Abstract
Rac GTPases are members of the Rho family regulating the actin cytoskeleton and implicated in neuronal development. Ubiquitous Rac1 and neuron-specific Rac3 GTPases are coexpressed in the developing mammalian brain. We used Cre-mediated conditional deletion of Rac1 in neurons combined with knockout of neuron-specific Rac3 to study the role of these GTPases in neural development. We found that lack of both genes causes motor behavioral defects, epilepsy, and premature death of mice. Deletion of either GTPase does not produce evident phenotypes. Double-knockout mice show specific defects in the development of the hippocampus. Selective impairment of the dorsal hilus of double-knockout animals is associated with alteration in the formation of the hippocampal circuitry. Axonal pathways to and from the dorsal hilus are affected because of the deficit of hilar mossy cells. Moreover, analysis of Rac function in hippocampal cultures shows that spine formation is strongly hampered only in neurons lacking both Rac proteins. These findings show for the first time that both Rac1 and Rac3 are important for the development of the nervous system, wherein they play complementary roles during late stages of neuronal and brain development.—Corbetta, S., Gualdoni, S., Ciceri, G., Monari, M., Zuccaro, E., Tybulewicz, V. L. J., de Curtis, I. Essential role of Rac1 and Rac3 GTPases in neuronal development.
Keywords: hippocampus, hilar mossy cells, dendritic spines, axons
RAC GTPASES ARE MEMBERS OF THE Rho family regulating actin dynamics (1). Evidence indicating the involvement of Rac proteins in different aspects of neuronal development includes formation of axon and dendrites, axonal navigation, and synaptogenesis (2, 3). There are three Rac genes in vertebrates encoding the Rac1, Rac2, and Rac3 proteins, sharing 88-92% sequence identity. Rac1 and Rac3 are coexpressed in neurons (4), and Rac2 is expressed in hematopoietic cells (5). Most studies on vertebrate neuronal development have focused on Rac1, whereas very few studies exist on Rac3, despite its neuronal coexpression with Rac1 (4).
Rac1 is ubiquitous, and Rac1-knockout mice are early embryonic lethal, because of the requirement for this GTPase in early development (6). To date, the analysis of transgenic mice expressing mutated forms of Rac1 has shown effects on axon terminals and dendritic spines in Purkinje cells (7). Although studies with mutants have been informative to start assembling a picture of how these proteins work to build the vertebrate nervous system, it is clear that conclusions from these studies must be taken with caution, because Rac1 mutants may interfere with the function of related GTPases. Recently, early conditional deletion (8) and knockdown in vitro (9) have confirmed a role of Rac1 in axon guidance in vivo and dendritogenesis in vitro, respectively.
Rac3 was first identified in humans (10) and in birds as Rac1B (11). The Rac3 protein is specifically expressed in the nervous system (11) and is developmentally regulated in avian and mouse brains, with a peak at times of intense neurite branching and synaptogenesis (12, 13). Rac3-knockout mice are viable (4), and recent findings indicate a role of Rac3 in cognitive development (14).
The relative contributions of Rac1 and Rac3 to the development of the vertebrate nervous system are unknown. Rac1 flox mice allowed the analysis of Rac1 function after conditional deletion of the gene in the immune system (15, 16) and in early brain development (8). On the other hand, the appearance of Rac3 first in vertebrates (3) suggests that this gene may play a role in the late development of brains of higher complexity. The possibility of functional interactions between Rac1 and Rac3 during the development of the vertebrate nervous system has never been addressed. By combining conditional mutagenesis of the Rac1 gene with the deletion of the Rac3 gene, we show here that both proteins are important for the late development of the brain and play key functions in later stages of neuronal maturation.
Materials and Methods
Mice and analysis of genomic DNA
Animal care was in accordance with institutional guidelines. SynI-Cre transgenic mice specifically expressing the viral Cre recombinase under the control of the synapsin I promoter in differentiated neurons (17), Rac1flox/flox (Rac1F/F) mice carrying floxed Rac1 alleles (16), Rac3−/− mice with deletion of the Rac3 gene (4), and ROSA26 mice (18) have been described previously.
For the generation of mice with the specific conditional deletion of Rac1 in neurons, Rac1F/F mice were bred to SynI-Cre transgenic animals. Offspring, Rac1F/+SynI-Cre mice, were crossed to Rac1F/F mice to obtain Rac1F/FRac3+/+SynI-Cre (Rac1N) mice carrying neuronal deletion of the Rac1 gene.
For the generation of mice with inactivation of both Rac1 and Rac3 in neurons, Rac3−/− mice were bred to Rac1F/F mice to generate Rac1F/+ Rac3−/+ mice. Rac1F/+ Rac3−/+ mice were intercrossed to generate Rac1F/F Rac3−/− mice (defined as Rac3KO). The Rac3−/− mice were mated to SynI-Cre mice to generate Rac3−/+SynI-Cre mice. These mice were crossed with Rac3−/− mice to obtain SynI-Cre/Rac3−/− mice, which were bred to Rac1F/F Rac3−/− mice to generate Rac1F/+ Rac3−/− SynI-Cre mice. Breeding of Rac1F/+ Rac3−/−SynI-Cre and Rac1F/FRac3−/− animals was finally used to produce Rac1F/FRac3−/− SynI-Cre mice (defined as Rac1N/Rac3KO), carrying deletions of both Rac3 and Rac1 genes in neurons. For experiments, Rac1F/F mice were crossed with Rac1F/+ SynI-Cre mice to obtain Rac1F/F and Rac1F/F SynI-Cre littermates, defined as WT and Rac1N, respectively, in the text; whereas Rac1F/F Rac3−/− mice were crossed with Rac1F/+SynI-Cre/Rac3−/− mice to obtain Rac1F/F/Rac3−/− and Rac1F/FSynI-Cre/Rac3−/− littermates, defined as Rac3KO and Rac1N/Rac3KO, respectively, in the text.
The genotypes were determined by polymerase chain reaction (PCR) on genomic DNA from tails or different organs. Briefly, tail or organ samples were incubated overnight in lysis buffer [50 mM Tris-Cl (pH 8), 100 mM EDTA, 100 mM NaCl, 1% SDS, and 1 mg/ml of proteinase K), followed by precipitation with isopropanol. Specific primers for PCR were F1 (5’-CATTTCTGTGGCGTCGCCAAC-3’) and R2 (5’-CACGCGGCCGAGCT GTGGTG-3’) for the Rac3 wildtype allele and R3 (5’-TTGCTGGTGTCCAGACCAAT-3’) from the lacZ gene for the Rac3 targeted allele (4). The three primers were used in a multiplex PCR with LA Taq (Takara Bio Inc., Otsu, Shiga, Japan) with the following amplification conditions: 1 min at 94°C, 30 cycles of 20 s at 98°C and 1 min at 66°C, and 10 min at 72°C at the end of the run. Amplification products were resolved on a 1.6% agarose gel. Primers Pr1 (5’-ATTTTGTGCCAAGGACAGTGACAAGCT-3’), Pr2 (5’-GAAGGAGAAGAAGCTGACTCCCATC-3’), and Pr3 (5’-CAGCCACAGGCAATGACAGATGTTC-3’) were used for the identification of the floxed and deleted Rac1 alleles. PCR analysis with these primers was performed with GoTaq polymerase (Promega, Madison, WI, USA) under the following amplification conditions: 5 min at 94°C, 30 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, followed by 7 min at 72°C. For the SynI-Cre transgene we used the primers Pr4 (5’-CCAG CACCAAAGGCGGGC-3’) and Pr5 (5’-TGCATCGACCGGTAATGCAG-3’) under the following conditions: 5 min at 94°C, 32 cycles of 30 s at 94°C, 30 s at 61°C, and 30 s at 72°C, followed by 7 min at 72°C.
Northern blot analysis
Total RNA was isolated from postnatal day (P) 13 brains and adult spleen with the RNeasy Midi kit (Qiagen, Milano, Italy). Northern blot analysis of total RNA (15 μg/lane) was performed as described previously (19). Blots were hybridized with a 378-bp PCR fragment corresponding to base pairs 181-558 of the translated Rac2 cDNA (amino acids 61-186) or with a 1.2-kb fragment corresponding to part of the 3’-untranslated region of the Rac1 cDNA. Hybridization took place in hybridization buffer supplemented with 32 P-labeled probes (1-2×106 cpm/ml) for 15 h at 65°C. After high-stringency washes at 65°C, X-ray films were exposed for 3-12 h to the hybridized filters.
Reflexological tests and epileptic behavior
Ten animals for each genotype (WT, Rac1N, Rac3KO, and Rac1N/Rac3KO) were tested for the following reflexological tests at P3, P6, P9, and P12 following a well-characterized protocol (20): righting reflex, negative geotaxis, cliff drop aversion, grasp reflex, and tail suspension. The responses in all tests except for the righting reflex were recorded using the following numerical scoring method: 0 (no response), 1 (weak response), and 2 (strong response). For the righting reflex, the maximal effect was graded as 3. The average scores for each group of mice were plotted for comparison. Animals were weighed before the tests. Statistical significance was assessed by the Student’s t test. Differences were considered significant at P < 0.05.
The behavioral manifestations of epileptic crises were classified by considering some of the parameters previously used to identify seizures (21), including running or bouncing, unilateral or bilateral forelimb and hind limb clonus plus falling, and tonic forelimb and hind limb extension. The classification was performed by an investigator unaware of the genotype of the animals.
Antibodies
The following antibodies and dilutions were used for biochemical and morphological analysis: anti-calretinin at 1:1000, and anti-tau-1 at 1:100 (Chemicon International, Temecula, CA, USA); anti-Cdc42 at 1:500 (Cell Signaling Technology, Danvers, MA, USA); anti-green fluorescent protein (GFP) at 1:400 (Molecular Probes Inc., Eugene, OR, USA); anti-Rac3 at 1:500 (4); anti-glutamate receptor subunits 2 and 3 (GluR2/3) at 1:100, anti-PSD95 at 1:500, anti-RhoA at 1:1000 and anti-Rac1 at 1:1000 (Upstate Biotechnology, Lake Placid, NY, USA); anti-zinc transporter-3 (ZnT-3) at 1:200 (gift of Richard Palmiter) (22); anti-neurofilament at 1:100 (Biomol International L.P., Plymouth Meeting, PA, USA); anti-synapsin I at 1:200 (gift of Flavia Valtorta, University Vita-Salute San Raffaele, Milan, Italy) (23); anti-active caspase-3 at 1:300 (BD Biosciences, San Jose, CA, USA); anti-VAMP2 at 1:500 (Synaptic System, Gõttingen, Germany); anti-glyceraldehyde-3-phosphate dehydrogenase (GADPH) at 1:5000 (Biogenesis Inc, Pool, UK); anti-Cre recombinase at 1:500 (Covance, Emeryville, CA, USA); anti-mitogen-activated protein 2 (MAP2) at 1:500 (Sigma-Aldrich, St. Louis, MO, USA); and anti-glial fibrillary acidic protein (GFAP) at 1:1 (gift of Virginia Lee, University of Pennsylvania School of Medicine, Philadelphia, PA, USA).
Biochemical analysis
Brains at the indicated developmental stages were extracted at 4°C with lysis buffer [1% Triton X-100, 150 mM NaCl, 1 mM sodium orthovanadate, 10 mM NaF, 20 mM Tris-Cl (pH 7.5), and protease inhibitors (Complete, EDTA-free, Protease Inhibitor Cocktail; Roche S.p.A., Milan, Italy)]. For immunoprecipitation, primary antibodies preadsorbed to 25 μl of protein A-Sepharose beads (Amersham Biosciences, Piscataway, NJ, USA) were added to lysates (2 mg of protein/immunoprecipitation) and incubated for 3 h at 4°C with rotation. Immunoprecipitates were washed 4 times with 0.5 ml of lysis buffer with 0.5% Triton X-100. Brain lysates (100 μg/lane) and immunoprecipitates were analyzed by SDS-PAGE and immunoblotting. After incubation with primary antibodies, filters were incubated with 0.2 μCi/ml of 125I-protein A or 125I-anti-mouse immunoglobulin (Amersham Biosciences), washed, and exposed to Amersham Hyperfilm-MP.
For the determination of the relative amounts of Rac3 and Rac1 in brain, duplicate aliquots of brain lysates from wild-type mice at P7, corresponding to the peak of Rac3 protein expression in brain (13), were immunoprecipitated with anti-Rac3 antibodies. Immunoprecipitates were blotted with either anti-Rac1 (recognizing both GTPases)- or anti-Rac3-specific antibodies. Blots were scanned, and the ratio between the values obtained with the two antibodies (anti-Rac3/anti-Rac1) was used to normalize with respect to Rac1 the values obtained by immunoblotting with anti-Rac3.
Histology, immunohistochemistry, and immunofluorescence
Between 4 and 10 mice/genotype were considered for each type of analysis. Mice were fixed under deep anesthesia by transcardial perfusion with 4% paraformaldehyde in phosphate-buffered saline (PBS). Brains were removed from skulls and postfixed overnight at 4°C. For histology, after fixation, organs were frozen or dehydrated and embedded in paraffin; 10-μm-thick sections were stained with cresyl violet or with hematoxylin and eosin, according to standard protocols.
For immunostaining, samples were washed with PBS, cryoprotected with sucrose in PBS, and frozen in optimal cutting temperature (O.C.T.) compound (VWR International Ltd., Poole, UK). For immunohistochemistry, 8- to 12-μm sections were blocked for 1 h with blocking buffer (0.1% Triton X-100, 10% goat serum, and 0.2 mg/ml bovine serum albumin in PBS), and incubated overnight at 4°C with primary antibodies. For immunoperoxidase staining, primary antibodies were detected by using a Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA, USA). Control sections were incubated with secondary antibodies only. Sections were analyzed with an Axio-plan2 microscope (Carl Zeiss MicroImaging GmbH, Jena, Germany).
For immunofluorescence, 8- to 20-μm-thick sections were blocked for 1 h in 10% serum and 0.1% Tween 20 in PBS for active caspase-3 or in 15% serum, 0.3% Triton X-100, 450 mM NaCl, and 20 mM sodium phosphate buffer (pH 7.4) for incubation with other antibodies. Sections were first incubated overnight at 4°C with primary antibodies, followed by incubation for 1.5-2 h at room temperature with fluorescently labeled secondary antibodies (Molecular Probes Inc.) or with tetramethylrhodamine B isothiocyanate-conjugated phalloidin (Sigma-Aldrich). Finally, sections were incubated with 4’,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) to detect the nuclei. Control sections were incubated with secondary antibodies only. Sections were analyzed with an UltraVIEW ERS spinning disk confocal microscope (PerkinElmer, Wellesley, MA, USA), a Bio-Rad MRC 1024 confocal microscope (Bio-Rad, Hercules, CA, USA), or a Zeiss Axiophot (Carl Zeiss MicroImaging GmbH).
Quantifications using ImageJ [U.S. National Institutes of Health (NIH), Bethesda, MD, USA] were performed on 7 to 8 sections from at least 3 to 4 different mice for each genotype and each experimental condition analyzed.
5-Bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) staining
SynI-Cre mice were mated with LacZ ROSA26 tester reporter mice (18). Double-heterozygous P13 and P8 mice were perfused under deep anesthesia with 4% paraformaldehyde in PBS. Brains were removed from skulls and immersed in fixative for 30 min at 4°C. Samples were washed with PBS, cryoprotected with 30% sucrose in PBS, and frozen in O.C.T. compound; 25-μm-thick sections were washed 3 times in 0.02% Nonidet P-40/PBS and stained in PBS with 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% Nonidet P-40, and 1 mg/ml X-gal at 37°C for 1.5 h. After 30 min in 4% paraformaldehyde in PBS at 4°C, sections were washed with PBS and mounted with Gelvatol.
Hippocampal neurons and transfections
Primary neuronal cultures were prepared from hippocampi of embryonic day (E) 17.5 mice from wild-type and mutant mice as described previously (24). Briefly, cells were plated onto poly (L-lysine)-coated glass coverslips (Sigma-Aldrich, Steinheim, Germany) in minimal essential medium (Life Technologies, Invitrogen Corp., Carlsbad, CA, USA) supplemented with 10% horse serum (Hyclone, Logan, UT, USA), 2 mM glutamine (BioWhittaker, Verviers, Belgium), and 3.3 mM glucose. Four hours after plating, coverslips were transferred into new dishes containing glia-conditioned hippocampal medium [minimal essential medium supplemented with 1% N2 supplement (Invitrogen, San Diego, CA, USA), 2 mM glutamine, 1 mM sodium pyruvate (Sigma-Aldrich), and 4 mM glucose]. Cells were incubated at 37°C in a 5% CO2 humidified atmosphere for the times indicated.
Neurons were either transfected with pEGFP-N1 plasmid (Clontech, Mountain View, CA, USA), or cotransfected with pEGFP-N1 and pEGFP-CRE plasmids (25) by using Lipo-fectamine 2000 (Invitrogen Corp.) in hippocampal medium. After fixation of cultures with 4% paraformaldehyde in 4% sucrose, 2 mM EGTA, and 120 mM sodium phosphate (pH 7.4), cells were permeabilized with 0.3% Triton X-100 and processed for immunofluorescence with the indicated antibodies. Primary antibodies were detected with Alexa Fluor 488/568-conjugated secondary antibodies (Molecular Probes). Images were captured with a Zeiss Axiophot epifluorescence microscope equipped with a C4742-95-12HR digital camera (Hamamatsu, Hamamatsu City, Japan).
Analysis of dendritic spine morphology
Neurons fixed at 14 days in vitro (DIV) were immunostained with anti-GFP and anti-PSD95 antibodies. For quantification, in each experiment 15-30 different 100-μm segments of primary and secondary dendrites from 7-10 neurons were analyzed for spines and protrusions. Two independent experiments were analyzed for each condition. GFP-positive dendritic protrusions were classified according to their morphology: mature spines including mushroom-shaped and stubby spines; other protrusions with no head, including short filopodia (<4 μm long), long filopodia (>4 μm long), and lamellipodia. The total protrusion density per 100 μm of dendrite was calculated by including all morphological classes of dendritic protrusions. At least 450 protrusions per condition per experiment have been considered for the quantification. Images were analyzed using Adobe Photoshop 8 (Adobe System, San Jose, CA, USA) and the public domain NIH ImageJ software. The reported values represent means ± SE. The data were analyzed by the Student’s t test (two-tailed distribution and two-sample unequal variance).
Results
Rac1N/Rac3KO mice are neurologically impaired
At the peak of expression, Rac3 represents ~8% of total Rac in brain (Fig. 1). Because Rac3 distribution is more restricted than that of Rac1, its expression in mouse neurons is likely to be higher than the average percentage in the whole brain. Rac1-null mice die before E9.5 (6), whereas Rac3-deficient mice are vital. To generate mice with Rac1 and Rac3 deletions in neurons, we used Rac3-null (Rac3KO) mice (4), mice with floxed Rac1 allele (Rac1flox) (16), and synapsin I Cre transgenic (SynI-Cre) mice expressing a Cre recombinase under the synapsin I promoter (17), to generate Rac1N (Rac1F/F SynI-Cre) and Rac1N/Rac3KO mice. SynI-Cre function is first detected at E12.5 (17) and is thus expected to remove Rac1 at late stages during neuronal development, when Rac3 expression in brain is high (4, 13).
Figure 1.
Relative amounts of Rac3 and Rac1 in P7 mouse brain. Duplicate samples from P7 brain lysates from wild-type animals were immunoprecipitated (2 mg protein lysate/immunoprecipitation) with anti-Rac3 antibody and blotted with either anti-Rac1 recognizing both Rac1 and Rac3 (top filter) or anti-Rac3-specific antibodies (bottom filter). Lane 1: 200 μg of lysate; lanes 2 and 4: 200 μg of unbound fractions after immunoprecipitation with preimmune or immune anti-Rac3 Ab, respectively; lanes 3 and 5: immunoprecipitation from 2 mg of lysate with preimmune or immune anti-Rac3 antibody, respectively. Quantification on blots (see Materials and Methods) from two independent experiments shows that Rac3 and Rac1 represent ~7.6 ± 0.4 and 92.4 ± 0.44%, respectively, of the total Rac in P7 mouse brain lysates.
We have analyzed the activity of the SynI-Cre transgene in SynI-Cre/ROSA26 tester mice (18). The trans-gene was widely active in neurons of developing brain (Fig. 2a). In the hippocampus, SynI-Cre was active in the CA3, hilus, and external granule cell layer of the dentate gyrus, whereas it was mostly undetectable in the CA1 and in the inner granule cell layer (Fig. 2b–d).
Figure 2. Activation of the SynI-Cre transgene.
X-Gal staining on P13 (a–c) and P8 (d) SynI-Cre/ROSA26 brain sections. In the hippocampus of P13 (b) and P8 mice (d), SynI-Cre expression is restricted mostly to the CA3 and dentate gyrus, including the hilus (c), and is absent in most cells of the CA1 region. Cb, cerebellum; Co, cortex; DG, dentate gyrus; Hi, hilus; Hp, hippocampus, Ob, olfactory bulb; Pn, pons; Th, thalamus. Scale bars = 500 μm (a); 200 μm (b, d); 100 μm (c).
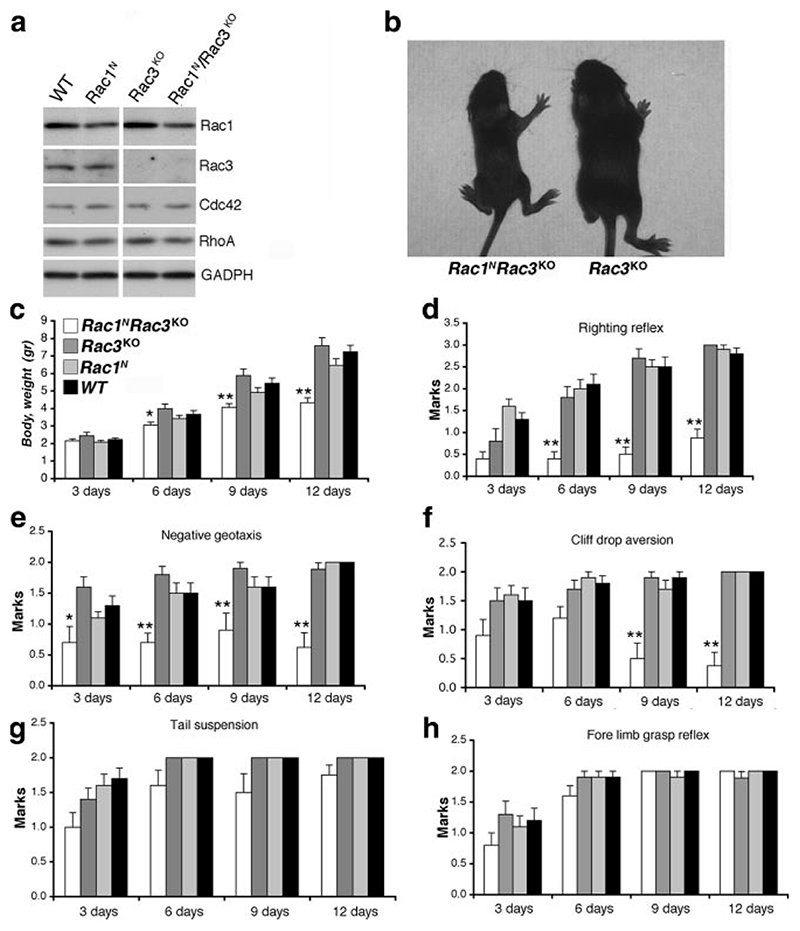
Rac1 deletion was confirmed by genomic PCR (Fig. 3a-c) and by the decrease of Rac1 protein (Fig. 4a) and transcript (Fig. 3d). Incomplete ablation of Rac1 in brain was due to retention of the protein in the glia and in the neurons in which SynI-Cre was not active (Fig. 2). As expected, Rac3 was absent in Rac3KO and Rac1N/Rac3KO brains (Fig. 4a). Rac1 and Rac3 were not up-regulated in the brains of Rac3KO and Rac1N mice, respectively (Fig. 4a) (4). Moreover, deletion of Rac1 and Rac3 was not compensated for by up-regulation of hematopoietic-specific Rac2 (Fig. 3d).
Figure 3. Production of Rac1N and Rac1N/Rac3KO mice.
a) Scheme of the Rac1 flox allele before (left) and after (right) Cre-mediated recombination. b, c) Genotyping by PCR on DNA from tail (b) and spinal cord and kidney (c) of mice with the indicated genotypes. DNAs were screened with primer (Pr) 2 and Pr3 to produce a 0.33-kb fragment from the floxed allele (F) and a 0.27-kb fragment from the wild-type allele (wt). The 0.17-kb PCR fragment from the deleted allele (del) was obtained with Pr1 and Pr3 from spinal cord. b) A 0.45-kb PCR fragment (Cre) was obtained in the SynI-Cre-positive transgenic mice with Pr4 and Pr5. PrF1 and PrR3 generate a 0.37-kb fragment from the Rac3 mutant allele (Rac3KO). Sequences of all primers are reported in Materials and Methods. d) Northern blot analysis on total RNA (15 μg/lane) hybridized with probes specific for Rac1 (left panels) or Rac2 (right panels; spleen as a positive control for Rac2). Bottom panels: gels used for blotting, stained with ethidium bromide.
Figure 4. Characterization of double-knockout mice.
a) Immunoblotting for the indicated antigens on P13 brain lysates. Rac1 is decreased in brains of Rac1N and Rac1N/Rac3KO (P<0.035) mice. Levels of Cdc42 and RhoA appear unaltered in lysates from mutant mice. b) Rac1N/Rac3KO (left) and Rac3KO (right) P13 mice. c-h) Body weight (c), righting reflex (d), negative geotaxis (e), cliff drop aversion (f), tail suspension (g), and forelimb grasp reflex (h) were assessed as described in Material and Methods. Bars and error bars represent mean ± se scores; 10 animals/group.*P < 0.05, **P < 0.005 vs. control littermates; Student’s t test.
Rac1N and Rac3KO mice developed normally, whereas Rac1N/Rac3KO mice were smaller in size (Fig. 4b, c). There were no differences in brain weight (P=0.41) between Rac1N/Rac3KO (0.31 g±0.023, n=5) and Rac3KO littermates (0.28 g±0.025, n=5), although Rac1N/Rac3KO mice revealed neurological abnormalities including decreased righting reflex, negative geotaxis, and cliff drop aversion (Fig. 4d-h). Moreover, the double-mutant mice developed spontaneous seizures (Supplemental Movie 1) and died around P13. Spontaneous seizures were evident in 25 of the 28 mice kept under observation, and they were particularly striking and prolonged at P12/P13, possibly contributing to the death of the animals around this age. For this reason, mice were sacrificed at P13 or earlier for the analyses performed in this study.
Rac1N/Rac3KO mice show specific defects in the dorsal hilus
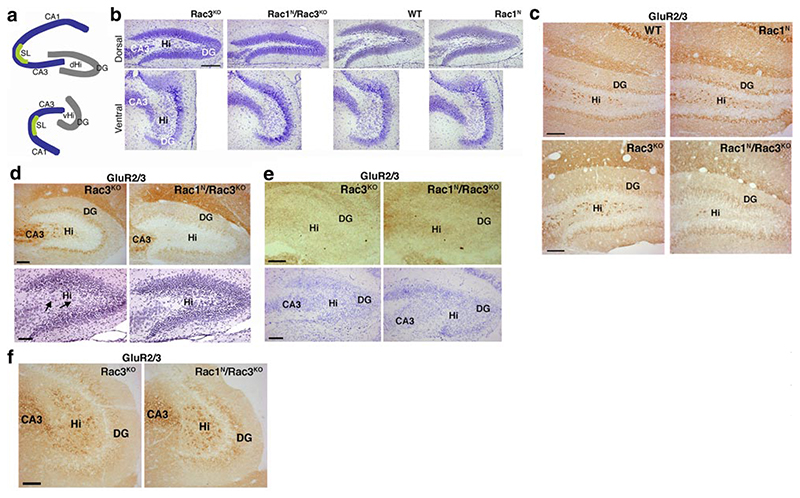
Analysis of the brains to look for anatomical correlates of the observed phenotypes revealed that the dorsal hippocampal hilus (Fig. 5a) was strikingly thinner in all P13 Rac1N/Rac3KO mice observed compared with that in WT and single mutant mice (Fig. 5b). The average reduction of the width of the dorsal hilus was 39.4 ± 3.8% compared with control Rac3KO littermates (n=9 Rac1N/Rac3KO, n=8 Rac3KO mice; P<0.0001). Thinning of the dorsal hilus was evident at different levels of the dorsal hippocampus of Rac1N/Rac3KO mice (unpublished results). The reduced thickness of the dorsal hilus was paralleled by a strong decrease in the number of large (Fig. 5b), GluR2/3-positive mossy cells (Fig. 5c). Hilar mossy cells represent the major excitatory neurons of the hilus, and they are involved in a range of physiological and pathological conditions (26, 27). The thinning of the dorsal hilus was evident also by staining for neurofilament and filamentous actin, whereas the representation of GFAP-positive non-neuronal cells was maintained in the hilus of double-knockout mice (S. Corbetta and I. de Curtis, unpublished observations). Similar effects were evident in P7 double-mutant mice (Fig. 5d), when GluR2/3-positive hilar mossy cells are first identifiable (28, 29), whereas no difference was observed at P4 (Fig. 5e), when GluR2/3-positive mossy cells are not detectable yet (29). Apoptosis could not explain the observed decrease in the numbers of GluR2/3-positive mossy cells in P7 and P13 Rac1N/Rac3KO mice, because it was equally negligible in the dorsal dentate area of double-knockout and control mice at P4 (Fig. 6a) and P7 (Fig. 6b). Moreover, in contrast to what was observed in the dorsal hilus, the thickness of the ventral hilus (Fig. 5b) and the presence of GluR2/3-positive mossy cells in this region of the hippocampus (Fig. 5f) were normal even in P13 Rac1N/Rac3KO animals.
Figure 5. Reduced numbers of mossy cells in Rac1N/Rac3KO dorsal hilus at P13 and P7.
a) Scheme of the dorsal and ventral hippocampus. DG, dentate gyrus; dHi, dorsal hilus; vHi, ventral hilus; SL, stratum lucidum. b) Nissl staining on sections of dorsal (top) and ventral (bottom) dentate gyrus from P13 Rac3KO, Rac1N/Rac3KO, WT, and Rac1N mice, showing reduced thickness of the dorsal hilus in the hippocampus of Rac1N/Rac3KO mice. c) Immunohistochemistry on P13 brain sections immunostained for GluR2/3. GluR2/3-positive cells are strongly reduced in the dentate hilus of P13 Rac1N/Rac3KO mice compared with Rac3KO, WT, and Rac1N mice. d) Immunostaining for GluR2/3-positive cells (top) and hematoxylin and eosin staining (bottom) on P7 brain sections. Large cells (arrows) are strongly reduced in the dorsal hilus of P7 Rac1N/Rac3KO mice compared with P7 Rac3KO mice. e) Immunostaining for GluR2/3 (top) and Nissl staining (bottom) of sections from P4 Rac3KO and Rac1N/Rac3KO mice. No differences are detectable in the hilus of P4 double-knockout mice compared with Rac3KO mice. GluR2/3-positive mossy cells are not detectable at P4. f) Sections from the ventral hippocampus of P13 mice immunostained for GluR2/3: the reduction in mossy cells is not evident in the ventral hilus of Rac1N/Rac3KO mice compared with control Rac3KO mice. Scale bars = 200 μm (b); 100 μm (c–f).
Figure 6. Apoptosis is not affected by deletion of Rac1 and Rac3 in the dorsal hippocampus.
Immunostaining with anti-active caspase-3 antibody (green) and DAPI (blue). a, b) P4 (a),and P7 (b) dorsal hippocampi. Caspase-positive apoptotic cells are rarely seen in Rac3KO and double-knockout mice. c) Sections of the interdigit region from the anterior limb of E13.5 wild-type mice were used as positive controls for apoptosis. DG, dentate gyrus; Hi, hilus. Scale bars = 100 μm.
Neuronal circuitry is affected in the dentate gyrus of Rac1N/Rac3KO mice
Because mossy cells are part of the circuitry that plays important regulatory functions in the hippocampus (27), we investigated the effects of mossy cell depletion on neuritogenesis and synaptogenesis in the dorsal hippocampus of Rac1N/Rac3KO mice. During brain development, the axons of mossy cells project to the proximal dendrites of granule cells (Fig. 7a) in the inner molecular layer of the ipsi- and contralateral dentate gyrus (30, 31). Calretinin-positive axonal projections of mossy cells (32) were markedly decreased in the inner molecular layer of the dorsal dentate gyrus of Rac1N/Rac3KO mice (Fig. 7b) as a consequence of the drastic bilateral reduction of mossy cells. Quantification showed that both the thickness and the density of the signal corresponding to the calretinin-positive projections were reduced in double-knockout mice with respect to control mice (Fig. 7c). The reduction of mossy cell axonal projections to the dorsal dentate gyrus was accompanied by the partial dispersion of calretinin-positive immature granule cells (32) at the border between the granule cell layer and the hilus of the dorsal hippocampus of double-mutant mice (Fig. 7b). On the other hand, mossy cell projections were normal in the ventral hippocampus of double-mutant mice (Fig. 7d).
Figure 7. Mossy cell axonal projections to dentate granule cells are reduced in the dorsal hilus of Rac1N/Rac3KO mice.
a) Scheme of major connections within the hippocampus involving hilar mossy cells and dentate granule cells. Granule cells send their axons (mossy fibers) both to mossy cells in the hilus and to pyramidal neurons in the CA3. Mossy cells are bidirectionally linked to granule cells by a positive feedback loop that is strategically placed between the entorhinal cortex and the hippocampal CA3 region. b) Immunofluorescence on sections of P13 brains stained for calretinin (green) that stains immature granule cells near the hilus (Hi), and mossy cell axonal projections to the inner molecular layer (IML). The calretinin-positive projections of mossy cells to the IML of the dorsal dentate gyrus (arrows) are strongly reduced in Rac1N/Rac3KO mice compared with Rac3KO mice. GCL, granule cell layer. c) Quantification of the thickness of the inner molecular layer positive for calretinin and of the density of the calretinin signal in the inner molecular layer of the dorsal hippocampi of Rac1N/Rac3KO (dKO) and Rac3KO mice. Values represent mean ± se percentages with respect to control Rac3KO mice (n=8) after normalization (Rac3KO mice = 100%). d) Photomicrographs of ventral hippocampus of Rac3KO and Rac1N/Rac3KO mice stained with antibodies for calretinin (green). In the ventral hilus, mossy cells show strong immunoreactivity for calretinin. Calretinin-positive axonal projections and mossy cells are not affected in the ventral hippocampus of Rac1N/Rac3KO mice. Scale bars = 100 μm.
A reduction of axons, dendrites, and synaptic terminals was evident within the dorsal hilus of P13 (Fig. 8) and P7 (not shown) Rac1N/Rac3KO mice. In contrast, dendrites and synaptic terminals were normal in the stratum lucidum of the CA3 both at P13 (Fig. 8c) and at P7 (not shown) and in the ventral hilus of Rac1N/Rac3KO animals (Fig. 8c). In particular, the innervation of mossy cells by collaterals of ZnT-3-positive mossy fibers (22), the axons of dentate granule cells, was strongly reduced in the dorsal hilus of Rac1N/Rac3KO mice, whereas mossy fiber projections from granule cells to the stratum lucidum of the dorsal CA3 pyramidal neurons were normal (Fig. 9a). Quantification on sections showed also that the densities of the label for ZnT-3, MAP2, and synapsin I were decreased in the dorsal hilus of Rac1N/Rac3KO mice compared with Rac3KO mice (Fig. 9b), indicating a decrease in the density of both axons and dendrites in this region. Therefore, both neuritogenesis and synaptogenesis were heavily disrupted in neurons of the dorsal dentate gyrus, which are known to mature postnatally (30).
Figure 8. Alteration of circuitry in the dorsal hilus of P13 Rac1N/Rac3KO mice.
a) Distribution of synapsin I (green), tau (red), and nuclear DAPI (blue) in P13 hippocampi. Reduced thickness of the dorsal hilus (Hi) in Rac1N/Rac3KO mice correlates with reduced tau-positive axons and synapsin I-positive presynaptic terminals in this region. b) Enlargements of hilus from respective panels shown in a. c) Hippocampal sections from Rac3KO and Rac1N/Rac3KO P13 mice immunostained for synapsin (green) and MAP2 (red). A specific reduction of synapsin-positive presynaptic terminals and MAP2-positive dendrites is evident in the dorsal hilus (dHi) of Rac1N/Rac3KO mice compared with control Rac3KO mice (left panels). No differences between Rac3KO and Rac1N/Rac3KO mice are observed in the stratum lucidum (SL) of the dorsal CA3 subfield (central panels) or in the ventral hilus (vHi) (right panels). Scale bars = 100 μm (a); 25 μm (b); 50 μm (c).
Figure 9. Mossy fiber projections from granule cells to mossy cells are strongly reduced in Rac1N/Rac3KO mice.
a) Hippocampi from Rac3KO (top panels) and Rac1N/Rac3KO (bottom panels) P13 mice were stained with anti-ZnT-3 (green), a marker for granule cell mossy fibers from the dentate gyrus (DG), and with DAPI (blue). ZnT-3-positive axonal terminals are strongly reduced in the dorsal hilus (dHi) of Rac1N/Rac3KO mice compared with control Rac3KO mice (right panels), but not in the stratum lucidum (SL) of the dorsal CA3 subfield (left panels). Scale bars = 100 μm. b) Quantification of density of ZnT-3, MAP2, and synapsin I signals in the dorsal hilus of Rac1N/Rac3KO (dKO) and Rac3KO mice. Values represent mean ± se percentages with respect to control Rac3KO mice (n=8 for ZnT-3 and MAP2; n=7 for synapsin I) after normalization (Rac3KO mice = 100%).
Rac1 and Rac3 are involved in dendritic spine formation
We have used well-established hippocampal cultures to further address the role of Rac1 and Rac3 in late stages of neuronal maturation and in dendritic spine formation in particular. These events are impaired by the expression of Rac1 mutants that cannot distinguish between Rac isoforms (7, 33). Because transgenic SynI-Cre was poorly effective in processing the Rac1-floxed allele in hippocampal cultures (Fig. 10a), we deleted the Rac1 gene by transfecting Rac1F/F and Rac1F/F/Rac3KO neurons with an enhanced green fluorescent protein (EGFP)-Cre expression vector (25). Comparison between EGFP-Cre-transfected and EGFP-transfected control neurons showed that the total numbers of small dendritic protrusions (including spines, filopodia, and lamellipodia) were significantly decreased after deletion of both Rac genes (−21%, P<0.00035). Postsynaptic dendritic spines were strongly reduced, whereas dendritic filopodia increased significantly in transfected neurons in which both Rac genes had been deleted (Fig. 10f, g) (P<0.00005). Deletion of Rac3 per se had no effects on spines (Fig. 10e, g), whereas deletion of Rac1 had only a weak effect on mushroomshaped spines (Fig. 10c, d, g) (P<0.0005). Thus, Rac1 and Rac3 play a synergistic role in the formation of dendritic spines.
Figure 10. Deletion of both Rac1 and Rac3 deeply affects spinogenesis.
a) Genomic DNA was extracted from the tail (T) of a Rac1F/+ SynI-Cre mouse, the spinal cord (SC) of a Rac1F/F SynI-Cre mouse, and 15 DIV hippocampal cultures obtained from two different Rac1F/+ SynI-Cre embryos (hc1 and hc2). DNAs were processed by PCR as detailed in Materials and Methods. PCR fragments are from floxed (F), wild-type (wt), and deleted (del) alleles. b) Hippocampal neurons from Rac1F/F mice were transfected at 4 DIV with EGFP and EGFP-Cre recombinase. Cotransfected cells fixed at 14 DIV were labeled with anti-GFP (green, cytoplasmic), anti-Cre (red, nuclear), and DAPI (blue). Same field is shown in both panels. Scale bar = 100 μm. c–f) Hippocampal neurons from Rac1F/F(c, d) or Rac1F/F/Rac3KO (e, f) mice were transfected at 4 DIV with GFP (c, e) or cotransfected with GFP-Cre and GFP (d, f). Cells were fixed at 14 DIV and stained with anti-GFP (green) and anti-PSD95 (red). Same fields are shown in the middle and bottom pictures of each panel. Scale bar = 5 μm. g) Quantification of the number of mature and immature spines on dendrites of hippocampal neurons, shown as average protrusion density (number/100 μm). Quantification was performed on ≥450 protrusions from 7-10 neurons per experiment per condition. Dendritic protrusions included mushroom and stubby spines, short (<4 μm) and long (>4 μm) filopodia, and lamellipodia. *P < 0.0005, **P < 0.00005 vs. respective control neurons; Student’s t test.
Discussion
We have shown that the GTPases Rac1 and Rac3 are synergistically critical for mammalian brain development, for which these proteins play complementary essential functions. In vivo, the deletion of the two genes in hippocampal neurons affects the development of the dentate gyrus, by hampering the formation of specific circuits involving hilar neurons and granule cells, two types of neurons that complete their differentiation postnatally (30, 34, 35). Furthermore, in hippocampal cultures the deletion of the two Rac genes leads to strong inhibition of dendritic spine development, in contrast to the deletion of either the Rac1 or Rac3 gene that causes minor or undetectable effects, respectively, on the development of spines. These findings not only reveal the functional importance of the two Rac genes in vertebrate neuronal development but also provide the first evidence that Rac1 and Rac3 contribute to the establishment of the hippocampal circuitry. The defects observed in the wiring of the hippocampus after the deletion of both genes may underlie the epileptic phenotype found in Rac1N/Rac3KO mice.
During development, Rac1 and Rac3 are coexpressed in hilar mossy cells, whereas only Rac1 is detected in granule cells (4). Thus, our data indicate that the loss of Rac1 and Rac3 in mossy cells causes a non-cell-autonomous defect in granule cells. The effects observed in the hippocampus of double-mutant mice are probably explained by the drastic decrease of mossy cells found in the dorsal hilus of Rac1N/Rac3KO mice. Mossy cells are believed to be important regulators of the signals arriving to the dentate gyrus from the cortex. Previous anatomic studies have indicated that these interneurons originate from the germinative zone immediately adjacent to the primordial dentate granule ventricular zone (36, 37). Impaired maturation and/or migration of these neurons represent likely explanations that await further characterization from studies on the poorly characterized ontogeny of these cells (29). The hypothesis of a nonautonomous defect in granule cells of Rac1N/Rac3KO mice is further supported by the finding that although mossy fiber collaterals to the hilus are not formed, granule cells of double-knockout mice can still extend mossy fibers to form synapses with another target within the hippocampus, the dendrites of the CA3 pyramidal neurons in the stratum lucidum. Moreover, the decreased innervation of the dorsal hilar region by granule cell axon collaterals correlates with the strongly reduced synapses observed in the hilus of double-mutant mice.
The analysis of Rac3-null mice and the comparative analysis presented in this study suggest that Rac1 and Rac3 are able to at least partially compensate for the lack of the other GTPase during neuronal development. On the other hand, the heavy neurological consequences of the double deletion of Rac1 and Rac3 in neurons show that both proteins contribute to the development of a functional nervous system in vertebrates.
Rac3 has strong homology to Rac1, with major sequence divergence at the carboxyl-terminal hypervariable region. It is likely that the gene for Rac3 did first appear in vertebrates, because an ortholog of Rac3 has not been found in less complex chordates (38), insects (39), or nematodes (40). In situ hybridization has confirmed that Rac3 is widely and specifically expressed in the developing nervous system of mice, where it is often but not always coexpressed with Rac1 (4). So far, with the exception of some endogenous Rac3 protein identified in primary lymphomas from Bcr/Abl P190 transgenic mice (41), the Rac3 protein has been detected exclusively in neural tissue. Because Rac3 shows highest and regulated expression during late brain development, with a peak at the time of intense neurite branching and synaptogenesis (12, 13), it is reasonable to hypothesize that this relatively new gene would contribute in the organization of the brain of more evolved animals by acting at later times during development.
So far, Rac1 has been considered the major if not only Rac GTPase required for vertebrate neuronal development, based mostly on the outcomes from studies using constitutively active or inactive Rac1 mutants. Although studies with these mutants have been essential to start assembling a picture of how Rac proteins may work to build the vertebrate nervous system, the results from these studies must be taken with caution before drawing final conclusions on the role of these GTPases in the vertebrate nervous systems.In fact, the possibility that the effects observed by expressing the Rac1 mutants might at least in part be due to interference also with Rac3 function in several neuronal populations cannot be ruled out. Therefore, the results reported here are fundamental to future work aiming to define the role of Rac GTPases in the development of diverse neuronal pathways of the central and peripheral nervous system, in which Rac1 and Rac3 are coexpressed. The impairment of hippocampal circuitry and the epileptic phenotype of double-mutant animals also suggest that these mice may be a valuable model to study the mechanisms of epileptogenesis.
In conclusion, the available evidence indicates that highly homologous Rac proteins perform complementary functions in the developing nervous system of vertebrates; both Rac1 and Rac3 need to be considered in future analysis of the function of Rac GTPases in neural development.
Footnotes
We thank Laura Croci, the personnel of the Alembic facility, Claudia Asperti, and Fabio Monzani for technical help. This work was supported by Telethon-Italy (Grant GGP05051) and by Fondazione Cariplo. We thank Jamey Marth (University of California, San Diego, CA, USA) for SynI-Cre mice; Louis Reichardt (University of California, San Francisco, CA, USA) for the pEGFP-Cre plasmid; and Virginia Lee (University of Pennsylvania School of Medicine, Philadelphia, PA, USA), Richard Palmiter (University of Washington School of Medicine, Seattle, WA, USA), and Flavia Valtorta (University Vita-Salute San Raffaele, Milan, Italy) for antibodies. We also thank Marco de Curtis, Jacopo Meldolesi, Anna Mondino, Flavia Valtorta, and Larry Wrabetz for helpful discussions.
References
- 1.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 2.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 3.De Curtis I. Functions of Rac GTPases during neuronal development. Dev Neurosci. 2008;30:47–58. doi: 10.1159/000109851. [DOI] [PubMed] [Google Scholar]
- 4.Corbetta S, Gualdoni S, Albertinazzi C, Paris S, Croci L, Consalez GG, de Curtis I. Generation and characterization of Rac3 knockout mice. Mol Cell Biol. 2005;25:5763–5776. doi: 10.1128/MCB.25.13.5763-5776.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Didsbury J, Weber RF, Bokoch GM, Evans T, Snyderman R. Rac, a novel ras-related family of proteins that are botulinum toxin substrates. J Biol Chem. 1989;264:16378–16382. [PubMed] [Google Scholar]
- 6.Sugihara K, Nakatsuji N, Nakamura K, Nakao K, Hashimoto R, Otani H, Sakagami H, Kondo H, Nozawa S, Aiba A, Katsuki M. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene. 1998;17:3427–3433. doi: 10.1038/sj.onc.1202595. [DOI] [PubMed] [Google Scholar]
- 7.Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Liao G, Waclaw RR, Burns KA, Linquist D, Campbell K, Zheng Y, Kuan CY. Rac1 controls the formation of midline commissures and the competency of tangential migration in ventral telencephalic neurons. J Neurosci. 2007;27:3884–3893. doi: 10.1523/JNEUROSCI.3509-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gualdoni S, Albertinazzi C, Corbetta S, Valtorta F, de Curtis I. Normal levels of Rac1 are important for dendritic but not axonal development in hippocampal neurons. Biol Cell. 2007;99:455–464. doi: 10.1042/BC20060119. [DOI] [PubMed] [Google Scholar]
- 10.Haataja L, Groffen J, Heisterkamp N. Characterization of RAC3, a novel member of the Rho family. J Biol Chem. 1997;272:20384–20388. doi: 10.1074/jbc.272.33.20384. [DOI] [PubMed] [Google Scholar]
- 11.Malosio ML, Gilardelli D, Paris S, Albertinazzi C, de Curtis I. Differential expression of distinct members of Rho family GTP-binding proteins during neuronal development: identification of Rac1B, a new neural-specific member of the family. J Neurosci. 1997;17:6717–6728. doi: 10.1523/JNEUROSCI.17-17-06717.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albertinazzi C, Gilardelli, Paris DS, Longhi R, de Curtis I. Overexpression of a neural-specific rho family GTPase, cRac1B, selectively induces enhanced neuritogenesis and neurite branching in primary neurons. J Cell Biol. 1998;142:815–825. doi: 10.1083/jcb.142.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolis A, Corbetta S, Cioce A, de Curtis I. Differential distribution of Rac1 and Rac3 GTPases in the developing mouse brain: implications for a role of Rac3 in Purkinje cell differentiation. Eur J Neurosci. 2003;18:2417–2424. doi: 10.1046/j.1460-9568.2003.02938.x. [DOI] [PubMed] [Google Scholar]
- 14.Corbetta S, D’Adamo P, Gualdoni S, Braschi C, Berardi N, de Curtis I. Hyperactivity and novelty-induced hyperreactivity in mice lacking Rac3. Behav Brain Res. 2008;186:246–255. doi: 10.1016/j.bbr.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Glogauer M, Marchal CC, Zhu F, Worku A, Clausen BE, Foerster I, Marks P, Downey GP, Dinauer M, Kwiatkowski DJ. Rac1 deletion in mouse neutrophils has selective effects on neutrophil functions. J Immunol. 2003;170:5652–5657. doi: 10.4049/jimmunol.170.11.5652. [DOI] [PubMed] [Google Scholar]
- 16.Walmsley MJ, Ooi SK, Reynolds LF, Smith SH, Ruf S, Mathiot A, Vanes L, Williams DA, Cancro MP, Tybulewicz VL. Critical roles for Rac1 and Rac2 GTPases in B cell development and signaling. Science. 2003;302:459–462. doi: 10.1126/science.1089709. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 19.Lehrach H, Diamond D, Wozney JM, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 20.Fox WM. Reflex-ontogeny and behavioural development of the mouse. Anim Behav. 1965;13:234–241. doi: 10.1016/0003-3472(65)90041-2. [DOI] [PubMed] [Google Scholar]
- 21.Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr Clin Neurophysiol. 1972;32:281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- 22.Palmiter RD, Cole TB, Quaife CJ, Findley SD. ZnT-3, a putative transporter of zinc into synaptic vesicles. Proc Natl Acad Sci U S A. 1996;93:14934–14939. doi: 10.1073/pnas.93.25.14934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valtorta F, Villa A, Jahn R, De Camilli P, Greengard P, Ceccarelli B. Localization of synapsin I at the frog neuromuscular junction. Neuroscience. 1988;24:593–603. doi: 10.1016/0306-4522(88)90353-3. [DOI] [PubMed] [Google Scholar]
- 24.Banker GA, Cowan WM. Rat hippocampal neurons in dispersed cell culture. Brain Res. 1977;126:397–425. doi: 10.1016/0006-8993(77)90594-7. [DOI] [PubMed] [Google Scholar]
- 25.Rico B, Beggs HE, Schahin-Reed D, Kimes N, Schmidt A, Reichardt LF. Control of axonal branching and synapse formation by focal adhesion kinase. Nat Neurosci. 2004;7:1059–1069. doi: 10.1038/nn1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sloviter RS. Decreased hippocampal inhibition and a selective loss of interneurons in experimental epilepsy. Science. 1987;235:73–76. doi: 10.1126/science.2879352. [DOI] [PubMed] [Google Scholar]
- 27.Ratzliff AH, Santhakumar V, Howard A, Soltesz I. Mossy cells in epilepsy: rigor mortis or vigor mortis? Trends Neurosci. 2002;25:140–144. doi: 10.1016/s0166-2236(00)02122-6. [DOI] [PubMed] [Google Scholar]
- 28.Fujise N, Kosaka T. Mossy cells in the mouse dentate gyrus: identification in the dorsal hilus and their distribution along the dorsoventral axis. Brain Res. 1999;816:500–511. doi: 10.1016/s0006-8993(98)01202-5. [DOI] [PubMed] [Google Scholar]
- 29.Li G, Berger O, Han SM, Paredes M, Wu NC, Pleasure SJ. Hilar mossy cells share developmental influences with dentate granule neurons. Dev Neurosci. 2008;30:255–261. doi: 10.1159/000110347. [DOI] [PubMed] [Google Scholar]
- 30.Ribak CE, Seress L, Amaral DG. The development, ultrastructure and synaptic connections of the mossy cells of the dentate gyrus. J Neurocytol. 1985;14:835–857. doi: 10.1007/BF01170832. [DOI] [PubMed] [Google Scholar]
- 31.Frotscher M, Seress L, Schwerdtfeger WK, Buhl E. The mossy cells of the fascia dentata: a comparative study of their fine structure and synaptic connections in rodents and primates. J Comp Neurol. 1991;312:145–163. doi: 10.1002/cne.903120111. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Fujise N, Kosaka T. Distribution of calretinin immunoreactivity in the mouse dentate gyrus. I. General description. Exp Brain Res. 1996;108:389–403. doi: 10.1007/BF00227262. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angevine JB., Jr Time of neuron origin in the hippocampal region: an autoradiographic study in the mouse. Exp Neurol Suppl. 1965;2:1–70. [PubMed] [Google Scholar]
- 35.Schlessinger AR, Cowan WM, Gottlieb DI. An autoradiographic study of the time of origin and the pattern of granule cell migration in the dentate gyrus of the rat. J Comp Neurol. 1975;159:149–175. doi: 10.1002/cne.901590202. [DOI] [PubMed] [Google Scholar]
- 36.Nowakowski RS, Rakic P. The mode of migration of neurons to the hippocampus: a Golgi and electron microscopic analysis in foetal rhesus monkey. J Neurocytol. 1979;8:697–718. doi: 10.1007/BF01206671. [DOI] [PubMed] [Google Scholar]
- 37.Nowakowski RS, Rakic P. The site of origin and route and rate of migration of neurons to the hippocampal region of the rhesus monkey. J Comp Neurol. 1981;196:129–154. doi: 10.1002/cne.901960110. [DOI] [PubMed] [Google Scholar]
- 38.Philips A, Blein M, Robert A, Chambon JP, Baghdiguian S, Weill M, Fort P. Ascidians as a vertebrate-like model organism for physiological studies of Rho GTPase signaling. Biol Cell. 2003;95:295–302. doi: 10.1016/s0248-4900(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 39.Hariharan IK, Hu KQ, Asha H, Quintanilla A, Ezzell RM, Settleman J. Characterization of rho GTPase family homologues in Drosophila melanogaster overexpressing Rhol in retinal cells causes a late developmental defect. EMBO J. 1995;14:292–302. doi: 10.1002/j.1460-2075.1995.tb07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–4488. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- 41.Cho YJ, Zhang B, Kaartinen V, Haataja L, de Curtis I, Groffen J, Heisterkamp N. Generation of Rac3 null mutant mice: role of Rac3 in Bcr/Abl-caused lymphoblastic leukemia. Mol Cell Biol. 2005;25:5777–5785. doi: 10.1128/MCB.25.13.5777-5785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]