Abstract
While apical growth in plants initiates upon seed germination, radial growth is only primed during early ontogenesis in procambium cells and activated later by the vascular cambium1. Although it is not known how radial growth is organized and regulated in plants, this system resembles the developmental competence observed in some animal systems, in which pre-existing patterns of developmental potential are established early on2,3. Here we show that the initiation of radial growth occurs around early protophloem sieve element (PSE) cell files of the root procambial tissue in Arabidopsis. In this domain cytokinin signalling promotes expression of a pair of novel mobile transcription factors, PHLOEM EARLY DOF (PEAR1, PEAR2) and their four homologs (DOF6, TMO6, OBP2 and HCA2), collectively called PEAR proteins. The PEAR proteins form a short-range concentration gradient peaking at PSE and activating gene expression that promotes radial growth. The expression and function of PEAR proteins are antagonized by well-established polarity transcription factors, HD-ZIP III4, whose expression is concentrated in the more internal domain of radially non-dividing procambial cells by the function of auxin and mobile miR165/166. The PEAR proteins locally promote transcription of their inhibitory HD-ZIP III genes, thereby establishing a negative feedback loop that forms a robust boundary demarking the zone of cell divisions. Taken together, we have established a network, in which the PEAR - HD-ZIP III module integrates spatial information of the hormonal domains and miRNA gradients during root procambial development, to provide adjacent zones of dividing and more quiescent cells as a foundation for further radial growth.
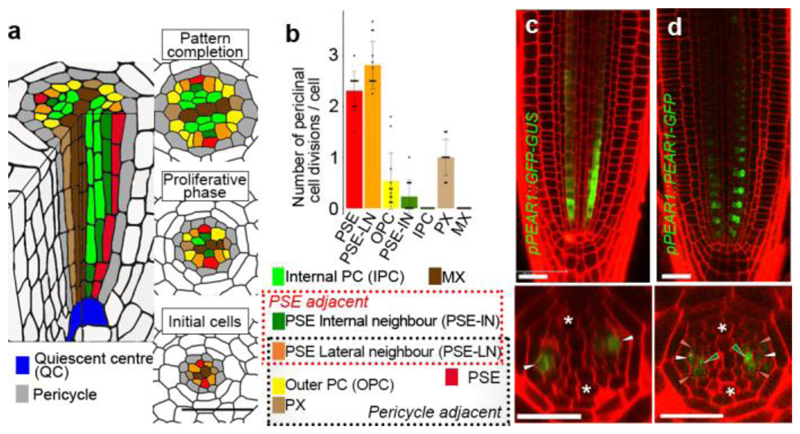
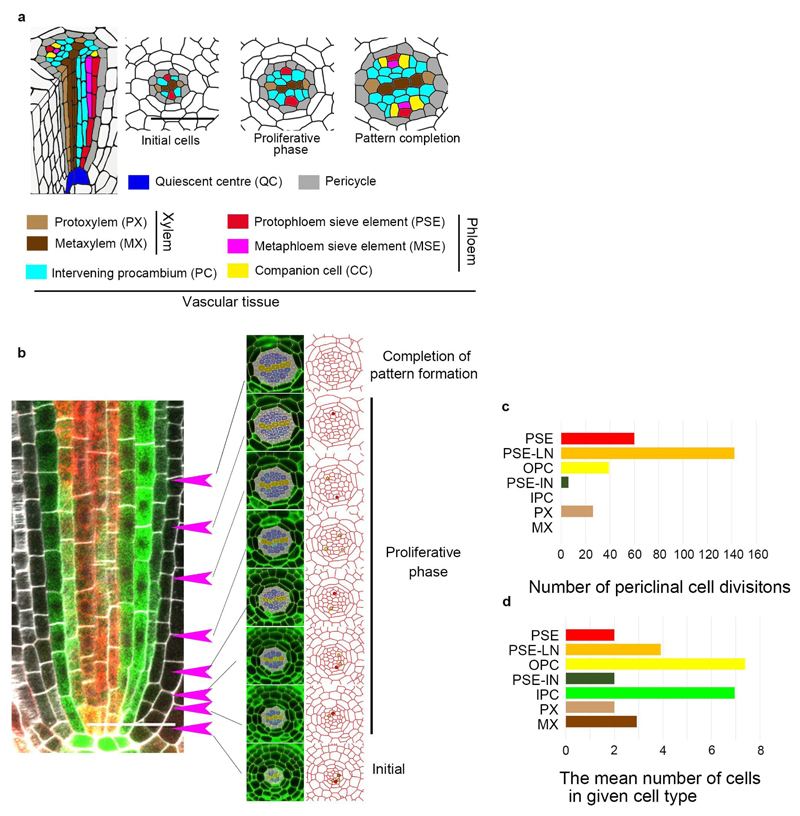
Cambial growth in plants is initiated within the procambial tissues of the apical meristems through periclinal (i.e. longitudinal) divisions associated with formation of the vascular tissues xylem and phloem1 (Extended Data Fig. 1a). It has been established that during procambial development in Arabidopsis roots there are distinct domains for high auxin and cytokinin signalling, which mark the regions for further development of xylem and phloem/procambium, respectively5–8. To accurately map the spatial distribution of the periclinal divisions, we established a new nomenclature for the root procambial cells, including PSE-lateral neighbours (PSE-LN) as cells directly contacting both PSE and the pericycle, the outer procambial cells (OPC) as procambial cells adjacent to the pericycle but not contacting PSE, and SE-internal neighbours (PSE-IN) as cells located internal to and directly contacting PSE (Fig. 1a). Both the PSE cell and PSE-LN showed higher activity of periclinal cell division than the OPC and PSE-IN (Fig. 1b, Extended Data Fig. 1b-d and Supplementary Information). We observed virtually no periclinal divisions in metaxylem (MX) and internal procambial cells (IPC) (Fig. 1b). Furthermore, blocking symplastic transport genetically9 between the PSE and the surrounding cells results in a dramatic reduction in the number of cell files, not only in PSE lineage but also in the PSE-LN lineage (Extended Data Fig. 2a-e). Thus, the proliferative activity in procambium is centred on and around PSE and may involve symplastic intercellular signals.
Figure 1. Periclinal cell divisions are centred around PSE, a domain highlighted by mobile PEAR transcription factors.
a, Schematic representation of procambial cells based on the position relative to PSE (red) and outer pericycle (gray). PSE neighbouring cells are classified as PSE lateral neighbour (PSE-LN, orange), a cell adjacent to both PSE and pericycle, or PSE internal neighbour (PSE-IN, dark-green), a cell adjacent to PSE but not pericycle. Intervening procambial cells are classified as outer PC (OPC, yellow), a procambial cell adjacent to pericycle, or internal PC (IPC), a procambial cell non-adjacent to pericycle. PX and MX represent protoxylem and metaxylem, respectively. b, Number of periclinal cell divisions in each cell during procambial development (273 division events in total from 13 independent roots, also see Supplementary Information). PSE and PSE-LN exhibited higher proliferative activity. Bar graphs represent mean. Error bars are s.d. Dots, individual data points. c, Expression of pPEAR1::GFP-GUS (n=17) exhibits a highly PSE-specific expression pattern in the vascular tissue, though a residual level of GFP signal is observed in PSE-IN, most likely due to the retention of fluorescent protein after the division of PSE. d, Expression of the translational fusion of PEAR1 to GFP (n=15). Fluorescent signal is observed not only in PSE but also in its neighbouring cells, including PSE-LN and PSE-IN. In c and d, n represents independent biological samples. White, dark-green, orange arrowheads and asterisks indicate PSE, PSE-IN, PSE-LN and protoxylem, respectively. Scale bars, 25 μm.
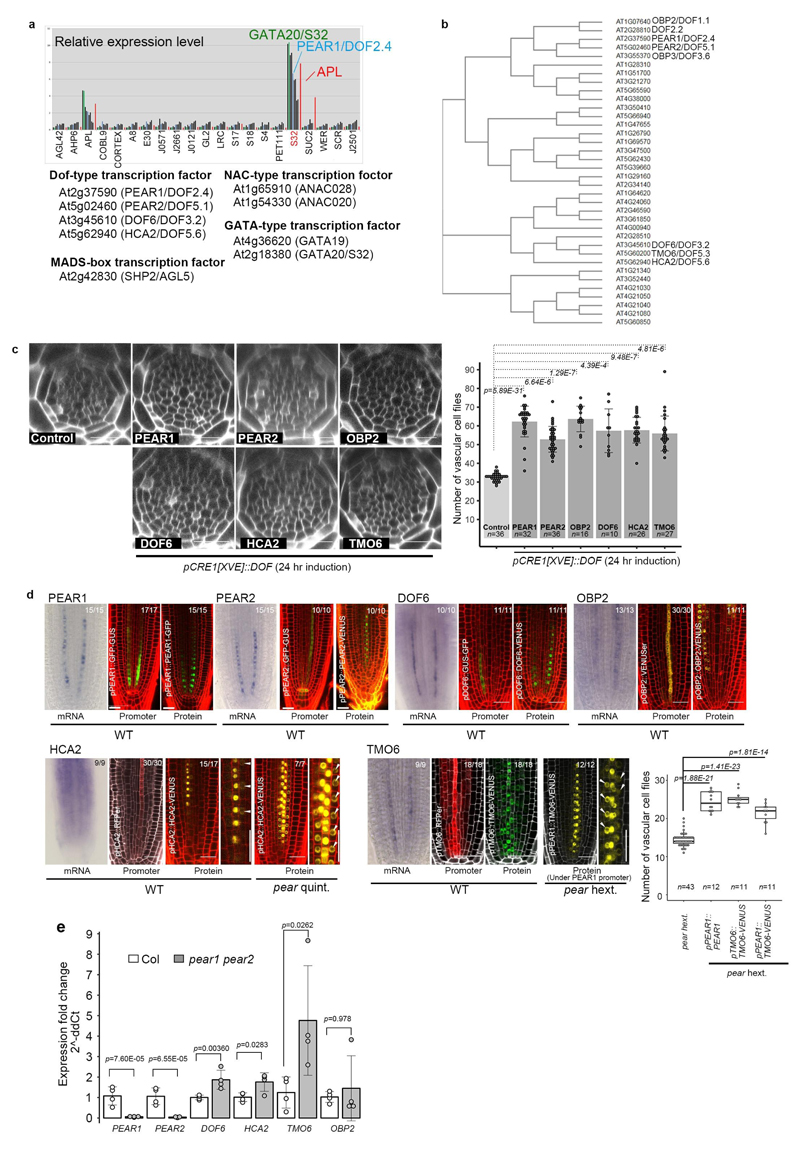
By searching in silico for transcription factors enriched in early PSE10 (Extended Data Fig. 3a), we identified a pair of DOF transcription factors11, PHLOEM-EARLY-DOF 1 (PEAR1)/DOF2.4 and PEAR2/DOF5.112 (Extended Data Fig. 3b). RNA in situ hybridization and transcriptional fusion constructs validated that both PEAR1 and PEAR2 are transcribed specifically in PSE cells (Fig. 1c and Extended Data Fig. 3d). However, fluorescent tagged versions of the PEAR proteins show localization not only in PSE but also in PSE neighbouring cells (PSE-LN and PSE-IN), indicating that these proteins move across short ranges via plasmodesmata (Fig. 1d, Extended Data Fig. 2f-g, 3d and 4a-d).
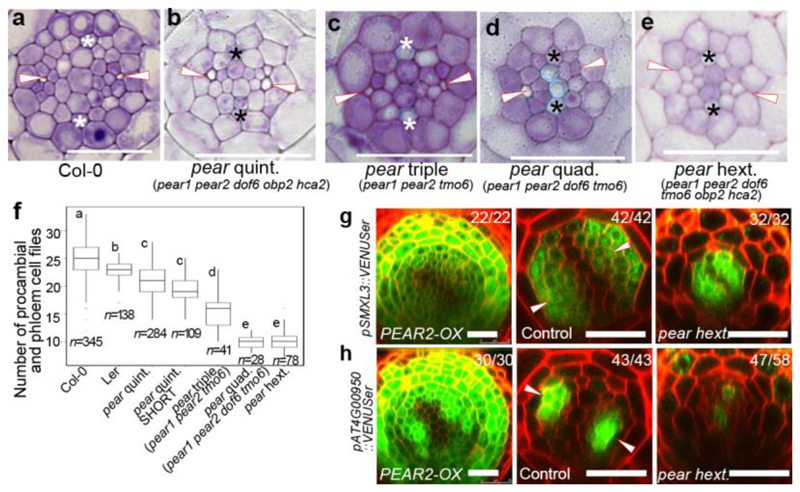
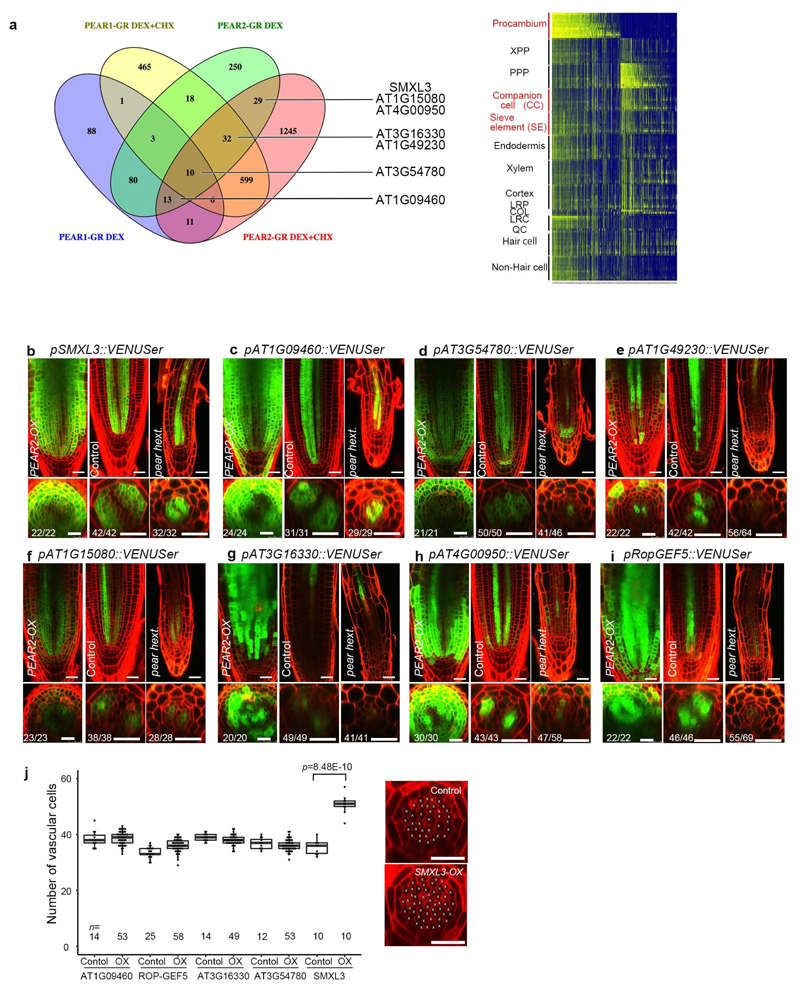
We next investigated whether the loss-of-function of these genes would lead to a phenotype corresponding to the one observed when symplastic transport is compromised (Extended Data Fig 2c). However, we did not find such phenotype in single or double mutants corresponding to PEAR1 and PEAR2 (Extended Data Fig. 5a and b). We subsequently identified DOF1.1/OBP213, DOF3.2/DOF614, DOF5.6/HCA215, and DOF5.3/TMO616 as additional PSE specific/abundantly expressed DOF genes with a broader gene product localization (Extended data Fig 3d). Furthermore, overexpression of any of these six loci results in an increased number of cell files (Extended Data Fig. 3c). In addition, we observed that DOF6, HCA2 and TMO6 are upregulated in pear1 pear2 double mutant apparently as a compensation response (Extended Data Fig. 3e, see also Supplementary Information). Among the several higher order combinatorial mutants involving all six genes, we found the pear1 pear2 tmo6 triple mutant to display reduced radial growth variably (Fig. 2a, c and f), while the corresponding three double mutants did not show this phenotype (Extended Data Fig. 5b). Furthermore, the pear1 pear2 dof6 tmo6 quadruple mutant results in all plants with further, uniformly reduced radial growth corresponding to the line with compromised symplastic trafficking (Fig. 2d, f and Extended Data Fig. 2c), indicating that these four mobile PEAR proteins play a major role in radial growth. In addition, the pear1 pear2 dof6 obp2 hca2 quintuple mutant resulted in a population of slowly elongating roots (around 30 per cent, n=300) with a reduction in radial growth (Fig. 2b and f), whereas the corresponding five quadruple mutants for the five genes did not display a strong phenotype (Extended Data Fig. 5b). The introduction of obp2 and hca2 mutations into the pear1 pear2 dof6 tmo6 quadruple background (resulting in the pear1 pear2 dof6 tmo6 obp2 hca2 hextuple mutant) did not result in further reduced radial growth (Fig. 2d-f), collectively suggesting a significant but minor contribution of OBP2 and HCA2. We were able to suppress the phenotype of the quintuple and/or hextuple mutants with all six genes (Extended Data Fig. 5c, d and see Supplementary Information). Collectively these data indicate that the mobile PEAR proteins redundantly control cell proliferation in and around PSE cells. Their effects are likely to be both cell autonomous and/or non-cell autonomous as several putatively direct target genes, including a central regulator of phloem formation SUPPRESSOR OF MAX2 1-LIKE3 (SMXL3)17, are expressed in both PSE and its surrounding cells (Fig. 2g-h and Extended Data Fig. 6, also see Supplementary Information). Moreover, ectopic expression of SMXL3 is sufficient to enhance periclinal cell divisions (Extended Data Fig. 6j).
Figure 2. PEAR genes activate periclinal cell division by controlling downstream genes in non-cell autonomous manner.
a-e, Cross-section of wild type (a), pear quintuple (pear1 pear2 dof6 obp2 hca2) (b), pear triple (pear1 pear2 tmo6) (c), pear quadruple (pear1 pear2 dof6 tmo6) (d) and pear hextuple (pear1 pear2 dof6 obp2 hca2 tmo6) (e), respectively. Each image is representative of independent biological samples analysed in f. f, Number of procambial and phloem cell files in wild type and pear combinatorial mutants. Values were calculated from root cross sections at the differentiation zone. Boxplot centres show median. For more information on boxplots, see Methods. Statistically significant differences between groups were tested using Tukey’s HSD test p<0.05. For individual P values, see Supplementary Table 3. n, independent biological samples. g-h, Expression of selected PEAR1/2 downstream genes in wild type, PEAR2 overexpression plant and pear hextuple mutant. SMXL3 is expressed in phloem and procambial tissue, whose expression is induced by PEAR2 overexpression, but not altered in pear hextuple. AT4G00950 gene is expressed in PSE and its neighbouring cells, whose expression is induced by PEAR2 overexpression and reduced in pear hextuple mutant. Number in each panel indicates samples with similar results of the total independent biological samples analysed. White arrowheads, PSE. Asterisks, protoxylem. Scale bars, 25 μm.
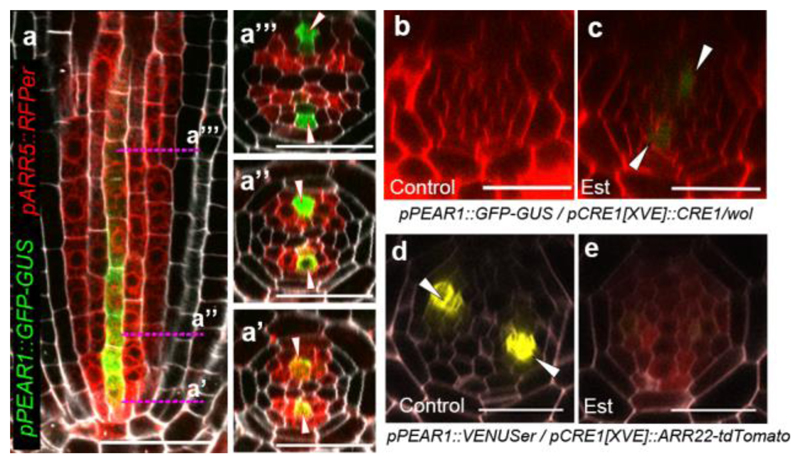
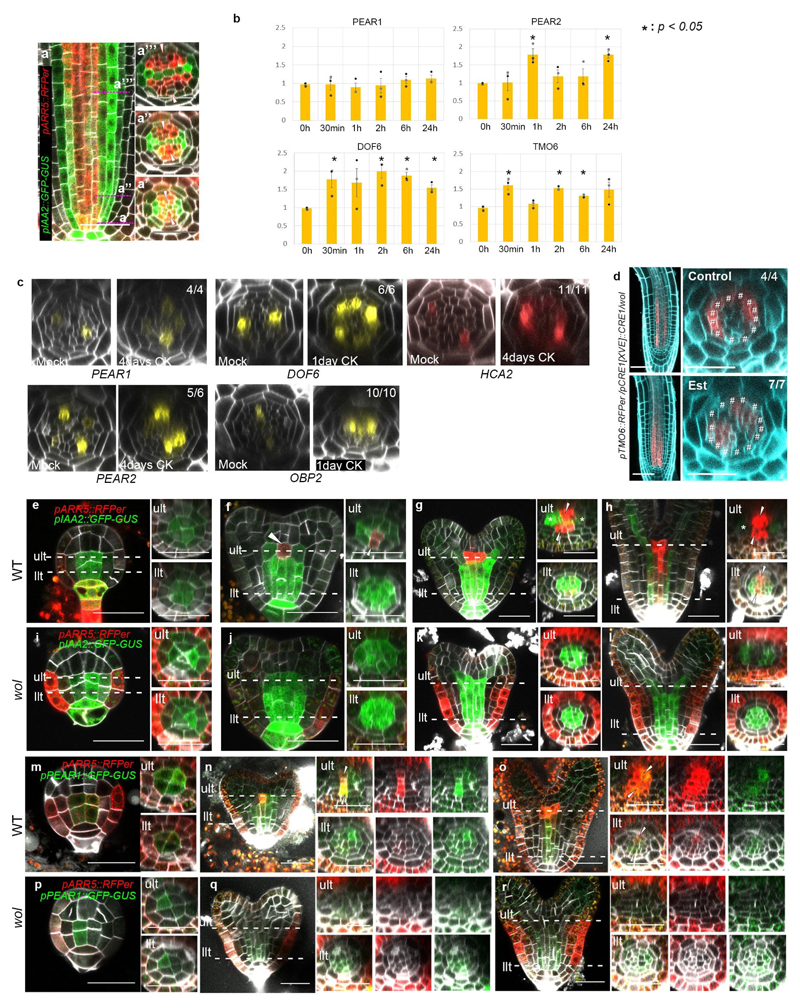
Earlier studies have highlighted cytokinins in regulating procambial cell proliferation6,8. During root development, cytokinin signalling reporter, pARR5::RFPer18 is initially activated and maintained in PSE and its surrounding procambial cells, later becoming concentrated in the procambial cells neighbouring to the xylem cells, while auxin response is maintained in xylem domain7,8 (Extended Data Fig. 7a, and see Supplementary Information). Cytokinin signalling reporter partially overlaps with the PEAR1 transcriptional domain (Fig. 3a). Exogenous cytokinin application rapidly increased the level of some of the PEAR transcripts (Extended Data Fig. 7b), and sustained cytokinin treatment resulted in a radial expansion of PEAR expression domains (Extended Data Fig. 7c). Conversely, both PEAR1 and TMO6 transcription were highly reduced in the procambial tissue of cytokinin signalling loss-of-function mutant wooden-leg (wol)5,19 (Fig. 3b and Extended Data Fig. 7d) and in plants overexpressing ARR2220, an inhibitor of cytokinin signalling (Fig. 3d and e). However, expression of both genes was restored by the induction of cytokinin signalling in wol (Fig. 3c and Extended Data Fig. 7d). In addition, we validated the requirement of cytokinin signalling for PEAR1 expression during embryogenesis (Extended Data Fig. 7e-r, and see Supplementary Information). Taken together, our results indicate that initiation of PEAR1 expression in early embryogenesis is independent of cytokinin signalling, but by the time the bisymmetric cytokinin pattern is formed at early heart stage, PEAR1 transcription is activated and maintained post-embryonically by cytokinins.
Figure 3. Cytokinin signalling triggers PEAR1 expression.
a, Expression of ARR5 and PEAR1 overlaps at the initial stage (a’) and early proliferative phase (a”), n=15. b-c, Transcription of PEAR1 in wol root, which is conditionally rescued by CRE1 induction (Est, estradiol treated). PEAR1 transcription is severely reduced in the condition with attenuated cytokinin response (b, n=6), and is restored after three days induction of CRE1 (c, arrowheads, n=6). d-e, PEAR1 transcription is down-regulated after 48 hours of ARR22 induction. n=5 and 7, respectively. In a-e, n represents independent biological samples. White arrowheads, PSE. Scale bars, 25 μm.
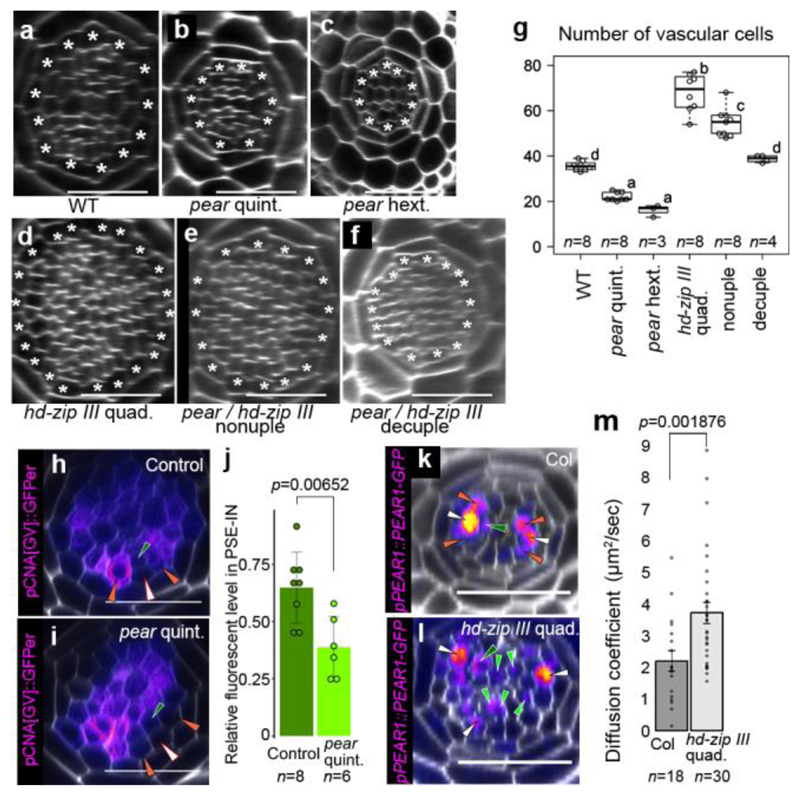
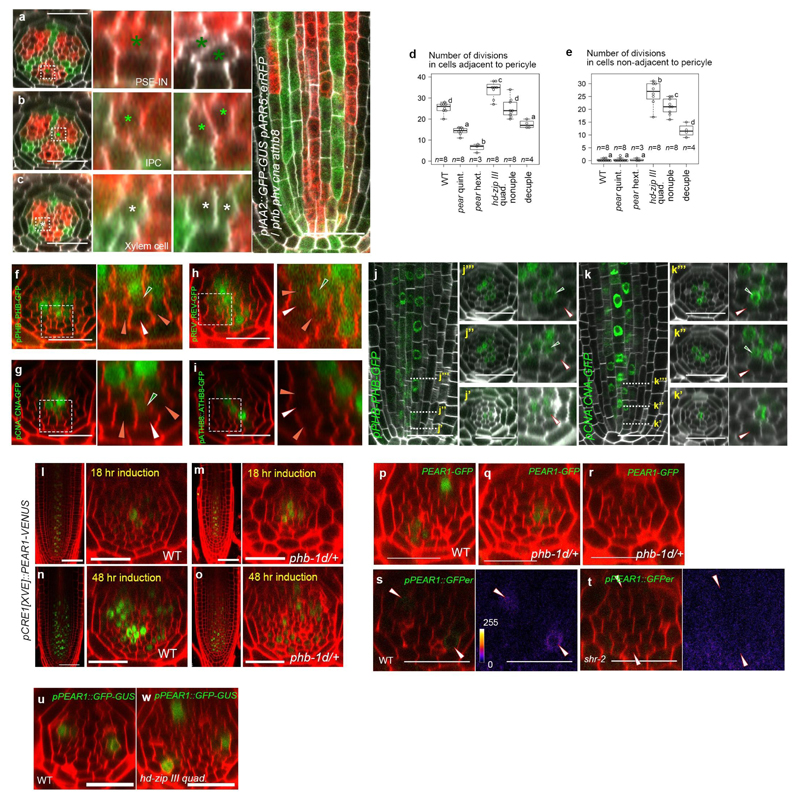
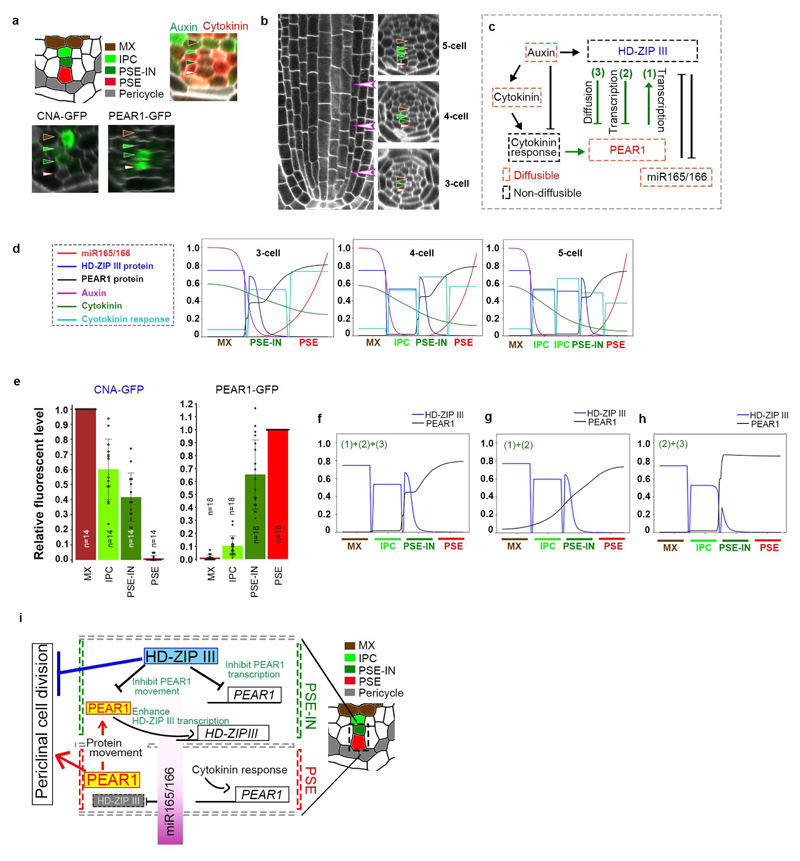
Almost no periclinal cell divisions were observed in the cells non-adjacent to the pericycle, including PSE-IN where both cytokinin response and PEAR protein are present (Fig. 1b-d and 3a), suggesting an inhibitory mechanism restricts PEAR function in the inner cells. We previously observed an increased cell number in the vascular tissue of quadruple loss-of-function mutant of the five Class III HOMEODOMAIN LEUCINE ZIPPER (HD-ZIP III) genes21,22 (Fig. 4a, d and g). These ectopic cell divisions occur in cells non-adjacent to the pericycle (Extended Data Fig. 8a-e). We observed high levels of three HD-ZIP III proteins, PHABULOSA (PHB), CORONA (CNA) and REVOLUTA (REV), in non-dividing procambial cells, IPC and PSE-IN, whereas their expression was absent in the actively dividing cells of the PSE and PSE-LN (Extended Data Fig. 8f-k). In this domain endodermal-derived mobile miR165/6 eliminates HD-ZIP III messenger RNA22,23, suggesting that HD-ZIP III inhibit periclinal cell divisions of PSE-IN by antagonizing the functions of co-localized PEAR proteins. This is further supported by our observation that overexpression of PEAR1 in the miRNA-resistant phb-1d mutant which has elevated levels of PHB22,23 is less effective than overexpressing PEAR1 in wild-type plants (Extended Data Fig. 8l-o). Hence, to sharpen the boundary between dividing and non-dividing cells, the expression pattern of both HD-ZIP III and PEAR proteins must be tightly controlled.
Figure 4. Antagonistic function of PEAR1 and HD-ZIP III sharpens the boundary between dividing and non-dividing cells.
a-f, An optical cross-section image of vascular tissue in wild type (a), pear quintuple (b), pear hextuple (c), hd-zip III quadruple (d), pear hd-zip III nonuple (e) and pear hd-zip III decuple mutant (f). Asterisks indicate pericycle cells. Each image is representative of independent biological samples analysed in g. g Quantification of vascular cell number. In the analysis of pear quintuple and pear hd-zip III nonuple, a population of slowly elongating roots is selected as described in Fig. 2f. h-i, Expression of CNA transcriptional reporter in the control (h, the heterozygous pear quintuple, n=4) and pear quintuple background (i, n=3). j,Fluorescent level in PSE-IN is significantly reduced in pear quintuple. n represents individual measurements across 4 or 3 independent biological samples, respectively. k-l, PEAR1-GFP localization in wild type (k, n=19) and hd-zip III quadruple mutant (l, n=17). PEAR1-GFP is broadly localized even in IPC in hd-zip III quadruple (l, light-green arrowheads). m, Average diffusion coefficient of PEAR1-GFP in wild-type and hd-zip III quadruple roots obtained by performing Raster Image Correlation Spectroscopy (RICS). In g-i and k-m, n represents independent biological samples. In g, boxplot centres show median. Statistically significant differences were tested using Tukey’s HSD test p<0.05. For individual P values, see Supplementary Table 3. In j and m, bar graphs represent mean. Error bars are s.d. (j) or s.e.m. (m). P values were calculated by two-sided Student’s t-test (j) or Mann–Whitney U test (m). Dots, individual data points. White, dark-green, orange, light-green arrowheads indicate PSE, PSE-IN, PSE-LN and IPC, respectively. Scale bars, 25 μm.
Auxin is known to promote the xylem associated HD-ZIP III transcription24,25. However, PHB, CNA and REV show broader expression of both transcriptional and translational reporters (Fig. 4h, Extended Data Fig. 8f-h and Extended Data Fig. 9), suggesting that other factors may enhance HD-ZIP III transcription in the peripheral region. Interestingly, we observed a significant reduction of CNA transcription in PSE-neighbouring cells in the pear quintuple background (Fig. 4h-j). In addition, PEAR1 overexpression enhanced the transcription of HD-ZIP III genes, especially in the central domain of vascular tissue (Extended Data Fig. 9). These data suggest that PEAR1 locally enhances HD-ZIP III transcription at PSE-neighbouring cells. As previous work has reported that PEAR1 has the potential to bind HD-ZIP III promoters26,27, it is possible that these interactions are direct. As HD-ZIP III and PEAR1 show complementary expression patterns, we explored whether HD-ZIP III could regulate PEAR1 transcription. PEAR1 expression was severely attenuated in mutants showing elevated levels of HD-ZIP III such as phb-1d and shr-222 (Extended Data Fig.8p-t). Together these data suggest a feedback loop between HD-ZIP III and PEAR1 transcription.
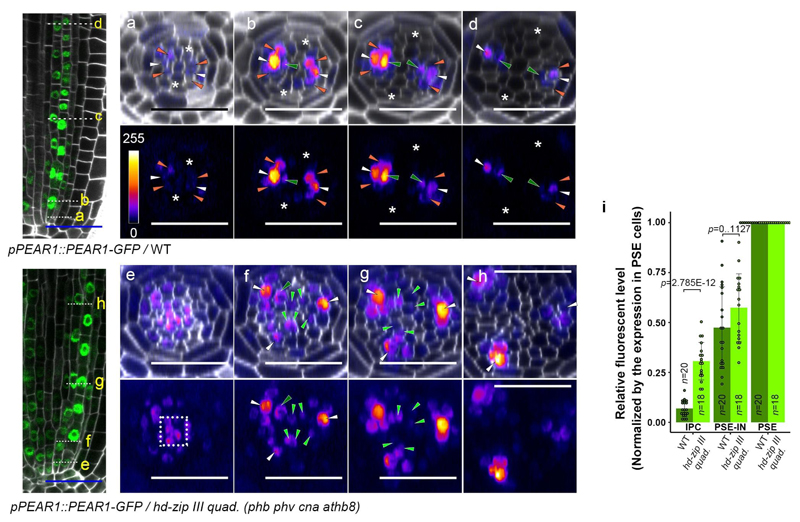
Furthermore, to examine a possible effect of the HD-ZIP III on the mobile PEAR1 proteins, we measured the diffusion coefficient and movement pattern of PEAR1-GFP in wild type and in the hd-zip III quadruple mutant where PSE is formed in a triarch arrangement but PEAR1 transcription is restricted to PSE as observed in wild type (Extended Data Fig. 8u and w). We observed that the diffusion coefficient of PEAR1-GFP is significantly higher and the protein moves further in the mutant compared to wild type (Fig. 4k-m and Extended Data Fig. 4). To understand the significance of this enhanced PEAR1 movement, we analysed the cell proliferation pattern of combinatorial pear1 pear2 dof6 obp2 hca2 phb phv cna athb8 nonuple and pear1 pear2 dof6 tmo6 obp2 hca2 phb phv cna athb8 decuple loss-of-function mutants. We found that these mutants showed a reduced number of periclinal cell divisions in the vascular cells both adjacent to and non-adjacent to the pericycle compared to the hd-zip III quadruple mutant (Fig. 4a-g and Extended Data Fig. 8d-e). This indicates that HD-ZIP III inhibit periclinal cell division partially through inhibiting PEAR1 movement to position the cell division zone around phloem.
In order to further conceptualize the observed interactions between PEAR and HD ZIP III and test the capacity of this network to generate sharp boundaries, we incorporated the PEAR factors into a spatially one dimensional network model with HD-ZIP III, miR165/6, auxin and cytokinin as defined in previous theoretical studies 8,28,29 (Supplementary Modelling Information). The model is defined on a line in one spatial dimension representing 3, 4 or 5 cells from the centre of the xylem axis to the outer edge of the PSE cell (Extended Data Fig. 10a-d and Supplementary Modelling Information). One particularly interesting aspect of the system is that the network involves dual negative feedback loops, in which HD-ZIP III transcription is activated by PEAR1 (Interaction (1) in Extended Data Fig.10c), while in turn both PEAR1 transcription and protein movement are inhibited by HD-ZIP III (Interaction (2) and (3), respectively in Extended Data Fig.10c). We ran simulations exploring the steady state patterns created in networks with the above interactions and in scenarios when one of the interactions was missing (Extended Data Fig. 10d-h). Based solely on two inputs imposed at the margins, auxin (xylem) and miR165/6 (outer margin), the model predicts the spatial distribution of cytokinin, as well as PEAR and HD-ZIP III proteins (Extended Data Fig. 10d-h). The version of the model incorporating all three interactions (i.e repressing both the transcription and movement of PEAR) results in the sharpest gradients of PEAR and HD-ZIP III proteins (Extended Data Fig. 10f) with both PEAR1 protein and HD-ZIP III localized within the PSE-IN, consistent with experimental observations (Extended Data Fig. 10e). To our knowledge this is the first report of a role for the dual regulation of both transcription and movement of a developmental regulator in sharpening boundaries.
Collectively our research has uncovered a regulatory network involving the dual regulation of gene transcription and protein movement, in which the spatial distribution of phytohormones and small RNA is decoded into the activity of two functionally antagonistic sets of transcription factors, PEAR and HD-ZIP III, during root procambial development (Extended Data Fig. 10i). The mobile PEAR factors promote cell proliferation around the two early protophloem sieve element cell files, which constitute two new organizers just proximal to the quiescent centre. These organizers surround a more quiescent central zone defined by the HD-ZIP III factors. In this way, the PEAR - HD-ZIP III module specifies a lateral meristem within an apical meristem and as such, forms a foundation for further cambial development30. Therefore, in the future it will be interesting to determine how extensively this procambial pathway also contributes to ontogenetically late processes such as wood and storage organ formation in the crop species.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment. Experiments were repeated at least twice. All experiments were repeated successfully.
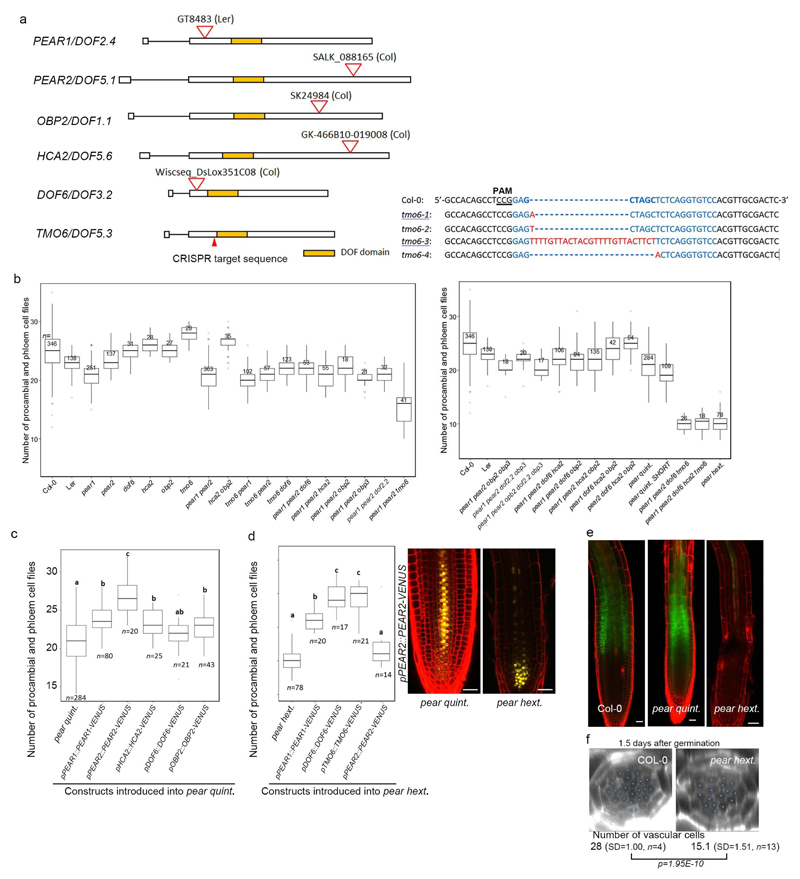
Plant materials and growth condition
Arabidopsis thaliana lines used in this study were either in Columbia or Landsberg erecta background. The following alleles were obtained from the publicly available collections: pear1 (CSHL_GT8483) in Ler, pear2 (SALK_088165) in Col-0, obp2 (SK24984) in Col-4, dof6 (Wiscseq_Ds_Llox351c08) in Col-0, hca2 (GK-466B10) in Col-0. Knock-out alleles of TMO6 were generated using CRISPR-Cas9 technology as previously described31. The following protospacer target sequence was selected as it had no predicted off-site targets and allowed screening via NheI restriction using the CRISPR-P web tool32. The Protospacer adjacent motif is underlined: GGACACCTGAGAGCTAGCTCCGG. Successful mutagenesis was confirmed via Sanger sequencing in plants of the T2 and T3 generation that no longer carried the Cas9 transgene. Four TMO6 mutant alleles were identified: tmo6-1 (+A), tmo6-2 (+T), tmo6-3 (deletion of 5 bp and at the same time insertion of 26 bp) and tmo6-4 (-5 bp) (Extended Data Fig. 5a). The alleles tmo6-1, -2, and -3 were found in the pear hextuple mutant and caused the pear hextuple phenotype, while tmo6-4 was found in the tmo6 single mutant, respectively. The genotyping primers for these mutants are listed in Supplementary Table 1. hd-zip III quadruple (phb phv cna athb8) was described previously21. Plant growth conditions were described previously5.
Histological analysis
Primary roots of vertically grown 4 to 5-day-old seedlings were used for histological analyses. For confocal imaging, root samples were stained with propidium iodide (PI), aniline blue (AB) or SCRI Renaissance 2200 (SR2200) (Renaissance Chemicals, UK). The method of PI and AB staining were described previously9,33. For SR2200 stain, root samples were fixed in SR2200 solution (4% paraformaldehyde, 0.1% (v/v) SR2200 in PBS buffer (pH7.4)). Then samples were washed with PBS buffer and transferred into the ClearSee solution34. Confocal imaging was performed on Leica TCS SP5, Leica TCS SP8, Leica TCS SP5 II HCS-A or Nikon C2 CLSM using a solid state blue laser (480nm) for GFP, a green laser (514nm) for VENUS, a lime laser (DPSS 561nm) for RFP and PI, and a UV laser (diode 405nm) for SR2200. Transverse plastic sections of root were performed as described previously5. For histological analyses of embryo, dissected embryos were mounted in SR2200 solution and visualized by the confocal microscopy.
Mapping of the position of periclinal cell divisions
A series of 2D confocal images of Arabidopsis root vascular tissue were recorded at 0.5 µm intervals using Nikon C2 CLSM or Leica TCS SP8. Cross section images in each developmental stage were created by ImageJ software from a series of 2D confocal images, and the cell segmentation was done using CellSeT35. For more information, see Supplementary Information.
Box plots
Box plots were created with standard box blot setting (the first and third quartiles, split by the median; whiskers extend to a maximum of 1.5× interquartile range (IQR) beyond the box.) Outliers are indicated as black dots.
DNA constructs and transgenic plants
Most of transgenic constructs were produced by using Gateway or multisite Gateway system (Invitrogen) as described previously18. To generate the transcriptional fusion constructs with GFP-GUS each promoter sequence was cloned into pDONR221 and fused to GFP-GUS coding sequence in the destination vector pBGWFS7 by normal LR reaction. For other transcriptional fusion constructs, including pPEAR1::VENUSer, pPEAR2::VENUSer, pAHA3::RFPer, pOBP2::VENUSer, pHCA2::RFPer, pTMO6::RFPer and pREV::RFPer and the transcriptional fusion constructs of PEAR1/PEAR2 downstream genes, each promoter was cloned into pDONRP4_P1R, and assembled with the coding sequence of fluorescent reporter (VENUSer or RFPer) and terminator into the destination vectors, pHm43GW (Hygromycin resistant), pBm43GW (Basta resistant) or by multisite Gateway system. To produce the transcriptional fusion constructs of HD-ZIP III, including PHB and CNA, each promoter was inserted upstream of the GAL4:VP16 (GV) coding region of pBIB-UAS-GFPer-NtADH5’-GV vector36. For most of the translational fusion constructs of PEAR genes, except for pPEAR1::PEAR1-GFP, each promoter was cloned into the first-box vector pDONRP4_P1R, and each coding sequence was cloned into vector pDONR221, thereafter each promoter and coding sequence were assembled with pDONR P2R_P3-terminator/reporter into pHm43GW, pBm43GW or pFR7mGW by multisite Gateway system18. To generate other translational fusion constructs, including pPEAR1::PEAR1-GFP, pCNA::CNA-GFP, pATHB8::ATHB-GFP and pREV::REV-GFP, each genomic fragment which contains promoter, coding and its 3’ region, was cloned into pAN19 vector. Then GFP coding sequence was fused to C-terminus of each coding sequence. Finally, each translational fusion sequence was inserted into the modified pBIN19 vector with Basta resistance23. For the overexpression construct, including PEAR genes and CRE1, the coding sequence of each genes was assembled with stele-specific estradiol-inducible promoter (pCRE1[XVE]) into pHm43GW or pBm43GW by the Multisite Gateway system described previously18. To construct pPEAR1[XVE]::icals3m, 1.5kb PEAR1 promoter was cloned into p1R4-ML:XVE vector18, and assembled with icals3m sequence into pBm43GW9. The primers for DNA construction and the list of plasmids are shown in Supplementary Table 1.
In situ hybridization
Amplified fragments of PEAR1, PEAR2 and OBP2 were cloned into pGEM-T Easy (Promega) vector and fragments of DOF6, HCA2, TMO6 into pCR-Bunt II-TOPO vector (Invitrogen) following manufacturer’s instructions. In order to obtain antisense probes, plasmids were first linearized by restriction enzyme treatment: MluI for PEAR1 and OBP2, ScaI for PEAR2, HindIII for TMO6 and DOF1, and XbaI for HCA2 were used. Linearized plasmids were digoxigenin (DIG) labelled using DIG RNA Labelling Kit (Roche) following manufacturer’s instructions. For PEAR1, OBP2, TMO6 and DOF1, T7 RNA polymerase and for PEAR2 and HCA2, SP6 RNA polymerase were used. mRNA detection on a whole-mount seedlings was performed as described37. Images were taken with Zeiss Axioimager microscope with either 20x or 40x objective.
Transcriptome analysis
Targets of PEAR1 and PEAR2 were identified by analysing transcriptional changes after dexamethasone (DEX) treatment of pRPS5A::PEAR1-GR and pRPS5A::PEAR2-GR. To identify putative direct targets, DEX treatment was also performed with cycloheximide (CHX), which inhibits protein synthesis and therefore activation of indirect targets. 3-day-old seedlings were grown on control medium and transferred to medium containing 10 μM DEX or 10 μM DEX and 10 μM CHX for 2h, after which root tips were collected and RNA extraction was performed. Total RNA (100 ng) was labelled using GeneChip WT PLUS Reagent Kit (Thermo Fisher Scientific) and hybridized to GeneChip Arabidopsis Gene 1.1 ST array plates (Affymetrix). Sample labelling, hybridization to chips, and image scanning were performed according to the manufacturer’s instructions. Microarray analysis was performed as previously described to yield significantly up-regulated genes (>1.0-fold; P < 0.05)8. Venn diagram of significantly up-regulated genes was made using Venny 2.1 on-line program (http://bioinfogp.cnb.csic.es/tools/venny_old/venny.php). Previously published root spatiotemporal expression data was used to make a heatmap to visualize predicted expression patterns of all PEAR1 and PEAR2 targets10. To have relative expression values for every gene in different root cell types and developmental stages, values for every gene were normalized based on its highest expression in one of the cell types. Heatmap was generated using R with gplots R-package38. The transcriptomics data files are submitted to GEO (accession number GSE115183).
Reporter analysis of PEAR1/2 downstream genes
When selecting genes for reporter analysis, putative direct targets were preferred. Significantly more direct targets were identified for PEAR2, and therefore those are overrepresented. Other considerations were how strongly they were upregulated, as well as their predicted expression pattern. Expression in early procambium or early phloem and procambium was preferred. AT1G49230, AT1G15080, AT3G16330, AT4G00950 and SMXL3 are putative direct targets of PEAR2 and with predicted expression in early phloem/procambium. AT3G54780 was chosen because it is a putative direct target of both PEAR1 and PEAR2, although no predicted expression data was available. AT1G09460, a direct target of PEAR2 and a target of PEAR1, was chosen because it was induced very strongly by both genes, although predicted to be expressed only very weakly in phloem/procambium.
Quantitative RT-PCR analysis
qRT-PCR analyses were performed as described previously39. Cytokinin treatment was done with 10μM 6-Benzylaminopurine (BA), and experiments were performed in three biological repeats and each of these with 3 technical repeats. RNA was extracted with the RNeasy kit (QIAGEN). Poly(dT) cDNA was prepared from 1 μg of total RNA with an iScript cDNA Synthesis Kit (Biorad) and analysed on a CFX384 Real-Time PCR detection system (BioRad) with iQ SYBR Green Supermix (BioRad) according to the manufacturer’s instructions. Expression levels were normalized to those of EEF1α and CDKA1;1. The primers are listed in Supplementary Table 1.
Phloem transport assay
The phloem translocation was judged by the transport and unloading of 5(6) Carboxyfluorascein diacetate (CFDA) as describe40. After application of the dye, plants were kept in agar plates and only placed on regular cover slips for imaging.
Raster image correlation spectroscopy (RICS)
To determine the rate of movement of GFP-labeled PEAR1 protein in wild type and hd-zip III quadruple (phb phv cna athb8) mutant background, Raster image correlation spectroscopy (RICS) was performed according to previous work41–43. Images were collected using a Zeiss 880 confocal microscope. Frames of 256x256 pixels were acquired using a raster scan with a dwell time of 8.19 μsec pixel -1 at a pixel size of either 100nm for 100 frames resulting in a line scan of 5.035ms. Diffusion coefficients were derived using the SimFCS software (https://www.lfd.uci.edu/globals/)44 from GFP-labeled PEAR1 vascular cells within the first 70 μm from the QC. Specifically, 18 observations from WT and 30 for the hd-zip III quadruple (phb phv cna athb8) mutant background were used for the RICS analysis using the SimFCS software The RICS algorithm by comparing the intensity fluctuations of one pixel to the fluctuations of the pixels next to it and the fluctuations of one pixel to itself over time, produces a spatio-temporal Auto Correlation Function (ACF) that captures the fluorescence dynamics of the particles in the volume44,45. The ACF is decomposed into two correlation functions that depend on ξ (the spatial lag in x) and ψ (the spatial lag in y). The first correlation function, S(ξ,ψ), calculates the spatio-temporal correlation due to the scanning of the microscope. The second correlation function, G(ξ,ψ), calculates the spatio-temporal correlation due to particles diffusing in the medium. The ACF, GS(ξ,ψ), takes both of these correlations into account by multiplying them: GS(ξ,ψ)= S(ξ,ψ)* G(ξ,ψ). The functions are constructed assuming that the distribution of fluorescence intensities follows a 3D Gaussian distribution. The decomposition of the ACF into two parts allows RICS to distinguish random, Brownian motion from diffusing particles in the medium45. The software fits the RICS-ACF using the pixel dwell time, pixel size, line scan and the Point Spread Function (PSF) bean waist of 0.241nm as previously obtained41 and returns the diffusion coefficient of the protein. The diffusion coefficient returned results in the ACF curve that best fits the data. Goodness of fit is determined by comparing the residuals to the amplitude of the ACF41–45.
Mathematical model
The mathematical model is formulated as a set of ordinary differential equations describing the set of interactions shown in Figure 4o, defined on a one-dimensional array of discrete spatial compartments representing a cross-section of root tissue. The spatial subdivisions may represent either cell or cell wall compartments, with multiple compartments per cell so that intracellular resolution is present within the model. Three, four or five cells are simulated, from the centre of the stele at the xylem axis to the edge of the stele where phloem is formed. The model is implemented as a single stand-alone text file using Python 2.7 plus the open source libraries Scipy, from which the 'odeint' function was used to solve the differential equations, and Matplotlib, which was used to plot the figures. See Supplementary Modelling Information for more details.
Extended Data
Extended Data Fig. 1. Quantification of periclinal cell division during procambial development.
a, Schematic representation of root vascular tissue of Arabidopsis. Procambial cells originate from their initial cells, and periclinal cell division increases the cell files during the proliferative phase, eventually resulting in a bisymmetric vascular pattern composed of a pair of phloem poles, which are separated from central xylem axis by intervening procambium. b, An example of mapping the position of periclinal cell divisions from the initial cells. From each position within the root vascular tissue (arrows), an optical cross-section image is constructed, and cells were segmented using CellSet. c, The number of periclinal cell divisions in each cell position (273 division events from 13 independent roots). d, The mean cell number in each category during procambial development. The number of events per cell in each group was calculated by diving the number of events by the mean cell number of each group during development (See Supplementary Information).
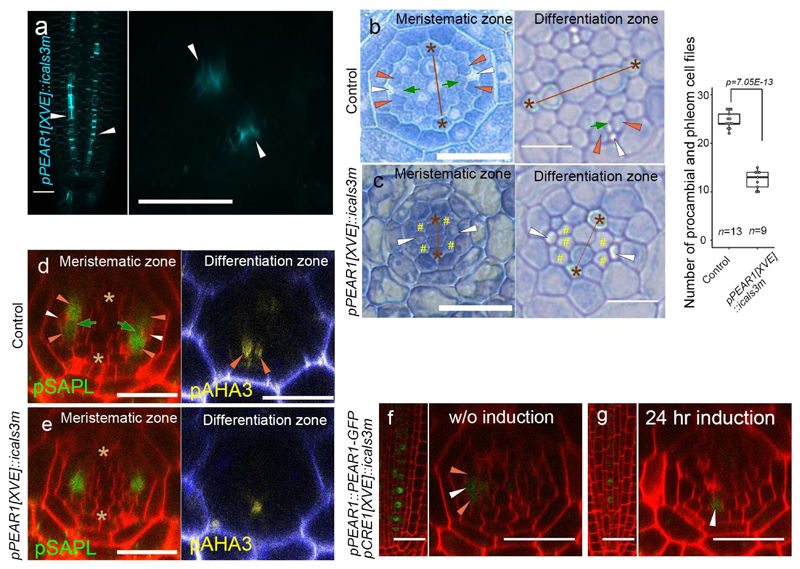
Extended Data Fig. 2. Inhibition of symplastic connection in early PSE results in the reduction of vascular cell number and in PSE-specific PEAR1-GFP localization.
a, Aniline blue stained primary root of pPEAR1[XVE]::icals3m after 24 hours of induction. Callose deposition occurs superficially in PSE cells (arrowheads, n=10). b-c, The vascular tissue of pPEAR1[XVE]::icals3m root, in non-induced (b, n=13) and after three days induction (c, n=9). In non-induced condition, PSE cells (white arrowheads) and their neighbouring cells, composed of MSE (dark-green arrows) and two lateral companion cells (orange arrowheads), are spatially separated from the xylem axis by intervening procambium. By contrast, after three days induction of callose deposition in PSE cells, only a single SE cell file is formed in each phloem pole (c, white arrowheads), and its neighbouring cells often touch the xylem axis (c, yellow hashtags). Number of procambial and phloem cell files is significantly reduced after three days induction. Boxplot centres show median. For more information on boxplots, see Methods. P value was calculated by two-sided Student’s t-test. d-e, Expression of Sister of APL (SAPL, AT3G12730) and ATPase 3 (AHA3, AT5G57350) in pPEAR1[XVE]::icals3m before (d, n=10 and 4, respectively) and after 24 hours of induction (e, n=10 and 4, respectively). SAPL is expressed in CC and MSE in meristematic zone, and AHA3 is expressed in differentiated CC (d). After induction, expression of these genes is restricted to a single cell file, indicating that symplastic cell communication between PSE and PSE-LN is required for the specification of PSE-neighbouring cell identity. f-g, PEAR1-GFP localization in pCRE1[XVE]::icals3m before (f, n=8) and after 24 hours of induction (g, n=7). PEAR1-GFP becomes specific to the PSE cell after the induction of callose deposition in whole vascular tissue, suggesting that PEAR1-GFP move in a short rage via plasmodesmata. White, orange and dark-green arrowheads indicate PSE, PSE-LN and PSE-IN, respectively. Asterisks indicate protoxylem (PX) cells. In a-g, n represents independent biological samples. Scale bars, 25 μm.
Extended Data Fig. 3. Identification of PEAR genes.
a, In silico analysis of the early phloem abundant transcription factors. Nine transcription factors are shown to be expressed abundantly in the early phloem cell (S32 fraction), containing four types of transcription factors, including DOF-type, MADS-box, NAC-type and GATA-type transcription factors. b, A phylogenetic tree of 36 Arabidopsis DOF transcription factors is drawn using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). c, Overexpression of PEAR genes, including PEAR1, PEAR2, OBP2, DOF6, HCA2 and TMO6, under the CRE1 inducible promoter enhances periclinal cell division in the vascular tissue. n represents independent biological samples. Bar graphs represent mean. Error bars are s.d. Dots, individual data points. P values were calculated by two-sided Student’s t-test. d, Expression of six PEAR genes, including PEAR1, PEAR2, DOF6, TMO6, OBP2 and HCA2, show similar expression patterns to PEAR1, in which both mRNA and transcriptional fusion reporter exhibit PSE-specific pattern with a broad protein localization. HCA2 translational fusion in wild-type background exhibits weak but detectable signal in PSE-neighbouring cells (arrows), and its expression level is enhanced in the pear quintuple mutant background. Though TMO6 mRNA is highly specific to PSE cells, its transcriptional fusion reporter shows a broad but PSE abundant expression pattern with a broad TMO6 protein localization. Mobility of TMO6 protein is more evident when TMO6-VENUS is expressed under PSE-specific PEAR1 promoter (pPEAR1::TMO6-VENUS) in pear hextuple. Number in each panel indicates samples with similar results of the total independent biological samples analysed. Boxplot centres show median. P value calculated by two-sided Student’s t-test. Dots, individual data points. e, A quantitative analysis of PEAR transcripts in pear1 pear2 double mutant background. Note that the level of transcripts of three PEAR genes, including TMO6, DOF6 and HCA2, is elevated in pear1 pear2 background, suggesting that a compensation mechanism would mask the effect of pear1 pear2 loss of function (also see Supplemental Information). Bar graphs represent mean. Error bars are s.d. Dots, individual data points. P values were calculated by two-sided Student’s t-test. Scale bars, 25 μm.
Extended Data Fig. 4. PEAR1-GFP localization during procambial development.
a-d, PEAR1-GFP localization in wild-type background (n=19, independent biological samples). The position of each optical section is indicated in the left panel showing the longitudinal section. At the position of the vascular initial cells, weak PEAR1-GFP signal is observed in PSE and neighbouring procambial cells but not in the xylem cells (a). During an early stage of the proliferative phase, the highest PEAR1-GFP signal is detected in the PSE, and substantial level of PEAR1-GFP signal is observed in PSE neighbouring cells, PSE-LN and PSE-IN (b, c) and its expression is maintained by the end of proliferation stage (d), indicating that the expression pattern of PEAR1-GFP is correlated with the domain having high proliferative activity, except for PSE-IN where almost no periclinal cell division is detected (see Fig. 1b). e-h, PEAR1-GFP localization in hd-zip III quadruple (phb phv cna athb8) mutant background (n=17, independent biological samples). The position of each optical section is indicated in left panel showing the longitudinal section. Broad localization of PEAR1-GFP is detected at the level of vascular initials. Central domain is highlighted with a dotted square (e). At the early stage of proliferative stage, fluorescent signal is detected in IPC cells (light-green arrowheads), as well as PSE and its neighbouring cells (f, g), and gradually becomes specific to PSE and its neighbours (h). i, Quantification of PEAR1-GFP signal in each cell type. Fluorescent intensity of PEAR1-GFP in IPC and PSE-IN cells during proliferative phase (b-c in wild-type, f-g in hd-zip III quadruple, respectively) was measured and normalized to the fluorescent intensity in PSE cells, confirming a broad distribution of PEAR1-GFP in hd-zip III quadruple. n represents individual measurements across 5 (wild type) or 4 (hd-zip III quadruple) independent biological samples, respectively. Bar graphs represent mean. Error bars are s.d. Dots, individual data points. P values were calculated by two-sided Student’s t-test. White, orange and dark-green and light-green arrowheads indicate PSE, PSE-LN, PSE-IN and IPC respectively. Scale bars represent 25 μm.
Extended Data Fig. 5. Loss of function of PEAR genes.
a, Organization of PEAR genes and CRISPR/Cas9-induced mutation in TMO6 locus. Deletions are denoted by dashes; insertions and a replacement are indicated by red letters. b, Quantification of phloem and procambium cell files in lower (left) and higher (right) order pear combinatorial mutants. Tukey’s HSD test is provided for all samples in Supplementary Table 3. c, pear quintuple mutant phenotype is suppressed by introduction of fluorescent-tagged PEAR proteins expressed under their native promoters. d, pear hextuple mutant phenotype is significantly suppressed by the introduction of PEAR1, DOF6 and TMO6 construct, but not PEAR2. In the pear hextuple background, PEAR2 expression is highly reduced in the vascular tissue. e, Phloem unloading assay in wild type, pear quintuple (with shortest roots) and pear hextuple (n=15, 8 and 15, respectively). Fluorescent CFDA dye is loaded on the cotyledon and imaged two hours after application (see Methods). Phloem transport and unloading is not changed in the shortest roots of the pear quintuple mutant strongly affected in the radial growth. pear hextuple shows defects in phloem transport. f, Phenotype of pear hextuple mutant at the early developmental stage (1.5 days after germination). The cell number in vascular tissue of pear hextuple is significantly reduced before the onset of phloem PSE differentiation and activation of the phloem transport (see Supplementary Information). P value was calculated by two-sided Student’s t-test. In b-f, n represents independent biological samples. In b-d, statistically significant differences between groups were tested using Tukey’s HSD test p<0.05. For individual P values, see Supplementary Table 3. Boxplot centres show median. For more information on boxplots, see Methods. Scale bars represent 25 μm.
Extended Data Fig. 6. Identification of genes acting downstream of PEAR.
a, Venn diagram showing the genes upregulated by overexpression of PEAR1 or PEAR2 with and without cycloheximide (chx). The analysis revealed 212 and 435 upregulated genes, in the respective experiments. Heatmap showing the predicted spatiotemporal expression patterns of all genes induced by PEAR1 or PEAR2. b-i, Expression patterns of eight selected genes responding to PEAR2 overexpression. In control conditions, all genes exhibit a broad expression pattern, in which five of them are transcribed both in phloem and procambial cells (b-d and i), and the rest of them are in PSE and its surrounding cells where PEAR proteins are accumulated abundantly (e-h). Whereas expression of SMXL3, AT1G09460 and AT1G15080 are maintained even in pear hextuple (b, c, f), the expression level of five genes are attenuated (d, e, g-i). Number in each panel indicates samples with similar results of the total independent biological samples analysed. j, Number of vascular cells after 3-days induction of overexpression of each PEAR downstream gene. In each case, several lines were analysed in parallel for a phenotypic change. Only SMXL3 overexpression can increase the vascular cell number (confirmed in three independent lines). Boxplot centres show median. For more information on boxplots, see Methods. n represents independent biological samples. Dots, individual data points. P value was calculated by two-sided Student’s t-test. Scale bars represent 25 μm.
Extended Data Fig. 7. Cytokinin controls PEAR expression.
a, Expression of auxin (pIAA2::GFP-GUS) and cytokinin (pARR5::RFPer) response genes (n=15). Auxin response is restricted to the xylem cells at initial stage (a’) and maintained during development (a”, a”’). High cytokinin response is activated initially and maintained in PSE and its neighbouring cell (a’, aa’), and later becomes concentrated into the intervening procambial cells flanking xylem cells (a”’). b, Exogenous cytokinin application rapidly promotes the transcript level of PEAR genes, including PEAR2, DOF6, and TMO6. Asterisks indicate significant (p<0.05) upregulation as determined by a two-sided t-test on three biological replicates. Bar graphs represent mean. Error bars are s.e.m. For individual P values, see Supplementary Table 3. c, Sustained cytokinin application leads to ectopic transcription of PEAR genes. The optical cross section images are obtained after 4 days (for PEAR1, PEAR2 and HCA2) or 1-day (DOF6 and OBP2) treatment of 1μM of BA. d, Conditional induction of CRE1 expression restores TMO6 transcription in wol root. In the absence of cytokinin response, TMO6 transcription is increased in pericycle and attenuated in the vascular tissue (Control, hashtags), and is restored in the vascular tissue after CRE1 induction (Est). e-l, Expression pattern of auxin (pIAA2::GFP-GUS) and cytokinin (pARR5::RFPer) response reporters during embryogenesis in wild type (e-h, n=29, 13, 13, 11, respectively) and wol (i-l, n=12, 8, 6, respectively). At globular stage, auxin response is activated among provascular cells both in wild type (e) and in wol (i). At early heart stage, cytokinin response is activated in cells positioned below shoot apical meristem (f, ult, arrowheads), and the stripe of cytokinin response domain is formed by mid heart stage (g, ult, arrowheads), simultaneously auxin response becomes concentrated in the cells proximal to the cotyledon (g, ult, asterisks), resulting in the bisymmetric hormonal response pattern. During the torpedo stage, cytokinin response domain reaches to llt (h, llt, arrowheads). In wol embryos, activation of cytokinin response in vascular tissue does not occur and a radial auxin response pattern is maintained (i-l). m-r, Expression of ARR5 and PEAR1 during embryogenesis. In the wild-type embryo (m-o, n=17, 25, 10, respectively), PEAR1 is broadly transcribed among provascular cells both in ult and llt with radial symmetric pattern at the globular stage (m). At the heart stage, PEAR1 transcription is enhanced in ult cells underneath the shoot apical meristem, which correlated with the activation of cytokinin response in this domain (n, arrowheads), and expression of both ARR5 and PEAR1 extends rootward and reaches to llt, becoming more concentrated within the cell files where phloem is specified post-embryonically (o, arrowheads). In wol embryos (p-r, n=19, 13, 13, respectively), PEAR1 transcription is initiated among provascular cells at the globular embryo stage (p) similar to wild type (m), but neither cytokinin response nor PEAR1 transcription occurs in ult at the heart stage (q), and PEAR1 expression is gradually attenuated by the torpedo stage (r). ult and llt represent upper- and lower tier, respectively. In a, e-r, n represents independent biological samples. In c and d, number in each panel indicates samples with similar results of the total independent biological samples analysed. Scale bars represent 25 μm.
Extended Data Fig. 8. HD-ZIP III restrict periclinal cell divisions during procambial development.
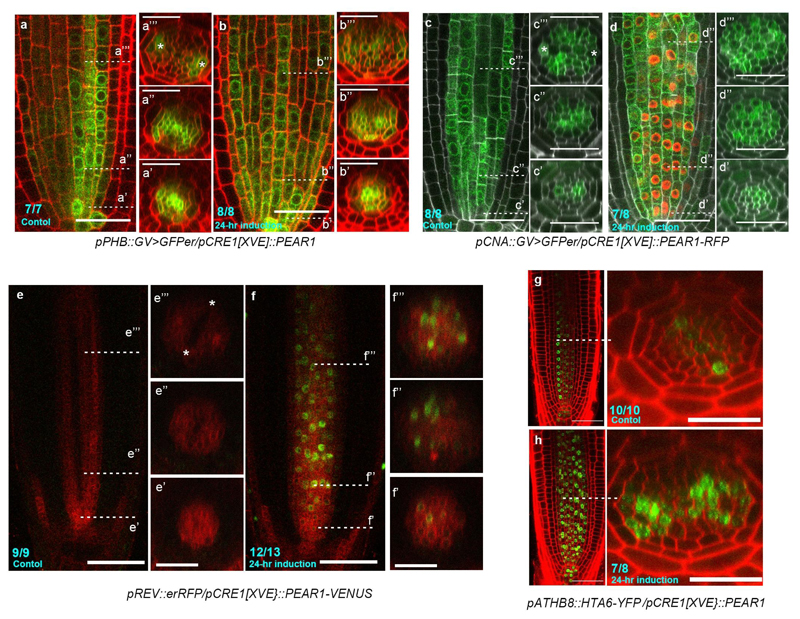
a-c, Periclinal cell divisions in the cells non-adjacent to pericycle, including PSE-IN (a), IPC (b) and xylem cell (c), occurs in hd-zip III quadruple (phb phv cna athb8). n=8. d-e, The number of periclinal cell divisions in cells adjacent (d) or non-adjacent to pericycle (e) in wild type, pear and hd-zip III combinatorial mutants. In the analysis of pear quintuple and pear hd-zip III nonuple, a population of slowly elongating roots is selected as described in Fig. 2f. Boxplot centres show median. Statistically significant differences between groups were tested using Tukey’s HSD test p<0.05. For individual P values, see Supplementary Table 3. f-i, Localization of PHB-GFP (f, n=12), CNA-GFP (g, n=10), REV-GFP (h, n=7) and ATHB8-GFP (i, n=5). j-k, Protein localization of PHB-GFP (j, n=7) and CNA-GFP (k, n=4) during procambial development. In the initial cells (j’, k’), both proteins are localized in metaxylem cells but not in PSE (white arrowheads). During the proliferation stage, PSE-IN (green arrowheads), which is produced by periclinal cell division in PSE, acquires the expression of both PHB-GFP (j”, j”’) and CNA (k”, k”’). l-o, Overexpression of PEAR1-VENUS under the CRE1 inducible promoter in wild type (l, n, n=5, 3, respectively) and heterozygous phb-1d background (phb-1d/+, m, o, n=5, 5, respectively). After 18 hours of induction, PEAR1-VENUS signal is detected in both backgrounds (l, m), however, enhanced periclinal divisions are only observed in wild type (l), and not in phb-1d (m). Longer induction of PEAR1 overexpression induces divisions even in phb-1d (n, o). p-r, pPEAR1-PEAR1-GFP expression is reduced in heterozygous phb-1d/+ background. Most of phb-1d heterozygotes exhibit a single PEAR1-GFP expressing pole (q, 72% n=11), and the expression of PEAR1-GFP is almost completely abolished in some roots of phb-1d (r, 18% n=11). s-t, The expression of pPEAR1::GFPer in wild type (s, n=10) and shr-2 (t, n=9). The fluorescent signal is below the limit of detection in shr-2. u-w, Expression of pPEAR1::GFP-GUS in WT (u, n=19) and hd-zip III quadruple mutant (w, n=11). In a-w, n represents independent biological samples. White, orange and dark-green arrowheads indicate PSE, PSE-LN, PSE-IN respectively. Scale bars represent 25 μm.
Extended Data Fig. 9. Overexpression of PEAR1 enhances the transcription of HD-ZIP III.
The transcription patterns of four HD-ZIP III, including PHB (a, b), CNA (c, d), REV (e, f) and ATHB8 (g, h) are visualized using their transcriptional fusion constructs. A longitudinal section is shown in the left panel, and the optical cross sections associated with this are shown in the right panels (the position of each section is indicated in the left panel). a-b, Transcription pattern of PHB (pPHB::GV>UAS::GFPer) in pCRE1[XVE]::PEAR1 plant before (a) and after 24 hours of induction of PEAR1 overexpression (b). PHB transcription is observed in whole vascular tissue at the initial and proliferative phase with peaks in xylem cells (a’), and its expression becomes concentrated into protoxylem cells (a” and a”’, asterisks indicate protoxylem cell). After the induction of PEAR1 overexpression, PHB expression in the central domain of the vascular tissue is maintained at the later stage, resulting in the radially symmetric PHB transcription pattern (b’ and b”). c-d, Transcription of CNA (pCNA::GV>UAS::GFPer) in pCRE1[XVE]::PEAR1-RFP plant before (c) and after 24 hours of induction (d). CNA transcription is observed mainly in xylem lineage at initial cells (c’), and becomes broader in whole vascular tissue, with peaks in procambial tissue, including PSE neighbouring cells (c”), and eventually its expression is gradually reduced in PSE and metaxylem, but is maintained in procambium, PSE neighbouring cells, as well as protoxylem cells (c”’). In a similar manner to PHB, CNA transcription in the central domain of the vascular tissue is maintained at the later stage when PEAR1-RFP is overexpressed (d”’). e-f, Transcription of REV (pREV::RFPer) in pCRE1[XVE]::PEAR1-VENUS plant before (e) and after 24 hours of induction (f). REV exhibits a distinct transcriptional pattern where its expression is initially uniform in vascular tissue (e’), and highest expression is localized in PSE, while decreasing towards xylem axis (e” and e”’). When PEAR1-VENUS is overexpressed (f), the transcription pattern of REV is also activated in the central domain of vascular tissue, resulting in the radial symmetric REV transcription pattern. g-h, The expression pattern of pATHB8::HTA6-YFP is highly specific to xylem cells (g), and its expression is enhanced after 24 hours of induction of PEAR1 overexpression with a broad expression domain (h). Number in each panel indicates samples with similar results of the total independent biological samples analysed.
Extended Data Fig. 10. The boundary between HD-ZIP III and PEAR proteins forms within the PSE-IN.
a, Summarizing results on pattern of auxin-cytokinin (data shown in Extended Data Fig. 7a), HD-ZIP III (CNA, data shown in Extended Data Fig. 8g and k) and PEAR (PEAR1, data shown in Fig. 1d and Extended Data Fig. 4a-d) in procambium. In the simulation, we simulate the concentration of HD-ZIP III and PEAR1 along the axis between metaxylem (MX, brown arrowheads) and PSE (white arrowheads). b, Summarizing result of procambial development. Number of cells between metaxylem and PSE increases during procambial development (data shown in Extended Data Fig. 1). Therefore, the model is defined as a line in one spatial dimension representing 3, 4 or 5 cells from the centre of the xylem axis to the outer edge of the PSE cell. c, The regulatory network embedded within each cell. Regulatory interactions shown using a black line have been published previously, whilst those using a green line are described for the first time here. d, Predicted concentration gradient of all elements in 3, 4 or 5 cells (from left to right). Within different root geometries corresponding to different growth stages in Arabidopsis, both PEAR1 and HD-ZIP III are co-localized in PSE-IN, forming a sharp concentration boundary within this cell. In f-h, only PEAR and HD-ZIP concentrations are shown, whilst all model components are shown here in all cases. e, Quantification of expression level of CNA-GFP and PEAR1-GFP at 4-cell region in wild-type background. n represents individual measurements across 3 (CNA-GFP) or 4 (PEAR1-GFP) independent biological samples, respectively. Bar graphs represent mean. Error bars are s.d. Dots, individual data points. f, Including all three interactions labelled in panel c in the model, results in the formation of sharp concentration gradients of PEAR1 (black line) and HD-ZIP III (blue line) with the boundary forming in the PSE-IN. g, In simulations where HD-ZIP III does not regulate PEAR1 diffusion (Interaction (3) in panel c), PEAR1 protein is predicted to spread into the procambium and metaxylem as shown in Fig. 4k-m and Extended Data Fig. 4. h, In simulations where PEAR1 does not activate HD-ZIP III transcription (Interaction (1) in panel c), the concentration of HD-ZIP III is reduced in the PSE-IN cell as shown in Fig. 4h-j. i, A regulatory mechanism forming the boundary between a dividing and a non-dividing cell during procambial development. White, orange, dark-green and light-green arrowheads indicate PSE, PSE-LN, PSE-IN and IPC. Scale bars, 25 μm.
Supplementary Material
This file contains a list of primers and plasmids used in this study.
This file contains individual P-values for Tukey’s HSD test (for Fig. 2f and Extended Data Fig. 5b-d) and for two-sided Student’s t-test (for Extended Data Fig. 7b). For Tukey’s HSD test, data contains: difference in mean values (diff), the lower end point of the interval (lwr), the upper end point of the interval (upr) and p-value after adjustment for the multiple comparisons (p adj).
This file contains a list of putative PEAR1/PEAR2 direct targets and their description.
Acknowledgements
We thank E. Scarpella, ABRC and NASC for materials, Katja Kainulainen, Karolina Blajecka, Mikko Herpola and Alessandro Mainardi for technical assistance, Natalie Clark for assisting with the scanning FCS analysis, Jenny Jansen for technical assistance with microarray hybridizations, Oleg Kambur and Lothar Kalmbach for assistance in generating the heatmap, and D. Weijers and T. Kakimoto for helpful discussions, A. Groenheide, E. Cornelissen, M. Chu, A. Korppoo and Alba Rodriguez Diez for technical support. This works was supported by Finnish Centre of Excellence in Molecular Biology of Primary Producers (Academy of Finland CoE program 2014-2019) decision #271832, the Gatsby Foundation [GAT3395/PR3)], University of Helsinki [award 799992091] and the European Research Council Advanced Investigator Grant SYMDEV [No. 323052] to Y. H., a NSF-BBSRC MCSB 1517058 to R.S. and Y.H., an ERC Consolidator grant (PLANTSTEMS), a Heisenberg Professorship of the German Research Foundation (DFG, GR2104/5-1) and the SFB 873 (DFG) to T.G., the Netherlands Organization for Scientific Research (NWO; VIDI-864.13.001) and The Research Foundation - Flanders (FWO; Odysseus II G0D0515N and 12D1815N) to W.S. and B.D.R., respectively, a JSPS postdoctoral fellowship for research abroad and JSPS KAKENHI Grant [17K15138] to S.M., Swiss National Science Foundation Postdoc Mobility Grant (P300P3_147894) to P.R., a JSPS Research Fellowship for Young Scientists and JSPS KAKENHI Grant [JP16J00131] to K.T., Bayer Science and Education Foundation, German Academic Exchange Service (DAAD) to B.B., The Finnish Academy of Science to J. O. H., Herchel Smith Postdoctoral Research Fellowship (Herchel Smith Fund) to S.O.
Footnotes
Code availability
The code for mathematical model is available on request.
Data availability
All lines and data supporting the findings of this study are available from the corresponding author upon request. The microarray data files are available at Gene Expression Omnibus (GEO) (accession no. GSE115183).
Author contributions
S.M., P.R. and I.S. contributed equally to this work. S.M. characterized the molecular interactions among PEAR and HD-ZIP III module. P.R. identified and quantified phenotype in the PEAR loss of function mutants with help of B.B. I.S. determined phloem specific DOFs and their downstream genes with input from B.D.R., W.S., M.B. and G. H. K.T. characterized PEAR-HD-ZIP III combinatorial mutants. B.B. generated tmo6 CRISPR mutants. J.H. performed in situ hybridization. N.M. and A.B. designed and performed computational modelling. H.H. produced CRE1 inducible line. S.O. assisted in the microarray experiments. K.H. and K.N. produced HD-ZIP III reporter lines. O.S. and A.P.M. provided the destination vector pSm43GW. R.S. and A.P.M. provided the pARR5::RFPer line. E.S.W., Y.K., T.G. and C.M. shared informative non-published data. R.S. analysed diffusion coefficient of PEAR1-GFP with P.R. B.D.R. and Y.H. participated in experimental design. S.M. and Y.H. wrote the manuscript and all authors commented on the manuscript. B.D.R. and Y.H. are co-corresponding authors.
References
- 1.Esau K. Vascular differentiation in plants. Holt, Rinehart and Winston; 1965. [Google Scholar]
- 2.Crick FH, Lawrence PA. Compartments and polyclones in insect development. Science. 1975;189:340–347. doi: 10.1126/science.806966. [DOI] [PubMed] [Google Scholar]
- 3.Dickson B, Sprenger F, Hafen E. Prepattern in the developing Drosophila eye revealed by an activated torso--sevenless chimeric receptor. Genes Dev. 1992;6:2327–2339. doi: 10.1101/gad.6.12a.2327. [DOI] [PubMed] [Google Scholar]
- 4.McConnell JR, et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 5.Mähönen AP, et al. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mähönen AP, et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 7.Bishopp A, et al. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol. 2011;21:917–926. doi: 10.1016/j.cub.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 8.De Rybel B, et al. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345 doi: 10.1126/science.1255215. 1255215. [DOI] [PubMed] [Google Scholar]
- 9.Vatén A, et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell. 2011;21:1144–1155. doi: 10.1016/j.devcel.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 11.Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7:555–560. doi: 10.1016/s1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- 12.Kim HS, et al. The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. Plant J. 2010;64:524–535. doi: 10.1111/j.1365-313X.2010.04346.x. [DOI] [PubMed] [Google Scholar]
- 13.Skirycz A, et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- 14.Rueda-Romero P, Barrero-Sicilia C, Gómez-Cadenas A, Carbonero P, Oñate-Sánchez L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J Exp Bot. 2012;63:1937–1949. doi: 10.1093/jxb/err388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, Qin G, Gu H, Qu LJ. Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell. 2009;21:3518–3534. doi: 10.1105/tpc.108.064139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlereth A, et al. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- 17.Wallner ES, et al. Strigolactone- and Karrikin-Independent SMXL Proteins Are Central Regulators of Phloem Formation. Curr Biol. 2017;27:1241–1247. doi: 10.1016/j.cub.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siligato R, et al. MultiSite Gateway-Compatible Cell Type-Specific Gene-Inducible System for Plants. Plant Physiol. 2016;170:627–641. doi: 10.1104/pp.15.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mähönen AP, et al. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol. 2006;16:1116–1122. doi: 10.1016/j.cub.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 20.Kiba T, Aoki K, Sakakibara H, Mizuno T. Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 2004;45:1063–1077. doi: 10.1093/pcp/pch128. [DOI] [PubMed] [Google Scholar]
- 21.Prigge MJ, et al. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlsbecker A, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyashima S, Koi S, Hashimoto T, Nakajima K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development. 2011;138:2303–2313. doi: 10.1242/dev.060491. [DOI] [PubMed] [Google Scholar]
- 24.Baima S, et al. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development. 1995;121:4171–4182. doi: 10.1242/dev.121.12.4171. [DOI] [PubMed] [Google Scholar]
- 25.Donner TJ, Sherr I, Scarpella E. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development. 2009;136:3235–3246. doi: 10.1242/dev.037028. [DOI] [PubMed] [Google Scholar]
- 26.O'Malley RC, et al. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 2016;166:1598. doi: 10.1016/j.cell.2016.08.063. [DOI] [PubMed] [Google Scholar]
- 27.Gaudinier A, et al. Enhanced Y1H assays for Arabidopsis. Nat Methods. 2011;8:1053–1055. doi: 10.1038/nmeth.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muraro D, et al. Integration of hormonal signaling networks and mobile microRNAs is required for vascular patterning in Arabidopsis roots. Proc Natl Acad Sci U S A. 2014;111:857–862. doi: 10.1073/pnas.1221766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellor N, et al. Theoretical approaches to understanding root vascular patterning: a consensus between recent models. J Exp Bot. 2017;68:5–16. doi: 10.1093/jxb/erw410. [DOI] [PubMed] [Google Scholar]
- 30.De Rybel B, Mähönen AP, Helariutta Y, Weijers D. Plant vascular development: from early specification to differentiation. Nat Rev Mol Cell Biol. 2016;17:30–40. doi: 10.1038/nrm.2015.6. [DOI] [PubMed] [Google Scholar]
- 31.Fauser F, Schiml S, Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014;79:348–359. doi: 10.1111/tpj.12554. [DOI] [PubMed] [Google Scholar]
- 32.Lei Y, et al. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant. 2014;7:1494–1496. doi: 10.1093/mp/ssu044. [DOI] [PubMed] [Google Scholar]
- 33.Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development. 2000;127:595–603. doi: 10.1242/dev.127.3.595. [DOI] [PubMed] [Google Scholar]
- 34.Kurihara D, Mizuta Y, Sato Y, Higashiyama T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 2015;142:4168–4179. doi: 10.1242/dev.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pound MP, French AP, Wells DM, Bennett MJ, Pridmore TP. CellSeT: novel software to extract and analyze structured networks of plant cells from confocal images. Plant Cell. 2012;24:1353–1361. doi: 10.1105/tpc.112.096289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashimoto K, Miyashima S, Sato-Nara K, Yamada T, Nakajima K. Functionally Diversified Members of the MIR165/6 Gene Family Regulate Ovule Morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2018;59:1017–1026. doi: 10.1093/pcp/pcy042. [DOI] [PubMed] [Google Scholar]
- 37.Hejátko J, et al. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat Protoc. 2006;1:1939–1946. doi: 10.1038/nprot.2006.333. [DOI] [PubMed] [Google Scholar]
- 38.Wickham H, Sievert C. ggplot2 : elegant graphics for data analysis. Second edition. Springer; 2016. [Google Scholar]
- 39.De Rybel B, et al. A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell. 2013;24:426–437. doi: 10.1016/j.devcel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Oparka KJ, Duckett CM, Prior DAM, Fisher DB. Real - time imaging of phloemun loading in the root tip of Arabidopsis. The Plant Journal. 1994;6:759–766. doi: 10.1046/j.1365-313X.1994.6050759.x. [DOI] [Google Scholar]
- 41.Clark NM, et al. Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. Elife. 2016;5 doi: 10.7554/eLife.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark NM, Sozzani R. Measuring Protein Movement, Oligomerization State, and Protein-Protein Interaction in Arabidopsis Roots Using Scanning Fluorescence Correlation Spectroscopy (Scanning FCS) Methods Mol Biol. 2017;1610:251–266. doi: 10.1007/978-1-4939-7003-2_16. [DOI] [PubMed] [Google Scholar]
- 43.O'Lexy R, et al. Exposure to heavy metal stress triggers changes in plasmodesmatal permeability via deposition and breakdown of callose. J Exp Bot. 2018;69:3715–3728. doi: 10.1093/jxb/ery171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Digman MA, et al. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys J. 2005;89:1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Digman MA, Gratton E. Analysis of diffusion and binding in cells using the RICS approach. Microsc Res Tech. 2009;72:323–332. doi: 10.1002/jemt.20655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains a list of primers and plasmids used in this study.
This file contains individual P-values for Tukey’s HSD test (for Fig. 2f and Extended Data Fig. 5b-d) and for two-sided Student’s t-test (for Extended Data Fig. 7b). For Tukey’s HSD test, data contains: difference in mean values (diff), the lower end point of the interval (lwr), the upper end point of the interval (upr) and p-value after adjustment for the multiple comparisons (p adj).
This file contains a list of putative PEAR1/PEAR2 direct targets and their description.