Abstract
Methods to deconvolve single-cell RNA sequencing data are necessary for samples containing a mixture of genotypes whether natural or experimentally combined. Multiplexing across donors is a popular experimental design which can avoid batch effects, reduce costs, and improve doublet detection. Using variants detected in the RNAseq reads, it is possible to assign cells to their donor of origin and to identify cross-genotype doublets that may have highly similar transcriptional profiles precluding detection by transcriptional profile. More subtle cross-genotype variant contamination can be used to estimate the amount of ambient RNA. Ambient RNA is caused by cell lysis prior to droplet partitioning and is an important confounder of scRNAseq analysis. Here we develop souporcell, a method to cluster cells using the genetic variants detected within the scRNAseq reads. We show that it achieves high accuracy on genotype clustering, doublet detection, and ambient RNA estimation as demonstrated across a range of challenging scenarios.
The ability to demultiplex mixtures of genotypes from droplet-based scRNAseq protocols, e.g. drop-seq1 or 10x Genomics2, is important because mixed sample scRNAseq is a powerful experimental design that reduces costs per donor, controls for technical batch effects3, and provides information on both cross-genotype doublets and the amount of ambient RNA in the experiment. While biochemical assays have been developed to enable multiplexing scRNAseq4,5, mixed genotype samples can be demultiplexed using the genetic variants available from the reads. Until recently, a genotype reference obtained via whole genome or exome sequencing has been required for each multiplexed individual prior to cell-sample categorization6. We present souporcell, a method to cluster cells by genotype, call doublet-cell barcodes, and infer the amount of ambient RNA in the experiment without the use of a genotype reference. We compare our method to demuxlet, the gold standard method that requires genotype information a priori, as well as two new tools that, like souporcell, do not require prior genetic information7,8. We show that souporcell not only outperforms these new methods, but also surpasses demuxlet on both cell assignment and doublet accuracy. Furthermore, souporcell explicitly models and estimates the amount of ambient RNA in the experiment, which is a major confounder of scRNAseq analysis with regard to both expression and genotype. Although a tool for ambient RNA quantification exists9, it requires prior knowledge in the form of one or more well expressed genes known to not be expressed in a particular cell type. Souporcell is freely available under the MIT open source license at https://github.com/wheaton5/souporcell.
Clustering model and data preprocessing
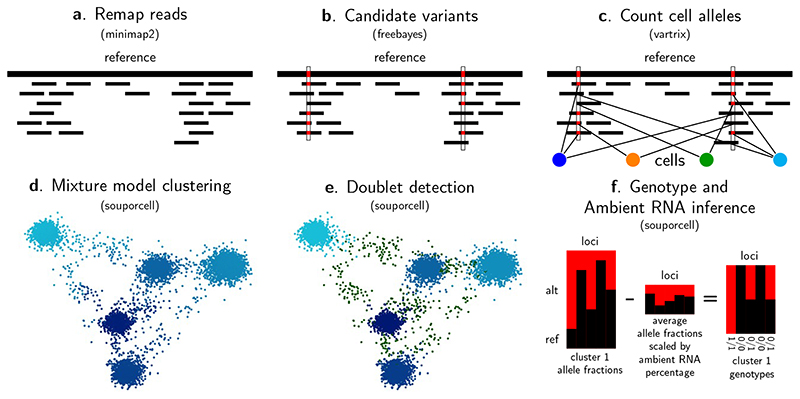
To cluster cells by genotype, we first must measure the allele information for each cell. To achieve the most accurate clustering, it is imperative that the variant calls and allele counts are measured accurately. While other tools start from the STAR aligned bam10 that is produced as part of running cellranger11, we have found several artifacts of the STAR alignments (methods) that are a significant source of false positive variants and reference bias. Instead, we remap the reads with minimap212 (Fig. 1a) which produces alignments more conducive to accurate variant calling. We call putative single nucleotide polymorphisms (SNPs) with freebayes13 (Fig. 1b). Next, we count alleles per cell with vartrix14(Fig. 1c) which avoids reference bias due to ambiguous support such as alignment end effects and corrects bases with duplicate reads via the UMI. If a source of reliable common variants is available, this can be used instead of the freebayes candidate variants.
Figure 1. souporcell overview.
a, We first remap the reads using minimap2 retaining the cell barcode and unique molecular identifier barcode for downstream use. b, We then call candidate variants using freebayes and c, count the allele support for each cell using vartrix. d, Using the cell allele support counts, we cluster the cells using sparse mixture model clustering (methods). e, Given the cluster allele counts, we categorize cells as doublets or singletons and excluding those doublets, f, we infer both the fraction of ambient RNA and the genotypes of each cluster (example for one cluster).
The clustering problem can be represented as a matrix X where each row represents a cell, each column represents a variant, and each element is the number of reads supporting each allele of the variant. We fit a mixture model with the cluster centers represented as the alternate allele fraction for each locus in the cluster. Because many clustering methods can easily get stuck in local optima, we cluster using a deterministic annealing variant of the expectation maximization algorithm15. This algorithm borrows ideas from statistical mechanics by treating the negative log probability of the data given the cluster centers as the energy state of the system and uses a temperature parameter that starts high and is slowly decreased to allow the solution to more often fall into the global optimal clustering. When the temperature parameter reaches 1.0, the loss function becomes the binomial density of the allele counts for the cluster center’s allele fractions (methods). The advantage of mixture model clustering over hard clustering is that cells can be partially assigned to multiple clusters, which naturally allows for both doublet cells and varying levels of ambient RNA (Fig. 1d). Having obtained the cluster centers, we identify doublet cell barcodes (Fig. 1e) by modeling a cell’s allele counts as being drawn from a beta-binomial distribution whose parameters are derived from either one or two clusters.
To identify the diploid genotypes of each cluster and the amount of ambient RNA (Fig. 1f), we assume that the allele counts for locus i of each cluster j are drawn from a binomial distribution with an alternative allele fraction of (1-ρ)fij+ρ*ai, where fij is 0, 0.5, or 1 (with a haploid mode limited to 0 and 1), ρ is a parameter representing the amount of ambient RNA and ai is the average allele fraction in the experiment. The ambient RNA shifts the observed allele fraction away from the underlying genotype allele fractions9. This model is implemented in the domain-specific language for probabilistic models, STAN16, and it solves for the maximum likelihood soup fraction with gradient descent.
There has been some concern in the community that it will be difficult to know which cluster corresponds to which individual after deconvolution with multiplexed scRNAseq experiments when genotypes are not known a priori. To address this, we propose an experimental design involving m overlapping mixtures for 2m-1 multiplexed individuals (Table 1). Each individual is assigned a binary number from 1 to 2m, where each bit corresponds to the inclusion (1) or exclusion (0) from each of the mixtures. This gives each individual a unique signature of inclusion/exclusion across the mixtures. Although each sample is in a different number of mixtures, the number of cells per experiment can be adjusted according to the number of mixtures that contain that sample. Souporcell provides a tool to match clusters from two experiments with shared samples (methods).
Table 1. Sample-cluster deconvolution experimental design.
| Mixture | 1 | 2 | 3 |
|---|---|---|---|
| Individual a | 0 | 0 | 1 |
| Individual b | 0 | 1 | 0 |
| Individual c | 0 | 1 | 1 |
| Individual d | 1 | 0 | 0 |
| Individual e | 1 | 0 | 1 |
| Individual f | 1 | 1 | 0 |
| Individual g | 1 | 1 | 1 |
| Mixture 1 | d | e | f | g |
| Mixture 2 | b | c | f | g |
| Mixture 3 | a | c | e | g |
This table outlines an experimental design of seven individuals with three overlapping mixtures to allow for clusters to be assigned to individuals. a, Shows the mapping of individuals to binary numbers where each digit of the binary number represents inclusion/exclusion from a mixture. b, The resulting mixtures.
Validation and Benchmarking
Currently, there are no good generative models available for batch effects, allele-specific expression, ambient RNA, and doublets in scRNAseq that can be used to generate in silico data for testing methods that cluster by genotype. To generate realistic data with known ground truth we sequenced five lines of induced pluripotent stem cells (iPSCs) from the Human iPSC initiative17 with the 10x Chromium single cell system, both individually and in a mixture of all five lines (with three replicates of the mixture). Each mixture contained 5-7,000 cells and ~25,000 UMIs per cell (Table S1). We first synthetically mixed 20% of the cells from the 5 individual samples while retaining their sample of origin. To make the synthetic mixture as close to real data as possible, we also simulated 6% doublets by switching all of the reads’ barcodes from one cell to that of another cell and 5% ambient RNA by randomly switching cell barcodes for 5% of the reads. A low dimensional representation of the expression matrix, E, reveals relatively little variation as expected since there is only one cell type present (Fig. 2a). Indeed, the most significant driver of expression appears to be the donor of origin, but the donor cells overlap in expression patterns and it is not possible to assign a donor to each cell based solely on expression patterns.
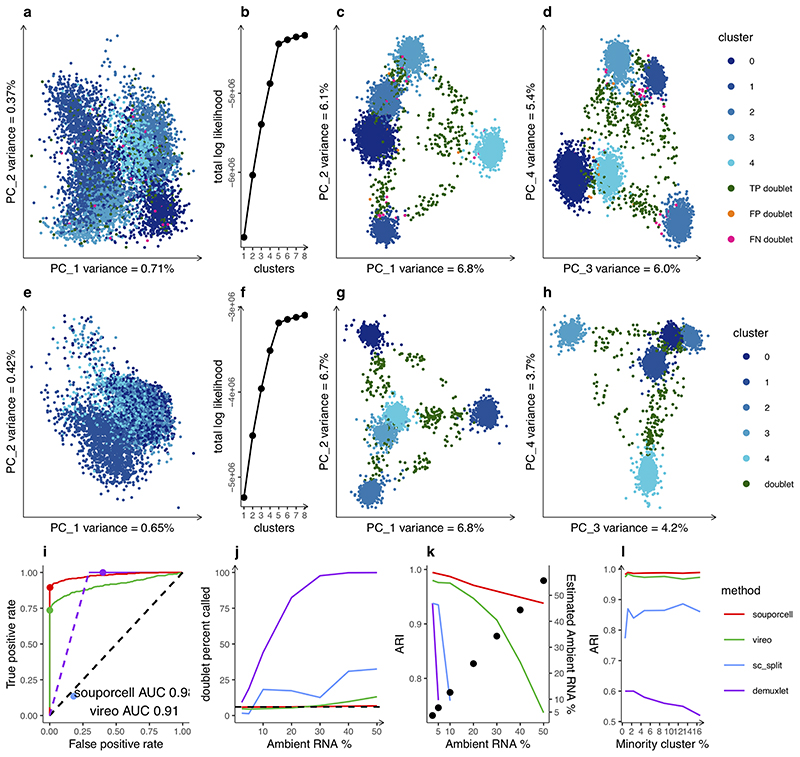
Figure 2. Evaluation of clustering accuracy.
a, Expression PCA of a synthetic mixture cells from five HipSci cells lines (n=7073 cells) with 5% ambient RNA and 6% doublets colored by known genotypes. Because these samples only contain one cell type, the largest remaining source of variation in the expression profile comes from the genotype, although the signal is not sufficient for accurate genotype clustering. b, Elbow plot of the number of clusters versus the total log likelihood showing a clear preference for the correct number of clusters (k=5). c and d, PCA of the normalized cell-by-cluster log likelihood matrix from souporcell (n=7073 cells). As this is a synthetic mixture in which we know the ground truth, we color by genotype clusters and highlight errors in orange (false positive doublets) and pink (false negative doublets). e, Expression PCA of a single replicate (see Fig. S1 for reps) of the experimental mixtures (n=4925 cells) colored by genotype clusters from souporcell. f, Elbow plot of the total log likelihood versus different numbers of clusters showing a clear preference for the correct number of clusters. g and h, PCA showing the first four PCs of the normalized cell-by-cluster log likelihood matrix colored by cluster (n=4925 cells). i, ROC curve of the doublet calls made by souporcell and vireo and a point estimate for scSplit (blue dot) for a synthetic mixture with 6% doublets 451/7073 and 10% ambient RNA. We show both the curves and the threshold chosen (points) for each tool. scSplit did not give a score so we simply show the point estimate. Demuxlet’s doublet probabilities were all 1.0 until the solid line starts, so we show a theoretical dotted line up to that point. j, Doublet call percentages for all tools on synthetic mixtures for varying amounts of ambient RNA versus the actual doublet rate (dotted line). k, Adjusted Rand Index (ARI) versus the known ground truth of synthetic mixtures with 6% doublets and a varying amount of ambient RNA. For levels >=10% ambient RNA, scSplit identified one of the singleton clusters as the doublet cluster, which means that the ARI was not clearly interpretable. Right y-axis vs points shows the estimated ambient RNA percent by souporcell versus the simulated ambient RNA percent. l, ARI of each tool on a synthetic mixture with 8% ambient RNA and 6% doublet rate with 1,000 cells per cluster for the first four clusters and a variable number of cells in the minority cluster (25-800 cells in the minority cluster).
We compared souporcell to vireo and scSplit, two other new tools that do not require prior genetic information. First, we ran variant calling and cell allele counting as recommended for each tool (methods). Using souporcell, we clustered cells by their genotypes, and evaluated the correct number of clusters through an elbow plot comparing the total log probability versus a varying number of clusters (Fig. 2b). The clustering output can be viewed as a matrix with cells as rows and clusters as columns with the values being the log likelihood of that cell versus the corresponding cluster. To visualize the five clusters identified by genotype we carried out a Principal Component Analysis (PCA) of the normalized log likelihood matrix, which reveals a clear separation of the clusters, with interspersed doublets (Fig. 2c and d). For these data souporcell assigned 6612/6622 singletons and 415/451 doublets correctly; four singletons were falsely labeled as a doublet, 35 doublets were misidentified as singletons, and one doublet and four singletons were unassigned. We carried out the same analysis for the three replicates of the experiment mixtures and show results for one (Fig. 2 row 2; see Fig. S1 for replicates). The expression PCA (Fig. 2e) and normalized cell-cluster loss PCA (Fig. 2g,h) of the experimental mixture were similar to the synthetic mixture indicating that the synthetic mixtures were an accurate approximation of real mixtures. To compare doublet detection between methods, we calculated a receiver-operator characteristic (ROC) curve of the doublet calls (Fig. 2i) on a synthetic mixture with 6% doublets and 10% ambient RNA that showed the area under the curve values of 0.98 and 0.91 for souporcell and vireo, respectively. We also show point estimates for the doublet threshold chosen. Demuxlet’s posterior doublet probability output did not have enough significant digits and is 1.0 until it starts varying with 27% false positives. The default doublet probability threshold for demuxlet gives nearly 40% false positive doublets.
Each of the five human iPSC lines has existing WGS data generated as part of the HipSci Project18. Therefore, for the experimentally mixed replicates, we compared each tool’s clustering to sample assignments obtained from demuxlet using genotypes available from the WGS. Demuxlet significantly overestimates doublets versus expectations based on the number of cells loaded11 (Table S2) especially as ambient RNA increases (Fig. 2j). Because we could not trust the doublet calls of demuxlet, we allowed scSplit, vireo, and souporcell to exclude their called doublets and then compared the remaining cells to demuxlet’s best single genotype assignment. The Adjusted Rand Index (ARI) of the remaining cell assignments versus demuxlet (Table S2) were 1.0 (fully concordant) for souporcell and vireo across the three replicates and an average of 0.97 for scSplit.
To evaluate the robustness of each tool across a range of parameters, we created synthetic mixtures of the five individual human iPSC scRNAseq experiments to test both the sensitivity to the ambient RNA level (Fig. 2j, k) and the ability to accurately assign cells to a cluster if it is much smaller than other clusters (Fig. 2m). For the ambient RNA experiment, we synthetically combined 20% of the cells from each of the five individual samples and simulated 6% intergenotypic doublets and a range of ambient RNA from 2.5%-50% representing realistic ranges previously reported9. We found that souporcell and vireo retain high accuracy with souporcell being more robust at accurately calling doublets in high ambient RNA cases (Fig. 2k). The ARI of scSplit and demuxlet suffered due to poor doublet detection. With these data we also show that souporcell is able to accurately estimate the amount of ambient RNA in the experiment (Fig. 2k). To test robustness to sample skew, e.g., one donor’s cells are underrepresented, we created a set of synthetic mixtures with 1,000 cells from each of four individual samples and 25-800 cells for the minority cluster including 8% ambient RNA and 6% doublets (Fig. 2m). We found that all tools performed well down to the minority cell cluster comprising only 1.2% (50 cells) of total cells (Fig. 2m), but only souporcell and vireo were able to correctly identify all minority sample singletons as their own cluster down to 0.6% of all cells. Again, demuxlet’s poor ARI was due primarily to extremely high levels of false positive doublets (Fig. 2l).
We then compared souporcell’s genotype and ambient RNA co-inference to vireo and scSplit versus the variants called from whole genome sequencing data. In scRNAseq data most variants have very low coverage per cluster compared to what would be generated from WGS data, thus the genotype accuracy is significantly lower than one would attain with genome sequencing. Nevertheless, souporcell surpasses both vireo and scSplit in genotype accuracy on a synthetically mixed sample with 6% doublets and 10% ambient RNA (Fig. S1i). The most common error mode for vireo and scSplit is calling homozygous reference loci as heterozygous variants (Fig. S1j) which is expected when ambient RNA is not accounted for, as it is not in these two tools.
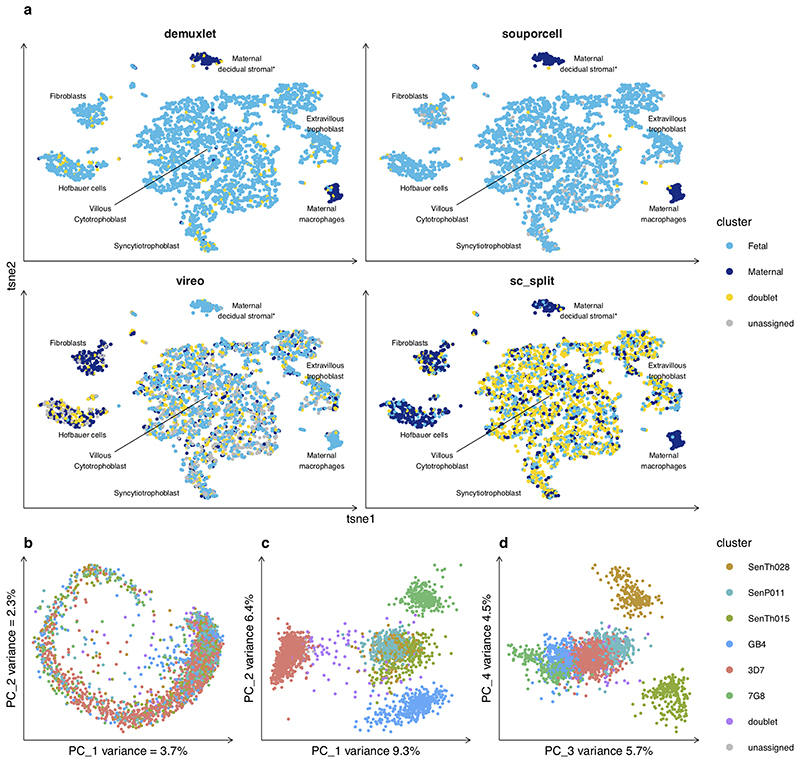
Next, we considered more challenging scenarios involving multiple cell types, widely varying numbers of cells per sample, and closely related genotypes. The decidua-placental interface plays an important role in pregnancy and birth, and is of importance to several diseases, including pre-eclampsia20. Recently, more than 70,000 cells were profiled by scRNAseq19 to explore the transcriptional landscape at this interface. The decidua is primarily composed of maternal cells with some invading fetal trophoblasts, while the placenta is largely composed of cells of fetal origin with the exception of maternal macrophages. In the study exploring this interface19, WGS from blood and placenta was used to genotype both mother and fetus, and demuxlet was used to assign cells to each individual. Here, we applied souporcell, vireo, and scSplit to two placental samples and one decidual sample from a single mother to determine if cellular origins could be established without reference genotypes. We show the expression t-SNE of a single placental sample labeled by cell type annotation19 and colored by genotype cluster as assigned by each method (Fig. 3a). While souporcell clusters agree with demuxlet and segregate with the expected cell type clusters, vireo and scSplit have major discordances with demuxlet. This is similar for the other samples tested (Fig. S2, Table S3). Comparing souporcell to demuxlet, there are 21 cells that demuxlet labels as maternal or fetal but which appear in the other individual’s cell type clusters. Based on the position of these cells in the expression t-SNE plot, it is most likely that these are errors in the demuxlet assignments that are not made by souporcell.
Figure 3. Application to challenging datasets.
a, Cell expression t-SNE plots of n=3,835 cells colored by each tool’s genotype assignments or clusters for placenta1 (other samples in Fig. S2). Cell phenotype clusters and cell genotype clusters co-segregate, with the majority of cell types being of fetal origin with the exception of maternal macrophages and *maternal decidual stromal cells, the latter of which (found only in one donor) were considered to be a non-placental artefact arising from the surgical procedure and were removed during data quality control in the original study19. We observe high concordance between souporcell and demuxlet (ARI 0.96) whereas vireo and scSplit have large discordances with ARI of 0 and 0.03 respectively. b, Expression PCA colored by genotype clusters for Plasmodium sample 1 (n=2608 cells) (other samples in Fig. S3) showing an even spread of genotypes throughout the asexual lifecycle. c and d, PCAs of first four PCs of souporcell’s normalized cell-by-cluster loss matrix showing good separation of each genotypic cluster (n=2608 cells).
We also tested souporcell on a non-human sample, the single-celled malaria parasite Plasmodium falciparum, for which single cell approaches are now used to explore natural infections21. Malaria infections often contain parasites from multiple different genetic backgrounds, and it is not possible to separate the strains prior to sequencing. These samples differ from human samples in a variety of ways; they are haploid when infecting humans, the genome is >80% A/T, and the transcriptome is only ~12 megabases (genome is ~23 Mb). We generated three datasets containing six genetically distinct strains of P. falciparum (methods) sampling 1893-2608 cells with median UMIs of ~1000. Analysis of the expression profile of one of these (see Fig. S3 for the others) reveals that the genotypes are distributed across the Plasmodium intra-erythrocytic cycle (Fig. 3b) while being well separated in normalized loss cluster space (Fig. 3c,d). The ARI for each method (Table S4) on the three Plasmodium data sets show superior performance for souporcell across the board, with scSplit suffering on all datasets and vireo performing poorly on one, which had an ARI versus demuxlet of 0.24. This sample was more difficult due to sample skew caused by a clonal expansion of one of the six strains.
Discussion
Here we have presented souporcell, a method for clustering scRNAseq cells by genotype using sparse mixture model clustering with explicit ambient RNA modeling. Our benchmarks show that souporcell can outperform all other currently available methods, including those that require genotypes a priori. Using more realistic and challenging test cases than previous studies, we show that souporcell is robust across a large range of parameters, and more so than any other currently available method. Moreover, souporcell is highly accurate for challenging datasets involving closely related maternal/fetal samples, and varying mixtures of Plasmodium falciparum strains. Limitations of souporcell include low signal to noise due to decreased UMI per cell and high numbers of donors causing increased local maxima. These issues are further explored in the supplementary note (Figs S1 and S5–7). Due to the advantages that mixtures give to scRNAseq experiments in ameliorating batch effects, improving doublet detection, and allowing for ambient RNA estimation, souporcell enables donor multiplexing designs to be used more easily than was previously possible, including in situations when no WGS or genotyping data are available. In addition to reducing cost and allowing for more complex and robust experimental designs, souporcell also enables valuable genotype information to be extracted and ambient RNA estimation at no additional cost.
Online Methods
Supplementary Methods
Remapping
We remap reads due to several different artifacts, described below. We first take the STAR aligned bam and create a fastq file from it using pysam and a custom python script (available at https://github.com/wheaton5/souporcell/renamer.py) while placing the UMI and cell barcode information in the read name for later use. We map these reads to the reference genome using minimap2 version 2.7-r654 with parameters -ax splice -t 8 -G50k -k 21 -w 11 --sr -A2 -B8 -O12,32 -E2,1 -r200 -p.5 -N20 - f1000,5000 -n2 -m20 -s40 -g2000 -2K50m --secondary=no, but have seen similar accuracy with the RNAseq aligner HiSat222. We resupply the cell barcode tags and UMI tags to the bam using pysam and a custom python script (available at https://github.com/wheaton5/souporcell/retag.py) and sort and index the bam file with samtools. All steps are now encapsulated into a simple pipeline script and provided as a singularity container for easy installation.
We identified three different artifacts introduced by the STAR alignments resulting in false positive variants as well as reference bias that causes reads that do not support the reference allele to appear as though they do. The first artifact is due to the way STAR handles spliced reads when the read does not match the reference well. STAR will take such a read and introduce multiple splice events that force it to fit the reference in a statistically spurious fashion. We have observed cigar strings such as 8M129384N12M50238N77M where the read matches one location well with 77 matches plus mismatches in one location. Instead of soft clipping the initial 20 bases that did not have a statistically significant alignment, STAR introduced two large splicing events to match very short regions (8 and 12 bases) to the reference. Due to the limitation of a read having a single mapping quality and these spliced reads being encoded as a single read object in the bam file, the variant callers will treat these spurious matches as high mapping quality. Consequently, for variants in these loci, the variant callers will count these reads as supporting the reference allele, thereby introducing reference bias and noise to the downstream clustering. This has been noted by others in the past, and GATK recommendations for variant calling on bulk RNAseq involve removing these regions of the alignments prior to variant calling23. The next set of artifacts is alignment parameter differences between STAR and aligners intended for variant calling. The second type of artifact we found was due to the soft clip penalty being higher in STAR and not being exposed as a parameter to the user. This leads to false positive variants due to the lack of soft clipping where other mappers would soft clip poorly matching read ends. The final issue is that the indel penalty relative to the mismatch penalty is much higher in STAR than other aligners. This causes the alignments to choose many mismatches over a single or few indels when possible and thus create false positive variants. This is a parameter which is exposed to the user but, the default makes the output of cellranger poorly suited for variant calling. For these reasons, we find it best to remap these reads with a mapper specifically tuned to genomic variant calling. We also have a -- skip_remap option and a --common_variants option as well as a --known_genotypes option. If using known variable sites such as from the 1k genomes project24 or if the genotypes of the donors are known, the remapping process is significantly less important. When provided with a known genotypes file, it initializes the cluster centers with the allele fractions corresponding to the known genotypes but then solves as normal from that point.
Variant Calling
Souporcell
Variant calling consists of two steps. First we identify candidate SNPs using freebayes (version v1.3.1-17-gaa2ace8) with parameters -iXu -C 2 -q 20 -n 3 -E 1 -m 30 --min-coverage 6 --max-coverage 100000 -- pooled-continuous. If one wished to use known common variant sites, one could skip this step and provide that vcf to the following step. In the second step we count alleles for each cell using the program vartrix (available at https://github.com/10XGenomics/vartrix (release version 1.1.3)) with parameters -- umi --mapq 30 --scoring-method coverage which gives us two sparse matrix outputs which represent the UMI allele counts per cell for each locus. For souporcell, we limit the loci considered for clustering to the ones with at least n cells (default =10) supporting each allele. For all human samples we used 10, but for the Plasmodium samples, we used n=4 due to the lower number of variants in the Plasmodium data. This provides us with fairly robust SNPs that have a good chance of aiding the clustering process.
Vireo
Vireo recommends running cellSNP (https://github.com/huangyh09/cellSNP version 0.1.6) on the STAR aligned bam with parameters --minMAF 0.1 --minCOUNT 100 limiting the analysis to loci with at least 100 UMIs and 10% minor allele fraction which are the settings we used throughout our analysis. For vireo donor clustering we use their R package, cardelino version 0.3.8.
scSplit
scSplit recommends using freebayes (the version we ran test on was v1.3.1-17-gaa2ace8) on the STAR aligned bam with parameters -iXu -C 2 -q 20 and then filtering for SNPs with a quality score >=30. We used bcftools for filtering with the command bcftools filter -e 'QUAL<30'. Then this vcf is used along with the matrix.py script in scSplit with the filtered vcf, the STAR aligned bam, and the cell barcode file as input to get the allele counts for each cell. For scSplit donor clustering we used git commit hash 52face6a4c1b291651bdf9b56328d168c7cb1fa6 cloned from master at https://github.com/jon-xu/scSplit on April 21, 2019.
Sparse mixture model clustering
Definitions
K: number of genotype clusters to be fixed at the outset. Lower case k will be used for indexing and referring to a specific cluster.
C: number of cells. Lower case c will be used for indexing and referring to a specific cell barcode. This barcode could have 0, 1, or more cells. It is important for some assumptions in this model that the majority of barcodes contain a single cell.
L: number of variant loci. Lower case l will be used to index and refer to a specific locus. We will assume only biallelic variants. Lc will be a list of loci with observed data in cell c.
A: Allele counts. Al,c is a vector of size 2 with the first number representing the number of reference alleles and the second representing the number of alt alleles seen at locus l in cell c.
□k,l: mixture parameter for allele fractions of cluster k at locus l. This is a real number representing the fraction of ref alleles in this cluster at this locus. We expect this to be near 1.0 (homozygous reference), 0.5 (heterozygous), or 0.0 (homozygous alt) but will be skewed from these values by noise, doublets, and ambient RNA.
T: temperature parameter for deterministic annealing process.
We define the likelihood of the data treating cells independently and marginalizing over the potential of each cell belonging to each cluster with a binomial likelihood for the alleles being drawn from the cluster center’s allele fraction.
Equation 1: Cluster model Likelihood function
We maximize this likelihood function using a deterministic annealing variant of the expectation maximization algorithm. The deterministic annealing approach adds a temperature parameter T which we initialize to 1/10th the average number of alleles expressed by each cell. At each temperature step we solve until convergence (total log likelihood change < 0.1). Each new temperature step the temperature is halved until <1 at which point we run a final step at T=1. We randomly initialize cluster centers and run this optimization 50 times by default and take the solution with the maximum total likelihood. At each temperature step, we define a temperature modified posterior for each cell belonging to each cluster as follows.
Equation 2: Deterministic annealing.
Which gives our maximization step according to the following equation.
Equation 3: Expectation Maximization update
Doublet detection
Definitions
Ak,l: Allele counts at locus l for all cells in cluster k according to the maximum probability cluster assignment from our clustering. This is a vector of size two with the ref and alt allele counts.
We treat the allele counts of each cell at each locus as random variables drawn from a beta-binomial distribution from either a single cluster or a pair of clusters. The beta-binomial is used to model our uncertainty in the binomial parameter p. For a single cluster the parameters are alpha = 1+alt counts and beta = 1+ref counts.
For the singleton case, we have
Equation 4: Singleton Likelihood
Where □ is the beta function and cluster i is the best fitting cluster for cell c.
The expected allele fractions of a doublet coming from cluster i, and cluster j is the average of the allele fractions of the two clusters. To obtain the pseudocounts needed to parameterize the beta-binomial, we use the total counts of the cluster with less coverage at this locus. That is,
Equations 5-6: Doublet beta-binomial parameters
The doublet probability given those conservative parameters becomes
Equation 7: Doublet likelihood
Where B is the beta function and α and β are the beta-binomial parameters described in Equations 5 and 6.
The posterior for each cell being a doublet is then given by
Equation 8: Doublet posterior
Where cluster i is the best fitting cluster for cell c and cluster j is the second-best fitting cluster for cell c and p(doublet) is the doublet prior. We allow the prior to be set by the user but have used an uninformed prior of 0.5 for all of our analysis.
We run the above process and remove doublet cells from the cluster allele counts repeatedly until we no longer find new doublets.
Genotype and ambient RNA co-inference
Definitions
ρ: mixture parameter representing the probability any given allele is arising from ambient RNA as opposed to from the cell associated with that barcode.
P: ploidy. We assume ploidy is limited to 1 or 2.
Al: total allele expression at locus l. This is again a vector of length 2 denoting the reference and alternative allele counts.
g: used to denote the number of copies of the reference allele. The expected reference allele rate without ambient RNA is g and g is an integer value ∈ [0..P]. Note that for biallelic variants and ploidy 1 or 2, g is sufficient to uniquely determine the genotype.
p(true): prior for variant being a true variant vs a false positive. The default is 0.9 which was the value used for all analyses.
Here, the proportion of ambient RNA in the system, ρ, is the only free parameter and we solve for it using maximum likelihood. The model treats each locus in each cluster as coming from one of three genotypes for diploid (0/0, 0/1, 1/1, here denoted by g = 0, 1, or 2) and two genotypes from haploid (0, 1). We treat each cluster as independent and each locus as independent, before marginalizing across the possible genotypes. The model also considers the possibility of the variant being a false positive. In this case, the variant will not segregate into distinct allele frequencies between different clusters and it will most likely not attain a value close to the standard allele frequencies expected from the diploid or haploid genotypes. Thus, we model the allele counts in each cluster as having come from a mixture of ambient RNA (an average allele fraction in the experiment) and from the cells in that cluster. The observed allele fractions are assumed to have been drawn from a binomial distribution with a probability that was skewed away from p = g/P by the level of ambient RNA ρ. Thus, the probability of the binomial from which the allele counts are drawn for true positive variants is the following.
Equation 9: True positive allele fraction
For a false positive the parameter is
Equation 10: False positive allele fraction
Thus, the full model is
Equation 11: Genotype and ambient RNA Likelihood function
We solve for ρ with gradient descent using the statistical modeling domain specific language STAN. Next, we calculate the posterior of the variant being a true positive for each of the three (or two in the haploid case) genotypes versus it being a false positive. The prior on variants being true positives can be set by the user, but defaults to 0.9 which is the value used in our analyses.
Human iPSC experiments
iPSC culture
Feeder-free iPSCs were obtained from the HipSci project18. Lines were thawed onto tissue culture-treated plates (Corning, 3516) coated with 5 μg/mL Vitronectin (rhVTN-N) (Gibco, A14700) using complete Essential 8 (E8) medium (StemCell Technologies, 05990) and 10 μM Rock inhibitor (Sigma, Y0503-1MG). Cells were propagated in E8 for 2 passages using 0.5 μM EDTA pH 8.0 (Invitrogen, 15575-038) for cell dissociation. Colonies were then dissociated into single cells using Accutase (Millipore, SCR005) and pooled in equal numbers, alongside individual lines, for one passage.
10x Single-cell 3’ RNA-seq
To create a single cell suspension, iPS cells were cultured as described above in six-well plates before being washed once with room temperature D-PBS (Gibco, 14190-144). The D-PBS was removed before adding 1 mL of Accutase (Millipore, SCR005). The cells were incubated at 37°C for seven minutes before adding 1 mL of E8 media. The cells were collected in a 15 mL Falcon tube and triturated three times with a 5 mL stripette to obtain a single cell suspension. To ensure no cell clumps remained, the cell suspension was passed through a 40 μm cell strainer. The cells were counted and the viability was assessed on a Countess automated cell counter (Life Technologies). GEMs (gel beads in emulsion) were created using the 10x Genomics Chromium™ Controller, according to the manufacturer’s protocol. All channels were loaded such that an estimated 10,000 cells were captured for GEM formation and successful library preparation. All samples were processed using a 10x Genomics Chromium™ Single Cell 3’ v2 kit (PN-120237), following the manufacturer’s instructions. Libraries were multiplexed and sequenced at a rate of one library per lane of a Hiseq 4000 (Illumina), acquiring 150 bp paired-end reads.
Synthetic mixtures
We generated synthetic mixtures with custom python scripts using pysam and numpy. We took all of the reads for a subset of cell barcodes from each of the individual experiments and combined them into a new dataset. We then simulated doublet formation by randomly choosing among the cell barcodes that we had already chosen for the mixture experiment and then chose a cross-genotype cell barcode with which to create a doublet. We then took all of the reads of one of those cell barcodes and changed their cell barcodes to that of the other cell. We also simulated ambient RNA by randomly changing a read’s cell barcodes to that of another cell barcode at a specified rate. The values of each of these parameters are described in the text and figure captions.
Demuxlet
We ran demuxlet git hash 85dca0a4d648d18e6b240a2298672394fe10c6e6 with default parameters except --field GT versus the cellranger bam, barcodes file, and vcf made by first downloading the exome bams from http://www.hipsci.org/ for each cell line, creating fastq files from them with samtools version 1.7 bam2fastq, then remapping to the cellranger reference with minimap2 version 2.7-r654 with parameters - ax sr, removing duplicates with samtools rmdup, and calling variants across the five bams with freebayes version v1.2.0-2-g29c4002-dirty with default parameters. Variants were then filtered with a custom python script using pyvcf such that the remaining variants be SNPs with QUAL >= 30.
Maternal/Fetal
We obtained two placental samples and one decidual sample from Vento et al19 at https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6701/samples/. The samples used were FCA7474065 (placenta1) (Fig. 3a), FCA747064 (placenta2 (Fig. S2b)), and FCA747063 (decidua1 (Fig. S2a) all from the same individual. We obtained the fastq files and ran cellranger version 2.1.1 on them with default parameters to obtain the bam and cell barcodes files which are the input to our system. We then ran souporcell, scSplit, and vireo on them with recommended settings for each tool as previously detailed and obtained the demuxlet calls used in Vento et al19. We ran souporcell, vireo, and scSplit on each of these and compared them to the demuxlet calls excluding the demuxlet doublet cells and the doublets called by each tool (Supp Table 3).
Plasmodium falciparum in vitro culturing and single cell analysis
P. falciparum strains were maintained in O+ blood in RPMI 1640 culture medium (GIBCO) supplemented with 25 mM HEPES (SIGMA), 10 mM D-Glucose (SIGMA), 50 mg/L hypoxanthine (SIGMA), and 10% human serum in a gas mix containing 5% O2, 5% CO2 and 90% N2. Human O+ erythrocytes were obtained from NHS Blood and Transplant, Cambridge, UK. All samples were anonymous. Plasmodium culture using erythrocytes and serum from human donors was approved by the NHS Cambridgeshire 4 Research Ethics Committee (REC reference 15/EE/0253) and the Wellcome Sanger Institute Human Materials and Data Management Committee. All P. falciparum clonal strains were obtained from MR4 (BEI resources): 3D7-HT-GFP (MRA-1029), 7G8 (MRA-152), GB4 (MRA-925), SenP011.02 (MRA-1176), SenTh015.04 (MRA-1181) and SenTh028.04 (MRA-1184). All strains were maintained in culture below 5% parasitemia for no less than 6 weeks without synchronization prior to the experiment in order to ensure maximum asynchronicity. Plasmodium1 pool was composed of 2 independently cultured flasks for each of the 6 strains. The Plasmodium1 pool was washed once in PBS, before resuspension in PBS at a concentration of 11,200 RBC/μl (corresponding to 479 parasites/μl). The Plasmodium2 pool was derived from an aliquot of the Plasmodium1 sample that had been resuspended in 200 μl of PBS and fixed with 800 μl of ice-cold methanol for 10 minutes on ice, before being washed twice in PBS and resuspended at 12,200 RBC/μl (corresponding 522 parasites/μl). The Plasmodium3 sample was derived from a mix of the 6 strains, grown in the same flask for 7 days and resuspended at 19,800 RBC/μl (corresponding to 960 parasites/μl). Hematocrits were established with a hemocytometer. Each cell suspension was loaded on one inlet of a 10x chromium chip according to manufacturer's instructions with a target recovery of 9000 cells per inlet. Chromium 10x v2 chemistry was used and libraries were prepared according to the manufacturer's instructions. Each 10x input library was sequenced on both lanes of a Hiseq 2500 Rapid Run using 75 bp paired-end sequencing.
Supplementary Material
Acknowledgments
We acknowledge the Wellcome Sanger Institute’s DNA Pipelines for construction of the 10x sequencing libraries. We thank Allan Muhwezi and Andrew Russell for assistance with parasite culture and 10x Single-cell 3’ RNA-seq respectively. In addition, we would like to thank Matthew Young for useful conversations about ambient RNA, Mirjana Efremova for providing information about the maternal/fetal data, and Katie Gray for assistance in interpreting the previously unannotated cluster. The Wellcome Sanger Institute is funded by the Wellcome Trust (grant 206194/Z/17/Z), which supports MKNL and MH. This work was supported by an MRC Career Development Award (G1100339) to MKNL. We would like to acknowledge the Wellcome Trust Sanger Institute as the source of the human induced pluripotent cell lines that were generated under the Human Induced Pluripotent Stem Cell Initiative funded by a grant from the Wellcome Trust and Medical Research Council, supported by the Wellcome Trust (WT098051) and the NIHR/Wellcome Trust Clinical Research Facility, and acknowledges Life Science Technologies Corporation as the provider of Cytotune (HipSci.org). The Cardiovascular Epidemiology Unit is supported by core funding from the UK Medical Research Council (MR/L003120/1), the British Heart Foundation (RG/13/13/30194; RG/18/13/33946) and the National Institute for Health Research [Cambridge Biomedical Research Centre at the Cambridge University Hospital’s NHS Foundation Trust]. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Footnotes
Author contributions: MKNL and MH conceived of the project. HH developed the methods, software, ran the tests and simulations, and created the figures. MKNL, MH, and HH wrote the manuscript with methods contributions from AT, AK, and MI. AT conducted the Plasmodium wet lab experiments. AK and MI conducted the HipSci cell line experiments. DG provided the HipSci cell lines and sequencing. RD provided feedback and guidance throughout the project.
Conflicts of interest: Haynes Heaton was a previous employee of 10x Genomics and holds shares in that company.
Data Availability
HipSci cell line data is available at ENA with accession numbers ERS2630499-ERS2630501 for the three replicates of the experimental mixture and ERS2630502-ERS2630507 for the individual cell lines of euts, nufh, babz, oaqd, and ieki respectively. This data is shown in Fig 2 and Supp Fig 1. Maternal Fetal data is available at https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6701/ with accession numbers FCA7474063-FCA7474065. This data is shown in Fig 3, Supp Fig 2. The plasmodium data is available on ENA with accessions ERS4280420, ERS4280419, and ERS4280421 for samples Plasmodium1-3 respectively. This data is shown in Fig 3, Supp Fig 3.
Code Availability
Souporcell is freely available under the MIT open source license at https://github.com/wheaton5/souporcell.
References
- 1.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng GXY, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 2016;34:303–311. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tung P-Y, et al. Batch effects and the effective design of single-cell gene expression studies. Sci Rep. 2017;7:39921. doi: 10.1038/srep39921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoeckius M, et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGinnis CS, et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods. 2019;16:619–626. doi: 10.1038/s41592-019-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang HM, et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2018;36:89–94. doi: 10.1038/nbt.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y, McCarthy DJ, Stegle O. Vireo: Bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol. 2019;20:273. doi: 10.1186/s13059-019-1865-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, et al. Genotype-free demultiplexing of pooled single-cell RNA-seq. Genome Biol. 2019;20:290. doi: 10.1186/s13059-019-1852-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young MD, Behjati S. SoupX removes ambient RNA contamination from droplet based single cell RNA sequencing data. bioRxiv. 2018 doi: 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv [q-bioGN] 2012 [Google Scholar]
- 14.Petti AA, et al. A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat Commun. 2019;10:3660. doi: 10.1038/s41467-019-11591-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda N, Nakano R. In: Advances in Neural Information Processing Systems 7. Tesauro G, Touretzky DS, Leen TK, editors. MIT Press; 1995. Deterministic Annealing Variant of the EM Algorithm; pp. 545–552. [Google Scholar]
- 16.Carpenter B, et al. Stan: A probabilistic programming language. J Stat Softw. 2017;76 doi: 10.18637/jss.v076.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Streeter I, et al. The human-induced pluripotent stem cell initiative—data resources for cellular genetics. Nucleic Acids Research. 2017;45:D691–D697. doi: 10.1093/nar/gkw928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilpinen H, et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vento-Tormo R, et al. Single-cell reconstruction of the early maternal—fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moffett A, Colucci F. Co-evolution of NK receptors and HLA ligands in humans is driven by reproduction. Immunol Rev. 2015;267:283–297. doi: 10.1111/imr.12323. [DOI] [PubMed] [Google Scholar]
- 21.Howick VM, et al. The Malaria Cell Atlas: a comprehensive reference of single parasite transcriptomes across the complete Plasmodium life cycle: file S1. doi: 10.1126/science.aaw2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirén J, Välimäki N, Mäkinen V. Indexing Graphs for Path Queries with Applications in Genome Research. IEEE/ACM Trans Comput Biol Bioinform. 2014;11:375–388. doi: 10.1109/TCBB.2013.2297101. [DOI] [PubMed] [Google Scholar]
- 23.Sahraeian SME, et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat Commun. 2017;8:59. doi: 10.1038/s41467-017-00050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
HipSci cell line data is available at ENA with accession numbers ERS2630499-ERS2630501 for the three replicates of the experimental mixture and ERS2630502-ERS2630507 for the individual cell lines of euts, nufh, babz, oaqd, and ieki respectively. This data is shown in Fig 2 and Supp Fig 1. Maternal Fetal data is available at https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6701/ with accession numbers FCA7474063-FCA7474065. This data is shown in Fig 3, Supp Fig 2. The plasmodium data is available on ENA with accessions ERS4280420, ERS4280419, and ERS4280421 for samples Plasmodium1-3 respectively. This data is shown in Fig 3, Supp Fig 3.
Souporcell is freely available under the MIT open source license at https://github.com/wheaton5/souporcell.