Abstract
Hotspot mutations of the oncogenes BRAF and NRas are the most common genetic alterations in cutaneous melanoma. Still, the nanoscale organization and signal coupling of these proteins remain incompletely understood, particularly upon expression of oncogenic NRas mutants. Here we employed single-molecule localization microscopy to study the nanoscale organization of NRas and BRAF at the plasma-membrane (PM) of melanoma cells. NRas and BRAF resided in self-clusters that did not associate well in resting cells. In EGF-activated cells, NRas clusters became more diffused while overall protein levels at the PM increased; thus allowing enhanced association of NRas and BRAF and downstream signaling. In multiple melanoma cell lines, mutant NRas resided in more pronounced self-clusters relative to WT NRas yet associated more with the clustered and more abundant BRAF. In cells resistant to trametinib, a clinical MEK inhibitor (MEKi), a similar co-clustering of NRas and BRAF was observed upon EGF activation. Strikingly, treatment of cells expressing mutant NRas with trametinib reversed the effect of mutant NRas expression by restoring the non-overlapping self-clusters of NRas and BRAF and by reducing their PM levels and elevated pERK levels caused by mutant NRas. Our results indicate a new mechanism for signal regulation of NRas in melanoma through its nanoscale dynamic organization and a new mechanism for MEKi function in melanoma cells carrying NRas mutations but lacking MEK mutations.
Keywords: Melanoma, Single molecule localization microscopy, Superresolution microscopy, Protein clustering
Introduction
The mitogenic pathway conveys growth signals from specific receptors (e.g. EGFR) into the cell and triggers its growth and division. This pathway is often described as a linear cascade of specific and synchronized molecular interactions (1). For instance mitogenic signals are conveyed from the growth receptors to Ras, RAF, MEK, and ERK. ERK then translocates to the nucleus, activates transcription and promotes cell growth (2). As a key step in this pathway, (H,K,N)-Ras and BRAF are recruited from the cell cytosol to its plasma membrane (PM), where these proteins get activated. Recent studies suggest that these proteins form transient complexes at the PM, together with additional proteins such as scaffolds, to convey the receptor signals (3). Indeed, NRas and BRAF have been described to form functional dimers and small (nanoscale) clusters at the PM (4). These proteins are then recycled in vesicles between the cell cytosol, its various organelles and the PM (5).
Multiple proteins involved in the mitogenic pathway undergo mutations that drive a variety of cancers by aberrant signaling, independent of external growth signals (6,7). Specifically, ~14-20% of melanomas carry mutations in NRas (8,9). Frequent mutations include G12D, G13D and Q61R (10). BRAF often carries the V600E activating mutation (11).
MEK inhibitors (MEKi) have become a prominent route for clinical treatment of melanoma patients, with some promising success rates (12). Still, the detailed dynamic organization of signaling complexes, nucleated by NRas or its oncogenic mutants on the PM, and their potential role in cell transformation and cancer progression remain poorly understood (13). Such understanding is critically needed for optimal targeting of the aberrant activity of these proteins in various cancers.
Here, we hypothesized that molecular complexes and clusters of NRas and BRAF at the PM could play a critical role in regulation of the mitogenic signal in health and disease. Thus, we employed single molecule localization microscopy (SMLM) to study NRas and BRAF at the PM of melanoma cells and in single molecule detail. Interestingly, NRas and BRAF resided in self-clusters that did not associate well in resting cells. In EGF-activated cells, these clusters became more diffused while overall protein levels at the PM increased; thus allowing enhanced association of NRas and BRAF and signaling downstream.
Mutant NRas (Q61R, G12D, G13D) resided in more pronounced self-clusters relative to WT NRas in multiple melanoma cell lines. Still, they could associate more with the clustered, and more abundant BRAF. Strikingly, treatment of such cells with trametinib, a clinical MEKi, reversed the effect of the mutant NRas expression. It restored the non-overlapping self-clusters of NRas and BRAF and reduced their PM levels. Treatment with trametinib could also reduce the elevated pERK levels, caused by the NRas mutants, to the levels of resting cells. Our results indicate a new mechanism for signal regulation of NRas in melanoma through its nanoscale dynamic organization. They also show a new mechanism for MEKi function in melanoma cells carrying NRas mutations, but lacking MEK mutations.
Materials and Methods
Cell lines
This study includes the following patient-derived melanoma cell lines (background mutations are in parentheses): 108T (NF1-H1366Q), A375 (BRAF-V600E), 72T (NRas-G12D), 74T (NRas-Q61R), 83T (NRas-G13D), 12T(NRas-Q61R). A subset of cell lines used in the study (108T, 72T, 74T, 83T and 12T) were derived from a panel of pathology-confirmed metastatic melanoma tumor resections collected from patients enrolled in IRB-approved clinical trials at the Surgery Branch of the National Cancer Institute. Pathology-confirmed melanoma cell lines were derived from mechanically or enzymatically dispersed tumor cells, which were then cultured in RPMI 1640 + 10% FBS at 37°C in 5% CO2 for 5-15 passages.
These cell lines were used in previous studies (14,15). They were originally derived from a panel of pathology-confirmed metastatic melanoma tumor resections collected from patients enrolled in IRB-approved clinical trials (with written informed consent) at the Surgery Branch of the National Cancer Institute. Pathology-confirmed melanoma cell lines were derived from mechanically or enzymatically dispersed tumor cells, which were then cultured in RPMI 1640 + 10% FBS at 37°C in 5% CO2 for 5-15 passages. Cell lines genotypes are indicated in Supplementary Table S1. All cell lines have been tested negative for mycoplasma.
Samples
In this work we constructed BRAF and NRas WT and three mutants G12D, G13D and Q61R conjugated to the photoactivatable fluorophores PAGFP and PAmCherry respectively by the gateway cloning method (16). All constructs containing photoactivatable fluorescent proteins (PAFPs) were cloned in pcDNA3 plasmids with a CMV promoter. The cloning of the fluorescent tags was performed at the N-terminus in order to prevent localization disruption. Constructs were validated by DNA sequencing. The 108T melanoma cells were transfected with PAmCherry-NRas (WT or mutants) and PAGFP-BRAF using Lipofetamin 3000 (L3000008, Invitrogen) for 48 h before EGF addition and fixation. Cell seeding and imaging was conducted on glass coverslips (#1.5 glass chambers, LabTek and Ibidi), and fixed with 2.4% Paraformaldehyde (PFA) for 30 min at 37°C. Trametinib (GSK1120212, Selleckchem, S2673) was used in a concentration of 10 nM for MEKi treatment of cells over 16 h.
For direct Stochastic Optical Reconstruction Microscopy (dSTORM) imaging, cells were seeded and fixed for 15 minutes after adding EGF. For permeabilization, we added 0.4 ml 0.1% Triton X-100 in PBS per well and incubated for 3-4 minutes. The cells were blocked by 2% normal goat serum in PFN (PBS + 10% serum + 0.02% sodium azide) for 30 minutes. For 0.5 million cells, we added 0.5 μg rabbit αhuman phsopho-BRAF (ab19283, Abcam) and mouse αhuman NRas (SC-31, L1115) as primary antibodies diluted in 2% normal goat serum in PFN, incubated for 60 minutes at RT and washed 3 times with PFN. Alexa647 (αrabbit, Invitrogen, A21244) and Atto488 (αmouse, Sigma, 62197) was added as secondary antibodies diluted (1/3000) in 2% normal goat serum in PFN, incubated for 45 minutes at RT and washed 3 times with PFN.
Western blot analysis
Proteins were extracted with RIPA buffer (R0278, Sigma) and Protease phosphatase inhibitor cocktail (PPC1010, Sigma). SDS-PAGE electrophoresis was performed using Novex WedgWell 4-12% gels (XP04120BOX, Invitrogen) and blotted on iBlot2 membranes (IB23001, Invitrogen). Western blot detection was performed with following antibodies: Anti-NRas (F155, Santa Cruz), Anti-ERK-P (ab201015, Abcam), Anti –ERK (4695, Cell Signaling), Anti-Tubulin (T6074, Sigma), Anti-ABCB5 (ab140667, Abcam).
The expression levels of proteins
In our study, we aimed to incorporate relatively low levels (~30%) of exogenous proteins relative to endogenous proteins. Individual expression levels of proteins were monitored by western blots (Fig. 1A) and imaging (throughout the study). Higher protein levels bear the risk of artificially promoting the mitogenic signaling of the cell. At these moderate levels of expression, exogenous proteins that are similar in sequence to the endogenous proteins (e.g. WT NRas and WT NRasPAmCherry) are expected to represent the behavior of the endogenous proteins by distributing randomly among the endogenous proteins (either as monomers or in clusters). Exogenous proteins that are different in sequence (e.g. NRas mutants carrying a fluorescent tag) could differ in their self-organization at the PM, and in their mutual interactions with other proteins (esp. BRAF, as in Figs. 3 and 5). Notably, in Fig. 7A,B, the moderate expression of mutant NRas was sufficient to cause upregulation of ERK, as measured by pERK, relative to the effect of expression of WT NRas.
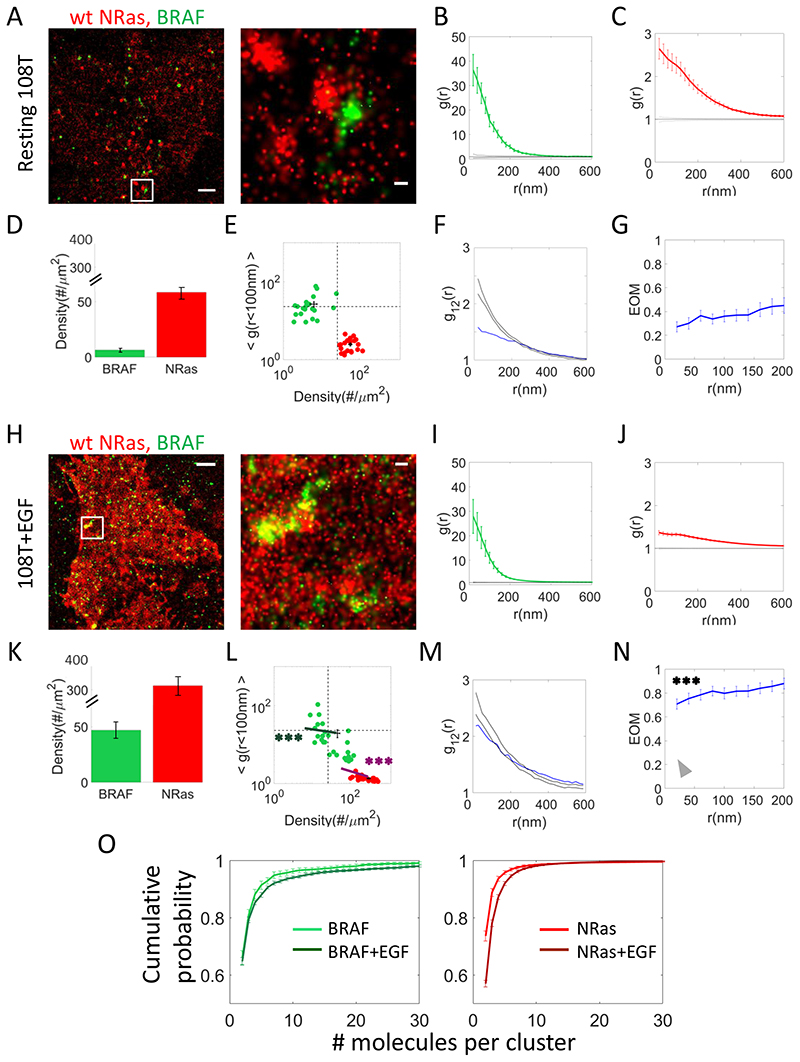
Fig. 1. NRas and BRAF reside and associate in clusters at the plasma membrane of melanoma cells that are altered after EGF activation.
(A) Two-colour PALM imaging of resting 108T melanoma cells expressing PAmCherryNRas and PAGFP-BRAF. Cells were seeded on the coverslip for 2 days before fixation. Shown is a representative cell (N=20). Bars – 2 μm (left) and 200 nm (right). (B) PCF of PAGFP-BRAF. (C) PCF of PAmCherry-NRas. (D) The density of BRAF and NRas at the PM. (E) A two dimensional map of self-clustering (value of g(0-100)) vs. protein density. Values are shown for individual cells as discs, either for BRAF (green) or NRas (red). Note that both axes are logarithmic. Dashed black lines are guidelines for comparison of results with additional measurements. (F) Bivariate PCF (BPCF) of BRAF and NRas (representative data shown for a single cell; N=20). (G) The extent of mixing (EOM) of BRAF and NRas (averaged over 20 cells) (See Analyses for further details). (H) Twocolour PALM imaging of 108T melanoma cells expressing PAmCherry-NRas and PAGFPBRAF. Cells were seeded on the coverslip for 2 days, and activated with EGF before fixation. Shown is a representative cell (N=27). Bars – 2 μm (left) and 200 nm (right). (I) PCF of PAGFP-BRAF. (J) PCF of PAmCherry-NRas. (K) The density of BRAF and NRas at the PM. (L) A two dimensional map of self-clustering (value of g(0)) vs. protein density. Data presentation is as in panel E. (M) Bivariate PCF (BPCF) of BRAF and NRas. (N) The EOM of BRAF and NRas. (O) Cumulative distributions of cluster sizes of (left) BRAF or (right) NRas. The distributions are presented in copy numbers, as quantified by a clustering algorithm applied on the PALM images (see Methods). Results are shown for resting and EGF-activated 108T cells. P-values relative to data in Fig. 1E: * < 0.05, ** < 0.01, *** < 0.005. P-values are summarized in Supplementary Table S4. Error bars in all relevant panels are SEM due to measurements on multiple cells.
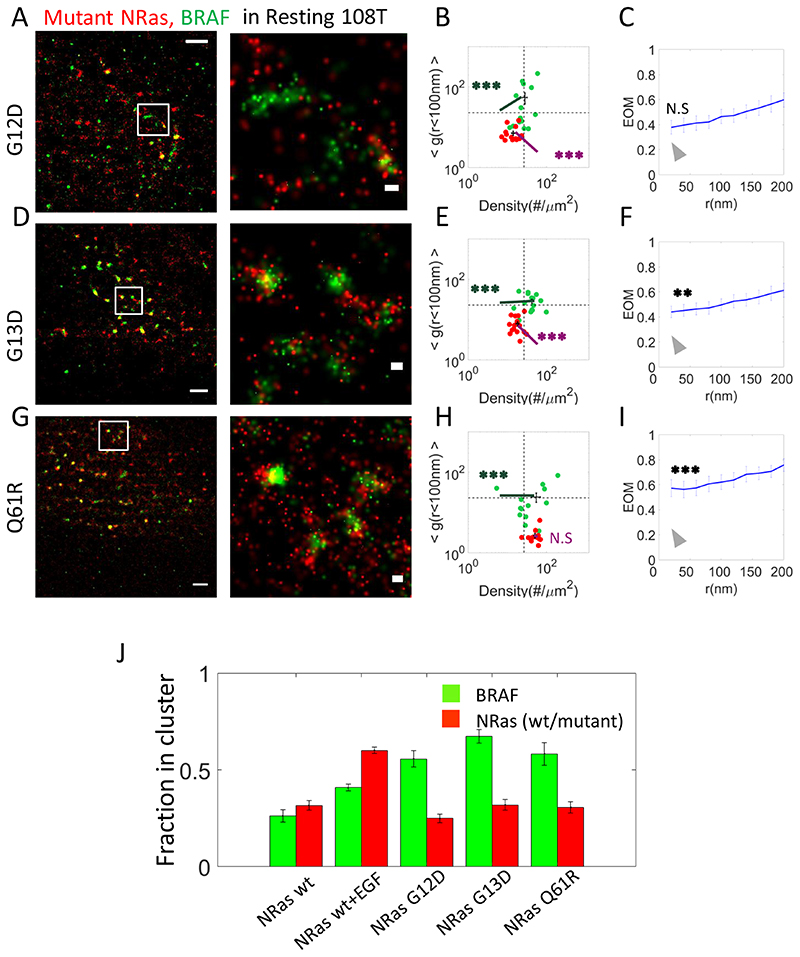
Fig. 3. Expression of NRas mutants enhance NRas clustering and BRAF recruitment.
(A,D,G) Two-colour PALM imaging of resting 108T melanoma cells expressing PAmCherry-NRas mutants (G12D, G13D and Q61R) and PAGFP-BRAF. Cells were seeded on the coverslip for 2 days before fixation. Shown are representative cell (N=14, N=15, N=13 for G12D, G13D and Q61R respectively). Bars – 2 μm (left) and 200 nm (right). (B,E,H) Two dimensional map of self-clustering (value of g(<100)) vs. protein density. Values are shown for individual cells as discs, either for BRAF (green) or NRas G12D (red). (C,F,I) The extent of mixing (EOM) of BRAF and NRas mutants (G12D, G13D and Q61R). (J) The fraction of either NRas or BRAF molecules found in clusters, under the conditions described in Figs. 1 and 3. Clustering analysis was performed using a clustering algorithm applied on the PALM images (see Methods, and as in Fig. 1O)..P-values relative to data in Fig. 1E: * < 0.05, ** < 0.01, *** < 0.005. P-values are summarized in Supplementary Table S4. Error bars in all relevant panels are SEM due to measurements on multiple cells.
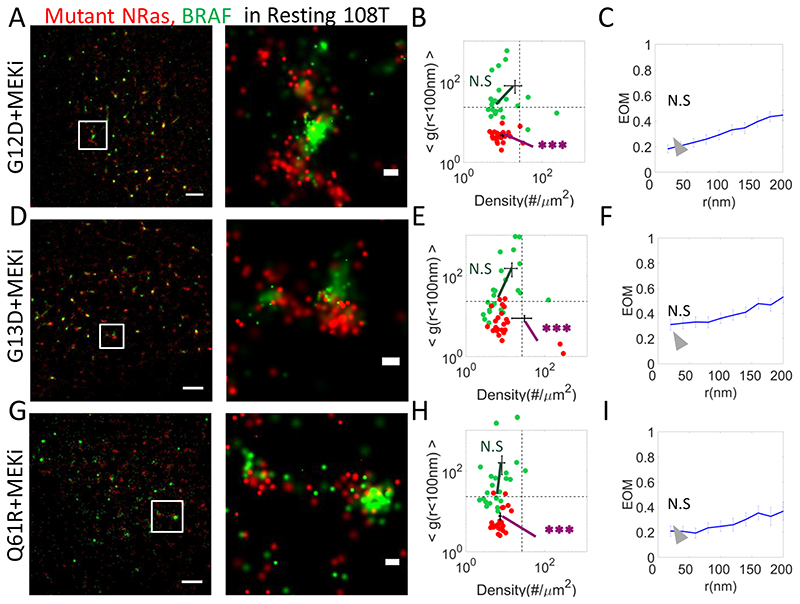
Fig. 5. Clinical inhibitors restore NRas-segregated BRAF clustering as in resting cells.
(A,D,G) Two-colour PALM imaging of resting 108T melanoma cells expressing PAmCherry-NRas mutants (G12D, G13, Q61R) and PAGFP-BRAF. Cells were seeded on the coverslip for 2 days and treated with MEKi 16h before fixation. Shown are representative cells (N=16, N=19, N=17). Bars – 2 μm (left) and 200 nm (right). (B,E,H) A two dimensional map of self-clustering (value of g(<100)) vs. protein density. Values are shown for individual cells as dots, either for BRAF (green) or mutant NRas (red). Note that both axes are logarithmic. Dashed black lines are guidelines for comparison of results with additional measurements. (C,F,I) The extent of mixing (EOM) of BRAF and mutant NRas (See Analyses in Methods). P-values relative to data in Fig. 1E: * < 0.05, ** < 0.01, *** < 0.005. P-values are summarized in Supplementary Table S4. Error bars in all relevant panels are SEM due to measurements on multiple cells.
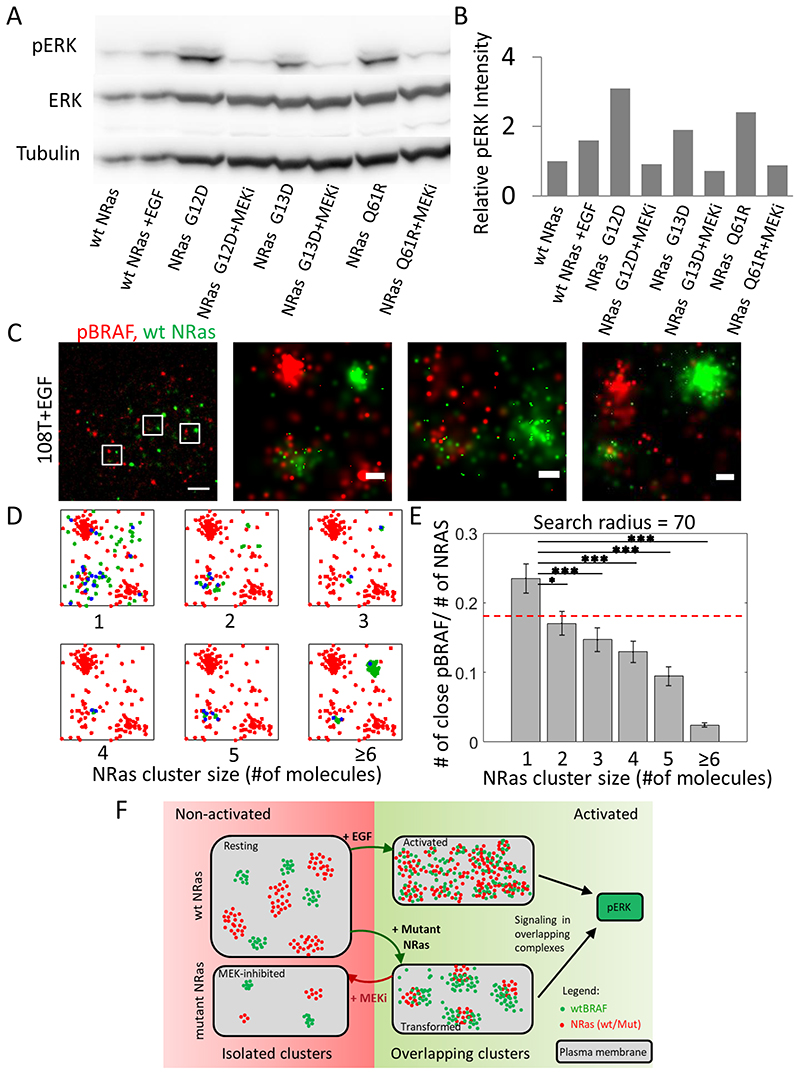
Fig. 7. MEK clinical inhibitor diminishes elevated ERK signaling by NRas mutants.
(A) Western blots of 108T cells transfected with either WT-NRas or mutant (G12D, G13D or Q61R) NRas. The cells were either resting or treated with EGF, or with MEKi. Shown are pERK, ERK and tubulin levels. (B) Relative pERK levels, normalized according to ERK levels, as quantified from the blot data in panel A. (C) Two colour dSTORM imaging of 108T cells. The cells were seeded on the coverslip for 2 days and treated with EGF 15 min before fixation. Endogenous NRas and pBRAF were tagged by Atto488 (green) and Alexa647 (red) respectively. Shown is a representative cell (N=22). Bars – 2 μm (left) and 200 nm (3 zoomed images on right). (D) The description of analyses that separates NRas molecules according to their self-cluster size (green points) and finds NRAS-associated pBRAF molecules for NRas in monomers or in clusters (NRas-pBRAF proximity of < 70 nm; blue points). In each plot, pBRAF molecules that were not associated with the specified NRas clusters are shown in red. (E) The ratio between the number of associated pBRAF to the number of NRas in each cluster size. Dashed red line is the average number of associated pBRAF across all cluster sizes. P-values in panel E: * < 0.05, ** < 0.01, *** < 0.005. (F) A model for NRas and BRAF organization and singling in clusters. The model shows segments of the PM of the cells, and the organization of NRas (red) and BRAF (green) within. The left PM cartoon represents resting conditions where BRAF and NRas reside in separated and pronounced clusters with small interaction. The top right PM cartoon represents activated conditions, in which BRAF and NRas become more diffused, their levels at the PM increase, and as a result - their interaction grows and signaling downstream is facilitated. The bottom right cartoon shows the effect of the expression of NRas oncogenic mutants. While mutant NRas become more clustered, they can efficiently recruit the more abundant BRAF and signal downstream. Treatment with MEKi reverses the molecular organization patterns of NRas and BRAF back to resting-like, and thus MEKi reduces the aberrant signaling by the oncogenic NRas mutants.
See further details in the SI regarding the expression levels of ABCB5 and the cell proliferation assay.
Antibody staining
For dSTORM imaging, cells expressing endogenous WT and mutant NRas were seeded and fixed on coverslips, as described above. For cell permeabilization, we added 0.4 ml 0.1% Triton X-100 in PBS per 1x1 cm2 well and incubated for 3-4 minutes. The cells were blocked by 2% normal goat serum in PFN (PBS + 10% serum + 0.02% sodium azide) for 30 minutes. Cells were next stained using a primary NRas antibody (L1115, Santa Cruz) and a secondary antibody stained with Alexa647 (company). This fluorophore was imaged using 647nm laser illumination at up to 100% (170mW in maximum). Direct-STORM buffer included GLOX (0.5 mg/mL glucose oxidase, 40 μg/mL catalase, 10% glucose), 10 mM 2-aminoethanethiol (MEA), in buffer that included 50 mM Tris and 10 mM NaCl with a pH of 8 (17). For the phosphorylation experiment, WT endogenous NRas and pBRAF were tagged by mouse anti-NRas (SC-31, L1115) and rabbit anti-pBRAF (ab19283, Abcam) as primary and Atto488 anti-mouse (αmouse, Sigma, 62197) and Alexa647 anti-rabbit (αrabbit, Invitrogen, A21244) as secondary respectively.
Imaging
We conducted two-colour photoactivated localization microscopy (PALM) imaging similar to previous studies (18), using a total internal reflection (TIRF) Nikon microscope with a CFI Apo TIRF X100 oil objective (NA 1.49, WD 0.12mm). PAGFP and PAmCherry were photoactivated using variable intensity (0.5-10% of 30mW in maximum) of 405nm laser illumination that was changed gradually from beginning of imaging to the end and alternate excitation using (50% of 90mW in maximum) 488nm laser excitation for PAGFP and (50% of 90mW in maximum) 561nm for PAmCherry unchanged during the imaging. Laser illumination at all wavelengths covered a circular area with a diameter of 80 μm at the sample. Movies of fixed cell imaging were acquired for up to 3000 frames at 13.1 fps of an EMCCD Ixon+ camera. The focus of the microscope was maintained throughout the imaging using the PerfectFocus system of the microscope.
Analyses
We used the ThunderSTORM software (19) to analyse PALM and dSTORM movies and generate images. Briefly, this software served to identify individual peaks and to assign them to individual molecules for rendering of the PALM or dSTORM images. The localization uncertainties of the different fluorophores used in our study peaked at ~20-30nm (Supplementary Fig. S1B). A distance threshold and a temporal gap were employed for peak grouping to account for possible molecular blinking (20). Temporal gaps were tested for each fluorophore separately to minimize possible over-counting of molecules.
In SMLM, single fluorophores can be typically detected over multiple frames till their photobleaching. Fluorophore blinking may cause absence of some detections from specific frames in the detection sequence. Such blinking and ‘off-frames’ may then lead to fluorophore overcounting, since single fluorophores could then be counted more than once (e.g. (21)). A standard approach to overcome this issue is to first characterize the statistics of ‘off-times’ for each fluorophore. Then, a threshold for maximal ‘allowed’ temporal gap between detections can be assigned (21–23)
Another potential issue that can lead to fluorophore overcounting is due to the spreading of the fluorophores’ localizations across their detected area (the point-spread function (PSF)) on the detector. To avoid both issues, a standard procedure in SMLM reconstruction is to group the detected peaks over space and time, and assign them to single fluorophores. We provide further details on this procedure in the SI, in Supplementary Fig. S2A–H and Grouping parameter values in Supplementary Table S2. These parameters were provided through dedicated filters during SMLM reconstruction using ThunderSTORM (19). The effect of selected grouping parameters on our results is shown in Supplementary Fig. S3A–D.
Drift compensation and channel registration were performed using dedicated algorithms in the ThunderSTORM software. Individual molecules were presented in corresponding to the probability density values of their fitted Gaussian. This scale was set for each species separately, according to its maximal probability density in the field (note that it should not be interpreted as the density of molecules in clusters).
Clustering and activated molecules selection analyses
To classify points into clusters we used the published algorithm DBSCAN using the Matlab function “rangesearch” (MathWorks) (24), with a distance threshold of <30 nm between molecules in same cluster. This algorithm served to generate the size distribution of molecular clusters in copy numbers (25). Histograms are shown in Fig. 1O as cumulative distributions to allow clearer comparison between results under various cell conditions. See further details in the SI on co-clustering analysis of NRas and BRAF (Fig. 6G).
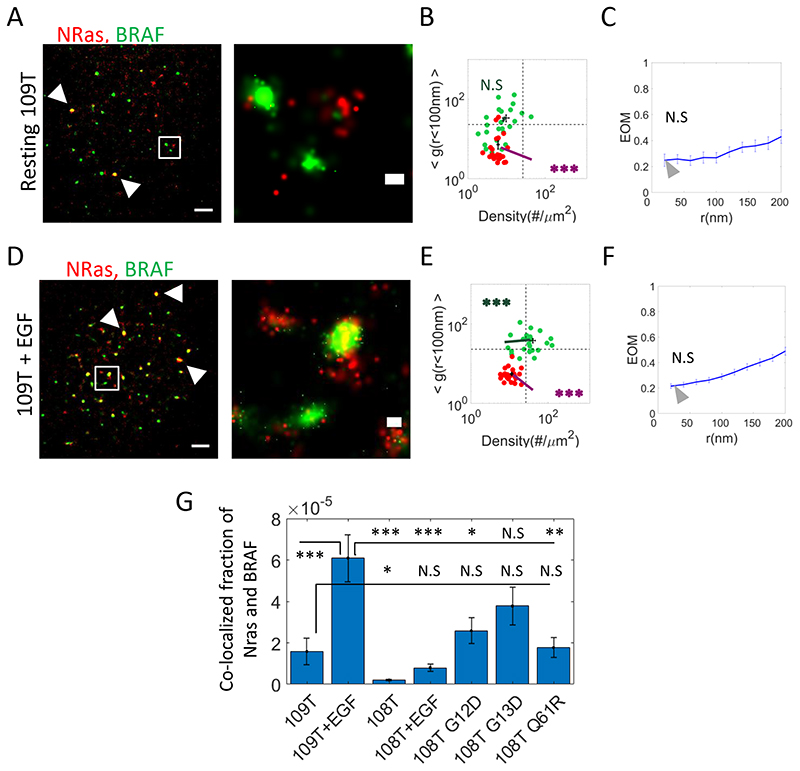
Fig. 6. NRas and BRAF molecular patterning change in resistant cells to MEKi relative to 108T cells.
(A,D) Two-colour PALM imaging of resting and EGF-activated 109T Trametinib-resistant melanoma cells, expressing PAmCherry-NRas and PAGFP-BRAF. Cells were seeded on the coverslip for 2 days before fixation. Shown is a representative cell (N=24 for resting cells and N=23 for EGF-activated cells). Bars – 2 μm (left) and 200 nm (right). (B,E) A two dimensional map of self-clustering (value of g(0-100)) vs. protein density. Values are shown for individual cells as discs, either for BRAF (green) or NRas (red). Note that both axes are logarithmic. Dashed black lines are guidelines for comparison of results with additional measurements. (C,F) The extent of mixing (EOM) of BRAF and NRas (See Analyses for further details). (G) The fraction of the pixels containing BRAF and NRas detections relative to NRas-containing pixels in 109T and in 108T melanoma cells expressing PAmCherry-NRas WT or mutants (G12D, G13, Q61R) and PAGFP-BRAF. P-values relative to data in Fig. 1E: * < 0.05, ** < 0.01, *** < 0.005. Error bars in all relevant panels are SEM due to measurements on multiple cells.
Modelling and simulations (Fig. 2)
see details in the Supplemental Materials and Methods and in Supplementary Table S3.
Second order statistics
A detailed description of the second order statistics used in this study has been published and extensively described (26–28). This description includes the univariate PCF, the bivariate PCF and extent of mixing (EOM). Briefly, Pair Correlation Function (PCF, denoted here also as g(r)) describes and quantifies in a point pattern how density varies as a function of distance from a reference particle/point. Usually PCF is normalized by the density of the sample, and was used in this normalized form throughout the study. The univariate PCF is used to explore a point pattern of a single species. It is further useful to compare the PCF results to a Poisson model that describes random placement of points across the field. This model results in a flat PCF, for which g(r)=1. Thus, higher values of the PSF (i.e. g(r)) indicate self-clustering.
For patterns with two species, Bivariate PCF (BPCF) statistics quantify the density of pattern 2 at distance r from an arbitrary point of pattern 1. In order to investigate whether or not two species (in a joint point pattern) are significantly interacting, we used the random labeling model. In this model, points of pattern 1 (n1) and points of pattern 2 (n2) distribute randomly at n1+ n2 fixed locations. Multiple Monte-Carlo simulations replicate 19 times the point patterns while randomly re-labeling the points (with the number of points from each species). The bivariate PCF of the original point pattern g12(r) is then compared to the bivariate PCFs of the simulations. We used the lowest and highest g12(r) of the different simulations as a 95% confidence interval for the acceptance or rejection of the model as a null hypothesis. Agreement of the data with the Random Labeling (RL) model indicates homogeneous mixing, and hence strong interaction (in a statistical sense) of the two species under study. Alternatively, a model of no interaction would result in a flat curve where g12(r)=1. The extent of mixing (EOM) represents the average of BPCF over multiple cells using 95% confidence interval due to the RL model. EOM is normalized between the values of +1 for perfect mixing and 0 for the model of No Interaction.
Statistical significance
In order to quantify the statistical significance of two-dimensional (density-g(r)) distributions we performed two-sample Kolmogorov-Smirnov using the function ‘kstest_2s_2d’ in Matlab. This function evaluates the difference between two vectors of 2-by-n given as an input. The result is a value between 0 and 1 (for none to perfect fit, respectively) (29). All significance values of the (density-g(r)) plots and of the EOM graphs are summarized in Supplementary Table S4.
In the rest of the analyses, p-values were calculated using Student’s T-test, assuming two-tailed distribution and samples with unequal variances.
Results
NRas and BRAF reside and associate in clusters at the plasma membrane of melanoma cells
To study the nanoscale organization of NRas and BRAF at the PM of melanoma cells, we turned to two-colour photoactivated localization microscopy (PALM) imaging. For that, NRas was tagged with the photoactivatable red fluorescent protein PAmCherry (30) (Supplementary Fig. S1A) and BRAF was tagged with the photoactivatable green fluorescent protein (PAGFP) (31). Two colour SMLM using these proteins has been verified (30), yielded localization uncertainties of ~20-30nm (Supplementary Fig. S1B), and used to study molecular organization for a variety of systems (e.g. (28,32)). In our current experiments, 108T melanoma cells (expressing WT BRAF and WT NRas) were transfected with these labelled proteins and seeded on coverslips, and fixed for imaging. The cells, in their resting state, were imaged in total internal reflection (TIRF) mode, in order to study the PM, while rejecting much of the background from the cell cytosol (Fig. 1A). Two colour PALM images of NRas and BRAF showed pronounced self-clusters of each of these proteins. We quantified the extent of relative self-clustering using univariate pair-correlation function (PCF, or g(r)) (26) (see details in the Analyses section of the Methods). For BRAF, the PCF curve averaged for multiple cells (Fig. 1B) indicated a level of significant self-clustering over 340nm (g(0-340nm)>1) and up to x36 higher than randomness (g(0-20)=36.3±6.4); compare green curve with flat black line of g(r)=1). NRas also showed significant self-clustering (Fig. 1C), however it was more diffused in comparison to BRAF clustering (g(0-20)= 2.64±0.23 and g(0-600nm)>1).
Protein abundance (i.e. density) is a second key characteristic of protein organization at PM, and is presented in Fig. 1D for NRas and BRAF. BRAF had a density of 6.4±1.3 molecules/μm2, while NRas had a significantly higher density, of 58.2±5.2 molecules/μm2 (p << 0.001).
To summarize these results here, and in the rest of the manuscript, we constructed a two dimensional map (Fig. 1E). In this map the y-axis represents the average values of g(<100), namely the extent of molecular self-clustering beyond randomness. The x-axis represents the protein density. These values are shown for individual cells as coloured discs, either for BRAF (green) or NRas (red). Note that both axes are logarithmic. The grids are guidelines that were defined for later comparison with additional measurements, as detailed in the rest of the study.
The colocalization between BRAF and NRas was quantified using the bivariate PCF statistics (BPCF (27); Fig. 1F; see Analyses). Briefly, through this statistics, the data is compared to two useful null hypotheses. The first is a model of no interaction between the proteins, resulting in a flat curve where g12(r)=1 ((26)). The second model is known as the ‘random labelling model’ and it indicates strong association of the interacting proteins in mutual clusters (i.e. molecular placement of both proteins is independent of their type). Our data indicate only partial mixing of NRas and BRAF, as experimental curve falls between the boarders of the two models. Normalizing the BPCF curves is useful for averaging the curves across multiple cells and results in our defined ‘Extent of mixing’ (EOM; Fig. 1G). Here, the model of no interaction is represented by EOM(r)=0, while homogeneous mixing (i.e. close association) is represented by EOM(r)=1. Again, the results where EOM(r) ranged between 0.2 and 0.4 indicated a relatively low colocalization of BRAF and NRAS.
NRas and BRAF show increased PM levels, more diffused organization and enhanced association upon EGF activation
Upon cell activation with EGF, NRas and BRAF get recruited to the PM (33). There, BRAF gets activated by active NRas, and further conveys the mitogenic signal downstream to MEK. To study the effect of MAPK pathway activation on the nanoscale organization of NRas and BRAF at the PM, we imaged melanoma (108T) cells using two-colour PALM after the addition of EGF to their medium (see Methods). As expected, the imaged cells demonstrated higher densities of NRas (by ~5 fold) and BRAF (by ~7 fold) as compared to resting cells (compare Figs. 1D,H). Both proteins seemed more diffused on average (yet not significantly), as found by PCF analyses for BRAF (compare g(0-20)=27.93±6.9 in Fig. 1I with g(0-20)=36.3±6.4 in Fig. 1B; p-value N.S.), and more so for NRas (compare g(0-20)=1.36±0.05 in Fig. 1J with g(0-20)= 2.64±0.23 in Fig. 1C; p=4.81E-05). These changes resulted in a significant shift of the data for BRAF and NRas in the clustering-density map (Fig. 1L; bold coloured lines originate from the coordinates of the averaged results of resting cells in Fig. 1E). Strikingly, BRAF and NRas now showed significantly increased mutual association in clusters, which approached homogeneous mixing in (BRAF) clusters (Fig. 1M,N; compare with the grey arrowhead that represent the EOM for resting cells expressing WT NRas in Fig. 1G).
Taken together, our results suggest a novel mechanism of signaling by NRas and BRAF in clusters. In resting cells, most of these proteins present at the PM reside in segregated clusters. Upon cell activation, NRas and BRAF clusters disintegrate, as their extent of relative self-clustering is reduced. The levels of these proteins increase. Thus, the more diffused organization of these proteins allows their close interaction and effective signaling downstream.
The dynamics of EGF-induced NRas and BRAF recruitment to the PM
We noted that NRas and BRAF resided at the PM in nanoscale clusters, both before and after EGF stimulation (Figs. 1B,C,I,J). Such clusters have been described before using immunogold EM (4) and PALM (34). Thus, we wanted to compare the size of these clusters in our data, in copy numbers of NRas and BRAF molecules. We show the cumulative probability of these proteins to reside in clusters of size 2-30mer. The distribution for both proteins is skewed (having no characteristic size). The majority (~70-80%) of the clusters were in dimers, another ~5-10% were trimers, with decreasing fractions of larger clusters (Fig 1L). The size-distributions shifted to larger clusters upon EGF-stimulation (darker green and red lines in Fig. 1O).
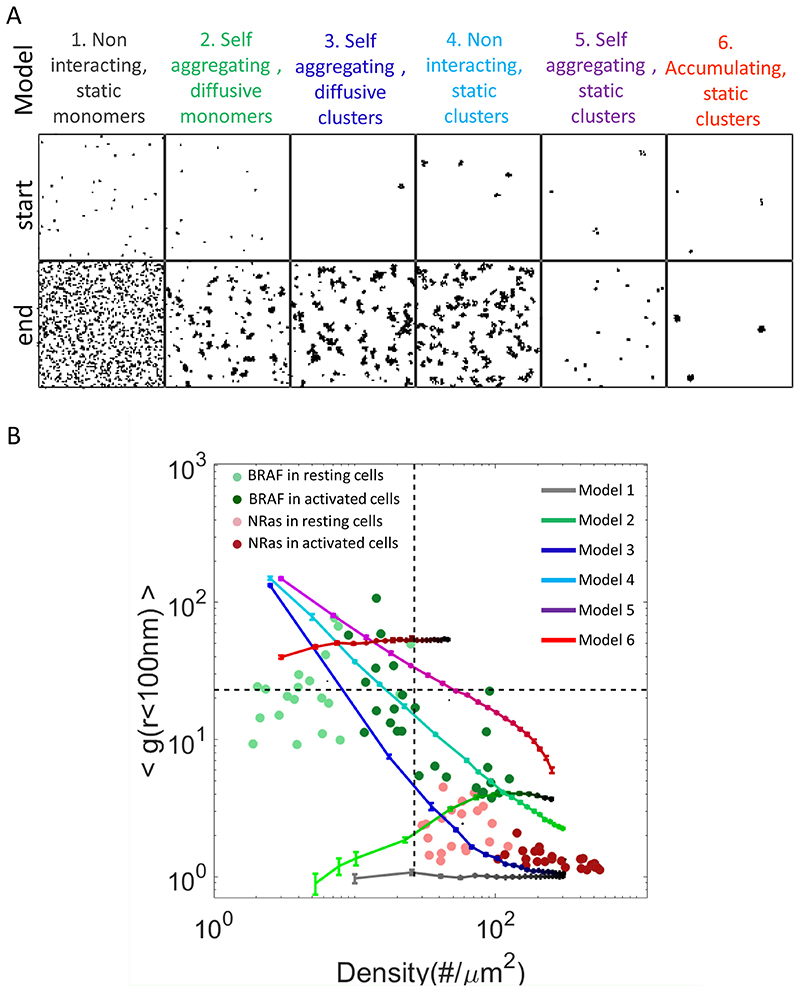
The recruitment of NRas and BRAF to the PM, upon EGF stimulation could follow various dynamic mechanisms. For instance, molecules could be delivered to the PM as monomers, or in vesicles. They could enrich pre-existing clusters or form new ones. The monomers and clusters could diffuse and interact at the PM, or remain stationary. Thus, we modelled such representative mechanisms (Fig. 2A; models 1-6), and simulated each model using Monte-Carlo simulation (with 20 repetitions for each model; see Methods). Initial conditions were chosen according to the measured densities in our PALM imaging experiments (Fig. 1). Representative initial and final patterns may serve for qualitative evaluation of the models (Fig. 2A; top and bottom rows, respectively). For quantitative evaluation, we analysed the clustering-density 2D plots, as for the experimental data (Figs. 1E,I, 2B). Cleary, models 1, 2 and 6 could be ruled out as their g(r)-density curves could not account for the experimental data of neither BRAF, nor NRas. Models 1 and 2 consider the arrival of proteins as monomers to the PM, while model 6 considers the arrival of the molecules into pre-existing, static clusters. Model 3 describes best the data for NRas, while model 4 describes best the data for BRAF. These models consider the arrival of molecules in clusters, and either allow their diffusion and aggregation at the PM (model 3) or does not allow diffusion and aggregation of the clusters (model 4). Still, model 5 cannot be ruled out for BRAF. This model considers arrival of molecules to the PM into new clusters that remain static. Thus, we conclude that self-aggregation in either dynamic or static clusters (model 3 for NRas and model 4 or 5 for BRAF) seem to be needed in order to account for our data.
Fig. 2. Clustering analyses of NRas and BRAF.
(A) The description of dynamic models that were simulated as Monte-Carlo reaction-diffusion processes in order to account for the organization of NRas and BRAF upon EGF activation. See further details on model parameters in Supplementary Table S3. Images include representative (top raw) starting and (bottom row) end patterns. (B) The g(0-100)-density map of the simulations due to the different models (1–6). Experimental results presented in Fig. 1 are overlaid for a convenient comparison with the simulation results.
Expression of NRas mutants results in enhanced NRas clustering and BRAF recruitment that allows signaling
We next aimed to study the organization of mutant NRas and BRAF at PM. Specifically, we studied the molecular organization in 108T cells overexpressing the melanoma-driving NRas mutants G12D (Fig. 3A–C,S1A), G13D (Fig. 3D–F,S1A) or Q61R (Fig. 3G–I,S1A). In resting cells, NRas levels at the PM were somewhat reduced, while BRAF levels were significantly elevated. Likewise, the extent of NRas self-clustering increased for G12D and G13D (Fig. 3A,B,D,E). Importantly, for all mutants, the colocalization of NRas and BRAF was increased in these resting cells (Fig. 3C,F,I; Supplementary Fig. S4A–F; clear significance was observed for the G13D and Q61R, while for G12D the increase was not significant). Thus, expression of NRas mutants lead to BRAF organization that resembles those of EGF-activated cells (compare results with Fig. 1E,L,N). Our results indicate that although mutant NRas becomes more clustered relative to WT NRas at the PM (Fig. 3J), the increased levels of BRAF at the PM could allow for its efficient mixing with the now more clustered NRas. Our results further suggest an unexplored mechanism of mutant NRas-enhanced signaling, by more efficient recruitment of BRAF.
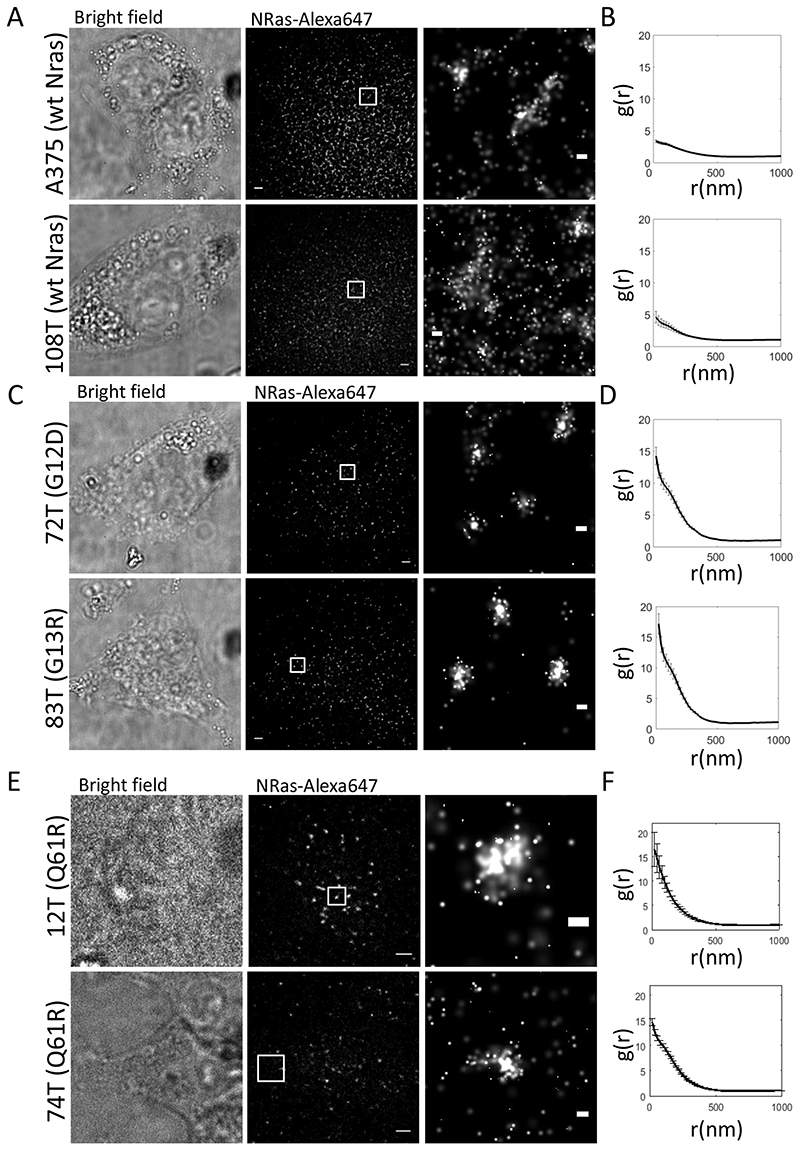
To validate that NRas mutants leads to increased self-clustering, we further imaged endogenous WT and mutant NRas proteins at the PM of multiple resting melanoma cell lines, A375 (WT NRas), 108T (WT NRas), 72T (G12D), 83T (G13R), 12T (Q61R) and 74T (Q61R). Imaging was conducted via direct Stochastic Optical Reconstruction Microscopy (dSTORM) (35) of NRas, labelled with mouse anti-NRas primary Ab and an anti-mouse secondary labelled with Alexa647. dSTORM imaging has a limited ability to quantify absolute molecular counts due to molecular blinking, antibody aggregation and the existence of multiple fluorophores on each antibody. Thus, we restrict our dSTORM-related analyses here (and later in the text) to a relative comparison of molecular clustering between the different cell lines. As before, WT NRas showed relatively low levels of self-clustering (Fig. 4A,B), while cell lines containing mutated NRas, demonstrated a significantly higher levels of NRas self-clustering (Fig. 4C,D,E,F). Thus, our previous of enhanced NRas self-clustering upon expression of NRas mutants does not depend on its overexpression, as needed for the PALM experiments. Notably, in contrast to the dSTORM assay, by PALM imaging we could directly visualize the mutant NRas protein and not average them with the WT proteins.
Fig. 4. Enhanced NRas clustering in multiple melanoma cell lines.
(A) Bright-field and Two-colour dSTORM imaging of resting melanoma cells expressing WT NRas (top and bottom rows: A375, 108T respectively). Cells were seeded on the coverslip for 2 days before fixation. Shown are representative cells (N=10, for each cell line). Bars – 2 μm (left) and 200 nm. (B) PCF of NRas for the data shown in A. (C) Brightfield and two-colour dSTORM imaging of resting melanoma cells expressing either NRas G12D (top rows, 72T), o NAS G13D (bottom row, 83T). Cells were treated as in A. Shown are representative cells (N=10, for each cell line). Bars – 2 μm (left) and 200 nm. (D) PCF of NRas for the data shown in C. (E) Brightfield and two-colour dSTORM imaging of resting melanoma cells expressing either NRas Q61R (top and bottom rows, 12T, 74T, respectively). Cells were treated as in A. Shown are representative cells (N=10, for each cell line). Bars – 2 μm (left) and 200 nm. (F) PCF of NRas for the data shown in E. Error bars in all relevant panels are SEM due to measurements on multiple cells.
Clinical MEK inhibitor restores segregation of NRas and BRAF clusters as in resting cells
Our model for NRas signaling at the PM suggests a linkage between NRas and BRAF activity to their dynamic and mutual organization at the PM. We also observe that mutant NRas tends to enhance association with BRAF and thus, promotes mitogenic signaling. Multiple clinical trials currently employ MEKi for reducing such aberrant signaling (36). Thus, we wondered whether MEK inhibitor could affect the organization of NRas (either WT or mutant) and BRAF at the PM. If so, the inhibitor might affect both the WT and oncogenic mutant versions of these proteins.
We started by imaging 108T cells overexpressing mutant NRas (namely G12D, D13D and Q61R) and BRAF after treatment of the cells with trametinib, MEKi (Fig. 5). These cells demonstrated enhanced clustering of both NRas and BRAF, relative to resting cells expressing WT NRas (compare Fig. 5A,D,G to Fig. 1A,E,G. See also and Supplementary Fig. S4B,D,F). NRas levels at the PM were significantly reduced (Supplementary Table S4), as for the cells that expressed mutant NRas without MEKi treatment (compare with Fig. 3B,E,H). In contrast, BRAF PM levels did not change significantly relative to the resting cells expressing WT NRas (compare Fig. 5B,E,H; green discs and Fig. 1E). Thus, the significant increase in BRAF levels caused by expression of NRas mutants (Fig. 3B,E,H) was cancelled by MEKi treatment.
Importantly, mutant NRas and BRAF were segregated from each other in the MEKi-treated cells (Fig. 5C,F,I), in contrast to the elevated colocalization that we saw in the absence of MEKi treatment (Fig. 3C,F,I).
We were interested if MEKi could also affect the nanoscale organization of NRas and BRAF in melanoma cells expressing WT NRas (Supplementary Fig. S5A–F). Similar to the results for cells expressing mutant NRas, we observed that the clustering extent of both proteins was enhanced (Supplementary Fig. S5A.B,D,E). NRas and BRAF also showed co-localization levels similar to those of resting cells (Supplementary Fig. S5C). Thus, MEKi could reverse the effect of the NRas mutants on the nanoscale organization of NRas and BRAF in cell expressing either WT or mutant NRas.
Taken together, these results suggest that trametinib is changing the organization and dynamics of NRas and BRAF on the plasma membrane. We suggest that this mechanism is the reduction of these proteins’ ability to interact and form mutual complexes at the PM. This mechanism seems to act in parallel to the proteins’ propensity for self-clustering (which seems to be their default state in resting cells).
Ras and BRAF in self-clusters associate upon EGF activation in MEKi-resistant cells
The continuous treatment of melanoma with MEKi often results in incurred resistance of the cells and cancer recurrence (37). Thus, we wanted to further test the possible relation of the nanoscale organization of NRas and BRAF in nanoclusters to MEKi resistance. For that, we imaged a melanoma cell line, 109T, which is significantly less sensitive to MEKi than 108T. This cell line has an IC50 of 804.7 nM, compared to the IC50 of 1.4 nM of 108T cells, and expresses the WT forms of NRas and BRAF (just like 108T cells).
In resting 109T cells, both NRas and BRAF remained in pronounced self-clusters (Fig. 6A B and Fig. S5G, H) with little mutual association (Fig. 6C). The density of NRas was significantly lower than in resting 108T cells (Fig. S5I, magenta line). However, upon EGF activation, NRas remained in self-clusters (Fig. 6D,E and Fig. S5J,K) -much like in cells expressing NRas mutants (Fig. 3A,D,G), and significantly different than in EGF-activated 108T cells (Fig. 1H). At the same time, the density of BRAF clusters grew significantly (Fig. 6E, green line and compare green bars in S5I, L). While the EOM between NRas and BRAF in these cells did not change relative to resting 109T cells (Fig. 6F), we could identify many more co-clusters of NRas and BRAF at the PM of these cells (Fig. 6D yellow clusters; compare with 6A). We quantified the fraction of the pixels containing BRAF and NRas detections (yellow pixels in Fig. 6D) relative to NRas -containing pixels (red pixels, respectively). We found that NRas and BRAF colocalization in mutual clusters was significantly higher in resting 109T cells than in 108T cells, and was comparable in extent to 108T cells expressing NRas mutants (Fig. 6G; note significance is relative to resting 109T cells in leftmost bar, and unless specified otherwise). Upon EGF activation, this fraction of co-clustered molecules increased ~4-fold relative to resting 109T cells, and significantly exceeded that of EGF-activated 108T cells and of 108T cells expressing NRas mutants.
We conclude that resistance to MEKi might involve a dynamic organization of NRas and BRAF in pronounced clusters at the plasma membrane that can come together upon EGF activation, as in cells expressing mutant NRas. This further suggests the existence of a shared mechanism of signaling regulation in NRas and BRAF clusters under conditions that mediate aggressive and persistent melanoma. Our study sets the stage for finding and characterizing such a mechanism.
Clinical MEK inhibitor diminishes ERK signaling of NRas mutants
To validate the signaling effects of EGF stimulation or mutant NRas overexpression on the mitogenic pathway in our experiments, we performed Western blots of pERK (and ERK) in the 108T cells (Fig. 7A). Indeed, the overexpression of mutant NRas (either G12D, G13D or Q61R) resulted in enhanced pERK levels, as compared to WT NRas resting cells (Fig. 7A,B). Moreover, pERK levels were also comparable or higher relative to EGF-activated, cells. As expected, MEKi treatment of the cells, transfected with mutant NRas, completely abrogated pERK levels. These results correlate between the nanoscale organization of BRAF and either WT or mutant NRas with the downstream activation of the mitogenic pathway.
Our results suggest a potentially new mechanism of MEKi function in melanoma cells carrying NRas mutations, but lacking MEK mutations. Thus, we further conducted a proliferation assay to make sure that the growth of these cells were affected by MEKi treatment. Indeed, for the tested cell lines, the number of cells that underwent MEKi treatment did not increase or even decreased over 96 hours while the number of cells without treatment increased by two- or three-fold during this time (Supplementary Fig. S6A). We further characterized the transformation state of our used cell lines, in comparison to a wide variety of other melanoma cells lines with various genetic backgrounds (Supplementary Table S1). We found that the 108T cell line showed elevated expression of the transformation marker ABCB5 (Supplementary Fig. S6B,C), which was above the average of 25 cell lines for the 108T cells, while 12T cells were slightly below average (Supplementary Fig. S6D). Thus, the cell lines used in this study (and esp. 108T) showed a transformed phenotype that was sensitive to MEKi treatment.
BRAF is more activated by monomeric NRas than clustered NRas
Next, we wanted to directly show that the clustered state of NRas can locally diminish its capability for signaling via phosphorylation of BRAF. For that, we performed two-color direct Stochastic Optical Reconstruction Microscopy (dSTORM) of phosphorylated BRAF (pBRAF) and NRas in the same cells and in single molecule detail. 108T cells were stimulated with EGF for 15 min, fixed and stained for NRas and pBRAF using primary antibodies and secondary antibodies stained with Atto488 and Alexa647 respectively. We found that NRas and pBRAF molecules showed both in self-clusters and in a diffused pattern (Fig. 7C, top; Supplementary Fig. S7A). We next distinguished the NRas molecules in either monomers or clusters of various sizes using the DBSCAN algorithm (25) (coded in Matlab). We then studied the abundance of pBRAF at NRas clusters, as a function of their size. Specifically, individual pBRAF molecules were assigned by their proximity (<70 nm) to NRas monomers and or to individual clusters (Fig. 7D). Next, we plotted the relative number of NRas-associated pBRAF in NRas clusters, as a function of their size (in copy number; Fig. 7E; absolute counts of NRas-proximal pBRAF are shown in Supplementary Fig. S7B). Strikingly, NRas monomers mixed significantly more with pBRAF molecules relative to NRas in self-clusters (Fig. 7E; bar for monomers is highest among all cluster-sizes, and bars get shorter with cluster size). Importantly, our results were insensitive to the chosen threshold of NRAF-pBRAF proximity (tested for thresholds of 35, 50, 70, 85 and 105nm; Supplementary Fig. S7C–F).
Our results indicate that clustered NRas can associate less with BRAF, and thus has a lower ability to signal downstream. On the other hand, diffusion and mixing of BRAF and NRas at the PM promote signaling and cell activation. Thus, our observed spatial redistribution of NRas and BRAF at the PM directly affects their signaling.
Discussion
In this study, we applied two-colour PALM to resolve the organization of NRas and BRAF at the PM of melanoma cells. Imaging showed that NRas and BRAF reside in pronounced clusters and with low interaction in resting 108T cells. EGF activation of the cells caused the increase of NRas and BRAF levels, along with their more diffused organization at the PM. This reorganization enabled their enhanced interaction and signaling. Expression of oncogenic mutants of NRas in resting cells led to a similar reorganization and enhanced signaling. MEKi treatment of cells led to reduced NRas protein levels at the PM. This treatment was also able to reverse the reorganization caused by NRas mutants upon their expression in such cells. We further found that NRas and BRAF colocalization in mutual clusters was significantly higher in resting MEKi-resistant (109T) cells relative to non-resistant (108T) cells, and was comparable in extent to such non-resistant cells expressing NRas mutants.
Taken together, our results indicate novel mechanisms for NRas-BRAF signaling at the PM through changes in their self and mutual organization (Fig. 7F). NRas and BRAF clusters should be more dispersed, along with the increase of protein levels, for enhanced NRas signaling upon EGF activation. While the expression of NRas oncogenic mutants increases NRas clustering, it also elevates the levels and dispersion of BRAF at the PM. This allows NRas mutants to efficiently recruit BRAF and productively signal downstream, as indicated by elevated pErk levels (Fig. 7A–E). Treatment with MEKi reverses BRAF levels and organization at the PM (the reverse shift in Fig. 7F). It results in NRas-segregated BRAF clusters, as in resting cells, and thus inhibits the signaling (Fig. 7A–E).
Traditionally, studies have addressed Ras and BRAF activity using various biochemical assays. Specifically, Western blotting and immunoprecipitation are insensitive to the exact organization of these proteins in clusters. Moreover, in such assays the results are averaged over many millions of cells and a staggering number of individual complexes. While diffraction-limited microscopy often achieves single cell resolution, it still averages the signal of thousands of protein complexes in each pixel (37).
More recently, high resolution imaging using immunogold-labeling TEM (IG-TEM) has revealed that Ras reside in small (5-8 mer) nanoclusters, with a size < 10nm (38–40). Notably, such labelling in TEM often suffers from poor labeling efficiency and multiple clustering artifacts (41). It also requires the ripping of the cells from a coverslip before labelling, which might alter the intact organization of the proteins under study (42). Such artifacts largely restrict the ability of this method to resolve molecular organization in intact cells.
In contrast, studies involving super-resolution microscopy have emphasized the role of Ras (43,44) and BRAF dimerization (6,45) (following biochemical studies (46)). Nan et al employed PALM and molecular complementation in their study. Still, these studies have also reported on larger scale self-clusters of the multiple Ras and RAF isoforms. Likewise, van Lengerich et al have shown EGF-induced clustering of the mitogenic receptors HER3 using dSTORM (47). Still, the mutual organization of NRas and BRAF has not been studied, nor were these proteins studied under the range of conditions studied here; esp. in human melanoma cells.
In contrast to these previous studies, our imaging approach has revealed relatively higher order clusters of NRas and BRAF. Both proteins demonstrated a size distribution of clusters from dimers to ~30 proteins (Fig. 10). Importantly, we could follow changes in this organization under various conditions, including EGF-induced cell activation, expression of oncogenic NRas mutations and after cell treatment with relevant clinical inhibitor. Mutant NRas resided in more pronounced self-clusters relative to WT NRas in multiple melanoma cell lines, yet could associate more with the clustered, and more abundant BRAF in 108T cells.
Importantly, cells treatment with the MEK-inhibitor trametinib had a pronounced effect on NRas and BRAF self-clustering and their spatial overlap. It diminished the density of these proteins at the PM, and increased their extent of self-clustering (Figs. 5,S3). MEKi treatment also restored the non-overlapping self-clusters of NRas and BRAF. Thus, MEKi treatment could restore NRas and BRAF PM levels and organization to their extent as in resting cells that express WT NRas. Since MEK acts downstream of NRas, this effect of its inhibitor exceeds the canonical mechanistic model by which this drug interferes with the catalytic site of MEK.
Taken together, our results raise the possibility of clinical treatment with MEKi of melanoma cells carrying NRas mutations, but lacking MEK mutations. Still, the underlying mechanisms of melanoma in treated patients seem to be complex. For instance, recent clinical updates report on gained resistance of melanoma cells to BRAF and MEK inhibitors in patients (48). Resistance mechanisms involve both genetic and epigenetic mechanisms. Mechanisms related to BRAF include the appearance of BRAF V600 mutations, gene amplification, splice variants, RAF-RAF dimerization, etc. (see (48,49) for review). The molecular mechanisms that facilitate drug resistance are of prominent clinical interest. Our results show that NRas and BRAF nanoscale patterning in MEKi-resistant cells generally resemble the organization of these molecules in cells that express oncogenic NRas mutations. This suggests a mechanism of NRas and BRAF patterning that is common to signal regulation in both cases of aggressive cancer progression due to either expression of melanoma-driver mutations or drug resistance.
Since MEKi treatment inhibits aberrant mitogenic signals downstream of BRAF, treatment of melanoma cells carrying NRas mutations, but lacking MEK mutations may still be effective in the face of such BRAF-dependent mechanisms (although such clinical treatment has not shown significant success so far) (50). Additional complications that are involved in cancer treatment with chemotherapy include the existence of residual dormant cancer cells and micro-metastases (49). Thus, our suggested MEKi treatment of melanoma cells carrying NRas mutations clearly requires careful testing and validation in model animals.
Supplementary Material
Statement of Significance.
Nanoscale dynamic organization of WT and mutant NRas relative to BRAF serves as a regulatory mechanism for NRas signaling and may be a viable therapeutic target for its sensitivity to MEKi.
Footnotes
Additional Information:
This research was supported by Grants no. 1417/13 and no. 1937/13 from the Israeli Science Foundation (E.S.). Y.S. is supported by the Israel Science Foundation grant No. 696/17, the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 754282), the ERC (CoG-770854), MRA (#622106), Rising Tide Foundation, Henry Chanoch Krenter Institute for Biomedical Imaging and Genomics, Estate of Alice Schwarz-Gardos, Estate of John Hunter, Knell Family, Peter and Patricia Gruber Award, and the Hamburger Family.
The authors declare no potential conflicts of interest
Contribution: E.S. supervised the research. E.S., A.H., R.S., J.S. and O.Y. designed the research; O.Y., R.S., J.S., and Y.R. performed research; O.Y., Y.R., and Y.N. analyzed the data; E.S., R.S. and A.H. wrote the paper.
Data availability
Data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Morrison DK. MAP kinase pathways. Cold Spring Harbor perspectives in biology. 2012:4. doi: 10.1101/cshperspect.a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keshet Y, Seger R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods in molecular biology. 2010;661:3–38. doi: 10.1007/978-1-60761-795-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. P Natl Acad Sci USA. 2005;102:15500–5. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hancock JF. Ras proteins: different signals from different locations. Nature reviews Molecular cell biology. 2003;4:373–84. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 5.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, et al. An Acylation Cycle Regulates Localization and Activity of Palmitoylated Ras Isoforms. Science. 2005;307:1746. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 6.Nan X, Collisson EA, Lewis S, Huang J, Tamguney TM, Liphardt JT, et al. Single-molecule superresolution imaging allows quantitative analysis of RAF multimer formation and signaling. Proc Natl Acad Sci U S A. 2013;110:18519–24. doi: 10.1073/pnas.1318188110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Luca A, Maiello MR, D’Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert opinion on therapeutic targets. 2012;16(Suppl 2):S17–27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 8.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome. Atlas N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazarian R, Shi HB, Wang Q, Kong XJ, Koya RC, Lee H, et al. Melanomas acquire resistance toB-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–U377. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friday BB, Adjei AA. Advances in targeting the Ras/Raf/MEK/Erk mitogen-activated protein kinase cascade with MEK inhibitors for cancer therapy. Clin Cancer Res. 2008;14:342–6. doi: 10.1158/1078-0432.CCR-07-4790. [DOI] [PubMed] [Google Scholar]
- 14.Philips MR, Der CJ. Seeing is believing: Ras dimers observed in live cells. P Natl Acad Sci USA. 2015;112:9793–4. doi: 10.1073/pnas.1511805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arafeh R, Qutob N, Emmanuel R, Keren-Paz A, Madore J, Elkahloun A, et al. Recurrent inactivating RASA2 mutations in melanoma. Nat Genet. 2015;47 doi: 10.1038/ng.3427. 1408-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alon M, Arafeh R, Sang Lee J, Madan S, Kalaora S, Nagler A, et al. CAPN1 is a novel binding partner and regulator of the tumor suppressor NF1 in melanoma. Oncotarget. 2018;9 doi: 10.18632/oncotarget.25805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzen F. Gateway (R) recombinational cloning: a biological operating system. Expert Opin Drug Dis. 2007;2:571–89. doi: 10.1517/17460441.2.4.571. [DOI] [PubMed] [Google Scholar]
- 18.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:1027–36. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman E, Barr V, Manley S, Patterson G, Balagopalan L, Akpan I, et al. Functional Nanoscale Organization of Signaling Molecules Downstream of the T Cell Antigen Receptor. Immunity. 2011;35:705–20. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ovesny M, Krizek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–90. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 22.Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Identification of clustering artifacts in photoactivated localization microscopy. Nature Methods. 2011;8:527–8. doi: 10.1038/nmeth.1627. [DOI] [PubMed] [Google Scholar]
- 23.Puchner EM, Walter JM, Kasper R, Huang B, Lim WA. Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory. P Natl Acad Sci USA. 2013;110:16015–20. doi: 10.1073/pnas.1309676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durisic N, Laparra-Cuervo L, Sandoval-Alvarez A, Borbely JS, Lakadamyali M. Single-molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate. Nature Methods. 2014;11:156–62. doi: 10.1038/nmeth.2784. [DOI] [PubMed] [Google Scholar]
- 25.Ester M, Kriegel HP, Sander Jorg, Xu Xiaowei. A density-based algorithm for discovering clusters a density-based algorithm for discovering clusters in large spatial databases with noise; KDD’96: Proceedings of the Second International Conference on Knowledge Discovery and Data Mining; 1996. pp. 226–31. [Google Scholar]
- 26.Wiegand T, Moloney KA. Rings, circles, and null-models for point pattern analysis in ecology. Oikos. 2004;104:209–29. [Google Scholar]
- 27.Sherman E, Barr VA, Samelson LE. Resolving multi-molecular protein interactions by photoactivated localization microscopy. Methods. 2013;59:261–9. doi: 10.1016/j.ymeth.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yakovian O, Schwarzer R, Sajman J, Neve-Oz Y, Razvag Y, Herrmann A, et al. Gp41 dynamically interacts with the TCR in the immune synapse and promotes early T cell activation. Scientific reports. 2018;8(1):9747. doi: 10.1038/s41598-018-28114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman E, Barr V, Manley S, Patterson G, Balagopalan L, Akpan I, et al. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity. 2011;35:705–20. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peacock JA. Two-Dimensional Goodness-of-Fit Testing in Astronomy. Mon Not R Astron Soc. 1983;202:615–27. [Google Scholar]
- 31.Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nature Methods. 2009;6(2):153–9. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–7. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 33.Sherman E, Barr VA, Merrill RK, Regan CK, Sommers CL, Samelson LE. Hierarchical nanostructure and synergy of multimolecular signalling complexes. Nature communications. 2016;7:12161. doi: 10.1038/ncomms12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson MF, Marais R. Ras protein signalling. Semin Immunol. 2000;12:63–73. doi: 10.1006/smim.2000.0208. [DOI] [PubMed] [Google Scholar]
- 35.Endesfelder U, Heilemann M. Direct Stochastic Optical Reconstruction Microscopy (dSTORM) Advanced Fluorescence Microscopy: Methods and Protocols. 2015;1251:262–75. doi: 10.1007/978-1-4939-2080-8_14. [DOI] [PubMed] [Google Scholar]
- 36.Akinleye A, Furqan M, Mukhi N, Ravella P, Liu DL. MEK and the inhibitors: from bench to bedside. J Hematol Oncol. 2013;6:27. doi: 10.1186/1756-8722-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman E, Barr V, Samelson LE. Super-resolution characterization of TCR-dependent signaling clusters. Immunol Rev. 2013;251:21–35. doi: 10.1111/imr.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Hancock JF. Ras nanoclusters: Versatile lipid-based signaling platforms. Biochimica et biophysica acta. 2015;1853:841–9. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Hancock JF, Prior IA. Electron microscopic imaging of Ras signaling domains. Methods. 2005;37:165–72. doi: 10.1016/j.ymeth.2005.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y, Hancock JF. Ras nanoclusters: Versatile lipid-based signaling platforms. Bba-Mol Cell Res. 2015;1853:841–9. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 41.Griffiths G, Lucocq JM. Antibodies for immunolabeling by light and electron microscopy: not for the faint hearted. Histochemistry and cell biology. 2014;142:347–60. doi: 10.1007/s00418-014-1263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung YH, Guo MY, Ma APY, Ng AMC, Djurisic AB, Degger N, et al. Transmission electron microscopy artifacts in characterization of the nanomaterial-cell interactions. Appl Microbiol Biot. 2017;101:5469–79. doi: 10.1007/s00253-017-8305-6. [DOI] [PubMed] [Google Scholar]
- 43.Nan XL, Tamgueney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, et al. Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway. P Natl Acad Sci USA. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen M, Peters A, Huang T, Nan XL. Ras Dimer Formation as a New Signaling Mechanism and Potential Cancer Therapeutic Target. Mini-Rev Med Chem. 2016;16:391–403. doi: 10.2174/1389557515666151001152212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nan XL, Collisson EA, Lewis S, Huang J, Tamguney TM, Liphardt JT, et al. Single-molecule superresolution imaging allows quantitative analysis of RAF multimer formation and signaling. P Natl Acad Sci USA. 2013;110:18519–24. doi: 10.1073/pnas.1318188110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freeman AK, Ritt DA, Morrison DK. The importance of Raf dimerization in cell signaling. Small GTPases. 2013;4:180–5. doi: 10.4161/sgtp.26117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Lengerich B, Agnew C, Puchner EM, Huang B, Jura N. EGF and NRG induce phosphorylation of HER3/ERBB3 by EGFR using distinct oligomeric mechanisms. P Natl Acad Sci USA. 2017;114:E2836–E45. doi: 10.1073/pnas.1617994114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kakadia S, Yarlagadda N, Awad R, Kundranda M, Niu JX, Naraev B, et al. Mechanisms of resistance to BRAF and MEK inhibitors and clinical update of US Food and Drug Administration-approved targeted therapy in advanced melanoma. Oncotargets Ther. 2018;11:7095–107. doi: 10.2147/OTT.S182721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merlino G, Herlyn M, Fisher DE, Bastian BC, Flaherty KT, Davies MA, et al. The state of melanoma: challenges and opportunities. Pigm Cell Melanoma R. 2016;29:404–16. doi: 10.1111/pcmr.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boespflug A, Caramel J, Dalle S, Thomas L. Treatment of NRAS-mutated advanced or metastatic melanoma: rationale, current trials and evidence to date. Ther Adv Med Oncol. 2017 Jul;9(7):481–492. doi: 10.1177/1758834017708160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available from the corresponding author upon reasonable request.