Abstract
Objectives
A high dairy protein intake in infancy, maternal pre-pregnancy BMI, and delivery mode are documented early programming factors that modulate the later risk of obesity and other health outcomes, but the mechanisms of action are not understood.
Methods
The Childhood Obesity Project is a European multicenter, double-blind, randomized clinical trial that enrolled healthy infants. Participating infants were either breastfed (BF) or randomized to receive higher (HP) or lower protein (LP) content formula in the first year of life. At the ages 5.5 years (n=276) and 8 years (n=232), we determined plasma metabolites by liquid chromatography tandem-mass-spectrometry of which 226 and 185 passed quality control at 5.5 years and 8 years, respectively. We assessed the effects of infant feeding, maternal pre-pregnancy BMI, smoking in pregnancy, delivery mode, parity, birth weight and length, and weight gain (0-24 months) on the metabolome at 5.5 and 8 years.
Results
At 5.5y, plasma alpha-ketoglutarate and the acylcarnitine/BCAA ratios tended to be higher in the HP than in the LP group, but no metabolite reached statistical significance (Pbonferroni>0.09). There were no group differences at 8y. Quantification of the impact of early programming factors revealed that the intervention group explained 0.6% of metabolome variance at both time points. Except for country of residence that explained 16% and 12% at 5.5y and 8y, respectively, none of the other factors explained considerably more variance than expected by chance.
Conclusions
Plasma metabolome was largely unaffected by feeding choice and other early programming factors and we could not prove the existence of a long term programming effect of the plasma metabolome.
Keywords: Metabolomics, high protein, alpha-ketoglutarate, BCAA, early programming
1. Introduction
The prevalence of non-communicable diseases such as obesity or diabetes has markedly increased in both high and low/low-medium income countries around the world 1.Therefore, increased attention is directed to the potential of prevention by early “programming”, a concept introduced by Günther Dörner in 1975 2 and expanded by Hales and Barker 3 and Lucas 4. The underlying concept is that certain environmental and nutritional exposures acting during limited time periods of developmental plasticity in early life lead to a lasting impact on later health and disease 5, 6. One example is the marked impact of a high protein intake in infancy, in excess of metabolic requirements, on plasma of insulin-releasing amino acids, which in turn stimulate the secretion of insulin and insulin-like growth factor I (IGF1), thereby inducing an increased weight gain during the first 2 y of life as well as increased adipogenic activity 7. A large European intervention study demonstrated that children with a conventionally high protein intake during the first year of life experience a greater weight gain during the first 2 years 8 and markedly higher obesity risk at early school age 9. The underlying mechanisms of these lasting programming effects are not fully understood. We aimed at exploring whether infant feeding and other early life factors induce lasting effects on the childhood metabolism. Metabolomics is the study of small molecules and metabolic intermediates, called metabolites, which are regarded as the result of the interaction of the genome, epigenome, transcriptome, proteome, and the environment 10, 11. The ensemble of all measurable metabolites is called metabolome. Using univariate and multivariate statistical approaches, we characterize and quantify the impact of (1) a higher protein infant formula and (2) other early programming factors, namely smoking during pregnancy, delivery mode, parity, maternal BMI, birth weight, birth length, and early weight gain, on the plasma metabolome of children aged 5.5 and 8 years.
2. Metarials and Methods
Study design
The data evaluated were collected as part of the European Childhood Obesity Project (CHOP), a double-blind, randomized, multicenter intervention trial conducted in five countries: Germany, Belgium, Italy, Poland, and Spain. Details of the design and results of the primary study of the first 2 y of life were reported previously 8, 9. Briefly, parents of healthy, singleton, term infants, who met the inclusion criteria, were invited to participate in a study on the effects of dietary protein on obesity and growth and assigned to a group of infants fully breastfed for at least 3 months (BF) or randomized double blind to infant and follow-on formulae with a higher or lower protein content in infant formula (HP and LP, respectively). They were randomized at an average age of 2 weeks but not later than 8 weeks. The cows’ milk protein content was 1.25 g/dL in the LP and 1.6 g/dL in the HP infant formulae, and 2.05 and 3.2 g/dL in the LP and HP follow-on formulae introduced after complementary feeding was introduced, respectively. An equal energy content was achieved by an addition of fat to the LP formulae. Complementary foods were chosen at the discretion of the families. At 4 months of age 30% of breastfed and 38% of formula-fed children had already introduced some complementary foods with an average intake of 170 kcal and 202 kcal per day, respectively. However, about 85% of all breastfed children were still breastfeeding at 6 months of age.
Ethics, consent and permissions
The study was approved by the ethics committees of all of the study centers (Belgium: Comitè d’Ethique Medicale de Centre Hospitalier Chretien Liege; No. OM87; Germany: Bayerische Landesärztekammer Ethik-Kommission, No. 02070; Italy: Azienda Ospedaliera San Paolo Comitato Etico, No 14/2002; Poland: Instytut Pomnik-Centrum Zdrowia Dziecka Komitet Etyczny, No 243/KE/2001; Spain: Comité ético de investigación clínica del Hospital Universitario de Tarragona Joan XXIII, Comité ético de investigación clínica del Hospital Universitario Sant Joan de Reus), and written informed parental consent was obtained for each infant (trial registration: ClinicalTrials.gov; identifier: NCT00338689). All research was performed in accordance with the Declaration of Helsinki.
Covariates & Early Programming Factors
We examined effects of infant feeding groups on the 5.5y and 8y metabolome, as well as effects of maternal pre-pregnancy BMI, maternal smoking during pregnancy, mode of delivery, birth weight and length, and early weight gain from birth to 24 months. Potential confounders considered throughout the analyses comprise age, sex, country of residence, and the highest educational level of either mother or father. Current BMI was considered as potential confounder for sensitivity purposes only as it is rather lying at the end of the programming pathway, thus not influencing the metabolites12.
Data on birth weight and length was obtained from the hospital records. All other anthropometric data were measured at visits to the study centers by repeatedly trained study personnel. We calculated the age- and sex-specific z-scores for weight based on the WHO growth standards 13, 14. We defined change in weight-for-age (WFA) as the absolute difference in the WFA z-scores (birth – 24 months). Data on pregnancy outcomes (maternal prepregnancy weight, gestational age at delivery, birth order, and delivery mode) were collected by questionnaire. Maternal height as well as the variable on parental educational level (according to ISCED) and smoking during pregnancy were assessed at the baseline visit. For more information see 8, 9.
Metabolomics analyses
Blood samples were collected and centrifuged and plasma samples frozen at -70°C (5.5y samples) and -80 oC (8y samples) at study centers, and shipped on dry ice to LMU Munich where they were stored at -80°C until analysis. Metabolites were measured by liquid chromatography tandem-mass-spectrometry. The detailed description of the measurements can be found in the appendix.
During the quantification processes, the samples of the study centers were randomly distributed across the batches. Samples of 5.5y and 8y were quantified separately. Each sample was measured once. The entire analytical process was post-processed by Analyst 1.5.1 and R (version 3.3.1). Quality control was based on six quality control samples per batch measured along with the study samples. If quality control samples in one batch had a coefficient of variation >0.3, we excluded the respective metabolite measurements in this batch from the analyses. Analytes of which we had measurements in less than half of the batches afterwards, were dropped completely from the analyses. The batch effect was removed by centering the concentrations around the overall median. Since concentrations at 5.5y and 8y might differ, quality control was performed separately on the 5.5y and 8y samples leading to slightly differing numbers of metabolites included in the respective analyses.
3. Calculation
All statistical analyses were performed using R (version 3.3.1). First, we screened the data graphically and excluded metabolite measurements that were distanced over 2SD from the nearest neighbor from the analysis. The focus of the analysis maternal diet a might be of early programming factors including a higher protein infant formula on the metabolic profile. We used two approaches to investigate the research question: first, we fit linear mixed effects models with a random intercept for study center of each metabolite on the respective early programming factor. Metabolite concentrations were log transformed (AA, polar lipids, NEFA, sums and ratios), and boxcox-transformed (TCA-metabolites and glutamine) to normalize distributions. We looked at the unadjusted models and the adjusted models (confounder: age, sex, and parental educational level). In a second step, we aimed at identifying and quantifying the variability of the metabolic profiles in terms of the feeding group and other early programming factors. We used the PC-PR2 method described by Fages et al 15. As this method required a complete case dataset, we restricted ourselves to metabolites with few missing values (Table A12). Briefly, a principal component analysis (PCA) is performed on the standardized (mean of 0 and standard deviation of 1) metabolomics data and each of the first k principal components, such that the first k principal components (PC) accounting for 80% variability, is regressed on the variable set of interest in a multivariable linear regression. Next to the early programming factors, age, sex, country of residence, and parental educational level were included as independent variables. Then, the overall Rpartial(q)2 quantifying the contribution of the independent variable q to the variation in metabolomic data is calculated as weighted mean of the Rpartial(I, q)2 of the k models using the proportions of explained variance of the i=1…k PCs as weights. To assess the likelihood that this Rpartial(q)2 was just observed by chance, we extended this method as follows: We bootstrapped each independent variable 500 times and repeated the PC-PR2 method using the original PC and the bootstrapped independent variable. Due to the bootstrapping of one side only, all associations found can be attributed to chance. We then calculated the 2.5% and 97.5% percentiles (referred to as 95%CIpercentile) of the 500 bootstrapped Rpartial(q)2. In case that the observed Rpartial(q)2 lay outside this range, we concluded the contribution of the independent variable was higher than one would expect by chance. We repeated this analysis stratified by sex. We also studied whether our finding of a breakpoint in BCAA catabolism infancy 16 could be replicated in later childhood. We used the same approach, which is descriptive analysis using locally-weighted polynomial regression (LOWESS) smoother and subsequently calculating piecewise linear mixed effect (LME) models with different breakpoints whose fit we compared by means of deviance.
4. Results
At 5.5y/8y of age, we obtained plasma samples from 291/249 formula-fed children of whom 276/232, respectively were fasted ≥6 hours at blood withdrawal. Twelve/fifteen of the non-fasted children were from Germany, one/zero from Belgium, and the information was missing for two/two children from Spain. All non-fasted children were excluded from the analyses. We also had plasma samples from 120/122 fasted previously breastfed children at 5.5y/8y, respectively. Characteristics of the dropouts are provided in Table A.2. Children with available blood samples had parents with higher educational status and their mothers smoked less often during pregnancy. Characteristics of the children included in the following analyses are shown in Table 1.
Table 1.
Number (%) or mean ± SD of the children with available blood samples at 5.5 years and 8 years of age. LP: randomized lower protein group; HP randomized higher protein group; BF: observational breastfed group.
| 5.5 years | 8 years | ||||
|---|---|---|---|---|---|
|
|
|
||||
| N | Number (%) or mean ± SD | N | Number (%) or mean ± SD | ||
| Feeding group | Breastfed (BF) | 396 | 120 (30%) | 354 | 122 (34%) |
| Lower protein (LP) | 139 (35%) | 116 (33%) | |||
| Higher protein (HP) | 137 (35%) | 116 (33%) | |||
| Sex | Female | 396 | 198 (50%) | 354 | 179 (51%) |
| Age at blood withdrawal (y) | 396 | 5.5 ± 0.07 | 354 | 8.1 ± 0.10 | |
| Country | Belgium | 396 | 61 (15%) | 354 | 59 (17%) |
| Italy | 143 (36%) | 75 (21%) | |||
| Poland | 70 (18%) | 92 (26%) | |||
| Spain | 122 (31%) | 128 (36%) | |||
| Highest parental educational level | No/Low | 395 | 42 (11%) | 353 | 37 (10%) |
| Middle | 213 (54%) | 175 (50%) | |||
| High | 140 (35%) | 141 (40%) | |||
| Prepregnancy BMI mother (kg/m2) | 377 | 23.7 ± 4.36 | 341 | 23.5 ± 4.15 | |
| Smoking during pregnancy | No | 396 | 317 (80%) | 353 | 287 (81%) |
| Yes | 79 (20%) | 66 (19%) | |||
| Delivery mode | Caesarean section | 393 | 89 (23%) | 353 | 80 (23%) |
| Forceps or Vacuum extraction | 38 (10%) | 37 (10%) | |||
| Spontaneous | 266 (68%) | 236 (67%) | |||
| Parity | 1 | 395 | 234 (59%) | 353 | 202 (57%) |
| 2 | 131 (33%) | 124 (35%) | |||
| 3 | 27 (7%) | 25 (7%) | |||
| 4+ | 3 (1%) | 2 (1%) | |||
| Birth weight (kg) | 396 | 3 293.3 ± 350.78 | 354 | 3 272.4 ± 334.76 | |
| Birth length (cm) | 394 | 50.4 ± 2.54 | 352 | 50.7 ± 2.75 | |
| Change in weight-for-age z-score (0-24mo) | 388 | 0.8 ± 0.99 | 340 | 0.8 ± 0.99 | |
| Height (cm) | 392 | 113.7 ± 4.51 | 353 | 129.5 ± 5.64 | |
| Weight (kg) | 393 | 20.7 ± 3.49 | 354 | 28.5 ± 6.21 | |
| BMI z Score | 391 | 0.3 ± 1.19 | 353 | 0.4 ± 1.27 | |
| Glucose levels (mg/dl) | 389 | 82.8 ± 6.86 | 348 | 83.5 ± 7.65 | |
| Insulin levels (µIU/ml) | 388 | 6.4 ± 3.10 | 350 | 8.8 ± 3.16 | |
In the 5.5y and 8y plasma, 22 and 21 amino acids (AA) did pass quality control, respectively. We furthermore quantified free carnitine, 41/38 acylcarnitines (Carn), 14/13 lysophosphatidylcholines (LPC), 33/19 diacyl-phosphatidylcholines (PCaa), 34/17 acylalkyl-phosphatidylcholines (PCae), and 31/15 sphingomyelins (SM) in the 5.5y/8y plasma samples, respectively, using flow-injection mass spectrometry. Non-esterified fatty acids (NEFA, count 35/45) and 15 / 16 organic acids, including metabolit the 5.5y and 8y samples leading to es of the tricarboxylic acid (TCA) cycle and keto-acids, were measured by LC-MS/MS. We calculated the sum of all AA, branched chain amino acids (BCAA), and LPC and 11 / 8 ratios depicting the first steps in BCAA catabolism.
Comparison of the metabolome across feeding groups
The results of the adjusted linear mixed effect models are shown in Figure 1. Concentrations and P-values of significantly different metabolites at the uncorrected α-threshold of 0.05 are given in Table 2. Neither at 5.5y nor 8y we found significant differences for any metabolite after correction for multiple testing.
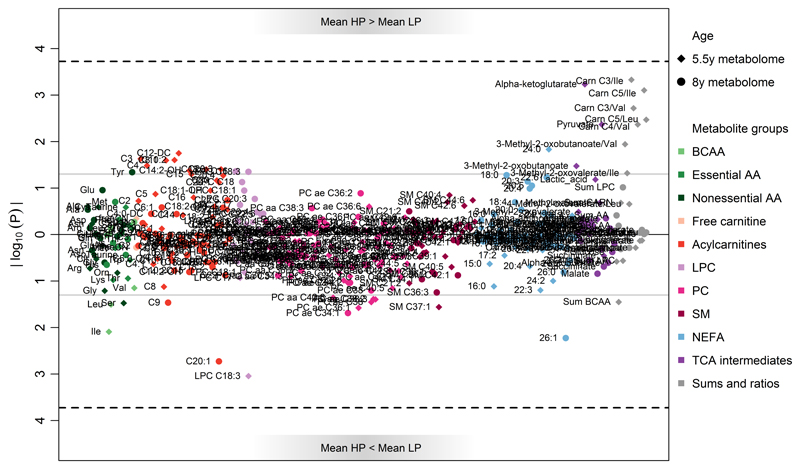
Figure 1.
Manhattan plot of the 5.5year (diamond) and 8year (circles) metabolite concentration differences between the children having received the lower protein (LP) and the higher protein (HP) infant formula. P-values are from linear mixed effects models adjusted for age at blood withdrawal, sex, and parental education. The grey solid line indicates the significance threshold of α=0.05, the black dashed line the Bonferroni corrected significance level of αBonferroni=0.00019. Abbreviations: AA, amino acids, LPC, lysophosphatidylcholine; PC, phosphatidylcholine; SM, sphingomyelins; NEFA, non-esterified acid; TCA, tricarboxylic acid cycle
Table 2.
Mean (SD) concentrations of metabolites that differed at the uncorrected significance level between the lower (LP), higher protein (HP) group or the breastfed (BF) infant at 5.5 and 8 years of age. Concentrations are in µmol/L. P values are from linear mixed effect (LME) models. We report the P from univariate and adjusted models as well as the uncorrected and Bonferroni corrected (PBonferroni) P values.
|
5.5y metabolome
| |||||||||||||||
| LP | HP | LP vs. HP | BF | BF vs. LP | BF vs. HP | ||||||||||
|
|
|
|
|||||||||||||
| (n=139) | (n=137) | Univariate LME | Adjusted† LME | (n=120) | Univariate LME | Adjusted† LME | Univariate LME | Adjusted† LME | |||||||
|
|
|
|
|
||||||||||||
| P | PBonferroni | P | PBonferroni | P | PBonferroni | P | PBonferroni | P | PBonferroni | P | PBonferroni | ||||
|
| |||||||||||||||
| Cys | 10 (4.18) | 11 (5.23) | 0.27 | 1 | 0.26 | 1 | 9.3 (5.46) | 0.024 | 1 | 0.028 | 1 | 0.013 | 1 | 0.017 | 1 |
| Ile | 62 (16.6) | 58 (17.6) | 0.012 | 1 | 0.008 | 1 | 58 (18) | 0.047 | 1 | 0.042 | 1 | 0.87 | 1 | 0.61 | 1 |
| Leu | 116 (25.7) | 111 (29.2) | 0.052 | 1 | 0.031 | 1 | 111 (29.8) | 0.07 | 1 | 0.051 | 1 | 0.84 | 1 | 0.96 | 1 |
| Ser | 91 (24) | 86 (24) | 0.041 | 1 | 0.034 | 1 | 85 (26.9) | 0.023 | 1 | 0.032 | 1 | 0.48 | 1 | 0.85 | 1 |
| Tyr | 62 (16.7) | 62 (16.9) | 0.74 | 1 | 0.74 | 1 | 59 (16.7) | 0.21 | 1 | 0.42 | 1 | 0.018 | 1 | 0.043 | 1 |
| freeCN | 41 (9.49) | 41 (9.7) | 0.3 | 1 | 0.45 | 1 | 37 (8.68) | 0.18 | 1 | 0.23 | 1 | 0.004 | 1 | 0.029 | 1 |
| Carn C3 | 0.33 (0.09) | 0.36 (0.1) | 0.01 | 1 | 0.023 | 1 | 0.32 (0.1) | 0.59 | 1 | 0.49 | 1 | 0.003 | 0.8 | 0.006 | 1 |
| Carn C4 | 0.22 (0.1) | 0.24 (0.1) | 0.021 | 1 | 0.033 | 1 | 0.19 (0.09) | 0.82 | 1 | 0.62 | 1 | 0.009 | 1 | 0.013 | 1 |
| Carn C5 | 0.11 (0.04) | 0.11 (0.04) | 0.11 | 1 | 0.13 | 1 | 0.1 (0.04) | 0.71 | 1 | 0.54 | 1 | 0.023 | 1 | 0.026 | 1 |
| Carn C6:1 | 0.013 (0.01) | 0.012 (0.005) | 0.92 | 1 | 0.75 | 1 | 0.01 (0.004) | 0.024 | 1 | 0.07 | 1 | 0.004 | 1 | 0.015 | 1 |
| Carn C8:1 | 0.071 (0.05) | 0.081 (0.06) | 0.023 | 1 | 0.025 | 1 | 0.071 (0.05) | 0.36 | 1 | 0.58 | 1 | 0.006 | 1 | 0.014 | 1 |
| Carn C10:2 | 0.059 (0.02) | 0.062 (0.02) | 0.025 | 1 | 0.025 | 1 | 0.055 (0.02) | 1 | 1 | 0.7 | 1 | 0.032 | 1 | 0.047 | 1 |
| Carn C12-DC | 0.15 (0.1) | 0.16 (0.09) | 0.021 | 1 | 0.018 | 1 | 0.12 (0.08) | 0.26 | 1 | 0.27 | 1 | <0.0001 | 0.045 | <0.0001 | 0.12 |
| Carn C14:2-OH | 0.024 (0.01) | 0.026 (0.01) | 0.055 | 1 | 0.042 | 1 | 0.024 (0.01) | 0.67 | 1 | 0.43 | 1 | 0.13 | 1 | 0.2 | 1 |
| Carn C16-OH | 0.018 (0.008) | 0.017 (0.008) | 0.74 | 1 | 0.63 | 1 | 0.014 (0.007) | 0.14 | 1 | 0.14 | 1 | 0.034 | 1 | 0.028 | 1 |
| Carn C18 | 0.044 (0.01) | 0.046 (0.02) | 0.044 | 1 | 0.04 | 1 | 0.038 (0.01) | 0.51 | 1 | 0.77 | 1 | 0.006 | 1 | 0.01 | 1 |
| Carn C20:1 | 0.00046 (2.1×10-4) | 0.00048 (2.0×10-4) | 0.09 | 1 | 0.08 | 1 | 0.00041 (1.7×10-4) | 0.95 | 1 | 0.91 | 1 | 0.041 | 1 | 0.035 | 1 |
| Carn C20:3 | 0.015 (0.007) | 0.016 (0.007) | 0.052 | 1 | 0.039 | 1 | 0.014 (0.006) | 0.44 | 1 | 0.33 | 1 | 0.46 | 1 | 0.53 | 1 |
| LPC C15 | 0.73 (0.4) | 0.73 (0.4) | 0.58 | 1 | 0.6 | 1 | 0.92 (0.4) | 0.034 | 1 | 0.035 | 1 | 0.01 | 1 | 0.052 | 1 |
| LPC C18:3 | 0.54 (0.3) | 0.46 (0.3) | 0.002 | 0.66 | 0.001 | 0.22 | 0.62 (0.4) | 0.89 | 1 | 0.75 | 1 | 0.1 | 1 | 0.049 | 1 |
| PC aa C28:2 | 0.099 (0.04) | 0.1 (0.05) | 0.99 | 1 | 0.96 | 1 | 0.11 (0.06) | 0.012 | 1 | 0.021 | 1 | 0.19 | 1 | 0.36 | 1 |
| PC aa C38:5 | 37 (10.6) | 37 (10.5) | 0.38 | 1 | 0.43 | 1 | 36 (10.6) | 0.06 | 1 | 0.049 | 1 | 0.35 | 1 | 0.26 | 1 |
| PC aa C42:6 | 0.67 (0.3) | 0.59 (0.3) | 0.049 | 1 | 0.044 | 1 | 0.57 (0.4) | 0.36 | 1 | 0.45 | 1 | 0.72 | 1 | 0.64 | 1 |
| PC ae C30:2 | 0.28 (0.1) | 0.27 (0.1) | 0.18 | 1 | 0.22 | 1 | 0.32 (0.1) | 0.021 | 1 | 0.017 | 1 | 0.053 | 1 | 0.08 | 1 |
| PC ae C36 | 1.3 (0.7) | 1.3 (0.6) | 0.73 | 1 | 0.78 | 1 | 1.4 (0.7) | 0.06 | 1 | 0.049 | 1 | 0.53 | 1 | 0.63 | 1 |
| PC ae C36:1 | 10 (6.16) | 9 (5.18) | 0.031 | 1 | 0.027 | 1 | 10 (6.63) | 0.77 | 1 | 0.6 | 1 | 0.32 | 1 | 0.4 | 1 |
| PC ae C36:6 | 0.57 (0.4) | 0.61 (0.4) | 0.19 | 1 | 0.25 | 1 | 0.53 (0.4) | 0.38 | 1 | 0.47 | 1 | 0.036 | 1 | 0.048 | 1 |
| PC ae C38 | 2.8 (1.56) | 2.4 (1.32) | 0.021 | 1 | 0.035 | 1 | 2.8 (1.66) | 0.81 | 1 | 0.73 | 1 | 0.025 | 1 | 0.1 | 1 |
| PC ae C38:2 | 5 (4.48) | 4.5 (4.28) | 0.07 | 1 | 0.041 | 1 | 4.7 (4.75) | 0.25 | 1 | 0.3 | 1 | 0.86 | 1 | 0.98 | 1 |
| PC ae C38:3 | 8.4 (5.17) | 7.6 (5.35) | 0.06 | 1 | 0.04 | 1 | 8 (4.23) | 0.77 | 1 | 0.9 | 1 | 0.08 | 1 | 0.09 | 1 |
| PC ae C42:4 | 1.4 (0.5) | 1.3 (0.4) | 0.18 | 1 | 0.12 | 1 | 1.1 (0.3) | 0.009 | 1 | 0.009 | 1 | 0.12 | 1 | 0.13 | 1 |
| SM C37:1 | 2.2 (1.3) | 1.8 (1.14) | 0.017 | 1 | 0.028 | 1 | 1.5 (0.9) | 0.8 | 1 | 0.79 | 1 | 0.74 | 1 | 0.9 | 1 |
| SM C42:6 | 2.2 (0.7) | 2.4 (0.9) | 0.32 | 1 | 0.24 | 1 | 2.3 (1.02) | 0.45 | 1 | 0.33 | 1 | 0.19 | 1 | 0.0498 | 1 |
| SM C43 | 0.44 (0.2) | 0.45 (0.2) | 0.52 | 1 | 0.71 | 1 | 0.39 (0.2) | 0.24 | 1 | 0.16 | 1 | 0.07 | 1 | 0.049 | 1 |
| NEFA 16:1 | 28 (18.7) | 25 (17.1) | 0.51 | 1 | 0.64 | 1 | 26 (19.3) | 0.054 | 1 | 0.047 | 1 | 0.28 | 1 | 0.19 | 1 |
| NEFA 24:0 | 0.2 (0.08) | 0.28 (0.2) | 0.007 | 1 | 0.015 | 1 | 0.2 (0.1) | 0.79 | 1 | 0.72 | 1 | 0.033 | 1 | 0.051 | 1 |
| NEFA 26:1 | 0.29 (0.2) | 0.26 (0.1) | 0.96 | 1 | 0.93 | 1 | 0.19 (0.1) | 0.016 | 1 | 0.03 | 1 | 0.008 | 1 | 0.012 | 1 |
| 3-Methyl-2-oxobutanoate | 15 (12.8) | 19 (14.8) | 0.041 | 1 | 0.034 | 1 | 16 (14.4) | 0.99 | 1 | 0.94 | 1 | 0.14 | 1 | 0.1 | 1 |
| Alpha-ketoglutarate | 5.9 (4.62) | 8 (5.94) | 0.001 | 0.27 | 0.001 | 0.14 | 7.5 (6.26) | 0.09 | 1 | 0.08 | 1 | 0.24 | 1 | 0.25 | 1 |
| Pyruvate | 188 (228) | 252 (258) | 0.011 | 1 | 0.004 | 1 | 228 (345) | 0.47 | 1 | 0.3 | 1 | 0.22 | 1 | 0.2 | 1 |
| Succininate | 5.2 (3.83) | 4.8 (2.5) | 0.36 | 1 | 0.35 | 1 | 5.7 (3.45) | 0.18 | 1 | 0.22 | 1 | 0.048 | 1 | 0.03 | 1 |
| Sum BCAA | 405 (93.7) | 388 (106) | 0.06 | 1 | 0.036 | 1 | 386 (100) | 0.07 | 1 | 0.044 | 1 | 0.83 | 1 | 0.97 | 1 |
| Ratio 3-Methyl-2-oxobutanoate/Val | 0.078 (0.08) | 0.11 (0.1) | 0.016 | 1 | 0.011 | 1 | 0.093 (0.1) | 0.66 | 1 | 0.67 | 1 | 0.13 | 1 | 0.11 | 1 |
| Ratio 3-Methyl-2-oxovalerate/Ile | 0.38 (0.3) | 0.46 (0.4) | 0.06 | 1 | 0.048 | 1 | 0.43 (0.3) | 0.4 | 1 | 0.6 | 1 | 0.72 | 1 | 0.55 | 1 |
| Ratio Carn C3/Ile | 0.0057 (0.002) | 0.0068 (0.003) | <0.0001 | 0.07 | <0.0001 | 0.11 | 0.0058 (0.002) | 0.35 | 1 | 0.39 | 1 | 0.015 | 1 | 0.011 | 1 |
| Ratio Carn C3/Val | 0.0015 (0.0005) | 0.0018 (0.0007) | 0.001 | 0.35 | 0.002 | 0.45 | 0.0015 (0.0006) | 0.46 | 1 | 0.43 | 1 | 0.021 | 1 | 0.024 | 1 |
| Ratio Carn C4/Val | 0.00099 (0.0005) | 0.0012 (0.0006) | 0.004 | 1 | 0.004 | 1 | 0.00094 (4.8×10-4) | 0.55 | 1 | 0.65 | 1 | 0.019 | 1 | 0.02 | 1 |
| Ratio Carn C5/Ile | 0.0019 (0.0007) | 0.0021 (0.0008) | 0.001 | 0.22 | 0.001 | 0.19 | 0.0019 (0.0007) | 0.35 | 1 | 0.48 | 1 | 0.017 | 1 | 0.01 | 1 |
| Ratio Carn C5/Leu | 0.00097 (3.6×10-4) | 0.0011 (4.1×10-4) | 0.004 | 0.97 | 0.003 | 0.8 | 0.00096 (3.6×10-4) | 0.49 | 1 | 0.61 | 1 | 0.02 | 1 | 0.018 | 1 |
| Ratio Carn C4/Val | 0.00099 (0.0005) | 0.0012 (0.0006) | 0.004 | 1 | 0.005 | 1 | 0.00094 (4.8×10-4) | 0.48 | 1 | 0.54 | 1 | 0.031 | 1 | 0.033 | 1 |
| Ratio Carn C5/Ile | 0.0019 (0.0007) | 0.0021 (0.0008) | 0.001 | 0.19 | 0.001 | 0.2 | 0.0019 (0.0007) | 0.22 | 1 | 0.28 | 1 | 0.051 | 1 | 0.03 | 1 |
| Ratio Carn C5/Leu | 0.00097 (3.6×10-4) | 0.0011 (4.1×10-4) | 0.003 | 0.86 | 0.004 | 0.84 | 0.00096 (3.6×10-4) | 0.3 | 1 | 0.37 | 1 | 0.07 | 1 | 0.06 | 1 |
|
| |||||||||||||||
|
8y metabolome
| |||||||||||||||
| LP | HP |
LP vs. HP
|
BF | BF vs. LP | BF vs. HP | ||||||||||
| Univariate LME | Adjusted† LME | Univariate LME | Adjusted† LME | Univariate LME | Adjusted† LME | ||||||||||
|
|
|
||||||||||||||
| (n=116) | (n=116) | P | PBonferroni | P | PBonferroni | (n=122) | P | PBonferroni | P | PBonferroni | P | PBonferroni | P | PBonferroni | |
|
| |||||||||||||||
| Asp | 7.5 (2.42) | 7.9 (3.43) | 0.42 | 1 | 0.49 | 1 | 8.3 (3.18) | 0.021 | 1 | 0.018 | 1 | 0.22 | 1 | 0.26 | 1 |
| Gln | 505 (136) | 502 (151) | 0.73 | 1 | 0.83 | 1 | 485 (165) | 0.07 | 1 | 0.046 | 1 | 0.09 | 1 | 0.04 | 1 |
| Glu | 89 (79.3) | 102 (95.3) | 0.08 | 1 | 0.11 | 1 | 117 (101) | 0.001 | 0.12 | 0.001 | 0.17 | 0.052 | 1 | 0.038 | 1 |
| Taurine | 51 (34.1) | 49 (29.9) | 0.68 | 1 | 0.36 | 1 | 42 (20.5) | 0.027 | 1 | 0.017 | 1 | 0.07 | 1 | 0.07 | 1 |
| Tyr | 64 (11.9) | 67 (12.6) | 0.08 | 1 | 0.046 | 1 | 65 (13.6) | 0.89 | 1 | 0.8 | 1 | 0.1 | 1 | 0.043 | 1 |
| Carn C3 | 0.62 (0.3) | 0.67 (0.4) | 0.83 | 1 | 0.95 | 1 | 0.57 (0.3) | 0.012 | 1 | 0.018 | 1 | 0.009 | 1 | 0.01 | 1 |
| Carn C3:1 | 0.04 (0.02) | 0.039 (0.03) | 0.9 | 1 | 0.88 | 1 | 0.031 (0.02) | 0.019 | 1 | 0.031 | 1 | 0.009 | 1 | 0.014 | 1 |
| Carn C6:1 | 0.021 (0.03) | 0.024 (0.03) | 0.26 | 1 | 0.26 | 1 | 0.035 (0.05) | 0.003 | 0.54 | 0.003 | 0.53 | 0.07 | 1 | 0.054 | 1 |
| Carn C9 | 0.03 (0.02) | 0.025 (0.01) | 0.038 | 1 | 0.034 | 1 | 0.031 (0.02) | 0.99 | 1 | 0.98 | 1 | 0.034 | 1 | 0.09 | 1 |
| Carn C18 | 0.03 (0.01) | 0.029 (0.01) | 0.23 | 1 | 0.26 | 1 | 0.028 (0.008) | 0.049 | 1 | 0.035 | 1 | 0.76 | 1 | 0.67 | 1 |
| Carn C18:2 | 0.063 (0.02) | 0.059 (0.02) | 0.06 | 1 | 0.2 | 1 | 0.055 (0.02) | 0.016 | 1 | 0.022 | 1 | 0.6 | 1 | 0.68 | 1 |
| Carn C18:2-OH | 0.0061 (0.003) | 0.0061 (0.004) | 0.39 | 1 | 0.26 | 1 | 0.0049 (0.003) | 0.016 | 1 | 0.021 | 1 | 0.32 | 1 | 0.46 | 1 |
| Carn C20:1 | 0.00035 (6.2×10-5) | 0.00032 (7.9×10-5) | 0.001 | 0.28 | 0.002 | 0.37 | 0.00033 (6.8×10-5) | 0.47 | 1 | 0.38 | 1 | 0.017 | 1 | 0.002 | 0.46 |
| Carn C20:4 | 0.00043 (1.5×10-4) | 0.00043 (1.4×10-4) | 0.46 | 1 | 0.26 | 1 | 0.0004 (1.6×10-4) | 0.21 | 1 | 0.2 | 1 | 0.023 | 1 | 0.03 | 1 |
| LPC C16 | 74 (35.9) | 84 (44.4) | 0.027 | 1 | 0.044 | 1 | 89 (50.9) | 0.001 | 0.13 | 0.001 | 0.15 | 0.17 | 1 | 0.11 | 1 |
| LPC C17 | 1.5 (0.8) | 1.6 (0.9) | 0.17 | 1 | 0.23 | 1 | 1.8 (1) | 0.001 | 0.11 | 0.002 | 0.4 | 0.011 | 1 | 0.016 | 1 |
| LPC C18 | 26 (13.2) | 29 (15.9) | 0.052 | 1 | 0.07 | 1 | 32 (18.5) | 0.001 | 0.16 | 0.001 | 0.27 | 0.14 | 1 | 0.12 | 1 |
| LPC C18:1 | 18 (5.95) | 20 (7.62) | 0.09 | 1 | 0.11 | 1 | 20 (6.91) | 0.006 | 1 | 0.008 | 1 | 0.42 | 1 | 0.36 | 1 |
| LPC C18:3 | 0.51 (0.2) | 0.61 (0.3) | 0.022 | 1 | 0.045 | 1 | 0.58 (0.3) | 0.66 | 1 | 0.59 | 1 | 0.14 | 1 | 0.26 | 1 |
| PC aa C32:1 | 14 (7.08) | 13 (5.57) | 0.71 | 1 | 0.84 | 1 | 13 (6.36) | 0.012 | 1 | 0.031 | 1 | 0.08 | 1 | 0.043 | 1 |
| PC aa C34:2 | 349 (78.9) | 346 (96.6) | 0.32 | 1 | 0.39 | 1 | 320 (86.1) | 0.004 | 0.79 | 0.008 | 1 | 0.11 | 1 | 0.08 | 1 |
| PC aa C36:2 | 257 (73.5) | 256 (86) | 0.22 | 1 | 0.33 | 1 | 243 (80) | 0.001 | 0.25 | 0.001 | 0.1 | 0.07 | 1 | 0.02 | 1 |
| PC aa C36:3 | 119 (25.7) | 115 (29.9) | 0.09 | 1 | 0.17 | 1 | 111 (25.7) | 0.004 | 0.68 | 0.003 | 0.65 | 0.25 | 1 | 0.15 | 1 |
| PC aa C38:4 | 103 (25) | 101 (33.2) | 0.45 | 1 | 0.32 | 1 | 97 (34.1) | 0.009 | 1 | 0.03 | 1 | 0.12 | 1 | 0.19 | 1 |
| PC aa C40:5 | 9.4 (3.2) | 9.7 (3.3) | 0.72 | 1 | 0.79 | 1 | 9 (3.28) | 0.22 | 1 | 0.16 | 1 | 0.08 | 1 | 0.034 | 1 |
| PC ae C34:1 | 12 (2.91) | 11 (3.43) | 0.014 | 1 | 0.02 | 1 | 12 (3.03) | 0.18 | 1 | 0.16 | 1 | 0.31 | 1 | 0.52 | 1 |
| PC ae C34:2 | 13 (4.18) | 13 (4.46) | 0.08 | 1 | 0.09 | 1 | 13 (4.45) | 0.025 | 1 | 0.035 | 1 | 0.6 | 1 | 0.54 | 1 |
| PC ae C36:1 | 8.1 (4.16) | 9.2 (6.63) | 0.44 | 1 | 0.42 | 1 | 12 (7.21) | 0.023 | 1 | 0.031 | 1 | 0.21 | 1 | 0.22 | 1 |
| PC ae C38:6 | 7.6 (2.39) | 7.3 (2.74) | 0.2 | 1 | 0.25 | 1 | 6.7 (2.63) | 0.005 | 0.9 | 0.005 | 0.99 | 0.25 | 1 | 0.28 | 1 |
| PC ae C40:4 | 3.6 (1.55) | 3.3 (1.49) | 0.58 | 1 | 0.58 | 1 | 4 (1.77) | 0.012 | 1 | 0.008 | 1 | 0.002 | 0.44 | 0.004 | 0.69 |
| SM C21:2 | 0.095 (0.06) | 0.1 (0.06) | 0.35 | 1 | 0.32 | 1 | 0.11 (0.06) | 0.038 | 1 | 0.048 | 1 | 0.048 | 1 | 0.0505 | 1 |
| SM C33:1 | 5.1 (1.62) | 5 (1.51) | 0.6 | 1 | 0.65 | 1 | 5.7 (1.67) | 0.002 | 0.38 | 0.014 | 1 | <0.0001 | 0.053 | 0.006 | 1 |
| SM C36:3 | 0.83 (0.3) | 0.74 (0.3) | 0.06 | 1 | 0.06 | 1 | 0.85 (0.3) | 0.43 | 1 | 0.54 | 1 | 0.006 | 1 | 0.038 | 1 |
| NEFA 20:4 | 7.1 (5.44) | 7.9 (5.78) | 0.07 | 1 | 0.1 | 1 | 8.2 (5.78) | 0.014 | 1 | 0.035 | 1 | 0.41 | 1 | 0.44 | 1 |
| NEFA 20:5 | 0.65 (0.6) | 0.69 (0.5) | 0.06 | 1 | 0.09 | 1 | 0.84 (0.6) | <0.0001 | 0.013 | 0.001 | 0.12 | 0.009 | 1 | 0.023 | 1 |
| NEFA 22:6 | 3.4 (1.77) | 3.7 (1.69) | 0.053 | 1 | 0.06 | 1 | 4.1 (2.01) | 0.002 | 0.29 | 0.012 | 1 | 0.11 | 1 | 0.37 | 1 |
| NEFA 26:1 | 0.18 (0.07) | 0.15 (0.06) | 0.004 | 0.83 | 0.006 | 1 | 0.15 (0.07) | 0.06 | 1 | 0.13 | 1 | 0.25 | 1 | 0.046 | 1 |
| Alpha-ketobutyrate | 15 (17.6) | 17 (23.9) | 0.21 | 1 | 0.25 | 1 | 18 (26.6) | 0.003 | 0.49 | 0.003 | 0.54 | 0.26 | 1 | 0.15 | 1 |
| Fructose-6-phosphate | 39 (11.4) | 40 (12.7) | 0.86 | 1 | 0.75 | 1 | 42 (13) | 0.016 | 1 | 0.11 | 1 | 0.013 | 1 | 0.02 | 1 |
| Fumarate | 0.69 (0.4) | 0.68 (0.3) | 0.58 | 1 | 0.65 | 1 | 0.61 (0.2) | 0.33 | 1 | 0.25 | 1 | 0.08 | 1 | 0.01 | 1 |
| Pyruvate | 137 (73.6) | 135 (74) | 0.28 | 1 | 0.49 | 1 | 102 (79.7) | 0.001 | 0.19 | 0.004 | 0.82 | 0.08 | 1 | 0.046 | 1 |
| Sum LPC | 165 (60.4) | 181 (71.3) | 0.07 | 1 | 0.1 | 1 | 187 (78.9) | 0.006 | 1 | 0.006 | 1 | 0.34 | 1 | 0.25 | 1 |
| Ratio Carn C3/Ile | 0.011 (0.005) | 0.012 (0.006) | 0.81 | 1 | 0.92 | 1 | 0.01 (0.006) | 0.006 | 1 | 0.008 | 1 | 0.004 | 0.88 | 0.005 | 0.92 |
| Ratio Carn C3/Val | 0.003 (0.001) | 0.003 (0.002) | 0.99 | 1 | 0.93 | 1 | 0.0027 (0.001) | 0.007 | 1 | 0.009 | 1 | 0.008 | 1 | 0.01 | 1 |
| Ratio Carn C5/Ile | 0.0032 (0.001) | 0.0033 (0.001) | 0.86 | 1 | 0.98 | 1 | 0.0031 (0.001) | 0.015 | 1 | 0.019 | 1 | 0.012 | 1 | 0.027 | 1 |
| Ratio Carn C5/Leu | 0.0016 (0.0006) | 0.0016 (0.0007) | 0.98 | 1 | 0.9 | 1 | 0.0015 (0.0007) | 0.015 | 1 | 0.021 | 1 | 0.015 | 1 | 0.031 | 1 |
Adjusted for age at blood withdrawal, sex, and parental education
Abbreviations: Carn, Acylcarnitine; lysoPC, lysophosphatidylcholine; NEFA, non-esterified acid; PC aa, diacyl-phosphatidylcholine; PC ae, acylalkyl-phosphatidylcholine; SM, sphingomyeline
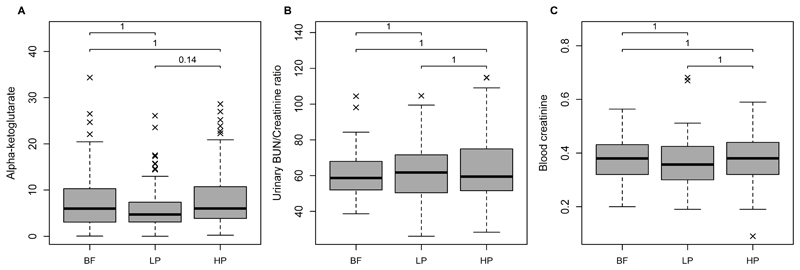
At 5.5y, alpha-ketoglutarate (αKG) was the metabolite differing most strongly between the intervention groups (uncorrected P [Puncorrected] = 0.001, Bonferroni corrected P [PBonferroni] = 0.14). Likewise elevated in the HP group were the TCA-intermediates pyruvate (Puncorrected = 0.004, PBonferroni=1) and 3-methyl-2-oxobutanoate (Puncorrected = 0.034, PBonferroni=1), and some ketoacid/BCAA and acylcarnitine/BCAA-ratios: 3-Methyl-2-oxobutanoate/Val, 3-Methyl-2-oxovalerate/Ile, Carn C3/Ile, Carn C3/Val, Carn C4/Val, Carn C5/Ile, and Carn C5/Leu were all higher in the HP group (Puncorrected from 0.001 to 0.042, PBonferroni>0.17). The mean BCAA concentrations were lower in the HP group but the difference did not reach statistical significance. Figure 2 displays the concentrations of αKG along with the urinary blood urea nitrogen (BUN)/creatinine ratio and the blood creatinine concentrations in the two intervention groups and the breastfed group. Only the comparison of αKG between the LP vs. HP groups reached a PBonferroni <0.15. The boxplots on αKG show furthermore that levels of αKG of the BF children are more similar to those of the HP children (mean: 5.9 µmol/L in LP; 8 µmol/L, 5.94 in HP; 7.5 µmol/L in BF). Results on the comparisons of the metabolite profile between the formula groups and the observational group of BF infants are shown in Figure A.2 and Table A.3. Twelve and 23 metabolites differed between BF vs. LP and BF vs. HP children, respectively. Some acylcarnitines species tended to be higher in HP than in BF children, whereas the sum of all acylcarnitines was not significantly different (Puncorrected = 0.17).
Figure 2.
Boxplots of the (A) 5.5year plasma alpha-ketoglutarate (αKG) concentrations (µmol/L), (B) the 5.5year urinary blood urea nitrogen (BUN)/creatinine ratio, and (C) 5.5year blood creatinine (mg/dl). Group differences were tested using linear mixed effects models adjusted for age at blood withdrawal, sex, and parental education. P values were Bonferroni corrected for the number of plasma metabolites & metabolite ratios (#: 240). BF, breastfed; LP, lower protein infant formula; HP, higher protein infant formula.
At the age of 8 y, there were less differences between LP and HP, and no metabolite or metabolite ratio differed significantly. LPC tended to be higher in HP vs LP, but only two LPC species reached the uncorrected α-threshold (LPC 16 and LPC 18:3: Puncorrected < 0.045). The sum of all LPC differed not significantly (Puncorrected = 0.1). Total LPC tended also to be lower in LP than in BF infants (sum of all LPC, Puncorrected = 0.006). Also at 8 years of age, no metabolite differed significantly between the BF and the HP or LP group. While LP children showed a similar metabolite profile as BF infants at 5.5y, they were more similar metabolite to HP children at 8y. Regarding the group of acylcarnitine/BCAA ratios which showed up at 5.5y, we observed a trend towards increased pyruvate and acylcarnitine/BCAA ratios in LP and HP children compared to BF children at 8y.
Quantification of the programming factors on the metabolome
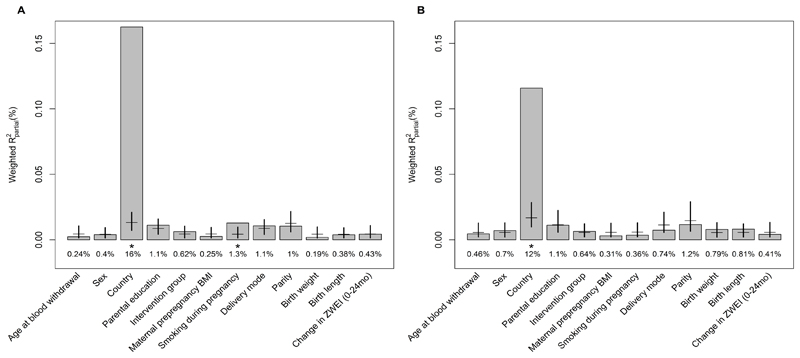
Figure 3 shows the contributions of feeding groups and other programming factors to the variations in metabolic profiles at 5.5 and 8y. Results on the LME regressing the metabolites on the programming factors are shown in Table A.4-A10.Together, all variables explained 22% and 16% of variance in the 5.5y and 8y metabolome, respectively. The variable explaining most of the variance was country of residence (Rpartial2 = 16% and 12% for 5.5y and 8y, respectively). Sex stratified analysis revealed that the country of residence explained more variance in girls (19% and 14% for 5.5y and 8y, respectively; compared to 14% and 11% in boys for 5.5y and 8y, respectively; Figure A.4). While in boys no other factor explained a significant proportion of variance in the metabolome, maternal smoking during pregnancy explained more variance than expected by chance in girls at 5.5y (Rpartial2 = 2.5% explained variability in the 5.5y metabolome; 95%CIpercentile 0.3% - 2.1). At 8y however, this effect was not seen any more (Rpartial2 = 0.8%; 95%CIpercentile 0.4% - 3%).
Figure 3.
Results of the PC-PR2 analysis showing the proportion of explained variance Rpartial2 of early programming factors and other selected variables in the (A) 5.5y metabolomics data of formula fed children, N=229; and the (B) 8y metabolomics data of formula fed children, N=180. Significance (*) is given if the observed Rpartial2 of the respective variable is outside of the bootstrapped percentile confidence interval under the assumption of chance.
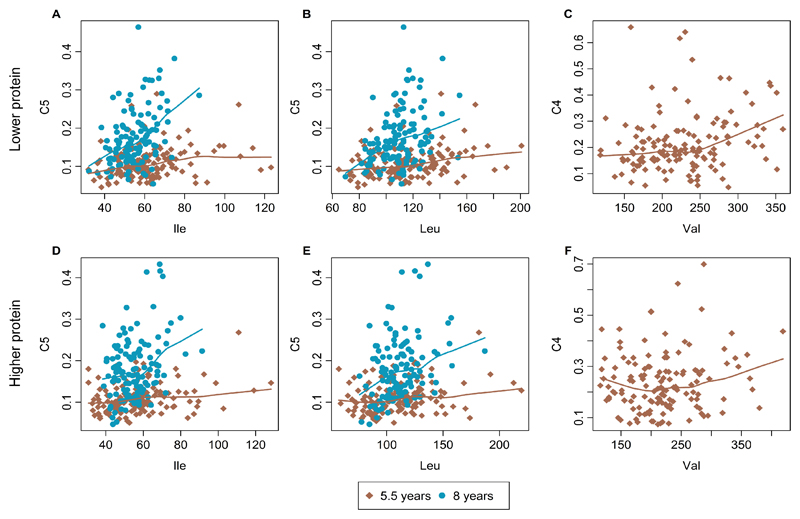
Breakpoint analysis
The scatterplots of the acylcarnitine/BCAA ratios with lowess smoothers for the 5.5y and the 8y data are shown in Figure 4. We did not find a breakpoint for any of the ratios, neither at any time point nor for any feeding group (P values for 5.5y: C5/Ile, 0.59 for LP and 0.14 for HP; C5/Leu, 0.55 for LP and 0.07 for HP; C4/Val, 0.26 for LP and 0.57 for HP; P values for 8y: C5/Ile, 0.80 for LP and 0.16 for HP; C5/Leu, 0.82 for LP and 0.46 for HP).
Figure 4.
Scatterplots with LOWESS lines to fit branched-chain amino acids (isoleucine, leucine, valine) versus their acylcarnitine degradation product in children having received a lower (A-C) and higher protein (D-F) infant formula milk. Concentrations are in micromoles. A and D, Isoleucine and acylcarnitine C5. B and E, Leucine and acylcarnitine C5. C and F, Valine and acylcarnitine C4.
5. Discussion
This randomized study shows that the impact of formula feeding in infancy with a different protein supply on the plasma metabolome of children aged 5.5 and 8 years is very small and not distinguishable from chance. Only a few metabolites were found to be affected, but there was no consistency between the 5.5y and 8y metabolome. Regarding the other early programming factors investigated, we found no indication for a programming effect. However, if we missed to include lifestyle factors that are associated to both, the childhood metabolome and the early programming factor, this result might suffer from the omitted-variable bias.
Differences in metabolome between HP and LP children
At 5.5y, the major differences between the infant feeding groups relate to alpha-ketoglutarate (αKG) and the ketoacid/BCAA and aclycarnitine/BCAA-ratios. While αKG is involved in the first step of BCAA catabolism, the ratios display single steps in the degradation pathway. αKG is also an intermediate in the tricarboxylic acid (TCA) cycle. However, neither the precursor nor the derivate of αKG, isocitrate and succinate, were different between the LP and HP group, thus we interpret our results as an indication of an altered BCAA catabolism: the elevated levels of αKG and the ratios might point towards increased BCAA degradation in HP children. The higher levels of pyruvate in HP might result from greater availability of acetyl-CoA from BCAA catabolism, which was reported to downregulate the decarboxylation of pyruvate to acetyl-CoA by pyruvate dehydrogenase 17.
αKG supplementation has been suggested to promote muscle synthesis 18–20. But our results on blood creatinine, which is the product of muscle creatinine catabolism and whose concentration depends on the subject’s muscle mass, do not suggest a higher protein catabolism in HP children as levels of creatinine were similar in LP and HP children. The elevated αKG and acylcarnitine/BCAA ratios might also result from of a higher protein intake in HP children. But the urinary BUN/creatinine ratio, which is an indicator for dietary protein intake 21, was similar in the two feeding groups.
At 8y, no metabolite or metabolite ratio was significantly different between HP and LP, and overall the impact of the infant feeding group on the metabolite profile was less strong. LPC tended to be higher in HP vs. LP. LPC are mainly formed by hydrolysis of the sn-2 fatty acid of phosphatidylcholine mediated primarily by lecithin-cholesterol acyltransferase (LCAT) 22. Several animal studies found low protein intake to decreased LCAT activity 23–25, but we have no indication of a difference in protein intake between groups in childhood, neither at 5.5y nor at 8y.
The one finding neglected so far complicates the interpretation of our results: Both the 5.5y levels of αKG as well as the 8y LPC levels in BF children were more similar to those of the HP children than those of the LP children. This is surprising as LP children were more similar to BF children with respect to early growth 8 and BMI at early school age 9. The randomization of the LP and HP group has been shown to be successful 8. Thus, we can exclude that our results are driven by different complementary foods. In contrast, the BF group was not randomized. Women who choose to breastfeed have usually a higher socioeconomic status 26. But also paternal factors such as income, unemployment, or paternity leave were found to influence the odds for breastfeeding 27. A comparison of the breastfed group to the formula groups may thus be severely biased due to confounding factors.
There are several possible explanations as to why we found no major effects of early nutritional and other programming factors on the 5.5y or 8y metabolome. One must consider that the plasma metabolome is a highly dynamic system which is influenced by many different factors, including current lifestyle and environment, day time 28–30, and gut microbiome 31, so that potential metabolic effects of early programming events might be masked by variation induced by other influencing factors. This possibility is supported by our findings that country of residence consistently explained a significant proportion of variance in the metabolomics data and that the effects of the formula milk were stronger at 5.5y than at 8y of age.
Breakpoint in BCAA catabolism
Previously, we have reported indications for saturation in BCAA catabolism in 6 month old children fed higher protein formula 16. At the later childhood ages studied here, no such breakpoints in the BCAA catabolism are detectable. A key reason could be the much lower protein intake per kg body weight in childhood as compared to the protein intake in infancy 32, which is also reflected by the much lower plasma BCAA concentrations at 5.5 and 8y as found previously in the 6 month old infants: the highest observed concentrations in children at 5.5y and 8y are below the previously determined breakpoint concentrations at 136 µmol/L (Ile) and 234 µmol/L (Leu) 16. The activity of BCKDH, which is the rate limiting step in BCAA catabolism 33, may increase with age. This hypothesis is supported by a study in rats whose liver BCKDH activity increased throughout the suckling period and reached adult levels at the weaning age 34. Data on humans is missing. For the interpretation of our results, we can thus only speculate on the increasing BCKDH activity. For the interpretation of our results, we can thus only speculate on the increasing BCKDH activity. This increased activity might be the reason why we do not find an indication for an exceed of the BCAA breakdown in healthy children aged 5-8 years who did not receive a special diet.
The effects of the other factors on the metabolome
The other programming factors studied were also not found to noticeably influence the plasma metabolome in childhood. Only maternal smoking during pregnancy accounted for more variability in the 5.5y metabolomics data than expected by chance. However, the effect was small, explaining only 1.4% of the variation and it was not seen at 8y. Sex stratified analyses indicated that smoking during pregnancy affected the metabolome at 5.5y of girls only. As the percent explained variability was still very small (2.5%) and only 0.4% percent points higher than what one would have been expected by chance, further studies are needed in order to verify if smoking during pregnancy affects girls to a larger extent.
Another interesting finding on differences between girls and boys is that of a larger effect of the country of residence on the metabolome in girls. In the overall studied population, country of residence explained the largest proportion of metabolite variation. This finding is in line with the results of Fages et al, the developers of the PC-PR2 method 15. The authors assigned 8% explained variability to country using 1H-NMR serum metabolomics measurements of adult individuals of the EPIC liver nested case–control study. Country of residence is certainly a proxy for many factors such as lifestyle, diet, and environment. The role of the single factors is by far not understood. A recent nutritional randomized, controlled, crossover trial highlights the difficult assessment of the effect of the diet on the metabolome and the high intra-individual differences 35, 36. Furthermore, although the same SOP were followed in all laboratories, the different laboratory conditions and personnel might as well add variation to the data. Thus, we cannot draw conclusion on what factors are comprised in the country effect and what is exactly exhibiting a higher effect on girls than on boys.
Birth weight is considered “a clinical outcome representative of the summation of exposures and insults that occurred in utero” 37. Many studies found a positive association between birth weight and later adiposity 38. A recent meta-analysis confirmed the higher obesity risk for high birth weight (>4000g) infants 39. In this present study, we did not find birth weight associated with the childhood metabolome, but we studied a rather healthy population without a high obesity prevalence and with little occurrence of extreme obesity. Of importance, this study included only infants born at full term with a birth weight appropriate for gestational age, which limits the range of exposures that might modulate later metabolome.
Strengths and limitations
The interpretation of our results must be carried out carefully. Statistically speaking, since the metabolomics measurements are a secondary outcome only and the study was not powered for this analysis, no statement on the null hypothesis is possible: we cannot conclusively deny the existence of a programming effect. Next to this limitation, we would like to discuss the problem that we could not adjust for all confounding factors such as lifestyle and diet. If these factors had an influence on both, the early programming variable and the childhood metabolome, we might have over- or underestimated the effect of the programming variables on the metabolites (the so-called ‘omitted-variable bias’). For instance, maternal diet might be very well associated with maternal pre-pregnancy BMI and the childhood metabolome. This problem can be solved by randomization since such factors are equally distributed across the intervention groups if randomization was successful. We have previously shown that parental and early infant characteristics did not differ between the randomized groups 8. Thus, in contrast to the early programming factors, we can assume that our results on the HP vs. LP randomized groups are not subject to confounding.
The big strength of this study is thus the long-term investigation of the effect of the randomized infant feeding group. Children from five European countries participated in this study. The intervention consisted of feeding isocaloric higher or lower protein infant formula for the first year of life. Using a high quality LC-MS/MS platform we determined highly specific quantitative measurements of plasma metabolite concentrations which cover a wide spectrum of metabolic pathways. We approached our research questions with both simple and advanced statistical methods and could not only characterize single effects but also quantify the global impact. A limitation is the considerable loss to follow up. However, dropout rates up to 6 years and reasons for dropout were similar in the two randomized feeding groups 9. Furthermore, even though we measured ~200 metabolites acting in different metabolic pathways, we cannot exclude that we may have missed molecules that might have been influenced by the intervention.
6. Conclusions
To our knowledge, this is the first study to investigate potential long-term effects of infant feeding on plasma metabolome at school age. We found trends towards a slightly altered metabolism in children having received a high protein infant formula. However, these trends were observed in the 5.5y metabolome only and were not replicated in the 8y metabolome. The strongest identified predictor of variance in metabolomics data was country of residence most likely reflecting the combined variation of many factors such as lifestyle, current dietary habits, and environmental factors. We cannot conclude that there is no programming effect nor can we exclude the possibility that potential modest early programming effects on the childhood plasma metabolome were masked by larger effect sizes of other influencing factors, such as current diet and lifestyle.
Supplementary Material
Acknowledgements
We gratefully thank Stephanie Winterstetter and Alexander Haag (Division of Metabolic and Nutritional Medicine, Dr von Hauner Children’s Hospital, University of Munich, Munich, Germany), who analysed the blood plasma samples.
Funding sources:
The authors’ work is financially supported in part by the Commission of the European Communities, Projects Early Nutrition (FP7-289346), DYNAHEALTH (H2020-633595) and LIFECYCLE (H2020-SC1-2016-RTD), and the European Research Council Advanced Grant META-GROWTH (ERC-2012-AdG 322605). No funding bodies had any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- AA
amino acids
- αKG
alpha-ketoglutarate
- BCAA
branched chain amino acids
- BF
breastfed
- BUN
urinary blood urea nitrogen
- Carn
acylcarnitines
- HP
higher protein formula
- LC-MS/MS
liquid chromatography mass spectrometry
- LCAT
lecithin-cholesterol acyltransferase
- LP
lower protein fsormula
- LPC
lysophosphatidylcholines
- NEFA
Non-esterified fatty acids
- PCA
principal component analysis
- PC
principal components
- PCaa
diacyl-phosphatidylcholines
- PCae
acyl-alkyl-phosphatidylcholines
- SM
sphingomyelines
- TCA
tricarboxylic acid cycle
- WFA
weight-for-age
Footnotes
Clinical Trial Registry number and website: ClinicalTrials.gov; identifier: NCT00338689; https://clinicaltrials.gov/ct2/show/NCT00338689;
Conflict of Interest
The authors declare that they have no competing interests with respect to this manuscript
References
- 1.Seidell JC, Halberstadt J. Obesity: The obesity epidemic in the USA - no end in sight? Nature reviews Endocrinology. 2016;12(9):499–500. doi: 10.1038/nrendo.2016.121. [DOI] [PubMed] [Google Scholar]
- 2.Dörner G. Anatomical neuroendocrinology. Karger Publishers; 1975. Perinatal hormone levels and brain organization; pp. 245–252. [Google Scholar]
- 3.Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35(7):595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 4.Lucas A. Programming by early nutrition in man. Ciba Found Symp. 1991;156:38–50. discussion 50-5. [PubMed] [Google Scholar]
- 5.Hellmuth C, Uhl O, Kirchberg FF, Grote V, Weber M, Rzehak P, et al. Effects of Early Nutrition on the Infant Metabolome. Nestle Nutrition Institute workshop series. 2016;85:89–100. doi: 10.1159/000439491. [DOI] [PubMed] [Google Scholar]
- 6.Koletzko B, Brands B, Grote V, Kirchberg FF, Prell C, Rzehak P, et al. Long-Term Health Impact of Early Nutrition: The Power of Programming. Annals of nutrition & metabolism. 2017;70(3):161–169. doi: 10.1159/000477781. [DOI] [PubMed] [Google Scholar]
- 7.Koletzko B, von Kries R, Closa R, Escribano J, Scaglioni S, Giovannini M, et al. Can infant feeding choices modulate later obesity risk? The American journal of clinical nutrition. 2009;89(5):1502S–1508S. doi: 10.3945/ajcn.2009.27113D. [DOI] [PubMed] [Google Scholar]
- 8.Koletzko B, von Kries R, Closa R, Escribano J, Scaglioni S, Giovannini M, et al. Lower protein in infant formula is associated with lower weight up to age 2 y: a randomized clinical trial. The American journal of clinical nutrition. 2009;89(6):1836–45. doi: 10.3945/ajcn.2008.27091. [DOI] [PubMed] [Google Scholar]
- 9.Weber M, Grote V, Closa-Monasterolo R, Escribano J, Langhendries JP, Dain E, et al. Lower protein content in infant formula reduces BMI and obesity risk at school age: follow-up of a randomized trial. The American journal of clinical nutrition. 2014;99(5):1041–51. doi: 10.3945/ajcn.113.064071. [DOI] [PubMed] [Google Scholar]
- 10.Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant molecular biology. 2002;48(1-2):155–71. [PubMed] [Google Scholar]
- 11.Nicholson JK. Global systems biology, personalized medicine and molecular epidemiology. Molecular systems biology. 2006;2(1) doi: 10.1038/msb4100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet. 2015;16(2):85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- 13.de Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J. Development of a WHO growth reference for school-aged children and adolescents. Bulletin of the World Health Organization. 2007;85(9):660–7. doi: 10.2471/BLT.07.043497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Onis M, Onyango AW, Van den Broeck J, Chumlea WC, Martorell R. Measurement and standardization protocols for anthropometry used in the construction of a new international growth reference. Food and nutrition bulletin. 2004;25(1 Suppl):S27–36. doi: 10.1177/15648265040251S104. [DOI] [PubMed] [Google Scholar]
- 15.Fages A, Ferrari P, Monni S, Dossus L, Floegel A, Mode N, et al. Investigating sources of variability in metabolomic data in the EPIC study: the Principal Component Partial R-square (PC-PR2) method. Metabolomics : Official journal of the Metabolomic Society. 2014;10(6):1074–1083. [Google Scholar]
- 16.Kirchberg FF, Harder U, Weber M, Grote V, Demmelmair H, Peissner W, et al. Dietary protein intake affects amino acid and acylcarnitine metabolism in infants aged 6 months. The Journal of clinical endocrinology and metabolism. 2015;100(1):149–58. doi: 10.1210/jc.2014-3157. [DOI] [PubMed] [Google Scholar]
- 17.Kerbey AL, Randle PJ, Cooper RH, Whitehouse S, Pask HT, Denton RM. Regulation of pyruvate dehydrogenase in rat heart. Mechanism of regulation of proportions of dephosphorylated and phosphorylated enzyme by oxidation of fatty acids and ketone bodies and of effects of diabetes: role of coenzyme A, acetyl-coenzyme A and reduced and oxidized nicotinamide-adenine dinucleotide. The Biochemical journal. 1976;154(2):327–48. doi: 10.1042/bj1540327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao K, Yin Y, Li X, Xi P, Wang J, Lei J, et al. Alpha-ketoglutarate inhibits glutamine degradation and enhances protein synthesis in intestinal porcine epithelial cells. Amino acids. 2012;42(6):2491–500. doi: 10.1007/s00726-011-1060-6. [DOI] [PubMed] [Google Scholar]
- 19.Hammarqvist F, Wernerman J, von der Decken A, Vinnars E. Alpha-ketoglutarate preserves protein synthesis and free glutamine in skeletal muscle after surgery. Surgery. 1991;109(1):28–36. [PubMed] [Google Scholar]
- 20.Cai X, Zhu C, Xu Y, Jing Y, Yuan Y, Wang L, et al. Alpha-ketoglutarate promotes skeletal muscle hypertrophy and protein synthesis through Akt/mTOR signaling pathways. Scientific reports. 2016;6 doi: 10.1038/srep26802. 26802. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Hosten AO. In: Clinical Methods: The History, Physical, and Laboratory Examinations. 3rd edn. Walker HK, Hall WD, Hurst JW, editors. Boston: 1990. BUN and Creatinine. [PubMed] [Google Scholar]
- 22.Jonas A. Lecithin cholesterol acyltransferase. Biochimica et biophysica acta. 2000;1529(1-3):245–56. doi: 10.1016/s1388-1981(00)00153-0. [DOI] [PubMed] [Google Scholar]
- 23.Park CS. Influence of dietary protein on blood cholesterol and related metabolites of growing calves. Journal of animal science. 1985;61(4):924–30. doi: 10.2527/jas1985.614924x. [DOI] [PubMed] [Google Scholar]
- 24.Yashiro M, Kimura S. Effect of voluntary exercise and dietary protein levels on serum lipoprotein distributions and lecithin: cholesterol acyltransferase (LCAT) activity of mice. Journal of nutritional science and vitaminology. 1980;26(1):59–69. doi: 10.3177/jnsv.26.59. [DOI] [PubMed] [Google Scholar]
- 25.Lamri MY, Meghelli-Bouchenak M, Boualga A, Belleville J, Prost J. Time course of changes in rat serum lecithin-cholesterol acyl-transferase activity and high-density-lipoprotein composition during the consumption of two different low-protein diets followed by a balanced diet. Nutrition. 1995;11(5):444–9. [PubMed] [Google Scholar]
- 26.Flacking R, Nyqvist KH, Ewald U. Effects of socioeconomic status on breastfeeding duration in mothers of preterm and term infants. European journal of public health. 2007;17(6):579–84. doi: 10.1093/eurpub/ckm019. [DOI] [PubMed] [Google Scholar]
- 27.Flacking R, Dykes F, Ewald U. The influence of fathers' socioeconomic status and paternity leave on breastfeeding duration: a population-based cohort study. Scandinavian journal of public health. 2010;38(4):337–43. doi: 10.1177/1403494810362002. [DOI] [PubMed] [Google Scholar]
- 28.Ang JE, Revell V, Mann A, Mantele S, Otway DT, Johnston JD, et al. Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography-mass spectrometry metabolomic approach. Chronobiology international. 2012;29(7):868–81. doi: 10.3109/07420528.2012.699122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bundy JG, Davey MP, Viant MR. Environmental metabolomics: a critical review and future perspectives. Metabolomics : Official journal of the Metabolomic Society. 2008;5(1):3. [Google Scholar]
- 30.Nicholson JK, Lindon JC, Holmes E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica; the fate of foreign compounds in biological systems. 1999;29(11):1181–9. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 31.Vernocchi P, Del Chierico F, Putignani L. Gut Microbiota Profiling: Metabolomics Based Approach to Unravel Compounds Affecting Human Health. Frontiers in microbiology. 2016;7:1144. doi: 10.3389/fmicb.2016.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Efsa Panel on Dietetic Products N, Allergies. Scientific Opinion on Dietary Reference Values for protein. EFSA Journal. 2012;10(2):2557. n/a. [Google Scholar]
- 33.Adeva-Andany MM, Lopez-Maside L, Donapetry-Garcia C, Fernandez-Fernandez C, Sixto-Leal C. Enzymes involved in branched-chain amino acid metabolism in humans. Amino acids. 2017;49(6):1005–1028. doi: 10.1007/s00726-017-2412-7. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y, Denne SC, Harris RA. Developmental pattern of branched-chain 2-oxo acid dehydrogenase complex in rat liver and heart. The Biochemical journal. 1993;290(Pt 2):395–9. doi: 10.1042/bj2900395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Perez I, Posma JM, Gibson R, Chambers ES, Hansen TH, Vestergaard H, et al. Objective assessment of dietary patterns by use of metabolic phenotyping: a randomised, controlled, crossover trial. Lancet Diabetes Endocrinol. 2017;5(3):184–195. doi: 10.1016/S2213-8587(16)30419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhupathiraju SN, Hu FB. One (small) step towards precision nutrition by use of metabolomics. Lancet Diabetes Endocrinol. 2017;5(3):154–155. doi: 10.1016/S2213-8587(17)30007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young BE, Johnson SL, Krebs NF. Biological determinants linking infant weight gain and child obesity: current knowledge and future directions. Advances in nutrition. 2012;3(5):675–86. doi: 10.3945/an.112.002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palatianou ME, Simos YV, Andronikou SK, Kiortsis DN. Long-term metabolic effects of high birth weight: a critical review of the literature. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme. 2014;46(13):911–20. doi: 10.1055/s-0034-1395561. [DOI] [PubMed] [Google Scholar]
- 39.Schellong K, Schulz S, Harder T, Plagemann A. Birth weight and long-term overweight risk: systematic review and a meta-analysis including 643,902 persons from 66 studies and 26 countries globally. PloS one. 2012;7(10):e47776. doi: 10.1371/journal.pone.0047776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.