Abstract
Neutrophils are critical to the generation of effective immune responses and for killing invading microbes. Paired immune receptors provide important mechanisms to modulate neutrophil activation thresholds and effector functions. Expression of the leukocyte immunoglobulin-like receptor (LILR)A6 (ILT8/CD85b) and LILRB3 (ILT5/CD85a) paired receptor system on human neutrophils has remained unclear due to the lack of specific molecular tools. Additionally, there is little known of their possible functions in neutrophil biology. The objective of this study was to characterise expression of LILRA6/LILRB3 receptors during human neutrophil differentiation and activation, and to assess their roles in modulating Fc receptor mediated effector functions. LILRB3, but not LILRA6, was detected in human neutrophil lysates following immunoprecipitation by mass spectrometry. We demonstrate high LILRB3 expression on the surface of resting neutrophils, and release from the surface following neutrophil activation. Surface expression was recapitulated in a human PLB-985 cell model of neutrophil-like differentiation. Continuous ligation of LILRB3 inhibited key IgA-mediated effector functions, including production of reactive oxygen species, phagocytic uptake and microbial killing. This suggests that LILRB3 provides an important checkpoint to control human neutrophil activation and their anti-microbial effector functions during resting and early-activation stages of the neutrophil life-cycle.
Introduction
Neutrophils are armed with an arsenal of strategies to kill invading microbes and to orchestrate inflammation (1). Upon infection, neutrophils are immediately activated and recruited to the site of infection where they detect and kill microbes through exposure to reactive oxygen species (ROS) and/or antimicrobial compounds following phagocytosis or degranulation (2). Additionally, neutrophils modulate immune responses by releasing cytokine and lipid mediators. However, if neutrophils are mobilized at the wrong time or place their potent effector functions and proinflammatory signals can damage the host and cause inappropriate inflammatory responses. Accordingly, neutrophils must be tightly regulated to avoid overreactive immune responses (3).
The activation threshold of neutrophils is tightly regulated to prevent inappropriate immune responses (4). Expression of inhibitory receptors and paired receptor systems provides major regulatory mechanisms (5, 6). However, though activation receptors including Fc receptors (FcRs), G protein-couple receptors (GPCRs) and Toll-like receptors (TLRs) have been studied extensively, there remains poor knowledge of the role of inhibitory and paired receptors in the regulatory process.
Paired receptors systems describe families of membrane receptors that share very similar extracellular regions but differ in their transmembrane regions, cytoplasmic regions and signalling capacity (6). Inhibitory receptors typically interact with self-proteins and provide mechanisms to raise activation thresholds and limit cell activity. Upon binding of ligands, immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in the cytoplasmic tails of inhibitory receptors become phosphorylated, leading to docking of SH2 domain containing tyrosine phosphatases, and suppression of downstream signalling cascades (7). In disease situations, the ligands of inhibitory receptors are often down-regulated or modified resulting in reduced inhibitory signals and a lowering of the activation threshold. Inhibitory receptors expressed on neutrophils include leukocyte-associated Ig-like receptor 1 (LAIR-1), signal inhibitory receptor on leukocytes 1 (SIRL-1), sialic acid-binding immunoglobulin-like lectin 5 (Siglec-5), and leukocyte immunoglobulin-like receptor (LILR)B2 (8, 9, 10, 11). The ITIM signalling of these receptors can counter-act activation of cells through immunoreceptor tyrosine-based activation motifs (ITAMs) signalling. Thus, inhibitory receptors can suppress FcR-mediated activation including signals from FcγRIIA (CD32a) and the Fc common γ chain cytoplasmic tail (utilised by FcγRIA or CD64, and FcαR or CD89) (4). Inhibitory receptors are often paired with activation receptor siblings that compete for the same ligands and regulate inhibitory signals (6). Siglec-14 possesses an almost identical ligand binding domain to Siglec-5, but associates with ITAM-bearing DAP12 adaptor protein and can activate immune cells (12). LAIR-2 is released in a soluble form from neutrophils and can bind the same ligand as LAIR-1 but with a higher affinity, thereby providing a mechanism to reduce LAIR-1 inhibitory signals (13). Consequently, paired receptors modify activation thresholds and help to generate balanced immune responses.
LILRA6 (ILT8/CD85b) and LILRB3 (ILT5/CD85a) are paired receptors expressed by myelomonocytic leukocytes (3, 14). LILRA6 and LILRB3 have high similarity in their extracellular domain and monoclonal antibodies generated against these receptors bind to neutrophils. Yet, as the antibodies are cross-reactive it has remained unclear which of these receptors is expressed in neutrophils and what their role is in neutrophil biology (5). LILRs are a family of 11 receptors with a powerful ability to modulate immune cell activity, function and phenotype (15, 16). LILRA, including LILRA6, possess short cytoplasmic tails, and signal through the ITAMs in the Fc common γ chain. In contrast, LILRB, including LILRB3, possess long cytoplasmic tails containing their own ITIMs. Consequently, binding of ligands to LILRAs or LILRBs can enhance or lower the immune activation threshold. The best-characterized LILRs on neutrophils are LILRA2 and LILRB2 which are demonstrated to induce neutrophil activation upon recognition of microbially cleaved immunoglobulins (17) and to modulate neutrophil phagocytosis and degranulation (9), respectively. Additionally, the mouse orthologue paired-immunoglobulin-like receptor (PIR)-B regulates respiratory burst in neutrophils (18). Characterisation of LILRA6 and LILRB3 in neutrophil biology has been hampered by a lack of LILRA6- and LILRB3-specific monoclonal antibodies (mAb) and because native ligands are unknown.
Antibodies opsonise pathogens to enhance the capacity of phagocytic cells to detect and destroy pathogens (3). IgG and IgA are the dominant antibody isotypes in serum and are recognised by FcγR’s or FcαR, respectively. Receptors expressed on neutrophils are FcγRI, FcγRIIIb and FcαR, that transduce signals through the ITAM of the receptor common γ chain, as well as FcγRIIa and FcγRIIb that transduce signals through their own ITAM and ITIM domains, respectively (3). Inhibitory receptors, including LILRB1, LILRB2 and LILRB4 (19,20), have been shown to inhibit IgG-mediated responses of phagocytes. Other inhibitory receptors, such as LAIR-1 and SIRL-1, inhibit IgA-mediated responses (21,22). Though the established model of ITIM-mediated inhibition of ITAM-driven activation suggests inhibitory receptors modulate both IgG and IgA-mediated functions, confirmatory studies are required. Notably, no studies have assessed the role of LILR to modulate IgA-mediated responses of phagocytes.
Here, we aimed to improve our understanding of LILRA6 and LILRB3 in neutrophil biology. We characterised their expression on resting neutrophils and during neutrophil activation. Next, we investigated their role in modulation of FcαR mediated effector functions, using mouse monoclonal antibodies with cross-linking and agonistic properties as a LILR stimulant. However, as mouse IgG can bind and stimulate human FcγR (23), we additionally used a human FcγR inhibitor called FLIPr-like in our assays to ensure Fab- but not Fc-mediated. This provided opportunities to probe LILR function and their capacity to modulate FcαR-mediated functions on neutrophils. We provide evidence that LILRB3 is expressed at the surface of resting and primed neutrophils, whereby it acts as a potent inhibitor of Fc receptor-mediated effector functions including reactive oxygen species (ROS) production, phagocytosis and killing of microbes. Expression can be recapitulated during differentiation of PLB-985 cells into a neutrophil-like cells, providing a model to improve understanding of the molecular mechanisms of LILRB3. Our data demonstrates that LILRB3 might be considered as the target of therapeutics to modify neutrophil functions and immune responses during disease situations.
Materials & Methods
Expression of recombinant LILR and IgA1 mAb
Signal peptides and extracellular domains of LILR were amplified from cDNA vectors (Table 1), and inserted into pcDNA3.4 vector. Forward primers contained an overhang for pcDNA3.4 vector, a BamHI restriction site, Kozak sequence and LILR-specific region. Reverse primers contained a overhang for pcDNA3.4 vector, a NotI restriction site, stop codon and 6xHis tag. PCR amplifications were performed with Phusion High Fidelty Taq polymerase, and thermocycling as follows:- 1 cycle (98°C for 2mins), 35 cycles (98°C 15 seconds, 62°C 30 seconds and 72°C 30 seconds), 1 cycle (72°C 10 minutes). rLILR was purified by affinity chromatography (ÄKTA Pure, GE Healthcare Life Sciences) using Nickel columns (GE Healthcare Life Sciences). Elutions were dialused into 50 mM Tris 300mM NaCl pH 8 at 4°C.
Table 1. LILR cDNA vectors and cloning vectors.
| cDNA vector | Primers |
|---|---|
| LILRA6 RDC0458 (R&D systems) |
Foward 5’CTCTAGAGGATCGAACCCTTGGATCCACCACCATGACCCCCACCCTCGCAGCCCTGCTCTGCCTAGGGCTGAGTCTGGGCCCCAGG3’ Reverse 5’CTAACCGGTAGGGATCGAACCCTTGCGGCCGCCTAGTGGTGATGGTGGTGATGGCGGATGAGATTCTCCACTGTG3’ |
| LILRB3 HG11978-M (Sino Biological) |
Foward 5’CTCTAGAGGATCGAACCCTTGGATCCACCACCATGACGCCCGCCCTCACAGCCCTGCTCTGCCTTGGGCTGAGTCTGGGCCCCAGGA3’ Reverse 5’CTAACCGGTAGGGATCGAACCCTTGCGGCCGCCTAGTGGTGATGGTGGTGATGCTCCAGGTATCTTCCCAGACCAGG3’ |
| LILRB1 ORF005932 (abmgood) |
Foward 5’CTCTAGAGGATCGAACCCTTGGATCCACCACCATGACCCCCATCCTCACGGTCCTG3’ Reverse 5’CTAACCGGTAGGGATCGAACCCTTGCGGCCGCCTAGTGGTGATGGTGGTGATGCGACCCAGTGGGGGTGAGGGGC3’ |
Vectors encoding anti-WTA-4497-IgA1 or anti-HSA-GA645-IgA1 were constructed by cloning gBlocks containing human IgA1 heavy (HC) and light chain (LC) constant regions into pcDNA3.4 vectors. gBlocks containing variable heavy (VH) and light chain (VL) sequences were cloned into pcDNA3.4-HCIg-hG1 and pcDNA3.4-LCIg-hk, as well as upstream KOZAK sequence and HAVT20 signal peptide, using NheI and BsiWI as the 3’cloning sites for VH and VL, respectively, to preserve the immunoglobulin heavy and kappa light chain amino acid sequence. VH and VL sequences were derived from previously described antibodies directed against staphylococcal wall teichoic acid GlcNAC-β (anti-WTA clone 4497; based on WO2014/193722)) and against human serum albumin (HSA) (anti-HSA clone CA465) (24, 25, 26). Antibodies were expressed as IgG1/Kappa in EXPI293F cells (Life Technologies) essentially as described before (27), and purified by affinity chromatography (ÄKTA Pure, GE Healthcare Life Sciences) using a Protein A column (GE Healthcare Life Sciences). Elutions into 0.1 M Citric acid pH 3.0 were dialysed against PBS at 4°C.
Recombinant (r)LILR and IgA1 were expresed in EXPI293F cells in EXPI media at 37°C with 5% CO2. 50 μg of vector was packaged into 250 μg PEI-MAX 40K (Polysciences, Inc) in 5 ml of Opti-MEM media (Gibco). After incubation at room temperature for 20 minutes, media was added to 50 ml of 2x106 cells/ml and cultured at 37°C with 5% CO2. Supernatants were harvested after 72 hours of culture, and dialysed against 50 mM Tris 300mM NaCl pH 8.
Antibodies
The following unlabelled antibodies were used in this study;- anti-LILRA6 (clone 921330, R&D Biosystems), anti-LILRB1 (clone GHI/75, ITK Diagnostics), anti-LILRB3 (clone 222821, R&D Biosystems), anti-CD89 (clone MIPa, BioRad), mouse IgG1 isotype (Southern Biotech), mouse IgG2a isotype (Southern Biotech). Antibodies conjugated with fluorescent labels used to measure receptor expression in this study are:- anti-mouse-IgG1-PE (Agilent), anti-CD35-PE (clone E11, BD Biosciences), anti-CD63-PE (clone 435, Immunotech), anti-CD11b-APC (BD biosciences), anti-CD66b-FITC (BD Biosciences), anti-CD62L-PE (BD biosciences) anti-CD63-FITC (Immunotech), anti-CD62L-FITC (BD biosciences), anti-CD16-FITC (BD biosciences), anti-CD32-FITC (BD biosciences), anti-CD64-FITC (MIBI) anti-CD89-PE (Serotec) and anti-CD32-PE (Serotec). Hamster anti-mouse-CD3e clone 145-2C11 (BD Biosciences) was used to test CD3ζ-mediated GFP expression in 2B4T cells. Antibodies used for Western blot assays were rabbit anti-LILRB3 pAb (Sino Biological), rabbit IgG and goat anti-rabbit-IgG-HRP (abcam).
Neutrophil isolation and activation
This study was carried out in accordance with the recommendations of METC-protocol 07-125/C approved on 1 March 2010 from the medical ethics committee of the University Medical Center Utrecht with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the 'medical ethics committee of the University Medical Center Utrecht'. Neutrophils were isolated by Ficoll/Histopaque centrifugation as previously described, and resuspended in RPMI1640 supplemented with 0.05% human serum albumin. Neutrophils were isolated on Ficoll/Histopaque gradients with > 98% purity and 99% viability. Secretory vesicle exocytosis was stimulated by incubation at 37°C for 60 minutes. Priming was stimulated by incubation with 10 μg/ml TNFα at 37°C for 60 minutes. Granule exocytosis was stimulated by incubation with 1μM fMLP and 10 μg/ml cytochalasin B for 60 minutes. In certain experiments, neutrophils were pre-incubated on ice with 5 μg/ml FLIPr-like to inhibit FcγR for 20 minutes (28).
Binding of mAb to Dynabeads or cells
Dynabeads (DB) or cells (35 μl of 5 x 106 cells/ml) were incubated with 5 μg/ml of mAb for 1 hr at 4°C. For detection of primary mAb, cells were incubated with 5 μg/ml of PE-conjugated goat anti-mouse-IgG for 1 hr at 4°C. Fluorescence was measured by flow cytometry. DB or cells were gated based on foward and side scatter plots. To produce LILR-coated DB, C-terminal His-tagged rLILR were attached to Dynabeads His-tag isolation & pulldown (ThermoFisher Scientific).
Immunoprecipitation of LILRA6/B3
3 x 107 cells were washed in ice-cold PBS, pelleted and lysed for 30 mins at 4°C using 500 μl of ice-cold 0.3% saponine containing protease inhibitors 1:1000 AEBSF and Leupeptin. After centrifugation for 15 minutes at 4°C, 100 μl of supernatants were incubated at 4°C with 25 μl of Dynabeads Protein A (DB) for Immunoprecipitation (ThermoFisher Scientific) for 2 hours. DB had previously been coated with anti-LILRA6, anti-LILRB3 or isotype controls following standard protocol. DB were washed 3 times in ice-cold lysis buffer, resuspended in sample buffer and heated to 95°C for 5 mins. DB lysates were seperated by SDS-PAGE, blotted onto membranes, probed with rabbit anti-LILRB3 pAb or rabbit IgG and detected with goat anti-rabbit-HRP.
On-bead digestion of immunoprecipitated LILRA6/B3 and mass spectrometry analysis
DB for immunoprecipitation were washed 3 times in ice-cold ultrapure water, resuspended in 50 μl reduction buffer containing 2% (w/v) sodium deoxycholate (SDC), 200mM Tris, 10mM tris(2-carboxyethyl)phosphine (TCEP) and heated in 95°C for 10 mins. Alkylation was done by adding 50μl of 60mM 2-chloroacetamide (CAA) at room temperature. 1μg of Chymotrypsin (Sigma-Aldrich) was added and digestion was performed at 37°C for 16 hours. The supernatant was kept after centrifugation at 20,000 g for 10 minutes at room temperature. SDC was removed by acidic precipitation with 0.5% trifluoroacetic acid (TFA). Peptides were cleaned by Oasis PRiME HLB 96 well plates, following manufacturer’s instructions, dried by SpeedVac and stored at -80°C until MS analysis.
Peptides were separated and analyzed using an Agilent 1290 Infinity HPLC system (Agilent Technologies, Waldbronn, Germany) coupled on-line to a Q-Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Reversed-phase separation was accomplished using a 100 μm inner diameter 2 cm trap column (in-house packed with ReproSil-Pur C18-AQ, 3 μm) (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) coupled to a 50 μm inner diameter 50 cm analytical column (in-house packed with Poroshell 120 EC-C18, 2.7 μm) (Agilent Technologies, Amstelveen, The Netherlands). Mobile-phase solvent A consisted of 0.1% formic acid (FA) in H2O, and mobile-phase solvent B consisted of 0.1% FA in 80% acetonitrile (ACN). Trapping was performed at a flow rate of 5 μl/min for 5 min with 0% B and peptides were eluted using a passive split flow of 300 nl/min for 80 min with 13% to 44% B over 65 min, 44% to 100% B over 3 min, 100% B for 1 min, 100% to 0% B over 1 min, and finally held at 0% B for 10 min. Mass spectra were collected in data-dependent acquisition (DDA) mode, automatically switching between MS and MS/MS. Full scans (from m/z 375 to 1,600) were acquired at a resolution of 60,000 and a target of 3e6 ions or a maximum injection time of 20 ms. Fragmentation was induced for the Top 15 peaks. Target peaks were isolated in a 1.4 Da isolation window and subjected to high-energy collision dissociation (HCD) with a normalized collision energy value of 27%. MS/MS spectra (from m/z 200 to 2,000) were acquired with a resolution of 30,000 using an AGC setting of 1e5 ions or a maximum injection time of 50 ms. Charge state screening was enabled, and precursors with unknown charge state or a charge state of 1 and >6 were excluded.
Raw shotgun LC-MS/MS data were searched with Proteome Discoverer (version 2.2, Thermo Scientific) using the Mascot search engine (version 2.6.1). Processing nodes included spectrum file reader, Minora feature detector (for LFQ), spectrum selector, Mascot and Percolator. Mascot searches were performed against a combined database of UniProt Swiss-Prot database: Homo sapiens (canonical and isoform) (September 2019, 20,417 entries) and manually checked variants of LILRA6/B3 (September 2019, 29 entries). Searches were performed with fixed Cys carbamidomethylation and variable Met oxidation and Ser/Thr phosphorylation of peptides. Chymotrypsin was chosen for cleavage specificity with a maximum of two missed cleavages allowed. The searches were performed using a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.05Da (HCD), followed by data filtering using Percolator, resulting in 1% false discovery rate (FDR). Mascot scores >20 were accepted. The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015561 and 10.6019/PXD015561 with the following login information: Username: reviewer73469@ebi.ac.uk; Password: spzXpdnf.
Detection of LILRB3 in cell supernatants
3 x 107 cells were pelleted. Supernatants were concentrated 10X on a 3 kDa ultra centrifugual filter column (Amicon). Supernatants were mixed 1:1 with 2X sample buffer, incubated at 95°C for 10 mins, seperated by SDS-PAGE and blotted onto membranes. After blocking, membranes were washed, probed with 1:5000 rabbit anti-LILRB3 pAb or rabbit IgG control, washed, probed with 1:10,000 goat anti-rabbit-IgG-HRP.
Culture and differentiation of PLB-985 cells
PLB-985 cells were cultured in RPM1640 supplemented with 10% FCS at 37°C with 5% CO2. To diffentiatie towards a neutrophil-like phenotype, PLB-985 cells were seeded at a density of 1 x 105 cells/ml, and cultured in the presence of 1.25% DMSO for up to 5 days (23, 29). To lyse DMSO-differentiated PLB-985 cells, 1 x 107 cells were washed in ice-cold PBS, pelleted and lysed for 30 mins at 4°C using 500 μl of ice-cold 0.3% saponine containing protease inhibitors 1:1000 AEBSF and Leupeptin. Immunoprecipitation of LILRB3 from PLB-985 cell lysates, and subsequent detection of LILRB3, was performed using the same methodologies as for neutrophils.
Construction of LILR reporter 2B4T cells
2B4 NFAT-GFP T cell reporter cells contain a NFAT-GFP construct composed of three tandem NFAT-binding sites fused with enhanced GFP cDNA (30). To express fusion proteins, DNA fragments containing the coding domain sequence (CDS) of the extracellular and transmembrane domains of LILRA6 and LILRB3 and the CD3ζ cytoplasmic tail were synthesized by Integrated DNA Technologies, and were ligated into a dual promoter lentiviral vector (BIC-PGK-Zeo-T2a-mAmetrine; RP172 derived from no.2025.pCCLsin.PPT.pA.CTE4⋅ scrT.eGFP.mCMV.hPGK.NG-FR.pre, as previously described) via Gibson reaction (31). Vectors containing correct inserts were confirmed by sequencing. The RP172 vector contains human EF1A promoter for potent expression of the cloned downstream gene and the selection marker for zeocin resistance. Lentiviral particles were created in HEK293T cells that were seeded at 6.25 x 104 cells/ml in RMPI1640 + 10% FCS+ 100 μg/ml penicillin and 100 μg/ml streptomycin and cultured at 37°C + 5% CO2 for 24 hours. 50 μl of RMPI1640 and TransIT-LT1 transfection reagent (MirusBio, USA) were mixed and incubated for 5 minutes at room temperature. Subsequently, 0.25 μg packaging mix (containing equal parts of pVSV-G, pMDL and pRSV vectors) and 0.25 μg of RP172 vector were added, mixed and incubated at room temperature for 20 minutes. This mixture was added to 1 ml of HEK293T cells and incubated at 37°C and 5% CO2 for 72 hours. 100 μl of the supernatant containing lentivirus particles was added to 500 μl of 1 x 105 cells/ml of 2B4T cells in RPMI 1640 supplemented with 10% FCS + 100 μg/ml penicillin, 100 μg/ml streptomycin and 8 μg/ml polybrene. After centrifugation at 2000 rpm for 90 minutes at 33°C, cells were supplemented with 500 μl of RPMI 1640 containing 10% FCS + 100 μg/ml penicillin and 100 μg/ml streptomycin and incubated at 37°C + 5% CO2. After 72 hours, cells were supplemented with an 5 ml of RPMI 1640 containing with 10% FCS, 100 μg/ml penicillin, 100 μg/ml streptomycin and 400 μg/ml zeocin in order to select for transfectants for 14 days. Cell cultures were formed and LILR expression was measured using anti-LILR mAb and flow cytometry analysis.
Induction of GFP expression in LILRCD3ζ reproter 2B4T cells
96-well plates were coated at 4°C overnight with 40 μl of 15 μg/ml anti-CD3ζ, anti-LILRA6, anti-LILRB3 or respective isotype control mAbs diluted in NaHCO3 buffer. 200 μl of 2B4T cells were seeded in 96-well plates at 2.5 x 105 cells/ml in RPMI 1640 containing 10% FCS, 100 μg/ml penicillin and 100 μg/ml streptomycin and cultured at 37°C + 5% CO2. After 18 hours, cells were resuspended in PBS and their fluorescence was measured by flow cytometry.
Measurement of neutrophil activation
White 96-well plates were coated at 4°C overnight with 40 μl of 5 μg/ml anti-CD89, anti-LILRB3, anti-LILRA6, mouse IgG1 isotype or mouse IgG2a isotype diluted in NaHCO3 buffer. Neutrophils were resuspended in IMDM + 0.05% HSA to 2.5 x 106 cells/ml. 50 μl of neutrophils were incubated on wells. After incubation at room temperature for 30 minutes, 100 μl of pre-warmed luminol balanced salt solution (LBSS; final concentration of luminol 220 μM) was added to wells after washing. The level of ROS production was determined as area under the curve, with background subtraction of neutrophils only, during 60 minutes.
Assessing modulation of IgA-mediated neutrophil activation
Clear 96-well plates were coated at 4°C overnight with 40 μl of 5 μg/ml anti-LILRA6 or mouse IgG1 isotype diluted in NaHCO3 buffer. Neutrophils were resuspended in IMDM + 0.05% HSA to 2.5 x 106 cells/ml. 50 μl of neutrophils were incubated on wells coated with anti-LILRA6, anti-LILRB3, mouse IgG1 or IgG2a mAb for 60 minutes at room temperature. After resuspension, neutrophils were transferred to wells of white plates that contained 100 μl of LBSS in order to measure luminescence. These wells had previously been coated with anti-CD89 or mouse IgG1 isotype control (40 μl of 5 μg/ml, incubated at 4°C overnight). The production of ROS was measured via luminescence detection at 2 minute intervals. The level of ROS production was determined as area under the curve, with background subtraction of neutrophils only, during 60 minutes.
IgA-mediated phagocytosis
Dynabeads M280 Streptavidin (DBs, ThermoFisher Scientific), which have a 2.8 μm diameter, were FITC-labelled by incubation for 30 mins in 500 μg/ml FITC. After washing, DBs were coated with biotinylated human serum albumin (100 μg/ml) following standard protocol. DBs were opsonised with anti-HSA-IgA1 or anti-WTA-IgA1 by incubation for 30 minutes at room temperature. Bound IgA was detected using 0.3 μg/ml anti-IgA-AF647 and flow cytometry. To measure phagocytic uptake, DBs were mixed at an multiplicity of infection (MOI) of 10:1 with 40 μl of 2.5 x106 neutrophils/ml, and incubated for 30 mins at 37°C with 200 rpm. Neutrophils were washed and fixed in 1% PFA. Fluorescence of neutrophils was measured by flow cytometry, and the % of FITC-positive neutrophils was measured. In both ROS production and phagocytosis experiments, neutrophils were pre-incubated for 60 minutes at 37°C + 5% CO2 on 96-well plates coated at 4°C overnight with anti-LILRA6 or mouse IgG1 isotype mAb.
IgA-mediated phagocytosis and killing of S. capitis
S. capitis ATCC 28740 and ATCC 28740-H strains were cultured in Tryptic Soy Broth (supplemented with 10 mg/ml chloramphenicol if required) at 37°C until absorbance at OD600 = 0.4. The S. capitis ATCC 29740-H carries the plasmid pRB474-tarFI2J2L2S which encodes the operon required for expression of polymer ribitol poly-phosphate (poly-RboP) wall teichoic acid (WTA) backbone with β-linked N-acetylglucosamine (β-O-GlcNAc) modifications (32). Thus, ATCC 29740-H is a hybrid strain expressing native WTA (poly-glycerolphosphate (poly-GroP) with α-O-N-acetylgalactosamine (GalNAc) modifications) and foreign WTA (poly-RboP and β-O-GlcNAc modifications). Bacteria were incubated with IgA1 mAb for 30 mins on ice. anti-hIgA-AF648 and flow cytometric analysis was used to detect surface bound IgA1. To FITC-label S. capitis, mid-logathrithmic phase cutures were incubated in 10 mg/ml FITC for 30 minutes on ice, and washed 5 times in PBS + 0.1% BSA. To assess IgA-dependent phagocytic uptake, FITC-labelled bacteria (5 μl of 3 x 108 CFU/ml) and neutrophils (45 μl of 3 x106 cells/ml) were mixed at an MOI of 10:1, were centrifuged at 500 × g for 3 mins to initiate the assay and were incubated for 30 mins at 37°C + 5% CO2 with 200 rpm. After washing, fluorescence of neutrophils was measured by flow cytometry.
To measure killing, bacteria (5 μl of 1 x 108 CFU/ml) and neutrophils (45 μl of 1 x107 cells/ml) were mixed at an MOI of 10:1 and incubated for 60 minutes at 37°C with 5% CO2. Reactions were stopped by addition of 200 μl of 0.3% ice-cold saponine and incubation on ice for 15 mins. CFU/ml were enumerated by serial dilution and culture of Brain Heart Infusion agar plates. All assays were performed in RPMI 1640 supplemented with 0.05% heat-inactivated human pooled serum. In both phagocytosis and killing experiments, neutrophils were pre-incubated for 60 minutes at 37°C on 96-well plates previously coated at 4°C overnight with anti-LILRA6 or mouse IgG1 isotype mAb. Data sets were tested for normal distributions using Shapiro-Wilk normality test, and analysed using parametric or non-parametric approaches as described in figure legends.
Results
LILRB3 is expressed by human neutrophils
We first assessed anti-LILRA6 and anti-LILRB3 mAb cross-reactivity. Recombinant extracellular domains of LILA6, B1 and B3 were expressed in a eukaryotic expression system. Both anti-LILRA6 and anti-LILRB3 bound to rLILRA6- and rLILRB3-, but not rLILRB1-, coated magnetic beads (Fig. 1A; 1B). This indicated anti-LILRA6 and anti-LILRB3 are cross-reactive. anti-LILRA6 bound better to rLILRA6 and anti-LILRB3 had enhanced binding to rLILRB3. This could be due to differences in affinity or glycosylation.
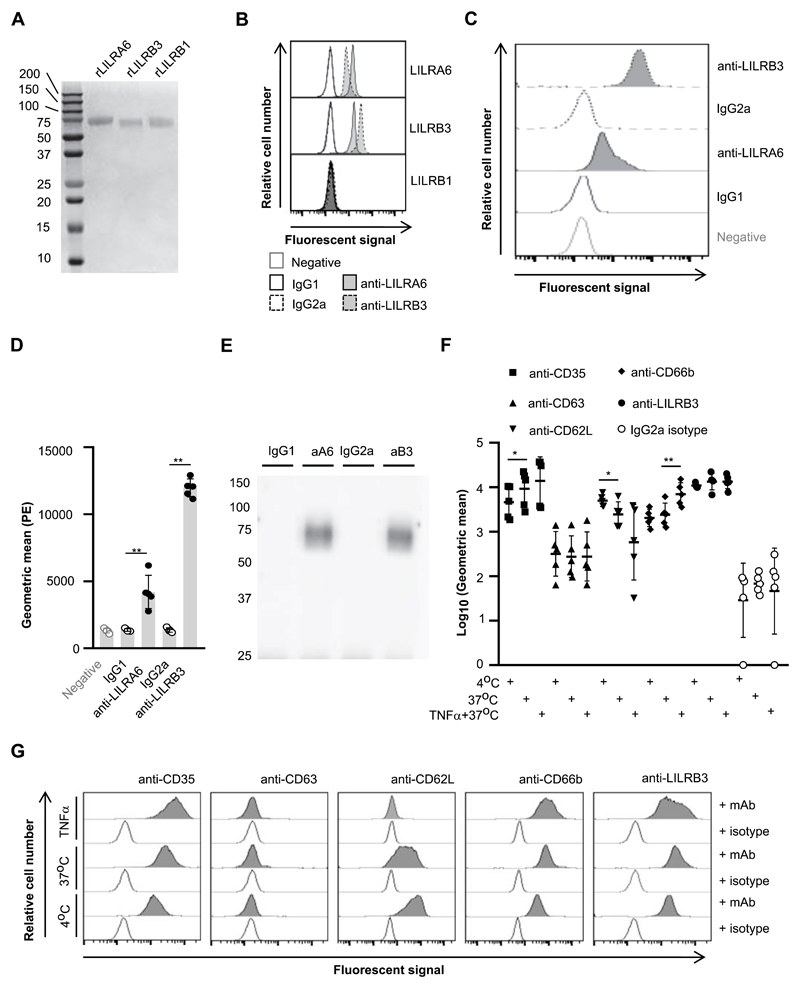
Figure 1. LILRB3 is expressed on the surface of resting neutrophils, and is down-regulated upon priming.
(A) Seperation of recombinant (r)LILRA6, LILRB3 and LILRB1 purified from eukaryotic expression system by SDS-PAGE. (B) Binding of anti-LILRA6, anti-LILRB3 and isotype control mAb to Dynabeads coated with rLILR. Representative flow cytomerty plots of n = 3 are shown. (C and D) Binding of anti-LILRA6, anti-LILRB3 and isotype control mAb to resting neutophils, using anti-IgG-PE as a seconday mAb. A representative experiment (C) and the integrated results from five separate experiments (D) were compared by Student t-test, where * = p < 0.05, ** = p < 0.01. (E) Immunoprecipitation of LILRA6 and/or LILRB3 from the surface of resting neutrophils. Neutrophil lysates were incubated with 5 μg/ml anti-LILRA6 (aA6), anti-LILRB3 (aB3), IgG1 or IgG2a (i) and DB protein G. Immunoprecipitated proteins were eluted from beads, seperated via SDS-PAGE, blotted onto membranes and detected using rabbit anti-LILRB3 pAb or rabbit IgG and goat anti-rabbit-IgG-HRP. Data is representative of n = 3 independent experiments. (F and G) Priming of neutrophils with TNFα does not alter LILRA6/B3 expression levels. Neutrophils were incubated at 4°C (resting), at 37°C (secretory vesicles exocytosed) or 37°C in the presence 5 μg/ml TNFα (primary granules exocytosed). Anti-IgG-PE used as a secondary mAb for anti-LILRB3 and IgG2a isotype control. Integrated results from five separate experiments (F) were compared by Student t-test, where * = p < 0.05, ** = p < 0.01. Data from a representative experiment are shown (G).
Next, we investigated LILRA6 and LILRB3 expression on primary neutrophils. Data from 5 healthy human donors was analyzed as geometric mean fluorescence in comparison to neutrophils stained with isotype control mAbs. Average geometric mean was 4729 and 1415 for anti-LILRA6 vs IgG1 isotype (p < 0.01), and 12190 vs 1459 for anti-LILRB3 and IgG2a isotype (p < 0.01) (Fig. 1C and 1D). LILRA6 (50 kDa) and LILRB3 (66.9 kDa) can be discriminated based on size due to differences in the length of their cytoplasmic tails. We immunoprecipitated receptors using neutrophil lysates in combination with anti-LILRA6, anti-LILRB3 or isotype control mAb and protein G coated beads. Immunoprecipitated proteins were separated by SDS-PAGE and blotted onto membranes. Using anti-LILRB3 pAb for probing, a single protein band of around 75 kDa was detected after immunoprecipitation with anti-LILRA6 and anti-LILRB3 (Fig 1E). As a control we confirmed that anti-LILRB3 pAb was able to detect both rLILRA6 and rLILRB3 by Western blotting (Supplementary Figure 1A). Collectively, the data suggested that one receptor, most likely LILRB3, was expressed on neutrophils at detectable levels. To confirm the identity of the protein, we performed mass spectrometry analysis of immunoprecipitated proteins from neutrophils lysates. A total of 21 LILRA6 or LILRB3 peptides were detected in samples immunoprecipitated using anti-LILRA6 and anti-LILRB3 mAb, which were absent in samples immunoprecipitated using control anti-IgG1 and anti-IgG2a mAb. Alignment of peptide sequences of all full-length LILRA6 and LILRB3 sequences in the UniProt database revealed 0 and 7 peptides were LILRA6- and LILRB3-specific respectively. Notably, peptides 5 (QRPAGAAETEPKDRGLL) and 6 (RRSSPAADVQEENLY) mapped to the cytoplasmic ITIM domain of LILRB3, and were found in samples from all 5 donors (Table 2; Supplementary Figure 1B). Therefore, mass spectrometry analysis indicated that neutrophils from 5/5 donors expressed LILRB3 and 0/5 donors expressed LILRA6. This is consistent with previous proteomic analysis revealing LILRB3 to be a major constituent of secretory granules (SVs) (33).
Table 2. Mass spectrometry detection of LILRA6 and LILRB3 in neutrophil lysates following immunoprecipitation.
Peptide sequences as follows 1 = QCGSDVGYNRF, 2 = TLQCGSDVGYNRF, 3 = DSQQLHSRGF, 4 = SSAGWSEPSDPLEMVMTGAY, 5 = QRPAGAAETEPKDRGLL, 6 = RRSSPAADVQEENLY, 7 = RLHKEGSPEPL
| Peptide | Donor 1 | Donor 2 | Donor 3 | Donor 4 | Donor 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Anti-A6 | Anti-B3 | Anti-A6 | Anti-B3 | Anti-A6 | Anti-B3 | Anti-A6 | Anti-B3 | Anti-A6 | Anti-B3 | |
| 1 | 7.45E+06 | 1.13E+06 | ||||||||
| 2 | 3.39E+07 | 7.79E+06 | ||||||||
| 3 | 5.09E+07 | 1.12E+07 | ||||||||
| 4 | 1.70E+06 | 4.28E+05 | ||||||||
| 5 | 7.17E+06 | 3.65E+06 | 1.04E+07 | 1.15E+07 | 3.03E+07 | 5.39E+07 | 2.36E+07 | 6.10E+07 | 7.70E+07 | 1.82E+07 |
| 6 | 3.42E+05 | 1.55E+06 | 3.09E+06 | 1.89E+06 | 2.34E+06 | 6.23E+06 | 1.17E+07 | 1.40E+06 | ||
| 7 | 4.27E+07 | 1.14E+07 | ||||||||
Immune receptors can also be expressed in secretory vesicles (SVs) or granules. We compared LILRB3 expression between resting neutrophils and those incubated at 37°C, which is enough to mobilize SV release (34). Incubation at 37°C induced up-regulation of SV marker CD35, but not granule marker CD63 (Fig. 1F & 1G). Additionally, CD62L was down-regulated. There was no significant change in LILRB3 expression (Fig. 1G). Incubating neutrophils with 5 μg/ml TNFα induced up-regulation of gelatinase granule marker CD66b, but no significant up- or down-regulation of CD63, CD35 nor CD62L (Fig. 1F& 1G). Mean LILRB3 surface content did not change upon TNFα priming. Individual flow cytometry plots suggested LILRB3 levels were more heterogenous following TNFα priming (Fig. 1G). This could mean LILRB3 is differentially regulated in sub-populations. Alternatively, there could have been simultaneous up- and down-regulation through TNFα priming associated mechanisms.
PLB-985 cell differentiation towards a neutrophil-like phenotype is associated with LILRB3 up-regulation
The expression of LILRB3 on the surface of resting neutrophils suggests LILRB3 may function during neutrophil differentiation. Recently, up-regulation of LILRA6/B3 transcripts was reported during differentiation of the promyeloid tumor cell line PLB-985 towards a neutrophil-like phenotype (29). We examined LILRA6/B3 expression during neutrophil-like differentiation of PLB-985 cells. Undifferentiated PLB-985 cells expressed CD32 and CD89 but not CD16 nor CD11b, as reported (Fig. 2A) (21, 29). Fluorescence of undifferentiated PLB-985 cells was elevated for anti-LILRB3 compared to IgG2a isotype (Fig. 2A; Supplementary Fig. 2A), suggestive that LILRA6/B3 are expressed at low basal levels on undifferentiated PLB-985 cells.
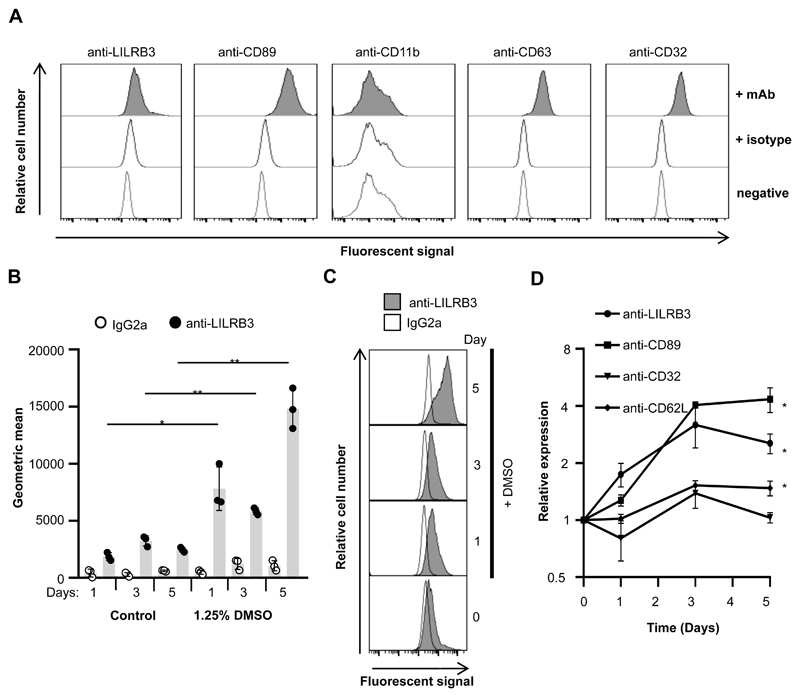
Figure 2. Differentiation of neutrophil-like PLB-985 like is associated with enhanced LILRA6/B3 surface expression.
(A) Undiffereniated PLB-985 cells express low levels of LILRA6/B3. PLB-985 cells were cultured in RPMI 1640 at 37°C with 5% CO2, and stained with primary antibodies. Anti-IgG-PE was used as a secondary mAb for anti-LILRB3 and IgG2a isotype control. A representative experiment from 3 sperate experiments is shown. (B, C and D) DMSO-differentiated PLB-985 cells expressed enhanced surface levels of LILRA6/B3. PLB-985 cells were cultured in the presence or absence of 1.25% DMSO at 37°C with 5% CO2 for up to 5 days. Cells were stained with mAb and fluouresence was measured by flow cytometry. Integrated results from three separate experiments (B) were compared by Student t-test, where * = p < 0.05, ** = p < 0.01. Data from a representative experiment are shown (C). Relative receptor expression on DMSO-differentiated PLB-985 populations to undifferentiated PLB-985 populations (D) was plotted, where Student t test was used to compare the relative receptor expression between day 0 and day 5.
Culture of PLB-985 cells with 1.25% DMSO for 5 days lead to increased CD89 and CD11b surface expression (Supplementary Fig. 2B), which was consistent with previous reports of neutrophil-like differentiation (22). LILRA6/B3 was up-regulated during differentiation with 1.25% DMSO (Fig. 2B, Fig. 2C), and after 5 days of differentiation there was a 2.54±0.3-fold up-regulation. This was concomitant with up-regulation of CD89 and CD62L (Fig. 2D).
To confirm expression of LILRA6 and/or LILRB3 in PLB-985 cells, we probed proteins immunoprecipitated from DMSO-differentiated PLB-985 cells by Western blot. Using rabbit anti-LILRB3 pAb for secondary detection, we found the presence of a single band at 75 kDa, most likely LILRB3, following immunoprecipitation with anti-LILRA6 and anti-LILRB3 mAb, but not respective isotype controls. We believe that the secondary antibody detected light and heavy chain Ig at band sizes 25 and 50 kDa, respectively. Thus, LILRB3 expression was up-regulated in PLB-985 cells cultured in the presence of 1.25% DMSO in comparison to control cells.
LILRB3 is down-regulated upon neutrophil activation
As mature neutrophils alter the composition of their surface receptors upon exposure to environmental stimuli, we next investigated the surface expression of LILRB3 during neutrophil activation. Fluorescence of neutrophils incubated at 37°C or 37°C in the presence of fMLP and cytochalasin B was normalised to fluorescence of neutrophils incubated at 4°C. Incubation at 37°C with fMLP and cytochalasin B lead to CD63 up-regulation, CD35 down-regulation, CD66b up-regulation and CD62L down-regulation (Fig. 3A, Fig. 3B), which are typical markers of neutrophil degranulation. Degranulation was associated with LILRB3 down-regulation (log10 geometric mean 4.12±0.18 vs 3.16±0.45, n = 5, p < 0.05). In addition, soluble LILRB3 was detected in supernatants of neutrophils incubated with fMLP and cytochalasin B but not buffer control (Fig. 3C).
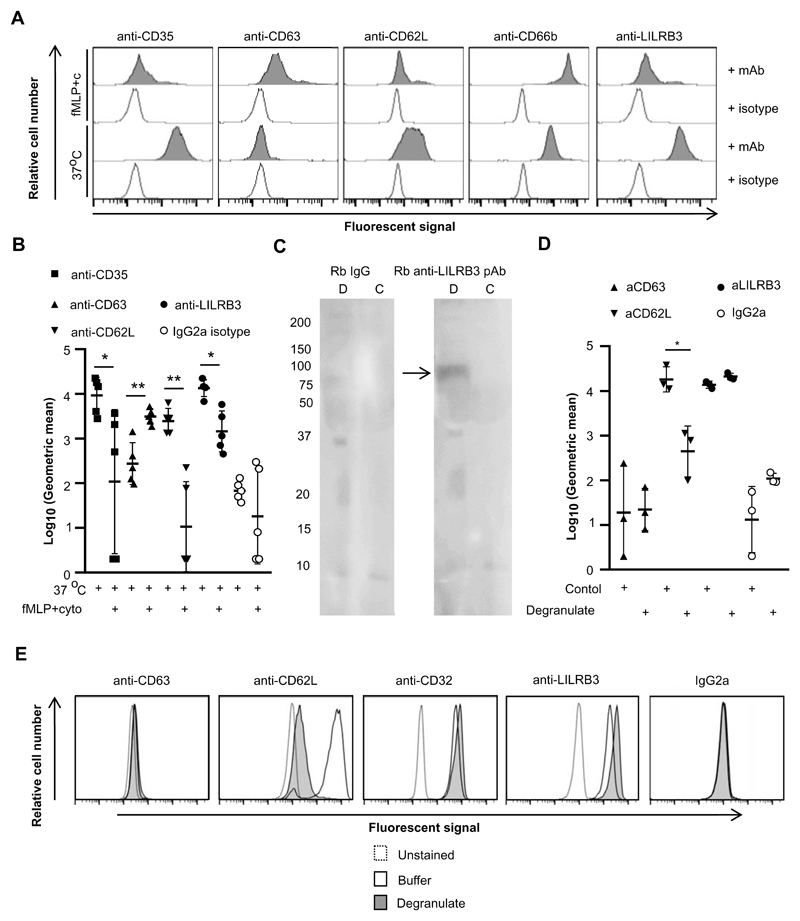
Figure 3. Neutrophil activation is associated with down-regulation of LILRB3 from the surface.
(A and B) Degranulation of neutrophils induced down-regulation of surface LILRB3 expression levels. Neutrophils were incubated at 37°C (secretory vesicles exocytosed) or 37°C in the presence of fMLP and cytochalsin B (granules exocytosed) and then stained for receptor expression. Anti-IgG-PE was used as a secondary mAb for anti-LILRB3 and IgG2a isotype control. A representative experiment (A) and the integrated results from five separate experiments (B) are shown. The MFI of neutrophil populations was normalized against unstained control neutrophils within each condition. Background level of receptor expression on neutrophils incubated at 37°C was normalized to 1 (37°C/37°C). Relative surface expression of receptors was calculated at 37°C in presence of fMLP and cytochalasin B in comparison to 37°C for granule release. (C) LILRB3 is detected in the supernatant of degranulated neutrophils. Supernates representing neutrophils degranulate (D; incubation in presence of fMLP and cytochalasin B at 37°C) or buffer control (C, incubation in 37°C) were seperated by SDS-PAGE, blotted onto membranes and probed with rabbit anti-LILRB3 pAb or rabbit IgG, and detected using goat anti-rabbit-IgG-HRP. Data representative of n = 3. (D and E) Proteases in neutrophil degranulate are not sufficient to induce down-regulation of surface LILRB3 expression. Neutrophils were incubated in the supernatant of previously degranulated neutrophils or control buffer for 1 hr at 37°C, and were then stained for receptor expression. Anti-IgG-PE used as a secondary mAb for anti-LILRB3 and IgG2a isotype control. Integrated results from three separate experiments (D) compared log10(Geometric mean fluorescence) values between neutrophils incubated at 37°C in degranulate or control buffer by Student t-test, where * = p < 0.05, ** = p < 0.01. Representative flow cytometry plots are shown (E).
Proteases contained in granules can cleave immune receptors, including CD62L, from the cell surface upon degranulation. To understand whether proteases released during degranulation cleaved LILRB3 from the neutrophil surface, we compared receptor expression on neutrophils incubated in the supernatant of previously degranulated neutrophils or a buffer control. Flow cytometry revealed no change in CD63 expression, indicating that the supernatant of degranulated neutrophils alone did not induce degranulation of resting neutrophils (Fig. 3D and 3E). The supernatant of degranulated neutrophils, but not buffer, induced down-regulation of CD62L. This indicated that proteases in the supernatant were active. However, LILRB3 was not down-regulated indicating that proteases contained within granules were not enough to release soluble LILRB3 (Fig. 3D and 3E).
Ligation of LILRB3 does not activate neutrophils
Inhibitory receptors can modify activation thresholds. 2B4 Nuclear Factor of Activated T cell–Green Fluorescent Protein (NFAT-GFP) reporter cells expressing chimeric proteins, consisting of extracellular and transmembrane domains of immune receptors fused to the CD3ζ cytoplasmic tail, have been utilised to identify agonists of immune receptors including LAIR-1 and LILR (35, 36). We developed 2B4T NFAT-GFP reporter cells to identify ligands that could efficiently cross-link LILRA6 and/or LILRB3 (Supplementary Figure 3A). Surface expression of chimeric proteins, composed of LILR extracellular and transmembrane domains fused to the CD3ζ cytoplasmic tail containing 3 x ITAM motifs, was detected in transduced cells (Supplementary Fig. 3B). GFP-expression was induced in 2B4T cell populations by stimulating native CD3 with anti-CD3 mAb (Supplementary Fig. 3C & 3D). Next, we tested agonistic effects of anti-LILR mAb (Fig. 4A and B). After incubation on anti-LILRA6 mAb coated wells, significantly more LILRA6CD3ζ–expressing 2B4T cells were GFP positive compared to control 2B4T cells. Likewise, anti-LILRB3 induced GFP production in a larger proportion of LILRB3CD3ζ–expressing 2B4T cells compared to control cells. Both anti-LILRA6 and anti-LILRB3 activated both LILRA6 and LILRB3 fusion proteins (Fig. 4A), indicating agonistic effects are cross-reactive. Notably, anti-LILRA6 was a better agonist of both receptors than anti-LILRB3. Differences in agonistic properties could be due to mAb-target affinities or glycosylation.
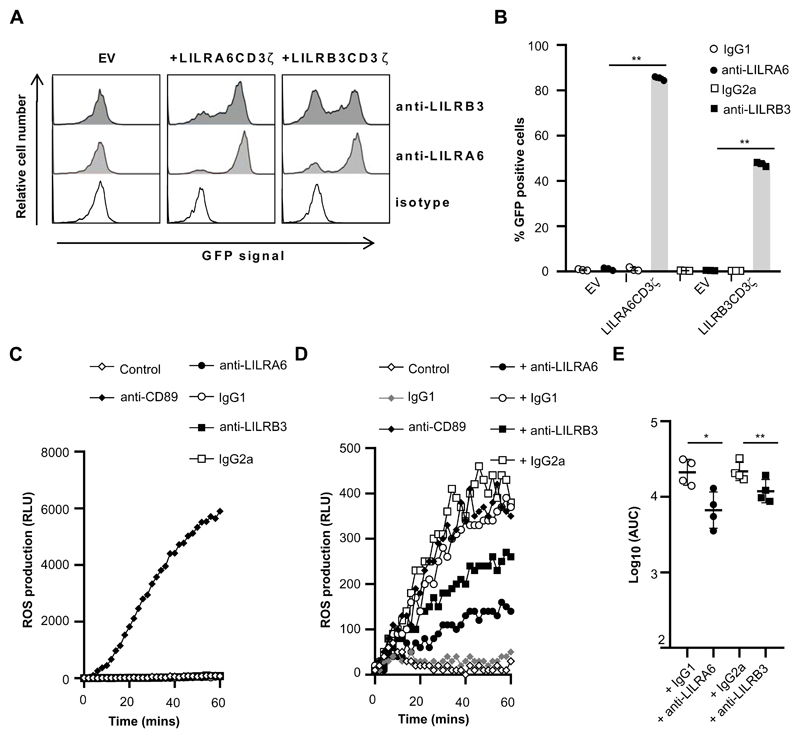
Figure 4. Cross-linking of LILRB3 suppresses FcαR mediated activation.
(A and B) GFP-expression in 2B4T cells expressing LILRCD3ζ (A6, B3) fusion proteins or empty vector (EV) control 2B4T cells following incubation at 37°C. Plates were previously coated with anti-LILRA6, anti-LILRB3 or isotype mAb. These mAb served as the agonist in these assays. Cells were incubated in coated wells for 18 hours, and GFP expression was measured by flow cytometry analysis. A representative experiment (A) and the integrated results from three separate experiments (B) were compared by Student t-test, where * = p < 0.05, ** = p < 0.01. (C) Cross-linking of LILRB3 does not induce ROS production by neutrophils. Neutrophils were incubated in the presence of 5 μg/ml FLIPr-like for 20 minutes, and then incubated on plates containing luminol. Plates were previously coated with anti-CD89, anti-LILRA6, anti-LILRB3 or isotype mAb. These mAb served as the agonist in these assays, and relative luminescence units (RLU) was measured over 60 minutes as an indicator of ROS production. (D and E) Cross-linking of LILRB3 suppresses CD89-mediated ROS production by neutrophils. Neutrophils (n = 4) were incubated on plates previously coated with anti-LILRA6, anti-LILRB3, IgG1 or IgG2a mAb. After 1 hour, neutrophils were stimulated through FcaR/CD89 by incubation on plates containing luminol that were previously coated with anti-CD89 or IgG1 isotype control. RLU was measured over 60 minutes as an indicator of ROS production. A representative experiment (D) and the integrated results from four separate experiments (E) were compared by Student t-test, where * = p < 0.05, ** = p < 0.01.
Neutrophils produce significant levels of ROS by cross-linking activation receptors including CD32 (FcγR2A), CD89 (FcαR) or LILRA2. We tested whether continuous ligation of LILRB3 could induce neutrophil activation and ROS production. Exposure of neutrophils to anti-CD89 mAb lead to increased luminescent signals and ROS production (Fig. 4C). Incubation of neutrophils on anti-LILRA6- or anti-LILRB3-coated wells did not induce enhanced luminescence signals (Fig. 4C). Data were consistent for neutrophils from 5 donors, indicative that LILRB3 cross-linking does not activate ROS production.
LILRB3 inhibits Fc receptor mediation activation
We aimed to test whether LILRB3 could modulate FcR-mediated ROS production using anti-LILRA6 and anti-LILRB3 mAb. Exposure of neutrophils to mouse IgG1 or IgG2a coated wells induced significant luminescence signals (Supplementary Fig. 4A). This is likely induced through cross-linking of human FcγR via the Fc region of mouse IgG. Neutrophils exposed to anti-LILRA6 IgG1, which is an agonist of LILRB3, produced significantly lower luminescent signals compared to neutrophils exposed to control IgG1 mAb (Supplementary Fig. 4B and 4C). Anti-LILRA6 treatment induced a 4.54-fold down-regulation of IgG1-mediated ROS production across donors, implying LILRB3 cross-linking suppresses FcγR-mediated activation.
Next, we investigated whether LILRB3 could modulate the activation of neutrophils through FcαR. To minimise mouse IgG background activation of human FcγR, we pre-treated neutrophils with FLIPr-like which can bind human FcγR and inhibit interactions with IgG (29). This ensured cross-linking of receptors (CD89 and LILRB3) via Fab regions of mAb, whilst limiting cross-linking of FcγR via Fc regions of mAb. Indeed, FLIPr-like inhibited activation of neutrophils via mouse IgG1 and IgG2a (Supplementary Figure 4A). Furthermore, ROS production induced by anti-CD89 mAb was reduced in the presence of FLIPr-like (Supplementary Figure 4D).
There was a significant reduction in FcαR-mediated ROS production in neutrophils pre-treated with anti-LILRA6 compared to IgG1 mAb (Fig. 4D & 4E). Likewise, anti-LILRB3 induced a significantly lower level of FcαR-mediated ROS production compared to IgG2a. Anti-LILRA6 induced a 3.22-fold decrease in FcαR-mediated ROS production compared to IgG1, whilst anti-LILRB3 induced a 1.70-fold decrease in FcαR-mediated ROS production compared to IgG2a. This indicated that LILRB3 could inhibit FcαR-mediated activation, and that anti-LILRA6 had a more potent effect than anti-LILRB3.
LILRB3 inhibits FcαR mediated phagocytosis
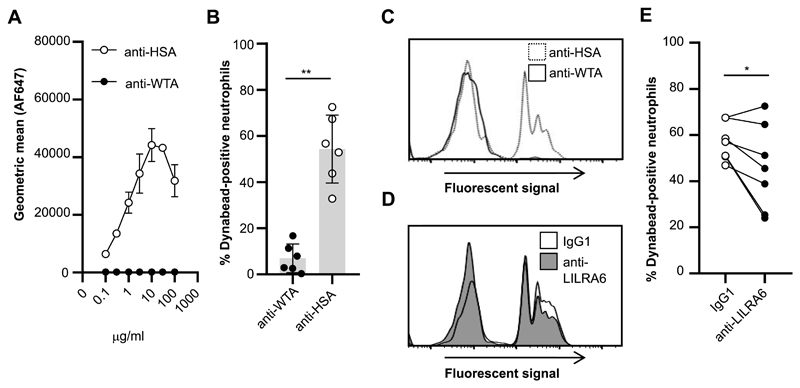
The finding that LILRB3 ligation inhibited FcαR-mediated activation suggested that LILRB3 may modulate a broader repertoire of antibody-mediated effector functions. We investigated whether LILRB3 ligation could modulate phagocytic uptake and microbial killing. The ability of LILRB3 to modulate IgA-mediated phagocytosis was tested using IgA1-opsonised microparticles. FITC-labelled dynabeads were coated with human serum albumin (HSA) and opsonised with anti-HSA-IgA1 or isotype control (Fig. 5A). There was a significant increase in % of fluorescent neutrophils incubated with anti-HSA-IgA1 opsonised dynabeads compared to isotype opsonised dynabeads (Fig. 5B and 5C). There was a significant reduction in phagocytic uptake of IgA1-coated dynabeads for neutrophils pre-incubated on anti-LILRA6 compared to IgG1- coated wells (Fig. 5D and 5E). This indicates LILRB3 stimulation can inhibit FcαR-mediated phagocytosis.
Figure 5. LILRB3 inhibits FcαR-mediated phagocytosis.
(A) Concentration-dependent binding of anti-HSA-IgA1 but not an isotype control to HSA-coated dynabeads, n = 3. (B and C) Phagocytic uptake of IgA-opsonised DB by neutrophils at an MOI of 10:1. Neutrophils were incubated in the presence of 5 μg/ml FLIPr-like for 20 minutes, and then incubated in the presence of IgA1-opsonised FITC-labelled dynabeads for 30 minutes at 37°C with 5% CO2. Fluorescence of neutrophils was measured by flow cytometry analysis. The % of fluorescent neutrophils was calculated for each neutrophil population exposed to IgA1-opsonised dynabeads. The integrated results from six separate donors (B) were compared by Student t-test, where * = p < 0.05, ** = p < 0.01, and representative flow cytometry plots are shown (C). (D and E) Continuous ligation of LILRB3 reduces phagocytic uptake of microparticles. Neutrophils were incubated in the presence of 5 μg/ml FLIPr-like for 20 minutes, and then incubated on anti-LILRA6 or IgG1 mAb coated plates. After 1 hour, neutrophils were incubated in the presence of IgA1-opsonised FITC-labelled dynabeads for 30 minutes at 37°C with 5% CO2. A representative experiment (D) and the integrated results from seven separate experiments (E), in which data from the same donor are linked, were compared by Student t-test, where * = p < 0.05, ** = p < 0.01.
LILRB3 inhibits FcαR-mediated antimicrobial effector functions
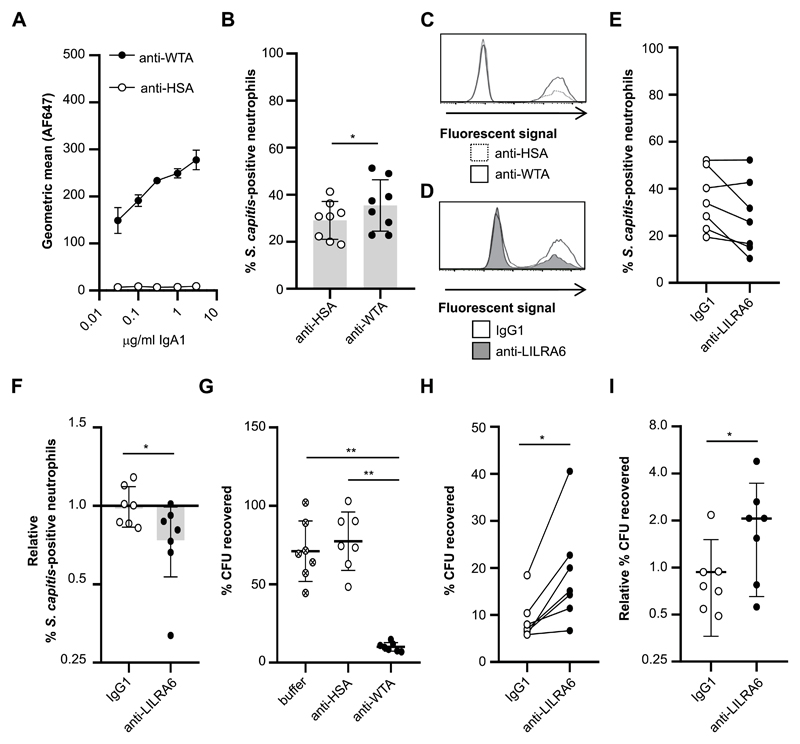
As appropriate levels of specific IgA can induce opsonisation, phagocytosis and killing of bacteria (37, 38), we assessed whether LILRB3 ligation can modulate IgA-mediated phagocytosis of live bacteria by neutrophils. Anti-WTA-4497 recognises β-linked N-acetylglucosamine (β-O-GlcNAc) modifications on the polymer ribitol poly-phosphate (poly-RboP) WTA backbone of S. aureus (24, 25, 32). Since S. aureus has evolved multiple mechanisms to evade neutrophil phagocytosis and killing (39), we utilised a S. capitis strain manipulated to express poly-RboP-WTA with β-O-GlcNAc (32). Anti-WTA-4497-IgA1 bound S. capitis ATCC 27840-H strain, but not wildtype strain ATCC 27840 (Fig. 6A; Supplementary Figure 4E & 4F). There was both IgA1-positive and IgA1-negative ATCC 27840-H populations in repeated experiments (Supplementary Figure 4E). 55.5% ± 4.17 and 1.83% ± 0.08 (n = 3, p < 0.01) of the S. capitis ATCC 27840-H populations were IgA1-positive when incubated with 3 μg/ml anti-WTA-4497-IgA1 or anti-HSA-IgA1, respectively. The incomplete opsonisation of the S. capitis population with anti-WTA-4497-IgA1 means there is a proportion of the bacterial population that cannot be phagocytosed through IgA-FcαR interactions. Nonetheless, there was enhanced IgA1 on S. capitis ATCC 27840-H bacteria opsonised with anti-WTA-4497-IgA1.
Figure 6. LILRB3 suppresses FcαR-mediated antimicrobial effector functions.
(A) Concentration-dependent binding of anti-WTA-IgA1 but not an isotype control to Staphylococcus capitis ATCC 27840-H strain. n = 3. (B and C) Phagocytic uptake of IgA-opsonised S. capitis by neutrophils at an MOI of 10:1. Neutrophils were incubated in the presence of 5 μg/ml FLIPr-like for 20 minutes, and then in the presence of anti-WTA-IgA1-opsonised FITC-labelled S. capitis for 30 minutes at 37°C with 5% CO2. Fluorescence of neutrophils was measured by flow cytometry analysis. The % of fluorescent neutrophils was calculated for each neutrophil population. The integrated results from eight separate donors (B) were compared by Student t-test, where * = p < 0.05, ** = p < 0.01, and representative flow cytometry plots are shown (C). (D, E and F) Continuous ligation of LILRB3 reduced phagocytic uptake of S. capitis at an MOI of 10:1. Neutrophils were incubated in the presence of 5 μg/ml FLIPr-like for 20 minutes, and then incubated on anti-LILRA6 or IgG1 coated plates. After 1 hour, neutrophils were incubated in the presence of anti-WTA-IgA1-opsonised FITC-labelled S. capitis for 30 minutes at 37°C with 5% CO2. Fluorescence of neutrophils was measured by flow cytometry analysis. The % of fluorescent neutrophils was calculated for each neutrophil population. A representative experiment (D) and the integrated results from seven separate donors (E), in which data from the same donor are linked, are shown. Relative % of fluorescent neutrophils (F) was calculated by normalizing values of against neutrophils pre-incubated on buffer control coated wells, and was compared by Student t-test, where * = p < 0.05, ** = p < 0.01. (G) Killing of IgA-opsonised S. capitis by neutrophils at a MOI of 1:1. Neutrophils were incubated in the presence of S. capitis opsonised with anti-WTA-IgA1 or anti-HSA-IgA or buffer control, for 60 minutes at 37°C with 5% CO2. Following neutrophil lysis, the % of CFU recovered at 60 minutes compared to 0 minutes was quantified by serial dilution and growth on BHI agar plates. Data from one donor was removed as an outlier using ROUT method. Data was analysed by Students t test, where * = p < 0.05, ** = p < 0.01. n = 7. (H and I) Continuous ligation of LILRB3 inhibited bacterial killing. Neutrophils were incubated on anti-LILRA6 or IgG1 coated plates for 1 hour prior to incubation for 60 minutes at 37°C with 5% CO2 in the presence of S. capitis opsonised with anti-WTA-IgA1 or anti-HSA-IgA or buffer control. After neutrophil lysis, the % of CFU recovered at 60 minutes compared to 0 minutes was quantified by serial dilution and growth on BHI agar plates. The integrated results from seven separate donors (H), in which data from the same donor are linked, were compared by Wilcoxon matched-pairs signed rank test, where * = p < 0.05, ** = p < 0.01. Relative % of recovered CFU (I) was calculated by normalizing values of neutrophils pre-incubated on anti-LILRA6 or IgG1 coated wells to buffer control, and compared Wilcoxon matched-pairs signed rank test, where * = p < 0.05, ** = p < 0.01.
We investigated phagocytosis of FITC-labelled S. capitis upon LILRB3 stimulation. A significantly higher proportion of neutrophils were fluorescent when S. capitis ATCC 27840-H was opsonised with anti-WTA-4497-IgA1 compared to isotype control (Fig. 6B, Fig. 6C). There was a strong trend, though no significant difference, for reduced phagocytic uptake of S. capitis during continuous ligation of LILRB3 compared to IgG1 treatment (Fig. 6D, Fig. 6E). When normalised for each donor, anti-LILRA6 induced significant reduction in the mean % fluorescent neutrophils compared to IgG1 treatment (Fig. 6F).
Finally, we investigated the impact of LILRB3 activation on microbial killing. Neutrophils were infected with S. capitis ATCC 27840-H at a MOI of 1 and the number of viable colony forming units (CFU) enumerated. Opsonisation with anti-WTA-4497-IgA1 lead to a significant decrease in the % of surviving bacteria compared to anti-HSA-IgA1 opsonisation or no opsonisation (Fig. 6G). Pre-incubation of neutrophils on anti-LILRA6 coated wells lead to a significant increase in % of recovered bacteria compared to IgG1 coated wells (Fig. 6H). There was a 2-fold increase in surviving bacteria when normalising data for each donor against buffer control coated wells (Fig. 6I). Our data argues LILRB3 can inhibit IgA-mediated anti-microbial effector functions.
Discussion
Knowledge of inhibitory receptors and paired receptor system properties has considerable implications for disease therapy (5). We aimed to characterize LILRA6 and LILRB3 in neutrophil biology. Our data provides robust evidence that LILRB3 is expressed at a high level on resting neutrophils, and that LILRB3 is released in a soluble form upon degranulation. Our data show that the LILRB3 is a potent inhibitor of FcR-mediated activation, ROS production, phagocytosis and microbial killing. The findings broaden our knowledge of the role of LILRB3 in human neutrophil biology.
Our data shows high expression of LILRB3 on resting neutrophils. Likewise, other paired receptors such as CEACAM1/3, Siglec5/14, and SIRPα/β are all expressed at high levels. As anti-LILRA6 and anti-LILRB3 mAb are cross-reactive, it has remained unclear which of these sibling receptors are expressed by neutrophils. Mass spectrometry analysis provided unequivocal evidence that LILRB3 is expressed on human neutrophils. Additionally, mass spectrometry analysis did not detect any evidence for LILRA6 expression. This suggests LILRA6 expression is not expressed on neutrophils, or expression is below detection limits.
Maturation, priming and activation can reshape the neutrophil surface. We detected no change in LILRB3 surface levels following SV exocytosis, though it is possible that LILRB3-positive SVs were mobilised during neutrophil purification. Our data suggests that priming with TNFα lead to heterogenous LILRB3 surface-expression levels, and that activation induces the release of LILRB3 from the surface. Though similar regulation is reported for the inhibitory receptor SIRL-1 (10), this contrasts other paired receptor systems including LAIR-1/-2, SIRPα/β and Siglec5/14 which are up-regulated upon activation (11, 40, 41). Thus, the LILRB3 and SIRL-1 systems are likely to regulate pre- and early-activation functions, whereas LAIR-1/-2, SIRPα/β and Siglec5/14 systems may regulate the termination of neutrophil functions. This also contrasts LILRB2 which is up-regulated upon degranulation (9).
Modulation of activation thresholds through inhibitory receptors is an important mechanism to control immune cell activity. Our data demonstrate that LILRB3 inhibits IgA-mediated oxidative burst, phagocytosis and microbial killing in vitro. IgA-opsonised bacteria have previously been shown to be effective at inducing ROS production but less effective at inducing phagocytosis (42). The data from our experiments indicate IgA opsonisation had a more profound enhancement on S. capitis killing compared to phagocytosis. This suggests that most microbes were killed through release of ROS and antimicrobial molecules. LILRB3 ligation was more effective at inhibiting IgA-induced ROS production than phagocytic uptake in our purified assays. In addition, LILRB3 ligation was more effective at inhibiting killing compared to phagocytosis of IgA-opsonised S. capitis. Collectively, this could suggest that IgA-opsonised bacteria are predominately killed through release of ROS and antimicrobial molecules, and that LILRB3 is an effective inhibitor of this mechanism.
Our studies utilised anti-LILR mAb as an agonist. Though our reporter system demonstrated agonistic properties of these mAb against LILRB3, 2B4 NFAT-GFP reporter systems are artificial. Therefore, additional resources need to be developed to identify agonists that activate LILR-bearing ITIM and ITAM, and to improve understanding of signal transduction pathways. In addition, further studies are required with appropriate controls to interrogate signalling mechanisms of LILRB3 in neutrophils.
Bacteria and bacterial products are sensed by phagocytes through pathogen recognition receptors (PRR) including TLRs, GPCRs and C-type lectin. Upon infection, PRR activation may induce the release of LILRB3 from the surface, allowing ROS production and intracellular killing of microbes at lower activation thresholds. The up-regulation of other inhibitory receptors upon activation and degranulation, such as LILRB2, LAIR-1, Siglec5 and CEACAM1, suggests they contribute to generating balanced immune responses to eliminate bacterial pathogens that do not have damaging effects to tissues.
We focused on investigating modulation of IgA-mediated responses. IgA is the most abundant antibody class at mucosal surfaces, and the second most abundant antibody class in serum. Cross-linking FcαR on neutrophils induces a variety of effector functions including phagocytosis, cytokine release, neutrophil extracellular trap (NET) release and ROS production. LAIR-1 and SIRL-1 have been shown to inhibit FcαR-mediated functions (21, 22). It is tempting to speculate that the LILRB3, LAIR-1 and SIRL-1 systems regulate IgA-mediated responses at mucosal surfaces and serum; though further studies are required. Paired receptor systems are known to regulate FcγR signalling. For example, LILRB1 and LILRB2 modulate FcγR responses on monocytes and/or neutrophils (9, 19). Given that FcαR and certain FcγR (FcγRIa and FcγRIIIa) signal through the Fc receptor common γ-chain, it is likely LILRB3 modulates both IgA and IgG mediated responses. However, there is a clear need for investigations to comprehensively assess whether paired receptor systems regulate both IgA and IgG mediated functions.
The PLB-985 cell line provides a useful model to study LILRA6 and LILRB3 in neutrophil biology, as they possess functions including ROS production, chemotaxis and phagocytosis. RNA-Seq analysis showed elevated expression of LILRA6 and LILRB3 genes during differentiation into neutrophil-like cells (29). We detected a 2.5-fold up-regulation in binding of anti-LILRB3 mAb to the surface of PLB-985 cells upon differentiation. Pull-down from DMSO-differentiated PLB-985 lysates detected a single band at 75 kDa in size, which was similar to that identified in pull-down from neutrophils and confirmed as LILRB3. This suggests LILRB3 is expressed in differentiated PLB-985 cells. This data supports the hypothesis that LILRB3 functions during neutrophil maturation and/or early activation phases. The PLB-985 cell system provides a valuable model for improving understanding LILRB3 in neutrophil biology, such as the identification of factors regulating LILRB3 expression and functions.
Identification of LILRB3 ligands is required to further elucidate a role in biology, and to provide novel molecular tools. Recently, LILRB3 is reported to bind the complement components C3b, iC3b, C3d and C4b (43). However, the functional consequences of these interactions remains uncharacterised. LILRB3 has also been shown to interact with a ligand associated with cytokeratin-8 on necrotic glandular epithelial cells (44). However, the specific ligand remains unidentified. LILRB3 could also have ligands and/or functions in a soluble form. This has been demonstrated for other LILR and immune receptors (13, 45).
Bacteria can directly interact with LILRs. This includes Staphylococcus aureus, Escherichia coli and Helicobacter pylori that can interact with LILRB3 and LILRB1 (36, 46), and Mycobacterium spp that can interact with LILRA1 and LILRB5. As these bacteria are opportunistic pathogens it is unclear whether LILRs have evolved as pathogen receptors, or whether bacteria have evolved to exploit LILRs. In either scenario the engagement of LILR by bacteria could have profound impacts on infection outcome.
In summary, the LILRB3 receptor is expressed at a high level on resting neutrophils and can inhibit FcR mediated functions and microbial killing. Rapid release of soluble LILRB3 upon activation suggests the LILRB3 may provide an important checkpoint to control activation and inflammation during neutrophil maturation. Further studies of LILRB3 inhibition of neutrophil development & functions are warranted. In addition, there is potential for development of immunomodulatory therapeutic approaches by targeting LILRB3.
Supplementary Material
Key points.
Neutrophils express LILRB3 which is released in a soluble form upon activation.
LILRB3 inhibits FcαR-mediated neutrophil activation and effector functions.
Acknowledgements
The authors would like to thank Nina van Sorge (UMC Utrecht) and Matevž Rumpret (UMC Utrecht) for advice, protocols and for providing PLB-985 cells and 2B4 NFAT-GFP T cells, and Sjors van der Lans (UMC Utrecht) for technical support.
Footnotes
Results incorporated in this standard received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement N° 700862. YZ was supported by a grant from the program of China Scholarships Council (201406170045). J.Z. and A.J.R.H. acknowledge support from the Netherlands Organization for Scientific Research (NWO) funding the Netherlands Proteomics Centre embedded in the roadmap facility X-Omics (project 184.034.019). J.Z additionally acknowledges support from the Chinese Scholarship Council (CSC).
References
- 1.Mócsai A. Diverse novel functions of neutrophils in immunity, inflammation, and beyond. J Exp Med. 2013;210:1283–1299. doi: 10.1084/jem.20122220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- 3.Futosi K, Fodor S, Mócsai A. Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int Immunopharmacol. 2013;17:638–650. doi: 10.1016/j.intimp.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Rees DJ, Szilagyi K, Kuijpers TW, Matlung HL, van den Berg TK. Immunoreceptors on neutrophils. Semin Immunol. 2016;28:94–108. doi: 10.1016/j.smim.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favier B. Regulation of neutrophil functions through inhibitory receptors: an emerging paradigm in health and disease. Immunol Rev. 2016;273:140–155. doi: 10.1111/imr.12457. [DOI] [PubMed] [Google Scholar]
- 6.Kuroki K, Furukawa A, Maenaka K. Molecular recognition of paired receptors in the immune system. Front Microbiol. 2012;3:429. doi: 10.3389/fmicb.2012.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steevels TA, Meyaard L. Immune inhibitory receptors: essential regulators of phagocyte function. Eur J Immunol. 2011;41:575–587. doi: 10.1002/eji.201041179. [DOI] [PubMed] [Google Scholar]
- 8.Avril T, Freeman SD, Attrill H, Clarke RG, Crocker PR. Siglec-5 (CD170) can mediate inhibitory signaling in the absence of immunoreceptor tyrosine-based inhibitory motif phosphorylation. J Biol Chem. 2005;280:19843–19851. doi: 10.1074/jbc.M502041200. [DOI] [PubMed] [Google Scholar]
- 9.Baudhuin J, Migraine J, Faivre V, Loumagne L, Lukaszewicz AC, Payen D, Favier B. Exocytosis acts as a modulator of the ILT4-mediated inhibition of neutrophil functions. Proc Natl Acad Sci U S A. 2013;110:17957–17962. doi: 10.1073/pnas.1221535110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steevels TA, Lebbink RJ, Westerlaken GH, Coffer PJ, Meyaard L. Signal inhibitory receptor on leukocytes-1 is a novel functional inhibitory immune receptor expressed on human phagocytes. J Immunol. 2010;184:4741–4748. doi: 10.4049/jimmunol.0902039. [DOI] [PubMed] [Google Scholar]
- 11.Verbrugge A, de Ruiter T, Geest C, Coffer PJ, Meyaard L. Differential expression of leukocyte-associated Ig-like receptor-1 during neutrophil differentiation and activation. J Leukoc Biol. 2006;79:828–836. doi: 10.1189/jlb.0705370. [DOI] [PubMed] [Google Scholar]
- 12.Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M. Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J. 2006;20:1964–1973. doi: 10.1096/fj.06-5800com. [DOI] [PubMed] [Google Scholar]
- 13.Lebbink RJ, van den Berg MC, de Ruiter T, Raynal N, van Roon JA, Lenting PJ, Jin B, Meyaard L. The soluble leukocyte-associated Ig-like receptor (LAIR)-2 antagonizes the collagen/LAIR-1inhibitory immune interaction. J Immunol. 2008;180:1662–1669. doi: 10.4049/jimmunol.180.3.1662. [DOI] [PubMed] [Google Scholar]
- 14.Tedla N, Gibson K, McNeil HP, Cosman D, Borges L, Arm JP. The co-expression of activating and inhibitory leukocyte immunoglobulin-like receptors in rheumatoid synovium. Am J Pathol. 2002;160:425–431. doi: 10.1016/S0002-9440(10)64861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown D, Trowsdale J, Allen R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue antigens. 2004;64:215–225. doi: 10.1111/j.0001-2815.2004.00290.x. [DOI] [PubMed] [Google Scholar]
- 16.Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol Rev. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- 17.Hirayasu K, Saito F, Suenaga T, Shida K, Arase N, Oikawa K, Yamaoka T, Murota H, Chibana H, Nakagawa I, Kubori T, et al. Microbially cleaved immunoglobulins are sensed by the innate immune receptor LILRA2. Nat Microbiol. 1:16054. doi: 10.1038/nmicrobiol.2016.54. [DOI] [PubMed] [Google Scholar]
- 18.Pereira S, Zhang H, Takai T, Lowell CA. The inhibitory receptor PIR-B negatively regulates neutrophil and macrophage integrin signaling. J Immunol. 2004;173:5757–5765. doi: 10.4049/jimmunol.173.9.5757. [DOI] [PubMed] [Google Scholar]
- 19.Fanger NA, Cosman D, Peterson L, Braddy SC, Maliszewski CR, Borges L. The MHC class I binding proteins LIR-1 and LIR-2 inhibit Fc receptor-mediated signaling in monocytes. Eur J Immunol. 1998;28:3423–34. doi: 10.1002/(SICI)1521-4141(199811)28:11<3423::AID-IMMU3423>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Lu HK, Rentero C, Raftery MJ, Borges L, Bryant K, Tedla N. Leukocyte Ig-like Receptor B4 (LILRB4) Is a Potent Inhibitor of FcγRI-mediated Monocyte Activation via Dephosphorylation of Multiple Kinases. J Biol Chem. 2009;284:34839–34848. doi: 10.1074/jbc.M109.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steevels TA, van Avondt K, Westerlaken GH, Stalpers F, Walk J, Bont L, Coffer PJ, Meyaard L. Signal inhibitory receptor on leukocytes-1 (SIRL-1) negatively regulates the oxidative burst in human phagocytes. Eur J Immunol. 2013;43:1297–1308. doi: 10.1002/eji.201242916. [DOI] [PubMed] [Google Scholar]
- 22.Olde Nordkamp MJ, van Eijk M, Urbanus RT, Bont L, Haagsman HP, Meyaard L. Leukocyte-associated Ig-like receptor-1 is a novel inhibitory receptor for surfactant protein D. J Leukoc Biol. 2014;96:105–111. doi: 10.1189/jlb.3AB0213-092RR. [DOI] [PubMed] [Google Scholar]
- 23.Lubeck MD, Steplewski Z, Baglia F, Klein MH, Dorrington KJ, Koprowski H. The interaction of murine IgG subclass proteins with human monocyte Fc receptors. J Immunol. 1985;135:1299–1304. [PubMed] [Google Scholar]
- 24.Fong R, Kajihara K, Chen M, Hotzel I, Mariathasan S, Hazenbos WLW, Lupardus PJ. Structural investigation of human S. aureus-targeting antibodies that bind wall teichoic acid. MAbs. 2018;10:979–991. doi: 10.1080/19420862.2018.1501252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehar SM, Pillow T, Xu M, Staben L, Kajihara KK, Vandlen R, DePalatis L, Raab H, Hazenbos WL, Morisaki JH, Kim J, et al. Novel antibody-antibiotic conjugate eliminates intracellular S aureus. Nature. 2015;527:323–328. doi: 10.1038/nature16057. [DOI] [PubMed] [Google Scholar]
- 26.Adams R, Griffin L, Compson JE, Jairaj M, Baker T, Ceska T, West S, Zaccheo O, Davé E, Lawson AD, Humphreys DP, et al. Extending the half-life of a fab fragment through generation of a humanized anti-human serum albumin Fv domain: An investigation into the correlation between affinity and serum half-life. MAbs. 2016;8:1336–1346. doi: 10.1080/19420862.2016.1185581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang XT, Sehlin D, Lannfelt L, Syvänen S, Hultqvist G. Efficient and inexpensive transient expression of multispecific multivalent antibodies in Expi293 cells. Biol Proced Online. 2017;19:11. doi: 10.1186/s12575-017-0060-7. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stemerding AM, Köhl J, Pandey MK, Kuipers A, Leusen JH, Boross P, Nederend M, Vidarsson G, Weersink AY, van de Winkel JG, van Kessel KP, et al. Staphylococcus aureus formyl peptide receptor-like 1 inhibitor (FLIPr) and its homologue FLIPr-like are potent FcγR antagonists that inhibit IgG-mediated effector functions. J Immunol. 2013;191:353–362. doi: 10.4049/jimmunol.1203243. [DOI] [PubMed] [Google Scholar]
- 29.Rincón E, Rocha-Gregg BL, Collins SR. A map of gene expression in neutrophil-like cell lines. BMC Genomics. 2018;19:573. doi: 10.1186/s12864-018-4957-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohtsuka M, Arase H, Takeuchi A, Yamasaki S, Shiina R, Suenaga T, Sakurai D, Yokosuka T, Arase N, Iwashima M, Kitamura T, et al. NFAM1, an immunoreceptor tyrosine-based activation motif-bearing molecule that regulates B cell development and signaling. Proc Natl Acad Sci U S A. 2004;101:8126–8131. doi: 10.1073/pnas.0401119101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Weijer ML, Bassik MC, Luteijn RD, Voorburg CM, Lohuis MA, Kremmer E, Hoeben RC, LeProust EM, Chen S, Hoelen H, Ressing ME, et al. A high-coverage shRNA screen identifies TMEM129 as an E3 ligase involved in ER-associated protein degradation. Nat Commun. 2014;5:3832. doi: 10.1038/ncomms4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winstel V, Liang C, Sanchez-Carballo P, Steglich M, Munar M, Bröker BM, Penadés JR, Nübel U, Holst O, Dandekar T, Peschel A, et al. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat Commun. 2013;4:2345. doi: 10.1038/ncomms3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rørvig S, Østergaard O, Heegaard NH, Borregaard N. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J Leukoc Biol. 2013;94:711–721. doi: 10.1189/jlb.1212619. [DOI] [PubMed] [Google Scholar]
- 34.Berger M, O'Shea J, Cross AS, Folks TM, Chused TM, Brown EJ, Frank MM. Human neutrophils increase expression of C3bi as well as C3b receptors upon activation. J Clin Invest. 1984;74:1566–1571. doi: 10.1172/JCI111572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebbink RJ, de Ruiter T, Adelmeijer J, Brenkman AB, van Helvoort JM, Koch M, Farndale RW, Lisman T, Sonnenberg A, Lenting PJ, Meyaard L. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J Exp Med. 2006;203:1419–1425. doi: 10.1084/jem.20052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hogan LE, Jones DC, Allen RL. Expression of the innate immune receptor LILRB5 on monocytes is associated with mycobacteria exposure. Sci Rep. 2016;6 doi: 10.1038/srep21780. 21780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janoff EN, Fasching C, Orenstein JM, Rubins JB, Opstad NL, Dalmasso AP. Killing of Streptococcus pneumoniae by capsular polysaccharide-specific polymeric IgA, complement, and phagocytes. J Clin Invest. 1999;104:1139–1147. doi: 10.1172/JCI6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorter A, Hiemstra PS, Leijh PC, van der Sluys ME, van den Barselaar MT, van Es LA, Daha MR. IgA- and secretory IgA-opsonized S. aureus induce a respiratory burst and phagocytosis by polymorphonuclear leucocytes. Immunology. 1987;61:303–309. [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy AJ, Lindsay JA. Staphylococcus aureus innate immune evasion is lineage-specific: a bioinfomatics study. Infect Genet Evol. 2013;19:7–14. doi: 10.1016/j.meegid.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Bühring HJ, Zen K, Burst SL, Schnell FJ, Williams IR, Parkos CA. Signal regulatory protein (SIRPalpha), a cellular ligand for CD47, regulates neutrophil transmigration. J Biol Chem. 2002;277:10028–10036. doi: 10.1074/jbc.M109720200. [DOI] [PubMed] [Google Scholar]
- 41.Erickson-Miller CL, Freeman SD, Hopson CB, D'Alessio KJ, Fischer EI, Kikly KK, Abrahamson JA, Holmes SD, King AG. Characterization of Siglec-5 (CD170) expression and functional activity of anti-Siglec-5 antibodies on human phagocytes. Exp Hematol. 2003;31:382–388. doi: 10.1016/s0301-472x(03)00046-8. [DOI] [PubMed] [Google Scholar]
- 42.Vidarsson G, van Der Pol WL, van Den Elsen JM, Vilé H, Jansen M, Duijs J, Morton HC, Boel E, Daha MR, Corthésy B, van De Winkel JG. Activity of human IgG and IgA subclasses in immune defense against Neisseria meningitidis serogroup B. J Immunol. 2001;166:6250–6256. doi: 10.4049/jimmunol.166.10.6250. 2001. [DOI] [PubMed] [Google Scholar]
- 43.Hofer J, Forster F, Isenman DE, Wahrmann M, Leitner J, Hölzl MA, Kovari JJ, Stockinger H, Böhmig GA, Steinberger P, Zlabinger GJ. Ig-like transcript 4 as a cellular receptor for soluble complement fragment C4d. FASEB J. 2016;30:1492–1503. doi: 10.1096/fj.15-275594. [DOI] [PubMed] [Google Scholar]
- 44.Jones DC, Hewitt CR, López-Álvarez MR, Jahnke M, Russell AI, Radjabova V, Trowsdale AR, Trowsdale J. Allele-specific recognition by LILRB3 and LILRA6 of a cytokeratin 8-associated ligand on necrotic glandular epithelial cells. Oncotarget. 2016;7:15618–15631. doi: 10.18632/oncotarget.6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An H, Chandra V, Piraino B, Borges L, Geczy C, McNeil HP, Bryant K, Tedla N. Soluble LILRA3, a potential natural antiinflammatory protein, is increased in patients with rheumatoid arthritis and is tightly regulated by interleukin 10, tumor necrosis factor-alpha, and interferon-gamma. J Rheumatol. 2010;37:1596–1606. doi: 10.3899/jrheum.091119. [DOI] [PubMed] [Google Scholar]
- 46.Nakayama M, Underhill DM, Petersen TW, Li B, Kitamura T, Takai T, Aderem A. Paired Ig-like receptors bind to bacteria and shape TLR-mediated cytokine production. J Immunol. 2007;178:4250–4259. doi: 10.4049/jimmunol.178.7.4250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.