig. 3c). Other cell types present in the tissue included a minor population of neural crest cells (SOX10+) and two clusters of post-mitotic neurons (STMN2+), emanating from two separate neurogenic branches from the MHB and hindbrain clusters (Fig. 3a-c and Supplementary Fig. 4a, d). Sub-clustering of these two STMN2+ clusters identified 10 different subtypes of neurons, deriving mainly from the hindbrain (SST+, VSX2+. GATA2+. ISL1+) but also from the dorsal MHB (TLX3+), diencephalon (BARHL1+) and forebrain (TRH+) (Supplementary Fig. 4e and Supplementary Table 1). We compared the day 14 data to a recently published scRNAseq dataset from the gastrulating mouse embryo at E8.517, and found very similar regional gene expression patterns in the neural tube cells of this dataset (Fig. 3d and Supplementary Fig. 4c). Integration with the mouse embryo dataset (E8.5) onto UMAP showed co-localisation of MiSTR cells with the neural regions of the embryo, whereas non-neural mouse clusters localised separately (Supplementary Fig. 4f). When integrating the MiSTR day 14 data with only relevant mouse neural tube clusters, we found the mouse neural cells segregated to co-localise with either rostral (OTX2+), caudal (HOXB1+), MB/MHB (PAX8+/FGF17+) or neural crest cells (SOX10+), indicating similarity in regionalised transcriptomes between the mouse neural tube and the MiSTR tissue (Supplementary Fig. 5a). In contrast, when integrating MiSTR data with published scRNAseq data from human pluripotent stem cell-derived neural organoids reported to give rise to midbrain and hindbrain neurons after extended culturing18, 19, we found co-localisation of OTX2+ forebrain progenitors, but could not detect cells expressing markers of midbrain, MHB or hindbrain progenitors in the organoid data (Supplementary Fig. 5b, c).
Figure 3. Single cell transcriptomics of dorsal and ventral MiSTR tissues.
(a, b) scRNAseq of 14 day dorsal MiSTR tissue (n=3 MiSTR tissues, 17683 cells) plotted in Uniform Manifold Approximation and Projection (UMAP) dimensions.
(a) Selected marker genes of neural patterning depict the rostro-caudal axis oriented left-to-right, revealed by the expression of genes marking the forebrain (FB), diencephalon (DiEn), midbrain (MB), midbrain-hindbrain boundary (MHB), and hindbrain (HB). Note the separate cluster of SOX10+ neural crest cells (NC) and a branch of STMN2+ neurons.
(b) Shared nearest neighbour (SNN) - based clusters calculated using Seurat. Cells are coloured according to their respective cluster and annotated based on literature and database search. dHB: dorsal Hindbrain, MHB: Midbrain-Hindbrain boundary, dTelenc: dorsal Telencephalon, cycl.: cycling, dDienc: dorsal Diencephalon, dMB: dorsal Midbrain. See also Supplementary Fig. 4a.
(c) RNA velocity analysis of day 14 dorsal MiSTR scRNAseq data (n=3 MiSTR tissues, 18241 cells), showing RNA trajectories away from the FGF17+ MHB cell cluster as well as towards postmitotic neurons (STMN2+), analysis performed in ScanPy.
(d) scRNAseq of E8.5 mouse embryonic stage plotted in UMAP dimensions showing selected marker genes of neural patterning, within the selected neural tube region (FB/MB/HB) highlighted in the overall map (top). See also Supplementary Fig. 4c. Plots were obtained from the scRNAseq map of mouse early development17, available at: https://marionilab.cruk.cam.ac.uk/MouseGastrulation2018.
(e, f) scRNAseq of 14 day ventral MiSTR tissue (n = 3 MiSTR tissues, 12669 cells) plotted in UMAP dimensions.
(e) Expression of genes marking the ventral forebrain (vFB), ventral midbrain (vMB) and floor plate (FP) revealed a rostro-caudal axis oriented right-to-left in the UMAP. This is in accordance with the distribution of hashtag barcodes marking each of the MiSTR regions (A-E). See also Supplementary Fig. 6a for additional gene expression patterns.
(f) SNN- based clusters calculated using Seurat. Cells are coloured according to their respective cluster and annotated based on literature search. HB FP: Hindbrain Floor Plate, MB FP: Midbrain Floor Plate, Hypothal. Prog.: Hypothalamic progenitors, cycl.: cycling, Dienc.: Diencephalic, vHB: ventral Hindbrain, vMB: ventral Midbrain, VLMCs: Vascular leptomeningeal Cells.
To assess if rostro-to-caudal organisation was also obtained at the single cell level in ventral MiSTR tissues and to link rostro-caudal fates with the physical location in the tissue, we performed scRNAseq on ventralised tissues using antibody-based cell hashing20 to discriminate cells coming from regions A-E. These data showed that ventral forebrain progenitors (SIX3+, SIX6+ and NKX2-1+) were derived exclusively from regions A and B whereas ventral midbrain progenitors (EN1+, LMX1A+, RSPO2+ and FOXA2+) were derived mainly from region C and hindbrain cells (NKX6-2+, PHOXB2+, HOXB2+) mainly from regions D and E (Fig. 3e, f and Supplementary Fig. 6a, b). In line with this, sub-clustering of neurons revealed ganglionic eminence neurons deriving from region A, hypothalamic neurons from regions A and B, ventral midbrain neurons from region C and ventral hindbrain neurons from regions D and E (Supplementary Fig. 6c). Strikingly, although the ventral MiSTR similar to dorsal MiSTR was clearly organised into forebrain, midbrain and hindbrain structures, no expression of FGF17 was detected in the ventral scRNAseq data (Supplementary Fig. 6a); consistent with the observation that FGF17 expression in the mouse brain is restricted to only the dorsal MHB (Supplementary Fig. 4b).
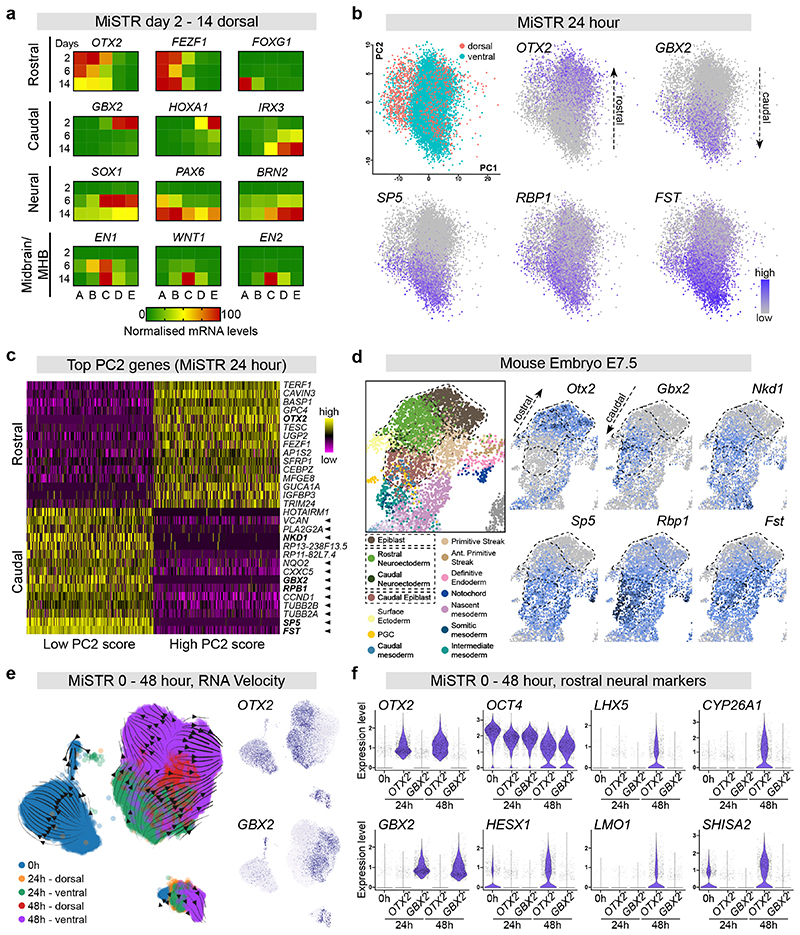
To investigate the temporal sequence of GSK3i-induced rostro-caudal organisation, we performed qRT-PCR analysis at days 2, 6 and 14 of differentiation. Although caudalising and neuralising factors were added simultaneously to the cultures, rostro-caudal organisation of the tissue was observed on day 2, before the appearance of neuroectodermal markers on day 6 (Fig. 4a and Supplementary Fig. 7a). The midbrain/MHB identity was first visible on day 6 as a peak of EN1 whereas other midbrain and MHB markers only appeared on day 14 of differentiation (Fig. 4a and Supplementary Fig. 7a).
Figure 4. Single cell transcriptomics of early MiSTR patterning and temporal dissection of MiSTR regionalisation.
(a) Heatmaps depicting temporal evolution of rostral, caudal, neural and midbrain/MHB markers (normalized expression) during dorsal (no SHH) MiSTR differentiation, at 2, 6 and 14 days under a 0 μM to 2 μM gradient, showing expression of early rostral and caudal markers (OTX2/FEZF1, GBX2/HOXA1) preceding expression of neural markers and midbrain/MHB markers. Note that the higher expression of GBX2 and HOXA1 on day 2 dims the rostro-caudal pattern which is also present on day 6 and 14 (see also corresponding line graphs and statistics in Supplementary Fig. 7a and Supplementary Table 2).
(b) PCA plot of scRNAseq data from 24-hour MiSTR tissue (n = 3 dorsal plus n = 3 ventral MiSTR tissues combined, total 9848 cells) showing clear segregation between OTX2+ and GBX2+ cells. Caudal cell identity was associated with activation of the WNT/β-catenin target genes SP5, RBP1 and FST, see also (c).
(c) Heatmap of the top 15 positively- and negatively loading genes on PC2 of 24 hour MiSTR tissue from the top 500 cells with lowest and highest PC2 scores (cells shown in columns). Cells of caudal fate (i.e. low PC2 score) showed high expression of genes identified as targets of the WNT/β-catenin pathway (marked by arrowheads, see also Supplementary Fig. 7b). Note that the earliest markers of rostral and caudal identity, OTX2 and GBX2, respectively are also identified as top loading genes on PC2. Genes highlighted in bold are plotted in d).
(d) scRNAseq data from the E7.5 mouse embryo (plotted in UMAP dimensions) shows similar rostro-caudal expression patterns for top-loading rostro-caudal genes identified in 24-hour MiSTR tissue. Regions associated with presumptive neural commitment are highlighted in dashed lines. See also Supplementary Fig. 8c. Note that genes marking rostro-caudal expression patterns in MiSTR at 24 hours are shared between epiblast and neuroectodermal cells in the mouse. Plots from mouse embryos were obtained from the scRNAseq map of mouse early development17. Note that the cluster annotated as “rostral neuroectoderm” contains both OTX2+ and GBX2+ cells, i.e. covering both presumptive forebrain, midbrain and hindbrain cells. Gene plots and annotations are available at: https://marionilab.cruk.cam.ac.uk/MouseGastrulation2018.
(e) RNA velocity analysis of integrated MiSTR scRNAseq data from 0 24 and 48 hours of differentiation (n=3 MiSTR tissues for each of the five conditions, 28598 cells combined), showing that cell differentiation trajectories are separated between OTX2+ and GBX2+ cells already from 24 hours of differentiation. Further note a clear directionality away from the OTX2+/GBX2+ expression boundary at 48 hours, indicating establishment of a molecular boundary effect between OTX2+ and GBX2+ cells. An unidentifiable mixed cluster at the bottom right, presumably resulting from technical artefacts, was disregarded for interpretation (see Methods).
(f) Violin plots showing expression levels of selected genes at 0 hours (7895 cells), and in the rostral (OTX2+) and caudal (GBX2+) MiSTR cell populations at 24 (8492 cells) and 48 hours (9705 cells). Each subset was downsampled to 1500 cells for visualization purposes. Note that presumptive markers CYP26A1, HESX1, LHX5, LMO1 and SHISA2 are enriched only in the OTX2+ population at 48 hours. These gene were confirmed also in the mouse to be specific for the rostral (OTX2+) neuroectoderm (see Supplementary Fig. 8c and d). See also Supplementary Table 2.
To further discern the temporal acquisition of rostro-caudal patterning and neural commitment, we performed scRNAseq of MiSTR tissue (dorsal and ventral) at 0, 24 and 48 hours of differentiation. Principal component analysis (PCA) showed a clear segregation of the OTX2- and GBX2-expressing cells already at 24 hours (Fig. 4b), and the majority of the caudal associated genes were identified as targets of the WNT/β-catenin pathway (Fig. 4c and Supplementary Fig. 7b), supporting that GSK3i-induced caudalisation was a result of canonical WNT signalling. These top caudal-associated genes in MiSTR at 24 hours accurately reflected the rostro-caudal expression gradient observed across the late epiblast and presumptive neurectoderm in the mouse at E7.517 (Fig. 4d). RNA velocity analysis on 0 – 48 hours MiSTR data showed separate cell differentiation trajectories from the OTX2+ and GBX2+ compartments, respectively. These OTX2+ versus GBX2+ trajectories emanated at 24 hours and were maintained as the cells proceeded differentiation to 48 hours, thus reinforcing that rostro-caudal identities were established at 24 hours (Fig. 4e). Genes associated with temporal maturation from 0-48 hours (PC1, Supplementary Fig. 8a, b) were shared markers of the presumptive neuroectoderm and late epiblast in mouse E7.5 (Supplementary Fig. 8c). In contrast, when analysing rostral and caudal cells separately (Fig. 4f), we could identify the first markers specific to the presumptive rostral neuroectodem at 48 hours (HESX1, LHX5, LMO1, SHISA2 and CYP26A1). These markers were confirmed in the mouse to be absent from the late epiblast and specifically expressed in the rostral (OTX2+) neurectoderm at E7.5-E7.75 (Supplementary Fig. 8c, d).
In summary, we report here for the first time an in vitro model for producing neural tissue with pre-defined and controllable rostro-caudal patterning equivalent to that of the early neural tube. In this model, we establish rostro-caudal patterning based exclusively on a single gradient of increasing WNT signalling in the absence of other morphogenic factors involved in neural tube patterning, such as WNT inhibitors21 and RA2. Such additional factors are likely important for refinement of boundaries and subdivision of regions and may be one reason the OTX2 boundaries in the MiSTR are not at the same sharpness as can be observed in vivo. Nonetheless, we show here that a WNT signalling gradient alone is sufficient to induce complex regionalisation of neural tissue with high reproducibility, extending from telencephalon over diencephalon and midbrain to anterior hindbrain and resulting in the formation of FGF-producing secondary organiser cells at the location of the MHB.
The rostro-caudal expression pattern of MiSTR tissues at 48 hours was similar to the expression pattern of the late epiblast and presumptive neuroectoderm in the mouse at E7.5. We could deduce from the model that rostro-caudal patterning was established prior to neural specification; something that has recently been confirmed for the segregation of spinal cord progenitors from anterior neural progenitors22, but which had not been shown for the segregation of hindbrain from forebrain neuroepithelium. With the model, we could deduce the first signs of a neural-specific transcription program emerging in the rostral cells, involving activation of HESX1, LHX5, SHISA2 and CYP26A1; factors which serve to inhibit WNT, FGF and RA signalling, respectively23–26, thereby likely safe-guarding rostral cells against caudalisation during neural plate formation. The caudal neuralising program appeared temporally delayed and the earliest molecular characteristics of this event remain to be determined. In summary, our study shows how a single morphogenic gradient can induce complex tissue regionalisation in vitro, and that the MiSTR model can be used as a reliable, predictable and accessible tool to investigate mechanisms of human rostro-caudal regionalisation and border formation.
Methods
hESC maintenance
Normally karyotyped, mycoplasma-free H9 and H1 hESCs (WiCell), RC17 hESCs (Roslin Cells) and hiPSC line C11 (generated at Lund University by J. Drouin-Ouellet and M. Parmar) were maintained on Matrigel (Corning) coated culture dishes in StemMACS iPS-Brew XF medium (Miltenyi Biotec), and passaged with EDTA (0.5 mM) once weekly.
Design and fabrication of the microfluidic gradient-generating cell culture system
The microfluidic cell culture system was developed as two main components, a) the microfluidic cell culture device, containing the hESC (connected by tubing to the external media-containing syringes and waste container), and b) a device holder with respective holding cassette (Fig. 1a and Supplementary Fig. 1a). The microfluidic device was fabricated in poly(dimethylsiloxane) (PDMS) using soft lithography27, comprising one bottom module with the Matrigel cavity (for cell seeding), and one top module, containing the microfluidic gradient generator and cell culture chamber (Fig 1a and Supplementary Fig. 1a, 1b). The serpentine channel gradient generator was designed to produce diffusive mixing of two inlet media under laminar flow in sequentially branching and recombining microfluidic channels11 (Fig. 1a; channel dimensions: 200 x 200 μm). In order to achieve sufficient spatial resolution to produce minimally overlapping rostro-caudal neural cell fates from forebrain to hindbrain, we designed the cell culture chamber of 20 x 10 x 0.5 mm (width x length x height). Furthermore, the length of the gradient (20mm) is approximately equivalent to the size of a post-conception week 8-10 human brain from forebrain to hindbrain.
The device holders and cassettes, as well as the injection moulds for the PDMS modules, were designed using CAD software (AutoCAD 2013/Fusion 360, Autodesk) and fabricated in plastic polyoxymethylene by micro-milling. The resulting design files (*.dxf) were exported to a CAD-CAM-CNC milling machine software (Galaad) to generate tool paths for the milling machine (ICP 4030, ISEL). The PDMS moulds were sealed with poly-(methyl-methacrylate) lids, each having an inlet and an outlet to allow for PDMS injection. PDMS (Sylgaard® 184 Silicone Elastomer Kit, Dow Corning), was mixed with a curer-to-base mass ratio of 1:10, and then degassed before injected into each mould, followed by curing at 60°C for 2h. The resulting solidified PDMS stamps were carefully peeled off and 1.25 mm diameter holes were punched out from the gradient generator PDMS stamp to allow for inlets and outlet tube connections to the device. The PDMS gradient generator and culture chamber stamps were aligned and bonded together28 to seal the microfluidics channels, thus forming the top PDMS module. Briefly, the two PDMS stamps were rolled onto a 10μm thin layer of uncured PDMS spun onto a glass, then aligned and assembled together. A final curing at 60°C for 2 hours ensured a robust bonding between the two stamps. One mm inner diameter silicone rubber tubes were glued to the inlets and the outlet of the top PDMS module using a silicone rubber glue (Elastosil E47, Wacker) enabling the connection of Teflon tubing from the syringes and to the outlet waste container. Inlet media were supplied with 10ml precision GasTight syringes (Hamilton - two syringes per device) mounted on low-pressure, high-precision, pulsating-free neMESYS pumps (Cetoni GmbH) to control the inlet flow. A total flow rate of 160 μl/h was applied (2x 80 μl/h from each syringe), which we calculated to produce minimal shear stress on the cells (see Supplementary Notes). The microfluidic device was sealed between a bottom holding cassette and a transparent polycarbonate lid, and held by the holder secured with screws (Fig. 1a and Supplementary Fig. 1a).
Quantitative measurement of gradient profile with fluorescein
The gradient profile in the cell culture chamber was measured using the fluorescence intensity of fluorescein (fluorescein sodium salt, FSS, Sigma-Aldrich) as a surrogate for GSK3i, since they have similar diffusion coefficients, calculated by the Stokes-Einstein equation29:
The entire cell culture chamber (20 x 10 mm) was imaged using an epifluorescence stereoscope (Olympus SZX) equipped with a cooled CCD camera (XC10, Olympus). Calibration images were acquired from eleven discrete dilutions of FSS in PBS (0μM, 0.05μM, 0.10μM…0.5μM). Each dilution was separately infused into the microfluidic device (both inlets) to obtain a stable fluorescent signal throughout the entire cell culture chamber. For each fluorescein dilution, 10 consecutive 14-bit grey-scale images of the fluorescence were obtained (cellSens Entry software, Olympus), before injecting PBS into the microfluidic device to remove all fluorescein residues. For acquisition of gradient images, 0.5μM fluorescein in PBS (= 100% concentration) and PBS only (= 0% concentration) were injected into the microfluidic device, and 10 images were acquired after 1 hour as a stationary fluorescein intensity gradient was established in the entire chamber. All images (calibration and gradient) were obtained using the same exposure and gain settings and subsequently processed, using an in-house developed image-processing algorithm in MATLAB (Mathworks, USA). Briefly, each set of 10 calibration images from the discrete concentrations (0%, 10%…, 100%) were averaged, assembled into a calibration stack, and an interpolation curve was made for all pixels sharing the same x and y coordinates in the stack. This procedure accommodated for any non-homogeneous illumination in the imaging set up. For each pixel in the averaged gradient image, the intensity was matched to the intensity of interpolation curve for the corresponding x,y pixel position in the calibration stack, thus converting the fluorescein pixel-intensity in the gradient image to a concentration value. As we calculated Péclet numbers >1 for both fluorescein and GSK3i, this indicated that their transport was mainly governed by convection and not diffusion. Hence, it was reasonably assumed that the GSK3i gradient profile was similarly linear and maintained across the cell culture chamber as was measured for FSS (Fig. 1b).
Assembly of the microfluidic cell culture system
Before each experiment, the bottom and top PDMS modules, the polycarbonate lid, the tubing and glass syringes were thoroughly cleaned with deionised water, and all were autoclaved separately. The holding cassette and device holder were sterilized by immersion in ethanol or by extensive ethanol wipe and dried at room temperature in sterile conditions – autoclaving of these parts was avoided to prevent any possible warping. The path from the glass syringes to the microfluidic serpentine was verified to be non-leaking and free of flow obstruction before each experiment.
All assembly steps were performed inside a cell culture flow hood, with previously sterilized MiSTR parts, to ensure sterility and prevent exposure to microparticles prone to clogging. On the day prior to initiation of differentiation (day −1), a matrix-bed was prepared by dispensing 200 μl of ice-cold Matrigel into the 1 mm deep cavity of the bottom PDMS module, placed inside the holding cassette (Supplementary Fig. 1a, b). Matrigel was allowed to gel at 37°C before cell seeding. Undifferentiated hESC colonies were detached using Accutase to yield a single-cell suspension, and 1.0-1.2*106 cells were seeded onto the Matrigel bed in 800μl of pluripotency medium (StemMACS iPS Brew XF) with 10 μM ROCK inhibitor (Y-27632). A sterile, temporary, custom-made PDMS wall was placed around the Matrigel cavity in the bottom PDMS module to contain the single-cell suspension, and the cells were cultured overnight at 37°. On day 0, the top PDMS module and attached inlet tubing, were first wetted with sterile Milli-Q water and bubbles flushed out from the whole media path. The same procedure was repeated 2 hours later, using a general usage syringe-pump (NE-4000, New Era Pump Systems), substituting the water with wash medium (DMEM/F12 supplemented with 5% knock-out serum replacement), while flushing out from the system any bubbles that may have formed. Once the full path (i.e. glass syringes, inlet tubing and microfluidic serpentine) was cleared of bubbles, the prepared bottom PDMS module with the seeded cells was brought from the incubator, the temporary PDMS wall was removed and the pluripotency medium was replaced wash medium. The bubble-cleared top PDMS module was then rapidly assembled onto the bottom PDMS module, both submerged in excess volume of wash medium to prevent any formation of bubbles in the device. The cassette holding the assembled device (with attached inlet tubing and media-filled glass syringes) was then inserted into the MiSTR holder and moved to a 37°C incubator. To minimise pump-related pressure differences, each pair (or two pairs) of media-containing syringes (0% and 100%) were mounted on a quadruple syringe holder attached on a single syringe pump module on the high-precision neMESYS pumps and the flow of differentiation media was initiated. The outlet tubing was connected to a sterile waste collector inside the incubator, located in a higher position than the MiSTR device to maintain a constant hydrostatic pressure in the outlet channels, thus minimising bubble nucleation.
Differentiation of hESCs in MiSTR
Differentiation in the custom-designed device was performed under a continuous flow (160 μl/h) of neural patterning medium (NPM) consisting of 1:1 mix of DMEM/F12 (Invitrogen) and NeuroMedium (Miltenyi Biotec), N2 supplement (1:200; Invitrogen), NeuroBrew-21 without vitamin A (1:100), supplemented with SB431542 (10 μM) and rh-Noggin (100 ng/ml) (all from Miltenyi Biotec). The NPM medium was devoid of Vitamin A (retinol), which is required for the synthesis of Retinoic acid. One syringe contained medium with added GSK3i, (100% side, CHIR99021, Miltenyi Biotec), the other without GSK3i (0%). For ventralising conditions, it was observed that the addition of SHH-C24II alone was not sufficient for efficient ventralisation in the MiSTR setup, although this has previously been shown to be sufficient in static cell culture conditions10. We hypothesised that this might be due to the absorption of SHH-C24II into the PDMS of the cell culture chamber. For efficient ventralisation, we therefore added a combination of SHH-C24II (200 ng/ml) and purmorphamine (0.5 μM, both from Miltenyi Biotec) to the medium of both inlet sides. Syringes were refilled with fresh medium every 2-4 days. On day 9 of differentiation, medium was changed to basal NPM medium without any added factors or GSK3i gradient, and kept at 160 μl/h until day 14 of differentiation. For 2 day Retinoic Acid (RA) gradient experiments, NPM was supplemented with SB431542 (10 μM) and rh-Noggin (100 ng/ml), and a gradient of 0 to 200nM of RA (Sigma-Aldrich). The differentiation protocol for 14 day hindbrain RA gradient experiments was adapted from a previous publication30: NPM was supplemented with SB431542 (10 μM) and rh-Noggin (100 ng/ml) (day 0 – 10), CHIR99021 (2 μM) (day 0-3), ventralizing factors (+SHH) as described above (day 3 – 14) and a 0 – 500nM gradient of RA from day 3 to day 14. See Supplementary Fig. 3c.
Immunostaining of MiSTR tissue samples
MiSTR tissue samples were fixed in 4% PFA for less than 1 hour, washed in PBS and embedded in OCT (Sakura) for cryosection, or stained as whole-mount (strips dissected along the longer axis under a stereoscope). Properly oriented frozen OCT-embedded samples were cryo-sectioned along the longer axis (i.e. along the gradient axis), and 14-18 μm thick sections collected on Superfrost Ultra Plus slides (ThermoFisher Scientific). For staining, sectioned and whole mount samples were washed in PBS and incubated for 1 hour in blocking buffer (PBS, 0,1% Triton X-100, 5% donkey serum), whole mounts with agitation. All antibodies were diluted in blocking buffer and incubated with samples overnight at 4°C. After incubation with primary antibodies, samples were washed extensively in PBS, and then incubated with species-matched, fluorophore-conjugated secondary antibodies (Jackson Laboratories) together with DAPI nuclei counter-stain. Whole mounts were mounted in Vectashield (Vector Laboratories), using coverslip spacers and sealed with nail polish. Cryosections were mounted in PVA-DABCO mounting media. See Supplementary Table 3 for list of primary antibodies.
Imaging
Images were obtained using a Leica DMI6000B epifluorescence system (20x 0.7 NA dry objective), or standard laser confocal system (Leica TCS SP8 or Zeiss LSM780) equipped with standard laser lines (405, 488, 561 and 633nm), using 10x 0.3 NA dry, 40x 1.3 NA oil-immersion or 63x 1.2 NA water-immersion objectives. Leica LAS X or Zeiss Zen Black software was used to perform tile scan imaging of large samples, which were stitched into one stack of images. Maximum projections of z-stacks were handled using Fiji (ImageJ, 1.52h). Uniform histogram corrections were performed using Adobe Photoshop CC, and figures were assembled using Adobe InDesign CC.
Quantitative real time reverse transcription PCR (qRT-PCR)
MiSTR tissue was sub-dissected into 5 equally sized samples using a custom-made standardised cutting tool. Tissue pieces were homogenised with QiaShredder column and RNA was isolated using RNeasy Micro kit (both from Qiagen). Reverse transcription was performed with random hexamer primers and Maxima First Strand cDNA Synthesis Kit (Thermo Scientific) using up to 1 μg of RNA from each sample. The cDNA was pipetted together with Sybr green Mastermix (Roche Life Sciences) and primers using the automated pipetting instrument Bravo (Agilent) or a liquid handler I-DOT (Dispendix), and samples were analysed by quantitative real-time PCR on a LightCycler 480 instrument (Roche Life Sciences) using a 2-step protocol with a 60°C annealing/elongation step. All qRT-PCR samples were run in technical duplicates, and the averaged Ct-values were used for calculations. Data are represented using the DDCt method. All fold changes are calculated as the average fold change relative to undifferentiated hESCs, based on 2 different housekeeping genes (ACTB and GAPDH). See Supplementary Table 4 for list of primers.
Quantification and statistical analysis
All qRT-PCR data (fold over undifferentiated hESC/hiPSC) was managed in Excel and statistically analysed using GraphPad Prism 7 software, p<0.05 was considered significant. For analysis between A-E regions within one condition and time point, one-way ANOVA was performed followed by a Tukey’s multiple comparison test between all regions. All datasets were tested for their normal distribution (Shapiro-Wilk test) and for equal variances (Brown-Forsythe test), and log transformed if needed before ANOVA. In case normal distribution or equal variance were not achieved, a non-parametric Kruskal-Wallis analysis was performed instead, followed by a Dunnett multiple comparison test. For analysis between dorsal and ventral conditions, and across time points, two-way ANOVA was performed on log transformed data followed by Sidak’s (dorsal vs ventral) or Tukey’s (day 2 vs. day 6 vs day 14) multiple comparison test. All multiple comparison tests were corrected using statistical hypothesis testing.
Rostral-caudal success exclusion criteria: Except for data presented in Fig. 1f, 1i, and Supplementary Fig. 1d, day 2-14 H9 MiSTR tissues that failed to pass the rostral-caudal success criterion were not used for analysis. Tissues failing this criterion accounted for 17.9% of all experiments (n=67, see Fig. 1j). Using relative mRNA expression data for OTX2 and GBX2 in each experiment, for regions A and E,
Location of OTX2-GBX2 border (Fig. 1k): arithmetic calculation between each neighbouring region, for each experiment using normalized OTX2 and GBX2 mRNA expression data,
Peak assessment (Fig. 2b): using the relative mRNA data for all regions A-E, value(s) scored above the 75 percentiles were considered the peak(s) in each experiment. For EN1 and WNT1, the peak difference to the average values (A-E) was 5960 fold ± 168.5 and 4852 fold ± 209.8 (mean ± SEM), respectively.
Single-cell RNA seq of MiSTR tissue
MiSTR tissue was divided into two halves along the gradient, one side used to collect the five A-E samples for mRNA analysis. The other side was dissociated into a single-cell suspension using Accutase (Thermofisher), for day 0, day 1 and day 2 samples, or with Neural dissociation kit (P) on a GentleMACS dissociator (both from Miltenyi Biotec) for day 14 samples. The cells were then frozen in CS10 CryoStor (Stemcell Technologies) and stored at −150°C until needed. In the meantime, each sample underwent mRNA analysis for quality control selection for scRNAseq. Cryopreserved cells, quality control cleared, were thawed, counted and mixed in equal numbers into a final single-cell suspension of 350 to 500 cells/μl solution in 0.01% BSA in PBS. For 0 to 48h tissues, cells from 3 biological MiSTR tissue replicates were pooled prior to sequencing. For day 14 tissues, the sequenced samples contained a mix of cells from regions A to E (n=1-3 biological replicates per region). A total of 9000-17000 cells were loaded per lane onto the 10X Chromium (10X Genomics) (see Supplementary Table 5) and cDNA libraries were generated according to manufacturer’s instructions. For day 14-ventral cells, each region from three biological replicates were thawed and pooled (in equal proportion) in 0.5% BSA in PBS, and incubated for 30 minutes with a unique cell hashing anti-human antibody (total 5 antibodies: TotalSeq A0251 – A0255; BioLegend). Further, cells from five regions were mixed in equal ratios and a total of 25000 cells were loaded on 10X lane using 10X V3 chemistry Kit. cDNA libraries (barcodes with hashtags and barcodes with mRNA expression) were produced according to manufacturer’s instructions. cDNA sequencing was performed on a NextSeq 500 (Illumina) according to the manufacturer’s instructions. Preliminary tests did not reveal significant differences between fresh and frozen cells (data not shown).
Processing, alignment and analysis of single-cell RNAseq data
Raw sequenced reads from day 0, 24-hour, 48-hour and day 14 dorsal tissues were processed by the 10X Genomics CellRanger2, while day 14 ventral (mRNA) reads were processed by 10X Genomics CellRanger 3.0 pipeline for alignment with the human reference genome (hg19) and generation of count matrices. Day 14 ventral hashtag reads were further processed by CITE-seq-count version 1.4.2 pipeline to generate hashtag count matrices.
Day 0, 24-hour and 48-hour analysis was performed using R version 3.5.0, and count matrices were then analyzed using the R package of Seurat version 2.3.4 for noise reduction PCA and data visualization. In order to remove experimental artifacts, cells with a gene count exceeding 2 times the median of genes per cell were identified as possible doublets and excluded from the analysis using the function FilterCells. In order to avoid biases towards biological processes related to cell metabolism, ribosomal and mitochondrial genes deleted from raw Seurat matrix using SubsetRow function of Seurat version 2.3.4. Similarly, cell cycle clustering-bias was mitigated by regressing out cell cycle genes (obtained elsewhere31) using the CellCycleScoring and the ScaleData functions from Seurat. Different MiSTR experiment matrices were integrated using the canonical correlation analysis implemented in Seurat version 2.3.4 based on the top 1000 highly variable genes from dataset, and further aligned using the AlignSubspace function. After integration of datasets, the highly variable genes were recalculated and used as a basis for Principle Component Analysis (PCA). Visualisation of gene expression was done on the PCA plots of the respective matrices.
Day 14-dorsal MiSTR, mouse gastrulation and pre-published organoids scRNAseq data analysis was performed using R package Seurat 3.0.2 on R version 3.6.1. Possible doublets (cell outliers with a gene count exceeding 2 times the median of genes per cell) and cells expressing more than 5% mitochondrial genes and 20% in case of day14 ventral multiplexed dataset were removed using Subset function of Seurat 3.0. To remove metabolism based bias ribosomal and mitochondrial genes were deleted from Seurat object and then further Seurat object was subjected to log normalization and cell cycle noise regression using NormalizeData, CellCycleScoring, ScaleData functions of Seurat 3.0 respectively. In order to compare mouse gastrulation scRNAseq data (downloaded from https://github.com/MarioniLab/EmbryoTimecourse2018/) with human cells based MISTR, the mouse gene names from raw data were mapped to human ortholog gene names using ensemble R package biomaRt_2.42.0 to create matrix containing human ortholog gene names. Further cell annotations from Marioni lab scRNAseq dataset were transferred to a new Seurat object. Day 14-dorsal tissue, mouse E8.5 cells, neural cells from mouse E8.5 and pre-published organoids scRNAseq datasets were integrated using an anchor based integrated strategy32 by implementing functions FindIntegrationAnchors and IntegrateData respectively on-to pre-processed datasets. In Supplementary Fig. 4f, visualisation of tissue clusters from the mouse dataset has been simplified compared to original annotations. Here, “Neural” is a compiled from the following clusters from the mouse E8.5 dataset: Neural crest, NMP, spinal cord, forebrain/midbrain/hindbrain and rostral neurectoderm, “Mesoderm” is compiled from: Caudal mesoderm, Exe mesoderm, Paraxial mesoderm, Intermediate mesoderm, Pharyngeal mesoderm, Somitic mesoderm, Mesenchyme, Cardiomyocytes and Allantois, “Endoderm” is compiled from: Def. Endoderm, Exe endoderm, Visceral endoderm and Parietal endoderm, “Blood/Endothelial” is compiled from: Blood progenitors 1/2, Erythroid 1/2/3, Endothelium and Haematoendothelial progenitors, “Non-neural, other” is compiled from Gut, PGC and notochord, “Surface ectoderm” represents surface ectoderm from the original dataset. For integration of MiSTR 14 days (dorsal) with relevant neural clusters from the mouse E8.5 dataset (Supplementary Fig. 5a), only clusters annotated as “forebrain/midbrain/hindbrain”, “rostral neurectoderm” and “Neural crest” from the mouse dataset were used.
Day 14-Ventral MiSTR cDNA library reads processing and different region cells demultiplexing was done by adding information from hashtag library as separate assay in main Seurat object and further implementing HTODemux function of Seurat 3.0. Clustering for day 14 dorsal (integrated) and ventral dataset was obtained by using modularity based Louvain clustering algorithm by running Seurat functions FindNeighbours and FindClusters, setting resolution parameter to 0.5. For visualization, Uniform Manifold Approximation and Projection (UMAP) plots were calculated using RunUMAP function of Seurat on the basis of pre-computed principle components.
Annotation of clusters was based on literature search and gene expression patterns from in situ hybridization data available from the Allen Institute for Brain Science (http://developingmouse.brain-map.org/) and from MGI Gene expression database (http://www.informatics.jax.org/expression.shtml), as well as comparison to gene expression patterns from the Mouse Brain Atlas (http://mousebrain.org).
RNA Velocity
For RNA velocity analysis Gene expression count matrixes including counts attributed unspliced RNA molecules have been obtained from the aligned reads using the Command Line Interface (CLI) developed as part of the Velocyto.py package16. Specifically, the tool velocyto run10x has been applied with default arguments and a gtf file corresponding to the human assembly GRCh37 has been passed as annotation. The output loom files comprise a four layered matrix one layer for spliced counts, one for unspliced, one for ambiguous counts and one summing all the counts. Further analysis and preprocessing was carried on using Python library SCANPY33 (version: https://github.com/theislab/scanpy/tree/dd5b6f759230fc33bd636f20d3e721dfbc1d5966)
As a part of our standard analysis, metabolism based bias was removed by deleting mitochondrial and ribosomal genes followed by normalization, log transformation and calculation of variable genes in loaded matrix by using pp.normalize_total and pp.log1p function respectively. In order to mitigate cell cycle effect, algorithm based on Seurat 3.2 (individual phase regression) was implemented using tl.score_genes_cell_cycle function followed by dataset integration based on MNN approach using pp.mnn_correct function. The clustering was based on Louvain clustering algorithm with the resolution set to 0.5 followed by visualization on UMAP space and then transferring UMAP coordinates for RNA velocity projections. RNA velocity was computed and visualized through the Python library scVelo Bergen. After reading the full dataset, preliminary data clean-up was carried using the function utilis.cleanup in order to remove unnecessary information from the loaded object. Filtering and normalization was performed using the function pp.filter_and_normalized with arguments min_shared_counts and n_top_genes respectively set to 10 and 3000. Analogously, possible doublets (cell outliers with a gene count exceeding 2.5 times the median of genes per cell) were excluded from the analysis using the function pp.filter_cells. Further pre-processing was carried using the function pp.moments, which computes moments for velocity estimation. Previously computed UMAP embedding information was then passed to the current object. Subsequently, gene-specific velocities were estimated using the function tl.velocity with the argument mode set to stochastic. The velocity graph was estimated using the function tl.velocity_graph and computed for the given embedding using the function tl.velocity_embedding. Finally, the results were visualized using the function pl.velocity_embedding_. Upon data visualization, a clearly separated cluster containing cells from all datasets (0, 24 and 48 hours) was evident in the bottom right corner of the plot. This cluster contained a mix of OTX2+ and GBX2+ cells and could not be attributed any biological meaning. In light of this, we speculate that this cluster resulted from technical artefacts and these cells were therefore disregarded during data interpretation.
Interrogation of gene expression in OTX2+ and GBX2+ cells from scRNAseq, 0-48 hours
MiSTR 0, 24-hour ventral, 48-hour dorsal and 48-hour ventral scRNAseq datasets were analysed with R version 3.5.3, Seurat version 3.1.2. The four individual samples were filtered for cells expressing gene counts under 5500, 6000, 4000 and 7000, respectively, and a mitochondrial gene expression under 7%, 10%, 5% and 8%, respectively. All samples were filtered for gene counts over 200. Each separate sample was processed using NormalizeData, FindVariableFeatures and ScaleData (with individual cell cycle regression) and integrated with batchelor version 1.2.4 fastMNN34 using 15 dimensions and automerge = TRUE. The integrated dataset was subset into 5 groups (0-hour, 24-hour OTX+, 24-hour GBX2+, 48-hour OTX+ and 48-hour GBX2+). OTX2+ and GBX2+ cells were defined as cells with an expression value > 0 for OTX2 and GBX2, respectively. For each group, positive markers were computed using FindMarkers (listed in Supplementary Table 1). All subsets were individually downsampled to 1500 cells for visualization with VlnPlot for selected markers.
hESC Mesodermal and Endodermal differentiation
To obtain positive controls for testing markers of mesodermal and endodermal differentiation (Supplementary Fig. 1c), H9 hESC were differentiated according to published protocols35, 36. In short, H9 hESCs were grow in pluripotency condition until full confluency and then changed to RMPI 1640 medium (Invitrogen) with 1:50 NeuroBrew-21 (Miltenyi Biotec), supplemented with 7.5 μM CHIR99021 (Miltenyi Biotec) for one day for Mesoderm induction35; or supplemented with 3 μM CHIR99021 on day 1, and with 100ng/ml of Activin A (Prepotech) for another 2 days, for Endoderm differentiation36.
Supplementary Material
Acknowledgements
This study was supported by the Novo Nordisk Foundation (NNF18OC0030286 to AK), The Lundbeck Foundation (R190-2014-3904 to THP), and the following to AK: Innovation Fund Denmark (BrainStem: 4108-00008A), the Strong Research Environment at Lund University Multipark, the Swedish Research Council (70862601/Bagadilico), The Crafoord Foundation, The Segerfalk Foundation, The Tore Nilsson Foundation, The Sven-Olof Janson Foundation and the Swedish Fund for Research Without Animal Experiments. The research leading to these results has received funding from the New York Stem Cell Foundation (MP), the European Research Council under the ERC Grant Agreement no. 30971 (MP), the Swedish Research Council (grant agreement 521-2012-5624, MP). The Novo Nordisk Foundation Center for Stem Cell Biology (DanStem) and the Novo Nordisk Foundation Center for Basic Metabolic Research (CBMR) are supported by Novo Nordisk Foundation grants (NNF17CC0027852 and NNF18CC0034900, respectively). MP is a New York Stem Cell Foundation Robertson Investigator. We thank Sol da Rocha Baez, Ingar Nilsson, Maria Madrona, Matias Heide Ankjær, Helle Kinggaard Lilja-Fischer (CBMR Single-cell Omics Platform), Helen Neil (DanStem Genomics Platform), Jutta Bulkescher (DanStem Imaging Platform) and Angeliki Meligkova (DanStem Stem Cell Culture Platform) for excellent technical and bioinformatics assistance and for use of instruments.
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Author Contributions
M.I., T.L., P.R., T.H.P., M.P., G.S.R and A.K.: Designed the study. M.I., P.R., G.S.R., P.A.K., D. M. R., G.B., K.L.E., O.K.M. and J.L.: Performed experiments. M.I., T.L., P.R. and A.K.: Wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests
Data availability
Single-Cell RNAseq data is deposited in the Gene Expression Omnibus (GEO) with accession number GSE135399. Single-Cell RNAseq code used for analysis is supplied as Supplementary Code in html and rmd files and can also be found on GitHub (https://github.com/kirkeby-lab).
References
- 1.Nordstrom U, Jessell TM, Edlund T. Progressive induction of caudal neural character by graded Wnt signaling. Nature neuroscience. 2002;5:525–532. doi: 10.1038/nn0602-854. [DOI] [PubMed] [Google Scholar]
- 2.Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci. 2005;6:553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- 3.Kiecker C, Lumsden A. The role of organizers in patterning the nervous system. Annu Rev Neurosci. 2012;35:347–367. doi: 10.1146/annurev-neuro-062111-150543. [DOI] [PubMed] [Google Scholar]
- 4.Ribes V, Briscoe J. Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harb Perspect Biol. 2009;1 doi: 10.1101/cshperspect.a002014. a002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meinhardt A, et al. 3D reconstitution of the patterned neural tube from embryonic stem cells. Stem Cell Reports. 2014;3:987–999. doi: 10.1016/j.stemcr.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demers CJ, et al. Development-on-chip: in vitro neural tube patterning with a microfluidic device. Development. 2016;143:1884–1892. doi: 10.1242/dev.126847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bagley JA, Reumann D, Bian S, Levi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birey F, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cederquist GY, et al. Specification of positional identity in forebrain organoids. Nat Biotechnol. 2019;37:436–444. doi: 10.1038/s41587-019-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkeby A, et al. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 2012;1:703–714. doi: 10.1016/j.celrep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Jeon NL, et al. Generation of Solution and Surface Gradients Using Microfluidic Systems. Langmuir. 2000;16:8311–8316. [Google Scholar]
- 12.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupe V, et al. In vivo functional analysis of the Hoxa-1 3’ retinoic acid response element (3’RARE) Development. 1997;124:399–410. doi: 10.1242/dev.124.2.399. [DOI] [PubMed] [Google Scholar]
- 14.Strate I, Min TH, Iliev D, Pera EM. Retinol dehydrogenase 10 is a feedback regulator of retinoic acid signalling during axis formation and patterning of the central nervous system. Development. 2009;136:461–472. doi: 10.1242/dev.024901. [DOI] [PubMed] [Google Scholar]
- 15.Yang L, et al. Analysis of FGF-dependent and FGF-independent pathways in otic placode induction. PLoS One. 2013;8:e55011. doi: 10.1371/journal.pone.0055011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Manno G, et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell. 2016;167:566–580 e519. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pijuan-Sala B, et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. 2019;566:490–495. doi: 10.1038/s41586-019-0933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quadrato G, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanton S, et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- 20.Stoeckius M, et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brafman D, Willert K. Wnt/beta-catenin signaling during early vertebrate neural development. Dev Neurobiol. 2017;77:1239–1259. doi: 10.1002/dneu.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metzis V, et al. Nervous System Regionalization Entails Axial Allocation before Neural Differentiation. Cell. 2018;175:1105–1118 e1117. doi: 10.1016/j.cell.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abu-Abed S, et al. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15:226–240. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andoniadou CL, et al. HESX1- and TCF3-mediated repression of Wnt/beta-catenin targets is required for normal development of the anterior forebrain. Development. 2011;138:4931–4942. doi: 10.1242/dev.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng G, Westerfield M. Lhx5 promotes forebrain development and activates transcription of secreted Wnt antagonists. Development. 2006;133:3191–3200. doi: 10.1242/dev.02485. [DOI] [PubMed] [Google Scholar]
- 26.Filipe M, Goncalves L, Bento M, Silva AC, Belo JA. Comparative expression of mouse and chicken Shisa homologues during early development. Dev Dyn. 2006;235:2567–2573. doi: 10.1002/dvdy.20862. [DOI] [PubMed] [Google Scholar]
- 27.Xia Y, Whitesides GM. Soft Lithography. Angewandte Chemie International Edition. 1998;37:550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 28.Satyanarayana S, Karnik RN, Majumdar A. Stamp-and-stick room-temperature bonding technique for microdevices. Journal of Microelectromechanical Systems. 2005;14:392–399. [Google Scholar]
- 29.Edward JT. Molecular volumes and the Stokes-Einstein equation. Journal of Chemical Education. 1970;47:261. [Google Scholar]
- 30.Maury Y, et al. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat Biotechnol. 2015;33:89–96. doi: 10.1038/nbt.3049. [DOI] [PubMed] [Google Scholar]
- 31.Nestorowa S, et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128:e20–31. doi: 10.1182/blood-2016-05-716480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kempf H, et al. Bulk cell density and Wnt/TGFbeta signalling regulate mesendodermal patterning of human pluripotent stem cells. Nat Commun. 2016;7 doi: 10.1038/ncomms13602. 13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Funa NS, et al. beta-Catenin Regulates Primitive Streak Induction through Collaborative Interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell. 2015;16:639–652. doi: 10.1016/j.stem.2015.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-Cell RNAseq data is deposited in the Gene Expression Omnibus (GEO) with accession number GSE135399. Single-Cell RNAseq code used for analysis is supplied as Supplementary Code in html and rmd files and can also be found on GitHub (https://github.com/kirkeby-lab).