Abstract
Pancreatic ductal adenocarcinoma is particularly metastatic, with dismal survival rates and few treatment options. Stiff fibrotic stroma is a hallmark of pancreatic tumours, but it is still unclear how stromal mechanosensing affects metastasis. Here, we show that mechanical changes in pancreatic cancer cell environment affects not only adhesion and migration, but also ATP/ADP and ATP/AMP ratios. Unbiased metabolomic analysis reveals the creatine phosphagen ATP recycling system as a major mechanosensitive target. This system depends on arginine flux through the urea cycle, and this is reflected by the increased incorporation of carbon and nitrogen from L-arginine into creatine, phospho-creatine and creatinine on stiff matrix. We identify creatine kinase-B (CKB) as a mechanosensitive transcriptional target of the Yes-associated protein (YAP), thus enhancing phosphocreatine. We further demonstrate a role of the creatine phosphagen system in invasive migration, chemotaxis and liver metastasis.
Introduction
Cell-ECM (extracellular matrix) interactions can be severely disrupted in fibrotic diseases and malignancy, where excessive ECM deposition and remodelling drive tumour progression and metastasis 1,2. In particular, pancreatic ductal adenocarcinoma (PDAC) is characterised by extensive matricellular fibrosis linked to aggressiveness 3. The most aggressive fibrotic forms are clustered molecularly into one subtype (squamous), which has the worst prognosis 4. ECM remodelling also increases stiffness and tissue tension, impacting cell behaviour through mechanotransduction, an important mediator of malignancy 5–7. The dense tumour stroma and poor vascularisation also limit nutrient and oxygen availability. Oncogenic mutations, such as KRasG12D impart metabolic flexibility to tumours, allowing aerobic glycolysis and conferring a survival advantage despite the hostile environment 8. Tumour cells adjust their metabolism, allowing survival despite limited nutrients, including scavenging of extracellular protein 9 and rewiring of glucose metabolism 10. The effects of stiff stroma and ECM remodelling on tumour cell behaviour are poorly understood but are crucial to deciphering why PDAC is so aggressive 11. Here, we explore connections between stromal stiffness and metabolism to gain insight into how energy production and consumption link to metastatic aggressiveness.
Adhesion to stiff matrix triggers integrin clustering leading to signaling. Specifically, the Yes associated protein 1 (YAP) component of the Hippo signalling pathway, is a major transcriptional pathway activated by mechanical stress and stiff ECM 12. The metabolic state of a cell impinges on YAP signalling, with nutrient starvation driving Hippo/YAP signalling, suggesting potentially promising anti-cancer therapeutic approaches13–16. However, it is unclear if the converse is true, e.g. does mechanosensing and thus YAP signalling affect the metabolic state of cells and if so, by what mechanisms?
Actin is an ATPase and ATP hydrolysis maintains polarity of actin filaments, such that the ATP-bound fast-growing end of filaments points toward a cellular membrane. This allows polarised polymerisation, creating force load against the plasma membrane17. Actin-generated force can move membrane vesicles and protrude the plasma membrane during migration and invasion through ECM. Polarised cells use microtubule and actin-based transport motors to cluster mitochondria near the leading edge to generate local ATP powering migration 18, 19. Additionally, localised mitochondria drive basement membrane invasion of the C. elegans gonadal anchor cell during development, bypassing matrix remodelling enzymes 20. Whether cancer cells employ similar mechanisms of localised energy generation to fuel migration and invasion remains to be explored.
In addition to localised mitochondrial ATP generation, cells also employ ATP recycling to generate and maintain local ATP gradients. Phosphorylated guanidine compounds, known as phosphagens, link to ATP hydrolysis and temporal buffering during dynamic energetic requirements. Among them, the phospho-creatine (pCr)/creatine kinase (CK) system (hereafter, referred to as the pCr-CK system) is one of the most well studied, perhaps due its extensive tissue distribution in vertebrates. The pCr-CK system operates in cell types with dynamic rates of energy turnover, such as neuronal, muscle and spermatozoa 21. The phosphagen system is based on the reversible phosphorylation of creatine to phosphocreatine, an energy storage molecule. In elongated cell systems like muscle and motile gametes, this system increases the free energy of ATP hydrolysis by steepening the gradient between local ATP and ADP. Additionally, it can act to overcome diffusion barriers between the source of ATP (mitochondria) and the site where ATP is needed (e.g. at the tail of the sperm) 21. This reaction is catalysed by creatine kinases and is directly coupled to ATP-ADP exchange22. The cytoplasmic creatine kinase B-type (CKB) could facilitate cell motility by locally providing ATP recycling at sites of actin remodelling 23. Increasing evidence also suggests the importance of the pCr-CK system in malignancies 24, 25. However, it is still unexplored whether this pathway could support cancer cell invasion or is actively involved in pancreatic tumorigenesis.
Here, we investigated the relationship between mechanosensing, motility and metabolic energy generation, and we discovered a dual effect of ECM mechanotransduction on ATP metabolism. Stiff substrata not only support ATP production through increased mitochondrial activity and oxidative phosphorylation, but also control ATP recycling through the pCr-CK system. In rigid environments, PDAC cells synthesise creatine and express CKB, engaging the phosphocreatine dependent ATP recycling mechanism. ECM mechanics regulate creatine phosphorylation through YAP-dependent expression of CKB. This correlates with elongation and polarisation of PDAC cells in response to migration on stiff ECM and supports a role of the pCr-CK system in delivering ATP to invasive protrusions. There, dynamic ATP recycling through the pCr-CK system supported actin dynamics and traction force generation, connecting actomyosin dynamics to energy recycling. The pCr-CK system promoted ECM invasion and metastasis, indicating a mechanism by which the stiff stroma of PDAC may drive aggressive invasive behaviour.
Results
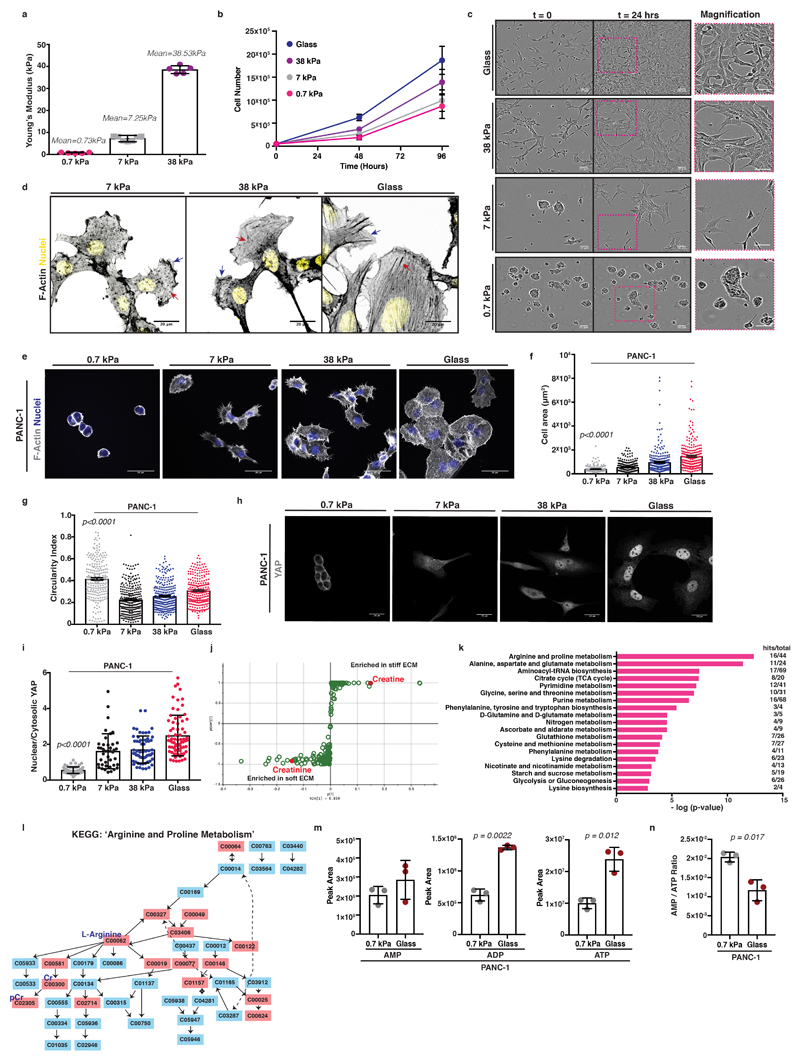
Matrix stiffness affects pancreatic cancer cell cytoskeletal organisation and shape
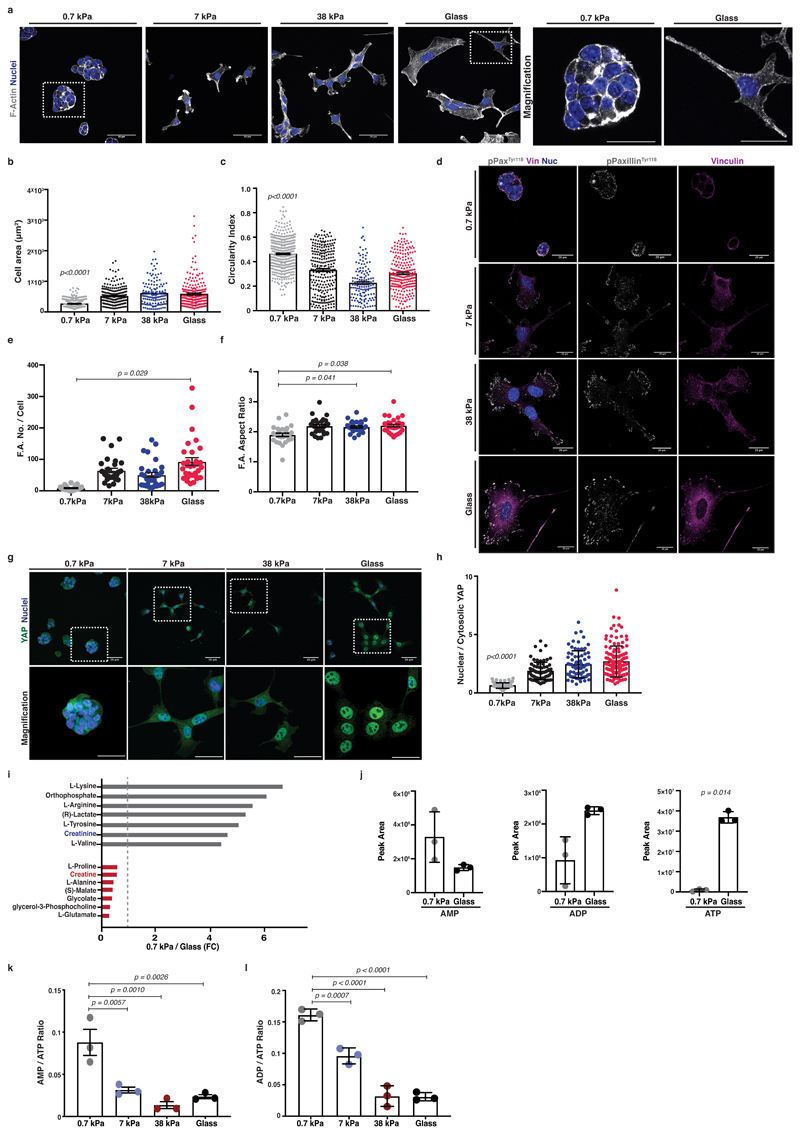
While it is evident that ECM mechanics influence the behaviour of normal epithelial cells 26, their role in transformed cells has been questioned 27. To probe mechanosensitivity of pancreatic cancer cells, we cultured murine KPC cells (pancreas tumour cells cultured from mice with Pdx1-Cre lox-stop-lox (LSL) KRasG12D and p53R172H 28) atop fibronectin containing hydrogels of increasing stiffness or on glass (Figure 1a-c and Extended Data 1a-c). Fibronectin is an important stromal component associated with pancreatic cancer metastasis 29. On surfaces ranging from glass (2-4 GPa), 38 kPa (stiff tumour) down to 0.7 kPa (soft, normal pancreas), cells showed a range of elongated and more spread morphologies with actin stress fibres (red arrows, Extended Data 1d) and lamellipodia (blue arrows, Extended Data 1d). However, on soft matrix of 0.7 kPa, cells appeared clustered and rounded (Figure 1a). As matrix stiffness decreased, cells spread over a smaller area and were more circular (Figure 1b, c). Cells were also less proliferative on softer matrix (Extended Data 1b), indicating robust mechanosensitivity. Additionally, focal adhesions (phospho-PaxillinY118 or Vinculin) decreased on softer substrata (Figure 1d-f). Specifically, phospho-PaxillinY118 positive focal complexes were decreased in number (Figure 1e) and diameter (aspect ratio) (Figure 1f). PANC-1 human PDAC cells behave similarly (Extended Data 1e-g). Thus, pancreatic cancer cells respond to stiffer matrix with increased F-actin organisation, focal adhesions and elongation.
Figure 1. Pancreatic cancer cells are mechanosensitive and untargeted metabolome profiling revealed major metabolic reprogramming with ECM mechanics.
Mouse KPC cells were cultured atop of 0.7-38kPa fibronectin-coated hydrogels and glass coverslips.
(a) Left; immunofluorescence of KPC cells cultured as indicated showing F-actin (grey) and nuclei (blue). Scale bars, 50μm. Right; Magnification of areas indicated by a dashed box. Scale bars, 25μm.
(b-c) Quantification of cell shape from (a) showing cell area (μm2) (b) and circularity index (c). Values are mean ± SEM from n=500 0.7kPa, n=257 7kPa, n=139 38kPa and n=225 glass, cells from 3 independent experiments. Statistical significance assessed by Kruskal-Wallis with Dunn’s multiple comparisons test.
(d) Immunofluorescence of KPC cells from (a) showing pPaxillinTyr118 (grey), Vinculin (magenta) and nuclei (blue). Scale bars, 20μm.
(e-f) Quantification of pPaxillinTyr118 positive particles from (d) showing number of focal adhesions per cell (e) and average aspect ratio of focal adhesions per cell (f). Values are mean ± SEM from n=27 0.7kPa, n=30 7kPa, n=30 38kPa and n=31 glass, cells from 3 independent experiments. Statistical significance assessed by two-tailed paired t-test on median values per experiment.
(g) Top; Immunofluorescence of KPC cells from (a) showing YAP (green) and nuclei (blue). Scale bars, 20μm. Bottom; Magnification of the areas indicated by a dashed box. Scale bars, 10μm.
(h) Quantification of (g) showing nuclear to cytosolic YAP ratio. Values are mean ± SD from n=84 0.7kPa, n=76 7kPa, n=74 38kPa and n=136 glass cells from 3 independent experiments. Statistical significance assessed by Kruskal-Wallis test with Dunn’s multiple comparisons test.
(i) Bar graph showing top hits of significantly altered (p<0.05) metabolites derived from untargeted global metabolome analysis plotted by their 0.7kPa/glass expression ratio (grey; enriched on 0.7 kPa, red; enriched on glass). Dotted line at x=1 indicates no difference.
(j) AMP, ADP and ATP nucleotide levels of KPC cells cultured as indicated. Values are mean ± SD from 3 biological replicates within the same day and representative of 3 independent experiments. Statistical significance assessed by two-tailed unpaired t-test with Welch’s correction.
(k-l) AMP to ATP (k) and ADP to ATP (l) ratios of KPC cells cultured as indicated. Values are mean ± SD from 3 biological replicates within the same day and representative from 3 independent experiments. Statistical significance assessed by one-way ANOVA.
YAP is a major transcriptional regulator of ECM mechanosensing 12 and is linked with tumorigenesis 30. We found that YAP accumulated in the nucleus with increasing stiffness in both mouse KPC (Figure 1g, h) and human PANC-1 cells (Extended Data 1h, i). In summary, pancreatic cancer cells sense matrix stiffness and respond by enhancing YAP nuclear localisation in addition to cytoskeletal and shape changes.
Matrix mechanics cause metabolic reprogramming in pancreatic cancer cells
We next asked how the changes of cytoskeletal organisation might also be accompanied by changes in the metabolic state of these cells. KRas mutations can drive metabolic plasticity 31 but the effect of mechanical stiffness of the microenvironment on this is unknown. Unbiased, untargeted global metabolome analysis of KPC cells in response to varying stiffness revealed major changes in metabolic reprogramming including arginine and creatinine enrichment on soft, 0.7 kPa environments (Figure 1i grey bars and Extended Data 1j), whereas proline and creatine were enriched on stiff substrata (Figure 1i red bars and Extended Data 1j). The Arginine and Proline KEGG metabolic pathways emerged from analysis as main metabolic cues tuned by ECM mechanics (Extended Data 1k, l). Since many of these metabolites can be key for ATP recycling, we further investigated the energetic states of these cells in response to mechanosensing. Indeed, cells sensing a stiff environment exhibited increased ADP and ATP levels (Figure 1j) as well as reduced AMP/ATP ratios, indicating enhanced ATP turnover rates (Figure 1k-l), changes also observed in human PANC-1 cells (Extended Data 1m, n). Thus, mechanosensing might affect the way pancreatic cancer cells manage their energy demands.
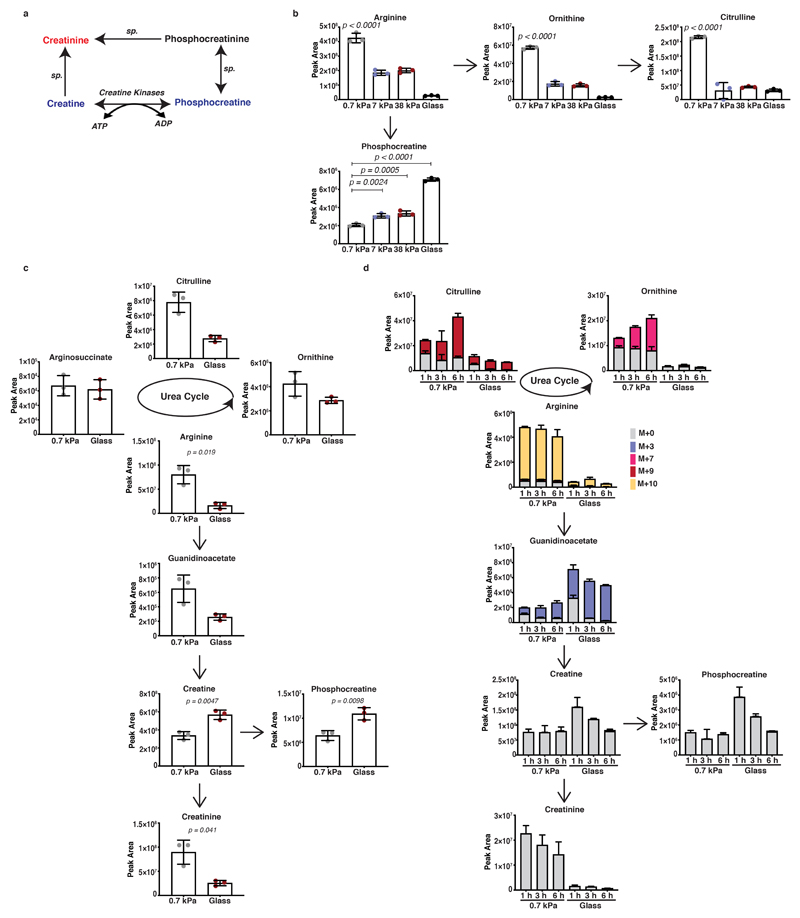
Matrix mechanics tune pancreatic cancer cell arginine metabolism and creatine biosynthesis
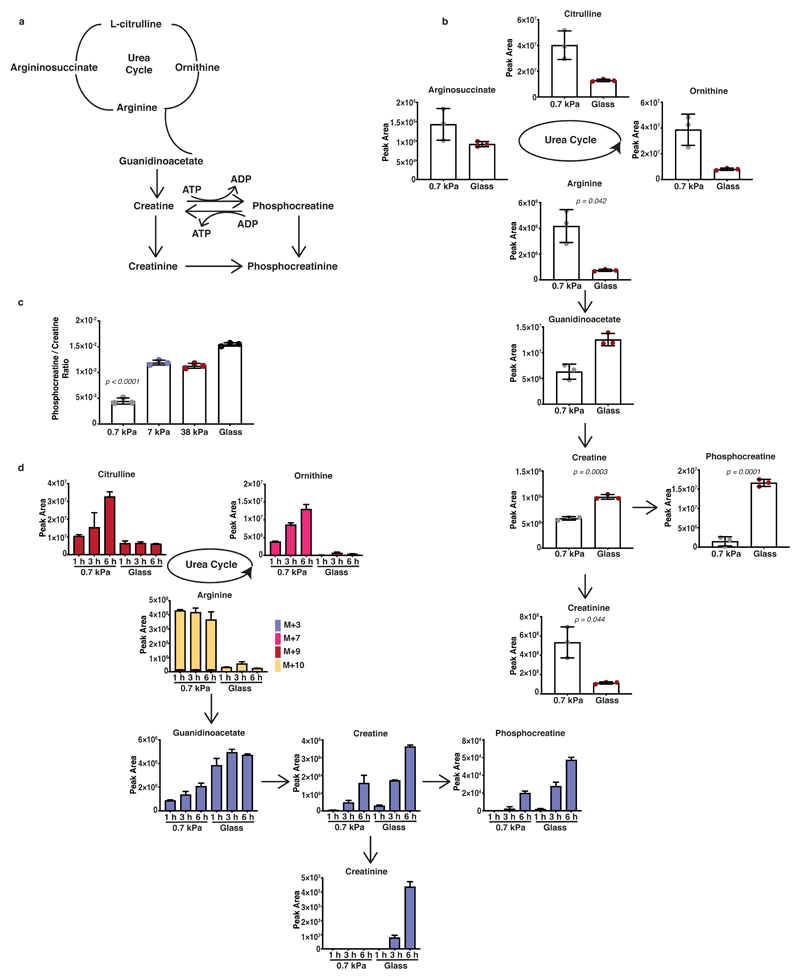
Since arginine metabolism through creatine synthesis could regulate ATP recycling (Figure 2a and Extended Data 2a), which provides energy for actin and myosin dynamics and thus cell motility, we further investigated the urea cycle and creatine biosynthetic pathways in response to mechanosensing. Targeted analysis of the intermediates of these pathways initially confirmed the observed reciprocal changes in creatinine (enriched on soft matrix) and creatine, phosphocreatine levels (enriched on stiff matrix) (Figure 2b and Extended Data 2b). Certainly, metabolite levels suggest that stiff environments favour increased entry of urea cycle metabolites into creatine synthesis and phosphorylation both in KPC (Figure 2b and Extended Data 2b) and human PANC-1 cells (Extended Data 2c). This response increased proportionally to ECM stiffness, with enhanced phosphocreatine/creatine ratio at 7 and 38 kPa, stiffness values relevant to tumour pathology 32 (Figure 2c). Our results so-far indicate that ECM stiffness could engage arginine metabolism towards creatine biosynthesis and that loss of this engagement upon soft, 0.7 kPa matrix, drives urea cycle activity. Carbon-13 (13C), nitrogen-15 (15N) tracer analysis of labelled carbon and nitrogen from arginine to urea cycle and creatine biosynthesis metabolites revealed increased labelling of urea cycle intermediates on softer substrata, whereas stiff environments displayed enhanced labelling of creatine pathway metabolites indicating a switch of arginine flux by ECM mechanics (Figure 2d and Extended Data 2d). Therefore, our data collectively suggest that stiffer matrix drives higher levels of ATP turnover and increased the activity of the phosphocreatine reaction, indicating engagement of an ATP recycling mechanism via the pCr-CK system. Cells shifted their metabolism in response to matrix stiffness, shunting L-arginine towards the creatine biosynthetic pathway, perhaps to meet the greater requirements for ATP recycling of stiff environments.
Figure 2. ECM stiffness directs creatine metabolism in PDAC cells.
(a) Schematic representation of the urea cycle, creatine biosynthesis and phosphocreatine-dependent ATP shuttle pathways.
(b) Urea cycle and creatine biosynthesis metabolic intermediates of KPC cells cultured on fibronectin-coated 0.7 kPa hydrogels and glass coverslips as indicated. Values are mean ± SD from 3 biological replicates within the same day and representative from 3 independent experiments. Statistical significance assessed by two-tailed unpaired t-test with Welch’s correction.
(c) Phosphocreatine to creatine ratio of KPC cells cultured on fibronectin-coated 0.7-38 kPa hydrogels and glass coverslips cultured as indicated. Values are mean ± SD from 3 biological replicates within the same day. Statistical significance assessed by one-way ANOVA.
(d) Bar plots showing labelled fraction of arginine-derived labelled carbon and nitrogen incorporation in urea cycle and creatine biosynthesis metabolites of KPC cells cultured as indicated and supplemented with L-arginine-13C615N4 for 1, 3 and 6 hours. Values are mean ±SD from 3 biological replicates within the same day.
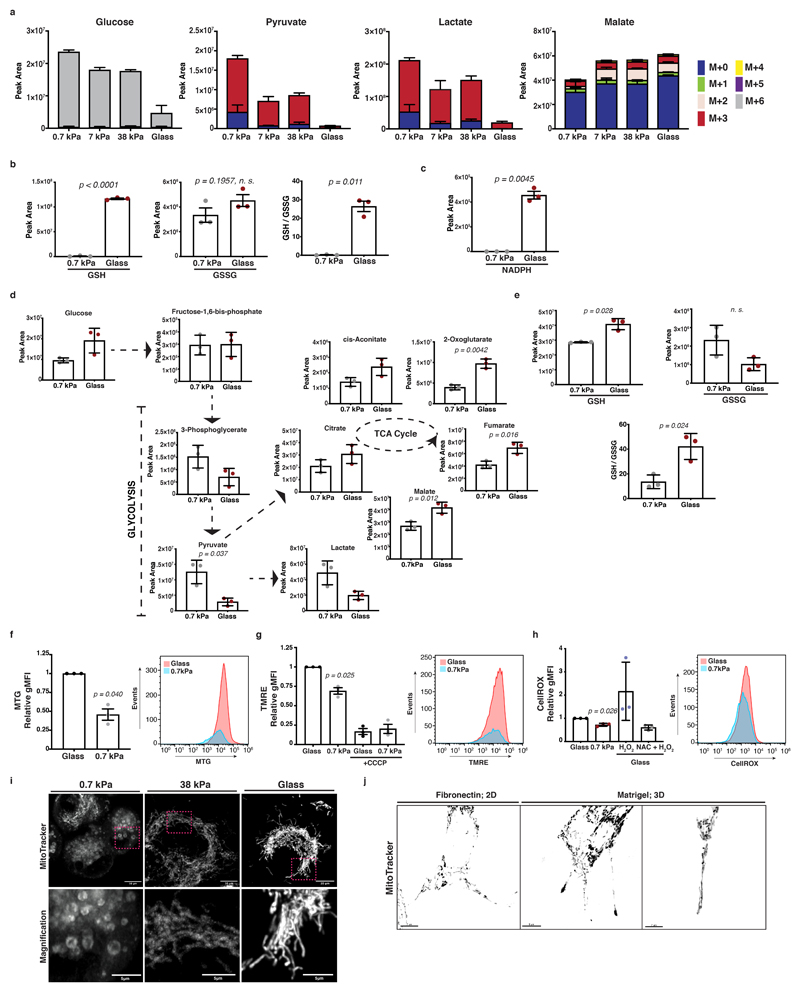
Matrix mechanics affect mitochondrial respiratory activity in pancreatic cancer cells
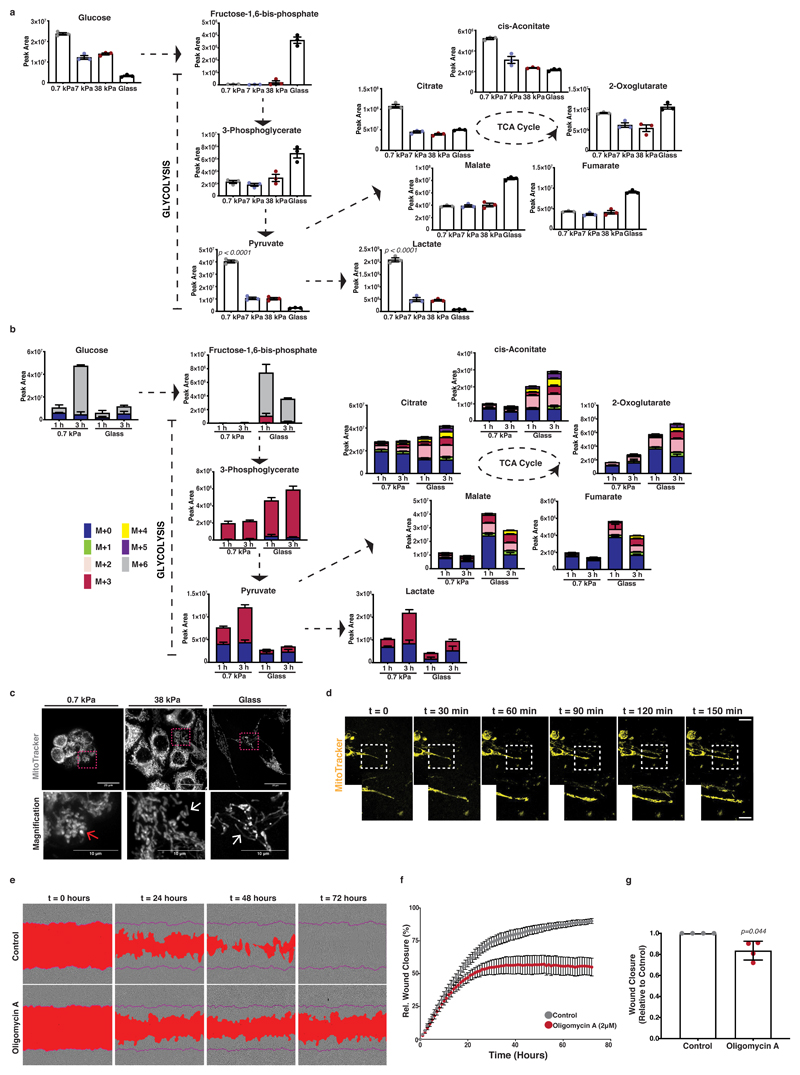
To further probe cellular bioenergetics in response to mechanosensing, we measured steady state levels of two major energy generating catabolic pathways, glycolysis and the TCA cycle. Oncogenic mutations confer a tendency toward aerobic glycolysis, as a means of maintaining metabolic plasticity in challenging environments 33. We observed accumulation of lactate and pyruvate in cells on soft matrix (Figure 3a), indicating higher levels of aerobic glycolysis. To analyse glucose flux, we performed a carbon-13 (13C) tracer analysis of metabolic intermediates of the glycolysis and TCA cycle pathways. This revealed increased glucose-derived labelling in pyruvate and lactate on softer substrates and increased TCA cycle labelling on stiffer substrates (Figure 3b and Extended Data 3a). To test whether stiff substrata favour oxidative phosphorylation activity, associated with increased reactive oxygen species (ROS), we assessed the redox state of our cells. Indeed, intermediates of the cellular antioxidant response (GSH, GSSG, GSH / GSSG ratio) (Extended Data 3b) and NADPH (Extended Data 3c) increased in stiff environments. Comparable differences in these pathways were also observed in human PANC-1 cells (Extended Data 3d, e). One possible explanation for this metabolic shift would be that mechanosensing affects mitochondrial integrity and homeostasis. Fluorescent labelling of mitochondria revealed more elongated and fused morphologies with increasing ECM rigidity in KPC cells (Figure 3c, Supplementary Video 1) that was accompanied by increased mitochondrial mass (Extended Data 3f), physiological mitochondrial membrane potential (Extended Data 3g) and ROS generation (Extended Data 3h) as measured using FACS analysis. Human PANC-1 cells also showed similar responses (Extended Data 3i). Thus, ECM mechanics could favour mitochondrial homeostasis and support total ATP production through oxidative phosphorylation activity. Since ECM stiffness is critical for the invasive transformation of PDAC cells, we assessed TCA cycle dependent ATP generation. By scratching a PDAC cell monolayer and embedding it in Matrigel matrix, we observed a remarkable enrichment of fused mitochondria along the pseudopods of ECM invading cells (Figure 3d and Extended Data 3j). When we treated cells with the mitochondrial ATP synthase and oxidative phosphorylation inhibitor, oligomycin A 34, we saw reduced cancer cell invasion (Figure 3e-g), indicating a functional importance of mitochondrial ATP production in invasive migration.
Figure 3. Mitochondrial dynamics and respiratory activity are induced by ECM mechanics in pancreatic cancer cells and support invasive behaviour.
In panels a-c, mouse KPC cells were cultured atop of 0.7-38kPa fibronectin-coated hydrogels and glass coverslips.
(a) Steady-state levels of glycolysis and TCA cycle intermediates of KPC cells cultured as indicated. Values are mean ± SD from 3 biological replicates within the same day. Statistical significance assessed by one-way ANOVA.
(b) Glucose-derived labelled carbon incorporation in glycolysis and TCA cycle intermediates of KPC cells cultured as indicated and supplemented with U-13C6-glucose for 1 and 3 hours. Values are mean ± SD from 3 biological replicates within the same day and representative from 3 independent experiments.
(c) Top; Maximum intensity projections of z-stack acquisitions of live KPC cells cultured as indicated showing labelled mitochondria (grey; MitoTracker). Scale bars, 20μm. Bottom; Magnification of the areas indicated by a dashed box. Scale bars, 10μm. White arrows highlight elongated and red arrows rounded mitochondria. Pictures representative of 3 independent experiments.
(d) Top; Representative pictures of KPC Matrigel-invading cells showing mitochondria labelling (yellow; Mitotracker) over 150 min. Scale bar, 20μm. Bottom right; Magnification of the areas indicated by a dashed box. Pictures representative of 3 independent experiments.
(e) Representative pictures of control or treated with oligomycin-A (2μM) PDAC-A cells invading 3D ECM. A mask over the wound area is annotated with red and a purple line indicates the initial wound area. Scale bars, 100μm. Pictures representative of 4 independent experiments.
(f) Wound closure over time from (e). Values are mean ± SD from 4 independent experiments. (g) Relative wound closure of (e) normalised at t1/2 wound closure of control. Values are mean ± SD from 4 independent experiments. Statistical significance assessed by two-tailed one-sample t-test on LN transformed values.
Collectively these data suggest that even in cells with identical genetic backgrounds (e.g. here KRasG12D; p53R172H), stiff ECM mechanics can induce specific alterations in pancreatic cancer cell metabolism, favouring ATP production through oxidative phosphorylation activity, which supports ECM invasion. However, since ATP is not actively stored in cells and displays high diffusion rates, we propose that ATP recycling through creatine biosynthesis and the phosphocreatine reaction is also crucial to mechanistically couple ECM mechanics with efficient ATP recycling during demanding activities like cancer invasion.
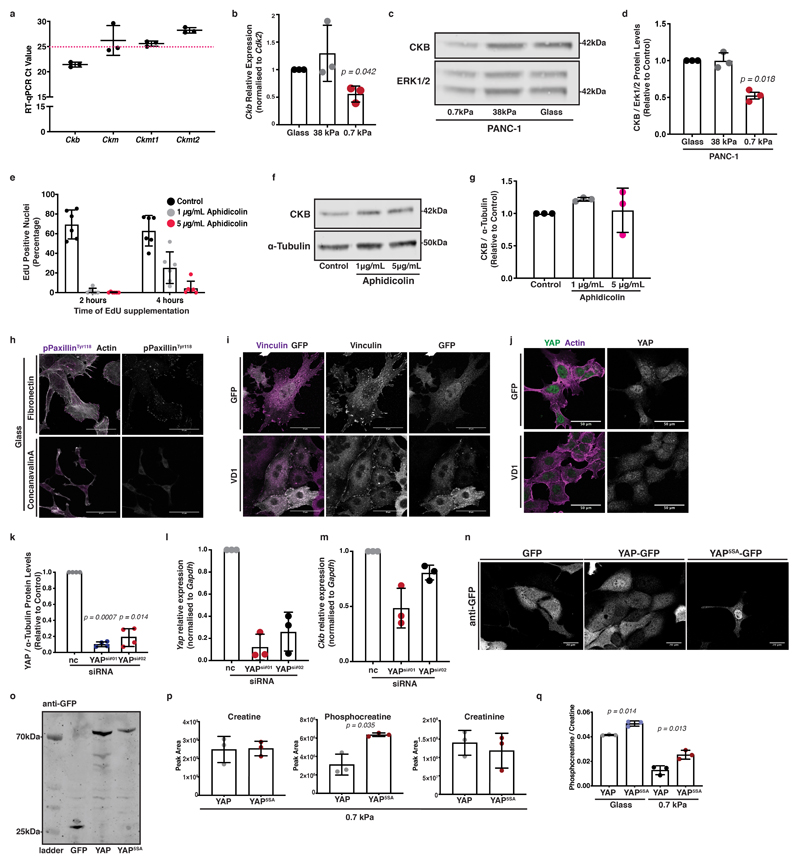
Creatine kinase B (CKB) is regulated by substrate adhesion and YAP nuclear localisation
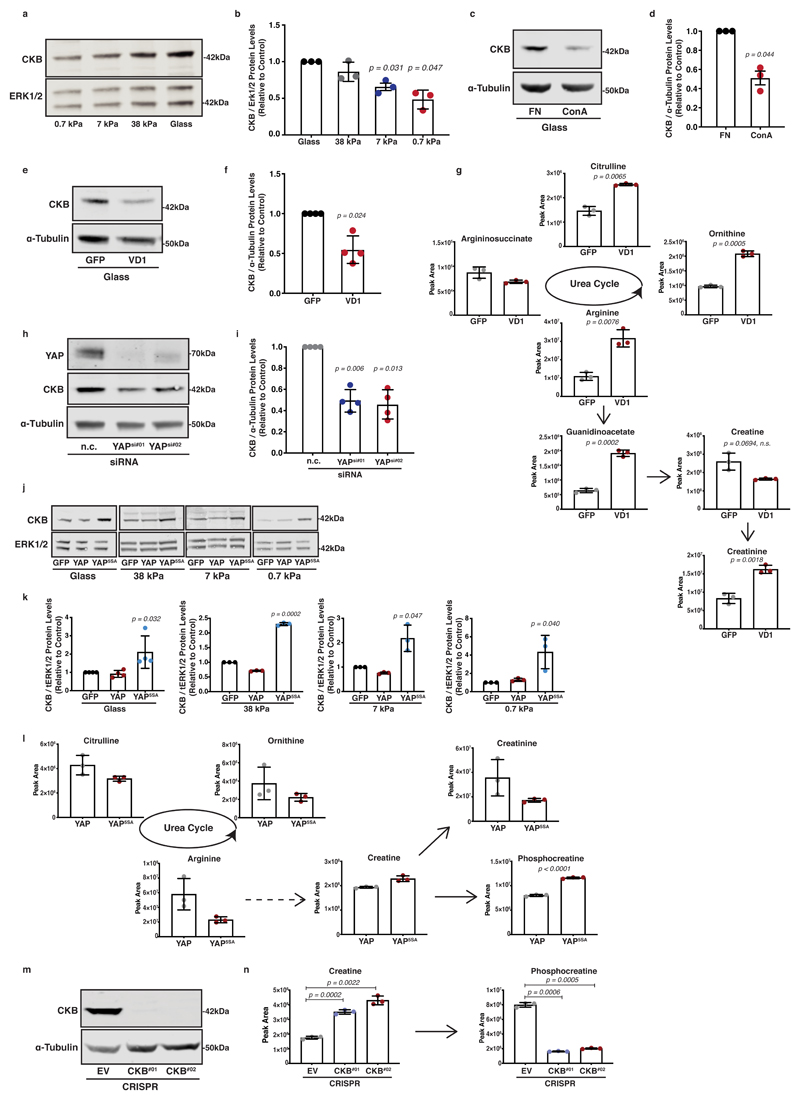
We next sought to understand whether ECM mechanics could be directly coupled to creatine biosynthesis and the phosphocreatine reaction. We hypothesised that loss of creatine phosphorylation on soft ECM could be the cause of phosphocreatine depletion and subsequent creatinine accumulation. We screened our cells for expression of creatine kinases, enzymes that catalyse the reversible phosphorylation of creatine that is coupled to ATP/ADP exchange. Among them, we detected significant expression of creatine kinase-b (Ckb) (Extended Data 4a). Interestingly, CKB protein and mRNA expression levels were elevated by increased matrix stiffness in both murine (Figure 4a-b and Extended Data 4b) and human PDAC cells (Extended Data 4c, d), but were not affected by aphidicolin, an inhibitor of proliferation (Extended Data 4e-g). Thus, CKB expression appears to be mechanosensitive, suggesting a possible mechanism by which ATP recycling is mechanoresponsive.
Figure 4. The phosphocreatine circuit depends on CKB in pancreatic cancer cells which is regulated by mechanosensing and YAP activity.
(a) KPC cells were cultured on 0.7-38kPa hydrogels and ECM-coated glass coverslips and immunoblotted for CKB and ERK1/2 (loading control). Pictures representative from 3 independent experiments.
(b) Densitometric quantification of (a). Values are mean ± SD and relative to control from 3 independent experiments.
(c) KPC cells were cultured on fibronectin (control) or concanavalin A (ConA) coated plates and immunoblotted for CKB and α-Tubulin (loading control). Pictures representative from 3 independent experiments.
(d) Densitometric quantification of (c). Values are mean ± SD and relative to control from 3 independent experiments.
(e) KPC cells expressing either GFP or VD1-GFP were immunoblotted for CKB and α-Tubulin (loading control). Pictures representative from 4 independent experiments.
(f) Densitometric quantification of (e). Values are mean ± SD from 4 independent experiments.
(g) Steady state levels of creatine biosynthesis intermediates of KPC cells from (e). Values are mean ± SD from 3 biological replicates within the same day. Statistical significance assessed by two-tailed Welch’s t-test.
(h) Control (nc) or YAP silenced (YAPsi#01, YAPsi#02) KPC cells were immunoblotted for YAP, CKB and α-Tubulin (loading control). Pictures representative from 4 independent experiments.
(i) Densitometric quantification of CKB expression from (h). Values are mean ± SD and relative to control from 4 independent experiments.
(j) KPC cells expressing GFP, GFP-YAP or GFP-YAP5SA were cultured as indicated and immunoblotted for CKB and ERK1/2 (loading control). Pictures representative from 3 independent experiments.
(k) Densitometric quantification of (j). Values are mean ± SD and relative to control from 4 (glass) and 3 independent experiments (0.7-38kPa).
(l) Steady state levels of creatine biosynthesis intermediates of KPC cells expressing either GFP-YAP or GFP-YAP5SA. Values are mean ± SD from 3 biological replicates within the same day.
(m) Control (EV) or CKB-depleted KPC cells (CKB #01, #02) were immunoblotted for CKB and α-Tubulin (loading control). Pictures representative of 3 independent experiments with similar results.
(n) Creatine and phosphocreatine levels of cells from (m). Values are mean ± SD from 3 biological replicates within the same day. Statistical significance assessed by two-tailed Welch’s t-test.
In b,d,f,i,k: statistical significance assessed by two-tailed one-sample t-test on LN transformed values.
Since mechanosensing depends on cell-ECM adhesion, we asked whether integrin-mediated adhesion could influence CKB expression. Concanavalin A (ConA) is a lectin that promotes adhesion in an integrin-independent way 35, so we used it to prevent the formation of focal complexes, while still permitting adhesion of cells to glass (Extended Data 4h). Surprisingly, cells downregulated CKB expression when plated on ConA-coated surfaces (Figure 4c, d), indicating a dependence of CKB expression specifically on integrin clustering and engagement. To further confirm the importance of focal adhesion formation in regulating CKB expression, we targeted vinculin with a dominant-negative small vinculin fragment, VD1, which blocks transmission of mechanical forces from integrins to actin cytoskeleton and therefore reduces nuclear YAP levels 26 (Extended Data 4i, j). Expression of this fragment reduced CKB protein levels (Figure 4e, f), indicating a tight connection between focal adhesion mechanosensing and CKB expression. The VD1 fragment also drove a reciprocal decrease of creatine and an increase of creatinine and urea cycle metabolite levels, recapitulating the creatine biosynthesis phenotype observed on soft substrates (Figure 4g).
As YAP is a major regulator of mechanosensitive transcription, we used siRNA-mediated silencing of YAP and found decreased CKB protein and mRNA expression levels (Figure 4h, i and Extended Data 4k-m). We also employed YAP5SA, a mutant where YAP nuclear localisation was enforced 36 (Extended Data 4n, o), which drove elevated CKB expression in all range of stiffnesses tested (Figure 4j, k), providing strong support for the idea that CKB is a YAP target in pancreatic cancer cells. Metabolomic analysis comparing KPC cells overexpressing YAP against those overexpressing YAP5SA, revealed phosphocreatine enrichment on both stiff (Figure 4l) and soft substrata (Extended Data 4p) increasing the phosphocreatine to creatine ratio (Extended Data 4q). Thus YAP drives CKB expression and engagement of the phosphocreatine reaction in PDAC cells, driving energy availability in a mechanosensitive way.
In line with this, CRISPR/Cas9 mediated depletion of CKB was sufficient to diminish phosphocreatine levels in pancreatic cancer cells (Figure 4m, n) suggesting that this response is specifically mediated through CKB in PDAC cells. Thus, PDAC cells appear not only to synthesise creatine with increasing stiffness, but also to carry the necessary machinery to engage the creatine/phosphocreatine reaction, a process influenced by integrin clustering, focal adhesion formation and YAP activity. Hence, the creatine phosphagen system is mechanoresponsive in PDAC cells.
The phosphagen system and CKB facilitates directed migration of pancreatic cancer cells
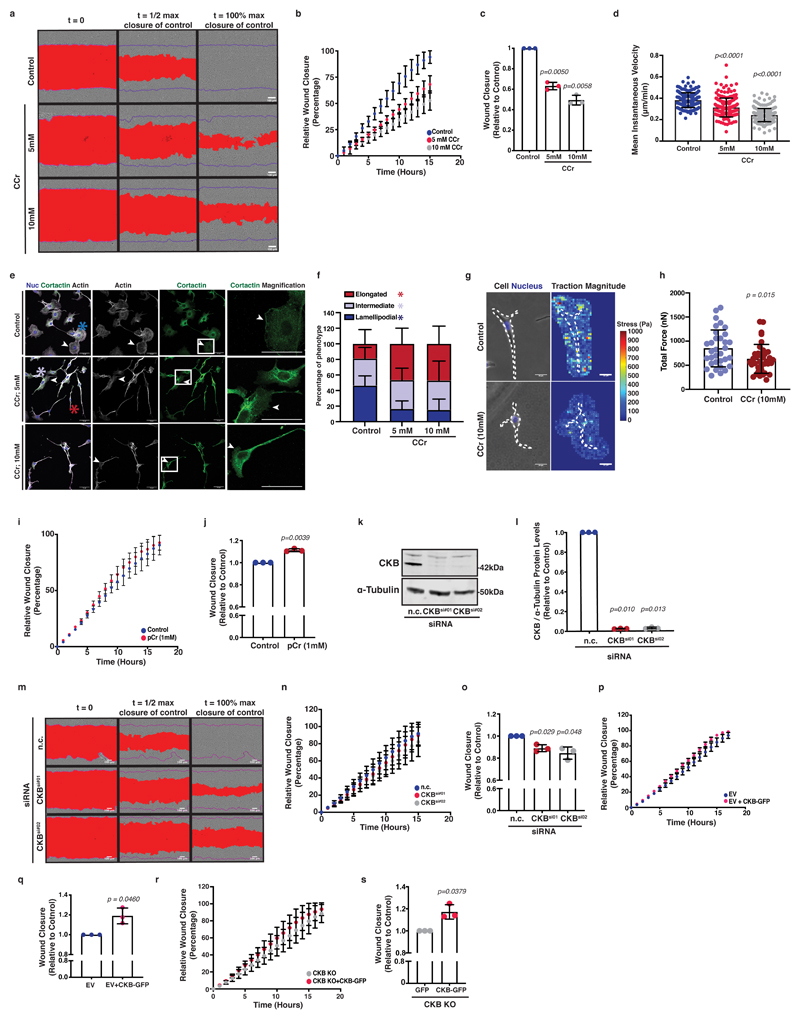
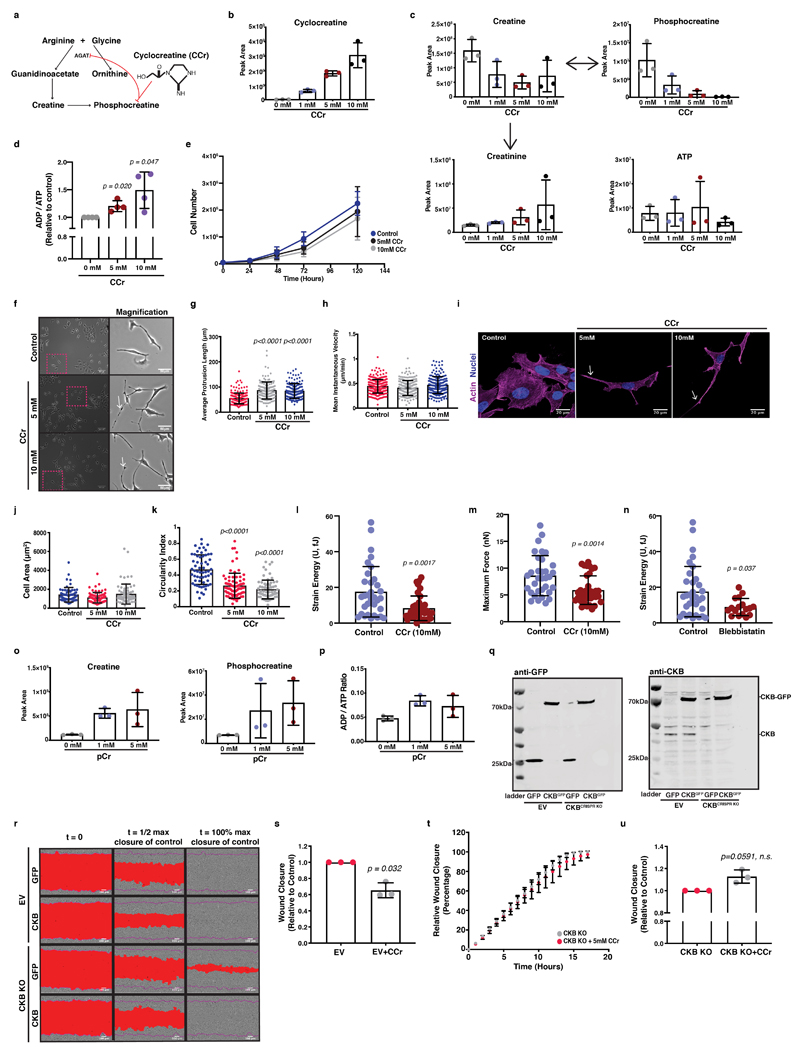
We reasoned that CKB activity and subsequently the pCr-CK system could metabolically support the migratory and invasive properties of cancer cells. The creatine phosphocircuit can be targeted using a creatine analogue, cyclocreatine (CCr; 1-carboxymethyl-2-iminoimidazolidine), which is a poor phosphate donor 22 (Extended Data 5a). Increasing concentrations of cyclocreatine diminished phosphocreatine levels and shifted the ADP/ATP ratios, with only a slight decrease in growth capacity after 72 hours (Extended Data 5b-e). Interestingly, cell migration speed and colonisation of a wounded monolayer were reduced in response to cyclocreatine, indicating a potential role in cell motility (Figure 5a-d; Supplementary Video 2). Cells at low confluence and supplemented with cyclocreatine, displayed formation of elongated, thin protrusions without affecting cell speed (Extended Data 5f-h; Supplementary Video 3); In line with this, circularity decreased, but not total cell area (Extended Data 5i-k). Cyclocreatine treatment of KPC cells cultured atop of collagen-I coated coverslips appeared elongated and displayed diminished lamellipodia (Figure 5e, f) suggesting a supportive role of the creatine phosphagen system toward actin and lamellipodia dynamics. We also hypothesised that this energy system could support actomyosin contractility and force generation, which we queried using traction force microscopy of KPC cells cultured atop fibronectin-coated stiff, 38 kPa, hydrogels. Cyclocreatine inhibited force generation, along with an overall deformation (strain) disruption (Figure 5g, h and Extended Data 5l-n). Thus, we implicate the pCr-CK system as a potential energetic linker between ECM mechanosensing and cellular mechanoresponses, with important implications for invasive migration.
Figure 5. Creatine homeostasis facilitates collective migration of pancreatic cancer cells.
(a) Pictures of control or CCr treated cells populating a wounded monolayer. Scale bars, 100μm.
(b-c) Wound closure over time (b) and relative closure at t1/2 of control from (a). Values are mean ± SD from 3 independent experiments.
(d) Cell velocity from (a). Values are mean ± SD from n=185 control, n=194 5mM and n=207 10mM CCr treated cells. Statistical significance assessed by Kruskal-Wallis with Dunn’s multiple comparisons test.
(e) Immunofluorescence of control or CCr treated cells, showing nuclei (blue), actin (grey) and cortactin (green). Arrows: lamellipodia. Asterisks: phenotype class; elongated (red), intermediate (grey) and lamellipodial (blue). Scale bars, 50μm.
(f) Percentage of phenotype from (e). Values are mean ± SD from 3 independent experiments. Statistical significance assessed by 2-way ANOVA; lamellipodial: control vs 5mM, p=0.0003; control vs 10mM, p<0.0001; elongated: control vs 5mM p=0.0011; control vs 10mM p=0.0006.
(g) Left; pictures of control or CCr treated cells showing cell body (grey) and nuclei (blue). Right; Traction fields of left panel. Scale bars, 20μm.
(h) Strain energy of (g). Values are mean ±SD from n=33 control and n=36 CCr treated cells from 3 independent experiments. Statistical significance assessed by two-tailed Mann-Whitney U test.
(i-j) Wound closure over time (i) and relative closure at t1/2 of control (j) from control or P-Cr treated cells populating a wounded monolayer. Values are mean ± SD from 3 independent experiments.
(k) Control (n.c.) or CKB silenced (CKBsi#01, CKBsi#02) cells were immunoblotted for CKB and α-Tubulin (loading control).
(l) Densitometric quantification of (k). Values are mean ± SD and relative to control from 3 independent experiments.
(m) Pictures of cells from (k) populating a wounded monolayer. Scale bars, 100μm.
(n-o) Wound closure over time (n) and relative closure at t1/2 of control (o) from (m). Values are mean ± SD from 3 independent experiments.
(p-s) Wound closure over time (p, r) and relative closure at t1/2 of control (q, s) from control (EV) or CKB-KO cells expressing GFP or CKB-GFP, populating a wounded monolayer. Values are mean ± SD from 3 independent experiments.
In c,j,l,o,q,s: statistical significance assessed by two-tailed one-sample t-test on LN transformed values.
Administration of phosphocreatine caused the reverse effect to cyclocreatine, increasing metabolites associated with ATP recycling (Extended Data 5o, p) and modestly, but reproducibly accelerating scratch wound migration (Figure 5i, j). Silencing CKB using siRNA (Figure 5k, l) induced delay of wound closure under the same setting (Figure 5m-o). In summary, creatine phosphorylation facilitates the energy requirements of pancreatic cancer cells for efficient migration. To validate this, we overexpressed a GFP-tagged construct of CKB in empty-vector (EV) control and CRISPR/Cas9-mediated CKB depleted cells (Extended Data 5q), accelerating migration in both wild-type control cells (Figure 5p, q and Extended Data 5r) and CKB-depleted cells (Figure 5r, s and Extended Data 5r). We also supplemented empty-vector (EV) control and CKB depleted cells with cyclocreatine without observing any migration defects in the latter (Extended Data 5s-u). Thus, when pancreatic cancer cells migrate collectively, such as in a wound healing assay, they rely on ATP recycling through the creatine-creatinine system and CKB activity to store and release energy through the reversible phosphocreatine reaction to feed into this pathway.
The phosphocreatine circuit facilitates pancreatic cancer cell ECM invasion and chemotaxis
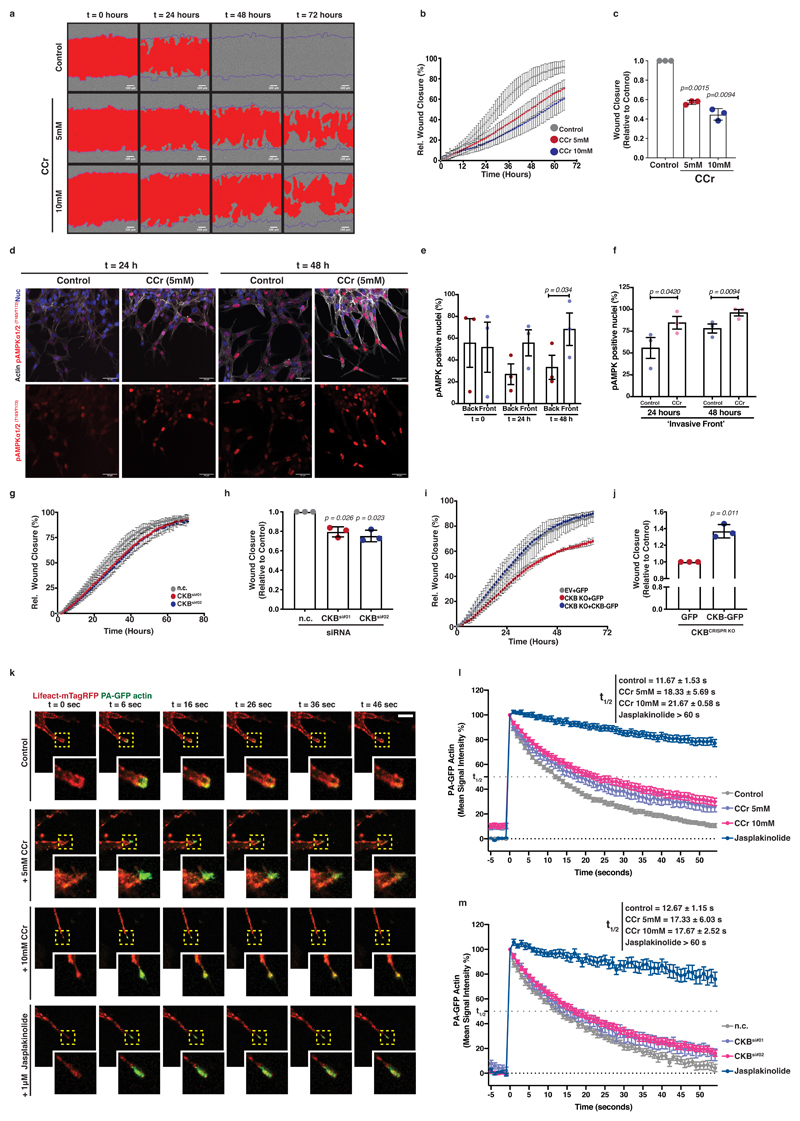
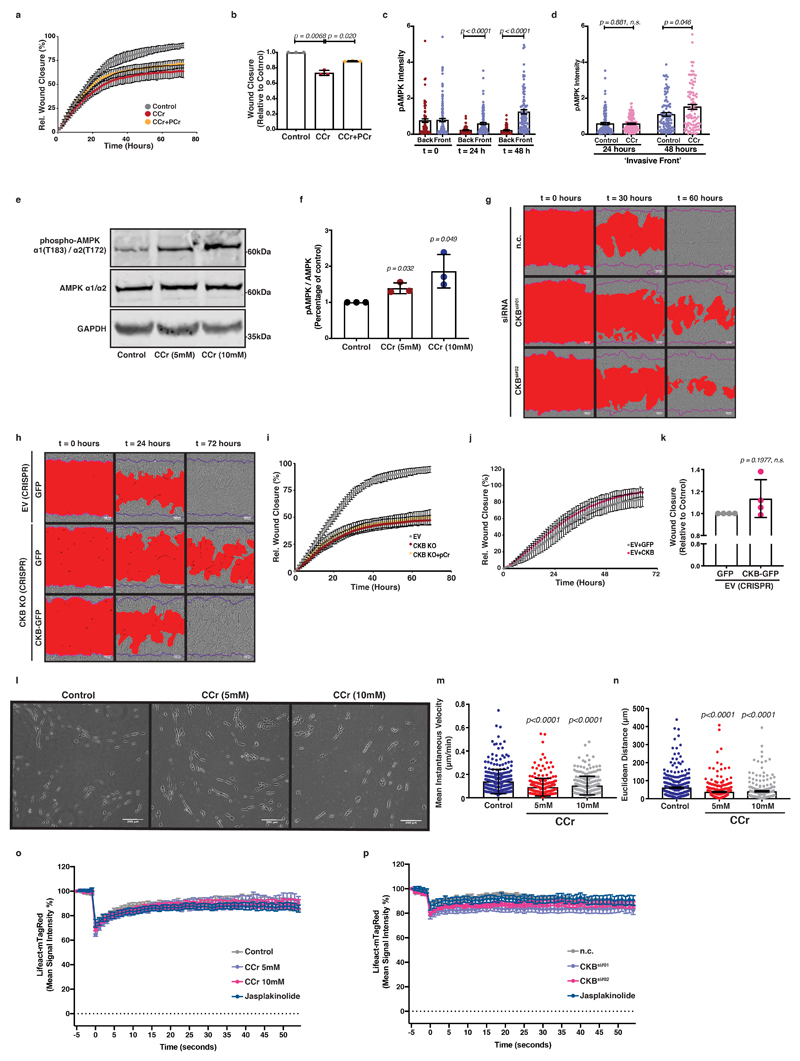
Invasion into ECM can be triggered by stiff substrates and requires dynamic pseudopods as well as directed chemotactic migration 37. Indeed, disruption of the phosphocreatine circuit with cyclocreatine resulted in major impairment of cancer cell invasion within a 3D, Matrigel-embedded, wounded cell monolayer (Figure 6a-c; Supplementary Video 4). Conversely, phosphocreatine supplementation accelerated ECM invasion of cyclocreatine-treated cells (Extended Data 6a, b). To assess whether loss of the pCr-CK system induced energetic stress during collective cell invasion, we visualised phospho-AMPK levels, a known sensor of cellular energetic stress 38, in control or cyclocreatine-treated Matrigel-embedded PDAC cell monolayers (Figure 6d). Interestingly, we observed phospho-AMPK activation in leader cells at invasive fronts following 24 hours of ECM invasion (Figure 6e and Extended Data 6c) that was significantly enhanced in cyclocreatine supplemented cells (Figure 6f and Extended Data 6d). Indeed, cyclocreatine also induced AMPK activation in PDAC cells in culture (Extended Data 6e, f). Thus, impairment of the phosphocreatine circuit opposed collective invasion. We tested the specific requirement for CKB expression during collective PDAC cell invasion. siRNA mediated CKB silencing delayed invasion of pancreatic cancer cells (Figure 6g, h and Extended Data 6g). Furthermore, CKB overexpression in CKB depleted cells accelerated cell invasion under the same setting, reverting the phenotype (Figure 6i, j and Extended Data 6h). Perhaps not surprisingly21, 22, phosphocreatine supplementation failed to rescue the invasion defect caused by CKB deletion (Extended Data 6i), highlighting not only that both the creatine kinase and the phosphate carrier are required for the phosphagen system to operate, but also that CKB is a major kinase mediator of the cytoplasmic phosphocreatine circuit in these PDAC cells. We noticed a modest trend for increased invasion of wild-type (EV) PDAC cells overexpressing GFP-CKB (Extended Data 6j, k) but these cells already have high levels of CKB and are highly invasive. We conclude that CKB expression and creatine phosphorylation promote efficient invasion of pancreatic cancer cells into the surrounding ECM.
Figure 6. Creatine homeostasis supports actin dynamics in invasive pseudopods of pancreatic cancer cells facilitating ECM invasion.
(a) Pictures of control or CCr treated cells invading 3D ECM. Scale bars, 100μm.
(b-c) Wound closure over time (b) and relative closure at t1/2 of control (c) from (a). Values are mean ± SD from 3 independent experiments.
(d) Immunofluorescence of cells from (a) showing actin (grey), nuclei (blue) and pAMPKα1T183/α2T172 (red) at the invasive front. Bottom; pAMPK channel (red) from top. Scale bars, 50 μm.
(e-f) Percentage of pAMPK positive nuclei over total nuclei at the ‘back’ and ‘front’ of control (e) and the ‘front’ of control or CCr treated cells (f) from (d). Values are mean ± SEM from 3 independent experiments. Statistical significance assessed by two-tailed paired t-test.
(g-h) Wound closure over time (g) and relative closure at t1/2 of control (h) from control (n.c.) or CKB silenced (CKBsi#01, CKBsi#02) cells invading 3D ECM. Values are mean ± SD from 3 independent experiments.
(i-j) Wound closure over time (i) and relative closure at t1/2 of control (j) from control (EV) or CKB-KO cells expressing GFP or CKB-GFP, invading 3D ECM. Values are mean ± SD from 3 independent experiments.
(k) Representative pictures showing PA-GFP-Actin (green) and LifeAct-mTagRed (red) in control, CCr or jasplakinolide treated cells photoactivated (t=6s) at the tips (yellow box, magnified) of pseudopods invading fibroblast-derived ECM. Scale bar, 10μm.
(l) Quantification of PA-GFP-Actin signal intensity from (k). Values are mean ±SEM from n=31 control, n=23 5mM CCr, n=35 10mM CCr and n=28 jasplakinolide treated cells from 3 independent experiments. Actin lifetime expressed as the t1/2 of intensity decay. t1/2 values are mean ± SD on average values per experiment.
(m) Quantification of PA-GFP-Actin signal intensity of control (nc) or CKB silenced (CKBsi#01, CKBsi#02) cells photoactivated at the tips of pseudopods invading fibroblast-derived ECM (as on (k)). Values are mean ±SEM from n=36 control (n.c.), n=31 CKBsi#01, n=38 CKBsi#02 and n=21 jasplakinolide treated cells from 3 independent experiments. Actin lifetime expressed as the t1/2 of intensity decay. t1/2 values are mean ± SD on average values per experiment.
In c,h,j: statistical significance assessed by two-tailed one-sample t-test on LN transformed values.
Furthermore, when cyclocreatine treated cells were cultured on 3D fibroblast-derived ECM at a low confluency, they displayed reduced cell speed and decreased directionality (Extended Data 6l-n). We have hypothesised that the CK/CP system could provide the required energy locally to power dynamic cytoskeletal remodelling. To test whether creatine-dependent ATP recycling may be important for actin dynamics within invading pseudopods, we measured actin dynamics using photoactivatible GFP-actin (PA-GFP-actin) at the pseudopod tips during invasion. The signal intensity of PA-GFP-actin was stabilised in pseudopods of CCr treated cells, indicating reduced actin treadmilling in response to disruption of the creatine phosphagen system (Figure 6k, l and Extended Data 6o, Supplementary Video 5). siRNA mediated CKB silencing caused an analogous response in actin dynamics (Figure 6m and Extended Data 6p). Therefore, the pCr-CK system is important for actin dynamics in invasive pseudopods, likely acting through local ATP recycling to maintain a steep gradient of high ATP and power dynamic actin turnover.
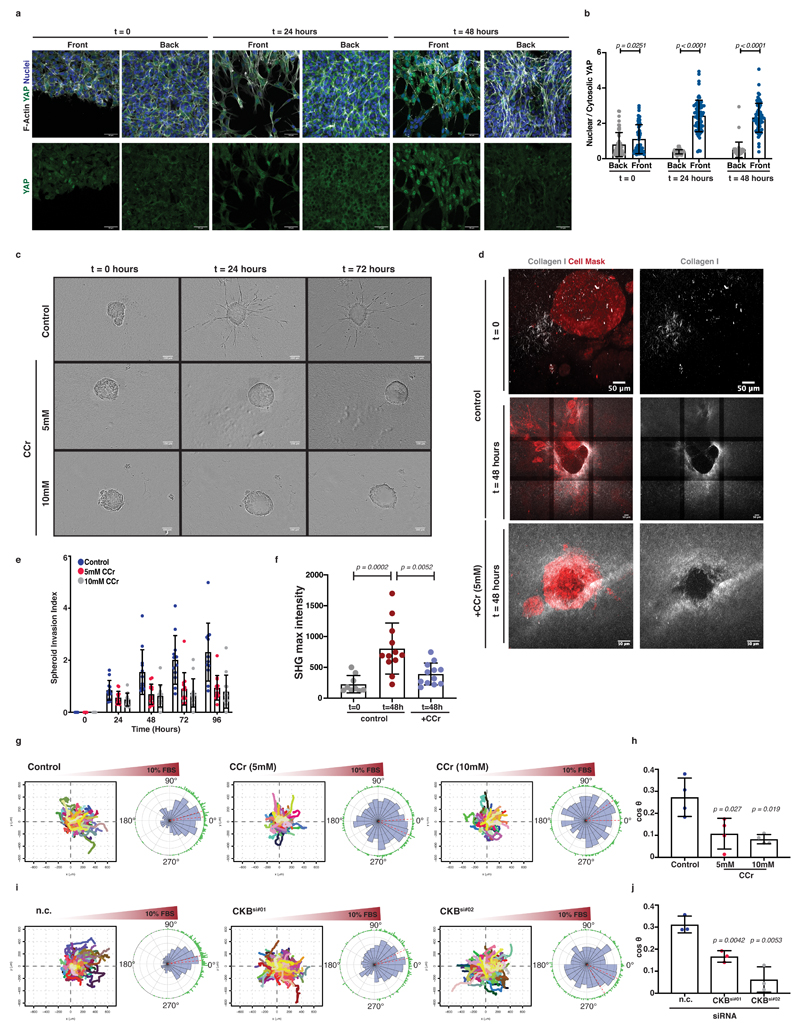
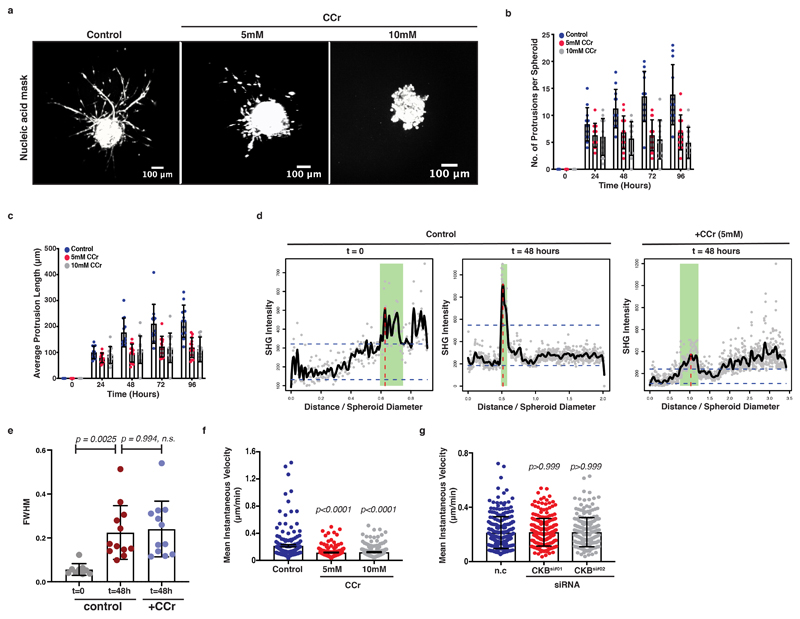
In addition to protrusion and actin dynamics, we proposed a role for the creatine phosphagen system in matrix remodelling and contractile function powering invasion. In agreement with higher tension sensing during invasion, PDAC leader cells at the invasive front displayed increased YAP nuclear localisation. On the contrary, nuclear YAP was remarkably absent from cells at the back of such ECM-embedded monolayers (Figure 7a, b). To test whether tension sensing and matrix remodelling was robustly dependent on the creatine phosphagen system in 3D, spheroids of KPC cells were embedded into a 3D Collagen I- Matrigel matrix, supplemented with CCr and invasion was monitored over a 96-hour period. Ccr effectively inhibited invasion (Figure 7c-e and Extended Data 7a-c; Supplementary Video 6). We also visualised collagen architecture under this setting, using second harmonic generation microscopy (SHG) which revealed that invading spheroids organise and enrich collagen I fibres in a surrounding ring (Figure 7d, f and Extended Data 7d,e). Such collagen accumulation around the spheroids could generate local ECM stiffening triggering their invasive transformation 39–41. Interestingly, CCr treatment was also accompanied by decreased accumulation of collagen I fibres around KPC spheroids (Figure 7d, f and Extended Data 7d, e).
Figure 7. Creatine homeostasis supports collagen remodelling and invasion of 3D PDAC spheroids and the chemotactic response of pancreatic cancer cells.
(a) Top; Immunofluorescence showing YAP (green), F-actin (grey) and nuclei (blue) at the ‘front’ and ‘back’ of ECM invading cells. Bottom; YAP channel (grey) from top. Scale bars, 50μm.
(b) Quantification of nuclear to cytosolic YAP ratio from (a). Values are mean ±SD from n=55 ‘back’, n=63 ‘front’ at t=0; n=51 ‘back’, n=73 ‘front’ at t=24h; n=44 ‘back’, n=69 ‘front’ at t=48h from 3 independent experiments. Statistical significance assessed by two-tailed Mann-Whitney test.
(c) Pictures of control or CCr-treated KPC spheroids invading Collagen I/Matrigel 3D ECM. Scale bars, 100μm.
(d) Left; Images showing cell mask (CellTracker; red) and Collagen I (SHG; grey) from (c). Right; Collagen-I channel (grey) from left. Scale bars 50 μm.
(e) Invasion index over time from (c). Values are mean ±SD from n=12 control, n=13 5mM and n=13 10mM CCr-treated spheroids from 3 independent experiments. Statistical significance assessed by two-tailed paired t-test on mean values per experiment; p-value on control vs 5mM at 48h is 0.0423, at 72h is 0.0066 and at 96h 0.0027. p-value on control vs 10mM at 72h is 0.0254 and at 96h is 0.0390.
(f) Max peak intensity of Collagen I (SHG) from (d). Each dot represents average value (from 6 plot profiles) per spheroid. Values are mean ±SD of n=9 t=0, n=12 t=48h and n=12 CCr-treated spheroids from 3 independent experiments. Statistical significance assessed by one-way ANOVA.
(g) Tracks of cells (spider plots) and circular rose plots of control or CCr-treated cells migrating towards a 10% FBS gradient.
(h) Chemotactic index (cosθ) of cells from (g). Values are mean ±SD from 4 independent experiments. Statistical significance assessed by two-tailed unpaired t-test with Welch’s correction on mean values per experiment.
(i) Tracks of cells (spider plots) and circular rose plots of control (n.c.) or CKB-silenced (CKBsi#01, CKBsi#02) cells migrating towards a 10% FBS gradient.
(j) Chemotactic index (cosθ) of cells from (i). Values are mean ±SD from 3 independent experiments. Statistical significance assessed by two-tailed unpaired t-test with Welch’s correction on mean values per experiment.
Since ECM invasion depends not only on matrix remodelling but also on chemotactic sensing of chemical cues and migrating towards them 42, we asked whether the creatine pathway could energetically support chemotaxis. Indeed, cyclocreatine supplementation (Figure 7g, h and Extended Data 7f; Supplementary Video 7) or CKB silencing (Figure 7i, j and Extended Data 7g) severely impaired the ability of pancreatic cancer cells to migrate towards a 10% serum stimulus in a chemotactic chamber. These results collectively suggest that CKB, through the pCr-CK system dependent ATP recycling, is a component of a major positive feedback loop driving ECM invasion.
CKB is overexpressed during PDAC progression and supports metastatic dissemination
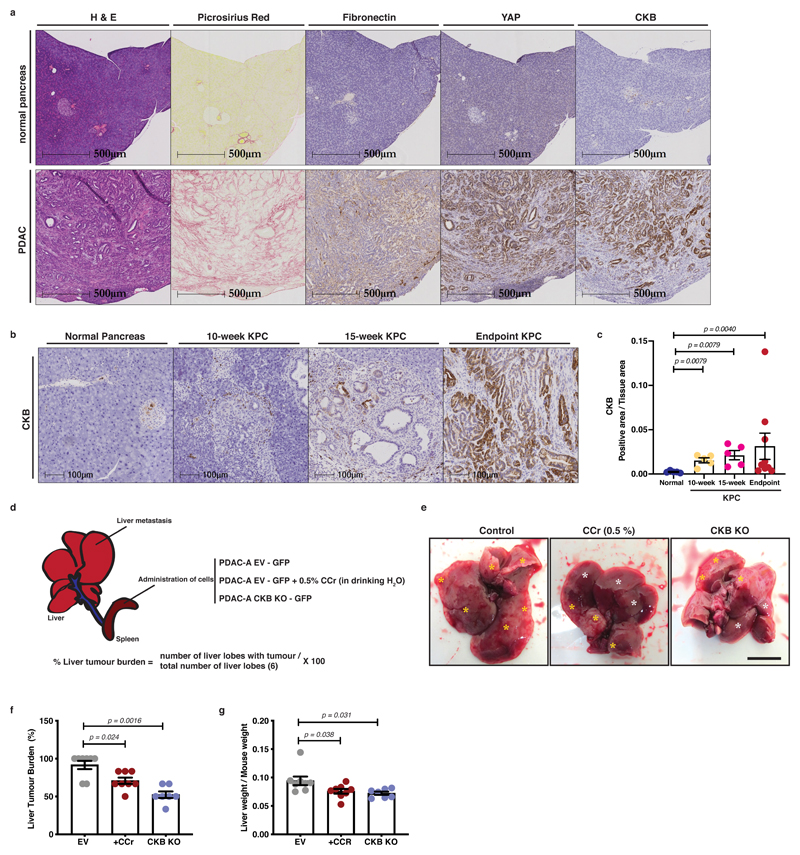
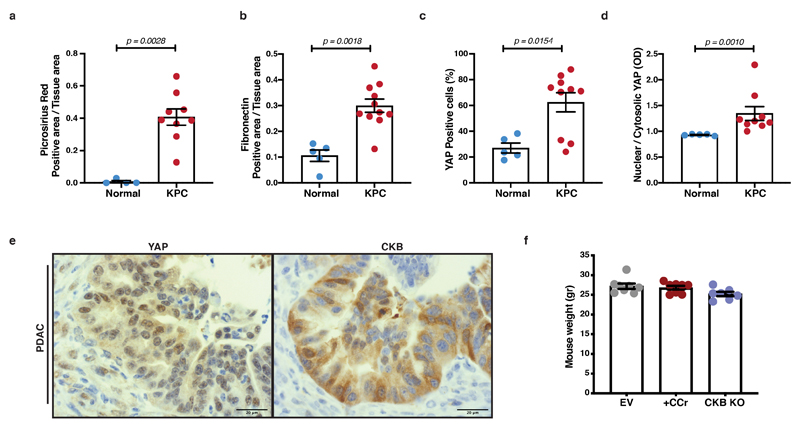
We hypothesised that the stiff matrix-rich stroma of PDAC tumours could increase CKB expression in vivo. We examined pancreata and tumours from the KPC mouse model of PDAC by immunohistochemistry, which revealed an increase of CKB signal in tumours. High levels of CKB also coincided with higher levels of total and nuclear YAP and stromal presence of Collagen I (Picrosirius red) and Fibronectin (Figure 8a and Extended Data 8a-e). Furthermore, while CKB appeared absent from normal ducts, we observed positive staining in pancreatic intraepithelial neoplasia (PanIN) lesions of KPC mice (10- and 15-week stages) indicating a potential role of the creatine phosphagen system in the progression towards invasive ductal adenocarcinomas (Figure 8b, c). It is well documented that human and mouse PDAC tumours show increased desmoplastic stroma and stiffness 32. We therefore propose that YAP signalling, by favouring CKB expression in response to stiff desmoplastic matrix, could facilitate the creatine-dependent energetic support of malignant cells in vivo.
Figure 8. CKB is expressed during PDAC progression and supports metastatic dissemination.
(a) Immunohistochemistry of pancreas from normal mice (Pdx1-Cre+;Kraswt/wt;p53wt/wt) and PDAC from KPC (Pdx1-Cre;LSLKrasG12D;LSLp53R172H) mice, showing H&E, Picrosirius Red, Fibronectin, YAP and CKB staining on serial sections. Representative pictures from 5 normal pancreata and 9 KPC mice. Scale bars, 500μm.
(b) Immunohistochemistry of normal pancreas and pancreas sections from 10-week, 15-week and endpoint PDAC from KPC mice showing CKB staining. Scale bars, 500μm.
(c) Quantification of CKB positive area per tissue area from (b). Values are mean ±SEM from n=5 normal pancreata, n=5 10-week, n=5 15-week and n=9 PDAC pancreata from KPC mice. Statistical significance assessed by two-tailed Mann-Whitney U-test comparing each KPC stage to normal pancreas.
(d) Graphical description of intrasplenic model and quantification strategy. Animals were injected with control (EV) or CKB-depleted (CKB-KO) cells. Control mice 1 week after surgery were randomly assigned to either normal diet or diet containing 0.5% CCr in drinking water. Liver tumour burden is defined as the percentage of liver lobes with tumour over the total number of liver lobes.
(e) Representative pictures of liver at time of dissection. Yellow asterisks indicate tumours, while white asterisks indicate tumour-free areas. Scale bar, 500μm.
(f) Liver tumour burden (%) of animals treated as indicated. Values are mean ±SEM from n=8 control (EV), n=8 EV+CCr and n=7 CKB-KO mice. Statistical significance assessed by two-tailed Mann-Whitney U-test comparing CCr treated to EV and CKB-KO to EV animals.
(g) Liver weight/mouse weight from (e, f). Values are mean ±SEM from n=8 control (EV), n=8 EV+CCr and n=7 CKB-KO mice. Statistical significance assessed by two-tailed Mann-Whitney U-test comparing CCr treated to EV and CKB-KO to EV animals.
To test whether the creatine phospho-circuit could support metastatic dissemination of PDAC cells in vivo, we used an orthotopic intrasplenic model, where tumour cells transit from spleen to seed metastasis in the liver 43, 44. Wild-type (EV) or CKB-depleted KPC cells were transplanted in the spleen of nude mice and after 7 days wild-type (EV) mice were randomly assigned to receive a normal water diet or diet containing cyclocreatine (0.5%) in drinking water. Following 21 days from surgery, mice were sacrificed and metastatic tumour burden in the liver was assessed (Figure 8d). Specifically, wild-type (EV) KPC cells generated metastatic tumours that extensively populated the surface of the liver lobes, while the metastatic burden was decreased with CCr supplementation or CKB-KO and tumours appeared more defined and localised (Figure 8e-g and Extended Data 8f).
These results collectively suggest that CKB, through the creatine phospho-circuit dependent ATP recycling mechanism, is a component of a major positive feedback loop of ECM mechanics, linking migration, mechanosensing and metabolism in pancreatic cancer, being simultaneously positively regulated by ECM force while it facilitates cell invasion and metastatic dissemination.
Discussion
Pancreatic cancer driver mutations, such as activated KRas, confer metabolic plasticity, allowing tumours to perform aerobic glycolysis and to consume alternative energy sources to overcome starvation 9. While the traditional Warburg effect considers tumours to rely on glucose consumption and aerobic glycolysis, tumours actually show considerable metabolic plasticity and the contributions of the microenvironment are increasingly appreciated8. In particular, pancreatic tumours also depend heavily on tricarboxylic acid (TCA) cycle activity, due to glucose starvation 45. We find that stromal stiffness drives the TCA cycle and may contribute to dependence on it, fueling PDAC aggressiveness. Mechanical properties of the ECM shape cell motility and signalling via adhesion and cytoskeletal coupling in normal fibroblasts 46. Additionally, pluripotent stem cells use mechanical cues during differentiation and for regulation of lipid metabolism 47. Furthermore, cancer cells and cancer-associated fibroblasts set up mutually beneficial feedback loops dependent on mechanosensing and YAP signalling, which involve scavenging scarce nutrients in the starved tumour microenvironment 48. However, direct connections between the energy needed for invasive cancer cell migration and mechanotransduction between cancer cells and the ECM are not well understood. We demonstrate here that ECM-derived mechanical cues and integrin-mediated mechanosensing are a major driver of metabolic plasticity for ATP production and recycling that fuels invasive migration of pancreatic cancer cells. Most importantly, we demonstrate that rigid ECM mechanics are an important regulator of arginine metabolism, directing arginine-derived carbon into creatine biosynthesis and away from the urea cycle. Urea cycle dysregulation is linked to tumorigenesis, favouring proliferation, mutagenesis and invasion and is associated with poor prognosis 49. Thus, deciphering the regulators of mechanosensing and arginine metabolism could highlight potential metabolic vulnerabilities of cancer cells.
Migrating cells localise their mitochondria towards leading protrusions, potentially as a mechanism to generate ATP locally to fuel local actin and myosin ATPase requirements 18, 19. Mitochondria positioning is regulated by the GTPase Miro1 in a complex with dynein or kinesins for transport along microtubules or with myo19 for shorter range transport along actin filaments 50. In vivo, localised mitochondrial recruitment to sites of invasion of the C. elegans anchor cell through basement membrane occurs during normal development 20. Anchor cell invasion resembles cancer cell invasion 51, 52 and here we find that pancreatic cancer cells localise their mitochondria into long protrusions when they invade, but also enhance ATP recycling via the creatine phosphagen system to maximise the efficiency of energy flow and to fuel invasion.
“The energy content of ATP is not fixed, but rather is a function of how far the ATP hydrolysis reaction is displaced from equilibrium…” 21. Quoted from Ellington21, highlights that the spatial concentration gradient of ATP in a cellular compartment is a crucial determinant of how much energy the cell can harness from the system. Thus, phosphagen systems, are highly conserved in evolution and facilitate transport of usable energy sources to places of great need as well as buffering ATP for rapid responses to changing energy demands 21. We find the main cytoplasmic creatine kinase, CKB, is upregulated in pancreatic cancer cells in response to stiff matrix, focal adhesion formation and translocation of YAP to the nucleus, implicating CKB as a YAP transcriptional target. CKB regulates the reversible phosphorylation of creatine to phosphocreatine which in turn transfers the phosphate to ADP to regenerate ATP. While CKB can facilitate actin and myosin-based migration in vitro of normal cells 23, we propose that CKB fuels the dynamic protrusion and migration of cancer cells that assume a migratory phenotype in association with rigid highly adhesive matrix.
Our results demonstrate that the phosphocreatine energy shuttle can metabolically support pancreatic cancer cell migration and invasion. Indeed, either by genetic manipulation of CKB or the chemical inhibitor CCr, pancreatic cancer cells showed defects in invasive migration. Specifically, CCr addition slowed actin turnover in invasive pseudopods, as well as impairing traction force generation, suggesting a direct connection of the phosphocreatine / creatine kinase system to actin and myosin dynamics in PDAC cells. Targeting the pCr-CK pathway also prevented chemotactic steering, a process requiring dynamic actin and myosin-dependent pseudopods and driving metastatic dissemination 53. More detailed studies may be needed to resolve whether CKB is important for pseudopod generation, splitting or stabilisation during chemotaxis. Nevertheless, our results propose that there is a mechano-dependent loop where a stiff ECM favours cellular invasion by controlling the regulation of the creatine phosphate shuttle and cytoplasmic ATP recycling.
While PCr administration could partially oppose the effects of CCr in vitro, PCr was unable to rescue CKB knockout in our hands. This is not surprising, since there is a need for both creatine phosphate and the creatine kinase for the phosphagen system to work 21, 22. This could also suggest an important non-redundant role for CKB in regeneration of ATP from PCr. Furthermore, CCr oral administration in nude mice transplanted intrasplenically with PDAC cells or CKB depletion from transplanted PDAC cells reduced metastatic liver tumour burden. This suggests an important role for the creatine phosphagen system as a metabolic supporter of PDAC cell metastasis.
Phosphocreatine shuttle activity supports the energetics of malignancies through activity of the mitochondrial compartment and CKMT1 24, 25. We propose that the cytoplasmic shuttle is also important to promote local energy gradients driving motility. In summary, our study suggests a mechanodependent regulation of ATP production and recycling that identify CKB as a potentially druggable target regulating ECM invasion and metastatic dissemination. Creatine is used as a nutritional supplement and in sports drinks, and cyclocreatine was highlighted as an inhibitor of mitochondrial CKMT1 to prevent growth of HER2+ breast cancer 25 as well as in acute myeloid leukemia 24. We suggest that the cytoplasmic CKB deserves further attention as a metabolic target for cancer dissemination.
Methods
Molecular cell biology methods and reagents
Primary murine KPC-PDAC and human PANC-1 cells were cultured in DMEM high glucose medium supplemented with FCS (10%), glutamine (2mM), sodium pyruvate (0.11 g x L-1), penicillin (10,000 units mL-1) and streptomycin (10,000 units mL-1). Cell lines were maintained at 37°C, under 5% CO2 and were tested multiple times over the study for mycoplasma contamination. Reagents were used as follows: Human Fibronectin Protein (1918-FN-02M, R&D Systems), Fibronectin bovine plasma (F1141, Sigma-Aldrich), Carbonyl cyanide 3-chlorophenylhydrazone (CCCP; C2759, Sigma-Aldrich), Concanavalin A from Canavalia ensiformis (L7647, Sigma-Aldrich), Aphidicolin from Nigrospora spaherica (A4487, Sigma-Aldrich), Corning Matrigel Basement Membrane Matrix (354234, Corning), Rat tail Collagen I (354249, Corning), Phosphocreatine disodium salt hydrate (P7936, Sigma-Aldrich), 2-Imino-1-imidazolidineacetic acid (Cyclocreatine; 377627, Sigma-Aldrich), 1-Carboxymethyl-2-iminoimidazolidine (Cyclocreatine; 57312, AstaTech), U-13C6-D-Glucose, 99% (CLM-1396-PK, Cambridge Isotope Laboratories / CK Isotopes, UK), Sigmacote (SL2, Sigma-Aldrich), sulfosuccinimidyl 6-(4’-azido-2’-nitrophenylamino)hexanoate (sulfo-SANPAH; 22589, Thermo Fisher Scientific), 3-(Acryloyloxy)propyltrimethoxysilane (L16400, Alfa Aesar), Acrylamide 40% Solution (A4058, Sigma-Aldrich), N,N’-methylene-bis-acrylamide 2% solution (M1533, Sigma-Aldrich), N,N,N’,N’-Tetramethylethylenediamine (TEMED; T9281, Sigma-Aldrich), Ammonium persulfate (A3678, Sigma-Aldrich), N-Acetyl-L-cysteine (A7250, Sigma-Aldrich), Ammonium hydroxide (221228, Sigma-Aldrich), Hydrogen peroxide (30%w/w; 31642, Sigma-Aldrich), tert-Butyl hydroperoxide solution (416665, Sigma-Aldrich), Puromycin dihydrochloride (A1113803, Thermo Fisher Scientific), Lipofectamine RNAiMAX (13778150, Thermo Fisher Scientific), Lipofectamine 2000 (11668019, Thermo Fisher Scientific), Lullaby (FLL73000, OZ Biosciences), Amaxa Cell Line Nucleofector Kit V (VCA-1003, Lonza), Oligomycin-A (75351-5MG, Sigma-Aldrich), L-Arginine:HCL (13C6, 99%; 15N4, 99%) (CNML-539-H-0.1 Cambridge Isotope Laboratories / CK Isotopes, UK), dialysed FBS (Gibco, 26400-044), SILAC DMEM Flex Media no glucose no phenol red (A2493901, Thermo Fisher Scientific), FluoSpheres™ Carboxylate-Modified Microspheres 0.2 μm red fluorescent (580/605) 2% solids (F8810, Thermo Fisher Scientific), blebbistatin (B0560, Sigma-Aldrich), jasplakinolide (2792, TOCRIS Bioscience), CellTracker Red CMTPX Dye (C34552, Thermo Fisher Scientific), Gelatin from cold water fish skin (G7041, Sigma-Aldrich), Saponin (47036, Sigma-Aldrich), NucBlue Live reagent (R37605, Thermo Fisher Scientific), Precision Red Advanced Protein Assay (ADV02-A; Cytoskeleton). For CRISPR/Cas9-mediated genome editing, the following sequences were cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 (gift from Feng Zhang, Addgene #62988) targeting mouse CKB: 5’-AAAGCCGCTCGGCGTGCACT-3’(CKBCRISPR#01) and 5’-ACGAGCATCCGGCCAGTCCC-3’ (CKBCRISPR#02). For siRNA mediated silencing the following sequences were used targeting mouse YAP, 5’- CTGGTCAAAGATACTTCTTAA-3’ (YAPsiRNA#01), 5’-ATGGAGAAGTTTACTACATAA-3’ (YAPsiRNA#02) and mouse CKB, 5’-CTGCCGCTTCCTAACTTATTA-3’ (CKBsiRNA#01) and 5’-AAGCTGTTGCTGACACTGAAA-3’ (CKBsiRNA#02). The following constructs were used, pEFGP-C1 (host lab), pEGFPC1/GgVcl1-258 (VD1, gift from Susan Craig, Addgene #46270), pEGFP-C3-hYAP1 (gift from Marius Sudol, Addgene #17843), LifeAct-mTagRFP (gift from M. Davidson, Addgene #54586), PA-GFP actin (gift from M. Davidson, Addgene #57121). YAP 5SA from a pCMV-flag YAP2 5SA plasmid (gift from Kunliang Guan, Addgene #27371) was introduced to the pEGFP-C1 vector, using PCR/restriction digest-based cloning. Using a pWZL Neo Myr Flag CKB plasmid (gift from William Hahn & Jean Zhao, Addgene #20466), CKB cDNA was isolated from the Myr Flag tag and introduced into pEGFP-C1 vector with PCR/restriction digest-based cloning. Reagents, DNA constructs and siRNA/CRISPR targeting sequences are also described at Supplementary Tables 1, 2 and 3 respectively. Antibodies and their validation are detailed in the Reporting Summary.
Transfection and genetic modulation of cells
Cells are described in the Reporting Summary and were typically transfected in suspension. Specifically 5 μg of DNA were transfected using Amaxa Cell Line Nucleofector kit (Lonza Bioscience) according to manufacturer’s instructions. To generate stable cell lines, stably transfected cells were selected with G-418 (G418S, Formedium) followed by fluorescence activated cell sorting (FACS). For FACS, gating was performed by cell size, live/dead and GFP-positive signal. For transfection of adhered cells, 1 μg of DNA was transfected with Lipofectamine 2000 reagent (Thermo Fisher Scientific) according to manufacturer’s instructions. For siRNA-mediated YAP silencing, YAP targeting oligos (25nM; Qiagen) or control siRNA (25nM; AllStars Negative Control siRNA, Qiagen) were transfected using the Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) according to manufacturer’s instructions. YAP-silenced cells were analysed 48h post-transfection. For siRNA-mediated CKB silencing, CKB targeting oligos (15nM; Qiagen) or control siRNA (15nM; AllStars Negative Control siRNA, Qiagen) were transfected using the Lullaby transfection reagent (OZ Biosciences) according to manufacturer’s instructions. Two-step siRNA delivery with 48h interval was performed and CKB-silenced cells were analysed 24h post-transfection. For CRISPR/Cas9-mediated knockout, single-guide RNA oligos were designed using the MIT CRISPR design tool (http://crispr.mit.edu/). Annealed oligonucleotides were cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0. Cells were transfected with 5μg of selected plasmid (control,empty vector (EV) or containing a guide RNA against CKB) using Amaxa Cell Line Nucleofector kit (Lonza Bioscience) according to manufacturer’s instructions. Stably transfected cells were selected using puromycin (2 μg mL-1). Plasmid vectors and siRNA/CRISPR targeting sequences are also described at Supplementary tables 2 and 3 respectively.
Polyacrylamide hydrogel preparation
Coverslips were cleaned in ultrasonic bath with MilliQ water for 30 min followed by 30 min ultrasonic bath with ethanol (100%). After rinsing with water, coverslips were RCA cleaned in a 5:1:1 solution of MilliQ water, hydrogen peroxide (30%w/w; 31642, Sigma-Aldrich) and ammonium hydroxide (221228, Sigma-Aldrich) for 10 min at 60°C.Then, coverslips were either submerged in Sigmacote (SL2, Sigma-Aldrich) for 5 min, washed with ethanol in ultrasonic bath for 15 min and dried with N2, to become water repellent, or were treated with 3-(Acryloyloxy)propyltrimethoxysilane (L16400, Alfa Aesar) in ethanol solution for 120 min and dried at 120°C for 60 min to activate stable interactions with polyacrylamide matrix. Polyacrylamide hydrogels were generated as described previously 54. Briefly 40% w/w acrylamide, 2% w/w bis-acrylamide, 1.5% w/w TEMED and 5% APS were mixed according to Supplementary Table 4 and the resulting solution was placed between a passivated and an activated coverslip. Following 30 min of polymerisation the two coverslips were separated and the polyacrylamide gel which was attached to the activated coverslip was washed and stored in sterile water at 4°C. For functionalization with ECM, polyacrylamide hydrogels were treated with 0.2 mg mL-1 sulfo-SANPAH solution in MilliQ water followed by UV irradiation (365 nm) for 10 min. Hydrogels were extensively washed with HEPES buffer (50mM, pH 8.5), incubated overnight with fibronectin (10μg mL-1) and washed extensively in PBS before use.
Atomic Force Microscopy (AFM)
Hydrogel elastic properties were measured by atomic force microscopy (AFM) using a Nanowizard 3 AFM model (JPK Instruments). Specifically, silicon spheres of 20 μm diameter were attached to tipless cantilevers with either a nominal spring constant of 0.03 N m-1 (Arrow TL1, NanoWorld) for soft substrata or with a nominal spring constant of 0.2 N m-1 (TL-CONT, NanoSensors, NanoWorld) for stiffer matrices. Mechanical measurements were performed into MilliQ water. The actual spring constant was calibrated by thermal tuning using the simple harmonic oscillator model. The Young’s Modulus was measured by recording the force displacement curves from mapping 1μm x 1μm areas. Three points near the gel centre were selected on each gel, separated at least by 20 μm between them. To compute Young’s elastic modulus (E), the Hertz model equation for spherical tips was fitted to the force-displacement curves using the JPK Data Processing software (version 4.2, JPK Instruments). An effective indentation of 500 nm and a Poisson’s ratio of 0.5 were applied.
Traction Force Microscopy (TFM)
Measurements of cellular traction forces were performed on fibronectin (10 μg mL-1) coated 38 kPa polyacrylamide hydrogels embedded with 0.2 μm red fluorescent carboxylate-modified microspheres (F8810; Thermo Fisher Scientific). Cells (10000 per cm2) were allowed to adhere for 16 hours before measurements either in full DMEM media or media supplemented with cyclocreatine (10 mM). Image acquisition was performed using an inverted EVOS FL Auto imaging system (Thermo Fisher Scientific) equipped with a PlanFL 40x/0.65 objective, before and after cell removal with trypsin (0.25%) treatment. Nuclei were labelled with NucBlue Live reagent (R37605, Thermo Fisher Scientific) and blebbistatin (B0560; Sigma) treatment (10μM, 60min) was used as a control of relaxed cellular traction forces. Image stacks before and after cell dissociation were processed in Fiji using the StackReg plugin 55 and bead displacements due to cell tractions and cell tractions were calculated with a previously published unconstrained Fourier-transform traction cytometry method 56. From the displacement field and traction force, the strain energy has been calculated.
Metabolite Extraction
Cells were plated in 6-well plates containing either hydrogels (32mm diameter) or glass coverslips of similar diameter and cultured in DMEM medium for 24 hours. Hydrogels were transferred into new 6-well plates, washed with PBS and metabolites were extracted with ice cold extraction buffer consisting of methanol, acetonitrile and water (50:30:20; all LCMS grade, Fisher Scientific). Lysis buffer volume added per well was adjusted to cell number, using cell count numbers from matching plates. For glucose isotope tracing experiments, media were replaced with fresh DMEM media containing U-13C6-D-Glucose, 99% (Cambridge Isotope Laboratories / CK Isotopes, UK) and after 60- and 180- min incubation period metabolites were extracted with ice cold extraction buffer as described earlier. For arginine isotope tracing experiments, following overnight growth, media were replaced with media containing L-Arginine:HCL (13C6, 99%; 15N4, 99%) (Cambridge Isotope Laboratories / CK Isotopes, UK) and after 1-, 3- and 6-hours of incubation, metabolites were extracted with ice cold extraction buffer as described earlier.
Liquid chromatography mass spectrometry (LCMS)
LCMS analysis was performed as described previously 57. Prepared samples were analysed on a LCMS platform consisting of an Accela 600 LC system and an Exactive mass spectrometer (Thermo Fisher Scientific). A Sequant ZIC-pHILIC column (4.6mm x 150mm, 5μm) (Merck) was used to separate the metabolites with the mobile phase mixed by A=20mM ammonium acetate in water and B=acetonitrile. A gradient program starting at 20% of A and linearly increasing to 80% at 30 min was used followed by washing (92% of A for 5 mins) and re-equilibration (20% of A for 10min) steps. The total run time of the method was 45 min. The LC stream was desolvated and ionised in the HESI probe. The Exactive mass spectrometer was operated in full scan mode over a mass range of 70–1,200 m/z at a resolution of 50,000 with polarity switching.
LCMS data processing and analysis
The LCMS raw data was converted into mzML files by using ProteoWizard and imported to MZMine 2.10 for peak extraction and sample alignment. Metabolite identification was conducted by matching exact m/z values (±5ppm) and retention times (±8%) of standards in a house-made database. For isotope tracing experiments a house-made database including all possible 13C and 15N isotopic m/z values of the relevant metabolites was used for the assignment of LCMS signals. The generated data in MZmine 2.10 was exported as .CSV files for further statistical analysis. For unbiased metabolomics SIMCA 14 (Umetrics AB, Sweden) was applied and the S-plot was made in orthogonal partial least squares-discrimination analysis (OPLS-DA). Pathway-enrichment analysis was performed using the MetaboAnalyst platform (https://www.metaboanalyst.ca) on the top metabolite hits identified by our unbiased analysis. For targeted metabolomics Microsoft Excel 2010 (Microsoft Office) was used for Student’s t test and calculation of fold-change.
Growth Curves
KPC cells (5x104 cells per well) were seeded in 12-well plates in triplicate wells and allowed to adhere overnight. Cells were counted each day for 5 days using a CASY Model TT Cell Counter (Innovartis, Roche Applied Science). Media were changed every 48 hours.
EdU Labelling of Cells
EdU labelling was performed using the Click-iT EdU Alexa Fluor 594 assay kit (C10354, Invitrogen) according to manufacturer’s instructions. Briefly, KPC cells were grown on glass coverslips for 16 hours either in normal DMEM-10% FBS media or media supplemented with aphidicolin (1 and 5 μg mL-1). Then EdU (10 μM) was supplemented for 2 and 4 hours. Following labelling, cells were washed in PBS, fixed in 4% PFA for 15 min and then permeabilised in 0.5% Triton X-100 solution in PBS for 20 min at room temperature. EdU detection was performed by incubating with Click-iT/Alexa Fluor 594 reaction cocktail and nuclei were counterstained with DAPI. Coverslips were mounted using ProLong Diamond antifade reagent (P36965, Invitrogen) and images were taken using a Zeiss 710 confocal microscope equipped with an Ec Plan Neofluar 20x/0.50 and a Ec Plan Neofluar 40x/1.30 objective. Nuclei were counted with Fiji software (ImageJ v2.0.0) using the Find Maxima command and percentage of EdU positive nuclei was calculated.
ECM coatings of coverslips
Round glass coverslips (typically 19mm diameter for immunofluorescence or 32mm diameter for protein isolation) were washed with Ethanol, oven-dried and then coated with either fibronectin (10μg mL-1) for 60min at room temperature or with Concanavalin A (ConA; 10 μg mL-1) for 16 hours at 4°C. Coated coverslips were washed in PBS and cells (2x104 cells per mL) were plated and cultured for 16 hours in DMEM-10% FBS.
Western blotting
Cells were lysed in radioimmunoprecipitation (RIPA) lysis buffer (150mM NaCl, 10mM Tris-HCl pH7.5, 1mM ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100, 0.1% SDS, 1X protease and phosphatase inhibitors (Roche)). Protein concentration was determined using Precision Red Advanced Protein Assay (Cytoskeleton, Inc.). Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membrane. Membranes were blocked and incubated for 16h in 4°C with one of the following antibodies: anti-YAP (1:1000; 14074, Cell Signaling Technology), anti-CKB (1:1000; ab151579, Abcam), anti-GFP (1:1000, 2955, Cell Signaling Technology), anti-ERK1/2 (1:1000; 9102, Cell Signaling Technology), anti-AMPKα (1:1000; 2532S, Cell Signaling Technology), anti-phospho-AMPKα1/α2 (T183/T172) (1:1000; ab23875, Abcam), anti-GAPDH (1:1000; MAB374, Millipore) and anti-αTubulin (1:3000; T6199, Sigma). Protein detection was achieved using Alexa-Fluor conjugated secondary antibodies and signal was imaged using the LI-COR Odyssey CLx (LI-COR Biosciences) system. All images were analysed using Image Studio Lite software, version 5.2.5. A list of antibodies used is also provided at Supplementary Table 5.
Immunofluorescence
Immunofluorescence was typically performed on 13- or 19-mm diameter coverslips or PAAm hydrogels with cells being seeded at low concentrations (1-2x104 cells per cm2). Media were aspirated and following a wash with 1x PBS, cells were fixed in 4% PFA for 10 min and then permeabilised with 0.1% Triton X-100 for 5min followed by 30 min incubation in blocking buffer (1%BSA/PBS). Cells were incubated for 60min with the following antibodies: anti-YAP (1:100; 14074, Cell Signaling Technology), anti-phosphoPaxillin (1:400; 2541, Cell Signaling Technology), anti-Vinculin (1:400; V9131, Sigma), anti-GFP (1:1000; ab13970, Abcam) and anti-Cortactin (1:200; 05-180, Millipore). Detection was performed using the following secondary antibodies: Alexa Fluor 488 donkey anti rabbit (1:500 dilution; A21206, Invitrogen), Alexa Fluor 594 donkey anti mouse (1:500 dilution; A21203, Invitrogen), Alexa Fluor 488 goat anti chicken (1:500 dilution; A11039, Invitrogen). Nuclei were visualised with DAPI (0.5μg mL-1; D1306, Invitrogen) and F-actin with Alexa Fluor 647 Phalloidin (1:100 dilution; A22287, Invitrogen) incubation along with secondary antibodies. Coverslips were mounted using ProLong Diamond antifade reagent (P36965, Invitrogen). Images were acquired using a Nikon A1R confocal microscope equipped with Nikon Apo 60x/1.40 and Plan Fluor 40x/1.30 objectives (Figure 1a,d, Extended Data 1d,e), a Zeiss 710 confocal microscope equipped with Ec Plan Neofluar 40x/1.30 and Plan-apochromat 63x/1.40 oil objectives (Figure 5e) or a Zeiss 880 Laser Scanning Microscope with Airyscan equipped with a Plan-Apochromat 63x/1.4 oil DIC M27 objective (Figure 1g, Extended Data 1h, Extended Data 4h-j,n, Extended Data 5i). All images were processed with Fiji software (ImageJ v2.0.0). A list of antibodies used is also provided at Supplementary Table 5.
Cell shape analysis on Figures 1b-c, Extended Data 1f-g were performed using Cell Profiler software (v3.0.0; CellProfiler), applying a mask for cell area based on F-actin staining and a mask for nuclei. Focal adhesion analysis on Figures 1e-f was performed using the Fiji software (ImageJ v2.0.0) and the Analyze particles function applying a minimum size threshold of 0.25μm2. YAP nuclear to cytosolic ratios on Figures 1h, Extended Data 1i, and 7b were calculated using the Fiji software (ImageJ v2.0.0) to quantify mean YAP fluorescence intensity on similar rectangular areas over and adjacent to the nucleus. Cell shape analysis on Extended Data 5j,k was performed by manually drawing around the cell perimeter based on F-actin staining and measuring the area and the shape descriptors using the Fiji software (ImageJ v2.0.0). Automated cell shape analysis on Figure 5f was performed using CellProfiler software (v3.1.8) to detect nuclei and cell shape (n ≥ 15 frames per condition over 3 experiments). Automated cell shape classification was performed using CellProfiler Analyst (v2.2.1) by training the software to recognise 3 distinct F-actin shapes (‘lamellipodial’, ‘intermediate’ and ‘elongated’) that were subsequently used to classify the dataset of F-actin images.
Live imaging of mitochondria
Cells were labelled using the MitoTracker Green (200nM; M7514, Thermo Fisher Scientific) reagent according to manufacturer’s instructions and imaged within a contained unit at 37°C and under 5% CO2 using a Zeiss 880 Laser Scanning Microscope with Airyscan and a Plan-Apochromat 63x/1.4 oil DIC M27 objective. Z-stack acquisitions at 1μm intervals were performed and 3D reconstructions were generated using the Imaris Image Analysis software (v.9.2.0; Bitplane). In addition, time series acquisitions were performed for 30mins at 1min intervals. Images were processed with Fiji software (ImageJ v2.0.0).
qRT-PCR
RNA was isolated from cells using the RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions and was used for cDNA synthesis with either DyNAmo cDNA synthesis kit (F-470L, Thermo Fisher Scientific) or Maxima First Strand cDNA synthesis kit (K1641, Thermo Fisher Scientific). Then qRT-PCR was performed using the DyNAmo HS SYBR Green qPCR kit (F410L, Thermo Fisher Scientific). PCR was performed on a C1000 Thermal Cycler (CFX96 Real time system, BioRad) as follows: 3min at 95ºC, 40-cycles of 20s 95°C, 30s 57°C, 30s 72°C and final 5min 72°C. Relative mRNA quantification was performed using the 2^-ddCT method for multiple genes. Primer sequences are provided at Supplementary Table 6.
Flow Cytometry
For quantifying mitochondrial mass and mitochondrial membrane potential, cells were labelled using the MitoTracker Green (200nM; M7514, Thermo Fisher Scientific) and TMRE-Mitochondrial Membrane Potential Assay Kit (100nM; ab113852, Abcam) respectively according to manufacturer’s instructions. Specifically, 2x105 cells were plated on 32mm diameter glass coverslips or PAAm hydrogels placed inside 6-well plates and grown for 16 hours at 37°C. MitoTracker Green (100 nM) and TMRE (100 nM) were added for 30 min. CCCP treatment (10μM; 30min) was used to deplete mitochondrial membrane potential. Live cells were collected and washed twice with PBS before flow cytometry. Cellular ROS were quantified using the CellROX Deep Red reagent (1μM; C10422, Thermo Fisher Scientific) according to manufacturer’s instructions. Tert-Butyl hydroperoxide treatment (100μM; 30min) and NAC administration (20mM, 60min) were used to positively and negatively control cellular ROS presence respectively. Specifically, 2x105 cells were plated on 32mm diameter glass coverslips or PAAm hydrogels placed inside 6-well plates and grown for 16 hours at 37°C. NAC (20mM) was administered for 60 min before addition of Tert-Butyl hydroperoxide treatment (100μM; 30min) and CellROX Deep Red reagent (1μM, 30 min). Live cells were collected and washed twice with PBS before flow cytometry. All measurements were in live cells using the Attune NxT (Thermo Fisher Scientific) flow cytometer. Data was analysed using the FlowJo software (v.10.5.3, FlowJo, LLC). DAPI (0.5 μg mL-1) was used to discriminate live/dead populations. Experimental design is provided in the Reporting Summary and gating strategies are provided in the Source Data.
ADP/ATP ratio bioluminescence assay
Cells (4x103) were plated in duplicate wells per condition on 96-well plates and after 24 hours of growth were supplemented with media containing 0, 5 or 10 mM cyclocreatine. Following 16 hours of cyclocreatine treatment, ADP / ATP ratio was assessed with a bioluminescent assay (Abcam, ab65313) according to manufacturer’s instructions. Luminescence was measured using a Spark multimode microplate reader (Tecan Trading AG).
Immunohistochemistry (IHC)
Immunohistochemical (IHC) staining for anti-CKB (ab108388, Abcam), anti-YAP (4912, Cell Signalling Technology) and anti-Fibronectin (A0245, Dako) was performed on 4 μm formalin fixed paraffin embedded (FFPE) sections which had previously been ovened at 60°C for 2 hours. IHC staining was performed on an Agilent Autostainer link48. The sections underwent manual dewaxing through xylene, graded alcohol and then washed in tap water before undergoing heat induced epitope retrieval (HIER). HIER was performed on an Agilent PT module where the 4 μm sections were heated to 98°C for 25 minutes in PT module 1 buffer (Thermo Fisher Scientific). After epitope retrieval sections were rinsed in Tris Buffered saline with Tween (Tbt) prior to being loaded onto the autostainer. Then sections underwent peroxidase blocking (Agilent), washed in Tbt before application of appropriate antibody at a previously optimised dilution (CKB, 1:200; YAP, 1:50; Fibronectin 1:600) for 40 minutes. The sections were then washed in Tbt before application of rabbit EnVision (Agilent) secondary antibody for 35 minutes. Sections were rinsed in Tbt before applying Liquid DAB (Agilent) for 10 minutes. The sections were then washed in water, counterstained with haematoxylin and coverslipped using DPX. The FFPE sections cut for IHC investigation were part of a set of serial sections where other serial sections were stained with Picrosirius red and H&E. A list of antibodies used is also provided at Supplementary Table 5.
Stained sections were scanned and analysed using the HALO Image Analysis Software (PerkinElmer). High magnification images were acquired using a Olympus BX51 microscope equipped with a UPlanFL N 40x/0.75 objective. A list of mice used for IHC staining is provided in Supplementary Table 7 and described in the Reporting Summary. All protocols and experiments were previously approved by the Animal Welfare and Ethical Review Body (AWERB) of the University of Glasgow and were accompanied by a UK Home Office project licence to the corresponding author PE494BE48.
Random Migration Assay
Cells were plated at low confluency onto fibronectin coated dishes and imaged for 16h using a Nikon TE2000 microscope with a Plan Fluor 10x/0.30 objective and equipped with a heated CO2 chamber. Images were analysed with Fiji software (ImageJ v2.0.0).
Scratch Wound Migration Assay
Confluent cell monolayers were generated by plating 3x105 cells per mL on 96-well Incucyte Imagelock plates for 16h. Wounding was performed using the 96-pin WoundMaker (Essen Bioscience) and images were acquired at 60min intervals for a period of 24 hours by the IncuCyte Zoom system (Essen Bioscience). Wound recovery was analysed with the automated Incucyte Zoom software (Essen Bioscience) providing relative wound density over time.
Wound ECM Invasion Assay
96-well Incucyte Imagelock plates were coated for 16h with Matrigel (100μg mL-1) at 37°C. Confluent cell monolayers were generated by plating 7x105 cells per mL for 4h. Then wells were wounded using the 96-pin WoundMaker (Essen Bioscience) and embedded in Matrigel (2.15 mg mL-1, diluted in DMEM-10%FBS). Following 60 min incubation at 37°C for 60min, DMEM-10%FBS or media containing appropriately diluted treatments (Cyclocreatine, Oligomycin A) were added to each well and images were acquired at 60min intervals for a period of 96 hours by the IncuCyte Zoom system (Essen Bioscience). Wound recovery was analysed with the automated Incucyte Zoom software (Essen Bioscience) providing relative wound density over time.
Immunolabelling of ECM-embedded cells
96-well plates were coated for 16h with Matrigel (100μg mL-1) at 37°C. Confluent cell monolayers were generated by plating 7x104 cells per well for 4h. Then wells were wounded using a sterile tip and were embedded in Matrigel (2.15 mg mL-1, diluted in DMEM-10% FBS). Following 60 min incubation at 37°C for 60min, DMEM-10% FBS or media containing appropriately diluted Cyclocreatine was added to each well. Wounded monolayers embedded in 3D ECM were washed once with 1x PBS and fixed with 4% PFA for 15 min at RT, either at t = 0, at t = 24 hours or at t = 48 hours following ECM setting. Immunofluorescence was then performed as previously described 58, 59. Specifically, following fixation, ECM-embedded cells were washed twice with PBS, blocked for 60 min in PFS buffer (PBS, 0.7% w/v fish skin gelatin (G7041; Sigma-Aldrich), 0.5% saponin (47036; Sigma-Aldrich)) and incubated with either anti-YAP (1:100; 14074, Cell Signaling Technology) or anti-phospho-AMPKα1/α2 (T183/T172) (1:100; ab23875, Abcam) primary antibodies diluted in PFS at 4°C for 16 hours with gentle shaking. Following 3 washes in PFS, ECM-embedded cells were incubated with Alexa Fluor 488 donkey anti rabbit (1:500 dilution; A21206, Invitrogen) secondary antibody and DAPI (0.5μg mL-1; D1306, Invitrogen) diluted in PFS for 60 min at RT and then washed twice in PFS and twice in PBS. Images were taken using a Nikon A1R confocal microscope equipped with a Plan Fluor 40x/1.30 objective. All images were processed with Fiji software (ImageJ v2.0.0). Phospho-AMPK intensity was calculated using the Fiji software (ImageJ v2.0.0) quantifying phospho-AMPK and DAPI mean fluorescence intensity over the nucleus. Values were exported in Excel software and phospho-AMPK mean fluorescence intensity for each cell was normalised over the respective DAPI mean fluorescence intensity.
Chemotaxis assay
Chemotaxis assays were performed as described previously 60. Briefly, cells were seeded onto fibronectin-coated coverslips and grown overnight in serum-free DMEM. Coverslips were mounted onto Insall chambers supplemented with 10%FBS as the chemoattractant. Imaging was performed with a Nikon TE2000-E time-lapse microscope using MetaMorph software for 48h at 30min intervals. Cells were tracked using the MTrackJ plugin in Fiji software (ImageJ v2.0.0). Spider plots, rose diagrams and circular statistics were performed using a custom R script (Rstudio, v.1.1.453).
3D Spheroid Invasion assay
Cells were plated into 96-well ultra-low attachment plates (Costar, Corning) at 500 cells per well in DMEM-10% FBS containing 1% (v/v) of Matrigel and allowed to form spheroids for 3 days. Then, plates were centrifuged (300 x g, 5 min) and 50μL of medium was removed, followed by incubation at 4°C for 20 minutes. Each spheroid was then overlaid with 50 μL of ice-cold collagen I - Matrigel (4mg/ml of each) solution. Plates were centrifuged (300 x g, 5 min) and incubated at 37°C for 60 min to allow matrix solidification. Then each spheroid was supplemented with 25 μl of either normal DMEM-10%FBS or media containing appropriately diluted Cyclocreatine. Spheroid growth was monitored over 5 days using an IncuCyte Zoom system (Essen Bioscience). To quantify ECM invasion, the number and the length of spheroid protrusions as well as spheroid diameter was quantified each 24 hours. Invasion index was defined as the ratio of the longest protrusions’ length (maximum invasion) over the spheroid diameter.
SHG imaging of 3D spheroids
KPC spheroids were stained with CellTracker Red CMTPX Dye (C34552, Thermo Fisher Scientific) for 60 min, embedded in Matrigel:Collagen I ECM and fixed with 4% PFA either at t=0 or at t= 48 hours following ECM introduction. Imaging was performed on a Nikon Eclipse TE2000-U inverted microscope with an Olympus long working distance 20x 0.95 NA water immersion objective. Excitation source was a Ti:Sapphire femtosecond pulsed laser (Cameleon, Coherent UK), irradiating at 80 MHz and tuned to a wavelength of 890 nm. A scan head specifically designed for multi-photon excitation was used (Trim-scope, LaVision Biotec, Germany). Band pass filters, 435/40 and 595/40, were used to filter the emission for the SHG and RFP channels respectively. Z-stack acquisitions were performed at 2μm steps covering an area around 300 μm deep in tiles of 500 μm by 500 μm regions containing the spheroids and the invasion area, if present. Subsequent analysis was performed in maximum intensity projections of z-stack acquisitions using Fiji software (ImageJ v2.0.0). Specifically, 6 SHG intensity profiles were generated around each spheroid (starting at the middle of the spheroid and ending at the end of the frame) and values were exported in Excel. SHG intensity values were plotted against the ratio of the distance from starting point divided by the spheroid diameter. Plot profile generation, identification of maximum SHG intensity peak (y max) and calculation of the full width at half maximum (FWHM, shaded green region on plots) was performed using a custom R script (Rstudio, v.1.1.453). Bar plots demonstrate average values (from 6 intensity profiles) per spheroid and were generated using Prism 8 (v8.2.0; GraphPad Software).
3D Cell-derived Matrix Invasion Assay
Gelatin-coated dishes were cross-linked with glutaraldehyde, quenched and equilibrated in DMEM containing 10% FBS. Human dermal fibroblasts were plated at near confluence and grown for 14 days in DMEM containing 10% FBS and ascorbic acid (50 μg mL-1). To produce decellularized matrices, cells were lysed using PBS containing 20mM NH4OH and 0.5% Triton X-100, and any nucleic acid residues were removed by incubation with DNaseI. Matrices were stored at 4°C in PBS containing calcium and magnesium and washed in PBS before cell seeding. Cells were plated at low confluency onto decellularized extracellular matrix derived from human fibroblasts and imaged for 16h using a Nikon TE2000 microscope with a Plan Fluor 10x/0.30 objective and equipped with a heated CO2 chamber. Images were analysed with Fiji software (ImageJ v2.0.0). Individual cells were tracked using the MTrackJ plugin in Fiji software (ImageJ v2.0.0).
Actin Photoactivation Microscopy
Photoactivation experiments were performed using a Zeiss 880 Laser Scanning Microscope with Airyscan and a Plan-Apochromat 63x/1.4 oil DIC M27 objective. PA-GFP-actin and LifeAct-mTagRFP were monitored with a 488 nm and a 561 nm laser respectively for 60 seconds with acquisitions every 1 sec. Photoactivation of PA-GFP-actin was performed with a 405-nm laser (power 33%, 0.67 μs pixel dwell time) irradiating a region of interest (ROI) at a pseudopod tip for each cell with a single pulse (0.41 μs pixel dwell time) at t=5.5 seconds. For cyclocreatine experiments 2×106 cells were transfected with 5μg PA-GFP-Actin and LifeAct-mTagRFP using Amaxa Cell Line Nucleofector kit (Lonza Bioscience) according to manufacturer’s instructions. 6 hours post-transfection, 5×104 cells were plated on 35mm glass-bottom dishes (P35G-1.5-14-C; MatTek Corporation) coated with fibroblast-derived ECM. Cells were treated with cyclocreatine (5mM, 10mM; 16h) or Jasplakinolide (1μM, 15 min) before imaging. For CKB silencing experiments, 5×104 cells were transfected with control (n.c.) siRNA (25nM; AllStars Negative Control siRNA, Qiagen) or CKB targeting oligos (25nM; Qiagen) using the Lipofectamine RNAiMAX transfection reagent (13778075, Thermo Fisher Scientific) according to manufacturer’s instructions. Following 24 hours of siRNA introduction, cells were transfected with 1μg PA-GFP-Actin and LifeAct-mTagRFP using Lipofectamine 2000 reagent (11668027, Thermo Fisher Scientific) according to manufacturer’s instructions. 6 hours post-transfection 5×104 cells were plated on 35mm glass-bottom dishes (P35G-1.5-14-C; MatTek Corporation) coated with fibroblast-derived ECM and imaging was performed following 16 hours incubation. Image analysis of PA-GFP-actin and LifeAct-mTagRFP intensity was performed with Zeiss ZEN software (ZEN 2.3 SP1 FP3 v. 14.0.17.29; Carl Zeiss Microscopy GmbH) and values were exported into Microsoft Excel for analysis. LifeAct-mTagRFP fluorescence intensity for each cell was calculated as a percentage of intensity at the intensity of the first acquisition (5 sec before photoactivation). PA-GFP-actin fluorescence intensity for each cell was initially normalised by the median of the intensity of the five first acquisitions before photoactivation and then as a percentage of intensity at the timepoint of photoactivation (t=0 sec). Fluorescent decay lifetime was calculated at t1/2 (=50% decay from t of photoactivation). Fluorescent decay curves of LifeAct-mTagRFP and PA-GFP-Actin intensities were generated using Prism 8 (v8.2.0; GraphPad Software).
In vivo PDAC Intrasplenic transplantation assay
Mice were maintained in the Animal House of the Beatson Institute according to UK Home Office regulations and in compliance with EU Directive 2010/63 and the UK Animals (Scientific Procedures) Act 1986. All protocols and experiments were previously approved by the Animal Welfare and Ethical Review Body (AWERB) of the University of Glasgow and were accompanied by a UK Home Office project licence to the corresponding author PE494BE48. Intrasplenic injection delivers tumor cells via the splenic vein that joins with the superior mesenteric vein to become the portal vein 43, 44. Following anaesthesia, a transverse incision in the left flank was made exposing the spleen and KPC cells (1x106) in 100 μl PBS were injected in the spleen of nude mice (CD-1nu, females, 6-week old; Charles River Laboratories, Wilmington, MA). Surgical clips were typically removed 7 days after surgery and mice were sacrificed 21 days after inoculation. Cyclocreatine (0.5%; 57312 AstaTech) was supplemented daily in drinking water and treatment was initiated 7 days following surgery and after clip removal. Mice were sacrificed 21 days following surgery and during dissection, the spleen and liver were typically removed and following weight measurements, the incidence of liver tumor development was examined.
Statistics and reproducibility
All datasets were analysed and plotted using Prism 8 (v8.2.0; GraphPad Software) unless otherwise stated. Differences between groups were tested for normal distribution and analysed using the appropriate statistical test, as mentioned in each figure legend. Error bars represent SD, unless otherwise stated. Statistical analysis and experimental design is also highlighted in the Reporting Summary.
Extended Data
Extended Data Fig. 1. Pancreatic cancer cells are mechanosensitive and untargeted metabolomic profiling reveals metabolic reprogramming via ECM mechanics.
Panels b-n, cells atop 0.7-38 kPa fibronectin-coated hydrogels or glass.
(a) Polyacrylamide hydrogel elasticity (kPa). Values area mean ±SD from n=5, 0.7kPa, n=7, 7kPa and n=5, 38kPa hydrogels from 3 independent preparations.
(b) Growth curves of KPC cells. Values are mean ±SEM from 3 independent experiments. Statistical significance assessed by two-tailed Mann-Whitney U test, on ‘glass vs 0.7kPa’ p=0.005 at 48h and p=0.014 at 96h, on ’38kPa vs 0.7kPa’ p=0.0086 at 48h and p=0.094 at 96h.
(c) Representative images of KPC cells. Scale bars, 50μm. Right panel; Magnification of areas indicated by a dashed box. Scale bars, 25μm.
(d) Immunofluorescence of KPC cells showing F-actin (black) and nuclei (gold). Scale bars, 20μm. Blue arrows lamellipodia and red arrows stress fibres.
(e) Immunofluorescence of PANC-1 cells showing F-actin (grey) and nuclei (blue). Scale bars, 50 μm.
(f-g) Quantification of (e) showing cell area (μm2) (f) and circularity index (g). Values are mean ±SEM from n=240, 0.7kPa, n=268, 7 kPa, n=276, 38kPa and n=256 glass cells from 3 independent experiments. Kruskal-Wallis with Dunn’s multiple comparisons test.
(h) Immunofluorescence of PANC-1 cells from (e) showing YAP (grey). Scale bars, 50μm.
(i) Quantification of (h) showing nuclear to cytosolic YAP ratio. Values are mean ±SD from n=66, 0.7kPa, n=41, 7kPa, n=61, 38kPa and n=72 on glass. Cells from 3 independent experiments. Kruskal-Wallis test with Dunn’s multiple comparisons test.
(j) σ-plot demonstrating metabolite enrichment on soft (0.7kPa, left) and stiff (glass, right) KPC cells.
(k) Bar graph pathway enrichment analysis of (j) clustered by -log(p-value). Untargetted analysis on n=3 ‘0.7kPa’ and n=3 ‘glass’ independent cultures on same day. Statistics: Fisher’s exact Test.
(l) ‘Arginine and Proline’ KEGG pathway from (k). Individual metabolites labelled by KEGG number (https://www.genome.jp/kegg/kegg3.html) and enriched metabolites highlighted (red). Cr: Creatine; pCr: Phosphocreatine.
(m-n) AMP, ADP and ATP levels (m) and AMP to ATP ratio (n) of PANC-1 cells. Values are mean ±SD from 3 biological replicates on same day. Two-tailed unpaired t-test with Welch’s correction.
Extended Data Fig. 2. ECM stiffness directs creatine metabolism in PDAC cells.
(a) Schematic representation of the phosphocreatine circuit. Red indicates metabolite enrichment on soft (0.7kPa) ECM, while blue indicates enrichment on stiff (glass) ECM.
(b) Urea cycle and creatine biosynthesis metabolic intermediates of KPC cells cultured on fibronectin-coated 0.7-38 kPa hydrogels and glass coverslips as indicated. Values are mean ± SD from 3 biological replicates within the same day. Statistical significance assessed by one-way ANOVA.
(c) Urea cycle and creatine biosynthesis metabolic intermediates of PANC-1 cells cultured as indicated. Values are mean ±SD from 3 biological replicates within the same day. Statistical significance assessed by two-tailed unpaired t-test with Welch’s correction.
(d) Arginine-derived labelled carbon and nitrogen incorporation in urea cycle and creatine biosynthesis metabolites of KPC cells cultured as indicated and supplemented with L-arginine-13C615N4 for 1, 3 and 6 hours. Values are mean ±SD from 3 biological replicates within the same day.
Extended Data Fig. 3. Mitochondrial dynamics and respiratory activity are induced by ECM mechanics in pancreatic cancer cells and support invasive behaviour.
In a-i, cells were cultured atop of 0.7-38kPa fibronectin-coated hydrogels and glass coverslips.
(a) Glucose-derived labelled carbon incorporation in glucose and TCA cycle intermediates of KPC cells as indicated and treated with U-13C6-glucose for 3 hours.
(b) GSH, GSSG levels and GSH/GSSG ratio of KPC cells cultured as indicated.
(c) NADPH levels of cells from (b).
(d) Glycolysis and TCA cycle metabolite levels of PANC-1 cells cultured as indicated.
(e) GSH, GSSG levels and GSH/GSSG ratio of PANC-1 cells from (d).
(f) Left; Mitochondrial mass (Mitotracker, MTG) of KPC cells as indicated. Values (gMFI) are mean ±SD relative to control (glass) from 3 independent experiments. Right; Representative histogram from left panel.
(g) Left; Mitochondrial membrane potential (TMRE) of KPC cells cultured as indicated. CCCP; negative control. Values (gMFI) are mean ±SD relative to control (glass) from 3 independent experiments. Right; Representative histogram from left panel.
(h) Left; Cellular ROS (CellROX) of KPC cells cultured as indicated. H2O2; positive, NAC+ H2O2; negative control. Values (gMFI) are mean ±SD relative to control (glass) from 3 independent experiments. Right; Representative histogram from left panel.
(i) Top; Maximum intensity projections of z-stack acquisitions of PANC-1 cells cultured as indicated showing labelled mitochondria. Scale bars, 10μm. Bottom; Magnification of areas indicated by a dashed box. Pictures representative of 2 independent experiments. Scale bars, 5μm.
(j) Maximum intensity projections of z-stack acquisitions of KPC cells cultured on fibronectin-coated dishes (left) or invading 3D ECM (middle, right) showing labelled mitochondria. Pictures representative of 3 independent experiments. Scale bars, 7μm (left, right) and 8μm (middle).
Values in a-e represent mean ±SD from 3 biological replicates performed on the same day.
In b,d,e: statistical significance assessed by two-tailed unpaired t-test with Welch’s correction.
In f,g,h: statistical significance assessed by two-tailed one-sample t-test on LN transformed values.
Extended Data Fig. 4. The phosphocreatine circuit depends on CKB in pancreatic cancer cells, which is regulated by mechanosensing and YAP activity.
(a) qRT-PCR of creatine kinases mRNA in KPC cells. Mean ±SD from 3 independent experiments.
(b) Ckb expression in KPC cells. Cdk2: normalisation control. Values mean ±SD and relative to control from 3 independent experiments. Two-tailed one-sample t-test on LN transformed values.
(c) PANC-1 cells immunoblotted for CKB and ERK1/2 (loading control).
(d) Densitometric quantification of (c). Mean ± SD and relative to control from 3 independent experiments. Two-tailed one-sample t-test on LN transformed values.
(e) % of EdU positive nuclei in KPC cells treated with aphidicolin. Mean ±SD from n=6 biological replicates and 2 independent experiments.
(f) KPC cells from (e) immunoblotted for CKB and α-Tubulin (loading control).
(g) Densitometric quantification of (f). Mean ±SD relative to control from 3 independent experiments.
(h) Immunofluorescence of KPC cells on fibronectin or concanavalin A, F-actin (grey) and pPaxillinTyr118 (magenta). Scale bars, 50μm.
(i) Immunofluorescence of KPC cells expressing GFP or VD1-GFP, Vinculin (magenta) and GFP (grey). Scale bars, 50μm.
(j) Immunofluorescence of cells from (i) showing YAP (Green) and F-actin (magenta). Scale bars, 50μm.
In h-j: pictures representative of 3 independent experiments.
(k) Densitometric quantification of YAP in YAP-silenced KPC cells. Mean ± SD and relative to control from 4 independent experiments. Two-tailed one-sample t-test on LN transformed values.
(l-m) Yap (l) and Ckb (m) expression from (k). Values are mean ± SD and relative to control from 3 independent experiments. Gapdh: normalisation control.
(n) Immunofluorescence of KPC cells expressing GFP, GFP-YAP or GFP-YAP5SA showing GFP (grey). Scale bars, 20μm.
(o) Cells from (n) were immunoblotted for GFP. Pictures in n-o representative of 3 independent experiments.
(p-q) Creatine pathway metabolites (p) and phosphocreatine/creatine ratio (q) of cells from (n) cultured on 0.7 kPa hydrogels. Values are mean ± SD from 3 biological replicates within the same day. Two-tailed unpaired t-test with Welch’s correction.
Extended Data Fig. 5. Creatine homeostasis facilitates collective migration of pancreatic cancer cells.
(a) CCr activity schematic.
(b) Ccr uptake by KPC cells. Values in b and c are mean ±SD from 3 biological replicates within the same day and representative of 2 independent experiments.
(c) Phosphocreatine circuit and ATP levels of (b).
(d) ADP to ATP ratio of (b). Mean ±SD from 4 independent experiments.
(e) Growth curves of (b). Mean ±SD from 3 independent experiments.
(f) Left; CCr-treated cells. Scale bars, 100μm. Right; Magnification of dashed boxes. White arrows; protrusions. Scale bars, 50μm.
(g) Average protrusion length from (f). Values are mean ±SD from n=206 control, n=145 5mM and n=142 10mM CCr-treated cells.
(h) Cell speed from (f). Mean ±SD from n=263 control, n=185 5mM and n=192 10mM CCr-treated cells from 3 independent experiments.
(i) Immunofluorescence of CCr-treated cells showing F-actin (magenta) and nuclei (blue). Scale bars, 20μm.
(j-k) Cell area (μm2) (j) and circularity index (k) from (i). Mean ±SD from n=60 control, n=78 5mM and n=69 10mM CCr-treated cells from 3 independent experiments.
(l-n) Total force (l), maximum force (m) of CCr-treated cells and strain energy of blebbistatin treated cells (n). Mean ±SD of n=33 control, n=36 CCr-treated and n=17 blebbistatin-treated cells from 3 independent experiments. Mann-Whitney U test.
(o) Creatine and phosphocreatine levels of control or pCr supplemented cells. Values in o and p are mean ±SD from 3 biological replicates within the same day.
(p) ADP/ATP ratio of (o).
(q) Control (EV) or CKBCRISPR KO cells expressing GFP or CKB-GFP were immunoblotted for GFP and CKB. Pictures representative of 3 independent experiments.
(r) Pictures of cells from (q) populating a wounded monolayer. Scale bars, 100μm.
(s) Relative wound closure of control or CCr-treated wild-type (EV) cells. Values in s, t, u are mean ±SD from 3 independent experiments.
(t-u) Relative wound closure (t) and relative closure at t1/2 of control (u) of control or CCr-treated CKBCRISPR KO cells.
In d,s,u: two-tailed one-sample t-test on LN transformed values.
In g,j,k: Kruskal-Wallis with Dunn’s multiple comparisons test.
Extended Data Fig. 6. Creatine homeostasis supports actin dynamics and ECM invasion of pancreatic cancer cells.
(a-b) Wound closure migration (a) and relative closure at t1/2 of control (b) from KPC cells invading 3D ECM as indicated. Mean ±SD from 3 independent experiments.
(c-d) pAMPK signal intensity at the ‘back’ and ‘front’ of control (c) and the ‘front’ of control or CCr-treated KPC monolayers (d) invading 3D ECM. Mean ±SEM from 3 independent experiments with n=84 ‘t=0,back’; n=113 ‘t=0,front’; n=63 ‘t=24h,back’; n=117 ‘t=24,front’; n=62 ‘t=48,back’; n=97 ‘t=48,front’; n=122 ‘t=24,CCr’; n=91 ‘t=48,CCr’ cells. Two-tailed paired t-test.
(e) Control or CCr-treated KPC cells immunoblotted for pAMPKα1T183/ α2T172, AMPKα1/α2 and GAPDH (loading control).
(f) Densitometric quantification of pAMPK/AMPK levels from (e). Mean ±SD and relative to control from 3 independent experiments.
(g) Pictures of control (nc) or CKB silenced (CKBsi#01, CKBsi#02) cells invading 3D ECM. Scale bars, 100μm.
(h) Control (EV) or CKB-depleted (CKB-KO) cells expressing GFP or CKB-GFP, invading 3D ECM. Scale bars, 100μm.
(i) Wound closure migration of control (EV), CKB-KO and CKB-KO P-Cr-treated cells invading 3D ECM. Mean ±SD from 2 independent experiments with 5 technical replicates.
(j-k) Wound closure over time (j) and relative closure at t1/2 of control (k) from control (EV) cells from (h). Values are mean ±SD from 4 independent experiments.
(l) Control or CCr-treated cells invading fibroblast-derived ECM. Scale bars, 200μm.
(m-n) Cell speed (m) and Euclidean distance (n) from (l). Values are mean ±SD from n=317 control, n=290 5mM and n=239 10mM CCr from 4 independent experiments. Kruskal-Wallis with Dunn’s multiple comparisons test.
(o-p) LifeAct-mTagRed signal intensity from actin photoactivation experiments; (o) control or CCr-treated cells. Mean ±SEM derived from n=31 control, n=23 5mM CCr, n=35 10mM CCr and n=28 jasplakinolide-treated cells from 3 independent experiments. (p) control (nc) or CKB-silenced (CKBsi#01, CKBsi#02) cells. Values are mean ±SEM derived from n=36 n.c., n=31 CKBsi#01, n=38 CKBsi#02 and n=21 jasplakinolide-treated cells from 3 independent experiments.
In b,f,k: two-tailed one-sample t-test on LN transformed values.
Extended Data Fig. 7. Creatine homeostasis supports collagen remodelling, invasion and the chemotactic response of pancreatic cancer cells.
(a) Representative 3D reconstructions of z-stack acquisitions showing nucleic acid labelling (grey) of control or CCr-treated spheroids after 96 hours of invasion within 3D ECM.
(b) Protrusion number per spheroid of control or CCr-treated KPC spheroids as indicated. Values are from n=12 control, n=13 5mM and n=13 10mM CCr-treated spheroids from 3 independent experiments.
(c) Average protrusion length (μm) of control or CCr-treated KPC spheroids as indicated. Values are from n=12 control, n=13 5mM and n=13 10mM CCr-treated spheroids from 3 independent experiments.
(d) Representative plot profiles of SHG intensity of control or CCr-treated KPC spheroids. Green-shaded area shows Full width half maximum (FWHM) of Collagen I (SHG) intensity.
(e) Full width half maximum (FWHM) of Collagen I (SHG) intensity from (d). Each dot represents average value (from 6 plot profiles) per spheroid. Values are mean ±SD of n=9 t=0, n=12 t=48h and n=12 CCr-treated spheroids from 3 independent experiments. Statistical significance assessed by one-way ANOVA.
(f) Cell speed of cells treated as indicated chemotaxing towards a 10% FBS gradient. Values are mean ± SEM from n=262 control, n=278 5mM and n=275 10mM CCr-treated cells from 4 independent experiments. Statistical significance assessed by Kruskal-Wallis with Dunn’s multiple comparisons test.
(g) Cell speed of control (n.c.) or CKB-silenced (CKBsi#01, CKBsi#02) cells chemotaxing towards a 10% FBS gradient. Values are mean ± SEM from n=241 control, n=252 CKBsi#01and n=232 CKBsi#02 cells from 3 independent experiments. Statistical significance assessed by Kruskal-Wallis with Dunn’s multiple comparisons test.
Extended Data Fig. 8. CKB is expressed during PDAC progression and supports metastatic dissemination.
(a) Quantification of PicroSirius Red positive area per tissue area from normal mice (Pdx1-Cre+;Kraswt/wt;p53wt/wt) and PDAC from KPC (Pdx1-Cre;LSLKrasG12D;LSLp53R172H) mice. Values are mean ±SEM from n=4 normal and n=9 PDAC pancreata. Statistical significance assessed by two-tailed Mann-Whitney U test.
(b) Quantification of Fibronectin positive area per tissue area from (a). Values are mean ±SEM from n=5 for normal and n=11 for PDAC pancreata. Statistical significance assessed by two-tailed Mann-Whitney U test.
(c-d) YAP positive cells (%) (c) and nuclear to cytosolic YAP ratio (d) from normal mice (Pdx1-Cre+;Kraswt/wt;p53wt/wt) and PDAC from KPC (Pdx1-Cre;LSLKrasG12D;LSLp53R172H) mice. Values are mean ±SEM from n=5 normal and n=10 (c),n=9 (d) PDAC pancreata. Statistical significance assessed by two-tailed Mann-Whitney U test.
(e) High magnification pictures of PDAC tissue sections from KPC mice showing YAP and CKB. Scale bars, 20μm. Representative of n=9 PDAC pancreata.
(f) Weight (gr) of animals at time of sacrifice treated as indicated from intrasplenic transplantation experiment. Values are mean ±SEM from n=8 control (EV), n=8 CCr-treated and n=7 CKB-KO mice.
Supplementary Material
Serum starved KPC cells were treated with cyclocreatine as indicated and let to chemotax towards a 10% FBS gradient using an Insall chamber set up. Cell migration was monitored for 48 hours with images acquired at 30 min intervals using a Nikon TE2000-E time-lapse microscope. Colour coded tracks indicate manual cell tracking quantification using the MTrackJ plugin in Fiji software (ImageJ v2.0.0). Scale bars, 50μm. Representative of 4 independent experiments.
KPC spheroids were treated with cyclocreatine as indicated, embedded in Matrigel-Collagen I matrix and monitored over a period of 96 hours with images acquired at 60 min intervals using the Incucyte System (Essen Bioscience). Scale bar, 200μm. Representative of 3 independent experiments
KPC cells were transfected with PA-GFP-Actin (green) and LifeAct-mTagRed (red), plated on fibroblast-derived ECM and treated with CCr or jasplakinolide as indicated. Cells were photoactivated (t=6s) at the tips of pseudopods invading fibroblast-derived ECM. Images were acquired at 1 sec intervals for a period of 60 sec. Scale bars, 10μm. Representative of 3 independent experiments
KPC cells were plated on Matrigel-coated plates to form a monolayer followed by monolayer wounding. Following embedding in Matrigel matrix and cyclocreatine supplementation as indicated, wound closure was monitored for 90 hours with images acquired at 60 min intervals using the Incucyte System (Essen Bioscience). Red mask indicates wound area and purple line marks the initial wound area. Scale bar, 100μm. Representative of 3 independent experiments
KPC cells were plated to form a monolayer and supplemented with cyclocreatine as indicated for 16 hours. Following monolayer wounding, wound closure was monitored for 24 hours with images acquired at 60 min intervals using the Incucyte System (Essen Bioscience). Red mask indicates wound area and purple line marks the initial wound area. Scale bar, 100μm. Representative of 3 independent experiments.
Mitochondria of live KPC cells cultured on glass or 0.7kPa hydrogels were labelled with Mitotracker Green (MTG) and imaging was performed using a Zeiss 880 confocal with Airyscan. Pictures were acquired at 60sec intervals for 30min total time. CCCP was administered 10 min before starting the imaging. Scale bars, 20μm. Representative of 3 independent experiments
Mitochondria of live KPC cells cultured on glass or 0.7kPa hydrogels were labelled with Mitotracker Green (MTG) and imaging was performed using a Zeiss 880 confocal with Airyscan. Pictures were acquired at 60sec intervals for 30min total time. CCCP was administered 10 min before starting the imaging. Scale bars, 20μm. Representative of 3 independent experiments
Acknowledgements
We acknowledge CRUK Beatson Institute Core Services and Advanced Technologies (C596/A17196) and especially Beatson Advanced Imaging Resource (BAIR). We thank S. Karim for KPC cell lines. We also thank C. Nixon and the Beatson Histology Facility, D. Bryant and his lab for advice, J. Murray for generating fibroblast-derived matrices and E. J. McGhee for assistance with SHG microscopy. We thank M. Neilson for assistance in analysis and statistical advice. We also thank T. Hamilton, C. Baxter and E. Onwubiko for helping with intrasplenic surgery. We thank T. Pompe (University of Leipzig) for providing analysis software for TFM experiments. We thank H. Spence and R.H. Insall for advice and discussion. VP is supported by a CRUK Glasgow Centre studentship (A18076) to MSS and LMM; LMM is supported by a CRUK core grant A15673; NP is supported by an MRC grant to LMM (MR/R017255/1). MSS is funded by an EPSRC Programme Grant (EP/P001114/1). MC is funded by an MRC UKRI/Rutherford Fund fellowship (MR/S005412/1). OM and TZ are funded by a Cancer Research UK Career Development Fellowship (C53309/A19702).
Footnotes
Data availability
Associated raw data is provided as Source Data Files associated with each main or Extended Data Figure. Unprocessed pictures of blots are provided in the Source Data. Original datasets, analyses and methodological details are available from the Source Data or from the corresponding author upon reasonable request. Information regarding experimental design and reagents can also be found in the Reporting Summary file.
Author contributions: VP performed most of the experiments and participated in conception and design, analysed data and drafted the manuscript with LMM. NP helped with experiments and offered advice. TZ performed liquid chromatography mass spectrometry metabolomics and data analysis. AJ performed all KPC mice work and provided tissue blocks from the KPC mice. MC performed traction force microscopy experiments. OM conceived and designed experiments, analysed data, edited the manuscript, and supervised TZ. MSS co-supervised VP, conceived and designed experiments and edited the manuscript. LMM conceived and designed experiments, co-supervised VP and drafted the manuscript with VP.
Competing Interests:
The authors declare no competing interests.
References
- 1.Papalazarou V, Salmeron-Sanchez M, Machesky LM. Tissue engineering the cancer microenvironment-challenges and opportunities. Biophys Rev. 2018;10:1695–1711. doi: 10.1007/s12551-018-0466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian C, et al. Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc Natl Acad Sci U S A. 2019 doi: 10.1073/pnas.1908626116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 5.Cukierman E, Bassi DE. Physico-mechanical aspects of extracellular matrix influences on tumorigenic behaviors. Semin Cancer Biol. 2010;20:139–145. doi: 10.1016/j.semcancer.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laklai H, et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat Med. 2016;22:497–505. doi: 10.1038/nm.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miroshnikova YA, et al. Tissue mechanics promote IDH1-dependent HIF1α–tenascin C feedback to regulate glioblastoma aggression. Nature Cell Biology. 2016;18:1336. doi: 10.1038/ncb3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyssiotis CA, Kimmelman AC. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017;27:863–875. doi: 10.1016/j.tcb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamphorst JJ, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vennin C, et al. Reshaping the Tumor Stroma for Treatment of Pancreatic Cancer. Gastroenterology. 2018;154:820–838. doi: 10.1053/j.gastro.2017.11.280. [DOI] [PubMed] [Google Scholar]
- 12.Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol. 2017;18:758–770. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mo JS, et al. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat Cell Biol. 2015;17:500–510. doi: 10.1038/ncb3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorrentino G, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, et al. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat Cell Biol. 2015;17:490–499. doi: 10.1038/ncb3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enzo E, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015;34:1349–1370. doi: 10.15252/embj.201490379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peskin CS, Odell GM, Oster GF. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophysical Journal. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunniff B, McKenzie AJ, Heintz NH, Howe AK. AMPK activity regulates trafficking of mitochondria to the leading edge during cell migration and matrix invasion. Mol Biol Cell. 2016;27:2662–2674. doi: 10.1091/mbc.E16-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuler MH, et al. Miro1-mediated mitochondrial positioning shapes intracellular energy gradients required for cell migration. Mol Biol Cell. 2017;28:2159–2169. doi: 10.1091/mbc.E16-10-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley LC, et al. Adaptive F-Actin Polymerization and Localized ATP Production Drive Basement Membrane Invasion in the Absence of MMPs. Dev Cell. 2019;48:313–328 e318. doi: 10.1016/j.devcel.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellington WR. Evolution and physiological roles of phosphagen systems. Annu Rev Physiol. 2001;63:289–325. doi: 10.1146/annurev.physiol.63.1.289. [DOI] [PubMed] [Google Scholar]
- 22.Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol Rev. 2000;80:1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- 23.Kuiper JW, et al. Local ATP generation by brain-type creatine kinase (CK-B) facilitates cell motility. PLoS One. 2009;4:e5030. doi: 10.1371/journal.pone.0005030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenouille N, et al. The creatine kinase pathway is a metabolic vulnerability in EVI1-positive acute myeloid leukemia. Nat Med. 2017;23:301–313. doi: 10.1038/nm.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurmi K, et al. Tyrosine Phosphorylation of Mitochondrial Creatine Kinase 1 Enhances a Druggable Tumor Energy Shuttle Pathway. Cell Metab. 2018 doi: 10.1016/j.cmet.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elosegui-Artola A, et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol. 2016;18:540–548. doi: 10.1038/ncb3336. [DOI] [PubMed] [Google Scholar]
- 27.Yang B, et al. Stopping Transformed Growth with Cytoskeletal Proteins: Turning a Devil into an Angel. bioRxiv. 2018 221176. [Google Scholar]
- 28.Morton JP, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proceedings of the National Academy of Sciences. 2010;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nature Cell Biology. 2015;17:816. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531:110–113. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice AJ, et al. Matrix stiffness induces epithelial-mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis. 2017;6:e352. doi: 10.1038/oncsis.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelletier M, Billingham LK, Ramaswamy M, Siegel RM. Extracellular flux analysis to monitor glycolytic rates and mitochondrial oxygen consumption. Methods Enzymol. 2014;542:125–149. doi: 10.1016/B978-0-12-416618-9.00007-8. [DOI] [PubMed] [Google Scholar]
- 35.Machesky LM, Hall A. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J Cell Biol. 1997;138:913–926. doi: 10.1083/jcb.138.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia D, Shaw RJ. AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Mol Cell. 2017;66:789–800. doi: 10.1016/j.molcel.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker AM, Bird D, Lang G, Cox TR, Erler JT. Lysyl oxidase enzymatic function increases stiffness to drive colorectal cancer progression through FAK. Oncogene. 2013;32:1863–1868. doi: 10.1038/onc.2012.202. [DOI] [PubMed] [Google Scholar]
- 40.Gkretsi V, Stylianopoulos T. Cell Adhesion and Matrix Stiffness: Coordinating Cancer Cell Invasion and Metastasis. Front Oncol. 2018;8:145. doi: 10.3389/fonc.2018.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker C, Mojares E, Del Rio Hernandez A. Role of Extracellular Matrix in Development and Cancer Progression. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nature Cell Biology. 2007;9:193. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 43.Rajendran S, et al. Murine bioluminescent hepatic tumour model. J Vis Exp. 2010 doi: 10.3791/1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soares KC, et al. A preclinical murine model of hepatic metastases. J Vis Exp. 2014 doi: 10.3791/51677. 51677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sousa CM, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prager-Khoutorsky M, et al. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- 47.Romani P, et al. Extracellular matrix mechanical cues regulate lipid metabolism through Lipin-1 and SREBP. Nat Cell Biol. 2019 doi: 10.1038/s41556-018-0270-5. [DOI] [PubMed] [Google Scholar]
- 48.Bertero T, et al. Tumor-Stroma Mechanics Coordinate Amino Acid Availability to Sustain Tumor Growth and Malignancy. Cell Metab. 2018 doi: 10.1016/j.cmet.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JS, et al. Urea Cycle Dysregulation Generates Clinically Relevant Genomic and Biochemical Signatures. Cell. 2018;174:1559–1570 e1522. doi: 10.1016/j.cell.2018.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez-Domenech G, et al. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 2018;37:321–336. doi: 10.15252/embj.201696380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hastie EL, Sherwood DR. A new front in cell invasion: The invadopodial membrane. Eur J Cell Biol. 2016;95:441–448. doi: 10.1016/j.ejcb.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherwood DR, Plastino J. Invading, Leading and Navigating Cells in Caenorhabditis elegans: Insights into Cell Movement in Vivo. Genetics. 2018;208:53–78. doi: 10.1534/genetics.117.300082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juin A, et al. N-WASP Control of LPAR1 Trafficking Establishes Response to Self-Generated LPA Gradients to Promote Pancreatic Cancer Cell Metastasis. Dev Cell. 2019;51:431–445 e437. doi: 10.1016/j.devcel.2019.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol. 2010;Chapter 10 doi: 10.1002/0471143030.cb1016s47. Unit 10 16. [DOI] [PubMed] [Google Scholar]
- 55.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 56.Muller C, Pompe T. Distinct impacts of substrate elasticity and ligand affinity on traction force evolution. Soft Matter. 2016;12:272–280. doi: 10.1039/c5sm01706h. [DOI] [PubMed] [Google Scholar]
- 57.Oakey LA, et al. Metabolic tracing reveals novel adaptations to skeletal muscle cell energy production pathways in response to NAD (+) depletion. Wellcome Open Res. 2018;3:147. doi: 10.12688/wellcomeopenres.14898.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bryant DM, et al. A molecular network for de novo generation of the apical surface and lumen. Nature Cell Biology. 2010;12:1035. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Román-Fernández Á, et al. The phospholipid PI(3,4)P2 is an apical identity determinant. Nature Communications. 2018;9 doi: 10.1038/s41467-018-07464-8. 5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Susanto O, Muinonen-Martin AJ, Nobis M, Insall RH. Visualizing Cancer Cell Chemotaxis and Invasion in 2D and 3D. Methods Mol Biol. 2016;1407:217–228. doi: 10.1007/978-1-4939-3480-5_17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Serum starved KPC cells were treated with cyclocreatine as indicated and let to chemotax towards a 10% FBS gradient using an Insall chamber set up. Cell migration was monitored for 48 hours with images acquired at 30 min intervals using a Nikon TE2000-E time-lapse microscope. Colour coded tracks indicate manual cell tracking quantification using the MTrackJ plugin in Fiji software (ImageJ v2.0.0). Scale bars, 50μm. Representative of 4 independent experiments.
KPC spheroids were treated with cyclocreatine as indicated, embedded in Matrigel-Collagen I matrix and monitored over a period of 96 hours with images acquired at 60 min intervals using the Incucyte System (Essen Bioscience). Scale bar, 200μm. Representative of 3 independent experiments
KPC cells were transfected with PA-GFP-Actin (green) and LifeAct-mTagRed (red), plated on fibroblast-derived ECM and treated with CCr or jasplakinolide as indicated. Cells were photoactivated (t=6s) at the tips of pseudopods invading fibroblast-derived ECM. Images were acquired at 1 sec intervals for a period of 60 sec. Scale bars, 10μm. Representative of 3 independent experiments
KPC cells were plated on Matrigel-coated plates to form a monolayer followed by monolayer wounding. Following embedding in Matrigel matrix and cyclocreatine supplementation as indicated, wound closure was monitored for 90 hours with images acquired at 60 min intervals using the Incucyte System (Essen Bioscience). Red mask indicates wound area and purple line marks the initial wound area. Scale bar, 100μm. Representative of 3 independent experiments
KPC cells were plated to form a monolayer and supplemented with cyclocreatine as indicated for 16 hours. Following monolayer wounding, wound closure was monitored for 24 hours with images acquired at 60 min intervals using the Incucyte System (Essen Bioscience). Red mask indicates wound area and purple line marks the initial wound area. Scale bar, 100μm. Representative of 3 independent experiments.
Mitochondria of live KPC cells cultured on glass or 0.7kPa hydrogels were labelled with Mitotracker Green (MTG) and imaging was performed using a Zeiss 880 confocal with Airyscan. Pictures were acquired at 60sec intervals for 30min total time. CCCP was administered 10 min before starting the imaging. Scale bars, 20μm. Representative of 3 independent experiments
Mitochondria of live KPC cells cultured on glass or 0.7kPa hydrogels were labelled with Mitotracker Green (MTG) and imaging was performed using a Zeiss 880 confocal with Airyscan. Pictures were acquired at 60sec intervals for 30min total time. CCCP was administered 10 min before starting the imaging. Scale bars, 20μm. Representative of 3 independent experiments