Abstract
Taxanes are standard therapy in clinical practice for metastatic breast cancer (BC), however, primary or acquired chemoresistance are a common cause of mortality. BC patient-derived xenografts (PDX) are powerful tools for the study of cancer biology and drug treatment response. Specific DNA-methylation patterns have been associated to different BC subtypes but its association with chemoresistance remains unstudied. Aiming to elucidate docetaxel-resistance mechanisms, we perform genome-wide DNA-methylation in BC PDX models, including luminal and triple-negative BC (TNBC) models sensitive to docetaxel, their matched models after emergence of chemoresistance and residual disease after short term docetaxel treatment. We found that DNA-methylation patterns from BC PDX models maintain the subtype-specific methylation patterns of clinical samples. Two main DNAmethylation clusters were found in TNBC PDX and remain stable during the emergence of docetaxel resistance; however, some genes/pathways were differentially methylated according to docetaxel response. A DNA-methylation signature of resistance able to segregate TNBC chemotherapy response was identified. Transcriptomic profiling of selected sensitive/resistant pairs and integrative analysis with methylation data demonstrated correlation between some differentially methylated and expressed genes in docetaxel-resistant TNBC PDX models. Multiple gene expression changes were found after the emergence of docetaxel resistance in TNBC. DNA-methylation and transcriptional changes identified between docetaxel-sensitive and resistant TNBC PDX models or residual disease may have predictive value for chemotherapy response in TNBC.
Keywords: TNBC (triple-negative breast cancer), PDX (patient-derived xenograft), chemoresistance, docetaxel, DNA-methylation, transcriptomics
Introduction
Combination of different chemotherapy regimens, together with targeted therapies are used for the treatment of distinct breast cancer subtypes (1). Despite initial responses, relapses or metastatic disease usually become treatment resistant in breast cancer patients (2), and account for the main cause of death in these patients: chemoresistant breast cancer has a 5-year survival rate of around 25% (2). Triple-negative breast cancer (TNBC), the most heterogeneous and aggressive subtype of breast cancer, has higher rates of response to neoadjuvant chemotherapy than other breast cancer subtypes, but resistance rapidly emerges (3). Taxanes are considered among the most active classes of compounds against TNBC and metastatic breast cancer (4). Docetaxel, a taxane chemotherapeutic agent used broadly for treatment of different cancer types, acts by binding to microtubules and avoiding tubulin subunits depolimerization, inducing cell cycle arrest, apoptosis and cell death (5). Several mechanisms have been suggested to confer docetaxel resistance for different cancer types, as prostate (6), gastric (7) and ovarian (8). In breast cancer, the best known chemoresistance mechanism is the modulation of drug efflux proteins (9). However, small molecule inhibitors of drug efflux proteins have being tested in patients with limited clinical success (9). Other molecular mechanisms of docetaxel resistance implying gene expression deregulation and epigenetic mechanisms have been proposed (10). Most studies have been performed in vitro using breast cancer cell lines (BCCLs), with limited predictive value in the clinics (11,12), or focusing on gene expression signatures of patient populations, where heterogeneity, particularly in TNBC is a major concern (13,14). Further insights to clarify the emergence of docetaxel resistance are urgently required. One important limitation is the availability of paired clinical samples before and after acquisition of resistance.
DNA-methylation-specific alterations are known hallmarks of human cancer (15,16). Several studies have also identified DNA-methylation signatures that can distinguish between breast cancer subtypes (17,18), and others that may be predictive of treatment response (19,20). DNA-methylation changes can contribute to cancer development through inactivation of tumor suppressor genes (BRCA1 and others) and, conversely, through activation of oncogenes (21). DNA-methylation has been also uncovered as an important clinical marker of drug response and resistance in breast cancer in some contexts (21,22).
Patient-derived xenograft (PDX) models have been proposed as a preferred tool for conducting basic and translational preclinical research, being biologically closer to patient tumors than cancer cell lines (11,12). Breast cancer PDX models maintain not only histopathological features from human tumors but also retain main genomic, transcriptomic and proteomic profiles and drug response (11,17). However, methylation patterns of breast cancer PDX models and their correlation with human breast tumors remain understudied. Multiple investigations support the relevance of PDX models as powerful preclinical tools for the study of drug resistance, but it is necessary to determine whether breast cancer methylation patterns are represented and maintained in PDX models to establish their suitability for preclinical methylation studies.
We have generated a panel of breast cancer PDX models comprising all histopathological subtypes (23). Mimicking the clinical scenario, TNBC PDX models respond to docetaxel while luminal PDX models proved to be resistant. However, after continuous in vivo exposure to docetaxel, initially sensitive TNBC PDX models become resistant (23). These paired sensitive and resistant TNBC PDX models, as well as the residual disease from sensitive TNBC PDX models, solve the gap between in vitro results and clinical samples and constitute powerful tools for the study of docetaxel chemoresistance.
We hypothesize that DNA-methylation and transcriptional changes contribute to the emergence of chemoresistance to docetaxel in TNBC patients. Here we performed genome-wide DNA-methylation in a panel of 12 breast cancer PDX models, corresponding residual disease after docetaxel treatment for the 9 TNBC, and paired chemoresistant-derived TNBC PDX tumors generated. Transcriptional profiles for selected docetaxel-sensitive/resistant TNBC PDX pairs were also obtained. We demonstrated that human breast cancer methylation patterns are conserved in breast cancer PDX models and can discriminate different subgroups within the heterogeneous TNBC subtype. Although global methylation is preserved during the emergence of chemoresistance, some gene-specific methylation changes are detected. Transcriptionally, multiple genes were differentially expressed between paired sensitive and resistant TNBC PDX tumors. Differentially methylated/expressed genes were associated to specific pathways. Similar pathways were found between methylation and expression analysis, suggesting that some gene expression changes are controlled by methylation during emergence of chemoresistance to docetaxel in TNBC PDX tumors, although most of the transcriptional changes identified were not related to methylation.
Materials and methods
Patient Characteristics and Generation of PDX
All human samples were obtained following institutional guidelines. Written informed consent for PDX generation and whole exome sequencing was obtained from all subjects and the study received approval from the corresponding institutional Ethics Committee in accordance with the Declaration of Helsinki. All research involving animals was performed in compliance with protocols approved by the Institutional Committees on Animal Care and adhering to European Union and international regulations. PDX models were generated as previously described (23). IDB PDX (TNBC IDB-01, IDB-02, IDB-09, luminal IDB-03 and IDB-04 and the HER2+ IDB-05), BCM PDX (BCM-4664, BCM-9161) and HCI-001 PDX models were generated by orthotopic transplantation of human fresh tumoral tissue or injection of metastatic cancer cells isolated from pleural effusions into the cleared mammary fat pad of immunodeficient mice, as described previously (23–25). VHIO models (VHIO-98, VHIO-127 and VHIO-270) were generated by subcutaneous implantation of primary tumor pieces or metastasis on the back of Foxn1nu mice as described in (26). VHIO, BCM and HCI PDX models were implanted in the intact fact pad of NOD.CB17-Prkdcscid/J mice of our animal facility where short term treatments with docetaxel (Residual Disease) and generation of resistant variants (IDB-09R, VHIO-98R and VHIO-127R) was performed as described (23). DNA-methylation profiles from paired sensitive and resistant TNBC PDX and/or residual disease previously described (23,27) and human samples of origin (when available) were obtained. All resistant TNBC PDX models used in this study were maintained under in vivo selective drug pressure and they were completely resistant [passage 3 of resistance for all model except VHIO-98R (passage 4 of resistance)]. Residual disease and resistant TNBC PDX tumors were collected between four and seven days after the last docetaxel treatment. Also, it is important to mention that IDB-02S model was partially sensitive, but from now on will be considered as sensitive to facilitate nomenclature.
DNA Extraction and RNAse Treatment
Total DNA from tissue was prepared with in house lysis buffer (100mM NaCl, 10mM Tris-Cl pH 8, 25mM EDTA). Frozen tumor tissues were fractionated using the POLYTRON® system PT 1200 E (Kinematica) and incubated 12-16 hours with 0.25% SDS (Invitrogen, 24730020), 0.25 mg/ml Proteinase K (Sigma Aldrich, P4850) and RNAse A (Sigma Aldrich, R5503) at 55°C in a termal block. For DNA purification, the homogenized sample was transferred to a phase lock gel heavy tube (VWR, 713-2538). Two steps of removal of proteins from nucleic acids using Phenol:Chloroform:Isoamyl Alcohol 25:24:1 (Sigma Aldrich, P3803) and three steps of nucleic acid washing with chloroform (VWR, 1024311000) were performed, centrifuging each time at 1500G during 5 minutes. After last spinning, remaining volume containing DNA was recovered and transferred to a solution containing 2.5x absolute ethanol (Merck Millipore, 1009832500) and 30 mM of sodium acetate pH 5.2 (Sigma Aldrich, S2889), mix and centrifuge at maximum for 5 minutes. Wash two times with 70% ethanol centrifuging at maximum for 5 minutes. Finally, resuspension in TE buffer (10 mM Tris-HCl, 1 mM disodium EDTA, pH 8.0) or ultrapure water (MilliQ purification system) was performed. All DNA samples were quantified by the fluorometric method (Quant-iT PicoGreen dsDNA Assay, Life Technologies, CA, USA), and assessed for purity by NanoDrop-1000 Spectrophotometer (Thermo Scientific, MA, USA) 260/280 and 260/230 ratio measurements. DNA integrity of samples was checked by electrophoresis in an agarose gel.
Bisulfite Conversion and Genome-Wide DNA-methylation Microarray
Five hundred nanograms of purified genomic DNA were treated with sodium bisulfite using the EZ DNA-methylation kit (Zymo Research, CA, USA). The incubation profile was 16 cycles at 95°C for 30 s, 50°C for 60 min and a final holding step at 4°C. 4 μl of bisulfite-converted DNA (50 ng/ul) were used for hybridization on Infinium Human Methylation 450K BeadChip, following the Illumina Infinium HD Methylation protocol. This consisted of a whole genome amplification step followed by enzymatic end-point fragmentation, precipitation and resuspension. The resuspended samples were hybridized on Human Methylation 450K BeadChips at 48°C for 16 h. Then unhybridized and non-specifically hybridized DNAs were washed away, followed by a single nucleotide extension using the hybridized bisulfite-treated DNA as a template. The nucleotides incorporated were labeled with biotin (ddCTP and ddGTP) and 2,4-dinitrophenol (DNP) (ddATP and ddTTP). After the single base extension, repeated rounds of staining were performed with a combination of antibodies that differentiated DNP and biotin by fixing them different fluorophores. Finally, the BeadChip was washed and protected in order to scan it.
The Illumina HiScan SQ scanner (Illumina, CA, USA) is a two-color laser (532 nm/660 nm) fluorescent scanner with a 0.375 μm spatial resolution capable of exciting the fluorophores generated during the staining step of the protocol. The intensities of the images were extracted and raw IDAT files were processed with Illumina’s GenomeStudio software and statistical analysis was done in R with minfi package. The methylation score for each CpG was represented as a β-value according to the fluorescent intensity ratio. β-values may take any value between 0 (non-methylated) and 1 (completely methylated) and they were used for all downstream analyses. All downstream analysis was conducted using the hg19/GRCh37 human genome assembly.
Methylation Clustering
Confirmation that samples from same donor were properly analysed, was assessed by confronting 59 SNP values obtained within the Illumina Infinium Human Methylation 450K BeadChip and the Illumina Infinium Human MethylationEPIC BeadChip microarray from the different samples. Scatter plots of β-values for PDX models and human samples of origin were produced to check if there was a degree of difference between the models. Afterwards, an analysis of methylation variance, comparing PDX models was applied to β-values obtaining the CpGs statistically significant (FDR < 0.01) taking into account a methylation difference between at least one of the groups equal or higher to 75%. An unsupervised heatmap representation of the PDX models was generated using those CpGs. Organizing the values by applying a hierarchical clustering method based on Manhattan distances aggregated by Ward’s linkage. An analysis of methylation variance revealed a minimum of 743 CpGs differentially methylated CpGs between TNBC vs luminal subtype from TCGA breast cancer patients, and then were applied to classify PDX models and breast cancer cell lines (BCCLs). Data from primary tumor samples were obtained from TCGA data portal (https://portal.gdc.cancer.gov/) while data from BCCLs were obtained from (28). DNAmethylation data from TCGA derives from Illumina Infinium Human Methylation 450K BeadChip platform which interrogates approximately 450,000 CpG sites in comparison with the EPIC microarray which interrogates 850,000 CpG sites approximately; around 90% of the Infinium Human Methylation 450K BeadChip microarray is still found in the Infinium MethylationEPIC BeadChip platform. Thus, data was merged to keep around 452,256 CpG sites present in both methylation platforms.
Unsupervised heatmap representation of the PDX models and 100 human mammary gland samples (from TCGA) was generated using randomly selected 1% of the CpG sites and organizing them by applying a hierarchical clustering method based on Manhattan distances aggregated by Ward’s linkage.
Pyrosequencing
Pyrosequencing assays were designed to analyze and validate the results obtained from the array under different scenarios. Sodium bisulfite modification of 1 μg of genomic DNA isolated from breast cancer PDX tumors was carried out with the EZ DNA-methylation Kit (Zymo Research Corporation) following the manufacturer’s protocol. Bisulfite-treated DNA was eluted in 30-μL volumes with 2 μL used for each PCR. The set of primers for PCR amplification and sequencing were designed with a specific program (PyroMark assay design version 2.0.01.15). Primer sequences were designed to hybridize with CpG-free sites to ensure methylation-independent amplification. PCR was performed with primers biotinylated to convert the PCR product to single-stranded DNA templates. We used the Vacuum Prep Tool (Biotage) to prepare single-stranded PCR products according to the manufacturer’s instructions. Pyrosequencing reactions and quantification of methylation were performed in a PyroMark Q96 MD version 2.0.6 (QIAGEN). Primers indicated below are in 5’ → 3’ direction.
Primers
| hSLC25A30 Forward | AGTTTTTATTGGTTTTTGTTAGTATTAGT |
| hSLC25A30 Reverse Biotinylated | TTCCCCAAATTTCTCTTCCACC |
| hSLC25A30 Forward PyroSeq | TGATAGTTTTAGATGGGGATA |
RNA Extraction and Gene Expression Microarray
Total RNA from tissue was prepared with Tripure Isolation Reagent (Roche). Frozen tumor tissues from sensitive and resistant IDB-01 and IDB-02 TNBC PDX models were fractionated using the POLYTRON® system PT 1200 E (Kinematica). Two-hundred-nanogram aliquots of total RNA were used for the production of fluorescent complementary RNA following the Two-Color Microarray-Based Gene Expression Analysis v. 6.5 (Agilent) protocol under manufacturer’s instructions. All samples were hybridized to the SurePrint G3 Human Gene Expression 8 × 60 K microarray (Agilent Technologies). The signal values were extracted using the Feature Extraction software (Agilent Technologies). After scanning and normalization processes, all the statistical treatment was realized under an R programming environment using Bioconductor’s package for gene expression analysis: Limma, RankProd, Marray, affy, pcaMethods, EMA y RamiGO.
Differentially expressed genes, after Limma analysis, were represented as a mean-centered gene expression graphs using web-based tool Morpheus from Broad Institute (29).
Correlation between Gene Expression and Methylation
To study the association between gene expression and DNA-methylation at gene level, data derived from both arrays was filtered to obtain the mean methylation value for each gene found in the expression array and annotated in the Infinium Human Methylation 450 BeadChip Array. In the case of methylation, CpG site probes falling on the promoter region of the known genes were considered, i.e., TSS1500, TSS200, 5’UTR, and 1st exon. Methylation beta values of CpG islands were averaged across CpG sites. A Pearson correlation test was performed for all the genes in the Expression Array, first correlating the genes intra models IDB-01 and IDB-02 and later calculating the correlation within the sensitive and resistant samples independently. Density plots were created with the correlation values per model or based on the sensitive/resistant conditions. Also, for those genes that were previously considered as differentially expressed (logFC >=1.5 and adj.P.Val <=0.05). A heatmap with the most differentially expressed genes was generated organizing the mean expression values by applying a clustering method based on Manhattan distances, and drawing its equivalent mean methylation values with its corresponding gene.
Statistical analysis and graphical representation were performed with R programming language (version 3.4 2017-04-21) and limma (version 3.30.13) and ggplot2 libraries. Significant differences between samples/models were assessed using Wilcoxon, Pearson or Chi-tests were appropriate with values of p<0.05 considered to be significant.
Gene Ontology/Pathway Analyses
To identify functional clusters of genes differentially methylated and/or differentially expressed, we performed Functional Annotation Clustering using DAVID (30) on the candidate genes obtained from sensitive/resistant pairs and sensitive/residual disease pairs comparisons.
GSEA Analysis
Gene Set Enrichment Analysis (GSEA) is freely available and is supported by the Broad Institute website (http://software.broadinstitute.org/gsea/index.jsp) and includes versions compatible with Java, R or Gene Pattern. All GSEA analyses presented here were performed using the R GSEA implementation.
DNAse Treatment and qRT-PCR
Prior to cDNA conversion, RNA was treated with DNA-free DNase I kit (Ambion, AM1906). cDNA was produced by reverse transcription using 1 μg of DNA-free RNA in a 35 μL reaction following TaqMan™ Reverse Transcription instructions (Applied Biosystems, N8080234). 20 ng/well of cDNA were used for the analysis performed in triplicate. Quantitative PCR was performed using the LightCycler® 480 SYBR green. Primer sequences are indicated below. Ct analysis was performed using LightCycler 480 software (Roche). All primers indicated below are in 5’ → 3’ direction.
| hEGFR Forward | CCTGTCTGGAAGTACGCAG |
| hEGFR Reverse | GCGATGGACGGGATCTTAGG |
| hERBB3 Forward | CCGCTTGACTCAGCTCACC |
| hERBB3 Reverse | CACGATGTCCCTCCAGTCAAT |
| hSLC25A30 Forward | ACTGCTGAGTGCGGTACATT |
| hSLC25A30 Reverse | GTCCTCTTGTCCCCTCTTGC |
| hPPiA Forward | ATGCTGGACCCAACACAAAT |
| hPPiA Reverse | TCTTTCACTTTGCCAAACACC |
Statistical Analyses
All data are expressed as mean ± SEM. Statistical comparison was performed by Student’s t test using GraphPad Prism version 5.04. p ≤ 0.05 was considered statistically significant. The statistical significance of difference between groups is expressed by asterisks: *0.01 < p < 0.05; **0.001 < p < 0.01; ***0.001 < p < 0.0001; ****p < 0.0001.
Results
DNA-methylation patterns of breast cancer PDX resemble human primary breast tumors
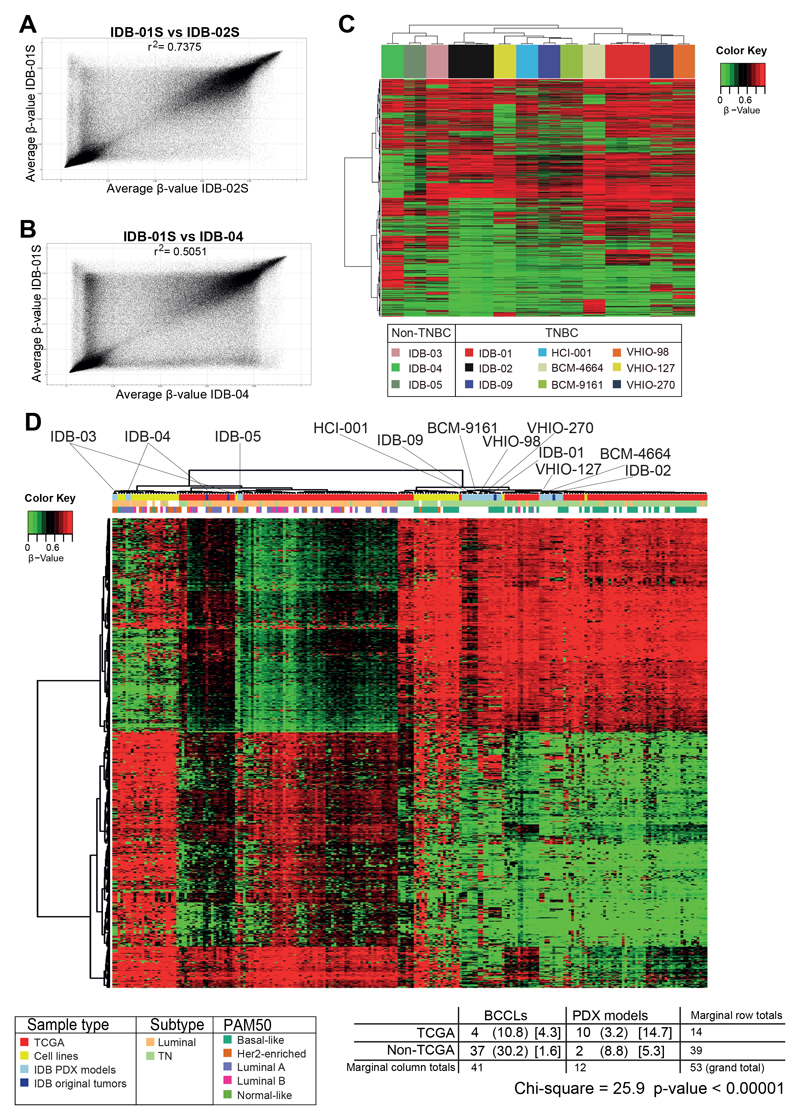
Hundreds of breast cancer PDX models have been established around the world (13), but whole-genome DNA-methylation analysis has not yet been reported. DNA-methylation status was analyzed for breast cancer PDX models and human tumors of origin when available. Methylation status was interrogated in 12 independent breast cancer PDX models previously generated (23–25) and four out of five human tumors of origin using the Infinium Human Methylation 450K and Infinium MethylationEPIC BeadChip Array from Illumina. The PDX models analysed include two luminal (IDB-03 and IDB-04), one ER+PR+HER2+ (IDB-05) and nine TNBC (IDB-01, IDB-02, IDB-09, HCI-001, BCM-4664, BCM-9161, VHIO-98, VHIO-127 and VHIO-270) all found to be sensitive to docetaxel (23, 27). Samples of residual disease after short term treatment with docetaxel for all sensitive TNBC PDX models and the matched PDX resistant variants (IDB-01R, IDB-02R, IDB-09R, VHIO-98R and VHIO-127R), generated after continuous docetaxel in vivo treatment were also included (23, 27) (Supplementary Figure S1A). The Infinium Human Methylation 450K BeadChip array contains 485,512 probes covering 99% of RefSeq genes. The Infinium MethylationEPIC BeadChip array contains over 850,000 probes, which cover more than 90% of the sites on the Infinium Human Methylation 450K BeadChip array, plus more than 350,000 novel CpGs at regions identified as potential enhancers in the FANTOM5 project (31). A SNP analysis was performed to validate the identity of the PDX models and the human original tumors from which they derived when available (Supplementary Figure S1B). To minimize differences due to intratumor biological variability, two/three biological replicates per condition were included in the analyses. A first approach comparing whole-genome DNA-methylation using all CpG sites β-values between sensitive TNBC PDX models (Figure 1A) or between a TNBC and a luminal PDX model (Figure 1B) revealed a higher correlation between breast cancer PDX models from the same subtype. Then, unsupervised analysis of most differentially methylated CpGs between breast cancer PDX models showed two major clusters that correlate with breast cancer subtypes: a cluster for TNBC PDX models and a cluster for non-TNBC PDX models (Figure 1C). Breast cancer PDX model IDB-03, which has lost estrogen and progesterone receptor expression (ER and PR, respectively), but expresses androgen receptor (17) (AR, data not shown) clusters with non-TNBC PDX models (Figure 1C). This result indicates that despite the loss of ER/PR in the breast cancer PDX model the global DNA-methylation profile is still luminal-like. Within the TNBC subgroup, segregation in two major DNA-methylation clusters is also observed (Figure 1C). One cluster contains the models IDB-02, VHIO-127 and BCM-9161, bearing BRCA1 germline mutations, as well as IDB-09 and HCI-001. A second cluster contains the non-BRCA1 mutated models IDB-01, VHIO-98, VHIO-270, and BCM-4664 (Figure 1C). Thus, segregation in two major DNA-methylation clusters based on hormone receptor expression observed in breast cancer patients (17) is maintained in our breast cancer PDX models, and two main TNBC methylations clusters, one of them enriched in BRCA1 mutated tumors, were found.
Fig. 1. Global DNA-methylation patterns of breast cancer PDX resemble breast cancer human samples.
A, B) Scatter plot of groupwise mean genome-wide DNA-methylation levels analyzed by GenomeStudio between two sensitive TNBC PDX models (A) or between a TNBC and a luminal PDX model (B). Correlations are indicated as r2. C) Unsupervised hierarchical clustering using 32.264 most differentially methylated CpGs between breast cancer PDX models. Methylation difference between at least one of the groups is equal or higher to 75% and p-value < 0.01. D) Supervised hierarchical clustering applying 743 selected CpGs that discriminate TNBC vs luminal subtype from TCGA breast cancer to BCCLs, breast cancer PDX models and human tumor samples of origin and TCGA clinical samples. Breast cancer PDX models are indicated. Chi-square and corresponding p-value indicating association between TCGA breast tumors and PDX models or BCCLs from the same subtype is indicated below.
Methylation patterns from breast cancer PDX models and human tumors of origin, TCGA breast cancer clinical samples (17) and breast cancer cell lines (BCCL) (28) were compared. A supervised cluster using the subtype methylation patterns extracted from TCGA revealed that each breast cancer PDX model and human tumor of origin was classified with human breast cancer samples from the same subtype (Figure 1D, Supplementary Table S1I). Ten out of the twelve breast cancer PDX models were mixed with TCGA breast tumors, while virtually all BCCLs classify separately from TCGA breast tumors (Figure 1D, chi square: 25.9 p value < 0.00001). This result demonstrates that subtype-specific global DNAmethylation from clinical breast cancer samples is conserved in breast cancer PDX models to a greater extent than in BCCLs. Distinctive DNA-methylation patterns for the different breast cancer subtypes that correlate with clinical implications have been previously described in a collection of clinical samples containing TNBC (EpiBasal) and non-TNBC (EpiLum), cohort 1 (32). A supervised hierarchical clustering of our breast cancer PDX models using these methylation signatures resulted in the classification of our TNBC (basal-like) PDX models as EpiBasal and the non-TNBC (luminal and triple-positive) PDX models as EpiLumB, including the IDB-03 model, despite the loss of ER/PR expression (Supplementary Figure S1C). This data supports the idea that the subtype-specific methylation patterns showed in primary breast tumors are maintained in breast cancer PDX models with clinical significance. Genome-wide DNA-methylation status from breast cancer PDX models and human mammary glands was compared (33,34), revealing a general hypomethylation pattern in breast cancer PDX models, while hypermethylation in some gene-specific CpG sites (Supplementary Figure S1D), which is also observed in original patient’s tumors. This result shows that variations in methylome between human mammary gland and human breast cancer are also conserved in breast cancer PDX models. Together these data reinforce the relevance of breast cancer PDX models to accelerate the translation into clinics of DNAmethylation studies.
Genome-wide DNA-methylation comparisons between paired sensitive and docetaxel resistant TNBC PDX models or residual disease evidences no global methylation changes
As DNA-methylation changes in BCCLs have also been shown to contribute to taxane chemoresistance (10,35), we investigated whether changes in DNA-methylation patterns could be associated with docetaxel resistance in TNBC PDX models. We analysed DNAmethylation patterns in paired sensitive and resistant (IDB-01S/R, IDB-02S/R, IDB-09S/R, VHIO-98S/R and VHIO-127S/R) and paired sensitive and residual disease (IDB-01S/RD, IDB-02S/RD, IDB-09S/RD, VHIO-98S/RD, VHIO-127S/RD, VHIO-270S/RD, HCI-001S/RD, BCM-4664S/RD, BCM-9161S/RD) TNBC PDX models.
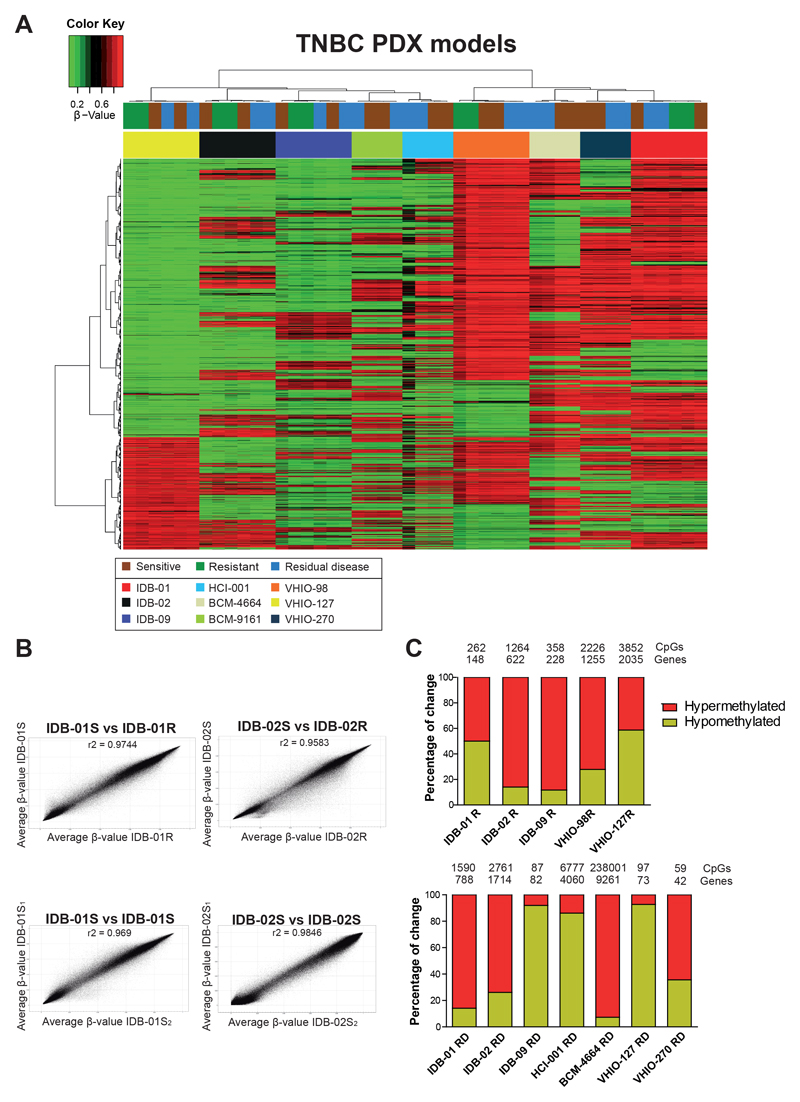
Unsupervised methylation cluster using the ten thousand most variable CpGs between all TNBC PDX models showed a clear separation between TNBC PDX models and no segregation based on docetaxel response (Figure 2A); however, within some models (VHIO-98, VHIO-127, VHIO-270 and BCM-4664) some separation between sensitive, resistant and residual disease was observed (Figure 2A). CpG site methylation levels were highly correlated between sensitive and resistant tumors from the same TNBC PDX model, to a similar extent to that achieved when two sensitive tumor replicates from the same TNBC PDX model were compared (Figure 2B, Supplementary Table S1A). Principal component analyses also revealed a similar distribution of models, IDB-02 and VHIO-127 showing overlapping positions, BCM-9161, HCI-001 being the closer ones to IDB-02, whereas BCM-4664, VHIO-98, VHIO-270 were closer to IDB-01S (Supplementary Figure S2A). IDB-01S and IDB-02S seem the most different ones according to this classification. This result suggests, on one hand, that methylation profile of residual disease is similarly close to that of resistant or sensitive tumors and, on the other hand, that most differences in DNAmethylation are due to intratumor heterogeneity, ruling out global methylation changes during the emergence of docetaxel resistance.
Fig. 2. Genome-wide DNA-methylation patterns between docetaxel sensitive, resistant and residual disease TNBC PDX models.
A) Unsupervised hierarchical methylation clustering using the 10.000 most variable CpGs between TNBC PDX models including sensitive (untreated), residual disease and resistant tumors. All resistant tumors were collected at passage three of treatment, one week after last DTX treatment (23,27). Sensitive tumors were collected from a similar passage than resistant ones, to discriminate DTX specific changes with those related with serial passages. Residual disease was collected after two to eight doses of docetaxel when tumors were shrinking B) Scatter plot of groupwise mean genome-wide DNA-methylation levels analyzed by GenomeStudio between sensitive and resistant pairs and between two different sensitive tumors from the same model. Correlations are indicated as r2. See also Supplementary Table S1A. C) Representation of differentially methylated sites indicating direction of change when comparing sensitive and resistant TNBC PDX tumors (top panel) or sensitive and residual disease (bottom panel). Total number of differentially methylated CpGs and genes for each TNBC PDX model are indicated. CpGs were selected by a methylation change higher than 30% and a standard deviation lower than 0.05 between samples from the same group. The number of differentially methylated CpG from BCM-9161 RD (0 CpG) and VHIO-98RD (2 CpG) was not enough to perform these analyses.
A comparison of whole genome DNA-methylation of sensitive and resistant groups based on CpG genomic region and CpG context revealed a genome-wide DNA-methylation increase in the promoter and CpG island context in the five chemoresistant TNBC PDX models (Supplementary Figure S2B-C). A similar genome-wide DNA-methylation increase was also observed in residual disease of most TNBC PDX models (Supplementary Figure S2B-C).
To identify key DNA-methylation changes associated with response to docetaxel, differentially methylated CpGs between sensitive and resistant/residual disease groups from all TNBC PDX models were extracted. Comparison of DNA-methylation at single base pair resolution using a minimum absolute methylation difference of 30% revealed high number of differentially methylated CpGs (hundreds to thousands) for some models (IDB-01, IDB-02, HCI-001, BCM-4664), and very few or none in others (BCM-9161, VHIO-98, VHIO-127) (Figure 2C, Supplementary Table S1B). Hypermethylation was found in some resistant TNBC PDX models (IDB-02R, IDB-09R, VHIO-98R) or residual disease (IDB-01RD, IDB-02RD, BCM-4664 or VHIO-270), but hypomethylation was also observed (IDB-09RD, HCI-001RD or VHIO-127RD) (Figure 2C).
Some of the differentially methylated sites observed in the resistant models or residual disease were located in the promoter and CpG islands, but most of the differentially methylated sites in the resistant models were located in intergenic regions and open sea sites; whereas in residual disease they were found in the body and open sea sites (Supplementary Figure S3A-B). Comparison of differentially methylated CpGs between resistant TNBC PDX models revealed little overlap between them (Supplementary Figure S3C). Comparisons of residual disease were done separately for the two clusters, but little overlap was also observed (Supplementary Figure S3D).
Analysis from Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were conducted on DAVID bioinformatics web-based tool (https://david.ncifcrf.gov/). This analysis revealed alterations in common pathways between several resistant TNBC PDX models, such as calcium signalling, plasma membrane, cell junction and cell adhesion. In residual disease of the BRCA1-enriched cluster main common pathways included calcium ion binding, cytoskeleton, but also GTPase activity, PI3K activity or Ras signalling pathway (Supplementary Table S1C-D).
Together these data suggest that emergence of chemoresistance to docetaxel in TNBC is not driven by global DNA-methylation changes nor single CpGs or genes between all TNBC PDX models, as no common differentially methylated CpGs or genes between TNBC PDX models were found. However, within each model methylation changes were observed in the docetaxel resistant-derived TNBC PDX models and residual disease.
DNA-methylation signature of resistance may predict response to chemotherapy in TNBC
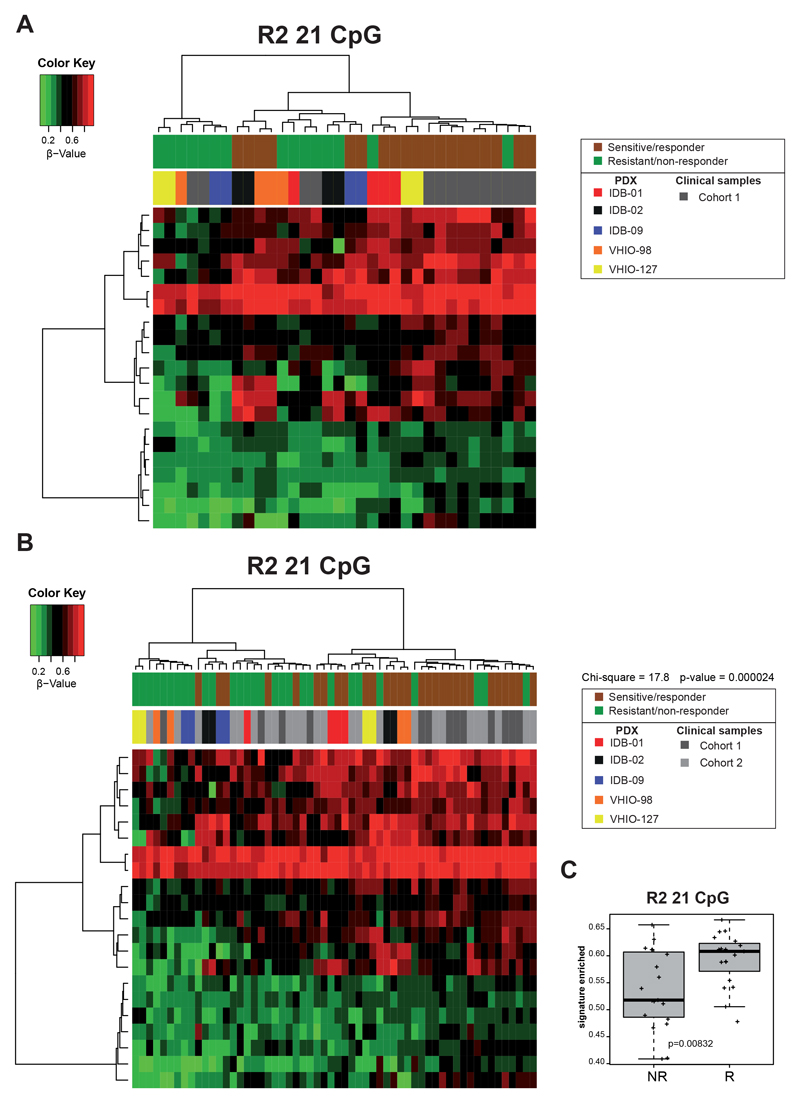
Next, we aimed to investigate the relevance of the DNA-methylation changes identified after short- or long-term treatment with docetaxel in our TNBC PDX models on basal-like clinical samples. We generated a methylation signature of docetaxel residual disease (RD1, 76 CpG) combining data from the 9 different pairs of S/RD TNBC PDX models and a methylation signature of docetaxel resistance (R1, 26 CpG) combining data from the 5 different pairs of S/R TNBC models analyzed (Supplementary Table S1E-F). Using the RD1 signature, TNBC PDX tumors do not longer cluster by model and two main clusters, one enriched in sensitive tumors and a second one enriched on residual disease were observed (Supplementary Figure S4A). Similarly, the R1 signature, segregate the PDX tumors in two clusters, one composed exclusively by resistant tumors, and a second one composed mainly by sensitive tumors (Supplementary Figure S4B).
We then tested these signatures in two independent TNBC clinical cohorts where DNAmethylation profiles were previously characterized (32,36). Cohort 1 contains samples from 14 basal-like breast cancer patients (9 alive/responders, 5 exitus/non responders, with no treatment information available) (32), and cohort 2 contains 24 samples (all collected before treatment) of TNBC breast cancer patients, treated neoadjuvantly with taxanes, alone or in combination with anthracyclines (10 responders, 14 non-responders) (36). When we apply the R1 and RD1 signatures to these clinical cohorts, PDX and clinical samples remain in different clusters, and the responders from the clinical cohort 2 were not segregated (Supplementary Figure S4C-D). However, the R1 signature, but not RD1, classifies the clinical samples from cohort 1 in 3 clusters: one of responders, one of non-responders and one enriched in responders (Supplementary Figure S4C-D). Then, we generated new signatures combining the clinical samples from cohort 1 with the S/RD and S/R PDX tumors (Supplementary Table S1G-H). In the resulting residual disease signature 2 (RD2, 478 CpG) there is a cluster composed mainly of residual disease PDX and a second cluster enriched in sensitive PDX and clinical responders (Supplementary Figure S5A). The same distribution is observed when the RD2 signature is applied to the clinical cohort 2 (Supplementary Figure S5B, chi square=9.31, p=0.002). The resulting resistant signature 2 (R2, 21 CpG) segregates clinical samples and PDX models in two clusters: one composed exclusively by resistant PDX/clinical non-responders and a second cluster divided in two subclusters, one enriched in resistant PDX/clinical non-responders and another enriched in sensitive PDX/clinical responders (Figure 3A). When this R2 signature is applied to the clinical cohort 2, for the first time all PDX and clinical samples are intermingled and subclassified by response to chemotherapy (Figure 3B). A cluster enriched in resistant PDX/clinical non-reponders and another enriched in sensitive PDX/clinical responders (chi square 17.8, p=0,00002) was shown. The mean methylation value of CpG from signature R2, but not RD2, could discriminate the clinical cohorts by chemotherapy response (Figure 3C and Supplementary Figure S5C). These results indicate that the R2 methylation signature may have predictive value to determine chemotherapy response to taxanes in TNBC patients.
Fig 3. DNA-methylation signatures as predictive tools for chemoresistance.
A, B) Supervised hierarchical clustering applying the R2 methylation signature: 21 CpGs that discriminate responders vs. non-responders (docetaxel sensitive vs resistant TNBC PDX (5 pairs) and alive vs exitus breast cancer patients from cohort 1) in these PDX/cohort 1 (A) and adding cohort 2 (responders/non-responders) (B). Differentially Methylated Positions were found using minfi in statistical software R, with a p-value of 0.001. Chi-square and corresponding p-value indicating segregation between responder and non-responders is indicated. C) Association of R2 methylation signature (mean CpG methylation values) with chemotherapy response in both clinical cohorts. Mean values, box and whiskers (min to max) and t test p-values are shown.
Poor association between gene-specific expression changes and DNA-methylation in TNBC PDX models
Next, we selected models IDB-01 and IDB-02, representative of each TNBC cluster for further analyses. An unsupervised methylation cluster using the ten thousand most variable CpGs between tumors within each TNBC PDX model was performed. Separation between sensitive and resistant samples for IDB-01 but not for the BRCA1-mutant IDB-02 was observed (Supplementary Figure S6A).
It is well known the machinery and relationship between epigenetic changes, that includes methylation, and gene expression control in cancer (37). To investigate whether changes in gene expression between sensitive and resistant tumors were controlled by methylation, whole gene expression microarray analyses were performed. Three paired sensitive and resistant tumors from each TNBC PDX model using SurePrint G3 Human Gene Expression Microarray v2 platform from Agilent were analyzed.
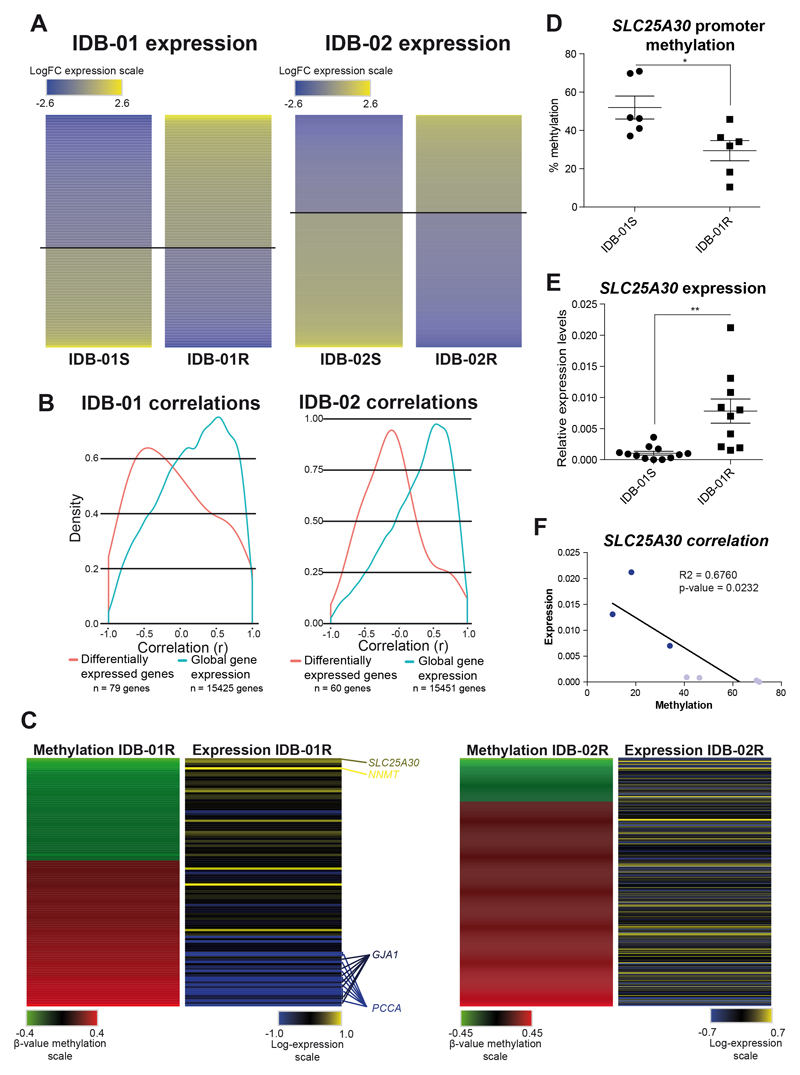
Pair-wise comparisons between sensitive and resistant IDB-01 and IDB-02 TNBC PDX models were performed using limma test, a complex ANOVA test that stabilizes the gene-specific variance estimates (38), to identify strongly, differentially expressed genes (adj. p-value < 0.05, fold change > |1.5|). A total of 702 genes in IDB-01, 304 downregulated and 398 overexpressed, and 769 genes in IDB-02, 445 downregulated and 324 overexpressed (Figure 4A, Supplementary Figure S6B and Supplementary Table S2A), were identified as differentially expressed genes in chemoresistant compared to chemosensitive tumors. A set of 35 genes, 7 in the same direction were identified as differentially expressed genes in common between both TNBC PDX models (Supplementary Figure S6B).
Fig. 4. Association between methylation and gene expression changes in sensitive and resistant TNBC PDX tumors.
A) Mean-centered gene expression of differentially expressed genes between sensitive and resistant tumors from IDB-01 and IDB-02 PDX model after limma test analysis. Genes are ordered vertically according to their expression. Overexpression (yellow) and underexpression (blue) are indicated. B) Density plot for correlation scores of methylation and gene expression for global, basal gene expression (blue line) and differentially expressed genes (red line) between sensitive and resistant IDB-01 PDX model. Number of genes used for correlations are indicated. C) Mean-centered methylation for each differentially promoter-methylated CpG and corresponding mean-centered gene expression between sensitive and resistant tumors from IDB-01 and IDB-02 PDX model. Genes are ordered vertically according to their methylation change. D, E) Mean methylation levels of 4 CpG islands from SLC25A30 promoter (D) and SLC25A30 mRNA expression levels relative to PPiA (E) in sensitive and resistant tumors from IDB-01 measured by pyrosequencing and qRT-PCR, respectively. Mean, SEM and statistical significance are represented. F) Correlation between methylation and gene expression in the same IDB-01 PDX tumors. IDB-01S tumors are indicated in light blue while IDB-01R tumors are indicated in dark blue. R-square and p-value indicating association between methylation and gene expression in IDB-01 TNBC PDX tumors are shown.
GSEA revealed that resistant samples from IDB-01 model showed enrichment in pathways involved in extracellular matrix receptor interaction, focal adhesion and cell differentiation, being the EGFR pathway the most enriched (Supplementary Table S2B). Differential mRNA expression levels were found by qRT-PCR from EGFR (Supplementary Figure S6C) which was increased in docetaxel resistant tumors, whereas expression of another partner of the pathway, ERBB3, significantly decreased (Supplementary Figure S6C). No different expression of HER2 was found between sensitive and resistant IDB-01 PDX tumors (23). Although NRG1, a ligand of the EGF receptor family, was differentially methylated in the gene body between sensitive and resistant tumors in both TNBC PDX models, its expression was below detection limits in both TNBC PDX models (Supplementary Table S1B and data not shown). In the case of IDB-02 TNBC PDX model, resistant tumors showed enrichment in genes associated to pathways of regulation of mitosis, replication and cell cycle according to the faster tumor growth found in chemoresistant tumors (23) (Supplementary Table S2B).
GO and KEGG term analyses of gene expression data for IDB-01R PDX model highlighted extracellular space as significant (Supplementary Table S2C), but other pathways were deregulated (mitochondrion, amino acid transport, regulation of nitric oxide). In IDB-02R PDX model, the BRCA1-mutant model, expression changes were related mainly with immune response pathways, but also extracellular space (as in IDB-01R) and matrix organization and cell adhesion pathways were differentially regulated (Supplementary Table S2C).
Correlations between promoter methylation (5’UTR, 1st exon, TSS200 and TSS1500 CpG gene regions) and global gene expression were extracted for IDB-01 and IDB-02 TNBC PDX models showing that there are mostly lineal positive correlations for both models [Figure 4B (blue lines), Supplementary Table S2D], indicating that global gene expression is not regulated by methylation. To characterize the significance of changes in DNA-methylation on gene expression during chemoresistance acquisition, a simple parametric test was applied to infer most differentially expressed genes between sensitive and resistant IDB-01 and IDB-02 TNBC PDX models and correlate it with methylation. Analysis revealed that most differentially expressed genes (adj. p-value < 0.05, LogFC > |1.5|) were negatively correlated with methylation in IDB-01 TNBC PDX model but not in IDB-02 [Figure 4B (red lines) and Supplementary Table S2D].
Conversely, the most differentially methylated CpGs in the promoter (5’UTR, 1st exon, TSS200 and TSS1500) (which are the most likely to affect expression), were selected and gene expression data for the corresponding transcripts was analyzed. As shown in Figure 4C and Supplementary Table S2D, expected negative associations between methylation and expression were obtained for several transcripts in IDB-01, including: the top CpG more strongly hypomethylated in resistant tumors, the solute carrier SLC25A30, and the methyltransferase NNMT, which has been involved in anthracycline resistance in breast cancer (39), both with just one differentially methylated CpG at promoter and showing increased gene expression in IDB-01R PDX model (Figure 4C and Supplementary Table S2A). Several CpG were found hypermethylated in the promoter of PCCA, a mitochondrial enzyme propionyl-CoA carboxylase, and GJA1, a gap junction gene that forms intercellular channels, and accordingly the corresponding genes were downregulated in resistant IDB-01R tumors (Figure 4C and Supplementary Table S2A). Associations between methylation and expression were virtually absent for the BRCA1-mutant model IDB-02 (Figure 4C and Supplementary Table S2D).
Pyrosequencing analysis confirmed that SLC25A30 promoter was differentially methylated, showing a hypomethylation pattern in IDB-01R (Figure 4D). As expected, analysis by qRTPCR showed higher mRNA expression levels of SLC25A30 in resistant IDB-01 TNBC PDX tumors (Figure 4E). Moreover, when we analyze methylation and expression data from same PDX tumor, a significant negative correlation was found (Figure 4F). SLC25A30 pertains to the family of solute carrier transporters and it appears to be consistently associated with ribosomal, mitochondrial and transport pathways (40) (Supplementary Table S2C). This data supports that SLC25A30 expression is controlled by promoter methylation and increased expression in resistant IDB-01 PDX could be associated to the emergence of chemoresistance.
Taken together, these results do not show a general negative correlation between methylation and expression in basal conditions, but some genes could be controlled by methylation during the emergence of resistance, at least in the IDB-01R model. Some clinically relevant pathways, as tyrosine kinase receptor pathway, have been unravelled using transcriptomic analyses of our paired sensitive and resistant TNBC PDX models, pointing out the reliability of breast cancer PDX models to study the emergence of resistance to chemotherapeutic drugs.
Discussion
Breast cancer PDX have demonstrated to resemble the heterogeneity, drug response, invasive capabilities and growth rates of human cancers better than established BCCLs (12,13). In this work we found that breast cancer PDX maintain the genome-wide DNA-methylation patterns and the segregation in two major DNA-methylation clusters based on breast cancer subtype observed in breast cancer patients (17). Moreover, our TNBC PDX models clustered in two main groups, one of them including all BRCA1 mutant models. It is known that tumors with BRCA1 mutations show multiple copy number changes, being most of these gains and losses of genomic DNA a very characteristic pattern of the BRCA1 tumors (41). Differences in copy number changes induce methylation detection changes (42); then distinct methylation patterns detected could be associated to changes in copy number. Previous studies found three distinct methylation clusters associated with survival within the TNBC subtype (43).
Global DNA-methylation profiles in PDX tumors are closer to primary tumors than those of BCCLs (44). Unlike in vitro cultured cell lines, which acquire accumulative epigenetic changes with passaging (45), breast cancer PDX models show a stable methylome even after continuous exposure to docetaxel. These results illustrate the superiority of breast cancer PDX models as compared to breast cancer cell lines as preclinical tools for methylation studies.
The high similarity in DNA-methylation patterns between docetaxel sensitive and resistant-derived or residual disease TNBC PDX models suggest that global DNA-methylation changes are not involved in the emergence of resistance to docetaxel in TNBC. However, a subset of differentially methylated CpGs, many of them in promoter regions, were found between sensitive and resistant-derived TNBC PDX models which could contribute to the emergence of resistance. Global DNA-methylation correlation values between sensitive and residual disease or resistant tumors were similar, which supports that most global methylation changes observed were due to intratumor variability.
The little overlap found in differential DNA-methylation and expression changes after docetaxel resistance emergence between TNBC models, reflects the high heterogeneity of the TNBC subtype, and the distinct biology of TNBC harbouring BRCA1 mutations.
Despite the heterogeneity, the resistance signature obtained combining data of all paired sensitive and resistant TNBC PDX models with a small cohort of TNBC clinical samples, was able to segregate a second cohort of TNBC patients as responders or non-responders to taxane-based therapy.
Global methylation and gene expression do not correlate in breast cancer PDX models, as observed in clinical samples (breast cancer, or other cancer types) (46), but for some genes, associations between differential CpG promoter methylation and gene expression were found, mainly in the IDB-01R model. The promoter of SLC25A30 is hypomethylated and the gene is overexpressed in IDB-01R PDX tumors, suggesting that methylation changes in SLC25A30 may be associated to the emergence of docetaxel resistance in some TNBC, but further evidences are required.
There are two main protein superfamilies of transporters: the ABC transporters and the SLC transporters (47). Some of these proteins, mainly ABC transporters have been associated with multidrug resistance in breast cancer (48). SLC25A30 pertains to the less studied SLC superfamily of transporters and it appears to be consistently associated with ribosomal, mitochondrial and transport pathways (34). Actually, mitochondrial dysfunction is a well-known mechanism of cancer chemoresistance (49).
Irrespectively of methylation, transcriptomic profiling revealed EGFR pathway as the most upregulated network during emergence of docetaxel resistance. Around 50% to 70% of TNBC/basal-like patients have shown to (over)express EGFR (50) and its expression has been associated with poor prognosis (51). EGFR and HER2 pathways have been associated with poor response to chemotherapy (9,52) and clinical combinations of taxanes plus TKR inhibitors are under current study for the treatment of TNBC patients (53) which will be supported by our findings.
We previously found that emergence of docetaxel resistance is accompanied by an increase in the CD49f+ population (23). Crosstalk and crossactivation between EGFR and CD49f has been described (54) and overexpression of both CD49f and EGFR genes has been associated with chemo and radioresistance in cancer (55) (56). New studies using EGFR inhibitors in combination with chemotherapy and other inhibitors, as dasatinib and mTOR/PI3K inhibitors show promising results (57).
In conclusion, our findings demonstrate that breast cancer PDX are suitable models for methylation studies elucidating mechanisms of drug resistance related to methylation and gene expression. Although the emergence of resistance to docetaxel is not driven by global methylation changes, we identified key differentially methylated genes that can contribute to chemoresistance in TNBC. Moreover, gene expression pathways associate with chemoresistance in the clinics are also observed using TNBC PDX models.
Supplementary Material
Implications.
Subtype-specific DNA-methylation patterns are maintained in BC PDX models. While no global methylation changes were found, we uncovered differentially DNA-methylated and expressed genes/pathways associated to the emergence of docetaxel resistance in TNBC.
Acknowledgements
We thank A Welm and Y De Rose for donation of the model HCI001 (TNBC) (24). We thank M Palafox, A Giro, G Boigues, the IDIBELL animal facility service, for technical support, L Alcaraz and D Amorós (Bioarray) for microarray analyses and members of the laboratory for useful discussions.
Funding
This work was supported by grants to E González-Suárez by the Spanish Ministerio de Ciencia, Innovación y Universidades, which is part of Agencia Estatal de Investigación (AEI), through the projects (SAF2008-01975, SAF2011-22893, SAF2014-55997-R, SAF2017-86117-R), and ISCIII PIE13/00022, co-funded by European Regional Development Fund. ERDF, a way to build Europe, by a Career Catalyst Grant from the Susan Komen Foundation CCR13262449. The laboratory of E Gonzalez-Suarez is funded by an ERC Consolidator grant LS4-682935. Aleix Prat laboratory is funded by the Instituto de Salud Carlos III-PI13/01718, by a Career Catalyst Grant from the Susan Komen Foundation, by Banco Bilbao Vizcaya Argentaria (BBVA) Foundation and by a Sociedad Española de Oncología Médica (SEOM) grant. JGM was recipient of a predoctoral grant from the MICINN. AC is recipient of a grant from the FI programme of the Secretariat for Universities and Research of the Department of Business and Knowledge of the Government of Catalonia. Grant co-funded by the European Social Fund (ESF) “ESF, “Investing in your future”. We thank CERCA Programme / Generalitat de Catalunya for institutional support. The PDX from VHIO were supported by a “GHD-pink” research support via the FERO Foundation to VS, Breast Cancer Research Foundation (BCRF-17-008). MT Lewis and L Dobrolecki are supported in part by the Breast Cancer Research Foundation, the Susan G. Komen Foundation, The V Foundation, NIH/NCI grant U54CA224076, BCM Breast Cancer SPORE P50 CA186784, BCM Cancer Center grant P30 CA125123, and a generous gift from the Korell family. ADL is funded by a contract “Juan Rodés” (JR17/00016) from ISCIII.
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. The original methylation and gene expression datasets generated during the current study are available in the GEO repository.
Methylation data can be retrieved from GSE110184: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110184 token: ebatmokernwxzgl
Expression data can be retrieved from GSE110153: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110153 token: gtajoqgurtkzjgx.
References
- 1.Rapoport BL, Demetriou GS, Moodley SD, Benn CA. When and How Do I Use Neoadjuvant Chemotherapy for Breast Cancer. Curr Treat Options Oncol. 2014;15:86–98. doi: 10.1007/s11864-013-0266-0. [DOI] [PubMed] [Google Scholar]
- 2.Cancer of the Breast (Female) - Cancer Stat Facts. [cited 2017 Jul 28]. [Internet]. Available from: https://seer.cancer.gov/statfacts/html/breast.html.
- 3.Liedtke C, Wang J, Tordai A, Symmans WF, Hortobagyi GN, Kiesel L, et al. Clinical evaluation of chemotherapy response predictors developed from breast cancer cell lines. Breast Cancer Res Treat. 2010;121:301–9. doi: 10.1007/s10549-009-0445-7. [DOI] [PubMed] [Google Scholar]
- 4.O’Shaughnessy J. Extending Survival with Chemotherapy in Metastatic Breast Cancer. The Oncologist. 2005;10:20–9. doi: 10.1634/theoncologist.10-90003-20. [DOI] [PubMed] [Google Scholar]
- 5.Herbst RS, Khuri FR. Mode of action of docetaxel - a basis for combination with novel anticancer agents. Cancer Treat Rev. 2003;29:407–15. doi: 10.1016/s0305-7372(03)00097-5. [DOI] [PubMed] [Google Scholar]
- 6.Lohiya V, Aragon-Ching JB, Sonpavde G. Role of Chemotherapy and Mechanisms of Resistance to Chemotherapy in Metastatic Castration-Resistant Prostate Cancer. Clin Med Insights Oncol. 2016;10:57–66. doi: 10.4137/CMO.S34535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubo T, Kawano Y, Himuro N, Sugita S, Sato Y, Ishikawa K, et al. BAK is a predictive and prognostic biomarker for the therapeutic effect of docetaxel treatment in patients with advanced gastric cancer. Gastric Cancer. 2016;19:827–38. doi: 10.1007/s10120-015-0557-1. [DOI] [PubMed] [Google Scholar]
- 8.Duran GE, Wang YC, Moisan F, Francisco EB, Sikic BI. Decreased levels of baseline and drug-induced tubulin polymerisation are hallmarks of resistance to taxanes in ovarian cancer cells and are associated with epithelial-to-mesenchymal transition. Br J Cancer. 2017;116:1318–28. doi: 10.1038/bjc.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray S, Briasoulis E, Linardou H, Bafaloukos D, Papadimitriou C. Taxane resistance in breast cancer: Mechanisms, predictive biomarkers and circumvention strategies. Cancer Treat Rev. 2012;38:890–903. doi: 10.1016/j.ctrv.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Kastl L, Brown I, Schofield AC. Altered DNA-methylation is associated with docetaxel resistance in human breast cancer cells. Int J Oncol. 2010;36:1235–41. doi: 10.3892/ijo_00000607. [DOI] [PubMed] [Google Scholar]
- 11.Byrne AT, Alférez DG, Amant F, Annibali D, Arribas J, Biankin AV, et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17:254–68. doi: 10.1038/nrc.2016.140. [DOI] [PubMed] [Google Scholar]
- 12.Dobrolecki LE, Airhart SD, Alferez DG, Aparicio S, Behbod F, Bentires-Alj M, et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016;35:547–73. doi: 10.1007/s10555-016-9653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Tham Y-L, et al. Patterns of resistance and incomplete response to docetaxel by gene expression profiling in breast cancer patients. J Clin Oncol Off J Am Soc Clin Oncol. 2005;23:1169–77. doi: 10.1200/JCO.2005.03.156. [DOI] [PubMed] [Google Scholar]
- 14.Wysocki PJ, Korski K, Lamperska K, Zaluski J, Mackiewicz A. Primary resistance to docetaxel-based chemotherapy in metastatic breast cancer patients correlates with a high frequency of BRCA1 mutations. Med Sci Monit Int Med J Exp Clin Res. 2008;14:SC7–10. [PubMed] [Google Scholar]
- 15.Pfeifer GP. Defining Driver DNA-methylation Changes in Human Cancer. [cited 2019 May 18];Int J Mol Sci. 2018 19 doi: 10.3390/ijms19041166. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteller M. Cancer epigenetics: DNA-methylation and chromatin alterations in human cancer. Adv Exp Med Biol. 2003;532:39–49. doi: 10.1007/978-1-4615-0081-0_5. [DOI] [PubMed] [Google Scholar]
- 17.Network TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fackler MJ, Umbricht C, Williams D, Argani P, Cruz L-A, Merino VF, et al. Genome-Wide Methylation Analysis Identifies Genes Specific to Breast Cancer Hormone Receptor Status and Risk of Recurrence. Cancer Res. 2011;71:6195–207. doi: 10.1158/0008-5472.CAN-11-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefansson OA, Villanueva A, Vidal A, Martí L, Esteller M. BRCA1 epigenetic inactivation predicts sensitivity to platinum-based chemotherapy in breast and ovarian cancer. Epigenetics. 2012;7:1225–9. doi: 10.4161/epi.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stone A, Cowley MJ, Valdes-Mora F, McCloy RA, Sergio CM, Gallego-Ortega D, et al. BCL-2 Hypermethylation Is a Potential Biomarker of Sensitivity to Antimitotic Chemotherapy in Endocrine-Resistant Breast Cancer. Mol Cancer Ther. 2013;12:1874–85. doi: 10.1158/1535-7163.MCT-13-0012. [DOI] [PubMed] [Google Scholar]
- 21.Stefansson OA, Esteller M. Epigenetic modifications in breast cancer and their role in personalized medicine. Am J Pathol. 2013;183:1052–63. doi: 10.1016/j.ajpath.2013.04.033. [DOI] [PubMed] [Google Scholar]
- 22.Győrffy B, Bottai G, Fleischer T, Munkácsy G, Budczies J, Paladini L, et al. Aberrant DNA-methylation impacts gene expression and prognosis in breast cancer subtypes. Int J Cancer. 2016;138:87–97. doi: 10.1002/ijc.29684. [DOI] [PubMed] [Google Scholar]
- 23.Gómez-Miragaya J, Palafox M, Paré L, Yoldi G, Ferrer I, Vila S, et al. Resistance to Taxanes in Triple-Negative Breast Cancer Associates with the Dynamics of a CD49f+ Tumor-Initiating Population. Stem Cell Rep. 2017;8:1392–407. doi: 10.1016/j.stemcr.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeRose YS, Wang G, Lin Y-C, Bernard PS, Buys SS, Ebbert MTW, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–20. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, et al. A Renewable Tissue Resource of Phenotypically Stable, Biologically and Ethnically Diverse, Patient-Derived Human Breast Cancer Xenograft Models. Cancer Res. 2013;73:4885–97. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN, et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2016;167:260–274.:e22. doi: 10.1016/j.cell.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gómez-Miragaya J, Navarro Diaz A, Tonda R, Beltran S, Palomero L, Palafox M, et al. Chromosome 12p amplification in triple-negative/BRCA1-mutated breast cancer associates with emergence of docetaxel resistance and carboplatin sensitivity. Cancer Res. 2019 doi: 10.1158/0008-5472.CAN-18-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–54. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morpheus. [cited 2017 Oct 16]. [Internet]. Available from: https://software.broadinstitute.org/morpheus/
- 30.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA-methylation profiling. [cited 2019 May 9];Genome Biol. 2016 17 doi: 10.1186/s13059-016-1066-1. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefansson OA, Moran S, Gomez A, Sayols S, Arribas-Jorba C, Sandoval J, et al. A DNA-methylation-based definition of biologically distinct breast cancer subtypes. Mol Oncol. 2015;9:555–68. doi: 10.1016/j.molonc.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA-methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–92. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 34.Narayan A, Ji W, Zhang XY, Marrogi A, Graff JR, Baylin SB, et al. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int J Cancer. 1998;77:833–8. doi: 10.1002/(sici)1097-0215(19980911)77:6<833::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 35.He D-X, Gu F, Gao F, Hao J, Gong D, Gu X-T, et al. Genome-wide profiles of methylation, microRNAs, and gene expression in chemoresistant breast cancer. Sci Rep. 2016;6:srep24706. doi: 10.1038/srep24706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pineda B, Diaz-Lagares A, Pérez-Fidalgo JA, Burgués O, González-Barrallo I, Crujeiras AB, et al. A two-gene epigenetic signature for the prediction of response to neoadjuvant chemotherapy in triple-negative breast cancer patients. [cited 2019 May 19];Clin Epigenetics. 2019 11 doi: 10.1186/s13148-019-0626-0. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perri F, Longo F, Giuliano M, Sabbatino F, Favia G, Ionna F, et al. Epigenetic control of gene expression: Potential implications for cancer treatment. Crit Rev Oncol Hematol. 2017;111:166–72. doi: 10.1016/j.critrevonc.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gehrmann ML, Fenselau C, Hathout Y. Highly altered protein expression profile in the adriamycin resistant MCF-7 cell line. J Proteome Res. 2004;3:403–9. doi: 10.1021/pr0340577. [DOI] [PubMed] [Google Scholar]
- 40.Haguenauer A, Raimbault S, Masscheleyn S, Gonzalez-Barroso M del M, Criscuolo F, Plamondon J, et al. A New Renal Mitochondrial Carrier, KMCP1, Is Up-regulated during Tubular Cell Regeneration and Induction of Antioxidant Enzymes. J Biol Chem. 2005;280:22036–43. doi: 10.1074/jbc.M412136200. [DOI] [PubMed] [Google Scholar]
- 41.Wessels LFA, van Welsem T, Hart AAM, van’t Veer LJ, Reinders MJT, Nederlof PM. Molecular Classification of Breast Carcinomas by Comparative Genomic Hybridization: a Specific Somatic Genetic Profile for BRCA1 Tumors. Cancer Res. 2002;62:7110–7. [PubMed] [Google Scholar]
- 42.Martin-Trujillo A, Vidal E, Monteagudo-Sánchez A, Sanchez-Delgado M, Moran S, Hernandez Mora JR, et al. Copy number rather than epigenetic alterations are the major dictator of imprinted methylation in tumors. [cited 2019 May 22];Nat Commun. 2017 8 doi: 10.1038/s41467-017-00639-9. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stirzaker C, Zotenko E, Song JZ, Qu W, Nair SS, Locke WJ, et al. Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nat Commun. 2015;6:5899. doi: 10.1038/ncomms6899. [DOI] [PubMed] [Google Scholar]
- 44.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, et al. Dynamic DNA-methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–67. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 46.Moarii M, Boeva V, Vert J-P, Reyal F. Changes in correlation between promoter methylation and gene expression in cancer. BMC Genomics. 2015;16:873. doi: 10.1186/s12864-015-1994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nigam SK. What do drug transporters really do. Nat Rev Drug Discov. 2015;14:29–44. doi: 10.1038/nrd4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kathawala RJ, Gupta P, Ashby CR, Chen Z-S. The modulation of ABC transporter-mediated multidrug resistance in cancer: a review of the past decade. Drug Resist Updat Rev Comment Antimicrob Anticancer Chemother. 2015;18:1–17. doi: 10.1016/j.drup.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Guerra F, Arbini AA, Moro L. Mitochondria and cancer chemoresistance. Biochim Biophys Acta. 2017;1858:686–99. doi: 10.1016/j.bbabio.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and Clinical Characterization of the Basal-Like Subtype of Invasive Breast Carcinoma. Clin Cancer Res. 2004;10:5367–74. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 51.Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ, Kim YJ, et al. High EGFR gene copy number predicts poor outcome in triple-negative breast cancer. Mod Pathol Off J U S Can Acad Pathol Inc. 2014;27:1212–22. doi: 10.1038/modpathol.2013.251. [DOI] [PubMed] [Google Scholar]
- 52.Abdelrahman AE, Rashed HE, Abdelgawad M, Abdelhamid MI. Prognostic impact of EGFR and cytokeratin 5/6 immunohistochemical expression in triple-negative breast cancer. Ann Diagn Pathol. 2017;28:43–53. doi: 10.1016/j.anndiagpath.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 53.Nabholtz JM, Abrial C, Mouret-Reynier MA, Dauplat MM, Weber B, Gligorov J, et al. Multicentric neoadjuvant phase II study of panitumumab combined with an anthracycline/taxane-based chemotherapy in operable triple-negative breast cancer: identification of biologically defined signatures predicting treatment impact. Ann Oncol Off J Eur Soc Med Oncol. 2014;25:1570–7. doi: 10.1093/annonc/mdu183. [DOI] [PubMed] [Google Scholar]
- 54.Yamada KM, Even-Ram S. Integrin regulation of growth factor receptors. Nat Cell Biol. 2002;4:E75–76. doi: 10.1038/ncb0402-e75. [DOI] [PubMed] [Google Scholar]
- 55.Ricci F, Fratelli M, Guffanti F, Porcu L, Spriano F, Dell’Anna T, et al. Patient-derived ovarian cancer xenografts re-growing after a cisplatinum treatment are less responsive to a second drug re-challenge: a new experimental setting to study response to therapy. Oncotarget. 2017;8:7441–51. doi: 10.18632/oncotarget.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song S, Honjo S, Jin J, Chang S-S, Scott AW, Chen Q, et al. The Hippo Coactivator YAP1 Mediates EGFR Overexpression and Confers Chemoresistance in Esophageal Cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2015;21:2580–90. doi: 10.1158/1078-0432.CCR-14-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costa R, Shah AN, Santa-Maria CA, Cruz MR, Mahalingam D, Carneiro BA, et al. Targeting Epidermal Growth Factor Receptor in triple negative breast cancer: New discoveries and practical insights for drug development. Cancer Treat Rev. 2017;53:111–9. doi: 10.1016/j.ctrv.2016.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The original methylation and gene expression datasets generated during the current study are available in the GEO repository.
Methylation data can be retrieved from GSE110184: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110184 token: ebatmokernwxzgl
Expression data can be retrieved from GSE110153: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110153 token: gtajoqgurtkzjgx.