Abstract
The objective of this study was to investigate the adjuvanticity of poly(I:C), a synthetic analogue of dsRNA capable of activating both TLR3 and RLRs, such as MDA-5 and RIG-I, as pathogen recognition receptors. While poly(I:C) is known to provoke a robust type I IFN, type III IFN, and Th1 cytokine response, its therapeutic use as a vaccine adjuvant is limited due to its vulnerability to nucleases and poor uptake by immune cells. To overcome these limitations, we encapsulated poly(I:C) into lipid nanoparticles (LNPs) containing an ionizable cationic lipid that can electrostatically interact with poly(I:C). We discovered that LNP-formulated poly(I:C) can trigger both lysosomal TLR3 and cytoplasmic RLRs, in vitro and in vivo, whereas poly(I:C) in an unformulated soluble form is only capable of triggering endosomal-localized TLR3. Administration of LNP-formulated poly(I:C) in mouse models led to efficient translocation to lymphoid tissue and concurrent innate immune activation following intramuscular (IM) administration, resulting in a significant increase in innate immune activation compared to unformulated soluble poly(I:C). When used as an adjuvant for recombinant full-length SARS-CoV-2 spike protein, LNP-formulated poly(I:C) elicited potent anti-spike antibody titers, surpassing those of unformulated soluble poly(I:C) by orders of magnitude. Moreover, prime and boost immunization with LNP-formulated poly(I:C) adjuvanted spike protein offered complete protection against a SARS-CoV-2 viral challenge in vivo, and serum from these mice was capable of significantly reducing viral infection in vitro. These findings highlight the potential of LNP-formulated poly(I:C) as a promising vaccine adjuvant that potently triggers both endosomal and cytoplasmic pathogen recognition receptors.
Introduction
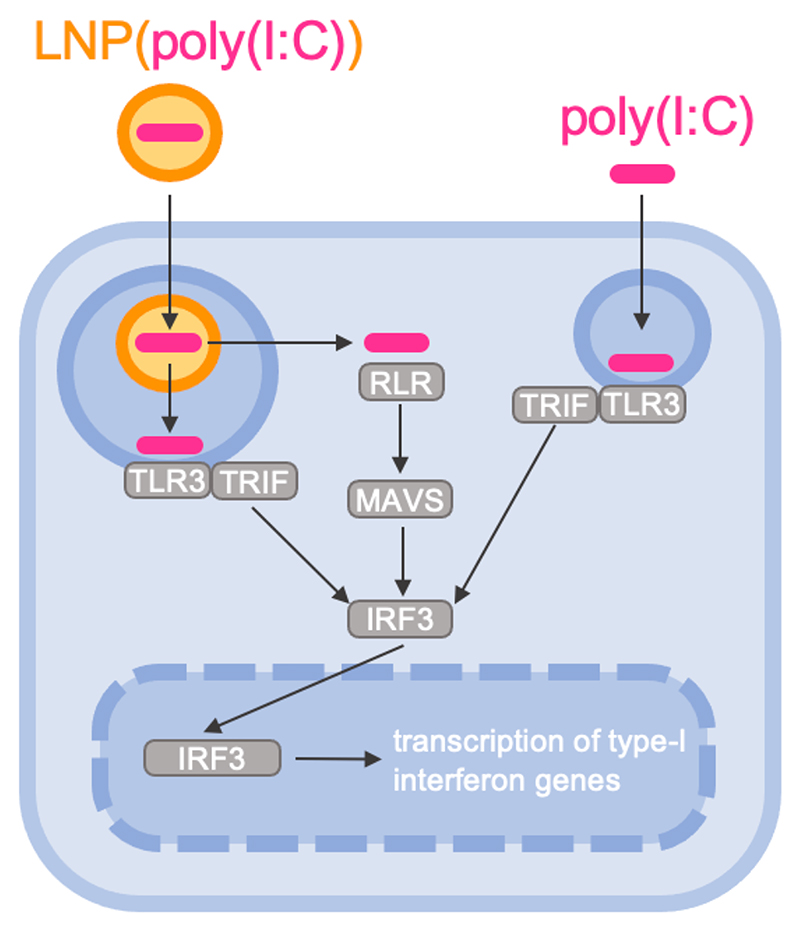
Recombinant protein and peptide subunit antigens are often weakly immunogenic and unable to mount protective humoral and cellular immune responses.[1] Therefore, most subunit antigens require co-formulation with adjuvants. Aluminum salts (alum) and oil-in-water emulsions are the most widely used adjuvants in vaccine formulations to date and some mainly elicit a T helper cell type-2 (Th2) skewed immune response.[2] Polyinosinic:polycytidylic acid (poly(I:C)) is a synthetic analogue of double stranded RNA (dsRNA) that can be recognized as a pathogen associated molecular pattern (PAMP) and can trigger several receptors in endosomes and cytoplasm of innate immune cells.[3] In the endosomes of antigen-presenting cells, poly(I:C) activates Toll Like Receptor 3 (TLR3) while in the cytoplasm poly(I:C) can trigger Retinoic acid-inducible gene-like receptors (RLRs) like Retinoic acid-inducible gene I (RIG-I) and the Melanoma Differentiation-associated 5 (MDA-5) receptors.[4] Innate immune responses following TLR3 activation rely on TIR-domain-containing adapter-inducing interferon-β (TRIF)-mediated signaling, whereas triggering of RLRs will result in RLR oligomerization, exposure of the N-terminal CARD domains and translocation to the mitochondria for further downstream signaling via mitochondrial antiviral signaling (MAVS) protein, resulting in the induction of strong type I and III interferon (IFN), which can promote type 1 cytokine responses (Figure 1).[4] This is of great relevance in the context of anti-viral and anti-cancer vaccination, by instigating robust levels of cytotoxic T cells and antibody isotypes with neutralizing and innate effector killing capacity.[4–6]
Figure 1. Simplified schematic representation of the signaling cascade in response to poly(I:C)-induced endosomal TLR3 and cytoplasmic RLR triggering.
In vivo, in soluble form, poly(I:C) is prone to degradation by nucleases, and poor uptake by antigen presenting cells in immune-inducing lymphoid tissues.[7] Furthermore, in its soluble form, poly(I:C) cannot reach into the cellular cytoplasm, and thus cannot efficiently activate RLRs. To improve its uptake by actively phagocytic cells such as dendritic cells and macrophages, poly(I:C) has been formulated into films,[8,9] hydrogels,[10] microparticles[11] and nanoparticles.[12,13] However, achieving cytoplasmic delivery remains very inefficient with most nanoparticles, especially in vivo.[14,15] Hence, the therapeutic potential of poly(I:C) is currently underdeveloped.
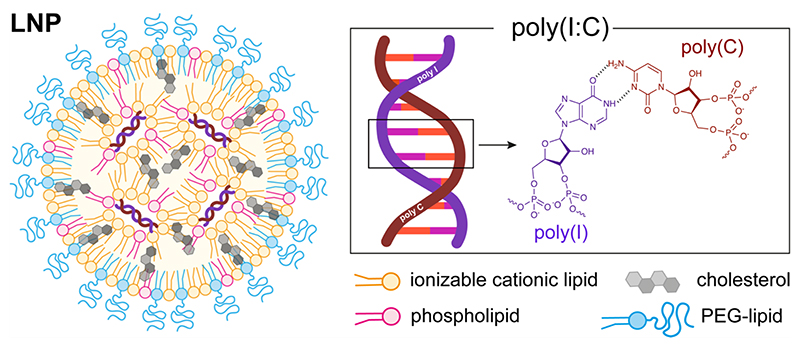
We hypothesized that encapsulating poly(I:C) into lipid nanoparticles[16,17] (LNP: Figure 2) through electrostatic complexation with an ionizable cationic lipid could overcome these limitations. LNPs are currently the most performant RNA delivery systems,[18–20] as demonstrated by the recent success of the mRNA LNP COVID-19 vaccines.[21] The LNPs used in this study are composed of an ionizable cationic lipid that, at low pH, can electrostatically complex with the anionic inosinic and cytidylic acid residues in the poly(I:C) backbone. LNPs protect nucleic acids from degradation in the extracellular medium and can be efficiently taken up by actively phagocytic cells in lymphoid tissues.[22] LNPs facilitate cytoplasmic delivery of a nucleic acid payload by destabilizing endosomal membranes. This is particularly relevant for accessing RLR, as these innate immune sensors are typically located in the cytoplasm of cells,[4] which is generally inaccessible to soluble, unformulated poly(I:C). Furthermore, nanoparticulate delivery systems mediate efficiently translocation to immune-inducing sites in lymphoid tissue,[11,23–28] which is of great relevance in the context of vaccine application.[29–35]
Figure 2. Schematic representation lipid nanoparticles (LNP) encapsulating poly(I:C).
Here we demonstrate that encapsulating poly(I:C) in LNP greatly enhances the innate immune response to poly(I:C), both in vitro and in vivo. While mere TLR3 triggering resulted in relatively poor adjuvanticity, combined TLR3 and RLR triggering induced robust innate and adaptive immune responses. Following intramuscular injection in mice, LNP-encapsulated poly(I:C) was delivered to dendritic cell (DC) subsets in draining lymph nodes and in the spleen, resulting in the activation of a broad range of myeloid and lymphoid immune cell subsets. Furthermore, when recombinant trimeric full-length spike protein of the SARS-CoV-2 virus was admixed with LNP-encapsulated poly(I:C), it induced robust antigen-specific neutralizing antibody responses superior to soluble poly(I:C) and generated protection against viral challenge in vivo.
Results and Discussion
Encapsulation of poly(I:C) in LNP
Poly(I:C) was encapsulated into LNP (Figure 2) using a rapid mixing method. An aqueous buffer (5 mM, pH 4) containing poly(I:C) was mixed vigorously with an ethanolic solution containing an ionizable lipid (S-Ac7-DOG; Figure S1 in the Supporting Information section), cholesterol, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and distearoyl-rac-glycerol-poly(ethylene glycol) (DSG-PEG; 2 kDa PEG). Cholesterol and DOPE served to facilitate nanoparticle formation and endosomal escape.[36] The PEG-lipid served for colloidal stabilization of the LNP. The reduction-sensitive ionizable lipid S-Ac7-DOG was recently developed in our laboratory as part of a combinatorial synthesis campaign aimed at identifying novel ionizable lipids for nucleic acid drug delivery.[37] ‘Low molecular weight` (LMW) poly(I:C), with a claimed size of 0.2 to 1 kilobases (kb), was used. We selected a molar ratio of ionizable lipid:cholesterol:DOPE:DSG-PEG of 50:38.5:10:1.5, based on established ratios used in mRNA delivery see Table S1 in the Supporting Information section for detailed composition).[36] LNP encapsulating poly(I:C), further referred to as LNP(poly(I:C)), were prepared at an N:P ration of 5:1 (N: molar fraction of ionizable amines in the ionizable lipid, P: molar fraction of anionic phosphate in poly(I:C)). LNP lacking poly(I:C), i.e., empty LNP, were prepared as a control.
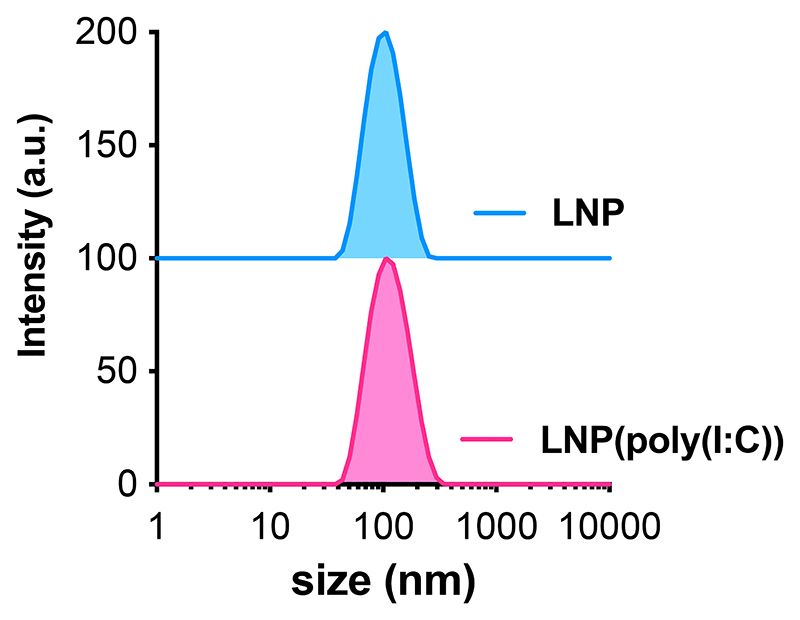
The size, electrophoretic mobility, and encapsulation efficiency of the LNP formulations were characterized using dynamic light scattering (DLS), electrophoretic mobility measurements, and a RiboGreen assay, as depicted in Figure 3 and Table 1. Both LNP(poly(I:C)) and empty LNP had a hydrodynamic diameter ranging from 90-110 nm, with a low polydispersity index (PDI) below 0.2, indicating a homogeneous population. RiboGreen assay confirmed complete encapsulation of poly(I:C) in the LNP. The zeta potential of both formulations was slightly positive at the physiological pH of 7.4. Notably, the zeta potential of nanoparticles is a measure of the electrostatic potential at the boundary between the dispersing medium and the “slipping plane” of the particle. Therefore, it assesses the properties of the LNP surface rather than the LNP core. The ionizable lipid, specifically S-Ac7-DOG, in our LNP formulations has a pK of 6.74[37] and thus will have reduced ionization at the physiological pH of 7.4, at which we measured the zeta potential of the LNP. We used an N:P ratio of 5:1. Consequently, the ionizable lipid is in excess and likely ensures encapsulation of poly(I:C) inside the core of the LNP. Therefore, the presence of poly(I:C) might not significantly alter the surface properties as it is shielded from the external environment. Moreover, the PEG (polyethylene glycol) coating can ‘shield’ the surface charge, further making the zeta potential of the LNP closer to neutral.
Figure 3. Intensity-based size distribution curves measured by DLS of LNP(poly(I:C)) and LNP formulations.
Table 1. Physicochemical characterization of LNP(poly(I:C)) and empty control LNP.
| z-average diameter (nm) | PDI | zeta-potential (mV) ± SD* | encapsulation efficiency (%) | |
|---|---|---|---|---|
| LNP(poly(I:C)) | 105 | 0.14 | 6.0 ± 4.9 | 100 |
| LNP | 96 | 0.10 | 3.5 ± 5.1 | / |
Measured in HEPES buffer pH 7.4.
In vitro cellular uptake and innate activation pathway of LNP(poly(I:C))
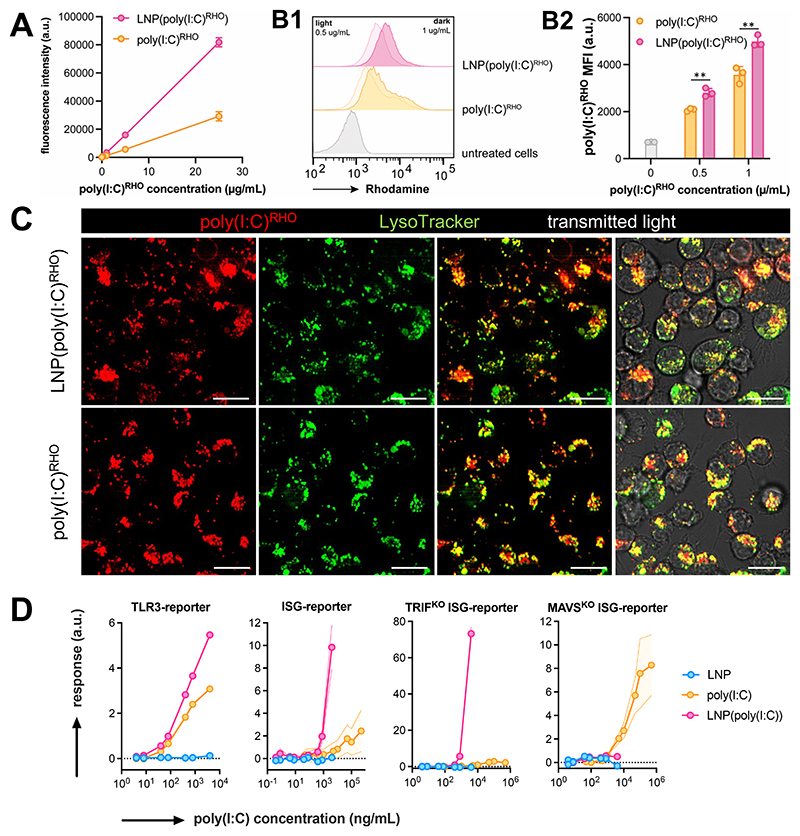
Fluorescently labeled rhodamine-conjugated poly(I:C) (poly(I:C)RHO) was employed to evaluate the uptake of LNP by antigen-presenting cells in vitro. Interestingly, the fluorescence emission intensity of poly(I:C)RHO was higher in LNP-formulated form than in unformulated form in solution (cfr. the experimental section for exact details how this was assessed). We then used the immortalized DC2.4 mouse dendritic cell line as a model for antigen-presenting cells.[38] The DC2.4 cells were treated with LNP(poly(I:C)RHO) and unformulated soluble poly(I:C)RHO for 12 hours, respectively, to assess their uptake efficiency. Flow cytometry analysis (Figure 4B) revealed that the uptake of LNP(poly(I:C)RHO) and poly(I:C)RHO) by DC2.4 cells was dose-dependent and was slightly higher in case of LNP(poly(I:C)RHO). However, the difference in fluorescence emission intensity between poly(I:C)RHO) and LNP(poly(I:C)RHO) induces a bias in the quantitative interpretation of these data. Confocal microscopy imaging (Figure 4C) confirmed the cellular internalization of both unformulated soluble poly(I:C)RHO and LNP(poly(I:C)RHO), showing a punctate intracellular pattern in cells. Counterstaining with LysoTracker demonstrated strong co-localization, suggesting that both unformulated soluble poly(I:C)RHO and LNP(poly(I:C)RHO) are primarily stored in intracellular acidic vesicles such as endosomes and lysosomes. It should be noted that endosomal escape with simultaneous cytoplasmic delivery of free poly(I:C)RHO is expected to happen only for a small fraction of LNP(poly(I:C)RHO), which makes it difficult to detect due to the strong signal from poly(I:C)RHO in endosomes.[39]
Figure 4. In vitro characterization of LNP(poly(I:C)).
(A) Fluorescence emission intensity (λex: 535 nm/λem: 670 nm) of poly(I:C)RHO and LNP(poly(I:C)RHO) (n=2, mean+SD). (B) Flow cytometry analysis of DC2.4 pulsed with LNP(poly(I:C)RHO) and unformulated soluble poly(I:C)RHO, (B1) histograms and (B2) corresponding MFI values. (n=3, mean ± SD ; t-test **: p < 0.01). (B) Confocal microscopy images of DC2.4 cells treated with LNP(poly(I:C)RHO) and poly(I:C)RHO. Cells were counterstained with LysoTrackerGreen. Scale bar represents 30 micron. (C) Innate immune activation by LNP(poly(I:C)) and poly(I:C) in TLR3- and ISG-reporter cell lines (n=6, mean ± SD). Note that a soluble, unformulated, poly(I:C) could be tested at concentrations that cannot be reached for LNP(poly(I:C)).
Next, we utilized a panel of commercially available reporter cell lines to examine the impact of LNP encapsulation on the ability of poly(I:C) to activate endosomal and cytoplasmic innate immune sensors (Figure 4D). HEK-Blue mTLR3 cells are HEK293 cells that have been genetically modified through co-transfection of the murine TLR3 gene and an inducible secreted embryonic alkaline phosphatase (SEAP) reporter gene. The SEAP gene is driven by the IFN-β minimal promoter fused to NF-κB and AP-1 binding sites. Activation of TLR3 by poly(I:C) triggers NF-κB and AP-1, leading to SEAP production, which can be quantified by a simple colorimetric assay. RAW-Lucia ISG cells are RAW264.7 macrophages that have been genetically engineered to carry an interferon regulatory factor (IRF)-inducible luciferase reporter construct. RAW264.7 cells express dsRNA RLRs. Activation of RLRs by poly(I:C) in the cytosol triggers IRF,[4] leading to luciferase production, which can be measured by bioluminescence detection. Additionally, we tested LNP(poly(I:C)) and poly(I:C) on RAW-Lucia ISG cells in which the signaling proteins involved in TLR3- and RLR-mediated innate activation, TRIF and MAVS, respectively,[4] were either knocked out or present. In TLR3 reporter cells, LNP encapsulation resulted in a significant increase in TLR3 activation by poly(I:C). In ISG reporter cells, LNP encapsulation led to a remarkable increase in ISG activation by poly(I:C). This response was maintained in TRIFKO ISG reporter cells, but was completely abolished in MAVSKO ISG reporter cells. Notably, all three ISG-reporter cell lines required high concentrations of unformulated, soluble poly(I:C) to induce a measurable response. Whereas the maximum concentration of poly(I:C) in TLR3-reporter cells was below 104 ng/mL, the ISG-reporter cells required a poly(I:C) concentration of up to 105–106 ng/mL. At such high concentrations, the samples containing LNP(poly(I:C)) induced severe cytotoxicity, making proper assessment at these levels unfeasible. Overall, our findings support the hypothesis that LNP encapsulation enhances the ability of poly(I:C) to strongly activate RIG-I and/or MDA-5, and it confirms that LNP can facilitate cytoplasmic delivery of poly(I:C).
LNP formulation strongly increases innate activation capacity of poly(I:C) in vivo
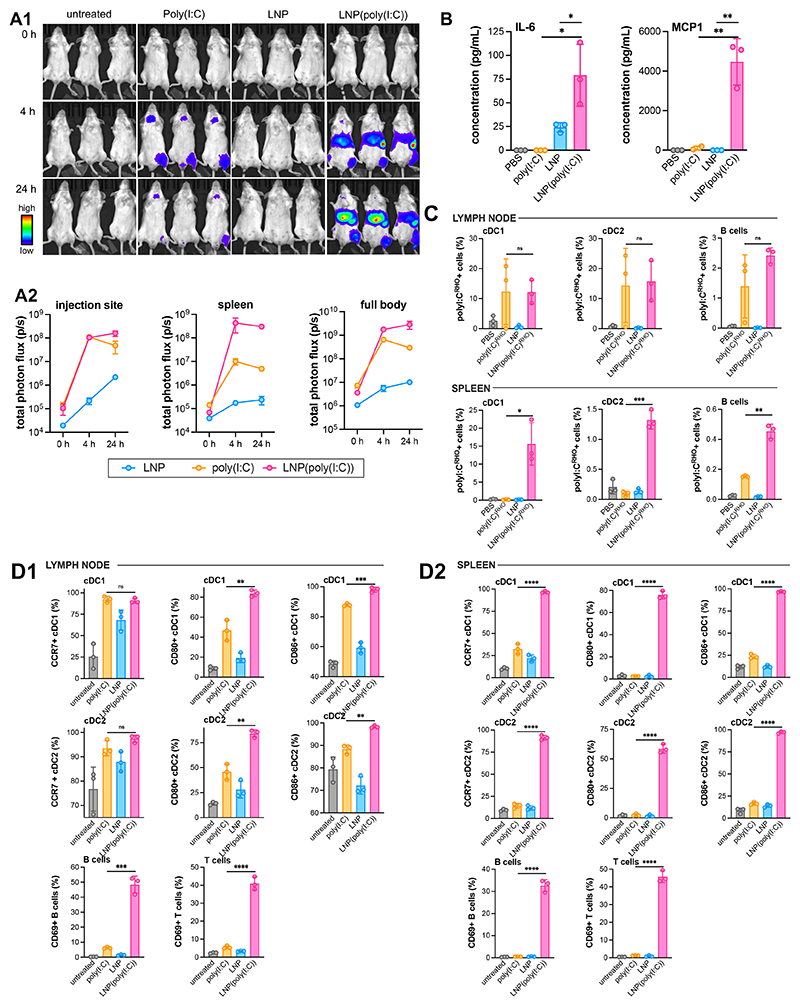
The influence of LNP formulation on the biodistribution and amplitude of the poly(I:C)-induced type I IFN response in vivo at the macroscopic tissue level was investigated using a IFN-β luciferase reporter mouse model (IFNβ+/Δβ-luc). This transgenic mouse model links the expression of IFN-β to the firefly luciferase gene, enabling spatiotemporal analysis of nanocarrier-mediated IFN-β induction through bioluminescence imaging.[40] IFNβ+/Δβ-luc mice were given an intramuscular injection into the quadriceps of an equivalent high dose of 25 μg poly(I:C) in unformulated soluble and LNP-formulated form. We deliberately selected a high dose to fully capture the effect of LNP formulation on the pharmacokinetic and pharmacodynamic profile of poly(I:C). Bioluminescence imaging was performed immediately upon injection, 4h, and 24h post-injection (Figure 5A). Unformulated soluble poly(I:C) induced IFN-β expression at the site of injection 4h post-injection, which declined strongly after 24h. IFN-β expression was also observed in the urinary tract and mouth, indicating renal secretion (and reingestion) of poly(I:C). LNP(poly(I:C)) induced a similar signal at the injection site 4h post-injection, but also induced strong IFN-β expression in the liver and spleen. The magnitude of the IFN-β expression further increased after 24h. Empty LNP did not induce a detectable response. To analyze cytokine levels in the blood, we performed Bio-Plex assays on cytokines (IL-1b, IL-4, IL-6, IL-10, IL-12(p70), IFN-γ, monocyte chemoattractant protein 1 (MCP1), TNF-α) at 6h post-injection (Figure 5B). MCP1 and IL-6 were increased in mice that received LNP(poly(I:C)), while cytokine levels in mice injected with unformulated soluble poly(I:C) remained below the detection level. Empty LNP also induced IL-6, but not MCP1, and IL-6 levels were lower than those induced by LNP(poly(I:C)). These findings are consistent with recent observations by Alameh et al., who reported on the ability of empty LNP to induce IL-6 in vivo in mice, suggesting that the adjuvant activity of empty LNP observed in an immunization setting may be attributed to its ability to induce IL-6.[41] The absence of a clear pro-inflammatory cytokine profile shortly after injection suggests that the LNP formulations are safe.
Figure 5. In vivo innate immune activation by LNP(poly(I:C)).
(A1) Bioluminescence imaging of BALB/c IFNβ+/Δβ-luc mice at 0 h, 4 h, 24 h after receiving intramuscular injection into the quadricep of a 25 μg dose of poly(I:C) in soluble unformulated and in LNP-formulated form. An equivalent dose of empty LNP was administered. (n = 3). (A2) Region of interest (ROI) analysis quantifying total flux (photons/second) of the full body, injection site, spleen of the corresponding samples (n = 3, mean±SD). (B) Bio-Plex analysis of cytokine levels in serum at 6 h post-injection (n=3, mean ± SD; t-test *: p < 0.05; **:p < 0.01). (C) Flow cytometry analysis of the uptake of poly(I:C)RHO in immune cell subsets in draining popliteal lymph node and the spleen at 24 h after receiving intramuscular injection into the quadricep of a 25 μg dose of poly(I:C)RHO in soluble unformulated and in LNP-formulated form. An equivalent dose of empty LNP was administered. (D) Flow cytometry analysis of immune cell activation and maturation in (D1) the draining popliteal lymph node and (D2) the spleen at 24 h after receiving intramuscular injection into the quadricep of a 25 μg dose of poly(I:C) in soluble unformulated and in LNP-formulated form. An equivalent dose of empty LNP was administered. (n = 3/group, mean ± SD; one-wat Anova *: p < 0.05; **:p < 0.01; ***:p < 0.001; ****:p < 0.0001).
We analyzed the effect of LNP formulation on the biodistribution of poly(I:C) (25 μg dose) on a cellular level by flow cytometry in the draining iliac lymph node and spleen at 24 h post IM injection in the quadricep. For fluorescent tracking, poly(I:C)RHO was used. In the draining lymph node, poly(I:C)RHO was detected in cDC1 and cDC2 dendritic cell subsets and, to a lesser extent, also in B cells. LNP formulation did not have a major impact on the delivery of poly(I:C)RHO to DCs (cDC1 and cDC2 DC subsets) and B cells (Figure 5C). In the spleen, by contrast, LNP(poly(I:C)RHO) showed, relative to unformulated soluble poly(I:C)RHO, a vast increase in delivery to cDC1 and cDC2 dendritic cells and to a lesser extent also to B cells.
The draining iliac lymph node and spleen were analyzed by flow cytometry, 24 hours post IM injection, to investigate the influence of LNP formulation on the cellular biodistribution of poly(I:C). Poly(I:C)RHO was used to enable fluorescent tracking. In the draining lymph node, poly(I:C)RHO was detected in cDC1 and cDC2 dendritic cell subsets, and to a lesser extent in B cells. The LNP formulation did not have a significant impact on the delivery of poly(I:C)RHO to DCs (cDC1 and cDC2 DC subsets) and B cells (Figure 5C). However, in the spleen, LNP(poly(I:C)RHO) showed enhanced delivery to cDC1 and cDC2 dendritic cells compared to unformulated soluble poly(I:C)RHO. Also, B cells received a higher dose of poly(I:C) when poly(I:C) was formulated in LNP.
Next, we analyzed the effect of LNP formulation on innate immune activation on a cellular level by flow cytometry in the draining iliac lymph node and spleen at 24 h post intramuscular injection. In the draining lymph node, both unformulated soluble poly(I:C) and LNP(poly(I:C)) induced upregulation of the maturation markers CCR7, CD80, and CD86 (Figure 5D1). LNP(poly(I:C)) and unformulated soluble poly(I:C) were equally effective in upregulating CCR7 expression. However, LNP(poly(I:C)) was more potent in upregulating CD80 and CD86 expression. Empty LNP induced intermediate maturation above background levels. In the draining lymph node, LNP(poly(I:C)) induced upregulation of the activation marker CD69 in B and T cells to a much higher extent than unformulated soluble poly(I:C). LNP did not induce activation of B and T cells. In the spleen, LNP(poly(I:C)) strongly induced maturation of cDC1 and cDC2 dendritic cells and induced strong activation of B and T cells (Figure 5D2). In the spleen, unformulated soluble poly(I:C) induced slight maturation of cDC1 and cDC2 dendritic cells but did not induce activation of B and T cells. Empty LNP had no effect on dendritic cells or B and T cells in the spleen. Taken together, our data provide clear evidence that the LNP formulation strongly amplifies the innate immune activation capacity of poly(I:C).
Uptake of LNP(poly(I:C)) by dendritic cells is likely to directly induce maturation of these cells. Our data showed that in the draining lymph node, there was no significant difference in the uptake of LNP(poly(I:C)) and unformulated soluble poly(I:C) by DCs. However, in the spleen, LNP(poly(I:C)) was delivered to DCs at a much higher rate than unformulated soluble poly(I:C). B and T cell activation in both the lymph node and spleen likely occurs as a bystander effect in response to cytokine secretion by dendritic cells or other TLR3- and RLR-expressing cells, such as muscle cells and epithelial cells that internalized LNP(poly(I:C)). The ability of LNP to deliver poly(I:C) to both endosomal vesicles (location of TLR3) and the cytoplasm (location of RLR) likely plays a major role in contributing to the strong innate immune activating capacity of LNP(poly(I:C)), as suggested by our in vitro data (Figure 4C). Moreover, the inherent properties of LNPs themselves, such as their ability to induce systemic levels of IL-6 and dendritic cell maturation in lymphoid tissue, might also contribute to the potency of LNP(poly(I:C)).
LNP formulation strongly alters the vaccine adjuvant properties of poly(I:C)
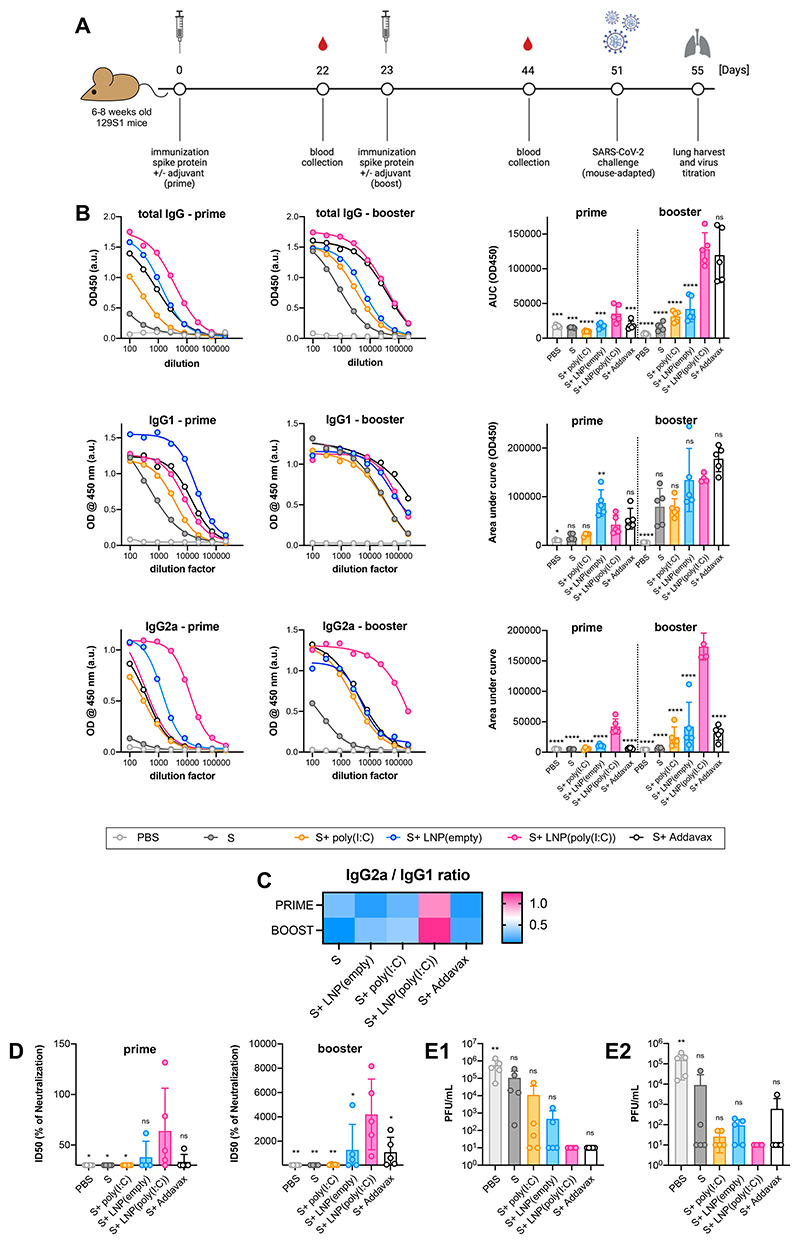
We investigated the adjuvant properties of LNP(poly(I:C)) by admixing them with recombinant full-length trimeric SARS-CoV-2[42] spike protein (further abbreviated as S protein). Well adjuvanted S protein is able to mount protective responses in mice as shown before by us and others.[43–45] 6-8 week old female 129S1 mice were immunized intramuscularly with a prime-boost schedule using 5 μg of recombinant S protein, either unadjuvanted or adjuvanted with LNP(poly(I:C)), LNP, or AddaVax, an MF59-like water-in-oil established control vaccine adjuvant. To limit the systemic immune activation, a 5-fold lower dose of poly(I:C) was used in this experiment. The experimental outline is depicted in Figure 6A, and blood was collected 22 days post-prime and 21 days post-boost vaccination for analysis of S protein-specific total IgG, IgG1, and IgG2a isotype/subclass titers by enzyme-linked immunosorbent assay (ELISA) (Figure 6B). Booster immunization increased antigen-specific antibody titers across all cohorts and isotype classes. The highest antigen-specific total IgG titers were measured in the LNP(poly(I:C)) adjuvanted cohorts. After prime, antigen-specific IgG1 titers were highest in mice immunized with antigen adjuvanted with empty LNP, whereas after boost, the Addavax-adjuvanted cohort had the highest IgG1 titers, confirming its type 2 skewing effect, and empty LNP and LNP(poly(I:C)) adjuvanted cohorts were on par. Antigen-specific IgG2a titers were greatly increased in LNP(poly(I:C)) adjuvanted cohorts, relative to all other cohorts, both after prime and boost. It is noteworthy that the adjuvanticity of soluble unformulated poly(I:C) was found to be very weak and barely increased antigen-specific antibody titers beyond those of non-adjuvanted antigen. Empty LNP also exhibited an adjuvant effect, which is in line with recent reports in the literature.[41] The high IgG2a/IgG1 (Figure 6C) ratio observed in LNP(poly(I:C)) adjuvanted cohorts suggests that LNP(poly(I:C)) skews towards a Th1 immune response and class switching towards an IgG2a isotype.
Figure 6. In vivo adjuvanticity of LNP(poly(I:C)).
(A) Experimental outline. (B) ELISA titers for (B1) IgG1, (B2) IgG2a, (B3) total IgG in mice sera collected 3 weeks post-prime (left) and post-boost (right) vaccination (graph represents area under the OD(450 nm) ELISA curve against serum dilutions for individual groups as well as Area under curve (AUC) calculated based on OD450 curves (mean ± SD) of the individual serum samples). (C) IgG1/IgG2a serum antibody ratio. (D) Half maximal inhibitory dilution ID50 of serum containing virus-neutralizing antibodies post-prime and post-boost vaccination (mean ± SD) in microneutralization assay using 350 tissue culture infectious dose 50 (TCID50) of USA-WA1/2020 SARS-CoV-2. The limit of detection was 30, corresponding to the lowest starting serum dilution used in microneutralization assays (E1) Viral lung titers and (E2) viral nasal turbinate titers represented as plaque-forming units (PFU)/mL (mean ± SD) after challenge with 10^5 PFU per animal of mouse-adapted SARS-CoV-2. The limit of detection was 67 PFU for plaque assays and the undetectable titers were given a value 10 (below detection limit) (n = 5, mean ± SD; one-way Anova *: p < 0.05; **:p < 0.01; ***:p < 0.001; ****:p < 0.0001).
We then investigated the capacity of sera from immunized mice to neutralize the ancestral USA-WA1/2020 virus, a Wuhan-like SARS-CoV-2 strain, in vitro using a microneutralization assay (Figure 6D). Sera collected post prime and post booster immunization were tested, and only serum from mice immunized with LNP(poly(I:C)) adjuvanted antigen induced viral neutralization beyond the background level from the post prime sera. All post-boost sera from mice immunized with adjuvanted antigen were able to neutralize the virus to some extent, and LNP(poly(I:C)), LNP, and AddaVax adjuvanted sera were on par. Immunization with unadjuvanted antigen or antigen adjuvanted with soluble unformulated poly(I:C) was unable to neutralize the virus in vitro, suggesting an absence of detectable neutralizing antibody titers in these groups post prime as well as post boost.
After booster immunization, mice received an intranasal dose of a mouse-adapted SARS-CoV-2 strain to determine the correlation between observed vaccine responses and protection from viral challenge. The mouse-adapted SARS-CoV-2 strain was derived from the ancestral Wuhan-like USA-WA1/2020 SARS-CoV-2 virus, antigenically matching the vaccine antigen, except for the N501Y mutation in its receptor binding domain, which allows the virus to bind to mouse ACE2 more efficiently, resulting in infection of laboratory strains of wild type mice.[46] Four days post challenge, lungs and nasal turbinates were harvested, and the residual viral load in these tissues was quantified by a plaque assay. Immunization with antigen adjuvanted with LNP(poly(I:C)) and AddaVax conferred sterilizing immunity, with plaque numbers below the detection limit in both lungs and nasal turbinates (Figure 6E). Unadjuvanted antigen and antigen adjuvanted with LNP and soluble unformulated poly(I:C) were unable to fully control viral infection.
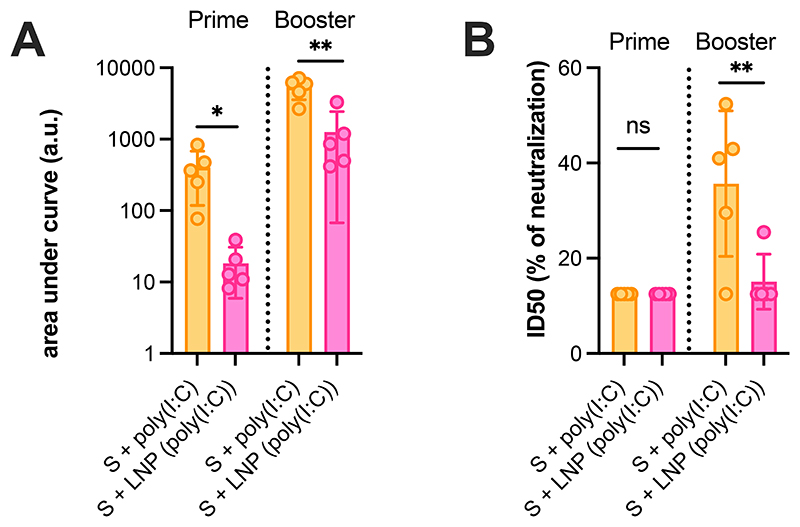
We repeated the vaccination study using trimeric recombinant SARS-CoV-2 spike protein in MAVS-/- mice. MAVS is an innate signaling adapter molecule that is involved in RLR signaling, but not TLR3 signaling. We observed that the dominant adjuvant effect of LNP(poly(I:C)) was abolished in MAVS-/- mice, with the highest ELISA binding titers observed in the group that received soluble poly(I:C) (Figure 7A). This was also reflected in the microneutralization assay, where the LNP(poly(I:C)) group had lower titers compared to the soluble poly(I:C) group, and overall low titers in MAVS-/- mice (Figure 7B). These results confirm our hypothesis that, also in vivo, induction of immunity by LNP(poly(I:C)) is primarily mediated by cytoplasmic RLR triggering, rather than TRL3 triggering.
Figure 7. In vivo adjuvanticity of LNP(poly(I:C)) is abrogated in MAVS-/- KO mice.
(A) ELISA titers for total IgG in MAVS-/- mice sera collected 3 weeks post-prime and post-boost vaccination. The graph represents area under curve (AUC) calculated based on OD(450 nm) curve against serum dilutions for individual animals (n=5, mean ± SD; one-way Anova *: p < 0.05; **:p < 0.01). (B) In vitro microneutralization titers observed in serum from vaccinated MAVS-/- mice (n=5, mean ± SD; one-way Anova **:p < 0.01) represented as ID50 of sera post-prime and post-boost vaccination (mean ± SD, one-way Anova) using 350TCID50 of USA-WA1/2020 SARS-CoV-2.
Conclusions
In this study, we investigated the encapsulation of the synthetic double-stranded RNA TLR3 agonist, poly(I:C), in LNPs via electrostatic interaction between the phosphate anions in the poly(I:C) backbone and a cationic amine of an ionizable lipid. Our results showed that LNP encapsulation maintained the capacity of poly(I:C) to trigger TLR3 in endosomes, while greatly amplifying its capacity to trigger RLRs by mediating delivery of poly(I:C) into the cytoplasm. This hypothesis is supported by literature data reporting in vitro experiments that show direct cytoplasmic delivery of poly(I:C) by transfection can activate RLRs and convert the TLR3-mediated induction of apoptosis into an immunoadjuvant effect driven by IFN-β production.[47] In vivo in mice, LNP formulation altered the pharmacokinetic profile of poly(I:C) upon intramuscular administration, resulting in vastly amplified innate immune activation in distal tissues, including liver and spleen. Immunization of mice with LNP(poly(I:C)) adjuvanted recombinant Spike protein from the SARS-CoV-2 virus yielded robust antigen-specific antibody titers, with LNP(poly(I:C)) vastly outperforming soluble unformulated poly(I:C), which exhibited rather weak adjuvant properties. Additionally, immune serum from mice receiving a prime and booster immunization of LNP(poly(I:C)) adjuvanted Spike protein strongly reduced viral infection in vitro, while prime and boost immunization of LNP(poly(I:C)) adjuvanted Spike protein fully protected against a mouse-adapted SARS-CoV-2 viral challenge in vivo.
We confirmed the importance of the innate signaling adapter molecule MAVS[4] for the adjuvant effect of LNP(poly(I:C)), although we were unable to discriminate the contributions of different RLRs (RIG-I or MDA5) to the observed adjuvant effect, as both signal through MAVS. Both RIG-I and MDA5 are known to be activated by poly(I:C) with the help of zinc finger protein ZCCHC3[48] but are also reported to have different preferential binding depending on molecular weight of the dsRNA. The efficiency of innate immune activation by Poly(I:C) depends on molecular mass of poly(I:C).[49] For these studies, we have used low molecular mass (LMM) poly(I:C). It has been suggested that MDA5 is mainly activated by high molecular weight (3kb) poly(I:C), whereas RIG-I would typically sense low molecular weight (<1.5kb) poly(I:C) as well as in vitro transcribed 5’ phosphorylated dsRNAs like the Sendai virus defective interfering RNA.[50–52] We anticipate that LNP(poly(I:C)) could find application in the context of vaccine design against viral infection and cancer, as well as in the context of cancer immunotherapy[53] to mount immunity against neo- and self-antigens. Overall, our findings suggest that LNP-formulation is a scalable and translational feasible approach that greatly amplifies the adjuvant properties of poly(I:C) by triggering both TLR3 and RLRs and altering its pharmacokinetic and pharmacodynamic properties in vivo. Notably, LNPs, including LNPs containing S-Ac7-DOG as an ionizable lipid,[54] have been amenable to lyophilization, which enables long-term storage and thereby facilitates their use in vaccine formulations.[55]
Experimental Section
Materials
Unless otherwise stated, all chemicals were purchased from Sigma Aldrich. Cell culture medium and supplements, PBS (1 x), penicillin/streptomycin (100 x), sodium pyruvate (100 x) and Quanti-iT RiboGreen RNA assay kit were purchased from Thermo Fischer. HEK-Blue-mTLR3 cells as well as HEK-Blue Detection, Normocin, poly(I:C) LMW, poly(I:C) LMW Rhodamine, AddaVax were obtained from Invivogen. DOPE, DSG-PEG2000 were purchased from Avanti Polar Lipids. Anti-mouse SARS-CoV-2 nucleoprotein and anti-mouse SARS-CoV-2 S protein antibodies were obtained from the Center for Therapeutic Antibody Development at the Icahn School of Medicine at Mount Sinai, New York. The secondary antibodies with conjugated HRP were obtained from Abcam.
Cell lines
The DC2.4 cell line was a kind gift from Dr. Kenneth Rock. (University of Massachusetts, Boston, USA). HEK-Blue mTLR3 cells, RAW-Lucia ISG cells, RAW-Lucia ISGO-KO-TRIF cells and RAW-Lucia ISGO-KO-MAVS cells were puchased from InvivoGen. Vero-E6, obtained from ATCC, and Vero-E6-TMPRSS2 cells (stably expressing TMPRSS2) were maintained in DMEM supplemented with 10% fetal bovine serum, 1X penicillin/streptomycin and 1X nonessential amino acids (Gibco) and further with puromycin for Vero-E6-TMPRSS2.
Mice
6–8-week-old 129S mice obtained from JAX Laboratories, MA. Mice were housed with food and water ad libitum in a specified pathogen-free animal facility at Icahn School of Medicine at Mount Sinai. Mice were vaccinated intramuscularly (50ul per hind leg per mouse) and infected via the intranasal route (10^5 PFU mouse adapted SARS-CoV-2 virus in 50μl PBS per mouse) under ketamine/xylazine anesthesia. All procedures were approved by the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee (IACUC-2013-1408). IPS-1 (MAVS-/-) deficient mice[56] were kindly provided by Dr. Matthias J. Schnell, Ph.D. Homozygous MAVS-/- were bred and housed in specific pathogen free (SPF) facilities maintained by the Center for Comparative Medicine and Surgery at the Icahn School of Medicine at Mount Sinai. All experiments were performed with sex-matched mice at 6–8 weeks of age. All studies were performed in accordance with the principles described by the Animal Welfare Act and the National Institutes of Health guidelines for the care and use of laboratory animals in biomedical research. The protocols for performing mice studies were reviewed and approved by Institutional Animal Care and Use committee (IACUC) at ISMMS.
Virus
The WA1-USA/2020 SARS-CoV-2 virus was obtained from BEI resources (NR-52281). The Mouse-adapted SARS-CoV-2 virus[46] as obtained after several serial passaging of WA1-USA/2020 SARS-CoV-2 virus in mice with different genetic backgrounds, at the Animal Biosafety level 3 (ABSL3) facility at Icahn school of Medicine at Mount Sinai. The virus was further propagated and titrated on Vero E6 cells and diluted appropriately in 1X PBS for intranasal infection in animals.
Instrumentation
Both DLS and ELS measurements were performed on a Zetasizer Nano ZS (Malvern Instruments Ltd., Malvern, U.K.) equipped with a HeNe laser (λ = 633 nm) and detection at scattering angle of 173°.
Flow cytometry analysis was performed on a BD LSRFortessa and BD FACSCanto. Data were processed using the FlowJo software package. Samples were prepared in 100 μL PBS and transferred to Eppendorf tubes prior to analysis.
Confocal microscopy images were taken via a Leica DMI6000B microscope (63 × 1.40 NA objective) coupled to an AndorDSD2 confocal scanner and a Zyla5.5 CMOS camera. Images were processed with ImageJ software package.
LNP formulation
Aqueous solutions of poly(I:C) were made by adding 250 μL of a poly(I:C) LMW stock solution (1 mg/mL in PBS) to 3.083 mL 5 mM acetate buffer (pH 4). Ethanol solutions (1.667 mL) consisted of S-Ac7-DOg, DOPE, cholesterol, DSG-PEG2000 (see Table S1 for LNP composition). LNPs were fabricated by solvent displacement, mixing rapidly with a vortex the ethanolic solution of lipids to the aqueous solution of poly(I:C). To remove ethanol, the formed LNP suspensions were dialyzed overnight with PBS using Slide-A-Lyzer cassettes (cut-off 3.5 kDa) (Thermo Fischer, U.S.A). Next, the LNP suspensions were concentrated by using an Amicon Ultra 10K Centrifugal Filter (MilliporeSigma, U.S.A.) (cut-off 3.5 kDa) to yield a poly(I:C) concentration of 0.5 mg/mL.
LNP(poly(I:C)RHO) formulation
Poly(I:C)RHO was formulated into LNP(poly(I:C)RHO) as mentioned above but not dialyzed. To serve as a proper control, poly(I:C)RHO was subjected to identical conditions (i.e., addition of ethanol, etc…). Fluorescence emission intensities of poly(I:C)RHO and LNP(poly(I:C)RHO), respectively, were recorded on an Ensight Multimode Microplate reader (PerkinElmer, U.S.A.) (λex = 530 nm λem = 560 nm).
DLS and ELS
100 μL of each sample was individually measured thrice via DLS. Cumulants analysis of the data gave the z-average and PDI (data provided as average). For ELS, each sample was diluted 10-fold in 5 mM HEPES buffer (pH 7.4) and was individually measured thrice. Zeta potential values were calculated based on Smoluchowski equation and were provided as average ± standard deviation.
Determination of encapsulation efficiency: RiboGreen assay
Quant-iT RiboGreen RNA Assay kit (Thermo Fisher, U.S.A.) was used to determine poly(I:C) encapsulation efficiency after LNP production. LNP samples were diluted in TE (Tris-EDTA) buffer to obtain a (theoretical) concentration of 1 μg/mL poly(I:C) (= working solutions). 50 μL of these working solutions were two-fold diluted in duplicate in a black 96-well plate with TE buffer. Similar samples were prepared with TE buffer supplemented with 2% Triton X-100 to lyse the LNPs. Next, 100 μL of RiboGreen solution (1:100 diluted RiboGreen reagent in TE buffer) was added to each well. Fluorescence was measured using an Ensight Multimode Microplate reader (PerkinElmer, U.S.A.) (λex = 485 nm λem = 528 nm). The resulting fluorescence values were subtracted by the fluorescence value of the reagent blank. The encapsulation efficiency (%) was calculated according to the equation , where Itotal is the fluorescence intensity of samples measured total poly(I:C) in presence of 2% Triton X-100 and Ifree is the fluorescence intensity of samples measured free or unencapsulated poly(I:C) in absence of 2% Triton X-100.
In vitro cellular uptake - Flow cytometry
DC2.4 cells were seeded in a 24-well plate at a concentration of 200 000 cells per well in 450 μL of culture medium and allowed to adhere overnight at 37°C (5% CO2). The cells were pulsed overnight, at 37°C (5% CO2), with 50 μL of poly(I:C)RHO and LNP(poly(I:C)RHO), respectively, resulting in a poly(I:C)RHO concentration of 0.5 μg/mL and 1 μg/mL, respectively. After overnight incubation, the culture medium was aspirated and washed with PBS. Next, 500 μL cell dissociation buffer was added and incubated for 15 min at 37°C (5% CO2) to detach the cells. The cell suspensions were transferred to Eppendorf tubes and centrifuged (5 min, 300 G, 4 °C). After aspiration of the supernatant, the cell pellets were resuspended in 200 μL PBS and analyzed using a BD LSRFortessa flow cytometer. Data were processed by FlowJo software package.
In vitro cellular uptake - Confocal microscopy
DC2.4 cells were seeded in WillCo-Dish glass bottom at a concentration of 50 000 cells in 180 μL culture medium and allowed to adhere overnight at 37°C (5% CO2). The cells were pulsed overnight with 20 μL of poly(I:C)-Rho LNP suspension (LNP(poly(I:C)-Rho)) in PBS (50 μg/mL poly(I:C) Rho) or soluble poly(I:C)-Rho (poly(I:C)-Rho) (50 μg/mL in PBS) at 37°C (5% CO2). After overnight incubation, the culture medium was aspirated and the cells were fixated with 4% PFA for 15 min at 37°C (5% CO2) followed by washing with PBS. A staining solution was prepared by adding 10 μL Hoechst (1 mg/mL stock solution in DMSO) to 2 mL PBS supplemented with 1% bovine serum albumin (BSA). 200 μL of this staining solution was added to the fixed cells and incubated for 40 min at 37°C (5% CO2). Finally, the samples were washed with PBS followed by confocal imaging using a Leica DMI6000B microscope (63 × 1.40 NA objective) coupled to an AndorDSD2 confocal scanner and a Zyla5.5 CMOS camera. Images were processed with ImageJ software package.
HEK-Blue mTLR3 innate immune activation assay
HEK-Blue mTLR3 cells were seeded in a flat-bottom 96-well plate at a concentration of 50 000 cells per well in 180 μL HEK-Blue detection medium (prepared per manufacturer instruction). The cells were pulsed overnight (6-16 h) with 20 μL of poly(I:C) LNP suspension (LNP(poly(I:C))), empty LNP (LNP(-)), soluble poly(I:C) or sterile endotoxine-free water (negative control) at 37°C (5% CO2) at concentrations of 3.333, 1.667, 0.333, 0.033, 0.003 μg/mL poly(I:C). After overnight incubation, SEAP levels were determined by measuring optical density at 620 nm using an Ensight Multimode Microplate reader (PerkinElmer, U.S.A.). Note: colorimetric quantification of the samples was obtained relative to the negative control and each concentration was performed in 5-fold.
Bio-Plex cytokine assay
Cytokines (IL-1b, IL-4, IL-6, IL-10, IL-12(p70), IFN-γ, MCP-1,TNF-α) in the mice serum samples were measured on a Luminex Bio-Plex suspension array system (Bio-Rad, U.S.A) according to the manufacturer’s instructions. Briefly, 4-fold diluted serum samples were added to magnetic capture beads (Bio-Rad) and incubated for 2 h at room temperature. After incubation, the beads were washed and a detection antibody (Bio-Rad) was added to the wells. After 1 h incubation the beads were washed again and incubated for 30 min with streptavidin-PE (Bio-Rad). Finally, the samples were measured by Bio-Plex 200 System (Bio-Rad). Cytokine concentrations were analyzed using standard curves and expressed as pg/mL.
Anti-SARS-CoV-2 S protein ELISA
Anti-SARS-CoV-2 S protein ELISA was performed to estimate S-specific antibody responses upon vaccination as described before.[43] Briefly, Maxisorp Nunc 96-well microtiter plates were coated with 50 μL per well of recombinant trimeric S protein, diluted to a concentration of 2 μg/mL in carbonate/bicarbonate buffer and incubated with 50μl per well overnight at 4°C. 50μl per well of three-fold serially diluted serum samples, starting from 1:100, were added to the antigen-coated plates followed by overnight incubation at 4°C. The plates were then washed in 1X PBS + 0.01% Tween20 and again incubated with appropriate horse-radish peroxidase (HRP)-conjugated secondary antibodies targeting total IgG, IgG1 or IgG2a antibodies at room temperature for 1 h. The plates were washed and developed with 100 μL of tetramethyl benzidine (TMB) substrate per well until blue color appeared. The reaction was terminated with 50 μL 1M H2SO4 and the absorbance was measured at 450 nm with 650 nm as a reference.
In vivo immune activation imaging
Luciferase reporter mice (IFNβ+/Δβ-luc) with a BALB/c background, aged 7-9 weeks, were housed in individual ventilated cages and given ad libitum access to food and water. 50 μL of poly(I:C) LNP suspension (LNP(poly(I:C))), empty LNP (LNP(-)), soluble poly(I:C) or PBS (untreated) were injected intramuscularly in the quadriceps (n=3) at an equivalent dose of 25 μg poly(I:C). 6 hours post-injection, blood was collected from mice for Bio-Plex cytokine analysis (see above). For in vivo imaging at the given time points (0, 4, 24 h), mice were injected subcutaneously with 200 μL D-luciferin and in vivo luminescence imaging was recorded 12 min later using the IVIS Lumina II imaging system. Local (injection site & spleen) luminescence and full body luminescence were quantified using the Living Image 4.4 software.
Analysis of in vivo lymphocyte targeting and activation
After the last time point (24 h) of the immune activation imaging (described above) luciferase reporter mice were sacrificed and iliac lymph nodes and spleens were isolated. Next, single cell suspensions were prepared from the dissected lymph nodes and spleens for flow cytometry analysis. Isolated lymph nodes and spleens were collected in ice cold PBS, smashed through 70 μm cell strainers, washed with PBS and stained for 30 min at 4°C with following primary labeled antibodies: CD3, CD20, CD11c, MHCII, CD86, CD80, CD40, CD69, CCR7. Live dead ratios were determined by staining with fixable dead/live staining and 123count ebeads were added to determine cellularity prior to analysis by a BD FACSCanto flow cytometer. Data were processed using the FlowJo software package.
In vivo cellular uptake by immune cell subsets
Luciferase reporter mice (IFNβ+/Δβ-luc) with a BALB/c background, aged 7-9 weeks, were housed in individual ventilated cages and given ad libitum access to food and water. 50 μL poly(I:C)-Rhodamine LNP suspension (LNP(poly(I:C)-Rho)), empty LNP (LNP(-)), soluble poly(I:C)-Rhodamine (soluble poly(I:C)-Rho) or PBS (untreated) were injected intramuscularly in the quadriceps (n=3) at an equivalent poly(I:C) dose of 25 μg. 24 hours post injection, mice were sacrificed and iliac lymph nodes and spleens were isolated. Next, single cell suspensions were prepared from the dissected iliac lymph nodes and spleens for analysis by flow cytometry. Isolated lymph nodes and spleens were collected in ice cold PBS, smashed through 70 μm cell strainers, washed with PBS and stained for 30 min at 4°C with DAPI and with the following primary labeled antibodies: CD11c, B220 and CD169. 123count ebeads were added to determine cellularity prior to analysis by a BD FACSCanto flow cytometer. Data were processed using the FlowJo software package.
Trimeric recombinant SARS-CoV-2 S protein vaccination
Trimeric full length recombinant S protein was produced as followed: only the ectodomain of the S protein (GenBank: MN908947.3) was cloned into a mammalian expression plasmid and the cleavage site was removed and stabilizing prolines were added at position 986 and 987 (24–26). A hexa-histidine tag as well as a T4 foldon trimerization domain was present in the plasmid for ease of purification. The S protein was expressed in 293F cells, using the ExpiFectamine 293 Transfection Kit (Thermo Fisher). Supernatant was collected on day 3 post transfection and Ni-NTA agarose (Qiagen) was used to purify the protein. S protein (5 μg/mouse) was used as such or mixed with adjuvant as described below and injected following a prime-boost schedule via the IM route with a BD 300 μL insulin syringe in the hamstring muscles of one hind leg (50 μL/mouse).
Adjuvants: soluble poly(I:C), LNP(poly(I:C)), LNP(-) and Addavax (0.2 mg/mL poly(I:C) or Addavax) were mixed 1:1 volume ratio with S protein (0.2 mg/mL) to yield an equivalent dose of 5 μg S protein and poly(I:C) per mouse.
In vitro microneutralization assay
To measure the neutralizing potential of SARS-CoV-2 vaccine-induced sera, an in vitro microneutralization assay was performed. Briefly, the S protein ± adjuvant-vaccinated mice sera were inactivated at 56°C for 30 min. Serum samples were serially diluted 3-fold starting from 1:30 dilution in infection medium (DMEM + 2% FBS + 1 x non-essential amino acids). The samples were incubated with 350 TCID50 of SARS-CoV-2 isolate USA-WA1/2020 (BEI resources; NR-52281) for 1 hour in an incubator at 37°C (5% CO2) and then transferred on pre-seeded Vero E6-TMPRSS2 cells in 96-well cell culture plates. The plates were incubated at 37°C for 48 hours and fixed in 4% formaldehyde. The cells were permeabilized with 0.1% Triton X-100 for 15 min at RT and washed three times with 1X PBS + 0.1% Tween 20 (PBST). The cells were then blocked in 5% milk in PBST for 1 hour at RT. After blocking, the cells wereincubated with anti-SARS-CoV-2 nucleoprotein and anti-SARS-CoV-2 S protein monoclonal antibodies, mixed in 1:1 volume ratio, for 1.5 hours at RT. The cells were washed again and incubated with HRP-conjugated anti-mouse IgG secondary antibody for 1 hour at room temperature followed by a brief PBS wash. Finally, 100 μL TMB substrate was added and incubated until blue color appeared and the reaction was terminated with 50 μL 1M H2SO4. Absorbance at 450nm was recorded and percentage inhibition calculated.
Lung Virus Titration
Plaque assays were performed to quantify and compare the replicating lung viral titers in vaccinated versus unvaccinated mice. Whole lungs and nasal turbinates were harvested from the mice four days post infection and homogenized in 500μl 1X PBS. After brief centrifugation, the tissue debris was discarded and the supernatant was 10-fold serially diluted starting from 1:10 dilution. Pre-seeded Vero-E6 cells were incubated with diluted lung and nasal turbinate homogenates for 1 hour at room temperature and then overlayed with 1 mL mixture of 2% oxoid agar and 2 x minimal essential medium (MEM) supplemented with 2% FBS. After 72 hours of incubation at 37°C (5% CO2) the plates were fixed in 4% formaldehyde, followed by immune-staining of infected cells with anti-mouse SARS-CoV-2 nucleoprotein and anti-mouse SARS-CoV-2 S protein monoclonal antibodies. After incubation with primary antibodies, horse radish peroxidase (HRP)-conjugated anti-mouse secondary antibody was added for 1 hour. Finally, the plaques were developed with TrueBlue substrate (KPL-Seracare). The final viral titers were calculated in terms of plaque forming units (PFU)/mL.
Supplementary Material
Acknowledgements
B.G.D.G. acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant n° 817938). We thank Daniel Flores, Marlene Espinoza, Jane Deng, and Ryan Camping for excellent administrative support, Richard Cadagan for technical support and Randy Albrecht for management and organization of the BSL3 facility. SARS-CoV-2 work in the M.S. laboratory is supported by NIH/NIAID R01AI160706 and NIH/NIDDK R01DK130425. S.C.P has received support from the Korea Health Industry Development Institute (KHIDI) for research under the Biomedical Global Talent Nurturing Program (HI22C2101). This study was also partly funded by CRIPT (Center for Research on Influenza Pathogenesis and Transmission), a NIH NIAID funded Center of Excellence for Influenza Research and Response (CEIRR, contract number 75N93021C00014), and by NIAID contract 75N93019C00046 to AG-S.
Footnotes
Conflicts of Interest
BGDG is co-inventor of a patent filing on the ionizable lipid used in this work for LNP formulation. This IP has been licensed to eTheRNA Immunotherapies. The M.S. laboratory has received unrelated funding support in sponsored research agreements from Phio Pharmaceuticals, 7Hills Pharma, ArgenX BV and Moderna. The A.G.-S. laboratory has received research support from GSK, Pfizer, Senhwa Biosciences, Kenall Manufacturing, Blade Therapeutics, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma, Applied Biological Laboratories and Merck, outside of the reported work. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Castlevax, Amovir, Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, CureLab Oncology, CureLab Veterinary, Synairgen, Paratus and Pfizer, outside of the reported work. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott and Astrazeneca. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York. S.C-C is inventor in patents associated with the use of viruses and/or immunotherapeutics for cancer. S.C-C has consulting agreements with the company Accurius.
References
- [1].Reed SG, Orr MT, Fox CB. Nat Med. 2013;19:1597–608. doi: 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- [2].Awate S, Babiuk LA, Mutwiri G. Front Immunol. 2013;4:114. doi: 10.3389/fimmu.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- [4].Rehwinkel J, Gack MU. Nat Rev Immunol. 2020;20(9):537–551. doi: 10.1038/s41577-020-0288-3. 2020 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ngoi SM, Tovey MG, Vella AT. J Immunol. 2008;181:7670–7680. doi: 10.4049/jimmunol.181.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, Casanova-Acebes M, Khudoynazarova M, Agudo J, Tung N, et al. Immunity. 2016;44:924–938. doi: 10.1016/j.immuni.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Martins KAO, Bavari S, Salazar AM. Expert Rev Vaccines. 2015;14:447–459. doi: 10.1586/14760584.2015.966085. [DOI] [PubMed] [Google Scholar]
- [8].Demuth PC, Min Y, Huang B, Kramer JA, Miller AD, Barouch DH, Hammond PT, Irvine DJ. Nat Mater. 2013;12(4):367–376. doi: 10.1038/nmat3550. 2013 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chiu Y-C, Gammon JM, Andorko JI, Tostanoski LH, Jewell CM. 2016;8:18722–18731. doi: 10.1021/acsami.6b06275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Song H, Huang P, Niu J, Shi G, Zhang C, Kong D, Wang W. Biomaterials. 2018;159:119–129. doi: 10.1016/j.biomaterials.2018.01.004. [DOI] [PubMed] [Google Scholar]
- [11].Jewell CM, Bustamante Lopez SC, Irvine DJ, López SCB, Irvine DJ. Proc Natl Acad Sci U S A. 2011;108:15745–50. doi: 10.1073/pnas.1105200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hafner AM, Corthésy B, Merkle HP. Adv Drug Deliv Rev. 2013;65:1386–1399. doi: 10.1016/j.addr.2013.05.013. [DOI] [PubMed] [Google Scholar]
- [13].Gale EC, Roth GA, Smith AAA, Alcántara-Hernández M, Idoyaga J, Appel EA. Adv Ther. 2020;3:2070001 [Google Scholar]
- [14].Morshedi Rad D, Alsadat Rad M, Razavi Bazaz S, Kashaninejad N, Jin D, Ebrahimi Warkiani M, Morshedi Rad D, Alsadat Rad M, Razavi Bazaz S, Kashaninejad N, et al. Adv Mater. 2021;33:2005363. doi: 10.1002/adma.202005363. [DOI] [PubMed] [Google Scholar]
- [15].Kauffman KJ, Webber MJ, Anderson DG. J Control Release. 2016;240:227–234. doi: 10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- [16].Hou X, Zaks T, Langer R, Dong Y. Nat Rev Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kulkarni JA, Witzigmann D, Chen S, Cullis PR, Van Der Meel R. Acc Chem Res. 2019;52:2435–2444. doi: 10.1021/acs.accounts.9b00368. [DOI] [PubMed] [Google Scholar]
- [18].Lokugamage MP, Vanover D, Beyersdorf J, Hatit MZC, Rotolo L, Echeverri ES, Peck HE, Ni H, Yoon JK, Kim YT, et al. Nat Biomed Eng. 2021;5(9):1059–1068. doi: 10.1038/s41551-021-00786-x. 2021 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ. Nat Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Miao L, Li L, Huang Y, Delcassian D, Chahal J, Han J, Shi Y, Sadtler K, Gao W, Lin J, et al. Nat Biotechnol. 2019;37:1174–1185. doi: 10.1038/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- [21].Jain S, Venkataraman A, Wechsler ME, Peppas NA. Adv Drug Deliv Rev. 2021;179:114000. doi: 10.1016/j.addr.2021.114000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nakamura T, Harashima H. Adv Drug Deliv Rev. 2020;167:78–88. doi: 10.1016/j.addr.2020.06.003. [DOI] [PubMed] [Google Scholar]
- [23].Chen Y, De Koker S, De Geest BG. Acc Chem Res. 2020;53(10):2055–2067. doi: 10.1021/acs.accounts.0c00260. [DOI] [PubMed] [Google Scholar]
- [24].Irvine DJ, Hanson MC, Rakhra K, Tokatlian T. Chem Rev. 2015;115:11109–11146. doi: 10.1021/acs.chemrev.5b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hanson MC, Crespo MP, Abraham W, Moynihan KD, Szeto GL, Chen SH, Melo MB, Mueller S, Irvine DJ. J Clin Invest. 2015;125:2532–46. doi: 10.1172/JCI79915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kuai R, Ochyl LJ, Bahjat KS, Schwendeman A, Moon JJ. Nat Mater. 2017;16:489–496. doi: 10.1038/nmat4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].De Koker S, Cui J, Vanparijs N, Albertazzi L, Grooten J, Caruso F, De Geest BG. Angew Chem Int Ed Engl. 2016;55:1334–1339. doi: 10.1002/anie.201508626. [DOI] [PubMed] [Google Scholar]
- [28].De Koker S, Naessens T, De Geest BG, Bogaert P, Demeester J, De Smedt S, Grooten J. J Immunol. 2010;184:203–211. doi: 10.4049/jimmunol.0803591. [DOI] [PubMed] [Google Scholar]
- [29].Reddy ST, van der Vlies AJ, Simeoni E, Angeli V, Randolph GJ, O’Neil CP, Lee LK, Swartz MA, Hubbell JA. Nat Biotechnol. 2007;25:1159–64. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- [30].Nembrini C, Stano A, Dane KY, Ballester M, van der Vlies AJ, Marsland BJ, Swartz MA, Hubbell JA. Proc Natl Acad Sci. 2011;108:E989–E997. doi: 10.1073/pnas.1104264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wilson JT, Keller S, Manganiello MJ, Cheng C, Lee C-C, Opara C, Convertine A, Stayton PS. ACS Nano. 2013;7:3912–3925. doi: 10.1021/nn305466z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, Ravindran R, Stewart S, Alam M, Kwissa M, et al. Nature. 2011;470:543–547. doi: 10.1038/nature09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Moon JJ, Suh H, Bershteyn A, Stephan MT, Liu H, Huang B, Sohail M, Luo S, Ho Um S, Khant H, et al. Nat Mater. 2011;10:243–251. doi: 10.1038/nmat2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu H, Moynihan KD, Zheng Y, Szeto GL, Li AV, Huang B, Van Egeren DS, Park C, Irvine DJ. Nature. 2014;507:519–522. doi: 10.1038/nature12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lynn GM, Sedlik C, Baharom F, Zhu Y, Ramirez-Valdez RA, Coble VL, Tobin K, Nichols SR, Itzkowitz Y, Zaidi N, et al. Nat Biotechnol. 2020;38:320–332. doi: 10.1038/s41587-019-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kulkarni JA, Witzigmann D, Leung J, Tam YYC, Cullis PR. Nanoscale. 2019;11:21733–21739. doi: 10.1039/c9nr09347h. [DOI] [PubMed] [Google Scholar]
- [37].manuscript in submission
- [38].Shen Z, Reznikoff G, Dranoff G, Rock K. J Immunol. 1997;158:2723–2730. [PubMed] [Google Scholar]
- [39].Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stöter M, et al. Nat Biotechnol. 2013;31(7):638–646. doi: 10.1038/nbt.2612. 2013 31. [DOI] [PubMed] [Google Scholar]
- [40].Nuhn L, Vanparijs N, De Beuckelaer A, Lybaert L, Verstraete G, Deswarte K, Lienenklaus S, Shukla NM, Salyer ACD, Lambrecht BN, et al. Proc Natl Acad Sci U S A. 2016;113:8098–8103. doi: 10.1073/pnas.1600816113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Alameh M-G, Tombácz I, Bettini E, Lederer K, Ndeupen S, Sittplangkoon C, Wilmore JR, Gaudette BT, Soliman OY, Pine M, et al. Immunity. 2021;54:2877–2892. doi: 10.1016/j.immuni.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ye T, Zhong A, García-Sastre A, Schotsaert M, De Geest BG. Angew Chemie. 2020;132:19045–19057. doi: 10.1002/anie.202008319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jangra S, De Vrieze J, Choi A, Rathnasinghe R, Laghlali G, Uvyn A, Van Herck S, Nuhn L, Deswarte K, Zhong Z, et al. Angew Chemie Int Ed. 2021;60:9467–9473. doi: 10.1002/anie.202015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rodrigues KA, Rodriguez-Aponte SA, Dalvie NC, Lee JH, Abraham W, Carnathan DG, Jimenez LE, Ngo JT, Chang JYH, Zhang Z, et al. Sci Adv. 2021;7:eabj6538. doi: 10.1126/sciadv.abj6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gale EC, Powell AE, Roth GA, Meany EL, Yan J, Ou BS, Grosskopf AK, Adamska J, Picece VCTM, d’Aquino AI, et al. Adv Mater. 2021;33:2104362. doi: 10.1002/adma.202104362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rathnasinghe R, Jangra S, Ye C, Cupic A, Singh G, Martínez-Romero C, Mulder LCF, Kehrer T, Yildiz S, Choi A, et al. Nat Commun. 2022;13:3921. doi: 10.1038/s41467-022-30763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Palchetti S, Starace D, De Cesaris P, Filippini A, Ziparo E, Riccioli A. J Biol Chem. 2015;290:5470–5483. doi: 10.1074/jbc.M114.601625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lian H, Zang R, Wei J, Luo W-W, Li S, H.-B. S. Correspondence Immunity. 2018;49:438–448. doi: 10.1016/j.immuni.2018.08.014. [DOI] [PubMed] [Google Scholar]
- [49].Zhou Y, Guo M, Wang X, Li J, Wang Y, Ye L, Dai M, Zhou L, Persidsky Y, Ho W. Innate Immun. 2013;19:184–192. doi: 10.1177/1753425912459975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Patel JR, Jain A, Chou YY, Baum A, Ha T, García-Sastre A. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- [52].Yoneyama M, Fujita T. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- [53].Bianchi F, Pretto S, Tagliabue E, Balsari A, Sfondrini L. Cancer Biol Ther. 2017;18:747–756. doi: 10.1080/15384047.2017.1373220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lamoot A, Lammens J, De Lombaerde E, Zhong Z, Gontsarik M, Chen Y, De Beer TRM, De Geest BG. Biomater Sci. 2023;11:4327–4334. doi: 10.1039/d2bm02031a. [DOI] [PubMed] [Google Scholar]
- [55].Kim B, Hosn RR, Remba T, Yun D, Li N, Abraham W, Melo MB, Cortes M, Li B, Zhang Y, et al. J Control Release. 2023;353:241–253. doi: 10.1016/j.jconrel.2022.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, et al. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.